Abstract
Species of the Lactobacillus acidophilus complex are generally considered to constitute most of the vaginal Lactobacillus flora, but the flora varies between studies. However, this may be due to difficulties in identifying the closely related species within the L. acidophilus complex by using traditional methods and to variations in the vaginal status of the participants. Two hundred two isolates from the vaginal fluids of 23 Swedish women without bacterial vaginosis, as defined by the criteria of Nugent et al. (R. P. Nugent, M. A. Krohn, and S. L. Hillier, J. Clin. Microbiol. 29:297-301, 1991), were typed by randomly amplified polymorphic DNA (RAPD) analysis and identified to the species level by temporal temperature gradient gel electrophoresis, multiplex PCR, and 16S ribosomal DNA sequencing. The vaginal flora of most participants was dominated by a single RAPD type, but five of them harbored two RAPD types representing two different species or strains. The most frequently occurring species were Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii. L. iners has not previously been reported as one of the predominant Lactobacillus species in the vagina.
The healthy human vagina is dominated by lactobacilli, which play an important role in protecting the host from urogenital infections (4, 17, 18). Furthermore, it is widely recognized that the microbial balance between lactobacilli as the dominating flora and other, mainly gram-negative anaerobes can be upset and frequently result in the syndrome of bacterial vaginosis (28). Thus, the microbial status of the healthy human vagina has to be defined in any study of vaginal lactobacilli. The clinical criteria of Amsel et al. (1) and/or the set of scores defined by Nugent et al. (19) for Gram-stained smears of vaginal fluid help to define the concept of bacterial vaginosis. These scores are generally considered to be useful in treating patients with bacterial vaginosis but can also be used to define a healthy vaginal status by excluding bacterial vaginosis and by combining the results with a short medical history and examination for sexually transmitted diseases.
A general opinion is that species of the Lactobacillus acidophilus complex constitute most of the healthy vaginal Lactobacillus flora (15). However, other species have been encountered, e.g., Lactobacillus fermentum, Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus jensenii, Lactobacillus casei, Lactobacillus delbrueckii, Lactobacillus vaginalis, and Lactobacillus salivarius (23, 26, 30). The group of organisms previously known as L. acidophilus was shown to be highly heterogeneous by Lauer et al. in 1980 (16). Subsequently, the group was divided into DNA homology groups A and B by Johnson et al. (11), to form the six separate species of the L. acidophilus complex: L. acidophilus (A1), Lactobacillus crispatus (A2), Lactobacillus amylovorus (A3), Lactobacillus gallinarum (A4), Lactobacillus gasseri (B1), and Lactobacillus johnsonii (B2) (8, 11). The species in the complex are obligately homofermentative and belong to the rRNA group L. acidophilus. L. jensenii, another common lactobacillus found in the vagina, belongs to the same phylogenetic group (25), and recently two new species have been added (9), i.e., Lactobacillus amylolyticus, isolated from beer (3), and Lactobacillus iners, from human sources (7).
The closely related species within the L. acidophilus complex are quite difficult and sometimes impossible to differentiate by phenotypic methods (13). In addition, the lack of reliable identification methods may account for some of the variation in the Lactobacillus flora of the vagina found in different studies (23, 30). Consequently, accurate genomic methods are needed in order to define the composition of the Lactobacillus flora in the vagina, not only for the treatment of infectious diseases but also to establish the normal Lactobacillus flora in different settings.
With this objective, we applied PCR-based methods for the identification of the Lactobacillus flora. Randomly amplified polymorphic DNA (RAPD) analysis (10, 22) was used to group the isolates; temporal temperature gradient gel electrophoresis (TTGE), where the 16S ribosomal DNA (rDNA) molecule is used, was used for identification to the species level (29); and multiplex PCR, where another area of the rDNA, namely, the intergenic spacer region between the 16S and the 23S rDNAs, is used for the identification (27), was also used.
MATERIALS AND METHODS
Bacterial strains.
Two hundred two isolates from the healthy vaginas of 23 Swedish women were typed and identified. All isolates were obligately homofermentative, as determined by using the 50CH system (API Systems, S. A., Montalieu Versieu, France) according to the manufacturer's instructions. As a key for the identification, 26 type and reference strains were used: L. acidophilus CCUG 5917T, Lactobacillus amylophilus CCUG 30137T, L. amylovorus CCUG 20531T, L. brevis CCUG 30670T, L. casei DSM 20011T, L. casei ATCC 334, L. casei subsp. pseudoplantarum DSM 20008, L. crispatus CCUG 30722T, Lactobacillus delbrueckii subsp. delbrueckii CCUG 34222T, L. delbrueckii subsp. lactis CCUG 31454T, L. fermentum ATCC 14931T, L. gallinarum CCUG 30724T, L. gasseri DSM 20243T, L. helveticus CCUG 30139T, L. iners CCUG 28746T, L. jensenii CCUG 35572T, L. johnsonii CCUG 30725T, Lactobacillus oris NCFB 2160T, Lactobacillus paracasei subsp. paracasei NCFB 151T, L. paracasei subsp. tolerans CCUG 34829T, L. plantarum ATCC 14917T, Lactobacillus rhamnosus DSM 20021T, Lactobacillus reuteri DSM 20016T, L. salivarius CCUG 31453T, Lactobacillus vaginalis CCUG 31452T, and Lactobacillus zeae DSM 20178 T.
Sampling.
Fifty-one women visiting the outpatient clinic, Department of Obstetrics and Gynaecology, and the antenatal care unit at the University Hospital, Linköping, Sweden, for planned gynecological examinations between June 1997 and May 1998 agreed to participate in the study. Most women came for their scheduled cervical screening. Samples were collected from the vaginal fluid with a cotton swab rolled over the upper third part of the lateral vaginal wall. The swabs were placed in modified Stuart medium (Copan transport medium; Venturi Transystem, Brescia, Italy) and sent to the laboratory, where they were cultured on the agar plates within 3 h. Another swab was similarly rolled over the same vaginal area and onto a glass slide. The slides were air dried, fixed by heat, and Gram stained. The slides were then assessed according to the criteria of Nugent et al. (19).
The specimens were vortexed for 5 s in 1 ml of phosphate-buffered saline (pH 7.2) and diluted in PBS to 1/100, 1/1,000, and 1/10,000. One milliliter of each dilution was spread onto horse blood agar (purchased from the Division of Clinical Microbiology, Faculty of Health Science, Linköping, Sweden), MRS (De Man Rogosa and Sharpe) agar (Oxoid, Basingstoke, Hampshire, United Kingdom), and Rogosa agar (Oxoid) plates. No cultivations on MRS agar were performed on the specimens from subjects A and B. All of the agar plates were incubated for 48 to 72 h in 10% CO2 and 5% O2. Lactobacilli from 23 women with normal vaginal flora according to the criteria of Nugent et al. (19) were included in the study.
One hundred eighty-six isolates with the ability to grow on Rogosa agar plates were randomly selected and studied, together with 16 isolates that grew only on horse blood agar.
All isolates and reference strains were cultured in Lactobacillus carrying medium (6) at 37°C in an anaerobic jar for 24 to 48 h prior to the preparation of the PCR template as previously described by Vásquez et al. (29). This template was used for all of the PCR-based analyses described below.
RAPD analysis.
Two hundred two isolates and the type strains were subjected to RAPD analysis. The primer used in the PCR amplification was a 9-mer with the sequence 3′-ACG CGC CCT-5′ (Scandinavian Gene Synthesis AB, Köping, Sweden). PCR amplification and agarose gel electrophoresis were performed as follows, as previously described by Quednau et al. (22). One microliter of PCR template was used in a PCR mixture containing PCR buffer with 1.5 mM MgCl2 (Roche Diagnostics GmbH, Mannheim, Germany), a 0.2 mM concentration of each nucleotide (Roche), and 2.5 units of Taq DNA polymerase (Roche). The final volume of the reaction mix was 50 μl, and it was covered with mineral oil (Perkin-Elmer, Norwalk, Conn.). The PCR amplification was conducted in a DNA thermal cycler (Perkin-Elmer) with the following temperature profile: 94°C for 45 s, 30°C for 120 s, and 72°C for 60 s for 4 cycles, followed by 94°C for 5 s, 36°C for 30 s (with an extension of 1 s for each cycle), and 72°C for 30 s for 26 cycles. The PCR session concluded with 72°C for 10 min, followed by cooling to 4°C. The PCR products were visualized by agarose gel electrophoresis, and photonegatives of RAPD gels were scanned with a Hewlett-Packard ScanJet 5300C into a computer at a resolution of 300 dpi. The gel images were then analyzed and grouped by GelCompar 4.2 (Applied Maths, Kortrijk, Belgium) with the Pearson product moment correlation coefficient (r) and the unweighted pair group method by arithmetic averages. The limit for the isolates that belonged to the same RAPD type was determined to 86% similarity.
TTGE.
Representative strains from each RAPD group were selected, PCR amplified with primers ENV1 and TTGE7-gc, and analyzed by TTGE, as described recently by Vásquez et al. (29). In total, 72 vaginal isolates and all type and reference strains were subjected to TTGE. A standard composed of TTGE products of strains that showed clear and bright bands at different positions was used to allow gel-to-gel comparison. Photonegatives of TTGE gels were scanned with a Hewlett-Packard ScanJet 5300C into a computer at a resolution of 300 dpi. After this, the gel images were analyzed by GelCompar 4.2 (Applied Maths) with the Dice coefficient (SD) and the unweighted pair group method by arithmetic averages.
Multiplex PCR.
Twenty-six vaginal isolates and all of the reference and type strains were assessed by multiplex PCR as previously described by Song et al. (27). PCR-G was used to group the lactobacilli with the primers Ldel-7, LU-1′, LU-3′, LU-5, and Lac-2. PCR II-1 with the primers Ljen-3, Laci-1, and 23-10C was applied for the identification of L. jensenii and L. acidophilus strains. Finally, PCR II-2 with primers Lcri-1, Lgas-1, Lcri-2, and Lgas-2 was performed to identify L. crispatus and L. gasseri strains. The PCR II-2 amplification was slightly modified by using a different annealing temperature (61°C instead of 65°C as described previously). The other modification to the protocol was that 1.25 or 1.5% agarose gel electrophoresis was used to analyze the amplicons instead of polyacrylamide gel electrophoresis.
16S rDNA sequencing.
Partial sequencing of the 16S rRNA gene was performed for strains with uncertain identity (Table 1). The target for identification was the first 900 nucleotides of the 5′ end of the gene with primers 593 and 390, which are complementary to universal regions U1 and U5, respectively (21). The PCR was performed in a Perkin-Elmer thermal cycler using the following temperature profile: 3 min at 96°C, 30 cycles consisting of 96°C for 15 s and 70°C for 2 min, and termination by cooling at 4°C. Following agarose gel electrophoresis, the PCR products were purified with a GFXPCR DNA and Gel Band Purification Kit according to the instructions of the manufacturer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). The purified PCR products were used in sequence reactions which were performed with the Thermo Sequenase Cy5 Dye Terminator Kit according to the instructions of the manufacturer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.), using a biotinylated primer (593-B) and the same temperature profile as described above. The sequencing reaction products were purified by applying streptavidin-coated superparamagnetic beads (Dynabeads M-280 Streptavidin; Dynal Biotech, Oslo, Norway) prior to loading the sequencing gel. Subsequent sequence determination was performed in an ALFexpressII instrument according to the instructions of the manufacturer (Amersham Pharmacia Biotech AB, Uppsala, Sweden).
TABLE 1.
Vaginal isolates derived from 23 women by the different genotypic approaches
| Subject | RAPD type | Species designation by:
|
||
|---|---|---|---|---|
| TTGEa | Multiplex PCRb | Sequencing | ||
| A | 15 | >1 bandc | L. crispatus | L. crispatus |
| B | 21 | L. jensenii | L. jensenii | NDd |
| 15 | >1 band | L. crispatus | L. crispatus | |
| C | 10 | L. crispatus | L. crispatus | ND |
| D | 4 | L. gasseri | L. gasseri | ND |
| E | 1 | L. gasseri | L. gasseri | ND |
| F | 3 | L. gasseri | L. gasseri | ND |
| Ge | 10 | >1 band | L. crispatus | L. crispatus |
| 12 | >1 band | L. crispatus | L. crispatus | |
| He | 9 | >1 band | L. crispatus | L. crispatus |
| 16 | >1 band | L. crispatus | L. crispatus | |
| I | 19 (L. iners) | L. iners | ND | ND |
| J | 6 (L. jensenii)f | L. jensenii | L. jensenii | ND |
| K | 17 | L. crispatus | L. crispatus | ND |
| L | 11 | L. gasseri | L. gasseri | ND |
| M | 5 | L. gasseri | L. gasseri | ND |
| 8 | L. crispatus | L. crispatus | ND | |
| N | 7 | >1 band | L. crispatus | L. crispatus |
| O | 19 (L. iners) | L. iners | ND | ND |
| P | 14 | L. gasseri | L. gasseri | ND |
| Q | 8 | >1 band | L. crispatus | L. crispatus |
| R | 6 (L. jensenii) | L. jensenii | L. jensenii | ND |
| S | 7 | >1 band | L. crispatus | L. crispatus |
| T | 19 (L. iners) | L. iners | ND | ND |
| U | 20 (L. iners) | L. iners | ND | ND |
| V | 10 | >1 band | L. crispatus | L. crispatus |
| 3 | L. jensenii | L. jensenii | ND | |
| W | 2 | L. gasseri | L. gasseri | ND |
See Fig. 1.
The isolates were grouped first by PCR-G and then identified to the species level by PCR II-1 and PCR II-2 (27).
The TTGE showed a pattern instead of a single band.
ND, not determined.
The subject showed two different L. crispatus strains.
The isolate has the same RAPD pattern as the type strain for L. jensenii.
RESULTS
Two hundred two isolates from 23 subjects with healthy vaginal mucosa were divided into 21 different RAPD types (Table 1). The RAPD types isolated from blood agar (subjects I, O, T, and U) showed no similarity with those of any of the isolates from Rogosa agar but had high similarity with the RAPD pattern from the type strain of L. iners (Table 1). No single subject was dominated by more than two RAPD types. In a majority of the individuals, only one RAPD type was found. Some of the RAPD types were recurrent; in addition, two subjects (G and H) harbored the same species but with different RAPD types (Table 1).
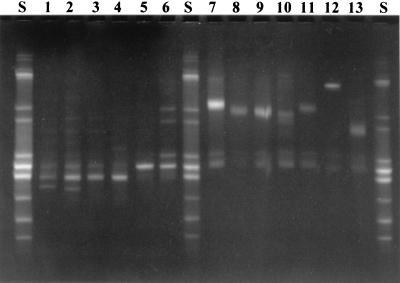
Most of the vaginal RAPD types could be identified to the species level by comparing the TTGE patterns of the isolates with those of the type strains (Table 1). The similarity in TTGE patterns within different isolates of the same RAPD type was usually high. Exceptions were the patterns for L. crispatus isolates A, B, G, H, N, Q, S, and V, which showed heterogeneity, and for L. gasseri isolates, which had one general TTGE pattern but showed a slight deviation in the position of the bands (Fig. 1 and Table 1).
FIG. 1.
TTGE patterns of representative vaginal Lactobacillus strains. Lanes S, standard. Lanes 1 to 6, L. crispatus strains: 1, 1B7; 2, 8R5; 3, 8R4; 4, 16B2; 5, 18 M7; 6, 26R6. Lanes 7 to 11, L. gasseri strains: 7, 5B1; 8, 6 M8; 9, 15R5; 10, 16R6; 11, 20 M47. Lane 12, L. jensenii strain 2RB5, Lane 13, L. iners strain 25B31.
The multiplex PCR enabled the identification to the species level of vaginal isolates belonging to L. crispatus, L. gasseri, and L. jensenii but not L. iners (Table 1).
L. crispatus isolates A, B, G, H, N, Q, S, and V, which showed heterogeneity in the TTGE, were sequenced and identified by using the BLAST program in GenBank.
Using the combined results of the different methods, the complete identification showed that L. crispatus, L. gasseri, L. iners, and L. jensenii were the most frequently occurring species in the healthy vaginas of 23 Swedish women (Table 1).
DISCUSSION
The predominant species of Lactobacillus found in the vaginas of healthy women as determined by reliable laboratory criteria (those of Nugent et al. [19]) have never been established before with the present genotypic approaches. RAPD analysis was used in this study to group the isolates and to observe the heterogeneity of the Lactobacillus flora of each subject. Most of the subjects had a very homogeneous flora dominated by a single RAPD type (Table 1). RAPD analysis has previously been used to differentiate separate strains of Lactobacillus spp. (10). However, comparing the strikingly unique RAPD patterns of the isolates that only could grow on blood agar (I, O, T, and U) with that of the type strain of L. iners directly identified this species (Table 1).
The different RAPD types were identified to the species level by TTGE by comparison of the TTGE profiles with those of the type strains. An exception where the identification by TTGE failed was a group of strains (A, B, G, H, N, Q, S, and V) that showed a high degree of heterogeneity in the TTGE patterns (Fig. 1 and Table 1). This could be due to heterogeneities in the 16S rRNA gene, a phenomenon previously described by Nübel et al. (20). Nevertheless, these isolates could be identified as L. crispatus, both by multiplex PCR and by 16S rDNA sequencing. The sequence data on the vaginal isolates of L. crispatus had high similarity with some newer sequence data reported by Kilic et al. (12).
In addition to the finding of L. iners, our results are in general agreement with other studies, showing a predominance of species within the L. acidophilus complex and the fact that only one or a few Lactobacillus species colonize the healthy human vagina. Antonio et al. (2) indicated L. crispatus, L. jensenii, and L. gasseri as three of the four most common species encountered in the vagina, applying whole-chromosomal DNA probes. Lachlak et al. (15) showed a dominance of group B of the L. acidophilus complex in asymptomatic women, but this was not the case in the present work, where L. crispatus was the predominant species of the complex.
L. iners is a rather newly described species (7). To our knowledge, L. iners has been reported only once before as being present in the vagina, by the same investigators who originally described the species (7). One explanation for the absence of L. iners in other studies could be that workers searching for lactobacilli do not usually examine isolates that can grow only on blood agar and not on typical Lactobacillus media such as Rogosa or MRS agar.
Species such as L. rhamnosus, L. fermentum, L. plantarum, and L. acidophilus have frequently been recovered from the vagina (23, 24, 30). None of them were found in the present study, and neither was the recently described species Lactobacillus fornicalis, which was isolated from the posterior fornix of the human vagina (5). The differences in Lactobacillus flora between studies may be attributed to a number of factors. One suggestion is that the intestinal lactobacilli differ in western countries and Japan (26), and the same may be true for the vagina (23). However, the difference in the Lactobacillus flora is unlikely to represent geographical variation alone. More likely explanations are variations in the way that samples are taken and treated, the vaginal status, or the fact that identification has often been based on phenotypic methods (18, 24). Phenotypic criteria are particularly unreliable for identification of members of the L. acidophilus complex and related species (13).
Grouping with RAPD and identification of representative isolates with TTGE and multiplex PCR were found to be a convenient way to identify the normal Lactobacillus flora of the vagina. Both TTGE and multiplex PCR are simpler to perform than 16S rDNA sequencing (14) or ribotyping (30).
REFERENCES
- 1.Amsel, R., P. A. Totten, C. A. Spiegel, K. C. Chen, D. Eschenbach, and K. K. Holmes. 1983. Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74:14-22. [DOI] [PubMed] [Google Scholar]
- 2.Antonio, M. A. D., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950-1956. [DOI] [PubMed] [Google Scholar]
- 3.Bohak, I., W. Back, L. Richter, M. Ehrmann, W. Ludwig, and K. H. Schleifer. 1998. Lactobacillus amylolyticus sp. nov., isolated from beer malt and beer wort. Syst. Appl. Microbiol. 21:360-364. [DOI] [PubMed] [Google Scholar]
- 4.Boris, S., J. E. Suárez, F. Vázquez, and C. Barbés. 1998. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 66:1985-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dicks, L. T. M., M. Silvester, P. A. Lawson, and M. D. Collins. 2000. Lactobacillus fornicalis sp. nov., isolated from the posterior fornix of the human vagina. Int. J. Syst. E vol. Microbiol. 50:1253-1258. [DOI] [PubMed] [Google Scholar]
- 6.Efthymiou, C., and C. A. Hansen. 1962. An antigenic analysis of Lactobacillus acidophilus. J. Infect. Dis. 110:258-267. [DOI] [PubMed] [Google Scholar]
- 7.Falsen, E., C. Pascual, B. Sjödén, M. Ohlén, and M. D. Collins. 1999. Phenotypic and phylogenetic characterisation of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int. J. Syst. Bacteriol. 49:217-221. [DOI] [PubMed] [Google Scholar]
- 8.Fujisawa, T., Y. Benno, T. Yaeshima, and T. Mitsuoka. 1992. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int. J. Syst. Bacteriol. 42:487-491. [DOI] [PubMed] [Google Scholar]
- 9.Gancheva, A., B. Pot, K. Vanhonacker, B. Hoste, and K. Kersters. 1999. A polyphasic approach towards the identification of strains belonging to Lactobacillus acidophilus and related species. Syst. Appl. Microbiol. 22:573-585. [DOI] [PubMed] [Google Scholar]
- 10.Johansson, M.-L., M. Quednau, G. Molin, and S. Ahrné. 1995. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett. Appl. Microbiol. 21:155-159. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. L., C. F. Phelps, C. S. Cummins, J. London, and F. Gasser. 1980. Taxonomy of the Lactobacillus acidophilus group. Int. J. Syst. Bacteriol. 30:53-68. [Google Scholar]
- 12.Kilic, A. O., S. I. Pavlova, S. Alpay, S. S. Kilic, and L. Tao. 2001. Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin. Diagn. Lab. Immunol. 8:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein, G., A. Pack, C. Bonaparte, and G. Reuter. 1998. Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 41:103-125. [DOI] [PubMed] [Google Scholar]
- 14.Kullen, M. J., R. B. Sanozky-Dawes, D. C. Crowell, and T. R. Klaenhammer. 2000. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511-516. [DOI] [PubMed] [Google Scholar]
- 15.Lachlak, N., E. Ageron, O. Zampatti, G. Michel, and P. A. D. Grimont. 1996. Composition of the Lactobacillus acidophilus complex isolated from vaginal flora. Microbiologica 19:123-132. [PubMed] [Google Scholar]
- 16.Lauer, E., C. Helming, and O. Kandler. 1980. Heterogeneity of the species Lactobacillus acidophilus (Moro) Hansen and Mocquot as revealed by biochemical characteristics and DNA-DNA hybridization. Zentbl. Bakteriol. Microbiol. Hyg. Abt. 1 Orig. C 1:150-168. [Google Scholar]
- 17.Martin, H. L., B. A. Richardson, P. M. Nyange, L. Lavreys, S. L. Hillier, B. Chohan, K. Mandaliya, J. O. Ndinya-Achola, J. Bwayo, and J. Kreiss. 1999. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J. Infect. Dis. 180:1863-1868. [DOI] [PubMed] [Google Scholar]
- 18.McLean, N. W., and I. J. Rosenstein. 2000. Characterisation and selection of a Lactobacillus species to re-colonise the vagina of women with recurrent bacterial vaginosis. J. Med. Microbiol. 49:543-552. [DOI] [PubMed] [Google Scholar]
- 19.Nugent, R. P., M. A. Krohn, and S. L. Hillier. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of Gram stain interpretation. J. Clin. Microbiol. 29:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersson, B. 1997. Direct solid-phase 16S rDNA sequencing: a tool in bacterial phylogeny. Ph.D. thesis. Royal Institute of Technology, Stockholm, Sweden.
- 22.Quednau, M., S. Ahrné, A. C. Petersson, and G. Molin. 1998. Identification of clinically important species of Enterococcus within 1 day with randomly amplified polymorphic DNA (RAPD). Curr. Microbiol. 36:332-336. [DOI] [PubMed] [Google Scholar]
- 23.Redondo-Lopez, V., R. L. Cook, and J. D. Sobel. 1990. Emerging role of Lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev. Infect. Dis. 12:856-872. [DOI] [PubMed] [Google Scholar]
- 24.Reid, G., J. A. McGroarty, L. Tomeczek, and A. W. Bruce. 1996. Identification and plasmid profiles of Lactobacillus species from the vagina of 100 healthy women. FEMS Immunol. Med. Microbiol. 15:23-26. [DOI] [PubMed] [Google Scholar]
- 25.Schleifer, K. H., and W. Ludwig. 1995. Phylogeny of the genus Lactobacillus and related genera. Syst. Appl. Microbiol. 18:461-467. [Google Scholar]
- 26.Song, Y.-L., N. Kato, Y. Matsumiya, C.-X. Liu, H. Kato, and K. Watanabe. 1999. Identification of and hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J. Clin. Microbiol. 37:3062-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, Y.-L., N. Kato, C.-X. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 28.Spiegel, C. A. 1991. Bacterial vaginosis. Clin. Microbiol. Rev. 4:485-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vásquez, A., S. Ahrné, B. Pettersson, and G. Molin. 2001. Temporal temperature gradient gel electrophoresis (TTGE) as a tool for identification of Lactobacillus casei, Lactobacillus paracasei, Lactobacillus zeae and Lactobacillus rhamnosus. Lett. Appl. Microbiol. 32:215-219. [DOI] [PubMed] [Google Scholar]
- 30.Zhong, W., K. Millsap, H. Bialkowska-Hobrzanska, and G. Reid. 1998. Differentiation of Lactobacillus species by molecular typing. Appl. Environ. Microbiol. 64:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]