Abstract
γ-Glutamyl aminopeptidase (GGT) activity is used as a specific marker for the identification of Neisseria meningitidis. We isolated from a healthy carrier and characterized an N. meningitidis isolate which lacked the activity due to the insertional mutation of the ggt gene, suggesting that naturally occurring N. meningitidis isolates do not always possess GGT activity.
Neisseria meningitidis is a gram-negative diplococcus pathogen that colonizes the nasopharynx. Selective media such as modified Thayer-Martin medium and New York City medium are usually used for the isolation of pathogenic neisseriae from swabs of the nasopharynx, where N. meningitidis, Neisseria gonorrhoeae, Neisseria lactamica, and sometimes Branhamella catarrhalis would exist. Further identification is performed on the basis of the biochemical properties listed in Table 1. Of the biochemical properties used for classification, γ-glutamyl aminopeptidase (EC 2.3.2.2; GGT) is considered a specific marker for N. meningitidis because its activity is positive in N. meningitidis but not in the other three species mentioned above (2). In fact, the detection of GGΤ activity is used by the Gonochek II enzymatic identification system (E-Y Laboratories Inc., San Mateo, Calif.) in the identification of N. meningitidis (1, 5, 6, 11). Here, we report the isolation and characterization of a natural isolate of N. meningitidis that is defective in GGT activity.
TABLE 1.
Biochemical properties of Neisseria strains
| Property | Result for straina
|
|
|---|---|---|
| NIID113 | NIID113/pHT198b | |
| Oxidase | + | + |
| Catalase | + | + |
| Fermentation of sugar | ||
| Glucose | + | + |
| Maltose | + | + |
| Sucrose | − | − |
| Levulose | − | − |
| Lactose | − | − |
| Nitrate reductase | − | − |
| Nitrite reductase | − | − |
| Production of polysaccharide | − | − |
| Gonochek II | ||
| Prolyl imonopeptidase | − | − |
| γ-Glutamyl aminopeptidase | − | + |
| β-Galactosidase | − | − |
+, reaction positive; −, reaction negative.
pHT198 is pHT128(9) carrying a wild-type ggt gene amplified with primers ggt-3 (GACTGCTGATGACATTAGCGG) and ggt-4 (GATTACTCACAATTTCCCCCTA) and using H44/76 chromosomal DNA as a template.
During an investigation of healthy carriers of N. meningitidis in Japan, N. meningitidis strain NIID113 was isolated from a 19-year-old healthy male by selection on modified Thayer-Martin medium using swabs of his nasopharynx. After confirming the oxidase-positive and gram-negative phenotypes, we examined the profile of biochemical properties of NIID113, such as sugar fermentation, which identified this strain as N. meningitidis (Table 1). The serogroup could not be specified because of the absence of agglutination with any kind of antisera (Difco), but it was suspected to be serogroup B based on a PCR method (8). In contrast to the results of the biochemical property assessment, the strain was identified as B. catarrhalis when it was assayed by the Gonochek II system (Table 1). The sole difference in the results of the Gonochek II assay from the criteria of biochemical properties for N. meningitidis was the absence of GGT activity. Consequently, we further studied the ggt gene.
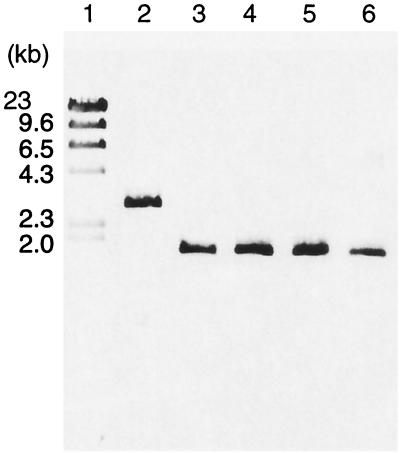
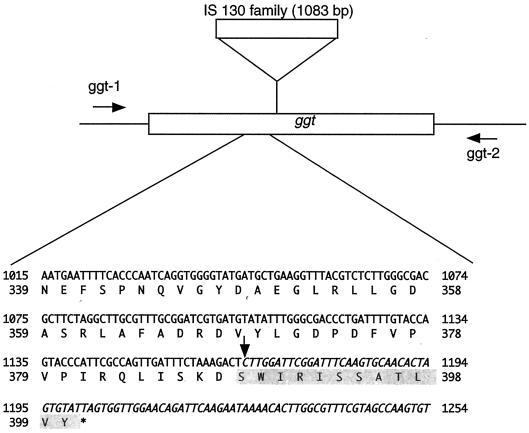
At first, to characterize a whole coding region of ggt, we tried to amplify a ggt allele by PCR with primers ggt-1 (ATGCCCTTGTATGGATCA) and ggt-2 (CTAATCACCCATCACTCGACCT) and ExTaq DNA polymerase (Takara Shuzo) and using purified chromosomal DNA as a template in GeneAmp PCR system 2400 (Applied Biosystems). Although a 1.8-kb fragment was amplified from H44/76, a typical N. meningitidis strain belonging to the electrophoretic type 5 complex isolated in Norway in the 1970s (kindly provided by L. O. Frøholm), a 2.9-kb fragment was amplified from NIID113 (Fig. 1). Amplified PCR products of the same size (1.8 kb) were observed among other GGT-positive clinical isolates of N. meningitidis (Fig. 1). This result suggests that the ggt gene of NIID113 was inactivated by an insertional mutation. To further analyze the ggt alleles, we performed sequencing with a BigDye Terminator Cycle Sequencing Ready Reaction kit (version 2.0; PE Biosystems) according to the manufacturer's protocol and using the PCR products as templates and an ABI PRISM 310 genetic analyzer (PE Biosystems). From the sequence data (DDBJ accession number AB084259), it was found that a 1,083-bp fragment of insertion sequence (IS) (DDBJ accession number AB076582) was inserted 1,165 bp downstream from the start codon of ggt, resulting in the loss of 206 C-terminal amino acid residues of GGT and the addition of 12 foreign amino acid residues (Fig. 2). The IS was identical to one that is widely distributed in the N. meningitidis genome (10). The ggt gene product, GGT, was not expressed in NIID113; this finding was confirmed by Western blotting with anti-GGT rabbit serum (data not shown). Furthermore, GGT activity was recovered by the introduction of pHT198, which carries a wild-type ggt gene on the pHT128 vector (9), into NIID113 (Table 1), suggesting that the defect in GGT activity was due to the insertional mutation of the ggt gene.
FIG. 1.
PCR amplification of the ggt gene from NIID113 (lane 2), H44/76 (lane 3), and other N. meningitidis clinical isolates, namely, NIID57 (lane 4), NIID99 (lane 5), and NIID100 (lane 6). Molecular mass markers are shown in lane 1.
FIG. 2.
Insertional site of IS in ggt. The vertical arrow behind the T residue (position 1165) indicates the insertional site of the IS. The IS sequence is shown in italics. Amino acid residues shaded in gray indicate the additional amino acid residues derived from the insertional sequence. An asterisk indicates the stop codon.
This is the first reported characterization of an N. meningitidis isolate from a healthy carrier with a deficiency in GGT activity resulting from an insertional mutation of the ggt gene. Since strains with carbohydrate degradation profiles that are inconsistent with the identification of Neisseria spp. have been occasionally found among Neisseria isolates (3, 4, 7), the detection of GGT activity would be a powerful and helpful method for identifying and differentiating Neisseria spp. However, researchers should carefully interpret a finding of GGT activity when the identification of N. meningitidis depends mainly on this characteristic, since the ggt gene in N. meningitidis strains can be inactivated by an insertional mutation in the strain's natural habitat, as shown by this study.
Nucleotide sequence accession numbers. The nucleotide sequences of ggt allele and IS in NIID113 have been deposited in the DDBJ/EMBL/Genbank nucleotide sequence database under accession numbers AB084259 and AB076582, respectively.
Acknowledgments
This work was supported by grants from the Ministry of Health, Welfare and Labor.
REFERENCES
- 1.Brown, J. D., and K. R. Thomas. 1985. Rapid enzyme system for the identification of pathogenic Neisseria spp. J. Clin. Microbiol. 21:857-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Amato, R. F., L. A. Eriquez, K. M. Tomfohrde, and E. Singerman. 1978. Rapid identification of Neisseria gonorrhoeae and Neisseria meningitidis by using enzymatic profiles. J. Clin. Microbiol. 7:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granato, P. A., R. Howard, B. Wilkinson, and J. Laser. 1980. Meningitis caused by maltose-negative variant of Neisseria meningitidis. J. Clin. Microbiol. 11:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoke, C., and N. A. Vedros. 1982. Characterization of atypical aerobic gram-negative cocci isolated from humans. J. Clin. Microbiol. 15:906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janda, W. M., M. G. Ulanday, M. Bohnhoff, and L. J. LeBeau. 1985. Evaluation of the RIM-N, Gonochek II, and Phadebact systems for the identification of pathogenic Neisseria spp. and Branhamella catarrhalis. J. Clin. Microbiol. 21:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philip, A., and G. C. Garton. 1985. Comparative evaluation of five commercial systems for the rapid identification of pathogenic Neisseria species. J. Clin. Microbiol. 22:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saez-Nieto, J. A., A. Fenoll, J. Vazquez, and J. Casal. 1982. Prevalence of maltose-negative Neisseria meningitidis variants during an epidemic period in Spain. J. Clin. Microbiol. 15:78-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taha, M. K. 2000. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J. Clin. Microbiol. 38:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi, H., and H. Watanabe. 2002. Broad-host-range vector of incompatibility group Q can work as a plasmid vector in Neisseria meningitidis: a new finding as a genetical tool. Microbiology 148:229-236. [DOI] [PubMed] [Google Scholar]
- 10.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 11.Welborn, P. P., C. T. Uyeda, and N. Ellison-Birang. 1984. Evaluation of Gonochek-II as a rapid identification system for pathogenic Neisseria species. J. Clin. Microbiol. 20:680-683. [DOI] [PMC free article] [PubMed] [Google Scholar]