Abstract
Current methods for producing cardiomyocytes from human induced pluripotent stem cells (hiPSCs) using 2D monolayer differentiation are often hampered by batch-to-batch variability and inefficient purification processes. Here, we introduce CM-AI, a novel artificial intelligence-guided laser cell processing platform designed for rapid, label-free purification of hiPSC-derived cardiomyocytes (hiPSC-CMs). This approach significantly reduces processing time without the need for chronic metabolic selection or antibody-based sorting. By integrating real-time cellular morphology analysis and targeted laser ablation, CM-AI selectively removes non-cardiomyocyte populations with high precision. This streamlined process preserves cardiomyocyte viability and function, offering a scalable and efficient solution for cardiac regenerative medicine, disease modeling, and drug discovery.
Subject terms: Stem-cell research, Cardiac regeneration, Stem-cell differentiation, Stem-cell biotechnology, Heart stem cells
AI based recognition of human stem cell derived cardiomyocytes combined with laser ablation of unwanted cells hastens cell manufacturing and improves cryopreservation viability.
Introduction
Human cardiomyocytes (CMs) can now be derived in virtually unlimited quantities using patient-specific human induced pluripotent stem cells (hiPSCs), enabling transformative advances in cardiac disease modeling, drug screening, and regenerative medicine1–3. 2D monolayer-based directed cardiac differentiation protocols have become widely adopted due to their relative simplicity and efficiency, with reported conversion yields of up to 95%4,5. However, differentiation efficiency is often inconsistent across batches (ranging from ~40 to 95%), necessitating downstream purification of CMs for most applications. Although 3D bioreactor and spinner flask-based formats show promise for large-scale hiPSC-CM production6, these approaches require complex engineering devices, and spinning action can produce detrimental shear stress on cells7. As such, 2D monolayer based hiPSC-CM production approaches remain the practical standard in most laboratories. Many 2D monolayer-based CM purification approaches have been developed for hiPSC-CMs over the years, but current approaches are not ideal for large-scale production and cryopreservation of healthy CMs with normal responses to cardioactive drugs8,9.
To address the need for purified hiPSC-CMs, a variety of methods have been explored. Fluorescent-based CM purification approaches have been employed with direct chemical labeling of mitochondria10, or via genetic modification of CMs to express green fluorescent protein (GFP) with subsequent fluorescent activated cell sorting (FACS)11. FACS-based enrichment of hiPSC-CMs, however, is not feasible for large-scale production due to significant loss of CMs and long processing times. Magnetic activated cell sorting (MACS) approaches have also been developed relying on cell-specific extracellular antigens that can be targeted with antibodies in order to purify hiPSC-CMs by positive or negative selection12–14. However, MACS-based sorting of hiPSC-CMs is not ideal for large-scale cell production owing to laborious multi-step cell processing procedures that can result in low cell yield and variable, user-dependent success rates15. The most commonly used CM purification approach to date for hiPSC-CM 2D monolayers is a metabolic selection media consisting of zero glucose and abundant lactate16. This metabolic selection media formulation relies on the unique CM ability to utilize lactate for energy production while most non-CMs, possessing fewer mitochondria, die. Although metabolic selection media is effective and popular for CM purification, it has been used historically to model myocardial ischemia in vitro17, and chronic long-term exposure ≥7 days produces 2D hiPSC-CMs possessing a heart failure-like functional and structural phenotype13,15. Thus there is a need for new methods of healthy hiPSC-CM production, purification, and processing in 2D monolayers. Production of healthy hiPSC-CMs is critical to the goals of recently enacted US legislation called the FDA Modernization Act 2.0/3.0, which is focused on the use of human-relevant in vitro approaches to replace animal testing for drug development and enhancement of the current clinical trial paradigm18,19.
Artificial intelligence (AI) is being implemented to enhance hiPSC research and cell processing20,21. Here, we introduce CM-AI, an AI-guided laser cell processing platform that enables rapid, label-free purification of hiPSC-CMs based on morphological features captured via phase-contrast imaging. For this, we utilized a cell processing device with deep machine learning capabilities from Kataoka Corporation (CPD-017, Kyoto, Japan), designed for laser-mediated ablation of unwanted cells in 2D monolayers as described before22. CM-AI provides an alternative hiPSC-CM purification and processing approach for production of healthy cardiomyocytes. Importantly, CM-AI enables shorter processing times compared to other hiPSC-CM processing protocols and can be executed by a wide range of users, enabling a larger number of lab personnel to contribute to cell production. We provide a careful validation of hiPSC-CM health, structure, cryopreservation, electrophysiological function, applications for 3D engineered heart tissue (3D EHT) generation, and drug responsiveness following CM-AI purification and processing.
Results and discussion
To train and validate the CM AI-based recognition of unpurified hiPSC-CMs in 2D monolayers, we utilized an established CRISPR/Cas9-engineered cell line with CM-specific GFP expression23. The TTN-GFP hiPSC cell line was differentiated to atrial chamber-specific cardiomyocytes using an established small molecule protocol24,25, and GFP+ CMs could be detected by day 8 of the protocol (Supplementary Fig. 1c, d). Training data was acquired between days 8 and 13 of atrial specific hiPSC-CM differentiation and included GFP images with corresponding overlaid phase contrast images for CM discrimination by CM-AI. Following training, CM-AI relies only on phase contrast images for CM identification, which enables CM-AI application to other hiPSC-CM cell lines including those without a genetic fluorescent tag. The use of CM-AI is a simple two-step process: Step 1. Image entire well, CM-AI detects cardiomyocytes and programs laser ablation of the non-myocyte population. Step 2. Following laser ablation, enriched hiPSC-CMs can be further maintained in culture and processed for downstream assays like electrophysiological screening. Step 1 requires 1 min for whole well image acquisition and AI recognition of the CM population, plus up to 7 min for laser ablation of the non-CM population in a single well, depending on the number of unwanted cells. Thus, CM-AI processing of a single six-well plate requires ~48 min in total. CM-AI also provides a quantitative report of the percentages of hiPSC-CMs (wanted cell population) and the non-myocytes (unwanted cell population) that are targeted for laser ablation in each well (Supplementary Figs. 2 and 3). CM-AI can also be programmed to process all six wells automatically, enabling the user to walk away while CPD-017 performs cell purifications. During CM-AI processing the plate of cells is housed within a built-in incubator (37 °C, 5% CO2) for stable maintenance of cell culture conditions. A Movie of contracting hiPSC-CMs following CM-AI and laser ablation of non-myocytes initially confirmed survival and health of the wanted CM cell population (Supplementary Movie 1).
For an initial validation, we quantified the hiPSC-CM GFP confluence area before and after laser ablation (Fig. 1c). On average, using CM-AI there was a 2.4% reduction of GFP area confluence, indicating a small loss of the CM population with processing. Importantly, following CM-AI processing and replating, the GFP confluence area represented greater than 85% of the total cell area per monolayer (Fig.1d). Further, using flow cytometry to count GFP+ cells we found that CM-AI purification was comparable to the purity achieved using the traditional metabolic media selection approach (Supplementary Fig. 4). hiPSC-CM purification following CM-AI processing was also validated by flow cytometry analysis using cardiac troponin T (cTnT) specific antibodies conjugated to Allophycocyanin (APC, Fig. 2). Using the TTN-GFP hiPSC-CM cell line, the average purity of three separate differentiations ranged from 91 to 93% cTnT+ cells (92.0 ± 1.0%, n = 3). Applying the CM-AI approach to non-labeled hiPSC-CMs was also successful (Fig.2b, c). Using a standard control hiPSC line without a genetic tag (19.9.11 hiPSC cell line), the average purity ranged from 86 to 99% cTnT+ cells (94.3 ± 7.2%, n = 3). Importantly, no expression of pluripotent stem cell genetic markers (NANOG and POU5F1, Supplementary Table 1) was detected by qRT-PCR in hiPSC-CM populations following CM-AI purification and processing.
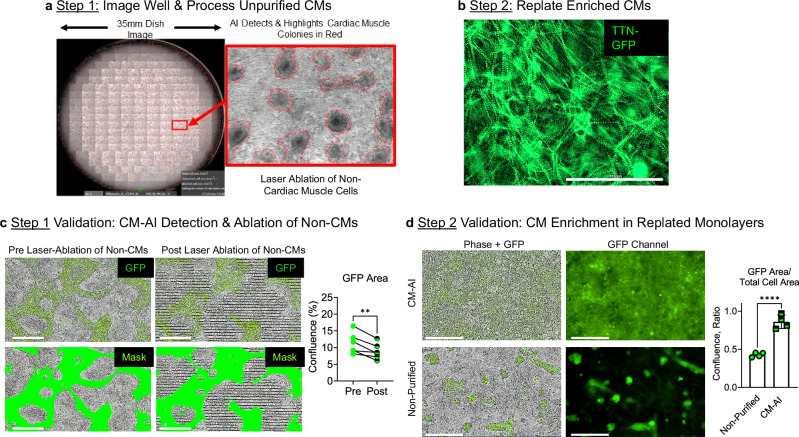
Fig. 1. CM-AI automated detection of CMs and laser ablation of non-myocytes in 2D monolayers.
a Step 1 in this hiPSC-CM enrichment process is to acquire a phase contrast image of a well of cardiac differentiation with AI detection of CM colonies (red). b Step 2 in this easy-to-use process is replating enriched hiPSC-CMs. c Phase contrast and fluorescent imaging of GFP+ CMs enrichment in 2D monolayers. Successful AI recognition of CMs in phase contrast images was validated using time-lapse imaging of monolayers pre-AI laser processing and post-AI laser processing. The overall GFP confluence did decrease significantly in six independent wells processed using CM-AI-laser ablation of the non-myocyte (GFP-) cell population (Pre GFP confluence = 11.23 ± 3.2%; Post GFP confluence = 8.88 ± 2.4%; n = 6 paired t-test, **P = 0.009). The green Mask was used for quantification of the GFP confluence area before and after CM-AI laser processing. d Validation of CM enrichment in replated monolayers. Following CM-AI laser processing and CM enrichment, CMs were replated on CELLvo™ MatrixPlus human ECM-coated plates for analysis. CM-AI laser-processed wells reached purity (>85%) levels significantly greater than unprocessed wells. (n = 4 monolayers per group; unpaired t-test; ****P < 0.0001).
Fig. 2. Validation of hiPSC-CM purity after CM-AI processing of two separate cell lines by flow cytometry.
a Flow cytometry plots of GFP + TTN-GFP hiPSC-CMs following CM-AI purification stained for cardiac troponin T (cTnT-APC). b Flow cytometry of untagged 19.9.11 hiPSC-CMs stained with cTnT-APC following CM-AI purification. c Summary of cTnT+ cell percentages from three independent differentiations using TTN-GFP and 19.9.11 hiPSC lines (n = 3 per line). Average purity was 92.0 ± 1.0% and 94.3 ± 7.2%, respectively. No residual expression of pluripotency markers (NANOG, POU5F1) detected post-CM-AI processing (qRT-PCR; Supplementary Table 1). Data are mean ± s.d.
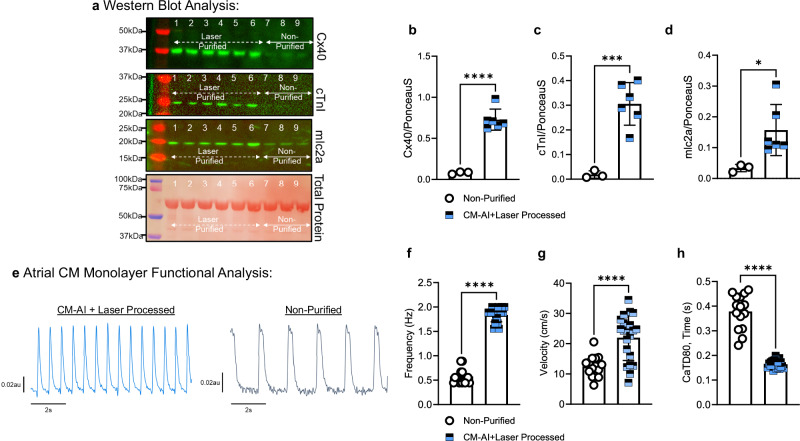
We analyzed the protein expression and function of CM-AI processed and replated atrial hiPSC-CM monolayers (Fig. 3). Western blot analysis revealed that atrial CM-specific protein expression was significantly enriched in CM-AI processed monolayers relative to non-purified control wells (Fig. 3a–d). For example, the cardiac atrial specific intercellular gap junction protein, connexin 40 (Cx40), was significantly enriched in CM-AI processed monolayers. Similarly, the CM-specific myofilament proteins, myosin light chain 2a (mlc2a) and cardiac troponin I (cTnI), were significantly enriched in CM-AI processed monolayers. To determine the impact of CM-AI purification on hiPSC-CM monolayer function, spontaneous pacemaker activations were recorded using a calcium-sensitive fluorophore as recently described25. CM-AI laser processed cells formed electrically connected monolayers with faster spontaneous beat rate, conduction velocity, and shorter calcium transient duration (CaTD80) than non-purified monolayers (Fig. 3e–h). Spontaneous activations and calcium transients of CM-AI purified and processed hiPSC-CM monolayers can be viewed in Supplementary Movie 2.
Fig. 3. Acute structural and electrical functional analysis of CM-AI purified hiPSC-CM monolayers.
a Western blot analysis of atrial-specific CM proteins in purified versus unpurified monolayers. CM-AI + Laser purified monolayers (n = 6, Lanes 1–6) showed significant enrichment of Cx40, cTnI, and mlc2a protein expression compared to time and batch-matched non-purified monolayers. b Cx40 protein expression was significantly enriched in laser-purified hiPSC-CM monolayers compared to non-purified monolayers (0.73 ± 0.13au; n = 6 vs. 0.08 ± 0.01au; n = 3; ****P < 0.0001, unpaired t-test). c cTnI protein expression was significantly greater in laser-purified monolayers compared to non-purified hiPSC-CM atrial monolayers (0.31 ± 0.09au; n = 6 vs. 0.02 ± 0.01au; n = 3; ***P = 0.0009, unpaired t-test). d mlc2a myofilament protein expression was significantly enriched in laser-purified hiPSC-CM monolayers compared to non-purified monolayers (0.16 ± 0.08au; n = 6 vs. 0.03 ± 0.01; n = 3; *P = 0.04, unpaired t-test). e Atrial hiPSC-CM monolayer function was quantified using calcium-sensitive dye (Calbryte 520AM, 5 μM) and high-resolution optical mapping. Representative spontaneous calcium flux traces from re-plated CM-AI + laser processed monolayers (blue) and replated non-purified monolayers (black). f CM-AI + laser processed monolayers had faster spontaneous beat rate compared to non-purified monolayers (1.84 ± 0.14 Hz; n = 21 monolayers vs. 0.58 ± 0.14 Hz; n = 16 monolayers; ****P < 0.0001, unpaired t-test). g Calcium wave propagation velocity was faster in CM-AI laser processed monolayers compared to non-purified monolayers (22.0 ± 7.6 cm/s; n = 21 monolayers vs. 12.5 ± 3.5 cm/s; n = 15 monolayers; ****P < 0.0001). h Calcium transient duration 80 (CaTD80) was significantly shorter in CM-AI laser processed monolayers compared to non-purified monolayers (0.164 ± 0.02 s; n = 21 vs. 0.379 ± 0.07 s; n = 15; ****P < 0.0001, unpaired t-test).
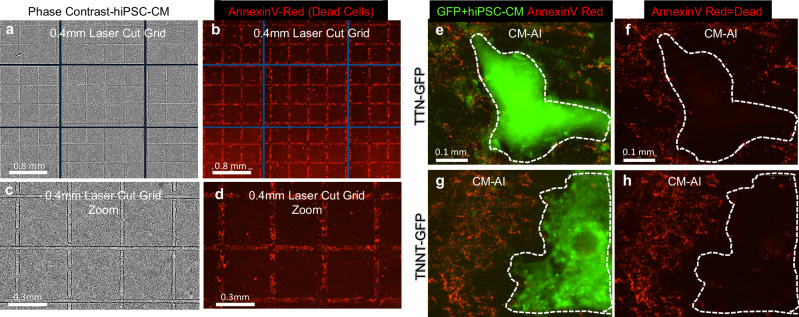
Next, we determined the health of hiPSC-CMs acutely following CM-AI and laser processing. AnnexinV reagent was utilized to assess cell health following laser irradiation using two different approaches. First, we tested the accuracy and precision of laser-induced cell killing by programming the laser to cut grids (0.4 mm squares, Fig. 4a–d) on an hiPSC-CM monolayer with subsequent AnnexinV staining to determine the location of dead cells. We found that cell death was limited to the grid lines of programmed laser irradiation (Fig. 4b, d). Second, we performed AnnexinV staining following CM-AI purification and processing using two separate GFP fluorescent tagged hiPSC-CM lines (Fig. 4e–h). In each experiment, AnnexinV-positive cells were restricted to the non-CM areas of 2D monolayers, indicating good health of the hiPSC-CM population. Further confirmation of laser ablation efficiency and health of remaining hiPSC-CMs was determined by optical mapping spontaneous calcium waves using a high-speed acquisition system as before (Fig.5)25,26. Following custom laser ablation in the shape of 2 block “Ms” on a 100% confluent hiPSC-CM monolayer, the remaining live hiPSC-CMs continued to contract rhythmically as a syncytium with the spontaneous calcium activation waves navigating around the laser-ablated regions (Supplementary Movie 3). This indicates functional integrity and health of the electrically and mechanically connected hiPSC-CMs monolayer regions outside of the laser-irradiated areas. In the future, custom patient-specific in vitro myocardial scarring diagnostic models may be implemented using this laser ablation approach for clinically translatable anti-arrhythmic medication screening and arrhythmia source predictions to guide radiofrequency ablation therapies.
Fig. 4. Precision and safety of CM-AI laser ablation.
a–d hiPSCs (19-9-11 hiPSC line) were plated as a monolayer on Kataoka Plates and differentiated to CMs. On day 7 hiPSC-CM monolayers were laser cut to make grids (0.4 mm2). a, c phase contrast images show where laser grids were made. b, d Staining with AnnexinV (red apoptosis reagent) shows dead cells are limited to the laser ablated areas. e–h AnnexinV (red) staining following CM-AI and laser processing to purify hiPSC-CMs using two different GFP-tagged cell lines. For each cell line tested, GFP+ CMs are outlined with white dotted line to enhance visualization in (f, h).
Fig. 5. Functional validation of CM-AI processed cardiomyocytes.
a–c Phase contrast images of custom programmed laser ablation test using a purified hiPSC-CM monolayer. The laser was programmed to kill cells in the shape of the block “M” × 2. d Spontaneous calcium activations were recorded using a calcium-sensitive probe (Calbryte520AM) and high-speed imaging system. Spontaneous activation waves navigated around the laser-ablated hiPSC-CMs and through the live hiPSC-CMs. Representative data shown from ≥3 replicates. Supplementary Movie 3 illustrates continuous contraction of live CM networks.
hiPSC-CM cryopreservation and recovery from thaw is critical for their implementation in drug discovery, cardiac safety screening, and to promote feasibility for “clinical trials in a dish” applications9,27. Using two separate hiPSC-CM lines, we tested the efficacy of cryopreserving atrial hiPSC-CMs produced using CM-AI (Figs.6–8). Post-thaw viability of hiPSC-CMs ranged from 62 to 74% (Fig.6a). Thawed hiPSC-CMs adherence and 2D confluent monolayer formation kinetics were measured using time-lapse microscopy (Fig.6b, c). For each hiPSC line tested, thawed hiPSC-CMs formed confluent monolayers with rhythmic contractions within 2.5 days from time of thaw. Following cryopreservation and replating, the atrial specific phenotype was maintained as verified by qRT-PCR analysis of the relative cardiac myosin isoform expression. We found MYH6 (fast, α-myosin) expression to be much greater than MYH7 (slow, β-myosin) gene expression as anticipated for healthy atrial myocardium of large animals including humans28,29. Cryopreserved hiPSC-CMs 2D monolayers responded to β-adrenergic stimulation (200 nM ISO) as expected with increased spontaneous beat rate and faster calcium impulse propagation velocity (Fig.6d–g). CM-AI laser processed and cryopreserved hiPSC-CMs were also used to test responsiveness to phosphodiesterase (PDE) inhibition using the PDE4 specific inhibitor, rolipram (Fig.7). Rolipram’s cardioactive effects are mediated by elevation of cAMP levels in hiPSC-CMs30,31, and as expected we found rolipram elevated hiPSC-CM beat rate, calcium transient amplitude, and impulse propagation velocity (Fig. 7c–g). Taken together, our results using ISO and rolipram suggest an intact and functional β-adrenergic signaling cascade in cryopreserved CM-AI processed atrial hiPSC-CMs.
Fig. 6. Post-thaw function and viability of cryopreserved CM-AI processed hiPSC-CMs.
a Post-thaw viability was similar between two hiPSC lines tested (TTN-GFP hiPSC-CM = 74.3 ± 7.4; 19.9.11 hiPSC-CM = 61.7 ± 2.9%, n = 3 separate batches per group). b Time-lapse imaging was used to quantify hiPSC-CM 2D monolayer formation kinetics following thaw, data is mean ± standard deviation at each time point. c Phase contrast and GFP images for each cell line tested in panel b on day 3.5 post thaw. d, e Representative calcium transient recordings from each hiPSC line at baseline and with isoproterenol (ISO, 200 nM) treatment. f Each cell line hiPSC-CMs responded to ISO with increased beat rate (TTN-GFP hiPSC-CM baseline = 68.9 ± 8.1; +ISO = 93.3 ± 10.3 bpm, n = 6) (19.9.11 hiPSC-CM baseline = 100.3 ± 15.6; +ISO = 130.0 ± 20.6bpm, n = 6). Paired t-test, ***P = 0.0001. g Each cell line hiPSC-CMs responded to 200 nM ISO with increased calcium wave propagation velocity (TTN-GFP hiPSC-CM baseline = 33.7 ± 5.9; +ISO = 46.9 ± 4.6 cm/s, n = 6) (19.9.11 hiPSC-CM baseline = 30.1 ± 3.4; +ISO = 37.2 ± 6.3 cm/s, n = 6). Paired t-test, ***P = 0.0003; **P = 0.04.
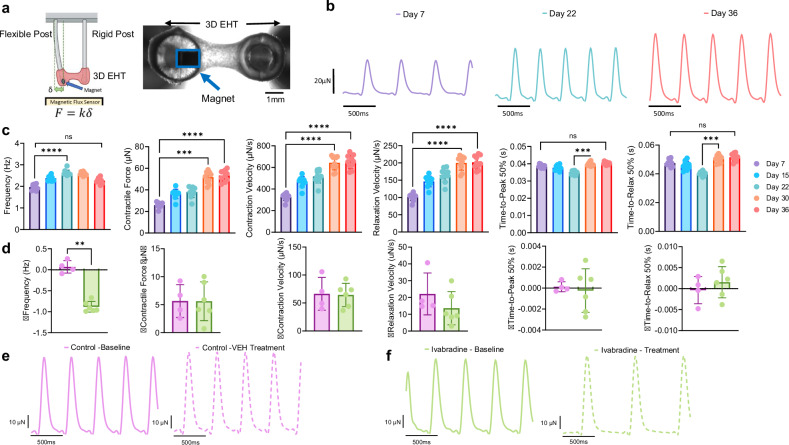
Fig. 8. Functional assessment of 3D engineered heart tissues (3D EHTs) derived from CM-AI-processed, cryopreserved hiPSC-derived atrial CMs.
aTop: Schematic illustration of the 3D EHT platform used to evaluate contractile function of CM-AI laser-processed hiPSC-derived atrial CMs. Right: Representative 10× brightfield image of a 3D EHT. The embedded magnet for force sensing is outlined in blue. Created in https://BioRender.comb Representative contractile force traces recorded from the same 3D EHT over time, demonstrating maturation of contractile performance. c Quantitative analysis of 3D EHT contractile function across multiple time points (Day 7, 22, and 36). Contraction frequency increased over time (Day 7 = 1.97 ± 0.13; Day 22 = 2.63 ± 0.12; Day 36 = 2.27 ± 0.12 Hz, n = 10). Peak contractile force increased over time (Day 7 = 25.9 ± 2.5; Day 22 = 38.0 ± 4.9; Day 36 = 53.3 ± 5.0 µN, n = 10). Contraction velocity increased over time (Day 7 = 327.3 ± 33.0; Day 22 = 516.4 ± 60.9; Day 36 = 650.5 ± 63.5 µN/s, n = 10). Relaxation velocity increased over time (Day 7 = 102.1 ± 10.6; Day 22 = 164.9 ± 19.7; Day 36 = 201.8 ± 18.9 µN/s, n = 10). Time to 50% peak of contraction (TTP 50%) increased from day 22 to 36 (Day 7 = 0.039 ± 0.0009; Day 22 = 0.034 ± 0.001; Day 36 = 0.040 ± 0.0008 s, n = 10). Time to 50% relaxation increased between days 22 and 36 (Day 7 = 0.048 ± 0.002; Day 22 = 0.040 ± 0.001; Day 36 = 0.051 ± 0.003 s, n = 10). All data represent repeated measurements from the same 10 3D EHTs at each time point. One way ANOVA; Friedman test; multiple comparisons; ****P < 0.0001; ***P = 0.0001. d Ivabradine treatment (0.3 nM) significantly reduced spontaneous beat rate (**P = 0.009, unpaired t-test and Kolmogorov–Smirnov test), while other contractile parameters remained unaffected. e Representative 3D EHT force traces at baseline and after vehicle control (VEH, dashed lines) treatment. f Representative force traces showing ivabradine treatment slowed the spontaneous beat rate of 3D EHTs as quantified in (d).
Fig. 7. Pharmacologic response of CM-AI cryopreserved hiPSC-CMs to PDE4 inhibition in 96-well high-throughput electrophysiology screen.
a Cryopreserved TTN-GFP atrial hiPSC-CMs phase contrast and GFP images, 20×. b Zoomed image of sarcomere structures in hiPSC-CMs. c Representative time-space plots indicate functional monolayer electrophysiological activations across the width of each well of the 96-well plate (9 mm diameter). Rolipram increased hiPSC-CM monolayer beat rate. d Representative calcium transient traces indicating the effect of rolipram to increase calcium transient amplitude. e Rolipram increased 2D monolayer beat rate (VEH = 80.0 ± 11.8; 1.0 µM Rolipram = 113.3 ± 33.0; 10.0 µM Rolipram = 129.2 ± 9.4 bpm, n = 8 per group). One way ANOVA, multiple comparisons, *P = 0.01; ***P = 0.0004. f Rolipram increased calcium transient amplitude (VEH = 0.53 ± 0.17; 1.0 µM Rolipram = 0.72 ± 0.40; 10.0 µM Rolipram=1.22 ± 0.50 ∆F/F0, n = 8 per group). One way ANOVA, multiple comparisons, **P = 0.004. g Rolipram increased calcium impulse conduction velocity (VEH = 23.5 ± 4.1; 1.0 µM Rolipram = 30.3 ± 17.6; 10.0 µM Rolipram = 47.9 ± 20.4 cm/s, n = 8 per group). One way ANOVA, Brown-Forsythe and Welch tests, multiple comparisons, *P = 0.02.
Finally, we tested the utility of cryopreserved CM-AI processed hiPSC-CMs for creating functional 3D engineered heart tissues (3D EHTs, Fig. 8). For these experiments cryopreserved hiPSC-CMs (TTN-GFP atrial hiPSC-CMs) were manufactured in Ann Arbor at the University of Michigan and then shipped to Baltimore laboratory at Johns Hopkins University where cells were thawed and cast into 3D EHTs (Fig. 8a). The cast 3D EHTs contracted spontaneously with normal rhythm (supplementary Movie 4) and contractile parameters were quantified (Supplementary Fig. 5). 3D EHT contractile function was quantified using magnetic force sensing; repeated magnetic force measurements were made over 36 days following thaw. Representative force traces and quantification indicate maturation of contractile performance over time (Fig. 8b, c). Here 3D EHTs response to ivabradine medication (0.3 nM, pacemaker current inhibitor) was tested. We found the expected ivabradine effect to reduce 3D EHT spontaneous beat rate (Fig. 8d, e) without altering contractility.
In summary, we have developed and validated an AI-guided laser purification method (CM-AI) that enables efficient, non-invasive, and scalable enrichment of hiPSC-derived cardiomyocytes. This technology eliminates the need for complex reagents, genetic modification, or cell-surface labeling, while delivering high-purity, functionally viable cardiomyocyte populations, compared to currently used antibody-based, fluorescent-based, and metabolic-based purification approaches. CM-AI enables hiPSC-CM purification as early as day 8 and up to day 13 of the cardiac differentiation timeline, while traditional approaches require at least 20 days of continuous culture prior to purification. CM-AI may be utilized on any hiPSC line and eliminates the need for MACS and FACS-based CM sorting methods that require complex fluidic systems, antibodies, and genetic modifications to generate highly purified hiPSC-CMs. Relative to the commonly utilized metabolic selection approach, CM-AI produces comparable numbers of hiPSC-CMs per six-well dish (Supplementary Fig. 6a) with a healthy phenotype (Supplementary Fig. 6b–d). The average yield of CMs per six-well plate is 20.1 million CMs using CM-AI, 8.2 million CMs using MACS, and 14.1 million CMs using the traditional metabolic selection approach. Thus, CM-AI produces sufficiently high numbers of CMs for downstream applications. CM-AI yield is less than recently described 2D roller bottle approaches which reported yield of up to 120 million CMs per 1-L bottle32. However, CM-AI yields of 1.75 million CMs/mL of media is comparable to recent 3D suspension based approaches that reported a CM yield of 1.2 million CMs/mL6. Importantly, CM-AI enables less experienced personnel to perform complex cell purification tasks with high efficiency in a fraction of the time. CM-AI and laser processing offers an alternative approach for hiPSC-CM production in 2D monolayers with successful cryopreservation for use in disease modeling, drug screening, and regenerative medicine. As a broadly applicable platform, CM-AI holds the potential to significantly streamline cardiac cell manufacturing workflows and to support personalized therapeutic development.
Methods
hiPSC cell culture maintenance and cardiac differentiation
hiPSCs were maintained and expanded as colonies in the undifferentiated, pluripotent state using standard protocols with approval from the University of Michigan HPSCRO (Human Pluripotent Stem Cell Research Oversight) Committee13,25,33,34. Two established genetically engineered hiPSC lines were utilized with CM-specific GFP expression: (1) TTN-GFP hiPSC line35, and (2) TNNT2-GFP hiPSC line (AICS-0037-172, Coriell Institute). One non-reporter hiPSC control line was used to determine utility of CM-AI for purification of non-labeled CMs (19.9.11 hiPSCs)36.
These hiPSCs were maintained as colonies in feeder-free conditions on Matrigel ECM in mTesr+ stem cell media (Stem Cell Technologies) with passaging every 7 days. Supplementary Fig. 1a, b shows the TTN-GFP hiPSC colonies and growth monitored using phase contrast time-lapse imaging (4×, Incucyte S3 Live Cell Imager, Sartorius). For cardiac-directed differentiation, hiPSCs were replated at a density of 1 × 106 cells per well of a six-well plate (Kataoka laser plates). Atrial cardiac directed differentiation of hiPSCs was executed as previously described24,25. GFP+ CMs were observed as early as day 8 of the differentiation timeline (Supplementary Fig. 1). On day 9–10 GFP+ CMs were observed with visible contractions. Non-myocytes were negative for GFP expression as expected (Fig. 1c). A total of five plates (30 wells) were utilized in this way for AI creation, training, and validation. Live cell imaging was used to determine GFP expression and for monitoring hiPSC maintenance (Incucyte S3, Sartorius, Ann Arbor, MI).
Cardiomyocyte-artificial intelligence (CM-AI) training and validation
For cardiomyocyte artificial intelligence (CM-AI) creation, we utilized the device software (Kataoka Cell Processor, CPD-017, Cell Processor Training, CpTr), which can create a classifier and training data. The learning data was created using phase contrast images (4×) and fluorescence images acquired with this device. It is important that training images be acquired on the same device that is used to implement CM-AI. The training data was labeled into three regions: CM region, non-CM region, and blank region. Based on the regions extracted by image processing from phase contrast images and fluorescence images, regions added and corrected manually by marking were used as training data. When performing image processing on a fluorescence image, we extracted the GFP portion and labelled the CM region by overlaying it with the corresponding phase contrast image. For CM-AI, we combined image processing and manual marking to create learning data equivalent to approximately three wells. U-Net, which is used for segmentation of biological images, was used to train the classifier of this device. We trained for 200 epochs and selected the classifier from the epochs that yielded results with the least loss on both training and validation data. CM-AI was used for automated detection of CMs in unpurified monolayers; with successful detection and marking of CMs within seconds of opening a CM well image (Fig. 1a). Validation of CM-AI was analyzed using standard phase contrast + GFP imaging before and after laser processing (Incucyte S3 Live Cell Analysis Instrument, Sartorius) throughout each well of six well dishes (n = 12 wells, 2 plates). GFP confluence area was calculated before and after CM-AI and laser cell processing and found to be minimally reduced following laser processing. Following CM-AI processing, hiPSC-CM monolayers were maintained for two days in metabolic selection media with daily washes (Hanks balanced salt solution with glucose) to further enhance CM enrichment. As a comparator, metabolic selection media were used as previously described2,16 with 10 days of chronic exposure to purify atrial CMs in parallel to the CM-AI + laser processing to enable direct comparison between the two purification approaches (Supplementary Figs. 4 and 6).
Flow cytometry
Flow cytometry was performed to assess the relative purity of CM culture purified with two different purification methods by quantification of percentage of GFP+ cells (Supplementary Fig. 4c). Briefly, cells 1 × 106 cells were fixed with cold 3% paraformaldehyde for 10 min on ice and resuspended in 1 × PBS (no Ca, Mg) 50 µL. Cells GFP+ CMs were captured by the Amnis ImageStreamX Mark II flow cytometer (Cytek Biosciences, USA). For GFP detection we used excitation laser at 488 nm and emission band at 480–560. The scatter plots were analyzed using the IDEAS 6.2 software. In other experiments (Fig.2), cTnT+ CMs were quantified by immunolabeling and FACS analysis as described before13. The gating strategy used to determine cTnT+ cell populations is displayed in Supplementary Fig. 7).
qRT-PCR analysis
For quantitative evaluation of mRNA expression, hiPSC-CM monolayers were replated on MatrixPlus human ECM coating as confluent monolayers in twelve-well plates as described before25. Expression of pluripotent stem cell markers was also evaluated with positive control RNA isolated from hiPSC maintenance cultures and appropriate negative controls. TaqMan probes (Applied Biosystems) were used for all reactions and are listed in Supplementary Table 1. Total cellular RNA was prepared using the RNAeasy Mini Kit (Qiagen) and cDNA was synthesized as described recently37. We calculated mRNA fold expression by the ΔΔCt method using the 18S rRNA as the housekeeping gene for normalization (Supplementary Fig. 6b). Every qPCR reaction was performed in triplicate and repeated using cDNA from at least three separate cardiomyocyte differentiation cultures. To determine relative myosin isoform gene expression (MYH6/MYH7) within samples, we used the ΔCt method. We found that average MYH6 ΔCt = 8.9 ± 0.53 and average MYH7 ΔCt = 17.6 ± 0.63, indicating that MYH6 » MYH7 gene expression in CM-AI processed atrial hiPSC-CMs as expected for healthy human cardiac tissue29.
hiPSC-CM protein expression analysis
CM-specific protein expression was analyzed by western blotting as described before13,38. Primary antibodies used include: Cx40 (ThermoFisher; Cat #36-4900), cTnI (Abcam; ab47003), and mlc2a (Synaptic Systems; #311011). Ponceau S total protein stain was applied to the nitrocellulose membrane prior to immunoblotting and images were used for normalization and quantification of protein expression. Protein band quantification was done by densitometry using ImageJ.
hiPSC-CM 2D monolayer functional analysis and drug responsiveness
Replated hiPSC-CM monolayer electrophysiological function was assessed using a calcium-sensitive fluorophore (CalBryte 520AM, AATBioquest) and high-resolution, high-throughput optical mapping device (100fps, CARTOX™, Fig. 3 &5-7) as recently described25,26. 96-well plates coated with CELLvo™ Matrix Plus human ECM were used to promote the maturation and adhesion of the cardiomyocytes. Spontaneous contractions were recorded in 10 s duration movies to quantify the beat rate, impulse conduction velocity, and calcium transient duration (CaTD80) using commercially available software (StemBioSys Optical Electrophysiology Analysis Tool). Isoproterenol (200 nM, Sigma Aldrich, I5627) was applied to 2D hiPSC-CM monolayers to determine the functionality of the β-adrenergic signaling system in CM-AI processed cells (Fig.6d–g). Rolipram (1, 10 µM, Enzo BML-PD175) was also applied to 2D hiPSC-CM monolayers to further probe the function of the β-adrenergic signaling pathway (Fig.7).
3D engineered heart tissue (EHT) casting and functional analysis
Atrial TTN GFP cardiomyocytes were differentiated, laser-purified, and cryopreserved at the University of Michigan and then shipped overnight to Johns Hopkins University. An estimated 17.3 × 106 CMs were removed from −80 °C and thawed in a 37 °C bead bath until completely thawed, less than 4 min. hiPSC-CMs were initially plated at 2D monolayers. Briefly, two six-well plates were coated with Geltrex (ThermoFisher, Geltrex™ LDEV-Free Reduced Growth Factor Basement Membrane Matrix, Catalog #: A1413201) in DMEM:F12 (ThermoFisher, DMEM/F-12, Catalog #:11320033) and incubated at 37 °C for at least 1 h. Each vial of CMs was dripped into RPMI/B27+ (ThermoFisher, RPMI 1640 Medium, HEPES, Catalog #: 22400089) and centrifuged to remove cryopreserving liquid. CMs were resuspended in RPMI/B27+ at a concentration of 75,000 CMs/mL and then plated onto the Geltrex plates at 1,500,000 CMs/well. CMs were incubated at 37 °C.
Once the CMs began spontaneously contracting, 3 days later, miniature 3D EHTs were cast as previously described39,40. CMs were lifted using 0.25% trypsin EDTA (ThermoFisher, Trypsin-EDTA (0.25%), phenol red, Catalog#: 25200056), and cardiac fibroblasts (Lonza, NHCF-V – Human Ventricular Cardiac Fibroblasts, Catalog #: CC-2904) were thawed following the protocol outlined above. Following centrifugation, CMs were resuspended at 8.3 × 106 CMs/mL of a proprietary media from Celogics, B27+, 5 g/L aminocaproic acid, 100 mg/L normocin, and ROCK inhibitor. Similarly, fibroblasts were resuspended in the same media at 2.5 × 106 fibroblasts/mL. Each tissue was cast onto the Curi Bio Mantarray Mini Kit posts and contains 500,000 CMs, 75,000 fibroblasts, and fibrinogen and thrombin solutions. Cast tissues were incubated at 37 °C for 80 min and then changed into fresh media supplemented with ROCK inhibitor. Tissues were changed into fresh media without ROCK inhibitor after 24 h and then changed every 2–3 days. A fluorescent Movie of spontaneously contracting TTN-GFP atrial 3D EHT was acquired using a Nikon microscope (Supplementary Movie 4, Nikon ECLIPSE Ti Inverted Microscope) at 4× magnification with a frame rate of 25 fps.
Spontaneous contractile measurements of the 3D EHTs were taken on the Mantarray Instrument on days 7, 15, 22, 30, and 36 (Fig. 8). The label-free recording of force using magnetic sensors enabled repeated measures of the same 3D EHTs over time. For drug responsiveness, additional contractile measurements were taken on day 42 and following 18-h treatment of 300 nM ivabradine (IVA). Data was extracted from the Mantarray as. h5 files which are analyzed by the CuriBio 3D Contractility Analysis software. Further analysis was completed in a custom MATLAB code. Confocal imaging completed at 10× on Leica SP8. Tissues that experienced significantly abnormally low contractile force or quiescence at any time point were removed from the data set as outliers. Graphing and statistical analysis were completed in GraphPad, column graphs presented as mean and standard deviation.
Statistics and reproducibility
Data was analyzed and statistics were performed using commercially available software (GraphPad Prism 10.0). Data are expressed as mean ± standard deviation, statistical tests used for each data set are included in figure legends along with P values, number of replicates (sample size), and indication of significance. Paired t-tests were used to compare a single set of matched measurements, such as in pre/post laser ablation analysis (Fig. 1c), pre/post ISO treatments (Fig.6f, g). Unpaired t-tests were used to compare the means of two independent or unrelated groups such as in the following figures: Figs.1d, 2c, 3b–d and f–h, 6a, 8d, and Supplementary Fig. 6b, d. Ordinary one way ANOVA analysis with Tukey’s multiple comparison test was utilized for comparison of three or more independent groups of data such as in: Fig.7e–g and Supplementary Figs. 4b and 6a. Repeated measures of 3D EHT force parameters were analyzed by one way ANOVA, Friedman test with Dunn’s Multiple Comparison test (Fig.8c).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Dr. Andre Monteiro da Rocha for guidance on flow cytometry data collection and analysis. This study was supported by the University of Michigan Frankel Cardiovascular Regeneration Core Laboratory and funding from the Frankel Cardiovascular Center. This work was supported by National Institutes of Health (NIH) UH3TR003271, R01HL164936, R01HL156947, UH3TR003519 (to D.-H. K.).
Author contributions
P.S., T.J.H., D.-H.K., P.K., and J.C. analyzed data and wrote the manuscript. T.J.H., J.M., P.S., and D.-H.K. conceived and designed the study. Y.K., M.Y., J.M. generated AI to detect cardiomyocytes and programmed laser ablation routine, T.J.H. and P.S. collected CM training image data. H.C., S.S., D.J., D.B., C.H., and T.J.H. collected images pre- and post-laser processing and performed and analyzed apoptosis experiments and data. J.A. and T.J.H. designed and analyzed experiments related to drug screening and optical recordings of electrophysiology. P.S. collected electrophysiology data and replated CMs for analysis in high-throughput plates. P.K. and D.-H.K. designed and analyzed experiments related to 3D engineered heart tissues. P.S. and M.M.N. collected and analyzed flow cytometry data. A.R. and L.V.D. procured genetically modified hiPSC lines and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks Eleonora Cianflone and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Daniele Torella and Christina Karlsson Rosenthal.
Data availability
Data, including AI training, electrophysiology, contractile function of 3D EHTs, flow cytometry, gene expression, and protein expression are available upon request from the corresponding authors. Data, including movie files, has also been deposited on Figshare with the following 10.6084/m9.figshare.2877141541.
Code availability
No custom code was used to generate or process the data described in the manuscript. Training data sets for CM-AI generation are available upon request from the corresponding authors.
Competing interests
T.J.H. declares employment at Greenstone Biosciences, consultant and scientific advisory board member of StemBioSys, Inc. D.-H.K. is a co-founder and scientific advisory board member of Curi Bio. M.M.N. is an employee of StemBioSys, Inc. Y.K., M.Y., and J.M. are employees of Kataoka Corporation. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Prakaimuk Saraithong, Peyton Krajcarski.
Contributor Information
Deok-Ho Kim, Email: dhkim@jhu.edu.
Todd J. Herron, Email: toddherr@stanford.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-025-08162-0.
References
- 1.Sharma, A., Wu, J. C. & Wu, S. M. Induced pluripotent stem cell-derived cardiomyocytes for cardiovascular disease modeling and drug screening. Stem Cell Res. Ther.4, 150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burridge, P. W. et al. Chemically defined generation of human cardiomyocytes. Nat. Methods11, 855–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, J. et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res.104, e30–e41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lian, X. et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA109, E1848–E1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, J. et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res.111, 1125–1136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prondzynski, M. et al. Efficient and reproducible generation of human iPSC-derived cardiomyocytes and cardiac organoids in stirred suspension systems. Nat. Commun.15, 5929 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghasemian, M. et al. Hydrodynamic characterization within a spinner flask and a rotary wall vessel for stem cell culture. Biochem. Eng. J.157, 107533 (2020). [Google Scholar]
- 8.Chirikian, O. et al. The effects of xeno-free cryopreservation on the contractile properties of human iPSC-derived cardiomyocytes. J. Mol. Cell. Cardiol.168, 107–114 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Zhang, J. Z. et al. Effects of cryopreservation on human induced pluripotent stem cell-derived cardiomyocytes for assessing drug safety response profiles. Stem Cell Rep.16, 168–181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hattori, F. et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods7, 61–66 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Bizy, A. et al. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res.11, 1335–1347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois, N. C. et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol.29, 1011–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, J. et al. In vitro model of ischemic heart failure using human induced pluripotent stem cell-derived cardiomyocytes. JCI Insight6, 10.1172/jci.insight.134368 (2021). [DOI] [PMC free article] [PubMed]
- 14.Tsujisaka, Y. et al. Purification of human iPSC-derived cells at large scale using microRNA switch and magnetic-activated cell sorting. Stem Cell Rep.17, 1772–1785 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossler, K. J. et al. Lactate and Immunomagnetic-purified hiPSC-derived cardiomyocytes generate comparable engineered cardiac tissue constructs. JCI Insight,10.1172/jci.insight.172168 (2023). [DOI] [PMC free article] [PubMed]
- 16.Tohyama, S. et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell12, 127–137 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Diaz, R. J. & Wilson, G. J. Studying ischemic preconditioning in isolated cardiomyocyte models. Cardiovasc Res.70, 286–296 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Zushin, P. H., Mukherjee, S. & Wu, J. C. FDA Modernization Act 2.0: transitioning beyond animal models with human cells, organoids, and AI/ML-based approaches. J. Clin. Investig.133, 10.1172/jci175824 (2023). [DOI] [PMC free article] [PubMed]
- 19.Carratt, S. A. et al. An industry perspective on the FDA Modernization Act 2.0/3.0: potential next steps for sponsors to reduce animal use in drug development. Toxicol. Sci.203, 28–34 (2024). [DOI] [PubMed] [Google Scholar]
- 20.Park, J. et al. Artificial intelligence-enabled quantitative phase imaging methods for life sciences. Nat Methods, 10.1038/s41592-023-02041-4 (2023). [DOI] [PubMed]
- 21.Vo, Q. D., Saito, Y., Ida, T., Nakamura, K. & Yuasa, S. The use of artificial intelligence in induced pluripotent stem cell-based technology over 10-year period: a systematic scoping review. PLoS ONE19, e0302537 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi, Y. et al. Automated adherent cell elimination by a high-speed laser mediated by a light-responsive polymer. Commun. Biol.1, 218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma, A. et al. CRISPR/Cas9-mediated fluorescent tagging of endogenous proteins in human pluripotent stem cells. Curr. Protoc. Hum. Genet96, 21.11.21–21.11.20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cyganek, L. et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight3, e99941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allan, A. et al. High-throughput longitudinal electrophysiology screening of mature chamber-specific hiPSC-CMs using optical mapping. iScience26, 107142 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monteiro da Rocha, A., Allan, A., Block, T., Creech, J. & Herron, T. J. High-Throughput Cardiotoxicity Screening Using Mature Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Monolayers. J Vis Exp, 10.3791/64364 (2023). [DOI] [PubMed]
- 27.van den Brink, L. et al. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Res.43, 101698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen, J. B. et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet.50, 1234–1239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiser, P. J., Portman, M. A., Ning, X.-H. & Moravec, C. S. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol.Heart Circ. Physiol.280, H1814–H1820 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Saleem, U. et al. Regulation of I(Ca, L) and force by PDEs in human-induced pluripotent stem cell-derived cardiomyocytes. Br. J. Pharm.177, 3036–3045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iqbal, Z. et al. Regulation of basal and norepinephrine-induced cAMP and ICa in hiPSC-cardiomyocytes: effects of culture conditions and comparison to adult human atrial cardiomyocytes. Cell. Signal.82, 109970 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Dhahri, W. et al. In vitro matured human pluripotent stem cell–derived cardiomyocytes form grafts with enhanced structure and function in injured hearts. Circulation145, 1412–1426 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Balistreri, M. et al. Effect of glucose on 3D cardiac microtissues derived from human induced pluripotent stem cells. Pediatr. Cardiol.38, 1575–1582 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Dow, R. et al. Bipolar patient specific in vitro diagnostic test reveals underlying cardiac arrhythmia phenotype caused by calcium channel genetic risk factor. Biol. Psychiatry Glob. Open Sci., 100296, 10.1016/j.bpsgos.2024.100296 (2024). [DOI] [PMC free article] [PubMed]
- 35.Sharma, A., Toepfer, C. N., Schmid, M., Garfinkel, A. C. & Seidman, C. E. Differentiation and contractile analysis of gfp-sarcomere reporter hiPSC-cardiomyocytes. Curr. Protoc. Hum. Genet.96, 21.12.21–21.12.12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, J. et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science324, 797–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jimenez-Vazquez, E. N. et al. SNTA1 gene rescues ion channel function and is antiarrhythmic in cardiomyocytes derived from induced pluripotent stem cells from muscular dystrophy patients. eLife11, e76576 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Block, T. et al. Human perinatal stem cell-derived extracellular matrix enables rapid maturation of hiPSC-CM structural and functional phenotypes. Sci. Rep.10, 19071–19071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mair, D. et al. Spaceflight-induced contractile and mitochondrial dysfunction in an automated heart-on-a-chip platform. Proc. Natl. Acad. Sci. USA121, e2404644121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsui, J. et al. Tunable electroconductive decellularized extracellular matrix hydrogels for engineering human cardiac microphysiological systems. Biomaterials272, 120764 (2021). [DOI] [PMC free article] [PubMed]
- 41.Herron. Todd Supplementary Data and Movies_COMMSBIO-24-1564B. figshare. Media. 10.6084/m9.figshare.28771415.v1 (2025).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Data, including AI training, electrophysiology, contractile function of 3D EHTs, flow cytometry, gene expression, and protein expression are available upon request from the corresponding authors. Data, including movie files, has also been deposited on Figshare with the following 10.6084/m9.figshare.2877141541.
No custom code was used to generate or process the data described in the manuscript. Training data sets for CM-AI generation are available upon request from the corresponding authors.