Abstract
Cyanobacteria may possess several enzymes that are directly involved in dihydrogen metabolism: nitrogenase(s) catalyzing the production of hydrogen concomitantly with the reduction of dinitrogen to ammonia, an uptake hydrogenase (encoded by hupSL) catalyzing the consumption of hydrogen produced by the nitrogenase, and a bidirectional hydrogenase (encoded by hoxFUYH) which has the capacity to both take up and produce hydrogen. This review summarizes our knowledge about cyanobacterial hydrogenases, focusing on recent progress since the first molecular information was published in 1995. It presents the molecular knowledge about cyanobacterial hupSL and hoxFUYH, their corresponding gene products, and their accessory genes before finishing with an applied aspect—the use of cyanobacteria in a biological, renewable production of the future energy carrier molecular hydrogen. In addition to scientific publications, information from three cyanobacterial genomes, the unicellular Synechocystis strain PCC 6803 and the filamentous heterocystous Anabaena strain PCC 7120 and Nostoc punctiforme (PCC 73102/ATCC 29133) is included.
INTRODUCTION
Cyanobacteria
Cyanobacteria (also called blue-green bacteria, blue-green algae, cyanophyceae, or cyanophytes) are a large and widespread group of photoautotrophic microorganisms, which originated, evolved, and diversified early in Earth's history. The earliest forms attributed to this group were found in sedimentary rocks formed 3.5 billion years ago, and it is commonly accepted that cyanobacteria played a crucial role in the Precambrian phase by contributing oxygen to the atmosphere (175).
At present, cyanobacteria are found in a wide range of habitats including aquatic (saltwater and freshwater), terrestrial, and extreme environments (like frigid lakes of the Antarctic or hot springs). Although called blue-green, cyanobacteria may display a variety of colors due to different combinations of the photosynthetic pigments chlorophyll a, carotenoids, and phycobiliproteins. All cyanobacteria combine the ability to perform an oxygenic photosynthesis (resembling that of chloroplasts) with typical prokaryotic features. The possession of chlorophyll a and the use of oxygenic photosynthesis distinguishes cyanobacteria from other photosynthetic bacteria, such as purple and green bacteria. Nevertheless, some cyanobacterial strains can perform anoxygenic photosynthesis by using hydrogen sulfide (H2S) as the electron donor. Many cyanobacteria can fix atmospheric dinitrogen (N2) (a capacity not possessed by any eukaryote) into ammonia (NH3), a form in which the nitrogen is further available for biological reactions. Although quite uniform in nutritional and metabolic respects, cyanobacteria are a morphologically diverse group with unicellular, filamentous, and colonial forms. Among certain filamentous cyanobacteria, there is some degree of cellular differentiation. Within the filament, vegetative cells may develop into structurally modified and functionally specialized cells: the akinetes (resting cells) or the heterocysts (cells specialized in nitrogen fixation). For more detailed information on cyanobacteria, see reference 218.
Unicellular and filamentous cyanobacteria can form symbioses with a wide diversity of hosts. In symbiosis, some cyanobionts perform both photosynthesis and nitrogen fixation while others exhibit only one of these properties (3, 22, 133, 161, 184). It is believed that ancestors of cyanobacteria evolved to become plastids after a long period of endosymbiosis. In biochemical and structural detail, cyanobacteria are especially similar to the chloroplasts of red algae (47, 104).
The taxonomy of cyanobacteria is still a controversial subject, with two prevailing approaches, the botanical approach (8-10, 102, 103) and the bacterial approach (167, 168, 190). Despite the differences, both botanists and bacteriologists divide the cyanobacteria into four or five major subgroups. In fact, the five sections recognized by Rippka et al. (167) coincide broadly with orders of other classifications: Chroococcales, Pleurocapsales, Oscillatoriales, Nostocales, and Stigonematales (for reviews on this subject, see references 218 and 219). At present, different molecular methods are being used and developed to infer phylogenetic relationships. However, the data available for cyanobacteria are still scarce (207, 219). Analysis of 16S rRNA sequences has given the most detailed hypothesis of the evolution of cyanobacteria (218, 219). A polyphasic taxonomy of cyanobacteria, integrating both phenotypic and genotypic characters is currently being assembled. A milestone in the study of cyanobacteria was the publication of the entire genome (3,573,470 bp) of the non-nitrogen-fixing unicellular cyanobacterium Synechocystis strain PCC 6803 (95, 144). A number of other cyanobacterial genome projects are being completed (see below).
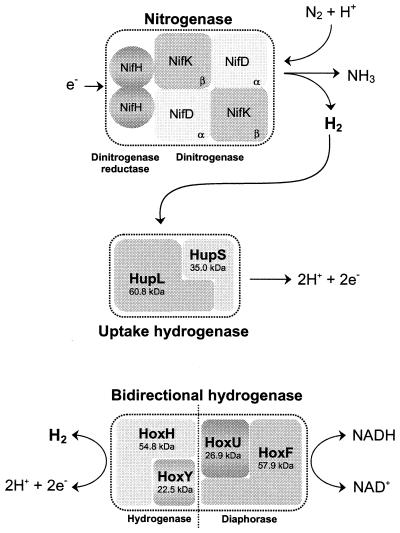
Cyanobacteria may possess several enzymes directly involved in hydrogen metabolism: nitrogenase(s) catalyzing the production of hydrogen (H2) concomitantly with the reduction of nitrogen to ammonia, an uptake hydrogenase catalyzing the consumption of hydrogen produced by the nitrogenase, and a bidirectional hydrogenase which has the capacity to both take up and produce hydrogen (Fig. 1) (13, 21, 58, 75, 87, 113, 115, 117).
FIG. 1.
Enzymes directly involved in hydrogen metabolism in cyanobacteria. While the uptake hydrogenase is present in all nitrogen-fixing strains tested so far, the bidirectional enzyme is distributed among both nitrogen-fixing and non-nitrogen-fixing cyanobacteria (although it is not a universal cyanobacterial enzyme) (192). The molecular masses indicated for the uptake hydrogenase subunits are mean values calculated from the deduced amino acid sequences of Anabaena strain PCC 7120 (39), Nostoc strain PCC 73102 (150), and A. variabilis ATCC 29413 (79), while the values for the subunits of the bidirectional enzyme are based on data exclusively from A. variabilis ATCC 29413 (173).
This review summarizes our knowledge about cyanobacterial hydrogenases. It is based on scientific publications and sequence information from the genome projects of the unicellular non-nitrogen-fixing Synechocystis strain PCC 6803 (95, 144) and the filamentous heterocystous strains Anabaena strain PCC 7120 (The Kazusa DNA Research Institute [http://www.kazusa.or.jp/cyano/anabaena/]) and Nostoc punctiforme (ATCC 29133, PCC 73102) (for preliminary sequence data, see the DOE Joint Genome Institute website [http://www.jgi.doe.gov/JGI_microbial/html/nostoc/nostoc_homepage.html]) (Fig. 2). In addition, genome projects are currently being performed using, e.g., unicellular cyanobacteria such as several Synechococcus strains (e.g., S. elongatus [http://www.kazusa.or.jp/en/] and Synechococcus strain WH8102 [http://spider.jgi-psf.org/JGI_microbial/html/synechococcus/synech_homepage.html]), Gloeobacter strain PCC 7421 (http://www.kazusa.or.jp/en/), and Microcystis aeruginosa (http://www.pasteur.fr/externe.html); the filamentous, nonheterocystous Spirulina platensis (http://hgc.igtp.ac.cn/); as well as the prochlorophyte Prochlorococcus marinus (http://www.jgi.doe.gov/JGI_microbial/html/prochlorococcus/prochlo_pickastrain.html). After introducing nitrogenases and hydrogenases, we focus on the two cyanobacterial hydrogenases, the uptake and the bidirectional enzymes, their regulation, and their accessory genes before finishing with an applied aspect—the use of cyanobacteria in the photoproduction of molecular hydrogen.
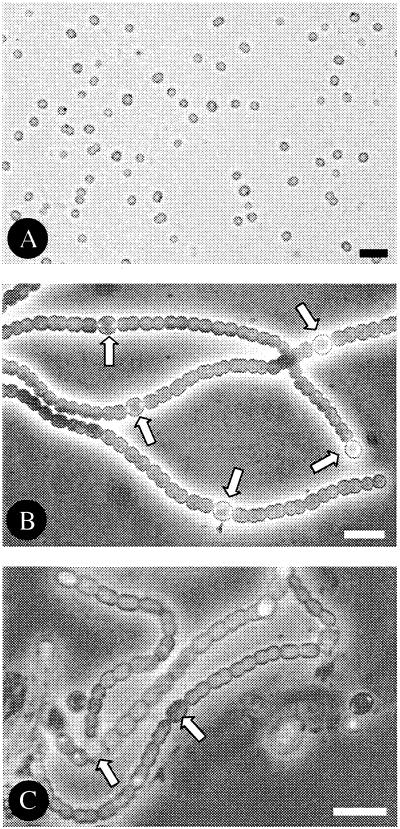
FIG. 2.
Microphotographs of the three cyanobacteria from which genome sequence information is included in the present review. (A) Synechocystis strain PCC 6803; (B) Anabaena strain PCC 7120; (C) Nostoc punctiforme PCC 73102/ATCC 29133. Note the presence of heterocysts (arrows) in the nitrogen-fixing cultures of Anabaena strain PCC 7120 and Nostoc strain PCC 73102. Bar, 10 μm.
Nitrogenases
Although nitrogenase is unquestionably a key enzyme for cyanobacterial hydrogen production, it is not subject of this review and so its properties will be discussed only briefly here.
The activity of nitrogenase, the enzymatic multiprotein complex for nitrogen fixation, is essential for the maintenance of the nitrogen cycle, since this element often limits the growth of organisms (35, 65, 158, 187). The diversity of prokaryotic microorganisms with the ability to fix nitrogen is in contrast to the remarkable conservation of the nitrogenase itself (36, 58, 80).
The nitrogenase complex consists of two proteins: the dinitrogenase (MoFe protein or protein I) and the dinitrogenase reductase (Fe protein or protein II), (Fig. 1). The dinitrogenase is an α2β2 heterotetramer of about 220 to 240 kDa, and the α and β subunits are encoded by nifD and nifK, respectively. The dinitrogenase reductase, encoded by nifH, is a homodimer of about 60 to 70 kDa and plays the specific role of mediating the transfer of electrons from the external electron donor (a ferredoxin or a flavodoxin) to the dinitrogenase (58, 127, 149). In addition to the three structural nif genes, many other genes are involved in the nitrogen fixation process and its regulation (for a review, see reference 26). Recently, following the completion of several cyanobacterial genome projects, it became evident that some strains contain multiple copies of certain nif genes; for example N. punctiforme (Nostoc strain PCC 73102) appears to have three copies of nifH (199; http://www.jgi.doe.gov/JGI_microbial/html/nostoc/nostoc_homepage.html).
The reduction of nitrogen to ammonia, catalyzed by nitrogenase, is a highly endergonic reaction requiring metabolic energy in the form of ATP. Moreover, this reaction is accompanied by an obligatory reduction of protons (H+) to hydrogen. Apparently, with a pure nitrogen atmosphere, at most 75% of the electrons (e−) would be allocated for nitrogen reduction and at least 25% would be allocated for proton reduction. Since two ATP molecules are required for each electron transferred from dinitrogenase reductase to dinitrogenase, the reaction requires a minimum of 16 ATP molecules until the dinitrogenase has accumulated enough electrons to reduce nitrogen to ammonia. The overall reaction can be written as follows:
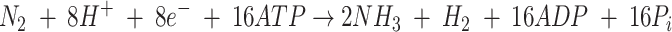 |
Nitrogenase is very oxygen labile; hence, all diazotrophs must protect the enzymatic complex from the deleterious effects of oxygen (O2). Cyanobacteria and prochlorophytes are the only prokaryotic organisms known to perform oxygenic photosynthesis (58, 129). Cyanobacteria have evolved different mechanisms and strategies, ranging from fixing nitrogen only under anoxic conditions to temporal or even spatial separation of nitrogen fixation and oxygen evolution, to protect their nitrogen-fixing machinery not only from atmospheric oxygen but also from the intracellularly generated oxygen (for reviews, see references 22, 26, 56, 140, 188, 220, and 221). Temporal separation between photosynthetic oxygen evolution and nitrogen fixation seems to be the most common strategy adopted by nonheterocystous cyanobacteria (92, 137, 165). Spatial separation of the two processes is achieved in many filamentous cyanobacteria by differentiation of vegetative cells into cells specialized in nitrogen fixation, i.e., the heterocysts. In the marine filamentous nonheterocystous Trichodesmium, a spatial separation probably occurs between the two processes without any obvious cellular differentiation; only a small fraction of cells within a colony or along the filament are capable of nitrogen fixation. In contrast to the irreversible changes occurring during heterocyst differentiation, those occurring in nonheterocystous cyanobacteria can be reversed (21, 44, 61, 140).
Some filamentous cyanobacteria are able to differentiate 5 to 10% of their vegetative cells into heterocysts (Fig 2). Heterocyst formation does not take place randomly within the filament. Recently, it was proposed that this process in Anabaena variabilis is not controlled by individual cells but by the entire filament (199). The filament senses the nitrogen status and responds by producing heterocyst differentiation signals. However, this does not exclude cell-to-cell signaling in the maintenance of heterocyst pattern (26, 199, 220), and it has been shown that the small diffusible peptide PatS and products of nitrogen fixation control heterocyst differentiation in Anabaena strain PCC 7120 (226, 227). The heterocyst provides a microanaerobic environment suitable for the functioning of nitrogenase since (i) it lacks photosystem II activity, (ii) it has a higher rate of respiratory oxygen consumption, and (iii) it is surrounded by a thick envelope that limits the diffusion of oxygen through the cell wall (56, 221). Heterocysts import carbohydrates and in return export glutamine to the vegetative cells. An interesting feature of heterocyst differentiation in cyanobacteria is the occurrence of developmentally regulated genome rearrangements (39-41, 67-69, 130, 134, 141, 142). These rearrangements occur late in heterocyst differentiation, at about the same time as the nitrogenase genes are transcribed. In vegetative cells of Anabaena strain PCC 7120, the fdxN, nifD, and hupL genes are interrupted by DNA elements that are excised, during heterocyst differentiation, by site-specific recombinases encoded by xisF, xisA, and xisC, respectively. In addition, the excision of the fdxN element requires the products of the two genes xisH and xisI (162).
Alternative nitrogenases.
In the heterocystous cyanobacterium A. variabilis ATCC 29413 several different nitrogenases have been identified and characterized (for details, see references 98, 99, 195, and 197 to 199).
(i) Mo-containing nitrogenases.
Two of the A. variabilis nitrogenases are molybdenum-dependent enzymes. The first one (the so-called conventional nitrogenase, encoded by the nif1 gene cluster) functions in heterocysts. The second enzyme (encoded by the nif2 gene cluster) functions strictly under anaerobic conditions in vegetative cells only (199).
(ii) V-containing nitrogenase.
The occurrence of a vanadium-containing nitrogenase was first reported by Kentemich et al. (98) and subsequently confirmed by Thiel (195). This enzyme is encoded by the vnfDGK gene cluster, which is transcribed in the absence of molybdenum and in which vnfDG are fused into a single gene (195).
(iii) Fe-containing nitrogenase.
A fourth nitrogenase is believed to be an iron enzyme similar to the one encoded by the anf gene cluster in Azotobacter vinelandii (23, 99).
Alternative nitrogenases have also been found in other cyanobacterial strains (23), and it is known that they differ from the conventional enzyme physically, chemically, and by their catalytic properties. One must bear in mind that all of the alternative enzymes investigated so far seem to allocate a higher proportion of electrons to the reduction of protons to hydrogen than does the conventional Mo enzyme complex. However, hydrogen produced by nitrogenase is generally taken up by an uptake hydrogenase, so that net hydrogen evolution by nitrogen-fixing cyanobacteria is barely observed, at least under aerobic conditions (7).
HYDROGENASES
The capacity of certain microorganisms to metabolize molecular hydrogen was discovered at the end of the 19th century (85) and later identified to be catalyzed by a hydrogenase (191). Since then hydrogenases have been observed and characterized in many microorganisms, including some algae, trichomonads, anaerobic ciliates, and chytrid fungi (4, 5, 59, 77, 86, 94, 100, 176, 222, 223, 225). The enzyme catalyzes the simplest of chemical reactions, the reversible reductive formation of hydrogen from protons and electrons:
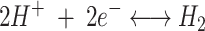 |
According to the metal composition of the active site, hydrogenases are classified into three major groups: NiFe, Fe, and metal-free hydrogenases (209).
The majority of hydrogenases are NiFe-containing enzymes, occurring in all bacterial classes (38, 159, 209). The first crystal structure of a NiFe hydrogenase, the representative heterodimeric soluble enzyme of Desulfovibrio gigas (6), was published in 1995 (212). This hydrogenase is involved in periplasmic H2 oxidation and is phylogentically related to other bacterial uptake hydrogenases (209). The binuclear Ni-containing active site in the large subunit contains an additional Fe ion (37, 60, 212). The nickel ion is ligated to four conserved cysteine residues (RxCGxCxxxH and DPCxxCxxH) at the N- and C-terminal regions, respectively, two of which are bridging the Fe and Ni ions. The Fe is also coordinated by two cyanide ions and one carbon monoxide molecule, two very uncommon biological ligands (18, 78). The hydrogen molecule is believed to access the active site through an identified hydrophobic channel (139), while conserved histidine and glutamate residues spanning from the active site to the molecule surface are responsible for the proton transfer (212). The transfer of electrons between the active site and the redox partner involves the participation of [FeS] clusters in the small subunit of NiFe hydrogenases. Sequence comparisons demonstrated that they possess several conserved cysteines, shown to participate in the formation of the [FeS] clusters. The heterodimeric NiFe hydrogenases are often directly linked to a variable diaphorase part containing additional prosthetic groups, e.g., [FeS] clusters, flavins, and cytochromes (6).
Cyanobacteria possess two functionally different NiFe hydrogenases, an uptake enzyme and a bidirectional enzyme. In 1995, structural genes encoding the latter enzyme were sequenced and characterized (173). These authors, to avoid confusion with the tritium exchange 42-kDa enzyme previously cloned (51-54), chose to call it a bidirectional hydrogenase, a name that is used throughout this review. The 42-kDa enzyme is commonly accepted to be an aminotransferase (228).
Cyanobacterial Uptake Hydrogenases
An uptake hydrogenase, with the evident function of catalyzing the consumption of the hydrogen produced by nitrogenase, has been found in all nitrogen-fixing cyanobacteria examined so far (87, 107, 192). Some physiological data, together with analysis of a mutant of the bidirectional hydrogenase gene hoxH, indicate that the unicellular non-nitrogen-fixing Synechococcus strain PCC 6301 (Anacystis nidulans) may also possess a hydrogen uptake enzyme (29, 48, 154-156). However, the presence of an uptake hydrogenase in non-nitrogen-fixing cyanobacteria is still a controversial subject, but the increasing availability of molecular data, particularly from genome projects, will help to clarify this question.
Since the small subunits of all known cyanobacterial uptake hydrogenases lack any N-terminal signal peptide (39, 79, 150) it has been suggested that the enzyme is localized on the cytoplasmic side of either the cytoplasmic or the thylakoid membrane (13, 209). In some filamentous strains, it is particularly expressed in the nitrogen-fixing heterocysts with little or no activity in the photosynthetic vegetative cells (see “Transcription of hupSL” below) (39, 87, 88, 152). The recycling of hydrogen produced by nitrogenase has been suggested to have at least three beneficial functions for the organism: (i) it provides the organism with ATP via the oxyhydrogen (Knallgas) reaction; (ii) it removes oxygen from nitrogenase, thereby protecting it from inactivation; and (iii) it supplies reducing equivalents (electrons) to nitrogenase and for other cell functions (32, 33, 90, 183, 216).
The uptake hydrogenase activity versus the bidirectional hydrogenase activity was characterized by Houchins and Burris (89) for Anabaena strain PCC 7120. In filaments, the uptake hydrogenase is resistant to atmospheric oxygen levels, but an inactivation that increased with the disruption of the filament was observed. The two hydrogenases differ in their thermal stability. Specifically, the uptake enzyme is more sensitive to high temperature, with a half-life of 12 min at 70°C, but less sensitive to competitive inhibition by carbon monoxide than is the bidirectional enzyme. Both enzymes have a low Km for hydrogen, but only the uptake hydrogenase activity was shown to be elicited by addition of hydrogen to the gas phase (48, 87, 116, 151, 169).
Biochemical and molecular data concerning filamentous heterocystous cyanobacteria point to the existence of at least two dissimilar subunits of about 60 and 35 kDa (39, 79, 88, 89, 112, 116, 150, 193). However, the exact subunit composition of cyanobacterial uptake hydrogenases and the molecular mass of the active holoenzyme are still unknown. It is premature to exclude the presence of additional subunits in the active form of the enzyme, since no active cyanobacterial uptake hydrogenase has yet been purified.
The cellular and subcellular localization of hydrogenases in cyanobacteria is unclear. It has been suggested that the enzyme is located in the thylakoid membranes of heterocysts (48), while others reported its presence in both the vegetative cells and heterocysts and a particular association with the cytoplasmic membrane (88, 116, 160). There is a diversity in the number of nitrogenases in a single nitrogen-fixing cyanobacterial strain, ranging from one in, e.g., Anabaena strain PCC 7120 and Nostoc strain PCC 73102 to at least three different nitrogenases in A. variabilis (98, 99, 195, 197, 198). However, since no study has addressed the question of putatively several uptake hydrogenases in a single strain, the pattern(s) of uptake hydrogenase cellular and subcellular localization might be different for different strains under different environmental conditions.
A strong correlation between the activity of an uptake hydrogenase and nitrogen fixation has been demonstrated in filamentous cyanobacteria (87, 107, 151, 221). In A. variabilis, exogenous hydrogen produces only a slight stimulatory effect on the in vivo hydrogen uptake during the early stages of nitrogenase induction (203). The authors suggest that the level of hydrogen evolved by nitrogenase is enough for the induction of hydrogenase synthesis. The temporary stimulation of hydrogen uptake in Nostoc strain PCC 73102 cultures incubated in darkness (i.e., cells with a much lower in vivo nitrogenase activity [126]) implies a strong correlation between the hydrogen uptake and nitrogenase activities and indicates that the addition of exogenous hydrogen alone is not enough to maintain the hydrogen uptake activity (151).
The biosynthesis of NiFe hydrogenases is closely related to Ni metabolism, both to form an active enzyme and at the transcriptional level (101, 135, 211, 212). Cyanobacterial hydrogenases seem to be no exception. In several strains the hydrogen uptake activity is dependent on the concentration of nickel in the growth medium (45, 46, 151, 152, 182, 202, 224).
Addition of organic carbon to the growth medium demonstrated that cells of Nostoc strain PCC 73102, grown either photo- or chemoheterotrophically, reach both higher nitrogenase and hydrogen uptake activities than do photoautotrophically grown cells (151). The maximal in vivo light-dependent hydrogen uptake activity occurred in cells grown heterotrophically in darkness, a condition that mimics symbiosis. This finding is in agreement with the suggested coupling between hydrogen consumption and an efficient nitrogen fixation in Nostoc sp. strain Cc and other symbiotic microorganisms (122, 124, 202). In contrast, autotrophically grown cells of a cyanobacterium isolated from Macrozamia communis have a higher level of both nitrogenase and hydrogenase activity than do heterotrophically grown cells (46). A rapid decrease in hydrogen uptake activity in chemoheterotrophically grown cultures of Anabaena cycadeae has been reported (105). Moreover, an inhibition of the uptake hydrogenase activity by the addition of glucose, with a consequent increase in hydrogen production by nitrogenase, was observed in the Gunnera-Nostoc symbiosis (123). This set of conflicting data is not easily explained. As already pointed out (124), the omission of nickel from the growth medium in some of the experiments might have influenced their results. Moreover, symbiotic cyanobacteria within coralloid roots of the cycads Cycas revoluta and Zamia furfuracea showed a significant in vivo hydrogen uptake (124).
Addition of the protein biosynthesis inhibitor chloramphenicol to Nostoc strain PCC 73102 cultures (151) inhibited the stimulation observed by nickel and organic carbon, suggesting a regulation at the synthesis and transcriptional level and not regulation of the activity of preexisting protein(s). A similar phenomenon has been reported for the filamentous nonheterocystous cyanobacterium Oscillatoria subbrevis (182). Based on midpoint potentials and inhibitor studies, the probable candidate for the initial electron acceptor of the uptake hydrogenase is plastoquinone (87). In A. variabilis, in vitro the uptake hydrogenase is activated by a reduced thioredoxin, leading to the proposal that in vivo the enzyme may be subjected to a form of redox control that also involves a thioredoxin (152, 185).
hupSL and the deduced proteins.
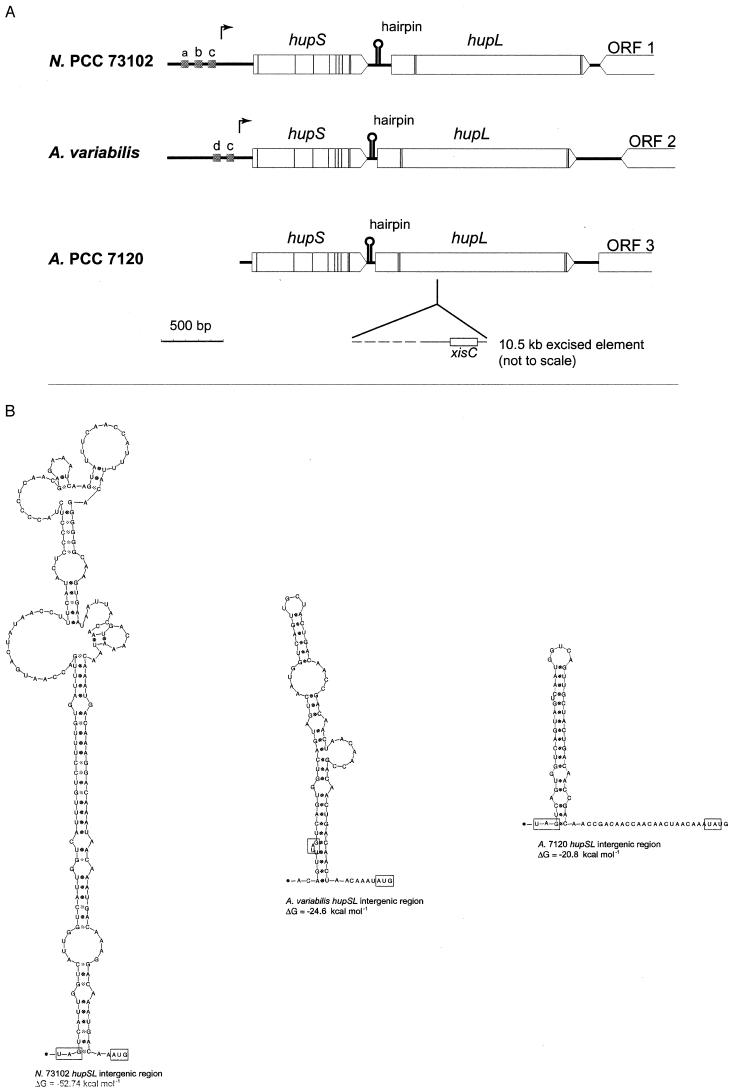
The first molecular data concerning cyanobacterial hydrogenases appeared in 1995. Carrasco et al. (39) described a novel developmental genome rearrangement for Anabaena strain PCC 7120, occurring in addition to the known nifD and fdxN rearrangements (see above), during the differentiation of a vegetative cell into a heterocyst. This third rearrangement occurs within a gene (hupL) encoding the large subunit of the uptake hydrogenase. The excision of a 10.5-kb DNA element occurs late during the heterocyst differentiation process, indicating that HupL in Anabaena strain PCC 7120 is expressed in heterocysts only. The excision occurs by a site-specific recombination, and the recombinase gene, xisC, was found 115 bp inside the right border of the excised element (Fig. 3A). Subsequently, the structural genes encoding both the small (hupS) and the large (hupL) subunits of the uptake hydrogenase have been sequenced and characterized in Anabaena strain PCC 7120 (www.kazusa.or.jp./cyano/anabaena), Nostoc strain PCC 73102 (150), and A. variabilis (79) (Fig. 3A).
FIG.3.
hupSL in the cyanobacteria Nostoc strain PCC 73102 (N. PCC 73102) (150), A. variabilis ATCC 29413 (79), and Anabaena strain PCC 7120 (A. PCC 7120) (39). (A) Thin vertical lines correspond to the nucleotide triplets encoding the conserved cysteine residues compared to HupSL in other organisms. Arrows show the positions of transcriptional start in Nostoc strain PCC 73102 and A. variabilis ATCC 29413. Grey boxes (a, b, c, and d) indicate the positions of putative promoter elements; a, NtcA-binding site; b, IHF-binding site; c, −10 promoter sequence; d, part of an FNR-binding site. Intergenic-sequence hairpin structures are also indicated (for details, see panel B), as well as positions and directions of putative ORFs, labeled 1 to 3, downstream of hupL. Note the presence of xisC and the subsequent rearrangement within hupL in Anabaena strain PCC 7120. (B) hupSL intergenic sequence hairpin structures. The stop codon of hupS and the start codon of hupL are boxed, and calculated ΔG values for each hairpin structure are shown below the respective structures (112).
All cyanobacterial hupSL genes sequenced so far are highly conserved, ranging from 83.8 to 95.1% nucleotide identities (Table 1), except that the occurrence of the hupL rearrangement/presence of xisC is not ubiquitous (17, 79, 192). They do, however, collectively differ significantly from the corresponding genes in other microorganisms, such as D. gigas (110), Bradyrhizobium japonicum (172), or Rhodobacter capsulatus (79, 108, 150). The deduced amino acid sequences of the cyanobacterial uptake hydrogenases all have >93% similarities to each other (Table 1; note the 99.7 and 99.6% amino acid similarity for HupS and HupL from Anabaena strain PCC 7120 and A. variabilis, respectively), whereas the corresponding percentage for Nostoc strain PCC 73102 HupL compared to D. gigas HupL is only 43%.
TABLE 1.
Sequence comparison of cyanobacterial hupSL and deduced amino acid sequencesa
| Strains | Nucleotide identity (%) |
Amino acid identity/similarity (%/%) |
||
|---|---|---|---|---|
| hupS | hupL | HupS | HupL | |
| Nostoc strain PCC 73102 vs A. variabilis | 84.4 | 83.8 | 88.8/93.8 | 91.1/95.1 |
| Anabaena strain PCC 7120 vs A. variabilis | 95.1 | 94.9 | 98.1/99.7 | 98.7/99.6 |
| Anabaena strain PCC 7120 vs Nostoc strain PCC 73102 | 84.2 | 85.0 | 88.8/93.8 | 90.6/95.1 |
HupS of cyanobacteria contains eight cysteine residues that clearly correspond to those involved in the formation of [FeS] clusters in D. gigas and a ninth cysteine whose position is only slightly different from that in other bacterial sequences (Fig. 3A; Table 2) (39, 79, 150, 212). The cyanobacterial small subunit also lacks the N-terminal twin-arginine signal peptide proposed to be involved in translocation of the protein, suggesting that the enzyme is not transported by the same machinery as in other organisms. Also missing in cyanobacterial HupS is the hydrophobic motif at the C terminus, but the role of this region in membrane anchoring is not clear since it is not present in all membrane-bound hydrogenases (122). The cyanobacterial HupL sequences contain putative Ni-binding sites characteristic for uptake hydrogenase large subunits at their N-terminal (FRGFE[I/V]ILRGKDPQAGLIVTPRIC62GIC65G[A/G]SH) and C-terminal (DPVEVGHVARSFDSC509LVC512TVHAH) ends (Table 2; Fig. 3A) (209), although at the C-terminal end the proline at position 508 is exchanged for a serine.
TABLE 2.
Positions and putative functions of conserved cysteine residues (Cys) in cyanobacterial uptake hydrogenases compared to those in Desulfovibrio gigas uptake hydrogenasea
It has been noted that some features of the cyanobacterial hupSL make it very similar to hupUV (hoxBC) involved in the regulation of the uptake hydrogenase in other microorganisms (R. capsulatus [50, 210], Ralstonia eutropha [109], and B. japonicum [24]), and as a consequence they may actually be encoding a regulatory hydrogenase rather than the uptake hydrogenase (150). One can point out that cyanobacterial HupS lacks the N-terminal signal peptide that is present in the small subunit of uptake hydrogenases from other microorganisms. However, the cyanobacterial HupL contains the C-terminal region, which is cleaved off during the maturation process of the protein, and an amino acid extension that is not present in HupV (150, 211); the N. punctiforme genome (http://www.jgi.doe.gov/JGI_microbia/html/nostoc/nostoc_homepage.html) does not contain any open reading frames (ORFs) similar to hupSL or hupUV, except for the already known hupSL. Low-stringency Southern hybridizations using Nostoc strain PCC 73102 and cloned hupSL (150) as the probe resulted in the identification of additional DNA fragments (115). However, recent experiments demonstrated that the noncoding region between hupS and hupL was the reason for the additional hybridization signal (112). Additionally, a hupL mutant of A. variabilis, cultivated under nitrogen-fixing conditions, exhibits significantly increased rates in hydrogen accumulation and produces three times more hydrogen than the wild type does (79). These results clearly demonstrate that the characterized cyanobacterial hupSL encode an uptake hydrogenase.
Generally, the structural genes encoding NiFe uptake hydrogenases are clustered in a similar physical organization forming a transcript unit, with hupS being located upstream of hupL. In several microorganisms, a third ORF, hupC, has been identified and is located directly downstream of the hupSL genes (83, 208, 211). It has been proposed that HupC, a b-type cytochrome, could play a role in mediating the electron transport to the terminal acceptor oxygen (43, 211). The cyanobacterial hupSL genes follow the general pattern (Fig. 3A) and are transcribed as a single transcript in A. variabilis ATCC 29413 (79) and in Nostoc strain PCC 73102 (112). In both organisms, as well as in Anabaena strain PCC 7120, ORFs have been detected downstream of the respective hupL genes, but none of them show any similarity to hupC and they are different in the three strains examined (Fig. 3A).
Transcription of hupSL.
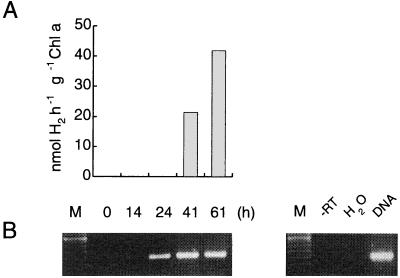
The first transcriptional study concerning cyanobacterial hupSL genes was performed with Anabaena strain PCC 7120, where non-nitrogen-fixing filaments of vegetative cells were transferred to nitrogen-fixing conditions. Reverse transcription-PCR (RT-PCR) demonstrated that hupL transcription coincides with heterocyst formation (39). Subsequent studies of Nostoc muscorum and A. variabilis ATCC 29413 (cyanobacteria without rearrangement of hupL; see “Genetic diversity of cyanobacterial hydrogenases” below) using RT-PCR and Northern blot analysis, respectively, confirmed the induction of a hupL transcript under nitrogen-fixing conditions only (17, 79). In N. muscorum, the induction of the transcript was followed by the appearance of an in vivo hydrogen uptake activity (Fig. 4) (17). In contrast to these results, a low level of hupL expression (detected by RT-PCR) has been found in A. variabilis ATCC 29413 vegetative cells grown with the addition of ammonia (28). Interestingly, when exposed to anaerobic conditions A. variabilis synthesizes a second Mo nitrogenase cellularly localized in the vegetative cells (199). The concomitant induction of the already identified uptake hydrogenase (79) or a possible alternative uptake hydrogenase deserves further studies. It was previously shown that ammonia-grown cells of A. variabilis have low but consistent in vivo hydrogen uptake, an activity that was attributed to the bidirectional enzyme (203). Moreover, some uptake hydrogenase activity could be detected in vegetative cells of Anabaena spp. grown microaerobically or anaerobically (88, 157).
FIG. 4.
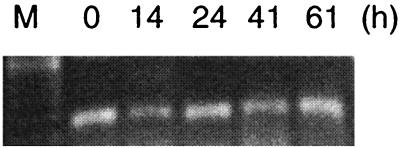
Induction of an in vivo light-dependent hydrogen uptake in Nostoc muscorum cells during a shift from non-nitrogen-fixing to nitrogen-fixing conditions. Ammonia-grown cells were transferred to nitrogen-fixing conditions (time = 0) and analyzed with a Clark-type electrode for the appearance of an in vivo light-dependent hydrogen uptake (A) and by RT-PCR for the presence of a hupL transcript (B) after 14, 24, 41, and 61 h (17). M, marker (100-bp DNA ladder). Controls (right panel) include only a PCR (-RT; all RNA samples, only t = 0 h shown, negative control), replacement of RNA with water (H2O, negative control), and use of genomic DNA instead of RNA (DNA, positive control).
The hupSL transcript in A. variabilis ATCC 29413 is 2.7 kb, and the transcription start site is located 103 bp upstream of the start codon, whereas in Nostoc strain PCC 73102 it is situated 259 bp upstream. Both start sites are preceded by putative −10 sequences, and putative promoter elements have been identified (Fig. 3A) (79, 112). In the A. variabilis sequence it is possible to identify half of a consensus FNR-binding sequence, and in Nostoc strain PCC 73102 there are possible integration host factor (IHF) and NtcA-binding sites (Fig. 3A) (79, 112). An FNR (fumarate nitrate reductase regulator)-binding site is involved in regulation of the Escherichia coli hyp operon (119), and the protein induces several operons during anaerobic growth (186). IHF is known to be involved in transcriptional activation of nif genes in purple bacteria (84), and the global cyanobacterial nitrogen regulator NtcA is necessary for heterocyst differentiation and expression of some of the genes involved in nitrogen fixation in Anabaena strain PCC 7120 (62, 82, 215).
The cyanobacterial hupSL genes differ from those of other microorganisms in being separated by longer intergenic regions (79, 150). These regions consist largely of 7-bp repeated sequences belonging to different families of short tandemly repeated repetitive sequences commonly found in heterocystous cyanobacteria (112, 131). Even though the specific short tandemly repeated repetitive sequences are not conserved, sequence analyses revealed that the possibility of a hairpin formation is a common feature, indicating a conserved two-dimensional structure rather than a specific sequence of the repeat itself (Fig. 3B) (112). The specific function(s) and origin of the repeats are not known (93, 131). One possible function of hairpins could be to increase the stability of the transcript. Another suggestion is that the hairpin structure may confer a translational coupling between the two structural genes (112).
Cyanobacterial Bidirectional Hydrogenases
The soluble or loosely membrane-associated cyanobacterial bidirectional hydrogenase is a common enzyme in both nitrogen-fixing and non-nitrogen-fixing cyanobacteria (75, 96, 97, 117, 177). However, recent data clearly demonstrated that, at least in nitrogen-fixing strains, it is not an universal enzyme and that strains missing the bidirectional enzyme have nothing obvious in common (192, 194). The Ni-containing bidirectional hydrogenase, partially purified and characterized from A. variabilis ATCC 29413 (178) differs from the uptake hydrogenase in its physical and catalytic properties (178, 202). In general, cyanobacterial bidirectional hydrogenases are characterized by their sensitivity to oxygen, thermotolerance, and high affinity to hydrogen (49, 87, 89, 163, 178). Methyl viologen, chemically reduced by dithionite, supports hydrogen evolution by all examined bidirectional hydrogenases and is usually used as an electron donor in the assay of the enzyme. Exposure to hydrogen during growth does not elicit additional activity of the bidirectional hydrogenase. Combined nitrogen in the growth medium also has little effect on the level of activity of the enzyme, suggesting that its function is independent of nitrogen fixation. Supporting this theory, Howarth and Codd (90) demonstrated that the biosynthesis of nitrogenase was not a prerequisite for the bidirectional hydrogenase biosynthesis and hydrogen evolution in several unicellular strains. The in vivo activity of the bidirectional enzyme in heterocystous strains increases considerably under anaerobic or microaerobic conditions (177), whereas in the unicellular non-nitrogen-fixing Gloeocapsa alpicola and Chroococcidiopsis thermalis the partial pressure of oxygen does not seem to have any significant influence (179, 180).
The physiological role of the bidirectional hydrogenase has been a matter of speculation and is still unclear. One possibility is that it functions as a mediator of the release of excess of reducing power in anaerobic environments. Kentemich et al. (97) and Schmitz et al. (173) reported that the low Km for hydrogen indicates that the enzyme generally operates in the direction of hydrogen uptake. Since the proton gradient in cyanobacteria is directed outward and the bidirectional hydrogenase has high affinity for hydrogen, they suggested that there may be a function in the oxidation of hydrogen at the periplasmic side and that the enzyme may allocate electrons to the respiratory chain (97, 173). Furthermore, it has been proposed that the enzyme functions as a valve for low-potential electrons generated during the light reaction of the photosynthesis, thus preventing the slowing of the electron transport chain under stress conditions (11). An incomplete version of respiratory complex I (11 subunits out of 14 strictly conserved in other prokaryotes like E. coli) was identified in cyanobacteria, and the bidirectional hydrogenase was suggested to fulfill the functions of the missing subunits (11, 12, 30, 174). However, other data do not support this theory (27, 64, 91, 189), including the fact that the bidirectional enzyme is not a universal cyanobacterial enzyme (192, 194). Since the bidirectional hydrogenase is absent in a significant set of strains, it seems unlikely that it plays a common, central role in cyanobacterial respiratory complex I, unless different cyanobacteria have adopted different strategies. Some of the conserved sequence motifs of the bidirectional hydrogenase are similar in the two corresponding complex I subunits, but apart from this there are only low sequence similarities (64). These authors proposed that the cyanobacterial and chloroplastidial complex, which both have 11 subunits in common with respiratory complex I, might work as a NADPH:plastoquinone oxidoreductase, possibly involved in the cyclic photosynthetic electron transport, and that the sequence similarities observed between the NADH dehydrogenase part of complex I and the bidirectional hydrogenase are due to a common ancestor. It should be pointed out that, at present, the existence of a respiratory 14-subunit complex I in cyanobacteria cannot be ruled out. Nostoc strain PCC 73102, a strain lacking the bidirectional hydrogenase (27, 194), respires at rates comparable to those of other cyanobacteria (27), and bidirectional hydrogenase-minus mutants of Synechocystis strain PCC 6803 exhibited growth and respiration rates comparable to those of the wild type (91). It is obvious that more data are necessary to clarify the function of the bidirectional hydrogenase in cyanobacteria.
hox genes and the deduced proteins.
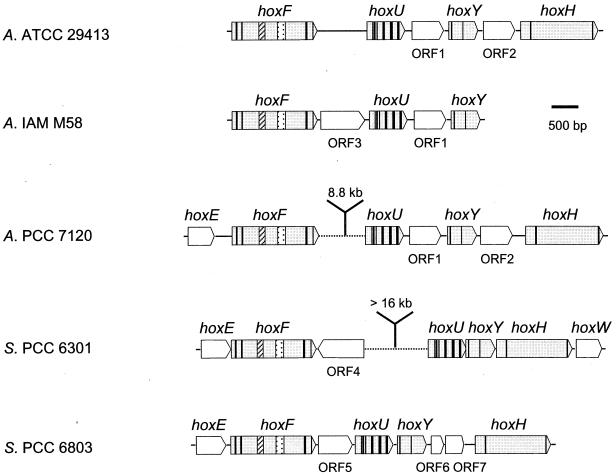
In 1995, Schmitz et al. (173) sequenced a set of structural genes (hox genes) encoding a bidirectional hydrogenase in A. variabilis ATCC 29413. These authors suggested that the bidirectional enzyme is a heterotetrameric enzyme consisting of a hydrogenase part (encoded by hoxYH) and a diaphorase part (encoded by hoxFU) (Fig. 1 and 5). In subsequent works, hox genes have been sequenced and characterized in the unicellular Synechocystis strain PCC 6803, Synechococcus strain PCC 6301 (12, 28-30, 95, 144, 174), and (partially) C. thermalis CALU 758 (178), as well as in the filamentous strains Anabaena strain PCC 7120 (www.kazusa.or.jp./cyano/anabaena) and (partially) A. variabilis IAM M58 (GenBank accession no. AB057405). The physical organizations of the structural genes encoding the bidirectional enzyme are similar (Fig. 5). In A. variabilis, Anabaena strain PCC 7120, Synechococcus strain PCC 6301, and Synechocystis strain PCC 6803, one or several additional ORFs have been identified between some of the structural genes. In A. variabilis IAM M58, an additional ORF is localized between hoxF and hoxU (Fig. 5). The intergenic distances between hoxF and hoxU in both Synechococcus strain PCC 6301 and Anabaena strain PCC 7120 are longer (at least 16 and approximately 9 kb, respectively) than in other strains (Fig. 5). An ORF positioned upstream of hoxF and termed hoxE has been identified and sequenced in Synechococcus strain PCC 6301 and Synechocystis strain PCC 6803, and is also present in Anabaena strain PCC 7120 (http://www.kazusa.or.jp./cyano/anabaena). It has been suggested to encode a third diaphorase subunit (28). Moreover, in Synechococcus strain PCC 6301, an ORF termed hoxW is localized downstream of hoxH, putatively encoding a protease (Fig. 5) (30).
FIG. 5.
Physical map of the genes encoding the bidirectional hydrogenase (hox) and additional ORFs (labeled 1 to 7, with identical numbers indicating homologous ORFs) in the cyanobacteria A. variabilis (two strains; A. ATCC 29413 [173] and A. IAM M58 [Gen. Bank accession no. AB057405]), Anabaena strain PCC 7120 (A. PCC 7120) (http://www.kazusa.or.jpn/cyano/anabaena), Synechococcus PCC 6301 (S. PCC 6301) (29, 30), and Synechocystis PCC 6803 (S. PCC 6803) (12, 95, 144). Vertical lines correspond to the positions of triplets encoding conserved cysteine residues compared to other microorganisms (for more details, see Table 4). ▨ and represent the NAD- and flavin mononucleotide-binding regions, respectively.
As with the uptake hydrogenase, molecular studies helped to clarify the picture of the subunit composition and molecular mass of the bidirectional hydrogenase. In agreement with the molecular data, previous work referred to large subunits of about 50 to 56 kDa in A. variabilis, Synechococcus strain PCC 6301, M. aeruginosa, and Spirulina plantensis (15, 96, 118) and to small subunits of about 17 kDa in A. variabilis and Synechococcus strain PCC 6301 (96). In A. variabilis, hoxH and hoxY encode predicted polypeptides of 54.8 and 22.5 kDa, respectively (173). Concerning the holoenzyme, earlier chromatographic studies reported an apparent molecular mass of 230 kDa for the hydrogenase of A. cylindrica (73). However, as pointed out by the authors, the enzyme was probably bound in a complex with other proteins. Considerably lower molecular masses (165 and 113 kDa) were reported for the bidirectional hydrogenase of Anabaena strain PCC 7120, with two-thirds of the activity being found in the 165-kDa peak (89). Native polyacrylamide gel electrophoresis followed by in vitro activity staining demonstrated the presence of a functional enzyme of about 118 kDa in induced cells of A. variabilis, Anabaena strain PCC 7120, and N. muscorum (177, 194). It is believed that in A. variabilis the four predicted polypeptides (Fig. 1) are assembled to form a tetrameric enzymatic complex, which is consistent with the molecular masses mentioned above.
The cellular and subcellular localization of the bidirectional hydrogenase is a topic that needs further investigation. Earlier work indicated that the enzyme is present in both the vegetative cells and heterocysts and that through gentle cell disruption procedures it was easily solubilized and thus located in the cytoplasm (73, 88, 89). However, other works suggest an association of the bidirectional hydrogenase with cell membranes. The enzyme of the unicellular Synechococcus strain PCC 6301 seems to be loosely associated with the cytoplasmic membrane (96, 97), whereas associations with the thylakoid membranes have been demonstrated in A. variabilis and Synechocystis strain PCC 6803 (11, 177).
Nucleotide sequence comparisons have shown that there is a reasonably high degree of homology between the hox genes of cyanobacteria and genes encoding the NAD+-reducing hydrogenase from the chemolithotrophic hydrogen-metabolizing bacterium R. eutropha as well as those encoding methyl viologen-reducing hydrogenases from species of the archaeal genera Methanobacterium, Methanococcus, and Methanothermus (173). Sequence comparisons between the bidirectional hydrogenase genes or deduced proteins of A. variabilis ATCC 29413, A. variabilis IAM M58, Anabaena strain PCC 7120, Synechococcus strain PCC 6301, and Synechocystis strain PCC 6803 reveal that they are highly homologous, with A. variabilis ATCC 29413 and Anabaena strain PCC 7120 being at least 95% identical when the deduced amino acid sequences of proteins are compared (Table 3). The two A. variabilis strains are less similar (78 to 88% identical), and all other comparisons are in the range of 58 to 71% identical amino acids of the deduced proteins, with the exception of 80 to 88% identical amino acids when comparing Anabaena strain PCC 7120 and A. variabilis IAM M58 (Table 3).
TABLE 3.
Sequence comparison of the structural genes hoxFUYH and the deduced amino acid sequences, of the cyanobacteria A. variabilis ATCC 29413, A. variabilis IAM M58, Anabaena strain PCC 7120, Synechococcus strain PCC 6301, and Synechocystis strain PCC 6803
| Strain | Gene or protein | Sequence comparisona with: |
|||
|---|---|---|---|---|---|
| A. variabilis IAM M58 | Anabaena strain PCC 7120 | Synechococcus strain PCC 6301 | Synechocystis strain PCC 6803 | ||
| A. variabilis ATCC 29413 | hoxFUYH | 78/82/76/—b | 93/94/91/94 | 63/62/60/63 | 65/64/67/67 |
| HoxFUYH | 82 (87)/88 (95)/78 (83)/— | 95 (96)/98 (99)/95 (96)/97 (98) | 62 (73)/63 (73)/60 (75)/64 (74) | 65 (76)/62 (75)/63 (77)/68 (77) | |
| A. variabilis IAM M58 | hoxFUYH | 79/82/76/— | 66/63/64/— | 67/66/65/— | |
| HoxFUYH | 81 (87)/89 (95)/80 (89)/— | 61 (72)/58 (71)/60 (75)/— | 67 (76)/61 (74)/62 (78)/— | ||
| Anabaena strain PCC 7120 | hoxFUYH | 63/77/63/63 | 66/65/67/66 | ||
| HoxFUYH | 60 (71)/61 (72)/60 (74)/65 (74) | 65 (76)/62 (75)/62 (76)/68 (77) | |||
| Synechococcus strain PCC 6301 | hoxFUYH | 64/66/63/66 | |||
| HoxFUYH | 59 (71)/70 (80)/62 (80)/71 (79) | ||||
Nucleotide identity (%) and amino acid identity (similarity) (%) are given for hoxF, hoxU, hoxY, and hoxH (and their proteins) separately.
—, not sequenced in A. variabilis IAM M58.
Some of the characteristics of the deduced proteins are summarized in Fig. 5 and Table 4. HoxH (the large subunit of the hydrogenase dimer) in cyanobacteria harbors six conserved cysteines, four matching the typical cysteine-containing N-terminal and C-terminal conserved motifs involved in the binding of nickel to the active site (174, 212). The unicellular strains contain an additional cysteine positioned 4 amino acids upstream of the C-terminal motif. In the small subunit of the hydrogenase dimer, HoxY, four cysteine residues, believed to be involved in coordinating a putative [4Fe4S] cluster (212), can be identified (Fig. 5; Table 4). The HoxU protein (the small subunit of the diaphorase moiety) contains several conserved cysteine residues putatively involved in the binding of [FeS] clusters. Two of the motifs are located at the N-terminal part of the protein; the first harbors four cysteines probably involved in binding a [2Fe2S] cluster, and the second harbors three cysteines residues putatively binding a [3Fe4S] or [4Fe4S] cluster (12, 174). The final conserved cysteine residue in the latter motif is preceded by an additional cysteine in the filamentous strains. Moreover, the C-terminal part of HoxU possesses two typical [4Fe4S] cluster-binding sites (Fig. 5; Table 4) (12, 174). The large subunit of the diaphorase moiety, HoxF, is generally less highly conserved in its N-terminal part. However, this region harbors two shorter, highly conserved cysteine-containing stretches with a longer nonconserved region in between, which is postulated to be involved in binding a [2Fe2S] cluster (Fig. 5; Table 4) (63). All five cyanobacterial strains contain another conserved cysteine directly upstream of this motif; the filamentous strains also harbor a cysteine immediately downstream. In the middle region of this subunit, typical glycine-rich binding sites for NAD (GxGxxGxxxG) and flavin mononucleotide (GxGxxxxGx10GxxG) can be identified (Fig. 5) (173). The five cyanobacterial HoxF proteins contain, in the NAD motif, another glycine following the second conserved one; in addition, in the two unicellular strains a third glycine immediately follows this additional glycine. The C-terminal end of HoxF contains a conserved motif with five cysteines, four of which correspond to the residues postulated to be involved in binding a [4Fe4S] cluster (Table 4) (170, 173). The fifth cysteine is positioned 9 amino acids upstream of the last conserved residue in this motif. In addition, a universal sixth cysteine preceding the conserved C-terminal motif is present in cyanobacterial HoxF.
TABLE 4.
Positions and putative functions of conserved cysteine residues (Cys) in cyanobacterial bidirectional hydrogenases compared to those in similar hydrogenases in other microorganisms
| Protein | Conserved residuesa | Function | Reference(s) |
|---|---|---|---|
| HoxF | Cys25, Cys30, Cys62, Cys66 | Binding of putative [2Fe2S] cluster | 12, 63, 174 |
| Cys453-463, Cys456-466, Cys459-469, Cys499-509 | Binding of putative [4Fe4S] cluster | 170, 173 | |
| HoxU | Cys36, Cys47, Cys50, Cys64 | Binding of putative [2Fe2S] cluster | 12, 174 |
| Cys100, Cys103, Cys109 | Binding of either putative [3Fe4S] or putative [4Fe4S] cluster | 12, 174 | |
| Cys148, Cys151, Cys154, Cys158, Cys192, Cys195, Cys198, Cys202 | Binding of two putative [4Fe4S] clusters | 12, 174 | |
| HoxY | Cys14, Cys17, Cys86-87, Cys150-153 | Binding of putative [4Fe4S] cluster | 212 |
| HoxH | Cys62, Cys65, Cys443-452, Cys446-455 | Binding of Ni | 159, 174, 212 |
The indicated intervals represent different positions in different strains.
Transcription of hox genes.
Transcriptional studies (using RT-PCR) indicate that the structural genes form a single transcriptional unit in A. variabilis ATCC 29413 (28). In contrast, in the unicellular Synechococcus strain PCC 6301 two transcripts were detected, the dicistronic hoxEF and the polycistronic hoxUYHWhypAB (28). Further promoter activities were identified in the hox locus by using β-galactosidase and bacterial luciferase as reporters (28). With the exception of a shift from non-nitrogen-fixing to nitrogen-fixing conditions, which was associated with no significant change in the transcript level of hoxH in N. muscorum (Fig. 6) (17), the influences of key environmental conditions on the transcription of the cyanobacterial hox genes remain to be examined.
FIG. 6.
Visualization of a hoxH transcript from N. muscorum during a shift from non-nitrogen-fixing to nitrogen-fixing conditions (the same experiment as described in the legend to Fig. 4). Transcripts from t = 0, 14, 24, 41, and 61 h after the transfer to nitrogen-fixing conditions are shown (17). M, marker (100-bp DNA ladder).
Genetic Diversity of Cyanobacterial Hydrogenases
The molecular diversity of cyanobacterial hydrogenases can be perceived from the results presented for a selected set of nitrogen-fixing Anabaena and Nostoc strains (Table 5). Although hup gene homologues are present in all strains examined, all kinds of combinations can be detected concerning the presence or absence of sequences homologous to xisC and to the bidirectional hydrogenase genes. Interestingly, at present there are no data supporting the existence of either multiple uptake hydrogenases or multiple bidirectional hydrogenases in a single cyanobacterial strain. It should be pointed out that it is not possible to establish any correlation between the presence or absence of the bidirectional hydrogenase and the occurrence of the gene responsible for the rearrangement of hupL. Similarly, there is no obvious connection to the different habitats from where the individual strains originally were isolated. The natural molecular variation of hydrogenases in different cyanobacteria is a field both to examine and to explore in an effort to understand the physiological function(s) as well as regulation of the respective enzyme and in an effort to identify and use the “correct genetic background” when constructing and examining a genetically engineered cyanobacterial strain.
TABLE 5.
Molecular diversity of hydrogenases in selected nitrogen-fixing Anabaena and Nostoc strainsa
| Group | Strainb | hup | xisC | hox | Reference(s) |
|---|---|---|---|---|---|
| A | Anabaena strain PCC 7120 | + | + | + | 39, 194 |
| Nostoc strain PCC 6314 | + | + | + | 192 | |
| Nostoc strain Mitsui 56111 | + | + | + | 192 | |
| B | A. variabilis ATCC 29413 | + | − | + | 17, 79, 173, 194 |
| N. muscorum CCAP 1453/12 | + | − | + | 17, 194 | |
| Nostoc strain Mitsui 91911 | + | − | + | 192 | |
| C | Nostoc strain Mitsui 38901 | + | + | − | 192 |
| Nostoc strain CYA 238 | + | + | − | 192 | |
| D | Nostoc strain PCC 73102 | + | − | − | 26, 150, 194 |
| Nostoc strain CYA 190 | + | − | − | 192 | |
| Nostoc strain ACOI 578 | + | − | − | 192 |
Diversity is demonstrated by the presence (+) or absence (−) of sequences homologous to hup (uptake hydrogenase), xisC (rearrangement in hupL in Anabaena strain PCC 7120), and hox (bidirectional hydrogenase) genes.
Selected strains are arranged in four groups: (A) strains containing hup, xisC, and hox; (B) strains containing hup and hox; (C) strains containing hup and xisC; and (D) strains containing only hup.
Accessory Genes
The maturation of nickel-containing enzymes, e.g., hydrogenases, ureases, and carbon monoxide dehydrogenases, is a complex process requiring accessory proteins (42, 81, 122, 127, 135, 211, 213). Initial work using E. coli revealed five ORFs, designated hypABCDE, affecting hydrogenases pleiotropically (119). At present, there is no information on the function of Hyp proteins in cyanobacteria. However, some of the homologous gene products of other bacteria (e.g., E. coli, R. eutropha, and B. japonicum) are well characterized, and in general homologous Hyp proteins fulfill similar functions in different organisms (summarized in Table 6) (42).
TABLE 6.
Conserved motifs and putative functions of putative cyanobacterial Hyp proteins
| Protein | Conserved motif(s) | Function(s) | References |
|---|---|---|---|
| HypA | Ni incorporation (putative) | 28, 71, 148, 201 | |
| Cx1-2Cx12-13Cx1-2C | [FeS] cluster binding or zinc finger motif, regulation | ||
| HypB | Ni incorporation | 28, 34, 71, 145, 147,148 | |
| N-terminal CxxCGCa | Protein stability | ||
| Histidine-rich N terminusa | Mobilization, carriage and storage of nickel | ||
| x2Nx2SSPGAGKTx27DAx6Gx44-48Px5Gx28x4TKxD | GTPase | ||
| HypC | Chaperone | 25, 121, 146 | |
| N-terminal MCL(G/A)(L/I)P | The cysteine interacts with the precursor of the large hydrogenase subunit and facilitates Ni insertion | ||
| HypD | Ni incorporation | 71, 146 | |
| CPVC | Potential metal binding | ||
| 5 conserved cysteines | Unknown | ||
| HypE | Purine derivate binding | 66, 71, 171 | |
| G(G/S)GGK(L/A)(M/S)x(Q/D)Lx55-62x(V/I)NDLA(V/M) (S/A)GaxP(L/R)x74-79GDxx(I/L)x(S/N)G(E/D)(L/I)G(N/R) HGx152-157(E/D)QLPRIC | Potential phosphoribosylformylglycinamidine cycloligase (AIR synthethase; EC 6.3.3.1) | ||
| HypF | CO/CN synthesis | 28, 146, 153 | |
| G(R/T)VQGVGFRx13G(D/W)V(C/N)Nx3G | Potential acylphosphatase | ||
| Cx2Cx18Cx2Cx24Cx2Cx18Cx2C | Two zinc fingers, transcriptional regulation | ||
| HHxAH | Histidine motif |
Not present in HypB′ of Synechocystis strain PCC 6803.
Organization and transcription of cyanobacterial hyp genes.
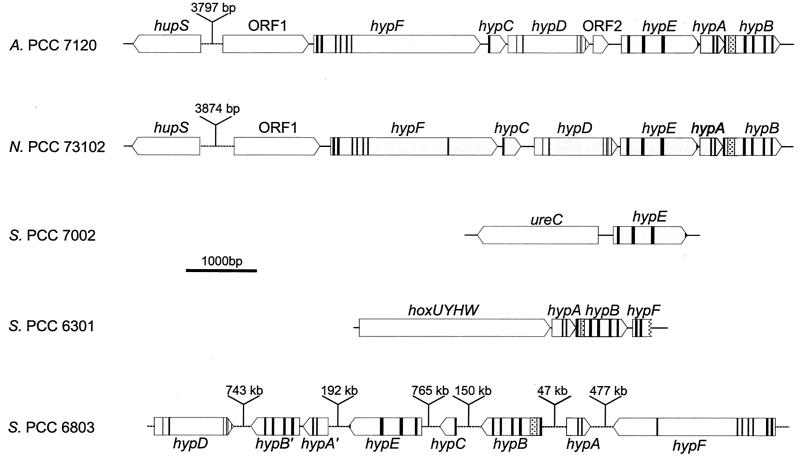
Although hydrogenase genes have been identified and characterized in several cyanobacteria (Tables 1 to 4 and 7), very little is known about the cyanobacterial hyp genes and the corresponding Hyp proteins (Fig. 7). Among the first hyp genes to be identified were hypABF in Synechococcus strain PCC 6301 (29) and hypB in Anabaena strain PCC 7120 (originally published as hupB [70]) (Fig. 7). The former are preceded by structural genes encoding the bidirectional hydrogenase of Synechococcus strain PCC 6301 (hoxUYH) and the putative protease HoxW, which might be involved in the maturation of HoxH (200) (Fig. 5 and 7). hypF was only partially sequenced. Transcriptional analysis (RT-PCR) revealed that hypA and hypB form a polycistronic transcript together with hoxUYHW, whereas hypF is part of a different transcript (28). hypB of Anabaena strain PCC 7120 is part of a gene cluster consisting of hypBAEDF (originally published as hupBAEDF [71]), in which hypA and hypB overlap by 10 bp (as in Nostoc strain PCC 73102 [74; http://jgi.doe.gov/JGI_microbial/html/nostoc/nostoc_homepage.html]). Analysis of the genome of Anabaena strain PCC 7120 (http://www.kazusa.or.jpn/cyano/anabaena) showed that hupSL and the hyp genes are approximately 3.8 kb apart and directed in opposite directions from each other (Fig. 7). Moreover, an ORF encoding a HypC homologue is located between hypF and hypD, and another ORF putatively encoding a 4-oxalocrotonate tautomerase homologue (217) is situated between hypD and hypE (Fig. 7). RT-PCR analysis revealed that hypB is expressed in heterocyst-induced but not in non-nitrogen-fixing cultures (71). A hyp gene cluster consisting of hypFCDEAB and an additional upstream ORF has been cloned from Nostoc strain PCC 73102 and sequenced (74). RT-PCR-based gene expression analysis showed that these genes form one transcript. Upstream of the transcriptional start site, putative binding sites for the global nitrogen regulator NtcA have been identified (74). A putative NtcA-binding site has also been identified upstream of the hupSL transcriptional start site (112), in agreement with the experimental evidence demonstrating that both the hyp and the hup operons are transcribed under nitrogen-fixing but not under non-nitrogen-fixing conditions (74). As in Anabaena strain PCC 7120, the hup operon is located upstream of the hyp operon (about 3.9 kb) and oriented in the opposite direction (Fig. 7). In Synechocystis strain PCC 6803, the hyp genes are scattered over the genome, with the exception of hypA′ and hypB′, which have the same direction and are 61 bp apart (Fig. 7). Two of the six hyp genes of Synechocystis strain PCC 6803, hypA and hypB, are duplicated (Fig. 7; Table 7) (95, 144). These are, at present, the only known duplications of cyanobacterial hyp genes, although duplication of hyp genes is not uncommon among bacteria. In addition, Synechocystis strain PCC 6803 contains an identified hupE homologue, an accessory gene with similarities to genes encoding both hydrogenase and urease accessory proteins (95, 144). A hypE homologue has been cloned and sequenced from Synechococcus strain PCC 7002 (171). The gene is located 213 bp upstream of ureC (encoding a putative urease subunit), but in the opposite direction (Fig. 7).
TABLE 7.
Summary of genes related to hydrogenases in cyanobacteria
| Strain | Accession no. (reference) of: |
||||||
|---|---|---|---|---|---|---|---|
| hupS | hupL | hoxE | hoxF | hoxU | hoxY | hoxH | |
| Unicellular | |||||||
| Synechococcus strain PCC 6301 | Y13471a (30) | Y13471 (30) | X97797 (29, 30) | X97797 (29, 30) | X97797 (29, 30) | ||
| Synechococcus strain PCC 7002 | |||||||
| Synechocystis strain PCC 6803 | NPa (11, 95, 144) | NP (11, 95, 144) | sll1220,b X97610 (12, 95, 144) | sll1221, X97610 (12, 95, 144) | sll1223, X97610 (12, 95, 144) | sll1224, X97610 (12, 95, 144) | sll1226, X97610 (12, 95, 144) |
| Filamentous heterocystous | |||||||
| A. variabilis ATCC 29413 | Y13216 (79) | Y13216 (79) | X79285 (173) | X79285 (173) | X79285 (173) | X79285 (173) | |
| A. variabilis IAM M58 | AB057405 | AB057405 | AB057405 | ||||
| Anabaena strain PCC 7120 | all0688c, U08013 (39) | all0687N/C, U08013 (39) | alr 0751 | alr0752 | alr0762 | alr0764 | alr0766 |
| N. punctiforme PCC 73102 (ATCC 29133) | c651/20, AF030525 (150) | c651/21, AF030525 (150) | NP (27)d | NP (27)d | NP (27)d | NP (194)d | NP (194)d |
| Accession no. (reference) of: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hoxW | hypA | hypA′ | hypB | hypB′ | hypC | hypD | hypE | hypF | ||||||||
| X97797 (29, 30) | X97797 (29, 30) | X97797 (29, 30) | X97797 (29, 30) | |||||||||||||
| AF035751 (171) | ||||||||||||||||
| slr1675 (95, 144) | sll1078 (95, 144) | Sll1432 (95, 144) | sll1079 (95, 144) | ssl3580 (95, 144) | slr1498 (95, 144) | sll1462 (95, 144) | sll0322 (95, 144) | |||||||||
| c286, AF006594 (71)d | c286, AF006594 (70, 71)d | c286d | c286, AF006594 (71)d | c286, AF006594 (71)d | c286d | |||||||||||
| c651/98, AF325724 (74)d | c651/99, AF325724 (74)d | c651/95, AF325829 (74)d | c651/96, AF325724 (74)d | c651/97, AF325724 (74)d | c651/94, AF325829 (74)d | |||||||||||
GenBank; http://www.ncbi.nlm.nih.gov/.
CyanoBase; http://www.kazusa.or.jp/cyano/cyano.html.
CyanoBase; http://www.kazusa.or.jp/cyano/anabaena/.
NP, not present.
FIG. 7.
Physical organization and conserved regions (vertical lines; for further explanations, see Table 6) of hitherto identified cyanobacterial hyp genes. The histidine-rich N terminals of hypB are indicated ( ). Anabaena strain PCC 7120 (A. PCC 7120) (70, 71; http://www.kazusa.or.jpn/cyano/anabaena), Nostoc PCC 73102 (N. PCC 73102) (74; http://www.jgi.doe.gov/JGI_microbial/html/nostoc/nostoc_homepage/html); Synechococcus strain PCC 7002 (S. PCC 7002) (171), Synechococcus strain PCC 6301 (S. PCC 6301) (29), and Synechocystis strain PCC 6803 (S. PCC 6803) (95, 144).
Interestingly, Anabaena strain PCC 7120 contains only one set of hyp genes, and the presence of both an uptake (39) and a bidirectional hydrogenase (1, 194) in this strain suggests that one copy is enough to fulfill the functions for both hydrogenases. Assuming that this is a general feature for cyanobacteria, the specific regulation of hyp genes in response to the differentially regulated hydrogenases in a single cyanobacterial strain deserves future attention.
CYANOBACTERIAL BIOHYDROGEN
As discussed above, two cyanobacterial enzymes are capable of hydrogen production: the nitrogenase(s) and the bidirectional hydrogenase. Individual strains may harbor both a bidirectional hydrogenase (although this is not a universal cyanobacterial enzyme [Table 5] [192]) and none to several nitrogenases, encoded by different structural genes. There is no report describing the existence of either several uptake hydrogenases or several bidirectional hydrogenases in a single cyanobacterial strain. An efficient photoconversion of water to hydrogen by cyanobacteria is certainly influenced by many other factors, and only an extensive knowledge of this field can lead to improvement of the rates of cyanobacterial hydrogen production. For example, it is known that immobilized cells produce more hydrogen than do free-living cultures (125) and that non-Mo-containing nitrogenases allocate more electrons to the production of hydrogen (206). The gas phase (214), the age and density of the culture, and the composition, pH, and temperature of the growth medium are also crucial to the final result. Most of the research on cyanobacterial hydrogen production has been carried out with nitrogen-fixing strains (31, 55, 106, 111, 166, 205, 206). In these organisms, the net hydrogen production is the result of hydrogen evolution catalyzed by nitrogenase and of hydrogen consumption catalyzed mainly by an uptake hydrogenase. Consequently, the production and selection of mutants deficient in hydrogen uptake activity is of great interest (76, 79, 115, 136). Moreover, the nitrogenase has a high ATP requirement and this lowers the potential solar energy conversion efficiencies considerably. On the other hand, the bidirectional hydrogenase requires much less metabolic energy but is extremely sensitive to oxygen (14, 15). For reviews on the potential, problems and prospects of hydrogen production by cyanobacteria, see references (19, 20, 72, 75, 113, 117, 120, 125, and 164).
Rates of Cyanobacterial Hydrogen Production
As discussed above, most of the research on rates of cyanobacterial hydrogen production has been carried out with nitrogen-fixing filamentous strains with typical values ranging from 0.17 to 4.2 nmol of hydrogen produced per μg of chlorophyll a per h (128). Interestingly, nitrogen-fixing cells of A. variabilis SPU 003 have the capacity to produce hydrogen mainly in darkness (181). Addition of various sugars stimulated the production of hydrogen, with mannose giving the highest rate, 5.58 nmol of hydrogen produced per mg dry weight per h (181). Since the hydrogen production is the result of hydrogen evolution catalyzed by nitrogenase and a hydrogen consumption catalyzed mainly by an uptake hydrogenase, the obvious improvements are to increase the hydrogen production by using alternative nitrogenases and by inhibiting the activity of the uptake hydrogenase. In a recent study, the hydrogen production by autotrophic, vanadium-grown cells (i.e., cells expressing the alternative vanadium-containing nitrogenase) of A. variabilis ATCC 29413 (wild type) and of its mutant PK84, impaired in the utilization of molecular hydrogen, was studied in a photobioreactor (204). The highest hydrogen production rates were observed in cultures grown at gradually increased irradiation. The wild-type strain evolved hydrogen only under a argon atmosphere, with the actual rate being as high as the potential rate (61% of oxygenic photosynthesis used for hydrogen production). In contrast, PK84 cells also produced hydrogen during growth under carbon dioxide-enriched air (13% of oxygenic photosynthesis used for hydrogen production, representing 33% of the potential rate) (204). A. variabilis PK84 has also been examined under simulated outdoor conditions (31). Use of a 4.35-liter automated helical tubular bioreactor and continuous cultivation for 2.5 months resulted in a maximum hydrogen evolution of 230 ml of hydrogen produced per 12-h light period at a growth density of 3.6 to 4.6 μg of chlorophyll a per ml of cell culture (31). Replacing air with argon doubled the rate of hydrogen evolution. Anoxygenic conditions over the dark periods had a negative effect on hydrogen production. Similarly, under aerobic outdoor conditions operated for 4 months, a maximum rate of 80 ml of hydrogen per reactor (4.35 liters) per hour was obtained from a batch culture on a bright day (57). The maximum efficiency of conversion of light to chemical energy of hydrogen was calculated to be 0.33 and 0.14% on a cloudy and a sunny day, respectively (57).
Strategies for Improving Cyanobacterial Strains for Photobiological Hydrogen Production
Thorough studies on hydrogen uptake and/or evolution have until now focused on only a few filamentous cyanobacteria, e.g., different Anabaena and Nostoc strains. However, other cyanobacteria, e.g., Oscillatoria, are able to fix nitrogen without forming heterocysts by using the strategy of temporal separation of the oxygen-sensitive nitrogen fixation and the oxygen-evolving photosynthesis. Such strains deserve a thorough examination of their hydrogen metabolism. Considering the versatility of cyanobacteria and their ability to survive under many different environmental conditions, more strains originating from different habitats must be studied with respect to their applicability in biohydrogen production. The genetic backgrounds (e.g., structural and accessory genes including mechanisms of regulation) of the selected strains and their overall capacities to be genetically modified should be determined. Of specific interest might be isolates originating from nitrogen-fixing associations. The situation of the symbiotic cyanobacteria is similar to that of steady-state cultures in bioreactors: the cells almost do not grow, they have a high metabolism in a restricted space, and they export at least one metabolite to the host. In addition, a heterotrophic capacity can be used during conversion of organic wastes into valuable, energy-rich compounds such as molecular hydrogen. Similarly, other cyanobacterial habitats mimicking these conditions, e.g., cyanobacterial mats, are also of great interest.
Several strategies are available for improving existing cyanobacterial strains for the biotechnological production of hydrogen. Inactivation of a gene encoding the uptake hydrogenase leads to a mutant that is not able to recycle the hydrogen evolved by the nitrogenase under nitrogen-fixing conditions. Consequently, hydrogen produced through the action of a nitrogenase will either be oxidized by the bidirectional hydrogenase or, if this enzyme is not present, be evolved from the cells. Recent experiments demonstrated that a nitrogen-fixing culture of an uptake-deficient mutant (a hup mutant generated by introducing a cassette of foreign DNA into hupL) of A. variabilis ATCC 29413 (a strain containing both an uptake hydrogenase and a bidirectional enzyme [Table 5]) produces molecular hydrogen (79). The absence of a bidirectional enzyme in cyanobacteria such as Nostoc strain PCC 73102 (Table 5) makes these strains interesting candidates for inactivation experiments. Identification and engineering of an oxygen-stable hydrogen-evolving hydrogenase might result in a photosynthesizing microorganism that evolves significant amounts of hydrogen. Interestingly, replacing Azotobacter vinelandii hydrogenase small-subunit cysteines with serines can create insensitivity to inhibition by oxygen and preferentially damage hydrogen oxidation over hydrogen evolution (132). Moreover, overproducing mutants might be obtained by providing genes encoding a selected hydrogenase on an expression vector. Coupling the genes to a strong promoter might lead to an increased amount of the hydrogenase and thus to increased levels of hydrogen produced by the organism. Similarly, overexpression might also be used to increase the nitrogenase activity. A thorough examination of the genes involved in the regulation of hydrogenase expression, e.g., hyp genes, might generate knowledge leading to further strategies for improving hydrogen production rates in cyanobacteria.
Genetic engineering has become increasingly important with the establishment of molecular biological tools and techniques for cyanobacteria. A few unicellular strains, including Synechococcus strains PCC 6301 and PCC 7942 and Synechocystis strain PCC 6803, are naturally transformable. Protocols and vector systems useful for the transfer of DNA into different cyanobacteria are available for nontransformable strains (196). These methods have been used with success in filamentous genera such as Anabaena and Nostoc, which are interesting candidates for future photobiotechnological applications (75, 76, 79). The introduction of the structural genes encoding a clostridial Fe hydrogenase into the unicellular cyanobacterium Synechococcus elongatus PCC 7942 and their subsequent transcription and translation into a functional hydrogen-evolving enzyme in a photosynthetic microorganism (16, 138) is interesting and deserves further studies.
Future Research and Development Directions and International Cooperation and Networks
The study of biohydrogen is a long-term approach to the development of knowledge and technologies aiming at producing molecular hydrogen from solar energy and water by using a renewable process. The present research and development activities within this field, with its long-term goal, can be promoted, stimulated, and made to advance at a higher rate through collaborations and networks. Besides some national biohydrogen programs and several individual projects, two major international initiatives can be recognized. (i) In the international program IEA (International Energy Agency; http://www.iea.org) Agreement of the Production and Utilization of Hydrogen, Annex 15 Photobiological Hydrogen Production, the main objectives are to investigate and develop processes and equipment for photobiological production of hydrogen by direct conversion of solar energy (114). The research is organized into four subtasks: light-driven hydrogen production by microalgae, maximization of photosynthetic efficiencies, hydrogen fermentations, and improvement of photobioreactor systems for hydrogen production. (ii) In the European program COST (European Cooperation in the Field of Scientific and Technical Research; http://www.belspo.be/cost/) 8.41 Biological and Biochemical Diversity of Hydrogen Metabolism, the main objective is to pool interrelated European expertise in order to understand the structural and molecular basis of the functions, as well as the factors that influence the activity and stability of hydrogenase enzymes.
ADDENDUM IN PROOF
The complete genomic sequence of Anabaena strain PCC 7120 was recently published by Kaneko et al. (T. Kaneko, Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata, DNA Res. 8:213, 227-253, 2001).
Acknowledgments
The present review is part of the IEA Agreement of the Production and Utilization of Hydrogen, Annex 15 Photobiological Hydrogen Production. Our research was financially supported by Fundação para a Ciência e Tecnologia (Portugal), Ångpanneföreningens Forskningsstiftelse (Sweden), the Swedish National Energy Administration (Statens Energimyndighet), and the Swedish Natural Science Research Council (NFR).
REFERENCES
- 1.Reference deleted.
- 2.Reference deleted.
- 3.Adams, D. G. 2000. Symbiotic interactions, p. 523-561. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Akhmanova, A., F. G. Voncken, K. M. Hosea, H. Harhangi, J. T. Keltjens, H. J. op den Camp, G. D. Vogels, and J. H. Hackstein. 1999. A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol. Microbiol. 32:1103-1114. [DOI] [PubMed] [Google Scholar]
- 5.Akhmanova, A., F. G. Voncken, T. van Alen, A. van Hoek, B. Boxma, G. D. Vogels, M. Veenhuis, and J. H. Hackstein. 1998. A hydrogenosome with a genome. Nature 396:527-528. [DOI] [PubMed] [Google Scholar]
- 6.Albracht, S. P. J. 1994. Nickel hydrogenases: in search of the active site. Biochim. Biophys. Acta 1188:167-204. [DOI] [PubMed] [Google Scholar]
- 7.Almon, H., and P. Böger. 1988. Nitrogen and hydrogen metabolism: induction and measurement. Methods Enzymol. 167:459-467. [Google Scholar]
- 8.Anagnostidis, K., and J. Komárek. 1985. Modern approach to the classification system of cyanophytes. 1. Introduction. Arch. Hydrobiol. Suppl. 71:291-302. [Google Scholar]
- 9.Anagnostidis, K., and J. Komárek. 1988. Modern approach to the classification system of cyanophytes. 3-Oscillatoriales. Arch. Hydrobiol. Suppl. 80:327-472. [Google Scholar]
- 10.Anagnostidis, K., and J. Komárek. 1990. Modern approach to the classification system of cyanophytes. 5-Stigonematales. Arch. Hydrobiol. Suppl. 86:1-73. [Google Scholar]
- 11.Appel, J., S. Phunpruch, and R. Schulz. 2000. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 173:333-338. [DOI] [PubMed] [Google Scholar]
- 12.Appel, J., and R. Schulz. 1996. Sequence analysis of an operon of NAD(P)-reducing nickel hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803 gives additional evidence for direct coupling of the enzyme to NADP(H)-dehydrogenase (complex I). Biochim. Biophys. Acta 1298:141-147. [DOI] [PubMed] [Google Scholar]
- 13.Appel, J., and R. Schulz. 1998. Hydrogen metabolism in organisms with oxygenic photosynthesis: hydrogenases as important regulatory devices for a proper redox poising? J. Photochem. Photobiol. Ser. B 47:1-11. [Google Scholar]
- 14.Asada, Y., and S. Kawamura. 1984. Hydrogen evolution by Microcystis aeruginosa in darkness. Agric. Biol. Chem. 48:2595-2596. [Google Scholar]
- 15.Asada, Y., S. Kawamura, and K.-K. Ho. 1987. Hydrogenase from the unicellular cyanobacterium Microcystis aeruginosa. Phytochemistry 26:637-640. [Google Scholar]
- 16.Asada, Y., Y. Koike, J. Schnackenberg, M. Miyake, I. Uemura, and J. Miyake. 2000. Heterologous expression of clostridial hydrogenase in the cyanobacterium Synechococcus PCC7942. Biochim. Biophys. Acta 1490:269-278. [DOI] [PubMed] [Google Scholar]
- 17.Axelsson, R., F. Oxelfelt, and P. Lindblad. 1999. Transcriptional regulation of Nostoc uptake hydrogenase. FEMS Microbiol. Lett. 170:77-81. [DOI] [PubMed] [Google Scholar]
- 18.Bagley, K. A., E. C. Duin, W. Roseboom, S. P. Albracht, and W. H. Woodruff. 1995. Infrared-detectable groups sense changes in charge density on the nickel center in hydrogenase from Chromatium vinosum. Biochemistry 34:5527-5535. [DOI] [PubMed] [Google Scholar]
- 19.Benemann, J. R. 1996. Hydrogen biotechnology: progress and prospects. Nat. Biotechnol. 14:1101-1103. [DOI] [PubMed] [Google Scholar]
- 20.Benemann, J. R. 1997. Feasibility analysis of photobiological hydrogen production. Int. J. Hydrogen Energy 22:979-987. [Google Scholar]
- 21.Bergman, B., J. R. Gallon, A. N. Rai, and L. J. Stal. 1997. N2 fixation by non-heterocystous cyanobacteria. FEMS Microbiol. Rev. 19:139-185. [Google Scholar]
- 22.Bergman, B., A. Matveyev, and U. Rasmussen. 1996. Chemical signalling in cyanobacterial-plant symbiosis. Trends Plant Sci. 6:191-197. [Google Scholar]
- 23.Bishop, P. E., and R. Premakumar. 1992. Alternative nitrogen fixation systems, p. 736-762. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 24.Black, L. K., and R. J. Maier. 1994. Sequences and characterization of hupU and hupV genes of Bradyrhizobium japonicum encoding a possible nickel-sensing complex involved in the hydrogenase expression. J. Bacteriol. 176:7102-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blokesch, M., A. Magalon, and A. Böck. 2001. Interplay between the specific chaperone-like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J. Bacteriol. 183:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böhme, H. 1998. Regulation of nitrogen fixation in heterocyst-forming cyanobacteria. Trends Plant Sci. 3:346-351. [Google Scholar]
- 27.Boison, G., H. Bothe, A. Hansel, and P. Lindblad. 1999. Evidence against a common use of the diaphorase subunits by the bidirectional hydrogenase and by the respiratory complex I in cyanobacteria. FEMS Microbiol. Lett. 174:159-165. [Google Scholar]
- 28.Boison, G., H. Bothe, and O. Schmitz. 2000. Transcriptional analysis of hydrogenase genes in the cyanobacteria Anacystis nidulans and Anabaena variabilis monitored by RT-PCR. Curr. Microbiol. 40:315-321. [DOI] [PubMed] [Google Scholar]
- 29.Boison, G., O. Schmitz, L. Mikheeva, S. Shestakov, and H. Bothe. 1996. Cloning, molecular analysis and insertional mutagenesis of the bidirectional hydrogenase genes from the cyanobacterium Anacystis nidulans. FEBS Lett. 394:153-158. [DOI] [PubMed] [Google Scholar]
- 30.Boison, G., O. Schmitz, B. Schmitz, and H. Bothe. 1998. Unusual gene arrangement of the bidirectional hydrogenase and functional analysis of its diaphorase subunit HoxU in respiration of the unicellular cyanobacterium Anacystis nidulans. Curr. Microbiol. 36:253-258. [DOI] [PubMed] [Google Scholar]
- 31.Borodin, V. B., A. A. Tsygankov, K. K. Rao, and D. O. Hall. 2000. Hydrogen production by Anabaena variabilis PK84 under simulated outdoor conditions. Biotechnol. Bioeng. 69:478-485. [DOI] [PubMed] [Google Scholar]
- 32.Bothe, H. 1982. Hydrogen production by algae. Experientia 38:59-64. [Google Scholar]
- 33.Bothe, H., J. Tennigkeit, and G. Eisbrenner. 1977. The utilization of molecular hydrogen by the blue-green alga Anabaena cylindrica. Arch. Microbiol. 114:43-49. [DOI] [PubMed] [Google Scholar]
- 34.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 35.Burris, R. H. 1991. Nitrogenases. J. Biol. Chem. 266:9339-9342. [PubMed] [Google Scholar]
- 36.Burris, R. H., and G. P. Roberts. 1993. Biological nitrogen fixation. Annu. Rev. Nutr. 13:317-335. [DOI] [PubMed] [Google Scholar]
- 37.Cammack, R. 1995. Redox enzymes. Splitting molecular hydrogen. Nature 373:556-557. [DOI] [PubMed] [Google Scholar]
- 38.Cammack, R. 1999. Hydrogenase sophistication. Nature 397:214-215. [DOI] [PubMed] [Google Scholar]
- 39.Carrasco, C. D., J. A. Buettner, and J. W. Golden. 1995. Programed DNA rearrangment of a cyanobacterial hupL gene in heterocysts. Proc. Natl. Acad. Sci. USA 92:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrasco, C. D., and J. W. Golden. 1995. Two heterocyst-specific DNA rearrangments of nif operons in Anabaena cylindrica and Nostoc sp. strain Mac. Microbiology 141:2479-2487. [DOI] [PubMed] [Google Scholar]
- 41.Carrasco, C. D., K. S. Ramaswamy, T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena xisF gene encodes a developmentally regulated site-specific recombinase. Genes Dev. 8:74-83. [DOI] [PubMed] [Google Scholar]
- 42.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 43.Cauvin, B., A. Colbeau, and P. M. Vignais. 1991. The hydrogenase structural operon in Rhodobacter capsulatus contains a third gene, hupM, necessary for the formation of a physiologically competent hydrogenase. Mol. Microbiol. 5:2519-2527. [DOI] [PubMed] [Google Scholar]
- 44.Cheng, J., C. R. Hipkin, and J. R. Gallon. 1999. Effects of inorganic nitrogen compounds on the activity and synthesis of nitrogenase in Gloeothece (Nägeli) sp. ATCC 27152. New Phytol. 141:61-70. [Google Scholar]
- 45.Daday, A., A. H. Mackerras, and G. D. Smith. 1985. The effect of nickel on hydrogen metabolism and nitrogen fixation in the cyanobacterium Anabaena cylindrica. J. Gen. Microbiol. 131:231-238. [Google Scholar]
- 46.Daday, A., and G. D. Smith. 1987. The hydrogenase-nitrogenase relationship in a symbiotic cyanobacterium isolated from Macrozamia communis L. Johnson. Aust. J. Plant Physiol. 14:319-324. [Google Scholar]
- 47.Douglas, S. E. 1994. Chloroplasts origins and evolution, p. 91-118. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 48.Eisbrenner, G., E. Distler, L. Floener, and H. Bothe. 1978. The occurrence of the hydrogenase in some blue-green algae. Arch. Microbiol. 118:177-184. [Google Scholar]
- 49.Eisbrenner, G., P. Roos, and H. Bothe. 1981. The number of hydrogenases in cyanobacteria. J. Gen. Microbiol. 125:383-390. [Google Scholar]
- 50.Elsen, S., A. Colbeau, J. Chabert, and P. M. Vignais. 1996. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 178:5174-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ewart, G. D., K. C. Reed, and G. D. Smith. 1990. Soluble hydrogenase of Anabaena cylindrica. Cloning and sequencing of a potential gene encoding the tritium exchange subunit. Eur. J. Biochem. 187:215-223. [DOI] [PubMed] [Google Scholar]
- 52.Ewart, G. D., and G. D. Smith. 1989. Purification and properties of soluble hydrogenase from the cyanobacterium Anabaena cylindrica. Arch. Biochem. Biophys. 268:327-337. [DOI] [PubMed] [Google Scholar]
- 53.Ewart, G. D., and G. D. Smith. 1989. An activity stain for the detection of cyanobacterial hydrogenase in polyacrylamide gels. Biochem. Int. 19:89-97. [Google Scholar]
- 54.Ewart, G. D., and G. D. Smith. 1989. Immunochemical analysis of the soluble hydrogenase from the cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 997:83-89. [DOI] [PubMed] [Google Scholar]
- 55.Famiglietti, M., A. Hochkoeppler, and P. L. Luisi. 1993. Surfactant-induced hydrogen production in cyanobacteria. Biotechnol. Bioeng. 42:1014-1018. [DOI] [PubMed] [Google Scholar]
- 56.Fay, P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fedorov, A. S., A. A. Tsygankov, K. K. Rao, and D. O. Hall. 2001. Production of hydrogen by an Anabaena variabilis mutant in a photobioreactor under aerobic outdoor conditions, p. 223-228. In J. Miyake, T. Matsunaga, and A. San Pietro (ed.), Biohydrogen II. Elsevier Science Ltd., Oxford, United Kingdom.
- 58.Flores, E., and A. Herrero. 1994. Molecular evolution and taxonomy of the cyanobacteria, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 59.Florin, L., A. Tsokoglou, and T. Happe. 2001. A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J. Biol. Chem. 276:6125-6132. [DOI] [PubMed] [Google Scholar]
- 60.Fontecilla-Camps, J. C. 1996. The active site of Ni-Fe hydrogenases: model chemistry and crystallographic results. J. Biol. Inorg. Chem. 1:91-98. [Google Scholar]
- 61.Fredriksson, C., and B. Bergman. 1997. Ultrastructural characterisation of cells specialized for nitrogen fixation in a non-heterocystous cyanobacterium Trichodesmium spp. Protoplasma 197:76-85. [Google Scholar]
- 62.Frias, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823-832. [DOI] [PubMed] [Google Scholar]
- 63.Friedrich, T., K. Steinmüller, and H. Weiss. 1995. The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 367:107-111. [DOI] [PubMed] [Google Scholar]
- 64.Friedrich, T., and H. Weiss. 1997. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J. Theor. Biol. 187:529-540. [DOI] [PubMed] [Google Scholar]
- 65.Gallon, J. R. 1992. Reconciling the incompatible: N2 fixation and O2. New Phytol. 122:571-609. [Google Scholar]
- 66.Garg, R. P., A. L. Menon, K. Jacobs, R. M. Robson, and R. L. Robson. 1994. The hypE gene completes the gene cluster for H2-oxidation in Azotobacter vinelandii. J. Mol. Biol. 236:390-396. [DOI] [PubMed] [Google Scholar]
- 67.Golden, J. W., C. D. Carrasco, M. E. Mulligan, G. J. Schneider, and R. Haselkorn. 1988. Deletion of a 55-kilobase-pair DNA element from the chromosome during heterocyst differentiation of Anabaena sp. strain PCC 7120. J. Bacteriol. 170:5034-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golden, J. W., M. E. Mulligan, and R. Haselkorn. 1987. Different recombination site specificity of two developmentally regulated genome rearrangements. Nature 327:526-529. [DOI] [PubMed] [Google Scholar]
- 69.Golden, J. W., S. J. Robinson, and R. Haselkorn. 1985. Rearrangement of nitrogen-fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature 314:419-423. [DOI] [PubMed] [Google Scholar]
- 70.Gubili, J., and D. Borthakur. 1996. The use of a PCR cloning and screening strategy to identify lambda clones containing the hupB gene of Anabaena sp. strain PCC 7120. J. Microbiol. Methods 27:175-182. [Google Scholar]
- 71.Gubili, J., and D. Borthakur. 1998. Organization of the hupDEAB genes within the hydrogenase gene cluster of Anabaena sp. strain PCC 7120. J. Appl. Phycol. 10:163-167. [Google Scholar]
- 72.Hall, D. O., S. A. Markov, Y. Watanbe, and K. K. Rao. 1995. The potential applications of cyanobacterial photosynthesis for clean technologies. Photosynth. Res. 46:159-167. [DOI] [PubMed] [Google Scholar]
- 73.Hallenbeck, P. C., and J. R. Benemann. 1978. Characterization and partial purification of the reversible hydrogenase of Anabaena cylindrica. FEBS Lett. 94:261-264. [Google Scholar]
- 74.Hansel, A., R. Axelsson, P. Lindberg, O. Troshina, R. Wünschiers, and P. Lindblad. 2001. Cloning and characterization of a hyp gene cluster in the filamentous cyanobacterium Nostoc sp. strain PCC 73102. FEMS Microbiol. Lett. 201:59-64. [DOI] [PubMed] [Google Scholar]
- 75.Hansel, A., and P. Lindblad. 1998. Towards optimization of cyanobacteria as biotechnologically relevant producers of molecular hydrogen, a clean and renewable energy source. Appl. Microbiol. Biotechnol. 50:153-160. [Google Scholar]
- 76.Hansel, A., F. Oxelfelt, and P. Lindblad. 1998. Construction of transconjugable plasmids for use in the insertion mutagenesis of Nostoc PCC 73102 uptake hydrogenase, p. 209-218. In O. R. Zaborsky, J. R. Benemann, T. Matsunaga, J. Miyake, and A. San Pietro (ed.), BioHydrogen. Plenum Press, New York, N.Y.
- 77.Happe, T., B. Mosler, and J. D. Naber. 1994. Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222:769-774. [DOI] [PubMed] [Google Scholar]
- 78.Happe, R. P., W. Roseboom, A. J. Pierik, S. P. Albracht, and K. A. Bagley. 1997. Biological activation of hydrogen. Nature 385:126.. [DOI] [PubMed] [Google Scholar]
- 79.Happe, T., K. Schütz, and H. Böhme. 2000. Transcriptional and mutational analysis of the uptake hydrogenase of the filamentous cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 182:1624-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haselkorn, R., and W. J. Buikema. 1992. Nitrogen fixation in cyanobacteria, p. 166-190. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 81.Hausinger, R. P. 1997. Metallocenter assembly in nickel-containing enzymes. J. Biol. Inorg. Chem. 2:279-286. [Google Scholar]
- 82.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hidalgo, E., J. M. Palacios, J. Murillo, and T. Ruiz-Argueso. 1992. Nucleotide sequence and characterization of four additional genes within the hydrogenase structural operon from Rhizobium leguminosarum bv. vicia. J. Bacteriol. 174:4130-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoover, T. R., E. Santero, S. Porter, and S. Kustu. 1990. The integration host factor stimulates interaction of RNA polymerase with NifA, the transcriptional activator for nitrogen fixation operons. Cell 63:11-22. [DOI] [PubMed] [Google Scholar]
- 85.Hoppe-Seyler, F. 1887. Die Methangärung der Essigsäure. Z. Phys. Chem. 11:561-568.
- 86.Horner, D. S., P. G. Foster, and T. M. Embley. 2000. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. E vol. 17:1695-1709. [DOI] [PubMed] [Google Scholar]
- 87.Houchins, J. P. 1984. The physiology and biochemistry of hydrogen metabolism in cyanobacteria. Biochim. Biophys. Acta 768:227-255. [Google Scholar]
- 88.Houchins, J. P., and R. H. Burris. 1981. Occurrence and localization of two distinct hydrogenases in the heterocystous cyanobacterium Anabaena sp. strain 7120. J. Bacteriol. 146:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Houchins, J. P., and R. H. Burris. 1981. Comparative characterization of two distinct hydrogenases from Anabaena sp. strain 7120. J. Bacteriol. 146:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Howarth, D. C., and G. A. Codd. 1985. The uptake and production of molecular hydrogen by unicellular cyanobacteria. J. Gen. Microbiol. 131:1561-1569. [Google Scholar]
- 91.Howitt, C. A., and W. F. J. Vermaas. 1999. Subunits of the NAD(P)-reducing nickel-containing hydrogenase do not act as part of the type-1 NAD(P)H-dehydrogenase in the cyanobacterium Synechocystis sp. PCC 6803, p. 595-601. In G. A. Peschek, W. Löffelhardt and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 92.Huang, T. C., R. F. Lin, M. K. Chu, and H. M. Chen. 1999. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. strain RF-1. Microbiology 145:743-753. [DOI] [PubMed] [Google Scholar]
- 93.Jackman, D. M., and M. E. Mulligan. 1995. Characterization of a nitrogen-fixation (nif) gene cluster from Anabaena azollae 1a shows that closely related cyanobacteria have highly variable but structured intergenic regions. Microbiology 141:2235-2244. [DOI] [PubMed] [Google Scholar]
- 94.Johnson, P. J., C. E. d'Oliveira, T. E. Gorrell, and M. Muller. 1990. Molecular analysis of the hydrogenosomal ferredoxin of the anaerobic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 87:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 30:185-209. [DOI] [PubMed] [Google Scholar]
- 96.Kentemich, T., M. Bahnweg, F. Mayer, and H. Bothe. 1989. Localization of the reversible hydrogenase in cyanobacteria. Z. Naturforsch. 44c:384-391.
- 97.Kentemich, T., M. Casper, and H. Bothe. 1991. The reversible hydrogenase in Anacystis nidulans is a component of the cytoplasmic membrane. Naturwissenschaften 78:559-560. [Google Scholar]
- 98.Kentemich, T., G. Danneberg, B. Hundeshagen, and H. Bothe. 1988. Evidence for the occurrence of the alternative, vanadium-containing nitrogenase in the cyanobacterium Anabaena variabilis. FEMS Microbiol. Lett. 51:19-24. [Google Scholar]
- 99.Kentemich, T., G. Haverkamp, and H. Bothe. 1991. The expression of a third nitrogenase in the cyanobacterium Anabaena variabilis. Z. Naturforsch. 46c:217-222.
- 100.Kessler, E. 1974. Hydrogenase, photoproduction and anaerobic growth, p. 456-473.In W. D. P. Stewart (ed.), Algal physiology and biochemistry. Blackwell Scientific, Oxford, United Kingdom.
- 101.Kim, H., C. Yu, and R. J. Maier. 1991. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J. Bacteriol. 173:3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Komárek, J., and K. Anagnostidis. 1986. Modern approach to the classification system of cyanophytes. 2-Chroococcales. Arch. Hydrobiol. Suppl. 73:157-226. [Google Scholar]
- 103.Komárek, J., and K. Anagnostidis. 1989. Modern approach to the classification system of cyanophytes. 4-Nostocales. Arch. Hydrobiol. Suppl. 82:247-345. [Google Scholar]
- 104.Kotani, H., and S. Tabata. 1998. Lessons from sequencing of the genome of a unicellular cyanobacterium, Synechocystis sp. PCC 6803. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:151-171. [DOI] [PubMed] [Google Scholar]
- 105.Kumar, A. P., B. T. V. V. Perraju, and H. N. Singh. 1986. Carbon nutrition and the regulation of uptake hydrogenase activity in free-living and symbiotic Anabaena cycadeae. New Phytol. 104:115-120. [DOI] [PubMed] [Google Scholar]
- 106.Kumazawa, S., and H. Asakawa. 1995. Simultaneous production of H2 and O2 in closed vessels by marine cyanobacterium Anabaena sp. TU37-1 under high-cell-density conditions. Biotechnol. Bioeng. 46:396-398. [DOI] [PubMed] [Google Scholar]
- 107.Lambert, G. R., and G. D. Smith. 1981. The hydrogen metabolism of cyanobacteria (blue-green algae). Biol. Rev. 56:589-660. [Google Scholar]
- 108.Leclerc, M., A. Colbeau, B. Cauvin, and P. M. Vignais. 1988. Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol. Gen. Genet. 214:97-107. [DOI] [PubMed] [Google Scholar]
- 109.Lenz, O., and B. Friedrich. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li, C., H. D. Jr. Peck, J. LeGall, and A. E. Przybyla. 1987. Cloning, characterization, and sequencing of the genes encoding the large and small subunits of the periplasmic [NiFe]hydrogenase of Desulfovibrio gigas. DNA 6:539-551. [DOI] [PubMed] [Google Scholar]
- 111.Lichtl, R. R., M. J. Bazin, and D. O. Hall. 1997. The biotechnology of hydrogen production by Nostoc flagelliforme grown under chemostat conditions. Appl. Microbiol. Biotechnol. 47:701-707. [Google Scholar]
- 112.Lindberg, P., A. Hansel, and P. Lindblad. 2000. hupS and hupL constitute a transcription unit in the cyanobacterium Nostoc sp. PCC 73102. Arch. Microbiol. 174:129-133. [DOI] [PubMed] [Google Scholar]
- 113.Lindblad, P. 1999. Cyanobacterial H2-metabolism: knowledge and potential/strategies for a photobiotechnological production of H2. Biotecnol. Apl. 16:141-144. [Google Scholar]
- 114.Lindblad, P., Y. Asada, J. Benemann, P. Hallenbeck, A. Melis, J. Miyake, M. Seibert, and O. Skulberg. 2000. IEA Hydrogen Agreement, Task 15: photobiological hydrogen production-an international collaboration, p. 56-59. In Z. Q. Mao, and T. N. Veziroglu (ed.), Hydrogen Energy Progress XIII. CICCST & IAHE, Beijing, China.
- 115.Lindblad, P., A. Hansel, F. Oxelfelt, P. Tamagnini, and O. Troshina. 1998. Nostoc PCC73102 and H2. Knowledge, research and biotechnological challenges, p. 53-63. In O. R. Zaborsky (ed.), BioHydrogen. Plenum Press, New York, N.Y.
- 116.Lindblad, P., and A. Sellstedt. 1990. Occurrence and localization of an uptake hydrogenase in the filamentous heterocystous cyanobacterium Nostoc PCC 73102. Protoplasma 159:9-15. [Google Scholar]
- 117.Lindblad, P., and P. Tamagnini. 2001. Cyanobacterial hydrogenases and biohydrogen: Present status and future potential, p. 143-169. In J. Miyake, T. Matsunaga, and A. San Pietro (ed.), Biohydrogen II. Elsevier Science Ltd., Oxford, United Kingdom.
- 118.Llama, M. J., J. L. Serra, K. K. Rao, and D. O. Hall. 1979. Isolation and characterization of the hydrogenase activity from the non-heterocystous cyanobacterium Spirulina maxima. FEBS Lett. 98:342-346. [DOI] [PubMed] [Google Scholar]
- 119.Lutz, S., A. Jacobi, V. Schlensog, R. Böhm, G. Sawers, and A. Böck. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenases isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 120.Madamwar, D., N. Garg, and V. Shah. 2000. Cyanobacterial hydrogen production. World J. Microbiol. Biotechnol. 16:757-767. [Google Scholar]
- 121.Magalon, A., and A. Böck. 2000. Dissection of the maturation reactions of the (NiFe) hydrogenase 3 from Escherichia coli taking place after nickel incorporation. FEBS Lett. 473:254-258. [DOI] [PubMed] [Google Scholar]
- 122.Maier, R. J., and E. W. Triplett. 1996. Toward more productive, efficient and competitive nitrogen-fixing symbiotic cyanobacteria. Crit. Rev. Plant. Sci. 15:191-234. [Google Scholar]
- 123.Man, H.-M., and W. B. Silvester. 1994. Interactions of H2 and carbon metabolism in moderating nitrogenase activity of the Gunnera/Nostoc symbiosis. Arch. Microbiol. 161:442-444. [Google Scholar]
- 124.Margheri, M. C., M. R. Tredici, G. Allotta, and L. Vagnoli. 1990. Heterotrophic metabolism and regulation of uptake hydrogenase activity in symbiotic cyanobacteria, p. 481-486. In M. Polsinelli, R. Materassi and M. Vincenzini (ed.), Developments in plant and soil sciences—biological nitrogen fixation. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 125.Markov, S. A., M. J. Bazin, and D. O. Hall. 1995. The potential of using cyanobacteria in photobioreactors for hydrogen production. Adv. Biochem. Eng. Biotechnol. 52:59-86. [Google Scholar]
- 126.Martel, A., E. Jansson, G. Garcia-Reina, and P. Lindblad. 1993. Ornithine cycle in Nostoc PCC 73102. Arginase, OCT and arginine deiminase, and the effects of addition of external arginine, ornithine, or citrulline. Arch. Microbiol. 159:506-511. [Google Scholar]
- 127.Masepohl, B., K. Schoelisch, K. Goerlitz, C. Kutzki, and H. Böhme. 1997. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Mol. Gen. Genet. 253:770-776. [DOI] [PubMed] [Google Scholar]
- 128.Masukawa, H., K. Nakamura, M. Mochimaru, and H. Sakurai. 2001. Photobiological hydrogen production and nitrogenase activity in some heterocystous cyanobacteria, p. 63-66. In J. Miyake, T. Matsunaga, and A. San Pietro (ed.), Biohydrogen II. Elsevier Science Ltd., Oxford, United Kingdom.
- 129.Matthijs, H. C. P., G. W. M. van der Staay, and L. R. Mur. 1994. Prochlorophytes: the other cyanobacteria? p. 49-64. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 130.Matveyev, A. V., E. Rutgers, E. Söderbäck, and B. Bergman. 1994. A novel genome rearrangement involved in heterocyst differentiation of the cyanobacterium Anabaena sp. PCC7120. FEMS Microbiol. Lett. 116:201-208. [Google Scholar]
- 131.Mazel, D., J. Houmard, A. M. Castets, and N. Tandeau de Marsac. 1990. Highly repetitive DNA sequences in cyanobacterial genomes. J. Bacteriol. 172:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McTavish, H., L. A. Sayavedra-Soto, and D. J. Arp. 1995. Substitution of Azotobacter vinelandii hydrogenase small-subunit cysteines by serines can create insensitivity to inhibition by O2 and preferentially damages H2 oxidation over H2 evolution. J. Bacteriol. 177:3960-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meeks, J. C. 1988. Symbiotic associations. Methods Enzymol. 167:113-125. [Google Scholar]
- 134.Meeks, J. C., E. L. Campbell and P. S. Bisen. 1994. Elements interrupting nitrogen fixation genes in cyanobacteria: presence and absence of a nifD element in clones of Nostoc sp. strain Mac. Microbiology 140:3225-3232. [Google Scholar]
- 135.Menon, N. K., J. Robbins, M. Der Vartanian, D. Patil, H. D. Peck, Jr., A. L. Menon, R. L. Robson, and A. E. Przybyla. 1993. Carboxy-terminal processing of the large subunit of [NiFe] hydrogenases. FEBS Lett. 331:91-95. [DOI] [PubMed] [Google Scholar]
- 136.Mikheeva, L. E., O. Schmitz, S. V. Shestakov, and H. Bothe. 1995. Mutants of the cyanobacterium Anabaena variabilis altered in hydrogenase activities. Z. Naturforsch. 50c:505-510.
- 137.Misra, H. S., and R. Tuli. 2000. Differential expression of photosynthesis and N2-fixation genes in the cyanobacterium Plectonema boryanum. Plant Physiol. 468:731-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miyake, M., J. Schnackenberg, C. Nakamura, Y. Asada, and J. Miyake. 2001. Molecular handling of hydrogenase, p. 205-219. In J. Miyake, T. Matsunaga, and A. San Pietro (ed.), Biohydrogen II. Elsevier Science Ltd., Oxford, United Kingdom.
- 139.Montet, Y., P. Amara, A. Volbeda, X. Vernede, E. C. Hatchikian, M. J. Field, M. Frey, and J. C. Fontecilla-Camps. 1997. Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics. Nat. Struct. Biol. 4:523-526. [DOI] [PubMed] [Google Scholar]
- 140.Mulholland, M. R., and D. G. Capone. 2000. The nitrogen physiology of the marine N2-fixing cyanobacteria Trichodesmium spp. Trends Plant Sci. 5:148-153. [DOI] [PubMed] [Google Scholar]
- 141.Mulligan, M. E., W. J. Buikema, and R. Haselkorn. 1988. Bacterial-type ferredoxin genes in the nitrogen fixation regions of the cyanobacterium Anabaena sp. strain PCC 7120 and Rhizobium meliloti. J. Bacteriol. 170:4406-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mulligan, M. E., and R. Haselkorn. 1989. Nitrogen-fixation (nif) genes of the cyanobacterium Anabaena sp. strain PCC 7120: the nifB-fdxN-nifS-nifU operon. J. Biol. Chem. 26:19200-19207. [PubMed] [Google Scholar]
- 143.Reference deleted.
- 144.Nakamura, Y., T. Kaneko, M. Hirosawa, N. Miyajima, and S. Tabata. 1998. CyanoBase, a www database containing the complete genome of Synechocystis sp. strain PCC 6803. Nucleic Acids Res. 20:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Olson, J. W., C. Fu, and R. J. Maier. 1997. The HypB protein from Bradyrhizobium japonicum can store nickel and is required for the nickel-dependent transcriptional regulation of hydrogenase. Mol. Microbiol. 24:119-128. [DOI] [PubMed] [Google Scholar]
- 146.Olson, J. W., and R. J. Maier. 1997. The sequences of hypF, hypC and hypD complete the hyp gene cluster required for hydrogenase activity in Bradyrhizobium japonicum. Gene 199:93-99. [DOI] [PubMed] [Google Scholar]
- 147.Olson, J. W., and R. J. Maier. 2000. Dual roles of Bradyrhizobium japonicum nickelin protein in nickel storage and GTP-dependent Ni mobilization. J. Bacteriol. 182:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 149.Orme-Johnson, W. H. 1992. Nitrogenase structure: where to now. Science 257:1639-1640. [DOI] [PubMed] [Google Scholar]
- 150.Oxelfelt, F., P. Tamagnini, and P. Lindblad. 1998. Hydrogen uptake in Nostoc sp. strain PCC 73102. Cloning and characterization of a hupSL homologue. Arch. Microbiol. 169:267-274. [DOI] [PubMed] [Google Scholar]
- 151.Oxelfelt, F., P. Tamagnini, R. Salema, and P. Lindblad. 1995. Hydrogen uptake in Nostoc strain PCC 73102: effects of nickel, hydrogen, carbon and nitrogen. Plant Physiol. Biochem. 33:617-623. [Google Scholar]
- 152.Papen, H., T. Kentemich, T. Schmülling, and H. Bothe. 1986. Hydrogenase activities in cyanobacteria. Biochimie 68:121-132. [DOI] [PubMed] [Google Scholar]
- 153.Paschos, A., R. S. Glass, and A. Böck. 2001. Carbamoylphosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett. 488:9-12. [DOI] [PubMed] [Google Scholar]
- 154.Peschek, G. A. 1979. Anaerobic hydrogenase activity in Anacystis nidulans. H2-dependent photoreduction and related reactions. Biochim. Biophys. Acta 548:187-202. [DOI] [PubMed] [Google Scholar]
- 155.Peschek, G. A. 1979. Aerobic hydrogenase activity in Anacystis nidulans. The oxyhydrogen reaction. Biochim. Biophys. Acta 548:203-215. [DOI] [PubMed] [Google Scholar]
- 156.Peschek, G. A. 1979. Evidence for two functionally distinct hydrogenases in Anacystis nidulans. Arch. Microbiol. 123:81-92. [Google Scholar]
- 157.Peterson, R. B., and C. P. Wolk. 1978. Localization of an uptake hydrogenase in Anabaena. Plant Physiol. 61:688-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Postgate, J. 1987. Nitrogen fixation. 2nd ed. New studies in biology. Edward Arnold Publishers, London, United Kingdom.
- 159.Przybyla, A. E., J. Robbins, N. Menon, and H. D. Peck, Jr. 1992. Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol. Rev. 88:109-136. [DOI] [PubMed] [Google Scholar]
- 160.Rai, A. N., M. Borthakur, E. Söderbäck, and B. Bergman. 1992. Immunogold localization of hydrogenase in the cyanobacterial-plant symbioses Peltigera canina, Anthoceros punctatus and Gunnera magellanica. Symbiosis 12:131-144. [Google Scholar]
- 161.Rai, A. N., E. Söderbäck, and B. Bergman. 2000. Cyanobacterium-plant symbioses. New Phytol. 147:449-481. [DOI] [PubMed] [Google Scholar]
- 162.Ramaswamy, K. S., C. D. Carrasco, T. Fatma, and J. W. Golden. 1997. Cell-type specificity of the Anabaena fdxN element rearrangement requires xisH and xisI. Mol. Microbiol. 23:1241-1249. [DOI] [PubMed] [Google Scholar]
- 163.Rao, K. K., and D. O. Hall. 1988. Hydrogenases: isolation and assay. Methods Enzymol. 167:501-509. [Google Scholar]
- 164.Rao, K. K., and D. O. Hall. 1996. Hydrogen production by cyanobacteria: potential, problems and prospects. J. Mar. Biotechnol. 4:10-15. [Google Scholar]
- 165.Reade, J. P., L. J. Dougherty, L. J. Rodgers, and J. R. Gallon. 1999. Synthesis and proteolytic degradation of nitrogenase in cultures of the unicellular cyanobacterium Gloeothece strain ATCC 27152. Microbiology 145:1749-1758. [DOI] [PubMed] [Google Scholar]
- 166.Reddy, P. M., H. Spiller, S. L. Albrecht, and K. T. Shanmugam. 1996. Photodissimilation of fructose to H2 and CO2 by a dinitrogen-fixing cyanobacterium, Anabaena variabilis. Appl. Environ. Microbiol. 62:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 168.Rippka, R., and M. Herdman. 1992. Pasteur culture collection of cyanobacterial strains in axenic culture. Catalogue & taxonomic handbook, vol. 1. Catalogue of strains. Institute Pasteur, Paris, France.
- 169.Rowell, P., and N. W. Kerby. 1991. Cyanobacteria and their symbionts, p. 373-407. In M. J. Dilworth, and A. R. Glenn (ed.), Biology and biochemistry of nitrogen fixation. Elsevier, Amsterdam, The Netherlands.
- 170.Runswick, M. J., R. B. Gennis, I. M. Pernley, and J. E. Walker. 1989. Mitochondrial NADH:ubiquinone reductase: complementary DNA sequence of the import precursor of the bovine 75-kDa subunit. Biochemistry 30:9697-9704. [DOI] [PubMed] [Google Scholar]
- 171.Sakamoto, T., V. B. Delgaizo, and D. A. Bryant. 1998. Growth on urea can trigger death and peroxidation of the cyanobacterium Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 64:2361-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sayavedra-Soto, L. A., G. K. Powell, H. J. Evans, and R. O. Morris. 1988. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc. Natl. Acad. Sci. USA 85:8395-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schmitz, O., G. Boison, R. Hilscher, B. Hundeshagen, W. Zimmer, F. Lottspeich, and H. Bothe. 1995. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur. J. Biochem. 233:266-276. [DOI] [PubMed] [Google Scholar]
- 174.Schmitz, O., and H. Bothe. 1996. The diaphorase subunit HoxU of the bidirectional hydrogenase as electron transferring protein in cyanobacterial respiration? Naturwissenschaften 83:525-527. [DOI] [PubMed] [Google Scholar]
- 175.Schopf, J. W. 2000. The fossil record: tracing the roots of the cyanobacterial lineage, p. 13-35. In B. A. Whitton, and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 176.Schulz, R. 1996. Hydrogenases and hydrogen production in eukaryotic organisms and cyanobacteria. J. Mar. Biotechnol. 4:16-22. [Google Scholar]
- 177.Serebriakova, L., N. A. Zorin, and P. Lindblad. 1994. Reversible hydrogenase in Anabaena variabilis ATCC 29413: presence and localization in non-N2-fixing cells. Arch. Microbiol. 161:140-144. [Google Scholar]
- 178.Serebryakova, L. T., M. Medina, N. A. Zorin, I. N. Gogotov, and R. Cammack. 1996. Reversible hydrogenase of Anabaena variabilis ATCC 29413: catalytic properties and characterization of redox centres. FEBS Lett. 383:79-82. [DOI] [PubMed] [Google Scholar]
- 179.Serebryakova, L. T., M. E. Sheremetieva, and P. Lindblad. 2000. H2-uptake and evolution in the unicellular cyanobacterium Chroococcidiopsis thermalis CALU 758. Plant Physiol. Biochem. 38:525-530. [Google Scholar]
- 180.Serebryakova, L. T., M. E. Sheremetieva, and A. A. Tsygankov. 1998. Reversible hydrogenase activity of Gloeocapsa alpicola in continuous culture. FEMS Microbiol. Lett. 166:89-94. [Google Scholar]
- 181.Shah, V., N. Gard, and D. Madamwar. 2001. Ultrastructure of the fresh water cyanobacterium Anabaena variabilis SPU 003 and its application for oxygen-free hydrogen production. FEMS Microbiol. Lett. 194:71-75. [DOI] [PubMed] [Google Scholar]
- 182.Shravan Kumar, C., and H. Polasa. 1991. Influence of nickel and copper on photobiological hydrogen production and uptake in Oscillatoria subbrevis strain 111. Proc. Indian Nat. Sci. Acad. B57:281-285.
- 183.Smith, G. D. 1990. Hydrogen metabolism in cyanobacteria, p. 131-143. In E. H. D. Kumar (ed.), Phycotalk 2. Rastogi & Co., Subhash Bazar, Meerut, India.
- 184.Smith, D. C., and A. E. Douglas. 1987. The biology of symbiosis. Edward Arnold Publishers, London, United Kingdom.
- 185.Spiller, H., G. Bookjans, and K. Shanmugan. 1983. Regulation of hydrogenase activity in vegetative cells of Anabaena variabilis. J. Bacteriol. 155:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 187.Stal, L. J. 1995. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131:1-32. [DOI] [PubMed] [Google Scholar]
- 188.Stal, L. J. 2000. Cyanobacterial mats and stromatolites, p. 61-120. In B. A. Whitton, and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 189.Stal, L. J., and R. Moezelaar. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179-211. [Google Scholar]
- 190.Stanier, R. Y., W. R. Sistrom, T. A. Hansen, B. A. Whitton, R. W. Castenholz, N. Pfennig, V. N. Gorlenko, E. N. Kondratieva, K. E. Eimhjellen, R. Whittenbury, R. L. Gherna, and H. G. Trüper. 1978. Proposal to place the nomenclature of the cyanobacteria (blue-green algae) under the rules of the International Code of Nomenclature of Bacteria. Int. J. Syst. Bacteriol. 28:335-336. [Google Scholar]
- 191.Stephenson, M., and L. H. Stickland. 1931. XXVII. Hydrogenase: a bacterial enzyme activating molecular hydrogen. I. The properties of the enzyme. Biochem. J. 25:205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tamagnini, P., J.-L. Costa, L. Almeida, M.-J. Oliveira, R. Salema, and P. Lindblad. 2000. Diversity of cyanobacterial hydrogenases, a molecular approach. Curr. Microbiol. 40:356-361. [DOI] [PubMed] [Google Scholar]
- 193.Tamagnini, P., F. Oxelfelt, R. Salema, and P. Lindblad. 1995. Immunological characterization of hydrogenases in the nitrogen-fixing Nostoc sp. strain PCC 73102. Curr. Microbiol. 31:102-107. [Google Scholar]
- 194.Tamagnini, P., O. Troshina, F. Oxelfelt, R. Salema, and P. Lindblad. 1997. Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme. Appl. Environ. Microbiol. 63:1801-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Thiel, T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 175:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Thiel, T. 1994. Genetic analysis of cyanobacteria, p. 581-611. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 197.Thiel, T., E. M. Lyons, and J. C. Erker. 1997. Characterization of genes for a second Mo-dependent nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 179:5222-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Thiel, T., E. M. Lyons, J. C. Erker, and A. Ernst. 1995. A second nitrogenase in vegetative cells of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 92:9358-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Thiel, T., and B. Pratte. 2001. Effect on heterocyst differentiation of nitrogen fixation in vegetative cells of the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 183:280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Thiemermann, S., J. Dernedde, M. Bernhard, W. Schroeder, C. Massanz, and B. Friedrich. 1996. Carboxyl-terminal processing of the cytoplasmic NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J. Bacteriol. 178:2368-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tibelius, K. H., L. Du, D. Tito, and F. Stejskal. 1993. The Azotobacter chroococcum hydrogenase gene cluster: sequences and genetic analysis of four accessory genes, hupA, hupB, hupY and hupC. Gene 127:53-61. [DOI] [PubMed] [Google Scholar]
- 202.Tredici, M. R., M. C. Margheri, R. De Philippis, and R. Materassi. 1990. The role of hydrogen metabolism in photoheterotrophic cultures of the cyanobacterium Nostoc sp. strain Cc isolated from Cycas circinalis L. J. Gen. Microbiol. 136:1009-1015. [Google Scholar]
- 203.Troshina, O. Y., L. T. Serebryakova, and P. Lindblad. 1996. Induction of H2-uptake and nitrogenase activities in the cyanobacterium Anabaena variabilis ATCC 29413: effects of hydrogen and organic substrate. Curr. Microbiol. 33:11-15. [DOI] [PubMed] [Google Scholar]
- 204.Tsygankov, A. A., V. B. Borodin, K. K. Rao, and D. O. Hall. 1999. H2 photoproduction by batch culture of Anabaena variabilis ATCC 29143 and its mutant PK84 in a photobioreactor. Biotechnol. Bioeng. 64:709-715. [DOI] [PubMed] [Google Scholar]
- 205.Tsygankov, A. A., L. T. Serebryakova, K. K. Rao, and D. O. Hall. 1998. Acetylene reduction and hydrogen photoproduction by wild-type and mutant strains of Anabaena at different CO2 and O2 concentrations. FEMS Microbiol. Lett. 167:13-17. [Google Scholar]
- 206.Tsygankov, A. A., L. T. Serebryakova, D. A. Sveshnikov, K. K. Rao, I. N. Gogotov, and D. O. Hall. 1998. Hydrogen photoproduction by three different nitrogenases in whole cells of Anabaena variabilis and the depedence on pH. Int. J. Hydrogen Energy 22:859-867. [Google Scholar]
- 207.Turner, S. 1997. Molecular systematics of oxygenic photosynthetic bacteria, p. 13-52. In D. Bhattacharya (ed.), Origins of algae and their plastids. Springer-Verlag, Vienna, Austria.
- 208.van Soom, C., J. Browaeys, C. Verreth, and J. Vanderleydan. 1993. Nucleotide sequence analysis of four genes, hupC, hupD, hupF, and hupG, downstream of the hydrogenase structural genes in Bradyrhizobium japonicum. J. Mol. Biol. 234:508-512. [DOI] [PubMed] [Google Scholar]
- 209.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 210.Vignais, P. M., B. Dimon, N. Zorin, A. Colbeau, and S. Elsen. 1997. HupUV proteins of Rhodobacter capsulatus can bind H2: evidence from the H-D exchange reaction. J. Bacteriol. 179:290-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vignais, P. M., and B. Toussaint. 1994. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch. Microbiol. 161:1-10. [DOI] [PubMed] [Google Scholar]
- 212.Volbeda, A., M.-H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 213.Voordouw, G. 1992. Evolution of hydrogenase genes. Adv. Inorg. Chem. 38:397-422. [Google Scholar]
- 214.Vyas, D., and H. D. Kumar. 1995. Nitrogen fixation and hydrogen uptake in four cyanobacteria. Int. J. Hydrogen Energy 20:163-168. [Google Scholar]
- 215.Wei, T. F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Weisshaar, H., and P. Böger. 1985. Pathways of hydrogen uptake in the cyanobacterium Nostoc muscorum. Arch. Microbiol. 142:349-353. [Google Scholar]
- 217.Whitman, C. P., B. A. Aird, B. A., W. R. Gillespie, W. R., and N. J. Stolowich. 1991. Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J. Am. Chem. Soc. 113:3154-3162. [Google Scholar]
- 218.Whitton, B. A., and M. Potts. 2000. Introduction to the cyanobacteria, p. 1-11. In B. A. Whitton, and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 219.Wilmotte, A. 1994. Molecular evolution and taxonomy of the cyanobacteria, p. 1-25. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 220.Wolk, C. P. 1996. Heterocyst formation. Annu. Rev. Genet. 30:59-78. [DOI] [PubMed] [Google Scholar]
- 221.Wolk, C. P., A. Ernest, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 222.Wünschiers, R., H. Senger, and R. Schulz. 2001. Electron pathways involved in H2-metabolism in the green alga Scenedesmus obliquus. Biochem. Biophys. Acta 1503:271-278. [DOI] [PubMed] [Google Scholar]
- 223.Wünschiers, R., K. Stangier, H. Senger, and R. Schulz. 2001. Molecular evidence for a Fe-hydrogenase in the green alga Scenedesmus obliquus. Curr. Microbiol. 42:353-360. [DOI] [PubMed] [Google Scholar]
- 224.Xiankong, Z., F. R. Tabita, and C. Van Baalen. 1984. Nickel control of hydrogen production and uptake in Anabaena spp. strains CA and 1F. J. Gen. Microbiol. 130:1815-1818. [Google Scholar]
- 225.Yarlett, N., C. G. Orpin, E. A. Munn, N. C. Yarlett, C. A. Greenwood. 1986. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 236:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yoon, H. S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 227.Yoon, H. S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zoysa, P. A., and C. J. Danpure. 1993. Possible evolutionary relationship between mammalian alanine:glyoxylate aminotransferase 1 and 42-kD subunit of cyanobacterial soluble hydrogenase. Mol. Biol. Evol. 10:704-706. [DOI] [PubMed] [Google Scholar]