Abstract
The regulatory logic of siderophore biosynthetic genes in bacteria involves the universal repressor Fur, which acts together with iron as a negative regulator. However in other bacteria, in addition to the Fur-mediated mechanism of regulation, there is a concurrent positive regulation of iron transport and siderophore biosynthetic genes that occurs under conditions of iron deprivation. Despite these regulatory differences the mechanisms of siderophore biosynthesis follow the same fundamental enzymatic logic, which involves a series of elongating acyl-S-enzyme intermediates on multimodular protein assembly lines: nonribosomal peptide synthetases (NRPS). A substantial variety of siderophore structures are produced from similar NRPS assembly lines, and variation can come in the choice of the phenolic acid selected as the N-cap, the tailoring of amino acid residues during chain elongation, the mode of chain termination, and the nature of the capturing nucleophile of the siderophore acyl chain being released. Of course the specific parts that get assembled in a given bacterium may reflect a combination of the inventory of biosynthetic and tailoring gene clusters available. This modular assembly logic can account for all known siderophores. The ability to mix and match domains within modules and to swap modules themselves is likely to be an ongoing process in combinatorial biosynthesis. NRPS evolution will try out new combinations of chain initiation, elongation and tailoring, and termination steps, possibly by genetic exchange with other microorganisms and/or within the same bacterium, to create new variants of iron-chelating siderophores that can fit a particular niche for the producer bacterium.
“Truth is never pure, and rarely simple.”
—Oscar Wilde
The Importance of Being Earnest, Act I
INTRODUCTION
Iron is an essential element for nearly all living systems; however, most of the iron in the biological fluids of vertebrates is found bound by transferrin, lactoferrin, and in red blood cells (12). Therefore, in establishing an infection, microorganisms depend heavily on their ability to use the host-complexed iron. A key feature which enables pathogenic bacteria to survive within the vertebrate host is the production of siderophores, iron-sequestering compounds, and the synthesis of their cognate transport systems, which are crucial in overcoming the nonspecific defense mechanisms of the host and allow for bacterial multiplication (10, 12, 24-30, 78, 79, 90).
The past few years have been a period of exceptional progress in the iron transport field and in the study of siderophores, during which the problem has moved from the stage of identification and cloning of the genes involved in the process of iron uptake to the assignment of biochemical functions to many of these genes, their linkage to specific pathways, and the visualization by X-ray diffraction analysis of the mechanisms of binding or acquisition of the ferric siderophore complex by the outer membrane protein receptors (10, 12, 26, 90). Progress in understanding how regulatory factors control expression of iron uptake genes has been a little slower, with a great deal of emphasis placed on the negative regulation by the universal repressor Fur, the common denominator in regulation of these iron uptake systems, which acts together with iron. Under iron-rich conditions, the Fe2+-Fur complexes bind to promoters containing a Fur box (Fur binding sequence) and repress transcription. Although Fur is required for transcriptional activation of some genes, this effect may be indirect (12, 90, 117, 131). Fur homologues have been identified in Escherichia coli and many other Gram-negative bacteria (12, 25). The Fur proteins of Vibrio anguillarum, Vibrio cholerae, and Vibrio vulnificus are very highly related, although they share only about 60% of their polynucleotide sequence with that of E. coli, while the Fur protein of Yersinia pestis is highly similar to Fur from E. coli and other enteric bacteria (25, 26, 83, 117, 132). However, the concept of regulation of iron uptake systems is becoming more complex with the recent findings of positive regulation of iron transport and siderophore biosynthetic genes, rather than only derepression mediated by the decrease of iron availability in the cell cytosol. The best-studied system is the ferric citrate system in E. coli, in which the FecI regulator has been demonstrated by Braun and Killmann to be an extracytoplasmic-function sigma factor acting in the initiation of transcription of the Fec operon (10). In V. anguillarum, Pseudomonas aeruginosa, and other microorganisms, positive regulation by a combination of positive regulatory proteins and the cognate siderophore has also been reported, and the mechanisms are under study (22, 26, 49, 50). Furthermore, it has also been demonstrated in V. anguillarum that negative control of iron uptake gene expression is not only due to repression of transcription initiation by a complex of Fur with iron (19) but also due to posttranscriptional regulation by antisense RNAs acting as a fine-tuning control mechanism (21, 26, 101). These regulatory strategies enable bacteria, via iron-sensing transcriptional activators and repressors, to turn on and off sets of genes that encode proteins for the assembly and export of siderophores and the specific uptake of the iron-siderophore complexes. Although a good deal of effort has been placed on the elucidation of how these various regulatory components of the iron uptake systems work on the control of expression of iron uptake systems in bacteria, there has also been a dramatic increase in research directed at understanding the molecular nature of siderophores and the genetics and enzymology of their biosynthesis, which are the topics of this review.
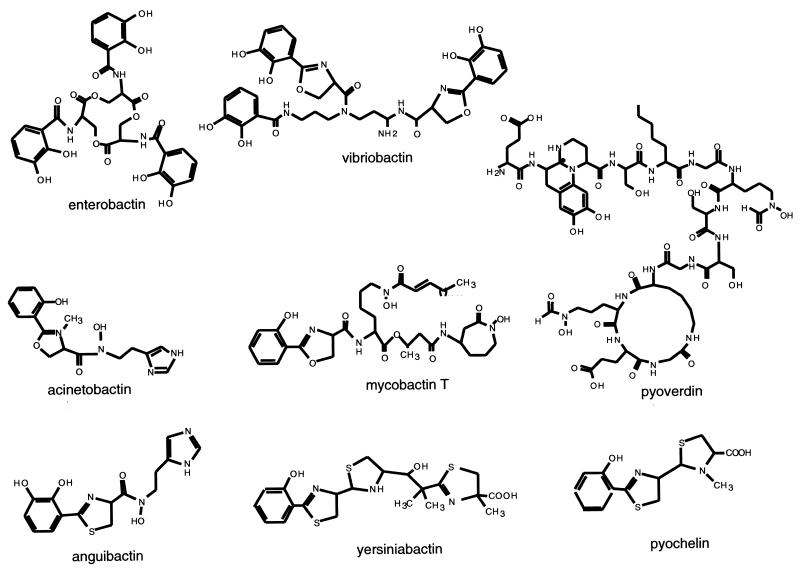
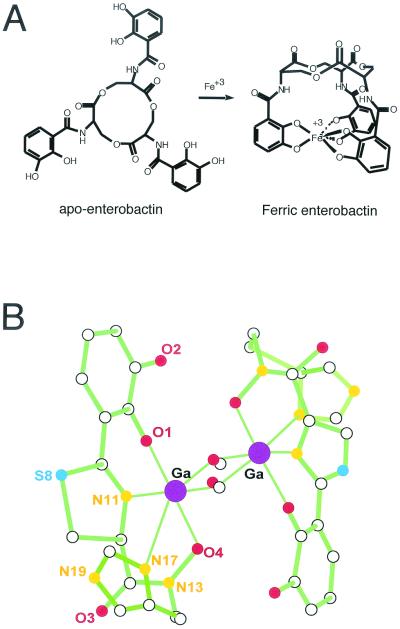
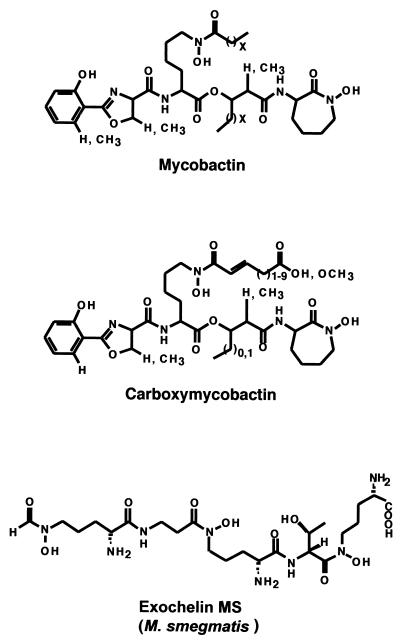
An all-encompassing, but possibly not entirely accurate, definition of siderophores is that they are small peptidic molecules, readily assembled by short, dedicated metabolic pathways, which contain side chains and functional groups that can provide a high-affinity set of ligands for coordination of ferric ions (25, 78, 79). Figure 1 shows the structural formulas for siderophores from various bacteria. There are three main types of iron-coordinating functional groups in siderophores. First, there are the N-hydroxy amino acid side chains as in anguibactin, with the oxygen atom as one of the ligands for Fe3+. Second, there are the adjacent hydroxyls of catechol rings, almost always derived from 2,3-dihydroxybenzoate (DHB), as represented in enterobactin, anguibactin, and acinetobactin. Variants may involve biosynthetic use of 2-hydroxybenzoate (salicylate) in place of 2,3-DHB, leading to phenolic moieties as iron ligands. The three catecholic side chains in E. coli enterobactin can provide a full hexadentate ligation set to ferric ion, accounting for the estimated Kd of 10−52 M that makes enterobactin such an astoundingly good scavenger for iron (78, 79) (Fig. 2A). Third, the nitrogen atoms of five-membered thiazoline and oxazoline rings, resulting from enzymatic cyclization of cysteinyl, seryl, or threonyl side chains, respectively, can also coordinate Fe 3+ (as is shown for the anguibactin siderophore complex with Ga3+ in Fig. 2B). This type of coordination is a common feature in the pyochelin, yersiniabactin, vibriobactin, anguibactin, and acinetobactin siderophores. The X-ray structure for the anguibactin-gallium complex also establishes the heterocyclic imine nitrogens as part of the iron coordination (3, 53) (Fig. 2B). Often the three iron-chelating functional groups are combined in the same siderophore, such as mycobactin and anguibactin, perhaps reflecting combinatorial biosynthetic assembly logic. In the case of anguibactin, the coordinating bonds are provided by an oxygen from the diphenolate group, an oxygen from the hydroxamate group, a nitrogen from the imidazole group, and a nitrogen from the thiazoline group. Two molecules of anguibactin, two of the metal ion and two of the solvent, complete the structure (Fig. 2B). Retrobiosynthesis of anguibactin,ω-N-hydroxy-ω-N}[2′-(2′,3′-dihydroxyphenyl) thiazolin-4′-yl]carboxyl}histamine, indicates that it is composed of one molecule of 2,3-dihydroxybenzoic acid (DHBA), one of l-cysteine, and one of N-hydroxy-histamine. Retrobiosynthesis of V. cholerae vibriobactin, N1-(2,3-dihydroxybenzoyl)-N5,N9-bis[2-(2,3-dihydroxyphenyl)-5-methyloxazolinyl-4-carboxamido]norspermidine, shows that it is composed of three molecules of DHBA, two of l-threonine, and one of the unusual polyamine norspermidine [bis(3-aminopropyl)amine] (54, 140, 141). The 2-(2-hydroxyphenyl)-thiazolinyl-thiazolidine-4 carboxylate in yersiniabactin (41-43, 45, 55, 56, 83, 110, 114, 115) and pyochelin (87, 93, 94) is reminiscent of anguibactin, while the two 2-(2,3-dihydroxyphenyl)-5-methyl-oxazoline-4-carboxamide groups in vibriobactin (57, 58) are reminiscent of the 2-(2-hydroxyphenyl)-oxazoline-4-carboxamide found in mycobactin from Mycobacterium tuberculosis (32, 86, 90, 124, 142, 143).
FIG. 1.
Structures of various bacterial siderophores: enterobactin from E. coli; vibriobactin from V. cholerae; acinetobactin from Acinetobacter calcoaceticus; mycobactin T from M. tuberculosis; pyoverdin and pyochelin from P. aeruginosa; anguibactin from V. anguillarum; and yersiniabactin from Y. pestis.
FIG. 2.
Spatial structures of enterobactin and anguibactin. (A) Enterobactin without and with Fe3+. Notice that the groups coordinating the iron are six oxygens from the three diphenolic groups. The complex requires one molecule of enterobactin. (B) X-ray structure of gallium anguibactin. This structure was determined from crystals obtained by complexing with Ga 3+ instead of Fe 3+ (53). Empty spheres: carbon (C); red spheres: oxygen (O); orange spheres: nitrogen (N); dark blue spheres: sulfur (S); purple spheres: gallium (Ga). Notice the coordination of two Ga3+ ions by two molecules of anguibactin using O-4 from the hydroxamate group, N-17 from the imidazole group, N-11 from the thiazoline group, and O-1 from the diphenol group. The complex requires two molecules of anguibactin and two molecules of methoxy since crystallization was carried out in a methanol solution. In biological fluids the complex will consist of two molecules of anguibactin, two Fe 3+ molecules, and two molecules of H2O.
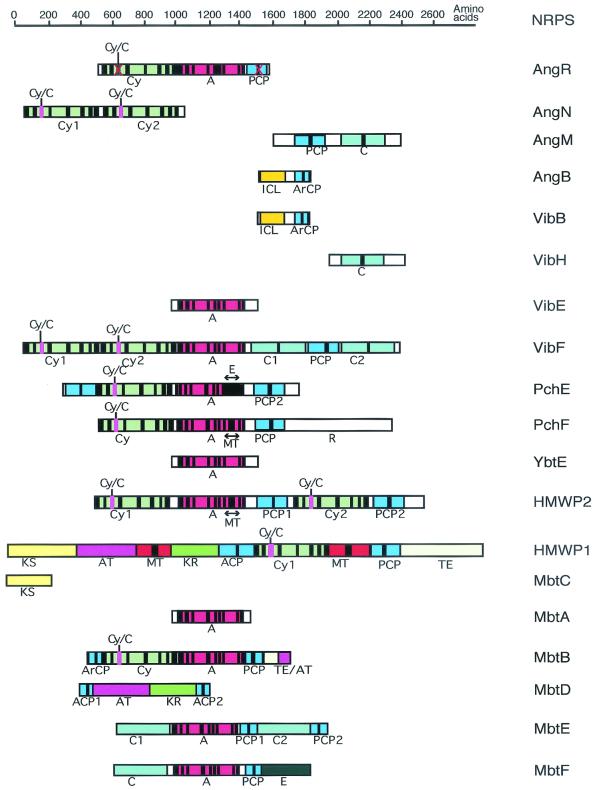
The peptidic backbone of siderophores, combined with the prevalence of nonproteinogenic amino acid units embedded in the iron chelators, suggested early on that these compounds would be nonribosomal peptides, and this has been validated in the past decade by the experiments carried out by the Walsh laboratory (39, 42, 56, 58, 86, 87, 96, 98, 106, 115) and by the microbial genome sequencing efforts, and thus many of the proteins involved in the biosynthesis of siderophores are nonribosomal peptide synthetases (NRPS). These type of enzymes were originally identified as catalyzing the syntheses of antibiotics and other substances in gram-positive bacteria (6, 47, 54, 59, 61, 63-70, 72, 103, 107, 116, 130). NRPS are multimodular enzymes that produce peptide products of a particular sequence without an RNA template. Instead, the order of monomeric amino acids activated and incorporated is specified by the order of NRPS domains (61, 70, 97, 106, 125, 127, 130). The elongating chains grow as a series of acyl-S-enzyme intermediates, tethered covalently to the NRPS assembly line via peptidyl carrier proteins domains (PCPs), that act as way stations for catalytic domains in the vicinity, to carry out chemical operations before the chain is translocated down to the next downstream carrier protein domain (54, 68, 125). The tethering thiol groups are posttranslationally introduced as phosphopantetheinyl arms (61, 65-67), and the NRPS assembly lines run by a multiple-thiol templating process (67, 70, 72). Nonribosomal peptides made by such enzymatic assembly lines include major antibiotics such as the penicillins and vancomycin, the immunosuppressant cyclosporine, and many of the siderophores.
An NRPS assembly line is comprised of autonomously folding domains, bundled together into functional modules to carry out steps of monomer selection and activation, chain elongation, and then chain termination. For example, the tripeptide aminoadipoyl-cysteinyl-valine (ACV) is produced by a 10-domain assembly line in a single enzyme of 480 kDa, while the undecapeptide cyclosporine is assembled by a 1.6-MDa single-subunit enzyme with 45 predicted domains (107, 130).
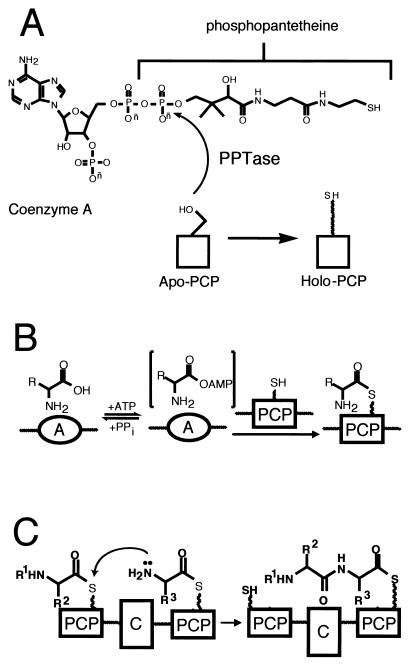
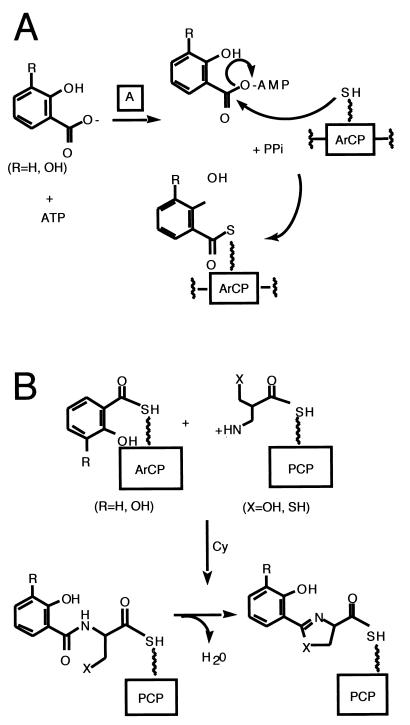
Each NRPS assembly line needs to be organized to carry out four kinds of catalytic operations. First, each of the PCP domains must be converted from the apo form to the holo form, bearing the phosphopantetheinyl arm (Fig. 3A). This is a posttranslational priming that must occur on each carrier protein way station for chain growth to proceed to full-length product. Typically, a dedicated priming enzyme, a phosphopantetheinyl transferase (PPTase) is encoded in the siderophore biosynthetic gene cluster (65). PPTases catalyze the attack of a side chain serine in a consensus sequence of the carrier protein domain on the pyrophosphate linkage of coenzyme A, transferring the phosphopantathenate (P-pant) moiety, yielding the phosphodiester linkage to the PCP serine side chain and releasing 3′,5′-ADP as soluble product (Fig. 3A).
FIG. 3.
Initiation of siderophore synthesis. (A) PPTase priming equation on apo PCP; (B) two-step A domain equation; (C) C domain equation.
Second, the assembly line must be able to select and activate monomers and incorporate them at particular positions in the growing chain. The carboxylic acids and amino acids to be activated are selected by adenylation (A) domains, operating with logic analogous to that of aminoacyl tRNA synthetases: the amino acid selected by the A domain active site is converted via attack on the cosubstrate ATP to the aminoacyl-AMP (Fig. 3B). This thermodynamically activated monomer is then transferred to the adjacent HS-pant-PCP domain (Fig. 3B), to covalently tether the aminoacyl moiety on the thiol way station and preserve the thermodynamic activation of the carboxyl groups as aminoacyl thioesters (aminoacyl-S-PCP).
The third unit operation of a biopolymer assembly line is chain elongation and is shown in Fig. 3C (16, 125, 131). In the NRPS superfamily of enzymes, siderophore chain growth is directional, from the most upstream, N-terminal PCP domains, to the most downstream, C-terminal PCP domains. Every peptide bond-forming step is tied to chain elongation and translocation. The peptide synthetase catalytic domains are termed condensation (C) domains; typically, a core elongation module of an NRPS assembly line has the three domains, C-A-PCP, as a functioning module. For an undecapeptide, e.g., cyclosporine, one would need 10 such elongation modules. The order of the elongation modules in the assembly line includes the ordering of the embedded A domains and thus encodes the selection of the amino acids to be incorporated. The actual peptide bond-forming step by each C domain involves an upstream peptidyl-S-PCP as the donor cosubstrate and the proximal downstream aminoacyl-S-PCP as the attacking, acceptor substrate. The translocated chain has grown by one peptide bond, is still tethered as a thermodynamically activated acyl-S-PCP, and is ready for another elongation cycle by the C domain of the next downstream module (Fig. 3C).
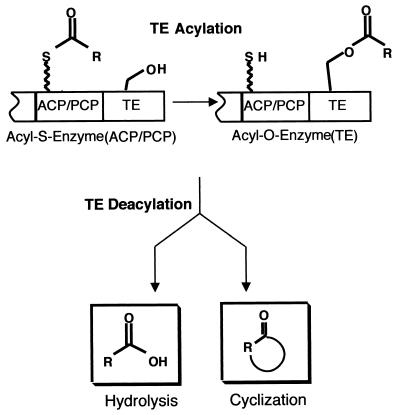
The fourth operation, shown in Fig. 4, is chain termination and release of the full-length nonribosomal peptide or siderophore (59). Because the peptidyl chains grow as a cascade of elongating acyl-S-enzyme intermediates, when the full-length chain arrives at the most-downstream PCP domain, it is still covalently docked and requires chemical cleavage from the enzyme assembly line. In the vast majority of NRPS the most-carboxy-terminal domain is a 30- to 35-kDa autonomously folding unit termed a thioesterase (TE), after the homologous function of such TEs in chain release in fatty acid synthase (FAS) and polyketide synthase (PKS) assembly lines (6, 122, 127). Chain release involves interdomain transfer from the PCP to the active site serine side chain in the active site of the TE to yield an acyl-O-TE intermediate (99). Figure 4 shows that this intermediate can experience two major fates: intermolecular hydrolysis, to release the free acid (e.g., the TE of ACV synthetase), or intramolecular capture by an OH or NH2 group in the peptidyl chain, to release a cyclic lactone or lactam (e.g., enterobactin or gramicidin).
FIG. 4.
Termination of siderophore synthesis.
As we examine features of the classes of siderophore synthetases discussed in detail below, there are a few general variations found in the siderophore synthetase family of NRPS assembly lines that are worth noting at the outset (see reference 88 for a recent overview).
First, with regard to chain initiation, the great bulk of bacterial siderophores so far identified contain an N-terminal modification of the peptide chain that usually derives from an aryl acid, such as salicylate and DHB noted above (Fig. 5A). These aryl acids serve as aryl-N-caps and may fulfill multiple functions. One could be analogous to the N-formylmethionine at the start of ribosomal protein biosynthesis in bacteria, imposing a directionality on chain growth by making the initiator amino acid only able to function as an N-acylated, peptide-like donor in the first chain elongation step. Another reason could be the incorporation of the phenolic or catecholic moiety that is one of the three common groups for ferric iron chelation. To install salicyl and DHB groups at the start of the assembly lines requires a dedicated A domain, for either salicyl-AMP or DHB-AMP formation (39, 40, 54, 86, 98, 99); in addition, the most-upstream carrier protein domain in siderophore assembly lines must be recognized for acylation by salicyl-AMP or DHB-AMP. This subset of PCP domains has been referred to as aryl carrier proteins (ArCPs) (127, 128).
FIG. 5.
Siderophore synthesis. Actions of A and ArCP (A) and Cy (C) domains.
Second, another feature prevalent in siderophores, and therefore encoded by or embedded in their siderophore synthetases, is the presence of thiazoline rings and oxazoline rings. We note below that these heterocycles are formed by variants of the condensation (C) domains, which have been termed cyclization (Cy) domains. Figure 5B shows that these domains first make the peptide linkages, using the downstream Cys-S-PCP, Ser-S-PCP, or Thr-S-PCP as attacking substrate. Then, before the chain can be translocated out of the vicinity, down to the next elongation module, the Cy domains catalyze the side chain cyclization and dehydration. The yersiniabactin (Ybt) synthetase and pyochelin (Pch) synthetase make tandem bis-heterocycles from two cysteine monomers (55, 87, 114, 115).
Third, in chain termination, siderophore synthetases show diverse catalytic propensity for siderophore chain transfer. We note the hydrolytic TE domains for Ybt and Pch synthetases and a remarkable cyclotrimerizing TE domain in the enterobactin (Ent) synthetase. For vibriobactin and anguibactin, the growing acyl chains are transferred out to soluble amine acceptors: norspermidine for vibriobactin and N-OH-histamine for anguibactin syntheses. In these chain-terminating steps, TE domains are not present in the assembly line, but rather C domains seem to be involved (55, 56, 128).
With these general concepts we turn to specific siderophore systems and discuss both the genetics of biosynthesis and the detailed biochemical mechanisms for the synthesis of these compounds.
ENTEROBACTIN
Introduction
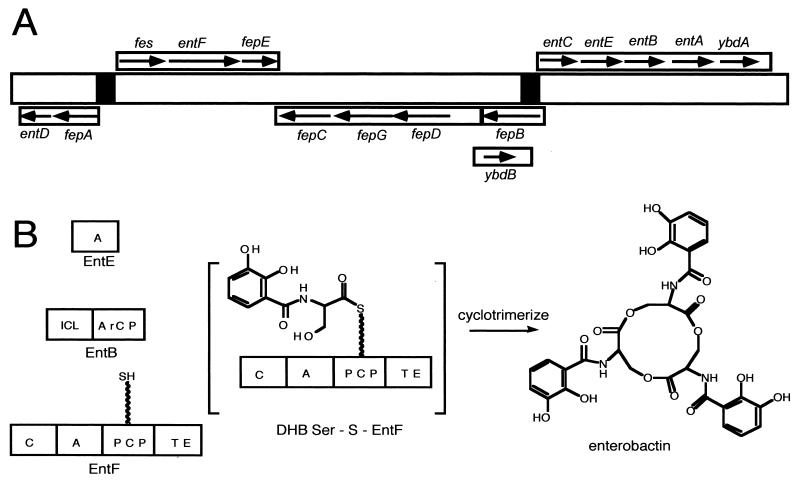
Enterobactin was first purified and characterized from Salmonella enterica serovar Typhimurium and E. coli culture supernatants (34, 77, 84, 90, 108). Figure 6A shows a schematic map of the enterobactin gene cluster, including the transcriptional units and the potential regulation sites. The initial stage for enterobactin synthesis requires the product of three genes: entC, which encodes an isochorismate synthetase; entB, which encodes 2,3-dihydro-2,3-dihydroxybenzoate synthetase; and entA, which encodes 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. The second stage deals with the synthesis of one molecule of enterobactin from three molecules each of 2,3-dihydroxybenzoic acid and l-serine. Products of the genes entD, entE, and entF and an assembly activity encoded in the carboxy terminus of entB catalyze this step. The cytosolic proteins EntE, EntF, and EntB, which are E. coli enzymes necessary for the final stage of enterobactin synthesis, are released by osmotic shock (8, 34, 52, 77). Consistent with the idea that cytoplasmic proteins found in “shockates” have an affinity for membranes, a small fraction of each was found in membrane preparations (34). It was further demonstrated that the enzymes were enriched in a minor membrane fraction of buoyant density intermediate between that of cytoplasmic and outer membranes, providing indirect support for the notion that these proteins may play a role in enterobactin excretion as well as synthesis. It is possible that enterobactin synthase is associated with a cytoplasmic membrane pump capable of excreting the siderophore. Enterobactin may be secreted directly from its site of synthesis through the envelope to the external milieu (8, 34).
FIG. 6.
Genetics and enzymology of enterobactin biosynthesis in E. coli. (A) Scheme of the enterobactin biosynthesis and transport genes. (B) Scheme of the synthetases encoded by the genes shown in panel A and the final product, enterobactin. ICL, isochorismate lyase activity.
Genetics
Transport of ferric enterobactin into the cell cytosol requires the products of at least five additional genes (34, 77, 90, 104). The enterobactin gene cluster includes 14 genes organized in six operons originating from three Fur-regulated bidirectional promoter-operator regions. One of these genes is fepA, which encodes the outer membrane protein receptor for complexes of ferric enterobactin. Another is fes, which is required for the intracellular release of iron from enterobactin. The other genes are fepB, fepC, fepD, fepE, and fepG, which intervene in the uptake of ferric enterobactin through the periplasm and the cytoplasmic membrane. In E. coli the enterobactin biosynthetic and transport genes are located in a large 22-kb genecluster, entD-fepA-fes-entD-fepE-fepC-fepG-fepD-fepB-entC-entE-entB-entA-ybdA, at 13 min on the E. coli chromosome (Fig. 6A) (M. Di Lorenzo, M. E. Tolmasky, S. Poppelaars, M. Nagasawa, S. Chai, and J. H. Crosa, submitted for publication). By using data obtained from gene fusions combined with DNA sequence analysis, it was concluded that four of the biosynthetic genes and ybdA were organized as an operon as entC-entE-entB-entA-open reading frame 15 (ORF15) (ybdB) (Fig. 6A). Primer extension analysis identified the transcription initiation site 55 nucleotides upstream of the entC translation initiation codon (52). A sequence similar to the consensus Fur box was found within this region. It is of interest that there is another operon, fepBDGC, which is initiated upstream of the entC gene but in the opposite strand (Fig. 6A). The −10 and −35 sequences for the two divergent promoters are located in the 103 bp that separate the initiation sites for these two mRNAs (52). A similar situation occurs on the other end of the enterobactin cluster, in which two divergent operons encode fepA-entD and fes-entF-fepE, respectively. Figure 6A shows that in this case the intergenic region also serves as the starting point for divergent mRNAs, although with more overlapping of the −10 and −35 promoter sequences and with only 18 bp separating the primary initiation sites.
Recently, overlapping and opposing promoter elements for the E. coli fepDGC operon and the ydbA gene (which encodes a 43-kDa cytoplasmic membrane protein) in the enterobactin cluster were also investigated by using a combination of site-directed mutagenesis and bidirectional transcriptional fusion vectors (23).
Compared to the E. coli consensus sequence both promoters were poorly conserved at the −35 sequence position. Mutations in either of the −35 sequences that resulted in higher transcription for one of the directions consistently reflected in a decrease of transcription in the opposite direction. A further consequence was a decrease in the iron regulation of that particular transcript. These studies indicated that there is a direct competition for RNA polymerase binding and that the expression levels of these promoters in addition possibly influenced by additional promoter sequence elements and accessory regulatory factors. Iron-mediated regulation of these two promoters via the Fur protein is likely to occur as a consequence of the relative promoter strengths and the position of two Fur-binding overlapping sequences which act as a compact regulatory region.
The mechanism involved in the regulation of the fepA-fes divergent promoters is influenced by the spatial distribution of this DNA region which allowed RNA polymerase to bind simultaneously the fes and fepA promoters and coordinately repress regardless of their different transcriptional activities. Comparison of runoff transcription patterns in the presence and absence of bound RNA polymerase indicated that in this divergent promoter system in which the fes promoter is stronger than the fepA promoter, there is a direct competition between the polymerase and regulators for overlapping target sites in the DNA.
The enterobactin genes are also found in other members of the Enterobacteriaceae such as Salmonella, Klebsiella, and Shigella species, as well as in Pseudomonas species. However, only 10% of the Shigella strains possess an active enterobactin iron transport system (25, 34).
Enzymology
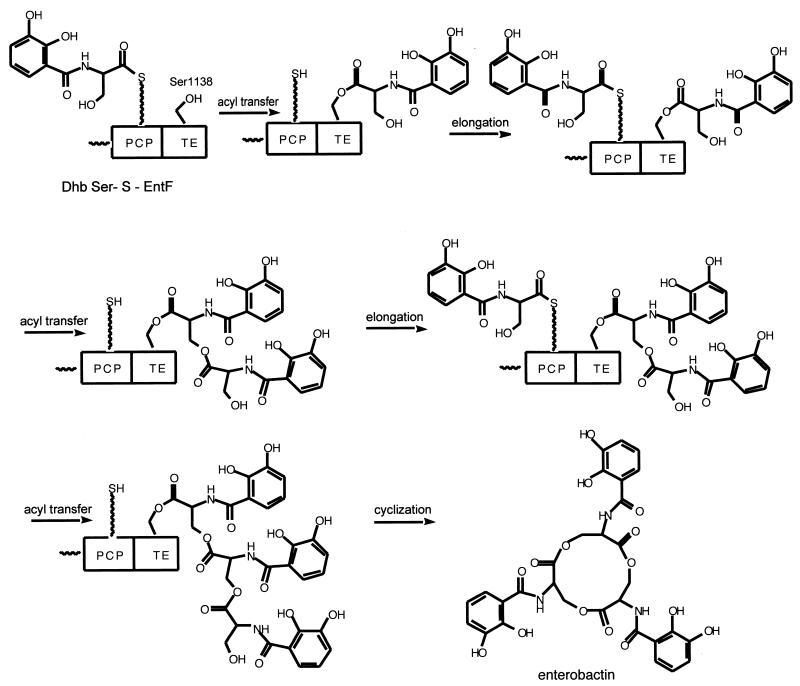
The enzymes that assemble enterobactin were among the first to be worked out for functional assays (92), substantially before bacterial genome sequences were available, and this knowledge led to the organizing concepts for the enterobactin system as a prototype for the enzymological approaches for all the other siderophores discussed below (39). A scheme of the assembly line is shown in Fig. 6B, and the detailed assembly line is indicated in Fig. 7. From these figures it is evident that three enzymes contribute seven domains. The EntE enzyme was first purified and characterized as a dihydroxybenzoyl-AMP ligase and found also to activate salicylate to salicyl-AMP reversibly, using the substrate-dependent 32PPi-ATP exchange assay that had been developed for detection of aminoacyl-AMP formation by aminoacyl tRNA synthetases. The DHB-AMP was formed at a rate of several hundred molecules per minute but released into free solution at a rate of only about 1 molecule min−1, consistent with kinetic sequestration of DHB-AMP in the EntE active site to slow down adventitious hydrolysis (92). The other three enzymes in the enterobactin synthetase assembly line are EntB, EntD, and EntF. The 140-kDa EntF protein was first purified; it was found to activate l-serine to seryl-AMP by the same 32PPi-ATP exchange assay, and substoichiometric amounts of phosphopantetheine were also detected in purified EntF by expression in a mutant blocked in EntD that incorporated radioactive beta alanine into the pantetheinyl prosthetic group of purified EntF (92, 98). To proceed further required the identification of the Ent assembly line-priming PPTase. The purification of the PPTase, termed holoacyl carrier protein synthase, ACPS, that acted on the apoACP subunit of the E. coli FAS (64) also demonstrated it would not prime the apo form of EntF and that therefore a second PCP-specific PPTase should exist in E. coli. This was predicted to be the EntD protein by homology analysis and experimentally validated when purified EntD was shown to be a PCP-specific PPTase (65). The fourth and last protein of the enterobactin synthetase system, EntB, was assigned its function by bioinformatic analysis that revealed this protein, harboring a known activity as an isochorismate lyase, involved in the pathway leading up to DHB (40) in its first 250 residues, had a C-terminal 100 residues that looked like an apoPCP domain. EntB was then phosphopantetheinylated with CoASH by EntD (40) to generate a second P-pant thiol way station in the Ent assembly line. The Walsh laboratory (39) later defined the EntB carrier protein as an ArCP domain since the holo EntB could be acylated with DHB via EntE action. The holoArCP on EntB is covalently loaded with DHB, while the holoPCP on EntF is auto-aminoacylated as a seryl-S-enzyme. The EntF C domain condenses DHB-S-ArCP with seryl-S-PCP, to yield a DHB-Ser-S chain on the PCP domain of EntF. Comparison of the DHB-Ser chain to the mature enterobactin, a DHB-Ser-cyclic trilactone, suggested that EntF must be able to cyclotrimerize the DHB-Ser-S-enzyme as the chain-terminating release step. The only remaining domain of EntF was the TE domain. With a mutant of the EntF TE domain's active site histidine, H1271A, the kcat for enterobactin was reduced 103-fold (106), and now a (DHB-Ser)2 dimer could be detected accumulating on the TE domain by mass spectrometric analysis of proteolytic fragments. This led to the explicit proposal, shown in Fig. 7, that the EntF TE can serve as a waiting room for the DHB-Ser acyl chain, protecting it from hydrolysis, while a second DHB-Ser chain builds up on the adjacent, upstream holoPCP domain as shown in the DHB-Ser-S-PCP, DHB-Ser-O-TE bisacyl enzyme intermediate. Attack by the side chain OH of the Ser moiety of the DHB-Ser-O-TE would yield the dimeric (DHB-Ser)2-O-TE acyl enzyme detected by mass spectrometry (106). Sequestration of this acyl enzyme would allow another DHB-Ser chain to arrive at the HS-Pant-PCP domain, and another interdomain acyl transfer would yield the (DHB-Ser)3-O-TE. Now this must achieve a conformation kinetically competent for intramolecular cyclization, to effect formation of the 12-ring trilactone and release the mature enterobactin siderophore. A cyclotrimerizing TE domain is a startling catalyst but is joined by a related TE, in the tyrocidine and gramicidin synthetase assembly lines, which, respectively, cyclize a decapeptidyl-O-TE (122) or cylcodimerize a pentapeptidyl-O-TE to the cyclic decapeptide antibiotic (61).
FIG. 7.
Enterobactin synthetase assembly line in E. coli.
The enterobactin synthetase component proteins EntE, -B, and -F have been examined for stable association by gel filtration analysis, without any indication of long-lived complexes. Also, in the reconstituted system of EntEBF-producing enterobactin at rates of 100 to 150 min−1 (40), the Kms for EntE and EntB indicate transient, readily associating and dissociating complexes. This three-protein assembly line is dynamic. Thus, the Ent system was the first siderophore synthetase to be characterized for P-Pant arms, for an ArCP domain in EntB, for identification of a dedicated PPTase for priming of the apoArCP and PCP domains, and for a cyclo-oligomerizing TE domain, all concepts that now have been generalized to other siderophore synthetases.
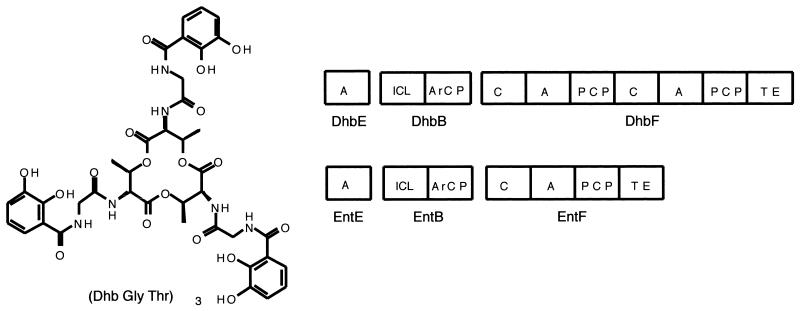
In that context, the genome sequencing of Bacillus subtilis indicated a cluster of related Ent genes, DhbB, -E, and -F, that on further examination were revealed to contain sequencing errors indicating premature ORF termination (69, 70). Reevaluation of these sequences identified the anticipated EntE and EntB homologs, but now the EntF homolog had an extra module, C-A-PCP, suggesting the B. subtilis homolog of enterobactin would be an aryl-N-capped dipeptide lactone (Fig. 8) (71). Sure enough, it has now been reported that (DHB-Gly-Thr)3 is the structure of the tricatecholic siderophore bacillobactin as shown in Fig. 8 (71).
FIG. 8.
Bacillobactin from B. subtilis. Comparison of the bacillobactin and enterobactin synthetases.
This finding validates the predictive logic for modular arrangement of catecholic siderophore synthetase assembly lines based on the EntF prototype and also validates the notion that algorithms for A domain selectivity (20, 109) have a robust success in predicting the amino acid to be activated by a novel A domain.
VIBRIOBACTIN
Introduction
The causative agent of cholera, V. cholerae, acquires iron via the vibriobactin-mediated iron transport system (12-15, 44, 129, 137-139). This bacterium can also use iron contained in heme or hemoglobin and also produces an iron-regulated hemolysin which may intervene in iron acquisition in vivo (51, 80, 81, 137-139). V. cholerae can use ferri-ferrichrome as well (95).
Vibriobactin is synthesized and secreted into the environment, where it binds ferric iron with high affinity. The ferri-vibriobactin complex is then transported into the cell by a process that requires the outer membrane receptor (113).
Unlike enterobactin, for which the biosynthetic and transport genes are located in a single genetic locus, vibriobactin genes are located in two separate gene clusters (34, 137-139). The genome of V. cholerae consists of two chromosomes (48, 123). Both vibriobactin system gene clusters are located on V. cholerae chromosome 1 (48, 123) but are separated by approximately 106 bp (13-15, 137-139). Chromosome 1 also contains most of the genes required for growth and pathogenicity of V. cholerae (123). This may reflect the central role of vibriobactin synthesis and utilization in the growth and survival of V. cholerae in at least one of its habitats.
Genetics
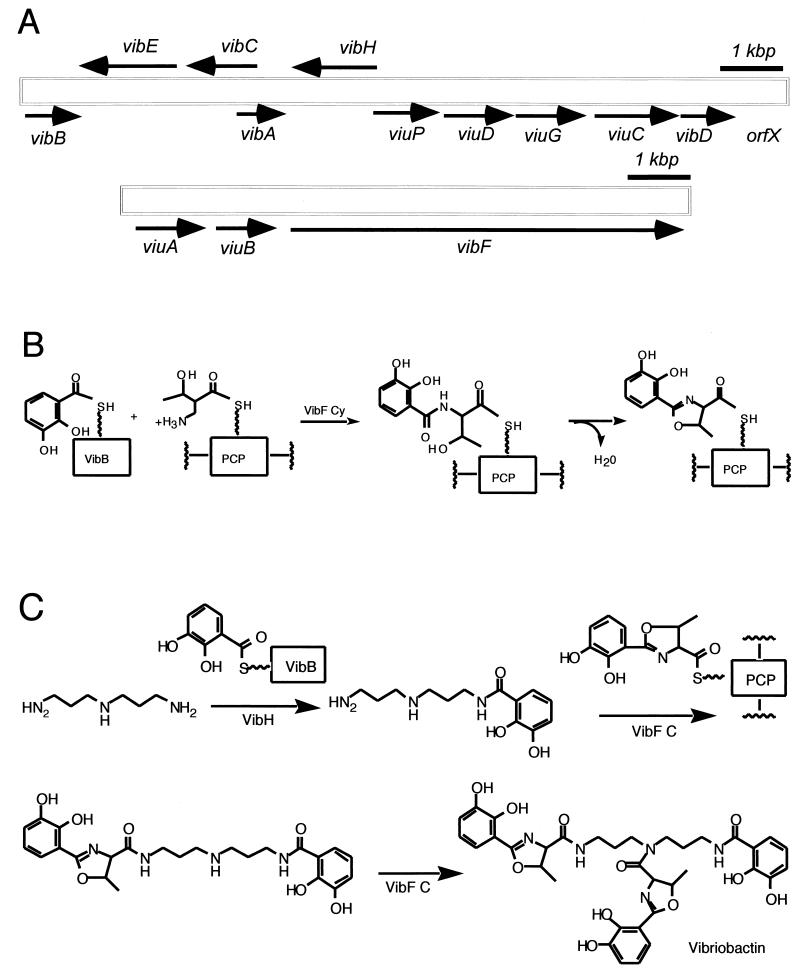
There are two regions of the V. cholerae chromosome that are concerned with vibriobactin-mediated iron uptake. Each cluster contains both biosynthetic genes and genes for vibriobactin utilization. Figure 9A shows that one of these clusters contains the vibriobactin transport and utilization genes viuA and viuB (13-15) and the biosynthetic gene vibF (13). The second region also shown in Fig. 9A includes the previously described genes for the synthesis of DHBA from chorismate (vibABC) and a gene for the activation of DHBA (vibE) (137-139). The region also contains genes for a periplasmic binding protein-dependent ABC transport system, which transports vibriobactin and enterobactin through the periplasm and across the inner membrane. It is unclear why the genes for vibriobactin synthesis and transport are divided into two genetic loci, but the separation of genes that usually map together has been observed for other iron acquisition systems in V. cholerae. For example, the heme receptor gene hutA maps at a distance from the other heme transport genes (80).
FIG. 9.
Genetics and enzymology of vibriobactin biosynthesis in V. cholerae. (A) Scheme of the vibriobactin biosynthesis and transport genes. (B and C) Vibriobactin assembly line. (B) Oxazoline formation; (C) Incorporation of norspermidine and completion.
Recently (137) two other vibriobactin biosynthetic genes were identified, vibD, which encodes a phosphopantetheinyl transferase, and vibH, located between vibA and viuP (Fig. 9A), which encodes a novel NRPS possessing only one condensation domain (137). Below we describe the genetic evidence suggesting their roles in vibriobactin synthesis. However, their roles became clear after the biochemical properties and in vitro synthesis of vibriobactin were studied, as shown in the next section. To determine whether vibD was required for vibriobactin biosynthesis, a vibD mutant was constructed by marker exchange (137) which was positive for the synthesis of catechols, indicating that the mutant had no defect in DHBA biosynthesis. The mutant, however, cross-fed V. cholerae mutants in vibB and vibA but it failed to stimulate the growth of the vibD and vibH mutant strains, indicating that it was not secreting vibriobactin. The vibriobactin synthesis defect in the vibD strain was only complemented by either vibD or entD encoded on a plasmid. Taken together, these data indicate that VibD is important for the assembly of vibriobactin, and on the basis of its sequence it could very well act to provide the phosphopantetheinyl transferase activity required for vibriobactin synthesis.
The investigators performed experiments similar to those carried out with vibD to determine whether vibH was also required for vibriobactin biosynthesis. Like the vibD mutant, the vibH mutant strain was positive for the synthesis of catechols, indicating that the conversion of chorismate to DHBA was not impaired. The vibH mutant did not cross-feed the vibD, vibB, or vibH mutants, indicating a defect in vibriobactin biosynthesis while it did cross-feed the vibA mutant, consistent with the ability of the vibH mutant to produce the catechol DHBA. A cloned copy of wild-type vibH in trans restored the ability to stimulate growth of each of the mutant strains. Therefore, VibH, like VibD, is required for the assembly of vibriobactin from DHBA, threonine, and norspermidine. The predicted VibH protein has a calculated molecular mass of 49.8 kDa and a predicted pI of 5.8. A BLAST search revealed that VibH has sequence homology with NRPS proteins, including B. subtilis DhbF (71, 97), E. coli EntF (92), and other proteins. Alignment of VibH with the best-characterized of the closely related proteins, EntF, revealed that the VibH protein aligns well with the first 452 amino acids of EntF. indicating that VibH contains only the condensation domain and no regions of homology to either an adenylation domain or a peptide carrier domain (54, 57). This unusual protein structure raises questions about the mechanism of action of VibH. Fortunately, considerable amount of work has already been carried out at the biochemical level, and now the mechanism of action of this protein is clearly understood as presented in the next section.
Enzymology
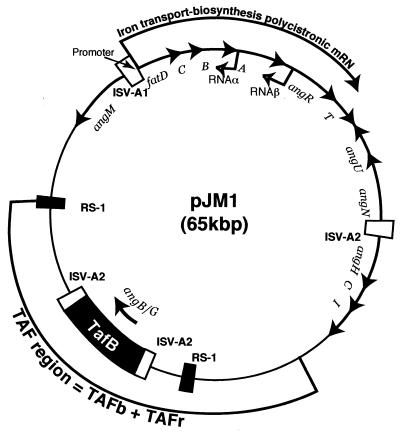
The two new enzymatic issues in the case of vibriobactin are how an oxazoline ring is formed and how the amide linkages are formed to the dihydroxyphenylthiazolinyl (DHPT) and dihydroxyphenyl oxazolinyl (DHPO) acyl groups. The VibE and VibB proteins are functional and structural analogs to the EntE and EntB enzymes noted previously for enterobactin assembly, acting to produce the DHB-S-pantetheinyl-VibB as aryl N-cap donor (Fig. 9B). This provides not only the catechol moiety in the two DHPO chains but also the DHB moiety that ends up on N6 of the norspermidine scaffold (57, 58). The VibF subunit has six predicted NRPS type domains, Cy-Cy-A-C-PCP-C, and has been purified to homogeneity after heterologous expression in E. coli (58). The A domain activates l-threonine and, as shown in Fig. 9B, attaches it to the HS-pant-PCP domain to yield the anticipated threonyl-S-PCP substrate for condensation/cyclization by one of the two Cy domains with DHB-S-VibB as donor. Figure 9B also shows that the DHB-Thr-S-VibF initial condensation product is then cyclized and dehydrated to give the HPTO-S-VibF intermediate. Figure 9C shows that the HPTO-S-VibF intermediate could be transferred to norspermidine as soluble substrate for one of VibF's two C domains, and indeed the transfer is observed. But the N6-acylated norspermidine DHB-N-norspermidine is 104 times more efficient in DHBO transfer by kcat and Km catalytic efficiency criteria, to yield the DHBO-N1-DHB-N6-norspermidine intermediate. In turn, the production of DHB-norspermidine is catalyzed by the fourth enzymatic subunit of the Vib assembly line, VibH, a free-standing C domain homolog that carries out amide bond formation like the reactions catalyzed by the C domains embedded in elongation modules of more conventional NRPS assembly lines. The DHBO-N1-DHB-N6-norspermidine is just one DHPO transfer step away from the mature vibriobactin. VibE, -B, and -F activate another molecule of DHB and threonine and condense them to the DHBO-S-VibF acyl enzyme, perhaps using the second of the tandem Cy domains in VibF, and then transfer this DHPO acyl chain, perhaps via the second C domain of VibF, to the uncapped N3 of the bisacyl norspermidine to yield the tris acylated vibriobactin (58) chelating groups, catechol, thiazoline, and N-OH, in one compact small molecular structure.
ANGUIBACTIN
Introduction
The bacterial fish pathogen V. anguillarum, a gram-negative polarly flagellated comma-shaped rod, is responsible for both marine and fresh water fish epizootics throughout the world (1). V. anguillarum causes a highly fatal hemorrhagic septicemic disease in salmonids and other fishes, including eels (1, 17, 46, 82, 89). The disease caused by V. anguillarum in salmon shows striking similarities to the septicemic disease in humans caused by V. vulnificus and Vibrio parahaemolyticus (1).
Genetics
The key feature which enables many pathogenic strains of V. anguillarum to survive within the vertebrate host in feral, as well as in intraperitoneal, experimental infections, is the possession of a 65-kb virulence plasmid, pJM1, shown in Fig. 10. This plasmid provides the bacteria with an iron-sequestering system that is crucial in overcoming the nonspecific defense mechanisms of the host (24-30, 119, 136). This system centers upon the synthesis of the siderophore anguibactin, an iron-scavenging compound, and subsequent transport of the ferric anguibactin complex into the cell cytosol via the cognate transport system proteins FatA, -B, -C, and -D (1-5, 60, 133, 136). Anguibactin is produced by the virulent strains of this bacterium in the host and in any other environment in which iron is chelated and thus not readily available (24-30, 134). The plasmid-encoded iron transport system and siderophore biosynthetic genes (shown in Fig. 10) are controlled by the concentration of available iron, via at least four plasmid-encoded regulators: two positive regulators, AngR (anguibactin system regulator) and TAF (transacting factor[s]); and two negative regulators, antisense RNAα and RNAβ (19, 21, 22, 25, 35, 100-102, 118-121). RNAα is very stable at high iron concentrations and acts at posttranscriptional level in the repression of fatB and fatA expression, while RNAβ, found under conditions of mild iron limitation, acts on the attenuation of expression of the angR gene in the iron transport-biosynthesis (ITB) operon (19, 102). Repression of the ITB operon at the transcriptional level requires the chromosomally encoded Fur protein (19, 117, 133). The promoter for the ITB operon (pITBO) was localized within a region ca. 300 bp upstream of fatD by primer extension and S1 mapping analysis (Fig. 10) (19, 126).
FIG. 10.
Genetic map of the pJM1 virulence plasmid of V. anguillarum highlighting those genes involved in the regulation, biosynthesis, and uptake of ferric anguibactin.
One of the other elements controlling expression of the ITB operon is AngR. Remarkably, this protein plays a role not only in the in the regulation of iron transport gene expression but also in anguibactin biosynthesis, and therefore virulence. The angR gene is encoded within a polycistronic message that includes the iron transport genes fatDCBA and the angT gene within the ITB operon (Fig. 10) (102, 119, 126, 132, 134). In addition, there is evidence that anguibactin itself enhances transcription of this operon, possibly independently of AngR and the TAF products (22). Recently, the TAF region was dissected into two subregions: TAFb, essential for anguibactin biosynthesis, and TAFr, associated with regulation of expression of the ITB operon. Within TAFb, two overlapping genes, angB and angG, were identified (131). These two genes are translated in frame. The ITB operon and other anguibactin biosynthetic genes located downstream are bracketed by the highly related ISV-A1 and IASV-A2 insertion sequences (Fig. 10), which are also highly related to the insertion sequences found flanking various thermostable direct hemolysin genes in V. parahaemolyticus, Vibrio mimicus, and non-O1 V. cholerae. This raises the possibility that some of these genes may have been acquired during evolution. A biosynthetic gene, angH, encoding a histidine decarboxylase, lies downstream of the transposase gene tnpB (Fig. 10). Insertions in angH are deficient in anguibactin biosynthesis and can be complemented with histamine (118), showing its involvement in anguibactin biosynthesis. Figure 10 also shows that there is a gene, angU, located downstream from angN and transcribed in the same orientation. Insertions in angU result in an anguibactin-deficient phenotype which can be complemented by the cloned angU gene (M. Di Lorenzo and J. H. Crosa, unpublished observations). These results indicate that angU is also essential for anguibactin biosynthesis. BLAST analysis demonstrated that the AngU protein shows 32% homology and 51% similarity with IucD, an oxidase involved in aerobactin biosynthesis in E. coli strains harboring the plasmid pColV K30 and 54% homology and 71% similarity to RhbE, an oxidase of Sinorhizobium melilotii. It is thus possible that angU is involved in the oxidation of histamine, resulting from AngH action, yielding N-hydroxy histamine.
Enzymology
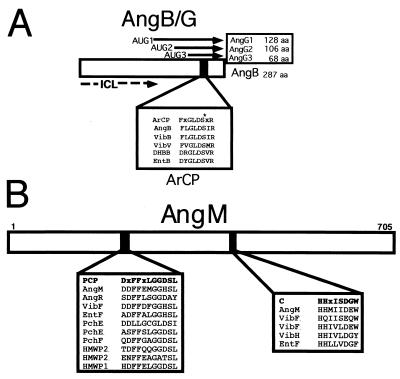
The enzymology of anguibactin biosynthesis is still under investigation; however, predictions can be made based on our knowledge of the structure of this siderophore and functions of potential biosynthetic genes inferred by homology studies. Figure 10 shows that the TAFb region consists of an apparent composite transposon, surrounded by ISAV2 repeated sequences, containing an ORF which is essential for anguibactin biosynthesis. This ORF was demonstrated to encode AngB, a 287-amino-acid polypeptide that shows significant homology to the EntB protein of E. coli and the VibB protein of V. cholerae (see Fig. 16A). AngB, which is essential for DHBA biosynthesis, is a precursor of anguibactin. Like EntB, the amino terminus of AngB possesses the isochorismate lyase activity (ICL in Fig. 11A), thereby explaining the need for this protein for the synthesis of DHBA (131). Analysis of mutations in the angB open reading frame provided evidence that in addition to angB, an overlapping gene, angG, exists at this locus and that it encodes three polypeptides which are in frame to the carboxy-terminal end of the 33-kDa AngB. Figure 11A shows that in addition to the DHBA synthesis function in the amino terminus (ICL) there is, at the carboxy terminus, an ArCP domain that is also present in the internal AngG polypeptides (132). This domain is where phosphopantetheinylation occurs at the serine residue. The P-Pant group acts as an acceptor of an activated aryl or amino acid group. ArCP domains intervene in assembly reactions during siderophore biosynthesis. By using site-directed mutagenesis a mutation at S248 was generated that leads to a complete abolishment of anguibactin production compared to the isogenic control. Yet DHBA production in this mutant was unaffected, further demonstrating the separability of the ICL and ArCP activities.
FIG. 16.
Pyochelin assembly line in P. aeruginosa.
FIG. 11.
Anguibactin biosynthesis in V. anguillarum. (A) AngB/G, provides the isochorismate lyase activity (ICL) and the ArCP for activated 2,3-DHBA. (B) AngM provides condensation and spacer domains.
Transposon insertions in the virulence plasmid identified another gene, angM, involved in anguibactin biosynthesis (33). Assessment of its sequence against the DNA database by BLAST analysis showed that AngM is highly homologous to V. cholerae VibF, required for vibriobactin biosynthesis (39% identity in a 287-amino-acid overlap; 52% similarity) (see Fig. 16). Although homologies exist in the PCP domain of HMPW1 and in the C domain of EntF, too many stretches of no homology reduce the overall percentage of similarity. Figure 11B shows that there are two domains in AngM that have extensive homology with the PCP and C domains, respectively, of NRPS. The PCP domain is the 4′ phosphopantetheine binding domain involved in thioester formation. The C motif is the elongation domain. No other sequence corresponding to an A domain, which is usually adjacent to the PCP domain, was identified in AngM.
To characterize the actual biosynthetic function of the angM gene, this gene was cloned in pBR325 (33). The cloned angM gene complemented the angM transposon mutant, restoring anguibactin production, underscoring the essential role played by this gene in anguibactin biosynthesis.
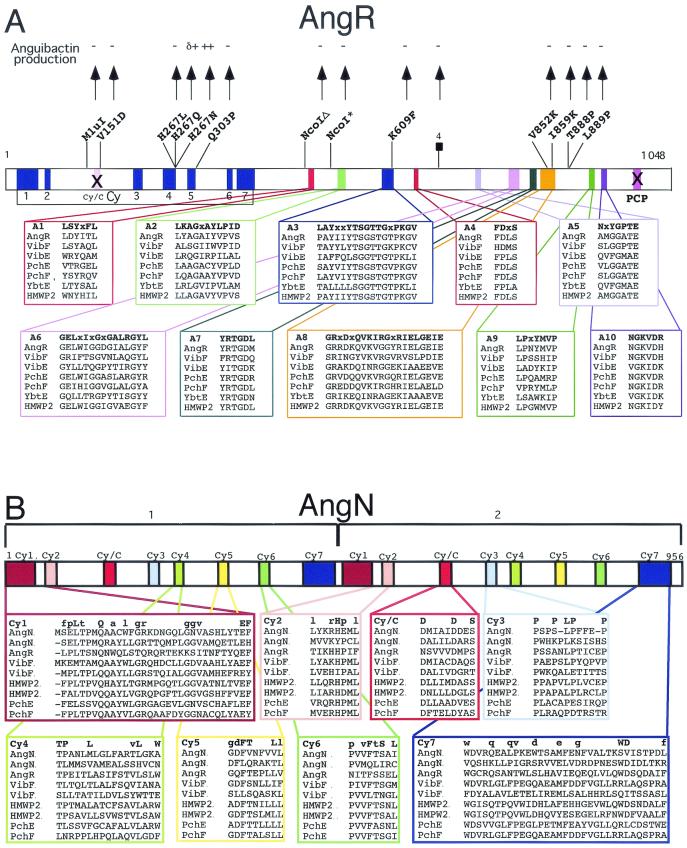
AngR is essential for both regulation and anguibactin biosynthesis (134). Figure 12A shows a scheme of the AngR amino acid sequence identifying the domain organization as Cy-A-PCP. The 10 subdomains of the A domain, A1 to A10, are conserved. The AngR Cy domains are compared to those from other NRPS in this figure as well as in Fig. 12B. There is also a PCP domain present in the carboxy terminus of AngR, which is compared to those from AngM and other NRPS in Fig. 11B. The PCP and Cy/C domains of AngR may not be functional, because an essential serine is replaced by alanine in the PCP domain, while the essential first aspartic acid is replaced by asparagine in the Cy/C subdomain, although the other seven Cy subdomains are very conserved. Transposition mutants within another region of the pJM1 plasmid resulted in the identification of the angN gene (Fig. 10). These mutants showed an anguibactin-deficient phenotype which could only be complemented with clones harboring the angN gene, demonstrating that it must play an essential role in anguibactin biosynthesis. Figure 10 shows that angN is located downstream of angR and is transcribed in the opposite orientation. Figure 12B shows that the predicted AngN protein has two Cy domains (AngN-1 and AngN-2) that are similar to each other (M. Di Lorenzo, C. T. Walsh, and J. H. Crosa, unpublished data). In Cy domains there are seven Cy subdomains and one Cy/C subdomain. The Cy1 to Cy7 subdomains are well conserved in the two Cy domains of AngN; however, nobody has yet demonstrated in any system whether Cy1 to Cy7 subdomains play a role in cyclization. Another gene, angT, which is located downstream of angR (Fig. 10) and is part of the ITB operon, is also essential in anguibactin biosynthesis, since its deletion leads to a 17-fold decrease in anguibactin production (133). AngT shows homology with TEs.
FIG. 12.
Anguibactin biosynthesis in V. anguillarum. (A) AngR may provide the adenylation domain to activate cysteine. (B) AngN may provide the cyclization domain(s) for thiazoline formation.
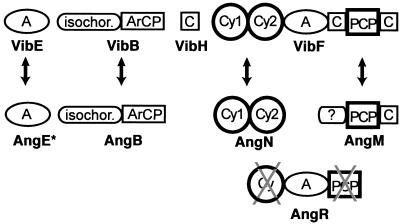
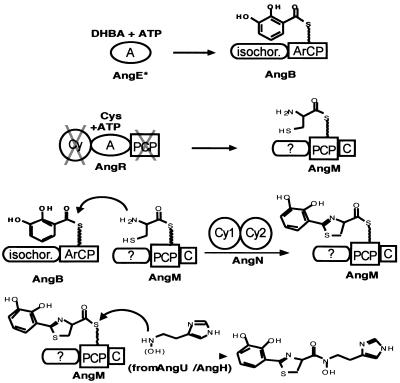
As seen in Fig. 1, the first two rings of the anguibactin structure are analogous to those in yersiniabactin and pyochelin, a phenolic-thiazoline moiety for iron chelation, but use dihydroxybenzoate rather than salicylate as the monomeric precursor, so the anguibactin has a catechol rather than a phenol substitution pattern. The right-hand half of anguibactin differs from Pch and Ybt in that this siderophore is not a free COOH siderophore but rather has the DHPT acyl group in amide linkage to an N-hydroxy-histamine. Formation of this amide is a distinguishing characteristic of the anguibactin assembly line. Analogously, the vibriobactin siderophore starts with a DHP ring connected to a five-ring heterocycle, but in this case an oxazoline rather than a thiazoline, yielding a DHPO two-ring moiety, derived from dihydroxybenzoate and threonine monomers. The DHPO acyl chain, like the DHPT acyl chain in anguibactin, is also in amide linkage. In this case the amine component is norspermidine, a six carbon N1,N3,N6-tri amine found in V. cholerae. Inspection of the vibriobactin and anguibactin synthetase subunits, shown to be necessary by genetic analysis, suggested the indicated order for the four Vib and four Ang subunits (Fig. 13). With the vibriobactin assembly line and enzymatic steps as framework, the anguibactin assembly line components can be ordered analogously for AngE (still not identified), -B, -N, -R, and -M, as shown in Fig. 14. The DHPT-S-Ang R acyl enzyme should be formed analogously and transferred to the N-hydroxyhistamine cosubstrate by the C domain of anguibactin. The anguibactin cluster contains two genes that are dedicated to conversion of the readily available amino acid histidine to the N-hydroxyhistamine dedicated cosubstrate, a histidine decarboxylase, AngH, and an N-oxygenase, AngU (reference 118 and unpublished observations). Experiments in our laboratories are currently being carried out to purify the anguibactin NRPS and synthesize anguibactin in vitro.
FIG. 13.
Comparison of vibriobactin and anguibactin NRPS. Crossed-out domains indicate that their sequence has mutations in crucial amino acid residues. AngE has not been identified as yet (also crossed out in the scheme).
FIG. 14.
Model for the anguibactin synthetase assembly line in V. anguillarum. The asterisk on AngE indicates that this protein has not yet been identified in V. anguillarum.
PYOCHELIN
Introduction
P. aeruginosa, an opportunistic pathogen, possesses many systems devoted to the transport of iron into the cell cytosol and can also utilize exogenous ferric enterobactin (31), a reflection of its ubiquity in the environment. This bacterium produces two siderophores: pyochelin and pyoverdin (also called pseudobactin), which are shown in Fig. 1 (7, 37, 74, 75, 93, 94, 105, 111, 112). The cognate protein receptors located in the outer membrane for ferric pyochelin, ferric pyoverdine, and ferric enterobactin have now been identified (7, 74, 75). The ferric enterobactin receptor, PfeA, with a molecular mass of 78 kDa, shows homology with the corresponding enterobactin receptor, FepA, of E. coli, and an activator for this gene, Pfe, has been identified (31). Recently another siderophore, pseudomonine, has been identified in Pseudomonas fluorescens as an oxazoline-containing, salicylic acid-based siderophore that, like anguibactin, includes a histamine moiety (73). This siderophore could have a role in plant disease suppression. It is of interest that the receptors for ferripyochelin and ferric pyoverdin are present at decreased levels in strains grown in the presence of enterobactin. Furthermore, production of the ferripyochelin receptor is similarly depressed in strains that are actively producing pyoverdine (85). Results from these investigators also indicate that siderophore production must also be similarly regulated, since pyochelin levels are very reduced in cultures of pyoverdin-producing strains compared to pyoverdin-deficient strains.
Genetics
The biosynthetic genes pchDCBA and pchEF, required for the formation of the siderophore pyochelin and its precursors salicylate and dihydroaeruginoate (Dha), are clustered with the pchR regulatory gene on the chromosome of P. aeruginosa. Salicylate is an intermediate in the biosynthetic pathway of pyochelin (93, 94). The 2.5-kb region upstream of the salicylate biosynthetic genes pchBA was found to contain two additional, contiguous genes, pchD and pchC, having the same orientation. The deduced amino acid sequence of the 60-kDa PchD protein was similar to those of the EntE protein (2,3-dihydroxybenzoate-AMP ligase) of E. coli and other adenylate-forming enzymes, suggesting that salicylate might be adenylated at the carboxyl group by PchD. The 28-kDa PchC protein showed similarities to TEs of prokaryotic and eukaryotic origin and might participate in the release of the product(s) formed from activated salicylate. Dha was identified in culture supernatants of iron-limited P. aeruginosa cells. Dha, which has antifungal properties, is thought to arise from the reaction of salicylate with cysteine, followed by the cyclization of cysteine (93). Insertion of the transcription and translation stop element omega (38) in the chromosomal pchD gene resulted in the abolishment of the production of Dha and pyochelin, suggesting that PchD-mediated activation of salicylate may be a common first step in the synthesis of both metabolites. Since the pchD::Ω insertion had a strong polar effect on the expression of the pchBA genes and salicylate synthesis, it was clear that the pchDCBA cluster must be part of a transcriptional unit (105). Underscoring this finding, a full-length pchDCBA transcript was detected in iron-limited cells of P. aeruginosa (105). Transcription of this operon started at tandemly arranged promoters, which overlapped with two Fur boxes and the promoter of the divergently transcribed pchR gene encoding an activator of pyochelin biosynthesis (49, 50).
There are three additional contiguous genes—pchG, pchH, and pchI—located downstream of the pchEF genes, probably forming a pchEFGHI operon (93, 94). The deduced amino acid sequences of PchH and PchI indicate that they possess features found in ATP binding cassette transport proteins with an export function. Moreover, PchG is a homolog of the Y. pestis and Yersinia enterocolitica proteins YbtU and Irp3, which are involved in the biosynthesis of yersiniabactin. A reductase function was attributed to PchG (93). A pchG null mutant resulted in the abolishment of pyochelin formation, whereas mutations in neither pchH nor pchI affected the production of salicylate, Dha, or pyochelin. If the pyochelin biosynthetic genes were expressed from a vector promoter, uncoupling them from Fur-mediated repression by iron- and PchR-dependent induction by pyochelin, in a P. aeruginosa mutant lacking the entire pyochelin biosynthetic gene cluster, the expressed pchDCBA and pchEFG genes were sufficient for salicylate, Dha, and pyochelin production (93). It was of interest that pyochelin formation was also obtained in E. coli with this clone as long as the E. coli entD gene, which provides a phosphopantetheinyl transferase necessary for PchE and PchF activation, was also concomitantly expressed.
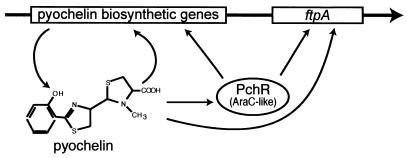
The pchR gene encodes a 31-kDaAraC-like regulatory protein which is required for production of the ferric pyochelin receptor (FptA) in response to iron limitation and to the presence of pyochelin. Heinrichs and Poole (49, 50) used transcriptional fusions fptA-lacZ and pchR-lacZ to study the regulation of gene expression for fptA and pchR. Their findings can be summarized as follows: inactivation of pchR by insertion of an Ω cartridge (38), with terminations signals for transcription and translation in both orientations, led to a dramatic decrease in the expression of fptA. However, insertional inactivation of pchR in a pyochelin-deficient background restored fptA expression to levels found in the pyochelin-proficient PchR-deficient strain. Therefore, it is apparent that PchR must repress fptA expression in the absence of pyochelin. This was further proved by noticing that the cloned pchR gene caused a fivefold decrease in expression of the fptA-lacZ fusion in E. coli. As in the case of pvdS, pchR expression was also repressed by iron. The data from this work indicated that PchR functions as both activator and repressor in controlling expression of fptA and pchR (49, 50). It is noteworthy that two partially conserved heptameric repeat sequences, CGAGGAA and CGTGGAT, were found upstream of the fptA −35 region. These sequences are also found upstream of the autoregulated pchR gene, suggesting that these sequences could function in PchR binding. A model of the regulation by PchR and pyochelin of pyochelin biosynthetic genes is shown in Fig. 15.
FIG. 15.
Model of regulation of the expression of pyochelin biosynthesis and uptake genes in P. aeruginosa.
It is possible that an interaction between ferric pyochelin and FptA initiates a signal transduction cascade which can lead to the release of an effector, which interacts with PchR to afford either activation or repression. Alternatively, there could be a direct interaction between PchR and the terminal element in the cascade. The attractive feature in this model is that the siderophore does not have to be transported inside the cell for transduction of the signal of successful iron chelation by pyochelin and concomitant need to enhance expression of the fptA gene. It is likely that FptA senses the level of pyochelin in the culture and can communicate it to PchR directly. Alternatively, the signal could be transduced by influencing pchR expression directly.
Enzymology
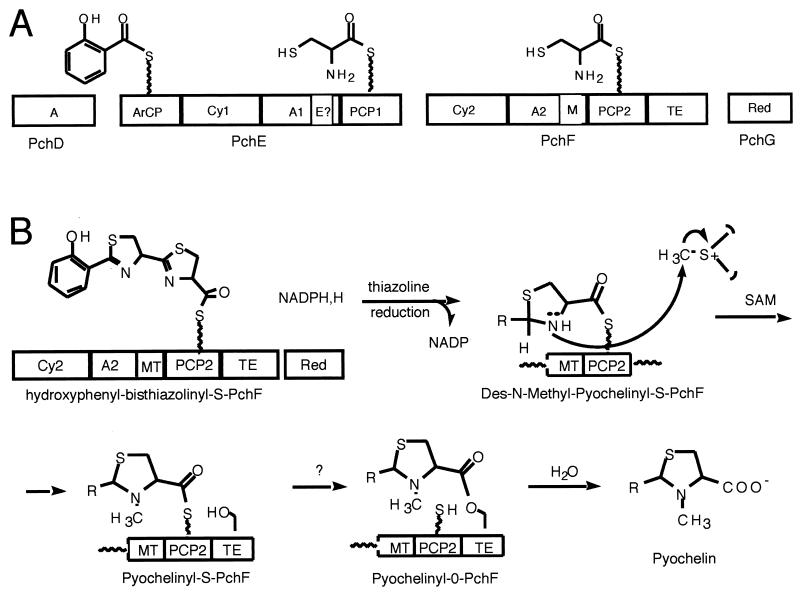
We focus here on the pyochelin system in which genetic analysis (87, 105) has indicated that the following pyochelin biosynthesis genes are necessary and sufficient for pyochelin formation: pchDEFG, which encode the products salicylate, ATP, two cysteines, NADPH, and S-adenosylmethionine (SAM) through to pyochelin. The steps involved in the pyochelin assembly line are shown in Fig. 16A and B. In this assembly line there are 10 domains rather than the 7 to make enterobactin and four notable chemical differences in the activity of constituent domains. First, the EntF C domain is replaced by Cy variants in both the PchE and PchF subunits such that the two cysteines activated by the PchE A domain and the PchF A domain become heterocyclized to a characteristic tandem. Second, there is a domain resembling a SAM-dependent methyltransferase (MT) embedded in the PchF subunit (Cy-A-MT-PCP-TE), and inspection of pyochelin suggests an N-methylation has occurred on a fully reduced thiazolidine ring (94). To convert a presumed initial tandem thiazoline-thiazoline to a thiazoline-thiazolidine ring system would require reduction of the C=N imine to the C—NH amine in pyochelin. The PchG subunit functions as such a thiazolinyl reductase (94). The fourth distinction from the Ent assembly line logic is that the PchF-TE is hydrolyzing rather than cyclizing, since pyochelin has a free COOH. An additional distinction is the use of salicylate as the aryl-N-cap rather than DHB. The PchAB genes are committed to salicylate biogenesis to provide sufficient monomer for siderophore assembly. When the growing chain gets to domain PCP2 in PchF, the third and most-downstream thiol way station, a hydroxyphenyl-thiazolinyl-thiazolinyl (HPTT)-S-enzyme has been constructed. This is released only very slowly by the TE domain. Instead, the HPTT-S-PchF acyl thioester enzyme lasts long enough to be acted on in trans by the PchG NADPH-dependent reductase, reducing the right-hand thiazolinyl ring to the fully reduced thiazolidinyl ring. In this oxidation state the ring nitrogen is more nucleophilic and is a substrate for methylation by the adjacent MT domain of PchF. The N-methylation converts a desmethyl-pyochelinyl-S-PchF to a pyochelinyl-S-PchF (94). It is presumed that now interdomain transfer of the mature Pch chain occurs, from pyochelinyl-S-PCP to pyochelinyl-O-TE, and that the TE domain is accessible to solvent water and hydrolytic release of mature pyochelin ensues. Again the four proteins of the Pch assembly line, like those of the Ent assembly line, seem to display rapid association and dissociation behavior, without a long-lived four protein complex.
The coordination site of pyochelin to ferric iron is not yet fully described but probably involves the phenolic-OH, the thiazoline, and perhaps the thiazolidine nitrogens and may, like anguibactin complexes (see Fig. 2), show a (pyochelin)2-Fe3+ stoichiometry.
YERSINIABACTIN
Introduction
The genus Yersinia contains at least 11 species, 3 of which are enteropathogenic for humans. Y. pestis is the agent of bubonic plague, while Y. enterocolitica causes a broad range of diseases ranging from acute bowel disease to extraintestinal manifestations such as reactive arthritis and uveitis. Plague, caused by Y. pestis, is a zoonotic disease affecting primarily rodents, and transmission between these hosts occurs via fleas. After a blood meal, Y. pestis grows in the digestive apparatus of the insect, blocking the proventricular valve between the midgut and esophagus. Regurgitation of infected blood can then occur into the mammal, and subsequent proliferation of Y. pestis cells occurs through the lymphatic system to the regional lymph nodes where they can multiply. The swollen lymph nodes or buboes define very clearly the name of the disease as bubonic pest. Spreading of the organism can now continue, reaching eventually quite high numbers in the liver, spleen, and other internal organs and finally resulting in septicemia. The cycle of this disease is completed when a flea from a host carries a bacteremic blood meal. Pneumonic plague due to infection of the lungs can lead to aerobic spreading of the disease in humans (83). A number of iron uptake systems have been identified in yersiniae, such as those involved in inorganic and heme transport. In toto, eight inorganic iron and two heme/hemoprotein transport systems have been described. In addition the siderophore yersiniabactin, which acts as a virulence factor for pathogenic yersinia strains growing in mice, is part of another iron transport system (83). Expression of pathogenicity by Yersinia requires the presence of a 70-kb pYV virulence plasmid that is found in high- and low-level-pathogenic strains (11, 83). Differences in mouse virulence seem to be chromosomally determined. Highly pathogenic strains possess a chromosomal cluster of iron-regulated genes designated the high-pathogenicity island (HPI). This island is absent in low-level-pathogenic or nonpathogenic strains and was found to be unstable in Yersinia strains. Its loss leads to a marked reduction in mouse virulence (11, 36, 83).
Genetics
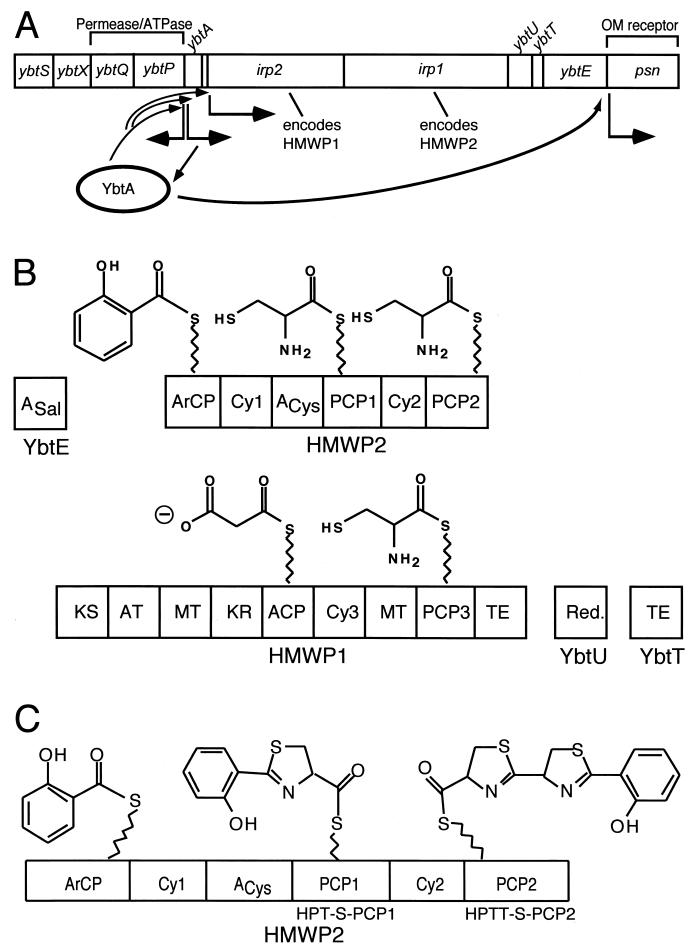
Figure 1 shows that the siderophore yersiniabactin contains phenolate, thiazoline, and thiazolidine rings. Yersiniabactin has an affinity for ferric iron of ∼4 × 1036 1/M and shows high structural similarity to siderophores produced by P. aeruginosa pyochelin and V. anguillarum anguibactin (11, 41-43, 45, 55, 56, 83, 110, 114, 115). The yersiniabactin biosynthetic, transport, and regulatory genes are encoded within an HPI possessed by virulent isolates of Y. pestis, Y. enterocolitica, and Yersinia pseudotuberculosis, as well as several types of pathogenic E. coli (18, 83). Yersiniabactin biosynthetic genes are arranged in four operons (Fig. 17A). These genes encode the following proteins: YbtE, high-molecular-weight proteins (HMWP) 1 and 2, YbtU, and YbtT. The pathogenicity island of Y. pestis is about 35 kb and is located within the 102-kb pgm locus, which is subject to high-frequency deletion (11, 18, 36, 41, 43, 83). The yersiniabactin system genes present in Y. pestis share almost 100% sequence identity with those of Y. enterocolitica (83). Fur boxes are found upstream of genes located somewhere else in the chromosome, such as yfeE, feoAB, and hasR, that are involved in other iron metabolic processes; expression of yfeE is not iron or Fur-regulated (S. W. Bearden and R. D. Perry, unpublished observations [cited in reference 83]). YbtA is an AraC-type regulator that activates transcription of the other ybt operons.
FIG. 17.
Genetics and enzymology of yersiniabactin biosynthesis in Y. pestis. (A) Scheme of the yersiniabactin biosynthesis and transport genes. (B and C) Schemes of the synthetases encoded by the genes shown in panel A and initiation steps in yersiniabactin biosynthesis.
Two proteins encoded by iron-repressible genes have been detected only in highly pathogenic Yersinia strains, being putatively located on the HPI: HMWP1 (260 kDa, encoded by irp1) and HMWP2 (190 kDa, encoded by irp2) (83). Inactivation of irp2 in Y. pseudotuberculosis results in a considerable reduction of mouse virulence (83). These proteins are important for siderophore yersiniabactin. The receptor of yersiniabactin, FyuA (so named for ferric yersiniabactin uptake), is a receptor with dual function: it is a receptor of the siderophore and a receptor of Y. pestis bacteriocin pesticin. Thus, highly pathogenic strains are pesticin sensitive (83). In Y. pestis, the fyuA gene, the irp2 gene, and the hms locus (encoding hemin storage) are located on a 102-kb fragment designated the pgm (pigmentation) locus. This fragment is flanked by insertion sequence element IS100 (83), which might be the leading cause of frequent deletions of the pgm locus. ybtA, a gene encoding a protein belonging to the AraC family of transcriptional regulators, was recently detected upstream the irp2 gene in Y. pestis. YbtA is believed to be a transcriptional activator of the yersiniabactin receptor and of the siderophore biosynthetic genes (83). An approximately 22-kb region of the pgm of Y. pestis encodes several iron-regulated proteins. This fragment may contain additional irp genes involved in siderophore synthesis, including irp1 (encoding HMWP1) (11, 83).
The ability to synthesize and take up the Yersinia siderophore yersiniabactin is a hallmark of the highly pathogenic Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica 1B. Four genes, irp1, irp3, irp4, and irp5, have been identified on a 13-kb chromosomal DNA fragment of Y. enterocolitica O8, WA-314. These genes, together with irp2, constitute the yersiniabactin biosynthetic gene cluster. The irp1 gene encodes the HMWP1 polypeptide with a predicted mass of 384.6 kDa. The first 3,000 bp of irp1 show similarity to the corresponding regions of the PKS genes of B. subtilis and Streptomyces antibioticus. The remaining part of irp1 is most similar to irp2, encoding HMWP2, which might be the reason for immunological cross-reactivity of the two polypeptides. Irp4 was found to have 41.7% similarity to TE-like protein of the anguibactin biosynthetic genes of V. anguillarum. Irp5 shows 41% similarity to EntE, the 2,3-dihydroxybenzoic acid-activating enzyme utilized in enterobactin synthesis of E. coli. Irp4 and Irp5 are nearly identical to YbtT and YbtE, recently identified in Y. pestis. irp3 has no similarity to any known gene. Inactivation of either irp1 or irp2 affects yersiniabactin synthesis. Mutations in irp1 or fyuA (encoding the yersiniabactin/pesticin receptor) result in downregulation of irp2 that can be upregulated by the addition of yersiniabactin while upregulation was achieved specifically by addition of yersiniabactin, which also suggesting autoregulation of genes involved in synthesis and uptake of yersiniabactin.
YbtA activates transcription of psn, ybtPQXS, HMWP1, HMWP2, and YbtE genes, possibly by activating transcription of the irp2 irp1 ybtUTE operon (Fig. 17A), while repressing transcription from its own promoter. YbtA binding sites are hypothesized to be present within the promoters of psn, ybtA, ybtPQXS, and irp2 irp1 ybtUTE as repeats, based on the fact that mutation of the repeat in psn caused loss of transcriptional activation (83). A psn mutation has no effect on the regulation activity of YbtA.
BLAST searches of the Y. pestis KIM10+ genome (UW Genome Project) have identified other potential regulatory genes: BarA (also termed AirS, for “attachment and iron regulation sensor”) in uropathogenic E. coli is a fused function sensor-response regulator with similarities to two-component systems. A mutation in barA/airS causes loss of siderophore production and expression of iron-repressible outer membrane proteins. Y. pestis KIM10+ has a homologue of barA/airS. Homologues of the S. enterica pmrAB genes are also present in Y. pestis KIM10+. This is of interest because PmrA and PmrB are the response regulator and sensor, respectively, of a two-component system in S. enterica that responds to extracellular iron. PmrA-PmrB appear to control genes required to avoid iron toxicity at high iron concentration.
Enzymology
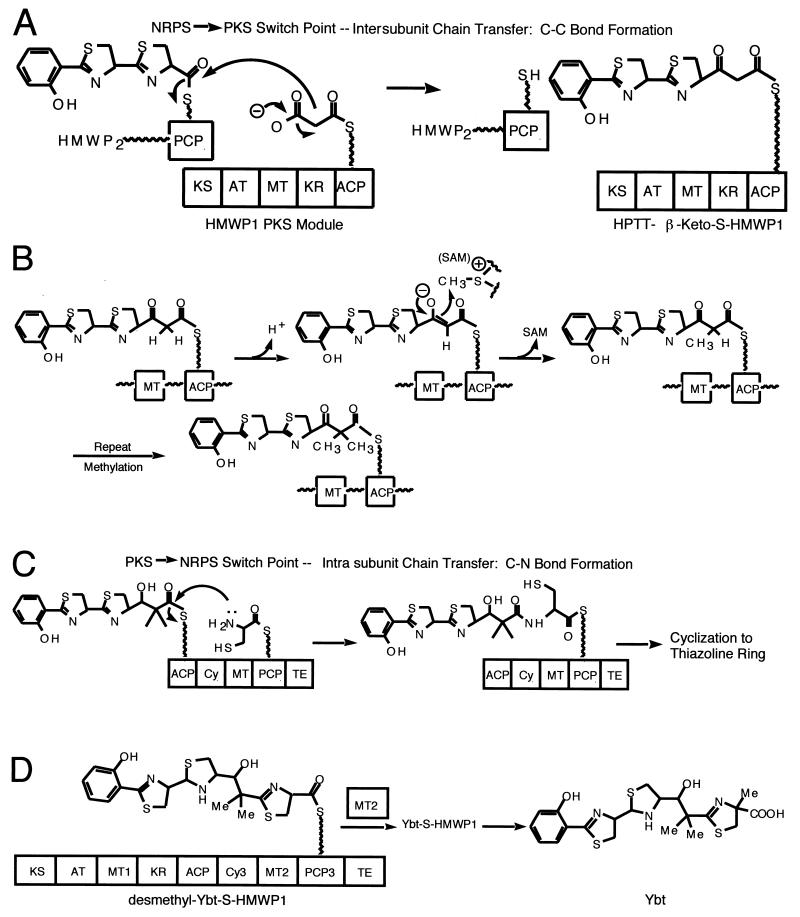
The yersiniabactin biosynthetic complex contains five proteins, YbtE, HMWP1 and HMWP2, and YbtU and YbtT (Figs. 17B and C). Comparison of the yersiniabactin and pyochelin structures shows an almost identical three ring (HPTT) moiety. This is essentially the full pyochelin molecule, while yersiniabactin has an additional portion, predicted to derive from a malonyl moiety and then a third five ring heterocycle. Where pyochelin has one N-methyl group, yersiniabactin has three C-methyl groups derived from SAM (41). Yersiniabactin is synthesized by a mixed PKS-NRPS strategy that features modular assembly of the siderophore from salicylate, a group derived from malonyl coenzyme A (malonyl-CoA), three molecules of cysteine, and three methyl groups. The yersiniabactin siderophore must therefore be assembled by a mixed system. The assembly lines for pyochelin and yersiniabactin are thus predicted to have equivalent domains in the same order. The first, the YbtE component, is, as anticipated, a salicyl-AMP ligase. The second domain should be an ArCP for salicyl tethering and this is the first domain of HMWP2. In total the 230-kDaprotein HMWP2 has six predicted domains: ArCP-Cy1-A-PCP1-Cy2-PCP2. Once primed, the three carrier proteins should be able to load salicyl, cysteinyl, and cysteinyl thioesters as shown (Fig. 17C), and the two Cy domains generate a Sal-Cys and then a Sal-Cys-Cys amide linkage, respectively. Because they are Cy domains, not just C domains, they should also be able to heterocyclize the Sal-Cys-S-PCP1 acyl enzyme intermediate to HPT-S-PCP1 and then the HPT-Cys-S-PCP2 to the HPTT-S-PCP2 enzyme (Fig. 17A and B) (76). Indeed all the predicted acyl- and aminoacyl-S-enzyme intermediates, including HPT and HPTT-S-enzyme forms have been detected by thiolytic release and HPLC and MS analysis (54, 55, 114). S to A point mutations in the ArCP, PCP1, and PCP2 carrier protein domains have been especially useful in leaving a given carrier protein domain in the inactive apo form and allowing acyl enzyme intermediates to build up at the holo carrier protein way stations that are upstream (55, 56).
Figure 18A shows that once the HPTT chain has reached the most-downstream carrier site on HMWP2, the chain is presumed to be transferred to the nine-domain, 350-kDa HMWP1 subunit and switch from NRPS-type assembly line modules to a PKS module that comprises the first five domains and 1,896 residues of HMWP1 (KS-AT-MT-KR-ACP), with predicted ketosynthase (KS), acyl transferase (AT), C-MT, ketoreductase (KR), and ACP domains. Four of the five domains in the PKS module are catalytic; the fifth, the ACP, is the first carrier protein way station on HMWP1. The intersubunit chain transfer between HMWP2 and HMWP1 would then be an NRPS-PKS switch point and generate in the elongating chain, a new C—C bond rather than the C—N bonds in NRPS modules. The transfer of the HPPT chain onto a decarboxylating malonyl-S-ACP in the PKS module would yield an HPTT-β-ketoacyl-S-ACP covalent acyl enzyme on HMWP1.
FIG. 18.
Enzymology of yersiniabactin biosynthesis in Y. pestis. (A) NRPS-PKS switch point; (B) methylation; (C) PKS-NRPS switch point; (D) methylation and termination.
The malonyl group will have been installed on the holo HS-ACP domain by catalytic malonyl transacylase activity of the AT domain with malonyl-CoA as substrate, a typical PKS module loading function. Three additional chemical transformations are proposed to occur to this acyl-S-ACP before it is translocated further downstream. One is thiazoline to thiazolidine reduction, analogous to the Pch biosynthetic pathway and there is a PchG homolog, YbtU (84), proposed by the Walsh laboratory to carry out the same thiazoline C=N to thiazolidine C—NH reduction. Then there are two C-methylations to be effected by the MT domain of the PKS module. It is very likely that these occur on the β-ketoacyl-S-ACP oxidation state since the required carbanion for tandem C-methylation would be kinetically and thermodynamically accessible by low energy enolization.
The last chemical step likely to be performed by the PKS module of HMWP1 is KR-mediated reduction of the dimethyl-β-ketoacyl-S-ACP to the β-hydroxyl group found in yersiniabactin (Fig. 18B). At this point all five domains of the PKS module will have carried out their ascribed functions and the siderophore growing chain can move to the last module, the last four domains of HMWP1 (residues 1896 to 3163), which comprise Cy3-MT-PCP3-TE (110). Inspection of the remaining tasks in siderophore assembly indicate addition of one more cysteine (Cys-S-PCP3), its heterocyclization (Cy3), C-methylation of the thiazoline (MT), and then hydrolytic release (TE). As shown in Fig. 18C the translocation of the chain from the ACP carrier protein site to the downstream PCP3 carrier protein site represents a second switch point of assembly line logic, this one from PKS back to NRPS, and the act of translocation is C-N bond formation by the Cy3 domain using the amino group of Cys-S-PCP3. The cysteinyl group is loaded onto PCP3 by the only A domain in the 16 domain, two subunit HMWP2-HMWP1 complex: the A domain back in the HMWP2 complex (Fig. 18D). When the chain has been translocated to PCPs and the third cysteinyl residue incorporated into yersiniabactin heterocyclized by Cy3 action, the penultimate intermediate is presumably the desmethyl yersiniabactin-S-PCP3 acyl enzyme. The Cα-H of this thiazolinyl-S-PCP3 is kinetically acidic, the Cα carbanion is readily accessible and is the presumed attacking species on the methyl group of SAM, catalyzed by the MT domain to yield the Ybt-S-PCP3 acyl enzyme, which is the mature acyl chain on the most downstream carrier protein domain (Fig. 18D).
All that is left is the chain termination step, in this case an intermolecular hydrolysis, effected by yersiniabactin chain transfer from PCP3 to the TE domain, where hydrolysis is rapid (114).
The final feature of the 16-domain yersiniabactin synthetase assembly line worth mention is indication of an editing function for hydrolytic removal of misacylated chains from the ACP and PCP3 carrier domains of the HMWP1 subunit. In a situation where a noncognate acyl-CoA, such as acetyl-CoA or benzoyl-CoA, is presented to the purified holo HMWP1 subunit in place of the cognate malonyl-CoA, the AT domain will misload. AT domains of PKS and FAS modules autoacylate on an active site serine and then transacylate to the HS-ACP partner domain. In place of the normal malonyl-S-ACP, misacylation will yield acetyl-S-ACP or benzoyl-S-ACP, and the assembly line will be blocked by a nonelongatable acyl chain. The HMWP1 subunit can edit the incorrect acyl chains to remove the roadblock by translocation of the acyl group from acyl-ACP down to HS-PCP3 and then to TE for hydrolysis. The net rates of hydrolysis were 172:50:1 for acetyl-:benzoyl-:malonyl-CoA, indication of a selective hydrolytic editing process that removes misacylated groups from the assembly line via a cascade of four acyl enzyme intermediates in the proof reading process (114).
MYCOBACTIN
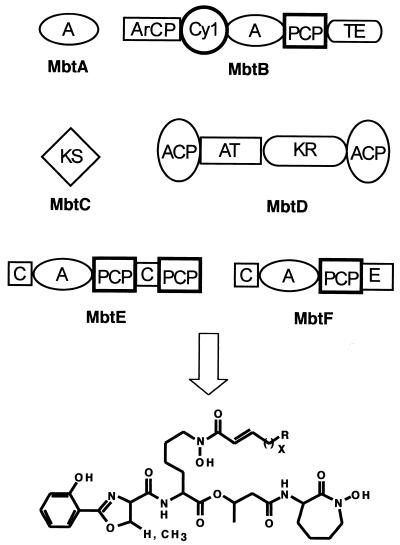
Introduction
Despite the availability of effective short-course chemotherapy (DOTS) and the Mycobacterium bovis Bacille Calmette-Guérin (BCG) vaccine, the tubercle bacillus continues to claim more lives than any other single infectious agent (90, 124). Recent years have seen increased incidence of tuberculosis in both developing and industrialized countries, the widespread emergence of drug-resistant strains,and a deadly synergy with the human immunodeficiency virus. The genus Mycobacterium includes, in addition to M. tuberculosis, the causative agent of tuberculosis, and Mycobacterium leprae, the causative agent of leprosy, other opportunistic pathogens that can cause disease in humans and animals. Members of the mycobacteria possess extra- and intracellular siderophores. Two secreted forms of exochelins, exochelin MS from Mycobacterium smegmatis and exochelin MN from Mycobacterium neoaurum, have been characterized, the first being a pentapeptide with three ornithinyl hydroxamates and the second being a hexapeptide, also with three ornithines, to reflect a short term carboxylic acid chain that in the mycobactin group is a longer acyl chain devoid of the terminal COOH moiety. Figure 19 shows structures of carboxymycobactins, exochelins, and mycobactin. The carboxymycobactins and exochelins are water soluble while the mycobactins are lipid soluble and thought to remain in mycobacterial cells (90). Solubilization of the external iron is effected by the extracellular siderophores, while mycobactin may play a role in providing a short-term iron storage before iron reaches the cytoplasm because of the presence of a thick lipophilic cell envelope (90, 124). In this review we focus on mycobactin, whose structures depend on the producing species, although they all share the same core nucleus and the property of being lipid soluble, intracellular siderophores with a mixed-type ferric iron chelation center (see Fig. 21).
FIG. 19.
Mycobacterial siderophores.
FIG. 21.
Mycobactin assembly line in M. tuberculosis. For mycobactin T, R is methyl (Me) and X is 14 to 17. For carboxymycobactins, R is COOH, COOMe and X is 1 to 5.
Genetics
An integrated map of the 4.4-Mb circular chromosome of this slow-growing pathogen has been established, and ordered libraries of cosmids and bacterial artificial chromosomes are available (32). The sequencing of the genome of M. tuberculosis revealed a cluster of 10 genes, mbtA to -J (32), that seemed likely to encode the enzymes for assembly of mycobactin and transport of iron (Fig. 20). In particular the six genes mbtA to mbtF could be analyzed as a 20-domain assembly line that has the mixed NRPS/PKS/NRPS order that could activate and elongate the monomers needed for mycobactin (86), including two polyketide synthetases (MbtC and MbtD) and three peptide synthetases (MbtB, MbtE, and MbtF); an isochorismate synthetase, MbtI, which provides salicylic acid; and an adenylating enzyme, MbtA, intervening in the activation of salicylic acid to be attached to MbtB, an aryl carrier protein. Furthermore the biosynthetic process also involves the activation of serine (or threonine) by means of the activity of a peptidyl carrier protein. The mbt operon may be responsible for the biosynthesis of the mycobacterial siderophores. The presence of only one nonribosomal peptide-synthesis system indicates that this pathway may generate both siderophores and that subsequent modification of a single-amino group of one lysine residue may account for the different physical properties and function of the siderophores. Using gene replacement techniques through homologous recombination, the mbtB was deleted and replaced with a hygromycin resistance cassette in the virulent strain of M. tuberculosis H37Rv. This mutant is restricted for growth under iron-limiting conditions, while it grows normally under iron-rich conditions. In addition to being defective in the production of all salicylate-derived products, the mutant was also impaired for growth in macrophage-like THP-1 cells, suggesting that siderophore production is required for virulence of M. tuberculosis. Recently, it was reported the cloning and characterization of a phosphopantetheinyl transferase from M. tuberculosis that modifies ArCP and PCP carrier protein domains in the mycobactin biosynthetic pathway (86).
FIG. 20.
Scheme of the mycobactin biosynthesis and transport genes.
Enzymology
As mentioned before the genes for exochelin MS have been identified (142, 143) but there appears to be one module too many and no in vitro characterization has yet occurred to resolve this issue. In the case of mycobactin, Fig. 21 shows that MbtA is an A domain subunit that would make salicyl-AMP. MbtB has two carrier protein domains, one for salicyl and one for serine/threonine covalent tethering, and a Cy domain to condense and cyclize to an HPO-S-MbtB acyl enzyme. The next protein in the assembly line should activate a lysine monomer, and either MbtE or MbtF could fulfill this NRPS-type elongation role. The next module should be a PKS module to provide a beta-hydroxy acyl moiety, and MbtD fits this requirement, with two ACP domains. The remaining of the two proteins MbtE and MbtF should activate the remaining lysine residue. There is an ORF that has homologies to lysine- and ornithine-N-oxygenases, for hydroxylation of both lysine residues. The cyclization to the seven-membered ring may be catalyzed by a variant TE or C domain. Finally the timing of acylation of the first N-OH-lysine is not yet clear nor is the specific AT partner enzyme identified.
SUMMARY AND CONCLUDING REMARKS
In those microorganisms in which it has been examined, the regulatory logic of genes involved in the synthesis of bacterial siderophores involves the universal repressor Fur, which acts together with the concentration of iron in the cell cytosol as a negative regulator. However, in some bacteria such as Yersinia sp., Pseudomonas sp., V. anguillarum, and other microorganisms, in addition to the Fur-mediated negative regulation there is a concurrent positive regulation of iron transport and siderophore biosynthetic genes that occurs under conditions of iron limitation. Thus, the chief regulatory logic of gene expression of iron transport-siderophore biosynthesis genes is microorganism dependent and could be simply negative control or a combination of both negative and positive mechanisms. The only operating regulatory mechanism in the case of the enterobactin and vibriobactin systems appears negative and mediated by Fur. However, the three divergent operons in the enterobactin system also show another fine mechanism of control that is dependent on the separation of the divergent promoters, what occurs at one promoter may affect the events occurring at the other. In the case of fepA-fes, the competition between the positive action of RNA polymerase and the negative action of Fur plays an important role on whether a gene will be turned on or off. RNA polymerase binding at the strongest fes promoter, which is very close to the weaker fepA promoter, does not allow the binding of Fur to the shared Fur box, and therefore the weaker fepA promoter is also expressed although always at a lower level than the expression of the strongest promoter.
Despite these regulatory differences, the mechanisms of siderophore all follow the same fundamental enzymatic logic: a series of elongating acyl-S-enzyme intermediates on multimodular protein assembly lines. These NRPS are the prokaryotic enzyme machinery that also turn out peptide antibiotics such as the precursors of penicillins, vancomycins, and pristinamycins. Each module in the NRPS assembly lines has a carrier protein domain primed with a phosphopantetheine arm that provides the terminal SH for tethering the growing siderophore/antibiotic chain. The other domains in each module are catalytic, carrying out local enzyme chemistry on the acyl chain before it is translocated to the next downstream module.
The monomer units selected for incorporation into the growing siderophore chain can vary depending on whether the monomer is utilized for chain initiation, chain elongation, or chain termination. Some monomers, such as salicylate and dihydroxybenzoate, are synthesized in the bacterium, and their biosynthetic genes are clustered and/or coregulated with the NRPS assembly line subunit genes. Other monomers are the amino acid constituents of the siderophore backbone [serine in enterobactin, glycine and threonine in (DHB-Gly-Thr)3, threonine in vibriobactin, cysteine in pyochelin, yersiniabactin, and anguibactin, and lysine in mycobactin] and are readily available in the producer cell.
During siderophore chain initiation and elongation steps the aryl acid and amino acid monomers get installed as thioesters on the thiol of the pantetheine arm of each module, with selection of the particular monomer by the A domain in each module. While this process serves to import the phenol and catechol chelating groups into the siderophore backbone, the other two chelating moieties, the N-hydroxy and the thiazoline/oxazoline moieties, are thought to be created from the amino acid moieties during chain elongation. This has been well established in pyochelin and yersiniabactin biogenesis for conversion of cysteinyl moieties in the elongating chain into thiazoline, and thiazolidine, rings. It is possible that lysine N6 hydroxylation occurs during chain elongation or instead on the free amino acid that is then selected and incorporated during chain growth, but that has not been established explicitly yet in either the anguibactin or mycobactin systems. Additional enzymatic tailoring steps can occur during siderophore chain elongation to modify functional properties of these iron-chelating natural products. The N-acylation of the mycobactins to increase their membrane solubility is one such modification, as is the thiazoline reduction and N-methylation that occurs in pyochelin chain maturation.
Siderophore chain termination occurs when the full-length acyl chain has reached the most-downstream carrier protein domain in the NRPS assembly line and is transferred to the chain-terminating TE domain. Hydrolysis can occur, producing the free carboxylate C terminus in siderophores such as pyochelin, vibriobactin, and mycobactin. Alternatively the acyl-O-TE intermediate can be sequestered from bulk solvent and be captured intramolecularly by a nucleophile in the siderophore chain. This is a serine side chain hydroxyl in the cyclotrimerization release in enterobactin, and there is corresponding capture by the threonine hydroxyl in the bacillobactin cyclic hexapeptide formation. A third alternative is transfer of the siderophore chain to an external nucleophile other than water. This can be to specific amine cosubstrates to make an amide linkage. The cosubstrate amine is histamine in anguibactin chain termination and norspermidine in vibriobactin chain termination, catalyzed by C-terminal condensation domains rather than TE domains in the anguibactin and vibrobactin synthetase assembly lines. The histamine may be hydroxylated before or after capture of the siderophore chain as that enzymology is not yet determined. In the vibriobactin case, the cosubstrate amine is DHB-N1-norspermidine and it is acylated twice at N5 and N9.
A substantial variety of siderophore structures are thus produced from similar NRPS assembly lines. Variation can come in the choice of phenolic acid selected as N-cap, the tailoring of amino acid residues during chain elongation, and the mode of chain termination and the nature of the capturing nucleophile of the siderophore acyl chain being released. The specific parts that get assembled and tailored in a given bacterium may reflect a combination of siderophore biosynthetic gene cluster inventory available, including the tailoring enzymes for heterocyclization or N-hydroxylation. This modular assembly logic can account for essentially all known siderophores, including several not explicitly discussed in this review, such as aerobactin, acinetobactin, and ornibactin.
Figure 22 summarizes the domains found in the NRPS that intervene in siderophore biosynthesis in the various bacteria discussed in this review. The comparison of these NRPS demonstrates very dramatically that the modular logic of siderophore NRPS must have been of the utmost importance in the evolution of these systems, especially in organisms that can obtain genetic information by conjugation and other DNA transfer mechanisms, including those mediated by conjugative transposons, and thus obtain horizontally additional genes for specific functions during the creation of new siderophore biosynthetic pathways. The ability to mix and match domains within modules and to swap modules themselves is likely to be ongoing in natural combinatorial biosynthesis. This NRPS evolution will try out new combinations of chain initiation, elongation and tailoring, and termination steps to create new variants of iron chelating siderophores that fit a particular niche for the producer bacterium.
FIG. 22.
Comparison of the domains of siderophore-NRPS from bacterial pathogens and E. coli.
The reader now must be wondering why is it that tris catechol arrays such as the versions found in enterobactin, bacillobactin, and vibriobactin, which provide very high affinity Fe(III) chelation, are not used universally in siderophore assembly. It is possible that the thiazoline/oxazoline and the N-hydroxy-Orn and N-hydroxy-Lys chelating groups offer some balance between lower affinity and trade off in energetics of biosynthesis, export, reuptake, and iron release. A curious finding in certain strains of E. coli and other enterobacteria causing extraintestinal infections brings into the picture factors that may have served to select against enterobactin-like siderophores during evolution. In addition to functional enterobactin biosynthesis and transport system genes, many virulent strains of enterobacteria responsible for extraintestinal infections also harbor the genes (chromosomal or plasmid-encoded) for the synthesis of the hydroxamate siderophore aerobactin and its cognate transport system for complexes with ferric iron (134). Strains in which the aerobactin system is modified to eliminate the synthesis of aerobactin and/or its transport system become avirulent, although they are still proficient in the production of enterobactin and its transport system. Therefore aerobactin, the siderophore with the dramatically lower affinity for iron (constant of association for iron is 1023 1/M) is essential for virulence while enterobactin, with the higher constant of association for iron (about 1052 1/M) does not play an important role in the ability of these bacteria to cause disease. Of course these values are derived for completely protonated ligands so that they have little relevance to physiological conditions; nevertheless, it is clear that aerobactin is thermodynamically inferior to enterobactin and even to transferrin, since the association constant of ferric transferrin is about 1036 1/M (27, 78, 79, 91). An investigation of the properties of these two siderophores led us first to an important difference: upon uptake, enterobactin is either enzymatically destroyed or esterified during intracellular release of the ferric ions and therefore is wasted, while iron is more readily released from aerobactin, which is then recycled to take up more iron (9), conferring a selective advantage for aerobactin. Furthermore, the relative low water solubility and chemical instability of enterobactin in serum could also favor aerobactin-mediated transport. It was also reported that aerobactin is in some way able to compartmentalize iron (134), suggesting that iron complexed to aerobactin may be channeled directly to be used for bacterial growth, while in the case of enterobactin, the assimilated iron appears to be contributed to an intracellular pool and the rate of its withdrawal for growth is probably concentration dependent (134). There are also kinetic and thermodynamic issues concerning the interaction of either of these two siderophores when they scavenge iron. Konopka et al. (62) demonstrated that aerobactin is more effective than enterobactin at equal molarity in delivering iron from transferrin to bacterial cells in human serum, and Williams and Carbonetti (134) found that aerobactin is more effective than enterobactin at complexing iron at very low concentrations of siderophore, a property which would confer advantage in the dynamic and fluid environments of the bloodstream and in the urinary tract, where bacterial extracellular products are continually diluted. In agreement with Konopka et al. (62), these investigators also reported that the presence of serum in the growth medium markedly reduced the effectiveness of enterobactin as a siderophore but had no significant influence on aerobactin-mediated iron assimilation. Other findings indicate that the vertebrate host, being exposed to the E. coli present in the normal flora, has developed antibodies against the enterobactin receptor, or even enterobactin itself, rendering it, or its receptor, inactive to interact (12, 96). Also, serum albumin has been reported to bind enterobactin, decreasing its effective concentration in serum and blocking enterobactin-mediated mobilization of transferrin iron. Therefore, the intrinsic relative inefficiency of enterobactin is likely to be exacerbated within an infected animal body by siderophore inactivation, due to its binding or that of its receptor to serum proteins. Another factor that may play a role in selecting aerobactin over enterobactin in these bacteria could also be the preferential expression of aerobactin biosynthetic genes under iron stress, possibly due to the differential expression of this operon compared to the enterobactin regulon.
It is therefore tempting to speculate that the disadvantages of enterobactin over aerobactin found in extraintestinal infections may also be found in other natural ecological niches: enterobactin and, possibly, other chemically related siderophores may not be adapted to the selective pressures encountered by most sideropohore-producing organisms, leading to the evolution of a whole array of microorganism- and/or niche-specific siderophore classes.
Acknowledgments
The work carried out in our laboratories and reported in this review was supported by NIH grants GM64600 and AI 19018 to J.H.C. and grants AI42738 and GM20011 to C.T.W.
REFERENCES
- 1.Actis, L. A., M. E. Tolmasky, and J. H. Crosa. 1999. Vibriosis, p. 523-557. In P. T. K. Woo and D. W. Bruno (ed.), Fish diseases and disorders, vol. 3. Viral, bacterial and fungal infections. Cab International Publishing, Wallingford, United Kingdom. [Google Scholar]
- 2.Actis, L. A., M. E. Tolmasky, D. H. Farrell, and J. H. Crosa. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 263:2853-2860. [PubMed] [Google Scholar]
- 3.Actis, L. A., W. Fish, J. H. Crosa, K. Kellerman, S. R. Ellenberger, F. M. Jauser, and J. Sanders-Loehr. 1986. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Actis, L. A., S. Potter, and J. H. Crosa. 1985. Iron regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 161:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1995. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17:197-204. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal, R., P. Caffrey, P. F. Leadley, C. J. Smith, and J. Staunton. 1995. The thioesterase of the erythromycin-producing polyketide synthase: mechanistic studies in vitro to investigate its mode of action and substrate specificity. J. Chem. Soc. Chem. Commun. 1519-1520.
- 7.Ankenbauer, R. G. 1992. Cloning of the outer membrane high-affinity Fe(III)-pyochelin receptor of Pseudomonas aeruginosa. J. Bacteriol. 174:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong, S. K., G. S. Pettis, L. J. Forrester, and M. A. McIntosh. 1989. The Escherichia coli enterobactin biosynthesis gene. entD: nucleotide sequence and membrane localization of its protein product. Mol. Microbiol. 3:757-766. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., C. Brazel-Faisst, and R. Schneider. 1984. Growth stimulation of Escherichia coli in serum by iron(III) aerobactin. Recycling of aerobactin. FEMS Microbiol. Lett. 21:99-103. [Google Scholar]
- 10.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 24104-24109. [DOI] [PubMed]
- 11.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 12.Bullen, J. J., and E. Griffiths. 1999. Iron binding proteins and host defense, p. 327-368. In J. J. Bullen and E. Griffiths (ed.), Iron and infection, 2nd ed. John Wiley and Sons Ltd., New York, N.Y.
- 13.Butterton, J. R., and S. B. Calderwood. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butterton, J. R., M. H. Choi, P. I. Watnick, P. A. Carroll, and S. B. Calderwood. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J. Bacteriol. 182:1731-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butterton, J. R., J. A. Stoebner, S. M. Payne, and S. B. Calderwood. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3729-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cane, D. E., C. Khosla, and C. Walsh. 1998. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282:63-68. [DOI] [PubMed] [Google Scholar]
- 17.Canestrini, G. 1893. La malattia dominante delle anguille. Atti Intituto Veneto Sci. Lett. Ed. Arti. 7:809-814. [Google Scholar]
- 18.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 19.Chai, S., T. Welch, and J. H. Crosa. 1998. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J. Biol. Chem. 273:33841-33847. [DOI] [PubMed] [Google Scholar]
- 20.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224 [DOI] [PubMed] [Google Scholar]
- 21.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:1885-1891. [DOI] [PubMed] [Google Scholar]
- 22.Chen, Q., A. M. Wertheimer, M. E. Tolmasky, and J. H. Crosa. 1996. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol. Microbiol. 22:127-134. [DOI] [PubMed] [Google Scholar]
- 23.Christoffersen, C., A. T. J. Brickman, I. Hook-Barnard, and M. A. McIntosh. 2001. Regulatory architecture of the iron-regulated fepD-ybdA bidirectional promoter region in Escherichia coli. J. Bacteriol. 183:2059-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosa, J. H. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature 284:566-568. [DOI] [PubMed] [Google Scholar]
- 25.Crosa, J. H. 1989. Genetics and molecular biology of siderophore mediated iron transport in bacteria. Microbiol. Rev. 53:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 67:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crosa, J. H. 1999. Molecular genetics of iron transport as a component of bacterial virulence, p. 255-288. In J. J. Bullen and E. Griffiths (ed.), Iron and infection, 2nd ed. John Wiley and Sons Ltd., New York, N.Y.
- 28.Crosa, J. H. 1984. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu. Rev. Microbiol. 38:69-89. [DOI] [PubMed] [Google Scholar]
- 29.Crosa, J. H., M. H. Schiewe, and S. Falkow. 1977. Evidence for plasmid contribution to the virulence of the fish pathogen Vibrio anguillarum. Infect. Immun. 27:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crosa, J. H., L. Hodges, and M. H. Schiewe. 1980. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect. Immun. 27:897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean, C. R., S. Neshat, and K. Poole. 1996. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeVoss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Earhart, C. F. 1996. Uptake and metabolism of iron and molybdenum, p. 1075-1090. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 35.Farrell, D. H., P. Mikesell, L. A. Actis, and J. H. Crosa. 1990. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P 22 cro and regulation by iron. Gene 86:45-51. [DOI] [PubMed] [Google Scholar]
- 36.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 37.Folschweiller, N., I. J. Schalk, H. Celia, B. Kieffer, M. A. Abdallah, and F. Pattus. 2000. The pyoverdin receptor FpvA, a TonB-dependent receptor involved in iron uptake by Pseudomonas aeruginosa. Mol. Membr. Biol. 17:123-133. [DOI] [PubMed] [Google Scholar]
- 38.Frey, J., and H. M. Krisch. 1985. Ω mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene 36:143-150. [DOI] [PubMed] [Google Scholar]
- 39.Gehring, A. M., K. A. Bradley, and C. T. Walsh. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495-8503. [DOI] [PubMed] [Google Scholar]
- 40.Gehring, A. M., I. Mori, and C. Walsh. 1998. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37:2648-2659. [DOI] [PubMed] [Google Scholar]
- 41.Gehring, A. M., E. DeMoll, J. D. Fetherstone, I. Mori, G. F. Mayhew, F. Blattner, C. Walsh, and R. Perry. 1998. Iron acquisition in plague: molecular logic in the biosynthesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 42.Gehring, A. M., I. Mori, R. D. Perry, and C. T. Walsh. 1998. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron chelating virulence factor of Yersinia pestis. Biochemistry. 37:11367-11650. [DOI] [PubMed] [Google Scholar]
- 43.Geoffroy, V. A., J. D. Fetherston, R. D. Perry. 2000. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 68:4452-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths, G. L., S. P. Sigel, S. M. Payne, and J. B. Neilands. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383-385. [PubMed] [Google Scholar]
- 45.Guilvout, I., O. Mercereau-Puijalon, S. Bonnefoy, A. Pugsley, and E. Carniel. 1993. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J. Bacteriol. 175:5488-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harbell, S. O., H. O. Hodgins, and M. H. Schiewe. 1979. Studies on the pathology of vibriosis in coho salmon. J. Fish Dis. 2:527-535. [Google Scholar]
- 47.Heaton, M. P. 1992. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J. Bacteriol. 174:4707-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heidelberg, J. F., et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinrichs, D. E., and K. Poole. 1993. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J. Bacteriol. 175:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henderson, D. P., and S. M. Payne. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 52.Hunt, M. D., G. S. Pettis, and M. A. McIntosh. 1994. Promoter and operator determinants for fur-mediated iron regulation in the bi-directional fepA-fes control region of the Escherichia coli enterobactin gene system. J. Bacteriol. 176:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jalal, M., D. Hossain, D. van der Helm, J. Sanders-Loehr, L. A. Actis, and J. H. Crosa. 1989. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J. Am. Chem. Soc. 111:292-296. [Google Scholar]
- 54.Keating, T., and C. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3:598-606. [DOI] [PubMed] [Google Scholar]
- 55.Keating, T., Z. Suo, D. Miller, and C. Walsh. 2000. Selectivity of the yersiniabactin synthetase adenylation domain in the two step process of amino acid activation and transfer to a holo-carrier protein. Biochemistry 39:2297-2306. [DOI] [PubMed] [Google Scholar]
- 56.Keating, T., D. Miller, and C. Walsh. 2000. Expression, purification, and characterization of the six domain 229 kDa HMWP2 subunit of yersiniabactin synthetase. Biochemistry 39:4729-4739. [DOI] [PubMed] [Google Scholar]
- 57.Keating, T., C. G. Marshall, and C. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthetase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 58.Keating, T., C. G. Marshall, and C. Walsh. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from Vib B, VibE, VibF, and VibH. Biochemistry 39:15522-15530. [DOI] [PubMed] [Google Scholar]
- 59.Keating, T., D. E. Ehmann, R. M. Kohli, C. G. Marshall, J. Trauger, and C. Walsh. 2001. Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. Chem. Biochem. 2:101-109. [DOI] [PubMed] [Google Scholar]
- 60.Koester, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 61.Kohli, R., J. Trauger, D. Schwarzer, M. Marahiel, and C. Walsh. 2001. Exploring the generality of peptide cyclization by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 40:7099-7108. [DOI] [PubMed] [Google Scholar]
- 62.Konopka, K., A. Bindereif, and J. B. Neilands. 1982. Aerobactin-mediated utilization of transferrin-iron. Biochemistry 21:6503-6508. [DOI] [PubMed] [Google Scholar]
- 63.Konz, D., and M. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R47. [DOI] [PubMed] [Google Scholar]
- 64.Lambalot, R., and C. Walsh. 1995. Cloning, oveproduction, and characterization of the E. coli holo-acyl carrier protein synthase. J. Biol. Chem. 270:24658-24661. [DOI] [PubMed] [Google Scholar]
- 65.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. Lacelle, M. A. Marahiel, R. Reid, C. Khosla, and C. Walsh. 1996. A new enzyme superfamily: the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 66.Li, J., R. Szittner, Z. S. Derewenda, and E. Meighan. 1996. Conversion of serine-114 to cysteine-114 and the role of the active site nucleophile in acyl transfer by myristoyl-ACP thioesterase from Vibrio harveyi. Biochemistry 35:9967-9973. [DOI] [PubMed] [Google Scholar]
- 67.Lipmann, F. 1980. Bacterial production of antibiotic peptides by thiol-linked synthesis on protein templates. Adv. Microb. Physiol. 21:227-266. [DOI] [PubMed] [Google Scholar]
- 68.Lipmann, F., W. Gevers, H. Kleinkauf, and R. Roskoski. 1971. Polypeptide synthesis on protein templates: the enzymatic synthesis of gramicidin S and tyrocidine. Adv. Enzymol. Relat. Areas Mol. Biol. 35:1-34. [DOI] [PubMed] [Google Scholar]
- 69.Marahiel, M. A. 1992. Multidomain enzymes involved in peptide synthesis. FEBS Lett. 307:40-43. [DOI] [PubMed] [Google Scholar]
- 70.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in non-ribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 71.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217 [DOI] [PubMed] [Google Scholar]
- 72.McDowell, R., and H. R. Morris. 1996. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 271:15428-15435. [DOI] [PubMed] [Google Scholar]
- 73.Mercado-Blanco, J., K. M. van der Drift, P. E. Olsson, J. E. Thomas-Oates, L. C. van Loon, and P. A. Bakker. 2001. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merriman, T. R., M. E. Merriman, and I. L. Lamont. 1995. Nucleotide sequence of pvdD, a pyoverdin biosynthesis gene from Pseudomonas aeruginosa: pvdD has similarity to peptide synthetases. J. Bacteriol. 177:252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer, J. M., A. Stintzi, and K. Poole. 1999. The ferripyoverdine receptor FpvA of Pseudomonas aeruginosa PAO1 recognizes the ferripyoverdines of P. aeruginosa PAO1 and P. fluorescens ATCC 13525. FEMS Microbiol. Lett. 170:145-150. [DOI] [PubMed] [Google Scholar]
- 76.Miller, D. A., and C. T. Walsh. 2001. Yersiniabactin synthetase: probing the recognition of carrier protein domains by the catalytic heterocyclization domains, Cy1 and Cy2, in the chain-initiation HWMP2 subunit. Biochemistry 40:5313-5321. [DOI] [PubMed] [Google Scholar]
- 77.Nahlik, M. S., T. J. Brickman, B. A. Ozenberger, and M. A. McIntosh. 1989. Nucleotide sequence and transcriptional organization of the Escherichia coli enterobactin biosynthesis cistrons entB and entA. J. Bacteriol. 171:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neilands, J. B. 1981. Microbial iron compounds. Annu. Rev. Biochem. 50:715-731. [DOI] [PubMed] [Google Scholar]
- 79.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 80.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of the two sets of tonB, exbB exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 81.O'Malley, S. M., S. L. Mouton, D. A. Occhino, M. T. Deanda, J. R. Rashidi, K. L. Fuson, C. E. Rashidi, M. Y. Mora, S. M. Payne, and D. P. Henderson. 1999. Comparison of the heme iron utilization systems of pathogenic vibrios. J. Bacteriol. 181:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O'Toole, R., D. L. Milton, P. Hoerstedt, and H. Wolf-Waltz. 1997. RpoN of the fish pathogen Vibrio anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology. 143:3849-3859. [DOI] [PubMed] [Google Scholar]
- 83.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pollack, J. R., and J. B. Neilands. 1970. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem. Biophys. Res. Commun. 39:989-992. [DOI] [PubMed] [Google Scholar]
- 85.Poole, K., S. Neshat, K. Krebes, and D. E. Heinrichs. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quadri, L. E. N., J. Sello, T. A. Keating, P. H. Weinreb, and C. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for the assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 87.Quadri, L. E. N., T. A. Keating, H. Patel, and C. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-2,4-bis-thazoline synthetase activity from PchD, PchE and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 88.Quadri, L. E. N. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol. Microbiol. 37:1-12. [DOI] [PubMed] [Google Scholar]
- 89.Ransom, D. P. 1978. Bacteriologic, immunologic, and pathologic studies of Vibrio sp. pathogenic to salmonids, p. 123. Ph.D. thesis. Oregon State University, Corvallis.
- 90.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 91.Raymond, T. K. N., and C. J. Carrano. 1979. Co-ordination chemistry of microbial iron transport. Accounts Chem. Res. 12:183-190. [Google Scholar]
- 92.Reichert, J., M. Sakitani, and C. Walsh. 1992. Characterization of EntF as a serine-activating enzyme. Protein Sci. 1:549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reimmann, C., L. Serino, M. Beyeler, and D. Haas. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135-3148. [DOI] [PubMed] [Google Scholar]
- 94.Reimmann, C., H. M. Patel, L. Serino, M. Barone, C. T. Walsh, and D. Haas. 2001. Essential PchG-dependent reduction in pyochelin biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 183:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rogers, M. B., J. A. Sexton, G. J. DeCastro, and S. B. Calderwood. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350-2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosenberg, H., and I. G. Young. 1974. Iron transport in the enteric bacteria, p. 67-82. In J. B. Neilands (ed.), Microbial iron metabolism: a comprehensive treatise. Academic Press, Inc., New York, N.Y.
- 97.Rowland, B. M., T. H. Grossman, M. S. Osburne, and H. W. Tabor. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178:119-123. [DOI] [PubMed] [Google Scholar]
- 98.Rusnak, F., W. S. Faraci, and C. Walsh. 1989. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound 2,3-dihydroxybenzoyl-adenylate product. Biochemistry 28:6827-6833. [DOI] [PubMed] [Google Scholar]
- 99.Rusnak, F., M. Skaitani, D. Drueckhammer, J. Reichert, and C. Walsh. 1991. Biosynthesis of the E. coli siderophore enterobactin: sequence of the EntF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry 30:2916-2927. [DOI] [PubMed] [Google Scholar]
- 100.Salinas, P., and J. H. Crosa. 1995. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene 160:17-23. [DOI] [PubMed] [Google Scholar]
- 101.Salinas, P., L. S. Waldbeser, and J. H. Crosa. 1993. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene 123:33-38. [DOI] [PubMed] [Google Scholar]
- 102.Salinas, P. C., M. E. Tolmasky, and J. H. Crosa. 1989. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc. Natl. Acad. Sci. USA 86:3529-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scholten, J. D., K. Chang, P. Babbit, H. Charest, M. Sylvestre, and D. Dunaway-Mariano. 1991. Novel enzymatic hydrolytic dehalogenation of a chlorinated aromatic. Science 253:182-185. [DOI] [PubMed] [Google Scholar]
- 104.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 105.Serino, L., C. Reimmann, P. Visca, M. Beyeler, V. Chiesa, and D. V. Haas. 1997. Biosynthesis of pyochelin and dihdroaeruginoic acid requires the iron-regulated pchDBCA operon in Pseudomonas aeruginosa. J. Bacteriol. 179:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shaw-Reid, C., N. Kelleher, H. Losey, A. Gehring, C. Berg, and C. Walsh. 1999. Assembly line enzymology by multimodular nonribosomal peptide synthetases: catalysis of both elongation and cyclolactonization by the thioesterase domain of E coli EntF. Chem. Biol. 6:385-400. [DOI] [PubMed] [Google Scholar]
- 107.Smith, D., J., A. J. Earl, and G. Turner. 1990. The multifunctional peptide synthetase performing the first step of penicillin biosynthesis in Penicillium chrysogenum is a 421,073 dalton protein similar to Bacillus brevis antibiotic synthetases. EMBO J. 9:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Staab, J. F., and C. F. Earhart. 1990. EntG activity of Escherichia coli enterobactin synthase. J. Bacteriol. 172:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 110.Stein, T., J. Vater, V. Kruft, B. Wittmann-Liebold, P. Franke, M. Panico, Z. Suo, C. Walsh, and D. Miller. 1999. Tandem heterocyclization activity of the multidomain 230 kDa HMWP2 subunit of Yersinia pestis yersiniabactin synthetase: interaction of the 1-1382 and 1383-2035 fragments. Biochemistry 38:14023-14035. [DOI] [PubMed] [Google Scholar]
- 111.Stintzi, A., P. Cornelis, D. Hohnadel, J. M. Meyer, C. Dean, K. Poole, S. Kourambas, and V. Krishnapillai. 1996. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 142:1181-1190. [DOI] [PubMed] [Google Scholar]
- 112.Stintzi, A., Z. Johnson, M. Stonehouse, U. Ochsner, J. M. Meyer, M. L. Vasil, and K. Poole. 1999. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J. Bacteriol. 13:4118-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stoebner, J. A., J. R. Butterton, S. B. Calderwood, and S. M. Payne. 1992. Identification of the vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suo, Z., H.-W. Chen, and C. Walsh. 2000. Acyl-CoA hydrolysis by the HMWP1 subunit of yersiniabactin synthetase: mutational evidence for a cascade of four acyl enzyme intermediates during hydrolytic editing. Proc. Natl. Acad. Sci. 97:14188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suo, Z., C. C. Tseng, and C. T. Walsh. 2001. Purification, priming, and catalytic acylation of carrier protein domains in the polyketide synthase and nonribosomal peptidyl synthetase modules of the HMWP1 subunit of yersiniabactin synthetase. Proc. Natl. Acad. Sci. USA 98:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Toh, H. 1991. Sequence analysis of the firefly luciferase family reveals a conservative sequence motif. Protein Seq. Data Anal. 4:111-117. [PubMed] [Google Scholar]
- 117.Tolmasky, M., A. M. Wertheimer, L. A. Actis, and J. H. Crosa. 1994. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J. Bacteriol. 176:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1994. A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1: histamine is a precursor in the biosynthesis of anguibactin. Mol. Microbiol. 15:87-95. [DOI] [PubMed] [Google Scholar]
- 119.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1988. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 160:860-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tolmasky, M. E., P. C. Salinas, L. A. Actis, and J. H. Crosa. 1988. Increased production of the siderophore anguibactin mediated by pJM1-like plasmids in Vibrio anguillarum. Infect. Immun. 56:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1993. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyper production of anguibactin. Infect. Immun. 61:3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trauger, J., R. Kohli, H. Mootz, M. Marahiel, and C. Walsh. 2000. Peptide cyclization catalyzed by the thioesterase domain of tyrocidine synthetase. Nature 407:215-218. [DOI] [PubMed] [Google Scholar]
- 123.Trucksis, M., J. Michalski, Y. K. Deng, and J. B. Kaper. 1998. The Vibrio cholerae genome contains two unique circular chromosomes. Proc. Natl. Acad. Sci. USA 95:14464-14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vergne, A. F., A. J. Waltz, and M. J. Miller. 2000. Iron chelators from mycobacteria (1954-1999) and potential therapeutic applications. Nat. Prod. Rep. 17:99-116. [DOI] [PubMed] [Google Scholar]
- 125.von Dohren, H., U. Keller, J. Vater, and R. Zocher. 1997. Nonribosomal biosynthesis of peptide antibiotics. Chem. Rev. 97:2675-2705. [DOI] [PubMed] [Google Scholar]
- 126.von Gabain, A., J. Belasco, J. Schottel, A. Chang, and S. Cohen. 1983. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc. Natl. Acad. Sci. USA 80:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walsh, C., A. M. Gehring, P. H. Weinreb, L. E. N. Quadri, and R. S. Flugel. 1997. Posttranslational modification of polyketide and nonribosomal peptide synthetases. Curr. Opin. Chem. Biol. 1:309-315. [DOI] [PubMed] [Google Scholar]
- 128.Walsh, C., H.-W. Chen, T. Keating, B. Hubbard, H. Losey, L. Luo, G. Marshall, D. Miller, and H. Patel. 2001. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on noribosomal peptide assembly lines. Curr. Opin. Chem. Biol. 5:525-534. [DOI] [PubMed] [Google Scholar]
- 129.Watnick, P. I., J. R. Butterton, and S. B. Calderwood. 1998. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes. irgA and irgB. Gene 209:65-70. [DOI] [PubMed] [Google Scholar]
- 130.Weber, G., K. Schorgendorfer, E. Schneider-Scherzer, and E. Leitner. 1994. The peptide synthetase catalyzing cyclosporin production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 26:12-25 [DOI] [PubMed] [Google Scholar]
- 131.Welch, T. J., S. Chai, and J. H. Crosa. 2000. The overlapping angB and angG genes are encoding within the trans-acting factor region of the virulence plasmid pJM1 in Vibrio anguillarum: essential role in anguibactin biosynthesis. J. Bacteriol. 182:6762-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wertheimer, A. M., M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1994. Structural and functional analyses of mutant Fur proteins with impaired regulatory function. J. Bacteriol. 176:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wertheimer, A. M., W. Verwej, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williams, P. H., and N. H. Carbonetti. 1986. Iron, siderophores, and the pursuit of virulence: independence of the aerobactin and enterobactin iron uptake systems in Escherichia coli. Infect. Immun. 51:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Witkowski, A., E. Witkowski, and S. Smith. 1994. Reengineering the specificity of a serine active-site enzyme. J. Biol. Chem. 269:379-383. [PubMed] [Google Scholar]
- 136.Wolf, M., and J. H. Crosa. 1986. Evidence for the role of a siderophore in promoting Vibrio anguillarum infections. J. Gen. Microbiol. 132:2949-2952. [DOI] [PubMed] [Google Scholar]
- 137.Wyckoff, E. E., S. L. Smith, and S. M. Payne. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 183:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wyckoff, E. E., A.-M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ABC transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yamamoto, S., M. A. R. Chowdhury, M. Kuroda, T. Nakano, Y. Koumoto, and S. Shinoda. 1991. Further study on polyamine compositions in Vibrionaceae. Can. J. Microbiol. 37:148-153. [DOI] [PubMed] [Google Scholar]
- 141.Yamamoto, S., S. Shinoda, M. Kawaguchi, K. Wakamatsu, and M. Makita. 1983. Polyamine distribution in Vibrionaceae: norspermidine as a general constituent of Vibrio species. Can. J. Microbiol. 29:724-728. [Google Scholar]
- 142.Yu, S., E. Fiss, and W. R. Jacobs, Jr. 1998. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180:4676-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhu, W., J. E. L Arcenaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two nonribosomal peptide synthetase genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]