Abstract
Fundamental features of microbial cellulose utilization are examined at successively higher levels of aggregation encompassing the structure and composition of cellulosic biomass, taxonomic diversity, cellulase enzyme systems, molecular biology of cellulase enzymes, physiology of cellulolytic microorganisms, ecological aspects of cellulase-degrading communities, and rate-limiting factors in nature. The methodological basis for studying microbial cellulose utilization is considered relative to quantification of cells and enzymes in the presence of solid substrates as well as apparatus and analysis for cellulose-grown continuous cultures. Quantitative description of cellulose hydrolysis is addressed with respect to adsorption of cellulase enzymes, rates of enzymatic hydrolysis, bioenergetics of microbial cellulose utilization, kinetics of microbial cellulose utilization, and contrasting features compared to soluble substrate kinetics. A biological perspective on processing cellulosic biomass is presented, including features of pretreated substrates and alternative process configurations. Organism development is considered for “consolidated bioprocessing” (CBP), in which the production of cellulolytic enzymes, hydrolysis of biomass, and fermentation of resulting sugars to desired products occur in one step. Two organism development strategies for CBP are examined: (i) improve product yield and tolerance in microorganisms able to utilize cellulose, or (ii) express a heterologous system for cellulose hydrolysis and utilization in microorganisms that exhibit high product yield and tolerance. A concluding discussion identifies unresolved issues pertaining to microbial cellulose utilization, suggests approaches by which such issues might be resolved, and contrasts a microbially oriented cellulose hydrolysis paradigm to the more conventional enzymatically oriented paradigm in both fundamental and applied contexts.
INTRODUCTION
Life on Earth depends on photosynthesis, which results in production of plant biomass having cellulose as the major component. The carbon cycle is closed primarily as a result of the action of cellulose-utilizing microorganisms present in soil and the guts of animals. Thus, microbial cellulose utilization is responsible for one of the largest material flows in the biosphere and is of interest in relation to analysis of carbon flux at both local and global scales. The importance of microbial cellulose utilization in natural environments is further enhanced by the status of ruminants as a major source of dietary protein. Finally, microbial cellulose utilization is also an integral component of widely used processes such as anaerobic digestion and composting.
Plant biomass is the only foreseeable sustainable source of fuels and materials available to humanity (410). Cellulosic materials are particularly attractive in this context because of their relatively low cost and plentiful supply. The central technological impediment to more widespread utilization of this important resource is the general absence of low-cost technology for overcoming the recalcitrance of cellulosic biomass. A promising strategy to overcome this impediment involves the production of cellulolytic enzymes, hydrolysis of biomass, and fermentation of resulting sugars to desired products in a single process step via a cellulolytic microorganism or consortium. Such “consolidated bioprocessing” (CBP) offers very large cost reductions if microorganisms can be developed that possess the required combination of substrate utilization and product formation properties (405).
Notwithstanding its importance in various contexts, fundamental understanding of microbial cellulose utilization is in many respects rudimentary. This is a result of the inherent complexity of microbial cellulose utilization as well as methodological challenges associated with its study. Understanding of cellulose hydrolysis can be approached at several levels of aggregation: components of cellulase enzyme systems, unfractionated cellulase systems, pure cultures of cellulolytic microorganisms, and mixed cultures of cellulolytic microorganisms. In general, our understanding is progressively less complete at more highly aggregated levels of study. Thus, although much remains to be elucidated at the level of enzyme components and the underlying genetics of such components, understanding of cellulose hydrolysis by unfractionated cellulase systems is still less complete, understanding of hydrolysis by pure cultures is more limited yet, and hydrolysis in multispecies cultures and mixed communities is least understood of all. There is a natural tendency for science to proceed over time toward a finer level of aggregation—e.g., from pathways to enzymes to genes—and this “reductionist” approach has yielded tremendous insights with respect to the life sciences generally and cellulose hydrolysis in particular. An alternative “integrative” approach, involving the development of an understanding of aggregated systems based on an understanding of their less aggregated components, is also a valid and important focus for scientific endeavor. With respect to cellulose hydrolysis, such integration is essential for research advances to be translated into advances in technological, ecological, and agricultural domains.
The great majority of cellulose hydrolysis research to date has focused on the genetics, structure, function, and interaction of components of cellulase enzyme systems. Several recent and comprehensive reviews address this large body of work (see “Cellulase enzyme systems” below). Whereas hydrolysis of cellulosic biomass has been approached in prior reviews and the research literature primarily as an enzymatic phenomenon, this review approaches the subject primarily as a microbial phenomenon. Thus, we intend our review to embody the integrative approach described in the previous paragraph.
The goals of this review are to collect and synthesize information from the literature on microbial cellulose utilization in both natural and technological contexts, to point out key unresolved issues, and to suggest approaches by which such issues can be addressed. In seeking to consider microbial cellulose utilization from an integrative perspective, we endeavor to consider a diversity of cellulolytic organisms and enzyme systems. This effort is, however, constrained by the information available, which is much more extensive for some types of systems and some levels of consideration than for others. Both aerobic and anaerobic organisms and enzymes are considered in our discussion of fundamentals (see “Fundamentals” below) and methodological aspects (see “Methodological basis for study” below). Our treatment of quantitative aspects of microbial cellulose utilization (see “Quantitative description of cellulose hydrolysis” below) of necessity focuses primarily on aerobic organisms and their enzymes. Information on anaerobic organisms and their enzymes is included in this section as possible, but is much more limited. In considering processing of cellulosic biomass (see “Processing of cellulosic biomass—a biological perspective” below) and organism development for consolidated bioprocessing (see “Organism development for consolidated bioprocessing” below), we focus on organisms producing reduced metabolic products via an effectively anaerobic metabolism because this is responsive to the needs, constraints, and opportunities associated with microbial conversion of cellulosic feedstocks (see“Processing of cellulosic biomass—a biological perspective” below). Literature pertaining to noncellulolytic organisms is included in cases where it provides important foundational understanding for topics involving cellulolytic organisms, as in the case of metabolic engineering of end product formation in cellulolytic anaerobes and expression of heterologous saccharolytic enzymes in noncellulolytic hosts (see “Organism development for consolidated bioprocessing” below). We conclude with a discussion of the genesis, status, and future direction of the microbial cellulose utilization field from both fundamental and biotechnological perspectives.
FUNDAMENTALS
Structure and Composition of Cellulosic Biomass
Cellulose, the most abundant component of plant biomass, is found in nature almost exclusively in plant cell walls, although it is produced by some animals (e.g., tunicates) and a few bacteria. Despite great differences in composition and in the anatomical structure of cell walls across plant taxa, a high cellulose content—typically in the range of approximately 35 to 50% of plant dry weight—is a unifying feature (410). In a few cases (notably cotton bolls), cellulose is present in a nearly pure state. In most cases, however, the cellulose fibers are embedded in a matrix of other structural biopolymers, primarily hemicelluloses and lignin, which comprise 20 to 35 and 5 to 30% of plant dry weight (410, 428, 707). Although these matrix interactions vary with plant cell type and with maturity (748), they are a dominant structural feature limiting the rate and extent of utilization of whole, untreated biomass materials. A detailed description of these interactions and the mechanisms by which they limit hydrolysis and utilization is beyond the scope of this paper and is the topic of several recent reviews (245, 749). The discussion below is focused primarily on cellulose itself, since it appears that—once stripped of the protective effects of other plant biopolymers—cellulose in native plant material shares many characteristics across plant taxa, including its potential for complete hydrolysis and utilization under the proper microbial and environmental conditions.
An important feature of cellulose, relatively unusual in the polysaccharide world, is its crystalline structure. Cellulose is synthesized in nature as individual molecules (linear chains of glucosyl residues) which undergo self-assembly at the site of biosynthesis (86). There is evidence that associated hemicelluloses regulate this aggregation process (19). Approximately 30 individual cellulose molecules are assembled into larger units known as elementary fibrils (protofibrils), which are packed into larger units called microfibrils, and these are in turn assembled into the familiar cellulose fibers.
The arrangement of individual chains within the elementary fibrils has largely been inferred from the fitting of X-ray diffraction data to statistical models that calculate structure based on minimum conformational energy. Individual models are a source of considerable controversy, even in terms of such fundamentals as the orientation of adjacent chains (parallel up versus parallel down) (354, 355, 510). Regardless of their orientation, the chains are stiffened by both intrachain and interchain hydrogen bonds. Adjacent sheets overlie one another and are held together (in cellulose I, the most abundant form of cellulose in nature) by weak intersheet van der Waals forces; despite the weakness of these interactions, their total effect over the many residues in the elementary fibril is considerable (538). The crystalline nature of cellulose implies a structural order in which all of the atoms are fixed in discrete positions with respect to one another. An important feature of the crystalline array is that the component molecules of individual microfibrils are packed sufficiently tightly to prevent penetration not only by enzymes but even by small molecules such as water.
Although cellulose forms a distinct crystalline structure, cellulose fibers in nature are not purely crystalline. The degree of departure from crystallinity is variable and has led to the notion of a “lateral order distribution” of crystallinity, which portrays a population of cellulose fibers in statistical terms as a continuum from purely crystalline to purely amorphous, with all degrees of order in between (427). In addition to the crystalline and amorphous regions, cellulose fibers contain various types of irregularities, such as kinks or twists of the microfibrils, or voids such as surface micropores, large pits, and capillaries (63, 127, 178, 428). The total surface area of a cellulose fiber is thus much greater than the surface area of an ideally smooth fiber of the same dimension. The net effect of structural heterogeneity within the fiber is that the fibers are at least partially hydrated by water when immersed in aqueous media, and some micropores and capillaries are sufficiently spacious to permit penetration by relatively large molecules—including, in some cases, cellulolytic enzymes (647, 648).
Purified celluloses used for studies of hydrolysis and microbial utilization vary considerably in fine structural features, and the choice of substrate for such studies undoubtedly affects the results obtained. Holocelluloses such as Solka Floc are produced by delignification of wood or other biomass materials. These materials contain substantial amounts of various hemicelluloses and often have a low bulk density suggestive of some swelling of cellulose fibers. Microcrystalline celluloses (e.g., Avicel and Sigmacell) are nearly pure cellulose, and the dilute-acid treatment used in their preparation removes both hemicelluloses and the more extensive amorphous regions of the cellulose fibers. Commercial microcrystalline celluloses differ primarily in particle size distribution, which (as indicated below) has significant implications for the rate of hydrolysis and utilization. Cellulose synthesized by the aerobic bacterium Acetobacter xylinum has been tremendously useful as a model system for studying cellulose biosynthesis, but has only been used for a few studies of microbial cellulose utilization. Like plant cellulose, bacterial cellulose is highly crystalline, but the two celluloses differ in the arrangement of glucosyl units within the unit cells of the crystallites (20), and genetic evidence suggests that the two celluloses are synthesized by enzymatic machinery that differs considerably at the molecular level (86). The two celluloses also differ substantially in rate of hydrolysis by fungal cellulases (246) and in rate of utilization by mixed ruminal bacteria (602, 731). The variable structural complexity of pure cellulose and the difficulty of working with insoluble substrates has led to the wide use of the highly soluble cellulose ether, carboxymethylcellulose (CMC), as a substrate for studies of endoglucanase production. Unfortunately, the use of CMC as an enzymatic substrate has weakened the meaning of the term “cellulolytic,” since many organisms that cannot degrade cellulose can hydrolyze CMC via mixed β-glucan enzymes (185). Because of the substituted nature of the hydrolytic products, relatively few microbes (including some fungi and Cellulomonas strains) can use CMC as a growth substrate.
Utilization of cellulosic biomass is more complex than is that of pure cellulose, not only because of the former's complex composition (i.e., presence of hemicelluloses and lignin) but also because of the diverse architecture of plant cells themselves. Plant tissues differ tremendously with respect to size and organization. Some plant cell types (e.g., mesophyll) have thin, poorly lignified walls that are easily degraded by polysaccharide-hydrolyzing enzymes. Others, like sclerenchyma, have thick cell walls and a highly lignified middle lamella separating cells from one another. These cell walls must be attacked from the inside (luminal) surface out through the secondary wall (as opposed to particles of pure cellulose, which are degraded from the outside inward). Thus, in addition to constraints imposed by the structure of cellulose itself, additional limitations are imposed by diffusion and transport of the cellulolytic agent to the site of attack. These constraints may severely limit utilization in some habitats (750).
Taxonomic Diversity
Until recently, hydrolysis and utilization of cellulose in amounts sufficient to provide usable energy to an organism were thought to be carried out exclusively by microorganisms. It now appears that some animal species, including termites and crayfish, produce their own cellulases, which differ substantially from those of their indigenous microflora (723), although the contribution of these enzymes to the nutrition of the animal is unclear. In examining the distribution of cellulolytic species across taxonomic groups, it is useful to consider microbial taxonomy based on phylogeny, rather than on a set of arbitrary morphological or biochemical characteristics as used in classical taxonomy. Current views of the evolutionary relatedness of organisms are based largely on phylogenetic trees constructed from measurements of sequence divergence among chronometric macromolecules, particularly small-subunit rRNAs (16S rRNA of procaryotes and 18S rRNA of eucaryotes [503, 752]). An inspection of these trees reveals that the ability to digest cellulose is widely distributed among many genera in the domain Bacteria and in the fungal groups within the domain Eucarya, although no cellulolytic members of domain Archaea have yet been identified. Within the eubacteria there is considerable concentration of cellulolytic capabilities among the predominantly aerobic order Actinomycetales (phylum Actinobacteria) and the anaerobic order Clostridiales (phylum Firmicutes). Fungal cellulose utilization is distributed across the entire kingdom, from the primitive, protist-like Chytridomycetes to the advanced Basidiomycetes.
The broad distribution of cellulolytic capability could suggest conservation of a cellulose-degrading capability acquired by a primordial ancestor early in evolutionary development; however, this would seem unlikely, given that the capacity for cellulose biosynthesis did not evolve until much later, with the development of algae, land plants and the bacterium A. xylinum. More likely is the convergent evolution toward a cellulolytic capability under the selective pressure of abundant cellulose availability following the development of cellulose biosynthesis. Evidence for such convergent evolution is discussed below (see “Molecular biology of cellulase enzymes”).
Fungi are well-known agents of decomposition of organic matter in general and of cellulosic substrates in particular (94, 462). Fungal taxonomy is based largely on the morphology of mycelia and reproductive structures during various stages of the fungal life cycle rather than on substrate utilization capability. Indeed, systematic characterization of growth substrates has not been carried out for many described fungal species. Therefore, it is currently unclear how broadly and deeply cellulolytic capability extends through the fungal world, and a consideration of the taxonomy of cellulolytic fungi may ultimately prove to be only a slightly narrower topic than consideration of fungal taxonomy in its entirety. Nevertheless, some generalizations can be made regarding the distribution of cellulolytic capabilities among these organisms.
A number of species of the most primitive group of fungi, the anaerobic Chytridomycetes, are well known for their ability to degrade cellulose in gastrointestinal tracts of ruminant animals. Although taxonomy of this group remains controversial (94), members of the order Neocallimastigales have been classified based on the morphology of their motile zoospores and vegetative thalli; they include the monocentric genera Neocallimastix, Piromyces, and Caecomyces and the polycentric genera Orpimomyces and Anaeromyces (376). Cellulolytic capability is also well represented among the remaining subdivisions of aerobic fungi. Within the approximately 700 species of Zygomycetes, only certain members of the genus Mucor have been shown to possess significant cellulolytic activity, although members of this genus are better known for their ability to utilize soluble substrates. By contrast, the much more diverse subdivisions Ascomycetes, Basidiomycetes, and Deuteromycetes (each of which number over 15,000 species [94]), contain large numbers of cellulolytic species. Members of genera that have received considerable study with respect to their cellulolytic enzymes and/or wood-degrading capability include Bulgaria, Chaetomium, and Helotium (Ascomycetes); Coriolus, Phanerochaete, Poria, Schizophyllum and Serpula (Basidiomycetes); and Aspergillus, Cladosporium, Fusarium, Geotrichum, Myrothecium, Paecilomyces, Penicillium, and Trichoderma (Deuteromycetes). For a more detailed consideration of fungal taxonomy and some of its unresolved issues, see reference 94.
When viewed through the lens of microbial physiology, the cellulolytic bacteria can be observed to comprise several diverse physiological groups (Table 1): (i) fermentative anaerobes, typically gram positive (Clostridium, Ruminococcus, and Caldicellulosiruptor) but containing a few gram-negative species, most of which are phylogenetically related to the Clostridium assemblage (Butyrivibrio and Acetivibrio) but some of which are not (Fibrobacter); (ii) aerobic gram-positive bacteria (Cellulomonas and Thermobifida); and (iii) aerobic gliding bacteria (Cytophaga, and Sporocytophaga). Generally, only a few species within each of the above-named genera are actively cellulolytic. The distribution of cellulolytic capability among organisms differing in oxygen relationship, temperature, and salt tolerance is a testament to the wide availability of cellulose across natural habitats. Complicating the taxonomic picture is the recent genomic evidence that the noncellulolytic solventogenic Clostridium acetobutylicum contains a complete cellulosomal gene cluster system that is not expressed, due in part to disabled promoter sequences (606). Examination of the rapidly expanding genomics database may reveal similar surprises in the future.
TABLE 1.
Major morphological features of cellulolytic bacteria
| Oxygen relationship | Genus | Representative speciesa | Gram reaction | Morphology | Growth temp.b | Resting state | Motility | Features of cellulase system | References |
|---|---|---|---|---|---|---|---|---|---|
| Aerobic | Acidothermus | A. cellulolyticus | + | Rod | Thermo | Noncomplexed, cell free | 48 | ||
| Bacillus | B. pumilis | + | Rod | Meso | Endospore | Flagellar | Noncomplexed, cell free | 220 | |
| Caldibacillus | C. cellovorans | + | Rod | Thermo | Endospore | Noncomplexed, cell free | 48 | ||
| Cellulomonasc | C. flavigena, C. uda | + | Rod | Thermo | None | Flagellar | Noncomplexed, cell free | 25, 26, 493 | |
| Cellvibrio | C. fulvus, C. gilvus | − | Curved rod | Meso | None | Flagellar | Noncomplexed, cell free | 612 | |
| Cytophaga | C. hutchinsonii | − | Rod | Meso | None | Gliding | Noncomplexed?, cell free? | 322, 384 | |
| Erwinia | C. carotovora | − | Rod | Meso | None | Flagellar | Noncomplexed, cell free | 29 | |
| Micromonospora | M. chalcae | + | Filamentous rod | Meso | Spored | Nonmotile | Noncomplexed, cell free | 200, 215 | |
| Pseudomonas | P. fluorescens var. cellulosa | − | Rod | Meso | None | Flagellar | Noncomplexed, cell free | 331 | |
| Sporocytophaga | S. myxococcoides | − | Rod | Meso | Spored | Gliding | Noncomplexed, cell free | 697 | |
| Rhodothermus | R. marinus | Rod | Thermo | 11, 48 | |||||
| Streptomyces | S. reticuli | + | Filamentous rod | Meso | Spored | Nonmotile | Noncomplexed, cell free | 715 | |
| Thermobifida | T. fusca | + | Filamentous rod | Thermo | Spored | Nonmotile | Noncomplexed, cell free | 777 | |
| Anaerobic | Acetivibrio | D. cellulolyticus | − | Curved rod | Meso | None | Nonmotile | Complexed | 327, 387, 589 |
| Anaerocellum | D. thermophilum | + | Rod | Thermo | None | Flagellar | Noncomplexed, cell free | 659 | |
| Butyrivibrio | B. fibrisolvens | + | Curved rod | Meso | None | Flagellar | Noncomplexed | 294 | |
| Caldicellulosiruptor | C. saccharolyticum | − | Rod | Thermo | None | Flagellar | Noncomplexed, cell free | 549 | |
| Clostridium | C. thermocellum, C. cellulolyticum | + | Rod | Thermo, meso | Endospore | Flagellar | Complexed, mostly cell boundc | 392, 415, 485, 532, 613 | |
| Eubacterium | E. cellulosolvens | + | Rod | Meso | None | Nonmotile | 699 | ||
| Fervidobacterium | F. islandicum | − | Rod | Thermo | None | Flagellar | 292 | ||
| Fibrobacter | F. succinogenes | − | Rod | Meso | None | Nonmotile | Complexed, cell bound | 88, 294, 463, 645 | |
| Halocella | H. cellulolytica | − | Rod | Meso | None | Flagellar | Noncomplexed, cell free | 622 | |
| Ruminococcus | R. albus, R. flavefaciens | + | Coccus | Meso | None | Nonmotile | Complexed, cell bound | 88, 294 | |
| Spirochaeta | S. thermophila | + | Spiral | Thermo | None | Noncomplexed, cell free | 8, 48 | ||
| Thermotoga | T. neapolitana | − | Rod | Thermo | 48 |
Not all strains of the indicated species are cellulolytic, and some less active or less studied cellulolytic species within these genera are not listed.
Meso, mesophilic; Thermo, themophilic.
Most strains can also grow anaerobically.
Unlike true endospores, these spores have only moderate resistance to environmental stress.
Among the bacteria, there is a distinct difference in cellulolytic strategy between the aerobic and anaerobic groups. With relatively few exceptions (549, 659), anaerobes degrade cellulose primarily via complexed cellulase systems exemplified by the well-characterized polycellulosome organelles of the thermophilic bacterium Clostridium thermocellum (606). Cellulolytic enzymes in C. thermocellum cultures are typically distributed both in the liquid phase and on the surface of the cells. However, several anaerobic species that utilize cellulose do not release measurable amounts of extracellular cellulase, and instead have localized their complexed cellulases directly on the surface of the cell or the cell-glycocalyx matrix. Most anaerobic cellulolytic species grow optimally on cellulose when attached to the substrate, and in at least a few species this adhesion appears to be obligate. Cellulolytic anaerobes resemble other fermentative anaerobes in that their cell yields are low, with the bulk of substrate being converted to various fermentation end products, including ethanol, organic acids, CO2, and H2.
Aerobic cellulose degraders, both bacterial and fungal, utilize cellulose through the production of substantial amounts of extracellular cellulase enzymes that are freely recoverable from culture supernatants (554, 606), although enzymes are occasionally present in complexes at the cell surface (67, 715). The individual enzymes often display strong synergy in the hydrolysis of cellulose. While many aerobic bacteria adhere to cellulose, physical contact between cells and cellulose does not appear to be necessary for cellulose hydrolysis. Kauri and Kushner (322) have shown that separating Cytophaga cells from cellulose via an agar layer or membrane filters appears to enhance cellulose utilization; they suggest that this separation may dilute hydrolytic products, thus relieving catabolite repression of enzyme synthesis. Aerobic cellulolytic bacteria and fungi produce high cell yields characteristic of aerobic respiratory growth, and this has led to considerable technological interest in producing microbial cell protein from waste cellulosic biomass (175, 567, 594, 623). In addition, many studies of aerobic cellulolytic microbes have focused on improving the yield and characteristics of cellulase enzymes. The physiology of the organisms themselves has received surprisingly little study, apart from studies on the effect of growth conditions on enzyme secretion (see, e.g., reference 236).
An interesting point suggested from Table 1 is that cellulose utilization generally proceeds via organisms that are either aerobic or anaerobic, but not both. Indeed, despite the wide distribution of facultatively anaerobic bacteria in general, members of the genus Cellulomonas are the sole reported facultatively anaerobic cellulose degraders (25, 26, 113, 150). Whether the general paucity of facultatively anaerobic groups is a consequence of a physiological or ecological incompatibility of two fundamentally different strategies for cellulose utilization employed by the two groups remains an interesting open question.
It is also notable that most aerobic cellulolytic bacterial species common in soil are classified within genera well known for secondary (non-growth-associated) metabolism, including the formation of distinct resting states (Bacillus, Micromonospora, and Thermobifida) and/or production of antibiotics (Bacillus and Micromonospora) and other secondary metabolites. While antibiotic production in cellulolytic species has not been systematically investigated, production of such compounds might provide additional selective fitness to compensate for their rather modest maximum growth rate on cellulose. An ability to form resting states relatively resistant to starvation or other environmental insult also provides a selective advantage in nature.
Cellulase Enzyme Systems
As noted in the discussion of structure and composition (see above), natural cellulosic substrates (primarily plant cell materials) are composed of heterogeneous intertwined polysaccharide chains with varying degrees of crystallinity, hemicelluloses and pectins, embedded in lignin. Microorganisms produce multiple enzymes to degrade plant cell materials, known as enzyme systems (722). Although this discussion focuses primarily on the action of hydrolytic enzyme systems on cellulose, it should be realized that such systems are also active on hemicellulose, and enzymes active specifically on hemicellulose are commonly coproduced by cellulolytic microorganisms. Prior reviews consider the complexed cellulases of anaerobic bacteria (31, 33, 36, 37, 38, 39, 40, 41, 43, 165, 166, 182, 383, 565, 606, 621), noncomplexed fungal and bacterial cellulases (122, 349, 579, 653, 672, 673, 756), cellulase structure and catalytic mechanisms (58, 136, 470, 624, 683, 722, 757, 758), cellulase (hydrolase) families (254, 255, 256, 258, 260), and biotechnological applications (52, 214, 501, 604, 605).
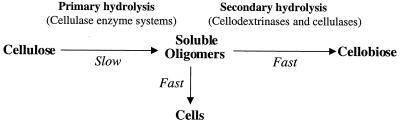
For microorganisms to hydrolyze and metabolize insoluble cellulose, extracellular cellulases must be produced that are either free or cell associated. The biochemical analysis of cellulase systems from aerobic and anaerobic bacteria and fungi has been comprehensively reviewed during the past two decades. Components of cellulase systems were first classified based on their mode of catalytic action and have more recently been classified based on structural properties (260). Three major types of enzymatic activities are found: (i) endoglucanases or 1,4-β-d-glucan-4-glucanohydrolases (EC 3.2.1.4), (ii) exoglucanases, including 1,4-β-d-glucan glucanohydrolases (also known as cellodextrinases) (EC 3.2.1.74) and 1,4-β-d-glucan cellobiohydrolases (cellobiohydrolases) (EC 3.2.1.91), and (iii) β-glucosidases or β-glucoside glucohydrolases (EC 3.2.1.21). Endoglucanases cut at random at internal amorphous sites in the cellulose polysaccharide chain, generating oligosaccharides of various lengths and consequently new chain ends. Exoglucanases act in a processive manner on the reducing or nonreducing ends of cellulose polysaccharide chains, liberating either glucose (glucanohydrolases) or cellobiose (cellobiohydrolase) as major products. Exoglucanases can also act on microcrystalline cellulose, presumably peeling cellulose chains from the microcrystalline structure (672). β-Glucosidases hydrolyze soluble cellodextrins and cellobiose to glucose (Fig. 1). Cellulases are distinguished from other glycoside hydrolases by their ability to hydrolyze β-1,4-glucosidic bonds between glucosyl residues. The enzymatic breakage of the β-1,4-glucosidic bonds in cellulose proceeds through an acid hydrolysis mechanism, using a proton donor and nucleophile or base. The hydrolysis products can either result in the inversion or retention (double replacement mechanism) of the anomeric configuration of carbon-1 at the reducing end (58, 751).
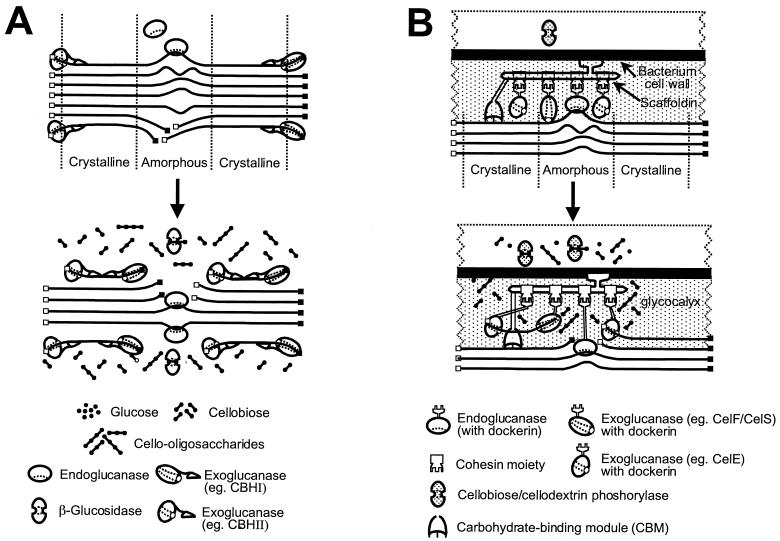
FIG. 1.
Schematic representation of the hydrolysis of amorphous and microcrystalline cellulose by noncomplexed (A) and complexed (B) cellulase systems. The solid squares represent reducing ends, and the open squares represent nonreducing ends. Amorphous and crystalline regions are indicated. Cellulose, enzymes, and hydrolytic products are not shown to scale.
The insoluble, recalcitrant nature of cellulose represents a challenge for cellulase systems. A general feature of most cellulases is a modular structure often including both catalytic and carbohydrate-binding modules (CBMs). The CBM effects binding to the cellulose surface, presumably to facilitate cellulose hydrolysis by bringing the catalytic domain in close proximity to the substrate, insoluble cellulose. The presence of CBMs is particularly important for the initiation and processivity of exoglucanases (673) (Fig. 1A). Revisiting the original model of cellulose degradation proposed by Reese et al. (563), a possible additional noncatalytic role for CBMs in cellulose hydrolysis was proposed: the “sloughing off” of cellulose fragments from cellulosic surfaces of, e.g., cotton fibers, thereby enhancing cellulose hydrolysis (161). Cellulase systems exhibit higher collective activity than the sum of the activities of individual enzymes, a phenomenon known as synergism. Four forms of synergism have been reported: (i) endo-exo synergy between endoglucanases and exoglucanases, (ii) exo-exo synergy between exoglucanases processing from the reducing and non-reducing ends of cellulose chains, (iii) synergy between exoglucanases and β-glucosidases that remove cellobiose (and cellodextrins) as end products of the first two enzymes, and (iv) intramolecular synergy between catalytic domains and CBMs (161, 672).
Cellulase systems are not merely an agglomeration of enzymes representing the three enzyme groups (endoglucanases, exoglucanases, and β-glucosidases, with or without CBMs), but rather act in a coordinated manner to efficiently hydrolyze cellulose. Microorganisms have adapted different approaches to effectively hydrolyze cellulose, naturally occurring in insoluble particles or imbedded within hemicellulose and lignin polymers (683). Cellulolytic filamentous fungi (and actinomycete bacteria) have the ability to penetrate cellulosic substrates through hyphal extensions, thus often presenting their cellulase systems in confined cavities within cellulosic particles (176). The production of “free” cellulases, with or without CBMs, may therefore suffice for the efficient hydrolysis of cellulose under these conditions. The enzymes in these cellulase systems do not form stable high-molecular weight complexes and therefore are called “noncomplexed” systems (Fig. 1A). By contrast, anaerobic bacteria lack the ability to effectively penetrate cellulosic material and perhaps had to find alternative mechanisms for degrading cellulose and gaining access to products of cellulose hydrolysis in the presence of competition from other microorganisms and with limited ATP available for cellulase synthesis. This could have led to the development of “complexed” cellulase systems (called “cellulosomes”), which position cellulase-producing cells at the site of hydrolysis, as observed for clostridia and ruminal bacteria (Fig. 1B). Noncomplexed cellulase systems are discussed first, highlighting the cellulase systems of the aerobic filamentous fungi Trichoderma reesei and Humicola insolens as well as aerobic actinomycetes belonging to the genera Cellulomonas and Thermobifida. The interesting multidomain cellulase systems of anaerobic hyperthermophilic bacteria are mentioned briefly. Thereafter the complexed cellulase systems of anaerobic Clostridium species, Ruminococcus species, and anaerobic fungi are considered.
Noncomplexed cellulase systems.
Cellulases from aerobic fungi have received more study than have those of any other physiological group, and fungal cellulases currently dominate the industrial applications of cellulases (235, 492, 614). In particular, the cellulase system of T. reesei (teleomorph:Hypocrea jecorina, initially called Trichoderma viride) has been the focus of research for 50 years (424, 561, 562, 563). T. reesei produces at least two exoglucanases (CBHI and CBHII), five endoglucanases (EGI, EGII, EGIII, EGIV, and EGV), and two β-glucosidases (BGLI and BGLII (358, 494, 664). Intensive efforts over several decades to enhance cellulase yields have resulted in strains that produce up to 0.33 g of protein/g of utilizable carbohydrate (177). The necessity for the two exoglucanases (cellobiohydrolases) has been attributed to their particular preferences for the reducing (CBHI) and nonreducing (CBHII) ends of cellulose chains of microcrystalline cellulose. This notion has also been supported by the exo-exo synergy observed between these two enzymes (259, 438, 489). Crystallography has elucidated the three-dimensional structures of the two cellobiohydrolases (163, 574). CBHI contains four surface loops that give rise to a tunnel with a length of 50 Å; CBHII contains two surface loops that give rise to a tunnel of 20 Å. These tunnels proved to be essential to the cellobiohydrolases for the processive cleavage of cellulose chains from the reducing or nonreducing ends. The three-dimensional (3-D) structure of CBHI confirmed that cellobiose is the major hydrolytic product as the cellulose chain passes through the tunnel. Occasionally, cellotriose or glucose is released during initial stages of hydrolysis (163). The structure of EGI (structurally related to CBHII) also has been resolved (345) to reveal the presence of shorter loops that create a groove rather than a tunnel. The groove presumably allows entry of the cellulose chain for subsequent cleavage. A similar groove was shown for the structure of EGIII (592), an endoglucanase that lacks a CBM.
Cellobiohydrolase activity is essential for the hydrolysis of microcrystalline cellulose. CBHI and CBHII are the principal components of the T. reesei cellulase system, representing 60 and 20%, respectively, of the total cellulase protein produced by the fungus on a mass basis (756). The important role of CBMs for both enzymes to ensure binding and processivity has been shown clearly (512). However, both the cellobiohydrolases are very slow at decreasing the degree of polymerization of cellulose. Endoglucanases are thought to be primarily responsible for decreasing degree of polymerization by internally cleaving cellulose chains at relatively amorphous regions, thereby generating new cellulose chain ends susceptible to the action of cellobiohydrolases (673). The need for five endoglucanase species in the T. reesei cellulase system has not been clearly explained, particularly considering that endoglucanases (with EGI and EGII as major species) represent less than 20% of the total cellulase protein of T. reesei. Synergism between endoglucanases and cellobiohydrolases has been shown for EGI (693), and EGII (437), and EGIII (489). However, synergism between endoglucanases has not been clearly demonstrated. Part of the problem may be that natural cellulosic substrates are not used for laboratory experiments due to their heterogeneous nature and the true functions of the different endoglucanases may not be observed on purified cellulose. It is noteworthy that some endoglucanases, such as EGI, have broad substrate specificity (e.g., xylanase activity [358]). The presence of CBMs is not essential for endoglucanase activity or for endo-exo synergism (592). Cellobiose, the major product of CBHI and CHBII activity, inhibits the activity of the cellobiohydrolases and endoglucanases (279, 437, 470).
The production of at least two β-glucosidases by T. reesei facilitates the hydrolysis of cellobiose and small oligosaccharides to glucose. Both BGLI and BGLII have been isolated from culture supernatants, but a large fraction of these enzymes remains cell wall bound (442, 690). The presence of β-glucosidases in close proximity to the fungal cell wall may limit loss of glucose to the environment following cellulose hydrolysis. T. reesei produces β-glucosidases at low levels compared to other fungi such as Aspergillus species (560). Furthermore, the β-glucosidases of T. reesei are subject to product (glucose) inhibition (102, 217, 417) whereas those of Aspergillus species are more glucose tolerant (138, 231, 724, 768). The levels of T. reesei β-glucosidase are presumably sufficient for growth on cellulose, but not sufficient for extensive in vitro saccharification of cellulose. T. reesei cellulase preparations, supplemented with Aspergillus β-glucosidase, are considered most often for cellulose saccharification on an industrial scale (560, 644).
The cellulase system of the thermophilic fungus H. insolens possesses a battery of enzymes that allows the efficient utilization of cellulose. The H. insolens cellulase system is homologous to the T. reesei system and also contains at least seven cellulases (two cellobiohydrolases [CBHI and CBHII] and five endoglucanases [EGI, EGII, EGIII, EGV, and EGVI]) (603). However, differences exist, such as the absence of a CBM in EGI of H. insolens. The enzymatic activity of the low-molecular-weight EGIII (also lacking a CBM) observed on different soluble cellulosic substrates was very low, and the natural function of this enzyme still remains unclear (603). Boisset et al. (65) studied the hydrolysis of bacterial microcrystalline cellulose (BMCC), using recombinant CBHI, CBHII, and EGV produced in Aspergillus oryzae, and elegantly showed that a mixture of the three enzymes allow efficient saccharification of crystalline cellulose. Moreover, optimal saccharification was observed when the mixture contained about 70 and 30% of total protein as CBHI and CBHII, respectively. Although the endoglucanase EGV was essential for efficient crystalline cellulose hydrolysis by either CBHI or CBHII, only 1 to 2% of the total protein was needed for maximum efficiency. The combination of all three enzymes yielded more than 50% microcrystalline cellulose hydrolysis. In comparison, the individual enzymes yielded less than 10% microcrystalline cellulose hydrolysis, whereas EGV plus CBHI, EGV plus CBHII, and CBHI plus CBHII yielded approximately 25, 14, and 33% hydrolysis from 3 g of bacterial microcrystalline cellulose (BMCC) per liter, respectively (65).
The white rot fungus Phanerochaete chrysosporium has been used as a model organism for lignocellulose degradation (78). P. chrysosporium produces complex arrays of cellulases, hemicellulases, and lignin-degrading enzymes for the efficient degradation of all three major components of plant cell walls: cellulose, hemicellulose, and lignin (79, 80, 118, 126, 698). Cellulose and hemicellulose degradation occur during primary metabolism, whereas lignin degradation is a secondary metabolic event triggered by limitation of carbon, nitrogen, or sulfur (80). P. chrysosporium produces a cellulase system with CBHII and six CBHI-like homologues, of which CBHI-4 is the major cellobiohydrolase (125, 698). Recently, a 28-kDa endoglucanase (EG28) lacking a CBM was isolated from P. chrysosporium (252). Synergism between the EG28 and the cellobiohydrolases was demonstrated, and it has been suggested that EG28 is homologous to EGIII of T. reesei and H. insolens. No other typical endoglucanase has been isolated from P. chrysosporium. However, Birch et al. (55) reported differential splicing in the CBM-encoding region of the cbh1.2 gene, depending on whether microcrystalline cellulose (Avicel) or amorphous cellulose (CMC) was used as the substrate. They proposed that differential splicing of the cbhI-like genes of P. chrysosporium could yield cellobiohydrolase and endoglucanase activity. Apart from cellobiohydrolases and possible endoglucanase activities, P. chrysosporium also produces cellobiose dehydrogenase that, in the presence of O2, oxidizes cellobiose to cellobionolactone, which reacts spontaneously with water to form cellobionic acid (251, 695). The biological function of cellobiose dehydrogenase has not been clarified, but its binding to microcrystalline cellulose and the enhancement of cellulose hydrolysis have been reported (28, 253). Cellobiose dehydrogenase may help generate hydroxyl radicals that could assist in lignin and cellulose depolymerization (251).
The best-studied species of cellulolytic aerobic bacteria belong to the genera Cellulomonas and Thermobifida (formerly Thermomonospora). Cellulomonas species are coryneform bacteria that produce at least six endoglucanases and at least one exoglucanase (Cex) (99). The individual cellulases of Cellulomonas resemble the cellulase systems of aerobic fungi and contain CBMs; however, cellulosome-like protuberant structures have been noted on Cellulomonas cells grown with cellulose and cellobiose as carbon sources (370, 714). The thermophilic filamentous bacterium Thermobifida fusca (formerly Thermomonospora fusca) is a major cellulose degrader in soil. Six cellulases, three endoglucanases (E1, E2, and E5), two exoglucanases (E3 and E6), and an unusual cellulase with both endoglucanase and exoglucanase activity (E4) have been isolated. The latter enzyme has high activity on BMCC and also exhibits synergism with both the other T. fusca endoglucanases and exoglucanases (304). The E4 enzyme also contains a family III CBM that assists the enzyme in processivity (303). Factorial optimization of the T. fusca cellulase system was undertaken, and the highest synergistic effect was shown with the addition of CBHI from T. reesei (335).
The thermophilic and hyperthermophilic procaryotes represent a unique group of microorganisms that grows at temperatures that may exceed 100°C. Several cellulolytic hyperthermophiles have been isolated during the past decade (48). Surprisingly, no cellulolytic thermophilic archaea have been described, although archaea that can grow on cellobiose and degrade other abundant polysaccharides, such as starch, chitin, and xylan, have been isolated (172, 656). Only two aerobic thermophilic bacteria have been described that produce cellulases: Acidothermus cellulolyticus (an actinomycete) and Rhodothermus (238, 584).
Complexed cellulase systems.
Microorganisms producing complexed cellulase systems (cellulosomes) are typically found in anaerobic environments, where they exist in consortia with other microorganisms, both cellulolytic and noncellulolytic. The cellulosome is thought to allow concerted enzyme activity in close proximity to the bacterial cell, enabling optimum synergism between the cellulases presented on the cellulosome. Concomitantly, the cellulosome also minimizes the distance over which cellulose hydrolysis products must diffuse, allowing efficient uptake of these oligosaccharides by the host cell (33, 606).
Cellulosomes are protuberances produced on the cell wall of cellulolytic bacteria when growing on cellulosic materials. These protuberances are stable enzyme complexes that are firmly bound to the bacterial cell wall but flexible enough to also bind tightly to microcrystalline cellulose. Cellulosomes from different clostridia (Clostridium thermocellum, Clostridium cellulolyticum, Clostridium cellulovorans, and Clostridium josui) and Ruminococcus species in the rumen have been studied in detail. The architecture of cellulosomes is similar among these organisms, although cellulosome composition varies from species to species. The cellulosome of the thermophilic C. thermocellum is discussed and briefly compared to those of the mesophilic C. cellulolyticum, C. cellulovorans, and R. albus (606).
The cellulosome structure of C. thermocellum was resolved through a combination of biochemical, immunochemical, ultrastructural, and genetic techniques (33). The cellulosome consists of a large noncatalytic scaffoldin protein (CipA) of 197 kDa that is multimodular and includes nine cohesins, four X-modules (hydrophilic modules), and a family III CBM. The scaffoldin is anchored to the cell wall via type II cohesin domains. A total of 22 catalytic modules, at least 9 of which exhibit endoglucanase activity (CelA, CelB, CelD, CelE, CelF, CelG, CelH, CelN, and CelP), 4 of which exhibit exoglucanase activity (CbhA, CelK, CelO, CelS), 5 of which exhibit hemicellulase activity (XynA, XynB, XynV, XynY, XynZ), 1 of which exhibits chitinase activity (ManA), and 1 of which exhibits lichenase activity (LicB), have dockerin moieties that can associate with the cohesins of the CipA protein to form the cellulosome. The assembly of the catalytic modules onto the scaffoldin, their composition, and their synergistic activity are still poorly understood. It is assumed that the cellulosome composition can vary and that the catalytic domains do not bind to specific cohesins (39). Preferred proximity relationships between specific catalytic domains cannot be excluded. The major exoglucanase, CelS, is always present in the cellulosome (466). CelS is a processive cellulase with a preference for microcrystalline or amorphous cellulose but not for CMC. CelS is thus defined as an exoglucanase and produces predominantly cellobiose with cellotriose as a minor product. Cellobiose acts as a strong inhibitor of CelS (356, 357). CelA is the major endoglucanase associated with the cellulosome (13, 606). Cellulosomes are remarkably stable, large complexes that can vary from 2 to 16 MDa and even up to 100 MDa in the case of polycellulosomes (39, 122, 606). The cellulosomes are extensively glycosylated (6 to 13% carbohydrate content), particularly on the scaffoldin moiety. The glycosyl groups may protect the cellulosome against proteases but may also play a role in cohesin-dockerin recognition (208).
Cellulosome preparations from C. thermocellum are very efficient at hydrolyzing microcrystalline cellulose (see “Rates of enzymatic hydrolysis” below). The high efficiency of the cellulosome has been attributed to (i) the correct ratio between catalytic domains that optimize synergism between them, (ii) appropriate spacing between the individual components to further favor synergism, and (iii) the presence of different enzymatic activities (cellulolytic or hemicellulolytic) in the cellulosome that can remove “physical hindrances” of other polysaccharides in heterogeneous plant cell materials.
Electron microscopy indicated that cellulosomes are compact “fist”-like structures that open when attaching to microcrystalline cellulose, allowing local spreading of the catalytic domains (Fig. 1B). Between the cellulosome and the cell wall is a stagnant region in which contact corridors and/or glycocalyces may be present, through which oligosaccharides remain in close proximity to the cell, restricting diffusion into the environment (31). Cellobiose and soluble cellodextrin transport are considered below (sec “Physiology of cellulolytic microorganisms”).
Cellulosome architecture in other clostridia is less complex. Taxonomically, the mesophilic C. cellulolyticum and C. josui belong, together with the thermophilic C. thermocellum, to group III within the Clostridiaceae. C. cellulovorans and C. acetobutylicum belong to group I within the Clostridiaceae and are more distant from C. thermocellum and C. cellulolyticum; however, the cellulosome components of C. cellulovorans are surprisingly similar to those of C. cellulolyticum. The cellulosome genes of these clostridia are clustered, and Tamaru et al. (665) suggested that C. cellulovorans could have acquired its cellulosome gene cluster through horizontal gene transfer from a common ancestor. The cellulosome of C. cellulolyticum is the best understood among mesophilic clostridia and is discussed as a model system here (43).
C. cellulolyticum cellulosomes vary from 600 kDa to about 16 MDa and, apart from the scaffoldin (CipC), may contain at least 13 distinct catalytic proteins. The CipC scaffoldin contains eight cohesins, two X-modules, and a family III CBM. As with C. thermocellum, exoglucanases form the major catalytic domains present in the cellulosome. CelE and CelF, exoglucanases (cellobiohydrolases) with opposite processivity, are always present in the C. cellulolyticum cellulosome (206, 513). The crystal structure of CelF revealed the presence of a tunnel (characteristic of processive exoglucanases); however, the tunnel may open into a cleft to allow endoglucanase-like entry of a cellulose chain in amorphous cellulose. CelF is thus considered a processive endoglucanase (514) with cellobiose as major product. Initially, small amounts of cellotriose are released, as observed for CelS of C. thermocellum. The ability of CelF to act on the interior of a cellulose chain may shed light on the question of how the cellulosome retains two processive enzymes attached to the scaffoldin and working in opposite directions.
ExgS is the major exoglucanase in the cellulosome of C. cellulovorans (166). ExgS is homologous to CelS of C. thermocellum and CelF of C. cellulolyticum (family 48 cellulases) and probably fulfills a similar function in the cellulosome. It is important to note that the cohesin-dockerin recognition is species specific (436). Fiérobe et al. (188) used this feature of C. thermocellum and C. cellulolyticum cellulosomes to engineer chimeric miniscaffoldins and chimeric catalytic domains, and they elegantly demonstrated two- to threefold synergism between the CelA endoglucanase and CelF exoglucanase of C. cellulolyticum when associated with miniscaffoldins. Determination of the genome sequence of the noncellulolytic C. acetobutylicum surprisingly revealed a cellulosome gene cluster (495). The noncellulolytic C. acetobutylicum can hydrolyze CMC but not amorphous or microcrystalline cellulose (621). It is tempting to speculate that C. acetobutylicum was once cellulolytic or that it fortuitously acquired the cellulosome gene cluster through horizontal gene transfer. Clostridium stercorarium is the only species from group III for which no cellulosome has been observed (606).
Ruminal bacteria of the genus Ruminococcus are phylogenetically related to, but do not fall within, the family Clostridiaceae. Recently, the presence of dockerin-like sequences in at least seven of the cellulase and xylanase genes of Ruminococcus flavefaciens and the production of 1.5-MDa cellulosome-like structures on the R. albus cell surface in the presence of cellobiose and organic acids (phenylacetic and phenylpropionic acid) suggested that Ruminococcus species indeed produce cellulosomes (162). A large protein of 250 kDa was isolated from R. albus cellulosomes, suggesting a possible large scaffoldin. The structure of the R. albus cellulosomes differs from that of the clostridia, suggesting an independent evolutionary path (336, 500). Fibrobacter succinogenes S85 is another efficient cellulolytic bacterium isolated from the rumen that, like the ruminococci, actively adheres to cellulose (184). Although the cellulases of F. succinogenes are cell associated, no cellulosome structures have been identified, and it would be interesting to know whether cellulose hydrolysis is mediated by cellulosomes in this actively cellulolytic anaerobe.
Anaerobic chytrid fungi are only found in the rumens of herbivorous animals (509) and produce highly active cellulases (68, 103, 745, 759). High-molecular-weight complexes with high affinity for microcrystalline cellulose have been isolated from Piromyces sp. strain E2. Conserved noncatalytic repeat peptide domains have been identified in cellulases and xylanases from Neocallimastix and Piromyces species and are thought to provide a docking function (180, 385). Recently, Steenbakkers et al. (639) used PCR primers based on DNA sequences that encode these 40-amino-acid cysteine-rich docking domains to recover the genes of several cellulosome-like components. Preliminary data indicate the presence of multiple scaffoldins; however they have not yet been isolated from culture fractions (639). Evidence is thus mounting that anaerobic fungi also utilize cellulosomes for hydrolysis of crystalline cellulose. Evolutionary convergence might have occurred between the anaerobic fungi and clostridia. However, the 40-amino-acid dockerin sequence of the anaerobic fungi differs significantly from those of the clostridia, suggesting independent development of the cellulosomes of anaerobic fungi.
Highly cellulolytic anaerobic hyperthermophiles are found in the genera Thermotoga (386) and Caldicellulosiruptor (549), and cellulases isolated from these organisms are often highly thermostable (66). A peculiar feature of the Caldicellulosiruptor hydrolases is the multidomain and multicatalytic nature of these “megazymes.” Many of these megazymes contain five or more domains, which can include a variety of cellulases, hemicellulases, and CBMs (48, 211). The megazymes differ in the number and position of catalytic domains and CBMs and could have evolved via domain shuffling. It is also tempting to speculate that the megazymes are primitive alternatives to operons, and could realize advantages associated with cellulosomes, such as facilitating synergism between different catalytic domains firmly attached to microcrystalline cellulose via multiple CBMs.
Glycoside hydrolase families.
Proteins are designated according to their substrate specificity, based on the guidelines of the International Union of Biochemistry and Molecular Biology (IUBMB). The cellulases are grouped with many of the hemicellulases and other polysaccharidases as O-glycoside hydrolases (EC 3.2.1.x). However, some of the auxiliary enzymes involved, particularly in hemicellulose hydrolysis, also belong to the group of glycosyltransferases (EC 2.4.1.x). Traditionally, the glycoside hydrolases and their genes were named at random. The classification of the glycoside hydrolases has become insufficient, with several thousand glycoside hydrolases identified during the last decade alone. An alternative classification of glycoside hydrolases into families was suggested based on amino acid sequence similarity (254). This classification has been updated several times (255, 256), but with the exponential growth in the number of glycoside hydrolases identified, Coutinho and Henrissat have begun to maintain and update the classification of glycoside hydrolases families at the Expasy server (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html) (124). Families were defined based on amino acid sequence similarities. There is usually a direct relationship between the amino acid sequence and the folding of an enzyme, and as the tertiary structures of many proteins were added, it became clear the families contain basic enzyme folds (257). At the latest update (26 July 2001), more than 5000 glycoside hydrolases were grouped into 86 families. Thus far, CBMs have been divided into 28 families.
Classification of glycoside hydrolases into structurally determined families provides valuable insights that extend and complement the functionally oriented IUBMB classification. The family classification scheme reflects the structural features of the enzymes, which are more informative than substrate specificity alone because the complete range of substrates is only rarely determined for individual enzymes. Once a 3-D structure in a family is known, it can be used to infer the structures of other members of the family. Tertiary structure, particularly at the active site, dictates the enzyme mechanism, and thus families also contain members whose enzyme mechanism is either inverting or retaining. Often enzymes contain multiple domains that belong to the glycoside hydrolase and glycosyltransferase groups. Classification into families defines the modules of such enzymes and resolves the contradiction about substrate specificity for multifunctional enzymes. The family classification also sheds light on the evolution of the glycoside hydrolases. Some families contain enzymes with different substrate specificities; for example, family 5 contains cellulases, xylanases, and mannanases. This suggests divergent evolution of a basic fold at the active site to accommodate different substrates. At the same time cellulases (hydrolyze β-1,4-glycosidic bonds) are found in several different families [families 5, 6, 7, 8, 9, (10), 12, 44, 45, 48, 61, and 74], suggesting convergent evolution of different folds resulting in the same substrate specificity. Some families are deeply rooted evolutionarily, such as family 9, which contains cellulases of bacteria (aerobic and anaerobic), fungi, plants (141), and animals (protozoa and termites [723]). In contrast, family 7 contains only fungal hydrolases whereas family 8 contains only bacterial hydrolases. Furthermore, cellulases from several families, and thus from different folds with either an inverting or retaining mechanism, are found in the same microorganism (for example, the C. thermocellum cellulosome contains endoglucanases and exoglucanases from families 5, 8, 9, and 48 [621]). Cellulases are thus a complex group of enzymes that appear to have evolved through convergence from a repertoire of basic folds. It is tempting to speculate that the pervasive diversity within the cellulase families reflects the heterogeneity of cellulose and associated polysaccharides within plant materials and diversity of niches where hydrolysis takes place. It might be also that nature has not yet fully optimized enzymes for the efficient hydrolysis of recalcitrant insoluble microcrystalline cellulose. In the era of microbial genomics, the large body of information obtained about sequence-structure relationships of existing members of the glycoside hydrolase families allows for the searching of putative glycoside hydrolases in cellulolytic and noncellulolytic microorganisms for which the genome sequence has been determined (258).
The classification of cellulases and other plant cell wall-hydrolyzing enzymes into families not only allows access to information on the structure, mechanism, and evolutionary origin but also structurally orders the ever-increasing list of newly identified hydrolases into functional groups. Henrissat et al. (260) recently proposed a new nomenclature for hydrolases in which the first three letters designate the preferred substrate, the next digits designate the glycoside hydrolase family, and the following capital letters indicate the order in which the enzymes were first reported. For example, the three enzymes CBHI, CBHII, and EGI of T. reesei are designated Cel7A (CBHI), Cel6A (CBHII), and Cel6B (EGI). When more than one catalytic domain is present, it is reflected in the name, such as Cel9A-Cel48A for the two catalytic domains of CelA of Caldocellulosiruptor saccharolyticus. However, researchers have still not completely embraced this new nomenclature. Two possible reasons for this could be (i) hesitance to let go of the established nomenclature and (ii) lack of substrate specificity, for instance the distinction between endoglucanase and exoglucanase/cellobiohydrolase activity. Because this discussion of cellulase systems and their components focuses primarily on catalytic functionality rather than structural relationships, the older, functionally based nomenclature is used below as we consider the molecular biology of cellulase enzymes.
Molecular Biology of Cellulase Enzymes
Regulation of cellulase production.
For T. reesei the production of enzymes for the utilization of complex substrates, such as cellulose, is induced only in the presence of the substrate (or products thereof) but suppressed when easily utilizable sugars, such as glucose, are available. Because cellulose is insoluble and cannot enter the cell, researchers have searched for the natural “inducer(s)” of cellulases (643). Seiboth et al. (609) used deletion mutants to examine the role of individual cellulase enzymes to produce the inducer of other cellulases. Deletion of either cbh2 or egl2 prevented the expression of other cellulase genes, suggesting that CBHII and EGII may play a role in the formation of the inducer. It is interesting that a Δcbh1 mutant increased the transcription of cbh2 more than twofold. Fowler and Brown (194) revealed that deletion of the bgl1 gene, which encodes the extracellular β-glucosidase BGL1, resulted in decreased endoglucanase activities and a lag in the transcription of cbh1, cbh2, egl1, and egl3, suggesting that a β-glucosidase may be partially responsible for formation of the inducer as well. As early as 1962, sophorose (β-1,2-glucobiose) was identified as a strong inducer of cellulases in T. reesei (423). It is assumed that sophorose is formed via the transglycosylation of cellobiose by a β-glucosidase, possibly BGLII of T. reesei (358, 691). However, it has not been demonstrated conclusively that sophorose is the natural inducer of cellulase production. Cellobiose, δ-cellobiose-1,5-lactone, and other oxidized products of cellulose hydrolysis, or even xylobiose resulting from xylan hydrolysis, have not been ruled out as the natural inducer(s). Moreover, the possibility that cellobiose functions as an inducer is more complex because at high levels it inhibits cellulase production (358). It should also be noted that not only the production of cellulases but also the production of hemicellulases is induced, presumably reflecting the intertwined occurrence of these polymers in nature (429).
Production of cellulases by T. reesei is regulated at the transcriptional level. Expression of the cellulase genes (cbh1, cbh2, egl1, egl2, and egl5) of T. reesei QM9414 is coordinated through transcription factors (296). The genes encoding the transcriptional factors ACEI (585) and ACEII (18) were identified based on their ability to bind to the T. reesei cbh1 promoter region and subsequently their DNA sequences were determined. ACEII is homologous to XlnR (700), a transcriptional activator identified in Aspergillus niger, and ACEII also stimulates the expression of cellulase and xylanase genes.
The general carbon catabolite repressor protein CRE1 represses the transcription of cellulase genes in T. reesei (295, 649, 663). The cellulase-hyperproducing T. reesei strain Rut C-30 has a cre1 mutation and still produces cellulases in the presence of glucose (295). The production of cellulases is repressed by CRE1 in the presence of glucose, but a basal level of production occurs in the absence of glucose (93). A link between catabolite repression and the energy status of the cell may exist. A study of four filamentous fungi revealed that extracellular cellulase was repressed at intracellular ATP concentrations above 10−7 mg/ml and that cyclic AMP (cAMP) played a role in derepression of enzyme synthesis (720). Basal levels of cellulase production presumably allow the production of the inducer through limited cellulose hydrolysis, which in turn mediates further induction of cellulase production. The mechanism by which sophorose or other inducers stimulate transcription through the transcriptional activators ACEI and ACEII is not clear yet.
Expression of the cellulase genes of T. fusca is also regulated at two levels: induction by cellobiose and catabolite repression in the presence of glucose (746). CelR represses cellulase production in the absence of cellulose or cellobiose. However, cellobiose acts as an inducer and inactivates CelR, thereby facilitating its dissociation from promoters allowing transcription of cellulase genes (635). Catabolite repression of cellulase genes occurs in the presence of glucose and may be regulated by cAMP levels, as indicated by studies done with Thermobifida curvata (747, 760). Various strains of Cellulomonas have been reported to produce high yields of cellulase on cellulosic substrates and lower yields on xylan, galactomannan, starch, and sugars (543). These data suggest that constitutive production of cellulases at basal levels occurs in the absence of glucose and that cellulase production is subjected to catabolite repression. Cellulosic substrates, as well as cellobiose and xylose, at moderate levels of 0.05 to 0.2 g/liter, serve as inducers for cellulase production (566).
Cellulosome formation in C. thermocellum occurs under carbon-limited conditions, with conflicting statements in the literature about whether induction is important (39, 458, 621). In cellobiose-grown C. thermocellum, celA, celD, and celF were detected during late exponential and early stationary phase, whereas celC occurred primarily in early stationary phase (455). Cellulase production is thus presumably down-regulated via catabolite repression. However, the composition of the cellulosome may be influenced by the carbon source used; for example, the major exoglucanase CelS is more prominent when cells are grown on cellulose than when they are grown on cellobiose (315, 458, 621). The cellulosome of C. cellulovorans is produced on cellulose but not on soluble carbohydrates such as glucose, fructose, cellobiose, or even CMC (60). However, C. cellulovorans grown on cellobiose and CMC does exhibit a high cellulase activity and transcription of cellulase genes. This suggests that cellulases may be produced on certain soluble carbohydrates, such as cellobiose, but that (poly)cellulosome assembly and detachment from the cell wall need some “triggering” from the presence of insoluble microcrystalline cellulose (60, 166, 665). Analysis of mRNA transcripts in the ruminal bacterium R. flavefaciens FD-1 has revealed somewhat contradictory results. Doerner et al. (164) have reported that the celA and celC genes were expressed constitutively while expression of the celB and celD was induced by cellulose. However, Wang et al. (721) have reported that the cellodextrinase celA and celE genes are both induced by cellulose.
Organization of cellulase genes.
The genes encoding cellulases are chromosomal in both bacteria and fungi. In the fungi, cellulase genes are usually randomly distributed over the genome, with each gene having its own transcription regulatory elements (683). Only in exceptional cases, such as for P. chrysosporium, are the three cellobiohydrolase-like genes clustered (126). A comparison of the promoter regions of cbh1, cbh2, eg1, and eg2 of T. reesei reesei revealed the presence of CRE1-binding sites through which catabolite repression is exerted (358). ACEI and ACEII activate transcription by binding to at least the cbh1 promoter region (18, 585).
In bacteria, the cellulase genes are either randomly distributed (e.g., in C. thermocellum [228]) or clustered on the genome (e.g., in C. cellulolyticum, C. cellulovorans, and C. acetobutylicum [42, 43]). The cellulase gene cluster of C. cellulovorans is approximately 22 kb in length and contains nine cellulosomal genes with a putative transposase gene in the 3′ flanking region. Similar arrangements have also been found in the chromosome of C. cellulolyticum and C. acetobutylicum, suggesting the presence a common bacterial ancestor to these mesophilic clostridia or the occurrence of transposon-mediated horizontal gene transfer events. Transcriptional terminators could be identified within these large gene clusters; however, promoter sequences have not yet been found (665).
Both cellulolytic bacteria and fungi (aerobic and anaerobic) primarily contain multidomain cellulases, with single-domain cellulases being the exception (e.g., EGIII of T. reesei and EG 28 of P. chrysosporium [252, 592]). The most common modular arrangements involve catalytic domains attached to CBMs through flexible linker-rich regions. The CBM module can be either at the N or C. terminus; the position is of little relevance when considering the tertiary structure of the protein. This arrangement is found predominantly in noncomplexed cellulase systems. The enzymes of complexed systems (anaerobic bacteria and fungi) are more diverse. Cellulosomal enzymes contain at least a catalytic domain linked to a dockerin. However, some enzymes contain multiple CBMs, a immunoglobulin-like domain (e.g., for CelE of C. cellulolyticum) (206), and a fibronectin type III domain (CbhA of C. thermocellum) (785). The most complex enzymes are those of the extremely thermophilic bacteria (48). The megazymes of the anaerobic hyperthermophile Caldicellulosiruptor isolate Tok7B.1 often have two catalytic domains, usually a cellulase and a hemicellulase domain (combinations from glycoside hydrolases families 5, 9, 10, 43, 44, and 48), linked through several CBM domains (211).
Gene duplication and horizontal gene transfer.
The large number of homologous cellulase genes observed within cellulolytic organisms, between related organisms, or between distant organisms within a niche environment, such as the rumen, suggest that chromosomal rearrangements and horizontal gene transfer contributed to the current rich repertoire of cellulase systems available. The presence of CBH1-like gene clusters in P. chrysosporium (126) and the highly homologous CelK and CbhA exoglucanases in C. thermocellum (786) suggests more recent gene duplication events. The formation of cellulases from the same family within a species but with different cellulase activity, such as EGI (Cel6B) and CBHII (Cel6A) of T. reesei, could represent more distant gene duplications, followed by substrate specificity divergence. The development of polyspecific families, such as the cellulases and hemicellulases in family 5, may represent common ancestor genes that underwent gene duplication followed by substantial divergence with regard to substrate specificity. Examples are the CelE (endoglucanase) and CelO (cellobiohydrolase) of C. thermocellum (621), as well as EGIII (endoglucanase) (587) and MANI (mannanase) in T. reesei (638). The different arrangement of catalytic domains and CBMs in the megazymes of the hyperthermophilic bacteria in all likelihood originated from intergenic domain shuffling through homologous or unequal crossover recombination events (48).
The role of horizontal gene transfer in the evolution of cellulase systems has been expected, but only recently has evidence of such events started to accumulate. The possibility that the cellulosomal gene cluster of C. cellulovorans could have been acquired through a transposase-mediated transfer event was discussed (665). The absence of introns in the glycoside hydrolase genes of the anaerobic fungi (in contrast to aerobic fungi, which contain introns in their glycoside hydrolase genes) raised suspicion that the anaerobic fungi acquired their genes from bacteria. Garcia-Vallvé et al. (203) systematically performed sequence homology analysis between the glycoside hydrolase protein and DNA sequences of the anaerobic fungi and the ruminal bacterium F. succinogenes. They also examined the G+C content and codon bias of the glycoside hydrolase genes of anaerobic fungi as well as the phylogenetic trees derived from the multialignment of orthologous sequences. Their analysis showed that the anaerobic fungi in all likelihood acquired the genes for cellulase systems from bacteria. The high microbial density in the rumen (1010 to 1011 bacteria per ml of ruminal fluid) and the consequent close proximity between ruminal bacteria and fungi, provide ideal conditions for horizontal gene transfer events to occur. Horizontal gene transfer has been demonstrated in the rumen (468, 483), suggesting genome plasticity in this niche environment that could also allow the anaerobic fungi to acquired new genes (430).
Physiology of Cellulolytic Microorganisms
Substrate preference.
The tremendous range of catabolic diversity among microorganisms is one of the distinguishing features of the microbial world. The range of this diversity varies widely among individual species, from highly specialized ones that can utilize only one or a few substrates as energy sources to highly versatile species that can utilize over 100 compounds as the sole carbon and energy source. In general, cellulolytic microbes lie near the specialist end of this continuum. They are primarily carbohydrate degraders and generally are unable to use proteins or lipids (or their components) as energy sources for growth. Cellulolytic microbes native to soil (e.g., the bacteria Cellulomonas and Cytophaga and most fungi) can generally utilize a variety of other carbohydrates in addition to cellulose (493, 543, 550). Anaerobic cellulolytic species (e.g., those of the genera Fibrobacter, Ruminococcus, and Clostridium) are more limited in their carbohydrate range, growing on cellulose and its hydrolytic products but often not on monosaccharides, oligosaccharides, and polysaccharides based on sugars other than glucose. C. thermocellum does not grow easily or well on glucose (486), and both C. thermocellum and R. albus use cellobiose in preference to glucose when both substrates are present (486, 680). A few cellulolytic anaerobic bacteria can utilize xylan (294). The specialist nature of the anaerobic cellulolytic microbes probably results mainly from the specialized enzymatic machinery for cellulose hydrolysis, the significant metabolic effort devoted to its synthesis, and other features peculiar to cellulose utilization. These characteristics, along with the high caloric value and natural abundance of cellulose itself, apply a significant selective pressure on microbes for its utilization—particularly if the organism develops a strategy for positioning itself in such a way as to gain earliest access to the products of cellulose hydrolysis. A specialist microbe, sufficiently well adapted to cellulose utilization, is unlikely to starve in any habitat (natural or man-made) receiving a periodic input of plant biomass.
Some of the more recently described anaerobic cellulolytic species (Anaerocellum thermophilum [659], C. saccharolyticus [549], and Halocella cellulolytica [622] display somewhat wider carbohydrate utilization spectra, with compounds such as starch and various monosaccharides variously reported to serve as substrates. There appears to be a tendency for a broader range of carbohydrate utilization in more extreme environments (thermophilic or halophilic), perhaps as a consequence of a smaller amount of cellulose input, possibly combined with the presence of fewer competing species in these habitats.
The nutrient requirements for growth of cellulolytic species include available nitrogen, phosphorus, and sulfur, plus standard macro- and microminerals and various vitamins. A few cellulolytic microbes have additional requirements (e.g., four- and five-carbon branched-chain volatile fatty acids in the case of the predominant ruminal cellulolytic bacteria). Although additional nutrients present in complex media (e.g., peptones and yeast extract) are not usually required, they often stimulate the growth of individual strains, sometimes dramatically.
Adhesion and formation of cellulose-enzyme-microbe complexes.
Catabolism of cellulose involves both enzymatic depolymerization of insoluble cellulose and cellular utilization of the hydrolytic products. There are two primary strategies for utilizing crystalline cellulose (Fig. 2). Aerobic bacteria and fungi do not adhere (or adhere only weakly) to cellulose, produce noncomplexed cellulases, and oxidize hydrolytic products to CO2 and water. Anaerobic bacteria and fungi display a greater tendency (or in some cases a requirement) to adhere to cellulose, produce primarily complexed cellulases exemplified by the cellulosome organelle, and produce a variety of fermentation end products. As noted below, these two strategies are not completely dichotomous, since various microbial species show different combinations of these characteristics.
FIG. 2.
General strategies of cellulose hydrolysis and utilization by aerobic (top) and anaerobic (bottom) bacteria. Cellulose, degradation products, and cellular features are not shown to scale. Some features of the alternate strategy type are utilized by one or more species. For example, the cellulase of the anaerobe Clostridium stercorarium is of the noncomplexed type, and members of the facultatively anaerobic Cellulomonas utilize an aerobic-type strategy for hydrolyzing cellulose but perform a mixed-acid fermentative catabolism of the hydrolytic products. Glycocalyces are the dominant means of adhesion among ruminal cellulolytic bacteria, but the importance of such structures in other anaerobic groups has not yet been systematically investigated. Refer to Fig. 1 for a more detailed comparison of complexed and noncomplexed systems at the enzymatic level.
Cellulose hydrolysis requires prior binding of enzymes to cellulose, either as an enzyme-substrate binary complex or as a cellulose-enzyme-microbe (CEM) ternary complex. Some cells with noncomplexed cellulase systems (e.g., Cellulomonas sp. strain NRCC 2406 [714]) show a tendency to adhere to cellulose, although such contact does not appear to be necessary for cellulose utilization (322). In the fungus T. reesei, considerable cellulase activity is located at the surface of growing hyphae (90), and growth on corncobs in a fluidized bed reactor has resulted in the formation of a biofilm ∼30 μm thick, which presumably maintains the contact of hyphae with cellulose (674). During degradation of plant cell walls, fungal hyphae confined in the intercellular spaces in plant cell walls are in close proximity to substrate even if they are not strictly “adhering,” and in this confined environment, loss of enzymes and hydrolytic products due to diffusion and convection is likely to be limited.
For anaerobic bacterial species, adhesion of cells to cellulose is much more common, and for some species it appears to be a requirement for rapid and efficient cellulose hydrolysis. In several species (e.g., C. thermocellum, C. cellulolyticum, and F. succinogenes), extracellular cellulolytic activity can be readily assayed in stationary-phase cultures, and culture supernatants have served as the starting point for the isolation of dozens of cellulase enzymes. Nevertheless, adhesion-defective mutants of C. thermocellum display substantially reduced cellulose hydrolysis and these mutants are often unstable, reverting to an adherent phenotype (32). In the early stages of batch culture growth, C. thermocellum (32, 485, 737, 741), and C. cellulolyticum (207) are found in close contact with cellulose, and adhesion of C. thermocellum has been explained by the presence of cellulose-binding modules within the cellulosome complex (32), which are anchored to the cell via a type II cohesin domain at the carboxyl terminus of the scaffoldin protein (see the discussion in reference 621).
Among the ruminal bacteria, adhesion appears to be even more pronounced than in the clostridia. F. succinogenes produces both a series of cellulose-binding proteins, some of which have endoglucanase activity (454), and a thin glycoprotein glycocalyx that results in strong adhesion to cellulose (360). As with C. thermocellum, adhesion-defective mutants of F. succinogenes S85 show little or no capacity to degrade cellulose and readily revert to the adherent phenotype (218, 453). R. flavefaciens and R. albus produce thick, tenacious glycocalyces (121, 734). For F. succinogenes and the ruminococci, inhibition of cell-cellulose adhesion or detachment of cells already adherent reduces or completely prevents cellulose utilization (360, 555, 732). Thus, adhesion seems to be an important strategy for cellulose utilization among anaerobic bacteria, and the CEM ternary complex is likely to be the major agent of cellulolysis.
The role of cell-free cellulases is less clear than that of cellulase present as part of CEM complexes. While cell-free enzymes may be capable of hydrolyzing cellulose in an in vitro assay, these enzymes must compete in microbial cultures with cell-adherent cellulases. During continuous growth of F. succinogenes (727) and R. flavefaciens (617) in cellulose-limited chemostats, both CMCase and Avicelase activities were essentially undetectable in culture supernatants despite high rates of cellulose removal from the culture. It is unlikely that the lack of cell-free cellulases was due to adsorption of extracellular cellulases by cellulose, because the surfaces of the cellulose particles were fully colonized by the bacteria and their glycocalyces, leaving no room for further adsorption of cell-free enzyme. Among cellulolytic anaerobes there are no experimental demonstrations to date that secreted cellulases make up a major part of cellulolytic activity of actively growing cultures, with the exception of Clostridium stercorarium (82, 606).
Adhesion, particularly when mediated via a sticky glycocalyx, can in theory impart a number of advantages to the cellulolytic microbe, including (i) providing a means of concentrating enzymes at the cellulose surface, (ii) permitting the cellulolytic organism first access to the oligomeric products of cellulose hydrolysis, (iii) protecting the ruminal microbe from the undesirable effects of protozoal grazing and attack by bacteriophages, and (iv) protecting hydrolytic enzymes from active ruminal proteases. This last potential advantage has recently been demonstrated experimentally (459). Overall, it is apparent that the potential benefits of adhesion clearly outweigh the additional expenditure of energy and biosynthetic precursors required for synthesis of glycocalyx or cellulose-binding factors.
The general mechanisms of adhesion at the point of contact to cellulose have recently been identified for several anaerobic cellulolytic species (453). These include (i) the cellulosome organelle, (ii) noncatalytic cellulose-binding proteins, (iii) glycosylated moieties of the bacterial glycocalyx or of specific binding proteins, and (iv) fimbriae or pilus-like structures. In some cases, the involvement of a specific molecule within these classes has been implicated, but in many cases the evidence is indirect and is based on the determination of gene sequences similar to those of adhesion components identified in other species. Individual species appear to employ several of the above mechanisms of adhesion. The rapid advances in our understanding of the mechanisms of adhesion by ruminal cellulolytic bacteria at the molecular level have been recently reviewed (453). While glycocalyces appear to be important for ruminal bacteria, additional research is needed to determine if they are important for, or are even produced by, other anaerobic bacteria. Recently, Desvaux et al. (157) have suggested that exopolysaccharide synthesis may represent a means of dissipation of excess carbon by C. cellulolyticum; perhaps glycocalyces have multiple functions for cellulolytic bacteria.
Uptake and phosphorylation of cellulose hydrolysis products.
The driving force for uptake of glucose and its oligomers appears to vary among cellulolytic species, although there is to date no evidence for involvement of classical phosphoenolpyruvate (PEP)-dependent phosphotransferase systems. Nongrowing cells of C. thermocellum display uptake of [14C]cellobiose and cellodextrins by a common, ATP-dependent system, while glucose enters via a separate mechanism that is also ATP dependent (651). Strobel (651) showed a sharp decline in the transport rate accompanying the addition of inhibitors that decreased intracellular ATP concentrations but not in response to inhibitors that abolished the proton motive force. These results are consistent with cellobiose and cellodextrin transport via an ATP-binding cassette protein (ABC protein), which is also a feature of the model for cellodextrin transport for C. cellulolyticum proposed by Desvaux et al. (156). The cellulolytic actinomycete bacterium Streptomyces reticuli produces an ABC protein that assists in the transport of cellobiose, cellotriose, and possibly other cellodextrins (601). An ABC protein associated with a xylan metabolism operon has been identified based on DNA sequence data in R. flavefaciens (21). By contrast, F. succinogenes utilizes a Na+ electrochemical gradient for uptake of glucose (195) and cellobiose (412), and the Na+ requirement for growth on cellulose suggests that Na+ may be the driving force for uptake of cellodextrins as well (740).
The relative importance of different soluble cellulose hydrolysis products (e.g., cellobiose and cellodextrins of various lengths) as intermediates taken up by cellulolytic bacteria is not in our view established. Microbial catabolism of glucose and cellobiose has received decades of attention, and its study predates the study of cellulose hydrolysis itself. By contrast, utilization of cellodextrins (defined here as oligomers larger than cellobiose and abbreviated here as Gn, where n is an integer≥ 3) has received little attention, perhaps because pure cellodextrins are commercially available only at a cost about 4 orders of magnitude greater than that of glucose. The cellodextrins G3 to G6 can be purified by column chromatography of mixed cellodextrins, which can be prepared by acid hydrolysis of pure cellulose (196, 447, 527). Yields of individual cellodextrins G3 to G6 vary depending on the conditions of hydrolysis, but they usually total 10 to 20% of the original amount of cellulose (447, 618). There is evidence that even purified cellodextrins contain trace amounts of chromatographically distinct carbohydrate oligomers of unknown structure (240).
Pure and mixed cellodextrins are excellent growth substrates for many carbohydrate-degrading bacteria, both cellulolytic and noncellulolytic. Most ruminal carbohydrate-fermenting bacteria that have been examined can grow on cellodextrin mixtures at maximum rates similar to that of cellobiose (G2 [575]). Three cellulolytic ruminal strains (F. succinogenes S85, R. flavefaciens FD-1, and R. albus 7) display Monod growth kinetics on individual cellodextrins (618). For all three strains, μmax shows little variation with chain length but S0.5μmax (the concentration permitting growth at half the maximal rate) declines with increasing chain length, most strongly for R. albus, less strongly for F. succinogenes, and only slightly for R. flavefaciens.
Cellulolytic anaerobes generally possess both an extracytoplasmic cellodextrinase that hydrolyzes cellodextrins
 |
and intracellular cellobiose and cellodextrin phosphorylases (CbP and CdP) that catalyze Pi-mediated (ATP-independent) phosphorolysis reactions:
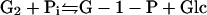 |
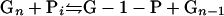 |
CbP is widely distributed in cellobiose-utilizing bacteria, both anaerobic (9, 10, 23, 249, 667, 738, 740) and aerobic (600), and the enzyme has even been found in the hyperthermophile Thermotoga neopolitana (769). The CbP from Cellvibrio gilvus is particularly interesting in that its wide glucosyl acceptor specificity has permitted the in vitro synthesis of numerous unusual di- and trisaccharides (670). By contrast, the distribution of CdP has not been systematically investigated, but the enzyme has been purified from C. thermocellum (615) and its presence has been inferred from labeling and metabolic studies with a number of cellulolytic anaerobes (740). One or more intracellular β-glucosidase enzymes that hydrolytically cleave cellobiose or cellodextrins to glucose have been detected in cellulolytic microbes (35, 297, 444, 446).
The simultaneous presence of extracellular cellodextrinase, intracellular CbP and CdP activities, and intracellular β-glucosidase in a variety of cellulolytic species suggests that cellodextrin and cellobiose metabolism potentially can occur by several processes: (i) extracytoplasmic hydrolysis with subsequent uptake and catabolism, (ii) direct uptake of followed by intracellular phosphorolytic cleavage and catabolism, and (iii) direct uptake followed by intracellular hydrolytic cleavage and catabolism. The relative importance of these alternatives is not well understood in cellulolytic bacteria with respect to metabolism of both cellodextrins and cellulose. This matter is important because of the potential for ATP production via the phosphorolytic cleavage of cellodextrins and cellobiose, as developed further below (see “Bioenergetics of microbial cellulose utilization”).
Knowledge of the relative extent of these reactions by growing cultures has been gained primarily through studies involving pure cellodextrins or mixtures of cellodextrins, rather than cellulose, at extracellular concentrations far in excess of those likely to occur in either natural or industrial environments. Although environmental concentrations have not been determined directly, measurements in the rumen indicate that concentrations of glucose (0.06 to 0.8 mM) (114, 320, 378, 537) are far higher than those of cellobiose (<0.02 mM) (321). Because the concentration of glucose is almost the same as the total concentration of reducing sugars, it is likely that cellodextrin concentrations are very low. In cell extracts of R. albus, the Vmax of phosphorolysis greatly exceeds that of hydrolysis (394), while the reverse is true for the noncellulolytic bacterium Prevotella ruminicola (393). It is interesting that activities of CbP in cell extracts of C. thermocellum are higher for cellulose-grown cells than for cellobiose-grown cells (10). Because CbP does not cleave G3 or higher oligomers and CdP does not cleave G2, the data suggest a major role for CbP during growth on cellulose even though its use provides less net ATP gain than does CdP. On the other hand, Russell (575) showed that cellodextrins are hydrolyzed extracellularly when added at millimolar concentrations to a cellobiose-grown culture. It is likely that the concentration of cellodextrins and the availability of other growth substrates (e.g., cellulose or cellobiose) are important in determining the fate of cellodextrins as well as the relative importance of phosphorolytic and hydrolytic cleavage. Given the widespread occurrence of phosphorolytic and hydrolytic routes for cellodextrin metabolism in cellulolytic microorganisms, the possibility that this apparent redundancy is of selective value bears consideration. Regulating the relative flux via these two routes may provide a means to adjust the rate of ATP supply in response to environmental factors (e.g., availability of substrate or nutrients).
As discussed in more detail below (see “Bioenergetics of microbial cellulose utilization”), cell yields increase markedly with increasing cellodextrin chain length in several cellulolytic organisms, and the cell yield on cellulose has recently been shown to be substantially higher than that on cellobiose in C. cellulolyticum. These data appear consistent with the following hypotheses: (i) phosphorolytic cleavage of β-glucosidic bonds occurs to a substantial extent relative to hydrolytic cleavage and (ii) a substantial fraction of carbohydrate utilized during growth on cellodextrins and cellulose is taken up directly as cellodextrins.
With respect to the second of these hypotheses, we note that it is very difficult to explain how the cell would derive any benefit (reflected in higher growth yields relative to that obtained on cellobiose) from growing on cellodextrins or cellulose unless cellodextrins are taken up by the cell without being first hydrolyzed to cellobiose. If further data support the notion that cellodextrins are an important or perhaps primary form of carbohydrate uptake in cells growing on cellulose, it will be of interest to reconcile this with understanding based on enzymatic studies. As depicted in Fig. 3, it is likely that enzymatic hydrolysis of oligomers is rapid relative to cellulose hydrolysis. Thus, in the absence of cells, oligomers would not be expected to accumulate and cellobiose would be the only apparent product of cellulose hydrolysis, as is commonly observed. However, the availability of cellodextrins at low concentrations does not preclude cells from competing with extracellular enzymes for these substrates, as suggested by cell yield data.
FIG. 3.
Hypothesis for the role of oligomers during microbially and enzymatically mediated cellulose hydrolysis.
Both CdP and CbP are reversible enzymes (Keq ≈ 4 [9, 487]) whose equilibrium constants in vitro actually favor intracellular synthesis of longer oligomers. Substantial losses of intracellular cellobiose and cellodextrins from the cell can occur via oligomer formation and secretion in growing cultures at high substrate concentrations (740). The physiological and ecological significance of this “cellodextrin efflux” is unclear, and it is possible that this process does not occur to a significant extent under substrate-limited microbial cellulose utilization. It has been proposed that cellodextrin efflux provides a means of feeding cellulose-nonadherent bacteria, both cellulolytic and noncellulolytic. While the benefits of cellulose-adherent cells feeding cellodextrins to nonadherent cells of the same species are intellectually attractive, the feeding of other species might seem counterproductive, were it not for the fact that some of these species are capable of cross-feeding other essential nutrients (e.g., branched-chain volatile fatty acids) required by some cellulolytic species (753). Moreover, cellodextrins may have currently unidentified regulatory functions important in controlling cellulolytic metabolism.
Equilibrium effects and reversibility may be manifested at microbial as well as enzymatic levels. In general, much less effort has been devoted to study of the effect of hydrolytic products on the kinetics of cellulose hydrolysis by growing cells than to the equivalent study of cell-free systems involving enzymes from both fungi (112, 279) and bacteria (see reference 458 for a review). Maglione et al. (416) noted the inhibitory effect of the cellobiose analog thiocellobiose on cellulose utilization by growing cultures of F. succinogenes S85, using fermentative production of succinate as a measure of substrate removal. These authors suggest that cellulose utilization in this organism is sensitive to feedback inhibition, and that microbial consumption of hydrolysis products is necessary for cellulolysis to proceed.
Fermentative catabolism and end products.
In strictly anaerobic, cellulolytic bacteria, G-1-P produced by the action of either CbP or CdP is metabolized to glucose-6-phosphate, the entry point to the Embden-Meyerhoff pathway of sugar catabolism. All of these species produce acetic acid and CO2 in substantial amounts, and individual species vary with respect to reduced products formed as a result of intracellular oxidation of reduced pyridine nucleotides. For Clostridium species and R. albus, ethanol and H2 are major reduced end products in pure culture, and acetyl coenzyme A (acetyl-CoA) is a key branch point associated with carbon flux to ethanol and acetate production (226, 486). The ruminal species F. succinogenes and R. flavefaciens produce large amounts of succinate, which in the rumen is converted by other bacteria to propionate, a gluconeogenic substrate for the ruminant host. Succinate production occurs via net fixation of CO2 by PEP carboxykinase, with subsequent conversion of oxaloacetate to malate and succinate (283, 448, 619). Lactate, produced in large amounts by many rapidly growing saccharolytic anaerobes, is generally not a major fermentation product in cellulolytic anaerobes, which have relatively low growth rates even on soluble sugars (207, 619). A notable exception is Anaerocellum thermophilum, the most rapidly growing of all cellulolytic bacteria, which produces lactate as the major end product of cellulose fermentation (659). The anaerobic chytridomycete fungus Neocallimastix frontalis produces ethanol, acetic acid, lactic acid, formic acid, H2, and CO2 as end products, while some other anaerobic fungi also produce succinic acid (676). In the anaerobic fungi, conversion of a pyruvate intermediate to acetate, CO2, and H2 occurs in a specialized organelle, the hydrogenosome, that contains pyruvate:ferredoxin oxidoreductase and hydrogenase (376). The reader should consult comprehensive reviews (396, 458, 569) for a more detailed consideration of the pathways by which anaerobes ferment carbohydrates.
The distribution of end products produced by the branched catabolic pathways of cellulose-fermenting anaerobes appears to be the result of control of metabolic flux at several levels: mass action effects involving the concentrations of intermediates, fermentation products, and electron carriers; enzyme activity; and enzyme synthesis. While experimental evidence supports the importance of the first two determinants, there is to date little evidence of transcriptional and translational control of end product formation in cellulolytic bacteria. The ruminal strains F. succinogenes S85 (727), R. albus 7 (516), and R. flavefaciens FD-1 (617) generally display relatively little change in the major fermentation end product ratios with changes in growth rate, pH, or nature of the carbohydrate source (cellulose or cellobiose), although R. flavefaciens does show a pronounced shift in minor end products (from H2 to formate) with increasing growth rate. C. cellulolyticum (156) and C. thermocellum (22) display an increase in the ratio of ethanol to acetate with increasing growth rate, and changes in end product ratios have been observed in response to the presence of yeast extract (226, 282, 519) in the medium. During growth of C. cellulolyticum in complex medium, NADH/NAD+ ratios as high as 57 have been reported, suggesting that catabolism is limited by the rate of NADH reoxidation (226, 519). Inability to regenerate NAD+ leads to metabolism of excess carbon via phosphoglucomutase, with the G-1-P ultimately used for cellodextrin synthesis and efflux, intracellular glycogen synthesis, and extracellular polysaccharide production (225, 227). The difficulty in reoxidizing NADH may explain why this species is the only mesophilic cellulolytic organism reported to produce lactate in excess of 0.5 mol per mol of hexose consumed (156).
High yields of solvents in lieu of organic acids have been attributed to high concentrations of H2, often associated with reduced agitation, for both cellulolytic (197, 369, 477) and noncellulolytic (170, 456) anaerobes. When incubated at 7 MPa under nitrogen, ethane, or propane, C. thermocellum exhibited an increased ratio of ethanol to acetate (47). Hydrogen removal also dramatically lowers ethanol yields in favor of acetate formation for cellulose fermentation by both C. thermocellum (737) and R. albus (518). Ethanol production in batch cultures of C. thermocellum was increased by the addition of acetate and lactate and decreased by the addition of ethanol (266). However, addition of ethanol, acetate, or lactate to the feed of xylose-limited continuous cultures of the noncellulolytic Thermoanaerobacterium (formerly Clostridium) thermosaccharolyticum resulted in no steady-state change in the ratio of ethanol to acidic end products (402). Removal of ethanol from T. thermosaccharolyticum continuous cultures by stripping a side stream withdrawn from the fermentor also resulted in no change in the ratio of ethanol to acidic end products (402). These results with T. thermosaccharolyticum suggest that factors in addition to extracellular product concentrations can play an important, and in some cases dominant, role in regulating fermentation product yields. Acetate formation is favored relative to lactate and ethanol formation for growth of C. cellulolyticum on cellobiose under carbon-limited compared to carbon-sufficient conditions (225, 226). With respect to noncellulolytic anaerobes, limitation by a nutrient other than the carbon source is a powerful modulator of solvent formation in C. acetobutylicum (443, 572), for which solvent formation is typically not growth associated. This, however, is not the case for T. thermosaccharolyticum (268), for which solvent formation is growth associated.
Utilization of cellobiose (and probably cellodextrins) by the facultatively anaerobic members of the genus Cellulomonas resembles that of strict anaerobes such as C. thermocellum: metabolism via intracellular CbP (25, 600), use of the Embden-Meyerhoff pathway (331), and production of ethanol and acetate as primary end products (113). The fact that cell yields of Cellulomonas fermentans were similar under both aerobic and anaerobic conditions (25) suggests that fermentative metabolism is the dominant pathway even in the presence of O2, although aerobic metabolism was accompanied by stationary-phase acetate oxidation without further increase in cell mass. Both acetate and low pH have been implicated in the inhibition of growth in mature Cellulomonas cultures (150).
Lack of a functional gene transfer system for most anaerobic cellulolytic bacteria has impeded the elucidation of the details of such physiological characteristics as cellodextrin utilization, cellulase synthesis and regulation, glycocalyx synthesis, and response to starvation. Sequencing of whole microbial genomes (currently in progress for C. thermocellum, F. succinogenes, and R. flavefaciens) will ultimately provide many insights on the genetic organization of the enzymes and associated components of the cellulolytic machinery.
Ecological Aspects of Cellulose-Degrading Communities
Different habitats in which cellulose is widely available, by their differing characteristics (water availability, oxygen availability, redox potential, temperature variability, and nutrient status) have fostered the development of cellulose utilization strategies that differ in enzyme architecture and presentation, rate and extent of cellulolysis, ancillary hydrolytic activities, fate of hydrolytic products, and interactions among cellulolytic and noncellulolytic microbes. Most (perhaps all) microbes thought to play a prominent role in cellulose hydrolysis in nature have evolved strategies that bring the cell close to the cellulose surface and give the cellulolytic organism “first access” to hydrolysis products.
In soils, cellulose is available primarily in the form of litter (dead plant material) that is relatively recalcitrant due to the high lignin content of terrestrial plants. A lack of fixed nitrogen and other nutrients may secondarily limit microbial growth, and the low moisture content of soils often favors the growth of fungi as the dominant cellulolytic biota (399). The fungal strategy for cellulolysis involves extracellular cellulases that work alongside lignolytic enzyme systems whose efficiency requires the continuous production of active oxygen species (e.g., by associated peroxidases). In one study, cellulolytic activity and the production of 14CO2 from [14C]cellulose decreased with soil depth (711), suggesting that cellulose utilization is largely an aerobic process, and the primary cellulolytic bacterial isolates were Cytophaga species, although Bacillus and Cellulomonas strains were also isolated. In composting cattle manure, filamentous bacteria from the genera Micromonospora, Cytophaga, and Sporocytophaga were the most numerically abundant isolates and fungi were much less abundant (215). As in any isolation study, the identity of uncultured species and their contribution to the biological process under study were unclear.
Because of its low lignin content, plant biomass produced in aquatic environments is typically degraded by bacteria, which are poor lignin degraders but are better adapted to an aquatic lifestyle than are fungi. The settling of plant detritus to the sediment layer establishes a localized zone of enrichment that may be anoxic as a result of microbial activity, resulting in a proliferation of anaerobic cellulolytic bacteria. Cellulolytic enzymes are presented more efficiently (e.g., as a polycellulosome complex by cells adhering to cellulose) to maximize biosynthetic economy and the capture of hydrolytic products. In poorly mixed aquatic habitats, hydrolysis of cellulose by secreted cellulases may be feasible, as suggested, for example, by the predominance of Cytophaga (known to actively secrete cellulases in culture) (322, 384, 421), in the removal of celluloses experimentally immersed in lake water and at sediment surfaces (275), or in removal of tulip poplar (Liriodendron tulipifera) leaves or algal detritus in streamwater (73).
Utilization of plant cell walls in the digestive tracts of herbivorous animals resembles in many respects that of anaerobic aquatic habitats, with the addition of a temporal constraint in the form of the physical passage of materials through the tract. In nonruminant animals, this rate of passage is too high to permit extensive fiber digestion by the slow-growing cellulolytic microflora; indeed, fiber is usually defined operationally in human nutrition as the organic portion of a feed that passes undigested through the tract. In ruminant animals, the retention time of plant fiber in the rumen is sufficiently long (48 h or more in some species) to allow the growth of a fibrolytic microbial population whose extensive fiber utilization contributes a major portion of the energy for the animal (707). The fibrolytic agents include both bacteria and Chytridomycete fungi. The bacteria are quantitatively more important, and in low-fiber diets the fungi are often absent (376). However, the fungi appear to enhance degradation via physical penetration and weakening of the plant cell walls (6, 7, 273). Both groups of microbes ultimately are destroyed by the acidic conditions in the ruminant's abomasum, a process aided (in the case of bacteria) by the production of lysozymes by abomasal tissue (305, 545). The amino acids released from microbial cell protein are absorbed in the small intestine, thus contributing to the protein nutrition of the ruminant.
In nature, cellulose utilization is carried out not by pure cultures of microorganisms but by multiple cellulolytic species coexisting with each other and with many noncellulolytic species. While cellulolytic species may compete directly for cellulose (104, 192, 193, 499, 616), both cellulolytic and noncellulolytic species can compete for cellodextrin products of cellulose hydrolysis, in cross-feeding of nutrients, and in production of inhibitory compounds. A number of examples of such interactions have been demonstrated in defined mixed cultures (Table 2).
TABLE 2.
Examples of interactions between cellulolytic and noncellulolytic microorganisms in defined mixed culture
| Cellulolytic organism | Noncellulolytic organism | Substrate | Cultivation method | Effects of coculturing | Reference |
|---|---|---|---|---|---|
| Clostridium thermocellum | Methanobacterium thermoautotrophicum | MCCa cellobiose | Batch, 60°C | Increased rate of cellulose degradation; end product shifts from ethanol to CH4 (via H2); interspiecies H2 transfer uncoupled at high growth rate on cellobiose | 737 |
| C. thermosaccharolyticum | Solka Floc | Batch, 60°C | 3-fold increase in ethanol production rate | 484 | |
| C. thermohydrosulfuricum | Steam-expl., aspen | 2-fold decrease in ethanol production rate; decreased soluble sugar accumulation | |||
| C. thermohydrosulfuricum | Cellulose | Increased rate of cellulose degradation; relief of yeast extract requirement through cross-feeding of vitamins and methionine | 467 | ||
| C. thermosaccharolyticum | Solka floc, corn stover | Batch, 60°C | Increased ethanol selectivity | 719 | |
| Fibrobacter succinogenes | Selenomonas ruminantium | Cellulose | Batch, 39°C | Utilization of cellulose fragments by S. ruminantium, conversion of succinate to propionate by S. ruminantium | 598 |
| Ruminococcus albus | Methanobrevibacter smithii | MCC | Continuous, 37°C | Methanogenesis via interspecies H2 transfer at all dilution rates; no measurable change in cellulose conversion or cell yield | 518 |
| F. succinogenes or Ruminococcus flavefaciens | Prevotella ruminicola | Orchardgrass or alfalfa | Batch, 38°C | Extent of cellulose digestion increased in some cocultures | 193 |
| F. succinogenes or Buytrivibrio fibrisolvens | Treponema bryantii | Barley straw | Batch, 39°C | Increase rate and extent of digestion of barley straw but not of pure cellulose | 361 |
| Trichoderma harzianum | Clostridium butyricum | Cellulose (undescribed) | Batch | Products of aerobic cellulose hydrolysis used by anaerobic, N2-fixing Clostridium | 712 |
| Cellulomonas flavigena | Azospirillum brasilense | Filter paper, wheat straw | Batch, 30°C | Physical association observed between cellulose degrader and N2-fixing Azospirillum | 239 |
| Cellulomonas flavigena | Xanthomonas sp. | Alkaline-treated bagasse | Continuous, 30-40°C | Higher μmax in coculture than in either monoculture | 540 |
MCC, microcrystalline cellulose.
Of particular interest in the context of improving the bioconversion of cellulosic biomass are reports of synergistic interactions among fibrolytic and nonfibrolytic bacteria grown on authentic plant cell wall material (192, 193, 451, 452, 719). In these examples, more complete utilization of cellulose and/or hemicelluloses sometimes has been observed, apparently by the simultaneous or sequential depolymerization of different, intimately associated cell wall polysaccharides. It should be noted, however, that almost all examples to date have been restricted to improved extent (rather than rate) of utilization.
The basis for improved fiber utilization in mixed cultures would seem unlikely to lie in the removal of either hydrolytic products (which are typically maintained at low concentrations even in monocultures) or fermentation end products (which may build to substantial concentrations but which are not particularly toxic or inhibitory to growth of the producing organism at the concentrations encountered in nature or in laboratory culture). Instead, it appears likely that removal of certain plant cell wall polysaccharides by one species or group of microbes may improve the accessibility of a second group to cellulose or to hemicelluloses. In this regard, it is interesting that some cellulolytic bacteria can actively depolymerize certain hemicelluloses (particularly xylans) and pectins but cannot effectively utilize the component monosaccharides and oligosaccharides, even in pure culture (i.e., in the absence of competition from noncellulolytic species [192]). Moreover, the soil bacterium Cellulomonas sp. strain ATCC 21399 synthesizes a suite of hydrolytic enzymes when grown on cellulose but not when grown on xylan, starch, or other substrates (543). This suggests that in nature these cellulolytic species utilize xylanases and pectinases primarily as tools to gain access to cellulose; i.e., they have sacrificed a reasonably abundant energy source (xylan or pectin) in exchange for an opportunity to exploit an even more abundant energy source (cellulose) that is utilizable by fewer competitors. Enzymatic cleavage of cell wall linkages between cinnamic acid and arabinoxylans by Ruminococcus albus and Butyrivibrio fibrisolvens has been proposed as another example of an activity whose primary function is to enhance accessibility of an organism to cellulose (435).
The ease with which cellulolytic microbes establish mutualistic interactions with noncellulolytic microbes has important implications in natural environments and in processes managed for human benefit. In pure culture, anaerobic bacteria that ferment cellulose generally produce a mixture of fermentation end products that include acetic acid, CO2, and reduced end products such as ethanol or succinate. In the presence of H2-consuming procaryotes, reducing equivalents are directed away from these reduced products and are instead used to reduce protons to H2 gas. The ultimate reduced product of the coculture depends on the nature of the H2-consuming symbiont (285, 449, 450, 518, 684, 728, 737): methane (from CO2-reducing methanogenic archaea), acetate (from CO2-reducing acetogenic bacteria), or H2S (from sulfate-reducing bacteria). The methanogenic process is widely exploited in the treatment of domestic wastes to produce a biogas used to offset the power costs of the treatment plant. In bioenergy scenarios that involve the direct conversion of cellulose by pure cultures to fuel ethanol (consolidated bioprocessing [CBP] [see “Process configurations” below]), a means must be provided to prevent contamination and growth by H2 consumers that would reduce the ethanol yield via competition for reducing equivalents.
Rate-Limiting Factors in Nature
What, then, determines the rate of conversion of cellulose fibers (containing many long chains of cellulose molecules) to individual, shorter chains that are more easily hydrolyzed? This question can be examined from the perspective of the cellulase enzymes themselves and from that of the microorganism responsible for synthesizing the enzyme and utilizing the hydrolytic products. For enzymatic hydrolysis of natural celluloses, several determinants of hydrolysis rate have been proposed, including crystallinity, degree of polymerization, particle size, pore volume, and accessible surface area (178). Evaluation of the relative importance of these fine-structure features in determining utilization is not straightforward. One of the consequences of the fine-structure variability of cellulose is that it is impossible to obtain a discrete population of particles with identical fine-structure features. Within any given cellulose sample, there is a great degree of variability in the size and shape of individual particles (718, 733). Measurements of fine-structure features such as particle size, crystallinity, or surface area yield average values for that population. Thus, these types of experiments are restricted to comparing measurements of hydrolysis or utilization among populations of particles and their average structural characteristics. Although we cannot obtain homogeneous populations of cellulose particles, we can obtain populations that differ sufficiently from one another to permit comparison with respect to their susceptibility to hydrolysis.
Even when considering a population of cellulose fibers, however, structure-utilization relationships are complicated by the interrelationships among the various structural features. The same structural discontinuities that contribute to increased pore volume, for example, also serve to lower the average degree of crystallinity. The unavoidable consequence of this fact is that it is difficult to alter (e.g., by physical or chemical treatments) one fine-structure feature without simultaneously altering others. Thus, as noted by several workers (202, 730), studies purporting to identify structural features that determine the rate of hydrolysis or utilization often have been overinterpreted, due to a failure to measure or consider changes in other structural determinants. Moreover, correlation is often interpreted as causation, which in at least some cases appears to be unfounded.
Crystallinity is widely regarded as a major determinant of cellulose hydrolysis at both enzymatic and microbial levels. Pretreatments of biomass that reduce crystallinity usually enhance the hydrolysis of cellulose by fungal cellulases (191, 595), but some pretreatments effective in enhancing hydrolysis have been reported to increase crystallinity (see, e.g., reference 1; additional references are given in “Pretreated substrates” below). Studies with pure celluloses indicate that amorphous celluloses are degraded 5 to 10 times more rapidly than are highly crystalline celluloses by both fungal enzymes (202) and ruminal bacteria (337, 729). On this basis, we would expect that crystallinity should increase during the course of cellulose hydrolysis as a result of a more rapid removal of amorphous material. Such measurements have yielded equivocal results. For the bacteria Cellulomonas (149), F. succinogenes (727), Clostridium cellulolyticum (156), and members of the ruminal microflora (205), no significant changes in relative crystallinity index (RCI; the most commmonly used estimate of degree of interchain hydrogen bonding) were observed during growth on cellulose. Similar observations have been reported with cell-free systems (see “Rates of enzymatic hydrolysis” below). Moreover, the correlation between RCI and rate of cellulose removal is relatively weak among celluloses of moderate to high crystallinity, in both enzymatic (95, 178, 191, 736) and whole-cell (139, 149, 733) systems. The disparity in the results from these studies may be partially explained by artifacts in the measurement of RCI. These measurements are typically performed by powder X-ray diffraction on dried material and can change greatly during recovery and drying of cellulose from biodegradation experiments or on suspension of dried cellulosic substrates in aqueous media (730).
Since cellulose hydrolysis is a surface phenomenon, available surface area is a potential determinant of hydrolytic rate, although there remains some debate about what constitutes the “available” surface area. Several studies have shown that the pore structure of cellulosic materials can accommodate particles of the size of a cellulolytic enzyme (223, 224, 648, 678, 736), and good correlation has been observed between total surface area (estimated from solute exclusion measurements and assumed pore geometries) (647) and the rate of substrate hydrolysis. Gama et al. (202), however, applying a modified solute exclusion technique to five different celluloses, have reported that cellulolytic enzymes do not penetrate the pore structure of purified celluloses. Moreover, these workers point out that effective cellulolysis requires synergism among several enzymes, the combined size of which is larger than the micropores that would accommodate a single enzyme. They conclude that “the external surface area, including the macropores, represents a measure of the effective contact area between cellulose and enzymes in the beginning of the reaction. However, this contact surface is not, in itself relevant to cellulose reactivity…because fragmentation can greatly increase accessible surface area” (202). This is an intriguing interpretation worthy of further study. Unfortunately, a meaningful quantitative relationship between surface area and the kinetics of cellulose hydrolysis did not emerge from their work, in which cellulose disappearance was reported only after a fixed incubation period (6 h). It is likely, however, that pore structure is a much more important determinant of hydrolysis in natural biomass materials than in purified celluloses, which have relatively smooth surfaces and lower porosity. On the whole, the relationship between surface area and hydrolytic rate in cell-free systems is rather equivocal.
By contrast, the relationship between available surface area and rate of cellulose digestion in whole-cell systems is strong across a variety of independent measurement techniques, suggesting that the available surface area is a more important determinant of rate of hydrolysis or utilization than is crystallinity. In mixed ruminal bacteria, where cellulolytic enzymes are retained primarily on the cell surface and the effective size of the catalytic unit is the size of the bacterial cell, it is clear that gross specific surface area (the external surface area excluding micropores) is an effective determinant of hydrolysis rate (416, 733). For F. succinogenes, the rate of succinate production from cellulose is directly proportional to surface area measured by absorption of Congo red dye (416). For mixed ruminal microbes, cellulose removal determined by weight loss is directly proportional to the gross specific surface area of the fiber, calculated from the measured dimensions of cellulose particles (733).
Even if hydrolytic cleavage of bonds is moderately rapid, cellulose hydrolysis may be impeded by the inability of enzymes to access additional substrate. This is likely to be due in part to most cellulose chains being buried within the microfibrils. Moreover, coverage of some surface chains by the “footprint” of enzyme molecules (particularly those in a complexed form) that covers many bonds (212) (see “Adsorption” below) physically restricts the binding of additional enzyme molecules to neighboring sites on the fiber. Finally, continued hydrolysis of cellulose requires both excision and removal of hydrolytic products from the site of attack to expose underlying cellulose chains to the enzymes. Walker et al. (718) have made direct measurements of the fragmentation of cellulose (i.e., the breakdown of cellulose into smaller particles, resulting from a separation of associated microfibrils or chains) resulting from the action of T. fusca cellulase and have shown that fragmentation precedes most of the release of reducing sugars. The importance of fragmentation is suggested from comparisons of the rate of weight loss of different cellulose allomorphs by pure cultures of ruminal cellulolytic bacteria and by mixed ruminal microflora (729). Allomorphs with virtually identical unit cell dimensions and similar particle sizes and RCI values showed considerable differences in rate of utilization, as measured by weight loss during bacterial growth. The most slowly utilized allomorphs (cellulose II and cellulose IIIII) were those that are thought to contain—in addition to the intrachain and interchain hydrogen bonds present in all crystalline allomorphs—hydrogen bonds between adjacent sheets, which would impede the fragmentation of microfibrils into individual chains. The relationship between fragmentation and cellulose hydrolysis has not been quantitatively addressed. Fragmentation would be expected to result in an increased reaction rate with time due to increased surface area. However, rates of cellulose hydrolysis by growing bacteria have been found to be constant or to decline with increasing substrate conversion (see “Kinetics of microbial cellulose utilization” below), consistent with the notion that factors other than increased surface area due to fragmentation are the most important in determining hydrolysis rates.
As a microbial process, cellulose utilization is subject to physical and chemical conditions in the environment. The effects of temperature are particularly dramatic. A comparison of maximum growth rate across cellulolytic species reveals a strong dependence on growth temperature (see “Kinetics of microbial cellulose utilization” below). Two other environmental parameters, pH and redox potential, affect the rate and extent of cellulose utilization. Regenerated celluloses (dyed cellophane strips) have been reported to be decomposed in both the water column and the sediment in acidifying lakes in Ontario (275). Rates of degradation (estimated from extraction of the dye from residual cellophane) were linear, suggesting that the process was limited by microbial activity rather than by the amount of substrate. Degradation rates were roughly similar under oxic and anoxic conditions, and no strong effects of pH on the rate of removal were observed, although the role of microenvironments on the process was not considered. By contrast, studies of leaf litter decomposition in aquatic environments have shown a clear inhibition of leaf litter mass removal under progressively acidic conditions (100, 198, 414). Inhibition of cellulose hydrolysis at low pH has also been observed in soil (280).
Effects of pH on cellulose utilization have also been noted in laboratory cultures. Anaerobic cellulolytic bacteria, like most fermentative microbes, grow within a fairly narrow pH range. In some habitats, pH fluctuations permit cellulose hydrolysis to occur at pH values below those supporting growth of the cellulolytic population. For example, substantial cellulose hydrolysis can occur by ruminal bacteria at pH below 6.0, once the bacteria have adhered to cellulose, synthesized a glycocalyx, and initiated bacterial growth at a higher pH (471). Nevertheless, we know of no cellulolytic anaerobe that grows (increases cell mass) at pH below 6.0, a fact that cannot currently be reconciled with observations that cellulose removal in some anaerobic mixed cultures is observed at pH as low as 4.5 (111) and has an optimum near pH 5.
METHODOLOGICAL BASIS FOR STUDY
Prior to the availability of molecular techniques, the central body of thought in microbial physiology was occupied with phenomena such as the rate of cell growth and substrate utilization, the overall stoichiometry of substrate utilization and product formation, cell yields and the thermodynamic efficiency of cell synthesis, substrate utilization for cell maintenance, synthesis of key catabolic enzymes in response to cultivation conditions (e.g., substrate availability and growth rate), cell lysis and death, and the extent of metabolic coupling. For microbial utilization of soluble substrates, a foundation of data involving such phenomena is available for a wide variety of organisms and the focus of physiological studies today is largely at the molecular level. However, this foundation is in general not available for microbial utilization of cellulose. In particular, many important studies remain to be done before our understanding of the physiology of microbial cellulose utilization is advanced to the level of the understanding of soluble substrate utilization in the mid-1970s. This situation has arisen largely because of methodological difficulties distinctive to this line of inquiry: quantification of cells and enzymes in the presence of solids, and substrate delivery for continuous culture.
Quantification of Cells and Enzymes in the Presence of Solids
For soluble substrates, cell concentration is most commonly measured directly via filtration and subsequent dry-weight determination or indirectly via light scattering or cell counting. Because of the presence of unutilized cellulose and/or insoluble lignin-rich residues (in the case of lignocellulosic substrates), application of these techniques to the study of microbial cellulose utilization is not practical. Performing cell counts would be feasible in cases where cells do not adhere to cellulose, but this is often not the case (see “Adhesion and formation of cellulose-enzyme-microbe complexes” above). More typically, performing cell counts would require enumerating cells adhered to cellulose particles in three dimensions as well as ensuring that a representative population of particles (e.g., with respect to size and extent of reaction) is considered. Accurate cell counts are normally not possible for cellulosic substrates by conventional methods.
Satisfactory methods are available for composite measurement of the mass of cells and cellulase (e.g., in a filter cake or pellet) involving cell lysis followed by analysis of total protein (119, 156, 291, 324, 457, 516) or nitrogen (407, 735). Compounds other than protein and nitrogen have been used in studies involving solid substrates as indicators of cell concentration, including phospholipids (190), DNA (244, 631), ATP (677), glucosamine (154), dehydrogenase activity (209), chitin (307), gene probes (640), and cell-specific antibodies (364). Metabolic measurements (e.g., of nutrient consumption, CO2 or heat evolution, and product formation) provide an additional class of methods that have been applied to the determination of cell concentration in the presence of solid substrates (98, 324). Such measurements are useful if the cell yield relative to the measured quantity is known with certainty from prior work, which is seldom the case for cellulolytic microorganisms. Techniques based on physical changes in the medium accompanying growth such as dielectric permittivity (242) and osmotic pressure (16) have been proposed, as have methods based on deconvolution of light-scattering data (323), but to date they have not found widespread applicability in the presence of cellulosic substrates. The reader is referred to Kennedy et al. (324) for a review of work related to measurement of cell mass in the presence of soluble substrates prior to 1992.
A significant set of questions related to microbial cellulose utilization require that cell and cellulase concentrations be determined independently of each other (411). Questions in this category include the determination of mass yields of cells and cellulase (grams per gram of substrate), the way these yields vary as a function of growth conditions, cell- and cellulase-specific hydrolysis rates (grams of cellulose hydrolyzed per gram of cells per hour or grams of cellulose per gram of cellulase per hour), the control of cellulase synthesis (e.g., in relation to different growth rates and substrates), and the ATP demand and “metabolic burden” associated with cellulase synthesis and growth on cellulose. Differential measurement of cell and cellulase concentrations is complicated because cellulase components or complexes are usually distributed among the culture broth, the cell surface, particulate biomass, and cellulose-enzyme-microbe complexes. In the frequent case where a significant fraction of the cellulase is retained on the cell surface, physical separation of cells and cellulase using described techniques is generally impractical. Because of these factors, few if any studies report quantitative data for the mass concentration of cells and cellulase accompanying microbial cellulose utilization.
Lynd and Zhang (411) have performed an analysis of measurement errors associated with various approaches for independently determining cell and cellulase concentrations. For cell concentration measurement, acceptable accuracy is expected when the concentration of a cell-specific component (e.g., DNA) is determined or when total protein concentration is determined in conjunction with a measurement specific to cellulase. Acceptable accuracy is not expected for most conditions if the cell concentration is based on either determination of simultaneous protein and nitrogen concentrations or on measurement of total-solids concentration in conjunction with a measurement specific to cellulase. For cellulase concentration measurement, acceptable accuracy is expected only when a measurement specific to cellulase is used and in general not when the cellulase concentration is based on the measurement of a component common to cellulase and cells such as protein, nitrogen, or dry weight. Enzyme-linked immunosorbent assay would appear to be a logical choice for a measurement specific to cellulase. This method has been used to detect and/or quantify concentrations of cellulase components (53, 189, 353, 491, 492) although thus far not to infer the overall concentration of cellulase enzymes.
Continuous Culture and Substrate Delivery
Continuous culture is the primary tool used to examine physiological characteristics of microorganisms in relation to growth rate. In addition, a variety of kinetic measurements can be made much more reliably—often by an order of magnitude or more—via replicated measurements of steady-state continuous cultures than derivative measurements made on transient batch cultures.
Quantities such as growth rate, hydrolysis rate, cell and cellulase yields, and the cell- and cellulase-specific hydrolysis rates can in principle be calculated from steady-state continuous culture data by using relatively simple equations (Table 3). For both soluble and insoluble cellulosic substrates, the growth rate may be arbitrarily set by varying the dilution rate (volumetic flow rate/working volume, units of inverse time). Based on mixing considerations only, at least three residence times are required following a change in input conditions (e.g., feed concentration or dilution rate) before a steady state is reached. Transient responses due to natural selection operative in the chemostat environment can require substantially longer periods before they are complete. At dilution rates typical of cellulose-grown chemostats, attainment of steady state usually requires several days and may require more than a week.
TABLE 3.
Equations for calculating parameters relevant to microbial cellulose utilization based on continuous culture data
| Parameter | Units | Symbol | Equationa |
|---|---|---|---|
| Growth rate | 1/h | μ |  |
| Hydrolysis rate | g of cellulose/liter/h | rS |  |
| Cell yield | g of cells produced/g of cellulose consumed | YX/S | 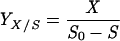 |
| Cellulase yield | g of cellulase produced/g of cellulose consumed | YE/S | 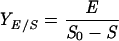 |
| Cell-specific hydrolysis rate | g of cellulose/g of cells/h | rSX | 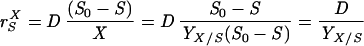 |
| Cellulase-specific hydrolysis rate | g of cellulose/g of cellulase/h | rSE | 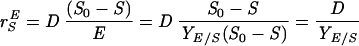 |
Symbol definitions: D, dilution rate (h−1); S0, feed cellulose concentration (g/liter); S, fermentor substrate concentration (g/liter); X, cell concentration (g/liter); E, cellulase concentration (g/liter).
Cellulose-grown continuous cultures are similar to continuous cultures grown on soluble substrates with respect to the concepts of balanced metabolism and substrate limitation. Once steady state has been achieved as indicated by constant values for extracellular variables (e.g., substrate, cell, and product concentrations) over time, chemostat cultures can be said to exist in a state of “balanced metabolism” in that their composition and the concentrations of intracellular metabolites are not changing significantly with time. Although this condition is similar to that exhibited by exponentially growing cells, steady-state chemostat cultures do not exhibit an exponential increase in cell concentration with time. The condition of substrate limitation, often desirable in chemostat studies, corresponds to the situation where an increase in the concentration of a given substrate (e.g., the carbon and energy source) would increase the rate of growth whereas an increase in the concentration of other substrates would have little or no such effect. As with soluble substrates, it is not necessary that the fermentor substrate concentration be zero in order for a culture to be cellulose limited.
Cellulose-fed chemostats differ from chemostats grown on soluble substrates with respect to the functional dependence of the effluent substrate concentration on the feed substrate concentration. Theoretical analysis based on either first-order or variable-order models for cellulose utilization kinetics (see, e.g., reference 633) indicate that the effluent cellulose concentration at a given dilution rate increases with increasing feed substrate concentrations. This phenomenon has also been observed experimentally for cellulose grown continuous cultures of both cellulolytic microorganisms (158, 407) and noncellulolytic microorganisms with added cellulase (632). By contrast, the effluent substrate concentration is the same regardless of the feed substrate concentration according to classical chemostat theory (630), and this result is routinely confirmed experimentally for soluble substrates.
Whereas many thousand papers report continuous cultivation of microorganisms on soluble substrates, there are but a few dozen papers involving continuous cultivation of microorganisms in submerged culture with insoluble cellulose as the growth substrate since Hobson (274) first considered this possibility. We suspect that the primary reason for this is that the apparatus for continuous culture on cellulosic substrates, and substrate delivery in particular, is more complex than for soluble substrates. The relatively long times required for continuous culture on cellulose and complications associated with cell concentration measurement may be additional contributing factors.
As summarized in Table 4, most studies of cellulose-grown chemostats have involved refined (often delignified) cellulosic substrates of a rather fine particle size (typically ≤50 μm) at a concentration between 5 and 25 g/liter. Substrate delivery has most commonly been achieved via a peristaltic pump, although other methods have been used. The range of applicability of peristaltic pumps for delivery of cellulosic substrates depends on the substrate particle size and slurry viscosity, solids concentration, and scale. In general, peristaltic pumps are not suitable for substrate delivery at high concentrations and/or large particle sizes.
TABLE 4.
Continuous culture using cellulosic substrates
| Substrate and organism(s) | Feed concn (g/liter) | Particle size (μm) | Substrate delivery | Feeding interval | Reference(s) |
|---|---|---|---|---|---|
| Pure cultures of cellulolytic micoorganisms | |||||
| Anaerobic | |||||
| Ball-milled filter paper; Ruminococcus flavefaciens, Fibrobacter succinogenes, Ruminococcus albus, Clostridium polysaccharolyticum | 1 | Pinch valve | Variable | 337, 338, 469 | |
| Avicel, pretreated wood; Clostridium thermocellum | 5-14 | 20, 250 | Peristaltic pump | <1 min | 407, 409 |
| Avicel; Ruminococcus albus, Methanobrevibacter smithiti | 11 | 20 | Recirculating loop and valve | 60 min | 450, 516-518 |
| Unspecified; thermophilic isolate | 10 | Peristaltic pump | 620 | ||
| Avicel; Piromyces sp. ± Methanobacterium formicum | 5 | 20 | Peristaltic pump | 30 min | 675 |
| Sigmacell; Ruminococcus flavefaciens, Fibrobacter succinogenes (±Selenomonas ruminantium or Streptococcus bovis) | 5 | 45 | Peristaltic pump with segmented gas/liquid delivery | <1 min | 104, 157, 158, 617, 727, 735, 740 |
| Aerobic | |||||
| Cerelose; Trichoderma viride (reesei) | 25 | Peristaltic pump | 84 | ||
| Avicel; Thermomonospora sp. strain N-35 | 5 | Peristaltic pump with recirculation loop | Continuous | 445 | |
| Solka Floc; Trichoderma reesei | 20 | 250 | |||
| Purified powdered cotton; Trichoderma reesei | 5 | 53 | Peristaltic pump | 210 | |
| Ball-milled wood; Trichoderma viride (reesei) | 5-11 | Peristaltic pump | 15 min | 535 | |
| Pure cultures of noncellulolytic microorganisms with added cellulase | |||||
| Delignified rice straw; Pichia stipitis | 10 | 286, 287 | |||
| Paper sludge; Saccharomyces cerevisiae | 100-200 | Solid | Piston pump | 8 h | Z. Fan and L. R. Lynd, submitted |
| Pretreated wood; Saccharomyces cerevisiae | 8-100 | 250 | Progressing cavity pump | 1 min | 632 |
| Nonsterile mixed cellulolytic cultures | |||||
| Avicel; cellulosic enrichment | 4-14 | 20, 50 | Pump (unspecified) | Intermittent | 111 |
| α-floc; anaerobic digester inoculum | 5-23 | Daily | 213 | ||
| Paper; thermophilic enrichment from compost | 10 | 2,000-3,000 | Daily | 344 |
In carrying out continuous culture on cellulosic substrates, care must be taken to ensure that the substrate is uniformly suspended in the feed reservoir, which is necessary for delivery of a representative and constant feed sample as the reservoir empties. Attention to the suspension of the substrate in the fermentor is also important. A nonuniform distribution of particles substantially complicates the interpretation of chemostat data, especially when it is not detected, although solids retention may be advantageous in an applied context. Uniform substrate dispersion in the feed reservoir can be verified by showing that the particulate substrate concentration in the reservoir is uniform with respect to height and time (as the medium is fed into the fermentor) and/or by showing that substrate concentrations are the same in the feed reservoir and in a subsample of the medium fed to the fermentor. Similarly, uniform substrate dispersion in the fermentor can be verified by showing that the substrate concentration is uniform with respect to height in the fermentor and/or that the substrate concentrations in the fermentor and fermentor effluent are the same. Sterilization of large laboratory vessels (e.g., 20-liter carboys) containing cellulosic substrates also presents potential complications. In particular, longer sterilization times are often required due to impeded convection in cellulose slurries.
QUANTITATIVE DESCRIPTION OF CELLULOSE HYDROLYSIS
Quantitative description of cellulose hydrolysis is of potential value in two contexts: (i) for structuring and testing our fundamental understanding and (ii) for designing and evaluating engineered systems based on quantitative models. Only fundamental aspects are considered in any detail in this section, although it is intended that the discussion be of value to those examining microbial cellulose utilization from applied perspectives as well.
Cellulase enzyme systems are composed of multiple proteins that interact by performing complementary functions related to cellulose hydrolysis and in some cases by forming multiprotein complexes (see “Cellulase enzyme systems” above). Because of such interactions, different behavior is exhibited when cellulase system components are present in combination as compared to when they are present in isolation. Cellulosic substrates are typically present as insoluble macroscopic particles with a distribution of sizes, shapes, and, in some cases, composition. Both cellulosic substrates occurring in nature and those resulting from pretreatment processes typically contain lignin, to which many cellulase components bind. Naturally occurring cellulosic substrates and some pretreated substrates also contain hemicellulose, which impedes access of cellulase components to 1,4-β-glucosidic bonds and may require for hydrolysis enzymatic activities distinct from those involved in cellulose degradation. Kinetic properties (e.g., adsorption capacity and affinity, substrate reactivity), chemical properties (fractional composition of different components), and physical properties (size, shape, density, and rigidity) generally vary over the course of hydrolysis. Cellulolytic organisms impact the rate of cellulose hydrolysis at least by determining the rate of cellulase production. Cellulase production by cellulolytic microorganisms is in turn determined by the interaction of multiple complex processes and variables of both an intracellular and extracellular nature. The presence of microorganisms in CEM complexes may impact cellulose hydrolysis with respect to features other than cellulase production (see “Kinetics of microbial cellulose utilization” above).
Given the above-listed enzymatic, substrate, and organismal properties, as well as the interactions among these properties, microbially mediated cellulose hydrolysis is an exceedingly complex phenomenon. The full extent of this complexity is not represented in any quantitative model proposed to date. All such models represent simplifications of the real situation, although this should not be taken as an impeachment of the utility of such models in either fundamental or applied contexts.
Depending on the purpose at hand, either a relatively simple or complex model may be sufficient. Within each subject area considered below, models are considered in a sequence roughly corresponding to their level of complexity. In several cases, we suggest structures for models that have not yet been developed in any detail. We do this with the hope of providing a framework helpful in identifying types of information required to develop a more complete understanding of microbial cellulose utilization at both the conceptual and quantitative levels. Care is taken in the discussion that follows to identify the enzymes and substrates used, and we suggest that caution is appropriate in generalizing results from one system to another.
Adsorption
Carbohydrate binding modules (CBMs) of cellulase enzymes readily adsorb to accessible sites on a cellulose-containing substrate particle to form a complex held together by specific, noncovalent interactive forces. Catalytic domains of cellulase system components may in some cases specifically adsorb to cellulose independently of CBMs, although this is generally thought to be less important than binding involving CBMs in the context of understanding and describing hydrolysis mediated by non-fractionated cellulase systems. Cellulase may also adsorb to lignin, which is thought to be nonspecific (506, 671). Formation of enzyme-cellulose complexes is a prerequisite for cellulose hydrolysis, and such complexes are a central feature of most conceptual and quantitative models for cellulose hydrolysis. Working with 26 cellulase preparations, 10 of them highly purified, Klyosov (347) showed a strong correlation between hydrolysis rates and values of the adsorption equilibrium constant.
Quantitative description of the adsorption of cellulase(s) to cellulose generally involves expressing the concentration of a cellulose-enzyme complex as a function of a vector of variables relevant to cellulase adsorption that describe the state of the system. In most adsorption models, such “state variables” include the total amount of cellulase present, the total amount of substrate present, substrate-specific and enzyme-specific parameters that impact adsorption (e.g., affinity and capacity), and variables that describe the physical and chemical environment (e.g., temperature and ionic strength). Experimental determination of the concentration of cellulose enzyme complex, [CE], is usually carried out by taking the difference between total cellulase present and unadsorbed cellulase, e.g. for a substrate containing only cellulose:
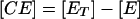 |
(1) |
where [ET] is the total concentration of binding sites on the enzyme and [E] is the concentration of binding sits on the enzyme not adsorbed to cellulose. Techniques for direct measurement of adsorbed enzyme would be desirable but are seldom employed (426).
Equilibrium is assumed in many adsorption models. The equilibrium assumption is often justified by the observation that the time required for adsorbed cellulase to reach a constant value is short relative to the time required for hydrolysis. Most studies find that adsorbed cellulase reaches a constant value in ≤90 mins, and many studies have found ≤30 min to be sufficient (74, 105, 332, 333, 334, 373, 508, 548, 625, 651, 652), whereas complete hydrolysis of cellulose usually requires a day or more. The simplest representation of adsorption equilibrium is via an equilibrium constant, Kd:
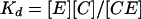 |
(2) |
where [C] is the concentration of accessible binding sites on cellulose not adsorbed to enzyme. Kd, [E], [C], and [CE] are taken here to have units of micromoles per liter. Other internally consistent units can also be used, and the use of units other than micromoles per liter for Kd is considered below. As an alternative to equilibrium models, some models (117, 488, 490) employ a dynamic description of adsorption such as
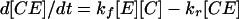 |
(3) |
where kr/kf = Kd.
Studies by Rabinovich et al. (described in reference 347) involving various cellulases and cellulose samples indicate that once a cellulase-cellulose complex is formed, the enzyme stays bound to the cellulose for a significant period (e.g., 30 min or more), during which hundreds of catalytic events occur. Surface diffusion rates of Cellulomonas fimi cellulase components on microcrystalline cellulose have been measured by Jervis et al. (312) and appear not to be rate limiting. Soluble sugars have no influence on adsorption behavior in cases where this has been examined (506, 507), although inhibition of adsorption by unidentified compounds in protein-extracted lucerne fibers has been reported (654).
Values for maximum binding capacities (enzyme per unit substrate) and affinity constants (1/Kd) are presented in Table 5 for both isolated cellulase components and cellulase mixtures and complexes as well as several substrates. It is notable that reported values for adsorption parameters for the same enzyme-substrate system differ by as much as an order of magnitude. The binding capacity of bacterial microcrystalline cellulose (BMCC) is higher than that of Avicel for both cases for which comparative data are available (T. fusca and T. reesei). The binding capacity of pretreated wood is similar to that of Avicel for the T. reesei system, but the C. thermocellum system exhibits a 19-fold-higher capacity for pretreated wood than for Avicel. The binding affinity (liters per gram of cellulase) for both Avicel and pretreated hardwood is the highest in Table 5 by over 100-fold for the C. thermocellum system, the only cellulase of the complexed type listed.
TABLE 5.
Summary of cellulase adsorption parametersa
| Organism, substrate and temp | Binding capacity
|
Binding affinity
|
Reference | |||
|---|---|---|---|---|---|---|
| mg/g | μmol/g | μmol/μmolb | Liters/g | Liters/μmol | ||
| Components | ||||||
| Cellulomonas fimi (30°C) | ||||||
| CenA BMCC | 144 | 3.1 | 5.0 × 10−4 | 41 | 1.89 | 212 |
| Cex BMCC | 184 | 3.6 | 5.8 × 10−4 | 33 | 1.71 | |
| Thermobifida fusca (50°C) | ||||||
| E3, Avicel | 26 | 0.4 | 6.5 × 10−5 | 3.1 | 0.20 | 70 |
| E3, BMCC | 741 | 11.4 | 1.8 × 10−3 | 1.5 | 0.10 | |
| E4, Avicel | 31 | 0.34 | 5.5 × 10−5 | 0.85 | 0.077 | |
| E4, BMCC | 875 | 9.7 | 1.6 × 10−3 | 0.49 | 0.044 | |
| E5, Avicel | 31 | 0.67 | 1.1 × 10−4 | 4.8 | 0.22 | |
| E5, BMCC | 556 | 12.0 | 1.9 × 10−3 | 2.8 | 0.13 | |
| Trichoderma reesei | ||||||
| CBHI, Avicel (4°C) | 48 | 0.74 | 1.2 × 10−4 | 0.69 | 0.044 | 439 |
| CBHI, Avicel (25°C) | 57 | 1.1 | 1.8 × 10−4 | 5.4 | 0.28 | 637 |
| CBHI, Avicel (25°C) | 15 | 0.29 | 4.7 × 10−5 | 682 | ||
| CBHI, Avicel (50°C) | 25 | 0.48 | 7.8 × 10−5 | 1.7 | 0.09 | 70 |
| CBHI/BMCC (50°C) | 239 | 4.6 | 7.5 × 10−4 | 5.4 | 0.28 | 70 |
| CBHII, Avicel (4°C) | 28 | 0.52 | 8.4 × 10−5 | 1.0 | 0.053 | 439 |
| CBHII, Avicel (25°C) | 11.3 | 0.24 | 3.9 × 10−5 | 682 | ||
| Trichoderma viride (30°C) | ||||||
| ExoI | 6.6 | 0.11 | 1.8 × 10−5 | 5.0 | 0.3 | 44 |
| ExoIII | 63 | 1.0 | 1.6 × 10−4 | 6.9 | 0.43 | |
| EndoI, Avicel | 130 | 2.5 | 4.0 × 10−4 | 0.88 | 0.04 | |
| EndoIII, Avicel | 26 | 0.45 | 7.3 × 10−5 | 12 | 0.68 | |
| EndoV, Avicel | 110 | 1.8 | 2.9 × 10−4 | 0.89 | 0.05 | |
| EndoVI, Avicel | 4.1 | 0.08 | 1.3 × 10−5 | 3.4 | 0.18 | |
| Multicomponent mixtures and complexes | ||||||
| Trichoderma reesei | ||||||
| Avicel (50°C) | 92 | 1.92 | 3.1 × 10−4 | 1.04 | 0.05 | 641 |
| Avicel (40°C) | 56 | 1.21 | 2.0 × 10−4 | 3.21 | 0.015 | 508 |
| Cellulose in pretreated woodc (40°C) | 81 | 1.68 | 2.7 × 10−4 | 1.82 | 0.087 | 506 |
| C. thermocellum | ||||||
| Avicel (60°C)d | 17.5 | 0.0083 | 1.4 × 10−5 | 246 | 517 | 50 |
| Pretreated wood(60°C)d,e | 317 | 0.15 | 2.5 × 10−5 | 344 | 722 | |
Values in bold are as reported; others are calculated.
Micromole of cellulase/micromole of β-glucosidic bond calculated using 6,173 μmol of β-glucosidic bonds/g of cellulose.
Dilute-acid-pretreated wood prepared at 220°C; an average molecular weight of 48,000 is assumed as in reference 641.
Calculated quantities based on a specific activity of 2.4 μmol/mg/min and a molecular mass of 2.1 × 106 Da.
Dilute-acid-pretreated wood prepared at 220°C.
Writing in 1988 in reference to adsorption of T. reesei cellulase to Avicel, Steiner et al. (641) made the observation that roughly 1 of 3,000 β-glucosidic bonds has the capacity to form an enzyme-substrate complex. The third column in Table 5 speaks to the generality of this important point. In particular, the ratio of enzyme-binding capacity per β-glucosidic bond present in the substrate is ≤2 × 10−3 for all enzyme-substrate combinations listed and ≤3 × 10−4 for enzymes most important in degrading crystalline cellulose (cellobiohydrolases and cellulosome-type complexes) adsorbed to Avicel or pretreated wood. These low values are probably due to limited accessibility associated with features of both the substrate and the enzyme. Substrate-associated inaccessbility involves β-glucosidic bonds that are inaccessible because they are covered by cellulose, hemicellulose, and/or lignin or because they are present in pores that are sufficiently small to prevent the passage of a cellulase molecule (223, 388, 464, 736). Enzyme-associated inaccessibility arises because the dimensions of cellulases and their binding domains greatly exceed the dimensions of the repeating cellobiose lattice unit on the cellulose surface (261), and so formation of an enzyme-substrate complex prevents additional complexes from being formed with β-glucosidic bonds covered by the bound cellulase. For example, Gilkes et al. (212) estimate that the number of cellobiose residues occupied by Cellulomonas fimi endoglucanases (CenA and CenX) in binding to BMCC is about 30. In addition to reduction of the binding capacity by a factor of ∼1/30 due to enzyme-associated inaccessibility, a further reduction in binding capacity by about 1/60 is necessary to result in the values listed in Table 5 for the overall ratio of cellulase-binding capacity as a fraction of β-glucosidic bonds for the C. fimi system [that is, (1/30) × (1/60) = 5.5 × 10−4]. It seems reasonable to assume that this further reduction is attributable to substrate-associated inaccessibility.
Whereas the ratio of enzyme-binding capacity to substrate reactive sites is ≤0.001 for most described cellulase-cellulose systems, this ratio is unity for most enzyme-catalyzed reactions involving soluble substrates. Because of this difference, it is unusual for substrate to be in excess during enzymatic hydrolysis of cellulose, although this is commonly the case for soluble substrates. Saturation of cellulosic substrates with cellulase is commonly observed in studies of cellulase adsorption (50, 71, 74, 437, 438, 439, 506, 681). Moreover, substrate is typically not in excess relative to cellulose-hydrolyzing activity (including cell-associated activity) in natural environments featuring microbial cellulose degradation (see “Uptake and phosphorylation of cellulose hydrolysis products” above). At cellulase loadings typical of engineered processes, it is commonly observed that free cellulase activity is present throughout the course of hydrolysis (24, 306, 506, 694, 773), which is indicative of accessible substrate sites not being in excess. As developed below and summarized in “Contrast to soluble substrates,” the relative rarity of substrate-excess conditions underlies several distinctive features of enzymatically mediated hydrolysis of cellulose compared to enzymatically mediated reactions involving soluble substrates.
Although usually not included in derivations for enzyme kinetics involving soluble substrates, it is important for cellulosic substrates to include a material balance on accessible cellulose binding sites:
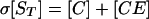 |
(4) |
where σ is the binding capacity of the substrate, corresponding to the density of accessible binding sites on cellulose to which enzyme could potentially bind (micromoles per gram), and [ST] is the concentration of substrate (grams per liter). The observation that substrate is usually not in excess during enzymatic hydrolysis of cellulose, discussed in the preceding paragraph, corresponds to the statement that [C] is in general not much higher than [CE]. Equation 4 may be substituted into equation 2 and solved for [CE] to give the Langmuir equation
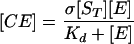 |
(5) |
A Langmuir equation of the general form of equation 5 is the most commonly used relationship to describe cellulase adsorption in the literature to date. It has been used to describe cellulase adsorption in studies involving individual components (44, 128, 332, 761), multicomponent noncomplexed systems (375, 506, 508, 716), and complexed systems (50) and for describing adsorption to lignocellulosic materials (50, 375, 506, 716), lignin (50, 506), and purified cellulose (44, 128, 332, 375, 508, 761).
Substitution of both enzyme and substrate material balances (equations 1 and 4) into the equilibrium constant (equation 2) gives a quadratic equation in [CE], as noted by several authors (71, 234, 641, 716):
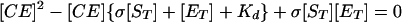 |
(6) |
Equation 6 has two roots, with the physically meaningful root being the one that satisfies the condition that 0 ≤ [CE] ≤ [ET] and σ[ST]. The values of [CE] given by the solution of equation exhibit the following important features. (i) Values for [CE] approach a constant value when either [ST] is increased and [ET] is held constant or [ET] is increased at constant [ST], representing saturation with either substrate or enzyme. Such dual saturation has been conclusively demonstrated in the literature for both noncomplexed (375, 641) and complexed (49) cellulases. (ii) The value of [ST] required to achieve an arbitrary extent of saturation in the concentration of [CE] (e.g., half the maximum concentration) is a function of [ET], with higher [ST] required at higher [ET]. This behavior is consistent with the adsorption study by Bernardez et al. (50) as well as the kinetic studies by Wald et al. (716), Lynd and Grethlein (406), and Bernardez et al. (49).
These two features differ sharply from the Michaelis-Menten model, which features saturation with substrate but not enzyme and a km that is independent of enzyme concentration. Indeed, it can readily be shown that in the limiting case where [ST] ≫ [ET], equation 6 reduces to the result implicit in the Michaelis-Menten equation:
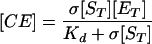 |
(7) |
However, the condition of excess substrate is uncommon for cellulose hydrolysis in most natural and engineered systems to which quantitative models might be applied, as discussed above. Thus, the Michaelis-Menten equation is for most purposes not useful for describing, understanding, or modeling cellulase enzyme systems acting on crystalline cellulose.
In cases where multiple cellulose-adsorbing components are present, description of adsorption behavior involves the simultaneous solution of several equations representing isotherms for the individual components (e.g., of the form of equation 6). Competitive adsorption among cellulase components has been described (366, 437, 578), and data have been interpreted in terms of some sites being common to different cellulase components while other sites are accessible only to a particular component (366, 578, 772). In some studies (332, 681), a positive interactive effect has been observed whereby more enzyme is adsorbed in the presence of multicomponent mixtures than when the components are present separately. Although the biochemical complexity of complexed cellulases is considerable, Bernardez et al. (49) have suggested that description and understanding of adsorption may be simplified relative to that in noncomplexed systems because of the dominance of adsorption via a single scaffoldin-like protein.
Cellulase enzyme components adsorb onto lignin as well as cellulose. Lignin acts as a competitive adsorbent for cellulase, with the result that rates of enzymatic hydrolysis are diminished by the presence of lignin even in the absence of any steric effect. Chernoglazov et al. (105) found that binding of cellulase to lignin can decrease the rate of hydrolysis by severalfold and can stop hydrolysis altogether before cellulose is exhausted. Sutcliffe and Saddler (657) found that β-glucosidase is adsorbed to lignin particularly strongly. Adsorption of unfractionated cellulase preparations from both T. reesei (506) and C. thermocellum (49) to dilute-acid-pretreated hardwood can be described by the Langmuir equation. For both of these systems, the fraction of cellulase adsorbed to cellulose rather than lignin approaches unity at low cellulase loadings but declines at higher cellulase loadings.
Notwithstanding the widespread use of the Langmuir equation and the fact that it is capable of describing some important general features of cellulase adsorption, the assumptions implicit in this equation are at best idealizations. In particular, binding sites on cellulosic substrates are thought to be nonuniform, and interactions among adsorbing molecules are thought to be common (437; also see below). To overcome these and other limitations, modifications of the Langmuir equation have been proposed, such as inclusion of two distinct types of binding sites (439, 637, 761) or a combined Langmuir-Freundlich model capable of describing negative or positive binding cooperativity (437, 439). Several studies have reported the formation of both loosely associated and tightly associated cellulase-cellulose complexes (346, 681). Suvajittanont et al. (658) have hypothesized that structural changes in cellulase enzymes occur upon cellulase adsorption.
The matter of adsorption reversibility, implicit in the Langmuir equation as well as in most dynamic representations of adsorption (e.g., equation 3), is far from clear. Release of bound cellulase is difficult to observe experimentally because of the high affinity of many cellulases for cellulose. Although careful studies with particular cellulases have clearly demonstrated fully reversible cellulase binding (72, 391), irreversible or less than fully reversible binding has been alluded to in recent (437, 658) as well as older (44, 489) studies.
Whereas most studies of cellulase adsorption have considered substrates that have not undergone appreciable reaction, the matter of adsorption behavior as the hydrolysis reaction proceeds has received some attention in the case of T. reesei cellulase preparations. Rapid initial adsorption followed by a steady return of cellulase to solution has been observed for both microcrystalline cellulose (461, 668) and pretreated lignocellulose (498, 506). An increase in the amount of adsorbed T. reesei cellulase preparation over the course of Avicel hydrolysis has also been reported (507). Ooshima et al. (506) found it justifiable to assume that the adsorption parameters for lignin present in mixed hardwood pretreated at 220°C did not change over the course of hydrolysis.
Quantitative description of the adsorption of microbial cells to cellulose-containing substrates involves expressing the concentration of cellulose-enzyme-microbe complexes, [CEM], as a function of variables relevant to microbial adsorption. This concentration could conceivably be defined in units reflecting the amount of substrate, enzyme, or microbial biomass involved in the complex. We suggest that is informative to define [CEM] in terms of enzyme units in order to facilitate comparison between the effectiveness of cellulase in microbial (involving CEM complexes) and cell-free (involving cellulase-enzyme complexes) contexts.
In contrast to the situation for cellulase adsorption, few quantitative data have been reported relative to the formation of CEM complexes. Cell adhesion has been associated with factors other than cellulase components expressed on the cell surface, including glycocalyx formation (see “Adhesion and formation of cellulose-enzyme-microbe complexes” above) and, in some species, the metabolic state of the cell (568). Thus, it does not appear promising to try to infer cell adhesion properties based solely on the adsorption properties of cellulase. The surface area accessible to cells probably excludes pores internal to substrate particles (733), although sites on such internal surfaces could presumably be accessed by cellulases released by an adherent cell. Saturation of barley straw with ruminal bacteria has been observed in the range of 23 or 33 mg of dry cells/g of straw, depending on the particular species considered (54). Batch growth curves of the rumen microorganisms R. flavefaciens and F. succinogenes (568), as well as C. cellulolyticum (207), exhibit a constant or increasing fraction of bound cells over the period during which growth and cellulose consumption occur. Based on this observation, description of microbial adsorption to cellulose in terms of a single affinity or desorption constant analogous to Kd appears to be unsatisfactory. This observation is also inconsistent with the hypothesis that the free cell concentration is zero until all available substrate sites are filled with excess cells accumulating in the medium thereafter.
Rates of Enzymatic Hydrolysis
Most models for the rate of enzymatic catalysis are based on the mathematical product of the concentration of the enzyme substrate complex and a proportionality factor relating this concentration to the reaction rate:
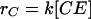 |
(8) |
where rC is the cellulose hydrolysis rate (substrate units/[volume × time]) and k is the rate constant, a proportionality factor between [CE] and rC (units as needed for dimensional consistency).
The previous section was concerned with how the concentration of the cellulose-cellulase complex(es) varies with respect to the relevant variables. This section is concerned primarily with the proportionality factor, k, which is a function of temperature and, in some models for cellulose hydrolysis, is also a function of additional state variables such as substrate conversion or enzyme age.
If there are multiple distinct enzyme-substrate complexes (e.g., in the case of different cellulase components), then one rate equation of the form of equation 8 must be written for each complex. In some models, multiple types of substrate sites are postulated; e.g., chain ends are differentiated from sites interior to the chain, which necessitates that one rate equation be written for each substrate type.
Models for the enzymatic hydrolysis of cellulose may also consider hydrolysis of one or more soluble oligosaccharides. The general form of the reaction rate equation for oligosaccharides, including cellobiose, is as follows: overall rate of reaction of Gj = rate Gj formed from cellulose hydrolysis + rate Gj formed from reaction of oligosaccharides of length >j − rate Gj reacts to form oligosaccharides with chain length <j, or
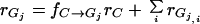 |
(9) |
where Gj is a soluble glucooligosaccharide of chain length j, rGj is the overall rate of formation of Gj (substrate units/volume/ time]), fC→Gj is the fraction of cellulose hydrolyzed directly to Gj, and rGj,i is the rate of formation of intermediate Gj by the ith reaction (+ if produced, − if consumed). If there are multiple soluble intermediates, one equation of the form of equation 9 must be written for each intermediate.
When the cellulase-cellulose complex is expressed in units of enzyme concentration, the parameter k represents the specific activity (rate/unit of enzyme) of the cellulase system. Table 6 presents data for specific activities for both multicomponent mixtures or complexes and components thereof with respect to Avicel, one of the most commonly used model substrates exhibiting a high degree of crystallinity; data for filter paper and pretreated birch are also included for the T. reesei system in order to facilitate comparison. It is important to note that specific activities measured during the initial stages of hydrolysis on a model substrate such as Avicel give an imperfect indication of the effectiveness of an enzyme preparation with respect to extended hydrolysis times and/or other substrates (3). The highest specific activities reported for multicomponent mixtures or complexes are higher than the highest reported specific activities for isolated components by 1.7-fold in the case of T. reesei (compare reference 174 and 773) and 159-fold in the case of C. thermocellum (compare references 5 and 465). The larger benefit of multiple protein components for the C. thermocellum system compared with T. reesei may be because most of the catalytically active components of the C. thermocellum cellulosome lack a cellulose-binding domain and are thus dependent on the noncatalytic dockerin protein for cellulose binding. The specific activity for CELA from Neocallimastix patriciarum is higher than that reported for the N. patriciarum complex and is by far the highest reported for an isolated component.
TABLE 6.
Specific hydrolysis rates for cellulosic substrates of high crystallinity in cell-free systems
| Organism | Cellulase preparation | Substrate and assay | Sp act (μmol/min/mg) | Reference | |
|---|---|---|---|---|---|
| Isolated component | |||||
| Clostridium cellulolyticum | CelA, expressed in E. coli and chromatographically purified | Avicel; 37°C, RS | 0.11 | 187 | |
| CelC, same | Avicel; 37°C | 0.017 | 186 | ||
| CelE, same | Avicel; 37°C | 0.06 | 206 | ||
| CelF, same | Avicel; 45°C | 0.17 | 564 | ||
| Clostridium thermocellum | Ion exchange-purified celA expressed in E. coli | Avicel; 60°C, RSa, 1 h | 0.083 | 607 | |
| Avicel (summarizes data for 8 components from various studies) | 0.0052-0.083 | 5 | |||
| Neocallimastix patriciarum | Immunoaffinity-purified CELA expressed in E. coli | Avicel; 40°C, RS, 30-60 min | 9.7 | 146 | |
| Crude recombinant CELA | Avicel; 40°C, RS, 30-60 min | 1.7 | 146 | ||
| Crude recombinant CELD | Avicel; 39°C | 0.18 | 767 | ||
| Ruminococcus flavefaciens | Chromatographically purified | Avicel; 39°C, RS | 0.049 | 249 | |
| Trichoderma reesei | Purified CBHI and CBHII | Avicel; 50°C, RS, 3 h | 0.014-0.027 | 682 | |
| Purified CBHI and CBHII | Avicel; 40°C, RS | 0.26-0.48 | 174 | ||
| Chromatographically purified, 5 components tested | Avicel; 30°C, RS, 20 h | 0.002-0.019 | 333 | ||
| Multicomponent mixtures and complexes | |||||
| Clostridum cellulolyticum | Partially purified cellulosome | Avicel | 0.2 | 199 | |
| Clostridium papyrosolvens | Size exclusion chromatography | Avicel | 0.037 | 539 | |
| Clostridium strain A11 | Affinity chromatography | Avicel; 34°C, RS (7) | 0.068 | 45 | |
| Clostridium thermocellum | Crude protein | Avicel, 60°C, 21 h | 0.0066 | 316 | |
| Size exclusion, affinity chromatography (2.1%) | Avicel | 0.1 | 350 | ||
| None (crude extracellular protein) | Avicel | 0.013 | 350 | ||
| Affinity digestion | Avicel; 60°C, RS | 13.2 | 465 | ||
| Affinity chromatography | Avicel; 60°C, RS | 6.0 | 465 | ||
| None (crude extracellular protein) | Avicel, 60°C, RS | 2.9 | 465 | ||
| Affinity digestion | Avicel; 60°C, RS 1 h | 2.4 | —b | ||
| Neocallimastix patriciarum | Crude protein | Avicel; 40°C, RS | 0.27 | 160 | |
| Piromyces strain E2 | Crude extracellular protein | Avicel; 40°C, RS | 0.26 | 160 | |
| Trichoderma reesei | Crude protein | Avicel; 37°C, 21 h | 0.000241 | 316 | |
| None (Celluclast, as received) | Avicel; 50°C, “initial rate” | 0.83 | 490 | ||
| None (crude broth) | Filter paper; 50°C, RS, 1 h | 0.6-0.8 | 177 | ||
| None (Celluclast, as received) | Petreated birch; 45°C, RS, 24 h | 1.0 | 773 | ||
| Chromatofocusing, reconstituted mixtures | Pretreated birch; 45°C, RS, 24 h | 0.34-1.16 | 773 |
RS, reducing sugars.
Y. Zhang and L. R. Lynd, submitted.
While some useful comparative observations can perhaps be made based on a compilation such as that presented in Table 6, a more obvious conclusions is that the results are highly variable even for the same organism and apparently similar cellulase preparations. Presumably this variability arises due to differences in assay conditions used by different investigators. In general, meaningful comparisons among different cellulase systems require side-by-side experiments in the same lab, which unfortunately are not common in the literature.
The idea that the specific activity of complexed cellulase systems is higher than that of noncomplexed cellulases, and T. reesei cellulases in particular, has often been mentioned in the literature (see, e.g., references 39, 160, 606, 621, 636, and 683). Most statements to this effect refer to the work of Johnson et al. (316) and/or Wood et al. (759), involving the cellulase systems of C. thermocellum and Neocallimastix frontalis, respectively. Johnson et al. reported a comparison of cellulase activity present in culture broth from C. thermocellum (0.2 mg of protein/ml) with that in reconstituted broth from T. reesei (9.5 mg of protein/ml). For both cotton and Avicel, rates of hydrolysis by C. thermocellum broth at 60°C were somewhat higher than rates of hydrolysis by T. reesei reconstituted broth at 50°C. These data have been interpreted to imply a 50-fold difference in specific activity (316, 683). The experimental design used in these experiments does not appear to eliminate the possibility that the reconstituted T. reesei broth contained cellulase at concentrations sufficiently high to saturate the substrate, which would result in a lower calculated specific activity. Further insight into this matter is provided by an unpublished experiment from Eric Johnson's thesis (reference 314, p. 63-65), which compared the hydrolysis of Avicel and phosphoric acid-swollen Avicel by broth dilutions from both organisms having the same amount of crude protein. In this experiment, the rate of Avicel hydrolysis was higher for C. thermocellum broth by a substantial margin (we estimate about sixfold based on initial rates). When combined with Johnson's estimates for the fraction of crude broth protein consisting of cellulase—30 to 35% for C. thermocellum and ≥85% for T. reesei—these data appear to imply that the specific activity of the C. thermocellum cellulase complex is about 15-fold higher than that of the T. reesei system. Remarkably, the rate of hydrolysis by the C. thermocellum system increased only slightly (we estimate about twofold based on initial rates) for hydrolysis of phosphoric-acid swollen cellulose compared to Avicel, whereas the hydrolysis by the T. reesei system was higher for phosphoric acid-swollen cellulose than for Avicel by over 50-fold.
Wood et al. (759) showed that an extracellular cellulase preparation from a coculture of the anaerobic fungus N. frontalis and the methanogenic Methanobrevibacter smithii exhibited roughly threefold-higher rates of cotton hydrolysis at 37°C than did a preparation from T. reesei at 50°C. However, an extracellular cellulase preparation from a pure culture of N. frontalis exhibited lower hydrolysis rates than did the T. reesei preparation. These comparative results were based on a consistent amount of endoglucanase activity (and not cellulase protein) for both systems. A subsequent study by Wilson and Wood (745) compared hydrolysis rates of cellulase preparations from C. thermocellum, N. frontalis, and T. reesei acting on cotton over a 10-fold range of protein concentrations. For all but the lowest protein concentrations tested, the specific activity of both complexed cellulases was higher than that of the noncomplexed T. reesei cellulase by roughly an order of magnitude. Solubilization was greater for the preparation from N. frontalis compared to the preparation from C. thermocellum for every protein concentration tested, although differences between these systems were twofold or less.
These results provide strong support for the notion that the specific activity of crystalline cellulose hydrolysis on a per-unit-cellulase-protein basis is higher for at least some complexed systems compared to the T. reesei system. The number of studies that speak directly to this point is, however, limited, and thus extensive data are not available with respect to variables such as sources of enzymes, substrates, enzyme preparation method, enzyme/substrate ratios, and extent of reaction. In light of this, the magnitude of the specific activity difference between complexed and noncomplexed system is uncertain, in our view, and would benefit from further study.
It is relevant to consider the hydrolytic activity of cellulase in comparison to other enzymes. The specific activity of T. reesei cellulase as measured by standard assays with crystalline cellulose as the substrate is about 100-fold lower than that of amylase (422). Klyosov (346) calculates kcat values, corresponding to k values in equation 8 when [CE] is expressed in moles, of 0.5 to 0.6 s−1 for T. reesei cellulase, 58 s−1, for amylase, and up to 100 to 1,000 s−1 for other hydrolases. The effectiveness of cellulase is further reduced because actual rates in extended hydrolysis are much lower than the initial rates measured in standard assays (see below). Because of these factors, the amount of cellulase required to achieve reasonable rates for practical applications can be substantial, e.g., 1.5 to 3% by mass of the initial amount of cellulose for the T. reesei cellulase system (422).
Notwithstanding the effect of lignin in terms of both sterically impeding access of enzymes to cellulose (see “Rate-limiting factors in nature” above) and acting as a competitive adsorbant (see the previous section), it is not correct to assume that higher rates will necessarily be observed on purified cellulosic substrates than on pretreated lignocellulose. For example, Bernardez et al. (49) compared initial hydrolysis rates for Avicel and dilute-acid-pretreated mixed hardwood using cellulase contained in cell-free broth from C. thermocellum. Pretreated wood was hydrolyzed up to 10-fold faster than Avicel at high enzyme loadings, consistent with the differences in enzyme adsorption capacity for these two substrates (Table 5).
The rate of enzyme-mediated hydrolysis of cellulose is inhibited by products of hydrolysis and is also potentially inhibited by fermentation products if hydrolysis and fermentation are carried out at the same time (see “Process configuration” below). As reviewed elsewhere (470, 533), cellulose hydrolysis is inhibited by cellobiose and to a much lesser extent by glucose for cellulase from both Trichoderma spp. and C. thermocellum. β-Glucosidase in T. reesei is highly sensitive to inhibition by glucose. Whether inhibition by soluble hydrolysis products is important for microbial cellulose utilization depends on whether such products accumulate in the microenvironments in which hydrolysis occurs, as discussed further below (see “Kinetics of microbial cellulose utilization”). Ethanol is less inhibitory to cellulose hydrolysis than is cellobiose by an order of magnitude in both Trichoderma spp. (533) and C. thermocellum (49).
Although initial rates are often used for biochemical characterization, it is of interest from both fundamental and applied perspectives to understand and describe the enzymatic hydrolysis of cellulose over the entire course of reaction—that is, over conversion values from 0 to 1, where conversion, χ, is defined by
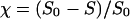 |
(10) |
where S0 is the substrate concentration at time t = 0 (batch) or entering (continuous) and S is the substrate concentration at time t > 0 (batch) or exiting (continuous). It may be noted that high values of χ are characteristic of most cellulose particles in both natural and engineered environments. A near-universal feature of cellulose hydrolysis observed in many studies over several decades is that the rate declines sharply as the reaction proceeds (corresponding to increasing values of χ) in a batch hydrolysis. Measurements of rate in conjunction with adsorbed enzyme (152, 498, 507) confirm that the phenomenon of declining rate with increasing conversion is observed on a specific (rate per adsorbed enzyme) as well as absolute basis.
Enzyme inactivation due to thermal effects (91, 117, 219), formation of an inactive enzyme-substrate (lignin) complex (234, 506, 657), and inhibition by hydrolysis products (91, 233, 377) have been implicated as important factors underlying the decreasing-rate phenomenon. However, it is significant to observe that this phenomenon has been documented in studies in which neither inactivation nor inhibition appears operative (693, 776).
Several studies have attributed declining rates of hydrolysis to a corresponding change in substrate reactivity. One subset of these studies postulates two types of cellulose that differ in their susceptibility to enzymatic attack (219, 289, 488, 536, 597, 716). While this “two-substrate” hypothesis cannot be rejected based on the literature to date, it also appears that the difference between the more reactive and less reactive substrate fractions is attributable primarily to factors other than crystallinity. If this difference were due to crystallinity, then cellulose crystallinity should increase over the course of reaction. However, relatively constant crystallinity over the course of enzymatic hydrolysis has been observed in studies involving a variety of cellulase systems (149, 178, 201, 374, 381, 547), although such crystallinity measurements may be due to artifacts (730).
A second subset of studies feature a continuous decline in substrate reactivity rather than two distinct substrate types. Working with pretreated poplar and the T. reesei system, Nutor and Converse (498) found that the rate of cellulose hydrolysis per adsorbed cellulase decreased monotonically by 1 to 2 orders of magnitude over the course of reaction. South et al. (633) used a conversion-dependent rate constant to represent the declining specific activity of the cellulase-cellulose complex over the course of simultaneous saccharification and fermentation of pretreated hardwood, using a commercial T. reesei cellulase preparation:
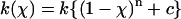 |
(11) |
The best-fit value of the exponent n was found to be 5.3, providing a further indication of the strong functional dependence of rate on conversion. South et al. tested but rejected a constant-reactivity model as an alternative to equation 11. Velkovska et al. (713) used equation 11 to model the rate of cellulose hydrolysis during growth of T. reesei on Solka Floc. A mean value for n of 6.1 gave the best fit to the data. The strong declines in specific hydrolysis rate observed by both South et al. (633) and Velkovska et al. (713) were obtained in the presence of a sugar-consuming micoorganism and at temperatures (37 and 28°C, respectively) unlikely to result in thermal denaturation of cellulase. Working with purified cellulase components from T. fusca, Zhang et al. (776) concluded that substrate heterogeneity causes the nonlinear kinetics exhibited during hydrolysis of filter paper whereas product inhibition and enzyme inactivation were rejected as explanations for this phenomenon. Product inhibition was ruled out because cellulase activity was not stimulated by β-glucosidase. Several lines of evidence supported a decline of substrate reactivity rather than enzyme deactivation, and a nearly threefold increase in the activation energy was observed for cellulose with conversion = 0.24 compared to conversion = 0.
Several groups have undertaken “restart” experiments wherein a partial hydrolysis is conducted, cellulase is removed, and the hydrolysis rate is measured on addition of new enzyme. As summarized in Table 7, results from restart experiments have been used as a basis to both confirm (488, 776) and reject (152, 233, 507) declining substrate reactivity as a primary factor responsible for the declining rate phenomenon. Gusakov et al. (233) explain declining rates in terms of deactivation of substrate-bound cellulase and product inhibition. The explanation of Ooshima et al. (507) is that synergistic interaction between cellulase components becomes less effective with increasing conversion. Väljamäe et al. (693) attribute the rate decline to steric hindrance due to nonproductive cellulose binding in combination with surface erosion. It may be noted that essentially all detailed investigations of the declining-rate phenomenon have been based on noncomplexed cellulases and that features of such systems may or may not be generalizable to other systems. Constant hydrolysis rates were observed under some conditions for the C. thermocellum cellulase system at conversion values up to 0.7 for pretreated wood and 0.4 for Avicel (406), although rates that decline with time in the usual fashion were observed under other conditions.
TABLE 7.
Summary of data from “restart” experiments
| Enzyme source (purification) | Substratea | Factors believed operative in uninterrupted hydrolyses
|
Declining rate observed upon restart? | Reference | |
|---|---|---|---|---|---|
| Inactivation | Inhibition | ||||
| Trichoderma longibrachiatum (unpurified) | CT cotton stalks | Yes | Yes | No | 233 |
| Trichoderma viride (unpurified) | Microcrystalline cellulose | No | Yesb | No | 507 |
| Celluclast (unpurified) | Microcrystalline cellulose | No | Yesb | Yes | 488 |
| Trichoderma reesei (unpurified) | Dilute-acid-pretreated MH | ? | ? | Slight | 152 |
| Thermobifida fusca (purified components) | Filter paper and BMCC | No | No | Yes | 776 |
| Trichoderma reesei (purified components) | BMCC | No | No | Yes | 693 |
BMCC, bacterial microcrystalline cellulose; CT, chemically treated; MH, mixed hardwood.
Product inhibition, although observed, was not deemed responsible for the declining rate observed.
For quantitative modeling of enzymatic hydrolysis of cellulose during simultaneous saccharification and fermentation in continuous well-mixed reactors, South et al. (633) found it necessary to integrate reaction rates over the time individual particles, or segments of the particle population, spend in the reactor according to the following equation:
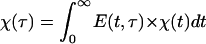 |
(12) |
where τ is the mean residence time, χ(τ) is the substrate conversion as a function of τ, E(t,τ) is the particle residence time distribution for a CSTR = e(t/τ)/τ, and 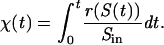
When parameters to equation 11 fit to batch hydrolysis data were used to predict steady-state continuous hydrolysis data using equation 12, essentially no increase in model error was observed. By contrast, if substrate reactivity was evaluated at the exit conversion, as is routinely done for analysis of soluble substrates in well-mixed continuous reactors, the model error was more than fivefold greater than that obtained using equation 12. These results were interpreted to mean that the reactivity of pretreated hardwood is a function of conversion as well as concentration under these conditions.
Bioenergetics of Microbial Cellulose Utilization
Distinctive features of microbial cellulose utilization compared to microbial utilization of monomeric sugars that are potentially important in the context of bioenergetics include (i) the metabolic burden represented by cellulase synthesis, (ii) potential net ATP gain as a result of phosphorolytic rather than hydrolytic cleavage of cellodextrins, (iii) bioenergetic demands for transport of cellulose hydrolysis products, and (iv) the metabolic burden represented by glycocalyx synthesis. The quantitative importance of these features are addressed in the following paragraphs; qualitative aspects are addressed above (see “Physiology of cellulolytic microorganisms”).
Several lines of evidence suggest that ATP allocation to cellulase synthesis is often a significant fraction of the ATP allocated to cell synthesis for both aerobic and anaerobic cellulolytic microorganisms. In particular, because specific activity is much lower for cellulase than for most other catabolic enzymes (see the previous section), the relative allocation of carbon and ATP to cellulase synthesis can be expected to be correspondingly larger for growth on cellulose as opposed to allocation of cellular resources to catabolic enzymes required for growth on soluble substrates. In the case of aerobic cellulase utilization, extensive data compiled by Esterbauer et al. (177) indicate that cellulase yields of 200 filter paper units (FPU) per g of cellulose and specific activities of 600 FPU/g of protein are typical of studies of extracellular cellulase production by T. reesei. These data imply a YE/S value of 0.33. Since it is unlikely that cell yield values substantially exceed these values, and since synthesis of 1 g of protein requires more ATP than synthesis of 1 g of cells (577), it may be inferred that the ATP allocated to cellulase synthesis is of a magnitude similar to that allocated to cell synthesis in T. reesei. For anaerobic growth on cellulose, modeling studies indicate that the rate of cellulose hydrolysis is maximized when about one-third of the discretionary ATP expenditure (excluding maintenance) is allocated to cellulase synthesis (710). This optimal ATP allocation is similar over a large range of values of assumed cellulase specific activity. Only limited data relevant to comparative evaluation of cell and cellulase yields are available, in part because of the difficulty of independently quantifying cell and cellulase production in many systems (see “Quantification of cells and enzymes is the presence of solid substrates” above).
Additional ATP beyond that obtained from catabolism of monosaccharides is potentially available to cellulolytic microorganisms as a result of the action of cellobiose phosphorylase (CbP) and cellodextrin phosphorylase (CdP) (discussed above [see “Uptake and phosphorylation of cellulolose hydrolysis products”]). The ATP gain accompanying phosphorolytic cleavage of a cellodextrin of length n is equal to n/(n − 1) and thus goes from zero for n = 1 to an asymptotic value of 1 as the cellodextrin length increases (Fig. 4). Evidence that the potential bioenergetic benefit of CdP and CbP is in fact realized to a significant extent comes from estimations of the cell yield based on measurement of optical density or protein synthesis. Observed cell yields are >30% higher on cellobiose than on glucose for C. thermocellum (650) and R. albus (680). In addition, cell yield increases of at least 25% have been observed to accompany growth on cellodextrins with chain lengths of >2 compared to cellobiose in the case of C. thermocellum (650) and R. albus (394). The true cell yield of C. cellulolyticum grown in continuous culture is 40% higher on Avicel than on cellobiose (156). In addition to these data for anaerobes, it may be noted that cellodextrin phophorylase activity has been detected in the aerobes Cellvibrio gilvus (293) and Cellulomonas fimi (600) and that increasing cell yield with increasing cellodextrin chain length has been reported for C. gilvus (612).
FIG. 4.
Effect of chain length (number of glucosyl units) on the ATP expenditure required per mole of glucosyl unit transported and on the potential ATP benefit per mole of glucosyl unit arising specifically from phosphorolytic activation via intracellular cellobiose phosphorylase and cellodextrin phosphorylase. The calculations assume a requirement for transport of 1 mol of ATP per mol of sugar regardless of chain length (see the text).
The bioenergetic cost of substrate transport is thought to be constant per transport event and thus independent of oligosaccharide chain length in E. coli (473). It has been proposed that this is also the case for C. thermocellum (650) and C. cellulolyticum (157). The ATP expenditure per hexose equivalent transported is equal to α/n, where α is the number of moles of ATP expended (directly or indirectly) per transport event and n is the cellodextrin chain length. As a result, this expenditure goes from α at n = 1 and to an asymptotic value of 0 as the cellodextrin chain length increases (Fig. 4). As discussed above (see “Uptake and phosphorylation of cellulose hydrolysis products”), ABC transport systems are thought to be used for cellodextrin transport by C. thermocellum and C. cellulolyticum but not F. succinogenes. Expenditure of 1 ATP per transport event, corresponding to α = 1, has been hypothesized for cellodextrin transport in C. thermocellum (650) and C. cellulolyticum (227); α = 2 has also been hypothesized for C. cellulolyticum (157). An alternating two-site mechanism involving one ATP hydrolyzed per transport event has been proposed based on detailed studies of both the LmrA ABC transporter in Lactococcus lactis (709) and the P-glycoprotein of humans (610). It has been hypothesized that this mechanism may also be applicable to other ABC transport systems (610, 709).
The magnitude of the potential bioenergetic benefit associated with growth on cellodextrins as opposed to glucose corresponds to the ATP gain accompanying phosphorolysis minus the difference between the ATP expenditure for monomer transport and the ATP expenditure for cellodextrin transport. This quantity may be visualized from Fig. 4 and may also be calculated as follows:
 |
(13) |
where f is the fraction of substrate cleaved phosphorolytically rather than hydrolytically, n is the cellodextrin chain length, and α is the number of moles of ATP expended (directly or indirectly) per transport event. By way of illustration, consider the hypothetical example of growth on cellopentaose (n = 5) relative to growth on glucose for the case where cellodextrin cleavage is entirely phosphorolytic (f = 1) and 1 ATP is expended per transport event (α = 1). For these values, the potential bioenergetic benefit for growth on cellopentaose relative to glucose, as calculated using equation 13, is 1.6 ATP/mol of hexose. Yet larger benefits result if α = 2 is assumed. While benefits of this magnitude are small compared to the ATP available from oxidative phosphorylation, they are large relative to the ATP available for anaerobic cellulolytic microorganisms with energy conservation via substrate-level phosphorylation. In particular, 1.6 ATP/mol of hexose corresponds to about a 50% increase in the total catabolic ATP available to a fermentative anaerobe (see “Metabolic engineering” below) and is also roughly commensurate with estimates for the ATP expended on cellulase synthesis, as discussed earlier in this subsection.
In comparison to growth on a monosaccharide, growth on cellulose affords opportunities for bioenergetic benefits via the action of phosphorylases and more efficient substrate transport but also incurs an additional bioenergetic cost for synthesis of cellulase and perhaps glycocalyces. As discussed above, available data are not inconsistent with the hypothesis that the magnitude of the bioenergetic benefit associated with growth on cellulose is equal to or in excess of the bioenergetic cost. Definitive testing of this hypothesis, however, requires further study.
Quantitative information on the composition and yields of glycocalyx formed by cellulolytic anaerobes is in general not sufficient to evaluate the bioenergetic cost of glycocalyx formation. A primary question in this context is the relative importance of de novo glycocalyx synthesis compared to nonspecific accretion. De novo synthesis as used here involves intracellular synthesis of glycocalyx components followed by secretion of these components into the cell's immediate surroundings. Accretion involves incorporation of glycocalyx components consisting of cellulose hydrolysis products that have not yet entered the cell. This distinction is potentially important with respect to bioenergetics because de novo-synthesized glycocalyx components are expected to exert an ATP demand whereas accreted components would not be expected to exert such a demand.
Lynd and Zhang (411) have proposed that the rates of ATP supply and demand during fully coupled microbial growth on cellulose may be described by an equation of the form
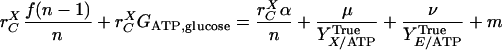 |
(14) |
where rCX is the moles of cellulose monomer hydrolyzed per gram of cells per hour, GATP,glucose is the moles of ATP from glycolysis and postpyruvate metabolism per mole of hexose, YX/ATPTrue is grams of cells per mole of ATP allocated to growth, YE/ATPTrue is grams of cellulase per mole of ATP allocated to cellulase synthesis, μ is the specific growth rate (reciprocal hours), ν is the specific cellulase synthesis rate (grams of cellulase per gram of cells per hour), and m is the maintenance ATP utilization rate (moles of ATP per gram of cells per hour). The first term in equation 14 represents the rate of ATP generation as a result of phosphorylytic cellodextrin cleavage, and the second term represents the rate of generation via glycolysis and postpyruvate metabolism. The third term, on the right of the equal sign, represents the rate of ATP expenditure for substrate transport. The fourth, fifth, and sixth terms represent rates of ATP expenditure for growth, cellulase synthesis, and maintenance, respectively. The first and fifth terms are specific to growth on cellulose, as are the magnitude and form of the third term. An additional term could be added for glycocalyx production if associated ATP demand were found to be significant.
Higher maintenance coefficients have been observed for glucose than for cellobiose in both C. thermocellum (650) and R. albus (680). At low growth rates (e.g., 0.02 h−1) of the cellulolytic Clostridium strain C7 on glucose, increased maintenance energy associated with elevated levels of cAMP and ppGpp was found to significantly reduce the rate of cell synthesis with little effect on the rate of exoenzyme synthesis (142). In F. succinogenes, glycogen storage and cycling appear to play a significant role in the energy economy of the cell and to be associated with sensitivity to cell lysis (738, 739). Glycogen formation and exopolysaccharide formation have also been noted in C. cellulolyticum grown on cellulose (157, 158). An important limitation of bioenergetically oriented studies of microbial cellulose utilization to date is that nearly all such studies have been carried out with soluble substrates. We attribute this primarily to the methodological challenges described above (see “Quantification of cells and enzymes in the presence of solid substrates”).
Kinetics of Microbial Cellulose Utilization
Cellulose hydrolysis limits the rate of microbial cellulose utilization under most conditions, as may be inferred from the observation that maximum growth rates on soluble sugars are usually several-fold faster than on crystalline cellulose. Consistent with hydrolysis being rate limiting, concentrations of soluble sugars resulting from cellulose hydrolysis are usually vanishingly small (≤0.5 mM or undetectable) in cellulose-fed systems that approach or achieve steady state. Such systems include chemostats involving both pure cultures (409, 516, 727) and mixed cultures (111) as well as the rumen (114, 320, 537). Soluble sugars are commonly observed to accumulate in batch cultures of cellulolytic bacteria, but this is in most cases probably due to continuing activity of cellulase enzymes after growth ceases (156).
The specific growth rate, μ, may be defined as
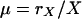 |
(15) |
where rX is the rate of cell formation (gram of cells per liter per hour) and X is the cell concentration. Microbial utilization of soluble substrates in batch cultures is often qualitatively described as having an “exponential” growth phase. However, an exponential increase in cell concentration with time is reasonable to expect only if the rate of increase of the cell concentration is proportional to the cell concentration, which requires that substrate be in excess and that substrate reactivity be constant through most of the batch growth curve. Although both of these conditions are met during the exponential phase of growth on a soluble substrate, it is unusual for either of them to be satisfied (let alone both of them) for growth on cellulosic substrates (see “Adsorption” and “Rates of enzymatic hydrolysis” above). Thus, there is little basis to expect exponential growth in batch cultures of bacteria growing on cellulose over any significant range of conversion values, and this has not been conclusively reported to our knowledge. The growth rate may be determined using equation 15 by estimating rx from differential batch data or, more reliably, from continuous culture data.
At the other extreme from exponential growth with cell concentration implicitly assumed to be limiting and substrate in excess, substrate concentration may be assumed to be limiting with cellulase and cells in excess.
In its simplest form, this latter assumption leads to first-order expressions for the rate of substrate utilization and growth in a batch culture such as equations 16 and 17:
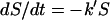 |
(16) |
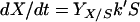 |
(17) |
where k′ is an empirical constant.
There is more experimental evidence to support cellulose hydrolysis being first-order in cellulose under at least some conditions as compared to the idea that microbes grow exponentially on cellulose.
It may be noted that these two possibilities are mutually exclusive. A first-order dependence of the rate of cellulose hydrolysis on cellulose concentration has been documented for pure and naturally occurring, but not pretreated, cellulosic substrates in batch cultures of mixed ruminal microorganisms (733) and anaerobic digester microflora (684). Such first-order dependence has also been observed in continuous pure cultures of R. flavefaciens (735) and R. albus (517) grown on microcrystalline cellulose and in continuous culture of T. reesei grown on ball-milled wood (535). However, the rate constant k′ has also been observed to decline with increasing conversion in microcrystalline cellulose-grown continuous cultures of R. flavefaciens (617) and F. succinogenes S85 (727) and in cultures of C. thermocellum grown on pretreated wood (407).
It is somewhat difficult to reconcile the observation of first-order hydrolysis kinetics for microbial cellulose utilization with known information about cellulose hydrolysis in cell-free systems (see “Rates of enzymatic hydrolysis” above). First, this would seem to require that the concentration of accessible substrate sites changes in proportion to the overall substrate concentration over the course of reaction, which has not been established in general. Second, first-order kinetics is not consistent with declining reactivity of cellulose with increasing conversion. In light of the significant evidence supporting such a decline in cell-free hydrolysis (see “Rates of enzymatic hydrolysis” above), it might be expected that this phenomenon would also be observed for microbial cellulose utilization. This, however, remains to be conclusively shown. Third, first-order kinetics are mechanistically defensible only under conditions where cellulase, or alternatively microbes with cellulase expressed on the cell surface, is entirely in excess. Conditions where neither substrate nor enzyme are in excess—that is, where the rate would increase if either the substrate or cellulase concentration were increased—are common for cell-free systems (see “Adsorption” above) and have also been documented for microbial cellulose utilization of pretreated substrates in continuous culture (407). Most if not all experimental data which show a first-order dependence of the cellulose hydrolysis rate with respect to cellulose concentration come from either nonpretreated substrates or model substrates such as Avicel, both of which have lower capacity to adsorb cellulase than is typical of pretreated substrates (50).
Table 8 presents values for the specific growth rate and first-order rate constant for microbial cellulose utilization. As with Tables 5 and 6, caution is appropriate when making comparative observations, since the data in Table 8 were not obtained under identical conditions with respect to apparatus and substrate. Subject to this potentially important limitation, available data are consistent with the following observations.
TABLE 8.
Kinetic parameters for microbial cellulose utilization
| Organism | Substrate, cultivation mode | Parameter value (h−1)
|
Reference | |
|---|---|---|---|---|
| Specific growth ratea | First-order rate constant | |||
| A. thermophilum | Microcrystalline cellulose, batch, 74°C | 0.4 (max, obs) | 659 | |
| Cellulomonas uda ATCC 21399 | Avicel, batch, 30°C | 0.027b | 148 | |
| Cellulomonas flavigena JC3 | Avicel, batch, 35°C | 0.006b | 139 | |
| C. cellulolyticumATCC 35319 | MN301, batch, 34°C | 0.057 (max, obs) | 157 | |
| C. cellulolyticum ATCC 35319 | MN301, continuous, 34°C | 0.083 (max, obs) | 0.05 | 157 |
| C. thermocellum ATCC 27405 | Avicel PH105, continuous, 60°C | 0.17 (max, obs) | 0.16b | 407 |
| C. thermocellum ATCC 27405 | Pretreated hardwood, continuous, 60°C | 0.13 (max, obs) | 407 | |
| F. succinogenes S85 | Sigmacell 20, continuous, 39°C | 0.076 (max, obs) | 0.07 | 727 |
| F. succinogenes S85 | Ball-milled filter paper, continuous, 39°C | 0.38 (max, obs) | 338 | |
| R. albus 8 | Avicel, continuous | 0.095 (max, obs)c | 0.05 | 517 |
| R. flavefaciens FD-1 | Sigmacell 20, continuous, 39°C | 0.10 (max, obs) | 0.08 | 617 |
| R. flavefaciens FD-1 | Ball-milled filter paper, continuous, 39°C | 0.51 (max, obs) | 338 | |
| Thermonospora sp. strain N-35 | Avicel, continuous, 55°C | 0.23 (max, extrap) | 445 | |
| T. reesei QM 9414 | Cotton, continuous, 28°C | 0.028 (max, obs) | 210 | |
| T. reesei Rut C30 | Solka Floc BW 200, 28°C | 0.077 (max, obs) | 250 | |
| T. reesei Rut C30 | Solka Floc 200, batch, 28°C | 0.125 (max, fitted) | 713 | |
| T. reesei QM 9414 | Ball milled cellulose, continuous, 30°C | 0.17 (max, extrap) | 0.1c | 535 |
| T. reesei QM9123 | Glucose, continuous, 30°C | 0.15 (max, extrap) | 84 | |
| Undefined acidogenic mixed culture from digester innoculum | Crystalline cellulose (20 μm), continuous, 35°C | 0.042 (max, obs) | 0.019c | 111 |
“Max, obs” corresponds to the reported growth rate for batch cultures studies and the highest reported steady-state dilution rate for continuous-culture studies. “Max, fitted” corresponds to the maximum growth rate observed in several runs with the value determined by parameter fitting. “Max, extrap” corresponds to the maximum growth rate extrapolated to zero conversion.
Rate constants calculated by Weimer (728) based on data from the references indicated.
Constant calculated for this work based on data from the references indicated.
(i) More reactive substrates such as ball-milled cellulose and Solka Floc support higher rates of growth and cellulose hydrolysis than do more recalcitrant substrates such as Avicel and cotton.
(ii) The three thermophilic cellulolytic microorganisms listed in the table, C. thermocellum, Thermomonospora sp. strain N-35, and A. thermophilum, exhibit substantially higher growth rates on cellulose than do any of the mesophiles listed on comparably reactive substrates. Available data exhibit a general trend of increasing growth rates on crystalline cellulose as a function of temperature, as shown in Fig. 5.
FIG. 5.
Relationship between growth temperature and maximum specific growth rate constant for aerobic (open circles) and anaerobic (solid circles) microorganisms grown on crystalline cellulose (r2 = 0.90). Data are from references 157, 210, 407, 445, 517, 617, 659, and 727.
(iii) Growth rates and first-order rate constants do not in general appear to be higher for aerobic cellulose utilizers (Thermomonospora spp., Cellulomonas spp., and T. reesei) than for anaerobic cellulose utilizers.
(iv) Although T. reesei is distinguished by being a prodigious producer of extracellular cellulase, there are no data to suggest that it exhibits unusually high rates of growth on cellulosic substrates. While not entirely conclusive, available data suggest that this organism grows rather slowly in submerged culture (which is not representative of its growth environment in nature) relative to other cellulolytic organisms on comparably reactive substrates.
It is surprising that the first-order rate constant for crystalline cellulose degradation by undefined acidogenic mixed cultures at mesophilic temperatures in the thorough study by Chyi and Dague (111) is generally lower than values for pure cultures of mesophiles growing on comparably reactive substrates.
The absence of an apparent difference between the specific growth rates mediated by anaerobes and aerobes, observation (iii) above, has implications with respect to the cell-specific cellulose hydrolysis rate, rSX (grams of cellulose per gram of cells per hour), which may be expressed as
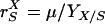 |
(18) |
Because cell yields of anaerobes growing on carbohydrates are typically severalfold lower than those for aerobes (577), it follows from equation 18 that if anaerobes and aerobes have similar growth rates on cellulose—as appears to be the case—the cell-specific cellulose hydrolysis rate is severalfold higher for anaerobes. This may also prove to be true with respect to the cellulase-specific hydrolysis rate mediated by CEM complexes in the presence of cellulolytic microbes, although available data do not speak directly to this point.
Models of microbial cellulose utilization that are more detailed than exponential growth or first-order substrate utilization, whether conceptual or quantitative, incorporate to various degrees our understanding embodied in models of adsorption of cellulase and/or cells (see “Adsorption” above), kinetics of enzymatic hydrolysis of cellulose (see “Rates of enzymatic hydrolysis” above), and bioenergetics (see “Bioenergetics of microbial cellulose utilization” above) and our understanding of relevant fundamentals (see “Fundamentals” above). For microbial cellulose utilization in general, the overall rate of cellulose hydrolysis is the sum of the rates due to hydrolysis associated with CE complexes and the rates due to hydrolysis associated with CEM complexes. For particular systems either CE-mediated or CEM-mediated hydrolysis may predominate. It would seem appropriate to allow for the possibility that the catalytic efficacy of cellulase in CE and CEM complexes may not be the same (discussed below). The overall rate of formation of an oligomer of length j is as considered above for enzymatic hydrolysis (equation 9) with the addition of microbial uptake. While separate substrate-enzyme and substrate-enzyme-microbe complexes might be hypothesized for oligosaccharides as well as for cellulose, this added complexity may not be warranted in light of the observation that oligosaccharide hydrolysis is usually not rate limiting. The overall rate of cell formation is the sum of the rates of uptake of oligomers of various lengths multiplied by the cell yield for that oligomer. Similarly, the overall rate of cellulase formation is the sum of the rates of uptake of oligomers of various lengths multiplied by the cellulase yield for that oligomer. The relative synthesis of cells and cellulase is determined by metabolic control mechanisms subject to bioenergetic constraints which are in part a function of the relative uptakes for oligomers of different lengths (see “Bioenergetics of microbial cellulose utilization” above). This line of thinking can be expressed symbolically by the following equations:
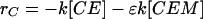 |
(19) |
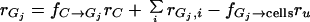 |
(20) |
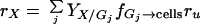 |
(21) |
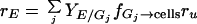 |
(22) |
where ɛ is an “enhancement factor” reflecting the ratio of the hydrolysis rate for a given amount of enzyme in a CEM complex to the hydrolysis rate of the same amount of enzyme in a CE complex, ru is the overall rate of microbial uptake of carbohydrates (substrate/volume/time), and rE is the rate of cellulase formation (enzyme/volume/time). Under conditions where oligomers do not accumulate, either because they are consumed as fast as they are formed or because the system is at steady state, the rate of cellulose hydrolysis, rC, is equal to the rate of carbohydrate uptake, ru. Equations 19 to 22 may be incorporated into reactor models by using an appropriate material balance, including for continuous systems consideration of the distribution of particle reactivities (see equation 12).
Both first-order substrate utilization and exponential growth models for microbial cellulose utilization are unstructured in the sense that the state of the biological phase (cells and other anabolites) is characterized by a single variable. By contrast, the framework represented by equations 19 through 22 is a structured model because the biophase is described by more than one variable: E and X. Models in which cells, enzymes, or both are described by more than one variable are also possible but will have features in common with equations 19 through 22.
Equations 19 though 22, together with the discussion in preceding sections, provide a framework for identifying key areas where understanding of microbial cellulose utilization is incomplete. Areas where the current state of understanding is particularly limiting in the context of quantitative description based on structured models include (i) adsorption of cells to cellulose; (ii) the relative effectiveness of cellulase when present in CEM complexes compared to CE complexes; (iii) the distribution of cellodextrin chain lengths taken up by cellulolytic microbes; and (iv) the yields of cells and cellulase, measured independently, and how these depend on growth conditions.
Enhanced effectiveness of cellulase enzymes when present as CEM complexes compared to CE complexes has been suggested in the literature. Such enhancement corresponds to ɛ > 1 in equation 19 and may be thought of as “enzyme-microbe synergy” by analogy to the “component-component synergy” that is well documented for cell-free cellulase systems (see “Cellulase enzyme systems” above). Reese (561) and Reese and Mandels (562) observed that rates of hydrolysis are substantially higher when mediated by growing cultures of T. reesei than are those for cell-free enzyme preparations even under conditions optimized for hydrolysis. It was hypothesized that the intimate association of hyphae with cellulose accounts for the higher hydrolysis rate in the growing culture (562). In the case of C. thermocellum, it has been proposed that cellulosome-containing protuberances may act as contact corridors channeling the diffusion of soluble degradation products from the cellulose fibers to the cells (39, 621).
With or without the formation of protuberances, the presence of cells on the surface of cellulose fibers can be expected to alleviate inhibition of cellulolysis by hydrolysis products. In the absence of cells located on the surface of cellulose fibers, transport of soluble hydrolysis products away from the fiber is driven by diffusion through the stagnant boundary layer surrounding a cellulose particle. Since diffusive transport requires a concentration gradient, the concentration of hydrolysis products at the fiber surface is higher than that in the bulk solution. In the presence of cells located on the surface of cellulose fibers, the concentration of hydrolysis products in the vicinity of the fiber will be lower due to both smaller diffusion distances and the locally high rate of uptake by adherent cells. This anticipated effect is over and above the decreased product inhibition observed in simultaneous saccharification and fermentation (SSF) with respect to separate hydrolysis and fermentation (SHF) (763), which is not associated with the presence of adherent cells in boundary layers. Alleviation of inhibition might be further enhanced if cells take up >G2 cellodextrins directly (see “Physiology of cellulolytic microorganisms” above), thus avoiding the formation of cellobiose and glucose, which are known inhibitors of cellulose hydrolysis (see “Rates of enzymatic hydrolysis” above). Recent studies have emphasized the importance of equilibrium and mass transfer effects in determining the rate of nonenzymatic hydrolysis of cellulose and hemicellulose. Torget et al. (685) have proposed that a highly structured diffusion-resistant water layer on the surface of cellulose fibers and the limited solubility of high-molecular-weight oligomers are important factors limiting hydrolysis rates. Both Torget et al. (685) and Jacobsen and Wyman (309) have hypothesized that removal of hydrolysis products and/or disruption of the structured water layer is important in accelerating hydrolysis rates. Such physical chemical factors are presumably operative during enzymatic hydrolysis as well and may play a role in determining the relative effectiveness of CEM and CE complexes. It may be noted that a first-order dependence on temperature is exhibited by both the growth rate on crystalline cellulose (Fig. 5) and the diffusion coefficient (56). The importance of mass transfer and solubility phenomena during microbial utilization of cellulose remains to be elucidated.
The extent of enzyme-microbe synergy has important implications in ecological and applied contexts but has not been evaluated in a quantitative fashion. At this point, about all that can be said is that there are several reasons to think that enzyme-microbe synergy may exist (corresponding to ɛ > 1) and little or no basis to assume that it does not (corresponding to ɛ = 1).
Descriptively adequate models are available which incorporate rates of cellulose hydrolysis and cell growth—including phenomena such as declining substrate reactivity, product inhibition, and batch or continuous reactors—provided that the cellulase loading is a model input (533, 599, 633, 710). However, no structured models are available describing microbial cellulose utilization in which YE/S is a variable model output, and the methodological basis for testing such a model is not established for the case where cellulase is expressed primarily on the cell surface. Development of such models represents a significant and achievable objective.
Beyond cellulose utilization by pure cultures, an additional level of complexity is introduced when multiple organisms are present either in defined cocultures or in undefined mixed cultures (discussed in “Ecological aspects of cellulose-degrading communities” above). There are few controlled studies that compare the rates of cellulose hydrolysis by pure cultures of cellulolytic bacteria with those observed for undefined mixed cultures, although such studies would be of interest.
Contrast to Soluble Substrates
Many if not most of the assumptions and conceptual cornerstones applicable to analysis of systems featuring enzymatically or cellularly mediated reaction of soluble substrates are not applicable to cellulosic substrates. This point is emphasized in Table 9.
TABLE 9.
Differences between biologically mediated reaction of cellulose and soluble carbohydrate substrates
| Soluble carbohydrates | Cellulose |
|---|---|
| Enzymatic reaction mechanism and kinetics | |
| Essentially all potential substrate-reactive sites are accessible | Only a small fraction of substrate-reactive sites are accessible |
| Substrate commonly in excess; hence, rate is proportional to [ET] | Excess substrate is uncommon; hence, at least partial rate saturation with increasing [ET] is usually encountered |
| Concentration is the only kinetically important substrate state variable | Conversion is an additional kinetically important substrate state variable in at least some systems |
| r/[CE] is a constant throughout the course of an irreversible reaction | r/[CE] decreases sharply over the course of the reaction |
| km, the substrate concentration at which half the maximum rate is observed, is a constant independent of [ET] | The substrate concentration at which half the maximum rate is observed increases with increasing [ET] |
| Cell growth, observable in batch culture | |
| Exponential growth, during which substrate is in excess and the concentration of cells is rate limiting, is readily observed | Exponential growth is typically not observed because substrate is typically not in excess and perhaps also because of declining substrate reactivity |
| Allocation of substrate between catabolism and cell synthesis is a key metabolic choice facing the cell | In addition to allocation between catabolism and anabolism, a second key metabolic choice involves allocation of carbon and energy between synthesis of cells and cellulase |
| ATP is synthesized via glycolysis and post-pyruvate metabolism (anaerobes) | Phosphorolytic cleavage of cellodextrins and cellobiose provides an additional potential route for synthesizing high-energy bonds |
| Cell growth, chemostat culture | |
| Substrate reactivity is equal to that leaving the fermentor | Substrate reactivity is equal to the weighted sum of particle reactivities integrated over the time each particle (or fraction of particles) spends in the fermentor |
| Steady-state substrate concentration is independent of the feed substrate concentration | Steady-state substrate concentration increases with increasing feed substrate concentration |
PROCESSING OF CELLULOSIC BIOMASS— A BIOLOGICAL PERSPECTIVE
Important distinguishing features of cellulosic biomass among potential feedstocks for biological processing include low purchase price, potential for supply on a very large scale, recalcitrance to reaction, and heterogeneous composition. For production of fuel and bulk chemicals, a feedstock having low cost and availability on a large scale is required and thus there is ample incentive to develop and apply technology that can cost effectively accommodate the recalcitrant, heterogeneous character of cellulosic biomass. For products with higher value and lower volume (e.g., fine chemicals or pharmaceuticals), there is much less incentive to use a low-cost feedstock, the scale of availability feedstock is not a significant issue, and feedstocks other than cellulosic biomass can be used which present fewer processing challenges. Because of these factors, it is natural in our view to focus on commodity products when considering biological processing of cellulosic feedstocks, and we do so here. The economics of cost-competitive commodity processes are dominated by feedstock cost and thus require high product yields (270, 401, 410, 763). As a result, there is a very strong incentive to conserve the reducing equivalents present in fermentable carbohydrate feedstocks, which is the defining feature of anaerobic metabolism. Biological production of commodity products from cellulosic biomass is thus likely to involve microbial metabolism that is effectively anaerobic, although development of microbes for use in such processes can in principle begin with either aerobes as well as anaerobes or facultative anaerobes. These ideas are developed more fully elsewhere (410).
We offer below a selective consideration of processing cellulosic biomass with an emphasis on biological aspects. More comprehensive treatments of this topic are available (131, 410, 614, 774).
Pretreated Substrates
Incubation of naturally occurring cellulosic materials in the presence of either pure cultures of cellulolytic microorganisms or cell-free enzyme preparations generally results in cellulose hydrolysis yields that are <20% of theoretical. As a result, process designs for biologically converting cellulosic materials nearly always include a pretreatment step. The properties of pretreated feedstocks are of central importance in defining objectives for developing organisms for use in biomass conversion processes (see “Organism development for consolidated bioprocessing” below).
The term “pretreatment” is widely used in the process engineering literature to refer to a process step which converts lignocellulosic biomass from its native form, in which it is recalcitrant to cellulase enzyme systems, into a form for which enzymatic hydrolysis is effective. We make no attempt here to provide a comprehensive review of the pretreatment field. Rather, the focus of this subsection is on describing the characteristics of pretreated materials as starting points for processing cellulosic substrates and for organism development pursuant to such processing. The reader is referred to more comprehensive work describing the diversity, mode of action, and evaluation of preatreatment processes (288, 309, 400, 434, 726).
Most of the β-glucosidic bonds in naturally occurring lignocellulosic materials are inaccessible to cellulase enzymes by virtue of the small size of the pores in the multicomponent spatially heterogeneous biomass matrix as well as enzyme-associated inaccessibility (see “Rates of enzymatis hydrolysis” above). In addition, cellulose in naturally occurring materials is closely associated with hemicellulose and other structural polysaccharides, and carbohydrate-rich microfibrils are surrounded by a lignin seal (see “Structure and composition of cellulosic biomass” above). Rendering lignocellulosic materials amenable to enzymatic hydrolysis thus involves overcoming both physical and chemical barriers. Compared to unpretreated materials, effectively pretreated lignocellulosic materials are generally characterized by increased surface area accessible to cellulase enzymes (porosity) and solubilization and/or redistribution of lignin. Increased porosity results from a combination of hemicellulose solubilization, lignin solubilization, and lignin redistribution. The relative importance of these factors differs greatly among different pretreatment processes.
Lignin redistribution is thought to explain why dilute acid and steam explosion are effective pretreatment processes although lignin is not removed (116). It is thought that lignin melts during pretreatment and coalesces upon cooling such that its properties are altered substantially (87, 116, 169, 666). The study by Ooshima et al. (506) involving the adsorption of T. reesei cellulase to dilute-acid-pretreated hardwood prepared at various temperatures provides an example of the effect of pretreatment on the state of lignin and enzyme accessibility. These investigators found that increasing the pretreatment temperature from 180 to 220°C was accompanied by an eightfold decrease in the adsorption capacity of lignin and a nearly sixfold increase in the adsorptive capacity of the cellulose.
As summarized in Table 10, proposed pretreatment processes include dilute acid, steam explosion at high solids concentration, “hydrothermal” processes, “organosolv” processes involving organic solvents in an aqueous medium, ammonia fiber explosion (“AFEX”), and strong alkali processes using a base such as NaOH or lime. Hydrothermal processes feature aqueous pretreatment with very low or zero concentration of added acid and either very low solids concentration and/or liquid flowing through a bed of solids. While Table 10 is not comprehensive, it does include most of the processes thought to be promising in the context of production of fuels and commodity chemicals from biomass. All these processes have been shown to produce, under appropriate conditions, pretreated fiber derived from herbaceous and/or hardwood-derived feedstocks that (i) retains nearly all of the cellulose present in the original material and (ii) allows close to theoretical yields upon enzymatic hydrolysis. The forms in which cellulose and particularly hemicellulose and lignin emerge from pretreatment are different for different processes, as summarized in Table 10.
TABLE 10.
Fate of biomass components for various pretreatment processes
| Process | Fate of biomass components under conditions leading to high cellulose digestibility
|
Reference(s) | ||
|---|---|---|---|---|
| Cellulose | Hemicellulose | Lignin | ||
| Dilute-acid pretreatment | Some depolymerization | 80-100% solubilization, primarily to monomers | Little or no solubilization, extensive redistribution | 223, 599, 671, 685 |
| Steam explosion at high solids concentration | Some depolymerization | 80-100% solubilization to a mixture of monomers oligomers, and degradation products | Little or no solubilization, extensive redistribution | 87, 248, 546 |
| Hydrothermal processes (see the text) | Some depolymerization | 80-100% solubilization, oligomers usually >50% | Partial solubilization (e.g., 20-50%) | 64, 372, 460b |
| Organic solvents with water | Substantial solubilization (varies but can be nearly complete) | Substantial solubilization (varies but can be nearly complete) | 110, 278 | |
| AFEX | Some decrystallization | Solubilization from 0 to 60% depending on moisture; ∼90% hydrolyzed to oligomers | Some solubilization (∼10-20%) | 132a |
| Sodium hydroxide pretreatment | Substantial swelling, type I → type II | Substantial solubilization (often >50%) | Substantial solublization (often >50%) | 726 |
| Lime pretreatment | Significant solubilization (to 30%) under some but not all conditions | Partial solubilization (∼40%) | 96, 319 | |
Also, B. E. Dale, personal communication.
Also, R. Torget, personal communication.
In the context of organism development for processing cellulosic biomass, several observations can be made based on the biomass pretreatment literature.
(i) The ability to hydrolyze cellulose is an essential organism development objective for biomass resulting from all pretreatment processes. For most pretreatments, it is further necessary that the cellulolytic enzyme system of the processing organism be effective against crystalline cellulose. Cellulose crystallinity does not decrease as a result of pretreatment of biomass by dilute acid (223, 348, 678), steam explosion (546, 581), or lime (97) under conditions resulting in high hydrolysis yields.
(ii) The ability to hydrolyze insoluble hemicellulose (as distinct from soluble hemicellulose hydrolysis products) is not an important organism development objective for pretreatment processes featuring essentially complete hemicellulose hydrolysis. Under conditions resulting in effective pretreatment, such processes include dilute-acid pretreatment (223, 686), steam explosion (87, 546), and hydrothermal pretreatment (64, 372, 460). Although strong-alkali pretreatment can be successful without entirely hydrolyzing hemicellulose (319), it has been observed that residual hemicellulose present after alkaline pretreatment is hydrolyzed by commercial cellulase preparations without the addition of enzymes expressly for hemicellulose hydrolysis (319, 408). Thus, even if insoluble hemicellulose is present after pretreatment, it may not be required that a processing organism produce hemicellulose-specific enzymes.
(iii) Conversion of all sugars derived from hemicellulose during pretreatment is a highly desired organism development objective. Utilization of hemicellulose-derived monomeric sugars, and xylose in particular, has received intensive investigation for over a decade (137, 237, 272, 299, 301, 310, 433, 775). With the exception of dilute-acid pretreatment, a substantial fraction of dissolved hemicellulose is present as oligomers for most processes in which associated hydrolysis products have been characterized, including steam explosion (248, 546), hydrothermal pretreatment (372), AFEX (B. Dale, personal communication) and countercurrent very-dilute-acid pretreatment (R. W. Torget, personal communication). Thus, for most pretreatment processes it is desirable that soluble oligomers originating from hemicellulose, in addition to monomeric sugars, be utilized.
(iv) To be of use in a practical process, a microorganism must remain metabolically active in the presence of inhibitory compounds generated during pretreatment with, at most, relatively low-cost detoxification measures taken. Such compounds arise from hydrolytic release of compounds present in unpretreated biomass (e.g., organic acids, extractives, and phenolics), reaction of carbohydrates and other solubilized components to form degradation products (e.g., furfural and hydroxymethyl furfural), and corrosion resulting in the release of inorganic ions (434). The amounts of inhibitors produced depend greatly on process conditions and configuration. The reader is referred to the recent review by Zaldivar et al. (774) and references therein for a comprehensive consideration of inhibition-related phenomena accompanying pretreatment.
(v) It is not necessary that organisms degrade lignin, although modifications that decrease cellulase binding to lignin are potentially of value.
Process Configurations
Four biologically mediated events occur in the course of converting cellulosic biomass into fuels and chemicals in processes featuring enzymatic hydrolysis: (i) cellulase production, (ii) hydrolysis of cellulose and (if present) other insoluble polysaccharides, (iii) fermentation of soluble cellulose hydrolysis products, and (iv) fermentation of soluble hemicellulose hydrolysis products. Alternative processing configurations can be categorized based on the degree to which these events are consolidated. As depicted in Table 11 separate hydrolysis and fermentation (SHF) involves four discrete process steps and as many as four different biocatalysts. Simultaneous saccharification and fermentation (SSF) consolidates hydrolysis and fermentation of cellulose hydrolysis products into one process step, with cellulase production and fermentation of hemicellulose hydrolysis products occurring in two additional discrete process steps. Simultaneous saccharification and cofermentation (SSCF) involves two process steps: cellulase production and a second step in which cellulose hydrolysis and fermentation of both cellulose and hemicellulose hydrolysis products occurs. In consolidated bioprocessing (CBP), cellulase production, hydrolysis, and fermentation of products of both cellulose and hemicellulose hydrolysis are accomplished in a single process step.
TABLE 11.
Evolution of biomass-processing schemes featuring enzymatic hydrolysisa
Each box represents a bioreactor (not to scale). See the text for definitions of abbreviations.
It may be noted that the term “consolidated bioprocessing (CBP)” is synonymous with direct microbial conversion (DMC) (282, 456, 719). In 1996 Lynd proposed that the term “consolidated bioprocessing” is preferable to “direct microbial conversion” (400). This was motivated by the observation that the CBP/DMC process configuration is no more direct than is SSF, since both configurations feature a fermentative step without a prior enzymatic hydrolysis step as in SHF. It is also the case that hydrolysis and fermentation in the CBP/DMC process configuration is no less simultaneous than in SSF. Rather than being “direct” or “simultaneous”, the main feature differentiating CBP from both SSF and SHF is the extent of consolidation, as indicated in Table 11.
Biomass processing technology has exhibited a trend toward increasing consolidation over time. This trend is most evident in the case of studies of the production of ethanol, which has received the most attention among biomass-derived fermentation products, but is likely to be applicable to other products as well. Prior to the mid-1980s, SHF was the most commonly considered process configuration. Since the studies by Wright and coworkers in 1988 (763, 764), SSF has been widely thought to have significant economic advantages over SHF. Simultaneous saccharification and cofermentation is currently viewed as an economically attractive near-term goal for process development (762), with potential cost benefits relative to SSF. The logical conclusion of this trend is CBP.
A process step dedicated to cellulase production is a feature of SHF, SSF, and SSCF but not of CBP. It is usually envisioned that dedicated cellulase production will be carried out by aerobic microorganisms. This is because of the much higher ATP yields, and correspondingly higher potential cellulase yields, of aerobic metabolism compared to anaerobic metabolism. By contrast, conversion of hydrolysis products to small molecules for use as fuels or bulk chemicals is likely to be carried out by microorganisms featuring a nonoxidative catabolism for all process configurations, as addressed at the beginning of this section.
Figure 6 contrasts metabolic aspects of process configurations featuring dedicated cellulase production (SHF, SSF, and SSCF) to CBP. In both configurations, the product(s) of interest is shown resulting from anaerobic catabolism. For dedicated cellulase production from the same feedstock used for fermentation, a fraction, F, of the feedstock is used to produce cellulase while the remainder (1 − F) of the feedstock is used to produce the desired product(s). Most design studies of SSF and SSCF configurations have reported values for F in the range of 0.03 to 0.05. Values of F in this range correspond to typical values for cellulase loading (10 to 15 FPU/g of cellulose hydrolyzed), together with higher-than-average yields for aerobic cellulase production (∼300 FPU/g of cellulose used to produce cellulase [177]), and result in reaction times in the range of 5 to 7 days (131, 762, 764). The optimum value of F from an economic viewpoint results from minimizing the sum of the cost of bioreactors in which cellulose hydrolysis occurs (which is high at low values of F, accompanied by low cellulase loading and long reaction times) and the cost and product yield loss and other costs associated with cellulase production (which are high at high values of F) (404). It is well known that higher cellulase loadings result in shorter reaction times than the economic optimum, but realizing such rates is not cost-effective because of the combined effect of the cost of cellulase production and the decreased product yield accompanying higher values of F.
FIG. 6.
Summary of material flows and bioenergetics for process configurations featuring dedicated cellulase production and consolidated bioprocessing.
A small fraction of the substrate processed, corresponding to F in Fig. 6, is available for cellulase synthesis for process configurations featuring dedicated cellulase production. However, cellulase can potentially be synthesized at high yield on a per-unit-substrate basis in a dedicated cellulase production configuration because of the large amount of ATP available from aerobic catabolism. For CBP, all of the substrate is available for cellulase synthesis, but potential cellulase yields on a per-unit-substrate basis are lower because of the smaller amount of ATP available from anaerobic catabolism.
Avoidance of the cost of dedicated cellulase production is the largest potential savings associated with CBP relative to current SSF technology. The value of such avoidance is equal to the cost of dedicated cellulase production, estimated at ≥$0.5/gal of ethanol produced by SSF, corresponding to about ∼$50/tonne of dry biomass hydrolyzed, in the comprehensive study by Hettenhaus and Glassner (267). A detailed process design study (405) also projected savings of this magnitude, with CBP reducing the overall processing costs by ca. twofold and the cost of biologically mediated process steps by ca. eightfold as compared to an SSF base case. Of prospective research-driven process improvements analyzed using process and cost models developed by the National Renewable Energy Laboratory, CBP has the largest potential cost savings (405, 766). The CBP processing concept is a potential breakthrough in low-cost processing of cellulosic biomass that is in principle applicable to production of any fermentation-derived product. However, this potential cannot be realized by organisms available today and requires the development of new and improved CBP-enabling organisms as addressed in the following section.
ORGANISM DEVELOPMENT FOR CONSOLIDATED BIOPROCESSING
Strategies
CBP requires a microbial culture that combines properties related to both substrate utilization and product formation. Desired substrate utilization properties include the production of a hydrolytic enzyme system allowing high rates of hydrolysis and utilization of resulting hydrolysis products under anaerobic conditions with a practical growth medium. Desired product formation properties include high product selectivity and concentrations. A cellulolytic culture with this combination of properties has not been described to date.
Development of microorganisms for cellulose conversion via CBP can be pursued according to two strategies. The native cellulolytic strategy involves naturally occurring cellulolytic microorganisms to improve product-related properties such as yield and tolerance. The recombinant cellulolytic strategy involves engineering noncellulolytic microorganisms that exhibit high product yields and tolerance so that they become able to utilize cellulose as a result of a heterologous cellulase system. Subsequent subsections review in detail the progress for each of these strategies with respect to developing cultures capable of utilizing cellulose in a CBP configuration. In the remainder of this subsection, we comment on strategic aspects related to hemicellulose utilization.
Over the last decade, the development of microorganisms capable of converting xylose and other hemicellulose-derived sugars to ethanol at high yields has been one of the most significant advances in the fields of both biomass conversion and metabolic engineering. Both of the strategies presented above with respect to cellulose conversion have been successfully applied to xylose conversion. The native-substrate utilization strategy is exemplified by the work of Ingram et al. with E. coli and Klebsiella oxytoca, in which organisms that naturally use hemicellulose-derived sugars were engineered to produce high product yields (299, 301). The recombinant substrate utilization strategy is exemplified by the work of Ho et al. (272, 273) and Hahn-Hägerdal et al. (237) with Saccharomyces cerevisiae, as well as that of Zhang and coworkers with Zymomonas mobilis (137, 775). In the work by these three groups, an organism with high product tolerance and yield was engineered so that it was able to use desired substrates. Engineering of microorganisms for utilization of soluble hemicellulose derivatives, extensively reviewed elsewhere (69, 272, 299, 310, 774), is rather advanced with several alternative organisms being considered for use in commercial processes. By contrast, the engineering of microorganisms for utilization of cellulose is in its nascent stages.
Consideration of the properties of pretreated feedstocks suggests that it is not necessarily required that organisms used in a CBP context possess an enzyme system specific to hydrolysis of insoluble hemicellulose (see “Pretreated substrates” above). Combining the ability to utilize cellulose and hemicellulose hydrolysis products into a single process step could be accomplished by a coculture of two compatible organisms, one with the ability to utilize cellulose and one with the ability to utilize pentose sugars. Such a coculture can be expected to be stable in light of each organism having a substrate which only it is able to use. Cocultures between cellulolytic and pentose-utilizing microbes are common in nature and to offer improved hydrolysis in the laboratory relative to pure cultures of cellulolytic bacteria (see “Ecological aspects of cellulose-degrading communities” above). Alternatively, the metabolic machinery required for cellulose utilization and utilization of hemicellulose sugars could be incorporated into a single organism. In such an arrangement, simultaneous utilization of both cellulose and pentoses would be desirable. This might be accomplished using a molecular approach by engineering the host organism so that it does not exhibit catabolite repression. A bioreactor-based approach is also promising in this context, since operating conditions may be chosen so that the concentrations of soluble hydrolysis products are maintained below levels triggering catabolite repression.
Native Cellulolytic Strategy
The native substrate utilization strategy for organism development pursuant to industrial conversion of cellulose via CBP can in principle begin with any cellulolytic microorganism. We restrict our attention here to cellulolytic microorganisms capable of anaerobic growth. It may be noted that use of a separate aerobic step for cell growth, as envisioned for processes involving Fusarium oxysporum (109) or Neurospora crassa (155), is not consistent with single-step processing as implicit in the CBP concept. We focus here on ethanol production because ethanol has been the main product considered in studies of CBP via the native cellulolytic strategy. Although organic acid production using naturally cellulolytic organisms has occasionally been proposed (382, 450, 556), the intolerance of described pure cultures of cellulolytic microorganisms to low pH appears to be a significant obstacle to this approach. Physiological features of cellulolytic bacteria are considered in some detail above (see “Physiology of cellulolytic microorganisms”) We consider here metabolic engineering of end product metabolism in solvent-forming anaerobes, tolerance to ethanol and other products, and genetic system development. At this time it is logical that work in these areas involving cellulolytic organisms should build on more advanced work involving noncellulolytic organisms. Selected results obtained with noncellulolytic organisms are therefore included in the discussion that follows.
Metabolic engineering.
Because of the high cost of feedstock in the production of fuels and commodity chemicals from biomass, a high yield (gram of product per gram of substrate) of saleable product(s) is essential from an economic point of view (see “Processing of cellulosic biomass—a biological perspective” above). Achieving high product yields is thus a primary focus of organism development via the native cellulolytic strategy.
Figure 7 presents the branched catabolism typical of ethanol-forming cellulolytic anaerobes from a conventional carbon-centered perspective beginning with monomeric sugars (shown here in unphosphorylated form). This perspective suggests targets for gene knockout to redirect carbon flux toward ethanol:lactate dehydrogenase, phosphotransacetylase, and acetate kinase. In all cellulolytic anaerobes for which data are available, pyruvate is catabolized via acetyl-CoA-yielding pyruvate:ferredoxin oxidoreductase rather than by acetaldehyde-yielding pyruvate decarboxylase or by pyruvate-formate lyase. Thus, regeneration of reduced electron carriers involves not only reduced NAD generated in the course of glycolysis but also reduced ferredoxin generated via the action of pyruvate dehydrogenase. Ferredoxin-NAD oxidoreductase is particularly important in this context. Fig. 7 presents an electron-centered perspective on fermentative metabolism. This perspective suggests that hydrogenase is a potential gene knockout target in addition to acetate kinase and phoshotransacetylase for redirecting flux away from acetic acid formation. As may be inferred from Fig. 7, it is not possible to produce acetate with regeneration of oxidized ferredoxin unless H2 is produced.
FIG. 7.
Catabolic reactions leading to the formation of various end products by ethanol-producing cellulolytic bacteria. 1, Lactate dehydrogenase; 2, pyruvate-ferredoxin oxidoreductase; 3, NADH-ferredoxin oxidoreductase; 4, hydrogenase; 5, phosphotransacetylase; 6, acetate kinase; 7, acetaldehyde dehydrogenase; 8, alcohol dehydrogenase; 9, glyceraldehyde-3-phosphate dehydrogenase. From references 396, 569, and 629.
Although substantial amounts of several organic end products are produced under most conditions by cellulolytic anaerobes, several lines of evidence suggest that organisms in this category are capable of metabolizing cellulose with near-exclusive production of a single end product. This is supported by (i) a priori consideration of biochemistry, (ii) experimental results featuring cellulolytic microorganisms producing high yields of a single fermentation product in response to mutation and selection and/or manipulation of the fermentor environment, and (iii) experimental results featuring noncellulolytic microorganisms producing high yields of a single product in response to mutation and selection as well as metabolic engineering.
It may be seen from Table 12 that the basic requirements of fermentative metabolism—generation of net ATP and regeneration of reduced electron carriers—can be met by producing ethanol, acetic acid, or lactic acid either singly or in combination.
TABLE 12.
Catabolic stoichiometry of ethanol, acetic acid, and lactic acid formation in cellulolytic anaerobes
| Step or enzyme | Reactiona |
|---|---|
| Glycolysis | Glucose + 2ADP + 2Pi + 2NAD+ → 2 Pyruvate− + 2 ATP + 2 NADH + 4H+ |
| Pyruvate:ferredoxin oxidoreductase | {MYE/G + MYA/G} {Pyruvate + CoASH + Fd(ox) → Acetyl-CoA + Fd(red) + CO2 + H+} |
| NADH:ferredoxin oxidoreductase | {MYE/G − MYA/G} {Fd(red) + NAD+ + 2H+ → Fd(ox) + NADH + H+} |
| Acetaldehyde dehydrogenase | MYE/G {Acetyl-CoA + NADH + H+ → Acetaldehyde + CoASH + NAD+} |
| Ethanol dehydrogenase | MYE/G {Acetaldehyde + NADH + H+ → Ethanol + NAD+} |
| Phosphotransacetylase | MYA/G {Acetyl-CoA + Pi → Acetyl phosphate + CoASH} |
| Acetate kinase | MYA/G {Acetyl phosphate + ADP → Acetate− + ATP + H+} |
| Hydrogenase | 2 MYA/G {Fd(red) + 2H+ → H2 + Fd(ox)} |
| Lactate dehydrogenase | MYL/G {Pyruvate− + NADH + H+ → Lactate− + NAD+} |
| Overall | Glucose + {2 + MYA/G} ADP + {2 + MYA/G}Pi → MYE/GEthanol + MYE/GEthanol + MYA/GAcetate + MYL/GLactate + {MYEt/Glu + MYA/G}CO2 + {MYL/G + MYA/G}H+ + 2MYA/GH2 + {2 + MYA/G}ATP |
MYE/G is the molar ethanol yield (moles of ethanol per mole of hexose);MYA/G is the molar acetic acid yield (moles of acetic acid per mole of hexose); MYL/G is the molar lactic acid yield (moles of lactic acid/mole of hexose). Note: MYE/G + MYA/G + MYL/G = 2.
Fermentation resulting in molar ratios of ethanol to organic acids exceeding 9:1 has been reported at low temperatures for the cellulolytic Clostridium saccharolyticum (477). Ethanol-to-organic-acid ratios of >5 have been obtained for strains of C. thermocellum developed using classical mutagenesis (719). The flexibility of product metabolism in cellulolytic anaerobes is underscored by the observation of molar ratios of ethanol to organic acids of <0.1 for C. thermocellum (737) and <0.15 for R. albus (518) in the presence of a hydrogen-consuming methanogen.
Ethanol-to-organic-acid ratios of >9 have been reported under favorable conditions for Thermoanaerobacter ethanolicus (742) and T. thermosaccharolyticum (268), which, although noncellulolytic, are thought to have end product metabolism generally similar to that of C. thermocellum. Classical mutagenesis techniques have been used to create strains capable of anaerobic growth with much-reduced activity of enzymes involved in product metabolism. Examples include mutants deficient in acetate kinase and/or phosphotransacetylase in the case of Thermoanaerobacter thermohydrosulfuricus (432), T. thermosaccharolyticum (573), and E. coli (232) and mutants deficient in lactate dehydrogenase in the case of E. coli (431).
During the 1980s, a substantial effort was devoted to strain isolation, optimization of culture conditions, and strain development via mutation and selection pursuant to fermenting cellulose and/or xylose with high ethanol yields (396, 401, 629, 719, 742). This work demonstrated very high ethanol yields a sufficient number of times in a sufficient number of organisms to provide substantial support for the biochemical and bioenergetic feasibility of fermenting cellulose with ethanol as the only significant organic end product. However, this effort did not result in robust strains that consistently produce ethanol at high yields under a broad range of conditions and in the hands of different investigators (401).
At this point, we believe that the most promising path along which to pursue the native cellulolytic organism development strategy is via metabolic engineering using molecularly based techniques. This, however, requires genetic systems by which to express foreign genes in cellulolytic anaerobes, which have in general not been available (discussed below). As such systems are developed, engineering of cellulolytic microorganisms for CBP will benefit from results obtained over the last 15 years pursuant to engineering of end product metabolism in noncellulolytic anaerobes. Examples of these results include enhancement of ethanol production in E. coli and K. oxytoca (reviewed by Ingram et al. [299, 301]), solvent production in C. acetobutylicum (reviewed by Mitchell [458]), and lactic acid production in yeasts (2, 147, 541). In these and other cases, metabolic flux is altered by blocking undesirable pathways, typically via homologous recombination-mediated “gene knockout” (59, 89, 221, 222, 359, 541, 744) and/or overexpression of genes associated with desirable pathways (2, 12, 17, 75, 147, 243, 441, 542). Gene knockout is more readily applicable than is overexpression for organisms with poorly developed genetic systems, is not subject to “saturation effects,” and can have the added benefit of irreversibility (183). Antisense RNA has also been used for altering catabolic flux in anaerobes (153).
Initial attempts to redirect flux commonly have unintended consequences, often as a result of either a metabolic imbalance involving either organic intermediates (159, 300) or electron carriers (277) or interactions with metabolic control systems (153). It is useful in our view to distinguish between fundamentally motivated studies that seek to use metabolic engineering to introduce a perturbation that is interesting to study and application-motivated studies that seek to achieve a given result. In the latter type of study, modifications beyond that undertaken initially are very often required to compensate for secondary effects accompanying the original metabolic manipulation. In the case of engineering E. coli to produce ethanol, for example, first-generation strains expressing pyruvate decarboxylase (76, 300) did not have sufficient alcohol dehydrogenase activity to function well at high sugar concentrations. This deficiency was subsequently corrected, and the performance improved (301). The success of engineering enteric bacteria for ethanol production and engineering yeast for lactic acid production suggests that it is reasonable to expect success in applied studies, given (i) a sustained effort commensurate with the complexity of the specific goal and (ii) that this goal is consistent with stoichiometric and bioenergetic constraints implicit in the metabolism of the host organism.
Growth inhibition by ethanol and other factors.
The ability or inability of cellulolytic anaerobes to utilize high concentrations of substrate(s) and produce a desired product at high concentration is important in the context of evaluating the potential of the native cellulolytic strategy in general and of specific host organisms in particular. Cessation of growth due to high ethanol concentrations is of particular interest for CBP. It is appropriate to give attention to growth limitation by factors other than ethanol because distinguishing between inhibition by ethanol and by other factors can be subtle, and because inhibitory factors other than ethanol are likely to accompany pretretreated feedstocks in real-world processes.
As reviewed elsewhere (298, 317), microorganisms are thought to be inhibited by ethanol as a result of end product inhibition of glycolytic enzymes and damage to the cell membrane. The ethanol tolerance of C. thermocellum has been investigated most extensively among cellulolytic anaerobes. Few data are available for other cellulolytic anaerobes, although such data would be of considerable interest. Most naturally occurring strains of C. thermocellum exhibit rather low ethanol tolerance; however, development of increased tolerance in response to exposure to ethanol has been described often for this organism (262, 660, 719) as well as for other thermophiles (30, 342, 397, 743). Thus, ethanol tolerance in C. thermocellum and noncellulolytic ethanol-producing thermophiles appears at a phenomenological level to be inducible rather than constitutive. As a result, rather different behavior may be observed depending on the extent to which organisms have been previously exposed to ethanol and transient responses to challenges with ethanol or another inhibitor are complete. Inhibition of C. thermocellum by ethanol has been attributed to a blockage in glycolysis associated with ethanol-induced changes in the cell membrane (263, 264, 265). Changes in membrane composition in response to ethanol have also been observed in other organisms (34, 167), including thermophiles (30, 318). Increased temperature, which, like ethanol, increases membrane fluidity, has been observed to markedly decrease ethanol tolerance in C. thermocellum (262, 362) as well as other organisms, both mesophilic (46, 290, 708) and thermophilic (14, 30).
Most current designs for processes involving enzymatically mediated hydrolysis of lignocellulosic substrates involve ethanol concentrations of ≤5% (by weight). Achieving ethanol concentrations substantially in excess of 5% (by weight) for such processes in the future is unlikely in light of constraints associated with slurry handling and because there is not a large incentive to do so in terms of economics or process energy requirements (400). There is, however, a substantial economic penalty associated with operation at ethanol concentrations lower than 4% (by weight) when conventional product recovery technology is used.
When considered in light of these processing requirements, the tolerance of selected strains of C. thermocellum appears to be sufficient with respect to added ethanol but not with respect to the maximum concentrations of ethanol produced in studies thus far. C. thermocellum strain SS22 (553) grows at added ethanol concentrations up to 64 g/liter, and strains A1 (660), C9 (262), and S7 (719) require added ethanol concentrations in the range of 27 to 50 g/liter to be 50% inhibited. Similar results have been reported for the xylose-utilizing noncellulolytic thermophile T. thermosaccharolyticum (30). Notwithstanding the rather high tolerance manifested with respect to added ethanol, the highest concentrations of ethanol produced by either of these organisms are ≤26 g/liter in studies to date (403, 596, 719).
Resolving the discrepancy between the apparent tolerance to added ethanol and the maximum concentration of ethanol produced for C. thermocellum and perhaps for other cellulolytic anaerobes is an important objective for the native cellulolytic CBP organism development strategy that will have a substantial bearing on its feasibility. A difference between the inhibitory effect of added and produced ethanol, in conjunction with different intracellular and extracellular ethanol concentrations, was proposed for yeast in studies prior to 1983 (34, 479, 497). However, subsequent unrebutted work has discounted this hypothesis for yeast (133, 134, 135, 229, 395), Z. mobilis (313), and E. coli (168). In light of these data and in particular biophysical analyses indicating that substantial differences between intracellular and extracellular ethanol concentrations across plasma membranes can exist for at most short periods (313, 395), it would appear unlikely that this discrepancy is due to a difference in the inhibitory effect of added and produced ethanol.
Working with T. thermosaccharolyticum, Lynd et al. (403) established that inhibition of continuous fermentation of high-feed xylose concentrations is due to salt resulting from base added for pH control and is not due to ethanol. Salt concentrations corresponding to those resulting from pH control also exert a strong inhibitory effect on cellulose fermentation by C. thermocellum (Y. Zhang and L. R. Lynd, unpublished data). These results are consistent with the hypotheses that (i) salt inhibition is responsible for the discrepancy observed in the literature between the tolerance to added ethanol and the maximum concentrations of ethanol produced by C. thermocellum and other thermophilic saccharolytic bacteria and (ii) metabolic engineering to reduce organic acid production may provide a means to increase not only ethanol yield but also apparent tolerance. Definitive testing of these hypotheses awaits further study. With respect to hypothesis (ii), it may be noted that metabolic engineering of E. coli to eliminate organic acid production allowed substantial increases in the concentrations of ethanol produced and substrate utilized compared to results obtained with nonengineered strains. These results are consistent with the observation that organic acids are more inhibitory than ethanol on a molar basis in this organism (301).
Inhibition of cellulolytic bacteria by compounds other than ethanol that are likely to occur in industrial processing environments has received relatively little attention, with most relevant studies involving C. thermocellum. Several studies have shown that C. thermocellum is capable of growing on pretreated lignocellulosic substrates that contain most or all of the lignin present prior to pretreatment (284, 409, 582), and in a direct comparison similar results were obtained for fermentation of Avicel and pretreated wood (407). Thus, the presence of insoluble lignin per se does not prevent high rates and extents of hydrolysis from being achieved (407). However, inhibition of C. thermocellum fermentation has been reported for hydrolysates resulting from autoclaved corn stover (22) or steam-exploded aspen (325, 326). Inhibition by pretreatment hydrolyzates is widely observed for noncellulolytic microbes as well (see “Pretreated substrates” above), and a systematic comparison of the relative sensitivity of cellulolytic and noncellulolytic anaerobes with respect to such inhibition has not been undertaken to our knowledge. Inhibition of C. thermocellum by acetate, both a common by-product of pretreatment and a fermentation product, has been investigated by Herrero et al. (265) with respect to fermentation of cellobiose. Unacclimated cells exhibited a 50% decrease in growth rate at near-neutral pH and an acetate concentration of 0.28 M. Acetate inhibition was attributed to decreasing the magnitude of the transmembrane proton motive force. Russell (576) has subsequently suggested anion accumulation as an alternative general explanation for the toxicity of organic acids.
An important but thus far unexplained phenomenon documented for both C. thermocellum (407) and C. cellulolyticum (158) is the decrease in substrate conversion with increasing feed substrate concentration in continuous culture. It is unlikely that this transition from cellulose-limited to cellulose-sufficient conditions is the result of the extracellular concentration of fermentation products, as indicated by experiments involving product addition as well as growth on cellobiose at high substrate concentrations. Nutrient concentrations appear to be sufficient in these studies as well. Possible explanations include (i) inhibition of cells, cellulase production, or the action of cellulase due to salt arising from organic acid production and addition of base to maintain pH control; (ii) accumulation of an intracellular metabolite to inhibitory levels; and (iii) a nutrient requirement specific to growth on cellulose. It is necessary to understand and ultimately overcome this limitation phenomenon in order to contemplate using naturally cellulolytic bacteria for industrial processes; this should be viewed as a key part of the more general issue of resolving the nature of growth limitation in these organisms.
Genetic system development.
Genetic system development has been reviewed with respect to noncellulolytic clostridia by Young et al. (771), Rood (571), Blaschek and White (62), and Phillips-Jones (534) and with respect to thermophiles by Mai and Wiegel (420). As presented by Mercenier and Chassy (440), primary choices associated with development of a genetic system include (i) the choice of strain or strains, (ii) the choice of DNA entry method, and (iii) the choice of transforming DNA. Development of transformation systems is in general a highly empirical endeavor, relying to a very large extent on repeated trials involving various combinations of strains, DNA entry methods, vectors, and conditions. Such development is particularly challenging in light of the large number of potentially important experimental variables involved and the related fact that negative results are very difficult to interpret.
It is widely observed that transformation is a strain-specific phenomenon and that what works in one strain often does not work in another. Screening a large number of strains has been a useful strategy in developing transformation systems in several organisms. For example, electrocompetent strains were obtained by screening 62 strains of Bacillus stearothermophilus (481) and 30 strains of Thiobacillus ferrooxidans (557). The presence or absence of restriction systems is an important host strain property in the context of genetic system development, and specific methylation to avoid restriction attack has been required to achieve significant transformation frequencies in several cases (101, 311, 441). However restriction systems have also been found to be absent in some gram-positive bacteria (62, 340). Electrotransformation has often been applied in conjunction with treatment to weaken the cell wall as a barrier to DNA uptake (173, 276, 418, 608) and in some cases such treatment appears to be required. Survival of a sufficient number of viable cells to detect a relatively infrequent transformation event is of the utmost importance when working with oxygen-sensitive anaerobes, and transformation of such organisms is routinely carried out in anaerobic glove boxes (771).
Natural competence is relatively rare and has not been described in any of the gram-positive anaerobes considered in the reviews cited at the beginning of this subsection; therefore, all techniques involving these organisms are based on artificially induced DNA uptake. Described protocols are based on DNA entry via protoplast transformation, chemically induced competence, conjugation, and electrotransformation. Protoplast transformation involves removing the cell wall (thought to be a barrier to DNA uptake in gram-positive bacteria), DNA uptake, and regeneration of normal walled cells (440). Such techniques were reported in 1984 for heat-treated cells of C. acetobutylicum (390) and in 1989 for B. stearothermophilus (765). Chemically induced competence (e.g., using CaCl2 or polyethylene glycol) has been widely used for gram-negative bacteria and was also used during the 1980s to develop transformation protocols for Clostridium thermohydrosulfuricum (now Thermoanaerobacter ethanolicus) (634), Streptococcus lactis (591), and Bacillus species (247, 662). Notwithstanding these reports, transformation of gram-positive bacteria based on protoplasts or chemically induced competence is increasingly viewed as cumbersome, difficult to reproduce, and generally less desirable than electrotransformation (458, 765). Conjugation has been used extensively for a few organisms such as C. beijerinckii, as reviewed by Young et al. (771), but is not widely used to transform gram-positive bacteria. Electrotransformation is increasingly common and is usually viewed as a preferred gene transfer method (440, 771). Electrotransformation systems were developed during the 1980s and 1990s in the case of noncellulolytic mesophilic clostridia including C. acetobutylicum, C. beijerinckii, C. perfringens, and at least five additional species (771), as well as T. thermosaccharolyticum (341) and Thermoanaerobacterium saccharolyticum (418, 419).
Vectors used in transforming gram-positive anaerobes include replicative plasmids, nonreplicative “suicide” vectors, phage-based vectors, and conjugative transposons. We comment further in this paragraph on the use of such vectors in noncellulolytic Clostridium species, reviewed by Phillips-Jones (534), Rood (571), and Young et al. (771), because of the significant number of cellulolytic anaerobes in this genus and because much progress has been made in this area in recent decades. “Shuttle vectors” based on replicative plasmids generally include replicons from both gram-positive and gram-negative bacteria as well as selectable markers suitable for use in both donor and recipient strains. Most of the replicons and markers used in Clostridium originate from other gram-positive genera, although this is not always the case. Suicide vectors designed for either single- or double (gene replacement)-crossover events can be used for gene knockout studies as discussed above (see “Metabolic engineering”). A filamentous virus-like particle from C. acetobutylicum NCIB 6444 has been isolated (329) and used to construct a replicative phagemid (330). Conjugative transposons with selectable antibiotic resistance markers may be used for gene isolation and characterization by insertional mutagenesis, as reviewed for clostridia by Young et al. (771).
Development of genetic systems for thermophilic cellulolytic bacteria, and thermophiles in general, involves additional considerations in addition to those relevant to such development for mesophiles. These considerations include the stability at elevated temperatures of antibiotics, genes coding for resistance to antibiotics or other phenotypic markers (418, 420), gene products conferring selectable properties, and plasmid replication. While it is important to incorporate these considerations into an experimental plan aimed at genetic system development, the literature suggests that they do not represent insurmountable obstacles in this context. Antibiotics and associated markers successfully used to select thermophilic transformants include chloramphenicol (151, 687), erythromycin (341), kanamycin (140, 418, 634, 687, 725), tetracycline (quoted in reference 420), and thiamphenicol (634). All of the markers used in these studies originated from mesophilic organisms. Replicons orginating from mesophilic organisms have been used in all but a few cases (140, 151). Investigation of antibiotic stability at elevated temperatures suggests that variables other than temperature have important impacts on stability (530) and that evaluation of antibiotic stability in the presence of specific “background” conditions to be used is advisable (420). Use of selective markers other than antibiotic resistance in thermophiles is of interest but has received relatively little study to date.
Heterologous expression of genes originating from cellulolytic bacteria in mesophilic hosts such as E. coli has been reported dozens if not hundreds of times. However, expression of foreign genes in cellulolytic organisms has been reported much less often and is in general in a nascent stage of development comparable to that for mesophilic Clostridium in the mid- to late 1980s.
In 1987, Tsoi et al. (687) investigated protoplast formation and regeneration for several strains identified as C. thermocellum and achieved transformation of one strain (F7) using pHV33 with selection based on kanamycin resistance and pMK419 with selection based on chloramphenicol resistance. In 1989, Kurose et al. (365) reported transformation of two thermophilic cellulolytic Clostridium strains using a shuttle vector conferring chloramphenicol resistance. In 1992, Cocconcelli et al. (115) reported electrotransformation of R. albus using pSC22 and pCK17. There have been no subsequent reports citing any of these three papers either by the groups responsible for them or by others.
More recently, several groups have attempted to transform C. thermocellum, thus far without success. C. thermocellum ATCC 27405 possesses an MboI-like (5′-GATC-3′ recognition sequence) restriction system that can be protected by the dam methylation system (340). Interestingly, this is also the case for four additional thermophilic, cellulolytic strains (511), suggesting that this restriction system may be a conserved feature among such strains. The occurrence of antibiotic sensitivity requisite for use of convenient selective markers appears widespread in thermophilic, cellulolytic anaerobes (511).
In 1998, conjugal transfer of transposon Tn 1545 from C. beijerinckii to Eubacterium cellulosolvens was reported by Anderson et al. (15). Recently, Jennert et al. (311) reported gene transfer to C. cellulolyticum ATCC 35319 via both conjugation and electrotransformation. Conjugative gene transfer was achieved using both pCTC1 with E. coli as the donor and Tn 1545 with Enterococcus faecalis as the donor. However, conjugal gene transfer frequencies were low, as also observed for E. cellulosolvens (15), and results were difficult to reproduce. Electroporation proved a more reliable method of gene transfer in the study by Jennert et al. (311), but only once incoming DNA protected from a restriction system recognizing the sequence 5′-CCGG-3′. More detailed studies of electrotransformation of C. cellulolyticum have been reported by Tardif et al. (669). Optimal conditions for electropermeabilization were determined based on measurement of ATP leakage (626). Transformants were obtained at field strengths of 7 or 7.5 kV cm−1 but not at other field strengths. Improved transformation frequencies were observed in liquid medium compared to solid medium, for which plating efficiencies were low. Plasmid DNAs including five different replicons were established with approximately equal efficiency (311), suggesting that proper methylation and optimization of electrotransformation conditions were in this case more critical than the source of transforming DNA.
Recombinant Cellulolytic Strategy
The recombinant cellulolytic strategy for organism development pursuant to cellulose conversion via CBP begins with noncellulolytic microorganisms having excellent product formation properties and involves heterologous expression of a functional cellulase system. Such heterologous expression has been undertaken for a variety of purposes with a variety of microorganisms. We focus here on studies aimed at, or at least anticipating, enablement of growth on cellulose. Thus, we do not, for example, catalogue the vast and important literature associated with cloning cellulase components in E. coli for the purpose of enzymological studies. Heterologous expression of cellulase pursuant to growth enablement has been investigated to date primarily in S. cerevisiae, enteric bacteria, and Z. mobilis. We focus on the body of work involving these three organisms because this encompasses the most advanced embodiment of the recombinant cellulolytic organism development strategy to date. Some work on heterologous cellulase expression has been undertaken with additional hosts (328, 419, 520), whose potential utility should not be dismissed. The possibility of restoring functional expression of cryptic cellulase genes in C. acetobutylicum (606) is also intriguing. General properties of yeast, enteric bacteria, and Z. mobilis as industrial biocatalysts, discussed elsewhere (774), are sufficiently established that an actively cellulolytic strain based on any of these hosts would probably find industrial application. We do not comment further on such properties here except to note that both the suitability of these organisms for use in industrial processes and the tools for genetic manipulation are much better established than is the case for naturally cellulolytic anaerobes (see “Native cellulolytic strategy” above). As discussed in the concluding discussion, these advantages must be weighed against the difficulty of enabling rapid growth on pretreated lignocellulose by organisms that are not naturally cellulolytic.
Before considering heterologous expression of cellulase enzyme systems in detail, it is worthwhile to first briefly examine the question of what components are, at a minimum, needed for a functional cellulase system in an organism that is not naturally cellulolytic. This question is approached with respect to pretreated substrates for which no hemicellulose-specific enzyme activities are required (see “Pretreated substrates” above). It should be recognized that the question of what constitutes a rudimentary saccharolytic enzyme system is considerably more complicated for nonpretreated (hemicellulose-containing) substrates and perhaps for some pretreated substrates as well.
For a noncomplexed cellulase system, the literature suggests that protein-specific hydrolysis rates will be lower if the following components are not present: a cellobiohydrolase attacking reducing ends, a cellobiohydrolase attacking nonreducing ends, an endoglucanase, and a β-glucosidase (either extracellular or cell associated). At least the cellobiohydrolases should have a CBM. For complexed cellulases, the following minimum set of components appear to be required: a scaffoldin protein with a CBM, at least two cohesins and a domain that binds to a cell wall-anchoring protein; a cell wall-anchoring protein; and at least one exoglucanase and an endoglucanase, both containing dockerins capable of binding to the scaffoldin protein. In addition, either a β-glucosidase or cellodextrin and cellobiose phosphorylases, together with the appropriate permeases, would be required.
Studies with reconstituted noncomplexed cellulase systems at roughly the levels of complexity indicated above mediate relatively rapid hydrolysis of crystalline cellulose in the case of both non-complexed (65) and complexed systems (188). Inclusion of cellulase components or other cellular features in addition to those listed may well be beneficial, and it is anticipated that the identity of such components will become more clear in the course of progress toward developing recombinant cellulolytic organisms.
Heterologous cellulase expression in bacteria.
(i) Zymomonas mobilis.
Several cellulase-encoding genes have been cloned and expressed in Z. mobilis with various degrees of success. Using a broad-host-range, mobilizable plasmid vector, the endoglucanase gene (eglX) from Pseudomonas fluorescens subsp. cellulosa was introduced into Z. mobilis (380). The transcription of the exlX gene was initiated from the CAT promoter present on the vector. The expression was rather poor since the CAT gene was insertionally inactivated during the cloning construction. This recombinant strain, however, produced the heterologous endoglucanase intracellularly throughout the growth phase independent of the glucose concentration in the medium (380). Similarly, introduction of the Bacillus subtilis endoglucanase into Z. mobilis also resulted in poor expression, and again no activity was obtained in the culture supernatant of the transformants (770).
In contrast to the P. fluorescens and B. subtilis genes, the endoglucanase gene (celZ) of Erwinia chrysanthemi was efficiently expressed in Z. mobilis (77). The specific activity of the Z. mobilis enzyme was comparable to that of the parent strain of E. chrysanthemi. Biosynthesis of CelZ was reported to occur during the exponential growth phase of Z. mobilis. Approximately 35% of the enzyme was released into the medium in the absence of detectable cell lysis. The endoglucanase appeared to be located in the periplasmic space of transformed Z. mobilis cells (77). Another cellulase gene that has been successfully expressed in Z. mobilis was cloned from Acetobacter xylinum (502). The CMCase gene from A. xylinum was efficiently expressed in Z. mobilis, and about 75% of the enzyme activity was detected in the periplasmic space. Few reports of heterologous cellulase expression in Z. mobilis have appeared in the last 5 years.
(ii) Enteric bacteria.
Two E. chrysanthemi endoglucanases, encoded by celY and celZ, and the A. xylinum cellulase gene have been expressed in both E. coli as well as the related enteric bacterium K. oxytoca (230, 502, 755, 779, 780, 781, 782). Initially the expression of celY in E. coli was poor due to promoter construction (230). With a low-copy-number vector, two E. coli glycolytic gene promoters (gap and eno) were tested and found to be less effective than the original E. chrysanthemi celZ promoter (782). However, by using a surrogate promoter from Z. mobilis, the expression of celZ in E. coli was increased sixfold. With this construct, large polar inclusion bodies were evident in the periplasmic space of the recombinant E. coli strain. Addition of the out genes from E. chrysanthemi caused a further increase in the production of CMCase activity and facilitated the secretion or release of more than 50% of the activity into the culure medium (782). The enhancement of expression of the E. chrysanthemi endoglucanase in E. coli achieved in the latter study is underlined by the fact that the total cellulase activity was estimated to represent 4 to 6% of the total cellular protein. The same cloning strategy in K. oxytoca resulted in a recombinant strain that produced CMCase activity equivalent to 5% of the total cellular protein (779).
In subsequent studies, Zhou and Ingram (780) found strong evidence for synergistic hydrolysis of CMC and acid-swollen cellulose by the two endoglucanases encoded by celZ and celY. CelY was reported to be unable to hydrolyze soluble cellooligosaccharides (cellotetraose and cellopentaose), but it hydrolyzed CMC to fragments averaging 10.7 glucosyl units (780). In contrast, CelZ hydrolyzed cellotetraose, cellopentaose, and amorphous cellulose to produce cellobiose and cellotriose as the major end products. CelZ hydrolyzed CMC to fragments averaging 3.6 glucosyl units. In combination, CelZ and CelY hydrolyzed CMC to smaller fragments having an average degree of polymerization of 2.3 hexose units (780). It was inferred that full synergy was obtained by sequential hydrolysis of CMC, with CelY acting first. This line of inquiry was then taken one step further by coexpressing the celY and celZ gene constructs in a K. oxytoca strain with the native ability to transport and metabolize cellobiose, thereby eliminating the need for supplemental β-glucosidase (778). In cellulose fermentation trials, Zhou et al. (778) found that most of the beneficial contribution could be attributed to CelY rather than CelZ. During the fermentation of crystalline cellulose with low levels of commercial cellulases of fungal origin, these recombinant cellulase-producing K. oxytoca strains produced up to 22% more ethanol than did the untransformed parental strain (778). Most recently, Zhou and Ingram (781) demonstrated growth and ethanol production on amorphous cellulose in the absence of added hydrolytic enzymes by a derivative of K. oxytoca M5A1 carrying chromosomally integrated copies of the Z. mobilis pdc and adhB genes for ethanol production and the celY and celZ endoglucanase genes of E. chrysanthemi. This recombinant strain, K. oxytoca SZ21, was reported to produce 20,000 U of endoglucanase activity per liter during fermentation. In combination with the native ability to metabolize cellobiose and cellotriose, this transgenic strain was able to ferment amorphous cellulose to ethanol with yields 58 to 76% of theoretical.
Heterologous cellulase expression in yeast.
The often-underestimated diversity of yeast species encompasses organisms with a broad range of properties that differ from S. cerevisiae and could be useful for CBP. However, S. cerevisiae has received the most attention with respect to heterologous cellulase expression as well as the production of ethanol and other commodity products (570).
(i) Endogenous saccharolytic enzymes of S. cerevisiae.
Although our central focus is the heterologous production of saccharolytic enzymes, we first consider endogenous saccharolytic enzymes produced by S. cerevisiae. This is of interest here because such enzymes may augment the effectiveness of heterologous cellulase components and because the expression of natural saccharolytic enzymes may provide clues to effective expression of recombinant enzymes. While industrial strains of S. cerevisiae are unable to ferment polysaccharides, certain strains produce saccharolytic enzymes with limited activity. These endogenous genes include the glucoamylase genes (STA1, STA2, STA3, and SGA1) of the diastatic strains of S. cerevisiae (544), the pectinase genes (PGU1 and PGL1) of Saccharomyces bayanus var. uvarum (61, 216), and the glucanase genes present in all S. cerevisiae strains (544).
The exo-β-1,3-glucanases produced by S. cerevisiae yield glucose as the end product (143, 472), whereas endo-β-1,3-glucanase releases a mixture of oligosaccharides with glucose as the minor product (482). Because β-1,3-glucan is the main structural polysaccharide responsible for the strength and rigidity of the yeast cell wall, β-1,3-glucanases have been suggested to play a role in important morphogenetic processes involving the controlled autolysis of β-1,3-glucan. During vegetative growth, several endo- and exo-1,3-β-glucanases are synthesized, some of which are secreted only to remain entrapped in the cell wall whereas others are released to the surrounding medium (181). In turn, the meiotic cycle leads to the induction of a new β-1,3-glucanase not present in vegetatively growing cells (120, 478). β-1,3-Glucanases possibly effect controlled cell wall hydrolysis during cell expansion, budding, conjugation, and sporulation. However, there is no direct evidence pointing to the involvement of these enzymes in particular functions during cytodifferentiation of the yeast cell (482).
Cloned and characterized β-1,3-glucanase genes from S. cerevisiae include EXG1, EXG2, BGL1, BGL2, SSG1, and SPR1 (343, 363, 478, 482, 593). From the restriction maps, nucleotide sequences, and chromosomal map positions it can be concluded that EXG1 is identical to BGL1 and that SSG1 is identical to SPR1. The chromosomal location of these four β-1,3-glucanase genes in S. cerevisiae is as follows: EXG1/BGL1 on chromosome XII, EXG2 on chromosome IV, BGL2 on chromosome IV, and SSG1/SPR1 on chromosome XV (120, 702). The EXG1 (BGL1) gene encodes a protein whose differential glycosylation accounts for the two main extracellular exo-1,3-β-glucanases (EXGI and EXGII), present in the culture medium of vegetatively growing cells (181, 371, 482). A second gene, EXG2, encodes a minor exo-1,3-β-glucanase (EXGIII) that is a high-molecular-weight protein exhibiting a high carbohydrate content but showing a significant degree of similarity in its protein fraction to that of the EXGI and EXGII exo-1,3-β-glucanases (371). A third gene, BGL2, encoding an endo-β-1,3-glucanase (472), has also been cloned (343). The EXG1 and EXG2 products are mainly secreted into the medium, whereas the BGL2 product is incorporated into the cell wall (472). A fourth gene, SSG1 (SPR1), encoding a sporulation-specific exo-β-1,3-glucanase, was cloned and characterized (478, 593). The predicted amino acid sequences of the polypeptides encoded by SSG1 and BGL2 are not homologous, whereas the SSG1, EXG1, and EXG2 gene products are highly homologous, differing mainly in their amino-terminal hydrophobic sequences (593). These leader sequences probably play a decisive role in directing these enzymes to particular subcellular locations, with the SSG1-encoded product remaining as an intracellular protein, presumably associated with the nascent ascospore envelope. The hydrophobic leader sequence of the SSG1-encoded protein fulfills the structural requirement for incorporation into a membranous system that allows its translocation into the ascospore wall (593). In view of the strong homology between the EXG1, EXG2, and SSG1 gene products and their different subcellular locations and timing of production, it is tempting to speculate a common evolutionary origin of the exo-β-1,3-glucanase genes with the acquisition of distinct structural and regulatory features that would allow them to respond to specific signals during the cell cycle (593). Further discussion of the expression, regulation, and function of these genes may be found elsewhere (544).
(ii) Expression of heterologous cellulase genes in S. cerevisiae.
Genes encoding cellulases have been cloned from various bacteria, filamentous fungi and plants and expressed in S. cerevisiae (Table 13). From Table 13 it is evident that a wide range of bacterial genes encoding cellulases have been cloned and expressed in S. cerevisiae. The endo-β-1,3-1,4-glucanase gene from Bacillus subtilis was expressed in S. cerevisiae under its own promoter and signal sequences (269, 628). The synthesis of high levels of β-glucanase in brewing yeast strains was achieved by placing this gene under the control of the S. cerevisiae ADH1 promoter on a high-copy-number 2μm-based plasmid vector (92). The fact that no extracellular endo-β-1,3-1,4-glucanase activity could be detected in cultures of S. cerevisiae was attributed to the inability of yeast to process the protein so as to promote secretion. However, even when the β-glucanase gene was fused to the ADH1P-MFα1S expression-secretion cassette, no extracellular enzyme activity could be detected in culture fluids of yeast transformants (704). By contrast, it was found that when the endo-β-1,4-glucanase gene (end1) from Butyrivibrio fibrisolvens was inserted into the same expression-secretion cassette (ADH1P-MFα1S-end1-TRP5T; designated END1), high levels of β-glucanase activity were secreted by laboratory strains of S. cerevisiae as well as wine and distillers' yeasts (701, 704).
TABLE 13.
Cloned cellulase genes expressed in S. cerevisiae
| Enzyme | Gene(s) | Donor | Reference(s) |
|---|---|---|---|
| Endo-β-glucanases | |||
| Endo-β-1,3-glucanase | bglH | Bacillus circulans | 480 |
| Endo-β-1,3-1,4-glucanase | Bacillus subtilis | 92 | |
| end1 | 269 | ||
| 628 | |||
| Endo-β-1,4-glucanase | end1 | Butyrivibrio fibrisolvens | 531, 701, 703 |
| Endoglucanase | Cellulomonas biazotea | 515 | |
| Endo-β-1,4-glucanase | cenA | Cellulomonas fimi | 627, 628, 754 |
| Endo-β-1,4-glucanase | Trichoderma longibrachiatum | 528, 529 | |
| Endo-β-glucanase | Clostridium thermocellum | 580 | |
| Endo-β-glucanase | Thermophilic bacterium | 281 | |
| Endo-β-1,3-glucanase | BGL2 | Saccharomyces cerevisiae | 343 |
| 472 | |||
| Endo-β-1,3-1,4-glucanase I (EGI) | (EGI) + egl1 | Trichoderma reesei | 4, 27, 522, 696 |
| Endo-β-1,3-1,4-glucanase III (EGIII) | egl1, egl3 egl4, egl5 | Trichoderma reesei | 522, 525, 586, 588, 655, 784 |
| FI-carboxymethylcellulase | Aspergillus aculeatus | 505, 688 | |
| Endo-β-1,3-1,4-glucanase | Barley | 308, 679 | |
| Endo-β-1,3-glucanase | Nicotiana plumbaginifolia | 144 | |
| Exo-β-glucanases | |||
| Exo-β-1,4-glucanase | cex | Cellulomonas fimi | 130 |
| Exo-β-1,3-glucanase I + II | EXG1/BGL1 | Saccharomyces cerevisiae | 482 |
| Exo-β-1,3-glucanase III | EXG2 | Saccharomyces cerevisiae | 343 |
| Cellobiohydrolases | |||
| Cellobiohydrolase | CEL3 | Agaricus bisporus | 108 |
| Cellobiohydrolase | cbh1 | Aspergillus aculeatus | 475, 476, 661 |
| Cellobiohydrolase | Clostridium thermocellum | 583, 689 | |
| Cellobiohydrolase | Penicillium janthinellum | 351 | |
| Cellobiohydrolase | CBH1 | Phanerochaete chrysosporium | 531, 703, 705 |
| Cellobiohydrolase | Thermophilic bacteria | 689 | |
| Cellobiohydrolase I (CBHI) | cbh1 | Trichoderma reesei | 4, 521, 696 |
| Cellobiohydrolase II (CBHII) | cbh2 | Trichoderma reesei | 4, 27 |
| 525, 783 | |||
| Endo/exoglucanases | |||
| Endo/exoglucanase | cel | Bacillus sp. strain DO4 | 107, 241 |
| Cellodextrinases | |||
| Cellodextrinase | celA | Ruminococcus flavefaciens | 531, 702, 705 |
| β-Glucosidases | |||
| β-Glucosidase | bgl1 | Aspergillus aculeatus | 661, 688 |
| β-Glucosidase | Aspergillus niger | 524 | |
| β-Glucosidase | bgl | Bacillus circulans | 106, 107 |
| β-Glucosidase | bglA, bglB, bglC | Cellulomonas biazotea | 550, 551 |
| β-Glucosidase | BGLN | Candida molischiana | 590 |
| β-Glucosidase | Candida pelliculosa | 352 | |
| β-Glucosidase | Kluyveromyces lactis | 558, 559 | |
| Cellobiase | BGL1 | Saccharomycopsis fibuligera | 413, 531, 705 |
| β-Glucosidase | BGL2 | Saccharomycopsis fibuligera | 413 |
The Bacillus circulans β-1,3-glucanase gene (bglH) has also been expressed in S. cerevisiae (480). Transcription of bglH was directed by the yeast GAL1 and SUC2 gene promoters, whereas secretion of β-glucanase was directed by the SUC2-encoded leader peptide. The presence of bacterial β-1,3-glucanase in the medium caused inhibition of yeast growth and cell expansion. Expression of SUC2-bglH in S. cerevisiae resulted in decreased cell size and expansion of vacuoles. The cause of these different symptoms was interpreted to be erosion of the β-1,3-glucan-containing cell wall by the exogenous enzyme and expression of bglH, leading to stress in the cells. The toxic effect of this bacterial β-1,3-glucanase in S. cerevisiae was evaded by prolonged culturing (15 days) at low temperature (16°C). Cho and Yoo (106) achieved high levels of expression when the B. circulans β-glucosidase gene (bgl) and the endo/exoglucanase gene (cel) from an unidentified Bacillus sp. (strain DO4) were inserted between the yeast ADH1 promoter and the PGK1 terminator sequences, with no apparent inhibitory effect on the growth of S. cerevisiae transformants.
Two endo-β-glucanase genes from thermophilic bacteria were cloned and expressed in S. cerevisiae without replacing the bacterial gene promoters and secretion signal sequences. The specific activity of the C. thermocellum enzyme synthesized in yeast was about 28% of that found in E. coli (580). By contrast, the specific activity of the endo-β-glucanase from an unidentified thermophilic anaerobe produced in S. cerevisiae was three- to sevenfold higher than in E. coli (281). The cellobiohydrolase gene from the latter bacterium was also expressed in S. cerevisiae (583). When this gene was placed under the control of the SUC2 promoter and invertase secretion signal, no cellobiohydrolase was secreted by the yeast transformant, suggesting that the translocation of the hybrid protein was influenced by the protein structure (689). When the SUC2 sequences were replaced by the STA1 promoter and glucoamylase secretion signal, the recombinant yeast secreted approximately 40% of the cellobiohydrolase synthesized into the culture medium. Alteration of the amino acid residues at the cleavage site resulted in a 3.5-fold increase in the total cellobiohydrolase production but did not affect the efficiency of secretion into the medium (689).
The endo-β-1,4-glucanase gene (cenA) from C. fimi was expressed in S. cerevisiae under the control of the yeast ADH1 and MEL1 (melibiase) gene promoters (627, 628). Secretion of the active enzyme by S. cerevisiae was greatly increased when the leader signal of the K1 killer toxin of melibiase was inserted immediately upstream of, and in frame with, the bacterial β-glucanase sequence. An S. cerevisiae strain expressing both the C. fimi cenA-encoded endo-β-1,4-glucanase (Eng) and the cex-encoded exo-β-glucanase (Exg) was able to saccharify filter paper and pretreated aspen wood chips in reaction mixtures that were supplemented with β-glucosidase (130, 754). In a similar study, the endoglucanase gene (515) and the β-glucosidase genes (551, 552) from Cellulomonas biazotea were also cloned and successfully expressed in S. cerevisiae.
In barley, approximately 75% of the endosperm cell wall consists of β-1,3-1,4-glucans. During germination as well as malting in the brewhouse, β-glucanase isoenzymes are secreted from the aleurone and scutellum and released into the endosperm. With the aim of constructing a glucanolytic brewing yeast strain, the β-1,3-1,4-glucanase gene from barley was cloned and expressed in S. cerevisiae (308). This gene was fused in frame to the signal sequences of the mouse α-amylase, yeast PHO5-encoded phosphatase, and SUC2-encoded invertase (504). These constructs were inserted behind the ADH1 and PGK1 promoters. The replacement of the ADH1 promoter with the PGK1 promoter resulted in a 20- to 100-fold increase in β-glucanase activity. The invertase leader peptide directed secretion of the β-1,3-1,4-glucanase more efficiently than did the leader peptides of α-amylase and phosphatase.
Regulated expression of the Nicotiana plumbaginifolia β-1,3-glucanase gene in S. cerevisiae resulted in a recombinant strain which can controllably lose some of its cell wall rigidity without lysis (144). This gene was expressed under the control of the yeast GAL1 promoter and the MFα1 secretion signal sequence. Production of β-1,3-glucanase by S. cerevisiae led to a strong growth inhibition by interfering with the cell wall growth from within the cell, and to a loss of up to 20% of some periplasmic enzymes as evidenced by the release of normally periplasmic-associated invertase.
Several cellulase genes from various fungi have also been expressed in S. cerevisiae. Five endo-β-1,4-glucanases (encoded by egl1, egl2, egl3, egl4, and egl5) and two cellobiohydrolases (CBHI and CBHII) from T. reesei were efficiently secreted into the culture medium by S. cerevisiae transformants (4, 27, 521, 522, 525, 586, 588, 655, 783, 784). Similar results were observed when the endo-β-1,4-glucanase gene from Trichoderma longibrachiatum was expressed in a wine yeast strain (528, 529). The cDNA copies of egl1, egl3, cbh1, and cbh2, carrying the T. reesei signal sequences, were expressed under the control of the yeast PGK1 and ENO1 gene promoters. These enzymes were not secreted efficiently until the late exponential or stationary growth phase, rendering the yeast cells larger and more irregular in shape (523). Both the endo-β-1,4-glucanases and cellobiohydrolases carrying the T. reesei leader peptides, efficiently entered the secretory pathway of S. cerevisiae, but they were produced in highly glycosylated forms and were heterogeneous in size. It would seem that these proteins had undergone extensive elongation of the outer mannose chains in the Golgi apparatus. However, these large and extensively glycosylated proteins were secreted efficiently and passed through the yeast cell wall to the culture medium. More than 80% of these cellulases were detected in the yeast culture medium. Most of the intracellular enzymes were located in the periplasmic space, and practically no enzyme was found soluble in the cytoplasm (523). Despite overglycosylation of the cellulases produced by S. cerevisiae, the specific activity of the yeast-made EGI was not significantly altered whereas a slight decrease in the specific activity of the yeast-made CBHII was observed in comparison with the native enzymes (27). The binding of the recombinant CBHII at a concentration of 30 μg/ml to crystalline cellulose was also decreased since only 50 to 70% of the yeast-made enzyme was bound to cellulose under conditions where 100% of the T. reesei enzyme was bound (521). This reduced ability to bind to the substrate was attributed to hyperglycosylation. However, the affinity of recombinant CBHII for crystalline cellulose was concentration dependent, indicating that the yeast-produced hyperglycosylated enzyme probably forms aggragates that bind less efficiently (523). CBHII still had greater activity than EGI against crystalline cellulose, whereas in the case of amorphous substrate the order was reversed (27). Evidence for synergism was obtained when mixtures of the two recombinant enzymes were used with a constant total protein dosage. Both yeast-produced enzymes were active against barley β-1,3-1,4-glucan but were inactive against β-1,3-1,6-glucan (laminarin). CBHII was inactive against xylan, whereas EGI exhibited considerably greater activity against insoluble, unsubstituted hardwood xylan than against amorphous cellulose. By comparison with two purified xylanases of T. reesei, the recombinant EGI produced xylooligosaccharides with longer mean chain length when acting on both substituted and unsubstituted xylan substrates (27).
Another fungal cellobiohydrolase gene that was cloned and expressed in S. cerevisiae originated from P. chrysosporium. A cDNA fragment encoding the cellobiohydrolase (cbh1-4) was amplified and cloned by PCR and expressed. The cbh1-4 gene was successfully expressed in S. cerevisiae under the control of the PGK1 promoter. The native P. chrysosporium signal sequence mediated secretion of cellobiohydrolase in S. cerevisiae. The construct was designated CBH1 (703). The CBH1-encoded activity in the recombinant S. cerevisiae was, however, rather low. By contrast, it was reported that, when the cellobiohydrolase gene (cel3) from Agaricus bisporus was fused to the yeast SUC2-encoded secretion signal, the yeast transformants showed enzymatic activity toward cellulose. However, long reaction times were required for degradation of CMC (108). Similar results were reported for the expression of a cellulase gene (resembling the P. chrysosporium cbh1-4 gene) from Penicillium janthinellum in S. cerevisiae (351).
In other work directed toward developing cellulolytic yeasts, a cDNA fragment encoding the FI-carboxymethylcellulase (FI-CMCase) of Aspergillus aculeatus was linked to the GAP (glyceraldehyde-3-phosphate dehydrogenase) promoter and transformed into S. cerevisiae (505). Production of FI-CMCase by S. cerevisiae was shown to be growth associated, but 92% of the total enzyme activity was not secreted into the medium. Cloning and expression of cellobiohydrolase (cbh1) and β-glucosidase (bgl1) genes of A. aculeatus in S. cerevisiae have been reported in a series of four papers (474, 476, 661, 688). It was found that by coexpressing the A. aculeatus cellobiohydrolase (cbh1) and β-glucosidase (bgl1) genes in S. cerevisiae the yeast transformants were able to hydrolyze up to 59% of added Avicel (661). Murai et al. (474, 475) succeeded in anchoring these enzymes on the cell surface of S. cerevisiae. The cellobiohydrolase and β-glucosidase genes, linked to the yeast glycoraldehyde-3-phosphate dehydrogenase promoter, were individually fused with the gene encoding the C-terminal half (320 amino acid residues from the C terminus) of the yeast α-agglutinin before they were introduced jointly into S. cerevisiae. These chimeric enzymes were delivered to the yeast cell surface by the secretion signal sequence of the native signal sequence of the A. aculeatus cellobiohydrolase and by the secretion signal sequence of the Rhizopus oryzae glucoamylase for β-glucosidase, respectively. The cellobiohydrolase and β-glucosidase activities were detected in the cell pellet fraction, not in the culture supernatant. The display of these enzymes on the yeast cell surface was confirmed by immunofluorescence microscopy, and yeast cells displaying these cell surface-anchored enzymes could grow on cellobiose or water-soluble cellooligosaccharides as the sole carbon source. This report by Murai et al. (475) demonstrated that the cell surface-engineered yeast with these “immobilized” cellobiohydrolase and β-glucosidase proteins could be endowed with the ability to assimilate cellooligosaccharides.
To enable S. cerevisiae to assimilate cellooligosaccharides, the Ruminococcus flavefaciens cellodextrinase gene (celA) was inserted between the yeast ADH1P-MFα1S expression-secretion cassette and the TRP5T terminator (702). The ADH1P-MFα1S-celA-TRP5T (designated CEL1) construct conferred to S. cerevisiae transformants an ability to synthesize and secrete cellodextrinase. This enzyme has a predominantly exo-type action on cellooligosaccharides, and cellobiose is the major end product of cellodextrin hydrolysis.
S. cerevisiae does not contain any β-glucosidase activity and seems to lack an uptake system for cellobiose. With the aim of constructing cellobiose-fermenting strains of S. cerevisiae, β-glucosidase genes were isolated from A. niger, Candida pelliculosa var. acetaetherius, S. fibuligera, and Kluyveromyces lactis (352, 413, 524, 558, 559). A laboratory strain of S. cerevisiae producing the K. lactis β-glucosidase intracellularly failed to grow on cellobiose. Growth could be achieved only if a strain permeable to the sugar derivative 5-bromo-4-chloro-3-indolyl-β-d galactoside (X-Gal) (and most probably to cellobiose) was used as a host (559). Approximately 80% of the C. pelliculosa β-glucosidase produced by S. cerevisiae was located in the periplasmic space (352). This periplasmic β-glucosidase activity facilitated growth on cellobiose. The β-glucosidase gene of Candida molischiana has also been cloned and successfully expressed in S. cerevisiae (590). In another instance, two β-glucosidase genes (BGL1 and BGL2) derived from S. fibuligera were expressed in S. cerevisiae (413). The substrate specificities of these two enzymes differed; the BGL1-encoded cellobiase hydrolyzed cellobiose efficiently, whereas the BGL2-encoded aryl-β-glucosidase did not. This finding is consistent with the observation that the S. cerevisiae transformant carrying the BGL1 fermented cellobiose to ethanol but the transformant carrying BGL2 did not.
Van Rensburg et al. (705) have introduced into S. cerevisiae genes chosen with the intention of expressing a rudimentary cellulase system: the B. fibrisolvens endo-β-1,4-glucanase (END1), the P. chrysosporium cellobiohydrolase (CBH1), the R. flavefaciens cellodextrinase (CEL1), and the S. fibuligera cellobiase (BGL1) gene constructs. The END1, CBH1, and CEL1 genes were inserted into yeast expression-secretion cassettes. Expression of END1, CBH1, and CEL1 was directed by the promoter sequences derived from the ADH2, PGK1, and ADH1 genes, respectively. In contrast, BGL1 was expressed under the control of its native promoter. Secretion of End1p and Cel1p was directed by the MFα1 signal sequence, whereas Cbh1p and Bgl1p were secreted using their native leader peptides. The construction of a fur1::ura3 S. cerevisiae strain allowed the autoselection of this multicopy URA3-based plasmid in rich medium. S. cerevisiae transformants secreting biologically active endo-β-1,4-glucanase, cellobiohydrolase, cellodextrinase, and cellobiase were able to hydrolyze various substrates including CMC, hydroxyethyl cellulose, laminarin, barley glucan, cellobiose, polypectate, birchwood xylan, and methyl-β-d-glucopyranoside.
In a later study the gene (cel) encoding the bifunctional endo/exoglucanase of a Bacillus sp. (241) was coexpressed with the β-glucosidase gene (bgl) of B. circulans (106). Using the δ sequences of the Ty1 retrotransposon as target sites for homologous recombination, the ADH1P-cel-PGK1T and ADH1P-bgl-PGK1T gene constructs were inserted at approximately 44 sites into the chromosomes of S. cerevisiae (106). When this δ-integrated recombinant S. cerevisiae strain was used in simultaneous saccharification and fermentation, an ethanol concentration of 2 wt% was obtained after 12 h from 50 g of microcrystalline cellulose per liter (107). Using this engineered strain expressing multiple copies of the bacterial endo/exoglucanase and β-glucosidase genes, a significantly reduced amount of commercial enzyme preparation was required during the SSF-based conversion of microcrystalline cellulose into ethanol.
Petersen et al. (531) took the concept of constructing a cellulose-degrading yeast one step further by engineering an S. cerevisiae strain for the degradation of four polysaccharides, i.e., starch, pectin, cellulose, and xylan (the main component of hemicellulose). This engineered S. cerevisiae strain contained the L. kononenkoae α-amylase gene (LKA1), the E. chrysanthemi pectate lyase gene (PEL5), the E. carotovora polygalacturonase gene (PEH1), the B. fibrisolvens endo-β-1,4-d-glucanase gene (END1), the P. chrysosporium cellobiohydrolase gene (CBH1), the S. cerevisiae exo-β-1,3-d-glucanase gene (EXG1), the S. fibuligera cellobiase gene (BGL1), and the A. niger endo-β-d-xylanase gene (XYN4) (531). This strain was able to grow on starch, pectate, and cellobiose, but the degradation of cellulose (Solka Floc and lichenan) and xylan was insignificant.
Expression of cellobiohydrolases (CBHs) in S. cerevisiae has been a particular focus of researchers in the field because of the vital role such enzymes play in degrading crystalline cellulose. Takada et al. (661) expressed the cbhI gene of A. aculeutus and used the resulting protein in conjunction with additional cellulases produced by S. cerevisiae to achieve up to 59% hydrolysis of Avicel. Cho and Yoo (106) reported measurable filter paper activity associated with production of an endo/exoglucanase originating from B. subtilis. Notwithstanding these notable studies, hydrolysis of high-crystallinity cellulose with enzyme preparations including CBHs produced by recombinant S. cerevisiae has not been widely reported, and has proved more challenging than has functional production of other classes of cellulase enzymes. Functional CBH expression represents at present a bottleneck to CBP organism development and to growth enablement (considered below) on crystalline cellulose in particular. Further progress in this area, including understanding the basis for both the successes and difficulties encountered in work to date, is an important goal for future research.
Another objective for current and future research on the development of CBP S. cerevisiae strains is the improvement of the secretory expression of the above-mentioned saccharolytic enzymes. High-level secretion of heterologous proteins, or native proteins for that matter, is not as readily achieved in S. cerevisiae as in some bacteria, fungi, or other yeast species (e.g., Pichia pastoris, Hansenula polymorpha, K. lactis, and Yarrowia lipolytica). Notwithstanding obstacles such as hyperglycosylation and hindered secretion due to the cell wall, there is an increasing number of examples of effective secretion of heterologous proteins by S. cerevisiae. Native secretion sequences have been found sufficient to effect proper posttranslational processing and secretion of functional proteins in the case of genes originating from fungal sources, including EgI, EgII, and Xyn2 of T. reesei (367, 368, 521); XynC, XlnA, and Man1 of Aspergillus (129, 398, 611); and a glucoamylase gene. In-frame fusions to the yeast MFα1S secretion sequence have been used to express in S. cerevisiae saccharolytic genes from bacteria, including the end1 gene of B. fibrosolvens, the cel1 gene of R. flavefaciens, the beg1 gene of B. subtilis, and the xlnD gene of A. niger (367, 368, 701, 702, 703). Although these proteins were often extensively glycosylated, they were still efficiently secreted through the yeast cell wall into the medium (367, 368). Secretion of a mannanase (Man1) of A. aculeatus was recently reported at levels corresponding to about 5% of cellular protein (611). Looking beyond saccharolytic enzymes, several mutant strains with a “supersecreting” phenotype showing substantially increased secretion of particular proteins have been isolated. For example, an ssc1 (pmr1) ssc2 double mutant secreted prochyomosin, bovine growth hormone, and scuPA at levels 5- to 50-fold higher than did nonmutated controls. Specific manipulations involving both the leader sequence and structural gene resulted in a substantial (up to 4.8-fold) impact on levels of secretion of single-chain proinsulin-like molecules into the culture supernatant (339).
In light of results such as these, we believe that it is reasonable to pursue expression in S. cerevisiae of cellulase at levels sufficient to enable growth on crystalline cellulose as required for CBP. At the same time, we acknowledge that achieving elevated secretion levels in this organism has to date often been a hit-or-miss proposition without a strong mechanistic basis. Integrated advancement of our fundamental understanding along with investigation of strategies to increase secretion levels is likely to be a particularly fruitful direction for future research in the context of organism development for CBP.
(iii) Growth on nonnative substrates by virtue of heterologous expression of saccharolytic enzymes.
Studies addressing growth enablement in liquid medium by virtue of heterologous expression of saccharolytic enzymes are listed in Table 14. Data for α-linked substrates (starch) are included as well as data for β-linked substrates (cellobiose, cellodextrins, and cellulose) because the field is at present more advanced with respect to starch compared to cellulose and its derivatives. Results from the single bacterial study targeting microbial utilization of starch are encouraging since modest growth and ethanol production from 40 g of starch per liter were achieved under anaerobic conditions. Growth of S. cerevisiae on starch, under aerobic conditions in most cases, has been demonstrated in several studies (145, 474, 531, 646). In a significant breakthrough, Birol et al. (57) reported the utilization of 100 g of starch per liter with production of 44 g of ethanol per liter and 8 g of cells per liter. Although ethanol and cell yields on starch were similar to those on glucose, the specific growth rate was nearly 10-fold lower on starch.
TABLE 14.
Summary of studies aimed at growth and fermentation enablement in liquid medium by virtue of heterologous expression of saccharolytic genes
| Substrate and host | Enzymes (genes) | Substrate | Growth or fermentationa | Reference |
|---|---|---|---|---|
| α-Linked substrates | ||||
| K. oxytoca | α-Amylase from Bacillus stearothermophilus, pullulanase from Thermoanaerobium brockii | Starch (40 g/liter) | Anaerobic fermentation, 15 g of ethanol per liter and 1.4 g of cells per liter | 171 |
| S. cerevisiae | Glucoamylase (GA1) | Soluble starch | CO2 release comparable to control with added amylase | 302 |
| α-Amylase (amyE), glucoamylase (glaA) | Corn starch (10 g/liter) | Final optical density of 2.0 | 145 | |
| α-Amylase (LKA1) | Soluble starch (20 g/liter) | Aerobic growth, final optical density >1.5 | 646 | |
| Glucoamylase from Rhizopus oryzae | Soluble starch (10 g/liter) | Aerobic growth with some ethanol production, final optical density >0.9 | 476 | |
| Amylopullulanase (LKA1), pectate lyase (PEL5), polygalacturonase (PEH1), endo-β-1,4-glucanase (END1), cellobiohydrolase (CBH1), exo-β-1,3-d-glucanase (EXG1), cellobiase (BGL1), and endo-β-d-xylanase (XYN4) | Starch | Aerobic growth, cell number > control | 531 | |
| α-Amylase (amyE), glucoamylase (glaA) | Soluble starch (100 g/liter) | Anaerobic growth, 44 g of ethanol per liter and 8 g of cells per liter | 57 | |
| β-Linked substrates | ||||
| K. oxytoca | Endoglucanases (celY and celZ) | Avicel | Anaerobic SSF with added cellulase, ethanol yields up to 22% more than control | 778 |
| Amorphous cellulose | Cellulose fermentation without added cellulase, ethanol yields 58-76% theoretical | 781 | ||
| S. cerevisiae | CMCase, β-glucosidase from Aspergillus aculeatus | Cellobiose (10 g/liter) | Aerobic growth, final optical density >1.5 | 476 |
| CMCase, β-glucosidase from Aspergillus aculeatus | Cellodextrins | Aerobic growth, cell number > control | 476 | |
| Amylopullulanase (LKA1), pectate lyase (PEL5), polygalacturonase (PEH1), endo-β-1,4-glucanase (END1), cellobiohydrolase (CBH1), exo-β-1,3-d-glucanase (EXG1), cellobiase (BGL1), and endo-β-d-xylanase (XYN4) | Cellobiose | Aerobic growth, cell number > control | 531 | |
| Endo/exoglucanse from Bacillus sp. strain DO4, β-glucosidase genes from Bacillus circulans | Cellodextrins | Growth, 2-fold higher cell mass and greater ethanol production compared to control | 107 | |
| Endo/exoglucanse from Bacillus sp. strain DO4, β-glucosidase genes from Bacillus circulans | Avicel | Anaerobic SSF, showed substantial enzyme production under aerobic and anaerobic conditions | 106 |
“Control” refers to a strain not expressing heterologous saccharolytic enzymes.
With respect to β-linked substrates, growth of recombinant S. cerevisiae on cellobiose (475, 531) and cellodextrins (107, 531) has been demonstrated. Anaerobic simultaneous saccharification and fermentation (SSF) of Avicel has also been investigated using an S. cerevisiae strain expressing endo/exo glucanase and β-glucosidase originating from Bacillus species. This strain produced filter paper activity under both aerobic and anaerobic conditions but was not shown to grow or produce ethanol in the absence of added cellulase (106). Anaerobic SSF of Avicel using K. oxytoca strains SZ21 and SZ22 expressing celY and celZ endoglucanases (see “Enteric bacteria” above) resulted in ethanol yields up to 22% higher than a control that did not express heterologous cellulases. However, no ethanol formation was observed in the absence of added cellulase (778). In a subsequent study (781), K. oxytoca strain SZ21 was shown to be capable of fermenting amorphous to cellulose to ethanol at yields 58 to 75% of theoretical in the absence of added cellulase. This represents the most advanced embodiment of cellulose processing via CBP by virtue of heterologous expression of saccharolytic enzymes.
As a result of a relatively small number of studies, all but one of which were conducted since 1995, significant progress has been made in the area of utilization of nonnative substrates by virtue of heterologous expression of saccharolytic enzymes. In the case of starch, the results of Birol et al. (57) are encouraging not only with respect to the feasibility of starch utilization, but also the with respect to the overall feasibility of enabling utilization of nonnative substrates. Increased rates of growth and hydrolysis as well as use of insoluble substrates would appear to be logical objectives for future work on starch utilization. Work aimed at microbial utilization of β-linked substrates has been undertaken more recently than work aimed at starch, with the first such studies appearing in 1998, and is less advanced. Growth on cellobiose and cellodextrins provides a point of departure that can be built upon in future work. It is desirable to build upon such results by examining benefits of endogenous cellulase production during SSF as well as anaerobic growth in the absence of cellulase enzymes, perhaps first on amorphous cellulose and then on crystalline cellulose. Recent work involving both enteric bacteria (778, 781) and yeast (106) represents significant first steps in this direction.
CONCLUDING DISCUSSION
Fundamentals
The last decade has seen marked advances in the depth and breadth of scientific understanding with respect to the structure, function, and genetics associated with the components of cellulase systems. Such advances include solving the 3-D structures of over two dozen cellulases, leading to a much better understanding of reaction mechanisms; the availability of many new protein sequences (300 in 1990, over 5,000 in 2001); meaningful new classification schemes based on structural features; and a better understanding of the regulation of cellulase genes, especially in fungal systems. Significant progress has also been made since 1990 with respect to understanding interactions among cellulase components. This includes a better understanding of synergistic interactions for an increasing number of noncomplexed cellulase systems, as well as a better and broader understanding of the structure and composition of cellulosomes. We expect that expanding knowledge of the molecular details of cellulose hydrolysis will continue at an accelerated pace during the coming decade. This expectation is supported by the progress made in the last decade, the powerful new tools that continue to become available, and the talent and dedication with which a substantial cadre of scientists is pursuing these issues.
Understanding cellulose hydrolysis as a microbial phenomenon builds on the foundation of knowledge pertaining to cellulose hydrolysis at a subcellular level but encompasses additional questions and lines of inquiry that are cellular in character. Such questions include those listed in Table 15.
TABLE 15.
Questions inherent to understanding microbial cellulose utilization most appropriately pursued by studies at a cellular level
| Physiological feature | Question(s) relative to growth on cellulose |
|---|---|
| Uptake of cellulose hydrolysis products | What is the distribution of oligoglucan chain lengths taken up by cellulolytic organisms? |
| What is the bioenergetic requirement for substrate transport? | |
| Metabolic control | What is the allocation of substrate carbon to cellulase and cell synthesis, how does this vary with growth conditions, and can such observations be reconciled with our understanding of gene expression at a subcellular level? |
| Bioenergetics | What is the allocation of ATP to cellulase and cell synthesis, and how does this vary with growth conditions? |
| What is the relative importance of phosphorolytic and hydrolytic cleavage of cellodextrins and/or cellobiose, does this depend on growth conditions, and, if so, how is it regulated? | |
| Can the rate of ATP production (e.g., due to phosphorolytic cleavage of oligomers, glycolysis, and post-pyruvate metabolism) be quantitatively reconciled with the rate of ATP consumption (e.g., due to substrate transport, cell synthesis, cellulase synthesis, maintenance, energy spilling, and perhaps other factors)? | |
| Considering all factors—including but not necessarily limited to ATP available from cellobextrin phosphorylation, ATP requirements for substrate transport, and the ATP required for cellulase synthesis—is the ATP available for growth of a microorganism growing on cellulose less than or greater than that available for growth of the same organism on soluble mono- or disaccharides? | |
| Cellulose-microbe adhesion and glycocalyx formation | What are the roles and relative importance of glycocalyces, cellulosomes, and other factors in microbial adhesion to cellulose, and how much does this vary among different cellulolytic microorganisms? |
| What is the relative prevalence, origin (e.g., accretion or de novo synthesis), and bioenergetic cost associated with synthesis and assembly of glycocalyx components? | |
| What function is served by the “protuberances” observed to form between cellulose and some cellulolytic microorganisms? | |
| Rates of microbial cellulose hydrolysis (pure cultures) | What are the comparative rates of cellulose hydrolysis by different microorganisms on both a cell- and cellulase-specific basis, and what factors are responsible for observed differences? |
| What is the relative effectiveness of cellulase systems in cellulose-enzyme-microbe complexes compared to cellulose-enzyme complexes? | |
| If cellulase-microbe synergy can be demonstrated, what is its mechanistic basis? | |
| Ecology and evolution | What are the ecological strategies and niches of various cellulolytic organisms, including interactions with other organisms, spatial relationship to the substrate (e.g., adhesion and penetration of pores by filaments), and the physical environment? |
| Can ecological strategies and niches be reconciled with cellular and subcellular features of the cellulolytic apparatus, and can the selective pressures leading to the evolution of such features be understood? | |
| In cases where cell-cell synergy can be demonstrated, what is its quantitative significance and mechanistic basis? |
The important body of work on microbial cellulose utilization undertaken to date is implicit in the framing of questions such as these and also gives hints to their answers. Still, investigation of the questions listed in Table 15 is in a nascent stage of development for most organisms.
Important tools for understanding microbial cellulose utilization have in many cases become available only recently or have not yet been developed. Such tools include systems that allow foreign genes to be expressed in cellulolytic microorganisms, which are established for the aerobic T. reesei (520) but not for most cellulolytic anaerobes. The recent development of an electrotransformation system for C. cellulolyticum makes possible new studies of microbial cellulose utilization using homologous recombination-mediated gene knockout. Such studies can be expected to yield exciting comparative results as similar systems become available for more cellulolytic microorganisms that are not currently transformable. Methods to independently quantify cells and cellulase can be expected to result in a second set of new insights, particularly in the areas of bioenergetics, metabolic control, and kinetics. New methods are required to fractionate and characterize glycocalyces and would be quite informative if developed. Continuous culture on cellulosic substrates is just now beginning to be applied in ways that give insights extending beyond summary description of substrate conversion and product formation, and it can be expected to yield rich insights in the coming years, especially when coupled with new analytical methods. Studies in which heterologous cellulase expression confers the ability to grow on nonnative substrates have begun to appear only in the last few years and represent an exciting frontier with the potential to become an important tool for fundamentally oriented investigations while also being relevant to applied goals (discussed subsequently).
The substantial potential of quantitative analysis to contribute to our understanding of cellulose hydrolysis at both subcellular and cellular levels has been realized to date to a very limited extent. In contexts such as specific activity of cellulases (Table 5) and adsorption (Table 6), it is at present often difficult to draw quantitative conclusions that extend beyond the reach of a particular study. This limitation may be addressed by paying more attention to methodological standardization and by undertaking more interspecific comparative studies under controlled conditions. These measures can be expected to shed light on several fundamental issues of considerable interest about which there is currently substantial uncertainty, the relative efficacy of complexed and noncomplexed cellulase systems being a case in point. Quantitative studies at different levels of aggregation (e.g., subcellular, pure culture, defined mixed culture, and undefined mixed culture) have great potential as a framework to test and develop our understanding but have seldom been undertaken.
Explanations for the features and molecular diversity of cellulase enzyme systems are logically sought in an understanding of the niches and adaptive strategies of the microorganisms in which these systems evolved. Conversely, results of molecular studies substantially enhance the depth and clarity with which the adaptive strategies of cellulolytic microorganisms can be understood. This potentially important complementarity can be more fully exploited in the future as understanding of microbial cellulose utilization advances.
Biotechnology
Several factors motivate the development of microorganisms possessing the properties required for cost-effective implementation of CBP in an industrial setting. The primary savings anticipated for CBP compared to other described process configurations featuring enzymatic hydrolysis result from elimination of costs associated with a process step dedicated to cellulase production. Other benefits which might be realized include higher product yields, higher rates, and improved stability of cultures and strains. Realization of these benefits is, however, by no means certain. Higher yields of fermentation products might be achieved because of the absence of oxidative metabolism in a CBP configuration. Higher rates might be achieved because of (i) the availability of all of the feedstock for cellulase production in CBP (compared to a small fraction of feedstock in other process configurations), (ii) the possibility of using high-specific-activity complexed cellulases in organisms developed for CBP, and (iii) cell-cellulase synergy. Improved culture stability may result for CBP as compared to processes featuring dedicated cellulase production and fermentation by noncellulolytic microbes because adherent cellulolytic bacteria used for CBP can be expected to outcompete many contaminants for products of cellulose hydrolysis. Moreover, selective pressure might maintain or improve high rates of microbial cellulose utilization in a CBP configuration whereas overproduction of cellulase in processes featuring dedicated cellulase production has negative selective value. In addition to its substantial cost reduction potential (see “Process configurations” above), it should be appreciated that CBP is in principle applicable to production of any fermentation product from cellulosic biomass.
The feasibility of CBP will be fully established only when a microorganism or microbial consortium is developed that satisfies the requirements discussed above (see “Strategies”). Short of such a definitive demonstration, analysis of the feasibility of CBP may be approached by considering data for naturally cellulolytic anaerobes or by using quantitative models. Nearly 90% of the cellulose in pretreated hardwood is hydrolyzed in 12 h by C. thermocellum grown in continuous culture at low feed substrate concentrations (407). Values for first-order rate constants for Avicel hydrolysis by cellulolytic anaerobes, 0.05 to 0.16 h−1 (Table 8), are consistent with reaction times of 44 to 14 h to achieve 90% cellulose hydrolysis. In light of these results, it may be inferred that the modest ATP available from anaerobic metabolism is sufficient to support the synthesis of both cells and cellulase at levels resulting in rather high rates of hydrolysis for highly crystalline cellulosic substrates. However, this has to date been demonstrated only at low substrate concentrations. Organisms engineered not to produce acetic acid, or which do not produce acetic acid naturally (e.g., Saccharomyces or Lactobacillus used as hosts for the recombinant cellulolytic strategy), will have somewhat lower GATP values than will naturally occurring cellulolytic anaerobes. Results from a model incorporating fermentative ATP generation, ATP expenditure for cellulase and cell synthesis, and cellulase kinetics indicate that acceptably fast hydrolysis rates are reasonable to expect with GATP = 2 ATP/mol (710). In particular, if a cellulase specific activity equal to that of T. reesei is assumed, then predicted reaction times are the same for CBP and current SSF configurations. At cellulase specific activities higher than that of T. reesei, predicted reaction times are shorter for CBP.
Organism development milestones for the native cellulolytic strategy are as follows. (i) Develop practical and reproducible genetic systems permitting heterologous gene expression and gene knockout in a variety of cellulolytic anaerobes. (ii) Unambiguously identify the factor(s) limiting growth under the conditions of interest, and establish that product tolerance is sufficient in a process context. (iii) Demonstrate that high product yields can be obtained in engineered strains. (iv) Demonstrate high-rate conversion of concentrated feedstocks (e.g., >50 g of carbohydrate per liter) by high-yielding strains. (v) Demonstrate that organisms can function adequately under conditions typical of an industrial environment, including tolerance to any inhibitors generated during pretreatment.
Genetic system development (milestone i) has to date been achieved only for C. cellulolyticum. The other milestones listed have not been achieved for naturally cellulolytic anaerobes. Successful realization of milestone i appears necessary to obtain high product yields in engineered strains (milestone iii) The feasibility of engineering high-yielding strains is supported by both stoichiometric considerations and successful metabolic engineering efforts involving similar end product metabolism (see “Metabolic engineering” above). A departure point for understanding product tolerance (milestone ii) is to determine the extent to which inhibition by salts resulting from organic acid production and addition of base for pH control is widespread. If it can be shown that such salt inhibition is a general explanation for the discrepancy between tolerance to ethanol and the maximum concentration of ethanol produced, this would have two important implications. First, the prospects for using naturally cellulolytic organisms in industrial CBP processes would be substantially improved. Second, It would be necessary to develop high-yielding strains producing little or no organic acids, and hence correspondingly low concentrations of salts resulting from pH neutralization, before high-rate conversion of concentrated feedstocks can be observed. That is, milestone iii would have to be achieved before milestone iv could be achieved.
Demonstrating that naturally cellulolytic organisms can function adequately in industrial environments (milestone v) is an important milestone that requires further study if it is to be achieved. With respect to pretreatment-generated inhibitors in particular, this milestone can be approached by identifying and developing an organism that exhibits tolerance to the inhibitors produced by a particular process or by identifying and developing a process that generates inhibitors that an organism can tolerate, or a combination of these. Understanding the mechanism of inhibitory effects and taking full advantage of organism's often considerable ability to evolve inhibitor resistance (e.g., in continuous culture) are likely to be important in this context.
Organism development milestones for the recombinant cellulolytic strategy are as follows. (i) Demonstrate that growth on noncrystalline cellulose is enabled and/or greater extents of hydrolysis are achieved in SSF of crystalline cellulose relative to a wild-type control. (ii) Demonstrate functional production and secretion of a variety of exoglucanases. (iii) Demonstrate an ability to grow on crystalline cellulose in the absence of added cellulases. (iv) Optimize cellulase expression and secretion based on a noncomplexed cellulase system to increase growth rates. (v) Express one or preferably several complexed cellulase systems (in different organisms) and compare their effectiveness to that of noncomplexed systems. (vi) Evaluate whether it is beneficial or necessary to incorporate additional features of naturally occurring cellulolytic bacteria.
Milestone i has been achieved for enteric bacteria and to some extent for S. cerevisiae (see “Growth on nonnative substrates by virtue of heterologous expression of saccharolytic enzymes” above). The other milestones listed have not been achieved. Functional expression of exoglucanase enzymes (milestone ii) has proved challenging in yeasts for reasons that are still not clear (see “Expression of heterologous cellulase genes in S. cerevisiae” above) and is very probably a prerequisite to demonstrating growth on crystalline cellulose in the absence of added enzymes (milestone iii). First-generation strains capable of such growth will probably grow quite slowly and will therefore benefit from improvement and optimization via a variety of approaches (milestone iv).
Anaerobic cellulolytic bacteria from nature meet the challenge of growth on a recalcitrant substrate and a very tight ATP budget by a collection of features that extend beyond the production of a functional cellulase system. These features include high-specific-activity cellulases of the complexed type, cell-substrate attachment, coupling of sugar phosphorylation to hydrolysis of β-glucosidic bonds (to at least some extent), glycocalyx formation, and (we think probably) energy-efficient substrate transport. For microorganisms expressing a heterologous cellulase system to achieve rates of growth and cellulose hydrolysis under anaerobic conditions, it may be advantageous or necessary for them to exhibit some or all of these features. Milestones v and vi reflect this perspective.
The two organism development strategies for CBP are associated with distinctive strengths and challenges. For the native cellulolytic strategy, organism development begins with organisms having highly evolved cellulose enzyme systems and metabolic features specific to cellulose that may be difficult to entirely replicate in an organism developed according to the recombinant cellulolytic strategy. However, the recombinant cellulolytic strategy begins with organisms having well-established properties related to robustness in industrial environments that may be difficult to replicate in an organism developed according to the native cellulolytic strategy. Although abstract arguments can be advanced about which strategy is more promising, our view is that such arguments cannot be conclusive on the basis of what we know today and that both strategies have merit and should be pursued. In support of this view, each strategy involves considerable uncertainties, different strategies may prove most advantageous for different products, organism development based on both native and recombinant substrate utilization strategies has proven successful for substrates other than cellulose (410), and the benefits of CBP are large enough to merit a parallel approach.
An added reason to pursue both the native and recombinant cellulolytic strategies is that each strategy has marked potential to inform the other. The recombinant strategy is informed by features of naturally cellulolytic bacteria specific to cellulose utilization. In addition, hydrolysis rates exhibited by naturally cellulolytic bacteria provide a performance standard for recombinant cellulolytic microbes. The native cellulolytic strategy is informed in the course of recreating functional cellulase systems one at a time pursuant to the recombinant cellulolytic strategy. In addition, product yields and tolerance exhibited by recombinant strategy host organisms—including S. cerevisiae, E. coli, and potentially others—represent a performance standard for cellulolytic microorganisms with engineered end product metabolism.
Both CBP organism development strategies involve very large challenges that will probably require a substantial sustained effort to overcome. However, the rewards for success in this endeavor are correspondingly large.
Alternative Cellulose Hydrolysis Paradigms
The vast majority of studies investigating cellulose hydrolysis and cellulase enzyme systems have proceeded within the context of an enzymatically oriented intellectual paradigm. In terms of fundamentals, this paradigm focuses on cellulose hydrolysis as primarily an enzymatic rather than microbial phenomenon. In terms of applications, the enzymatic paradigm anticipates processes featuring production of cellulase in a step separate from that in which the cellulosic feedstock is hydrolyzed for the purpose of conversion to a desired product. This paradigm is clearly manifested in statements accompanying the early work of pioneers in the field. For example, Sternberg (642) wrote in a 1976 statement attributed to Mandels and Weber (425): “Thousands of microorganisms have the ability to grow on cellulose. Although many of these grow quire rapidly only a few produce extracellular cellulases capable of converting crystalline cellulose to glucose in vitro.” Reese and Mandels (562) wrote in 1971: “The dream of cellulase investigators is to develop a commercially feasible process for converting waste cellulose to glucose. The recent successful commercial practice of enzymatically converting starch to glucose gives new hope to this dream. The chief problem is obtaining sufficiently active enzymes and highly reactive substrates so that relatively high sugar concentrations can be obtained in a reasonable time. The sugars produced can be used as a source of glucose, they can be converted to protein, or to fat, by feeding them to appropriate microorganisms. The sugars could also be used for production of alcohol.”
In response to the needs of the enzymatic paradigm, research focused primarily on microorganisms that actively secrete cellulases. Since higher levels of cellulase secretion are observed in aerobic microorganisms than in anaerobes, it was logical for studies inspired by the enzymatic paradigm to focus on cellulase production using aerobes as well as their noncomplexed cellulase enzyme systems.
An alternative microbially oriented paradigm considers cellulose hydrolysis as a microbial phenomenon and anticipates processes in a CBP configuration featuring cellulase production, cellulose hydrolysis, and fermentation in a single step. This microbial paradigm naturally leads to an emphasis on different fundamental issues, organisms, cellulase systems, and applied milestones compared to those of the enzymatic paradigm. Advancement of the microbial paradigm is fostered by investigating microbes, as opposed to enzymes, that rapidly hydrolyze cellulose. Issues associated with microbial cellulose utilization (see, e.g., Table 15) are thus central components of the body of fundamental knowledge underlying the microbial paradigm, whereas such issues are more peripheral relative to the enzymatic paradigm. It is logical in the context of the microbial paradigm to focus on anaerobic microorganisms and their distinctive complexed cellulase systems since the production of desired reduced product(s) is a required feature of processes configured according to the CBP concept. Thus, while T. reesei is exceptionally well suited to the needs of processes based on the enzymatic paradigm, it is ill suited with respect to the needs of the microbial paradigm in light of its unremarkable cellulose hydrolysis rates (see “Kinetics of microbial cellulose utilization” above) and production of water and CO2 as metabolic products. Applied milestones associated with advancing CBP by either the native or recombinant cellulolytic strategies (see above) have little overlap with milestones for the enzymatic paradigm.
A focus on the enzymatic paradigm was responsive to the tools available when pursuant studies were initiated nearly a half century ago. At that time, an industrial process could be imagined based on hydrolysis using cellulases recovered from an actively secreting organism such as T. reesei followed by high-yield fermentation of the resulting sugars to desired products by using available microorganisms such as yeast or lactic acid bacteria. By contrast, the development of microorganisms for CBP via strategies such as metabolic engineering of end product formation in naturally cellulolytic anaerobes or heterologous expression of cellulase enzymes could not have been imagined.
Today, biotechnology is central to progress toward applied objectives within the context of both the microbial and enzymatic cellulose hydrolysis paradigms. For the microbial paradigm, achieving the organism development milestones we foresee pursuant to both the native and recombinant cellulolytic strategies (see above) will be based to a very large extent on the successful application of biotechnological tools. For the enzymatic paradigm, cellulases with higher specific activity than current commercial preparations are highly desirable and are likely to be required (614). Developing such cellulases can be approached either by using new heterologous expression systems for cellulases that naturally have high specific activity or by using protein engineering to create new, improved enzyme systems. Notwithstanding the substantial differences between the microbial and enzymatic paradigms outlined in the preceding paragraphs, it is possible that results from work undertaken within both these paradigms may be incorporated into advanced technology in a convergent manner. For example, high-specific-activity cellulases could in principle be incorporated into CBP-enabling microorganisms.
In the decades since commercially feasible cellulose conversion processes featuring enzymatic hydrolysis were first envisioned by Reese, Mandels, and other pioneers in the field, sustainable resource supply, energy security, and global climate change have emerged as dominant issues affecting the well-being of humankind. The motivation for realizing this vision has thus increased substantially even as the magnitude of the challenges involved has become more apparent. In spite of the great effort that has been devoted to the field, there exist today biotechnological approaches to developing practical processes for the conversion of cellulose to fuels and commodity chemicals that are both promising and relatively unexplored. An important subset of such approaches involves microbial cellulose utilization.
Acknowledgments
L.R.L., W.V.Z., and I.S.P. were partially supported by cooperative agreement DE-FC36-00GO10591 from the U.S. Department of Energy; L.R.L. was also partially supported by grant 00-35504-9495 from the U.S. Department of Agriculture.
We are grateful to Bruce Dale and Robert Torget for personal communications cited herein. We also thank Wil Konigs, James Russell, Herb Strobel, Charles Wyman, and Yiheng Zhang for useful discussions.
REFERENCES
- 1.Acebal, C., M. P. Castillón, P. Estrada, I. Mata, E. Costa, J. Aguedo, D. Romero, and F. Jimenez. 1986. Enhanced cellulase production from Trichoderma reesei QM9414 on physically pretreated wheat straw. Appl. Microbiol. Biotechnol. 24:218-223. [Google Scholar]
- 2.Adachi, E., M. Torigoe, M. Sugiyama, J. Nikawa, and K. Shimizu. 1998. Modification of metabolic pathways of Saccharomyces cerevisiae by the expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH value. J. Ferment. Bioeng. 86:284-289. [Google Scholar]
- 3.Adney, W. S., C. I. Erhman, J. O. Baker, S. R. Thomas, and M. E. Himmel. 1994. Cellulase assays: methods from empirical mathematical models. ACS Symp. Ser. 566:218-235. [Google Scholar]
- 4.Aho, S., and M. Paloheimo. 1990. The conserved terminal region of Trichoderma reesei cellulases forms a strong antigenic epitope for polyclonal antibodies. Biochim. Biophys. Acta. 1087:137-141. [DOI] [PubMed] [Google Scholar]
- 5.Ahsan, M. M., M. Matsumoto, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Purification and characterization of the family J catalytic domain derived from the Clostridium thermocellum endoglucanase CelJ. Biosci. Biotechnol. Biochem. 61:427-431. [DOI] [PubMed] [Google Scholar]
- 6.Akin, D. E., W. S. Borneman, and C. E. Lyon. 1990. Degradation of leaf blades and stems by monocentric and polycentric isolates of ruminal fungi. Anim. Feed Sci. Technol. 31:205-221. [Google Scholar]
- 7.Akin, D. E., C. E. Lyon, W. R. Windham, and L. L. Rigsby. 1989. Physical degradation of lignified stem tissues by ruminal fungi. Appl. Environ. Microbiol. 55:611-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksenova, H. Y., F. A. Rainey, P. H. Janssen, G. A. Zavarzin, and H. W. Morgan. 1992. Spirochaeta thermophila, new species, an obligately anaerobic, polysaccharolytic, extremely thermophilic bacterium. Int. J. Syst. Bacteriol. 42:175-177. [Google Scholar]
- 9.Alexander, J. K. 1961. Characteristics of cellobiose phosphorylase. J. Bacteriol. 81:903-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander, J. K. 1972. Cellobiose phosphorylase from Clostridium thermocellum. Methods Enzymol. 28:944-948. [Google Scholar]
- 11.Alfredsson, G. A., J. K. Kristjánsson, S. Hjorleifsdóttir, and K. O. Stetter. 1988. Rhodothermus marinus, new genus new species, a thermophilic, halophilic bacterium from submarine hot springs in Iceland. Microbiology 134:299-306. [Google Scholar]
- 12.Alterthum, F., and L. O. Ingram. 1989. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl. Environ. Microbiol. 55:1943-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alzari, P. M., H. Souchon, and R. Domínguez. 1996. The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265-275. [DOI] [PubMed] [Google Scholar]
- 14.Amartey, S. A., D. J. Leak, B. S. Hartley. 1991. Effects of temperature and medium composition on the ethanol tolerance of Bacillus stearothermophilus LLD-15. Biotechnol. Lett. 13:627-632. [Google Scholar]
- 15.Anderson, K. L., Megehee, J. A., and V. H. Varel. 1998. Conjugal transfer of transposon TN 1545 into the cellulolytic bacterium Eubacterium cellulosolvens. Lett. Appl. Microbiol. 26:35-37. [DOI] [PubMed] [Google Scholar]
- 16.Aoyagi, H., M. Uemura, O. Hiruta, H. Takebe, and H. Tanaka. 1995. Estimation of microbial cell concentration in suspension culture by the osmotic pressure measurement of culture broth. Biotechnol. Tech. 9:429-434. [Google Scholar]
- 17.Aristidou, A. A., K.-Y. San, and G. N. Bennett. 1994. Modification of central metabolic pathway in Escherichia coli to reduce acetate accumulation by heterologous expression of the Bacillus subtilis acetolactate synthase gene. Biotechnol. Bioeng. 44:944-951. [DOI] [PubMed] [Google Scholar]
- 18.Aro, N., A. Saloheimo, M. Ilmén, and M. Penttilä. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 19.Atalla, R. H., J. M. Hackney, I. Uhlin, and N. S. Thompson. 1993. Hemicelluloses as structure regulators in the aggregation of native cellulose. Int. J. Biol. Macromol. 15:109-112. [DOI] [PubMed] [Google Scholar]
- 20.Atalla, R. H., and D. L. Vanderhart. 1984. Native cellulose: a composite of two distinct crystalline forms. Science 223:283-285. [DOI] [PubMed] [Google Scholar]
- 21.Aurilia, V., J. C. Martin, K. P. Scott, D. K. Mercer, M. E. A. Johnston, and H. J. Flint. 2000. Organisation and variable incidence of genes concerned with the utilization of xylans in the rumen cellulolytic bacterium Ruminococcus flavefaciens. Anaerobe 6:333-340. [Google Scholar]
- 22.Avgerinos, G. C., and D. I. C. Wang. 1983. Selective solvent delignification for fermentation enhancement. Biotechnol. Bioeng. 25:67-83. [DOI] [PubMed] [Google Scholar]
- 23.Ayers, W. A. 1959. Phosphorolysis and synthesis of cellobiose by cell extracts from Ruminococcus flavefaciens. J. Biol. Chem. 234:2819-2822. [PubMed] [Google Scholar]
- 24.Bader, J., K.-H. Bellgardt, A. Singh, P. K. R. Kumar, and K. Schügerl. 1992. Modeling and simulation of cellulase adsorption and recycling during enzymatic hydrolysis of cellulosic materials. Bioprocess Eng. 7:235-240. [Google Scholar]
- 25.Bagnara, C., C. Gaudin, and J. P. Bélaïch. 1987. Physiological properties of Cellulomoma fermentans, a mesophilic cellulolytic bacterium. Appl. Microbiol. Biotechnol. 26:170-176. [Google Scholar]
- 26.Bagnara, C., R. Toci, C. Gaudin, and J. P. Bélaïch. 1985. Isolation and characterization of a cellulolytic microorganism, Cellulomonas fermentans, sp. nov. Int. J. Syst. Bacteriol. 35:502-507. [Google Scholar]
- 27.Bailey, M. J., M. Siika-aho, A. Valkeajärvi, and M. E. Penttilä. 1993. Hydrolytic properties of two cellulases of Trichoderma reesei expressed in yeast. Biotechnol. Appl. Biochem. 17:65-76. [PubMed] [Google Scholar]
- 28.Bao, W. and V. Renganathan. 1992. Cellobiose oxidase of Phanerochaete chrysosporium enhances crystalline cellulose degradation by cellulases. FEBS Lett. 302:77-80. [DOI] [PubMed] [Google Scholar]
- 29.Barras, F., F. van Gijsegem, and A. K. Chatterjee. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32:201-234. [Google Scholar]
- 30.Baskaran, S., H.-J. Ahn and L. R. Lynd. 1995. Investigation of the ethanol tolerance of Clostridium thermosaccharolyticum in continuous culture. Biotechnol. Prog. 11:276-281. [Google Scholar]
- 31.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. [DOI] [PubMed] [Google Scholar]
- 32.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:379-386. [DOI] [PubMed] [Google Scholar]
- 34.Beavan, M. J., C. Charpentier, A. H. Rose. 1982. Production and tolerance of ethanol in relation to phopholipid fatty-acyl composition in Saccharomyces cerevisiae. J. Gen. Microbiol. 128:1447-1455. [Google Scholar]
- 35.Bedino, S., G. Testore, and F. Obert. 1985. Comparative study of glucosidases from the thermophilic fungus Thermoascus aurantiacus Miehe—purification and characterization of intracellular beta-glucosidase. Ital. J. Biochem. 34:341-355. [PubMed] [Google Scholar]
- 36.Béguin, P. 1990. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 44:219-248. [DOI] [PubMed] [Google Scholar]
- 37.Béguin, P., and P. M. Alzari. 1998. The cellulosome of Clostridium thermocellum. Biochem. Soc. Trans. 26:178-185. [DOI] [PubMed] [Google Scholar]
- 38.Béguin, P., and J.-P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 39.Béguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 40.Béguin, P., J. Millet, and J.-P. Aubert. 1992. Cellulose degradation by Clostridium thermocellum: from manure to molecular biology. FEMS Microbiol. Lett. 100:523-528. [DOI] [PubMed] [Google Scholar]
- 41.Béguin, P., J. Millet, S. Chauvaux, S. Salamitou, K. Tokatlidis, J. Navas, T. Fujino, M. Lemaire, O. Raynaud, M.-K. Daniel, and J.-P. Aubert. 1992. Bacterial cellulases. Biochem. Soc. Trans. 20:42-46. [DOI] [PubMed] [Google Scholar]
- 42.Bélaïch, J.-P., A. Bélaïch, H.-P. Fierobe, L. Gal, C. Gaudin, S. Pagés, C. Reverbel-Leroy, and C. Tardif. 1999. The cellulolytic system of Clostridium cellulolyticum, p. 479-487. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers, Tokyo, Japan.
- 43.Bélaïch, J.-P., C. Tardif, A. Bélaïch, and C. Gaudin. 1997. The cellulolytic system of Clostridium cellulolyticum. J. Biotechnol. 57:3-14. [DOI] [PubMed] [Google Scholar]
- 44.Beldman, G., A. G. J. Voragen, F. M. Rombouts, M. F. Searle-van Leeuwen, and W. Pilnik. 1987. Adsorption and kinetic behavior of purified endoglucanases and exoglucanases from Trichoderma viride. Biotechnol. Bioeng. 30:251-257. [DOI] [PubMed] [Google Scholar]
- 45.Benoit, L., C. Cailliez, A. Géhin, J. Thirion, G. Raval, and H. Petitdemange. 1995. Carboxymethylcellulase and avicelase activities from a cellulolytic Clostridium strain A11. Curr. Microbiol. 30:305-312. [DOI] [PubMed] [Google Scholar]
- 46.Benschoter, A. S., and L. O. Ingram. 1986. Thermal tolerance of Zymomonas mobilis: temperature-induced changes in membrane composition. Appl. Environ. Microbiol. 51:1278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berberich, J. A., B. L. Knutson, H. J. Strobel, S. Tarhan, S. E. Nokes, and K. A. Dawson. 2000. Product selectivity shifts in Clostridium thermocellum in the presence of compressed solvents. Ind. Eng. Chem. Res. 39:4500-4505. [Google Scholar]
- 48.Bergquist, P. L., M. D. Gibbs, D. D. Morris, V. S. J. Te'o, D. J. Saul, and H. W. Morgan. 1999. Molecular diversity of thermophilic cellulolytic and hemicellulolytic bacteria. FEMS Microbiol. Ecol. 28:99-110. [Google Scholar]
- 49.Bernardez, T. D., K. A. Lyford, and L. R. Lynd. 1994. Kinetics of the extracellular cellulases of Clostridium thermocellum on pretreated mixed hardwood and Avicel. Appl. Microbiol. Biotechnol. 41:620-625. [Google Scholar]
- 50.Bernardez, T. D., K. Lyford, D. A. Hogsett, and L. R. Lynd. 1993. Adsorption of Clostridium thermocellum cellulases onto pretreated mixed hardwood, Avicel, and lignin. Biotechnol. Bioeng. 42:899-907. [DOI] [PubMed] [Google Scholar]
- 51.Bernier, R., and F. Stutzenberger. 1987. Preferential utilization of cellobiose by Thermomonospora curvata. Appl. Environ. Microbiol. 53:1743-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhat, M. K. 2000. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18:355-383. [DOI] [PubMed] [Google Scholar]
- 53.Bhat, S., R. A. Hutson, E. Owen, and M. K. Bhat. 1997. Determination of immunological homology between cellulosome subunits and cloned endoglucanases and xylanases of Clostridium thermocellum. Anaerobe 3:347-352. [DOI] [PubMed] [Google Scholar]
- 54.Bhat, S., R. J. Wallace, and E. R. Ørskov. 1990. Adhesion of cellulolytic ruminal bacteria to barley straw. Appl. Environ. Microbiol. 56:2698-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birch, P. R. J., P. F. G. Sims, and P. Broda. 1995. Substrate-dependent differential splicing of introns in the regions encoding the cellulose binding domains of two exocellobiohydrolase I-like genes in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 61:3741-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bird, R. B., W. E. Stewart, E. N. Lightfoot. 1960. Transport phenomena. John Wiley & Sons, Inc., New York, N.Y.
- 57.Birol, G. Z. I. Onsan, B. Kirdar, S. G. Oliver. 1998. Ethanol production and fermentation characteristics of recombinant Saccharomyces cerevisiae strains grown on starch. Enzyme Microb. Technol. 22:672-677. [Google Scholar]
- 58.Birsan, C., P. Johnson, M. Joshi, A. MacLeod, L. McIntosh, V. Monem, M. Nitz, D. R. Rose, D. Tull, W. W. Wakarchuck, Q. Wang, R. A. J. Warren, A. White, and S. G. Withers. 1998. Mechanisms of cellulases and xylanases. Biochem. Soc. Trans. 26:156-160. [DOI] [PubMed] [Google Scholar]
- 59.Biswas, I., A. Gruss, S. D. Erlich, and E. Maguin. 1993. High efficiency gene inactivation and replacement system for Gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blair, B. G., and K. L. Anderson. 1999. Regulation of cellulose-inducible structures of Clostridium cellulovorans. Can. J. Microbiol. 45:242-249. [DOI] [PubMed] [Google Scholar]
- 61.Blanco, P, C. Sieiro, A. Diaz, and T. G. Villa. 1994. Production and partial characterization of an endopolygalacturonase from Saccharomyces cerevisiae. Can. J. Microbiol. 40:974-977. [DOI] [PubMed] [Google Scholar]
- 62.Blaschek, H. P., and B. A. White. 1995. Genetic systems development in the clostridia. FEMS Microbiol. Rev. 17:349-356. [Google Scholar]
- 63.Blouin, F. A., L. F. Martin, and S. P. Rowland. 1970. Gel permeation properties of cellulose. III. Measurement of pore structure of unmodified and of mercerized cottons in fibrous form. Textile Res. J. 40:809-813. [Google Scholar]
- 64.Bobleter, O. 1994. Hydrothermal degradation of polymers derived from plants. Prog. Polym. Sci. 19:797-841. [Google Scholar]
- 65.Boisset, C., C. Petrequin, H. Chanzy, B. Henrissat, and M. Schülein. 2001. Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 72:339-345. [DOI] [PubMed] [Google Scholar]
- 66.Bok, J.-D., D. A. Yernool, and D. E. Eveleigh. 1998. Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neopolitana. Appl. Environ. Microbiol. 64:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bond, K., and F. Stutzenberger. 1989. A note on the localization of cellulosome formation in Thermomonospora curvata. J. Appl. Bacteriol. 67:605-609. [Google Scholar]
- 68.Borneman, W. S., D. E. Akin, and L. G. Ljungdahl. 1989. Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microbiol. 55:1066-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bothast, R. J., N. N. Nichols, and B. S. Dien. 1999. Fermentations with new recombinant microorganisms. Biotechnol. Prog. 15:867-875. [DOI] [PubMed] [Google Scholar]
- 70.Bothwell, M. K., S. D. Daughhetee, G. Y. Chaua, D. B. Wilson, and L. P. Walker. 1997. Binding capacities for Thermomonospora fusca E3, E4, and E5, the E3 binding domain, and Trichoderma reesei CBHI on Avicel and bacterial microcrystalline cellulose. Biores. Technol. 60:169-178. [Google Scholar]
- 71.Bothwell, M. K., and L. P. Walker. 1995. Evaluation of parameter estimation methods for estimating cellulase binding constants. Biores. Technol. 53:21-29. [Google Scholar]
- 72.Bothwell, M. K., D. B. Wilson, D. C. Irwin, and L. P. Walker. 1997. Binding reversibility and surface exchange of Thermomonospora fusca E3 and E5 and Trichoderma reesei CBHI. Enzyme Microb. Technol. 20:411-417. [Google Scholar]
- 73.Bott, T. L., and I. A. Kaplan. 1991. Selection of surrogates for a genetically engineered microorganism with cellulolytic capability for ecological studies in streams. Can. J. Microbiol. 37:848-857. [Google Scholar]
- 74.Boussaid, A., and J. N. Saddler. 1999. Adsorption and activity profiles of cellulases during the hydrolysis of two Douglas fir pulps. Enzyme Microb. Technol. 24:138-143. [Google Scholar]
- 75.Boynton, Z. L., G. N. Bennett, and F. B. Rudolph. 1996. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 62:2758-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brau, B., and H. Sahm. 1986. Cloning and expression of the structural gene for pyruvate decarboxylase of Zymomonas mobilis in Escherichia coli. Arch. Microbiol. 144:296-301. [Google Scholar]
- 77.Brestic-Goachet, N., P. Gunasekaran, B. Cami, and J. C. Baratti. 1989. transfer and expression of an Erwinia chrysanthemi cellulase gene in Zymomonas mobilis. J. Gen. Microbiol. 135:893-902. [Google Scholar]
- 78.Broda, P., P. Birch, P. Brooks, J. L. Copa-Patino, M. L. Sinnott, C. Tempelaars, Q. Wang, A. Wyatt, and P. Sims. 1994. Phanerochaete chrysosporium and its natural substrate. FEMS Microbiol. Rev. 13:189-196. [DOI] [PubMed] [Google Scholar]
- 79.Broda, P., P. R. J. Birch, P. R. Brooks, and P. F. G. Sims. 1995. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl. Environ. Microbiol. 61:2358-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broda, P., P. R. J. Birch, P. R. Brooks, and P. F. G. Sims. 1996. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol. Microbiol. 19:923-932. [DOI] [PubMed] [Google Scholar]
- 81.Bronnenmeier, K., K. P. Rücknagel, and W. L. Staudenbauer. 1991. Purification and properties of a novel type of exo-1,4-β-glucanase (Avicelase II) from the cellulolytic thermophile Clostridium stercorarium. Eur. J. Biochem. 200:379-385. [DOI] [PubMed] [Google Scholar]
- 82.Bronnenmeier, K. and W. L. Staudenbauer. 1988. Resolution of Clostridium stercorarium cellulase by fast protein liquid chromatography (FPLC). Appl. Microbiol. Biotechnol. 27:432-436. [Google Scholar]
- 83.Bronnenmeier, K., and W. L. Staudenbauer. 1990. Cellulose hydrolysis by a highly thermal stable endo-1,4-β-glucanase (Avicelase I) from Clostridium stercorarium. Enzyme Microb. Technol. 12:431-436 [Google Scholar]
- 84.Brown, D. E., and M. A. Zainudeen. 1977. Growth kinetics and cellulase biosynthesis in continuous culture of Trichoderma viride. Biotechnol. Bioeng. 14:941-958. [DOI] [PubMed] [Google Scholar]
- 85.Brown, M. A., and M. D. Levine. 1993. Assessment of costs and benefits of flexible and alternative fuel use in the U.S. transportation sector. Technical report 11. Evaluation of a wood-to-ethanol process. DOE/EP-0004. U.S. Department of Energy, Washington, D.C.
- 86.Brown, R. M., Jr., and I. M. Saxena. 2000. Cellulose biosynthesis: a model for understanding the assembly of biopolymers. Plant Physiol. Biochem. 38:57-67. [Google Scholar]
- 87.Brownell, H. H., and J. N. Saddler. 1987. Steam pretreatment of lignocellulosic material for enhanced enzymatic hydrolysis. Biotechnol. Bioeng. 29:228-235. [DOI] [PubMed] [Google Scholar]
- 88.Bryant, M. P. 1959. Bacterial species of the rumen. Bacteriol. Rev. 23:125-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bunch, P. K., F. Mat-Jan, N. Lee, and D. P. Clark. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 90.Busto, M. D., N. Ortega, and M. Perez-Mateos. 1996. Location, kinetics and stability of cellulases induced in Trichoderma reesei cultures. Biores. Technol. 57:187-192. [Google Scholar]
- 91.Caminal, G., J. López-Santin, and C. Sola. 1985. Kinetic modeling of the enzymatic hydrolysis of pretreated cellulose. Biotechnol. Bioeng. 27:1282-1290. [DOI] [PubMed] [Google Scholar]
- 92.Cantwell, B., G. Brazil, N. Murphy, and D. J. McConnell. 1986. Comparison of expression of the endo-β-1,3-1,4-glucanase gene from Bacillus subtilis in Saccharomyces cerevisiae from the CYC1 and ADH1 promoters. Curr. Genet. 11:65-70. [DOI] [PubMed] [Google Scholar]
- 93.Carle-Urioste, J. C., J. Escobar-Vera, S. El-Gogary, F. Henrique-Silva, E. Torigoi, O. Crivellaro, A. Herrera-Estrella, and H. El-Dorry. 1997. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 272:10169-10174. [DOI] [PubMed] [Google Scholar]
- 94.Carlile, M. J., and S. C. Watkinson. 1997. The fungi, p. 269-275. Academic Press, New York, N.Y.
- 95.Caulfield, D. F., and W. E. Moore. 1974. Effect of varying crystallinity of cellulose on enzymatic hydrolysis. Wood Sci. 6:375-377. [Google Scholar]
- 96.Chang, V. S., B. Burr, and M. T. Holtzapple. 1997. Lime pretreatment of switchgrass. Appl. Biochem. Biotechnol. 63-65:3-19. [DOI] [PubMed] [Google Scholar]
- 97.Chang, V. S., and M. T. Holtzapple. 2000. Fundamental factors affecting biomass enzymatic reactivity. Appl. Biochem. Biotechnol. 84-86:5-37. [DOI] [PubMed] [Google Scholar]
- 98.Chattaway, T., A. L. Demain, and G. Stephanopoulos. 1992. Use of various measurements for biomass estimation. Biotechnol. Prog. 8:81-84. [Google Scholar]
- 99.Chaudhary, P., N. N. Kumar, and D. N. Deobagkar. 1997. The glucanases of Cellulomonas. Biotechnol. Adv. 15:315-331. [DOI] [PubMed] [Google Scholar]
- 100.Chauvet, E., and J. Mercé. 1988. Aquatic hyphomycetes: their role in the decomposition of leaf-litter. Rev. Sci. Eau 1:203-216. [Google Scholar]
- 101.Chen, C.-K., C. M. Boucle, and H. P. Blaschek. 1996. Factors involved in the transformation of previously non-transformable Clostridium perfringens type B. FEMS Microbiol. Lett. 140:185-191. [DOI] [PubMed] [Google Scholar]
- 102.Chen, H. Z., M. Hayn, and H. Esterbauer. 1992. Purification and characterization of two extracellular β-glucosidases from Trichoderma reesei. Biochim. Biophys. Acta 1121:54-60. [DOI] [PubMed] [Google Scholar]
- 103.Chen, H. Z., X. L. Li, and L. G. Ljungdahl. 1997. Sequencing of a 1,3-1,4-beta-d-glucanase (lichenase) from the anaerobic fungus Orpinomyces strain PC-2: properties of the enzyme expressed in Escherichia coli and evidence that the gene has a bacterial origin. J. Bacteriol. 179:6028-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen, J., and P. J. Weimer. 2001. Competition among three predominant ruminal cellulolytic bacteria in the absence or presence of non-cellulolytic bacteria. Microbiology 147:21-30. [DOI] [PubMed] [Google Scholar]
- 105.Chernoglazov, V. M., O. V. Ermolova, and A. A. Klyosov. 1988. Adsorption of high-purity endo-1,4-β-glucanases from Trichoderma reesei on components of lignocellulosic materials: cellulose, lignin, and xylan. Enzyme Microb. Technol. 10:503-507. [Google Scholar]
- 106.Cho, K. M., and Y. J. Yoo. 1999. Novel SSF process for ethanol production from microcrystalline cellulose using the δ-integrated recombinant yeast, Saccharomyces cerevisiae L2612δGC. J. Microbiol. Biotechnol. 9:340-345. [Google Scholar]
- 107.Cho, K. M., Y. J. Yoo, and H. S. Kang. 1999. δ-Integration of endo/exo-glucanase and β-glucosidase genes into the yeast chromosomes for direct conversion of cellulose to ethanol. Enzyme Microb. Technol. 25:23-30. [Google Scholar]
- 108.Chow, C. M., E. Yague, S. Raguz, D. A. Wood, and C. F. Thurston. 1994. The Cel3 gene of Agaricus bisporus codes for a modular cellulase and is transcriptionally regulated by the carbon source. Appl. Environ. Microbiol. 60:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Christakopoulos, P., B. J. Macris, and D. Kekos. 1989. Direct fermentation of cellulose to ethanol by Fusarium oxysporum. Enzyme Microb. Technol. 11:236-239. [Google Scholar]
- 110.Chum, H. L., D. K. Johnson, S. Black, J. Baker, K. Grohmann, K. V. Sarkanen, K. Wallace, H. A. Schroeder. 1988. Organosolv pretreatment for enzymatic hydrolysis of poplars. I. Enzyme hydrolysis of cellulosic residues. Biotechnol. Bioeng. 31:643-649. [DOI] [PubMed] [Google Scholar]
- 111.Chyi, Y. T., and R. R. Dague. 1994. Effects of particulate size in anaerobic acidogenesis using cellulase as a sole carbon source. Water Environ. Res. 66:670-678. [Google Scholar]
- 112.Claeyssens, M., and G. Aerts. 1992. Characterization of cellulolytic activities in commercial Trichoderma reesei preparations: an approach using small, chromogenic substrates. Biores. Technol. 39:143-146. [Google Scholar]
- 113.Clemmer, J. E., and C. L. Tseng. 1986. Identification of the major anaerobic end products of Cellulomonas sp. (ATCC 21399). Biotechnol. Lett. 8:823-826. [Google Scholar]
- 114.Clinquart, A., C. Van Eenaeme, I. Dufrasne, M. Gielen, and L. Istasse. 1995. Soya oil in the diet of growing-fattening bulls. I. Effects on animal performance and carcass composition. J. Anim. Physiol. Anim. Nutr. 74:9-14. [Google Scholar]
- 115.Cocconcelli, P. S., E. Ferrari, F. Rossi, and V. Bottazzi. 1992. Plasmid transformation of Ruminococcus albus by means of high-voltage electroporation. FEMS Microbiol. Lett. 94:203-208. [DOI] [PubMed] [Google Scholar]
- 116.Converse, A. O. 1993. Substrate factors limiting enzymatic hydrolysis, p. 93-106. In J. N. Saddler (ed.) Bioconversion of forest and agricultural plant residues. CAB International, Wallinford, Conn.
- 117.Converse, A. O., R. Matsuno, M Tanaka, M. Taniguchi. 1988. A model of enzyme adsorption and hydrolysis of microcrystalline cellulose with slow deactivation of the adsorbed enzyme. Biotechnol. Bioeng. 32:38-45. [DOI] [PubMed] [Google Scholar]
- 118.Copa-Patiño, J. L., G. K. Young, and P. Broda. 1993. Production and initial characterisation of the xylan-degrading system of Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 40:69-76. [Google Scholar]
- 119.Cordova-López, J., M. Gutiérrez-Rojas, S. Huerta, G. Saucedo-Castañeda, and E. Favela-Torres. 1996. Biomass estimation of Aspergillus niger growing on real and model supports in solid state fermentation. Biotechnol. Tech. 10:1-6. [Google Scholar]
- 120.Correa, J., C. R. Vazquez de Aldana, P. San Segundo, and F. Del Rey. 1992. Genetic mapping of 1,3-β-glucanase-encoding genes in Saccharomyces cerevisiae. Curr. Genet. 22:283-288. [DOI] [PubMed] [Google Scholar]
- 121.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 122.Coughlan, M. P. 1990. Cellulose degradation by fungi, p. 1-35. In W.M. Fogarty and C.T. Kelly (ed.), Microbial enzymes and biotechnology, 2nd ed. Elsevier Applied Science, London, United Kingdom.
- 123.Coughlan, M. P., K. Hon-Nami, H. Hon-Nami, L. Ljungdahl, J. J. Paulin, and W. E. Rigsby. 1985. The cellulolytic enzyme complex of Clostridium thermocellum is very large. Biochem. Biophys. Res. Commun. 130:904-909. [DOI] [PubMed] [Google Scholar]
- 124.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23 In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation, Uni Publishers Co., Tokyo, Japan.
- 125.Covert, S. F., A. Vanden Wymelenberg, and D. Cullen. 1992. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 58:2168-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Covert, S. F., J. Bolduc, and D. Cullen. 1992. Genomic organization of a cellulase gene family in Phanerochaete chrysosporium. Curr. Genet. 22:407-413. [DOI] [PubMed] [Google Scholar]
- 127.Cowling, E. B. 1975. Physical and chemical constraints in the hydrolysis of cellulose and lignocellulosic materials. Biotechnol. Bioeng. Symp. 5:163-181. [PubMed] [Google Scholar]
- 128.Creagh, A. L., E. Ong, E. Jervis, D. G. Kilburn, and C. A. Haynes. 1996. Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. USA 93:12229-12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crous, J. M., I. S. Pretorius, and W. H. van Zyl. 1995. Cloning and expression of an Aspergillus kawachii endo-1,4-β-xylanase gene in Saccharomyces cerevisiae. Curr. Gen. 28:467-473. [DOI] [PubMed] [Google Scholar]
- 130.Curry, C., N. Gilkes, G. O'Neill, R. C. Miller, Jr., and N. Skipper. 1988. Expression and secretion of a Cellulomonas fimi exoglucanase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 54:476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dale, B. E. 1999. Biobased industrial products: priorities for research and commercialization. National Research Council, National Academy Press, Washington D.C. [PubMed]
- 132.Dale, B. E., and M. J. Moriera. 1982. A freeze-explosion technique for increasing cellulose hydrolysis. Biotechnol. Bioeng. Symp. Ser. 12:31-43. [Google Scholar]
- 133.D'Amore, T., C. J. Panchal, I. Russell, G. G. Stewart. 1988. Osmotic pressure effects and intracellular accumulation of ethanol in yeast during fermentation. J. Ind. Microbiol. 2:365-372. [Google Scholar]
- 134.Dasari, G., F. Roddick, M. A. Connor, N. B. Pamment. 1983. Factors affecting the estimation of intracellular ethanol concentrations. Biotechnol. Lett. 5:715-720. [Google Scholar]
- 135.Dasari, G., M. A. Worth, M. A. Connor, N. B. Pamment. 1990. Reasons for the apparent difference in the effects of produced and added ethanol on culture viability during rapid fermentations by Saccharomyces cerevisiae. Biotechnol. Bioeng. 35:109-122. [DOI] [PubMed] [Google Scholar]
- 136.Davies, G. J. 1998. Structural studies on cellulases. Biochem. Soc. Trans. 26:167-173. [DOI] [PubMed] [Google Scholar]
- 137.Deanda, K., M. Zhang, C. Eddy, S. Picataggio. 1996. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl. Environ. Microbiol. 62:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Decker, C. H., J. Visser, and P. Schreier. 2000. β-Glucosidases from five black Aspergillus species: study of their physico-chemical and biocatalytic properties. J. Agric. Food Chem. 48:4929-4936. [DOI] [PubMed] [Google Scholar]
- 139.De Coninck-Chosson, J. 1988. Aerobic degradation of cellulose and adsorption properties of cellulases in Cellulomonas uda JC3: effects of crystallinity of substrate. Biotechnol. Bioeng. 31:495-501. [DOI] [PubMed] [Google Scholar]
- 140.De Grado, M., P. Castan, and J. Berenguer. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241-245. [DOI] [PubMed] [Google Scholar]
- 141.Del Campillo, E. 1999. Multiple endo-1,4-β-d-glucanase (cellulase) genes in Arabidopsis. Curr. Top. Dev. Biol. 46:39-61. [DOI] [PubMed] [Google Scholar]
- 142.D'Elia, J., and W. Chesbro. 1992. Maintenance energy demand affects biomass synthesis but not cellulase production by a mesophilic Clostridium. J. Ind. Microbiol. 10:123-133. [Google Scholar]
- 143.Delrey, F., T. G. Villa, T. Santos, I. García-Acha, and C. Nombela. 1982. Purification and partial characterization of a new, sporulation specific, exo-β-glucanase from Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 105:1347-1353. [DOI] [PubMed] [Google Scholar]
- 144.Demolder, J., M. de Backer, W. Fiers, and R. Contreras. 1993. Phenotypic effects in Saccharomyces cerevisiae after regulated expression of β-(1,3)-glucanase from Nicotiana plumbaginifolia. J. Biotechnol. 27:295-305. [DOI] [PubMed] [Google Scholar]
- 145.De Moraes, L. M. P., S. Astolfi, and G. Oliver. 1995. Development of yeast strains for the efficient utilization of starch: evaluation of constructs that express α-amylase and glucoamylase separately or as bifunctional fusion proteins. Appl. Microbiol. Biotechnol. 43:1067-1076. [DOI] [PubMed] [Google Scholar]
- 146.Denman, S., G.-P. Xue, and B. Patel. 1996. Characterization of a Neocallimastix patriciarum cellulase cDNA (celA) homologous to Trichoderma reesei cellobiohydrolase II. Appl. Environ. Microbiol. 62:1889-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dequin, S., E. Baptista, and P. Barre. 1999. Acidification of grape musts by Saccharomyces cerevisiae wine yeast strains genetically engineered to produce lactic acid. Am. J. Enol. Vitic. 50:45-50. [Google Scholar]
- 148.Dermoun, Z., and J. P. Bélaïch. 1985. Microcalorimetric study of cellulose degradation by Cellulomonas uda ATCC 21399. Biotechnol. Bioeng. 27:1005-1011. [DOI] [PubMed] [Google Scholar]
- 149.Dermoun, Z., and J. P. Bélaïch. 1988. Crystalline index change in cellulose during aerobic and anaerobic Cellulomonas uda growth. Appl. Microbiol. Biotechnol. 27:399-404. [Google Scholar]
- 150.Dermoun, Z., and J. P. Bélaïch. 1988. Effects of cellobiose concentration on the anaerobic growth of Cellulomonas uda. Biotechnol. Lett. 10:365-368. [Google Scholar]
- 151.De Rossi, E., P. Brigidi, N. E. Welker, G. Riccardi, and D. Matteuzzi. 1994. New shuttle vector for cloning in Bacillus stearothermophilus. Res. Microbiol. 145:579-583. [DOI] [PubMed] [Google Scholar]
- 152.Desai, S. G., and A. O. Converse. 1997. Substrate reactivity as a function of the extent of reaction in the enzymatic hydrolysis of lignocellulose. Biotechnol. Bioeng. 56:650-655. [DOI] [PubMed] [Google Scholar]
- 153.Desai, R. P., and E. T. Papoutsakis. 1999. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Desgranges, C., C. Vergoignan, M. Georges, and A. Durand. 1991. Biomass estimation in solid state fermentation. Appl. Biochem. Biotechnol. 35:200-205. [Google Scholar]
- 155.Deshpande, V., S. Keskar, C. Mishra, and M. Rao. 1986. Direct conversion of cellulose/hemicellulose to ethanol by Neurospora crassa. Enzyme Microb. Technol. 8:149-152. [Google Scholar]
- 156.Desvaux, M., E. Guedon, and H. Petitdemange. 2000. Cellulose catabolism by Clostridium cellulolyticum growing in batch culture on defined medium. Appl. Environ. Microbiol. 66:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Carbon flux distribution and kinetics of cellulose fermentation in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. J. Bacteriol. 183:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Kinetics and metabolism of cellulose degradation at high substrate concentrations in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. Appl. Environ. Microbiol. 67:3837-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Diaz-Ricci, J. C., L. C. Regan, and J. E. Bailey. 1991. Effect of alteration of the acetic acid synthesis pathway on the fermentation pattern of Escherichia coli. Biotechnol. Bioeng. 38:1318-1324. [DOI] [PubMed] [Google Scholar]
- 160.Dijkerman, R., H. J. N. Op den Camp, and C. van der Drift. 1996. Cultivation of anaerobic fungi in a 10-l fermenter system for the production of (hemi)cellulolytic enzymes. Appl. Microbiol. Biotechnol. 46:85-91. [Google Scholar]
- 161.Din, N., H. G. Damude, N. R. Gilkes, R. C. Miller, R. A. J. Warren, and D. G. Kilburn. 1994. C1-Cx revisited: intramolecular synergism in a cellulase. Proc. Natl. Acad. Sci. USA 91:11383-11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ding, S. Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Divne, C., J. Ståhlberg, T. Reinikainen, L. Ruohonen, G. Pettersson, J. K. C. Knowles, T. T. Teeri, and T. A. Jones. 1994. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265:524-528. [DOI] [PubMed] [Google Scholar]
- 164.Doerner, K. C., G. T. Howard, R. I. Mackie, and B. A. White. 1992. β-Glucanase expression in Ruminococcus flavefaciens FD-1. FEMS Microbiol. Lett. 93:147-153. [Google Scholar]
- 165.Doi, R. H., M. Goldstein, S. Hashida, J. S. Park, and M. Takagi. 1994. The Clostridium cellulovorans cellulosome. Crit. Rev. Microbiol. 20:87-93. [DOI] [PubMed] [Google Scholar]
- 166.Doi, R. H., J.-S. Park, C.-C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 167.Dombek, K. M., and L. O. Ingram. 1984. Effects of ethanol on the Escherichia coli plasma membrane. J. Bacteriol. 157:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dombek, K. M., and L. O. Ingram. 1986. Magnesium limitation and its role in apparent toxicity of ethanol during yeast fermentation. Appl. Environ. Microbiol. 52:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Donaldson, L. A., K. K. Y. Wong, and K. L. Mackie. 1988. Ultrastructure of steam-exploded wood. Wood Sci. Technol. 22:103-114. [Google Scholar]
- 170.Doremus, M. G., J. C. Linden, A. R. Moreira. 1985. Agitation and pressure effects on acetone-butanol fermentation. Biotechnol. Bioeng. 27:852-860. [DOI] [PubMed] [Google Scholar]
- 171.Dos Santos, V. L., E. F. Araújo, E. G. de Barros, and W. V. Guimãraes. 1999. Fermentation of starch by Klebsiella oxytoca P2, containing plasmids with α-amylase and pullulanase genes. Biotechnol. Bioeng. 65:673-676. [DOI] [PubMed] [Google Scholar]
- 172.Driskill, L. E., K. Kusy, M. W. Bauer, and R. M. Kelly. 1999. Relationship between glycosyl hydrolase inventory and growth physiology of the hyperthermophile Pyrococcus furiosus on carbohydrate-based media. Appl. Environ. Microbiol. 65:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dunny, G. M., L. N. Lee, and D. J. Le Blanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.El Gogary, S., A. Leite, O. Crivellaro, D. E. Eveleigh, and H. El-Dorry. 1989. Mechanism by which cellulose triggers cellobiohydrolase I gene expression in Trichoderma reesei. Proc. Natl. Acad. Sci. USA 86:6138-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.El-Nawwi, S. A., and A. A. El-Kader. 1996. Production of single-cell protein and cellulase from sugarcane bagasse: effect of culture factors. Biomass Bioenerg. 11:361-364. [Google Scholar]
- 176.Eriksson, K. E. L., R. A. Blanchette, and P. Ander. 1990. Microbial and enzymatic degradation of wood and wood components, Springer-Verlag, New York, N.Y.
- 177.Esterbauer, H., W. Steiner, I. Labudova, A. Hermann, and M. Hayn. 1991. Production of Trichoderma cellulase in laboratory and pilot scale. Biores. Technol. 36:51-65. [Google Scholar]
- 178.Fan, L. T., Y.-H. Lee, and D. H. Beardmore. 1980. Mechanism of the enzymatic hydrolysis of cellulose: effects of major structural features of cellulose on enzymatic hydrolysis. Biotechnol. Bioeng. 22:177-199. [Google Scholar]
- 179.Fan, L. T., Y.-H. Lee, and D. H. Beardmore. 1980. Major chemical and physical features of cellulose materials as substrates for enzymatic hydrolysis. Adv. Biochem. Eng. 14:101-117. [Google Scholar]
- 180.Fanutti, C., T. Ponyi, G. W. Black, G. P. Hazlewood, and H. J. Gilbert. 1995. The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J. Biol. Chem. 270:29314-29322. [DOI] [PubMed] [Google Scholar]
- 181.Farkas, V., P. Biely, and S. Baue. 1973. Extracellular β-glucanases of the yeast,Saccharomyces cerevisiae. Biochim. Biophys. Acta. 321:246-455. [DOI] [PubMed] [Google Scholar]
- 182.Felix, C. R., and L. G. Ljungdahl. 1993. The cellulosome: the exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 183.Fell, D. A. 1998. Increasing the flux in metabolic pathways: a metabolic control analysis perspective. Biotechnol. Bioeng. 58:121-124. [DOI] [PubMed] [Google Scholar]
- 184.Fields, M. W., S. Mallik, and J. B. Russell. 2000. Fibrobacter succinogenes S85 ferments ball-milled cellulose as fast as cellobiose until cellulose surface area is limiting. Appl. Microbiol. Biotechnol. 54:570-574. [DOI] [PubMed] [Google Scholar]
- 185.Fields, M. W., J. B. Russell, and D. B. Wilson. 1998. The role of ruminal carboxymethylcellulases in the degradation of β-glucans from cereal grain. FEMS Microbiol. Ecol. 27:261-268. [Google Scholar]
- 186.Fiérobe, H.-P., C. Bagnara-Tardif, C. Gaudin, F. Guerlesquin, P. Sauvé, A. Bélaïch, and J.-P. Bélaïch. 1993. Purification and characterization of endo-glucanase C from Clostridium cellulolyticum. Catalytic comparison with endonuclease A. Eur. J. Biochem. 217:557-565. [DOI] [PubMed] [Google Scholar]
- 187.Fiérobe, H.-P., C. Gaudin, A. Bélaïch, M. Loutfi, E. Faure, C. Bagnara, D. Baty and J.-P. Bélaïch. 1991. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173:7956-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fiérobe, H.-P., A. Mechaly, C. Tardif, A. Bélaïch, R. Lamed, Y. Shoham, J.-P. Bélaïch, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 189.Fillingham, I. J., P. A. Kroon, G. Williamson, H. J. Gilbert, and G. P. Hazlewood. 1999. A modular cinnamoyl ester hydrolase from the anaerobic fungus Piromyces equi acts synergistically with xylanase and is part of a multiprotein cellulose-binding cellulase-hemicellulase complex. Biochem. J. 343:215-224. [PMC free article] [PubMed] [Google Scholar]
- 190.Findlay, R. H., G. M. King, and L. Watling. 1989. Efficacy of phospholipid analysis in determining microbial biomass from sediments. Appl. Environ. Microbiol. 55:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Focher, B., A. Marzetti, M. Cattaneo, P. L. Beltrame, and P. Carniti. 1981. Effects of structural features of cotton cellulose on enzymatic hydrolysis. J. Appl. Polym. Sci. 26:1989-1999. [Google Scholar]
- 192.Fondevila, M., and B. A. Dehority. 1994. Degradation and utilization of forage hemicellulose by rumen bacteria, singly and in coculture or added sequentially. J. Appl. Bacteriol. 77:541-548. [DOI] [PubMed] [Google Scholar]
- 193.Fondevila, M., and B. A. Dehority. 1996. Interactions between Fibrobacter succinogenes, Prevotella ruminicola, and Ruminococcus flavefaciens in the digestion of cellulose from forages. J. Anim. Sci. 74:678-684. [DOI] [PubMed] [Google Scholar]
- 194.Fowler, T., and R. D. Brown, Jr. 1992. The bgl1 gene encoding extracellular β-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 6:3225-3235. [DOI] [PubMed] [Google Scholar]
- 195.Franklund, C. V., and T. L. Glass. 1987. Glucose uptake by the cellulolytic ruminal anaerobe Bacteroides succinogenes. J. Bacteriol. 169:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Freer, S. N., and R. W. Detroy. 1982. Direct fermentation of cellodextrins to ethanol by Candida wickerhamii and C. lusitaniae. Biotechnol. Lett. 4:453-458. [Google Scholar]
- 197.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Friberg, F., C. Otto, and B. S. Svensson. 1980. Effects of acidification dynamics of allochthonous leaf material and benthic invertebrate communities in running water, p. 304-305. In D. Drablos and A. Tollan (ed.), Ecological impact of acid precipitation. Sur Nedbors Virkning Pa Skog og Fisk, Oslo, Norway.
- 199.Gal, L., S. Pagès, C. Gaudin, A. Bélaïch, C. Reverbel-Leroy, C. Tardif, and J.-P. Bélaïch. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gallagher, J., A. Winters, N. Barron, L. McHale, and A. P. McHale. 1996. Production of cellulase and beta-glucosidase activity during growth of the actinomycete Micromonospora chalcae on cellulose-containing media. Biotechnol. Lett. 18:537-540. [Google Scholar]
- 201.Gama, F. M., and M. Mota. 1997. Enzymatic hydrolysis of cellulose (I): relationship between kinetics and physico-chemical parameters. Biocatalysis Biotrans. 15:221-236. [Google Scholar]
- 202.Gama, F. M., J. A. Teixeira, and M. Mota. 1994. Cellulose morphology and enzymatic reactivity: a modified solute exclusion technique. Biotechonol. Bioeng. 43:381-387. [DOI] [PubMed] [Google Scholar]
- 203.Garcia-Vallvé, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17:352-361. [DOI] [PubMed] [Google Scholar]
- 204.Gardner, R. M., K. C. Doerner, and B. A. White. 1987. Purification and characterization of an exo-β-1,4-glucanase from Ruminococcus flavefaciens FD-1. J. Bacteriol. 169:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Garleb, K. A., L. D. Borquin, J. T. Hsu, G. W. Wagner, S. J. Schmidt, and G. C. Fahey, Jr. 1991. Isolation and chemical analysis of nonfermented fiber fractions of oat hulls and cottonseed hulls. J. Anim. Sci. 69:1255-1271 [DOI] [PubMed] [Google Scholar]
- 206.Gaudin, C., A. Bélaïch, S. Champ, and J.-P. Bélaïch. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gelhaye, E., H. Petitdemange, and R. Gay. 1993. Adhesion and growth rate of Clostridium cellulolyticum ATCC 35319 on crystalline cellulose. J. Bacteriol. 175:3452-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gerwig, G. J., J. P. Kamerling, J. F. G. Vliegenthart, E. Morag, R. Lamed, and E. A. Bayer. 1993. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J. Biol. Chem. 268:26956-26960. [PubMed] [Google Scholar]
- 209.Ghaly, A. E., R. Kok, and J. M. Ingrahm. 1989. Growth rate determination of heterogeneous microbial population in swine manure. Appl. Biochem. Biotechnol. 22:59-78. [DOI] [PubMed] [Google Scholar]
- 210.Ghose, T. K., and V. Sahai. 1979. Production of cellulases by Trichoderma reesei QM 9414 in fed-batch and continuous-flow culture with cell recycle. Biotechnol. Bioeng. 21:283-296. [DOI] [PubMed] [Google Scholar]
- 211.Gibbs, M. D., R. A. Reeves, G. K. Farrington, P. Anderson, D. P. Williams, and P. L. Bergquist. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40:333-340. [DOI] [PubMed] [Google Scholar]
- 212.Gilkes, N. R., E. Jervis, B. Henrissat, B. Tekant, R. C. Miller, R. A. J. Warren, and D. G. Kilburn. 1992. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J. Biol. Chem. 267:6743-6749. [PubMed] [Google Scholar]
- 213.Girard, P., J. M. Scharer, and M. Moo-Young. 1986. Two-stage anaerobic digestion for the treatment of cellulosic wastes. Chem. Eng. J. 33:B1-B10. [Google Scholar]
- 214.Glick, B. R., and J. J. Pasternak. 1989. Isolation, characterization and manipulation of cellulase genes. Biotechnol. Adv. 7:361-386. [DOI] [PubMed] [Google Scholar]
- 215.Godden, B., and M. J. Penninckx. 1984. Identification and evolution of the cellulolytic microflora present during composting of cattle manure: on the role of actinomycetes sp. Ann. Microbiol. 135B:69-78. [DOI] [PubMed]
- 216.Gognies, S, A. Gainvors, M. Aigle, and A. Belarbi. 1999. Cloning, sequence analysis and overexpression of a Saccharomyces cerevisiae endopolygalacturonase-encoding gene (PGLI). Yeast 15:11-21. [DOI] [PubMed] [Google Scholar]
- 217.Gong, C. S., M. R. Ladisch, and G. T. Tsao. 1977. Cellobiase from Trichoderma viride: purification, properties, kinetics, and mechanism. Biotechnol. Bioeng. 19:959-981. [DOI] [PubMed] [Google Scholar]
- 218.Gong, J. H., and C. W. Forsberg. 1989. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 55:3039-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.González, G., G. Caminal, C. de Mas, and J. López-Santin. 1989. A kinetic model for pretreated wheat straw saccharification by cellulase. J. Chem. Technol. Biotechnol. 44:275-288. [Google Scholar]
- 220.Gordon, R. E., W. C. Haynes, and C. Hor-Nay Pang. 1973. The genus Bacillus. Agriculture handbook 427. Agricultural Research Service, US. Department of Agriculture, Washington, D.C.
- 221.Green, E. M., and G. N. Bennett. 1996. Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl. Biochem. Biotechnol. 57-58:213-221. [DOI] [PubMed] [Google Scholar]
- 222.Green, E. M., Z. L. Boynton, L. M. Harris, F. B. Rudolph, E. T. Papoutsakis, and G. N. Bennett. 1996. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142:2079-2086. [DOI] [PubMed] [Google Scholar]
- 223.Grethlein, H. E. 1985. The effect of pore size distribution on the rate of enzymatic hydrolysis of cellulosic substrates. Bio/Technology 2:155-160. [Google Scholar]
- 224.Grous, W. R., A. O. Converse, and H. E. Grethlein. 1986. Effect of steam explosion pretreatment on pore size and enzymatic hydrolysis of poplar (Populus tremuloides). Enzyme Microb. Technol. 8:274-280 [Google Scholar]
- 225.Guedon, E., M. Desvaux, and H. Petitdemange. 2000. Kinetic analysis of Clostridium cellulolyticum carbohydrate metabolism: importance of glucose-1-phosphate and glucose-6-phosphate branch points for distribution of carbon fluxes inside and outside cells as revealed by steady-state continuous culture. J. Bacteriol. 182:2010-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Guedon, E., S. Payot, M. Desvaux, and H. Petitdemange. 1999. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J. Bacteriol. 181:3262-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Guedon, E., S. Payot, M. Desvaux, and H. Petitdemange. 2000. Relationships between cellobiose catabolism, enzyme levels, and metabolic intermediates in Clostridium cellulolyticum grown in a synthetic medium. Biotechnol. Bioeng. 67:327-335. [DOI] [PubMed] [Google Scholar]
- 228.Guglielmi, G., and P. Béguin. 1998. Cellulase and hemicellulase genes of Clostridium thermocellum from five independent collections contain few overlaps and are widely scattered across the chromosome. FEMS Microbiol. Lett. 161:209-215. [DOI] [PubMed] [Google Scholar]
- 229.Guijarro, J. M., and R. Lagunas. 1984. Saccharomyces cerevisiae does not accumulate ethanol against a concentration gradient. J. Bacteriol. 160:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Guiseppi, A., J. L. Aymeric, B. Cami, F. Barras, and N. Creuzet. 1991. Sequence analysis of the cellulase-encoding celY gene of Erwinia chrysanthemi: a possible case of interspecies gene transfer. Gene 106:109-114. [DOI] [PubMed] [Google Scholar]
- 231.Gunata, Z., and M. J. Vallier. 1999. Production of a highly glucose-tolerant extracellular β-glucosidase by three Aspergillus strains. Biotechnol. Lett. 21:219-223. [Google Scholar]
- 232.Gupta, S., and D. P. Clark. 1989. Escherichia coli derivatives lacking both alcohol dehydrogenase and phosphotransacetylase grow anaerobically by lactate fermentation. J. Bacteriol. 171:3650-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gusakov, A. V., A. P. Sinitsyn, and A. A. Klyosov. 1985. Kinetics of the enzymatic hydrolysis of cellulose. 1. A mathematical model for a batch reactor process. Enzyme Microb. Technol. 7:346-352. [Google Scholar]
- 234.Gusakov, A. V., and A. P. Sinitsyn. 1992. A theoretical analysis of cellulase product inhibition: effect of cellulase binding constant, enzyme/substrate ratio, and β-glucosidase activity on the inhibition pattern. Biotechnol. Bioeng. 40:663-671. [DOI] [PubMed] [Google Scholar]
- 235.Gusakov, A. V., A. P. Sinitsyn, J. A. Manenkova, and O. V. Protas. 1992. Enzymatic saccharification of industrial and agricultural lignocellulosic wastes—main features of the process. Appl. Biochem. Biotechnol. 34-35:625-637. [Google Scholar]
- 236.Haggett, K. D., W. Y. Choi, and N. W. Dunn. 1978. Mutants of Cellulomonas which produce increased levels of β-glucosidase. Eur. J. Appl. Microbiol. Biotechnol. 6:189-191. [Google Scholar]
- 237.Hahn-Hägerdal, B., F. Wahlbom, M. Gárdonyi, W. H. van Zyl, R. R. Cordero Otero, and L. Jönsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose fermentation—a review. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 238.Halldórsdóttir, S., E. T. Thórólfsdóttir, R. Spilliaert, M. Johansson, S. H. Thórbjarnardóttir, A. Palsdóttir, G. O. Hreggvidsson, J. K. Kristjánsson, O. Holst, and G. Eggertsson. 1998. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulase of glycosyl hydrolase family 12. Appl. Microbiol. Biotechnol. 49:277-284. [DOI] [PubMed] [Google Scholar]
- 239.Halsall, D. M., and D. J. Goodchild. 1986. Nitrogen fixation associated with development and localization of mixed populations of Cellulomonas sp. and Azospirillum brasilense grown on cellulose or wheat straw. Appl. Environ. Microbiol. 51:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hamacher, K., G. Schmid, H. Sahm, and C. Wandrey. 1985. Structural heterogeneity of cellooligomers homogeneous according to high-resolution size-exclusion chromatography. J. Chromatogr. 319:311-318. [Google Scholar]
- 241.Han, S. J., Y. J. Yoo, and H. S. Kang. 1995. Characterization of bifunctional cellulase and its structural gene. J. Biol. Chem. 270:26012-26019. [DOI] [PubMed] [Google Scholar]
- 242.Harris, C. M., R. W. Todd, S. J. Bungard, R. W. Lovitt, J. G. Morris, and D. B. Kell. 1987. Dielectric permittivity of microbial suspensions at radio frequencies: a novel method for the real-time estimation of micobial biomass. Enzyme Microb. Technol. 9:181-186. [Google Scholar]
- 243.Harris, L. M., R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol. Bioeng. 67:1-11. [PubMed] [Google Scholar]
- 244.Hashimoto, S., M. Fujita, and R. A. Baccay. 1982. Biomass determination in the anaerobic digestion of night soil. J. Ferment. Technol. 60:51-54. [Google Scholar]
- 245.Hatfield, R. D., J. Ralph, and J. H. Grabber. 1999. Cell wall structural foundations: molecular basis for improving forage digestibility. Crop Sci. 39:27-37. [Google Scholar]
- 246.Hayashi, N., J. Sugiyama, T. Okano, and M. Ishihara. 1997. The enzymatic susceptibility of cellulose microfibrils of the algal-bacterial type and the cotton-ramie type. Carbohydr. Res. 305:261-269. [Google Scholar]
- 247.Heierson, A., R. Landen, A. Lovgren, G. Dalhammar, and H. G. Boman. 1987. Transformation of vegetative cells of Bacillus thuringiensis by plasmid DNA. J. Bacteriol. 169:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Heitz, M., E. Capek-Menard, P. G. Koeberle, J. Gagné, E. Chornet, R. P. Overend, J. D. Taylor, and E. Yu. 1991. Fractionation of Populus tremuloides at the pilot plant scale: optimization of steam pretreatment conditions using the Stake II technology. Biores. Technol. 35:23-32. [Google Scholar]
- 249.Helaszek, C. T., and B. A. White. 1991. Cellobiose uptake and metabolism by Ruminococcus flavefaciens. Appl. Environ. Microbiol. 57:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hendy, N. A., C. R. Wilke, and H. W. Blanch. 1984. Enhanced cellulase production in fed-batch cuture of Trichoderma reesei C30. Enzyme Microb. Technol. 6:73-77. [Google Scholar]
- 251.Henriksson, G., G. Johansson, and G. Pettersson. 2000. A critical review of cellobiose dehydrogenases. J. Biotechnol. 78:93-113. [DOI] [PubMed] [Google Scholar]
- 252.Henriksson, G., A. Nutt, H. Henriksson, B. Pettersson, J. Stahlberg, G. Johansson, and G. Pettersson. 1999. Endoglucanase 28 (Cel12A), a new Phanerochaete chrysosporium cellulase. Eur. J. Biochem. 259:88-95. [DOI] [PubMed] [Google Scholar]
- 253.Henriksson, G., A. Salumets, C. Divne, and G. Pettersson. 1997. Studies of cellulose binding by cellobiose dehydrogenase and a comparison with cellobiohydrolase 1. Biochem. J. 324:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Henrissat, B., I. Callebaut, S. Fabrega, P. Lehn, J. P. Mornon, and G. Davies. 1995. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. USA 92:7090-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Henrissat, B., and G. J. Davies. 2000. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124:1515-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Henrissat, B., H. Driguez, C. Viet, and M. Schülein. 1985. Synergism of cellulases from Trichoderma reesei in the degadation of cellulose. Bio/Technology 3:722-726. [Google Scholar]
- 260.Henrissat, B., T. T. Teeri, and R. A. J. Warren. 1998. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 425:352-354. [DOI] [PubMed] [Google Scholar]
- 261.Henrissat, B., B. Vigny, A. Buleon, and S. Perez. 1988. Possible adsorption sites of cellulases on crystalline cellulose. FEBS Lett. 231:177-182. [Google Scholar]
- 262.Herrero, A. A., and R. F. Gomez. 1980. Development of ethanol tolerance in Clostridium thermocellum: effect of growth temperature. Appl. Environ. Microbiol. 40:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Herrero, A. A., R. F. Gomez, and M. F. Roberts. 1982. Ethanol-induced changes in the membrane lipid composition of Clostridium thermocellum. Biochem. Biophys. Acta 693:195-204. [DOI] [PubMed] [Google Scholar]
- 264.Herrero, A. A., R. F. Gomez, and M. F. Roberts. 1985. 31P NMR studies of Clostridium thermocellum mechanism of end product inhibition by ethanol. J. Biol. Chem. 260:7442-7451. [PubMed] [Google Scholar]
- 265.Herrero, A. A., R. F. Gomez, B. Snedecor, C. J. Tolman, and M. F. Roberts. 1985. Growth inhibition of Clostridium thermocellum by carboxylic acids: a mechanism based on uncoupling by weak acids. Appl. Microbiol. Biotechnol. 22:53-62. [Google Scholar]
- 266.Herrero-Molina, A. A. 1981. The physiology of Clostridium thermocellum in relation to its energy metabolism. Ph.D. thesis, Massachusetts Institute of Technology, Cambridge.
- 267.Hettenhaus, J., and D. Glassner. 1997. Cellulase assessment for biomass hydrolysis. National Renewable Energy Laboratory, Golden, Colo.
- 268.Hill, P. W., T. R. Klapatch, and L. R. Lynd. 1993. Bioenergetics and end-product regulation of Clostridium thermosaccharolyticum in response to nutrient limitation. Biotechnol. Bioeng. 43:873-883. [DOI] [PubMed] [Google Scholar]
- 269.Hinchliffe, E., and W. G. Box. 1984. Expression of the cloned endo-1,3-1,4-β-glucanase gene of Bacillus subtilis in Saccharomyces cerevisiae. Curr. Genet. 8:471-475. [DOI] [PubMed] [Google Scholar]
- 270.Hinman, N. D., J. Schell, C. J. Riley, P. W. Bergeron, and P. J. Walter. 1992. Preliminary estimate of the cost of ethanol production for SSF technology. Appl. Biochem. Biotechnol. 34-35:639-649. [Google Scholar]
- 271.Ho, N. W. Y., Z. Chen, and A. P. Brainard. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 64:1852-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ho, N. W. Y., Z. Chen, A. Brainard, and M. Sedlak. 1999. Successful design and development of genetically-engineered Saccharomyces yeasts for effective cofermentation of glucose and xylose from cellulosic biomass to fuel ethanol. Adv. Biochem. Eng. Biotechnol. 65:164-192. [DOI] [PubMed] [Google Scholar]
- 273.Ho, Y. W., and N. Abdullah. 1999. The role of rumen fungi in fibre digestion: Review. Asian-Australas. J. Anim. Sci. 12:104-112. [Google Scholar]
- 274.Hobson, P. N. 1965. Continuous culture of rumen bacteria: apparatus. J. Gen. Microbiol. 38:161-166. [DOI] [PubMed] [Google Scholar]
- 275.Hoeniger, J. M. 1985. Microbial decomposition of cellulose in acidfying lakes of south-central Ontario. Appl. Environ. Microbiol. 50:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp cremoris grown with glycine in osmotically-stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hols, P., A. Ramos, J. Hugenholtz, J. Delcour, W. M. de Vos, H. Santos, and M. Kleerebezem. 1999. Acetate utilization in Lactococcus lactis deficient in lactate dehydrogenase: a rescue pathway for maintaining redox balance. J. Bacteriol. 181:5521-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Holtzapple, M. T., and A. E. Humphrey. 1984. The effect of organosolv pretreatment on the enzymatic hydrolysis of poplar. Biotechnol. Bioeng. 26:670-676. [DOI] [PubMed] [Google Scholar]
- 279.Holtzapple, M., M. Cognata, Y. Shu, and C. Hendrickson. 1990. Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol. Bioeng. 36:275-287. [DOI] [PubMed] [Google Scholar]
- 280.Holub, Z., A. Simonovicova, and V. Banasova. 1993. The influence of acidification on some chemical and microbiological properties of soil, those determining plant viability. Biologia (Bratislava) 48:671-675. [Google Scholar]
- 281.Honda, H., S. Iijima, and T. Kobayashi. 1988. Cloning and expression in Saccharomyces cerevisiae of an endo-β-glucanase gene from a thermophilic cellulolytic anaerobe. Appl. Microbiol. Biotechnol. 28:57-58. [Google Scholar]
- 282.Hon-Nami, K., S. Goto, M. Tomita, Y. Takagi, K. Sekine, E. Okuma, S. Yonemura, K. Sato, and T. Saiki. 1988. Direct microbial conversion of cellulose to ethanol by a new isolate, Clostridium thermocellum I-1-B. p. 71-76. In Proceedings of the 8th International Symposium on Alcohol Fuels. New Energy and Industrial Development Corporation, Tokyo, Japan.
- 283.Hopgood, M. F., and D. J. Walker. 1969. Succinic acid production by rumen bacteria. III. Enzymic studies on the formation of succinate by Rumincoccus flavefaciens. Aust. J. Biol. Sci. 22:1413-1424. [Google Scholar]
- 284.Hörmeyer, P. Tailliez, J. Millet, H. Girard, G. Bonn, O. Bobleter, and J.-P. Aubert. 1988. Ethanol production by Clostridium thermocellum grown on hydrothermally and organosolv-pretreated lignocellulosic materials. Appl. Microbiol. Biotechnol. 29:528-535. [Google Scholar]
- 285.Horvan, B., F. Rieu-Lesme, G. Fonty, and P. Gouet. 1996. In vitro interaction between rumen H2-producing cellulolytic microorganisms and H2-utilizing acetogenic and sulfate-reducing methanogenic bacteria. Anaerobe 2:175-180. [Google Scholar]
- 286.Hoshino, K., T. Sasakura, K. Sugai, M. Taniguchi, and M. Fujii. 1994. Production of ethanol from reclaimed paper using a combination of a reversibly soluble-autoprecipitating cellulase and flocculating yeast cells. J. Chem. Eng. Jpn. 27:260-262. [Google Scholar]
- 287.Hoshino, K., H. Yamasaki, C. Chida, S. Morohashi, T. Sasakura, T. M. Taniguchi, and M. Fujii. 1997. Continuous simultaneous saccharification and fermentation of delignified rice straw by a combination of two reversibly soluble-autoprecipitating enzymes and pentose-fermenting yeast cells. J. Chem. Eng. Jpn. 30:30-37. [Google Scholar]
- 288.Hsu, T.-A. 1996. Pretreatment of biomass, p. 179-212. In C. E. Wyman (ed.) Handbook on biomass ethanol. Taylor and Francis, Washington, D.C.
- 289.Huang, A. A. 1975. Enzymatic hydrolysis of cellulose to sugar. Biotechnol. Bioeng. Symp. 5:245-252. [PubMed] [Google Scholar]
- 290.Huang, S.-Y., and J. C. Chen. 1988. Analysis of the kinetics of ethanol fermentation with Zymomonas mobilis considering temperature effect. Enzyme Microb. Technol. 10:431-439. [Google Scholar]
- 291.Huang, T.-L., Y. W. Han, and C. D. Callihan. 1971. Application of the Lowry method for determination of cell concentration in fermentation of waste cellulosics. J. Ferment. Technol. 49:574-576. [Google Scholar]
- 292.Huber, R., C. R. Woese, T. Langworthy, J. K. Kristjánsson, and K. O. Stetter. 1990. Fervidobacterium islandicum, new species, a new extremely thermophilic eubacterium belonging to the “Thermotogales.” Arch. Microbiol. 154:105-111. [Google Scholar]
- 293.Hulcher, F. H., and K. W. King. 1957. Disaccharide preference for an aerobic cellulolytic bacterium, Cellvibrio gilvus n.sp. J. Bacteriol. 76:565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hungate, R. E. 1966. The rumen and its microbes. Academic Press, Inc., New York, N.Y.
- 295.Ilmén, M., M. L. Onnela, S. Klemsdal, S. Keränen, and M. Penttilä. 1996. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 253:303-314. [DOI] [PubMed] [Google Scholar]
- 296.Ilmén, M., A. Saloheimo, M.-L. Onnela, and M. E. Penttilä. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Inglin, M., B. A. Feinberg, and J. R. Loewenberg. 1980. Partial purification and characterization of a new intracellular β-glucosidase of Trichoderma reesei. Biochem. J. 185:515-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ingram, L. O. 1990. Ethanol tolerance in bacteria. Crit. Rev. Biotechnol. 9:305-319. [DOI] [PubMed] [Google Scholar]
- 299.Ingram, L. O., H. C. Aldrich, A. C. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Unverwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. D. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 300.Ingram, L. O., and D. P. Clark. July1991. Ethanol production using engineered mutant E. coli. U.S. patent 5,028,539.
- 301.Ingram, L. O., P. F. Gomez, X. Lai, M. Moniruzzaman, B. E. Wood, L. P. Yomano, and S. W. York. 1998. Metabolic engineering of bacteria for ethanol production. Biotechnol. Bioeng. 58:204-214. [DOI] [PubMed] [Google Scholar]
- 302.Innis, M. A., M. J. Holland, P. C. McCabe, G. E. Cole, V. P. Wittman, R. Tal, W. K. Watt, D. H. Gelfand, J. P. Holland, and J. H. Meade. 1985. Expression, glycosylation, and secretion of an Aspergillus glucoamylase by Saccharomyces cerevisiae. Science 228:21-26. [DOI] [PubMed] [Google Scholar]
- 303.Irwin, D., D. H. Shin, S. Zhang, B. K. Barr, J. Sakon, P. A. Karplus, and D. B. Wilson. 1998. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Irwin, D. C., M. Spezio, L. P. Walker, and D. B. Wilson. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 305.Irwin, D. M., and A. C. Wilson. 1990. Concerted evolution of ruminant stomach lysozymes: characterization of a lysozyme complementary DNA clones from sheep and deer. J. Biol. Chem. 265:4944-4952. [PubMed] [Google Scholar]
- 306.Ishihara, M., S. Uemura, N. Hayashi, J. Jellison, and K. Shimizu. 1991. Adsorption and desorption of cellulase components during enzymatic hydrolysis of steamed shirakamba (Betula platyphylla Skatchev) wood. J. Ferment. Bioeng. 72:96-100. [Google Scholar]
- 307.Ito, K., A. Kimizuka, H. Okazaki, and S. Kobayashi. 1989. Mycelial distribution in rice koji. J. Ferment. Bioeng. 68:7-13. [Google Scholar]
- 308.Jackson, E. A., G. M. Ballance, and K. K. Thomsen. 1986. Construction of a yeast vector directing the synthesis and release of barley (1-3,1-4)-β-glucanase. Carlsberg Res. Commun. 51:445-458. [Google Scholar]
- 309.Jacobsen, S. E., and C. E. Wyman. 2000. Cellulose and hemicellulose hydrolysis models for application to current and novel pretreatment processes. Appl. Biochem. Biotechnol. 84-86:81-96. [DOI] [PubMed] [Google Scholar]
- 310.Jeffries, T. W., and N.-Q. Shi. 1999. Genetic engineering for improved xylose fermentation by yeasts. Adv. Biochem. Eng. Biotechnol. 65:117-161. [DOI] [PubMed] [Google Scholar]
- 311.Jennert, K. C. B., C. Tardif, D. I. Young, and M. Young. 2000. Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 146:3071-3080. [DOI] [PubMed] [Google Scholar]
- 312.Jervis, E. J., C. A. Haynes, and D. G. Kilburn. 1997. Surface diffusion of cellulases and their isolated binding domains on cellulose. J. Biol. Chem. 272:24016-24023. [DOI] [PubMed] [Google Scholar]
- 313.Jobses, I. M. L., and J. A. Roels. 1986. The inhibition of the maximum specific growth and fermentation rate of Zymomonas mobilis by ethanol. Biotechnol. Bioeng. 28:554-563. [DOI] [PubMed] [Google Scholar]
- 314.Johnson, E. A. 1983. Regulation of cellulase activity and synthesis in Clostridium thermocellum. Ph.D. thesis. Massachusetts Institute of Technology, Cambridge.
- 315.Johnson, E. A., F. Bouchot, and A. L. Demain. 1985. Regulation of cellulase formation in Clostridium thermocellum. J. Gen. Microbiol. 131:2303-2308. [Google Scholar]
- 316.Johnson, E. A., M. Sakajoh, G. Halliwell, A. Madia, and A. L. Demain. 1982. Saccharification of complex cellulosic substrates by the cellulase system from C. thermocellum. Appl. Environ. Microbiol. 43:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Jones, R. P. 1989. Biological principles for the effects of ethanol. Enzyme Microb. Technol. 11:130-153. [Google Scholar]
- 318.Jung, S. H., J. G. Zeikus, and R. I. Hollingsworth. 1994. A new family of very long chain α,ω-dicarboxylic acids is a major structural fatty acyl component of the membrane lipids of Thermoanaerobacter ethanolicus 39E. J. Lipid Res. 35:1057-1065. [PubMed] [Google Scholar]
- 319.Kaar, W. E., and M. T. Holtzapple. 2000. Using lime pretreatment to facilitate the enzymatic hydrolysis of corn stover. Biomass Bioenerg. 18:189-199. [Google Scholar]
- 320.Kajikawa, H., M. Amari, and S. Masaki. 1997. Glucose transport by mixed ruminal bacteria from a cow. Appl. Environ. Microbiol. 63:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kajikawa, H., and S. Masaki. 1999. Cellobiose transport by mixed ruminal bacteria from a cow. Appl. Environ. Microbiol. 65:2565-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kauri, T., and D. J. Kushner. 1985. Role of contact in bacterial degradation of cellulose. FEMS Microbiol. Ecol. 31:301-306. [Google Scholar]
- 323.Kennedy, M. J., M. S. Thakur, D. I. C. Wang, and G. N. Stephanopoulos. 1992. Estimating cell concentration in the presence of suspended solids: a light-scattering technique. Biotechnol. Bioeng. 40:875-888. [DOI] [PubMed] [Google Scholar]
- 324.Kennedy, M. J., M. S. Thakur, D. I. C. Wang, and G. N. Stephanopoulos. 1992. Techniques for the estimation of cell concentration in the presence of suspended solids. Biotechnol. Prog. 8:375-381. [DOI] [PubMed] [Google Scholar]
- 325.Khan, A. W. 1988. Factors affecting the utilization of steam- and explosion-decompressed aspen wood by cellulolytic anaerobes. Biomass 15:269-280. [Google Scholar]
- 326.Khan, A. W., M. Asther, and C. Giuliano. 1984. Utilization of steam- and explosion-decompressed aspen wood by some anaerobes. J. Ferment. Technol. 62:335-339. [Google Scholar]
- 327.Khan, A. W., E. Meek, L. C. Sowden, and J. R. Colvin. 1994. Emendation of genus Acetivibrio and description of Acetivibrio cellulosolvens, new species, of nonmotile cellulolytic mesophile. Int. J. Syst. Bacteriol. 34:410-422. [Google Scholar]
- 328.Kim, A. Y., G. T. Attwood, S. M. Holt, B. A. White, and H. P. Blaschek. 1994. Heterologous expression of endo-β-1,4-glucanase from Clostridium cellulovorans in Clostridium acetobutylicum ATCC 824 following transformation of the engB gene. Appl. Environ. Microbiol. 60:337-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kim, A. Y., and H. P. Blaschek. 1991. Isolation and characterization of a filamentous viruslike particle from Clostridium acetobutylicum NCIB 6444. J. Bacteriol. 173:530-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kim, A. Y., and H. P. Blaschek. 1993. Construction and characterization of a phage-plasmid hybrid (Phagemid), pCAK1, containing the replicative form of viruslike particle CAK1 isolated from Clostridium acetobutylicum NCIB 6444. J. Bacteriol. 175:3838-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kim, B. H. 1987. Carbohydrate catabolism in cellulolytic strains of Cellulomonas, Pseudomonas, and Nocardia. Korean J. Microbiol. 25:28-33. [Google Scholar]
- 332.Kim, D. W., Y. H. Jang, and Y. K. Jeong. 1998. Adsorption kinetics and behaviour of two cellobiohydrolases from Trichoderma reesei on microcrystalline cellulose. Biotechnol. Appl. Biochem. 27:97-102. [Google Scholar]
- 333.Kim, D. W., Y. K. Jeong, Y. H. Jang, and J. K. Lee. 1994. Purification and characterization of endoglucanase and exoglucanase components from Trichoderma viride. J. Ferment. Bioeng. 77:363-369. [Google Scholar]
- 334.Kim, D. W., T. S. Kim, Y. K. Jeong, and J. K. Lee. 1992. Adsorption kinetics and behaviors of cellulase components on microcrystalline cellulose. J. Ferment. Bioeng. 73:461-466. [Google Scholar]
- 335.Kim, E., D. C. Irwin, L. P. Walker, and D. B. Wilson. 1998. Factorial optimization of a six-cellulase mixture. Biotechnol. Bioeng. 58:494-501. [DOI] [PubMed] [Google Scholar]
- 336.Kirby, J., J. C. Martin, A. S. Daniel, and H. J. Flint. 1997. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol. Lett. 149:213-219. [DOI] [PubMed] [Google Scholar]
- 337.Kistner, A., and J. H. Kornelius. 1990. A small-scale, three-vessel continuous culture system for quantitative studies of plant fibre degradation by anaerobic bacteria. J. Microbiol. Methods 12:173-182. [Google Scholar]
- 338.Kistner, A., J. H. Kornelius, and G. S. Miller. 1983. Kinetic measurements on bacterial cultures growing on fibres. S. Afr. J. Anim. Sci. 13:217-220. [Google Scholar]
- 339.Kjeldsen, T. 2000. Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol. 54:277-286. [DOI] [PubMed] [Google Scholar]
- 340.Klapatch, T. R., A. L. Demain, and L. R. Lynd. 1996. Restriction endonuclease activity in Clostridium thermocellum and Clostridium thermosaccharolyticum Appl. Microbiol. Biotechnol. 45:127-131. [DOI] [PubMed] [Google Scholar]
- 341.Klapatch, T. R., M. L. Guerinot, and L. R. Lynd. 1996. Electrotransformation of Clostridium thermosaccharolyticum. J. Ind. Microbiol. 16:342-347. [DOI] [PubMed] [Google Scholar]
- 342.Klapatch, T. R., D. A. L. Hogsett, S. Baskaran, S. Pal, and L. R. Lynd. 1994. Organism development and characterization for ethanol production using thermophilic bacteria. Appl. Biochem. Biotechnol. 45-46:209-223. [Google Scholar]
- 343.Klebl, F., and W. Tanner. 1989. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 171:6259-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Kleijntjens, R. H., P. A. de Boks, and K. C. A. M. Luyben. 1986. A continuous thermophilic cellulose fermentation in an upflow reactor by a Clostridium thermocellum-containing mixed culture. Biotechnol. Lett. 8:667-672. [Google Scholar]
- 345.Kleywegt, G. J., J. Y. Zou, C. Divne, G. J. Davies, I. Sinning, J. Stahlberg, T. Reinikainen, M. Srisodsuk, T. T. Teeri, and T. A. Jones. 1997. The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J. Mol. Biol. 272:383-397. [DOI] [PubMed] [Google Scholar]
- 346.Klyosov, A. A. 1988. Cellulases of the third generation, p. 97-99. In J.-P. Aubert, P. Beguin, and J. Millet (ed.), Biochemistry and genetics of cellulose degradation. Academic Press, Ltd., London, United Kingdom.
- 347.Klyosov, A. A. 1990. Trends in biochemistry and enzymology of cellulose degradation. Biochemistry 29:10577-10585. [DOI] [PubMed] [Google Scholar]
- 348.Knappert, D. R., H. E. Grethlein, and A. O. Converse. 1981. Partial acid hydrolysis of poplar wood as a pretreatment for enzymatic hydrolysis. Biotechnol. Bioeng. Symp. Ser. 11:66-77. [Google Scholar]
- 349.Knowles, J., P. Lehtovaara, and T. Teeri. 1987. Cellulase families and their genes. Trends Biotechnol. 5:255-261. [Google Scholar]
- 350.Kobayashi, T., M. P. M. Romaniec, U. Fauth, and A. L. Demain. 1990. Subcellulosome preparation with high cellulase activity from Clostridium thermocellum. Appl. Environ. Microbiol. 56:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Koch, A., C. T. O. Weigel, and G. Schulz. 1993. Cloning, sequencing, and heterologous expression of a cellulase encoding cDNA (cbh1) from Penicillium janthinellum. Gene 124:57-65. [DOI] [PubMed] [Google Scholar]
- 352.Kohchi, C., and A. Toh-e. 1986. Cloning of Candida pelliculosa β-glucosidase gene and its expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 203:89-94. [DOI] [PubMed] [Google Scholar]
- 353.Kolbe, J., and C. P. Kubicek. 1990. Quantification and identification of the main components of the Trichoderma cellulase complex with monoclonal antibodies using an enzyme-linked immunosorbent (ELISA) assay. Appl. Microbiol. Biotechnol. 34:26-30. [DOI] [PubMed] [Google Scholar]
- 354.Koyama, M., W. Helbert, T. Imai, J. Sugiyama, and B. Henrissat. 1997. Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proc. Nat. Acad. Sci. USA 94:9091-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kroon-Batenburg, L. M. J., and J. Kroon. 1997. The crystal and molecular structures of cellulose I and II. Glycoconj. J. 14:677-690. [DOI] [PubMed] [Google Scholar]
- 356.Kruus, K., A. Andreacchi, W. K. Wang, and J. H. D. Wu. 1995. Product inhibition of the recombinant CelS, an exoglucanase component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 44:399-404. [DOI] [PubMed] [Google Scholar]
- 357.Kruus, K., W. K. Wang, J. Ching, and J. H. D. Wu. 1995. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 177:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Kubicek, C. P., and M. E. Penttilä. 1998. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma, p. 49-72. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2 ed. Taylor & Francis Ltd., London, United Kingdom.
- 359.Kubo, Y., H. Takagi, and S. Nakamori. 2000. Effect of gene disruption of succinate dehydrogenase on succinate production in a sake yeast strain. J. Biosci. Bioeng. 90:619-624. [DOI] [PubMed] [Google Scholar]
- 360.Kudo, H., K.-J. Cheng, and J. W. Costerton. 1987. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can. J. Microbiol. 33:267-271. [DOI] [PubMed] [Google Scholar]
- 361.Kudo, H., K.-J. Cheng, and J. W. Costerton. 1987. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can. J. Microbiol. 33:244-248. [DOI] [PubMed] [Google Scholar]
- 362.Kundu, S., T. K. Ghose, and S. N. Mukhopadhyay. 1983. Bioconversion of cellulose into ethanol by Clostridium thermocellum—product inhibition. Biotechnol. Bioeng. 25:1109-126. [DOI] [PubMed] [Google Scholar]
- 363.Kuranda, M. J., and P. W. Robbins. 1987. Cloning and heterologous expression of glycosidase genes from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:2585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kurane, R., T. Suzuki, Y. Takahara, N. Kurita, and M. Miyaji. 1979. Application of fluorescent antibody staining technique to trace the microorganism inoculated in biological treatment systems. Agric. Biol. Chem. 43:2093-2098. [Google Scholar]
- 365.Kurose, N., T. Miyazaki, T. Kakimoto, J. Yagyu, M. Uchida, A. Obayashi, and Y. Murooka. 1989. Isolation of plasmids from thermophilic clostridia and construction of shuttle vectors in Escherichia coli and cellulolytic bacteria. J. Ferment. Bioeng. 68:371-374. [Google Scholar]
- 366.Kyriacou, A., R. J. Neufeld, and C. R. MacKenzie. 1989. Reversibility and competition in the adsorption of Trichoderma reesei cellulase components. Biotechnol. Bioeng. 33:631-637. [DOI] [PubMed] [Google Scholar]
- 367.La Grange, D. C., I. M. Claeyssens, I. S. Pretorius, and W. H. van Zyl. 2001. Degradation of xylan to d-xylose by recombinant Saccharomyces cerevisiae coexpressing the Aspergillus niger xylosidase (xlnD) and the Trichoderma reesei xylanase II (xyn2) genes. Appl. Environ. Microbiol. 67:5512-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.La Grange, D. C., I. S. Pretorius, and W. H. van Zyl. 1996. Expression of a Trichoderma reesei β-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 62:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Lamed, R. J., J. H. Lobos, and T. M. Su. 1988. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl. Environ. Microbiol. 54:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Larriba, G. 1993. Translocation of proteins across the membrane of the endoplasmic reticulum: a place for Saccharomyces cerevisiae. Yeast 9:441-463. [DOI] [PubMed] [Google Scholar]
- 372.Laser, M., D. Schulman, S. G. Allen, J. Lichwa, M. J. Antal, and L. R. Lynd. 2002. A comparison of liquid hot water and steam treatments of sugar cane bagasse for bioconversion to ethanol. Biores. Technol. 81:33-44. [DOI] [PubMed] [Google Scholar]
- 373.Lee, N. E., and J. Woodward. 1989. Kinetics of the adsorption of Trichoderma reesei C30 cellulase to DEAE-Macrosorb. J. Biotechnol. 11:75-82. [Google Scholar]
- 374.Lee, S. B., I. H. Kim, D. D. Y. Ryu, and H. Taguchi. 1983. Structural properties of cellulose and cellulase reaction mechanism. Biotechnol. Bioeng. 25:33-51. [DOI] [PubMed] [Google Scholar]
- 375.Lee, S. B., H. S. Shin, D. D. Y. Ryu, and M. Mandels. 1982. Adsorption of cellulase on cellulose: effect on physicochemical properties of cellulose on adsorption and rate of hydrolysis. Biotechnol. Bioeng. 24:2137-2153. [DOI] [PubMed] [Google Scholar]
- 376.Lee, S. S., J. K. Ha, H. S. Kang, T. McAllister, and K.-J. Cheng. 1997. Overview of energy metabolism, substrate utilization and fermentation characteristics of ruminal anaerobic fungi. Korean J. Anim. Nutr. Feedstuffs 21:295-314. [Google Scholar]
- 377.Lee, Y.-H., and L. T. Fan. 1983. Kinetic studies of enzymatic hydrolysis of insoluble cellulose. II. Analysis of extended hydrolysis times. Biotechnol. Bioeng. 25:939-966. [DOI] [PubMed] [Google Scholar]
- 378.Leedle, J. A. Z., M. L. Coe, and R. A. Frey. 1995. Evaluation of health and ruminal variables during adaptation to grain-based diets in beef cattle. Am. J. Vet. Res. 56:885-892. [PubMed] [Google Scholar]
- 379.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Lejeune, A., D. E. Eveleigh, and C. Colson. 1988. Expression of an endoglucanase gene of Pseudomonas fluorescens var. cellulosa in Zymomonas mobilis. FEMS Microbiol. Lett. 49:363-366. [Google Scholar]
- 381.Lenz, J., H. Esterbauer, W. Sattler, J. Schurz, and E. Wrentschur. 1990. Changes of structure and morphology of regenerated cellulose caused by acid and enzymatic hydrolysis. J. Appl. Polym Sci. 41:1315-1326. [Google Scholar]
- 382.Le Ruyet, P., H. C. Dubourguier, and G. Albagnac. 1984. Homoacetogenic fermentation of cellulose by a coculture of Clostridium thermocellum and Acetogenium kivui. Appl. Environ. Microbiol. 48:893-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Leschine, S. B. 1995. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 49:399-426. [DOI] [PubMed] [Google Scholar]
- 384.Li, X., and P. Gao. 1997. Isolation and partial properties of cellulose-decomposing strain of Cytophaga sp. LX-7 from soil. J. Appl. Microbiol. 82:73-80. [Google Scholar]
- 385.Li, X.-L., H. Z. Chen, and L. G. Ljungdahl. 1997. Monocentric and polycentric anaerobic fungi produce structurally related cellulases and xylanases. Appl. Environ. Microbiol. 63:628-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Liebl, W. 2001. Cellulolytic enzymes from Thermotoga species. Methods Enzymol. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 387.Lin, C., J. W. Urbance, and D. A. Stahl. 1994. Acetivibrio cellulolyticus and Bacteroides cellulosolvens are members of the greater clostridial assemblage. FEMS Microbiol. Lett. 124:151-155. [DOI] [PubMed] [Google Scholar]
- 388.Lin, J. K., M. R. Ladisch, J. A. Patterson, and C. H. Noller. 1987. Structural properties of cellulose and cellulase reaction mechanism. Biotechnol. Bioeng. 29:976-981. [Google Scholar]
- 389.Lin, K. W., M. R. Ladisch, D. M. Schaefer, C. H. Noller, V. Lechtenberg, and G. T. Tsao. 1981. Review on effect of pretreatment on digestibility of cellulosic materials. AIChE Symp. Ser. 207:102-106. [Google Scholar]
- 390.Lin, Y. L., and H. P. Blaschek. 1984. Transformation of heat-treated Clostridium acetobutylicum protoplasts with pUB110 plasmid DNA. Appl. Environ. Microbiol. 48:737-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Linder, M., and T. T. Teeri. 1996. The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc. Natl. Acad. Sci. USA 93:12251-12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Ljungdahl, L. G., F. Bryant, L. Carriera, T. Saiki, and J. Wiegel. 1981. Some aspects of thermophilic and extreme thermophilic microorganisms, p. 397-419. In A. Holleander (ed.), Trends in the biology of fermentations for fuels and chemicals. Plenum Press, New York, N.Y.
- 393.Lou, J., K. A. Dawson, and H. J. Strobel. 1996. Role of phosphorolytic cleavage in cellobiose and cellodextrin metabolism by the ruminal bacterium Prevotella ruminicola. Appl. Environ. Microbiol. 62:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Lou, J. R., K. A. Dawson, and H. J. Stobel. 1997. Cellobiose and cellodextrin metabolism by the ruminal bacterium Ruminococcus albus. Appl. Environ. Microbiol. 35:221-227. [DOI] [PubMed] [Google Scholar]
- 395.Loureiro, V., and H. G. Ferreira. 1983. On the intracellular accumulation of ethanol in yeast. Biotechnol. Bioeng. 25:2263-2269. [DOI] [PubMed] [Google Scholar]
- 396.Lovitt, R. W., B. H. Kim, G.-J. Shen, and J. G. Zeikus. 1989. Solvent production by micoorganisms. Crit. Rev. Biotechnol. 7:107-186. [Google Scholar]
- 397.Lovitt, R. W., R. Longin, and J. G. Zeikus. 1984. Ethanol production by thermophilic bacteria: physiological comparison of solvent effects on parent and alcohol-tolerant strains of Clostridium thermohydrosulfuricum. Appl. Environ. Microbiol. 48:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Luttig, M., I. S. Pretorius, and W. H. van Zyl. 1997. Cloning of two β-xylanase-encoding genes from Aspergillus niger and their expression in Saccharomyces cerevisiae. Biotechnol. Lett. 19:411-415. [Google Scholar]
- 399.Lynch, J. M. 1988. The terrestrial environment, p. 103-131. In J. M. Lynch and J. E. Hobbie (ed.), Microorganisms in action: concepts and applications in microbial ecology, 2nd ed., Blackwell Scientific Publishers, Oxford, United Kingdom.
- 400.Lynd, L. R. 1996. Overview and evaluation of fuel ethanol production from cellulosic biomass: technology, economics, the environment, and policy. Annu. Rev. Energy Environ. 21:403-465. [Google Scholar]
- 401.Lynd, L. R. 1989. Ethanol production from lignocellulosic substrates using thermophilic bacteria: critical evaluation of potential and review, Adv. Biochem. Eng. Biotechnol. 38:1-52. [Google Scholar]
- 402.Lynd, L. R., H.-J. Ahn, G. Anderson, P. W. Hill, D. S. Kersey, T. Klapatch. 1991. Thermophilic ethanol production: investigation of ethanol yield and tolerance in continuous culture. Appl. Biochem. Biotechnol. 28/29:549-570. [Google Scholar]
- 403.Lynd, L. R., S. Baskaran, and S. Casten. 2001. Salt accumulation associated with KOH added for pH control, and not ethanol, limits growth of Thermoanaerobacterium thermosaccharolyticum HG-8 at elevated feed xylose concentrations in continuous culture. Biotech. Prog. 17:118-125. [DOI] [PubMed] [Google Scholar]
- 404.Lynd, L. R., J. H. Cushman, R. J. Nichols, and C. E. Wyman. 1991. Fuel ethanol from cellulosic biomass. Science 251:1318-1323. [DOI] [PubMed] [Google Scholar]
- 405.Lynd, L. R., R. T. Elander, and C. E. Wyman. 1996. Likely features and costs of mature biomass ethanol technology. Appl. Biochem. Biotechnol. 57-58:741-761. [Google Scholar]
- 406.Lynd, L. R., and H. E. Grethlein. 1987. Hydrolysis of dilute acid pretreated mixed hardwood and purified microcrystalline cellulose by cell-free broth from Clostridium thermocellum. Biotechnol. Bioeng. 29:92-100. [DOI] [PubMed] [Google Scholar]
- 407.Lynd, L. R., H. E. Grethlein, and R. H. Wolkin. 1989. Fermentation of cellulosic substrates in batch and continuous culture by Clostridium thermocellum. Appl. Environ. Microbiol. 55:3131-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Lynd, L. R., K. Lyford, C. R. South, G. P. van Walsum, and K. Levenson. 2001. Evaluation of paper sludges for amenability to enzymatic hydrolysis and conversion to ethanol. Tappi J. 82:1-19. [Google Scholar]
- 409.Lynd, L. R., R. H. Wolkin, and H. E. Grethlein. 1987. Continuous fermentation of pretreated hardwood and Avicel by Clostridium thermocellum. Biotechnol. Bioeng. Symp. Ser. 17:265-274. [Google Scholar]
- 410.Lynd, L. R., C. E. Wyman, and T. U. Gerngross. 1999. Biocommodity engineering. Biotechnol. Prog. 15:777-793. [DOI] [PubMed] [Google Scholar]
- 411.Lynd, L. R., and Y. Zhang. 2002. Quantitative determination of cellulase concentration as distinct from cell concentration in studies of microbial cellulose utilization: analytical framework and methodological approach. Biotechnol. Bioeng. 77:467-475. [DOI] [PubMed] [Google Scholar]
- 412.Maas, L. K., and T. L. Glass. 1991. Cellobiose uptake by the cellulolytic ruminal anaerobe Fibrobacter (Bacteroides) succinogenes. Can. J. Microbiol. 37:141-147. [DOI] [PubMed] [Google Scholar]
- 413.Machida, M., I. Ohtsuki, S. Fukui, and I. Yamashita. 1988. Nucleotide sequence of Saccharomycopsis fibuligera genes for extracellular β-glucosidases as expressed in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 54:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Mackay, R. J., and K. E. Kersey. 1985. A preliminary study of aquatic insect communities and leaf decomposition in acid streams near Dorset, Ontario. Hydrobiologia 122:3-11. [Google Scholar]
- 415.Madden, R. H., M. J. Bryder, and N. J. Poole. 1982. Purification and isolation of an anaerobic, cellulolytic, bacterium, Clostridium papyrosolvens sp. nov. Int. J. Syst. Bacteriol. 32:87-91. [Google Scholar]
- 416.Maglione, G., J. B. Russell, and D. B. Wilson. 1997. Kinetics of cellulose digestion by Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 63:665-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Maguire, R. J. 1977. Kinetics of the hydrolysis of cellobiose and p-nitrophenyl-β-d-glucoside by cellobiase of Trichoderma viride. Can. J. Biochem. 55:19-26. [DOI] [PubMed] [Google Scholar]
- 418.Mai, V., W. W. Lorenz, and J. Wiegel. 1997. Transformation of Thermoaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol. Lett. 148:163-167. [Google Scholar]
- 419.Mai, V., and J. Wiegel. 2000. Advances in development of a genetic system for Thermoanaerobacterium spp.: expression of genes encoding hydrolytic enzymes, development of a second shuttle vector, and integration of genes into the chromosome. Appl. Environ. Microbiol. 66:4817-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Mai, V., and J. Wiegel. 1999. Recombinant DNA applications in thermophiles, p. 511-519. In A. L. Demain and J. E. Davis (ed. in chief) Manual of industrial microbiology and biotechnology, 2nd ed. ASM Press, Washington, D.C.
- 421.Malek, M. A., N. A. Chowdhury, Q. M. Youssouf, and N. Choudhury. 1988. Bacterial cellulases and saccharification of lignocellulosic materials. Enzyme Microb. Technol. 10:750-753. [Google Scholar]
- 422.Mandels, M. 1985. Applications of cellulases. Biochem. Soc. Trans. 13:414-416. [DOI] [PubMed] [Google Scholar]
- 423.Mandels, M., F. W. Parrish, and E. T. Reese. 1962. Sophorose as an inducer of cellulase in Trichoderma reesei. J. Bacteriol. 83:400-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Mandels, M., and E. T. Reese. 1957. Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. J. Bacteriol. 73:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Mandels, M., and J. Weber. 1969. The production of cellulases. Adv. Chem. Ser. 95:391-413. [Google Scholar]
- 426.Mansfield, S. D., C. Mooney, and J. N. Saddler. 1999. Substrate and enzyme characteristics that limit cellulose hydrolysis. Biotechnol. Prog. 15:804-816. [DOI] [PubMed] [Google Scholar]
- 427.Marchessault, R. H., and J. A. Howsmon. 1957. Experimental evaluation of the lateral order distribution in cellulose. Text. Res. J. 27:30-41. [Google Scholar]
- 428.Marchessault, R. H., and P. R. Sundararajan. 1993. Cellulose, p. 11-95 In G. O. Aspinall (ed.), The polysaccharides, vol. 2. Academic Press, Inc., New York, N.Y.
- 429.Margolles-Clark, E., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 430.Martin, W. 1999. Mosaic bacterial chromosomes: a challenge on route to a tree of genomes. Bioessays 21:99-104. [DOI] [PubMed] [Google Scholar]
- 431.Mat-Jan, F., K. Y. Alam, and D. P. Clark. 1989. Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J. Bacteriol. 171:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Mayer, M. A. G., K. Bronnenmeier, W. H. Schwarz, C. Schertler, and W. L. Staudenbauer. 1995. Isolation and properties of acetate kinase- and phosphotransacetylase-negative mutants of Thermoanaerobacter thermohydrosulfuricus. Microbiology 141:2891-2896. [Google Scholar]
- 433.McMillan, J. D. 1994. Conversion of hemicellulose hydrolyzates to ethanol. ACS Symp. Ser. 566:411-437. [Google Scholar]
- 434.McMillan, J. D. 1994. Pretreatment of lignocellulosic biomass. ACS Symp. Ser. 566:292-324. [Google Scholar]
- 435.McSweeney, C. S., A, Dulieu, and R. Bunch. 1998. Butyrivibrio spp. and other xylanolytic microorganisms from the rumen have cinnamoyl esterase activity. Anaerobe 4:57-65. [DOI] [PubMed] [Google Scholar]
- 436.Mechaly, A., H. P. Fiérobe, A. Bélaïch, J. P. Bélaïch, R. Lamed, Y. Shoham, and E. A. Bayer. 2001. Cohesin-dockerin interaction in cellulosome assembly: a single hydroxyl group of a dockerin domain distinguishes between nonrecognition and high affinity recognition. J. Biol. Chem. 276:9883-9888. [DOI] [PubMed] [Google Scholar]
- 437.Medve, J., J. Karlsson, D. Lee, and F. Tjerneld. 1998. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59:621-634. [PubMed] [Google Scholar]
- 438.Medve, J., J. Ståhlberg, and F. Tjerneld. 1994. Adsorption and synergism of cellobiohydrolase I and cellobiohydrolase II of Trichoderma reesei during hydrolysis of microcrystalline cellulose. Biotechnol. Bioeng. 44:1064-1073. [DOI] [PubMed] [Google Scholar]
- 439.Medve, J., J. Ståhlberg, and F. Tjerneld. 1997. Isotherms for adsorption of cellobiohydrolase I and II from Trichoderma reesei on microcrystalline cellulose. Appl. Biochem. Biotechnol. 66:39-56. [DOI] [PubMed] [Google Scholar]
- 440.Mercenier, A., and B. M. Chassy. 1988. Strategies for the development of bacterial transformation systems. Biochimie 70:503-517. [DOI] [PubMed] [Google Scholar]
- 441.Mermelstein, L., and E. T. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T 1 methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Messner, R., K. Hagspiel, and C. P. Kubicek. 1990. Isolation of a β-glucosidase-binding and activating polysaccharide from cell walls of Trichoderma reesei. Arch. Microbiol. 154:150-155. [Google Scholar]
- 443.Meyer, C. L., and E. T. Papoutsakis. 1989. Increased levels of ATP and NADH are associated with increased solvent production in continuous cultures of Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 30:450-459. [Google Scholar]
- 444.Meyer, H. P., and G. Canevascini. 1981. Separation and some properties of two intracellular β-glucosidases of Sporotrichum thermophile. Appl. Environ. Microbiol. 41:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Meyer, H. P., and A. E. Humphrey. 1982. Cellulase production in continuous culture with the mutant Thermomonospora sp. N-35. Chem. Eng. Commun. 19:149-156. [Google Scholar]
- 446.Mihoc, A., and D. Kluepfel. 1990. Purification and characterization of beta-glucosidase from Streptomyces lividans 66. Can. J. Microbiol. 36:53-56. [Google Scholar]
- 447.Miller, G. L., J. Dean, and R. Blum. 1960. A study of methods for preparing oligosaccharides from cellulose. Arch. Biochem. Biophys. 91:21-26. [Google Scholar]
- 448.Miller, T. L. 1978. The pathway of formation of acetate and succinate from pyruvate by Bacteroides succinogenes. Arch. Microbiol. 117:145-152. [DOI] [PubMed] [Google Scholar]
- 449.Miller, T. L., E. Currenti, and M. J. Wolin. 2000. Anaerobic bioconversion of cellulose by Ruminococcus albus, Methanobrevibacter smithii, and Methanosarcina barkeri. Appl. Microbiol. Biotechnol. 54:494-498. [DOI] [PubMed] [Google Scholar]
- 450.Miller, T. L., and M. J. Wolin. 1995. Bioconversion of cellulose to acetate with pure cultures of Ruminococcus albus and a hydrogen-using acetogen. Appl. Environ. Microbiol. 61:3832-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Miron, J., and D. Ben-Ghedalia. 1992. The degradation and utilization of wheat-straw cell-wall monosaccharide components by defined ruminal cellulolytic bacteria. Appl. Microbiol. Biotechnol. 38:432-437. [Google Scholar]
- 452.Miron, J., and D. Ben-Ghedalia. 1993. Digestion of structural polysaccharides from Panicum and vetch hays by the rumen bacterial strains Fibrobacter succinogenes BL2 and Butyrivibrio fibrisolvens D1. Appl. Microbiol. Biotechnol. 39:756-759. [DOI] [PubMed] [Google Scholar]
- 453.Miron, J., D. Ben-Ghedalia, and M. Morrison. 2001. Adhesion mechanisms of rumen cellulolytic bacteria. J. Dairy Sci. 84:1294-1309. [DOI] [PubMed] [Google Scholar]
- 454.Miron, J., and C. W. Forsberg. 1999. Characterisation of cellulose-binding proteins that are involved in the adhesion mechanism of Fibrobacter intestinalis DR7. Appl. Microbiol. Biotechnol. 51:491-497. [DOI] [PubMed] [Google Scholar]
- 455.Mishra, S., P. Béguin, and J.-P. Aubert. 1991. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J. Bacteriol. 173:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Mistry, F. R., and C. L. Cooney. 1989. Production of ethanol by Clostridium thermosaccharolyticum. I. Effect of cell recycle and environmental parameters. Biotechnol. Bioeng. 34:1295-1304. [DOI] [PubMed] [Google Scholar]
- 457.Mitchell, D. A., E. Gumbira-Sa'it, P. F. Greenfield, and H. W. Doelle. 1991. Protein measurement in solid-state fermentation. Biotechnol. Tech. 5:437-442. [DOI] [PubMed] [Google Scholar]
- 458.Mitchell, W. J. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microbiol. Physiol. 39:31-130. [DOI] [PubMed] [Google Scholar]
- 459.Miwa, T., S. Takagi, and T. Hino. 2000. Degradation of the cellulase of ruminal bacteria by the protease of coexisting bacteria in vitro. Anim. Sci. Jpn. 71:157-163. [Google Scholar]
- 460.Mok, W. S.-L., and M. J. Antal. 1992. Uncatalyzed solvolysis of whole biomass hemicellulose by hot compressed liquid water. Ind. Eng. Chem. Res. 31:1157-1161. [Google Scholar]
- 461.Moloney, A., and M. P. Coughlan. 1983. Sorption of Talaromyces emersonii cellulase on cellulosic substrates. Biotechnol. Bioeng. 25:271-280. [DOI] [PubMed] [Google Scholar]
- 462.Montegut, D., N. Indictor, and R. J. Koestler. 1991. Fungal deterioration of cellulosic textiles: a review. Int. Biodeterior. 28:209-226. [Google Scholar]
- 463.Montgomery, L., B. A. Flesher, and D. A. Stahl. 1982. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Bacteriol. 38:430-435. [Google Scholar]
- 464.Mooney, C. A., S. D. Mansfield, M. G. Touhy, and J. N. Saddler. 1998. The effect of initial pore volume and lignin content on the enzymatic hydrolysis of softwoods. Biores. Technol. 64:113-119. [Google Scholar]
- 465.Morag, E., E. A. Bayer, and R. Lamed. 1992. Affinity digestion for the near-total recovery of purified cellulosome from Clostridium thermocellum. Enzyme Microb. Technol. 14:289-292. [Google Scholar]
- 466.Morag, E., E. A. Bayer, G. P. Hazlewood, H. J. Gilbert, and R. Lamed. 1993. Cellulase S-S (CelS) is synonymous with major cellobiohydrolase (subunit S8) from the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 43:147-151. [DOI] [PubMed] [Google Scholar]
- 467.Mori, Y. 1995. Nutritional interdependence between Thermoanaerobacter thermohydrosulfuricus and Clostridium thermocellum. Arch. Microbiol. 164:152-154. [Google Scholar]
- 468.Morrison, M. 1996. Do ruminal bacteria exchange genetic material? J. Dairy Sci. 79:1476-1486. [DOI] [PubMed] [Google Scholar]
- 469.Morrison, M., R. I. Mackie, and A. Kistner. 1990. 3-Phenylpropanoic acid improves the affinity for Ruminococcus albus for cellulose in continuous culture. Appl. Environ. Microbiol. 56:3220-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Mosier, N. S., P. Hall, C. M. Ladisch, and M. R. Ladisch. 1999. Reaction kinetics, molecular action, and mechanisms of cellulolytic proteins. Adv. Biochem. Eng. Biotechnol. 65:23-40. [DOI] [PubMed] [Google Scholar]
- 471.Mouriño, F. M., R. Akkarawongsa, and P. J. Weimer. 2001. Initial pH as a determinant of cellulose digestion rate by mixed ruminal microorganisms in vitro. J. Dairy Sci. 84:848-859. [DOI] [PubMed] [Google Scholar]
- 472.Mrsa, V., F. Klebl, and W. Tanner. 1993. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-1,3-glucanase. J. Bacteriol. 175:2102-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Muir, M., L. Williams, and T. Ferenci. 1985. Influence of transport energization on the growth yield of Escherichia coli. J. Bacteriol. 163:1237-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Murai, T., M. Ueda, H. Atomi, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Kawaguchi, M. Arai, and A. Tanaka. 1997. Genetic immobilization of cellulase on the cell surface of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 48:499-503. [DOI] [PubMed] [Google Scholar]
- 475.Murai, T., M. Ueda, T. Kawaguchi, M. Arai, and A. Tanaka. 1998. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl. Environ. Microbiol. 64:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Murai, T., M. Ueda, M. Yamamura, H. Atomi, Y. Shibasaki, N. Kamasawa, M. Osumi, T. Amachi, and A. Tanaka. 1997. Construction of a starch-utilizing yeast by cell surface engineering. Appl. Environ. Microbiol. 63:1362-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Murray, W. D., and A. W. Khan. 1983. Ethanol production by a newly isolated anaerobe, Clostridium saccharolyticum. Effects of culture medium and growth conditions. Can. J. Microbiol. 29:342-347. [Google Scholar]
- 478.Muthukumar, G., S. H. Suhng, P. T. Magee, R. D. Jewell, and D. A. Primerano. 1993. The Saccharomyces cerevisiae SPR1 gene encodes a sporulation-specific exo-1,3-β-glucanase which contributes to ascospore thermoresistance. J. Bacteriol. 175:386-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Nagodawithana, T. W., and K. H. Steinkraus. 1976. Influence of rate of ethanol production and accumulation on viability of Saccharomyces cerevisiae in rapid fermentation. Appl. Environ. Microbiol. 31:158-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Nakajima, H., K. Noguchi, M. Yamamoto, R. Aono, and K. Horikoshi. 1993. Expression of an 87-kD-β-1,3-glucanase of Bacillus circulans IAM1165 in Saccharomyces cerevisiae by low-temperature incubation. Biosci. Biotechnol. Biochem. 57:2039-2042. [DOI] [PubMed] [Google Scholar]
- 481.Narumi, I., K. Sawakami, S. Nakamotoa, N. Nakayama, T. Yanagisawa, N. Takahashi, and H. Kihara. 1992. A newly-isolated Bacillus stearothermophilus K1041 and its transformation by electroporation. Biotechnol. Tech. 6:83-86. [Google Scholar]
- 482.Nebreda, A. R., T. G. Villa, J. R. Villanueva, and F. Del Rey. 1986. Cloning of genes related to exo-β-glucanase production in Saccharomyces cerevisiae: characterization of an exo-β-glucanase structural gene. Gene 47:245-259. [DOI] [PubMed] [Google Scholar]
- 483.Netherwood, T., R. Bowden, P. Harrison, A. G. O'Donnell, D. S. Parker, and H. J. Gilbert. 1999. Gene transfer in the gastrointestinal tract. Appl. Environ. Microbiol. 65:5139-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Ng, T. K., A. Ben-Bassat, and J. G. Zeikus. 1981. Ethanol production by thermophilic bacteria: fermentation of cellulosic substrates by cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl. Environ. Microbiol. 41:1337-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Ng, T. K., P. J. Weimer, and J. G. Zeikus. 1977. Cellulolytic and physiological properties of Clostridium thermocellum. Arch. Microbiol. 114:1-7. [DOI] [PubMed] [Google Scholar]
- 486.Ng, T. K., and J. G. Zeikus. 1982. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J. Bacteriol. 150:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Ng, T. K., and J. G. Zeikus. 1986. Synthesis of 14C-cellobiose with Clostidium thermocellum cellobiose phosphorylase. Appl. Environ. Microbiol. 52:902-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Nidetzky, B., and W. Steiner. 1993. A new approach for modeling cellulase-cellulose adsorption and the kinetics of the enzymatic hydrolysis of microcrystalline cellulose. Biotechnol. Bioeng. 42:469-479. [DOI] [PubMed] [Google Scholar]
- 489.Nidetzky, B., W. Steiner, M. Hayn, and M. Claeyssens. 1994. Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interactions. Biochem. J. 298:705-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Nidetzky, B., W. Zachariae, G. Gercken, M. Hayn, and W. Steiner. 1994. Hydrolysis of cellooligosaccharides by Trichoderma reesei cellobiohydrolases: Experimental data and kinetic modeling. Enzyme Microb. Technol. 16:43-52. [Google Scholar]
- 491.Nieves, R. A., Y.-C. Chou, M. E. Himmel, and S. R. Thomas. 1995. Quantification of Acidothermus cellulolyticus E1 endoglucanase and Thermomonospora fusca E3 exoglucanase using enzyme-linked immunosorbent assay (ELISA). Appl. Biochem. Biotechnol. 51-52:211-223. [Google Scholar]
- 492.Nieves, R. A., C. I. Ehrman, W. S. Adney, R. T. Elander, and M. E. Himmel. 1998. Technical communication: survey and analysis of commercial cellulase preparations suitable for biomass conversion to ethanol. World J. Microbiol. Biotechnol. 14:301-304. [Google Scholar]
- 493.Nishise, H., A. Ogawa, Y. Tani, and H. Yamada. 1985. Studies on microbial glycerol dehydrogenase. 4: glycerol dehydrogenase and glycerol dissimilation in Cellulomonas sp. NT3060. Agric. Biol. Chem. 49:629-636. [Google Scholar]
- 494.Nogawa, M., M. Goto, H. Okada, and Y. Morikawa. 2001. l-Sorbose induces cellulase gene trancription in the cellulolytic fungus Trichoderma reesei. Curr. Genet. 38:329-334. [DOI] [PubMed] [Google Scholar]
- 495.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, G. Gibson, M. Hong, J. Dubois, D. Qiu, J. Hitti, T. Aldredge, M. Ayers, R. Bashirzadeh, H. Bochner, M. Boivin, S. Bross, D. Bush, C. Butler, A. Caron, A. Caruso, R. Cook, P. Daggett, C. Deloughery, J. Egan, D. Ellston, M. Engelstein, J. Ezedi, K. Gilbert, A. Goyal, J. Guerin, T. Ho, K. Holtham, P. Joseph, P. Keagle, J. Kozlovsky, M. LaPlante, G. LeBlanc, W. Lumm, A. Majeski, S. McDougall, P. Mank, J. I. Mao, D. Nocco, D. Patwell, J. Phillips, B. Pothier, S. Prabhakar, P. Richterich, P. Rice, D. Rosetti, M. Rossetti, M. Rubenfield, M. Sachdeva, P. Snell, R. Spadafora, L. Spitzer, G. Shimer, H. U. Thomann, R. Vicaire, K. Wall, Y. Wang, K. Weinstock, P. Lai, A. Wonsey, Q. Xu, L. Zhang, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Norkrans, B. 1950. Influence of cellulolytic enzymes from Hymenocetes on cellulose preparations of different crystallinity. Physiol. Plant. 3:75-81. [Google Scholar]
- 497.Novak, M., P. Strehaiano, M. Moreno, G. Goma. 1981. Alcoholic fermentation—on the inhibitory effect of ethanol. Biotechnol. Bioeng. 23:201-211. [Google Scholar]
- 498.Nutor, J. R. K., and A. O. Converse. 1991. The effect of enzyme and substrate levels on the specific hydrolysis rate of pretreated poplar wood. Appl. Biochem. Biotechnol. 28-29:757-772. [Google Scholar]
- 499.Odenyo, A. A., R. I. Mackie, D. A. Stahl, and B. A. White. 1994. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: pure culture studies with cellulose and alkaline hydrogen peroxide-treated wheat straw. Appl. Environ. Microbiol. 60:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Ohara, H., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci. Biotechnol. Biochem. 64:254-260. [DOI] [PubMed] [Google Scholar]
- 501.Ohmiya, K., K. Sakka, S. Karita, and T. Kimura. 1997. Structure of cellulases and their applications. Biotechnol. Genet. Eng. Rev. 14:365-414. [DOI] [PubMed] [Google Scholar]
- 502.Okamoto, T., S. Yamano, H. Ikeaga, and K. Nakmura. 1994. Cloning of the Acetobacter xylinum cellulase gene and its expression in Escherichia coli and Zymomonas mobilis. Appl. Microbiol. Biotechnol. 42:563-568. [DOI] [PubMed] [Google Scholar]
- 503.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Olsen, O., and K. K. Thomsen. 1989. Processing and secretion of barley (1-3,1-4)-β-glucanase in yeast. Carlsberg Res. Commun. 54:29-39. [DOI] [PubMed] [Google Scholar]
- 505.Ooi, T., K. Minamiguchi, T. Kawaguchi, H. Okada, S. Murao, and M. Arai. 1994. Expression of the cellulase (FI-CMCase) gene of Aspergillus aculeatus in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 58:954-956. [DOI] [PubMed] [Google Scholar]
- 506.Ooshima, H., D. S. Burns, and A. O. Converse. 1990. Adsorption of cellulase from Trichoderma reesei on cellulose and lignaceous residue in wood pretreated by dilute sulfuric acid with explosive decompression. Biotechnol. Bioeng. 36:446-452. [DOI] [PubMed] [Google Scholar]
- 507.Ooshima, H., M. Kurakake, J. Kato, and Y. Harano. 1991. Enzyme activity of cellulase adsorbed on cellulose and its change during hydrolysis. Appl. Biochem. Biotechnol. 31:253-266. [DOI] [PubMed] [Google Scholar]
- 508.Ooshima, H., M. Sakata, and Y. Harano. 1983. Adsorption of cellulase from Trichoderma viride on cellulose. Biotechnol. Bioeng. 25:3103-3114. [DOI] [PubMed] [Google Scholar]
- 509.Orpin, C. G. 1977. The occurrence of chitin in the cell walls of rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 99:215-218. [DOI] [PubMed] [Google Scholar]
- 510.O'Sullivan, A. C. 1997. Cellulose: the structure slowly unravels. Cellulose 4:173-207. [Google Scholar]
- 511.Ozkan, M. S., G. Desai, Y. Zhang, D. M. Stevenson, J. Beane, M. L. Guerinot, and L. R. Lynd. 2002. Characterization of thirteen newly-isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J. Ind. Microbiol. Biotechnol. 27:275-280. [DOI] [PubMed] [Google Scholar]
- 512.Palonen, H., M. Tenkanen, and M. Linder. 1999. Dynamic interaction of Trichoderma reesei cellobiohydrolases Ce16A and Ce17A and cellulose at equilibrium and during hydrolysis. Appl. Environ. Microbiol. 65:5229-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Parsiegla, G., M. Juy, C. Reverbel-Leroy, C. Tardif, J. P. Bélaïch, H. Driguez, and R. Haser. 1998. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 Å resolution. EMBO J. 17:5551-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Parsiegla, G., C. Reverbel-Leroy, C. Tardif, J. P. Bélaïch, H. Driguez, and R. Haser. 2000. Crystal structures of the cellulase Ce148F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39:11238-11246. [DOI] [PubMed] [Google Scholar]
- 515.Parvez, S., M. I. Rajoka, F. Fariha, and K. A. Malik. 1994. Cloning of endoglucanase genes from Cellulomonas biazotea into E. coli and S. cervisiae using shuttle vector Yep24. Folia Microbiol. 39:251-254. [DOI] [PubMed] [Google Scholar]
- 516.Pavlostathis, S. G., T. L. Miller, and M. J. Wolin. 1988. Fermentation of insoluble cellulose by continuous cultures of Ruminococcus albus. Appl. Environ. Microbiol. 54:2655-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Pavlostathis, S. G., T. L. Miller, M. J. Wolin. 1988. Kinetics of insoluble cellulose fermentation by continuous cultures of Ruminococcus albus. Appl. Environ. Microbiol. 54:2660-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Pavlostathis, S. G., T. L. Miller, M. J. Wolin. 1990. Cellulose fermentation by continuous cultures of Ruminococcus albus and Methanobrevibacter smithii. Appl. Microbiol. Biotechnol. 33:109-116. [Google Scholar]
- 519.Payot, S., E. Guedon, C. Cailliez, E. Gelhaye, and H. Petitdemange. 1998. Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased NADH reoxidation as a factor limiting growth. Microbiology 144:375-384. [DOI] [PubMed] [Google Scholar]
- 520.Penttilä, M. 1998. Heterologous protein production in Trichoderma, p. 365-382 In G. E. Harman and C. P. Kubicek (ed.) Trichoderma and Gliocladium. Taylor and Francis Ltd., London, United Kingdom.
- 521.Penttilä, M. E., L. André, P. Lehtovaara, M. Bailey, T. T. Teeri, and J. K. C. Knowles. 1988. Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene 63:103-112. [DOI] [PubMed] [Google Scholar]
- 522.Penttilä, M. E., L. André, M. Saloheimo, P. Lehtovaara, and J. K. C. Knowles. 1987. Expression of two Trichoderma reesei endoglucanases in the yeast Saccharomyces cerevisiae. Yeast 3:175-185. [DOI] [PubMed] [Google Scholar]
- 523.Penttilä, M., P. Lehtovaara, and J. Knowies. 1989. Cellulolytic yeast and their applications, p. 247-267. In P. J. Barr, A. J. Brake, and P. Valenzuela (ed.), Yeast genetic engineering. Butterworth, Stoneham, Mass.
- 524.Penttilä, M. E., K. M. H. Nevalainen, A. Raynal, and J. K. C. Knowles. 1984. Cloning of Aspergillus niger genes in yeast. Expression of the gene encoding Aspergillus β-glucosidase. Mol. Gen. Genet. 194:494-499. [Google Scholar]
- 525.Penttilä, M. E., M. L. Suihko, U. Lehtinen, and J. Knowles. 1987. Construction of brewer's yeasts secreting fungal endo-β-glucanase. Curr. Genet. 12:413-420. [Google Scholar]
- 526.Percy, A., H. Ono, and K. Hayashi. 1998. Acceptor specificity of cellobiose phosphorylase from Cellvibrio gilvus: synthesis of three branched trisaccharides. Carbohydr. Res. 308:423-429. [DOI] [PubMed] [Google Scholar]
- 527.Pereira, A. N., M. Mobedshahi, and M. R. Ladisch. 1988. Preparation of cellodextrins. Methods Enzymol. 160:26-38. [Google Scholar]
- 528.Pérez-González, J. A., L. H. De Graaff, J. Visser, and D. Ramon. 1996. Molecular cloning and expression in Saccahromyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl. Environ. Microbiol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Pérez-González, J. A., R. González, A. Querol, J. Sendra, and D. Ramon. 1993. Construction of a recombinant wine yeast strain expressing β-(1,4)-endoglucanase and its use in microvinification processes. Appl. Environ. Microbiol. 59:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Peteranderl, R., E. B. Shotts, and J. Wiegel. 1990. Stability of antibiotics under growth conditions for thermophilic anaerobes. Appl. Environ. Microbiol. 56:1981-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Petersen, S. H., W. H. Van Zyl, and I. S. Pretorius. 1998. Development of a polysaccharide degrading strain of Saccharomyces cerevisiae. Biotechnol. Tech. 12:615-619. [Google Scholar]
- 532.Petitdemange, E., F. Caillet, J. Giallo, and C. Gaudin. 1984. Clostridium cellulolyticum sp. nov., a cellulolyltic mesophilic species from decayed grass. Int. J. Syst. Bacteriol. 34:155-159. [Google Scholar]
- 533.Philippidis, G. P., T. K. Smith, and C. E. Wyman. 1993. Study of the enzymatic hydrolysis of cellulose for production of fuel ethanol by the simultaneous saccharification and fermentation process. Biotechnol. Bioeng. 41:846-853. [DOI] [PubMed] [Google Scholar]
- 534.Phillips-Jones, M. K. 1995. Introduction of recombinant DNA into Clostridium spp. Methods Mol. Biol. 47:227-235. [DOI] [PubMed] [Google Scholar]
- 535.Pietersen, N. 1977. Continuous cultivation of Trichoderma viride on cellulose. Biotechnol. Bioeng. 19:337-348. [Google Scholar]
- 536.Pietersen, N., and E. W. Ross. 1979. Mathematical model for enzymatic hydrolysis and fermentation of cellulose by Trichoderma. Biotechnol. Bioeng. 21:997-1017. [Google Scholar]
- 537.Piwonka, E. J., J. L. Firkins, and B. L. Hull. 1994. Digestion in the rumen and total tract of forage-based diets with starch or dextrose supplements fed to Holstein heifers. J. Dairy Sci. 77:1570-1579. [DOI] [PubMed] [Google Scholar]
- 538.Pizzi, A., and N. Eaton. 1985. The structure of cellulose by conformational analysis. 2. The cellulose polymer chain. J. Macromol. Sci. Chem. 22:105-137. [Google Scholar]
- 539.Pohlschröder, M., S. B. Leschine, and E. Canale-Parola. 1994. Multicomplex cellulase-xylanase system of Clostridium papyrosolvens C7. J. Bacteriol. 176:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Ponce-Noyola, T., and M. de la Torre. 1993. Interactions of a mixed culture composed of Cellulomonas flavigena and Xanthomonas sp. growing in continuous culture on sugar cane bagasse. Appl. Microbiol. Biotechnol. 40:531-534. [Google Scholar]
- 541.Porro, D., M. M. Bianchi, L. Brambilla, R. Menghini, D. Bolzani, V. Carrera, J. Lievense, C.-L. Liu, B. M. Ranzi, L. Frontali, and L. Alberghina. 1999. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl. Environ. Microbiol. 65:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Porro, D., L. Brambilla, B. M. Ranzi, E. Martegani, and L. Alberghina. 1995. Development of metabolically-engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol. Prog. 11:294-298. [DOI] [PubMed] [Google Scholar]
- 543.Poulsen, O. M., and L. W. Petersen. 1988. Growth of Cellulomonas sp. ATCC 21399 on different polysaccharides as sole carbon source induction of extracellular enzymes. Appl. Microbiol. Biotechnol. 29:480-484. [Google Scholar]
- 544.Pretorius, I. S. 1997. Utilization of polysaccharides by Saccharomyces cerevisiae, p. 435-458. In F. K. Zimmermann and K. D. Entian (ed.), Yeast sugar metabolism—1997. Technomic Publishing Company, Lancaster, Pa.
- 545.Prieur, D. J. 1986. Tissue specific deficiency of lysozyme in ruminants. Comp. Biochem. Physiol. Ser. B. 85:349-354. [DOI] [PubMed] [Google Scholar]
- 546.Puls, J., K. Poutanen, H. U. Körner, and L. Viikari. 1985. Biotechnological utilization of wood carbohydrates after steaming pretreatment. Appl. Microbiol. Biotechnol. 22:416-423. [Google Scholar]
- 547.Puls, J., and T. M. Wood. 1991. The degradation pattern of cellulose by extracellular cellulases of aerobic and anaerobic microorganisms. Biores. Technol. 36:15-19. [Google Scholar]
- 548.Rabinovich, M. L., V. M. Chernoglazov, and A. A. Klesov. 1983. Isoenzymes of endoglucanase in cellulase complexes: different affinity for cellulose and different role in the hydrolysis of an insoluble substrate. Biochemistry 48:321-329. [Google Scholar]
- 549.Rainey, F. A., A. M. Donnison, P. H. Janssen, D. Saul, A. Rodrigo, P. L. Bergquist, R. M. Daniel, E. Stackebrandt, and H. W. Morgan. 1994. Description of Caldicellulosiruptor saccharolyticus gen. nov., sp. nov: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol. Lett. 120:263-266. [DOI] [PubMed] [Google Scholar]
- 550.Rajoka, M. I., and K. A. Malik. 1997. Cellulase production by Cellulomonas biazotea cultured in media containing different cellulosic substrates. Biores. Technol. 59:21-27. [Google Scholar]
- 551.Rajoka, M. I., S. Parvez, and K. A. Malik. 1992. Cloning of structural genes for β-glucosidase from Cellulomonas biazotea into E. coli and Saccharomyces cerevisiae using shuttle vector pBLU-D. Biotechnol. Lett. 14:1001-1006. [Google Scholar]
- 552.Rajoka, M. I., A. Bashir, S. R. A. Hussein, M. T. Gauri, S. Parvez, and K. A. Malik. 1998. Cloning and expression of β-glucosidase genes in Escherichia coli and Saccharomyces cerevisiae using shuttle vector pYES 2.0. Folia Microbiol. 43:129-135. [DOI] [PubMed] [Google Scholar]
- 553.Rani, K. S., and G. Seenayya. 1999. High ethanol tolerance of new isolates of Clostridium thermocellum strains SS21 and SS22. World J. Microbiol. Biotechnol. 15:173-178. [DOI] [PubMed] [Google Scholar]
- 554.Rapp, P., and A. Beerman. 1991. Bacterial cellulases. p. 535-595. In C. H. Haigler and P. J. Weimer (ed.), Biosynthesis and biodegradation of cellulose. Marcel Dekker, Inc., New York, N.Y.
- 555.Rasmussen, M. A., R. B. Hespell, B. A. White, and R. J. Bothast. 1988. Inhibitory effects of methylcellulose on cellulose degradation by Ruminococcus flavefaciens. Appl. Environ. Microbiol. 54:890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Ravinder, T., B. Ramesh, G. Seenayya, and G. Reddy. 2000. Fermentative production of acetic acid from various pure and natural cellulolosic materials by Clostridium lentocellum SG6. World J. Microbiol. Biotechnol. 16:507-512. [Google Scholar]
- 557.Rawlings, D. E., and T. Kusano. 1994. Molecular genetics of Thiobacillus ferrooxidans. Microbiol. Rev. 58:39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Raynal, A., C. Gerbaud, M. C. Francingues, and M. Guérineau. 1987. Sequence and transcription of the β-glucosidase gene of Kluyveromyces fragilis cloned in Saccharomyces cerevisiae. Curr. Genet. 12:175-184. [DOI] [PubMed] [Google Scholar]
- 559.Raynal, A., and M. Guérineau. 1984. Cloning and expression of the structural gene for β-glucosidase of Kluyvermyces fragilis in Escherichia coli and Saccharomyces cerevisiae. Mol. Gen. Genet. 195:108-115. [DOI] [PubMed] [Google Scholar]
- 560.Reczey, K., A. Brumbauer, M. Bollok, Z. Szengyel, and G. Zacchi. 1998. Use of hemicellulose hydrolysate for β-glucosidase fermentation. Appl. Biochem. Biotechnol. 70-72:225-235. [DOI] [PubMed] [Google Scholar]
- 561.Reese, E. T. 1956. A microbiological process report: enymatic hydrolysis of cellulose. Appl. Microbiol. 4:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Reese, E. T., and M. Mandels. 1971. Enzymatic degradation. p. 1079-1094. In N. M. Bikales and L. Segal (ed.), Cellulose and cellulose derivatives. Wiley Interscience, New York, N.Y.
- 563.Reese, E. T., R. G. H. Sui, and H. S. Levinson. 1950. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J. Bacteriol. 59:485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Reverbel-Leroy, C., S. Pagès, A. Bélaïch, J.-P. Bélaïch, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Robson, L. M., and G. H. Chambliss. 1989. Cellulases of bacterial origin. Enzyme Microb. Technol. 11:626-644. [Google Scholar]
- 566.Rodríguez, H., F. Alea, and E. Kyslíkova. 1996. Regulation of cellulolytic activity in Cellulomonas sp. IIbc. Bioresource Technol. 55:79-82. [Google Scholar]
- 567.Rodríguez, H., and R. Gallardo. 1993. Single cell protein production from bagasse pith by a mixed bacterial culture. Acta Biotechnol. 13:141-149. [Google Scholar]
- 568.Roger, V., G. Fonty, S. Komisarczuk-Bony, and P. Gouet. 1990. Effects of physicochemical factors on the adhesion to cellulose avicel of the ruminal bacteria Ruminococcus flavefaciens and Fibrobacter succinogenes subsp. succinogenes. Appl. Environ. Microbiol. 56:3081-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Rogers, P., and G. Gottschalk. 1993. Biochemistry and regulation of acid and solvent production in clostridia. p. 25-50. In D. R. Woods (ed.), The clostridia and biotechnology. Butterworth-Heinemann, Stoneham, Mass.
- 570.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene expression in yeast: a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 571.Rood, J. I. 1997. Genetic analysis in Clostridium perfringens, p. 67-72. In J. I. Rood, B. A. McClane, and J. G. Songer (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Ltd., London, United Kingdom.
- 572.Roos, J. W., J. K. McLaughlin, and E. T. Papoutsakis. 1985. The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol. Bioeng. 27:681-694. [DOI] [PubMed] [Google Scholar]
- 573.Rothstein, D. M. 1986. Clostridium thermosaccharolyticum strain deficient in acetate production. J. Bacteriol. 165:319-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Rouvinen, J., T. Bergfors, T. Teeri, J. K. Knowles, and T. A. Jones. 1990. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249:380-386. [DOI] [PubMed] [Google Scholar]
- 575.Russell, J. B. 1985. Fermentation of cellodextrins by cellulolytic and non-cellulolytic rumen bacteria. Appl. Environ. Microbiol. 49:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Russell, J. B. 1992. Another explanation for the toxicity of fermenting acids at low pH: anion accumulation vs uncoupling. J. Appl. Biotechnol. 73:363-370. [Google Scholar]
- 577.Russell, J. B., and G. M. Cook. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59:48-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Ryu, D. D. Y., C. Kim, and M. Mandels. 1984. Competitive adsorption of cellulase components and its significance in a synergistic mechanism. Biotechnol. Bioeng. 26:488-496. [DOI] [PubMed] [Google Scholar]
- 579.Ryu, D. D. Y., and M. Mandels. 1980. Cellulases: biosynthesis and applications. Enzyme Microb. Technol. 2:91-102. [Google Scholar]
- 580.Sacco, M., J. Millet, and J. P. Aubert. 1984. Cloning and expression in Saccharomyces cerevisiae of a cellulase gene from Clostridium thermocellum. Ann. Microbiol. (Inst. Pasteur) 135A:485-488. [DOI] [PubMed] [Google Scholar]
- 581.Saddler, J. N., H. H. Brownell, L. P. Clermont, and N. Levitin. 1982. Enzymatic hydrolysis of cellulose and various pretreated wood fractions. Biotechnol. Bioeng. 24:1389-1402. [DOI] [PubMed] [Google Scholar]
- 582.Saddler, J. N., and M. K. H. Chan. 1984. Conversion of pretreated lignocellulosic substrates to ethanol by Clostridium thermocellum in monoculture and co-culture with Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. Can. J. Microbiol. 30:212-220. [Google Scholar]
- 583.Saito, T., T. Suzuki, A. Hayashi, H. Honda, M. Taya, S. Iijima, and T. Kobayashi. 1990. Expression of a thermostable cellulase gene from a thermophilic anaerobe in Saccharomyces cerevisiae. J. Ferment. Bioeng. 69:282-286. [Google Scholar]
- 584.Sakon, J., W. S. Adney, M. E. Himmel, S. R. Thomas, and P. A. Karplus. 1996. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry 35:10648-10660. [DOI] [PubMed] [Google Scholar]
- 585.Saloheimo, A., N. Aro, M. Ilmén, and M. Penttilä. 2000. Isolation of the ace1 gene encoding a Cys2-His2 transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817-5825. [DOI] [PubMed] [Google Scholar]
- 586.Saloheimo, A., B. Henrissat, A.-M. Hoffrén, O. Teleman, and M. Pentillä. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 587.Saloheimo, M., P. Lehtovaara, M. Penttilä, T. T. Teeri, J. Ståhlberg, G. Johansson, G. Pettersson, M. Claeyssens, P. Tomme, and J. K. C. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei the characterization of both gene and enzyme. Gene 63:11-21. [DOI] [PubMed] [Google Scholar]
- 588.Saloheimo, M., T. Nakari-Setälä, M. Tenkanen, and M. Penttilä. 1997. cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in Yeast. Eur. J. Biochem. 249:584-591. [DOI] [PubMed] [Google Scholar]
- 589.Sanchez, C. R., P. C. Schvartz, and B. H. Ramos. 1999. Growth and endoglucanase activity of Acetivibrio cellulolyticus grown in three different cellulosic substrates. Rev. Microbiol. 30:310-314. [Google Scholar]
- 590.Sánchez-Torres, P., L. González-Candelas, and D. Ramón. 1998. Heterologous expression of a Candida molischiana anthocyanin-β-glucosidase in a wine yeast strain. J. Agric. Food Chem. 46:354-360. [DOI] [PubMed] [Google Scholar]
- 591.Sanders, M. E., and M. A. Nicholson. 1987. A method for genetic transformation of nonprotoplasted Streptococcus lactis. Appl. Environ. Microbiol. 53:1730-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Sandgren, M., A. Shaw, T. H. Ropp, S. Wu, R. Bott, A. D. Cameron, J. Ståhlberg, C. Mitchinson, and T. A. Jones. 2000. The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 Å resolution. J. Mol. Biol. 308:295-310. [DOI] [PubMed] [Google Scholar]
- 593.San-Segundo, P., J. Correa, C. R. Vázquez de Aldana, and F. Del Rey. 1993. SSG1, a gene encoding a sporulation-specific 1,3-β-glucanase in Saccharomyces cerevisiae. J. Bacteriol. 175:3823-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Sarikaya, A., and M. R. Ladisch. 1999. Solid-state fermentation of lignocellulosic plant residues from Brassica napus by Pleurotus ostreatus. Appl. Biochem. Biotechnol. 82:1-15. [DOI] [PubMed] [Google Scholar]
- 595.Sasaki, T., T. Tanaka, N. Nanbu, Y. Sato, and K. Kainuma. 1979. Correlation between x-ray diffraction measurements of cellulose crystal structure and the susceptibility to microbial cellulase. Biotechnol. Bioeng. 21:1031-1042. [Google Scholar]
- 596.Sato, K., M. Tomita, S. Yonemura, S. Goto, K. Sekine, E. Okuma, Y. Takagi, K. Hon-Nami, and T. Saiki. 1993. Characterization and ethanol hyperproduction by Clostridium thermocellum I-1-B. Biosci. Biotechnol. Biochem. 57:2116-2121. [Google Scholar]
- 597.Sattler, W., H. Esterbauer, O. Glatter, and W. Steiner. 1989. The effect of enzyme concentration on the rate of the hydrolysis of cellulose. Biotechnol. Bioeng. 33:1221-1234. [DOI] [PubMed] [Google Scholar]
- 598.Scheifinger, C. C., and M. J. Wolin. 1973. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 26:789-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Schell, D. J., M. F. Ruth, and M. P. Tucker. 1999. Modeling the enzymatic hydrolysis of dilute acid-pretreated Douglas fir. Appl. Biochem. Biotechnol. 77-79:67-81. [Google Scholar]
- 600.Schimz, K. L., B. Broll, and B. John. 1983. Cellobiose phosphorylase (EC 2.4.1.20) of Cellulomonas: occurrence, induction, and its role in cellobiose metabolism. Arch. Microbiol. 135:241-249. [Google Scholar]
- 601.Schloesser, A., T. Aldekamp, and H. Schrempf. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 190:127-132. [DOI] [PubMed] [Google Scholar]
- 602.Schofield, P., R. E. Pitt, and A. N. Pell. 1994. Kinetics of fiber digestion from in vitro gas production. J. Anim. Sci. 72:2980-2991. [DOI] [PubMed] [Google Scholar]
- 603.Schülein, M. 1997. Enzymatic properties of cellulases from Humicola insolens. J. Biotechnol. 57:71-81. [DOI] [PubMed] [Google Scholar]
- 604.Schülein, M. 1998. Kinetics of fungal cellulases. Biochem. Soc. Trans. 26:164-167. [DOI] [PubMed] [Google Scholar]
- 605.Schülein, M. 2000. Protein engineering of cellulases. Biochim. Biophys. Acta 1543:239-252. [DOI] [PubMed] [Google Scholar]
- 606.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 607.Schwarz, W. H., F. Gräbnitz, and W. L. Staudenbauer. 1986. Properties of Clostridium thermocellum endoglucanase produced in Escherichia coli. Appl. Environ. Microbiol. 51:1293-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Scott, P. T., and J. I. Rood. 1989. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene 82:327-333. [DOI] [PubMed] [Google Scholar]
- 609.Seiboth, B., S. Hakola, R. L. Mach, P. Suominen, and C. P. Kubicek. 1997. Role of four major cellulases in triggering cellulase gene expression in Trichoderma reesei. J. Bacteriol. 179:5318-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Senoir, A. E., M. K. Al-Shawi, and I. L. Urbatsch. 1995. The catalytic cycle of P-glycoprotein. FEBS Lett. 377:285-289. [DOI] [PubMed] [Google Scholar]
- 611.Setati, M. E., P. Ademark, W. H. van Zyl, B. Hahn-Hägerdal, and H. Stålbrand. 2001. Expression of the Aspergillus aculeatus endo-β-1,4-mannanase encoding gene (man1) in Saccharomyces cerevisiae and characterisation of the recombinant enzyme. Protein Expression Purif. 21:105-114. [DOI] [PubMed] [Google Scholar]
- 612.Shafer, M. L., and K. W. King. 1965. Utilization of cellulose oligosaccharides by Cellvibrio gilvus. J. Bacteriol. 89:113-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Sharma, V. K., and J. C. Hagen. 1995. Isolation and characterization of Clostridium hobsonii comb. nov. Biores. Technol. 51:61-74. [Google Scholar]
- 614.Sheehan, J., and M. Himmel. 1999. Enzymes, energy, and the environment: a strategic perspective on the U.S. Department of Energy's research and development activities for bioethanol. Biotechnol. Prog. 15:817-827. [DOI] [PubMed] [Google Scholar]
- 615.Sheth, K., and J. K. Alexander. 1969. Purification and properties of β-1,4-oligoglucan: orthophosphate glucosyltransferase from Clostridium thermocellum. J. Biol. Chem. 244:457-464. [PubMed] [Google Scholar]
- 616.Shi, Y., C. L. Odt, and P. J. Weimer. 1997. Competition for cellulose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 63:734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Shi, Y., and P. J. Weimer. 1992. Response surface analysis of the effects of pH and dilution rate on Ruminococcus flavefaciens FD-1 in cellulose-fed continuous culture. Appl. Environ. Microbiol. 58:2583-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Shi, Y., and P. J. Weimer. 1996. Utilization of individual cellodextrins by three predominant ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 62:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Shi, Y., P. J. Weimer, and J. Ralph. 1997. Formation of formate and hydrogen, and flux of reducing equivalents and carbon in Ruminococcus flavefaciens FD-1. Antonie Leeuwenhoek 72:101-109. [DOI] [PubMed] [Google Scholar]
- 620.Shimizu, T., K. Kudo, H. Tanaka, and Y. Nasu. 1992. Cellulose digestion by an extremely thermophilic anaerobic bacterium. Osaka Soc. Ferment. Technol. 70:443-449. [Google Scholar]
- 621.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 622.Simankova, M. V., N. A. Chernych, G. A. Osipov, and G. A. Zavarzin. 1993. Halocella cellulolytica, gen. nov. sp. nov., a new obligately anaerobic, halophilic, cellulolytic bacterium. Syst. Appl. Microbiol. 16:385-389. [Google Scholar]
- 623.Singh, A., A. B. Abidi, A. K. Agarwal, and N. S. Darmwal. 1991. Single cell protein production by Aspergillus niger and its evaluation. Zentbl. Mikrobiol. 146:181-184. [PubMed] [Google Scholar]
- 624.Singh, A., and K. Hayashi. 1995. Microbial cellulases: protein architecture, molecular properties and biosynthesis. Adv. Appl. Microbiol. 40:1-44. [DOI] [PubMed] [Google Scholar]
- 625.Singh, A., P. K. R. Kumar, and K. Schugerl. 1991. Adsorption and reuse of cellulases during saccharification of cellulosic materials. J. Biotechnol. 18:205-212. [Google Scholar]
- 626.Sixou, S., N. Eynard, J. M. Exoubas, E. Werner, and J. Teissié. 1991. Optimized conditions for electrotransformation of bacteria are related to the extent of electropermiabilization. Biochim. Biophys. Acta 1088:135-138. [DOI] [PubMed] [Google Scholar]
- 627.Skipper, N. A., R. P. Bozzato, D. Vetter, R. W. Davies, R. Wong, and J. E. Hopper. 1987. Use of the melibiase promoter and signal peptide to express a bacterial cellulase from yeast, p. 137-148 In G. G. Stewart, I. Russell, R. D. Klein, and R. R. Hiebsch (ed.). Biological research on industrial yeasts. vol. 1. CRC Press, Inc., Boca Raton, Fla.
- 628.Skipper, N., M. Sutherland, R. W. Davies, D. Kilburn, R. C. Miller, A. Warren, and R. Wong. 1985. Secretion of a bacterial cellulase by yeast. Science 230:958-961. [DOI] [PubMed] [Google Scholar]
- 629.Slapack, G. E., I. Russell, and G. G. Stewart. 1987. Thermophilic microbes in ethanol production. CRC Press, Inc., Boca Raton, Fla.
- 630.Slater, J. H. 1988. Microbial populations and community dynamics, p. 51-74. In J. M. Lynch and J. E. Hobbie (ed.), Micro-organisms in action: concepts and applications in microbial ecology, 2nd ed. Blackwell Scientific Publishers, Oxford, United Kingdom.
- 631.Solomon, B. O., L. E. Erickson, and S. S. Yang. 1983. Estimation of biomass concentration in the presence of solids for the purpose of parameter estimation. Biotechnol. Bioeng. 25:2469-2477. [DOI] [PubMed] [Google Scholar]
- 632.South, C. R., D. A. Hogsett, and L. R. Lynd. 1993. Continuous fermentation of cellulosic biomass to ethanol. Appl. Biochem. Biotechnol. 39-40:587-600. [Google Scholar]
- 633.South, C. R., D. A. L. Hogsett, and L. R. Lynd. 1995. Modeling simultaneous saccharification and fermentation of lignocellulose to ethanol in batch and continuous reactors. Enzyme Microb. Technol. 17:797-803. [Google Scholar]
- 634.Soutschek-Bauer, E., L. Hartl, and W. L. Staudenbauer. 1985. Transformation of Clostridium thermohydrosulfuricum DSM 586 with plasmid DNA. Biotechnol. Lett. 7:705-710. [Google Scholar]
- 635.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of celR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 636.Srinivasan, K., M. Murakami, Y. Nakashimada, and N, Nishio. 2001. Efficient production of cellulolytic and xylanolytic enzymes by the rumen anaerobic fungus, Neocallimastix frontalis, in a repeated batch culture. J. Biosci. Bioeng. 91:153-158. [DOI] [PubMed] [Google Scholar]
- 637.Ståhlberg, J., G. Johansson, and G. Petterson. 1991. A new model for enzymatic hydrolysis of cellulose based on the two-domain structure of cellobiohydrolase-1. Bio/Technology 9:286-290. [Google Scholar]
- 638.Stålbrand, H., A. Saloheimo, J. Vehmaanpera, B. Henrissat, and M. Penttilä. 1995. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Steenbakkers, P. J. M., X. L. Li, E. A. Ximenes, J. G. Arts, H. Z. Chen, L. G. Ljungdahl, and H. J. M. Op den Camp. 2001. Noncatalytic docking domains of cellulosomes of anaerobic fungi. J. Bacteriol. 183:5325-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Steffan, R. J., A. Breen, R. M. Atlas, and G. S. Sayler. 1989. Application of gene probe methods for monitoring specific microbial populations in freshwater ecosystems. Can. J. Microbiol. 35:681-685. [Google Scholar]
- 641.Steiner, W., W. Sattler, and H. Esterbauer. 1988. Adsorption of Trichoderma reesei cellulase on cellulose: experimental data and their analysis by different equations. Biotechnol. Bioeng. 32:853-865. [DOI] [PubMed] [Google Scholar]
- 642.Sternberg, D. 1976. Production of cellulase by Trichoderma. Biotechnol. Bioeng. Symp. Ser. 6:35-53. [PubMed] [Google Scholar]
- 643.Sternberg, D., and G. R. Mandels. 1980. Regulation of the cellulolytic system in Trichoderma reesei by sophorose: induction of cellulase and repression of β-glucosidase. J. Bacteriol. 144:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Sternberg, D., P. Vijayakumar, and E. T. Reese. 1977. β-Glucosidase: microbial production and effect on enzymatic hydrolysis of cellulose. Can. J. Microbiol. 23:139-147. [DOI] [PubMed] [Google Scholar]
- 645.Stewart, C. S., and H. J. Flint. 1989. Bacteroides (Fibrobacter) succinogenes, a cellulolytic anaerobic bacterium from the gastrointestinal tract. Appl. Microbiol. Biotechnol. 30:433-439. [Google Scholar]
- 646.Steyn, A. J. C. J. Marmur, and I. S. Pretorius. 1995. Cloning, sequence analysis and expression in yeasts of a cDNA containing a Lipomyces kononenkoae alpha-amylase-encoding gene. Gene (Amsterdam) 166:65-71. [DOI] [PubMed] [Google Scholar]
- 647.Stone, J. E., and A. M. Scallan. 1968. A structural model for the cell wall of water-swollen wood pulp fibres based on their accessibility to macromolecules. Cellulose Chem. Technol. 3:343-358. [Google Scholar]
- 648.Stone, J. E., A. M. Scallan, E. Donefer, and E. Ahlgren. 1969. Digestibility as a simple function of a molecule of similar size to a cellulase enzyme. Adv. Chem. Ser. 95:219-241. [Google Scholar]
- 649.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS. Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 650.Strobel, H. J. 1995. Growth of the thermophilic bacterium Clostridium thermocellum in continuous culture. Curr. Microbiol. 31:210-214. [Google Scholar]
- 651.Strobel, H. J., F. C. Caldwell, and K. A. Dawson. 1995. Carbohydate transport by the anaerobic thermophilic Clostridium thermocellum LQRI. Appl. Environ. Microbiol. 61:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Stutzenberger, F. 1987. Selective adsorption of endoglucanases from Thermomonospora curvata on protein-extracted lucerne fibers. Lett. Appl. Microbiol. 5:1-4. [Google Scholar]
- 653.Stutzenberger, F. 1990. Bacterial cellulases, p. 37-70. In W. M. Fogarty and C. T. Kelly (ed.), Microbial enzymes and biotechnology, 2nd ed. Elsevier Applied Science, London, United Kingdom.
- 654.Stutzenberger, F., and G. Lintz. 1986. Hydrolysis products inhibit adsorption of Trichoderma reesei C30 cellulases to protein-extracted lucerne fibres. Enzyme Microb. Technol. 8:341-344. [Google Scholar]
- 655.Suihko, M. L., U. Lehtinen, B. Zurbriggen, A. Vilpola, J. Knowles, and M. Pentillä. 1991. Construction and analysis of recombinant glucanolytic brewer's yeast strains. Appl. Microbiol. Biotechnol. 35:781-787. [Google Scholar]
- 656.Sunna, A., M. Moracci, M. Rossi, and G. Antranikian. 1997. Glycosyl hydrolases from hyperthermophiles. Extremophiles 1:2-13. [DOI] [PubMed] [Google Scholar]
- 657.Sutcliffe, R., and J. N. Saddler. 1986. The role of lignin in the adsorption of cellulases during enzymatic treatment of lignocellulosic material. Biotechnol. Bioeng. Symp. Ser. 17:749-762. [Google Scholar]
- 658.Suvajittanont, W., J. McGuire, and M. K. Bothwell. 2000. Adsorption of Thermomonospora fusca E5 cellulase on silanized silica. Biotechnol. Bioeng. 67:12-18. [DOI] [PubMed] [Google Scholar]
- 659.Svetlichnyi, V. A., T. P. Svetlichnaya, N. A. Chernykh, and G. A. Zavarzin. 1990. Anaerocellum thermophilum, new genus new species, an extremely thermophilic cellulolytic eubacterium isolated from hot springs in the valley of geysers (Russian SFSR, USSR). Mikrobiologiya 59:598-604. [Google Scholar]
- 660.Taillez, P., S. H. Girard, R. Longin, P. Beguin, and J. Millet. 1989. Cellulose fermentation by an asporogenous mutant and an ethanol-tolerant mutant of Clostridium thermocellum. Appl. Environ. Microbiol. 55:203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Takada, G., T. Kawaguchi, J. Sumitani, and M. Arai. 1998. Expression of Aspergillus aculeatus No. F-50 cellobiohydrolase I (cbh1) and β-glucosidase 1 (bgl1) genes by Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 62:1615-1618. [DOI] [PubMed] [Google Scholar]
- 662.Takahashi, W., H. Yamagata, K. Yamaguchi, N. Tsukagoshi, and S. Udaka. 1983. Genetic transformation of Bacillus brevis 47, a protein-secreting bacterium, by plasmid DNA. J. Bacteriol. 156:1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Takashima, S., H. Iikura, A. Nakamura, H. Masaki, and T. Uozumi. 1996. Analysis of Cre1 binding sites in the Trichoderma reesei cbh1 upstream region. FEMS Microbiol. Lett. 145:361-366. [DOI] [PubMed] [Google Scholar]
- 664.Takashima, S., A. Nakamura, M. Hidaka, H. Masaki, and T. Uozumi. 1999. Molecular cloning and expression of the novel fungal beta-glucosidase genes from Humicola grisea and Trichoderma reesei. J. Biochem. 125:728-736. [DOI] [PubMed] [Google Scholar]
- 665.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Tanahashi, M., S. Takada, T. Aoki, T. Goto, T. Higuchi, and S. Hanai. 1983. Characterization of explosion wood. 1. Structure and physical properties. Wood Res. 69:36-51. [Google Scholar]
- 667.Tanaka, K., T. Kawaguchi, Y. Imada, T. Ooi, and M. Arai. 1995. Purification and properties of cellobiose phosphorylase from Clostridium thermocellum. J. Ferment. Bioeng. 79:212-216. [Google Scholar]
- 668.Tanaka, M., H. Nakamura, M. Taniguchi, T. Morita, R. Matsuno, and T. Kamikubo. 1986. Elucidation of adsorption processes of cellulases during hydrolysis of crystalline cellulose. Appl. Microbiol. Biotechnol. 23:263-268. [Google Scholar]
- 669.Tardif, C., H. MaÂmar, M. Balfin, and J. P. Belaich. 2001. Electrotransformation studies in Clostridium cellulolyticum. J. Ind. Microbiol. Biotechnol. 16:1-4. [DOI] [PubMed] [Google Scholar]
- 670.Tariq, M. A., and K. Hayashi. 1995. Synthesis of three heterodisaccharides, 4-O-β-glucopyranosyl-6-deoxy-d-glucose, 4-O-β-d-glucopyranosyl-d-mannosamine, and 4-O-β-d-glucopyranosyl-d-mannose, and confirmation of their structures by 13C-NMR and MS. Biochem. Biophys. Res. Commun. 214:568-575. [DOI] [PubMed] [Google Scholar]
- 671.Tatsumoto, K., J. O. Baker, M. P. Tucker, K. K. Oh, A. Mohagheghi, K. Grohmann, and M. E. Himmel. 1988. Digestion of pretreated aspen substrates: hydrolysis rates and adsorptive loss of cellulase enzymes. Appl. Biochem. Biotechnol. 18:159-174. [Google Scholar]
- 672.Teeri, T. T. 1997. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15:160-167. [Google Scholar]
- 673.Teeri, T. T., A. Koivula, M. Linder, G. Wohlfahrt, C. Divne, and T. A. Jones. 1998. Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose? Biochem. Soc. Trans. 26:173-178. [DOI] [PubMed] [Google Scholar]
- 674.Tengerdy, R. P., W. H. Rho, and A. M. Mohagheghi. 1991. Liquid fluidized bed starter culture of Trichoderma reesei for cellulase production. Appl. Biochem. Biotechnol. 27:195-204. [Google Scholar]
- 675.Teunissen, M. J., R. J. S. Baerends, R. A. G. Knelissen, H. J. Op den Camp, and H. J. Knelissen. 1992. A semi-continuous culture system for production of cellulolytic and xylanolytic enzymes by the anaerobic fungus Piromyces sp. strain E2. Appl. Microbiol. Biotechnol. 38:28-33. [Google Scholar]
- 676.Teunissen, M. J., and H. J. Op den Camp. 1993. Anaerobic fungi and their cellulolytic and xylanolytic enzymes. Antonie Leeuwenhoek 63:63-76. [DOI] [PubMed] [Google Scholar]
- 677.Thierry, A., and R. Chicheportiche. 1988. Use of ATP bioluminescence measurements for the estimation of biomass during biological humification. Appl. Microbiol. Biotechnol. 28:199-202. [Google Scholar]
- 678.Thompson, D. N., H. C. Chen, and H. E. Grethlein. 1992. Comparison of pretreatment methods on the basis of available surface area. Biores. Technol. 39:155-163. [Google Scholar]
- 679.Thomsen, K. K., E. A. Jackson, and K. Brenner. 1988. Genetic engineering of yeast: construction of strains that degrade β-glucans with aid of a barley gene. ASBC J. 46:31-36. [Google Scholar]
- 680.Thurston, B., K. A. Dawson, and H. J. Strobel. 1993. Cellobiose versus glucose utilization by the ruminal bacterium Ruminococcus albus. Appl. Environ. Microbiol. 59:2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Tomme, P., V. Heriban, and M. Claeyssens. 1990. Adsorption of two cellobiohydrolases from Trichoderma reesei to Avicel: evidence for “exo-exo” synergism and possible “loose complex” formation. Biotechnol. Lett. 12:525-530. [Google Scholar]
- 682.Tomme, P., H. Van Tilbeurgh, G. Pettersson, J. Van Damme, J. Vanderkerckhove, J. Knowles, T. Teeri, and M. Claeyssens. 1988. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 170:575-581. [DOI] [PubMed] [Google Scholar]
- 683.Tomme, P., R. A. J. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 684.Tong, X. G., L. H. Smith, and P. L. McCarty. 1990. Methane fermentation of selected lignocellulosic materials. Biomass 21:239-255. [Google Scholar]
- 685.Torget, R. W., J. S. Kim, and Y. Y. Lee. 2000. Fundamental aspects of dilute acid hydrolysis/fractionation kinetics of hardwood carbohydrates. 1. Cellulose hydrolysis. Ind. Eng. Chem. Res. 39:2817-2825. [Google Scholar]
- 686.Torget, R. W., P. Walter, M. Himmel, and K. Grohmann. 1991. Dilute-acid pretreatment of corn residues and short-rotation woody crops. Appl. Biochem. Biotechnol. 28-29:75-86. [Google Scholar]
- 687.Tsoi, T. V., N. A. Chuvil'skaya, Y. Y. Atakishieva, T. D. Dzhavakhishvili, V. K. Akimenko, and A. M. Boronin. 1987. Clostridium thermocellum—a new object of genetic investigations. Mol. Genet. Mikrobiol. Virusol. 11:18-23. [PubMed] [Google Scholar]
- 688.Ueda, M., and A. Tanaka. 2000. Cell surface engineering of yeast: construction of arming yeast with biocatalyst. J. Biosci. Bioeng. 90:125-136. [PubMed] [Google Scholar]
- 689.Uozumi, N., A. Hayashi, T. Ito, A. Patthra, I. Yamashita, S. Iijima, and T. Kobayashi. 1993. Secretion of thermophilic bacterial cellobiohydrolase in Saccharomyces cerevisiae. J. Ferment. Bioeng. 75:399-404. [Google Scholar]
- 690.Usami, S., K. Kirimura, M. Imura, and S. Morikawa. 1990. Cellular localization of the constitutive β-glucosidase in Trichoderma viride. J. Ferment. Bioeng. 70:185-187. [Google Scholar]
- 691.Vaheri, M., M. Leisola, and V. Kaupinnen. 1979. Transglycosylation products of cellulase system of Trichoderma reesei. Biotechnol. Lett. 1:41-46. [Google Scholar]
- 692.Väljamäe, P., V. Sild, A. Nutt, G. Pettersson, and G. Johansson. 1999. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 266:327-334. [DOI] [PubMed] [Google Scholar]
- 693.Väljamäe, P., V. Sild, G. Pettersson, and G. Johansson. 1998. The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface—erosion model. Eur. J. Biochem. 253:469-475. [DOI] [PubMed] [Google Scholar]
- 694.Vallander, L., and K. E. Eriksson. 1987. Enzyme recirculation in saccharification of lignocellusic materials. Enzyme Microb. Technol. 9:714-720. [Google Scholar]
- 695.Vallim, M. A., B. J. H. Janse, J. Gaskell, A. A. Pizzirani-Kleiner, and D. Cullen. 1998. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl. Environ. Microbiol. 64:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Van Arsdell, J. N., S. Kwok, V. L. Schweickart, M. B. Ladner, D. H. Gelfand, and M. A. Innis. 1987. Cloning, characterization and expression in Saccharomyces cerevisiae of endoglucanase I from Trichoderma reesei. Bio/technology 5:60-64. [Google Scholar]
- 697.Vance, I., C. M. Topham, S. L. Blayden, and J. Tampion. 1980. Extracellular cellulase production by Sporocytophaga myxococcoides NCIB 8639. J. Gen. Microbiol. 117:235-242. [Google Scholar]
- 698.Vanden Wymelenberg, A., S. Covert, and D. Cullen. 1993. Identification of the gene encoding the major cellobiohydrolase of the white rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 59:3492-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Van Gylswyk, N. O., and J. J. T. K. Van der Toon. 1986. Description and designation of a neotype strain of Eubacterium cellulosolvens (Cillobacterium cellulosolvens). Int. J. Syst. Bacteriol. 36:275-277. [Google Scholar]
- 700.Van Peij, N. N. M. E., M. M. C. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Van Rensburg, P., W. H. van Zyl, and I. S. Pretorius. 1994. Expression of the Butyrivibrio fibrisolvens endo-β-1,4-glucanase gene together with the Erwinia pectate lyase and polygalacturonase genes in Saccharomyces cerevisiae. Curr. Genet. 27:17-22. [DOI] [PubMed] [Google Scholar]
- 702.Van Rensburg, P., W. H. van Zyl, and I. S. Pretorius. 1995. Expression of the Ruminococcus flavefaciens cellodextrinase gene in Saccharomyces cerevisiae. Biotechnol. Lett. 17:481-486. [Google Scholar]
- 703.Van Rensburg, P., W. H. van Zyl, and I. S. Pretorius. 1996. Co-expression of a Phanerochaete chrysosporium cellobiohydrolase gene and a Butyrivibrio fibrisolvens Endo-β-1,4-glucanase gene in Saccharomyces cerevisiae. Curr. Genet. 30:246-250. [DOI] [PubMed] [Google Scholar]
- 704.Van Rensburg, P., W. H. van Zyl, and I. S. Pretorius. 1997. Over-expression of the Saccharomyces cerevisiae exo-β-1,3-glucanase gene together with the Bacillus subtilis endo-β-1,3-1,4-glucanase gene and the Butyrivibrio fibrisolvens endo-β-1,4-glucanase gene in yeast. J. Biotechnol. 55:43-53. [DOI] [PubMed] [Google Scholar]
- 705.Van Rensburg, P., W. H. van Zyl, and I. S. Pretorius. 1998. Engineering yeast for efficient cellulose degradation. Yeast 14:67-76. [DOI] [PubMed] [Google Scholar]
- 706.Van Soest, P. J. 1973. The uniformity and nutritive availability of cellulose. Fed. Proc. 32:1804-1808. [PubMed] [Google Scholar]
- 707.Van Soest, P. J. 1994. Nutritional ecology of the ruminant, 2nd ed. Cornell University Press, Ithaca, N.Y.,
- 708.Van Uden, N. 1985. Ethanol toxicity and ethanol tolerance in yeasts. Annu. Rep. Ferment. Proc. 8:1-58. [Google Scholar]
- 709.Van Veen, H. W., A. Margolles, M. Müller, C. F. Higgins, and W. N. Konings. 2000. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 19:2503-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Van Walsum, G. P., L. R. Lynd. 1998. Allocation of ATP to synthesis of cells and hydrolytic enzymes in cellulolytic fermentative microorganisms: bioenergetics, kinetics, and bioprocessing. Biotechnol. Bioeng. 58:316-320. [PubMed] [Google Scholar]
- 711.Vardavakis, E. 1989. Seasonal fluctuations of aerobic cellulolytic bacteria, and cellulase and respiratory activities in a soil profile under a forest. Plant Soil 115:145-150. [Google Scholar]
- 712.Veal, D. A., and J. M. Lynch. 1987. Associative cellulolysis and nitrogen fixation by co-cultures of Trichoderma harzianum and Clostridium butyricum: the effects of ammonium nitrogen on these processes. J. Appl. Bacteriol. 63:245-253. [Google Scholar]
- 713.Velkovska, S., M. R. Marten, and D. F. Ollis. 1997. Kinetic model for batch cellulase production by Trichoderma reesei RUT C30. J. Biotechnol. 54:83-94. [DOI] [PubMed] [Google Scholar]
- 714.Vladut-Talor, M., T. Kauri, and D. J. Kushner. 1986. Effects of cellulose on growth, enzyme production and ultrastructure of a Cellulomonas species. Arch. Microbiol. 144:191-195. [Google Scholar]
- 715.Wachinger, G., K. Bronnenmeier, W. L. Staudenbauer, and H. Schrempf. 1989. Identification of mycelium-associated cellulase from Streptomyces reticuli. Appl. Environ. Microbiol. 55:2653-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Wald, S., C. R. Wilke, and H. W. Blanch. 1984. Kinetics of the enzymatic hydrolysis of celluose. Biotechnol. Bioeng. 26:221-230. [DOI] [PubMed] [Google Scholar]
- 717.Waldo, D. R., L. W. Smith, and E. L. Cox. 1972. Model of cellulose disappearance from the rumen. J. Dairy Sci. 55:125-129. [DOI] [PubMed] [Google Scholar]
- 718.Walker, L. P., D. B. Wilson, and D. C. Irwin. 1990. Measuring fragmentation of cellulose by Thermomonospora fusca cellulase. Enzyme Microb. Technol. 12:378-386. [Google Scholar]
- 719.Wang, D. I. C., G. C. Avgerinos, I. Biocic, S. D. Wang, and H. Y. Fang. 1983. Ethanol from cellulosic biomass. Philos. Trans. R. Soc. Lond. Ser. B 300:323-333. [Google Scholar]
- 720.Wang, D. Z., Y. Zu, and P. Gao. 1996. Studies of the regulation of cellulase systems by ATP and cAMP in mycelial fungi. Weishengwu Xuebao 36:12-18. [Google Scholar]
- 721.Wang, W. Y., S. J. Reid, and J. A. Thomson. 1993. Transcriptional regulation of an endoglucanase and a cellodextrinase gene in Ruminococcus flavefaciens FD-1. J. Gen. Microbiol. 139:1219-1226. [DOI] [PubMed] [Google Scholar]
- 722.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 723.Watanabe, H., and G. Tokuda. 2001. Animal cellulases. Cell. Mol. Life Sci. 58:1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Watanabe, T., T. Sato, S. Yoshioka, T. Koshijima, and M. Kuwahara. 1992. Purification and properties of Aspergillus niger β-glucosidase. Eur. J. Biochem. 209:651-659. [DOI] [PubMed] [Google Scholar]
- 725.Wayne, J., and S. Y. Xu. 1997. Identification of a thermophilic plasmid origin and its cloning within a new Thermus-E. coli shuttle vector. Gene 195:321-328. [DOI] [PubMed] [Google Scholar]
- 726.Weil, J., P. J. Westgate, K. Kohlmann, and M. R. Ladisch. 1994. Cellulose pretreatments of lignocellulosic substrates. Enzyme Microb. Technol. 16:1002-1004. [DOI] [PubMed] [Google Scholar]
- 727.Weimer, P. J. 1993. Effects of dilution rate and pH on the ruminal celluloytic bacterium Fibrobacter succinogenes S85 in cellulose fed continuous culture. Arch. Microbiol. 160:288-294. [DOI] [PubMed] [Google Scholar]
- 728.Weimer, P. J. 1996. Why don't ruminal bacteria digest cellulose faster? J. Dairy Sci. 79:1496-1502. [DOI] [PubMed] [Google Scholar]
- 729.Weimer, P. J., A. D. French, and T. A. Calamari. 1991. Differential fermentation of cellulose allomorphs by ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 57:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Weimer, P. J., J. M. Hackney, and A. D. French. 1995. Effects of chemical treatments and heating on the crystallinity of cellulose and their implications for evaluating the effect of crystallinity on cellulose biodegradation. Biotechnol. Bioeng. 48:169-178. [DOI] [PubMed] [Google Scholar]
- 731.Weimer, P. J., J. M. Hackney, H.-J. G. Jung, and R. D. Hatfield. 2000. Fermentation of a bacterial cellulose/xylan composite by mixed ruminal microflora: implications for the role of polysaccharide matrix interactions in plant cell wall biodegradability. J. Agric. Food Chem. 48:1727-1733. [DOI] [PubMed] [Google Scholar]
- 732.Weimer, P. J., R. D. Hatfield, and D. R. Buxton. 1993. Inhibition of ruminal cellulose fermentation by extracts of the perennial legume cicer milkvetch (Astragalus cicer). Appl. Environ. Microbiol. 59:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Weimer, P. J., J. M. Lopez-Guisa, and A. D. French. 1990. Effect of cellulose fine structure on kinetics of its digestion by mixed ruminal microorganisms invitro. Appl. Environ. Microbiol. 56:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Weimer, P. J., and C. L. Odt. 1995. Cellulose degradation by ruminal microbes: physiological and hydrolytic diversity among ruminal cellulolytic bacteria. ACS Symp. Ser. 618:291-304. [Google Scholar]
- 735.Weimer, P. J., Y. Shi, and C. L. Odt. 1991. A segmented gasl/liquid delivery system for continuous culture of microorganisms on insoluble substrates and its use for growth of Ruminococcus flavefaciens on cellulose. Appl. Microbiol. Biotechnol. 36:178-183. [Google Scholar]
- 736.Weimer, P. J., and W. M. Weston. 1985. Relationship between the fine structure of native cellulose and cellulose degradability by the cellulase complexes of Trichoderma reesei and Clostridium thermocellum. Biotechnol. Bioeng. 27:1540-1547. [DOI] [PubMed] [Google Scholar]
- 737.Weimer, P. J., and J. G. Zeikus. 1977. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence and presence of Methanobacterium thermoautotrophicum. Appl. Environ. Microbiol. 33:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738.Wells, J. E., and J. B. Russell. 1994. The endogenous metabolism of Fibrobacter succinogenes and its relation to cellobiose transport, viability, and cellulose digestion. Appl. Microbiol. Biotechnol. 41:471-476. [Google Scholar]
- 739.Wells, J. E., and J. B. Russell. 1996. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl. Environ. Microbiol. 62:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Wells, J. E., J. B. Russell, Y. Shi, and P. J. Weimer. 1995. Cellodextrin efflux by the cellulolytic ruminal bacterium Fibrobacter succinogenes and its potential role in the growth of non-adherent bacteria. Appl. Environ. Microbiol. 61:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Wiegel, J., and M. Dykstra. 1984. Clostridium thermocellum: adhesion and sporulation while adhered to cellulose and hemicellulose. Appl. Microbiol. Biotechnol. 20:59-65. [Google Scholar]
- 742.Wiegel, J., and L. J. Ljungdahl. 1981. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic anaerobic bacterium. Arch. Microbiol. 128:343-348. [Google Scholar]
- 743.Wiegel, J., and L. G. Ljungdahl. 1986. The importance of the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 43:147-151. [DOI] [PubMed] [Google Scholar]
- 744.Wilkinson, S. R., and M. Young. 1994. Targeted integration of genes into the Clostridium acetobutylicum chromosome. Microbiology 140:89-95. [Google Scholar]
- 745.Wilson, C. A., and T. M. Wood. 1992. The anaerobic fungus Neocallimastix frontalis: isolation and properties of a cellulosome-type enzyme fraction with the capacity to solubilize hydrogen-bond-ordered cellulose. Appl. Microbiol. Biotechnol. 37:125-129. [Google Scholar]
- 746.Wilson, D. B. 1992. Biochemistry and genetics of actinomycete cellulases. Crit. Rev. Biotechnol. 12:45-63. [DOI] [PubMed] [Google Scholar]
- 747.Wilson, D. B., and D. C. Irwin. 1999. Genetics and properties of cellulases. Adv. Biochem. Eng. Biotechnol. 65:1-21. [Google Scholar]
- 748.Wilson, J. R. 1993. Organization of forage plant tissues, p. 1-32. In H. G. Jung, D. R. Buxton, R. D. Hatfield, and J. Ralph (ed.), Forage cell wall structure and digestibility. American Society of Agronomy—Crop Science Society of America—Soil Science Society of America, Madison, Wisc.
- 749.Wilson, J. R., and R. D. Hatfield. 1997. Structural and chemical changes of cell wall types during stem development: consequences for fibre degradation by rumen microflora. Aust. J. Agric. Res. 48:165-180. [Google Scholar]
- 750.Wilson, J. R., and D. R. Mertens. 1995. Cell wall accessibility and cell structure limitations to microbial digestion of forage. Crop Sci. 35:251-259. [Google Scholar]
- 751.Withers, S. G. 2001. Mechanisms of glycosyl transferases and hydrolyses. Carbohydr. Polym. 44:325-337. [Google Scholar]
- 752.Woese, C. R. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 97:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Wolin, M. J. 1990. Rumen fermentation: biochemical interactions between the populations of a microbial community, p. 237-251. In D. E. Akin et al. Microbial and plant opportunities to improve lignocellulose utilization by ruminants. Elsevier Science Publications, New York, N.Y.
- 754.Wong, W. K. R., C. Curry, H. R. S. Parekh, M. Wayman, R. W. Davies, D. G. Kilburn, and N. Skipper. 1988. Wood hydrolysis by Cellulomonas fimi endoglucanase and exoglucanase coexpressed as secreted enzymes in Saccharomyces cerevisiae. Bio/technology 6:713-719. [Google Scholar]
- 755.Wood, B. E., and L. O. Ingram. 1992. Ethanol production from cellobiose, amorphous cellulose and crystalline cellulose by recombinant Klebsiella oxytoca containing chromosomally integrated Zymomonas mobilis genes for ethanol production and plasmids expressing thermostable cellulase genes from Clostridium thermocellum. Appl. Environ. Microbiol. 58:2103-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Wood, T. M. 1992. Fungal cellulases. Biochem. Soc. Trans. 20:46-53. [DOI] [PubMed] [Google Scholar]
- 757.Wood, T. M., and V. Garcia-Campayo. 1990. Enzymology of cellulose degradation. Biodegradation. 1:147-161. [Google Scholar]
- 758.Wood, T. M., and V. Garcia-Campayo. 1994. Enzymes and mechanisms involved in microbial cellulolysis, p. 197-231. In C. Ratledge (ed.), Biochemistry of microbial degradation. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 759.Wood, T. M., C. A. Wilson, S. I. McCrae, and K. N. Joblin. 1986. A highly active extracellular cellulase from the anaerobic rumen fungus Neocallimastix frontalis. FEMS Microbiol. Lett. 34:37-40. [Google Scholar]
- 760.Wood, W. E., D. G. Neubauer, and F. J. Stutzenberger. 1984. Cyclic AMP levels during induction and repression of cellulase biosynthesis in Thermomonospora curvata. J. Bacteriol. 160:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Woodward, J., M. K. Hayes, and N. E. Lee. 1988. Hydrolysis of cellulose by saturating and non-saturating concentrations of cellulase: implications for synergism. Bio/Technology 6:301-304. [Google Scholar]
- 762.Wooley, R., M. Ruth, D. Glassner, and J. Sheehan. 1999. Process design and costing of bioethanol technology: a tool for determining the status and direction of research and development. Biotechnol. Prog. 15:794-803. [DOI] [PubMed] [Google Scholar]
- 763.Wright, J. D. 1988. Ethanol from biomass by enzymatic hydrolysis. Chem. Eng. Prog. 84:62-74. [Google Scholar]
- 764.Wright, J. D., C. E. Wyman, and K. Grohmann. 1988. Simulateous saccharification and fermentation of lignocellulose: process evaluation. Appl. Biochem. Biotechnol. 18:75-89. [Google Scholar]
- 765.Wu, L., and N. E. Welker. 1989. Protoplast transformation of Bacillus stearothermophilus NUB36 by plasmid DNA. J. Gen. Microbiol. 135:1315-1324. [DOI] [PubMed] [Google Scholar]
- 766.Wyman, C. E. 1999. Biomass ethanol: technical progress, opportunities, and commercial challenges. Annu. Rev. Energy Environ. 24:189-226. [Google Scholar]
- 767.Xue, G.-P., K. S. Gobius, and C. G. Orpin. 1992. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarum encoding three multifunctional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J. Gen. Microbiol. 138:2397-2403. [DOI] [PubMed] [Google Scholar]
- 768.Yan, T. R., and C. L. Lin. 1997. Purification and characterization of a glucose-tolerant β-glucosidase from Aspergillus niger CCRC 31494. Biosci. Biotechnol. Biochem. 61:965-970. [DOI] [PubMed] [Google Scholar]
- 769.Yernool, D. A., J. K. McCarthy, D. E. Eveleigh, and J. D. Bok. 2000. Cloning and characterization of the glucooligosaccharide catabolic pathway β-glucan glucohydrolase and cellobiose phosphorylase in the marine hyperthermophile Thermotoga neapolitana. J. Bacteriol. 182:5172-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Yoon, K. H., S. H. Park, and M. Y. Pack. 1988. Transfer of Bacillus subtilis endo-β-1,4-glucanase gene into Zymomonas anaerobia. Biotechnol. Lett. 10:213-216. [Google Scholar]
- 771.Young, D. I., V. J. Evans, J. R. Jefferies, K. C. B. Jennert, Z. E. V. Phillips, A. Ravagnani, and M. Young. 1999. Genetic methods in clostridia. Methods Microbiol. 29:191-207. [Google Scholar]
- 772.Yu, A. H. C., D. Lee, and J. N. Saddler. 1995. Adsorption and desorption of cellulase components during the hydrolysis of a steam-exploded birch substrate. Biotechnol. Appl. Biochem. 21:203-216. [Google Scholar]
- 773.Yu, A. H. C., and J. N. Saddler. 1995. Identification of essential cellulase components in the hydrolysis of steam-exploded birch substrate. Biotechnol. Appl. Biochem. 21:185-202. [Google Scholar]
- 774.Zaldivar, J., J. Nielsen, and L. Olsson. 2001. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 56:17-34. [DOI] [PubMed] [Google Scholar]
- 775.Zhang, M., C. Eddy, K. Deanda, M. Finkelstein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]
- 776.Zhang, S., D. E. Wolfgang, and D. B. Wilson. 1999. Substrate heterogeneity causes the nonlinear kinetics of insoluble cellulose hydrolysis. Biotechnol. Bioeng. 66:35-41. [DOI] [PubMed] [Google Scholar]
- 777.Zhang, Z., Y. Wang, and J. Ruan. 1998. Reclassification of Thermomonospora and Microtetraspora. Int. J. Syst. Bacteriol. 48:411-422. [DOI] [PubMed] [Google Scholar]
- 778.Zhou, S. F. C. Davis, and L. O. Ingram. 2001. Gene integration and expression and extracellular secretion of Erwinia chrysanthemi endoglucanase CelY (celY) and CelZ (celZ) in ethanologenic Klebsiella oxytoca P2. Appl. Environ. Microbiol. 67:6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Zhou, S., and L. O. Ingram. 1999. Engineering endoglucanase-secreting strains of ethanologenic Klebsiella oxytoca P2. J. Ind. Microbiol. Biotechnol. 22:600-607. [DOI] [PubMed] [Google Scholar]
- 780.Zhou, S., and L. O. Ingram. 2000. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J. Bacteriol. 182:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Zhou, S., and L. O. Ingram. 2001. Simultaneous saccharification and fermentation of amorphous cellulose to ethanol by recombinant Klebsiella oxytoca SZ21 without supplemental cellulase. Biotechnol. Lett. 23:1455-1462. [Google Scholar]
- 782.Zhou, S., L. P. Yomano, A. Z. Saleh, F. C. Davis, H. C. Aldrich, and L. O. Ingram. 1999. Enhancement of expression and apparent secretion of Erwinia chrysanthemi endoglucanase (encoded by celZ) in Escherichia coli B. Appl. Environ. Microbiol. 65:2439-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Zurbriggen, B., D. J. Bailey, M. E. Penttilä, K. Poutanen, and M. Linko. 1990. Pilot scale production of a heterologous Trichoderma reesei cellulase by Saccharomyces cerevisiae. J. Biotechnol. 13:267-278. [DOI] [PubMed] [Google Scholar]
- 784.Zurbriggen, B. D., M. E. Penttilä, L. Viikari, and M. J. Bailey. 1991. Pilot scale production of a Trichoderma reesei endo-β-glucanase by brewer's yeast. J. Biotechnol. 17:133-146. [DOI] [PubMed] [Google Scholar]
- 785.Zverlov, V. V., G. A. Velikodvorskaya, W. H. Schwarz, K. Bronnenmeier, J. Kellermann, and W. L. Staudenbauer. 1998. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J. Bacteriol. 180:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Zverlov, V. V., G. A. Velikodvorskaya, W. H. Schwarz, J. Kellermann, and W. L. Staudenbauer. 1999. Duplicated Clostridium thermocellum cellobiohydrolase gene encoding cellulosomal subunits S3 and S5. Appl. Microbiol. Biotechnol. 51:852-859. [DOI] [PubMed] [Google Scholar]