Abstract
Current X-ray diffraction and cryoelectron microscopic data of ribosomes of eubacteria have shed considerable light on the molecular mechanisms of translation. Structural studies of the protein factors that activate ribosomes also point to many common features in the primary sequence and tertiary structure of these proteins. The reconstitution of the complex apparatus of translation has also revealed new information important to the mechanisms. Surprisingly, the latter approach has uncovered a number of proteins whose sequence and/or structure and function are conserved in all cells, indicating that the mechanisms are indeed conserved. The possible mechanisms of a new initiation factor and two elongation factors are discussed in this context.
INTRODUCTION
There is overwhelming evidence that the genetic code is, with relatively minor variations, conserved throughout species. It is thus fully expected that the apparatus that deciphers this code should be correspondingly conserved. The intermediates and the products of the translation reaction are conserved, giving further credence to this idea. Current X-ray diffraction analysis of the eubacterial ribosomes at 5.5-Å resolution has resulted in the direct visualization of the tRNA substrates in the ribosomal A, P, and E sites (27, 204). Since the sites are conserved (173), this argues for conservation of the mechanisms in translation as well.
The ribosomal proteins, the tRNAs, and the aminoacyl-tRNA synthetases have remarkably conserved structures throughout the species, as have the protein factors that are involved in initiation, elongation, and termination (149, 150). The only apparent exception to the fact that all of the components of translation are conserved is the initiation reaction (113). The initiation processes are functionally similar in all cells, but the components of the reactions appear to differ between archaebacteria/eukaryotes and the eubacteria. Thus, three factors are assumed sufficient for initiation in eubacteria, IF1, IF2, and IF3. In contrast, eukaryotic initiation involves additional proteins, many of which are made up of several subunits (83, 84). This observation implies that the evolution of these processes differed, an issue that has been elegantly addressed by Kyrpides and Woese (113).
Here, we review observations that several initiation factors and one elongation factor of Escherichia coli, first isolated as required to reconstitute translation (55, 58, 65, 74), bear a surprising degree of similarity in structure and function to the eukaryotic initiation factors eIF4A and eIF5A and to the elongation factor eEF3, previously described as a fungus-specific protein. The possible mechanism of action of these proteins is discussed after a brief review of the initiation and elongation reactions. Reviews are cited whenever possible.
TRANSLATIONAL INITIATION
In all cells, translation initiation establishes the reading phase of the genetic code. The start codon in the mRNA interacts with a special methionyl-tRNA (fMet-tRNAfMet or Met-tRNAi) in the peptidyl donor center of the small subunit of ribosomes. The start codon is usually AUG or GUG; rarely, other codons such as UUG, AUU, and AUA can also initiate synthesis (70, 76, 185). Start codons are ambiguous since they also specify insertion of methionine, valine, or other amino acids at internal positions. These triplets also occur out of phase in the genetic transcript. Therefore, features of the mRNA, apart from the start codon, are necessary to phase translation of the genetic transcript. Several reviews are available (70, 76, 185).
In most eubacteria, a second determinant of specificity is an mRNA polypurine tract 5′ of the start triplet. This sequence complements the 3′ end of the 16S rRNA of the 30S subunit and is called the Shine-Dalgarno (SD) sequence (176). If the SD sequence is truncated or deleted in an mRNA, normal ribosomes cannot translate the encoded proteins (88). However, if the 16S rRNA is altered so as to anneal to these defective mRNAs, translation is possible (88). Biochemical, genetic, and statistical data emphasize the importance of this mRNA-16S rRNA interaction but indicate that the SD sequence is not always necessary and is often not sufficient to specify the start of protein synthesis (70, 76, 185). Other regions of the 16S rRNA, i.e., bases 1471 to 1480 and 458 to 466, have been proposed to bind the mRNA 3′ or 5′ of the initiation codon (184). These sequences occur in stem-loop structures that are not exposed on the 30S subunit (184). Mutations in the 16S rRNA complementary to the mRNA downstream sequences do not significantly alter synthesis (147, 160). Deletion of these “downstream” elements is necessary to establish their function (147).
Early studies of the binding properties of the bacteriophage Qβ-A protein ribosome-binding site and analogues of this sequence indicated the following. (i) The intact start triplet is strictly required to bind the mRNA to the ribosome in the presence or in the absence of the fMet-tRNAfMet. (ii) The upstream SD signal confers stability to mRNA binding. (iii) The number and the type of bases between the SD signal and the start triplet affect initiation efficiency (60, 139). Identical requirements have been reported for a variety of mRNAs (71, 80-82, 161). It is thought that the SD sequence in most transcripts may help transiently to anchor the mRNA on the ribosome, allowing for the kinetic selection of the start triplet in closer proximity to the ribosomal P site (76, 77).
It has been suggested that an extended anticodon of the fMet-tRNAfMet (i.e., bases 3′ or 5′ of the tRNA anticodon that base pair with bases 5′ or 3′ of the mRNA start codon) enhances the rate of formation and the stability of initiation complexes (60, 63, 80, 123). The bases 5′ of AUG markedly modulate initiation complex formation (60). In addition, the start codon itself is frequently found embedded in stretches of longer sequence homology to the anticodon loop (62). Mutations that disrupt the potential complementarity of the extended anticodon with the mRNA reduce translational rates, suggesting that they may be involved in the AUG recognition process. Examples of such features are given in reference 62. Similar results have been obtained with point mutations in the start triplet of the lacZ gene (89, 132).
mRNA secondary structure can affect translational initiation (40, 80, 118, 196). In some cases, the secondary structure masks initiation sites, e.g., AUG in the case of the bacteriophage R17 or MS2 RNA replicase or the SD sequence in the case of the lamB protein of E. coli (40, 70, 80, 185). In other cases, disruption of the secondary structure increases the ability of the ribosomes to recognize correct (and a few incorrect) initiation sites (118). Mutations that disrupt the secondary structure of ribosome-binding sites increase the expression of various genes or exhibit polar effects, presumably by exposing or creating initiation signals (70, 76, 79, 118, 185). The expression of the MS2 coat protein gene depends quantitatively on the thermodynamic stability of the secondary structures neighboring the AUG start codon (40).
Computer-assisted phylogenetic sequence comparisons and mutational studies suggest that the coding regions of several hundred mRNAs fold into a uniform, almost periodic pattern of secondary structure. In contrast, sequences 5′ of start codons exhibit a relative lack of potential secondary structure (61). Many mutations 5′ of the start site of prokaryotic and eukaryotic mRNAs were found to impair initiation by sequestering the start triplet in secondary structures (70, 71, 111). These studies suggest that the differences in mRNA secondary structure of the start and of the coding sequences, particularly the transition in such structures, may also be an important determinant of initiation site selection (53, 61). This, in turn, implies that an active process may be required to unwind such structures so that elongation can proceed.
Eukaryotic Initiation
There are many similarities in the process of initiation across all organisms. The similarities and differences that underlie the eukaryotic and eubacterial processes have been reviewed in detail by Hershey and Merrick (83, 84). Briefly, the start codon, the use of a unique initiator tRNA to decode it, and many aspects of the recognition mechanisms are similar, if not identical, in all cells. Thus, ribosomes must be dissociated in the presence of the ubiquitous initiation factor, IF3. A preinitiation complex that harbors the initiator tRNA on the small subunit occurs in both eukaryotic and prokaryotic cells (76, 84, 111). However, marked differences occur, which reflect the fact that the translation of eubacterial transcripts is coupled to transcription. In eukaryotes, complex mechanisms exist to splice and modify the ends of the transcripts prior to transporting them from the nucleus into the cytoplasm of the cell. In contrast to the eubacterial mRNAs, most eukaryotic mRNAs are monocistronic. However, eukaryotic ribosomes have the ability to recognize the internal ribosome entry sites of several mRNAs through a special mechanism (91, 94).
An important difference in the mechanisms of initiation of eukaryotic and eubacterial cells reflects the fact that eukaryotic mRNAs are usually modified by a 5′-terminal 7mG cap structure (111) and have a 3′ poly(A) extension (95). A special mechanism exists for recognizing the 5′ cap structure. In similar fashion to eubacteria, the eukaryotic initiation codon is flanked by sequences that favor a specific consensus. Thus, an A is favored 3 bases 5′ of the AUG, although many transcripts have a G at this position. Also, a unique G is usually found 3′ of the start codon. The consensus sequence, i.e., bases near the 5′ and 3′ borders of AUG, is known to be important in start site recognition. Mutation of a nonstart AUG to the proper sequence context results in initiation of a new protein at that site (111).
The eukaryotic start site is generally devoid of stable secondary structures, as is the case for the eubacterial mRNAs. However, the length of the region that is free of secondary structure is smaller in the eukaryotic transcripts, probably reflecting the relative shorter length of the eukaryotic start site (53). Secondary structures that occlude recognition of the 7mG cap impair initiation (111). In general, recognition of the 5′ terminus of the mRNA and of the 7mG cap structure precedes binding of the 40S subunit to the mRNA. These interactions, in turn, precede scanning of the transcript up to the start codon (111).
More proteins are involved in the process of initiation in eukaryotic cells than in eubacteria, and several of these proteins are polymeric, amplifying the possibilities for translation regulation. The genes encoding initiation proteins have been cloned and sequenced, and the structures of most of the encoded proteins have been determined (84). The details of their structure and function have been reviewed recently (84).
Initiation Reaction
Initiation in all cells requires the conversion of the 70S (80S) ribosome into 30S (40S) and 50S (60S) subunits (102). In eubacteria, the reaction may occur via two pathways in which the 30S initiation factor complex can either interact first with the mRNA or with fMet-tRNA (76, 77). Formation of the 30S-mRNA-fMet-tRNA complex requires binding of the IF2-GTP-fMet-tRNA into the P site. Without initiation factors, elongator tRNAs compete successfully for P-site occupancy (59, 81, 82). EF3 prevents the association of ribosomal subunits and impedes the codon-specific attachment of elongator tRNAs (81, 82). IF2 is a latent GTPase that may initiate these proofreading events (76). IF2 may discriminate initiator from elongator tRNAs by recognizing the acceptor end of the tRNA. In contrast, IF3 appears to select the anticodon stem-loop of the initiator tRNA (81, 82). IF1 stimulates the action of IF2 and IF3 by binding to the A site of the 30S subunit (76). Thus, the initiation factors guarantee the exclusive occupancy of the P site by fMet-tRNA. The initiation factors are probably not involved in mRNA binding but instead affect the position of the mRNA on the ribosome. Thus, mRNA cross-links to the r-proteins S1, S3, S5, S14, and S21. Initiation factors shift the position of the cross-links concurrent with the accommodation of the mRNA into the proper site and the displacement of the elongator tRNAs from this site (76, 77). The binding of the eubacterial IF3 results in a conformational change in the ribosomes which promotes the subsequent binding of the mRNA to the 30S subunit (76, 102). Cryoelectron microscope (cryo-EM) data of the complex of IF3 with the ribosome are consistent with this view (124).
The structures of IF1 and IF3 have been determined (76). IF1 and IF2 form a complex that resembles the structure of domains IV and V of the EFG-GDP complex (19). Domains IV and V of the EFG-GDP complex, in turn, resemble the EFTu ternary complex and have been postulated to mimic the structure of tRNA. Such structural similarity also appears to aid binding of the EFTu-GTP-aminoacyl-tRNA complex to the ribosomal A site. There is considerable evidence, apart from this structural resemblance, that the initiation factors ensure the stable entrance of the initiator RNA into the ribosomal P site (80, 81).
Two forms of IF2 and IF3 occur in E. coli (169). It is possible that these forms act on alternate pathways for initiation in which tRNAi interacts first with the 30S particle or with the 30S-mRNA complex (76, 77, 196).
Similarly, the eukaryotic proteins, eIF3 and eIF1A, bind to the 40S subunit and prevent the association of the 60S subunit. Although the exact mechanism is not known, it is suggested that subunit antiassociation may occur by an allosteric transition (84), by steric hindrance, or perhaps by preventing the displacement of the eIF2-GTP-Met-tRNA from the 40S by the 60S subunit (84). In addition, other factors, eIF6 and eIF3A, have been reported to impede the association of the subunits by binding to the 60S particle, thus further preventing an inappropriate joining of the subunits (84). The exact mechanism of eIF6 action has not yet been established.
One expects that a function such as the joining of the ribosomal subunits should remain a conserved process. In eubacteria, the junction of the ribosomal subunits is composed mostly of rRNA sequences, and it is assumed that these base pair with the 50S particle in some fashion. However, it appears that the process requires a protein, the “rescue” factor, to join properly in a functional complex devoid of IF3 (66). Direct evidence that the “rescue protein” competes with the eubacterial IF3 to foster 70S subunit formation suggests that this protein is indeed the counterpart of IF5. A more precise assignment depends on determining the structures of these proteins. What is clear is that the joining of the ribosomal subunits is also dependent on common functions carried out by nonribosomal proteins. For the case of the eubacterial ribosomes, the “rescue” factor may help unmask or align the rRNA sequences in the 30S and 50S subunits which may help join these particles.
IF1A has a significant (21%) sequence identity to the eubacterial factor IF1 and acts in analogous fashion to this protein. On the other hand, eIF3 is composed of 11 subunits and thus differs from the single eubacterial protein that displays a similar mechanism. The ternary complex that is formed, Met-tRNAi-GTP-eIF2 in eukaryotes, is catalyzed by eIF2, although this protein is composed of three subunits; the γ subunit binds the Met-tRNAi and GTP. The sequence of the eubacterial IF2 is, however, not related to the eIF2. The comparable protein is instead eIF5B (84, 113), which is a ribosome-dependent GTPase, as is the eubacterial IF2.
In eukaryotic cells, the binding of the mRNA to the tRNAi-ribosome complex requires factors that are not represented in eubacterial cells or have not yet been identified. Among these factors are (i) eIF2A, a protein that acts only with AUG in formation of the tRNAi-40S-AUG complex; (ii) eIF2B, a protein that stimulates the exchange of GDP for GTP in eIF2—the phosphorylation of eIF2B by a variety of means is involved in a unique series of reactions that are the targets for a negative regulation of translation; (iii) eIF3A and eIF6, which prevent subunit association by binding to the 60S particle; and (iv) the 7mG cap, which occurs as a singular feature of most eukaryotic mRNAs—a special mechanism is used to recognize the cap structure. The eukaryotic eIF4F is composed of a 25-kDa cap-binding protein designated eIF4E, a 220-kDa protein (eIF4G) that acts as chaperone for the action of eIF3, eIF4A, eIF4B, and eIF4E. The eIF4F complex unwinds the secondary structure of the mRNAs 5′ proximal to the 7mG cap. This protein complex acts as an RNA helicase and is obligatory for translation in reticulocytes (84). For a summary of the nomenclature and function of the initiation factors, see Table 1 (see also reference 84).
TABLE 1.
Initiation factors
| Eukaryotes | Archaea | Eubacteria | Ribosomal function |
|---|---|---|---|
| eIF1 | + | AUG recognition; binding of 40S subunit to 5′ end of mRNA | |
| eIF1A | + | IF1 | tRNAi binding; subunit antiassociation |
| eIF2α | + | Affects eIF2B binding by phosphorylation | |
| eIF2β | + | Binds GTP, helps recognize P-eIF2B, eIF5 | |
| eIF2γ | + | IF2 (α and β), Sel B | GTP-dependent tRNAi binding to small subunit |
| eIF2A | ? | IF2β? | SeCys-tRNA binding, GTPase |
| eIF2Bα, eIF2Bβ, eIF2Bδ | I, II, III | Helps recognize P-eIF2 | |
| eIF2Bγ, eIF2Bβɛ | Guanine nucleotide exchange (GEF) for eIF2 activity | ||
| eIF3 | − | IF3 (α and β) | Subunit antiassociation; proofreading for tRNAi |
| eIF3A | + | Binds to eIF2B, eIF5; subunit antiassociation; binds to 60S subunit | |
| eIF4A | + | IF4A (W2) | ATP-dependent helicase; unwinding of mRNA required for inititation |
| eIF4B | − | Binds RNA; stimulates processive helicase activity of eIF4A | |
| eIF4E | − | Recognizes mG cap of 7mRNA | |
| eIF4G | − | Chaperone for eIF4A, eIF4B, eIF4E, and eIF3 | |
| eIF4H | − | Binds RNA; stimulates helicase activity of eIF4A | |
| eIF5 | − | “Rescue”? | Stimulates eIF2 GTPase on 40S subunit; subunit joining |
| eIF5A | + | IF5A (EF-P) | Stimulates synthesis of first peptide bond |
| eIF5B | + | IF2 (α and β) | Mediates GTP hydrolysis with eIF2 on 40S, subunit joining |
| eIF6 | ? | Mediates subunit antiassociation, binds to 60S subunit |
Function of the RNA Helicases in Initiation
eIF4A is the only protein in the eIF4F complex that possesses an intrinsic ATP-dependent RNA helicase activity. The eIF4A protein is the prototype of the DEA(D/H) family of helicases that exhibit either RNA-RNA or RNA-DNA specificity (84, 126). This helicase activity is, however, nonprocessive and bidirectional and acts to unwind only 3 to 5 bases from the mRNA termini (84, 126). The eIF4A homodimer has a weak intrinsic helicase activity, which is enhanced by association with other proteins of the eIF4F complex. Of these proteins, eIF4B can stimulate the processive activity of eIF4A when in solution or when bound to eIF4F (126). eIF4A can, in turn, enhance the activity of eIF4F (84). A new protein, eIF4H, appears to act in the same fashion as eIF4B (84).
Kinetic studies of the eIF4A protein indicate that the affinity of the enzyme is modulated by ATP · Mg2+ and ADP · Mg2+ such that the complex of the enzyme with the nucleoside triphosphate is much higher than that for the nucleoside diphosphate (120). The enzyme appears to bind RNA in a random fashion, and the hydrolysis of ATP is not required for eIF4A to bind to single-stranded RNA. The presence or absence of phosphate and the bound nucleoside has been postulated to act as a switch that modulates the structure of the enzyme and its ability to interact with single-stranded RNA. The binding and hydrolysis produce cyclical conformational changes in the enzyme that alter its affinity for the single-stranded substrate so that the energy of ATP hydrolysis can be converted into work (120).
The eIF4A protein dissociates faster from the single-stranded RNA substrate than it can hydrolyze ATP. Thus, the protein probably does not act alone as a processive helicase. Clearly, the helicase activity may depend on the binding of the eIF4B protein (84, 126). The structure of the complex between eIF4A and eIF4B has not yet been determined. Nevertheless, it is likely that the eIF4B or perhaps the newly described eIF4H, which acts in the same fashion as eIF4B, could cause a conformational change that forms a tighter structure around the RNA-binding site. The higher affinity of the eIF4B for the RNA may cause the molecule to transiently dissociate from the complex, enabling the movement of the RNA substrate. This movement is likely to be coupled to the hydrolysis of ATP through the cyclical conformational changes described above.
In the case of eukaryotic initiation, it appears necessary that the ribosomes scan from the 5′ terminus to the position of the start codon of most mRNAs (111). It has been postulated that the intrinsic helicase activity of eIF4A could act as a “clamp” on the 5′-terminal region of the mRNA. ATP, which is necessary for scanning, could be hydrolyzed and result in the opening and closing of the active site dependent on the cycles of ATP hydrolysis (111). The merit of this idea depends, in part, in proving definitely whether scanning occurs by the systematic and unidirectional movement of the ribosome relative to the mRNA 5′ terminus (111). The discovery that eukaryotic ribosomes can initiate at internal sites suggests that scanning is not restricted to starting at the 5′ terminus of the mRNA (94).
A provocative idea has been put forth to explain the results of introducing a start codon 3′ or 5′ to the termination codon of the coat protein of MS2 or fr bacteriophages (1). The lysis protein, which occurs within the coat protein-coding region, is not expressed unless the upstream coat gene is translated. The lysis start site was positioned by mutation 3′ or 5′ of the termination codon of the coat protein. It was observed that if the coat protein termination codon occurred 3′ of the two competent lysis start sites, the 3′ start site was selected, whereas if the termination occurred 5′ of both of the inserted starts, the 5′ start site was utilized (1). These results suggest that ribosomes scan the mRNA in both directions after they terminate translation until they encounter a functional start site (1). Other early experiments by Sarabhai and Brenner (172) had proposed that ribosomes drift from the termination site to the restart site of the rIIB gene of bacteriophage T4.
Thus, a common mechanism may underlie the reinitiation of synthesis in eubacteria and the internal entry site of eukaryotic ribosomes. It also appears that scanning of the eukaryotic mRNAs need not be restricted to recognition of the 5′ start site of the transcript.
Structure of eIF4A
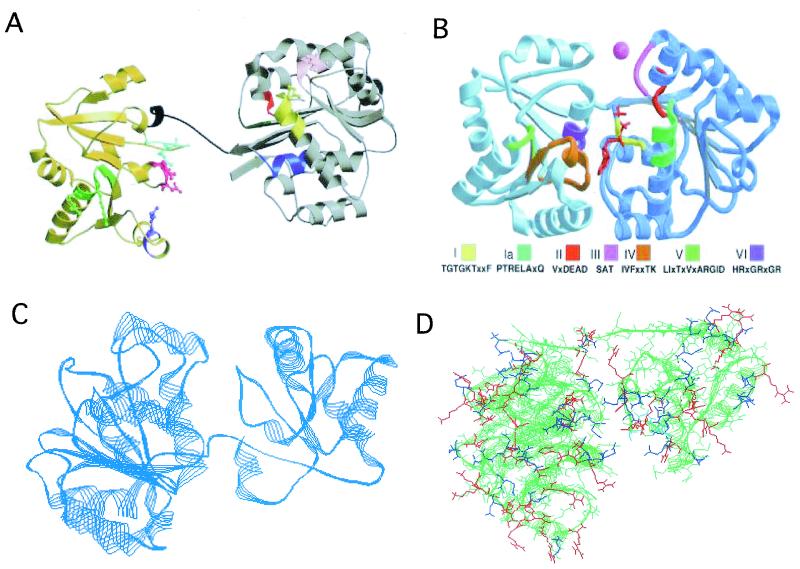
The crystal structure of the full-length eIF4A at 2.8 Å resolution has revealed some features that bear on the mechanism of this helicase (9, 25). The model of the molecule derived from these studies is shown in Fig. 1A. The molecule has a dumbbell structure which is composed of two domains joined by an extended 11-residue linker. The molecule is about 80 Å long, and the linker is about 18 Å long. The amino-terminal domain (residues 1 to 223) of eIF4A exhibits an α-β domain which appears to have an identical folding topology and tertiary structure as that found in other helicases and in the RecA protein (25).
FIG. 1.
(A) Ribbon representation of the structure of the full-length eIF4A. The amino-terminal end is shown in brown, and the carboxyl-terminal end is shown in gold. The connecting flexible II residue linker is shown in black. The following domains are colored as indicated: the N-terminal domain, which harbors motif I, which has the Walker A motif, ASQSGTGKT (residues 65 to 72), blue; the Ia motif, PTRELA (residues 97 to 102), yellow; the GG motif (residues 125 to 126), orange; the TPGR (residues 145 to 148), pink; motif II, which harbors the Walker B DEAD motif (residues 169 to 172), red; and motif III, which harbors SAT (residues 200 to 202), green; the C-terminal motif IV, VIFCNTRR (residues 263 to 270), green; the conserved R motif, arginine-298, purple; the RGID motif in motif V (residues 321 to 324), magenta; and the HRIGRGGR (residues 345 to 352) of motif VI, cyan. (Reprinted from reference 25 with permission of the publisher.) (B) Ribbon representation of the structure of the full-length aIF4A from the archaebacterial M. jannaschii derived from the X-ray diffraction data at 1.8-Å resolution. The C-terminal end of the molecule is shown to the left of the figure. The color designation of domains I to IV is given below the figure. (Reprinted from reference 186 with permission of the publisher.) (C) Ribbon representation of the computer-derived structure of the eubacterial IF4A (W2) protein. Approximately 10% of the amino acid sequence of the N terminus of the protein is missing. The E. coli K-12 protein was patterned after the archaebacterial structure in panel B. The C-terminal end of the molecule is shown to the left of the figure. (D) Surface representation of aIF4A of M. jannaschii with the relative distribution of charged amino acid residues. Positively charged residues are shown in blue, and negatively charged residues are shown in red. The molecule is shown with the N-terminal domain to the left and the C-terminal domain to the right. The figure shows that the C-terminal end of the molecule and the bottom of the N-terminal end and the linker that joins the two domains are basic. This figure was generated by the Swiss Plot program.
The C-terminal end of eIF4A is disordered (Fig. 1A). This may permit the linker domain to be relatively flexible so that the eIF4A might be a distended molecule. The size of the single-stranded RNA-binding site is about 15 nucleotides per eIF4A monomer, compatible with a predicted extended structure (25). Mutations at the C-terminal end are known which reduce the ATP hydrolysis rate (16). Since ATP is bound by the N terminus, these observations are consistent with an extended structure. It has been suggested that ATP binding by eIF4A alters the conformation of the protein that results in a compact structure in which the two domains interact directly with each other (25).
The two domains that comprise the “dumbbell” structure of eIF4A occur in other helicases, as do the conserved helicase motifs. This is not surprising, since all these enzymes interact with ATP and with polynucleotides. The compact structure suggested by the work of Caruthers et al. (25) also occurs in other helicases whose structures have been determined. It has been noted, however, that the eukaryotic enzyme has a weak helicase activity which is strongly potentiated by other proteins of the eIF4F complex. These proteins may, in fact, stabilize a more compact structure of the eIF4A helicase.
The structures of several helicases, PcrA DNA helicase, hepatitis C virus RNA helicase, and Uvr DNA helicase, have been compared (25). These models have common features that may be relevant to the mechanism of eIF4A action. These include an extended single-stranded oligonucleotide binding region within domains 1 and 2. Motifs I and II of domain 1 of the molecule are juxtaposed with motifs V and VI of domain 2, as shown in Fig. 1A for the eIF4A structure. The orientation of the domains may differ somewhat in the different helicases.
The features of the eIF4A structure have been interpreted to suggest the following. (i) Several groups in the amino and carboxyl termini of the molecule which occur along the interface harbor the bound single-stranded oligonucleotide. Motif IV, which harbors the oligonucleotide-binding motif, the “QXXR” sequence, is specific to the DEA(D/H) box helicases and may directly interact with oligonucleotides. (ii) Several amino acid side chains in motifs V and VI contact the ATP-binding site and the DEAD motif of the amino-terminal domain. This arrangement suggests a role in coupling interactions with ATP or ADP to conformational changes in the protein (25). Mutations in the three arginines in motif VI of the mouse eIF4A reduce the ATPase activity of the protein, supporting this suggestion. (iii) The His-345 of eIF4A is conserved in this position in all helicases and forms a salt bridge with the last aspartic acid residue of the DEAD motif. The DNA helicases studied have altered residues at this (DEXX) site. It is clear that all helicases have structurally conserved motifs and regions that vary depending broadly on their substrate specificity.
The structure of aIF4A from Methanococcus jannaschii has been determined by X-ray diffraction at the 1.8-Å resolution (186). The structure harbors two domains and bears a striking resemblance to the “dumbbell” structure of the eukaryotic protein. The structure also has a relatively long linker joining the two domains (Fig. 1B).
The computer-derived structure of 90% of the eubacterial IF4A (W2) sequence (Fig. 1C) reveals that the structure indeed resembles the “dumbbell” feature of the eIF4A and aIF4A proteins. The C-terminal end of the eubacterial protein appears larger than the corresponding domain of the eIF4A. The linker between the two domains of the molecule and the relatively disordered carboxyl-terminal end suggests that this molecule might also be flexible as is the eIF4A structure.
Thus far, it is unclear whether helicases catalyze the unwinding of RNA and DNA structures through a “rolling” model which requires that the subunits of the proteins bind alternatively to single- and double-stranded RNA or DNA or by an “inchworm” model which requires binding of the helicase to single-stranded regions of RNA or DNA. The “rolling” model is best visualized if the helicases are dimers when complexed to their substrates. However, this is not always the case. Crystal structures of the RNA or DNA helicase complex may resolve this important mechanistic issue as well as the problem of where the RNA or DNA binds to the proteins. As a first approximation, the charge distribution of the archaebacterial aIF4A (W2) is such that relatively more basic groups occur on the surface of the C-terminal end. Some basic residues also occur on the interface on the N-terminal domain of the molecule (Fig. 1D). Thus, it is possible that the RNA substrates bind to the C-terminal surfaces and to the N-terminal junction. The RNA could unwind by the relative movement of the domains as proposed for the eukaryotic eIF4A.
Eubacterial IF4A (W2) Protein
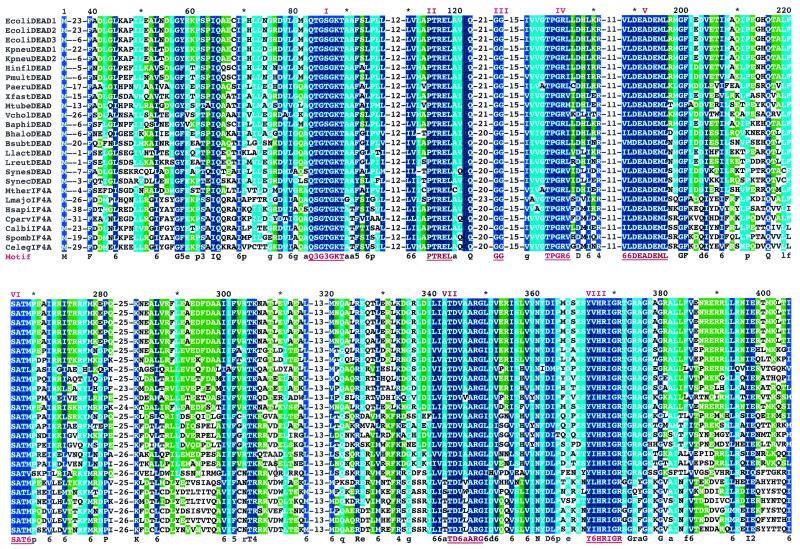
Recently a gene, deaD, has been shown to be equivalent to the gene encoding eIF4A of eukaryotic cells (122, 192). The E. coli protein harbors 87% amino acid sequence similarity to the eukaryotic eIF4A. Highly conserved motifs of representative sequences of eubacterial, archaeal, and eukaryotic proteins are given in Fig. 2. High-identity regions follow residue 39 of these proteins and occur throughout the molecules. The C-terminal ends of the proteins vary considerably in length. Clearly, the sequences of the IF4A (W2) protein are highly conserved. The sequences of the eukaryotic proteins are also conserved but appear to have diverged from those of the eubacterial and archaebacterial proteins. Three genes occur in E. coli, which differ principally in the length of the N-terminal sequences of the proteins. (Different forms of eIF4A have also been reported [84].) However, the C-terminal ends of the E. coli proteins are longer than those of the representative species of Caenorhabditis elegans, Candida albicans, and Methanococcus thermoautotrophicum. A marked region of identity, LDEADMLXXGF, underlies all of the sequences examined.
FIG. 2.
Alignment of amino acid sequence of the conserved domains of eIF4A and IF4A (W2) proteins from representative species of eubacteria and archaebacteria. Identical residues within the conserved domains are shown in blue, and highly conserved residues are shown in light blue and green. The sequences were aligned using the Swiss Plot program.
The eubacterial gene encoding the IF4A (W2) was originally isolated as a multicopy suppressor of a temperature-sensitive mutation in the ribosomal protein S2 (192). The S2 protein, in turn, is required for the assembly of several other proteins, particularly S1, which has a well-known role in the initiation reaction and which is also an RNA helix-destabilizing protein (187, 188).
Study of the helicase activity of the eubacterial protein indicates that it acts as a helix-destabilizing protein (122). Unlike the eukaryotic protein, the IF4 (W2) protein unwinds double-stranded RNA past the 3 to 5 residues that are acted upon by the eukaryotic protein (100, 122) as a result of interactions with eIF4B. Thus, an eIF4B-like protein may not be required for the processive action of the eubacterial enzyme.
The sequence of the IF4A (W2) protein has a motif (HRIGRXXR) that is involved in binding RNA and in hydrolysis of ATP (41). Unlike the eukaryotic protein, however, the eubacterial protein does not hydrolyze ATP in the presence of several polynucleotides (122). Since the protein can stimulate the melting of an RNA duplex in the absence of added ATP, the IF4A (W2) protein from eubacteria may be a helix-destabilizing protein rather than an ATP-dependent RNA helicase as is the case for the eukaryotic eIF4A (100, 122).
Many different functions have been attributed to the IF4A (W2) protein. An elegant genetic study in which the host RNA polymerase was replaced by more efficient T7 polymerase indicated that the β-galactosidase yield was markedly decreased but that the overexpression of the DEAD box, IF4A (W2), stabilized the yield of the mRNA and increased the amount of the encoded proteins. Several mRNA transcripts behaved in similar fashion, and it was suggested that the protein stabilizes mRNA structures (92). Binding of IF4A (W2) to ribosomes promotes initiation, which, in turn, could stabilize the mRNAs in question (122).
The eubacterial IF4A (W2) may exert certain interesting modes of regulating synthesis. The eubacterial protein is strongly induced under cold stress (100). A 15-fold increase in the activity is found upon a shift in temperature from 0 to 15°C (100). The protein induced by cold stress occurs bound to fully assembled 70S ribosomes.
The W2 protein stimulates synthesis programmed by templates that harbor secondary structures but is not required for the synthesis of polypeptides programmed by mRNAs devoid of secondary structures, e.g., poly(rU) (55). The N-terminal sequence of the W2 protein was found to be identical to that of the E. coli deaD gene product. The deaD gene product, in turn, has 54% identity and 87% sequence similarity to a eukaryotic factor, eIF4A, known to be involved in unwinding the secondary structure of mRNAs that impede the formation of initiation complexes. Polyvalent anti-eIF4A antibody cross-reacts with W2 (122).
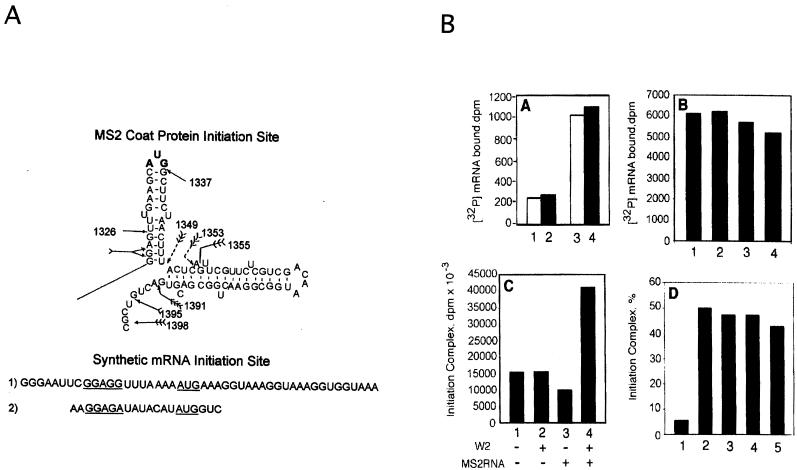
To test if W2, in its putative role as an RNA helix-destabilizing factor, affects initiation per se, initiation was studied in a defined in vitro system. Two types of mRNA templates (each harboring the known determinants of initiation) were studied. On the one hand, synthetic mRNAs that bore little secondary structure and, on the other, the highly structured native mRNA were examined to assess what effect, if any, W2 might have on initiation. Initiation complexes using unstructured mRNAs are readily formed, and W2 does not appear to enhance their formation. Initiation complex formation using the structured MS2 RNA template, on the other hand, was markedly stimulated by the addition of W2 (Fig. 3). Since the unstructured mRNA harbored the SD region and a suitably spaced AUG start codon, it is unlikely that the W2 protein is involved in the recognition of the SD sequence. The protein could conceivably be involved in unwinding a secondary structure that masks the SD sequence, but this is unlikely because ribosomes can unmask stronger secondary structures which sequester SD sequences (162).
FIG. 3.
(A) Secondary structure of the initiation site of the MS2 RNA coat protein cistron, sequence of the start site of the 200-bp unstructured mRNAs, and sequence of the start site of the unstructured 20-bp mRNA. (B) Ribosome-mRNA complexes with unstructured mRNAs or with the structured MS2 RNA. (A) W2 binds unstructured mRNAs (lanes 1 and 2, 20 and 200 bp of [32P]mRNA; lanes 4 and 5, 20 and 200 bp of [32P]mRNA plus 0.2 μg of W2). (B) Binding of 200 bp of unstructured [32P]mRNA to ribosomes with 0, 0.1, 0.2, and 0.4 μg of W2 (lanes 1 to 4). (C) Ternary initiation complexes, f[35S]Met-tRNA-ribosome-MS2 RNA, require W2. (D) Ternary initiation complexes, fMet[35S]Met-tRNA-ribosome-unstructured mRNA, do not require W2 (lane 1, no mRNA; lane 2, 200 bp of mRNA; lane 3, 200 bp of mRNA plus 0.1 μg of W2; lane 4, 20 bp of mRNA; lane 5, 20 bp of mRNA plus 0.1 μg of W2). See reference 122 for experimental details.
An important result is that the formation of the ternary initiation complex, fMet-tRNA-30S-mRNA, is markedly dependent on the IF4A (W2) helicase, indicating that the protein acts as an essential intermediate in the initiation reaction (122) (Fig. 3). It is possible that the IF4A (W2) protein unwinds structures that occur in the start or in the coding region so as to effect proper accommodation of the initiation complex. Thus, the IF4A (W2) protein might be involved in scanning for formation of the proper ternary complex. Although more evidence is needed to ascertain this, it is clear that the IF4A (W2) protein is an important factor for initiation on structured mRNA sites.
PEPTIDE BOND SYNTHESIS
Peptidyl Transferase
It is well established that the large subunit of the ribosome harbors the catalytic components responsible for peptide bond synthesis. The peptidyl transferase of the 50S subunit catalyzes a number of displacement reactions of the general type RCO- X + B → RCO-B + X, where X is OR′ and B is either water or a nucleophilic reagent of the type R′′-OH, R′′-NH2, or R′′-SH. Further, peptidyl transferase promotes the hydrolysis of esters in the presence of the release proteins, RF1 and RF2, and transesterification in the presence of OH donors (140). During peptide bond synthesis at physiological pH, the charge on the amino group of the incoming aminoacyl-tRNA is positive. A partial positive charge is also present on the carboxylate group where the nascent polypeptide chain is esterified to tRNA. A histidine group in an essential 50S protein, e.g., L2, has been proposed to act as a proton sink to neutralize these charges (33).
The crystal structure of the 50S subunit at the 2.4-Å resolution has revealed that the peptidyl transferase domain is protein free. The active site of this region has been cocrystallized with analogs of the CCA amino acids which act as inhibitors of peptide bond synthesis and a truncated aminoacyl-tRNA (141). These substrates bind to the A and P sites. The nucleotides that bind these substrate analogs are highly conserved within domain V (167). The residue that may be involved in catalysis is A2451, whose N-3 is about 3 Å from the phosphoramide oxygen of the bound peptide bond synthesis inhibitor and about 4 Å from the amide nitrogen of the peptide bond being formed (141). The unusual pKa of A2451, which may be the essential catalytic base, derives in part from the fact it can form hydrogen bonds with G2447, which in turn interacts with a buried phosphate (135). Further, A2451 is essential for peptide bond synthesis (73). It has been suggested that the α-amino group of the aminoacyl-tRNA attacks the carboxyl carbon that acylates the 3′ hydroxyl group of the peptidyl-tRNA, resulting in the formation of an oxyanion intermediate at the carboxyl atom. The oxyanion dissociates to add an additional amino acid to the nascent protein chain esterified to the tRNA and bound to the A site. The deacyl-tRNA remains in the P site (141). Thus, it appears that the peptidyl transferase is a ribozyme and that the proteins L2, L3, and L4 help form the locus where A2451 can form the essential oxyanion (141). The region indeed catalyzes peptide bond synthesis as the reaction products are reported to cocrystallize at this site and result in a pre-translocation intermediate (173a). Mutants of A2451 are conditionally lethal but retain some peptidyl transferase activity in vitro. Further, mutants in G2447 are viable and have little or no effect or synthesis (154, 191a). The residual activity of the peptidyl transferase in the A2451 mutants might be compensated by perhaps another functional group in the rRNA. Alternatively, it is possible that the intermediate only enhances the rate of peptide bond synthesis. Substrate binding may be affected by the A2451 mutants, suggesting that this elegant mechanism remains to be proven.
rRNA-tRNA Interactions and the Peptidyl Transferase Active Center
Genetic evidence, experiments involving the use of antibiotics, and the results of photocross-linking studies of aminoacyl-tRNA analogues (reviewed in references 167 and 181) have implicated the highly conserved domain V of 23S rRNA and, in particular, the central loop of this domain as the essential player in peptide bond formation. Indeed, A2451, which has been implicated to be involved in catalysis, occurs in domain V (141). Mutations that confer resistance to antibiotics that impair peptide bond synthesis map within the exposed loops of domain V (167, 180, 182).
Chemical footprinting has also identified nucleotides protected from chemical attack which are bound to the P, A, or E sites of the 23S rRNA within the sites depicted in the secondary structure of domain V (170, 182). Protection of the E site-bound tRNA is restricted to the 50S particle, whereas protection of the P and A site-bound tRNAs is scattered throughout other ribosomal domains on the 50S and 30S particles (73).
Mutations in G2585 of the 23S rRNA exhibit effects far away from the site where the peptidyl-tRNA substrate binds, suggesting that conformational changes affect the site of peptide bond formation. Indeed, a large number of mutations selected randomly on residues 2493 to 2606 in the central loop of domain V affect peptide bond formation (reviewed in reference 167). Thus, domain V, which binds the tRNA substrates, is also very likely to be involved in catalysis of peptide bond synthesis. The function of this center appears to be affected by distal rRNA elements.
Limited Capacity of Peptidyl Transferase To Synthesize Certain Dipeptides
In spite of the rapid progress in understanding the mechanism of peptide bond synthesis, little has been done to understand the basis for the observation that in vitro the peptidyl transferase can efficiently synthesize peptide bonds only with certain aminoacylated substrates (reviewed in reference 189). The ribozyme-catalyzed reaction also polymerizes different aminoacyl substrates with widely different reaction rates, suggesting that this may be a property of RNA-catalyzed reactions (205). Certain antibiotics, such as chloramphenicol, inhibit peptide bond synthesis with some aminoacyl-tRNA template combinations and not others. In vitro, the peptidyl transferase activity of various aminoacylated substrates depends on the identity of their side chains (189). A number of researchers have reported, for example, that aminoacyl-tRNAs bearing large aromatic side chains are active while several other aminoacyl-tRNAs are much less reactive (189). In contrast, synthesis of proteins in vivo does not show such a preference (189).
Reconstitution studies indicate that the assembled peptidyl transferase of the 70S ribosome catalyzes peptide bond synthesis at a higher rate than does the peptidyl transferase of the 50S subunit but does not efficiently form peptide bonds with most amino acids (69). A soluble protein, EFP, stimulates peptide bond synthesis from several aminoacyl-tRNAs by the peptidyl transferase (52, 69) and restores this specificity. The general properties of the EFP protein are discussed below.
efp Gene in Eubacteria
The E. coli efp gene was cloned, sequenced, and mapped to 94.3 min on the chromosome (6). The gene has been overexpressed in E. coli, and the overexpressed protein has been crystallized (5). EFP bears no obvious amino acid sequence similarities to any other translation factor (5, 6). Interruption of the efp gene encoding EFP is lethal to the cell and results in abrupt cessation of protein synthesis, specifically in an impairment of peptide bond formation (7). Thus, the efp gene is essential for cell growth and for viability. The efp gene is highly conserved in eubacteria, in archaea, and also in eukaryotes where the protein it encodes is called aIF5A and eIF5A, respectively (113). A number of bacteria harbor genes whose sequences are nearly identical to that of the efp gene. Thus, the efp gene in the Haemophilus influenzae genome encodes a protein that has 54% identity and 87% similarity to the E. coli EFP protein. In these cases, the N-terminal end of the EFP exhibits similarity to the ribosomal proteins L18 and L27, which are among the proteins that cross-link to puromycin and stimulate peptide bond synthesis reconstituted from 23S rRNA and proteins such as L2, L3, and L4 (175). The smallest bacterial genome, Mycoplasma genitalium, maintains a copy of the efp gene. Translation factor aIF5A of the archaebacterial Methanococcus jannaschii has the same conserved motifs throughout the N- and C-terminal regions of the prokaryotic EFP. Examples of eubacterial sequences are shown in Fig. 4.
FIG. 4.
Alignment of amino acid sequences of the EFP proteins from eubacterial, archaebacterial, and eukaryotic sources. The Lys-31 motif in the eubacterial sequences bears an unusual modification. Lys-54 of the eukaryotic sequences is modified by hypusine. Identical residues in the eukaryotic sequences are shown in yellow, sequences unique to eubacterial sequences are shown in blue, and conserved residues are shown in green.
Eukaryotic eIF5A
A number of activities similar to that of EFP have been detected in yeast, archaebacteria, and eukaryotic cells and their corresponding proteins are called eIF5A (8, 178). eIF5A was named on the basis of the observations that the protein stimulates fMet-puromycin synthesis but does not promote polyphenylalanine synthesis directed by poly(rU) templates (178). In mammalian cells, eIF5A occurs in both the nucleus and the cytoplasm. The eukaryotic protein may play an additional role in regulating the export of mRNA as well as in protein synthesis (11, 12). eIF5A is a cofactor of the Rev transactivator protein of human immunodeficiency virus type 1 and of the Rex protein of human T-cell leukemia virus type 1 which mediates the translocation of viral mRNAs from the nucleus into the cytoplasm (11). Thus, it is possible that eIF5A might interact with a nuclear export system. Indeed, misssense mutants of the gene encoding eIF5A completely block Rev translocation, demonstrating the requirement for eIF5A in Rev- mediated nuclear export (11).
The altered expression of eIF5A is correlated with several disease states. Thus, the expression of the protein is elevated in human immunodeficiency virus type 1-infected patients. Hypusine [Nɛ-(4-amino-2-hydroxybutyl)-l-lysine] formation is also significantly elevated in Ras oncogene-transfected mouse cells (31). In contrast, human carcinoma cells treated with alpha-2 interferon exhibit a reduced level of hypusine synthesis as well as an increase in the production of epidermal growth factor at the tumor cell surface, which decreases the proliferation of the tumor cells (24).
A requirement for eIF5A in cell proliferation is well established. Thus, deletion of the eIF5A gene in Saccharomyces cerevisiae reduces protein synthesis (174) and depletion of eIF5A in budding yeast arrests cells at the G1 stage of growth and results in the production of nonviable spores (104).
The genes encoding these proteins have been sequenced. The eIF5A sequences have the strongest region of homology near the site of hypusine modification that is essential to the function of these proteins (151). Interestingly, eIF5A is the only protein that has been reported to be modified by hypusine. S. cerevisiae has two essential genes that encode eIF5A; they exhibit 26% identity in a 108-amino-acid sequence near Lys-54 of the eIF5A, where the hypusine modification occurs. The homologous region in the E. coli EFP near Lys-31 is modified by an unusual group of 148 ± 1 Da (molecular mass measured by mass spectrometric analysis). This group differs from hypusine but may also be essential for the activity and stability of the native protein (H. Aoki, M. Yaguchi, D. Watson, M. Pearson, and M. C. Ganoza, unpublished observations). The Lys-31 region has a special motif, VKPGK, that is conserved in many different bacteria and in the aIF5A of the archaebacterial M. jannaschii.
The EFP proteins have three highly conserved motifs. It is clear that a significant proportion of the total amino acids are similar in the eukaryotic and prokaryotic proteins (Fig. 4). Domain I, containing the longest contiguous stretch of conserved amino acids, harbors the Lys-31 modified residue in the E. coli EFP protein. Each domain appears to have highly conserved amino acid residues unique to eubacterial EFP proteins (blue in Fig. 4). The eukaryotic sequences also have unique motifs (yellow) which are interspaced between the other conserved domains of the proteins. The structure of the eubacterial Staphylococcus aureus proteins derived by X-ray diffraction at the 1.9-Å resolution (T. E. Benson, M. C. McCroskey, J. I. Cialdella, G. Choi, and J. D. Pearson, Proc. Second Symp. Struct. Aspects. Protein Synthesis, p. 15, 2000) has three main domains, whereas the archaeal and eukaryotic proteins have two such domains (108). The presence of this third domain in the eubacterial sequences creates a protein fold that suggests that EFP might function as a tRNA mimic (Benson et al., Abstract) This difference, particularly the absence of the third domain, may be important in eukaryotic mRNA transport and/or stabilization.
Several eukaryotic genes that encode eIF5A encode fewer than 190 residues (Fig. 4). This in itself explains the gaps observed at, for example, the C terminus at residues 124 to 136. The eubacterial EFP sequences have a well-conserved C-terminal domain that does not occur in archaebacteria or eukaryotes, suggesting a functional difference in these proteins. Nevertheless, it is clear that many amino acids are similar in the eukaryotic and eubacterial proteins. Amino acids within similar domains are divergent between the bacterial and eukaryotic sequences, but there seems to be a pattern of conservation of discrete, perhaps critical, amino acids in all these proteins. Study of the phylogenetic relationships between the prokaryotic and eukaryotic sequences suggests that these proteins coevolved with the organism in question, emphasizing their essential character. Thus, the genes encoding the eIF5A and EFP proteins from different species are highly conserved, but more evidence is needed to establish whether they represent orthologous functions.
Structure of aIF5A
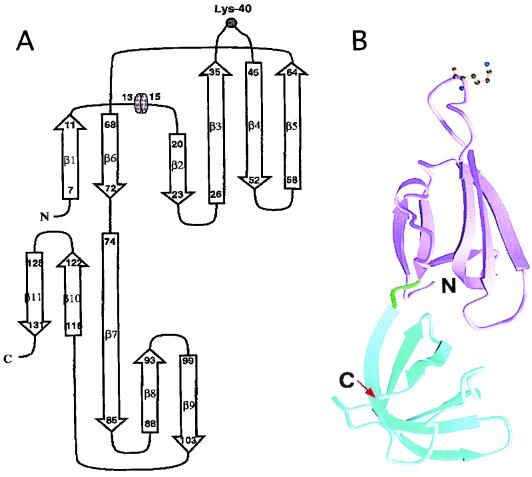
EFP has 84% sequence similarity to the M. jannaschii aIF5A protein, which has been crystallized and whose structure has been solved at the 1.8-Å resolution (108). The deduced structure has two different domains. The highly conserved residues that harbor hypusine in aIF5A, near the middle of the molecule, form a loop at the end of the long molecule. The crystal structure of the M. jannaschii aIF5A shows that the protein is made of two β-sheet domains arranged in an elongated structure that is about 63 Å long and 26 Å wide (108). Figure 5 shows the arrangement of domain I, which has the β1 to β6 strands and the helix. Domain II (residues 74 to 132) harbors strands β7 to β11. The hypusine site of modification occurs in a long loop between strands β3 and β4. Domain II resembles the oligonucleotide-binding fold (OB fold [134]) found in the E. coli cold shock proteins, the Staphylococcus nuclease, the N-terminal end of the yeast Asp-tRNA synthetase, and the eubacterial initiation factor IF1. The structure of aIF5A suggests that the protein may bind to nucleic acids. The OB fold that occurs in aIF5A and in initiation factor 1 is involved in binding the former protein to the ribosomal A site (129).
FIG. 5.
(A) Ribbon diagram and representative sketch of the structure of aIF5A from M. jannaschii. The ribbon structure was derived from X-ray diffraction patterns at 1.8-Å resolution. The sketch (A) shows the eleven β sheets in two domains of the molecule joined by flexible links that contain the Lys-54 site of hypusine modification. (Reprinted from reference 108 with permission of the publisher.) (B) Ribbon structure of the eIF5A, showing the two domains of the molecule linked by the flexible linker that harbors the hypusine residue. The molecule bears opposite charges in the C- and N-terminal ends.
The N-terminal domain (I) of aIF5A is acidic, whereas the C-terminal domain (II) is basic. Thus, the structure of the protein is highly polar. These features and the flexible hinge between the domains of the molecule are likely to play important functions in its mode of action. The X-ray diffraction-derived structure of aIF5A from the archaebacterial Pyrobaculum aerophilum at 1.75-Å resolution shows that these features are conserved (152).
Possible Role of EFP in the Peptidyl Transferase Reaction
An E. coli cell has 800 to 900 copies of EFP or about 0.1 to 0.2 copy per ribosome, suggesting that this protein may function catalytically (4). Biochemical studies have not revealed any effect of EFP on the initiation reaction. Further, antibiotics that inhibit initiation or translocation had no effect on the EFP-stimulated synthesis of peptide bonds. In contrast, EFP enhanced the inhibition of peptide bond formation imparted by chloramphenicol and lincomycin, suggesting the EFP somehow facilitated or stabilized the interaction of these agents with the peptidyl transferase active center (5).
Ribosome reconstitution experiments showed that L16, or its first 47-amino-acid N-terminal fragment, was required for the EFP-mediated peptide bond synthesis whereas L11, L15, and L7/L12 were not, suggesting that EFP operates at a different ribosomal site than used by most other translation factors (64) (Fig. 6).
FIG. 6.
(A) Requirement for EFP and L16 in peptide bond synthesis by reconstituted 70S ribosomes (○, no EFP; ▪, with 2 μg of EFP); the cores were reconstituted with L16 (▴), and then EFP was added (•). (B) Synthesis of dipeptides from fMet-tRNA and 5′ CCA amino acids; association constants in the presence or absence of EFP as a function of the length of the amino acid side chain. Data are from references 52 and 64. The sizes of the amino acid side chains are derived from X-ray diffraction data.
The interaction between the EFP protein and the native ribosome and ribosomal subunits is of particular interest in elucidating the exact role and function of EFP. The results of primer extension experiments provide clues to the nature of the in vitro associations between the rRNA molecule and the EFP protein. A selected number of bases on 16S and 23S rRNA are protected from the chemical probes by the assembly of the ribosome and the EFP complex. In several regions of the rRNA molecules, assembly causes enhanced reactivity of specific bases, which are interpreted to be the result of protein-dependent conformational changes in the rRNA (130).
Previous studies have demonstrated that streptomycin markedly inhibits the EFP-mediated synthesis of peptide bonds (5). The footprints observed on contact of EFP with the 30S subunit are near one of the regions where streptomycin binds to the 16S rRNA of the particle. This is within the decoding center of the ribosome, which is engaged in the mRNA-tRNA, codon-anticodon interactions. The bases that are enhanced by interactions of EFP with the 30S subunit are in the vicinity of the S1 and S5 proteins, which also bind within the decoding center of the ribosome.
EFP also interacts with domain II of the 50S subunit (H. Aoki, J. Lang, A. Lang, and M. C. Ganoza, submitted for publication). Domain II also has binding sites for both EFTu and EFG. The stimulation of peptide bond synthesis by EFP does not require EFG. However, EFP may influence the activity of EFTu. As the incoming aminoacyl-tRNA-GTP-EFTu complex first approaches the A site, it is too far away from the aminoacyl-tRNA in the P site to form a peptide bond (27, 128). Consequently, EFTu accommodates the incoming aminoacyl-tRNA to a position where a peptide bond can be formed. The hydrolysis of GTP that results on binding of EFTu-GTP-aminoacyl-tRNA to the ribosome is essential for binding the aminoacyl-tRNA substrates to the peptidyl transferase active center. EFP could accelerate the ribosomal GTPase activity of EFTu so as to promote the approximation of the aminoacyl-tRNAs into the peptidyl transferase active site.
Interactions between EFP and the 23S rRNA on domain V are of especial interest because EFP has been predicted to enhance elongation by interacting with the peptidyl transferase center. Indeed, the protection analysis demonstrates that EFP is in contact with the A site of domain V of the 23S rRNA. In binding directly to the A site, EFP could directly increase the affinity of the incoming aminoacyl-tRNA (22, 181). The binding of EFP to the A site also enhances the reactivity of bases in the 50S E site (Aoki et al., submitted). These interactions would increase the rate of efficiency of translation, as EFP is known to do (52).
Since the EFP-binding domain is very near the site of EFTu and EFG binding on both the 30S and 50S subunits (Aoki et al., submitted), it is likely that the binding site may span both subunits. The oligonucleotide-binding domain of the EFP protein could bind the aminoacyl-tRNA while the hydrophobic residues near the central loop of the molecule might interact with the peptidyl transferase center.
The contacts observed on the 30S at G577 and A579 are near the 530 loop that forms part of the binding site for EFTu (156) as well as one of the streptomycin-binding sites (180). The proposed elongated structure of the protein could facilitate interactions with both subunits that may enable the proper approximation of the tRNA substrates required for peptide bond synthesis. We suggest that EFP functions by interacting with the A site of the 30S subunit as well as with the A site of the peptide bond-forming center of the large ribosomal subunit.
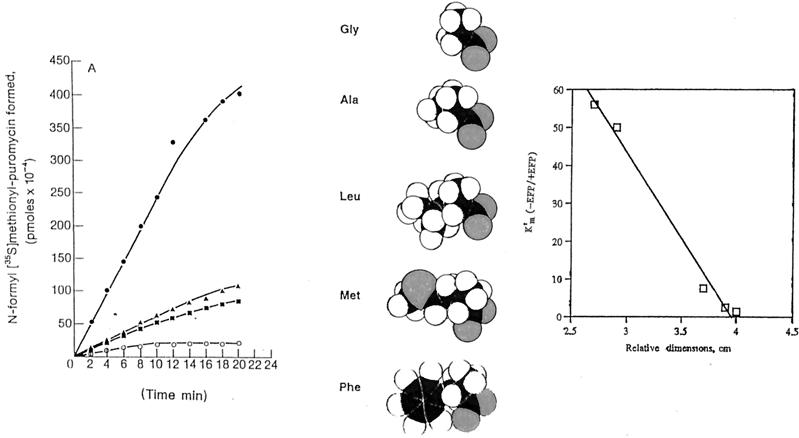
EFP stimulates the initial rate of translation programmed by certain synthetic templates when synthesis is initiated by N-blocked aminoacyl-tRNAs (52). EFP could function in the formation of the first peptide bond and, more probably, could promote the synthesis of certain dipeptides during subsequent elongation. The later idea is consistent with the fact that EFP occurs bound to 70S ribosomes as well as to polyribosomes. EFP was first isolated as a factor that stimulated the translation of a natural mRNA [in contrast to that of simple, synthetic messages such as poly(rU)]. Subsequently, purified EFP was shown to stimulate peptide bond formation by the ribosome in the puromycin reaction model when the concentration of puromycin was decreased (68). Several aminoacyl 5′ CCA fragments were synthesized and tested as acceptors in peptide bond formation, using fMet-tRNA, bound to 70S ribosomes, as donor. These studies showed that the intrinsic ability of the ribosome to catalyze the synthesis of certain dipeptides was extremely poor while other dipeptides were efficiently synthesized. Notably, EFP stimulated dipeptide synthesis in the cases of those aminoacyl acceptors that are poor substrates for the intrinsic peptidyl transferase. For example, fMet-Gly synthesis on ribosomes was stimulated 50- to 60-fold by the addition of EFP whereas fMet-Phe synthesis was barely affected. Significant stimulation of the synthesis of fMet-Ala, fMet-Leu, fMet-Val, fMet-Lys, and fMet-Met also occurred on addition of the EFP protein (52, 69) (Fig. 6). These results suggest that the side chain of the aminoacyl acceptors is essential for the stimulation of synthesis and the amino acid charge is not necessarily the feature that is recognized by the EFP protein. As a first approximation, the ability of EFP to stimulate peptide bond synthesis correlates inversely with the relative size of the amino acid side chain of the aminoacyl acceptor (52).
Molecular modeling of the peptide bond synthesis reaction between puromycin and an analogue of the peptidyl-tRNA indicates that the bulky dimethoxy benzene ring in the puromycin structure allows the molecule to bend in a U shape (158). In such a conformation, puromycin makes a perfect steric fit with the NH2 group of the peptidyl-tRNA, which facilitates the formation of a peptide bond. Alterations of the puromycin structure that distort the U shape of the molecule, such as the substitution of a Gly residue at this position, drastically reduce the reaction rate (reviewed in reference 189). This suggests that the side chains of the amino acids are important to the reaction rate and prompts the suggestion that the EFP protein may directly or indirectly promote the approximation of aminoacyl-tRNAs bearing small amino acid side chains into the active center of the peptidyl transferase. The fact that little, if any, peptide bond synthesis occurs with acceptors that have small side chains in the absence of EFP could explain the requirement for this protein in vivo, where initiation of many protein-encoding reading frames commonly begins with such amino acids.
The observation that EFP, a nonribosomal protein, can restore the ability of the peptidyl transferase to polymerize amino acids that are otherwise poor substrates points to the structural and/or dynamic complexity of this reaction. IF2 and EFTu are known to bind to the 3′ terminus of the initiator or aminoacyl-tRNA, respectively, making it unlikely that EFP also interacts directly with the tRNA 3′ terminus. On the other hand, elements regulating the active center of the peptidyl transferase may span a larger portion of the ribosome, as suggested by mutagenesis studies (73, 167). Reconstitution studies show that several 50S subunit proteins (apart from L2, L3, and L4) significantly stimulate peptide bond synthesis (45, 175). Experiments involving antibiotics that interact with peptidyl transferase indicate that the active center can exist in a number of functional conformers (167). Given such plasticity, we suggest that EFP may act to restructure the active site in a way that promotes interaction between the peptidyl transferase and its less favorable aminoacyl-tRNA substrates. Thus, EFP may be viewed as a critical regulatory molecule for peptide bond formation during chain elongation.
PROTEIN CHAIN ELONGATION
Protein chain elongation entails a cycle consisting of alignment of aminoacyl-tRNAs by their specific codons in mRNA (decoding), peptide bond synthesis, and movement of mRNA relative to the ribosome (translocation). The eubacterial proteins, EFTu, EFTs, and EFG (eEF1α, eEF1βγ, and eEF2 of eukaryotes), facilitate these processes on ribosomes. Reviews are available (22, 36, 116, 144, 146, 202).
Decoding
The first elongator tRNA is attached to the ribosomal complex as a GTP-EFTu-aminoacyl-tRNA intermediate, which accelerates the rate of binding of the aminoacyl-tRNA moiety to the mRNA-programmed ribosome. Hydrolysis of GTP, promoted by L7/L12 of the 50S subunit, is required before a peptide bond can be made. The energy of GTP cleavage may be expended to proofread near-cognate aminoacyl-tRNAs and/or to allow for the alignment of the correct codon-anticodon interaction. After GTP hydrolysis, EFTu-GDP leaves the ribosome and a peptide bond can be formed. EFTs catalyzes the exchange of GDP in EFTu-GDP with free GTP. Fungi have another elongation factor, eEF3, which is a ribosome-dependent ATPase (29). This reaction is analogous to that first discovered in E. coli and called W, which ejects tRNAs from 70S ribosomes (57, 58, 74). (The name of the W protein [a truncated version of RbbA] was changed to reflect the fact that the protein is a ribosome-bound ATPase.) Indeed, it has been shown subsequently (using heteropolymeric mRNA analogues) that eEF3 also promotes the ejection of tRNAs from ribosomes (194). The genes encoding the elongation factors have been cloned and sequenced (15, 42). The structures of EFTu-GDP, EFTu-GTP-aminoacyl-tRNA, and EFG-GDP have been determined (2, 10, 37, 142).
Translocation
Following peptide bond formation with the peptidyl-tRNA, the tRNA must be moved or translocated from the ribosomal A site to the P site. Peptide bond synthesis by the peptidyl transferase of the 50S particle also leaves the deacyl-tRNA in the P site of the ribosome (173). Thus, after or during translocation, the deacyl-tRNA must also be translocated from the P site to the E site (73, 163).
The movement (translocation) of the nascent peptidyl-tRNA from the A site to the P site is inherent to the ribosome (183) but is catalyzed by elongation factor EFG and GTP (183). This movement is substantial, involving changes in the order of 50 Å at the elbow of the tRNA during each step of the elongation cycle. The tRNA substrates must remain bound to the ribosome during this process in order to maintain the reading frame. The energy to drive this mechanism comes largely from hydrolysis of GTP that results from contact of the EFG-GTP complex with the ribosome (73, 165, 202). Presumably, GTP hydrolysis allows the release of EFG from ribosomes (73, 165, 202). The results of RNase protection assays and site-directed mRNA cross-linking data, before and after addition of EFG, indicate that translocation moves the mRNA by 3 nucleotides (13, 73, 128, 165, 202).
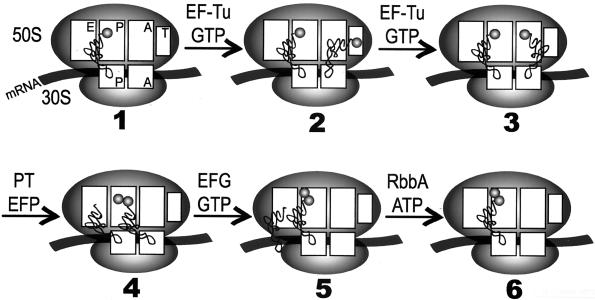
Chemical footprinting revealed that tRNAs bound in various partial elongation reactions protect specific bases on rRNA from chemical probes (13, 73, 165, 202). The principal observations are that the 3′ and 5′ ends of the tRNAs are in different states in the two ribosomal subunits, such that a single tRNA can be found on the 30S A site and on the 50S P site. These positions are termed the “hybrid” sites of the particle (128). These footprinting, chemical protection experiments of the translation reaction programmed by poly(rU) revealed that the tRNA, particularly the 3′ terminus of the molecule, undergoes considerable movement during each step of the elongation cycle (128). The EFTu-GTP-aminoacyl-tRNA complex does not bind to the ribosomal A site directly (128, 164). The tRNA footprints suggest that this initial contact occurs with a hybrid A/T site in which the anticodon of the tRNA and the mRNA on the 30S subunit remain fixed and that the 3′ terminus of the tRNA does not bind directly to the A site but to the A/T site of the 50S particle. Following GTP hydrolysis, the aminoacyl-tRNA is moved from the A/T site into the ribosomal A site (A/A hybrid site equivalent). Peptide bond synthesis follows after ejection of the EFTu-GDP complex from the ribosome, leaving deacyl-tRNA in the P site and the peptidyl-tRNA in the A site. The EFG-GTP complex binds at this stage of the process. The hybrid site model and the site of EFTu and EFG action are shown in Fig. 7. We also posit a possible site of action for EFP and RbbA in this scheme. The action of EFG may be preceded by movement of the deacyl-tRNA from the P site to the E site (163). This reaction is stimulated strongly by the RbbA protein, but it is not certain whether the RbbA protein ejects tRNAs only from the E site after peptide bond synthesis or from the P site also (57). In the former case, EFG may mediate translocation both of the deacyl-tRNA from the P site to the E site and of the peptidyl-tRNA from the A site to the P site.
FIG. 7.
Requirement for RbbA and EFP in MS2 programmed synthesis reconstituted from homogeneous proteins. Data are from reference 52.
Several experiments agree with the hybrid model. Using fluorescence energy transfer, Hardesty and colleagues (148) reported that the photolabeled tRNA moves more than 20 Å after peptide bond synthesis, a result in full accord with the prediction of the hybrid model. However, a powerful new labeling method which is able to score for tRNA interactions in the pre- and posttranslocational states did not detect all of the changes predicted by the hybrid model (38). Spahn and Nierhaus have proposed an alternate model for the reaction which assumes that the ribosomes have a movable site that carries the substrates during the pre- to posttranslocation process (181). Recent cryo-EM analysis of the translocating ribosome gave results consistent with features of both models (46).
There is evidence that EFTu enters the ribosome at a site different from the A site that binds the aminoacyl-tRNA. EFTu binds to ribosomes that already carry an aminoacyl-tRNA (114, 164). There is also EM evidence that the binding site for EFTu differs from that of other ribosomal sites (114). The crystal structure of the 70S ribosome has revealed the position of the three tRNA-binding sites and the contiguous site for binding of elongation factors (27). We refer to this site as the T site. EFP acts in concert with the peptidyl transferase to bring about peptide bond synthesis with a variety of aminoacyl-tRNA acceptors. Peptide bond formation results in a large rearrangement of the ribosome, as predicted by the hybrid model and by the data obtained with the fluorescent energy probes. We propose an active mechanism, in which the W (RbbA) protein is required for the ejection of the tRNAs that must occur after peptide bond synthesis. Also, EFTu may be actively removed from the ribosome prior to this step. The binding of EFG could bring about translocation, and the hydrolysis of GTP could occur as a result of the EFG-ribosome interaction. The similarity in the tertiary structure of EFTu-aminoacyl-tRNA and EFG (both have domains that resemble tRNA [202]) may be important to bind the factors to the T site. The T site is probably near the L7/L12 protein dimers that act with EFTu and EFG on the ribosome. The ribosome itself could bring about the translocation reaction on interaction with EFG (183, 202).
The translocation mediated by EFG occurs by contact of the G domain of the molecule with 23S rRNA bases, which include the α-sarcin loop on domain VI. This contact results in molecular rearrangements of EFG which apparently change the conformation of the ribosome to an unlocked state. The resulting loosening of the tRNA contacts may be coupled to movement in regions of the ribosome, which could, in turn, move the tRNA relative to the mRNA (202). A disruption of the contacts made by domain 4 of EFG would reestablish the locked state of the ribosome, and EFG would be released (202).
It is imperative that the translocation reaction overcome the energy barrier required to move the substrates while maintaining the codon-anticodon interaction. The footprinting data of Moazed and Noller (128) suggest that the peptidyl-tRNA should remain anchored on the 50S particle. This ensures that the peptidyl-tRNA remains stably bound to the ribosome. Recent data suggest that the 30S subunit is able to effect translocation in the presence of EFG (101). One possibility is that codon-anticodon interactions occur in a subsite of the 30S subunit that is protected from factor interactions and is regulated by the P-site-bound substrates. This, in turn, suggests that part of the 30S subunit may move relative to the 50S particle. The head and body of the 30S subunit are held together by a single-stranded RNA that could enable such movement. Three-dimensional cryo-EM maps of the E. coli ribosomes have revealed that the two subunits rotate relative to each other on binding of EFG to the ribosome. This effects a ratchet-like movement which opens the mRNA channel. After GTP hydrolysis, this rotation is followed by the advance of the mRNA-tRNA2 complex. The original position of the subunits is reestablished after EFG-GDP is released from the ribosomes (46). This model is compatible with the footprinting data of Moazed and Noller (128) and with the model first proposed by Woese (203) that the structure of the anticodon stem-loop of tRNAs permits a reciprocating ratchet-like movement of the tRNA-mRNA complex (46). One of the proteins, required for reconstitution of synthesis, the RbbA protein, is a ribosome-dependent ATPase (105, 106, 107). The RbbA protein has features in common with EF3 of yeast (29, 30, 110). The possible mode of action of these proteins in chain elongation is discussed below.
Elongation Factor 3 of Fungi
In fungi, particularly the yeast S. cerevisiae, an additional factor beyond EFTu and EFG (eEF1α and eEF2 in eukaryotes) is needed for the elongation phase of translation. The discovery of this protein, elongation factor 3 (EF3), was based on the observation that, in vitro, yeast ribosomes were not able to synthesize polyphenylalanine from a poly(rU) template without EF3 and ATP (39, 177).
The gene encoding EF3 in S. cerevisiae, yef3, was cloned from a genomic library (171). The gene is present in a single copy, but recent analysis of the genomic sequence database of S. cerevisiae has shown that a second gene may encode EF3 (K. Chakraburtty, personal communication). This second gene would code for a protein with high homology but no identity to EF3. EF3 has a molecular mass of 115.7 kDa.
EF3 was determined to be a ribosome-dependent ATPase, using yeast ribosomes (30, 127). Analysis of the amino acid sequence of EF3 revealed an ATP-/GTP-binding motif, GX4GK(S/T) (3, 41, 103). A second motif present in EF3 is found only among ATP-binding proteins (198). Together, the two sequences constitute what is called the ATP-binding cassette (ABC) (3). ABC proteins make up the largest known orthologous group of proteins, many of which are membrane transporters.
EF3-homologous proteins have been found in the fungi Candida albicans and Pneumocystis carinii (42, 131, 204). Immunoblot analysis of extracts from higher eukaryotic species using anti-EF3 antibody have not revealed any immunologically cross-reacting proteins. However, ribosomes of higher eukaryotes show significant ATPase activity, which may play a similar role in protein synthesis to that of EF3 (44, 109, 110).
The role of EF3 in protein synthesis seems to be to facilitate the binding of aminoacyl-tRNA to the A site of the ribosome. In yeast, EF1α is the homologue of the eubacterial EFTu. When EF1α is present at catalytic amounts, EF3 stimulates the EF1α-mediated binding of aminoacyl-tRNA to the A site (30, 103, 195). It also influences the binding of deacyl-tRNA to the E site and stimulates the exchange of labeled deacyl-tRNA bound to the E site of yeast ribosomes (29) independent of EF1α (EFTu).
Eubacterial EF3
Ribosomes are inert in the absence of several protein factors that generally adhere loosely to either the 30S subunit (initiation factors IF1, IF2, and IF3) or to 70S ribosomes (elongation factors EFTu and EFG and termination factors RF1, RF2, and RF3).
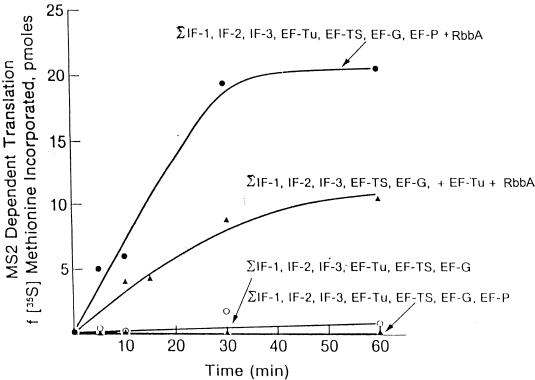
Reconstitution of synthesis using all of the homogeneous initiation, elongation, and termination factors has revealed that they do not suffice to promote the synthesis of peptides directed by native mRNA templates (55, 58, 65, 74). One of the proteins discussed above, EFP, is required for peptide bond synthesis by 70S ribosomes in the absence of organic solvents generally used to facilitate the interaction of the tRNA substrate analogues with the peptidyl transferase of the 50S subunit (52, 69). The second protein is the W2 (IF4A) helix-destabilizing protein also discussed above. A third required protein, now called RbbA (for “ribosome-bound ATPase”), was discovered during attempts to explain the ATP requirement in the transfer of fully aminoacylated-tRNAs into peptides whose synthesis was directed by the coat protein of MS2 RNA (57, 58, 74). The stimulation of synthesis directed by MS2 RNA and homogeneous initiation and elongation factors requires EFP and RbbA, as shown in Fig. 8. The RbbA protein accounts for the bulk of the ATPase activity associated with 70S ribosomes and 30S subunits (106). The gene, yhih, was cloned from E. coli genomic DNA. A His-encoding tag was inserted into the N-terminal sequence of the gene, and the RbbA protein was purified to homogeneity (105, 106, 107).
FIG. 8.
Hybrid state model for the translational elongation cycle (128). tRNA-binding sites on the 50S and 30S subunits are represented by the upper and lower rectangles, respectively. The 50S subunit harbors A (aminoacyl), P (peptidyl), and E (exit) sites, and the 30S subunit has only A and P sites. tRNAs are represented by vertical bars, and amino acids are represented by small circles. mRNA is represented as a line bound to the 30S subunit. The directional movement of the acylated and deacylated tRNAs through the ribosome, catalyzed by elongation factors EFTu and EFG, during the translational cycle is indicated, as are the proposed stimulation of the peptidyl transferase by EFP and tRNA ejection by RbbA. PT, peptidyl transfer.
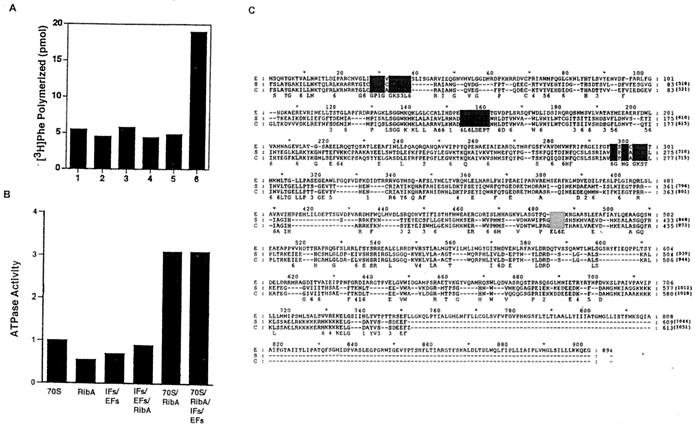
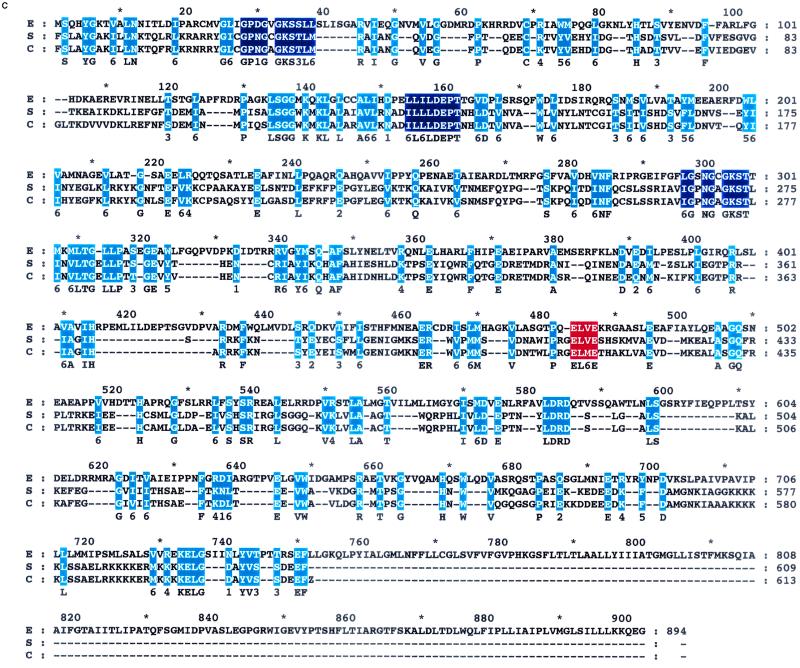
Several properties of the RbbA protein, including its ability to cross-react with antibodies specific to the yeast EF3, suggested a similarity in these proteins. As shown in Fig. 9, the predicted amino acid sequence of RbbA contains the ATP/GTP-binding motif GX4GK(S/T) (106). The amino acid sequence contains two of these motifs. Another duplicated motif in the amino acid sequence is found only among ATP-binding proteins (3, 198). The two domains together constitute the ABC (3, 198). ABCs are most common among membrane transporters of all species, but they are also part of the amino acid sequence of the yeast translation factor, EF3. The identification of the ATP-binding domains suggests that RbbA is an ATPase, as opposed to the notion that RbbA simply activates some intrinsic ATPase activity of the ribosome.
FIG. 9.
(A) Requirement for RbbA in synthesis with pure elongation factors. Lane 1, EFTu, EFG, GTP (EF); lane 2, EF plus AMPPcP; lane 3, EF plus ATP; lane 4, EF plus RbbA; lane 5, EF plus RbbA plus AMPPcP; lane 6, EF plus RbbA plus ATP. (B) Ribosome-dependent ATPase activity of RbbA. Data are from references 105 to 107. (C) Predicted amino acid sequence of RbbA. The predicted amino acid sequence of RbbA based on open reading frame yhih sequenced by Blattner et al. (15) is shown as blue-highlighted letters representing amino acids that are part of the ABC. Blue-highlighted letters represent amino acids that make up the aminoacyl-tRNA synthetase motif shared with the yeast EF3. Yellow-highlighted letters represent amino acids that are identical or very similar in the S. cerevisiae (S), C. albicans EF3 (C), and E. coli (E) RbbA proteins. The sequences were aligned with the Swiss Plot program.
Even though RbbA cross-reacts with antibody against EF3, no extensive homology outside of the ABC domains could be found between the amino acid sequences of RbbA and EF3. There is 22% identity and 40% similarity between RbbA and the C-terminal half of EF3. The only other motif RbbA and EF3 share is a motif common among aminoacyl-tRNA synthetases. This amino acid motif is ELVES in EF3. In RbbA, the motif is ELVEK (Fig. 9), compared to ELVEQ and ELLEK in prolyl-tRNA and valyl-tRNA synthetases, respectively. No other translation factor (or ABC transporter) analyzed contains this motif. In every case, there is predicted to be an amphiphilic α-helical domain near the motif. The presence of the motif in RbbA may indicate a possible interaction of RbbA with tRNA, but the motif may simply act as a general RNA recognition motif as well.
Several antibiotics have been examined as potential inhibitors of the RbbA protein. Antibiotics that impair translocation and peptide bond synthesis do not inhibit the ribosome-dependent activity of RbbA. In contrast, hygromycin B and streptomycin inhibit both poly(rU)-directed polyphenylalanine synthesis and the ATPase activity of 70S ribosomes. The two antibiotics exhibit similar synthesis inhibition curves, but hygromycin B promotes a stronger inhibition of the ribosome-bound ATPase activity (106, 107). RbbA accounts for much of the observed ATPase activity of 70S ribosomes as well as 30S subunits. Hygromycin B also displaces RbbA from the ribosome. Hygromycin B protects base 1494 on the E. coli 30S subunit (180). Mutants resistant to hygromycin B in Tetrahymena map to base 1495 of the corresponding 30S subunit (182a). Hygromycin B must cause changes in the ribosomal particle that specifically affect the ribosomal ATPase activity.
The binding site of RbbA to the 30S subunit has been studied by use of Traut's reagent, 2-iminothiolane (107, 193), and the cross-linked products were separated by two-dimensional diagonal electrophoresis and identified with anti-EF3 and with anti-S1 antibodies. The largest cross-links observed migrate above 200 kDa in a protein complex composed of RbbA and S1.
Possible Function of EF3
The exact function of RbbA in protein synthesis has not yet been determined. However, several properties of RbbA can be taken into consideration when suggesting models for its possible role in protein synthesis. First, RbbA has a site for the binding of elongation factor Tu (107). In the binding experiments, EFTu was present as EFTu-GDP. This is the form of EFTu that is released from the ribosome after aminoacyl-tRNA is bound to the A site of the ribosome. EFTu binds mainly to the 50S subunit of the ribosome, but it has been shown that the 530 loop of 16S rRNA also mediates the binding of the EFTu ternary complex to the ribosome. A G-to-A mutation at base 530 reduced EFTu ternary-complex binding to the ribosome (156). Thus, EFTu may bind to the 30S subunit at this region. Powers and Noller (156) placed the 530 loop directly in the decoding site of the ribosome, and Van Ryk and Dahlberg (197) demonstrated that a single base mutation in the 530 loop significantly affected translational fidelity.
Second, RbbA is found bound to both 70S ribosomes and 30S subunits. RbbA also binds specifically to 16S rRNA (106), suggesting that 16S rRNA nucleotides make up part of the binding site on the 30S subunit. RbbA enhances the chemical and enzymatic reactivity of A889 and G890 in the free 16S rRNA and protects A925 (105, 106, 107). These bases are probably near contact sites for RbbA and neighbor a poly(G) stretch in 16S rRNA, G885 to G888. In the crystal structure of the 30S subunit, A925 occurs immediately above the A889-G890 helix switch. This region of 16S rRNA has been demonstrated by Lodmell and Dahlberg (119) to exist in two different conformations. Conformation A results in hyperaccurate ribosomes; the A site becomes very stringent for the binding of cognate aminoacyl-tRNAs. In conformation B, the ribosomes become prone to errors and the A site becomes more open to aminoacyl-tRNAs.
The 16S rRNA of wild-type ribosomes exists in both conformations. Lodmell and Dahlberg (119) suggested that the 30S ribosomal proteins, S5 and S12, probably facilitate this conformational switch that affects the binding of tRNA at both the A and P sites because these two proteins have been implicated in the ribosomal hyperaccurate and ram phenotypes.
RbbA binds strongly to poly(G) sequences (105, 106, 107). G885 to G888 are conserved among all small-subunit rRNAs (119); thus, this poly(G) stretch in 16S rRNA is likely to be essential for ribosome function. Based on binding experiments with 16S rRNA, RbbA may bind to this region of the rRNA, and the poly(G) stretch is probably also a binding site for RbbA. Both RNase T1 and DEP preferentially modify unpaired bases. Conformation B (error prone) leaves these two bases unpaired. The second conformation, A (hyperaccurate), pairs them with U911 and C910. By binding to A925 and enhancing the reactivity of A889 and G890, RbbA may drive the conformation of the rRNA toward the unpairing of A889 and G890 during the reaction.
In the structural models of the 30S subunit (23), for the two regions, G869 occurs just above the G889 helix switch (23) and A925 occurs in the neck region of the 30S subunit at the point of the mRNA entry (23). Thus, it is possible that RbbA promotes mRNA movement on the 30S subunit by acting through the 16S rRNA G889-G890 helix switch.
In the tertiary model of 16S rRNA presented by Brimacombe et al. (18), the 530 loop and G885 to G890 are brought near each other by a site linked to ribosomal protein S5. S5 is implicated in ram mutations and may mediate the conformational switch described above (119). It is possible that the two regions are linked such that conformational changes in one region trigger changes in the other. Since RbbA binds EFTu, the interaction between the two molecules may trigger suspected conformational changes in 16S rRNA during the decoding process.
The 900 region of 16S rRNA also has been implicated in 50S-30S subunit joining. Merryman et al. (125) demonstrated that bases 892 to 896 and bases 900 to 902 are protected when 30S subunits interact with 50S subunits. This region of 16S rRNA is also near to or interacts with ribosomal proteins S5, S8, and S16 (18, 119).
RbbA is cross-linked to 30S ribosomal subunit protein S1 in the 30S subunit (107). S1, in turn, has been cross-linked to A892 of 16S rRNA (146). The neutron-determined protein map of the 30S subunit (23) and its X-ray diffraction patterns (32) show how RbbA may fit within the subunit. The S5, S8, and S16 proteins also closely neighbor the 900 region of 16S rRNA. This provides additional evidence that RbbA binds to this region of 16S rRNA. S1 is thought to be an RNA-dependent helicase that helps recruit mRNA to the 30S subunit (187, 188). It is positioned in the decoding center of the ribosome (187, 188).
The final consideration is the action of the antibiotic hygromycin B, which binds to the 30S subunit and protects bases A1408 and G1494 of 16S rRNA (180, 182). These bases also lie in the decoding center of the ribosome (27, 200). Hygromycin B affects translation at two different steps of elongation. First, hygromycin B, like other aminoglycosides, induces misreading (180). This is an effect due to the distortion of the A site (decoding center). Hygromycin B also affects the translocation process (180). The mRNA is often mistranslocated (i.e., moved more or less than the 3 bases) in the presence of hygromycin B. This also causes misreading at the A site.
Hygromycin B inhibits at least 75 to 80% of the ATPase activity of 70S ribosomes (107). Functionally similar antibiotics like streptomycin, which gives a footprint at 16S rRNA base 902 (180), and neomycin exhibit less effect (25 and 10%, respectively) on the ATPase activity. Hygromycin B was also capable of releasing RbbA from the ribosome (106, 107). This could be due to either direct competition for a binding site or, more probably, based on comparative binding-site data, hygromycin B-induced conformational changes within the ribosome. Of interest, however, is the work by Wilms et al. (200) that demonstrated cross-linking between the 900 region (RbbA region) and position 1468 (hygromycin B region) of 16S rRNA. The most recent X-ray crystallography maps of the 70S ribosome also show a close interaction between the two rRNA regions at the decoding center (27).
The above considerations suggest a role for RbbA in the decoding process of protein synthesis. In all models of translation, it is imperative that the codon-anticodon interactions of the tRNAs and mRNA remain intact during the translocation event. The footprinting data of Moazed and Noller (128) suggest that the peptidyl-tRNA remains tightly bound to the P site of the 50S subunit before the translocation event. The peptidyl-tRNA anticodon end is in the 30S subunit A site (the hybrid A/P site). Since the tRNA is firmly anchored on the ribosome, this suggests that the 30S subunit is moved with respect to the 50S subunit. The head and the body of the 30S subunit are held together by a single-stranded region of rRNA that could facilitate this movement (128). RbbA binds near this site. Since RbbA is a 30S subunit ATPase and has affinity for EFTu-GDP, it is possible that RbbA functions by facilitating the release of EFTu-GDP from the ribosome. This would, in turn, also facilitate the release of deacyl-tRNA from the E site through allosteric interactions (181), and codon-anticodon interactions could be maintained throughout.
RbbA exhibits activity that is similar to that of yeast translation factor EF3. Both are ATPases that can be stimulated by ribosomes, and both stimulate polyphenylalanine synthesis in vitro (106, 107). Both EF3 and RbbA are a strict requirement for in vitro synthesis. The main difference between the two proteins appears to lie in their affinities for ribosomes of fungal and bacterial origins. Recent work by Chakraburtty's group suggests that EF3 functions by removing deacylated tRNA from the E site, as does the RbbA (W) protein (57, 58, 74, 194). According to the “allosteric three-site” model of elongation, this would also increase aminoacyl-tRNA binding at the A site (181). EF3 was shown previously to facilitate EF1α (EFTu)-dependent binding of aminoacyl-tRNA to the A site (29).
Also of some interest may be the fact that EF3 of fungi has an E. coli ribosomal protein S5-homologous domain in its N-terminal region (29). This region has no homology to RbbA, and the yeast S5 homologue, S4, does not contain the S5 motif of EF3. It may be interesting to see if RbbA and S5 constitute an equivalent of EF3. What seems clear is that RbbA and EF3 have enough properties in common to suggest that they participate in a common function in translation.
TRANSLATIONAL TERMINATION
Any one of the three triplets, UAA, UAG, or UGA, when present in the reading frame, codes for termination of synthesis. In eubacteria, two proteins, RF-1 and RF-2, are required to recognize the termination signal and to stimulate hydrolysis of the nascent peptidyl-tRNA by the peptidyl transferase of the 50S subunit. A third protein, RF-3, acts with GTP to eject these factors from the ribosome. RF1 and RF2 were first identified during early attempts to reconstitute synthesis from pure initiation and elongation factors (51, 54). Several reviews are available (26, 34, 35, 191).
In vivo and in vitro observations suggest that UGA can be read as termination, tryptophan, cysteine, or selenocysteine (67, 121). Also, UAA, UAG, or UGA can be suppressed without loss of viability (67). This misreading is necessary for the synthesis of some essential proteins (191). There is genetic and biochemical evidence that the code to terminate synthesis is longer than a single triplet and that, in certain cases, sequences in the mRNA either 5′ or 3′ of these codons may influence the decision of termination or insertion of an amino acid (suppression) (67, 191). Suppression is used for the synthesis of many bacteriophage and retroviral proteins. It has been proposed that the C-terminal end of the nascent peptide, which is specified by the two codons at the 5′ side of UGA, markedly influences this decision by promoting termination (14).
The specificity of the termination reaction was first studied using synthetic polymers bearing UAA in different contexts. The polymers were analyzed by nuclear magnetic resonance spectroscopy and for their ability to promote RF-1- or RF-2-mediated termination. A model was proposed based on these data, explaining the effect of context. Recognition of UAA depends, at least in part, on the nature of the bases surrounding the termination codons. A loosely stacked conformation of UAA favors termination, whereas nucleosides which encourage strong base stacking restrict release. A survey of the sequences that flank stop codons has verified their nonrandom nature and has shown the preference for UAAU and UAAG as efficient stop signals in E. coli and eukaryotic mRNAs, respectively (21, 56, 155). Also, a strong base bias has been observed 5′ of the nonsense triplets (21, 56). Thus, the normal signal for termination may be determined by a longer sequence in the mRNA than merely the termination codon.
The conserved protein motifs of RF1 and RF2 have been proposed to resemble a tRNA-like structure which also occurs in the EFTu-GTP-aminoacyl-tRNA complex and underlies the EFG-GDP structure and that of the RRF protein (19, 93a, 136, 137) (see below). Omnipotent mutations are known which alter the specificity of RF2, resulting in a molecule that recognizes all three nonsense codons (93). Interchanging the mutated site between RF1 (SPF) and RF2 (PAT) alters their nonsense codon specificity (136). A tripeptide (SPF) in RF1 and RF2 confers stop codon specificity in a hybrid RF1-RF2 protein (93). It has been suggested that the RFs interact directly with nonsense codons in mRNA because the release factors cross-link to the stop codons in mRNA (155). All this argues that the factors indeed participate in nonsense codon recognition.
The crystal structure of the E. coli RF2 has been determined at 1.8-Å resolution. The tripeptide motif (SPF), proposed to be involved in nonsense codon recognition, and the conserved GGQ motif, thought to be involved with the peptidyl transferase center, occur in loops of an L-shaped structure that occur about 23 Å apart. Thus, if the structural model of the recombinant protein indeed represents the native structure, it is unlikely that the same molecule recognizes the stop codon and promotes hydrolysis of the completed protein of the peptidyl-tRNA intermediate (197). It has been suggested that RF2 achieves the ability to stimulate peptidyl-tRNA hydrolysis only by functioning in close concert with rRNA. However, recent cryo-EM data suggest a more L-like shape for the structure of the RF2-ribosome-bound complex, implying that RF2 can decode stop codons and promote hydrolysis of the nascent chain on the ribosome (M. Ehrenberg, personal communication).
Interactions between the stop signal and defined nucleotides in 16S rRNA have been emphasized by mutations that affect RF recognition (7). Indeed, suppressors of UGA that occur by mutation of 16S rRNA have been identified (7). Other suppressors, e.g., G1093A in the 23S rRNA, are UGA specific and thus also affect termination. It is possible that the GTPase center that harbors this suppressor affects the binding of RF2 to the ribosome. Other suppressors that are codon dependent have been isolated in regions of rRNAs that could possibly alter the conformation of the RF-binding sites. RF1, on the other hand, may bind in a somewhat different position which overlaps the LI-binding site (191). It is possible that the mutations in the GTPase center and on the 16S rRNA that suppress nonsense codons are important for proper termination. Indeed, other suppressors of rRNA affect termination (133).
The hydrolytic center is likely to be in domain V of the 23S rRNA, which is the peptidyl transferase domain. Early experiments suggest this, and the peptidyl transferase of the 50S subunit promotes peptidyl-tRNA hydrolysis in the absence of the release factors with suitable nucleophiles.
Eukaryotic cells have one release factor, eRF1, that promotes termination with all three nonsense codons and recognizes tetranucleotides that harbor any of the three nonsense codons (48, 49). The structure of the eRF1 is highly conserved in eukaryotic cells and harbors a GGQ sequence motif that occurs in all the species (50). Mutations in the glycine residues of this GGQ motif obliterate peptidyl-tRNA hydrolysis (50). However, mutations of the Gln residue only partly restrict in vitro termination (175a).
The structure of eRF1 has been determined by X-ray diffraction (179). The molecule has three domains that fold in an L-type structure that resembles a tRNA molecule. Domains 1, 2, and 3 correspond to the anticodon loop, the aminoacyl acceptor stem, and the T stem of tRNA, respectively (179). In the tRNA-like structure of the protein, the “anticodon” end of the molecule can bind a tetranucleotide harboring a termination codon (179). Mutations at this site are lethal and result in improper codon recognition (179). Other mutations most commonly occur by suppression of nonsense codons due to improper functioning of a release factor. These mutations result in insertion of an amino acid at the site of the nonsense codon.
The eRF1 structure has a highly conserved shallow groove on the surface of domain 1, which is thought to be involved in stop codon recognition (137). Mutations in the eRF1 groove (anticodon-like region) result in nonsense suppression (137). The eRF molecule also harbors a site that may be used in activating the esterase activity of the peptidyl transferase (50). The groove is about 80 Å from the GGQ motif of the molecule, which may be involved in peptidyl-tRNA hydrolysis (50). Thus, it is thought that eRF1 binds to the A site of the ribosome, where the GGQ motif bound to water can effect a nucleophilic attack on the ester of the peptidyl-tRNA bound to the P site, resulting in hydrolysis of the peptidyl-tRNA harboring the completed protein (50). The conservation of the GGQ motif suggests that a similar mechanism might underlie termination in all cells, in spite of sequence and structural differences in the release factor of the eukaryotic and eubacterial source. However, it is likely that the recognition mechanism is more complex than this hypothesis implies.
The third eubacterial release factor, RF3, acts to unbind the RFs in a series of reactions (204b). RF3 stably binds GDP and acts as a guanine exchange factor on the ribosome-RF complex. Hydrolysis of peptidyl-tRNA by the RFs allows RF3 to bind GTP, which in turn induces the dissociation of RF1 and RF2 from the ribosome. After GTP hydrolysis, RF3 is also released from the ribosomes (204b).
A second eukaryotic factor, eRF3, stimulates the eRF1 activity as well as the GTPase activity of ribosomes in the presence of eRF1 and GTP (48, 49). eRF3 has features in common with the eubacterial protein (112) but has little sequence similarity to this protein. The C-terminal sequence of eRF3 is similar to EF1A and harbors a GTPase domain (49, 137). The N-terminal domain of the molecule appears dispensable, whereas the C-terminal end is essential for the function of the molecule.
In M. genitalium, the smallest known bacterial species, RF2 has been dispensed with but a copy of RF1 has been maintained. In this case, the UGA codon has been assigned to Gln (47). Similarly, only RF1 occurs in mammalian and S. cerevisiae mitochondria (115, 153). In these cases the UGA codon has been reassigned as Trp.
In eubacteria, the disassembly of the termination complex involves GTP and two factors, RRF (for “ribosome recycling factor”) and EFG (86, 87, 99). In the absence of RRF, ribosomes scan the mRNA and reinitiate synthesis downstream of the stop codon (97). The RRF gene also increases translational fidelity, but this mechanism has not been fully investigated (98). The RRF protein is essential for cell viability (99).
Archaebacteria appear to be the only group of organisms lacking an RRF homologue. In eukaryotes, RRF is a chloroplastic (168) or mitochondrial (A. Kaji et al., personal communication) protein. The mitochondrial RRF homologue is essential for mitochondrial maintenance (Kaji et al., personal communication). A eukaryotic cytoplasmic homologue of RRF has not been found. Instead, it has been postulated that the cytoplasmic ribosomes bind the 5′ and 3′ termini of the mRNA together through a poly(A)-binding protein which, in turn, forms part of the eIF4F protein complex (94, 95). The interaction of the 3′ and 5′ termini of the mRNA in this complex is thought to facilitate reinitiation events (95).
CONCLUDING COMMENTS
Tables 1 and 2 contain a summary of the nomenclature and of the general functions of each initiation, elongation and termination factor from eubacterial and from archaebacterial and eukaryotic sources. It is immediately apparent that the factors and the functions that they promote are highly conserved. There are a few important exceptions, and part of this review has dealt with their consideration.
TABLE 2.
Elongation and termination factors
| Eukaryotes | Archaea | Eubacteria | Ribosomal function |
|---|---|---|---|
| eEF1A | + | EFTu | GTP-dependent binding of aminoacyl-tRNA to 70S_(80S); proofreading for noncognate tRNAs |
| eEF1βγ | + | EFTs | Exchanges GDP for GTP in EFTu |
| eEF2 | + | EFG | Translocation of peptidyl-tRNA; ribosome-dependent GTPase |
| eEF3 | ? | RbbA (W2) | Ribosome-dependent ATPase; tRNA ejection from ribosomes |
| eRF1 | + | RF1 | Nonsense (UAA, UAG) codon recognition, ribosome-dependent hydrolysis of peptidyl-tRNA |
| − | RF2 | Nonsense codon (UAA, UGA) recognition, ribosome-dependent hydolysis of peptidyl-tRNA | |
| eRF3 | − | RF3 | GTP-dependent recycling of RFs |
| mt RRFa | − | RPF | Reinitiation; unbinding of mRNA after termination |
The RRF of eukaryotes occurs in mitochondria (mt) but probably not in the cytoplasm of the cell.
The initiation reactions have conserved most of the critical proteins. Where functions differ, subtle changes, such as the modification of the protein factors or the acquisition of additional functions, e.g., the recognition of the 7mG-cap structure, may have evolved for a tighter regulation of the process.
One is struck by the complexity of the eukaryotic initiation factors. One of the most obvious examples of this complexity is the eIF3 protein, which has 11 subunits. Interestingly, many of these subunits have regulatory functions and form complexes with other initiation factors (84). Here, as with other multimeric proteins, one recalls the parallel in the proposed evolution of the ribosome. The finding of discrete units composed, e.g., of 5.8S rRNA suggests that perhaps contemporary ribosomes evolved from ancient component parts (72, 138). For the proteins, gene duplication would have the advantage of enabling individual member of the aggregates to be regulated by a variety of diverse mechanisms.
Many of the differences in the initiation factors from eubacterial and archaebacterial-eukaryotic sources relate to “recycling” functions. Thus, the equivalent of EFTs of eubacteria does not always occur in eukaryotic cells (84). Similarly, the initiation factors eIF2Bγ and eIF2Bɛ are not represented in eubacteria and, like EFTs, are involved in recycling guanine nucleotides; i.e., they possess GEF activity. One is tempted to speculate that the small size of the eubacterial cell effectively increases the concentration of its components, perhaps rendering these functions dispensable. On the other hand, proteins such as eIF4G, which act as a chaperone for the assembly of the initiation complexes on the 5′ terminus of the eukaryotic ribosomes, may be required to ensure that the concentration of 7mG-cap recognition factor is sufficiently high to target the mRNA start site.
The triplets that code for termination and initiation are affected strongly by the context of the neighboring bases. This also may be an evolutionary advantage since it enables termination to occur or to be suppressed so as to increase the number of proteins that can be synthesized from a single reading frame. The fact that a protein can be made from a methionine codon in the coding region by a single mutation (111) implies that the ambiguous nature of the AUG start codon can potentially be used to enlarge the number of proteins that can be synthesized from a single gene.
Finally, the flexible nature of start and stop codons suggests that alternate mechanisms may be used to decipher such codons. This may ensure that these processes occur in spite of changes in the proteins that regulate their action.
A summary of the elongation and termination factors is given in Table 2. Clearly, with minor exceptions, the elongation process is dependent on factors whose structures have been conserved. It is evident that the functions attributed to EF3 are not restricted to fungi. The RbbA protein of E. coli has significant features in its structure and function which strongly suggest that it acts in similar fashion to EF3. Similarly, eukaryotic cells harbor a tightly associated ATPase activity that may act in similar fashion in translation. The major activities involved in decoding, translocation, and proofreading appear to have been maintained throughout evolution.
The termination reaction has also conserved its vital proteins. In this case, the function of the two eubacterial termination proteins has been harnessed by one larger eukaryotic protein able to perform both recognition functions. The fact that a single mutation in the eubacterial RF2 protein alters the coding properties of the protein so that it now recognizes all three rather than two nonsense codons underlies the functional resemblance of these proteins. Also, the conserved residue in the terminal tip of the tRNA-like “L” structure that occurs in these proteins might be essential to their function, particularly to their ability to hydrolyze the completed peptidyl-tRNA. The eubacterial and eukaryotic proteins have little sequence homology; however, they appear to fold in a tRNA-like structure with domains that are compatible with their action in codon recognition and in promoting ester hydrolysis by the peptidyl transferase. The dimensions of the eubacterial RF2, however, are not strictly compatible with this notion but suggest, instead, that the binding of the RF to the rRNA may be important to its recognition function. The cryo-EM-detected structures appear compatible with this view.
There is at present scant evidence that the restart mechanism used in eubacteria also occurs in eukaryotic cells. Perhaps the juxtapositioning of the mRNA termini in eukaryotic cells bypasses the more complex restart mechanism used by eubacterial cells.
An emerging principle about the structure of the translation factors is their ability to mimic tRNA-like shapes. This was first observed for the elongation factor EFG-GDP and the ternary complex of EFTu-GTP-aminoacyl-tRNA. Other tRNA-like structures are found in RF2 of E. coli, although the putative “anticodon” ends of the protein and the CCA terminus are not really able to simultaneously decipher stop at the 30S A site and chain cleavage by the peptidyl transferase. It is noteworthy, however, that the molecules can be made to fit both rRNA sites, given contacts with the 16S and 23S rRNA. Finally, the RRF protein also mimics a tRNA, and, surprisingly, the S. aureus EFP structure can be superimposed on the L shape of the tRNAPhe structure. Arguments have been made that the structure of the initiation factors and their complexes may also fit such a pattern, but this remains unproven. Whether this interesting insight holds up in eukaryotic cells is unknown. It is known that eRF1 does resemble a tRNA structure.
There is much evidence that proteins evolved from ancestral proteins. It appears that some proteins also mimic the structure of tRNAs. It has been speculated that the protein synthetic machinery, and perhaps a primitive molecular version of it, had RNA that evolved to recognize RNA—that some proteins perhaps replaced the functions of RNA by mimicking and by recognizing RNA and that perhaps, during these transitions, proteins eventually evolved to recognize these proteins (31a). The comparison of the eukaryotic, archaeal, and eubacterial translation systems seems replete with examples of each of the above types of schemes, although clearly this is quite speculative at present.
Most of the proteins, apart from the initiation factors 1, 2, and 3 and the elongation factors EFTu, EFTs, and EFG or the RRF factor, were discovered by reconstituting synthesis in vitro by using pure initiation and elongation proteins and E. coli ribosomes programmed with MS2 RNA (51, 54, 57, 58, 74). These and genetic experiments indicate that the proteins described here are essential for synthesis. However, the reconstituted in vitro systems are not nearly as efficient as the live cells (57, 58, 65, 74, 175b). Thus, it is possible that additional eubacterial proteins are yet to be discovered. Nonetheless, it is evident that the translation reactions are for more highly conserved among the species than was heretofore suspected.
Acknowledgments
We are grateful to many colleagues for discussion of this work, particularly Andrew J. Becker, Knud Nierhaus, Kalpana Chakraburtty, Akira Kaji, Al Dahlberg, and James Friesen. We are also grateful for the critical comments of John Xu and Martin Lovman and for the excellent work of Jay Lu, Kathleen Dekany, Andrea Long, and Sally Adams. We thank Lizhu Ke, Jennifer Yang, Ivona Kodieradski, Nada Barraclough, and Cris Cunningham (Meackle) for excellent technical help over the years.
We thank the Medical Research Council of Canada, the Natural Sciences and Engineering Council of Canada, and the Pharmacia Corporation for financial support.
Footnotes
This work is dedicated to the memory of Clelia H. Finney.
REFERENCES
- 1.Adhin, M. R., and J. van Duin. 1990. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 213:811-818. [DOI] [PubMed] [Google Scholar]
- 2.Ævarsson, A., E. Brazhnikov, M. Garber, J. Zheltonosova, Y. Chirgadze, S. al-Karadaghi, L. A. Svensson, and A. Liljas. 1994. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 13:3669-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, G. F., C. S. Mimura, S. R. Holbrook, and V. Shyamala. 1992. Traffic ATPases: a superfamily of transport proteins operating from Escherichia coli to humans. Adv. Enzymol. Relat. Areas Mol. Biol. 65:1-47. [DOI] [PubMed] [Google Scholar]
- 4.An, G., B. R. Glick, J. D. Friesen, and M. C. Ganoza. 1980. Identification and quantification of elongation factor EFP in E. coli. Can. J. Biochem. 97:23-28. [Google Scholar]
- 5.Aoki, H., S. L. Adams, D. Watson, M. Yaguchi, I. Kozieradzki, and M. C. Ganoza. 1997. Molecular characterization of the efp gene product involved in a peptidyl transferase reaction. Biochimie 79:7-11. [DOI] [PubMed] [Google Scholar]
- 6.Aoki, H., S. L. Adams, D. G. Chung, M. Yaguchi, S. E. Chuang, and M. C. Ganoza. 1991. Cloning, sequencing and overexpression of the gene for prokaryotic factor EFP involved in peptide bond synthesis. Nucleic Acids Res. 19:6215-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki, H., K. Dekany, S. L. Adams, and M. C. Ganoza. 1997. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J. Biol. Chem. 272:32254-32259. [DOI] [PubMed] [Google Scholar]
- 7a.Arkov, A. L., and E. J. Murgola. 1999. Ribosomal RNAs in translation termination: facts and hypothesis. Biochemistry 64:1354-1359. [PubMed] [Google Scholar]
- 8.Bartig, D., K. Lemkemeier, J. Frank, F. Lottspeich, and F. Klink. 1992. The Archaebacteria hypusine-containing protein. Eur. J. Biochem. 204:751-758. [DOI] [PubMed] [Google Scholar]
- 9.Benz, H. L., H. Trachsel, and U. Baumann. 1999. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae—the prototype of the DEAD box protein family. Structure 7:671-679. [DOI] [PubMed] [Google Scholar]
- 10.Berchtold, H., L. Reshetnikova, C. O. Reiser, N. K. Schirmer, M. Sprinzl, and R. Hilgenfeld. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365:126-132. (Erratum, 365: 368.) [DOI] [PubMed] [Google Scholar]
- 11.Bevec, D., H. Jaksche, M. Oft, T. Wöhl, M. Himmelspach, A. Pacher, M. Schebesta, K. Koettnitz, M. Dobrovnik, R. Csonga, F. Lottspeich, and J. Hauber. 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of Rev cofactor eIF5A. Science 271:1858-1860. [DOI] [PubMed] [Google Scholar]
- 12.Bevec, D., H. Klier, W. Holter, E. Tschachler, P. Valent, F. Lottspeich, T. Baumruker, and J. Hauber. 1994. Induced expression of the hypusine containing protein eukaryotic initiation factor 5A in activated human T lymphocytes. Proc. Natl. Acad. Sci. USA 91:10829-10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyer, D., E. Skripkin, J. Wadzack, and K. H. Nierhaus. 1994. How the ribosome moves along the mRNA during protein synthesis. J. Biol. Chem. 269:30713-30717. [PubMed] [Google Scholar]
- 14.Bjornsson, A., S. Mottagui-Tabar, and L. A. Isaksson. 1996. Structure of the C-terminal end of the nascent peptide influences translational termination. EMBO J. 15:1696-1704. [PMC free article] [PubMed] [Google Scholar]
- 15.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 16.Blum, S., S. R. Schmid, A. Pause, P. Buser, P. Linder, N. Sonenberg, and H. Trachsel. 1992. ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci USA 89:7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borowski, C., M. V. Rodnina, and W. Wintermeyer. 1996. Truncated elongation factor G lacking the G domain promotes translocation of the 3′ end but not of the anticodon domain of peptidyl-tRNA. Proc. Natl. Acad. Sci. USA 93:4202-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brimacombe, R., J. Atmadja, W. Stiege, and D. Schuler. 1998. A detailed model of the three-dimensional structure of Escherichia coli 16S ribosomal RNA in situ in the 30S subunit. J. Mol. Biol. 199:115-136. [DOI] [PubMed] [Google Scholar]
- 19.Brock, S., K. Szkaradkiewiez, and M. Spernzl. 1998. Initiation factors and protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol. 29:409-417. [DOI] [PubMed] [Google Scholar]
- 20.Broderson, D. E., W. H. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, hygromycin B on 30S ribosomal subunits. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 21.Buckingham, K., D. G. Chung, T. Neilson, and M. C. Ganoza. 1987. Recognition of translational termination signals. Biochim. Biophys. Acta 909:92-98. [DOI] [PubMed] [Google Scholar]
- 22.Burkhardt, N., R. Junemann, C. M. Spahn, and K. H. Nierhaus. 1998. Ribosomal tRNA binding sites: three-site models of translation. Crit. Rev. Biochem. Mol. Biol. 33:95-149. [DOI] [PubMed] [Google Scholar]
- 23.Capel, M. S., D. M. Engelman, B. R. Freeborn, M. Kjeldgaard, J. A. Langer, V. Ramakrishnan, D. G. Schindler, D. K. Schneider, B. P. Schoenborn, I. Y. Sillers, et al. 1987. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science 238:1403-1406. [DOI] [PubMed] [Google Scholar]
- 24.Caraglia, H., A. Passeggio, S. Beninati, A. Leardi, L. Nicolini, S. Improta, A. Pinto, A. R. Bianco, P. Tagliaferri, and A. Abbruzzese. 1997. Interferon alpha 2 recombinant epidermal growth factor modulates proliferation and hypusine synthesis in human epidermoid cancer KB cells. Biochem. J. 324:737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caruthers, J. M., E. R. Johnson, and D. B. McKay. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA 97:13080-13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caskey, T., E. Scolnick, R. Tompkins, J. Goldstein, and G. Milman. 1970. Peptide chain termination; codon, protein factor, and ribosomal requirements. Cold Spring Harbor Symp. Quant. Biol. 34:479-488. [DOI] [PubMed] [Google Scholar]
- 27.Cate, J. H., M. M. Yusupov, G. Z. Yusupova, T. N. Earnest, and H. F. Noller. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285:2095-2104. [DOI] [PubMed] [Google Scholar]
- 28.Cech, T. R., A. J. Zaug, and P. Brogowski. 1981. In vitro splicing of the ribosomal RNA precursor of Tetrahymema: involvement of a guanosine nucleotide in the excision of the intervining sequence. Cell 27:487-496. [DOI] [PubMed] [Google Scholar]
- 29.Chakraburtty, K., and F. J. Triana-Alonso. 1998. Yeast elongation factor 3: structure and function. Biol. Chem. 379:831-840. [DOI] [PubMed] [Google Scholar]
- 30.Chakraburtty, K., and A. Kamath. 1998. Protein synthesis in yeast. Int. J. Biochem. 20:581-590. [DOI] [PubMed] [Google Scholar]
- 31.Chen, Z. P., and K. Y. Chen. 1997. Marked elevation of hypusine formation activity on eukaryotic initiation factor 5A in V-HA-RAS transformed mouse NIH3T3 cells. Cancer Lett. 115:235-241. [DOI] [PubMed] [Google Scholar]
- 31a.Clark, B. F. C., S. Thirup, M. Kjeldgaard, and J. Nyborg. 1999. Structural information for explaining the molecular mechanism of protein biosynthesis. FEBS Lett. 452:41-46. [DOI] [PubMed] [Google Scholar]
- 32.Clemens, W. M., Jr., J. L. May, B. T. Wimberly, J. P. McCutcheon, M. Capel, and V. Ramakrishnan. 1999. Structure of bacterial 30S subunit at 5.5Å resolution. Nature 400:833-840. [DOI] [PubMed] [Google Scholar]
- 33.Cooperman, B. S., T. Wooten, D. P. Romero, and R. R. Traut. 1995. Histidine 229 in protein L2 is apparently essential for 50S peptidyl transferase activity. Biochem. Cell Biol. 73:1087-1094. [DOI] [PubMed] [Google Scholar]
- 34.Craigen, W. J., and C. T. Caskey. 1986. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322:273-275. [DOI] [PubMed] [Google Scholar]
- 35.Craigen, W. J., C. C. Lee, and C. T. Caskey. 1990. Recent advances in peptide chain termination. Mol. Microbiol. 4:861-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czworkowski, J., and P. B. Moore. 1996. The elongation phase of protein synthesis. Prog. Nucleic Acid Res. Mol. Biol. 54:293-332. [DOI] [PubMed] [Google Scholar]
- 37.Czworkowski, J., J. Wang, T. A. Steitz, and P. B. Moore. 1994. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 13:3661-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dabrowski, M., C. M. Spahn, M. A. Schafer, S. Patzke, and K. H. Nierhaus. 1998. Protection patterns of tRNAs do not change during ribosomal translocation. J. Biol. Chem. 273:32793-32800. [DOI] [PubMed] [Google Scholar]
- 39.Dasmahapatra, B., and K. Chakraburtty. 1981. Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 256:9999-10004. [PubMed] [Google Scholar]
- 40.deSmit, M. H., and J. van Duin. 1990. Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acids Res. Mol. Biol. 38:1-35. [DOI] [PubMed] [Google Scholar]
- 41.Dever, T. E., M. J. Glynias, and W. C. Merrick. 1987. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. USA 84:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Domenico, B. J., J. Lupisella, M. Sandbaken, and K. Chakraburtty. 1992. Isolation and sequence analysis of the gene encoding translation elongation factor 3 from Candida albicans. Yeast 8:337-352. [DOI] [PubMed] [Google Scholar]
- 43.Egebjerg, J., N. Larsen, and R. Garrett. 1990. Structural map of 23 rRNA, p. 168-179. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 44.El'skaya, A. G., A. I. Serebryanik, and G. V. Ovcharenko. 1995. Stimulation of yeast EF3 factor by mammalian ribosomes. Ukr. Biokhim. Zh. 67:28-32. [PubMed] [Google Scholar]
- 45.Franceschi, F. J., and K. H. Nierhaus. 1990. Ribosomal proteins L15 and L16 are late assembly proteins of the large ribosomal subunit. J. Biol. Chem. 265:16676-16682. [PubMed] [Google Scholar]
- 46.Frank, J., and K. R. Agrawal. 2000. A ratchet-like inter-subunit reorganization of the ribosomes during translocation. Nature 406:318-322. [DOI] [PubMed] [Google Scholar]
- 47.Fraser, C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 48.Frolova, L., X. Le Goff, H. H. Rasmussen, S. Cheperegin, G. Drugeon, M. Kress, I. Arman, A. L. Haenni, L. E. Celis, M. Philippe, J. Justesen, and L. Kisselev. 1994. A highly conserved eukaryotic protein family possessing properties of a polypeptide chain release factor. Nature 372:701-703. [DOI] [PubMed] [Google Scholar]
- 49.Frolova, L., S. LeGoff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependant guanosine triphosphatase. RNA 4:334-341. [PMC free article] [PubMed] [Google Scholar]
- 50.Frolova, L., R. Y. Tsivkowvskii, G. F. Sivolobova, N. Y. Oparina, O. I. Serpinsky, V. M. Blinov, S. I. Tatkov, and L. L. Kisselev. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganoza, M. C. 1966. Polypeptide chain termination in cell-free extracts of E. coli. Cold Spring Harbor Symp. Quant. Biol. 31:273-278. [DOI] [PubMed] [Google Scholar]
- 52.Ganoza, M. C., and H. Aoki. 2000. Peptide bond synthesis: function of the efp gene product. J. Biol. Chem. 381:553-559. [DOI] [PubMed] [Google Scholar]
- 53.Ganoza, M. C., and B. G. Louis. 1993. Potential secondary structure at the translational start domain of eukaryotic and prokaryotic mRNAs. Biochimie 76:428-439. [DOI] [PubMed] [Google Scholar]
- 54.Ganoza, M. C., and T. N. Nakamoto. 1996. Studies on the mechanism of polypeptide chain termination in cell-free extracts of E. coli. Proc. Natl. Acad. Sci. USA 55:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ganoza, M. C., and J. L. Fox. 1974. Isolation of a soluble factor needed for protein synthesis with various messenger ribonucleic acids other than poly(U). J. Biol. Chem. 249:1037-1048. [PubMed] [Google Scholar]
- 56.Ganoza, M. C., K. Buckingham, P. Hader, and N. Nielson. 1984. Effect of base sequence on in vitro protein-chain termination. J. Biol. Chem. 259:14101-14104. [PubMed] [Google Scholar]
- 57.Ganoza, M. C., C. Cunningham, and R. M. Green. 1995. A new factor from Escherichia coli affects translocation of mRNA. J. Biol. Chem. 270:26377-26381. [DOI] [PubMed] [Google Scholar]
- 58.Ganoza, M. C., C. Cunningham, and R. M. Green. 1985. Isolation and point of action of a factor from Escherichia coli required to reconstruct translation. Proc. Natl. Acad. Sci. USA 82:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganoza, M. C., C. C. Cunningham, D. G. Chung, and T. Nielson. 1991. A proposed role for IF3 and EFT in maintaining the specificity of prokaryotic initiation complex formation. Mol. Biol. Rep. 15:33-38. [DOI] [PubMed] [Google Scholar]
- 60.Ganoza, M. C., A. Fraser, and T. Neilson. 1978. Nucleotides contiguous to AUG affect translation initiation. Biochemistry 17:2769-2775. [DOI] [PubMed] [Google Scholar]
- 61.Ganoza, M. C., E. C. Kofoid, P. Marliere, and B. G. Louis. 1987. Potential secondary structure at translational initiation sites. Nucleic Acids Res. 15:345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganoza, M. C., P. Marliere, E. C. Kofoid, and B. G. Louis. 1985. Initiator tRNA may recognize more than the initiation codon in mRNA: a model for translational initiation. Proc. Natl. Acad. Sci. USA 82:4587-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ganoza, M. C., P. Sullivan, C. C. Cunningham, E. C. Kofoid, and T. Neilson. 1982. Effect of bases contiguous to AUG on translation initiation. J. Biol. Chem. 257:8228-8232. [PubMed] [Google Scholar]
- 64.Ganoza, M. C., N. Zahid, and R. Baxter. 1985. Stimulation of peptidyl transferase reactions by a soluble protein. Eur. J. Biochem. 146:287-294. [DOI] [PubMed] [Google Scholar]
- 65.Ganoza, M. C. 1977. The Ayerst Award Lecture 1976. Novel factors in protein biosynthesis Can. J. Biochem. 55:267-281. [DOI] [PubMed] [Google Scholar]
- 66.Ganoza, M. C., H. Aoki, I. Kozieradzki, and I. Schwartz. 1993. Reconstitution of translation. Evidence for the involvement of the rescue protein in the association/dissociation of ribosomal subunits. Eur. J. Biochem. 217:839-847. [DOI] [PubMed] [Google Scholar]
- 66a.Ganoza, M. C., and M. C. Kiel. 2001. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 45:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garen, A. 1968. Sense and nonsense in the genetic code. Science 160:149-159. [DOI] [PubMed] [Google Scholar]
- 68.Glick, B. R., and M. C. Ganoza. 1975. Identification of a soluble protein that stimulates peptide bond synthesis. Proc. Natl. Acad. Sci. USA 72:4257-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glick, B. R., S. Chladek, and M. C. Ganoza. 1979. Peptide bond formation stimulated by protein synthesis factor EFP depends on the aminoacyl moiety of the acceptor. Eur. J. Biochem. 97:23-28. [DOI] [PubMed] [Google Scholar]
- 70.Gold, L. 1988. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 57:199-233. [DOI] [PubMed] [Google Scholar]
- 71.Gold, L., D. Pribnow, T. Schneider, S. Shinedling, B. S. Singer, and G. Stormo. 1981. Translation initiation in procaryotes. Annu. Rev. Microbiol. 35:265-403. [DOI] [PubMed] [Google Scholar]
- 72.Gray, M. W. and M. N. Schnare. 1996. Evaluation of rRNA gene organization, p. 49-69. In R. A. Zimmerman and A. Dahlberg (ed.), Ribosomal RNA structure, evolution, processing, and function in protein biosynthesis. CRC Press, Inc., Boca Raton, Fla.
- 73.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66:679-716. [DOI] [PubMed] [Google Scholar]
- 74.Green, R. H., R. B. Glick, and M. C. Ganoza. 1985. Requirements for in vitro reconstruction of protein synthesis. Biochem. Biophys. Res. Commun. 126:792-798. [DOI] [PubMed] [Google Scholar]
- 75.Grenizmann, G. P., J. Kelly, S. Laalami, M. Shuda, M. A. Firpo, Y. Cenatiempo, and A. Kaji. 1998. Release factor RF3 GTPase activity acts in disassembly of the ribosome termination complex. RNA 4:973-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gualerzi, C. O., L. Brandi, E. Caserta, A. La Teara, R. Spurio, J. Tomsic, and C. L. Pon. 2000. Translation initiation in bacteria, p. 447-494. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome. Structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 77.Gualerzi, C. V., A. L. Teana, R. Spurio, M. A. Canonaco, H. Severini, and C. L. Pon. 1990. Initiation of protein synthesis in prokaryotes: recognition of mRNA by ribosomes and molecular basis for the function of initiation factors, p. 281-291. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 78.Guerrier-Takada, C., K. Gardiner, T. Marsh, N. Pace, and S. Altman. 1983. The RNA moiety of ribonuclease P is the catalytic subunity of the enzyme. Cell 27:849-857. [DOI] [PubMed] [Google Scholar]
- 79.Hall, M. N., J. Gabay, M. Debarbouille, and M. Schwartz. 1982. A role for mRNA secondary structure in the control of translation initiation. Nature 295:616-618. [DOI] [PubMed] [Google Scholar]
- 80.Hartz, D., D. S. McPheeters, and L. Gold. 1991. Influence of mRNA determinants on translation initiation in Escherichia coli. J. Mol. Biol. 218:83-97. [DOI] [PubMed] [Google Scholar]
- 81.Hartz, D., D. S. McPheeters, and L. Gold. 1989. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 3:1899-1912. [DOI] [PubMed] [Google Scholar]
- 82.Hartz, D., D. S. McPheeters, and L. Gold. 1990. From polynucleotide to natural mRNA translation initiation: function of Escherichia coli initiation factors, p. 275-280. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 83.Hershey, J. W. B. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717-755. [DOI] [PubMed] [Google Scholar]
- 84.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 85.Higgins, C. F., I. D. Hiles, G. P. Salmond, D. R. Gill, J. A. Downie, I. J. Evans, I. B. Holland, L. Gray, S. D. Buckel, A. W. Bell, et al. 1986. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323:448-450. [DOI] [PubMed] [Google Scholar]
- 86.Hirashima, A., and A. Kaji. 1972. Factor-dependent release of ribosomes from messenger RNA—requirement for two heat-stable factors. J. Mol. Biol. 65:43. [DOI] [PubMed] [Google Scholar]
- 87.Hirashima, A., and A. Kaji. 1973. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 248:7580. [PubMed] [Google Scholar]
- 88.Hui, A., and H. A. de Boer. 1987. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:4762-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hui, A., J. Hayflick, K. Dinkenspiel, and H. A. DeBoer. 1984. Mutagenesis of the three bases preceding the start codon of the β-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 3:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hyde, S. C., P. Emsley, M. J. Hartshorn, M. M. Mimmack, U. Gileadi, S. R. Pearce, M. P. Gallagher, D. R. Gill, R. E. Hubbard, and C. F. Higgins. 1990. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346:362-365. [DOI] [PubMed] [Google Scholar]
- 91.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iost, I., M. Dreyfus, and P. Linder. 1994. mRNAs can be stabilized by DEAD-box proteins. Nature 372:193-196. [DOI] [PubMed] [Google Scholar]
- 93.Ito, K., M. Uno, and Y. Nakamura. 1998. Single amino acid substitution in prokaryotic release factor 2 permits it to terminate translation at all three stop codons. Proc. Natl. Acad. Sci. USA 95:8165-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93a.Ito, K., M. Uno, and Y. Nakamura. 2000. A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature 403:680-684. [DOI] [PubMed] [Google Scholar]
- 93b.Ito, K., K. Ebihara, M. Uno, and Y. Nakamura. 1996. Conserved motifs in prokaryotic and eukaryotic release factors: tRNA-protein mimicry hypothesis. Proc. Natl. Acad. Sci. USA 93:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jackson, W. F., and A. Kaminski. 1985. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 95.Jacobson, A. 1996. Poly(A) metabolism and translation: the closed-loop model, p. 451-480. In J. W. B. Harshey et al. (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 96.Janosi, L., H. Hara, S. Zhang, and A. Kaji. 1996. Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys. 32:121-201. [DOI] [PubMed] [Google Scholar]
- 97.Janosi, L., S. Mottagui-Tabar, L. A. Isaksson, Y. Sekine, E. Ohtsubo, S. Zhang, S. Goon, S. Nelken, M. Shuda, and A. Kaji. 1998. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 17:1141-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Janosi, L., L. Ricker, and A. Kaji. 1996. Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie 78:959. [DOI] [PubMed] [Google Scholar]
- 99.Janosi, L., I. Shimizu, and A. Kaji. 1994. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl. Acad. Sci. USA 91:4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones, P. G., M. Mitta, Y. Kim, W. Yiang, and M. Inouye. 1996. Cold shock induces a major ribosome-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joseph, S., and H. F. Noller. 1998. EFG catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 17:3478-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaempfer, R. 1972. Initiation factor IF-3: a specific inhibitor of ribosomal subunit association. J. Mol. Biol. 71:583-598. [DOI] [PubMed] [Google Scholar]
- 103.Kamath, A., and K. Chakraburtty. 1989. Role of yeast elongation factor 3 in the elongation cycle. J. Biol. Chem. 264:15423-15428. [PubMed] [Google Scholar]
- 104.Kang, H., and J. W. B. Hershey. 1994. Effect of initiation factor eIF5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 269:3934-3940. [PubMed] [Google Scholar]
- 105.Kiel, M. 1999. Identification and characterization of an Escherichia coli ribosomal ATPase. Ph.D. thesis. University of Toronto Press, Toronto, Canada.
- 106.Kiel, M. C., H. Aoki, and M. C. Ganoza. 1999. Identification of a ribosomal ATPase in Escherichia coli cells. Biochemie 81:1097-1103. [DOI] [PubMed] [Google Scholar]
- 107.Kiel, M. C., and M. C. Ganoza. 2000. Functional interactions of an Escherichia coli ribosomal ATPase. Eur. J. Biol. Chem. 268:278-286. [DOI] [PubMed] [Google Scholar]
- 108.Kim, K. K., L. W. Hung, H. Yokota, R. Kim, and S. H. Kim. 1998. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at the 1.8Å resolution. Proc. Natl. Acad. Sci. USA 95:10419-10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kovalchuke, O., and K. Chakraburtty. 1994. Comparative analysis of ribosome-associated adenosinetriphosphatase (ATPase) from pig liver and the ATPase of elongation factor 3 from Saccharomyces cerevisiae. Eur. J. Biochem. 226:133-140. [DOI] [PubMed] [Google Scholar]
- 110.Kovalchuke, O., J. Ziehler, and K. Chakraburtty. 1995. Comparative analysis of ATPase of yeast elongation factor 3 and ATPase associated with Tetrahymena ribosomes. Biochemie 77:713-718. [DOI] [PubMed] [Google Scholar]
- 111.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 112.Kuni, M., K. Ito, J. Moffat, K. Matsuma, K. McCaughan, T. Nobukuni, W. Tate, and Y. Nakamura. 1994. Identification of the prfC gene, which encodes peptide release factor 3 of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5798-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kyrpides, N. C., and C. R. Woese. 1998. Universally conserved initiation factors. Proc. Natl. Acad. Sci. USA 95:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lake, J. 1979. Ribosome structure and function, p. 207-236. In G. Chamliss, G. R. Craven, J. Davies, K. Davis, I. Kahan, and M. Nomura (ed.), Ribosomes. Structure, function and genetics. University Park Press, Baltimore, Md.
- 115.Lee, C. C., K. M. Trimms, C. N. A. Trotman, and W. P. Tate. 1987. Isolation of a rat mitochondrial release factor. Accommodation of the changed genetic code for termination. J. Biol. Chem. 262:3548-3552. [PubMed] [Google Scholar]
- 116.Liljas, A. 1990. Some structural aspects of elongation, p. 309-317. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Wagner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 117.Reference deleted.
- 118.Lodish, H. F. 1970. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J. Mol. Biol. 50:689-702. [DOI] [PubMed] [Google Scholar]
- 119.Lodmell, J. S., and A. E. Dahlberg. 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277:1262-1267. [DOI] [PubMed] [Google Scholar]
- 120.Lorsch, J. R., and D. Herschlag. 1998. The DEAD-box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37:2180-2193. [DOI] [PubMed] [Google Scholar]
- 121.Low, S. C., and M. J. Berry. 1996. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biol. Sci. 21:203-208. [PubMed] [Google Scholar]
- 122.Lu, J., H. Aoki, and M. C. Ganoza. 1999. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int. J. Biochem. Cell Biol. 31:215-229. [DOI] [PubMed] [Google Scholar]
- 123.Manderschied, U., S. Bertram, and H. G. Gassen. 1978. Initiator-tRNA recognized a tetranucleotide codon during the 30S initiation complex formation. FEBS Lett. 90:162-166. [DOI] [PubMed] [Google Scholar]
- 124.McCutcheon, J. P., R. K. Agrawal, S. M. Phillips, R. A. Grassucci, S. E. Gerchman, W. M. Clemons, Jr., V. Ramakrishnan, and J. Frank. 1999. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. USA 96:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Merryman, C., D. Moazed, J. McWhirter, and H. F. Noller. 1999. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285:97-105. [DOI] [PubMed] [Google Scholar]
- 126.Methot, N., A. Pause, J. W. B. Hershey, and N. Sonenberg. 1994. The translation initiation factor eIF4B contains an RNA binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 14:2307-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miyazaki, M., M. Uritani, and H. Kagiyama. 1988. The yeast peptide elongation factor 3 (EF3) carries an active site of ATP hydrolysis which can interact with various nucleoside triphosphates in the absence of ribosomes. J. Biochem. (Tokyo) 104:445-450. [DOI] [PubMed] [Google Scholar]
- 128.Moazed, D., and H. F. Noller. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342:142-148. [DOI] [PubMed] [Google Scholar]
- 129.Moazed, D., R. R. Samaha, C. O. Gualerzi, and H. F. Noller. 1995. Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol. 248:207-210. [DOI] [PubMed] [Google Scholar]
- 130.Moazed, D., S. Stern, and H. F. Noller. 1986. Rapid chemical probing of conformations in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 187:399-417. [DOI] [PubMed] [Google Scholar]
- 131.Morimyo, M., K. Mita, T. Higashi, K. Sugaya, E. Hongo, M. Ajimura, S. Sasanuma, T. Nohata, T. Kimura, H. Inoue, Y. Ishihara, and S. Koike. 1997. Schizosaccharomyces pombe. Tanpakushitsu Kakusan Koso 42(17 Suppl.):2920-2926. [PubMed] [Google Scholar]
- 132.Munson, L. M., G. D. Stormo, and R. L. Niece. 1984. lacZ translation initiation mutations. J. Mol. Biol. 177:663-683. [DOI] [PubMed] [Google Scholar]
- 133.Murgola, A. J., A. L. Arkov, N. S. Chernyaeva, K. O. F. Hedenstierna, and F. T. Pagel. 2000. rRNA functional sites and structures for peptide chain termination, p. 509-518. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 134.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muth, G. W., S. A. Ortoleva-Donnelly, and S. A. Strobel. 2000. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289:947-950. [DOI] [PubMed] [Google Scholar]
- 136.Nakamura, Y., and K. Ito. 1998. How protein reads the stop codon and terminates translation. Genes Cells 3:265-278. [DOI] [PubMed] [Google Scholar]
- 137.Nakamura, Y., Y. Kawazu, M. Uno, K. Yoshimura, and K. Ito. 2000. Genetic probes to bacterial release factors: tRNA mimicry hypothesis and beyond, p. 519-526. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 138.Nazar, R. N. 1980. A 5.8S rRNA-like sequence in prokaryotic 23S rRNA. FEBS Lett. 119:212-214. [DOI] [PubMed] [Google Scholar]
- 139.Neilson, T., E. C. Kofoid, and M. C. Ganoza. 1980. Association of a synthetic precistronic region with 70S ribosomes is enhanced by an intact initiation triplet and a sequence complementary to the 3′-terminus of 16S rRNA. Nucleic Acids Res. Symp. Ser. 7:313-323. [PubMed] [Google Scholar]
- 140.Nierhaus, K. H., M. Schulze, and B. C. Cooperman. 1980. Molecular mechanisms of the ribosomal peptidyl transferase. Biol. Chem. Int. 1:185-192. [Google Scholar]
- 141.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 142.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. F. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EFTu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 143.Noller, H. F. 1984. Structure of ribosomal RNA. Annu. Rev. Biochem. 53:119-162. [DOI] [PubMed] [Google Scholar]
- 144.Noller, H. F. 1991. Ribosomal RNA and translation. Annu. Rev. Biochem 60:191-227. [DOI] [PubMed] [Google Scholar]
- 145.Noller, H. F., V. Hoffarth, and L. Zimniak. 1992. Unusual resistance of peptidyl tranferase to protein extraction procedures. Science 256:1416-1419. [DOI] [PubMed] [Google Scholar]
- 146.Noller, H. F., D. Moazed, S. Stern, T. Powers, P. N. Allen, J. M. Robertson, B. Weiser, and K. Triman. 1990. Structure of rRNA and its functional interactions in translation, p. 73-92. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 147.O'Connor, M., T. Asai, C. L. Squires, and A. E. Dahlberg. 1999. Enhancement of translation by the downstream box does not involve mRNA-rRNA base pairing. Proc. Natl. Acad. Sci. USA 96:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Odom, O. W., W. D. Picking, and B. Hardesty. 1990. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry 29:10734-10744. [DOI] [PubMed] [Google Scholar]
- 149.Olsen, G., and C. R. Woese. 1997. Archaeal genomics: an overview. Cell 89:991-994. [DOI] [PubMed] [Google Scholar]
- 150.Ouzounis, C., and N. Kyrpides. 1996. The emergence of major cellular processes in evolution. FEBS Lett. 390:119-123. [DOI] [PubMed] [Google Scholar]
- 151.Park, M. H., E. C. Wolff, and J. E. Folk. 1993. Hypusine: its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors 4:95-104. [PubMed] [Google Scholar]
- 152.Peat, T. S., J. Newman, G. S. Waldo, J. Berendzen, and T. C. Terwilliger. 1998. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.7Å resolution. Structure 6:1207-1214. [DOI] [PubMed] [Google Scholar]
- 153.Pel, H. J., C. Maat, M. Rep, and L. A. Grivell. 1992. The yeast nucleic gene MRF1 encodes a mitochondrial peptide chain release fetor and cures several mitochondrial RNA splicing defects. Nucleic Acids Res. 20:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Piccirilli, J. A., T. S. McConnell, A. J. Zaug, H. F. Noller, and T. R. Cech. 1992. Aminoacyl esterase activity of the Tetrahymena ribozyme. Science 256:1420-1424. [DOI] [PubMed] [Google Scholar]
- 154a.Polacek, N., M. Gaynor, A. Yassin, and A. S. Mankin. 2001. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411:498-501. [DOI] [PubMed] [Google Scholar]
- 155.Poole, E. S., C. M. Brown, and W. P. Tate. 1995. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 14:151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155a.Poole, E. S., L. L. Major, S. A. Mannering, and W. P. Tate. 1998. Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res. 26:954-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Powers, T., and H. F. Noller. 1994. The 530 loop of 16S rRNA: a signal to EFTu? Trends Genet. 10:27-31. [DOI] [PubMed] [Google Scholar]
- 157.Prescott, C., L. Krabben, and K. H. Nierhaus. 1991. Ribosomes containing the C-1054 mutation in E. coli 16S rRNA act as suppressors of all three nonsense codons. Nucleic Acids Res. 19:5281-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raache, I. D. 1971. Stereochemistry of the puromycin reaction. Biochem. Biophys. Res. Commun. 43:168-173. [DOI] [PubMed] [Google Scholar]
- 159.Raué, H. A., W. Musters, C. A. Rutgers, J. Van Riet, and R. J. Planta. 1990. rRNA: from structure to function, p. 217-235. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 160.Resch, A., K. Tedin, A. Grundling, A. Mundlein, and U. Blasi. 1996. Downstream box-anti downstream box interactions are dispensable for translation initiation of leaderless mRNAs. EMBO J. 15:4740-4748. [PMC free article] [PubMed] [Google Scholar]
- 161.Ringquist, S., S. Shinedling, D. Barrick, L. Green, Binkley, G. D. Stormo, and L. Gold. 1992. Translation initiation in Escherichia coli: sequences within the ribosome binding site. Mol. Microbiol. 6:1219-1229. [DOI] [PubMed] [Google Scholar]
- 162.Ringquist, S., T. Jones, E. E. Snyder, T. Gibson, I. Boni, and L. Gold. 1995. High-affinity RNA ligands to Escherichia coli ribosomes and ribosomal protein S1: comparison of natural and unnatural binding sites. Biochemistry 34:3640-3648. [DOI] [PubMed] [Google Scholar]
- 163.Robertson, J. M., and W. Wintermeyer. 1987. Mechanism of ribosomal translocation. tRNA binds transiently to an exit site before leaving the ribosome during translocation. J. Mol. Biol. 195:525-540. [DOI] [PubMed] [Google Scholar]
- 164.Rodnina, M. V., T. Pape, R. Fricke, L. Kuhn, and W. Wintermeyer. 1996. Initial binding of the elongation factor Tu∗GTP∗aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem. 271:646-652. [DOI] [PubMed] [Google Scholar]
- 165.Rodnina, M. V., A. Savelsbergh, V. I. Katunin, and W. Wintermeyer. 1997. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385:37-41. [DOI] [PubMed] [Google Scholar]
- 166.Rodnina, M. V., A. I. Serebryanik, G. V. Ovcharenko, and A. V. El'Skaya. 1994. ATPase strongly bound to higher eukaryotic ribosomes. Eur. J. Biochem. 225:305-310. [DOI] [PubMed] [Google Scholar]
- 167.Rodriguez-Fonseca, D., R. Amils, and R. A. Garrett. 1995. Fine structure of the peptidyl transferase centre on 23S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol. 247:224-235. [DOI] [PubMed] [Google Scholar]
- 168.Rolland, N., L. Janosi, M. A. Block, A. Shuda, E. Teyssier, C. Miege, C. Cheniclet, J. Carde, A. Kaji, and J. Joyard. 1999. Plant ribosome recycling factor homologue is a chloroplastic protein and is bacterial in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl. Acad. Sci. USA 96:5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sacerdot, C., G. Vachon, S. Laalami, F. Morel-Deville, Y. Cenatiempo, and M. Grunberg-Manago. 1992. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational codons for IF2 beta. J. Mol. Biol. 225:67-80. [DOI] [PubMed] [Google Scholar]
- 170.Samaha, R. R., R. Green, and M. F. Noller. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377:309-314. [DOI] [PubMed] [Google Scholar]
- 171.Sandbaken, M. G., J. A. Lupisella, B. DiDomenico, and K. Chakraburtty. 1990. Protein synthesis in yeast. Structural and functional analysis of the gene encoding elongation factor 3. J. Biol. Chem. 265:15838-15844. [PubMed] [Google Scholar]
- 172.Sarabhai, A., and S. Brenner. 1967. A mutant which reinitiates the polypeptide chain after chain termination. J. Mol. Biol. 27:145-162. [DOI] [PubMed] [Google Scholar]
- 173.Saruyama, H., and K. H. Nierhaus. 1986. Evidence that the three-site model for the ribosomal elongation cycle is also valid in the archaebacterium Halobacterium halobium. Mol. Gen. Genet. 204:221-228. [Google Scholar]
- 173a.Schmeing, T. M., A. C. Seila, J. L. Hansen, B. Freeborn, J. K. Soukup, S. A. Scaringe, S. A. Strobel, P. B. Moore, and T. A. Steitz. 2002. A pre-translocation intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9:225-230. [DOI] [PubMed] [Google Scholar]
- 174.Schnier, J., H. G. Schwelberger, Z. Smit-McBride, H. A. Kang, and J. W. B. Hershey. 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3105-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schulze, H., and K. H. Nierhaus. 1982. Minimal set of ribosomal components for reconstitution of the peptidyl transferase activity. EBMO J. 1:609-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175a.Seit Nebi, A., L. Frolova, N. Ivanova, A. Poltaraus, and L. Kisselev. 2001. Substitutions of the glutamine residue in ubiquitous GGQ tripeptide in human eRF1 do not entirely abolish the release factor activity. Mol. Biol. 34:849-900. [PubMed] [Google Scholar]
- 175b.Shimuzu, Y., A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa, T. Ueda. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19:751-755. [DOI] [PubMed] [Google Scholar]
- 176.Shine, J., and I. Dalgarno. 1975. Determinants of cistron specificity in bacterial ribosomes. Nature 254:34-38. [DOI] [PubMed] [Google Scholar]
- 177.Skogerson, L., and D. Engelhardt. 1977. Dissimilarity in protein chain elongation factor requirements between yeast and rat liver ribosomes. J. Biol. Chem. 252:1471-1475. [PubMed] [Google Scholar]
- 178.Smit-McBride, Z., T. E. Deve, W. C. Merrick, and J. W. B. Hershey. 1989. Protein synthesis initiation factor eIF-4D (eIF-5A). J. Biol. Chem. 264:1578-1583. [PubMed] [Google Scholar]
- 179.Song, H., P. Mugnier, A. K. Das, H. M. Webb, D. R. Evans, M. F. Tuite, B. A. Hemmings, and D. Barford. 2000. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100:311-321. [DOI] [PubMed] [Google Scholar]
- 180.Spahn, C., and C. Prescott. 1997. Throwing a spanner in the works: antibiotics and the translation apparatus. J. Mol. Med. 74:423-439. [DOI] [PubMed] [Google Scholar]
- 181.Spahn, C. M., and K. H. Nierhaus. 1998. Models of the elongation cycle: an evaluation. Biol. Chem. 379:753-772. [DOI] [PubMed] [Google Scholar]
- 182.Spahn, C. M. T., M. A. Spahn, M. A. Shäfer, A. A. Krayevsky, and K. H. Nierhaus. 1996. Conserved nucleotides of 23S rRNA located at the ribosomal peptidyl transferase center. J. Biol. Chem. 271:32857-32862. [DOI] [PubMed] [Google Scholar]
- 182a.Spangler, E. A., and E. H. Blackburn. 1985. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics puromycin and hygromycin. J. Biol. Chem. 260:6334-6340. [PubMed] [Google Scholar]
- 183.Spirin, A. S. 1985. Ribosomal translocation: facts and models. Prog. Nucleic Acid Res. Mol. Biol. 32:75-114. [DOI] [PubMed] [Google Scholar]
- 184.Sprengart, M. L., E. Fuchs, and A. G. Porter. 1996. The downstream box: an efficient and independent tranlation initiation signal in Escherichia coli. EMBO J. 15:660-674. [PMC free article] [PubMed] [Google Scholar]
- 185.Steitz, J. A. 1979. Genetic signals and nucleotide sequence in messenger RNA, p. 349-399. In R. F. Goldberger (ed.), Biological regulation and development. Plenum Press, New York, N.Y.
- 186.Story, R. M., H. Li, and J. N. Abelson. 2001. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 98:1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Subramanian, A. R. 1984. Structure and function of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28:101-142. [DOI] [PubMed] [Google Scholar]
- 188.Subramanian, A. R. 1984. Structure and function of the largest Escherichia coli ribosomal protein. Trends Biochem. Sci. 9:491-494. [Google Scholar]
- 189.Symons, R. H., R. J. Harris, P. Greenwell, D. J. Eckermann, and E. F. Vanin. 1978. The use of puromycin analogs and related compounds to probe the active centre of peptidyl transferase on Escherichia coli ribosomes. Bioorg. Chem. 4(Suppl.):409-439. [Google Scholar]
- 190.Taniguchi, T., and C. Weissman. 1978. Site-directed mutations in the initiator region of the bacteriophage Qβ coat cistron and their effect on ribosome binding. J. Mol. Biol. 118:533-565. [Google Scholar]
- 191.Tate, W. P. and C. M. Brown. 1992. Perspectives in biochemistry. Translational terminations “stop” for protein synthesis or “pause” for regulation of gene expression. Biochemistry 31:2443-2450. [DOI] [PubMed] [Google Scholar]
- 191a.Thompson, J., D. F. Kim, M. O'Connor, K. R. Lieberman, M. A. Bayfield, S. T. Gregory, R. Green, H. F. Noller, and A. E. Dahlberg. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. USA 98:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Toone, M. W., K. E. Rudd, and J. D. Friesen. 1991. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress mutations in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 173:3291-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Traut, R. R., J. M. Lambert, G. Boileau, and J. W. Kenny. 1980. Protein topography of Escherichia coli ribosomal subunits as inferred from protein crosslinking, p. 89-110. In G. Chambliss, G. R. Craven, J. Davies, K. Davis, L. Kahn, and M. Nomura (ed.), Ribosomes. Structure, function and genetics. University Park Press, Baltimore, Md.
- 194.Triana-Alonso, F. J., K. Chakraburtty, and K. H. Nierhaus. 1995. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270:20473-20478. [DOI] [PubMed] [Google Scholar]
- 195.Uritami, M., and M. Miyazaki. 1988. Role of yeast peptide elongation factor 3 (EF3) at the AA-tRNA binding steps. J. Biochem. (Tokyo) 104:118-126. [DOI] [PubMed] [Google Scholar]
- 196.van Duin, J., G. P. Overbeek, and C. Backendorf. 1980. Functional recognition of phage RNA by 30S ribosomal subunits in the absence of initiator tRNA. Eur. J. Biochem. 110:593-597. [DOI] [PubMed] [Google Scholar]
- 197.Van Ryk, D. L., and A. E. Dahlberg. 1995. Structural changes in the 530 loop of Escherichia coli 16S RNA in mutants with impaired translational fidelity. Nucleic Acids Res. 23:3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197a.Vestgaard, B., L. van Bich, G. R. Andersen, J. Nyborg, R. H. Buckingham, and M. Kjeldgaard. 2001. Bacterial polypeptide release factor RF2 is structurally distinct from eRF1. Mol. Cell 8:1375-1382. [DOI] [PubMed] [Google Scholar]
- 198.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding motif. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Welch, E. M., W. Wang, and S. W. Peltz. 2000. Translation termination: it is not the end of the story, p. 467-485. In N. Sonenberg, J. W. H. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 200.Wilms, C., J. W. Noah, D. Zhong, and P. Wollenzien. 1997. Exact determination of UV crosslinks in 16S ribosomal RNA in 30S ribosomal subunits. RNA 3:602-612. [PMC free article] [PubMed] [Google Scholar]
- 201.Wilson, D. N., M. E. Dalphin, H. J. Pel, L. L. Major, J. B. Mansell, and W. P. Tate. 2000. Factor-mediated termination of protein synthesis: a welcome return to the mainstream of translation, p. 495-508. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 202.Wilson, K. S., and H. F. Noller. 1998. Molecular movement inside the translational engine. Cell 92:337-349. [DOI] [PubMed] [Google Scholar]
- 203.Woese, C. 1970. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature 226:817-820. [DOI] [PubMed] [Google Scholar]
- 204.Ypma-Wong, M. F., W. A. Fonzi, and P. S. Sypherd. 1992. Fungus-specific translation elongation factor 3 gene, is present in Pneumocystis carinii. Infect. Immun. 60:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204a.Yusupov, M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 204b.Zavialov, A. V., R. H. Buckingham, and M. Ehrenberg. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107:115-124. [DOI] [PubMed] [Google Scholar]
- 205.Zhang, B., and T. R. Cech. 1997. Peptide bond formation by in vitro selected ribozymes. Nature 390:96-100. [DOI] [PubMed] [Google Scholar]