Abstract
Filamentous fungi are unique organisms—rivaled only by actinomycetes and plants—in producing a wide range of natural products called secondary metabolites. These compounds are very diverse in structure and perform functions that are not always known. However, most secondary metabolites are produced after the fungus has completed its initial growth phase and is beginning a stage of development represented by the formation of spores. In this review, we describe secondary metabolites produced by fungi that act as sporogenic factors to influence fungal development, are required for spore viability, or are produced at a time in the life cycle that coincides with development. We describe environmental and genetic factors that can influence the production of secondary metabolites. In the case of the filamentous fungus Aspergillus nidulans, we review the only described work that genetically links the sporulation of this fungus to the production of the mycotoxin sterigmatocystin through a shared G-protein signaling pathway.
INTRODUCTION
Fungi are remarkable organisms that readily produce a wide range of natural products often called secondary metabolites. In this review we will use the terms interchangeably. In many cases, the benefit these compounds confer on the organism is unknown (21). However, interest in these compounds is considerable, as many natural products are of medical, industrial and/or agricultural importance. Some natural products are deleterious (e.g., mycotoxins), while others are beneficial (e.g., antibiotics) to humankind (35). Although it has long been noted that biosynthesis of natural products is usually associated with cell differentiation or development, and in fact most secondary metabolites are produced by organisms that exhibit filamentous growth and have a relatively complex morphology, until recently the mechanism of this connection was not clear. A critical advance in this regard was the establishment of a G-protein-mediated growth pathway in Aspergillus nidulans that regulates both asexual sporulation and natural product biosynthesis (55). Since then, several studies have provided insight into other molecules and pathways that link chemical and morphological differentiation processes in fungi. The focus of this review is to provide an overview of the research establishing this connection, and this review covers the progress made to date in elucidating relationships between natural product metabolism and fungal development. We conclude with speculation as to why such a relationship could be of value to the organism.
SECONDARY METABOLISM IS ASSOCIATED WITH DEVELOPMENTAL PROCESSES
Secondary metabolism is commonly associated with sporulation processes in microorganisms (56, 77, 113), including fungi (21, 103). Secondary metabolites associated with sporulation can be placed into three broad categories: (i) metabolites that activate sporulation (for example, the linoleic acid-derived compounds produced by A. nidulans [23, 26, 27, 79]), (ii) pigments required for sporulation structures (for example melanins required for the formation or integrity of both sexual and asexual spores and overwintering bodies [6, 63]), and (iii) toxic metabolites secreted by growing colonies at the approximate time of sporulation (for example the biosynthesis of some deleterious natural products, such as mycotoxins [55, 120]). These and other examples of secondary metabolites that fit into these categories are shown in Table 1.
TABLE 1.
Fungal secondary metabolites associated with sporulation processes and development
| Secondary metabolite | Producing fungus | Association with development | Reference(s) |
|---|---|---|---|
| Linoleic-acid derived psi factor | Aspergillus nidulans | Induces sporulation; affects ratio of asexual to sexual spore development | 23, 26, 27, 79 |
| Zearalenone | Fusarium graminearum | Induces sporulation; enhances perithecial formation | 125 |
| Butyrolactone I | Aspergillus terreus | Induces sporulation and lovastatin production | 102 |
| Melanin | Colletotrichum lagenarium | Associated with appressorial formation | 116 |
| Melanin | Alternaria alternata | UV protection of spore | 63 |
| Melanin | Cochliobolus heterotrophus | Required for spore survival | 72 |
| Spore pigment | Aspergillus fumigatus | Required for virulence | 121 |
| Mycotoxins | Aspergillus spp. | Produced after sporulation | 12, 49, 98 |
| Patulin | Penicillium urticae | Antibiotic; produced after sporulation | 103 |
Secondary Metabolites That Are Sporogenic Factors
Secondary metabolite production usually commences late in the growth of the microbe, often upon entering the stationary or resting phase (21). In early observations it was noted that the environmental conditions required for sporulation and secondary metabolism were often similar and were more stringent than those for pure vegetative growth (21, 51, 103). Although it was once thought that natural products were essential for sporulation, there are many examples of fungal strains that still sporulate but are deficient in secondary metabolite production, for example, Penicillium urticae patulin mutants (103) and A. nidulans sterigmatocystin mutants (106). In addition, some secondary metabolites such as brevianamides A and B produced by Penicillium brevicompactum appear only after conidiation has commenced (14). It is possible, however, that some natural products have subtle effects on sporulation, as recent studies of A. nidulans sterigmatocystin mutants indicate that they exhibit a decrease in asexual spore production not detectable by the unaided eye (97, 108).
Some secondary metabolites have easily observable effects on morphological differentiation in fungi. Several studies show that compounds excreted by mycelium can induce asexual and sexual sporulation in other fungi; this phenomenon operates across species and genera (51, 88). In most cases, these compounds have not been identified but are presumed to be natural products produced as the mycelia ages. Other natural products affecting fungal development are better characterized. Fusarium graminearum produces an estrogenic mycotoxin called zearalenone that enhances perithecial production in F. graminearum. The addition of dichloros, an inhibitor of zearalenone synthesis, inhibits the sexual development of this fungus (125). Butyrolactone I, an inhibitor of eukaryotic cyclin-dependent kinases produced by Aspergillus terreus, increases hyphal branching, sporulation, and production of another secondary metabolite, lovastatin, in this fungus (102). Butyrolactone-containing molecules act as self-regulating factors in some bacteria and control many biological functions such as antibiotic and virulence factor production (33). In A. nidulans, endogenous oleic acid- and linoleic acid-derived molecules called psi factor govern the ratio of asexual to sexual spores in this organism (23, 26).
Pigments
Some fungal pigments are natural products associated with developmental structures, the most common being melanins. Melanins are generally dark brown pigments formed by oxidative polymerization of phenolic compounds and are synthesized during spore formation for deposition in the cell wall. Melanin biosynthesis has been well studied in pathogenic fungi, where the pigment not only contributes to the survival of the fungal spore by protecting against damaging UV light but is also an important virulence factor.
In Colletotrichum lagenarium, melanin biosynthesis has been associated with the formation of appressoria (116). Appressoria are infection structures required for host penetration, and any impairment in their formation can reduce virulence. In Alternaria alternata, melanin deposition is also involved in spore development. Disruption of the A. alternata melanin biosynthetic gene brm2 dramatically decreases melanin production in this fungus. The conidia produced are reduced in diameter and are more sensitive to UV light than the wild type (63). In another fungus, the maize pathogen Cochliobolus heterotrophus, fitness studies of albino spore mutants in a greenhouse indicated that melanin production is required for survival of this fungus (72).
The effect of melanin biosynthesis on the virulence of fungal human pathogens has also been studied. In Aspergillus fumigatus, a cause of invasive aspergillosis in immunocompromised patients, spore pigment is a virulence factor. Disruption of the alb1 gene, which encodes a putative polyketide synthase, creates a pigmentless conidial phenotype and leads to a significant reduction in fungal infection of a murine model (121).
Mycotoxins
The most deleterious of natural products, in terms of health effects, are the mycotoxins. A relationship between mycotoxin production and sporulation has been documented in several mycotoxigenic genera. For example, in Aspergillus parasiticus certain chemicals that inhibit sporulation have also been shown to inhibit the production of aflatoxin (98). In A. parasiticus and A. nidulans chemical inhibition of polyamine biosynthesis inhibits sporulation and aflatoxin and sterigmatocystin production, respectively (49). Several papers have shown that Aspergillus mutants deficient in sporulation are also unable to produce aflatoxin (see reference 12 and references therein). In 1977, it was shown that Claviceps purpurea mutants lacking the ability to form conidia on agar plates also produce less toxic alkaloids (93). In the same year, Sekiguchi and Gaucher (103) reported that P. urticae NRRL 2159A mutants blocked at an early stage of conidiation produced markedly less patulin under growth conditions which normally supported patulin biosynthesis. This study suggests an indirect effect of sporulation on patulin biosynthesis (although patulin biosynthetic mutations have no effect on conidiation [103]). In Fusarium, the case for the existence of a genetic connection between sporulation and mycotoxin production has recently been identified. A mutation in the Fusarium verticillioides FCC1 gene results in reduced sporulation and reduced fumonisin B1 biosynthesis (104). Also, introducing a dominant-activated allele of an A. nidulans α G-protein into Fusarium sporotrichioides produces strains with elevated production of T-2 mycotoxin but reduced conidium formation (115).
REVIEW OF ASPERGILLUS DEVELOPMENT AND MYCOTOXIN PRODUCTION
Aspergillus Developmental Structures
To fully understand the relationship between sporulation and mycotoxin biosynthesis, we will now briefly review these developmental processes in Aspergillus with a focus on A. nidulans. Most Aspergillus spp. propagate solely by asexual spores called conidia (e.g., Aspergillus flavus and A. parasiticus), while other species produce both conidia and sexual spores called ascospores (e.g., A. nidulans, teleomorph: Emericella nidulans). A. nidulans conidia are formed on specialized complex structures called conidiophores, while ascospores are produced inside spherical sexual fruiting bodies called cleistothecia. Most asexual species, including A. flavus and A. parasiticus, form resistant structures called sclerotia. For Aspergillus species that are problematic opportunistic pathogens of plants and animals (including humans), conidia serve as the major source of inoculum.
The formation of conidiophores begins with a stalk that extends from a thick-walled foot cell. The tip of the stalk begins to swell, forming a vesicle. From the vesicle, cells called sterigmata are formed (31, 81, 86), and chains of conidia originate from the sterigmata. Genetic and molecular studies of conidial reproduction in A. nidulans identified a gene, brlA (31), that encodes a transcriptional regulator proposed to govern the activation of developmental genes at the time of vesicle formation (2, 3). Loss-of-function brlA mutations result in no conidiation and abnormally long conidiophore stalks (17, 58) and do not affect sterigmatocystin production in A. nidulans (49). However, forced expression of brlA from an inducible promoter in vegetative cells causes immediate activation of conidiation-specific genes, resulting in conidiophore production (8). The effect of brlA overexpression on sterigmatocystin production has not been investigated. Both conidiation and sterigmatocystin biosynthesis are coregulated, at a step prior to brlA expression, by a common signal transduction pathway. This regulatory pathway will be the subject of our review in sections below.
Aflatoxin and Sterigmatocystin Biosynthesis
Since the discovery of aflatoxin and its deleterious properties to humans and animals, efforts have been directed toward the understanding of the molecular mechanisms leading to its biosynthesis. The last decade has seen significant progress in characterizing the genes required for aflatoxin biosynthesis in A. flavus and A. parasiticus and sterigmatocystin biosynthesis in A. nidulans (sterigmatocystin is the penultimate precursor in the aflatoxin biosynthetic pathway).
Genes required for the synthesis of aflatoxin and sterigmatocystin are well conserved between aspergilli and are located in large gene clusters (18, 120, 130). The relative order and transcriptional direction of some of the homologous gene pairs though are not conserved (64). Thus far, most of the genes in the respective clusters have been shown to encode enzymes required for toxin biosynthesis (64). At least two genes, aflR and aflJ, play a regulatory role (80, 91, 131). Several studies of aflR have demonstrated it encodes a sterigmatocystin/aflatoxin pathway-specific transcription factor (29, 119, 126, 131). aflR deletion mutants in all three Aspergillus spp. do not express biosynthetic genes in the sterigmatocystin-aflatoxin cluster nor do they produce sterigmatocystin or aflatoxin (29, 92, 131). Loss of aflR does not stop spore or sclerotial production but does affect the numbers of spores (97, 108) and sclerotia (25, 120) formed. The function of aflJ is not well defined, but disruption of aflJ in A. flavus results in a failure to produce aflatoxin (80).
Environmental Factors That Affect Mycotoxin Production and Development in Aspergillus spp.
Some environmental factors affect mycotoxin production, conidiation, and cleistothecial and sclerotial production concurrently. Physical parameters affecting mycotoxin and/or spore production in Aspergillus spp. include temperature (42), availability of an air-surface interface (50), and pH (20, 32). Nutritional factors such as carbon source and nitrogen source can also affect both mycotoxin production and morphological differentiation (65). Additionally, some compounds present in seeds commonly infected by Aspergillus species can influence both toxin production and fungal development (22, 24, 48, 135).
Effects of pH
Although aflatoxin and sterigmatocystin production, like that of penicillin, appears to be influenced by growth medium pH, studies addressing pH regulation of these mycotoxins have produced complex and at times contradictory results. Cotty (32) determined a link between aflatoxin production and sclerotial morphogenesis based on changes of both chemical and morphological differentiation in response to pH. At pH 4.0 or below, sclerotial production is reduced by 50% in A. flavus while aflatoxin production is maximal (32). However, Buchanan and Ayres (20) concluded from their work that the initial pH of growth medium was not an important determinant for mycotoxin production. In fact, the effect of pH on aflatoxin biosynthesis is dependent on the composition of growth media. Keller et al. (65) showed that A. nidulans and A. parasiticus produced less mycotoxin as the pH of the growth medium increased. In addition, pH-sensing mutants of A. nidulans exhibit both sporulation (94, 118) and sterigmatocystin production aberrations (94). Further research is necessary to elucidate the complex interactions between pH and other environmental factors that influence morphological and chemical differentiation in Aspergillus.
Effects of Carbon and Nitrogen Source
Availability and type of carbon and nitrogen source affect sterigmatocystin and aflatoxin production. Simple sugars such as glucose, fructose, sucrose, and sorbitol as sole carbon sources support high fungal growth, sporulation, and aflatoxin production (1, 19, 59, 75, 76). In contrast, peptone (1, 19, 42, 75) and the more-complex sugars like galactose, xylose, mannitol, and lactose (59) do not support aflatoxin production well.
The choice of nitrogen source used in the growth medium can have different effects on sterigmatocystin and aflatoxin production in different Aspergillus species. Nitrate as the nitrogen source has been shown by some groups to repress the synthesis of aflatoxin intermediates in A. parasiticus (59, 85) but enhance sterigmatocystin production in A. nidulans (42). Feng and Leonard (42) also observed no sterigmatocystin production in ammonium-containing media. Other studies (65, 82) indicate sterigmatocystin and aflatoxin production increases in ammonium-based media and decreases in nitrate-based medium. Nitrogen source influences not only mycotoxin production but also the formation of developmental structures in Aspergillus spp. Studies in which A. flavus is grown on agar media containing either nitrate or ammonium as the sole nitrogen source have shown that development of sclerotia occurs on nitrate but not on ammonium (13).
Effects of Host Seed Constituents
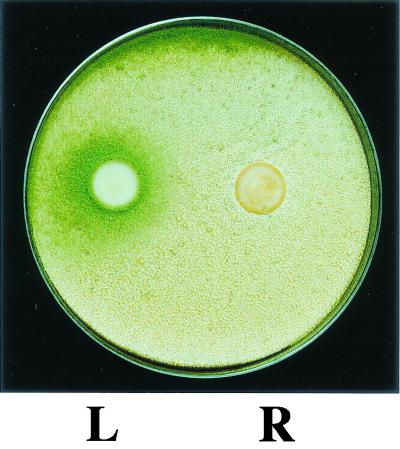
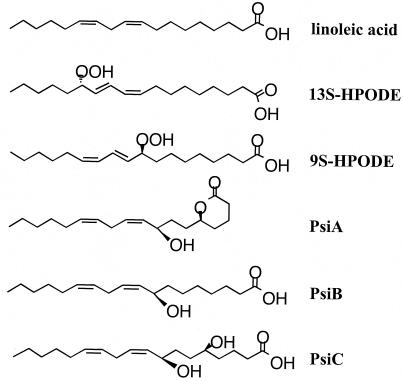
In A. nidulans, linoleic acid is metabolized into a series of sporogenic molecules called psi factors (27). Calvo et al. (24) have demonstrated that linoleic acid and the hydroperoxylinoleic acid derivatives from seeds increase asexual spore production in A. flavus, A. parasiticus, and A. nidulans. Figure 1 demonstrates how the addition of 13S-hydroperoxylinoleic acid to growth medium can stimulate conidiation of A. flavus. The seed fatty acids closely resemble psi factors in structure (Fig. 2). As discussed earlier, psi factors are fatty acid-derived compounds synthesized endogenously by Aspergillus (27) that influence the development of this fungus. The results presented above suggest that the sporogenic effect of seed fatty acids could take place through interference and/or mimicking of psi factor.
FIG. 1.
Sporogenic effect of linoleic acid-derived compounds on Aspergillus. Conidia of A. flavus were spread evenly over YGT medium (26) at a concentration of 106 spores/ml. Discs containing 1 mg of 13S-hydroperoxylinoleic acid (L) or solvent control (R) were placed on the surface, and the plate was incubated at 29°C for 2 days. Conidium production is induced by 13S-hydroperoxylinoleic acid (24).
FIG. 2.
Structures of linoleic acid and linoleic acid-derived molecules from seed-, 13S-hydroperoxylinoleic acid (13S-HPODE)- and 9S-HPODE-, and linoleic acid-derived sporogenic molecules from A. nidulans, psiA, psiB, and psiC.
Linoleic acid also supports aflatoxin production, whereas the hydroperoxylinoleic acid derivatives and downstream breakdown products are reported to inhibit (22, 36, 48, 127) or stimulate (22, 34, 89) aflatoxin production. This differential effect on aflatoxin production might be associated with the placement of functional groups on the carbon backbone of fatty acid derivatives. For example, Burow et al. (22) have shown specifically that treatment of Aspergillus with 13S-hydroperoxylinoleic acid decreases the accumulation of aflatoxin biosynthetic gene transcripts. On the other hand, 9S-hydroperoxylinoleic acid lengthens the time aflatoxin and sterigmatocystin transcripts accumulate.
Many other plant compounds have been reported to affect fungal growth and aflatoxin production. For a review of these metabolites, see reference 134.
SIGNAL TRANSDUCTION PATHWAY
The previous section demonstrates that interactions between sporulation and mycotoxin production in relation to environmental factors are not understood at a genetic level. Presumably, the Aspergillus fungal cell receives many developmental and mycotoxin-stimulating signals from the environment. The detection and integration of these signals leading to a comprehensive response by the fungus could involve many genetic elements. However, once these signals are transmitted into the fungal cell, it appears that a common signal transduction pathway is partially responsible for tying fungal development to natural product biosynthesis. A major contribution to the elucidation of this genetic connection between fungal development and biosynthesis of natural products was made in the study of Aspergillus developmental mutants first described by Bennett and collaborators (10-12), in which they observed that conidial mutants also lost the ability to produce aflatoxin. Further studies by Kale et al. (60, 61) revealed that these A. flavus and A. parasiticus developmental mutants also lack aflR and aflatoxin enzymatic gene transcripts and fail to accumulate other aflatoxin pathway intermediates. A similar phenotype was found in developmental A. nidulans mutants called “fluffy” (37, 78, 117, 123). Hicks et al. (55) demonstrated that some of these A. nidulans developmental mutants are also deficient in sterigmatocystin production. These findings led to the discovery of a signal transduction pathway regulating both conidiation and sterigmatocystin-aflatoxin biosynthesis.
Overview of Signal Transduction in Fungi
In Saccharomyces cerevisiae, several signaling pathways operate to regulate filamentous growth and differentiation (71). One prominent pathway involves components of the mitogen-activated protein (MAP) kinase pathway that is activated in response to pheromones and nutrition availability. In haploid S. cerevisiae cells, the MAP kinase pathway mediates the response to pheromones during mating (71). In diploid cells, the same signaling pathway is used to stimulate filamentous growth in response to nitrogen limitation and other environmental signals (46). A second well-studied signal transduction pathway in S. cerevisiae operates through nutrient sensing and functions in parallel to the MAP kinase pathway. It involves a heterotrimeric G-protein-coupled receptor, adenylate cyclase, cyclic AMP (cAMP) signaling molecules, and protein kinase A (PKA) and regulates pseudohyphal growth (71). Upstream of PKA are at least two other small, monomeric GTP-binding proteins called Ras1 and Ras2 (71). Ras1 and Ras2 regulate cAMP production by interacting with adenylate cyclase, and Ras2 regulates the MAP kinase pathway during filamentous growth. In Schizosaccharomyces pombe, a Ras1 homolog plays a role in MAP kinase pathway activation but not in regulating cAMP production (71).
Some proteins acting downstream of PKA in the G-protein signaling pathway also interact with components of the MAP kinase pathway (66). For example, MAP kinase and cAMP signaling pathways both activate expression of the FLO11 gene. This gene encodes a cell surface protein required for pseudohyphal formation in S. cerevisiae (101). Other studies of S. cerevisiae indicate that cAMP-dependent protein kinases might positively regulate signaling in the MAP kinase cascade (83).
Protein kinase signaling pathways have also been examined in some pathogenic fungi (see reference 71 for a recent review). Briefly, in Ustilago maydis, cAMP signaling appears to control the switch between budding and filamentous growth. This switch is essential for the ability of the fungus to cause diseases of crops (47). cAMP signaling is also involved in appressorial formation in Magnaporthe grisea, a key step in the infection process of rice (see reference 66 and references therein). An α or β G-protein subunit of the human pathogen Cryptococcus neoformans regulates mating, virulence, and melanin biosynthesis (6). From these examples it is clear that the G-protein signaling pathway is essential for fungal growth and infection.
A G-Protein/cAMP/PKA Signaling Pathway Connects Sporulation and Sterigmatocystin-Aflatoxin Production in Aspergillus spp.
Characterization of fluffy mutants in Aspergillus G-protein signaling.
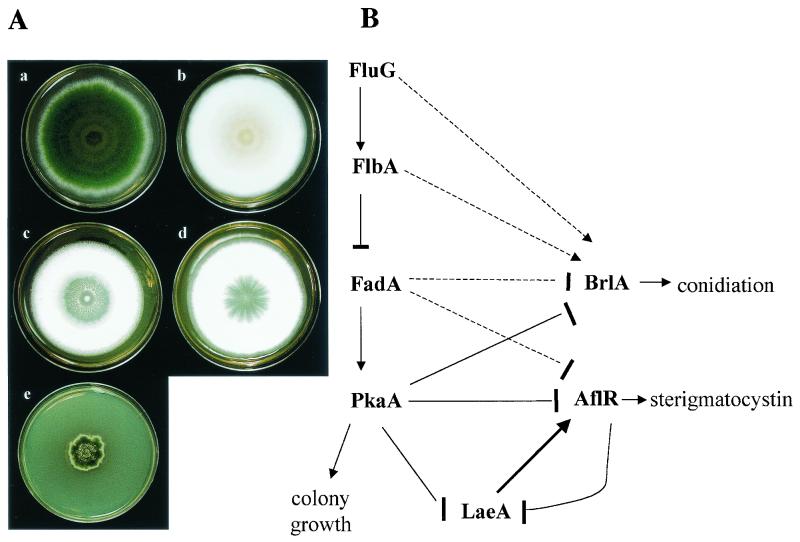
Insight into an Aspergillus G-protein signal transduction pathway originated by complementation of single-locus asexual developmental mutants called fluffy. Six fluffy loci were initially identified that were the result of recessive mutations in the fluG, flbA, flbB, flbC, flbD, and flbE genes. Fluffy mutants have similar phenotypes in being deficient in conidiophore formation and brlA expression (4, 5, 37, 70, 123). Two of these genes, fluG and flbA, were later found to regulate both asexual development and sterigmatocystin production (55), with fluG believed to act upstream of flbA (Fig. 3). fluG is involved in the synthesis of an extracellular low-molecular-weight diffusible factor that activates a pathway leading to conidiation and sterigmatocystin production (69). FluG loss-of-function mutations (e.g., ΔfluG, Table 2 and Fig. 3) present an aconidial, non-sterigmatocystin-producing phenotype (55). Overexpression of fluG in submerged culture was sufficient to induce brlA expression and subsequent conidiation (68). However, fluG overexpression was not sufficient to induce precocious sterigmatocystin production, indicating that FluG-independent regulatory elements are required in sterigmatocystin gene cluster activation (55).
FIG. 3.
(A) Phenotype effect of signaling mutants. Strains shown are wild-type A. nidulans (a), ΔfluG (b), ΔflbA (c), fadAG42R (d), and ΔpkaA (e). (B) Proposed model of G-protein signal transduction regulating sterigmatocystin production and sporulation. Arrows indicate positive regulation, and blocked arrows indicate negative regulation. Solid lines indicate known pathways. Dashed lines indicate postulated pathways. Abbreviations: FluG, early acting developmental regulator; FlbA, regulator of G-protein signaling; FadA, α subunit of G-protein; PkaA, catalytic subunit of PKA; LaeA, AflR regulator; AflR, sterigmatocystin-aflatoxin-specific transcription factor; BrlA, conidiation-specific transcription factor (55, 106).
TABLE 2.
Alleles having effects on sporulation and/or sterigmatocystin production.
| Allele(s) | Function | Mutation effect | Reference(s) |
|---|---|---|---|
| aflR | Pathway-specific regulator of stc gene expression | 91 | |
| ΔaflR | Loss of sterigmatocystin-aflatoxin production | 29, 92, 131 | |
| Affects conidiation and sclerotial development | 25, 97, 108, 120 | ||
| OEaflR | Early and increased mycotoxin production | 106 | |
| brlA | Pathway-specific regulator of conidiogenesis genes | 31 | |
| ΔbrlA | Abolishment of conidiation | 17, 58 | |
| OEbrlA | Early onset of conidiation, including sporulation in submerged liquid culture; no effect on sterigmatocystin production | 8 | |
| fluG | Involved in synthesis of extracellular, diffusible factor required for conidiation | 69 | |
| ΔfluG | Aconidial; no sterigmatocystin production | 55 | |
| OEfluG | Activates conidiation; early sterigmatocystin production observed in submerged liquid culture but not on plates | 55 | |
| flbA | Encodes RGS-containing protein; active FlbA inactivates G-protein pathway | 70 | |
| ΔflbA | Aconidial; no sterigmatocystin production | 55 | |
| OEflbA | Premature sporulation, early sterigmatocystin production | 55 | |
| fadA | α-Subunit of heterotrimeric G protein | 133 | |
| ΔfadA and fadAG203R | Premature conidiation; early sterigmatocystin production | 55 | |
| fadAG42R | Aconidial; no sterigmatocystin production | 55 | |
| pkaA | cAMP-dependent PKA | 106 | |
| ΔpkaA | Results in hypersporulation; aberrant sterigmatocystin production | 106 | |
| OE pkaA | Decreased conidiation; no sterigmatocystin production | 106 | |
| ΔpkaA ΔflbA | Double deletion mutant; partially restores some sporulation; restores sterigmatocystin production | 106 | |
| ΔpkaA fadAG42R | Partially restores sporulation but not sterigmatocystin production | 106 | |
| sfaD | β-Subunit of heterotrimeric G-protein | 99 | |
| ΔsfaD | Increased sporulation | 105 |
Δ indicates the gene has been deleted. OE indicates the gene is overexpressed. fadAG203R is the dominant negative fadA allele. fadAG42R is the dominant activated fadA allele.
FlbA loss-of-function mutants exhibit a phenotype similar to those of ΔfluG strains, and in addition, the mycelium starts to autolyze after several days of growth (55) (Table 2 and Fig. 3). The FlbA protein was found to contain a regulator of G-protein signaling (RGS) domain (70), suggesting that ΔflbA strains harbor a constitutively active G-signaling pathway responsible for the aconidial, non-sterigmatocystin-producing phenotype (Fig. 3). This was supported by the identification of another mutant, fadAG42R, presenting the same phenotype as ΔflbA. fadA is an α subunit of a heterotrimeric G protein, putatively the target for the RGS domain of flbA (133). When bound to GTP, FadA is in an active form that favors vegetative growth (Fig. 3). Developmental activation leading to sporulation and sterigmatocystin production presumably requires FlbA to enhance the intrinsic GTPase activity of FadA. The fadAG42R mutant is a dominant-activated form of FadA unable to hydrolyze GTP (55) (Table 2 and Fig. 3). Mutants overexpressing flbA or mutants carrying a fadA dominant negative allele where FadA is inactive, e.g., fadAG203R, show premature sterigmatocystin production and conidiation (55). These results support a model in which FlbA negatively regulates the FadA signaling pathway that blocks conidiation and sterigmatocystin biosynthesis (Fig. 3).
Loss-of-function flbA or fadAG42R dominant activating mutations block sterigmatocystin production and conidiation through inhibition of the expression of the sterigmatocystin gene transcription factor aflR and the asexual developmental transcription factor brlA (55, 133). Conidiation can be rescued in loss-of-function flbA mutants by forced brlA overexpression (J. Hicks and N. P. Keller, unpublished data). However, aflR overexpression failed to reestablish sterigmatocystin production in a ΔflbA background (K. Shimizu, J. Hicks, and N. P. Keller, unpublished data). These results suggest differential downstream regulation in the signal transduction pathway for brlA and aflR.
Role of pkaA, which encodes a PKA catalytic subunit.
The understanding of the mechanism by which FlbA and FadA regulate conidiation and sterigmatocystin production was recently extended by characterization of a cAMP-dependent protein kinase catalytic subunit (pkaA) (106). Deletion of pkaA leads to hyperconidiation and has little effect on sterigmatocystin production, while pkaA overexpression inhibits brlA and aflR expression and, concomitantly, conidiation and mycotoxin production—the latter being a phenotype resembling ΔflbA and dominant-activated fadAG42R mutants (106) (Table 2 and Fig. 3). Deletion of pkaA restores conidiation in the ΔflbA and fadAG42R backgrounds but restores sterigmatocystin production only in the ΔflbA but not fadAG42R background (106). These data suggest that FlbA and FadA inhibition of conidiation is mediated in part through PkaA and that FlbA has a FadA-independent, PkaA-dependent role in regulating sterigmatocystin production. In addition to transcriptionally regulating brlA and aflR, PkaA also regulates AflR and perhaps BrlA posttranscriptionally (106). Recent studies suggest that PkaA negatively regulates AflR activity through phosphorylation (J. Hicks, K. Shimizu, and N. P. Keller, Abstr. XXI Fungal Genet. Conf., p. 144, 2001). Transcriptional regulation of AflR by PkaA might be mediated by LaeA, a protein required for aflR expression, and negatively regulated by PkaA (J. W. Bok and N. P. Keller, Abstr. XXI Fungal Genet. Conf., p. 144, 2001) (Fig. 3).
Other Components of the Signal Transduction Pathway Controlling A. nidulans Development
With regards to Aspergillus development, several other signaling proteins have been identified that are involved in sporulation and colony formation. One protein, RasA, is a homolog of the yeast Ras proteins. The levels of activated RasA protein in fungal cells regulate the development of A. nidulans (111). High levels of active RasA result in spores that cannot produce germ tubes, whereas intermediate levels lead to uncontrolled vegetative growth similar to the ΔflbA, fadAG42R, and pkaA overexpression phenotypes. Preliminary data suggest that activated Ras protein also inhibits sterigmatocystin production (K. Shimizu and N. P. Keller, unpublished data). In S. cerevisiae but not S. pombe, Ras activity is partially mediated through PKA, and efforts are under way to determine if Ras inhibition of development and sterigmatocystin production in A. nidulans requires PkaA activity.
Recently, Fillinger et al. (43) isolated cyaA, which encodes adenylate cyclase, from A. nidulans (43). Deletion of cyaA severely affects spore germination, causing a delay in germ tube emergence, but the effect on sterigmatocystin production is unknown.
The sfaD gene of A. nidulans encodes the β-subunit of the heterotrimeric G-protein complex (99). sfaD deletion mutants exhibit conidiation in submerged cultures, similar to the phenotype described for strains containing a dominant negative allele of fadA (i.e., fadAG203R), but unlike in fadAG203R strains, sterigmatocystin production is reduced in the ΔsfaD mutant under these conditions (Shimizu and Keller, unpublished) (Table 2). In addition, constitutive active FadA can cause proliferative growth in the absence of sfaD. Also, introduction of ΔsfaD into a ΔflbA background can restore sporulation (99, 132). These results suggest that SfaD and FadA are both positive growth regulators with partially overlapping functions and that FlbA controls both proteins.
Conservation of FadA Regulation of Aflatoxin Production in Other Aspergillus spp.
Proteins of the G-protein signaling pathway of A. nidulans appear to have functional counterparts in A. parasiticus and A. flavus. Hicks et al. (55) transformed the A. nidulans fadAG42R dominant active allele into a norsolorinic acid (e.g., the first stable intermediate in the aflatoxin pathway)-accumulating strain of A. parasiticus. The resultant strain did not produce norsolorinic acid and had the same fluffy phenotype as an A. nidulans fadAG42R mutant (55). S. C. Sim and N. P. Keller (unpublished data) obtained an aconidial, null aflatoxin phenotype in A. flavus strains transformed with the fadAG42R allele.
Signal Transduction Regulation of Sporulation and Secondary Metabolism in Other Organisms
Antibiotics.
The best-known example of a genetic connection between antibiotic production and morphological development is found in the filamentous bacteria Streptomyces spp., in which common elements regulating sporulation and antibiotic production have been reported (56). For example, the translation regulator bldA (67) encodes a tRNA essential to translate the ATT leucine codon, which is scarce in Streptomyces. Deletion of this gene results in reduction of spore production and failure to synthesize antibiotics, without affecting growth. In A. nidulans, a relationship between conidiation and the production of the antibiotic penicillin has also recently been described (115). The dominant activating fadAG42R allele, which represses conidiation and sterigmatocystin biosynthesis, stimulates the expression of ipnA (an enzymatic gene of the A. nidulans penicillin gene cluster) and stimulates concomitant penicillin biosynthesis (115).
Cyclopiazonic acid.
Several strains of A. flavus produce the mycotoxin cyclopiazonic acid as well as aflatoxin. Introduction of the fadAG42R allele into A. flavus inhibits the production of both mycotoxins (Sim and Keller, unpublished).
Trichothecenes.
Introduction of the A. nidulans fadAG42R allele into the fungus F. sporotrichioides generated strains with reduced conidial production and increased trichothecene production (115). Expression of the fadAG42R allele reduced Fusarium spore production 50 to 95% and resulted in delayed radial growth compared with the wild type.
Pigments.
In Cryphonectria parasitica, the chestnut-blight fungus, deletion of cpg1, which encodes the α-subunit of a heterotrimeric G protein, leads to a marked reduction in fungal growth rate and spore production and a loss of virulence. Reduced pigmentation is also observed (45). The β-subunit of a G protein, Cpg2, has also been identified in this fungus. Its deletion also decreases virulence, conidiation, and pigment production (62).
OTHER GENETIC FACTORS COUPLING DEVELOPMENT AND SECONDARY METABOLISM
CCAAT Binding Protein Complex
The A. nidulans CCAAT binding protein complex PENR1 has been reported to regulate both development and penicillin production (74, 112). PENR1 positively regulates the expression of two penicillin biosynthetic genes, ipnA and aatA. Deletion of hapB, hapC, and hapE genes, which encode PENR1 complex components, resulted in identical phenotypes characterized by low levels of ipnA and aatA expression as well as a slow-growth phenotype and a reduction in conidiation (74, 87, 112).
WD Protein Regulation
A. nidulans rcoA encodes a member of the WD protein family. These proteins contain WD repeats, conserved sequence motifs usually ending with Trp-Asp (WD) (110). Members of this family have a role in signal transduction pathways and/or nuclear scaffolding complexes and/or act as global regulators involved in repression of a broad number of genes (84, 122). Several WD proteins have been found to be important in fungal development. In S. pombe, Frz1 is required for spore production (9). In Neurospora crassa, RCO1 is involved in the control of vegetative growth, sexual reproduction, and asexual development (128). S. cerevisiae Tup1p is the best-known example of a fungal WD global regulator. Tup1p forms a complex with other proteins and interacts with histone H3 and H4, stabilizing the nucleosomes on the gene promoters and hence blocking access by the RNA polymerase II complex (38).
RcoA in A. nidulans (53) has closest similarity to N. crassa RCO1, and the respective mutants share some similar aberrance in growth. A. nidulans ΔrcoA strains are deficient in both sterigmatocystin and asexual spore production (53). brlA and aflR transcription is greatly suppressed in these strains, but fluG, flbA, and fadA expression is not affected, thus suggesting that the regulatory role of RcoA may be independent of the G-protein signaling pathway.
Polyamine Pathway
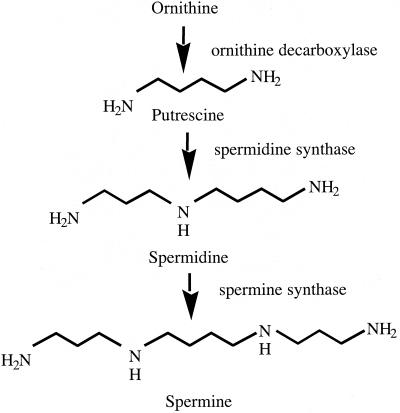
Polyamines are small aliphatic molecules important for normal cell growth and development in a wide range of organisms (100). The most common polyamines are spermidine, spermine, and putrescine, and their biosynthetic relationship is shown in Fig. 4. Rajam and Galston (96) showed that inhibitors of polyamine biosynthesis reduced mycelial growth when applied to cultures of some phytopathogenic fungi, demonstrating the importance of these molecules for fungal development. Guzman-de-Peña et al. (49) described a positive correlation between polyamine biosynthesis and asexual development, sexual development, and sterigmatocystin production. Diamino butanone, an inhibitor of ornithine decarboxylase (the enzyme required for putrescine biosynthesis), blocked the formation of aerial mycelium, asexual and sexual sporulation, and sterigmatocystin biosynthesis in A. nidulans and asexual sporulation and aflatoxin biosynthesis in A. parasiticus (50). Supplementation of exogenous putrescine could partially overcome the repressive effects of the drug. In A. nidulans, spermidine is essential for growth, sporulation, and mycotoxin production, since deletion of the spdA gene, which encodes a spermidine synthase, is lethal (J. Yuan, J. W. Bok, D. Guzman-de-Peña, and N. P. Keller, Mol. Microbiol., in press). Supplementing media with spermidine restores growth and allows asexual sporulation and sterigmatocystin production to resume (Jin et al., submitted).
FIG. 4.
Polyamine biosynthetic pathway.
pacC
In an earlier section we stated that pH affected fungal development and sterigmatocystin and aflatoxin production in Aspergillus spp. in a complex fashion. Keller et al. (65) hypothesized that control of aflatoxin and sterigmatocystin gene expression in response to pH could be mediated via the global transcriptional regulator PacC (118). At acidic pHs, the PacC protein is in an inactive conformation and is unable to bind target sites. At alkaline pHs, the PacC protein is cleaved (40), producing an active form. This activated PacC isoform can then bind to the promoters of target genes, activating the expression of alkaline expressed genes, and repressing the expression of acid expressed genes. This suggested that PacC might be a negative regulator of aflatoxin-sterigmatocystin gene expression, as these genes were more highly expressed at acidic pH (65). Ehrlich et al. (39) showed that the promoter region of the A. parasiticus aflR contains at least one PacC binding site and several promoters of stc genes in A. nidulans also contain putative PacC binding sites (65), thus allowing for the possibility of aflatoxin-sterigmatocystin regulation by PacC. Besides possibly regulating aflatoxin-sterigmatocystin production (65), PacC is also a regulator of genes involved in penicillin production (41, 118). At elevated pHs, PacC activates expression of the isopenicillin N synthase gene, which is involved in penicillin biosynthesis. In Penicillium chrysogenum, PacC also activates the expression of the isopenicillin N synthase homologue, pcbC (114). In addition, PacC regulates morphogenesis. Deletion of the entire pacC coding region results in reduction of growth and conidiation (118). Truncated pacC alleles are also deficient in conidiation (94).
areA
As previously mentioned, Cotty (32) studied the influence of the nitrogen source on development and aflatoxin production in A. flavus and determined a link between nitrogen source, aflatoxin production, and sclerotial morphogenesis. The influence of nitrogen source on sclerotial production at the molecular level is still unclear. However, the repression of aflatoxin production by nitrate might be partially explained by AreA-mediated regulation. AreA is a positive-acting repressor of genes involved in nitrogen metabolism. This GATA factor recognizes specific sequences in the promoter regions of genes under its control (124). areA has been recently cloned in A. parasiticus, and it has been shown in electrophoretic mobility shift assays that AreA binds the region between aflR and aflJ in this fungus (30). This finding suggests that areA participates in the nitrate-mediated negative regulation of gene transcription for aflatoxin biosynthesis in A. parasiticus. Liu and Chu (75) showed that aflR expression in A. parasiticus was inhibited by nitrate. This inhibition can be overcome by overexpression of aflR (28). Nitrate might also regulate aflatoxin biosynthesis posttranscriptionally. Flaherty and Payne (44) overexpressed aflR in nitrate and media lacking nitrate. Although aflatoxin transcripts were detected in both cases, aflatoxin biosynthesis inhibition in the presence of nitrate was not alleviated. This result indicates that nitrate regulates mycotoxin production not only transcriptionally but also posttranscriptionally.
C-Type Cyclins
Fumonisins are a group of mycotoxins produced by F. verticillioides. One of the genes involved in fumonisin biosynthesis, FUM5, encodes a polyketide synthase and has recently been characterized (95). Recently, evidence connecting fumonisin production and sporulation has been reported (104). A regulator gene of the fumonisin pathway has been found, FCC1, which encodes a putative polypeptide with similarities to C-type cyclins of S. cerevisiae (73, 104). Cyclins are essential activating subunits of cyclin-dependent kinases and include a large family of proteins with conserved function in many organisms (7). The FCC1 mutant does not express FUM5 and therefore does not produce fumonisin B1. Additionally, the FCC1 mutant shows reduced asexual spore production when grown on cracked corn. However, the effect of FCC1 mutation on fumonisin production and development was shown to be pH dependent, as the wild-type phenotype was partially recovered in low-pH media.
CONCLUSIONS
It has been noted since the earliest days of fungal manipulation that many species of filamentous fungi readily synthesize complex compounds that are putatively helpful but not necessary for survival and whose production is presumably costly to maintain. Furthermore, production is often linked to fungal development. Some compounds might function as virulence factors, or their presence could give a competitive edge to the producing organism or enhance the survivability of spores. Some secondary metabolites stimulate sporulation and therefore influence the development of the producing organism and neighboring members of the same species, perhaps enhancing the fitness of a community of related species. Natural products are often produced late in fungal development, and their biosynthesis is complex. This complexity is due to a number of factors that affect secondary metabolite production. These include (i) the influence of a number of external and internal factors on natural product biosynthesis, (ii) the involvement of many sequential enzymatic reactions required for converting primary building blocks into natural products, (iii) tight regulation of natural product enzymatic gene expression by one or more transcriptional activators, (iv) close association of natural product biosynthesis with primary metabolism, and (v) close association of natural products with later stages of fungal development, particularly sporulation. Furthermore, the genes required for biosynthesis of some natural products are clustered, perhaps as a consequence of these factors. Gene clusters contain all or most of the genes required for natural product biosynthesis, and logic suggests that their maintenance could only be selected for if the final natural product conferred some advantage to the producing organism. Moreover, natural product biosynthetic gene clusters can be conserved between organisms, for example the sterigmatocystin-aflatoxin biosynthetic gene cluster in several Aspergillus spp.
As discussed in the preceding sections, some natural products directly enhance sporulation of the producing organisms. Relatively few of these compounds have been described. Other secondary metabolites function to protect the spore; for example, melanin is a protectant from UV damage. Most natural products play no obvious roles in sporulation or spore protection but are secreted into the environment at a time in the life cycle of the fungus that corresponds with sporulation. However, some of the compounds that are excreted into the environment could have subtle effects on the organism that are not immediately obvious. For example, although sterigmatocystin was long thought to have no effect on the producing fungus, recent studies (97, 108) have demonstrated an effect of sterigmatocystin on Aspergillus spore production.
Aspergillus spp. produce mycotoxins at a time that coincides with spore development. In A. nidulans, these processes are regulated by G-protein signaling pathway components (55, 106, 133). Inactivation of a G α-subunit, FadA, leads to concomitant sporulation and mycotoxin production. Some of the G-protein signaling regulatory elements described for A. nidulans are conserved among other filamentous fungi (6, 16, 55, 62, 66, 115, 116, 129). However, they can have different or even opposite roles in regulating the biosynthesis of natural products (115). As Tag and collaborators reported, the fadAG42R allele negatively regulates sterigmatocystin production but positively regulates trichothecene and penicillin production (115). Why should the same G-protein signaling pathway oppositely regulate two or more different natural products? The particular example of opposite regulation of sterigmatocystin and penicillin production in A. nidulans by the same signaling pathway allows us to speculate on the necessity of differential regulation when we consider that these two metabolites are also oppositely regulated by pH (penicillin is favored in alkali environments, and sterigmatocystin is favored in acidic environments). Although penicillin is destroyed at low pHa, aflatoxin and sterigmatocystin are very stable molecules not affected by pH extremes. All three of these molecules are complex and incur a large energy cost. Aflatoxin is known to be toxic to insects (90), while penicillin is a renowned bactericidal antibiotic. One could speculate that in their natural environment, Aspergillus spp. face more competition from bacteria in alkaline soils, while acidic soils are generally less populous in bacteria. Therefore, penicillin is produced during fungal development in alkaline environments to destroy its bacterial competition, while sterigmatocystin and aflatoxin secretion and accumulation in mycelial and sclerotial tissue in acidic environments provide protection against insects. Penicillin and sterigmatocystin biosynthesis is temporal in nature, with penicillin biosynthetic gene transcription occurring earlier than stc gene transcription (115). This would ensure that penicillin is secreted into the environment first to kill off fast-growing prokaryotes, allowing the fungus to establish itself in the community. Later, penicillin production ceases and sterigmatocystin is produced to protect against eukaryotic competitors.
The G-protein signaling pathway operates in most organisms and appears to be a conserved mechanism linking external stimuli to a coordinated response by the organism. In Aspergillus, homologs of familiar proteins identified in other organisms, such as an RGS domain protein, α- and β-subunits of heterotrimeric G proteins, and cAMP-dependent PKA, are involved in controlling genes involved in conidiation (via brlA expression) and mycotoxin production (via aflR expression). Although most of the work on the G-protein signaling pathway linking mycotoxin production and sporulation has been done using Aspergillus species, particularly A. nidulans, evidence indicates the presence of a similar pathway regulating tricothecene production in Fusarium (115). The G-protein signaling pathway is an appealing mechanism to link fungal development with natural product metabolism for the following reasons: environmental influences on sporulation and sterigmatocystin and aflatoxin production in Aspergillus spp. can be very complex and contradictory, particularly with regards to pH. If multiple signals for development and mycotoxin production are received external to the cell by different receptors that sense pH, sugar, and nitrogen content of the environment, a system is required to integrate these signals and communicate a coherent message to the nucleus, leading to a coordinated response. Because of the ubiquitous nature of the G-protein signaling pathway in higher organisms, it seems likely that Aspergillus has coopted this pathway to link sporulation and development with mycotoxin production. We speculate that the same mechanism could operate in other fungi that excrete natural products at the onset of sporulation. However, this signaling pathway might not be the only mechanism to coordinate natural product biosynthesis and sporulation. In liquid shake cultures, Aspergillus does not sporulate but still produces mycotoxins. Furthermore, some linoleic acid-derived seed compounds can promote sporulation but inhibit aflatoxin gene transcription as described below.
Aspergillus spp. produce mycotoxins during the onset of sporulation. These compounds might influence the fitness of the producing organism but are not considered sporulating factors per se. However, this genus also secretes linoleic acid-derived molecules, termed psi factors, which do influence asexual and sexual development of the fungus (27). In addition, some plant unsaturated fatty acids have been shown to influence asexual and sexual development (24) and mycotoxin production (22) in this fungus. It is interesting to speculate why Aspergillus spp. produce sporulating molecules that resemble host seed constituents. Perhaps the production of secondary metabolites that mimic the effect of oilseed compounds on fungal growth and development represents a mechanism for survival by the fungi when it is not feeding on a host seed. For A. nidulans, a soil organism, the production of linoleic acid-derived psi factors required for development could indicate it evolved from oilseed-infesting aspergilli.
Aflatoxin and sterigmatocystin production in Aspergillus spp. has been extensively studied due to the deleterious effect of the mycotoxin on human health. While medical investigations have focused on understanding the mechanisms of aflatoxin toxicity and carcinogenicity in humans, these effects of sterigmatocystin and aflatoxin on human health are a consequence of ingesting infected crops and are not the primary function of these compounds. Why these fungi produce sterigmatocystin and aflatoxin; what effect, if any, these compounds have on fungal development; and what ecological role the mycotoxins play are complex questions. In A. nidulans, mutants that produce no sterigmatocystin or accumulate different intermediates of this mycotoxin have been shown in corn and medium-based experiments to be less fit than wild-type strains (97, 108), as defined by reduced sporulation. The further one proceeds along the sterigmatocystin pathway, the more fit the organism becomes. Therefore, these results suggest sterigmatocystin can increase the competitive fitness of the organism.
In recent years, great progress has been made in the discovery of signaling pathways that connect fungal development with natural product biosynthesis. However, much remains to be learned. The complexity of these regulatory networks, with multiple target sites and interconnections with other regulatory mechanisms, makes their full elucidation a challenging task. Emerging technologies should be brought to bear on this puzzle. A. nidulans has a large number of mutants available to address these issues. Furthermore, most of the A. nidulans genome has been sequenced and is available (Cereon Inc.). An illuminating set of experiments could involve microarray analysis of genes involved in the G-protein signaling pathway, fatty acid metabolism (as unsaturated fatty acids influence development), acetate metabolism (acetate is derived from the catabolism of fatty acids and is the primary building block for aflatoxin and sterigmatocystin), and sterigmatocystin biosynthesis. Information obtained from the model organism A. nidulans could lead to an understanding of how these regulatory pathways directly or indirectly control fungal development and the biosynthesis of natural products in other microorganisms. This will open up new broad and exciting fields of applications in which the production of beneficial natural products could be enhanced and the production of those with deleterious effects could be reduced or eliminated.
Acknowledgments
A. M. Calvo and R. A. Wilson made equal contributions to this work.
REFERENCES
- 1.Abdollahi, A., and R. L. Buchanan. 1981. Regulation of aflatoxin biosynthesis: characterization of glucose as an apparent inducer of aflatoxin production. J. Food Sci. 46:143-146. [Google Scholar]
- 2.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 3.Adams, T. H., H. Deising, and W. E. Timberlake. 1990. brlA requires both zinc fingers to induce development. Mol. Cell. Biol. 10:1815-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams, T. H., W. A. Hide, L. N. Yager, and B. N. Lee. 1992. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12:3827-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams, T. H., and J.-H. Yu. 1998. Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans. Curr. Opin. Microbiol. 1:674-677. [DOI] [PubMed] [Google Scholar]
- 6.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews, B., and V. Measday. 1998. The cyclin family of budding yeasts: abundant use of a good idea. Trends Genet. 14:66-72. [DOI] [PubMed] [Google Scholar]
- 8.Andrianopoulos, A., and W. E. Timberlake. 1991. ATTS, a new and conserved DNA binding domain. Plant Cell 3:747-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asakawa, H., K. Kitamura, and C. Shimoda. 2001. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol. Genet. Genom. 265:424-435. [DOI] [PubMed] [Google Scholar]
- 10.Bennett, J. W. 1981. Loss of norsolorinic acid and aflatoxin production by a mutant of Aspergillus parasiticus. J. Gen. Microbiol. 124:429-432. [Google Scholar]
- 11.Bennett, J. W., P. M. Leong, S. Kruger, and D. Keyes. 1986. Sclerotial and low aflatoxigenic morphological variants from haploid and diploid Aspergillus parasiticus. Experientia 42:841-851. [Google Scholar]
- 12.Bennett, J. W., and K. E. Papa. 1988. The aflatoxigenic Aspergillus, p. 264-280. In D. S. Ingram and P. A. Williams (ed.), Genetics of plant pathology, vol. 6. Academic Press, London, United Kingdom.
- 13.Bennett, J. W., P. L. Rubin, L. S. Lee, and P. N. Chen. 1979. Influence of trace elements and nitrogen source on versicolorin production by a mutant strain of Aspergillus parasiticus. Mycopathologia 69:161-166. [DOI] [PubMed] [Google Scholar]
- 14.Bird, B. A., A. T. Remaley, and I. M. Campbell. 1981. Brevianamides A and B are formed only after conidiation has begun in solid cultures of Penicillium brevicompactum. Appl. Environ. Microbiol. 42:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Borgia, P. T., C. L. Dodge, L. E. Eagleton, and T. H. Adams. 1994. Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans. FEMS Microbiol. Lett. 122:227-232. [DOI] [PubMed] [Google Scholar]
- 17.Boylan, M. T., P. M. Mirabito, C. E. Willett, C. R. Zimmerman, and W. E. Timberlake. 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 7:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, D., J.-H. Yu, H. Kelkar, M. Fernandes, T. Nesbitt, N. P. Keller, T. H. Adams, and T. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchanan, R. L., and H. G. Stahl. 1984. Ability of various carbon sources to induce and support aflatoxin biosynthesis by Aspergillus parasiticus. J. Food Safety 6:271-279. [Google Scholar]
- 20.Buchanan, R. L., Jr., and J. C. Ayres. 1975. Effects of initial pH on aflatoxin production. Appl. Microbiol. 30:1050-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu'Lock, J. D. 1961. Intermediary metabolism and antibiotic synthesis. Adv. Appl. Microbiol. 3:293-342. [DOI] [PubMed] [Google Scholar]
- 22.Burow, G. B., T. C. Nesbitt, J. Dunlap, and N. P. Keller. 1997. Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant-Microbe Interact. 10:380-387. [Google Scholar]
- 23.Calvo, A. M., H. W. Gardner, and N. P. Keller. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276:20766-20774. [DOI] [PubMed] [Google Scholar]
- 24.Calvo, A. M., L. L. Hinze, H. W. Gardner, and N. P. Keller. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65:3668-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cary, J. W., K. C. Erlich, M. Wright, P. K. Chang, and D. Bhatnagar. 2000. Generation of aflR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:680-684. [DOI] [PubMed] [Google Scholar]
- 26.Champe, S. P., and A. A. E. El-Zayat. 1989. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171:3982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champe, S. P., P. Rao, and A. Chang. 1987. An endogenous inducer of sexual development in Aspergillus nidulans. J. Gen. Microbiol. 133:1383-1388. [DOI] [PubMed] [Google Scholar]
- 28.Chang, P.-K., K. C. Ehrlich, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl. Environ. Microbiol. 61:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang, P.-K., J. W. Cary, D. Bhatnagar, T. E. Cleveland, J. W. Bennett, J. E. Linz, C. P. Woloshuk, and G. A. Payne. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang, P.-K., J. Yu, D. Bhatnagar, and T. E. Cleveland. 2000. Characterization of the Aspergillus parasiticus major nitrogen regulatory gene, areA. Biochim. Biophys. Acta 1491:263-266. [DOI] [PubMed] [Google Scholar]
- 31.Clutterbuck, A. J. 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotty, P. 1988. Aflatoxin and sclerotial production by Aspergillus flavus: influence of pH. Phytopathology 78:1250-1253. [Google Scholar]
- 33.Davies, D. G., M. R. Parsek, B. H. Pearson, J. W. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 34.De Luca, C., M. Picardo, A. Breathnach, and S. Passi. 1996. Lipoperoxidase activity of Pityrosporum: characterisation of by-products and possible role in pityriasis versicolor. Exp. Dermatol. 5:49-56. [DOI] [PubMed] [Google Scholar]
- 35.Demain, A. L., and A. Fang. 2000. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 69:1-39. [DOI] [PubMed] [Google Scholar]
- 36.Doehlet, D. C., D. T. Wicklow, and H. W. Gardner. 1993. Evidence implicating the lipoxygenase pathway in providing resistance to soybean against Aspergillus flavus. Phytopathology 183:1473-1477. [Google Scholar]
- 37.Dorn, G. L. 1970. Genetic and morphological properties of undifferentiated and invasive variants of Aspergillus nidulans. Genetics 66:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich, K. C., J. W. Cary, and B. G. Montalbano. 1999. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Biochim. Biophys. Acta 1444:412-417. [DOI] [PubMed] [Google Scholar]
- 40.Espeso, E. A., T. Roncal, E. Diez, L. Rainbow, E. Bignell, J. Alvaro, T. Suarez, S. H. Denison, J. Tilburn, H. N. Arst, Jr., and M. A. Peñalva. 2000. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 19:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Espeso, E. A., J. Tilburn, H. N. J. Arst, Jr., and M. A. Peñalva. 1993. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 12:3947-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng, G. H., and T. J. Leonard. 1998. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 64:2275-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fillinger, S., M.-K. Chaveroche, K. Shimizu, N. P. Keller, and C. d'Enfert. 2002. cAMP and ras signaling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 44.Flaherty, J. E., and G. A. Payne. 1997. Overexpression of aflR leads to upregulation of pathway gene expression and increased aflatoxin production in Aspergillus flavus. Appl. Environ. Microbiol. 63:3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 93:14122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavrias, V., A. Andrianopoulos, C. J. Gimeno, and W. W. Timberlake. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255-1263. [DOI] [PubMed] [Google Scholar]
- 47.Gold, S., D. Gillian, K. Barrett, and J. W. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8:2805-2816. [DOI] [PubMed] [Google Scholar]
- 48.Goodrich-Tanrikulu, M., N. Mahoney, and S. B. Rodriquez. 1995. The plant growth regulator methyl jasmonate inhibits aflatoxin production by Aspergillus flavus. Microbiology 141:2831-2837. [DOI] [PubMed] [Google Scholar]
- 49.Guzman-de-Peña, D., J. Aguirre, and J. Ruiz-Herrera. 1998. Correlation between the regulation of sterigmatocystin biosynthesis and asexual and sexual sporulation in Emericella nidulans. Antonie Leeuwenhoek 73:199-205. [DOI] [PubMed] [Google Scholar]
- 50.Guzman-de-Peña, D., and J. Ruiz-Herrera. 1997. Relationship between aflatoxin biosynthesis and sporulation in Aspergillus parasiticus. Fungal Genet. Biol. 21:198-205. [DOI] [PubMed] [Google Scholar]
- 51.Hadley, G., and C. E. Harrold. 1958. The sporulation of Penicillium notatum westling in submerged liquid cultures. J. Exp. Bot. 9:418-428. [Google Scholar]
- 52.Reference deleted.
- 53.Hicks, J., R. A. Lockington, J. Strauss, D. Dieringer, C. P. Kubicek, J. Kelly, and N. P. Keller. 2001. RcoA has pleiotropic effects on Aspergillus nidulans cellular development. Mol. Microbiol. 39:1482-1493. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.Hicks, J., J.-H. Yu, N. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G-alpha protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hopwood, D. A. 1988. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc. R. Soc. Lond. B 235:121-138. [DOI] [PubMed] [Google Scholar]
- 57.Reference deleted.
- 58.Johnstone, I. L., S. G. Hughes, and A. J. Clutterbuck. 1985. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 4:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kacholz, T., and A. L. Demain. 1983. Nitrate repression of averufin and aflatoxin biosynthesis. J. Nat. Prod. 46:499-506. [Google Scholar]
- 60.Kale, S. P., D. Bhatnagar, and J. W. Bennett. 1994. Isolation and characterization of morphological variants of Aspergillus parasiticus. Mycol. Res. 98:645-652. [DOI] [PubMed] [Google Scholar]
- 61.Kale, S. P., J. W. Cary, D. Bhatnagar, and J. W. Bennett. 1996. Characterization of experimentally induced, nonaflatoxigenic variant strains of Aspergillus parasiticus. Appl. Environ. Microbiol. 62:3399-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasahara, S., and D. L. Nuss. 1997. Targeted disruption of a fungal G-protein beta subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 63.Kawamura, C., T. Tsujimoto, and T. Tsuge. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 64.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 65.Keller, N. P., C. Nesbitt, B. Sarr, T. D. Phillips, and G. B. Burow. 1997. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology 87:643-648. [DOI] [PubMed] [Google Scholar]
- 66.Kronstad, J., A. De Maria, D. Funnell, R. D. Laidlaw, N. Lee, M. M. de Sa, and M. Ramesh. 1998. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch. Microbiol. 170:395-404. [DOI] [PubMed] [Google Scholar]
- 67.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 68.Lee, B. N., and T. H. Adams. 1996. FluG and FlbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA beta activation. EMBO J. 15:299-309. [PMC free article] [PubMed] [Google Scholar]
- 69.Lee, B. N., and T. H. Adams. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal. Genes Dev. 8:641-651. [DOI] [PubMed] [Google Scholar]
- 70.Lee, B. N., and T. H. Adams. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14:323-334. [DOI] [PubMed] [Google Scholar]
- 71.Lengeler, K. B., R. C. Davidson, C. D'souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascade regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leonard, K. J. 1977. Virulence, temperature optima, and competitive abilities of isolines of races T and O of Bipolaris maydis. Phytopathology 67:1273-1279. [Google Scholar]
- 73.Liao, S. M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. Van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 74.Litzka, O., K. Then Bergh, and A. A. Brakhage. 1996. The Aspergillus nidulans penicillin-biosynthesis gene aat (penDE) is controlled by a CCAAT-containing DNA element. Eur. J. Biochem. 238:675-682. [DOI] [PubMed] [Google Scholar]
- 75.Liu, B.-H., and F. S. Chu. 1998. Regulation of aflR and its product, AflR, associated with aflatoxin biosynthesis. Appl. Environ Microbiol. 64:3718-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luchese, R. H., and W. F. Harrigan. 1993. Biosynthesis of aflatoxin—the role of nutritional factors. J. Appl. Bacteriol. 74:5-14. [DOI] [PubMed] [Google Scholar]
- 77.Mapleston, R. A., M. J. Stone, and D. H. Williams. 1992. The evolutionary role of secondary metabolites. Gene 115:151-157. [DOI] [PubMed] [Google Scholar]
- 78.Martinelli, S. D., and A. J. Clutterbuck. 1971. A quantitative survey of conidiation mutants in Aspergillus nidulans. J. Gen. Microbiol. 69:261-268. [DOI] [PubMed] [Google Scholar]
- 79.Mazur, P., K. Nakanishi, A. A. E. El-Zayat, and S. P. Champe. 1991. Structure and synthesis of sporogenic psi factors from Aspergillus nidulans. J. Chem. Soc. Chem. Commun. 20:1486-1487. [Google Scholar]
- 80.Meyers, D. M., G. Obrian, W. L. Du, D. Bhatnagar, and G. A. Payne. 1998. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 64:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mims, C. W., E. A. Richardson, and W. E. Timberlake. 1988. Ultrastructural analysis of conidiophore development in the fungus Aspergillus nidulans using freeze-substitution. Protoplasma 44:132-141. [Google Scholar]
- 82.Morrice, J., D. A. Mackenzie, A. J. Parr, and D. B. Archer. 1998. Isolation and characterisation of the acetyl-CoA carboxylase gene from Aspergillus nidulans. Curr. Genet. 34:379-385. [DOI] [PubMed] [Google Scholar]
- 83.Mösch, H.-U., E. Kubler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neer, E. J., C. J. Schmidt, R. Nambudripad, and T. F. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371:297-300. [DOI] [PubMed] [Google Scholar]
- 85.Niehaus, W. G. J., and W. Jiang. 1989. Nitrate induces enzymes of the mannitol cycle and suppresses versicolorin synthesis in Aspergillus parasiticus. Mycopathologia 107:131-139. [DOI] [PubMed] [Google Scholar]
- 86.Oliver, P. T. P. 1972. Conidiophore and spore development in Aspergillus nidulans. J. Gen. Microbiol. 73:45-54. [DOI] [PubMed] [Google Scholar]
- 87.Papagiannopoulos, P., A. Andrianopoulos, J. A. Sharp, M. A. Davis, and M. J. Hynes. 1996. the hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol. Gen. Genet. 251:412-421. [DOI] [PubMed] [Google Scholar]
- 88.Park, D., and P. M. Robinson. 1969. Sporulation in Geotrichum candidum. Br. Mycol. Soc. 52:213-222. [Google Scholar]
- 89.Passi, S., M. Nazzaro-Porro, C. Fanelli, A. A. Fabbri, and P. Fasella. 1984. Role of lipoperoxidation in aflatoxin production. Appl. Microbiol. Biotechnol. 19:186-190. [Google Scholar]
- 90.Payne, G. A. 1992. Aflatoxin in maize. Crit. Rev. Plant Sci. 10:423-440. [Google Scholar]
- 91.Payne, G. A., and M. P. Brown. 1998. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36:329-362. [DOI] [PubMed] [Google Scholar]
- 92.Payne, G. A., G. J. Nystrom, D. Bhatnagar, T. E. Cleveland, and C. P. Woloshuk. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pazoutova, S., V. Pokorny, and Z. Rehacek. 1977. The relationship between conidiation and alkaloid production in saprophytic strains of Claviceps purpurea. Can. J. Microbiol. 23:1182-1187. [DOI] [PubMed] [Google Scholar]
- 94.Piñero, D. P. 1999. Isolation and characterization of the Aspergillus parasiticus pacC gene. Master's thesis. Office of Graduate Studies of Texas A & M University, College Station.
- 95.Proctor, R. H., A. E. Desjardins, and R. D. Plattner. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27:100-112. [DOI] [PubMed] [Google Scholar]
- 96.Rajam, M. V., and A. W. Galston. 1985. The effects of some polyamine biosynthetic inhibitors on growth and morphology of phytopathogenic fungi. Plant Cell Physiol. 26:683-692. [DOI] [PubMed]
- 97.Ramaswamy, A. 2002. Ecological analysis of secondary metabolite production in Aspergillus spp. Master's thesis. Office of Graduate Studies of Texas A & M University, College Station.
- 98.Reiß, J. 1982. Development of Aspergillus parasiticus and formation of aflatoxin B1 under the influence of conidiogenesis affecting compounds. Arch. Microbiol. 133:236-238. [DOI] [PubMed] [Google Scholar]
- 99.Rosén, S., J.-H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruiz-Herrera, J. 1994. Polyamines, DNA methylation, and fungal differentiation. Crit. Rev. Microbiol. 20:143-150. [DOI] [PubMed] [Google Scholar]
- 101.Rupp, S., E. Summers, H.-J. Jo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schimmel, T. G., A. D. Coffman, and S. J. Parsons. 1998. Effect of butyrolactone I on the producing fungus, Aspergillus terreus. Appl. Environ. Microbiol. 64:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sekiguchi, J., and G. M. Gaucher. 1977. Conidiogenesis and secondary metabolism in Penicillium urticae. Appl. Environ. Microbiol. 33:147-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shim, W.-B., and C. P. Woloshuk. 2001. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl. Environ. Microbiol. 67:1607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reference deleted.
- 106.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reference deleted.
- 108.Sim, S. C. 2001. Characterization of genes in the sterigmatocystin gene cluster and their role in fitness of Aspergillus nidulans. Master’s thesis. Office of Graduate Studies of Texas A & M University, College Station.
- 109.Reference deleted.
- 110.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 111.Som, T., and V. S. R. Kolaparthi. 1994. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol. Cell. Biol. 14:5333-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Steidl, S., P. Papagiannopoulos, O. Litzka, A. Andrianopoulos, M. A. Davis, A. A. Brakhage, and M. J. Hynes. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol. Cell. Biol. 19:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stone, M. J., and D. H. Williams. 1992. On the evolution of functional secondary metabolites (natural products). Mol. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 114.Suarez, T., and M. A. Peñalva. 1996. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol. Microbiol. 20:529-540. [DOI] [PubMed] [Google Scholar]
- 115.Tag, A., J. Hicks, G. Garifullina, C. Ake, T. D. Phillips, M. Beremand, and N. P. Keller. 2000. G-protein signalling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38:658-665. [DOI] [PubMed] [Google Scholar]
- 116.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMKI regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13:374-383. [DOI] [PubMed] [Google Scholar]
- 117.Tamame, M., F. Antequera, J. R. Villanueva, and T. Santos. 1983. High-frequency conversion of a “fluffy” developmental phenotype in Aspergillus spp. by 5-azacytidine treatment: evidence for involvement of a single nuclear gene. Mol. Cell. Biol. 3:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Peñalva, and H. N. J. Arst. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 120.Trail, F., N. Mahanti, and J. Linz. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755-765. [DOI] [PubMed] [Google Scholar]
- 121.Tsai, H.-F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van der Voorn, L., and H. L. Ploegh. 1992. The WD-40 repeats. FEBS Lett. 307:131-134. [DOI] [PubMed] [Google Scholar]
- 123.Wieser, J., B. N. Lee, J. W. Fondon, and T. H. Adams. 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27:62-69. [DOI] [PubMed] [Google Scholar]
- 124.Wilson, R. A., and H. N. Arst. 1998. Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 62:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wolf, J. C., and C. J. Mirocha. 1973. Regulation of sexual reproduction in Gibberella zeae (Fusarium roseum ‘Graminearum’) by F-2 (zearalenone). Can. J. Microbiol. 19:725-734. [DOI] [PubMed] [Google Scholar]
- 126.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wright, M. S., D. M. Greene-McDowelle, H. J. Zeringue, Jr., D. Bhatnagar, and T. E. Cleveland. 2000. Effects of volatile aldehydes from Aspergillus-resistant varieties of corn on Aspergillus parasiticus growth and aflatoxin biosynthesis. Toxicon 38:1215-1223. [DOI] [PubMed] [Google Scholar]
- 128.Yamashiro, C. T., D. J. Ebbole, B., R. E. Brown, C. Bourland, L. Madi, and C. Yanofsky. 1996. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a G1 causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu, J., D. Bhatnagar, and T. E. Cleveland. 2000. Cloning of a sugar utilization gene cluster in Aspergillus parasiticus. Biochim. Biophys. Acta 1493:211-214. [DOI] [PubMed] [Google Scholar]
- 131.Yu, J.-H., R. A. E. Butchko, M. Fernandes, N. P. Keller, T. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]
- 132.Yu, J.-H., S. Rosén, and T. H. Adams. 1999. Extragenic suppressors of loss-of-function mutations in the Aspergillus FlbA regulator of G-protein signaling domain protein. Genetics 151:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu, J.-H., J. Weiser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]
- 134.Zaika, L. L., and R. L. Buchanan. 1987. Review of compounds affecting the biosynthesis or bio-regulation of aflatoxins. J. Food Prot. 50:691-708. [DOI] [PubMed] [Google Scholar]
- 135.Zeringue, H. J., Jr. 2000. Identification and effects of maize silk volatiles on cultures of Aspergillus nidulans. J. Agric. Food Chem. 48:921-925. [DOI] [PubMed] [Google Scholar]