Abstract
The σS (RpoS) subunit of RNA polymerase is the master regulator of the general stress response in Escherichia coli and related bacteria. While rapidly growing cells contain very little σS, exposure to many different stress conditions results in rapid and strong σS induction. Consequently, transcription of numerous σS-dependent genes is activated, many of which encode gene products with stress-protective functions. Multiple signal integration in the control of the cellular σS level is achieved by rpoS transcriptional and translational control as well as by regulated σS proteolysis, with various stress conditions differentially affecting these levels of σS control. Thus, a reduced growth rate results in increased rpoS transcription whereas high osmolarity, low temperature, acidic pH, and some late-log-phase signals stimulate the translation of already present rpoS mRNA. In addition, carbon starvation, high osmolarity, acidic pH, and high temperature result in stabilization of σS, which, under nonstress conditions, is degraded with a half-life of one to several minutes. Important cis-regulatory determinants as well as trans-acting regulatory factors involved at all levels of σS regulation have been identified. rpoS translation is controlled by several proteins (Hfq and HU) and small regulatory RNAs that probably affect the secondary structure of rpoS mRNA. For σS proteolysis, the response regulator RssB is essential. RssB is a specific direct σS recognition factor, whose affinity for σS is modulated by phosphorylation of its receiver domain. RssB delivers σS to the ClpXP protease, where σS is unfolded and completely degraded. This review summarizes our current knowledge about the molecular functions and interactions of these components and tries to establish a framework for further research on the mode of multiple signal input into this complex regulatory system.
INTRODUCTION
σS, or RpoS, is a sigma subunit of RNA polymerase in Escherichia coli that is induced and can partially replace the vegetative sigma factor σ70 (RpoD) under many stress conditions. As a consequence, transcription of numerous σS-dependent genes is activated (for reviews, see references 75, 77, 79, 124, and 125). Consistent with the multiple functions of the σS regulon, the rpoS gene was discovered independently and named accordingly by several groups (recently summarized in references 79 and 105). It was identified as a gene involved in near-UV resistance (nuv) (216); as a regulator for the katE-encoded catalase HPII (katF) (126, 186), exonuclease III (xthA) [186], and acidic phosphatase (appR) (211); and, finally, as a starvation-inducible gene encoding a central regulator for stationary phase-inducible genes (csi-2) (113). Only then was it recognized that all the previous studies had described alleles of the same gene (113, 212), which codes for a sigma factor (152, 156, 209). Because of its crucial role in stationary phase or under stress conditions, the name σS or RpoS was proposed (113). In addition, the term σ38 is used sometimes (although the molecular mass of σS deviates from 38 kDa in various species and even in some E. coli strains). The rpoS gene has also been identified in other enteric and related bacteria. At present, it seems that σS occurs in the γ branch of the proteobacteria, i.e., in a group of gram-negative bacteria that includes many species with special importance for humans because of their pathogenic or beneficial potential. With minor variations, the general function of σS in these bacteria appears to be similar to that in E. coli (summarized in reference 79).
In more recent studies, it was demonstrated that σS and σS-dependent genes not only are induced in stationary phase but actually respond to many different stress conditions (76, 82, 121, 144, 148). Therefore, σS is now seen as the master regulator of the general stress response, which is triggered by many different stress signals, is often (though not always) accompanied by a reduction or cessation of growth, and provides the cells with the ability to survive the actual stress as well as additional stresses not yet encountered (cross-protection). This is in pronounced contrast to specific stress responses, which are triggered by a single stress signal and result in the induction of proteins that allow cells to cope with this specific stress situation only. While specific stress responses tend to eliminate the stress agent and/or to mediate repair of cellular damage that has already occurred, the general stress response renders cells broadly stress resistant in such a way that damage is avoided rather than needing to be repaired (for a recent review of different bacterial stress responses, see reference 203).
The major function of the general stress response is thus preventive, which is clearly reflected in the σS-dependent multiple stress resistance observed with starved or otherwise stressed cells (82, 113, 136) (for a recent review of the general stress response that also includes physiological aspects, see reference 79). Accordingly, the majority of the more than 70 σS-dependent genes known so far confer resistance against oxidative stress, near-UV irradiation, potentially lethal heat shocks, hyperosmolarity, acidic pH, ethanol, and probably other stresses yet to be identified. Additional σS-controlled gene products generate changes in the cell envelope and overall morphology (stressed E. coli cells tend to become smaller and ovoid). Metabolism is also affected by σS-controlled genes, consistent with σS being important under conditions where cells switch from a metabolism directed toward maximal growth to a maintenance metabolism. σS also controls genes mediating programmed cell death in stationary phase, which may increase the chances for survival for a bacterial population under extreme stress by sacrificing a fraction of the population in order to provide nutrients for the remaining surviving cells (22). Finally, a number of virulence genes in pathogenic enteric bacteria have been found to be under σS control, consistent with the notion that host organisms provide stressful environments for invading pathogens (recently summarized in reference 80). However, even though numerous σS-dependent genes have been identified (see references 77, 79, and 125 for recent compilations), many more such genes will probably be found in the future. Moreover, the functions of the genes known so far are incompletely understood. Even after more than 10 years of intensive research on σS, much remains to be learned about the physiology of the σS-mediated response. The same is true for the regulatory interdependencies within the large regulatory network directed by σS. By contrast, the basic mechanisms of regulation of σS itself are now reasonably well understood and are the subject of this review (for recent minireviews on the same subject, see references 81 and 125). Since by far most of the relevant work has been done with E. coli and Salmonella, the systems referred to in this review are those described for these enteric bacteria if not otherwise mentioned. Whereas the physiological function of σS is comparable in all species where it has been discovered to date, there are significant differences between enteric bacteria and pseudomonads in the regulation of σS that are outlined specifically.
THE PROBLEM OF MULTIPLE STRESS SIGNAL INTEGRATION
Complex and physiologically far-reaching bacterial responses often use a single master regulator at the interface of upstream signal processing and downstream regulatory mechanisms. In the general stress response of E. coli, σS plays the role of this top-level master regulator. Other examples of such central regulators are σB in the general stress response of various gram-positive bacteria (172) and the response regulator Spo0A in sporulation initiation of Bacillus subtilis (202). The master regulators serve as the decisive information processing units, which connect complex signaling networks with downstream regulatory cascades or networks that ultimately control the expression of numerous structural genes associated with a response. These regulatory networks exhibit a hierarchical and modular structure; i.e., they can be subdivided into lower-level smaller modules that are under the control of secondary regulators, which also allow specific signal input at such lower and more confined levels. A master regulator may also commit the cell to a certain complex developmental program, with specific temporal and spatial control being exerted by secondary regulalors.
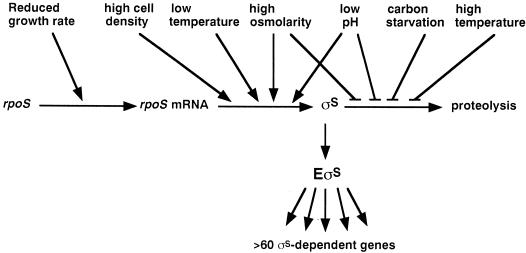
Depending on the type of master regulator (sigma factor, two-component response regulator, etc.) as well as on whether its cellular level or its activity (or both) is the decisive parameter, signal transduction and integration upstream of the central regulator can be very different. In the general stress response of E. coli, the decisive parameter is the cellular level of σS (however, as described below, there is now some initial evidence that σS may also be subject to some sort of activity control). σS levels increase in response to starvation for carbon, nitrogen, or phosphate sources as well as for amino acids. This leads to entry into stationary phase, i.e., a complete cessation of growth, but σS can also be induced by a partial reduction of the growth rate (64, 89, 92, 113, 114, 157, 210). Additional inducing conditions are hyperosmolarity (148), nonoptimally high (144) or low (199) temperature, acidic pH (18, 121), high cell density (114), and probably other environmental stress situations.
How can so many different stress signals be integrated toward a single parameter, i.e., the intracellular σS concentration? Do they generate a common intracellular signal? Initially it seemed that a reduction or cessation of growth might be such a signal. However, σS is also induced in late exponential phase (provided that a certain cell density is reached) without any change of growth rate (114). In response to the classical heat shock procedure (i.e., a shift from 30 to 42°C), σS is induced even though growth is accelerated (144). On the basis of more recent studies, it has become clear that the concept of a unifying intracellular σS-inducing signal cannot be correct, simply because different environmental signals affect different levels and therefore completely different processes in the regulation of σS.
Regulation of σS occurs at nearly every theoretically possible level (Fig. 1). rpoS transcription is stimulated by controlled downshifts in growth rate in a chemostat (157, 210) as well as by continuous reduction in growth rate which results in an inversely correlated increase in rpoS transcription (5- to 10-fold) (113, 114). By contrast, abrupt cessation of growth, as for example, in response to sudden glucose starvation, only weakly increases rpoS transcription (less than twofold) (113, 114). rpoS translation, i.e., the rate of translation of already existing rpoS mRNA, is stimulated (i) by high osmolarity (hyperosmotic shift rapidly activates translation more than fivefold, but also continuous growth at high osmolarity has clear effects) (148), (ii) during growth at moderately low temperatures (e.g., at 20°C) (199), (iii) on reaching a certain cell density (approximately 1 × 108 to 2 × 108 cells ml−1) during growth in minimal glucose medium (114), and (iv) in response to a pH downshift from pH 7 to pH 5 in rich medium (G. Kampmann and R. Hengge-Aronis, unpublished data).
FIG. 1.
Various levels of σS regulation are differentially affected by various stress conditions. An increase of the cellular σS level can be obtained either by stimulating σS synthesis at the levels of rpoS transcription or rpoS mRNA translation or by inhibiting σS proteolysis (which under nonstress conditions is extraordinarily rapid). The most rapid and strongest reaction can be achieved by a combination of these processes (as observed, e.g., on hyperosmotic or pH shifts). For further details, see the text.
Besides this multifaceted regulation of σS synthesis, there is also control of σS degradation. In cells growing on minimal medium, σS (which is produced at a low but measurable rate) is degraded with a half-life between 1 min and a few minutes (114, 144, 148, 208). However, in response to stresses such as starvation (114, 208), shift to hyperosmolarity (148), the classical heat shock (144), or pH downshift to pH 5 (18), σS proteolysis is considerably reduced or even completely inhibited. As a consequence, σS rapidly accumulates in the cell. The kinetics of this stabilization can be very rapid (on hyperosmotic shift, σS is strongly stabilized within a few minutes [148]) or can take somewhat more time (as, e.g., after heat shock [144]). This again indicates that even in cases where the same level of σS regulation is affected by different stress conditions, the regulatory mechanisms involved are likely to be different.
In recent years, considerable progress has been made in identifying (i) cis-acting regulatory regions at the DNA, mRNA, or protein levels; (ii) trans-acting factors such as regulatory proteins or regulatory RNAs; and (iii) additional large or small molecules that modulate σS regulation at these different levels. In general, the closer to the rpoS gene, its mRNA, or the σS protein itself these factors act, the better understood is their molecular mechanism of action. By contrast, the upper parts of the corresponding signal transduction pathways have remained more elusive. It is, however, already clear that these pathways do not operate independently and in parallel until they finally converge to influence, e.g., rpoS mRNA secondary structure or the activity of a specific σS recognition factor for proteolysis. Rather, these pathways are highly interconnected, such that specific stress conditions can influence the cellular σS concentration by multiple mechanisms. Moreover, specific components of these pathways also control each other. Thus, σS is controlled by a complex signal transduction network whose redundancy, additiveness, and internal feedback regulatory loops are crucial for its admirable signal-integrative power (and probably its nonlinear behavior) but at the same time have made life difficult for researchers trying to elucidate these pathways.
REGULATION OF rpoS TRANSCRIPTION
Soon after σS had been recognized as a stationary-phase regulator that was itself induced in stationary phase (113), it became clear that much of its regulation was due to posttranscriptional mechanisms (114, 127, 135). Although σS protein levels are very low in exponentially growing cells, relatively high levels of rpoS mRNA are present and do not seem to change in response to several stresses that actually result in strongly elevated σS protein levels. Therefore, attention has focused on the control of rpoS translation and σS proteolysis (see below) whereas rpoS transcription has remained insufficiently characterized. Nevertheless, transcriptional regulation of rpoS occurs, e.g., during the gradual decrease in growth rate when cells grow in rich medium and finally enter stationary phase. Under these conditions, rpoS transcription is activated approximately 5- to 10-fold (113, 114, 135, 153, 190, 208).
Promoters Contributing to rpoS Transcription
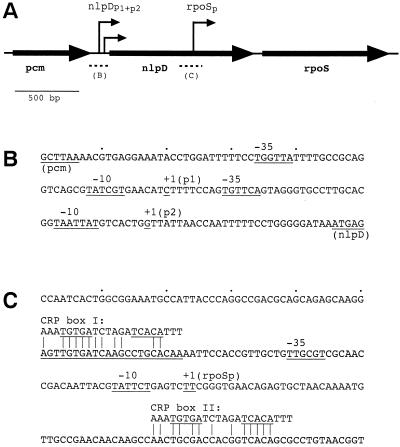
Several promoters are involved in rpoS transcription (Fig. 2). Two transcripts can be detected by Northern analysis (8). Polycistronic nlpD-rpoS mRNA originates from two closely spaced promoters (nlpDp1 and nlpDp2) upstream of the nlpD gene (112, 115), which encodes a lipoprotein of unknown function (87, 115). Another promoter (rpoSp) is located within the nlpD gene and produces a monocistronic rpoS mRNA with an unusually long nontranslated 5′ region of 567 nucleotides (112, 208). Studies with transcriptional fusions that included a 5′ deletion analysis indicated that this transcript is the major rpoS mRNA (112). Moreover, rpoSp accounts for activation of transcription in Luria broth-grown cells during entry into stationary phase (112, 208). The NlpD protein is not stationary phase induced, which indicates that the nlpD promoters are not growth phase regulated (115).
FIG. 2.
Transcriptional control regions upstream of the rpoS gene. (A) The nlpD-rpoS operon is located at 61.76 min on the E. coli chromosome, where it is trancribed in counterclockwise direction. (B) The operon promoters (nlpDp1 and nlpDp2) contribute to basal expression of rpoS but are not regulated by growth rate or growth phase (115). (C) The major rpoS promoter (rpoSp) is located within the nlpD gene, is flanked by two putative cAMP-CRP binding sites (CRP box I and II), and is subject to stationary-phase induction when cells are grown on rich medium (112). Broken lines in panel A indicate the relative positions of the sequences shown in panels B and C.
In other enteric bacteria, Vibrionaceae members, and pseudomonads studied so far, the nlpD and rpoS genes are always linked, which suggests similar transcriptional regulation to that in E. coli. Downstream of rpoS, however, variations are quite common and even occur between different E. coli strains (28, 37, 83). However, nlpD can occur alone in bacteria that do not possess an rpoS gene, such as Haemophilus influenzae (55).
Trans-Acting Factors Controlling rpoS Transcription
cAMP-CRP and EIIA(Glc).
In strains carrying mutations in cya (encoding adenylate cyclase) or crp (encoding the cyclic AMP [cAMP] receptor protein [CRP], also called the catabolite activator protein), σS levels and the activities of transcriptional rpoS::lacZ fusions are already high in exponential phase, indicating that cAMP-CRP is a negative regulator of rpoS transcription. In the cya mutant, this phenotype can be reversed by the addition of external cAMP (113, 114). However, the mode of action of cAMP-CRP in rpoS transcriptional control seems to depend on the growth phase. Recent evidence indicates that during entry into stationary phase, cAMP-CRP positively controls rpoS transcription (F. Scheller and R. Hengge-Aronis, unpublished results). This may resolve an apparent contradiction between the above-mentioned observation of high log-phase levels of σS in cya or crp mutants (114) and the finding that certain rpoS::lacZ fusions show reduced expression in a cya background (135).
Two putative cAMP-CRP binding sites are present upstream and downstream of rpoSp (Fig. 2), and the role of these potential binding sites is currently under investigation. Whereas the location of the upstream cAMP-CRP box is similar to that in the lac promoter and corresponds to a classical activator position at a class I promoter (31), the location of the second cAMP-CRP box downstream of the transcriptional start site may suggest an inhibitory action. In addition, cAMP-CRP may also have an indirect effect on rpoS expression, since cya or crp mutants exhibit a reduced growth rate, which in turn can affect rpoS transcription, as mentioned above.
Adenylate cyclase activity is positively modulated by the crr-encoded EIIA(Glc), which is the soluble part of the glucose-specific EII component of the phosphotransferase system for solute uptake. Consistent with this, a crr mutation results in elevated σS levels during exponential phase, which reflects increased rpoS transcription as well as increased rpoS translation. The former can be suppressed by cAMP addition, indicating that EIIA(Glc) affects rpoS transcription through its modulation of adenylate cyclase activity (217). Phenotypically, the crr mutant thus seems to mimic the log-phase behavior of the cya mutant.
Polyamines stimulate adenylate cyclase expression at the level of translational initiation, and at the same time they lead to 2.3- and 4-fold increases of the cellular levels of σS and σ28, respectively (235). This polyamine-induced upregulation of σS may mimic the positive effect of cAMP-CRP on rpoS transcription during entry into stationary phase.
The GacS-GacA two-component and Las-Rhl quorum-sensing systems in pseudomonads.
GacA is a two-component response regulator in various Pseudomonas species that has long been known to positively affect the production of secondary products such as antibiotics, toxins, and lytic exoenzymes during entry into stationary phase (117, 123, 178). The cognate histidine sensor kinase is the GacS (LemA) protein (85, 178). What is actually sensed by GacS has not been clarified. Homologs of GacA are present in Salmonella (SirA, which controls certain virulence genes [93]) and E. coli (YecB or UvrY [see below]). The GacS-GacA two-component system is at the top of a regulatory cascade that controls the LasI-LasR quorum-sensing system, which in turn regulates a second quorum-sensing system, RhlI-RhlR (110, 167, 176). These quorum-sensing systems are crucial for the control of virulence factors, exoenzymes, and stress-protective proteins as well as for the formation of biofilms (62, 74, 111, 159, 232); for summaries of quorum-sensing systems, see references 61 and 231).
Mutations in gacA or gacS also result in a more than 80% reduction of σS levels and equally reduced expression of a transcriptional rpoS::lacZ fusion specifically during transition into stationary phase (227). Whether this regulation of rpoS by the Gac system is direct or indirect has not been demonstrated. There are also conflicting data about how rpoS is linked to the Las-Rhl cascade. An earlier study had indicated that rpoS is under positive control of both the Las and Rhl systems (110). More recently, however, rpoS was found not to be affected by rhl mutations; in contrast, rhlI is upregulated in rpoS mutants, indicating that σS is a negative regulator of the Rhl system (228). This fits with the observation that pyocyanin and pyoverdin are overproduced in rpoS mutants (205), since these virulence-associated factors are under positive control of the Rhl system (27).
In contrast to the situation in Pseudomonas, the GacA homolog YecB (UvrY) in E. coli appears not to be involved in the control of rpoS expression. Even though YecB overproduction stimulated the expression of transcriptional and translational rpoS::lacZ fusions and resulted in a twofold-higher level of σS specifically during transition into stationary phase, a yecB::cat knockout mutation did not alter the expression of these rpoS::lacZ fusion, nor were σS levels affected (Kampmann and Hengge-Aronis, unpublished). While σS plays similar physiological roles in E. coli and Pseudomonas (188, 205), differential control by YecB-GacA may reflect the different environmental conditions characteristic of the natural habitats of these bacteria.
BarA, a histidine sensor kinase in search of a response regulator.
The E. coli homolog of GacS is a hybrid sensor kinase called BarA (40% identity and 59% overall similarity at the amino acid level to P. aeruginosa GacS). BarA was previously found as a multicopy suppressor of an envZ mutation; i.e., when present at high levels, BarA is able to cross-phosphorylate the response regulator OmpR and thereby activate porin synthesis (88, 154). Under the name AirS, BarA has also been identified as a virulence factor in uropathogenic E. coli (241). There is evidence that BarA plays a positive role in rpoS expression. A strain with a lacZ insertion in the chromosomal copy of barA, which was originally isolated as a hydrogen peroxide-sensitive mutant, exhibits reduced levels of σS (150, 151). In the mutant, rpoS mRNA levels were reduced during exponential phase but were normal in stationary phase. Therefore, BarA was suggested as a positive regulator of rpoS transcription (150). By specifically affecting rpoS mRNA levels in exponential phase, this control may determine the range within which σS levels can be modulated by posttranscriptional control mechanisms in response to various stress conditions.
The homology to the GacS-GacA system, as well as recent biochemical data (166), suggests that BarA is a cognate sensor kinase for YecB. Therefore, it seems surprising that BarA but not YecB (see above) is involved in σS control. However, it is possible that BarA acts on more than one response regulator with an unknown target response regulator being involved in rpoS control. BarA (GacS) belongs to the complex “built-in phosphorelay” sensor kinases in which sensor, transmitter, receiver, and histidine-containing phosphotransfer domains are combined in a single polypeptide chain. In view of their multiple interactions, phosphorelay components seem especially adequate for establishing such phosphotransfer networks.
PsrA in pseudomonads: a TetR-like regulator.
A search for insertional mutations that downregulated the expression of a transcriptional rpoS::lacZ fusion in P. putida yielded a mutant defective in a gene termed psrA (for “Pseudomonas sigma regulator”), which encodes a TetR repressor-like regulatory protein. psrA is required for increased rpoS transcription during entry into stationary phase and is negatively autoregulated (104). Whether this control of rpoS is direct or indirect is currently unknown.
Role for Polyphosphate in rpoS Regulation
Inorganic polyphosphate occurs in most microorganisms and often accumulates in stationary phase or under other stress conditions (106, 107). In E. coli, the actual polyphosphate level is the result of a balance between synthesis (catalyzed by polyphosphate kinase, encoded by ppk) and degradation (catalyzed by exopolyphosphatases, encoded by ppx and gppA) (106). Polyphosphate accumulation is positively affected by the “alarmone” guanosine 3′,5′-bispyrophosphate (ppGpp; see below), which seems to inhibit the ppx-encoded exopolyphosphatase (108). Polyphosphate stimulates Lon-mediated degradation of ribosomal proteins; i.e., it may be crucial for gaining access to intracellular amino acid pools under conditions of sudden carbon or amino acid starvation (109).
Polyphosphate-free ppk mutants are multiple stress sensitive and impaired in stationary phase survival (36, 175). Consistent with these phenotypes, σS levels as well as the expression of a transcriptional rpoS::lacZ fusion are reduced in a strain that overproduces yeast exopolyphosphatase and is therefore depleted of polyphosphate (195). These findings indicate that polyphosphate somehow stimulates rpoS transcription and thereby contributes to stationary-phase induction of rpoS (which in rich media is partly due to increased rpoS transcription). However, polyphosphate fails to stimulate rpoS transcription in vitro and therefore may exert an indirect influence in vivo (195).
Small Molecules That Influence rpoS Transcription
ppGpp.
ppGpp levels in E. coli strongly increase in response to amino acid limitation (triggering the stringent response) or starvation for carbon, nitrogen, and phosphorus sources. Amino acid limitation causes a rise in the cellular level of uncharged tRNA, which is sensed by the ribosome-associated RelA protein (ppGpp synthase I). Under other starvation conditions, ppGpp synthesis is mediated by SpoT (ppGpp synthase II). SpoT is also the degrading enzyme. Only relA spoT double mutants are completely devoid of ppGpp (32).
Such ppGpp-free mutants contain strongly reduced σS levels. Glucose and phosphate starvation, but not amino acid limitation, still induce σS in these mutants (albeit to lower levels than in the wild type). On the other hand, σS accumulation can be triggered by artificially stimulating ppGpp accumulation (64).
ppGpp affects rpoS transcription, as demonstrated with transcriptional rpoS::lacZ fusions (112). However, ppGpp does not seem to specifically target the promoters involved in rpoS transcription, since a transcriptional rpoS::lacZ fusion construct, in which these natural promoters were deleted (and basal expression was due to vector-dependent transcriptional readthrough activity), exhibited similarly reduced expression as the promoter-carrying construct in a ppGpp-free genetic background. It was therefore proposed that in the case of rpoS, ppGpp may affect transcriptional elongation or transcript stability rather than transcriptional initiation (112). In the absence of ppGpp, starvation may result in an uncoupling of transcription and translation, which may lead to increased premature termination, as demonstrated for lacZ mRNA (52, 219, 220).
It is also unclear whether this ppGpp effect is direct or indirect. An increase in the cellular ppGpp content results in the accumulation of polyphosphate, which also stimulates rpoS transcription by an unknown mechanism (see above). It is therefore conceivable that ppGpp acts indirectly via polyphosphate. The finding that a polyphosphate-depleted strain is not impaired in ppGpp accumulation but contains strongly reduced σS levels is consistent with such an indirect mode of action (195). Clearly more research is required to elucidate these relationships at the molecular level.
Is rpoS expression controlled by quorum sensing?
Sometimes high cell density in a bacterial population turns out to be the inducing signal for “stationary phase-inducible” genes. The classical “quorum-sensing” system is the lux system in Vibrio fischeri, where a membrane-permeable acylated homoserine lactone (acylated HSL, the “autoinducer”) is produced by LuxI and accumulates in the medium. Beyond a certain threshold concentration, the autoinducer binds to and activates LuxR, which stimulates the expression of the luxI and luxR genes themselves and the luciferase structural genes (60, 61, 187). Numerous bacterial species contain homologs of the LuxI-LuxR pair, which control a wide variety of output functions (recently summarized in reference 231). Other types of quorum-sensing systems use different kinds of inducing molecules; e.g., gram-positive species use small peptides in general. A hallmark of all these systems is their inducibility on addition of conditioned medium, i.e., spent supernatant obtained from a culture grown to relatively high cell density, which contains the inducing molecule in sufficient concentration.
With respect to rpoS induction by conditioned medium, conflicting data have been reported. Such induction (approximately fourfold) was observed for a transcriptional rpoS::lacZ fusion, and acetate was proposed as the inducing agent (190). With a different rpoS::lacZ operon fusion present in multiple copies, fourfold induction was also found with conditioned Luria broth medium (153), but when the same fusion was present in single copy in the chromosome, induction by spent medium was reduced to a mere 1.6-fold (197). In another study, a transcriptional rpoS::lacZ fusion was found to be completely unaffected by conditioned medium (63). With a set of single-copy transcriptional and translational rpoS::lacZ fusions (114), very little if any induction was obtained, no matter whether conditioned rich or minimal medium was used, and even spent medium freshly prepared in parallel with the induction experiments (to compensate for potential instability of a putative inducer) had little effect on rpoS expression levels (D. Traulsen and R. Hengge-Aronis, unpublished results). It therefore seems that quorum sensing mediated by some excreted medium component does not play a significant role in the regulation of rpoS in E. coli. Therefore, translational induction of rpoS beyond a certain cell density (114) may also be connected to some metabolic alterations rather than to quorum sensing mediated by an excreted substance (see below). The finding that rpoS itself is probably not or is only weakly controlled by quorum sensing does not preclude certain σS-transcribed genes from being subject to such regulation, which may affect the promoters of these genes directly (11, 197, 206).
The E. coli genome sequence (23) does not reveal obvious homologs of genes encoding known acyl-HSL synthases of the LuxI and AinS families (17, 61, 66). There is, however, a LuxR-related protein, SdiA, which may respond to an unidentified acyl-HSL (63, 197). Expression of SdiA itself, as well as the activity of a known target promoter, ftsQp2, responds negatively to conditioned (E. coli) medium, which may mean that the potential of E. coli to respond to an acyl-HSL via SdiA (and thereby activate cell division genes) is downregulated in stationary phase.rpoS, however, does not seem to be under the control of SdiA (63).
Homoserine lactone and homocysteine thiolactone.
Nonacylated HSL has been implicated in rpoS control in E. coli. It was reported that a thrA metL lysC mutant, which is deficient early in the branched pathway that leads to biosynthesis of lysine, methionine, threonine, and isoleucine, had reduced σS levels, which apparently could be suppressed by exogenously adding HSL (at concentrations up to 1 mM). This suppression was weaker in the presence of multiple copies of RspA. Such overexpression also reduced stationary-phase expression of an rpoS::lacZ fusion (which apparently was a transcriptional fusion). Therefore, it was hypothesized that HSL is an inducer for rpoS expression and that RspA may be involved in the degradation of HSL (86). At the time this work was published, this seemed to be in line with quorum-sensing studies that demonstrated the role of acylated HSLs in gene regulation. However, it is now known that free HSL is not the precursor for acylated HSLs that serve as autoinducers in LuxI-LuxR-like systems (72, 165) but, rather, plays the role of an intracellular metabolite. Moreover, there is good evidence against quorum sensing affecting rpoS regulation (see above).
More recently it was found that HSL (up to 1 mM) added to wild-type E. coli did not induce rpoS (197), and the RspA overproduction effect on rpoS is now considered nonphysiological (67). Nevertheless, the idea that RspA may degrade HSL is consistent with the recent observation that RspA-overproducing strains indeed seem to have increased homoserine and decreased HSL levels (U. Sauer, personal communication). Unexpectedly, these strains actually show elevated σS levels, which would not seem consistent with HSL being an inducer for rpoS (226).
Another recent study (67) implicates specifically the methionine biosynthesis pathway in σS control. A metE mutant, which is deficient for conversion of homocysteine to methionine and therefore accumulates homocysteine thiolactone (HCTL), exhibits increased σS levels. Moreover, an asd mutant, which is deficient earlier in methionine biosynthesis (and therefore also in homocysteine and HCTL formation), shows decreased σS levels, and this phenotype could be suppressed by exogenous HCTL (1 mM) (67). During entry into stationary phase in minimal glucose medium, a 2.5-fold accumulation of HCTL was observed in wild-type cells (67). Under these conditions, however, there is little if any activation of rpoS transcription, but σS accumulation is due to posttranscriptional control (114; also see below). Unfortunately, HCTL effects were demonstrated by assaying for σS protein only, and so the level of control affected remains open to speculation (67).
In summary, these studies (67, 86) have demonstrated that amino acid biosynthetic pathways, in particular the branch that leads to methionine (with HCTL as the putative effector), can influence the expression of rpoS. It has not been clarified, however, whether this effect is at the level of rpoS transcription, and the underlying molecular mechanism remains unknown.
Acetate and other weak acids.
In an early study that reported the induction of a transcriptional rpoS::lacZ fusion in spent culture supernatant, the fermentation product acetate (used at 40 mM) was found to activate rpoS expression (190). In another study, however, acetate did not have an effect on rpoS expression (153). The studies agreed that benzoate (10 or 25 mM) has an inducing effect, and it was concluded that this may be so in general for weak acids with a pKa of 4.8 to 4.9 (40 mM propionate was also found to be effective) (153, 190). However, a translational rpoS::lacZ fusion did not show this induction by weak acids (190). One has to take into account, however, that these studies were performed before the advent of limitless precise PCR cloning, and therefore reporter gene fusions had to be constructed in rather complicated ways and often remained incompletely characterized. To finally settle the issue of weak acids in rpoS control, these experiments would have to be repeated and expanded with the precisely characterized systems available today.
Recent genome-wide analyses have shown that addition of acetate to buffered medium results in the activation of various σS-dependent genes and proteins, but unfortunately the cellular concentration and regulation of σS itself were not studied under these conditions (7, 100). Similar conditions resulted in increased synthesis of σS in Salmonella, but the underlying control mechanisms seemed at least in part posttranscriptional (39).
Cellular NADH-to-NAD+ ratio.
Experiments with a transcriptional rpoS::luxAB fusion in a nuoG mutant background (which is defective in a subunit of NADH dehydrogenase) suggested that a high NADH-to-NAD+ ratio somehow downregulates rpoS transcription. Consistent with this proposal, rpoS transcription is low under oxygen-limited (microaerobic) growth conditions, where NADH levels should increase due to the scarcity of oxygen as an electron acceptor for respiration (194). The mechanistic basis of this effect is unclear.
REGULATION OF rpoS TRANSLATION
Initial evidence for posttranscriptional regulation of rpoS was provided by clearly different patterns of expression of transcriptional and translational rpoS::lacZ fusions (114, 127, 135). Translational rpoS::lacZ fusions can actually reflect regulation of rpoS translation as well as of σS proteolysis (114, 148, 192). The latter can be excluded by using translational fusions that contain fewer than 173 N-terminal codons of rpoS, since an essential proteolytic recognition element is located at and around K173 in σS (20; also see below).
Translation of rpoS mRNA is stimulated by a shift to hyperosmolarity (114, 148), by low temperature (199), by a shift to acidic pH (pH 5; Kampmann and Hengge-Aronis, unpublished), or during late exponential phase when a growing culture reaches a certain cell density (114). After the onset of carbon starvation, i.e., on entry into stationary phase, rpoS translation is reduced again, and further increases in σS levels are then due to inhibition of σS degradation, as described below (114).
Role of rpoS mRNA Secondary Structure
There are two species of rpoS mRNA of clearly different lengths (the locations of relevant promoters are given in Fig. 2). Polycistronic nlpD-rpoS mRNA can have two different 5′ ends since there are two closely spaced promoters upstream of nlpD (115). Monocistronic rpoS mRNA originates from rpoSp within the nlpD gene and contains an unusually long nontranslated 5′ region of 567 nucleotides (112, 208). This leader sequence is functionally important, since 5′ deletions in it reduce rpoS expression (38).
Even under conditions where σS protein is hardly detectable, cells produce fair amounts of rpoS mRNA, which seems to remain constant under the translation-inducing conditions mentioned above (8, 147). It is generally believed that control of the rate of translation of already existing complete rpoS mRNA is based on an mRNA secondary structure in which the translational initiation region (TIR) is based paired and therefore not sufficiently accessible to ribosomes (under noninducing conditions). Certain stress signals are hypothesized to trigger changes in this mRNA secondary structure that allow more frequent translational initiation. However, the actual appearance of this rpoS mRNA structure is still largely a matter of speculation.
Theoretical predictions generated with the MFOLD computer program (using complete or partial rpoS mRNA sequences) indicate that approximately 340 nucleotides at the 5′ end of rpoS mRNA fold into an very stable and complex cruciform-type structure (Traulsen and Hengge-Aronis, unpublished). Further downstream, the putative structures are somewhat less stable and the TIR has the potential to fold into two energetically almost equivalent principal structures. One is characterized by a large hairpin that includes the Shine-Dalgarno sequence. There is genetic evidence against this structure playing a role in rpoS translational control (S. Bouché and R. Hengge-Aronis, unpublished results). In the second putative structure, the region around the Shine-Dalgarno sequence is partially base paired to an “internal antisense” region located further upstream, with a relatively long and probably internally structured intervening sequence. There are, however, several theoretical possibilities for the exact location of the “internal upstream antisense” region (Traulsen and Hengge-Aronis, unpublished). Several variations of this second theoretical structure have been published (30, 38, 120, 131), and it is generally believed that this structure may come close to the in vivo reality under noninducing conditions. The only preliminary experimental evidence that such an “internal upstream antisense” structure is in principle correct is provided by two different complementary double point mutations, which showed wild-type expression levels (although one double mutant altered the regulatory pattern, in particular Hfq dependency [see below]) (30, 38). However, the exact details of the in vivo rpoS mRNA secondary structure still await experimental clarification.
In fact, the problem of the correct in vivo rpoS mRNA secondary structure is more complex than, e.g., in the related case of rpoH mRNA, which encodes the heat shock sigma factor σ32.rpoH mRNA folds into a translationally incompetent secondary structure also involving an internal antisense element, which opens up upon heat shock, resulting in a directly temperature-triggered translational induction of rpoH (summarized in reference 236). This process does not involve any regulatory proteins (143), and the experimentally demonstrated rpoH mRNA secondary structure is the one theoretically predicted (142). In rpoS mRNA, however, several proteins and small regulatory RNAs are positively or negatively involved in translational control, and at least some of these can directly bind to rpoS mRNA in vitro (see below). Therefore, theoretical calculations or in vitro structural probing based on rpoS mRNA alone is likely to yield incorrect or at least incomplete results. It seems that the only way of settling the issue of the correct rpoS mRNA secondary structure and its dynamics may be in vivo structural probing. Wild-type strains under different conditions as well as various mutants with cis- or trans-regulatory defects in rpoS translation will have to be tested in such experiments. However, in view of the technical difficulties of such an endeavor, especially with a large mRNA with complex and semistable secondary structure, it is not surprising that such data have yet to be reported for rpoS mRNA.
trans-Acting Factors Involved in rpoS Translation
The RNA binding protein Hfq (HF-I).
More than 30 years ago, the Hfq protein was identified as a host factor (host factor I [HF-I]) essential for replication of phage Qβ RNA (56, 57). Hfq acts as an accessory component of Qβ replicase that binds to several sites in Qβ RNA including the 3′ end (14, 139, 193). Hfq is required for initiating replication specifically of the Qβ RNA plus strand, probaby by affecting the secondary structure at its 3′ end (191). The role of the ribosome-associated (45) Hfq protein in E. coli physiology, however, remained enigmatic until an hfq mutant was observed to have a very pleiotropic phenotype (214), which resembles the phenotype of an rpoS mutant (147). This led to the discovery that Hfq is required for efficient rpoS translation (29, 146). While this can explain the pleiotropy of hfq mutants, Hfq also has physiological functions that are independent of σS (147). In particular, it stimulates the degradation of ompA, miaA, mutS, and its own mRNA (215, 221, 222). Hfq is a 11.2-kDa oligomer-forming protein (57). While this review was under revision, Hfq was reported to form hexameric rings homologous to eukaryotic Sm and Lsm proteins, which occur in the spliceosome and play various roles in mRNA processing (140, 240).
The molecular function of Hfq in rpoS translation is still relatively speculative. Epistasis experiments, where hfq mutations were combined to other mutations or overproduction constructs that affect rpoS translation indicated that Hfq is probably directly involved in translation initiation; i.e., it acts close to or at the level of rpoS mRNA (130, 146, 200, 217, 238). rpoS mRNA coimmunoprecipitates with Hfq in cellular extracts (238). Hfq also binds with high affinity to several sites in a large 5′ fragment of rpoS mRNA synthesized in vitro, which is predicted to fold into the same secondary structure as the wild-type mRNA (Traulsen and Hengge-Aronis, unpublished). A 5′ deletion analysis of rpoS mRNA indicated that regions relatively far upstream of the TIR are important for translational stimulation by Hfq (38). Thus, Hfq binds rpoS mRNA, just as it is able to bind Qβ RNA. However, Hfq does not show similarity to RNA helicases. This makes an active processive unfolding activity unlikely. Alternatively, by binding to a few crucial positions of rpoS mRNA, Hfq may affect the equilibrium between possible alternative secondary structures that are differentially productive for translational initiation. Thus, Hfq may stabilize a semistable rpoS mRNA secondary structure, which can easily open up when some additional stimulating factor is induced or activated (e.g., HU protein or DsrA-RNA, [Fig. 3; see below]). Yet another possibility is that Hfq does not necessarily affect rpoS mRNA secondary structure (although this would not be excluded) but acts like a “platform” bound to rpoS mRNA that recruits additional factors involved in rpoS translational control. The finding that single potentially base-pair-disrupting point mutations in the TIR or in the region likely to be base paired to the TIR result in increased rpoS translation and reduced Hfq dependence (30) appears consistent with both of these putative mechanisms of Hfq action.
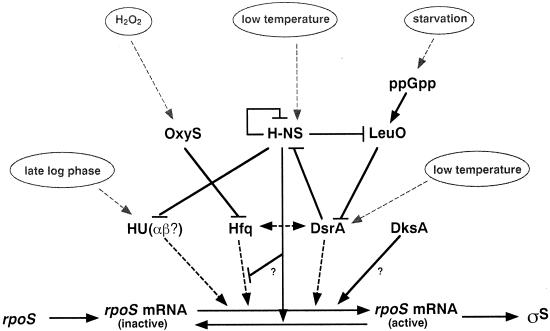
FIG. 3.
The rpoS translational control network. rpoS mRNA is thought to occur in at least two different conformations, one being a more closed structure with the translation initiation region base paired to an upstream internal antisense element, and the other being a more open and translationally competent structure. The translation-stimulating factors Hfq, HU, and DsrA RNA can bind to rpoS mRNA (indicated by broken heavy lines) and together probably drive it into the translationally competent structure. The other components shown are likely to act more indirectly (for further details, see the text).
It was reported that Hfq can also bind to DsrA RNA (200), which is a small regulatory RNA partially complementary to rpoS mRNA that stimulates rpoS translation above all at low temperature (see below for details). Therefore, it was suggested that Hfq may influence DsrA action by forming an active DsrA-Hfq complex and/or by altering DsrA structure (200). However, at 37°C an hfq mutation reduces rpoS translation much more than a dsrA mutation does (146, 199), indicating that Hfq does not act exclusively through DsrA. A hypothetical model consistent with all data available would be that Hfq bound to rpoS mRNA recruits DsrA into a ternary complex (in which secondary structures of rpoS mRNA and/or of DsrA could also be altered) and thereby facilitate translational stimulation by DsrA. In this complex, several Hfq molecules or oligomers bound to different sites on rpoS mRNA could be present. During revision of this review, ternary-complex formation with Hfq was also reported for flhA mRNA and OxyS RNA (240) as well as for galK mRNA and spot 42 RNA (140). Both studies came to the conclusion that the role of Hfq (and perhaps of Sm and LSm proteins in general) is to facilitate specific RNA-RNA interaction. Thus, Hfq could stimulate any process dependent on such RNA-RNA interactions.
If so, stress signal input into rpoS translational control would not necessarily be via a control of the activity or the level of Hfq itself. In fact, hfq mutants show overall reduced activity of translational rpoS::lacZ fusions or rates of σS synthesis (as measured in pulse-labeling experiments), but regulation by stress signals, e.g., by hyperosmotic shift, is not abolished (146). This suggests that stress signals affect the cellular concentrations or activities of the specifically translation-activating or inhibiting components (e.g., DsrA and OxyS) that can join the rpoS mRNA-Hfq complex. These components indeed exhibit pronounced regulation (see below).
HU: a nucleoid protein that also stimulates rpoS translation.
Protein HU is a major protein component of the bacterial nucleoid. It affects overall nucleoid structure and topology but also participates in specific gene regulation, DNA recombination, and DNA repair (155). In addition, HU is required for optimal survival during prolonged starvation (35). In members of the Enterobacteriaceae and Vibrionaceae, two homologous subunits (HUα and HUβ [encoded by hupA and hupB, respectively]) contribute to the formation of active HU protein (158). During growth, the HUα2 homodimer is abundant, whereas during late exponential phase, HUβ is induced and HUαβ heterodimers are formed in E. coli (35). An HU-deficient hupAB double mutant exhibits strongly reduced σS levels because of reduced rpoS translation (12).
In vitro, HU binds with high affinity to a small rpoS mRNA fragment (150 nucleotides covering the TIR and the upstream antisense region probably base paired to the TIR) (12) as well as to a larger fragment (covering more than 700 nucleotides starting from the original mRNA 5′ end) that also binds Hfq (Traulsen and Hengge-Aronis, unpublished). As a DNA binding protein, HU has a strong preference for nicked or cruciform DNA (94). Thus, HU may preferentially recognize secondary-structure elements, such as pronounced bends or kinks, which also occur in RNA secondary structure. HU may directly alter the rpoS mRNA secondary structure, but it is unknown how this effects relates to that of Hfq or of other components that affect rpoS translation (Fig. 3).
Since the induction of HUβ (which is under the negative control of the nucleoid protein FIS [34]) correlates with stimulation of rpoS translation during late exponential growth phase, and specifically since the HUαβ heterodimer is required for stationary phase survival (35), it is tempting to speculate that the heterodimer is the form of HU involved in rpoS translation. However, this hypothesis has yet to be tested experimentally. Nevertheless, phylogenetically, the occurrence of an HUαβ heterodimer correlates with the occurrence of σS (with the exception of the Pseudomonas group, but regulation of σS is significantly different in several aspects in this group).
H-NS and StpA: histone-like proteins acting as RNA chaperones?
H-NS is an abundant histone-like protein with functions in nucleoid organization as well as in gene regulation, where in nearly all cases it acts as a repressor or silencer that can form large nucleoprotein complexes. StpA is a closely related paralog of H-NS with similar properties (although it seems more efficient as an RNA chaperone). Just as with HUα and HUβ, homo- as well as heterooligomers are formed by H-NS and StpA (for reviews, see references 9, 43, and 230).
H-NS-deficient mutants exhibit strongly increased σS levels, which in exponential phase are already similar to those reached by the wild-type only in stationary phase or under other stress conditions (15, 234). In these hns mutants, the rate of rpoS translation is enhanced and proteolysis of σS is strongly reduced or even abolished (234). The slow growth and genetic instability typical of hns mutants are at least partially connected to these abnormally high σS levels, since they can be suppressed by mutations in rpoS (15).
Mechanistically, it is still unclear how H-NS downregulates rpoS translation, but there are a number of possibilities for direct or indirect influences (Fig. 3). Since H-NS can bind to RNA (although high-affinity specific binding has not yet been demonstrated [40, 48]), it may directly interact with rpoS mRNA and affect its secondary structure, perhaps in a transient way as an RNA chaperone. H-NS may also counteract the effects of positive regulators of rpoS translation such as Hfq and/or HU. These possibilities are not mutually exclusive, since the positively acting factors and H-NS may have opposite effects on the equilibrium between two rpoS mRNA conformations that can be translated with different efficiencies (Fig. 3). Consistent with H-NS counteracting Hfq, H-NS deficiency has no effect on rpoS translation in a hfq mutant background (146). H-NS and HU in general seem to play antagonistic roles, e.g., in determining DNA supercoiling (44) or in the expression of certain genes such as ompF (41, 164). Thus, it is also possible that H-NS inhibits rpoS translation by affecting the cellular level of HU or by directly counteracting the stimulatory effect of HU on rpoS translation.
Another candidate for promoting rpoS translation is the H-NS homolog StpA. Several studies have shown that StpA levels are significantly lower than H-NS levels (201, 239), although one report gives approximately equal numbers of H-NS and StpA molecules per cell (10). It seems clear, however, that H-NS and StpA regulate each other negatively at the level of transcription. Therefore, an hns mutant should have an increased cellular concentration of StpA (201, 239). Moreover, StpA is upregulated after a hyperosmotic shift (58). Thus, increased StpA levels appear to correlate with increased rpoS translation. Since StpA can act as a RNA chaperone (40), it was tempting to speculate that it may stimulate rpoS translation. However, high-log-phase levels of σS in an hns mutant were not suppressed by introducing an stpA mutation, and also osmotic induction of rpoS translation was normal in a stpA mutant (Bouché and Hengge-Aronis, unpublished). Therefore, under these conditions, StpA does not seem to play a role in rpoS translation. This, however, does not exclude a potential involvement of StpA under different conditions or in other genetic backgrounds.
Role of small regulatory RNAs in rpoS translation: DsrA, OxyS, and RprA.
Several small regulatory RNAs with important fine-tuning functions in complex regulatory circuits have been identified in E. coli (summarized in reference 1), and three very recently published studies suggest that small regulatory RNAs in E. coli are much more common and significant than previously thought (6, 179, 224). rpoS translation seems to be an especially prominent target for such regulation, with three regulatory RNAs having been found so far. While DsrA and RprA promote rpoS translation, OxyS has an inhibitory function.
DsrA was originally identified as a multicopy suppressor of H-NS-mediated silencing of the rcsA gene in E. coli (198) and was then found to be essential for increased rpoS translation at low temperature (199). DsrA is a stable 87-nucleotide RNA that folds into a three-stem-loop structure (119, 131). A region covering most of stem-loop 1 and the following single-stranded part of DsrA is complementary to an upstream “antisense element” in rpoS mRNA that is assumed to base pair with the TIR region, suggesting that DsrA functions by an “anti-antisense” mechanism that disrupts intramolecular basepairing in rpoS mRNA (119, 120, 131). DsrA plays only a minor role for rpoS translation in cells grown at 37 or 42°C yet becomes the major stimulating factor at 30°C and especially at 20°C (199). The basis of low-temperature translational induction of σS is the clearly enhanced transcription of dsrA as well as a sixfold-increased stability of DsrA at low temperature (177). DsrA and Hfq were recently reported to interact specifically, and Hfq was suggested to stabilize DsrA as well as to alter its secondary structure in a way that promotes association with rpoS mRNA (200). Whether the formation of such a binary complex facilitates DsrA action on rpoS mRNA or whether Hfq already bound to rpoS mRNA (as described above) recruits DsrA, the result is likely to be a ternary complex (see also above). Hfq may affect the secondary structure of both RNAs such that they optimally interact, and with all partners involved interacting with each other, the complex is probably relatively stable. As a result, the formation of an “open” conformation at the TIR of rpoS mRNA that allows ribosome entry would be facilitated (Fig. 3).
Besides rpoS mRNA, DsrA has at least one other target, hns mRNA. While DsrA was initially thought to act like a conventional antisense RNA interfering with hns translation initiation (120), it now seems likely that a region corresponding to unfolded stem-loop 2 of DsrA forms a coaxial stack with two regions in hns mRNA. Negative regulation of hns expression is a consequence of more efficient degradation of hns mRNA within this complex (118). DsrA is predicted to form similar complexes with argR and ilvIH mRNAs, but an involvement of DsrA in the regulation of these genes has not yet been demonstrated (118).
Multifunctionality exerted by different regions may be common in small regulatory RNAs, since it has also been observed for OxyS. OxyS is a 109-nucleotide regulatory RNA that folds into a similar secondary structure to that of DsrA. As a member of the OxyR regulon, OxyS is induced by oxidative stress (hydrogen peroxide) and acts as a pleiotropic regulator (2). Small regions located in loops 1 and 3 of OxyS control translation of fhlA (which encodes a transcriptional activator) by forming a “kissing complex” with two sites of fhlA mRNA, one of which contains the Shine-Dalgarno sequence (3, 5). By contrast, the rather long A-rich single-stranded region between stem-loops 2 and 3 of OxyS is involved in negative regulation of rpoS translation, although this part of OxyS does not show significant sequence complementarity to rpoS mRNA. Coimmunoprecipitation experiments indicate that OxyS binds to Hfq protein. Thus, OxyS may sequester Hfq or form a translationally incompetent ternary complex with Hfq and rpoS mRNA (238) (Fig. 3). OxyS-mediated translational repression of rpoS may be a fine-tuning mechanism to avoid redundant overinduction of oxidative-stress protective genes (katG, gorA, and dps) that are under dual positive control of OxyR/σ70 and σS. It may also prevent the uneconomical induction of the large multifunctional σS regulon under conditions where the cell has to cope with oxidative stress only, i.e., a situation that can be managed by the stress-specific OxyR-mediated response alone.
The third small regulatory RNA involved in rpoS translational control, RprA, was found as a multicopy suppressor for a dsrA mutation (130). In the dsrA mutant background, an rprA null mutation also reduces hyperosmotic stimulation of rpoS translation. However, in the presence of DsrA, neither RprA overproduction nor its knockout seems to affect rpoS expression. Thus, RprA clearly has the potential to stimulate rpoS translation, but the physiological conditions under which this becomes relevant are unknown. The rprA promoter is under positive control of RcsB, a response regulator that activates capsule synthesis (unpublished evidence mentioned in reference 130). RprA exhibits some sequence complementarity to the upstream “antisense” element that basepairs with the TIR of rpoS mRNA and may thus act similarly to DsrA (M. Majdalani and S. Gottesman, personal communication.
The LysR-like regulator LeuO: a repressor for dsrA expression.
LeuO is a LysR-like regulator (189), which is strongly repressed by H-NS in growing E. coli cells (101). Overproduction of LeuO (either from a multicopy plasmid or in a mutant that carries a Tn 10 transposon immediately upstream of leuO with pout of the transposase gene reading into leuO) reduces rpoS translation, especially at low temperature. This effect is entirely dependent on the presence of DsrA, and LeuO was shown to repress dsrA transcription (101). This regulation is direct since LeuO binding sites have recently been identified in the dsrA promoter region (177). A leuO knockout mutation, however, does not affect increased rpoS translation during late exponential phase or in response to high osmolarity or low temperature. This is not entirely surprising, since under these conditions, leuO expression is repressed or even “silenced” by H-NS (101). However, during entry into stationary phase, leuO is induced in a ppGpp-dependent manner (50). This ppGpp-mediated activation may be indirect, since leuO expression is subject to a “promoter relay” activation mechanism that involves the surrounding ilvIH and leuABCD operons (33, 51). As a consequence, LeuO probably downregulates DsrA in stationary phase. While this may alter the expression of other targets of DsrA, the σS level is not affected (E. Klauck and R. Hengge-Aronis, unpublished results), probably because other σS-inducing mechanisms compensate for the reduced levels of DsrA. When all these results are taken together, the physiological role of LeuO is far from clear. However, a hint may come from the “cryptic” bgl operon, where LeuO can antagonize H-NS-mediated (and under certain conditions also σS-dependent) “silencing” (160, 218). Interestingly, the bgl operon becomes expressed in a mammalian host (97). It is thus conceivable that LeuO plays an important regulatory role in a host environment.
DnaK and DksA: a link to heat shock and chaperones.
The heat shock chaperone DnaK, as well as a protein termed DksA (originally identified as a DnaK suppressor [95]), has been implicated in rpoS translation. A dnaK mutant exhibits a stationary-phase-specific multiple-stress-sensitive phenotype very similar to that observed for rpoS mutants (180, 181). This correlates with reduced σS levels in starving dnaK mutant cells (144, 181). Part of this effect is due to reduced rpoS translation, since it can also be seen with RpoS::LacZ hybrid proteins that are not subject to proteolysis. The mechanism behind this effect remains unknown, but the overproduction of the heat shock sigma factor σ32 in the dnaK mutant does not play a role, since a suppressor mutation that reduces the σ32 level and/or activity does not suppress the dnaK effect on σS (144).
DksA is a putative zinc binding protein with similarity to the transcriptional activator TraR (59) and other prokaryotic and eukaryotic regulators (103). The basis of dnaK suppression by multiple copies of dksA is still unclear, but it was suggested that production of some stress response factors might be involved (16). This is consistent with the more recent finding that dksA mutations in Salmonella exhibit reduced σS induction in stationary phase and after a shift to acidic pH. Work with rpoS::lacZ translational fusions indicated that DksA affects rpoS translation by some not yet characterized mechanism (225). In P. aeruginosa, overexpression of DksA inhibits the expression of rhlI, rhlAB, and lasB (26). This would be in line with a repressing effect of σS on the rhl system (228). However, additional data suggest that this effect of DksA overproduction is not due to upregulation of σS alone (26).
EIIA(Glc): a link to the carbon source and energy supply.
A crr mutant, which is defective in the glucose-specific PTS component EIIA(Glc), contains strongly elevated σS levels. Both transcriptional and posttranscriptional effects contribute to this phenotype (217). Higher expression of a transcriptional rpoS::lacZ fusion is fully suppressed by cAMP addition, indicating that the effect reflects stimulation of adenylate cyclase by EIIA(Glc) and negative control of rpoS transcription by cAMP-CRP (see above). However, high σS levels and increased activity of a translational rpoS::lacZ fusion are not fully suppressed by cAMP addition, nor does this effect of crr disruption disappear in a rssB mutant background, where σS is not degraded. Thus, EIIA(Glc) obviously downregulates rpoS translation by some uncharacterized and perhaps indirect mechanism. Moreover, the phosphorylated form of EIIA(Glc) is required for this activity. However, external addition of glucose, which is known to drastically decrease the level of phosphorylated EIIA(Glc) (207), does not result in σS induction (217). In the absence of phosphotransferase system-mediated glucose uptake, however, phosphorylation of EIIA(Glc) reflects the intracellular phosphoenolpyruvate-to-pyruvate ratio (84). Negative regulation of σS by EIIA(Glc) may thus be a function of this ratio, which depends on the nature of the carbon source and the energy supply in general (217).
The cold shock domain proteins CspC and CspE.
CspC and CspE belong to the CspA cold shock protein family in E. coli, although they are expressed at 37°C and are not temperature regulated (169). Overproduction of these two RNA binding proteins strongly stabilizes and thereby increases the cellular level of rpoS mRNA. Whether this is a direct or indirect effect is currently unknown. Such high rpoS mRNA levels are assumed to translate into higher σS levels, since the σS-dependent genes osmY, dps, proP, and katG are significantly activated. Conversely, a cspC cspE double mutant exhibits reduced osmotic induction of osmY and dps (168). Unfortunately, rpoS mRNA levels were not determined in the osmotic shift experiment, and in general the rates of σS synthesis and the cellular σS level were not monitored directly in this CspC-CspE study. It was previously reported that a shift to high osmolarity does not increase the rpoS mRNA level (146), which would not be consistent with rpoS mRNA-stabilizing factors playing a major role in osmotic regulation of rpoS. Therefore, it is possible that the CspC and CspE effects on osmY and dps expression are direct and do not always reflect the regulation of rpoS (168).
A Small Molecule That Influences rpoS Translation: UDP-Glucose
UDP-glucose has been implicated in σS regulation, since several mutants with defects in central carbon metabolism that result in UDP-glucose deficiency exhibit increased σS levels during exponential growth (24). These defects can be in phosphoglucose isomerase (encoded by pgi), with the mutant growing on fructose, as well as phosphoglucomutase (pgm) or UDP-glucose pyrophosphorylase (galU), with the latter two mutants growing on glucose. Glucose and galactose given in trace amounts to the pgi and pgm mutants, respectively, rapidly replenish the internal UDP-glucose pool and in parallele result in a rapid decrease of σS levels (24). More recent work with transcriptional and translational rpoS::lacZ fusions and direct pulse-chase measurements of σS synthesis and degradation indicate that UDP-glucose specifically affects rpoS translation. Moreover, enhanced σS levels in a galU mutation are observed only with an intact hfq gene, which suggests that UDP-glucose directly or indirectly interferes with Hfq function in rpoS translation (A. Muffler and R. Hengge-Aronis, unpublished results). However, the molecular mechanism of UDP-glucose action has yet to be clarified, and it is also unknown whether the cellular UDP-glucose level changes in response to any stress signals that affect rpoS translation.
rpoS Translational Control Network and Stress Signal Input
When all the regulatory factors involved in rpoS translation are considered together, a highly intertwined network characterized by positive and negative feedback regulation emerges (Fig. 3). The regulatory output of this network under different physiological conditions is difficult to predict, especially when changing environmental conditions affect the cellular levels of indirectly acting and multiply connected components such as H-NS or LeuO. DsrA is obviously a central player, since it affects the two global regulators σS and H-NS, with the latter in turn downregulating σS. Thus, DsrA seems to have a dual positive effect on rpoS translation, one direct and the other indirect via H-NS. DsrA, H-NS, and LeuO also seem to form a negative feedback loop (Fig. 3). The physiological function of this regulatory loop, i.e., its behavior and consequences for rpoS translation when external stress signals affect the level of single components in the loop, is currently not clear.
The complexity of the rpoS translational control network makes it difficult to define stress signal input. The only σS-inducing condition, for which the underlying signal transduction mechanism seems pretty straightforward, is a shift to low temperature (around 20°C). This treatment clearly induces DsrA RNA (177), which in turn has a direct positive effect on rpoS translation as described above. Whether low-temperature induction of H-NS (116) plays any role in rpoS regulation is unclear. Starvation induction of LeuO (50) may be relevant only at low temperature, since LeuO acts by repressing DsrA RNA (101). So far, however, different stress conditions have not been studied in combination. Late-exponential-phase induction of rpoS translation (114) correlates with the induction of HUβ (35), but whether this reflects a causal relationship is a matter of speculation. Finally, the intracellular signal that is triggered by osmotic upshift and stimulates rpoS translation more than fivefold within a few minutes (148) is completely elusive.
As complex as the translational control network may be, it is even further interconnected to the networks that control rpoS transcription and σS stability. If ppGpp stimulates LeuO expression under starvation conditions (50), it may at the same time be a strong positive regulator of rpoS transcription (see above) and a negative regulator of rpoS translation (Fig. 3). The physiological function of this multiple role of ppGpp is currently not clear. Its purpose may be to avoid nonappropriate overexpression of σS under conditions of combined stresses, e.g., in response to starvation at low temperature. Also, EIIA(Glc) affects transcription (by controlling adenylate cyclase activity) as well as rpoS translation. H-NS, on the other hand, represses rpoS translation and at the same time keeps σS protein levels down by somehow stimulating σS turnover (see below). At present, it still seems appropriate and helpful to treat the different levels of σS control as separate “regulatory modules.” In the somewhat longer run, however, their interconnection will have to be taken into account.
REGULATION OF σS PROTEOLYSIS
rpoS transcription as well as translation can be stimulated under certain stress conditions, but even in cells that grow in the relative absence of stress, there is a certain basal rate of σS synthesis. However, the cellular σS level remains low because of rapid degradation (114, 208). During growth in rich medium, rpoS transcription is very low and the steady-state σS level is usually at or below the limit of detection, which makes quantitative analyses of σS synthesis and proteolysis difficult. During growth in minimal medium, however, rates of σS synthesis and turnover can be determined by pulse-chase experiments and σS levels can be quantified by immunoblot analysis. Under these conditions, the σS half-life is between 1 min and several minutes (depending on the carbon source) (114, 144, 148, 192, 208). This rapid turnover sets the stage for various stress conditions affecting σS levels by modulating the rate of σS proteolysis. In general, it seems that relatively threatening stress conditions tend to affect σS degradation, maybe because this allows the most rapid reaction. These stresses include sudden carbon starvation (114, 208), osmotic upshift (148), and shift to acidic pH (18), which result in σS stabilization within a few minutes. On the other hand, the classical heat shock procedure, i.e., a shift from 30 to 42°C, results in a more moderate increase in σS half-life, which takes approximately 20 min to develop (144).
σS Degradation by the Complex ATP-Dependent ClpXP Protease
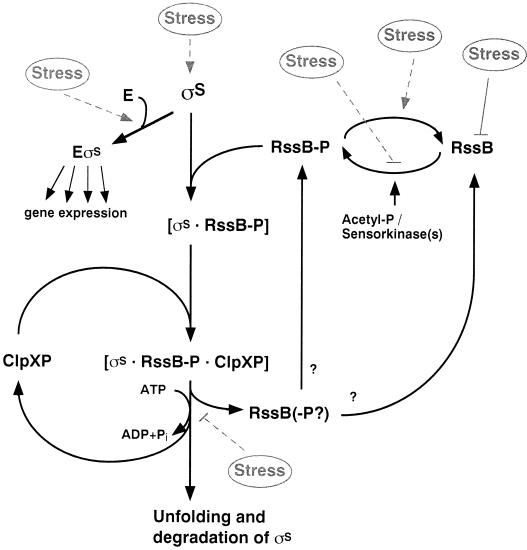
The ClpXP protease is responsible for σS degradation. ClpXP is a barrel-shaped processive protease consisting of two six-subunit rings of the ATP-hydrolyzing ClpX chaperone, which play the role of substrate-discriminating and unfolding gatekeepers to the inner proteolytic chamber formed by two seven-subunit rings of ClpP (69, 99, 223). Mutations in clpP as well as in clpX result in stabilization of σS (192). Since the clpP and clpX genes constitute an operon (68, 133), the clpP phenotype could in principle have been due to polarity on clpX, but the inability to suppress σS stability in the clpP mutant by providing clpX in trans confirmed that the entire ClpXP complex is required for σS proteolysis (Muffler and Hengge-Aronis, unpublished). Recently, it has been possible to reconstitute σS degradation in vitro, and these experiments have defined ClpXP as well as a specific recognition factor (see below) as essential and sufficient for the basic process of σS proteolysis (243). σS degradation by ClpXP is complete; i.e., no stable degradation products have been observed.
The Response Regulator RssB: a σS Recognition Factor with Phosphorylation-Modulated Affinity
In contrast to other ClpXP substrates, σS cannot be recognized by ClpXP alone, as demonstrated both in vivo and in vitro (145, 243). Rather, a specific recognition factor, the RssB protein (also termed SprE, MviA, or ExpM in different bacterial species), is required (4, 18, 145, 170). A mutation in rssB results in the stabilization of σS (and also of otherwise unstable RpoS::LacZ hybrid proteins) and therefore in elevated σS levels in exponential phase (145, 170). RssB belongs to the two-component response regulator family of proteins, whose activity is modulated by phosphorylation of a conserved aspartyl residue in the N-terminal receiver domain (D58 in RssB). In vitro experiments have shown that phosphorylated RssB directly interacts with σS (20). Phosphorylation as well as σS binding in vitro is lost with RssB variants, in which D58 is replaced by other amino acids, consistent with strains carrying the same mutations exhibiting high levels of stable σS (21, 25, 102). RssB is essential for σS degradation in vitro (243) and may be specific for σS, since turnover of another ClpXP substrate, λO protein, does not depend on RssB (242, 243). In conclusion, the response regulator RssB is an essential, specific, and direct σS recognition factor, whose affinity for σS and therefore whose activity in σS proteolysis are modulated by phosphorylation of its receiver domain.
Like most response regulators, RssB consists of at least two domains, the N-terminal receiver and a C-terminal output domain (the latter could also be more than a single domain). The unique role of RssB in proteolysis is reflected in a unique output domain(s) without similarity to any other protein of known function. In certain response regulators, the output domain alone is mechanistically responsible for the molecular function (most often in transcription initiation), with the receiver domain imposing regulation by phosphorylation-modulated intramolecular inhibition (42, 70). In other cases, phosphorylation of the receiver domain actively contributes to the output function, e.g., by stimulating oligomerization (53, 233) or by exposing an interactive surface in the receiver itself (138, 237). RssB belongs to the latter class, since the isolated N- and C-terminal domains of RssB are functionally inactive in vitro and in vivo; i.e., the N-terminal receiver domain plays an active and positive role in RssB function (102). The molecular details of the RssB-σS interaction remain to be elucidated, but there is evidence that RssB, unlike many other response regulators, does not dimerize or oligomerize on phosphorylation and/or σS binding and that the RssB-σS complex exhibits 1:1 stoichiometry (102).
The cellular concentration of RssB (which in growing cells is around the limit of detection) is the limiting factor for the rate of σS proteolysis in vivo. This means that RssB can be titrated by increased σS synthesis (174). This mechanism can be exploited for stress-induced stabilization of σS (see below). On the other hand, cells have to continuously adjust the expression of RssB to σS in order to maintain σS proteolysis during growth despite controlled or accidential variations in the rate of σS synthesis. This is achieved by a homeostatic feedback coupling that is provided by rssB transcription being dependent on σS (185; Pruteanu and Hengge-Aronis, submitted). These two reports, however, do not agree on the location of the σS-dependent promoter, since Ruiz et al. (185) invoke a promoter just upstream of rssB, which was not found by Pruteanu and Hengge-Aronis (174), who provide evidence that rssB transcription is driven exclusively from the σS-controlled rssAB operon promoter. σS control of rssB expression also results in indirect negative autoregulation of rpoS as well as of rssB, since σS stimulates the expression of a factor, RssB, that initiates σS disappearance (174).
The Turnover Element: the RssB Binding Site within σS
Unlike many other proteolysis substrates, which feature recognition sequences or elements at or close to the N or C termini (96, 229), σS was found to contain a “turnover element” somewhere in the middle of its sequence. Initial evidence for such a proteolysis-promoting element came from the analysis of RpoS::LacZ hybrid proteins carrying N-terminal σS fragments of different lengths. Whereas relatively short hybrid proteins were stable and yielded high β-galactosidase activities in log phase, extending the σS part beyond a certain region resulted in hybrid proteins that were subject to the same regulated turnover as σS itself and yielded low β-galactosidase activities (148, 192). These studies roughly mapped the turnover element somewhere in or downstream of region 2.4 (which is involved in recognition of the −10 promoter element). Consistent with σS and σ70 recognizing the same −10 consensus (19, 47, 81), there is extreme amino acid similarity of these two sigmas up to the end of region 2.4. Just beyond this point, however, the sequences diverge. Reasoning that only σS is unstable and therefore should contain the turnover element, a number of amino acids in this region of σS, which clearly differ from those in σ70, were replaced by the latter ones. This identified K173 as an absolutely crucial amino acid for σS proteolysis. A single point mutation, K173E, eliminates rapid σS proteolysis (20). Single mutations in E174 or V177 also enhance the σS half-life two- and threefold, respectively (20). In conclusion, K173 is a core amino acid of the turnover element. Moreover, K173 is also crucial for promoter recognition in the extended −10 part of a promoter (specifically of a C in position −13) (19), and this part of σS or σ70 is now termed region 2.5 (13, 19).
In vitro experiments with the σS(K173E) variant demonstrated that K173 is essential for interaction with RssB. In other words, the turnover element around K173 represents the binding site for RssB (or an essential part thereof) (20). Unfortunately, there is no experimentally determined structural information for region 2.5 of sigma factors (a known partial structure of σ70 ends with region 2.4 [132]), but the sequence between V172 and K188 is strongly predicted to be in an α-helical conformation. The double role of K173 in RssB binding and in interaction with the extended −10 promoter region also means that K173 must be surface exposed, no matter whether σS is in the RNA polymerase complex or not. In an initial attempt to estimate the extension of the binding site for RssB, the amino acids predicted to form the α-helix in region 2.5 were N-terminally fused to β-galactosidase and were found to be sufficient for RssB binding in vivo as well as in vitro (A. Stüdemann, E. Klauck, and R. Hengge-Aronis, unpublished results). This means that a relatively small part of σS, most probably just one α-helix, is sufficient for interaction with RssB, whereas on the other side, the entire RssB protein (or at least more than a single domain) is required for binding. This situation is reminiscent of protein binding by the DnaK chaperone, where a small target sequence with one crucial amino acid in the substrate protein is bound in a pocket formed by DnaK (134, 184, 244). One may speculate that a comparable mechanism operates for σS binding to RssB, with formation of a high-affinity binding cavity in RssB being dependent on phosphorylation of its receiver domain.
Initiation of σS Proteolysis: the RssB Cycle
Once bound to RssB, σS is transferred to the ClpXP protease, where, like other Clp protease substrates, it is unfolded and completely degraded by an ATP hydrolysis-dependent mechanism (Fig. 4). A ternary complex between σS, RssB, and ClpX and a quaternary complex also involving ClpP have been observed in vitro (243). RssB is then released from the complex, as indicated by in vitro as well as in vivo data (102, 243). Thus, RssB is recycled and plays a catalytic role in the initiation of σS degradation. Taking into account a σS half-life of 1.5 min (114), the cellular σS-to-RssB ratio of approximately 20:1 (21), and the fact that RssB remains a monomer in the σS-RssB complex (102), it has been estimated that a single molecule of RssB can initiate the degradation of at least six or seven molecules of σS per minute (in cells growing in minimal glucose medium). The real number may actually be somewhat higher, since this estimation was based on the degradation of fully synthesized σS molecules only (as visible in pulse-chase and immunoprecipitation experiments). In addition, however, nascent σS polypeptide chains can probably enter the degradation pathway as soon as they are long enough to contain the recognition site for RssB (102).
FIG. 4.
Role of RssB-ClpXP and putative signal input in the σS recognition and degradation pathway. The response regulator RssB is an essential, specific, and direct σS recognition factor. RssB delivers σS to the ClpXP protease, where σS is unfolded and completely degraded whereas RssB is released. σS binding requires RssB phosphorylation, but it is unclear whether the catalytic cycle of RssB involves obligatory dephosphorylation during release and subsequent rephosphorylation. Stress signals may affect (i) the phosphorylation of RssB and therefore RssB-σS complex formation; (ii) the cellular level of RssB (which in growing cells is rate limiting for σS proteolysis); (iii) the synthesis of σS such that RssB becomes titrated on σS overproduction; (iv) σS association with RNA polymerase core enzyme, which protects against binding by RssB; and (v) the function of the ClpXP protease itself (see the text for details). However, the molecular details of the stress signal input pathways involved are still largely unknown.
So far, it is unknown whether RssB is dephosphorylated during its catalytic cycle. Dephosphorylation could be a convenient mechanism for RssB release. Contact with ClpXP may stimulate an RssB autophosphatase activity, which in purified RssB alone would be cryptic (spontaneous in vitro dephosphorylation occurs with a half-life of more than 1 h) (102). Alternatively, σS may lose its affinity for RssB during its unfolding and initial transfer into ClpP, which would alter the conformation of σS in and around region 2.5, whose integrity is required for RssB interaction (20). Dephosphorylation of RssB during its catalytic cycle would imply rephosphorylation as an obligatory part of this cycle (Fig. 4), which has interesting regulatory implications (see below).
Signal Integration in the Control of σS Proteolysis
Defining the linkages between stress signal transduction pathways and the σS recognition and degradation pathway remains a challenge for future studies, mainly because these signal transduction pathways themthelves have yet to be elucidated. However, it is becoming apparant that the basic RssB-ClpXP system has the potential to act like a multiple-signal-integration machinery. Theoretically, this system provides a wide range of possibilities for downregulating the rate of σS degradation by stress signal transduction pathways (Fig. 4). In the presence of preliminary data only, the existence of these mechanims remains speculative at present, but the following theoretical discussion may provide useful hypotheses for future work.
With RssB being a response regulator, it is reasonable to expect that some stresses will affect the phosphorylation state of RssB. Unfortunately, this could not yet be demonstrated directly in vivo, since the cellular RssB level is at the limit of (and sometimes below) detection in immunoblot experiments (21, 141). So far, no cognate sensor kinase for RssB has been identified and the E. coli genome sequence does not provide any obvious candidate. Acetyl phosphate seems to contribute to RssB phosphorylation, since acetyl phosphate-free pta-ackA mutants exhibit longer σS half-lifes; however, since σS proteolysis is not completely abolished in these mutants, at least one additional phosphoryl donor for RssB is likely to exist (25). It therefore seems possible that RssB is phosphorylated by “cross talk” from other sensor kinases, consistent with phosphorylated RssB (and therefore rapid σS recognition and degradation) representing the “default” state of the system in the absence of stress. Environmental stress would then trigger some mechanism that actively dephosphorylates RssB. However, a specific RssB phosphatase (or a sensor kinase that switches to RssB phosphatase activity) still awaits identification.
An interesting variation on this theme becomes possible if RssB is obligatorily dephosphorylated during its release from the complex with σS and ClpXP (Fig. 4). In this case, a specific stress-activated phosphatase may be dispensable, and the entire regulation of phosphorylation and dephosphorylation of RssB could be mediated by one (or several) cross-talking sensor kinases that would then have to be inhibited by certain stresses. However, signal input flexibility and precision would certainly be higher if the system actively controls phosphorylation as well as dephosphorylation, which could be differentially targeted by different stress conditions.
In cells growing in minimal medium, there is a finely tuned balance between σS synthesis and proteolysis. There is evidence that the cellular level of RssB is the rate-limiting factor for σS proteolysis in vivo (174). Accordingly, a sudden strong increase in σS synthesis results in σS stabilization because of titration of RssB. This is observed on artificial induction of σS synthesis ( 174), or on osmotic upshift or pH downshift, where the rate of σS synthesis increases severalfold within a few minutes (146, 148; Kampmann and Hengge-Aronis, unpublished). Therefore, osmotically triggered or pH-triggered stabilization of σS may in part be a passive consequence of the stimulation of rpoS mRNA translation (see above).
With RssB being limiting for σS proteolysis, it is theoretically also possible that some sort of stress may result in a reduction of RssB levels rather than of RssB activity. However, starvation leads to a moderate increase in the cellular RssB concentration (21, 185; Pruteanu and Hengge-Aronis, submitted), whereas osmotic upshift has no effect (21). Also, for some other stresses known to affect σS proteolysis, alterations in RssB levels were not observed (M. Pruteanu and R. Hengge-Aronis, unpublished results).
Association of σS with RssB or RNA polymerase core enzyme seems mutually exclusive. Core enzyme protects σS against degradation in vitro and, at equimolar concentrations with RssB, can even actively displace RssB from the σS-RssB complex (243). This suggests that any factors that in vivo may disfavor σ70 in its competion with σS for core polymerase or that may somehow directly stimulate σS holoenzyme formation would also contribute to σS stabilization. Such σS-activating and -stabilizing factors have not yet been unequivocally identified, but there are reasons to postulate their existence (see below).
Finally, there is indirect in vivo evidence that a σS-RssB complex is still formed in carbon-starved cells (21), which indicates that under these conditions, inhibition of some activity in the unfolding and degradation pathway downstream from σS-RssB binding contributes to stabilization of σS. In that respect, it may be relevant that the ClpX level is likely to be reduced due to growth stage-specific clpPX mRNA processing in stationary-phase cells (122).
In summary, this extraordinary potential for multiple signal integration in the RssB-ClpXP system can explain why so many different stress signals can finally result in the same phenomenon, i.e., σS stabilization. In the future, it will have to be worked out which stresses act by which of the mechanisms outlined above. An increased flexibility and fine-adaptive power would be achieved if certain stresses used different combinations of these mechanisms.
Additional Factors with Uncharacterized Molecular Functions in σS Turnover
It is obvious from the previous section that signal integration in the control of σS recognition and degradation is highly complex and that therefore probably several, if not many, components involved are still missing from our picture. Unfortunately, however, it may be predicted that in a system that integrates many signal input pathways (which may also be interconnected!), a mutation in a single signal-transducing component probably produces a minor or even no phenotype unless the mutant is tested under very specific conditions. This may explain why mutant searches using screens that reflect σS degradation have not yielded mutations in novel genes with clear-cut phenotypes other than in rssB, clpP, or clpX (49; F. Reindl, E. Kampmann, and R. Hengge-Aronis, unpublished results). However, there is circumstantial evidence that certain genes somehow contribute to the control of σS proteolysis.
RssA.
The rssA gene is located upstream of rssB, and the two genes constitute an operon with a single promoter upstream of rssA (174). The N-terminal part of RssA belongs to a family of putative serine esterases of unclear physiological functions that have been conserved from bacteria to humans (128). RssA deficiency, as well as overproduction, was observed to have minor but reproducible effects on the cellular σS levels (G. Kampmann, M. Marquardt, and R. Hengge-Aronis, unpublished results). So far, it is not clear whether RssA acts directly or indirectly and what its actual biochemical function is. RssA may be involved in RssB dephosphorylation under some conditions, but alternative explanations are at present not excluded.
The histone-like protein H-NS.
hns mutants exhibit abnormally high σS levels in exponential phase (15, 234). Although H-NS is known as an abundant nucleoid-associated protein that represses or even silences the transcription of numerous genes (230), its effect in the control of σS is posttranscriptional, with rpoS translation being stimulated and σS proteolysis being strongly reduced (15, 234). It seems likely that H-NS indirectly affects σS degradation by controlling the expression of some other regulatory factor. The stability of σS in the hns mutant is certainly not due to a lack of RssB; rather, the cellular RssB level even seems slightly increased in the mutant (102), consistent with σS control of rssB expression (174, 185). Alternatively, H-NS may regulate some component involved in the control of RssB activity, such as, phosphorylation, but the component(s) still awaits identification.
The LysR homolog LrhA.
In stationary-phase cells, the outer membrane porin OmpF is downregulated by a σS-dependent mechanism (170). Overexpression of LrhA, a regulator of the LysR family with hitherto unknown function, was found to suppress this phenotype by reducing σS levels, whereas an lrhA null mutation had the opposite phenotype. Epistasis experiments with rssB mutants have shown that LrhA affects σS proteolysis. The lrhA effect was still observed in a strain in which rssB was expressed from a nonnative promoter. Therefore, it was speculated that LrhA somehow affects the activity of RssB, i.e., its phosphorylation state (65). However, LrhA belongs to a family of transcriptional regulators, and therefore it is likely to control the expression level of some other factor, which may then play a direct role in of RssB activation. Since stress-induced alterations in the rate of σS proteolysis are too rapid to involve transcriptional induction of some factor, which in turn affects RssB activity, LrhA is unlikely to be part of a stress signal transduction pathway that controls σS proteolysis. A homolog of LrhA, HexA, is known in Erwinia carotovora, where it controls genes involved in exoenzyme synthesis, plant virulence, and motility (73, 149).
The DnaK chaperone.
In stationary phase, dnaK mutants show reduced σS content and exhibit a pleiotropic phenotype very similar to that of an rpoS mutant (144, 180, 181). As outlined above, part of this effect is due to reduced rpoS translation. In addition, however, various lines of evidence indicate that dnaK mutants are partially defective for σS stabilization in starved cells (144, 181). However, the mechanistic link between the DnaK chaperone and the σS recognition and degradation system (Fig. 4) has not been identified. It is interesting that the DnaK system plays opposite roles in controlling the proteolysis of σS and of the heat shock sigma factor σ32: whereas DnaK stabilizes σS, it is crucial for the degradation of σ32. DnaK may also play a role in heat shock stabilization of σS, which is a relatively slow process (taking up to 20 min) that more or less correlates with the accumulation of the heat shock protein DnaK (144). By contrast, the extremely rapid and transient heat shock stabilization of σ32 is due to titration of the DnaK chaperone by suddenly accumulating denatured proteins (236).
It is possible that DnaK plays a role in starvation sensing. Sudden carbon source starvation is almost immediately followed by a strong reduction in overall protein biosynthesis and consequently in reduced levels of newly synthesized but not yet natively folded polypeptides, some of which are DnaK substrates. Thus, more DnaK may be available to somehow protect σS from degradation (144). At present it is unknown whether only DnaK or the entire DnaK chaperone machine (also including DnaJ and GrpE) is involved in σS protection from proteolysis.
A Small Molecule That Affects σS Proteolysis: Acetyl Phosphate
During growth on glucose as a carbon source, acetyl phosphate is produced by phosphotransacetylase (encoded by the pta gene) from acetyl coenzyme A. Acetate kinase (ackA) then uses acetyl phosphate and ADP to produce ATP and acetate, which is excreted (46, 173). An acetyl phosphate-free pta-ackA mutant exhibits an approximately twofold-increased σS half-life, and since acetyl phosphate is an excellent phosphoryl donor for RssB in vitro, a similar in vivo function has been postulated (25). With a few exceptions (98, 173, 196), this effect is in contrast to findings with most other response regulators, where in vivo phosphotransfer from acetyl phosphate usually cannot be detected unless the cognate sensor kinase (which in the absence of its specific stimulus often acts as the response regulator phosphatase) is eliminated by mutation (137). Thus, for most response regulators, phosphorylation with acetyl phosphate is possible in vitro but does not play a physiological role. For σS proteolysis, however, it seems to be physiologically relevant. Nevertheless, acetyl phosphate cannot be the only phosphodonor for RssB, because σS turnover is not completely abolished in the pta-ackA mutant. Also, the observation that the pta-ackA mutation has a minor (and under some conditions no) effect on the σS level (25, 39) indicates that acetyl phosphate plays a rather subtle role in the combination of all the influences that together determine the actual σS level (in practice this means that in order to see such an effect, it is not sufficient to measure the σS level, an all-integrative parameter, but that σS degradation has to be directly assayed, e.g., by pulse-chase labeling).
Recently, the effects of acetate addition (at neutral pH) on genome-wide gene expression have been investigated. More than two dozen acetate-inducible genes (7) or proteins (100) have been identified, many of which are σS controlled. This effect is specific for acetate since formate produced largely opposite effects (100). In one of these studies, acetyl phosphate was excluded as the direct inducer, since a pta-ackA mutant exhibited constitutively high levels of the same proteins. Therefore, a high acetyl coenzyme A level was proposed to be the inducing signal (100). Unfortunately, σS itself and its different levels of control were not studied in those acetate-treated cells (see above for discussion of a putative effect of acetate on rpoS transcription).
REGULATION OF σS ACTIVITY
σS activity in transcriptional initiation requires its association with RNA polymerase core enzyme. However, of all the sigma factors of E. coli, σS is the one with the lowest affinity for the core enzyme in vitro (129). Moreover, even in stationary phase, the cellular level of σS does not exceed approximately one-third of the cellular level of σ70 (89, 92). Given these basic data and the general competion of sigma factors for core, one wonders how σS can recruit RNA polymerase core to any significant extent in vivo and activate the expression of genes at all. It may be that the putative anti-σ70 factor Rsd (90, 91) shifts the balance somewhat in favor of σS, but, given the relatively low cellular level of Rsd (91), this effect cannot be expected to quantitatively eliminate σ70 activity. Moreover, some stationary-phase-induced genes are expressed by σ70-containing RNA polymerase (77). It is conceivable that covalent modification (163) or ppGpp binding (213) of core polymerase may improve the interaction with σS. In addition, it is tempting to speculate that at least under stress conditions, where σS is induced, some unidentified factor(s) may exist that stimulates its interaction with core polymerase.
In Vivo Evidence for Regulation of σS Activity
The crl gene product stimulates the expression of curli fimbriae (162), which are involved in cell-cell aggregation (182, 183) and adhesion to eukaryotic cells (71, 162). Synthesis of curli (with the subunits encoded by csgAB) is also dependent on σS (8, 161). More recently, it was found that the role of Crl is not curli specific but that Crl has a stimulatory effect on the expression of a number of σS-activated genes. Also, for negative effects of σS (e.g., on the expression of OmpF or in a not further clarified negative autoregulation of σS itself), Crl seems to play a synergistic role (171). However, Crl does not downregulate σS itself (if anything, σS levels increase in a crl mutant, because the above-mentioned negative feedback in σS control is relieved). Moreover, Crl does not seem to be a DNA binding protein since it does not contain any known DNA binding motif, nor could (nonspecific) binding to DNA cellulose be observed. Therefore, it was proposed that Crl may activate σS, perhaps by modulating the σS association with RNA polymerase core (171). This is certainly an attractive hypothesis, but a direct demonstration of such a function is still missing. If Crl stimulates σS-core interaction in stationary phase, it could be expected to directly interact with σS and/or core, and it should also contribute to σS stabilization (since association with core protects σS from RssB binding, as outlined above), but these specific hypotheses have not yet been tested.
There are specific conditions where high cellular levels of σS do not result in high expression of σS-dependent genes, i.e., where σS levels and activities do not appear to correlate. This seems to be the case when σS is artificially overproduced during exponential growth (e.g., from an isopropyl-β-d-thiogalactopyranoside [IPTG]-inducible promoter [R. Lange and R. Hengge-Aronis, unpublished results]). Another such situation is provided by the classic glucose-lactose diauxie experiment (54). σS is degraded with a half-life of 2 min during the first growth phase on glucose. During the lag phase, σS is completely stabilized, resulting in the accumulation of σS as well as mRNA of the σS-dependent osmY gene. When the cells then start to grow on lactose, the σS half-life remains relatively high (more than 20 min) and σS levels therefore decrease only slowly. However, no osmY mRNA can be detected during the end of the lag phase and during growth on lactose (54). This observation is formally reminiscent of the inactivation of σ32 during temperature downshift, where σ32 levels decrease much more slowly than expression levels of σ32-dependent heat shock genes (204).
It is tempting to speculate that under such conditions, some factor(s) necessary for σS activation may be missing or inactive (or some inactivating factor may be abundant). However, such effects have to be interpreted with caution, because some σS-dependent genes require additional regulatory factors besides σS (78), which may not be present or active under the specific conditions studied.
In summary, there is initial although not conclusive evidence that activity of σS, i.e., probably σS association with RNA polymerase core in competition with other sigma factors, is regulated. At present, it can only be speculated that core enzyme modification (e.g., by ppGpp binding) or additional proteins (e.g., Crl) could be involved. One of the problems, however, is to experimentally distinguish this activation of σS from activation of σS-dependent transcription by some conventional regulatory mechanism. A clear answer probably requires in vitro transcription experiments that would allow the effects on sigma factor competition to be separated from “normal” activation of transcription.
The Response Regulator RssB Can Act Like an Anti-Sigma Factor for σS
In wild-type cells, RssB binding to σS is the first step in σS delivery to the ClpXP protease. However, in clp mutants as well as in stationary-phase cells engineered to contain a slightly increased RssB-to-σS ratio, RssB binding to σS results in σS inhibition as a transcription initiation factor; i.e., reduced expression of σS-dependent genes can be observed (21, 141, 242). This suggested that in the absence of ClpXP, or under conditions where σS degradation is inhibited at the protease level, RssB can in principle act like an anti-sigma factor, e.g., by interfering with σS-core polymerase association (21). Recent in vitro data indicate that the association of σS with RssB is indeed mutually exclusive with σS-core association (243).
Thus, in principle, RssB has the potential to function as an anti-sigma factor for σS. Are there any conditions where this is physiologically relevant? Under the best-studied conditions (growth in minimal medium with various carbon sources), any binding of σS to RssB results in rapid degradation of σS. Moreover, RssB is present at clearly substoichiometric concentrations (21). However, two scenarios where σS inhibition by RssB could be physiologically relevant are at least conceivable (21). First, earlier in evolution, RssB may have been a stress-regulated anti-σS factor (originally produced in stoichiometric amounts with σS) before it was recruited by the proteolysis machinery to serve as a specific recognition factor with a catalytic function. Second, there may be unidentified conditions where (i) significant levels of σS are present, (ii) RssB may be upregulated, and (iii) ClpXP may be less active or downregulated. Although RssB is moderately upregulated in stationary phase (21, 174, 185), further studies of rssB regulation have so far not produced evidence that there are any strongly RssB-inducing conditions under which RssB could function as an anti-σS factor (Pruteanu and Hengge-Aronis, unpublished). Thus, the evolutionary scenario of a change in RssB function from anti-σS factor to σS proteolysis recognition factor seems more likely.
CONCLUSIONS AND PERSPECTIVES
With an ever-increasing number of factors that contribute to rpoS transcription and translation as well as to σS proteolysis, σS now appears to be an E. coli protein with one of the most complex regulation systems. Nevertheless, a relatively clear picture of the basic regulatory mechanisms, at least in posttranscriptional regulation, has emerged recently. The basic control of rpoS translation uses rpoS mRNA secondary structure, the Hfq and HU proteins, and small RNAs such as the DsrA mRNA. The core σS degradation machinery clearly consists of the ClpXP protease and the phosphorylation-modulated σS recognition factor RssB. Beyond this, however, numerous questions have yet to be answered.
Above all, the way in which the multiple signals that control σS are integrated remains largely unexplored. Translational control of rpoS involves a plethora of components (Fig. 3), but what are their molecular functions and interplay? How does late log phase, high osmolarity, or shift to acid pH affect the rate of rpoS translation? In σS proteolysis, the way in which the RssB-ClpXP system functions allows us to predict the overall functions of the “missing” components that have yet to be identified (Fig. 4). These include factors that, in response to certain stress conditions, affect (i) RssB phosphorylation, (ii) ClpXP activity in general or its ability to specifically degrade σS, or (iii) σS association with core RNA polymerase. The latter also indicates a link between the control of σS proteolysis and activity. In general, the different levels of σS control do not operate independently from each other, but components like H-NS or EIIA(G1c), which affect more than one level of control, may play a coordinating role.
There is growing evidence for complex connections between σS regulation and other regulatory circuits. These include a linkage to oxidative stress that operates via OxyS RNA; to the CRP regulon, catabolite repression, and inducer exclusion that uses cAMP-CRP and EIIA(Glc); or to the heat shock response which involves the DnaK chaperone. These connections are certainly relevant under multiple simultaneous stress conditions, which is probably a more natural situation than the carefully controlled single-stress situations usually studied in the laboratory.
With all the currently available information taken together, we appear to be approaching a situation where the σS regulatory network is becoming so complex that quantitative (mathematical) analysis and simulation may become helpful in really understanding its inherent overall potential and actual behavior under different conditions. With many regulatory components and their basic biochemical functions now identified, such analysis seems feasible. Since σS is connected to many other crucial regulatory modules in the cell, it may even provide a good starting point for a future quantitative analysis of the entire cellular regulatory network.
Acknowledgments
I thank numerous colleagues for communicating results prior to publication and the present and past members of my laboratory who have contributed to establishing the knowledge summarized in this review.
Research in my laboratory has been generously funded by the Deutsche Forschungsgemeinschaft (Priority Program “Regulatory Networks in Bacteria”; SFB 156-A8; Gottfried-Wilhelm-Leibniz Program), the State of Baden-Württemberg (Landesforschungspreis), and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Altuvia, S., and E. G. H. Wagner. 2000. Switching on and off with RNA. Proc. Natl. Acad. Sci. USA 97:9824-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia, S., D. Weinstein-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: roles as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia, S., A. Zhang, L. Argaman, A. Tiwari, and G. Storz. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17:6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, R. A., E. T. Palva, and M. Pirhonen. 1999. The response regulator ExpM is essential for the virulence of Erwinia carotovora subsp. carotovora and acts negatively on the sigma factor RpoS (σS). Mol. Plant-Microbe Interact. 12:575-584. [DOI] [PubMed] [Google Scholar]
- 5.Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300:1101-1112. [DOI] [PubMed] [Google Scholar]
- 6.Argaman, L., R. Herschber, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 7.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnqvist, A., A. Olsén, and S. Normark. 1994. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 9.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 10.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baca-DeLancey, R. R., M. M. T. South, X. Ding, and P. N. Rather. 1999.Escherichia coli genes regulated by cell-to-cell signalling. Proc. Natl. Acad. Sci. USA 96: 4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balandina, A., L. Claret, R. Hengge-Aronis, and J. Rouvière-Yaniv. 2001. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 39:1069-1079. [DOI] [PubMed] [Google Scholar]
- 13.Barne, K. A., J. A. Bown, S. J. W. Busby, and S. D. Minchin. 1997. Region 2.5. of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the “extended-10” motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Qβ plus und minus strand RNAs by Qβ replicase. J. Mol. Biol. 232:512-521. [DOI] [PubMed] [Google Scholar]
- 15.Barth, M., C. Marschall, A. Muffler, D. Fischer, and R. Hengge-Aronis. 1995. A role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of Prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 18.Bearson, S. M. D., W. H. Benjamin Jr., W. E. Swords, and J. W. Foster. 1996. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 178:2572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoterσS-dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 20.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker, G., E. Klauck, and R. Hengge-Aronis. 2000. The response regulator RssB, a recognition factor for σS proteolysis in Escherichia coli, can act like an anti-σS factor. Mol. Microbiol. 35:657-666. [DOI] [PubMed] [Google Scholar]
- 22.Bishop, R. E., B. K. Leskiw, R. S. Hodges, C. M. Kay, and J. H. Weiner. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J. Mol. Biol. 280:583-596. [DOI] [PubMed] [Google Scholar]
- 23.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. ColladoVides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 24.Böhringer, J., D. Fischer, G. Mosler, and R. Hengge-Aronis. 1995. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 177:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouché, S., E. Klauck, D. Fischer, M. Lucassen, K. Jung, and R. Hengge-Aronis. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 27:787-795. [DOI] [PubMed] [Google Scholar]
- 26.Branny, P., J. P. Pearson, E. C. Pesci, T. Köhler, B. H. Iglewski, and C. van Delden. 2001. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J. Bacteriol. 183:1531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, E. W., J. E. LeClerc, B. Li, W. L. Payne, and T. A. Cebula. 2001. Phylogenetic evidence for horizontal transfer of mutS alleles among naturally occurring Escherichia coli strains. J. Bacteriol. 183:1631-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, L., and T. Elliott. 1997. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 32.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt,(ed), R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechtes and H. E. Umberger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 33.Chen, C. C., M. Fang, A. Majumder, and H.-Y. Wu. 2001. A 72-base pair AT-rich DNA sequence functions as a bacterial gene silencer. J. Biol. Chem. 276:9478-9485. [DOI] [PubMed] [Google Scholar]
- 34.Claret, L., and J. Rouvière-Yaniv. 1996. Regulation of HU alpha and HU beta by CRP and FIS in Escherichia coli. J. Mol. Biol. 263:126-139. [DOI] [PubMed] [Google Scholar]
- 35.Claret, L., and J. Rouvière-Yaniv. 1997. Variation in HU composition during growth of E. coli: the heterodimer is required for long term survival. J. Mol. Biol. 273:93-104. [DOI] [PubMed] [Google Scholar]
- 36.Crooke, E., M. Akiyama, N. N. Rao, and A. Kornberg. 1994. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 269:6290-6295. [PubMed] [Google Scholar]
- 37.Culham, D. E., and J. M. Wood. 2000. An Escherichia coli reference collection group B2- and uropathogen-associated polymorphism in the rpoS-mutS region of the E. coli chromosome. J. Bacteriol. 182:6272-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunning, C., and T. Elliott. 1999. RpoS synthesis is growth rate regulated in Salmonella typhimurium but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J. Bacteriol. 181:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cusick, M. E., and M. Belfort. 1998. Domain structure and RNA annealing activity of the Escherichia coli regulatory protein StpA. Mol. Microbiol. 28:847-857. [DOI] [PubMed] [Google Scholar]
- 41.Deighan, P., A. Free, and C. J. Dorman. 2000. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 38:126-139. [DOI] [PubMed] [Google Scholar]
- 42.Djordjevic, S., P. N. Goudreau, Q. P. Xu, A. M. Stock, and A. H. West. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorman, C. J., J. C. D. Hinton, and A. Free. 1999. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7:124-128. [DOI] [PubMed] [Google Scholar]
- 44.Dri, A.-M., P. L. Moreau, and J. Rouvière-Yaniv. 1992. Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene 120:11-16. [DOI] [PubMed] [Google Scholar]
- 45.DuBow, M., T. Ryan, R. A. Young, and T. Blumenthal. 1977. Host factor for coliphage Qβ RNA replication: presence in prokaryotes and association with the 30S ribosomal subunit in Escherichia coli. Mol. Gen. Genet. 153:39-43. [DOI] [PubMed] [Google Scholar]
- 46.el-Mansi, E. M., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 47.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure forσS-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 48.Falconi, M., M. T. Gualtieri, A. La Teana, M. A. Losse, and C. L. Pon. 1988. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15 kDa Escherichia coli DNA-binding protein H-NS. Mol. Microbiol. 2:323-329. [DOI] [PubMed] [Google Scholar]
- 49.Fang, F. C., C.-Y. Chen, D. G. Guiney, and Y. Xu. 1996. Identification of σS-regulated genes in Salmonella typhimurium: complementary regulatory interactions betweenσS and cyclic AMP receptor protein. J. Bacteriol. 178:5112-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang, M., A. Majumder, K.-J. Tsai, and H.-Y. Wu. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276:64-70. [DOI] [PubMed] [Google Scholar]
- 51.Fang, M., and H.-Y. Wu. 1998. A promoter relay mechanism for sequential gene activation. J. Bacteriol. 180:626-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Faxén, M., and L. A. Isaksson. 1994. Functional interactions between translation, transcription and ppGpp in growing Escherichia coli. Biochim. Biophys. Acta. 1219:425-434. [DOI] [PubMed] [Google Scholar]
- 53.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 14:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer, D., A. Teich, P. Neubauer, and R. Hengge-Aronis. 1998. The general stress sigma factor σS of Escherichia coli is induced during diauxic shift from glucose to lactose. J. Bacteriol. 180:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. Mckenney, G. Sutton, W. Fitzhugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. Mcdonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 56.Franze de Fernandez, M. T., L. Eoyang, and J. T. August. 1968. Factor fraction required for the synthesis of bacteriophage Qβ RNA. Nature (London) 219:588-590. [DOI] [PubMed] [Google Scholar]
- 57.Franze de Fernandez, M. T., W. S. Hayward, and J. T. August. 1972. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 247:824-831. [PubMed] [Google Scholar]
- 58.Free, A., and C. J. Dorman. 1997. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frost, L., K. Ippen-Ihler, and M. Skurray. 1995. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 61.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the luxR-luxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gambello, M. J., S. Kaye, and B. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.García-Lara, J., L. H. Shang, and L. I. Rothfield. 1996. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J. Bacteriol. 178:2742-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σS is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator SprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilson, L., A. Kuo, and P. V. Dunlap. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177:6946-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodrich-Blair, H., and R. Kolter. 2000. Homocysteine thiolactone is a positive effector of σS levels in Escherichia coli. FEMS Microbiol. Lett. 185:117-121. [DOI] [PubMed] [Google Scholar]
- 68.Gottesman, S., W. P. Clark, V. de Crecy-Lagard, and M. R. Maurizi. 1993. ClpX, an alternative subunit for the ATP-dependent Clp proteoase of Escherichia coli: sequence and in vivo activities. J. Biol. Chem. 268:22618-22626. [PubMed] [Google Scholar]
- 69.Grimaud, R., M. Kessel, B. Beuron, A. C. Steven, and M. R. Maurizi. 1998. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273:12476-12481. [DOI] [PubMed] [Google Scholar]
- 70.Grimsley, J. K., R. B. Tjalkens, M. A. Strauch, T. H. Bird, G. B. Spiegelman, Z. Hostomsky, J. M. Whiteley, and J. A. Hoch. 1994. Subunit composition and domain structure of the Spo0A sporulation transcription factor of Bacillus subtilis. J. Biol. Chem. 269:16977-16982. [PubMed] [Google Scholar]
- 71.Hammar, M., A. Arnquist, Z. Bian, A. Olsén, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 72.Hanzelka, B. L., M. R. Parsek, D. L. Val, P. V. Dunlap, J. E. Cronan, and E. P. Greenberg. 1999. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J. Bacteriol. 181:5766-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harris, S. J., Y. Shih, S. D. Bentley, and G. P. C. Salmond. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence factors. Mol. Microbiol. 28:705-717. [DOI] [PubMed] [Google Scholar]
- 74.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. H. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 75.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in stationary phase gene regulation in Escherichia coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 76.Hengge-Aronis, R. 1996. Back to log phase:σS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 77.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaschter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 78.Hengge-Aronis, R. 1999. Interplay of global regulators in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 79.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178.In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 80.Hengge-Aronis, R. 2000. A role for the σS subunit of RNA polymerase in the regulation of virulence genes. Adv. Exp. Med. Biol. 485:85-93. [DOI] [PubMed] [Google Scholar]
- 81.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 82.Hengge-Aronis, R., R. Lange, N. Henneberg, and D. Fischer. 1993. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 175:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herbelin, C. J., S. C. Chirillo, K. A. Melnick, and T. S. Whittam. 2000. Gene conservation and loss in the mutS-rpoS genomic region of pathogenic Escherichia coli. J. Bacteriol. 182:5381-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hogema, B. M., J. C. Arents, R. Bader, K. Eijkemans, H. Yoshida, H. Takahashi, H. Alba, and P. W. Postma. 1998. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIA(Glc). Mol. Microbiol. 30:487-498. [DOI] [PubMed] [Google Scholar]
- 85.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huisman, G. W., and R. Kolter. 1994. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science 265:537-539. [DOI] [PubMed] [Google Scholar]
- 87.Ichikawa, J. K., C. Li, J. Fu, and S. Clarke. 1994. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J. Bacteriol. 176:1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishige, K., S. Nagasawa, S. Tokishita, and T. Mizuno. 1994. A novel device of bacterial signal transducers. EMBO J. 13:5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jishage, M., and A. Ishihama. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major σ subunit of RNA polymerase. Proc. Natl. Adac. Sci. USA 95:4953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jishage, M., and A. Ishihama. 1999. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 181:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnston, C., D. A. Pegues, C. J. Hueck, C. A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 94.Kamashev, D., A. Balandina, and J. Rouvière-Yaniv. 1999. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 18:5434-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang, P. J., and E. A. Craig. 1990. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 172:2055-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karzai, A. W., E. D. Roch, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 97.Khan, M. A., and R. E. Isaacson. 1998. In vivo expression of the β-glucoside (bgl) operon of Escherichia coli occurs in mouse liver. J. Bacteriol. 180:4746-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim, S.-B., B.-S. Shin, S.-K. Choi, C.-K. Kim, and S.-H. Park. 2001. Involvement of acetyl phosphate in the in vivo activation of the response regulator ComA in Bacillus subtilis. FEMS Microbiol. Lett. 195:179-183. [DOI] [PubMed] [Google Scholar]
- 99.Kim, Y. I., R. E. Burton, B. M. Burton, R. T. Sauer, and T. A. Baker. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5:639-648. [DOI] [PubMed] [Google Scholar]
- 100.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klauck, E., J. Böhringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 102.Klauck, E., M. Lingnau, and R. Hengge-Aronis. 2001. Role of the response regulator RssB in σS recognition and initiation of σS proteolysis in Escherichia coli. Mol. Microbiol. 40:1381-1390. [DOI] [PubMed] [Google Scholar]
- 103.Klug, A., and J. Schwabe. 1995. Protein motifs. 5. Zinc fingers. FASEB J. 9:597-604. [PubMed] [Google Scholar]
- 104.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kolter, R. 1999. Growth in studying the cessation of growth. J. Bacteriol. 181:697-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic poylphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 107.Kulaev, I. S., and V. M. Vagabov. 1983. Polyphosphate metabolism in micro-organisms. Adv. Microbiol. Physiol. 24:83-171. [DOI] [PubMed] [Google Scholar]
- 108.Kuroda, A., H. Murphy, M. Cashel, and A. Kornberg. 1997. Guanosine tetra-and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240-21243. [DOI] [PubMed] [Google Scholar]
- 109.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 110.Lafiti, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 111.Lafiti, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. A. B. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 112.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA-polymerase in Escherichia coli. J. Bacteriol. 177:4676-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 114.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA-polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 115.Lange, R., and R. Hengge-Aronis. 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13:733-743. [DOI] [PubMed] [Google Scholar]
- 116.La Teana, A., A. Brandi, M. Falconi, R. Spurio, C. L. Pon, and C. O. Gualerzi. 1991. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. USA 88:10907-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acac. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 119.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interaction at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma factor σS (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 122.Li, C., Y. P. Tao, and L. D. Simon. 2000. Expression of different-size transcripts from the clpP-clpX operon of Escherichia coli during carbon deprivation. J. Bacteriol. 182:6630-6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liao, C. H., D. E. McCallus, and W. F. Fett. 1994. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol. Plant-Microbe Interact. 7:391-400. [DOI] [PubMed] [Google Scholar]
- 124.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor σS (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 125.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 126.Loewen, P. C., and B. L. Triggs. 1984. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J. Bacteriol. 160:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Loewen, P. C., I. von Ossowski, J. Switala, and M. R. Mulvey. 1993. KatF (σS) synthesis in Escherichia coli is subject to posttranscriptional regulation. J. Bacteriol. 175:2150-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lush, M. J., Y. Li, D. J. Read, A. C. Willis, and P. Glynn. 1998. Neuropathy target esterase and a homologous Drosophila neurodegeneration-associated mutant protein contain a novel domain conserved from bacteria to man. Biochem. J. 332:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 131.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanims, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malhotra, A., E. Severinova, and S. A. Darst. 1996. Crystal structure of a σ70 subunit fragment from E. coli RNA polymerase. Cell 87:127-136. [DOI] [PubMed] [Google Scholar]
- 133.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumphrey, B.Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease in Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 134.Mayer, M. P., H. Schröder, S. Rüdiger, K. Paal, T. Laufen, and B. Bukau. 2000. Multipstep mechanism os substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7:586-593. [DOI] [PubMed] [Google Scholar]
- 135.McCann, M. P., C. D. Fraley, and A. Matin. 1993. The putative σ factor KatF is regulated posttranscriptionally during carbon starvation. J. Bacteriol. 175:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative σ factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173:4188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 138.McEvoy, M. M., A. Bren, M. Eisenbach, and F. W. Dahlquist. 1999. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J. Mol. Biol. 289:1423-1433. [DOI] [PubMed] [Google Scholar]
- 139.Miranda, G., D. Schuppli, I. Barrera, C. Hausherr, J. M. Sogo, and H. Weber. 1996. Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: role of RNA interaction mediated by ribosomal protein S1 and host factor. J. Mol. Biol. 267:1089-1103. [DOI] [PubMed] [Google Scholar]
- 140.Mϕller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq, a bacterial Sm-like rotein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 141.Moreno, M., J. P. Audia, S. M. D. Bearson, C. Webb, and J. W. Foster. 2000. Regulation of sigma-S degradation in Salmonella enterica var typhimurium: in vivo interactions between sigma-S, the response regulator MviA (RssB) and ClpX. J. Mol. Microbiol. Biotechnol. 2:245-254. [PubMed] [Google Scholar]
- 142.Morita, M., M. Kanemori, H. Yanagi, and T. Yura. 1999. Heat-induced synthesis of σ32 in Escherichia coli: structural and functional dissection of rpoH mRNA secondary structure. J. Bacteriol. 181:401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morita, M. T., Y. Tanaka, T. Kodama, Y. Kyogoku, H. Yanagi, and T. Yura. 1999. Translational induction of heat shock transcriptional factor σ32: evidence for a built-in RNA thermosensor. Genes Dev. 13:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Muffler, A., M. Barth, C. Marschall, and R. Hengge-Aronis. 1997. Heat shock regulation of σS turnover: a role for DnaK and relationship between stress responses mediated by σS and σ32 in Escherichia coli. J. Bacteriol. 179:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 146.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for the translational regulation of rpoS in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 147.Muffler, A., D. D. Traulsen, D. Fischer, R. Lange, and R. Hengge-Aronis. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Muffler, A., D. D. Traulsen, R. Lange, and R. Hengge-Aronis. 1996. Posttranscriptional osmotic regulation of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 178:1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mukherjee, A., Y. Cui, W. Ma, Y. Liu, and A. K. Chatterjee. 2000. hexA of Erwinia carotovora spp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator or extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Environ. Microbiol. 2:203-215. [DOI] [PubMed] [Google Scholar]
- 150.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 151.Mukhopadhyay, S., and H. E. Schellhorn. 1997. Identification and characterization of hydrogen peroxide-sensitive mutants of Escherichia coli: genes that require. J. Bacteriol. 179:330-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mulvey, M. R., and P. C. Loewen. 1989. Nucleotide sequence of katF of Escherichia coli suggest KatF protein is a novel σ transcription factor. Nucleic Acids Res. 17:9979-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mulvey, M. R., J. Switala, A. Borys, and P. C. Loewen. 1990. Regulation of transcription of katE and katF in Escherichia coli. J. Bacteriol. 172:6713-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nagasawa, S., S. Tokishita, H. Aiba, and T. Mizuno. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6:799-807. [DOI] [PubMed] [Google Scholar]
- 155.Nash, H. A. 1996. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies, p. 149-179. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia Coli. R. G. Landes Co., Austin, Tex.
- 156.Nguyen, L. H., D. B. Jensen, N. E. Thompson, D. R. Gentry, and R. R. Burgess. 1993. In vitro functional characterization of overproduced Escherichia colikatF/rpoS gene product. Biochemistry 32:11112-11117. [DOI] [PubMed] [Google Scholar]
- 157.Notley, L., and T. Ferenci. 1996. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli. J. Bacteriol. 178:1465-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oberto, J., and J. Rouvière-Yaniv. 2001. Does the parallel evolution pattern between the replication-segregation proteins and HU have a biological significance? Biochimie 83:61-66. [DOI] [PubMed] [Google Scholar]
- 159.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ohta, T., C. Ueguchi, and T. Mizuno. 1999. rpoS function is essential for bgl silencing caused by C-terminally truncated H-NS in Escherichia coli. J. Bacteriol. 181:6278-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Olsén, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin binding curli in Escherichia coli. Mol. Microbiol. 7:523-536. [DOI] [PubMed] [Google Scholar]
- 162.Olsén, A., A. Jonsson, and S. Normark. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652-655. [DOI] [PubMed] [Google Scholar]
- 163.Ozaki, M., A. Wada, N. Fujita, and A. Ishihama. 1991. Growth phase-dependent modification of RNA polymerase in Escherichia coli. Mol. Gen. Genet. 230:17-23. [DOI] [PubMed] [Google Scholar]
- 164.Painbeni, E., M. Caroff, and J. Rouvière-Yaniv. 1997. Alterations of the outer membrane composition in Escherichia coli laching the histone-like protein HU. Proc. Natl. Acad. Sci. USA 94:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pernestig, A.-K., Ö. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 167.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Phadtare, S., and M. Inouye. 2001. Role CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Phadtare, S., K. Yamanaka, and M. Inouye. 2000. The cold shock response, p. 33-45. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 170.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator, SprE, controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 172.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 173.Prüβ, B. M., and A. J. Wolfe. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973-984. [DOI] [PubMed] [Google Scholar]
- 174.Pruteanu, M., and R. Hengge-Aronis. The cellular level of the recognition factor RssB is rate-limiting for σS proteolysis: implications for RssB regulation and signal transduction in σS turnover in Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed]
- 175.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance can survival of stationary- phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Reimmann, C., M. Beyeler, A. Lafiti, H. Winteler, M. Foglino, A. Lasdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserin lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 177.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of rpoS: temperature regulation of dsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rich, J. J., T. G. Kinscherf, T. Kitten, and D. K. Willis. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 176:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rivas, E., R. J. Klein, T. A. Jones, and S. R. Eddy. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 11:1369-1373. [DOI] [PubMed] [Google Scholar]
- 180.Rockabrand, D., T. Arthur, G. Korinek, K. Livers, and P. Blum. 1995. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J. Bacteriol. 177:3695-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurum strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 184.Rüdiger, S., M. P. Mayer, J. Schneider-Mergener, and B. Bukau. 2000. Modulation of substrate specificity of the DnaK chaperone by alteration of a hydrobphobic arch. J. Mol. Biol. 304:245-251. [DOI] [PubMed] [Google Scholar]
- 185.Ruiz, N., C. N. Peterson, and T. J. Silhavy. 2001. RpoS-dependent transcriptional control of sprE: regulatory feedback loop. J. Bacteriol. 183:5974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sak, B. D., A. Eisenstark, and D. Touati. 1989. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF product. Proc. Natl. Acad. Sci. USA 86:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Salmond, G. P. C., B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1995. The bacterial “enigma”: cracking the code of cell-cell communication. Mol. Microbiol. 16:615-624. [DOI] [PubMed] [Google Scholar]
- 188.Sarniguet, A., J. Kraus, M. D. Henkels, A. M. Muehlchen, and J. E. Loper. 1995. The sigma factor σS affects antibiotic and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. USA 92:12255-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 190.Schellhorn, H. E., and V. L. Stones. 1992. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J. Bacteriol. 174:4769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schuppli, D., G. Miranda, H. C. Tsui, M. E. Winkler, J. M. Sogo, and H. Weber. 1997. Altered 3′-terminal RNA structure in phage Qbeta adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. USA 94:10239-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schweder, T., K.-H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σS) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Senear, A. W., and J. A. Steitz. 1976. Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J. Biol. Chem. 251:1902-1912. [PubMed] [Google Scholar]
- 194.Sevcik, M., A. Sebková, J. Volf, and I. Rychlík. 2001. Transcription of arcA and rpoS during growth of Salmonella typhimurium under aerobic and microaerobic conditions. Microbiology 147:701-708. [DOI] [PubMed] [Google Scholar]
- 195.Shiba, T., K. Tsutsumi, H. Yano, Y. Ihara, A. Kameda, K. Tanaka, H. Takahashi, M. Munekata, N. N. Rao, and A. Kornberg. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acac. Sci. USA 94:11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sitnikov, D. M., J. B. Schineller, and T. O. Baldwin. 1996. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. USA 93:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sledjeski, D., and S. Gottesman. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:2003-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in E. coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 200.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sondén, B., and B.-E. Uhlin. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15:4970-4980. [PMC free article] [PubMed] [Google Scholar]
- 202.Sonenshein, A. L. 2000. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 3:561-566. [DOI] [PubMed] [Google Scholar]
- 203.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress responses. ASM Press, Washington, D.C.
- 204.Straus, D. B., W. A. Walter, and C. A. Gross. 1989. The activity of σ32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes Dev. 3:2003-2010. [DOI] [PubMed] [Google Scholar]
- 205.Suh, S.-J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Takahashi, H., T. Inada, P. Postma, and H. Aiba. 1998. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIAGlc, the glucose-specific phosphotransferase protein, in Escherichia coli. Mol. Gen. Genet. 259:317-326. [DOI] [PubMed] [Google Scholar]
- 208.Takayanagi, Y., K. Tanaka, and H. Takahashi. 1994. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol. Gen. Genet. 243:525-531. [DOI] [PubMed] [Google Scholar]
- 209.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, σ38, is a second principal sigma factor of RNA polymerase in stationary phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Teich, A., S. Meyer, H. Y. Lin, L. Andersson, S. O. Enfors, and P. Neubauer. 1999. Growth rate related concentration changes of the starvation response regulators σS and ppGpp in glucose-limited fed-batch and continuous cultures of Escherichia coli. Biotechnol. Prog. 15:123-129. [DOI] [PubMed] [Google Scholar]
- 211.Touati, E., E. Dassa, and P. L. Boquet. 1986. Pleiotropic mutations in appR reduce pH 2.5 acid phosphatase expression and restore succinate untilization in CRP-deficient strains of Escherichia coli. Mol. Gen. Genet. 202:257-264. [DOI] [PubMed] [Google Scholar]
- 212.Touati, E., E. Dassa, J. Dassa, P. L. Boquet, and D. Touati. 1991. Are appR and katF the same Escherichia coli gene encoding a new sigma transcription initiation factor? Res. Microbiol. 142:29-36. [DOI] [PubMed] [Google Scholar]
- 213.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem. 276:1220-1225. [DOI] [PubMed] [Google Scholar]
- 214.Tsui, H.-C. T., H.-C. L. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 215.Tsui, H. C. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tuveson, R. W., and R. B. Jonas. 1979. Genetic control of near-UV (300-400 nm) sensitivity independent of the recA gene in strains of Escherichia coli K12. Photochem. Photobiol. 30:667-676. [DOI] [PubMed] [Google Scholar]
- 217.Ueguchi, C., N. Misonou, and T. Mizuno. 2001. Negative control of rpoS expression by phosphoenolpyruvate: carbohydrate phosphotransferase system in Escherichia coli. J. Bacteriol. 183:520-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ueguchi, C., T. Ohta, C. Seto, T. Suzuki, and T. Mizuno. 1998. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 180:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vogel, U., and K. F. Jensen. 1994. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176:2807-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vogel, U., M. Sϕrensen, S. Pedersen, K. F. Jensen, and M. Kilstrup. 1992. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol. Microbiol. 6:2191-2200. [DOI] [PubMed] [Google Scholar]
- 221.Vytvytska, O., J. S. Jakobsen, G. Balcunatie, J. S. Andersen, M. Baccarini, and A. von Gabain. 1998. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. USA 95:14118-14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Vytvytska, O., I. Moll, V. R. Kaberdin, A. von Gabain, and U. Blasi. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14:1109-1118. [PMC free article] [PubMed] [Google Scholar]
- 223.Wang, J. M., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 224.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 226.Weikert, C., F. Canonaco, U. Sauer, and J. E. Bailey. 2000. Overexpression of RspAB improves recombinant protein production in Escherichia coli. Metab. Eng. 4:293-299. [DOI] [PubMed] [Google Scholar]
- 227.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The Two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 230.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175-185. [DOI] [PubMed] [Google Scholar]
- 231.Winans, S. C., and J. Zhu. 2000. The role of cell-cell communication in confronting the limitations and opportunities of high population densities, p. 261-272. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 232.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. C. Salmond, B. W. Bycroft, A. Lazdunski, G. S. A. B. Stewart, and P. Williams. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wyman, C., I. Rombel, A. K. North, C. Bustamante, and S. Kustu. 1997. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science 275:1658-1661. [DOI] [PubMed] [Google Scholar]
- 234.Yamashino, T., C. Ueguchi, and T. Mizuno. 1995. Quantitative control of the stationary phase-specific sigma factor, σS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 14:594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yoshida, M., K. Kashiwagi, G. Kawai, A. Ishihama, and K. Igarashi. 2001. Polyamine enhancement of the synthesis of adenylate cyclase at the translational level and the consequential stimulation of the synthesis of RNA polymerase σ28 subunit. J. Biol. Chem. 276:16289-16295. [DOI] [PubMed] [Google Scholar]
- 236.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 237.Zapf, J., U. Sen, Madhusudan, J. A. Hoch, and K. I. Varughese. 2000. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 8:851-862. [DOI] [PubMed] [Google Scholar]
- 238.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang, A., S. Rimsky, M. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:1-20. [DOI] [PubMed] [Google Scholar]
- 241.Zhang, J. P., and S. Normark. 1996. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science 273:1234-1236. [DOI] [PubMed] [Google Scholar]
- 242.Zhou, A. N., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhu, X. T., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata, M. E. Gottesman, and W. A. Hendrickson. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]