Abstract
Human oral bacteria interact with their environment by attaching to surfaces and establishing mixed-species communities. As each bacterial cell attaches, it forms a new surface to which other cells can adhere. Adherence and community development are spatiotemporal; such order requires communication. The discovery of soluble signals, such as autoinducer-2, that may be exchanged within multispecies communities to convey information between organisms has emerged as a new research direction. Direct-contact signals, such as adhesins and receptors, that elicit changes in gene expression after cell-cell contact and biofilm growth are also an active research area. Considering that the majority of oral bacteria are organized in dense three-dimensional biofilms on teeth, confocal microscopy and fluorescently labeled probes provide valuable approaches for investigating the architecture of these organized communities in situ. Oral biofilms are readily accessible to microbiologists and are excellent model systems for studies of microbial communication. One attractive model system is a saliva-coated flowcell with oral bacterial biofilms growing on saliva as the sole nutrient source; an intergeneric mutualism is discussed. Several oral bacterial species are amenable to genetic manipulation for molecular characterization of communication both among bacteria and between bacteria and the host. A successful search for genes critical for mixed-species community organization will be accomplished only when it is conducted with mixed-species communities.
INTRODUCTION
Communication is a key element in successful organizations. The bacteria on human teeth and oral mucosa have developed the means by which to communicate and thereby form successful organizations. These bacteria have coevolved with their host to establish a highly sophisticated relationship in which both pathogenic and mutualistic bacteria coexist in homeostasis. The fact that human oral bacteria are not found outside the mouth except as pathogens elsewhere in the body (51) points to the importance of this relationship. Communication among microorganisms is essential for initial colonization and subsequent biofilm formation on the enamel surfaces of teeth and requires physical contact between colonizing bacteria and between the bacteria and their host. Without retention on the tooth surface, the bacteria are swallowed with the saliva. Through retention, these bacteria can form organized, intimate, multispecies communities referred to as dental plaque.
Sequential changes in populations of bacteria associated with tooth eruption (20, 21, 102, 138) as well as with caries development (53) and periodontal disease states (109, 136) are known. Temporal changes in populations of bacteria on tooth surfaces after professional cleaning are ordered and sequential (92, 114, 115). Such sequential changes must occur through attachment and growth of different bacterial species. With the attachment of each new cell type, a nascent surface is presented for the attachment of other kinds of bacteria, resulting in a progression of nascent surfaces and concomitant changes in species diversity (79, 137). Such coordination indicates communication. In the absence of communication, these orderly changes would be random. Due to the dynamics of growth and adherence, the bacterial populations in the oral cavity are constantly changing, even during the intervals between normal daily oral hygiene treatments. It is unlikely that the various species within oral biofilms function as independent, discrete constituents; rather, these organisms function as a coordinated community that uses intra- and interspecies communication.
For the past 40 years, pure cultures of oral bacteria have been isolated from supragingival and subgingival dental plaque removed from healthy and diseased sites. The numbers and variety of bacteria obtained from many clinical conditions have been catalogued (10, 19, 72, 109, 137). Estimates of the bacterial species diversity in the oral cavity, based on both culture-dependent methods (109, 136) and culture-independent methods (83, 123), indicate about 500 species. About 415 species are estimated to be present in subgingival plaque (123), and many of these are also found in supragingival plaque. Cultured species account for about 60% of the organisms identified by molecular methods, indicating that the oral cavity is an environment where most species can be studied by routine culture methods. This is distinct from other environments where less than 1% of the clones obtained by molecular methods represent cultured species (2, 44, 54, 118). Thus, dental plaque is one of the best-described mixed-species bacterial communities. Because it is easily accessible, it is convenient to study this complex model system.
Most of the cultured species of oral bacteria have been tested for their ability to physically interact with and adhere to different species, and all display specific recognition patterns with their respective partner cells (154). This recognition between genetically distinct cells in suspension and resultant clumping is called coaggregation (72). Recognition between a suspended cell type and one already attached to a substratum is termed coadhesion (9). These interactions often appear to be mediated by complementary protein-adhesin and saccharide-receptor components on the two cell types (22, 27, 28, 143, 154). In many coaggregates, galactosides are competitive inhibitors of coaggregation, indicating a likely lectin-carbohydrate recognition between cognate molecules on the two cell surfaces (27, 78, 103, 154). These properties offer microbial ecologists and molecular biologists opportunities to explore the mechanisms of cell-cell recognition and their role in fostering bacterial biofilm communities in dental plaque.
Model systems useful for the study of mixed-species communities include biofilms on substrata (31, 155, 156) and planktonic communities grown in chemostats (11, 12). One model that has been employed to study oral bacterial colonization in vivo is a retrievable enamel chip worn by human volunteers (92, 115, 121). An in vitro model consisting of a flowcell with saliva-coated surfaces has offered an excellent platform for studying the adherence and growth of oral bacteria (75, 76, 119, 120). Community organization in the in vivo enamel chip and the in vitro flowcell model systems can be investigated by using confocal laser microscopy.
Avenues of communication among these ever-changing populations are likely to include metabolite exchange (57), cell-cell recognition (73, 120), genetic exchange (89), and signaling molecules produced by the host (82) and by other bacteria (15, 26, 48). A concert of communication methods probably occurs to establish transitory community organizations. The species participating in these oral communities compete and cooperate en route to establishing a climax community that may encompass all of the more than 500 known species. Overall, little is known about the communication mechanisms used by oral bacteria to establish these communities and to prevent disappearance from their habitat. This review focuses on these mechanisms, with the greatest attention devoted to cell-cell recognition and autoinducer-2, a signaling molecule produced by many bacteria, including oral bacteria. This review does not cover the equally exciting area of bacterium-host interactions.
In this review, our definition of communication among oral bacteria is not limited to effects of transcriptional activators, chemotactic signals, or two-component regulatory elements. Rather, here we include the view of bacteria responding to their environment. The first response in a biofilm is attachment to a surface, and this is communication. If adherence were nonspecific, then all bacteria could attach to oral surfaces, and this does not occur. After adherence, bacteria form multispecies communities. As in nonmicrobial ecosystems, some species are more successful at the beginning and other species are more successful at the end of community evolution; the timing of these successful opportunities may well relate to sequential modifications of the environment by the community. The methods by which these bacteria communicate to accomplish ordered multispecies communities is an active area of research, and we discuss some of the progress made in understanding communication among oral bacteria.
SPATIOTEMPORAL MODEL OF ORAL BACTERIAL COLONIZATION
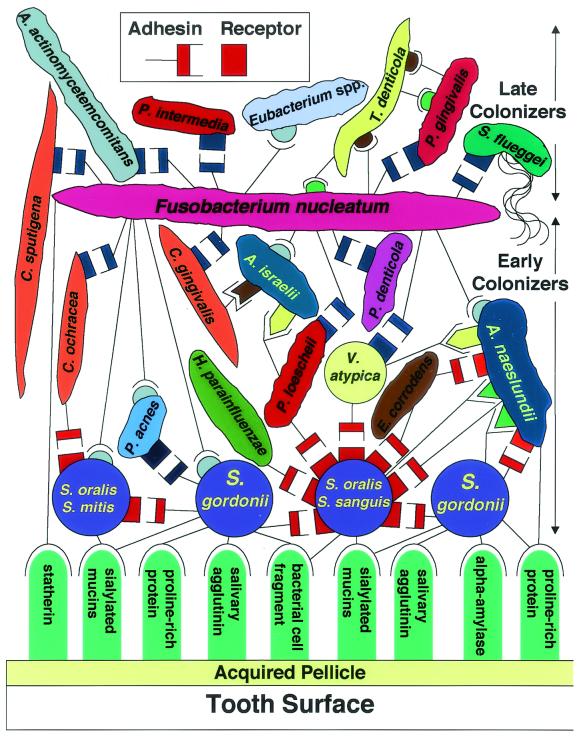
Development of the oral microbial community involves competition as well as cooperation among the 500 species that compose this community. A few of those oral species are shown in Fig. 1 in a diagram illustrating competition and cooperation among early and late colonizers of the tooth surface. The acquired pellicle, which is composed of a variety of host-derived molecules, coats the enamel surface within minutes after professional cleaning and is a source of receptors recognized by the primary colonizers of dental plaque. These receptors include mucins, agglutinins, proline-rich proteins, phosphate-rich proteins such as statherin, and enzymes such as alpha-amylase (Fig. 1, blue-green columns, bottom). Each is a known receptor for particular oral species.
FIG. 1.
Spatiotemporal model of oral bacterial colonization, showing recognition of salivary pellicle receptors by early colonizing bacteria and coaggregations between early colonizers, fusobacteria, and late colonizers of the tooth surface. Each coaggregation depicted is known to occur in a pairwise test. Collectively, these interactions are proposed to represent development of dental plaque and are redrawn from Kolenbrander and London (79). Starting at the bottom, primary colonizers bind via adhesins (round-tipped black line symbols) to complementary salivary receptors (blue-green vertical round-topped columns) in the acquired pellicle coating the tooth surface. Secondary colonizers bind to previously bound bacteria. Sequential binding results in the appearance of nascent surfaces that bridge with the next coaggregating partner cell. Several kinds of coaggregations are shown as complementary sets of symbols of different shapes. One set is depicted in the box at the top. Proposed adhesins (symbols with a stem) represent cell surface components that are heat inactivated (cell suspension heated to 85°C for 30 min) and protease sensitive; their complementary receptors (symbols without a stem) are unaffected by heat or protease. Identical symbols represent components that are functionally similar but may not be structurally identical. Rectangular symbols represent lactose-inhibitable coaggregations. Other symbols represent components that have no known inhibitor. The bacterial strains shown are Actinobacillus actinomycetemcomitans, Actinomyces israelii, Actinomyces naeslundii, Capnocytophaga gingivalis, Capnocytophaga ochracea, Capnocytophaga sputigena, Eikenella corrodens, Eubacterium spp., Fusobacterium nucleatum, Haemophilus parainfluenzae, Porphyromonas gingivalis, Prevotella denticola, Prevotella intermedia, Prevotella loescheii, Propionibacterium acnes, Selenomonas flueggei, Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, Streptococcus sanguis, Treponema spp., and Veillonella atypica.
Streptococci constitute 60 to 90% of the bacteria that colonize the teeth in the first 4 h after professional cleaning (115). Other early colonizers include Actinomyces spp., Capnocytophaga spp., Eikenella spp., Haemophilus spp., Prevotella spp., Propionibacterium spp., and Veillonella spp. Many of the physical interactions that occur between the organisms of this community are known (72, 73, 154), and some are depicted in Fig. 1. The complementary symbols depict physical interactions known to occur between a pair of species. The different shapes and colors of the complementary symbols in Fig. 1 represent potentially distinct coaggregations. Rectangle-shaped symbols of any color represent lactose-inhibitable coaggregations, which are prevalent among oral bacteria (27, 72). Those of the same color represent functionally similar but not identical coaggregations. For example, the red rectangle symbols on the purple circle representing Streptococcus oralis and Streptococcus sanguis indicate a receptor polysaccharide named 1 Gn with the structure →PO4− →6GalNAcα1→3Rhaβ1→4Glcβ1→6Galfβ1→6GalNAcβ1→3Galα1→)n (28). The GalNAcβ1→3Gal receptor site in 1 Gn is recognized by functionally similar adhesins on Streptococcus gordonii, Haemophilus parainfluenzae, Prevotella loescheii, Veillonella atypica, Eikenella corrodens, and Actinomyces naeslundii. These adhesins are of various molecular sizes, and the species bearing the adhesins compete with each other for binding to the receptor polysaccharide (72, 73, 154). Thus, it is postulated that coaggregation and coadherence are integral to communication between species and help to establish patterns of spatiotemporal development.
Early Colonizers
Viridans streptococci (Fig. 1, bottom, purple circles), especially S. gordonii, are ideal model organisms to study because they are early colonizers, they coaggregate with a variety of oral bacteria, they bind to several host molecules, and many are genetically transformable. Streptococcus is the only genus of oral bacteria that demonstrates extensive intrageneric coaggregation as well as intergeneric coaggregation (73, 78). The ability to bind to other early colonizers and to host molecules (Fig. 1, lower half) may confer an advantage on these viridans streptococci in establishing early dental plaque. S. gordonii binds to salivary agglutinin glycoproteins by SspA/B (37), to alpha-amylase by AbpA (14, 132), and to the α2-3-linked sialic acid termini of O-linked oligosaccharides of host glycoconjugates by Hsa (142). Hsa has 113 serine-rich dodecapeptide repeats (142) and may be the sialic acid-binding adhesin involved in utilization of sialic acid as a nutrient, as has been shown for several viridans streptococci (17).
S. gordonii binds to acidic proline-rich proteins (PRPs) via the ProGln termini of PRPs (52). Acidic PRPs account for 25 to 30% of the total proteins in saliva; they regulate calcium phosphate and hydroxyapatite crystal equilibrium and thus contribute to stabilizing tooth integrity. They are encoded by host loci PRH1 and PRH2 and comprise five variants of 150 or 171 residues (63). One of these, acidic PRP-1, has been studied in some detail and may serve as a nutrient source for early colonizers. S. gordonii rapidly degrades the 150-amino-acid protein into two detectable peptides, the 105-amino-acid peptide derived from the amino-terminal region and a peptide corresponding to the 40 C-terminal amino acids, which are further degraded to oligopeptides (88). The pentapeptide expected to result from the intervening region was not detected, but a synthetic peptide of the same sequence (Arg106Gly107Arg108Pro109Gln110) caused detachment of A. naeslundii from PRP-1-coated latex beads, suggesting a mechanism for competition with other early colonizers (88). Thus, PRPs are not only receptors for binding bacteria (Fig. 1), they are also a ready nutrient source with an ecological bonus of modulating the adherence of potentially competing bacteria such as A. naeslundii (Fig. 1, lower right) at the tooth surface.
Oral bacteria capable of binding to a receptor and subsequently utilizing the receptor as a nutrient are suited to this task because they have diverse mechanisms for attachment. They can bind to other host receptors or to other bacteria while degrading the PRP nutrient. The numerous interactions between streptococci, host molecules, and other early colonizers demonstrate multifactorial communication between bacteria and their environment.
Early-colonizing bacteria have been shown to regulate gene expression in response to a saliva-containing environment. In saliva, S. gordonii DL1 increases expression of sspA/B (39), encoding surface proteins that bind to salivary agglutinin (37), an interaction depicted at the bottom of Fig. 1. Thus, streptococci suspended in saliva may bind to salivary agglutinin and elicit enhanced expression of sspA/B. Several other saliva-regulated genes include S. gordonii hppA, encoding an oligopeptide-binding lipoprotein, a homologue of the Streptococcus mitis glucose kinase gene gki, and a homologue of Lactococcus lactis clpE, encoding a member of the Clp protease family (39). These saliva-regulated proteins represent only a small assortment of the expected total proteins regulated by environmental conditions. They do, however, indicate that S. gordonii responds, at the transcriptional level, to elements from its natural oral environment.
Considering that each of the approximately 500 species in the oral bacterial community (Fig. 1) has such a potential for gene regulation in response to host-produced molecules and physical interactions with other bacteria, the complexity of possible interactions within the oral environment and the number of opportunities for cell-to-cell communication become daunting.
F. nucleatum and Late Colonizers
The bacterial species shown in Fig. 1 are placed in either of two general categories, early colonizers or late colonizers. This placement is based on the species of bacteria identified in dental plaque during temporal sampling after oral hygiene procedures were conducted (109, 115, 137). Fusobacterium nucleatum, however, is unusual and is intentionally placed at the border between early and late colonizers (Fig. 1) for the following reasons. First, F. nucleatum is the most numerous gram-negative species in healthy sites, and its numbers increase markedly in periodontally diseased sites (109). It is always present whenever Treponema denticola and Porphyromonas gingivalis are also present, suggesting that its presence predates that of the other two species and may be required for their colonization (137). Second, F. nucleatum coaggregates with all of the early colonizers and the late colonizers (4, 77). The bacteria representing early colonizers coaggregate with only a specific set of other early colonizers but not with all of them and generally not with any of the late colonizers (73, 79, 154). Although all the late colonizers coaggregate with F. nucleatum, they generally do not coaggregate with each other. A few exceptions, such as T. denticola coaggregating with P. gingivalis, have been reported (158) (Fig. 1, top right). Thus, F. nucleatum acts as a bridge between early and late colonizers, which may partially explain why fusobacteria are so numerous in samples from both healthy and diseased sites.
The cognate receptors (Fig. 1, blue rectangles) on the surface of the partners of F. nucleatum display functional similarity. They are depicted as competing for binding with the same F. nucleatum adhesins (Fig. 1, upper half). This idea is based on the observation that spontaneous mutants of F. nucleatum which were selected solely on the basis of being unable to coaggregate with P. gingivalis had also lost the ability to coaggregate with all of the lactose-inhibitable coaggregation partners but retained the ability to coaggregate with other partners (4). Just as these receptors exhibit functional similarity, adhesins recognizing the same 1 Gn receptor polysaccharide on S. oralis and S. sanguis (Fig. 1, lower half) exhibit functional similarity. The lactose-inhibitable coaggregations (Fig. 1, blue rectangles) between F. nucleatum and its partners appear to be mediated by the same galactose-binding adhesin that mediates attachment of F. nucleatum to mammalian cells, including human buccal epithelial cells and gingival and periodontal ligament fibroblasts (152). A mutant of F. nucleatum with no galactose-binding activity does not bind to eukaryotic cells. An independent study reported galactose-inhibitable binding and invasion of human gingival epithelial cells by F. nucleatum (59). These results attribute great potential significance to galactose-sensitive adhesins for initiating communication between early and late colonizers as well as with their host.
Finally, in addition to interactions with oral bacteria and host cells, F. nucleatum interacts with and binds host-derived molecules, such as plasminogen (33). F. nucleatum is generally nonproteolytic, but organisms that coexist with it, such as P. gingivalis, are highly proteolytic and can activate fusobacterium-bound plasminogen to form fusobacterium-bound plasmin, a plasma serine protease (33). Acquisition of proteolytic ability on its cell surface confers on the fusobacteria a new metabolic property, the ability to process potential peptide signals in the community. These peptides may be used as nutrients by fusobacteria or by other biofilm residents. F. nucleatum also induces expression of β-defensin 2, a small cationic peptide produced by mucosal epithelial cells (82). P. gingivalis does not elicit β-defensin 2 production, suggesting a distinction between these two important oral bacteria and their role in stimulating innate immune responses (82). Although F. nucleatum is often considered a periodontal pathogen, it may instead contribute to maintaining homeostasis and improving host defense against true pathogens.
Cooperation and Competition
In addition to fusobacteria acting as the principal coaggregation bridge between early and late colonizers, bridging among early colonizers is also possible. Some of these coaggregation bridges are shown in Fig. 1. For example, coaggregation between P. loescheii and S. oralis is lactose inhibitable (Fig. 1, red rectangle), and coaggregation between P. loescheii and Actinomyces israelii is lactose noninhibitable (Fig. 1, yellow obelisk). S. oralis is not able to coaggregate with A. israelii; therefore, P. loescheii acts as a bridge of coaggregation. Both A. israelii and P. loescheii coaggregate with F. nucleatum, which coaggregates with all the late colonizers.
Coaggregation bridges are mechanisms of cooperation because they bring together two species that are not coaggregation partners. Such bridges may be critical for temporary retention of bacteria on a nascent surface and may facilitate eventual bacterial colonization of the biofilm. The bridges are distinct from competition, which occurs when multiple species compete for binding to the same receptor. Competitive and cooperative mechanisms may be central to successful mixed-species colonization as well as the proper succession of genera known to occur on teeth in both health and disease.
COMMUNITY ARCHITECTURE AND METABOLIC COMMUNICATION
In Vitro
Experimental approaches towards understanding human oral communities often start with in vitro studies and simple model systems. Several model biofilm systems have been developed, and most involve the flow of nutrients over a surface to which bacteria are attached (155, 156). Others employ a static support, such as a hydroxyapatite disk immersed in growth medium with gentle shaking (58). The substratum, nutrient medium, and oral bacteria chosen for study vary considerably among these models. Results from each model system contribute to our understanding of the growth and activity of bacteria on oral surfaces.
One system used in our laboratory is the flowcell, based on a design by Palmer and Caldwell (119), in which a microscope slide and coverslip are separated by a silicone rubber gasket that forms two channels (75). Sterile saliva is used as the sole nutrient source and is used to coat the glass before addition of bacteria to the flowcell. During a static period, bacteria bind to the saliva conditioning film, after which saliva flow is initiated, leading to the formation of a biofilm.
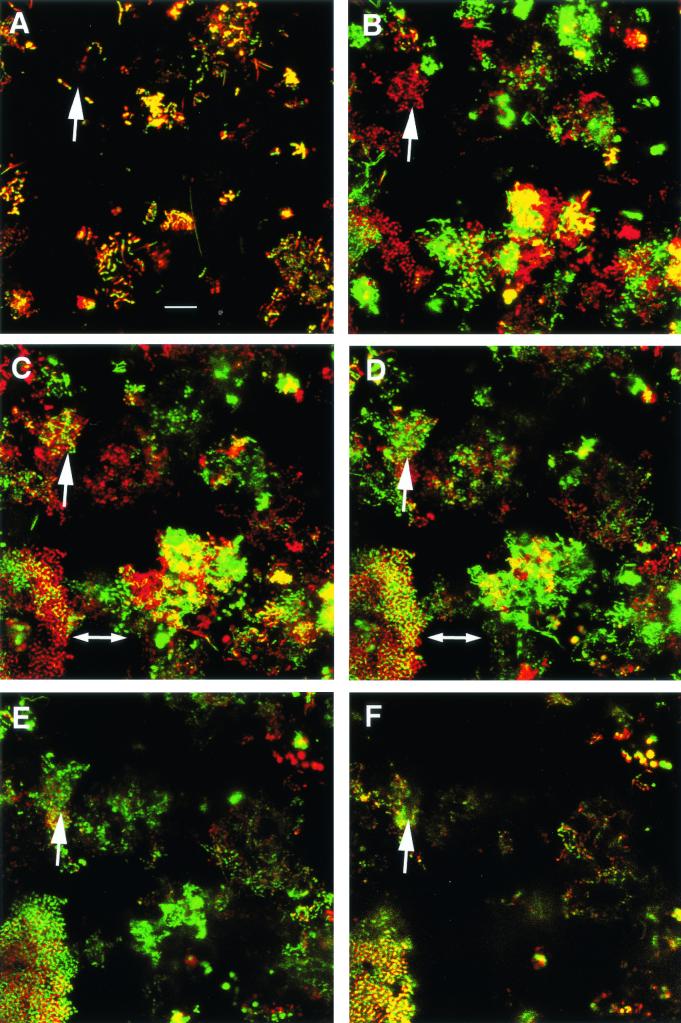
Inoculation of the flowcell with whole, unfiltered saliva followed by laminar flow of sterile saliva overnight results in a bacterial biofilm community attached to the saliva-coated substratum and extending towards the lumen (Fig. 2). After 18 h of growth, microcolonies consisting of different bacterial morphotypes are found in juxtaposition, suggesting construction of mixed-species communities. The spaces between these microcolonies indicate that communication by diffusible small molecules would require transmission over large distances and through a large volume of saliva compared to the cell-to-cell distances within a microcolony. The presence of several distinct cellular morphologies at the substratum (Fig. 2A) clearly indicates that many of the attached cells are already arranged as mixed-species communities.
FIG. 2.
Human oral biofilm formed in vitro with a saliva inoculum and using sterile saliva as its sole source of nutrient. The 25-μm-thick biofilm was grown overnight suspended from the underside of the coverslip of a flowcell with saliva flowing through once at 0.2 ml per min. Bacterial juxtaposition and biofilm architecture were imaged by confocal scanning laser microscopy after staining the cells with Live/Dead stain (Molecular Probes, Eugene, Oreg.). The color of the cells is from the red (propidium iodide; damaged or permeable cell membrane) and green (SYTO 9; healthy cell) fluorescent stains. Colocalization of both fluorophores results in yellow staining. Confocal scanning laser microscopy acquires optical sections through the biofilm; each optical section is 0.5 μm thick. The entire biofilm is represented in six images (A to F). Panel A is the 0.5-μm optical section at the substratum and shows the biofilm footprint. Panel F is the top 0.5 μm of the biofilm where it projects into the lumen of the flowcell. The other four projection images contain eight sections per projection and show the 4-μm-thick regions from 4 to 8 μm from the substratum (B), 8 to 12 μm from the substratum (C), 12 to 16 μm from the substratum (D), and 16 to 20 μm from the substratum (E). Regions indicated by arrows are described in the text. Bar, 10 μm. Microscopic observations and image acquisition were performed on a TCS 4D system (Leica Lasertechnik GmbH, Heidelberg, Germany).
Many initial attachments occur by only a few cells (Fig. 2A, arrow), compared to the contiguous, more voluminous colony mass extending toward the lumen (Fig. 2B, C, D, E, and F, arrows). This feature is characteristic of multispecies biofilm growth in laminar flow conditions as used in Fig. 2 and in certain monospecies biofilms (119). Under these conditions, initial colonization on the substratum is followed by axial growth and accumulation toward the lumen. In laminar flow, little shear force occurs at the substratum because salivary flow velocity is greatly reduced at the substratum compared to that in the lumen bulk phase. Accordingly, delivery of nutrient is higher in the faster-flowing region and may account for the greater biovolume found distant from the substratum. The larger cell masses may contact each other (Fig. 2C, double-headed arrow) and thus facilitate communication within the large surrounding void. Increasing separation between contacted communities (Fig. 2D, double-headed arrow) may accompany progression of the biofilm towards the lumen (Fig. 2E and F).
The apparent diversity and range of interactions that seem to occur in mixed-species communities (as shown in Fig. 2) help explain why seemingly healthy and active multispecies communities are capable of growth solely on saliva (34). The participants in this multispecies biofilm must signal each other in numerous ways from initial colonization through the various physiological stages en route to a mature biofilm community. It is important to consider that the laterally and axially (from top to bottom) organized communities are not composed of cells exhibiting just one cellular morphology. Rather, each of these adjacent communities may comprise many species. In fact, cells of nearly identical morphology may be different species.
Because of this complexity, such communities are difficult to study without highly specific tools or probes. Probes that are fluorescent permit imaging by confocal laser microscopy and can be used to examine spatiotemporal relationships. Several types have been used with success, including green fluorescent protein (5, 60), fluorescently labeled antibodies to surface antigens (5, 75, 120), and fluorescently labeled 16S rRNA-targeted probes (110).
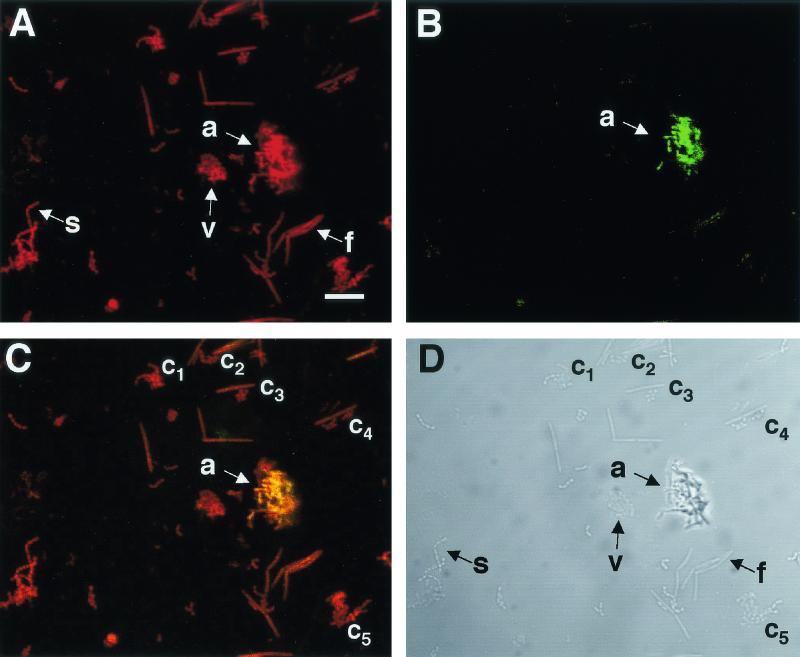
Each bacterial species has a unique DNA sequence encoding the 16S subunit rRNA (16S rDNA). Modern bacterial phylogenetic schemes are based on the 16S rDNA sequence, and signature sequences unique to a particular species can be used as nucleotide probes for fluorescence in situ hybridization (FISH) to localize species in their natural habitat. Probe specificity can be evaluated by blot surveys against RNA from a range of oral species (122) or by mixing a selected group of oral bacterial species and examining by FISH the specificity of the probe in whole cells by confocal microscopy (Fig. 3). In Fig. 3, the probe was targeted to the 16S rRNA of Actinomyces serovar WVA963 strain PK1259 and was specific for actinomyces cells (Fig. 3B, green cells). This mixture of cells also contained fusobacteria (spindle-shaped cells), streptococci (spherical cells in chains), and veillonellae (smaller spherical cells in clumps). Four oral bacterial genera are represented here, and each coaggregates with the other three.
FIG. 3.
Four-genus mixture of oral bacteria stained with a specific probe by FISH and the nonspecific nucleic acid stain SYTO 59. (A) Confocal micrograph of a field of cells stained with the nucleic acid stain SYTO 59. The field of cells contains the following species: Actinomyces serovar WVA963 strain PK1259 (a), F. nucleatum PK1594 (f), S. gordonii DL1 (s), and Veillonella atypica PK1910 (v). (B) Confocal micrograph of the same field showing the location of the fluorescein isothiocyanate-labeled actinomyces-specific probe. The image demonstrates that the probe interacts only with actinomyces cells (a). (C) Overlay of confocal micrographs (A and B), demonstrating the specificity of the actinomyces probe. Areas of colocalization of fluorescein isothiocyanate and SYTO-59 markers appear yellow. The yellow actinomyces cells are in contact with other cells seen at the edges of the actinomyces cluster. Coaggregations (c1 to c5) of different species within the mixed culture are also visible. (D) Differential interference contrast image of the field of cells using transmitted light. The distinct morphologies of the various cells in the mixed culture are visible. Bar, 10 μm. Microscopic observations and image acquisition were performed on a TCS 4D system (Leica Lasertechnik GmbH, Heidelberg, Germany).
Several coaggregates were visible after briefly mixing the four species to prepare this specimen (Fig. 3). The large coaggregate (Fig. 3C, arrow), consisting of actinomyces (yellow) and other cells (red), clearly shows the juxtapositioning of distinct cell types. Juxtapositioning is also seen in the examples of other coaggregations (Fig. 3C and D, c1-5), but the identity of the spherical cells cannot be unequivocally determined from morphology alone. The specificity of nucleic acid probes makes them particularly useful in identifying bacteria in situ, and FISH would be an excellent method for identifying the organisms shown in Fig. 2. Thus, specific 16S rRNA-targeted probes are useful for identifying oral bacteria within dental plaque without disrupting community architecture. The combination of FISH with autoradiography can provide additional information on the metabolism of these cells in complex communities by coupling identification of species with uptake of radiolabeled substrates (87, 117).
Using fluorescently conjugated antibodies raised against cell surface epitopes to probe biofilms for the location of particular organisms has the advantage of detecting a functional surface component in a complex biofilm. The disadvantage of this technique is the extensive characterization of the antibody required to determine its specificity. In some situations, even highly specific monoclonal antibodies react with an antigen, such as phosphorylcholine, which is found on several genera of oral bacteria (55). Such broad cross-reactivity may be useful for detecting cells expressing the antigen but does not permit identification of species in a mixed-species community. However, in other situations, it is likely that a functional surface receptor polysaccharide or cognate adhesin is found only on certain species and thus would map both a function and taxonomy. We have used both antibodies and green fluorescent protein to identify bacteria in dual-species biofilms in vitro (5, 120). A summary of this study follows.
When unamended sterile saliva is the nutrient source for monoculture biofilms, S. gordonii DL1 is capable of growth, whereas S. oralis 34 and A. naeslundii T14V are unable to grow (120). Each of these species coaggregates with the other two, and we tested the possibility that coaggregation between S. gordonii and either S. oralis or A. naeslundii in a biofilm might enhance communication with and growth of the latter two species. In pairwise combination, S. gordonii grew independently of the other two organisms; neither A. naeslundii nor S. oralis grew to any significant level. Rather, it seems that S. oralis and A. naeslundii were simply retained in coculture with S. gordonii. However, when S. oralis and A. naeslundii, the two species that failed to grow independently on saliva, were introduced sequentially into a flowcell, they coaggregated and grew luxuriantly (120). Moreover, the amount of growth exhibited in this mutualistic interaction was much greater than the independent growth by S. gordonii. These data provide a dramatic example of the consequences of mutualism and suggest that independent growth may not be the most advantageous strategy in complex communities.
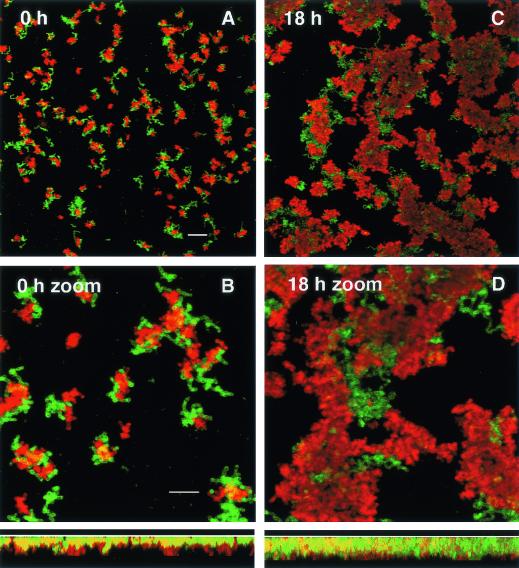
Keeping in mind that both A. naeslundii and S. oralis are capable of binding to receptors in the salivary conditioning film, sequential addition of these two species to the flowcell could result in random and independent adherence to the saliva-coated surface. However, immediately after the unbound cells are removed by salivary flow, nearly all of the cells of the second species, A. naeslundii, were coadherent with the previously bound S. oralis cells (Fig. 4A). A preference for coadherence clearly occurs, because the two species are seen in juxtaposition rather than in a random distribution over the substratum. Examination of the coadherence in the Z dimension (Fig. 4, Z section below panel B) reveals that the red-stained cells (actinomyces) are predominantly localized on top of the green-stained cells (streptococci), confirming the binding of the actinomyces to the already adherent streptococci. After overnight saliva flow, extensive growth of both species is obvious (Fig. 4C and D). The biomass of both species increased laterally as well as axially, and the biofilm appeared denser (Fig. 4, Z section below panel D). Thus, retention through coadherence appears to lead to cooperative growth on saliva for these two species.
FIG. 4.
Representative confocal scanning laser microscopy images of a two-species biofilm formed by S. oralis 34 and A. naeslundii T14V. Maximum projection images were taken at 0 h (A and B) and 18 h (C and D) of salivary flow. S. oralis 34 was incubated statically in a saliva-coated flowcell for 15 min before initiation of salivary flow at 0.2 ml/min for 15 min. A. naeslundii T14V was then added and incubated statically for 15 min, and salivary flow was resumed for 15 min (equals time zero). S. oralis 34 cells were labeled with rabbit anti-S. oralis 34 serum (gift of J. Cisar), followed by indodicarbocyanine-conjugated goat anti-rabbit immunoglobulin antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.); indodicarbocyanine fluorescence is presented in green. A. naeslundii T14V was labeled with a mouse monoclonal antibody against type 1 fimbriae (gift of J. Cisar), followed by indocarbocyanine-conjugated goat anti-mouse immunoglobulin antibody (Jackson ImmunoResearch Laboratories); indocarbocyanine fluorescence is presented in red. A. naeslundii cells are frequently located in direct proximity to S. oralis cells. After 18 h of saliva flow, growth of both genera is apparent. Dimensions of the regions displayed are 250 μm by 250 μm (x-y perspectives; A and C) and 83 μm by 83 μm (x-y perspectives; B and D; 3× zoom of the center portion of the upper panels). Rotation of the maximum projection to display the x-z perspective (83 μm by 10 μm) is shown below panels B and D. The substratum position in the x-z perspective is indicated by the white line. Bars, 20 μm (A and C) and 10 μm (B and D). Microscopic observations and image acquisition were performed on a TCS 4D system (Leica Lasertechnik GmbH, Heidelberg, Germany).
Even simple retention of a species without growth in a biofilm can be of critical in vivo significance. Retention is the principal requirement for growth in a flowing environment such as the oral cavity. It allows communication signals synthesized after cell contact to enter the nascent environment and thereby encourage development of other, more favorable interbacterial relationships. For example, for the species discussed above (Fig. 4) (120), retention of S. oralis in an S. gordonii-dominated biofilm could enable S. oralis to wait for introduction of a more favorable partner, such as A. naeslundii, to join the biofilm community. Coadherence of A. naeslundii to S. oralis could then result in luxuriant growth of the partners. In addition, rapid growth of S. oralis and A. naeslundii could diminish growth of S. gordonii through competition for the same nutrient. Indeed, it is known that S. oralis is a dominant early colonizer of the tooth surface, while S. gordonii is present in lower numbers (114, 115). The ability to form advantageous partnerships and, by comparison, the ability to grow independently represent two of the physiological communication strategies that may occur in developing early dental plaque communities.
Another form of communication among oral bacteria in dental plaque is the exchange of free DNA from lysed cells to transformation-competent cells. Transformation of S. gordonii has been shown to occur in vitro with plasmid DNA in human saliva (104). The half-life for transforming activity of both plasmid and chromosomal DNA is only 5.7 s in saliva in the mouth, but 5.7 s is sufficient for DNA uptake and protection from salivary nucleases (105). Natural genetic transformation has been reported for many oral streptococci in planktonic cultures and occurs by induction of genetic competence through a competence-stimulating peptide (CSP) signaling system (61, 97). Competence in these bacteria is a quorum-sensing phenomenon, where the quorum size required may be species and strain specific. The products of at least six genes, comAB, comX, and comCDE, are involved in CSP signaling. CSP is encoded by comC and is usually found in a genetic locus with comD and comE, which encode a histidine kinase and a response regulator, respectively. A mutation in comD was identified in a screen of S. gordonii mutants with biofilm formation defects (94). Thus, it appears that oral streptococci in biofilms may communicate by a quorum-sensing CSP signaling system.
Cvitkovitch and coworkers discovered the quorum-sensing CSP signaling system in Streptococcus mutans and showed that this system is essential for genetic competence in S. mutans (89) and that it is involved in biofilm formation (91). Knockout mutants defective in comC, comD, comE, and comX were constructed and compared with the wild type. All of the mutants formed altered biofilms. Either exogenous addition of CSP or complementation of mutant comC with a plasmid containing wild-type comC restored the wild-type biofilm phenotype to the comC mutant (91). However, neither addition of CSP nor complementation showed any effect on the biofilms formed by the other com mutants. This quorum-sensing system also functions to regulate acid tolerance in S. mutans biofilms formed in vitro (90). Densely packed cells are more resistant to killing by low pH than are cells in less populated regions of the biofilm (90).
Accompanying the discovery of the CSP system is the evidence that biofilm growth of the streptococcus greatly enhances both competence induction and uptake and integration of a variety of plasmid and chromosomal donor DNAs (89). With biofilm cells, transformation frequencies 10 to 600 times the rate seen with planktonic cells were observed and were maximal after 8 to 16 h of growth. Lysed cells in the biofilm could act as donors of chromosomal DNA, indicating that communities of living and dead cells may share information through genetic exchange. DNA could also act as a nutrient source for the growing biofilm (46), or possibly, extracellular DNA may contribute to initial establishment of oral bacterial biofilms in a manner similar to that recently shown for biofilms formed by a nonoral bacterium (153).
In another example, transfer of native conjugative transposon Tn916-like elements encoding tetracycline resistance (Tetr) from one oral streptococcal species to another in a model biofilm was recently reported (126, 127). The significance of this finding is twofold: (i) tetracycline is used to treat periodontal disease, and Tetr elements circulating within dental plaque would thwart treatment as well as potentially transfer Tetr to transient residents on their way to other body sites, and (ii) DNA transfer favors the possibility of genetic manipulation of these organisms and will thus facilitate molecular characterization of communication in biofilms. Thus, significant progress has been made in answering the question of whether DNA is transferred among bacteria in densely packed environments such as dental plaque.
Imaging by conventional confocal microscopy is difficult with thick specimens. Two-photon excitation microscopy is a potentially valuable tool for resolution of cell shapes at depths greater than 100 μm. Two-photon excitation microscopy coupled with fluorescence lifetime imaging microscopy has been used to study pH gradients in biofilms formed by a consortium of 10 species of oral bacteria (150). Pulses of 14 mM sucrose were supplied to the consortium to elicit acid production. Fluorescence decay of carboxyfluorescein was used to examine pH within the biofilm. Distinct microzonal variations in pH and sharp pH gradients were observed, indicating spatial heterogeneity of response to the same metabolic stimulus. This observation provides evidence for architectural organization of consortia and metabolism within the biofilm. Distinct microbial communities occur within the larger biofilm architecture and include a range of individual responses. Cells at the border of a niche are situated between cells that are responding by producing acid from sucrose and cells outside the niche responding in a different way. Thus, consortia can be understood to consist of physiologically heterogeneous species that respond differently to a given external stimulus. For example, the response may be growth, which may protect other members if the growing cells modify the local environment.
Metabolic Communication
The examples of metabolic communication discussed here are limited to interactions in which at least one organism benefits. This arena of metabolic communications among oral bacteria has been reviewed extensively (57, 73, 84, 100). Beneficial interactions may occur through the excretion of a metabolite by one organism that can be used as a nutrient by a different organism or through the breakdown of a substrate by the extracellular enzymatic activity of one organism that creates biologically available substrates for different organisms. An example of the latter enzymatic activity is sequential hydrolysis of a complex glycoprotein by several bacteria acting in sequence on the product of a previous bacterium's action, as has been shown for oral streptococci (18).
Within the oral cavity, bacteria form multispecies communities that are distinguishable primarily by their location (supragingival versus subgingival versus epithelial). The subgingival community has the highest species richness and the greatest capacity for pathogenic outcome, such as periodontal tissue destruction. In an examination of cocultures of putative periodontal pathogens, such as P. gingivalis and T. denticola, cocultures produced more biomass than was observed in the respective monocultures; most of the coculture biomass was in the form of cell aggregates, and the coculture was transferable over at least five successive inoculations (56).
Cell-free supernatants from monocultures were tested for the ability to stimulate growth of the companion organism. The supernatant from cultures of P. gingivalis could increase the growth of T. denticola and vice versa. Succinate was found in the T. denticola supernatant and was utilized by P. gingivalis monocultures, as determined by lowered concentrations of succinate during growth. The P. gingivalis supernatant contained six fatty acids, including isobutyric acid, and although none of those acids was removed by T. denticola monocultures, isobutyric acid was found to stimulate growth of T. denticola monocultures when added as a supplement at a concentration lower than that at which it occurred in the P. gingivalis supernatant (56). These results imply that cross-feeding between P. gingivalis and T. denticola occurs and that T. denticola requires only minor amounts of P. gingivalis-produced isobutyric acid for maximal growth stimulation.
About 60 oral species of Treponema have been identified (123), and spirochetes constitute a large percentage of the total oral bacterial numbers. Accordingly, a large T. denticola population could benefit greatly through interaction with a small P. gingivalis population. In a separate study, a stimulatory effect of P. gingivalis supernatant on T. denticola growth was attributed to proteinaceous substances (111); this study did not examine the inverse interaction. Thus, synergistic interactions between P. gingivalis and other anaerobic bacteria such as oral spirochetes yielded increased growth and may contribute to increased virulence of these potential periodontal pathogens.
To investigate the outcome of multispecies interactions on virulence, P. gingivalis and F. nucleatum were tested in a murine subcutaneous lesion model. Simultaneous injection of these bacteria at the same site resulted in larger lesions and higher morbidity than did injections of a single species (40). In a concurrent study, this synergistic effect was shown to occur even when each bacterial strain was injected separately on opposite sides of the same animal (45). Thus, this polymicrobial infection demonstrated cooperativity in virulence outcome.
Veillonellae are anaerobic gram-negative cocci that exist in supra- and subgingival plaque communities, where they make up 1 to 5% of the total cultivatable anaerobic bacteria (80). Veillonellae can utilize short-chain organic acids, especially lactate, for growth. These organic acids are excreted by streptococci during growth on sugars and are the basis for the metabolic communication documented in vitro (106) and in vivo in gnotobiotic rats (107, 149). Also, it has been shown in vivo that veillonellae are not capable of colonizing the tooth surface without streptococci as metabolic partners and that larger populations of veillonellae develop in coculture with streptococci that recognize them as a coaggregation partner than in coculture with streptococci with which they do not coaggregate (101).
The conversion of the lactic acid formed by streptococci to less potent acids, such as acetic acid, by veillonellae has been assumed to reduce caries susceptibility in the host, although little experimental evidence supports this hypothesis. Instead, a molecular study suggests that veillonellae are present together with streptococci in carious lesions (8).
While this molecular approach does not establish causality, it does indicate the types of bacteria present at specific sites in health and in disease. Such studies have identified reproducible bacterial communities found at particular disease sites (137), and thus, they identified bacterial populations that may arise through interaction with one another. Studies of biofilm architecture at these sites together with studies of the physiological consequences of that architecture will be required to sort out the occurrence and nature of metabolic communication.
In Vivo
Significant advances have been made in clinical research directed at understanding the architecture of supragingival and subgingival dental plaque in situ. From the five reports discussed here, a picture of dental plaque architecture is emerging.
Supragingival plaque is formed on the outwardly visible enamel surface of teeth. It has been studied in situ by bonding to teeth a device consisting of an enamel piece with an adherent nylon ring (157). After removal from the tooth surface, the supragingival plaque biofilm formed within the nylon ring on the enamel substratum is examined by confocal scanning laser microscopy. The voids observed among the adherent biomass (157) appear to be filled with fluid, as has been reported for several in vitro biofilms composed of one or only a few species (32, 86, 159). On the basis of these observations, voids may serve as avenues for metabolic communication as well as for signaling molecules and antimicrobial compounds.
One method used to study the diffusion of molecules and the efficacy of antimicrobials in undisturbed supragingival plaque was to measure penetration of differential stains into plaque formed in vivo. In these studies, a bovine dentin disk was bonded to an acrylic appliance that was worn by a human subject (159). The disk contained three parallel 200-μm-wide grooves, which were about 500 μm deep and served as the location for plaque accumulation. Upon removal of the appliance, the disks were broken in half along the middle groove. One half was covered with 0.2% chlorhexidine for 1 min; the other half was the control. After treatment, both halves were rinsed with saline, stained with a mixture of ethidium bromide and fluorescein diacetate, and viewed by confocal scanning laser microscopy. Ethidium bromide, a red fluorescent nucleic acid stain, permeates only cells with damaged cell membranes, and fluorescein diacetate penetrates all cells but is nonfluorescent until the green fluorescein moiety is freed by intracellular esterases.
The biofilms grew up to 65 μm thick, and the plaque structure appeared diffuse, with morphologically heterogeneous cell shapes. Both stains penetrated the entire thickness, indicating that there was no barrier to small molecules in these plaque biofilms. In contrast, as measured by cell death (ethidium fluorescence), the antimicrobial chlorhexidine had an effect primarily near the lumen (159). Only the cells near the lumen may have been killed because (i) cells located deeper are more resistant to killing, (ii) antimicrobial action is titrated by contact with surface-exposed biomass and thus the dose of antimicrobial received deeper in the biofilm is ineffectual, or (iii) biofilms may not be indiscriminately open to all molecules throughout their thickness. Furthermore, as plaque biofilm community composition changes, the gating and selectivity of diffusible molecules may also change in response to communication signals. Biofilms may select and gate putative communication signals, and the biofilm inhabitants may temporally regulate such gating. Additional studies to determine the selective porosity of biofilms to signaling molecules will help answer this question.
To study subgingival plaque, oral biofilms formed below the gum line, researchers either use a model system or extract teeth and examine the periodontal region directly. One model system involves three different materials placed into periodontal pockets of patients with rapidly progressing clinical periodontitis (151). A membrane of polytetrafluoroethylene (a material used to cover superficial defects after surgery that can be colonized by plaque bacteria), gold foil (for scanning electron microscopy), and dentin were used. Periodontal pocket depths were approximately 8 mm, and the materials were held in place for 3 to 6 days, yielding biofilms 40 to 45 μm thick.
Analyses were conducted by confocal scanning laser microscopy with fluorescently labeled 16S rRNA-directed oligonucleotide probes and by electron microscopy. Probes specific for Treponema spp. and general bacterial (EUB338 [1]) probes were used to detail the architecture of the periodontal biofilm in situ. The deepest zones of the pockets were colonized principally by spirochetes and gram-negative bacteria, whereas shallow regions contained mostly gram-positive cocci (151). The composition of the carrier material had little influence on colonization. Complex communities of interspersed, morphologically distinct cell types were commonly observed, as were characteristic organized arrangements of mixed cell types, such as rosettes consisting of a gram-positive central bacterium surrounded by gram-negative rods.
The rosettes and other mixed-cell-type arrangements seen in situ bear striking similarity to those seen in vitro during coaggregation studies (74) and in other in situ electron microscopy studies (69, 93). Species-specific FISH probes allowed this first unequivocal demonstration of the spatial arrangement of spirochetes in situ within the periodontal biofilm (151). As additional species-specific oligonucleotide probes are designed, the entire architecture of dental plaque in situ can be elucidated.
In another study examining spatial relationships, Noiri et al. (112) extracted teeth from sites with periodontal pocket depths of approximately 8 mm from patients with advanced adult periodontitis. Intact periodontal pockets were preserved, and the localization of five species, Prevotella nigrescens, F. nucleatum, T. denticola, Eikenella corrodens, and Actinobacillus actinomycetemcomitans, was investigated by immunohistochemistry. Localization of bacteria within the periodontal pocket was analyzed by separating the pocket into nine zones. Vertical zones extended from the gingival margin to the deep pocket and were categorized as shallow, middle, and deep. Horizontal zones, from the tooth surface to the pocket epithelial surface, were called tooth attached, unattached, and epithelium associated.
The results of the immunohistochemistry revealed that P. nigrescens is located at the epithelium-associated plaque area in the middle pocket zone. The middle and deep pocket zones of the unattached area were preferentially colonized by F. nucleatum and T. denticola, but these two bacteria could be found in all zones except one or two of the shallow zones. E. corrodens was located primarily in tooth-attached plaque zones, whereas A. actinomycetemcomitans was found infrequently and only in the middle, unattached plaque zone. F. nucleatum was found in samples from 9 of 15 patients, whereas the other species were found in six or fewer samples. In an earlier study, this research group showed that an antiserum reactive against actinomyces detected cells predominantly in the tooth-attached, shallow and middle zones (113).
Thus, it appears that each oral bacterial species colonizes preferred sites within the subgingival plaque architecture. This selective colonization suggests that species organize into communities through communication with their host and with other species that occupy the host's substrata.
SOLUBLE-SIGNAL COMMUNICATION AMONG ORAL BACTERIA
Transfer of genes by competence-inducing pathways is one of the most-studied forms of communication by oral bacteria. This method of communication is mediated by competence-stimulating peptides (CSPs) (62), which are small, cationic peptides of 14 to 23 amino acid residues that are produced by at least 10 species of oral streptococci (62, 89). Since several reviews and book chapters have discussed the important role of CSPs (62, 97, 108), this method of oral bacterial communication will not be discussed in detail here. Likewise, readers are referred to other reports and reviews describing the roles of sex pheromones and their inhibitor peptides in the well-described mating system of Enterococcus faecalis (3, 29, 30).
Signaling by soluble molecules also occurs between Streptococcus salivarius and Streptococcus pyogenes by the lantibiotic peptides that modulate their own and interspecies lantibiotic synthesis and that may be a mechanism for controlling susceptible streptococci in the human oral cavity (148). Many oral bacteria produce bacteriocin peptides; some are lantibiotic peptides, and others are nonlantibiotic peptides (65, 125). These peptides are thought to influence the ecology of these bacteria, but their mechanism of communicating this influence is unknown. One investigation was conducted with several oral streptococci known to coaggregate with each other (147). One strain, S. gordonii DL1 (Challis), produced a bacteriocin, and five strains were sensitive to the bacteriocin, but neither the production of nor sensitivity to the bacteriocin could be correlated to the ability to coaggregate, suggesting that cell-cell attachment and bacteriocin production are not related.
Physical communication is ubiquitous among oral bacteria in that all of the human oral bacteria tested coaggregate with at least one other genetically distinct species (73). Recently, several examples of production of a quorum-sensing molecule called autoinducer-2 (AI-2) have been reported in oral bacteria (15, 26, 48, 49). We will propose some ideas on possible methods of communication involving AI-2 in a mixed-species community.
Autoinducer-2
Methods of communication among genetically identical cells are likely to be different from the communication signals exchanged among species. AI-2 is proposed to be a signal mediating messages among different species in a mixed-species community (108, 133), distinguishing it from the family of acyl homoserine lactones, typified by autoinducer-1, that regulate gene expression in genetically identical cells (6, 50). No evidence of acyl homoserine lactone production by oral bacteria has been found, suggesting that oral bacterial species do not use acyl homoserine lactones for signaling (49, 154). In contrast, luxS, the gene encoding the enzyme essential for production of AI-2, is present in several genera of oral bacteria (15, 26, 48, 49). AI-2 was discovered in the marine bacterium Vibrio harveyi, in which it is a signal molecule that regulates bioluminescence (7). A set of bioluminescent reporter strains of V. harveyi were constructed (139, 140) and have been used by many investigators to assay for the production of AI-2 by other bacteria. Through these studies, AI-2 has been implicated in gene regulation in several species besides V. harveyi, including Streptococcus pyogenes (98), Escherichia coli (35), Salmonella enterica serovar Typhimurium (141), and two human oral bacteria, A. actinomycetemcomitans (48) and P. gingivalis (15, 26).
Signaling by AI-2 to produce light in V. harveyi is accomplished through binding of AI-2 to LuxP, followed by a phosphorylation cascade (25). The structure of AI-2 bound to its primary receptor LuxP in V. harveyi was recently determined to be a furanosyl borate diester (25). LuxP is an AI-2 sensor protein and is located in the periplasmic space of V. harveyi. AI-2 is produced from S-adenosylmethionine through several steps, including the required enzymatic conversion of the intermediate S-ribosylhomocysteine by LuxS to 4,5-dihydroxy-2,3-pentanedione, which is unstable and is predicted to cyclize spontaneously (133, 134) into a variety of molecules called pro-AI-2 before forming a mature AI-2-LuxP complex (25). The site of borate addition to form AI-2 is unknown and may be in the AI-2-producing cell, outside the cell, or in the recipient cell (25). AI-2 and conserved luxS have been found in more than 30 bacterial species (108, 140), including both gram-positive and gram-negative organisms. Considering the broad representation of luxS among bacteria, AI-2 has been proposed to be a universal interspecies signal (108, 133), but this role has yet to be demonstrated in an ecologically relevant consortium of bacteria.
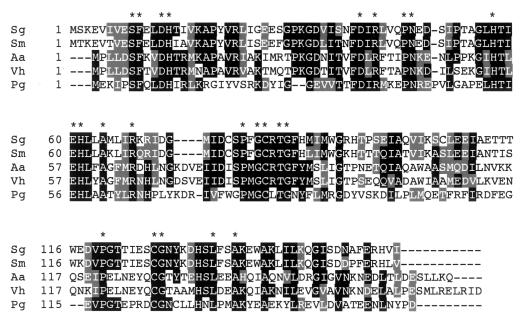
Alignment of LuxS sequences from 26 organisms revealed 23 invariant amino acid residues (64). An alignment of the amino acid sequences deduced from the luxS genes of four oral bacteria, A. actinomycetemcomitans, P. gingivalis, S. gordonii, and Streptococcus mutans, as well as V. harveyi shows that all 23 invariant amino acids are present (Fig. 5). The streptococcal LuxS sequences are 83% identical to each other and 30 to 40% identical to the other three sequences, demonstrating that the gram-positive streptococcal enzymes are more closely related to each other than to enzymes from the three gram-negative species in this alignment. The A. actinomycetemcomitans LuxS sequence is 72% identical to V. harveyi LuxS but only 31% identical to the LuxS sequence of P. gingivalis, which is 29% identical to V. harveyi LuxS. Finding luxS in both gram-positive and gram-negative strains of oral bacteria suggests a role for AI-2 in mixed-species communities such as dental plaque.
FIG. 5.
Alignment of the deduced amino acid sequences of luxS from S. gordonii DL1 (Sg; accession number AY081773), S. mutans UA159 (Sm; www.genome.ou.edu/smutans.html), A. actinomycetemcomitans HK1651 (Aa; www.genome.ou.edu/act.html), V. harveyi BB120 (Vh; accession no. AF120098), and P. gingivalis W83 (Pg; www.tigr.org). Consensus of at least 50% identical amino acid residues is denoted by black boxes; conserved amino acid substitutions are highlighted with gray boxes. Asterisks are placed above the 23 amino acid residues that were shown to be invariant in 26 LuxS sequences aligned by Hilgers and Ludwig (64).
In a survey of 16 species of oral bacteria, F. nucleatum, P. gingivalis, and Prevotella intermedia produced high levels of AI-2 under the conditions tested (49). Another periodontal bacterium, A. actinomycetemcomitans, was subsequently reported to produce AI-2 (48). These AI-2-producing species are all involved in the polymicrobial infection known as periodontal disease. Additional oral species are being surveyed for production of AI-2 with the expectation that AI-2-mediated communication may play a central role in gene regulation in mixed-species communities.
Two oral species, P. gingivalis and A. actinomycetemcomitans, were examined for regulation of gene expression by AI-2 (15, 26, 48). Based on the V. harveyi reporter assay, conditioned medium from these two oral bacteria elicited a response that was approximately 10 and 20%, respectively, of that of the V. harveyi positive control (26, 48). As measured by reverse transcription-PCR, two genes relevant to hemin acquisition were reported to be induced in a P. gingivalis luxS mutant, and three other genes were repressed, indicating a response to luxS inactivation (26). Expression levels of genes that were altered in the P. gingivalis mutant were also affected by exposure of the mutant to conditioned medium from a recombinant E. coli strain harboring the A. actinomycetemcomitans luxS gene on a plasmid (48). Additionally, as measured by reverse transcription-PCR, exposure of A. actinomycetemcomitans to conditioned medium from the recombinant AI-2-producing E. coli strain described above resulted in increased transcription of afuA, which encodes a periplasmic protein involved in iron transport (48). As measured by a cytotoxicity assay, leukotoxin production was enhanced in A. actinomycetemcomitans cultures exposed to conditioned medium from the parent strain or from the recombinant E. coli strain described above (48).
In a separate study (15), it was reported that a P. gingivalis luxS mutant produced approximately 30% less Lys-gingipain and about 45% less Arg-gingipain, two cysteine proteases that are synthesized at high cell densities. While the regulation of genes involved in iron acquisition, protease synthesis, and leukotoxin production appears to be influenced by the presence of AI-2 in P. gingivalis and A. actinomycetemcomitans, it is not yet clear whether the differential regulation of these genes affects the virulence of these oral pathogens in vivo. Nonetheless, the pairing of P. gingivalis and A. actinomycetemcomitans has provided a starting point for demonstrating that AI-2 may serve as an interspecies signal (108, 133) in two organisms that coexist in the same ecological niche.
Modeling Oral Bacterial Mixed-Species Communication by AI-2
A LuxP homologue was found in A. actinomycetemcomitans (48) but not in P. gingivalis (26). This leaves open the possibility that oral bacteria may have alternate binding proteins for AI-2 that may be like the periplasmic ribose-binding proteins of E. coli and S. enterica serovar Typhimurium (7) and a recently discovered transporter of AI-2 in S. enterica serovar Typhimurium (141), which are all homologous to LuxP of V. harveyi. In accordance with the concept of alternative binding proteins, it is possible that a variety of cognate fits of pro-AI-2 molecules and binding proteins may occur within a mixed-species community. The binding affinity of each pro-AI-2/binding protein pair may be characteristic of a species and thus permit numerous potential cognate fits within mixed-species communities. Thus, by using their respective cognate binding proteins, many species could simultaneously sample a mixture of pro-AI-2 in their microenvironment. Likewise, a mixture of pro-AI-2 in the microenvironment may be sampled differently by different bacteria because the concentration of pro-AI-2 molecules is critical for specificity in regulating gene expression. Thus, the capacity of AI-2 to serve as a universal signal within a mixed-species bacterial community may stem from individual organisms' abilities to bind and respond to unique molecules within the environmental pool of pro-AI-2.
It is challenging to postulate attractive mechanisms of mixed-species communication by the proposed universal interspecies signal AI-2. First, some but not necessarily all of the species in the community must express an active LuxS, since it is required for synthesizing pro-AI-2. Second, to be universal, all the species must respond to pro-AI-2. Third, as culture supernatants from a variety of genera elicit light production in the appropriate V. harveyi reporter strains, at least some of the extracellular pro-AI-2 molecules produced by different species must have identical structures. However, not all of the pro-AI-2 molecules need to be identical. Thus, a mixture of pro-AI-2 molecules may be a significant factor in the ability of an organism to compete by sensing, selecting, and responding to a particular isomer within the blend of pro-AI-2 molecules. Analogs of AI-2 such as 4-hydroxy-5-methyl-furanone were found to stimulate the Vibrio reporter strain to produce light when added at 1,000-fold the concentration of AI-2 required for light production (134). This demonstrates that, although less efficient, the LuxP of V. harveyi will bind and respond to molecules other than naturally synthesized pro-AI-2. Assuming that several and perhaps all of the species in a mixed-species community such as dental plaque are producing pro-AI-2, the species-specific mode of communication may be based simply on correctly identifying the proper isomer in the pro-AI-2 signal blend, resulting in an advantageous gene regulation response.
Sorting and responding to pro-AI-2 may occur by mechanisms similar to those used by other biological kingdoms. Only a narrow window of signal concentration may elicit a response, as has been found for insect pheromones released and received by members of the order Lepidoptera (130, 131). Coevolution of emitters and receivers of insect pheromones occurs, and communication between the correct males and females must happen in a background of many signals specific for other species. Too much or too little pheromone elicits no response by the potential mate. Too constant an emission gets no response. Pheromones may be of the same general chemical structures but in different blends, which are specific for the correct mating. By having several chemicals in the blend, the combinations of the chemicals can yield an almost unlimited variety of blends, and each species of Lepidoptera may emit and respond to only one blend. In a similar way, the combinations of pro-AI-2 molecules formed by members of a bacterial community may be sorted and received uniquely for each species within a mixed-species community such as dental plaque.
Other factors in a microenvironment may modulate the detection and response to a blend of pro-AI-2 signals. As has been reported for distinct pH gradients caused by juxtapositioning of a variety of species (150), other chemical or physical gradients may form with certain adjacent species in a microenvironment. These conditions may influence the production of pro-AI-2 or the blend of pro-AI-2 in the extracellular pool. For example, in S. enterica serovar Typhimurium, increased AI-2 production occurs in high-osmolarity and low-pH environments, and a protein synthesis-dependent degradation of the signal occurs in low-osmolarity conditions (139). A prominent role for acidic environments has long been known in dental plaque, where acid-producing bacteria cause caries and consequently pose a constant threat to oral health. The acidic microenvironment present in dental plaque may be essential for proper response to chemical communication with AI-2. The physical attachment of cells of different species may cause contact-induced gene regulation, perhaps in all coaggregating cells, and result in altered production of and response to a mixture of pro-AI-2 molecules.
No matter what mechanism mixed-species communities use for AI-2 signaling, progress in understanding the role of AI-2 in complex microbial communities will come when experiments are conducted with mixed-species communities.
DIRECT-CONTACT SIGNAL: ANTIGENS I AND II
The antigen I/II (Ag I/II) family of surface proteins is represented on many species of human oral streptococci (Table 1). They contain the LPXTG motif recognized by gram-positive sortase enzymes, which tether this class of proteins to the peptidoglycan layer (47, 135). These large polypeptides range from about 1,450 to 1,570 amino acid residues and possess seven discrete, well-conserved structural domains. These multifunctional proteins bind to other bacteria, host tissue, salivary conditioning film on teeth, and soluble molecules. Thus, the Ag I/II polypeptides are excellent candidates for mediating the adherence of streptococci to surfaces and providing an opportunity to communicate with other residents of the environment. A complete description of these properties and the immunogenicity of the proteins is found in two reviews (66, 67). Some of the recent progress in defining a role for Ag I/II proteins in promoting the dominance of streptococci in dental plaque will be discussed here.
TABLE 1.
Ag I/II homologues of oral streptococci
| Organism | Protein(s) | Reference(s) |
|---|---|---|
| S. constellatus | —a | 24, 99 |
| S. cricetus | PAa | 144 |
| S. gordonii | SspA, SspB | 37 |
| S. intermedius | — | 24, 124 |
| S. mitis | — | 23 |
| S. mutans | SpaP, Pac, Protein I/IIf | 71, 116 |
| S. oralis | — | 16 |
| S. sobrinus | SpaA, PAg | 85, 146 |
—, homologue with no designation.
Considering that Ag I/II proteins on different streptococcal species contain well-conserved structural domains, it is not surprising that they also exhibit overlapping functions, such as binding to human salivary glycoproteins. However, they do possess specific functions as well, which arise from the activity of distinct structural domains. Small changes in the primary sequence can have profound changes in the functional character of Ag I/II proteins. For example, in SspB, an Ag I/II homologue in S. gordonii, a region spanning residues 1167 to 1250 is required for adherence to a coaggregation partner, P. gingivalis (13). Changing two residues, Asn1182 to Gly and Val1185 to Pro, yields a protein predicted to possess secondary structure similar to that of SpaP, an Ag I/II homologue in S. mutans, which does not coaggregate with P. gingivalis. Recombinant strains of S. gordonii bearing the modified proteins lost the ability to coaggregate with P. gingivalis (38), confirming that this region of these two homologues differs functionally as well as structurally.
In contrast, an example of functional similarity is that the Ag I/II proteins of both S. mutans and S. gordonii bind to human collagen type I, a major component of dentin, and facilitate invasion of human root dentinal tubules by these species (95). Testing the multifunctional capabilities of SspB was accomplished by comparing the abilities of S. gordonii and S. mutans to bind to human collagen type I and to coaggregate with P. gingivalis. Both abilities were required for Streptococcus-Porphyromonas coinvasion of dentinal tubules (96). P. gingivalis was able to invade dentinal tubules when cocultured with S. gordonii bearing SspA and SspB. An S. gordonii sspA/B mutant deficient in collagen binding did not support coinvasion with P. gingivalis; S. mutans bearing SpaP invaded human root dentinal tubules but did not support coinvasion with P. gingivalis. The results of these studies illustrate discrete functions embodied within the Ag I/II structures and the relevance of surface adhesins to the colonization of indigenous environments by bacteria.
Another discrete function of Ag I/II proteins is the ability to mediate coaggregation with actinomyces partner cells. S. gordonii strains DL1 and M5 each have two Ag I/II paralogs, SspA and SspB, which are involved in coaggregation with A. naeslundii. The genes encoding these proteins are arranged in tandem on the chromosomes of these strains and, in strain DL1, are known to exhibit different patterns of regulation in response to environmental conditions (43). Furthermore, it appears that sspB transcription is positively regulated by the sspA gene product, suggesting an additional role for SspA (42).
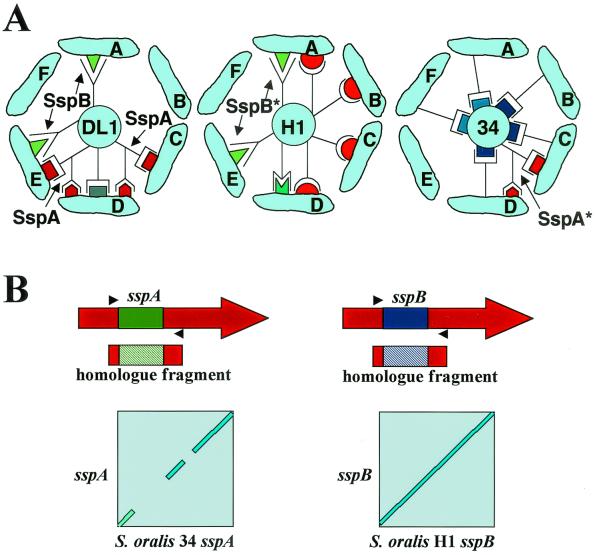
Earlier surveys of coaggregations between streptococci and actinomyces yielded six coaggregation groups of actinomyces, representing the range of coaggregations observed with approximately 90% of the 300 human oral A. naeslundii strains tested (41, 73). Recent results focused on S. gordonii DL1 have shown that SspA and SspB are adhesins critical for coaggregation with four of the six actinomyces coaggregation groups (41) (Fig. 6A). Others have shown that inactivation of sspA of S. gordonii DL1 resulted in a loss of greater than 50% of the wild-type ability to coaggregate with A. naeslundii T14V (coaggregation group A) and A. naeslundii PK606 (coaggregation group D) (68).
FIG. 6.
(A) Involvement of Ag I/II family members in Streptococcus-Actinomyces coaggregations. The diagram shows coaggregations between S. gordonii DL1, S. oralis H1, and S. oralis 34 (representatives of streptococcal coaggregation groups 1, 2, and 3, respectively) with six actinomyces coaggregation groups (groups A, B, C, D, E, and F) (72). Actinomyces coaggregation groups are symbolized by oblong cellular shapes containing letters representing the groups and surround the three circles representing streptococcal strains DL1, H1, and 34. The model of S. gordonii DL1 coaggregation is based on identification of the independent coaggregation functions of SspA and SspB of S. gordonii DL1 (41). The models for S. oralis strains H1 and 34 are based on pairwise coaggregations between actinomyces and streptococci, lactose-inhibitable coaggregations, and loss of certain coaggregation functions by spontaneous mutants (72). The complementary symbols shown here are described in the legend to Fig. 1. *, putative protein designations. (B) Graphic representation of sspA and sspB (accession number U40027), with divergent regions shown in green and purple and gene fragments amplified from S. oralis 34 and S. oralis H1 encoding Ag I/II homologues. The locations of primers (solid arrowheads) used to amplify the homologous fragments from S. oralis H1 (purple-stippled rectangle) and S. oralis 34 (green-stippled rectangle) are shown. Blast2 alignments (145), shown below, reveal that the divergent regions of S. gordonii sspB and the S. oralis H1 homologue are conserved over the length of this region (conserved sequences appear as a straight line at a 45o angle). However, alignment of the sspA divergent region and the corresponding region from the S. oralis 34 homologue reveals gaps in the sequence identity. The similarities and differences in these sequences may be responsible for the coaggregations with A. naeslundii strains shown in panel A. See text for more detail.
Construction of sspB insertion mutants of both S. gordonii DL1, and an isogenic sspA mutant allowed the first study of the independent functions of SspA and SspB (41). SspA exhibited two coaggregation-specific functions; it participated in lactose-inhibitable and lactose-noninhibitable interactions (Fig. 6A, cognate symbols complementary to red rectangle and red obelisk, respectively). Mutation of sspA resulted in changes in coaggregation with three of the four actinomyces coaggregation groups. For example, the sspA mutant was unable to coaggregate with actinomyces coaggregation group C (Fig. 6A). SspB mediated only lactose-noninhibitable coaggregations (Fig. 6A, cognate symbol complementary to yellow-green triangle). The sspB mutant was unable to coaggregate with actinomyces coaggregation group A, and its coaggregation with actinomyces coaggregation group E was changed from lactose noninhibitable to lactose inhibitable. A change from lactose noninhibitable to lactose inhibitable is possible because the former coaggregation masks the latter in the presence of lactose. The effects of the mutations were additive in the sspAB double mutant; however, it retained a single previously unrecognized lactose-inhibitable coaggregation with A. naeslundii PK606 (coaggregation group D) (Fig. 6A, cognate symbol complementary to gray rectangle). Thus, SspA and SspB appear to mediate all coaggregations between S. gordonii DL1 and the actinomyces coaggregation groups except the lactose-inhibitable coaggregation with actinomyces coaggregation group D.
SspA and SspB show a high level of sequence identity in S. gordonii strains M5 and DL1 (37). The complete sequences of sspA and sspB from S. gordonii DL1 have recently been determined (accession number U40027). Comparison of the C-terminal half of the deduced sequences of SspA and SspB shows that they are 98% identical, and the sequences of the N-terminal third of the polypeptides are 96% identical. The intervening divergent regions are only 37% identical. Comparison of these sequences to the corresponding sequences of strain M5 indicates 93% identity between the SspA sequences and 95% identity between the SspB sequences. Thus, the genetic organization of these two genes as well as the sequence similarity both between the paralogs in strains M5 and DL1 and between the orthologs of strains M5 and DL1 indicate close relationships.
This broad range of functions attributed to SspA and SspB suggested that perhaps other streptococci might also employ one or both of these proteins in mediating coaggregations with actinomyces. Two examples are depicted along with the coaggregation model of S. gordonii DL1 with the six actinomyces coaggregation groups A to F (Fig. 6A). Both S. oralis H1 and S. oralis 34 express a surface protein that cross-reacts with antiserum (gift of D. Demuth) recognizing both SspA and SspB. However, S. oralis H1 and S. oralis 34 exhibit patterns of coaggregation with actinomyces that are distinct from that of S. gordonii DL1 (Fig. 6A).
By using primers designed to hybridize with conserved sequence flanking the divergent region of both sspA and sspB of S. gordonii DL1, genomic DNA from S. oralis strains H1 and 34 was used as the template to PCR amplify the divergent region of the Ag I/II genes from these strains (Fig. 6B). Sequence analysis of the resultant PCR products revealed that the S. oralis 34 divergent region was 86% identical to sspA, while the S. oralis H1 divergent region was 93% identical to the corresponding region of sspB. Alignment (145) of the divergent region and short flanking conserved regions of S. gordonii DL1 sspA with the sspA homologue fragment from S. oralis 34 shows large gaps in sequence identity (Fig. 6B, left graph). In comparison, alignment of the sspB homologue fragment from S. oralis H1 with S. gordonii DL1 sspB contains no gaps (Fig. 6B, right graph).
Considering this identity of S. gordonii DL1 sspB and the S. oralis H1 homologue, it is not surprising that both S. gordonii DL1 and S. oralis H1 exhibit the same SspB-mediated interactions with actinomyces (Fig. 6A, left and center, respectively). Accordingly, comparison of streptococci bearing the less similar SspA homologues revealed distinct coaggregation patterns in S. gordonii DL1 and S. oralis 34 (Fig. 6A, left and right, respectively). Apparently, changes predicted in the S. oralis 34 SspA homologue are sufficient to prevent recognition of the lactose-sensitive receptor on actinomyces coaggregation group E. Examination of SspA and SspB homologues in these streptococcal strains indicates that these proteins are integral to the adherence abilities of oral streptococci and are structurally and functionally distinct.
Besides mediating adherence to actinomyces, these two surface proteins bind salivary agglutinin glycoprotein (36, 68). This may represent one of the many mechanisms that streptococci employ to adhere to salivary conditioning film on enamel surfaces. The outcome of S. gordonii DL1 binding to saliva was investigated by random arbitrarily primed PCR. Such studies showed that exposure of S. gordonii DL1 to saliva resulted in induction of sspA/B expression (39). Thus, bathing cells of S. gordonii DL1 in saliva, a condition expected to occur in the oral cavity, causes them to synthesize surface adhesins that bind salivary receptors. One hypothesis is that such binding would encourage removal of the streptococcal cells by agglutination and swallowing. Another hypothesis is that such binding promotes attachment to surfaces through salivary receptor bridging between SspA/B-bearing planktonic cells and already attached bacteria. In fact, coating streptococci with saliva may enhance binding to surfaces and prevent disappearance from the mouth by swallowing.
Consistent with the second hypothesis, we have shown that suspending streptococci in saliva does not reduce their ability to coaggregate with actinomyces (81) or with other streptococci (78). Also, our results with streptococci and actinomyces growing in biofilms on saliva-conditioned glass surfaces with saliva as the sole nutrient source indicate that these bacteria adhere and grow luxuriantly (120). Presumably, the ability to communicate under these conditions is enhanced by being coated with saliva, because the habitat in which these bacteria coevolved with their host is bathed in saliva.
FUTURE DIRECTIONS
In plaque, as in most natural biofilm ecosystems, spatial heterogeneity exists with respect to species location in the axial and lateral dimensions (see Fig. 2); the spatial organization changes with time as a result of interspecies communication and gene expression. Elucidating the architecture of dental plaque at the species level is possible with the use of confocal microscopy coupled with appropriate probes based on 16S rDNA sequences and antibodies that recognize cell surface epitopes of oral bacteria. Probe specificity can be tested in biofilm models in vitro. Temporal dental plaque studies can be conducted in situ. The development of fluorescence-based reporters that function in an anaerobic environment needs to be encouraged. When these probes become available, they will promote study of gene regulation in anaerobic periodontal biofilm communities that include human oral spirochetes. Gene regulation induced by cell-cell contact is underinvestigated; the development of reporters based on green fluorescent protein will facilitate future studies. Current approaches such as in vivo expression technology, DNA microarrays, and proteomics provide the platforms for large-scale analysis of gene expression.
The genomes of F. nucleatum (70), S. mutans (B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Lin, S. Li, S. Kenton, H. Lai, J. D. White, R. E. McLaughlin, M. McShan, D. Ajdic, and J. Ferretti, Streptococcus mutans Genome Sequencing Project, 26 March 2002 posting date; www.genome.ou.edu/smutans.html), P. gingivalis (http://www.tigr.org), and A. actinomycetemcomitans (B. A. Roe, F. Z. Najar, S. Clifton, T. Ducey, L. Lewis, and S. W. Dyer, Actinobacillus Genome Sequencing Project, 26 March 2002 posting date; www.genome.ou.edu/act.html) have been sequenced, and at least 10 other oral bacterial genomes are being sequenced. Investigation in silico will offer great opportunity to identify specific genes hypothesized to be relevant to intraspecies and interspecies communication, such as comC and luxS, respectively. Indeed, searching in silico yielded luxS of P. gingivalis (26), A. actinomycetemcomitans (48), and S. pyogenes (98), as well as comC of S. mutans (89).
Subtle regulation of gene expression in any organism within a community may lead to significant changes in the organism's ability to participate in community activities, such as use of community-formed metabolic end products as nutrients. Known progressive changes in species composition in natural communities with respect to time, nutrient availability, disease state, or other environmental conditions alert us to consider that a mutation in a relevant gene may cause only a subtle change that is critical in contributing to an organism's success in a community. Such subtle changes may be controlled by the regulation of multiple integrated pathways. Results from future research will build a better understanding of how small changes cause large shifts in population composition and metabolic output of mixed-species communities.
Furthermore, inactivation of a gene involved in mixed-species community formation may cause a subtle variation in an organism's phenotype only during critical transitions in population composition and have no effect on population composition before or after the transition. Dental plaque on human tooth enamel undergoes population transitions each day between oral hygiene regimens. Identifying genes that are critical for an organism's success in this environment may be possible not by screening mutants for absence of growth but rather by investigating subtle changes in an organism's participation in the community.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. [DOI] [PubMed] [Google Scholar]
- 5.Aspiras, M. B., K. M. Kazmerzak, P. E. Kolenbrander, R. McNab, N. Hardegen, and H. F. Jenkinson. 2000. Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro. Appl. Environ. Microbiol. 66:4074-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 8.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos, R., H. C. van der Mei, J. M. Meinders, and H. J. Busscher. 1994. A quantitative method to study co-adhesion of microorganisms in a parallel plate flow chamber: basic principles of the analysis. J. Microbiol. Methods 20:289-305. [Google Scholar]
- 10.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med.. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 11.Bradshaw, D. J., and P. D. Marsh. 1999. Use of continuous flow techniques in modeling dental plaque biofilms. Methods Enzymol. 310:279-296. [DOI] [PubMed] [Google Scholar]
- 12.Bradshaw, D. J., P. D. Marsh, G. K. Watson, and C. Allison. 1998. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66:4729-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, A. E., J. D. Rogers, E. M. Haase, P. M. Zelasko, and F. A. Scannapieco. 1999. Prevalence of the amylase-binding protein A gene (abpA) in oral streptococci. J. Clin. Microbiol. 37:4081-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 16.Burnie, J. P., W. Brooks, M. Donohoe, S. Hodgetts, A. al-Ghamdi, and R. C. Matthews. 1996. Defining antibody targets in Streptococcus oralis infection. Infect. Immun. 64:1600-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers, H. L., K. A. Homer, and D. Beighton. 1996. Utilization of sialic acid by viridans streptococci. J. Dent. Res. 75:1564-1571. [DOI] [PubMed] [Google Scholar]
- 18.Byers, H. L., E. Tarelli, K. A. Homer, and D. Beighton. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469-479. [DOI] [PubMed] [Google Scholar]
- 19.Carlsson, J. 2000. Growth and nutrition as ecological factors, p. 67-130. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymondham, United Kingdom
- 20.Carlsson, J., H. Grahnen, G. Jonsson, and S. Wikner. 1970. Early establishment of Streptococcus salivarius in the mouth of infants. J. Dent. Res. 49:415-418. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson, J., H. Grahnen, G. Jonsson, and S. Wikner. 1970. Establishment of Streptococcus sanguis in the mouths of infants. Arch. Oral Biol. 15:1143-1148. [DOI] [PubMed] [Google Scholar]
- 22.Cassels, F. J., C. V. Hughes, and J. L. Nauss. 1995. Adhesin receptors of human oral bacteria and modeling of putative adhesin-binding domains. J. Ind. Microbiol. 15:176-185. [DOI] [PubMed] [Google Scholar]
- 23.Chan, A., P. G. Egland, and P. E. Kolenbrander. 2002. Novel antigen I/II family members: adhesins of human oral streptococci. J. Dent. Res. (Special Issue A) 81:A287. [Google Scholar]
- 24.Chatenay-Rivauday, C., I. Yamodo, M. A. Sciotti, N. Troffer-Charlier, J. P. Klein, and J. A. Ogier. 2000. TNF-alpha release by monocytic THP-1 cells through cross-linking of the extended V-region of the oral streptococcal protein I/II. J. Leukoc. Biol. 67:81-89. [DOI] [PubMed] [Google Scholar]
- 25.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 26.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cisar, J. O., A. L. Sandberg, C. Abeygunawardana, G. P. Reddy, and C. A. Bush. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5:655-662. [DOI] [PubMed] [Google Scholar]
- 28.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 47-65. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 30.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 31.Costerton, J. W. 2000. Biofilms in the new millennium: musings from a peak in Xanadu, p. 329-344. In D. G. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom
- 32.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 33.Darenfed, H., D. Grenier, and D. Mayrand. 1999. Acquisition of plasmin activity by Fusobacterium nucleatum subsp. nucleatum and potential contribution to tissue destruction during periodontitis. Infect. Immun. 67:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong, M. H., and J. S. van der Hoeven. 1987. The growth of oral bacteria on saliva. J. Dent. Res. 66:498-505. [DOI] [PubMed] [Google Scholar]
- 35.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demuth, D. R., P. Berthold, P. S. Leboy, E. E. Golub, C. A. Davis, and D. Malamud. 1989. Saliva-mediated aggregation of Enterococcus faecalis transformed with a Streptococcus sanguis gene encoding the SSP-5 surface antigen. Infect. Immun. 57:1470-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 38.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dû, L. D., and P. E. Kolenbrander. 2000. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect. Immun. 68:4834-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersole, J. L., F. Feuille, L. Kesavalu, and S. C. Holt. 1997. Host modulation of tissue destruction caused by periodontopathogens: effects on a mixed microbial infection composed of Porphyromonas gingivalis and Fusobacterium nucleatum. Microb. Pathog. 23:23-32. [DOI] [PubMed] [Google Scholar]
- 41.Egland, P. G., L. D. Dû, and P. E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Sabaeny, A., D. R. Demuth, and R. J. Lamont. 2001. Regulation of Streptococcus gordonii sspB by the sspA gene product. Infect. Immun. 69:6520-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Sabaeny, A., D. R. Demuth, Y. Park, and R. J. Lamont. 2000. Environmental conditions modulate the expression of the sspA and sspB genes in Streptococcus gordonii. Microb. Pathog. 29:101-113. [DOI] [PubMed] [Google Scholar]
- 44.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feuille, F., J. L. Ebersole, L. Kesavalu, M. J. Stepfen, and S. C. Holt. 1996. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 64:2094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 48.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gendron, R., D. Grenier, and L. Maheu-Robert. 2000. The oral cavity as a reservoir of bacterial pathogens for focal infections. Microbes Infect. 2:897-906. [DOI] [PubMed] [Google Scholar]
- 52.Gibbons, R. J., D. I. Hay, and D. H. Schlesinger. 1991. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect. Immun. 59:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibbons, R. J., and J. van Houte. 1975. Dental caries. Annu. Rev. Med. 26:121-136. [DOI] [PubMed] [Google Scholar]
- 54.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 55.Gmür, R., T. Thurnheer, and B. Guggenheim. 1999. Dominant cross-reactive antibodies generated during the response to a variety of oral bacterial species detect phosphorylcholine. J. Dent. Res. 78:77-85. [DOI] [PubMed] [Google Scholar]
- 56.Grenier, D., and D. Mayrand. 1986. Nutritional relationships between oral bacteria. Infect. Immun. 53:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grenier, D., and D. Mayrand. 2000. Periodontitis as an ecological imbalance, p. 275-310. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymondham, United Kingdom.
- 58.Guggenheim, M., S. Shapiro, R. Gmür, and B. Guggenheim. 2001. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 67:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han, Y. W., W. Shi, G. T. Huang, S. Kinder-Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of green fluorescent protein fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 61.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 62.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-19. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 63.Hay, D. I., A. Bennick, D. H. Schlesinger, K. Minaguchi, G. Madapallimattam, and S. K. Schluckebier. 1988. The primary structures of six human salivary acidic proline-rich proteins (PRP-1, PRP-2, PRP-3, PRP-4, PIF-s and PIF-f). Biochem. J. 255:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilgers, M. T., and M. L. Ludwig. 2001. Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proc. Natl. Acad. Sci. USA 98:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 67.Jenkinson, H. F., and R. J. Lamont. 1997. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 8:175-200. [DOI] [PubMed] [Google Scholar]
- 68.Jenkinson, H. F., S. D. Terry, R. McNab, and G. W. Tannock. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones, S. J. 1972. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch. Oral Biol. 17:613-616. [DOI] [PubMed] [Google Scholar]
- 70.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelly, C., P. Evans, L. Bergmeier, S. F. Lee, A. Progulske-Fox, A. C. Harris, A. Aitken, A. S. Bleiweis, and T. Lehner. 1989. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 258:127-132. [DOI] [PubMed] [Google Scholar]
- 72.Kolenbrander, P. E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu. Rev. Microbiol. 42:627-656. [DOI] [PubMed] [Google Scholar]
- 73.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 74.Kolenbrander, P. E., and R. N. Andersen. 1988. Intergeneric rosettes: sequestered surface recognition among human periodontal bacteria. Appl. Environ. Microbiol. 54:1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolenbrander, P. E., R. N. Andersen, K. Kazmerzak, R. Wu, and R. J. Palmer, Jr. 1999. Spatial organization of oral bacteria in biofilms. Methods Enzymol. 310:322-332. [DOI] [PubMed] [Google Scholar]
- 76.Kolenbrander, P. E., R. N. Andersen, K. M. Kazmerzak, and R. J. Palmer, Jr. 2000. Coaggregation and coadhesion in oral biofilms, p. 65-85. In D. G. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom
- 77.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix. Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ. Microbiol. 56:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kolenbrander, P. E., and J. London. 1993. Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol. 175:3247-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolenbrander, P. E., and L. V. Moore. 1992. The Genus Veillonella, p. 2034-2047. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 2. Springer-Verlag, New York, N.Y.
- 81.Kolenbrander, P. E., and C. S. Phucas. 1984. Effect of saliva on coaggregation of oral Actinomyces and Streptococcus species. Infect. Immun. 44:228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuramitsu, H., and R. P. Ellen. 2000. Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Norfolk, England.
- 85.LaPolla, R. J., J. A. Haron, C. G. Kelly, W. R. Taylor, C. Bohart, M. Hendricks, J. P. Pyati, R. T. Graff, J. K. Ma, and T. Lehner. 1991. Sequence and structural analysis of surface protein antigen I/II (SpaA) of Streptococcus sobrinus. Infect. Immun. 59:2677-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li, T., P. Bratt, A. P. Jonsson, M. Ryberg, I. Johansson, W. J. Griffiths, T. Bergman, and N. Strömberg. 2000. Possible release of an ArgGlyArgProGln pentapeptide with innate immunity properties from acidic proline-rich proteins by proteolytic activity in commensal Streptococcus and Actinomyces species. Infect. Immun. 68:5425-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li, Y.-H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li, Y.-H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liljemark, W. F., C. G. Bloomquist, C. L. Bandt, B. L. Pihlstrom, J. E. Hinrichs, and L. F. Wolff. 1993. Comparison of the distribution of Actinomyces in dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease. Oral Microbiol. Immunol. 8:5-15. [DOI] [PubMed] [Google Scholar]
- 93.Listgarten, M. A., H. Mayo, and M. Amsterdam. 1973. Ultrastructure of the attachment device between coccal and filamentous microorganisms in “corn cob” formations of dental plaque. Arch. Oral Biol. 18:651-656. [DOI] [PubMed] [Google Scholar]
- 94.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love, R. M., M. D. McMillan, and H. F. Jenkinson. 1997. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect. Immun. 65:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Love, R. M., M. D. McMillan, Y. Park, and H. F. Jenkinson. 2000. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordonii depends upon binding specificity of streptococcal antigen I/II adhesin. Infect. Immun. 68:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lunsford, R. D. 1998. Streptococcal transformation: essential features and applications of a natural gene exchange system. Plasmid 39:10-20. [DOI] [PubMed] [Google Scholar]
- 98.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 99.Ma, J. K., C. G. Kelly, G. Munro, R. A. Whiley, and T. Lehner. 1991. Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect. Immun. 59:2686-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayrand, D., and D. Grenier. 1998. Bacterial interactions in periodontal diseases. Bull. Inst. Pasteur 96:125-133. [Google Scholar]
- 101.McBride, B. C., and J. S. van der Hoeven. 1981. Role of interbacterial adherence in colonization of the oral cavities of gnotobiotic rats infected with Streptococcus mutans and Veillonella alcalescens. Infect. Immun. 33:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCarthy, C., M. L. Snyder, and R. B. Parker. 1965. The indigenous oral flora of man. I. The newborn to the one-year-old infant. Arch. Oral Biol. 10:61-70. [DOI] [PubMed] [Google Scholar]
- 103.McIntire, F. C., A. E. Vatter, J. Baros, and J. Arnold. 1978. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect. Immun. 21:978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mercer, D. K., K. P. Scott, W. A. Bruce-Johnson, L. A. Glover, and H. J. Flint. 1999. Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva. Appl. Environ. Microbiol. 65:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mercer, D. K., K. P. Scott, C. M. Melville, L. A. Glover, and H. J. Flint. 2001. Transformation of an oral bacterium via chromosomal integration of free DNA in the presence of human saliva. FEMS Microbiol. Lett. 200:163-167. [DOI] [PubMed] [Google Scholar]
- 106.Mikx, F. H., and J. S. van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20:407-410. [DOI] [PubMed] [Google Scholar]
- 107.Mikx, F. H., J. S. van der Hoeven, K. G. Konig, A. J. Plasschaert, and B. Guggenheim. 1972. Establishment of defined microbial ecosystems in germ-free rats. I. The effect of the interactions of Streptococcus mutans or Streptococcus sanguis with Veillonella alcalescens on plaque formation and caries activity. Caries Res. 6:211-223. [DOI] [PubMed] [Google Scholar]
- 108.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 109.Moore, W. E. C., and L. V. H. Moore. 1994. The bacteria of periodontal diseases. Periodontology 5:2000.66-77. [DOI] [PubMed] [Google Scholar]
- 110.Moter, A., and U. B. Göbel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 111.Nilius, A. M., S. C. Spencer, and L. G. Simonson. 1993. Stimulation of in vitro growth of Treponema denticola by extracellular growth factors produced by Porphyromonas gingivalis. J. Dent. Res. 72:1027-1031. [DOI] [PubMed] [Google Scholar]
- 112.Noiri, Y., L. Li, and S. Ebisu. 2001. The localization of periodontal-disease-associated bacteria in human periodontal pockets. J. Dent. Res. 80:1930-1934. [DOI] [PubMed] [Google Scholar]
- 113.Noiri, Y., K. Ozaki, H. Nakae, T. Matsuo, and S. Ebisu. 1997. An immunohistochemical study on the localization of Porphyromonas gingivalis, Campylobacter rectus and Actinomyces viscosus in human periodontal pockets. J. Periodontal Res. 32:598-607. [DOI] [PubMed] [Google Scholar]
- 114.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 115.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 116.Okahashi, N., C. Sasakawa, M. Yoshikawa, S. Hamada, and T. Koga. 1989. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol. Microbiol. 3:673-678. [DOI] [PubMed] [Google Scholar]
- 117.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 119.Palmer, R. J., Jr., and D. E. Calwell. 1995. A flowcell for the study of plaque removal and regrowth. J. Microbiol. Methods 24:171-182. [Google Scholar]
- 120.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Palmer, R. J., Jr., R. Wu, S. Gordon, C. G. Bloomquist, W. F. Liljemark, M. Kilian, and P. E. Kolenbrander. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337:393-403. [DOI] [PubMed] [Google Scholar]
- 122.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 123.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Petersen, F. C., S. Pasco, J. Ogier, J. P. Klein, S. Assev, and A. A. Scheie. 2001. Expression and functional properties of the Streptococcus intermedius surface protein antigen I/II. Infect. Immun. 69:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roberts, A. P., G. Cheah, D. Ready, J. Pratten, M. Wilson, and P. Mullany. 2001. Transfer of Tn916-like elements in microcosm dental plaques. Antimicrob. Agents Chemother. 45:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Roberts, A. P., J. Pratten, M. Wilson, and P. Mullany. 1999. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 177:63-66. [DOI] [PubMed] [Google Scholar]
- 128.Reference deleted.
- 129.Reference deleted.
- 130.Roelofs, W. L. 1995. Chemistry of sex attraction. Proc. Natl. Acad. Sci. USA 92:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Roelofs, W. L., and R. A. Jurenka. 1996. Biosynthetic enzymes regulating ratios of sex pheromone components in female redbanded leafroller moths. Bioorg. Med. Chem. 4:461-466. [DOI] [PubMed] [Google Scholar]
- 132.Rogers, J. D., R. J. Palmer, Jr., P. E. Kolenbrander, and F. A. Scannapieco. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 134.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 135.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 136.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontology 5:2000.7-25. [DOI] [PubMed] [Google Scholar]
- 137.Socransky, S. S., A. D. Haffajee, C. M. A., C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 138.Socransky, S. S., and S. D. Manganiello. 1971. The oral microbiota of man from birth to senility. J. Periodontol. 42:485-496. [DOI] [PubMed] [Google Scholar]
- 139.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 140.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 142.Takahashi, Y., K. Konishi, J. O. Cisar, and M. Yoshikawa. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Takahashi, Y., A. L. Sandberg, S. Ruhl, J. Muller, and J. O. Cisar. 1997. A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to alpha2-3-linked sialic acid-containing receptors. Infect. Immun. 65:5042-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tamura, H., T. Kikuchi, R. Shirato, and H. Kato. 2001. Cloning and DNA sequencing of the surface protein antigen I/II (PAa) of Streptococcus cricetus. FEMS Microbiol. Lett. 196:251-256. [DOI] [PubMed] [Google Scholar]
- 145.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 146.Tokuda, M., N. Okahashi, I. Takahashi, M. Nakai, S. Nagaoka, M. Kawagoe, and T. Koga. 1991. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infect. Immun. 59:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tompkins, G. R., M. A. Peavey, K. R. Birchmeier, and J. R. Tagg. 1997. Bacteriocin production and sensitivity among coaggregating and noncoaggregating oral streptococci. Oral Microbiol. Immunol. 12:98-105. [DOI] [PubMed] [Google Scholar]
- 148.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van der Hoeven, J. S., A. I. Toorop, and F. H. Mikx. 1978. Symbiotic relationship of Veillonella alcalescens and Streptococcus mutans in dental plaque in gnotobiotic rats. Caries Res. 12:142-147. [DOI] [PubMed] [Google Scholar]
- 150.Vroom, J. M., K. J. De Grauw, H. C. Gerritsen, D. J. Bradshaw, P. D. Marsh, G. K. Watson, J. J. Birmingham, and C. Allison. 1999. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl. Environ. Microbiol. 65:3502-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wecke, J., T. Kersten, K. Madela, A. Moter, U. B. Göbel, A. Friedmann, and J. Bernimoulin. 2000. A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol. Lett. 191:95-101. [DOI] [PubMed] [Google Scholar]
- 152.Weiss, E. I., B. Shaniztki, M. Dotan, N. Ganeshkumar, P. E. Kolenbrander, and Z. Metzger. 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol. Immunol. 15:371-377. [DOI] [PubMed] [Google Scholar]
- 153.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 154.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 155.Wilson, M. 1999. Use of constant depth film fermentor in studies of biofilms of oral bacteria. Methods Enzymol. 310:264-279. [DOI] [PubMed] [Google Scholar]
- 156.Wimpenny, J. 2000. An overview of biofilms as functional communities, p. 1-24. In D. G. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom
- 157.Wood, S. R., J. Kirkham, P. D. Marsh, R. C. Shore, B. Nattress, and C. Robinson. 2000. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J. Dent. Res. 79:21-27. [DOI] [PubMed] [Google Scholar]
- 158.Yao, E. S., R. J. Lamont, S. P. Leu, and A. Weinberg. 1996. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol. Immunol. 11:35-41. [DOI] [PubMed] [Google Scholar]
- 159.Zaura-Arite, E., J. van Marle, and J. M. ten Cate. 2001. Conofocal microscopy study of undisturbed and chlorhexidine-treated dental biofilm. J. Dent. Res. 80:1436-1440. [DOI] [PubMed] [Google Scholar]