Abstract
Purpose
Although IDH-mutant astrocytomas exhibit more favorable survival outcomes compared to their IDH-wildtype counterparts, therapeutic failure in recurrent cases persists as a significant clinical challenge. The objective of this study is to identify genes associated with recurrence in IDH-mutant astrocytoma and to elucidate their expression pattern, biological functions, and prognostic value.
Methods
RNA-sequencing data of patients with IDH-mutant astrocytoma were collected from 96 cases in the Chinese Glioma Genome Atlas (CGGA) database, 150 cases in CGGA2019 and 222 cases in The Cancer Genome Atlas (TCGA). Differentially expressed genes (DEGs) were identified using unpaired t-tests between recurrent and primary IDH-mutant astrocytoma. GO and KEGG analyses were performed to analyze these DEGs. Pearson correlation analysis was employed to assess the correlation between High mobility group AT-hook 2 (HMGA2) and genes associated with cell invasion and extracellular matrix components. Kaplan–Meier analyses and univariate and multivariate Cox regression analyses were conducted to assess the prognosis.
Results
HMGA2 was highly expressed in patients with recurrent IDH-mutant astrocytoma in comparison to those with primary IDH-mutant astrocytoma. Patients with higher HMGA2 expression are more likely to have high-grade gliomas and to be in the O6-methylguanine-DNA methyltransferase promoter (MGMTp) methylation group. Functional enrichment and correlation analyses revealed that HMGA2 is closely related to extracellular matrix content and cell migration and invasion ability. HMGA2 is an independent prognostic factor associated with poor prognosis in patients with IDH-mutant astrocytoma.
Conclusions
HMGA2 was highly expressed in recurrent IDH-mutant astrocytoma, with higher expression levels associated with increased cell migration and invasion abilities. HMGA2 has the potential to serve as a biomarker for poor prognosis and may represent an effective therapeutic target in the treatment of IDH-mutant astrocytoma.
Keywords: Astrocytoma, IDH-mutant, Recurrent tumors, HMGA2, Cell invasion
Introduction
Diffuse glioma is the most frequent primary malignant tumor of the central nervous system (CNS) in adults, with a high incidence rate, recurrence rate, and an unfavorable prognosis [18, 23]. The most recent 2021 World Health Organization (WHO) Central Nervous System (CNS) classification system categorizes adult-type diffuse gliomas into three types based on histological diagnosis and molecular markers: (1) astrocytoma, isocitrate dehydrogenase (IDH)-mutant, (2) oligodendroglioma, IDH-mutant and 1p/19q-codeleted, and (3) glioblastoma, IDH-wildtype. IDH-mutant astrocytoma, characterized by IDH1/2 mutations without 1p/19q codeletion, encompasses WHO grades 2–4 and exhibits a better prognosis compared to wild-type gliomas despite its malignant potential [7, 27]. However, the therapeutic landscape for recurrent cases remains challenging, as traditional treatments (surgery, radiotherapy, and temozolomide) often fail to achieve durable responses. Recent advances in targeted therapy have emerged: The FDA approved vorasidenib, a dual IDH1/2 inhibitor, for recurrent IDH1-mutated gliomas based on the INDIGO trial [6, 16]. This randomized Phase III study demonstrated improved progression-free survival (PFS) and time to the next anticancer intervention (TTNI) versus the placebo group, marking a significant milestone in the management of IDH-mutant gliomas. While these developments hold promise for improving outcomes, the identification of additional therapeutic targets remains critical to enhance survival benefits in these patients.
Differential expression analyses of RNA-seq datasets from the Chinese Glioma Genome Atlas (CGGA) and CCGA2019 revealed that the high mobility group AT-hook 2 (HMGA2) gene is commonly upregulated in recurrent IDH-mutant astrocytomas compared to the primary group. HMGA2, a member of the high mobility group superfamily, is a nuclear non-histone architectural transcription factor and functions by regulating the transcription of target genes. Previous research has demonstrated the importance of HMGA2 in various biological processes, including the cell cycle process, apoptosis, DNA damage and repair, angiogenesis, epithelial-mesenchymal transition, metastasis, and drug resistance [1, 21, 26]. In addition, HMGA2 is also considered a stem cell marker that is hardly detected in mature cells but can be re-expressed in most malignant tumors [13, 15]. Nevertheless, the precise function of HMGA2 in IDH-mutant astrocytomas remains uncertain, and further investigation is necessary.
In the present study, HMGA2 is found to be enriched in high-grade IDH-mutant astrocytoma and the O6-methylguanine-DNA methyltransferase promoter (MGMTp) group. Functional clustering analyses and correlation analyses revealed that HMGA2 is involved in modulating invasion ability and the extracellular matrix components. Finally, survival analyses indicated that HMGA2 is an independent prognostic factor for poor survival in patients with IDH-mutant astrocytoma. The above evidence suggested the crucial role of HMGA2 in the metastasis and recurrence of IDH-mutant astrocytoma, which indicates its potential to be a biomarker for relapse diagnosis and be applied in the IDH-mutant clinical treatment.
Methods
Samples collection and data acquisition
The mRNA sequencing data and corresponding clinical information were derived from the CGGA portal website (http://www.cgga.org.cn/, Dataset ID: mRNAseq_325 and mRNAseqg_693, gene expression unit: FPKM), and The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/, gene expression unit: FPKM). A total of 468 patients diagnosed with IDH-mutant astrocytoma according to the 2021 WHO CNS classification were collected, including 96 from CGGA, 150 from CGGA2019, and 222 from TCGA datasets.
Volcano plot
Unpaired t-tests were performed to analyze the differentially expressed genes (DEGs) between recurrent and primary IDH-mutant astrocytoma using Microsoft Excel (v2021). |log2 (fold change) | of > 1 and a false discovery rate (FDR) control (adjusted P < 0.05) were set as the cutoff. The “ggplot2” package was used to generate volcano plots to visually depict the genes exhibiting noteworthy alterations, highlighted in either red or blue.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysis
Pearson correlation analysis was conducted to select the top 500 genes most closely associated with HMGA2. These genes were subsequently subjected to GO and KEGG enrichment analysis using the DAVID database (v6.8) (https://DAVIDncifcrf.gov/). The official gene symbol was used as the identifier for Homo sapiens. The visualization was conducted using the R software.
Correlation analysis and survival analysis
The correlations between HMGA2 and cell invasion, as well as extracellular matrix components, were calculated by Pearson correlation analyses. Survival analyses were performed using Kaplan–Meier survival curves and univariate and multivariate Cox proportional hazard models, which were carried out with SPSS statistical software (version 26.0; IBM, Armonk, NY, USA). In the Kaplan–Meier analysis, patients with IDH-mutant astrocytoma were stratified into high-expression and low-expression groups using cohort-specific optimal cutoffs (CGGA: 0.03; CGGA2019: 0.02; TCGA: 1.6015). Patients with missing information were excluded from the analysis.
Statistical analysis
The statistical analysis and data visualization were conducted utilizing the R programming language (version 4.3.1, available at https://www.r-project.org/). GraphPad Prism software (version 9.5), IBM SPSS Statistics (version 26.0), and Microsoft Office 2021 were employed for data manipulation and visualization. An unpaired t-test was conducted to investigate the disparities between the two groups, considering the distribution of the data. A p-value of less than 0.05 was considered statistically significant.
Results
Significant upregulation of HMGA2 in patients with recurrent IDH-mutant astrocytoma
DEGs were identified between recurrent and primary IDH-mutant astrocytoma, and a total of 705 DEGs were screened, including 520 upregulated and 185 downregulated genes, from the CGGA database (Fig. 1A). In the CGGA2019 database, 602 DEGs were obtained and 474 genes were upregulated while 128 genes were downregulated (Fig. 1B). By intersecting the DEGs identified in the CGGA and CGGA2019 databases, we obtained 26 overlapping genes that were consistently upregulated in recurrent IDH-mutant astrocytomas (Figs. 1C and D). Among these 26 genes, HMGA2 exhibited the highest fold changes in both datasets (CGGA: fold change = 5.204, P = 0.003; CGGA2019: fold change = 6.163, P = 0.005). Notably, HMGA2 has been implicated in tumor recurrence-related processes such as cell migration, invasion, and extracellular matrix remodeling [1, 26], making it a promising candidate for further investigation.
Fig. 1.
Differentially expressed genes (DEGs) between primary and recurrent IDH-mutant astrocytoma. Volcano plots of DEGs in (A) CGGA and (B) CGGA2019 with the cut-offs of |log2 (fold change) | value of > 1 and false discovery rate (FDR) control (adjusted P < 0.05). HMGA2 was upregulated in recurrent IDH-mutant astrocytoma in both (C) CGGA and (D) CGGA2019 datasets
HMGA2 was closely related to clinicopathological characteristics in IDH-mutant astrocytoma
We next investigated the correlation between HMGA2 and clinic pathological features in IDH-mutant astrocytoma. As shown in Fig. 2A, patients with higher HMGA2 are more likely to have high-grade gliomas (WHO Grade 3 and 4) in the CGGA database. These findings were independently validated in the CGGA2019 and TCGA databases (Figs. 2B and C), indicating a link between HMGA2 expression and the malignant progression of IDH-mutant astrocytoma. Furthermore, as methylation of MGMTp is a predictor of increased temozolomide (TMZ) sensitivity [5], we analyzed the relationship between HMGA2 and MGMTp methylation. The results revealed enrichment of HMGA2 in the MGMTp methylation group in the CGGA database, whereas no relationship was observed in the CGGA2019 and TCGA datasets (Figs. 2D-F).
Fig. 2.
The association between HMGA2 expression with clinicopathological characteristics. The expression of HMGA2 in recurrent IDH-mutant astrocytoma with different WHO grades in (A) CGGA, (B) CGGA2019, and (C) TCGA databases. The expression of HMGA2 was higher in the MGMTp methylation group in the (D) CGGA dataset, but not in the (E) CGGA2019 and (F) TCGA datasets
HMGA2 is associated with extracellular matrix and cell migration
Pearson correlation analysis (| R |> 0.5, P < 0.05) was used to screen the genes most related to HMGA2 in the CGGA, CGGA2019, and TCGA databases, and the corresponding gene functions were subsequently analyzed by GO and KEGG. Regarding the biological processes (BP), genes associated with HMGA2 are mainly engaged in extracellular matrix organization, collagen fibril organization, and positive regulation of cell migration (Figs. 3A, E, I). With respect to cellular components (CC), these genes are primarily associated with extracellular matrices, extracellular regions, and extracellular spaces (Figs. 3B, F, J). For molecular functions (MF), these genes are involved in extracellular matrix structural constituent, collagen binding, and fibronectin binding (Figs. 3C, G, K). In addition, KEGG pathway analyses were conducted, which revealed pathways including extracellular matrix (ECM)-receptor interaction (hsa04512), PI3 K-Akt signaling pathway (hsa04151), and focal adhesion (hsa04510) (Figs. 3D, H, L). The aforementioned results indicate that HMGA2 may contribute to the recurrence of IDH-mutant astrocytoma by enhancing cell migration and invasion capabilities.
Fig. 3.
The potential functions of HMGA2 in IDH-mutant astrocytoma. GO analyses of the top 500 HMGA2 related genes in IDH-mutant astrocytoma, including (A, E, and I) biological processes (BP), (B, F, and J) cellular component (CC), and (C, G, and K) molecular function (MF) in CGGA, CGGA2019, and TCGA databases. KEGG analysis of HMGA2 in the (D) CGGA, (H) CGGA2019, and (L) TCGA datasets
Higher expression of HMGA2 indicates greater invasive ability in IDH-mutant astrocytoma
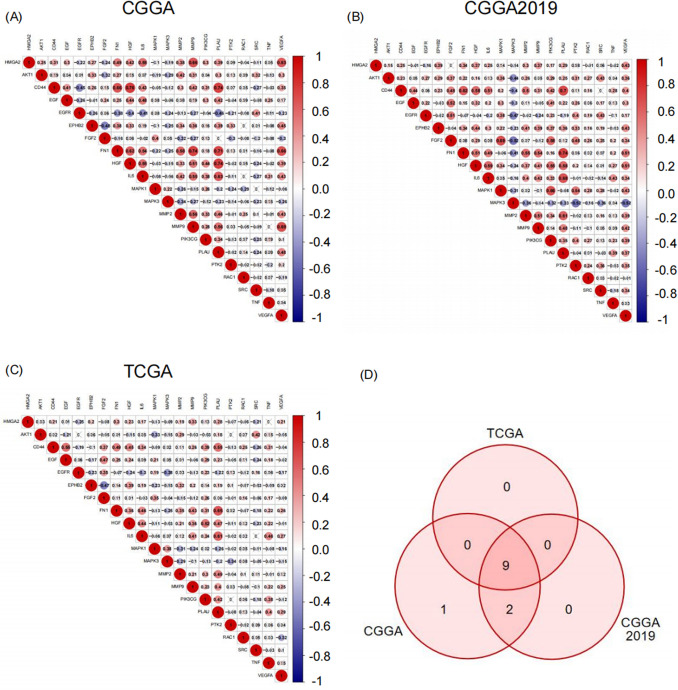
Correlation analyses further revealed that HMGA2 expression was positively correlated with most of the genes associated with cell motility and invasive potential, with the most significant correlations observed with MMP9 (Matrix Metalloproteinase-9), IL-6 (Interleukin-6), and VEGFA (Vascular Endothelial Growth Factor A) in the CGGA, CGGA2019, and TCGA databases (Figs. 4A-C). Consequently, nine common genes were identified to be positively correlated with HMGA2 in all three databases. These findings indicated a correlation between high HMGA2 expression and cell invasion in IDH-mutated astrocytoma (Fig. 4D).
Fig. 4.
The relationship between HMGA2 with cell migration and invasion abilities. Correlation matrix between HMGA2 and genes associated with cell migration and invasion in (A) CGGA, (B) CGGA2019, and (C) TCGA databases. The red circle indicates a positive correlation and the blue circle indicates a negative correlation. Grey “ × ” indicates no significant correlation. (D) Venn diagram illustrating the overlapping genes that exhibited a positive correlation with HMGA2 in all three databases
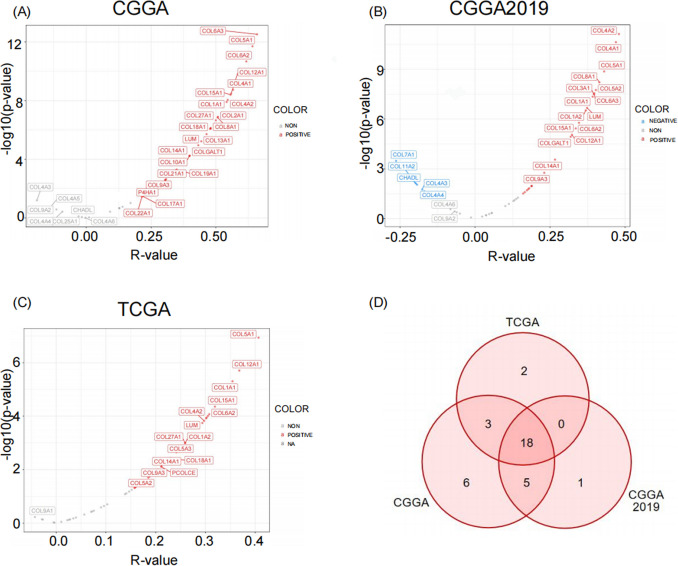
The expression of HMGA2 is closely related to the expression of extracellular matrix components
Tumor invasion is thought to begin with tumor cells adhering to the extracellular matrix. Therefore, correlation analyses were conducted, revealing a significant positive correlation between HMGA2 and the expression of most extracellular matrix components in CGGA, CGGA2019, and TCGA (Figs. 5A-C). This suggests that HMGA2 may have a role in cell invasiveness in IDH-mutant astrocytoma through the regulation of the extracellular matrix.
Fig. 5.
Correlation analysis between HMGA2 and extracellular matrix. Pearson correlation analyses between HMGA2 and genes associated with extracellular matrix in (A) CGGA, (B) CGGA2019, and (C)TCGA databases. Blue indicates a significant negative correlation, red indicates a significant positive correlation, and gray indicates no significant correlation. (D) Venn diagram illustrating the common genes that were correlated with HMGA2 in the CGGA, CGGA2019, and TCGA databases
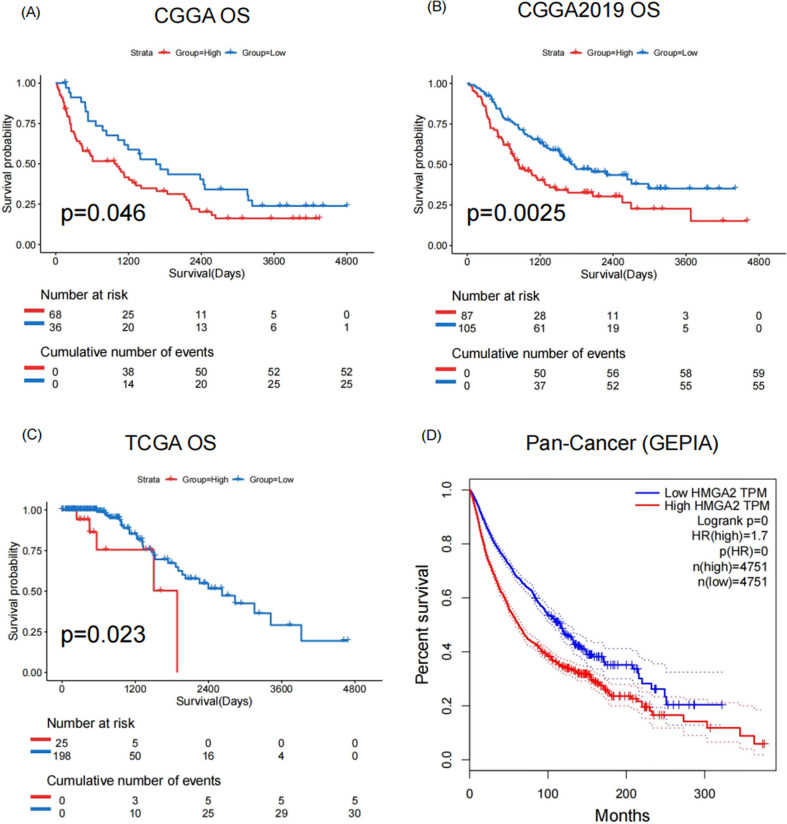
HMGA2 is an independent adverse prognostic factor for overall survival (OS) in patients with IDH-mutant astrocytoma.
The Kaplan–Meier curve shows that patients with higher expression of HMGA2 had a shorter OS in IDH-mutant astrocytoma in CGGA, CGGA2019, and TCGA datasets (Figs. 6A-C). This finding was also observed in the HMGA2 high-expression group in the pan-cancer survival analysis based on the GEPIA (Gene Expression Profiling Interactive Analysis) database (Fig. 6D). Furthermore, the univariate Cox analyses of HMGA2, age, gender, MGMTp status, and WHO grade, as well as the subsequent multivariate Cox regression, confirmed that HMGA2 and WHO grade were independent prognostic factors for OS in all three databases (Tables 1, 2, 3). These results suggested that HMGA2 may be a promising independent prognostic predictor in IDH-mutant astrocytoma.
Fig. 6.
Prognosis analysis of HMGA2 in IDH-mutant astrocytoma. (A) Kaplan–Meier survival curve for overall survival (OS) in patients with high and low HMGA2 expression in (A) CGGA, (B) CGGA2019 and (C) TCGA databases, which stratified by optimal cutoff value (CGGA: 0.03; CGGA2019: 0.02; TCGA: 1.6015). (D) The pan-cancer survival analysis of HMGA2 in the GEPIA database
Table 1.
Univariate and multivariate analysis of prognostic parameters in CGGA(OS)
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| HMGA2 |
1.821 (1.300–2.552) |
< 0.0001 |
1.457 (1.033–2.054) |
0.032 |
| Age at Diagnosis |
1.001 (0.974–1.029) |
0.919 | ||
| Gender |
0.766 (0.489–1.199) |
0.243 | ||
| MGMTp Status |
1.401 (0.834–2.354) |
0.203 | ||
| WHO Grade |
2.251 (1.716–2.952) |
< 0.0001 |
2.183 (1.654–2.880) |
< 0.0001 |
Table 2.
Univariate and multivariate analysis of prognostic parameters in CGGA2019(OS)
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| HMGA2 |
1.753 (1.217–2.537) |
0.003 |
1.541 (1.057–2.247) |
0.025 |
| Age at Diagnosis |
1.003 (0.982–1.023) |
0.810 | ||
| Gender |
1.002 (0.680–1.477) |
0.990 | ||
| MGMTp Status |
0.918 (0.607–1.388) |
0.685 | ||
| WHO Grade |
2.632 (1.985–3.490) |
< 0.0001 |
2.505 (1.892–3.318) |
< 0.0001 |
Table 3.
Univariate and multivariate analysis of prognostic parameters in TCGA (OS)
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| HMGA2 |
1.682 (1.215–2.329) |
0.002 |
1.637 (1.167–2.296) |
0.004 |
| Age at Diagnosis |
1.024 (0.994–1.055) |
0.121 | ||
| Gender |
0.853 (0.436–1.670) |
0.643 | ||
| MGMTp Status |
1.107 (0.385–3.180) |
0.851 | ||
| WHO Grade |
1.909 (1.081–3.369) |
0.026 |
1.730 (0.997–3.001) |
0.051 |
Discussion
Following the latest 2021 WHO CNS classification system, IDH status has become one of the fundamental factors associated with survival outcomes in glioma [25]. Accordingly, adult diffuse gliomas can be assigned into three types: astrocytoma, IDH-mutant; Oligodendroglioma, IDH-mutant and 1p/19q-deficient; and Glioblastoma, IDH-wildtype. Due to the general epigenetic silencing of the glycolysis pathway, IDH-mutant gliomas grow more slowly than their wild-type counterpart and therefore predict a relatively favorable prognosis. However, it is commonly observed that IDH-mutant astrocytoma may experience a recurrence, leading to the infiltrating of normal brain tissue and becoming an incurable malignancy if untreated [12, 19]. Consequently, the identification of efficacious therapeutic targets for recurrent IDH-mutant astrocytoma is of paramount importance for the prolongation of survival and the improvement of quality of life in patients.
HMGA2 has recently been identified as a critical factor implicated in the process of tumor metastasis in various malignancies, including rectal [17], ovarian [24], and breast cancers [11]. This study further demonstrated the close relationship between HMGA2 and the recurrence of IDH-mutant astrocytoma by utilizing public datasets comprising extensive clinical samples and employing molecular pathology analyses. The results suggests that HMGA2 may be a potential therapeutic target in the recurrence of IDH-mutant astrocytoma. In this study, we first analyzed the DEGs between recurrent and primary IDH-mutant astrocytoma in CGGA and CGGA2019 databases and found significant upregulation of HMGA2 in both datasets. Then, in line with the previous study [3], increased expression of HMGA2 was found to be correlated with a higher tumor grade, which further corroborates the involvement of HMGA2 in the recurrence of IDH-mutant astrocytoma. Furthermore, the upregulation of HMGA2 was found to be associated with MGMTp methylation status in our research. As TMZ sensitivity is closely linked to the methylation status of the MGMT promoter [3], it can be postulated that HMGA2 may be a predictive factor for TMZ therapy in IDH-mutant astrocytoma.
Previous studies have demonstrated that HMGA2 leads to tumor progression through various mechanisms. Firstly, HMGA2 affects the expression of genes involved in cell proliferation, cell cycle, and survival [2, 10, 11]. Secondly, HMGA2 can inhibit the expression of B-cell lymphoma-2 (Bcl-2) to inhibit cell apoptosis and promote tumor proliferation [4, 8, 9, 14]. Thirdly, HMGA2 regulates the DNA repair process by influencing DNA repair proteins, thereby enhancing tumor cell survival. Other mechanisms include involvement in the phenotype of cancer stem cells, promotion of vascularization during cancer development, increased epithelial-to-mesenchymal transition (EMT), and metastasis, and so on [20, 22]. In the present study, we observed a positive correlation between HMGA2 expression and genes associated with cell invasion and the extracellular matrix, suggesting that HMGA2 may contribute to the recurrence of IDH-mutant astrocytoma by influencing the extracellular matrix, cell migration, and invasion. Furthermore, patients with high expression of HMGA2 exhibited a significantly shorter OS period, indicating that HMGA2 may have both predictive and prognostic value in IDH-mutant astrocytoma.
Nevertheless, this study is constrained by several limitations. First, the analyses were based on public datasets, and further experiments, including cell line studies and animal experiments, will be required to verify the results. Second, the absence of an established pharmaceutical agent for the selective targeting of HMGA2 necessitates further research efforts for the development of a clinical treatment.
In conclusion, a bioinformatics analysis was conducted to investigate the role and impact of HMGA2 in IDH-mutant astrocytoma. The findings contribute to the understanding of the mechanisms underlying the recurrence of IDH-mutant astrocytoma and identify HMGA2 as a potential therapeutic target for IDH-mutant astrocytoma.
Abbreviations
- CNS
Central Nervous System
- WHO
World Health Organization
- HMGA2
High mobility group AT-hook 2
- MGMT
O6-methylguanine-DNA methyltransferase
- IDH
Isocitrate Dehydrogenases
- PFS
Progression-free survival
- TTNI
Time to the next anticancer intervention
- CGGA
Chinese Glioma Genome Atlas
- TCGA
The Cancer Genome Atlas
- GO
Gene Ontology
- BP
Biological Processes
- CC
Cellular Components
- MF
Molecular Functions
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DEGs
Differentially expressed genes
- ECM
Extracellular Matrix
- MMP9
Matrix Metalloproteinase-9
- IL-6
Interleukin-6
- VEGFA
Vascular Endothelial Growth Factor A
- GEPIA
GeneExpression Profiling Interactive Analysis
- TMZ
Temozolomide
- Bcl-2
B-cell lymphoma-2
- EMT
Epithelial-to-mesenchymal transition
Authors’ contributions
ZG and LYW are responsible for the study design. LYW, SYL and ZG are responsible for data extraction and analysis. LYW and ZYL are responsible for writing the manuscript. ZG is responsible for the revision of the draft. All authors approved the final draft.
Funding
This study was supported by The Science & Technolngy Development Fund of Tianjin Education Commission for Higher Education [Grant number: 2022 KJ147].
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga, http://www.cgga.org.cn/
Declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Tianjin University of Traditional Chinese Medicine. The study was conducted according to the principles of the Declaration of Helsinki.
Patient consent for publication
Patient consent for publication was not required for this study.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Luying Wan and Ziyi Liu contributed equally to this work.
References
- 1.Esmailzadeh S et al (2017) siRNA-Mediated silencing of HMGA2 induces apoptosis and cell cycle arrest in human colorectal carcinoma. J Gastrointest Cancer 48(2):156–163 [DOI] [PubMed] [Google Scholar]
- 2.Gao X et al (2017) HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer 8(5):501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horbinski C et al (2022) Clinical implications of the 2021 edition of the WHO classification of central nervous system tumours. Nat Rev Neurol 18(9): 515–529 [DOI] [PubMed]
- 4.Kaur H et al (2015) The chromatin-modifying protein HMGA2 promotes atypical teratoid/rhabdoid cell tumorigenicity. J Neuropathol Exp Neurol 74(2):177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb YN (2024) Vorasidenib: First approval. Drugs 84(10):1325–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z et al (2022) HMGA2-Snai2 axis regulates tumorigenicity and stemness of head and neck squamous cell carcinoma. Exp Cell Res 418(1):113271 [DOI] [PubMed] [Google Scholar]
- 7.Liu H et al (2014) RKIP inhibits gastric cancer cell survival and invasion by regulating the expression of HMGA2 and OPN. Tumour Biol 35(12):11949–11958 [DOI] [PubMed] [Google Scholar]
- 8.Louis DN et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malek A et al (2008) HMGA2 gene is a promising target for ovarian cancer silencing therapy. Int J Cancer 123(2):348–356 [DOI] [PubMed] [Google Scholar]
- 10.Mansouri A et al (2019) MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol 21(2):167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansoori B et al (2016) HMGI-C suppressing induces P53/caspase9 axis to regulate apoptosis in breast adenocarcinoma cells. Cell Cycle 15(19):2585–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansoori B et al (2021) HMGA2 as a critical regulator in cancer development. Genes (Basel) 12(2) [DOI] [PMC free article] [PubMed]
- 13.Mansoori B et al (2021) HMGA2 supports cancer hallmarks in triple-negative breast cancer. Cancers (Basel) 13(20) [DOI] [PMC free article] [PubMed]
- 14.Mansoori B et al (2020) Overexpression of HMGA2 in breast cancer promotes cell proliferation, migration, invasion and stemness. Expert Opin Ther Targets 1–11 [DOI] [PubMed]
- 15.Mellinghoff IK et al (2023) Vorasidenib in IDH1- or IDH2-Mutant low-grade glioma. N Engl J Med 389(7):589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JJ et al (2023) Isocitrate dehydrogenase (IDH) mutant gliomas: A Society for Neuro-Oncology (SNO) consensus review on diagnosis, management, and future directions. Neuro Oncol 25(1):4–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu Y et al (2019) miR-16 regulates proliferation and apoptosis of pituitary adenoma cells by inhibiting HMGA2. Oncol Lett 17(2):2491–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrom QT et al (2022) CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro Oncol 24(Suppl 5):v1–v95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pentimalli F et al (2018) Retraction: Suppression of HMGA2 protein synthesis could be a tool for the therapy of well differentiated Liposarcomas overexpressing HMGA2. Cancer Res 78(24):6909 [DOI] [PubMed] [Google Scholar]
- 20.Rautajoki KJ et al (2023) Genomic characterization of IDH-mutant astrocytoma progression to grade 4 in the treatment setting. Acta Neuropathol Commun 11(1):176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X et al (2015) A novel anti-proliferative role of HMGA2 in induction of apoptosis through caspase 2 in primary human fibroblast cells. Biosci Rep 35(1) [DOI] [PMC free article] [PubMed]
- 22.Tan N et al (2019) Reactive oxygen species metabolism-based prediction model and drug for patients with recurrent glioblastoma. Aging (Albany NY) 11(23):11010–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X et al (2011) Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res 17(8):2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weller M et al (2021) EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 18(3):170–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J et al (2016) Elevated HMGA2 expression is associated with cancer aggressiveness and predicts poor outcome in breast cancer. Cancer Lett 376(2):284–292 [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Wei JJ (2013) HMGA2 and high-grade serous ovarian carcinoma. J Mol Med (Berl) 91(10):1155–1165 [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Mo Q, Wang X (2019) Oncological role of HMGA2 (Review). Int J Oncol 55(4):775–788 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga, http://www.cgga.org.cn/