Abstract
Background
A common sequence polymorphism at codon 72 of the p53 gene encoding either arginine or proline was recently shown to be functionally relevant for apoptosis induction in vitro. In B-type chronic lymphocytic leukemia (B-CLL), p53 gene mutations occur in a subset of patients and are associated with impaired survival and drug resistance. Here, we address the functional relevance of the codon 72 single nucleotide (SNP) polymorphism for cell death sensitivity following exposure to clinically employed cytotoxic drugs and γ-irradiation.
Methods
138 B-CLL samples were analysed by SSCP-PCR and sequencing for single nucleotide polymorphism at codon 72 of the p53 gene. The in vitro cytotoxicity assay (DiSC-assay) was performed with 7 drugs (chlorambucil, mafosfamide, fludarabine phosphate, methylprednisolone, doxorubicin, vincristine) or γ-irradiation.
Results
Of the138 B-CLL samples, 9 samples were homozygous for proline (Pro/Pro), 78 samples homozygous for arginine (Arg/Arg), and 49 samples heterozygous (Arg/Pro). No differences were found for patient survival and cell death triggered by 7 cytotoxic drugs or γ-irradiation.
Conclusion
These data indicate that polymorphic variants of p53 codon 72 are not clinically relevant for apoptosis induction or patient survival in B-CLL.
Background
The tumor suppressor gene p53 plays a central role in the induction of cell cycle arrest, senescence and apoptosis [1-5]. The polyproline domain (PP domain) of p53 spanning amino acids 62–91 is involved in apoptosis induction and facilitates transactivation of pro-apoptotic genes by p53 [6].
Located in this PP domain is at codon 72 a common single nucleotide polymorphism (SNP), resulting in either a proline residue (p53Pro) or an arginine residue (p53Arg). Thus, each individual inherits a p53 genotype that can be heterozygous (Arg/Pro) or homozygous for either arginine (Arg/Arg) or proline (Pro/Pro). The polymorphism is balanced, varies with latitude and race, and is maintained at different allelic frequencies across the population [7]. These two SNPs appear to be different both biochemically and biologically [8-11]. Differences in apoptosis susceptibility to cytotoxic drugs were described [12,13], and the response and survival to radiochemotherapy in clinical samples of squamous cell carcinomas was found to be increased in case the arginine allele is retained [13].
Chronic lymphocytic B-cell lymphoma is still an incurable disease and may be addressed as a disease of intrinsic apoptosis deficiency. It is a disease where the mutational status of the p53 gene is linked to patient survival [14] (and references herein). We therefore asked whether the codon 72 polymorphism is of clinical relevance for in vitro resistance to cytotoxic drugs, γ-irradiation and patient prognosis.
Methods
Patients
Samples from 138 B-CLL patients (99 male, 39 female, age 63.2 ± 0.5 (mean ± SEM) were analysed. Peripheral blood was analysed for drug sensitivities using fresh cells. The same samples were analysed for mutations in the p53 DNA binding domain and of the p53 codon 72 SNP, using snap frozen cells from the same specimens. Only patients with high peripheral blood leukocyte count were included in this analysis (median WBC 120.8/nl, range 20.7–1262.2/nl). The Binet stage was A in 29 cases, B in 24 cases, A/B in 15 cases (due to insufficient information on clinical lymph node status) and C in 62. Of the 138 patients, 80 were pretreated (58%) with one to six drug regimens (mean number of pretreatments for these 80 patients ± SEM: 1.93 ± 1.005). Median survival was 30.1 months; median follow up for the 17 censored patients was 97.9 months. This study was performed in accordance with local ethical standards and the declaration of Helsinki.
Analysis of p53 codon 72 polymorphism
SNP in the p53 codon 72 were analysed by genomic SSCP-PCR analysis and DNA sequencing. The primers were CGG ACG ATA TTG AAC AAT GG (sense) and CGT TTT CTG GGA AGG GAG AG (antisense), resulting in a PCR product of 167 bp. A standard PCR reaction in 50 μl with a final concentration of 0.6 mM MgCl2 was performed (40 cycles, annealing temperature 56°C). PCR products were denatured and separated on a nondenaturating 10% polyacrylamide gel at 500 V and 50 mA for 2 h at 22°C and analysed by silver staining [15,16]. Sequence polymorphism was confirmed by DNA sequencing, as described [14]. A result was obtained in 136 samples (98.6%). Mutations in the DNA-binding domain of the p53 gene (exon 5 to 8) has been analyzed previously by the use of SSCP-PCR and sequencing as described in detail elsewhere [14].
Chemosensitivity assay
B-CLL cells were exposed to cytotoxic drugs (chlorambucil, mafosfamide, fludarabine phosphate, methylprednisolone, doxorubicin, vincristine) or γ-irradiation (2 Gy) and cultured for 92 h. Percentages of drug-induced cell death and LD90 doses were determined by the use of a standardized morphometric test, the DiSC assay [17,18].
Data analysis
For intervariable assessment, the non-parametric Mann-Whitney U-test (for 2 groups) or the Kruskal-Wallis test (for 3 groups) or the χ2-test or Fisher's exact test for categorical parameters were applied. Overall survival was estimated by the Kaplan-Meier product-limit method. For the chemosensitivity data, LC90 doses were determined by calculating the log dose at which the fitted survival probability was equal to 0.1. LC90 values were logged (base 10) before calculation of mean and SEM, as described [19].
Results
Codon 72 SNP and clinical data
Of the 138 samples analysed, 136 could be amplified by genomic PCR : 78 samples were Arg/Arg, 9 samples were Pro/Pro, and 49 samples were Arg/Pro. Patients age and gender was not different in the subgroups (63.3 ± 1.07 years; male/female 55/23 in Arg/Arg, 65.2 ± 2.7 years, male/female 6/3 in Pro/Pro, 63.2 ± 1.5; male/female 37/12 in Pro/Arg). In the Binet's stage, the distribution was: stage A: 18 Arg/Arg, 1 Pro/Pro, 10 Pro/Arg; stage B: 10 Arg/Arg, 1 Pro/Pro, 13 Pro/Arg; stage C: 37 Arg/Arg, 6 Pro/Pro, 18 Pro/Arg; stage A or B: 9 Arg/Arg, 1 Pro/Pro, 5 Pro/Arg (p = 0.6).
78 patients were pretreated with a median of 2 treatment regimens. There was, however, no association between pretreatment status and codon 72 SNP (p = 0.6).
Codon 72 SNP and IgVH and p53 gene mutational analysis
In 113 samples, the IgVH-gene status was known, and in all 136 cases the p53 genotype was known [14]. Concerning the IgVH-gene status, of the hypermutated samples, 19 were Arg/Arg, 0 Pro/Pro, 10 Pro/Arg and of the pre-germinal center samples, 50 were Arg/Arg, 6 Pro/Pro, 28 Pro/Arg (p = 0.3). Concerning the p53 genotype of the B-CLL samples, of the 22 p53-mutated samples, 17 were Arg/Arg, 2 Pro/Pro, 3 Pro/Arg and of the 114 p53-wildtype samples, 61 were Arg/Arg, 7 Pro/Pro, 46 Pro/Arg (p = 0.06).
Codon 72 SNP and survival
There was no difference in overall survival for the 3 different groups (p = 0.885) (Fig. 1B). This was also true when only the p53 wild type samples were analysed (p = 0.46).
Figure 1.
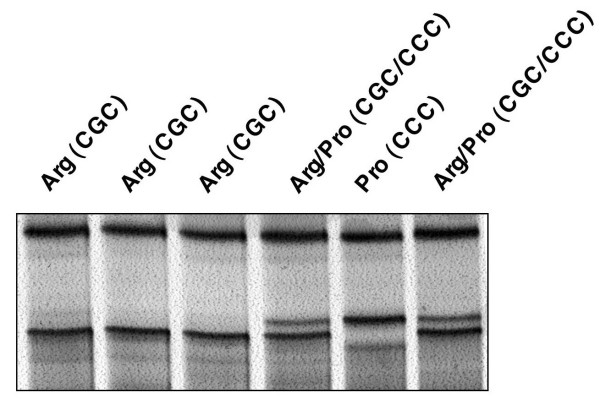
Analysis of p53 codon 72 SNP in B-CLL. Silver-stained polyacrylamide gel of 6 B-CLL samples analysed by SSCP-PCR for the codon 72 SNP. Sequencing of the 138 B-CLL samples confirmed the polymorphisms in p53 gene, codon 72 (exon 4): 6.6% of the samples were homozygous for proline (Pro/Pro), 57.4% were homozygous for arginine (Arg/Arg), and 36% were heterozygous with both an arginine and a proline allele (Arg/Pro).
Codon 72 SNP and cell death sensitivity
For none of the tested drugs (chlorambucil, mafosfamide, doxorubicine, vincristine, fludarabine, caldribine and methylprednisolone), a correlation to the p53 codon 72 SNP was seen. This was the case irrespective of the p53 mutational status (Table 1). For γ-irradiation, there was a trend (p = 0.099) for better survival of the B-CLL cells when the sample is homozygous for proline. This finding is in accordance with a recent report on response rates and survival of head and neck carcinoma patients treated with radiochemotherapy where patients with a homozygous proline genotype had an impaired outcome to treatment [13]. This association of the Pro/Pro genotype failed, however, to attain statistical significance. This, together with the lack of correlation with patient survival indicates a rather limited clinical relevance of such codon 72 polymorphisms.
Table 1.
Resistance to ionising irradiation and cytotoxic drugs in relation to the p53 codon 72 status
| all patients | p53 wild type | |||||||
| p53 codon 72 Arg/Arg | p53 codon 72 Pro/Pro | p53 codon 72 Arg/Pro | p | p53 codon 72 Arg/Arg | p53 codon 72 Pro/Pro | p53 codon 72 Arg/Pro | p | |
| γ-irradiation (% surviving cells) |
36.3 ± 4.7 (n = 49) |
50.0 ± 10.1 (n = 7) |
24.2 ± 4.8 (n = 26) |
0.099 | 30.6 ± 4.8 (n = 39) |
54.7 ± 13.4 (n = 5) |
22.9 ± 4.9 (n = 25) |
0.14 |
| log10 LC90 chlorambucil | 0.73 ± 0.07 (n = 78) |
0.76 ± 0.19 (n = 9) |
0.62 ± 0.08 (n = 49) |
0.63 | 0.6 ± 0.08 (n = 61) |
0.76 ± 0.22 (n = 7) |
0.57 ± 0.08 (n = 46) |
0.70 |
| log10 LC90 mafosfamide | 0.32 ± 0.05 (n = 75) |
0.37 ± 0.12 (n = 9) |
0.21 ± 0.05 (n = 47) |
0.39 | 0.3 ± 0.06 (n = 59) |
0.38 ± 0.16 (n = 7) |
0.21 ± 0.05 (n = 44) |
0.42 |
| log10 LC90 fludarabine | 0.18 ± 0.08 (n = 78) |
0.13 ± 0.18 (n = 9) |
-0.32 ± 0.1 (n = 49) |
0.32 | 0.11 ± 0.1 (n = 61) |
0.22 ± 0.23 (n = 7) |
-0.06 ± 0.01 (n = 46) |
0.28 |
| log10 LC90 cladribine | -0.76 ± 0.1 (n = 78) |
-0.85 ± 0.18 (n = 9) |
-1.0 ± 0.11 (n = 49) |
0.10 | -0.89 ± 0.1 (n = 61) |
-0.79 ± 0.22 (n = 7) |
-1.04 ± 0.11 (n = 46) |
0.22 |
| log10 LC90 vincristine | 0.13 ± 0.1 (n = 55) |
0.01 ± 0.27 (n = 8) |
-0.22 ± 0.11 (n = 39) |
0.71 | -0.14 ± 0.11 (n = 40) |
0.05 ± 0.33 (n = 6) |
-0.25 ± 0.11 (n = 36) |
0.59 |
| log10 LC90 doxorubicin | -0.55 ± 0.04 (n = 78) |
-0.52 ± 0.06 (n = 9) |
-0.61 ± 0.05 (n = 49) |
0.64 | -0.55 ± 0.05 (n = 61) |
-0.49 ± 0.07 (n = 7) |
-0.63 ± 0.05 (n = 46) |
0.35 |
| log10 LC90 methylprednisolone | 0.99 ± 0.14 (n = 78) |
1.36 ± 0.45 (n = 9) |
0.95 ± 0.18 (n = 48) |
0.70 | 0.99 ± 0.16 (n = 61) |
1.52 ± 0.47 (n = 7) |
0.95 ± 0.18 (n = 45) |
0.57 |
For the cytostatic drugs (chlorambucil, mafosfamide, fludarabine, cladribine, vincristine, doxorubicin or methylprednisolone) log10 LC90 concentrations were compared. In case of ionizing γ-irradiation with a fixed dose of 2 Gy, percentages of surviving cells were compared. p53 WT: wild type p53 gene. Mean +/- SEM is given. Statistical significance was calculated by means of the Kruskal-Wallis test.
Discussion
Recently, a sequence polymorphism at codon 72 the p53 gene (exon 4) encoding either arginine (CGC) or proline (CCC) was suggested to result in a drastically altered biological and biochemical behaviour of p53 in vitro. Compared to the proline encoding allele, the arginine allele appeared to trigger a more pronounced apoptosis response, whereas the proline allele induced significantly more G1 arrest [8,10,13].
The question of clinical relevance was addressed in a few case control studies. There, the proline allele was associated with urothelial [20], thyroid [21,22], and colorectal carcinomas [23] and chronic myeloid leukemia [24], whereas homozygosity for arginine was associated with advanced lung cancer [25]. Concerning patient outcome, in Italian breast cancer patients, the retention of an arginine allele was correlated with a reduction of survival in one study [26]. Another study addressed the clinical relevance of this SNP for treatment response in solid tumors. There, the homozygous proline genotype was correlated with impaired response to radiochemotherapy and reduced survival in head and neck carcinoma [13]. Furthermore, p53 protein encoded by the arginine allele appears to be more susceptible to HPV-E6 protein-induced degradation [27]. Although many studies investigated the relationship of the allelic distribution and susceptibility to HPV-associated cervical carcinoma, two recent meta-analyses could not establish such a correlation with the p53 gene codon 72 SNP [28,29].
In B-CLL, the p53 gene is known to be of clinical relevance concerning survival and treatment response [14]. We therefore investigated the potential clinical relevance of the p53 gene codon 72 SNP in a cohort of 138 samples from patients with B-CLL with respect to survival and drug sensitivity.
The distribution of the proline and arginine alleles in the B-CLL samples was, however, not different of frequency distributions observed in the general population [7]. Moreover, we found no correlation between codon 72 p53 SNP and p53 Exon 5–8 mutations or the IgVH hypermutation status or clinical Binet stage. Likewise, survival was not different in the p53 codon 72 SNP subgroups. Furthermore, no correlation with in vitro drug sensitivities was seen. The trend for reduced sensitivity to irradiation in case of homozygosity for proline is in accordance with a previous report in head and neck cancer [7], but fails to reach statistical significance. These data indicate that the codon 72 SNPs of p53 have per se no clinical relevance, at least in B-CLL. This may indicate either that B-CLL differs significantly from other tumors with regard to the regulation of cell death induced by cytotoxic anticancer therapies or that positive reports from other studies in clinical samples are false positive, eg. due to small sample size.
Conclusion
Although p53 gene mutations in the DNA-binding domain are clinically relevant for patients with B-CLL, the p53 gene codon 72 SNP was not found to be of clinical relevance in patients with B-CLL, neither for patient survival nor for ex vivo sensitivity for cytotoxic drugs and γ-irradiation.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
IS compiled the study, carried out the p53 genetic analysis, performed the statistical analysis and drafted the manuscript. AB carried out the cell death assays and provided the clinical samples and data, MH analysed the IgH mutational status, BD participated in the design of the study, PTD participated in the design of the study and drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Figure 2.
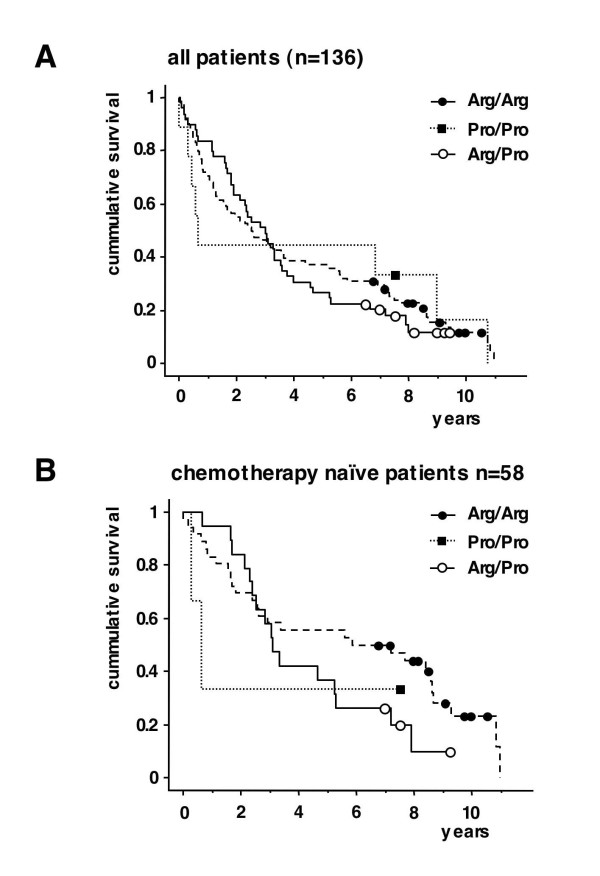
Kaplan-Meier-survival estimates for overall survival for B-CLL patients with an Arg/Arg, Pro/Pro and Arg/Pro p53 codon 72 genotype. A: All patients. Squares and circles indicate censored patients. Log rang test p = 0.9 B: Subgroup of chemotherapy naïve patients ("no pretreatment") (n = 58). Squares and circles indicate censored patients. Log rang test p = 0.3.
Acknowledgments
Acknowledgements
We thank Sylvia Scheele, Alison Burlton, Philip Bell, and David Head for expert technical assistance. The following drug companies kindly supplied drugs: Asta (mafosfamide), Schering (fludarabine). This work was funded in part by the following grants: "Schwerpunktprogramm Apoptose" by the Deutsche Krebshilfe and by the Deutsche Forschungsgemeischaft (grants to IS and PTD).
Contributor Information
Isrid Sturm, Email: isrid.sturm@charite.de.
Andrew G Bosanquet, Email: agb@caltri.org.
Michael Hummel, Email: michael.hummel@charite.de.
Bernd Dörken, Email: bernd.dorken@charite.de.
Peter T Daniel, Email: pdaniel@mdc-berlin.de.
References
- Hemmati PG, Gillissen B, von Haefen C, Wendt J, Starck L, Güner D, Dörken B, Daniel PT. Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene. 2002;21:3149–3161. doi: 10.1038/sj.onc.1205458. [DOI] [PubMed] [Google Scholar]
- Hemmati PG, Normand G, Verdoodt B, von Haefen C, Hasenjager A, Güner D, Wendt J, Dörken B, Daniel PT. Loss of p21 disrupts p14ARF-induced G1 cell cycle arrest but augments p14ARF-induced apoptosis in human carcinoma cells. Oncogene. 2005;24:4114–4128. doi: 10.1038/sj.onc.1208579. [DOI] [PubMed] [Google Scholar]
- Mrozek A, Petrowsky H, Sturm I, Kraus J, Hermann S, Hauptmann S, Lorenz M, Dörken B, Daniel PT. Combined p53/Bax mutation results in extremely poor prognosis in gastric carcinoma with low microsatellite instability. Cell Death Differ. 2003;10:461–467. doi: 10.1038/sj.cdd.4401193. [DOI] [PubMed] [Google Scholar]
- Normand G, Hemmati PG, Verdoodt B, von Haefen C, Wendt J, Güner D, May E, Dörken B, Daniel PT. p14ARF induces G2 cell cycle arrest in p53- and p21-deficient cells by down-regulating p34cdc2 kinase activity. J Biol Chem. 2005;280:7118–7130. doi: 10.1074/jbc.M412330200. [DOI] [PubMed] [Google Scholar]
- Rau B, Sturm I, Lage H, Berger S, Schneider U, Hauptmann S, Wust P, Riess H, Schlag PM, Dörken B, Daniel PT. Dynamic expression profile of p21WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003;21:3391–3401. doi: 10.1200/JCO.2003.07.077. [DOI] [PubMed] [Google Scholar]
- Baptiste N, Friedlander P, Chen X, Prives C. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene. 2002;21:9–21. doi: 10.1038/sj.onc.1205015. [DOI] [PubMed] [Google Scholar]
- Sjalander A, Birgander R, Kivela A, Beckman G. p53 polymorphisms and haplotypes in different ethnic groups. Hum Hered. 1995;45:144–149. doi: 10.1159/000154275. [DOI] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O´Nicons J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, Vousden KH, Allday MJ, Gusterson B, Ikawa S, Hinds PW, Crook T, Kaelin WGJ. A common polymorphism acts as an intragenic modifier of mutant p53 behavior. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Salvioli S, Barbi C, Mishto M, Trapassi C, Gemelli C, Storci G, Olivieri F, Monti D, Franceschi C. p53 codon 72 genotype affects apoptosis by cytosine arabinoside in blood leukocytes. Biochem Biophys Res Commun. 2002;299:539–541. doi: 10.1016/S0006-291X(02)02691-8. [DOI] [PubMed] [Google Scholar]
- Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X, Crook T. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–3337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- Sturm I, Bosanquet AG, Hermann S, Güner D, Dörken B, Daniel PT. Mutation of p53 and consecutive selective drug resistance in B-CLL occurs as a consequence of prior DNA-damaging chemotherapy. Cell Death Differ. 2003;10:477–484. doi: 10.1038/sj.cdd.4401194. [DOI] [PubMed] [Google Scholar]
- Sturm I, Köhne CH, Wolff G, Petrowsky H, Hillebrand T, Hauptmann S, Lorenz M, Dörken B, Daniel PT. Analysis of the p53/BAX pathway in colorectal cancer: low BAX is a negative prognostic factor in patients with resected liver metastases. J Clin Oncol. 1999;17:1364–1374. doi: 10.1200/JCO.1999.17.5.1364. [DOI] [PubMed] [Google Scholar]
- Sturm I, Petrowsky H, Volz R, Lorenz M, Radetzki S, Hillebrand T, Wolff G, Hauptmann S, Dörken B, Daniel PT. Analysis of p53/BAX/p16(ink4a/CDKN2) in esophageal squamous cell carcinoma: high BAX and p16(ink4a/CDKN2) identifies patients with good prognosis. J Clin Oncol. 2001;19:2272–2281. doi: 10.1200/JCO.2001.19.8.2272. [DOI] [PubMed] [Google Scholar]
- Bosanquet AG, Bell PB. Enhanced ex vivo drug sensitivity testing of chronic lymphocytic leukaemia using refined DiSC assay methodology. Leuk Res. 1996;20:143–153. doi: 10.1016/0145-2126(95)00127-1. [DOI] [PubMed] [Google Scholar]
- Bosanquet AG, Bell PB. Novel ex vivo analysis of nonclassical, pleiotropic drug resistance and collateral sensitivity induced by therapy provides a rationale for treatment strategies in chronic lymphocytic leukemia. Blood. 1996;87:1962–1971. [PubMed] [Google Scholar]
- Bosanquet AG, Bosanquet MI. Ex vivo assessment of drug response by differential staining cytotoxicity (DiSC) assay suggests a biological basis for equality of chemotherapy irrespective of age for patients with chronic lymphocytic leukaemia. Leukemia. 2000;14:712–715. doi: 10.1038/sj.leu.2401727. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Tsukino H, Nakao H, Imai H, Katoh T. p53 Codon 72 polymorphism and urothelial cancer risk. Cancer Lett. 2003;189:77–83. doi: 10.1016/S0304-3835(02)00518-9. [DOI] [PubMed] [Google Scholar]
- Granja F, Morari J, Morari EC, Correa LA, Assumpcao LV, Ward LS. Proline homozygosity in codon 72 of p53 is a factor of susceptibility for thyroid cancer. Cancer Lett. 2004;210:151–157. doi: 10.1016/j.canlet.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Boltze C, Roessner A, Landt O, Szibor R, Peters B, Schneider-Stock R. Homozygous proline at codon 72 of p53 as a potential risk factor favoring the development of undifferentiated thyroid carcinoma. Int J Oncol. 2002;21:1151–1154. [PubMed] [Google Scholar]
- Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutierrez-Enriquez S, Hall J, Guino E, Peinado MA, Capella G, Canzian F. A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene. 2004;23:1954–1956. doi: 10.1038/sj.onc.1207305. [DOI] [PubMed] [Google Scholar]
- Bergamaschi G, Merante S, Orlandi E, Galli A, Bernasconi P, Cazzola M. TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica. 2004;89:868–869. [PubMed] [Google Scholar]
- Papadakis ED, Soulitzis N, Spandidos DA. Association of p53 codon 72 polymorphism with advanced lung cancer: the Arg allele is preferentially retained in tumours arising in Arg/Pro germline heterozygotes. Br J Cancer. 2002;87:1013–1018. doi: 10.1038/sj.bjc.6600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, Marzi E, Trapassi C, Storci G, Olivieri F, Franceschi C. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res. 2003;9:4860–4864. [PubMed] [Google Scholar]
- Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: a meta-analysis review. Cancer Epidemiol Biomarkers Prev. 2004;13:11–22. doi: 10.1158/1055-9965.epi-083-3. [DOI] [PubMed] [Google Scholar]
- Jee SH, Won SY, Yun JE, Lee JE, Park JS, Ji SS. Polymorphism p53 codon-72 and invasive cervical cancer: a meta-analysis. Int J Gynaecol Obstet. 2004;85:301–308. doi: 10.1016/j.ijgo.2003.08.017. [DOI] [PubMed] [Google Scholar]


