Abstract
Background
Breast cancer ranks as the foremost cause of cancer-related mortality among women globally. Timely diagnosis stands as the most effective approach in mitigating breast cancer mortality rates. There exists a close relationship between immune processes and tumorigenesis. This study aims to elucidate the immune mechanisms and potential biomarkers associated with breast cancer using bioinformatics techniques.
Objective
Initially, differentially expressed genes were identified through consensus analysis of invasive breast cancer (BRCA) samples sourced from The Cancer Genome Atlas (TCGA) database, focusing on immunotherapy response. Subsequently, protein–protein interaction (PPI) networks, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were employed to refine the selection of potential genes. Lastly, expression and prognostic analyses of hub genes were conducted to identify reliable key genes, with a focus on CCL19. Immunohistochemistry (IHC) was employed to assess the differential expression of CCL19 in both tumor and adjacent breast tissue samples. Additionally, protein correlation analysis, signaling pathway analysis, immune infiltration analysis, gene co-expression analysis, and drug sensitivity analysis of CCL19 were conducted to investigate its pathological clinical features and potential biological functions.
Results
CCL19 expression exhibited a significant increase in breast cancer. Elevated CCL19 expression correlates with advanced tumor stage and indicates a favorable prognosis in breast cancer. CCL19 expression correlates with the abundance of diverse tumor-infiltrating immune cells (TIICs). CCL19 exhibits a positive correlation with the expression of the majority of immune-related genes. Enrichment analysis revealed the involvement of CCL19 in immune-related pathways and the PI3K-Akt signaling pathway. These findings suggest that CCL19 may influence breast cancer prognosis through immune infiltration. Patients exhibiting high CCL19 expression demonstrated more favorable responses to immunotherapy.
Conclusion
Breast cancer demonstrates overexpression of CCL19. CCL19 holds promise as a biomarker for forecasting breast cancer prognosis and as a potential therapeutic target.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-025-02715-9.
Keywords: CCL19, Breast cancer, Biomarker, Immune infiltration
Introduction
Breast cancer ranks as the most prevalent malignancy among women globally. The incidence of breast cancer has been rising in recent years. In 2020, roughly 2.3 million new cases of breast cancer were diagnosed globally among women. Breast cancer is the leading cause of cancer-related mortality among women, representing a significant threat to women’s health [1]. It is widely recognized that tumor cells are regulated by the tumor microenvironment (TME). The interactions between tumor cells and the TME complicate the process of tumorigenesis, making it challenging to elucidate [2]. Thus, a comprehensive investigation into the expression levels of cancer-related genes is crucial for the diagnosis, treatment, and prognosis of breast cancer.
CCL19, located on human chromosome 17, belongs to the chemokine family. CCL19 is produced by lymph nodes, the spleen, and dendritic cells and exhibits abundant expression in the thymus and lymph nodes [3]. CCL19 plays a significant role in immune response, particularly in the transportation of T cells, NK cells, and dendritic cells (DCs). CCL19 mediates tumor progression via its ligand CCR7 [4]. A growing body of literature has documented differential expression of CCL19 in various malignant tumors, including gastric cancer, lung cancer, cervical cancer, and colorectal cancer [5–8]. This suggests that CCL19 may play a potential role in tumorigenesis. CCL19 has been shown to suppress tumors in breast, gastric, and colorectal cancer (CRC) [5, 7, 9]. Conversely, overexpression of CCL19 has been associated with tumor-promoting effects in small cell lung cancer (SCLC) and cervical cancer [6, 8]. Several studies have identified CCL19 as a potential therapeutic target for cancer. Injection of CCL19 into mouse models of both lung cancer and colorectal cancer has led to tumor shrinkage [5, 10]. However, research on the specific mechanisms and therapeutic applications of CCL19 in breast cancer remains limited.
Consequently, we employed a range of methodologies, including gene expression analysis, protein correlation analysis, signaling pathway analysis, prognostic analysis, gene co-expression analysis, and drug sensitivity analysis, to comprehensively investigate the role of CCL19 in breast cancer. The expression of CCL19 was analyzed in conjunction with tumor-infiltrating lymphocytes (TILs) and immunomodulatory factors. Our results indicate a close association between CCL19 and breast cancer immunity. CCL19 may serve as a significant predictive biomarker for breast cancer prognosis and could represent a novel therapeutic target.
Methods
Ethical approval
This study was approved by the institutional research ethics committee of the Second Affiliated Hospital of Anhui Medical University (Approval No. AHMU2 ND-202178). All 101 patients were pathologically confirmed breast cancer patients who treated at the Second Affiliated Hospital of Anhui Medical University from January 2017 to December 2021. Patients with comorbidity of other malignancies were excluded. All selected patients had no prior chemotherapy, radiotherapy, or targeted therapy.
Data collection
GSE108474 data were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm). The GSE103668 (Triple Negative Breast cancer) dataset was used to predict immunotherapy response. The transcribed values are transformed log2 using the"limma"package in R. All data involved in this study were obtained and applied in accordance with the database Publication Guidelines and data access policies.
Identification of DEGs
21 breast cancer samples from GSE108474 data were divided into two groups based on whether there was a response to immunotherapy. “Limma” of R software was used to determine the differentially expressed genes (DEGs) between different clusters [11]. Adjusted p value < 0.05, |logFc|> 1 was defined as the screening threshold for differential mRNA expression. The heatmaps were all visualized by the R package heatmap. The SRTING database (cn.string-db.org) was used to construct the PPI network. Data obtained from the SRTING database were used to identify 20 hub genes with the cytoHubba plug-in of Cytoscape software [12] and combined into a visual network. To further confirm the potential functions of DEGs, enrichment analysis was performed. The “Cluster Profiler” package in R was used to analyze the GO function and the KEGG pathway [13].
CCL19 expression analysis
Gene expression data and clinical information of breast cancer were downloaded from TCGA. Additional pan-cancer datasets based on the GEO cohort were obtained from the GENT2 database. The differences of CCL19 mRNA expression levels of tumor tissues and normal tissues among various tumor types was analyzed. Next, the clinical data downloaded were used to divide the samples into two groups based on the expression level of CCL19. The differences in clinical data between the two groups were analyzed separately.
Prognosis analysis
The data from TCGA database was used to assess the difference in survival between the CCL19 high group and CCL19 low group. Overall survival (OS) and disease free survival (DFS) were used to assess the prognostic value of CCL19 in breast cancer. Multivariate Cox regression analysis was performed using the"survival"and"rms"packages in R software. The analysis included factors such as CCL19, age, gender, and tumor stage to construct a Cox proportional hazards model. The"regplot"package was used to visualize the model as a nomogram, and a calibration curve for the prediction model was generated simultaneously.
Genetic mutation analysis
The genetic mutation characteristics of CCL19 were studied through the CBioPortal database (www.cBioPortal.org) [14]. Further, based on the CCL19 expression data from TCGA, the correlation of tumor mutational burden (TMB)/microsatellite instability (MSI) and CCL19 expression was described by using Spearman analysis. The above figure was implemented by the R package “ggstatsplot”.
Immune infiltration analysis
CIBERSORT [15] was used to calculate the degree of immune cell infiltration in the BRCA queue in TCGA. Wilcoxon signed rank test was applied to analyze the differences in tumor infiltrating immune cells between different groups. The ESTIMATE algorithm was used to calculate the immune and matrix scores between two groups [16]. We use TIMER 2.0 database (timer.cistrome.org) to study the association of CCL19 expression with immune cell infiltration [17]. Depmap (https://depmap.org/portal/) was used to obtain genome information of breast cancer cell lines and sensitivity to genetic and small molecule perturbations.
Gene co-expression analysis
Oncomine database (www.oncomine.org) is a tumor microarray database, which was used for gene co-expression analysis of CCL19 in this study. CCL19 was subjected to gene co-expression analysis including immune activator, immunosuppressant, chemokine, chemokine receptor and MHC genes through the TIBIS database (cis.hku.hk/TISIDB/index.php).
PPI network and gene enrichment analysis
SRTING database (cn.string-db.org) was used to conduct an online search for the mutual aid relationship of CCL19 proteins [18]. The obtained results were visualized with Cytoscape [12]. The R package"enrich plot"was used to study the enrichment of CCL19 in signaling pathways. GO and KEGG were used to assess the potential function of CCL19 through R package ‘clusterProfiler’. The R package"ggplot2"was used for plotting.
Drug susceptibility analysis
Considering the role of immune checkpoint inhibitors in immunotherapy, the correlation between CCL19 and immune checkpoints was analyzed. Furthermore, the response of immunotherapy between the low expression and high expression CCL19 groups was predicted. The immunophenotype score (IPS) reflects the efficacy of immunotherapy. The higher the IPS score, the stronger the immunogenicity. The IPS of TCGA-BRCA patients was derived from the Cancer Immunome Atlas (TCIA) (https://TCIA.at/home).
Genomics of Cancer Drug Sensitivity (GDSC) (www.cancerrxgene.org), the largest pharmacogenomics database, was used to predict chemotherapy response of 1101 breast cancer samples from TCGA [19]. The prediction process is implemented by the R package “pRRophetic” [20]. The mRNA expression data and drug sensitivity data were combined and subjected to Pearson correlation analysis to obtain the correlation of CCL19 expression withIC50 value of the drugs. P-values were adjusted by false discovery rate (FDR).
IHC
Specimens from patients were immersed in methanol solution and embedded in paraffin. Anti-CCL19 (1:200; Abcam) antibody and Anti-CD8 (1:300; Abcam) antibody were used for immunohistochemical staining. Paraffin blocks were cut into 3 µm slices and placed in a 57 °C environment for 90 min to dry. Slices were placed in xylene and a range of concentrations of alcohol. Slices were subsequently boiled in a citrate solution. Endogenous hydrogen peroxide activity was blocked by hydrogen peroxide (0.3%). Slices were incubated with primary antibody overnight at 4 °C. After removal, wash three with phosphate-buffered saline (PBS) and incubate with secondary antibody for 30 min at 37 °C. Sections were stained in 3,3-diaminobenzidine tetrahydrochloride (DAB) and hematoxylin. The staining results were scored by two pathologists, who were unaware of the sample's information. Positive reactions were defined as brown staining. The IHC score is calculated by the staining intensity (0, negative; 1, weak; 2, moderate; 3, strong) multiplied by the positive area (0, less than 5%; 1, 5% to 25%; 2, 26% to 50%; 3, 51% to 75%; 4, greater than 75%). The full score is 12 points, where 0–7 points are considered low expression and 8–12 points are considered high expression [21].
Cell culture
Human breast cancer cell lines (MCF-7 and MDA-MB-231) were bought from the American Type Culture Collection (ATCC). All cells were cultured in DMEM medium with 10% fetal bovine serum and 100 × penicillin–streptomycin solution. All cells were cultured at 5% CO2 and 37 °C. Recombinant Human CCL19 (JN0318, Baiaoleibo, Beijing) was added to the cell line and continued to culture.
Cell proliferation assay
Breast cancer cells were inoculated into 96-well plates. Cell viability was calculated by CCK-8 system after 24, 48 and 72 h. The optical density (OD) value of each hole was measured at 450 nm (BioTek USA).
Colony formation assay
Breast cancer cells were cultured in 6-well plates for 2 weeks. Culture 500 cells per well until colony formation. The formed colonies were fixed with 4% paraformaldehyde (PFA) for 30 min and stained with 0.5% crystal violet for 5 min. The number of clones formed (more than 50 cells per clone) was manually measured and photographed.
Transwell
For invasion assay, the top chamber was pre-coated with Matrigel. Next, the cells were added culturing in a serum-free medium. In the meantime, RPMI-1640 medium with 10% FBS was put in the lower chamber. After that, cells that remained in the upper chamber were removed. Invaded cells were finally stained with crystal violet and observed. For migration assay, the upper chamber was not coated with Matrigel, and the remaining procedures were similar to those in the invasion assay.
Statistical analysis
All the above statistical analysis and R packages were performed using R software (version 4.0.2) (www.Rproject.org) for statistical analysis. P < 0.05 indicates statistical significance.
Results
Identification of DEGs
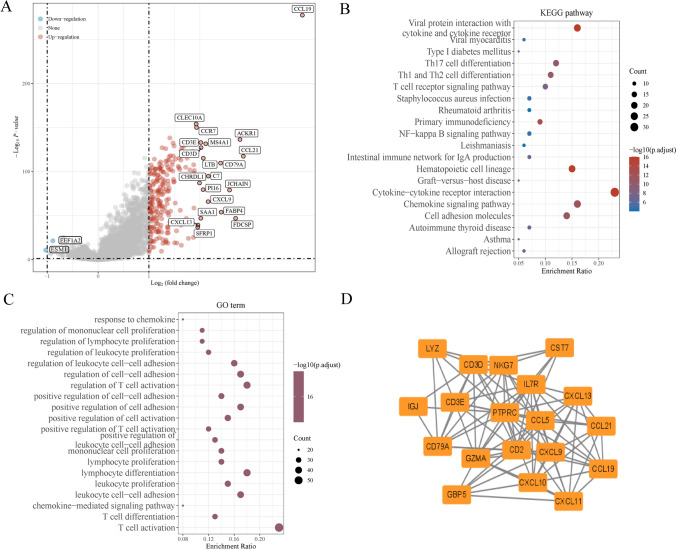
The flow chart of this study is shown in Fig. 1. BRCA samples from GSE103668 were divided into two groups by the response to immunotherapy. 583 DEGs in total were obtained by screening the two groups, of which 581 genes were up-regulated and 2 genes were down-regulated (Fig. 2A). To further investigate the biological functions and underlying mechanisms of DEGs in breast cancer, GO enrichment analysis and KEGG pathway analysis were used. GO analysis showed that DEGs were mainly enriched in negative regulation of immune system process, regulation of cell–cell adhesion, leukocyte cell–cell adhesion, and T cell activation (Fig. 2B). KEGG analysis showed that DEGs were mainly involved in chemokine signaling pathway, cytokine-cytokine receptor interaction, and cell adhesion molecules (Fig. 2C).
Fig. 1.
Flowchart of this study
Fig. 2.
Analysis of DEGs based on immunotherapy response. A DEGs volcano map analysis; B KEGG pathway of DEGs; C GO analysis of DEGs; D Hub gene was selected from PPI network through Cytoscape plug-in Cytohubba
The Expression Level and Prognostic Value of the Hub Gene
.DEGs PPI network graphs are constructed. Cytoscape's plugin cytoHubba was used to seek for the most important nodes, and finally PTPRC, CST7, C1S, CD2, IL7R, CD3E, CD3D, GZMA, CCL5, LYZ, CXCL9, NKG7, GBP5, CXCL11, CD79 A, CXCL10, JCHAIN, CCL19, CCL21, CXCL13 were identified as hub genes (Fig. 2D). To determine the diagnostic significance and prognostic value of hub genes, the GEPIA database was used to validate the expression (Supplementary Fig. 1) and prognosis (Supplementary Fig. 2) of 20 genes in breast cancer. The above results show that CCL19 is differentially expressed in breast cancer tissues. Moreover, the expression of CCL19 is related to the OS of patients. Previous studies have shown that CCL19 exists in the tumor microenvironment of breast cancer and plays an important role [3]. Therefore, CCL19 was selected as the hub gene in this study.
Expression of CCL19 in tumor tissue
First, the mRNA expression level of CCL19 in different tumor tissues was analyzed. Evaluation of CCL19 mRNA expression in pan-cancer by TCGA database showed that CCL19 expression was lower in bladder Urothelial Carcinoma (BLCA), cholangio carcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and Neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), thyroid carcinoma (THCA), uterine Corpus Endometrial Carcinoma (UCEC), and higher in BRCA (Fig. 3A). Data from the GEO database also support the above results (Fig. 3B). Furthermore, analysis of differential expression of CCL19 based on cancer and cancer adjacent tissue samples in the TCGA database showed that CCL19 expression was significantly higher in breast cancer tumor tissues compared with normal tissues (Fig. 3C). CCL19 is differentially expressed between different molecular types of breast cancer, including triple-negative, HER2-overexpressing, Luminal A, and Luminal B (Fig. 3D). CCL19 expression was assessed in different cancer stages, and CCL19 expression was statistically different between tumor stages I and II, II and III, and II and V (Fig. 3E). The data were divided into two groups according to the expression level of CCL19, and the differences in clinical data between the two groups were analyzed. There were significant differences in age, tumor stage, and T classification between the two groups (Fig. 3F).
Fig. 3.
Differential expression of CCL19. A Expression of CCL19 in different tumors from TCGA database; B Expression of CCL19 in different tumors from GEO database; C Expression of CCL19 in tumor tissues and normal tissues of breast cancer from TCGA database; D Expression of CCL19 in different subtypes of breast cancer from TCGA database; E Expression of CCL19 in different stages of breast cancer from TCGA database; F Correlation analysis between CCL19 and different pathological features in breast cancer
Prognosis Value of CCL19 in breast cancer
.To further inspect the relationship between CCL19 expression and breast cancer prognosis and survival, we divided breast cancer samples into high and low expression group according to the median value of CCL19 expression level. By analyzing survival data with the Kaplan–Meier curve, we noticed that CCL19 expression level was significantly associated with better OS (Fig. 4A, B).
Fig. 4.
Survival analysis curve of CCL19 in breast cancer. A, B OS and DFS in breast cancer patients through Kaplan–Meier curve; C Univariate Cox regression analysis of CCL19 in breast cancer; D Multivariate Cox regression analysis of CCL19 in breast cancer; E The prognostic nomogram with CCL19 and clinical variables; F The 1-, 3- and 5-year calibration plots demonstrated the performance of the nomogram
Univariate and multivariate Cox regression analyses were conducted to determine independent prognostic factors. Univariate Cox regression analysis showed that age, stage, T, N, and CCL19 were closely related to survival rate (Fig. 4C). The above-mentioned influencing factors were incorporated into the multi-factor COX analysis. The results showed that CCL19 (P = 0.017, HR = 0.911, 95%CI 0.844–0.983) was also significantly correlated with survival rate (Fig. 4D). In summary, the expression of the CCL19 can be preliminarily determined that the volume of CCL19 is related to the prognosis of patients. A prognostic nomogram including CCL19 was constructed, which also included age, gender, TNM, Grade, and Stage. Among them, age has a higher influence, followed by CCL19 expression (Fig. 4E). The calibration curve obtained simultaneously with the nomogram. The curves of 1-year, 3-year, and 5-year prediction values are basically distributed along the diagonal line (Fig. 4F), indicating that the model has good predictive performance.
Genomic mutation of CCL19 in breast cancer
Since cancer is associated with Genomic mutations, we explored genomic alterations in CCL19. CCL19 had different alterations frequencies in different breast cancer subtypes, including 1.39% (breast invasive ductal carcinoma) and 0.84% (breast mixed ductal and lobular carcinoma) (Supplementary Fig. 3 A). The"Mutations"modules of the cBioPortal database showed that the alterations frequency of CCL19 was 1.1%, including amplification, deep deletion, and missense mutation (Supplementary Fig. 3B). In total, only one sample of CCL19 had a missense mutation. Supplementary Fig. 3 C shows the location of the alterations in CCL19. Amplification is the most common type of CCL19 alterations. We further explored the relationship between CCL19 expression and TMB (tumor mutational burden) and MSI (microsatellite instability) of breast cancer in TCGA. We noticed that the expression of CCL19 was negatively correlated with TMB and MSI (Supplementary Fig. 3D and E).
PPI network and enrichment analysis of CCL19 in breast cancer
The genes significantly associated with CCL19 were identified through the dataset from TCGA (Supplementary Table 1). Ten co-expressed genes significantly positively or negatively correlated with CCL19 were shown in the circular graph (Fig. 5A). According to the median expression level of CCL19, breast cancer samples were divided into high and low expression group. Next, we compared the DEG analysis between the two groups. | log2 (multiple change) |> 1, and adjusted for p < 0.05. As the result, 1386 genes were identified as DEG (Supplementary Table S2). The top 50 upregulated genes and top 50 downregulated genes were shown in the heatmap (Fig. 5B).
Fig. 5.
A Co-expressed genes significantly associated with CCL19 were shown as correlation circle plots. Red, positive correlation; green, negative correlation; B The top 50 upregulated DEGs and top 50 downregulated DEGs were displayed in the heatmap. Red, upregulated genes; blue, downregulated genes; C The circle graph of enrichment analysis; D The signaling pathways that were CCL19-related differentially expression genes activated in breast cancer through GSEA
The obtained genes were subjected to GO and KEGG analysis using the “clusterProfiler” package. The gene set with the highest credibility was selected to draw a ring diagram (Fig. 5C). The interaction between CCL19 and its functional related proteins was shown in Supplementary Fig. 4 A-B. GO analysis found that CCL19 is involved in Leukocyte mediated immunity, positive regulation of cell activation, and antigen binding (Supplementary Fig. 5 A, Supplementary Table 3). The results of KEGG showed that CCL19 expression was associated with Cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine-receptor and Primary immunity (Supplementary Fig. 5B, Supplementary Table 4). GSEA was used to determine the signaling pathway of different activated CCL19 related genes in breast cancer. The results showed that the immune related gene set was significantly enriched in CCL19 related DEG (Fig. 5D).
The Relationship between CCL19 Expression and Immune Cell Infiltration in breast cancer
CIBERSORT calculation was used to determine the proportion of 22 immune cells infiltrated in the tissues of each breast cancer patient. The sample was divided into high and low expression groups according to the expression level of the target gene CCL19. The “limma” package was used to compare the differences in immune cells between the high and low expression groups. Figure 6A shows the distribution of immune infiltrating cells in the two groups. The correlation between CCL19 expression and immune cell expression was visualized as a Lolipop plot (Fig. 6B). The high expression group of CCL19 showed an increase in CD8 + T cells and M1 macrophages (Fig. 6C, D). The expression of CCL19 was significantly negatively correlated with M0 macrophages and M2 macrophages (Fig. 6E, F).
Fig. 6.
A The proportion of 22 immune cells infiltrating in low and high CCL19 expression groups; B Relationships between the expression of CCL19 and 22 types of immune infiltration cells; C–F The relationships between the expression of CCL19 and CD8 + Tcells, M0 macrophages, M1 macrophages and M2 macrophages; G ESTIMATE algorithm was used to investigate the correlation between the two groups in Immune scores and Stromal scores
The correlation between CCL19 expression and immune cell infiltration in pan cancer of the TIMER database is shown in Supplementary Fig. 6. XCELL, TIMER, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, and EPIC were integrated to compare the correlation between different groups of immune cells, indicating differences in immune status between low and high CCL19 expression groups (Supplementary Fig. 7 A). After adjusting the purity of the tumor, further analysis of the correlation between CCL19 and the abundance of cancer immune cells revealed that CCL19 expression was negatively correlated with B cells, CD4-T naïve cells, CD8-T cells, CD4-T memory, CD4-T effector memory and Myeloid DC cells (Supplementary Fig. 7B-G). Next, estimation algorithms were used to investigate the correlation immune score and matrix score between the two groups. We found that the immune score and matrix score of the high CCL19 expression group were higher than those of the low CCL19 expression group (Fig. 6G), which proved that the expression level of CCL19 could affect the immune activity of breast TME in breast cancer.
To understand the effect of CCL19 knockout on the viability of different breast cancer cells, we explored the CCL19 loss screening from Depmap. The Gene Effects value of CCL19 in breast cancer cell lines is less than 0, which means that CCL19 will inhibit cell proliferation (Supplement Table 5).
Analysis of co‑expression genes associated with CCL19 in breast cancer
The gene co-expression analysis module method of the Oncomine database was used to analyze the CCL19 mRNA of TCGA breast cancer patients. As can be seen from Supplementary Fig. 8 A, the number of genes significantly positively correlated with CCL19 was higher than those of negative correlation. Supplementary Fig. 8B, C shows top 50 significant genes positively and negatively associated respectively. The results show that CCL19 is highly correlated with SELP and KLRB1, reflecting that CCL19 may take part in in tumor development by mediating activated endothelial cells, inhibiting NK cells, and activating acid sphingomyelinase [22–24].
The TIBIS database provided us with the co-expression relationship of CCL19 with immune activator, immunosuppressant, chemokine, chemokine receptor and MHC genes. The heatmap generated by the co-expression analysis indicated that a large number of immune-related genes were significantly positively correlated with the expression of CCL19 in breast cancer (Supplementary Fig. 8D–H).
Correlation between CCL19 expression and drug sensitivity in breast cancer
To explore the relationship between CCL19 expression and common immune checkpoint-related genes, the expression of immune checkpoint-related genes in the tumor sample sequencing data was extracted for correlation testing. The correlation test filter was set to P-value < 0.001. The heat map showed the positive regulatory relationship between CCL19 and multiple immune checkpoints, including LAIR1, ICOS, PDCD1 and TIGIT (Fig. 7A, B).
Fig. 7.
A, B The association between ICIs and CCL19. C–F The correlation between immunophenoscore and different CCL19 expression groups. G–I IC50 value of KIN001-102, tak-715 and 17-AAG
Integrate the immunotherapy effect scoring files in TCIA with the sorted TCGA sequencing data according to the sample ID. According to the expression of CCL19, the patients were divided into two groups: high and low expression. The “limma” package was used to compare the immunotherapy scores between the high and low expression groups. The “ggpurb” package was used to express the final difference comparison results into violin plots. The results of immunophenotypic analysis showed that the group with high CCL19 expression exhibited higher IPS, indicating that patients with high CCL19 expression exhibited greater positive responses to immunotherapy (Fig. 7C–F).
IC50 (semi-inhibitory concentration) is an important indicator for evaluating drug efficacy or sample treatment response and was used to analyze the sensitivity of patients with different CCL19 expression levels to drugs. The results showed that patients with high CCL19 expression had lower IC50 values for TAK-715 and KIN001-102(Fig. 7G, H). In contrast, patients with high CCL19 expression had higher IC50 values for 17-AAG (Fig. 7I).
Validation of the expression, prognosis, and immune-Related exploration of CCL19 through clinical samples
IHC staining was performed on the cancer tissue and normal tissue samples from hospital patients. All staining steps were carried out in strict accordance with the standard procedure. The results showed that CCL19 was darker brown in tumor tissue, which represented higher expression (Fig. 8A, B). For hospital patients, we compared the differential expression of CCL19 in different groups. Breast cancer patients from hospitals were grouped based on age, nuclear grade, tumor stage, and molecular type (Table 1). Neither OS nor RFS based on clinical samples were found to be significantly associated (Supplementary Fig. 9). Thanks to early tumor stage and good treatment, breast cancer patients from our hospital have fewer recurrences and deaths within five years. In order to draw more accurate conclusions, it is necessary to further expand the research sample size in the future.
Fig. 8.
Expression of CCL19 in breast cancer patients from hospitals. A IHC images of CCL19 in tumor and adjacent normal tissues; B IHC score of CCL19 expression in tumor and adjacent tissues; C IHC images of CD8,PD-1 and PD-L1 expression in groups with CCL19 high and low expression in breast cancer samples
Table 1.
Correction between CCL19 expression and clinical characteristics of patients with BRCA (n = 101)
| CLINICOPATHOLOGICAL Characteristics | Number | Low expression |
High expression | P-value |
|---|---|---|---|---|
| Age (years) | 0.456 | |||
| ≤ 55 | 48 | 20 | 28 | |
| > 55 | 53 | 26 | 27 | |
| Stage | 0.036 | |||
| Stage I | 34 | 10 | 24 | |
| Stage II | 57 | 29 | 28 | |
| Stage III | 10 | 7 | 3 | |
| T classification | 0.327 | |||
| T1 | 41 | 15 | 26 | |
| T2 | 58 | 30 | 28 | |
| T3 | 2 | 1 | 1 | |
| N classification | 0.053 | |||
| N0 | 76 | 29 | 47 | |
| N1 | 15 | 10 | 5 | |
| N2 | 8 | 5 | 3 | |
| N3 | 2 | 2 | 0 | |
| Grade | 0.034 | |||
| 1 | 18 | 9 | 9 | |
| 2 | 75 | 30 | 45 | |
| 3 | 8 | 7 | 1 | |
| Subtype | 0.506 | |||
| Luminal A | 18 | 9 | 9 | |
| Luminal B | 59 | 24 | 35 | |
| HER-2 + | 15 | 7 | 8 | |
| Basal | 9 | 6 | 3 | |
| ER status | 0.147 | |||
| Positive | 28 | 16 | 12 | |
| Negative | 73 | 30 | 43 | |
| PR status | 0.298 | |||
| Positive | 32 | 17 | 15 | |
| Negative | 69 | 29 | 40 | |
| HER2 status | 0.277 | |||
| Positive | 65 | 27 | 38 | |
| Negative | 36 | 19 | 17 | |
| Ki-67 ststus | 0.326 | |||
| ≤ 14 | 19 | 9 | 10 | |
| > 15 | 82 | 37 | 45 | |
To explore the correlation between CCL19 and immunotherapy, IHC expression levels of CCL19 and CD8 were calculated. We found that CCL19 expression was significantly correlated with the expression levels of CD8, PD-1 and PD-L1 (Fig. 8C).
CCL11 inhibits the proliferation, migration, and invasion of BC cells.
Next, we analyzed the biological function of CCL19 in breast cancer through cellular experiments. CCK-8 and colony formation experiments showed that the proliferation and colony formation of BC cells were inhibited in vitro after the addition of CCL19 (Fig. 9A–E). To further evaluate the role of CCL11 in BC cell progression, we investigated the migration and invasion of cells adding CCL11BC. Transwell experiment showed that the addition of CCL11 significantly reduced the invasion and migration ability of BC cells (Fig. 9F–H).
Fig. 9.
CCL19 inhibits malignant behavior of breast cancer cell lines. A, B After adding CCL19, the proliferation capacity of breast cancer cells was measured by CCK-8; C–E After adding CCL19, the proliferation and viability of breast cancer cells were detected by plate cloning; F–H Transwell assay evaluated the migration and invasion ability of breast cancer cells after adding CCL19
Discussion
In recent decades, significant improvements in breast cancer survival rates have been observed [25]. Nevertheless, the fundamental mechanisms driving breast cancer tumorigenesis remain only partially understood. Interactions between tumor cells and diverse components of the tumor microenvironment contribute to the overall complexity of cancer [26]. Recent advancements in genomic research have significantly enhanced the understanding of breast cancer diagnosis and treatment. Genetic data, gene activation pathways, and gene-mediated functions are critical to understanding and treating cancer [27–29]. Tumors undergo transitions between elimination, equilibrium, and escape as the status of the immune system fluctuates [30, 31]. Hence, immune-related genes play a crucial role in tumor progression, garnering increasing attention in breast cancer diagnosis and treatment [32–34]. This study classified 21 breast cancer samples from the GSE108474 dataset into two groups based on immunotherapy responses, followed by differential expression gene (DEG) screening within the clusters. Then, DEG differentially expressed in the two clusters was screened. Key hub genes identified include PTPRC, CST7, C1S, CD2, IL7R, CD3E, CD3D, GZMA, CCL5, LYZ, CXCL9, NKG7, GBP5, CXCL11, CD79 A, CXCL10, JCHAIN, CCL19, CCL7, CCL21, and CXCL13. Considering the expression and prognostic implications of the central gene in BRCA, CCL19 was identified as the key gene for this study.
The CCL19 gene, situated at 9p13 on human chromosome 17, belongs to the CC chemokine family [3, 35]. CCL19 is commonly viewed as a homeostatic chemokine that regulates immune cell trafficking, thereby contributing to cancer development. CCL19 promotes lymphocyte homing and lymph node metastasis of tumor cells through its interaction with the CCR7 receptor [4]. CCL19 exhibits a dual role in tumorigenesis. First, CCL19 suppresses tumors by inducing anti-tumor immune responses and inhibiting angiogenesis. Zhuoqing Xu et al. concluded through cell experiments and animal experiments that CCL19 can inhibit CRC angiogenesis by promoting the expression of miR-206 [5]. High expression of CCL19 significantly inhibited the proliferation of GC cells and tumor growth in vitro and in vivo by up regulating the CCR7/AIM2 pathway [7]. Conversely, CCL19 also plays a role in promoting tumor progression by inducing tumor cell proliferation and metastasis. Qian Liu et al. found that CCL19 is linked to lymph node metastasis in small cell lung cancer patients and promotes tumor metastasis by impairing CD8 + T cell function [8, 34]. The knockout of CCL19 attenuates tumor cell proliferation in cervical cancer [6].
CCL19 demonstrates both anti-tumor and pro-tumor activities in different contexts of breast cancer. CCL19 has the potential to mediate strong anti-tumor responses in the body. The CCL19/CCR7 axis has been identified as regulating EMT and mediating tumor cell invasion and migration through AKT signaling pathway in breast cancer. Hossein Hozhabri et al. found that the overexpression of CCL19 in breast cancer patients was associated with better OS and RFS [36]. CCL19 can be combined and cleared by CX-CKR, which promotes breast cancer cell invasion and metastasis [37]. CCL19 induces NK cells and inhibits tumor growth when breast cancer tumor-bearing mice are injected with CCL19 [38, 39]. However, Xu et al. found that CCL19 administration in vitro increased the invasion, migration and EMT of breast cancer cells. CCL19 facilitates the secretion of IL-6 and IL-1β by DC cells, which promote tumor growth in triple-negative breast cancer (TNBC), leading to tumor invasion and worse prognosis in patients [40]. Recent research has identified CCL19 as a potential prognostic biomarker that shows significant correlation with TILs in breast cancer [41]. Sangeetha Prabhakaran et al. identified 12 chemokine gene expression characteristics, including CCL19, capable of distinguishing breast cancer with heterotopic lymph node-like structures rich in tumor location, and predicting more favorable long-term outcomes [42].
This study revealed a significant upregulation of CCL19 in breast cancer tumor tissue, which was positively correlated with patient prognosis. Data from the TCGA and GEPIA databases indicated a significant association between CCL19 mRNA expression levels, cancer stage, and tumor subtype in breast cancer patients. IHC results from hospital patients further corroborated this finding. In addition, the mutation rate of CCL19 in Invasive ductal carcinoma patients was 1.39%. Amplification represents the most common type of CCL19 mutation in breast cancer. CCL19 expression exhibited a negative correlation with MSI and TMB in breast cancer. The current study identified a significant correlation between CCL19 expression and the infiltration levels of various TILs in the tumor microenvironment. These findings support the hypothesis that CCL19 regulates tumor progression by modulating the trafficking of immune cells, including DCs, T cells and NK cells. Research has demonstrated that CCL19 induces maturation and migration of DCs while reducing apoptosis [43]. CCL19 indirectly increases T cell proliferation by promoting the production of IL-12, TNFα and IL-1β by DC cells (20). CCL19 can increase the proliferation of NK cells triggered by IL-2 (20,21). CCL19 augments T cell responses by recruiting and facilitating their steady-state transport along a concentration gradient of CCL19 [44]. This study identified a positive correlation between CCL19 and the density of infiltrating CD8 + T cells in the TME within our cohort, suggesting a potential role of CCL19 in regulating breast cancer progression through CD8 + T cells. Functional network analysis suggested that CCL19 may contribute to breast cancer progression by engaging in signaling pathways such as the PI3 K-Akt pathway and Human Papillomavirus infection. Peptides derived from Human Endogenous Retroviruses (HERV) have been shown to stimulate CCL19 production in pancreatic cancer cells via the PI3 K pathway [45]. CCL19 enhances Akt phosphorylation and upregulates Bcl-2 protein expression in HNSCC cells. Elevated Bcl-2 expression inhibits cisplatin-induced apoptosis, implying a significant role of CCL19 in chemoresistance [46].
Chemotherapy is widely recognized as a potent inducer of apoptosis. Tumor cells evade apoptosis by downregulating pro-apoptotic pathways, upregulating anti-apoptotic signals, or inducing false apoptotic signals [47]. Hence, evaluating the sensitivity of breast cancer patients with varying CCL19 expression levels to chemotherapy is imperative. The results demonstrated a significant correlation between CCL19 expression levels and the IC50 values of TAK-715, KIN001-102, and 17-AAG. Research has demonstrated that direct intratumoral injection of CCL19 significantly reduces tumor volume in a mouse model of lung cancer [10, 48]. Xu et al. observed significant inhibition of intratumoral angiogenesis following treatment with CCL19 in both colon cancer cells and mice [5]. Co-injection of CCL19 and cytosine-phosphorothioguanine oligonucleotides (CpG ODN) into tumor sites in a mouse model led to substantial tumor shrinkage [49]. Treatment with CCL19 markedly upregulated Bcl-xL and Bcl-2 proteins in breast cancer cells, thereby countering anoikis [50]. The combination of CCL19-encoding plasmid DNA (CCL19 pDNA) and PD-L1 inhibitor (BMS-1) significantly inhibited tumor development..
In this study, we employed multiple bioinformatics approaches to screen for hub genes in BRCA samples, identifying CCL19 as the most prominent gene. Although our findings are promising, several limitations remain in this study. First, the survival analysis results are limited by the generally favorable prognosis associated with breast cancer. In our cohort, no significant differences in OS and RFS were observed among groups with varying levels of CCL19 expression. A longer follow-up period is required to thoroughly assess the impact of CCL19 on patient prognosis. Second, the precise mechanisms and pathways through which CCL19 influences breast cancer progression remain unclear. Finally, while we evaluated the drug sensitivity of CCL19 using the GDSC database, further experimental validation is necessary. Additional experiments are essential for further verification. Addressing these limitations will guide future research directions.
In conclusion, our study identifies CCL19 as a promising biomarker for breast cancer. However, the mechanisms underlying CCL19's role in regulating tumor progression in breast cancer remain poorly understood. Thus, future research will focus on elucidating the genetic axis of CCL19 in breast cancer.
Supplementary Information
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
XC contributed to conception and design of the study. YW organized the database. LY and XW performed the statistical analysis. XC and ZD wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research is supported by Anhui Provincial Health Research Project (Grant no. AHWJ2024BAh30024).
Data availability
Data is provided within the manuscript or supplementary information files. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Institutional Research Ethics Committee of The Second Affiliated Hospital of Anhui Medical University (Hefei, China), and written informed consent was obtained from all patients.
The accordance statements
All experimental methods in this study were conducted in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221: 107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gowhari Shabgah A, et al. Does CCL19 act as a double-edged sword in cancer development? Clin Exp Immunol. 2022;207(2):164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salem A, et al. CCR7 as a therapeutic target in Cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1): 188499. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z, et al. CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1α/VEGF-A pathway in colorectal cancer. Cell Death Dis. 2018;9(10):974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, et al. Increased CCL19 expression is associated with progression in cervical cancer. Oncotarget. 2017;8(43):73817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou R, et al. CCL19 suppresses gastric cancer cell proliferation, migration, and invasion through the CCL19/CCR7/AIM2 pathway. Hum Cell. 2020;33(4):1120–32. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, et al. CCL19 associates with lymph node metastasis and inferior prognosis in patients with small cell lung cancer. Lung Cancer. 2021;162:194–202. [DOI] [PubMed] [Google Scholar]
- 9.Kim BJ, et al. Lymphoidal chemokine CCL19 promoted the heterogeneity of the breast tumor cell motility within a 3D microenvironment revealed by a Lévy distribution analysis. Integr Biol (Camb). 2020;12(1):12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillinger S, et al. CCL19 reduces tumour burden in a model of advanced lung cancer. Br J Cancer. 2006;94(7):1029–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi L, et al. Comprehensive analysis of the PD-L1 and immune infiltrates of m(6)A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids. 2020;21:299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509-w514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605-d612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9(9): e107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M, et al. SPOCK1/SIX1axis promotes breast cancer progression by activating AKT/mTOR signaling. Aging (Albany NY). 2020;13(1):1032–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleshkan VV, et al. Transcription of the KLRB1 gene is suppressed in human cancer tissues. Mol Gen Mikrobiol Virusol. 2007;4:3–7. [PubMed] [Google Scholar]
- 23.Wyrożemski Ł, Qiao SW. Immunobiology and conflicting roles of the human CD161 receptor in T cells. Scand J Immunol. 2021;94(3): e13090. [DOI] [PubMed] [Google Scholar]
- 24.Avan A, et al. AKT1 and SELP polymorphisms predict the risk of developing cachexia in pancreatic cancer patients. PLoS ONE. 2014;9(9): e108057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel RL, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. [DOI] [PubMed] [Google Scholar]
- 26.Arneth B. Tumor microenvironment. Medicina (Kaunas). 2019;56(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker-Smith TL, Peck J. Genetic and genomic advances in breast cancer diagnosis and treatment. Nurs Womens Health. 2019;23(6):518–25. [DOI] [PubMed] [Google Scholar]
- 28.López-Otín C, et al. Meta-hallmarks of aging and cancer. Cell Metab. 2023;35(1):12–35. [DOI] [PubMed] [Google Scholar]
- 29.Satake H, et al. Radiomics in breast MRI: current progress toward clinical application in the era of artificial intelligence. Radiol Med. 2022;127(1):39–56. [DOI] [PubMed] [Google Scholar]
- 30.Candeias SM, Gaipl US. The immune system in cancer prevention, development and therapy. Anticancer Agents Med Chem. 2016;16(1):101–7. [DOI] [PubMed] [Google Scholar]
- 31.Wu M, et al. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J Hematol Oncol. 2022;15(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badiu DC, et al. Modulation of immune system-strategy in the treatment of breast cancer. In Vivo. 2021;35(5):2889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, et al. Inflammation and cancer: paradoxical roles in tumorigenesis and implications in immunotherapies. Genes Dis. 2023;10(1):151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, et al. Tumor-targeting intravenous lipid emulsion of paclitaxel: characteristics, stability, toxicity, and toxicokinetics. J Pharm Anal. 2022;12(6):901–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z, et al. CCL5/CCR5 axis in human diseases and related treatments. Genes Dis. 2022;9(1):12–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hozhabri H, et al. A comprehensive bioinformatics analysis to identify potential prognostic biomarkers among CC and CXC chemokines in breast cancer. Sci Rep. 2022;12(1):10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng LY, et al. Involvement of a novel chemokine decoy receptor CCX-CKR in breast cancer growth, metastasis and patient survival. Clin Cancer Res. 2009;15(9):2962–70. [DOI] [PubMed] [Google Scholar]
- 38.Braun SE, et al. The CC chemokine CK beta-11/MIP-3 beta/ELC/Exodus 3 mediates tumor rejection of murine breast cancer cells through NK cells. J Immunol. 2000;164(8):4025–31. [DOI] [PubMed] [Google Scholar]
- 39.Yin J, et al. Epigenetic modulation of antitumor immunity and immunotherapy response in breast cancer: biological mechanisms and clinical implications. Front Immunol. 2023;14:1325615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang H, et al. Human breast cancer-derived soluble factors facilitate CCL19-induced chemotaxis of human dendritic cells. Sci Rep. 2016;6:30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, et al. CCL19 has potential to be a potential prognostic biomarker and a modulator of tumor immune microenvironment (TIME) of breast cancer: a comprehensive analysis based on TCGA database. Aging (Albany NY). 2022;14(9):4158–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prabhakaran S, et al. Evaluation of invasive breast cancer samples using a 12-chemokine gene expression score: correlation with clinical outcomes. Breast Cancer Res. 2017;19(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hjortø GM, et al. Differential CCR7 targeting in dendritic cells by three naturally occurring CC-chemokines. Front Immunol. 2016;7:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Y, et al. CCL19 enhances CD8(+) T-cell responses and accelerates HBV clearance. J Gastroenterol. 2021;56(8):769–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudo-Saito C, et al. Induction of immunoregulatory CD271+ cells by metastatic tumor cells that express human endogenous retrovirus H. Cancer Res. 2014;74(5):1361–70. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, et al. Autocrine and paracrine chemokine receptor 7 activation in head and neck cancer: implications for therapy. J Natl Cancer Inst. 2008;100(7):502–12. [DOI] [PubMed] [Google Scholar]
- 47.Pfeffer CM, Singh ATK. Apoptosis: a target for anticancer therapy. Int J Mol Sci. 2018;19(2):448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hillinger S, et al. EBV-induced molecule 1 ligand chemokine (ELC/CCL19) promotes IFN-gamma-dependent antitumor responses in a lung cancer model. J Immunol. 2003;171(12):6457–65. [DOI] [PubMed] [Google Scholar]
- 49.Oh SM, Oh K, Lee DS. Intratumoral administration of secondary lymphoid chemokine and unmethylated cytosine-phosphorothioate-guanine oligodeoxynucleotide synergistically inhibits tumor growth in vivo. J Korean Med Sci. 2011;26(10):1270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su ML, et al. Inhibition of chemokine (C–C motif) receptor 7 sialylation suppresses CCL19-stimulated proliferation, invasion and anti-anoikis. PLoS ONE. 2014;9(6): e98823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.