Abstract
The hepatitis C virus (HCV)-specific CD4+ T-cell response against nonstructural proteins is strongly associated with successful viral clearance during acute hepatitis C. To further develop these observations into peptide-based vaccines and clinical immunomonitoring tools like HLA class II tetramers, a detailed characterization of immunodominant CD4+ T-cell epitopes is required. We studied peripheral blood mononuclear cells from 20 patients with acute hepatitis C using 83 overlapping 20-mer peptides covering the NS3 helicase and NS4. Eight peptides were recognized by ≥40% of patients, and specific CD4+ T-cell clones were obtained for seven of these and three additional, subdominant epitopes. Mapping of minimal stimulatory sequences defined epitopes of 8 to 13 amino acids in length, but optimal T-cell stimulation was observed with 10- to 15-mers. While some epitopes were presented by different HLA molecules, others were presented by only a single HLA class II molecule, which has implications for patient selection in clinical trials of peptide-based immunotherapies. In conclusion, using two different approaches we identified and characterized a set of CD4+ T-cell epitopes in the HCV NS3-NS4 region which are immunodominant in patients achieving transient or persistent viral control. This information allows the construction of a valuable panel of HCV-specific HLA class II tetramers for further study of CD4+ T-cell responses in chronic hepatitis C. The finding of immunodominant epitopes with very constrained HLA restriction has implications for patient selection in clinical trials of peptide-based immunotherapies.
Chronic hepatitis C is a global health problem with an estimated 170 million chronically infected patients (1). The prevalence of chronic hepatitis C is between 0.5 and 3% in Europe and North America, where it is a leading cause of chronic liver disease, cirrhosis, and hepatocellular carcinoma. About 50% of patients do not respond to current treatment modalities, and there is no prophylactic vaccine. Spontaneous viral clearance is observed only in acute hepatitis C, and understanding the hepatitis C virus (HCV)-specific immune response that induces permanent viral clearance may lead to the rational development of prophylactic or therapeutic vaccines against hepatitis C. Most knowledge about the correlates of protective immune responses in humans comes from the study of patients with acute symptomatic hepatitis C (4, 7, 12, 14, 16-18). Forty to fifty percent of these patients achieve spontaneous viral clearance, which is associated with a strong and maintained HCV-specific CD4+ and CD8+ T-cell response. Among the HCV-specific CD4+ T-cell responses, the reactivity against nonstructural HCV proteins like NS3 and NS4 seems to be most prominent (4, 7, 14). Several studies have identified immunogenic peptides, mainly within HCV core and the nonstructural proteins (2, 3, 5, 8-11, 15, 19). In fact, more than 60% of the entire NS3-NS4 sequence has been reported to contain CD4+ T-cell epitopes that are recognized by certain individuals, also reflecting the high polymorphism of the HLA class II genes. An overview of previously published CD4+ T-cell epitopes is available at the HCV immunology database of the Los Alamos National Laboratory (http://hcv.lanl.gov/content/immuno/tables/helper_summary.html), and epitopes that have been identified using CD4+ T-cell lines or clones are summarized in the supplemental material for this paper (see Table S1 in the supplemental material). Nevertheless, in most cases the epitopes have not been mapped to the minimal T-cell-stimulating sequence nor has HLA restriction been defined at a T-cell clonal level. However, a detailed knowledge of immunodominant CD4+ T-cell epitopes including the optimal length of the peptide for T-cell stimulation and the spectrum of HLA restriction is necessary for the development of HLA class II tetramers, to define the role of viral escape mutations, and eventually for the design of peptide-based immunotherapies.
In this paper we identified and characterized 10 immunodominant CD4+ T-cell epitopes within the NS3 helicase, NS4A, and NS4B region by a combined approach: first, we measured the proliferative responses of peripheral blood mononuclear cells (PBMC) of 20 patients with acute hepatitis C to a set of 83 individual 20-mer peptides. Then, a large panel of HCV-specific CD4+ T-cell clones was isolated and expanded from the blood of patients with acute hepatitis C by stimulation with recombinant HCV antigens. The T-cell clones were characterized with regard to fine specificity and HLA restriction. A very high rate of concordance in the recognition of epitopes was found between the two approaches, suggesting that the epitopes thus identified represent naturally processed immunodominant epitopes which are promising candidates for peptide-based vaccines and for the development of HLA class II tetramers.
MATERIALS AND METHODS
Patients.
NS3- and NS4-specific T-cell clones were isolated from seven patients with acute hepatitis C infection. In 17 patients with acute hepatitis C infection and in three patients who previously cleared the virus spontaneously a peptide screening (20-mers covering the region of amino acids [aa] 1207 to 2014) was performed by proliferation assay (Table 1). The diagnosis of acute hepatitis C was based on the following criteria: (i) elevated serum alanine aminotransferase levels, at least 20 times above the upper limit of normal; (ii) documented seroconversion of HCV antibodies; (iii) positive PCR for HCV RNA in at least the first serum sample; (iv) history of sudden onset of liver disease in previously healthy persons; and (v) exclusion of other possible causes of acute hepatitis (e.g., autoimmune liver disease, viral hepatitis, etc.).
TABLE 1.
Clinical characteristics of patients with acute hepatitis Ca
| Patient no. | Sex | Age (yr) | HCV genotype | Time after onset (mo) | HCV RNA | Outcome | HLA-DRB1 | No. of peptide epitopes | CD4+ T-cell clones (specificity) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 17 | 3 | 0.4 | + | Self limited | *0101, *0404 | 4 | |
| 2 | F | 40 | NA | 0.5 | − | Self limited | *0101, *0701 | 36 | p37, p64 |
| 3 | M | 59 | 1a | 0.7 | + | Self limited | DR3, DR15 | 20 | |
| 4 | M | 60 | 3 | 1.0 | − | Self limited | DR2, DR10 | 0 | |
| 5 | F | 43 | 1b | 1.0 | − | Self limited | DR2, DR11 | 14 | |
| 6 | M | 39 | 1a | 1.3 | − | Self limited | *0301, *1601 | 1 | p52 |
| 7 | F | 25 | 1b | 1.4 | − | Self limited | *1501 | 19 | p22b,c, p42 |
| 8 | F | 36 | 1b | 2.3 | + | Self limited | *0401, *1501 | 14 | |
| 9 | F | 22 | 4 | 2.4 | − | Self limited | *0301, *1501 | 4 | |
| 10 | F | 56 | 1b | 5.3 | − | Self limited | *0701, *1501 | 16 | |
| 11 | M | 20 | 1b | 7.8 | − | Self limited | *0101, *1104 | 4 | |
| 12 | M | 46 | 1b | 13.7 | − | Self limited | *0101, *1501 | 0 | p24, p64c, p71c |
| 13 | F | 29 | 3 | 66.2 | − | Self limited | *0102, *1104 | 12 | p60, p62, p64 |
| 14 | M | 48 | NA | 66.7 | − | Self limited | *1104, *1502 | 24 | p5b,c, p22b |
| 15 | F | 24 | NA | 0.6 | NA | Unknown | *0401, *1301 | 10 | |
| 16 | F | 34 | 1a | 0.7 | 2,000 | Chronic | DR2, DR13 | 13 | |
| 17 | M | 39 | 3 | 1.5 | − | Chronic | DR2, DR10 | 7 | |
| 18 | F | 40 | NA | 2.8 | 350 | Chronic | *0701, *1501 | 24 | |
| 19 | F | 23 | 1b | 3.5 | − | Chronic | *0102, *1501 | 2 | p22 |
| 20 | F | 33 | 1b | 8.3 | − | Chronic | DR1, DR10 | 10 | |
| 21 | M | 1b | Chronic | *0101, *1301 | p49 | ||||
| 22 | M | 3 | Chronic | *0402, *1101 | p61 |
Abbreviations: M, male; F, female; NA, not available.
CD4+ T-cell clones from these patients have been published previously by our group (3).
Of these specificities, more than one clone could be isolated from the patient.
HLA-DRB1* typing.
Low-resolution DRB1* typing was performed by sequence-specific oligonucleotide hybridization using the Biotest ELPHA DR low-resolution plates (Biotest, Dreieich, Germany) in a Tecan Genesis RMP robotic hybridization system (Tecan, Crailsheim, Germany) according to the manufacturer's instructions.
High-resolution DRB1* typing was obtained by group-specific PCR and subsequent DNA sequencing (for details and primer sequences see Table S2 in the supplemental material).
HCV proteins and peptides.
The following fragments of HCV proteins were purchased from Microgen Inc. (Munich, Germany): NS3 (aa 1007 to 1534) and NS4 (aa 1616 to 1863), expressed in Escherichia coli and purified by ion-exchange chromatography followed by preparative sodium dodecyl sulfate gel electrophoresis. In addition, recombinant HCV proteins comprising the NS3 and NS4 region of the HCV polyprotein (c33c = aa 1192 to 1457, c100 = aa 1569 to 1931, and c200 = aa 1192 to 1931) were kindly provided by M. Houghton (Chiron, Emeryville, CA). These proteins were expressed as COOH-terminal fusion proteins with human superoxide dismutase in yeast (Saccharomyces cerevisiae). Proteins were >90% pure.
Eighty-three overlapping peptides (20-mers) covering the region of aa 1207 to 2014 were synthesized by Chiron Mimotopes (Clayton, Australia). Truncated peptides covering aa 1535 to 1554, aa 1585 to 1604, aa 1655 to 1674, aa 1685 to 1704, aa 1765 to 1784, aa 1785 to 1804, aa 1805 to 1824, and aa 1875 to 1894 were synthesized by Chiron Mimotopes (Clayton, Australia) and EMC (Microcollections, Tübingen, Germany). Lyophilized peptides were reconstituted at 20 mg/ml in dimethyl sulfoxide (Roth, Karlsruhe, Germany) and were diluted to 1 mg/ml in RPMI 1640 medium (Biochrom, Berlin, Germany).
Proliferation assay.
PBMC were isolated on Ficoll-Hypaque gradients (Biochrom, Berlin, Germany) and washed four times in phosphate-buffered saline. PBMC (5 × 104/well) were incubated in 96-well U-bottom plates (TPP, Trasadingen, Switzerland) for 5 days in the presence of HCV proteins (2 μg/ml; c33c, c100, or c200) or synthesized peptides (10 μg/ml) in 150 μl of tissue culture medium (RPMI 1640 medium; Biochrom, Berlin, Germany) containing 2 mM l-glutamine, 1 mM sodium pyruvate (Serva, Heidelberg, Germany), 100 U of penicillin (Biochrom, Berlin, Germany) per ml, 100 μg of streptomycin (Biochrom, Berlin, Germany) per ml, and 5% human AB serum (Pan, Aidenbach, Germany). Cultures were labeled by incubation for 16 h with 2 μCi of [3H]thymidine (specific activity, 80 mCi/mmol; Amersham, Little Chalfont, United Kingdom). The cells were collected and washed on filters (Dunn, Asbach, Germany) by using a cell harvester (Skatron, Sterling, VA), and the amount of radiolabel incorporated into DNA was estimated with a beta counter (LKB/Pharmacia, Uppsala, Sweden). Triplicate cultures were assayed routinely, and the results are expressed as mean counts per minute. The stimulation index was calculated as the ratio of counts per minute obtained in the presence of antigen to that obtained without antigen. A stimulation index of >3, corresponding to the mean + 2 standard deviations of 10 healthy, HCV-negative controls, was considered significant
Clone cells (1 × 104 to 3 × 104) were incubated with antigen-presenting cells (autologous PBMC or HLA-DR-matched PBMC) in the presence of HCV protein (1 mg/ml) or synthesized peptides (10 μg/ml). After 3 to 5 days cultures were labeled by incubation for 16 h with 2 μCi of [3H]thymidine (specific activity, 80 mCi/mmol; Amersham, Little Chalfont, United Kingdom). Subsequently, the proliferation assay was performed as described above.
Generation of T-cell clones and specificity testing.
PBMC (5 × 104/well) of seven patients with acute hepatitis C (patient characteristics are listed in Table 1) were incubated in 96-well U-bottom plates (TPP, Trasadingen, Switzerland) in the presence of HCV proteins (2 μg/ml) in 150 μl of tissue culture medium. On day 6, recombinant interleukin 2 (IL-2) was added to a final concentration of 200 U/ml (IL-2 was a gift of A. de Baey, Basel Institute for Immunology). On day 10 cells were cloned at 0.5 cells/well in the presence of 3 × 104 irradiated PBMC per well, 200 U of IL-2 per ml, and 2 μg of phytohemagglutinin (Murex Biotech Ltd., Kent, United Kingdom) per ml. After 3 to 5 weeks clones were tested for specificity to HCV antigens. Clone cells (104 to 105) were added to 3 × 104 autologous or HLA-DR-matched, irradiated PBMC with and without 1 μg/ml HCV protein or 10 μg/ml HCV peptide and cultured for 5 days. Then the proliferation assay was performed as described above.
For expansion, T-cell clones were stimulated every 2 to 5 weeks with irradiated allogeneic PBMC, 200 U/ml IL-2, and 2 μg/ml phytohemagglutinin.
Determination of HLA restriction.
For determination of HLA restriction proliferation assays were performed in the presence or absence of anti-HLA class II antibodies anti-DR (Becton Dickinson, San Jose, CA), anti-DP (Becton Dickinson, San Jose, CA), and anti-DQ (Becton Dickinson, San Jose, CA). Autologous or HLA-DR-matched PBMC were pulsed with the HCV protein for 24 h, and then the antigens were washed off. After irradiation of the PBMC the clone cells and the antibody (10 μl) were added. After identification of the presenting class II molecule, fine analysis was performed using the following partially matched, homozygous, lymphoblastoid cell lines as antigen-presenting cells: KAS116 (DRA1*0101, DRB1*0101, DRB6*0101, DQA1*0101, DQB1*0501, DPA1*0201, and DPB1*1301), JESTHOM (DRA1*0101, DRB1*0101, DRB6*0101, DQA1*0101, DQB1*0501, DPA1*01, and DPB1*0401), PMG075 (DRA1*0101, DRB1*0102, DRB6*0101, DQA1*0101, DQB1*0501, DPA1*01/0201, and DPB1*0301/0401), SCHU (DRA1*0102, DRB1*1501, DRB5*0101, DRB6*0201, DQA1*0102, DQB1*0602, DPA1*01, and DPB1*0402), DUCAF (DRA1*0101, DRB1*0301, DRB3*0202, DQA1*0501, DQB1*0201, DPA1*01, and DPB1*0202), RSH (DRA1*0102, DRB1*0302, DRB3*0101, DQA1*0401, DQB1*0402, DPA1*02022/0301, and DPB1*0101/0401), SPO010 (DRA1*0101, DRB1*1101, DRB3*0202, DQA1*0102, DQB1*0502, DPA1*01, and DPB1*02012), BM21 (DRA1*0101, DRB1*1101, DRB3*0202, DQA1*0501, DQB1*0301, DPA1*0201, and DPB1*1001), CB6B (DRA1*0101, DRB1*1301, DRB3*0202, DQA1*0103, DQB1*0603, DPA1*02021, and DPB1*1901), HO301 (DRA1*0102, DRB1*1302, DRB3*0301, DQA1*0102, DQB1*0605, DPA1*0201, and DPB1*0501), MOU (DRA1*0101, DRB1*07, DRB4*0101, DQA1*0201, DQB1*0201, DPA1*01, and DPB1*02012), PITOUT (DRA1*0101, DRB1*07, DRB4*0101, DQA1*0201, DQB1*0201, DPA1*01, and DPB1*0401). Fine analysis was performed with a fluorescence-activated cell sorting-based test system as described previously. In brief, a total of 1 × 104 to 3 × 104 clone cells were incubated with 3 × 104 antigen-pulsed antigen-presenting cells in 96-well V-bottom plates for 16 h, washed, incubated with CD25-fluorescein isothiocyanate (Immunotech, Marseille, France) and CD4 TC (Caltag, Burlingame, CA) for 30 min at 4°C, and washed again. Fluorescence was measured with a FACScan cell sorter (Becton Dickinson). A gate was set for CD4+ T cells, and binding of CD25 antibodies was expressed as median fluorescence intensity.
RESULTS
Patients selected for PBMC stimulation with overlapping peptides.
Fresh PBMC of 20 patients with current or past acute hepatitis C who had mounted a CD4+ T-cell response against recombinant NS3 or NS4 antigens before were tested with a set of 83 20-mer peptides, overlapping by 10 amino acids, and covering amino acid positions 1207 to 2014 of a genotype 1b HCV strain. In 13 patients, the assay was performed within the first 3 months of clinical onset of acute hepatitis C, in two patients it was performed between month 3 and month 6, and in five patients it was performed during follow-up after viral clearance (Table 1). Fourteen patients had a self-limited course of acute hepatitis C, five developed chronic hepatitis C, and one patient was lost to follow-up. Of the patients with acute self-limited hepatitis C tested in the early phase, three were still HCV RNA positive whereas the remainder were already HCV PCR negative. Of the five chronically evolving patients three were HCV PCR negative at the time of testing but relapsed later and the other two had low viral loads of 2,000 and 350 IU/ml, so that all T-cell responses measured in this assay correspond to a phase of viral control (Table 1 shows more details). Importantly, in all chronically evolving patients, the NS3-NS4-specific T-cell proliferation was lost upon viral relapse (data not shown), emphasizing the association with sustained HCV-specific CD4+ T-cell responses and long-term viral clearance.
Results of PBMC stimulation with overlapping peptides.
Eighteen of 20 patients had a significant proliferative CD4+ T-cell response to at least one of the NS3-NS4-derived peptides at the time of testing. In representative experiments, depletion of CD4+ T cells led to complete abrogation of antigen-specific proliferation (data not shown), suggesting that the 20-mer peptides preferentially stimulated CD4+ T cells. The patients responded to a mean of 11.7 different epitopes (range, 0 to 36), and only 11 peptides were never recognized by any of the patients.
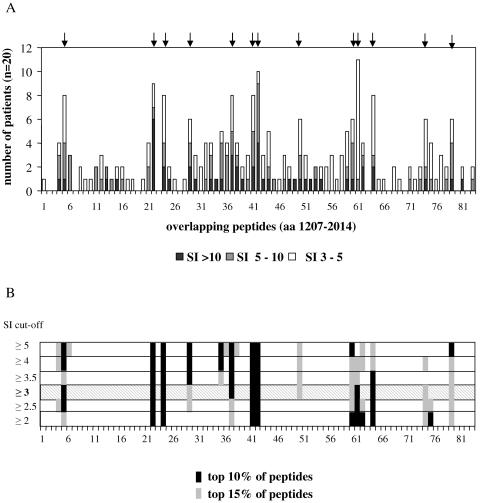
Importantly, however, 13 peptides were recognized by 30% or more of the patients, of which eight peptides were recognized by at least 40% (Fig. 1). The amino acid positions and the sequences of the peptides are summarized in Table 2.
FIG. 1.
Proliferation assay with 83 peptides covering NS3 helicase and NS4. PBMC of 20 patients with current or past acute hepatitis C were tested with a set of 83 20-mer peptides, overlapping by 10 amino acids, and covering amino acid positions 1207 to 2014 of a genotype 1b HCV strain by proliferation assay. The results are shown as numbers of significant reactions (i.e., stimulation index [SI] of >3) per peptide (A). The peptides which were recognized by 30% or more of the 20 patients are marked by arrows. If the cutoff for significance is varied from >2 to >5, only minor changes are observed in the hierarchy of immunodominant epitopes (B).
TABLE 2.
T-cell epitopes recognized by PBMC of at least 30% of patients
| No. | Amino acid position | Sequence (genotype 1b) | Frequency of recognition [no. (%)] | Specific CD4+ T-cell clones |
|---|---|---|---|---|
| p5 | 1242-1261 | AAYAAQGYKVLVLNPSVAAT | 8 (40) | (+)a |
| p22 | 1388-1407 | GRHLIFCHSKKKCDELATKL | 9 (45) | + |
| p24 | 1406-1425 | KLVAMGINAVAYYRGLDVSV | 8 (40) | + |
| p29 | 1450-1469 | SVIDCNTCVTQTVDFSLDPT | 6 (30) | (+)a |
| p37 | 1535-1554 | TTVRLRAYMNTPGLPVCQDH | 8 (40) | + |
| p41 | 1575-1594 | QTKQSGENLPYLVAYQATVC | 8 (40) | + |
| p42 | 1585-1604 | YLVAYQATVCARAQAPPPSW | 10 (50) | + |
| p50 | 1665-1674 | GVLAALAAYCLSTGCVVIVG | 6 (30) | − |
| p60 | 1765-1784 | MWNFISGIQYLAGLSTLPGN | 6 (30) | + |
| p61 | 1775-1794 | LAGLSTLPGNPAIASLMAFT | 11 (55) | + |
| p64 | 1805-1824 | QTLLFNILGGWVAAQLAAPG | 8 (40) | + |
| p74 | 1905-1924 | HVGPGEGAVQWMNRLIAFAS | 6 (30) | − |
| p79 | 1955-1974 | QLLRRLHQWISSECTTPCSG | 6 (30) | − |
CD4+ T-cell clones specific for this epitope have previously been characterized and published by our group (3).
NS3-NS4-specific CD4+ T-cell clones.
NS3-NS4-specific CD4+ T-cell clones were generated by stimulating PBMC with recombinant antigen (aa 1192 to 1972) and subsequent cloning by limiting dilution. Specific clones were isolated from 7 of the 20 patients who were also studied in the peptide assay and two additional patients with acute hepatitis C (Table 1). CD4+ T-cell clones against 11 epitopes were generated, one of which (p22) has been partially characterized by us before. Importantly, the T-cell clones recognized 8 of the 13 immunodominant epitopes identified by the peptide library in PBMC. For seven of the eight most immunodominant epitopes at least one specific CD4+ T-cell clone could be isolated.
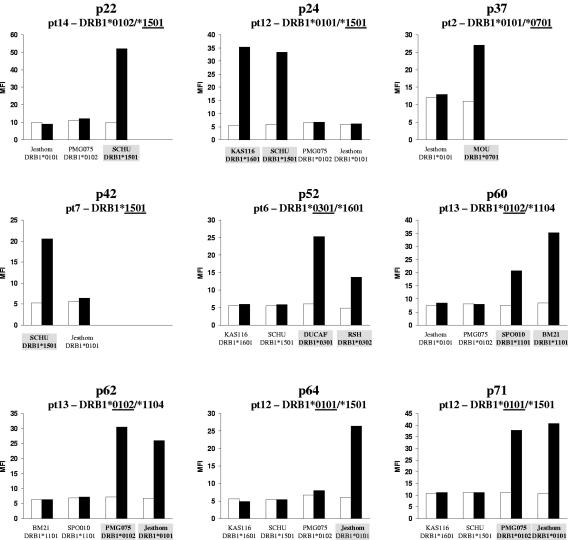
To define the HLA restriction, first inhibition with HLA-DR, -DP, and -DQ antibodies was performed and revealed HLA-DR for all epitopes. Subsequently, CD4+ T-cell clones were stimulated in the presence of partially matched homozygous lymphoblastoid cell lines (Fig. 2). For all but two (p49 and p61) the HLA restriction of specific CD4+ T-cell clones could be determined. Three T-cell clones were HLA-DRB1*0101 restricted (specific for p62, p64, and p71), three T-cell clones were restricted by HLA-DRB1*1501 (specific for p22, p24, and p42), and one T-cell clone each was restricted by HLA-DRB1*0301 (p52), HLA-DRB1*0701 (p37), and HLA-DRB1*1104 (p60), respectively (Table 3). All HLA-DRB1*0101-restricted T-cell clones were cross-reactive with HLA-DRB1*0102, irrespective of whether the donor of the T cells was HLA-DRB1*0101 (patients 2 and 12) or *0102 (patient 13). Similarly, for the HLA-DRB1*1501-restricted epitopes, a patient with HLA-DRB1*1502 (patient 14) responded to all three epitopes, suggesting that the minimal differences between these HLA-DR alleles do not play a major role for T-cell recognition.
FIG. 2.
HLA restriction of NS3/4-specific CD4+ T-cell clones. NS3/4-specific CD4+ T-cell clones were incubated with partially matched homozygous lymphoblastoid cell lines carrying one of the patient's HLA-DR alleles, in the presence of recombinant NS3/4 protein (black bars) or medium control (empty bars). After 16 h activation of specific CD4+ T-cell clones was determined by fluorescence-activated cell sorting for CD25 expression and shown as mean fluorescence intensity (MFI).
TABLE 3.
Mapping of the minimal, stimulatory sequence within CD4+ T-cell epitopes
| No. | aa | Patient(s) | Source | Minimal/optimal epitopea | HLA restriction |
|---|---|---|---|---|---|
| p22 | 1388-1407 | 14, 19 | Clones | GRHLIFCHSKRKCDELATKL | DRB1*1501/DRB5*0501 |
| 14 | PBMC | GRHLIFCHSKR | DRB1*1502/*1104 | ||
| p24 | 1406-1425 | 12 | Clone | KLVAMGINAVAYYRGLDVSV | DRB1*1501 |
| PBMC | |||||
| p37 | 1535-1554 | 2 | Clone | TTVRLRAYMNTPGLPVCQDH | DRB1*0701 |
| 2 | PBMC | AYMNTPGLPV | DRB1*0101/*0701 | ||
| 14 | PBMC | LRAYMNTPGLPV | DRB1*1502/*1104 | ||
| p42 | 1585-1604 | 7 | Clone | YLVAYQATVCARAQAPPPSW | DRB1*1501 |
| 3 | PBMC | YLVAYQATVC | DRB1*0301/*1501 | ||
| 2 | PBMC | NLPYLVAYQATVCAR | DRB1*0101/*0701 | ||
| p49 | 1655-1674 | 21 | Clone | VVTSTWVLVGGVLAALAAYC | DRB1*0101/*1301 |
| 2 | PBMC | TSTWVLVGGVLAA | DRB1*0101/*0701 | ||
| p52 | 1685-1704 | 6 | Clone | RVVLSGKPAIIPDREVLYRE | DRB1*0301 |
| 3 | PBMC | IIPDREVLYR | DRB1*0301/*1501 | ||
| p60 | 1765-1784 | 13 | Clone | MWNFISGIQYLAGLSTLPGN | DRB1*1104 |
| 14 | PBMC | YLAGLSTLPG | DRB1*1502/*1104 | ||
| p61 | 1775-1794 | 22 | Clone | LAGLSTLPGNPAIASLMAFT | DRB1*0402/*1101 |
| p62 | 1785-1804 | 13 | Clone | PAIASLMAFTAAVTSPLTTS | DRB1*0102 |
| 14 | PBMC | PAIASLMAFTA | DRB1*1502/*1104 | ||
| p64 | 1805-1824 | 2, 12, 13 | Clones | QTLLFNILGGWVAAQLAAPG | DRB1*0101, *0102 |
| 21 | PBMC | FNILGGWVAAQL | DRB1*0101/*1301 | ||
| 2 | PBMC | FNILGGWVA | DRB1*0101/*0701 | ||
| p71 | 1875-1894 | 12 | Clone | STEDLVNLLPAILSPGALVV | DRB1*0101 |
| 2 | PBMC | NLLPAIL | DRB1*0101, *0701 |
Boldface amino acids are required for optimal T-cell stimulation; the underlined amino acids represent the minimal epitope that induces significant T-cell stimulation.
Obviously, individual T-cell clones may not necessarily be representative for the total T-cell response against a particular epitope in the same or different patients and some CD4+ T-cell epitopes have promiscuous HLA binding characteristics. To study this further, we compared the proliferative PBMC response in the 20 patients with respect to the presence or absence of the HLA-DR allele which was identified by the CD4+ T-cell clone (Fig. 3). For the HLA-DRB1*1501/DRB5*0101-restricted epitopes p22 and p24, a significant proliferation was present in 7 and 8 of 13 HLA-DR15 patients, respectively, but in only one of seven HLA-DR15-negative patients (P < 0.05). For p42, two HLA-DR15-negative patients also showed a significant proliferative PBMC response, suggesting that other HLA class II alleles can also present this epitope or an overlapping epitope contained in the 20-mer peptide. For the peptides p37, p60, and p64, HLA restriction seemed to be less constrained, although the HLA restriction identified by the CD4+ T-cell clone in each case was the dominant restriction element. Surprisingly, p64 was recognized by 4 of 13 HLA-DR1-negative patients, although CD4+ T-cell clones from three different patients were exclusively HLA-DR1 restricted. This implies that p64 is presented by HLA class II molecules other than HLA-DR1 but that it may be particularly immunodominant in HLA-DR1-positive individuals.
FIG. 3.
Analysis of patient HLA-DRB1 and PBMC response to immunodominant epitopes. For six peptides the proliferative PBMC responses in 20 patients were analyzed with respect to the presence or absence of the HLA-DR allele which was identified as presenting the HLA molecule by CD4+ T-cell clones (x axis). The stimulation index of each patient is represented by the black dots (y axis). The numbers of patients with positive PBMC responses to the epitope in relation to the total numbers of patients with the respective HLA allele are shown above the panels. For p22 and p24, a significant proliferation was significantly associated with the presence of the HLA-DRB1*15 alleles (**, P < 0.01; *, P < 0.05).
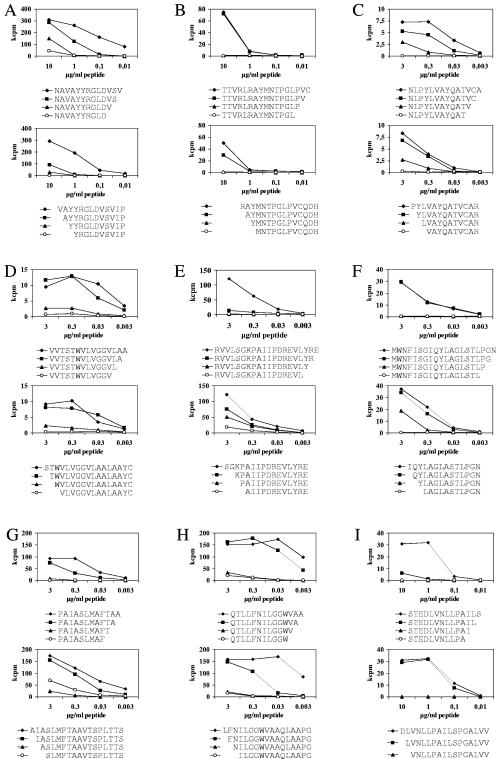
Mapping of minimal CD4+ T-cell epitopes.
For nine epitopes we synthesized N-terminal and C-terminal stepwise truncated peptide variants (up to 25 variants per epitope). Specific CD4+ T-cell clones were first tested against the complete set of truncated variants at high peptide concentrations (10 μg/ml). Subsequently, the four peptides neighboring the position where stimulatory capacity was lost were selected, and the CD4+ T-cell clones were retested over a broad range of different peptide concentrations (Fig. 4). Although the loss of stimulatory activity sometimes depended on addition or removal of a single amino acid, more frequently a gradual loss of stimulatory activity, over one to three amino acid positions, was observed. We therefore distinguished optimal stimulatory peptides, which yielded the strongest T-cell proliferation and were between 10 and 15 amino acids long, from minimal stimulatory peptides, which were the shortest fragments still able to elicit a significant proliferative CD4+ T-cell response and which were between 8 and 13 amino acids long (Table 3).
FIG. 4.
Mapping of minimal epitopes of HCV-specific CD4+ T-cell clones. Nine sets of synthesized N-terminal and C-terminal stepwise truncated peptide variants (up to 25 variants per epitope) were synthesized, and specific CD4+ T-cell clones were tested against the complete set of truncated variants. The four peptides neighboring the position where stimulatory capacity was lost were selected, and nine CD4+ T-cell clones (A to I) were stimulated with the peptides over a broad range of concentrations (x axis). The results are shown as counts per minute (y axis). Minimal stimulatory sequences were between 8 and 13 amino acids in length, but optimal T-cell stimulation was usually observed with 10- to 15-mers.
Similar to the HLA restriction, minimal stimulatory sequences may vary for different T-cell clones although they are still specific for the same epitope. Although PBMC contain polyclonal T cells potentially recognizing different, overlapping epitopes within a given sequence, we attempted to repeat the epitope mapping with PBMC from different patients. Since stimulation indices for PBMC are usually much lower than those for T-cell clones, which may compromise the correct interpretation of the data, only PBMC from patients with very strong reactivities to the given epitope were selected, limiting the analysis to seven epitopes, since for two epitopes no appropriate donor was available. Importantly, the T-cell clones and the PBMC yielded very similar epitopes which in most cases differed by no more than one or two amino acids at the N or C terminus. Even for the epitopes p37 and p62, for which no HLA-matched PBMC donor with strong reactivity was available, the recognized minimal epitopes were almost identical. This suggests that for many epitopes there is a limited variability of the minimal stimulatory sequence within a given epitope, even if the epitope is presented by a different HLA class II molecule.
DISCUSSION
The detailed characterization of immunodominant CD4+ T-cell epitopes is prerequisite (i) to the rational design of peptide-based, T-cell-targeted immunotherapies; (ii) for the synthesis of HLA class II tetramers as an important new tool for the monitoring of natural and vaccine-induced immune responses; and (iii) for a better understanding of the relevance of viral variability and immune escape.
Since several reports have suggested that the NS3-NS4 region is immunodominant for CD4+ T cells (4, 7, 14), we have focused our study on an approximately 800-amino-acid stretch from position 1207 to amino acid 2014, covering the NS3 helicase, NS4A, and NS4B. Using overlapping 20-mer peptides to stimulate PBMC directly ex vivo, we identified 13 peptides that were recognized by ≥30% of patients with acute or resolved hepatitis C. Of these, eight peptides were also recognized by specific CD4+ T-cell clones in independent cloning procedures. Moreover, looking at the eight peptides that were recognized by ≥40% of patients, seven could also be confirmed by CD4+ T-cell clones. Surprisingly, one immunodominant epitope against which no T-cell clones were raised was the epitope aa 1248 to 1261. In previous studies, this epitope was recognized by NS3-specific CD4+ T-cell lines and clones from four of five (3) and eight of eight (11) patients, respectively. In both studies, however, all but one patient responding to aa 1248 to 1261 carried either the HLA-DRB1*11/DQB1*0301 haplotype or the HLA-DRB1*0401 allele. Unintentionally, these HLA class II alleles were present in only one of eight patients who were selected for T-cell cloning, which may explain the lack of T-cell clones specific for this epitope in the current study.
Presentation of viral epitopes to CD4+ T cells critically depends on the HLA class II molecule involved. Since the HLA class II gene locus is the most polymorphic gene locus of the human genome, with several hundred different alleles, a complete mapping of CD4+ T-cell epitopes that can be immunogenic in the context of a certain HLA class II molecule is elusive. The aim of this study was therefore to identify epitopes that, first, are presented by common HLA class II alleles and, second, are frequently recognized in the group of patients carrying the particular allele. Only such epitopes can be reasonable candidates to be included in T-cell-targeted immunotherapies and obviously only for these epitopes is construction of HLA class II tetramers likely to be meaningful if applied to the analysis of CD4+ T-cell kinetics. In this study, the HLA restrictions could be unequivocally identified for nine T-cell epitopes and all were restricted by HLA class II alleles that occur at frequencies of 10 to 30% in most ethnicities (http://www.ncbi.nlm.nih.gov/projects/mhc/ihwg.fcgi?cmd=page&page=AnthroMain). Importantly, five epitopes (p22, p24, p37, p42, and p64) were recognized by more than 50% of patients carrying the appropriate HLA class II allele in a single proliferation assay. CD4+ T-cell epitopes have been described as being rather promiscuous with regard to HLA class II binding (13), and this has also been shown for several HCV-derived epitopes, e.g., epitope 1248 to 1261 (3). However, in our study, recognition of the epitopes p22, p24, and p42 was virtually restricted to the group of patients carrying HLA-DRB1*1501. Based on these results, an NS4-specific HLA-DR1 tetramer could be synthesized, which stained specific CD4+ T-cell clones and successfully identified specific CD4+ T cells in PBMC from patients with acute hepatitis C at a frequency of up to 0.7% of CD4+ T cells (M. Lucas et al., 11th International Symposium on Hepatitis C and Related Viruses, Heidelberg, Germany, 3 to 7 October 2004).
HCV is a highly variable virus, frequently circulating as a complex mixture of quasispecies (6). All immunological techniques so far make use of peptides or recombinant proteins to identify specific T cells, and consequently only T cells specific for, or cross-reacting with, the chosen sequence will be found. In this study we have used genotype 1b-derived peptides and genotype 1a-derived recombinant antigens and found high agreement between T-cell responses. In fact, if the minimal epitopes are analyzed for genetic variability, 8 of 10 epitopes are highly conserved between genotypes (see Table S3 in the supplemental material) and also within a given genotype (see Table S4 in the supplemental material), with >94% of all published genotype 1b sequences being identical. The notable exception is p52, which is located within NS4A, which is extremely variable with 26 different genotype 1b variants in the database (http://hcvpub.ibcp.fr), none of which occurs more frequently than 16.6%. Therefore, in the case of p52, the fact that only 1 of 20 patients responded in our PBMC assay may significantly underestimate the immunogenicity of this epitope. Accordingly, other, more variable epitopes may exist that have escaped detection due to sequence variability. Those will become detectable only with reagents synthesized according to the individual patient's infecting HCV isolate(s), which will be possible only in selected cases. From a practical point of view, however, the conserved epitopes are certainly the preferred candidates for use in peptide-based vaccines and for the synthesis of HLA class II tetramers.
In conclusion, we identified a set of CD4+ T-cell epitopes within the NS3-NS4 region which are immunodominant in patients achieving transient or persistent viral control. The majority of the epitopes are highly conserved and are recognized in the context of common HLA-DR alleles, so that these epitopes—in combination with appropriate CD8+ T-cell epitopes—may be principal candidates for use in trials of peptide-based immunotherapy to treat chronic hepatitis C.
Supplementary Material
Acknowledgments
We thank Jutta Döhrmann, Carmen Amsel, and Daniela Novak for excellent technical assistance.
This work was supported by the European Commission (QLK2-CT-2002-01329), the HepNet (Kompetenznetz Hepatitis, Teilprojekt 10.2.1), and the Wilhelm-Sander-Stiftung (2003.044.1). This work is part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance supported by a grant (LSHM-CT-2004-503359) from the Priority 1 “Life Sciences, Genomics and Biotechnology for Health” program in the 6th Framework Programme of the EU.
J. T. Gerlach and A. Ulsenheimer contributed equally to this work.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alter, M. J. 1999. Hepatitis C virus infection in the United States. J. Hepatol. 31:88-91. [DOI] [PubMed] [Google Scholar]
- 2.Day, C. L., G. M. Lauer, G. K. Robbins, B. McGovern, A. G. Wurcel, R. T. Gandhi, R. T. Chung, and B. D. Walker. 2002. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J. Virol. 76:12584-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diepolder, H. M., J. T. Gerlach, R. Zachoval, R. M. Hoffmann, M. C. Jung, E. A. Wierenga, S. Scholz, T. Santantonio, M. Houghton, S. Southwood, A. Sette, and G. R. Pape. 1997. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J. Virol. 71:6011-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, E. A. Wierenga, T. Santantonio, M. C. Jung, D. Eichenlaub, and G. R. Pape. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 346:1006-1007. [DOI] [PubMed] [Google Scholar]
- 5.Eckels, D. D., H. Zhou, T. H. Bian, and H. Wang. 1999. Identification of antigenic escape variants in an immunodominant epitope of hepatitis C virus. Int. Immunol. 11:577-583. [DOI] [PubMed] [Google Scholar]
- 6.Farci, P., and R. H. Purcell. 2000. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 20:103-126. [PubMed] [Google Scholar]
- 7.Gerlach, J. T., H. M. Diepolder, M. C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 8.Godkin, A., N. Jeanguet, M. Thursz, P. Openshaw, and H. Thomas. 2001. Characterization of novel HLA-DR11-restricted HCV epitopes reveals both qualitative and quantitative differences in HCV-specific CD4+ T cell responses in chronically infected and non-viremic patients. Eur. J. Immunol. 31:1438-1446. [DOI] [PubMed] [Google Scholar]
- 9.Harcourt, G. C., M. Lucas, I. Sheridan, E. Barnes, R. Phillips, and P. Klenerman. 2004. Longitudinal mapping of protective CD4+ T cell responses against HCV: analysis of fluctuating dominant and subdominant HLA-DR11 restricted epitopes. J. Viral Hepat. 11:324-331. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann, R. M., H. M. Diepolder, R. Zachoval, F. M. Zwiebel, M. C. Jung, S. Scholz, H. Nitschko, G. Riethmuller, and G. R. Pape. 1995. Mapping of immunodominant CD4+ T lymphocyte epitopes of hepatitis C virus antigens and their relevance during the course of chronic infection. Hepatology 21:632-638. [PubMed] [Google Scholar]
- 11.Lamonaca, V., G. Missale, S. Urbani, M. Pilli, C. Boni, C. Mori, A. Sette, M. Massari, S. Southwood, R. Bertoni, A. Valli, F. Fiaccadori, and C. Ferrari. 1999. Conserved hepatitis C virus sequences are highly immunogenic for CD4+ T cells: implications for vaccine development. Hepatology 30:1088-1098. [DOI] [PubMed] [Google Scholar]
- 12.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lund, O., M. Nielsen, C. Kesmir, A. G. Petersen, C. Lundegaards, P. Worning, C. Sylvester-Hvid, K. Lamberth, G. Roder, S. Justesen, S. Buus, and S. Brunak. 2004. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 55:797-810. [DOI] [PubMed] [Google Scholar]
- 14.Missale, G., R. Bertoni, V. Lamonaca, A. Valli, M. Massari, C. Mori, M. G. Rumi, M. Houghton, F. Fiaccadori, and C. Ferrari. 1996. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J. Clin. Investig. 98:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penna, A., G. Missale, V. Lamonaca, M. Pilli, C. Mori, P. Zanelli, A. Cavalli, G. Elia, and C. Ferrari. 2002. Intrahepatic and circulating HLA class II-restricted, hepatitis C virus-specific T cells: functional characterization in patients with chronic hepatitis C. Hepatology 35:1225-1236. [DOI] [PubMed] [Google Scholar]
- 16.Rahman, F., T. Heller, Y. Sobao, E. Mizukoshi, M. Nascimbeni, H. Alter, S. Herrine, J. Hoofnagle, T. J. Liang, and B. Rehermann. 2004. Effects of antiviral therapy on the cellular immune response in acute hepatitis C. Hepatology 40:87-97. [DOI] [PubMed] [Google Scholar]
- 17.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai, S. L., Y. F. Liaw, M. H. Chen, C. Y. Huang, and G. C. Kuo. 1997. Detection of type 2-like T-helper cells in hepatitis C virus infection: implications for hepatitis C virus chronicity. Hepatology 25:449-458. [DOI] [PubMed] [Google Scholar]
- 19.Wertheimer, A. M., C. Miner, D. M. Lewinsohn, A. W. Sasaki, E. Kaufman, and H. R. Rosen. 2003. Novel CD4+ and CD8+ T-cell determinants within the NS3 protein in subjects with spontaneously resolved HCV infection. Hepatology 37:577-589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.