Abstract
The capsid of infectious bursal disease virus (IBDV), a nonenveloped virus of the family Birnaviridae, has a T=13l icosahedral shell constituted by a single protein, VP2, and several disordered peptides, all derived from the precursor pVP2. In this study, we show that two of the peptides, pep11 and pep46, control virus assembly and cell entry. Deletion of pep11 or even simple substitution of most of its residues blocks the capsid morphogenesis. Removal of pep46 also prevents capsid assembly but leads to the formation of subviral particles formed by unprocessed VP2 species. Fitting with the VP2 atomic model into three-dimensional reconstructions of these particles demonstrates that the presence of uncleaved pep46 causes a steric hindrance at the vertices, blocking fivefold axis formation. Mutagenesis of the pVP2 maturation sites confirms that C terminus processing is necessary for VP2 to acquire the correct icosahedral architecture. All peptides present on virions are accessible to proteases or biochemical labeling. One of them, pep46, is shown to induce large structural rearrangements in liposomes and to destabilize target membranes, demonstrating its implication in cell entry.
Birnaviruses are nonenveloped double-stranded RNA (dsRNA) viruses that include economically important animal pathogens (12). Like all dsRNA viruses, the genomes of birnaviruses need to remain hidden throughout the viral life cycle, avoiding detection by the host defense mechanisms. Viral transcription and replication occur within the viral particle. Consequently, birnaviruses have to cross the target cell membrane in order to reach the nucleotide pool of the cytoplasm, where they initiate their replication cycle. To insure viral translocation into the cell, most dsRNA viruses exhibit a capsid constituted by several concentric protein shells (19, 24, 29). The most external shell is generally lost during the cell entry process and is thought to be involved in a local destabilization of the target membrane (5). The remaining inner shell, generally referred to as the “core,” constitutes the viral transcription machinery (18). In contrast to these viruses, birnavirus particles possess a single-layer capsid, which is competent for both transcription (6, 31) and membrane translocation. The molecular mechanisms governing the crossing of single-shell dsRNA viruses through the barrier represented by a target cell membrane are currently not understood.
Birnaviruses display an icosahedral architecture of triangulation T=13 laevo (3, 28). The crystal structures of an avian birnavirus, the infectious bursal disease virus (IBDV), and of its capsid protein, VP2, were recently determined to a resolution of 7 and 3Å, respectively. Fitting the atomic model of the capsid protein into the virion electron density map revealed that VP2 is the only component of the virus icosahedral capsid. Surprisingly, all other proteins (VP1 and VP3) and peptides present in the viral particle are not visible in the icosahedral symmetric shell (10).
The major components of the birnavirus particle arise from the proteolytic processing of the polyprotein pVP2-VP4-VP3 (referred herein as IBDA polyprotein), encoded by one of the two genomic segments (segment A). VP4 is the viral protease that cleaves the polyprotein, releasing proteins pVP2 and VP3 within the infected cell (1). In the case of IBDV, subsequent serial cleavage at the C terminus of pVP2 by VP4 gives rise to the mature VP2 protein and four peptides composed of 46, 7, and 11 amino acids (aa) (pep46, pep7a, pep7b, and pep11; see Fig. 1), all associated with the virus particle (11). VP1, the viral RNA-dependent RNA polymerase, is covalently linked to the two genomic segments and is encoded by segment B. Free VP1 molecules are also present within viral particles (12).
FIG. 1.
Schematic representation of IBDA polyprotein mutants in the pVP2 C-terminal domain and resulting assemblies. At the top of the figure, the IBDA polyprotein and the polyprotein fused at its C terminus with the GFP via a 7-amino-acid linker are represented. The sequences of the structural peptides pep46, pep7a, pep7b, and pep11, deriving from the processing of the C-terminal domain of pVP2, are reported according to the single-letter code. Numbering refers to the amino acid position in the polyprotein. Deletions and substitutions were engineered at the C terminus of pVP2. The deleted amino acids are indicated by asterisk and the substitutions by the amino acid encoded. The 30 central amino acids of pep46 are represented by three dots. The four types of assembly (T=13 VLP, pVP2 tubes, pVP2 irregular particles, and VP2* subviral particles) generated by the mutants and purified by density gradients are indicated on the left. Representative samples collected from the bands present in the gradients were negatively stained with 2% uranyl acetate and are shown at the top right of the figure.
Birnavirus particle morphogenesis is a complex process controlled by interactions of VP3 with both pVP2 and VP1 (8, 21, 22, 27). Expression of the pVP2-VP4-VP3 polyprotein gene in the absence of VP1 results in the assembly of tubes displaying helical symmetry and composed of unprocessed pVP2 (9, 23). While expression of the gene segment corresponding to VP2 (441 amino acids) in the absence of other viral components leads to the formation of T=1 subviral particles (4), expression of the VP2 precursor, pVP2 (512 amino acids), leads to the formation of irregular particles (9). Coexpression of the polyprotein and VP1 genes leads to the sequential maturation of pVP2 and the concomitant pVP2/VP2/VP3/VP1 assembly into T=13 virus-like particles (VLP) that are morphologically identical to the virions (8, 22, 28). Whereas the VLP shell is constituted by 260 VP2 trimers, the subviral particles contain only 20 trimers. VLP formation requires the negative charges at the C terminus of VP3 to be screened by VP1, as in the virus particle, or by any exogenous peptide fused at its C terminus or simply deleted (8, 9). Whereas VP3 plays a pivotal role in viral morphogenesis, it is not closely associated with VP2 in the virus particle (11), and the VP2/VP3 stoichiometry varies for different birnaviruses (14). Moreover, during viral assembly, the maturation of pVP2 gives rise to peptides with functions as yet unknown.
This study shows that peptide pep46 controls the morphogenesis of the viral particles formed by VP2. We also demonstrate that peptide pep11 controls the maturation of pVP2, in particular allowing the release of pep46 from VP2. Furthermore, the three-dimensional (3D) structure of VP2 particles indicates that pep46, when unreleased from VP2, blocks the assembly of the fivefold axis by steric hindrance during capsid assembly. Surprisingly, three of the four peptides were found to be accessible to biochemical labeling on the virion, demonstrating their localization at the external surface of the particle. In addition to controlling viral morphogenesis, it was also found that pep46 interacts with liposomes and destabilizes the target cell membrane, strongly suggesting an additional role in the cell entry process. Consequently, these results show that birnaviruses exhibit a unique strategy to control both assembly and cell entry based on the biophysical properties of a 46-amino-acid peptide endowed with dual functions.
MATERIALS AND METHODS
Plasmid and recombinant baculovirus constructs.
Plasmid pFBΔIBDA-GFP, encoding the IBDA polyprotein fused at its C terminus to the green fluorescent protein (GFP) was previously described (9). Mutations were previously introduced by using the Pfu DNA polymerase with the QuikChange site-directed mutagenesis kit (Stratagene) in the plasmid pT7-A-HDR (11). The DNA stretches harboring the mutations (deletions or substitutions) and defined by NarI (nucleotide 902) and NruI (nucleotide 1689) restriction sites were excised and subcloned into pFBΔIBDA-GFP, restricted by NarI and NruI. pFBΔIBDA-GFP mutants were validated by sequence analysis using adequate primers. The resulting plasmids were used to generate the recombinant baculoviruses. Briefly, the pFBΔIBDA-GFP derivatives were transformed into DH10Bac competent cells that contain the bacmid. Colonies containing recombinant bacmids were identified by disruption of the lacZα gene. High-molecular-weight DNA was prepared from selected colonies and used to transfect Sf9 cells with lipofectin. Recombinant baculovirus were generated by standard procedures, and high-titer viral stocks of the recombinant baculoviruses (108 PFU/ml) were prepared.
Preparation of protein assembly specimens.
Sf9 cells were infected at a multiplicity of infection higher than 5 PFU/ml in the presence of protease inhibitors leupeptin (0.5 μg/ml) and aprotinin (1 μg/ml), collected 100 h postinfection after addition of the same inhibitors at the same concentrations, and then treated with Freon 113. Purification was carried out by density gradient centrifugation in a CsCl solution. The concentration of protein in the purified suspension was estimated by the method of Bradford using bovine serum albumin as the standard and UV spectrophotometry at 280 nm.
Electron microscopy.
Specimens were prepared from the appropriate CsCl gradient fraction containing the different assembled forms by desalting through Micro Bio-Spin chromatography columns (Bio-Rad) equilibrated with a buffer containing 50 mM Tris, pH 7.4, and 150 mM NaCl. Samples of the suspensions were applied to an air glow-discharged carbon-coated grid and stained with a 2% uranyl acetate aqueous solution. Electron cryomicroscopy was performed as previously described (15). Grids were observed in a CM12 electron microscope (Philips) operated at 80 kV.
Three-dimensional reconstructions.
Micrographs were scanned with a Nikon Coolscan 8000 at a resolution of 4,000 dpi, which corresponds to a pixel size of 1.8 Å. Individual particles were selected from micrographs using the program ×3d (7). Every center of mass was then calculated after reducing the resolution to 15Å and smoothing the resulting map using a Wang radius of 5Å (34).
The three-dimensional reconstruction was made using the Rico suite of programs (25). This protocol consists of three main steps: preparation of the data, search of the views of the individual projections, and the building of the final reconstruction. First, the images were Fourier-Bessel transformed and sampled on 500 pixels in the 0.0- to 0.1-Å−1 range. Bessel functions up to order 200 were calculated. Second, for each image and all possible view angles in 3-degree steps, low-resolution icosahedral reconstructions were calculated using the 150 first-sampling points (0.03 Å−1) and including only up to 30 Bessel functions. Each of these reconstructions was then projected and correlated to the original image. The view parameters of the best-correlated reconstruction were taken as a reference, and the view parameters of the rest of the images were redetermined and refined taking the reference into account. The center of each projection was also refined. No model is thus necessary in this reconstruction method. Another cycle of view/center refinement was carried out at higher resolution using order 50 in Bessel functions and 250 pixels (0.05 Å−1).
The third step consists in building the final reconstruction from the oriented and centered images. For this, the complete Fourier-Bessel expansion was used up to a resolution of 15Å. This procedure gave a map for each of the three micrographs. These were independently corrected for C-terminal fragment (CTF) effects. The zeros of the transfer function were searched as described by Conway et al. (7). Phase-flip corrections were calculated for each group of images. To minimize the introduction of noise in the reconstructions, data near the zeros of the CTF functions were eliminated. These three CTF-corrected Fourier transforms were mixed and softened using a 5-Å Wang filter (35).
Fitting of the atomic model into the virus-like particle reconstruction.
The complete virus capsid of IBDV (10) or the 13 VP2 monomers contained in the asymmetric unit were fitted in the virus-like particle reconstruction using the program URO (26). All the figures of maps and protein coordinates were rendered using the PyMOL molecular graphics system (DeLano Scientific LLC, San Carlos, CA; http://www.pymol.org).
Protein and peptide biochemical analyses. (i) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses.
Samples were loaded on a 10% polyacrylamide gel, and proteins were revealed by Coomassie blue staining.
(ii) Western blot analyses.
Gels were transferred on Immobilon membranes (Millipore) for 1 h at 50 V. Proteins were visualized using dilutions of ascites fluids and relevant secondary antibodies conjugated to alkaline phosphatase (Biosys) and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate as the substrate.
(iii) Biotinylation and trypsin digestion of virions.
Virus particles (0.5 μg/μl) were resuspended in phosphate-buffered saline (PBS) and incubated 1 hour at room temperature in 1 mM EZ-Link sulfo-NHS-LC-biotin [sulfosuccinimidyl-6-(biotinamido)hexanoate; Pierce] to add a biotin group to lysines and N-terminal amine. Virus was desalted using Microspin G25 columns (Amersham) before matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis. Trypsin was added in virus suspensions to a protein/enzyme mass ratio of 200/1 at 37°C. Generated peptides were analyzed by MALDI-TOF after 1 min of digestion.
(iv) MALDI-TOF analysis.
The sample was then allowed to dry at room temperature before addition of 0.5 μl of the matrix solution. This dried-droplet sampling method was employed using a freshly prepared solution at 3 mg/ml of 1-cyano-4-hydroxycinnamic acid matrix in 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluororacetic acid. For acquisition, the accelerating voltage used was 20 kV. Peptide spectra were recorded in positive-reflector mode and with a delayed extraction of 130 ns and a 62% grid voltage. To analyze some peptides, spectra were recorded by the positive linear method with a delayed extraction of 160 ns and a 62% grid voltage. The spectra were calibrated using an external calibration sample composed of Des-Arg 1 bradykinin [(M + H)+ = 904.468 Da], human angiotensin I [(M + H)+ = 1,296.685 Da], neurotensin [(M + H)+ = 1,672.917 Da], melittin from bee venom [(M + H)+ = 2,845.762 Da], and bovine insulin B chain disulfonate [(M + H)+ = 3,494.651 Da].
Peptide synthesis.
The peptide pep46 was obtained by automated solid-phase synthesis using the 9-fluorenylmethoxy carbonyl strategy and then purified by reversed-phase high-performance liquid chromatography using procedures already reported for the production of other proteins (13). The protein was analyzed by mass spectrometry and confirmed to have purity higher than 98%.
Peptide activity.
The primary hepatocellular carcinoma epithelial chicken LMH, the quail pectoralis “fibrosarcoma” QT6, the LSCC-BK3 chicken B-lymphoid, and the human HeLa and HEp-2 cell lines were grown in RPMI medium supplemented with 10% fetal calf serum, 1% chicken serum, 2 mM glutamine, and 1 mM sodium pyruvate complemented with tryptose phosphate broth (Gibco-BRL) for QT6 cells. The three chicken cell lines are permissive to IBDV.
(i) Permeabilization activity.
The activity of the peptides was studied by incubation of live cells in suspension or subconfluent monolayers on coverslips with peptides (20 to 0.5 μM in RPMI culture medium without calf serum). After 1 h of incubation at 37°C, a 50-μl aliquot of cell supernatant was clarified at 3,000 rpm for 3 min and diluted four times in RPMI medium. Cell lactate dehydrogenase (LDH) release was measured using the Cytotox 96 nonradioactive kit (Promega), as described by the manufacturer. Briefly, 50 μl was incubated with 50 μl of reactant for 30 min in darkness. Reactions were then stopped by addition of 50 μl of 1 M acetic acid. Optical density (OD) was then measured at 490 nm. The percentage of LDH release was determined by the formula 100 × (OD -OD0)/(ODmax − OD0). OD0 and ODmax correspond to the ODs associated with untreated cells and Triton X-100-treated cells, respectively. OD represents the optical density of cells incubated with peptides. The permeabilizing activity of the peptides was also measured at a concentration of 10 μM at different times postincubation. The assays were performed as described above with the exception that the cells were rinsed and incubated at 4°C or in the presence of 0.5% (wt/vol) sodium azide in RPMI medium at 37°C.
(ii) Hemolytic activity.
Avian or sheep erythrocytes (50 μl of blood) were pelleted at 3,000 rpm for 3 min, rinsed with 1× PBS, and resuspended for 1 h in 0.3 ml PBS with various peptide concentrations at 4 or 37°C. After centrifugation, the optical density of the supernatants was measured at 405 nm/540 nm.
RESULTS
Virus assembly and proteolytic maturation of the IBDV capsid protein precursor pVP2 are concomitant. In the absence of VP1, the formation of VLP requires an exogenous sequence such as that of GFP to be fused at the C terminus of VP3 (9). To explore the relationship between capsid formation and pVP2 maturation, we constructed 17 baculoviruses expressing IBDA-GFP mutants characterized by substitutions or deletions in the pVP2 C-terminal domain (Fig. 1). Validation of the constructs was carried out by in vitro expression and Western blot analysis (not shown). For most mutants, primary processing generating pVP2, VP4, and VP3-GFP was effective. However, as previously found (20), mutation of the pVP2-VP4 junction site reduced the yield of pVP2 release (data not shown). Wild-type and mutated proteins were expressed and the resulting assemblies purified by isopycnic centrifugation.
pep11 controls the maturation of pVP2 and is essential to viral assembly.
Density gradients of IBDA-GFP fusion polyprotein deleted from pep11 (IBDA-GFPΔpep11) display no band, demonstrating that pep11 is essential to the capsid assembly process (Fig. 1 and 2A). To better define the contribution of pep11 to capsid morphogenesis, we constructed eight baculoviruses expressing the IBDA-GFP polyprotein containing one alanine substitution at positions varying from 503 to 510 (Fig. 1). Wild-type and mutated polyproteins were expressed in the baculovirus-insect cell system and the resulting assemblies purified (Fig. 2A and C). Because VP3 is fused to GFP, its presence is revealed by fluorescence properties. The density gradients corresponding to two mutants (IBDA-GFP-S503A and IBDA-GFP-T510A) show a band that is fluorescent under UV illumination (not shown). While one gradient (IBDA-GFP-I506A) shows no band at all, all gradients of all other central pep11 substitution mutants display nonfluorescent bands. Electron microscopy of negatively stained preparations shows that the fluorescent bands (mutants S503A and T510A) contain VLP. In contrast, observation of the nonfluorescent bands reveals tubes (mutants G504A and Q508A) or small irregular particles (mutants R505A, R507A, and L509A). As expected, no particular assembly could be detected for mutant I506A. SDS-PAGE associated with Western blot analysis shows that the major components of the VLP are VP3-GFP and VP2 (Fig. 2B and D). Maturation intermediates of pVP2 are also visible. The other assemblies, tubes and small irregular particles, are mainly constituted by pVP2, demonstrating that a single mutation in pep11 is sufficient to block VLP assembly, pVP2 processing, and VP3 recruitment. These findings indicate that pep11 and most of its residues control the VP2 assembly process by regulation of the proteolytic maturation of pVP2, suggesting that the processing of certain pVP2 molecules precedes VLP assembly.
FIG. 2.
Biochemical and electron microscopy analyses of the structures generated by expression of IBDA-GFP polyproteins modified at the C terminus of pVP2. (A) After an overnight isopycnic centrifugation, each gradient corresponding to one of the four deletion mutants was illuminated with a white (left) or UV (right) light and photographed. A band is visible for mutants Δpep46, Δpep7a, and Δpep7b. The bands associated with Δpep7a and Δpep7b are fluorescent, in contrast to the Δpep46 band. Samples collected from the bands were concentrated and negatively stained with 2% uranyl acetate solution. (B) Material extracted from each band shown in panel A was analyzed by SDS-PAGE followed by Coomassie blue staining (left) or Western blot analyses with anti-pVP2/VP2 or anti-VP3 monoclonal antibodies (right). (C) Gradients corresponding to substitution mutants in the pep11 domain were photographed. Concentrated samples were processed for electron microscopy analysis. (D) Material extracted from each band shown in panel C was analyzed by SDS-PAGE followed by Western blot analyses with anti-pVP2/VP2 or anti-VP3 monoclonal antibodies.
pep46 controls the geometry of VP2 assemblies.
The deletion of pep46 enables the formation of another type of assembly. The density gradient of IBDA-GFPΔpep46 shows a nonfluorescent band (Fig. 1 and 2A). Electron microscopy of negatively stained preparations reveals the presence of spherical particles with a diameter of about 25 nm (Fig. 2A). SDS-PAGE associated with Western blot analysis shows that the main component of the spherical particles is a protein with an apparent molecular mass of 41 kDa (a value slightly larger than that of mature VP2, 38 kDa), which is reactive with an anti-VP2/pVP2 antibody (Fig. 2B). Henceforth, this high-molecular-weight version of VP2 will be referred as VP2*. Consistent with the absence of fluorescence, no VP3-GFP was identified in the VP2* preparations.
To better characterize VP2*, we precisely determined its molecular weight by mass spectrometry analysis and compared it to those of pVP2 and VP2. The average mass of VP2* was found to be 49,477 Da. This value matches the results (49,404 Da) of the subtraction of the pep46 mass (4,874 Da) from the theoretical average mass of pVP2 (54,278 Da), taking into account the mass of the water molecule (18 Da) that is generated during peptidic bond formation. VP2* is thus constituted by VP2 (aa 1 to 441 of the polyprotein) elongated by pep7a, pep7b, and pep11 (aa 487 to 512 of the polyprotein). When observed by electron microscopy, the VP2* particles (Fig. 3) appear to have a structure similar to the T=1 subviral particles obtained when only VP2 is expressed (4).
FIG. 3.
Combined electron cryomicroscopy and X-ray crystallography studies of VP2 and VP2* subviral particles. (a) Image of VP2 (left) and VP2* (right) embedded in ice. No major structural difference can be observed. (b) 3D reconstruction of VP2 (blue) and VP2* (green). In red, the atomic model of the VP2 subviral particle is shown at a resolution limited to 15 Å. The upper row shows the reconstructions seen from the top down the threefold axis. The lower row represents a central section through the reconstructions. (c) Result of the fit of the atomic model of IBDV VP2 into the VP2 and VP2* subviral particle reconstructions. The reconstruction electron densities of VP2 (blue) fit well with the VP2 atomic model, which is represented as a red ribbon. At the fivefold axis, the internal cavity is filled in the case of VP2*. Because the C terminus of VP2 points towards this cavity, we can conclude that the cavity is occupied by the C terminus of VP2*.
At this point, we can conclude that pep46 and pep11 are essential to IBDV capsid assembly. The fact that point mutants in the central part of pep11 have an efficient primary pVP2-VP4 processing but do not assemble shows that the presence of pep11 is a prerequisite for both tube and VLP formation. Indeed, pep11 seems essential to the release of pep46 from VP2. Finally, pep46 controls the geometry of the VP2 assemblies: its presence or absence leads to the formation of T=13 icosahedral VLP or T=1 subviral particles, respectively.
To understand the control of the geometrical switch leading to objects having T=13 or T=1 icosahedral symmetries, we determined the structure of the VP2* subviral particle.
Structure and comparison of the VP2* and VP2 particles.
Figure 3a shows VP2* and VP2 icosahedral particles embedded in vitreous ice. The three-dimensional structure was calculated with a recently developed 3D reconstruction method based on symmetry-adapted functions (24). Figure 3b illustrates the reconstructions at a resolution of about 15 Å calculated with 172 and 99 particles of VP2* and VP2, respectively. The reconstructions demonstrate that VP2* and VP2 particles exhibit dodecahedral geometries and indeed have similar structures. However, they display a significant structural difference in an internal domain located near the fivefold axes: a cavity present in VP2 particles is filled in their VP2* homologues. This shows that the additional residues present in VP2* are internal and located near the fivefold axes.
Fitting the atomic model of VP2 into the VP2 and VP2* subviral particle reconstructions confirms that the C terminus of VP2 is located next to the internal cavity at the fivefold axes (Fig. 3c). The cavity present in VP2 subviral particles is completely filled by the extra 25 amino acids (pep7a plus pep7b plus pep11) of VP2*. These observations strongly suggest that unreleased pep46, which is 46 amino acids long, cannot fit into the cavity, thus resulting in steric hindrance to the formation of the fivefold axis required for the assembly of icosahedral particles. This accords with the fact that pVP2 forms various assemblies, tubes with helical symmetry and irregular particles, that do not all display fivefold symmetry.
Icosahedral-particle formation requires the release of pep46.
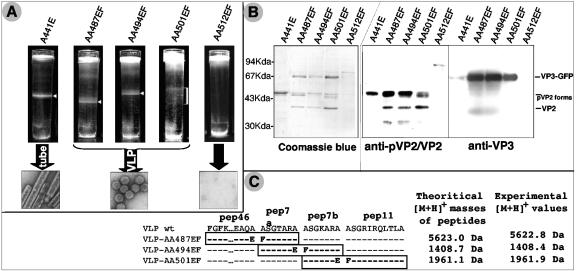
To confirm that the presence of pep46 results in a steric hindrance at the fivefold axes in icosahedral particles, several baculoviruses driving the expression of the polyprotein IBDA-GFP mutated at the pVP2 cleavage sites were constructed (Fig. 1). The five mutants, A441E, AA487EF, AA494EF, AA501EF, and AA512EF, gave rise to a polyprotein with a proteolytic maturation of pVP2 blocked at the mutated positions 441-442, 487-488, 494-495, 501-502, and 512-513, respectively. Polyprotein mutants were expressed and the resulting assemblies purified. As shown in Fig. 4A, a band is visible in density gradients for all mutants except for the one affecting the pVP2-VP4 junction (AA512EF). This last mutant does not lead to high-molecular-weight assemblies. Electron microscopy of A441E samples reveals tubes with a diameter of about 55 nm, similar to pVP2 tubes (9, 23). In contrast, observation of the AA487EF, AA494EF, and AA501EF bands reveals the presence of VLP with a diameter of about 60 nm. These particles are indistinguishable from VLP-GFP and viral particles (9). SDS-PAGE associated with Western blot analysis shows that the main component of the tubes is unprocessed pVP2, revealing that cleavage at the VP2-pep46 junction is required to allow cleavages at the peptide junctions pep46-pep7a, pep7a-pep7b, and pep7b-pep11 (Fig. 4B). On the contrary, the major components of the VLP are the VP3-GFP fusion protein and the mature form VP2. Different maturation forms of pVP2 are also visible in the various VLP, and the pVP2 processing concords with the engineered mutations: proteolytic cleavage is blocked only at the mutated sites.
FIG. 4.
Analysis of the structures generated by expression of IBDA-GFP polyproteins modified at the pVP2 cleavage sites. (A) After density centrifugation, each gradient corresponding to the five mutants was photographed. Gradients contain either no or one band. Samples collected from the bands were concentrated and negatively stained with 2% uranyl acetate solution. A block in assembly was evidenced when the pVP2-VP4 primary cleavage site was mutated. Tubes were assembled when the VP2-pep46 cleavage was mutated. (B) Material extracted from each band shown in panel A was analyzed by SDS-PAGE followed by Coomassie blue staining (left) or Western blot analyses with anti-pVP2/VP2 or anti-VP3 monoclonal antibodies (right). (C) Characterization of the peptides present in the VLP by mass spectrometry analysis. Theoretical and experimental masses of the mutated peptides present in the VLP are shown. The masses corresponding to the fused peptides pep46-pep7a, pep7a-pep7b, and pep7b-pep11 were evidenced in the VLP AA487EF, AA494EF, and AA501EF, respectively.
In conclusion, these substitution mutants demonstrate that two proteolytic cleavages at the VP2-pep46 and pVP2-VP4 junctions are critical to the formation of VLP.
pep7a and pep7b are not essential to viral assembly.
The processing of pVP2 was further analyzed on the mutated VLP (AA487EF, AA494EF, and AA501EF) by mass spectrometry. We previously showed that pep46, pep7a, pep7b, and pep11, with theoretical masses of 4,874.7, 633.3, 660.4, and 1,185.7 Da, respectively, are present in IBDA-GFP VLP (11). As expected, mutation of a cleavage site withdraws one generated peptide and increases the mass of one of the peptides (Fig. 4C). Three heavier peptides with [M + H]+ masses of 5,622.8, 1,408.4, and 1,961.9 Da were detected in VLP AA487EF, AA494EF, and AA501EF, respectively. These peptide masses match the theoretical masses of the expected fused peptides. These results show that, when the cleavage sites defining pep7a and pep7b are mutated, the pVP2 processing is modified but still remains effective and leads to the formation of VLP.
To verify that pep7a and pep7b play a minor role in viral assembly, we constructed two deletion mutants, IBDA-GFPΔpep7a and IBDA-GFPΔpep7b, that were expressed, extracted, and purified. For both mutants, the gradient shows a band that is fluorescent under UV illumination (Fig. 1 and 2A). In both cases, electron microscopy of negatively stained preparations reveals VLP with a diameter of about 65 nm. These particles are similar to VLP-GFP. All these particles have the same protein composition and are characterized by efficient pVP2 maturation (Fig. 2B). These results confirmed that pep7a and pep7b are not essential for capsid assembly.
The key role of pep46 and pep11 in viral assembly suggests that they are associated with the viral capsid components. The structure of IBDV virions determined at a resolution of 7 Å indicates that the icosahedral capsid is constituted by only a single component, VP2. Hence, the peptides released during the pVP2 maturation process do not follow the icosahedral symmetry. To assess their physical relationship with the VP2 shell, we tested their accessibility by biochemical labeling and mild trypsin digestions on purified virions.
Peptide accessibility.
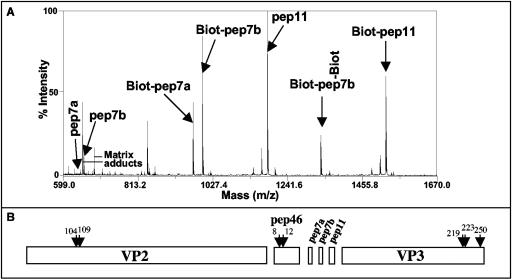
To investigate the presence of the four peptides (pep46, pep7a, pep7b, and pep11) at the viral surface, virions were biotinylated and labeled peptides analyzed by mass spectrometry. Figure 5A shows that the three small peptides, pep7a, pep7b, and pep11, were indeed accessible to labeling. Due perhaps to the low number of lysines in pep46 (one), no biotinylation of pep46 was observed (not shown). Although it contains 14 lysines, the internal protein VP3 is not labeled (not shown). The accessibility of pep46 in the viral particles was further analyzed. Figure 5B shows that VP2, pep46, and the C terminus of VP3 are accessible to mild trypsin digestion of the virus particles. Cleavage products revealing cleavages after lysines 8 and 12 in pep46 were identified. The digestion sites of VP2 are localized at positions 104 and 109, very close to the SDE loop located at the fivefold and the quasisixfold axes of the capsid (10). These observations suggest that all the peptides are located near the fivefold and quasisixfold axes of the viral particles, while VP3 is in the internal compartment defined by VP2. The three smaller peptides (pep7a, pep7b, and pep11) display high accessibility, suggesting that they lie at the external surface of the virus particle.
FIG. 5.
Peptide accessibility to biochemical labeling and protease. (A) IBDV particles were submitted to biotinylation before mass spectrometry analysis. A mass/charge window ranging from 599 to 1,670 is presented. The peptides pep7a, pep7b, and pep11 with their biotinylated forms were observed. Because of the presence of a lysine in the pep7a sequence, two biotinylated forms of pep7b were detected. pep46 and VP3 were not biotinylated in these experimental conditions. (B) IBDV particles were submitted to trypsin digestion, and the generated peptides were analyzed by mass spectrometry. The peptides were assigned to the primary sequences of the main structural components of the virions VP2 and VP3 and the peptides pep46, pep7a, pep7b, and pep11. The numbers and arrows indicate amino acid positions that are located immediately upstream the cleavage sites. The results presented were obtained after 1 minute of coincubation of the virus with trypsin. Cleaved products of pep46 were also observed for longer coincubation times.
The sequence analysis of the peptides of IBDV and other birnavirus homologues shows that pep46 has a remarkably hydrophobic N terminus that is followed by a predicted amphipathic domain (11). All these observations suggest that pep46, in addition to its role in the virus assembly pathway, may also interact with membranes and contribute into the entry of the virus into the target cell. To test this hypothesis, we studied the effect of pep46 on liposomes and cell membranes.
Pep46 destabilizes liposomes and cell membranes.
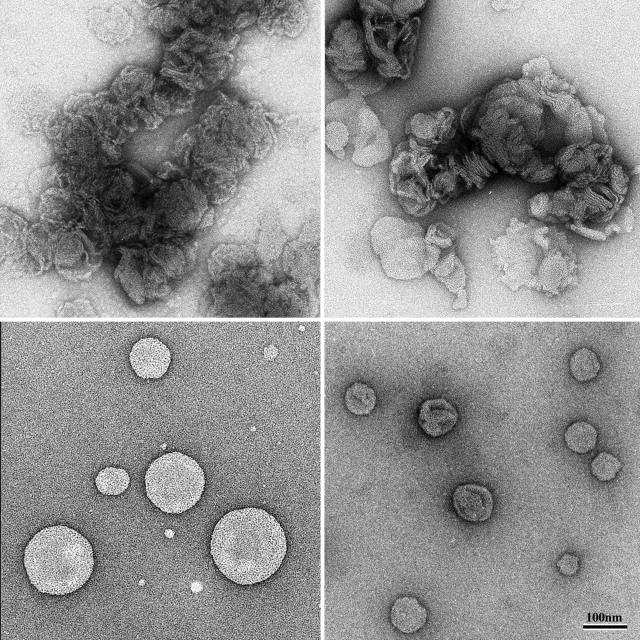
Figure 6 (lower left panel) shows a liposome preparation. The liposomes have a typical spherical appearance with an apparent diameter of typical values in the range of 150 to 400 nm. Incubation with pep46 leads to a spectacular liposome aggregation, characterized by large planar surfaces between interacting membranes (Fig. 6, upper left panel). It is difficult to assess if the liposomes have been disrupted. Melittin is known to induce large membrane changes (16) resulting in hemolysis when incubated with erythrocytes (34). The effect of pep46 on liposomes is similar to the one induced by melittin (Fig. 6, upper right panel). In contrast, peptides pep7a, pep7b, and pep11 have no effect on liposomes (shown for pep11 in Fig. 6, lower right panel).
FIG. 6.
Images of liposomes in the presence of different peptides. pep46, upper left; melittin, upper right; pep11, lower right; untreated liposomes, lower left. Unlike pep11, pep46 and melittin induce large structural changes in liposomes.
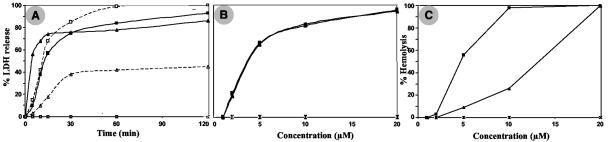
The drastic effects observed on synthetic lipid liposomes are also found on biological membranes. When pep46 is added to various cells (IBDV-permissive cell lines, LSCC-BK3, LMH, and QT6; IBDV nonpermissive cell lines, HEP-2 and HeLa), lactate dehydrogenase is released from the cytoplasm (not shown). In all cells, and as exemplified for LMH cells in Fig. 7, the greater the incubation period (Fig. 7A) and the peptide concentration (Fig. 7B), the greater the effect. Once again, the magnitude of the effect of pep46 is similar to that observed with melittin: 80% of the total lactate dehydrogenase is released in 30 min. However, pep46 and melittin behave differently in response to temperature changes—pep46 has less effect at 4°C than at 37°C—suggesting that the rigidity of the aliphatic chains constituting the membranes is an important factor contributing to the action of the peptide (Fig. 7A). The other IBDV peptides have no effect on cells when used separately or combined. Finally, incubation of IBDV peptides with erythrocytes confirms these findings. Pep46 and melittin induce erythrocyte hemolysis contrary to other IBDV peptides (Fig. 7C).
FIG. 7.
Cell membrane permeabilization activity of the viral peptides. (A and B) Peptide activity was quantified by measuring the LDH released from LMH cells and expressed as a percentage of the total cellular LDH. (A) The LDH released in the culture cell medium was measured after 1 hour of incubation with pep46 (at 37°C [▴] or 4°C [▵]) or mellitin (at 37°C [▪] and 4°C [□]) at a concentration of 10 μM. No release was observed for pep7a (⋄) and pep11 (×) at 37 or 4°C. (B) The activities of pep46 (▴) and mellitin (▪) on cells display the same concentration dependence. Again, no activity could be observed for pep11 (×) at 37°C. (C) Hemolysis activity of the peptides. In contrast to pep11 (×), pep46 (▴) and melittin (▪) display hemolysis activity. Note, however, that the concentration dependence is different in the two cases.
DISCUSSION
While assembly and maturation of the IBDV capsid protein pVP2/VP2 were known to be concomitant (8, 9), we hereby demonstrate that these two processes are interdependent. The autoproteolytic processing of the IBDA polyprotein, giving rise to pVP2, VP4, and VP3, is carried out by VP4 via its protease activity. In addition to VP4, the subsequent pVP2 processing requires the presence of VP1 and VP3. This requirement acts in favor of the existence of a large quaternary maturation complex formed by VP1, pVP2, VP3, and VP4. While VP3 and VP1 have been shown to interact (32), promoting the assembly of the VP2 viral capsid (8, 22), affinity between other proteins of the complex has not to date been evidenced (33). This observation suggests that the affinity between proteins mainly depends on the formation of the multiprotein complex formed around pVP2. In addition, as far as the assembly is concerned, VP1 can be replaced in the multimeric maturation complex by an unrelated exogenous sequence fused at the C terminus of VP3, suggesting that VP1 needs merely to be present in the vicinity of VP3 in order to screen the negative charges of the VP3 C terminus (9). The association of pVP2 with VP3 can be considered as a chemical equilibrium reaction favoring the dissociation of the species. VP1 in the virus or any peptide interacting with the C terminus of VP3 in the VLP will displace the equilibrium towards the pVP2-bound VP3 complex and thus favor the formation of the maturation complex (8). Similarly, overexpression of VP3 modifies the equilibrium in favor of the VP3-bound pVP2 and of the maturation complex (27). In all cases, the maturation complex appears to be formed through weak and nonspecific interactions, suggesting that cellular constituents or different physicochemical conditions may substitute some components of the maturation complex. Concordant to this conclusion, it has been shown that VLP are assembled in the vaccinia virus expression system without requiring VP1 (17) in contrast to the observations in the baculovirus/Sf9 system.
All assemblies appear to be controlled by the maturation complex. When it has a high protease activity, VLP are efficiently produced. When its enzymatic activity is reduced, for instance when VP1 is missing, tubes essentially made of pVP2 are generated. The absence of maturation activity prevents the formation of ordered objects and leads to the assembly of pVP2 into irregular particles. Self-assembly of VP2 does not require any processing and leads to the formation of T=1 subviral particles (4).
The formation of the VLPs requires pVP2 to be cleaved by the maturation complex at two processing sites: the VP2-pep46 and the pVP2-VP4 junctions. Other cleavages within the maturation complex (at pep46-pep7a, pep7a-pep7b, and pep7b-pep11 junctions) do not seem to participate in the capsid formation process. The expression of a polyprotein deleted from pep7a or pep7b even gives rise to VLP. In agreement with these results, reverse genetics shows that pep7a and pep7b are not essential for virus rescue (11). On the contrary, pep11 and pep46 are crucial determinants of the viral assembly process. The sequence of pep11 is so critical that most of its substitutions block capsid morphogenesis as well as cleavage at the VP2-pep46 junction. These results suggest that pep11 activates either the VP4 protease or, more probably, allows the VP2-pep46 junction to adopt the right conformation to be cleaved. In addition, the processing of pVP2 starts at its C terminus and extends towards its N terminus; the VP2-pep46 junction is thus the last one to be cleaved. Mutation of the VP2-pep46 site prevents any processing of pVP2, suggesting that the maturation complex, in particular in the pep11 and pep46 domains, has indeed a chaperone activity, allowing the processing of pVP2.
The peptide pep46 controls the geometry and size of the synthesized viral particle. While its presence results in the formation of T=13 icosahedral particles, its deletion leads to the formation of smaller T=1 subviral particles constituted by VP2*, which correspond to VP2 fused with peptides pep7a, pep7b, and pep11. The structures of both VP2* and VP2 assemblies were determined at a resolution of about 15 Å. The comparison of the calculated map allows the identification of the pVP2-specific domain in the structure. It is located in an internal cavity close to the fivefold axis of the VP2 assembly. This cavity is present in both the cryomicroscopy reconstruction and the X-ray electron density map of VP2 particles (10). The 25 additional amino acids present in VP2* fill the cavity, showing that the 46 amino acids of pep46 represent a steric hindrance to the formation of the fivefold axes in the viral particles. The fact that some of the peptides are accessible to biochemical labeling also demonstrates that the C terminus of pVP2 points towards the exterior of the particles and represents a steric hindrance to the formation of the fivefold axes. These observations imply that, at the fivefold axes, the proteolytic cleavage at the VP2-pep46 junction precedes the formation of the capsid. Due to packing differences, the presence of pep46 appears possible at the quasisixfold axes. Accordingly, the pVP2 protein is able to assemble in rigid tubes, forming a hexagonal lattice. At this point, it can also be argued that pep46 is essential to the formation of the quasisixfold axes as well as the assembly of a large-diameter particle. The fact that the pVP2 protein is fully processed in the viral particles, even though VP4 is not a structural protein, suggests that the maturation at the quasisixfold axis occurs after assembly.
Although the peptides were not visualized on the surface of the IBDV particle by X-ray crystallography (10), they are accessible to mild trypsin digestion or to biotinylation, demonstrating that they are located either at the external surface of the viral particle (pep7a, pep7b, and pep11) or underneath the surface at the fivefold or quasisixfold axes (pep46). Sequence analysis shows that pep46 possesses amphiphilic properties, suggesting significant membrane affinity. Indeed, pep46 strongly interacts with liposomes and induces the release of cytosolic enzymes from cultured cells and hemolysis of erythrocytes. In fact, pep46 has a similar effect to melittin on both liposomes and cellular membranes.
The peptide pep46 appears to possess a dual function. It controls the geometry of the viral particle and has drastic effects on membranes, strongly suggesting that it participates in viral entry. This dual function can be roughly schematized. When pep46 is unreleased from VP2, it prevents the formation of fivefold axes. After cleavage, pep46 is located near the surface of the particle at the fivefold and quasisixfold axes, ready to participate in virus cell entry. The properties of pep46 suggest a viral entry mechanism similar to the one advanced for rotavirus, another group of dsRNA viruses (30). A peptide, pep46 for IBDV, a VP7 proteolytic fragment for rotaviruses, destabilizes the cellular membrane, allowing the entry of the viral transcription-competent particle into the cytoplasm of the host cell. While entry is triggered by the calcium concentration variation and associated release of VP7 for rotaviruses, the entry signal dissociating pep46 from the viral surface remains to be identified for birnaviruses. It is plausible to conclude that a cellular receptor induces conformational changes for VP2, changing its affinity for pep46, thus triggering membrane-pep46 interactions.
The properties of pep46 also appear to be similar to those of the gamma peptide of nodavirus, another family of nonenveloped virus. The gamma peptide (44 residues) alters the cell membrane structure and thus mediates the translocation of the positive-strand genomic RNA (2). The function similarities between these two peptides are surprising, taking into account the fact that they arise from such different viral families: single- versus double-stranded RNA viruses. Indeed, they confirm the structural similarities of their capsid proteins and reinforce the evolutionary link proposed between single- and double-stranded RNA viruses (10).
Acknowledgments
We thank J.-F. Bouquet and F.-X. Legros (Mérial) for providing the 1C6 monoclonal antibody (mab) specific for pVP2/VP2 and IBDV virus, N. Eterradossi (AFSSA) for the mab 20 specific for VP3, and Félix Rey (CNRS) for critical reading of the manuscript.
B.D. and J.L. acknowledge support from the INRA and CNRS, the CNRS programs PCV and “Dynamique et réactivité des assemblages biologiques,” the ACI “Microbiologie” from the French MRT, and the EU COST action 839. C.C. was funded by a French MRT fellowship. M.G. was funded by an INRA-Région Ile de France fellowship. J.P. acknowledges EMBO (ALTF 230/2002) and the EU (HPMF-CTF-2002-01805) for fellowships.
REFERENCES
- 1.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. R. Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. [DOI] [PubMed] [Google Scholar]
- 3.Bottcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caston, J. R., J. L. Martinez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodriguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandran, K., and M. L. Nibert. 2003. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 11:374-382. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. 1975. Ribonucleic acid polymerase activity in purified infectious pancreatic necrosis virus of trout. Biochem. Biophys. Res. Commun. 62:689-695. [DOI] [PubMed] [Google Scholar]
- 7.Conway, J. F., B. L. Trus, F. P. Booy, W. W. Newcomb, J. C. Brown, and A. C. Steven. 1996. Visualization of three-dimensional density maps reconstructed from cryoelectron micrographs of viral capsids. J. Struct. Biol. 116:200-208. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier, C., J. Lepault, B. Da Costa, and B. Delmas. 2004. The last C-terminal residue of VP3, glutamic acid 257, controls capsid assembly of infectious bursal disease virus. J. Virol. 78:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals new structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 11.Da Costa, B., C. Chevalier, C. Henry, J. C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas, B., F. S. B. Kibenge, J. C. Leong, E. Mundt, V. N. Vakharia, and J. L. Wu. 2004. Birnaviridae, p. 561-569. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy, VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 13.de Rocquigny, H., D. Ficheux, C. Gabus, M. C. Fournie-Zaluski, J. L. Darlix, and B. P. Roques. 1991. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 180:1010-1018. [DOI] [PubMed] [Google Scholar]
- 14.Dobos, P., B. J. Hill, R. Hallett, D. T. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 16.Dufourcq, J., J.-F. Faucon, G. Fourche, J.-L. Dasseux, M. Le Maire, and T. Gulik-Krzywicki. 1986. Morphological changes of phosphatidylcholine bilayers induced by melittin: vesicularization, fusion, discoidal particles. Biochim. Biophys. Acta 859:33-48. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Arias, A., C. Risco, S. Martinez, J. P. Albar, and J. F. Rodriguez. 1998. Expression of ORF A1 of infectious bursal disease virus results in the formation of virus-like particles. J. Gen. Virol. 79:1047-1054. [DOI] [PubMed] [Google Scholar]
- 18.Gillies, S., S. Bullivant, and A. R. Bellamy. 1971. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science 174:694-696. [DOI] [PubMed] [Google Scholar]
- 19.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 20.Lejal, N., B. Da Costa, J.-F. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 21.Lombardo, E., A. Maraver, J. R. Castón, J. Rivera, A. Fernandez-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maraver, A., A. Ona, F. Abaitua, D. Gonzalez, R. Clemente, J. A. Ruiz-Diaz, J. R. Caston, F. Pazos, and J. F. Rodriguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Torrecuadrada, J. L., J. R. Caston, M. Castro, J. L. Carrascosa, J. F. Rodriguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa, A., N. Miyazaki, J. Taka, H. Naitow, A. Ogawa, Z. Fujimoto, H. Mizuno, T. Higashi, Y. Watanabe, T. Omura, R. H. Cheng, and T. Tsukihara. 2003. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure 11:1227-1238. [DOI] [PubMed] [Google Scholar]
- 25.Navaza, J. 2003. On the three-dimensional reconstruction of icosahedral particles. J. Struct. Biol. 144:13-23. [DOI] [PubMed] [Google Scholar]
- 26.Navaza, J., J. Lepault, F. A. Rey, C. Alvarez-Rua, and J. Borge. 2002. On the fitting of model electron densities into EM reconstructions: a reciprocal-space formulation. Acta Crystallogr. D Biol. Crystallogr. 58:1820-1825. [DOI] [PubMed] [Google Scholar]
- 27.Ona, A., D. Luque, F. Abaitua, A. Maraver, J. R. Caston, and J. F. Rodriguez. 2004. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322:135-142. [DOI] [PubMed] [Google Scholar]
- 28.Pous, J., C. Chevalier, M. Ouldali, J. Navaza, B. Delmas, and J. Lepault. 2005. Structure of birnavirus-like particles determined by combined electron cryo-microscopy and X-ray crystallography. J. Gen. Virol. 86:2339-2346. [DOI] [PubMed] [Google Scholar]
- 29.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 A resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz, M. C., J. Cohen, and F. Michelangeli. 2000. Role of Ca2+ in the replication and pathogenesis of rotavirus and other viral infections. Cell Calcium 28:137-149. [DOI] [PubMed] [Google Scholar]
- 31.Spies, U., H. Muller, and H. Becht. 1987. Properties of RNA polymerase activity associated with infectious bursal disease virus and characterization of its reaction products. Virus Res. 8:127-140. [DOI] [PubMed] [Google Scholar]
- 32.Tacken, M. G., B. P. Peeters, A. A. Thomas, P. J. Rottier, and H. J. Boot. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J. Virol. 76:11301-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 34.Tosteson, M. T., S. J. Holmes, M. Razin, and D. C. Tosteson. 1985. Melittin lysis of red cells. J. Membr. Biol. 87:35-44. [DOI] [PubMed] [Google Scholar]
- 35.Wang, B. C. 1985. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 115:90-112. [DOI] [PubMed] [Google Scholar]