Abstract
We examine biochemical characteristics of the herpes simplex virus (HSV) tegument protein VP22 by gel filtration, glycerol sedimentation, and chemical cross-linking experiments and use time course radiolabeling and immunoprecipitation assays to analyze its synthesis and interaction with other infected-cell proteins. VP22 was expressed as a delayed early protein with optimal synthesis requiring DNA replication. In immunoprecipitation assays, VP22 was found in association with several additional proteins including VP16 and a kinase activity likely to be that of UL13. Furthermore, in sizing chromatography experiments, VP22 was present in several higher-order complexes in infected cells. From gel filtration analysis the major form of VP22 migrated with a molecular mass of approximately 160 kDa, consistent with its presence as a tetramer, or a dimer complexed with other proteins, with a fraction of the protein migrating at larger molecular mass. In vitro-synthesized VP22 sedimented in a size range consistent with a mixture of tetramers and dimers. Short N- or C-terminal deletions resulted in migration almost exclusively as dimers, indicating that VP22, in the absence of additional virus-encoded proteins, could form higher-order assemblies, most likely tetramers, but that both N-and C-terminal determinants were required for stabilizing such assemblies. Consistent with this we found that isolated proteins encompassing either the N-terminal or C-terminal region of VP22 sedimented as dimers, and that the purified C-terminal domain could be cross-linked into dimeric structures. These results are discussed with regard to possible virus and host interactions involved in VP22 recruitment into virus particles.
The herpesvirus tegument is a proteinaceous compartment of the virion localized between the capsid and the envelope (7). For herpes simplex virus (HSV) the tegument accounts for approximately 50% of the volume of the virion and more than 15 virus-encoded proteins have been reported to be recruited to it (27). The stoichiometries of assembled tegument proteins differ greatly. In HSV, VP22, encoded by the UL49 gene (18), is among the most abundant of the tegument proteins being present at about 1,500 to 2,000 molecules per virion (21, 24).
VP22 has a molecular mass of 38 kDa and is a basic, phosphorylated protein that is well conserved throughout the alphaherpesvirus subfamily. A number of studies, particularly cell biology oriented studies on localization and compartmentalization, have begun to shed light on its role in virus infection. We and others have examined the subcellular localization of VP22 by immunofluorescence (2, 12, 29) and more recently, VP22 fused to the green fluorescent protein has been used to investigate the localization and trafficking of VP22 in live infected cells (15, 22). Green fluorescent protein-VP22 localizes predominantly to the cytoplasm of infected cells and translocates from the cytoplasm to the nucleus during cell division, most likely via an association with mitotic chromatin (13, 15). Different conclusions on the localization of VP22, indicating a predominantly nuclear localization have been reached from results of immunofluorescence studies in fixed cells for HSV (34) and for pseudorabies virus (9). The precise reasons for the differences in conclusions regarding localization from the different types of studies are not clear. VP22 expressed in isolation in transient transfection assays also has the capacity to bind, reorganize, and stabilize cellular microtubules in a manner similar to that demonstrated for cellular microtubule-associated proteins (14), leading to the proposal for a VP22-microtubule interaction that may occur in HSV-1-infected cells.
VP22 is subject to posttranslational modification, including ADP-ribosylation (1) and phosphorylation. Multiple phosphorylated forms of VP22 are present within infected cells together with a nonphosphorylated form present in approximately equivalent amounts (16, 17, 20, 34). Interestingly, it appears that the nonphosphorylated form of VP22 is specifically incorporated into assembling virions, suggesting that the mechanism(s) involved in tegument assembly in some way differentiates between these two species of VP22 (9, 17, 29, 34). It has also been recently suggested that phosphorylation may be involved in the dissociation of VP22 from the tegument upon virus entry into the cell (30).
Examination of the precise role of VP22 by analysis of the phenotype of deletion mutants in the context of virus infection have not produced unified conclusions. A virus containing a deleted variant of VP22 was constructed and this variant incorporated extremely reduced amounts of the protein (33). While the variant grew with approximately wild-type efficiency in culture, the fact that a null variant was not isolated indicated that some residual VP22 function may be critical. The isolation of complete null mutants in bovine herpesvirus (25) and recently in pseudorabies virus (9) has demonstrated that VP22 is not essential for replication of those viruses in culture, while in contrast other studies have indicated that the corresponding gene is required for replication of Marek's disease virus (10). Recent results demonstrate that for HSV, VP22 is not absolutely essential for replication in culture but viruses lacking VP22 display a cell type defect in replication and an altered profile of virion-assembled proteins (11).
Despite this information, however, to date there have been relatively few studies reporting biochemical characterization of VP22, and any complete understanding of the role and detailed mechanisms of assembly and involvement in virus-host interactions will require such analyses in addition to imaging studies of its localization. In this work we use time course radiolabeling and immunoprecipitation to analyze the synthesis of VP22 in HSV-1-infected cells and its interaction with additional infected-cell proteins. We also employ gel filtration and glycerol sedimentation to examine VP22 protein complexes in virus-infected cells, or of the isolated protein synthesized in vitro. VP22 from virus-infected cells was present in multiple higher-order forms with the major soluble form migrating with a molecular mass of approximately 160 kDa, consistent with its presence as a tetramer or potentially a dimer associated with other species. A fraction of VP22 migrated with a molecular mass of approximately 290 kDa, consistent with a larger complex formed with additional virus proteins or a higher-order oligomer. VP22 expressed in isolation by in vitro synthesis migrated with a sedimentation profile consistent with a mixture of tetramers and dimers. Interestingly, both N-terminal and C-terminal regions of VP22 migrated as dimers, and cross-linking studies provided supportive evidence for dimer formation of at least the C-terminal subregion. We propose a model for interactions which could account for higher-order interactions with virus and host components.
MATERIALS AND METHODS
Cell culture and virus infection.
HeLa cells (human epithelial cervical carcinoma cell line) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum. For virus infection, cells in 35-mm cluster dishes or 100-mm plates, were infected with HSV-1 strain 17 at a multiplicity of infection of 5 or 10 or mock infected. After 1 h incubation, the inoculum was removed and replaced with DMEM supplemented with 2% fetal calf serum. For radiolabeling with [35S]methionine, cells were infected in 35-mm wells and the medium was replaced at different times postinfection with labeling medium consisting of DMEM (lacking methionine), 2 mM l-glutamine, 2% fetal calf serum, and 20 μCi/ml [35S]methionine. For infections in the presence of the DNA synthesis inhibitor phosphonoacetic acid, the medium was replaced immediately after removal of the inoculum with DMEM supplemented with 2% fetal calf serum and 400 μg/ml phosphonoacetic acid (Sigma).
At different times during infection the cells were harvested, washed, and either disrupted directly in sodium dodecyl sulfate (SDS) lysis buffer for a total lysate, or soluble extracts were made using a high-salt extraction buffer. The production of whole-cell soluble extract was essentially as described before (36). Briefly, infected-cell pellets were frozen and then resuspended by rapid pipetting in one packed cell volume of high-salt buffer (400 mM NaCl, 20 mM HEPES [pH 7.9], 1 mM EDTA, 5% vol/vol glycerol, 1 mM dithiothreitol, plus the following cocktail of protease inhibitors; 1 mM phenylmethylsulfonyl fluoride, 0.22 mM tosyl lysyl chloromethyl ketone (TLCK), 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 1 μg/ml pepstatin A). The extracts were clarified by centrifugation at 12,000 × g for 30 min at 4°C.
Immunoprecipitation and in vitro phosphorylation assays.
Soluble extracts from infected cells were diluted 1:10 in 0.2 M high-salt buffer (200 mM NaCl, 20 mM HEPES [pH 7.9], 1 mM EDTA, 5% vol/vol glycerol, 1 mM dithiothreitol, 0.1% vol/vol Nonidet-P40, containing the protease inhibitor cocktail described above). For immunoprecipitations of in vitro-translated products, 10 μl of the appropriate samples were similarly diluted. The diluted extracts were clarified by centrifugation at 12,000 × g for 10 min at 4°C, antibody was added, and the samples wereincubated at 4°C for 3 h followed by addition of 25 μl protein A beads (Pharmacia) for a further 30 min. Nonspecifically bound proteins were removed by washing the beads 4 times with 40 volumes of 0.2 M high-salt buffer. The immunoprecipitated proteins were then used either for in vitro phosphorylation assays, or solubilized in 10 μl 4x SDS lysis buffer for separation by polyacrylamide gel electrophoresis (PAGE).
For the in vitro phosphorylation assays, following the final washes above, the protein A beads were washed once with 50 volumes of kinase buffer (50 mM NaCl, 20 mM MgCl2, 50 mM Tris [pH 7.5], 4 mM dithiothreitol, and protease inhibitor cocktail). Following addition of 1 μl of [γ-32P]ATP, the beads containing the immunoprecipitated proteins were incubated for 15 min at 37°C. The proteins were then solubilized in 10 μl 4x SDS lysis buffer and separated by PAGE.
RNA isolation.
HeLa cells were mock infected or infected with 10 PFU per cell of HSV-1 (18) in the presence or absence of 400 μg/ml of phosphonoacetic acid. At different time points cells were washed with phosphate-buffered saline, trypsinized, pelleted, and suspended on ice in 50 mM Tris-HCl, pH 8, 140 mM NaCl, 1.5 mM MgCl2 and NP-40 (0.5%). Cytoplasmic RNA was extracted and purified using the RNeasy mini kit (QIAGEN), including a DNase step on the column, which was performed using the manufacturer's instructions. Equal amounts of RNA from each sample were subjected to one-step reverse transcription-PCR by using VP22-specific primers to amplify VP22. The primers for VP22 were TTCGTCCAGTACGACGAGTCGGATTA and ATGGTGGTGATGCCAAGGAGTTCGT. PCR products were separated on a 1% agarose gel, stained with ethidium bromide, and visualized by UV trans-illumination. Reverse images are shown.
Vectors for in vitro transcription/translation.
For in vitro transcription/translation of full-length VP22, the plasmid pGCM22b was first constructed by inserting the BglII fragment from pGE109, containing the complete UL49 open reading frame (15), into the BamHI site of the commercial vector pGEM3Zf(+) (Promega).
The construction of N- and C-terminally truncated VP22 variants was based on the plasmid pUL49ep and related plasmid pUL49-4 (24). The plasmid pUL49-4 contains the entire UL49 open reading frame engineered with a BamHI site preceding the start codon and a HindIII site following the stop codon. A series of plasmids containing linker insertions at different positions was made. Oligonucleotides containing a BglII site were first inserted into the RsaI site (at codon 60), the HincII site (at codon 159), the MscII site (at codon 172), the SmaI site (at codon 192), the NaeII site (at codon 194), and the HincII site (at codon 267), to create pUL49-4 ins60, ins159, ins172, ins192, ins194, and ins267, respectively.
pUL49-4 del(119-192) was constructed by removing a SmaI fragment, containing codons 119 to 192, from pUL49-4 and inserting the same oligonucleotide. Carboxy-terminal deletions of VP22 were based on fragments from these plasmids inserted into pGCM25, which provides stop codons in three frames. Thus, plasmids pGCM28, pGCM29, and pGCM31 were constructed by inserting the BamHI/BglII fragments from pUL49-4 ins192, ins159, and ins267, respectively, into BamHI-digested pGCM25. This created expression vectors for VP22 codons 1 to 192, 1 to 159, and 1 to 267, respectively.
Vectors for in vitro transcription/translation of the amino-terminal deletions of VP22 were based on pGCM32 and pGCM33, which provided start codons in two different frames downstream of the T7 promoter. Plasmids pGCM32 and pGCM33 were constructed by inserting the annealed oligonucleotides 5′-AATTCATGGGCG-3′ and 5′-GATCCGCCCATG-3′, and 5′-AATTCATGGGCGG-3′, 5′-GATCCCGCCCATG-3′, respectively, into EcoRI/BamHI-digested pGEM3Zf(+). This resulted in the addition of a start codon (underlined above) downstream of the T7 promoter with the BamHI site in two different reading frames relative to the start codon. Plasmids pGCM35 and pGCM37 were then constructed by inserting the BglII/HindIII fragments from pUL49-4 ins60 and ins172, respectively, into BamHI/HindIII-digested pGCM32. This created expression vectors for VP22 codons 60 to 301 and 172 to 301, respectively. Plasmid pGCM36 was constructed by inserting the BglII/HindIII fragment from pUL49-4 ins159, into BamHI/HindIII-digested pGCM33, creating an expression vector for VP22 codons 159 to 301.
Production of 35S-labeled in vitro-synthesized proteins (TnT protein).
In vitro transcription/translation reactions were performed using the Promega TNT coupled rabbit reticulocyte lysate system according to the manufacturers instructions. Each 50 μl reaction contained 1 μg of plasmid DNA, 40 units RNasin (Promega), 0.2 mM amino acid mixture minus methionine, 40 μCi [35S]methionine (Amersham), 1 μl TNT T7 polymerase, 2 μl TNT reaction buffer, and 25 μl TNT rabbit reticulocyte lysate. The reaction mixtures were incubated for 90 min at 30°C, and stored at −20°C. For the purpose of clarity, all in vitro-transcribed/translated 35S-labeled products are referred to as TnT products.
Size measurements.
For analysis of higher-order assemblies in infected-cell extracts or of in vitro-synthesized VP22, we used both size exclusion chromatography and glycerol gradient sedimentation, from which the sedimentation coefficient (in Svedberg units) is calculated. From these measurements the apparent size of a protein or protein assembly can be calculated.
Size exclusion chromatography.
Size exclusion chromatography was performed using a Superdex 200 HR 10/30 column (Pharmacia). The column was equilibrated with 3 column volumes (72 ml) of GF-150 or GF-500 buffer containing 20 mM Tris-HCl (pH 8.0), 20% vol/vol glycerol, plus the protease inhibitor cocktail, and 150 mM or 500 mM NaCl. Infected-cell extracts (50 μl, 1.25 × 106 cell equivalents) were first dialyzed against GF-150 or GF-500 buffer, mixed with an equal volume of the corresponding buffer, and then clarified by centrifugation at 12,000 × g for 10 min at 4°C, before loading onto the column. For in vitro-synthesized samples, 50 μl of TnT product were used and processed identically. Fractions (0.5 ml) were collected, and 50 μl of each fraction was separated by SDS-PAGE. Radiolabeled bands in the gels were quantitated using a Molecular Dynamics PhosphorImager. The values for each fraction are expressed as a percentage of the maximum value obtained for each run, and plotted versus elution volume. The elution profile of known molecular mass markers (Pharmacia) were analyzed in GF-150 buffer.
Glycerol gradient centrifugation.
Size analysis by glycerol gradient centrifugation was performed in 12 ml polyallomer tubes. A 0.2 ml 60% glycerol cushion was made at the bottom of each tube in buffer (50 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol, and the protease inhibitor cocktail) containing 150 mM NaCl (GL-150 buffer) or 500 mM NaCl (GL-500 buffer). Onto this cushion, a 5% to 20% glycerol gradient was formed (total 11.2 ml), in either GL-150 or GL-500 buffer. The gradients were loaded with 100 μl of infected-cell extract (2.5 × 105 cell equivalents), or 100 μl TnT product and centrifuged at 39,000 rpm, for 28 h at 4°C (Sorvall TFT 41.1 rotor). Fractions were collected using an automatic drop counter set to 15 drops per fraction (approximately 400 μl per fraction) and 50 μl of each fraction was separated by SDS-PAGE. Molecular mass sedimentation markers were fractionated in parallel using GF-150 buffer.
Calculation of the Stokes radius using size exclusion chromatography.
The Kav the classical method of describing the elution position of a protein from size exclusion chromatography is calculated as Kav = (Ve − V0)/(Vt − V0), where Ve is the peak elution volume of a protein, V0 is the void volume of the column, and Vt is the total volume of the column. All the elution profiles in this work are shown in terms of elution volume (Ve, in ml). The Kav value of the peak elution of a protein is a function of its Stokes radius. Since Ve is linearly proportional to Kav, a similar function exists between Ve and Stokes radius. Having demonstrated a linear relation between the Ve and the Stokes radii of known protein standards (data not shown), a calibration equation was used for calculating the Stokes radii from Ve.
Calculation of the sedimentation coefficient using glycerol gradient sedimentation.
Protein standards were sedimented in duplicate gradients and their peak positions measured by SDS-PAGE analysis of the fractions. A standard curve was derived by plotting the sedimentation coefficients for known standards, e.g., aldolase 7.3 S, bovine serum albumin 4.3 S versus peak elution. The linear regression equation (data not shown) from this standard curve was used for calculating the sedimentation coefficients of VP22 or VP16 species.
Western blotting.
Following separation by SDS-PAGE, proteins were electrophoretically transferred onto nitrocellulose membranes. The membranes were blocked for 1 h in phosphate-buffered saline containing 5% wt/vol nonfat dried milk, 0.2% Tween 20 and then processed by standard methods. Primary antibodies were anti-VP22 mouse monoclonal antibody P43 (1:100), anti-VP22 rabbit polyclonal serum AGV30 (1:5,000), and anti-VP16 mouse monoclonal antibody LP1 (1:1,000). Secondary antibodies, depending on the primary antibody were: horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Bio-Rad), horseradish peroxidase-conjugated goat anti-rabbit (Bio-Rad), and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin M (μ-chain specific, Sigma). Secondary antibodies were diluted 1/2,000 for detection by diaminobenzidine or 1/4,000 for detection by chemiluminescence.
Cross-linking experiments.
VP22 159-301 was expressed as a His-tagged product and purified to homogeneity by nickel-chelating and cation exchange chromatography exactly as previously described (31). Protein samples (12 μg) were incubated with either of various homobifunctional chemical cross-linking agents bis(sulfosuccinimidyl)suberate (BS3), ethylene glycolbis(sulfosuccinimidylsuccinate) (sulfo-EGS), or disulfosuccinimidyl tartrate (sulfo-DST) according to manufacturers standard protocol (Pierce). The reagents chosen are reactive towards primary amines, but vary in the length of the spacer chains between functional groups: sulfo-DST (6.4 Å), BS3 (11.4 Å), and sulfo-EGS (16.1 Å). In brief, protein samples were incubated with increasing concentrations (60, 150, and 300 μM) of the cross-linkers in citrate buffer on ice for 2 h. The reactions were quenched by the addition of 50 mM Tris for 30 min on ice, and samples then denatured in SDS lysis buffer and separated by SDS-PAGE under reducing conditions.
RESULTS
Expression of VP22 during HSV-1 infection.
The accumulation of HSV-1 VP22 protein and rate of synthesis were analyzed by Western blot analysis or pulse-labeling analysis using anti-VP22 specific polyclonal antiserum AGV30 (14) or the anti-VP22 monoclonal antibody P43 (18). HeLa cells were infected with HSV-1 (multiplicity of infection, 10) or mock infected, and pulse-labeled with [35S]methionine at different times postinfection. The cells were lysed, total cell extracts were prepared and separated by SDS-PAGE, and the gels were subjected to autoradiography (Fig. 1a). To identify bands corresponding to VP22 and as a comparison VP16, samples were electroblotted and the membranes were subjected to autoradiography or probed with monoclonal antibodies to detect VP22 (P43) and VP16 (LP1). An example showing the analysis for one of the later time points is shown in Fig. 1b. Alignment of the VP16 and VP22 bands detected by immunoblotting with the 35S-labeled profile on the same blot allowed assignment of the bands corresponding to VP16 and VP22 in the pulse-labeling time course (Fig. 1a, arrows).
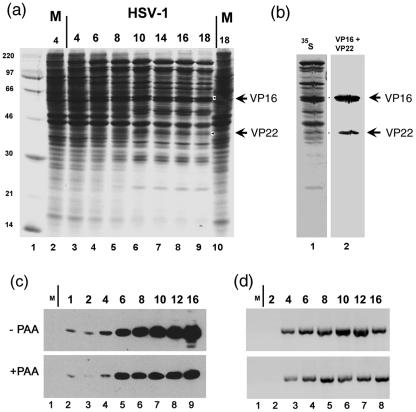
FIG. 1.
Time course of VP22 and VP16 accumulation. (a) After infection (HSV-1, multiplicity of infection of 10) or mock infection (M), HeLa cells were pulse-labeled with 20 μCi/ml [35S]methionine for the times shown above each lane (hours after infection or mock infection, labeling interval 2 h). Total cell lysates (5 × 104 cell equivalents) from infected (lanes 3 to 9) or mock-infected cells (lanes 2 and 10) were then separated by SDS-PAGE and the gels were exposed for autoradiography. (b) In parallel a duplicate gel containing samples from the 16-h time point was electroblotted and probed with anti-VP22 (P43) and anti-VP16 (LP1) antibodies. Detection of VP22 and VP16 by chemiluminescence is shown in lane 2, while the corresponding autoradiogram is shown in lane 1. The bands corresponding to VP22 and VP16 were then assigned and are indicated by arrows in panels a and b. The migration of 14C-labeled mass markers is shown to the left of panel a. (c) Accumulation of VP22 in the presence and absence of phosphonoacetic acid. Western blot analysis of the accumulation of VP22 in the presence and absence of phosphonoacetic acid (PAA). Total cell lysates of infected or mock-infected cells (lane 1), harvested at the times indicated above each lane after infection in the absence or presence of 400 μg/ml phosphonoacetic acid, were separated by SDS-PAGE, electroblotted, and probed with anti-VP22 antibody (AGV30). Detection was by chemiluminescence. (d) Cells were infected in the presence and absence of PAA as before, RNA isolated at the times indicated, and VP22-specific RNA amplified by RT-PCR using specific primers. The results are consistent with those obtained by analyzing protein synthesis and show progressive increase of VP22 RNA in the absence of PAA compared to a plateau at 6 to 8 h in the presence of PAA.
A novel labeled band corresponding to VP16 could be readily detected by 6 h postinfection while that for VP22 was more clearly detected slightly later, between 8 and 10 h (Fig. 1a, lanes 3 to 9). It is difficult to judge the relative kinetics of VP22 expression compared to VP16 by crude total accumulation (whether in pulse-labeling or Western blotting) and such kinetics showed minor differences in absolute rates over the course of several experiments. Nonetheless semiquantitative analysis of the corresponding bands by PhosphorImager analysis indicated that VP16 was consistently synthesized in greater amounts (three- to fourfold) than VP22, taking into account a 3:1 ratio of methionine content between VP16 and VP22.
To examine more qualitative aspects of the relative kinetics of VP22 we examined accumulation in the presence and absence of phosphonoacetic acid, an inhibitor of HSV DNA synthesis. Cells were infected in the presence and absence of phosphonoacetic acid (400 μg/ml), harvested at different times after infection and total lysates separated by SDS-PAGE, electroblotted and probed with anti-VP22 antibody (AGV30). In the absence of phosphonoacetic acid, VP22 was detectable in this case at 1 h postinfection, increasing by 4 h, with a progressive accumulation during the remainder of the infection (Fig. 1c, lanes 2 to 9). In the presence of phosphonoacetic acid, VP22 was still detected at 1 h postinfection with accumulation until 6 h postinfection to a level similar to that observed in the absence of the inhibitor. From 6 h onward, there was little further increase in the accumulation of VP22 in the presence of phosphonoacetic acid. Consistent results were obtained when analyzing VP22 expression at the level of RNA accumulation (Fig. 1d), showing a progressive increase in RNA levels in the absence of PAA, compared to a plateau from approximately 6 h onward in the presence of PAA. Thus, VP22 exhibits early-late kinetics, requiring viral DNA replication for maximal expression.
Expression of VP22 was further analyzed by immunoprecipitation from soluble extracts produced from infected-cells labeled with [35S]methionine at different times postinfection as above (Fig. 2). The amounts of VP22 labeled at each time point were also quantified by PhosphoImager analysis (data not shown). De novo expression of VP22 could be detected by 2 to 4 h postinfection and increased until 14 to 16 h postinfection, followed by a decrease in rate of synthesis at the late times of infection. Only a minor background of nonspecifically bound species from mock-infected cells was observed (lanes 2), and the immunoprecipitation analysis demonstrated the coprecipitation with VP22 of several specific infected-cell proteins (Fig. 2a, arrows). Of the VP22-associated proteins, the 65-kDa protein was the most prominent, with peak binding to VP22 at around 8 to 14 h postinfection (Fig. 2a, lanes 6 to 8). We also observed 57-kDa and 47-kDa proteins coprecipitating with VP22 with peak binding in the same time intervals, a 150-kDa protein which appeared more abundant slightly later, 14 to 18 h postinfection (Fig. 2a, lanes 9 to 10), and two other VP22-associated proteins of 25 kDa and 80 kDa, also observed at late times of infection. The differences in timing of the peak appearance of these VP22-interacting species may reflect a qualitative temporal difference in interaction, with perhaps the 80-kDa and 25-kDa species associating in a distinct fashion later than the others, but without information on the total abundance of the interacting species it is presently difficult to make such a conclusion. However, what is clear is that a number of specific infected-cell proteins associated with VP22.
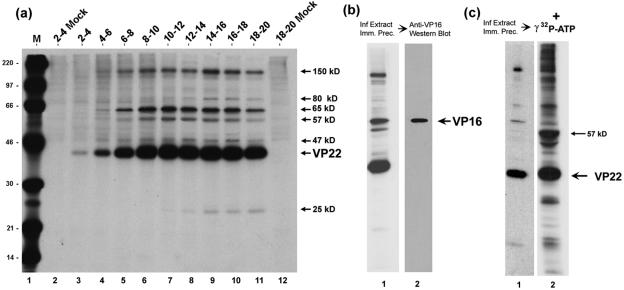
FIG. 2.
Analysis of VP22 interacting species by immunoprecipitation. (a) HeLa cells were infected (multiplicity of infection, 5) or mock-infected and pulse-labeled with 20 μCi/ml [35S]methionine for the times shown above each lane (hours). Soluble extracts were prepared and VP22 was immunoprecipitated with anti-VP22 antibody AGV30. Equivalent samples were separated by SDS-PAGE and exposed for autoradiography. The position of VP22 and the VP22-associated proteins is shown to the right of the panel. Lane M, 14C-labeled protein size markers with masses shown to the left of the panel in kDa. (b) VP22 was immunoprecipitated with AGV30 from infected-cell extracts labeled with 20 μCi/ml [35S]methionine from 12 to 16 h postinfection. The samples were separated and exposed for autoradiography as before. A duplicate gel was electroblotted onto nitrocellulose and the blot probed with anti-VP16 monoclonal antibody (LP1). VP16 was detected by Western blotting of the VP22 immunoprecipitate and comigrated with the 65-kDa band detected by autoradiography. (c) A kinase activity immunoprecipitates with VP22. Autoradiograph of the immunoprecipitation of VP22 from infected-cell extracts performed as in panel a. Duplicate immunoprecipitation reactions were equilibrated in kinase buffer and [γ-32P]ATP. Following incubation at 37°C for 15 min the proteins were solubilized in SDS lysis buffer, separated on SDS-PAGE, and exposed for autoradiography of 32-P labeled bands. The positions of VP22 and the major in vitro phosphorylation product are shown the right of the panel.
VP16 is the 65-kDa VP22-associated protein.
The 65-kDa protein which coprecipitated with VP22 is of the correct size for VP16, and we have earlier demonstrated in reciprocal experiments that a population of VP22 was bound to VP16 when the latter was immunoaffinity purified from infected cells (12). To confirm that the 65-kDa band was indeed VP16, infected cell extracts immunoprecipitated with anti-VP22 antibody were subsequently probed by Western blotting with anti-VP16 monoclonal antibody (Fig. 2b, lane 2). VP16 was detected specifically in the immunoprecipitation from infected-cell extracts, comigrating as expected with the labeled 65-kDa band.
A kinase activity associates with VP22.
In considering the apparent molecular weights of the other VP22-associated proteins, a candidate for the 57-kDa VP22-associated protein is UL13, which like VP22 is assembled within the tegument of HSV virions (6, 32). UL13 is a kinase which has been implicated in both virus-induced nuclear kinase activity and tegument associated kinase activity (6), and previous analysis of phosphorylation in vitro also showed that VP22 was a target for UL13 (5). Unfortunately, the available antibody against UL13 does not detect the protein in Western blot analysis. Alternatively, to examine whether a kinase activity was coprecipitated with VP22, we performed in vitro phosphorylation assays following immunoprecipitation of VP22 from infected or mock-infected cell extracts (Fig. 2c). Anti-VP22-immunoprecipitated samples (35S-labeled profile, Fig. 2c, lane 1) were incubated in a kinase buffer together with [γ-32P]-ATP. Of the two main 32P-labeled bands (Fig. 2c, arrows) one clearly comigrated with VP22, while the other migrated with the 57-kDa species. Judged by the 35S-labeled profile, this latter species was present in relatively minor amounts yet represented, after VP22, the next most abundant phosphorylated species. While these results do not demonstrate that the 57-kDa band is UL13, they do demonstrate firstly that a kinase activity coprecipitates with VP22, secondly that VP22 is a substrate for this kinase, and that the 57-kDa band which coprecipitates with VP22 is also a substrate for this activity and likely the most efficient substrate.
Of the other species which coprecipitated with VP22, a candidate for the 80-kDa VP22-associated protein is VP13/14, which has an apparent molecular mass of ca. 82 kDa and is also a tegument-associated protein, the product of the UL47 gene. Using the anti-UL47 polyclonal antibody (R220) we have shown that the 80-kDa VP22-associated protein comigrates with the VP13/14 proteins (data not shown). However, we have attempted but have been unable to demonstrate a reciprocal VP22 coprecipitation using the anti-UL47 antibody in immunoprecipitation assays. Precise identification of these bands and the additional less prominent VP22-associated species will be pursued elsewhere and the remainder of this work concerns further biochemical characterization of the nature of VP22 in infected cells and reconstruction assays.
Biochemical analysis of VP22 in infected-cell extracts.
VP22 and VP16 are among the most abundant components of the HSV virion, and as shown here and previously (12) interact with one another. We next wished to examine whether any higher-order assemblies of either component could be demonstrated in soluble extracts from HSV infected cells. To that end HeLa cells were infected with HSV-1 at a multiplicity of infection of 10, labeled with [35S]methionine from 12 to 16 h postinfection, and soluble extracts were prepared and fractionated by size exclusion or by sedimentation analyses.
Size exclusion chromatography of the extracts was performed using a Superdex 200 HR 10/30 column which had been calibrated with a range of markers. Following fractionation the samples were first analyzed by SDS-PAGE and autoradiography to give an estimate of separation of the total labeled cell proteins (Fig. 3a). The overall protein profile from this fractionation procedure indicates that the infected-cell proteins were reasonably well separated, with infected-cell-specific bands ranging from the 30-kDa up to the 600-kDa size range. The same fractions were then analyzed by Western blot analysis using the anti-VP22 antibody (Fig. 3b). The migration of VP16 was also analyzed and the positions of relative peak elution for both proteins were calculated by scanning using a PhosphorImager and plotting the percent maximal value against the elution volume, Ve (Fig. 3b).
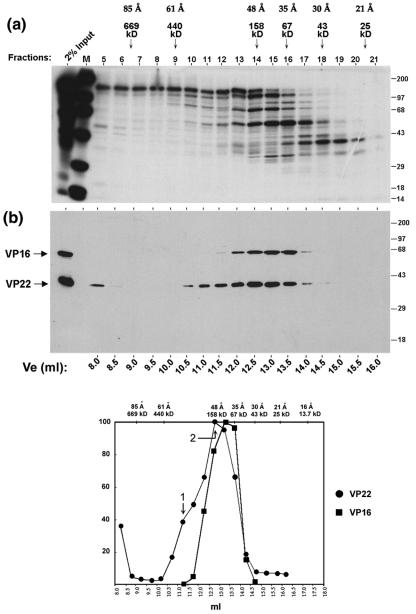
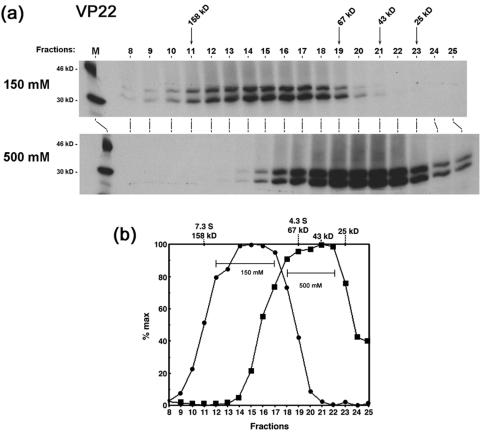
FIG. 3.
VP22 forms high molecular complexes in infected cells. (a) Autoradiograph of the elution profile from size exclusion chromatography of infected-cell extracts. High-salt extract prepared from HeLa cells infected with 5 PFU/cell of HSV-1 and labeled with 20 μCi of [35S]methionine from 12 to 16 h postinfection was fractionated by size exclusion chromatography as described in Materials and Methods using G-150 (150 mM salt buffer). One-tenth (50 μl) of each fraction (fractions 5 to 21) was separated in an SDS-PAGE gel and subjected to autoradiography. Lane M shows 14C-labeled protein standards and their positions are shown to the right of each panel. The first lane shows 2% of the input extract used in the separation analysis. The peak elution positions of the protein standards used to calibrate the size exclusion column are shown on the top of the panel along with their Stokes radii. (b) Western blot analysis of the fractions using AGV30 and LP1. A duplicate gel to that shown in panel a was electroblotted onto nitrocellulose and probed with anti-VP22 (AGV30) or anti-VP16 (LP1) antibodies. The elution volume of each fraction is shown at the bottom of the panel. The positions of VP16 and VP22 are shown to the left of the panel. (c) The elution profiles of VP22 (solid circles) and VP16 (solid squares) were measured by PhosphoImager densitometry analysis of the scanned blot and plotted as a percentage of the maximal peak versus elution volume. Peaks are numbered as discussed in the text. The elution positions of protein standards used in size exclusion chromatography are shown on the top of the graph along with their Stokes radii.
A very minor amount of VP22 (Fig. 3b, fraction 5) eluted close to the void volume and was likely to represent a minor amount of aggregated protein. The bulk of VP22 eluted within the column between fractions 10 and 17 (Ve, 10.5 to 14 ml) with a leading shoulder indicating the possible presence of two forms of VP22 (Fig. 3b, peaks 1 and 2). Peak 2 was the major form of VP22, with an estimated size of approximately 160 kDa (Stokes radius of 48 Å). Although difficult to precisely assign a peak elution for peak 1 due to overlap with peak 2, we estimated a shoulder peak at 11.25 ml corresponding to a size of approximately 290 kDa (Stokes radius of 57 Å). In the same samples, VP16 eluted with a more symmetric elution profile, indicating that it was a single species with an estimated size from size exclusion chromatography of 108 kDa and a Stokes radius of 41 Å (Fig. 3b). We could not resolve the VP22 peaks 1 and 2 better by size exclusion chromatography and further characterization was carried out using glycerol gradient sedimentation analysis.
For glycerol gradient sedimentation, the same extracts were separated in 5 to 25% glycerol gradients. To examine whether the higher-order species of VP22 were affected by salt concentration we also performed the sedimentation analysis at both physiological salt concentration (150 mM NaCl) and elevated salt concentration (500 mM NaCl) as indicated (Fig. 4a and b). As in size exclusion chromatography, VP22 migrated with a broad profile across the fractions, while VP16 exhibited a much narrower profile and sedimented at a lower size range (Fig. 4a). At physiological salt concentration, a minor amount of VP22 (Fig. 4, fraction 1) sedimented close to the bottom of the gradient and was likely due to aggregated species.
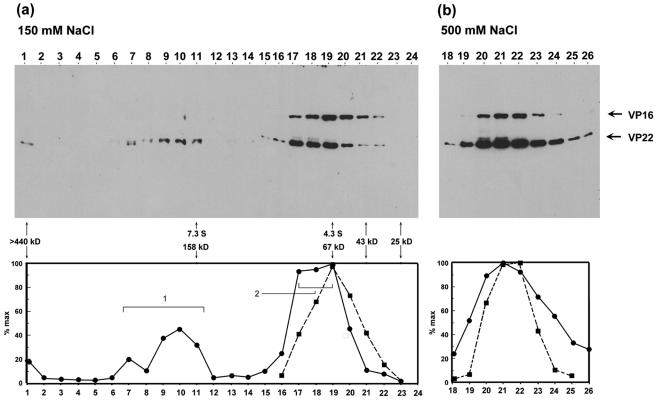
FIG. 4.
Glycerol gradient sedimentation analysis of VP22 and VP16. Extracts from infected HeLa cells (multiplicity of infection, 5) prepared as before were analyzed by glycerol gradient sedimentation in 150 mM salt buffer (panel a) or 500 mM salt buffer (panel b), as described in Materials and Methods. Fractions were collected from the bottom of the gradient and aliquots were separated by SDS-PAGE and electroblotted onto nitrocellulose and the sedimentation profiles of VP22 and VP16 gels were determined after probing with anti-VP22 and anti-VP16 antibodies. The abundance of VP22 and VP16 was measured and the profiles of VP22 (solid circles) and VP16 (solid squares) were plotted as percentages of the maximal peak versus fraction number. Peaks for VP22 are numbered as discussed in the text. The peak elution positions of the protein standards used in sedimentation analysis are shown on the graph along with their sedimentation coefficients.
Two peaks of VP22 were observed within the gradients. Peak 1 (fractions 7 to 11) had a sedimentation coefficient in the range 7.3 S to 9 S, corresponding to a size of between 160 kDa and 200 kDa. The second form, peak 2, fractionated as a separate peak (peak fractions 17 to 19), with a sedimentation coefficient in the range 4.3 S to 5.3 S, corresponding to an approximate size of between 67 kDa and 90 kDa, which may also represent more than one species. We note that during repeat analyses of these fractions, VP22 peak 1 was initially as abundant as peak 2, but upon storage and freeze-thawing of samples progressively diminished with greater accumulation of peak 2. We believe this reflects instability of multimeric forms in peak 1 and possible dissociation into peak 2. VP16, in the same gradient, eluted as a more well defined symmetric peak (sedimentation coefficient 4.3 S, corresponding to a size of 67 kDa).
To examine the salt sensitivity of the observed VP22 species, sedimentation was repeated at 500 mM NaCl. Under these conditions, no VP22 peak 1 was detected and almost all the protein was now found migrating in lower-molecular-weight complexes between fractions 19 and 24. (We note that although VP16 migrated in a position consistent with a largely monomeric form at 0.15 M NaCl, its position had shifted marginally at 0.5 M NaCl.) Although VP22 peak 1 was no longer observed at 0.5 M NaCl, nevertheless the bulk of VP22 eluted at a position at least as large as that of VP16. One interpretation consistent with these results is that this form of VP22 represents a salt-stable dimer of the protein, while the larger forms represent higher-order homo-oligomers, or assemblies with additional proteins, which dissociate to VP22 dimers at elevated salt concentrations (see below).
Table 1 summarizes the results from both size exclusion chromatography and glycerol gradient sedimentation analysis of VP22 and VP16 from infected cells. Two forms of VP22 (peaks 1 and 2) exhibit different size in each type of analysis. The less abundant peak detected (peak 1), was approximately 290 kDa by size exclusion chromatography and 167 to 200 kDa by glycerol gradient sedimentation, while the most abundant species, peak 2, was 158 kDa by size exclusion chromatography and 67 to 90 kDa by glycerol gradient sedimentation.
TABLE 1.
Summary of size analyses of VP22 and VP16 in infected-cell extractsa
| Protein | Peak | Size exclusive, Å (kDa) | Glycerol gradient, kDa (S) |
|---|---|---|---|
| VP22 | 1 | 57 (∼290) | 160-200 (7.3-9.0) |
| 2 | 48 (158) | 67-90 (4.3-5.3) | |
| VP16 | 1 | 41 (108) | 67 (4.3) |
Summary of the calculation of Stokes radius, estimated from size exclusion chromatography together with the corresponding apparent mass. Sedimentation coefficients, calculated from glycerol gradient sedimentation, are given together with size from this analysis. Major peaks 1 and 2 for VP22 correspond to those discussed in the text.
As in any such analysis, this difference in the estimated size of each species by the different biochemical methods may be due to the fact that the elution profile of a protein in size exclusion chromatography is sensitive to the shape, while in glycerol gradient sedimentation elution is largely independent of shape. It is possible that VP22 is not globular and this could explain the difference in its estimated size from size exclusion chromatography and glycerol gradient sedimentation. However, it is also possible that the 167- to 200-kDa peak seen by sedimentation does indeed correspond to the larger 290-kDa peak by size exclusion but that this assembly is more stable in gel filtration and dissociates to the broader 167- to 200-kDa species during sedimentation in gradients. Similarly it is possible that the 158-kDa peak 2 seen by size exclusion is less stable in sedimentation dissociating to the 67-kDa to 90-kDa peak 2 observed in the gradients.
Formally, since the actual makeup and stoichiometry of each peak are not known, it is not possible to conclusively state that the oligomeric species observed in size exclusion chromatography are the same as those observed in glycerol gradient sedimentation. We note that in an attempt to examine this, VP22 was immunoprecipitated from each of the gradient fractions and the immunoprecipitation products were then exposed for autoradiography to qualitatively examine the protein profiles of the complexes. However, the results were inconclusive as there was too little protein in the immunoprecipitations from the fractions to be detected by autoradiography (data not shown). More precise structural composition of the VP22 assemblies will require larger-scale preparative analyses rather than the analytical-scale analyses carried out here.
Nevertheless we can conclude from these results that VP22 exists in more than one higher-order assembly in infected cells, and that a likely minimal size for the bulk of the protein ranges from approximately 160 kDa (determined from size exclusion) to 67 to 90 kDa (from sedimentation). This is significantly larger than monomeric VP22. Moreover the results indicate the formation of at least one further higher-order assembly, of approximately 290 kDa (from size exclusion) or 167 kDa (from sedimentation).
Biochemical analysis of in vitro-synthesized VP22.
To examine further biochemical aspects of VP22, and in particular whether it existed in higher-order complexes in the absence of the other infected-cell proteins, we next performed similar sedimentation and size exclusion analyses using in vitro-synthesized [35S]methionine-labeled VP22. We also examined deletion variants of VP22, lacking N- or C-terminal regions. For ease of reference to discriminate in the text between infected-cell VP22 and in vitro-synthesized VP22, the latter has been termed VP22(TnT).
Glycerol gradient sedimentation of VP22(TnT).
Glycerol gradient sedimentation was performed on VP22(TnT) at either physiological (150 mM) or high-salt (500 mM) concentrations in order to observe any salt-induced effects. Following sedimentation the fractions were separated on SDS-PAGE gels and exposed for autoradiography. Each fraction was quantified by PhosphorImager analysis and plotted as a percentage of the maximum versus the fraction number. At 150 mM salt, VP22(TnT) sedimented as a broad peak (Fig. 5a). The major species (fractions 12 to 17) sedimented at 5.4 S to 6.9 S, corresponding to sizes of approximately 100 kDa to 146 kDa. At the high salt concentration, VP22(TnT) sedimented as a distinctly smaller though still relatively diffuse peak, ranging from 3.2 S to 4.7 S, corresponding to 34 kDa to 79 kDa, with the majority of the protein in the upper region of this range.
FIG. 5.
Glycerol gradient sedimentation analysis of in vitro-synthesized VP22. (a) Autoradiograph of the elution profile of in vitro-synthesized VP22 following glycerol gradient sedimentation analysis at 150 mM and 500 mM salt, as indicated. VP22 was expressed in vitro in a coupled transcription-translation system as described in Materials and Methods and the expressed product, VP22(TnT), subjected to sedimentation analysis exactly as before at a final concentration of either 150 mM or 500 mM NaCl. Aliquots of each of the fractions were separated by SDS-PAGE and the gels were subjected to autoradiography. Lane M shows 14C-labeled markers. The elution positions of the mass protein standards are shown on the top of the panel. (b) The level of VP22(TnT) in each of the fractions in the gels in panel a was measured using a PhosphorImager analysis and the intensities were plotted as a percentage of maximal fraction versus fraction number.
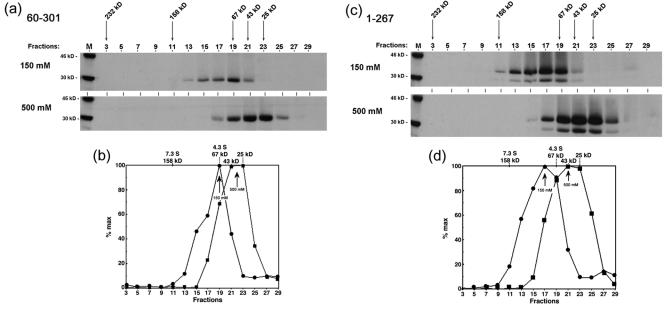
We note the presence in SDS gels of a slightly smaller product of VP22(TnT) which was observed also with the variant VP22.1-267, but not with VP22.60-301 (see below). This could be due to inefficient initiation or proteolytic clippage of the N-terminal region. It is possible that the appearance of these slightly shorter products could be incorporated into hetero-oligomers with the full-length versions, though this does not alter the main conclusions. Thus, since the predicted mass of VP22 is 32.2 kDa, our results are most consistent with the interpretation that at physiological salt concentration, the bulk of VP22(TnT) sedimented as a tetramer, perhaps with a population of dimers, and that at the high-salt concentration the protein dissociates into a salt-stable dimer, together with a population of monomers.
The sedimentation profiles of VP22.60 to 301(TnT) and VP22.1-267(TnT) at 150 mM and 500 mM salt are shown in Fig. 6. (For space considerations we have not included all data and graphical representations.) The main conclusion from this analysis was that deletion of the N-terminal 60 residues had a significant effect on VP22 migration such that at the physiological salt concentration, the bulk of VP22.60-301(TnT) sedimented as a peak of 4.3 S corresponding to a size of 68 kDa, possibly with a minor population of approximately 110 kDa (Fig. 6b). At the higher salt concentration, the main species now sedimented with a size of 33 kDa (3.2 S). The linear mass of VP22.60-301(TnT) is 25.3 kDa. Therefore, the results of this analysis are most consistent with the interpretation that deletion of the N-terminal region of VP22 resulted in destabilization, such that at normal salt concentration VP22.60-301(TnT) sedimented mainly as a dimer (rather than the main distribution as tetramers of intact VP22), and that at elevated salt VP22.60-301(TnT) was mainly monomeric.
FIG. 6.
Effect on VP22 sedimentation of deletion of N-terminal and C-terminal residues. Autoradiograph of the elution profile of VP22.60-301(TnT) and VP22.1-267(TnT) following glycerol gradient sedimentation analysis at 150 mM and 500 mM salt. Synthesis, sedimentation, and analysis were as described for Fig. 7.
This same analysis was performed on VP22.1-267(TnT). The results indicate that deletion of the extreme C-terminal residues also resulted in destabilization of VP22 (Fig. 6c,d). Thus, at 150 mM salt VP22.1-267(TnT) sedimented with a different profile to that of intact VP22, with a size of 90 kDa (5.1 S). Increased salt concentration also affected sedimentation such that at 500 mM NaCl, VP22.1-267(TnT) sedimented at approximately 40 kDa (3.6 S). Since the linear mass of VP22.1-267(TnT) is 28.6 kDa, these results are again most consistent with the presence of a dimer at normal salt concentration and monomer at 500 mM salt.
The results of the glycerol gradient sedimentation of VP22 and the amino- and carboxy-terminal deletions are summarized in Table 2. The results indicate that the in vitro-synthesized VP22 exists in a broad distribution most consistent with a mixture of mainly tetramers and dimers. It is possible that under certain buffer conditions VP22 could be stabilized so that it quantitatively migrated as a tetramer, or it may be that some additional cofactor (viral or cellular) could be required for full stabilization into a tetrameric configuration. But the results are reasonably consistent with those from infected-cell lysates. Furthermore the results indicate that the N- and C-terminal ends of VP22 are involved in promoting higher-order assembly, such that their loss results in much reduced formation of the tetrameric form with mainly dimeric forms observed, and destabilization to monomers at high salt.
TABLE 2.
Summary of sedimentation analysis of in vitro-synthesized VP22 and deletion variantsa
| Protein (size, kDa) | Peak | S | Size (kDa)
|
|
|---|---|---|---|---|
| 150 mM NaCl | 500 mM NaCl | |||
| VP22 (32.2) | Major | 5.4-6.9 | 101-146 | 34-79 |
| VP22.60-301 (25.3) | Minor | ∼5.8 | ∼113 | 34 |
| Major | 4.3 | 68 | 34 | |
| VP22.1-267 (28.6) | Minor | ∼6.6 | ∼134 | 40 |
| Major | 5.1 | 90 | ||
Summary of the sedimentation profiles of in vitro-synthesized VP22 and deletion variants. Sedimentation was performed at either 150 mM or 500 mM NaCl. The presence of minor species is discussed in the text. Only size is given for the results at 500 mM NaCl. The predicted size for each species as calculated from the linear sequence of amino acids is given below each variant in the left-hand column.
Dimerization of both the N-terminal and C-terminal products of VP22.
We also performed size exclusion chromatography on in vitro-synthesized VP22 variants encompassing the N-and C-terminal sections of the protein. These variants included VP22.1-192(TnT), 1-159(TnT), 159-301(TnT) and 172-301(TnT) and an internal deletion variant lacking residues 119 to 192, termed del(119-192)(TnT). Each variant was subjected to size exclusion chromatography, fractions were collected, aliquots were separated by SDS-PAGE and migration was measured using a PhosphorImager and plotted as a percentage of the maximum versus the elution volume. Rather than show the individual gels and quantitation from these analyses, the main observations are summarized in Table 3.
TABLE 3.
Summary of size exclusion chromatography of in vitro-synthesized VP22 and deletion variantsa
| VP22 protein (kDa) | Stokes radius (Å) | Size (kDa) in 150 mM NaCl | Oligomer |
|---|---|---|---|
| 159-301 (15.5) | 28 | 31 | Dimer |
| 172-301 (14.2) | 28 | 31 | Dimer |
| 1-267 (26.6) | 35 | 67 | Dimer |
| 1-192 (20.3) | 28 | 41 | Dimer |
| 1-159 (16.8) | 25 | 32 | Dimer |
| del(119-192 24.5) | 19 | 20 | Monomer |
Summary of the calculation of Stokes radius, estimated from size exclusion chromatography together with the corresponding size of in vitro-synthesized VP22 variants. The size as calculated from the linear sequence of amino acids is given below each variant in the left-hand column and the likely oligomerization form in the right-hand column.
VP22.1-192(TnT) and 1-159(TnT) eluted as single peaks of 41 kDa and 32 kDa, respectively. Since these proteins have linear masses of 20.3 kDa and 16.8 kDa, respectively, their elution profiles are consistent with dimer formation. Similarly VP22.159-301(TnT) and VP22.172-301(TnT) also eluted as a single species, both peaking with a size of approximately 31 kDa. The linear masses of these variants are 15.5 kDa and 14.2 kDa, respectively, and again the results are consistent with migration as dimer species. The internal deletion variant del(119-192)(TnT) has a linear mass of 24.5 kDa, and migrated with a size of 20 kDa, in this case indicating that it was likely to be largely monomeric.
Cross-linking dimers of the conserved core of VP22.
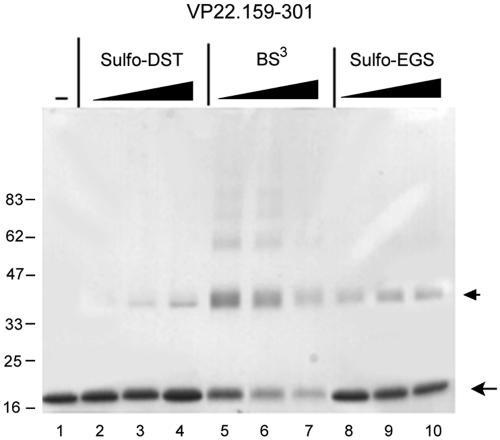
In the attempt to provide independent evidence for dimer formation by subregions of VP22, we expressed the variant VP22.159-301 in bacteria and purified the species to homogeneity. While intact VP22 did not express well in bacteria, this subregion was readily amenable to expression and purification as previously described (31). Purified VP22.159-301, devoid of any contaminating species as judged by SDS-PAGE (Fig. 7, lane 1) was incubated in the absence or presence of increasing concentrations of the homo-bifunctional cross-linking agents bis-(sulfosuccinimidyl)-suberate (BS3), ethylene glycol bis-(sulfosuccinimidyl)-succinate (sulfo-EGS), or disulfosuccinimidyl-tartrate (sulfo-DST). The reagents are reactive towards primary amines and vary in the length of the spacer chains between functional groups: sulfo-DST (6.4 Å), BS3 (11.4 Å), and sulfo-EGS (16.1 Å). After 15 min the reactions were quenched, and the samples were denatured in SDS loading buffer and analyzed by SDS-PAGE.
FIG. 7.
Cross-linking of VP22.159-301. VP22.159-301 was expressed and purified exactly as previously described (31). Protein samples (12 μg) were incubated with the homobifunctional chemical cross-linking agents disulfosuccinimidyl tartrate (sulfo-DST, lanes 2 to 4), bis(sulfosuccinimidyl)suberate (BS3, lanes 5 to 7), or ethylene glycolbis(sulfosuccinimidylsuccinate) (sulfo-EGS, lanes 8 to 9), in increasing concentrations (60, 150, and 300 μM) of the cross-linkers in citrate buffer on ice for 2 h. The reactions were quenched by the addition of 50 mM Tris for 30 min on ice, and samples were then denatured in SDS lysis buffer and separated by SDS-PAGE under reducing conditions. The positions of the VP22.159-301 in the absence of cross-linker (lane 1) and the major cross-linked species are indicated by arrows.
The single 18-kDa species of VP22.159-301 was observed in the absence of cross-linker (Fig. 7, lane 1). Both sulfo-DST (Fig. 7, lanes 2 to 4) and sulfo-EGS (Fig. 7, lanes 8 to 10) promoted the appearance of an additional species migrating at ca. 38 kDa, consistent with the cross-linking of VP22.159-301 dimers. Sulfo-EGS appeared to cross-link more efficiently than sulfo-DST, which could be consistent with its longer spacer arm (16.1Å versus 6.4 Å). Increasing the concentration of either of these cross-linkers did not result in the appearance of higher-order species. Interestingly the cross-linker BS3, while inducing the formation of stable dimers, also resulted in the appearance of lesser amounts of higher-order species (Fig. 7, lanes 5 to 7). The linker length of BS3 is shorter than that of sulfo-EGS, and while this may be a quantitative effect we did not detect the presence of higher-order species when using sulfo-EGS at higher concentrations. It may be that BS3 has a particular linker length which more readily reveals additional less abundant multimers of VP22.159-301.
We note that similar attempts to provide independent evidence for dimer formation of the isolated N terminus have proved difficult due to poor expression of this product in bacteria. Nevertheless, at least for VP22.159-301, the results together with those from size fractionation of the in vitro-synthesized VP22, provide very strong evidence for dimer formation of VP22.159-301 and support the proposal for dimer formation of the N-terminal region. This conclusion is also consistent with the destabilizing effect of deletion of the N-terminal region (i.e., 60 to 301) on formation of higher-order VP22 assemblies, likely to be tetramers.
DISCUSSION
VP22, encoded by the UL49 gene of HSV-1 (18) is among the most abundant of the tegument proteins being present at about 1,500 to 2,000 molecules per virion (21, 24). Recent results demonstrate that for HSV, while not absolutely essential for replication in culture, viruses lacking VP22 display a cell type defect in replication and an altered profile of virion-assembled proteins (11). A number of studies have begun to shed light on VP22 localization and compartmentalization during virus infection (2, 9, 13-15, 22, 23, 26, 28, 29, 34, 35). VP22 has been reported to interact with membranes in infected cells (3) and, potentially in a related fashion, to bind to the cytoplasmic tail of the glycoprotein gD (4). Interactions between VP22 and glycoproteins E and M have been reported in PRV (19). However, there is comparatively little data on the biochemical analysis of VP22 itself. Any complete understanding of, for example, how tegument proteins are recruited or nucleate virion assembly, or indeed self assemble (see below), and of the details of their functions within cells will require biochemical and biophysical characterization.
Here we report characterization of VP22 expression and initial biochemical analysis by gel filtration, glycerol sedimentation and chemical cross-linking. We could find no reports of kinetic pulse-labeling experiments of total infected protein synthesis which assigned VP22 based on comigration with an immunologically identified band and performed such analysis to give an approximation of the relative abundance of VP22 synthesized in infected cells. While such analysis has limitations, we estimate that VP22 is synthesized in significantly lower amounts than VP16, the other major tegument protein in HSV. Since these proteins are both major components and recruited in approximately similar abundance (7, 21, 24), this analysis supports the proposal that there may be a mechanism(s) for selective tegument recruitment. For VP22 it has further been proposed that phosphorylation may be involved, with the hypophosphorylated form specifically assembled into virions (17).
In coprecipitation assays a number of infected-cell proteins were detected in VP22 immunoprecipitates, indicating that at least a population of VP22 is complexed with other species. We identified one of these proteins as VP16, consistent with previous results showing in reciprocal experiments that VP22 copurified with VP16 (12). We also demonstrated a 57-kDa species interacting with VP22, which we believe is likely to be the UL13 protein kinase, and indeed a kinase activity which phosporylates VP22 was present. This interpretation is consistent with previous results showing that virions contain a kinase, lacking in a UL13 −ve strain, which phosphorylates VP22 (5), and also that while VP22 is phosphorylated by a cellular kinase (16, 17), its phosphorylation is stimulated by UL13 (20). While phosphorylation of VP22 and association with UL13 were not the main aims of this analysis, it is possible that the coprecipitation of VP22 with VP16, the kinase/UL13, the 80-kDa, and other species reflects the existence of a macromolecular complex with a role in either phosphorylation or assembly (see below). Further analysis in this work concentrated upon size fractionation of VP22 both in infected cells and of the protein in isolation, particularly with a view to the possibility of higher-order complexes of the protein which reflect its role in assembly.
At physiological salt concentrations using both techniques, VP22 was present in several high-molecular-weight forms in infected-cell extracts. By size exclusion chromatography the main species of VP22 had a size of approximately 160 kDa with a leading fraction indicating the presence of a larger population (Fig. 3, Table 1). By sedimentation analysis the main population (Fig. 4, peak 2) fractionated with a size of 67 to 90 kDa, with a distinct well-separated population migrating at ca. 167 to 200 kDa (Fig. 4, peak 1). To help determine the nature of these VP22 species, we examined the size profile of the protein in isolation. Numerous attempts to express and purify the intact protein in e.g., bacterial expression systems failed and we utilized in vitro-synthesized VP22 using reticulocyte lysates as a source. While imperfect, this allowed us to perform analytical-scale experiments on the protein; and the conclusions were reinforced by cross-linking analysis of the isolated, purified C-terminal region of VP22, which is expressed well in bacteria (31).
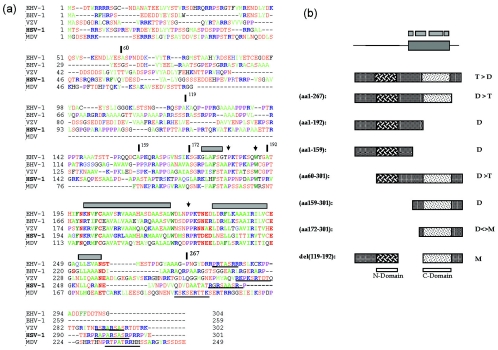
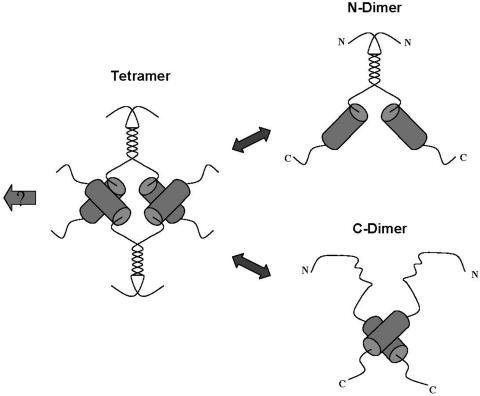
Thus, VP22 synthesized in vitro fractionated in a pattern most consistent with a mixture of dimers and tetramers at physiological salt concentration, dissociating into mainly dimers at the high salt concentration. The results from the biochemical analysis of the deletions are consistent with this hypothesis and point to the existence of two dimerization domains (see summary in Fig. 8b). Cross-linking studies performed on the purified C-terminal region further support the proposal for its ability to form dimers. The minimal amino-terminal domain seems to be contained within residues 60 to 119 while the carboxy-terminal domain could be refined to within residues 172 to 267, although residues beyond 267 are required for full activity. We propose that each domain can homo-dimerize, and these dimers interact to form tetramers. Each domain seems to be able to function somewhat independently, but deletion at the termini does abrogate tetramer formation. A model of VP22 oligomerization based on these results is illustrated in Fig. 9.
FIG. 8.
Conservation of a core domain and summary of VP22 oligomerization studies. (a) Sequences of VP22 homologues from equine herpesvirus type 1 (EHV-1), bovine herpesvirus type 1 (BHV-1), varicella-zoster virus (VZV), HSV-1, and Marek's disease virus (MDV) were aligned using the Clustal multiple-sequence alignment algorithm. The alignment was imported into the Genedoc program and alignments were optimized using information from all pairwise alignments. For ease of interpretation, residues have been color coded: A, F, I, L, M, V, W, and Y indicated in green; D, E, N, S, and T are in red; R and K are in blue; and P is in purple. Numbers relating to the boundaries of several HSV constructs used in this work are indicated with vertical bars above the sequence, while regions predicted to adopt secondary structure are indicated by gray boxes. A potential conserved motif towards the C-terminal end is underlined. (b) Summary of the oligomerization state of VP22 and various subregions. The top schematic indicate the conserved core with regions predicted from computer algorithms to adopt potential alpha-helical structure shown above. The preponderance of tetramers (T) and dimers (D) and monomers (M) is indicated for each VP22 species. Shading in two regions indicates the conclusion that VP22 contains two dimerization interfaces (N) and (C) as discussed in the text.
FIG. 9.
Model of VP22 oligomerization states. Based on the results from size fractionation experiments and analysis of subregions of the protein, VP22 is illustrated as a tetramer, having dimerization interfaces within the N-terminal and C-terminal regions. Dissociation of the tetramer leads to dimers, which for the full-length protein could formally be composed of dimers held together by interactions at either interface. The conserved core region and interface at the N terminus are not designed to indicate a particular structure, though computer algorithms predict alpha-helix formation within the C-terminal domain of the protein, as discussed in the text. The arrow to the left indicates the possibility that VP22 tetramers are further recruited into higher-order complexes, as discussed in the text.
In light of these data, the results from infected-cell extracts suggest that peak 1 (with a size of 167 kDa) may be composed of tetramers. Similarly, one of the forms in peak 2 could be a dimer of approximate 80 to 90 kDa. Given the association between VP16 and VP22 reported previously (12) and demonstrated here in reciprocal immunoprecipitation experiments, one question we wished to address was the possible identification of large molecular complexes containing these two proteins in size fractionation analysis. However, the highest-molecular-mass forms of VP22 in either sedimentation or size fractionation did not contain detectable VP16. It remains possible that additional, less-stable larger complexes containing VP22 and VP16 exist but dissociate, and indeed the largest forms of VP22 were unstable after glycerol gradient fractionation.
While further attempts are in progress using different isolation and buffer conditions in an attempt to stabilize any such complexes, it is also as likely that the native VP22 peak 1 form is not associated with VP16 and thus is an independent oligomer or an assembly with components other than VP16. On the other hand VP22 peak 2 migrated in an overlapping pattern with VP16 by both size exclusion and sedimentation analysis. It was evident, particularly from the sedimentation analysis, that VP22 peak 2 was larger than VP16, and it is possible that the higher size range within this peak, i.e., around 90 to 100 kDa, could contain a complex of VP16 and a monomer (perhaps dimer) of VP22. We attempted to precipitate each of the fractions after size fractionation to determine the profile of associated proteins and whether it altered across the size range but the analytical-scale experiments reported here were not sensitive enough. However, in transfected cells expressing the proteins in the absence of other virus proteins, VP22 and VP16 clearly form some type of VP22-VP16 complex (12), and bacterially expressed VP16 and VP22.159-301 also coprecipitate (data not shown). Future analysis of infected cells on a more preparative scale and the use of purified proteins should therefore help characterize the size and stoichiometry of VP16-VP22 complexes.
Analysis of the sequence alignments of the VP22 homologues in other alphaviruses reveals some interesting features pertinent to these studies and warrants some speculation (Fig. 8a). While there are short sections of homology within the N-terminal region, a major feature is the presence of a well conserved 100-residue section conserved across the family corresponding in HSV-1 approximately to residues 160 to 260. Further analysis using secondary-structure prediction algorithms confine probabilities of secondary structure to within this region, indicating a series of potential alpha helices (Fig. 8a, gray boxes) linked by coils or turns. The presence of, for example, a completely conserved asparagine (HSV residue 197) and tryptophan (residue 221) is consistent with the preference for these in capping and terminating alpha helices, and they are followed by conserved proline residues indicative of a tight turn into the next helix.
Whether or not the actual VP22 structure reflects the prediction algorithms, it seems likely that the ability to express and purify region 159 to 301 is related to its ability to form an isolated folded structure, which as proposed above forms dimers. Furthermore we have previously shown that addition of short negatively charged molecules (phosphodiester, or phorothioate oligonucleotides) catalyzes the ability of this core domain to assemble into a particle which has a regular spherical shape (31). Further work is in progress to examine whether this can be recapitulated with negatively charged peptides and whether this activity reflects any aspect of recruitment and assembly into the tegument. e.g., interaction with the negatively charged tail of VP16 (12). While there is clearly redundancy in the requirement for tegument proteins, we believe it is likely that the ability of major tegument proteins such as VP22 to homo-oligomerize, or to be effectively cross-linked by virtue of interactions with other tegument proteins will be important features in their recruitment into virions. This study provides a useful starting point for consideration of those features of VP22.
REFERENCES
- 1.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 2.Blouin, A., and J. A. Blaho. 2001. Assessment of the subcellular localization of the herpes simplex virus structural protein VP22 in the absence of other viral gene products. Virus Res. 81:57-68. [DOI] [PubMed] [Google Scholar]
- 3.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope gD binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 5.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, C., A. J. Davison, A. Dolan, M. C. Frame, D. J. McGeoch, D. M. Meredith, H. W. Moss, and A. C. Orr. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303-311. [DOI] [PubMed] [Google Scholar]
- 7.Dargan, D. 1986. The structure and assembly of herpesviruses, p. 359-437. In J. R. Harris and W. Horne (ed.), Electron microscopy of proteins, vol. 5:VIral structure. Academic Press, New York, N.Y. [Google Scholar]
- 8.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., W. Hafezi, A. Whitely, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-endocing gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, G., D. O'Reilly, and P. O'Hare. 1999. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J. Virol. 73:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, G. D., and D. M. Meredith. 1992. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J. Gen. Virol. 73:723-726. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiss, B. J., J. E. Tavis, L. M. Metzger, D. A. Leib, and L. A. Morrison. 2001. Temporal regulation of herpes simplex virus type 2 VP22 expression and phosphorylation. J. Virol. 75:10721-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of Type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotsakis, A., L. E. Pomeranz, A. Blouin, and J. A. Blaho. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J. Virol. 75:8697-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60-68. [DOI] [PubMed] [Google Scholar]
- 25.Liand, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL9 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, A., P. O'Hare, J. McLauchlan, and G. Elliott. 2002. Herpes simplex virus tegument protein VP22 contains overlapping domains for cytoplasmic localization, microtubule interaction, and chromatin binding. J. Virol. 76:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 30.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Normand, N., H. van Leeuwen, and P. O'Hare. 2001. Particle formation by a conserved domain of the herpes simplex virus protein VP22 facilitating protein and nucleic acid delivery. J. Biol. Chem. 276:15042-15050. [DOI] [PubMed] [Google Scholar]
- 32.Overton, H. A., D. J. McMillan, L. S. Klavinskis, L. Hope, A. J. Ritchie, and P. Wong-kai-in. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 33.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Leeuwen, H., G. Elliott, and P. O'Hare. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 76:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, C. 1984. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature (London) 309:229-234. [DOI] [PubMed] [Google Scholar]