Abstract
Type B leukemogenic virus is a variant of mouse mammary tumor virus (MMTV) that causes thymic lymphomas rather than mammary tumors in mice. We demonstrate that conversion of a mammotropic MMTV to a T-cell-tropic virus requires two alterations in the long terminal repeat: (i) acquisition of a T-cell-specific enhancer and (ii) loss of transcriptional repression through deletion of negative regulatory elements (NREs) or by suppression of NRE activity after appropriate positioning of the enhancer.
Mouse mammary tumor virus (MMTV) is an oncogenic retrovirus that induces mammary tumors in mice (8), whereas a T-cell-tropic variant, type B leukemogenic virus (TBLV), causes thymic lymphomas (1). Compared to the MMTV long terminal repeat (LTR), the TBLV LTR has a 443-bp deletion that eliminates a negative regulatory element (NRE) (2, 3, 9-11) and a 124-bp substitution that generates a triplicated element, which serves as a T-cell enhancer (12). The deletion and substitution truncate the superantigen (sag) gene (2) to eliminate the C-terminal interaction with the T-cell receptor (TCR) (22, 24). All T-cell-tropic MMTV variants isolated from thymic lymphomas harbor partial or complete NRE deletions, and some reportedly also contain T-cell enhancers (7, 13, 21). The MMTV NRE has been shown to repress viral expression in a number of cell types, including lymphoid cells (3, 11, 19, 25), and mutation of a SATB1-binding site within the NRE increases MMTV LTR-reporter gene expression in the lymphoid tissues of transgenic mice relative to that in the mammary gland (3, 11).
To determine the contribution of the MMTV LTR to viral disease specificity, various mutant proviruses have been constructed by our laboratory and others. Viruses produced using an MMTV molecular clone, HYB-MTV (20) (Fig. 1A), which contained the entire LTR from thymotropic MMTVs MA and DL8 (23) or a 440-bp segment (encompassing the deleted and substituted region) of the TBLV LTR (14) induced thymic lymphomas instead of mammary tumors. These studies suggested that cis-acting LTR elements determine viral tumorigenicity since TBLV Sag is dispensable for lymphomagenesis in BALB/cJ mice (14). To distinguish the relative contributions of the NRE and the T-cell enhancer to MMTV-induced disease, we analyzed additional mutants.
FIG. 1.
Construction and characterization of mutant MMTV LTRs. (A) Diagram of the hybrid proviral clone of MMTV, HYB-MTV. The 5′ portion of the provirus, including the LTR, is derived from the endogenous MMTV strain Mtv1 (white), while the 3′ portion, including the LTR, is derived from the exogenous C3H MMTV (gray). The coiled extension at the 3′ end of the provirus denotes the flanking cellular DNA at the site of proviral integration. The recognition sites of restriction enzymes used in this study are also shown. (B) Diagram of the MMTV, TBLV, and mutant MMTV LTRs. The arrow on the MMTV LTR shows the transcription start site at the U3-R border. sag coding potential is shown as a black (full-length Sag) or gray (truncated Sag) box under each LTR. (C) Transcriptional efficiencies of wild-type MMTV, mutant MMTV, and TBLV LTRs in transient-transfection assays of Jurkat T cells. Luciferase (LUC) activity is reported in light units normalized for DNA uptake as measured by cotransfection with the Renilla luciferase expression plasmid, pRL-TK. LUC activities from the TBLV and mutant MMTV LTRs are depicted relative to that from the MMTV LTR (assigned a value of 1). Standard deviations from the means of triplicate assays are shown. (D) Transcriptional efficiencies of wild-type MMTV, mutant MMTV, and TBLV LTRs in HC11 mouse mammary cells. HC11 cells were grown in the absence (white bars) or presence (gray bars) of 10−6 M DEX for 24 h prior to assays. LUC activity is reported in light units normalized for DNA uptake as measured by cotransfection with the pRL-TK reporter plasmid. LUC activities from the TBLV and mutant MMTV LTRs are depicted relative to that from the MMTV LTR in the absence of DEX (assigned a value of 1). Standard deviations from the means of triplicate assays are shown. Significance was determined by Student's t test.
The TBLV T-cell enhancer masks NRE-mediated transcriptional repression.
Two different MMTV LTRs were engineered by insertion of the TBLV T-cell enhancer (E) at the StuI site upstream of the NRE (MTVStuE LTR), similar to its position in the TBLV LTR (2), or at the AflII site downstream of the NRE (MTVAflE LTR) (Fig. 1B). In both cases, the NRE remained intact. Enhancer insertion truncated the sag gene in the MTVStuE LTR but not the MTVAflE LTR. The third mutant LTR, MTVΔNRE, has been described previously (12) and lacks the entire NRE and the flanking T-cell-enhancer sequences (Fig. 1B).
The mutant LTRs were linked to the firefly luciferase gene and used in transient-transfection assays of human Jurkat T cells. Wild-type TBLV and MTVAflE LTRs demonstrated similar transcriptional strengths that were ca. 700-fold higher than that of the MMTV LTR (Fig. 1C). As previously demonstrated, deletion of the NRE alone (MTVΔNRE) resulted in a 3-fold increase in reporter gene activity (12) (data not shown), whereas insertion of the TBLV T-cell enhancer upstream of the NRE (MTVStuE) resulted in a 250-fold elevation of luciferase activity (Fig. 1C). These results indicate that the placement of the TBLV enhancer between the MMTV NRE and the LTR promoter abolishes NRE activity in Jurkat T cells.
Since the TBLV enhancer contains a triplication of the hormone-responsive element (12), we also analyzed the transcriptional activity of the mutant LTRs in mouse mammary cells expressing functional glucocorticoid receptors. All LTRs, except for that of MTVΔNRE, had statistically similar transcriptional efficiencies after transient transfections of HC11 cells grown in the absence of a glucocorticoid, dexamethasone (DEX), suggesting that T-cell enhancer addition (12) conferred no transcriptional advantage in mammary cells (Fig. 1D). As anticipated, the wild-type MMTV LTR gave a fivefold increase in reporter activity in the presence of DEX. Glucocorticoid induction of the MTVΔNRE LTR was not statistically significant (P > 0.25), presumably because NRE deletion elevates basal MMTV transcription approximately fourfold (Fig. 1D). Enhanced glucocorticoid induction of the TBLV, MTVAflE, and MTVStuE LTRs may be attributed to altered spacing and triplication of a hormone-responsive element within the TBLV enhancer (2). However, the overall expression from the MTVStuE LTR appeared to be diminished compared to that from the MTVAflE LTR, both of which contained the NRE, suggesting that the NRE may serve as a position-dependent insulator (4). Placement of the TBLV enhancer downstream of the NRE (MTVAflE) may provide the same effect as NRE deletion (TBLV).
Lymphomagenic MMTVs require NRE loss and T-cell enhancer acquisition.
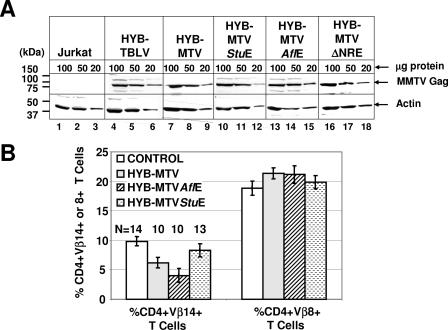
AvrII-to-SstI segments of the mutant MMTV LTRs were replaced in the corresponding region of the HYB-MTV 3′ LTR (Fig. 1A). Jurkat T cells stably expressing wild-type (Jurkat/HYB-MTV) or mutant viruses (Jurkat/HYB-MTVStuE, Jurkat/HYB-MTVAflE, and Jurkat/HYB-MTVΔNRE) were isolated. Comparable MMTV Gag expression levels of transfected cells were verified by Western blotting (Fig. 2A) prior to intraperitoneal inoculation of 2 × 107 cells into weanling BALB/cJ mice. Sag-reactive CD4+ Vβ14+ T cells were deleted at 3 months postinoculation with Jurkat/HYB-MTV and Jurkat/HYB-MTVAflE (Fig. 2B), while nonreactive CD4+ Vβ8+ T cells remained unaffected. The significantly faster T-cell deletion in HYB-MTVAflE-infected mice might be due to the higher viral loads resulting from enhanced viral transcription and replication in T cells (Fig. 1C). Deletion of Sag-reactive T cells was not detectable in mice injected with HYB-MTVStuE transfectants, consistent with our previous data that truncation of Sag by more than a few amino acids is incompatible with T-cell deletion (22).
FIG. 2.
Protein expression and Sag-mediated deletion induced by wild-type MMTV and mutant viruses. (A) Western blot analysis comparing viral Gag expression levels in Jurkat T cells expressing wild-type (Jurkat/HYB-MTV) or mutant (Jurkat/HYB-MTVStuE, Jurkat/HYB-MTVAflE, and Jurkat/HYB-MTVΔNRE) MMTV proviruses. Stably transfected Jurkat cells were derived by electroporation with the appropriate proviral constructs carrying the hygromycin resistance cassette, followed by growth selection in 250 μg/ml hygromycin. Three different amounts (100, 50, and 20 μg) of whole-cell lysates were analyzed on 8% sodium dodecyl sulfate-containing polyacrylamide gels. The Gag precursor (arrow) was detected with an MMTV capsid-specific monoclonal antibody (17) using a protocol described previously (15). The same amounts of cellular lysates were incubated with antibodies specific for actin as a control for protein loading. (B) Flow cytometric analysis of Sag-mediated peripheral deletion of T cells. Injected mice were analyzed at 3 months after injection with Jurkat T cells stably expressing wild-type or mutant MMTV proviruses. CD4+ Vβ14+ and CD4+ Vβ8+ peripheral T cells were detected using phycoerythrin-conjugated anti-mouse CD4 monoclonal antibody (RM4-5) and fluorescein-conjugated anti-mouse TCR Vβ14 (14-2) or Vβ8.1 and 8.2 (MR5-2) monoclonal antibodies from Pharmingen (San Diego, Calif.). The cells were analyzed using a FACSCalibur (Becton Dickinson, Franklin Lakes, N.J.) and CELLQuest software. The percentages of TCR Vβ+ cells in the gated CD4+ T-cell populations were calculated. The number of animals (N) analyzed from each group is indicated above each bar. The mean percentage (±standard deviation) of CD4+ TCR Vβ+ T cells in each group is shown.
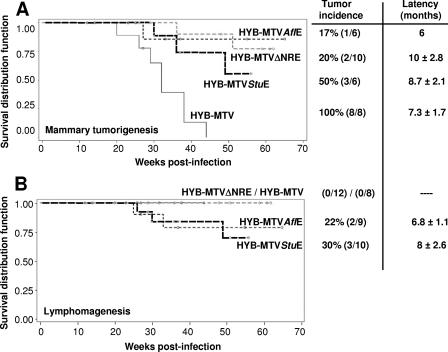
HYB-MTV induced mammary tumors with an incidence of 100% and an average latency of 7.3 ± 1.7 months. HYB-MTVΔNRE also induced only mammary tumors in 20% of the inoculated female mice within 10 ± 2.8 months, indicating that NRE deletion alone is insufficient for lymphomagenesis. Our previous data indicated that HYB-TBLV induced exclusively T-cell lymphomas, with an incidence of 50% and an average latency of 5.7 ± 1.5 months (14). However, 50% of HYB-MTVStuE-infected mice developed mammary tumors within 8.7 ± 2.1 months, whereas 30% developed thymic lymphomas within 8 ± 2.6 months. One animal infected with HYB-MTVStuE (mouse 4) developed both tumor types. HYB-MTVAflE infection induced a 17% incidence of mammary tumors and a 22% incidence of thymic lymphomas, with latencies of 6 months and 6.8 ± 1.1 months, respectively. Kaplan-Meier plots followed by Wilcoxon or log rank tests indicated that the mammary tumor incidence for each of the mutants was significantly different than that for wild-type HYB-MTV (Fig. 3A), but the difference among the various mutants was not statistically significant. Lymphomas have never been observed after infection with HYB-MTV (Fig. 3B), even after the inoculation of much larger numbers of animals (22, 26). The lower incidence of mammary and T-cell tumors induced by MTVStuE viruses may be due to a suboptimal arrangement of cis-acting sequences in the LTRs, leading to lower transcriptional activity (Fig. 1C) and favoring recombination with endogenous MMTVs. The lower incidence and latency of tumors induced by MTVAflE did not lead to lower transcriptional activity or recombination with endogenous MMTVs; however, Sag-mediated deletion by this virus was significantly faster statistically than that by HYB-MTV (Fig. 2B). Thus, altered tumorigenesis by MTVAflE might result from the deletion of T cells required for dissemination of the virus or tumor targets, as suggested by Papiernik et al. (16).
FIG. 3.
Kaplan-Meier plots of incidence of mammary tumors and thymic lymphomas induced by wild-type and mutant MMTV proviruses. Female mice were caged to maintain continuous breeding and lactation. Mice that died from nontumor causes (e.g., birthing problems) before the average latency of tumor induction were excluded from the study. (A) Development of mammary tumors. Numbers in parentheses indicate the numbers of animals with tumors/total numbers of injected females. The circles indicate one or more animals that died without any detectable tumors. (B) Development of thymic lymphomas. Numbers in parentheses indicate numbers of animals with tumors/total numbers of injected males and females. One mouse injected with HYB-MTVStuE developed both a mammary tumor and a thymic lymphoma after 11 months of latency.
To determine the LTR structure of newly integrated proviruses, PstI-digested tumor DNA was analyzed by Southern blotting using probes specific for either the MMTV LTR (Fig. 4A) or the NRE (Fig. 4B). Most integrated proviruses in HYB-MTVAflE-induced lymphomas contained the NRE (Fig. 4A and B, lanes 5 and 7). Furthermore, these LTRs retained their original size, indicative of an intact T-cell enhancer with a 62-bp triplication. Shorter LTRs might harbor fewer enhancer copies or a partial deletion of the NRE. Growth of TBLV-induced lymphomas appears to select proviral integrations with suboptimal numbers of LTR enhancer repeats to modulate oncogene expression (6). LTRs of predominant acquired proviruses in HYB-MTVStuE-induced lymphomas lacked the entire NRE and were detected only by the MMTV LTR probe (Fig. 4A and B, lanes 10 and 11). The reduced size of these LTRs is consistent with complete NRE deletion and retention of the T-cell enhancer. NRE-lacking recombinants were not observed in HYB-MTVStuE-induced mammary cancer (Fig. 4A and B, lanes 8, 9, and 12), even in an animal that developed both tumor types (compare lanes 11 and 12). The presence of the T-cell enhancer in lymphomagenic HYB-MTVAflE and HYB-MTVStuE viruses was verified by reverse transcription-PCR, cloning, and sequencing (data not shown).
FIG. 4.
LTR structure of acquired proviruses in wild-type and mutant MMTV-induced mammary tumors (MT) and thymic lymphomas (TL). (A) Southern blotting of tumor DNA using the full-length MMTV LTR probe. (B) Southern blotting of tumor DNA with the MMTV NRE probe (StuI-to-AflII fragment of the MMTV LTR). Genomic DNA was digested with PstI, separated on 0.8% agarose gels, transferred to nitrocellulose, and hybridized as previously described (9). Southern blotting of genomic DNA from an uninfected BALB/cJ mouse liver (LI) was used to distinguish the bands originating from the three endogenous Mtvs in the BALB/cJ strain. White asterisks (lanes 5, 7, 10, 11, and 13) indicate the LTR fragments of predominant acquired proviruses in virus-induced tumors.
These results suggest that two events are required for MMTV-induced lymphomagenesis: loss of NRE-mediated transcriptional repression and acquisition of a T-cell enhancer. Selection of proviruses lacking the NRE in HYB-MTVStuE-induced lymphomas may be driven by the incomplete suppression of NRE activity by the upstream T-cell enhancer, as observed in transient-transfection assays in T cells. HYB-MTVAflE is not subject to this selection pressure, since the T-cell enhancer located downstream of the NRE appears to mask transcriptional repression. The ability of HYB-MTVAflE to induce lymphomas also indicates that Sag truncation is dispensable for this process, in agreement with our previous data (14). Interestingly, the LTRs of acquired proviruses in the HYB-MTVΔNRE-induced mammary tumor had reacquired the NRE, suggesting that the presence of this region might favor mammary tumorigenesis (Fig. 4A and B, lane 13). Furthermore, the low tumor incidence of HYB-MTVΔNRE might be dependent on the appearance of such recombinants.
In conclusion, development of lymphomagenic MMTVs requires alterations of transcriptional regulatory elements in the LTR. Loss of NRE activity and T-cell enhancer acquisition allow increased viral expression in T cells, resulting in a higher probability of insertional mutagenesis. The viral enhancer also may be critical for transcriptional upregulation of selected cellular oncogenes, such as c-myc and rorc, in T cells (5, 6, 18).
Acknowledgments
We acknowledge helpful suggestions from the Dudley lab.
We also acknowledge support from NIH grants P01 CA77760 and R01 CA34780.
REFERENCES
- 1.Ball, J. K., and G. A. Dekaban. 1987. Characterization of early molecular biological events associated with thymic lymphoma induction following infection with a thymotropic type-B retrovirus. Virology 161:357-365. [DOI] [PubMed] [Google Scholar]
- 2.Ball, J. K., H. Diggelmann, G. A. Dekaban, G. F. Grossi, R. Semmler, P. A. Waight, and R. F. Fletcher. 1988. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J. Virol. 62:2985-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramblett, D., C.-L. L. Hsu, M. Lozano, K. Earnest, C. Fabritius, and J. Dudley. 1995. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J. Virol. 69:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brasset, E., and C. Vaury. 2005. Insulators are fundamental components of the eukaryotic genomes. Heredity 94:571-576. [DOI] [PubMed] [Google Scholar]
- 5.Broussard, D. R., M. M. Lozano, and J. P. Dudley. 2004. Rorγ (Rorc) is a common integration site in type B leukemogenic virus-induced T-cell lymphomas. J. Virol. 78:4943-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broussard, D. R., J. A. Mertz, M. Lozano, and J. P. Dudley. 2002. Selection for c-myc integration sites in polyclonal T-cell lymphomas. J. Virol. 76:2087-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley, J., and R. Risser. 1984. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J. Virol. 49:92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley, J. P. 1999. Mouse mammary tumor virus, p. 965-972. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology. Academic Press Ltd., London, United Kingdom.
- 9.Hsu, C.-L. L., C. Fabritius, and J. Dudley. 1988. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J. Virol. 62:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. W., P. G. Moffitt, K. L. Morley, and D. O. Peterson. 1991. Multipartite structure of a negative regulatory element associated with a steroid hormone-inducible promoter. J. Biol. Chem. 266:24101-24108. [PubMed] [Google Scholar]
- 11.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mertz, J. A., F. Mustafa, S. Meyers, and J. P. Dudley. 2001. Type B leukemogenic virus has a T-cell-specific enhancer that binds AML-1. J. Virol. 75:2174-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalides, R., E. Wagenaar, and P. Weijers. 1985. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol. Cell. Biol. 5:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mustafa, F., S. Bhadra, D. Johnston, M. Lozano, and J. P. Dudley. 2003. The type B leukemogenic virus truncated superantigen is dispensable for T-cell lymphomagenesis. J. Virol. 77:3866-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mustafa, F., M. Lozano, and J. P. Dudley. 2000. C3H mouse mammary tumor virus superantigen function requires a splice donor site in the envelope gene. J. Virol. 74:9431-9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papiernik, M., A. C. Wache, C. Pontoux, and B. Nabarra. 1997. Massive mammary gland infection and pregnancy-dependent mammary tumor development in mice infected neonatally with mouse mammary tumor virus (SW) but not in mice infected as adults, despite a dramatic local response. Eur. J. Immunol. 27:2145-2151. [DOI] [PubMed] [Google Scholar]
- 17.Purdy, A., L. Case, M. Duvall, M. Overstrom-Coleman, N. Monnier, A. Chervonsky, and T. Golovkina. 2003. Unique resistance of I/LnJ mice to a retrovirus is due to sustained interferon gamma-dependent production of virus-neutralizing antibodies. J. Exp. Med. 197:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajan, L., D. Broussard, M. Lozano, C. G. Lee, C. A. Kozak, and J. P. Dudley. 2000. The c-myc locus is a common integration site in type B retrovirus-induced T-cell lymphomas. J. Virol. 74:2466-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross, S. R., C.-L. L. Hsu, Y. Choi, E. Mok, and J. P. Dudley. 1990. Negative regulation in correct tissue-specific expression of mouse mammary tumor virus in transgenic mice. Mol. Cell. Biol. 10:5822-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shackleford, G. M., and H. E. Varmus. 1988. Construction of a clonable, infectious, and tumorigenic mouse mammary tumor virus provirus and a derivative genetic vector. Proc. Natl. Acad. Sci. USA 85:9655-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theunissen, H. J. M., M. Paardekooper, L. J. Maduro, R. J. A. M. Michalides, and R. Nusse. 1989. Phorbol ester-inducible T-cell-specific expression of variant mouse mammary tumor virus long terminal repeats. J. Virol. 63:3466-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrona, T. J., M. Lozano, A. A. Binhazim, and J. P. Dudley. 1998. Mutational and functional analysis of the C-terminal region of the C3H mouse mammary tumor virus superantigen. J. Virol. 72:4746-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanagawa, S.-I., K. Kakimi, H. Tanaka, A. Murakami, Y. Nakagawa, Y. Kubo, Y. Yamada, H. Hiai, K. Kuribayashi, T. Masuda, and A. Ishimoto. 1993. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J. Virol. 67:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yazdanbakhsh, K., C. G. Park, G. M. Winslow, and Y. Choi. 1993. Direct evidence for the role of COOH terminus of mouse mammary tumor virus superantigen in determining T cell receptor Vβ specificity. J. Exp. Med. 178:737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu, Q., U. Maitra, D. Johnston, M. Lozano, and J. P. Dudley. 2004. The homeodomain protein CDP regulates mammary-specific gene transcription and tumorigenesis. Mol. Cell. Biol. 24:4810-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]