Abstract
We have engineered monomeric gp120 in such a way as to favorably present the conserved epitope for the broadly neutralizing antibody b12 while lowering the exposure of epitopes recognized by some weakly neutralizing and nonneutralizing antibodies. The work presented here describes the immune response in rabbits immunized with two prototype, engineered gp120s to explore the relationship between antigenicity and immunogenicity for these mutants. The GDMR gp120 mutant (residues 473 to 476 on gp120 altered from GDMR to AAAA) has a series of substitutions on the edge of the CD4 binding site (CD4bs), and the mCHO gp120 mutant has seven extra glycans relative to the wild-type protein. Importantly, serum mapping showed that both mutants did not elicit antibodies against a number of epitopes that had been targeted for dampening. The sera from rabbits immunized with the GDMR gp120 mutant neutralized some primary viruses at levels somewhat better than the wild-type gp120 immune sera as a result of an increased elicitation of anti-V3 antibodies. Unlike wild-type gp120 immune sera, GDMR gp120 immune sera failed to neutralize HXBc2, a T-cell line adapted (TCLA) virus. This was associated with loss of CD4bs/CD4-induced antibodies that neutralize TCLA but not primary viruses. The mCHO gp120 immune sera did not neutralize primary viruses to any significant degree, reflecting the masking of epitopes of even weakly neutralizing antibodies without eliciting b12-like antibodies. These results show that antibody responses to multiple epitopes on gp120 can be dampened. More precise focusing to a neutralizing epitope will likely require several iterations comparing antigenicity and immunogenicity of engineered proteins.
The failure of monomeric gp120 to prevent human immunodeficiency virus (HIV) infection in human efficacy trials has fueled the pursuit of new approaches for eliciting broadly neutralizing antibodies. Monomeric gp120 elicits neutralizing antibodies against HIV strains adapted to grow in laboratory tissue culture but not against primary isolates of HIV circulating in populations. This observation, initially made more than a decade ago, began to focus attention on the structural biology of gp120 and gp41 (the HIV envelope spike) and on understanding the interaction of HIV with the few known antibodies that can neutralize a broad range of strains. Such antibodies have been detected both in sera from HIV-infected people and as broadly neutralizing monoclonal antibodies (MAbs).
A small panel of broadly neutralizing MAbs isolated from HIV-infected people has helped identify conserved regions of gp120 that can be targeted by a next-generation HIV vaccine. These MAbs include the human MAb b12, recognizing an epitope overlapping the CD4 binding site (CD4bs) of gp120 (9, 44); 2G12, recognizing a conserved cluster of oligomannose chains on gp120 (10, 42, 46, 51); and 447-52D, recognizing a conserved motif at the tip of the V3 loop (12, 16, 18, 48). Studies of the broadly neutralizing MAbs suggest that their neutralizing capacity is associated with the ability to bind to functional envelope (Env) on the virus but does not correlate with binding to isolated gp120 (41, 45). Therefore, failure of an immunogen to elicit broad neutralizing antibodies is interpreted as a failure to elicit antibodies with reasonable binding affinities for conserved regions of functional Env. The presence of epitopes recognized by broadly neutralizing antibodies on monomeric gp120 argues that it is still a potential template for HIV type 1 (HIV-1) vaccine design, although not in its native unmodified form. Monomeric gp120 generally elicits antibodies that are overwhelmingly directed to epitopes that are not well presented on the Env trimer of primary viruses and are nonneutralizing or weakly neutralizing.
Two different approaches at modifying Env molecules and presenting neutralizing epitopes more favorably are being explored. The first approach focuses on the use of modified gp120, gp140, or gp160 glycoproteins. For example, various envelope glycoproteins have been generated in which the variable loops have been deleted, with the aim of increasing the exposure of neutralizing epitopes in the CD4bs and CD4-induced (CD4i) site (23, 26). Unfortunately, to date this method has failed to elicit the desired level of neutralizing antibodies capable of recognizing their cognate epitopes on wild-type virus. However, in one study, gp140 oligomers were generated with a partially deleted V2 loop and shown to elicit antibodies that can neutralize the homologous wild-type virus (3). Another study in which V1/V2 loops were deleted along with shortening of the V3 stem showed elicitation of antibodies with higher potency than wild-type or only the V1/V2 deletion mutant (59). In other studies, partially deglycosylated recombinant gp160 or recombinant viruses expressing gp120 glycosylation-deficient mutants have been generated (6, 40). This method, too, has problems because antibodies to the exposed epitopes fail to recognize wild-type antigen. A recent study by Kang et al. (22) showed that partial deletion of variable loops with removal of three to five glycosylations helped expose epitopes for neutralizing antibodies favorably on gp140. However, this is only an antigenicity study and is yet to be evaluated as a means to improve immunogenicity. In other studies, fusion intermediates in which gp120 or gp140 is covalently cross-linked to CD4 have been used as immunogens (14). The antibodies elicited were able to neutralize some primary viruses, but it is unclear whether these antibodies are gp120 or CD4 directed. In one study, gp120 was constrained by cross-linking it to antibody A32 (25). However, the A32-gp120 did not elicit better neutralizing antibodies than gp120. One report also discusses stabilizing the conformation of monomeric gp120 to mimic the CD4 bound conformation by the introduction of Phe-43 cavity-filling mutation such as S375W (55).
The second approach focuses on the preservation or reconstruction of the trimeric envelope spike. The essential concept is that immunizing with a close mimic of the functional trimer will improve the chances of eliciting neutralizing antibodies. Soluble gp140 trimers are being used as the prototype immunogen (3, 7, 13, 19, 53, 58). Soluble gp140 trimers containing the ectodomain of gp41 covalently linked to gp120 have been generated by fusing GCN4 trimerization domains or T4 bacteriophage fibritin trimeric motifs to the C terminus of soluble, uncleaved gp140 glycoproteins (56, 57). In a slightly different approach proteoliposomes have been generated containing native, trimeric uncleaved gp160ΔCT (where ΔCT indicates deletion of the cytoplasmic tail) glycoproteins (20). These trimers have similar but not identical antigenic profiles, as probed by MAb reactivity, to Env expressed on infected cells. Immunization with HIV-1 isolate YU2 gp140-GCN4 trimers was shown to be more efficient at eliciting neutralizing antibodies than YU2 gp120 monomers (7, 19, 58). One limitation of this approach is that the soluble trimer is presented as an uncleaved form, and this may limit the ability of the recombinant trimer to fully mimic the native trimer (33). In a study aimed at circumventing the problems with uncleaved gp140, cysteine residues have been incorporated into gp120 and gp41. This prevents dissociation of the two subunits through the formation of an intersubunit disulfide bridge upon expression of cleaved gp140 (4). However, a mutation (I559P) that destabilizes the six-helix bundle formation or expression on a membrane setting was needed for the expression of this molecule in a stable trimeric form (43). In a very recent report, a group has used formaldehyde-treated and heat-inactivated HIV-1 virus particles for immunization. This method was used to present to the immune system a native Env spike in a membrane-bound and cleaved form. The results are encouraging, and the immune sera showed a slightly broader range of neutralization potential (37). Most of the modified Env molecules discussed above are currently being used in immunogenicity studies in small animal models.
Can we engineer monomeric gp120 to suppress responses to unwanted epitopes and enhance responses to epitopes such as b12? Recent work in our laboratory has helped give a clearer picture of the binding site of the broadly neutralizing b12 antibody on gp120 (34, 44) and has led to two strategies to engineer gp120 to achieve this goal. The first is alanine scanning mutagenesis that helped generate variants that favored b12 binding relative to weakly neutralizing anti-CD4bs antibodies. The quadruple alanine mutant GDMR gp120 (residues 473 to 476 on gp120 altered from GDMR to AAAA) showed b12 binding to be favored relative to anti-CD4bs antibodies such as b6 or F105. The second strategy is hyperglycosylation of monomeric gp120 where undesired epitopes (i.e., those that induce weakly neutralizing and nonneutralizing antibodies) are masked using the selective incorporation of N-linked glycans (35). This mechanism is similar to that used by HIV itself, in which glycans shield antigenic determinants from neutralizing antibody recognition. The mutant mCHO gp120 with seven extra sugars fails to bind weakly neutralizing CD4bs antibodies while still binding the broadly neutralizing b12 antibody. Another related mutant, mCHO-GDMR gp120, fails to bind the V3 loop-specific antibodies 447-52D and 19b and the V1/V2 loop-specific antibodies G3-4 and G3-136, indicating masking of some portions of these variable regions. We presumed that the extra glycosylations were responsible for the loss of binding by the loop antibodies, but this was further investigated here. An earlier report had shown that the addition of an N-glycan to the V3 loop could reduce the antibody response to part of the loop (15).
The work presented in this paper deals with immunizing rabbits with our first generation of engineered gp120 mutants, GDMR gp120 and mCHO gp120, and the characterization of sera to explore the relationship between antigenicity and immunogenicity.
MATERIALS AND METHODS
Materials.
The monoclonal antibodies used in this study were 17b, 48d, A32, C11, 19b, and 39F, kindly provided by J. Robinson (31, 32, 47, 50, 54); 447-52D and 697-D, kindly provided by M. Gorny and Susan Zolla-Pazner (12, 16-18); 8.22.2, kindly provided by A. Pinter (21); 2G12, kindly provided by H. Katinger (10, 42, 46, 51); F425-B4e8, kindly provided by L. Cavacini (11); and HIV-IG, kindly provided by J. Mascola. MAbs b12, b6, b3, and 4KG5 were produced in-house (1, 2, 8, 9, 60); D7324 was purchased from Cliniqa (Fallbrook, CA), and sCD4 was purchased from Progenics (NY). Plasmid pSVIIIexE7pA-HXBc2 was a gift from J. Sodroski. The JR-FL, JR-CSF, ADA (all HIV-1 isolates), and Env genes were cloned into the same plasmid as described previously. pCAGGS-SF162 was kindly provided by J. Binley. The following reagents were obtained from the AIDS Research and Reference Reagent Program: F105 (contributed by M. Posner and L. Cavacini) (38, 39), pNL4-3.Luc.R−E− (contributed by Nathaniel Landau), U87.CD4.CCR5 cells, U87.CD4.CXCR4 (contributed by H. Deng and D. Littman), and HIV-1 consensus subtype B Env (15-mer) peptides (complete set of 210 peptides).
ELISAs.
Enzyme-linked immunosorbent assay (ELISA) plate wells were coated overnight at 4°C with 50 μl of sheep anti-gp120 antibody (D7324) at 5 μg/ml in phosphate-buffered saline (PBS). Wells were washed four times (with 200 μl each time) with PBS containing 0.05% Tween 20 using an automated plate washer (Molecular Devices) and blocked with 3% bovine serum albumin (BSA) for 1 h at room temperature (RT). After the BSA was aspirated, gp120 (at 1 μg/ml) (wild-type gp120, GDMR gp120, or mCHO gp120) was added to the wells in dilution buffer (PBS containing 1% BSA and 0.02% Tween) and allowed to incubate for 2 h at RT. (The wild-type gp120, GDMR-gp120, and mCHO-gp120 used for this capture ELISA were produced and purified as described below. The final concentration of the protein was determined by a bicinchoninic acid protein assay [Pierce]). The wells were washed four times, and primary antibodies were incubated for 1 h at RT, at a starting concentration of 2.5 to 10 μg/ml, and diluted fourfold in dilution buffer. The wells were washed four times, and secondary antibody goat anti-human immunoglobulin G (IgG) F(ab′)2 horseradish peroxidase (HRP; Pierce) diluted 1:1,000 in dilution buffer (in the case of human Abs) or anti-hemagglutinin HRP high-affinity antibody (3F10) diluted 1:1,000 (in the case of 4KG5-hemagglutinin single-chain fragment-variable antibody) was added and incubated for 1 h at RT. The plates were washed four times and developed by adding 50 μl of TMB (3,3′, 5,5′-tetramethylbenzidine) solution according to the manufacturer's instructions (Pierce). The HRP reaction was stopped by adding 50 μl of H2SO4 (2 M), and the plates were read at an optical density at 450 nm (OD450) on a microplate reader (Molecular Devices).
An ELISA to determine serum binding to homologous and heterologous Envs was performed as described above (with purified vaccinia-produced protein) except that instead of primary antibodies, rabbit sera at 1/2,000 diluted twofold (total of eight dilutions) was incubated for 1 h 30 min at RT. The wells were washed four times, and 50 μl of goat anti-rabbit IgG F(ab′)2 (Pierce) conjugated to HRP at a 1:1,000 dilution was added, and the plate was incubated for 1 h at RT and developed as described above.
An ELISA to determine serum binding titers was performed by coating microwells with 50 μl (1 μg/ml) of wild-type gp120JR-FL (produced in CHO cells; kindly provided by W. Olson and N. Schulke, Progenics) overnight at 4°C. The procedure then followed that of serum binding to homologous and heterologous Envs, as described above.
Protein production using the vaccinia system and purification using lectin.
Glycoproteins were produced in HEK 293T cells using recombinant vaccinia viruses engineered to express the designated protein as previously described (13a.). Cells were infected at a multiplicity of infection of 5 for 1 h then washed twice with PBS, and medium was replaced with Dulbecco's modified Eagle's medium, penicillin-streptomycin, without serum, and incubated at 37°C for 48 h. Supernatants were clarified by low-speed centrifugation and filtration through a 0.22-μm-pore-size filter. Triton X-100 (0.1%) was added to inactivate residual vaccinia. Secreted gp120 was purified from the supernatant by column chromatography using Galanthus nivalis lectin cross-linked to agarose beads (Vector Laboratories, Burlingame, Calif.). Chromatography was performed on a Bio-Rad LP system with a 2 ml/min flow rate. The column was equilibrated with morpholineethanesulfonic acid (MES; 20 mM MES, 130 mM NaCl, 10 mM CaCl2, pH 7.0) buffer, and supernatant was applied, washed with MES buffer containing 500 mM NaCl, followed by MES buffer, and eluted in MES buffer containing 750 mM methyl manno-pyranoside. The eluate was concentrated and exchanged into MES buffer by centrifugal filtration, and the final concentration was determined using a bicinchoninic acid protein assay (Pierce, Rockford, IL). The proteins were assayed with silver staining and by Western blotting.
Rabbit immunization.
New Zealand White female rabbits, weighing 3 to 5 kg, three per immunogen, were immunized with purified protein. Prebleeds were taken prior to immunization to be used as controls. Each rabbit was injected with 120 μg of protein in 1 ml of 1× Ribi adjuvant (R-730 [monophosphoryl lipid A, synthetic trehalose dicorynomycolate, cell wall skeleton]; Corixa) subcutaneously (0.5 ml), intramuscularly (0.1 ml at two sites), and intradermally (0.05 ml at six sites) in the case of the wild-type and GDMR rabbits. The mCHO rabbits were immunized in a similar manner except that the route of injection for the 1 ml of protein (in Ribi) was subcutaneous. The boosts were carried out at 4-week intervals, and bleeds were taken 10 days postinjection. A total of five boosts were carried out. Animal housing and immunization procedures adhered to the protocols of the Institutional Animal Care and Use Committee.
Neutralization assay and V3 peptide inhibition.
In this assay, virus competent for a single-round replication was produced by cotransfection of 293T cells with plasmids pSVIIIexE7-JR-FL, -JR-CSF, -ADA, or -HXBc2, or pCAGGS-SF162 or vesicular stomatitis virus glycoprotein and pNL4-3.luc.R−E−. The degree of virus neutralization by antibody was determined by measuring luciferase activity. Briefly, 3 × 104 U87.CD4.CCR5 or CXCR4 (for HXBc2) cells in 200 μl of medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 1 μg of puromycin/ml, 300 μl of G418/ml, glutamine, and penicillin-streptomycin) were added to microplate wells (96-well flat bottom; Corning Inc., Corning, N.Y.) and incubated for 24 h at 37°C in 5% CO2. Sixty microliters of medium containing an amount of virus previously determined to yield 100,000 relative light units was mixed with 60 μl of sera diluted 1:2 twofold and incubated for 2 h at 37°C in 5% CO2. After 2 h, the medium was aspirated from the plates with U87 cells, and 100 μl of serum and virus was added and the culture was incubated 2 h at 37°C in 5% CO2. After the incubation, serum and virus were aspirated, and the wells were washed with 100 μl of medium twice; 200 μl of fresh medium was added, and the plates were incubated for a further 3 days. On the third day, medium was aspirated from wells, and 60 μl of luciferase cell culture lysis reagent (Promega, Madison, Wis.) was added. The wells were scraped, and the lysate was mixed by pipetting. The plate was centrifuged at 1,800 × g for 10 min at 4°C. Twenty microliters was transferred to an opaque assay plate (Corning), luciferase reagent (Promega) was added, and the luciferase activity was measured on a luminometer (Orion, Berthold Detection Systems). Serum and virus were incubated for only 2 h with cells as previously described (58) since this was found to give the least nonspecific activity from serum (especially effects of enhancement). In addition, each immune serum sample was always compared to the activity measured with the given prebleed serum diluted in the same fashion to control for nonspecific neutralization activity.
V3 antibody-specific neutralization of primary virus was assayed by a V3 peptide inhibition assay developed by J. Mascola (19). We used a 23-mer YU2 V3 peptide (SynPep, Dublin, CA) to probe the neutralization of SF162 and ADA viruses. The peptide was used at a final concentration of 30 μg/ml and incubated with serum diluted 1:8 for 30 min at 37°C in 5% CO2. Virus was added, and the mixture was incubated for a further 2 h at 37°C in 5% CO2. This mixture was then added to the cells, and the assay was carried out as described above. Negative controls were set up in which peptide was incubated with virus only, or peptide was incubated with nonimmune sera, and this in turn was compared to virus neutralization in the absence of any peptide. The JR-FL V3 (23-mer) peptide used at ViroLogic Inc. was synthesized at the Scripps Research Institute (core facility).
Serum competition ELISAs.
(i) The serum competition assay is similar to one described by Moore and Ho (29). In our assay, serum was initially titrated against wild-type gp120 immobilized on microwell plates. In order to help equalize the levels of polyclonal antibodies specific to gp120 between different serum samples, serum was initially titrated against gp120, and the last serum dilution that gave an OD reading of 1.9 was determined. This fixed dilution (varying from ∼1 in 10,000 to 20,000 for the different sera) was subsequently used to compete increasing concentrations of known human monoclonal antibodies. For the competition ELISA, microwell plates were coated with 1 μg/ml gp120JR-FL (Progenics) in PBS overnight at 4°C. Plates were washed and blocked with 3% BSA for 1 h 30 min at RT. BSA was aspirated and two monoclonal antibodies per group (b12 and 15e, b6 and F105, 447-52D and 19b, F425-B4e8 and 39F, and C11 and A32) each at a starting concentration of 5 μg/ml and diluted fourfold, was added to the wells, and the plates were incubated for 3 h at RT. Antibody solution was aspirated, and the serum diluted at a fixed concentration determined previously (as described above), was added, after which the plate was incubated for a further 1 h 30 min. Plates were washed and goat-anti rabbit IgG Fc fragment (with minimal reactivity to human serum proteins) (Jackson ImmunoResearch) conjugated to alkaline phosphatase (AP) diluted at 1:1,000 was added for 1 h at RT. Plates were washed and developed by adding 50 μl of AP substrate, prepared by adding one tablet of disodium p-nitrophenyl phosphate (Sigma) to 5 ml of AP staining buffer, pH 9.6, and the OD405 was measured. The percent change in serum binding was calculated as follows: (OD at which serum binding plateaus in the presence of increasing concentration of human MAb/OD at which serum binds at a fixed concentration in the absence of MAb) × 100. Therefore, percent inhibition of polyclonal serum binding to gp120 by a human MAb equals 100 minus the percent change in serum binding (calculated from the formula shown above).
(ii) The ELISA format for competition with biotinylated antibodies was set up as described above. Briefly, 1 μg/ml of gp120JR-FL was captured with sheep anti-gp120 antibody for 2 h at RT. It was then washed, and polyclonal sera were added at a single concentration of a 1/100 dilution for 2 h at RT. Excess solution was aspirated, and biotinylated MAb (10 to 5 μg/ml) diluted fourfold were added for 1 h 30 min at RT. Plates were washed, and streptavidin conjugated to alkaline phosphatase (Vector Laboratory), diluted 1:300, was added, and the plates were incubated for 1 h at RT. Plates were washed and developed with AP substrate, and the OD was measured at 405 nm.
Serum binding to linear peptides.
A linear peptide set (115 peptides; 15 mers) based on the consensus clade B sequence was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The soluble peptides were dissolved in PBS, and insoluble peptides were dissolved in 10% or 50% dimethyl sulfoxide. Microwell plates were coated at a concentration of 4 nmol per well in 50 μl of coating buffer (23) (15 mM Na2CO3, 35 mM NaHCO3, 3 mM NaN3, pH 9.6) overnight at 4°C. The following day the plates were washed (PBS containing 0.05% Tween 20) and blocked with 3% BSA for 1 h at RT. BSA was aspirated, and 50 μl of serum diluted 1/100 in dilution buffer (PBS containing 1% BSA and 0.02% Tween) was added and incubated for 1 h at RT. The wells were washed, and goat anti-rabbit IgG Fc fragment (Jackson ImmunoResearch) conjugated to AP, diluted 1:1,000 in dilution buffer, was added for 1 h at RT. Plates were washed again and developed with AP substrate, and the OD was read at 405 nm.
RESULTS
Antigenic profiles of purified GDMR gp120 and mCHO gp120 compared to wild-type gp120.
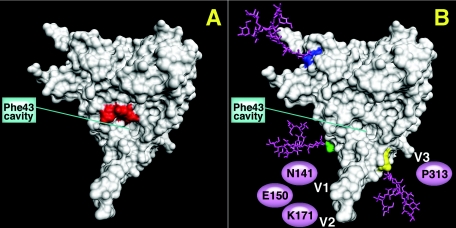
The engineered gp120 proteins used in our previous reports (35, 36) were made by transient transfection of 293T cells and ELISAs performed with cell supernatants to determine antigenicity. All immunogens were based on a scaffold of codon-optimized JR-FL gp120 sequence. One mutant, GDMR gp120 (where residues 473 to 476 were mutated to alanine) (Fig. 1A) was previously shown to favorably bind b12 compared to other weakly neutralizing CD4bs antibodies such as b6, b3, and F105 (35). It also did not bind CD4-IgG2 or CD4-induced (CD4i) antibodies (48d and 17b). The second mutant, mCHO gp120 (Fig. 1B), is a hyperglycosylated gp120 that has seven extra sugar attachment motifs (NXT), three on the core (at positions H92, Q114, and I423) and four on the variable loops (on V1-N141, E150, V2-K171, and V3-P313). The binding affinities of CD4bs antibodies such as F105, b6, b3, F91, and 15e were significantly reduced for the mCHO gp120 mutant relative to wild-type gp120 (35). However, mCHO gp120 bound b12, albeit with slightly lowered affinity compared to wild-type gp120. A32, a C1-C4 antibody, also failed to bind. V1/V2 and V3 loop-specific antibody binding was not tested with the mCHO gp120 mutant in our previous report.
FIG. 1.
gp120 core structure with spatial location of the V1/V2/V3 loops representing mutant GDMR gp120 (A), where residues 473 to 476 (red) lining the CD4 Phe-43 binding cavity were mutated to alanine. (B) Mutant mCHO gp120 (CD4 binding face) with sugars (pink) attached on the core to residues H92 (dark blue), Q114 (green), and I423 (yellow). The putative location of V1 and V2 loops with three sugars at position N141, E150, and K171 and the putative location of the V3 loop with one sugar at position P313 are also represented. The monomer structure is from the Protein Data Bank of HXBc2 gp120 by Kwong et al. (24). The images were made using the PMV program by C. Corbaci. R. Cardoso attached the coordinates for the sugars at residues H92, Q114, and I423 on the gp120 core representing the extra glycans on the core of mCHO gp120 mutant.
For immunization, purified protein was needed in larger amounts than readily available from transient transfections, and therefore the protein was produced using the vaccinia virus system in 293T cells and purified using the Galanthus nivalis lectin (described in Material and Methods). The purified proteins were used in ELISA to ascertain the antigenicity of the vaccinia-produced proteins before being used for immunization. Results of antibody binding studies with the GDMR gp120 mutant compared to wild-type gp120 are shown in Table 1. The CD4bs antibodies b6, F105, and b3, as well as the CD4i antibodies 17b and 48d, failed to bind GDMR gp120. The binding of the CD4i antibodies did not improve in the presence of saturating amounts of soluble CD4. A32, a C1-C4 antibody, and C11, a C1-C5 antibody, also bound very poorly. However, GDMR gp120 mutant bound the broadly neutralizing antibody b12 similarly to wild-type gp120. Antibodies to the V3 loop such as 447-52D, 19b, F425-B4e8, and 39F bound with similar apparent affinity as wild-type gp120 to the GDMR gp120 mutant. The antibody 4KG5, a single-chain fragment-variable antibody that recognizes a discontinuous conformational epitope involving V1/V2 and V3 loops (60), bound with 10-fold lower apparent affinity to GDMR gp120 compared to wild-type gp120. This was unexpected since residues 473 to 476 (GDMR) are not thought to play a direct role in the binding of 4KG5. Of the antibodies specific to the V2 loop, 8.22.2 (21), which recognizes a linear epitope, bound to the GDMR gp120 mutant with apparent affinity similar to the wild-type gp120, but 697-D (17), which recognizes a conformational epitope, bound with reduced affinity. Taken together, the failure of the GDMR gp120 mutant to recognize CD4i antibodies and the reduced affinity for 4KG5 and 697-D MAbs are consistent with some degree of conformational rearrangement involving the V1/V2 and V3 loops in the mutant protein.
TABLE 1.
Antigenicity of GDMR and mCHO gp120 mutants compared to wild-type gp120a
| Antibody | JR-FL gp120 antigen binding ratio
|
||
|---|---|---|---|
| Wild-type | GDMR | mCHO | |
| CD4bs | |||
| b12 | 1.00 | 1.00 | 0.5 |
| b6 | 1.00 | 0.00 | 0.12 |
| Fab b3 | 1.00 | 0.00 | 0.05 |
| F105 | 1.00 | 0.00 | 0.00 |
| CD4-IgG2 | 1.00 | 0.00 | 0.00 |
| CD4i | |||
| 48d | 1.00 | 0.00 | 0.00 |
| 48d plus CD4 | 1.00 | 0.00 | 0.00 |
| 17b | 1.00 | 0.00 | 0.00 |
| 17b plus CD4 | 1.00 | 0.00 | 0.00 |
| Sugar, 2G12 | 1.00 | 0.8 | 0.6 |
| V3 loop | |||
| 447-52D | 1.00 | 1.00 | 0.00 |
| 19b | 1.00 | 1.00 | 0.00 |
| F425-B4e8 | 1.00 | 1.00 | 0.1 |
| 39F | 1.00 | 1.00 | 0.8 |
| V2 loop | |||
| 697-D | 1.00 | 0.23 | 0.01 |
| 8.22.2 | 1.00 | 1.00 | 0.01 |
| V1/V2/V3, 4KG5 | 1.00 | 0.08 | 0.00 |
| C1-C4, A32 | 1.00 | 0.05 | 0.00 |
| C1-C5, C11 | 1.00 | 0.1 | 0.00 |
| Serum, HIVIG | 1.00 | 0.3 | 0.05 |
Ratios of apparent antibody binding affinities (K50s) to the mutant are shown in comparison to wild-type gp120. Boldface indicates that the binding affinity of the antibody for the mutant is less than 25% of wild-type gp120.
The antigenicity of the purified vaccinia-produced mCHO gp120 protein compared to wild-type gp120 is shown in Table 1. HIV-IG, a polyclonal IgG preparation from HIV-1-infected humans, bound poorly to this mutant. Antibody b12 bound better than b6 or b3, albeit with somewhat lower apparent affinity than to wild-type gp120. The mCHO gp120 protein did not bind F105 or CD4-IgG2. In our previous report, the masking effect by glycosylation of the variable loops was shown with a variant of mCHO gp120, mCHO-GDMR gp120 mutant (with the four alanines at 473 to 476). Since the presence of the GDMR mutation (described above) in mCHO-GDMR gp120 might have changed the orientation of the loops, we repeated ELISAs on mCHO gp120 with antibodies to the variable loops. Antibodies to the crown of the V3 loop such as 447-52D and 19b failed to bind, while F425-B4e8 binding was reduced, and an antibody to the stem 39F bound well. This might indicate a masking effect by the sugar on the crown of the V3 loop inhibiting binding of crown-specific V3 loop antibodies but not those to the stem of the V3 loop. Antibodies to the V2 loop, 697-D and 8.22.2, failed to bind the mCHO-gp120 mutant, and this might indicate masking of the corresponding epitopes by the three extra sugars on the V1/V2 loops of the mutant.
Immunization and serum binding to homologous and heterologous Env gp120s.
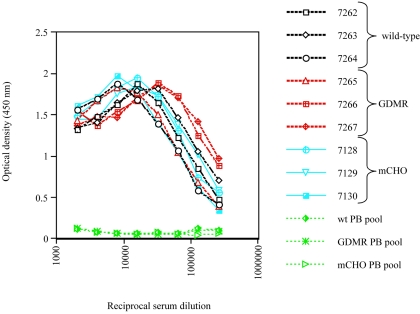
Three rabbits per wild-type gp120, GDMR gp120, or mCHO gp120 immunogen group were immunized with 120 μg of purified vaccinia-produced protein. Booster injections were performed at 4-week intervals, and bleeds were taken 10 days postinjection. A total of five booster injections were carried out. Serum binding titers were determined against wild-type gp120JR-FL coated directly on plates. Half-maximal serum binding titers were determined from anti-gp120 binding curves of fourth bleed sera. Titers ranged from 7 × 104 to 25 × 104 (Fig. 2 and Table 2). All three groups had comparatively similar serum binding titers; the extra glycosylation in the mCHO gp120 mutant group did not alter the overall anti-gp120 serum binding titers. The fourth bleed sera for all three immunogen groups were taken for further analysis, and assays described hereafter will pertain to fourth bleed sera unless otherwise indicated.
FIG. 2.
ELISA of the binding to wild-type gp120 of sera from rabbits immunized with wild-type (black) (rabbits 7262 to 7264), GDMR (red) (rabbits 7265 to 7267) and mCHO gp120s (blue) (rabbits 7128 to 7130). Pooled prebleed sera (green) from each group were used as negative controls. Half-maximal serum binding titers were calculated based on the graphs and are presented in Table 2.
TABLE 2.
Neutralization of pseudotyped HIV-1 by gp120 antigen-immunized rabbit seraa
| Rabbit no. | Antigen JRFL gp120 and mutants | Half-maximal serum binding titers to wt gp120 | Serum dilution at 50% neutralization
|
|||||
|---|---|---|---|---|---|---|---|---|
| SF162 | JR-FL | JR-CSF | ADA | HXBc2 (TCLA) | VSV-G control | |||
| 7262 | Wild-type | 10 × 104 | 1:16 | 1:8 | −b | − | 1:32 | − |
| 7263 | Wild-type | 18 × 104 | 1:32 | − | − | 1:8 | 1:64 | − |
| 7264 | Wild-type | 7.5 × 104 | 1:32 | 1:4 | − | − | 1:32 | − |
| 7265 | GDMR | 8 × 104 | 1:32 | 1:4 | 1:4 | 1:16 | − | − |
| 7266 | GDMR | 25 × 104 | 1:128 | − | 1:4 | 1:4 | − | − |
| 7267 | GDMR | 30 × 104 | 1:128 | 1:4 | 1:8 | 1:32 | − | − |
| 7128 | mCHO | 10 × 104 | − | − | − | − | − | − |
| 7129 | mCHO | 15 × 104 | 1:16 | − | − | − | − | − |
| 7130 | mCHO | 7 × 104 | − | − | − | − | − | − |
wt, wild-type; VSV-G, vesicular stomatitis virus glycoprotein.
−, lower than 50% neutralization by the respective serum at the lowest serum dilution tested (1:4).
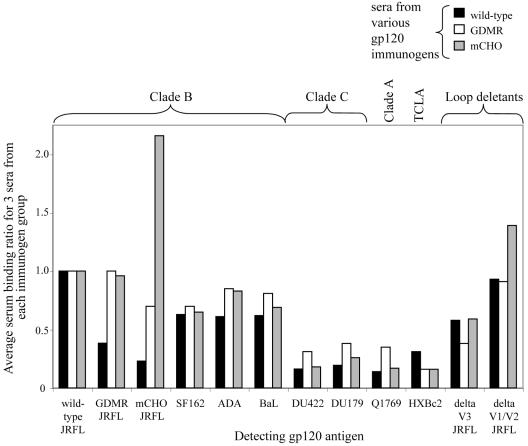
In order to determine the serum binding specificity to homologous and heterologous Envs, purified gp120s were captured with sheep anti-gp120 antibody (D7324) in a sandwich ELISA. The half-maximal serum binding titer for each individual serum against different Env gp120s was determined, and from that the binding ratio was calculated with respect to the serum binding to wild-type gp120JR-FL (the homologous strain). The average binding ratios for three rabbit sera (per immunogen) against different captured Env gp120s are shown in Fig. 3. The key observations are the following. (i) The sera from rabbits immunized with wild-type and GDMR and mCHO gp120s bind wild-type gp120JR-FL with similar apparent affinities as shown previously by serum titration.(ii) Sera from all immunogen groups exhibited good cross-reactivity to different clade B gp120s (SF162, ADA, and BaL) but the levels were lower when binding to gp120 from other clades (clade C-DU422 and DU179; clade A-Q1769) and also to T-cell line-adapted (TCLA) HXBc2 gp120. (iii) Sera from all three groups showed a strong V3-reactive antibody component as suggested by reduced binding to V3 loop deleted gp120JR-FL. (iv) Sera from rabbits immunized with wild-type gp120 bind GDMR gp120 and mCHO gp120 with reduced apparent affinity, suggesting that a significant percentage of the sera contains antibodies directed against CD4bs, CD4i, and C1-C4/C5 (discontinuous), antibodies that fail to bind these mutants. (v) Sera raised against mCHO gp120 bind the immunizing antigen somewhat better than to wild-type gp120, indicating that it elicits antibodies against neo-epitopes created in the presence of the extra glycans that are not present on wild-type gp120. (vi)Sera raised against wild-type gp120 and GDMR gp120 did not show the presence of a large V1/V2-reactive antibody component as determined by binding to V1/V2 loop deleted gp120JR-FL. However, sera from mCHO gp120-immunized rabbits bound the V1/V2 deletion gp120 mutant with higher apparent affinity compared to wild-type gp120 (when used as the detecting antigen), and this may explain the epitope specificity of some of the antibodies raised by immunization with the mCHO gp120 mutant that did not react with wild-type gp120.
FIG. 3.
ELISA binding of sera from wild-type (black), GDMR (white), and mCHO (gray) gp120-immunized rabbits to wild-type and mutant gp120s and to different gp120s from clades A, B, and C, HXBc2 (T-cell line adapted), V1/V2 loop-deleted (JR-FL gp120),and V3 loop-deleted (JR-FL gp120) gp120. The half-maximal serum binding titer (K50) for each individual serum against different gp120s was determined, and from that the binding ratio was calculated with respect to the serum binding to wild-type JR-FL gp120 (the homologous Env) (K50 of seruma binding to gp120x/K50 of seruma binding to wild-type gp120, where a represents one particular serum and x represents one of the different gp120s listed above). Each bar represents the average binding ratio for three rabbit sera per immunogen group (y axis) to different detecting gp120s (x axis).
Neutralization of virus by wild-type gp120-, GDMR gp120-, and mCHO gp120-immunized rabbit sera.
Neutralization assays were performed on immune sera (Table 2). Neutralization was evaluated against primary clade B CCR5-dependent viruses: SF162, a highly sensitive virus, and the moderately resistant viruses ADA, JR-CSF and JR-FL. We also checked for neutralization of HXBc2, a TCLA virus (Table 2). Sera from GDMR gp120-immunized animals appeared to be overall somewhat more effective in neutralizing the primary viruses than sera from wild-type gp120-immunized animals. Sera from mCHO gp120-immunized animals did not significantly neutralize the primary viruses. The GDMR gp120 immune sera showed the highest titers against the sensitive SF162 virus (1:128 for two out of three animals) and showed consistent neutralization of JR-CSF (titers 1:4 to 1:8) and ADA (titers of 1:4 to 1:32). The neutralization potential of wild-type gp120 and GDMR gp120 immune sera was also measured against a panel of different viruses at ViroLogic Inc. (5) (data not shown). In this case, some moderate and resistant primary clade B viruses, 1196, 92US712, 92HT593 and 92BR020, failed to be neutralized by any of the immune sera at the lowest dilution tested (1:20). GDMR gp120 immune sera neutralized SF162 virus at a titer four times higher than wild-type gp120 immune sera, as seen in our in-house assay. Both wild-type gp120 and GDMR gp120 immune sera failed to neutralize JR-FL and JR-CSF viruses at a 1:20 serum dilution.
Sera from both GDMR gp120- and mCHO gp120-immunized rabbits failed to neutralize the TCLA virus HXBc2, while wild-type gp120 immune sera was effective. There was thus an inversion of the activities of GDMR gp120 and wild-type gp120 immune sera for the TCLA virus compared to the primary viruses. This was also shown for another TCLA virus, NL4-3, where two out of the three GDMR gp120 immune sera failed to neutralize the virus (assay performed at ViroLogic Inc., [5]) (data not shown). The results are consistent with the importance of CD4bs and/or CD4i antibodies in neutralization of TCLA viruses. Such antibodies are typically very weak in neutralizing primary viruses and should not generally be elicited by GDMR gp120.
V3 peptide-based neutralization inhibition studies were performed to determine the contribution of anti-V3 antibodies to the neutralizing activity in the sera (Table 3). The V3 peptide used was a 23-amino-acid YU2 V3 sequence that has two amino acids different from the homologous JR-FL V3 sequence. The V3 peptide effectively inhibited neutralization of SF162 and ADA by the GDMR gp120 immune sera by approximately 70% and 60%, respectively. The peptide inhibited SF162 neutralization by wild-type gp120 immune sera by 50%. In an independent pseudotyped virus neutralization assay (performed at ViroLogic Inc., [5]), JR-FL V3 peptide (homologous sequence to the immunizing antigen) was used in neutralization inhibition of SF162 virus, and a similar reduction in neutralization was observed for both wild-type gp120 and GDMR gp120 immune sera (Table 3). This result suggests that the improved neutralization activity seen in the case of GDMR gp120 immune sera is due to a considerable degree to the presence of higher levels of V3 loop-specific antibodies. On the other hand, unlike GDMR gp120 immune sera, wild-type gp120 immune sera neutralized the TCLA virus NL4-3, and this was not inhibited in the presence of JR-FL V3 peptide in two of the three wild-type gp120 immune sera and inhibited only slightly in the third rabbit sera (Table 3). This result indicates that antibodies other than those against the V3 loop mediate the neutralization of TCLA viruses by wild-type gp120 immune sera.
TABLE 3.
Percentage reduction in neutralization by the wild-type or GDMR gp120 immune sera in the presence of YU2 or JR-FL V3 peptide (23 mer)a
| Animal and antigen (JR-FL gp120 background) | % Reduction in neutralization of virus
|
|||
|---|---|---|---|---|
| Primary SF162 virus with YU2 V3 peptide | Primary ADA virus with YU2 V3 peptide | Primary SF162 virus with JR-FL V3 peptide | TCLA NL43 virus with JR-FL V3 peptide | |
| 7262 WT | 47 | − | 50 | 0 |
| 7263 WT | 38 | − | 24 | 0 |
| 7264 WT | 61 | − | 52 | 6 |
| 7265 GDMR | 70 | 72 | 97 | − |
| 7266 GDMR | 87 | − | 99 | − |
| 7267 GDMR | 72 | 52 | 82 | ND |
Dash (−) indicates lower than 50% neutralization of a virus by the respective serum. WT, wild type; ND, not determined.
Mapping serum antibody responses using human monoclonal antibodies to inhibit binding of wild-type gp120, GDMR gp120, and mCHO gp120 immune sera.
We performed assays to map serum responses in order to explore the relationship between immunogenicity and antigenicity. This may in the future allow for improvement in the design of similar antigens. The assay used was similar to one described previously (29), where prebound monoclonal antibodies to wild-type gp120 were used to inhibit the binding of sera from HIV-1-infected patients. In the earlier assay, HIV-1-positive sera were directly labeled with alkaline phosphatase and used at a fixed concentration to compete unlabeled monoclonal antibodies. In our case, prelabeling was not necessary as human monoclonal antibodies were used to inhibit the binding of rabbit polyclonal sera (to wild-type gp120), and these were detected with anti-rabbit secondary antibody (with minimal reactivity to human serum proteins). Since the polyclonal serum competition with monoclonal antibodies was not very sensitive against antibodies produced in smaller fractions of the total serum response (such as CD4bs antibodies), we used two monoclonal antibodies per group to target a broader epitope footprint on gp120.
The majority, approximately 40 to 50%, of antibodies in the serum in all gp120-immunized groups were V3 loop-specific antibodies, as measured by inhibition of serum binding with human monoclonal antibodies to the V3 loop (447-52D, 19b, F425-B4e8, and 39F). Antibodies in the polyclonal sera targeted to the crown of the V3 loop were measured by inhibition in the presence of the V3 crown-specific antibodies 447-52D and 19b, and those to the stem of V3 loop were measured by inhibition in the presence of 39F (an antibody to the N terminus of V3 stem; J. Robinson, unpublished observations) and F425-B4e8 (characterized as a V3 stem-specific antibody [11], but recent observations indicate that the antibody is susceptible to changes in V3 crown residues). The competition assays were also performed with each antibody individually (447-52D, 19b, 39F, and F425-B4e8), with the result that the percentage inhibition was the same as when they were used in pairs, with the exception of 19b, which was lower. The GDMR gp120 immune sera exhibited an interesting pattern, with an overall increase in antibodies to both the crown (20%) and the stem (30%) of the V3 loop compared to wild-type gp120 immune sera (Fig. 4). This result suggests an improved exposure of epitopes both in the crown and stem of the V3 loop in the GDMR gp120 mutant and also supports the observation made through antigenicity studies of some reorientation of the variable loops. However, it is important to note that the crown and stem of the V3 loop are not completely independent, and there will be many anti-V3 antibodies that have footprints overlapping both epitopes. Two out of the three sera from rabbits immunized with mCHO gp120 also showed similarly enhanced levels of V3 loop specific antibodies. Of note, Garrity et al. (15) reported previously that a single glycosylation on the GPGR V3 crown epitope can lead to increased antibody response to epitopes on either side of the crown.
FIG. 4.
Competition ELISA between immune rabbit sera and MAbs for binding to gp120. Shown here are values obtained from competition ELISA in which wild-type gp120- and GDMR gp120 immune sera binding to wild-type gp120 is inhibited in the presence of human MAbs specific to the V3 loop (447-52D, F425-B4e8, and 39F). Percentage of V3 loop antibodies in the total polyclonal sera was determined by percent inhibition of serum binding to wild-type gp120 (as described in Materials and Methods) and is shown on the y axis. The percentages shown on the graph for wild-type gp120 and GDMR gp120 mutant are the averages of values obtained for three rabbits per group.
Responses to the CD4bs by any of the immunogens were relatively weak. Two of three wild-type gp120 immune sera had around 10% CD4bs antibodies as detected by inhibition of serum binding in the presence of antibodies b6 and F105, but this value dropped to undetectable levels in all three sera from GDMR gp120-immunized rabbits (data not shown). One of three sera from mCHO gp120-immunized rabbits also had around 10% of the total serum antibodies inhibited by b6 and F105.
Mapping the serum antibody responses by biotinylated MAb competition.
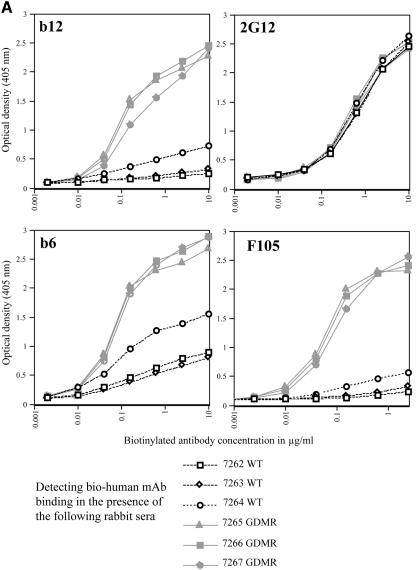
Polyclonal serum competition assays, where changes in serum antibody binding are detected in the presence of known MAbs (as described above), are inefficient when the antibodies being detected are close to 10% of the total antibody response or lower. Therefore, as an alternative, we set up assays with biotinylated human MAbs in the presence of rabbit sera to detect directly the inhibition of MAb binding to monomeric gp120 (27, 52). In this assay, biotinylated human MAb was titrated in the presence of a fixed concentration of polyclonal rabbit sera (1/100 dilution), and the human MAb binding was determined with streptavidin-alkaline phosphatase. The data for wild-type gp120 and GDMR gp120 immune sera competed with several human MAbs are shown in the form of graphs in Fig. 5. Table 4 tabulates the results from competition assays against a panel of human MAbs by wild-type gp120-, GDMR gp120-, and mCHO gp120-immunized rabbit sera. Described below is the simplest interpretation of the results with the bio-competition assay, where inhibition of binding by polyclonal serum of a bio-MAb is taken as the absence of a similar antibody in the serum. However, it is important to add that many epitopes can overlap. In addition, there are indirect effects where binding to one epitope can reduce the binding of an antibody in a completely different epitope (30). Unlike sera from rabbits immunized with wild-type gp120, weakly neutralizing CD4bs antibodies with specificities similar to b6 and F105 were almost undetectable in the sera from rabbits immunized with GDMR gp120 and mCHO gp120. This result is in accordance with the design of the mutants. The GDMR gp120 and mCHO gp120 mutants were also designed to elicit b12-like antibodies; however, this outcome was poorly represented in the immune sera as detected by biotinylated b12 antibody competition. The 2G12 competition assay did not show any difference between the different sera. This is a good control for the assay that shows absence of binding inhibition of 2G12 antibody by polyclonal serum antibodies raised against different gp120s, since such antibodies (to glycan clusters recognized by 2G12) are not normally elicited upon gp120 immunization. CD4i activity was determined in the presence of sCD4. There was a clear decrease in the levels of CD4i antibodies (48d and 17b) in the GDMR gp120 and mCHO gp120 immune sera. This result is also in accordance with the antigenic profiles exhibited by the gp120 mutants. Lower levels of A32-like antibodies were found in the sera from rabbits immunized with mCHO gp120 than in the case of GDMR gp120-immunized rabbits, which in turn were lower than sera raised from rabbits immunized with wild-type gp120. This observation also corresponds to antigenicity profiles that were previously shown with these mutants. Antibodies like C11 against a discontinuous C1-C5 epitope were also expressed at lower levels in the GDMR gp120 and mCHO gp120 immune sera than in the sera raised from the wild-type gp120 immunization. Both mutants had shown reduced recognition by C11 in antigenicity studies.
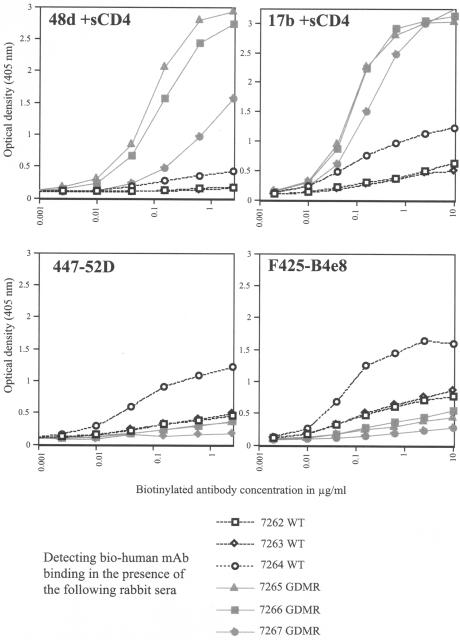
FIG. 5.
Competition ELISAs between biotinylated MAbs and immune rabbit sera. The x axis represents the concentration of biotinylated human MAb (μg/ml). The y axis represents the binding of biotinylated MAb detected with streptavidin-alkaline phosphatase at OD405. Biotinylated monoclonal antibody binding was inhibited in the presence of a single dilution (1/100) of wild-type gp120- or GDMR gp120 immune sera. The wild-type gp120-immunized rabbit sera (rabbits 7262 to 7264) inhibiting the binding of the respective bio-MAbs are shown as black lines; and the GDMR gp120-immunized rabbit sera (rabbits 7265 to 7267) inhibiting the binding of the respective bio-MAbs are shown as gray lines. (A) Inhibition of bio-b12 or bio-2G12 (top graphs) and bio-b6 or bio-F105 (bottom graphs). (B) Inhibition of bio-48d or bio-17b (in the presence of 2 μg/ml, sCD4) (top graphs) and bio-447-52D or bio-F425-B4e8 (bottom graphs).
TABLE 4.
Biotinylated human MAb binding inhibition to monomeric gp120 in the presence of wild-type, GDMR, and mCHO gp120 immune sera
| Rabbit no. and gp120 immunogen | Binding by antibodya
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4bs
|
Sugar
|
CD4i
|
C1-C4 region
|
V3 loop
|
V2 loop
|
C1-C5 region
|
|||||
| b12 | b6 | F105 | 2G12 | 48d +CD4 | 17b +CD4 | A32 +CD4 | 447-52D | F425-B4e8 | 697-D | C11 | |
| 7262 WT | 0.0 | 0.2 | 0.0 | 0.7 | 0.0 | 0.0 | 0.2 | 0.0 | 0.5 | 2 | 0.0 |
| 7263 WT | 0.0 | 0.11 | 0.0 | 0.7 | 0.0 | 0.0 | 0.2 | 0.0 | 0.5 | 1.5 | 0.0 |
| 7264 WT | 0.13 | 0.4 | 0.5 | 0.7 | 0.0 | 0.6 | 0.4 | 1.00 | 0.8 | 1.5 | 0.15 |
| Average WT | 0.04 | 0.24 | 0.17 | 0.7 | 0.0 | 0.2 | 0.27 | 0.33 | 0.6 | 1.7 | 0.05 |
| 7265 GDMR | 0.55 | 0.67 | 1.0 | 0.75 | 0.6 | 1.0 | 0.8 | 0.0 | 0.0 | 1.5 | 0.5 |
| 7266 GDMR | 0.5 | 1.0 | 0.83 | 0.75 | 0.4 | 1.0 | 0.3 | 0.0 | 0.0 | 2.0 | 0.8 |
| 7267 GDMR | 0.5 | 0.67 | 0.63 | 0.75 | 0.13 | 0.3 | 0.8 | 0.0 | 0.0 | 1.0 | 0.3 |
| Average GDMR | 0.52 | 0.8 | 0.82 | 0.75 | 0.4 | 0.77 | 0.63 | 0.0 | 0.0 | 1.5 | 0.53 |
| 7128 mCHO | 0.7 | 1.0 | 0.83 | 1.0 | 0.9 | 1.0 | 0.9 | 0.8 | 0.5 | 1.0 | 0.4 |
| 7129 mCHO | 0.7 | 1.0 | 1.0 | 1.0 | 0.6 | 1.0 | 1.0 | 0.5 | 0.5 | 1.5 | 0.5 |
| 7130 mCHO | 1.0 | 0.89 | 1.0 | 1.0 | 0.7 | 1.0 | 1.0 | 1.0 | 1.0 | 1.2 | 0.4 |
| Average mCHO | 0.8 | 0.96 | 0.94 | 1.0 | 0.73 | 1.0 | 0.97 | 0.77 | 0.67 | 1.23 | 0.47 |
| Prebleed | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Inhibiting antibody (2μg/ml)b | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
The half-maximal binding of bio-MAbs in the presence of immune sera was compared to binding of the bio-MAb in the presence of nonimmunized rabbit (prebleed) sera. The bio-MAb binding in the presence of nonimmune sera is given a value of 1.0 and inhibition of bio-MAb antibody in the presence of unbiotinylated MAb at 2 μg/ml is given a value of 0.0. WT, wild type.
5 μg/ml of C11 was used as inhibiting Ab.
The 447-52D-like antibodies to the crown of the V3 loop were found in abundance in all three GDMR gp120 immune sera, and this was also the case with two of the three wild-type gp120 immune sera (Fig. 5B). Antibody F425-B4e8 was also found to be inhibited better by the sera from rabbits immunized with GDMR gp120 than by wild-type gp120 or mCHO gp120 immune sera (Table 4), indicating that F425-B4e8-like antibodies were more abundant in the GDMR gp120 immune sera. This result is in accordance with the increased levels of antibodies to both the crown and stem of V3 loop seen in the previous assay with the GDMR gp120 immune sera. In contrast, sera from rabbits immunized with mCHO gp120 had comparatively lower levels of 447-52D-like antibodies. Since the earlier studies showed an abundance of V3 loop antibodies, it appears that these may be directed to the stem rather than the tip of the loop.
Antibodies against a conformational epitope in the V2 loop defined by the MAb 697-D were not present in wild-type gp120, GDMR gp120, or mCHO gp120 immune sera, as determined by the absence of inhibition of binding of this antibody to gp120.
Mapping of serum responses using linear peptides.
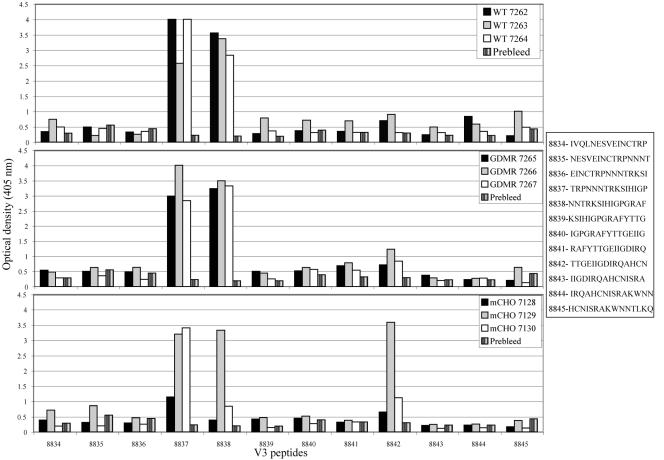
A whole clade B gp120 consensus sequence comprising 115 15-mer overlapping peptides was scanned for binding differences between wild-type gp120, GDMR gp120, and mCHO gp120 immune sera (constant dilution of 1/100). The ELISA binding data for all 115 peptides is not shown (only those of the V3 peptides) (Fig. 6) as the overall binding pattern did not vary between the different groups, and the occasional differences are explained below. Antibodies binding linear peptides in the C1 region were found in all three groups of sera including the mCHO gp120 mutant that had two extra sugar attachment motifs in this region. Two of the three sera from GDMR gp120-immunized rabbits showed increased levels of antibodies to the V2 loop sequence ALFYKLDVVPIDNDN; this was also the case with sera from all three mCHO gp120-immunized rabbits. The sera raised against GDMR gp120 showed increased levels of antibodies against some linear peptides in the C2 region different from the level seen with sera from rabbits immunized with wild-type gp120. The V3 response in sera from both wild-type gp120- and GDMR gp120-immunized rabbits was restricted to just two peptides, TRPNNNTRKSIHIGP and NNTRKSIHIGPGRAF (Fig. 6). However, two of the three sera raised against mCHO gp120 had lower levels of antibodies against the second peptide. This is in accordance with the results described above that showed that antibodies to the crown of the V3 loop were in lower abundance in the mCHO gp120 immune sera compared to wild-type gp120 or GDMR gp120 immune sera. Both wild-type gp120 and mCHO gp120 immune sera showed no detectable antibody response to the V1 peptides where the extra sugars were incorporated at positions 141 and 150 onto the mCHO gp120 mutant. All three sera from mCHO gp120-immunized rabbits had a large response against the C4 peptide VGKAMYAPPIRGQIR, which was not the case with sera raised against wild-type or GDMR gp120s. Overall, the sera from both wild-type gp120- and GDMR gp120-immunized rabbits did not show any large differences in recognition of linear peptides. The differences with regards to CD4bs, CD4i, and C1-C4/C5 gp120 core antibodies seen with GDMR gp120 and mCHO gp120 immune sera compared to sera from wild-type gp120-immunized rabbits (in the competition ELISAs described earlier) appears to be mainly due to changes in levels of conformational antibodies directed against discontinuous epitopes and not due to antibodies raised against linear epitopes.
FIG. 6.
ELISA of serum binding to linear overlapping V3 peptides. The sequences of peptides are shown in the right-hand panel with peptide numbers as given by National Institutes of Health AIDS Reagent Repository. The upper panel represents three (rabbits 7262 to 7264) wild-type sera, middle panel represents three (rabbits 7265 to 7267) GDMR sera, and bottom panel represents three (rabbits 7128 to 7130) mCHO sera. The y axis represents binding measured at OD405
DISCUSSION
In this report, we have described a set of immunogenicity studies on first generation monomeric gp120 mutants that have been engineered toward eliciting b12-like antibodies. The main aims of our experiments were twofold: first, to obtain information on the immunogenicity of the mutants; second, to map serum responses to explore the relationship between antigenicity and immunogenicity. Our findings are being used to improve the design and construction of further mutants.
A key observation from this study is that engineering of gp120 can lead to a targeted reduction in the elicitation of multiple nonneutralizing and weakly neutralizing antibody specificities. The vaccinia-produced GDMR gp120, where residues 473 to 476 were mutated to alanine, mirrored the antigenicity profile previously established (35, 36) for 293T cell-derived GDMR gp120, and in addition further antigenic properties were defined. Our findings can be summarized as follows: (i) CD4bs antibodies, b6, b3, and F105 that are weakly neutralizing failed to bind, while binding affinity for the broadly neutralizing antibody b12 remained the same as wild-type gp120; (ii) CD4i antibodies failed to bind the GDMR gp120 mutant; (iii) C1-C4/C5 gp120 antibodies bound GDMR gp120 with lowered affinity compared to wild-type gp120; (iv) V3 loop antibodies bound the GDMR gp120 mutant with similar affinities as wild-type gp120; (v) a V2 loop antibody (8.22.2) to a linear epitope bound the GDMR gp120 mutant with similar apparent affinity as wild-type gp120 but the antibody (697-D) to a conformational V2 epitope bound with lower affinity; (vi) a V1/V2/V3 loop-dependent antibody, 4KG5, exhibited lower binding to the GDMR gp120 mutant compared to wild-type gp120. Overall, the difference in antibody binding affinities for GDMR gp120 mutant by antibodies to the variable loops that are conformation dependent suggested a degree of conformational rearrangements in the V1/V2 and V3 variable loops. In addition, the lack of recognition of the GDMR gp120 mutant by CD4i antibodies is likely linked to a change in the orientation of the variable loops.
Mapping of the serum responses to the GDMR gp120 mutant by competition studies showed reduced levels of CD4bs and CD4i antibodies compared to the sera from rabbits immunized with wild-type gp120. These observations are consistent with the antigenicity profiles exhibited by the GDMR gp120 mutant. A large proportion of the total serum antibodies in the GDMR gp120-immunized rabbits were specific to the V3 loop. It has been shown previously that sera raised from humans vaccinated with gp120 contain a large proportion of antibodies directed against the V3 loop (27, 29). Notably, there was a 20 to 30% increase in the percentage of V3 loop antibodies in the sera from rabbits immunized with GDMR gp120 compared to sera of wild-type gp120-immunized rabbits. This result is consistent with the notion that some rearrangements of the V1/V2 and/or V3 loops in the GDMR gp120 mutant produced increased epitope exposure and improved immunogenicity of V3 loop epitopes. There is also a possibility that the increased levels of V3 loop antibodies to the GDMR gp120 mutant may be a compensatory response due to dampening of the elicitation of CD4bs and CD4i antibodies. The immunogenicity results for the GDMR gp120 mutant are encouraging in that the serum antibody responses correspond fairly well to the antigenic profiles exhibited by the mutant.
The GDMR gp120 immune sera exhibited a somewhat improved neutralizing activity against a few primary viruses compared to sera from wild-type gp120-immunized rabbits. We sought to investigate whether this could be attributed to the elicitation of b12-like antibodies as envisaged for the GDMR gp120 mutant. V3 peptide competition studies showed that most (70 to 90%) of the neutralization, where present, could be attributed to V3 loop antibodies. It would seem that changes in the conformation and/or exposure of the V3 loop as a result of the GDMR mutation increases the titer of a neutralizing antibody response that has some degree of cross-reactivity. Furthermore, the inability of the GDMR gp120 immune sera to neutralize the TCLA viruses HXBc2 and NL4-3 argues against the induction of b12-like antibodies since both these viruses are very sensitive to b12 neutralization and, indeed, to CD4bs antibody neutralization in general (5, 28, 49).
Immunization with wild-type gp120 or GDMR gp120 had opposite effects on the relative ability of the resulting sera to neutralize primary and TCLA viruses. Wild-type gp120 immune sera neutralized the TCLA viruses HXBc2 and NL4-3, presumably reflecting the elicitation of antibodies such as CD4bs and CD4i, antibodies that effectively neutralize such viruses. The V3 loop of HXBc2 and NL4-3 is notably different from that of the immunizing JR-FL isolate, and neutralization titers were not significantly affected by the presence of JR-FL V3 peptide. It is therefore highly unlikely that neutralization of the TCLA virus NL4-3 by the wild-type gp120 immune sera was mediated by V3 loop-specific antibodies to linear epitopes. In contrast, the GDMR gp120 immune sera did not neutralize HXBc2 or NL4-3 virus, as opposed to its ability to neutralize some primary viruses. This is consistent with the loss of CD4bs and CD4i antibodies in the corresponding sera. The pattern of these results clearly indicates that neutralization of TCLA virus is not necessarily predictive of neutralization of primary viruses; i.e., one can have primary virus neutralization in the absence of TCLA neutralization. Therefore, the use of TCLA neutralization as a sole prescreen or first stage evaluation of immune sera is probably not advisable.
The mCHO gp120 mutant was designed to eliminate weakly neutralizing and nonneutralizing antibodies in the CD4bs, CD4i, C1-C4/C5, and V1/V2 loops and part of the V3 loop while still maintaining binding to antibody b12. The mCHO gp120 mutant with seven extra glycosylation moieties still gave a robust serum antibody binding titer by ELISA, similar to the sera from wild-type gp120-immunized rabbits. Mapping sera by competition studies showed that mCHO gp120 elicited lower levels of CD4bs, CD4i, and C1-C4/C5 conformational antibodies than wild-type gp120 as envisaged by its design. This is an interesting observation that shows that antibody responses to three major gp120 core epitopes can be dampened by the presence of only three extra glycans.
Wild-type gp120 immune sera did not elicit detectable levels of conformational V2 loop-specific antibodies as determined by the lack of serum inhibition of biotinylated 697-D antibody and this was also the case with mCHO gp120 immune sera. However there were some antibodies against linear V2 peptides both in the wild-type gp120 and mCHO gp120 immune sera. It is therefore difficult to speculate whether the extra sugar on the V2 loop necessarily masked responses to the V2 loop in the mCHO gp120 mutant since such antibodies were not elicited in great numbers by wild-type gp120 immunization either. However, an improved serum binding response by the mCHO gp120 immune sera was seen to a linear 15-mer peptide in the V2 loop, the C terminus of the K171 residue where an extra sugar was incorporated.
Sera from mCHO gp120-immunized rabbits did not produce antibodies that bind to the crown of the V3 loop as determined by the lack of inhibition of biotinylated 447-52D antibody binding in the presence of mCHO gp120 immune sera compared to sera from wild-type gp120- or GDMR gp120-immunized rabbits. In addition, two out of the three mCHO gp120 immune sera also failed to bind one of the two V3 crown-specific 15-mer peptides (NNTRKSIHIGPGRAF), against which both wild-type gp120 and GDMR gp120 immune sera bound the best. The reasons for this could be either the mutation of the GPGR sequence or the effective masking of the V3 crown by the extra sugar moiety. The lowered levels of V3 crown-specific antibodies in the mCHO gp120 immune sera is in accordance with the lack of recognition of the mutant by V3 crown antibodies 447-52D and 19b in antigenicity studies. However, in a different setup of the competition ELISA, where inhibition of the mutant immune serum binding to wild-type gp120 is detected in the presence of human monoclonal antibodies, mCHO gp120 immune serum binding was effectively inhibited in the presence of monoclonal antibodies to the V3 loop. The results indicate that there is still a significant proportion of V3 loop antibodies in the mCHO gp120 immune sera to parts of the V3 loop other than the crown. This result is consistent with the antigenicity studies that showed the mCHO gp120 mutant bound well to the V3 stem-specific antibody 39F. It is, however, difficult to say conclusively whether the masking of the crown of the V3 loop or the mutation in the GPGR sequence caused the reduction in antibodies (like 447-52D) specific to the crown. In a report by Garrity et al. (15), a complete block in the antibody response against the V3 loop as a whole was seen only when there were three to four extra sugar attachment motifs and at least two extra glycans on the V3 loop, and in that case the response shifted to the immunodominant V1 loop. Therefore, in order to completely mask the V3 loop, additional glycosylations may be needed. Similarly, to prevent a shift in antibody response to other variable loops, extra sugars may also be required to fully mask the V1/V2 loops. In light of this, we have recently modified the mCHO gp120 mutant further by the incorporation of several additional glycosylation sites in the V2 and V3 loops (36).
The increased levels of antibodies to a 15-mer V2 loop peptide in the mCHO gp120 immune sera in contrast to wild-type gp120 immune sera raised some concern about the possible generation of neo-epitopes within the mCHO gp120 mutant. Therefore, the mCHO gp120 immune sera were tested for the presence of antibodies to such putative epitopes by using serum antibody binding ELISAs. Immune sera from mCHO gp120 bound to itself somewhat better than to wild-type gp120 and also to gp120 with a deletion of the V1/V2 loop, indicating that some of the serum antibodies from mCHO gp120-immunized rabbits recognized epitopes that were not well exposed on wild-type gp120. Mapping of mCHO gp120 immune sera using linear peptides spanning the whole gp120 sequence (115, 15-mer, overlapping peptides) detected improved recognition by all three rabbit sera from mCHO gp120 immunization of chiefly two 15-mer peptides, one in the C terminus of the V2 loop and other in the conserved C4 region of gp120, in contrast to wild-type gp120 immune sera. Therefore, clearly the mCHO gp120 mutant elicits some neo-epitope antibodies, but the levels are not overwhelming, and, more importantly, most serum antibodies raised against this mutant recognized wild-type gp120 as well as immune sera from wild-type gp120-immunized rabbits.
The mCHO gp120 immune sera exhibited a loss of neutralization efficiency compared to wild-type gp120. This is probably due to the lowered levels of weakly neutralizing CD4bs and CD4i antibodies and to reduced levels of V3 crown-specific antibodies that are an important fraction of the neutralizing antibody repertoire in the immune sera from monomeric gp120. The sera from rabbits immunized with wild-type gp120 elicited CD4bs antibodies as detected by competition assays with bio-b12, bio-b6, or bio-F105. However, the wild-type gp120 immune sera (rabbits 7262, 7263, and 7264) failed to neutralize antibody b12-sensitive viruses such as HIV-1 strains JR-FL, JR-CSF, or ADA to any significant degree. This suggests that b12-like antibodies are not elicited following wild-type gp120 immunization. We had hoped that, unlike wild-type gp120, the engineered gp120s designed to expose the b12 epitope would elicit b12-like antibodies. In the case of the mCHO gp120, the failure to elicit b12-like antibodies might be due to the lowered exposure of the b12 epitope as determined by the reduced affinity for b12 antibody binding. As indicated above, we have sought to address this problem by the engineering of mutants with improved affinity for b12 (36) that may prove superior in eliciting b12-like antibodies.
In most studies, where a number of potential HIV-1 antigens are being tested for immunogenicity in small animal models, testing for serum titers and testing for neutralizing antibody response are the regular assays of choice (3, 7, 53, 58). Serum mapping studies aimed at determining the specificities of antibodies raised by an antigen are not examined to any great extent. Competition assays between monoclonal antibodies and polyclonal sera do not always give direct answers regarding the different types of antibodies present in polyclonal sera. This is also true when monoclonal antibodies are competed against other monoclonal antibodies, where decreases in binding can be observed when one antibody binds to gp120 even though the second antibody does not bind to precisely the same epitope (30). Mapping polyclonal immune serum is not straightforward but vital and provides answers as to the success of a strategy to modify HIV-1 Env to elicit neutralizing antibodies.
To summarize, we have immunized with engineered monomeric gp120 that favorably presents the conserved epitope for the broadly neutralizing antibody b12 while lowering the exposure of the weakly neutralizing and nonneutralizing immunodominant epitopes. Two strategies were used, one where residues were changed on the gp120 core such that a selection of weakly neutralizing antibodies fail to bind (GDMR gp120 mutant). The second was to mask immunodominant loops, the CD4-induced site and the nonneutralizing face of gp120 with carbohydrate moieties (mCHO gp120 mutant). One significant finding in this report is that through design, it is possible to reduce elicitation of unwanted antibodies to a multiplicity of epitopes. However, we have not yet been able to focus the response to the desired b12 epitope, and this will likely require further engineering of gp120.
Acknowledgments
We thank Christina Corbaci and Rosa Cardoso for help with making Fig. 1. We thank Louis Altamura and Tsiri Agbenyega from R. Dom's laboratory for technical support. We thank Charlotte Berndt for technical support. We thank Michael Zwick for helpful discussions with the manuscript.
We acknowledge support from grants from the National Institutes of Health (grants AI33292 and AI060425 to D.R.B.), the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium, the Pendleton Trust Foundation, American Foundation for AIDS Research (amfAR grant 106436-34-RFVA to R.P), Elizabeth Glaser Pediatric AIDS Foundation (grant 77497 to M.L), and Public Health Service Grant from National Institutes of Health (grant F32 AI10577 to B.P).
REFERENCES
- 1.Barbas, C. F., III, E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, E. Norrby, and D. R. Burton. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolmstedt, A., S. Sjolander, J. E. Hansen, L. Akerblom, A. Hemming, S. L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 7.Bower, J. F., X. Yang, J. Sodroski, and T. M. Ross. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 78:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R., C. F. Barbas III, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 11.Cavacini, L., M. Duval, L. Song, R. Sangster, S. H. Xiang, J. Sodroski, and M. Posner. 2003. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17:685-689. [DOI] [PubMed] [Google Scholar]
- 12.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 1998. Generation of recombinant vaccinia viruses, UNIT 16.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley and Sons, New York, N.Y.
- 14.Fouts, T., K. Godfrey, K. Bobb, D. Montefiori, C. V. Hanson, V. S. Kalyanaraman, A. DeVico, and R. Pal. 2002. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. USA 99:11842-11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrity, R. R., G. Rimmelzwaan, A. Minassian, W. P. Tsai, G. Lin, J. J. de Jong, J. Goudsmit, and P. L. Nara. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279-289. [PubMed] [Google Scholar]
- 16.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundner, C., Y. Li, M. Louder, J. Mascola, X. Yang, J. Sodroski, and R. Wyatt. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 331:33-46. [DOI] [PubMed] [Google Scholar]
- 20.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, Y., W. J. Honnen, C. P. Krachmarov, M. Burkhart, S. C. Kayman, J. Corvalan, and A. Pinter. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J. Immunol. 169:595-605. [DOI] [PubMed] [Google Scholar]
- 22.Kang, S. M., F. S. Quan, C. Huang, L. Guo, L. Ye, C. Yang, and R. W. Compans. 2005. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 331:20-32. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. B., D. P. Han, C. Cao, and M. W. Cho. 2003. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology 305:124-137. [DOI] [PubMed] [Google Scholar]
- 24.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, H. X., S. M. Alam, J. R. Mascola, J. Robinson, B. Ma, D. C. Montefiori, M. Rhein, L. L. Sutherland, R. Scearce, and B. F. Haynes. 2004. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 78:5270-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, S., R. Wyatt, J. F. Richmond, F. Mustafa, S. Wang, J. Weng, D. C. Montefiori, J. Sodroski, and H. L. Robinson. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res. Hum. Retrovir. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 27.Montefiori, D. C., B. S. Graham, J. Zhou, R. A. Bucco, D. H. Schwartz, L. A. Cavacini, and M. R. Posner. 1993. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. NIH-NIAID AIDS Vaccine Clinical Trials Network. J. Clin. Investig. 92:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. Kessler II, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, J. P., R. L. Willey, G. K. Lewis, J. Robinson, and J. Sodroski. 1994. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 68:6836-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancera, M., and R. Wyatt. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145-156. [DOI] [PubMed] [Google Scholar]
- 34.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2004. Improved design of an antigen with enhanced specificity for the broadly HIV-neutralizing antibody b12. Protein Eng. Des. Sel. 17:749-758. [DOI] [PubMed] [Google Scholar]
- 37.Poon, B., J. T. Safrit, H. McClure, C. Kitchen, J. F. Hsu, V. Gudeman, C. Petropoulos, T. Wrin, I. S. Chen, and K. Grovit-Ferbas. 2005. Induction of humoral immune responses following vaccination with envelope-containing, formaldehyde-treated, thermally inactivated human immunodeficiency virus type 1. J. Virol. 79:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7-14. [PubMed] [Google Scholar]
- 39.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 40.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas, 3rd, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 45.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, C. F., Jr., S. Silver, A. T. Profy, S. D. Putney, A. Langlois, K. Weinhold, and J. E. Robinson. 1990. Human monoclonal antibody that recognizes the V3 region of human immunodeficiency virus gp120 and neutralizes the human T-lymphotropic virus type IIIMN strain. Proc. Natl. Acad. Sci. USA 87:8597-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12:193-204. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turbica, I., M. Posner, C. Bruck, and F. Barin. 1995. Simple enzyme immunoassay for titration of antibodies to the CD4-binding site of human immunodeficiency virus type 1 gp120. J. Clin. Microbiol. 33:3319-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, Z. Y., B. K. Chakrabarti, L. Xu, B. Welcher, W. P. Kong, K. Leung, A. Panet, J. R. Mascola, and G. J. Nabel. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwick, M. B., R. Kelleher, R. Jensen, A. F. Labrijn, M. Wang, G. V. Quinnan, Jr., P. W. Parren, and D. R. Burton. 2003. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 77:6965-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]