Abstract
We used experimental infection of rhesus macaques with envelope gp120 V3 loop isogenic simian-human immunodeficiency virus (SHIV) molecular clones to more clearly define the impact of human immunodeficiency virus type 1 coreceptor usage in target cell selectivity and the rates of CD4+-T-cell depletion. Functional assays demonstrate that substitution of the V3 loop of the pathogenic CXCR4-tropic (X4) SHIVSF33A2 molecular clone with the corresponding sequences from the CCR5-tropic (R5) SHIVSF162P3 isolate resulted in a switch of coreceptor usage from CXCR4 to CCR5. The resultant R5 clone, designated SHIVSF33A2(V3), is replication competent in vivo, infecting two of two macaques by intravenous inoculation with peak viremia that is comparable to that seen in monkeys infected with X4-SHIVSF33A2. But while primary infection with the X4 clone was accompanied by rapid and significant loss of peripheral and secondary lymphoid CD4+ T lymphocytes, infection with R5-SHIVSF33A2(V3) led to only a modest and transient loss. However, substantial depletion of intestinal CD4+ T cells was observed in R5-SHIVSF33A2(V3)-infected macaques. Moreover, naïve T cells that expressed high levels of CXCR4 were rapidly depleted in X4-SHIVSF33A2-infected macaques, whereas R5-SHIVSF33A2(V3) infection mainly affected memory T cells that expressed CCR5. These findings in a unique isogenic system illustrate that coreceptor usage is the principal determinant of tissue and target cell specificity of the virus in vivo and dictates the dynamics of CD4+-T-cell depletion during SHIV infection.
Since the discovery of the chemokine receptors CCR5 and CXCR4 as the major coreceptors (CoRs) for entry of human immunodeficiency virus type 1 (HIV-1), mounting evidence indicates that coreceptor expression and usage play crucial roles in viral transmission, persistence, and pathogenesis (28). Most HIV-1 strains transmitted between humans use CCR5 as their coreceptor (R5 strains). With progression to disease, variants that use CXCR4 (X4 viruses) emerge in about 50% of infected individuals and are associated with a more rapid rate of peripheral CD4+-T-cell loss (33). We and others have used infection of nonhuman primates with simian-human immunodeficiency virus (SHIV) expressing the envelopes of R5 and X4 HIV-1 strains as a model system to study the impact of coreceptor usage in HIV-1 infection and AIDS pathogenesis (4). In infection of rhesus macaques (RM) with R5 and X4 pathogenic SHIVs, SHIVSF162P3 and SHIVSF33A, respectively, we previously reported a slower rate of peripheral CD4+-T-cell depletion in R5-SHIVSF162P3-infected RM that recapitulated the progression of infection seen in humans (13, 15). In contrast, infection with pathogenic X4-SHIVSF33A resulted in rapid and precipitous decline in peripheral and lymphoid CD4+ T cells, mirroring infection with X4 isolates (15, 16, 22). However, analysis of lamina propria lymphocytes (LPLs) purified from jejunal tissues revealed that R5-SHIVSF162P3 caused severe depletion of mucosal CD4+ T cells within 2 weeks of infection, whereas the loss of mucosal CD4+ T cells in X4-SHIVSF33A-infected macaques was more gradual (15). Different tissue sites of replication and CD4+-T-cell depletion patterns in macaques infected with CXCR4- and CCR5-tropic viruses have also been reported previously by others (30, 31), leading to the suggestion that target cell availability as influenced by differential coreceptor expression and viral tropism as dictated by coreceptor preference have profound consequences on clinical disease. Nevertheless, the strains used in all these studies were independent isolates, with variants that differed in genetic sequences and, hence, in properties other than host cell tropism which might have contributed to the differences seen in the pathogenic sequela. For instance, a major (7 out of 10 clones) and a minor (1 out of 10 clones) variant, as determined by V1 to V5 env sequences, that differed in their entry efficiency and neutralization susceptibility were present in the R5-SHIVSF162P3 isolate we used (17). Furthermore, while both X4-SHIVSF33A and R5-SHIVSF162P3 showed neutralization resistance, the X4 virus appeared to be more fusogenic and cytopathic in vitro (6, 7, 18). A confirmation that early targeting of different T-cell populations and different tissues by X4 and R5 viruses is due solely to the coreceptors used could greatly benefit from infection of RM with variants that differ only in their coreceptor binding site sequences.
For HIV-1, differential usage of coreceptor in vitro has been shown to depend on the charge and/or structure of the V3 loop of envelope gp120 (2, 8). Towards the objective of constructing isogenic SHIVs that differ only in coreceptor preference, the V3 loop of SHIVSF33A2, a molecular clone that displays the infection characteristics of the parental pathogenic X4-SHIVSF33A isolate in RM (14), was replaced with the corresponding region of the R5-SHIVSF162P3 isolate (18). The function and coreceptor choice of the resulting chimeric envelope glycoprotein were determined. Furthermore, infection of RM with virus expressing the chimeric V3 envelope, designated SHIVSF33A2(V3), was characterized and compared to infection with the X4 isogenic SHIVSF33A2 molecular clone.
MATERIALS AND METHODS
Cells.
Human osteosarcoma cells expressing CD4 and either CCR5 (HOS.CD4.CCR5) or CXCR4 (HOS.CD4.CXCR4) were kind gifts from N. Landau (Salk Institute, La Jolla, CA) and were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 0.5 μg/ml of puromycin (Sigma-Aldrich, St. Louis, MO), and penicillin-streptomycin. 293T cells used for transfection were cultured in Dulbecco's modified Eagle's medium with 10% FBS and penicillin-streptomycin. Human peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll-Hypaque gradient purification followed by stimulation with 3 μg/ml of phytohemagglutinin A (Sigma-Aldrich) in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 U/ml of interleukin-2 (Chiron Corp., Emeryville, CA).
Plasmid constructs and virus production.
PCR-based overlapping extension methodology was employed to replace the V3 loop of X4-SHIVSF33A2 env gp120 with that of R5-SHIVSF162P3. Briefly, a HincII-to-HindIII fragment of Env33A2 (6) (nucleotides 1249 to 2386 [GenBank accession number M38427]) was subcloned into pBlueScript KS II(+) vector to serve as the template for PCR amplifications. The inner amplification primers used were SH5 (5′-GGG GAA AGC ATT TTA TGC AAC AGG AGA CAT AAT AGG AGA TAT AAG ACA AGC ACA TTG TAA CAT TAG TAG AGC-3′) and SH6 (5′-GCA TAA AAT GCT TTC CCC GGT CCT ATA GGT ATA CTT TTT CTT GTA TTG TTA TTG GGT CTT GTA C-3′), which encompassed the env gp120 V3 sequence of R5-SHIVSF162P3 (in italics, with overlapping regions underlined) flanked by the C2 and C3 sequences of X4-SHIVSF33A2. The outer primers used were T3 and T7. The amplified fragment was completely sequenced for verification, and an envelope expression vector containing the V3 chimeric env, Env33A2(V3), was constructed by subcloning the HincII-to-HindIII-amplified fragment back into Env33A2 (Fig. 1). The same fragment was ligated into the corresponding regions of SHIVSF33A2 3′ DNA (14) to generate the 3′ genome of the V3 isogenic clone SHIVSF33A2(V3). Env162P3 (18)-, Env33A2-, and Env33A2(V3)-pseudotyped luciferase reporter viruses were prepared by cotransfecting 293T cells with the NL4.3-Luc-E−R− vector and the corresponding Env expression plasmid using the DMRIE-C reagent (Invitrogen). Reporter viruses were harvested 72 h posttransfection and quantified for p24gag antigen content (Beckman Coulter, Fullerton, CA). SHIV was obtained by cotransfection of 293T cells with SIVmac239 5′ and SHIVSF33A2 or SHIVSF33A2(V3) 3′ hemigenomes followed by cocultivation with phytohemagglutinin A-stimulated PBMCs as previously described (23). Recovered viruses were amplified and propagated in human PBMCs, and culture supernatants were collected for viral p27gag antigen determination (Coulter Corporation, Miami, FL) and for assessment of the 50% tissue culture infectious dose in human PBMCs.
FIG. 1.
Comparison of the V3 loop sequence of Env33A2 and Env33A2(V3). Chimeric Env was constructed by replacing the V3 loop of X4-SHIVSF33A2 gp120 (white) with the corresponding sequences of R5-SHIVSF162P3 (black) as described in Materials and Methods.
Entry and blocking assays.
A total of 5 × 103 HOS.CD4.CCR5 or HOS.CD4.CXCR4 cells were seeded in 96-well plates 24 h before use. Cells were infected with 5 ng p24 equivalent of the indicated pseudotyped viruses in the presence of 2 μg/ml of polybrene followed by incubation for 72 h at 37°C. At the end of the incubation period, cells were harvested, lysed, and processed according to the manufacturer's instructions (Promega, Madison, WI). Entry, as quantified by luciferase activity, was measured with an MLX microtiter plate luminometer (Dynex Technologies, Inc., Chantilly, VA). For the blocking assay, 5 × 105 PBMCs were pretreated with various concentrations of AOP-RANTES (a generous gift of Oliver Hartley, University of Zurich) for 1 h before infection. Percent blocking was calculated by the amount of entry in the presence of AOP-RANTES relative to that in the absence of AOP-RANTES.
Animal infections.
All infections were carried out in adult RM (Macaca mulatta) individually housed at the Tulane National Primate Research Center in compliance with the Guide for the Care and use of Laboratory Animals (28a). Animals were confirmed to be serologically negative for simian type D retrovirus, simian immunodeficiency virus (SIV), and simian T-cell lymphotrophic virus prior to infection. Whole blood lymphocytes were sampled at designated time intervals. Plasma viremia was quantified by branched DNA analysis (Bayer Diagnotics, Emeryville, CA), and absolute CD4+ and CD8+ cell counts were monitored by TruCount (BD Biosciences, Palo Alto, CA). Biopsy samples were obtained by surgery at 2 and 8 weeks postinfection (wpi) for the SHIVSF33A2(V3)-infected animals and at 2 and 9 wpi for the SHIVSF33A2-inoculated animals. The percentages of CD4+ T cells in the biopsy samples were analyzed by flow cytometry (FACScalibur) using CD3-fluorescein isothiocyanate (FITC), CD4-phycoerythrin (PE), and CD8-peridinin chlorophyll protein antibodies. Further phenotyping of peripheral and lymphoid CD4+ T cells was performed by using CD28-allophycocyanin, CD95-PE or -FITC, CCR5-PE, and CXCR4-PE. Except for CD3-FITC (BioSource, Camarillo, CA), all antibodies were obtained from BD Biosciences.
RESULTS
The V3 loop of X4-SHIVSF33A2 dictates CoR usage.
The 35-amino-acid V3 domains of the X4-SHIVSF33A2 molecular clone and the R5-SHIVSF162P3 isolate differ in 10 amino acids (Fig. 1), with the overall charge of this region being +7 for SHIVSF33A2 and +5 for SHIVSF162P3, values that are consistent with those reported previously for other X4 and R5 HIV-1 strains (10). In single-round infectivity assays, we found that contrary to Env33A2, the chimeric V3 envelope Env33A2(V3) was not able to mediate entry into HOS.CD4 cells expressing CXCR4. Instead, Env33A2(V3), similar to Env162P3, used CCR5 as the CoR (Fig. 2A). To confirm the coreceptor usage of Env33A2(V3), blocking of entry into human PBMCs with AOP-RANTES, an amino terminus-modified form of the CCR5 natural ligand RANTES (36), was assessed. Results showed that entry of virus expressing Env33A2(V3) was efficiently blocked by AOP-RANTES in a dose-dependent manner, while virus expressing Env33A2 was not affected, even at a concentration of 250 nM (Fig. 2B). In fact, a slight increase in Env33A2-mediated infection in the presence of high concentrations of AOP-RANTES (>50 nM) was observed, consistent with reports of stimulation of X4 virus replication by CC chemokines (9, 19). Taken together, the data showed that Env33A2(V3) is functional and that the substitution of the V3 loop of Env33A2 with that of Env162P3 resulted in a switch in coreceptor usage from CXCR4 to CCR5.
FIG. 2.
In vitro properties of Env33A2(V3). (A) Relative entry of luciferase reporter viruses expressing Env33A2, Env162P3, and Env33A2(V3) into HOS.CD4.CXCR4 (black bars) and HOS.CD4.CCR5 (white bars) cells. (B) CCR5 usage of reporter viruses expressing Env33A2(V3) (▵) was verified by blocking entry into human PBMCs with the modified CCR5 ligand AOP-RANTES. Viruses pseudotyped with Env33A2 (▪) served as negative controls. All data were the means of triplicates.
SHIVSF33A2(V3) is replication competent in RM.
Env33A2(V3) was then used to replace the corresponding region of X4-SHIVSF33A2 to construct the chimeric virus SHIVSF33A2(V3). The two clones differ only in the V3 loop, with an overall identity of 99.88%. Two macaques each were inoculated intravenously with 103 50% tissue culture infectious doses of R5-SHIVSF33A2(V3) (macaques BT35 and CF17) or X4-SHIVSF33A2 (macaques CA05 and CA06). All four animals were infected, with peak viremia of 106 to 108 RNA copies/ml plasma at 2 to 3 wpi and with kinetics of virus replication slightly faster in the X4-SHIVSF33A2-infected macaques than in the R5-SHIVSF33A2(V3)-infected macaques (Fig. 3A and B). Replication of both viruses declined thereafter, with a postpeak viral load of 102 to 103 RNA copies/ml plasma at 20 wpi.
FIG. 3.
Virologic and immunologic measurements in SHIV-infected RM. (A) Animals CA05 and CA06 inoculated with X4-SHIVSF33A2 and (B) animals BT35 and CF17 inoculated with R5-SHIVSF33A2(V3). Viremia (RNA copies/ml [solid lines]) and absolute CD4+-T-cell counts (dashed lines) of peripheral blood were monitored during the first 20 weeks of infection.
The dynamics of CD4+-T-cell loss differ in R5-SHIVSF33A2(V3)- and X4-SHIVSF33A2-infected macaques.
We next examined the impact of X4 and R5 clone infection on the CD4+-T-cell compartment. A drop in peripheral CD4+ T cells, a feature that is characteristic of infection with the pathogenic isolate R5-SHIVSF162P3 and with SIV, was observed in both R5-SHIVSF33A2(V3)-infected animals during peak viremia (2 to 3 wpi). The levels of circulating CD4+ T lymphocytes fluctuated thereafter but rebounded close to preinfection values by 8 wpi (Fig. 3B). In contrast, X4-SHIVSF33A2 infection led to a rapid and more sustained loss of peripheral CD4+ T cells (Fig. 3A).
To further examine the impact of CoR usage on the dynamics of the CD4+-T-cell compartment, the percentages of CD4+ T cells in various lymphoid and nonlymphoid tissues during peak (2 to 3 wpi) and postpeak (8 to 9 wpi) infection were analyzed (Fig. 4). No significant loss in CD4+ T cells was noted in the draining and peripheral lymph nodes (LNs) of the two R5-SHIVSF33A2(V3)-infected animals during the peak or postpeak infection period (Fig. 4B). Despite a viral load that is comparable to that of the R5-SHIVSF33A2(V3)-infected animals, however, a 30 to 50% loss of CD4+ T cells was found in all lymphoid tissues of CA05 examined at 9 wpi (Fig. 4A). In CA06, the X4-SHIVSF33A2-infected animal with a log higher peak viremia, massive depletion of CD4+ T cells was seen in both the draining and systemic LNs within 2 weeks of infection, with some degree of restoration noted in these secondary LNs by 9 wpi.
FIG. 4.
Percentage of CD4+ T lymphocytes in various lymphoid and nonlymphoid compartments of (A) X4-SHIVSF33A2- and (B) R5-SHIVSF33A2(V3)-infected RM. The percentages of CD4+ T cells in the inguinal (Ing), iliac (Ili), mesenteric (Mes), and colonic (Col) lymph nodes, BM, and LPL from the jejunum were analyzed before (white bars), during peak (2 to 3 wpi [gray bars]), and postpeak (8 to 9 wpi [black bar]) infection. Note that the mesenteric LN of CA06 was not available for analyses. Dashed bars show percentages of CD4+ T cells in the same compartments of uninfected control macaques for reference when preinfection values for the infected macaques were not available. Error bars show standard errors of values from two to three uninfected animals.
Contrary to what was observed in the secondary lymphoid compartments, a 60% and 55% drop in CD4+ T cells in the bone marrow (BM) and mucosal tissues such as the lamina propria of the gut (LPL), respectively, in CF17 was present at 8 wpi (Fig. 4B). A similar degree of postpeak CD4+-T-cell loss was also noted in the BM and LPL in the other R5-SHIVSF33A2(V3)-infected monkey, BT35. For X4-SHIVSF33A2 infection, a decrease in BM CD4+ T cells (25%) was seen in CA05 at 2 wpi, but dramatic depletion was already present in CA06. While the gut remained relatively intact in both X4 clone-infected macaques at 2 wpi, substantial CD4+-T-cell depletion (60%) was detected in CA06 by 9 wpi (Fig. 4A), while CA05 displayed a more modest loss (25%). The targeting of BM by the X4 and R5 isogenic SHIV molecular clones is in agreement with reports that CD34+ progenitor cells could be infected by HIV-1 variants with different phenotypes (34, 41). Furthermore, although the dynamics of CD4+-T-cell loss among the two X4-SHIVSF33A2-infected macaques varies due to a difference in viral load, the data show that by switching the coreceptor usage of this clone virus through replacement of its V3 loop, the site of early virus replication changes. While the X4 clone preferentially targets and depletes cells in peripheral blood and secondary LNs within the first few weeks of infection, the R5 clone targets mucosal tissue sites. The preferential depletion of CD4+ T cells in the gut and not the LNs of the R5 clone-infected animals during primary infection is consistent with observations made in HIV-1-infected humans (5, 11, 27), SIV-infected macaques (21, 25, 26, 37, 39), and animals infected with the pathogenic R5-SHIVSF162P3 isolate (15).
T-cell-subset distribution and CoR expression varied in lymphoid and mucosal tissues of RM.
CXCR4 and CCR5 are differentially expressed on naïve and memory CD4+ T cells, the two major T-cell subpopulations that can be distinguished in macaque cells by surface staining for the CD28 and CD95 antigens (32). To determine whether the differential compartmentalization of the X4 and R5 clones during primary infection is influenced by the availability and susceptibility of their target cells, the frequency of the two T-cell subsets in various lymphoid and mucosal tissues and CoR expression on their surface were determined (Fig. 5). Consistent with previous reports (1, 3, 20, 35, 38, 40), naïve (CD95low CD28high) CD4+ T lymphocytes were found in abundance in the periphery and LNs but were rare in the gut. More than 95% of cells in this latter compartment were memory cells (CD95high CD28high or CD95high CD28low) (Fig. 5A). Virtually all the naïve cells were positive for CXCR4 and negative for CCR5 (Fig. 5B), while cells of the memory phenotype expressed both receptors, with higher CCR5 expression on memory cells in the gut than in the periphery and secondary lymphoid compartments (Fig. 5C). Within the macaque host, therefore, there is greater representation of CXCR4-positive cells in peripheral blood and LNs, while the majority of CCR5-expressing cells reside in mucosal effector sites. This, then, explains the early preferential targeting of the X4 clone to the periphery and LN compartments and the R5 clone to the gut-associated lymphoid tissue since the greatest number of their respective target cells reside in these compartments.
FIG. 5.
T-cell subset distribution (A) and CoR expression in naïve (B) and memory (C) T cells in various lymphoid and nonlymphoid tissues of uninfected control RM. Lymphocytes from peripheral blood (PBMC), lymphoid (inguinal [Ing] and mesenteric [Mes]) and mucosal (LPL) tissues, and BM were prepared from two to three uninfected macaques, and a CD4+ small lymphocyte gate was used for phenotyping of CD28 and CD95 surface expression. CXCR4 and CCR5 expression levels on naïve and memory T cells, the two major CD4+-T-lymphocyte subsets defined by CD28/CD95 costaining, were further determined. Error bars show standard errors of data.
Differential target cell selectivity in R5-SHIVSF33A2(V3)- and X4-SHIVSF33A2-infected macaques.
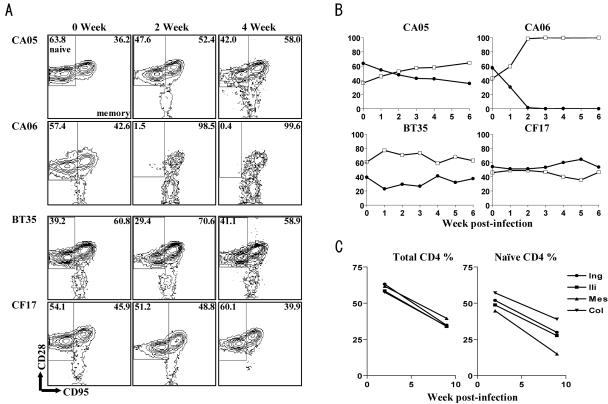
To further establish CoR specificity of the two clones in vivo and to assess the impact of CoR preference on target cell selectivity, changes in the phenotype (Fig. 6) as well as chemokine receptor expression (Fig. 7) in circulating and lymphoid CD4+ T cells were determined. As anticipated, the X4 clone selectively targeted and depleted naïve CD4+ T cells in peripheral blood. The percentage of this T-cell subpopulation dropped from a baseline value of 63.8% to 42% within 2 to 4 weeks of infection in CA05 (Fig. 6A) and further declined by 6 wpi (Fig. 6B). A more drastic loss of naïve T cells, from 57.4% to 1.5% within 2 weeks of infection, was seen in CA06, the monkey with the higher peak viremia (Fig. 6A). By 6 wpi, >99.9% of circulating CD4+ T cells in CA06 were cells of the memory phenotype (Fig. 6B). Similar preferential early targeting of naïve T cells was also seen in the LNs, where loss of this T-cell subset accounted for the total drop in percentage of CD4+ T cells (Fig. 6C). For animals infected with the R5-SHIVSF33A2(V3) clone, a rather modest (∼15%) but specific loss of memory cells was observed in CF17 at 2 to 4 wpi (Fig. 6A), and this further decreased at 5 wpi before rebounding close to preinfection values by 6 wpi (Fig. 6B). In BT35, an increase in the memory T-cell subset, similar to that reported for SIV infection (25), was seen at 1 wpi, and this was followed by a gradual decline.
FIG. 6.
Different subsets of CD4+ T cells are targeted by X4-SHIVSF33A and R5-SHIVSF33A2(V3). PBMCs and lymph node cells collected at the indicated times postinfection from SHIVSF33A2- and SHIVSF33A2(V3)-infected RM were phenotyped for CD3, CD4, CD28, and CD95 expression. (A) Contour plots depict differential depletion of naïve (CD95low CD28high) and memory (CD95high CD28low/high) CD4+ T cells in early X4 (CA05 and CA06) and R5 (BT35 and CF17) SHIV clone infections, respectively. (B) Changes in the levels of naïve (•) and memory (□) CD4+ T cells in blood during the first 6 weeks of virus infection with X4 and R5 SHIV molecular clones. (C) Changes in total percentages of CD4+ T cells as well as percentages of naïve cells in various lymphoid tissues of macaque CA05.
FIG. 7.
CXCR4- and CCR5-positive cells are selectively depleted by X4-SHIVSF33A2 and R5-SHIVSF33A2(V3), respectively. Data were generated by gating through lymphocytes and then gating through CD4+ cells. After initial gating on naïve (CD95low CD28high) and memory (CD95high CD28low/high) CD4+ lymphocytes, individual samples were analyzed for CXCR4 and CCR5 expression. Percentages of gated naïve and memory CD4+ T cells in (A) X4 and (B) R5 clone-infected RM that express CXCR4 or CCR5 are shown.
When changes in chemokine receptor expression on circulating CD4+ T lymphocytes were examined, results showed that naïve cells expressing high levels of CXCR4 were selectively eliminated in early X4-SHIVSF33A2 infection, followed or accompanied by depletion of CXCR4+ memory cells (Fig. 7A). The difference in the tempo of depletion of the two CXCR4+-T-cell subpopulations was more evident in CA06, where a complete loss of naïve CXCR4+ cells compared to a 40% drop in CXCR4+ memory cells was present at 3 wpi. In contrast, a transient drop in CCR5+ memory T lymphocytes was seen in both R5-SHIVSF33A2(V3)-infected macaques at 3 wpi, and this was accompanied by a corresponding increase in the percentage of CXCR4+ memory T cells (Fig. 7B). By 5 wpi, however, a rebound in the percentage of CCR5+ memory cells was noted. The selective destruction of CXCR4+ lymphocytes by the X4 clone and CCR5+ lymphocytes by the R5 clone further demonstrates their CoR specificity in vivo and shows that in addition to the extent of virus replication, the dynamics of CD4+-T-cell loss are influenced by the CoR usage of the inoculating virus, the level of CoR expression on target cells, and the representation of these cells in the various tissue compartments.
DISCUSSION
The use of SHIV isogenic clones that differ only in their CoR binding sequences for infection of nonhuman primates provides an ideally suited model to study the impact of HIV-1 coreceptor usage on the composition and dynamics of the CD4+-T-cell compartment. We report here the generation and characterization of such viruses. We show that the substitution of the V3 loop of the X4-SHIVSF33A2 molecular clone with the corresponding sequences of the R5-SHIVSF162P3 isolate alone was sufficient to alter its coreceptor preference in vitro and its replication characteristics in vivo. Differences in the sites of early virus replication as well as the rates of CD4+-T cell-loss in the periphery, LNs, and mucosal tissue compartments were seen in RM infected with the X4 compared to the R5 isogenic SHIV molecular clones. Infection with X4-SHIVSF33A2 resulted in a rapid drop, massively in the case of CA06, in CD4+-T-cell numbers in peripheral blood and secondary LNs during the period of peak virus production (Fig. 3 and 4). This loss of CD4+ T cells in the peripheral tissues preceded depletion of CD4+ T cells in the intestinal mucosa of CA06, the X4-SHIVSF33A2-infected macaque with the highest viral burden. In contrast, loss of CD4+ T cells in the blood of both R5-SHIVSF33A2(V3)-infected macaques was modest and transient, with little or no depletion seen in the secondary lymph nodes during primary infection. However, lower percentages of CD4+ T cells were seen in the gut-associated lymphoid tissue, consistent with preferential depletion in mucosal effector sites by the R5 clone. These observations of differential sites of early virus replication in animals infected with the X4 and R5 isogenic clones are consistent with previous findings in animals infected with the pathogenic X4-SHIVSF33A and R5-SHIVSF162P3 isolates (15) and demonstrate that coreceptor utilization, and not other undetermined strain-related differences, is the major determinant for this difference.
We showed not only differential early targeting of various tissue compartments in X4 and R5 clone-infected macaques but also preferential depletion of particular CD4+-T-cell subsets within these compartments. Naïve T cells were dramatically depleted in X4-SHIVSF33A2-infected macaques, whereas the memory T cells were targeted by R5-SHIVSF33A2(V3) (Fig. 6 and 7). These findings of differences in tissue and T-cell targeting by X4 and R5 viruses can, to a large extent, be explained by differential expression of their cognate receptors on the two major CD4+-T-cell subsets and the relative tissue distribution of these cell populations in vivo. CCR5 expression is largely restricted to memory T cells that are infrequent in peripheral blood and LNs but prevalent in tissue effector sites such as intestinal lamina propria (Fig. 5). Accordingly, CD4+-T-cell loss occurs predominantly in the gut of R5-SHIVSF33A2(V3)-infected macaques, with only modest depletion seen in peripheral blood (Fig. 3) and no depletion seen in the secondary lymphoid tissue compartments (Fig. 4). In contrast, CXCR4 is expressed on memory as well as naïve CD4+ T cells in peripheral blood and all lymphoid tissues examined (Fig. 5). In this regard, the observation that the loss of CD4+ T cells in the peripheral tissues was followed by elimination of CD4+ T cells in the intestinal mucosa of CA06 indicates that there is no absolute restriction in the sites of CD4+-T-cell depletion induced by SHIV infection but that the rate and extent of T-cell loss in these various tissue compartments are influenced by the total number of susceptible target cells that are available within these sites. Conceivably, the faster kinetics of replication seen for the X4 isogenic clone are due to the larger pool of susceptible target cells available for this virus (Fig. 3A and 5). Furthermore, the greater frequency and abundance of CXCR4 expression on naïve cells compared to memory cells can explain the early targeting of this T-cell subset by X4-SHIVSF33A2. The effects of infection by CXCR4- and CCR5-using viruses on naïve and memory CD4 subsets have recently been described using X4-SHIV and SIV (29-31). Our use of an isogenic system, showing early depletion of CXCR4+ naïve cells by the X4-SHIVSF33A2 molecular clone and CCR5+ memory cells by the R5-SHIVSF33A2(V3) clone (Fig. 7), confirms and extends findings in those other reports, demonstrating that coreceptor preference is the principal basis of this selectivity.
In a prior study, one of two macaques inoculated intravenously with a cell-free X4-SHIVSF33A2 molecular clone suffered precipitous peripheral CD4+-T-cell loss and progression to simian AIDS within 30 wpi, while the other displayed a less severe but sustained depletion of CD4+ T cells and controlled its infection (14). The variability in the level of viremia and kinetics and extent of cell destruction seen in these and the present two macaques infected with the X4-SHIVSF33A2 molecular clone contrasts with the patterns observed in animals infected with the uncloned X4-SHIVSF33A strain, suggesting that the clonal virus is less pathogenic than the viral isolate. But the possibility of individual host variability cannot be excluded. Additional studies in a larger group of animals are required to address this. Despite these variations, however, the data show that the pattern of CD4+-T-cell loss induced by X4-SHIVSF33A2 clone infection is dramatically altered through modifications of its V3 loop. Furthermore, the availability of these X4 and R5 isogenic SHIV molecular clones for infection of macaques is expected to enhance our understanding of the role of tropism in transmission, persistence, and pathogenesis of HIV-1 infection. For example, coinfection of RM with these viruses should allow us to determine whether cell tropism alone dictates the R5 dominance we observed in macaques dually infected with the pathogenic isolates X4-SHIVSF33A and R5-SHIVSF162P3 (12). Additionally, since the sites of replication and immune cells targeted by the clones differ, questions of interest will be whether the type and/or extent of selective immune pressures exerted by the shared host on the two viruses will differ as well. An examination of sequence changes in the envelope and/or other genomic regions of viruses recovered from isogenic X4- and R5-SHIV-infected macaques over time, coupled with antiviral immune measurements, may provide critical insights into these questions. Last, although coreceptor switch has rarely been reported in SIV-infected macaques (24, 31), infection with the R5-SHIVSF33A2(V3) clone, a virus generated on the backbone of an X4 clone with only 10 amino acid changes in the V3 loop, may provide a unique opportunity to investigate whether reversion to an X4 phenotype can occur in infection of RM.
In summary, although the number of animals used is limited, this proof-of-concept study using an isogenic system clearly shows that the course of infection with the X4-SHIVSF33A2 molecular clone is markedly altered by changing its coreceptor choice through replacement of its V3 loop sequences with those of an R5 strain. Coreceptor targeting of specific CD4+-T-cell subpopulations coupled with the susceptibility and distribution of the target cells contribute to the differing rates and sites of CD4+-T-cell depletion seen in X4-SHIVSF33A2- and R5-SHIVSF33A2(V3)-infected macaques. Further studies with these two clones in larger, more systematic studies are expected to provide important information on the role of tropism in HIV-1 infection and disease induction.
Acknowledgments
We thank Lisa Chakrabarti and Viviana Simon for critical comments and Peter Lopez for help with fluorescence-activated cell sorter analyses.
This research was supported by grants from the U.S. National Institutes of Health (AI46980, AI41945, and CA72822).
REFERENCES
- 1.Anton, P. A., J. Elliott, M. A. Poles, I. M. McGowan, J. Matud, L. E. Hultin, K. Grovit-Ferbas, C. R. Mackay, I. S. Y. Chen, and J. V. Giorgi. 2000. Enhanced levels of functional HIV-1 co-receptors on human mucosal T cells demonstrated using intestinal biopsy tissue. AIDS 14:1761-1765. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Bleul, C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogers, W. M., C. Cheng-Mayer, and R. C. Montelaro. 2000. Developments in preclinical AIDS vaccine efficacy models. AIDS 14(Suppl. 3):S141-S151. [PubMed] [Google Scholar]
- 5.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti, L. A., T. Ivanovic, and C. Cheng-Mayer. 2002. Properties of the surface envelope glycoprotein associated with virulence of simian-human immunodeficiency virus SHIVSF33A molecular clones. J. Virol. 76:1588-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 9.Dolei, A., A. Biolchini, C. Serra, S. Curreli, E. Gomes, and F. Dianzani. 1998. Increased replication of T-cell-tropic HIV strains and CXC-chemokine receptor-4 induction in T cells treated with macrophage inflammatory protein (MIP)-1alpha, MIP-1beta and RANTES beta-chemokines. AIDS 12:183-190. [DOI] [PubMed] [Google Scholar]
- 10.Fouchier, R. M., M. Groenink, A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guadalupe, M., E. Reay, S. Sankaran, T. Prindiville, J. Flamm, A. McNeil, and S. Dandekar. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77:11708-11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harouse, J. M., C. Buckner, A. Gettie, R. Fuller, R. Bohm, J. Blanchard, and C. Cheng-Mayer. 2003. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc. Natl. Acad. Sci. USA 100:10977-10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harouse, J. M., A. Gettie, T. Eshetu, R. C. Tan, R. Bohm, J. Blanchard, G. Baskin, and C. Cheng-Mayer. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIVSF162P3. J. Virol. 75:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harouse, J. M., A. Gettie, R. C. Tan, T. Eshetu, M. Ratterree, J. Blanchard, and C. Cheng-Mayer. 2001. Pathogenic determinants of the mucosally transmissible CXCR4-specific SHIVSF33A2 map to env region. J. Acquir. Immune Defic. Syndr. 27:222-228. [DOI] [PubMed] [Google Scholar]
- 15.Harouse, J. M., A. Gettie, R. C. H. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 16.Harouse, J. M., R. C. Tan, A. Gettie, P. Dailey, P. A. Marx, P. A. Luciw, and C. Cheng-Mayer. 1998. Mucosal transmission of pathogenic CXCR4-utilizing SHIVSF33A variants in rhesus macaques. Virology 248:95-107. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, M., C. Buckner, J. Harouse, A. Gettie, J. Blanchard, J. E. Robinson, and C. Cheng-Mayer. 2003. Antigenic variations in the CD4 induced sites of the CCR5-tropic, pathogenic SHIVSF162P3 gp120 variants. J. Med. Primatol. 32:211-217. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, M., J. M. Harouse, A. Gettie, C. Buckner, J. Blanchard, and C. Cheng-Mayer. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIVSF162P3 maps to envelope gp120. J. Virol. 77:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinter, A., A. Catanzaro, J. Monaco, M. Ruiz, J. Justement, S. Moir, J. Arthos, A. Oliva, L. Ehler, S. Mizell, R. Jackson, M. Ostrowski, J. Hoxie, R. Offord, and A. S. Fauci. 1998. CC-chemokines enhance the replication of T-tropic strains of HIV-1 in CD4(+) T cells: role of signal transduction. Proc. Natl. Acad. Sci. USA 95:11880-11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel, E. J., J. Boisvert, K. Murphy, M. A. Vierra, M. C. Genovese, A. J. Wardlaw, H. B. Greenberg, M. R. Hodge, L. Wu, E. C. Butcher, and J. J. Campbell. 2002. Expression of the chemokine receptors CCR4, CCR5, and CXCR3 by human tissue-infiltrating lymphocytes. Am. J. Pathol. 160:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148-1152. [DOI] [PubMed] [Google Scholar]
- 22.Luciw, P., C. Mandell, S. Himathongkham, J. Li, T. Low, K. Schmidt, K. Shaw, and C. Cheng-Mayer. 1999. Fatal immunopathogenesis by SIV/HIV-1 (SHIV) containing a variant form of the HIV-1SF33 env gene in juvenile and newborn rhesus macaques. Virology 263:112-127. [DOI] [PubMed] [Google Scholar]
- 23.Luciw, P. A., E. Pratt-Lowe, K. E. S. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. USA 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marx, P. A., and Z. Chen. 1998. The function of simian chemokine receptors in the replication of SIV. Semin. Immunol. 10:215-223. [DOI] [PubMed] [Google Scholar]
- 25.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093-1097. [DOI] [PubMed] [Google Scholar]
- 26.Mattapallil, J. J., Z. Smit-McBride, M. McChesney, and S. Dandekar. 1998. Intestinal intraepithelial lymphocytes are primed for gamma interferon and MIP-1β expression and display antiviral cytotoxic activity despite severe CD4+ T-cell depletion in primary SIV infection. J. Virol. 72:6421-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. P., S. G. Kitchen, P. Pugach, and J. A. Zack. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 20:111-126. [DOI] [PubMed] [Google Scholar]
- 28a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 29.Nishimura, Y., C. R. Brown, J. J. Mattapallil, T. Igarashi, A. Buckler-White, B. A. Lafont, V. M. Hirsch, M. Roederer, and M. A. Martin. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. USA 102:8000-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura, Y., T. Igarashi, O. K. Donau, A. Buckler-White, C. Buckler, B. A. Lafont, R. M. Goeken, S. Goldstein, V. M. Hirsch, and M. A. Martin. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. USA 101:12324-12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picker, L. J., S. I. Hagen, R. Lum, E. F. Reed-Inderbitzin, L. M. Daly, A. W. Sylwester, J. M. Walker, D. C. Siess, M. Piatak, Jr., C. Wang, D. B. Allison, V. C. Maino, J. D. Lifson, T. Kodama, and M. K. Axthelm. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29-43. [DOI] [PubMed] [Google Scholar]
- 33.Rowland-Jones, S. L. 2003. Timeline: AIDS pathogenesis: what have two decades of HIV research taught us? Nat. Rev. Immunol. 3:343-348. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz, M. E., C. Cicala, J. Arthos, A. Kinter, A. T. Catanzaro, J. Adelsberger, K. L. Holmes, O. J. Cohen, and A. S. Fauci. 1998. Peripheral blood-derived CD34+ progenitor cells: CXC chemokine receptor 4 and CC chemokine receptor 5 expression and infection by HIV. J. Immunol. 161:4169-4176. [PubMed] [Google Scholar]
- 35.Sallusto, F., D. Lenig, C. R. Mackay, and A. Lanzavecchia. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simmons, G., P. R. Clapham, L. Picard, R. E. Offord, M. M. Rosenkilde, T. W. Schwartz, R. Buser, T. N. C. Wells, and A. E. Proudfoot. 1997. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science 276:276-279. [DOI] [PubMed] [Google Scholar]
- 37.Smit-McBride, Z., J. J. Mattapallil, M. McChesney, D. Ferrick, and S. Dandekar. 1998. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4+ T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72:6646-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vajdy, M., R. Veazey, I. Tham, C. deBakker, S. Westmoreland, M. Neutra, and A. Lackner. 2001. Early immunologic events in mucosal and systemic lymphoid tissues after intrarectal inoculation with simian immunodeficiency virus. J. Infect. Dis. 184:1007-1014. [DOI] [PubMed] [Google Scholar]
- 39.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 40.Veazey, R. S., K. G. Mansfield, I. C. Tham, A. C. Carville, D. E. Shvetz, A. E. Forand, and A. A. Lackner. 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 74:11001-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiser, B., H. Burger, P. Campbell, S. Donelan, and J. Mladenovic. 1996. HIV type 1 RNA expression in bone marrows of patients with a spectrum of disease. AIDS Res. Hum. Retrovir. 12:1551-1558. [DOI] [PubMed] [Google Scholar]