Abstract
Simian immunodeficiency virus of sooty mangabeys (SIVsmm) is recognized as the progenitor of human immunodeficiency virus type 2 (HIV-2) and has been transmitted to humans on multiple occasions, yet the epidemiology and genetic diversity of SIVsmm infection in wild-living populations remain largely unknown. Here, we report the first molecular epidemiological survey of SIVsmm in a community of ∼120 free-ranging sooty mangabeys in the Taï Forest, Côte d'Ivoire. Fecal samples (n = 39) were collected from 35 habituated animals (27 females and 8 males) and tested for SIVsmm virion RNA (vRNA). Viral gag (800 bp) and/or env (490 bp) sequences were amplified from 11 different individuals (eight females and three males). Based on the sensitivity of fecal vRNA detection and the numbers of samples analyzed, the prevalence of SIVsmm infection was estimated to be 59% (95% confidence interval, 0.35 to 0.88). Behavioral data collected from this community indicated that SIVsmm infection occurred preferentially in high-ranking females. Phylogenetic analysis of gag and env sequences revealed an extraordinary degree of genetic diversity, including evidence for frequent recombination events in both the recent and distant past. Some sooty mangabeys harbored near-identical viruses (<2% interstrain distance), indicating epidemiologically linked infections. These transmissions were identified by microsatellite analyses to involve both related (mother/daughter) and unrelated individuals, thus providing evidence for vertical and horizontal transmission in the wild. Finally, evolutionary tree analyses revealed significant clustering of the Taï SIVsmm strains with five of the eight recognized groups of HIV-2, including the epidemic groups A and B, thus pointing to a likely geographic origin of these human infections in the eastern part of the sooty mangabey range.
Simian immunodeficiency viruses (SIVs) have been identified in no fewer than 36 different nonhuman primate species in subsaharan Africa (8); however, only two SIV strains, SIVcpz from central chimpanzees (Pan troglodytes troglodytes) and SIVsmm from sooty mangabeys (Cercocebus atys atys), are known to have crossed the species barrier, generating human immunodeficiency virus type 1 (HIV-1) and HIV-2, respectively. Although SIVcpz and SIVsmm have each been transmitted to humans on multiple occasions (30), their subsequent spread within human populations has been quite variable. In the case of HIV-1, infections with groups N and O have been largely confined to Gabon and Cameroon (4, 5, 42), while group M has spread pandemically (51, 52). In the case of HIV-2, groups A and B have infected substantial numbers of people in west Africa (17, 23, 34, 54, 79), while groups C to H have each been identified only in single individuals (14, 18, 22, 76). Circumstances that favored cross-species transmission and subsequent amplification of only a subset of these viruses are unknown but have implications for potential future SIV transmissions.
While much has been learned about SIVcpz infection of wild chimpanzees (63, 64, 75), less is known about the prevalence, distribution, and natural history of SIVsmm in wild sooty mangabeys. Studies of captive animals housed primarily in U.S. primate centers have suggested that SIVsmm infection is common and frequently exceeds 50% prevalence in sexually mature adults (3, 21). Animals were found to harbor substantially divergent SIVsmm strains which most likely were introduced by naturally infected founder animals imported from west Africa (3, 45). Interestingly, nearly all SIVsmm-infected sooty mangabeys in captivity maintain normal CD4+-T-cell counts and fail to develop immunodeficiency despite life-long infection and high-level virus replication (11, 60, 68). This observation of nonpathogenic infection is starkly different from that of rhesus macaques experimentally infected with the same virus that experience rapid CD4+ cell loss and clinical progression to AIDS and death (12, 68, 69). SIVsmm infection has also been documented in wild-caught sooty mangabeys in west Africa, primarily in animals hunted for bushmeat (2) or kept as household pets (13, 47). One study conducted in 1996 in Sierra Leone employed a trap-and-release strategy, and although SIVsmm infections were identified, this strategy failed to sample the majority of targeted animals (13). Here, we utilize an alternative, noninvasive approach to determine the prevalence of SIVsmm infection in wild-living sooty mangabeys and characterize the phylogenetic relationships of field strains of this virus. This approach involves the direct amplification of SIVsmm viral RNA from fecal samples by reverse transcription-PCR (RT-PCR) analysis (43).
Sooty mangabeys live in the primary and secondary forests of coastal west Africa in a range that once extended from the Casamance River in Senegal to the Sassandra/Nzo River system in Côte d'Ivoire (26, 38) (Fig. 1). Because of hunting and habitat destruction, sooty mangabeys in Senegal, Guinea Bissau, and parts of Guinea are now largely extinct; however, they are still numerous in Sierra Leone, Liberia, and western Côte d'Ivoire (2, 13, 56, 59). Within this natural range, by far, the most SIVsmm strains (82%) have been identified in Sierra Leone (Fig. 1 and Table 1, and references therein). One additional strain was isolated from a household pet in Liberia (SIVsmmLIB1) (13, 47), and two others (SIVsmmCI2 and SIVsmmCI8) were recovered from sooty mangabeys captured in western Côte d'Ivoire and housed in the Abidjan Zoo, Côte d'Ivoire (50). This paucity of SIVsmm strains of defined geographic origin has hampered attempts to determine the phylogeography of SIVsmm and to identify the lineages that have given rise to epidemic HIV-2. In fact, of the eight known groups of HIV-2, only three have been traced back to a likely point of origin. HIV-2 groups E and F viruses are closely related to SIVsmm strains from Sierra Leone, and the single HIV-2 group D strain is closely related to the Liberian SIVsmmLIB1 strain (13). In each case, phylogenetic and epidemiological data strongly suggested cross-species transmission in a particular location (14, 22, 30), but similar geographic linkages have not been found for the other HIV-2 groups, including the more prevalent groups A and B, thus leaving their geographic origin open to question.
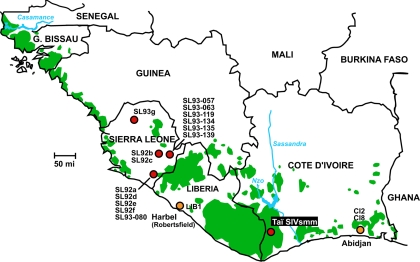
FIG. 1.
Historical range of the sooty mangabey (C. atys atys) from south of the Casamance River in Senegal to the Nzo/Sassandra River systems in Côte d'Ivoire. The geographic origin of previously reported SIVsmm strains from Sierra Leone, Liberia, and western Côte d'Ivoire is shown in relation to the location of the Taï field site. Red dots indicate known capture locations of SIVsmm-infected sooty mangabeys, while yellow dots indicate sites where infected sooty mangabeys were identified in captivity (CI2 and CI8 were captured in western Côte d'Ivoire, close to the Liberian border). Green areas depict remaining coastal forest areas (http://www.globalforestwatch.org). The range of the white-crowned mangabey (C. atys lunulatus) extends from east of the Sassandra River to the Volta River in Ghana (26, 38). Country boundaries and major cities are indicated.
TABLE 1.
SIVsmm strains of known geographic origin
| SIVsmm strain | Status | Yra | Ageb | Sexc | Countryd | Locale | Diagnosisf | Sequenceg | Surveyh | Virus isolation | No. of infected animals/no. tested (%)h | Reference or source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL92b | wild born | 1992 | Juvenile | M | Sierra Leone | Panguma | Plasma Ab | FL | Household pet | Yes | 2/66 (3) | 10, 13 |
| SL92c | wild born | 1992 | Juvenile | M | Sierra Leone | Panguma | Plasma Ab | P (gag, env) | Household pet | No | 2/66 (3) | 13 |
| SL92a | wild living | 1992 | Adult | M | Sierra Leone | Gombu | Plasma Ab | P (gag, env) | Trapped SM | Yes | 4/10 (40) | 13 |
| SL92d | wild living | 1992 | Adult | F | Sierra Leone | Gombu | Plasma Ab | P (gag, env) | Trapped SM | No | 4/10 (40) | 13 |
| SL92e | wild living | 1992 | Adult | F | Sierra Leone | Gombu | Plasma Ab | P (gag, env) | Trapped SM | No | 4/10 (40) | 13 |
| SL92f | wild living | 1992 | Adult | F | Sierra Leone | Gombu | Plasma Ab | P (gag, env) | Trapped SM | No | 4/10 (40) | 13 |
| SL93-080 | wild born | 1992 | Juvenile | M | Sierra Leone | Gombu | Spleen DNA | P (gag, pol, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-057 | wild born | 1992 | Adult | F | Sierra Leone | Bombowo | Spleen DNA | P (gag, pol, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-063 | wild born | 1992 | Adult | M | Sierra Leone | Bombowo | Spleen DNA | P (gag, pol, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-119 | wild born | 1993 | Adult | M | Sierra Leone | Bombowo | Spleen DNA | P (gag, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-134 | wild born | 1993 | Adult | M | Sierra Leone | Bombowo | Spleen DNA | P (gag, pol, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-135 | wild born | 1993 | Juvenile | M | Sierra Leone | Bombowo | Spleen DNA | P (gag, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93-139 | wild born | 1993 | Juvenile | F | Sierra Leone | Bombowo | Spleen DNA | P (pol, env) | Bushmeat market | No | 7/12 (58) | 2 |
| SL93g | wild living | 1993 | NA | NA | Sierra Leone | Mafuri | Fecal vRNA | P (env) | Nonhabituated SMi | No | 1/61 (2)j | 45 |
| LIB1 | wild born | 1989 | Juvenile | F | Liberia | Robertsfield | Plasma Ab | P (gag, pol, env) | Household pet | Yes | 2/25 (8) | 13, 47 |
| CI2 | wild born | 1979 | Juvenile | NA | Côte d'Ivoire | NAe | Plasma Ab | P (gag, env) | Captive SM (Abidjan Zoo) | Yes | 3/6 (50) | 50; this study |
| CI8 | wild born | 1981 | Juvenile | NA | Côte d'Ivoire | NAe | Plasma Ab | P (gag, env) | Captive SM (Abidjan Zoo) | Yes | 3/6 (50) | 50; this study |
| Taï1 | wild living | 2002 | Juvenile | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59)k | This study |
| Taï13 | wild living | 2002 | Adult | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï17 | wild living | 2002 | Juvenile | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï22 | wild living | 2002 | Adult | M | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï23 | wild living | 2002 | Adult | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï29 | wild living | 2002 | Adult | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï31 | wild living | 2002 | Adult | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï32 | wild living | 2002 | Adult | F | Cote d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï33 | wild living | 2002 | Adult | M | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï35 | wild living | 2002 | Adult | M | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag, env) | Habituated SM | No | 11/35 (59) | This study |
| Taï37 | wild living | 2002 | Adult | F | Côte d'Ivoire | Taï Forest | Fecal vRNA | P (gag) | Habituated SM | No | 11/35 (59) | This study |
Year of analysis of captive sooty mangabeys and year of SIVsmm detection in wild-living sooty mangabeys.
Estimated age at the time of analysis. Adult, >6 years; juvenile, 2 to 5 years; NA, not available.
M, male; F, female; NA, not available.
Country of capture or home range of wild sooty mangabeys.
CI2 and CI8 were captured in western Côte d'Ivoire close to the Liberian border (exact location is unknown).
Test performed to diagnose SIVsmm infection. Ab, antibody.
SIVsmm sequence information available. FL, full-length sequence; P, partial sequence.
Sooty mangabey population surveyed. SM, sooty mangabey.
Number of infected sooty mangabeys per total number screened in study in which the infected sooty mangabey was identified.
Fecal samples were not preserved in RNAlater, and their species origin was not genetically confirmed.
Prevalence was calculated for the entire habituated community (n = 120).
One of the last remaining contiguous forest blocks in coastal west Africa extends from eastern Liberia to western Côte d'Ivoire (Fig. 1). To investigate whether SIVsmm is endemic in this area, we employed noninvasive detection methods to conduct a molecular epidemiological survey of ∼120 sooty mangabeys in the Taï National Forest in southwestern Côte d'Ivoire. This community has been studied extensively since 1997 (48, 56-58) and is habituated to the presence of human observers. The Taï field site thus provided a unique opportunity to investigate SIVsmm infection in wild-living sooty mangabeys whose social relationships and behavioral characteristics are well established. Using RT-PCR amplification of fecal virion RNA and microsatellite analyses of putative transmission pairs, we determined the prevalence of SIVsmm infection, molecularly characterized 11 new SIVsmm strains, and investigated the routes of natural SIVsmm transmission in the wild.
MATERIALS AND METHODS
Study site and sample collection.
The present study was carried out in a group of habituated sooty mangabeys (n ∼ 120) in the Taï National Park, Côte d'Ivoire (6°20′N to 5°10′N and 4°20′W to 6°50′W) (56-58). Individual group members were identified based on facial features and gait and cross-referenced with a photographic record of the entire group. Fecal samples (10 to 20 g) were collected into 50-ml conical tubes filled with 20 ml of RNAlater (Ambion, Austin, TX) under direct observation, with the respective animal's name and time and date of collection recorded on the tube. Samples were stored at the base camp for 5 to 60 days at ambient temperature (26 to 28°C) and subsequently shipped to the University of Alabama at Birmingham. Upon receipt, samples were stored at −80°C.
Nucleic acid extraction from sooty mangabey fecal samples.
Total RNA was extracted from RNAlater-preserved fecal specimens using the RNAqueous Midi-Kit (Ambion; Austin, TX) as previously described (63-65). Briefly, ∼1 ml of the 1:1 stool-RNAlater suspension was mixed with 6 ml of lysis buffer, clarified by centrifugation, and passed through a nucleic acid-binding filter. Total nucleic acids were eluted in 1 ml of elution buffer, and RNA was selectively precipitated using LiCl. The pelleted sample was then washed with 70% ethanol, air dried, and resuspended in 50 μl RNA storage solution (Ambion, Austin, TX). Fecal DNA was extracted using the QIAamp Stool DNA Mini kit (QIAGEN, Valencia, CA). Briefly, 400 μl of fecal RNAlater mixture was resuspended in stool lysis buffer, clarified, and passed through a DNA-binding column. Bound DNA was eluted in 150 μl storage solution.
Amplification of SIVsmm sequences from fecal RNA.
RT-PCR amplification of fecal virion RNA was performed as previously described by using two sets of primers specific for SIVsmm gag and env sequences (43). These genomic regions were chosen since the great majority of HIV-2 and SIVsmm strains in the Los Alamos HIV Sequence Database have been sequenced in these same regions, thus allowing for comprehensive phylogenetic analyses (40). cDNA was synthesized using the R1 primer followed by two rounds of PCR using primers F1/R1 and F2/R2, respectively. The gag primers included SM-gag-F1 (5′-TGGGAGATGGGCGCGAGAAACTCCGTC-3′), SM-gag-R1 (5′-ATCAGCAGTGTCTGTGTCATCCAATT-3′), SM-gag-F2 (5′-AGGGAAAAAAGCAGATGAATTAGAA-3′), and SM-gag-R2 (5′-GCTCTTGTAGAAYCTATCTACATA-3′). The env (gp41 region) primers included SM-gp41-F1 (5′-GCTACGGCAGGTTCTGCA ATGGG-3′), SM-gp41-R1 (5′-CTGGTCCTTGCGGATATGGATCTG-3′), SM-gp41-F2 (5′-GCTGTCCGCTCAGTCCCGGACTTT-3′), and SM-gp41-R2 (5′-GGAGGAGAACACTGGCCTATA-3′). RT-PCR products from gag (800 bp) and env (490 bp) regions were purified (QIAGEN) and directly sequenced using the inner (F2/R2) primers.
Nucleotide sequence accession numbers. GenBank accession numbers for the sequences determined in this study are AY835434 to AY835453).
Microsatellite analyses.
Fecal samples that yielded near-identical viral sequences were tested for possible misidentification by microsatellite analyses (9, 15, 19, 49). For allele sizing, the forward primer was labeled with a fluorescent dye (HEX or FAM). Both reverse and forward primers were purchased as custom primers (MapPairs; Invitrogen, Carlsbad, CA). Fecal DNA was subjected to PCR amplification in a total volume of 20 μl containing 10 μl 2× AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA), 7.5 μg bovine serum albumin (Ambion, Austin, TX), and 4 pmol of each primer. PCRs were performed in duplicate with corresponding negative (distilled water) and positive (DNA of the person performing the microsatellite typing) controls included. Samples were subjected to a step-down PCR corresponding to an initial 10-min denaturation step at 95°C followed by 12 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s with decreasing annealing temperatures of 0.5°C per cycle, followed by 45 cycles with an annealing temperature of 54°C for 30 s and a final elongation step for 10 min at 72°C. For each sample, PCR products from all four alleles were pooled, and 1 μl of the pooled sample was mixed with 0.5 μl of a ROX-labeled molecular weight ladder (Applied Biosystems, Foster City, CA), 1 μl loading dye, and 2.5 μl formaldehyde. The mixture was denatured at 95°C for 5 min and placed on ice; 1.5 μl was subsequently loaded onto a 6% GenePage Plus polyacrylamide gel (Amresco, Solon, OH) and subjected to electrophoresis at 3,000 V for 3.2 h at 51°C using an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA). Samples were sized using GeneScan software (Applied Biosystems, Foster City, CA) using a molecular weight curve generated from the size standard using the Local Southern algorithm included in the GeneScan software. All PCRs were performed in duplicate. Sequences from sooty mangabeys whose genotype appeared homozygous were amplified a minimum of seven times (using increasing quantities of genomic DNA) to exclude allelic dropout.
Phylogenetic analysis.
Deduced amino acid sequences of amplified SIVsmm gag and env (gp41) fragments were aligned using CLUSTAL W (72) with representatives of the SIVsmm/HIV-2 lineage from the Los Alamos HIV/SIV Sequence Database (GenBank accession numbers were as follows: for HIV-2A/60415K, L33076; HIV-2A/CAM2, D00835; HIV-2B/UC1, L07625; HIV-2B/EHO, U27200; HIV-2C/2238, L33077 and M87128; HIV-2D/F0784, L33083 and M87074; HIV-2E/PA, L33087; HIV-2F/93SL2, U75441 and U75440; HIV-2G/Abt96, AF208027; HIV-2H/96FR12034, AY530889; SIVsmmCI2, AY932816 and X78508; SIVsmmCI8, AY932818 and X78509; SIVsmmSL92a, U48818 and U17646; SIVsmmSL92b, U48811 and U48819; SIVsmmSL92c, U48812 and U48820; SIVsmmSL92d, U48813 and U48821; SIVsmmSL92e, U48814 and U48822; SIVsmmSL92f, U48815 and U48823; SIVsmmSL93-057, AY864793 and AY864791; SIVsmmSL93-063, AY864794 and AY864792; SIVsmmSL93-080, AY864795 and AY864790; SIVsmmSL93-119, AY864796 and AY864786; SIVsmmSL93-134, AY864797 and AY864787; SIVsmmSL93-135, AY864798 and AY864788; SIVsmmSL93-139, AY864789; SIVmne, M32741; SIVsmmPBj, L03295; SIVstm, M83293; SIVsmmBro85, U48817; SIVsmmLIB1, U48816 and U48824; SIVsmmM951, AY158984 and AY159610; SIVsmmD177, AY158974 and AY159618; SIVsmmM935, AY158970; SIVsmmG992, AY158971 and AY159612; SIVsmmF098, AY158982 and AY159619; SIVsmmFr85, AF041989; SIVsmmFr66, AF041987). The amino acids were then replaced by their corresponding codons, yielding alignments of 618 nucleotides for gag and 372 nucleotides for env (gp41), respectively. Trees were inferred by neighbor-joining (62), maximum-likelihood (20), and Bayesian (78) methods. Neighbor-joining trees were implemented in CLUSTAL W (72) using Kimura's correction (37) and 1,000 bootstrap replicates. Maximum-likelihood trees were implemented in PHYML (27) with 100 bootstrap replicates, and the Bayesian method was implemented in MrBayes (33) with 5 million (gp41) and 10 million (gag) generations. Both likelihood methods used the HKY model (31) with gamma-distributed rates at sites (77). Bayesian likelihoods and parameters were examined with the Tracer program (Tracer v1.2 [http://evolve.zoo.ox.ac.uk/software.html/id=tracer]), and all estimated sample sizes were above 100.
SIVsmm prevalence calculations.
In a previous study of captive sooty mangabeys, we determined the sensitivity of SIVsmm virion RNA detection in fecal samples to be 50% (95% confidence interval, 0.32 to 0.69) (43). We thus used this assay sensitivity to calculate the prevalence of SIVsmm infection in the Taï Forest community. For each sooty mangabey, the probability that it would be detected as being infected, if it was truly infected, was calculated, taking into consideration the number of specimens analyzed (one or two samples) as well as the test sensitivity. SIVsmm prevalence was then estimated as the number of sooty mangabeys that tested positive divided by the number of sooty mangabeys expected to test positive (given the assay sensitivity), assuming that all individuals were positive. For these calculations, it was assumed that detection of viral RNA in multiple fecal samples collected from the same individual was not correlated. Upper and lower confidence limits were calculated based on the number of sooty mangabeys in the community which could be infected and yet would have a greater than 5% chance of showing as few or as many infected sooty mangabeys as were detected, respectively.
Statistical analyses.
The agonistic and affiliative interactions among female sooty mangabeys in the Taï Forest have previously been reported (56-58). Correlations between dominance rank and fecal viral RNA positivity were assessed using a Mann-Whitney U test. Associations were considered significant at a P value of <0.05 (all tests were two tailed).
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences determined in this study are AY835434 to AY835453.
RESULTS
Prevalence of SIVsmm infection in the Taï sooty mangabey community.
To determine the prevalence of SIVsmm infection in the wild, we collected fecal samples from 35 members of a large group (n ∼ 120) of free-ranging sooty mangabeys in the Taï National Park, Côte d'Ivoire. The group's home range, which comprises about 6 to 8 square kilometers, is located near the western border of the park, approximately 20 km southeast of the town of Taï (Fig. 1), and overlaps with that of four or five other nonhabituated sooty mangabey communities. During the collection period from June to August 2002, the study group consisted of 6 to 10 adult (>6 years old) males, 34 adult (>6 years old) females, 54 juveniles (2 to 5 years old), and 22 infants (0 to 1 year old). Thirty-nine fecal samples were collected from 27 females and 8 males representing 30 adults and 5 juveniles (Table 2 and Fig. 2). Thus, approximately 68% of the adult and 9% of the juvenile population were sampled; no samples were obtained from infants.
TABLE 2.
Noninvasive SIVsmm survey of a habituated community of free-ranging sooty mangabeys in Taï Forest, Côte d'Ivoire
| Dominance ranka | Sooty mangabey | Sexb | Age groupc | Age (yr) | Comments | No. of infants | Date of sample collection (mo/day/yr) | No. of vRNA-positive fecal sample/no. tested | SIVsmm strain |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Sana | F | Adult | >10 | Likely mother of Louise and Felicite | Multiparous | 7/10/02 | 1/1 | Taï31 |
| 2 | Felicite | F | Adult | 7-9 | Likely daughter of Sana | 1 | 6/4/02 | 1/1 | Taï29 |
| 3 | Louise | F | Adult | 8-11 | Likely daughter of Sana | 2 | 6/4/02 | 0/1 | |
| 5 | Foulard | F | Adult | >6 | At least 2 | 7/23/02; 7/31/02 | 0/2 | ||
| 6 | Teddy | F | Juvenile | 4-4.5 | 0 | 1/19/02 | 0/1 | ||
| 7 | Diamond | F | Adult | >10 | Multiparous | 6/18/02; 6/4/02 | 2/2 | Taï11; Taï32 | |
| 8 | Regina | F | Juvenile | 4-4.5 | Daughter of Rita | 0 | 8/3/02 | 1/1 | Taï1 |
| 10 | Rita | F | Adult | >10 | Mother of Regina | Multiparous | 6/4/02 | 1/1 | Taï23 |
| 11 | Ema | F | Adult | >10 | Multiparous | 6/8/02 | 0/1 | ||
| 13 | Mandy | F | Adult | >10 | Multiparous | 7/14/02 | 0/1 | ||
| 14 | Kina | F | Juvenile | 5-6 | 0 | 6/7/02; 7/13/02 | 2/2 | Taï3; Taï17 | |
| 17 | Bijou | F | Adult | >10 | Multiparous | 7/3/02 | 0/1 | ||
| 18 | Tina | F | Adult | >10 | Multiparous | 7/3/02 | 0/1 | ||
| 19 | Agathe | F | Adult | >6 | 1-2 | 6/8/02 | 1/1 (gag only) | Taï37 | |
| 20.5 | Yoko | F | Adult | 5-6 | 0 | 6/4/02 | 0/1 | ||
| 22 | Olga | F | Adult | >10 | Multiparous | 6/4/02 | 1/1 | Taï13 | |
| 25 | Sita | F | Adult | >10 | Multiparous | 7/30/02 | 0/1 | ||
| 27 | Farah | F | Adult | >10 | Multiparous | 7/14/02 | 0/1 | ||
| 28 | Cana | F | Adult | >10 | Multiparous | 6/9/02 | 0/1 | ||
| 29 | Belle | F | Adult | 6-7.5 | 1 | 7/22/02 | 0/1 | ||
| 30 | Gisa | F | Adult | >12 | Multiparous | 6/7/02 | 0/1 | ||
| 31 | Ronda | F | Adult | >10 | Multiparous | 6/6/02 | 0/1 | ||
| 32 | Sora | F | Adult | >10 | Multiparous | 6/23/02 | 0/1 | ||
| 33.5 | Cora | F | Adult | 6-8 | 2 | 6/11/02 | 0/1 | ||
| 35 | Jeune | F | Adult | >6 | 1 | 7/29/02 | 0/1 | ||
| 36.5 | Vivianne | F | Adult | >6 | Multiparous | 6/11/02 | 0/1 | ||
| 36.5 | Anne | F | Adult | >6 | Multiparous | 7/29/02 | 0/1 | ||
| Arno | M | Adult | 7-10 | High-ranking, long-term member | 6/22/02 | 0/1 | |||
| Simon | M | Adult | >10 | Low-ranking, long-term member | 7/6/02 | 0/1 | |||
| Marlon | M | Adult | >10 | Mid-ranking, long-term member | 6/10/02; 7/7/02 | 1/2 | Taï33 | ||
| Steve | M | Adult | >10 | Mid-ranking, long-term member | 7/24/02 | 1/1 | Taï22 | ||
| Awa | M | Juvenile | 5-7 | Long-term member | 8/4/02 | 0/1 | |||
| Christophe | M | Adult | 7-8 | Occasional visitor | 7/7/02 | 0/1 | |||
| Sam | M | Adult | >10 | Mid-ranking, long-term member | 6/27/02 | 1/1 | Taï35 | ||
| Fetisch | M | Juvenile | 5-7 | Long-term member | 8/3/02 | 0/1 |
FIG. 2.
Male (left) and subadult female (right) sooty mangabeys from the Taï Forest community.
RNA was extracted from all fecal samples and subjected to RT-PCR analysis using two sets of consensus primers designed to amplify an 800-bp fragment in gag and a 490-bp fragment in the gp41 region of env. This analysis identified 11 sooty mangabeys that were fecal virion RNA (vRNA) positive using one or both primer sets (Table 2). Sequence analysis of the respective amplification products confirmed infection with genetically diverse SIVsmm strains (see below). Fecal vRNA was detected in 8 of 27 (30%) females and in 3 of 8 (38%) males. All vRNA-positive males were sexually mature adults, while two of eight vRNA-positive females were juveniles. Two of the latter were daughters (Regina and Felicite) of vRNA-positive mothers (Sana and Rita); however, a second daughter (Louise) of one of these mothers (Sana) was fecal vRNA negative (Table 2).
We previously determined the sensitivity of SIVsmm virion RNA detection in fecal samples from infected sooty mangabeys to be 50% (95% confidence interval, 0.32 to 0.69) (43). We thus used this estimate to calculate the prevalence of SIVsmm infection in the Taï community. Taking into consideration the assay sensitivity as well as the numbers of samples actually analyzed, we estimated this prevalence to be 59% (95% confidence interval, 0.35 to 0.88).
Genetic diversity of SIVsmm in the Taï sooty mangabey community.
To determine the relationships of the newly identified Taï viruses to each other and to previously characterized SIVsmm strains, we constructed phylogenetic trees from amplified gag and env (gp41) nucleotide sequences using a Bayesian likelihood approach. In the absence of suitably closely related outgroups, the two trees were midpoint rooted to maximize their similarity (Fig. 3). Inspection of these trees yielded several unexpected results. The most striking feature was the extent of topological differences between the gag and env (gp41) trees (Fig. 3), indicating extensive recombination within the SIVsmm radiation. For example, in the gag region, Taï32 grouped with Taï1 and Taï23; however, in the env tree, Taï32 clustered closely with Taï29 and Taï31. Similarly, in the gag tree, Taï13 was most closely related to HIV-2 group G (Abt96); however, in the env tree, Taï13 clustered closely with Taï22 and Taï33. In both instances, mosaic branches fell near the tips of the trees, indicating recombination in the very recent past. However, discordant branching was also evident deeper in the trees. For example, HIV-2 groups A and B clustered together in the gag tree but were separated by intervening SIVsmm sequences in the env tree. Similarly, SIVsmm strains from Sierra Leone were interspersed with SIVsmm from Côte d'Ivoire in the gag tree, but the two groups clustered separately in the env tree. These examples provide evidence for recombination having occurred at all levels in the tree. Since recombination requires that two different viruses infect the same animal, coinfection and/or superinfection must be a relatively frequent occurrence in wild-living sooty mangabeys.
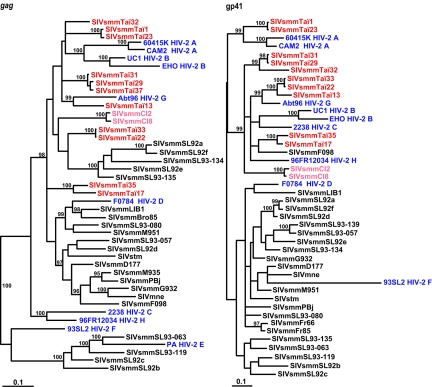
FIG. 3.
Phylogenetic relationships of SIVsmm from the Taï Forest to previously characterized SIVsmm and HIV-2 strains in gag and env regions. SIVsmm from Côte d'Ivoire is highlighted in red (CI2 and CI8 from captive sooty mangabeys are shown in pink); the eight groups of HIV-2 are shown in blue. The trees were inferred by the Bayesian method; numbers on branches are percent posterior probabilities (values of 95% and above are shown). The scale bars indicate 0.1 substitutions per site.
Inspection of gag and env trees also revealed an unusual clustering of the single representative of HIV-2 group F (93SL2) (14). In both genomic regions, 93SL2 was highly divergent from all other SIVsmm and HIV-2 strains. In the gag tree, it formed a distinct lineage that clustered independently from other viruses, whereas in the env tree, it clustered closely but with a long terminal branch with SIVs from a pig-tailed macaque (SIVmne) and a captive sooty mangabey (SIVsmmD177) (Fig. 3). In env trees constructed by the neighbor-joining method, 93SL2 did not fall into this same group but formed a lineage distinct from that of all other SIVsmm/HIV-2 viruses (not shown); however, this is most simply explained by “long-branch attraction,” an artifact of this tree-building method (46). Inspection of the 93SL2 sequence did not reveal any obvious sign of G-to-A (or other) hypermutation, and its removal from the Bayesian analyses did not change the topology for the other strains in any significant way. Thus, it seems clear that 93SL2 is mosaic and that the sooty mangabey virus that gave rise to this human infection was also recombinant. Whether the long branch in the env tree reflects accelerated evolution following cross-species transmission requires further study.
The different clades of HIV-2 were originally termed “subtypes” but have since been renamed “groups,” in analogy with HIV-1 (18), because several groups appear to have resulted from separate cross-species transmission events. While it has long been recognized that the rare groups of HIV-2 must have been generated by independent transfers of SIVsmm (14, 22, 23), it has not been clear whether the two epidemic groups, groups A and B, were the result of one or two transmission events (30). This is because none of the known SIVsmm strains were particularly closely related to either group. This is still the case in the gag tree, where HIV-2 groups A and B cluster together (Fig. 3); however, in the env tree, HIV-2 group A clusters closely (and with high posterior probability) with a lineage of Taï Forest viruses (Taï1 and Taï23), while HIV-2 group B is most closely related to the single HIV-2 group C strain (2238) within a larger clade comprising other Taï viruses as well as HIV-2 groups G and H. This provides the strongest evidence yet that HIV-2 groups A and B arose from separate cross-species transmission events.
While SIVsmm strains from Sierra Leone and Côte d'Ivoire form interspersed lineages in the gag tree, they cluster separately in the env tree (Fig. 3). This difference in topology must be the result of recombination. However, it seems unlikely that the observed dichotomy of strains from the two distinct geographic regions (as depicted in the env tree) was caused by random events. To investigate this further, we constructed a second set of trees intended to clarify the underlying topologies. This was accomplished by excluding probable recombinants as well as other problematic SIVsmm and HIV-2 sequences. Specifically, we excluded HIV-2 group E (no available gp41 sequence), HIV-2 group F (likely mosaic), HIV-2 group D and SIVsmmLIB1 (likely mosaic), most SIV strains from captive sooty mangabeys and macaques in U.S. primate centers (unknown geographic origin), and a group of SIVsmm strains (SL92a, SL92d, SL92e, SL92f, SL93-080, SL93-134, SL93-135, SL93-139, and SL93-057) from Sierra Leone (likely mosaic). The resulting trees (Fig. 4) looked considerably more consistent with respect to their topologies (the green arrows indicate where the excluded Sierra Leonean SIVsmm sequences would join, indicating that they indeed comprise recombinants). Although there were still differences between the gag and env trees (e.g., the relative position of Taï1 and Taï23), these differences were not as pervasive as in the full data set. Instead, the new trees emphasize the phylogeographic division between SIVsmm strains from Sierra Leone and Côte d'Ivoire and in addition clearly show that HIV-2 groups A, B, C, G, and H are more closely related to SIVsmm strains from Côte d'Ivoire than those from Sierra Leone. Together, these data support an origin of HIV-2 groups A, B, C, G, and H in the eastern part of the sooty mangabey range.
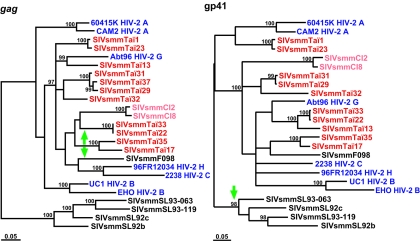
FIG. 4.
Phylogenetic relationships of SIVsmm from the Taï Forest to previously characterized SIVsmm and HIV-2 strains in gag and env regions, with probable recombinant sequences excluded (see text for details). SIVsmm from Côte d'Ivoire is highlighted in red, while the eight groups of HIV-2 are shown in blue. The green arrows indicate where the excluded Sierra Leonean SIVsmm sequences would join (in the gag tree, SIVsmm strains SL92a, SL92e, SL92f, SL93-134, and SL93-135 join on the Taï33/Taï22 branch, whereas the other excluded strains join on the F098 branch; in the env tree, all excluded sequences join the Sierra Leonean SIVsmm cluster). The trees were inferred as described in the legend of Fig. 3.
Epidemiologically linked infections in the Taï sooty mangabey community.
Phylogenetic analyses also identified several subsets of Taï viruses that differed in less than 2% of their gag and env nucleotide sequences, strongly suggesting that they represented epidemiologically linked infections (Fig. 3 and 4). These putative transmission cases (which were confirmed in multiple independent amplification reactions) involved the following animals: Rita (Taï23) and Regina (Taï1); Sana (Taï31), Felicite (Taï29), and Agathe (Taï37; gag sequences only); and Steve (Taï22) and Marlon (Taï33), respectively. Observational studies indicated that Rita and Regina represented a known mother/daughter pair. Sana and Felicite were suspected to represent a mother/daughter pair based on nursing and grooming behavior. No observational data were available for possible familial relationships of Agathe, Steve, or Marlon.
To confirm the authenticity of these putative transmission cases, we extracted fecal DNA and PCR amplified six highly polymorphic microsatellite loci (D18s536, D2s433, D2s1333, D9s905, D2s1326, and D4s243) originally identified in the human genome and more recently used for paternity studies in chimpanzees and baboons (9, 15, 19, 49). The selected loci comprised two alleles (paternally and maternally inherited), which consisted of tetranucleotide repeats of variable length flanked by conserved genomic sequences (amplicon sizes for the six loci ranged from 157 to 165 bp, 186 to 210 bp, 278 to 306 bp, 283 to 291 bp, 242 to 271 bp, and 160 to 172 bp, respectively). Heterozygous loci were confirmed in a second amplification reaction, while homozygous loci were amplified a minimum of seven times to exclude stochastic amplification of only one of two possible alleles (allelic dropout) (49). Although the frequency of these alleles in the larger Taï community has not been determined, the selected loci provided sufficient genetic information to exclude sample misidentification and to examine suspected familial relationships. The results are summarized in Table 3. Each member of a putative transmission pair differed from the other member(s) of that pair (or group) in at least one allele at three of the six microsatellite loci analyzed (Table 3). Thus, the fecal samples containing near-identical SIVsmm sequences were not inadvertently collected from the same individuals but were derived from bona fide transmission cases.
TABLE 3.
Microsatellite analysis of epidemiologically linked SIVsmm infections
| Transmission group/pair | Sooty mangabey | Sexa | SIVsmm strain | Length of amplification product (bp)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D18s536 | D2s433 | D2s1333 | D9s905 | D2s1326 | D4s243 | ||||||||||
| 1 | Sana | F | Taï31 | 161c | 161c | 190 | 210 | 282 | 302 | 283 | 287 | 246 | 271 | 160 | 172 |
| 1 | Felicite | F | Taï29 | 161 | 165 | 190 | 194 | 294 | 302 | 283c | 283c | 267 | 271 | 168 | 172 |
| 1 | Agathe | F | Taï37 | 157 | 161 | 206 | 210 | 278 | 298 | 283 | 291 | 242 | 246 | 168 | 172 |
| 2 | Rita | F | Taï23 | 157 | 165 | 186 | 206 | 278 | 298 | 283 | 291 | 242 | 250 | 164c | 164c |
| 2 | Regina | F | Taï1 | 157 | 165 | 202 | 206 | 278 | 282 | 283 | 291 | 242 | 271 | 164 | 168 |
| 3 | Steve | M | Taï22 | 161 | 165 | 190c | 190c | 278 | 282 | 291c | 291c | 263 | 271 | 168 | 172 |
| 3 | Marlon | M | Taï33 | 157 | 161 | 186c | 186c | 302 | 306 | 283c | 283c | 267 | 271 | 160 | 164 |
M, male; F, female.
Microsatellite loci were amplified from fecal DNA; two alleles per locus are shown.
Homozygous loci were amplified a minimum of seven times to exclude allelic dropout.
Table 3 also shows that Rita and Regina, as well as Sana and Felicite, each share one allele at all six microsatellite loci. To determine the likelihood that they represented mother/daughter pairs, we used the proportion of homozygous loci to estimate the mean microsatellite allele frequency at 16.7% (Table 3). A likelihood ratio test showed that both Rita and Regina, as well as Sana and Felicite, were more than 40 times more likely to represent mother/daughter pairs than unrelated individuals. By contrast, Steve and Marlon shared no alleles at four of the six microsatellite loci (Table 3), and a likelihood ratio test showed that these two males were more than 200 times more likely to represent unrelated individuals than siblings. Finally, microsatellite analyses excluded Agathe and Felicite to represent full siblings despite their closely related viruses (locus D2s1333 excluded Sana from being Agathe's mother). Thus, near-identical SIVsmm sequences were identified in both related and unrelated individuals, suggesting vertical as well as horizontal routes of SIVsmm transmission.
Dominance rank and SIVsmm infection in the Taï sooty mangabey community.
Female sooty mangabeys in the Taï Forest have been the subjects of several behavioral studies. Specifically, dominance relationships, kin-based alliances, and social rank have been investigated in both adult and juvenile females (56-58). Dominance rank was defined according to the direction of submissive behavior (avoid, yield, and supplant [for definitions, see reference 56]). A linear dominance hierarchy was constructed on the basis of 3,690 observed agonistic interactions. If a relationship between individuals was unclear, an average rank was assigned. The probability that the observed linearity resulted from a random process had a P value of <0.0001. As shown in Table 1, fecal samples were collected from females ranging in dominance rank from 1 (alpha female) to 36.5. This provided an opportunity to examine whether there was an association between SIVsmm infection and social rank. Figure 5 depicts the results of this analysis. There was a statistically significant excess of higher-ranking females among the SIVsmm fecal RNA-positive individuals (Mann-Whitney U test; two-tailed, P < 0.02]).
FIG. 5.

Female sooty mangabey dominance rank and SIVsmm infection. SIVsmm vRNA-positive and -negative individuals are shown, with their relative dominance rank indicated by filled circles (also see Table 2).
DISCUSSION
Wild-living sooty mangabeys represent a substantial SIV reservoir.
Although sooty mangabeys are the recognized source of HIV-2, no study to date has examined how frequently SIVsmm infection occurs in wild-living communities, how the virus is transmitted in these settings, and whether the natural history of SIVsmm infection in wild-living animals differs from that of captive populations. Sooty mangabeys are still abundant in Sierra Leone, Liberia, and western Côte d'Ivoire, where they continue to be hunted for bushmeat (26, 38, 59). Since multiple cross-species transfers of SIVsmm have been documented, the magnitude of the existing SIVsmm reservoir is of public health interest, yet the prevalence of SIVsmm infection in wild-living communities remains largely unknown. Trapping of free-ranging animals is inefficient (13), and bushmeat market surveys have provided information for only a small number of animals (2). Thus, alternative screening approaches were needed to determine the extent of the current SIVsmm reservoir.
In this study, we conducted the first molecular epidemiological survey of sooty mangabeys in their natural habitat. The Taï community is unique in several aspects. First, it represents the only community of free-ranging sooty mangabeys that has been habituated to the presence of human observers (56-58). Second, it is located in an area of the sooty mangabey range that has not previously been surveyed for SIVsmm infection (Fig. 1 and Table 1). Third, this wild community exhibits behavioral characteristics that may be of epidemiological importance. Unlike captive animals (28), the Taï sooty mangabeys exhibit a social system in which alliances and affiliations are strongly influenced by kinship, and female juveniles frequently acquire the rank and social status of their mothers (56-58). By testing the majority of adult individuals as well as some juveniles, we identified the prevalence of SIVsmm infection in this community to be 59% (95% confidence interval, 0.35 to 0.88). Although biased toward an older age group (since no infants were sampled), this estimate is very similar to prevalence rates previously reported for captive (58%) and wild-caught (57%) sooty mangabeys (2, 21). Thus, the infection levels of wild-living sooty mangabeys approach those of wild-living African green monkeys (1, 7, 35, 53) and mandrills (70), which are among the most commonly infected primate species known to date. Moreover, the finding of SIVsmm at still another location within the sooty mangabey range supports the notion that this infection is uniformly distributed and widespread (Fig. 1). The low infection rates reported for pet sooty mangabeys (13) are unlikely to be due to regional differences in SIVsmm prevalence (Table 1); rather, vertical transmission is likely to account for only a fraction of naturally occurring infections (most pets are acquired as infants at a very young age). Finally, the finding of highly divergent as well as recombinant virus strains within the home range of a single sooty mangabey group supports the notion that the natural history of SIVsmm is one of efficient virus spread both within and between wild communities.
The widespread infection of wild sooty mangabeys also raises questions concerning the infection status of a closely related subspecies, the white-crowned mangabey (Cercocebus atys lunulatus), whose habitat is immediately adjacent to that of the sooty mangabey and extends from the eastern shores of the Nzo/Sassandra system in Côte d'Ivoire eastward to the Volta River in Ghana (26, 38). White-crowned mangabeys are not known to be naturally infected with SIV; however, only very few individuals have been tested (60). Moreover, white-crowned mangabeys are susceptible to SIV infection in captivity, as demonstrated by the finding of SIVver infections (from east African vervet monkeys) in white-crowned mangabeys housed in a Kenyan primate center (73). Since the two subspecies appear to interbreed where their habitats overlap (25), it will be important to test whether free-ranging white-crowned mangabeys in west Africa harbor SIVsmm (or any other SIV). Such studies would not only address whether white-crowned mangabeys possibly contribute to the current SIVsmm reservoir but would also shed light on the question of when sooty mangabeys and the related red-capped mangabeys (Cercocebus torquatus) first acquired their SIV infections (6).
Routes of SIVsmm transmission in wild-living sooty mangabeys.
Phylogenetic analysis of the Taï viruses identified three sets of closely related infections, which were shown by microsatellite analyses to represent bona fide transmission cases (Table 3). Two of these involved mother/daughter pairs, suggesting vertical transmission. Mother-to-infant transmission is the most likely explanation for the closely related viruses infecting Rita and Regina, since Regina was a juvenile at the time of testing (Table 2). Vertical transmission may also have caused Felicite's infection; however, since Sana and Felicite were both sexually mature adults at the time of sampling and had given birth to one or more offspring (Table 2), transmission from a common sexual partner cannot formally be excluded. Human infants acquire HIV-1 infection either during the course of pregnancy (in utero) or labor/delivery (intrapartum) or through infected breast milk (postpartum) (66). The near-identical viruses in Rita and Regina thus suggest similar transmission scenarios in wild-living sooty mangabeys, although no conclusions can be drawn concerning perinatal or postnatal infection.
Although there is evidence for vertical transmission, sexual acquisition of SIVsmm is likely a more frequent occurrence in wild-living communities. In our study, only 2 of 11 SIVsmm-infected animals were juveniles, consistent with the notion that SIVsmm infection, like SIVagm infection, increases with age and sexual maturity (21, 53). Moreover, the finding of near-identical SIVsmm strains in two seemingly unrelated males, Steve and Marlon, is best explained by infection through a common sexual partner, since adult males do not normally associate with each other. The fact that such a partner was not identified does not argue against this possibility, since not all adult females were sampled and since the sensitivity of fecal virion RNA detection is only 50%. A common sexual partner may also explain the unexpectedly close relationship of Agathe's (Taï37) and Felicite's (Taï29) virus in the gag tree (Fig. 3), since these two females were excluded to represent full siblings (Table 3). It should be noted that sooty mangabeys exhibit female philopatry, meaning that females remain within their natal groups, while males disperse. Male invasions are common during the mating season (C. Benneton, F. Range, and R. Noë, unpublished data), and roving males may represent an efficient means of spreading SIVsmm between communities. This may also explain the extensive genetic diversity observed within single communities (13; Fig. 3).
Finally, horizontal transmission could also occur through biting or fighting, since injuries, including open wounds, are frequently observed, especially during the mating season, when aggressive interactions among adult males increase. Open wounds are cleaned by licking, and this process can involve many different individuals. Several studies have shown that primates are highly susceptible to SIV infection by the oral route (61). For example, atraumatic application (via a cotton swab) of infectious SIVmac to the back of the tongue was sufficient to infect not only juvenile but also adult macaques (71). Given that sooty mangabeys have particularly high systemic viral loads, ranging between 104 to 106 viral RNA copies per ml of plasma (12, 60, 68), it is conceivable that oral exposure to infectious SIVsmm in the context of wound cleaning or the extraction of blood-sucking parasites represents still another route of infection. Such a scenario could explain the finding of near-identical viruses in animals who do not constitute full siblings or direct mating partners (e.g., Agathe and Felicite). Future studies will need to determine to what extent vertical, sexual, and other potential routes of transmission contribute to the maintenance of SIVsmm infection in the wild.
SIVsmm infection seems nonpathogenic in its natural host.
In contrast to HIV-infected humans (or SIVmac-infected macaques), SIVsmm-infected captive sooty mangabeys do not generally exhibit signs of immunodeficiency, CD4-T-cell loss, or opportunistic infections, despite continuous virus replication and high systemic viral loads (11, 12, 32, 36, 60, 68, 69). The mechanisms underlying this nonpathogenic phenotype are not understood; however, recent studies indicate that SIVsmm-infected sooty mangabeys maintain T-lymphocyte populations and manifest far lower levels of immune activation than what is typically observed for pathogenic SIVmac and HIV-1 infections (11, 68). It has thus been proposed that this attenuated immune activation is a key factor in avoiding immunopathology and progression to AIDS. Nonetheless, there are anecdotal reports of AIDS and opportunistic infections in SIVsmm-infected senescent sooty mangabeys (44), indicating that under certain circumstances, SIVsmm infection may be pathogenic even for its natural host. Since environmental factors could contribute to disease manifestation, we examined the Taï records for evidence of adverse health effects associated with SIVsmm infection.
Within sooty mangabey communities, both males and females form linear dominance hierarchies, with adult males outranking all other individuals (28, 29). Females acquire ranks that are very similar to those of their mothers, at least until they reach maturity (56-58). Interestingly, we found a statistically significant association between high female dominance rank and fecal viral RNA positivity (Fig. 5). High social rank in female sooty mangabeys is associated with greater foraging efficiency (high-ranking females have greater access to food) and lower social stress (high-ranking females are less likely to be attacked) (56-58). Moreover, high-ranking females are believed to have a lower infant mortality and thus a larger number of living offspring. Finally, the dominance hierarchy is usually quite stable. In the Taï community, there were no rank changes among adult females between 1998 and 2002, and this was also true for the relative ranks of 14 of 16 juvenile females during 2001 to 2002 (58). Thus, SIVsmm infection does not seem to negatively impact female sooty mangabey reproductive success. Moreover, fecal RNA detection correlates with plasma viral loads (43), suggesting that high-ranking female sooty mangabeys may also have higher systemic viral loads. Although social rank is not a direct indicator for physical fitness and health, the correlation of fecal vRNA positivity and social rank would suggest that even in the wild, SIVsmm infection is generally not pathogenic for its natural host.
The reason for an apparently greater prevalence of SIVsmm infection among high-ranking females is not known. It is possible that high-ranking females copulate more frequently and/or mate with a greater number of male partners. However, preliminary observational data from the Taï community suggest that the number of observed copulations involving high- and low-ranking females is the same when controlled for the number of menstrual cycles. Thus, it is presently unclear whether high-ranking females are, in fact, more frequently exposed to SIVsmm through mating activities. If this were the case, then our data would suggest that female promiscuity, a strategy commonly believed to increase survival by guarding against infanticide by males (74; C. Fruteau, F. Range, and R. Noë, unpublished), is associated with a cost: a higher likelihood of acquiring sexually transmitted infections. Alternatively, the association between social rank and SIVsmm infection could be due to other (nonsexual) factors. As discussed above, the great majority of affiliative relationships among the Taï females, including grooming and other interactions, involve individuals of similar rank. Moreover, high-ranking females are groomed more frequently than low-ranking females (67). If certain types of grooming behaviors (e.g., wound cleaning and extraction of parasites) increase the risk of SIVsmm acquisition, this could explain the observed predominance of SIVsmm infection among high-ranking females.
Tracing the geographic origin of epidemic HIV-2.
The phylogenetic analysis of the newly derived Taï viruses yielded three important findings. First, the unprecedented number of SIVsmm recombinants identified within a single wild-living community suggests that super- or coinfection may be more frequent in sooty mangabeys than in other SIV-infected primates. Second, the interspersion of SIVsmm sequences with HIV-2 groups A and B now provides direct evidence that these human infections were generated by two independent transmission events and that two such transfers gave rise to epidemically spreading HIV-2. Third, the close phylogenetic relationships of the Taï viruses with HIV-2 groups A, B, C, G, and H suggest geographic links between five of the eight HIV-2 groups and SIVsmm strains from Côte d'Ivoire.
The single example of HIV-2 group G (Abt96) was recovered from a blood donor in Côte d'Ivoire (76). In both the gag and env trees, Abt96 clusters with SIVsmm strains from the Taï Forest, and this relationship has also been confirmed for larger sequence fragments in the gag gene (not shown). Thus, the transmission event that gave rise to HIV-2 group G most likely occurred in Côte d'Ivoire, echoing previous examples of rare HIV-2 groups being most closely related to SIVsmm strains from the same locality (14, 30). Two other epidemiological “dead-end” infections, HIV-1 groups C and H, are also more closely related to SIVsmm strains from Côte d'Ivoire than to those from Sierra Leone. The single representative of group H, 96FR12034, was originally identified in an AIDS patient who sought medical treatment in France (18). However, the patient's history indicated that he was a native of Côte d'Ivoire, from a town close to the Liberian border (18). The single representative of HIV-2 group C, 2238, was identified in a Liberian agricultural worker whose home village is unknown (22, 23). Again, the close phylogenetic relationships of HIV-2 groups C and H with Taï SIVsmm strains suggest a recent geographic origin of these human infections within western Côte d'Ivoire or eastern Liberia.
Genetic analysis of the Taï viruses also revealed a close association with the two groups of epidemic HIV-2. This is not surprising for HIV-2 group B, since the great majority of group B infections have been identified in Côte d'Ivoire and neighboring countries to the east (34, 54). By contrast, HIV-2 group A is prevalent throughout west Africa (17, 23, 51). Much of the previous literature has focused on a possible origin of HIV-2 group A towards the west of the sooty mangabey range because of the very high prevalence of HIV-2 group A in rural Guinea Bissau, which is believed to have resulted from the social, cultural, and health care changes associated with the war of independence in the late 1960s (41, 55). However, the clustering of HIV-2 group A with SIVsmm strains from the Taï Forest, particularly evident in env, suggests that group A, as well as group B, originated in the east of the sooty mangabey range. In this context, it should be noted that HIV-2 was present in rural Côte d'Ivoire as early as 1965 (39). Moreover, the area with the most pronounced clinical manifestation of a newly introduced pathogen often does not coincide with the location of the natural animal reservoir. A case in point is the HIV-1 group M epidemic, which was first recognized as a clinical entity in east Africa, yet the chimpanzee subspecies that gave rise to this HIV-1 group is located in west central Africa (16, 24, 30). HIV-2 group A thus seems to be another example of a zoonotic pathogen whose area of greatest clinical impact does not coincide with the location of the original cross-species transfer.
Despite the observed relationships between HIV-2 groups A, B, C, G, and H and SIVsmm from Côte d'Ivoire, the phylogeography of SIVsmm is not entirely straightforward. While Fig. 4 shows a clear phylogeographic division between SIVsmm strains from Sierra Leone and Côte d'Ivoire, there is also a group of recombinant Sierra Leonian SIVsmm strains that clusters with other Sierra Leonean viruses in the env tree but joins the radiation of Côte d'Ivoire SIVsmm at two different positions in the gag tree (green arrows in Fig. 4). This implies that SIVsmm strains from these different clades must have “met” (i.e., coinfected wild sooty mangabeys) at some point in the past, presumably at an intervening location. In this context, the molecular epidemiology of SIVsmm in Liberia, a country which lies between Côte d'Ivoire and Sierra Leone, would be of particular interest (Fig. 1); unfortunately, no such information exists. Despite these gaps in knowledge, the close relationships of HIV-2 groups A and B with SIVsmm from the Taï Forest provide the best evidence yet for a geographic origin of epidemic HIV-2 in the eastern part of the sooty mangabey range.
Human zoonotic risk.
To what extent SIVsmm still poses a risk for human infection is not known but could be considerable, given the magnitude of the existing SIVsmm reservoir. Interestingly, a seroepidemiological survey of 10,000 rural villagers in northern Sierra Leone conducted in 1997 failed to identify evidence of ongoing SIVsmm transmission, despite epidemiological evidence of continuous exposure to SIVsmm (14). By contrast, we report here links between sooty mangabey viruses from Côte d'Ivoire and five of the eight known HIV-2 groups, including the two most prevalent ones. These data thus raise the possibility that virus, host, and/or environmental factors may have been (and may remain) more favorable for cross-species transfers in the eastern part of the sooty mangabey range than elsewhere in west Africa. This hypothesis is testable. Given the extent of SIVsmm genetic information now available, it should be possible to develop molecular-based assays capable of detecting and distinguishing new SIVsmm transmissions. Such surveys should allow determination of whether SIVsmm is still transmitted to humans and, if so, whether there are geographic hotspots for such transmission events.
Acknowledgments
We thank G. Silvestri, S. Staprans, A. Kaur, and P. Johnson for unpublished SIVsmm viral load data in captive sooty mangabeys; A. Vandamme and R. Camacho for helpful discussions; R. Pého for sample collection and field work; the CSRS and the staff of the CRE research station (Taï National Park) for logistical support; R. Mereddy and M. Salazar for expert technical assistance; and W. J. Abbott for artwork and manuscript preparation. We also thank the Ministère de la Recherche Scientifique and the Ministère de l'Agriculture et des Ressources Animales of Côte d'Ivoire for permission to conduct research in the Taï National Park.
This work was supported in part by grants from the National Institutes of Health (RO1 AI 058715, RO1 AI 50529, N01 AI 85338, and P20 AI 27767), the Max Planck Institute Seewiesen (Abteilung Wicker), and the Howard Hughes Medical Institute. F.R. was supported by a grant from the German government (DAAD Doktorandenstipendiums im Rahmen des gemeinsamen Hochschulsonder-programms III von Bund und Ländern).
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apetrei, C., M. J. Metzger, D. Richardson, B. Ling, P. T. Telfer, P. Reed, D. L. Robertson, and P. A. Marx. 2005. Detection and partial characterization of simian immunodeficiency virus SIVsm strains from bush meat samples from rural Sierra Leone. J. Virol. 79:2631-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apetrei, C., A. Kaur, N. W. Lerche, M. J. Metzger, I. Pandrea, J. Hardcastle, S. Fakelstein, R. Bohm, J. Kohler, V. Traina-Dorge, T. Williams, S. Strapans, G. Plauche, R. S. Veazey, H. M. McClure, A. A. Lackner, B. Gormus, D. L. Robertson, and P. A. Marx. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79:8991-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayouba, A., S. Souquieres, B. Njinku, P. M. Martin, M. C. Muller-Trutwin, P. Roques, F. Barre-Sinoussi, P. Mauclere, F. Simon, and E. Nerrienet. 2000. HIV-1 group N among HIV-1-seropositive individuals in Cameroon. AIDS 14:2623-2625. [DOI] [PubMed] [Google Scholar]
- 5.Ayouba, A., P. Mauclere, P. M. Martin, P. Cunin, J. Mfoupouendoun, B. Njinku, S. Souquieres, and F. Simon. 2001. HIV-1 group O infection in Cameroon, 1986-1998. Emerg. Infect. Dis. 7:466-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. [DOI] [PubMed] [Google Scholar]
- 8.Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi-Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J. Virol. 78:7748-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchan, J. C., S. C. Alberts, J. B. Silk, and J. Altmann. 2003. True paternal care in a multi-male primate society. Nature 425:179-181. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti, L. A., A. Luckey, and P. A. Marx. 2001. A divergent simian immunodeficiency virus from sooty mangabey with an atypical Tat-TAR structure. AIDS Res. Hum. Retrovir. 17:1155-1165. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti, L. A. 2004. The paradox of simian immunodeficiency virus infection in sooty mangabeys: active viral replication without disease progression. Frontiers Biosci. 9:521-539. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Z., P. Telfier, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Constable, J. L., M. V. Ashley, J. Goodall, and A. E. Pusey. 2001. Noninvasive paternity assignment in Gombe chimpanzees. Mol. Ecol. 10:1279-1300. [DOI] [PubMed] [Google Scholar]
- 16.Corbet, S., M. C. Müller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damond, F., C. Apetrei, D. L. Robertson, S. Souquiere, A. Lepretre, S. Matheron, J. C. Plantier, F. Brun-Vezinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infecting patients living in France. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 18.Damond, F., M. Worobey, P. Campa, I. Farfara, G. Colin, S. Matheron, F. Brun-Vezinet, D. L. Robertson, and F. Simon. 2004. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res. Hum. Retrovir. 20:666-672. [DOI] [PubMed] [Google Scholar]
- 19.Ely, J. J., D. L. Gonzalez, A. Reeves-Daniel, and W. H. Stone. 1998. Individual identification and paternity determination in chimpanzees (Pan troglodytes) using human short tandem repeat (STR) markers. Int. J. Primatol. 19:255-269. [Google Scholar]
- 20.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 21.Fultz, P. N., T. P. Gordon, D. C. Anderson, and H. M. McClure. 1990. Prevalence of natural infection with simian immunodeficiency virus and simian T-cell leukemia virus type I in a breeding colony of sooty mangabey monkeys. AIDS 4:619-625. [DOI] [PubMed] [Google Scholar]
- 22.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 23.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 25.Groves, C. P. 1978. Phylogenetic and population systematics of the mangabeys (Primates: Cercopithecoidea). Primates 19:1-34. [Google Scholar]
- 26.Groves, C. P. 2001. Primate taxonomy. Smithsonian series in comparative evolution biology. Smithsonian Institution Press, Washington, D.C.
- 27.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 28.Gust, D. A., and T. P. Gordon. 1994. The absence of a matrilineally based dominance system in sooty mangabeys, Cercocebus torquatus atys. Anim. Behav. 47:589-594. [Google Scholar]
- 29.Gust, D. A. 1995. Moving up the dominance hierarchy in young sooty mangabeys. Anim. Behav. 50:15-21. [Google Scholar]
- 30.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch, V. M. 2004. What can natural infection of African monkeys with simian immunodeficiency virus tell us about the pathogenesis of AIDS? AIDS Rev. 6:40-53. [PubMed] [Google Scholar]
- 33.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 34.Ishikawa, K., W. Janssens, J. S. Banor, T. Shinno, J. Piedade, T. Sata, W. K. Ampofo, J. A. Brandful, Y. Koyanagi, N. Yamamoto, W. F. Canas-Ferreira, Y. Adu-Sarkodie, and T. Kurata. 2001. Genetic analysis of HIV type 2 from Ghana and Guinea-Bissau, West Africa. AIDS Res. Hum. Retrovir. 17:1661-1663. [DOI] [PubMed] [Google Scholar]
- 35.Jolly, C., J. E. Phillips-Conroy, T. R. Turner, S. Broussard, and J. S. Allan. 1996. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops). J. Med. Primatol. 25:78-83. [DOI] [PubMed] [Google Scholar]
- 36.Kaur, A., L. Alexander, S. I. Staprans, L. Denekamp, C. L. Hale, H. M. McClure, M. B. Feinberg, R. C. Desrosiers, and R. P. Johnson. 2001. Emergence of cytotoxic T lymphocyte escape mutations in nonpathogenic simian immunodeficiency virus infection. Eur. J. Immunol. 31:3207-3217. [DOI] [PubMed] [Google Scholar]
- 37.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 38.Kingdon, J. 1997. The Kingdon field guide to African mammals. Princeton University Press, Princeton, N.J.
- 39.Le Guenno, B. 1989. HIV1 and HIV2: two ancient viruses for a new disease? Trans. R. Soc. Trop. Med. Hyg. 83:847. [DOI] [PubMed] [Google Scholar]
- 40.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, B. Wolinsky, and B. Korber. 2003. HIV Sequence Compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 41.Lemey, P., O. G. Pybus, B. Wang, N. K. Saksena, M. Salemi, and A. M. Vandamme. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. USA 100:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemey, P., O. G. Pybus, A. Rambaut, A. J. Drummond, D. L. Robertson, P. Roques, M. Worobey, and A. M. Vandamme. 2004. The molecular population genetics of HIV-1 group O. Genetics 167:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling, B., M. L. Santiago, S. Meleth, B. Gormus, H. M. McClure, C. Apetrei, B. H. Hahn, and P. A. Marx. 2003. Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma. J. Virol. 77:2214-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling, B., C. Apetrei, I. Pandrea, R. S. Veazey, A. A. Lackner, B. Gormus, and P. A. Marx. 2004. Classic AIDS in a sooty mangabey after an 18-year natural infection. J. Virol. 78:8902-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling, B., P. Telfer, P. Reed, D. L. Robertson, and P. A. Marx. 2004. A link between SIVsm in sooty mangabeys (SM) in wild-living monkeys in Sierra Leone and SIVsm in an American-based SM colony. AIDS Res. Hum. Retrovir. 20:1348-1351. [DOI] [PubMed] [Google Scholar]
- 46.Markine-Goriaynoff, N., J. P. Georgin, M. Goltz, W. Zimmermann, H. Broll, H. M. Wamwayi, P. P. Pastoret, P. M. Sharp, and A. Vanderplasschen. 2003. The core 2 β-1,6-N-acetylglucosaminyltransferase-mucin encoded by bovine herpesvirus 4 was acquired from an ancestor of the African buffalo. J. Virol. 77:1784-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marx, P. A., Y. Li, N. W. Lerche, S. Sutjipto, A. Gettie, J. A. Yee, B. H. Brotman, A. M. Prince, A. Hanson, R. G. Webster, and R. C. Desrosiers. 1991. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a west African pet sooty mangabey. J. Virol. 65:4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGraw, W. S., and R. Bshary. 2002. Association of terrestrial mangabeys (Cercocebus atys) with arboreal monkeys: experimental evidence for the effects of reduced ground predator pressure on habitat use. Int. J. Primatol. 23:311-325. [Google Scholar]
- 49.Morin, P. A., K. E. Chambers, C. Boesch, and L. Vigilant. 2001. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol. Ecol. 10:1835-1844. [DOI] [PubMed] [Google Scholar]
- 50.Peeters, M., W. Janssens, K. Fransen, J. Brandful, L. Heyndrickx, K. Koffi, E. Delaporte, P. Piot, G. M. Gershy-Damet, and G. van der Groen. 1994. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Côte d'Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res. Hum. Retrovir. 10:1289-1294. [DOI] [PubMed] [Google Scholar]
- 51.Peeters, M., C. Toure-Kane, and J. N. Nkengasong. 2003. Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS 17:2547-2560. [DOI] [PubMed] [Google Scholar]
- 52.Perrin, L., L. Kaiser, and S. Yerly. 2003. Travel and the spread of HIV-1 genetic variants. Lancet Infect. Dis. 3:22-27. [DOI] [PubMed] [Google Scholar]
- 53.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primatol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 54.Pieniazek, D., D. Ellenberger, L. M. Janini, A. C. Ramos, J. Nkengasong, M. Sassan-Morokro, D. J. Hu, I. M. Coulibally, E. Ekpini, C. Bandea, A. Tanuri, A. E. Greenberg, S. Z. Wiktor, and M. A. Rayfield. 1999. Predominance of human immunodeficiency virus type 2 subtype B in Abidjan, Ivory Coast. AIDS Res. Hum. Retrovir. 15:603-608. [DOI] [PubMed] [Google Scholar]
- 55.Poulsen, A. G., P. Aaby, H. Jensen, and F. Dias. 2000. Risk factors for HIV-2 seropositivity among older people in Guinea-Bissau. A search for the early history of HIV-2 infection. Scand. J. Infect. Dis. 32:169-175. [DOI] [PubMed] [Google Scholar]
- 56.Range, F., and R. Noe. 2002. Familiarity and dominance relations among female sooty mangabeys in the Taï National Park. Am. J. Primatol. 56:137-153. [DOI] [PubMed] [Google Scholar]
- 57.Range, F., and R. Noe. 2005. Can simple rules account for the pattern of triadic interactions in juvenile and adult female sooty mangabeys? Anim. Behav. 69:445-452. [Google Scholar]
- 58.Range, F. Social behaviour of free-ranging juvenile sooty mangabeys (Cercocebus torquatus atys). Behav. Ecol. Sociobiol., in press.
- 59.Refisch, J., and I. Koné. 2005. Impact of commercial hunting on monkey populations in the Taï region, Côte d'Ivoire. Biotropica 37:136-144. [Google Scholar]
- 60.Rey-Cuille, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruprecht, R. M., T. W. Baba, V. Liska, N. B. Ray, L. N. Martin, M. Murphey-Corb, T. A. Rizvi, B. J. Bernacky, M. E. Keeling, H. M. McClure, and J. Andersen. 1999. Oral transmission of primate lentiviruses. J. Infect. Dis. 179(Suppl. 3):S408-S412. [DOI] [PubMed] [Google Scholar]
- 62.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 63.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 64.Santiago, M. L., M. Lukasik, S. Kamenya, Y. Li, F. Bibollet-Ruche, E. Bailes, M. N. Muller, M. Emery, D. A. Goldenberg, J. S. Lwanga, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, D. P. Watts, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, J. F. Brookfield, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santiago, M. L., F. Bibollet-Ruche, N. Gross-Camp, A. C. Majewski, M. Masozera, I. Munanura, B. A. Kaplin, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Noninvasive detection of simian immunodeficiency virus infection in a wild-living L'Hoest's monkey (Cercopithecus Ihoesti). AIDS Res. Hum. Retrovir. 19:1163-1166. [DOI] [PubMed] [Google Scholar]
- 66.Scarlatti, G. 2004. Mother-to-child transmission of HIV-1: advances and controversies of the twentieth centuries. AIDS Rev. 6:67-78. [PubMed] [Google Scholar]
- 67.Schino, G. 2001. Grooming, competition and social rank among female primates: a meta-analysis. Anim. Behav. 62:265-271. [Google Scholar]
- 68.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 69.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souquière, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclère, P. A. Marx, F. Barré-Sinoussi, B. H. Hahn, M. C. Müller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stahl-Hennig, C., R. M. Steinman, K. Tenner-Racz, M. Pope, N. Stolte, K. Matz-Rensing, G. Grobschupff, B. Raschdorff, G. Hunsmann, and P. Racz. 1999. Rapid infection of oral mucosal-associated lymphoid tissue with simian immunodeficiency virus. Science 285:1261-1265. [DOI] [PubMed] [Google Scholar]
- 72.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomonaga, K., J. Katahira, M. Fukasawa, M. A. Hassan, M. Kawamura, H. Akari, T. Miura, T. Goto, M. Nakai, M. Suleman, M. Isahakia, and M. Hayami. 1993. Isolation and characterization of simian immunodeficiency virus from African white-crowned mangabey monkeys (Cercocebus torquatus lunulatus). Arch. Virol. 129:77-92. [DOI] [PubMed] [Google Scholar]
- 74.Van Schaik, C. P., and C. H. Janson (ed.). 2000. Infanticide by males and its implications. Cambridge University Press, Cambridge, United Kingdom.
- 75.Worobey, M., M. L. Santiago, B. F. Keele, J. B. N. Ndjango, J. B. Joy, B. L. Labama, D. Dhed'a, A. Rambaut, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2004. Origin of AIDS: contaminated polio vaccine theory refuted. Nature 428:820. [DOI] [PubMed] [Google Scholar]
- 76.Yamaguchi, J., S. G. Devare, and C. A. Brennan. 2000. Identification of a new HIV-2 subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res. Hum. Retrovir. 16:925-930. [DOI] [PubMed] [Google Scholar]
- 77.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306-314. [DOI] [PubMed] [Google Scholar]
- 78.Yang, Z., and B. Rannala. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 14:717-724. [DOI] [PubMed] [Google Scholar]
- 79.Zeh, C., D. Pieniazek, S. M. Agwale, K. E. Robbins, L. Odama, N. Sani-Gwarzo, M. S. Gboun, U. S. Inyang, T. M. Folks, C. Wambebe, and M. L. Kalish. 2005. Nigerian HIV type 2 subtype A and B from heterotypic HIV type 1 and HIV type 2 or monotypic HIV type 2 infections. AIDS Res. Hum. Retrovir. 21:17-27. [DOI] [PubMed] [Google Scholar]