Abstract
Virions of porcine reproductive and respiratory syndrome virus (PRRSV) contain six membrane proteins: the major proteins GP5 and M and the minor proteins GP2a, E, GP3, and GP4. Here, we studied the envelope protein requirements for PRRSV particle formation and infectivity using full-length cDNA clones in which the genes encoding the membrane proteins were disrupted by site-directed mutagenesis. By transfection of RNAs transcribed from these cDNAs into BHK-21 cells and analysis of the culture medium using ultracentrifugation, radioimmunoprecipitation, and real-time reverse transcription-PCR, we observed that the production of viral particles is dependent on both major envelope proteins; no particles were released when either the GP5 or the M protein was absent. In contrast, particle production was not dependent on the minor envelope proteins. Remarkably, in the absence of any one of the latter proteins, the incorporation of all other minor envelope proteins was affected, indicating that these proteins interact with each other and are assembled into virions as a multimeric complex. Independent evidence for such complexes was obtained by coexpression of the minor envelope proteins in BHK-21 cells using a Semliki Forest virus expression system. By analyzing the maturation of their N-linked oligosaccharides, we found that the glycoproteins were each retained in the endoplasmic reticulum unless expressed together, in which case they were collectively transported through the Golgi complex to the plasma membrane and were even detected in the extracellular medium. As the PRRSV particles lacking the minor envelope proteins are not infectious, we hypothesize that the virion surface structures formed by these proteins function in viral entry by mediating receptor binding and/or virus-cell fusion.
Porcine reproductive and respiratory syndrome virus (PRRSV) belongs to the family of Arteriviridae, which also comprises Equine arteritis virus (EAV), Lactate dehydrogenase-elevating virus (LDV), and Simian hemorrhagic fever virus (24, 42). This family belongs to the order of Nidovirales, together with the Coronaviridae, Toroviridae, and Roniviridae (2, 3). Arteriviruses are enveloped RNA viruses that contain a positive-strand RNA genome and synthesize a 3′ nested set of six or eight subgenomic RNAs (sgRNAs) that encode the structural proteins. Analyses of purified virions of PRRSV have indicated that they are composed of seven proteins, i.e., four envelope glycoproteins named GP2a (encoded by open reading frame 2a [ORF2a]), GP3 (ORF3), GP4 (ORF4), and GP5 (ORF5); a nonglycosylated membrane protein M (ORF6); the nucleocapsid protein N (ORF7); and a nonglycosylated envelope protein, E, that is expressed from a second ORF (ORF2b) entirely contained within ORF2 (22, 25, 35, 48). The 29- to 30-kDa GP2a and 31- to 35-kDa GP4 proteins are both putative class I integral membrane proteins with an N-terminal signal sequence and a C-terminal membrane anchor, containing two and four predicted N-glycosylation sites, respectively (22). The E protein is a small 10-kDa protein with a potential N-terminal N-myristoylation site and a casein kinase II phosphorylation site, a central hydrophobic domain, and a hydrophilic C terminus containing a cluster of basic residues (32). The E protein was reported to be the predominant product of sgRNA2 (48). The 45- to 50-kDa GP3 protein is highly glycosylated potentially by seven N-linked oligosaccharides and has a single N-terminal hydrophobic domain (22). Whereas the GP3 protein was shown to be incorporated into virions of European-type strains (35), for North American strains, its structural nature has been questioned (11, 19). For EAV, GP2b, GP3, and GP4 have been reported to exist as covalently linked heterotrimeric complexes (45). Furthermore, incorporation of these glycoproteins and of the E protein in EAV particles was shown to be interdependent (43). The GP5 and M proteins exist as disulfide-linked heterodimers in PRRSV particles (20). The membrane topologies of these proteins have not yet been accurately determined, but by analogy with LDV (8), the M protein is assumed to be a class III integral membrane protein with an N-terminal ectodomain of only 16 amino acids followed by three membrane-spanning domains and a C-terminal endodomain of about 78 residues (22). The GP5 protein is assumed to consist of an N-terminal signal sequence of approximately 30 amino acids, followed by an ectodomain of approximately 35 residues with a variable number of potential N-glycosylation sites, a long hydrophobic region of about 60 residues that is presumed to span the membrane one to three times, and a hydrophilic C terminus of approximately 70 amino acids (22).
Little is known about the structural requirements for PRRSV virion formation and infectivity. Several studies point towards an important role for the GP5 and M proteins in PRRSV assembly (38, 39), but nothing is yet known about the function of the minor structural proteins. Recently, Welch and coauthors (40) demonstrated that the ORF2a/b or ORF4 products are essential for a North American strain of PRRSV; mutant genomes from which these ORFs had been deleted did not generate infectivity for reasons that were not investigated. In the case of EAV, each of the structural proteins was shown to be required for the production of infectious progeny virus (26). It appeared that the homologous GP5 and M proteins are essential for virus assembly, in contrast to the minor envelope proteins, which are dispensable for virus assembly but not for EAV infectivity (43).
The aim of the present study was to determine the membrane protein requirements for PPRSV virion formation and infectivity using a collection of infectious cDNA clones in which the correct expression of each of these proteins was individually disabled. The results indicated that while the major proteins are essential for particle formation, the minor proteins are essential for virus infectivity. Furthermore, the results indicated that the minor proteins are incorporated into virions as a multimeric complex. Independent confirmation of this interaction between these proteins was indeed obtained by their coexpression in cells using a Semliki Forest virus (SFV) expression system which showed that the glycoproteins are retained in the endoplasmic reticulum (ER) unless all are expressed together.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
BHK-21 cells were grown in Glasgow minimal essential medium (Gibco BRL) complemented with 5% fetal bovine serum (FBS), 10% tryptose phosphate broth, 20 mM HEPES (pH 7.4), 2 mM glutamine, 100 U ml−1 penicillin, and 100 U ml−1 streptomycin. Porcine alveolar lung macrophages (PAMs) were maintained in MCA-RPMI 1640 medium (Gibco BRL) containing 5% FBS, 500 U ml−1 penicillin, and 500 U ml−1 streptomycin (42). LV4.2.1, a derivative of the original Lelystad virus (LV) Ter Huurne strain, which has been adapted for growth on MA-104 cells and derivatives thereof (35), was propagated in MARC-145 cells (16). The following antibodies were used for immunoprecipitation of PRRSV structural proteins: polyclonal rabbit serum p690, raised against a Lelystad virus-specific peptide consisting of amino acids 64 to 78 of the GP2a protein (22); monoclonal antibody (MAb) WBE2, directed against the GP3 protein (6); MAb 122.1, directed against amino acids 40 to 79 of the GP4 protein (35); MAb 126.3, directed against an epitope on the ectodomain of the M protein (39); and MAb 122.17, directed against the PRRSV N protein (35). MAb 3AH9, raised against amino acids 170 to 201 of the GP5 protein (30), was used to detect the expression of the GP5 protein in an immunoperoxidase monolayer assay (IPMA). MAb1, directed against the bovine herpesvirus type 1 gE/gI protein complex, was used as a control in the affinity purification assays (36).
Generation of full-length gene knockout constructs and complementing expression vectors.
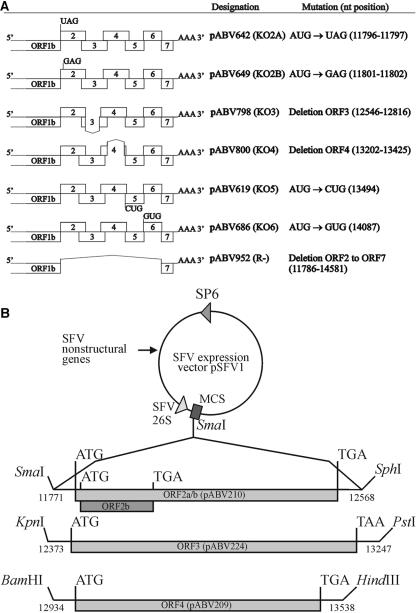
Using site-directed PCR mutagenesis (Table 1) and shuttle plasmids, mutant full-length PRRSV cDNA clones were generated in which the ORFs were disrupted either by mutating their translation initiation codons (KO2a, KO2b, KO5, and KO6), or by deleting the ORF from just after the termination codon of the preceding ORF to 40 nucleotides 5′ of the transcription-regulating sequence of the subsequent ORF (KO3 and KO4) (Table 1 and Fig. 1A). Mutations were introduced into pABV437, the PacI mutant of the genome-length cDNA clone of LV (21).
TABLE 1.
Primers used to generate the mutant full-length cDNAs and the pCIneo expression constructs containing ORFs 2a and 2b of PRRSV LV
| Primer designationa | Primer sequence (5′→3′)b | Nucleotide positionc | Purpose |
|---|---|---|---|
| LV183 (+) | GCCGTGAATTCGGGGTGTAGCAATGGGG | 11779 | KO2a |
| LV25 (−) | AATCGGATCCTGGTCAGCTCGAATGATGTG | 12529 | KO2a and KO2b |
| LV189A (+) | GCCGTGAATTCGGGGTGATGCAGTGGGGTCAC | 11779 | KO2b |
| LV250 (−) | ACTGGGCCCTCAGCTCGAATGATG | 12531 | KO3 |
| LV253 (−) | ACTGGGCCCTTATCGTGATGTACTGGGGAGTAC | 13178 | KO4 |
| LV90 (+) | TCGACTTGTCGCTAGTATGC | 10798 | KO3, KO4, and R− |
| LV186 (+) | TGTTCGCCATTCTCTTGGCAATCTGAGATGT | 13472 | KO5 |
| LV38 (−) | AATCGGATCCTCTGGTTTTTACCGGCC | 14600 | KO5 |
| LV96 (+) | CCAACTGCAGACCGGATGTCC | 12025 | KO6 |
| LV187 (−) | ATCGTCTAGGCCTCCCACTGCTCAGCC | 14078 | KO6 |
| LV407 (−) | AGGCGGCAGCCGGGGCTTGCCTAGCTCTACCTGC | 11737 | R− |
| LV408 (−) | ATGTTAACGACGCGTCGTCACGGCACTTGCCCAGGCGGCAGCCG | 11759 | R− |
| LV271 (+) | GCTCTAGAGCCGCCGCCATGCAATGGGGTCACTGTGG | 11796 | pCIneo-ORF2a; incorporation of XbaI plus consensus Kozak sequence |
| LV278 (−) | ATAGTTTAGCGGCCGCTCAGCTCGAATGATGTGTTGC | 12525 | pCIneo-ORF2a; incorporation of NotI |
| LV272 (+) | GCTCTAGAGCCGCCGCCATGGGGTCACTGTGGAG | 11801 | pCIneo-ORF2b; incorporation of XbaI plus consensus Kozak sequence |
| LV279 (−) | ATAGTTTAGCGGCCGCTCATAGGACCTTCGATAGTTCG | 12013 | pCIneo-ORF2b; incorporation of NotI |
Positive-sense primers and negative-sense primers are marked by a + and −, respectively.
The restriction sites are indicated by single underlining, and the consensus Kozak sequence is indicated by double underlining. The mutated translation initiation sites are indicated in boldface type.
The position of each primer with respect to the nucleotide sequence of PRRSV LV (GenBank accession no. M96262).
FIG. 1.
Composition of the PRRSV constructs. (A) Design of the PRRSV knockout constructs. Depicted are the point mutations of the initiation codons of ORF2a (KO2a), ORF2b (KO2b), ORF5 (KO5), and ORF6 (KO6); the deletions in ORF3 (KO3) and ORF4 (KO4); and the deletion of ORF2 to ORF7 (R−). The names of the constructs and the nucleotide (nt) positions of the mutations are shown on the right. (B) Design of the recombinant SFV constructs. Restriction enzyme sites, their positions in the nucleotide sequence of PRRSV LV (GenBank accession no. M96262), and the names of the constructs providing the genes (pABV) are indicated.
Construct pABV952 was used as a negative control; this construct contains a deletion from just after the termination codon of PRRSV ORF1ab to the transcription-regulating sequence of ORF7 (Table 1 and Fig. 1A) and consequently lacks an essential RNA replication signal located in ORF6 (M. V. Kroese et al., unpublished results). This construct will be referred to below as “R−” (replication negative).
To produce the lacking proteins in trans, sequences encoding these proteins were cloned into the pCIneo mammalian expression vector (Promega). For this purpose, ORFs 2a and 2b of pABV437 were amplified using the respective oligonucleotides located upstream and downstream of the respective ORFs (Table 1). The nucleotide sequences directly upstream of the initiator codons of the ORFs were modified according to a Kozak consensus sequence. In addition, the restriction sites XbaI and NotI were added upstream and downstream of the ORFs, respectively. The PCR fragments were digested with XbaI and NotI and ligated into the similarly digested pCIneo vector. This resulted in plasmids pABV782 and pABV783, containing ORFs 2a and 2b, respectively. All PCR-generated plasmids were verified by sequencing of the entire insert.
Expression of GP2a, E, GP3, and GP4 by recombinant Semliki Forest virus.
To study the interdependence of the minor glycoproteins GP2a, GP3, and GP4 for intracellular transport, these proteins were expressed by the SFV expression system as developed by Liljestrom and Garoff (18). ORF3 of LV was cloned in expression vector pSFV1 (18) as described previously for ORF2d (pABV262) (25) and ORF4 (pABV265) (23). For the design of the recombinant SFV constructs, see Fig. 1B. Briefly, ORF3 was excised from plasmid pABV224 (22) with KpnI and PstI, treated with Klenow polymerase, and ligated into the SmaI site of expression vector pSFV1, which had been dephosphorylated using calf intestine phosphatase. This resulted in plasmid pABV267. Plasmid pABV267 turned out to contain two complete copies of ORF3 in the same orientation but was observed to express the GP3 protein correctly. Recombinant SFV was prepared according to the methods described previously by Liljestrom and Garoff (18).
In vitro transcription of RNA, electroporation of BHK-21 cells, and inoculation of PAMs.
The in vitro generation of full-length PRRSV transcripts and the electroporation of BHK-21 C13 cells to transfect these transcripts have been previously described (47). To examine the viability of the PRRSV knockout mutants, the transfected BHK-21 cells were fixed 24 h posttransfection, and immunostaining (IPMA [see below]) was performed with an antibody to the N protein. For analysis of infectious progeny produced by the BHK-21 cells, the culture supernatants of the transfected cells were used to inoculate PAMs (passage 1 [p1]). After incubation for 24 h at 37°C, these cells were also fixed for IPMA, and the culture supernatants were inoculated once again onto new PAMs (p2), followed by an IPMA on the cells 24 h later.
Rescue of PRRSV knockout mutants.
In vitro-synthesized RNA transcripts of the mutant full-length cDNAs and complementing pCIneo cDNAs were cotransfected by electroporation into BHK-21 cells which were subsequently incubated under standard conditions. To study whether complementation of the gene knockout mutants had occurred, the culture medium was harvested and two passages on PAMs were performed as described above.
IPMA.
Immunostaining of BHK-21 cells and of PAMs was performed with the aforementioned MAbs against the GP3, GP4, GP5, M, and N proteins by the method described previously by Wensvoort et al. (41). Expression of the GP2a and E proteins could not be investigated in the IPMA since reactive antibodies are currently unavailable.
Metabolic labeling of cells and virus.
Twelve hours after transfection of BHK-21 cells with the full-length PRRSV RNAs, the cells were washed three times with starvation medium (MEM-E without l-methionine, l-cysteine, and l-glutamine [ICN]) and subsequently incubated with the same medium supplemented with 1.5 mg ml−1 methionine and 24 mg ml−1 cysteine, 1% l-glutamine, 100 U ml−1 penicillin, 100 U ml−1 streptomycin, and 5% dialyzed FBS. After 1 hour, 157.5 mCi Tran[35S] label (ICN) was added, and the cells were further incubated for 5 h at 37°C. After clarification of the culture supernatants at 6,000 rpm for 10 min at 4°C with a Hermle ZK380, the labeled viruses were pelleted through a 0.5 M sucrose cushion (in 20 mM Tris-HCl, pH 7.6, 100 mM NaCl, and 1 mM EDTA) by ultracentrifugation (Beckman R65 rotor at 48,000 rpm for 3 h at 4°C). The labeling of LV virions propagated in MARC-145 cells and the purification of labeled extracellular virions by sucrose cushion have been previously described (25).
Pulse-chase labeling of BHK-21 cells infected with recombinant SFVs.
The pulse-chase labeling of BHK-21 cells infected with recombinant SFVs has been described previously (25). Briefly, BHK-21 cells were infected with one or more recombinant SFVs at a multiplicity of infection (MOI) of 5 each. At 18 h postinfection (p.i.), the cells were starved for methionine for 30 min followed by a pulse with l-[35S]methionine for 30 min. Subsequently, the cells were chased with 5 mM nonradioactive l-methionine for 0, 150, and 300 min or for the time periods indicated in the legends to Fig. 4 to 6.
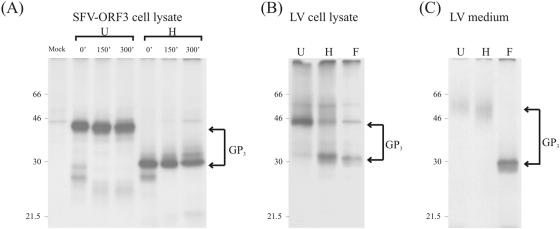
FIG. 4.
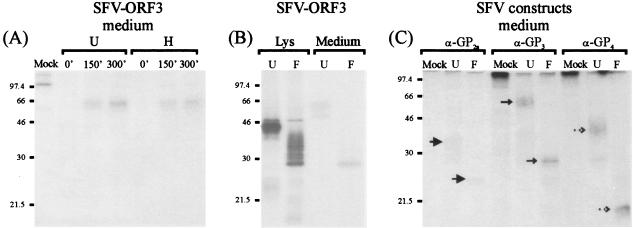
Immunoprecipitation analysis of GP3 expressed by SFV-ORF3 or by LV. (A) GP3 was immunoprecipitated with anti-GP3 (α-GP3) from lysates of BHK-21 cells infected with SFV-ORF3 at an MOI of 5. At 18 h p.i., the cells were starved for methionine for 30 min and subsequently labeled with l-[35S]methionine for 30 min. The cells were then chased with 5 mM nonradioactive l-methionine for 0, 150, and 300 min. (B) GP3 was immunoprecipitated with α-GP3 from lysates of MARC-145 cells infected with LV4.2.1. Cells were infected with LV4.2.1 at an MOI of 2, and at 18 h p.i., they were labeled with l-[35S]methionine for 30 min and chased for 300 min. (C) GP3 was immunoprecipitated with α-GP3 from extracellular LV4.2.1 virions labeled and purified by sedimentation through a sucrose cushion as described previously (25). Immunoprecipitates were treated with EndoH (H) or PNGaseF (F) or were left untreated (U).
FIG. 6.
Analysis of GP2a, GP3, and GP4 released into the medium of BHK-21 cells. (A) Immunoprecipitation of the GP3 protein from the medium of BHK-21 cells infected with SFV-ORF3. Cells were pulse-chase labeled as described in the legend to Fig. 4A. Immunoprecipitates were treated with EndoH (H) or left untreated (U). (B) Immunoprecipitation of the GP3 protein from the cell lysate (Lys) and medium of BHK-21 cells infected with SFV-ORF3. The cells were pulse-labeled, followed by a chase of 300 min. Immunoprecipitates were treated with PNGase F (F) or left untreated (U). (C) Immunoprecipitation of GP2a, GP3, and GP4 proteins from the medium of BHK-21 cells coinfected with SFV-ORF2a, SFV-ORF3, and SFV-ORF4. At 4 h p.i., the cells were pulse-labeled, followed by a chase of 300 min. The medium was harvested, and the GP2a, GP3, and GP4 proteins were subjected to immunoprecipitation with anti-GP2a (α-GP2a), anti-GP3 (α-GP3), or anti-GP4 (α-GP4) antibodies. Precipitates were either treated with PNGaseF (F) or left untreated (U). The arrows indicate the respective proteins.
Affinity purification of viral particles.
To isolate viral particles using affinity purification, the virus pellets were resuspended in phosphate-buffered saline (PBS) supplemented with protease inhibitors (Pefabloc-Sc [25 mg ml−1], pepstatin [0.7 mg ml−1], aprotinin [1 mg ml−1], and leupeptin [0.5 mg ml−1]). Virus particle suspensions were incubated overnight at 4°C in the presence of the appropriate antibody. The next day, 3 mg protein A Sepharose was added to each sample, and the suspensions were incubated at 4°C for another 2 h. The immune complexes formed were washed three times with PBS, resuspended in 30 ml of 2× NuPAGE LDS sample buffer supplemented with 50 mM dithiothreitol, and heated for 5 min at 100°C. To confirm the specificity of the affinity purification, control experiments were performed without antibodies and with MAb1, which is directed against the bovine herpesvirus type 1 gE/gI complex (36).
Radioimmunoprecipitations.
To isolate radiolabeled viral proteins, lysates of cells or of virus pellets were prepared in PBS-TDS (PBS containing 1 g sodium dodecyl sulfate [SDS], 10 ml Triton X-100, and 5 g sodium desoxycholate per liter) supplemented with protease inhibitors. The lysates were incubated overnight at 4°C in the presence of the appropriate antibody. The next day, 3 mg protein A Sepharose was added to each sample, and the samples were incubated at 4°C for another 2 h. The immune complexes were washed three times with PBS-TDS, resuspended in 30 ml of 2× NuPAGE LDS sample buffer supplemented with 50 mM dithiothreitol, and heated for 5 min at 100°C.
Endoglycosidase treatment.
Endoglycosidase treatments were performed with Endo-β-N-acetyl-d-glycosaminidase H (EndoH; New England Biolabs) and with peptide N-glycosidase F (PNGaseF; New England Biolabs) according to the manufacturer's instructions.
Gel electrophoresis.
Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using a 12% polyacrylamide gel (NuPAGE). Gels were fixed for at least 1 h in 40% (vol/vol) methanol and 10% (vol/vol) acetic acid and dried on Whatman 3MM paper, and radioactivity was visualized by phosphorimaging (STORM-840). Intensities of the protein bands were quantified using ImageQuant 5.1 image analysis software.
RNA isolation and RNA quantification.
To investigate the relative amounts of viral genomic RNA (as a measure of total viral particles), a real-time reverse transcription (RT)-PCR was performed using the Light Cycler (Roche) as previously described (37).
Statistical analysis.
Overall equality of the amounts of viral particles produced by wild-type and mutant viruses, each expressed as relative amounts of RNA as determined by RT-PCR, was tested using the Kruskal-Wallis test. Interesting contrasts between groups of viruses or between different viruses were tested using the permutation test. Significance levels were set at 5%. Statistical analyses were performed with StatXact-5 for Windows (Cytel Software Corporation, 2001).
RESULTS
Requirement of the PRRSV envelope protein genes for infectious virus production.
To determine the requirement of each of the PRRSV envelope proteins for the production of infectious progeny, we generated full-length cDNA clones in which the genes encoding the envelope proteins were disabled, one at a time, either by mutation of their translation initiation codon or by deletion of a major part of the ORF (Fig. 1A) (KO2a, -2b, -3, -4, -5, and -6). BHK-21 cells were transfected with in vitro RNA transcripts of the gene knockout constructs and immunostained 24 h later with an antibody to the N protein. As judged from the expression of the N protein, which for each KO mutant was similar to that of the wild-type (wt) control, neither genome replication nor mRNA transcription appeared to be affected in the gene knockout constructs. BHK-21 cells are nonpermissive to PRRSV; hence, production of infectious virus cannot be determined by monitoring the spread of the infection through the transfected culture. To investigate the presence of infectious progeny virus released by the BHK-21 cells, the supernatants of these cultures were inoculated onto PAMs, the natural host cells of PRRSV. The results of the immunoassays performed 24 h p.i. revealed that no or only few cells had become infected with any of the mutants. In a typical experiment, up to approximately 0.1% of the cells stained positive compared to about 20% in the case of the wt. When the culture supernatants of these PAMs (p1) were passaged once more onto fresh cells (p2), none of the cells stained positive except for the cells inoculated with the wt control. These observations are similar to those described previously in comparable experiments with EAV gene knockout viruses (26, 49). From our results, we conclude that each of the PRRSV envelope proteins is essential for infectious virus production.
Membrane protein requirements for viral particle formation.
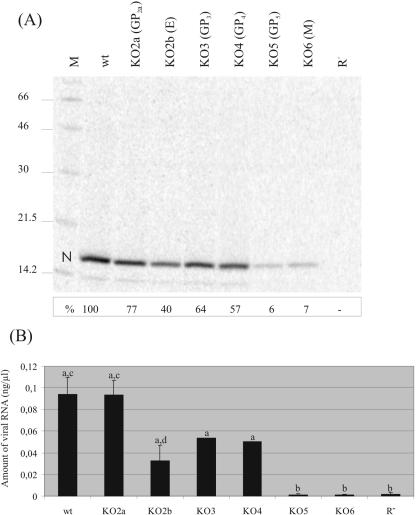
Knowing the importance of each of the envelope proteins for the virus, we subsequently investigated whether any of these mutations affected the formation of viral particles. BHK-21 cells transfected with in vitro transcripts of the different full-length constructs were labeled in parallel with Tran[35S] label. Particles released into the culture supernatant were concentrated by sedimentation through a sucrose cushion. The pellets were dissolved in PBS-TDS and subjected to immunoprecipitation with an antibody to the N protein. SDS-PAGE of the immunoprecipitates showed that the gene knockout constructs not expressing (functional forms of) the GP2a, E, GP3, or GP4 protein were still capable of producing noninfectious viral particles as judged by the appearance of the N protein (Fig. 2A). However, in the absence of the GP5 or the M protein, only traces of the nucleocapsid protein were observed (approximately 6% of the wt), implying that GP5 and M are both required for virion formation. The low amounts of N protein detected in the culture medium most likely represent intracellular N protein (nucleocapsids) released due to cell lysis.
FIG. 2.
Presence of virus-like particles in transfection supernatants. (A) Immunoprecipitation analysis of recombinant viruses. BHK-21 cells were transfected with in vitro transcripts of the full-length constructs and labeled with Tran[35S] label. Virus-like particles in the culture supernatants of the transfected cells were concentrated by sedimentation through a sucrose cushion. Pellets were dissolved in PBS-TDS and subjected to immunoprecipitation with a MAb directed against the N protein. The immunoprecipitated proteins were analyzed in an SDS-12% polyacrylamide gel. “N” indicates the position of the nucleocapsid protein. At the left, the positions and sizes (kilodaltons) of the molecular mass marker proteins run in parallel (lane M) are shown. BHK-21 cells transfected with replication-negative mutant R− were used as a negative control. The intensities of the N-protein bands were quantified using ImageQuant5.1 image analysis software. The values (in percentages of wt) are shown below the respective N-protein bands. (B) Quantification of relative amounts of viral RNA. BHK-21 cells were transfected with full-length RNA transcripts. At 24 h posttransfection, culture supernatants were harvested, viral RNA was isolated, and a real-time RT-PCR was performed. Shown are the mean values of two (KO3 and KO4) or four (wt, KO2a, KO2b, KO5, KO6, and R−) experiments. A statistically significant difference (P ≤ 0.05) was observed between wt/KO2a/KO2b/KO3/KO4 jointly on the one hand and KO5/KO6/R− jointly on the other hand (“a” versus “b”). In addition, a statistically significant difference (P ≤ 0.05) was observed between viral RNA levels obtained for wt and KO2a compared to KO2b (“c” versus “d”).
Rescue of PRRSV gene knockout mutants with constructs expressing the lacking protein.
To verify whether the inability of our gene knockout constructs to produce infectious progeny was indeed due to our intended mutations rather than to changes unintentionally introduced in the full-length cDNAs, we performed complementation experiments using plasmids expressing the missing proteins. For this purpose, we transfected BHK-21 cells either with in vitro transcripts of full-length construct KO2a or KO2b alone or together with pCIneo constructs expressing the GP2a or the E protein, respectively. An immunostaining experiment performed 24 h posttransfection with MAb 122.17 showed that N protein expression induced by the gene knockout constructs was comparable to that by the wt transcript (Table 2, passage p0 BHK-21). As expected, no virus replication was observed after inoculation of PAMs with the supernatants of single-transfected BHK-21 cells (Table 2, p1 PAMs). On the other hand, inoculation of PAMs with the culture supernatants of the double-transfected BHK-21 cells resulted in infection as judged from the immunopositive cells (Table 2, p1), indicating that the failure of our knockout mutants to produce infectious progeny virus was solely caused by the gene disruptions. When a second passage (Table 2, p2) was performed, no virus replication was observed except for the wt control, confirming that the infectious progeny consisted of complemented virus particles only capable of a single round of replication. Our observations are in agreement with data published very recently by Welch et al. (40) showing that infectious PRRSV lacking ORF2 or ORF4 can be rescued by growth in complementing cell lines providing the missing protein in trans. Similar complementation was reported for EAV using cell lines expressing the minor envelope proteins (49).
TABLE 2.
Rescue of PRRSV gene knockout mutants with constructs expressing the lacking protein
| Mutant | Complementa | p0 BHK-21b | p1 PAMsb | p2 PAMsb |
|---|---|---|---|---|
| Wild type | — | +++ | +++ | +++ |
| KO2A | — | +++ | − | − |
| KO2A | pCIneo-ORF2a | +++ | + | − |
| KO2A | SFV-ORF2 | +++ | + | − |
| KO2B | — | +++ | − | − |
| KO2B | pCIneo-ORF2b | +++ | + | − |
| KO2B | SFV-ORF2 | +++ | + | − |
Expression constructs and recombinant SFV used for in trans complementation of the full-length gene knockout constructs. —, no complement added.
Virus replication was analyzed by IPMA at 24 h posttransfection (BHK-21) or postinfection (PAMs) using MAb 122.17 directed against the N protein. The degree of positive staining is indicated by the number of plusses (+ to +++); no staining is indicated by a minus (−).
Release of viral RNA from cells transfected with mutant viral RNA transcripts.
To characterize the virus-like particles produced in the absence of the GP2a, E, GP3, or GP4 protein, we analyzed whether the particles present in the culture supernatants of transfected cells contained viral RNA by using real-time RT-PCR. A clear distinction was observed between viral RNA levels obtained for mutants KO2a, KO2b, KO3, and KO4 compared to those for KO5 and KO6 (Fig. 2B). Whereas the amounts of RNA detected in the culture supernatants of mutant viruses KO2a, KO2b, KO3, and KO4 were in the same order of magnitude as that of the wt, the viral RNA levels of KO5 and KO6 were dramatically reduced (1% of the wt) (Fig. 2B), consistent with the immunoprecipitation results. Closer inspection of the relative amounts of viral RNA produced by the KO2a, KO2b, KO3, and KO4 mutants revealed that while the amount of viral RNA obtained for the KO2a mutant was similar to that of wt virus, it was significantly reduced by about two-thirds in the case of mutant KO2b. Viruses KO3 and KO4 also produced lower levels of viral RNA (50% of the wt), but these amounts were statistically not significantly different from those of the wt. The results are consistent with the interpretation that no viral particles are produced if either the GP5 or the M protein is absent but that particle formation is only marginally affected by the lack of any of the other envelope proteins.
Virion envelope composition in the absence of GP2a, E, GP3, or GP4.
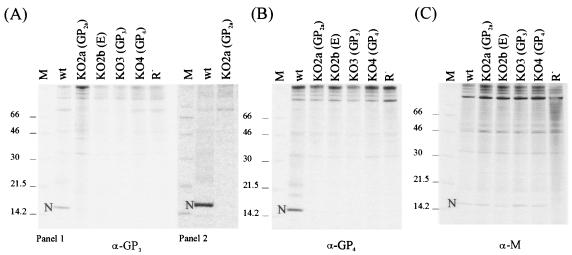
Next, we analyzed the composition of the noninfectious viral particles produced in the absence of one of the minor envelope proteins. As these proteins are extremely difficult to detect in PRRSV preparations due to their low abundance, we used an affinity purification assay to indirectly detect their incorporation into virions using the presence of the N protein as a readout parameter. Intact virus particles released into transfected cell culture supernatants were affinity purified by incubation with MAbs directed against epitopes in the ectodomains of the proteins followed by collection of the particles using protein A-Sepharose. Purification of wild-type particles with either the anti-GP3 or the anti-GP4 antibody revealed an intense N-protein band on SDS-PAGE (Fig. 3A and B). In contrast, application of the procedure to KO2a, KO2b, KO3, and KO4 using the same antibodies did not reveal any detectable N protein (Fig. 3A and B). Note that the faint band at the level of the N protein in the experiment depicted in Fig. 3A (panel 1) was not seen in other experiments (Fig. 3A, panel 2). In parallel, a control experiment was performed with an anti-M antibody to ascertain the production of enveloped viral particles (Fig. 3C). N-protein bands were seen for all constructs, confirming that such particles had indeed been produced. These results indicated that the incorporation of GP3 and GP4 into virus particles requires simultaneous expression of all four minor envelope proteins, suggesting that these proteins are assembled as a multimeric complex. Unfortunately, no reactive antibodies were available for GP2a and E. Thus, given the undetectable low amounts of radioactivity of the different minor proteins in the viral particles, an analysis of the effects of the different gene knockouts on the incorporation of the other minor glycoproteins was not feasible.
FIG. 3.
Composition of virus-like particles in the absence of minor envelope proteins. Radiolabeled virus-like particles were generated and processed as described in the legend to Fig. 2. After sedimentation, virus-like particles were resuspended in PBS and subjected to affinity purification. (A) Purification of particles with MAb anti-GP3 (α-GP3). Panels 1 and 2 show the results of two representative experiments. (B) Purification of particles with MAb anti-GP4 (α-GP4). (C) Purification of particles with MAb anti-M (α-M).
Analysis of putative interactions between the minor envelope proteins.
The results obtained so far point to interactions between the minor proteins as an essential requirement for their particle assembly. To allow an alternative study of these interactions, we completed a set of SFV recombinants by generating SFV-ORF3 (Fig. 1B). The constructs SFV-ORF2 (25) and SFV-ORF4 (23, 25) have already been described. SFV-ORF2 contains the coding sequences for the GP2a protein as well as those for the small E protein. To establish whether the E protein is indeed expressed from this vector, we included it in our complementation experiments. As shown in Table 2, cotransfection of BHK-21 cells with in vitro RNA transcripts of SFV-ORF2 and of full-length construct KO2a or KO2b resulted in successful complementation. These observations indicated that both the GP2a protein and the E protein are correctly expressed from the alphavirus vector, rendering our collection of SFV vectors suitable for studying interactions between the minor proteins.
When the SFV-ORF2 and SFV-ORF4 vectors were separately expressed in BHK-21 cells, the GP2a and GP4 glycoproteins were apparently retained in the ER as their N-glycans remained endoglycosidase H (EndoH) sensitive (25; J. J. M. Meulenberg, unpublished results). Led by this observation, we hypothesized that the minor glycoproteins might acquire transport competence only when assembled into a multimeric complex. We therefore studied their interaction by assessing their ability to be transported through the Golgi complex when coexpressed in different combinations in BHK-21 cells. Transport competence was assessed by analyzing their sensitivity to EndoH and peptide N-glycosidase F (PNGaseF). We started by checking our newly obtained SFV-ORF3 vector and analyzed the independent expression and transport of the GP3 protein in BHK-21 cells. The protein immunoprecipitated from radiolabeled cell lysates migrated in gel at approximately 45 kDa. EndoH treatment reduced the apparent molecular mass to about 29 kDa, consistent with the removal of possibly seven N-linked glycans (Fig. 4A). The GP2a and GP4 proteins have previously been shown to become EndoH resistant over time when expressed in cells infected with LV, being fully EndoH resistant in the extracellular virus particles (23, 25). Analysis of the GP3 protein in LV-infected MARC-145 cells indicated that the GP3 protein behaved slightly different (Fig. 4B and C). Even after a chase time of 300 min, the glycoprotein immunoprecipitated from the infected cell lysate was still largely immature in its oligosaccharides. The GP3 protein appeared as a discrete band of approximately 45 kDa that was reduced to about 29 kDa after treatment with EndoH and PNGaseF (Fig. 4B), similar to the independently expressed protein (Fig. 4A). However, as for GP2d and GP4, in extracellular virions, only the mature, EndoH-resistant form of the GP3 protein was detected (Fig. 4C). Here, the glycoprotein appeared as a fuzzy band of approximately 50 to 60 kDa, presumably due to the extensive and heterogeneous maturation of its N-linked glycans. The apparent size of the virion-derived GP3 was reduced to 29 kDa by removal of the oligosaccharides with PNGaseF, confirming the identity of the GP3 protein (Fig. 4C).
Interactions between GP2a, GP3, and GP4 are required for transport to the Golgi.
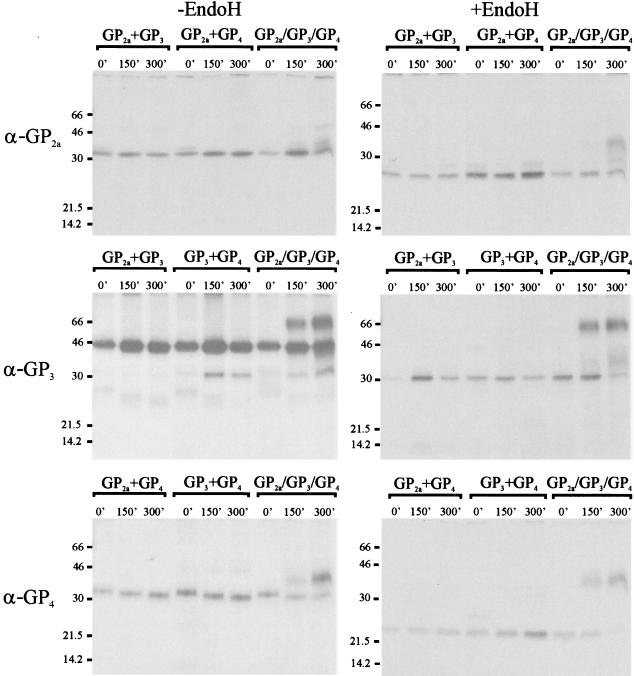
Next, we studied the transport behavior of the minor structural proteins when coexpressed. BHK-21 cells were coinfected with different combinations of the recombinant SFV viruses at an MOI of approximately 5 each to ensure that most cells were infected by two or three different viruses. The degree of infection (90 to 100%) was confirmed in an immunostaining assay (data not shown). Cells were pulse-labeled for 30 min, chased for up to 300 min, and lysed, after which the glycoproteins were immunoprecipitated, and the maturation state of their glycans was analyzed using EndoH. It appeared that in the three possible combinations where two glycoproteins (and the E protein) were coexpressed none of the proteins became EndoH resistant (Fig. 5). Also, when we replaced one of the three recombinant SFV viruses by SFV-ORF5, no maturation was observed for any of the glycoproteins involved (data not shown). Only when GP2a plus E, GP3, and GP4 were expressed together did the majority of the proteins become EndoH resistant after a chase period of 300 min (Fig. 5), indicating that simultaneous expression of all minor glycoproteins (and presumably of the E protein) is essential for their transport. The size of the EndoH-sensitive form of the GP4 protein seemed to decrease slightly during the chase period, irrespective of the presence of other glycoproteins. As the size of the deglycosylated backbone of GP4 did not change, the observed decrease in size might be due to some intermediate changes (trimming) of the N-glycan moieties.
FIG. 5.
EndoH sensitivities of coexpressed GP2a, GP3, and GP4 proteins. BHK-21 cells were coinfected with different combinations of SFV-ORF2a, SFV-ORF3, and SFV-ORF4 as indicated above the lanes. The cells were pulse-chase labeled as described in the legend to Fig. 4A. The cells were lysed, and the GP2a, GP3, and GP4 proteins were subjected to immunoprecipitation with anti-GP2a (α-GP2a), anti-GP3 (α-GP3), or anti-GP4 (α-GP4) antibodies. Precipitates were either left untreated (−EndoH; left-hand panels) or treated with EndoH (+EndoH; right-hand panels).
Interaction of GP2a, GP3, and GP4 leads to secretion of proteins into the medium.
To investigate whether the interactions between the GP2a, GP3, and GP4 proteins resulted in release of the proteins into the extracellular medium, we analyzed the culture supernatants of BHK-21 cells infected with the individual SFV recombinants or coinfected with all three SFV recombinant viruses. Individual expression of the minor glycoproteins did not lead to any detectable GP2a or GP4 in the medium of SFV-ORF2- or SFV-ORF4-infected BHK-21 cells after a 300-min chase (data not shown). In contrast, a small amount of GP3 was precipitated from the medium of BHK-21 cells infected with SFV-ORF3 only (Fig. 6A). Treatment of the immunoprecipitated GP3 protein with EndoH showed that this secreted product was EndoH resistant (Fig. 6A). After treatment with PNGaseF, the size of GP3 was similar to that of the fully deglycosylated form obtained from the cell lysate (Fig. 6B). Analysis of the culture supernatant of BHK-21 cells coinfected with SFV-ORFs 2, 3, and 4 showed that, besides GP3, GP2a and GP4 were also now detected in the culture medium after a chase period of 300 min (Fig. 6C). The GP2a and GP4 proteins were both resistant to EndoH (data not shown) and sensitive to PNGaseF (Fig. 6C). To check whether the appearance of GP2a, GP3, and GP4 in the extracellular medium was not due to lysis of the cells, we performed a control experiment in which we subjected the medium of BHK-21 cells infected with SFV-ORF4 to immunoprecipitation with anti-GP4. No GP4 was detected (data not shown). Hence, we conclude that coexpression of GP2a/E, GP3, and GP4 resulted in transport of these proteins from the ER through the Golgi complex followed by their release into the culture medium.
DISCUSSION
In the present study, we investigated the structural requirements for PRRSV virion formation and infectivity using full-length cDNA clones in which the genes encoding the membrane proteins were disrupted by site-directed mutagenesis. From our results, we draw three main conclusions. First of all, each of the membrane proteins is essential for the production of infectious virus. Second, the GP5 and M protein are both crucial for particle formation, consistent with a role as the fundamental building blocks of the viral envelope. Third, while not required for particle assembly, the minor glycoproteins GP2a, GP3, and GP4, and probably also the minor envelope protein E, interact with each other. This interaction is likely to be of critical importance for their incorporation as heteromultimeric complexes into PRRSV particles and essential for rendering these particles infectious.
Until recently, the GP5 protein and, to a lesser extent, the M protein were considered to have their main function in the targeting of arteriviruses to and their entry into cells. This view changed drastically after Dobbe et al. (5) demonstrated that replacing the ectodomain of the EAV GP5 by that of the homologous proteins of PRRSV and LDV in the context of a full-length cDNA clone did not alter the tropism of the resulting chimeric viruses. Similarly, replacement of the ectodomain of the M protein of PRRSV by that of other arteriviruses also did not change viral tropism (39). While a role of these proteins in cell entry can still not be ruled out (in fact, data by Delputte et al. [4] demonstrate a contribution to PRRSV host cell attachment through their interaction with a heparin-like receptor), the great abundance of GP5-M heterodimers in the viral envelope argues for a primary function of these proteins in viral structure and assembly. The dramatic effect on particle production that we observed here when the synthesis of either of the proteins was disabled strongly supports this view. The disulfide bond between GP5 and M presumably occurs between cysteine residues at positions 50 and 8, respectively (38). Mutating either one of these residues resulted in a complete block of particle production (38), indicating that the covalent association of GP5 and M is crucial for assembly. The same holds true for EAV where the homologous GP5 and M proteins were also found to be required for particle formation (43) and where the essential requirement for heterodimerization between GP5 and M proteins was also shown by mutagenesis of the cysteine residues involved (31).
PRRSV particle assembly is not dependent on the minor proteins. Analysis of viral protein and RNA in the culture media of cells transfected with single gene knockout constructs revealed that in all four cases, RNA-containing particles were released. Except when the E protein was lacking, which led to a significant decrease, the amounts of particles produced were not much lower than the amount of virus obtained with the wild-type construct. Interestingly, the absence of any of the four proteins appeared to affect the incorporation of all three others. Thus, no GP3 was detected in particles when the expression of either GP2a, or E, or GP4 was prevented, and no detectable GP4 was incorporated in the absence of GP2a, E, and GP3. Similar observations were made with EAV. However, though strongly reduced, some E protein continued to be incorporated into EAV particles in the absence of each of the minor glycoproteins (43), a feature we could not establish for PRRSV due to the lack of an E-specific antibody.
The interdependence of the minor PRRSV envelope proteins for their incorporation into virions strongly indicated that these proteins have to associate in infected cells to form heteromultimeric complexes prior to or during their assembly into virions. The low abundance of these proteins in viral particles prevented the direct demonstration of such complexes. Indirect evidence was, however, obtained by using a biological feature, i.e., intracellular transport, as an indicator for their interaction. By analyzing the maturation state of the N-linked oligosaccharides, we observed that the minor glycoproteins were each retained in the ER region of the cell when expressed individually or together with one of the others. They acquired transport competence, however, when they were all coexpressed together, in which case they were collectively carried through the Golgi complex to the plasma membrane and even detected in the extracellular medium. Since GP2a, GP3, and GP4 are envelope proteins, it is most likely that they associated in membranous structures, presumably in the form of particles. Although the identification of these structures was beyond the scope of this study, we performed an electron microscopic analysis of BHK-21 cells in which GP2a, GP3, and GP4 had been coexpressed and of the medium of these cells, but this did not reveal any virus-like particles.
As the ORF encoding the E protein is fully contained within the sequence encoding GP2a, our experiments do not allow conclusions about the specific contribution of the E protein to the interaction process. Our results are consistent with those reported recently for EAV. Here, the occurrence of a covalently linked GP2b/GP3/GP4 complex in virions could be directly demonstrated. Interestingly, while the disulfide linkage between the EAV GP2b and GP4 proteins appeared to be formed intracellularly, the covalent association of GP3, presumably by linking to GP4 (46), occurs after the assembled virus has been released from the infected cell (45). Whether or not the proteins in the PRRSV complex are also covalently linked is presently unclear. In this respect, it is of note that the cysteine residue (Cys102), which in GP2b of EAV is supposedly responsible for the formation of the cystine bridge with GP4 (46), is not conserved in the GP2a protein of PRRSV (10).
As argued above, the GP5-M heterodimers are unlikely to play a predominant role in arterivirus targeting and/or entry. Hence, this function should be attributed to the minor proteins. As these proteins occur in particles as heteromultimeric complexes, they probably constitute the virion-exposed structure that mediates binding of the virion to the primary receptor, which has not yet been identified for any arterivirus except for PRRSV, where a macrophage-restricted sialoadhesin was found to be involved (34). PRRSV binding to sialoadhesin leads to clathrin-mediated internalization of the virion into an endosome. A subsequent drop in pH in the endosome is required for PRRSV replication (17, 28). For other viruses such as orthomyxo- and alphaviruses (14, 29), it has been observed that an acidic pH triggers a conformational change in the fusion protein(s) that ultimately leads to the fusion of the viral and endosomal membranes and the delivery of the nucleocapsid into the cell's cytoplasm. If the arterivirus minor protein complex would indeed be involved in this fusion process, its unique nature and composition would point to a fusion mechanism that is significantly different from the recently described class I and class II mechanisms that drive the fusion of, for instance, influenza viruses (7, 33) and flaviviruses (13), respectively.
The GP3 protein seems to take a special position among the arterivirus glycoproteins. While its membrane topology has not been resolved and remains speculative (12, 44), its virion structural nature is also still controversial. For LDV, the GP3 protein was reported to be a nonstructural, soluble glycoprotein that is secreted from infected cells (9). In contrast, the GP3 protein of EAV was clearly demonstrated to be incorporated into virions (44). For PRRSV, both situations have been described. Thus, the GP3 protein of the prototype European strain LV was found to be incorporated into virions (35), whereas that of North American strains was observed to occur as a secreted protein (sGP3) (11, 19). In the present study, we provided new evidence for the incorporation of the GP3 protein into LV particles, but additionally, we demonstrated the independent secretion of a substantial amount of GP3 by cells expressing the protein from an alphavirus vector. This dual nature of GP3 is reminiscent of glycoproteins of several other viruses such as the glycoproteins G (gG) of alphaherpesviruses (1, 27) and the Erns glycoprotein of BVDV (15). The gG protein of alphaherpesviruses is found in the virus envelope and, in some alphaherpesviruses, is also secreted after proteolytic processing. Recently, these secreted forms of gG were reported to have chemokine binding activity, thereby inhibiting the biological activity of these chemokines in vitro (1). The BVDV Erns protein, in addition to being a virion protein, is also secreted from infected cells into the extracellular environment. This secreted form was recently shown to block the double-stranded RNA interferon-inducing signal (15). Whether the secreted PRRSV GP3 protein is also of physiological relevance remains to be established.
Acknowledgments
We are grateful to Joop de Bree for performing the statistical analyses, to Renate Hakze-van der Honing for performing the real-time RT-PCR experiments, and to Lisette Cornelissen for helpful discussions.
REFERENCES
- 1.Bryant, N. A., N. Davis-Poynter, A. Vanderplasschen, and A. Alcami. 2003. Glycoprotein G isoforms from some alphaherpesviruses function as broad-spectrum chemokine binding proteins. EMBO J. 22:833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 3.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 4.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparin-like receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobbe, J. C., Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288:283-294. [DOI] [PubMed] [Google Scholar]
- 6.Drew, T. W., J. P. Lowings, and F. Yapp. 1997. Variation in open reading frames 3, 4 and 7 among porcine reproductive and respiratory syndrome virus isolates in the UK. Vet. Microbiol. 55:209-221. [DOI] [PubMed] [Google Scholar]
- 7.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. [DOI] [PubMed] [Google Scholar]
- 8.Faaberg, K. S., and P. G. Plagemann. 1995. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology 212:512-525. [DOI] [PubMed] [Google Scholar]
- 9.Faaberg, K. S., and P. G. Plagemann. 1997. ORF 3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology 227:245-251. [DOI] [PubMed] [Google Scholar]
- 10.Godeny, E. K., A. A. de Vries, X. C. Wang, S. L. Smith, and R. J. de Groot. 1998. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 72:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonin, P., H. Mardassi, C. A. Gagnon, B. Massie, and S. Dea. 1998. A nonstructural and antigenic glycoprotein is encoded by ORF3 of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 143:1927-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedges, J. F., U. B. Balasuriya, and N. J. MacLachlan. 1999. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology 264:92-98. [DOI] [PubMed] [Google Scholar]
- 13.Heinz, F. X., and S. L. Allison. 2003. Flavivirus structure and membrane fusion. Adv. Virus Res. 59:63-97. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 15.Iqbal, M., E. Poole, S. Goodbourn, and J. W. McCauley. 2004. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J. Virol. 78:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 17.Kreutz, L. C., and M. R. Ackermann. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res. 42:137-147. [DOI] [PubMed] [Google Scholar]
- 18.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (New York) 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 19.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardassi, H., B. Massie, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98-112. [DOI] [PubMed] [Google Scholar]
- 21.Meulenberg, J. J., J. N. Bos-de Ruijter, G. Wensvoort, and R. J. Moormann. 1998. An infectious cDNA clone of porcine reproductive and respiratory syndrome virus. Adv. Exp. Med. Biol. 440:199-206. [DOI] [PubMed] [Google Scholar]
- 22.Meulenberg, J. J., A. Petersen-den Besten, E. P. de Kluyver, R. J. Moormann, W. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meulenberg, J. J., A. P. van Nieuwstadt, A. van Essen-Zandbergen, and J. P. Langeveld. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 71:6061-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meulenberg, J. J. M., M. M. Hulst, E. J. de Meijer, P. L. J. M. Moonen, A. den Besten, E. P. de Kluyver, G. Wensvoort, and R. J. M. Moormann. 1994. Lelystad virus belongs to a new virus family, comprising Lactate dehydrogenase-elevating virus, Equine arteritis virus, and Simian hemorrhagic fever virus. Arch. Virol. Suppl. 9:441-448. [DOI] [PubMed] [Google Scholar]
- 25.Meulenberg, J. J. M., and A. Petersen-den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44-51. [DOI] [PubMed] [Google Scholar]
- 26.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 27.Murata, T., F. Goshima, H. Takakuwa, and Y. Nishiyama. 2002. Excretion of herpes simplex virus type 2 glycoprotein D into the culture medium. J. Gen. Virol. 83:2791-2795. [DOI] [PubMed] [Google Scholar]
- 28.Nauwynck, H. J., X. Duan, H. W. Favoreel, P. van Oostveldt, and M. B. Pensaert. 1999. Entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages via receptor-mediated endocytosis. J. Gen. Virol. 80:297-305. [DOI] [PubMed] [Google Scholar]
- 29.Peisajovich, S. G., and Y. Shai. 2003. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta 1614:122-129. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez, M. J., J. Sarraseca, J. Fominaya, E. Cortes, A. Sanz, and J. I. Casal. 2001. Identification of an immunodominant epitope in the C terminus of glycoprotein 5 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 82:995-999. [DOI] [PubMed] [Google Scholar]
- 31.Snijder, E. J., J. C. Dobbe, and W. J. M. Spaan. 2003. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 77:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. Raamsman, and A. A. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegmann, T. 2000. Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion. Traffic 1:598-604. [DOI] [PubMed] [Google Scholar]
- 34.Vanderheijden, N., P. L. Delputte, H. W. Favoreel, J. Vandekerckhove, J. van Damme, P. A. van Woensel, and H. J. Nauwynck. 2003. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophages. J. Virol. 77:8207-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Nieuwstadt, A. P., J. J. Meulenberg, A. van Essen-Zandbergen, A. Petersen-den Besten, R. J. Bende, R. J. Moormann, and G. Wensvoort. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Oirschot, J. T., M. J. Kaashoek, M. A. Maris-Veldhuis, K. Weerdmeester, and F. A. Rijsewijk. 1997. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J. Virol. Methods 67:23-34. [DOI] [PubMed] [Google Scholar]
- 37.van Rijn, P. A., G. J. Wellenberg, R. Hakze-van der Honing, L. Jacobs, P. L. Moonen, and H. Feisma. 2004. Detection of economically important viruses in boar semen by quantitative RealTime PCR technology. J. Virol. Methods 120:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verheije, M. H. 2002. Genetic engineering of the porcine reproductive and respiratory syndrome virus: exploration of avenues towards a vaccine, p. 95-104. PhD thesis. Utrecht University, Utrecht, The Netherlands.
- 39.Verheije, M. H., T. J. Welting, H. T. Jansen, P. J. Rottier, and J. J. Meulenberg. 2002. Chimeric arteriviruses generated by swapping of the M protein ectodomain rule out a role of this domain in viral targeting. Virology 303:364-373. [DOI] [PubMed] [Google Scholar]
- 40.Welch, S.-K., R. Jolie, D. S. Pearce, W. D. Koertje, E. Fuog, S. L. Shields, D. Yoo, and J. G. Calvert. 2004. Construction and evaluation of genetically engineered replication-defective porcine reproductive and respiratory syndrome virus vaccine candidates. Vet. Immunol. Immunopathol. 102:277-290. [DOI] [PubMed] [Google Scholar]
- 41.Wensvoort, G., C. Terpstra, J. Boonstra, M. Bloemraad, and D. van Zaane. 1986. Production of monoclonal antibodies against swine fever virus and their use in laboratory diagnosis. Vet. Microbiol. 12:101-108. [DOI] [PubMed] [Google Scholar]
- 42.Wensvoort, G., C. Terpstra, J. M. A. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, F. Wagenaar, J. M. Broekhuijsen, P. L. J. M. Moonen, T. Zetstra, E. A. de Boer, H. J. Tibben, M. F. de Jong, P. van 't Veld, G. J. R. Groenland, J. A. van Gennep, M. T. Voets, J. H. M. Verheijden, and J. Braamskamp. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 43.Wieringa, R., A. A. de Vries, J. van der Meulen, G. J. Godeke, J. J. Onderwater, H. van Tol, H. K. Koerten, A. M. Mommaas, E. J. Snijder, and P. J. Rottier. 2004. Structural protein requirements in equine arteritis virus assembly. J. Virol. 78:13019-13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieringa, R., A. A. de Vries, M. J. Raamsman, and P. J. Rottier. 2002.Characterization of two new structural glycoproteins, GP3 and GP4, of equine arteritis virus. J. Virol. 76:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wieringa, R., A. A. de Vries, and P. J. Rottier. 2003. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 77:6216-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wieringa, R., A. A. F. de Vries, S. M. Post, and P. J. M. Rottier. 2003. Intra- and intermolecular disulfide bonds of the GP2b glycoprotein of equine arteritis virus: relevance for virus assembly and infectivity. J. Virol. 77:12996-13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wissink, E. H., M. V. Kroese, J. G. Maneschijn-Bonsing, J. J. Meulenberg, P. A. van Rijn, F. A. Rijsewijk, and P. J. Rottier. 2004. Significance of the oligosaccharides of the porcine reproductive and respiratory syndrome virus glycoproteins GP2a and GP5 for infectious virus production. J. Gen. Virol. 85:3715-3723. [DOI] [PubMed] [Google Scholar]
- 48.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]
- 49.Zevenhoven-Dobbe, J. C., S. Greve, H. van Tol, W. J. M. Spaan, and E. J. Snijder. 2004. Rescue of disabled infectious single-cycle (DISC) equine arteritis virus by using complementing cell lines that express minor structural glycoproteins. J. Gen. Virol. 85:3709-3714. [DOI] [PubMed] [Google Scholar]