Abstract
Background
Klebsiella pneumoniae producing extended-spectrum β-lactamase (ESBL) colonizing and transmitting in the intestine, especially in children, have significant public health implications. Investigating antibiotic resistance, antibiotic resistance genes (ARGs), virulence factor genes (VFGs), and genetic relationships may help us to explore the characteristics and differences of ESBL-positive K. pneumoniae in children with and without diarrhea.
Methods
After selecting and pairing, 26 pairs of 52 ESBL-positive K. pneumoniae strains were isolated from 323 children with diarrhea and 393 children without diarrhea. Antimicrobial susceptibility test and whole genome sequencing were performed to explore antibiotic resistance, ARGs, and VFGs. The genetic relationship was explored by conducting a maximum likelihood phylogenetic tree and investigating plasmid and sequence type (ST).
Results
All strains showed resistance to cephalosporins, with ESBL-producing genes widely carried (98.1%). Carbapenem-resistant K. pneumoniae (CRKP) were found in both groups. Hypervirulent K. pneumoniae (hvKP) were isolated from children with diarrhea carrying iucA on plasmid. The emergence of ST5670 CRKP and ST2108 hvKP highlighted the necessity for close monitoring of community-acquired K. pneumoniae.
Conclusions
Severe drug resistance was found among ESBL-positive K. pneumoniae strains isolated from children with and without diarrhea. Attention must be paid to ESBL-positive K. pneumoniae colonized in the intestine of children, and pathogen and ARG monitoring in children should be strengthened, even in healthy people.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13099-025-00700-9.
Keywords: ESBL, Klebsiella pneumoniae, Diarrhea, Children
Background
Diarrhea remains a leading cause of mortality in children globally. Over the past two decades, the incidence of diarrhea has remained high, with nearly 1.7 billion cases of childhood diarrheal disease, and 500,000 children under five lose their lives due to diarrhea every year in the world [1, 2]. In China, community-acquired diarrhea is highly prevalent among children, and the high cumulative burden of diarrhea will raise the risk of stunting [3–5]. Bacteria account for 60.0–85.0% of the pathogens in diarrheal cases, and the antibiotic selection for clinical treatment depends on the severity and duration of the infection [6]. Third-generation cephalosporins are commonly employed for treating severe diarrhea caused by Enterobacteriaceae bacteria [7–9]. However, the overuse of antibiotics has accelerated the emergence of antimicrobial resistance (AMR), compromising the efficacy of antibiotics in the treatment of bacterial diarrhea [10]. Bacteria that produce extended-spectrum β-lactamase (ESBL) could develop resistance to cephalosporins and carbapenems, which are major antibiotics in clinical practice [11, 12]. Klebsiella pneumoniae, a human commensal and opportunistic pathogen, is one of causative agents of diarrhea infections, particularly among children with weakened immunity, causing a wide range of nosocomial and community-acquired infections worldwide [13–15]. Though K. pneumoniae is not the main cause pathogen of diarrhea, diarrhea caused by this pathogen has been reported, especially in China. From 2018 to 2021, in a certain district of Beijing, the total detection rate of K. pneumoniae in diarrhea cases was 10.43% (115/1103), of which 66 cases were infected by K. pneumoniae only [16]. Over four years, 322 dirrhea infants caused by K. pneumoniae were discovered in Zhengzhou children’s hospital [17]. ESBL-producing K. pneumoniae, one of the most common and severe multidrug-resistant (MDR) pathogens in hospitals, can cause nosocomial infection outbreaks [15, 18, 19]. Carbapenem is an important antibiotic used in clinical practice, and resistance to carbapenem in K. pneumoniae poses a major threat to public health [20–24]. ESBL-positive K. pneumoniae has been found to be widely carried in hospitals and communities across Europe, Africa, Asia, and other regions [25–27]. ESBL-producing K. pneumoniae constitutes a considerable public health challenge owing to its resistance to a broad spectrum of antibiotics, particularly beta-lactams such as penicillins and cephalosporins [15]. This multidrug resistance (MDR) complicates treatment options, frequently leaving carbapenems as one of the few viable alternatives, although there is a rising incidence of carbapenem-resistant strains. K. pneumoniae is a common etiological agent of healthcare-associated infections, including pneumonia, bloodstream infections, and urinary tract infections, particularly among immunocompromised individuals [24]. The increasing prevalence of ESBL-producing strains highlights the critical need for enhanced antibiotic stewardship, robust infection control measures, and the advancement of novel therapeutic strategies to address the challenges posed by multidrug-resistant pathogens [19, 20]. Hospitalized patients worldwide have been found to have high prevalences of hypervirulent K. pneumoniae (hvKP), multidrug-resistant K. pneumoniae (MDR-KP), and carbapenem-resistant K. pneumoniae (CRKP) [28, 29]. The high prevalence of MDR strains and the plasmid-mediated dissemination of resistance genes found in K. pneumoniae pose significant therapeutic challenges for immunocompromised children under 5 years old [14]. This has led to previously unheard of public health issues, including the emergence of Carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP).
To characterize and compare ESBL-positive K. pneumoniae isolates obtained from paired children with and without diarrhea, an individually matched case-control study was conducted, filtering out 52 paired strains from a total of 716 diarrhea and non-diarrhea samples. And a systematic investigation was conducted to assess antibiotic resistance, identify antibiotic resistance genes (ARGs), detect virulence factor genes (VFGs), determine plasmid incompatibility (Inc) types, and analyze the genetic relationship among strains.
Methods
Sample collection and bacteria isolation
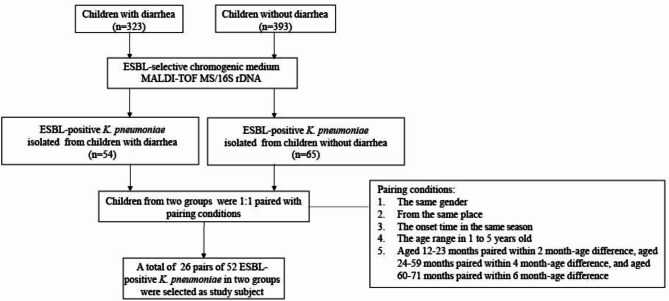
In order to mitigate potential information biases, children’s information and clinical information were collected by standardized trained doctors using a standardized case registration form. Children with diarrhea were all community-acquired cases, characterized by more than three abnormal defecations within a 24-hour period (inclusive). The diarrheal cases were children with mild diarrhea who scored ≤ 10 on the Vesikari 20-point scale, not having bacteremia [30]. Children without diarrhea were either recruited from the community or enrolled in the same hospital as the cases who were hospitalized for reasons other than diarrhea and did not have a history of diarrhea within four weeks. Children were excluded if they were underlying chronic metabolic diseases or if they had received antibiotic treatment within four weeks before enrollment. A total of 716 fresh stool samples from 323 children with diarrhea and 393 children without diarrhea were collected. Children studied were living in the Yangtze River Delta region. The stool samples were cultured on an ESBL-selective chromogenic medium (CHROMagar, France) to select ESBL-producing strains. K. pneumoniae identification was confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Billerica, MA, USA); those that could not be identified by MALDI-TOF MS were identified by 16 S rDNA sequencing. Totally, 54 and 65 ESBL-producing K. pneumoniae strains were isolated from children with and without diarrhea, respectively. Children with diarrhea who were isolating ESBL-positive K. pneumoniae were individually 1:1 matched with children without diarrhea according to children’s age, gender, living region, and onset time. The children were paired with the same gender and from the same city, and the onset time was in the same season. Careful balancing was also conducted for factors such as family conditions, hygiene practices, and nutritional conditions to minimize potential bias. The ages of the children ranged from 1 to 5 years old, and to ensure precise age pairing between children with and without diarrhea, we adhered to a specific age grouping to ensure comparable age distributions between groups and minimize the potential influence of age-related differences on our results [31]. Children aged 12–23 months were paired within a 2-month age difference, aged 24–59 months were paired within a 4-month age difference, and aged 60–71 months were paired within a 6-month age difference (Fig. 1). A total of 26 pairs of children were selected, and 52 ESBL-positive K. pneumoniae strains isolated from them were served as our study subjects.
Fig. 1.
Flowchart for determining two groups of extended-spectrum β-lactamase (ESBL) positive Klebsiella pneumoniae isolates
Antimicrobial susceptibility testing
The antimicrobial susceptibility test (AST) was performed on the ESBL-positive K. pneumoniae isolates using broth microdilution and the Phoenix NMIC-413 AST panel for 25 antibiotics: cefazolin, cefepime, cefoxitin, ceftazidime, ceftriaxone, cefuroxime, amoxicillin-clavulanate, ampicillin-sulbactam, piperacillin-tazobactam, aztreonam ertapenem, imipenem, meropenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, tetracycline, tigecycline, minocycline, colistin, nitrofurantoin, chloramphenicol, and trimethoprim-sulfamethoxazole. The drug resistance of isolates to antibiotics was identified according to the minimum inhibitory concentration with reference to the Clinical and Laboratory Standards Institute (CLSI) criteria M100, 34th Edition. Escherichia coli ATCC 25,922 was used as a quality control strain [32].
DNA extraction and genome sequencing
DNA was extracted using a Wizard Genomic DNA Extraction Kit (Promega, Madison, WI, USA). Paired-end libraries with insert sizes of ~150 bp were constructed using MGIEasy Fragmentase DNA Library Prep module and sequenced on MGI200 platform. Raw reads were filtered using fastp, and the high-quality reads were de novo assembled using SPAdes (version 3.5.0), with additional options of ‘-careful -k 33,55,77’. QUAST (version 5.2.0) was used to evaluate the quality of assemblies, and those with too many contigs or too low N50 values were re-sequenced [32]. The assemblies of 52 K. pneumoniae sequenced in this study were uploaded to GenBank under the project PRJNA1175053 (Supplementary Table S1).
ARGs, VFGs, and plasmid identification
ARGs were identified using Abricate (version 0.8) and compared with the ResFinder database (version 4.1.11). VFGs were identified using Kleborate (version 2.3.2). HvKP was considered since it harbors at least one hypervirulence gene, such as iroB, iucA, peg-344, rmpA, and rmpA2 [33]. The distributions of genes on chromosomes or plasmids were identified by Plasmer (version 0.1). Plasmid replicons were identified with the PlasmidFinder database (version 2022-03-09) using blastn (version 2.10.0).
Phylogenetic relationship analysis
Multi-locus sequence type was determined in silico using Institut Pasteur MLST databases and software (https://bigsdb.pasteur.fr/klebsiella/). New alleles and sequence type (ST) were submitted and approved. Publicly available genomes of 142 ST15, two ST14, and four ST25 K. pneumoniae strains isolated from China were downloaded from GenBank (Supplementary Table S2). A maximum likelihood (ML) phylogenetic tree was inferred using parsnp (version 1.7.2), and pairwise single nucleotide polymorphism (SNP) distances in core genome alignments were identified using pairsnp (version 0.1.0). Isolates with ≤ 21 SNPs were considered to have a genetic relationship [34]. The tree and corresponding metadata information were visualized using iTOL (https://itol.embl.de/).
Statistical analysis
Clustering analysis of AMR phenotypes and ARGs was performed using the OmicStudio tools (https://www.omicstudio.cn/tool). The distribution of phenotypes, ARGs, VFGs, and Inc types in two groups was compared using McNemar tests or Fisher’s exact tests. The number of ARGs distributed on plasmid or chromosome in two groups were compared using a paired-samples T-test, and the comparison of AMR patterns was conducted using non-parametric tests. The Kappa index was used to analyze the consistency between the resistance genotypes and phenotypes with a value range of 0–1 [35]. SPSS (version 21) was used for statistical analyses. Statistical significance was defined as P < 0.05.
Results
Antibiotic resistance features revealed by AST: equally severe in children with and without diarrhea
The AST of all isolates to 25 antimicrobial agents across 10 drug classes revealed high resistance rates to cephalosporins (100.0%), tetracyclines (73.1%), sulfonamides (67.3%), and quinolones (30.8%). Resistance to minocycline was significantly higher in isolates from children without diarrhea (34.62%), compared to those in children with diarrhea (11.5%) (P < 0.05) (Fig. 2).
Fig. 2.
The heatmap and cluster analysis of antibiotic resistance (left) and carrying antibiotic resistant genes (ARGs) (right) were conducted, with the bar chart on the top showing the resistance rate (left) and the carrying rate of ARGs (right) in two groups. The statistical difference was marked in red at the top of the bars. The strains primarily aggregated into three primary clades according to antibiotic-resistant distribution (left) and also clustered in three distinct branches by ARGs (right). Clusters of phenotypes were matched to the clusters of ARGs and connected in different color lines [Heatmap of antibiotic resistance for 25 antibiotic agents including cefazolin (CFZ), cefepime (FEP), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefuroxime (CXM), amoxicillin-clavulanate (AMC), ampicillin/sulbactam (SAM), piperacillin-tazobactam (TZP), aztreonam (ATM), ertapenem (EIP), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin (GEN), tobramycin (TOB), ciprofloxacin (CIP), levofloxacin (LVX), tetracycline (TET), tigecycline (TGC), minocycline (MIN), colistin (COL), nitrofurantoin (NIT), chloramphenicol (CHL), and trimethoprim-sulfamethoxazole (SXT)]
Among K. pneumoniae strains isolated from children with diarrhea, the highest resistance rates were observed for cefazolin (96.2%), followed by cefuroxime (88.5%) and ceftriaxone (88.5%); all strains showed sensitivity to cefoxitin. One CRKP strain was identified, exhibiting resistance to ertapenem. The resistant rates of quinolone antibiotics varied, with the rates from 15.4% (ciprofloxacin) to 26.9% (levofloxacin). All strains tested were sensitive to tigecycline, with 11.5% of the strains were resistant to minocycline and 73.1% to tetracycline. All strains exhibited sensitivity to colistin.
Similarly, the strains isolated from children without diarrhea exhibited high resistance to multiple antibiotics. Resistance to cefazolin (100.0%), cefuroxime (96.2%), and ceftriaxone (92.3%) was prevalent, although all strains remained susceptible to cefoxitin. One strain belonged to CRKP, displaying resistance to ertapenem, imipenem, and meropenem. Among quinolones, the resistant rates of ciprofloxacin and levofloxacin were at 19.2% and 23.1%, respectively. The isolates showed high resistance to tetracycline (73.1%) and minocycline (34.6%), while all isolates were sensitive to tigecycline. The strains were also sensitive to colistin.
The prevalence of MDR ESBL-positive K. pneumoniae was 88.5% in children with diarrhea and 78.8% in children without diarrhea. The most common MDR pattern was cephalosporins—tetracyclines—trimethoprim-sulfamethoxazole. No significant differences were found in MDR prevalence or MDR patterns between the two groups (P > 0.05). All CRKP strains were classified as MDR.
ARG identification highlighted widespread carriage of ESBL-producing genes
A total of 85 ARG types spanning ten classes were identified among all ESBL-positive K. pneumoniae strains, with aph(3”)-Ib, aph-Id, and qnrB91 carrying rates in children with diarrhea higher than in children without diarrhea (P < 0.05). ESBL-producing genes were widely carried (98.1%), with CTX-M being the most common type. Gene blaCTX-M-14 was detected in 36.5% of strains, while blaCTX-M-15 was 18.4%. Unique genes identified in strains from children with diarrhea included blaCTX-M-24, blaLEN, blaSHV-185, and blaSHV-33, along with carbapenem-resistance genes blaIMP-4. In contrast, strains from children without diarrhea uniquely carried blaTEM-1A, blaCTX-M-27, blaCTX-M-65, blaCTX-M-99, and blaNDM-5. The most common quinolone resistant genes were OqxA (76.93%) and qnrS1 (51.93%), while the isolates in children without diarrhea specially harbored qnrB1, qnrB52, and qnrB6.
The clustering results of resistance phenotypes were highly consistent with ARG-based genotype clustering results, with the majority converging into three distinct branches (Fig. 2). The consistency between resistance genotypes and phenotypes for each antibiotic in the two groups was evaluated using the Kappa index value (Supplementary Table S3). In children with diarrhea, consistency ranged from 0.272 (chloramphenicol) to 1.000 (amoxicillin/clavulanate), while in children without diarrhea, it ranged from 0.293 (trimethoprim-sulfamethoxazole) to 0.898 (tetracycline).
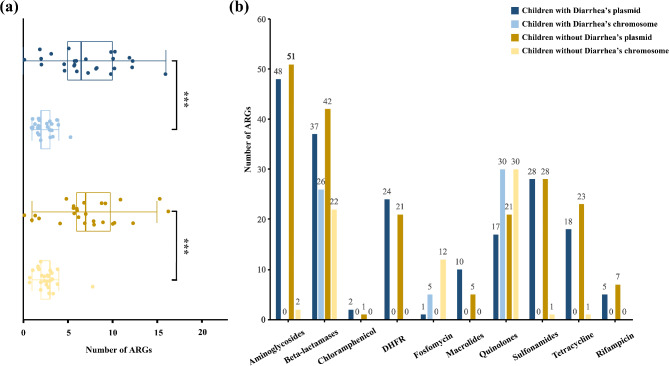
Plasmid prediction analysis indicated that the number of ARGs located on plasmids (1–16, average: 7.16) was significantly higher than those located on chromosomes (1–5, average: 2.52) in both groups (P < 0.05) (Fig. 3a). However, there were no statistically significant differences in the distribution of each type of ARGs between the two groups (P > 0.05) (Fig. 3b). Aminoglycoside, macrolide, sulfonamide, tetracycline, chloramphenicol, rifampicin, and trimethoprimresistant genes were predominantly identified on plasmids, whereas fosfomycin resistance genes were primarily located on chromosomes. Quinolone and β-lactam resistance genes were distributed both plasmids and chromosomes. Specifically, qnr-type quinolone resistance genes were located on plasmids, while Oqx were predominantly identified on chromosomes. For β-lactamase genes, CTX-M, TEM, DHA, and LAP types were mainly identified on plasmids, while SHV, OKP, and LEN were mainly located on chromosomes. The carbapenem-resistance genes blaIMP-4 was plasmid-borne, and blaNDM-5 was carried by an IncX3 plasmid.
Fig. 3.
The distribution of ARGs of two groups on plasmid and chromosome was showed. (a) The number of ARGs carried by each strain was shown in children with and without diarrhea distributed on plasmid or chromosome, and there was no difference between groups (P > 0.05), but a significant difference can be found on distributing positions (P < 0.05); (b) The distribution of ARGs for each type of antibiotics was illustrated in two groups. Different ARGs had varying preferences for chromosomes or plasmids. Aminoglycoside, macrolide, sulfonamide, tetracycline, chloramphenicol, rifampicin, and trimethoprim resistant genes were predominantly found on plasmids, whereas fosfomycin resistance genes were primarily located on chromosomes. Quinolone and β-lactam resistance genes were distributed on both plasmids and chromosomes, exhibiting distinct subtype variations
VFGs identification detected HvKP carrying IucA in children with diarrhea
Seven strains carried concerning VFGs and had the probability to synthesize pathogenic factors, with four found in children with diarrhea (15.4%) and three in children without diarrhea (11.5%). Notably, two strains in children with diarrhea carrying iucA were classified as hvKP (3.8%), with iucA located on the plasmid IncFIB. Additionally, yersiniabactin-producing virulence gene cluster, dispersed on chromosome, was identified in two strains from children with diarrhea (7.7%) and three strains from children without diarrhea (11.5%).
Plasmid analysis identified incfib as the main type of plasmid carrying HvKP VFGs
A total of 14 plasmid Inc types were identified in 52 isolates. Antibiotic resistance plasmids were carried by 51 strains (98.1%), virulence plasmids were carried by two strains (7.7%), and plasmids with both were carried by two strains, though virulence genes and ARGs were not present on the same plasmid. IncFIB (78.8%) was the most prevalent Inc type, followed by repB (28.8%) and IncFII (26.9%) (Supplementary Figure S1). No statistically significant differences were observed in plasmid types between the two groups (P > 0.05). Several kinds of ARGs, aadA2, dfrA12, sul1, and blaCTX-M-14, were located on the IncFIB plasmid, while ESBL-producing genes blaTEM-1B and blaCTX-M-15 were identified on the IncFII plasmid.
Phylogenetic analysis revealed the most common ST belonged to clone group 15
Six novel sequence types (STs) were submitted and named from ST6903 to ST6908. Among the 52 isolates, 45 different STs were observed, with clone group 15 containing ST15 and ST14 being the most prevalent (Supplementary Figure S2). ML phylogenetic tree and pairwise SNP distances based on core genome alignments demonstrated that ST15 strain obtained in our study clustered mainly with CRKP strains, and may share a possible genetic relationship with a strain isolated from a patient in Hunan in the same year (SNP = 58) (Fig. 4a). Two strains of ST14 in our study clustered with two ST14 CRKP strains isolated in China; however, the high SNP distance (SNP ≥ 1670) suggested no direct genetic relationship among them (Fig. 4b). The hvKP strains were identified as ST2108 and ST25, while the CRKP strains belonged to ST5670 and ST20. The ST25 hvKP strain in our study may be genetically distant from other ST25 hvKP strains in China (SNP ≥ 349) (Fig. 4c).
Fig. 4.
Maximum likelihood (ML) phylogenetic analysis of strains. (a) One strain of ST15 from our study clustered with 142 ST15 K. pneumoniae strains, most of which were carbapenem-resistant. The strain from our study showed a potential genetic relationship with a strain isolated from a patient in Hunan in the same year (SNP = 58); (b) Two ST14 strains from our study were clustered with two carbapenem-resistant K. pneumoniae (CRKP) ST14 strains isolated from China. The high SNP distance (SNP ≥ 1670) indicated no direct genetic relationship among them; (c) One ST25 strain from our study clustered with four K. pneumoniae strains of ST25 from China. The strain isolated in our study and three strains in Genbank were hypervirulent K. pneumoniae (hvKP), with no CRKP identified. The ST25 hvKP strain in our study may be genetically distant from other hvKP ST25 strains (SNP ≥ 349); (d) All strains from our study were clustered in two branches. The heatmap of antibiotic resistance, carried ARGs, and carried vital virulent genes were followed
Phylogenetic relationship between strains isolated from children with and without diarrhea
Based on the SNPs in the core genomes, ML phylogenetic analysis revealed that all isolates formed two distinct branches; CRKP (1/2, 50.0%) and hvKP (2/2,100.0%) were in clade B. Two ST14 strains isolated from children with diarrhea exhibited a small SNP distance (SNP = 12), suggesting a potential genetic relationship. Two ST585 strains isolated from children with and without diarrhea showed no SNP differences (SNP = 0) and the onset time were close (in five days), indicating a possible epidemiological relationship (Fig. 4d).
Discussion
ESBL-producing K. pneumoniae drives nosocomial and community-acquired infections among children worldwide [14, 36]. This study revealed the characteristics and difference of resistance, ARGs and VFGs, plasmid Inc type, sequence type, and the genetic relationship of paired ESBL-positive K. pneumoniae strains isolated from children with and without diarrhea.
All isolated strains in this study were resistant to cephalosporins, with 90.4% resistance to ceftriaxone, a third-generation cephalosporin. This finding aligns with the high ceftriaxone resistance rate (96.8%) reported for ESBL-producing K. pneumoniae isolated from hospitals in China [37]. In addition, almost all strains carried ESBL-producing genes (98.1%). The CTX-M enzymes are the most common ESBL type in strains isolated from hospital-associated and community-acquired infections, with CTX-M-15 dominating worldwide, followed by CTX-M-14 [38]. CTX-M-15-producing genes are mainly carried by the IncF plasmid group, including IncFII [39]. In our study, CTX-M enzymes were also predominant, with CTX-M-14 (36.5%) and CTX-M-15 (18.4%) being the most frequently detected. We also found that CTX-M-15 was carried by IncFII plasmids, the major Inc type of plasmid in our study. The rate of resistance, MDR and ARGs carried is unexpectedly high in both children with and without diarrhea, and this issue could be a result of the extensive use of antibiotics and mobile genetic elements transmission. The MDR rate and the proportion of strains co-resistant to both β-lactam and quinolone were slightly higher in children with diarrhea, and the resistance to tobramycin was only found in children with diarrhea. While in children without diarrhea, the resistance to imipenem and meropenem was found, and the resistance to minocycline was higher (P < 0.05) than children with diarrhea. Antibiotic resistant K. pneumoniae is more likely to survive and colonize in intestinal tract under the selective pressure of antibiotic treatment, even making it difficult to treat clinically. Therefore, it is imperative to monitor resistance patterns, the resistance transmission, and clinical antibiotic usage, while also prioritizing surveillance of ESBL-positive K. pneumoniae prevalence in both children with and without diarrhea [14, 40, 41]. Based on the “One Health” approach, the high rates of drug resistance and widespread carriage of ARGs should be concerned not only in children with diarrhea, but also in healthy children.
ST11 and ST15 are major STs in China and worldwide, with ST15 emerging as a high-risk clone frequently causing hospital outbreaks and the dominant sequence type of CRKP bloodstream infections in northeast China [42, 43]. While ST11 was not detected in our study, ST15 strain resistant to third-generation cephalosporins and carried SHV-28 on the chromosome was isolated from healthy children and mainly clustered with CRKP strains isolated from China. Some strains of ST15 mediating carbapenem resistance, and many carrying virulence plasmids, indicated that ST15 as important clone group promotes the global spread of resistance genes and virulence genes [44]. Therefore, it is necessary to strengthen the monitoring of ST15 in China.
A high prevalence of CRKP has been increasingly reported in hospitalized patients worldwide [28, 45]. In this study, the prevalence of CRKP was relatively low (3.8%), below the resistance rate (10.0%) reported by the national bacteria drug resistance monitoring system [46]. Notably, CRKP was not only detected in children with diarrhea but also in healthy children. Moreover, a novel ST5670 CRKP was first identified worldwide, isolated from children with diarrhea. The carbapenem gene blaNDM-5 carried by plasmid IncX3 was identified in healthy children. Plasmid IncX3, known for its strong cross-species transmission potential [47, 48], may facilitate the spread of blaNDM-5 among strains.
Recently, the World Health Organization issued its latest report warning that hvKP has spread worldwide [49]. The carrying rate of hvKP in healthy people was from 4 to 5.19% [50]. In this study, two hvKP strains carrying VFGs iucA on plasmid IncFIB were isolated from children with diarrhea. Other hypervirulent genes iro and iuc repoted could also be carried and transmitted broadly by plasmid IncFIB in hvKP [51]. ARGs, ESBL-producing genes blaSHV and blaCTX-M, were also found in hvKP and were carried on chromosomes and plasmids, respectively. The VFGs and ARGs were carried by plasmids harbored in hvKP, and the horizontal transfer of virulence plasmid across strains may be one of the reasons for diarrhea [52]. The ST2108 hvKP was found in China for the first time. Thus, it’s also critical to keep an eye on the spread of enteric pathogens and virulent plasmids in diarrheal patients, especially in pediatric populations. If children are infected by hvKP, the CRKP strain colonized in the gut may acquire the plasmid carrying iucA, which may facilitate iron acquisition and promote gut inflammation, allowing it to develop into hv-CRKP and exacerbating mucosal damage in immunocompromised hosts [33]. It is critical to control the colonization and dissemination of K. pneumoniae, especially CRKP, hvKP, and even hv-CRKP.
Due to the relatively small sample size and the single-center design of this study, the generality of the findings to other populations may be limited. To address this, the multi-center study will be conducted in the future.
Conclusions
We conducted a comprehensive analysis of the drug resistance phenotypes, genomic characteristics, and genetic relationships of ESBL-positive K. pneumoniae isolated from children with and without diarrhea. The findings highlight the carriage of ARGs and VFGs in ESBL-positive K. pneumoniae are serious not only in children with diarrhea but also in those without diarrhea, underscoring the need for enhanced surveillance of drug-resistant bacteria and ARGs in children, including healthy individuals. Improved monitoring of pathogenic bacteria and ARGs in both diarrheal and healthy pediatric populations is essential to mitigate the spread of resistant and hypervirulent strains.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- ESBL
Extended-spectrum β-lactamase
- ARG
Antibiotic-resistance gene
- VFG
Virulence factor gene
- ST
Sequence type
- CRKP
Carbapenem-resistant K. pneumoniae
- hvKP
Hypervirulent K. pneumoniae
- AMR
Antimicrobial resistance
- MDR
Multidrug-resistant
- MDR-KP
Multidrug-resistant K. pneumoniae
- hv-CRKP
Carbapenem-resistant hypervirulent K. pneumoniae
- Inc
Incompatibility
- MALDI-TOF MS
Matrix-assisted laser desorption ionization-time of flight mass spectrometry
- AST
Antimicrobial susceptibility test
- ML
Maximum likelihood
- SNP
Single nucleotide polymorphism
Author contributions
Y. Z.: Conceptualization, Methodology, Investigation, Writing-Original Draft, Visualization. M. W.: Investigation, Methodology, Validation, Writing-Review and Editing. Z. L.: Methodology, Validation, Writing-Review and Editing. Y. P.: Visualization, Supervision. Y. Y.: Visualization, Supervision. X. L.: Investigation, Methodology. Z. L.: Visualization, Supervision. B. K.: Resources, Supervision, Funding Acquisition. M. Z.: Conceptualization, Resources. X. L.: Conceptualization, Resources, Supervision, Writing-Review and Editing, Funding Acquisition. All authors have reviewed and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (grant numbers: 2022YFC2303900, 2024YFC3406305), and the major projects of the National Natural Science Foundation of China (grant numbers: 22193064).
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request. The assemblies of 52 K. pneumoniae sequenced in this study were uploaded to GenBank under the project PRJNA1175053.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by Ethical Committee of National Institute for communicable disease control and prevention (protocol code ICDC-2017003, 3 Jul. 2017) (protocol code ICDC-2017004, 3 Jul. 2017).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mei Zeng, Email: zengmeigao@aliyun.com.
Xin Lu, Email: luxin@icdc.cn.
References
- 1.Ali HA, Nuh AM, Abdi HA, et al. Multilevel analysis of prevalence and determinants of diarrhea among under-five children in Somalia: insights from the Somalia demographic and health survey 2020. BMC Public Health. 2025;25(1):202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Diarrhoeal disease 2024 [Available from: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease
- 3.Dal Canto G, Artaiga THE, Mohamed AI et al. Amoebic dysentery complicated by hypovolemic shock and Sepsis in an infant with severe acute malnutrition: A case report. Microorganisms. 2023;11(1). [DOI] [PMC free article] [PubMed]
- 4.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and National life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388(10053):1459–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Checkley W, Buckley G, Gilman RH, et al. Multi-country analysis of the effects of diarrhoea on childhood stunting. Int J Epidemiol. 2008;37(4):816–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries RM, Linscott AJ. Practical guidance for clinical microbiology laboratories: diagnosis of bacterial gastroenteritis. Clin Microbiol Rev. 2015;28(1):3–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gramundi I, Albornoz E, Boutureira M, et al. Characterization of third generation cephalosporin-resistant Escherichia coli clinical isolates from Ushuaia, Argentina. Rev Argent Microbiol. 2023;55(1):43–8. [DOI] [PubMed] [Google Scholar]
- 8.Ingle DJ, Levine MM, Kotloff KL, et al. Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa. Nat Microbiol. 2018;3(9):1063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jomehzadeh N, Ahmadi K, Nasiri Z. Evaluation of biofilm formation and antibiotic resistance pattern in Extended-Spectrum β-Lactamase-Producing Escherichia coli strains. Biomedical Biotechnol Res J (BBRJ). 2022;6(2):175–9. [Google Scholar]
- 10.Tribble DR. Antibiotic therapy for acute watery diarrhea and dysentery. Mil Med. 2017;182(S2):17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamma PD, Aitken SL, Bonomo RA, et al. Infectious diseases society of America 2022 guidance on the treatment of Extended-Spectrum β-lactamase producing enterobacterales (ESBL-E), Carbapenem-Resistant enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat resistance (DTR-P. aeruginosa). Clin Infect Dis. 2022;75(2):187–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jomehzadeh N, Ahmadi K, Shaabaninejad H, et al. Plasmid-mediated AmpC β-Lactamase gene analysis in Klebsiella Pneumoniae clinical isolates. Biomedical Biotechnol Res J (BBRJ). 2022;6(4):582–5. [Google Scholar]
- 13.Prokesch BC, TeKippe M, Kim J, et al. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2016;16(9):e190–5. [DOI] [PubMed] [Google Scholar]
- 14.Akenten CW, Khan NA, Mbwana J, et al. Carriage of ESBL-producing Klebsiella pneumoniae and Escherichia coli among children in rural Ghana: a cross-sectional study. Antimicrob Resist Infect Control. 2023;12(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jomehzadeh N, Ahmadi K, Bahmanshiri MA. Investigation of plasmid-mediated quinolone resistance genes among clinical isolates of Klebsiella pneumoniae in Southwest Iran. J Clin Lab Anal. 2022;36(7):e24342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Zhao J, Liu C, et al. Pathogenic and molecular characteristics of Klebsiella pneumoniae in fecal samples from diarrhea cases in a district of Beijing in 2018–2021. Chin J Zoonoses. 2024;40(8):745–957. [Google Scholar]
- 17.Wang XH, Zhou WH, Su J. Observation of occurance of infant diarrhea caused by KIebsiella pneumoniae. J Med Forum. 2005;26(12):24–5. [Google Scholar]
- 18.Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185–8. [DOI] [PubMed] [Google Scholar]
- 19.Jomehzadeh N, Saki M, Ahmadi K, et al. The prevalence of plasmid-mediated quinolone resistance genes among Escherichia coli strains isolated from urinary tract infections in Southwest Iran. Mol Biol Rep. 2022;49(5):3757–63. [DOI] [PubMed] [Google Scholar]
- 20.Jomehzadeh N, Jahangirimehr F, Chegeni SA. Virulence-associated genes analysis of carbapenemase-producing Escherichia coli isolates. PLoS ONE. 2022;17(5):e0266787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian X, Zheng X, Sun Y, et al. Molecular mechanisms and epidemiology of Carbapenem-Resistant Escherichia coli isolated from Chinese patients during 2002–2017. Infect Drug Resist. 2020;13:501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z, Liu L, Ye L, et al. Genomic insight into the distribution and genetic environment of bla(IMP-4) in clinical carbapenem-resistant Klebsiella pneumoniae strains in China. Microbiol Res. 2023;275:127468. [DOI] [PubMed] [Google Scholar]
- 23.Budia-Silva M, Kostyanev T, Ayala-Montaño S, et al. International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nat Commun. 2024;15(1):5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jomehzadeh N, Rahimzadeh M, Ahmadi B. Molecular detection of extended-spectrum β-lactamase- and carbapenemase-producing Klebsiella pneumoniae isolates in Southwest Iran. Trop Med Int Health. 2024;29(10):875–81. [DOI] [PubMed] [Google Scholar]
- 25.Ballén V, Gabasa Y, Ratia C, et al. Antibiotic resistance and virulence profiles of Klebsiella pneumoniae strains isolated from different clinical sources. Front Cell Infect Microbiol. 2021;11:738223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hijazi SM, Fawzi MA, Ali FM, et al. Prevalence and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in healthy children and associated risk factors. Ann Clin Microbiol Antimicrob. 2016;15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng L, Liu Z, Liu C, et al. The distribution characteristics of global blaOXA-carrying Klebsiella pneumoniae. BMC Infect Dis. 2023;23(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chia JH, Chu C, Su LH, et al. Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M beta-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol. 2005;43(9):4486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hala S, Malaikah M, Huang J, et al. The emergence of highly resistant and hypervirulent Klebsiella pneumoniae CC14 clone in a tertiary hospital over 8 years. Genome Med. 2024;16(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Huang Y, Xue P, et al. A duplex droplet digital PCR assay for Salmonella and Shigella and its application in diarrheal and non-diarrheal samples. Int J Infect Dis. 2022;120:210–6. [DOI] [PubMed] [Google Scholar]
- 31.Kotloff KL, Blackwelder WC, Nasrin D, et al. The global enteric multicenter study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis. 2012;55(Suppl 4):S232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X, Luo M, Wang M, et al. High carriage and possible hidden spread of multidrug-resistant Salmonella among asymptomatic workers in Yulin, China. Nat Commun. 2024;15(1):10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo TA, Olson R, Fang CT, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56(9). [DOI] [PMC free article] [PubMed]
- 34.David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy-Zane MS, Pyle L. Reliability of the anterior humeral line index compared with the Gartland classification for posteriorly hinged Supracondylar humerus fractures. Orthopedics. 2018;41(4):e502–5. [DOI] [PubMed] [Google Scholar]
- 36.Le T, Wang L, Zeng C, et al. Clinical and Microbiological characteristics of nosocomial, healthcare-associated, and community-acquired Klebsiella pneumoniae infections in Guangzhou, China. Antimicrob Resist Infect Control. 2021;10(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Zhou K, Zheng B, et al. High prevalence of ESBL-Producing Klebsiella pneumoniae causing Community-Onset infections in China. Front Microbiol. 2016;7:1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castanheira M, Simner PJ, Bradford PA. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob Resist. 2021;3(3):dlab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol. 2013;39(1):79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo QX, Lu P, Chen YB et al. ESKAPE in China: epidemiology and characteristics of antibiotic resistance. Emerg Microbes Infec. 2024;13(1). [DOI] [PMC free article] [PubMed]
- 41.Li H, Yan SJ, Li DD, et al. Trends and patterns of outpatient and inpatient antibiotic use in China’s hospitals: data from the center for antibacterial surveillance, 2012-16. J Antimicrob Chemoth. 2019;74(6):1731–40. [DOI] [PubMed] [Google Scholar]
- 42.Feng L, Zhang M, Fan Z. Population genomic analysis of clinical ST15 Klebsiella pneumoniae strains in China. Front Microbiol. 2023;14:1272173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Y, Yang Y, Feng Y, et al. Prevalence and clonal diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal infections: A systematic review of 128 articles across 30 countries. PLoS Med. 2023;20(6):e1004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y, Jiang T, He X, et al. Global phylogeography and genomic epidemiology of Carbapenem-Resistant bla(OXA-232)-Carrying Klebsiella pneumoniae sequence type 15 lineage. Emerg Infect Dis. 2023;29(11):2246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Jiang X, Yang T, et al. Genomic epidemiology of Carbapenemase-producing Klebsiella pneumoniae in China. Genomics Proteom Bioinf. 2022;20(6):1154–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.the national monitoring network. 2022 national bacteria bacterial drug resistance monitoring report (briefly) 2023 [Available from: https://www.carss.cn/Report/Details/917
- 47.Findlay J, Poirel L, Kessler J, et al. New Delhi Metallo-β-Lactamase-Producing enterobacterales bacteria, Switzerland, 2019–2020. Emerg Infect Dis. 2021;27(10):2628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Yuan S, Xuan L, et al. Emergence of a novel sequence type carbapenem-resistant hypervirulent Klebsiella pneumoniae ST6417 harboring bla(NDM-5) on the lncX3 plasmid. Microbiol Spectr. 2024;12(10):e0098424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Health Organization. Disease Outbreak News; Antimicrobial Resistance, Hypervirulent Klebsiella pneumoniae - Global situation 2024 [Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON527
- 50.Yang J, Li Y, Tang N, et al. The human gut serves as a reservoir of hypervirulent Klebsiella pneumoniae. Gut Microbes. 2022;14(1):2114739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam MMC, Wyres KL, Judd LM, et al. Tracking key virulence loci encoding aerobactin and Salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y, Wu CY, Wang BJ et al. Characterization difference of typical KL1, KL2 and ST11-KL64 hypervirulent and carbapenem-resistant Klebsiella pneumoniae. Drug Resist Update. 2023;67. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request. The assemblies of 52 K. pneumoniae sequenced in this study were uploaded to GenBank under the project PRJNA1175053.