Abstract
Feeding and reproduction are known to be closely correlated with each other, and the seasonal breeders show breeding season-dependent feeding behavior. However, most model animals do not have definite breeding seasonality, and the mechanisms for such feeding behavior remain unclear. Here, we focused on female medaka (Oryzias latipes); they show breeding season-dependent feeding behavior, and their condition of breeding season can be experimentally controlled by day-length. We first demonstrated that, among previously reported feeding-related peptides (neuropeptides involved in feeding), agouti-related peptide 1 (agrp1) and neuropeptide y b (npyb) show higher brain expression under the breeding condition than under the non-breeding one. Combined with analysis of agrp1 knockout medaka, we obtained results to suggest that long day-induced sexually mature condition, especially ovarian estrogenic signals, increase the expressions of agrp1 in the brain, which results in increased food intake to promote reproduction. Our findings advance the understanding of neural mechanisms of feeding behavior for reproductive success.
Research organism: Other
Introduction
Feeding behavior is essential to animals for their survival and reproduction and is known to be modulated by various internal and external factors: nutritional status, sexual maturity, temperature, seasonality, etc. This behavior is known to be closely correlated with reproduction (Kauffman and Rissman, 2004), which is an essential biological activity important for the animal life. Previous studies reported that nutritional-state modulates reproductive behaviors and functions (Chen et al., 2006; Amirjani et al., 2019; Volk et al., 2017; Lynn et al., 2010). For example, musk shrews show defective sexual behavior under fasted conditions (Temple and Rissman, 2000). In addition, not a few studies demonstrated that fasting-induced low energy condition suppresses reproduction (Evans and Anderson, 2012; Kalra and Kalra, 1996; Kirkwood et al., 1987; Merry and Holehan, 1979; Hasebe et al., 2016). Thus, it has been well investigated how nutritional status resulting from feeding modulates reproduction. On the other hand, it has been reported that some animals change their feeding behavior during the breeding season. For instance, the black seabream migrates to the shallow water during the breeding season (Tsuyuki, 2018; Kawai et al., 2020) where they can get more food, and the white-tailed deer spends more time for feeding under reproductive status (Stone et al., 2017). Such a close relationship between reproduction and feeding is thought to be important for biological fitness. However, the regulatory mechanisms for breeding season-dependent feeding behavior are still unknown. One possible reason is that most of the model animals appear to have lost the well-defined breeding season. Although the mammalian models, mice and rats, and teleost model zebrafish, have reproductive cycles of about 4–5 days (Nilsson et al., 2015; Peute et al., 1978), they do not have definite breeding seasonality. Thus, the mechanisms for long-term changes in feeding behavior according to the breeding season have not yet been studied in detail.
Here, as a model animal for the seasonal breeder, we used a teleost fish, medaka (Oryzias latipes). Medaka is a useful model animal, whose reproductive status can be experimentally controlled by day-length (Robinson and Rugh, 1943; Egami, 1954) while keeping an appropriate temperature constant. In the long-day (LD) condition (14 h light/10 h dark), female medaka becomes reproductive and regularly spawns every day, while it becomes non-reproductive in the short-day (SD) condition (10 h light/14 h dark). In other words, LD or SD condition can induce breeding or non-breeding season of female medaka, respectively. Thus, medaka enables us to analyze the mechanisms of breeding season-dependent feeding behavior without consideration for possible changes in metabolism and gene expressions due to the changes in ambient temperature, which means medaka is suitable for this study.
Regulatory mechanisms of feeding behavior have mainly been analyzed in mammals. These studies reported that some neuropeptides, such as agouti-related peptide (AgRP) and neuropeptide Y (NPY), are involved in the control of feeding and called ‘feeding-related peptides’ as key molecules for the regulation of feeding behavior (Hahn et al., 1998; Aponte et al., 2011; Krashes et al., 2011; Andermann and Lowell, 2017). Teleosts have also been thought to possess a regulatory mechanism for feeding similar to mammals. In fact, expression of homologous genes coding for feeding-related peptides have been reported (Rønnestad et al., 2017; Conde-Sieira and Soengas, 2016). On the other hand, although administration of some of them have been suggested to induce feeding behavior in teleosts as well (Rønnestad et al., 2017), their functions in feeding behaviors still remain unclear.
In the present study, to understand mechanisms of breeding season-dependent feeding behavior, we focused on female medaka, which clearly show seasonal changes in breeding conditions by day length (Mitani et al., 2010; Kanda et al., 2008) under the fixed appropriate temperature. We first quantified changes in feeding behavior according to the breeding states and found that female medaka under the condition of breeding season (LD) eat more than those under the condition of non-breeding (SD). Therefore, we searched for genes that show breeding state-dependent changes in expression and found some candidates for feeding-related peptides in medaka. We then analyzed expressions of the candidate genes by using RNA-seq, in situ hybridization (ISH), and RT-qPCR, and analyzed phenotypes of gene knockout medaka. These results led us to conclude that AgRP1 plays a key role in the breeding season-dependent changes in feeding behavior via ovarian estrogenic signals.
Results
Feeding behavior of female medaka is upregulated in the breeding season
To analyze food intake of male and female medaka in breeding/non-breeding seasons, we first established a method for measuring food intake in medaka. In brief, we placed medaka in a white cup, fed brine shrimp to medaka in all-you-can-eat style for 10 min, and counted the leftover brine shrimp in the cup with a ‘shrimp-counter’ system (called Japanese ‘Wanko-soba’-like method, Figure 1—source code 1 and Figure 1—figure supplement 1). We used this system to analyze food intake of male and female medaka under the breeding condition equivalent to that in the breeding season (kept under LD condition) or under the non-breeding condition equivalent to that in non-breeding season (SD condition) (Figure 1A; Kanda et al., 2008). We found that female medaka under the breeding (LD) condition eat more than those under the non-breeding (SD) condition (Figure 1B; p=0.02519). In contrast to female, in males there was no significant difference in food intake between the breeding and non-breeding condition (Figure 1C; p=0.6540). Since these results demonstrated that females, not males, show breeding season-dependent feeding behavior, we focused only on female medaka in the following analyses on neuronal mechanism for breeding season-dependent feeding behavior. Next, to examine which gene products modulate feeding behavior of female medaka in the breeding season, we performed mRNA-sequencing (RNA-seq) using the whole brain of female medaka in breeding condition (LD) and non-breeding condition (SD) (Figure 1—figure supplement 2A). Overall, 1025 genes showed significantly different expression between LD and SD female medaka. Figure 1—figure supplement 2B shows a heat map of representative genes mainly related to neuroendocrine system, which were differently expressed between LD and SD females. Among the conventional candidate feeding-related neuropeptides, we identified two kinds of neuropeptides, agrp1 and npyb, both of which showed higher expression in LD than in SD (Figure 1—figure supplement 2C and D). Both AgRP and NPY are known to have orexigenic effects mainly in mice (Schwartz et al., 2000; Andermann and Lowell, 2017). Therefore, in the subsequent analyses, we focused on agrp1 and npyb as candidate genes that modulate breeding season-dependent feeding behavior in female medaka.
Figure 1. Reproductive female medaka show larger amount of food intake.
(A) Light conditions for breeding and non-breeding status. (B) Food intake (10 min) of female medaka in long day (LD) (orange; n=7) and SD (blue; n=7) conditions, normalized to the amount of artemia eaten by medaka in LD (breeding) condition (p=0.02519, U=42.5). (C) Food intake (10 min) of male medaka in LD (orange; n=7) and short day (SD) (blue; n=7) conditions, normalized to the amount of artemia eaten by medaka in LD (breeding) condition (p=0.6540, U=20.5). Mann–Whitney U test, *p<0.05. n.s., not significant.

Figure 1—figure supplement 1. 'Wanko-soba’ method for calculation of the amount of food intake of fish.

Figure 1—figure supplement 2. Whole-brain gene expression in long day (LD) (n=3) and short day (SD) (n=3) female medaka.

AgRP1, NPYa, and NPYb may be the ‘feeding-related peptides’ in female medaka
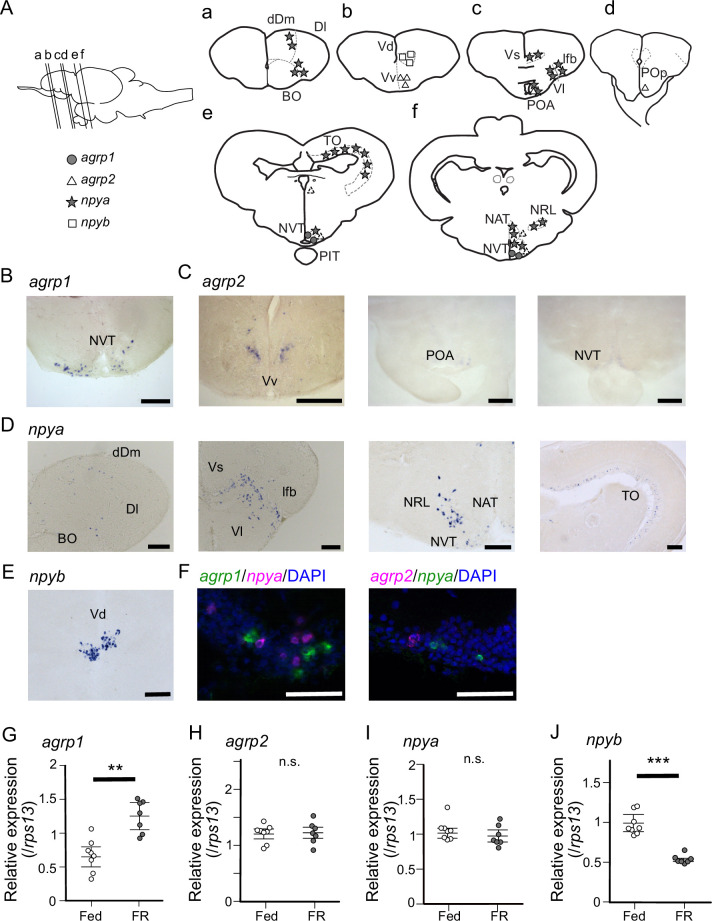
Medaka has two agrp paralogues, agrp1 and agrp2, and two npy paralogues, npya and npyb, which arose from third round whole genome duplication early in the teleost lineage (Liu et al., 2019; Sundström et al., 2008). Therefore, we next examined the anatomical distribution of neurons expressing agrp1, npyb, and their paralogs in the female brain by in situ hybridization (ISH). We found that agrp1- and npyb-expressing neurons are distributed in local brain regions (Figure 2A, B and E), while npya- and agrp2- expressing neurons are more widely distributed (Figure 2A, C and D) in the brain. The agrp1 neurons were distributed in the nucleus ventralis tuberis (NVT) of the hypothalamus (Figure 2B), while agrp2 neurons were expressed in the telencephalon and in the hypothalamus (Figure 2C). On the other hand, npya neurons were distributed more widely from telencephalon to hypothalamus (Figure 2D). npyb neurons were distributed locally in the nucleus ventralis telencephali pars dorsalis (Vd) of the telencephalon (Figure 2E).
Figure 2. agrp1- and npyb-expressing neurons are distributed locally, while npya- and agrp2-expressing neurons are distributed more widely in the brain.
(A) Illustration of the lateral view of medaka brain and distributions of cell bodies expressing each agrp or npy gene. The oblique lines labeled with (a–e) indicate the level of the frontal sections in (a–e). (a–e) Illustrations of frontal sections showing the distribution of neurons expressing each gene. The localization of neurons is indicated in the right half of the illustrations. BO, bulbus olfactorius; dDm, dorsal region of area dorsalis telencephali pars medialis; Dl, area dorsalis telencephali pars lateralis; lfb, lateral forebrain bundle; NAT, nucleus anterior tuberis; NRL, nucleus recessus lateralis; NVT, nucleus ventralis tuberis; PIT, pituitary; POA, area preoptica; POp, nucleus preopticus pars paravocellularis; TO, tectum opticum; Vd, area ventralis telencephali pars dorsalis; Vl, area ventralis telencephali pars lateralis; Vs, area ventralis telencephali pars supracommissuralis; Vv, area ventralis telencephali pars ventralis; agrp1 (gray circle), agrp2 (open triangle: high expression, dotted triangle: low expression), npya (star), npyb (open square). (B) agrp1-expressing neurons are localized in NVT. Scale bar: 100 μm. (C) agrp2-expressing neurons are observed in Vv, POA, and NVT. Scale bar: 100 μm. (D) npya-expressing neurons are distributed in dDm, Dl, BO, Vs, Vl, lfb, NRL, NAT, NVT, and TO. Scale bar: 100 μm. (E) npyb-expressing neurons are localized in Vd. Scale bar: 100 μm. (F) Left: agrp1 (green) and npya (magenta) are distributed in NVT, but the two genes are not co-expressed. Right: agrp2 (magenta) and npya (green) are distributed in NVT, but the two genes are not co-expressed. Scale bars: 50 μm. (G–J) agrp and npy expressions in the whole brain of female medaka under normally fed condition (Fed; white; n=7) or 2-week food restricted (FR; gray; n=8). (G) agrp1 (p=0.001243, U=2), (H) agrp2 (p=0.9551, U=29), (I) npya (p=0.2319, U=39), and (J) npyb (p=0.0003108, U=56). Mann–Whitney U test, **p<0.01, ***p<0.001. n.s., not significant.
Figure 2—figure supplement 1. The number of npya-expressing neurons and the expression of agrp2 in the hypothalamus of fed and food-restricted (FR) medaka.
In mice, agrp is known to be only expressed in the hypothalamus and mostly co-expressed with npy (Hahn et al., 1998), and these AgRP/NPY neurons are known to regulate mammal feeding behavior (Shutter et al., 1997; Broberger et al., 1998; Ollmann et al., 1997; Takahashi and Cone, 2005). In medaka, on the other hand, agrp1 signals were not observed in npya neurons (Figure 2F, left), although the both genes were expressed in the hypothalamus. In addition, agrp2 signals were not observed in hypothalamic npya neurons (Figure 2F, right), either. These results suggest that AgRP and NPY are not co-expressed in medaka. Since AgRP and NPY of medaka showed different expressing patterns compared with other animals such as mice, we examined whether they act as modulators of feeding. We divided female medaka in LD condition into two groups; one group was kept under normally fed condition (Fed), and the other was kept under 2-week food restricted condition (FR). We then analyzed whole-brain expressions of these four genes. RT-qPCR analysis demonstrated that 2-week FR increased the expression of agrp1 (Figure 2G; p=0.001243) but decreased that of npyb (Figure 2J; p=0.0003108), suggesting that the two peptides are involved in feeding in an opposite manner. On the other hand, agrp2 did not significantly change their expressions between Fed and FR conditions (Figure 2H; p=0.9551). Although npya did not significantly change their expressions between Fed and FR conditions (Figure 2I; p=0.2319), it may be possible that npya expression changed in a specific brain region, since npya neurons are widely distributed in various brain regions as described above (Figure 2D). Since it is suggested that NPY released from hypothalamic npy-expressing neurons controls food intake in mice (Kohno and Yada, 2012), we also examined the npya-expression in medaka hypothalamus by ISH. We counted npya-expressing neurons in each hypothalamic region and compared them between Fed and FR female medaka (Figure 2—figure supplement 1). We found that the total number of npya-expressing neurons in the hypothalamus was significantly larger in Fed compared with FR (Figure 2—figure supplement 1A; p=0.02857). Here, significant increase in cell number was observed in nucleus recessus lateralis (NRL) (p=0.02857) and nucleus anterior tuberis (NAT) (p=0.02857), but not in NVT (p=0.1143) (Figure 2—figure supplement 1B–D). On the other hand, the expression of agrp2 did not show remarkable difference in the hypothalamus under food restriction or not (Figure 2—figure supplement 1E). Thus, the results suggest that AgRP1, NPYa, and NPYb may be the ‘feeding-related peptides’ in female medaka.
Both agrp1 and npyb show higher expression levels in LD than in SD female medaka
To further examine the result of RNA-sequencing (Figure 1—figure supplement 2), we compared expression of agrp1 and npyb between the female medaka under the breeding condition (LD) and those under the non-breeding condition (SD) using ISH and whole-brain RT-qPCR (Figure 3). First, we performed whole-brain RT-qPCR and found that the expression level of agrp1 was higher in LD than in SD female (Figure 3A, p=0.001865). In ISH experiments, we observed larger number of agrp1-expressing neurons in LD than in SD females (Figure 3B and C; p=0.02828, Figure 3—figure supplement 1). Since the expression level of agrp1 was higher in LD than that in SD (Figure 3A), higher expression of agrp1 under the breeding condition may be due to the increase in the number of neurons expressing agrp1. On the other hand, npyb expression in RT-qPCR was significantly higher in LD than that in SD (Figure 3D; p=0.0001554), although ISH analysis demonstrated that npyb-expressing cell number was not significantly different between LD and SD (Figure 3E; p=0.4206). These results suggest that the expression level for each neuron increased in LD compared with SD. Thus, higher expression of npyb under the breeding condition may be due to the increase of expressions in each neuron expressing npyb.
Figure 3. agrp1 and npyb show higher expression levels in long day (LD) than in short day (SD) female.
(A) agrp1 expression in the whole brain of LD (orange; n=8) and SD (blue; n=8) female medaka (p=0.001865, U=60). (B) In situ hybridization (ISH) of agrp1-expressing neurons in LD and SD female medaka. Scale bar: 100 μm. (C) The number of neurons expressing agrp1 in LD (orange; n=5) and SD (blue; n=5) (p=0.02828, U=23). (D) npyb expression in the whole brain of LD (orange; n=8) and SD (blue; n=8) female medaka (p=0.0001554, U=64). (E) The number of neurons expressing npyb in LD (orange; n=5) and SD (blue; n=5) (p=0.4206, U=17). The upper, middle, and lower bars show the third quartile, median, and the first quartile, respectively. Mann–Whitney U test, *p<0.05, **p<0.01, ***p<0.001. n.s., not significant.
Figure 3—figure supplement 1. Time course of the number of neurons showing in situ hybridization (ISH) signals for agrp1.
In juvenile female medaka, expression levels of neither agrp1 nor npyb show significant change according to the day-length
The results thus far indicates that expressions of agrp1 and npyb are upregulated in female medaka under the condition of breeding season. Since the breeding/non-breeding state is experimentally controlled by day-length (LD/SD) in the present study, we examined which of the two factors, day-length itself or substance(s) from LD-induced mature ovary, regulates the expression of agrp1 and npyb. Here, we used sexually immature juvenile medaka and compared their whole-brain expressions of agrp1 and npyb under LD/SD conditions using RT-qPCR (Figure 4). We found that expression levels of neither agrp1 nor npyb show significant difference between LD and SD (Figure 4A [p=0.4179] and Figure 4B [p=0.3357]). Furthermore, food intake of juvenile female was not different between LD and SD (Figure 4—figure supplement 1; p=0.7197). These results suggest that neither of them is regulated directly by day-length itself. Instead, the gene expression is suggested to be regulated by LD-induced sexual maturity.
Figure 4. In juvenile female medaka, expression levels of neither agrp1 nor npyb show significant change according to the day-length.
(A) agrp1 expression in the brain of juvenile female medaka (p=0.4179, U=21). (B) npyb expression in the brain of juvenile female medaka (p=0.3357, U=19). Long day (LD): orange, n=8; short day (SD): blue, n=7. The upper, middle, and lower bars show the third quartile, median, and the first quartile, respectively. Mann–Whitney U test. n.s., not significant.
Figure 4—figure supplement 1. Food intake of juvenile female does not show significant change according to the day-length.
Estrogen, which is released from mature ovary, may affect the expression of agrp1
Among various factors associated with ovarian maturity, estrogens are known to be abundantly released from mature ovary and play important roles in reproductive readiness, sexual behavior, and so on Jennings and de Lecea, 2020; Naftolin et al., 2007; Adachi et al., 2007; Clarkson and Herbison, 2009; Wintermantel et al., 2006; Micevych and Meisel, 2017; Melo and Ramsdell, 2001. Among the ovarian estrogens, 17β-estradiol (E2) is the major factor important for reproduction (Kanda et al., 2011; Kelly and Qiu, 2010), and the blood E2 concentration of LD-conditioned female medaka is higher than those of SD (Ikegami et al., 2022). Thus, we hypothesized that E2 regulates the expression of agrp1 and npyb under the condition of breeding season. We analyzed the expression of agrp1 and npyb in sham-operated (Sham), ovariectomized (OVX, fish with surgical ablation of the ovary), and OVX medaka with E2-administration (OVX+E) (Figure 5A and B). The OVX medaka were allowed to survive at least for 2 weeks to clear the endogenous E2 (Kayo et al., 2020), and spawning of all the Sham medaka were confirmed for three consecutive days. By using whole-brain RT-qPCR, we found that OVX induces significantly lowered agrp1 expression than Sham, and OVX+E shows a tendency to recover agrp1 expression compared with OVX (Figure 5A; Sham vs OVX: p=0.04310, OVX vs OVX+E: p=0.05790, Sham vs OVX+E: p=0.2000), which suggests that the ovarian E2 regulates agrp1 expression. On the other hand, the expression levels of npyb did not show significant differences among the three groups (Figure 5B; Sham vs OVX: p=0.1386, OVX vs OVX+E: p=0.9991, Sham vs OVX+E: p=0.08120). In addition, food intake of OVX female was not significantly different between LD and SD (Figure 5—figure supplement 1; p=0.7308), which suggests that ovarian signal may be important for breeding season-dependent feeding behavior. Therefore, we focused more on the estrogenic regulation of agrp1 expression.
Figure 5. Estrogen, which is secreted from mature ovary, may affect the expression of agrp1.
(A) Relative expression of agrp1 in Sham (white; n=6), OVX (ovariectomized medaka, gray; n=6), and OVX+E (OVX medaka kept in the tank containing E2, yellow; n=7). Sham vs OVX: p=0.04310, OVX vs OVX+E: p=0.05790, Sham vs OVX+E: p=0.2000. (B) Relative expression of npyb in Sham, OVX, and OVX+E. Sham vs OVX: p=0.1386, OVX vs OVX+E: p=0.9991, Sham vs OVX+E: p=0.08120. The upper, middle, and lower bars show the third quartile, median, and the first quartile, respectively. Steel–Dwass test, *p<0.05. n.s., not significant. (C) Photographs of brain slices after experiments of double in situ hybridization (agrp1 [green] and estrogen receptors [magenta]). (D) Expanded photograph of agrp1 and esr2a co-expressing neurons (white arrowhead). Scale bars: 50 µm.

Figure 5—figure supplement 1. Food intake of OVX female does not show significant change according to the day-length.

Estrogens act mainly by interacting with estrogen receptors (Chen et al., 2022). Medaka has three kinds of estrogen receptors; esr1, esr2a, and esr2b (Tohyama et al., 2016; Kinoshita et al., 2009), and all of them have been reported to be expressed in NVT (Zempo et al., 2013), in which agrp1 was also expressed (Figure 2B). Therefore, we examined co-expression of these esr genes and agrp1. As shown in Figure 5C and D, esr2a signal was clearly co-expressed in some agrp1-expressing neurons, which strongly suggests that E2 affects the expression of agrp1 via esr2a in those neurons of NVT.
agrp1−/− female medaka show a decrease in food intake and in the number of fertilized eggs
Our present experimental evidence thus far highlights the importance of agrp1 as the factor modulating the season-dependent feeding behavior in medaka. To analyze the function of AgRP1 in medaka, we generated knockout medaka of agrp1 (agrp1−/−) by using CRISPR/Cas9. The designed CRISPR guide RNA cleaved targeted sites of agrp1 (exon3, Figure 6—figure supplement 1A), and we obtained agrp1−/− medaka, which has lots of amino acid changes in functional site for AgRP1 (Figure 6—figure supplement 1B). In agrp1−/− brain, AgRP1 immunoreactive signals, which were observed in WT, were not found (Figure 6—figure supplement 1C). These suggested that agrp1−/− possess nonfunctional AgRP. As for phenotype of the knockout, the agrp1−/− female medaka appeared skinny and the body weight was significantly lower than that of agrp1+/+ (Figure 6A; body weight: p=0.04113; abdominal length: p=0.002165). In addition, abdominal height of agrp1−/− was also smaller than that of agrp1+/+, while the body length was not significantly different (Figure 6A; body length: p=0.3939). We next analyzed food intake of agrp1−/− female medaka in LD condition (breeding). As shown in Figure 6B (p=0.004329) and Figure 6—figure supplement 2 (agrp1+/+ vs agrp1−/+: p=0.5470, agrp1+/+ vs agrp1−/−: p=0.009353, agrp1−/+ vs agrp1−/−: p=0.01234), we found that LD agrp1−/− female medaka eat less than agrp1+/+. Then, we kept agrp1−/− medaka in LD or SD condition and compared their food intake. In contrast to agrp1+/+, agrp1−/− in LD condition did not show a significant increase in food intake compared with SD (Figure 6C; p=0.5953). We also examined whether loss of AgRP1 affects reproductive function. Whereas the agrp1−/− females were fertile, the pairs of agrp1−/− female and agrp1+/+ male resulted in fewer spawned eggs than agrp1+/+ pairs (Figure 6D; p=0.008658). In addition, the ovarian size of agrp1−/− appeared to be smaller than agrp1+/+ (Figure 6E, left). In particular, since relative ovarian weight normalized by body weight (gonadosomatic index [GSI]) of agrp1−/− female tended to be marginally smaller than agrp1+/+ (Figure 6E, right; p=0.06494), the smaller body size of agrp1−/− (Figure 6A) may drastically affect ovarian morphology. Since the number of spawned eggs was decreased in agrp1−/− female, we analyzed gene expressions of gonadotropins which should affect ovarian maturation. Oocyte maturation and ovulation are known to be regulated by gonadotropins, follicular-stimulating hormone (FSH) and luteinizing hormone (LH). As shown in Figure 6F, agrp1−/− females showed lower levels of expression of gonadotropin genes (fshb and lhb; lhb: p=0.008658; fshb: p=0.02597), which suggests that loss of function of agrp1 impaired breeding season-dependent feeding behavior and led to attenuation of reproductive functions, especially the decrease in number of spawned eggs in the breeding season.
Figure 6. agrp1−/− female medaka show decrease in food intake and the number of fertilized eggs.
(A) Lateral views of representative agrp1+/+ and agrp1−/− female medaka (left), and body weight, body length, and abdominal height (right) of agrp1+/+ (white; n=6) and agrp1−/− (gray; n=6). All the fish are 4-month-old adult medaka. Scale bar: 1 cm. body weight: p=0.04113, U=31; body length: p=0.3939, U=24; abdominal height: p=0.002165, U=36. (B) Food intake (10 min) of agrp1+/+ (white; n=6) and agrp1−/− (gray; n=6) female medaka. Each amount of food intake is normalized by the average number of that of agrp1+/+ medaka (p=0.004329, U=35). (C) Food intake (10 min) of LD agrp1−/− (white; n=9) and SD agrp1−/− (gray; n=7) female medaka. Each amount of food intake is normalized by the average number of that of LD agrp1−/− medaka (p=0.5953, U=37). (D) The number of eggs spawned by agrp1+/+ (white; n=6) and agrp1−/− (gray; n=5) female medaka in a day. Each female was paired with a wildtype male (p=0.008658, U=28.5). (E) Photograph of ovary in representative agrp1+/+ and agrp1−/− female (left) and the gonado-somatic index (GSI, right). n=6 of each group (p=0.06494, U=30). Scale bar: 1 mm. (F) Expression of gonadotropin genes (lhb and fshb) in the pituitary of agrp1+/+ (white; n=6) and agrp1−/− (gray; n=6) female medaka. lhb: (p=0.008658, U=34); fshb: (p=0.02597, U=32). The upper, middle, and lower bars show the third quartile, median, and the first quartile, respectively. Mann–Whitney U test, *p<0.05, **p<0.01. n.s., not significant.

Figure 6—figure supplement 1. Mutation site of agrp1 knockout medaka.

Figure 6—figure supplement 2. Food intake of WT, agrp1 hetero, and homo knockout medaka.

Discussion
In the present study, we took advantage of female medaka, which clearly shows breeding season-dependent feeding behavior and found that neuropeptides, agrp1 and npyb, show higher expression under the breeding condition than under the non-breeding condition. We also obtained results to suggest that the expression of both agrp1 and npyb changes depending on nutritional status of female medaka. In addition, ovariectomy and E2 administration changed expression of agrp1 but not npyb, suggesting that increased release of ovarian E2 in the breeding season upregulates the agrp1 expression, which results in the facilitation of female feeding behavior. Finally, loss-of-function mutation of AgRP1 decreased the amount of food intake and the number of spawned eggs. The present results suggest that breeding season-dependent feeding behavior can be modulated by the increased expression of agrp1 upregulated by increased release of ovarian estrogen in the breeding season (Figure 7). To date, not a few previous reports have shown the influence of nutritional status on reproduction (Evans and Anderson, 2012; Kalra and Kalra, 1996; Kirkwood et al., 1987; Merry and Holehan, 1979; Hasebe et al., 2016). On the other hand, although seasonal breeders have been reported to show changes in feeding behavior during the breeding season (Tsuyuki, 2018; Kawai et al., 2020), its neuroendocrine mechanisms have largely remained enigmatic. Our present results may provide a neuroendocrinological model for the mechanisms that play a key role in the control of breeding season-dependent feeding behavior in teleosts.
Figure 7. Illustration of mechanisms of breeding season-dependent feeding behavior in medaka suggested by the present study.

(A) Long day (LD) condition in the breeding season induces ovarian maturation, which facilitates release of estrogen (E2) from the mature ovary. (B) High concentration of serum E2 increases agrp1 expression in the brain via the estrogen receptors, especially, esr2a. (C) Higher expression of AgRP1 is suggested to activate neural circuitry for feeding, which leads to an increase in food intake and egg spawning of female medaka in the breeding season.
Feeding-related peptides AgRP and NPY in medaka
Here, we demonstrated that female medaka eat more under the condition of breeding season (Figure 1B). Various kinds of neuropeptides in the brain have been suggested to control feeding, and these are generally called ‘feeding-related peptides’ (Funahashi et al., 2003). In the present study, we first used a seasonally breeding model teleost medaka and searched for the ‘feeding-related peptides’ involved in seasonal feeding behavior. A whole-brain RNA-seq analysis using female medaka under the breeding condition (LD) and non-breeding condition (SD) revealed two kinds of feeding-related peptides, agrp1 and npyb, which show different expression levels between LD and SD (Figure 1—figure supplement 2). In mammals, AgRP and NPY are known to have orexigenic function and are co-expressed in hypothalamic neurons (Schwartz et al., 2000; Hahn et al., 1998). Previous studies in mammals (Schwartz et al., 2000; Andermann and Lowell, 2017; Muroi and Ishii, 2016) have suggested neural mechanisms of appetite including functions of AgRP and NPY. However, such mechanisms in non-mammalian vertebrates such as teleosts (Rønnestad et al., 2017; Blanco and Soengas, 2021) have not yet been clarified. Our present study using medaka has shown possible functions of AgRP and NPY in teleost feeding behavior, especially in a breeding season-dependent manner.
Our present study using medaka showed that female medaka express agrp1 in hypothalamus, and food restriction increases the agrp1 expression (Figure 2B and G). It has been reported that leptin receptor-knockout medaka show higher food intake and higher expressions of agrp1 and npya than wild type, whereas the expression of agrp2 and npyb remained to be analyzed (Chisada et al., 2014). Zebrafish has also been used as a model animal in teleosts. In zebrafish, food restriction increased agrp1 (Song et al., 2003; Opazo et al., 2018) expression, and transgenic overexpression of agrp1 caused gain of body weight (Song and Cone, 2007), as in mammals (Graham et al., 1997; Adam et al., 2002; Hahn et al., 1998; Ilnytska and Argyropoulos, 2008). It has also been reported that agrp1 knockout zebrafish eat less than the wild type (Shainer et al., 2019), although loss of AgRP in mice showed little effect on food intake (Qian et al., 2002). Our present results and these previous studies strongly support that agrp1 regulates feeding and may act as an orexigenic factor in teleosts. On the other hand, agrp2 neurons were distributed mainly in telencephalon, and its weak expressions were also observed in the POA and the hypothalamus (Figure 2C), which is different from the results in zebrafish (Shainer et al., 2017). In the present study, food restriction did not remarkably affect the agrp2 expression in medaka (Figure 2, Figure 2—figure supplement 1E), and AgRP2 in zebrafish is suggested to play an important role in stress response, not feeding (Shainer et al., 2019). Thus, it is highly probable that agrp2 is involved in functions other than feeding in female medaka.
Furthermore, we demonstrated that npya is expressed in multiple brain regions including hypothalamus in medaka (Figure 2D), which is similar to mammals (Gray and Morley, 1986) and zebrafish (Yokobori et al., 2012; Jeong et al., 2018). We also showed that npya is not co-expressed with agrp1 nor agrp2 (Figure 2F) as in the zebrafish (Jeong et al., 2018), suggesting that the relationship between NPY and AgRP of teleosts may be different from that of mammals, in which most of the agrp-expressing neurons co-express npy (Hahn et al., 1998). Moreover, the present study also showed that npyb expression is localized in telencephalon (Figure 2E), which is similar to the previous report using tiger puffer (Kamijo et al., 2011). Previous studies of NPY in zebrafish showed that zebrafish has only one type of NPY (NPYa) (Söderberg et al., 2000; Larsson et al., 2009) and has lost NPYb during evolution. Like in mammals (Marks et al., 1992; Clark et al., 1984; Glenn Stanley et al., 1986; Marks and Waite, 1997; Baldock et al., 2009), food restriction in zebrafish increased the npya expression in the hypothalamus (Song et al., 2003; Opazo et al., 2018), and intracerebroventricular administration of NPYa increased food intake (Yokobori et al., 2012). Although these zebrafish studies suggest that NPYa may increase food intake, it is still debatable since body weight was not significantly different between npya knockout zebrafish and wild type (Shiozaki et al., 2020). Interestingly, by analyzing both npya and npyb expression in medaka of different nutritional conditions, we found that food restriction decreased the npya-expressing cell number in the hypothalamus (Figure 2—figure supplement 1) and npyb expression level (Figure 2J). These changes in npya and npyb expressions are not consistent with previous studies using other conventional model animals described above (Yokobori et al., 2012; Marks et al., 1992; Clark et al., 1984; Glenn Stanley et al., 1986; Marks and Waite, 1997; Baldock et al., 2009). The present study may suggest that the function of npy may be different among teleosts. In addition, npyb expression was increased under the breeding condition (LD), while LD female showed increase in food intake (Figure 3F). Thus, decreased expression of npyb by food restriction (Figure 2J) may suggest that the change in npyb expression reflects nutritional condition in medaka. Thus, future study of npya and npyb functions in the control of feeding will be necessary.
As described above, we found that agrp1, npya, and npyb change expression levels in response to nutritional status. Among these three genes, we suggest that agrp1 most probably affects relatively long-term feeding in the breeding season, which agrees well with the recent studies in mice showing the function of AgRP as a long-term orexigenic factor. In mice, it has been reported that intracerebroventricular administration of AgRP increases food intake for 1 week (Hagan et al., 2000), and stimulation of receptors expressed in AgRP neurons triggers AgRP release, leading to an increase in food intake for 3 days (Nakajima et al., 2016). The present study also suggests a long-term (seasonal) orexigenic effect of AgRP in teleosts and may also provide an important insight into the understanding of common regulatory mechanisms of feeding by AgRP among various animal species.
High concentration of E2 in the breeding season facilitates agrp1 expression
Our results suggest that agrp1 and npyb show higher expressions under the breeding condition (LD) (Figure 3), but the experiments using juvenile female medaka (Figure 4) showed that expression levels of these two genes do not change according to the day-length itself but to the LD-induced sexual maturity. In addition, the present results indicate that the ovarian estrogen E2 upregulates agrp1 expression mainly via the estrogen receptors esr2a that are co-expressed in some population of agrp1 neurons in the hypothalamic nucleus NVT (Figure 5). Since LD female medaka (breeding) shows high blood concentration of E2 (Ikegami et al., 2022), this pathway may be important for breeding season-dependent feeding behavior. Especially, in teleost, main egg protein for nutrition is vitellogenin, whose expression is also facilitated by E2 (Tohyama et al., 2017). Taken together, it is suggested that E2 may synchronously regulate amount of food intake and female-specific reproductive signals (vitellogenin production and oocyte maturation), which plays a key role in reproductive success in oviparous animals.
In mammals, previous studies have reported on inconsistent effects of ovary and E2 on feeding. Ablation of ovary caused suppression of food intake in mice (Yu et al., 2020), whereas it caused no change in rats (Roesch, 2006). On the other hand, administration of E2 decreased food intake in both mice (Yu et al., 2020) and rats (Roesch, 2006). It should be noted that these laboratory rodents only exhibit short estrous cyclicity and have lost breeding seasonality, and the blood E2 concentration drastically changes in a few days (Nilsson et al., 2015). Thus, it is possible that the control mechanisms of feeding may be different between animals with short estrous cyclicity and those with breeding seasonality.
Furthermore, the present study suggests different control mechanisms of feeding between the animals with breeding seasonally and those without. Here, we showed that E2 directly modulates agrp1 expression via esr2a receptors co-expressed in the agrp1 neurons (Figure 5C), while in mice, AgRP/NPY neurons are reported to be suppressed by E2 indirectly via esr1 (erα)-expressing Kiss1 neurons located in the hypothalamic arcuate nucleus (Qiu et al., 2018; Dubois et al., 2016; Yang et al., 2017). On the other hand, in medaka, expressions of all kinds of estrogen receptors are reported to be localized in NVT (Zempo et al., 2013), in which agrp1 expression is also localized (Figure 2B). In addition, esr2a has been reported to be involved in the feedback regulation of follicle-stimulating hormone in the pituitary and in the development of oviduct, and esr2a knockout females are completely infertile (Kayo et al., 2019). Our hypothesis that estrogen signaling via esr2a affects agrp1 expression may highlight another important function of esr2a for reproduction, while a possibility still remains that esr1- and esr2b-expressing neurons also affect agrp1 expression indirectly.
AgRP1 changes feeding behavior depending on LD-induced sexual maturity, which causes the increase in food intake in the breeding season
Since our results thus far indicate the importance of agrp1, which shows upregulated expression directly stimulated by the ovarian E2 in the breeding season, we examined phenotypes of agrp1 knockout (agrp1−/−) medaka. We found that agrp1−/− medaka under the condition of breeding season eat less (Figure 6B) and spawn a smaller number of eggs (Figure 6D) than WT. Furthermore, the agrp1−/− females did not show significant difference of food intake in LD and SD (Figure 6C). These results strengthen our hypothesis that agrp1 is involved in the increased food intake in the breeding season. Furthermore, agrp1−/− female displayed light body weight (Figure 6A), accompanied by smaller ovary (Figure 6E) and low level of expression of gonadotropins, fshb and lhb (Figure 6F), which are considered to have caused smaller number of spawned eggs. All of these results support our hypothesis that AgRP1 plays an important role in the breeding season-dependent feeding behavior, which culminates in normal reproduction.
In summary, by using a seasonal breeder medaka, we found evidence to suggest that long day-length facilitates ovarian maturation and E2 release, which upregulates agrp1 expression of hypothalamic neurons to activate neural circuitry for feeding behavior and boost oocyte maturation. We propose that this kind of positive feedback control may be important for animals that spawn many eggs every day in the breeding season (Figure 7); medaka needs plenty of food for the production of many eggs. In other words, the metabolic costs of producing eggs on a daily basis in medaka would inevitably require increased food intake. Indeed, previous study showed a need for high food intake for reproduction (Hasebe et al., 2016). It is expected that future studies will elucidate whether or not the present findings in medaka are applicable to other seasonal breeders as well.
Materials and methods
Animals
Female and male wild-type d-rR medaka (O. latipes) and agrp1 knockout (agrp1−/−) medaka were maintained in pairs or shoals at 27℃. Fish were fed three times a day with brine shrimp and flake food (Otohime B-2; San-u Fish Farm, Osaka, Japan). Their reproductive status was controlled by day-length (LD [14 h light/10 h dark]: reproductive, SD [10 h light/14 h dark]: non-reproductive). The light-on time was 8:00 AM. We used juvenile medaka (~5 weeks after fertilization) and adult medaka (>3 months after fertilization). Female medaka, which spawned at least three consecutive days, were used as reproductive ones. For the analysis of the effect of food restriction, reproductive and non-reproductive female medaka were fasted for 14 days or 10 days. Note that all medaka survived after food restriction. For the analyses of food intake, we food-restricted medaka for 6 h after 10 min feeding in the morning and sampled their whole brains for subsequent experiments. Food-restricted medaka were sampled at the same time as the other fed medaka. All experiments and fish maintenance were conducted in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan) and the protocols approved by the Animal Care and Use Committee of Graduate School of Science, the University of Tokyo (permission number, 17-1, 20-6), and the Animal Care and Use Committee of Graduate School of Agriculture, Tokyo University of Agriculture and Technology (permission number, R05-15, R06-27).
Food intake assay
Each 6 h food-restricted medaka was put into a white cup with 100 mL breeding water and was habituated for 5 min. Then, we fed medaka by application of 200 μL aliquots of food water containing brine shrimp in all-you-can-eat style and serve another aliquot once done with it, which is repeated N times (like the Japanese ‘Wanko soba’; so-called Japanese ‘Wanko soba’ method). Then, 10 min after the start, we stopped feeding medaka and placed a magnetic bar to stir the breeding water so that the shrimp concentration will be constant. Then, we collected 10 mL aliquot from the experimental cup by using a micro pipette and transferred it to a conical tube. The conical tube was frozen overnight, and the leftover brine shrimp sunk in the bottom were counted by ‘shrimp-counter’. We counted the number of brine shrimp in the 200 μL solution three times before and after the experiments, and the average number was used. The food intake was calculated as follows.
(Food Intake) = (The average number of brine shrimp in the solution) * (number of aliquots, N)
- (number of leftover brine shrimp sunk in the bottom) * 10
Food intake was normalized by the average of LD or WT medaka.
‘Shrimp-counter’ system
The number of shrimps in the solution was counted using OpenCV3 library (Intel, Santa Clara, CA) run under a Python script (Figure 1—source code 1). This script was run under Anaconda 4.4.0 for Windows running Python 3.5.
RNA-sequencing
We collected two whole brains of LD or SD female in one tube (note that pituitary was confirmed not to be included) and extracted total RNA by using NucleoSpin RNA Plus kit (MACHEREY-NAGEL, Düren, Germany). cDNA was obtained by KAPA Stranded mRNA-Seq Kit (Kapa Biosystems, Inc, Wilmington, MA) and KAPA Library preparation kit (Kapa Biosystems, Inc). Then, it was applied to a next-generation sequencer Hiseq 2500 (Illumina, San Diego, CA), following the standard protocol of Illumina system. We selected the candidate genes judging from transcripts per million (TPM) for expression value in the obtained data using CLC Genomics Workbench. We made volcano plot using R (R Development Core Team, 2023) and RStudio (2023) and colored dots, which indicate p-value <0.05 and |log FC|>1. In addition, we made a heatmap of genes related to neuroendocrine system using DESeq2 (Love et al., 2014).
Histological analysis of the distribution of agrp- and npy-neurons in the brain
To analyze the distribution of agrp- and npy-expressing neurons, we performed ISH for agrp and npy on frozen sections of reproductive medaka. In brief, female medaka was anesthetized (FA100, Bussan Animal Health Co, Ltd, Osaka, Japan), and its brain was picked up and fixed with 4% paraformaldehyde (PFA)/PBS. In analyses on agrp2 expression, we performed perfusion-fixation by using 4% PFA/PBS. After incubation with 30% sucrose/PBS, brains were embedded in 5% low melting agar/20% sucrose/PBS and sectioned at a thickness of 25 µm. The sections were hybridized with agrp1 (ENSORLG00000000398, 177 bases), agrp2 (ENSORLG00000029106, 303 bases), npya (ENSORLG00000004649, 288 bases) and npyb (ENSORLG00000007880, 288 bases)-specific digoxigenin (DIG)-labeled RNA probes and performed nitro blue tetrazolium (NBT)/ 5-bromo-4-chloro-3-indolyl-phosphate color-reaction (BCIP) after wash and incubation with anti-digoxigenin antibody (Cat# 11093274910; Roche; RRID:AB_514497) as previously reported (Zempo et al., 2013). Photographs were taken with a digital camera (DFC310FX; Leica Microsystems, Wetzlar, Germany) attached to an upright microscope (DM5000B; Leica Microsystems).
Histological analysis of agrp1-, npy-, and estrogen receptor (esr)-expressing neurons
To examine whether agrp1-expressing neurons co-express npya and esr, we prepared agrp1 fluorescein-labeled RNA probe and carried out double ISH as previously reported (Umatani et al., 2022). esr DIG-labeled probes were kindly given by Dr. Kayo (Kyoto Univ.), and we used npya DIG-labeled probe described in the previous paragraph. In brief, we made brain sections as described above and applied both agrp1 fluorescein-labeled and each DIG-labeled RNA probes. Signals for npya, esr1, esr2a, and esr2b were visualized by incubation with anti-digoxigenin antibody (Cat# 11207733910; Roche; RRID:AB_514500) and TSA Plus Cy3 System (TSA-Plus Cyanine 3 system, Cat# NEL744001KT, Akoya Biosciences, Marlborough, MA). After inactivation of Cy3 system by 3% H2O2, we applied peroxidase-conjugated anti-fluorescein antibody (Cat# 11426346910, Roche; RRID:AB_840257) on sections and performed TSA Plus biotin system (Cat# NEL749A001KT, Akoya Biosciences). Then, signals for agrp1 were visualized by Alexa 488 conjugated streptavidin (Cat# S11223, Invitrogen). For counter-staining of cell nuclei, DAPI in PBS was applied on section. On the other hand, to examine whether agrp2-expressing neurons co-express npya, we used npya fluorescein-labeled RNA probe and agrp2 DIG- labeled one. Double ISH of them was performed according to the same method described above. Fluorescent images were acquired with a confocal laser-scanning microscope (AXR, Nikon, Tokyo, Japan) using excitation and emission wavelengths of 405 nm and 429–474 nm for DAPI, 488 nm and 512–526 nm for Alexa 488, and 561 nm and 571–625 nm for Cy3, respectively. These were photographed at the Tokyo University of Agriculture and Technology for Smart Core facility Promotion Organization.
Quantitative real-time polymerase chain reaction (RT-qPCR)
A whole brain or a pituitary was collected from each medaka and total RNA was extracted by using FastGene RNA basic kit (Nippon Genetics Co, Ltd) according to the manufacturer’s instructions. For the juvenile medaka, we checked their sex as previously reported, and two samples of the same sex were mixed and used as one sample. Total RNA samples were reverse transcribed by FastGene cDNA synthesis 5×ReadyMix OdT according to the manufacturer’s instructions. For the analyses of the brain, 1 μL of cDNA diluted with 10-fold MQ was mixed with KAPA SYBR Fast qPCR kit (Kapa Biosystems, Inc) and amplified with Lightcycler96 [Roche; 95℃ 150 s (95℃ 10 s, 60℃ 10 s, 72℃ 15 s)×45 cycles]. For the analysis of the pituitary, 1 μL of cDNA diluted with fivefold MQ was mixed with KAPA SYBR Fast qPCR kit and amplified with Lightcycler96 [Roche; 95℃ 150 s (95℃ 10 s, 60℃ 10 s, 72℃ 10 s)×45 cycles]. The data was normalized by housekeeping gene, ribosomal protein s13 (rps13). Primer sequences were as follows:
AgRP1 RT-PCR F1 CCAATTTCCAGTCACCGAAG
AgRP1 RT-PCR R1 CTGGGTCCAACACAGAATCA
AgRP2 RT-PCR F1 TTGTTGTGCTTCTTGCTGCT
AgRP2 RT-PCR R1 ACAGAGCTCCAAACGGTGTC
NPYa SE CTCATCACAAGACAGAGGTATGGG
NPYa AS GGGTTGTAACTTGACTGTGGAAGTG
NPYb SE CTGCCTGCTCCTCTGTTTTTTCTC
NPYb AS CACAGTGTCTGGGTTGTCTCTCTTTC
qPCR FSHb Fw new TGGAGATCTACAGGCGTCGGTAC
qPCR FSHb Rv new AGCTCTCCACAGGGATGCTG
qPCR LHb Fw new AGGGTATGTGACTGACGGATCCAC
qPCR LHb Rv new TGCCTTACCAAGGACCCCTTGATG
RPS13 SE GTGTTCCCACTTGGCTCAAGC
RPS13 AS CACCAATTTGAGAGGGAGTGAGAC
Sham operations, ovariectomy, and E2 administration
Ovariectomy and E2 administration were performed according to a previous study (Kayo et al., 2020). Briefly, reproductive female medaka were anesthetized with 0.02% MS-222 (Sigma-Aldrich, St. Louis, MO) and their ovaries were excised via intraperitoneal operation. Sham operation group was anesthetized, received an abdominal incision without removing the ovaries, and received skin suture by using a silk thread. After checking that all Sham females spawn, we prepared three tanks; two tanks contained 7–8 OVX medaka, and one tank contained Sham medaka in 2 L breeding water in it. We dissolved β-estradiol 1.4 mg in 1 mL EtOH (E2 stock) and dispensed 2 μL of E2 stock or the same amount of 100% ethanol for the control tank. Ethanol or E2-containing water were changed every day. After the steroid treatment for 5 days, the medaka were anesthetized, and their whole brains were collected for RT-qPCR analysis.
Generation of agrp1 KO medaka lines
We generated agrp1 KO medaka lines by using CRISPR/Cas9. Cas9 mRNA and tracer RNA were purchased from Integrated DNA Technologies (IDT, Coralville, IA). The guide RNA sequence for digestion by CRISPR/Cas9 complex was ‘CCTCACCAGCAGTCCTGCCTGG’.
Mixture of Cas9 protein, tracer RNA, CRISPR RNA, GFP mRNA diluted with PBS and 0.02% phenol red (final concentration: Cas9 protein; 500 ng/μL, tracer RNA; 100 ng/μL, CRISPR RNA; 50 ng/μL, GFP mRNA; 5 ng/μL) was injected into the cytoplasm of one- or two-cell-stage embryos (F0). To obtain homozygous transgenic offspring, the carriers were crossed with each other.
Measurement of body size and ovary size
We took photographs of fish bodies from the lateral side by using a digital camera MC120HD (Leica) and calculated the abdominal and body length using ImageJ. For the analysis of body length, we measured the length from the mouth to the base of the tail. GSI was calculated as ovary weight/ body weight * 100.
Immunohistochemistry using AgRP1 antibody
We made brain sections of WT and agrp1 KO as described in the ‘Histological analysis of the distribution of agrp- and npy-neurons in the brain’. After washing with PBST two times for 10 min, sections were incubated with AgRP1 antibody (1:1000; rabbit polyclonal AgRP [83-132] amide [human], Cat# H-003-53; RRID:AB_2313908, Phoenix Pharmaceutical, Burlingame, CA)/0% goat serum/PBS overnight. On the next day, slides were washed with PBST and incubated with anti-rabbit biotinylated goat antibody (1:200; Cat#BA-1000; Vector Laboratories, Burlingame, CA) for 1 h. Then we applied Alexa 488 conjugated streptavidin (1:500, Cat#S11223, Invitrogen) and DAPI (1:2000; Dojindo Laboratories, Kumamoto, Japan). Photographs were taken with a digital camera (Leica Microsystems) attached to an upright microscope (Leica Microsystems).
Statistics
For the statistical analysis, we used Kyplot 5.0 software (Kyence, Osaka, Japan) or R software (R Development Core Team, 2023) with RStudio (version 2023.06.0+421). For the comparison of the TPM, we used Student’s t-test. For the comparison of agrp1 and npyb expressions in OVX and E2-administrated medaka, Steel–Dwass test was used for multiple comparison of the expression level. In the other experiments, we used Mann–Whitney U test. In all statistical analysis, significance levels were described as follows: *p<0.05, **p<0.01, and ***p<0.001.
Code availability
The code of ‘Shrimp-counter’ system is available in the present study in Figure 1—source code 1.
Acknowledgements
We thank Drs. Mikoto Nakajo (Osaka Med and Pharm Univ), Soma Tomihara (Hiroshima Univ), and Kana Ikegami (Kitasato Univ) for helpful discussion. We also thank Dr. Daichi Kayo (Kyoto Univ.) for his kind supply of esr DIG-labeled probes. In addition, we deeply appreciate Dr. Hiroyuki Takeda (U Tokyo) for his continued kind support and encouragement during our experiments. We also thank Dr. Yutaka Miura (Tokyo Univ of Agri and Tech) for his kind support of our experiments. As for the help of animal care, we thank Ms. Hisako Kohno, Miho Kyokuwa, Hiroko Tsukamoto, and Risa Nakaba. We thank Ms. Maiko Matsuda (Vesper studio) for her kind gift of medaka illustrations to us. The present study was supported by JSPS KAKENHI (JP21J20864 and JP22KJ0597 to YT, JP26221104 and JP21K06262 to YO, JP20H03071 to CU), and MEXT Initiative for Realizing Diversity in the Research Environment (Leadership training for women) (CU).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Chie Umatani, Email: chie@go.tuat.ac.jp.
Kristin Tessmar-Raible, University of Vienna, Austria.
Kate M Wassum, University of California, Los Angeles, United States.
Funding Information
This paper was supported by the following grants:
Japan Society for the Promotion of Science JP21J20864 to Yurika Tagui.
Japan Society for the Promotion of Science JP26221104 to Yoshitaka Oka.
Japan Society for the Promotion of Science JP20H03071 to Chie Umatani.
Ministry of Education, Culture, Sports, Science and Technology MEXT Initiative for Realizing Diversity in the Research Environment to Chie Umatani.
Japan Society for the Promotion of Science JP22KJ0597 to Yurika Tagui.
Japan Society for the Promotion of Science JP21K06262 to Yoshitaka Oka.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing - original draft, Writing – review and editing.
Data curation, Validation, Investigation, Methodology.
Data curation, Validation, Investigation, Methodology.
Data curation, Validation, Investigation, Visualization.
Software, Methodology.
Software, Supervision, Funding acquisition, Investigation, Methodology, Writing – review and editing.
Resources, Supervision, Funding acquisition, Writing – review and editing.
Resources, Supervision, Funding acquisition, Writing – review and editing.
Conceptualization, Resources, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing - original draft, Project administration, Writing – review and editing.
Ethics
All experiments and fish maintenance were conducted in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan) and the protocols approved by the Animal Care and Use Committee of Graduate School of Science, the University of Tokyo (Permission number, 17-1, 20-6) and the Animal Care and Use Committee of Graduate School of Agriculture, Tokyo University of Agriculture and Technology (Permission number, R05-15, R06-27).
Additional files
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files; source data files have been provided for Figures 1–6.
References
- Adachi S, Yamada S, Takatsu Y, Matsui H, Kinoshita M, Takase K, Sugiura H, Ohtaki T, Matsumoto H, Uenoyama Y, Tsukamura H, Inoue K, Maeda KI. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. The Journal of Reproduction and Development. 2007;53:367–378. doi: 10.1262/jrd.18146. [DOI] [PubMed] [Google Scholar]
- Adam CL, Archer ZA, Findlay PA, Thomas L, Marie M. Hypothalamic gene expression in sheep for cocaine- and amphetamine-regulated transcript, pro-opiomelanocortin, neuropeptide Y, agouti-related peptide and leptin receptor and responses to negative energy balance. Neuroendocrinology. 2002;75:250–256. doi: 10.1159/000054716. [DOI] [PubMed] [Google Scholar]
- Amirjani S, Asemi Z, Bazarganipour F, Aramesh S, Allan H, Sayadi M, Tabatabaei MS, Mohamadian Z, Zabti F, Iranpak N, Heydarzadeh A, Taghavi SA, Badehnoosh B, Khashavi Z. Dietary intake and lifestyle behaviour in different phenotypes of polycystic ovarian syndrome: a case-control study. Journal of Human Nutrition and Dietetics. 2019;32:413–421. doi: 10.1111/jhn.12646. [DOI] [PubMed] [Google Scholar]
- Andermann ML, Lowell BB. Toward a wiring diagram understanding of appetite control. Neuron. 2017;95:757–778. doi: 10.1016/j.neuron.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature Neuroscience. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock PA, Lee NJ, Driessler F, Lin S, Allison S, Stehrer B, Lin E-JD, Zhang L, Enriquez RF, Wong IPL, McDonald MM, During M, Pierroz DD, Slack K, Shi YC, Yulyaningsih E, Aljanova A, Little DG, Ferrari SL, Sainsbury A, Eisman JA, Herzog H. Neuropeptide Y knockout mice reveal a central role of NPY in the coordination of bone mass to body weight. PLOS ONE. 2009;4:e8415. doi: 10.1371/journal.pone.0008415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco AM, Soengas JL. Leptin signalling in teleost fish with emphasis in food intake regulation. Molecular and Cellular Endocrinology. 2021;526:111209. doi: 10.1016/j.mce.2021.111209. [DOI] [PubMed] [Google Scholar]
- Broberger C, De Lecea L, Sutcliffe JG, Hökfelt T. Hypocretin/orexin- and melanin-concentrating hormone-expressing cells form distinct populations in the rodent lateral hypothalamus: relationship to the neuropeptide Y and agouti gene-related protein systems. The Journal of Comparative Neurology. 1998;402:460–474. [PubMed] [Google Scholar]
- Chen SE, McMurtry JP, Walzem RL. Overfeeding-induced ovarian dysfunction in broiler breeder hens is associated with lipotoxicity. Poultry Science. 2006;85:70–81. doi: 10.1093/ps/85.1.70. [DOI] [PubMed] [Google Scholar]
- Chen P, Li B, Ou-Yang L. Role of estrogen receptors in health and disease. Frontiers in Endocrinology. 2022;13:839005. doi: 10.3389/fendo.2022.839005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisada S, Kurokawa T, Murashita K, Rønnestad I, Taniguchi Y, Toyoda A, Sakaki Y, Takeda S, Yoshiura Y. Leptin receptor-deficient (knockout) medaka, Oryzias latipes, show chronical up-regulated levels of orexigenic neuropeptides, elevated food intake and stage specific effects on growth and fat allocation. General and Comparative Endocrinology. 2014;195:9–20. [PubMed] [Google Scholar]
- Clark JT, Kalra PS, Crowley WR, Kalra SP. Neuropeptide Y and human pancreatic polypeptide stimulate feeding behavior in rats. Endocrinology. 1984;115:427–429. doi: 10.1210/endo-115-1-427. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE. Oestrogen, kisspeptin, GPR54 and the pre-ovulatory luteinising hormone surge. Journal of Neuroendocrinology. 2009;21:305–311. doi: 10.1111/j.1365-2826.2009.01835.x. [DOI] [PubMed] [Google Scholar]
- Conde-Sieira M, Soengas JL. Nutrient sensing systems in fish: impact on food intake regulation and energy homeostasis. Frontiers in Neuroscience. 2016;10:603. doi: 10.3389/fnins.2016.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois SL, Wolfe A, Radovick S, Boehm U, Levine JE. Estradiol restrains prepubertal gonadotropin secretion in female mice via activation of ERα in kisspeptin neurons. Endocrinology. 2016;157:1546–1554. doi: 10.1210/en.2015-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egami N. Effect of artificial photoperiodicity on time of oviposition in the fish, Oryzias latipes. Annot Zool Japonenses. 1954;27:57–62. [Google Scholar]
- Evans JJ, Anderson GM. Balancing ovulation and anovulation: integration of the reproductive and energy balance axes by neuropeptides. Human Reproduction Update. 2012;18:313–332. doi: 10.1093/humupd/dms004. [DOI] [PubMed] [Google Scholar]
- Funahashi H, Takenoya F, Guan JL, Kageyama H, Yada T, Shioda S. Hypothalamic neuronal networks and feeding-related peptides involved in the regulation of feeding. Anatomical Science International. 2003;78:123–138. doi: 10.1046/j.0022-7722.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- Glenn Stanley B, Kyrkouli SE, Lampert S, Leibowitz SF. Neuropeptide Y chronically injected into the hypothalamus: A powerful neurochemical inducer of hyperphagia and obesity. Peptides. 1986;7:1189–1192. doi: 10.1016/0196-9781(86)90149-X. [DOI] [PubMed] [Google Scholar]
- Graham M, Shutter JR, Sarmiento U, Sarosi I, Stark KL. Overexpression of Agrt leads to obesity in transgenic mice. Nature Genetics. 1997;17:273–274. doi: 10.1038/ng1197-273. [DOI] [PubMed] [Google Scholar]
- Gray TS, Morley JE. Neuropeptide Y: anatomical distribution and possible function in mammalian nervous system. Life Sciences. 1986;38:389–401. doi: 10.1016/0024-3205(86)90061-5. [DOI] [PubMed] [Google Scholar]
- Hagan MM, Rushing PA, Pritchard LM, Schwartz MW, Strack AM, Van Der Ploeg LH, Woods SC, Seeley RJ. Long-term orexigenic effects of AgRP-(83---132) involve mechanisms other than melanocortin receptor blockade. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2000;279:R47–R52. doi: 10.1152/ajpregu.2000.279.1.R47. [DOI] [PubMed] [Google Scholar]
- Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nature Neuroscience. 1998;1:271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- Hasebe M, Kanda S, Oka Y. Female-specific glucose sensitivity of GnRH1 neurons leads to sexually dimorphic inhibition of reproduction in medaka. Endocrinology. 2016;157:4318–4329. doi: 10.1210/en.2016-1352. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Kajihara S, Umatani C, Nakajo M, Kanda S, Oka Y. Estrogen upregulates the firing activity of hypothalamic gonadotropin-releasing hormone (GnRH1) neurons in the evening in female medaka. Journal of Neuroendocrinology. 2022;34:e13101. doi: 10.1111/jne.13101. [DOI] [PubMed] [Google Scholar]
- Ilnytska O, Argyropoulos G. The role of the Agouti-Related Protein in energy balance regulation. Cellular and Molecular Life Sciences. 2008;65:2721–2731. doi: 10.1007/s00018-008-8104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings KJ, de Lecea L. Neural and hormonal control of sexual behavior. Endocrinology. 2020;161:bqaa150. doi: 10.1210/endocr/bqaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong I, Kim E, Kim S, Kim HK, Lee DW, Seong JY, Park HC. mRNA expression and metabolic regulation of npy and agrp1/2 in the zebrafish brain. Neuroscience Letters. 2018;668:73–79. doi: 10.1016/j.neulet.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Kalra SP, Kalra PS. Nutritional infertility: the role of the interconnected hypothalamic neuropeptide Y-galanin-opioid network. Frontiers in Neuroendocrinology. 1996;17:371–401. doi: 10.1006/frne.1996.0010. [DOI] [PubMed] [Google Scholar]
- Kamijo M, Kojima K, Maruyama K, Konno N, Motohashi E, Ikegami T, Uchiyama M, Shioda S, Ando H, Matsuda K. Neuropeptide Y in tiger puffer (Takifugu rubripes): distribution, cloning, characterization, and mRNA expression responses to prandial condition. Zoological Science. 2011;28:882–890. doi: 10.2108/zsj.28.882. [DOI] [PubMed] [Google Scholar]
- Kanda S, Akazome Y, Matsunaga T, Yamamoto N, Yamada S, Tsukamura H, Maeda Ki, Oka Y. Identification of KiSS-1 product kisspeptin and steroid-sensitive sexually dimorphic kisspeptin neurons in medaka (Oryzias latipes) Endocrinology. 2008;149:2467–2476. doi: 10.1210/en.2007-1503. [DOI] [PubMed] [Google Scholar]
- Kanda S, Okubo K, Oka Y. Differential regulation of the luteinizing hormone genes in teleosts and tetrapods due to their distinct genomic environments--insights into gonadotropin beta subunit evolution. General and Comparative Endocrinology. 2011;173:253–258. doi: 10.1016/j.ygcen.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, Rissman EF. A critical role for the evolutionarily conserved gonadotropin-releasing hormone II: mediation of energy status and female sexual behavior. Endocrinology. 2004;145:3639–3646. doi: 10.1210/en.2004-0148. [DOI] [PubMed] [Google Scholar]
- Kawai K, Fujita H, Sanchez G, Furusawa S, Umino T. Estimating the spawning season of black sea bream Acanthopagrus schlegelii in Hiroshima Bay, Japan, from temporal variation in egg density. Fisheries Science. 2020;86:645–653. doi: 10.1007/s12562-020-01433-1. [DOI] [Google Scholar]
- Kayo D, Zempo B, Tomihara S, Oka Y, Kanda S. Gene knockout analysis reveals essentiality of estrogen receptor β1 (Esr2a) for female reproduction in medaka. Scientific Reports. 2019;9:8868. doi: 10.1038/s41598-019-45373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayo D, Oka Y, Kanda S. Examination of methods for manipulating serum 17β-Estradiol (E2) levels by analysis of blood E2 concentration in medaka (Oryzias latipes) General and Comparative Endocrinology. 2020;285:113272. doi: 10.1016/j.ygcen.2019.113272. [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Qiu J. Estrogen signaling in hypothalamic circuits controlling reproduction. Brain Research. 2010;1364:44–52. doi: 10.1016/j.brainres.2010.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Murata K, Naruse K, Tanaka M. Medaka: Biology, Management, and Experimental Protocols. John Wiley & Sons; 2009. [DOI] [Google Scholar]
- Kirkwood RN, Cumming DC, Aherne FX. Nutrition and puberty in the female. The Proceedings of the Nutrition Society. 1987;46:177–192. doi: 10.1079/pns19870026. [DOI] [PubMed] [Google Scholar]
- Kohno D, Yada T. Arcuate NPY neurons sense and integrate peripheral metabolic signals to control feeding. Neuropeptides. 2012;46:315–319. doi: 10.1016/j.npep.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, Lowell BB. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. The Journal of Clinical Investigation. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson TA, Tay BH, Sundström G, Fredriksson R, Brenner S, Larhammar D, Venkatesh B. Neuropeptide Y-family peptides and receptors in the elephant shark, Callorhinchus milii confirm gene duplications before the gnathostome radiation. Genomics. 2009;93:254–260. doi: 10.1016/j.ygeno.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Liu R, Kinoshita M, Adolfi MC, Schartl M. Analysis of the role of the Mc4r system in development, growth, and puberty of medaka. Frontiers in Endocrinology. 2019;10:213. doi: 10.3389/fendo.2019.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn SE, Stamplis TB, Barrington WT, Weida N, Hudak CA. Food, stress, and reproduction: short-term fasting alters endocrine physiology and reproductive behavior in the zebra finch. Hormones and Behavior. 2010;58:214–222. doi: 10.1016/j.yhbeh.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Marks JL, Li M, Schwartz M, Porte D, Baskin DG. Effect of fasting on regional levels of neuropeptide Y mRNA and insulin receptors in the rat hypothalamus: An autoradiographic study. Molecular and Cellular Neurosciences. 1992;3:199–205. doi: 10.1016/1044-7431(92)90039-5. [DOI] [PubMed] [Google Scholar]
- Marks JL, Waite K. Intracerebroventricular neuropeptide Y acutely influences glucose metabolism and insulin sensitivity in the rat. Journal of Neuroendocrinology. 1997;9:99–103. doi: 10.1046/j.1365-2826.1997.00554.x. [DOI] [PubMed] [Google Scholar]
- Melo AC, Ramsdell JS. Sexual dimorphism of brain aromatase activity in medaka: induction of a female phenotype by estradiol. Environmental Health Perspectives. 2001;109:257–264. doi: 10.1289/ehp.01109257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry BJ, Holehan AM. Onset of puberty and duration of fertility in rats fed a restricted diet. Reproduction. 1979;57:253–259. doi: 10.1530/jrf.0.0570253. [DOI] [PubMed] [Google Scholar]
- Micevych PE, Meisel RL. Integrating neural circuits controlling female sexual behavior. Frontiers in Systems Neuroscience. 2017;11:42. doi: 10.3389/fnsys.2017.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani Y, Kanda S, Akazome Y, Zempo B, Oka Y. Hypothalamic Kiss1 but not Kiss2 neurons are involved in estrogen feedback in medaka (Oryzias latipes) Endocrinology. 2010;151:1751–1759. doi: 10.1210/en.2009-1174. [DOI] [PubMed] [Google Scholar]
- Muroi Y, Ishii T. A novel neuropeptide Y neuronal pathway linking energy state and reproductive behavior. Neuropeptides. 2016;59:1–8. doi: 10.1016/j.npep.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Naftolin F, Garcia-Segura LM, Horvath TL, Zsarnovszky A, Demir N, Fadiel A, Leranth C, Vondracek-Klepper S, Lewis C, Chang A, Parducz A. Estrogen-induced hypothalamic synaptic plasticity and pituitary sensitization in the control of the estrogen-induced gonadotrophin surge. Reproductive Sciences. 2007;14:101–116. doi: 10.1177/1933719107301059. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Cui Z, Li C, Meister J, Cui Y, Fu O, Smith AS, Jain S, Lowell BB, Krashes MJ, Wess J. Gs-coupled GPCR signalling in AgRP neurons triggers sustained increase in food intake. Nature Communications. 2016;7:10268. doi: 10.1038/ncomms10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson ME, Vandenput L, Tivesten Å, Norlén AK, Lagerquist MK, Windahl SH, Börjesson AE, Farman HH, Poutanen M, Benrick A, Maliqueo M, Stener-Victorin E, Ryberg H, Ohlsson C. Measurement of a comprehensive sex steroid profile in rodent serum by high-sensitive gas chromatography-tandem mass spectrometry. Endocrinology. 2015;156:2492–2502. doi: 10.1210/en.2014-1890. [DOI] [PubMed] [Google Scholar]
- Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- Opazo R, Plaza-Parrochia F, Cardoso dos Santos GR, Carneiro GRA, Sardela VF, Romero J, Valladares L. Fasting upregulates npy, agrp, and ghsr without increasing ghrelin levels in zebrafish (Danio rerio) larvae. Frontiers in Physiology. 2018;9:1901. doi: 10.3389/fphys.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peute J, van der Gaag MA, Lambert JG. Ultrastructure and lipid content of the liver of the zebrafish, Brachy Danio rerio, related to vitellogenin synthesis. Cell and Tissue Research. 1978;186:297–308. doi: 10.1007/BF00225539. [DOI] [PubMed] [Google Scholar]
- Qian S, Chen H, Weingarth D, Trumbauer ME, Novi DE, Guan X, Yu H, Shen Z, Feng Y, Frazier E, Chen A, Camacho RE, Shearman LP, Gopal-Truter S, MacNeil DJ, Van der Ploeg LHT, Marsh DJ. Neither agouti-related protein nor neuropeptide Y is critically required for the regulation of energy homeostasis in mice. Molecular and Cellular Biology. 2002;22:5027–5035. doi: 10.1128/MCB.22.14.5027-5035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Rivera HM, Bosch MA, Padilla SL, Stincic TL, Palmiter RD, Kelly MJ, Rønnekleiv OK. Estrogenic-dependent glutamatergic neurotransmission from kisspeptin neurons governs feeding circuits in females. eLife. 2018;7:e35656. doi: 10.7554/eLife.35656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . Vienna, Austria: R Foundation for Statistical Computing; 2023. https://www.R-project.org/ [Google Scholar]
- Robinson EJ, Rugh R. The reproductive processes of the fish, oryzias latipes. The Biological Bulletin. 1943;84:115–125. doi: 10.2307/1538054. [DOI] [Google Scholar]
- Roesch DM. Effects of selective estrogen receptor agonists on food intake and body weight gain in rats. Physiology & Behavior. 2006;87:39–44. doi: 10.1016/j.physbeh.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Rønnestad I, Gomes AS, Murashita K, Angotzi R, Jönsson E, Volkoff H. Appetite-controlling endocrine systems in teleosts. Frontiers in Endocrinology. 2017;8:73. doi: 10.3389/fendo.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Shainer I, Buchshtab A, Hawkins TA, Wilson SW, Cone RD, Gothilf Y. Novel hypophysiotropic AgRP2 neurons and pineal cells revealed by BAC transgenesis in zebrafish. Scientific Reports. 2017;7:44777. doi: 10.1038/srep44777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shainer I, Michel M, Marquart GD, Bhandiwad AA, Zmora N, Ben-Moshe Livne Z, Zohar Y, Hazak A, Mazon Y, Förster D, Hollander-Cohen L, Cone RD, Burgess HA, Gothilf Y. Agouti-related protein 2 is a new player in the teleost stress response system. Current Biology. 2019;29:2009–2019. doi: 10.1016/j.cub.2019.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Kawabe M, Karasuyama K, Kurachi T, Hayashi A, Ataka K, Iwai H, Takeno H, Hayasaka O, Kotani T, Komatsu M, Inui A. Neuropeptide Y deficiency induces anxiety-like behaviours in zebrafish (Danio rerio) Scientific Reports. 2020;10:5913. doi: 10.1038/s41598-020-62699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutter JR, Graham M, Kinsey AC, Scully S, Lüthy R, Stark KL. Hypothalamic expression of ART, a novel gene related to agouti, is up-regulated in obese and diabetic mutant mice. Genes & Development. 1997;11:593–602. doi: 10.1101/gad.11.5.593. [DOI] [PubMed] [Google Scholar]
- Söderberg C, Wraith A, Ringvall M, Yan YL, Postlethwait JH, Brodin L, Larhammar D. Zebrafish genes for neuropeptide Y and peptide YY reveal origin by chromosome duplication from an ancestral gene linked to the homeobox cluster. Journal of Neurochemistry. 2000;75:908–918. doi: 10.1046/j.1471-4159.2000.0750908.x. [DOI] [PubMed] [Google Scholar]
- Song Y, Golling G, Thacker TL, Cone RD. Agouti-related protein (AGRP) is conserved and regulated by metabolic state in the zebrafish, Danio rerio. Endocrine. 2003;22:257–265. doi: 10.1385/ENDO:22:3:257. [DOI] [PubMed] [Google Scholar]
- Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB Journal. 2007;21:2042–2049. doi: 10.1096/fj.06-7503com. [DOI] [PubMed] [Google Scholar]
- Stone DB, Cherry MJ, Martin JA, Cohen BS, Miller KV. Breeding chronology and social interactions affect ungulate foraging behavior at a concentrated food resource. PLOS ONE. 2017;12:e0178477. doi: 10.1371/journal.pone.0178477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström G, Larsson TA, Brenner S, Venkatesh B, Larhammar D. Evolution of the neuropeptide Y family: new genes by chromosome duplications in early vertebrates and in teleost fishes. General and Comparative Endocrinology. 2008;155:705–716. doi: 10.1016/j.ygcen.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Takahashi KA, Cone RD. Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology. 2005;146:1043–1047. doi: 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- Temple JL, Rissman EF. Brief refeeding restores reproductive readiness in food-restricted female musk shrews (Suncus murinus) Hormones and Behavior. 2000;38:21–28. doi: 10.1006/hbeh.2000.1596. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Miyagawa S, Lange A, Ogino Y, Mizutani T, Ihara M, Tanaka H, Tatarazako N, Kobayashi T, Tyler CR, Iguchi T. Evolution of estrogen receptors in ray-finned fish and their comparative responses to estrogenic substances. The Journal of Steroid Biochemistry and Molecular Biology. 2016;158:189–197. doi: 10.1016/j.jsbmb.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Ogino Y, Lange A, Myosho T, Kobayashi T, Hirano Y, Yamada G, Sato T, Tatarazako N, Tyler CR, Iguchi T, Miyagawa S. Establishment of estrogen receptor 1 (ESR1)‐knockout medaka: ESR 1 is dispensable for sexual development and reproduction in medaka, Oryzias latipes. Development, Growth & Differentiation. 2017;59:552–561. doi: 10.1111/dgd.12386. [DOI] [PubMed] [Google Scholar]
- Tsuyuki ATU. Assessment of ichthyofauna at oyster rafts in Hiroshima Bay, Japan, using underwater video cameras. Aquaculture Science. 2018;66:267–274. [Google Scholar]
- Umatani C, Yoshida N, Yamamoto E, Akazome Y, Mori Y, Kanda S, Okubo K, Oka Y. Co-existing Neuropeptide FF and gonadotropin-releasing hormone 3 coordinately modulate male sexual behavior. Endocrinology. 2022;163:bqab261. doi: 10.1210/endocr/bqab261. [DOI] [PubMed] [Google Scholar]
- Volk KM, Pogrebna VV, Roberts JA, Zachry JE, Blythe SN, Toporikova N. High-fat, high-sugar diet disrupts the preovulatory hormone surge and induces cystic ovaries in cycling female rats. Journal of the Endocrine Society. 2017;1:1488–1505. doi: 10.1210/js.2017-00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, Korach KS, Greiner E, Pérez CA, Schütz G, Herbison AE. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Stires H, Belden WJ, Roepke TA. The arcuate estrogen-regulated transcriptome: estrogen response element-dependent and -independent signaling of ERα in female mice. Endocrinology. 2017;158:612–626. doi: 10.1210/en.2016-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori E, Azuma M, Nishiguchi R, Kang KS, Kamijo M, Uchiyama M, Matsuda K. Neuropeptide Y stimulates food intake in the Zebrafish, Danio rerio. Journal of Neuroendocrinology. 2012;24:766–773. doi: 10.1111/j.1365-2826.2012.02281.x. [DOI] [PubMed] [Google Scholar]
- Yu K, He Y, Hyseni I, Pei Z, Yang Y, Xu P, Cai X, Liu H, Qu N, Liu H, He Y, Yu M, Liang C, Yang T, Wang J, Gourdy P, Arnal JF, Lenfant F, Xu Y, Wang C. 17β-estradiol promotes acute refeeding in hungry mice via membrane-initiated ERα signaling. Molecular Metabolism. 2020;42:101053. doi: 10.1016/j.molmet.2020.101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempo B, Kanda S, Okubo K, Akazome Y, Oka Y. Anatomical distribution of sex steroid hormone receptors in the brain of female medaka. The Journal of Comparative Neurology. 2013;521:1760–1780. doi: 10.1002/cne.23255. [DOI] [PubMed] [Google Scholar]