Abstract
Background
GPR56/ADGRG1 is an adhesion G protein-coupled receptor involved in cell-matrix interactions and metastasis of human melanoma cells. Previously, we demonstrated that GPR56 activation in melanoma cells triggers Gα12/13-RhoA signaling, leading to increased IL-6 production and enhanced cell migration. Yet little is known of the downstream signaling effectors and their specific roles in regulating melanoma cellular phenotypes.
Results
In this study, we show that GPR56 activation induces Rho-ROCK-MLC and JAK-STAT3 signaling, which temporally and differentially drive amoeboid-like morphology and IL-6 upregulation. Interestingly, GPR56-induced JAK-STAT3 activation is partially regulated by Rho-ROCK-MLC signaling but not vice versa. Moreover, receptor auto-proteolysis modulates the magnitude of GPR56-mediated signaling, and its unique intracellular regions contribute to the selective regulation of unique signaling pathways and associated cellular phenotypes.
Conclusion
Our findings reveal complex GPR56-mediated biased signaling through the Rho-ROCK-MLC and JAK-STAT3 pathways, highlighting these networks as potential therapeutic targets for modulating distinct tumorigenic phenotypes in human melanoma cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-025-02267-z.
Keywords: Adhesion GPCR, IL-6, Cytoskeletal remodelling, GPR56, Melanoma, Signalling
Introduction
G protein-coupled receptors (GPCRs) play an important role in cellular communication during tumor development, acting variably as either tumor promoters or suppressors [1, 2]. The adhesion-class GPCRs (aGPCRs) represent the 2nd largest GPCR family in humans and have been increasingly implicated either pro-tumorigenic or anti-tumorigenic roles. Indeed, several aGPCRs have been identified as mutated or aberrantly expressed across diverse cancer types [3–6]. GPR56/ADGRG1 is a cancer-associated aGPCR first identified in human melanoma cell lines, where its expression was found to inversely correlated with the metastatic potential of tumor cells [7, 8]. Tissue transglutaminase (TG2) was later revealed as the extracellular matrix ligand for GPR56 in melanoma tissues and the interaction between TG2 and GPR56 was found to suppress melanoma cell growth, angiogenesis, and metastasis [9–12]. Recently, a low GPR56 expression-associated transcriptomic signature was specifically linked to the mesenchymal phenotypes of various cancer types [13]. These findings suggest that GPR56 may function as a potential tumor suppressor by partially inhibiting epithelial-mesenchymal transition.
Conversely, GPR56 was shown to enhance cell growth and/or drug resistance of colorectal, non-small-cell lung carcinoma, and epithelial ovarian tumors hence suggesting a plausible oncogenic function [14–20]. Moreover, Pabst et al. identified GPR56 as a novel surface marker of leukemia stem cell (LSC) for the majority of acute myeloid leukemia (AML) [21]. GPR56 expression evidently contributes to AML development and higher GPR56 expression levels not only correlate with the LSC gene signature but are also associated with high-risk AML subgroups, poor clinical outcome, and drug resistance [22–25]. In the immune system, GPR56 was revealed by us as an inhibitory receptor of human natural killer cells, thus a potential immune checkpoint molecule [26]. Bilemjian et al. recently detected prominent GPR56 expression in various tumor-infiltrating lymphocyte (TIL) subsets, with effector memory and central memory CD8+ T cells exhibiting the highest levels of GPR56 expression [27]. Importantly, GPR56 expression in TILs was further upregulated following T-cell receptor activation. Additionally, overexpression of GPR56 greatly impaired T cell migration, suggesting that GPR56 may function as an immune checkpoint in specific T cell populations [27]. Finally, animal studies investigating the role of GPR56 in endogenous cancer progression across various tumor models have yielded mixed results, highlighting the complex and context-dependent role of GPR56 in tumorigenesis [28]. In short, the functional role of GPR56 in tumor development appears to be dependent on cell type and/or tumor stage-specific contexts.
The divergent roles of GPR56 in tumorigenesis are likely due to its complex protein structure, multiple binding partners, diverse receptor isoforms, and varied activation/signaling mechanisms [29]. As an archetypal aGPCR, the extracellular region of GPR56 extends markedly from the heptahelical transmembrane (7TM) moiety, and consists of a pentraxin/laminin/neurexin/sex-hormone-binding-globulin-like (PLL) domain and a GPCR autoproteolysis-inducing (GAIN) domain [30]. Furthermore, GPR56 is fully processed by the characteristic auto-proteolytic cleavage at the GPCR proteolysis site (GPS) and expressed as a bipartite complex on the membrane [31]. Hence, GPR56 signaling is thought to be mediated primarily by the well-accepted tethered agonism mechanism in which receptor activation is triggered by the newly-exposed agonistic Stachel peptide of the C-terminal fragment (CTF) following the dissociation of the N-terminal fragment (NTF) [32–34]. Nevertheless, alternative activation mechanisms including the Stachel-independent and NTF-CTF non-dissociation modes have also been proposed for GPR56 [35–37]. Besides TG2, a diverse range of binding partners and ligands for GPR56 has been identified, including collagen III, laminin, progastrin, CD9/CD81, heparin, and phosphatidylserine [29]. Furthermore, extensive alternative RNA splicing results in the production of at least five distinct GPR56 receptor isoforms, each exhibiting different signaling activities and/or intensities [38]. Most critically, interactions between unique GPR56 isoforms and their ligands have been shown to mediate tissue- and cell type-specific functions [39–41].
Given its disparate tumorigenic roles, there is an increasing need to better understand the cancer type-specific regulation of GPR56-mediated signaling. To date, most studies on GPR56 signaling have focused on the activation of the Gα12/13-RhoA axis; however, other signaling pathways, including Gαi, Gαq, Gβγ, β-arrestin, and mTOR have also been implicated [29, 42]. We have shown previously that GPR56 activation in human melanoma cells by an immobilized agonistic CG4 monoclonal antibody (mAb) triggered Gα12/13-RhoA signaling, promoting IL-6 upregulation and cell migration [42, 43]. Herein, we demonstrate that GPR56 activation elicits Rho-ROCK-MLC and JAK-STAT3 signaling cascades downstream of Gα12/13 in melanoma cells to promote amoeboid-like morphological change and increased IL-6 production. Interestingly, GPS auto-proteolysis plays a role in regulating the magnitude of GPR56-mediated signaling. Furthermore, we uncover a GPR56 isoform-dependent modulation of signaling activities and cellular phenotypes, which are partially regulated by distinct regions of the intracellular loop (ICL) and cytoplasmic tail. In conclusion, our findings offer new insights into GPR56-mediated biased signaling and its associated tumorigenic phenotypes in human melanoma cells.
Materials and methods
Reagents and antibodies
All chemicals and reagents were purchased from Sigma (MO, USA) and Invitrogen (MA, USA) unless otherwise specified. Signalling inhibitors used include Rho-GTPase inhibitor, Rhosin hydrochloride (Catalogue No. 5003)(Tocris Bioscience, Bristol, UK); ROCK inhibitors, Y27632 (#sc-281642) and H1152 (#sc-203592) (Santa Cruz, TX, USA); myosin II inhibitor, (±)-Blebbistatin (#sc-203532) (Santa Cruz, TX, USA); STAT3 inhibitors, STAT3 inhibitor V (STAT3i, #sc-202818)(Santa Cruz, TX, USA) and LLL12 (#1792-5)(BioVision, CA, USA); JAK1/2 inhibitors, JAK inhibitor I (JAKi, #sc-204021) and AZD1480 (#sc-364735)(Santa Cruz, TX, USA). Antibodies (Abs) used for signalling western blot analysis, including anti-MLCII (#3672), anti-pMLCII (#3671), anti-JAK2 (#3230), anti-pJAK2 (#3771), anti-STAT3 (#9145), and anti-pSTAT3 (#9145) were all obtained from Cell Signalling Technology, Inc. (MA, USA). GPR56-specific CG2, CG3, and CG4 monoclonal antibodies (mAbs) were developed in-house as described previously [43]. Mouse isotype control IgG1 (Clone 11711) was from R&D System (MN, USA).
Cell culture
All culture media were supplemented with 10% heat inactivated fetal calf serum (FCS), 2 mM L-glutamine, 50 IU/mL penicillin and 50 µg/mL streptomycin. All cell lines were incubated at 37 °C in a 5% CO2, 95% humidity incubator. Human melanoma cell lines including A375 (CRL-1619), A2058 (CRL-11147), C32 (CRL-1585), MeWo (HTB-65), SK-MEL-5 (HTB-70), and RPMI-7951 (HTB-60) were obtained from ATCC (VA, USA). A375 and A2058 cells were cultured in the Dulbecco’s Modified Eagle Medium (DMEM), whereas C32, SK-MEL5, RPMI-7951, and MeWo cells were cultured in Minimum Essential Media (MEM). For cell treatment with signalling inhibitors, Rhosin (60 µM), Y27632 (60 µM), H1152 (20 µM), (±)-Blebbistatin (25 µM), STAT3i (5 µM), LLL12 (10 µM), JAKi (10 µM), and AZD1480 (20 µM) were used unless otherwise specified.
Establishment of GPR56-knock out melanoma cell lines
A2058 and MeWo cell lines were subjected to CRISPR/Cas9-mediated human GPR56-specific gene silencing according to the standard procedures. Briefly, cells were co-transfected with the GPR56-specific guide RNA-producing pRGEN-Human-GPR56-U6-SG-1 construct along with the pRGEN-Cas9-CMV and pHRS-Human-GPR56-1 plasmids (ToolGen, Seoul, South Korea). Transfected cells were selected in medium containing hygromycin (100 µg/mL, Invitrogen) for several days. The established stable cells were subjected to fluorescence-activated cell sorting (FACS) and western blotting analyses to confirm GPR56 gene silencing effects using GPR56-specific mAbs.
Establishment of cell lines stably expressing the wild-type, alternatively-spliced, and truncated GPR56 receptor isoforms
Stable A375 melanoma cells over-expressing distinct GPR56 isoforms or truncated GPR56 variants were generated using the pFB-Neo retroviral transduction system (Agilent Technologies Inc., CA, USA) as described previously [43, 44]. In brief, the cDNAs of desired GPR56 variants were generated by the TOOLSite-Directed Mutagenesis Kit (BIOTOOLS Co., Ltd., Taipei, Taiwan) for the T383A mutant or amplified by PCR using appropriate primers (summarized in Table S1) and cloned into the pFB-Neo vector, which was then co-transfected with the pVPack and pVPackGP vectors into HEK-293T cells. Virion-containing supernatant was collected 48 h post-transfection and used to infect A375 cells in the presence of polybrene (8 µg/mL). Infected cells were cultured in DMEM containing G418 (1 mg/mL) for ~ 10–14 days. The specific expression of distinct GPR56 isoforms in stable A375 cells was verified by FACS and western blotting.
GPR56 activation and phenotypic analyses of melanoma cells
Unless otherwise specified, GPR56 activation was induced by receptor ligation using immobilized GPR56-specific CG4 mAb as described previously [43]. CG3 mAb and mouse IgG1 isotype were routinely included as negative controls. Briefly, melanoma cells (1 × 106 cells/well) were cultured in 6-well plates pre-coated without or with CG3, CG4, or control Ab (10 µg/mL). Cell morphology was examined and recorded under inverted microscopy. The degree of cell shape changes was determined by counting the ratio of cells with fully-spread mesenchymal-like versus round-shaped amoeboid-like morphology in 3 different fields. The cell length was calculated using the ImageJ software (NIH, USA). For the analysis of IL-6 production, conditioned medium (CM) was collected at indicated time points, spun by centrifugation at 6,000 g for 20 min at 4 °C and stored at -80 °C until use. IL-6 concentration was determined using a Human IL-6 Matched ELISA Pair Set (Sino Biological, China) according to the manufacturer’s protocols.
Confocal and high-resolution image analyses
CG3 and CG4 mAbs were added on poly-lysine coated coverslips overnight at room temperature (RT). Cells (1 × 105 cells/well) were incubated on the Ab-coated coverslips for 6 h at 37 °C, followed by fixation in 4% paraformaldehyde/PBS for 20 min at 4 °C. Cells were blocked and permeabilized in blocking buffer containing 0.1% saponin for 30 min at 4 °C. Permeabilized cells were incubated with phalloidin (1:200) for 1 h at 4 °C. Finally, cells were stained by Hoechst in PBS (1:1000) and then mounted with mounting gel (20% glycerol in PBS) on the slides. The expression pattern of actin filaments was observed using ZEISS LSM780 (Germany). For the high-resolution cell image analysis, cells (5 × 105 cells/dish) were seeded onto the µ-Dish35mm, high imaging dish (ibidi, Germany) pre-coated with CG3 or CG4 mAb. Cell morphologies were observed and recorded using Holo-Tomography 3D Cell Explorer (NanoLive, Switzerland).
FACS and Western blotting analyses
FACS analysis of surface GPR56 expression in melanoma cells was carried out as described previously [45]. Briefly, cells were harvested and fixed in 4% paraformaldehyde/PBS for 10 min at 4 °C. Fixed cells were incubated for 1 h in cold blocking buffer (1% BSA/5% serum of 2nd Ab/PBS) and subsequently incubated with anti-GPR56 CG2 mAb (2 µg/mL) in blocking buffer for 1 h before washing. Cells were incubated for 1 h with fluorochrome-conjugated goat anti-mouse IgG in blocking buffer (1:200), washed, and followed by analysis using FACScan flow cytometer (BD Biosciences). For western blotting analysis, cells were lysed and protein lysates were separated in 10% SDS-PAGE gels, blotted, and probed with the primary Ab (pre-determined concentration) and HRP-conjugated 2nd Ab (1: 500) as described [43]. The blotting results were photographed using BioSpectrum 610 Imaging System (UVP, Upland, CA, USA). For the signalling analysis, GPR56 activation was alternatively induced by first incubating cells (2 × 106 cells/mL) in suspension with CG4 mAb (10 µg/mL) for 30 min at 4 °C, washed, and followed by Ab ligation with the goat anti-mouse IgG (5 µg/mL) at 37 °C in serum-free medium. Reaction was stopped at different time points of incubation as indicated and cell lysates were isolated immediately. Total cell lysates were separated in SDS-PAGE gels and analysed using the following primary Abs: anti-MLC II (1:1000), and anti-pMLC II (1:1000), anti-JAK2 and anti-pJAK2 (1:1000), anti-STAT3 (1:200), and anti-pSTAT3 (1:2000). The reaction was detected with HRP-conjugated 2nd Abs (1:2500) and ECL reagents. As a protein loading control, GAPDH and β-actin were probed by mouse anti-GAPDH (#60004, Proteintech, IL, USA) and anti-actin Ab (clone C4/MAB-1501, Merck).
RhoA activation assay
The G-LISA® RhoA Activation Assay Biochem kit (# BK124, Cytoskeleton, Inc., Denver, CO, USA) was used to detect active, GTP-bound RhoA in cell lysates according to the protocols suggested by the manufacturer. Briefly, cell lysates (0.5 mg/mL, 50 µL/well) were added into wells of the RhoA-GTP binding 96-well plate for 30 min at 4 °C on a cold orbital microplate shaker (200 rpm). After extensive washes with wash buffer at RT, wells were filled with the antigen presenting buffer (200 µL/well) for exactly 2 min at RT. Wells were washed extensively before incubating with pre-diluted anti-RhoA Ab (50 µL/well) for 45 min at RT on an orbital microplate shaker (200–400 rpm). Following thorough washes, HRP-labelled 2nd Ab (1:62.5, 50 µL/well) was added into wells for 45 min incubation at RT. Wells were washed again before incubating with HRP detection reagent (50 µL/well) for 10 min at 37 °C. The reaction was terminated by adding HRP stop buffer (50 µL/well) and the reaction signal was determined by measuring absorbance at 490 nm.
Cell migration and invasion assays
Cell migration and invasion analyses were performed using 96-well Cell Migration/Chemotaxis Assay (#ab235673) and Cell Invasion Assay (#ab235697) Kits (Abcam Limited.), respectively. Briefly, cells were serum-starved for 24 h and re-suspended at a density of 1 × 106 cells/mL in serum-free medium containing 5% BSA. Plates were prepared before the cell migration and invasion assays according to the manufactures’ protocol. For cell invasion assay, the matrigel invasion chamber was pre-incubated without or with CG4/IgG1 mAb (5 µg/mL) for 1 h. Cells (100 µL/well) were treated without or with indicated mAbs (5 µg/mL) and placed in the upper chamber equipped with an 8-µm pore filter membrane. The bottom chambers were filled with medium without or with 10% FCS. As a positive control, the migration inducer solution supplied by the kit was added in the lower chambers. The plates were kept in a 37 °C incubator for 16 h. Migrated cells were dissociated from the bottom of filter membrane using 100 µL/well of the mix solution (100 µL of Cell Invasion Dye in 1 mL of Cell Dissociation Solution) at 37 °C for 60 min. Fluorescence intensity was measured in a fluorescence plate reader (Molecular Devices) with a Em/Ex = 530/590 nm filter set.
Time-lapse analysis of cell migration
To monitor the movement of live cells, 2.5 × 104 A375/GPR56-S1 cells were stained with a microtracker (#M7512, Invitrogen) and seeded in a Corning BioCoat™ Matrigel Invasion Chamber (Corning) pre-incubated without or with CG4 mAb (5 µg/mL) for 1 h. Cells were placed in the Tokai Hit stage top incubation system (TOKAI HIT Co. Ltd.), which precisely controls temperature, humidity, and CO2 levels. For the time-lapse analysis of cell movement, cells were imaged every hour for 24 h using a Nikon Eclipse Ji confocal microscope. Images were captured under a focused laser beam with 3% power and a 1.0 AU pinhole using an S Plan Fluor ELWD 20x ELWD N.A. 0.45 objective. All images of live cells were deconvoluted and denoised with NIS-elements v6.1 (Nikon) and analyzed using a 3D object tracker to determine the movement of migrating cells in XYZ coordinates. The Kolmogorov–Smirnov test was used for the statistical analysis of cell movement.
Statistical analysis
Unless indicated otherwise, the data set of at least three independent experiments done in triplicate was included for statistical analysis, which was performed using the student t test or one-way ANOVA analysis by Prism 5 software (GraphPad Software Inc., La Jolla, California, USA). The results were shown as means ± standard error of the mean (SEM). The statistical significance of p value was set at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
GPR56 activation induces increased IL-6 production and amoeboid-like morphology in melanoma cells
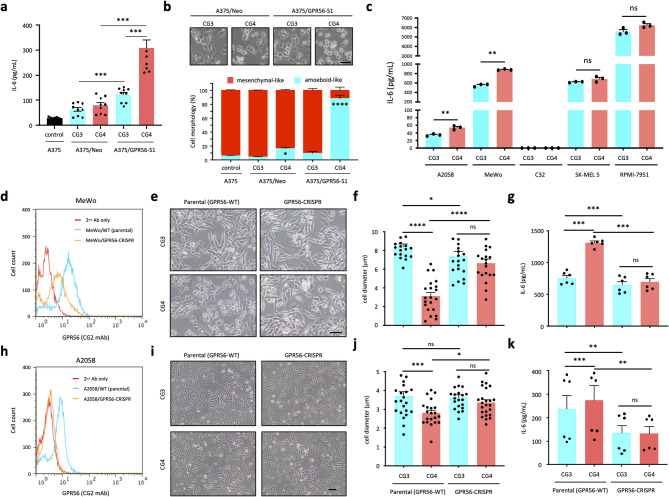
In line with our previous findings, stable A375 human melanoma cells over-expressing GPR56 splicing variant 1 (A375/GPR56-S1) produced significantly more IL-6 when stimulated with immobilized activating CG4 versus none-activating control CG3 mAb (Fig. 1a) [43]. In fact, in direct comparison to A375/Neo cells that expressed a low level of endogenous GPR56, A375/GPR56-S1 cells consistently secreted more IL-6 even when incubated with CG3 (Fig. 1a). These results suggest that forced expression of GPR56-S1 in A375 cells alone leads to constitutive receptor activation and elevated IL-6 production, which is further exacerbated by the agonistic CG4 mAb. In parallel, distinct cell morphologies were clearly noted in the two cell groups. Specifically, a large percentage (88.291 ± 4.731%) of A375/GPR56-S1 cells displayed a round-shaped amoeboid-like morphology when seeded in CG4-coated plates. By contrast, only ~ 16% (16.174 ± 1.273%) of CG4-stimulated A375/Neo cells exhibited the same morphology. On the other hand, the majority of A375/Neo (95.785 ± 0.557%) and A375/GPR56-S1 (90.515 ± 2.510%) cells showed a fully-spread mesenchymal-like morphology when cultured in the uncoated control or CG3-coated plates (Fig. 1b). Notably, no apparent difference in cell adhesion was observed in A375/GPR56-S1 cells cultured on plates coated with CG3 and CG4. Additionally, CG4-induced amoeboid-like cells remained adherent to the culture plates without any noticeable cell loss or death during long-term culture (data not shown). Furthermore, the morphological changes induced by CG4 were sustained even when A375/GPR56-S1 cells were cultured in the presence of various extracellular matrix proteins, such as fibronectin, collagen-I, and collagen-III. This observation underscores the role of CG4-induced GPR56 activation in regulating cellular morphology (Fig. S1a). In conclusion, CG4-elicited GPR56 activation in A375 cells drives both IL-6 upregulation and distinct morphological changes.
Fig. 1.
GPR56 activation induces IL-6 up-regulation and morphological change in human melanoma cell lines. (a) Analysis of IL-6 levels in the supernatants of A375/Neo and A375/GPR56-S1 cells incubated with immobilized CG3 and CG4 mAbs for 24 h. Untreated parental A375 cells were included as a negative control (n = 9). (b) Analysis of cell morphologies of A375/Neo and A375/GPR56-S1 cells cultured for 24 h on plates coated with CG3 and CG4 mAbs. Images in the top panel showed morphologies of mAb-treated cells, while the plot in the lower panel represented the percentage of elongated mesenchymal-like vs. round-shaped amoeboid-like cells from 3 random fields per group (n = 3). (c) Analysis of IL-6 levels in the supernatants of A2058, MeWo, C32, SK-MEL5, and RPMI-7951 melanoma cell lines incubated with immobilized CG3 and CG4 mAbs for 24 h (n = 3). (d, h) FACS analysis of surface GPR56 expression levels in parental and GPR56-CRISPR MeWo (d) and A2058 (h) cells as indicated. (e, f, I, j) Morphological changes of parental and GPR56-CRISPR MeWo (e, f) and A2058 (i, j) cells incubated with immobilized CG3 and CG4 mAbs were shown as microscopy images (e, i) and size changes in cell diameter (f, j) at 24 h of culture (f, n = 21; j, n = 24). Analysis of IL-6 levels in the supernatants of parental and GPR56-CRISPR MeWo (g) and A2058 (k) cells incubated with immobilized CG3 and CG4 mAbs for 24 h (g, n = 6; k, n = 6). Scale bar, 50 μm. Data are presented as means ± SEM from independent experiments performed in triplicate. ns, non-significant
These two distinct phenotypes captured our interest, prompting us to investigate whether they are commonly triggered by GPR56 activation in human melanoma cells. To this end, we analyzed five additional melanoma cell lines—A2058, MeWo, C32, SK-MEL-5, and RPMI-7951—that express different levels of endogenous GPR56. Consistently, varying degrees of CG4-induced morphological change were observed across all five cell lines, with the extent of these changes correlating roughly to their respective GPR56 expression levels (Fig. S1b, c). Once again, no obvious shape change was noted when cells were cultured on non-activating CG3-coated plates. Finally, IL-6 production was upregulated significantly by A2058 and MeWo cells upon CG4 but not CG3 stimulation. Of note, C32 cells did not produce any IL-6, while SK-MEL-5 and RPMI-7951 cells secreted high levels of IL-6 constitutively whether untreated or treated with CG3 and CG4 (Fig. 1c and Fig. S1c).
Next, CRISPR/Cas9-mediated GPR56 gene disruption was performed in A2058 and MeWo cells to verify the specific role of endogenous GPR56 in regulating cell morphology and IL-6 production. As expected, the two distinct CG4-induced phenotypes were significantly attenuated in A2058/GPR56-CRISPR and MeWo/GPR56-CRISPR cells compared to their parental counterparts (Fig. 1d-k). In truth, the two GPR56-CRISPR cell lines produced significantly less IL-6 than their parental cells, even when incubated with CG3 (Fig. 1g, k). Collectively, these results indicate that GPR56 activation in melanoma cells generally leads to upregulated IL-6 production and morphological changes in a manner dependent on GPR56 expression levels.
Amoeboid-like morphology and IL-6 upregulation are two unique phenotypes temporally induced by GPR56-mediated signaling in melanoma cells
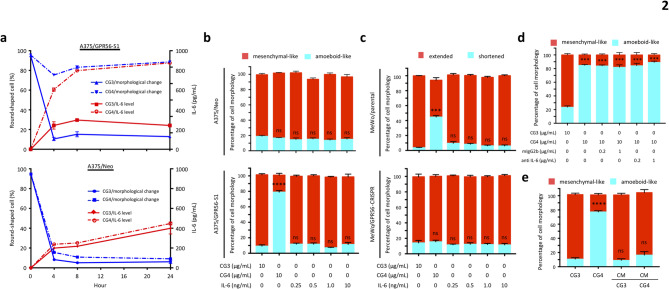
IL-6 is a pro-inflammatory cytokine with both pro- and anti-tumor functions, including modulation of cellular morphology [46, 47]. This prompted us to wonder whether CG4-induced amoeboid-like morphology is a direct, primary outcome of GPR56-mediated signaling, or if it is a secondary effect driven by IL-6 (or other soluble factors). To verify this, we first conducted a time-course phenotypic analysis of CG4-stimulated A375/GPR56-S1 cells. This revealed significant morphological changes at 2 h of incubation and increased IL-6 production at 4 h. Both phenotypes peaked around 8 h of culture and persisted for at least 24 h (Fig. 2a and Fig. S2a, b). Importantly, similar results were also observed in MeWo cells (Fig. S2c, d), indicating that GPR56-induced morphological change generally takes place prior to IL-6 upregulation, probably due to the suppression of cell spreading, i.e. anti-spreading.
Fig. 2.
Amoeboid-like morphology and IL-6 up-regulation are two independent melanoma cell phenotypes induced by GPR56 activation. (a) Time course analyses of indicated phenotypical changes of A375/Neo and A375/GPR56-S1 cells stimulated with the immobilized CG4 mAb. CG3 was used as a negative control (n = 3). (b, c) Analysis of the effect of exogenous IL-6 on cell morphologies of A375 (b) and MeWo (c) cells as indicated. Cells incubated with immobilized CG3 and CG4 mAbs were included as controls (n = 4). (d) Analysis of the effect of a functional blocking anti IL-6 mAb on CG4-induced amoeboid-like morphology of A375/GPR56-S1 cells. A mouse IgG2b isotype was included as a negative control (n = 3). (e) Analysis of the effect of conditioned medium (CM) of CG3-/CG4-treated A375/GPR56-S1 cells on cell morphology of A375/GPR56-S1 cells (n = 3). Data are presented as means ± SEM from independent experiments performed in triplicate. ns, non-significant
Since cell anti-spreading occurs shortly after cellular contact with immobilized CG4, when IL-6 levels are still low, secreted IL-6 is unlikely to be responsible for this phenotype. In fact, the addition of exogenous IL-6, even at high concentrations, failed to induce cell rounding in A375 and MeWo cells (Fig. 2b, c). Incubation of A375/GPR56-S1 cells with a functional blocking anti-IL-6 mAb did not impede CG4-induced anti-spreading (Fig. 2d). Finally, no obvious shape changes were noted when cells were cultured in conditioned medium collected from A375/GPR56-S1 cells stimulated with immobilized CG4 or CG3 mAb (Fig. 2e). Thus, we conclude that anti-spreading and IL-6 upregulation are two distinct phenotypes temporally induced by GPR56-mediated signaling in melanoma cells.
The RHO-ROCK-MLC and JAK-STAT3 signaling pathways differentially regulate GPR56-elicited amoeboid-like morphology and IL-6 upregulation in melanoma cells
The Gα12/13-RhoA axis has been identified as the predominant signaling pathway triggered by GPR56 activation [31, 41, 48, 49]. Rho-associated protein kinase (ROCK) is the principal downstream effector of small Rho GTPases and Rho-ROCK signaling is known to regulate cell morphology, polarity, and migration via dynamic cytoskeletal remodeling [50, 51]. Of special interest, ROCK activation enhances the phosphorylation of myosin light chain (MLC), which facilitates and reinforces actin-myosin II interactions, ultimately driving increased cellular contractility and an amoeboid-like morphology [52–54]. Moreover, Rho-ROCK signaling has been implicated in the regulation of IL-6 expression in several cell types [55, 56].
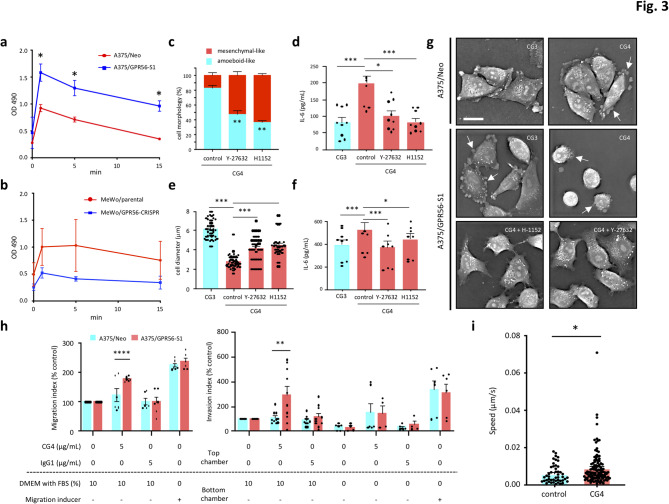
We therefore investigated the role of Rho-ROCK signaling in GPR56-mediated phenotypic manifestations. In line with earlier reports, A375/GPR56-S1 cells produced significantly higher levels of active RhoA-GTP than did A375/Neo cells soon after receptor ligation by CG4, starting at 1 min and lasted for at least 15 min (Fig. 3a). Likewise, more active RhoA-GTP was produced in parental MeWo cells than MeWo/GPR56-CRISPR cells upon CG4 stimulation (Fig. 3b). Consistently, incubation of A375/GPR56-S1 and MeWo cells with two different ROCK inhibitors, Y-27,632 and H-1152, dramatically attenuated GPR56-elicited morphological changes and IL-6 upregulation (Fig. 3c-f and Fig. S3a, b). A closer examination of the anti-spreading phenotype revealed pronounced membrane blebbing in CG4-treated A375/GPR56-S1 cells, which were effectively “rescued” upon treatment with ROCK inhibitors (Fig. 3g). In truth, cell blebbing was also noted in some CG4-treated A375/Neo and CG3-treated A375/GPR56-S1 cells even though these cells mostly displayed a mesenchymal-like morphology (Fig. 3g). In summary, these results establish a clear connection between GPR56-mediated Rho-ROCK signaling, cytoskeletal remodeling, and IL-6 upregulation.
Fig. 3.
GPR56-elicited RHO-ROCK-MLC signalling pathway promotes amoeboid-like morphology, IL-6 up-regulation, and cell migration and invasion in melanoma cells. (a, b) Analyses of active RhoA-GTP levels in A375 (a) and MeWo (b) cells incubated with immobilized CG4 mAb at indicated time points (n = 3). (c-f) Analyses of the effect of two ROCK inhibitors, Y-27,632 and H1152, on CG4-induced amoeboid-like morphology and IL-6 up-regulation of A375/GPR56-S1 cells (c, d) and MeWo cells (e, f) (c, n = 3; d, n = 9; e, n = 50; f, n = 9). (g) High-resolution image analyses of cell morphologies of CG4-treated A375/GPR56-S1 cells in the absence or presence of ROCK inhibitors at 6 h of culture. A375/Neo cells and CG3 mAb treatment were included as negative controls. Scale bar, 20 μm. White arrows indicate cell blebbing. (h) Cell migration and invasion assays of A375/Neo and A375/GPR56-S1 cells under different treatment conditions. Migrated cells were stained and measured by Ext.530/Emi.590 (n = 3). Data were analysed using two-way ANOVA Tukey’s multiple comparisons test. Data are presented as means ± SEM from independent experiments performed in triplicate. (i) Time-lapse cell migration assay of A375/GPR56-S1 cells treated without or with CG4 (5 µg/mL). In total, 48 control cell movements and 120 CG4-treated cell movements were analysed. Data were analysed using the Kolmogorov–Smirnov test. *p < 0.05
Given that Rho/ROCK-regulated actomyosin contractility promotes cell blebbing, amoeboid morphology, and increased cell motility, we next investigated the effects of CG4 treatment on melanoma cell migration and invasion. Indeed, A375/GPR56-S1 cells exhibited significantly enhanced cell migration and invasion in response to 10% FBS chemoattractant after CG4 incubation, compared to A375/Neo cells (Fig. 3h). In contrast, no similar effects were observed in cells treated with the IgG1 control. Notably, the time-lapse cell migration assay revealed a significantly increased migration speed in A375/GPR56-S1 cells following CG4 stimulation compared to the control (Fig. 3i). These findings align remarkably well with the amoeboid-like phenotype observed in CG4-activated A375/GPR56-S1 cells.
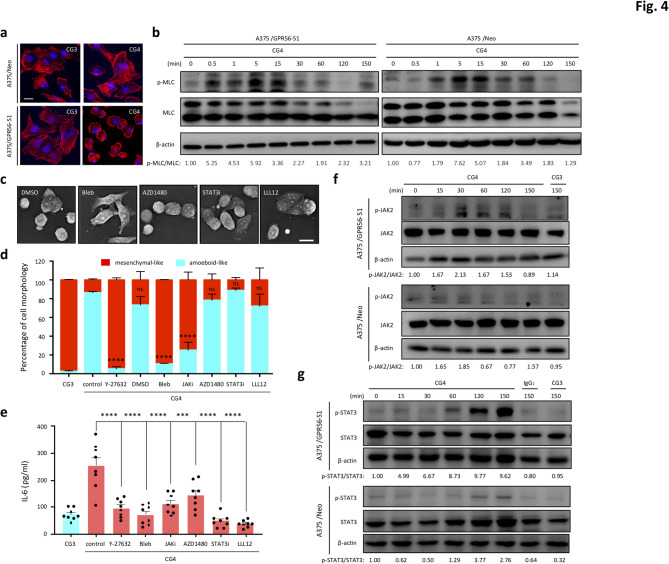
Consistently, confocal imaging of CG4-treated A375/GPR56-S1 cells revealed shortened, peripherally enriched F-actin patterns characteristic of amoeboid-like cells. Conversely, extensive actin-rich membrane protrusions were observed in CG3-treated A375/GPR56-S1 and control A375/Neo mesenchymal-like cells (Fig. 4a). As expected, CG4 stimulation induced significantly-increased and more rapid MLC phosphorylation in A375/GPR56-S1 cells compared to A375/Neo cells (Fig. 4b and Fig. S4a). Likewise, MeWo/parental cells exhibited elevated MLC phosphorylation relative to MeWo/GPR56-CRSIPR cells under CG4 treatment (Fig. S4b). Conceivably, incubation of A375/GPR56-S1 cells with ROCK inhibitors significantly reduced CG4-induced MLC phosphorylation, which aligns with their inhibitory effects on amoeboid-like morphologies and IL-6 upregulation (Fig. S4c, Fig. 4d-e). We next treated cells with blebbistatin, a myosin ATPase-specific inhibitor that destabilizes actomyosin filaments [57]. As expected, CG4-induced cell blebbing and amoeboid-like morphologies were abolished in cells treated with blebbistatin, as well as in those treated with ROCK inhibitors (Figs. 3g and 4c-d and Fig. S3a). Interestingly, CG4-induced IL-6 upregulation was also dramatically reduced in the presence of blebbistatin (Fig. 4e). Together, these results confirm that GPR56-induced anti-spreading and increased IL-6 production in melanoma cells are largely regulated by the Rho-ROCK-MLC signaling pathway.
Fig. 4.
Differential regulation of amoeboid-like morphology and IL-6 up-regulation by biased RHO-ROCK-MLC and JAK-STAT3 signalling pathways. (a) Confocal fluorescence analysis of F-actin (red) staining patterns in A375/Neo and A375/GPR56-S1 cells incubated with immobilized CG3 and CG4 mAb at 6 h of culture. Cell nucleus was marked by Hoechst staining (blue). Scale bar, 10 μm. (b) Western blotting analysis of p-MLC and total MLC levels of A375/Neo and A375/GPR56-S1 cells incubated with CG4 mAb at indicated time points. (c) High-resolution image analyses of cell morphologies of CG4-treated A375/GPR56-S1 cells at 6 h of culture in the absence or presence of signalling inhibitors as indicated. DMSO was used as a negative control. Scale bar, 20 μm. (d, e) Morphological analysis (d, n = 3) and ELISA analysis of IL-6 levels (e, n = 8) of CG4-treated A375/GPR56-S1 cells in the absence or presence of signalling inhibitors as indicated. Data are presented as means ± SEM from independent experiments performed in triplicate. ns, non-significant. (f, g) Western blotting analyses of p-JAK2 and total JAK levels (f) and p-STAT3 and total STAT3 levels (g) of A375/Neo and A375/GPR56-S1 cells incubated with CG4 mAb at indicated time points. CG3 and an IgG1 isotype were used as negative controls. Probing with the anti β-actin Ab was used to show the equal loading of lysate samples
STAT3 is the primary cytoplasmic effector of the IL-6/IL-6 receptor (IL-6R) signaling axis and the active STAT3 transcription factor is known to enhance the transcription and biosynthesis of IL-6, hence resulting in a positive feedback regulation [46, 47]. Importantly, a direct link of the Rho-ROCK signaling pathway to STAT3 activation and a close interrelationship between actomyosin contractility and the JAK-STAT3 pathway have been established previously [58, 59]. We hence investigated the potential role of JAKs and STAT3 in GPR56-induced IL-6 upregulation and anti-spreading. As shown, JAK2 and STAT3 phosphorylation was indeed much increased in A375/GPR56-S1 cells than in A375/Neo cells when stimulated by CG4 (Fig. 4f, g and Fig. S4d). Similar results were also noted in MeWo/parental cells versus MeWo/GPR56-CRSIPR cells (Fig. S4b, e). Intriguingly, while incubation of cells with inhibitors of JAK and STAT3 markedly inhibited GPR56-induced IL-6 upregulation, their effects on cell morphology were marginal at best (Fig. 4c-e). Specifically, only the reversible, cell-permeable JAK inhibitor I (JAKi), but not another small-molecule JAK inhibitor (AZD1480) nor the STAT3 inhibitors (STAT3i and LLL12) showed a significant inhibitory effect on CG4-induced amoeboid-like morphology in A375/GPR56-S1 cells (Fig. 4d). The significant inhibitory effect of JAKi, but not AZD1480, on CG4-induced morphological changes may be attributed to the distinct activities of these two inhibitors on different JAKs and intracellular kinases. Nevertheless, these results suggest that the JAK-STAT3 signaling pathway is crucial for IL-6 upregulation, but plays a minimal role in the morphological changes of melanoma cells elicited by GPR56 activation.
Temporal regulation and cross-talk of the GPR56-induced ROCK-MLC and JAK-STAT3 signaling pathways in melanoma cells
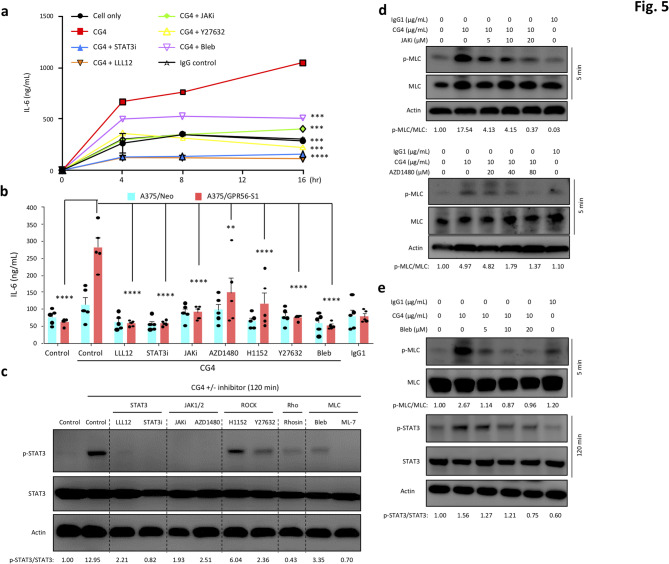
The distinct roles of Rho-ROCK-MLC and JAK-STAT3 signaling pathways in regulating GPR56-induced anti-spreading and IL-6 upregulation suggest a potential temporal relationship and cross-talk between the two pathways. Of note, enhanced phosphorylation of MLC, JAK2, and STAT3 was detected in a temporal manner and peaked at different time points (pMLC at 0.5–5 min, pJAK2 at 15–60 min, and pSTAT3 at 60–120 min) in CG4-reated A375/GPR56-S1 and MeWo cells (Fig. 4b, f, g and Fig. S4f). Importantly, we found that GPR56-induced IL-6 upregulation was inhibited by all signaling inhibitors tested as early as 4 h after culture and persisted for up to 16 h (Fig. 5a, b). In contrast, GPR56-induced amoeboid-like morphology observed at 2 h of incubation was attenuated by ROCK-MLC inhibitors, but not by most JAK/STAT3 signaling inhibitors (Fig. 4d). These results strongly suggest that GPR56-elicited JAK-STAT3 signaling is partly modulated by the Rho-ROCK-MLC pathway, but it seems unlikely that the reverse is true.
Fig. 5.
Involvement of the RHO-ROCK-MLC and JAK-STAT3 signalling pathways in GPR56-induced IL-6 up-regulation in melanoma cells. (a) Time course analyses of CG4-induced IL-6 up-regulation of A375/GPR56-S1 cells cultured in the absence or presence of signalling inhibitors as indicated. (b) Analyses of CG4-induced IL-6 up-regulation of A375/Neo and A375/GPR56-S1 cells cultured for 20 h in the absence or presence of signalling inhibitors as indicated (n = 5). Data are presented as means ± SEM from independent experiments performed in triplicate. (c-e) Western blotting analysis of p-MLC and total MLC, p-STAT3 and total STAT3 levels of CG4-stimulated A375/GPR56-S1 cells in the absence or presence of signalling inhibitors as indicated. Cells alone and cells incubated with an immobilized IgG1 were included as negative controls
To decipher the temporal relationship and possible cross-talk of the Rho-ROCK-MLC and JAK-STAT3 signaling axes induced by GPR56, we analyzed CG4-induced MLC and STAT3 activation in the presence of various signaling inhibitors at different time points. Indeed, while inhibitors of JAKs and STAT3 abolished STAT3 phosphorylation detected at 120 min, their effects on MLC phosphorylation at 5 min was relatively weak. Of note, higher concentrations of JAK and STAT3 inhibitors did show some inhibitory effects on MLC phosphorylation (Fig. 5c, d and Fig. S4g). By contrast, inhibitors of the Rho-ROCK-MLC pathway all lead to a dramatic retardation of MLC phosphorylation at 5 min and a partial reduction of STAT3 phosphorylation at 120 min (Fig. 5c, e and Fig. S4c). We hypothesize that the diminished but persistent STAT3 phosphorylation observed in the presence of ROCK/MLC inhibitors likely results from the independent positive feedback signaling of the IL-6 receptor, activated by basal IL-6 constitutively produced by melanoma cells. These results indicate that the GPR56-triggered Rho-ROCK-MLC pathway not only regulates cytoskeletal organization and cell morphology, but also positively modulates the JAK/STAT3/IL-6 pathway, which plays a minor role in regulating ROCK-MLC signaling.
GPS auto-proteolysis modulates the magnitude of GPR56-elicited signaling
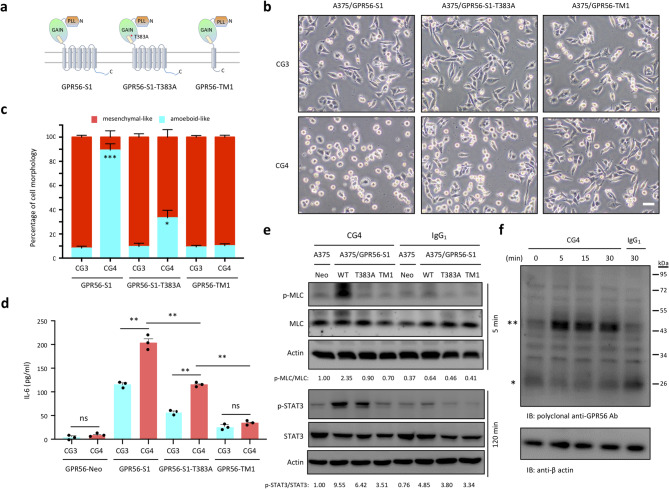
We have shown previously that GPR56-induced IL-6 upregulation is dependent on its GPS auto-proteolysis and 7TM region [43]. To determine whether the same is true for the anti-spreading phenotype, we compared the morphology of A375/GPR56-S1, A375/GPR56-S1-T383A, and A375/GPR56-TM1 cells in response to CG4 stimulation. The T383A mutant is a GPS cleavage-deficient receptor, while GPR56-TM1 represents GPR56 receptor containing only the first TM (Fig. 6a and Fig. S5a). Comparable GPR56 expression was detected in these stable cells by flow cytometry and western blot analyses using CG4 mAb and rabbit polyclonal anti-serum against the cytoplasmic C-terminal peptide of GPR56. Importantly, these analyses not only verified that the respective GPR56 receptor variants were properly processed as expected, but also identified monomeric and dimeric forms of GPR56-CTF in lysates from A375/GPR56-S1 cells, as reported previously (Fig. S5a, b) [31].
Fig. 6.
The role of GPS auto-proteolysis and 7TM in regulating GPR56-mediated signalling. (a) Schematic diagrams depict the various recombinant GPR56 receptors analyzed in the study, including the wild-type (S1), GPS cleavage-deficient (S1-T383A), and the first TM only-containing variant (GPR56-TM1). (b, c) Analysis of morphological changes of A375/GPR56-S1, A375/GPR56-S1-T383A and A375/GPR56-TM1 cells incubated with immobilized CG3 and CG4 mAbs at 24 h of culture. Scale bar, 20 μm. The data in (c) represented the percentage of elongated mesenchymal-like cells and round-shaped amoeboid-like cells from 3 different fields (100 total cells per field) of images shown in (b). (d) Analysis of IL-6 levels in the supernatants of A375/Neo, A375/GPR56-S1, A375/GPR56-S1-T383A, and A375/GPR56-TM1 cells incubated with immobilized CG3 and CG4 mAbs for 24 h (n = 3). Data are presented as means ± SEM from independent experiments performed in triplicate. ns, non-significant. (e) Western blotting analysis of p-MLC and total MLC, p-STAT3 and total STAT3 levels of indicated stable A375 cell lines incubated with CG4 or IgG1 at 5 and 120 min, respectively. A375/Neo cells were included as a negative control. (f) Western blotting analysis of the GPR56-CTF subunit in lysates of A375/GPR56-S1 cells following the binding of CG4 mAb at different time points. Blots were probed by a rabbit polyclonal Ab against a GPR56 cytoplasmic peptide sequence. * and ** denote the monomeric and dimeric forms of GPR56-CTF, respectively
Surprisingly, we found that ~ 35% (33.300 ± 10.763%) of A375/GPR56-S1-T383A cells displayed amoeboid-like morphology when incubated with CG4, but remained predominantly mesenchymal appearance (90.434 ± 4.801%) upon CG3 treatment. Conversely, the majority of A375/GPR56-TM1 cells manifested the mesenchymal-like morphology whether stimulated by CG3 (90.825 ± 2.383%) or CG4 (89.791 ± 2.802%)(Fig. 6b, c). Likewise, IL-6 levels produced by A375/GPR56-S1-T383A cells were significantly higher than those of A375/GPR56-TM1 cells, but lower than those from A375/GPR56-S1 cells upon CG4 stimulation (Fig. 6d). Consistent with these phenotypic effects, the levels of pMLC and pSTAT3 were lower in CG4-treated A375/GPR56-S1-T383A cells in comparison to those of A375/GPR56-S1 cells. Meanwhile, no noticeable signaling events were observed in A375/GPR56-TM1 cells under the same treatment conditions (Fig. 6e).
Altogether, these results affirm that an intact 7TM region is absolutely required for GPR56-mediated signaling. However, unexpectedly, the GPS-unprocessed variant appears capable of inducing partial signaling, leading to minor phenotypic changes upon activation. This supports the concept of allosteric regulation in the activation and signaling of GPS cleavage-deficient aGPCRs [60]. Therefore, we conclude that GPS proteolysis is involved in modulating the magnitude of GPR56-mediated signaling (full verse partial), probably by regulating the extent to which the agonistic Stachel peptide is exposed.
GPR56-elicited signaling is regulated differentially by unique regions/residues of its 7TM moiety
The 7TM moiety plays an essential role in regulating the signaling output of GPCRs by determining the selective coupling of receptor to specific G proteins and signal transducers, as well as influencing other protein modifications, including phosphorylation, ubiquitination, and dimerization [61–63]. Intriguingly, increased signals of the dimeric GPR56-CTF form were detected soon after CG4-induced GPR56 activation (Fig. 6f). This result strongly suggests that GPR56-CTF adapts a predominantly homo-dimeric conformation following CG4 stimulation, likely due to structural and/or molecular modifications of the 7TM domain.
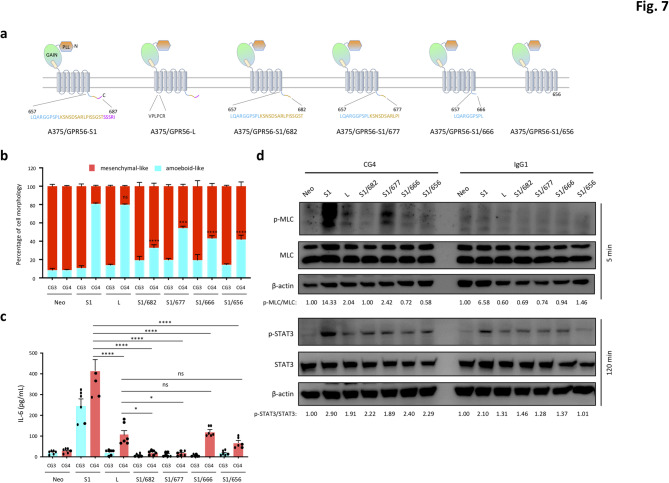
To dissect the role of the 7TM region in regulating GPR56-mediated signaling, we looked closer at the GPR56 isoforms with different lengths of the 7TM domain. Previous studies have shown that the long-form GPR56 variant (GPR56-L) contains 6 additional amino acid residues in the first ICL and induces a weaker signaling activity in comparison to the GPR56-S1 variant, suggesting a regulatory role for the first ICL in GPR56-mediated signaling [38]. In addition, the C-terminal tail region of GPR56-7TM domain contains numerous Ser/Thr residues, which serve as potential phosphorylation sites and may act as regulatory determinants for distinct signaling pathways.
We therefore established stable A375 cells expressing the GPR56-L isoform (A375/GPR56-L) and GPR56-S1 variants with different truncated C-terminal ends. In total, 4 C-terminally truncated GPR56-S1 variants, including S1/682, S1/677, S1/666, and S1/656, which obliterated specific Ser/Thr-rich sequences were generated (Fig. 7a, S6a). As demonstrated, these stable A375 cell lines all displayed similar mesenchymal-like morphology when cultured on uncoated and CG3-coated culture plates (Fig. 7b, S6b). Interestingly, divergent effects on cell morphology and IL-6 production were noted in distinct stable cell lines upon CG4 stimulation. As shown, the majority (~ 83%) of A375/GPR56-L cells exhibited an amoeboid-like morphology, similar to A375/GPR56-S1 cells. However, IL-6 production by A375/GPR56-L cells was significantly lower compared to A375/GPR56-S1 cells (Fig. 7b, c). By contrast, amoeboid-like morphology was identified in approximately 30–40% of stable A375 cells expressing the GPR56-S1/682, S1/677, S1/666, and S1/656 variants, all of which produced significantly lower or basal levels of IL-6 following CG4 incubation (Fig. 7b, c).
Fig. 7.
Differential regulation of GPR56-elicited signalling by unique regions/residues of the 7TM moiety. (a) Schematic diagrams depict the various GPR56 receptor variants analyzed in the study, including the wild-type short form 1 (S1), long form (L), and C-terminally truncated GPR56-S1/682, S1/677, S1/666, and S1/656 receptors. (b, c) Analysis of morphological changes (b) and secreted IL-6 levels (c) of stable A375 cells expressing indicated GPR56 receptor variants incubated with immobilized CG3 and CG4 mAbs at 24 h (n = 5). Data are presented as means ± SEM from independent experiments performed in triplicate. ns, non-significant. (d) Western blotting analysis of p-MLC and total MLC, p-STAT3 and total STAT3 levels of indicated stable A375 cell lines incubated with CG4 or IgG1 at 5 and 120 min, respectively
In keeping with these cellular phenotypes, western blotting analyses revealed that A375/GPR56-L cells exhibit significant MLC phosphorylation, but only weak STAT3 activation upon CG4 activation, compared to cells expressing the GPR56-S1 isoform (Fig. 7d). Conversely, aside from a strong MLC phosphorylation signal detected in CG4-activated A375/GPR56-S1/677 cells, none of the other GPR56-truncated variants induced significant MLC or STAT3 activation under the same incubation conditions (Fig. 7d). In conclusion, the divergent signaling and phenotypical manifestations induced by GPR56-L and C-terminally truncated GPR56-S1 variants suggest isoform-specific biased signaling of GPR56, regulated differentially, in part, by its ICL and cytoplasmic sequences. Specifically, the length and/or sequence of the 1st ICL region, along with the distinct C-terminal Ser/Thr-rich sub-regions, appear to play unique regulatory roles in modulating GPR56-mediated ROCK-MLC and JAK-STAT3 signaling pathways.
Discussion
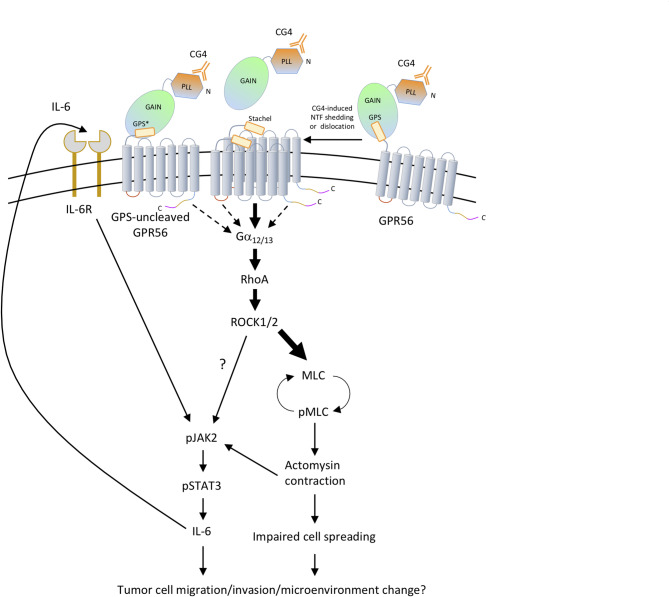
Mutated and dysregulated GPCRs are well-known drivers for cancer progression. Thus, cancer-associated GPCRs represent one of favorable biomarkers and pharmacological targets of anti-cancer therapy [64–67]. In recent years, the role of aGPCRs in tumorigenesis has gained increasing attention, with several of these receptors being considered promising targets of targeted cancer therapy [6, 68, 69]. In this study, we dissected the GPR56-induced signaling pathways and their associated tumorigenic phenotypes in human melanoma cells. Our findings demonstrate that GPR56 activation triggers Rho-ROCK-MLC signaling downstream of Gα12/13, leading to the promotion of amoeboid-like morphology and upregulation of IL-6 (Figs. 1, 2, 3 and 4). Interestingly, GPR56-induced IL-6 production relies heavily on the JAK2-STAT3 pathway, which is partially regulated by, and likely functions downstream of, the Rho-ROCK-MLC pathway. Conversely, the JAK2-STAT3 pathway plays only a minor role in influencing the ROCK-MLC pathway and the associated morphological change (Figs. 5 and 8). Amoeboid-like morphology and IL-6 are well-established pro-tumorigenic factors involved in cell migration, invasion, and immune modulation of tumor microenvironment (TME). These findings suggest that GPR56 likely plays a role in the mesenchymal-to-amoeboid transition and TME modification in melanoma. Moreover, the Rho/ROCK and JAK/STAT signaling pathways, both critical in cancer pathogenesis, have been explored as potential therapeutic targets [70, 71]. Our results provide valuable insights into potential signaling-intervention points for addressing GPR56-modulated tumorigenic phenotypes in melanoma (Fig. 8).
Fig. 8.
The proposed model of GPR56-mediated signaling in human melanoma cell. The binding of the CG4 mAb to the PLL domain of GPR56 induces GPR56-NTF shedding and formation of dimeric GPR56-CTF, which triggers Gα12/13 coupling and RhoA/ROCK activation. MLC phosphorylation is induced immediately downstream of ROCK and leads to cytoskeletal rearrangement, actomyosin contraction, and amoeboid-like morphology. Additionally, JAK2/STAT3 signaling is activated either directly by ROCK or pMLC-mediated actomyosin contraction to induce IL-6 production, which acts via an autocrine mechanism through the IL-6 receptor (IL-6R) to activate the baseline JAK2/STAT3 signaling activity. GPS cleavage-deficient GPR56 induces a lower signaling output than the fully-cleaved WT receptor, while differential signaling activities are elicited by GPR56 variants with different ICL or cytoplasmic tail sequences
The role of GPS cleavage in aGPCR activation has been a topic of considerable debate. While this highly conserved proteolytic modification implies a shared functional importance across most aGPCRs, some aGPCRs remain as uncleaved full-length proteins, suggesting alternative GPS cleavage-independent activation mechanisms. Although the tethered agonism model posits that GPS proteolysis and subsequent NTF-CTF separation are prerequisites for aGPCR activation via the newly exposed Stachel peptide, recent findings have indicated that aGPCRs can also be activated without NTF-CTF dissociation or GPS cleavage, as proposed by the allosteric activation/inhibition models [32, 33, 60]. Our previous results demonstrate that immobilized CG4 mAb activates GPR56 by promoting NTF-CTF dissociation in a CD9/CD81-dependent manner, aligning with the tethered agonism model of aGPCRs [43]. However, a more detailed functional analysis of the GPS-uncleaved GPR56 variant revealed partial signaling activities that led to minor phenotypic changes (Fig. 6). This new observation supports the allosteric regulation model of aGPCR activation, suggesting that the availability of the agonistic Stachel peptide, regulated by GPS proteolysis and/or NTF-CTF interaction, influences the magnitude of receptor activation (full versus partial).
The revelation of differential Rho-ROCK-MLC and JAK2-STAT3 signaling induced by distinct GPR56 isoforms is particularly intriguing, as it strongly suggests the presence of novel biased signaling. Biased agonism in GPCRs was initially proposed to explain how the same receptor can activate different signaling pathways when bound to distinct ligands. This occurs through the selective interaction of ligand-induced receptor conformations with specific signaling transducers, such as G proteins or β-arrestins [72–74]. In addition to biased ligands, later research has broadened the understanding of GPCR biased signaling to include other contributing factors, such as biased receptors, system bias, location bias, and even specific subdomains of bias [72, 75]. Our results indicate that GPR56-induced biased signaling occurs downstream of Gα12/13 and is partially regulated by its ICL and cytoplasmic sub-regions. Variations in the sequence or length of these regions likely contribute to the adaptation of distinct receptor conformations or modifications, driving differential signaling outcomes (Figs. 7 and 8). In this regard, it is noteworthy that CG4-induced activation leads to the increased formation of homo-dimeric GPR56-CTF (Fig. 6f), as oligomeric GPCR structures are known to produce signaling outputs distinct from those of monomeric GPCRs [74, 75]. Therefore, distinct GPR56 isoforms, differentially expressed within a given cell population or tissue, may elicit biased signaling upon ligand binding and activation. Coincidentally, this aligns with recent findings on cell type-specific alternatively spliced latrophilin 3/Adgrl3 variants, which control the biased coupling of Gαs or Gα12/13 to regulate synapse formation in the mouse brain [76]. Critically, the distinct roles of specific cytoplasmic Ser/Thr-rich sub-regions in regulating GPR56-mediated signaling suggest the involvement of post-translational modifications, such as phosphorylation, which are known to modulate the selective interaction of GPCRs with unique signaling effectors. Future studies will be crucial to explore the role of these potential molecular modifications in biased signaling across different GPR56 isoforms.
In addition to the numerous cellular ligands and binding partners identified to date, various GPR56-specific reagents, including Abs, monobodies, and small molecules, have been developed for its functional analysis [11, 18, 37, 41, 77–79]. Moreover, the preclinical anti-tumor efficacy of a GPR56-specific Ab-drug conjugate has recently been evaluated in colorectal cancer models [80]. Intriguingly, these GPR56-targeting molecules induce a variety of effects, including cell activation, adhesion, and migration, partly through shear force-induced NTF shedding, cytoskeletal remodeling, or receptor internalization. Notably, these cellular responses appear to be mediated by distinct G protein-dependent or -independent signaling pathways, depending on the binding regions and/or affinities of the GPR56-targeting molecules. Altogether, these findings emphasize the need for comprehensive signaling studies prior to the therapeutic use of GPR56-targeting agonists, antagonists, and inhibitors. Indeed, a recent study by Luo et al. demonstrated that targeting unique GPR56-mediated signaling in hepatocellular carcinoma with specific inhibitors led to promising results in inhibiting the in vivo metastasis of hepatocellular carcinoma cells [81]. Future strategies aimed at targeting the GPR56-induced signaling networks identified in this study may yield similar therapeutic benefits in the treatment of human melanomas.
Conclusions
In summary, we reveal the intricate biased Rho-ROCK-MLC and JAK-STAT3 signaling networks induced by CG4-mediated GPR56 activation in human melanoma cells that lead to amoeboid-like morphology and up-regulated IL-6 production. Our findings provide novel insights into the role of specific signaling effectors as potential targets to regulate GPR56-modulated tumorigenic phenotypes of melanoma cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Gin-Wen Chang for the critical reading of the manuscript. The authors acknowledge the technical assistance from the Microscopy and Instrumentation Centers, Chang Gung University and the technical and instrumental support from Lin Trading Co., Ltd., Taiwan.
Author contributions
KYH, KFN, KYI, YFC, CMH, WCY, MS, and HHL designed the experiments, analysed, and interpreted the data. KYH, KFN, KYI, YCC, YFC, CMH, WCY, HYC, and TCC performed experiments. KYH, KFN, KYI, and HHL wrote the manuscript. HHL supervised the project. KYH, KFN, and HHL obtained funding.
Funding
This study was supported by grants from Chang Gung Memorial Hospital to KFN (CMRPG3M0461 and CMRPG3M1941) and to HHL (CMRPD1M0033 and CMRPD1M0323) and the National Science and Technology Council (NSTC), Taiwan to KYH (NSTC-108-2811-B-182-507) and HHL (NSTC-110-2320-B-182-024, NSTC-113-2918-I-182-001 and NSTC-113-2320-B-182-009).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Competing financial interests
The authors declare no competing financial interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kuan-Yeh Huang, Kwai-Fong Ng and Kuan-Yu I contributed equally to this work.
References
- 1.Wu V, Yeerna H, Nohata N, Chiou J, Harismendy O, Raimondi F, Inoue A, Russell RB, Tamayo P, Gutkind JS. Illuminating the Onco-GPCRome: novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J Biol Chem. 2019;294:11062–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. [DOI] [PubMed] [Google Scholar]
- 3.Aust G. Adhesion-GPCRS in tumorigenesis. Adv Exp Med Biol. 2010;706:109–20. [DOI] [PubMed] [Google Scholar]
- 4.Lin HH. Adhesion family of G protein-coupled receptors and cancer. Chang Gung Med J. 2012;35:15–27. [DOI] [PubMed] [Google Scholar]
- 5.Lala T, Hall RA. Adhesion G protein-coupled receptors: structure, signaling, physiology, and pathophysiology. Physiol Rev. 2022;102:1587–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gad AA, Balenga N. The emerging role of adhesion GPCRs in Cancer. ACS Pharmacol Translational Sci. 2020;3:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng KF, Chen TC, Stacey M, Lin HH. Role of ADGRG1/GPR56 in Tumor Progression. Cells. 2021;10. [DOI] [PMC free article] [PubMed]
- 8.Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett. 1999;446:292–8. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci USA. 2006;103:9023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Chen G, Mohanty S, Scott G, Fazal F, Rahman A, Begum S, Hynes RO, Xu L. GPR56 regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011;71:5558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Friedland S, Corson N, Xu L. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 2014;74:1022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Xu L. GPR56 in cancer progression: current status and future perspective. Future Oncol. 2012;8:431–40. [DOI] [PubMed] [Google Scholar]
- 13.Moreno M, Pedrosa L, Pare L, Pineda E, Bejarano L, Martinez J, Balasubramaniyan V, Ezhilarasan R, Kallarackal N, Kim SH, Wang J, Audia A, Conroy S, Marin M, Ribalta T, Pujol T, Herreros A, Tortosa A, Mira H, Alonso MM, Gomez-Manzano C, Graus F, Sulman EP, Piao X, Nakano I, Prat A. Bhat, K. P. & de la Iglesia, N. GPR56/ADGRG1 Inhibits Mesenchymal Differentiation and Radioresistance in Glioblastoma. Cell reports. 2017;21:2183–2197. [DOI] [PubMed]
- 14.Ji B, Feng Y, Sun Y, Ji D, Qian W, Zhang Z, Wang Q, Zhang Y, Zhang C, Sun Y. GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelialmesenchymal transition through PI3K/AKT signaling activation. Oncol Rep. 2018;40:1885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kausar T, Sharma R, Hasan MR, Tripathi SC, Saraya A, Chattopadhyay TK, Gupta SD, Ralhan R. Clinical significance of GPR56, transglutaminase 2, and NF-kappaB in esophageal squamous cell carcinoma. Cancer Invest. 2011;29:42–8. [DOI] [PubMed] [Google Scholar]
- 16.Ke N, Sundaram R, Liu G, Chionis J, Fan W, Rogers C, Awad T, Grifman M, Yu D, Wong-Staal F, Li QX. Orphan G protein-coupled receptor GPR56 plays a role in cell transformation and tumorigenesis involving the cell adhesion pathway. Mol Cancer Ther. 2007;6:1840–50. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Huang Z, Yang W, Li Z, Xing S, Li H, Hu B, Li P. Expression of orphan GPR56 correlates with tumor progression in human epithelial ovarian cancer. Neoplasma. 2017;64:32–9. [DOI] [PubMed] [Google Scholar]
- 18.Shashidhar S, Lorente G, Nagavarapu U, Nelson A, Kuo J, Cummins J, Nikolich K, Urfer R, Foehr ED. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24:1673–82. [DOI] [PubMed] [Google Scholar]
- 19.Sud N, Sharma R, Ray R, Chattopadhyay TK, Ralhan R. Differential expression of G-protein coupled receptor 56 in human esophageal squamous cell carcinoma. Cancer Lett. 2006;233:265–70. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Chatterjee T, Godoy C, Wu L, Liu QJ, Carmon KS. GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-Mediated mechanism. Mol Cancer Res. 2019;17:2196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabst C, Bergeron A, Lavallee VP, Yeh J, Gendron P, Norddahl GL, Krosl J, Boivin I, Deneault E, Simard J, Imren S, Boucher G, Eppert K, Herold T, Bohlander SK, Humphries K, Lemieux S, Hebert J, Sauvageau G, Barabe F. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127:2018–27. [DOI] [PubMed] [Google Scholar]
- 22.Bargal SA, Rafiee R, Crews KR, Wu H, Cao X, Rubnitz JE, Ribeiro RC, Downing JR, Pounds SB, Lamba JK. Genome-wide association analysis identifies SNPs predictive of in vitro leukemic cell sensitivity to cytarabine in pediatric AML. Oncotarget. 2018;9:34859–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daga S, Rosenberger A, Quehenberger F, Krisper N, Prietl B, Reinisch A, Zebisch A, Sill H, Wolfler A. High GPR56 surface expression correlates with a leukemic stem cell gene signature in CD34-positive AML. Cancer Med. 2019;8:1771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daria D, Kirsten N, Muranyi A, Mulaw M, Ihme S, Kechter A, Hollnagel M, Bullinger L, Dohner K, Dohner H, Feuring-Buske M, Buske C. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia. 2016;30:1734–41. [DOI] [PubMed] [Google Scholar]
- 25.Jentzsch M, Bill M, Grimm J, Schulz J, Schuhmann L, Brauer D, Goldmann K, Wilke F, Franke GN, Behre G, Ponisch W, Vucinic V, Niederwieser D, Platzbecker U, Schwind S. High expression of the stem cell marker GPR56 at diagnosis identifies acute myeloid leukemia patients at higher relapse risk after allogeneic stem cell transplantation in context with the CD34+/CD38- population. Haematologica. 2020;105:e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang GW, Hsiao CC, Peng YM, Vieira Braga FA, Kragten NA, Remmerswaal EB, van de Garde MD, Straussberg R, Konig GM, Kostenis E, Knauper V, Meyaard L, van Lier RA, van Gisbergen KP, Lin HH, Hamann J. The adhesion G Protein-Coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 2016;15:1757–70. [DOI] [PubMed] [Google Scholar]
- 27.Bilemjian V, Vlaming MR, Freile A, Huls J, De Bruyn G, M., Bremer E. (2022) The novel immune checkpoint GPR56 is expressed on Tumor-Infiltrating lymphocytes and selectively upregulated upon TCR signaling. Cancers (Basel). 14. [DOI] [PMC free article] [PubMed]
- 28.Xu L, Begum S, Barry M, Crowley D, Yang L, Bronson RT, Hynes RO. GPR56 plays varying roles in endogenous cancer progression. Clin Exp Metastasis. 2010;27:241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh AK, Lin HH. The role of GPR56/ADGRG1 in health and disease. Biomedical J. 2021;44:534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzman GS, Ackerman SD, Ding C, Koide A, Leon K, Luo R, Stoveken HM, Fernandez CG, Tall GG, Piao X, Monk KR, Koide S, Arac D. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 2016;91:1292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang NY, Hsiao CC, Huang YS, Chen HY, Hsieh IJ, Chang GW, Lin HH. Disease-associated GPR56 mutations cause bilateral frontoparietal polymicrogyria via multiple mechanisms. J Biol Chem. 2011;286:14215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebscher I, Schon J, Petersen SC, Fischer L, Auerbach N, Demberg LM, Mogha A, Coster M, Simon KU, Rothemund S, Monk KR, Schoneberg T. A tethered agonist within the ectodomain activates the adhesion G Protein-Coupled receptors GPR126 and GPR133. Cell Rep. 2015;10:1021. [DOI] [PubMed] [Google Scholar]
- 33.Stoveken HM, Hajduczok AG, Xu L, Tall GG. Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc Natl Acad Sci USA. 2015;112:6194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu B, Luo R, Jin P, Li T, Oak HC, Giera S, Monk KR, Lak P, Shoichet BK, Piao X. GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J Biol Chem. 2019;294:19246–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem. 2011;286:28914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishore A, Purcell RH, Nassiri-Toosi Z, Hall RA. Stalk-dependent and Stalk-independent signaling by the adhesion G Protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J Biol Chem. 2016;291:3385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzman GS, Zhang S, Gupta A, Koide A, Koide S, Arac D. Stachel-independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc Natl Acad Sci USA. 2017;114:10095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JE, Han JM, Park CR, Shin KJ, Ahn C, Seong JY, Hwang JI. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J Cancer Res Clin Oncol. 2010;136:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Chiou B, Gilman CK, Luo R, Koshi T, Yu D, Oak HC, Giera S, Johnson-Venkatesh E, Muthukumar AK, Stevens B, Umemori H, Piao X. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020;39:e104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci USA. 2011;108:12925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeung J, Adili R, Stringham EN, Luo R, Vizurraga A, Rosselli-Murai LK, Stoveken HM, Yu M, Piao X, Holinstat M, Tall GG. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc Natl Acad Sci USA. 2020;117:28275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang KY, Lin HH. The activation and signaling mechanisms of GPR56/ADGRG1 in melanoma cell. Front Oncol. 2018;8:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang NY, Peng YM, Juang HH, Chen TC, Pan HL, Chang GW, Lin HH. GPR56/ADGRG1 activation promotes melanoma cell migration via NTF dissociation and CTF-Mediated Galpha12/13/RhoA signaling. J Invest Dermatology. 2017;137:727–36. [DOI] [PubMed] [Google Scholar]
- 44.Chiang NY, Chang GW, Huang YS, Peng YM, Hsiao CC, Kuo ML, Lin HH. Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J Cell Sci. 2016;129:2156–69. [DOI] [PubMed] [Google Scholar]
- 45.Peng YM, van de Garde MD, Cheng KF, Baars PA, Remmerswaal EB, van Lier RA, Mackay CR, Lin HH, Hamann J. Specific expression of GPR56 by human cytotoxic lymphocytes. J Leukoc Biol. 2011;90:735–40. [DOI] [PubMed] [Google Scholar]
- 46.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Reviews Clin Oncol. 2018;15:234–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. 2018;18:773–89. [DOI] [PubMed] [Google Scholar]
- 48.Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–78. [DOI] [PubMed] [Google Scholar]
- 49.Ackerman SD, Garcia C, Piao X, Gutmann DH, Monk KR. The adhesion GPCR Gpr56 regulates oligodendrocyte development via interactions with Galpha12/13 and RhoA. Nat Commun. 2015;6:6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK Inhibition for cancer therapy. EMBO Rep. 2012;13:900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loirand G. Rho kinases in health and disease: from basic science to translational research. Pharmacol Rev. 2015;67:1074–95. [DOI] [PubMed] [Google Scholar]
- 52.Agarwal P, Zaidel-Bar R. Principles of actomyosin regulation in vivo. Trends Cell Biol. 2019;29:150–63. [DOI] [PubMed] [Google Scholar]
- 53.Orgaz JL, Crosas-Molist E, Sadok A, Perdrix-Rosell A, Maiques O, Rodriguez-Hernandez I, Monger J, Mele S, Georgouli M, Bridgeman V, Karagiannis P, Lee R, Pandya P, Boehme L, Wallberg F, Tape C, Karagiannis SN, Malanchi I, Sanz-Moreno V. Myosin II reactivation and cytoskeletal remodeling as a hallmark and a vulnerability in melanoma therapy resistance. Cancer Cell. 2020;37:85–103. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graziani V, Rodriguez-Hernandez I, Maiques O, Sanz-Moreno V. The amoeboid state as part of the epithelial-to-mesenchymal transition programme. Trends Cell Biol. 2022;32:228–42. [DOI] [PubMed] [Google Scholar]
- 55.Ma W, Sze KM, Chan LK, Lee JM, Wei LL, Wong CM, Lee TK, Wong CC, Ng IO. RhoE/ROCK2 regulates chemoresistance through NF-kappaB/IL-6/ STAT3 signaling in hepatocellular carcinoma. Oncotarget. 2016;7:41445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goyal P, Brunnert D, Ehrhardt J, Bredow M, Piccenini S, Zygmunt M. Cytokine IL-6 secretion by trophoblasts regulated via sphingosine-1-phosphate receptor 2 involving Rho/Rho-kinase and Rac1 signaling pathways. Mol Hum Reprod. 2013;19:528–38. [DOI] [PubMed] [Google Scholar]
- 57.Zhao FQ, Padron R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz-Moreno V, Gaggioli C, Yeo M, Albrengues J, Wallberg F, Viros A, Hooper S, Mitter R, Feral CC, Cook M, Larkin J, Marais R, Meneguzzi G, Sahai E, Marshall CJ. ROCK and JAK1 signaling cooperate to control actomyosin contractility in tumor cells and stroma. Cancer Cell. 2011;20:229–45. [DOI] [PubMed] [Google Scholar]
- 59.Orgaz JL, Pandya P, Dalmeida R, Karagiannis P, Sanchez-Laorden B, Viros A, Albrengues J, Nestle FO, Ridley AJ, Gaggioli C, Marais R, Karagiannis SN, Sanz-Moreno V. Diverse matrix metalloproteinase functions regulate cancer amoeboid migration. Nat Commun. 2014;5:4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vizurraga A, Adhikari R, Yeung J, Yu M, Tall GG. Mechanisms of adhesion G protein-coupled receptor activation. J Biol Chem. 2020;295:14065–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flock T, Hauser AS, Lund N, Gloriam DE, Balaji S, Babu MM. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z, Yang F, Zhang D, Liu Z, Lin A, Liu C, Xiao P, Yu X, Sun JP. Phosphorylation of G Protein-Coupled receptors: from the barcode hypothesis to the flute model. Mol Pharmacol. 2017;92:201–10. [DOI] [PubMed] [Google Scholar]
- 63.Gurevich VV, Gurevich EV. The structural basis of the arrestin binding to GPCRs. Molecular Cellular Endocrinology. 2019;484:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discovery. 2011;10:47–60. [DOI] [PubMed] [Google Scholar]
- 65.Insel PA, Sriram K, Wiley SZ, Wilderman A, Katakia T, McCann T, Yokouchi H, Zhang L, Corriden R, Liu D, Feigin ME, French RP, Lowy AM, Murray F. GPCRomics: GPCR expression in Cancer cells and tumors identifies new, potential biomarkers and therapeutic targets. Front Pharmacol. 2018;9:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schoneberg T, Liebscher I. Mutations in G Protein-Coupled Receptors: Mechanisms, Pathophysiology and Potential Therapeutic Approaches. Pharmacol Rev. 2021;89–119. [DOI] [PubMed]
- 67.Chaudhary PK, Kim S. (2021) An Insight into GPCR and G-Proteins as Cancer Drivers. Cells. 2021;10. [DOI] [PMC free article] [PubMed]
- 68.Haubner S, Mansilla-Soto J, Nataraj S, Kogel F, Chang Q, de Stanchina E, Lopez M, Ng MR, Fraser K, Subklewe M, Park JH, Wang X, Riviere I, Sadelain M. Cooperative CAR targeting to selectively eliminate AML and minimize escape. Cancer Cell. 2023;41:1871–91. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ravn-Boess N, Roy N, Hattori T, Bready D, Donaldson H, Lawson C, Lapierre C, Korman A, Rodrick T, Liu E, Frenster JD, Stephan G, Wilcox J, Corrado AD, Cai J, Ronnen R, Wang S, Haddock S, Sabio Ortiz J, Mishkit O, Khodadadi-Jamayran A, Tsirigos A, Fenyo D, Zagzag D, Drube J, Hoffmann C, Perna F, Jones DR, Possemato R, Koide A, Koide S, Park CY, Placantonakis DG. The expression profile and tumorigenic mechanisms of CD97 (ADGRE5) in glioblastoma render it a targetable vulnerability. Cell Rep. 2023;42:113374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barcelo J, Samain R, Sanz-Moreno V. Preclinical to clinical utility of ROCK inhibitors in cancer. Trends cancer. 2023;9:250–63. [DOI] [PubMed] [Google Scholar]
- 71.Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113:365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discovery. 2018;17:243–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wootten D, Christopoulos A, Marti-Solano M, Babu MM, Sexton PM. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat Rev Mol Cell Biol. 2018;19:638–53. [DOI] [PubMed] [Google Scholar]
- 74.Wang W, Qiao Y, Li Z. New insights into modes of GPCR activation. Trends Pharmacol Sci. 2018;39:367–86. [DOI] [PubMed] [Google Scholar]
- 75.Eiger DS, Pham U, Gardner J, Hicks C, Rajagopal S. GPCR systems pharmacology: a different perspective on the development of biased therapeutics. The Am J Physiology-cell Physiology. 2022;322:C887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang S, DeLeon C, Sun W, Quake SR, Roth BL, Sudhof TC. Alternative splicing of latrophilin-3 controls synapse formation. Nature. 2024;626:128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohta S, Sakaguchi S, Kobayashi Y, Mizuno N, Tago K, Itoh H. Agonistic antibodies reveal the function of GPR56 in human glioma U87-MG cells. Biol Pharm Bull. 2015;38:594–600. [DOI] [PubMed] [Google Scholar]
- 78.Chatterjee T, Zhang S, Posey TA, Jacob J, Wu L, Yu W, Francisco LE, Liu QJ, Carmon KS. Anti-GPR56 monoclonal antibody potentiates GPR56-mediated Src-Fak signaling to modulate cell adhesion. J Biol Chem. 2021;296:100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vizurraga AL, Robertson MJ, Yu M, Skiniotis G, Tall GG. Hexahydroquinoline derivatives are selective agonists for the adhesion G Protein-Coupled receptor ADGRG1/GPR56. Mol Pharmacol. 2023;104:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacob J, Francisco LE, Chatterjee T, Liang Z, Subramanian S, Liu QJ, Rowe JH, Carmon KS. An antibody-drug conjugate targeting GPR56 demonstrates efficacy in preclinical models of colorectal cancer. Br J Cancer. 2023;128:1592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luo Y, Lu J, Lei Z, Rao D, Wang T, Fu C, Zhu H, Zhang Z, Liao Z, Liang H, Huang W. GPR56 facilitates hepatocellular carcinoma metastasis by promoting the TGF-beta signaling pathway. Cell Death Dis. 2024;15:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.