Abstract
Hepatitis E virus (HEV), a human plus-stranded RNA virus, contains three open reading frames (ORF). Of these, ORF1 encodes the viral nonstructural polyprotein, ORF2 encodes the major capsid protein, and ORF3 codes for a phosphoprotein of undefined function. Recently, using the yeast two-hybrid system to screen a human cDNA liver library, we have isolated and characterized AMBP (α1-microglobulin/bikunin precursor), which specifically interacts with the ORF3 protein of HEV. The ORF3 protein expedites the processing and secretion of α1-microglobulin. When checked individually for interaction, the second processed protein from AMBP, bikunin, strongly interacted with the full-length ORF3 protein. This protein-protein interaction has been validated by immunoprecipitation in both COS-1 and Huh7 cells and by His6 pull-down assays. In dual-labeling immunofluorescent staining, followed by fluorescence microscopy of transfected human liver cells, ORF3 colocalized with endogenously expressed bikunin. Finally, a 41-amino-acid C-terminal region of ORF3 has been found to be responsible for interacting with bikunin. The importance of this virus-host protein-protein interaction, with reference to the viral life cycle, has been discussed.
Hepatitis E is an acute disease endemic in many countries throughout developing parts of the world, in particular on the continents of Africa and Asia, where it causes epidemics and sporadic infections. The causative agent, hepatitis E virus (HEV), is transmitted via the fecal-oral route, predominantly through contaminated water (7, 10, 25, 26). HEV is a plus-stranded RNA virus with a 7.2-kb genome containing three open reading frames (ORF), ORF1, ORF2, and ORF3, encoding three different proteins (20, 32, 35). ORF1 (5,079 bp) is at the 5′ end of the genome and is predicted to code for the putative nonstructural proteins with sequences homologous to those encoding viral methyltransferases, proteases, helicases, and RNA-dependent RNA polymerases (1, 20, 27, 35). In the absence of a reliable in vitro culture system for HEV, fundamental studies on its replication and expression strategy have not been undertaken. ORF2 and ORF3 have been expressed in Escherichia coli, animal cells, baculovirus, and yeast and in vitro in a coupled transcription-translation system (12, 15, 17, 22, 23). ORF2, the major capsid protein for HEV, is synthesized as a precursor and is processed through signal sequence cleavage into the mature protein, which is capable of self-association (18, 36). ORF3 encodes a small 13.5-kDa phosphoprotein that is expressed intracellularly, associates with the cytoskeleton, and shows no major processing (3, 42). The ORF3 protein dimerizes using a 43-amino-acid region, interacts with SH3 domains, and activates mitogen-activated protein kinase (21, 37). The phosphorylated form of the ORF3 protein has also been shown to interact with the nonglycosylated form of the major capsid protein, ORF2 (38). These properties of ORF3 clearly indicate that this protein may have multiple roles in HEV pathogenesis. Recently, through a yeast two-hybrid screen, a liver-specific protein, α1-microglobulin/bikunin precursor (AMBP), had been isolated as an interaction partner for ORF3. Also, detailed studies showed that ORF3 expedited the processing and export of one of the processed proteins, α1-microglobulin (α1m), from the hepatocyte (39).
In this study, we have focused on the second processed protein of AMBP, called bikunin. Bikunin is a Kunitz-type serine protease inhibitor that is secreted from liver cells in free and bound forms (8). Using yeast two-hybrid techniques, we have studied protein-protein interactions of this protein and found it to be a strong interaction partner for the ORF3 protein. The ORF3-bikunin interaction results were further confirmed using coupled transcription-translation His6 binding assays and immunoprecipitation in both COS-1 and Huh7 cells. Further, our studies using dual-labeling immunofluorescent staining followed by fluorescence microscopy in human liver cells showed colocalization of the ORF3 protein with bikunin. Finer mapping of this interaction revealed that a 41-amino-acid C-terminal region of the ORF3 protein was responsible for binding with bikunin. The biological significance of the ORF3-bikunin interaction in viral pathogenesis is discussed.
Bikunin strongly interacts with the ORF3 protein.
In order to identify the cellular interacting host partners of the ORF3 protein, a yeast two-hybrid screen using a human liver cDNA library was conducted, from which AMBP was isolated (39). The AMBP gene codes for two different unrelated proteins, α1m and bikunin. The full-length AMBP cDNA referred to as R352, a gift from Jean-Philippe Salier, was used as a template to PCR-clone bikunin in frame into the yeast two-hybrid and other cloning vectors used in this study (Table 1).
TABLE 1.
Yeast strains, plasmids and recombinant plasmid constructs used in this study
| Strain or plasmid construct | Description or reference |
|---|---|
| Yeast strains | |
| Y190 | MATatrp1-901 his3 leu2-3,112 ura3-52 ade2 gal4 gal80URA3::GAL-lacZ LYS2::GAL-HIS3 |
| PJ69.4a | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δgal80Δ Lys2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ |
| PJ69.4α | MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δgal180Δ Lys2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ |
| Plasmids | |
| pGAD424/pACT2 | GAL4 AD vector [GAL4(768-881)]; LEU2, 2μm, Ampr |
| pGBT9/pAS2/pAS2-1 | GAL4 DNA-BD vector [GAL4(1-147)]; TRP1,2 |
| pGBT9-ORF3 | Tyagi et al. (37) |
| pGAD424-ORF3 | Tyagi et al. (37) |
| pAS2-1-ORF3 | Tyagi et al. (39) |
| pAS2-ORF3 | Tyagi et al. (38) |
| pGBT9-ORF3 1-57 | pGBT9-ORF3 digested with BstEII and BamHI and religated |
| pGBT9-ORF3 57-123 | pGBT9-ORF3 digested with BstEII and EcoRI and religated |
| pGBT9-ORF3 1-81 | pGBT9-ORF3 digested with EagI and BamHI and religated |
| PGBT9-ORF3 83-123 | pGBT9-ORF3 digested with EagI and EcoRI and religated |
| pAS2-R352 | Tyagi et al. (39) |
| pACT2-bikunin | PCR-amplified and cloned from pACT2-AMBP using SmaI and BamHI sites (underlined) for forward (5′-CCCATGGCCCCGGGGATGGAAAACTTCAATA-T3′) and reverse (5′CGGGATCCTGCAGCAGCGCTCCGGACTCTCG3′), respectively |
| pSG-R352 | Tyagi et al. (39) |
| pSG-AMBP | Tyagi et al. (39) |
| pMT-bikunin | pACT2-Bikunin digested using SmaI and EcoRI sites, fragment ligated in pMT |
| pSG-bikunin | pACT2-Bikunin digested using SmaI and BamHI sites, fragment ligated in pSG |
| pSG-AD-ORF3 83-123 | pGBT9-ORF3 83-123 digested with HinDIII fragment ligated in pSG |
| pSG-ORF3 | Jameel et al. (15) |
| pMT-ORF3 | Jameel et al. (15) |
| pSG-His6ORF3 | Zafrullah et al. (42) |
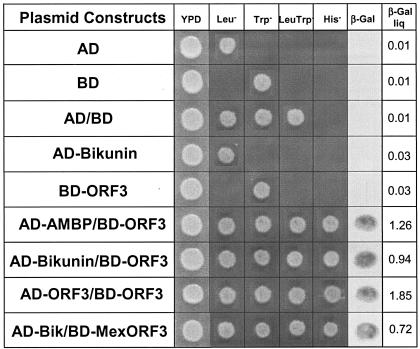
All constructs were verified by restriction digestion and sequencing. DNA manipulations were carried out as described by Sambrook et al. (29). Saccharomyces cerevisiae Y190 (MATa trp1-901 his3 leu2-3,112 ura3-52 ade2 gal4 gal80URA3::GAL-lacZ LYS2::GAL-HIS3) cells were cotransformed with activation domain (AD)-bikunin fusion protein along with binding domain (BD)-ORF3 protein. Strain Y190 contains integrated copies of both HIS3 and lacZ reporter genes under the control of GAL4 binding sites. Cotransformants were isolated and tested for His+ prototrophy by growth on synthetic dextrose medium lacking Trp, Leu, and His (SDTrp−Leu−His− dropout medium) and β-galactosidase activity on filter and liquid assays. The results of the two-hybrid assay are shown in Fig. 1. The yeast extract-peptone-dextrose (YPD) plate showed unrestricted growth of all transformants. Neither of the two plasmids singly was able to induce HIS3 or lacZ expression in yeast. Single transformants, the yeast host strain, and the cotransformants were plated on all the restrictive-medium plates. Only transformants that possessed the BD plasmid or constructs containing it grew on SDTrp− plates, whereas only transformants containing the AD plasmid or constructs derived from it grew on SDLeu− plates. The transformants containing both BD-ORF3 and AD-bikunin were able to grow on SDTrp−Leu−His− plates. The C-terminal region of ORF3 is highly conserved in all HEV strains, except in the Mexican strain (14). The ORF3 from the Mexican strain of HEV (BD-Mex ORF3) was also tested for interaction with AD-bikunin and showed growth on the SDTrp−Leu−His− plates. The second reporter gene (lacZ) tested positive for expression in a filter lift assay, resulting in a blue color for the positive cotransformants. The full-length AMBP, isolated from the human cDNA liver library (39), and the ORF3-ORF3 (37) interacting cotransformants were used as positive controls in the assay.
FIG. 1.
Yeast two-hybrid analysis showing ORF3 protein interaction with full-length bikunin. Cotransformants showing ORF3-ORF3 (19) and ORF3-AMBP (39) interactions were used as positive controls. All appropriate negative and positive controls are shown. Growth on YPD (nonselective) and selective media are shown. Leu−, SDLeu− medium; Trp−, SDTrp− medium; LeuTrp−, SDLeu−Trp− medium; His−, SDLeu−Trp−His− medium; β-Gal, results from the β-galactosidase filter assay; MexORF3, ORF3 from Mexican strain of HEV. Liquid-β-galactosidase (β-Gal liq) assay results are also shown. Single transformants and cotransformants were analyzed in a liquid-β-galactosidase assay and compared with each other. Values are given in arbitrary units. The numbers represent the means of results from five independent transformants. Y190 corresponds to the untransformed host strain. Transformants with more than one plasmid are separated by a slash.
The liquid-galactosidase assay was conducted to determine the strength of the ORF3-bikunin interaction. The assay was conducted using the substrate chlorophenol red-β-d-galactopyranoside (CPRG) as described previously (5). Relative enzymatic activity was determined in five independent transformants from each group. The relative liquid-β-galactosidase-assay unit measurements of the four positive clones, namely, AD-AMBP/AD-ORF3 (1.26 units), AD-Bikunin/BD-ORF3 (0.94 units), AD-Bikunin/BD-Mex ORF3 (0.72 units), and AD-ORF3/BD-ORF3 (1.85 units), were found to be comparable.
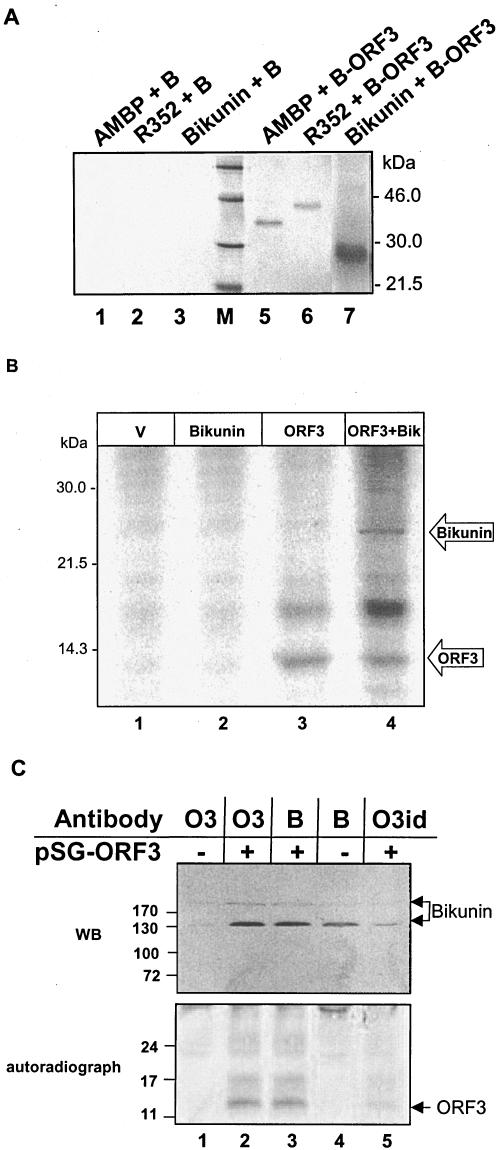
To further confirm our full-length interactions, we used an in vitro approach. The full-length ORF3 gene was cloned with an N-terminal His6 tag into the pSG vector (Table 1). The His6-ORF3 protein was expressed using the rabbit reticulocyte lysate-coupled transcription-translation system as described previously (37). In separate reactions, [35S]methionine was used to radiolabel proteins expressed from pSG-AMBP, pSG-R352, and pSG-bikunin (Table 1). Nonradioactive His6-ORF3 was then bound to charged Ni- nitrilotriacetic acid (Ni-NTA) beads, washed with phosphate-buffered saline three times, and then equally aliquoted into three tubes. Radiolabeled AMBP, R352, and bikunin were then added to these tubes, each containing nonradioactive immobilized ORF3. In separate control experiments, approximately equal amounts of each radioactive protein were added to tubes with charged Ni-NTA beads in the absence of ORF3. After 4 h of incubation at 4°C with gentle shaking, the beads were washed three times with phosphate-buffered saline to remove all unbound protein. The samples were boiled in sodium dodecyl sulfate (SDS) loading buffer for 4 min and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). The results of this experiment are shown in Fig. 2A. The control experiments showed clearly that none of the proteins bound to the beads in the absence of ORF3. Both AMBP (Fig. 2A, lane 5) and R352 (Fig. 2A, lane 6) were clearly visible on the autoradiogram, showing good binding to ORF3 in vitro. A band representing bikunin (Fig. 2A, lane 7) was also clearly visible.
FIG. 2.
In vitro interaction of the ORF3 protein with bikunin. (A) Nonradioactive His6-ORF3 was expressed in vitro and bound to Ni-NTA beads (represented by “+ B” in lane labels). [35S]methionine-radiolabeled AMBP, full-length AMBP (R352), and bikunin proteins were synthesized using an in vitro-coupled transcription-translation system and tested for binding with immobilized ORF3 protein. As a control experiment, these proteins were also bound to Ni-NTA beads alone. Lane M represents molecular mass markers. (B) Coimmunoprecipitation of bikunin and the ORF3 protein. COS-1 cells expressing ORF3 plus bikunin (ORF3+Bik) were immunoprecipitated using ORF3 antibodies (lane 4). Lane 3, ORF3 expression; lane 2, binding control for bikunin using ORF3 antibodies; lane 1, vector control. (C) Association of the bikunin and ORF3 proteins in Huh7 human hepatoma cells. Cells transfected with pSG (lanes 1 and 4) or pSG ORF3 (lanes 2, 3, and 5) were labeled with [35S]cysteine-methionine promix and immunoprecipitated with antibodies specific for ORF3 (lanes 1 and 2) or bikunin (lanes 3 and 4) or immunodepleted with anti-ORF3 antibody (lane 5). Aliquots of the samples were resolved by 6% SDS-PAGE and immunoblotted with antibikunin antibody (upper panel) or were resolved by 15% SDS-PAGE, the gel was dried, and bands were detected by autoradiography (lower panel). Numbers to the left of the gels represent molecular mass markers in kilodaltons. O3, B, and O3id denote antibodies for ORF3, bikunin, and ORF3 immunodepletion, respectively. WB, Western blot.
To confirm the bikunin-ORF3 interaction, we further studied it in animal cells. Mammalian COS-1 cells were transiently transfected with pMT-ORF3 alone or along with pMT-bikunin (Table 1) as described previously (38). [35S]methionine-labeled cell lysates were immunoprecipitated using ORF3 polyclonal antibodies. Cotransfection of COS-1 cells with pMT-ORF3 and pMT-bikunin, followed by immunoprecipitation with anti-ORF3 antibodies, showed both of the full-length proteins (Fig. 2B, lane 4). ORF3 antibody did not cross-react with bikunin (Fig. 2B, lane 1), whereas single transfection of pMT-ORF3 showed the expression of ORF3 with anti-ORF3 antibodies (Fig. 2B, lane 3). Association of bikunin with ORF3 was further confirmed using Huh7 human hepatoma cells, which produce bikunin as an endogenous protein. Cells expressing ORF3 protein were checked for their ability to coimmunoprecipitate bikunin. As shown in Fig. 2C, ORF3 was capable of associating with processed bikunin (upper panel). Two bands of approximately 240 kDa and 150 kDa were observed by immunoblotting with antibikunin antibody (which most probably represent IαI [comprising H1 plus H2 plus bikunin] and IαLI [comprising H2 plus bikunin] or PαI [comprising H3 plus bikunin]) (28). The significantly diminished intensity of the IαI band in comparison to that of IαLI/PαI may be due to decreased formation of the tertiary conjugate in Huh7 cells. The lower gel shows the expression of ORF3 in the same set of samples as a control. One set of samples was immunodepleted with anti-ORF3 antibody to show the specificity of interaction (Fig. 2C, lane 5).
Cellular colocalization of bikunin with ORF3.
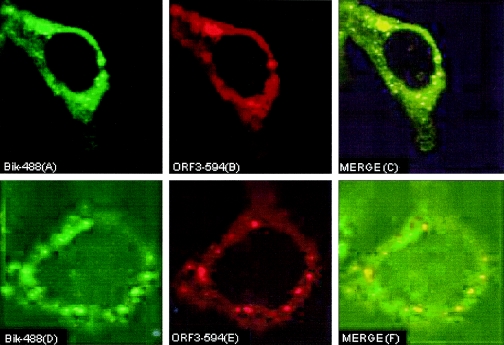
To validate the bikunin-ORF3 interaction in the physiological environment, we decided to study the colocalization pattern of the two proteins. We have recently shown that ORF3 localizes in the cytoplasm as well as in the endoplasmic reticulum and Golgi apparatus (39). The AMBP protein is processed in the Golgi compartment to yield α1m and bikunin. In an effort to understand the cellular localization of bikunin, we used Huh7 human hepatoma cells, which express bikunin endogenously. These cells were subjected to immunofluorescent staining followed by fluorescence microscopy, as described previously (38). To study colocalization of ORF3 with bikunin, dual-labeling experiments were conducted (Fig. 3). In these experiments, Huh7 cells were transiently transformed with pMT-ORF3. For immunofluorescence staining, the primary antibodies used were monoclonal anti-ORF3 at a 1:200 dilution and polyclonal antibikunin at a 1:500 dilution. For these experiments, the conjugated secondary antibodies (1:1,000 dilution) used were goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG, coupled to either Alexa 594 (red) or Alexa 488 (green) dye (Molecular Probes, Eugene, OR).
FIG. 3.
Colocalization of bikunin with ORF3 in liver cells. Huh7 cells transiently transfected with pMT-ORF3 were doubly labeled with polyclonal antibikunin and monoclonal anti-ORF3, followed by the Alexa 488 or Alexa 594 conjugated anti-rabbit IgG or anti-mouse IgG antibodies, respectively. Separate images showing bikunin distribution (A and D), ORF3 distribution (B and E), and a merger of both (C and F) were acquired. Colocalizations are shown in yellow in the merge panels.
The distribution of recombinant ORF3, as observed previously (21, 38, 39), was cytoplasmic (Fig. 3B and E). Bikunin showed cytoplasmic distribution with distinct aggregates forming in the cytoplasm (Fig. 3A and D). Bikunin also showed colocalization with ORF3 in the cytoplasm, as indicated by the yellow color in the merged images (Fig. 3C and F), which coincided with aggregates of bikunin. These results confirmed that the two proteins colocalize in the cellular environment of a hepatocyte.
The C-terminal 41-amino-acid region of the ORF3 protein is responsible for interacting with bikunin.
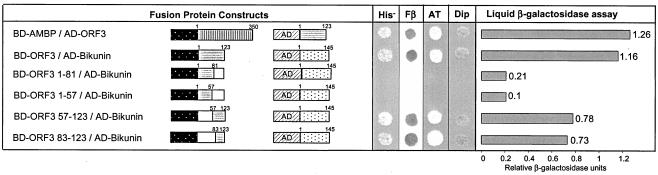
In order to map the interaction domain of ORF3, various deletion constructs (Table 1) were used. These deletion constructs of ORF3 were cotransformed with full-length bikunin (Fig. 4). While BD-ORF3 1-57 and BD-ORF3 1-81 did not show His+ prototrophy with AD-bikunin, BD-ORF3 57-123 was able to turn on the reporter gene in the yeast two-hybrid system. Further, BD-ORF3 83-123, a smaller subset of the amino acid 57 to 123 region, also showed reporter gene activity with AD-bikunin, thereby further narrowing down the interaction domain. The quantitative liquid-β-galactosidase assay revealed that the strength of the interaction of the BD-ORF3 57-123/AD-bikunin cotransformants and BD-ORF3 83-123/AD-bikunin cotransformants remained unaltered and were less than those of the full-length AD-bikunin/BD-ORF3. On the other hand, BD-ORF3 1-57/AD-bikunin cotransformants and BD-ORF3 1-81/AD-bikunin cotransformants showed negligible β-galactosidase activity. Thus, the deletion mapping results clearly showed that the ORF3 protein C-terminal region (amino acids 83 to 123) was largely responsible for the ORF3-bikunin interaction.
FIG. 4.
Mapping of the interaction domain for ORF3 using the yeast two-hybrid system. AD regions (represented by slanting lines) were cloned in-frame with AMBP (vertical lines), bikunin (black dots on a white background), and ORF3 (horizontal lines). BD regions (white dots on a black background) were cloned in-frame with AMBP and ORF3. Open boxes represent regions that were deleted from the wild-type sequences from ORF3. The numbers above the boxes represent the first and last nucleotides of the regions included in the deletion constructs. His−, growth on SDTrp−Leu−His− medium; Fβ, results from the β-galactosidase filter assay (only blue filters are visible in shades of gray); AT, growth on SDTrp−Leu−His− medium with 50 mM 3-amino-1,2,3-trizole; Dip, growth of diploids tested through the genetic two-hybrid approach. The bar graph and numbers represent relative β-galactosidase activities from the liquid-β-galactosidase assay.
HEV cannot be cultured routinely, although it has recently been propagated in primary macaque hepatocytes (33, 34) and a virus resembling HEV has been cultured in A549 cells (13). As a result, studies on HEV protein synthesis, processing, and assembly have been limited to heterologous gene expression systems. Recently, we have shown that ORF3 interacts with AMBP and accelerates the export of processed α1m from the hepatocytes, indicating its possible role in downregulation of a localized immune response in the microenvironment around the infected liver cell (39). In this study, we have examined the second processed protein that emerges from the processing of AMBP and found that it interacts strongly with ORF3. This protein is a liver-specific Kunitz-type serine protease inhibitor called bikunin. The interaction of ORF3 and bikunin is stronger than the ORF3-α1m interaction. This finding led to various interesting aspects of this study, including our ability to carefully establish the domain in the ORF3 protein that is responsible for this interaction. We were able to show that the interaction domain was located in the C-terminal region of ORF3 and was beyond the previously described phosphorylation domain for this protein. Hence, phosphorylation does not play a role in the ORF3-bikunin interaction. Although the bikunin-ORF3 interaction is independent of phosphorylation, the interaction domain of ORF3 overlaps with the ORF3 dimerization domain (amino acids 81 to 123) (37) and SH3 binding domain (amino acids 75 to 113) (21). This region has also been shown to carry an immunodominant epitope, antibodies to which are universally present in infected humans and animals (24). Moreover, this region is highly conserved in all HEV strains sequenced to date (2, 4, 24, 32, 35, 41) except the Mexican strain (14). However, our experiments with the ORF3 from the Mexican strain of HEV indicated that despite the variability in the sequence, the Mexican strain ORF3 still interacts with bikunin in the two-hybrid system. Our current data, along with previous observations, clearly suggest that the C-terminal region of ORF3 is a multifunctional domain. Strategic overlapping of these interaction domains may be a possible mechanism to ensure regulation of these interactions. Steric hindrance may ensure only one interaction, or a combination of them, at a given time. Alternatively, these interactions might be far removed from each other, due to their cellular localization and different expression time points during the life cycle of the virus, and hence each may bear no relation to the other whatsoever.
Huh7 human liver cells expressing AMBP endogenously were stained using antibikunin antibodies. Using anti-α1m antibodies, liver cells showed staining in the cytoplasm, primarily in the perinuclear region, which is characteristic of proteins translocating to the endoplasmic reticulum (39). AMBP gets processed in this region (trans-Golgi) to α1m and bikunin, which in turn get transported in free or bound form out of the hepatocyte (8). In similar experiments, when liver cells were stained with antibikunin antibody, distinct aggregates in the cytoplasm were observed. Through dual-staining immunofluorescent microscopy of liver cells endogenously expressing AMBP, bikunin was shown to colocalize with the ORF3 protein.
Bikunin is a 145-amino-acid serine protease inhibitor and is maintained at high levels in blood serum and urine; however, it has been detected in other tissues as well. In urine it exists in free form, while in blood serum both free and IαI-complexed forms are present (28). Bikunin belongs to the Kunitz family of protease inhibitors, by virtue of two tandemly arranged Kunitz-type domains, and is active against a broad range of enzymes that include trypsin, chymotrypsin, plasmin, leukocyte elastase, cathepsin B, and cathepsin H (9, 16, 31). Bikunin provides the IαI family members with their protease inhibitory capacity, and the IαI family has been implicated in inflammatory responses (28).
In cancers, administration of bikunin may block tumor cell invasion by a direct inhibition of tumor cell-associated plasmin activity, as well as by inhibiting urokinase-type plasminogen activator expression at the gene and protein levels, possibly through suppression of CD44 dimerization and/or the mitogen-activated protein kinase signaling cascade (19). In addition, bikunin has been shown to be capable of neutralizing granzyme K (40). Granzymes, along with perforins, are major components of cytosolic granules that play an important role in T-cell- and natural killer cell-mediated cytotoxicity against virally infected host cells (6, 30, 31). Other pharmaceutical effects of bikunin, like suppression of pancreatitis, colitis, and arthritis, also point towards bikunin as an immunosuppressive protein (11). Our previous studies showing enhanced secretion of the AMBP processed protein, α1m, in the presence of a viral protein (ORF3) and now its interaction with bikunin indicate that an immunosuppressive environment is being created around the infected liver cell. A future study of viral pathogenesis will reveal if the virus uses both of the processed proteins of AMBP to its advantage.
Acknowledgments
We gratefully acknowledge the generous gifts of the yeast two-hybrid vectors and strains from Stephen Elledge, the PJ69-4a and PJ69-4α strains from Philip James, the R352 gene and antibodies against bikunin from Jean-Philippe Salier, and the ORF3 gene and antibodies to the ORF3 protein from Shahid Jameel. The laboratory assistance of Ravinder Kumar and Purnima Kumar is greatly appreciated. Confocal microscopy, a Wellcome Trust laboratory, and help from Preeti Malik, Shahid Jameel, and Chetan Chitnis are gratefully acknowledged.
This work was supported by internal funds from ICGEB.
REFERENCES
- 1.Ansari, I. H., S. K. Nanda, H. Durgapal, S. Agrawal, S. K. Mohanty, D. Gupta, S. Jameel, and S. K. Panda. 2000. Cloning, sequencing, and expression of hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1). J. Med. Virol. 60:275-283. [PubMed] [Google Scholar]
- 2.Aye, T. T., T. Uchida, X.-Z. Ma, F. Iida, T. Shikata, H. Zhuang, and K. M. Win. 1992. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986-1988) of China. Nucleic Acids Res. 20:3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aye, T. T., T. Uchida, X. Ma, F. Iida, T. Shikata, H. Zhuang, and K. M. Win. 1992. Sequence comparison of the capsid region of hepatitis E viruses isolated from Myanmar and China. Microbiol. Immunol. 36:615-621. [DOI] [PubMed] [Google Scholar]
- 4.Aye, T. T., T. Uchida, X. Ma, F. Iida, T. Shikata, M. Ichikawa, T. Rikihisa, and K. Win. 1993. Sequence and gene structure of the hepatitis E virus isolated from Myanmar. Virus Genes 7:95-109. [DOI] [PubMed] [Google Scholar]
- 5.Bai, C., and S. J. Elledge. 1996. Gene identification using the yeast two-hybrid system. Methods Enzymol. 273:331-347. [DOI] [PubMed] [Google Scholar]
- 6.Berke, G. 1995. The CTL's kiss of death. Cell 81:9-12. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, D. W. 1990. Enterically-transmitted non-A, non-B hepatitis. Br. Med. Bull. 46:442-461. [DOI] [PubMed] [Google Scholar]
- 8.Bratt, T., T. Cedervall, and B. Akerstrom. 1994. Processing and secretion of rat alpha 1-microglobulin-bikunin expressed in eukaryotic cell lines. FEBS Lett. 354:57-61. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L., S. J. T. Mao, and W. J. Larsen. 1992. Identification of a factor in fetal bovine serum that stabilizes the cumulus extracellular matrix. A role for a member of the inter-alpha-trypsin inhibitor family. J. Biol. Chem. 267:12380-12386. [PubMed] [Google Scholar]
- 10.Emerson, S. U., and R. H. Purcell. 2001. Recombinant vaccines for hepatitis E. Trends Mol. Med. 7:462-466. [DOI] [PubMed] [Google Scholar]
- 11.Fries, E., and A. M. Blom. 2000. Bikunin—not just a plasma proteinase inhibitor. Int. J. Biochem. Cell Biol. 32:125-137. [DOI] [PubMed] [Google Scholar]
- 12.He, J., A. W. Tam, P. O. Yarbough, G. R. Reyes, and M. Carl. 1993. Expression and diagnostic utility of hepatitis E virus putative structural proteins expressed in insect cells. J. Clin. Microbiol. 31:2167-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, R., N. Nakazono, K. Ishii, D. Li, O. Kawamata, R. Kawaguchi, and Y. Tsukada. 1995. Existing variations on the gene structure of hepatitis E virus strains from some regions of China. J. Med. Virol. 47:299-302. [DOI] [PubMed] [Google Scholar]
- 14.Huang, R. T., D. R. Li, L. I. Wei, X. R. Huang, X. T. Yuan, and X. Tian. 1992. Isolation and identification of hepatitis E virus in Xinjiang, China. J. Gen. Virol. 73:1143-1148. [DOI] [PubMed] [Google Scholar]
- 15.Jameel, S., M. Zafrullah, M. H. Ozdener, and S. K. Panda. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 70:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaumeyer, J. F., J. O. Polazzi, and M. P. Kotick. 1986. The mRNA for a proteinase inhibitor related to the HI-30 domain of inter-alpha-trypsin inhibitor also encodes alpha-1-microglobulin (protein HC). Nucleic Acids Res. 14:7839-7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khudyakov, Y. E., M. O. Favorov, D. L. Jue, T. K. Hine, and H. A. Fields. 1994. Immunodominant antigenic regions in a structural protein of the hepatitis E virus. Virology 198:390-393. [DOI] [PubMed] [Google Scholar]
- 18.Khudyakov, Y. E., N. S. Khudyakova, H. A. Fields, D. Jue, C. Starling, M. O. Favorov, K. Krawczynski, L. Polish, E. Mast, and H. Margolis. 1993. Epitope mapping in proteins of hepatitis E virus. Virology 194:89-96. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, H., M. Suzuki, Y. Hirashima, and T. Terao. 2003. Bikunin is a Kunitz-type protease inhibitor predominantly found in human amniotic fluid. Biol. Chem. 384:749-754. [DOI] [PubMed] [Google Scholar]
- 20.Koonin, E. V., A. E. Gorbalenya, M. A. Purdy, M. N. Rozanov, G. R. Reyes, and D. W. Bradley. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-stranded RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 89:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and D. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389-42400. [DOI] [PubMed] [Google Scholar]
- 22.Lal, S. K., P. Tulasiram, and S. Jameel. 1997. Expression and characterization of the hepatitis E virus ORF3 protein in the methylotrophic yeast, Pichia pastoris. Gene 190:63-67. [DOI] [PubMed] [Google Scholar]
- 23.Panda, S. K., I. H. Ansari, H. Durgapal, S. Agrawal, and S. Jameel. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 74:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panda, S. K., S. K. Nanda, M. Zafrullah, I.-H. Ansari, M. H. Ozdener, and S. Jameel. 1995. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J. Clin. Microbiol. 33:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell, R. H., and S. U. Emerson. 2000. Hepatitis E virus infection. Lancet 355:578. [DOI] [PubMed] [Google Scholar]
- 26.Purcell, R. H., and J. R. Ticehurst. 1988. Enterically transmitted non-A, non-B hepatitis: epidemiology and clinical characteristics, p. 131-137. In A. J. Zukerman (ed.), Viral hepatitis and liver disease. Alan R. Liss, Inc., New York, N.Y.
- 27.Reyes, G. R., C. C. Huang, A. W. Tam, and M. A. Purdy. 1993. Molecular organization and replication of hepatitis E virus (HEV). Arch. Virol. 7:15-25. [DOI] [PubMed] [Google Scholar]
- 28.Salier, J. P., P. Rouet, G. Raguenez, and M. Daveau. 1996. The inter-α-inhibitor family: from structure to regulation. Biochem. J. 315:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Shresta, S., C. T. Pham, D. A. Thomas, T. A. Graubert, and T. J. Ley. 1998. How do cytotoxic lymphocytes kill their targets? Curr. Opin. Immunol. 10:581-587. [DOI] [PubMed] [Google Scholar]
- 31.Sjoberg, E. M., and E. Fries. 1990. One of the major sulphated proteins secreted by rat hepatocytes contains low-sulphated chondroitin sulphate. Biochem. J. 272:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tam, A. W., M. M. Smith, M. E. Guerra, C. C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam, A. W., R. White, E. Reed, M. Short, Y. Zhang, T. R. Fuerst, and R. E. Lanford. 1996. In vitro propagation and production of hepatitis E virus from in vivo-infected primary macaque hepatocytes. Virology 215:1-9. [DOI] [PubMed] [Google Scholar]
- 34.Tam, A. W., R. White, P. O. Yarbough, B. J. Murphy, C. P. McAtee, R. E. Lanford, and T. R. Fuerst. 1997. In vitro infection and replication of hepatitis E virus in primary cynomolgus macaque hepatocytes. Virology 238:94-102. [DOI] [PubMed] [Google Scholar]
- 35.Tsarev, S. A., S. U. Emerson, G. R. Rees, T. S. Tsareva, L. J. Letgers, I. A. Malik, M. Iqbal, and R. H. Purcell. 1992. Characterization of a prototype strain of hepatitis E virus. Proc. Natl. Acad. Sci. USA 89:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyagi, S., S. Jameel, and S. K. Lal. 2001. A yeast two-hybrid study on self-association of the ORF2 protein of hepatitis E virus. Biochem. Biophys. Res. Commun. 284:614-621. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi, S., S. Jameel, and S. K. Lal. 2001. Self-association and mapping of the interaction domain of hepatitis E virus ORF3 protein. J. Virol. 75:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyagi, S., H. Korkaya, M. Zafrullah, S. Jameel, and S. K. Lal. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759-22767. [DOI] [PubMed] [Google Scholar]
- 39.Tyagi, S., M. Surjit, A. Kar-Roy, S. Jameel, and S. K. Lal. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor α1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308-29319. [DOI] [PubMed] [Google Scholar]
- 40.Wilharm, E., M. A. A. Parry, R. Friebel, H. Tschesche, G. Matschiner, C. P. Sommerhoff, and D. E. Jenne. 1999. Generation of catalytically active granzyme K from Escherichia coli inclusion bodies and identification of efficient granzyme K inhibitors in human plasma. J. Biol. Chem. 274:27331-27337. [DOI] [PubMed] [Google Scholar]
- 41.Yin, S., R. H. Purcell, and S. U. Emerson. 1994. A new Chinese isolate of hepatitis E virus: comparison with strains recovered from different geographical regions. Virus Genes 9:23-32. [DOI] [PubMed] [Google Scholar]
- 42.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]