Abstract
Enhancers, as distal cis-regulatory elements in the genome, have a pivotal influence on orchestrating precise gene expression. Enhancer RNAs (eRNAs), transcribed from active enhancer regions, are increasingly recognized as key regulators of transcription. N6-methyladenosine (m6A), the most plentiful internal modification in eukaryotic mRNAs, has garnered significant research interest in recent years. With advancements in high-throughput sequencing technologies, it has been established that m6A modifications are also present on eRNAs. An accumulative body of evidence demonstrates that aberrant enhancers, eRNAs, and m6A modifications are intimately connected with carcinoma onset, progression, invasion, metastasis, treatment response, drug resistance, and prognosis. However, the underlying molecular mechanisms governing m6A modification of eRNAs in cancer remain elusive. Here, we review and synthesize current understanding of the regulatory roles of enhancers, eRNAs, and m6A modifications in cancer. Furthermore, we investigate the possible roles of eRNAs m6A modification in tumorigenesis based on existing literature, offering novel perspectives and directions for future research on epigenetic regulatory mechanisms in cancer cells.
Keywords: Epigenetics, Enhancer, eRNAs, m6A modifications, Cancer
Introduction
The mechanisms regulating gene expression in higher eukaryotes are extremely complex, with different genes being activated in a cell-type-specific manner, resulting in cell types with specialised morphology and function [1]. Enhancers are DNA sequences consisting of 200–1500 base pairs that function in regulating gene expression, influencing cell identity, and regulating cell-type-specific gene expression [2]. The human genome contains a significantly higher number of enhancers compared to genes [3], with one enhancer capable of regulating multiple genes, while a single gene can be influenced by numerous enhancers [4]. However, aberrant enhancer regulation can lead to dysregulation of gene transcription, ultimately contributing to tumorigenesis and progression. Enhancer remodelling, altered enhancer activity, and even transcription factor (TF) dysregulation, which indirectly alters enhancer activity, can be key factors in tumourigenesis.
In the activated state, the enhancer attracts TFs, recruits RNA polymerase II (RNA Pol II), and transcribes to produce eRNAs. eRNAs are widely present in human cells and possess multiple biological functions. In recent years, increasing evidence has revealed that eRNAs can regulate the expression of proximal or distal target genes through a variety of mechanisms [5]. Furthermore, eRNAs can contribute to cancer development by altering gene transcription, as well as protein-RNA interactions to regulate oncogenes, tumour suppressor genes, and cancer signalling pathways [6]. In terms of potential clinical applications, eRNAs are of great value for further research as cancer-related biomarkers and therapeutic targets.
m6A modification is a methylation modification that occurs on RNA, accounting for about 60% of methylation modifications [7]. m6A methylation is primarily regulated by three types of proteins: writing proteins, which catalyse the formation of m6A methylation; erasing proteins, which demethylate RNA; and reading proteins, which recognise m6A methylation sites [8]. The m6A modification plays a crucial role in diverse cellular processes and is closely associated with cancer development [9]. For example, dysregulated m6A modifications significantly influence glycolysis and tumour progression in hepatocellular carcinoma (HCC) [10]. Moreover, studies have demonstrated that targeted modulation of m6A can enhance anti-tumour therapy efficacy in gastric cancer (GC) [11].
With the advancement of new technologies, m6A modification on eRNAs has been gradually uncovered. Several studies have investigated how m6A modification affects eRNAs, including their stability and transcriptional activation, as well as the possible involvement of m6A-related eRNAs in cancer. In this review, we describe the characterisation of enhancers and eRNAs, their mechanisms of action in transcriptional activation, and their association with cancer. Furthermore, we extensively explore the pathophysiological utility of m6A modification on eRNAs and comprehensively analyse its implications in cancer, offering new perspectives on the epigenetic regulatory mechanisms within cancer cells.
Enhancer and enhancer RNAs
Enhancer
Enhancers, which are non-coding sequences in the genome, have the ability to activate or increase the transcription of linked genes in specific cell types. With the advancement of genomic research, our understanding of enhancers has become more comprehensive. Enhancers are typically characterised by the following features: (1) They can function independently of the direction, distance, and location of the target gene. Generally, enhancers are located in intergenic and intronic regions, with some found in exonic regions. The distance from the target gene can vary from a few hundred bases to several thousand bases. For instance, the distance between the mouse SHH gene and its enhancer ZRS sequence exceeds 1 million base pairs [12, 13]. (2) Enhancers contain specific DNA sequences that allow TF binding. These sequences primarily consist of dense clusters of transcription factor binding sites (TFBs) that bind to cell type-specific TFs, co-regulatory factors, structural proteins, chromatin modifiers, and some enzymes [1]. (3) They are typically tissue-specific or cell type-specific, as demonstrated by the first mammalian cell enhancer found in the intron region of the immunoglobulin (Ig) heavy chain gene [14]. (4) Enhancers are characterised by a lack of nucleosomes and heightened sensitivity to nucleases, such as DNase I [15]. (5) Enhancers are evolutionarily conserved in both sequence and function [16]. Based on these characteristics, approximately 400,000 potential enhancers have been identified in the mammalian genome [17].
Genes can be regulated by either a single enhancer or multiple enhancer elements, which may cluster together to form super-enhancers. Super-enhancers cover larger genomic regions and contain higher concentrations of TFs, mediator coactivators, and histone markers linked to transcriptional activity compared to regular enhancers [18]. In addition to super-enhancers, two specific types of enhancers exist: shadow enhancers and stretch enhancers. Shadow enhancers are essentially a set of enhancers that influence a shared target gene, resulting in overlapping expression patterns in both space and time [19]. Stretch enhancers, identified through computational analysis of type 2 diabetes data, are broad regions of enhancers larger than 3 kb. These enhancers also increase gene expression and function in a cell type-specific manner [20] (Table 1).
Table 1.
Comparison of super-enhancers, shadow enhancers, and stretch enhancers
| Feature | Super-enhancers | Shadow enhancers | Stretch enhancers | References |
|---|---|---|---|---|
| Genomic organization | Large enhancer clusters formed by multiple neighboring ordinary enhancers | A set of enhancers on the genome that regulate the same gene |
Large genomic regions characterized by enhancers |
[20–23] |
| Typical size | 8-20 kb | — | Larger than 3 kb | [20–22] |
| Regulatory function | Powerful activation of cellular identity genes | Gene expression stability | Directing cell-specific gene expression | [20–22, 24, 25] |
| Relevance to cancer | Driving aberrant activation of oncogenes to maintain tumor properties | Redundant regulation ensures high expression of oncogenes | Activation of distal oncogenes | [19, 20, 26–29] |
The enhancer can be activated by specific histone modifications. During transcriptional activation, the enhancer recruits two key complexes: the Mll3/Mll4/COMPASS complex and the p300/CBP complex. The former has methyltransferase activity, which mediates the monomethylation of histone H3 lysine 4 (H3K4me1), while the latter has acetyltransferase activity and mediates acetylation of histone H3 lysine 27 (H3K27ac). Upon labelling with H3K4me1 or H3K27ac, the enhancer becomes active. Therefore, H3K4me1 and H3K27ac can serve as markers for enhancers in chromatin immunoprecipitation sequencing analyses and can be used for the identification of enhancers in the genome [30, 31]. Enhancers exist in multiple states beyond the activated state, including primed, poised, and inactive states. Activated enhancers feature histone modifications and open chromatin, whereas enhancers in the primed or poised states lack H3K27ac modifications, and silent enhancers exhibit few epigenetic signatures [32]. As cis-acting elements, enhancers can also be modified by methylation, such as the addition of a methyl group at position 5 of the cytosine ring (5-methylcytosine, 5mC). Studies have shown that active enhancers have lower levels of 5mC compared to silent enhancers, though the precise role of 5mC in regulating enhancer activity remains unclear [33].
Before an enhancer can activate a promoter for transcription, it must first establish a link with the promoter to facilitate the exchange of information or material. Several models have been proposed to explain how enhancers communicate with promoters, such as the sliding model, the linking model, the contact model, the action-at-a-distance model, and the kissing model. However, the exact mechanism of enhancer-promoter interaction is still not fully understood [13, 34–37]. Regardless of the interaction mechanism, once enhancers and promoters interact, transcription is activated. Mechanistic models of enhancer-activated transcription can be categorised into four types: (1) enhancer-recruited protein complexes are transferred directly from the enhancer to the promoter; (2) enhancer-mediated protein complexes are recruited to the promoter; (3) enhancers accumulate a high concentration of TFs, coactivators, and chromatin remodelling agents near the promoter, increasing the likelihood of transcriptional activation at the promoter; (4) enhancer-mediated post-translational modification of the promoter [38]. The relationship between enhancers and promoters, and the mechanics of enhancer-activated transcription, remains a challenging area of research that requires further investigation for a deeper understanding.
Enhancer RNAs
In 2010, two research teams independently observed differences in the transcription of enhancers in neurons and macrophages, leading to the discovery of eRNAs. This discovery expanded the non-coding RNA (ncRNA) family and prompted the scientific community to explore eRNAs and their role in transcriptional regulation. To date, it is estimated that humans possess between 40,000 and 65,000 eRNAs [39]. Most eRNAs contain a 5’cap structure but lack a 3’polyadenylation (polyA) modification, typically being no longer than 2 kb, and are bi-directionally transcribed from active enhancers. A small proportion of eRNAs are transcribed unidirectionally from active enhancers; these are relatively long, have both 3’polyA modifications and 5’cap structures, and are generally more stable [40–42]. However, not all enhancers are capable of producing eRNAs. Enhancers that generate eRNAs possess the following characteristics: (1) histone modifications (such as H3K4me1 and H3K27ac) or the presence of certain histone variants (e.g., H2AZ); (2) they are located in open chromatin regions; (3) they are bound by transcriptional coactivators; (4) they exhibit low levels of DNA methylation [43]. Additionally, super-enhancers, which were mentioned earlier, can also be transcribed, and transcription from these super-enhancers can generate larger amounts of eRNAs [44].
eRNAs, along with mRNAs and long non-coding RNAs (lncRNAs), belong to the ncRNA category, sharing similarities and notable differences. mRNAs and lncRNAs are longer, more stable, and typically undergo splicing and 3′ polyA modification, unlike most eRNAs. However, some unidirectional eRNAs transcripts do undergo splicing and 3′ polyA modifications. Studies have shown that mRNAs and lncRNAs are 90–100 times more stable than eRNAs [45]. eRNAs are rapidly degraded by nuclear RNA exosome complexes after synthesis, and this short lifespan complicates their sequencing. To detect eRNAs, techniques such as global run-on sequencing (GRO-seq) and its derivatives, such as precise nuclear run-on sequencing (PRO-seq), have been employed. Researchers have refined methods to detect eRNAs based on their 5’-cap structure, including PRO-cap and 5’GRO-seq. Additionally, nanopore direct RNA sequencing has been applied in the analysis of cellular mRNAs, non-coding RNAs (ncRNAs), and the detection of various RNA viruses [46], and single-molecule imaging has made breakthroughs in revealing the translation and decay of mRNAs [47], potentially applicable to eRNAs analysis through further technical advancements.
eRNAs are generated in enhancer regions marked by high levels of H3K4me1 and low levels of H3K4me3 histone modifications, in contrast to lncRNAs, which are produced in promoter regions with high H3K4me3 modifications. The active enhancer regions producing eRNAs are associated with high levels of serine-5-phosphorylated RNA Pol II, whereas mRNA-producing promoter regions are linked to serine-2-phosphorylated RNA Pol II [48]. Interestingly, it was found that eRNAs transcribed from super-enhancers can function similarly to lncRNAs. Thus, eRNAs and lncRNAs exhibit functional similarities, leading to the hypothesis that lncRNAs may have evolved from eRNAs, stabilised and acquired functionality over evolutionary time. However, this hypothesis requires further investigation to verify. Additionally, eRNAs expression is positively correlated with the active enhancer marker H3K27ac, which is commonly used to mark active enhancers. Notably, eRNAs themselves serve as a more precise indicator of active enhancers, aiding the identification of new enhancers [49].
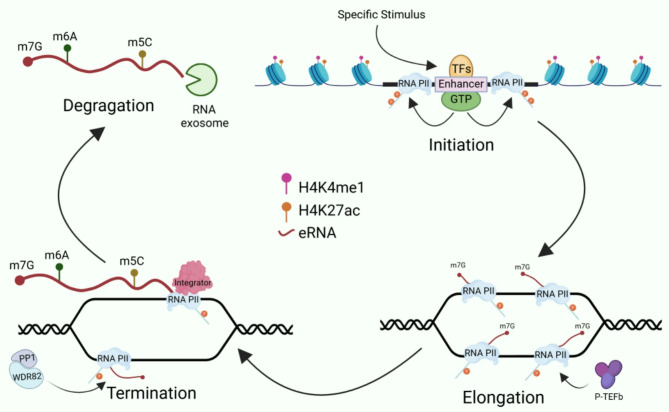
Typically, eRNAs production occurs prior to the transcription of the corresponding gene and involves three key steps. In the first step, TFs and coactivators are recruited. Upon specific stimuli, TFs and coactivators are recruited to interact with the enhancer region, promoting nucleosome remodelling and recruiting additional TFs, cofactors, and complexes (e.g., P300/CBP) [50]. In the second step, histone modification occurs at the enhancer site, with H3K27 acetylation mediated by P300/CBP, which promotes chromatin accessibility, further opening the enhancer region and recruiting RNA Pol II and bromodomain-containing protein 4 (BRD4). In the third step, eRNAs are transcribed, elongated, and processed. BRD4 regulates the recruitment of transcription initiation factors and the formation of transcription mediator complexes, promoting the elongation of RNA Pol II at highly acetylated enhancers [51]. Estrogen receptor-α (ERα), a central TF, associates with estrogen-responsive enhancers in MCF-7 breast cancer cells and selectively recruits a prominent complex of other DNA-binding TFs, ultimately forming a large complex known as MegaTrans, which is essential for eRNAs transcription [52]. However, it remains unclear whether this complexity exists in other cell types. Furthermore, it has been found that eRNAs transcriptional pausing and elongation are mediated by Spt5 and the positive transcription elongation factor b (P-TEFb) complex [53]. Additionally, Integrator, a large 12-subunit complex with nucleic acid endonuclease activity, mediates eRNAs transcription termination by interacting with the Pol II C-terminal domain (CTD) and cleaving the 3′ end of the primary eRNAs transcript [54]. Once released from chromatin, nascent eRNAs are degraded by nuclear RNA exosome complexes in a 3′-5′ manner. The enzymatic activity of these complexes is supported by two proteins, Rrp44 (Dis3) and Rrp6 (Exosc10). During eRNAs production, harmful DNA/RNA hybrids, such as R-loops, may form, where eRNAs are enzymatically modified to prevent the formation of base pairs, significantly impacting genomic stability. In knockout cells for the Dis3 and Exosc10 genes, R-loop structures in the transcribed regions of eRNAs were significantly increased, indicating that nuclear RNA exosome complexes may play a role in degrading detrimental secondary structures during eRNAs formation [55]. Methylation modifications, including m6A and m5C (5-methylcytosine), have also been detected on eRNAs, influencing their stability and metabolism. At the initial stages of eRNAs transcription, the cap-binding complex (CBC) associates with eRNAs to form the m7G (N7-methylguanosine) cap at the 5′ end (Fig. 1).
Fig. 1.
A diagram illustrates eRNAs transcription process, including modifications and degradation. Following the preliminary attachment of general transcription factors (GTFs) and other TFs in response to specific signals, RNA Pol II is brought to the enhancer to start bidirectional transcription. The enhancer region is rich in H3K4me1 and H3K27ac modifications. The C-terminal domain (CTD) of Pol II in the enhancer region is primarily phosphorylated at the Tyr1 and Ser5 sites, which regulate the pausing and elongation of eRNAs transcription. Integrators can mediate the transcription termination of eRNAs. WDR82 (WD repeat domain 82) and PP1 (phosphatase 1) are engaged in the early ending of eRNAs transcription. Once eRNAs are released from chromatin, they are degraded by nuclear RNA exosome complexes in a 3’-5’ manner. Various RNA modifications, maintaining m5C, m6A, and m7G, are frequently present in eRNAs and affect their stability
The function of eRNAs was initially controversial. Some researchers argued that, due to their low abundance and short half-life, eRNAs were merely “noise” in the chromatin environment opened by enhancers and by-products of gene transcription. However, more studies have refuted this claim. eRNAs are not only markers of active enhancers, but also crucial regulators of transcription. eRNAs play the following roles in transcriptional regulation: (1) regulation of chromatin remodelling. eRNAs recruit Cohesin and bind to its components, SMC3 and RAD21, to regulate chromatin accessibility at the target promoter. Conversely, depletion of eRNAs decreases chromatin accessibility at enhancers and promoters [56]. eRNAs also stabilise enhancer-promoter loops by directly interacting with chromatin loop factors, such as MED12 [57]. (2) Regulation of histone modifications. eRNAs interact with CBP, stimulating its catalytic activity and increasing histone acetylation at enhancers [58]. eRNAs can also activate p300 and induce H3K27 acetylation at enhancers [59]. Similarly, eRNAs interacting with the EZH2 subunit antagonise its nucleosome-binding activity and inhibit the methyltransferase activity of PRC2, thus reducing the deposition of inhibitory H3K27me3 [60]. (3) Recruitment of TFs and co-regulators, thereby modulating their activity. eRNAs can capture TF YY1 and increase its local concentration at transcription sites [61]. eRNAs also interact directly with BRD4 to amplify the interaction between H3K27ac and BRD4, thus maintaining enhancer activity [62]. Recent studies have found that eRNAs capture dissociated TFs through weak RNA-mediated interactions, promoting their recombination with regulatory elements [63]. (4) Direct participation in or interference with transcription, enhancing transcription elongation. eRNAs can interact with the positive elongation factor (P-TEFb) to enhance transcription. In neurons, eRNAs act as a decoy for the negative elongation factor (NELF) after inducing the immediate early gene (IEG), luring it away from the IEG and converting the paused RNA Pol II into productive elongation [64]. Furthermore, eRNAs regulate the proper loading of target sites by CAAA bundles, which control the intermediate heterogeneous cytosolic nuclear ribonucleoprotein L (hnRNPL) [65].
Enhancer and enhancer RNAs in cancer
Enhancer in cancer
Enhancers function as cis-regulatory elements that elevate gene expression by cooperating with promoters to orchestrate transcriptional programmes. With growing research into enhancer biology, it is now well-established that enhancer dysregulation—manifesting as loss of native enhancers, emergence of de novo enhancers, or aberrant enhancer activation/inhibition—plays a pivotal role in tumorigenesis. The disruption of enhancer function in cancer can be broadly attributed to three primary categories: genetic alterations, epigenetic reprogramming, and TF dysregulation.
Firstly, genetic alterations can directly influence enhancer activity. These include copy number variations, chromosomal translocations, amplifications, insertions, deletions, and both intra- and inter-chromosomal inversions. Single nucleotide polymorphisms (SNPs) and structural variants in enhancer regions are now recognised as key contributors to cancer risk. Genome-wide association studies (GWAS) have revealed that a significant proportion of cancer-associated variants—including those linked to lung and breast cancers—are enriched within enhancer regions [66]. For instance, a point mutation within the enhancer region of the *TERT* gene enhances TF binding, leading to elevated telomerase activity and uncontrolled proliferation of tumour cells [67].
In a genome-wide CRISPR interference (CRISPRi) screen, researchers identified 66 enhancers within the melanoma genome associated with highly recurrent regions (HRRs), which appear to function as tumour-suppressive elements. These were stratified into four categories using a latent block model: (1) enhancers frequently deleted in patients, (2) enhancers containing recurrent mutations at functional loci, (3) enhancers with sparse mutations, and (4) enhancers with high-level duplications in proximity to oncogenes. Among these, 21 enhancers exhibited copy number gains and 17 showed heterozygous deletions. Functionally, these disrupted enhancers were implicated in the EGFR signalling pathway, central carbon metabolism, and the AMPK signalling cascade, collectively contributing to tumour progression [68]. Similarly, chromatin structure can be altered by enhancer-associated structural variants; in osteosarcoma, abnormal chromatin looping between the *MYC* enhancer and promoter augments oncogenic transcription by increasing spatial proximity [69]. Hence, enhancer amplification, hijacking, or mutation is critical to cancer initiation and progression.
Secondly, alterations in epigenetic modifications can also affect enhancer function, such as aberrant DNA methylation and modifications like histone methylation and acetylation. Aberrant hypermethylation of enhancers associated with tumour suppressor genes, such as CDKN2A, can result in gene silencing, whereas hypomethylation of enhancers linked to oncogenes, such as ERα in breast cancer (BC), facilitates the binding of TFs [70, 71]. TP63 has been associated with poor prognosis in prostate cancer (PCa) due to its elevated expression. The methylation level of enhancers in the epithelium of prostate basal cells decreases when TP63 is expressed, but it may be responsible for the enhanced hydroxymethylation of cytosine (hmC) at certain enhancers by TP63. However, not all CpG sites related to TP63 undergo demethylation when TP63 is expressed [72]. In castration-resistant prostate cancer (CRPC), inhibition of lysine-specific demethylase 1 (LSD1/KDM1A) blocks the demethylation of the AR pioneer factor FOXA1, which then recruits BRD4 to chromatin, disrupting FOXA1’s global binding and reducing enhancer accessibility. Genetic and epigenetic alterations occur simultaneously in most cancer cells [73].
In addition, abnormalities in TFs, such as mutations, aberrant expression, chimerism, altered function, altered stability, or even crosstalk between TFs, can also cause changes in enhancer activity and thus affect cancer. The overexpression of FOXA1 in BC remodels ER signalling pathway enhancers by recruiting the MLL3 complex, promoting cell proliferation [74]. Consistent with this, ΔNp63 recruits p300 to restructure the enhancer landscape in lung adenocarcinoma (LUAD), driving squamous differentiation phenotypes [75]. Knockdown of forkhead box protein D2 (FOXD2) in colonoid organs downregulates the expression of more than 200 genes, leading to the loss of colorectal identity and causing colorectal carcinogenesis. Mechanistically, FOXD2 opens chromatin and recruits K3K4 monomethyltransferase (MLL4) to introduce histone modifications, and enhancers and promoters further form differential chromatin loops to regulate chromatin structure, re-edit enhancers, and induce apoptosis [76]. In acute myeloid leukaemia, an in-frame mutation in the C-terminal region of the enhancer binding protein alpha (C/EBPα) alters its basic leucine zipper structural domain, producing a C/EBPα C-terminal mutant (C/EBPα-Cm). The resulting mutant has a reduced capability to bind with DNA, causing decreased enhancer activity of the ULBP2/5/6 genes and reduced gene expression [77]. This ultimately allows tumour cells to evade natural killer cell-mediated cell lysis.
Abnormal enhancer activity is intricately linked to tumour drug resistance, sensitivity to targeted therapy, and response to chemotherapy. Aberrant enhancer activation can stabilise the continuous expression of oncogenes and maintain survival signalling pathways in tumour cells, thereby reducing the cytotoxic effects of conventional chemotherapy or targeted therapy. For instance, the nuclear matrix binding protein SATB1 mediates the formation of an enhancer-promoter loop of the BCL2 gene, upregulating the expression of the anti-apoptotic protein BCL2 and leading to tumour cell resistance to apoptosis-inducing drugs such as cisplatin [78]. Therefore, individual classification based on enhancer genetic variations can provide new biomarkers for predicting treatment responses and guide drug selection. Thus, the enhancer regulatory network plays a “double-edged sword” role in cancer treatment responses: its abnormal activation can drive drug resistance, while targeted intervention may restore treatment sensitivity.
Enhancer-targeted therapeutic strategies can be categorised as follows: (1) development of small-molecule inhibitors targeting enhancers: the BET inhibitor JQ1 suppresses cancer stemness by disrupting enhancer architecture and downregulating BRD4-dependent stemness-related genes. Clinically, combining JQ1 with cisplatin has shown promise in enhancing chemosensitivity and improving patient survival outcomes [79]. Beyond JQ1, other enhancer-targeting agents, including the CDK7 inhibitor THZ1 and the CDK8/CDK19 dual inhibitor cortistatin A, have demonstrated efficacy in inhibiting tumour progression [80, 81]. Furthermore, clinical trials of BET inhibitors in haematologic malignancies have validated the feasibility of enhancer-targeted therapies. (2) Antisense Oligonucleotides (ASOs): ASOs are chemically synthesised nucleic acid analogues [82]. Modified ASOs can precisely downregulate enhancer-derived noncoding RNAs by disrupting enhancer-promoter looping [83]. Therapeutic ASOs exhibit high specificity and the ability to modulate enhancers that are traditionally considered “undruggable”. However, efficient delivery of ASOs to target tissues remains a major challenge for clinical translation [84]. (3) CRISPR/Cas9 genome editing systems: the CRISPR/Cas9 deletion system enables functional studies of enhancers by directly removing specific enhancer regions [85]. Advanced epigenome-editing tools, such as enCRISPRa (enhancer-targeted CRISPR activation) and enCRISPRi(enhancer-targeted CRISPR interference), can remodel epigenetic modifications at enhancers to either activate or suppress their transcriptional activity and downstream target genes [86]. Collectively, small-molecule compounds and ASOs targeting enhancer elements offer potential to restore enhancer homeostasis, modulate eRNAs levels, and thus interfere with tumour progression.
Tumour-specific hyperactive enhancers are frequently enriched at oncogenic loci (e.g., MYC, BCL2), and distinct enhancer signatures across tumour subtypes provide a foundation for precision targeting. However, several practical challenges must be addressed: (1) the vast complexity and functional redundancy of enhancer networks, coupled with high interindividual heterogeneity, complicate therapeutic targeting; (2) the blood-brain barrier penetration capacity of enhancer-targeted inhibitors requires optimisation for central nervous system malignancies.
In terms of potential risks, enhancer-targeted therapies carry inherent risks due to the involvement of enhancers in normal physiological processes. For instance, small-molecule inhibitors may exhibit off-target effects by simultaneously modulating multiple enhancers. BET inhibitors, while suppressing oncogenic enhancers, may inadvertently activate immune-related pathways, triggering excessive inflammatory responses [87]. Additionally, genes regulated by multiple enhancers (e.g., redundant enhancers) may resist single-enhancer inhibition, whereas over-editing could disrupt cellular homeostasis. Enhancer-targeted interventions might also perturb chromatin architecture, affecting non-target genes. For example, targeting the MYC enhancer could dysregulate adjacent noncoding RNAs like PVT1, leading to genomic instability [88]. Despite their transformative potential, the risks of enhancer-targeted strategies stem from the complexity of enhancer regulatory networks and insufficient technological specificity. Future efforts should integrate multi-omics data and machine learning models to predict critical enhancers, coupled with AI-driven design of next-generation inhibitors. Balancing innovation with risk mitigation will be essential to advance precision medicine sustainably.
Enhancer RNA in cancer
eRNAs play an indispensable role in enhancer activation as well as transcriptional regulation. Genetic alterations and epigenetic modifications in the enhancer region can significantly influence eRNAs expression. Growing evidence indicates that abnormal eRNAs expression can drive tumour progression.
eRNAs are crucial in the regulation of gene expression through various mechanisms. In cancer, they can either enhance the transcription of oncogenes or suppress the activity of tumour suppressor genes. For example, insulin receptor substrate 2 (IRS2), an oncogene located on chromosome 13, is essential for driving cell proliferation, clonogenic survival, and anti-apoptotic activity in oral squamous cell carcinoma (OSCC). The IRS2-linked enhancer RNA (IRS2e), located adjacent to the IRS2 locus, exhibits elevated expression in OSCC and is associated with unfavourable clinical outcomes [89]. Additionally, research has shown that CCAT1-L, an enhancer RNA embedded in a super-enhancer near the MYC locus, is expressed from a genomic region 515 kb upstream of MYC in human colorectal tumours [90]. Functionally, this long non-coding RNA (lncRNA) facilitates the formation of long-range chromatin loops, thereby modulating MYC transcription. Interestingly, the architecture of these loops undergoes remodelling during cancer progression, leading to dysregulated gene expression. In renal cancer cells, the level of eRNA p53BER2 is lower compared to normal tissues. Silencing p53BER2 mitigates the cytotoxic effects of nutlin-3 in TP53-WT cell lines, as well as the G1-phase blockade and senescence in these cells [91]. Furthermore, additional experiments have indicated that p53BER2 can induce cell-cycle arrest and DNA repair by interacting with BRCA2. Overall, eRNAs regulate oncogenes and tumour suppressors through various transcriptional, epigenetic, and structural mechanisms. Comprehensive studies combining eRNA profiling with functional assays are necessary to better understand their roles in cancer development and treatment.
In addition to regulating tumour-related genes, eRNAs are involved in aberrant signalling cascade response pathways in tumours. CCAT1 is a cancer-associated eRNA targeted downstream of TP63 and SOX2 in squamous cell carcinoma (SCC). TP63 and SOX2 co-occupy the promoter and super-enhancer of CCAT1 to activate its transcription in SCC cells, and CCAT1 subsequently forms complexes with TP63 and SOX2. Silencing CCAT1 in SCC significantly impairs cancer cell viability, clonogenic ability, and reduces tumour volume. Further studies found that CCAT1 interacts with SOX2 and TP63 to mediate EGFR expression by binding with its enhancer, consequently activating the MEK/ERK1/2 and PI3K/AKT signalling pathways [92].
Because eRNAs are associated with immune escape and treatment resistance in cancer cells, they can be clinically used as markers of tumour treatment responsiveness and resistance. The eRNA LINC02257 is notably linked to poor prognosis in colorectal adenocarcinoma patients. Abnormal LINC02257 expression alters the tumour immune microenvironment across different tumour categories [93]. Additionally, the relocation of proto-oncogenes to transcriptionally active regions is a common chromosomal rearrangement in cancer. For example, the transfer of the proto-oncogene MYC to the immunoglobulin (IGH) heavy chain motif occurs in 85% of Burkitt lymphoma cases, leading to increased MYC proto-oncogene expression. The IGH/MYC translocation results in overexpression of the eRNA AL928768.3. Overexpression of AL928768.3 increases lymphoma cell growth and resistance to chemotherapy, whereas no such resistance is observed in B-cell lines without this translocation. Targeting AL928768.3 to reduce resistance in Burkitt’s lymphoma may become a common therapeutic approach in the clinic [94].
Furthermore, eRNAs can serve as therapeutic targets and potential biomarkers for diagnosis and prognosis. The proto-oncogene MYC is a key driver in leukemogenesis and AML (Acute myeloid leukemia) progression, while the orphan nuclear receptor NR4A1, a tumour suppressor frequently silenced in AML patients, directly binds to MYC super-enhancers, eliminating the necessary coactivators to disrupt AML-selective MYC super-enhancer activity, thus inhibiting tumour cell growth both in vivo and in vitro [95]. To better predict the prognosis of cervical adenocarcinoma (CESC) patients, researchers identified 7936 eRNAs that were distinctly expressed in CESC patients through bioinformatic analysis and further identified a prognostic model composed of eight eRNAs. The 8-eRNA model demonstrates higher accuracy and stability and is an independent prognostic indicator compared to clinicopathological classification alone [96]. The construction of the 8-eRNA model provides important insights for exploring novel therapeutic targets for cervical cancer patients. As of now, many eRNAs in cancer cells remain untapped, and we anticipate further research to uncover the relationship between eRNAs and cancer.
In conclusion, eRNAs emerge as both dynamic biomarkers and functional executors of enhancer activity, playing pivotal roles in oncogenesis, metastatic progression, and therapeutic resistance. Mechanistically, eRNAs directly regulate cell cycle-associated genes to drive tumour cell proliferation and survival. Notably, Li et al. demonstrated that among 626 cancer-associated eRNAs, a significant subset (including CCND1 and MYC targets) exhibit potent pro-proliferative effects, with experimental eRNAs suppression leading to marked downregulation of these oncogenes and subsequent suppression of PCa growth [97]. Importantly, eRNA quantitative trait loci (eRNA-QTLs) show spatial colocalisation with cancer risk variants, establishing their regulatory significance in tumourigenesis. Beyond primary tumour growth, eRNAs orchestrate metastatic programmes through microenvironmental remodelling. This is exemplified in BC models where eRNA-mediated activation of epithelial-mesenchymal transition (EMT) genes promotes invasive phenotypes [98]. Furthermore, eRNAs drive therapeutic resistance via multimodal regulation of drug metabolism enzymes (e.g., cytochrome P450 family) and survival signalling cascades. Illustrating this paradigm, Shi et al. have characterised the dual functionality of super-enhancer-derived LINC02454 in modulating temozolomide (TMZ) sensitivity through epigenetic and post-transcriptional mechanisms in glioma [99]. These collective findings position eRNA-targeted therapies as a transformative frontier in oncology. Strategic modulation of eRNAs networks may overcome current limitations in precision oncology approaches, particularly for treatment-refractory malignancies.
RNA m6A modification
RNA m6A modification
Post-transcriptional modifications, a crucial domain of epigenetics, have come out as key regulators of numerous physiological and pathological processes. According to MODOMICS, over 170 distinct chemical modifications of RNA had been established in living organisms by the end of 2023 [100]. Among these, N6-methyladenosine (m6A) stands out as the most prevailing, abundant, and evolutionarily conserved internal co-transcriptional modification in eukaryotic RNAs. As a dynamically reversible process, RNA m6A modification is catalyzed by writer proteins, removed by erasers, and recognized by reader proteins, predominantly accumulating near termination codons, the 3’ untranslated region (3’UTR), and internal exons. At the molecular level, m6A influences almost every dimension of RNA metabolism, including mRNA translation, degradation, splicing, folding, nuclear export, stabilization, and decay. Moreover, m6A modifications play integral roles in diverse biological processes, such as transcriptional regulation, intracellular signaling, and the DNA damage response [101–103]. In the last several years, accumulating researches have demonstrated that RNA m6A modification further modulates tumor development by regulating tumor cell metabolism, underscoring its significance in cancer biology.
Regulators of m6A (writer, eraser, reader)
Writer
The most extensively studied m6A-modifying complex is the m6A methyltransferase complex, which comprises methyltransferase-like protein 3 (METTL3), methyltransferase-like protein 14 (METTL14), and the cofactor Wilms Tumor 1 Associated Protein (WTAP). Each of these components plays a crucial role in the enzymatic process. In recent years, additional accessory proteins participating in m6A modification have been discovered, maintaining methyltransferase-like protein 16 (METTL16), methyltransferase-like protein 5 (METTL5), human methyltransferase (ZCCHC4), RNA-binding motif protein 15 (RBM15), and Vir-like m6A methyltransferase-associated protein (VIRMA/KIAA1429). These proteins contribute to m6A deposition on various structured RNAs, such as U6 snRNA and 28 S rRNA, and, under certain conditions, within mRNA introns [104–108].
Eraser
m6A erasers function as demethylases, removing m6A modifications via oxidative reactions that require ferrous iron (Fe²⁺) as a cofactor and α-ketoglutarate as a co-substrate. The finding of the first m6A eraser, fat mass and obesity-associated protein (FTO), by Professor He Chuan in 2011 provided compelling evidence that m6A modifications are dynamically reversible, leading to a surge in m6A research. The second major m6A eraser, alkylation repair homolog 5 (ALKBH5), was discovered in 2013. Like FTO, ALKBH5 belongs to the α-ketoglutarate-dependent dioxygenase family. Aside from FTO and ALKBH5, other erasers such as ALKBH1 and ALKBH3 have been identified. ALKBH1 is involved in tRNA demethylation [109], whereas ALKBH3 functions as an m1A demethylase (N1-methyladenosine), acting on both m1A residues in tRNA and m6A modifications in RNA [110, 111].
Reader
m6A-modified readers encompass a varied range of proteins, containing the YTH domain-containing family (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2), the insulin-like growth factor 2 mRNA-binding protein family (IGF2BP1, IGF2BP2, IGF2BP3), the HNRNP family (HNRNPA2B1, HNRNPC, HNRNPG), eukaryotic translation initiation factors (eIF3, eIF4E, eIF4G), and embryonic lethal abnormal visual-like protein 1 (ELAVL1). Recently, a novel m6A reader, proline-rich coiled-coil 2 A (Prrc2a), has also been identified. These readers regulate downstream RNA metabolism either by indirectly binding RNA through m6A-induced structural changes or by directly recognizing and interacting with m6A-modified sites. Their functions influence various cellular processes, maintaining RNA splicing, nuclear export, stability, and translation, thereby playing critical roles in disease development (Table 2).
Table 2.
Functions of m6A regulators in RNA metabolism
| Type | m6A regulators | Function | References |
|---|---|---|---|
| Writers | METTL3 | Catalyzes m6A modification | [112] |
| METTL14 | A core subunit of the m6A methyltransferase forms a heterodimer with METTL3 to facilitate m6A modification | [112] | |
| WTAP | The regulatory subunit of the m6A methyltransferase and recruits METTL3 and METTL14 into the nuclear speckles | [113] | |
| METTL16 | Accountable for m6A modification of U6 snRNA, snRNA and other lncRNAs | [104] | |
| METTL5 | Accountable for m6A modification of 18 S rRNA | [105] | |
| ZCCHC4 | Accountable for m6A modification of 28 S rRNA | [106] | |
| RBM15 | Binds the m6A complex and recruits it to a special RNA site | [114] | |
| VIRMA/KIAA1429 | Directs the methyltransferase components to specific RNA region | [115] | |
| Erasers | FTO | Demethylates m6A, also has activity towards m6Am and m1A | [116, 117] |
| ALKBH5 | Principally demethylates m6A | [118] | |
| ALKBH1 | Acting as a tRNA demethylase by removing N(1)-methyladenine, which regulates translation initiation and elongation | [109] | |
| ALKBH3 | tRNAs demethylase | [111] | |
| Readers | YTHDF1 | Enhances mRNA translation | [119] |
| YTHDF2 | Enhances mRNA degradation | [119] | |
| YTHDF3 | Prompts translation and degradation by interacting with YTHDF1 and YTHDF2 | [120] | |
| YTHDC1 | Leads to RNA splicing and export | [121] | |
| YTHDC2 | Improves the translation efficiency of target mRNA | [121] | |
| IGF2BP1/2/3 | Enhances the stability and translation of mRNA | [122] | |
| HNRNPA2B1 | Spurs primary microRNA processing | [123] | |
| eIF3 | Promotes mRNA translation | [124] | |
| ELAVL1 | RNA-binding protein | [125] | |
| Prrc2a | Recognizes m6A modification and associates with myelin formation and meiosis | [126, 127] |
Cross talk between m6A modification and other posttranscriptional modifications
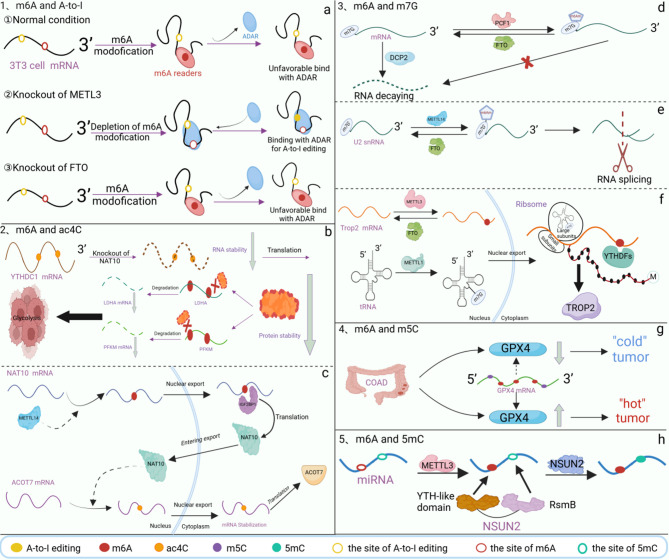
Post-transcriptional modification represents an essential regulatory mechanism in gene expression, involving a complex interplay of various processes. The interaction between m6A modification and other RNA modifications in both physiological and pathological contexts has been increasingly recognized; however, research in this field remains in its early stages. As early as 2018, genome-wide analyses revealed that m6A modification negatively regulates adenosine-to-inosine (A-to-I) RNA editing. Adenosine deaminases acting on RNA (ADARs) catalyze the conversion of adenosine to inosine in double-stranded RNA substrates. In mouse 3T3 cells, knockdown of METTL3 contributed to an enhanced A-to-I editing, whereas depletion of FTO resulted in its downregulation. Despite these observations, the precise mechanism by which m6A influences A-to-I editing within the same transcript remains unclear. It is hypothesized that m6A-modified RNAs may exhibit reduced binding affinity for ADAR, or alternatively, that the presence of m6A-bound reader proteins may sterically hinder subsequent A-to-I editing [128]. N-acetyltransferase 10 (NAT10) is the only enzyme identified to catalyze N4-acetylcytosine (ac4C) modification [129]. In osteosarcoma, NAT10 knockdown impairs the translation of the m6A reader YTHDC1. Through disrupting this pathway, NAT10 depletion reduces osteosarcoma cell growth, migration, and invasion while increasing global m6A levels [130]. Beyond acetylation and A-to-I editing, m6A also exhibits crosstalk with various methylation modifications. Under normal physiological conditions, decapping enzyme 2 (DCP2) regulates mRNA degradation when the 5’ cap is modified by m7G [131]. If the first nucleotide following the m7G cap is 2’-O-dimethyladenosine (Am), this Am residue can be further methylated by phosphorylated CTD-interacting factor 1 (PCIF1), forming the m6Am modification. FTO is capable of removing this m6Am mark, thereby maintaining its dynamic reversibility. Notably, the presence of an m7G cap adjacent to m6Am protects the transcript from DCP2-mediated decapping, underscoring the functional coordination between these modifications [132]. m6Am modification has also been detected within the interior regions of U2 small nuclear RNA (snRNA) alongside m7G-modified caps. The cooperative action of m6Am and m7G modifications has an essential impact on regulating RNA splicing [133]. In bladder cancer (BCa), trophoblast cell surface antigen 2 (TROP2) is highly upregulated. METTL3-mediated m6A modification at the 3’UTR of TROP2 mRNA enhances its translation, while METTL1-mediated m7G modification in tRNA improves translation efficiency. These modifications act synergistically to increase TROP2 expression, thereby promoting BCa progression [134]. Interestingly, IGF2BP family proteins recognize both m6A and m7G modifications within mRNAs in cancer cells. Notably, the spatial distribution of m6A and m7G is similar when bound by IGF2BP, but their regulatory effects on mRNA stability differ. IGF2BP-mediated recognition of m7G promotes mRNA degradation, whereas recognition of m6A enhances mRNA stability. This suggests a potential functional interplay between these modifications, as more than half of m7G-modified mRNAs were found to be simultaneously modified by m6A [135]. 5mC methylation is a well-characterized epigenetic modification in mammalian DNA. Remarkably, single-molecule quantum sequencing of miR-200c-5p—a microRNA derived from colorectal cancer (DLD-1) cells—revealed simultaneous m6A and 5mC modifications. Further studies demonstrated that RNA 5mC methylation is catalyzed by NOP2/Sun RNA methyltransferase 2 (NSUN2), an enzyme possessing two distinct structural domains: a YTH-like domain responsible for recognizing m6A, and an RsmB domain that catalyzes cytosine methylation. As a result, m6A modifications at positions #7 and #11 in miR-200c-5p are consistently associated with 5mC modification at position #13, highlighting the coordinated regulatory interplay between these two modifications [136] (Fig. 2).
Fig. 2.
The crosstalk between m6A modification and other posttranscriptional modifications. (a) There is a negative regulatory effect of m6A modification on A-to-I editing. Knockdown of METTL3 in mouse 3T3 cells upregulates A-to-I while knockdown of FTO downregulates A-to-I. (b) Knockout of NAT10, a major enzyme for ac4C acetylation, reduces the stability of YTHDC1, rendering the m6A sites on PFKM and LDHA mRNA unrecognized and ultimately positively regulating glycolysis in osteosarcoma cells. (c) METTL14 facilitates the m6A modification of NAT10 mRNA, while IGF2BP1 interacts with the m6A site on NAT10 mRNA to enhance its translation, resulting in NAT10 production. NAT10 mediates the ac4C modification of ACOT7 mRNA, promoting its translation to form ACOT7. (d) Catalyzes m6Am modification near the m7G cap, which increases resistance to DCP2-mediated decapping. (e) METTL14 installs m6Am modification at an internal site in U2 snRNA, which culminates in influencing global alternative RNA splicing. (f) METTL3 regulates m6A modification at the 3’-UTR in TROP2 mRNA to enhance translation of TROP2 mRNA, and METTL1 mediates m7G modification in tRNA to improve mRNA translation efficiency, which synergistically increase TROP2 expression to promote bladder cancer cell development. (g) Both m6A and m5C modifications are present in GPX4 mRNA, and both modifications work together to maintain redox homeostasis in COAD. (h) METTL3 mediates m6A modification of miRNAs, and NSUN2 recognizes m6A through the YTH-like structure domain and mediates 5mC methylation modification through the RsmB structural domain
The relationship between enhancer and m6A modification
Disruptions in epigenetic regulation play a pivotal role in numerous physiological and pathological conditions, with particularly profound implications for cancer development. Two key components in the field of epigenetics are m6A RNA methylation and enhancers. m6A is widely recognised as a post-transcriptional modification, while enhancers primarily regulate gene transcription. However, the relationship between m6A and enhancers remains unclear. Some potential effects of m6A on enhancers have been documented. In the context of acute myeloid leukaemia, the establishment of active enhancer elements potently induces IGF2BP2 and IGF2BP3 expression. These RNA-binding proteins, in turn, stabilise m6A-modified DDX21 transcripts. Functionally, DDX21 orchestrates an oncogenic programme by recruiting TF YBX1 to activate ULK1 expression, ultimately promoting leukemogenesis through enhanced proliferation and apoptosis resistance [137]. In clear cell renal cell carcinoma (ccRCC), the m6A eraser enzyme FTO regulates bromodomain-containing protein 9 (BRD9) expression by demethylating its mRNA to enhance stability. Importantly, the TF SOX17 mediates BRD9 recruitment to de novo enhancers that control key oncogenic networks, facilitating ccRCC progression [138]. Notably, we identified a METTL14-m6A-BPTF regulatory axis in renal cell carcinoma metastasis. While m6A modification destabilised BPTF mRNA, BPTF expression remained crucial for promoting lung metastasis. METTL14 deficiency led to BPTF accumulation, which subsequently altered the enhancer repertoire to reinforce oncogenic pathways and form active enhancers that reprogrammed cellular metabolism towards glycolysis. This work uncovers previously unrecognised functional links between RNA epigenetics and chromatin remodelling in cancer progression [139]. In conclusion, these results provide greater insight into the connection between enhancers and m6A. Nevertheless, only a limited number of studies have verified that m6A plays a role in enhancer regulation. It is still uncertain whether enhancers influence m6A in a mechanistic way, and further research is required to shed light on this issue.
RNA m6A modification in cancer
Numerous studies have demonstrated that m6A modification and its regulatory proteins exert significant influence over cancer initiation, progression, and prognosis. This section summarises the key roles of m6A modifications across various cancers (see Table 3).
Table 3.
The roles of m6A regulators in cancer
| Cancer types | m6A regulators | Expression | Targets | Function | Role in cancer | References |
|---|---|---|---|---|---|---|
| AML | IGF2BP2 | Upregulation | MYC/GPT2/SLC1A5 |
Prompting cell growth and proliferation Hindering cell differentiation and apoptosis |
Oncogene | [144] |
| Glioblastoma | ALKBH5 | Upregulation | VEGFA/CHK1/CD73 | Prompting cell migration, invasion, and resistance to radiotherapy | Oncogene | [146–148] |
| SRSF7 | Upregulation | PBK | Enhancing cell proliferation and migration | Oncogene | [149] | |
| LC | METTL3 | Upregulation | circIGF2BP3/DCP2 | Prompting cell immune escape and resistance to chemotherapy | Oncogene | [151, 153] |
| IGF2BP3 | Upregulation | COX6B2 | Prompting cell resistance to chemotherapy | Oncogene | [154] | |
| FTO | Downregulation | ESR1 | Inhibiting cell growth and metastasis | Anti-oncogene | [152] | |
| YTHDF1/IGF2BP3 | Upregulation | ESR1 | Prompting cell growth and metastasis | Oncogene | [152] | |
| ESCA | METTL3 | Upregulation | MYC | Promoting cell proliferation, invasion, metastasis, and immune escape | Oncogene | [157, 158] |
| GC | METTL3/IGF2BP2 | Upregulation | PABPC1/miRNA-118/STAT5A | Prompting cell proliferation and migration | Oncogene | [159–161] |
| YTHDF1 | Upregulation | IFN-γ | Inhibiting cell growth and metastasis | Oncogene | [164] | |
| YTHDF2 | Upregulation | CyclinD1 | Prompting cell growth, invasion, and resistance to radiotherapy and chemotherapy | Oncogene | [165] | |
| YTHDF3 | Upregulation | lncRNAs/miRNAs/mRNA | Prompting cell growth and invasion | Oncogene | [166] | |
| HCC | METTL3/METTL14 | Upregulation | ACLY, SCD1 | Prompting cell growth and invasion | Oncogene | [167] |
| WTAP/IGF2BP3 | Upregulation | circCCAR1 | Prompting cell growth, metastasis, resistance to chemotherapy, and immunosuppression | Oncogene | [170] | |
| YTHDF1 | Upregulation | ATG2A | Prompting cell growth and metastasis | Oncogene | [234] | |
| PC | WTAP/IGF2BP3 | Upregulation | GBE1 | Promoting cell proliferation and stemness | Oncogene | [175] |
| CRC | YTHDF2 | Downregulation | STEAP3-AS1 |
Inhibiting cell growth, proliferation and metastasis |
Anti-oncogene | [178] |
| METTL3 | Upregulation | POU6F2-AS1 | Promoting cell growth and invasion | Oncogene | [179] | |
| METTL16/IGF2BP1 | Upregulation | SOGA1 | Promoting cell growth and proliferation | Oncogene | [180] | |
| METTL17 | Upregulation | LRPPRC/MSSUs | Promoting cell growth, proliferation and invasion | Oncogene | [181] | |
| ALKBH5 | Upregulation | AXIN2 | Promoting cellular immunosuppression | Oncogene | [182] | |
| RCC | HNRNPC | Upregulation | PPAP2B | Prompting cell proliferation and metastasis | Oncogene | [183] |
| METTL14 | Upregulation | TRAF1 | Prompting cell resistance to chemotherapy | Oncogene | [184] | |
| ALKBH5 | Upregulation | AURKB | Prompting cell growth, proliferation, invasion | Oncogene | [185] | |
| BCa | RBM15/METTL3 | Upregulation | ENO1 | Promoting cell growth | Oncogene | [186] |
| WTAP | Upregulation | circ0008399 | Promoting cell resistance to chemotherapy | Oncogene | [187] | |
| YTHDF2 | Upregulation | DHCR7 | Promoting cell invasion and metastasis | Oncogene | [188] | |
| FTO | Upregulation | MALAT1 | Promoting cell growth and proliferation | Oncogene | [189] | |
| PCa | YTHDC1 | Upregulation | SLC12A5 | Prompting cell metastasis, and resistance to chemotherapy | Oncogene | [190] |
| YTHDF3 | Upregulation | SPOP/NXK3.1/TWIST1/SNAI2 | Prompting cell growth and proliferation | Oncogene | [191] | |
| METTL3 | Upregulation | circRBM33 | Prompting cell growth, invasion, and resistance to chemotherapy | Oncogene | [192] | |
| RBM15 | Upregulation | FTO-IT1 | Prompting cell growth and proliferation | Oncogene | [193] | |
| FTO | Downregulation | DDIT4 | Inhibiting cell growth and invasion | Anti-oncogene | [194] | |
| BC | METTL3/YTHDF2 | Upregulation | LATS1 | Prompting cell growth and invasion | Oncogene | [195] |
| ALKBH5 | Upregulation | FOXO1 | Prompting cell stemness, and resistance to chemotherapy | Oncogene | [196] | |
| IGF2BP2 | Upregulation | CDK6 | Prompting cell growth and proliferation | Oncogene | [197] | |
| hnRNPA2B1 | Upregulation | ALYREF/NXF1 | Prompting cell stemness and proliferation | Oncogene | [198] | |
| OC | METTL3 | Upregulation | circPLPP4 | Prompting cell resistance to chemotherapy | Oncogene | [199] |
| ALKBH5 | Upregulation | ITGB1/FSH | Prompting cell proliferation and metastasis | Oncogene | [200, 201] | |
| YTHDC1 | Downregulation | PIK3R1 | Prompting cell growth and proliferation | Anti-oncogene | [202] | |
| CC | ALKBH5 | Downregulation | circCCDC134 | Prompting cell proliferation and metastasis | Anti-oncogene | [203] |
| HNRNPC | Upregulation | FOXM1 | Prompting cell invasion and metastasis | Oncogene | [204] | |
| METTL3 | Upregulation | HSPA9 | Prompting malignant transformation of cells | Oncogene | [205] | |
| EC | WTAP | Downregulation | EGR1 | Hindering cell growth, and resistance to chemotherapy | Anti-oncogene | [207] |
| METTL3 | Downregulation | NLRC5 | Hindering cell proliferation and metastasis | Anti-oncogene | [208] | |
| OSCC | METTL14 | Upregulation | CALD1 | Promoting cell growth and metastasis | Oncogene | [209] |
| osteosarcoma | METTL14 | Upregulation | MN1 | Prompting cell growth and resistance to chemotherapy | Oncogene | [210] |
| melanoma | METTL14 | Upregulation | RUNX2 | Prompting cell growth and metastasis | Oncogene | [211] |
| pitNET | FTO | Upregulation | DSP | Prompting cell growth and resistance to chemotherapy | Oncogene | [212] |
| NPC | WTAP | Upregulation | DIAPH1-AS1 | Prompting cell growth and metastasis | Oncogene | [213] |
| Laryngeal cancer | METTL3 | Upregulation | HOXA10-AS | Prompting cell proliferation and oxidative resistance | Oncogene | [216] |
| HSCC | METTL3 | Upregulation | circCUX1 | Prompting cell resistance to radiotherapy | Oncogene | [217] |
| seminoma | METTL3 | Upregulation | TFAP2C | Prompting cell resistance to chemotherapy | Oncogene | [218] |
AML acute myeloid leukemia, LC lung cancer, ESCA esophageal cancer, GC gastric cancer, HCC hepatocellular carcinoma, PC pancreatic cancer, CRC colorectal cancer, RCC renal cell carcinoma, BCa bladder cancer, PCa prostate cancer, BC breast cancer, OC ovarian cancer, CC cervical cancer, EC endometrial cancer, OSCC oral squamous cell carcinoma, NPC nasopharyngeal carcinoma, pitNET pituitary neuroendocrine tumors, HSCC hypopharyngeal squamous cell carcinoma
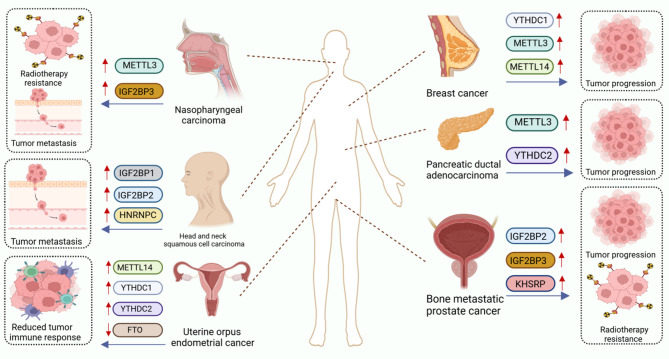
Acute myeloid leukemia
AML, the most prevalent form of adult acute leukaemia, is an aggressive haematological malignancy characterised by the clonal expansion of immature myeloid cells, impaired differentiation of myeloid progenitors, and infiltration of malignant clones into the bone marrow, peripheral blood, and other tissues. These pathological processes lead to uncontrolled proliferation, blocked differentiation, and disrupted haematopoiesis. The development of AML has been linked to dysregulation of m6A regulators including METTL3, METTL14, ALKBH5, YTHDF1, YTHDF2, and IGF2BP2 [140–144]. Among these, IGF2BP2 expression is significantly elevated in AML, particularly in leukaemia stem cells (LSCs) and leukaemia-initiating cells (LICs), and is strongly associated with poor prognosis. IGF2BP2 enhances the expression of MYC, GPT2, and SLC1A5 in an m6A-dependent manner, promoting glutamine uptake and metabolism. This leads to improved mRNA stability and enhanced translation, fuelling the tricarboxylic acid (TCA) cycle and accelerating AML cell proliferation. These findings suggest that IGF2BP2 inhibitors may hold therapeutic promise in future AML treatment [144].
Glioblastoma
Glioblastoma is the most common and lethal primary brain tumour, known for its invasive growth, extensive vascularisation, and neurodestructive behaviour. Expression levels of YTHDF family members and METTL14 are elevated in glioblastoma tissues compared to normal brain tissue. In particular, YTHDF2 and YTHDF3 are markedly upregulated in high-grade gliomas [145]. ALKBH5 is also highly expressed in gliomas, and its knockdown has been shown to reduce angiogenesis both in vitro and in vivo [146]. Previous research has linked ALKBH5 with glioma cell invasion, as well as radioresistance and chemoresistance [147, 148]. Furthermore, ALKBH5 expression correlates with glioma malignancy and patient prognosis, positioning it as a potential clinical biomarker [146]. Additionally, SRSF7 (Serine/Arginine-Rich Splicing Factor 7), a novel m6A regulator, has been found to promote glioma cell proliferation and migration. It does so by recruiting the methyltransferase complex to selectively enhance m6A methylation near its RNA binding sites, which are often located in genes associated with cell growth and motility [149]. These findings provide important insights into the development of glioma-targeted therapies.
Lung cancer
Lung cancer (LC) is one of the most aggressive malignancies worldwide and accounts for approximately 18% of all cancer-related deaths. Compared to adjacent normal tissues, expression of METTL3, METTL16, YTHDF1, and IGF2BP3 is elevated in LC [150–152], while FTO expression is typically reduced [152]. A circular RNA derived from IGF2BP3, known as circIGF2BP3, is significantly overexpressed in non-small cell lung cancer (NSCLC). This circRNA (circular RNA) is generated via back-splicing of exons 4 and 13 and undergoes m6A modification mediated by METTL3, with YTHDC1 facilitating its circularisation. CircIGF2BP3 impairs CD8⁺ T cell responses and promotes tumour immune evasion [153]. In addition, METTL3 enhances m6A methylation of DCP2, leading to its degradation and the promotion of mitochondrial autophagy, which in turn contributes to chemoresistance in NSCLC [151]. Recent findings indicate that IGF2BP3 also promotes metabolic reprogramming, thereby contributing to acquired drug resistance [154]. FTO expression is significantly lower in NSCLC tumour tissues compared to adjacent non-cancerous tissues. One of FTO’s targets is ESR1 (oestrogen receptor alpha). YTHDF1 and IGF2BP3 each recognise distinct m6A sites on ESR1 mRNA, enhancing its stability and promoting tumour growth. Overexpression of FTO leads to reduced m6A methylation of ESR1 mRNA. These insights into the FTO–YTHDF1–IGF2BP3–ESR1 axis present new opportunities for NSCLC treatment [152]. However, studies on ALKBH5 in NSCLC have yielded conflicting results. Some research suggests ALKBH5 overexpression promotes NSCLC progression, while other studies indicate that elevated ALKBH5 levels inhibit tumour cell proliferation and promote apoptosis [155, 156]. These discrepancies highlight the need for further investigation into the context-dependent role of ALKBH5 in LC.
Esophageal cancer
Oesophageal cancer (ESCA) is a common and highly fatal malignancy of the digestive tract. Recent studies have revealed a positive correlation between Fusobacterium nucleatum (Fn) infection and METTL3 expression in ESCA tissues, with Fn significantly upregulating METTL3. c-Myc mRNA has been identified as a downstream target of METTL3, and the m6A reader YTHDF1 binds to the m6A site on c-Myc mRNA, enhancing its stability and thereby increasing c-Myc expression, which promotes ESCA cell invasion and metastasis [157]. Serine hydroxymethyltransferase 2 (SHMT2), a key mitochondrial enzyme involved in one-carbon unit metabolism, is significantly elevated in ESCA tissues and associated with poor prognosis. METTL3, ALKBH5, and FTO mediate the m6A modification of c-Myc mRNA, and SHMT2 further regulates this modification via the one-carbon metabolic pathway by increasing intracellular SAM (S-adenosylmethionine) levels, ultimately promoting c-Myc expression and facilitating immune escape [158]. Thus, targeting the one-carbon unit metabolic pathway may represent a promising therapeutic strategy for ESCA.
Gastric cancer
GC is the fifth most prevalent malignancy worldwide. Both METTL3 and its downstream target STAT5A act as oncogenes in GC. Helicobacter pylori infection has been shown to further elevate the expression of METTL3 and STAT5A, which correlates with poor patient prognosis. The reader protein IGF2BP2 binds to the m6A-modified site on STAT5A mRNA, enhancing its stability and protein expression, thereby increasing the invasive potential of GC cells [159]. In addition, METTL3 can promote gastric carcinogenesis by facilitating EMT and by binding to non-m6A-modified mRNAs [160, 161]. Although ALKBH5 has been explored for its role in GC cell invasion and progression [162], its expression is significantly lower in GC tissues compared to normal tissues and is further reduced in patients with distant metastases [163]. This inconsistency has led to debate regarding the role of ALKBH5 in GC, necessitating further research. Moreover, engineered small extracellular vesicles have been developed to deliver siRNA targeting YTHDF1, effectively reducing its expression and thereby inhibiting GC progression and metastasis, offering a novel therapeutic avenue [164]. YTHDF2 has also been shown to promote GC cell proliferation both in vitro and in vivo, as well as to increase resistance to radiotherapy [165]. Similarly, YTHDF3 enhances GC cell invasiveness by activating the PI3K/AKT pathway and modulating the immune microenvironment. Its upregulation is often associated with poor clinical outcomes [166].
Hepatocellular carcinoma
HCC is the second leading cause of cancer-related death globally. In HCC cells, overexpression of METTL3 and METTL14 leads to m6A-mediated stabilisation of oncogenic transcripts, thereby accelerating tumour progression [167]. CircRNAs also play critical roles in HCC. For example, circGPR137B is significantly downregulated in HCC and forms a feedback loop with miR-4739 and FTO, collectively suppressing metastasis [168]. Paradoxically, FTO is upregulated in HCC and contributes to tumour proliferation. Targeting FTO with inhibitors has shown potential to suppress HCC growth and enhance chemosensitivity [169]. Additionally, EIF4A3 promotes the generation of circCCAR1, whose stability is enhanced by WTAP-mediated m6A modification via IGF2BP3 interaction. Elevated levels of circCCAR1 impair CD8⁺ T-cell function, facilitating HCC progression and metastasis [170]. Multi-omics analyses have also revealed that NOTCH1 is a direct downstream target of YTHDF1, which binds to the m6A-modified NOTCH1 mRNA, enhancing its stability and translation. Inhibiting YTHDF1 reduces cancer cell stemness and improves therapeutic responses, making it a promising target for HCC treatment [171].
Pancreatic cancer
Pancreatic cancer (PC) is the most lethal gastrointestinal malignancy, known for its insidious onset, rapid progression, and dismal prognosis. METTL3 has been found to promote PC cell proliferation, whereas METTL16 inhibits PC cell metastasis and FTO reduces drug resistance [172–174]. The m6A modification of GBE1 (Glucan branching enzyme 1) mRNA by WTAP and IGF2BP3 enhances its stability and expression, correlating with poor prognosis in PC [175]. Additionally, DDIT4-AS1, a downstream target of ALKBH5, is upregulated via ALKBH5-mediated m6A demethylation, increasing stemness and chemoresistance [176]. Under hypoxic conditions, ALKBH5 decreases global m6A levels in PC cells, thereby enhancing glycolysis. YTHDF2 recognises m6A-modified HDAC4 (histone deacetylase type 4) mRNA, stabilising its expression. HDAC4, in turn, regulates the acetylation of HIF1α, a direct target of ALKBH5. This establishes a positive feedback loop involving ALKBH5/HDAC4/HIF1α, which drives glycolytic metabolism and promotes PC cell migration under hypoxia [177]. This regulatory axis may offer a therapeutic opportunity for hypoxia-targeted PC treatment.
Colorectal cancer
Colorectal cancer (CRC) is one of the most common malignancies globally, ranking third in incidence and second in mortality. The relationship between m6A modification and CRC progression has been well established. The antisense lncRNA STEAP3-AS1, induced by hypoxia, promotes CRC cell growth and migration, correlating with poor prognosis. STEAP3-AS1 and YTHDF2 compete for binding, disrupting the interaction between YTHDF2 and STEAP3 mRNA. This competition protects STEAP3 mRNA from m6A-mediated degradation, thereby enhancing the expression of STEAP3 protein to support CRC progression [178]. Another long non-coding RNA, POU6F2-AS1, is also upregulated in CRC. METTL3-induced m6A modification of POU6F2-AS1 promotes CRC cell proliferation and adipogenesis [179]. METTL16 expression correlates with the inhibition of SOGA1 (suppressor of glucose by autophagy) and PDK4 (pyruvate dehydrogenase kinase 4). METTL16 enhances SOGA1 expression by binding to IGF2BP1, leading to the upregulation of PDK4, which subsequently promotes glycolytic reprogramming and accelerates CRC progression [180]. METTL17 expression is elevated in CRC patients, and its depletion increases sensitivity to ferroptosis while inhibiting CRC cell proliferation, migration, invasion, and tumour growth in xenograft models [181]. ALKBH5 interacts with and demethylates AXIN2, a Wnt repressor, leading to the degradation of IGF2BP1 and promoting immunosuppression, thus fostering CRC development. Targeting ALKBH5 may provide therapeutic benefits for CRC patients [182].
Renal cell carcinoma
Among urologic malignancies, renal cell carcinoma (RCC) has a comparatively high mortality rate, and its incidence continues to rise. Many studies have linked m6A regulators to the development of RCC. However, it has now been discovered that METTL3, the IGF2BP family, and FTO all appear to play conflicting roles in RCC, with even the function of the same m6A regulator differing depending on the RCC subtype. CircPPAP2B was found to be overexpressed in highly aggressive RCC cells and associated with poor prognosis. Furthermore, it was demonstrated that circPPAP2B interacts with HNRNPC in an m6A-dependent manner and promotes HNRNPC nuclear translocation. On-degradable ubiquitination of HNRNPC leads to subcellular re-localisation, which causes metastasis of RCC [183]. Tumour necrosis factor receptor-associated factor TRAF1 expression is up-regulated in sunitinib-resistant RCC cells. METTL14 regulates the mRNA stability of TRAF1 in an IGF2BP2-dependent manner to maintain sunitinib resistance [184]. ALKBH5, which is highly expressed in RCC, stabilises the AURKB mRNA (encoding aurora kinase B) in an m6A-dependent manner to promote RCC cell growth and invasion [185]. Therefore, targeting METTL14 and ALKBH5 signalling pathways could offer a promising therapeutic avenue for RCC.
Bladder cancer
BCa is the second most common genitourinary carcinoma. Recurrence, metastasis, and drug resistance are three key concerns of BCa that need to be addressed. METTL3, METTL14, and WTAP are elevated in BCa compared to adjacent normal tissues. The RBM15/METTL3 complex mediates the m6A modification of ENO1 mRNA, and the oncogene ENO1 stimulates the growth of BCa cells. YTHDF1 recognises this m6A modification and accelerates the effectiveness of ENO1 protein translation, thus promoting the progression of BCa [186]. Binding of WTAP to circ0008399 speeds up the establishment of the m6A methyltransferase complex, improving circ0008399’s mRNA stability and triggering the expression of TNFα-inducible protein 3 (TNFAIP3). TNFAIP3 overexpression decreases BCa’s chemosensitivity [187]. BCa invasion in vitro and metastasis in vivo were facilitated by the upregulation of 7-dehydrocholesterol reductase (DHCR7) in BCa tissues. Decreased mRNA degradation due to YTHDF2-mediated increased DHCR7 in BCa induced the cAMP/protein kinase A/FAK pathway, greatly enhancing the invasive and metastatic capability of BCa, leading to poor prognosis [188]. Notably, the m6A eraser FTO functions as an oncogenic factor in BCa, as well as m6A writers. It has been shown that FTO catalyses the demethylation of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), regulates microRNA miR-384 and MAL T-cell differentiation protein 2 (MAL2) expression, and ultimately promotes the progression of BCa [189]. Thus, whether some crosstalks between m6A erasers and writers occur in BCa, causing co-carcinogenesis, remains unclear.
Prostate cancer
PCa is the second most prevalent malignant tumour, which can become more challenging to treat after a few years if it develops into CRPC. The dysregulation of m6A modification is strongly linked to PCa progression. SLC12A5, a neuron-specific potassium chloride co-transporter, can form a complex with YTHDC1 and then upregulate the TF HOXB13 to promote PCa progression [190]. In CRPC, YTHDF3 expression is upregulated and is associated with poor survival and high-grade metastasis. YTHDF3 promotes the RNA degradation of SPOP and NXK3.1 but stabilises the RNA expression of m6A-dependent TWIST1 and SNAI2, ultimately promoting the proliferation of CRPC cells [191]. The m6A-modified circRBM33 could bind to FMR1 and form a binary complex. The knockdown of METTL3 disrupted the formation of the complex. The circRBM33 in the complex can stimulate PCa cells’ mitochondrial respiration, while FMR1 can regulate cellular respiration. When these two work together, they can accelerate the proliferation of PCa cells and promote invasion [192]. FTO-IT1 is a lncRNA that is negatively associated with patient survival and is elevated in anti-androgenic and chemotherapy-resistant PCa. FTO-IT1 can directly interact with RMB15 to hinder the m6A methyltransferase complex, which subsequently decreases the mRNA m6A levels and stability of a subset of p53 target genes, ultimately resulting in increased cancer cell growth [193]. Besides, FTO can regulate the EMT in PCa cells, which is linked to bone metastasis [194].
Breast cancer
BC is the most prevalent carcinoma and the leading cause of cancer-related mortality in women. m6A regulators are essential in the development and treatment of BC. LAST1 is identified as an oncogene, with its m6A modification mediated by METTL3. YTHDF2 recognises the m6A modification in LAST1 mRNA, resulting in reduced stability. By increasing LATS1 mRNA expression and activating YAP/TAZ in the Hippo pathway, the knockdown of METTL3 or overexpression of YTHDF2 can prevent the onset of BC [195]. ALKBH5 expression was increased in BC patients and positively correlated with poor prognosis and chemotherapy resistance. ALKBH5 promotes m6A demethylation of FOXO1 mRNA and enhances the stability of FOXO1 mRNA in DOX-resistant BC cells, increasing the resistance of BC cells to chemotherapy [196]. Cell cycle protein-dependent kinase 6 (CDK6) is a direct target of IGF2BP2 in BC cells. IGF2BP2 directly regulates CDK6 in an m6A-dependent manner to accelerate BC progression [197]. Knockdown of HnRNPA2B1 or inhibition of m6A-tagged mRNA export can cause maintenance of nuclear m6A-tagged mRNAs linked to stemness maintenance. This would inhibit BC cell self-renewal and effectively improve the efficacy of DOX treatment [198]. Consequently, inhibition of m6A-tagged mRNA and its nuclear export may be a prospective therapeutic approach.
Ovarian cancer
Ovarian cancer (OC) is one of the three most common gynaecologic malignancies, and patients often have poor prognosis due to the lack of effective screening strategies. m6A modification has an impact on OC’s development, migration, invasion, and drug resistance. m6A can modify circPLPP4 and increase RNA stability, resulting in elevated circPLPP4 in OC cells. CircPLPP4 can act as a microRNA sponge to sequester miR-136, which further competitively up-regulates the expression of phosphatidylinositol-3-kinase regulatory subunit 1 (PIK3R1), leading to an increase in drug resistance in OC cells. Targeting m6A-modified circPLPP4 may be a prospective therapeutic target for OC-resistant patients [199]. Integrin subunit β1 (ITGB1) is a downstream target of ALKBH5 regulation. High expression of ALKBH5 demethylates m6A modifications in ITGB1 mRNA, which in turn prevents YTHDF2 from mediating the m6A-dependent degradation of this mRNA. This results in increased ITGB1 expression and enhanced phosphorylation of focal adhesion kinase (FAK) and Src proto-oncogene, ultimately promoting lymph node metastasis of OC [200]. Moreover, follicle stimulating hormone (FSH) was also able to promote epithelial-mesenchymal transition in OC cells, leading to OC metastasis in an m6A-dependent manner [201]. YTHDC1 is an oncogene in OC progression, and overexpression of YTHDC1 hindered OC advancement both in vivo and in vitro, but YTHDC1 expression was down-regulated in OC patients [202]. More investigation is required to explore whether YTHDC1 could be a novel therapeutic avenue for OC.
Cervical cancer
Cervical cancer (CC) is the fourth most common gynaecological tumour worldwide and is primarily induced by persistent infection with high-risk human papillomavirus (HPV). Drug resistance, metastasis, and CC cell proliferation are all closely related to m6A modifications. It has been found that, except for ALKBH5, whose expression is down-regulated in CC and exerts an anti-tumour effect, all other m6A modifiers are up-regulated in expression and exert an oncogenic effect. CircCCDC134, a cyclic RNA with m6A modification, stimulates CC cell growth and invasion both in vivo and in vitro. ALKBH5 has the capacity to mediate the demethylation of circCCDC134 to decrease its stability, while YTHDF2 recognises the m6A on circCCDC134 in an m6A-dependent manner and improves its stability. As ALKBH5 expression was reduced in CC and YTHDF2 was increased in CC, they synergistically enhanced circCCDC134 levels and accelerated CC progression [203]. HNRNPC significantly decreased the patients’ overall survival by recognising the m6A site on FOXM1 RNA (oncogene) in an m6A-dependent manner and controlling the isoform conversion of FOXM1 to encourage lymph node metastasis in CC [204]. Expression of the CC-derived exosome Mortalin was increased in plasma exosomes isolated from CC patients. m6A methylation in the 3’UTR of METTL3-catalysed HSPA9 mRNA conferred higher mRNA stability and enhanced translational efficiency to Mortalin [205]. Overexpression of Mortalin, in turn, facilitated proliferation, migration, and invasion of CC cells.
Endometrial cancer
Endometrial cancer (EC) is a highly prevalent gynaecologic carcinoma with increasing morbidity and mortality worldwide. m6A modifications can interact with circRNAs to influence the growth, invasion, and migration of EC cells. CircCHD7 connects with IGF2BP2 in an m6A-dependent manner by increasing mRNA stability and increases the expression of the downstream target gene, platelet-derived growth factor receptor β (PDGFRB), which ultimately enhances EC cell proliferation and hampers apoptosis [206]. Reduced levels of m6A modification and WTAP in EC patients correlate with the stemness of endometrial cancer stem cells (ECSC). The discovery of the WTAP/EGR1/PTEN axis offers a novel therapeutic strategy for EC [207]. Low METTL3 levels were related to poor prognosis in EC patients, while METTL3 expression was positively correlated with CD8 levels. Increased METTL3 in patients raises the ratio of CD8 + T cells and impedes EC progression, whereas METTL3 depletion has the opposite effect. The NLR family CARD structural domain containing 5 (NLRC5) is a target for METTL3-mediated m6A modification, which prevents its degradation through a YTHDF2-dependent mechanism. Elevated METTL3 levels enhance immunosurveillance and reduce EC immune evasion, suggesting that METTL3 and YTHDF2 could serve as diagnostic and prognostic factors for EC [208].
Other cancers
There is a connection between m6A modification and tumourigenesis in major systems of the body. Here, we present an overview of other cancers that have been comparatively less studied in relation to m6A. Research has revealed that m6A modifications on mRNAs and regulators of m6A are closely associated with the progression of various cancers. For example, in OSCC, METTL14 is involved in OSCC cell growth and metastasis by regulating the m6A level of CALD1 mRNA [209]. Additionally, METTL14 mediates m6A modification on MN1 mRNA to promote osteosarcoma progression and drug resistance [210], m6A modification on RUNX2 mRNA, mediated by METTL14, prompts migration and invasion of melanoma cells [211]. In pituitary neuroendocrine tumours, elevated levels of FTO cause a decline in the m6A level of DSP mRNA, which increases resistance to tumour therapy [212]. In nasopharyngeal carcinoma (NPC), WTAP mediates m6A modification of the lncRNA DIAPH1-AS1, enhancing its stability and promoting NPC metastasis [213]. In thyroid cancer and lymphoma, m6A-related lncRNAs (LINC02471, DOCK9-DT) can be useful biomarkers [214, 215]. Additionally, m6A modifications in various RNA types across different cancers can lead to distinct forms of tumour resistance. In laryngeal cancer, HOXA10-AS sequesters miR-29b-3p to function as a competitive endogenous RNA, enhancing tumour cell oxidative resistance and accelerating laryngeal cancer progression [216]. In hypopharyngeal squamous cell carcinoma, METTL3 mediates the m6A modification of circCUX1 to increase its expression and enhance cancer cell resistance to radiotherapy [217]. In seminoma, METTL3 acts as a TF-activated enhancer binding protein 2C mRNA (TFAP2C) m6A writer, increasing the stability of TFAP2C mRNA, which subsequently causes enhanced DNA repair-related gene expression, including WEE1 and BRCA1, ultimately resulting in cancer cells becoming resistant to cisplatin chemotherapy [218].
m6A modification exhibits dual regulatory roles in cancer phenotypes. First, m6A can drive pro-tumorigenic effects by activating key oncogenic pathways, promoting immune evasion, and reprogramming metabolic processes. For instance, m6A methylation of EphA2 and VEGFA enhances vasculogenic mimicry in CRC through activation of the PI3K/AKT and ERK1/2 signalling pathways [219]. Additionally, METTL14 modulates mitochondrial metabolism-related genes to enhance tumour cell energy acquisition [220]. Conversely, m6A modifications can exert anti-tumorigenic effects by suppressing malignant transformation and enhancing chemosensitivity. For example, m6A modification-mediated lncRNA TP53TG1 inhibits GC progression by regulating CIP2A stability [221]. The m6A modification of TGFB2 triggers a reconfiguration of lipid metabolism, which subsequently contributes to the development of gemcitabine resistance in pancreatic ductal adenocarcinoma (PDAC) [222]. Notably, the same regulatory factors may display opposing roles across cancer types. FTO promotes oncogene expression in AML but suppresses tumour progression in gliomas, a divergence potentially linked to tissue-specific signalling pathways or microenvironmental interactions [223, 224]. In chemotherapy resistance, m6A modifications may either promote drug resistance by regulating resistance-associated genes, such as METTL3 upregulation, which induces cisplatin resistance in OC [225]. In immunotherapy, m6A modifications regulate immune checkpoints by modulating PD-1/PD-L1 expression. YTHDF1 enhances PD-L1 mRNA translation to facilitate immune evasion in PCa [226]. Furthermore, ALKBH5 deficiency promotes M1-polarised macrophage infiltration, improving immunotherapy outcomes [155]. The dual roles of m6A modifications and their microenvironment-dependent effects underscore the need for personalised intervention strategies in cancer therapy.
Strategies targeting m6A mRNA can be categorised as follows: (1) targeting m6A regulatory factors. For instance, YTHDF2 has been shown to effectively regulate leukaemia cell proliferation [227], while targeting METTL3 holds potential efficacy in improving gemcitabine therapy for PC [228]. (2) Utilising RCas9 technology to modify m6A sites, enabling precise alteration of specific m6A loci [229]. (3) Combination therapies integrating conventional tumour treatments with novel epigenetic drugs. m6A mRNA regulators have been implicated in tumour radioresistance and demonstrated significant clinical utility when combined with kinase inhibitors (KIs), 5-fluorouracil (5-FU), gemcitabine, anthracyclines, and cisplatin [230]. Moreover, the integration of m6A mRNA regulation with immune checkpoint blockade has been recognised as a promising therapeutic approach for the treatment of malignancies [231].
Currently, no FDA-approved m6A-targeted anticancer drugs are available. The METTL3 inhibitor STC-15 is undergoing Phase I clinical trials (NCT05584111) for various malignancies, including lung, hepatic, bone, colorectal, cutaneous, breast cancers, and malignant melanoma [232]. CCS1 (Bisantrene), a specific FTO-targeting agent that inhibits m6A demethylase activity, has entered Phase II clinical trials (NCT04989335) in combination with fludarabine and clofarabine for relapsed or refractory AML [233]. These developments indicate that clinical trials of m6A-targeted therapies remain in their preliminary stages.
Several challenges and potential risks persist in developing m6A-targeted cancer therapeutics: firstly, current pharmacological strategies demonstrate insufficient target discrimination. Small-molecule inhibitors targeting m6A-associated enzymes might interact with other structurally similar enzymes or molecular pathways, leading to unintended consequences. Antisense oligonucleotide (ASO) approaches carry inherent risks of sequence-dependent off-target binding, which could inadvertently disrupt RNA processing mechanisms and compromise transcriptomic fidelity. Secondly, differential m6A modification patterns between tumour and normal tissues may result in unexpected toxicities. Additionally, dynamic changes in m6A modification during disease progression and tumour microenvironment evolution complicate therapeutic outcome prediction. Thirdly, potential side effects such as immunosuppression or hepatotoxicity require careful evaluation. Most current research remains at the preclinical stage, presenting significant challenges in clinical translation. Overall, while m6A targeting shows therapeutic promise, overcoming these obstacles necessitates further research to develop more specific, efficient, and safer therapeutic strategies.
m6A modification of enhancer RNAs
The revelation that a large number of eRNAs are produced within the human genome has transformed our perception of enhancers, indicating that they can act not just as DNA but also as regulatory RNAs [235]. Pathological samples show a significant production of eRNAs, and mutations linked to diseases often coincide with enhancers that display eRNAs transcription, underscoring the potential involvement of enhancer transcription or eRNAs transcripts in disease development [236].
Among the more than 170 types of cellular RNA modifications, N6-adenosine methylation (m6A) stands out as the most common and important post-transcriptional modification in eukaryotic cells [237]. Recent findings indicate that m6A modification is widespread in eRNAs and may affect their stability, localisation, and function [101, 238]. However, the molecular function of m6A has mainly been explored in more abundant mRNA and post-transcriptional regulatory processes, leaving the role of m6A-modified eRNAs in transcriptional regulation largely unexplored. Additional research is needed to investigate the impact of other post-transcriptional modifications on eRNAs.
m6A modification of enhancer RNAs detection
eRNAs are generated during the transcription process and are known to be unstable [239]. Most eRNAs have half-lives of under 10 min, making them difficult to capture in steady-state RNA samples [240]. Therefore, more sophisticated and sensitive sequencing technologies are necessary to accurately analyse m6A modifications on eRNAs. Recent advancements in m6A sequencing technology offer new methods to explore the post-transcriptional regulation of eRNAs. Detection techniques can be categorised into two types: antibody-dependent and antibody-independent (Table 4). Despite several improvements in this technology in recent years, including the availability of commercial m6A MeRIP kits, the localisation of m6A is less precise, typically spanning peak regions of about 50–200 base pairs that may include multiple DRAC motifs, and there is a risk of false-positive enrichment in areas of gene alternative splicing [241]. Furthermore, the need for several dozen to hundreds of micrograms (µg) of total RNA limits the application of m6A epitranscriptome profiling in clinical contexts, as most patient samples yield only a few micrograms of total RNA. Therefore, it is crucial to develop an m6A profiling method suitable for low-input RNA samples to better understand m6A dynamics in various diseases.
Table 4.
The advantages and limitations of m6A detection methods
| m6A detection methods | Advantages | Limitations | References | |
|---|---|---|---|---|
| Anti-body dependent | MeRIP-seq |
1.Simple to perform 2.Good universality and wide applicability |
1.False-positive 2.False-negative 3.Limited resolution |
[248] |
| m6A-seq2 |
1.Merge samples for IP 2.Reduce the technical variability |
1.False-positive | [249] | |
| miCLIP-seq | 1.Single-base resolution | 1.Unable to quantify and locate | [250] | |
| MINT-Seq | 1.Recognize m6A on nascent RNA | 1.False-positive | [242] | |
| Anti-body independent | m6A-SEAL-seq | 1.High sensitivity and specificity |
1.Tedious and time-consuming 2.Unable to achieve single-base resolution |
[251] |
| m6A-SAC-seq |
1.Single-base resolution 2.Test of small samples |
1.Based on an enzyme 2.Sequence perference |
[252] | |
| Nanopore direct RNA sequencing | 1.Single-base resolution |
1.High cost and low accuracy 2.Require high RNA quality and quantity |
[253] | |
| GLORI-seq | 1.Absolute quantification of single-base resolution |
1. High cost 2. The calculation complexity is high |
[254] | |
| DART-seq | 1.High sensitivity | 1.Low accuracy | [255] | |
Recently, researchers have employed Methylation Inscribed Nascent Transcript Sequencing (MINT-Seq), a new sequencing technique that effectively enriches and quantifies m6A distribution on nascent RNA, leading to the identification of several novel m6A sites on introns [242]. Additionally, a recent study introduced a method for identifying m6A sites across the transcriptome in single cells, uncovering unexpected variability in the presence and abundance of m6A sites among individual cells and highlighting differentially methylated mRNAs throughout the cell cycle [243]. Furthermore, researchers have created CRISPR/dCas13 (Rx)-mediated m6A editors by combining them with RNA methyltransferase GhMTA (TME) or demethylase GhALKBH10 (TDE) to induce specific m6A site demethylation or methylation. This demonstrates that these m6A editors can be used to investigate the effects of specific m6A modifications [244]. Although high-throughput sequencing technologies have facilitated the swift accumulation of data regarding numerous m6A sites, these methodologies remain susceptible to generating false-positive or false-negative outcomes due to inconsistencies in the sequencing process or technical complications. Consequently, it is imperative to seek more reliable approaches for the detection of m6A modifications.
Most eRNAs degrade so rapidly that they are challenging to detect, and current sequencing techniques struggle to capture their entire length. However, recent studies utilising sequencing-based nascent RNA methods have introduced effective tools for identifying RNA synthesis and unstable transcripts, such as eRNAs. Techniques like NET-seq, GRO-seq, and TT-seq have emerged [240, 245–247]. These nascent RNA sequencing approaches have opened new possibilities for capturing eRNAs sequences. In the future, researchers may integrate these nascent RNA sequencing techniques with m6A detection methods to address the rapid degradation of eRNAs, thereby enabling more precise identification of m6A modifications on eRNAs.
m6A modification of enhancer RNAs in cancer
In 2019, researchers employed methylated RNA immunoprecipitation sequencing (MeRIP-Seq) to profile the m6A landscape across the entire transcriptome in 21 human fetal tissue samples, identifying 1,908 enhancer-derived lncRNAs containing m6A modifications [256]. Notably, the pattern of m6A deposition on eRNAs closely resembled that observed in mRNAs, both conforming to the conserved RRACH motif [257]. Since this discovery, successive researches have discovered the impact of m6A modifications on eRNAs stability. For instance, in mouse embryonic stem cells (mESCs), m6A methylation was investigated to promote eRNAs degradation. Knockdown of METTL3 or YTHDC1 contributed to reduced m6A levels on eRNAs as well as a subsequent increase in eRNAs abundance. Mechanistically, METTL3 functions as an m6A writer, catalyzing m6A modifications on eRNAs, while YTHDC1, an m6A reader, facilitates eRNAs decay through nuclear exosome-mediated degradation. Consequently, depletion of either METTL3 or YTHDC1 leads to eRNAs stabilization. Additionally, stabilized eRNAs actively recruit TFs, such as YY1 and CBP/EP300, to maintain an open chromatin state while simultaneously displacing repressive complexes like PRC2. This process ultimately enhances chromatin accessibility and activates the transcription of downstream genes in an m6A-dependent manner, inducing a positive feedback loop [258]. These findings illustrate that m6A modifications act as a regulatory switch controlling eRNAs abundance, stability, and chromatin dynamics, thereby influencing gene transcription. Conversely, an independent discovery in BC cell line MCF-7 reported the opposite effect—m6A modifications were found to protect eRNAs from degradation. In METTL3-knockdown MCF-7 cells, eRNAs production were significantly reduced, and premature termination of transcription was observed due to enhanced recruitment of the Integrator complex, which mediates RNA cleavage. Interestingly, loss of METTL3 caused a marked enhancement in the nucleic acid endonuclease subunit (INTS11) at enhancer regions. However, there is a lack of clarity about how Integrator is recruited to target genes and how its nucleic acid endonuclease activity is regulated. Mechanistically, METTL3-mediated m6A modifications appeared to impair Integrator recruitment by stabilizing RNA polymerase II (Pol II). Additionally, m6A modifications facilitated the recruitment of hnRNPG and YTHDC1, protecting eRNAs from endonucleolytic cleavage [259]. Thus, these findings suggest that m6A modifications, along with m6A readers, synergistically enhance eRNAs stability in certain cellular contexts. However, the strength of antagonism between METTL3 and Integrator may not fully explain the ability of METTL3 to positively regulate transcription. Thus, the m6A deposited by METTL3 on nascent eRNAs may also play other regulatory roles in transcription and/or post-transcription. In summary, the conflicting conclusions drawn from these two studies are likely attributed to differences in cellular environments—mESCs versus MCF-7 cells—highlighting the context-dependent nature of m6A regulation. This discrepancy emphasizes the demand for further research to elucidate the precise impact of m6A modifications on eRNAs stability across different cell types. Notably, it is well validated that m6A has a pivotal impact on promoting mRNA degradation in cytoplasmic lysates. However, its effects on various RNA species appear to vary relying on the stage of the RNA life cycle or cellular context. Furthermore, a recent study challenged the long-standing notion that m6A modifications primarily accelerate mRNA degradation in coding regions. Instead, the study demonstrated that m6A modifications in the 3’UTR stabilize mRNAs more efficiently than those in coding regions [260]. This suggests that m6A modifications exert differential regulatory effects depending on their position within the transcript. However, whether this principle extends to eRNAs remains to be determined and requires further investigation.
In addition to regulating eRNAs stability, m6A modifications have also been implicated in cancer development, progression, invasion, and metastasis. For instance, in breast cancer MCF-7 cells, m6A modifications are widely distributed on eRNAs, and eRNAs harboring m6A tags tend to be longer, more stable, and highly cell-type specific. These modifications serve as essential recognition sites for the m6A reader protein YTHDC1, which directly activates enhancers and induces eRNAs transcription. Furthermore, YTHDC1 binding to eRNAs facilitates chromatin condensation, engages in crosstalk with transcriptional coactivators (BRD4), and enhances enhancer activity, ultimately promoting oncogene expression and breast cancer progression. Thus, m6A modifications on eRNAs play a coordinated role with reader proteins in enhancer activation and gene transcriptional regulation, extending their influence beyond eRNAs function alone [101]. Interestingly, while previous findings in mESCs indicated that YTHDC1 depletion increased eRNAs stability, this study in MCF-7 cells reported that YTHDC1 depletion did not affect eRNAs stability but rather hampered enhancer function and gene transcription. This distinction may be attributed to differences in cell types or variations in experimental methodologies, though the precise underlying mechanism remains unclear [258]. A similar regulatory role of m6A-modified eRNAs has been observed in PDAC. Super-enhancer-associated RNAs (seRNAs) harboring m6A modifications were found to be longer and enriched with RRACH motifs compared to their non-m6A-modified counterparts. Notably, chromatin regions associated with highly m6A-modified seRNAs exhibited significantly increased H3K4me3 levels. Mechanistically, cofilin-1 (CFL1), a filament-severing protein, was established as a cofactor of METTL3 that interacts with its zinc finger domain, promoting METTL3 nuclear co-localization and facilitating localized chromatin modifications that drive oncogene expression. Additionally, YTHDC2, an m6A reader of seRNAs, was found to interact with the histone methyltransferase mixed-lineage leukemia protein 1 (MLL1/KMT2A) to cooperatively enhance localized H3K4me3 deposition and promote PDAC progression. Notably, silencing METTL3 or YTHDC2 significantly suppressed the malignant phenotype of PDAC. These findings suggest that m6A modifications of seRNAs, mediated by METTL3 and CFL1, are identified by YTHDC2, which subsequently employs MLL1 to enhance chromatin accessibility and drive oncogene transcription [261, 262]. Thus, the CFL1-YTHDC2-MLL1 axis plays an important role in the epigenetic regulation of chromatin remodeling during PDAC development. In uterine corpus endometrial cancer (UCEC), IGFBP7 antisense RNA1 (IGFBP7-AS1) functions as a critical eRNA regulating IGFBP7, a member of the IGFBP gene family. IGFBP7-AS1 and IGFBP7 were substantially lower in UCEC tissues than in normal tissues, and reduced IGFBP7-AS1 expression was linked to poor clinical outcomes. Further analysis disclosed a link between IGFBP7-AS1 expression and key m6A regulators. In addition to FTO, declined expression of METTL14, WTAP, RBM15, YTHDF1, YTHDF2, YTHDC1, YTHDC2, and HNRNPC led to an increase in IGFBP7-AS1 expression. Notably, patients with high IGFBP7-AS1 expression exhibited elevated basophil and immune cell infiltration, as well as improved responses to immunotherapy. Mechanistically, IGFBP7-AS1 may improve the tumor immune microenvironment and affect immune lymphocytes such as T cells, mast cells, macrophages M1 and M2, thereby inducing protective immunity in UCEC. These findings suggest that increased m6A modifications on IGFBP7-AS1 lead to its downregulation, contributing to poorer clinical outcomes in UCEC patients. However, the association between m6A modifications and eRNAs expression levels is unclear, perhaps by affecting the stability of eRNAs. In UCEC, m6A affects eRNAs stability possibly by recruiting m6A reading proteins to regulate degradation or stabilization, or by disrupting the local secondary structure of eRNAs to expose or hide nuclease cleavage sites, or by regulating the post-transcriptional processing of eRNAs, which no study has yet been able to demonstrate [263]. Surprisingly, a recent study revealed that enhancer-driven eRNAs can indirectly prevent m6A site recognition on mRNAs, thereby accelerating HCC progression. Super-enhancer (SE) profiling in HCC cell lines identified a novel lncRNA, LINC01089, which was markedly elevated in HCC tissues and in relation to poor patient prognosis. While LINC01089 overexpression had no influence on HCC cell growth, it considerably boosted EMT, migration, invasion, and metastasis. Mechanistically, LINC01089 was found to interact with the RRM1 domain (aa71-149) of hnRNPM, promoting exon 3 skipping in DIAPH3 mRNA during alternative splicing. Exon 3 of DIAPH3 harbors an m6A site recognized by IGF2BP3, which enhances DIAPH3 mRNA stability. Consequently, exon 3 skipping leads to decreased DIAPH3 mRNA stability, resulting in elevated levels of phosphorylated ERK (pERK), phosphorylated Elk1 (pElk1), and Snail, thereby driving HCC metastasis. The depletion of LINC01089 resulted in elevated levels of DIAPH3 protein, suppressed the ERK/Elk1/Snail signaling pathway, and ultimately impeded the process of epithelial-mesenchymal transition in HCC cells [264].
In addition to their role in cancer progression, m6A-modified eRNAs have also been implicated in cancer drug resistance and hold potential as clinical biomarkers. One notable example is SUCLG2-AS1, a super-enhancer (SE)-derived lncRNA that is highly expressed in NPC tissues. This lncRNA harbors 14 highly conserved m6A modification sites, and its elevated expression has been correlated with poorer prognosis and reduced survival rates in NPC patients. Mechanistically, the m6A reader IGF2BP3 identifies the METTL3-mediated m6A modifications on SUCLG2-AS1, preventing its degradation and thereby enhancing its stability. Additionally, knockdown of SUCLG2-AS1 significantly reduced SOX2 protein levels in NPC cells. Furthermore, CTCF (CCCTC-binding factor), a multifunctional TF, was found to be recruited to the SOX2 promoter and enhancer regions by SUCLG2-AS1. Through long-range chromatin looping, SUCLG2-AS1 facilitated SOX2 transcriptional activation, leading to SOX2 accumulation and subsequent NPC metastasis and radioresistance. These findings suggest that m6A-modified and stabilized SUCLG2-AS1 enhances SOX2 transcription by recruiting CTCF, thereby regulating the interaction between the super-enhancer and SOX2 promoter regions. Given its oncogenic role, targeting the m6A modification sites on SUCLG2-AS1 to reduce its stability may provide a novel therapeutic method for NPC [265]. In bone-metastatic prostate cancer (mPCa), MLXIPe has been identified as a tumor-specific eRNA modified by m6A. m6A levels of MLXIPe were considerably elevated in mPCa tissues compared to primary PCa and adjacent non-tumor tissues. Importantly, increased MLXIPe m6A levels were linked to poorer survival in mPCa patients, and m6A-modified MLXIPe was specifically enriched in bone-metastatic lesions. Further analysis revealed that PSMD9, a gene linked to mPCa progression, was a downstream target of MLXIPe. Notably, PSMD9 expression was correlated with lower recurrence-free survival following radiotherapy, and its 5’UTR contained m6Am modifications. Mechanistically, KHSRP, an RNA-binding protein that serves as a dual reader of m6A and m6Am, was found to regulate both MLXIPe and PSMD9. Specifically, KHSRP utilized its KH1/2 domain to recognize m6Am on PSMD9 mRNA and its KH3/4 domain to recognize m6A modifications on MLXIPe. This dual recognition mechanism stabilized both MLXIPe and PSMD9 transcripts. Knockdown of XRN2, a highly active 5’ > 3’ ribonuclease, led to increased PSMD9 mRNA stability. Additionally, MLXIPe was shown to prevent XRN2-mediated degradation of PSMD9 mRNA via m6A modification and the KHSRP complex, ultimately leading to increased PSMD9 expression and enhanced production of DNA repair proteins. As a result, m6A-modified MLXIPe contributed to radiotherapy resistance in mPCa cells. Silencing the m6A sites on MLXIPe markedly reduced radiotherapy resistance, thereby restoring the sensitivity of mPCa cells to radiotherapy. These findings underscore the potential of targeting m6A modifications on MLXIPe as a strategy for overcoming radiotherapy resistance in mPCa [238]. Another study identified five m6A-associated eRNAs that were significantly connected with prognosis in patients with head and neck squamous cell carcinoma (HNSCC). These eRNAs were utilized to establish a prognostic risk model according to their expression and m6A modification status. The model included the following five eRNAs: AC005562.1, COPDA1, LINC00271, MIAT, and MIR4435-2HG. Functional enrichment analysis suggested that the m6A-associated eRNAs risk model was involved in tumor progression and immune-related biological pathways. Notably, the expression levels of COPDA1 and MIAT were positively correlated with the infiltration rates of naïve B cells, CD8 + T cells, and activated CD4 + memory T cells, whereas their expression was inversely correlated with MIR4435-2HG levels. Compared to normal tissues, MIAT and MIR4435-2HG were significantly upregulated in HNSCC. Interestingly, while high MIAT expression was in association with enhanced tumor immune cell infiltration and improved anti-tumor immunity, resulting in a better prognosis, elevated MIR4435-2HG expression was associated with worsened disease progression and increased tumor aggressiveness. Further functional assays demonstrated that silencing MIR4435-2HG suppressed cancer cell migration and invasion, further supporting its oncogenic role in HNSCC. However, the roles of the remaining three eRNAs (AC005562.1, COPDA1, and LINC00271) in HNSCC remain unclear and require further investigation. Taken together, these findings indicate that the 5m6A-associated eRNAs risk model has potential as a predictive tool for patient survival and response to immunotherapy. By leveraging this risk model, clinicians may be able to identify high-risk HNSCC patients and develop personalized immunotherapy or targeted therapy strategies based on their m6A-associated eRNA profiles [266] (Fig. 3).
Fig. 3.
The role of eRNAs m6A in cancer. The regulators of eRNAs m6A are engaged in the progression of numerous cancers, including BC, PDAC, mPCa, NPC, HNSCC and UCEC
In clinical diagnostics, eRNAs m6A modifications demonstrate unique advantages over traditional biomarkers, including high sensitivity, robust specificity, and multi-omics correlations. The development of a “liquid biopsy 2.0” system—integrating Visium HD spatial transcriptomics, eRNA m6A profiling, cfDNA methylation, protein biomarkers (e.g., PD-L1), and nanopore sequencing—holds significant potential. Within this framework, deep neural networks (DNNs, e.g., Transformer models) can be trained to automatically decipher modification signatures in nanopore sequencing signals for AI-assisted interpretation, while rapid detection of cell-free RNA may be achieved through microfluidic chips and CRISPR-Cas13a systems. For future therapeutic strategies targeting eRNAs m6A modifications, several directions hold promise: firstly, small-molecule inhibitors could be designed by targeting the m6A writer complex or blocking reader protein functions, such as developing METTL3 allosteric inhibitors and YTHDF1/2-specific antagonistic peptides. Secondly, RNA-targeted therapies may include precision editing of m6A modification patterns to reduce off-target effects. Thirdly, combination therapeutic strategies, such as PARP inhibitors combined with METTL3 inhibitors or immune checkpoint inhibitors paired with YTHDF2 antagonists, warrant exploration. In summary, eRNAs m6A modifications serve as a critical nexus linking epigenetic regulation and transcriptional programmes, offering novel perspectives for cancer diagnosis and treatment. It is anticipated that through the integration of multi-omics approaches and cross-disciplinary applications of nanotechnology, targeting eRNAs m6A modifications will enable a comprehensive clinical workflow—from early screening to targeted intervention—ultimately improving patient survival outcomes and advancing the field of precision oncology.
Conclusions and future directions
Despite accounting for approximately 98% of the genome, the precise function of enhancers—one of the most significant classes of non-coding regulatory factors—remains largely elusive. Within the last couple of years, advances in high-throughput sequencing technologies have given deeper perception into the mechanisms by which enhancers drive cell-type-specific gene expression. However, these discoveries have simultaneously led to a more fluid and nuanced definition of enhancers. The identification of hundreds of thousands of enhancer-like regulatory elements that function through mechanisms distinct from classical enhancers has prompted a reconsideration of how enhancers should be defined. As a cutting-edge technological approach, crispr-based editing has been applied to identify novel enhancer elements, but challenges remain in applications across the genome and in multiple cell types. Perhaps in the future it will be possible to combine crispr-based editing with single-cell sequencing to reveal more in-depth functional mechanisms of enhancers or enhancer RNAs [267]. Moving forward, the refinement and integration of enhancer identification methodologies will be essential to distinguishing enhancer-like elements from classical enhancers. A more comprehensive understanding of their molecular mechanisms will not only contribute to redefining enhancers but also enhance our ability to interpret their regulatory roles. Furthermore, the mechanisms governing enhancer-promoter interactions and the precise processes by which enhancers activate transcription remain largely unresolved. In addition, enhancer dysregulation has emerged as a critical area of focus in cancer research. Structural variations, epigenetic modifications, and alterations in enhancer activity have been implicated in oncogene overexpression and tumor suppressor gene inactivation. However, the full repertoire of enhancers involved in tumorigenesis has yet to be elucidated. Key questions persist regarding the regulatory influence of known enhancers on their target genes, the extent of shared and distinct biological effects of enhancer dysregulation across different tumor types, and the feasibility of targeting enhancers for therapeutic intervention. Addressing these challenges will be essential for leveraging enhancer biology in cancer diagnosis and treatment.
An accumulating number of researches has underlined the pivotal impact of m6A RNA modification on cancer. Although m6A-related studies are still in their early stages, multiple studies strongly demonstrate that dysregulation of m6A regulators is closely linked to key oncogenic processes, maintaining cancer cell proliferation, invasion, metastasis, and drug resistance. Interestingly, some m6A regulators display dual roles, acting as both oncogenes and tumor suppressors in different cancer types. For instance, METTL3 promotes tumorigenesis in LC, ESCA, GC, HCC, CRC, PCa, BCa, BC, OC, CC, laryngeal cancer, hypopharyngeal squamous cell carcinoma, and seminoma, yet exerts tumor-suppressive effects in EC. Similarly, FTO has been investigated to enhance tumor advancement in BCa and pituitary neuroendocrine tumors while playing a tumor-suppressive role in LC and PCa. More strikingly, certain regulators display dual roles within the same cancer type. ALKBH5, for example, acts as both an oncogene and tumor suppressor in LC, GC, and PC, while FTO exhibits similar paradoxical effects in HCC. Beyond their roles in tumorigenesis, m6A modifications also mediate various forms of therapy resistance. For instance, METTL3 has been implicated in oxidative stress resistance in laryngeal cancer, radioresistance in hypopharyngeal squamous cell carcinoma, and chemoresistance in seminoma. These observations raise several fundamental questions: Why can both m6A writers and erasers be dysregulated within the same cancer type, contributing to comparable oncogenic or tumor-suppressive effects? Why do the same regulatory factors exert opposing roles across different cancers? How can a single regulator display dual functions within the same tumor type? What determines the ability of an m6A regulator to mediate distinct forms of therapy resistance across different malignancies? The underlying molecular mechanisms remain unclear, potentially involving variations in the tumor microenvironment (TME) or complex epigenetic crosstalk. Further investigation is required to elucidate these mechanisms. Additionally, m6A regulators have demonstrated potential as biomarkers for cancer diagnosis, prognosis, and targeted therapy. METTL3, FTO, ALKBH5, and YTHDF1/2/3 have been identified as prognostic biomarkers for multiple cancers, while METTL3 and FTO also serve as diagnostic markers. YTHDF1/2, METTL17, ALKBH5, and YTHDC1 have emerged as promising therapeutic targets, with interfering RNA against YTHDF1 successfully synthesized to inhibit GC progression. However, the translation of such therapeutic strategies into clinical applications remains in its infancy, with significant gaps in preclinical and clinical validation. Given the extensive regulatory influence of m6A on gene expression, its therapeutic potential must be carefully assessed to determine possible side effects and clinical viability. Furthermore, our findings highlight an intricate interplay between m6A RNA modifications and other epigenetic modifications, including DNA methylation, histone modifications, and RNA demethylation. This interplay has been implicated in pivotal biological processes such as RNA splicing and decay, significantly impacting the progression of osteosarcoma, OC, BCa, and CRC. However, it remains unclear whether these interactions are cancer-type specific or represent a more universal mechanism. Additionally, whether m6A modifications engage in broader crosstalk with other DNA and histone modifications in cancer progression remains an open question, lacking systematic study. Future research should address these pressing questions, refining our understanding of m6A modifications and their broader implications in oncogenesis. By integrating advanced methodologies and exploring novel regulatory networks, we can unlock new dimensions of cancer epigenetics and pave the way for more effective diagnostic and therapeutic strategies.
The number of eRNAs in the human transcriptome is extraordinarily large, and their functions are largely dependent on enhancers. However, due to their inherent instability and low abundance, there is an urgent need to develop more sensitive sequencing technologies beyond GRO-seq, PRO-seq, and ChIP-seq to enable more precise identification of eRNAs. Advancements in this area will offer deeper perspectives into the functional roles of eRNAs. While eRNAs are known to enhance enhancer activity by facilitating the formation of enhancer-promoter loops, the mechanisms by which they regulate the expression of distal genes remain unclear. eRNAs have a dual impact on cancer, contributing to both oncogene activation and tumor suppressor regulation while also participating in signaling cascades. However, these represent only a fraction of the functional mechanisms underlying cancer-associated eRNAs, and further investigation is needed to illuminate additional regulatory pathways. Moreover, in oncogenic signaling pathways, eRNAs are not only subject to regulation but also act as key regulatory elements themselves. The precise molecular mechanisms governing these interactions remain largely undefined. Additionally, eRNAs have been implicated in immune evasion and therapeutic resistance and have demonstrated potential as biomarkers for cancer diagnosis, prognosis, and even therapeutic targeting. However, several significant challenges hinder their clinical application. The complexity of obtaining human tissue samples, the difficulty of in vivo functional validation, and the potential toxicity and limitations of eRNA-based therapies necessitate extensive foundational research before transitioning to clinical trials. Our findings indicate that m6A-modified eRNAs tend to be longer in certain cancer cells, but whether differences exist in the transcription and function of long versus short eRNAs remains an open question. Furthermore, we strongly suspect that m6A-mediated regulation of eRNAs stability is highly context-dependent. In breast cancer MCF7 cells, m6A modifications did not significantly alter eRNAs stability, whereas in NPC, m6A modifications increased eRNAs stability, and in mESCs, m6A modifications led to eRNAs destabilization. However, a definitive conclusion regarding the influence of different cellular environments on m6A-mediated eRNAs stability has yet to be reached. Another critical yet unresolved question is whether m6A modifications exert differential effects on eRNAs stability depending on their specific modification sites. Existing evidence suggests that m6A modifications predominantly influence eRNAs stability, causing the activation of cancer-related genes, which subsequently drives cancer cell proliferation, invasion, metastasis, and drug resistance. Additionally, eRNAs can interfere with the recognition of m6A sites in mRNAs, thereby altering mRNA stability and subsequently promoting cancer metastasis. To explore the therapeutic potential of m6A-modified eRNAs, future investigation should concentrate on characterizing their advanced molecular structures to assist the exploration of more precise targeting strategies. Although the influence of m6A modifications on eRNA-mediated carcinoma advancement is of significant biological importance, current research in this area remains limited, highlighting the need for further comprehensive investigations. Beyond m6A, other RNA modifications, such as m1A, m5C, and 5-hydroxymethylcytosine (5hmC), have also been reported in eRNAs. However, the involvement of these modifications in cancer remains poorly understood and will require extensive long-term studies.
In conclusion, an in-depth investigation into the m6A modifications of eRNAs has the potential to reshape our understanding of cancer biology. The intricate interplay between enhancers, eRNAs, and m6A modifications reveals a complex yet promising frontier in epigenetics. Future research should adopt innovative, multidimensional, and interdisciplinary approaches to explore this field further. We anticipate that these efforts will lead to the development of precise epigenetic-targeted cancer therapies in the near future.
Acknowledgements
Abbreviations
- eRNAs
Enhancer RNAs
- m6A
N6-methyladenosine
- TF
Transcription factor
- TFBs
Transcription factor binding sites
- Ig
Immunoglobulin
- H3K4me1
Histone H3 lysine 4
- H3K27ac
Histone H3 lysine 27
- 5mC
5-methylcytosine
- ncRNAs
Non-coding RNAs
- lncRNA
Long non-coding RNAs
- miRNAs
microRNAs
- polyA
3’polyadenylation
- GRO-seq
Global run-on sequencing
- PRO-seq
Precise nuclear run-on sequencing
- BRD4
Bromodomain-containing protein 4
- ERα
Estrogen receptor-α
- P-TEFb
Positive transcription elongation factor b
- CTD
C-terminal domain
- WDR82
WD repeat domain 82
- PP1
Phosphatase 1
- CBC
Cap binding complex
- NELF
Negative elongation factor
- IEG
Immediate early gene
- hnRNPL
Heterogeneous cytosolic nuclear ribonucleoprotein L
- GTFs
General transcription factors
- CRISPRi
CRISPR interference
- HRRs
Highly recurrent regions
- hmC
Hydroxymethylation of cytosine
- LSD1/KDM1A
Lysine-specific demethylase 1
- FOXD2
Forkhead box protein D2
- C/EBPα
Enhancer binding protein alpha
- ULBP2
UL16-binding protein 2
- C/EBPα-Cm
C/EBPα C-terminal mutant
- OSCC
Oral squamous cell carcinoma
- HNSCC
Head and neck squamous cell carcinoma
- IRS2e
IRS2-linked enhancer RNA
- SCC
Squamous cell carcinoma
- AKT/PI3K
Phosphatidylinositol 3-kinase
- MEK1/2
Mitogen-activated protein kinase kinase 1/2
- ERK1/2
Extracellular signal-regulated kinase 1/2
- TME
Tumor microenvironment
- IGH
Immunoglobulin heavy chain
- CESC
Cervical adenocarcinoma
- TNM
Tumor-lymph node-metastasis
- METTL3
Methyltransferase-like protein 3
- METTL14
Methyltransferase-like protein 14
- WTAP
Wilms tumor 1 associated protein
- SAM
S-adenosylmethionine
- RRACH
Recognizing specific RNA sequences
- METTL16
Methyltransferase-like protein 16
- METTL5
Methyltransferase-like protein 5
- ZCCHC4
Human methyltransferase
- RBM15
RNA-binding motif protein 15
- VIRMA/KIAA1429
Vir-like m6A methyltransferase-associated protein
- 3'UTR
3’ untranslated region
- Fe²⁺
Ferrous iron
- FTO
Fat mass and obesity-associated protein
- ALKBH5
Alkylation repair homolog 5
- HCC
Hepatocellular carcinoma
- EMT
Epithelial-mesenchymal transition
- HNRNP
Heterogeneous nuclear ribonucleoprotein
- ELAVL1
Embryonic lethal abnormal visual-like protein 1
- circRNA
Circular RNA
- Prrc2a
Proline-rich coiled-coil 2 A
- A-to-I
Adenosine-to-inosine
- ADARs
Adenosine deaminases acting on RNA
- NAT10
N-acetyltransferase 10
- ac4C
N4-acetylcytosine
- PFKM
Phosphofructokinase
- LDHA
Lactate dehydrogenase A
- ACOT7
Acyl-coenzyme A thioesterase 7
- DCP2
Decapping enzyme 2
- m7G
N7-methylguanosine
- PCIF1
Phosphorylated CTD-interacting factor 1
- TROP2
Trophoblast cell surface antigen 2
- COAD
Colon adenocarcinoma
- hnRNA
Heterogeneous nuclear RNA
- GPX4
Glutathione peroxidase 4
- m5C
5-methylcytosine
- NSUN2
NOP2/Sun RNA methyltransferase 2
- AML
Acute myeloid leukemia
- LSCs
Leukemia stem cells
- SRSF7
Serine/arginine-rich splicing factor 7
- LC
Lung cancer
- NSCLC
Non-small cell lung cancer
- LUAD
Lung adenocarcinoma
- ESR1
Estrogen receptorα
- ESCA
Esophageal cancer
- Fn
Fusobacterium nucleatum
- GC
Gastric cancer
- PC
Pancreatic cancer
- GBE1
Glucan branching enzyme 1
- HDAC4
Histone deacetylase type 4
- CRC
Colorectal cancer
- SOGA1
Suppressor of glucose by autophagy
- PDK4
Pyruvate dehydrogenase kinase 4
- RCC
Renal cell carcinoma
- AURKB
Encoding aurora kinase B
- BCa
Bladder cancer
- TNFAIP3
TNFα-inducible protein3
- DHCR7
7-dehydrocholesterol reductas
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- MAL2
Mal T-cell differentiation protein 2
- PCa
Prostate cancer
- CRPC
Castration-resistant prostate cancer
- BC
Breast cancer
- CDK6
Cell cycle protein-dependent kinase 6
- ALYREF
Aly/REF export factor
- NXF1
Nuclear RNA export factor 1
- OC
Ovarian cancer
- PIK3R1
Phosphatidylinositol-3-kinase regulatory subunit 1
- ITGB1
Integrin subunit β1
- FAK
Focal adhesion kinase
- FSH
Follicle stimulating hormone
- CC
Cervical cancer
- HPV
Human papillomavirus
- EC
Endometrial cancer
- PDGFRB
Platelet-derived growth factor receptorβ
- ECSC
Endometrial cancer stem cells
- EGR1
Early growth response factor-1
- NLRC5
NLR family CARD structural domain containing 5
- NPC
nasopharyngeal carcinoma
- TFAP2C
TF-activated enhancer binding protein 2 C
- MeRIP-Seq
Methylated RNA immunoprecipitation sequencing
- MINT-Seq
Methylation-inscribed nascent transcript sequencing
- mESCs
Mouse embryonic stem cells
- INTS11
Nucleic acid endonuclease subunit
- Pol II
RNA polymerase II
- PDAC
Pancreatic ductal adenocarcinoma
- seRNAs
Super-enhancer-associated RNAs
- CFL1
Cofilin-1
- MLL1/KMT2A
Mixed-lineage leukemia protein 1
- UCEC
Uterine corpus endometrial cancer
- CTCF
CCCTC-binding factor
- mPCa
Bone-metastatic prostate cancer
- m1A
N1-methyladenosine
- 5hmC
5-hydroxymethylcytosine
- enCRISPRi
Enhancer-targeted CRISPR interference
- SNPs
Single nucleotide polymorphisms
- GWAS
Genome-wide association studies
- ASOs
Antisense Oligonucleotides
- enCRISPRa
Enhancer-targeted CRISPR activation
- IRS2
Insulin receptor substrate 2
- eRNA-QTLs
eRNA quantitative trait loci
- TMZ
Temozolomide
- ccRCC
Clear cell renal cell carcinoma
- BRD9
Bromodomain-containing protein 9
- KIs
Kinase inhibitors
- 5-FU
5-fluorouracil
Author contributions
Conception and design of the review article: Y. H., L. M. and M. L. Interpretation of relevant literature and manuscript writing: J. S. and M. Y. The author(s) read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32300465 and No. 82260493), Natural Science Foundation of Xinjiang Uygur Autonomous Region (Grant No. 2022D01C725) and Xinjiang Key Laboratory of Molecular Biology for Endemic Diseases (XJDFB2024G02).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet. 2018;50:1296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis MW, Li S, Franco HL. Transcriptional control by enhancers and enhancer RNAs. Transcription. 2019;10:171–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Xiong F, Li W. Enhancer RNAs in cancer: regulation, mechanisms and therapeutic potential. RNA Biol. 2020;17:1550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, Dolan K, Zhu X, Hubert N, Tao Y, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep. 2018;25:1816–e18281814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Li S, Shen L, Li J, Zhang D, Yu J, Huang L, Liu N, Lu H, Xu M. M6A-modified BFSP1 induces aerobic Glycolysis to promote liver cancer growth and metastasis through upregulating Tropomodulin 4. Mol Biomed. 2025;6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Zhang X, Liu C, Wu Y, Wen Y, Zheng R, Xu C, Tian J, Peng Q, Zheng X, et al. Engineering a nano-drug delivery system to regulate m6A modification and enhance immunotherapy in gastric cancer. Acta Biomater. 2025;191:412–27. [DOI] [PubMed] [Google Scholar]
- 12.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. [DOI] [PubMed] [Google Scholar]
- 13.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–55. [DOI] [PubMed] [Google Scholar]
- 14.Neuberger MS. Expression and regulation of Immunoglobulin heavy chain gene transfected into lymphoid cells. Embo J. 1983;2:1373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–53. [DOI] [PubMed] [Google Scholar]
- 16.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An integrated encyclopedia. Of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du M, Stitzinger SH, Spille JH, Cho WK, Lee C, Hijaz M, Quintana A. Cissé, II: direct observation of a condensate effect on super-enhancer controlled gene bursting. Cell. 2024;187:331–e344317. [DOI] [PubMed] [Google Scholar]
- 19.Kvon EZ, Waymack R, Gad M, Wunderlich Z. Enhancer redundancy in development and disease. Nat Rev Genet. 2021;22:324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. Chromatin stretch enhancer States drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator Establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney PH, Shrestha B, Xiong J, Zhang T, Rushlow CA. Shadow enhancers modulate distinct transcriptional parameters that differentially effect downstream patterning events. Development. 2022;149. [DOI] [PMC free article] [PubMed]
- 25.Cannavò E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, Ciglar L, Korbel JO, Furlong EE. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol. 2016;26:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bal E, Kumar R, Hadigol M, Holmes AB, Hilton LK, Loh JW, Dreval K, Wong JCH, Vlasevska S, Corinaldesi C, et al. Author correction: Super-enhancer hypermutation alters oncogene expression in B cell lymphoma. Nature. 2022;611:E2. [DOI] [PubMed] [Google Scholar]
- 27.Kvon EZ, Waymack R, Gad M, Wunderlich Z. Author correction: enhancer redundancy in development and disease. Nat Rev Genet. 2021;22:337. [DOI] [PubMed] [Google Scholar]
- 28.Gaynor L, Singh H, Tie G, Badarinath K, Madha S, Mancini A, Bhattacharya S, Hoshino M, de Sauvage FJ, Murata K, et al. Crypt density and recruited enhancers underlie intestinal tumour initiation. Nature. 2025;640:231–39. [DOI] [PMC free article] [PubMed]
- 29.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective Inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubo N, Chen PB, Hu R, Ye Z, Sasaki H, Ren B. H3K4me1 facilitates promoter-enhancer interactions and gene activation during embryonic stem cell differentiation. Mol Cell. 2024;84:1742–e17521745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Wu X, Xie W. Activation, decommissioning, and dememorization: enhancers in a life cycle. Trends Biochem Sci. 2023;48:673–88. [DOI] [PubMed] [Google Scholar]
- 33.Angeloni A, Bogdanovic O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019;63:707–15. [DOI] [PubMed] [Google Scholar]
- 34.Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V. Cisse, II: mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang L, Cao C, Ji L, Cai Z, Wang D, Ye R, Chen J, Yu X, Zhou J, Bai Z, et al. Complementary Alu sequences mediate enhancer-promoter selectivity. Nature. 2023;619:868–75. [DOI] [PubMed] [Google Scholar]
- 36.Barshad G, Lewis JJ, Chivu AG, Abuhashem A, Krietenstein N, Rice EJ, Ma Y, Wang Z, Rando OJ, Hadjantonakis AK, Danko CG. RNA polymerase II dynamics shape enhancer-promoter interactions. Nat Genet. 2023;55:1370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, Snetkova V, Bower G, Jacinto S, Clock B, Dizehchi A, Barozzi I, Mannion BJ, Alcaina-Caro A, Lopez-Rios J, et al. Increased enhancer-promoter interactions during developmental enhancer activation in mammals. Nat Genet. 2024;56:675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang JH, Hansen AS. Enhancer selectivity in space and time: from enhancer-promoter interactions to promoter activation. Nat Rev Mol Cell Biol. 2024;25:574–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drabløs F, Lennartsson A, Rönnerblad M, Hrydziuszko O, Vitezic M, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by Establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Z, Li W. Enhancer RNA: what we know and what we can achieve. Cell Prolif. 2022;55:e13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song P, Han R, Yang F. Super enhancer LncRNAs: a novel hallmark in cancer. Cell Commun Signal. 2024;22:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain M, Abu-Shumays R, Olsen HE, Akeson M. Advances in nanopore direct RNA sequencing. Nat Methods. 2022;19:1160–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dave P, Roth G, Griesbach E, Mateju D, Hochstoeger T, Chao JA. Single-molecule imaging reveals translation-dependent destabilization of mRNAs. Mol Cell. 2023;83:589–e606586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–63. [DOI] [PubMed] [Google Scholar]
- 49.Carullo NVN, Phillips Iii RA, Simon RC, Soto SAR, Hinds JE, Salisbury AJ, Revanna JS, Bunner KD, Ianov L, Sultan FA, et al. Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res. 2020;48:9550–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu E, Zemke NR, Berk AJ. Promoter-specific changes in initiation, elongation, and homeostasis of histone H3 acetylation during CBP/p300 Inhibition. Elife. 2021;10. [DOI] [PMC free article] [PubMed]
- 52.Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014;159:358–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, Adelman K. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018;32:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–76. [DOI] [PubMed] [Google Scholar]
- 55.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MC, An O, Li Z, Yeoh AEJ, Fullwood MJ, et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood. 2019;134:239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168:135–e149122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou TY, Kraus WL. Analysis of estrogen-regulated enhancer RNAs identifies a functional motif required for enhancer assembly and gene expression. Cell Rep. 2022;39:110944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, Lauberth SM. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol. 2018;25:687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oksuz O, Henninger JE, Warneford-Thomson R, Zheng MM, Erb H, Vancura A, Overholt KJ, Hawken SW, Banani SF, Lauman R, et al. Transcription factors interact with RNA to regulate genes. Mol Cell. 2023;83:2449–e24632413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi L, Li S, Maurer K, Zhang Z, Petri M, Sullivan KE. Enhancer RNA and NFκB-dependent P300 regulation of ADAMDEC1. Mol Immunol. 2018;103:312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y, Zhou J, He L, Li Y, Yuan J, Sun K, Chen X, Bao X, Esteban MA, Sun H, Wang H. MyoD induced enhancer RNA interacts with HnRNPL to activate target gene transcription during myogenic differentiation. Nat Commun. 2019;10:5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ying P, Chen C, Lu Z, Chen S, Zhang M, Cai Y, Zhang F, Huang J, Fan L, Ning C, et al. Genome-wide enhancer-gene regulatory maps link causal variants to target genes underlying human cancer risk. Nat Commun. 2023;14:5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Min J, Shay JW. TERT promoter mutations enhance telomerase activation by Long-Range chromatin interactions. Cancer Discov. 2016;6:1212–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Luo M, Liang Q, Zhao K, Hu Y, Wang W, Feng X, Hu B, Teng J, You T, et al. Landscape of enhancer disruption and functional screen in melanoma cells. Genome Biol. 2023;24:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.See YX, Chen K, Fullwood MJ. MYC overexpression leads to increased chromatin interactions at super-enhancers and MYC binding sites. Genome Res. 2022;32:629–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arya AK, Bhadada SK, Singh P, Sachdeva N, Saikia UN, Dahiya D, Behera A, Bhansali A, Rao SD. Promoter hypermethylation inactivates CDKN2A, CDKN2B and RASSF1A genes in sporadic parathyroid adenomas. Sci Rep. 2017;7:3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siersbæk R, Scabia V, Nagarajan S, Chernukhin I, Papachristou EK, Broome R, Johnston SJ, Joosten SEP, Green AR, Kumar S, et al. IL6/STAT3 signaling hijacks Estrogen receptor α enhancers to drive breast Cancer metastasis. Cancer Cell. 2020;38:412–e423419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sultanov R, Mulyukina A, Zubkova O, Fedoseeva A, Bogomazova A, Klimina K, Larin A, Zatsepin T, Prikazchikova T, Lukina M, et al. TP63-TRIM29 axis regulates enhancer methylation and chromosomal instability in prostate cancer. Epigenetics Chromatin. 2024;17:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li M, Liu M, Han W, Wang Z, Han D, Patalano S, Macoska JA, Balk SP, He HH, Corey E, et al. LSD1 Inhibition disrupts Super-Enhancer-Driven oncogenic transcriptional programs in Castration-Resistant prostate Cancer. Cancer Res. 2023;83:1684–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 2016;17:2715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Napoli M, Wu SJ, Gore BL, Abbas HA, Lee K, Checker R, Dhar S, Rajapakshe K, Tan AC, Lee MG, et al. ∆Np63 regulates a common landscape of enhancer associated genes in non-small cell lung cancer. Nat Commun. 2022;13:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim HM, Kang B, Park S, Park H, Kim CJ, Lee H, Yoo M, Kweon MN, Im SH, Kim TI, Roh TY. Forkhead box protein D2 suppresses colorectal cancer by reprogramming enhancer interactions. Nucleic Acids Res. 2023;51:6143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu M, Du M, Yu J, Qian Z, Gao Y, Pan W, Zhao X, Wang M, Li H, Zheng J, et al. CEBPA mutants down-regulate AML cell susceptibility to NK-mediated Lysis by disruption of the expression of NKG2D ligands, which can be restored by LSD1 Inhibition. Oncoimmunology. 2022;11:2016158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Y, Wang Z, Sun L, Shao L, Yang N, Yu D, Zhang X, Han X, Sun Y. SATB1 mediates Long-Range chromatin interactions: A dual regulator of Anti-Apoptotic BCL2 and Pro-Apoptotic NOXA genes. PLoS ONE. 2015;10:e0139170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yokoyama Y, Zhu H, Lee JH, Kossenkov AV, Wu SY, Wickramasinghe JM, Yin X, Palozola KC, Gardini A, Showe LC, et al. BET inhibitors suppress ALDH activity by targeting ALDH1A1 Super-Enhancer in ovarian Cancer. Cancer Res. 2016;76:6320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen CH, Yang N, Zhang Y, Ding J, Zhang W, Liu R, Liu W, Chen C. Inhibition of super enhancer downregulates the expression of KLF5 in basal-like breast cancers. Int J Biol Sci. 2019;15:1733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36:945–61. [DOI] [PubMed] [Google Scholar]
- 82.Seth PP, Tanowitz M, Bennett CF. Selective tissue targeting of synthetic nucleic acid drugs. J Clin Invest. 2019;129:915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blinka S, Reimer MH Jr., Pulakanti K, Rao S. Super-Enhancers at the Nanog locus differentially regulate neighboring Pluripotency-Associated genes. Cell Rep. 2016;17:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu L, Zhou W, Lin L, Chen A, Feng J, Qu X, Zhang H, Yue J. Delivery of therapeutic oligonucleotides in nanoscale. Bioact Mater. 2022;7:292–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moorthy SD, Davidson S, Shchuka VM, Singh G, Malek-Gilani N, Langroudi L, Martchenko A, So V, Macpherson NN, Mitchell JA. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res. 2017;27:246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li K, Liu Y, Cao H, Zhang Y, Gu Z, Liu X, Yu A, Kaphle P, Dickerson KE, Ni M, Xu J. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat Commun. 2020;11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marchant V, Trionfetti F, Tejedor-Santamaria L, Rayego-Mateos S, Rotili D, Bontempi G, Domenici A, Menè P, Mai A, Martín-Cleary C, et al. BET protein inhibitor JQ1 ameliorates experimental peritoneal damage by Inhibition of inflammation and oxidative stress. Antioxid (Basel). 2023;12. [DOI] [PMC free article] [PubMed]
- 88.Shigeyasu K, Toden S, Ozawa T, Matsuyama T, Nagasaka T, Ishikawa T, Sahoo D, Ghosh P, Uetake H, Fujiwara T, Goel A. The PVT1 LncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol Cancer. 2020;19:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan L, Wang J, Guo J, Jia R. Enhancer RNA IRS2e is essential for IRS2 expression and the oncogenic properties in oral squamous cell carcinoma. Carcinogenesis. 2023;44:119–28. [DOI] [PubMed] [Google Scholar]
- 90.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L LncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–35. [DOI] [PubMed] [Google Scholar]
- 92.Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L, Xiao JF, Li CQ, Huang ML, Ding LW, et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao J, Liu Y, Yi J, Liu X. LINC02257, an enhancer RNA of prognostic value in Colon adenocarcinoma, correlates with Multi-Omics Immunotherapy-Related analysis in 33 cancers. Front Mol Biosci. 2021;8:646786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stasevich EM, Uvarova AN, Murashko MM, Khabusheva ER, Sheetikov SA, Prassolov VS, Kuprash DV, Demin DE, Schwartz AM. Enhancer RNA AL928768.3 from the IGH locus regulates MYC expression and controls the proliferation and chemoresistance of Burkitt lymphoma cells with IGH/MYC translocation. Int J Mol Sci. 2022;23. [DOI] [PMC free article] [PubMed]
- 95.Call SG, Duren RP, Panigrahi AK, Nguyen L, Freire PR, Grimm SL, Coarfa C, Conneely OM. Targeting oncogenic super enhancers in MYC-Dependent AML using a small molecule activator of NR4A nuclear receptors. Sci Rep. 2020;10:2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang L, Zhang J, Songyang Z, Xiong Y. Identification and validation of eRNA as a prognostic Indicator for cervical Cancer. Biology (Basel). 2024;13. [DOI] [PMC free article] [PubMed]
- 97.Chen W, Wang Z, Wang Y, Lin J, Chen S, Chen H, Ma X, Zou X, Li X, Qin Y, et al. Enhancer RNA Transcriptome-Wide association study reveals a distinctive class of Pan-Cancer susceptibility eRNAs. Adv Sci (Weinh). 2025;12:e2411974. [DOI] [PMC free article] [PubMed]
- 98.Liao H, Wang H, Zheng R, Yu Y, Zhang Y, Lv L, Zhang B, Chen J. LncRNA CARMN suppresses EMT through inhibiting transcription of MMP2 activated by DHX9 in breast cancer. Cell Signal. 2024;113:110943. [DOI] [PubMed] [Google Scholar]
- 99.Shi T, Guo D, Zheng Y, Wang W, Bi J, He A, Fan S, Su G, Zhao X, Zhao Z, et al. Bivalent activity of super-enhancer RNA LINC02454 controls 3D chromatin structure and regulates glioma sensitivity to Temozolomide. Cell Death Dis. 2024;15:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cappannini A, Ray A, Purta E, Mukherjee S, Boccaletto P, Moafinejad SN, Lechner A, Barchet C, Klaholz Bruno P, Stefaniak F, Bujnicki JM. MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2024;52:D239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, Moroz-Omori EV, Shao J, Zhu X, Bolt MJ, et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81:3368–e33853369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yin J, Ding F, Cheng Z, Ge X, Li Y, Zeng A, Zhang J, Yan W, Shi Z, Qian X, et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling. Cell Death Dis. 2023;14:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu F, Wei J, Cui X, Yu C, Ni W, Bungert J, Wu L, He C, Qian Z. Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 2021;49:5779–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ignatova VV, Stolz P, Kaiser S, Gustafsson TH, Lastres PR, Sanz-Moreno A, Cho YL, Amarie OV, Aguilar-Pimentel A, Klein-Rodewald T, et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020;34:715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M, Lafontaine DLJ. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O’Donohue MF, Guzmán UH, Małecki JM, Wu J, Kirpekar F, et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ren W, Lu J, Huang M, Gao L, Li D, Wang GG, Song J. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat Commun. 2019;10:5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016;167:1897. [DOI] [PubMed] [Google Scholar]
- 110.Esteve-Puig R, Climent F, Piñeyro D, Domingo-Domènech E, Davalos V, Encuentra M, Rea A, Espejo-Herrera N, Soler M, Lopez M, et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood. 2021;137:994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang L, Duan HC, Paduch M, Hu J, Zhang C, Mu Y, Lin H, He C, Kossiakoff AA, Jia G, Zhang L. The molecular basis of human ALKBH3 mediated RNA N(1) -methyladenosine (m(1) A) demethylation. Angew Chem Int Ed Engl. 2024;63:e202313900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park SH, Ju JS, Woo H, Yun HJ, Lee SB, Kim SH, Győrffy B, Kim EJ, Kim H, Han HD, et al. The m(6)A writer RBM15 drives the growth of triple-negative breast cancer cells through the stimulation of Serine and glycine metabolism. Exp Mol Med. 2024;56:1373–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123–41. [DOI] [PubMed] [Google Scholar]
- 116.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Zhang H, Wang H. N(1)-methyladenosine modification in cancer biology: current status and future perspectives. Comput Struct Biotechnol J. 2022;20:6578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Toh JDW, Crossley SWM, Bruemmer KJ, Ge EJ, He D, Iovan DA, Chang CJ. Distinct RNA N-demethylation pathways catalyzed by nonheme iron ALKBH5 and FTO enzymes enable regulation of formaldehyde release rates. Proc Natl Acad Sci U S A. 2020;117:25284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, Chang YH, Jiang T, Wang YZ. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang Y, Jiang X, Chen J, Liu L, Liu G, Sun K, Liu W, Zhu X, Guan Q. The m(6)A reader YTHDC2 maintains visual function and retinal photoreceptor survival through modulating translation of PPEF2 and PDE6B. J Genet Genomics. 2024;51:208–21. [DOI] [PubMed] [Google Scholar]
- 122.Zhang H, Sun Y, Wang Z, Huang X, Tang L, Jiang K, Jin X. ZDHHC20-mediated S-palmitoylation of YTHDF3 stabilizes MYC mRNA to promote pancreatic cancer progression. Nat Commun. 2024;15:4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang J, Zhang J, Liu H, Meng L, Gao X, Zhao Y, Wang C, Gao X, Fan A, Cao T, et al. N6-methyladenosine reader hnRNPA2B1 recognizes and stabilizes NEAT1 to confer chemoresistance in gastric cancer. Cancer Commun (Lond). 2024;44:469–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang J, Shin MK, Park J, Hwang HJ, Locker N, Ahn J, Kim D, Baek D, Park Y, Lee Y, et al. An interaction between eIF4A3 and eIF3g drives the internal initiation of translation. Nucleic Acids Res. 2023;51:10950–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andrade D, Mehta M, Griffith J, Oh S, Corbin J, Babu A, De S, Chen A, Zhao YD, Husain S, et al. HuR reduces Radiation-Induced DNA damage by enhancing expression of ARID1A. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed]
- 126.Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tan X, Zheng C, Zhuang Y, Jin P, Wang F. The m6A reader PRRC2A is essential for meiosis I completion during spermatogenesis. Nat Commun. 2023;14:1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiang JF, Yang Q, Liu CX, Wu M, Chen LL, Yang L. N(6)-Methyladenosines modulate A-to-I RNA editing. Mol Cell. 2018;69:126–e135126. [DOI] [PubMed] [Google Scholar]
- 129.Xie R, Cheng L, Huang M, Huang L, Chen Z, Zhang Q, Li H, Lu J, Wang H, Zhou Q, et al. NAT10 drives cisplatin chemoresistance by enhancing ac4C-Associated DNA repair in bladder Cancer. Cancer Res. 2023;83:1666–83. [DOI] [PubMed] [Google Scholar]
- 130.Mei Z, Shen Z, Pu J, Liu Q, Liu G, He X, Wang Y, Yue J, Ge S, Li T, et al. NAT10 mediated ac4C acetylation driven m(6)A modification via involvement of YTHDC1-LDHA/PFKM regulates Glycolysis and promotes osteosarcoma. Cell Commun Signal. 2024;22:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu H, Li L, Chen KM, Homolka D, Gos P, Fleury-Olela F, McCarthy AA, Pillai RS. Decapping enzyme NUDT12 partners with BLMH for cytoplasmic surveillance of NAD-Capped RNAs. Cell Rep. 2019;29:4422–e44344413. [DOI] [PubMed] [Google Scholar]
- 132.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. Reversible methylation of m(6)A(m) in the 5’ cap controls mRNA stability. Nature. 2017;541:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen H, Gu L, Orellana EA, Wang Y, Guo J, Liu Q, Wang L, Shen Z, Wu H, Gregory RI, et al. METTL4 is an SnRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020;30:544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen C, Chao Y, Zhang C, Hu W, Huang Y, Lv Y, Liu B, Ji D, Liu M, Yang B, et al. TROP2 translation mediated by dual m(6)A/m(7)G RNA modifications promotes bladder cancer development. Cancer Lett. 2023;566:216246. [DOI] [PubMed] [Google Scholar]
- 135.Liu C, Dou X, Zhao Y, Zhang L, Zhang L, Dai Q, Liu J, Wu T, Xiao Y, He C. IGF2BP3 promotes mRNA degradation through internal m(7)G modification. Nat Commun. 2024;15:7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ohshiro T, Konno M, Asai A, Komoto Y, Yamagata A, Doki Y, Eguchi H, Ofusa K, Taniguchi M, Ishii H. Single-molecule RNA sequencing for simultaneous detection of m6A and 5mC. Sci Rep. 2021;11:19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao Y, Zhou Y, Qian Y, Wei W, Lin X, Mao S, Sun J, Jin J. m(6)A-dependent upregulation of DDX21 by super-enhancer-driven IGF2BP2 and IGF2BP3 facilitates progression of acute myeloid leukaemia. Clin Transl Med. 2024;14:e1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang C, Chen L, Lou W, Su J, Huang J, Liu A, Xu Y, He H, Gao Y, Xu D, Li Q. Aberrant activation of m6A demethylase FTO renders HIF2α(low/-) clear cell renal cell carcinoma sensitive to BRD9 inhibitors. Sci Transl Med. 2021;13:eabf6045. [DOI] [PubMed] [Google Scholar]
- 139.Zhang C, Chen L, Liu Y, Huang J, Liu A, Xu Y, Shen Y, He H, Xu D. Downregulated METTL14 accumulates BPTF that reinforces super-enhancers and distal lung metastasis via glycolytic reprogramming in renal cell carcinoma. Theranostics. 2021;11:3676–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li M, Ye J, Xia Y, Li M, Li G, Hu X, Su X, Wang D, Zhao X, Lu F, et al. METTL3 mediates chemoresistance by enhancing AML homing and engraftment via ITGA4. Leukemia. 2022;36:2586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang M, Xie Z, Tan Y, Wu Y, Wang M, Zhang P, Yuan Y, Li J. METTL14-mediated N6-methyladenosine modification of TCP1 mRNA promotes acute myeloid leukemia progression. Cell Signal. 2024;122:111304. [DOI] [PubMed] [Google Scholar]
- 142.Li R, Wu X, Xue K, Feng D, Li J, Li J. RNA demethylase ALKBH5 promotes tumorigenesis of t (8;21) acute myeloid leukemia via ITPA m6A modification. Biomark Res. 2023;11:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hong YG, Yang Z, Chen Y, Liu T, Zheng Y, Zhou C, Wu GC, Chen Y, Xia J, Wen R, et al. The RNA m6A reader YTHDF1 is required for acute myeloid leukemia progression. Cancer Res. 2023;83:845–60. [DOI] [PubMed] [Google Scholar]
- 144.Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, Zhou K, Li W, Hu J, Fu C, et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40:1566–e15821510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11:480–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fan Y, Yan D, Ma L, Liu X, Luo G, Hu Y, Kou X. ALKBH5 is a prognostic factor and promotes the angiogenesis of glioblastoma. Sci Rep. 2024;14:1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kowalski-Chauvel A, Lacore MG, Arnauduc F, Delmas C, Toulas C, Cohen-Jonathan-Moyal E, Seva C. The m6A RNA demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers (Basel). 2020;13. [DOI] [PMC free article] [PubMed]
- 148.Malacrida A, Di Domizio A, Bentivegna A, Cislaghi G, Messuti E, Tabano SM, Giussani C, Zuliani V, Rivara M, Nicolini G. MV1035 overcomes Temozolomide resistance in Patient-Derived glioblastoma stem cell lines. Biology (Basel). 2022;11. [DOI] [PMC free article] [PubMed]
- 149.Cun Y, An S, Zheng H, Lan J, Chen W, Luo W, Yao C, Li X, Huang X, Sun X, et al. Specific regulation of m(6)A by SRSF7 promotes the progression of glioblastoma. Genomics Proteom Bioinf. 2023;21:707–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xie YX, Wang L, Zhou ZH, Liu WJ, Wang W, Yang JH, He ML, Qiu JG, Jiang BH. m(6)A RNA methyltransferase METTL16 induces Cr(VI) carcinogenesis and lung cancer development through glutamine biosynthesis and GLUL expression. J Hazard Mater. 2024;480:136093. [DOI] [PubMed] [Google Scholar]
- 151.Sun Y, Shen W, Hu S, Lyu Q, Wang Q, Wei T, Zhu W, Zhang J. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. J Exp Clin Cancer Res. 2023;42:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu X, Qiu S, Zeng B, Huang Y, Wang X, Li F, Yang Y, Cao L, Zhang X, Wang J, Ma L. N(6)-methyladenosine demethyltransferase FTO mediated m(6)A modification of Estrogen receptor alpha in non-small cell lung cancer tumorigenesis. Oncogene. 2024;43:1288–302. [DOI] [PubMed] [Google Scholar]
- 153.Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lin Z, Li J, Zhang J, Feng W, Lu J, Ma X, Ding W, Ouyang S, Lu J, Yue P, et al. Metabolic reprogramming driven by IGF2BP3 promotes acquired resistance to EGFR inhibitors in Non–Small cell lung Cancer. Cancer Res. 2023;83:2187–207. [DOI] [PubMed] [Google Scholar]
- 155.Hua X, Xu Q, Wu R, Sun W, Gu Y, Zhu S, Liu X, Lv T, Song Y. ALKBH5 promotes non-small cell lung cancer progression and susceptibility to anti-PD-L1 therapy by modulating interactions between tumor and macrophages. J Exp Clin Cancer Res. 2024;43:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang Z, Lin G, Hong Y, Weng L, Zhu K, Zhuang W. High expression of AlkB homolog 5 suppresses the progression of non-small cell lung cancer by facilitating ferroptosis through m6A demethylation of SLC7A11. Environ Toxicol. 2024;39:4035–46. [DOI] [PubMed] [Google Scholar]
- 157.Guo S, Chen F, Li L, Dou S, Li Q, Huang Y, Li Z, Liu W, Zhang G. Intracellular Fusobacterium nucleatum infection increases METTL3-mediated m6A methylation to promote the metastasis of esophageal squamous cell carcinoma. J Adv Res. 2024;61:165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Qiao Z, Li Y, Cheng Y, Li S, Liu S. SHMT2 regulates esophageal cancer cell progression and immune escape by mediating m6A modification of c-myc. Cell Biosci. 2023;13:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zang Y, Tian Z, Wang D, Li Y, Zhang W, Ma C, Liao Z, Gao W, Qian L, Xu X, et al. METTL3-mediated N(6)-methyladenosine modification of STAT5A promotes gastric cancer progression by regulating KLF4. Oncogene. 2024;43:2338–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wei X, Huo Y, Pi J, Gao Y, Rao S, He M, Wei Q, Song P, Chen Y, Lu D, et al. METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol. 2022;24:1278–90. [DOI] [PubMed] [Google Scholar]
- 161.Liu T, Feng YL, Wang RY, Yang S, Ge YL, Zhang TY, Li J, Li CY, Ruan Y, Luo B, Liang GY. Long-term MNNG exposure promotes gastric carcinogenesis by activating METTL3/m6A/miR1184 axis-mediated epithelial-mesenchymal transition. Sci Total Environ. 2024;913:169752. [DOI] [PubMed] [Google Scholar]
- 162.Wang Q, Huang Y, Jiang M, Tang Y, Wang Q, Bai L, Yu C, Yang X, Ding K, Wang W, et al. The demethylase ALKBH5 mediates ZKSCAN3 expression through the m(6)A modification to activate VEGFA transcription and thus participates in MNNG-induced gastric cancer progression. J Hazard Mater. 2024;473:134690. [DOI] [PubMed] [Google Scholar]
- 163.Hu Y, Gong C, Li Z, Liu J, Chen Y, Huang Y, Luo Q, Wang S, Hou Y, Yang S, Xiao Y. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol Cancer. 2022;21:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.You Q, Wang F, Du R, Pi J, Wang H, Huo Y, Liu J, Wang C, Yu J, Yang Y, Zhu L. m(6) A reader YTHDF1-Targeting engineered small extracellular vesicles for gastric Cancer therapy via epigenetic and immune regulation. Adv Mater. 2023;35:e2204910. [DOI] [PubMed] [Google Scholar]
- 165.Yang J, Chen Y, He Y, Da M. YTHDF2 promotes gastric cancer progression and enhances chemoradiotherapy resistance. Drug Dev Res. 2024;85:e22179. [DOI] [PubMed] [Google Scholar]
- 166.Yu Y, Meng LL, Chen XY, Fan HN, Chen M, Zhang J, Zhu JS. m(6)A reader YTHDF3 is associated with clinical prognosis, related RNA signatures and immunosuppression in gastric cancer. Cell Signal. 2023;108:110699. [DOI] [PubMed] [Google Scholar]
- 167.Yang Y, Cai J, Yang X, Wang K, Sun K, Yang Z, Zhang L, Yang L, Gu C, Huang X, et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol Ther. 2022;30:2342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu L, Gu M, Ma J, Wang Y, Li M, Wang H, Yin X, Li X. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. 2022;21:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen A, Zhang VX, Zhang Q, Sze KM, Tian L, Huang H, Wang X, Lee E, Lu J, Lyu X, et al. Targeting the oncogenic m6A demethylase FTO suppresses tumourigenesis and potentiates immune response in hepatocellular carcinoma. Gut. 2024;74:90–102. [DOI] [PMC free article] [PubMed]
- 170.Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, Jiang J, Wang L, Mang Y, Gao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang X, Su T, Wu Y, Cai Y, Wang L, Liang C, Zhou L, Wang S, Li XX, Peng S, et al. N6-Methyladenosine reader YTHDF1 promotes stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. 2024;84:827–40. [DOI] [PubMed] [Google Scholar]
- 172.Chen J, Zhang H, Xiu C, Gao C, Wu S, Bai J, Shen Q, Yin T. METTL3 promotes pancreatic cancer proliferation and stemness by increasing stability of ID2 mRNA in a m6A-dependent manner. Cancer Lett. 2023;565:216222. [DOI] [PubMed] [Google Scholar]
- 173.Yi T, Wang C, Ye X, Lin J, Lin C, Qin F, Yang W, Ye Y, Ning D, Lan J, et al. METTL16 inhibits pancreatic cancer proliferation and metastasis by promoting MROH8 RNA stability and inhibiting CAPN2 expression. Int J Surg. 2024;110:7701–19. [DOI] [PMC free article] [PubMed]
- 174.Lin K, Zhou E, Shi T, Zhang S, Zhang J, Zheng Z, Pan Y, Gao W, Yu Y. m6A eraser FTO impairs gemcitabine resistance in pancreatic cancer through influencing NEDD4 mRNA stability by regulating the PTEN/PI3K/AKT pathway. J Exp Clin Cancer Res. 2023;42:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jin W, Yao Y, Fu Y, Lei X, Fu W, Lu Q, Tong X, Xu Q, Su W, Hu X. WTAP/IGF2BP3-mediated GBE1 expression accelerates the proliferation and enhances stemness in pancreatic cancer cells via upregulating c-Myc. Cell Mol Biol Lett. 2024;29:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang Y, Liu X, Wang Y, Lai S, Wang Z, Yang Y, Liu W, Wang H, Tang B. The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol Cancer. 2022;21:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu X, Feng M, Hao X, Gao Z, Wu Z, Wang Y, Du L, Wang C. m6A methylation regulates hypoxia-induced pancreatic cancer glycolytic metabolism through ALKBH5-HDAC4-HIF1α positive feedback loop. Oncogene. 2023;42:2047–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M, Zhang Z, Chen Y, Xie N, Gao W, et al. Hypoxia-induced LncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m(6)A-mediated degradation of STEAP3 mRNA. Mol Cancer. 2022;21:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jiang T, Qi J, Xue Z, Liu B, Liu J, Hu Q, Li Y, Ren J, Song H, Xu Y, et al. The m(6)A modification mediated-lncRNA POU6F2-AS1 reprograms fatty acid metabolism and facilitates the growth of colorectal cancer via upregulation of FASN. Mol Cancer. 2024;23:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wei W, Zhang ZY, Shi B, Cai Y, Zhang HS, Sun CL, Fei YF, Zhong W, Zhang S, Wang C, et al. METTL16 promotes glycolytic metabolism reprogramming and colorectal cancer progression. J Exp Clin Cancer Res. 2023;42:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li H, Yu K, Hu H, Zhang X, Zeng S, Li J, Dong X, Deng X, Zhang J, Zhang Y. METTL17 coordinates ferroptosis and tumorigenesis by regulating mitochondrial translation in colorectal cancer. Redox Biol. 2024;71:103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhai J, Chen H, Wong CC, Peng Y, Gou H, Zhang J, Pan Y, Chen D, Lin Y, Wang S, et al. ALKBH5 drives immune suppression via targeting AXIN2 to promote colorectal Cancer and is a target for boosting immunotherapy. Gastroenterology. 2023;165:445–62. [DOI] [PubMed] [Google Scholar]
- 183.Zheng Z, Zeng X, Zhu Y, Leng M, Zhang Z, Wang Q, Liu X, Zeng S, Xiao Y, Hu C, et al. CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis. Mol Cancer. 2024;23:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen Y, Lu Z, Qi C, Yu C, Li Y, Huan W, Wang R, Luo W, Shen D, Ding L, et al. N(6)-methyladenosine-modified TRAF1 promotes Sunitinib resistance by regulating apoptosis and angiogenesis in a METTL14-dependent manner in renal cell carcinoma. Mol Cancer. 2022;21:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang X, Wang F, Wang Z, Yang X, Yu H, Si S, Lu J, Zhou Z, Lu Q, Wang Z, Yang H. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m(6)A-dependent manner. Ann Transl Med. 2020;8:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shen C, Liu J, Xie F, Yu Y, Ma X, Hu D, Liu C, Wang Y. N6-Methyladenosine enhances the translation of ENO1 to promote the progression of bladder cancer by inhibiting PCNA ubiquitination. Cancer Lett. 2024;595:217002. [DOI] [PubMed] [Google Scholar]
- 187.Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, Zhong H, Jiang Y, Zhang X, Jiang G. Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder Cancer. Cancer Res. 2021;81:6142–56. [DOI] [PubMed] [Google Scholar]
- 188.Zeng Y, Luo Y, Zhao K, Liu S, Wu K, Wu Y, Du K, Pan W, Dai Y, Liu Y, et al. m6A-Mediated induction of 7-Dehydrocholesterol reductase stimulates cholesterol synthesis and cAMP signaling to promote bladder Cancer metastasis. Cancer Res. 2024;84:3402–18. [DOI] [PubMed] [Google Scholar]
- 189.Tao L, Mu X, Chen H, Jin D, Zhang R, Zhao Y, Fan J, Cao M, Zhou Z. FTO modifies the m6A level of MALAT and promotes bladder cancer progression. Clin Transl Med. 2021;11:e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yuan S, He SH, Li LY, Xi S, Weng H, Zhang JH, Wang DQ, Guo MM, Zhang H, Wang SY, et al. A potassium-chloride co-transporter promotes tumor progression and castration resistance of prostate cancer through m(6)A reader YTHDC1. Cell Death Dis. 2023;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Duan J, Fan D, Chen P, Xiang J, Xie X, Peng Y, Bai J, Li T, Li Y, Song H, et al. YTHDF3 regulates the degradation and stability of m6A-Enriched transcripts to facilitate the progression of Castration-Resistant prostate Cancer. J Pineal Res. 2024;76:e13003. [DOI] [PubMed] [Google Scholar]
- 192.Zhong C, Long Z, Yang T, Wang S, Zhong W, Hu F, Teoh JY, Lu J, Mao X. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int J Biol Sci. 2023;19:1543–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang J, Wei J, Sun R, Sheng H, Yin K, Pan Y, Jimenez R, Chen S, Cui XL, Zou Z, et al. A LncRNA from the FTO locus acts as a suppressor of the m(6)A writer complex and p53 tumor suppression signaling. Mol Cell. 2023;83:2692–e27082697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao Y, Hu X, Yu H, Sun H, Zhang L, Shao C. The FTO mediated N6-Methyladenosine modification of DDIT4 regulation with tumorigenesis and metastasis in prostate Cancer. Res (Wash D C). 2024;7:0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xu Y, Song M, Hong Z, Chen W, Zhang Q, Zhou J, Yang C, He Z, Yu J, Peng X, et al. The N6-methyladenosine METTL3 regulates tumorigenesis and Glycolysis by mediating m6A methylation of the tumor suppressor LATS1 in breast cancer. J Exp Clin Cancer Res. 2023;42:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu X, Li P, Huang Y, Li H, Liu X, Du Y, Lin X, Chen D, Liu H, Zhou Y. M(6)A demethylase ALKBH5 regulates FOXO1 mRNA stability and chemoresistance in triple-negative breast cancer. Redox Biol. 2024;69:102993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xia T, Dai XY, Sang MY, Zhang X, Xu F, Wu J, Shi L, Wei JF, Ding Q. IGF2BP2 drives cell cycle progression in Triple-Negative breast Cancer by recruiting EIF4A1 to promote the m6A-Modified CDK6 translation initiation process. Adv Sci (Weinh). 2024;11:e2305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jin T, Yang L, Chang C, Luo H, Wang R, Gan Y, Sun Y, Guo Y, Tang R, Chen S, et al. HnRNPA2B1 isgylation regulates m6A-Tagged mRNA selective export via ALYREF/NXF1 complex to foster breast Cancer development. Adv Sci (Weinh). 2024;11:e2307639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li H, Lin R, Zhang Y, Zhu Y, Huang S, Lan J, Lu N, Xie C, He S, Zhang W. N6-methyladenosine-modified circPLPP4 sustains cisplatin resistance in ovarian cancer cells via PIK3R1 upregulation. Mol Cancer. 2024;23:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sun R, Yuan L, Jiang Y, Wan Y, Ma X, Yang J, Sun G, Zhou S, Wang H, Qiu J, et al. ALKBH5 activates FAK signaling through m6A demethylation in ITGB1 mRNA and enhances tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer. Theranostics. 2023;13:833–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu X, Zhuang X, Yu H, Li P, Li X, Lin H, Teoh JP, Chen Y, Yang Y, Cheng Y, et al. FSH induces EMT in ovarian cancer via ALKBH5-regulated snail m6A demethylation. Theranostics. 2024;14:2151–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang X, Chen Q, Bing Z, Zhou S, Xu Z, Hou Y, Zhao Y, Zhao S, Wang T. Low expression of m6A reader YTHDC1 promotes progression of ovarian cancer via PIK3R1/STAT3/GANAB axis. Int J Biol Sci. 2023;19:4672–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liang L, Zhu Y, Li J, Zeng J, Wu L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J Exp Clin Cancer Res. 2022;41:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu YY, Xia M, Chen ZB, Liao YD, Zhang CY, Yuan L, Pan YW, Huang H, Lu HW, Yao SZ. HNRNPC mediates lymphatic metastasis of cervical cancer through m6A-dependent alternative splicing of FOXM1. Cell Death Dis. 2024;15:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ao K, Yin M, Lyu X, Xiao Y, Chen X, Zhong S, Wen X, Yuan J, Ye M, Zhang J, et al. METTL3-mediated HSPA9 m6A modification promotes malignant transformation and inhibits cellular senescence by regulating Exosomal Mortalin protein in cervical cancer. Cancer Lett. 2024;587:216658. [DOI] [PubMed] [Google Scholar]
- 206.Shi R, Zhao R, Shen Y, Wei S, Zhang T, Zhang J, Shu W, Cheng S, Teng H, Wang H. IGF2BP2-modified circular RNA circCHD7 promotes endometrial cancer progression via stabilizing PDGFRB and activating JAK/STAT signaling pathway. Cancer Gene Ther. 2024;31:1221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang B, Wang Y, Wang W, Wang Z, Zhang Y, Pan X, Wen X, Leng H, Guo J, Ma XX. WTAP/IGF2BP3 mediated m6A modification of the EGR1/PTEN axis regulates the malignant phenotypes of endometrial cancer stem cells. J Exp Clin Cancer Res. 2024;43:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhan L, Zhang J, Zhang JH, Liu XJ, Guo B, Chen JH, Tang ZH, Wang WY, Wang QY, Wei B, Cao YX. METTL3 facilitates immunosurveillance by inhibiting YTHDF2-mediated NLRC5 mRNA degradation in endometrial cancer. Biomark Res. 2023;11:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen R, Zhang S, Li H, Yang M, Lu Y, Zhang X. METTL14 promotes oral squamous cell carcinoma progression by regulating the mRNA and m6A levels of CALD1. J Environ Pathol Toxicol Oncol. 2023;42:71–81. [DOI] [PubMed] [Google Scholar]
- 210.Li HB, Huang G, Tu J, Lv DM, Jin QL, Chen JK, Zou YT, Lee DF, Shen JN, Xie XB. METTL14-mediated epitranscriptome modification of MN1 mRNA promote tumorigenicity and all-trans-retinoic acid resistance in osteosarcoma. EBioMedicine. 2022;82:104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhang X, Zhang X, Liu T, Zhang Z, Piao C, Ning H. METTL14 promotes migration and invasion of choroidal melanoma by targeting RUNX2 mRNA via m6A modification. J Cell Mol Med. 2022;26:5602–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zou Y, Bao X, Li D, Ye Z, Xiang R, Yang Y, Zhu Z, Chen Z, Zeng L, Xue C, et al. FTO-mediated DSP m(6)A demethylation promotes an aggressive subtype of growth hormone-secreting pituitary neuroendocrine tumors. Mol Cancer. 2024;23:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Li ZX, Zheng ZQ, Yang PY, Lin L, Zhou GQ, Lv JW, Zhang LL, Chen F, Li YQ, Wu CF, et al. WTAP-mediated m(6)A modification of LncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022;29:1137–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen D, Zhao H, Guo Z, Dong Z, Yu Y, Zheng J, Ma Y, Sun H, Zhang Q, Zhang J, et al. Identification of m6A-related LncRNAs LINC02471 and DOCK9-DT as potential biomarkers for thyroid cancer. Int Immunopharmacol. 2024;133:112050. [DOI] [PubMed] [Google Scholar]
- 215.Yang Q, Lu Y, Du A. m6A-related LncRNAs as potential biomarkers and the LncRNA ELFN1-AS1/miR-182-5p/BCL-2 regulatory axis in diffuse large B-cell lymphoma. J Cell Mol Med. 2024;28:e18046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao K, Chen L, Xie Y, Ren N, Li J, Zhai X, Zheng S, Liu K, Wang C, Qiu Q, et al. m6A/HOXA10-AS/ITGA6 axis aggravates oxidative resistance and malignant progression of laryngeal squamous cell carcinoma through regulating Notch and Keap1/Nrf2 pathways. Cancer Lett. 2024;587:216735. [DOI] [PubMed] [Google Scholar]
- 217.Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, Fan Y. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wei J, Yin Y, Zhou J, Chen H, Peng J, Yang J, Tang Y. METTL3 potentiates resistance to cisplatin through m(6) A modification of TFAP2C in seminoma. J Cell Mol Med. 2020;24:11366–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu X, He H, Zhang F, Hu X, Bi F, Li K, Yu H, Zhao Y, Teng X, Li J, et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kahl M, Xu Z, Arumugam S, Edens B, Fischietti M, Zhu AC, Platanias LC, He C, Zhuang X, Ma YC. m6A RNA methylation regulates mitochondrial function. Hum Mol Genet. 2024;33:969–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Fang D, Ou X, Sun K, Zhou X, Li Y, Shi P, Zhao Z, He Y, Peng J, Xu J. m6A modification-mediated LncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci. 2022;113:4135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ma MJ, Shi YH, Liu ZD, Zhu YQ, Zhao GY, Ye JY, Li FX, Huang XT, Wang XY, Wang JQ, et al. N6-methyladenosine modified TGFB2 triggers lipid metabolism reprogramming to confer pancreatic ductal adenocarcinoma gemcitabine resistance. Oncogene. 2024;43:2405–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-Methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tao B, Huang X, Shi J, Liu J, Li S, Xu C, Zhong J, Wan L, Feng B, Li B. FTO interacts with FOXO3a to enhance its transcriptional activity and inhibits aggression in gliomas. Signal Transduct Target Ther. 2020;5:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cui S. METTL3-mediated m6A modification of Lnc RNA RHPN1-AS1 enhances cisplatin resistance in ovarian cancer by activating PI3K/AKT pathway. J Clin Lab Anal. 2022;36:e24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang Y, Jin P, Wang X. N(6)-methyladenosine regulator YTHDF1 represses the CD8 + T cell-mediated antitumor immunity and ferroptosis in prostate cancer via m(6)A/PD-L1 manner. Apoptosis. 2024;29:142–53. [DOI] [PubMed] [Google Scholar]
- 227.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises Cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–e148136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–9. [DOI] [PubMed] [Google Scholar]
- 229.Rau K, Rösner L, Rentmeister A. Sequence-specific m(6)A demethylation in RNA by FTO fused to RCas9. RNA. 2019;25:1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhuang H, Yu B, Tao D, Xu X, Xu Y, Wang J, Jiao Y, Wang L. The role of m6A methylation in therapy resistance in cancer. Mol Cancer. 2023;22:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, Agrawal K, Gonzalez GM, Wang Y, Patel SP, Rana TM. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A. 2020;117:20159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wu Z, Smith AR, Qian Z, Zheng G. Patent landscape of small molecule inhibitors of METTL3 (2020-present). Expert Opin Ther Pat. 2024;1–16. [DOI] [PMC free article] [PubMed]
- 233.Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, et al. Targeting FTO suppresses Cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–e9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, Wang X. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–34. [DOI] [PubMed] [Google Scholar]
- 236.Chen H, Li C, Peng X, Zhou Z, Weinstein JN, Liang H. A Pan-Cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018;173:386–e399312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible M⁶A RNA methylation. Nat Rev Genet. 2014;15:293–306. [DOI] [PubMed] [Google Scholar]
- 238.Zhao Y, Wen S, Li H, Pan CW, Wei Y, Huang T, Li Z, Yang Y, Fan S, Zhang Y. Enhancer RNA promotes resistance to radiotherapy in bone-metastatic prostate cancer by m(6)A modification. Theranostics. 2023;13:596–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–23. [DOI] [PubMed] [Google Scholar]
- 240.Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, Cramer P. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–8. [DOI] [PubMed] [Google Scholar]
- 241.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–89. [DOI] [PubMed] [Google Scholar]
- 242.Xiong F, Wang R, Lee JH, Li S, Chen SF, Liao Z, Hasani LA, Nguyen PT, Zhu X, Krakowiak J, et al. RNA m(6)A modification orchestrates a LINE-1-host interaction that facilitates Retrotransposition and contributes to long gene vulnerability. Cell Res. 2021;31:861–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tegowski M, Flamand MN, Meyer KD. scDART-seq reveals distinct m(6)A signatures and mRNA methylation heterogeneity in single cells. Mol Cell. 2022;82:868–e878810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yu L, Alariqi M, Li B, Hussain A, Zhou H, Wang Q, Wang F, Wang G, Zhu X, Hui F, et al. CRISPR/dCas13(Rx) derived RNA N(6)-methyladenosine (m(6)A) dynamic modification in plant. Adv Sci (Weinh). 2024;11:e2401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. [DOI] [PubMed] [Google Scholar]
- 249.Dierks D, Garcia-Campos MA, Uzonyi A, Safra M, Edelheit S, Rossi A, Sideri T, Varier RA, Brandis A, Stelzer Y, et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat Methods. 2021;18:1060–7. [DOI] [PubMed] [Google Scholar]
- 250.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang Y, Xiao Y, Dong S, Yu Q, Jia G. Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat Chem Biol. 2020;16:896–903. [DOI] [PubMed] [Google Scholar]
- 252.Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, Harada BT, Dai Q, Wei J, Zhang L, et al. m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol. 2022;40:1210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gao Y, Liu X, Wu B, Wang H, Xi F, Kohnen MV, Reddy ASN, Gu L. Quantitative profiling of N(6)-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using nanopore direct RNA sequencing. Genome Biol. 2021;22:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Liu C, Sun H, Yi Y, Shen W, Li K, Xiao Y, Li F, Li Y, Hou Y, Lu B, et al. Absolute quantification of single-base m(6)A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol. 2023;41:355–66. [DOI] [PubMed] [Google Scholar]
- 255.Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Xiao S, Cao S, Huang Q, Xia L, Deng M, Yang M, Jia G, Liu X, Shi J, Wang W, et al. The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat Cell Biol. 2019;21:651–61. [DOI] [PubMed] [Google Scholar]
- 257.Wang K, Peng J, Yi C. The m(6)A consensus motif provides a paradigm of epitranscriptomic studies. Biochemistry. 2021;60:3410–2. [DOI] [PubMed] [Google Scholar]
- 258.Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, Zhao S, Shen B, Gao Y, Han D, He C. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xu W, He C, Kaye EG, Li J, Mu M, Nelson GM, Dong L, Wang J, Wu F, Shi YG, et al. Dynamic control of chromatin-associated m(6)A methylation regulates nascent RNA synthesis. Mol Cell. 2022;82:1156–e11681157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhou Y, Ćorović M, Hoch-Kraft P, Meiser N, Mesitov M, Körtel N, Back H, Naarmann-de Vries IS, Katti K, Obrdlík A, et al. m6A sites in the coding region trigger translation-dependent mRNA decay. Mol Cell. 2024;84:4576–93.e4512. [DOI] [PubMed]
- 261.m(6)A methylation of super-enhancer RNAs facilitates chromatin accessibility. Nat Genet. 2023;55:2037–8. [DOI] [PubMed] [Google Scholar]
- 262.Li R, Zhao H, Huang X, Zhang J, Bai R, Zhuang L, Wen S, Wu S, Zhou Q, Li M, et al. Super-enhancer RNA m(6)A promotes local chromatin accessibility and oncogene transcription in pancreatic ductal adenocarcinoma. Nat Genet. 2023;55:2224–34. [DOI] [PubMed] [Google Scholar]
- 263.Liu J, Yin J, Wang Y, Cai L, Geng R, Du M, Zhong Z, Ni S, Huang X, Yu H, Bai J. A comprehensive prognostic and immune analysis of enhancer RNA identifies IGFBP7-AS1 as a novel prognostic biomarker in uterine Corpus endometrial carcinoma. Biol Proced Online. 2022;24:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Su T, Zhang N, Wang T, Zeng J, Li W, Han L, Yang M. Super Enhancer-Regulated LncRNA LINC01089 induces alternative splicing of DIAPH3 to drive hepatocellular carcinoma metastasis. Cancer Res. 2023;83:4080–94. [DOI] [PubMed] [Google Scholar]
- 265.Hu X, Wu J, Feng Y, Ma H, Zhang E, Zhang C, Sun Q, Wang T, Ge Y, Zong D, et al. METTL3-stabilized super enhancers-lncRNA SUCLG2-AS1 mediates the formation of a long-range chromatin loop between enhancers and promoters of SOX2 in metastasis and radiosensitivity of nasopharyngeal carcinoma. Clin Transl Med. 2023;13:e1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cai H, Liang J, Jiang Y, Tan R, Hou C, Hou J. Integrative analysis of N6-Methyladenosine-Related enhancer RNAs identifies distinct prognosis and tumor immune Micro-Environment patterns in head and neck squamous cell carcinoma. Cancers (Basel). 2022;14. [DOI] [PMC free article] [PubMed]
- 267.Klein JC, Chen W, Gasperini M, Shendure J. Identifying novel enhancer elements with CRISPR-Based screens. ACS Chem Biol. 2018;13:326–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.