Abstract
Assembly of an infectious retrovirus requires the incorporation of the envelope glycoprotein complex during the process of particle budding. We have recently demonstrated that amino acid substitutions of a tyrosine residue in the cytoplasmic domain block glycoprotein incorporation into budding Mason-Pfizer monkey virus (M-PMV) particles and abrogate infectivity (C. Song, S. R. Dubay, and E. Hunter, J. Virol. 77:5192-5200, 2003). To investigate the contribution of other amino acids in the cytoplasmic domain to the process of glycoprotein incorporation, we introduced alanine-scanning mutations into this region of the transmembrane protein. The effects of the mutations on glycoprotein biosynthesis and function, as well as on virus infectivity, have been examined. Mutation of two cytoplasmic residues, valine 20 and histidine 21, inhibits viral protease-mediated cleavage of the cytoplasmic domain that is observed during virion maturation, but the mutant virions show only moderately reduced infectivity. We also demonstrate that the cytoplasmic domain of the M-PMV contains three amino acid residues that are absolutely essential for incorporation of glycoprotein into virions. In addition to the previously identified tyrosine at residue 22, an isoleucine at position 18 and a leucine at position 25 each mediate the process of incorporation and efficient release of virions. While isoleucine 18 may be involved in direct interactions with immature capsids, antibody uptake studies showed that leucine 25 and tyrosine 22 are part of an efficient internalization signal in the cytoplasmic domain of the M-PMV glycoprotein. These results demonstrate that the cytoplasmic domain of M-PMV Env, in part through its YXXL-mediated endocytosis and intracellular trafficking signals, plays a critical role in the incorporation of glycoprotein into virions.
The envelope (Env) glycoprotein of Mason-Pfizer monkey virus (M-PMV), like those of other retroviruses, is synthesized on the rough endoplasmic reticulum (ER) and is cotranslationally glycosylated and inserted into the lumen of the ER (5-8, 30). Shortly after synthesis, the glycosylated precursor is assembled into trimers, a process which is thought to be required for transport of Env from the ER to the Golgi complex (2, 20). It is then cleaved by a cellular protease into two subunits, gp70 (SU) and gp22 (TM), in a late compartment of the Golgi complex (26). The oligomeric, noncovalently associated gp70 and gp22 complexes are then transported to the plasma membrane, where they are incorporated into budding virions (10, 68). The SU glycoprotein is responsible for receptor binding, whereas the TM glycoprotein is responsible for anchoring the SU protein at the surface of infected cells or the viral membrane. The TM glycoprotein also mediates virus-cell membrane fusion during viral entry as well as cell-cell fusion via a fusion peptide and heptad repeat motifs located at the extracellular domain (2, 18, 35, 69, 74, 76). This fusion process also is influenced by the cytoplasmic domain of the TM glycoprotein as demonstrated previously (9, 13, 19, 36, 44, 55, 68, 70).
As is observed with murine leukemia virus (MuLV) and Gibbon ape leukemia virus, but unlike most other retroviruses, a viral protease-mediated maturational cleavage of the TM cytoplasmic domain occurs following virus release, which results in conversion of gp22 into gp20 (9, 10, 13, 55, 67). Based on cytoplasmic domain truncation mutants, this maturational cleavage of the cytoplasmic domain appears to dramatically increase the fusion activity of the TM proteins and results in the loss of 17 amino acids from the carboxy terminus of the cytoplasmic domain (9, 68).
The incorporation of glycoprotein into budding virions is essential for the formation of an infectious virus particle, since retrovirus Env proteins play important roles in receptor binding and membrane fusion. In the case of the alphaviruses, an interaction between the cytoplasmic domain of the spike glycoprotein and the virus nucleocapsid has been demonstrated directly and is absolutely required for virus budding (1, 23, 72, 87). For retroviruses, which do not require glycoprotein expression for virus assembly and release, the nature of capsid-envelope interactions is less well defined. In M-PMV, the glycoprotein appears to play an important role in intracellular transport of assembled capsids to the plasma membrane, and mutations that interfere with Env incorporation also decrease the efficiency of virus release (64, 65, 68). In contrast, Rous sarcoma virus, which encodes a glycoprotein lacking a cytoplasmic domain, can efficiently assemble and infect cells (53). In the case of Moloney MuLV, some deletion mutations in the cytoplasmic domain of the TM protein decrease infectivity without reducing glycoprotein incorporation (33). Evidence derived from Env and Gag mutagenesis and pseudotyping studies of human immunodeficiency virus type 1 (HIV-1) has accumulated both for and against the existence of a specific interaction between the TM cytoplasmic domain and matrix (MA) domain of Gag (15, 17, 51, 63). However, in recent studies, it was demonstrated that mutations in HIV-1 MA that block the incorporation of full-length HIV-1 Env into virions do not affect the incorporation of heterologous retroviral Env glycoprotein with short cytoplasmic domains or HIV-1 Env mutants containing large truncations in the cytoplasmic domain (21, 22, 46). Additional studies have shown that cytoplasmic mutations that block incorporation could be reverted by mutations in MA (45). These findings imply that the incorporation of Env glycoprotein with long cytoplasmic domains depends on specific interaction between sequences in the cytoplasmic domain of the TM protein and those within MA, whereas the incorporation of a glycoprotein with a short cytoplasmic domain does not (3, 21, 29, 45, 50, 79).
Previously, we demonstrated that the mutations in a tyrosine residue at position 22 in the cytoplasmic domain block glycoprotein incorporation into budding M-PMV virions and abrogate infectivity (68). The incorporation function of the tyrosine mutants could be rescued, but only partially, by the aromatic amino acid tryptophan. In this study we utilized alanine-scanning mutagenesis of amino acid residues 2 to 33 within the cytoplasmic domain, which are highly conserved among D-type retroviruses, to investigate the contribution of this region to Env incorporation. The accompanying paper (70) describes the effects of these mutations on the fusogenicity of Env expressed in the absence of Gag and protease. Here, we examined the effects of these substitutions on Env biosynthesis in the context of a proviral M-PMV expression construct and examined the impact of the mutations on Env incorporation into virions and on virus infectivity. These studies indicate that in addition to the previously described tyrosine, two additional amino acids play a critical role in incorporation of the glycoprotein into budding virions and two other amino acids play an important role in the maturational cleavage of the cytoplasmic domain of the TM protein.
MATERIALS AND METHODS
DNAs, cell culture, and transfections.
The pMTΔE vector (72) is an M-PMV proviral expression vector in which the tat gene of HIV-1 has replaced the env gene of M-PMV. Each of the mutated sequences described in the accompanying publication (70) were excised from the pTMT vectors by digestion with EcoRI and BlpI, and the fragment was inserted into the proviral vector pSARM4, which has unique EcoRI-BlpI sites (69). All the mutations were confirmed by DNA sequencing.
COS-1 cells were obtained from the American Type Culture Collection. The HOS-CD4/LTR-hGFP (GHOST) cell line, which expresses green fluorescent protein (GFP) under the control of an HIV long terminal repeat (LTR) was obtained through the AIDS Reference and Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, and was originally contributed by Vineet N. KewalRamani and Dan R. Littman (11, 43). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (Sigma), 10 U of penicillin G sodium/ml, and 10 μg of streptomycin sulfate/ml (Pen-Strep; GibcoBRL). The HOS-CD4/LTR-hGFP cells were additionally maintained in medium containing hygromycin and G418 (Geneticin) as recommended by the contributor. Each mutant DNA was transfected into COS-1 cells by using FuGENE6 (Roche Molecular Biochemicals).
Glycoprotein incorporation assay.
COS-1 cells in 100-mm-diameter plates were transfected with 5 μg of the molecular clone pSARM4 containing either the wild-type (WT) or mutant env gene. Two days after transfection, the cells were labeled with 500 μCi of [3H]leucine (Perkin-Elmer, NEN) in 0.8 ml of leucine-deficient DMEM. The cells were labeled for 90 min, at which time the label was removed, complete DMEM containing 10% fetal bovine serum was added, and incubation was continued for 6 h. The supernatant was collected and, following removal of the cell debris by filtering through a 0.45-μm-pore-size filter, the supernatant was loaded onto a 25% (wt/vol) sucrose cushion in phosphate-buffered saline (PBS) and centrifuged for 30 min in a TLA 100.3 rotor (Beckman) at 100,000 rpm. The pellet was resuspended in lysis buffer B (1% Triton X-100, 15% sodium deoxycholate, 0.15 M NaCl, 0.1% sodium dodecyl sulfate [SDS], 0.05 M Tris, pH 7.5), and M-PMV viral proteins were immunoprecipitated with goat anti-M-PMV antiserum. Immunoprecipitates were analyzed by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography.
Cleavage of peptides by M-PMV PR in vitro.
The cleavage of peptides derived from M-PMV cytoplasmic domain sequences was assayed by evaluation of the cleavage products by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Vydac C18 RP column in a methanol-H2O system (61). The peptides used in this study were synthesized by the solid-phase method, purified by RP-HPLC, and analyzed by amino acid analysis and mass spectrometry. The standard reaction conditions for the cleavage of peptides were as follows: 50 mM sodium acetate, pH 5.3, 0.3 M NaCl, 0.05% β-mercaptoethanol, 6 mM EDTA, 330 μM peptide, and 0.6 μM protease in 120 μl, overnight at 37°C. The peptide products generated by protease (PR) were characterized by amino acid composition analysis. In experiments for determination of kinetic constants Km and kcat, the concentration of peptides was varied between 25 and 1,500 μM and the reaction mixture was incubated for 1 h.
Single-round virus infectivity assay.
The single-round infectivity of M-PMV molecular clones was determined as described before (69). Briefly, COS-1 cells in 100-mm-diameter plates were cotransfected with both the glycoprotein expression vector pTMT, containing either the wild-type or the mutant env gene, and pMTΔE. At 48 h after cotransfection, culture supernatants were collected and filtered though a 0.45-μm-pore-size filter. Relative levels of reverse transcriptase (RT) activity were determined for each sample as previously described (12), and the levels of RT were normalized by dilution with complete medium. The normalized supernatants were used to infect HOS-CD4/LTR-hGFP cells with 15 μg/ml Polybrene in duplicate. After 48 h of incubation, cells were washed twice with deficient PBS and 500 μl of PBS-1 mM EDTA was added to resuspend the cells. The resuspended cells were analyzed for the expression of GFP by flow cytometry.
Antibody uptake analysis.
All immunofluorescence-based antibody uptake analyses were done as described previously with some modifications (49). The glycoprotein expression vector pTMT containing either wild-type or mutant env was transfected into COS-1 cells grown on glass coverslips. To determine relative levels of Env surface expression, unfixed cells on the coverslips were washed two times with cold PBS, incubated with goat anti-M-PMV for 30 min on ice, and then washed three times with PBS. They were fixed with methanol-acetic acid (95:5 [vol/vol]) for 30 min at −20°C, washed once in PBS, and then stained with fluorescein isothiocyanate-labeled rabbit anti-goat immunoglobulin G (Molecular Probes) for 30 min at room temperature with rocking. In order to compare levels of internalization of wild-type and mutant Env, COS-1 cells grown on coverslips were washed in PBS at 48 h posttransfection. Cells were then incubated with goat anti-M-PMV at 4°C for 30 min and washed once with PBS. These cells were then incubated at 37°C for 15 min or 30 min with 100 μl of DMEM. Chloroquine (100 μM) was added to prevent lysosomal degradation of endocytosed Env-antibody complexes. Surface and endocytosed goat anti-M-PMV antibody was detected using fluorescein isothiocyanate-labeled rabbit anti-goat immunoglobulin G as described above. All the samples were observed and photographed with an Olympus IX 70 microscope.
RESULTS
Mutant glycoprotein incorporation into virions.
Previously, we demonstrated that amino acid substitutions into Y22 blocked incorporation of glycoprotein into budding virions (68). To test if the alanine-scanning cytoplasmic domain substitutions affected glycoprotein incorporation into virions, each pSARM4 clone, containing either the wild-type or mutated env gene, was transfected into COS-1 cells and, at 48 h posttransfection, the cells were labeled with [3H]leucine for 90 min. Following a 6-h chase in complete medium, virus particles in the supernatants were filtered, pelleted through a 25% sucrose cushion, and then immunoprecipitated as described in Materials and Methods.
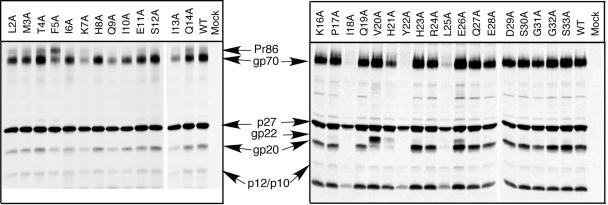
After a 6-h pulse-chase labeling period, the mature products of the env gene (gp70 and gp20) and the gag gene (p27 and p10/p120) can be observed in the viral pellet for the wild type and most of the mutants (Fig. 1). Interestingly, alanine substitutions at amino acid residues I18 and L25, as well as the previously characterized Y22A mutation, block glycoprotein incorporation into virions, as evidenced by the absence of gp70 and gp20 in the virus pellets. In contrast, V20A mutant virions incorporate normal levels of Env proteins, but the amino acid substitution inhibits the viral protease-mediated cleavage of the cytoplasmic residue, so that the bulk of this TM protein is in the form of the uncleaved gp22. The H21A mutation appears to incompletely inhibit both incorporation of glycoprotein into virions and cleavage of the TM protein.
FIG. 1.
Incorporation of mutant glycoprotein into virions. Alanine mutations were substituted for every residue of the cytoplasmic domain of the TM protein from leucine at position 2 to serine at position 33, as described elsewhere (70). Virus-containing supernatants from metabolically labeled COS-1 cells transfected with either wild-type or mutant pSARM4 constructs were centrifuged through a 25% sucrose cushion. The virus pellets were recovered, immunoprecipitated with goat anti-M-PMV serum, and analyzed by SDS-PAGE. The mutant designation is shown above each lane, and the positions of the viral proteins are indicated in the middle.
Consistent with our observations in Env-expressing cells, where cleavage of Pr86 was inhibited and almost equal amounts of gp70 and Pr86 were found at the cell surface (70), the F5A mutant virions incorporate almost equal amounts of precursor glycoprotein and gp70. Similarly, the I13A mutant consistently exhibited lower amounts of gp70 glycoprotein on the plasma membrane when expressed in the context of a glycoprotein expression vector (70), and it was also incorporated at lower levels into virus particles. The K7A and Q9A mutant virions also incorporated decreased levels of glycoprotein into budding virions in the context of a proviral clone. These two mutations increase the fusogenicity of the glycoprotein in cell-cell fusion assays.
As we observed previously for the Y22 substitution mutants that were deficient in Env incorporation (68), the amounts of virions released from cells transfected with both the I18A and the L25A mutants were decreased significantly from that observed with wild type or mutants that incorporated Env at wild-type levels.
Cleavage of peptides by M-PMV PR in vitro.
Both the V20A and the H21A mutations appeared to inhibit cleavage of the TM protein. In order to directly determine the inhibitory effect of these mutations on cleavage by the M-PMV PR, peptides that represented residues 18 to 27 of the cytoplasmic domain, and which contain the region critical for maturational cleavage, were synthesized. One peptide represented the wild-type sequence, while the other two incorporated either the V20A or the H21A substitution. Peptides were cleaved for 1 h at pH 5.3 as described in Materials and Methods. Determination of the kcat/Km ratio gives a measurement of the catalytic efficiency, and it allows one to compare the sensitivities of the peptides with different mutations to the M-PMV PR.
As expected, cleavage of the peptide containing the wild-type sequence by the M-PMV PR occurred between the first histidine and the tyrosine (data not shown). While cleavage occurred at the same site for the mutant peptides, cleavage was severely impaired. The kcat/Km ratio of the wild-type peptide was approximately 19 times higher than those of the peptides with mutations at V20A or H21A, indicating that the mutations introduced into these sites of the cytoplasmic domain decrease their hydrolysis significantly and in this way block maturational cleavage by the viral PR (Table 1). Although the peptides with the V20A or H21A mutation were cleaved at a similar low efficiency (kcat/Km), the maximum rate of hydrolysis (kcat) for the peptide with the H21A mutation was about three times higher than the peptide with V20A mutation, while the peptide with the V20A mutation had a three-times-higher substrate affinity (Km) than the H21A peptide (Table 1). These results indicate that while the underlying mechanisms for the inhibitory effect on cleavage of these mutations are different, both effectively block cleavage of the cytoplasmic domain by the viral PR in virions.
TABLE 1.
Cleavage of gp22-derived peptides
| Mutation | Peptidesa | kcat (s−1) | Km (mM) | kcat/Km (s−1/mM) |
|---|---|---|---|---|
| WT | IQVHYHRLEQ | 0.43 | 0.136 | 3.15 |
| V20A | IQAHYHRLEQ | 0.053 | 0.299 | 0.18 |
| H21A | IQVAYHRLEQ | 0.175 | 1.076 | 0.17 |
Peptides were cleaved by M-PMV PR at pH 5.3 in 0.05 M sodium acetate containing 0.3 M NaCl, 0.05% β-mercaptoethanol. The mutation is shown in boldface in each of the mutant peptides.
Analysis of single-round infectivity by GFP expression.
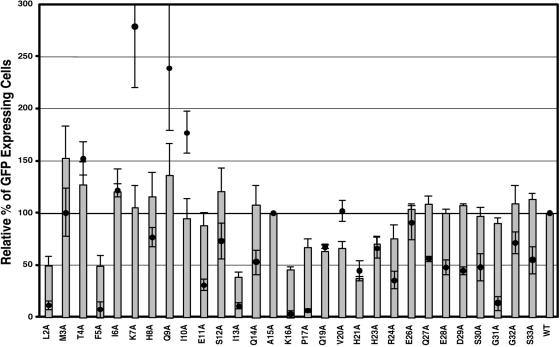
In order to analyze the effect of the mutations introduced into the cytoplasmic domain on virus infectivity, a complementation assay was used. In this single-round infection assay the mutant Envs were assayed for their ability to complement the pMTΔE vector, which encodes the tat gene of HIV-1 in place of the M-PMV env gene. The pTMT and pMTΔE vectors were cotransfected into COS-1 cells, and 2 days after transfection the culture supernatants were assayed for RT activity as described previously. Equivalent amounts of RT-containing supernatant were used to infect GHOST cells, and 2 days postinfection the cells were analyzed by flow cytometry for the number of the GFP-positive cells. As a negative control, the assay was also performed by transfecting COS-1 cells with either the pMTΔE vector or pTMT vector alone, to measure the background GFP expression level. Because the I18A, Y22A, and L25A mutants did not incorporate glycoprotein at detectable levels, we did not measure the infectivities of these mutants (Fig. 2).
FIG. 2.
Viral entry and single-round infectivity assay. COS-1 cells were cotransfected with pTMT and pMTΔEnv expression vectors as described in Materials and Methods. At 48 h posttransfection, culture medium from cells expressing virus was filtered and normalized for reverse transcriptase activity. The normalized medium was used to infect HOS-CD4/LTR-hGFP cells. The number of GFP-expressing cells was quantitated by fluorescence-activated cell sorter analysis. The mean percentage (± the standard deviation) of GFP-expressing cells relative to wild type from three independent experiments is shown for each of the mutants (gray bars). Superimposed on the infectivity data are the fusogenicity data for each mutant in the context of a full-length Env protein expression vector (70) (filled circles).
None of the mutant Env proteins was completely defective at mediating virus entry. The most defective mutants (L2A, F5A, I13A, K16A, and P17A) retained approximately 40% of the infectivity of virus containing the wild-type Env. This contrasts with results from fusion assays with the unprocessed Env protein alone, in which these mutants were highly defective for fusion (70) (Fig. 2). This was also observed for the bulk of the mutations (G31A in particular) that are C-terminal to the maturational cleavage site within the TM protein. These mutant viruses, for the most part, exhibit infectivity equivalent to wild-type virus. This argues strongly that the detrimental effects of these downstream mutations are reversible, are only observed in Env proteins with a full-length gp22 protein, and are lost on cytoplasmic domain cleavage. The E11A and P17A mutations with low fusogenicity (31% and 8%) in the context of Env alone also mediate efficient infectivity (90% and 73%) similar to that of the wild-type virus, suggesting that even for these more-membrane-proximal mutations, CT cleavage modulates the effect of the alanine substitution. Interestingly, the T4A, K7A, Q9A, and I10A mutants, which showed increased levels of fusogenicity in the cell-cell fusion assay (1.5- to 2.75-fold that of wild type), with the Q9A and I10A mutants inducing large syncytium formation in the context of full-length Env protein, only mediate wild-type levels of infectivity.
Virions containing the V20A mutation are infectious in this single-cycle assay. The facts that this mutant is as fusogenic as the wild-type Env when expressed from the pTMT vector alone and mediates moderate levels (60%) of infectivity even though maturational cleavage of the TM protein is blocked in the virus argue that complete protease mediated-cleavage of the cytoplasmic domain is not essential for virus-cell fusion. The H21A mutant, which partially blocks incorporation and maturational cleavage of the glycoprotein, has an intermediate level of infectivity. Three mutants, Q19A, H23A, and R24A, which have lower levels of infectivity compared with the wild type, are located in the region encompassing the residues (I18-L25) required for incorporation, suggesting that a subtle change in Env-MA interactions might be the basis for the reduced biological activity.
Effects of mutations in cytoplasmic residues on endocytosis of the M-PMV Env protein.
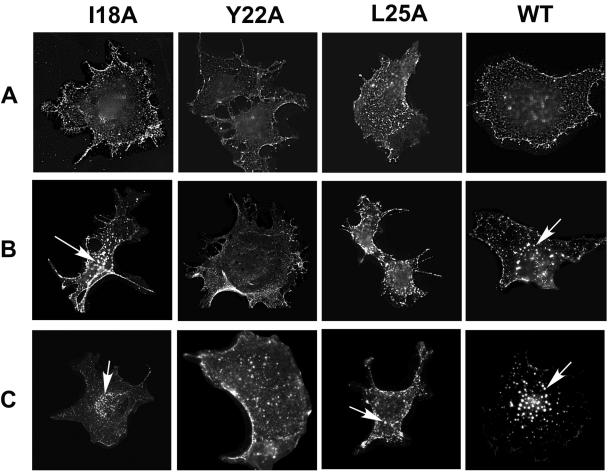
To corroborate our previous surface biotinylation data, which indicated that the I18A, Y22A, and L25A mutant viral glycoproteins are expressed on the cell surface (70), and to determine if these three mutations that block Env incorporation also play a role in the endocytosis of the M-PMV Env protein, we employed an antibody uptake assay. COS-1 cells grown on glass coverslips were transfected with pTMT vectors encoding either the wild-type or mutant env genes. Three sets of transfected COS-1 cells were incubated with goat anti-M-PMV antiserum for 30 min on ice. One set of COS-1 cells was fixed to detect steady-state Env expression on the cell surface, and the other two sets of cells were then shifted to 37°C in the presence of chloroquine for an additional 15 min and 30 min, respectively, to measure internalization of surface-bound antibodies.
In the steady-state surface staining, the wild-type and the all the other mutant glycoproteins tested showed a similar surface staining pattern (Fig. 3A). An analysis of the uptake of the surface-bound goat anti-M-PMV antibody showed dramatic differences in the manner by which the different glycoproteins were internalized. After a 15-min period of antibody uptake, the wild-type and I18A Env showed reduced surface immunofluorescence and the appearance of brightly staining vesicular structures (Fig. 3B). By 30 min the bulk of the immunofluorescent staining was localized to these perinuclear vesicular structures (Fig. 3C). In contrast, the Y22A- and L25A-expressing cells showed extensive plasma membrane staining and only a few fluorescent endocytic vesicles even after 30 min of incubation at 37°C (Fig. 3C). These data indicate that the tyrosine residue at position 22 and leucine 25 of the cytoplasmic domain of M-PMV Env both play a key role in the endocytosis of the glycoprotein, as well as in the incorporation of Env into virions.
FIG. 3.
Antibody uptake mediated by wild-type (WT) and incorporation-defective mutant glycoproteins in COS-1 cells. (A) Steady-state surface expression of wild-type and mutant env gene products was detected after incubation of the unfixed, pTMT vector-transfected COS-1 cells on ice with goat anti-M-PMV antibody followed by methanol-acetic acid fixation and secondary antibody incubation. (B and C) Internalization of wild-type and mutant Env proteins from the plasma membrane to the prelysosomal vesicles was visualized by incubating Env-expressing cells with goat anti-M-PMV antibody in the presence of chloroquine at 37°C for either 15 min (B) or for 30 min (C); the cells were then fixed and stained as for panel A. Arrows indicate internalized glycoproteins.
Biological effects of double mutations of Q9A and V20A.
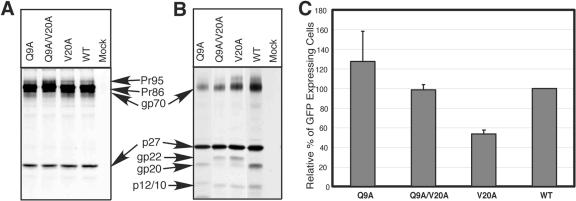
Previously, we described that the Q9A mutation in the cytoplasmic domain of the TM protein induces massive syncytium formation in the cell-cell fusion assay in the absence of cytoplasmic domain cleavage of Env (70). The V20A mutation blocks maturational cleavage of the cytoplasmic domain of the TM protein in the context of the proviral clone and, despite exhibiting wild-type fusion activity in the context of the full-length Env protein, mediates a reduced level (60%) of infectivity in the context of virus. We therefore asked whether the highly fusogenic Q9A mutation could compensate for the inhibitory effect of V20A. The Q9A/V20A mutant env gene was cloned into the M-PMV infectious molecular clone pSARM4, and the effects of the double mutations on glycoprotein incorporation into virions, maturational cleavage, and infectivity were tested as described above.
The Q9A/V20A mutation, like the Q9A substitution, resulted in decreased levels of Env into virions and, like the V20A mutation, inhibited maturational cleavage of the TM protein (Fig. 4A and B). Nevertheless, this double mutant still induced massive syncytium formation in the context of the Env protein alone or when expressed from the proviral clone (data not shown), and in an infectivity assay the Q9A/V20A double mutant had a comparable level of infectivity as the wild type (Fig. 4C). Taken together, these data indicate that the inhibitory effect of the V20A mutation on infectivity could be suppressed by introducing a second site mutation, Q9A, into the cytoplasmic domain of the M-PMV TM protein.
FIG. 4.
Expression of Q9/V20 mutants in the context of the pSARM4 provirus. WT and mutant proviruses were transfected into COS-1 cells and viral proteins were metabolically labeled as described in Materials and Methods. (A) Cell lysates were immunoprecipitated with goat anti-M-PMV serum and analyzed by SDS-PAGE. The mutant designation is shown above each lane. (B) Virus-containing supernatants from metabolically labeled COS-1 cells transfected with either wild-type or mutant pSARM4 constructs were centrifuged through a 25% sucrose cushion. The virus pellets were recovered, immunoprecipitated with goat anti-M-PMV serum, and analyzed by SDS-PAGE. The positions of the viral bands are indicated. (C) Single-round infectivity assay. COS-1 cells were cotransfected with pTMT and pMTΔEnv expression vectors as described in Materials and Methods. At 48 h posttransfection, culture medium from cells expressing virus was filtered and normalized for reverse transcriptase activity. Normalized medium was used to infect HOS-CD4/LTR-hGFP cells. Infectivity was quantitated by fluorescence-activated cell sorter analysis of the number of GFP-expressing cells.
DISCUSSION
We previously demonstrated that C-terminal truncation of the cytoplasmic domain of the TM glycoprotein and substitution mutations in the Y22 residue in this domain block incorporation of the glycoprotein into budding virions and abrogate infectivity (9, 68). In this study, using alanine-scanning mutagenesis, we have investigated the possibility that additional amino acids in the cytoplasmic domain of the TM protein are involved in the incorporation and function of the viral env gene products.
The cytoplasmic domains of several viral glycoproteins have been shown to play important roles in specifying the site of virus assembly and their own incorporation into virions (51, 63, 64, 85). In the case of HIV-1, the cytoplasmic domain has been implicated in the pathogenic effects of the virus in cell culture (34, 77) and has been shown to play an important role in the incorporation of glycoprotein into virions (16, 22, 24, 29, 31, 37, 39, 46, 50, 60, 79). Studies with both HIV and simian immunodeficiency virus have shown that certain mutations in the matrix domain of the Gag precursors block incorporation of the Env protein into virions during budding (22, 24, 38, 40, 45, 50, 75). In alphaviruses, the cytoplasmic domain of the spike glycoprotein has been shown to interact with a hydrophobic cavity created by aromatic amino acids in the nucleocapsid protein, thereby enabling the incorporation of the spike protein and facilitating virus budding (1, 72).
The betaretroviruses, exemplified in this study by M-PMV, employ a morphogenic process that is distinct from that of the other retroviruses that assemble at the plasma membrane. Immature, preassembled intracytoplasmic capsids of M-PMV migrate from their sites of assembly within the cytoplasm to the plasma membrane (28, 57-59, 62, 65, 71, 82), and previous studies from this laboratory have shown that this process is dependent on Env and vesicular transport (64, 65). These data argues that the initial interactions between the cytoplasmic domain of the TM protein and the MA domain of Gag occur intracellularly and may influence the incorporation of Env during budding from the plasma membrane.
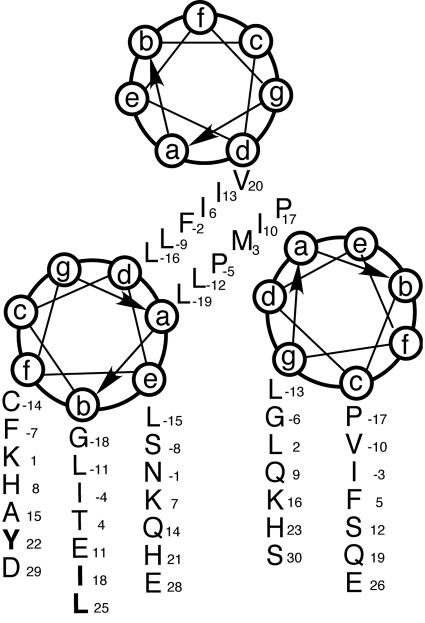
In order to investigate the ability of the mutated glycoproteins to be incorporated into virus, the mutated env genes were cloned into an infectious molecular clone, pSARM4. The results of these studies showed that mutation of key residues, I18 and L25, as well as the previously reported Y22 resulted in the abrogation of glycoprotein incorporation into virions. Structure predictions of the cytoplasmic domain of the M-PMV TM protein indicate that there is a high potential for the membrane-spanning domain and the N-terminal half of this region to form an alpha-helical coiled-coil structure. Secondary structure predictions of the cytoplasmic domain coupled with the positions of the three amino acid residues (I18, Y22, and L25) would place all three on the same face (position b and f) of the helix (Fig. 5). Thus, in the Env trimer, these residues would face outward and be in a position to interact with each monomer of an MA trimer as well as components of the cellular endocytosis machinery.
FIG. 5.
Helical wheel presentation of the membrane-spanning domain and the N-terminal 30 residues of the cytoplasmic domain of the M-PMV TM protein. Amino acid residues comprising the membrane-spanning domain are designated by negative numbers, and those that are critical for glycoprotein incorporation are shown in bold.
Previously, our laboratory demonstrated that the Env protein colocalizes with Gag at a pericentriolar assembly site and that it must be transported to the pericentriolar recycling endosome in order to initiate export of the immature capsids from this site (64, 65). Trafficking of the viral glycoproteins to the recycling endosomes can occur by retrieval from the cell surface through endocytosis or directly from the Golgi complex (42). These previous experiments argue strongly that endocytic trafficking of M-PMV Env glycoprotein is required for the efficient cytoplasmic transport of the capsids. Interestingly, out of the three amino acid residues (I18, Y22, and L25) identified as critical for the glycoprotein incorporation, the Y22 and L25 amino acids are part of a well-conserved tyrosine-based endocytosis signal motif—YXXL (16, 31, 48, 49, 68, 75). Antibody uptake assays demonstrated that substitution of Y22 or L25 with alanine abrogated internalization of the glycoprotein (Fig. 3) and that in the absence of this internalization both intracellular transport of Gag and incorporation of Env were inhibited. Because wild-type levels of Env are found on the plasma membrane for both the Y22A and L25A mutants, it is clear that simply being present at the plasma membrane is insufficient for incorporation, and it appears that the intracellular interactions we have observed between immature capsids and Env are critical for directing the capsids to Env-enriched membrane regions, where incorporation can occur. Env-containing recycling endosomes, cotransported with capsids to the plasma membrane, might be the source of such membranes, and it is likely that Y22 and L25 play a critical role in this intracellular trafficking program. Indeed, in the absence of such a mechanism it is difficult to explain how Env can be excluded from the Y22A and L25A mutant virions.
It is not clear whether Y22 and L25 play any role other than trafficking of Env to the correct intracellular compartment and ultimately the correct membrane domain for incorporation, since their function in endocytosis precedes any potential interaction with immature capsids. However, I18 does not affect endocytosis of Env and is predicted to be located on the outer face of the helical coiled-coil predicted for this region. Mutation of this residue to alanine also interferes with intracellular transport of immature capsids as well as Env incorporation and so is most likely to be involved in direct interactions with the matrix domain of immature capsids. Second-site reversion experiments similar to those carried out with HIV (45) should allow us to confirm this hypothesis.
It is becoming evident for other retroviruses that the plasma membrane is not the only place where Env and Gag interact. Recently, HIV-1, which encodes several highly conserved endocytosis motifs in the cytoplasmic domain, was shown to bud directly into a compartment of the late endosome known as the multivesicular body in both primary monocyte-derived macrophages and tissue culture cell lines (47, 52, 54, 66). Proline-rich motifs, called late domains, have been identified in the Gag proteins of a number of retroviruses (4, 25, 78, 80), rhabdoviruses (32), and filoviruses (27, 84) and appear to play a critical role in viral budding. In M-PMV, release of viral particles from the plasma membrane requires interactions between a late domain (PPPY motif) and a NEDD4-related component of the ESCRT machinery (14, 41, 73, 81, 83). Thus, trafficking of Env and Gag through endosomal compartments could also provide spatial access to the cell-encoded protein complexes that play a critical role in the final stages of viral budding. Taken together, the data presented here strongly suggest that the endocytosis of M-PMV Env is intricately interconnected with the incorporation of glycoprotein into virions and efficient cytoplasmic transport of the immature capsids for virus budding.
Alanine substitution of two other amino acid residues, valine at position 20 (V20A) and histidine at position 21 (H21A), inhibited viral protease-mediated cleavage of the glycoprotein during the maturational process. These residues represent the P1 (H21) and P2 (V20) positions of the protease cleavage site within the cytoplasmic domain. The analysis of in vitro cleavage of peptides corresponding to this region demonstrated that both substitutions inhibit protease cleavage at this site by reducing catalytic efficiency almost 20-fold. The V20A mutant showed high fusogenicity as a full-length glycoprotein, and though this mutation prevented cleavage of the cytoplasmic domain when expressed in the context of the provirus, it still retained approximately 60% of the infectivity of wild-type virus. Construction of a Q9A/V20A double mutant restored infectivity to that of the wild type. These observations argue that complete protease-mediated cleavage of the TM protein is not essential for virus-cell fusion. However, neither V20A nor H21A block TM cleavage completely, and we cannot rule out the possibility that the presence of one cleaved TM protein in the TM trimers is sufficient to activate fusogenicity, as has been shown for MuLV (56, 88). In MuLV, mutation of the protease cleavage site did not affect Env incorporation but did reduce virus infectivity (33, 55, 86). It will be interesting to determine whether other substitutions within V20 or H21, which block cleavage completely, interfere with M-PMV infectivity.
Some of the Env mutations (L2A, F5A, I13A, K16A, P17A, and G31A) resulted in very low levels of fusogenicity when expressed as full-length glycoproteins but exerted only a minor effect on infectivity in the context of the virus. The fusion inhibitory effects of the G31A mutation would be expected to be lost in the context of mature virus, since this region of the TM is absent following maturational cleavage. Less expected, however, was the increase in biological activity observed with the other mutants, which exhibited infectivities of about 40% to 60% that of wild type in the context of virions. These results would argue that the maturational cleavage releases structural constraints that are present in the full-length TM protein.
In conclusion, the results of the study presented here demonstrate that the cytoplasmic domain of M-PMV Env plays a multifunctional role in virus replication. The identification of additional amino acid residues that are critical for the incorporation of the M-PMV glycoprotein into budding virions points to specific protein-protein interactions within the cell as an essential prerequisite for this process. Further analyses aimed at defining the nature of this molecular interaction at the level of both Env and Gag will be essential for understanding the cell biology of M-PMV assembly.
Acknowledgments
We thank Susan Dubay for her valuable advice on experiments and Tshana Thomas for excellent technical assistance.
This work was supported by grant CA-27834 from the National Institutes of Health to E.H. and by grant IAA4055304 from the Grant Agency of the Academy of Sciences of the Czech Republic to I.P. Immunofluorescence analysis was performed at the High Resolution Image Facility. Fluorescence-activated cell sorter analysis was performed at the Flow Cytometry Core Facilities of the University of Alabama at Birmingham Center for AIDS Research (supported by NIH grant P30-AI-27767).
REFERENCES
- 1.Barth, B. U., M. Suomalainen, P. Liljestrom, and H. Garoff. 1992. Alphavirus assembly and entry: role of the cytoplasmic tail of the E1 spike subunit. J. Virol. 66:7560-7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, H. B., S. P. Tucker, S. R. Kar, S. A. McPherson, D. T. McPherson, J. W. Dubay, J. Lebowitz, R. W. Compans, and E. Hunter. 1995. Oligomerization of the hydrophobic heptad repeat of gp41. J. Virol. 69:2745-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot, G., K. Janvier, S. Le Panse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for Env incorporation into virions and infectivity. J. Virol. 77:6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. (Erratum, 78:4383, 2004.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradac, J., and E. Hunter. 1986. Polypeptides of Mason-Pfizer monkey virus. II. Synthesis and processing of the env gene products. Virology 150:491-502. [DOI] [PubMed] [Google Scholar]
- 6.Bradac, J. A., and E. Hunter. 1986. Polypeptides of Mason-Pfizer monkey virus. III. Translational order of proteins on the gag and env gene specified precursor polypeptides. Virology 150:503-508. [DOI] [PubMed] [Google Scholar]
- 7.Brody, B. A., and E. Hunter. 1992. Mutations within the env gene of Mason-Pfizer monkey virus: effects on protein transport and SU-TM association. J. Virol. 66:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brody, B. A., M. G. Kimball, and E. Hunter. 1994. Mutations within the transmembrane glycoprotein of Mason-Pfizer monkey virus: loss of SU-TM association and effects on infectivity. Virology 202:673-683. [DOI] [PubMed] [Google Scholar]
- 9.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, B. A., S. S. Rhee, M. A. Sommerfelt, and E. Hunter. 1992. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc. Natl. Acad. Sci. USA 89:3443-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. R. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee, S., J. Bradac, and E. Hunter. 1985. A rapid screening procedure for the isolation of nonconditional replication mutants of Mason-Pfizer monkey virus: identification of a mutant defective in pol. Virology 141:65-76. [DOI] [PubMed] [Google Scholar]
- 13.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conibear, E. 2002. An ESCRT into the endosome. Mol. Cell 10:215-216. [DOI] [PubMed] [Google Scholar]
- 15.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 16.Delamarre, L., C. Pique, A. R. Rosenberg, V. Blot, M. P. Grange, I. Le Blanc, and M. C. Dokhelar. 1999. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J. Virol. 73:9659-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubay, J. W., S. R. Dubay, H. J. Shin, and E. Hunter. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J. Virol. 69:4675-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubay, J. W., S. J. Roberts, B. Brody, and E. Hunter. 1992. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J. Virol. 66:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Einfeld, D., and E. Hunter. 1988. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. USA 85:8688-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez, S. A., A. Burny, and J. L. Affranchino. 1996. Identification of domains in the simian immunodeficiency virus matrix protein essential for assembly and envelope glycoprotein incorporation. J. Virol. 70:6384-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H. D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 27.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong, S., G. Choi, S. Park, A. S. Chung, E. Hunter, and S. S. Rhee. 2001. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 75:2526-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hourioux, C., D. Brand, P. Y. Sizaret, F. Lemiale, S. Lebigot, F. Barin, and P. Roingeard. 2000. Identification of the glycoprotein 41(TM) cytoplasmic tail domains of human immunodeficiency virus type 1 that interact with Pr55Gag particles. AIDS Res. Hum. Retrovir. 16:1141-1147. [DOI] [PubMed] [Google Scholar]
- 30.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 31.Inabe, K., M. Nishizawa, S. Tajima, K. Ikuta, and Y. Aida. 1999. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 73:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 78:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalia, V., S. Sarkar, P. Gupta, and R. C. Montelaro. 2003. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J. Virol. 77:3634-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao, S. M., E. D. Miller, and L. Su. 2001. A leucine zipper motif in the cytoplasmic domain of gp41 is required for HIV-1 replication and pathogenesis in vivo. Virology 289:208-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, F. J., N. Manel, Y. Boublik, J. L. Battini, and M. Sitbon. 2003. Human T-cell leukemia virus type 1 envelope-mediated syncytium formation can be activated in resistant mammalian cell lines by a carboxy-terminal truncation of the envelope cytoplasmic domain. J. Virol. 77:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodge, R., J. P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mammano, F., E. Kondo, J. Sodroski, A. Bukovsky, and H. G. Gottlinger. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 69:3824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manrique, J. M., C. C. Celma, J. L. Affranchino, E. Hunter, and S. A. Gonzalez. 2001. Small variations in the length of the cytoplasmic domain of the simian immunodeficiency virus transmembrane protein drastically affect envelope incorporation and virus entry. AIDS Res. Hum. Retrovir. 17:1615-1624. [DOI] [PubMed] [Google Scholar]
- 40.Manrique, J. M., C. C. Celma, E. Hunter, J. L. Affranchino, and S. A. Gonzalez. 2003. Positive and negative modulation of virus infectivity and envelope glycoprotein incorporation into virions by amino acid substitutions at the N terminus of the simian immunodeficiency virus matrix protein. J. Virol. 77:10881-10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxfield, F. R., and T. E. McGraw. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121-132. [DOI] [PubMed] [Google Scholar]
- 43.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347-52354. [DOI] [PubMed] [Google Scholar]
- 48.Novakovic, S., E. T. Sawai, and K. Radke. 2004. Dileucine and YXXL motifs in the cytoplasmic tail of the bovine leukemia virus transmembrane envelope protein affect protein expression on the cell surface. J. Virol. 78:8301-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochsenbauer, C., S. R. Dubay, and E. Hunter. 2000. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol. Cell. Biol. 20:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owens, R. J., J. W. Dubay, E. Hunter, and R. W. Compans. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 88:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez, L. G., G. L. Davis, and E. Hunter. 1987. Mutants of the Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J. Virol. 61:2981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 55.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rein, A., C. Yang, J. A. Haynes, J. Mirro, and R. W. Compans. 1998. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J. Virol. 72:3432-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee, S. S., H. X. Hui, and E. Hunter. 1990. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J. Virol. 64:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhee, S. S., and E. Hunter. 1991. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport and membrane association of a capsid. EMBO. J. 10:535-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee, S. S., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 60.Ritter, G. D., Jr., G. Yamshchikov, S. J. Cohen, and M. J. Mulligan. 1996. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J. Virol. 70:2669-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rumlova, M., T. Ruml, J. Pohl, and I. Pichova. 2003. Specific in vitro cleavage of Mason-Pfizer monkey virus capsid protein: evidence for a potential role of retroviral protease in early stages of infection. Virology 310:310-318. [DOI] [PubMed] [Google Scholar]
- 62.Sakalian, M., and E. Hunter. 1999. Separate assembly and transport domains within the Gag precursor of Mason-Pfizer monkey virus. J. Virol. 73:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salzwedel, K., J. T. West, Jr., M. J. Mulligan, and E. Hunter. 1998. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J. Virol. 72:7523-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 65.Sfakianos, J. N., R. A. LaCasse, and E. Hunter. 2003. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 4:660-670. [DOI] [PubMed] [Google Scholar]
- 66.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 67.Sommerfelt, M. A., S. R. Petteway, Jr., G. B. Dreyer, and E. Hunter. 1992. Effect of retroviral proteinase inhibitors on Mason-Pfizer monkey virus maturation and transmembrane glycoprotein cleavage. J. Virol. 66:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song, C., S. R. Dubay, and E. Hunter. 2003. A tyrosine motif in the cytoplasmic domain of Mason-Pfizer monkey virus is essential for the incorporation of glycoprotein into virions. J. Virol. 77:5192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song, C., and E. Hunter. 2003. Variable sensitivity to substitutions in the N-terminal heptad repeat of Mason-Pfizer monkey virus transmembrane protein. J. Virol. 77:7779-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song, C., K. Micoli, and E. Hunter. 2005. Activity of the Mason-Pfizer monkey virus fusion protein is modulated by single amino acids in the cytoplasmic tail. J. Virol. 79:11569-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stansell, E., E. Tytler, M. R. Walter, and E. Hunter. 2004. An early stage of Mason-Pfizer monkey virus budding is regulated by the hydrophobicity of the Gag matrix domain core. J. Virol. 78:5023-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suomalainen, M., P. Liljestrom, and H. Garoff. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 74.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wild, C., J. W. Dubay, T. Greenwell, T. Baird, Jr., T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilk, T., T. Pfeiffer, and V. Bosch. 1992. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology 189:167-177. [DOI] [PubMed] [Google Scholar]
- 78.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yasuda, J., and E. Hunter. 2000. Role of matrix protein in the type D retrovirus replication cycle: importance of the arginine residue at position 55. Virology 268:533-538. [DOI] [PubMed] [Google Scholar]
- 83.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu, X., X. Yuan, Z. Matsuda, T. H. Lee, and M. Essex. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J. Virol. 66:4966-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zavorotinskaya, T., and L. M. Albritton. 1999. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. J. Virol. 73:5621-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao, H., B. Lindqvist, H. Garoff, C. H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO. J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao, Y., L. Zhu, C. A. Benedict, D. Chen, W. F. Anderson, and P. M. Cannon. 1998. Functional domains in the retroviral transmembrane protein. J. Virol. 72:5392-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]