Abstract
Massive numbers of palm civets were culled to remove sources for the reemergence of severe acute respiratory syndrome (SARS) in Guangdong Province, China, in January 2004, following SARS coronavirus detection in market animals. The virus was identified in all 91 palm civets and 15 raccoon dogs of animal market origin sampled prior to culling, but not in 1,107 palm civets later sampled at 25 farms, spread over 12 provinces, which were claimed to be the source of traded animals. Twenty-seven novel signature variation residues (SNVs) were identified on the spike gene and were analyzed for their phylogenetic relationships, based on 17 sequences obtained from animals in our study and from other published studies. Analysis indicated that the virus in palm civets at the live-animal market had evolved to infect humans. The evolutionary starting point was a prototype group consisting of three viral sequences of animal origin. Initially, seven SNV sites caused six amino acid changes, at positions 147, 228, 240, 479, 821, and 1080 of the spike protein, to generate low-pathogenicity viruses. One of these was linked to the first SARS patient in the 2003-2004 period. A further 14 SNVs caused 11 amino acid residue changes, at positions 360, 462, 472, 480, 487, 609, 613, 665, 743, 765, and 1163. The resulting high-pathogenicity groups were responsible for infections during the so-called early-phase epidemic of 2003. Finally, the remaining six SNVs caused four amino acid changes, at positions 227, 244, 344, and 778, which resulted in the group of viruses responsible for the global epidemic.
Severe acute respiratory syndrome (SARS), caused by a novel SARS-associated coronavirus (SARS-CoV), initially emerged in southern China in late 2002 and quickly spread worldwide, threatening public health and socioeconomic stability. Since the first reported isolation of a SARS-CoV-like virus from palm civets in an animal market in Guangdong, speculation about the origin of SARS virus has largely focused on the animal species in this region (6). When SARS reemerged in Guangdong in the winter of 2003-2004, provincial officials took aggressive action, ordering a sweep through farms and food markets to destroy any animals that might harbor a SARS-CoV-like virus (16). This strategy was based on research showing that critical genetic sequences are almost identical in viruses sequenced from humans and palm civets, in particular masked palm civets and raccoon dogs (16, 28). However, the dynamics and natural distribution of SARS-CoV-like virus in palm civets from various geographical regions remain unclear (28). The study of the SARS-CoV-like virus in palm civet populations in the Xinyuan animal market and on farms in various geographical regions is thus likely to provide important clues to understanding the transmission, persistence, and evolution of this deadly viral pathogen and to help in the control and prevention of future outbreaks of SARS (28).
MATERIALS AND METHODS
Specimen collection.
Triplicate rectal and throat swabs were taken from palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides) at Xinyuan animal market, Guangzhou, China, in January 2004, prior to culling commencement. Ninety-one civets and 15 raccoon dogs were randomly selected from 18 vendors with booths located in four blocks dedicated to the sale of civets and raccoon dogs. We also collected environmental specimens from these blocks, including 5 animal-cage swabs, 10 cash-table swabs, and 9 wall swabs. Specimens from farm-based animals in other Chinese provinces were collected between January and September 2004. Generally, two farms in each province were sampled. The criteria for selecting farms to sample included that they used to sell animals from a booth at Xinyuan animal market or that they claimed to previously trade >80% of their animals to Guangdong province. In total, 1,107 palm civets from 25 farms in 12 provinces were sampled during 2004 (Fig. 1; see Fig. S1 in the supplemental material). Sterile sampling techniques were practiced to ensure that cross-contamination between animals did not occur (6). Hair samples and photographs of each animal were also taken for further identification or phylogenetic studies, if required. The specimens were suspended in viral transport medium and transported on dry ice to a laboratory within 48 h.
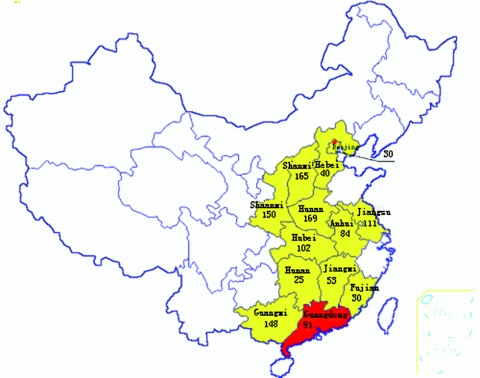
FIG. 1.
Geographic locations and numbers of palm civets sampled in China in 2004.
Real-time and nested RT-PCR amplification.
We used a single-step real-time reverse transcription-PCR (RT-PCR) diagnostic kit (PG Biotech Co. Ltd., Shenzhen, China), with the assay carried out on a Roche Light Cycler instrument. Manual nested RT-PCR was performed for further confirmation. Positive (plasmid provided by the manufacturer) and negative (medium used for sampling) controls were included in each run, and all possible precautions to prevent cross-contamination were taken. A negative control was incorporated for every five nested PCRs to monitor cross-contamination. Only specimens which tested positive for both the N (nucleocapsid protein) and P (replicase) genes of SARS-CoV-like virus by nested RT-PCR were scored as positive.
Quantitative analysis of viral loads in specimens.
The viral loads in throat and rectal swabs from civets were determined using a SARS virus fluorescence quantitative PCR diagnostic kit (Da An Bioengineering Co. Ltd., Guangzhou, China). Each PCR run included a 10-fold dilution series of positive SARS-CoV genomic template controls (for standard curve construction; supplied by the manufacturer), a no-template control for the extraction step to monitor possible contamination during sample processing, and a no-template control for the PCR amplification step. Real-time PCR was performed on an ABI 7000 real-time detection system, with 5 μl of cDNA added to each 50-μl reaction mixture, and the results were analyzed with software provided by the manufacturer (30).
DNA sequencing and analysis.
DNA sequences were determined directly from primary specimens. PCR products were purified from an agarose gel and sequenced at the Beijing Genome Research Center, China, using a 373A DNA sequencer (Applied Biosystems, Foster City, Calif.) and a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems). All sequences were confirmed by sequencing both strands. Comparative analyses of spike (S) gene sequences and protein amino acid sequence predictions were performed using the maximum likelihood methods of PAUP (version 4.0) (7). Published S gene sequences from other viruses were obtained from the GenBank database.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the viral sequences determined for this study, as well as published sequences used in phylogenetic analyses, are provided in Table 1.
TABLE 1.
SARS-CoV-like viruses detected in market animals and patients
| Straina | Source | Location and date of sampling | Proposed group affiliation | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| A030G | Raccoon dog | Animal market, Guangzhou, January 2004 | Prototype | AY687357 | This study |
| A031G | Raccoon dog | Animal market, Guangzhou, January 2004 | Prototype | AY687358 | This study |
| A022G* | Civet | Animal market, Guangzhou, January 2004 | Prototype | AY686863 | This study |
| B029G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY687361 | This study |
| C025G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY687370 | This study |
| A001G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY687354 | This study |
| Civet007G* | Civet | Restaurant TDL, Guangzhou, January 2004 | Low pathogenicity | AY572034 | This study |
| C013G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY687365 | This study |
| C029G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686872 | This study |
| B033G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686862 | This study |
| B040G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686864 | This study |
| C017G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686867 | This study |
| B039G* | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686864 | This study |
| Civet020G* | Civet | Restaurant TDL, Guangzhou, January 2004 | Low pathogenicity | AY572038 | This study |
| A013G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686855 | This study |
| C019G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686869 | This study |
| C014G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686866 | This study |
| C028G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686871 | This study |
| GD03T0013 | Patient | Guangzhou, December 2003 | Low pathogenicity | AY304486 | This study |
| B012G | Civet | Animal market, Guangzhou, January 2004 | Low pathogenicity | AY686869 | This study |
| SZ3* | Civet | Market, Shenzhen, May 2003 | High pathogenicity | AY525636 | 6 |
| SZ16* | Raccoon dog | Market, Shenzhen, May 2003 | High pathogenicity | AY304488 | 6 |
| GD01* | Patient | Guangdong, February 2003 | High pathogenicity | AY278489 | 18 |
| HGZ8L1-A* | Patient | Guangzhou, early phase of epidemic | High pathogenicity | AY394981 | 4 |
| ZS-A* | Patient | Zhongshan, early phase of epidemic | High pathogenicity | AY394997 | 4 |
| ZS-B* | Patient | Zhongshan, early phase of epidemic | High pathogenicity | AY394996 | 4 |
| ZS-C* | Patient | Zhongshan, early phase of epidemic | High pathogenicity | AY395003 | 4 |
| GZ60* | Patient | Guangzhou, February 2003 | High pathogenicity | AY304491 | 6 |
| HGZ8L1-B* | Patient | Guangzhou, February 2003 | High pathogenicity | AY394982 | 4 |
| HKU-36871* | Patient | Hong Kong, March 2003 | High pathogenicity | AY304492 | 20 |
| BJ01* | Patient | Beijing, March 2003 | High pathogenicity | AY278488 | 2 |
| BJ03* | Patient | Beijing, March 2003 | High pathogenicity | AY278490 | 2 |
| CUHK-W1* | Patient | Hong Kong, March 2003 | High pathogenicity | AY278554 | 26 |
| HKU-39849* | Patient | Hong Kong, March 2003 | Epidemic | AY278491 | 32 |
| HKU-65806* | Patient | Hong Kong, March 2003 | Epidemic | AY304493 | 6 |
| CUHK-Su10* | Patient | Amoy Gardens, Hong Kong, March 2003 | Epidemic | AY282752 | 3 |
| Fra* | Patient | Frankfurt, March 2003 | Epidemic | AY310120 | 25 |
| GZ50* | Patient | Guangzhou, March 2003 | Epidemic | AY304495 | 6 |
| Tor2* | Patient | Toronto, March 2003 | Epidemic | AY274119 | 22 |
| Urbani* | Patient | Vietnam, March 2003 | Epidemic | AY278741 | 19 |
*, whole genome sequences were obtained.
RESULTS
Prevalence of SARS-CoV-like virus in palm civets at a live-animal market.
Both real-time and nested RT-PCR for the N and P genes showed that all of the palm civets and raccoon dogs sampled at the Xinyuan animal market prior to culling in January 2004 were positive for a SARS-CoV-like virus. Eighty-four of the 91 palm civets tested positive with both rectal and throat swabs. The other seven palm civets tested positive with throat or rectal swabs only. Of the 15 raccoon dogs, 12 tested positive with both throat and rectal swabs, while 3 tested positive with throat swabs only. Of the 24 environmental specimens, 22 tested positive, including 4 of 4 animal-cage swabs, 8 of 9 cash-table swabs, and 10 of 11 wall swabs that were taken from 16 booths spread over the four blocks of the market where the animals were sampled. For all specimens, the nested RT-PCR results were confirmed by sequencing the PCR fragments of the N and P genes. In addition, 15 S gene and 2 whole-genome sequences of SARS-CoV-like virus were directly determined from palm civet and raccoon dog samples taken from the market. A 29-nucleotide sequence that is deleted in human SARS-CoV was detectable in all of these virus sequences (6).
Investigation of geographic origin of palm civets harboring SARS-CoV-like virus.
To trace the possible geographical origin of SARS-CoV-like virus, we sampled 1,107 palm civets from the provinces the market vendors claimed that market animals had been traded from (Fig. 1; see Fig. S1 in the supplemental material). These provinces included Anhui, Beijing, Fujian, Guangxi, Henan, Hebei, Hubei, Hunan, Jiangsu, Jiangxi, Shanxi, and Shaanxi (Fig. 1). Surprisingly, for the period of January to September 2004, all of the 1,107 civets sampled in other parts of China tested negative for SARS-CoV-like virus (see Fig. S1 in the supplemental material).
Quantitative analysis of SARS-CoV-like virus in palm civets at Xinyuan animal market.
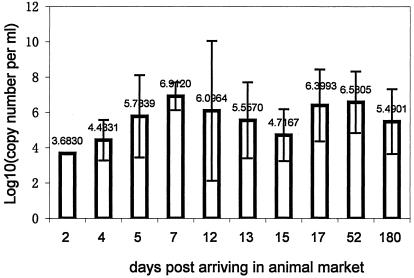
It appeared that the majority of the tested palm civets, if not all, were not infected at farms but most likely were infected at the Xinyuan animal market by other palm civets or as yet unidentified animals harboring the virus. To test this hypothesis, the viral loads in rectal swabs from 76 of the 91 palm civets sampled at the market were quantitatively measured using a real-time PCR assay designed to estimate viral copy numbers in clinical specimens (15, 17). The viral load detected in a rectal swab from one palm civet which had been at the market for 2 days was 103.68. Five palm civets that had been at the market for 4 days had virus loads averaging 104.43. Virus loads peaked on day 7 (106.9) (Fig. 2). Due to the massive clearing action implemented by the local government to remove infection sources, each animal at the Xinyuan animal market could only be sampled once. The palm civets which tested negative for SARS-CoV-like virus in rectal swabs but positive in throat swabs had been at the market for 2, 20 (n = 2), or more than 60 days (n = 2). While it was not common for animals to remain in the market for extended periods of time, there is a possibility that some animals might become reinfected if the market is heavily contaminated.
FIG. 2.
Quantitative measurement of SARS-CoV-like virus in rectal swabs taken from palm civets after their arrival at an animal market, determined by fluorescent real-time RT-PCR. Bars are labeled with the mean viral load for each time point. Error bars represent standard deviations for triplicate determinations.
SNV residue and phylogenetic analysis of S gene sequences.
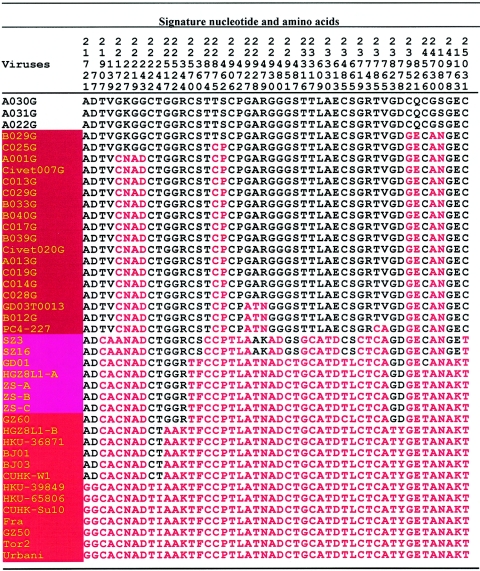
When we compiled the complete spike (S) gene sequences from the 15 market palm civets together with those obtained from 2 raccoon dogs and 2 palm civets captured at a restaurant in January 2004 (M. Wang et al., submitted), recently published sequences from an animal market, 11 virus sequences from the so-called early-phase epidemic, and 7 isolate sequences from the late-phase 2003 epidemic, 27 signature nucleotide variation (SNV) residues were identified (Fig. 3) (4, 21). Of these, 4 were synonymous and 23 were nonsynonymous mutations. Analysis of the 27 SNV residues revealed that one palm civet-derived S gene and two S genes from raccoon dogs had none of the 27 SNVs mutated, representing the prototype virus. All 27 SNV residues from representative isolates responsible for the epidemic of 2003 carried mutations, including HKU-39849, HKU65806, CUHK-Su10, Fra, GZ50, Tor2, and Urbani (Table 1) (6, 9, 13, 17). Isolates from the so-called early- and middle-phase epidemic had 21, 22, or 25 SNV mutations. These included virus sequences from Guangzhou, Hong Kong, Beijing, and other Chinese provinces, namely, GD01, HGZ8L1-A, ZS-A, ZS-B, ZS-C, GZ60, HGZ8L1-B, HKU-36871, BJ01, BJ03, and CUHK-W1 (4, 5). Two viruses of animal origin, SZ3 and SZ16, had mutations in 17 of the 27 residues (6). It should be noted that the S gene sequence (GD03T0013) determined from the first SARS patient identified in winter 2003 had mutations in only 8 of the 27 SNV residues (4). An isogenic strain with the same SNV pattern, B012G, was detected in a palm civet from the Xinyuan animal market (Table 1; Fig. 3 to 5).
FIG. 3.
Signature nucleotide and amino acid variation analysis of spike protein.
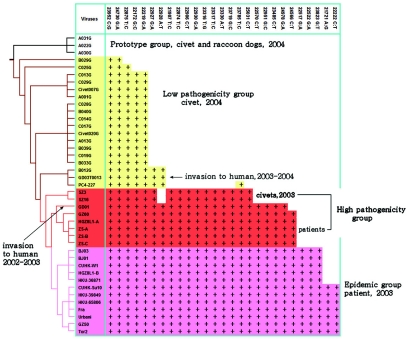
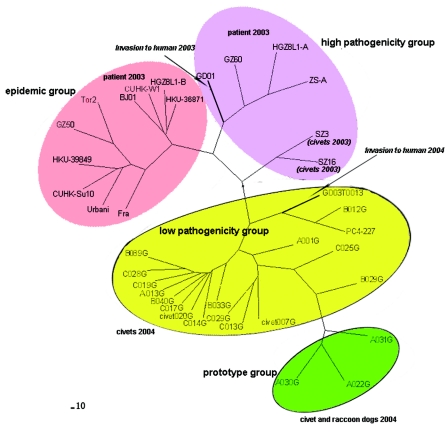
FIG. 5.
The evolution process and signature nucleotide variation of SARS-CoV-like virus, based on spike gene sequences. The phylogenetic tree was constructed using the maximum likelihood methods of PAUP, with the raccoon dog sequence A031G used as the root. The viruses of the prototype group, the low-pathogenicity group, the high-pathogenicity group, and the epidemic group and their SNV patterns are unshaded or shaded in yellow, red, or pink, respectively. Information on various sequences is detailed in Table 1.
To determine the phylogenetic relationships of the SARS-CoV-like viruses, S gene sequences obtained from primary specimens were aligned and analyzed by PAUP software using maximum likelihood. The data indicated that the SARS-CoV-like virus originated from animal virus sequences such as A022G, A030, and A031, of which two were from raccoon dogs and one was from a palm civet (Fig. 3 to 5). When using the spike gene sequences of the raccoon dog strain A031G as the original prototype, the evolutionary process of the virus could be observed. The gradual signature nucleotide variation in various virus sequences matched with the evolution process (Fig. 5). Phylogenetic analysis with whole-genome sequences of SARS-CoV-like virus gave similar results to those obtained with S gene sequences (see Fig. S2 in the supplemental material) (4, 5). The data also indicated that the virus responsible for the first SARS patient in the winter of 2003 (GD03T0013) was identical to a virus sequence from the market (B012G) (Fig. 3 to 5). This patient might thus have caught the virus from palm civets from the Xinyuan animal market. An additional 40 SNV residues were identified when whole-genome sequences of viral RNAs from palm civets were compared with those from patients (see Fig. S3 in the supplemental material). Thirty of the SNVs were located on the gene encoding polyprotein (orf1ab), with others located on genes for the uncharacterized proteins Orf 3 (four SNVs), M protein (two SNVs), ORF 11, ORF 13, and nucleocapsid protein.
DISCUSSION
Palm civets in Guangdong province were first culled during May 2003, when captive wildlife was considered a potential infectious source. These animals reentered Xinyuan animal markets at the end of August 2003, after the ban on selling certain species of farmed wildlife in China was lifted (6). The reappearance of SARS-CoV-like virus in the Xinyuan animal market in winter 2003 represented a significant new risk to humans throughout the world. We demonstrated that the SARS-CoV-like virus was overwhelmingly prevalent in palm civets and raccoon dogs in the market itself (100% of sampled animals were positive for virus). The continued presence of SARS-CoV-like virus in the Xinyuan animal market thus provided a pool where the virus may have persisted and evolved over extended periods of time (4, 28).
Since 75 of the 91 palm civets sampled at Xinyuan market were claimed to have been traded from other provinces, it was reasonable to speculate that the virus was brought into the market by animals infected during the transportation period or on farms. However, SARS-CoV-like virus was not detected in 1,107 palm civets examined from 25 farms in 12 provinces of China which were sampled between January and September 2004 (Fig. 1; see Fig. S1 in the supplemental material). Of the 25 farms, 4 were sampled twice and 1 was sampled three times. Note that we traced market civets back to a farmer who directly distributed animals to markets. His 17 palm civets sampled at the Xinyuan animal market all tested positive for SARS-CoV-like virus. However, all of the palm civets (n = 169) at his farm in Henan province tested negative for SARS-CoV-like virus by the same PCR method. This finding strongly suggested that the palm civets were infected at the market by other palm civets or by other animals harboring the virus rather than at the farm. A recently published serological study detailed a very similar observation (27). While all of 47 civet serum samples collected in early June 2003 from two farms in other provinces were negative for SARS-CoV neutralizing antibodies, 14 of 18 samples obtained from the Xinyuan animal market in January 2004 tested positive. In addition, four animals from a farm in Shanwei, Guangdong, showed low levels of neutralizing antibodies; however, it was claimed that most animals from this farm were obtained from various markets within Guangdong province. It was suggested that the animals had possibly been exposed to SARS-CoV prior to their arrival on the farm (27).
If the animals were infected at the market, then new arrivals should possess a relatively low or nil viral load, and the viral load should increase in a similar manner to that observed during the experimental housing of uninfected animals with an infected animal. Martina and coworkers reported that when noninfected cats and ferrets were housed with an infected animal, transmission occurred, and that viral titers increased gradually after 2 days, peaking on days 6 to 8 posthousing (14). In the present study, animals started shedding virus as early as 2 days after arrival. Note that there were no data for day 1 and that only one animal was observed at the 2-day time point. An average of 104.43 viral copies per ml of specimen was observed for six palm civets which had been at the market for 4 days. The peak virus load was observed in animals which had been at the market for 7 days; this declined by day 15 (Fig. 2). The observation that the transmission of virus to the palm civets at the market reflects the pattern observed for the experimental transmission of SARS-CoV-like virus between laboratory animals indicates that both of these situations may mirror natural virus transmission. When palm civets were experimentally infected, the viral genome was detected by RT-PCR in throat and anal swabs from 3 to 18 days postinoculation (29). Although the virus is rapidly cleared from experimentally infected mice, hamsters, cats, and ferrets, it is possible that it persists in market civets for a longer period of time. Viral genomic RNA could be detected in the spleens and lymph nodes of experimentally infected palm civets for up to 34 or 35 days postinoculation (29). This apparent persistent infection of SARS-CoV-like virus in palm civets supports the idea that they are a major animal reservoir of infection.
It seems that palm civets are extremely susceptible to SARS-CoV and that the Xinyuan animal market was likely the source of infection, where the virus was amplified, circulated, excreted though the respiratory and intestinal tracts of palm civets, and subsequently disseminated to cause sporadic disease in humans. After the palm civets and other highly suspect animals were cleared from the market in January 2004, no further natural human infections were reported. The initial introduction of the virus to the market might have been via a raccoon dog, a palm civet, another as yet unidentified animal, or a combination of these. The zoological biodiversity of the Xinyuan animal market was large, including live donkeys, calves, goats, sheep, piglets, American minks, raccoon dogs, farmed foxes, hog badgers, porcupines, nutria, guinea pigs, rabbits, and birds. Animals were presented in small wire cages piled atop one another, which highly favors the transmission of any pathogens present. The mixing of wild and domestic animals of various species and geographic origins likely further increased the probability of pathogen transmission. Continuing studies should be conducted with a view to monitoring the possible reemergence of virus at the Xinyuan animal market and tracing the original animal reservoir.
Nucleotide signature variation has been observed in various SARS-CoV-like virus sequences and has been put forward as a genetic tool to define distinct genotypes, to trace the sources of infection, and to understand the evolution of the SARS-CoV-like virus. However, all of the data reported so far have focused on human virus sequences, for which only a few SNV residues have been defined (4, 5, 20, 26, 31). Based on SNV patterns, phylogenetic analysis, and the strain-related clinical and epidemiological records available, we divided the SARS-CoV-like viruses into the following four groups: the prototype group, the low-pathogenicity group, the high-pathogenicity group, and the epidemic group (Fig. 3 to 5; Table 2). The prototype group included three virus sequences, two from raccoon dogs and one from a palm civet. We designated it as such because none of the 27 SNV residues of the spike gene were mutated in those virus sequences compared with human isolates (Fig. 3 to 5). In addition, when one of these (A031G) was used as a root to construct a phylogenetic tree using maximum likelihood methods, the three sequences seemed to form a single group from which the other SARS-CoV viruses evolved (Fig. 4 and 5). None of the viruses in this group has been detected in humans, suggesting that they only infect animals.
TABLE 2.
Virological, clinical, and epidemiological features of SARS-CoV groupsa
| Group | Origin | Detection in phases of 2003 epidemic
|
Spike gene analysis result
|
Human infection | Clinical features
|
Epidemiological features
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early | Middle | Late | No. of SNVs mutated | Formation of distinct phylogenetic group(s) | Clinical presentation | Close contact infection | Contact history with patient | Contact history with animal(s) | Responsible for large outbreak | |||
| Prototype | Palm civet, raccoon dog | NA | NA | NA | None | Yes | No | NA | NA | NA | NA | NA |
| Low pathogenicity | Palm civet, patient | NA | NA | NA | 2-7 | Yes | Yes | Mild | None identified | No | Yes | No |
| High pathogenicity | Patient | Yes | No | No | 17-22 | Yes | Yes | Severe | Yes | Yes | Yes | No |
| Epidemic | Patient | No | Yes | Yes | 25-27 | Yes | Yes | Severe | Yes | Yes | No | Yes |
NA, not applicable.
FIG. 4.
Phylogenetic analysis of SARS-CoV-like virus based on S gene sequences using the maximum likelihood methods of PAUP. A bootstrap analysis with 1,000 replicates was used to measure the support of the resulting topologies. The prototype, low-pathogenicity, high-pathogenicity, and epidemic groups of viruses are shaded with various colors. The years and host species of the determined virus sequences are provided. Detailed information regarding those sequences is listed in Table 1.
The low-pathogenicity group included most of the S gene sequences obtained in this study, as well as those responsible for the sporadic infections of winter 2003-2004 and recently published sequences from market palm civets (21). It was designated the low-pathogenicity group for several reasons: first, SNV residues 2 to 7 of the spike gene were mutated; second, it formed a distinct group on the constructed phylogenetic tree; third, the most mutated sequence (GD03T0013, with 7 SNVs) had caused human infection in the 2003-2004 period; and finally, and most importantly, infected patients had only mild clinical presentations and low infectivity, with none of the 113 close contacts of the four patients becoming infected (Table 2) (4, 12). It seems that although members of the low-pathogenicity group can infect both humans and animals, they cannot cause an outbreak, since the SNVs critical for SARS-CoV binding to receptors of Vero cells had not been extensively mutated (23, 24).
The high-pathogenicity group included isolates involved in the so-called early phase of the 2003 epidemic. Based on epidemiological investigations, the 2003 epidemic was divided into early, middle, and late phases by a group of Chinese scientists (4). The early phase was defined as the period from 16 November 2002 to 30 January 2003. It featured 11 index cases that had arisen locally in the absence of any contact history, identified from different geographical locations within Guangdong province (4). Virus isolates or sequences in this group were mainly from patients with contact histories traceable to some of the earliest independent cases in Guangzhou and not those isolated from the late-phase epidemic of 2003. These viruses had 16 to 22 of the 27 SNVs mutated, and they formed a separate group on the phylogenetic tree (Fig. 3 to 5). One of them, GD01, had a 29-nucleotide sequence that was suggested to be a marker for animal isolates that is deleted when viruses adapt to humans (18). Isolates from palm civets and raccoon dogs obtained in 2003 were also included in the high-pathogenicity group, although they have been detected in neither patients nor palm civets from other markets so far. Therefore, this group of viruses may reflect natural virus evolution to human adaptation and may cause severe clinical presentations.
Viruses with 25 or all of the 27 SNV residues mutated on the spike gene were assigned to the epidemic group, since all of them were involved in large outbreaks and in the worldwide epidemic, including BJ01 (Beijing), TW1 (Taiwan), Urbani (Vietnam), TOR-2 (Canada), and HKU-39849 and CUHK-W1 (Hong Kong) (3, 13, 19, 31, 32). These viruses were dominant in the middle and late phases of the 2003 epidemic. The middle phase began on 31 January 2003, when the first major SARS outbreak occurred in a hospital; a superspread event was identified and was associated with >130 primary and secondary infections. The late phase began with the Hotel M outbreak on 21 February 2003 (4). Isolate-related clinical and epidemiological information suggested that viruses in this group were responsible for severe clinical presentations, high infectivity, many close-contact infections, a large number of deaths, large outbreaks, and/or the worldwide epidemic. When adult palm civets were experimentally infected, the epidemic group isolate SARS-CoV BJ01 caused higher average body temperatures and stimulated slightly stronger antibody production than the high-pathogenicity virus GD01, which contains the signature 29-nucleotide sequence (29). It should be noted that body temperature is the only measurable clinical symptom presented by infected civets and other nonhuman animals.
Through the study of the accumulation of SNV residue mutations and phylogenetic analysis data of the S gene, we can observe SARS-CoV-like virus microevolutionary events as they unfold (Fig. 3 to 5). The evolution event seems to have been initiated at nucleotides 23952 (C→G) and 24730 (G→A), resulting in changes at amino acid residues 821 and 1080 in the S2 region of the S protein. The S2 domain contains heptad repeats 1 and 2 (HR1 and HR2). Target cell-bound S protein molecules might change their conformation by associating via HR1 and HR2 interactions to form an oligomeric structure, leading to the fusion of viral and target cell membranes (8). Five additional SNV residue changes resulted in four amino acid substitutions, at residues 147, 228, 240, and 479, of the S protein. In the second stage of evolution, the virus had an additional 14 SNV residues mutated, causing 11 amino acid changes (positions 609, 462, 360, 472, 480, 487, 613, 665, 743, 765, and 1163; Fig. 3 to 5). In the third stage, the remaining six SNV residues were mutated, resulting in four amino acid changes, at positions 344, 778, 227, and 244 (Fig. 3 to 5). Fourteen of the 15 amino acid changes in the second and third stages occurred in the S1 region. It has recently been reported that binding of the S1 domain to its receptor, angiotensin-converting enzyme 2 (ACE2), on host cells is responsible for SARS-CoV entry into cells. Sui and colleagues have suggested that the N-terminal 261 to 672 amino acids of the S protein could compete with soluble ACE2 for an association with the S1 domain (23). Li et al. generated two sets of truncated fragments of the spike protein and identified a fragment of 259 amino acids (amino acids 441 to 700) as the major immunodominant epitope. They speculated that amino acids 1 to 510 of the S glycoprotein represent a unique domain containing the receptor-binding site, analogous to the S1 subunit of other coronaviruses (11). These data suggest that the first 590 amino acids of the SARS-CoV S protein are required for interaction with the surfaces of Vero E6 cells.
SNV residue analysis also provided insights into the mechanism that the SARS-CoV-like virus utilizes to cross the species barrier and cause disease in humans (1). The difference between GD03T0013, the virus sequence determined from the first SARS patient in 2003-2004, and the most closely related animal isolate from a palm civet, C028G, a member of the low-pathogenicity group, is a single amino acid substitution (from K or R to N) at position 479 of the spike protein (Fig. 3) (4). The amino acid variations between GZ01 and SZ3 or SZ16 are at positions 360, 487, 665, and 1163 of the S protein (Fig. 3) (4, 5). These amino acid substitutions may play a critical role in the ability of the SARS-CoV-like virus to jump from animal hosts to humans and cause severe disease (Fig. 4 and 5) (6, 10, 20). However, this hypothesis remains to be confirmed experimentally.
Song et al. have recently reported that SARS-CoV evolves at a relatively constant neutral rate in both humans and palm civets, based on neutral mutation rate analysis of five major coding sequences (orf1ab, S, E, M, and N) of virus sequences from palm civets (2003 and 2004) and human patients (2004) (21). When a rooted phylogenetic tree was constructed, virus sequences obtained from palm civets in 2003 and 2004 were not in the same primary transmission lineage. Therefore, the 2002-2003 epidemic and the 2003-2004 episodes were caused by two independent viral transmissions from animals to humans (21). There is no evidence that SARS-CoV is still circulating in the human population, but it appears to continue to circulate in animals, particularly in palm civets and raccoon dogs (H. Q. Jing, X. G. Jiang, H. Zhen, H. X. Wang, Z. G. Cui, D. Jin, Y. C. Xiao, M. Y. Yan, B. Kan, and J. Xu, unpublished data). Our observations suggest that when SARS-CoV-like virus arrives at an animal market, the majority of palm civets, if not all, will become infected, and that the virus will evolve rapidly in animals to cause disease. Therefore, it is critical to identify the original animal reservoir to remove the continuing threat of SARS.
Supplementary Material
Acknowledgments
This work was supported by a special grant for identifying the animal reservoir of the SARS-CoV-like virus (2003AA208407 to J.G.X.) provided by the Ministry of Science and Technology of China and by a grant from the U.S. National Institutes of Health (NIH U19 AI51915).
We are thankful for the technical support provided by employees of the Beijing Human Genomics Center, China.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Antia, R., R. R. Regoes, J. C. Koella, and C. T. Bergstrom. 2003. The role of evolution in the emergence of infectious diseases. Nature 426:658-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, S., E. Qin, Z. Xu, W. Li, J. Wang, Y. Hu, Y. Liu, S. Duan, J. Hu, Y. Han, J. Xu, Y. Li, Y. Yi, Y. Zhou, W. Lin, H. Xu, R. Li, Z. Zhang, H. Sun, J. Zhu, M. Yu, B. Fan, Q. Wu, W. Lin, L. Tang, B. Yang, G. Li, W. Peng, W. Li, T. Jiang, Y. Deng, B. Liu, J. Shi, Y. Deng, W. Wei, H. Liu, Z. Tong, F. Zhang, Y. Zhang, C. Wang, Y. Li, J. Ye, Y. Gan, J. Ji, X. Li, X. Tian, F. Lu, G. Tan, R. Yang, B. Liu, S. Liu, S. Li, J. Wang, J. Wang, W. Cao, J. Yu, X. Dong, and H. Yang. 2003. Complete genome sequences of the SARS-CoV: the BJ group (isolates BJ01-BJ04). Genomics Proteomics Bioinformatics 1:180-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chim, S. S., S. K. Tsui, K. C. Chan, T. C. Au, E. C. Hung, Y. K. Tong, R. W. Chiu, E. K. Ng, P. K. Chan, C. M. Chu, J. J. Sung, J. S. Tam, K. P. Fung, M. M. Waye, C. Y. Lee, K. Y. Yuen, and Y. M. Lo. 2003. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet 362:1807-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. [DOI] [PubMed] [Google Scholar]
- 5.Guan, Y., J. S. Peiris, B. Zheng, L. L. Poon, K. H. Chan, F. Y. Zeng, C. W. Chan, M. N. Chan, J. D. Chen, K. Y. Chow, C. C. Hon, K. H. Hui, J. Li, V. Y. Li, Y. Wang, S. W. Leung, K. Y. Yuen, and F. C. Leung. 2004. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet 363:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann, L. M., I. Hotzel, W. P. Cheevers, K. P. On Top, G. S. Lewis, and D. P. Knowles. 2004. Seven new ovine progressive pneumonia virus (OPPV) field isolates from Dubois Idaho sheep comprise part of OPPV clade II based on surface envelope glycoprotein (SU) sequences. Virus Res. 102:215-220. [DOI] [PubMed] [Google Scholar]
- 8.Kliger, Y., and E. Y. Levanon. 2003. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiol. 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 10.Li, L., Z. Wang, Y. Lu, Q. Bao, S. Chen, N. Wu, S. Cheng, J. Weng, Y. Zhang, J. Yan, L. Mei, X. Wang, H. Zhu, Y. Yu, M. Zhang, M. Li, J. Yao, Q. Lu, P. Yao, X. Bo, J. Wo, S. Wang, and S. Hu. 2003. Severe acute respiratory syndrome-associated coronavirus genotype and its characterization. Chin. Med. J. 116:1288-1292. [PubMed] [Google Scholar]
- 11.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, G., Q. Chen, J. Xu, Y. Liu, W. Lim, J. S. Peiris, L. J. Anderson, L. Ruan, H. Li, B. Kan, B. Di, P. Cheng, K. H. Chan, D. D. Erdman, S. Gu, X. Yan, W. Liang, D. Zhou, L. Haynes, S. Duan, X. Zhang, H. Zheng, Y. Gao, S. Tong, D. Li, L. Fang, P. Qin, and W. Xu. 2004. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg. Infect. Dis. 10:1774-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 14.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, A. G. Van, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng, E. K., D. S. Hui, K. C. Chan, E. C. Hung, R. W. Chiu, N. Lee, A. Wu, S. S. Chim, Y. K. Tong, J. J. Sung, J. S. Tam, and Y. M. Lo. 2003. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin. Chem. 49:1976-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Normile, D. 2004. Infectious diseases. Viral DNA match spurs China's civet roundup 31. Science 303:292. [DOI] [PubMed] [Google Scholar]
- 17.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin, E., X. He, W. Tian, Y. Liu, W. Li, J. Wen, J. Wang, B. Fan, Q. Wu, G. Chang, W. Cao, Z. Xu, R. Yang, J. Wang, M. Yu, Y. Li, J. Xu, B. Si, Y. Hu, W. Peng, L. Tang, T. Jiang, J. Shi, J. Ji, Y. Zhang, J. Ye, C. Wang, Y. Han, J. Zhou, Y. Deng, X. Li, J. Hu, C. Wang, C. Yan, Q. Zhang, J. Bao, G. Li, W. Chen, L. Fang, C. Li, M. Lei, D. Li, W. Tong, X. Tian, J. Wang, B. Zhang, H. Zhang, Y. Zhang, H. Zhao, X. Zhang, S. Li, X. Cheng, X. Zhang, B. Liu, C. Zeng, S. Li, X. Tan, S. Liu, W. Dong, J. Wang, G. K. Wong, J. Yu, J. Wang, Q. Zhu, and H. Yang. 2003. A genome sequence of novel SARS-CoV isolates: the genotype, GD-Ins29, leads to a hypothesis of viral transmission in South China 2. Genomics Proteomics Bioinformatics 1:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 20.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song, H. D., C. C. Tu, G. W. Zhang, S. Y. Wang, K. Zheng, L. C. Lei, Q. X. Chen, Y. W. Gao, H. Q. Zhou, H. Xiang, H. J. Zheng, S. W. Chern, F. Cheng, C. M. Pan, H. Xuan, S. J. Chen, H. M. Luo, D. H. Zhou, Y. F. Liu, J. F. He, P. Z. Qin, L. H. Li, Y. Q. Ren, W. J. Liang, Y. D. Yu, L. Anderson, M. Wang, R. H. Xu, X. W. Wu, H. Y. Zheng, J. D. Chen, G. Liang, Y. Gao, M. Liao, L. Fang, L. Y. Jiang, H. Li, F. Chen, B. Di, L. J. He, J. Y. Lin, S. Tong, X. Kong, L. Du, P. Hao, H. Tang, A. Bernini, X. J. Yu, O. Spiga, Z. M. Guo, H. Y. Pan, W. Z. He, J. C. Manuguerra, A. Fontanet, A. Danchin, N. Niccolai, Y. X. Li, C. I. Wu, and G. P. Zhao. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 102:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadler, K., V. Masignani, M. Eickmann, S. Becker, S. Abrignani, H. D. Klenk, and R. Rappuoli. 2003. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 1:209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui, J., W. Li, A. Murakami, A. Tamin, L. J. Matthews, S. K. Wong, M. J. Moore, A. S. Tallarico, M. Olurinde, H. Choe, L. J. Anderson, W. J. Bellini, M. Farzan, and W. A. Marasco. 2004. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 101:2536-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sui, J., W. Li, A. Roberts, L. J. Matthews, A. Murakami, L. Vogel, S. K. Wong, K. Subbarao, M. Farzan, and W. A. Marasco. 2005. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 79:5900-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 26.Tsui, S. K., S. S. Chim, and Y. M. Lo. 2003. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N. Engl. J. Med. 349:187-188. [DOI] [PubMed] [Google Scholar]
- 27.Tu, C., G. Crameri, X. Kong, J. Chen, Y. Sun, M. Yu, H. Xiang, X. Xia, S. Liu, T. Ren, Y. Yu, B. T. Eaton, H. Xuan, and L. F. Wang. 2004. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 10:2244-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster, R. G. 2004. Wet markets—a continuing source of severe acute respiratory syndrome and influenza? Lancet 363:234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, D., C. Tu, C. Xin, H. Xuan, Q. Meng, Y. Liu, Y. Yu, Y. Guan, Y. Jiang, X. Yin, G. Crameri, M. Wang, C. Li, S. Liu, M. Liao, L. Feng, H. Xiang, J. Sun, J. Chen, Y. Sun, S. Gu, N. Liu, D. Fu, B. T. Eaton, L. F. Wang, and X. Kong. 2005. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 79:2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, X., G. Cheng, B. Di, A. Yin, Y. He, M. Wang, X. Zhou, L. He, K. Luo, and L. Du. 2003. Establishment of a fluorescent polymerase chain reaction method for the detection of the SARS-associated coronavirus and its clinical application. Chin. Med. J. 116:988-990. [PubMed] [Google Scholar]
- 31.Yeh, S. H., H. Y. Wang, C. Y. Tsai, C. L. Kao, J. Y. Yang, H. W. Liu, I. J. Su, S. F. Tsai, D. S. Chen, and P. J. Chen. 2004. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA 101:2542-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng, F. Y., C. W. Chan, M. N. Chan, J. D. Chen, K. Y. Chow, C. C. Hon, K. H. Hui, J. Li, V. Y. Li, C. Y. Wang, P. Y. Wang, Y. Guan, B. Zheng, L. L. Poon, K. H. Chan, K. Y. Yuen, J. S. Peiris, and F. C. Leung. 2003. The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39). Exp. Biol. Med. (Maywood) 228:866-873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.