Abstract
Exposed seronegative individuals (ES) with persistent high-risk sexual behavior may be less susceptible to human immunodeficiency virus type 1 (HIV-1) infection because they carry the chemokine receptor (CR) gene alleles CCR5 open reading frame (ORF) Δ32, CCR5 promoter −2459G, or CCR2 ORF 64I (CCR2-64I), all of which have been found to diminish HIV-1 infectivity and/or disease progression. To investigate this, we determined the haplotypes for these three genetic loci in 93 ES and 247 low-risk control individuals. To test if protective haplotypes exert their effect by modulating CR expression, we measured the protein expression of CCR5 and CXCR4 on circulating CD4+ T cells and CD14+ monocytes in 71 ES and 92 controls. To avoid investigator bias, the analysis was performed without knowledge of each subject's risk and genotype. The CCR5 −2459G allele was significantly enriched in ES Caucasian men, who constituted the majority (84%) of the ES cohort, compared to the control Caucasian men (P = 0.02). This increase was mostly attributable to a higher frequency of the −2459 A/G versus the −2459 A/A genotype in individuals heterozygous for the Δ32 allele (P = 0.012). No protective influence of the CCR2-64I allele was observed. The haplotypes CCR5 ORF Δ32/CCR5 −2459A (in complete linkage disequilibrium) and CCR5 ORF wt/CCR5 −2459G had a cumulative negative effect on the expression of CCR5, since we measured significantly reduced CCR5 densities on both T-helper cells and monocytes only when both haplotypes were present. Densities of CCR5 on lymphocytes and monocytes were correlated (r = 0.59; P < 0.0001), indicating concordance of CCR5 expression patterns across different cell types. We conclude that the CCR5 ORF Δ32/wt-CCR5 −2459 A/G genotype combination offers an advantage in resisting sexual HIV-1 transmission and that this effect is mediated by a relative paucity of CCR5 on potential target cells of HIV-1.
Human immunodeficiency virus type 1 (HIV-1) requires expression of CD4 in conjunction with a chemokine coreceptor, most frequently CCR5 or CXCR4, to productively infect target cells. Since the discovery of these coreceptors, it has become well established that alterations in their gene expression and function can impact HIV-1 disease progression. For example, inheritance of the chemokine receptor (CR) polymorphisms CCR5 open reading frame (ORF) Δ32, CCR5 promoter −2459 A-to-G, and CCR2 ORF 190 G-to-A (CCR2-64I) are associated with delayed development of the AIDS in HIV-1-infected patients (4, 5, 9, 14, 16, 17, 20, 21, 29, 33, 35, 36, 38, 39, 45, 53, 54). Moreover, individuals homozygous for the CCR5-Δ32 allele, comprising about 1% of the Caucasian population, have strongly reduced susceptibility to R5-dependent HIV-1 infection (7, 26, 49). However, it is clear that the infrequent inheritance of homozygosity for the CCR5-Δ32 allele cannot account for the majority of persons worldwide who are repeatedly exposed to HIV-1 by high-risk activities but remain seronegative and uninfected.
In this study, we investigated the contributions of the CCR5 −2459G and CCR2-64I polymorphisms, as well as heterozygosity for CCR5-Δ32, to protection against HIV-1 infection in a well-defined exposed seronegative (ES) longitudinal cohort (8, 12) in contrast to a low-risk uninfected control group. Whereas CCR5 ORF Δ32/Δ32 homozygosity occurs in just 1% of Caucasians, the CCR5 −2459 A/G plus CCR5 ORF Δ32/wt or CCR2 ORF 64I/wt genotype combinations comprise approximately 5% each. However, the effects of these two genotype combinations on the susceptibility to HIV-1 infection in vivo have not been conclusively determined (6, 7, 13, 14, 17, 22, 27, 30, 31, 37, 49, 53, 55, 61). Here, we have compared the frequencies of the CCR5 ORF wt/Δ32, CCR5 −2459A/G, and CCR2 ORF 64V/I alleles and allele combinations between the ES and the low-risk control group. In addition, we determined the impact of inheriting specific haplotypic combinations of these alleles in our cohorts on the expression of CCR5 on HIV-1 target cells. Our results indicate that inheritance of the CCR5 ORF Δ32/wt plus −2459 A/G genotype combination occurs more commonly in ES than in low-risk control individuals and that these genotypes together may act to reduce target cell susceptibility to HIV-1.
MATERIALS AND METHODS
Study populations.
Ninety-three healthy HIV-1-seronegative individuals ≥18 years old who reported high-risk sexual activity with known HIV-1-infected partners were recruited within metropolitan Seattle for longitudinal study at the Fred Hutchinson Cancer Research Center HIV-1 Vaccine Trials Unit. Enrollment criteria were unprotected sexual intercourse with a known HIV-1-infected person ≥6 times in the previous 6 months or an average of twice weekly ≥4 months within 2 years of enrollment. Prior receipt of an HIV-1 vaccine was an exclusion criterion. Volunteers entering the study were designated ES. A medical history, physical examination, complete blood cell count, T-cell subset analysis, and HIV-1 testing were done at the screening visit to confirm eligibility. HIV-1 infection was excluded by the following tests done at the screening visit and study day 0: HIV-1/2 enzyme-linked immunosorbent assay (ELISA) and Western blot (50), HIV-1 plasma RNA reverse transcription-PCR (Amplicor HIV-1 Monitor; Roche Molecular Systems) (19), and peripheral blood mononuclear cell (PBMC) DNA PCR (41). HIV-1/2 ELISA and Western blot were repeated every 3 months, and HIV-1 PBMC DNA PCR was performed again 4 weeks and 8 weeks after enrollment and then repeated yearly. Clinicians performed HIV-1 risk reduction counseling during each visit. The volunteers completed a questionnaire concerning risk behavior and provided blood during visits scheduled every month over the initial 3 months and every 3 to 6 months thereafter.
A control group of 247 HIV-1-seronegative individuals ≥18 years old reporting HIV low-risk sexual activities were also recruited. Each ES and control volunteer provided written consent prior to enrollment. All aspects of the study were approved by the Institutional Review Board at the University of Washington and the Fred Hutchinson Cancer Research Center.
Genotyping of genetic polymorphisms.
PBMC were isolated by Ficoll-Hypaque density centrifugation, and PCR and DNA restriction fragment length polymorphism analyses were performed as previously described for genotyping of the CCR5 ORF Δ32 (43), CCR5 promoter −2459G (23), and CCR2 ORF 64I (37) polymorphisms (25). The forward primer for CCR2 ORF 64I was slightly modified from the original publication to 5′-TTTTGTGGGCAACATGATGG-3′ to achieve clearer separation of the bands on the gel that are characteristic of the wild type (wt) or the 64I allele.
Flow cytometric analysis of CCR5 and CXCR4.
In preliminary experiments, the saturating concentrations of the anti-CCR5 and anti-CXCR4 monoclonal antibodies were determined (2.5 μg/ml) and used throughout the study. PBMC were incubated with either anti-CCR5 (clone 2D7), anti-CXCR4 (clone 12G5), or immunoglobulin G2a (IgG2a) isotype control (all BD Pharmingen) in phosphate-buffered saline supplemented with 1% bovine serum albumin and 0.1% sodium azide (fluorescence-activated cell sorting [FACS] buffer) for 30 min at 4°C. After two washes in FACS buffer, cells were incubated in fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG Fc F(ab′)2 (1:100; Qifikit; DAKO) for 20 min at 4°C. Cells were washed twice, and a final incubation step was performed with a combination of either phycoerythrin (PE)-conjugated anti-CD3 and allophycocyanin (APC)-conjugated anti-CD4 or anti-CD14-PE and anti-CD8-APC (all BD Pharmingen). 7-Aminoactinomycin D (7-AAD; Sigma) (1 μg/ml) was added during this step to mark dead cells. Two final washes were performed in FACS buffer containing 10 μg/ml actinomycin D (Sigma) to prevent leakage of 7-AAD from dead cells. Cells were fixed in 2% paraformaldehyde containing 5 μg/ml actinomycin D and analyzed on a Calibur flow cytometer (Becton Dickinson) within 5 days after staining. 7-AAD+ dead cells were excluded from the analysis, and the percentages of CCR5- and CXCR4-expressing CD3+ CD4+-T-helper lymphocytes and CD14+ monocytes were determined.
The receptor densities of CCR5 and CXCR4 per cell were quantified by comparing FITC staining of cells to a standard curve, established by analysis of a mixture of five calibrated bead populations coated with five different quantities of immunoglobulins/bead (Qifikit). Beads were incubated with the FITC-conjugated goat anti-mouse IgG Fc F(ab′)2 in parallel to the samples and acquired with identical fluorescence gain and compensation settings. The fluorescence intensity of each bead population was plotted against its designated number of immunoglobulins/bead, and the five data points were interpolated by linear regression. CD3+ CD4+-T-lymphocyte and CD14+ monocyte populations were defined as CCR5+ or CXCR4+ by comparison to the corresponding isotype control, and the regression formula was used to convert the geometric mean of the fluorescence intensity of these CCR5+ or CXCR4+ cell populations to the mean density of receptors per cell.
In vitro HIV-1 infection of CD4+ T lymphoblasts.
The R5-tropic molecular clone HIV-1JR-CSF was used for infectivity studies, grown from the proviral plasmid pYK-JR-CSF (3) (NIH AIDS Research and Reference Reagent Program) by calcium phosphate-mediated transfection of HEK293T cells. Two days after transfection, the cell-free virus supernatant was collected, and the virus titer was determined on multinuclear activation of galactosidase indicator (MAGI) cells (58) transduced with CCR5, which were kindly provided by Michael Emerman (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
Cryopreserved PBMC from ES and control individuals were allowed to rest overnight and then were depleted twice of CD8+ T lymphocytes by incubation with anti-CD8 antibody-coated beads and magnetic column depletion (Miltenyi). CD4-enriched cells were stimulated for 3 days with 1.5 μg/ml phytohemagglutinin (PHA; Remel) in HEPES-buffered RPMI 1640 supplemented with 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM l-glutamine (all Gibco), and 10% heat-inactivated fetal bovine serum (Gemini Bio-Products) (R-10). Following stimulation, nonadherent cells were removed, washed twice, and resuspended in R-10 containing 100 U/ml interleukin-2 (Chiron) (R10-100). PHA-stimulated CD4+-enriched lymphoblasts were incubated with HIV-1JR-CSF at multiplicities of infection (MOI) of 0.003 and 0.006 for 4 h at 37°C. Cells were washed three times and plated in quadruplicates for each MOI into a 96-well round-bottom tissue culture plate at 2 × 105 cells and 200 μl R10-100 per well. On days 3 and 5, 100 μl of medium was replaced. Seven days after infection, supernatants were harvested for HIV-1 p24 determination by ELISA (Perkin-Elmer).
Statistical analysis and specimen blinding.
Proportions of individuals harboring a specific genotype or genotype combination were compared between ES and control individuals by the chi-square test.
Flow cytometric analysis was done without knowledge of the risk assignment of the samples, and data were unblinded only upon completion of the study. In some samples, very low percentages of positive cells were observed, which indicated low receptor expression levels. However, the scarcity of positive cells in these cases did not permit an accurate determination of the receptor density. This problem occurred when the percentage of positive cells was <1%; for the statistical analysis, these samples were assigned a density equal to the mean density of all other values in the group − 2 standard deviations. Among the 163 specimens that were evaluated by flow cytometry, this occurred once each for CCR5 and CXCR4 expression on CD4+ T cells and 19 times and 23 times for CCR5 and CXCR4 expression on monocytes, respectively.
Percentages and densities of CCR5 and CXCR4 expression were compared between multiple groups harboring different genotype combinations using the Kruskal-Wallis statistic. If a significant difference was detected by the Kruskal-Wallis test (KW) (P ≤ 0.05), then individual two groups were compared by Dunn's multiple comparison and Mann-Whitney test. If a difference was significant by the Mann-Whitney test (P ≤ 0.05), but not by the Dunn's test (P > 0.05), which adjusts for multiple comparisons, it was considered a trend. Correlations between lymphocyte densities and either monocyte densities or lymphoblast infectivities were tested by Spearman correlation.
RESULTS
Relative resistance to HIV-1 infection in exposed seronegative individuals with continuing high-risk behavior.
The ES study population consisted of 93 individuals with an identified HIV-1-infected sexual partner who met the enrollment criteria and had no evidence of HIV-1 infection by serology and PCR testing at the time of recruitment. Seventy-eight (84%) reported unprotected anal or vaginal intercourse six times or more during the 6 months prior to enrollment, and 11 (12%) engaged in these activities a minimum of twice weekly for ≥4 months within the past 2 years. The median age at study entry was 37.4 years (range, 20.3 to 60.2 years). Eighty-eight ES were Caucasian (95%), 3 ES were Hispanic, 1 ES was African-American, and 1 ES was American Indian/Alaska Native. Eighty-one ES were male (87%), and of these, 76 (82% of all) ES were men having sex with men (MSM); 12 ES were female (13%). Eleven men and one woman also acknowledged injection drug use either at screening or during follow-up.
The ES individuals were followed for a median of 102.6 weeks. The majority (66%) reported unprotected sexual intercourse with known HIV-1-infected partners during the first 6 months of the study, and 47% followed for ≥1 year continued this practice (Table 1). In addition, approximately one-third reported high-risk sexual activity with partners of unknown HIV-1 status over the course of the study (Table 1). The total number of sexual partners also remained stable over the study period (Table 1). Additional details of high-risk sexual activities were previously described among 46 MSM within this cohort (12).
TABLE 1.
High-risk activities of 93 exposed seronegative individuals
| Parameter | No. of individuals |
|---|---|
| Frequency of unprotected intercourse | |
| With known HIV-1-infected partner since enrollment | |
| During first 6 mo of study (n = 93) | |
| ≥6 | 37 |
| 1-5 | 24 |
| 0 | 32 |
| During last 6 mo of study (n = 75)a | |
| ≥6 | 21 |
| 1-5 | 14 |
| 0 | 40 |
| With partner of unknown HIV-1 status since enrollment | |
| During first 6 mo of study (n = 93) | |
| ≥6 | 22 |
| 1-5 | 12 |
| 0 | 59 |
| During last 6 mo of study (n = 75)a | |
| ≥6 | 13 |
| 1-5 | 12 |
| 0 | 50 |
| No. of sexual partners | |
| During first 6 mo of study (n = 93) | |
| ≥5 | 44 |
| 2-4 | 12 |
| 1 | 32 |
| 0 | 5 |
| During last 6 mo of study (n = 75)a | |
| ≥5 | 35 |
| 2-4 | 7 |
| 1 | 33 |
| 0 | 0 |
Excludes individuals who were observed for <1 year.
Eleven of the 93 ES seroconverted, with an observed seroincidence rate of 3.29 HIV-1 infections per 100 person years (95% confidence interval [CI], 1.35, 5.24). All 11 seroconverters were MSM, which corresponds to 3.94 HIV-1 infections per 100 person years for all 76 MSM in the ES cohort (95% CI, 1.61, 6.26). We observed a lower seroincidence rate in the ES MSM in contrast to MSM reporting unprotected intercourse, regardless of partner HIV-1 serostatus, who enrolled in the Seattle HIVNET Vaccine Preparedness Study (7.68; 95% CI, 4.13, 14.27) (51). Thus, although the ES persistently engaged in high-risk sexual activities, there was a trend toward lower HIV-1 susceptibility during the period of our study than in the contemporaneous Seattle HIVNET MSM cohort.
Increased frequency of the CCR5 −2459 A/G genotype in CCR5 ORF Δ32/wt ES.
We hypothesized that maintenance of HIV-1 seronegativity in our ES cohort could be attributed to an increased prevalence of protective genetic polymorphisms in HIV-1-binding chemokine receptors. To explore this, we compared the CCR5 promoter −2459G, the CCR5-Δ32, and the CCR2-64I allele frequencies in the 93 ES and a control population consisting of 247 persons reporting low-risk sexual activities. The overall CCR5-Δ32 allele frequencies were 14.5% in ES and 9.1% in the controls (P = 0.041; Table 2). This difference was attributable to the higher percentage of Caucasians in our ES cohort (96.7% versus 74.9% in the controls; P < 0.0001), particularly since the CCR5-Δ32 allele frequencies did not differ significantly between ES and controls when only Caucasians (15.0% versus 11.9%; P = 0.308) or Caucasian men (16.0% versus 14.4%; P = 0.673; Table 2) were compared. The CCR2-64I allele frequencies were similar in ES and controls (7.0% versus 9.1%; P = 0.378; Table 2), as well as when stratified by Caucasian ethnicity (6.1% versus 8.4%; P = 0.348) and Caucasian men (4.5% versus 7.2%; P = 0.28; Table 2).
TABLE 2.
Allele frequencies of CCR5 ORF wt/Δ32, CCR5 −2459 A/G, and CCR2 ORF 64V/I in ES and normal-risk control individuals
| Alleles | No. with allele
|
|||||
|---|---|---|---|---|---|---|
| All genders and ethnicities
|
Caucasian men
|
|||||
| ES (n = 93) | Control (n = 247) | Pa | ES (n = 78) | Control (n = 104) | Pa | |
| CCR5 ORF | ||||||
| wt | 159 | 449 | 0.041 | 131 | 178 | 0.673 |
| Δ32 | 27 | 45 | 25 | 30 | ||
| CCR2 ORF | ||||||
| 64V | 173 | 449 | 0.378 | 149 | 193 | 0.28 |
| 64I | 13 | 45 | 7 | 15 | ||
| CCR5 −2459 | ||||||
| A | 100 | 265 | 0.978 | 85 | 130 | 0.124 |
| G | 86 | 229 | 71 | 78 | ||
Chi-square test.
The CCR5 −2459G allele frequencies did not differ significantly between ES and controls when all individuals (46.2% versus 46.4%; P = 0.978; Table 2) or all Caucasians (46.7% versus 42.2%; P = 0.318) were analyzed. However, a trend toward a higher frequency of the protective CCR5 −2459G allele in ES Caucasian men was observed (45.5% versus 37.5%; P = 0.124; Table 2). Since the majority of the ES were Caucasian men (84% versus 42% in the controls), we focused our subsequent analyses of CCR5 −2459 genotypes on Caucasian men from the ES and the control populations. The two genotypes containing one or two CCR5 −2459G alleles, −2459 A/G and −2459 G/G, were more frequent in ES than in control Caucasian men (74.4% versus 57.7%; P = 0.02; Table 3). This increase was reflected primarily by the predominance of the CCR5 −2459 A/G genotype among CCR5 Δ32/wt heterozygote ES subjects. In this case, 74% of Δ32/wt ES subjects carried the −2459 A/G genotype, compared to 37% of Δ32/wt controls (P = 0.012; Table 3). Of note, due to complete linkage disequilibrium between Δ32 and −2459A (35, 39), Δ32/wt heterozygotes cannot carry the −2459 G/G genotype. In contrast, among CCR5 wt/wt individuals, the CCR5 −2459 A/G and −2459 G/G genotypes were not significantly more frequent in ES subjects than in controls (78.6% versus 66.2%; P = 0.122; Table 3). Thus, in comparison to the controls, ES Caucasian men who carried the CCR5-Δ32 allele on one chromosome were more likely to carry the CCR5 −2459G allele on the other chromosome. This observation suggests that the combined CCR5 ORF Δ32/wt plus −2459 A/G genotype confers an advantage in resisting HIV-1 infection.
TABLE 3.
Genotype frequencies of CCR5 ORF Δ32 and CCR5 −2459 in Caucasian men
| Genotype
|
No. with genotype
|
Pa | ||
|---|---|---|---|---|
| CCR5 ORF | CCR5 −2459 | ES (n = 78) | Control (n = 104) | |
| All | A/A | 20 | 44 | 0.02 |
| A/G + G/G | 58 | 60 | ||
| wt/wt | A/A | 12 | 25 | 0.122 |
| A/G + G/G | 44 | 49 | ||
| Δ32/wt | A/Ab | 5 | 19 | 0.012 |
| A/Gb | 14 | 11 | ||
| Δ32/Δ32 | A/Ab | 3 | 0 | NAc |
Chi-square test.
Δ32 is in complete linkage disequilibrium with −2459A.
NA, not applicable.
Association of the combined CCR5 ORF Δ32/wt plus −2459 A/G genotype with lower CCR5 densities on circulating CD4+ T cells and CD14+ monocytes.
The protective effect of the combined CCR5 ORF Δ32/wt plus −2459 A/G genotype may be due to lower expression levels of CCR5 on the surface of mucosal target cells encountered by HIV-1 during sexual transmission (18, 48). Because we were unable to obtain rectal or vaginal tissues in most of our study participants, we assessed PBMC as a surrogate to investigate this hypothesis. By flow cytometry, we determined the percentages of CCR5-expressing CD4+ T cells and CD14+ monocytes as well as their receptor surface densities in 71 ES subjects and 92 controls.
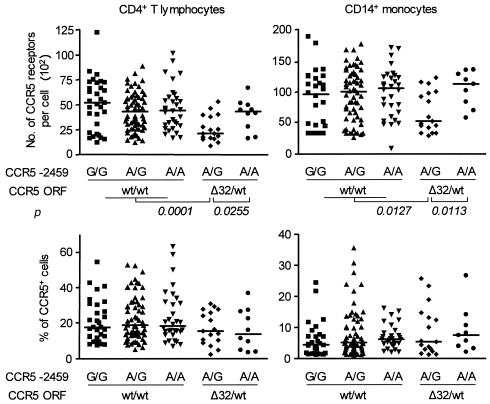
Among all genotype combinations, CCR5 ORF Δ32/wt plus −2459 A/G was associated with lower densities of CCR5 on CD4+ T cells (KW, P = 0.0005) and monocytes (KW, P = 0.0206) that expressed this receptor, but had no effect on the percentage of cells expressing CCR5 (Fig. 1). For CD4+ T cells, CCR5 densities ranged from 854 to 5,284 (median, 2,087) in Δ32/wt plus −2459 A/G individuals, compared to 1,589 to 6,654 (median, 4,296) in Δ32/wt plus −2459 A/A individuals (Mann-Whitney, P = 0.026; Dunn, P > 0.05) and 1,198 to 12,422 (median, 4,490) in all CCR5 ORF wt/wt individuals (Mann-Whitney, P = 0.0001; Dunn, P < 0.001). For CD14+ monocytes, CCR5 densities ranged from 2,948 to 12,266 (median, 5,293) in Δ32/wt plus −2459 A/G individuals, compared to 5,838 to 13,548 (median, 11,241) in Δ32/wt plus −2459 A/A individuals (Mann-Whitney, P = 0.0113; Dunn, P < 0.05) and 918 to 18,863 (median, 10,182) in all CCR5 ORF wt/wt individuals (Mann-Whitney, P = 0.0127; Dunn, P < 0.05).
FIG. 1.
Association between CCR5 ORF plus −2459 genotype combinations and CCR5 expression. Stored peripheral blood CD4+ T cells and CD14+ monocytes from 163 individuals were blinded and analyzed by FACS for the mean number of CCR5 surface receptors per CCR5+ cell and the percentage of CCR5-expressing cells. CCR5 genotypes were determined by restriction fragment length polymorphism analysis. Stratification of CCR5 densities and percentages by CCR5 ORF plus −2459 genotype combinations was performed after completion of all FACS analyses. Horizontal bars represent population medians. P values were determined by Mann-Whitney test. wt, wild type.
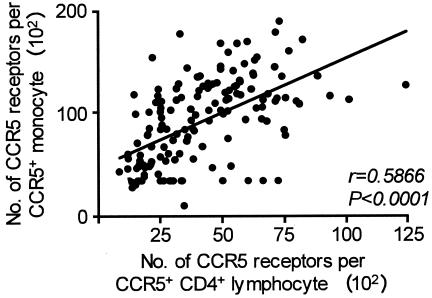
The CCR5 ORF Δ32/wt or −2459 A/G genotype individually had no significant effect on CCR5 densities on the HIV-1 target cells. Likewise, the CCR2-64I allele independently or in the various haplotypic combinations with the other two genomic loci had no influence on CCR5 expression (data not shown). The CCR5 densities on circulating CD4+ T cells and monocytes were positively correlated (Spearman correlation, r = 0.5866; P < 0.0001; Fig. 2). We did not observe a trend toward higher or lower expression of CXCR4 in ES versus controls or between any of the genotypes, with the exception of negligible percentages and densities of CXCR4 on monocytes from Δ32/Δ32 persons (mean percentage of CXCR4+ monocytes, 0.54%; n = 3). Thus, the combined CCR5 ORF Δ32/wt plus −2459 A/G genotype, present in increased frequencies in the ES Caucasian men, is associated with lower expression levels of CCR5 on both circulating CD4+ T cells and CD14+ monocytes.
FIG. 2.
Correlation between T-helper cell and monocyte surface CCR5 densities. Peripheral blood CD4+ T cells and CD14+ monocytes from 163 individuals were analyzed by FACS for the mean number of CCR5 surface receptors per CCR5+ cell. Line fit was done by linear regression. r, Spearman correlation coefficient.
Peripheral CD4+ T lymphoblasts from ORF Δ32/wt plus −2459 A/G individuals do not exhibit increased resistance to direct in vitro infection.
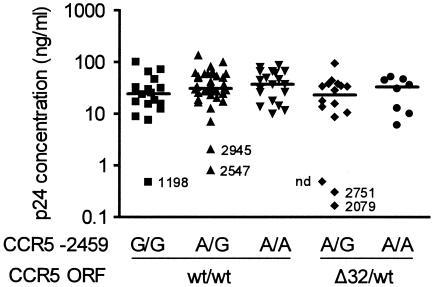
We postulated that the lower expression levels of CCR5 observed in CCR5 ORF Δ32/wt plus −2459 A/G individuals decreased the susceptibility of target cells to infection with R5-tropic HIV-1 strains (1, 18, 24, 44, 48). To screen for this effect, we infected PHA-stimulated CD4+ T lymphoblasts from 77 ES and 25 control individuals with HIV-1JR-CSF and stratified infectivity levels by CCR5 ORF plus −2459 genotype combinations. In preliminary experiments in seven low-risk control donor cells, we determined MOI of 0.003 and 0.006 as optimal viral inocula to detect gradually increasing Gag p24 production over time in cell culture supernatants and with a common peak production at day 7 after infection. For both infecting doses, no overall difference in infectivity was observed between CCR5 ORF Δ32/wt plus −2459 A/G persons and persons harboring other genotype combinations (Fig. 3). Likewise, we did not observe a correlation between the infectivity of CD4+ lymphoblasts and CCR5 densities on circulating T-helper cells obtained from the same individuals at different venipuncture dates (Spearman, r = 0.1585; P = 0.1577; data not shown). However, six donors had markedly lower infectivity levels than the others (Fig. 3). All six possessed at least one CCR5 −2459G allele, and three of the six (50%) harbored the CCR5 ORF Δ32/wt plus −2459 A/G genotype combination. Five of the six donor cells with lower infectivity were also tested for CCR5 expression densities on CD4+ T cells, and these were all below the low 33rd percentile. In addition, lack of CCR5 expression in the three individuals homozygous for the CCR5-Δ32 allele correlated with absence of productive in vitro infection (data not shown). We conclude that the observed relative in vivo resistance to HIV-1 infection of the CCR5 ORF Δ32/wt plus −2459 A/G genotype and its associated decrease in CCR5 expression are not reflected by the in vitro infectivity of PHA-activated CD4+ T lymphoblasts.
FIG. 3.
HIV-1 infectivity of CD4+ lymphoblasts. CD4-selected, PHA-stimulated lymphoblasts from 102 individuals were infected with HIV-1JR-CSF at MOI of 0.006 (not shown) and 0.003 (shown). p24 concentrations in culture supernatants were determined 7 days after infection. Stratification of p24 values by CCR5 genotype combinations was performed after completion of all infection assays. Numbers next to symbols are the mean number of CCR5 surface receptors per cell measured on peripheral blood CCR5+ CD4+ T cells from the same individuals but obtained at different clinic visits from those used to obtain the cells used for the infectivity assays. nd, not done.
DISCUSSION
In contrast to CCR5-Δ32 homozygosity, which confers near 100% protection against HIV-1 infection (7, 26, 49), the benefit of carrying just one Δ32 allele has not been well defined. Published data demonstrate a disease-retarding effect of the CCR5 Δ32/wt genotype in HIV-1-infected individuals (5, 17, 38, 39, 45) but are conflicting as to whether Δ32 heterozygosity is associated with relative resistance to HIV-1 transmission (7, 13, 14, 31, 37, 49, 61). Our findings here demonstrate that a single Δ32 allele exerts a protective effect against viral transmission only if it occurs combined with the −2459G allele in the CCR5 promoter region (33, 35) on the other chromosome. Significantly lower CCR5 expression in these CCR5 Δ32/wt plus −2459 A/G individuals as compared to other genotype combinations suggests that decreased CCR5 expression is the factor limiting viral transmission. In contrast, CCR2 ORF 64I carrier status was not associated with resistance to infection, which is in keeping with previously published data (53), or with diminished CCR5 expression.
The observed enrichment of the CCR5 −2459 A/G genotype within our Δ32/wt Caucasian ES MSM also led to a significant increase of the overall frequency of the CCR5 Δ32/wt plus −2459 A/G genotype combination across the whole ES cohort when compared to all control individuals (P = 0.0478). This was mostly attributable to the markedly increased percentage of Caucasians in our ES cohort (95% in ES versus 75% in controls), since the Δ32 allele is relatively common in Caucasians (∼10%), but rare in other ethnicities (26, 28, 34, 49). It is possible that Caucasians became overrepresented in our ES cohort because they more often than other ethnicities display a relative resistance to infection. Our study shows that part of this ethnic advantage may be related to the increased prevalence of the CCR5 Δ32/wt plus −2459 A/G genotype combination in Caucasians compared to other ethnic groups. For example, among the 65 non-Caucasians in our two cohorts, only one Pacific Islander was a Δ32 heterozygote. Such a low frequency of the Δ32 allele in non-Caucasians is in agreement with other reports (26, 34, 49), and the CCR5 Δ32/wt plus −2459 A/G genotype combination therefore plays a miniscule role at best for resistance to infection in non-Caucasian cohorts, such as exposed seronegative female commercial sex workers in Africa (46).
An enrichment of the CCR5 Δ32/wt plus −2459 A/G combination would be expected in Caucasian ES versus Caucasian controls. We observed 17.9% CCR5 Δ32/wt plus −2459 A/G individuals in ES versus 10.6% in control Caucasians. Although this difference was not statistically significant (P = 0.215), when we compared our ES cohort to much larger Caucasian control cohorts that were reported by others, the enrichment of the CCR5 Δ32/wt plus −2459 A/G genotype combination in our ES cohort was apparent: Gonzales et al. (10), n = 959 and P = 0.0211; the Multicenter AIDS Cohort (MACS) results shown by Tang et al. (55), n = 469 and P = 0.0829; McDermott et al. (35) n = 347 and P = 0.0199; and the three control cohorts combined, n = 1,775 and P = 0.0213. Similar to our ES cohort, Tang et al. have reported a trend for an enrichment of the CCR5 Δ32/wt plus −2459 A/G genotype in the 90 most highly exposed and persistently HIV-1-seronegative men (HEPS) in the MACS cohort (55). If the HEPS and our ES cohort are combined (n = 168) and compared to the combined control cohorts, the level of significance increases further (P = 0.0095). This underscores that the CCR5 Δ32/wt plus −2459 A/G genotype combination is associated with a protective effect against HIV-1 transmission.
Both CCR5-Δ32 heterozygosity and the −2459G allele have been separately linked to lower CCR5 expression on PBMC (1, 2, 40, 48, 52, 57, 59), and the −2459G allele has also been shown to display lower CCR5 promoter activity by in vitro reporter gene analysis (35). In our study, CCR5-Δ32 heterozygosity or −2459G carrier status did not independently affect CCR5 expression. However, the combined CCR5 ORF Δ32/wt and CCR5 −2459 A/G genotype was clearly associated with decreased CCR5 densities on peripheral blood CD4+ T cells and CD14+ monocytes, indicating that both genotypes act cumulatively to restrict CCR5 expression. Of note, the highest CCR5 densities in the CCR5 Δ32/wt plus −2459 A/G group were around the median CCR5 expression measured for the other genotype combinations. This may reflect the finding by Tang et al. that CCR5 Δ32/wt plus −2459 A/G individuals could be further subdivided into two groups with distinct sets of additional genetic polymorphisms, only one of which was associated with resisting infection (55).
CCR5 densities on T cells and monocytes were positively correlated, indicating that trends toward high or low CCR5 expression existed across different cell types in each individual. This suggests that the relative CCR5 expression levels on peripheral blood CD4+ T cells and CD14+ monocytes may be extrapolated to leukocyte populations in other compartments. Thus, in CCR5 ORF Δ32/wt plus −2459 A/G individuals, HIV-1 transmission may be hindered by a relative paucity of CCR5 on mucosal target cells. Several studies have established a relationship between CCR5 cell surface density in various non-mucosal cell populations and susceptibility to infection (24, 40, 44, 45, 56, 60). It would be interesting to actually confirm that mucosal target cells such as intraepithelial Langerhans cells (LCs) express lower CCR5 levels and are less susceptible to infection in CCR5 ORF Δ32/wt plus −2459 A/G individuals. In an elegant study, Kawamura et al. have reported that skin-derived LCs from ORF Δ32/wt plus −2459 A/G individuals were markedly less susceptible to HIV-1 than LCs from ORF Δ32/wt plus −2459 A/A individuals (18). This finding most likely reflects a difference in CCR5 expression between ORF Δ32/wt −2459 A/G and −2459 A/A individuals—as we have seen in our cohorts—although the authors did not evaluate the CCR5 densities of the isolated LCs. Arguably, cell numbers obtained from mucosal or skin tissue samples are mostly too low to perform both receptor density and infectivity studies. Our infectivity experiments with CD4+ lymphoblasts, however, indicate that results obtained with surrogate target cells need to be interpreted with caution. Using PHA-stimulated peripheral blood CD4+ T lymphocytes for screening, we could not detect a difference in infectivity levels between ORF Δ32/wt −2459 A/G and −2459 A/A individuals. It is likely that in vitro hyperactivation overcame the differences that existed in CCR5 promoter activity before stimulation with the mitogen, and infectivity levels in the two groups were therefore equal. This finding also emphasizes the possibility that relative resistance to infection conferred by specific CCR5 genotype combinations can be overcome by cellular activation, as may be the case during sexually transmitted disease-triggered mucosal inflammation (15, 32, 42, 47).
In conclusion, our results indicate that some individuals who practice high-risk sexual behavior may resist infection because they possess the CCR5 Δ32/wt plus −2459 A/G genotype combination, which is associated with relatively weak CCR5 expression. This suggests that a single Δ32 allele can exert a protective effect against viral transmission only if it pairs with the −2459G allele in the CCR5 promoter region of the other chromosome. Despite the enrichment of the CCR5 Δ32/wt plus −2459 A/G genotype combination in our ES cohort, the majority of ES individuals do not carry it and additional mechanisms of resistance, such as the dose of CCR5-binding immune response genes (11), therefore have to be considered.
Acknowledgments
We thank Jean Lee for patient recruitment, Chunhui Wang for help with genotyping, Micky Moerbe for help with data analysis, Ted Gooley for review of the statistical methods, and Marta Bull for editing of the manuscript.
This work was supported by National Institutes of Health grants AI 51980 (F.H.), AI 27757 (The University of Washington CFAR New Investigator Award to F.H. and Clinical Research Core Award to H.L.), AI 45402 (T.Z.), AI 49109 (T.Z.), AI 56994 (T.Z.), and AI 35605 (M.J.M.) and a Burroughs Wellcome Clinical Scientist Award (M.J.M).
REFERENCES
- 1.Agrawal, L., X. Lu, J. Qingwen, Z. VanHorn-Ali, I. V. Nicolescu, D. H. McDermott, P. M. Murphy, and G. Alkhatib. 2004. Role for CCR5Δ32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J. Virol. 78:2277-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benkirane, M., D. Y. Jin, R. F. Chun, R. A. Koup, and K. T. Jeang. 1997. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5delta32. J. Biol. Chem. 272:30603-30606. [DOI] [PubMed] [Google Scholar]
- 3.Cann, A. J., J. A. Zack, A. S. Go, S. J. Arrigo, Y. Koyanagi, P. L. Green, S. Pang, and I. S. Y. Chen. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clegg, A. O., L. J. Ashton, R. A. Biti, P. Badhwar, P. Williamson, J. M. Kaldor, G. J. Stewart et al. 2000. CCR5 promoter polymorphisms, CCR5 59029A and CCR5 59353C, are under represented in HIV-1-infected long-term non-progressors. AIDS 14:103-108. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, O. J., M. Vaccarezza, G. K. Lam, B. F. Baird, K. Wildt, P. M. Murphy, P. A. Zimmerman, T. B. Nutman, C. H. Fox, S. Hoover, J. Adelsberger, M. Baseler, J. Arthos, R. T. Davey, Jr., R. L. Dewar, J. Metcalf, D. J. Schwartzentruber, J. M. Orenstein, S. Buchbinder, A. J. Saah, R. Detels, J. Phair, C. Rinaldo, J. B. Margolick, A. S. Fauci et al. 1997. Heterozygosity for a defective gene for CC chemokine receptor 5 is not the sole determinant for the immunologic and virologic phenotype of HIV-infected long-term nonprogressors. J. Clin. Investig. 100:1581-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contopoulos-Ioannidis, D. G., T. R. O'Brien, J. J. Goedert, P. S. Rosenberg, and J. P. Ioannidis. 2003. Effect of CCR5-delta32 heterozygosity on the risk of perinatal HIV-1 infection: a meta-analysis. J. Acquir. Immune. Defic. Syndr. 32:70-76. [DOI] [PubMed] [Google Scholar]
- 7.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien et al.. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 8.Goh, W. C., J. Markee, R. E. Akridge, M. Meldorf, L. Musey, T. Karchmer, M. Krone, A. Collier, L. Corey, M. Emerman, and M. J. McElrath. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548-557. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, E., M. Bamshad, N. Sato, S. Mummidi, R. Dhanda, G. Catano, S. Cabrera, M. McBride, X. H. Cao, G. Merrill, P. O'Connell, D. W. Bowden, B. I. Freedman, S. A. Anderson, E. A. Walter, J. S. Evans, K. T. Stephan, R. A. Clark, S. Tyagi, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 1999. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. USA 96:12004-12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez, G. Catano, R. J. Nibbs, B. I. Freedman, M. P. Quinones, M. J. Bamshad, K. K. Murthy, B. H. Rovin, W. Bradley, R. A. Clark, S. A. Anderson, R. J. O'Connell, B. K. Agan, S. S. Ahuja, R. Bologna, L. Sen, M. J. Dolan, and S. K. Ahuja. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434-1440. [DOI] [PubMed] [Google Scholar]
- 12.Hladik, F., A. Desbien, J. Lang, L. Wang, Y. Ding, S. Holte, A. Wilson, Y. Xu, M. Moerbe, S. Schmechel, and M. J. McElrath. 2003. Most highly exposed seronegative men lack HIV-1-specific, IFN-gamma-secreting T cells. J. Immunol. 171:2671-2683. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, T. L., R. R. MacGregor, H. Burger, R. Mick, R. W. Doms, and R. G. Collman. 1997. CCR5 genotypes in sexually active couples discordant for human immunodeficiency virus type 1 infection status. J. Infect. Dis. 176:1093-1096. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 15.Humphreys, T. L., C. T. Schnizlein-Bick, B. P. Katz, L. A. Baldridge, A. F. Hood, R. A. Hromas, and S. M. Spinola. 2002. Evolution of the cutaneous immune response to experimental Haemophilus ducreyi infection and its relevance to HIV-1 acquisition. J. Immunol. 169:6316-6323. [DOI] [PubMed] [Google Scholar]
- 16.Ioannidis, J. P., D. G. Contopoulos-Ioannidis, P. S. Rosenberg, J. J. Goedert, A. De Rossi, T. Espanol, L. Frenkel, M. J. Mayaux, M. L. Newell, S. G. Pahwa, C. Rousseau, G. Scarlatti, S. Sei, L. Sen, and T. R. O'Brien. 2003. Effects of CCR5-delta32 and CCR2-64I alleles on disease progression of perinatally HIV-1-infected children: an international meta-analysis. AIDS 17:1631-1638. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis, J. P., P. S. Rosenberg, J. J. Goedert, L. J. Ashton, T. L. Benfield, S. P. Buchbinder, R. A. Coutinho, J. Eugen-Olsen, T. Gallart, T. L. Katzenstein, L. G. Kostrikis, H. Kuipers, L. G. Louie, S. A. Mallal, J. B. Margolick, O. P. Martinez, L. Meyer, N. L. Michael, E. Operskalski, G. Pantaleo, G. P. Rizzardi, H. Schuitemaker, H. W. Sheppard, G. J. Stewart, I. D. Theodorou, H. Ullum, E. Vicenzi, D. Vlahov, D. Wilkinson, C. Workman, J. F. Zagury, and T. R. O'Brien. 2001. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann. Intern. Med. 135:782-795. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura, T., F. O. Gulden, M. Sugaya, D. T. McNamara, D. L. Borris, M. M. Lederman, J. M. Orenstein, P. A. Zimmerman, and A. Blauvelt. 2003. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. USA 100:8401-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khadir, A., F. Coutl'ee, P. Saint Antoine, C. Olivier, H. Voyer, and A. Kessous Elbaz. 1995. Clinical evaluation of Amplicor HIV-1 test for detection of human immunodeficiency virus type 1 proviral DNA in peripheral blood mononuclear cells. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:257-263. [PubMed] [Google Scholar]
- 20.Knudsen, T. B., T. B. Kristiansen, T. L. Katzenstein, and J. Eugen-Olsen. 2001. Adverse effect of the CCR5 promoter −2459A allele on HIV-1 disease progression. J. Med. Virol. 65:441-444. [PubMed] [Google Scholar]
- 21.Kostrikis, L. G., Y. Huang, J. P. Moore, S. M. Wolinsky, L. Zhang, Y. Guo, L. Deutsch, J. Phair, A. U. Neumann, and D. D. Ho. 1998. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 4:350-353. [DOI] [PubMed] [Google Scholar]
- 22.Kostrikis, L. G., A. U. Neumann, B. Thomson, B. T. Korber, P. McHardy, R. Karanicolas, L. Deutsch, Y. Huang, J. F. Lew, K. McIntosh, H. Pollack, W. Borkowsky, H. M. L. Spiegel, P. Palumbo, J. Oleske, A. Bardeguez, K. Luzuriaga, J. Sullivan, S. M. Wolinsky, R. A. Koup, D. D. Ho, and J. P. Moore. 1999. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J. Virol. 73:10264-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristiansen, T. B., T. B. Knudsen, S. Ohlendorff, and J. Eugen-Olsen. 2001. A new multiplex PCR strategy for the simultaneous determination of four genetic polymorphisms affecting HIV-1 disease progression. J. Immunol. Methods 252:147-151. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Y. L., C. Mettling, P. Portales, J. Reynes, J. Clot, and P. Corbeau. 2002. Cell surface CCR5 density determines the postentry efficiency of R5 HIV-1 infection. Proc. Natl. Acad. Sci. USA 99:15590-15595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, H., Y. Hwangbo, S. Holte, J. Lee, C. Wang, N. Kaupp, H. Zhu, C. Celum, L. Corey, M. J. McElrath, and T. Zhu. 2004. Analysis of genetic polymorphisms in CCR5, CCR2, stromal cell-derived factor-1, RANTES, and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin in seronegative individuals repeatedly exposed to HIV-1. J. Infect. Dis. 190:1055-1058. [DOI] [PubMed] [Google Scholar]
- 26.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 27.Louisirirotchanakul, S., H. Liu, A. Roongpisuthipong, E. E. Nakayama, Y. Takebe, T. Shioda, and C. Wasi. 2002. Genetic analysis of HIV-1 discordant couples in Thailand: association of CCR2 64I homozygosity with HIV-1-negative status. J. Acquir. Immune Defic. Syndr. 29:314-315. [DOI] [PubMed] [Google Scholar]
- 28.Lucotte, G. 2002. Frequencies of 32 base pair deletion of the (Delta 32) allele of the CCR5 HIV-1 co-receptor gene in Caucasians: a comparative analysis. Infect. Genet. Evol. 1:201-205. [DOI] [PubMed] [Google Scholar]
- 29.Mangano, A., E. Gonzalez, R. Dhanda, G. Catano, M. Bamshad, A. Bock, R. Duggirala, K. Williams, S. Mummidi, R. A. Clark, S. S. Ahuja, M. J. Dolan, R. Bologna, L. Sen, and S. K. Ahuja. 2001. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J. Infect. Dis. 183:1574-1585. [DOI] [PubMed] [Google Scholar]
- 30.Mangano, A., J. Kopka, M. Batalla, R. Bologna, and L. Sen. 2000. Protective effect of CCR2-64I and not of CCR5-delta32 and SDF1-3′A in pediatric HIV-1 infection. J. Acquir. Immune. Defic. Syndr. 23:52-57. [DOI] [PubMed] [Google Scholar]
- 31.Marmor, M., H. W. Sheppard, D. Donnell, S. Bozeman, C. Celum, S. Buchbinder, B. Koblin, and G. R. Seage III. 2001. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J. Acquir. Immune Defic. Syndr. 27:472-481. [DOI] [PubMed] [Google Scholar]
- 32.Martin, H. L., Jr., P. M. Nyange, B. A. Richardson, L. Lavreys, K. Mandaliya, D. J. Jackson, J. O. Ndinya-Achola, and J. Kreiss. 1998. Hormonal contraception, sexually transmitted diseases, and risk of heterosexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 178:1053-1059. [DOI] [PubMed] [Google Scholar]
- 33.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 34.Martinson, J. J., N. H. Chapman, D. C. Rees, Y. T. Liu, and J. B. Clegg. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 16:100-103. [DOI] [PubMed] [Google Scholar]
- 35.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, and P. M. Murphy. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352:866-870. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, L., M. Magierowska, J. B. Hubert, C. Rouzioux, C. Deveau, F. Sanson, P. Debre, J. F. Delfraissy, I. Theodorou et al. 1997. Early protective effect of CCR-5 delta 32 heterozygosity on HIV-1 disease progression: relationship with viral load. AIDS 11:F73-F78. [DOI] [PubMed] [Google Scholar]
- 37.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. W. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338-340. [DOI] [PubMed] [Google Scholar]
- 38.Mulherin, S. A., T. R. O'Brien, J. P. Ioannidis, J. J. Goedert, S. P. Buchbinder, R. A. Coutinho, B. D. Jamieson, L. Meyer, N. L. Michael, G. Pantaleo, G. P. Rizzardi, H. Schuitemaker, H. W. Sheppard, I. D. Theodorou, D. Vlahov, and P. S. Rosenberg. 2003. Effects of CCR5-Delta32 and CCR2-64I alleles on HIV-1 disease progression: the protection varies with duration of infection. AIDS 17:377-387. [DOI] [PubMed] [Google Scholar]
- 39.Mummidi, S., S. S. Ahuja, E. Gonzalez, S. A. Anderson, E. N. Santiago, K. T. Stephan, F. E. Craig, P. O'Connell, V. Tryon, R. A. Clark, M. J. Dolan, and S. K. Ahuja. 1998. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat. Med. 4:786-793. [DOI] [PubMed] [Google Scholar]
- 40.Ometto, L., M. Zanchetta, A. Cabrelle, G. Esposito, M. Mainardi, L. Chieco-Bianchi, and A. De Rossi. 1999. Restriction of HIV type 1 infection in macrophages heterozygous for a deletion in the CC-chemokine receptor 5 gene. AIDS Res. Hum. Retrovir. 15:1441-1452. [DOI] [PubMed] [Google Scholar]
- 41.Paxton, W. B., R. W. Coombs, M. J. McElrath, M. C. Keefer, J. Hughes, F. Sinangil, D. Chernoff, L. Demeter, B. Williams, L. Corey et al. 1997. Longitudinal analysis of quantitative virologic measures in human immunodeficiency virus-infected subjects with > or = 400 CD4 lymphocytes: implications for applying measurements to individual patients. J. Infect. Dis. 175:247-254. [DOI] [PubMed] [Google Scholar]
- 42.Plummer, F. A., J. N. Simonsen, D. W. Cameron, J. O. Ndinya-Achola, J. K. Kreiss, M. N. Gakinya, P. Waiyaki, M. Cheang, P. Piot, A. R. Ronald et al. 1991. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 163:233-239. [DOI] [PubMed] [Google Scholar]
- 43.Quillent, C., E. Oberlin, J. Braun, D. Rousset, G. Gonzalez-Canali, P. Metais, L. Montagnier, J. L. Virelizier, F. Arenzana-Seisdedos, and A. Beretta. 1998. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 351:14-18. [DOI] [PubMed] [Google Scholar]
- 44.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynes, J., P. Portales, M. Segondy, V. Baillat, P. Andre, B. Reant, O. Avinens, G. Couderc, M. Benkirane, J. Clot, J. F. Eliaou, and P. Corbeau. 2000. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 181:927-932. [DOI] [PubMed] [Google Scholar]
- 46.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Royce, R. A., A. Sena, W. Cates, Jr., and M. S. Cohen. 1997. Sexual transmission of HIV. N. Engl. J. Med. 336:1072-1078. [DOI] [PubMed] [Google Scholar]
- 48.Salkowitz, J. R., S. E. Bruse, H. Meyerson, H. Valdez, D. E. Mosier, C. V. Harding, P. A. Zimmerman, and M. M. Lederman. 2003. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin. Immunol. 108:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 50.Schacker, T., A. C. Collier, J. Hughes, T. Shea, and L. Corey. 1996. Clinical and epidemiologic features of primary HIV infection. Ann. Intern. Med. 125:257-264. [DOI] [PubMed] [Google Scholar]
- 51.Seage, G. R., III, S. E. Holte, D. Metzger, B. A. Koblin, M. Gross, C. Celum, M. Marmor, G. Woody, K. H. Mayer, C. Stevens, F. N. Judson, D. McKirnan, A. Sheon, S. Self, S. P. Buchbinder et al. 2001. Are US populations appropriate for trials of human immunodeficiency virus vaccine? Am. J. Epidemiol. 153:619-627. [DOI] [PubMed] [Google Scholar]
- 52.Shieh, B., Y. E. Liau, P. S. Hsieh, Y. P. Yan, S. T. Wang, and C. Li. 2000. Influence of nucleotide polymorphisms in the CCR2 gene and the CCR5 promoter on the expression of cell surface CCR5 and CXCR4. Int. Immunol. 12:1311-1318. [DOI] [PubMed] [Google Scholar]
- 53.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, and S. J. O'Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 54.Tang, J., C. Rivers, E. Karita, C. Costello, S. Allen, P. N. Fultz, E. E. Schoenbaum, and R. A. Kaslow. 1999. Allelic variants of human beta-chemokine receptor 5 (CCR5) promoter: evolutionary relationships and predictable associations with HIV-1 disease progression. Genes Immun. 1:20-27. [DOI] [PubMed] [Google Scholar]
- 55.Tang, J., B. Shelton, N. J. Makhatadze, Y. Zhang, M. Schaen, L. G. Louie, J. J. Goedert, E. C. Seaberg, J. B. Margolick, J. Mellors, and R. A. Kaslow. 2002. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J. Virol. 76:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuttle, D. L., J. K. Harrison, C. Anders, J. W. Sleasman, and M. M. Goodenow. 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 72:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatesan, S., A. Petrovic, D. I. Van Ryk, M. Locati, D. Weissman, and P. M. Murphy. 2002. Reduced cell surface expression of CCR5 in CCR5Delta 32 heterozygotes is mediated by gene dosage, rather than by receptor sequestration. J. Biol. Chem. 277:2287-2301. [DOI] [PubMed] [Google Scholar]
- 58.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression patterns correlate with infectability by macrophage-tropic HIV-1 in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zucker-Franklin, D., M. Fraig, and G. Grusky. 1995. Interaction of human immunodeficiency virus type 1, human T-cell leukemia/lymphoma virus type I (HTLV-I), and HTLV-II with in vitro-generated dendritic cells. Clin. Diagn. Lab. Immunol. 2:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]