Abstract
Using a C-terminal domain (PCT) of the measles virus (MV) phosphoprotein (P protein) as bait in a yeast two-hybrid screen, a cDNA identical to the recently described human p53-induced-RING-H2 (hPIRH2) cDNA was isolated. A glutathione S-transferase-hPIRH2 fusion protein expressed in bacteria was able to pull down P protein when mixed with an extract from P-expressing HeLa cells in vitro, and myc-tagged hPIRH2 could be reciprocally coimmunoprecipitated with MV P protein from human cells. Additionally, immunoprecipitation experiments demonstrated that hPIRH2-myc, MV P, and nucleocapsid (N) proteins form a ternary complex. The hPIRH2 binding site was mapped to the C-terminal X domain region of the P protein by using a yeast two-hybrid assay. The PCT binding site was mapped on hPIRH2 by using a novel yeast two-hybrid tagged PCR approach and by coimmunoprecipitation of hPIRH2 cysteine mutants and mouse/human PIRH2 chimeras. The hPIRH2 C terminus could mediate the interaction with MV P which was favored by the RING-H2 motif. When coexpressed with an enhanced green fluorescent protein-tagged hPIRH2 protein, MV P alone or in a complex with MV N was able to redistribute hPIRH2 to outside the nucleus, within intracellular aggregates. Finally, MV P efficiently stabilized hPIRH2-myc expression and prevented its ubiquitination in vivo but had no effect on the stability or ubiquitination of an alternative ubiquitin E3 ligase, Mdm2. Thus, MV P protein is the first protein from a pathogen that is able to specifically interact with and stabilize the ubiquitin E3 ligase hPIRH2 by preventing its ubiquitination.
Measles virus (MV) is responsible for an acute childhood disease that, in 2002, infected almost 40 million infants and caused 0.8 million deaths (42). It is an enveloped virus belonging to the Mononegavirales order, the Paramyxoviridae family, and the Morbillivirus genus. The genome is a nonsegmented, negative-strand RNA. It comprises six genes, the genes for nucleoprotein (N) and phosphoprotein (P protein) and matrix (M), fusion (F), hemagglutinin (H), and large (L; polymerase) proteins, in that order, flanked by two short sequences, the leader (Le) and the trailer (Tr), which contain the genome and antigenome promoters, respectively. The MV RNA-dependent RNA polymerase comprising the L and P proteins uses a ribonucleoprotein template composed of the genomic RNA in complex with the N protein. The P protein from Paramyxoviridae acts as a bridge between L and ribonucleoprotein and has partially overlapping binding sites for both L (40) and N proteins. P has multiple functions: (i) it binds to the N-terminal domain of L (7), (ii) it acts as a molecular chaperone and stabilizes the L protein (20), (iii) it places the polymerase complex on the nucleocapsid, (iv) it allows viral transcription and replication, and (v) it prevents the illegitimate encapsidation of cellular RNA by keeping N in a monomeric N° state (22). P is a modular protein with two distinct functional domains, an acidic and intrinsically disordered N-terminal domain (PNT) and a C-terminal domain (PCT) (24). PNT binds to the conserved N-terminal domain of N (NCORE), while PCT binds to the C-terminal domain of N (NTAIL) (7, 19). PNT, which is dispensable for transcription, acts as the chaperone for N° and is required for replication (8, 19, 45). PCT is the most conserved sequence within the Paramyxoviridae family and contains all the regions required for viral transcription. PCT can self-oligomerize in tetramers (19, 44). PCT binding to NTAIL involves the induced helix folding of the N α-MORE region to the compact three-α-helix bundle of the PCT C-terminal X domain (XD) (23, 28, 29, 32). NTAIL protrudes from the surface of the nucleocapsid core made of self-assembled NCORE bound to viral RNA (1, 24, 25).
MV infection of human host cells results from a largely unknown complex interplay between viral and cellular activities. While viral entry through MV H binding to CD46 or CD150 cellular receptors and F-mediated virus envelope fusion to the plasma membrane have been thoroughly investigated (see reference 16 for a review), the cellular mechanisms involved in viral replication, assembly, and budding remain largely unknown. From the cellular point of view, MV infection activates the interferon pathway and induces cytopathic effects, including syncytium formation, cell cycle arrest, and apoptosis (see reference 16 for a review). The physiological consequences of MV infection on the immune system have been intensively investigated because of the MV immunological paradox which associates the induction of a profound cellular immunodepression with a lifelong protective immunity (see reference 16 for a review).
Herein, we report the identification of the first cellular partner of the MV polymerase cofactor P protein, a ubiquitin E3 ligase which has been recently described for mice as androgen receptor N-terminus-interacting protein (ARNIP) and for humans as p53-induced-RING-H2 (PIRH2) protein (2, 30, 31).
MATERIALS AND METHODS
Antibodies, cells, and virus.
The following antibodies were used: anti-MV P homemade rabbit polyclonal antibody 371171 (7) and mouse monoclonal antibody (MAb) 49.21 (7, 36), anti-MV-F Y503 MAb, anti-MV-N MAb cl120 (17), anti-myc MAb 9E10, anti-hemagglutinin (HA) MAb (clone HA-7) (Sigma), anti-Sendai virus P polyclonal Ab PCV1193, and anti-Mdm2 MAb 2A10. Human epithelial HeLa and 293T cells were grown as described previously (47). The Hallé strain of MV and the recombinant vaccinia viruses vv-P (41) and vv-T7 (14), encoding MV P and T7 polymerase, respectively, were amplified as described previously (47).
Plasmid constructs.
The hybrids BD- and AD-MV P, PNT P(1-230), and PCT(231-507) and N proteins subcloned into pGBKT7 or pGADT7 (Clontech) have been described previously (7). Different deletion mutants of PCT, according to modular structure prediction (24, 32), were subcloned in frame with the GAL4-BD domain in pGBKT7. pcDNA3-myc-hPIRH2 was constructed by subcloning mutants into a homemade pcDNA3-myc vector (myc-hPIRH2). pEGFP-hPIRH2, pEGFP-C145/8S, pcDNA4.1-hPIRH2-myc, and point mutant derivatives C102S-myc, C122S-myc, C145/8S-myc, C164S-myc, and C172S-myc have been described previously (31). Mouse PIRH2 (mPIRH2)-myc and chimeric human N-terminal/mouse C-terminal (hN/mC) and mouse N-terminal/human C-terminal (mN/hC) hybrids were amplified using pcDNA3-myc-mARNIP (2) and pcDNA4.1-hPIRH2-myc as templates, respectively, and then subcloned into pcDNA4.1. Sendai virus P protein (Psev) was amplified from pTM1/P (a kind gift from D. Kolakofsky) and subcloned into psc6. psc6-P, psc6-N (37), and pCG-EGFP-P, encoding MV P, N, and enhanced green fluorescent protein (EGFP)-P hybrid proteins (9), respectively, were kindly provided by R. Cattaneo. Glutathione S-transferase (GST)-hPIRH2 (30), pCMV-Mdm2, and HA-ubiquitin (Ub) in pTM123 were kindly provided by A. Puisieux and E. Decroly.
Yeast two-hybrid screening.
BD-PCT was used as a bait protein to screen a human HeLa MATCHMAKER cDNA library cloned into a pGADT7-Rec vector (Clontech) according to the manufacturer's protocol. Positive plasmids were isolated from several independent yeast clones and identified by DNA sequencing to carry full-length human PIRH2 cDNA (AD-hPIRH2). The specificity of the interaction was confirmed by retransforming AD-hPIRH2 into Y187 yeast cells and remating with BD-PCT expressed in AH109 yeast cells, according to a previously reported procedure (7). AD-P, AD-PNT, pGBKT7 (BD), and BD-N were used as controls. Experiments were repeated three times for consistency.
Binding site mapping by Y2H-TPCR screening.
To identify the P binding site on hPIRH2, we adapted a tagged random primer PCR for the screening of cDNA fragment libraries in a yeast two-hybrid assay called Y2H-TPCR (Fig. 1). Full-length hPIRH2 cDNA was amplified using AD-hPIRH2 as the template with specific primers for hPIRH2. The PCR amplicon was purified and used as a template for random PCR amplification of hPIRH2 cDNA fragments using primer A (GACCATGATTACGCCGAATTCNNNNNNNNNNNNNNN) that contains an EcoRI site (italics). Following PCR, random PCR amplicons were purified (Amersham) to remove excess primer A. Amplicons served as PCR templates for further amplification using primer B (GACCATGATTACGCCGAATTC). Amplicons with sizes of ∼250 to 300 bp were purified and subcloned in frame with the C terminus of AD in pGADT7 with EcoRI. This library of hPIRH2 fragments was amplified in Escherichia coli DH5α. The hPIRH2 library was cotransformed with BD-PCT into AH109 yeast using Yeastmaker Yeast Transformation System 2 (Clontech) and streaked onto SD/−Trp/−Leu and SD/−Trp/−Leu/His/−Ade (QDO) plates. Clones growing on QDO-α-galactosidose (α-Gal) plates were recovered by transformation into bacteria, and inserts were sequenced.
FIG. 1.
Principle of Y2H-TPCR screening assay used to determine the P binding site on hPIRH2. Primer A comprises 15 specific nucleotides (GACCATGATTACGCC) noted as short lines at the 5′ end and 15 random nucleotides at the 3′ end (narrow arrow, N15) linked by the EcoRI site in the middle (E, GAATTC). Primer B contains only 15 specific nucleotides with the EcoRI link at the 3′ end.
GST pull-down assay.
Recombinant proteins were expressed and purified as described previously (30) with minor modifications. GST-hPIRH2 and GST were expressed in E. coli BL21(DE3) (Novagen) and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside overnight at room temperature. After cell lysis and sonication, recombinant proteins were affinity purified using glutathione Sepharose 4 fast-flow beads (Amersham Pharmacia Biotech). Approximately 10 μg recombinant proteins or GST alone with beads was added to 100 μl of vv-P-infected HeLa cell lysates and incubated in lysis buffer (50 mM Tris pH 8.0, 5 mM EDTA, 150 mM NaCl, and 0.5% NP-40) for 3 h with agitation. Beads were recovered and washed five times with washing buffer (5% sucrose, 5.0 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.5 M NaCl, and 1% NP-40) by centrifugation at 3,000 rpm for 5 min. Samples were boiled in Laemmli buffer prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using anti-P MAb.
Cell transfection and infection.
Recombinant proteins and small interfering RNA (siRNA) were expressed by transient transfection. HeLa cells were transfected using Lipofectin reagent (Invitrogen). To increase the protein expression level, cells were infected for 1 h with vv-T7 (13) at a multiplicity of infection (MOI) of 5, washed, and then transfected. 293T cells were transfected with calcium phosphate (Sigma) unless otherwise indicated. In some experiments, 5 μM MG132 (Peptide International, Inc.) was added to the medium the day before cell harvesting. Cells were harvested 48 h posttransfection.
To obtain cell extracts expressing MV P protein, HeLa cells were infected with recombinant vv-P for 1 h at an MOI of 2 and then washed and incubated in fresh medium for 24 h before cell harvesting.
To investigate the effects of hPIRH2 overexpression on MV infection, cells were infected 1 day after transfection. 293T cells, seeded the day before, were infected with MV at an MOI of 1, incubated for 2 h at 37°C, and then washed once and incubated in culture medium in the presence of 20 μg/ml of the fusion-inhibitory peptide Z-d-Phe-l-Phe-Gly (38) to prevent syncytium formation.
Coimmunoprecipitation.
HeLa or 293T cells were washed once with ice-cold phosphate-buffered saline (PBS) and lysed in 1 ml of lysis buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 150 mM NaCl, and 0.5% NP-40) on ice and then further disrupted by passage through a needle. Cell debris were pelleted by centrifugation, and supernatants were precleared with 20 μl of protein G Sepharose 4 Fast Flow (Amersham Pharmacia Biotech) for 2 h. Supernatants were incubated with primary antibody for 2 h at 4°C with rotation. Next, the mixture was centrifuged, and supernatants were mixed with 20 μl of protein G beads. After overnight incubation at 4°C, beads were collected and washed five times in washing buffer (5% sucrose, 5.0 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.5 M NaCl, and 1% NP-40) and once with PBS. Bound proteins were eluted from beads by boiling them with Laemmli buffer and separated by SDS-PAGE before Western blotting.
In vivo ubiquitination assay.
293T cells were transfected with plasmids encoding N, P, Psev, hPIRH2, C145/8S, mPIRH2, Mdm2, and HA-tagged ubiquitin either alone or in combination. Cells were collected 48 h later, and lysates were subjected to Western blotting and immunoprecipitation (IP) as described above. The total DNA level was kept constant by the addition of the empty vector.
Immunofluorescence.
HeLa cells were plated onto glass coverslips and transfected with different plasmids, as indicated in the legend to Fig. 7. After 48 h, cells were washed with PBS, fixed in 4% paraformaldehyde, washed three times, and permeabilized with 0.3% Triton X-100. After three washes, cells were incubated with the anti-N MAb cl120 and/or anti-P rabbit polyclonal serum for 1 h. After three further washes, cells were incubated with rhodamine-conjugated anti-mouse immunoglobulin G or Alexa 546-conjugated anti-rabbit immunoglobulin G for 1 h. Nuclei were labeled using TO-PRO-3 (Molecular Probes). Cells were analyzed by fluorescence confocal microscopy.
FIG. 7.
MV N protein alters the localization of hPIRH2 via interaction with MV P. HeLa cells were transfected with plasmids as indicated and stained with anti-N MAb cl120 and rhodamine-conjugated secondary antibodies (A, B, and C) and anti-P polyclonal P (D, E, and F) and Alexa 546-conjugated secondary antibodies. Nuclei were stained with TO-PRO-3. Merged signals are shown in the right panels.
siRNA.
pSUPER coding for siRNA targeting hPIRH2 mRNA (si-hPIRH2) was constructed as described previously (5) using two synthetic complementary oligonucleotides (5′-gat ccc c TGG ACG ATC CAC TGT TCA G ttc aag aga CTG AAC AGT GGA TCG TCC A ttt ttg gaa a-3′ and 5′-agc ttt tcc aaa aa TGG ACG ATC CAC TGT TCA G tct ctt gaa CTG AAC AGT GGA TCG TCC A ggg-3′; uppercase indicates sequences that are complementary to the target gene; lowercase indicates flanking sequences). siRNA against GFP (si-GFP) (15) was kindly provided by Frederic Iseni. To evaluate si-hPIRH2 efficiency, 293T cells were cotransfected with si-hPIRH2 and pEGFP-C145/8S, si-GFP and pEGFP-C145/8S, si-hPIRH2 and C145/8S-myc, or si-GFP and C145/8S-myc. Cell lysates were analyzed by assessing fluorescence or Western blotting using anti-GFP and anti-myc MAbs, respectively.
RESULTS
Interaction of human PIRH2 with the C-terminal region of MV P protein.
To identify cellular partner(s) of MV P protein, a yeast two-hybrid screening of a human HeLa cDNA library was performed. Full-length P and PNT fused in frame with the GAL4-BD proved unsuitable because they transactivated yeast reporter genes. When PCT was used as bait, nine clones grew on QDO-α-Gal plates, and three clones corresponded to the same cDNA that was identical to the published sequence of hPIRH2 (2, 30). The protein, termed AD-hPIRH2, was in frame with the GAL4-AD domain and covered full-length hPIRH2. The specificity of the interaction was further investigated by using this plasmid to retransform Y187 yeast cells and by repeating the mating with BD-PCT in AH109 yeast cells. AD-hPIRH2 strongly interacted with BD-PCT, but it did not interact with BD or BD-N. Likewise, BD-PCT did not interact with AD-PNT (data not shown).
In vitro interaction of hPIRH2 with MV P.
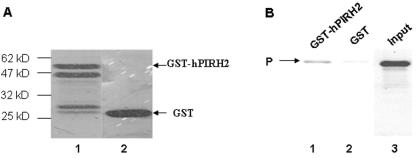
To investigate whether hPIRH2 interacts with full-length P, we performed an in vitro GST pull-down experiment with full-length GST-fused hPIRH2 expressed in bacteria. The GST-hPIRH2 fusion produced a protein of the expected size with some proteolytically cleaved products (Fig. 2A). When mixed with an MV P-containing cell lysate from HeLa cells infected with vv-P, GST-hPIRH2, but not GST alone, was able to pull down MV P (Fig. 2B). However, the interaction appeared to be weak, possibly due to C-terminal degradation of GST-hPIRH2.
FIG. 2.
In vitro interaction of P and PIRH2 in a GST pull-down assay. (A) hPIRH2 was expressed as a GST fusion protein, affinity purified, resolved by SDS-PAGE, and stained with Coomassie blue (lane 1). GST protein was similarly purified (lane 2). (B) A total of 10 μg GST or GST-hPIRH2, immobilized on glutathione-Sepharose, was used for the binding of HeLa cell lysates infected by vv-P. Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and by immunoblotting with MAb 49.21. The input lane represents 1/10 of the protein used in the binding reaction.
hPIRH2 interacts with full-length P in vivo.
Interaction between hPIRH2 and P was next studied in human cells by coimmunoprecipitation. After transfection of pcDNA3-myc-hPIRH2 in HeLa cells, the expression level of myc-hPIRH2 was very low when detected by anti-myc antibody, unlike another hybrid construct (pcDNA3-myc-IRF3) detected by the same MAb, suggesting that hPIRH2 might have a short half-life (data not shown) as previously reported (31). Fortunately, by expressing T7 polymerase using vv-T7 infection in HeLa cells, the expression level of myc-hPIRH2 was largely increased (data not shown). Whole extracts from HeLa cells transfected with pcDNA3-myc-hPIRH2 and/or psc6-P and infected with vv-T7 were subjected to immunoprecipitation. Reciprocal specific co-IP of myc-hPIRH2 and P proteins was observed by either anti-P (Fig. 3A) or anti-myc MAb (Fig. 3B). Similarly, MV P protein could also be coimmunoprecipitated with an alternatively tagged form of hPIRH2, HA-hPIRH2 (not shown). To exclude an artifact due to the presence of a tag at the hPIRH2 N terminus or vv-T7 infection, hPIRH2-myc, in which the myc tag was fused at the C terminus of hPIRH2, was coexpressed with P in 293T cells without vv-T7 infection. P could also be successfully coimmunoprecipitated (Fig. 3C) using anti-myc MAb. Additionally, the PCT domain could also be coimmunoprecipitated with HA-hPIRH2 (data not shown). When the homologous Psev was coexpressed with hPIRH2-myc in 293T cells, it was unable to be coimmunoprecipitated by the anti-myc MAb (Fig. 3D). Taken together, we conclude that P and hPIRH2 physically and specifically interact in vivo.
FIG. 3.
In vivo interaction of P and hPIRH2. (A to C) Coimmunoprecipitation of MV P protein with myc-hPIRH2 (A and B) and hPIRH2-myc (C) in HeLa (A and B) and 293T (C) cells using P (A)- or myc (B and C)-specific antibodies. Antibody specificity is shown by the lack of immunoprecipitated material (lanes 1). Left panels show protein expression in whole-cell extracts of input samples. (D) Inability of Psev and hPIRH2-myc to be coimmunoprecipitated in 293T cells (lane 2, right panel). The controls are the same as those in panels A to C. Lane 0 on the left panel shows the lack of nonspecific labeling after immunoblotting. Proteins were expressed by transfection with the corresponding expression vectors (A to D) without (C and D) or with (A and B) infection with vv-T7. In experiments illustrated in panels A and B, MG132 proteasome inhibitor was added for 8 h before cell harvesting. α, anti.
Interaction of PCT with hPIRH2 is mediated by its extreme C-terminal XD region and favored by the PCT oligomerization domain.
To identify the region of PCT that interacts with hPIRH2, we next tested a series of PCT deletion mutants, generated according to structure prediction (24, 32), by using the yeast mating assay. Only constructs containing the XD resulted in the activation of the yeast reporter genes (Fig. 4A). Moreover, from the level of α-galactosidase expression, the levels of interaction of BD-PCT-Spacer and BD-PCT with AD-hPIRH2 were of similar magnitudes and much stronger than those observed with the BD-PX, BD-PXS, BD-PDL BD-XDL, and BD-XD fragments. The latter fragments lack the P(304-375) PMD domain that we demonstrated in Fig. 4A to be the oligomerization domain of MV P, as predicted (32). Indeed, only PMD-containing fragments were able to interact with AD-PCT (Fig. 4A). Since all constructs were expressed and were of their expected molecular masses (Fig. 4B), the lack of interaction of PMD-containing fragments in the absence of XD suggests that PMD does not bind to hPIRH2 but that PCT oligomerization increases its avidity to hPIRH2.
FIG. 4.
XD mediates MV P binding to hPIRH2. (A) Ability of PCT and deletion mutants (numbers correspond to amino acid sequence) fused to BD to bind AD-hPIRH2 in the yeast mating assay. AD-PNT and AD-PCT were used as negative and positive controls, respectively. (B) Expression of various deletion mutant proteins in yeast strain AH109. Proteins were immunoblotted with rabbit 371171 antibody against P. Arrowheads indicate the positions of mutant proteins.
MV N, MV P, and hPIRH2 form a ternary complex in vivo.
Within MV PCT, the XD binds to the αMORE region of NTAIL (23, 29). The ability of hPIRH2 and N to compete for binding to XD and/or to produce a ternary complex with MV P protein was investigated by transfection and co-IP analysis with human 293T cells. As expected, the anti-N MAb specifically pulled down P with N in the absence or presence of myc-hPIRH2 (Fig. 5, right panel). Furthermore, the anti-N MAb efficiently coimmunoprecipitated hPIRH2-myc only when the three proteins were coexpressed (Fig. 5). It should be noted that when expressed, the amount of each protein was roughly the same between samples. Thus, P can act as a bridge between hPIRH2 and N, and these three proteins can form a stable ternary complex in vivo.
FIG. 5.
Coimmunoprecipitation of hPIRH2, MV P, and MV N proteins expressed in human cells. Western blot analysis of proteins from whole-cell lysates and anti-P immunoprecipitations from 293T cells transfected with various combinations of hPIRH2-myc, MV P, and MV N proteins, as indicated. α, anti.
Mapping of the PCT interaction site on hPIRH2 using a novel Y2H-TPCR assay.
By using a novel Y2H-TPCR assay established in our laboratory, a library of random hPIRH2 PCR fragments subcloned downstream from Gal4-AD was screened for interaction against BD-PCT. This assay is derived from a random PCR-based screening for soluble domains using green fluorescent protein (18, 26). We extended its use to screen for interaction domains by fusing it with the GAL4-AD domain. Sixteen clones grew in selective medium following hPIRH2 deletion library transformation into BD-PCT-containing yeast cells. Ten clones corresponded to hPIRH2 sequences in frame with BD (Table 1), with five clones coding for the same hPIRH2(157-261) C-terminal fragment. When assessed after subcloning and cotransformation with BD-PCT and control constructs, only clones containing this last fragment (157-261) showed specific interactions, although to a weak extent. Indeed, the cotransformed yeasts grew primarily only on the less stringent, selective SD/−Trp/−Leu/−His medium (Table 1). However, the clones selected on SD/− Trp/−Leu/−His medium were then able to subsequently grow on highly selective QDO-α-Gal medium and became blue, indicating protein-protein interactions. When we coexpressed the HA-hPIRH2(157-261) fragment with P protein in HeLa cells, the anti-HA MAb failed to coimmunoprecipitate MV P protein (data not shown). Our interpretation of these data is that the hPIRH2(157-261) fragment encompasses a P binding site that mediates only a weak protein-protein interaction and that there may possibly be another P binding site which failed to be identified by our Y2D-TPCR assay.
TABLE 1.
The PCT interaction site maps within the C-terminal part of PIRH2 as determined by a Y2H-TPCR assaya
| Construct | Growth onb:
|
||
|---|---|---|---|
| SD/−Trp/−Leu (two plasmids) | SD/−Trp/−Leu/−His (single selective gene) | SD/−Trp/−Leu/−His/−Ade (two selective genes) | |
| AD + BD-PCT | + | − | − |
| AD-PIRH2 + BD | + | − | − |
| AD-PIRH2 + BD-PCT | + | + | + |
| AD-(12-106) + BD-PCT | + | − | − |
| AD-(69-106) + BD-PCT | + | − | − |
| AD-(69-160) + BD-PCT | + | − | − |
| AD-(157-261) + BD-PCT | + | + | − |
| AD-(157-261) + BD | + | − | − |
Plasmids encoding putative positive clones were cotransformed with BD-PCT and selected on media with increasing stringency as indicated. After 7 days, clones were assessed for growth (AD, pGADT7 empty vector; BD7 pGBKT7 empty vector; AD-PIRH27 full-length PIRH2 fused to the AD domain).
+, clone growth; −, no growth.
Both hPIRH2 C-terminal and RING-H2 regions mediate interactions with MV P.
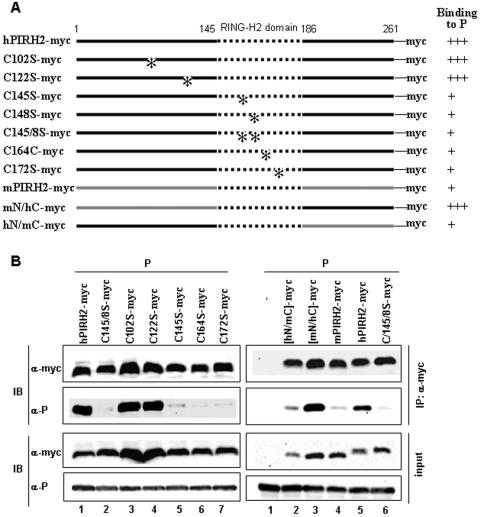
We postulated that the conserved cysteine-rich RING-H2 PIRH2(145-186) domain, which is the active site of ubiquitin E3 ligase activity of PIRH2 (30), may participate in hPIRH2 binding to P. Single and double mutations in conserved cysteines within the RING domain disrupt its structure and abolish the E3 ligase activity (2). The ability of P to bind a series of hPIRH2-myc mutants with conserved cysteines altered to serines within (C145S, C148S, C145/8S, C164S, and C172S) and outside (C102S and C122S) the RING domain (Fig. 6A) was investigated. After immunoprecipitation of hPIRH2 from transfected 293T cells, only the two point mutants outside the RING domain were found to co-IP with P as efficiently as their wild-type counterpart (Fig. 6B, left panel). All point mutants within the RING domain essentially lost their ability to bind to P. These differences were not related to major differences in protein expression levels (Fig. 6B, left panel). Thus, the hPIRH2 RING-H2 domain is involved in the interaction with MV P protein in whole cells.
FIG. 6.
Both RING motif and C-terminal regions of hPIRH2 protein mediate binding to MV P protein. The region of PIRH2 binding to P was analyzed in coimmunoprecipitation assays. (A) Schematic representation of the sequences of hPIRH2 (black lines), mPIRH2 (gray lines), and the conserved RING motif (dotted lines). *, Cys-to-Ser mutations. (B) Western blot analysis of the proteins from transfected 293T whole-cell lysates (input, lower panels) and from anti-myc immunoprecipitations (IP, upper panels). The experiment was carried out as described in the legend for Fig. 3D. α, anti.
hPIRH2 and mPIRH2 are highly homologous and differ by only 18 and 7 residues within the N- and C-terminal regions, respectively, flanking the fully conserved PIRH2(145-186) RING-H2 motif. This led us to test the ability of mPIRH2-myc to interact with MV P in 293T cells. Only small amounts of P protein could be pulled down together with mPIRH2-myc by using anti-myc MAb (Fig. 6B, right panel). We next created two mouse and human chimeric proteins with reciprocal exchanges of their N- and C-terminal regions, hN/mC-myc and mN/hC-myc. After coexpression with P in 293T cells and IP with anti-myc MAb, only the mN/hC chimera was as efficient as hPIRH2-myc in coimmunoprecipitating MV P protein (Fig. 6B, right panel). Thus, hPIRH2 binding to P likely involves two domains, the RING-H2 motif and the C-terminal hPIRH2 (187-261) moiety.
P protein affects subcellular localization of hPIRH2.
It has previously been demonstrated that hPIRH2 subcellular localization might be critical in its regulation (31). To assess whether MV proteins might affect hPIRH2 activities in this way, full-length EGFP-hPIRH2, EGFP-C145/8S, and control EGFP were used to study hPIRH2 cellular localization in HeLa cells by confocal microscopy. These three proteins exhibited the same diffuse nuclear and cytoplasmic fluorescence (data not shown). MV N protein exhibited aggregation with a speckled distribution (Fig. 7A to C), while MV P protein displayed a diffuse cytoplasmic distribution (data not shown). Coexpression of MV N protein did not change the broad distribution of any EGFP proteins (Fig. 7A to C), and N protein distribution was unchanged upon EGFP-hPIRH2 expression. In contrast, coexpression of MV P, which is cytoplasmic and excluded from the nucleus, restricted distribution of EGFP-hPIRH2 to the cytoplasm and prevented its nuclear localization but did not change the distribution of EGFP or EGFP-C145/8S (data not shown). As expected (22, 46), P protein became aggregated in part and colocalized with the now exclusively cytoplasmic speckled distribution of N protein when these proteins were coexpressed (Fig. 7D to F and data not shown) (22). Furthermore, both EGFP-hPIRH2 and EGFP-C145/8S, but not EGFP, colocalized with P aggregated into the N-rich-punctuated areas (Fig. 7D, E, and F, respectively). Altogether, these data showed that EGFP-hPIRH2 is colocalized in vivo with P and aggregated by N protein, in agreement with its ability to form ternary complexes which can be coimmunoprecipitated. The ability of MV P to sequester, in part, the RING mutant EGFP-C145/8S into N-rich areas and its inability to prevent some nuclear localization of EGFP-C145/8S (Fig. 7F) are indicative of a weak residual interaction likely mediated by the C terminus of hPIRH2, in agreement with the small amount of P which could be coimmunoprecipitated with this hPIRH2 mutant.
P protein is partly unstable and degraded by the proteasome but does not behave as a substrate for hPIRH2.
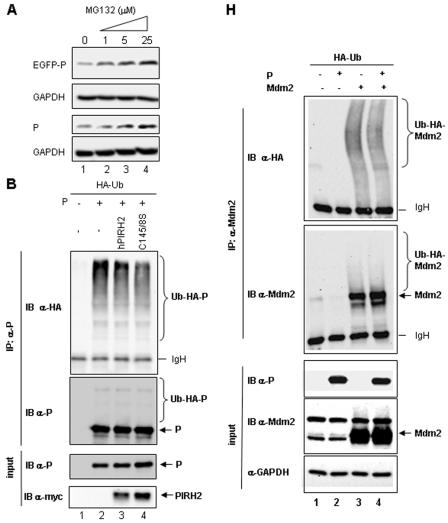
We next assessed whether MV P protein is a substrate for PIRH2-mediated ubiquitination and proteasome degradation. When an EGFP-P hybrid molecule or P protein was transiently expressed, it was further stabilized in a dose-dependent manner in the presence of MG132, whereas cellular GAPDH levels were unaltered (Fig. 8A). Accordingly, in the presence of MG132, polyubiquitinated P was detected after anti-P immunoprecipitation using anti-HA MAb, which was poorly recognized by the anti-P MAb (Fig. 8B, lane 2). The coexpression of hPIRH2-myc or the C145/8S-myc mutant changed neither the expression nor the ubiquitination level of P protein (Fig. 8B, lanes 3 and 4). Thus, P protein is ubiquitinated and degraded by the proteasome but is unlikely to represent a substrate for hPIRH2.
FIG. 8.
MV P does not act as a substrate for PIRH2, but it stabilizes hPIRH2 and inhibits its ubiquitination. (A) P protein is partly unstable and degraded by the proteasome. HeLa cells were transfected for 48 h with pCG-EGFP-P or psc6-P, and various concentrations of MG132 were added 24 h before cell harvesting. (B) P protein is ubiquitinated but does not behave as a substrate for exogenous hPIRH2-myc. 293T cells were transfected with plasmids, as indicated, in the presence of MG132. After 48 h, cell lysates were immunoprecipitated using anti-P polyclonal Ab and analyzed by Western blotting with anti-P and anti-HA MAb. (C) P (lower panel) stabilizes hPIRH2-myc (upper panel) in vivo. 293T cells were transfected as indicated with MG132 or cycloheximide where detailed. GAPDH was used as a protein loading control. (D) In the context of MV infection, the ability of P to stabilize hPIRH2-myc is undetectable. 293T cells were transfected with hPIRH2-myc, and 24 h later, they were infected with MV and harvested 48 h after infection. Cells were treated, where indicated, with 5 μM MG132 for 24 h before harvesting. Lysates were analyzed by Western blotting for N, P, and hPIRH2 proteins. (E) P prevents in vivo ubiquitination of hPIRH2. 293T cells were transfected with plasmids, as indicated, and treated with 5 μM MG132. Anti-myc immunoprecipitations were analyzed by Western blotting with anti-HA MAb or anti-myc MAb. Protein contents of the cell lysates prior to immunoprecipitation were determined by using anti-myc, anti-P, and anti-GAPDH antibodies, as indicated. Ubx1 and Ubx2 likely indicate mono- and diubiquitinated forms of PIRH2-myc. (F) MV P inhibition of hPIRH2 ubiquitination is dose dependent and is unaffected by either MV N protein or P protein from Sendai virus. Psev (lane 3), MV P and N proteins (lane 4), or increasing amounts of P (lanes 6 to 9) were coexpressed with Ub-HA and hPIRH2-myc. Cell extracts were probed, by Western blotting, using anti-myc, anti-Psev, anti-MV N, anti-MV P, or anti-GAPDH MAb (top to bottom panels, respectively). (G) Abrogation of PIRH2 expression and ubiquitination by si-PIRH2. 293T cells were transfected as indicated, and cell lysates were subjected to Western blotting with anti-myc, anti-P, or anti-GAPDH antibodies. (H) Inability of MV P to inhibit the in vivo ubiquitination of Mdm2. HA-epitope-tagged Ub was coexpressed with P (lanes 2 and 4) and/or Mdm2 (lanes 3 and 4) in 293T cells. Two days after transfection, Mdm2 was immunoprecipitated, and the immunoprecipitated material was probed with anti-HA MAb and anti-Mdm2 MAb (upper two panels). Expression of MV P, Mdm2, and GAPDH in total cell extracts (inputs) was also analyzed by Western blotting (lower panels). α, anti.
P stabilizes hPIRH2 protein by inhibiting its ubiquitination in vivo.
hPIRH2 has a short half-life and is a target for proteasome-mediated degradation (31). Accordingly, after transfection into 293T cells, the hPIRH2-myc protein was almost undetectable by Western blotting, unless the cells were pretreated with the proteasomal inhibitor MG132 (Fig. 8C), whereas the level of GAPDH protein remained unaltered. For this reason, cells transfected with hPIRH2 were treated with MG132 in some previous experiments (see the legend for Fig. 3). Proteasomal degradation of hPIRH2 also depends upon an intact RING-H2 motif, since the C145/8S-myc double mutant was efficiently expressed with or without MG132 (data not shown). Given that we have shown that MV proteins can regulate hPIRH2 cellular localization, MV P protein was further assessed for its effects on hPIRH2 stability in the absence of the proteasome inhibitor. P coexpression largely increased hPIRH2-myc protein abundance to levels comparable to those observed following MG132 treatment (Fig. 8C, lanes 3 and 4). To ascertain that this effect was at the posttranslational level and not at the transcriptional level, the experiment was also performed in the presence of cycloheximide to inhibit de novo protein synthesis. A short (4-h) pretreatment with cycloheximide (50 μg/ml) prior to cell harvesting did not affect the stabilization of hPIRH2 by MV P (Fig. 8C, lane 6). Western blotting confirmed P protein expression where appropriate (Fig. 8C, lower panel). An effect similar to that of MV P protein on the stability of EGFP-hPIRH2 protein was observed, whereas P protein did not change EGFP levels (data not shown). Thus, hPIRH2 is specifically stabilized in the presence of MV P.
We next asked whether P protein that is expressed during MV infection is able to stabilize exogenous hPIRH2-myc or if the overexpression of hPIRH2 can affect MV replication. Surprisingly, hPIRH2-myc was very poorly detected in MV-infected cells (Fig. 8D). The low level was due to the inability of the virally expressed P to stabilize most of the hPIRH2-myc because, in the presence of MG132, a large amount of hPIRH2 was detected (Fig. 8D, lane 2). Furthermore, the levels of virally expressed P and N proteins were insensitive to the coexpression of hPIRH2-myc, whether it was stabilized by MG132 treatment or not (Fig. 8D, compare lanes 1 and 2, and data not shown).
Ubiquitination of hPIRH2 has previously been demonstrated to regulate hPIRH2 at the posttranslational level (31). The data showing stabilization of hPIRH2 upon P expression led us to investigate whether P might affect ubiquitination of hPIRH2. HA-Ub was coexpressed by transfection with hPIRH2-myc, C145/8S-myc, or mPIRH2-myc with or without P in the presence of MG132. Western blotting of total lysates from cells coexpressing HA-Ub and hPIRH2-myc, detected with anti-myc MAb, revealed numerous higher-molecular-weight protein bands, in addition to hPIRH2-myc protein (Fig. 8E, input panel, lane 3), likely representing polyubiquitinylated hPIRH2-myc. Indeed, following IP with anti-myc MAb, the same bands (except the one corresponding to nonubiquitinated hPIRH2-myc) were detected by using the anti-HA MAb (Fig. 8E, IP:α-myc, top panel, lane 3). Interestingly, in the whole-cell extract, two bands have the expected molecular weights of mono- and diubiquitinated hPIRH2-myc proteins (Fig. 8E, input panel, lane 3, Ubx1 and Ubx2). Furthermore, although they were undetectable in the whole-cell extracts, ubiquitinated forms of mPIRH2-myc and C145/8S-myc were also detected in the anti-myc immunoprecipitated material (Fig. 8E, IP:α-myc, top panel, lanes 5 and 7, respectively). This suggests that relatively small portions of total C145/8-myc and mPIRH2-myc are subjected to polyubiquitination.
When MV P was coexpressed with PIRH2 protein, it readily abolished hPIRH2 ubiquitination (Fig. 8E) without changing the expression level of the unmodified hPIRH2 (Fig. 8E, input panel). This effect was specific, since it was not observed after coexpression of Psev or MV N proteins which failed to bind to hPIRH2 (Fig. 8F), and it was dose dependent (Fig. 8F). Additionally, MV N protein, which forms ternary complexes with P and hPIRH2, did not alter the capacity of MV P to inhibit hPIRH2 ubiquitination (Fig. 8F).
To further ascertain that the ladder of protein bands detected by the anti-myc antibody in whole-cell extracts corresponds to ubiquitinated hPIRH2-myc and not other proteins, the ubiquitination assay was repeated in the presence of si-hPIRH2, introduced by cotransfection. This siRNA efficiently eliminated the expression of exogenous hPIRH2-myc (Fig. 8G, anti-myc panel). The silencing effect was specific since siRNA targeting GFP had no effect, and neither siRNA altered endogenous GAPDH (Fig. 8G). Interestingly, coexpression of MV P with hPIRH2-myc similarly abolished the ladder of (ubiquitinated) protein bands but did not affect the level of hPIRH2-myc expression (Fig. 8G). These data were further confirmed in coimmunoprecipitation experiments (data not shown). We thus conclude that P protein expression efficiently inhibits the in vivo ubiquitination of hPIRH2-myc, C145/8S-myc, and mPIRH2-myc.
Since the RING motif of hPIRH2 is involved in its interaction with MV P protein, we asked whether P may also inhibit another structurally related E3 ligase. We chose Mdm2, since PIRH2 and Mdm2 act on the same p53 substrate. After cotransfection with P, Mdm2 was abundantly expressed, but little ubiquitinated Mdm2 could be detected in total cell lysates, and ubiquitin-conjugated Mdm2 could be detected only after immunoprecipitation with anti-Mdm2 MAb (Fig. 8H). Furthermore, levels of ubiquitinated Mdm2 were not changed when MV P protein was coexpressed (Fig. 8H, top panel).
Taking all the data together, we conclude that MV P inhibits hPIRH2 ubiquitination via its specific interaction with this ubiquitin E3 ligase. As a consequence, MV P protein enhances the stability of hPIRH2 by preventing its degradation by the proteasome.
DISCUSSION
We have identified hPIRH2 as the first human cellular partner of MV P protein. PIRH2 was recently identified in mice as an ARNIP and in humans as hPIRH2 (2, 30). This protein was further characterized as interacting with the histone acetylase TIP60, which stabilizes and partially retains hPIRH2 within the nucleolus (31). PIRH2 belongs to the class of E3 ubiquitin ligases with a RING-H2 motif (2, 30). The only known substrate for hPIRH2-mediated ubiquitination is the p53 transactivator (30), which is a potent cell cycle inhibitor and apoptosis inducer (see reference 48 for a review).
MV P protein binds to hPIRH2 in vitro and in vivo as demonstrated by GST pull-down, reciprocal coimmunoprecipitation, and colocalization experiments. This binding is likely specific because the closely related P protein from Sendai virus cannot be coimmunoprecipitated with hPIRH2 or mPIRH2 (data not shown). In addition, the reciprocal binding sites were mapped to the X domain of MV P and to the hPIRH2(145-186) RING-H2 domain and the hPIRH2(187-261) C-terminal domain. In the Y2H assay, the interaction of XD with hPIRH2 was enhanced in the presence of PMD, a coiled-coil motif which was predicted, and demonstrated here, to be the oligomerization domain of MV P, which, by analogy with Sendai virus P protein, should assemble into tetramers (24, 44). This suggests that hPIRH2 may also make oligomers. Indeed, the structurally related Mdm2 E3 ligase is a dimer (43). Since both PCT and P can be coimmunoprecipitated with hPIRH2, the natively unfolded PNT fragment (24) is unlikely to be involved in the P-hPIRH2 interaction. The crystal structure of the XD reveals a stable domain composed of three α-helices arranged in a three-helix bundle (23). XD is responsible for the induced folding of NTAIL, the natively unfolded domain of MV N protein (23, 32). It binds to the α-helix N(477-505) region, termed αMORE, so as to make a four-helix bundle. Rather surprisingly, we found that N, P, and hPIRH2 can make ternary complexes which can be coimmunoprecipitated. Furthermore, the three proteins colocalized in vivo with aggregated forms of N protein. However, when expressed in the context of a viral infection, stabilization of exogenous hPIRH2-myc by P was not detected. It should be stressed that during MV infection, there is a large excess of N protein over P protein because of the mRNA gradient transcription (6, 35) and the accumulation of nucleocapsids. Furthermore, N°P complexes, which mainly involve PNT binding to NCORE (8, 19), are very short-lived (35), possibly leaving most P engaged in interactions involving PCT-to-NTAIL binding. Altogether, these data strongly suggest that hPIRH2 and N proteins do compete with each other for binding to the P XD region. Since P protein is a tetramer, according to the structure of the homologous Sendai virus P protein (44), we predict that hPIRH2 and NTAIL associate separately with P monomers which might belong to the same P tetramer. Although Paramyxovirinae PCT structures are highly conserved and share several common features, Psev, a mouse pathogen, binds to neither hPIRH2 nor mPIRH2 despite having XD folding identical to that of MV (3).
The C-terminal hPIRH2(157-261) region, which lacks half of the RING-H2 domain, was identified as containing a P binding site by using our novel Y2H-TPCR assay. These data were confirmed in coimmunoprecipitation experiments using mouse/human hybrid molecules.
Two lines of evidence suggest that the RING finger motif plays a role in hPIRH2 binding to P: (i) mPIRH2, which has the same RING motif sequence as hPIRH2, can bind to P protein; and (ii) an hPIRH2 mutant within the RING-H2 motif almost abolishes hPIRH2 binding. RING motifs of other proteins, including Mdm2, can mediate protein-protein interactions (4, 33, 39). Alternatively, since an isolated RING domain can have self-oligomerized properties (27), the RING motif may act indirectly by mediating hPIRH2 oligomerization, as it does for Mdm2 (43), and thus may stabilize the interaction with P tetramers. In this latter case, the weak but significant mPIRH2 binding to P would be mediated by the C-terminal region, albeit with a lower affinity because of the seven-residue difference with its human counterpart or because it makes heterologous oligomers with the limited amount of endogenous hPIRH2. The fact that, in a colocalization assay, MV N protein can still aggregate the EGFP-C145/8S mutant via a P-mediated bridge favors the C terminus of hPIRH2 as the primary P binding site. How the C terminus of hPIRH2 and possibly the RING-H2 motif interact with XD remains to be unraveled, and cocrystallization attempts are under way.
mPirh2 ubiquitinates p53, promoting its degradation by the proteasome (30). Although P is stabilized in part in the presence of MG132 and is ubiquitinated, the expression of exogenous hPIRH2 does not enhance P ubiquitination and degradation, suggesting that P is not a substrate for hPIRH2. As described previously (31), we found that exogenously expressed hPIRH2 is unstable and ubiquitinated in cells. MV P protein stabilizes hPIRH2 at the posttranslational level and efficiently prevents hPIRH2 ubiquitination. The cellular protein TIP60 binds to hPIRH2 and, like MV P, stabilizes hPIRH2 but by a different mechanism, as it partly sequesters hPIRH2 into the nucleolus (31). The mechanism by which P protein inhibits ubiquitination of hPIRH2, but not the related Mdm2 E3 ligase, cannot be directly inferred from our data. We speculate the following. (i) The ubiquitination of the hPIRH2-C145/8S mutant, which occurs at a weaker level but is also inhibited by P, indicates that the RING motif favors the ubiquitination and that the ubiquitination may occur outside the RING motif. Interestingly, the hPIRH2 C terminus contains a weak putative PEST sequence (amino acids 198 to 228), which could be the primary ubiquitination site (31). In this case, P may act by shielding the ubiquitination site, thereby stabilizing hPIRH2. (ii) Like Mdm2 (12), hPIRH2 may mediate its own ubiquitination and degradation through its RING motif. Indeed, the compact RING finger structure interacts with an E2 ubiquitin-conjugating enzyme(s) (2) and facilitates ubiquitination of bound substrates (see reference 48 for a review). The hPIRH2-C145/8S protein with a double mutation within the RING motif can be ubiquitinated in cells. These data are in agreement with a previous observation and were thought to be indicative of the inability of hPIRH2 to mediate its own ubiquitination (31). Indeed, this mutant is stabilized because it escapes proteasome-mediated destruction. The RING motif was thought to not be required for ubiquitination of PIRH2 but to be involved in its recruitment by the proteasome (31). However, we cannot exclude the possibility that ubiquitination of the C145/8S mutant, which has its E3 ligase activity disabled (2), is performed by endogenous hPIRH2. Indeed, the P protein may act as a direct inhibitor of the E3 ligase of hPIRH2. This hypothesis is currently under investigation by testing the ability of MV P to inhibit PIRH2-mediated p53 ubiquitination. (iii) Finally, P may favor a yet-to-be-identified deubiquitination pathway, although MV P sequence alignment does not highlight any significant homology with known deubiquitinases, such as HAUSP.
Does hPIRH2 play a role in MV replication? Despite our best attempts, we have been unable to find any detectable effect of overexpressing exogenous hPIRH2 on MV infection, whatever the follow-up of the infection (including the percentage of MV glycoprotein-expressing cells or the protein expression levels of different MV genes determined by Western blotting). However, the effect of hPIRH2 may be cell type specific, since (i) MV replication can infect many different cell types, including lymphocytic, epithelial, dendritic, thymic, and neuronal cells; and (ii) hPIRH2 expression likely varies according to the cell type in vivo, as indicated by the data available for its mouse counterpart (2). Alternatively, hPIRH2 may target one of the other MV proteins, such as the L protein for ubiquitination and proteosomal degradation. Indeed, P protein has been described as a chaperone for the L protein, which is very unstable in cells (20).
Reciprocally, MV may interfere with the cellular function of hPIRH2. hPIRH2 is one of four cellular E3 ligases with RING motifs known to mediate ubiquitination of p53 for proteasome destruction (30). Interestingly, MV infection induces cell cycle inhibition with accumulation of lymphocytes in G0/G1 phase (34) and apoptosis of many cell types (10), although the underlying mechanisms are not well understood. Whether MV P interference with hPIRH2 activity is involved in these processes, possibly by blocking E3 ligase activity of hPIRH2 and increasing p53 levels, is currently under investigation. Indeed, the silencing of mPIRH2 induces a p53-mediated cell cycle arrest in G1 phase (30). Interestingly, a p53-mediated induction of apoptosis has been described for Sendai virus (21) and many other viruses (11).
In summary, we report the first viral protein, MV phosphoprotein, that is able to interfere with the biology of the human E3 ubiquitin ligase PIRH2, one of the regulators of the p53 pathway.
Acknowledgments
This work was supported in part financially by the Medical Research Council (grant identification no. 63468) and the Commission of the European Communities specific RTD program “Quality of Life and Management of Living Resources” (QLK2-CT2001-01225).
This work does not necessarily reflect the Commission's views and in no way anticipates the Commission's future policy in this area.
We thank L. Beitel, R. Leng, R. Cattaneo, R. Vigne, E. Decroly, T. F. Wild, R. Drillien, F. Iseni, B. Moss, R. Agami, A. Puisieux, S. Longhi, and D. Kolakofsky for providing useful reagents and acknowledge the use of the confocal microscopy facilities of the University Ctμ and the invaluable help of Béatrice Burdin and Sandra Garvi-Balvay. We are greatly indebted to Carine Soriano for her excellent technical help in setting up the Y2H-TPCR assay and to Sébastien Plumet for performing useful reverse transcriptase QPCR to monitor RNA levels by using the CeCIL university facility center.
REFERENCES
- 1.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 2.Beitel, L. K., Y. A. Elhaji, R. Lumbroso, S. S. Wing, V. Panet-Raymond, B. Gottlieb, L. Pinsky, and M. A. Trifiro. 2002. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29:41-60. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard, L., N. Tarbouriech, M. Blackledge, P. Timmins, W. P. Burmeister, R. W. Ruigrok, and D. Marion. 2004. Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 319:201-211. [DOI] [PubMed] [Google Scholar]
- 4.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M., J. C. Cortay, and D. Gerlier. 2003. Measles virus protein interactions in yeast: new findings and caveats. Virus Res. 98:123-129. [DOI] [PubMed] [Google Scholar]
- 8.Curran, J., J.-B. Marq, and D. Kolakofsky. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 69:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaux, P., and R. Cattaneo. 2004. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J. Virol. 78:11632-11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esolen, L. M., S. W. Park, J. M. Hardwick, and D. E. Griffin. 1995. Apoptosis as a cause of death in measles virus-infected cells. J. Virol. 69:3955-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 12.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., J.-B. Marq, F. Iseni, S. Martin, and D. Kolakofsky. 2004. A short peptide at the amino terminus of the Sendai virus C protein acts as an independent element that induces STAT1 instability. J. Virol. 78:8799-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlier, D., H. Valentin, D. Laine, C. Rabourdin-Combe, and C. Servet-Delprat. 2005. Subversion of the immune system by measles virus: a model for the intricate interplay between a virus and the human immune system. In M. Oldstone & P. Lachman (ed.), Microbial subversion of host immunity. Horizon Scientific Press, Caister Academic Press, Norfolk, United Kingdom.
- 17.Giraudon, P., M. F. Jacquier, and T. F. Wild. 1988. Antigenic analysis of African measles virus field isolates: identification and localisation of one conserved and two variable epitope sites on the NP protein. Virus Res. 10:137-152. [DOI] [PubMed] [Google Scholar]
- 18.Grothues, D., C. R. Cantor, and C. L. Smith. 1993. PCR amplification of megabase DNA with tagged random primers (T-PCR). Nucleic Acids Res. 21:1321-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 20.Horikami, S. M., S. Smallwood, B. Bankamp, and S. A. Moyer. 1994. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205:540-545. [DOI] [PubMed] [Google Scholar]
- 21.Huang, S., L. K. Qu, and A. E. Koromilas. 2004. Induction of p53-dependent apoptosis in HCT116 tumor cells by RNA viruses and possible implications in virus-mediated oncolysis. Cell Cycle 3:1043-1045. [PubMed] [Google Scholar]
- 22.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 23.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 24.Karlin, D., F. Ferron, B. Canard, and S. Longhi. 2003. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84:3239-3252. [DOI] [PubMed] [Google Scholar]
- 25.Karlin, D., S. Longhi, and B. Canard. 2002. Substitution of two residues in the measles virus nucleoprotein results in an impaired self-association. Virology 302:420-432. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki, M., and F. Inagaki. 2001. Random PCR-based screening for soluble domains using green fluorescent protein. Biochem. Biophys. Res. Commun. 280:842-844. [DOI] [PubMed] [Google Scholar]
- 27.Kentsis, A., R. E. Gordon, and K. L. Borden. 2002. Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA 99:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston, R. L., W. A. Baase, and L. S. Gay. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kingston, R. L., D. J. Hamel, L. S. Gay, F. W. Dahlquist, and B. W. Matthews. 2004. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. USA 101:8301-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 31.Logan, I. R., V. Sapountzi, L. Gaughan, D. E. Neal, and C. N. Robson. 2004. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J. Biol. Chem. 279:11696-11704. [DOI] [PubMed] [Google Scholar]
- 32.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638-18648. [DOI] [PubMed] [Google Scholar]
- 33.Meza, J. E., P. S. Brzovic, M. C. King, and R. E. Klevit. 1999. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 274:5659-5665. [DOI] [PubMed] [Google Scholar]
- 34.Naniche, D., S. I. Reed, and M. B. Oldstone. 1999. Cell cycle arrest during measles virus infection: a G0-like block leads to suppression of retinoblastoma protein expression. J. Virol. 73:1894-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plumet, S., W. P. Duprex, and D. Gerlier. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 79:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pothier, P., and J. B. Bour. 1997. Applications des anticorps monoclonaux en virologie. Med. Mal. Infect. 11:592-597. [Google Scholar]
- 37.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson, C. D., and P. W. Choppin. 1983. Oligopeptides that specifically inhibit membrane fusion by paramyxoviruses: studies on the site of action. Virology 131:518-532. [DOI] [PubMed] [Google Scholar]
- 39.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 40.Smallwood, S., K. W. Ryan, and S. A. Moyer. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154-163. [DOI] [PubMed] [Google Scholar]
- 41.Spehner, D., R. Drillien, and P. M. Howley. 1997. The assembly of the measles virus nucleoprotein into nucleocapsid-like particles is modulated by the phosphoprotein. Virology 232:260-268. [DOI] [PubMed] [Google Scholar]
- 42.Stein, C. E., M. Birmingham, M. Kurian, P. Duclos, and P. Strebel. 2003. The global burden of measles in the year 2000—a model that uses country-specific indicators. J. Infect. Dis. 187:S8-14. [DOI] [PubMed] [Google Scholar]
- 43.Tanimura, S., S. Ohtsuka, K. Mitsui, K. Shirouzu, A. Yoshimura, and M. Ohtsubo. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447:5-9. [DOI] [PubMed] [Google Scholar]
- 44.Tarbouriech, N., J. Curran, R. W. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 45.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. D. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent, S., D. Spehner, S. Manie, R. Delorme, R. Drillien, and D. Gerlier. 1999. Inefficient measles virus budding in murine L.CD46 fibroblasts. Virology 265:185-195. [DOI] [PubMed] [Google Scholar]
- 47.Vincent, S., I. Tigaud, H. Schneider, C. J. Buchholz, Y. Yanagi, and D. Gerlier. 2002. Restriction of measles virus RNA synthesis by a mouse host cell line: trans-complementation by polymerase components or a human cellular factor(s). J. Virol. 76:6121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, Y., C. C. Li, and A. M. Weissman. 2004. Regulating the p53 system through ubiquitination. Oncogene 23:2096-2106. [DOI] [PubMed] [Google Scholar]