Abstract
Primate lentiviruses have narrow host ranges, due in part to their sensitivities to mammalian intracellular antiviral factors such as APOBEC3G and TRIM5α. Despite the protection provided by this innate immune system, retroviruses are able to transfer between species where they can cause disease. This is true for sooty mangabey simian immunodeficiency virus, which has transferred to humans as HIV-2 and to rhesus macaques as SIVmac, where it causes AIDS. Here we examine the sensitivities of the closely related HIV-2 and SIVmac to restriction by TRIM5α. We show that rhesus TRIM5α can restrict HIV-2 but not the closely related SIVmac. SIVmac has not completely escaped TRIM5α, as shown by its sensitivity to distantly related TRIM5α from the New World squirrel monkey. Squirrel monkey TRIM5α blocks SIVmac infection after DNA synthesis and is not saturable with restriction-sensitive virus-like particles. We map the determinant for TRIM5α sensitivity to the structure in the capsid protein that recruits CypA into HIV-1 virions. We also make an SIV, mutated at this site, which bypasses restriction in all cells tested.
Retroviruses are not strictly species specific and are able to jump, or zoonose, from one species to another. The recent transfers of HIV-1 and HIV-2 into the human population and the resultant AIDS pandemic underline the threat to human health posed by such retroviral zoonotic transfers. It is therefore of interest to consider the molecular mechanisms of the species barriers that protect against zoonotic retroviral infection and to identify the antiviral factors involved. Recently, two classes of antiretroviral factors have been described as an intrinsic immune system (3). The first comprises members of the Cem15/APOBEC3G family of deaminase enzymes (29). They are packaged into sensitive virions and catalyze the deamination of the viral DNA, leading to degradation, mutation, and loss of infectivity (18). This has led primate lentiviruses to evolve vif, which recruits APOBEC3G to the proteasome in an infected cell (10, 20, 30). A second distinct class of inhibitory activities is exemplified by the TRIM5 alleles of humans (Ref1) and monkeys (Lv1) (2, 7, 11, 13, 16, 21, 24, 32, 34, 38). TRIM5α blocks infection early after viral entry, before the establishment of a provirus. Several TRIM5 alleles, notably those from human and Agm, are able to block infection by both lentiviruses and gamma retroviruses. Rhesus macaque Lv1/TRIM5α and APOBEC3G both contribute to the inability of HIV-1 to infect and cause disease in rhesus macaques, the most valuable primate model for AIDS. Other antiviral activities such as ZAP (8) and Lv2 (28) suggest that innate or intrinsic antiviral immune systems will be complex and diverse.
TRIM5 is member of the tripartite motif-containing family of proteins (25). The tripartite motif comprises a RING domain, encoding two atypical zinc fingers, one or two B-box domains, and a coiled-coil domain. The B-box and coiled-coil domains mediate protein-protein interactions between TRIM family members (25). Some TRIM proteins, including TRIM5α, additionally contain a B30.2 or SPRY domain. While the longest splice variant, TRIM5α, restricts retroviral infection, the shorter TRIM5γ acts as a dominant negative when overexpressed and restores restricted infectivity (32). The SPRY domain encodes antiviral specificity (26, 33, 39), and the TRIM5∂ RING domain has been shown to act as an E3 ligase for autoubiquitination in vitro (37), suggesting that ubiquitination of incoming viral capsid might underlie the antiviral process.
Here we characterize the differential restriction of two closely related retroviruses, HIV-2 and SIVmac, by TRIM5α alleles from Old World rhesus macaques and New World squirrel monkeys. Both of these viruses are products of zoonosis from sooty mangabeys. We show that squirrel monkey TRIM5α is uniquely active after viral DNA synthesis and is unsaturable with sensitive virus-like particles. We define the viral TRIM5α-sensitive motif in the capsid protein, in the loop equivalent to the HIV-1 cyclophilin A binding loop.
MATERIALS AND METHODS
Abbreviations.
SIV, simian immunodeficiency virus; SIVmac, SIV from macaques; HIV-1, human immunodeficiency virus type 1; VLP, virus-like particles; siRNA, small interfering RNA; CA, capsid; CypA, cyclophilin A; FACS, fluorescence-activated cell sorting; GFP, green fluorescent protein; RFP, red fluorescent protein; ELISA, enzyme-linked immunosorbent assay; RT, reverse transcriptase; VSV-G, vesicular stomatitis virus glycoprotein; Agm, African green monkey; MLV, murine leukemia virus.
Cell lines, viral vectors, infection assays, and quantitative PCR.
Cell lines TE671, 293T, FRhk4, CRFK, and pindak have been described previously (11, 16). VSV-G-pseudotyped SIVmac vectors (22) were prepared by FuGENE-6 transfection of 293T cells as described previously (2). HIV-2 vectors were produced as described above by use of the HIV-2 RODA gag-pol expression vector pSVR∂NB (9) and an HIV-2 vector encoding GFP derived from pSVR∂NBPuro∂H (9) by replacing the SV40 puro element with a cytomegalovirus immediate early promoter driving GFP expression from plasmid pCNCG (2) at the SalI site. Infection assays were performed in six-well plates containing 105 cells per well. Infected cells expressing GFP were enumerated by fluorescence-activated cell sorting (Becton Dickinson) 48 h after infection. Viral reverse transcriptase activity was measured by reverse transcriptase ELISA (Cavidi Tech, Uppsala, Sweden). TaqMan quantitative PCR to measure viral DNA synthesis was performed as previously described using primers/probe specific to GFP as described previously (2).
Expression of TRIM5α in CRFK cells.
mRNA was purified from rhesus macaque FRhK4 cells and squirrel monkey pindak cells by using TRIzol reagent (Invitrogen), and cDNA was synthesized by using SuperScript (Invitrogen). The cDNA was used as a template for PCR using primers fwd 5′-CAGACGAATTCCACCATGGCTTCTGGAATCCTG-3′ and rev 5′-GGACGTTCGAAATAGAAAGAAGGGAGACAGC-3′; EcoRI and Csp45I sites are underlined. TRIM5α PCR products were cloned into the MLV expression vector CXCR, a derivative of CNCG, between the EcoRI and the Csp45I sites such that it was expressed under the control of the MLV long terminal repeat in infected cells (16). This virus also encodes RFP (Clontech), also expressed in infected cells. TRIM5α- and RFP-encoding vectors were prepared by 293T transfection using NB-tropic Moloney MLV core and a VSV-G envelope as described previously (16). CRFK cells were transduced with vector encoding TRIM5α and RFP. Single cell clones were isolated by limiting dilution and positive clones identified by red fluorescence microscopy. Positive clones were infected with GFP-encoding virus and, 48 h after that, assayed for green fluorescence by FACS as previously described (16). Rhesus TRIM5α sequence was verified by DNA sequencing. Squirrel monkey TRIM5α was determined by sequencing three independent clones. DNA analysis was performed with DNACowboy (Blue Tractor Software, Malltraeth, Wales, United Kingdom).
Disruption of TRIM5α expression with siRNA.
Disruption of squirrel monkey TRIM5α expression by siRNA was performed by transfecting cells with siRNA oligonucleotides (QIAGEN) using Oligofectamine (Invitrogen) as described previously (16). The siRNA oligonucleotide sequence to squirrel monkey was GUUGGAAGCUGACGUCAGA. siRNA to rhesus and human TRIM5 was delivered using a lentiviral vector based on pHR′SINcPPT-SGW (1), referred to here as CSGW. CSGW was modified for siRNA expression as follows. The human U6 promoter was amplified by PCR from the human cell line HBL-6 by using primers forward 5′-GGGGCTGCAGAAGGTCGGGCAGGAAGAGGGCCTATTTCCC-3′ and reverse 5′-TATCGTATGCATGCATGCAAAAAATCTAGAGAAGCGTCGACGGTGTTTCGTCCTTTCCACAAG-3′. It was cloned into self-ligated pGEM-T-Easy (Promega) by use of PstI and NsiI sites. Synthetic oligonucleotides 5′-TCGAATTCAGGATCCTGAGCTCGTGCA-3′ and 5′-CGAGCTCAGGATCCTGAATTCGATGCA-3′ were then annealed and inserted into the NsiI site. The resulting plasmid was named pGEM-U6-LINKER. Synthetic oligonucleotides 5′-CTTAGGGAGGTCAGGTTGAGTCTTCGGGCTCAACTTGACCTCCCTGAGCTTTTT-3′ and 5′-CTAGAAAAAGCTCAGGGAGGTCAAGTTGAGCCCGAAGACTCAACCTGACCTCCCTAAG-3′, designed according to the siRNA sequence described by Stremlau et al. (32), were annealed and ligated to SalI-digested, mung bean nuclease blunt-ended, XbaI-digested pGEM-U6-LINKER. The resulting plasmid contains an U6-shRNA siRNA cassette, which was cloned into CSGW by use of EcoRI. This vector was packaged into VSV-G-pseudotyped HIV-1 virus by transfection of 293T cells as described above.
Forty-eight h after siRNA transfection or infection with siRNA-encoding vector, cells were infected with GFP-encoding retroviral vectors. Green-fluorescing, infected cells were enumerated by FACS (Becton Dickinson) 48 h after this.
Mutagenesis.
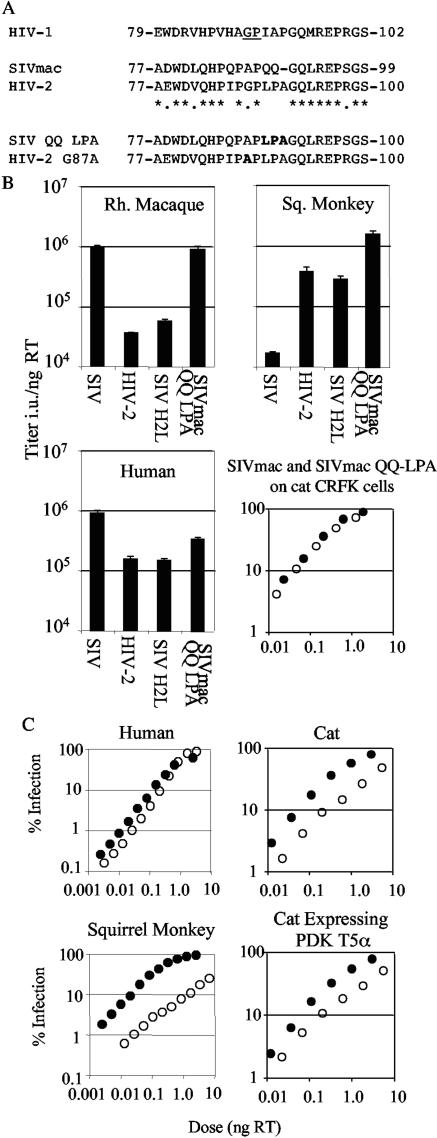
Mutants of SIVmac packaging plasmid pSIV3+, G87A and QQ89-90LPA, were made by using the QuikChange protocol (Stratagene). Oligonucleotide sequences are available on request. SIV3+ containing HIV-2 residues CA77 to -107 was made by PCR using HIV-2 packaging plasmid pSVR∂NB as a template and oligonucleotides fwd CAATGAGGAAGCTGCAGAATGGGATG and rev CTACTGTACTAGTTGTCCCTGCTATG containing the underlined PstI and SpeI sites. This fragment, cut PstI and SpeI, was used to replace the corresponding fragment in pSIV3+. This mutagenesis effectively alters SIVmac residues 78 to 98 into residue 77, and 99 to 107 are conserved (see Fig. 5). All constructs were sequenced. Sequences were analyzed and primers designed using DNACowboy (Blue Tractor Software, Malltraeth, Wales, United Kingdom).
FIG. 5.
Mutational analysis reveals that TRIM5α sensitivity determinants lie in the capsid region equivalent to the cyclophilin A binding loop of HIV-1. (A) Partial capsid sequences of HIV-1, HIV-2, SIVmac, and SIVmac mutants. CA residue numbers are marked, mutated residues are in bold, and alignment symbols refer to SIVmac and HIV-2. The G-P motif responsible for recruiting CypA into HIV-1 cores is underlined. (B) Titers of SIVmac, HIV-2, SIVmac encoding HIV-2 CA residues 78 to 98 (SIV H2L), and SIVmac containing HIV-2 residues 89 to 91 (SIVmac QQ LPA) were determined on human TE671 cells, rhesus macaque FRhK4 cells, and squirrel monkey pindak cells. Errors are standard errors of the mean from two independent experiments performed with two independent virus prepara-tions. Virus doses to determine titer infected between 1 and 15% of target cells. Also shown are titrations of SIVmac GFP (filled symbols) and SIVmac QQ-LPA (open symbols) on feline CRFK cells. (C) Serial dilutions of wild-type HIV-2 GFP (filled symbols) and HIV-2 G87A (open symbols) GFP were titrated onto human TE671 cells, squirrel monkey pindak cells, feline CRFK cells, and feline CRFK cells expressing squirrel monkey TRIM5. Viral doses were determined by RT ELISA. Plots are representative of two independent experiments performed with two independent viral preparations. Viral titers, in infectious units per nanogram of reverse transcriptase (Titer i.u./ng RT), were measured by ELISA.
Nucleotide sequence accession number.
Squirrel monkey TRIM5α has been assigned GenBank accession number AY928202.
RESULTS
Human, rhesus macaque, and squirrel monkey cell lines are differentially permissive to HIV-2 and SIVmac infection.
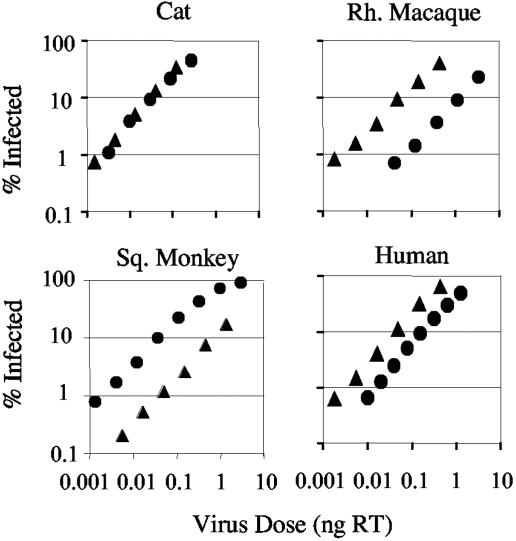
We sought cell lines that had different permissivities to the closely related primate lentiviruses HIV-2 and SIVmac. We titrated HIV-2 GFP and SIVmac GFP onto human (TE671), cat (CRFK), rhesus macaque (FRhK4), and squirrel monkey (pindak) cells. Viral doses were equalized by assaying reverse transcriptase activity. While the titers of HIV-2 GFP and SIVmac GFP are similar on cat cells, the titer of HIV-2 GFP on rhesus cells and the titer of SIVmac GFP on squirrel monkey cells are restricted by around 1 order of magnitude (Fig. 1). In addition, the HIV-2 GFP titer is slightly reduced on human cells compared to SIVmac GFP titer.
FIG. 1.
Titration of HIV-2 and SIVmac-GFP onto cells from cat, rhesus macaque, squirrel monkey, and human. Threefold serial dilutions of HIV-2 GFP (•) and SIVmac GFP (▴) were titrated onto cells, and GFP expression was measured 48 h later. Virus doses were measured by reverse transcriptase ELISA. Results are representative of three repetitions.
TRIM5-specific siRNA rescues infectivity of restricted virus in rhesus and squirrel monkey cells.
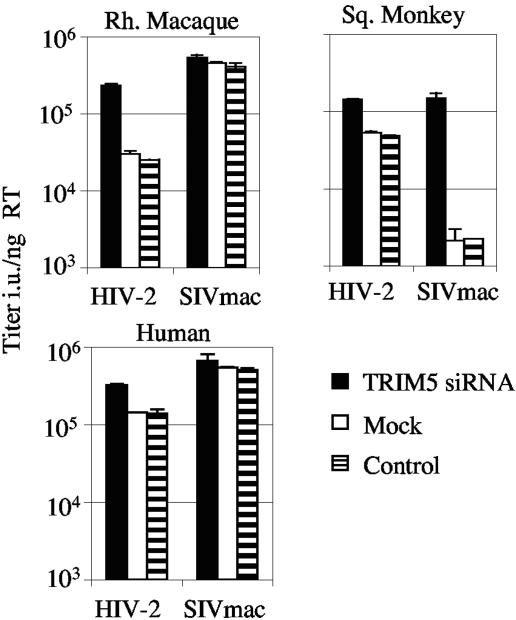
To examine the role of TRIM5α in the species-specific infectivities of HIV-2 and SIVmac on rhesus macaque and squirrel monkey cells, we used siRNA. Reducing TRIM5α expression restores the infectivity of restricted HIV-2 GFP in rhesus cells but does not affect the titer of unrestricted SIVmac GFP. Furthermore, the introduction of TRIM5α siRNA into squirrel monkey cells significantly restores the infectivity of restricted SIVmac GFP and slightly increases the infectivity of HIV-2 GFP. These data indicate that the restriction factor TRIM5α is essential for the reduced infectivity of HIV-2 GFP in cells from rhesus macaque and for the reduced infectivity of SIVmac GFP in squirrel monkey cells. The weak restriction of HIV-2 in human cells is also partially abrogated by a reduction of TRIM5 expression.
Expression of human, rhesus, or squirrel monkey TRIM5α in permissive feline cells renders them nonpermissive for restricted retrovirus.
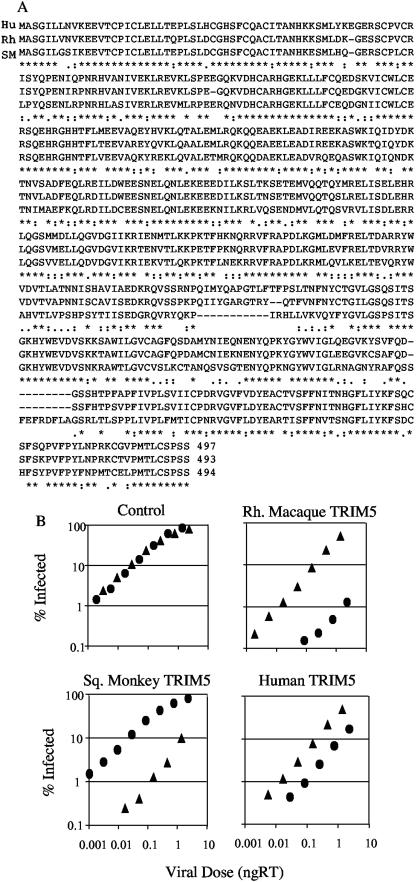
To confirm the role of TRIM5α in restriction of HIV-2 and SIVmac GFP, we cloned the rhesus macaque and squirrel monkey TRIM5α genes from rhesus FRhK4 cells and squirrel monkey pindak cells by using PCR (see Fig. 3A). The TRIM5α sequence from rhesus FRhK4 cells is identical to the previously described rhesus TRIM5α sequence obtained from rhesus primary cells. The squirrel monkey sequence is closely related to that of rhesus TRIM5α but has significant differences in the C-terminal SPRY domain (see Fig. 3A) (32). We note that the region of squirrel monkey TRIM5α homologous to that containing residue R332 in human T5α, recently shown to control the inverse restriction of HIV-1 by human and rhesus TRIM5αs (39), is absent in the squirrel monkey sequence. There is also an insertion of nine residues in the squirrel monkey SPRY domain. The fact that the SPRY domain largely controls human, rhesus, and Agm TRIM5α antiviral specificity suggests that these differences are likely to be important to the antiviral specificity of squirrel monkey TRIM5α (31, 39).
FIG.3.
Expression of rhesus, squirrel monkey, or human TRIM5α in permissive feline cells renders them nonpermissive for restricted retrovirus. (A) Human (Hu), rhesus (Rh), and squirrel monkey (SM) TRIM5α sequences are aligned. (B) Cat cells were left untransduced as a control or were transduced with retroviral vector encoding rhesus macaque TRIM5α, squirrel monkey TRIM5α, or human TRIM5α, and TRIM5α-positive clones were isolated by limiting dilution. Serial dilutions of HIV-2 GFP (•) or SIVmac GFP (▴) were then titrated onto the TRIM5α-positive cells and the percent infected cells determined by FACS. Viral doses were measured by RT ELISA. Results are representative of three repetitions.
We derived feline CRFK cell lines stably expressing rhesus and squirrel monkey TRIM5α as described previously (16). CRFK cells expressing human TRIM5α have been described previously (16). We titrated HIV-2 GFP and SIVmac GFP on unmodified and TRIM5α-expressing CRFK cells (see Fig. 3B). Unmodified cells are equally permissive to both viruses, whereas expression of human TRIM5α renders them able to weakly restrict SIVmac and more strongly restrict HIV-2. The expression of rhesus TRIM5α reduces the titer of HIV-2 GFP by around 100-fold and that of SIVmac GFP by around fivefold. The expression of squirrel monkey TRIM5α reduces the titer of SIVmac GFP by around 100-fold but has no effect on the titer of HIV-2 GFP. These observations are consistent with the notion that the human, rhesus macaque, and squirrel monkey TRIM5α alleles are responsible for the differences in HIV-2 and SIVmac titers on cell lines derived from these species. The stronger restriction of HIV-2 compared to endogenous TRIM5α expression in human cells (compare Fig. 1 and 2 to Fig. 3) and the additional restriction of SIVmac by human TRIM5α is likely to be due to TRIM5 overexpression. Overexpression of restriction factors has previously been described as leading to restriction of previously insensitive virus (4, 16).
FIG. 2.
TRIM5-specific siRNA rescues infectivity of restricted virus in rhesus, squirrel monkey, and human cells. Rhesus macaque FRhK4 cells, squirrel monkey pindak cells, or human TE671 cells were left untreated (striped bars) or were mock treated (open bars) or treated with siRNA to TRIM5 (solid bars). Forty-eight h later, cells were infected with equal doses (determined by RT ELISA) of HIV-2 GFP or SIVmac GFP as shown. Doses were chosen such that the percentages of infected cells were between 0.5 and 2%. Errors are standard errors of the mean of two independent infections by two independent virus preparations. Viral titers, in infectious units per nanogram of reverse transcriptase (Titer i.u./ng RT), were measured by ELISA.
Restriction by squirrel monkey TRIM5 is after DNA synthesis and is not saturable with sensitive virus-like particles.
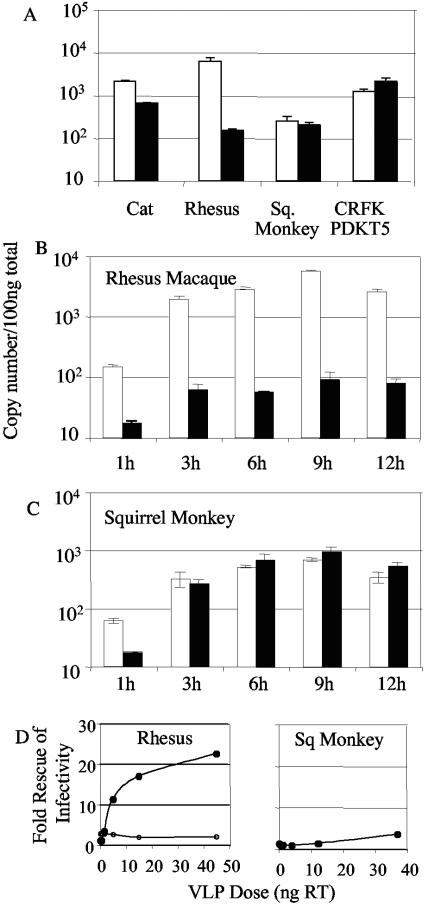
All examples of TRIM5-mediated innate immunity to retroviral infection so far described are mediated early after viral entry, before significant reverse transcription has occurred (2, 7, 11, 13, 16, 21, 24, 32, 34, 38). In order to test whether this is the case for the restriction of HIV-2 by rhesus TRIM5α and for that of SIVmac by squirrel monkey TRIM5α, we measured the amount of viral DNA 6 hours after infection by equivalent doses of HIV-2 and SIVmac GFP by using quantitative TaqMan PCR. CRFK cells were equivalently infected by equal doses of HIV-2 and SIVmac (Fig. 1), but SIVmac DNA synthesis was slightly higher (Fig. 4A). HIV-2 DNA synthesis was reduced by 100-fold in rhesus monkey cells in comparison to that of SIVmac. The differences in DNA synthesis between HIV-2 and SIVmac correlate with the differences in the observed titers (Fig. 1). Surprisingly, levels of DNA synthesis by HIV-2 and SIVmac are very close 6 h after the infection of squirrel monkey cells (Fig. 4A). We therefore performed a time course experiment, measuring viral DNA at fixed points after the infection of rhesus macaque (Fig. 4B) and squirrel monkey (Fig. 4C) cells by HIV-2 GFP and SIVmac GFP. These data show that the amount of unrestricted SIVmac DNA is always higher than that of restricted HIV-2 DNA in rhesus cells at each time point, indicating that the block to HIV-2 infection is at or before DNA synthesis. Conversely, after the infection of squirrel monkey cells, both restricted SIVmac and unrestricted HIV-2 GFP synthesize similar amounts of DNA, with a single exception at the earliest time point at 1 hour, at which the restricted SIVmac makes more DNA than the unrestricted virus, HIV-2. These data indicate that squirrel monkey TRIM5α blocks SIVmac infectivity without significantly affecting DNA synthesis. We then tested whether the later block to infection mediated by squirrel monkey TRIM5α was conserved in feline CRFK cells expressing squirrel monkey TRIM5α. We compared DNA synthesis by HIV-2 GFP and that by SIVmac GFP 6 h after infection. The result indicates that the timing of squirrel monkey TRIM5α restriction is conserved when the TRIM5α is exogenously expressed in feline cells (Fig. 4A).
FIG. 4.
Restriction by squirrel monkey TRIM5α is after DNA synthesis and is not saturable with restriction-sensitive virus-like particles. (A) Cells from species indicated, i.e., cat CRFK, rhesus FRhK4, squirrel (Sq.) monkey pindak, or cat CRFK expressing squirrel monkey TRIM5α (CRFK PDKT5), were infected with equal doses of SIVmac GFP (open bars) and HIV-2 (solid bars) determined by RT ELISA. Six h postinfection, total DNA was purified and 100 ng subjected to quantitative PCR for the GFP target. This experiment was repeated as described above, except that infections were performed on (B) rhesus macaque FRhK4 cells or (C) squirrel monkey pindak cells, and DNA was purified from parallel samples at the time points indicated. Errors are standard errors of the mean of two independent infections performed in parallel, and data are representative of two repetitions using independent virus preparations. (D) A fixed dose of HIV-2 GFP (filled symbols) or SIVmac GFP (open symbols) was used to infect rhesus macaque FRhK4 cells in the presence or absence of a serial dilution of HIV-1 VLP. SIVmac GFP was used to infect squirrel monkey pindak cells in the presence or absence of a serial dilution of SIVmac VLP. The n-fold increase in viral titer in the presence of the VLP is plotted as a function of VLP dose as measured by RT ELISA. The fixed doses of GFP-encoding viruses were chosen to infect between 0.5 and 2% of the target cells.
All TRIM5-mediated restrictions of retroviral infection found thus far have been shown to be saturable by exposure of cells to restriction-sensitive VLP (2, 7, 21, 35). We imagine that the VLP enter the cells, titrate the available TRIM5, and render the cells permissive to restricted virus. Saturation of TRIM5α in rhesus FRhK4 cells with HIV-1 VLP increases the titer of restricted HIV-2 GFP by more than 20 times (Fig. 4D). Unrestricted SIVmac titer remains unaffected. However, restriction of SIVmac GFP in squirrel monkey cells is not saturated with VLP. At high doses of SIVmac VLP, we were unable to rescue infectivity by more than around threefold (Fig. 4D). This is despite the fact that the highest VLP dose of ∼40 ng RT represents a multiplicity of infection of at least 5 on these cells and will infect all of the cells, according to the data shown in Fig. 1. This implies that the inability of the VLP to saturate is not explained by an insufficient amount of VLP in these experiments.
Mutational analysis reveals that TRIM5α sensitivity determinants lie in the capsid region equivalent to the cyclophilin A binding loop of HIV-1.
In order to map the region of the capsid responsible for the differential sensitivity of the closely related HIV-2 and SIVmac to TRIM5α restriction, we performed a mutational analysis. We reasoned that swapping sections of capsid between closely related viruses would be tolerated without a loss in fitness that could be misinterpreted as sensitivity to restriction. We replaced a section of the SIV capsid with the equivalent region from the HIV-2 capsid. HIV-2 residues CA78 to CA98 replaced residues CA78 to CA97 in SIVmac (Fig. 5A). This includes the region equivalent to the CypA binding loop of the HIV-1 capsid. The titer of the resulting SIVmac-based SIV H2L was determined on human TE671, rhesus FRhK4, and squirrel monkey pindak cells (Fig. 5B). Sensitivity to human, rhesus, and squirrel monkey TRIM5α was determined by this part of the capsid, and the transfer of CA78 to 98 from HIV-2 to SIVmac transferred HIV-2's sensitivity to human and rhesus TRIM5αs and its insensitivity to squirrel monkey TRIM5α.
In order to further map the TRIM5α-sensitive motif, we mutated SIVmac CA residues 89 to 90, QQ, to LPA, the equivalent residues in HIV-2 (Fig. 5A), and titrated this virus on rhesus, squirrel monkey, and human cell lines (Fig. 5B). Interestingly, this mutation had no effect on the titer on rhesus cells but restored infectivity for squirrel monkey cells. These data indicate that sensitivity to TRIM5α is controlled by residues in this exposed loop and that the sensitivity to human and rhesus TRIM5α can be separated from the sensitivity to squirrel monkey TRIM5α. We also compared the titer of wild-type SIVmac and SIVmac QQ-LPA on CRFK cells (Fig. 5B). The titers of the wild type and the mutant virus are the same on three unrestrictive cell types, i.e., rhesus, human, and feline, indicating that this mutant has not become nonspecifically high titer.
An inspection of the HIV-2 CA sequence (Fig. 5A) and comparison with the equivalent region of HIV-1 revealed a glycine-proline motif at CA87 to -88 in HIV-2. The recruitment of CypA into the HIV-1 capsid by such a G-P motif has been implicated in HIV-1's sensitivity to TRIM5α restriction (15, 17, 36). We therefore mutated CA A87G in the SIVmac capsid to create the G-P motif found in HIV-2. We also mutated HIV-2 G87A to destroy the motif in HIV-2. Generating a G-P motif in SIVmac had no significant effect on the titer of SIVmac in human, rhesus macaque, or squirrel monkey cells (data not shown). However, destroying the G-P motif in HIV-2 reduced the titer of HIV-2 G87A in a species-specific way (Fig. 5C). HIV-2 G87A titer is almost equal to that of the wild-type virus in human and is reduced by three times in feline cells. However, it is specifically and strongly reduced in squirrel monkey cells. This does not appear to be due to an acquired sensitivity to squirrel monkey TRIM5α, as evidenced by similar titers of mutant and wild-type virus in feline CRFK cells and feline CRFK cells expressing squirrel monkey TRIM5α (Fig. 5C). Furthermore, reduction of TRIM5 expression in squirrel monkey cells with siRNA does not significantly increase the titer of HIV-2 G87A (data not shown). The reduction in titer in squirrel monkey cells is accompanied by an atypical nonlinear titration curve. These observations suggest that HIV-2 G87A has acquired either sensitivity to another antiviral factor or a reduced ability to interact with required factors specifically in squirrel monkey cells. The titers of these mutants are not influenced by cyclosporine A treatment, indicating an independence from cyclophilin A (data not shown). We also note that cyclosporine A treatment had no effect on the infectivity of either HIV-2 or SIVmac in human cells in spreading infection assays (5).
DISCUSSION
The recent descriptions of TRIM5α as an important mediator of retroviral permissivity in primates has enabled us to examine the TRIM5α sensitivities of closely related zoonotic retroviruses that have been able to transfer between primates. We find that, not surprisingly, rhesus TRIM5α is unable to restrict infection by SIVmac, which causes an AIDS-like disease in infected rhesus macaques (19). This virus has not completely escaped TRIM5α sensitivity, however, as shown by its restriction by a distantly related TRIM5α protein from squirrel monkeys. This observation explains the low titer of SIVmac on squirrel monkey cells (14). HIV-2, however, which is very closely related to SIVmac, is strongly restricted by rhesus TRIM5α, suggesting that these two viruses may have been under different selection pressures in their transfer to humans and rhesus monkeys, respectively. HIV-2 restriction in rhesus cells is likely to explain the difficulty of infecting rhesus macaques with this virus. The successful infection of rhesus macaques has been achieved, but only after the selection of particular animals based on the ability of SIVmac to replicate in their peripheral blood lymphocytes (6). Overexpression of TRIM5α, shown in Fig. 3, reveals that TRIM5 antiviral specificity broadens when TRIM5 is overexpressed. Similar observations have been described for the murine restriction factor Fv1 (4). These experiments reveal significant antiviral activity when TRIM5α protein levels are high within a cell, suggesting that differences in TRIM5α expression levels may account for differing abilities of individuals or cells within an individual to replicate a particular virus. HIV-2 is weakly restricted by human TRIM5α, particularly when overexpressed, and we speculate that TRIM5α might contribute to the lower pathogenicity of HIV-2 than of HIV-1, a virus that is clearly not restricted by human TRIM5α.
The cloning and sequencing of the squirrel monkey TRIM5α allele reveals that it is closely related to human and rhesus macaque TRIM5α and, unlike the owl monkey TRIM-Cyp protein (23, 27), has an intact SPRY domain. Measurement of viral DNA synthesis after infection reveals that squirrel monkey TRIM5α-restricted virus, SIVmac, is able to synthesize as much DNA as the unrestricted virus is, even when TRIM5α is exogenously expressed in feline cells. This is reminiscent of restriction by the murine gag-like restriction factor Fv1 and shows for the first time that strong restriction, after DNA synthesis, is not unique to the gag-like Fv1 protein. In contrast to Fv1 restriction, restriction by squirrel monkey TRIM5α is not readily saturable with sensitive VLP even at multiplicities of infection that saturate restriction in rhesus macaque cells. Although a previous report has demonstrated a partial saturation of SIVmac restriction in squirrel monkey cells, these authors also concluded that partial rescue of restricted infectivity with very high doses of VLP was unique to these cells (7). It therefore remains unclear whether squirrel monkey TRIM5α has an atypical, unsaturable, post-reverse transcription mechanism of restriction or whether such differences are caused by subtle changes in factors such as intracellular localization.
In the case of TRIM5αs from rhesus macaque, squirrel monkey, and owl monkey, and probably for other TRIM5αs, it appears that a major site of interaction between TRIM5 and retroviral capsid is at the exposed loop responsible for packaging CypA into HIV-1 virions. Neither HIV-2 nor SIV recruit CypA into their cores, and drugs that block CA-CypA interactions do not affect the titers of these viruses (5). Transferring a region, equivalent to the HIV-1 CypA binding sequence, of the HIV-2 CA gene to SIVmac is able to confer the specific restriction sensitivity or insensitivity of HIV-2 to SIVmac. Furthermore, small changes within this exposed loop can render SIVmac insensitive to TRIM5α. Currently available retroviral capsid crystal structures, including HIV-1, equine infectious anemia virus, and human T-cell leukemia virus, all include an exposed loop similar to the HIV-1 CypA binding structure. Such conservation might suggest that this structure is required for a host-virus interaction unrelated to CypA and consequently is an attractive target for TRIM5α. This idea might be supported by the specific loss of titer of HIV-2 G87A in squirrel monkey cells as well as its nonlinear titration curve. Such observations could be explained by the G87A mutation altering the ability of the capsid to interact with both positive and negative factors.
The effectiveness of an antiviral factor will depend on the ease with which a virus can escape its activity through mutation. We have shown that small changes in SIVmac CA can render the virus insensitive to restriction by rhesus and squirrel monkey TRIM5α. Single amino acid changes also mediate the escape of MLV-N from both human and Agm TRIM5α as well as the escape of HIV-1 from restriction by owl monkey TRIM-cyp (23, 27, 34, 36). Conversely, point mutations in human and rhesus TRIM5α broaden antiviral specificity to block viruses insensitive to the wild-type proteins (33, 39). While these experiments may suggest that virus can easily escape TRIM5, the effectiveness of innate immunity will depend on several classes of antiviral pathways acting simultaneously and perhaps in concert with the adaptive immune system, for example, with the antiviral activity of cytotoxic T cells reactive against gag (15). We therefore suspect that it may be more difficult for a virus to escape TRIM5α activity through changing CA sequence than CA mutagenesis experiments in vitro might suggest. Moreover, recent work has provided phylogenetic evidence that TRIM5α is positively selected, suggesting that virus and TRIM5α might evolve rapidly in competition, an effect referred to as the Red Queen effect, where simultaneous rapid evolution retains the status quo (26).
An HIV-1 mutant completely insensitive to rhesus macaque TRIM5α would improve animal models of AIDS, allowing the use of HIV-1 rather than a virus based predominantly on SIVmac. Such a virus has remained elusive despite a number of mutational analyses (12, 15). A rhesus-tropic HIV-1 would have to be modified to encode a vif protein active against rhesus APOBEC3G, but this is feasible. Our observations suggest that an HIV-1 insensitive to rhesus TRIM5α might be attainable and further efforts to make such a virus are worthwhile. Furthermore, a better understanding of host-virus interactions is likely to be valuable in the generation of more-effective vectors for gene delivery and for the identification of targets for antiviral therapeutics.
Acknowledgments
We thank Paul Bieniasz for reagents and sharing unpublished data, Ben Webb and Yasu Takeuchi for helpful advice, and Jonathan Scammel, Andrew Lever, and Francois Loic Cosset for reagents.
This work was supported by the Wellcome Trust, a UCL Graduate School Fellowship to Z.K., and a Medical Research Council Ph.D. studentship to S.J.W.
REFERENCES
- 1.Bainbridge, J. W., C. Stephens, K. Parsley, C. Demaison, A. Halfyard, A. J. Thrasher, and R. R. Ali. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene. Ther. 8:1665-1668. [DOI] [PubMed] [Google Scholar]
- 2.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 4.Bock, M., K. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro, B. A., M. Nepomuceno, N. W. Lerche, J. W. Eichberg, and J. A. Levy. 1991. Persistent infection of baboons and rhesus monkeys with different strains of HIV-2. Virology 184:219-226. [DOI] [PubMed] [Google Scholar]
- 7.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 11.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda, Y., L. Ylinen, M. Kahar-Bador, and G. J. Towers. 2004. The influence of gag on human immunodeficiency virus type 1 species-specific tropism. J. Virol. 78:11816-11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 19.Letvin, N. L., M. D. Daniel, P. K. Sehgal, R. C. Desrosiers, R. D. Hunt, L. M. Waldron, J. J. MacKey, D. K. Schmidt, L. V. Chalifoux, and N. W. King. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71-73. [DOI] [PubMed] [Google Scholar]
- 20.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 21.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 23.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573.15243629 [Google Scholar]
- 28.Schmitz, C., D. Marchant, S. J. Neil, K. Aubin, S. Reuter, M. T. Dittmar, and A. McKnight. 2004. Lv2, a novel postentry restriction, is mediated by both capsid and envelope. J. Virol. 78:2006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 30.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 31.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 33.Stremlau, M., M. J. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299.11027299 [Google Scholar]
- 35.Towers, G., M. Collins, and Y. Takeuchi. 2002. Abrogation of Ref1 restriction in human cells. J. Virol. 76:2548-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 37.Xu, L., L. Yang, P. K. Moitra, K. Hashimoto, P. Rallabhandi, S. Kaul, G. Meroni, J. P. Jensen, A. M. Weissman, and P. D'Arpa. 2003. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp. Cell Res. 288:84-93. [DOI] [PubMed] [Google Scholar]
- 38.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]