Abstract
Picornavirus RNA replication involves the specific synthesis of negative-strand intermediates followed by an accumulation of positive-strand viral RNA in the presence of a multitude of cellular mRNAs. Previously, in an effort to identify cis-acting elements required for initiation of negative-strand RNA synthesis, we deleted the entire 3′ noncoding regions from human rhinovirus and poliovirus genomic RNAs. These deletion mutation transcripts displayed a severe delay in RNA accumulation following transfection of HeLa cells. Interestingly, in subsequent infection of HeLa cells, the deletion-mutant poliovirus displayed only a moderate deficiency in RNA synthesis. These data suggested that the delay in the production of cytopathic effects after transfection may have been due to an RNA replication defect overcome by the accumulation of a compensatory mutation(s) generated during initial rounds of RNA synthesis. In this study, we have sequenced the entire genome of the deletion-mutant virus and found only two nucleotide changes from the parental clone. Transfection analysis of these sequence variants revealed that the sequence changes did not provide compensatory functions for the 3′ noncoding region deletion mutation replication defect. Further examination of the deletion mutant phenotype revealed that the severe replication defect following RNA transfection is due, in part, to nonviral terminal sequences present in the in vitro-derived deletion mutation transcripts. Our data suggest that poliovirus RNA harboring a complete 3′ noncoding region deletion mutation is infectious (not merely quasi-infectious).
In the first step of poliovirus (PV) RNA replication, viral and host cell proteins form a membranous replication complex in which positive-strand viral RNA is used as the template to generate full-length, negative-strand RNA. The viral RNA polymerase (3Dpol) has been shown to be necessary and sufficient for elongation of an oligo(U)-primed, polyadenylated RNA template in vitro (17). Furthermore, the viral polymerase can nonspecifically generate the reverse complement of nonviral polyadenylated RNAs when provided with oligo(U) (13, 50). Because the replication complex discriminates between the biochemically similar 3′ ends of cellular polyadenylated mRNAs and the polyadenylated PV genome, a cis-acting RNA replication element is thought to be required for template selection and the proper assembly of the replication initiation complex.
Within the family Picornaviridae, several RNA elements involved in genomic replication have been described. A stem-loop structure required for viral RNA replication was identified within the capsid-coding sequence of human rhinovirus 14 (35, 36). This replication element, termed cre, is located about 2.3 kb downstream from the 5′ end and more than 4 kb from the 3′ end of the genome. Elements with similar structures have been described in a number of other picornaviruses, each containing a conserved AAACA motif (21, 22, 33, 46). In PV1, the cre is located in the coding region of the nonstructural protein 2C. Additional studies have shown that the in vitro uridylylation of the viral protein VPg is greatly stimulated by the presence of the PV1 cre (46). The uridylylated form of VPg has long been thought to act as a protein primer for both positive-strand and negative-strand RNA synthesis, suggesting that the cre is necessary for the uridylylation of VPg during a PV infection.
Another replication element is found in enterovirus sequences corresponding to nucleotides 1 to ∼100 of the positive-strand genomic RNA (3, 74). This element is confined within stem-loop I of the PV 5′ NCR (noncoding region) (24) and has a cloverleaf-like structure (3). This RNA structure binds viral protein 3CD and host protein PCBP, forming a ribonucleoprotein complex (2, 19, 45). Mutational analysis of the cloverleaf RNA suggests that the ribonucleoprotein complex is required for RNA replication (2, 54, 56) by acting either in cis for negative-strand synthesis (7, 20) or in trans for positive-strand synthesis (3). In addition, a ribonucleoprotein complex has also been shown to form at the 3′ end of the complementary negative-strand RNA (9, 54). Further complicating the analysis, mutations of the cloverleaf structure have resulted in defects in cap-independent translation of the poliovirus genome (45, 60). It has also been suggested that the 5′ cloverleaf ribonucleoprotein complex is involved in RNA stability (40), which would indirectly account for the apparent role in both replication and translation. Whether direct or indirect, the mechanism by which the PV 5′ cloverleaf contributes to genomic replication has not been fully elucidated.
These findings highlight several important aspects of replication elements in picornaviruses. First, conserved sequences and structures involved in RNA replication need not be merely targeting signals that recruit the RNA genome to the replication complex but can be functional elements in any number of essential processes in the replication cycle. Second, functional RNA structure elements may be contained in the positive strand, the negative strand, or both strands of complementary RNAs. In addition, elements involved in RNA replication need not be close (in terms of linear sequence) to the sites of RNA replication initiation. Finally, picornavirus genomes contain multiple RNA replication element sequences, with each one likely to have a unique function.
For several reasons, the PV 3′ NCR is a logical candidate to encode a cis-acting replication element that is required for the initiation of negative-strand RNA synthesis. Binding of the PV 3′ NCR to components within the replication complex could be responsible for bringing the 3′ poly(A) tract into close proximity to the 3Dpol to initiate negative-strand synthesis. Furthermore, while the PV 3′ NCR is presumably not subject to the selective pressure of codon maintenance, there is a high degree of sequence conservation among the genomic 3′ NCRs of the three PV serotypes. Such sequence conservation suggests that a strong selection pressure, like the preservation of a cis-acting replication element, is important in maintaining the primary structure (and therefore secondary structure and/or tertiary structure) of this region of the viral genome. Finally, it is likely that a conserved RNA motif is required for the replication complex to discriminate between PV genomic RNA and the multitude of cellular mRNAs. While there are at least two cis-acting replication elements in the PV1 genome, there is no evidence that either of these elements serves to target the replication complex.
Several reports have proposed that RNA structures within the PV 3′ NCR function in PV replication. Jacobson et al. proposed a pseudoknot structure that involves sequences from the 3Dpol gene and the 3′ NCR (28). Disruption of this proposed structure by an 8-nucleotide insertion gave rise to a mutant virus with a temperature-sensitive phenotype; however, insertions of 2 or 10 nucleotides at the same site produced viruses with wild-type growth phenotypes (57). A more extensive study designed to disrupt the putative pseudoknot structure also yielded anomalous infectivities of transfected mutated cDNAs (47). Alternatively, Pilipenko et al. proposed the existence of two stem-loops (X and Y) within the 3′ NCR of poliovirus (48). The existence of these structures would preclude the formation of the pseudoknot structure. Furthermore, a proposed tertiary “kissing” interaction involving base pairing between the loop regions of stem-loops X and Y was suggested (48, 49). Transfection of transcripts containing mutations in either loop region resulted in recovered virus that contained reversion mutations or second-site compensatory mutations predicted to reestablish the “kissing” interaction. However, not all of the recovered viruses had mutations that could reestablish this interaction, and the authors suggested the possible existence of unidentified second-site compensatory mutations. While these studies suggested a role for the PV 3′ NCR in genome replication, a clear mechanistic description and a precise definition of a minimal cis-acting replication element have yet to emerge.
To more clearly define the minimal boundaries of and requirements for a cis-acting replication element within the PV 3′ NCR, we previously reported the deletion of the entire 3′ NCR from a full-length PV cDNA construct (pT7PVΔ3′NCR) (63). Surprisingly, transfections of transcripts harboring this complete PV 3′ NCR deletion mutation (Δ3′ NCR PV1) demonstrated that, while the deletion mutation was deleterious to RNA replication, the lesion was not lethal. When deletion mutation transcripts were transfected into HeLa cells, there was a long delay before the detection of cytopathic effects (CPE) and RNA accumulation. We showed that the replication delay was not due to a defect in the accumulation of viral translation products. In contrast, virus recovered from these transfections had only a moderate RNA replication defect. The recovered viruses maintained the complete 3′ NCR deletion mutation and were able to accumulate to levels 1 log10 lower than wild type. Further analysis of the recovered virus suggested that there was a defect in the initiation of positive-strand RNA synthesis (8). The long delay before the abrupt appearance of newly synthesized viral RNA after transfection with pT7PVΔ3′NCR-derived RNA, coupled with the almost-wild-type phenotype of third-passage Δ3′ NCR PV1, suggested that a severe RNA replication defect was overcome by the accumulation of a compensatory mutation(s) generated during initial rounds of viral RNA replication.
In an attempt to identify compensatory mutations responsible for the replication phenotype of Δ3′ NCR PV, we sequenced the entire genome of a plaque-purified isolate (4-1 M). Furthermore, to understand how identified mutations affect PV genome replication in the absence of a 3′ NCR, we reconstructed the pT7PVΔ3′NCR full-length cDNA by reverse transcription-PCR (RT-PCR) amplification of subgenomic segments of the Δ3′ NCR PV1 genome. Here we show that a second-site compensatory mutation is not required to achieve nearly wild-type RNA replication of the deletion mutation RNA. We also show that there are no significant differences in the kinetics of translation or in the stability of the deletion mutation transcripts compared to transcripts corresponding to wild-type poliovirus genomes. In addition, we found that removing the covalently linked viral protein VPg does not affect the infectivity of deletion mutant viral RNA. However, establishing an authentic viral polyadenylated terminus increases the specific infectivity of deletion mutation transcripts, suggesting that the PV 3′ NCR may be involved in site selection for initiation of negative-strand RNA synthesis.
MATERIALS AND METHODS
Virus stocks and plaque assay.
Δ3′ NCR PV1 mutant virus stocks were plaque isolated and passaged in HeLa (R19) monolayers as described previously (11). HeLa cell monolayers were infected with serial dilutions of virus and overlaid with semisolid minimal essential medium (MEM) containing 0.45% agarose. Well-isolated plaques were harvested. Virus was twice plaque purified and passaged three times in HeLa (R19) monolayers at 37°C to amplify the virus stocks. Plaque assays were performed as described previously (11).
One-step growth cycle analysis.
One-step growth analysis was performed on HeLa (R19) monolayers in 60-mm plates at 37°C as described previously (64). HeLa cell monolayers were infected at a multiplicity of infection (MOI) of 15 for 30 min at room temperature in 60-mm plates and washed with phosphate-buffered saline, followed by the addition of 3 ml MEM plus 8% newborn calf serum with 1% nonessential amino acids. Infected-cell monolayers were harvested and virus was released from the cells by five freeze/thaw cycles, and the titer of the clarified supernatant was determined by plaque assays.
Total cellular RNA extraction.
Total cytoplasmic RNA for viral genome sequencing and for use in cDNA synthesis was harvested by the Nonidet P-40, sodium dodecyl sulfate (SDS) lysis method, modified after Campos and Villarreal (10). HeLa cell monolayers were infected at an MOI of 15. At 5 to 6 h postinfection, cells were harvested and collected by centrifugation. Cells were lysed by adding Nonidet P-40-TSM buffer (0.15 M NaCl, 5 mM MgCl2, 10 mM Tris-HCl [pH 7.6], 2% [vol/vol] Nonidet P-40). The lysate was clarified by centrifugation, and the cytoplasmic fraction was equilibrated with an equal volume of RNA extraction buffer (8 M urea, 2% SDS, 20 mM EDTA, 0.3 M NaCl, 50 mM Tris, pH 8.0). Total cytoplasmic RNA was phenol-chloroform extracted, ethanol (EtOH) precipitated, and resuspended in diethyl pyrocarbonate (DEPC)-treated H2O.
Plasmids.
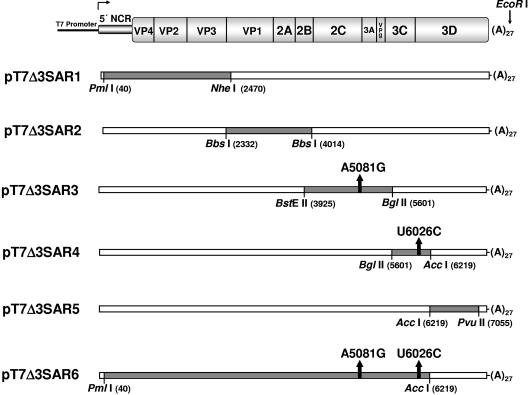
A T7 bacteriophage promoter-containing plasmid encoding a genome length PV1 cDNA (pT7-PV1) (23) and a PV1 genome harboring a complete 3′ NCR deletion (pT7PVΔ3′NCR) (63) have been described previously. Synthesis of cDNA was performed as described previously (15). Plasmids containing suballelic reconstructions pT7Δ3SAR1, -2, -3, -4, and -5 were generated by RT-PCR amplification of Δ3′ NCR PV1 (4-1 M) viral RNA genomic segments from extracted total cytoplasmic RNA using primer sets (PV genomic position is indicated in brackets) T2(+) [12 to 41]/PV2365(−) [2365 to 2384], DBPV2184(+) [2184 to 2203]/JT2C4169(−) [4169 to 4188], DBPV2992(+) [2992 to 3011]/L453(−) [5629 to 5647], PV2B(+) [3995 to 4015]/PV3D3(−) [6233 to 6252], and L449(+) [6012 to 6026]/S7181(−) [7181 to 7197], respectively. The cDNAs were cloned into the pT7PVΔ3′NCR construct using the restriction endonuclease sites indicated in Fig. 2. Plasmid pT7Δ3SAR6 was generated by combining the NheI-to-BstEII DNA fragment of pT7Δ3SAR2, the BstEII-to-BglII DNA fragment of pT7Δ3SAR3, and the BglII-to-AccI DNA fragment of pT7Δ3SAR4 with the vector containing the AccI-to-NheI DNA fragment of pT7Δ3SAR1.
FIG. 2.
Schematic map of suballelic reconstructions. The figure shows how the full-length deletion mutation cDNA, pT7PVΔ3′NCR, was reconstructed with cDNA fragments of the third-passage Δ3′ NCR PV1 (4-1 M) RNA. Suballelic portions of the genome were reverse transcribed, PCR amplified, and cloned into appropriate restriction sites of pT7PVΔ3′NCR. The segments are displayed as the shaded areas, and the restriction endonucleases (and restriction site locations) used in the cloning of these suballelic reconstructions are labeled. The arrows within the shaded genomic segments indicate the location and nucleotide changes of Δ3′ NCR PV1 (4-1 M) RNA from the parental clone. Each plasmid was linearized using restriction endonuclease EcoRI and was under the transcriptional control of a T7 bacteriophage promoter element. A genomic map of PV is displayed at the top of the panel for reference.
pT7PVΔ3′MluI was generated by inserting a PCR product from the 3′ end of pT7PVΔ3′NCR. The PCR product had an MluI site positioned at the 3′ end of the poly(A) tract and was cloned into BglII-to-MluI sites of the previously described clone pT7PV1(MluI) (68). The poly(A) tract PCR product was longer than the template, extending the poly(A) tract to contain 51 adenosine residues. pT7RzPVMluI and pT7RzΔ3′MluI were generated by cloning the StuI-to-PinAI site of prib(+)Rluc (27) into pT7PV1(MluI) and pT7PVΔ3′MluI, respectively. pT7RzΔ3′BcgI was generated by the introduction of a BcgI linker into the MluI site of pT7RzΔ3′MluI.
Genomic sequence.
cDNAs generated for the construction of pT7Δ3SAR1, -2, -3, -4, and -5 were generated in duplicate to account for possible errors introduced by the DNA polymerases in the RT-PCRs. These same cDNAs were used for sequencing the Δ3′ NCR PV1 (4-1 M) genome. The 5′ end (nucleotides ∼1 to 100) of the Δ3′ NCR PV1 (4-1 M) RNA genome was sequenced directly as described previously (11). The 3′-terminal sequences of the viral genome were determined via asymmetric RT-PCR and were reported previously (63). Sequences between ∼7100 and ∼7300 were sequenced from an RT-PCR-generated cDNA using the primer set PV3C(+) [5480 to 5501]/PV7350(−) [7350 −7369].
RNA transfection and specific-infectivity analysis.
HeLa cell transfections were carried out with either in vitro-derived transcripts or purified virion RNA as described previously (11). Viral RNA was prepared by extraction of sucrose gradient-purified poliovirus as previously described (14, 26). HeLa cell monolayers in 60-mm plates were transfected with 200 μl of a mixture consisting of RNA (at various concentrations), 1 mg/ml DEAE-dextran, and TS buffer (137 mM NaCl, 4.4 mM KCl, 0.7 mM Na2HPO4, 25 mM Tris, 0.5 mM MgCl2, 0.68 mM CaCl2, adjusted to pH 7.45). Transfection mixtures were added to each monolayer for 30 to 40 min at room temperature. The mixtures were removed, and the monolayers were washed once with MEM plus 8% newborn calf serum with 1% nonessential amino acids. Transfected cells were then overlaid with either MEM plus 8% newborn calf serum with 1% nonessential amino acids or with semisolid 0.45% agarose media to determine specific infectivities. The transfected cells were incubated at 37°C for 2 to 4 days. Monolayers were fixed with 10% trichloroacetic acid and stained with crystal violet for plaque analysis when appropriate.
Extended poly(A) tracts were synthesized in reaction mixtures containing in vitro-derived RNAs, poly(A) polymerase reaction buffer (50 mM KCl, 0.7 mM MnCl2, 0.2 mM EDTA, 100 μg/ml acetylated bovine serum albumin, 10% glycerol, 20 mM Tris-HCl, pH 7.0), 0.5 μM ATP, and recombinant yeast poly(A) polymerase (600 U; USB). Reaction mixtures were incubated for 20 min at 37°C, and reactions were terminated by the addition of 10 mM EDTA and incubation on ice. Quantity and integrity of RNAs were determined by 1% agarose gel separation, ethidium bromide staining, and comparison to virion RNAs of known concentration.
In vitro RNA stability.
In vitro-derived 32P-labeled transcripts were generated as described previously (53). Stability of radiolabeled transcripts was determined in a 30-μl mixture of 67% HeLa S-10 translation-replication extract (see below), radiolabeled RNAs (1.5 × 104 cpm/reaction), and DEPC-H2O (to 30 μl) in the presence or absence of 0.2 μg of cycloheximide. Reaction mixtures were incubated at 37°C, and 5-μl portions were removed at specific times. Reactions were stopped by the addition of 250 μl of a buffered SDS solution (0.5% SDS, 1 mM EDTA, 100 mM NaCl, 10 mM Tris-HCl, pH 7.5). RNA from these reactions was phenol-chloroform extracted twice, precipitated with EtOH, and pelleted at 15,000 × g for 20 min. RNA pellets were washed twice with 70% EtOH, dried, and resuspended in 40 μl of DEPC-treated H2O. The RNAs from each reaction were separated by electrophoresis on a 1.1% agarose gel, stained with ethidium bromide, and visualized by UV illumination. The gels were dried, and the radiolabeled RNAs were detected and quantified by phosphorimager analysis (Molecular Dynamics, Sunnyvale, California).
In vitro translation.
Extracts were prepared as described by Barton et al. (5) with the following minor differences (64): (i) HeLa cells were resuspended in a volume equal to that of the cell pellet using hypotonic buffer, and (ii) the initiation factor preparation used to supplement the translation reactions contained glycerol to a final concentration of 10%. In vitro translation reaction mixtures were incubated with in vitro-transcribed RNA from pT7-PV1 or pT7PVΔ3′NCR or with purified virion PV1 RNA in the presence of 15 μCi [35S]methionine and 2 mM guanidine-HCl.
Computer-predicted RNA secondary structures.
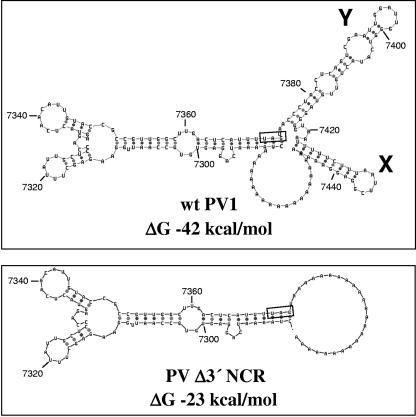
Secondary structures for 3′-terminal sequences starting at position 7287 of the PV1 genome were determined using Mfold, version 3.1 (34, 75), under default settings (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/). Two minimum-free-energy structures were generated for each sequence, and the structures that maintained stem-loop Y are shown in Fig. 7.
FIG. 7.
Computer-modeled secondary structures of PV 3′ RNA sequences with and without a 3′ NCR. RNA sequences corresponding to nucleotide positions 7287 through 7441 of wild-type PV1 or nucleotide positions 7287 through 7373 of Δ3′ NCR PV1 include 24 A residues of the 3′ poly(A) tract. Minimum-free-energy values are indicated below each structure. The first stop codon is boxed in each structure, and proposed stem-loop structures X and Y of the 3′ NCR are indicated on the wild-type structure.
RESULTS
Single-cycle growth analysis of virus accumulation.
In a previous report, we had demonstrated that in vitro-derived PV transcripts harboring a complete 3′ NCR deletion mutation were infectious following transfection of HeLa cells (63). Following transfection, a long delay preceded the abrupt appearance of genomic RNA as determined by Northern blot analysis. Synchronous with the detection of genomic RNA was the appearance of CPE. Virus was harvested following these transfections, and sequence analysis revealed that there were no changes at or near the deletion site. A high-titer virus stock was generated by isolating virus through two rounds of plaque purification followed by three passages in HeLa cells. This virus isolate, called Δ3′ NCR PV1 (4-1 M), was sequenced to verify the stability of the deletion mutation. Subsequent infections with Δ3′ NCR PV1 (4-1 M) showed that the virus was capable of achieving nearly wild-type levels of genomic RNA replication. Therefore, this virus was thought to contain one or more second-site compensatory mutations.
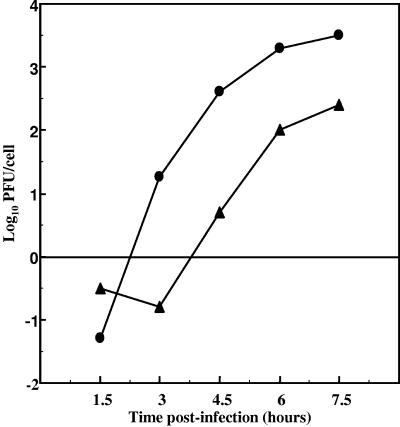
To further characterize the growth kinetics and maximum accumulation levels of Δ3′ NCR PV1 (4-1 M), we performed a synchronized single-cycle growth analysis by infecting HeLa cell monolayers at a high MOI with either wild-type virus or deletion-mutant virus. We harvested the cells at specific times after infection, measured the accumulation of virus, and expressed the viral titer as PFU/cell (Fig. 1). The single-cycle growth curve in Fig. 1 shows that the mutant virus accumulated at a slightly slower rate than wild-type virus. In addition, maximum mutant virus accumulation was less than that of wild type by ∼1 log10 unit. These data could account for the small-plaque phenotype we observed on HeLa cell monolayers (8). Our data are consistent with the slightly delayed kinetics of deletion-mutant RNA accumulation following infection that we had observed previously (63). In contrast, virus replication kinetics following transfection with deletion mutation transcripts display a very significant (∼30- to 50-h) delay in the accumulation of virus compared to wild-type PV1 transcripts. Here we report that, following viral infection, an ∼1-h delay in deletion mutant virus replication kinetics was observed compared to wild-type PV1, representing a significant increase in the kinetics of viral replication and a nearly wild-type phenotype of deletion mutant virus. Because we had shown that the high-titer stock of Δ3′ NCR PV1 maintained the complete 3′ NCR deletion, these data suggested that the virus may have undergone nucleotide changes in the genome outside of the 3′ NCR that resulted in an increased replicative capacity.
FIG. 1.
Single-cycle growth analysis of PV1 and Δ3′ NCR PV1. HeLa cell monolayers were infected with either wild-type PV1 (•) or Δ3′ NCR PV1 (4-1 M) (▴) at an MOI of 15 and incubated at 37°C, and cells were harvested at 1.5, 3, 4.5, 6, and 7.5 h postinfection. The virus titer was determined by plaque assays.
Complete genomic nucleotide sequence of Δ3′ NCR PV1 (4-1 M).
To identify putative second-site mutations responsible for the increase in deletion-mutant virus replication, we determined the nucleotide sequence of the entire genome of the deletion-mutant virus and compared this sequence to that of the parental pT7PVΔ3′NCR clone (29, 63). Changes from the parental sequence were subsequently analyzed for the ability to confer increased specific activity consistent with the growth properties of virus recovered after transfection of the transcripts with a complete 3′ NCR deletion. The virus stock used for sequence analysis, Δ3′ NCR PV1 (4-1 M), was generated by two rounds of plaque purification followed by three passages of virus amplification. RNA was extracted from purified deletion-mutant virus, and segments of the RNA genome were RT-PCR amplified in two parallel reactions (Fig. 2; see Materials and Methods). The resulting cDNAs include nucleotide sequences representing the entire genome except the 5′-terminal sequences, which were sequenced directly. We identified a mutation (A5081G) resulting in an amino acid change (I1447V) in the carboxy terminus of the 2C protein (amino acid 320 of 2C). This amino acid change is adjacent to a putative RNA binding domain (52). The sequence from independent plaque-purified virus 2-1 S was wild type at this position. In contrast, the sequence from secondary plaque-purified virus 4-1 S, which shares a common primary-plaque lineage with 4-1 M, contained the same A5081G mutation, suggesting that this nucleotide change was isolated in the first round of plaque purification. Finally, another sequence change (U6026C) resulted in an amino acid mutation (Y1762H) near the amino terminus of the 3Dpol (amino acid 14 of 3Dpol). This amino acid change is only four residues from a previously described compensatory mutation (N18Y in 3Dpol) in a chimeric poliovirus containing a mutated 3′ NCR of HRV14 (38). Virus stock 4-1 S was wild type at this location. Because 4-1 S shares a primary plaque lineage with 4-1 M, we speculate that this mutation was not present in the primary isolate but must have originated in the second round of plaque purification. Neither the sequence change in the 2C gene nor the sequence change in the 3D gene was conserved among the isolates. These data suggested that there was not a common reversion strategy and that multiple independent reversion mutations could have arisen to compensate for the loss of the entire PV 3′ NCR.
Suballelic reconstruction of the deletion mutation cDNA.
To determine if a particular mutation, or the pair of mutations, could confer greater infectivity upon deletion mutation transcripts, pT7PVΔ3′NCR was reconstructed with suballelic segments from the Δ3′ NCR PV1 (4-1 M) virus genome (Fig. 2). Suballelic cDNAs were generated by RT-PCR amplification of five separate segments of the third-passage Δ3′ NCR PV1 (4-1 M) genome. The cohort of these suballelic segments represents the entire genome except for the very 5′ and 3′ termini. The mutations identified by complete genomic nucleotide sequencing (A5081G and U6026C) were individually contained within two separate suballelic reconstructions, pT7Δ3SAR3 and pT7Δ3SAR4, respectively. Compared to the original 3′ NCR deletion construct, transcripts that contained a segment of the genome possessing a compensatory mutation might be expected to have faster growth kinetics, as evidenced by the production of CPE at an earlier time posttransfection than observed following transfection with the original pT7PVΔ3′NCR-derived transcripts.
HeLa cell transfections were carried out with in vitro-derived transcripts produced by T7 RNA polymerase from each of the individual suballelic reconstructed cDNAs. None of the reconstructions displayed a rescue phenotype after transfection (defined by a reduced time to observed CPE) compared to transfection carried out with pT7PVΔ3′NCR transcripts (data not shown). In addition, when transcripts containing both mutations A5081G and U6026C (pT7Δ3SAR6; Fig. 2) were used in this transfection analysis, no rescue phenotype was observed (data not shown).
Specific-infectivity analysis of virion and in vitro-derived RNAs.
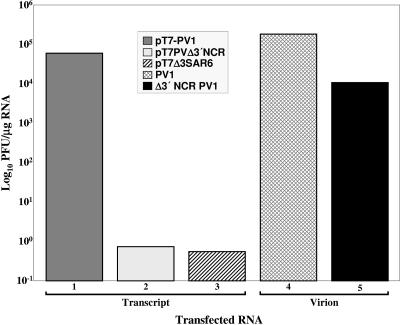
To provide a quantitative assessment of the putative rescue functions of pT7Δ3SAR6, we compared its specific infectivity to that of RNAs isolated from virions of both Δ3′ NCR PV1 (4-1 M) and PV1. Since the Δ3′ NCR PV RNAs were recovered from progeny virions, these RNAs should be infectious and contain all necessary compensatory elements required for nearly wild-type replication. We determined the specific infectivity (PFU/μg RNA) of wild-type pT7-PV1 transcripts, purified wild-type PV1 virion RNA, pT7PVΔ3′NCR transcripts, pT7Δ3SAR6 transcripts, and purified Δ3′ NCR PV1 virion RNA (Fig. 3). As expected, pT7-PV1 transcripts had approximately a 10-fold-lower specific infectivity than those of purified wild-type PV virion RNA (Fig. 3, compare bars 1 and 4) (25, 27, 66). The specific infectivity of either the pT7PVΔ3′NCR or pT7Δ3SAR6 transcripts was greater than 4 log10 units lower than that of wild-type transcripts. In contrast, the specific infectivity of purified deletion-mutant virion RNA was greater than 104 PFU/μg RNA. The difference in the specific infectivities of virion RNA from the wild-type and deletion-mutant viruses is consistent with the single-cycle growth data obtained for these virus isolates (Fig. 1). These data demonstrate that, although the pT7Δ3SAR6 reconstruction contains a majority of the Δ3′ NCR PV1 (4-1 M) genome sequence, including mutations A5081G and U6026C, a rescue phenotype was not detected.
FIG. 3.
Specific-infectivity analysis of wild-type and 3′ NCR-deleted RNAs in HeLa cell monolayers. Serial dilutions of RNA were transfected into HeLa (R19) monolayers via DEAE-dextran, and specific infectivities of RNAs were determined by plaque assay. RNAs were derived from in vitro transcription reactions (bars 1 to 3) or from purified RNA from sucrose gradient-isolated virions (bars 4 and 5). Specific infectivities represent the averages of multiple experiments performed in duplicate. Virion RNA concentration was determined by spectrophotometer readings of optical density at 260 nm, and in vitro-derived transcript RNA was quantitated by ethidium bromide staining and comparison against a poliovirus RNA standard in a 1% agarose gel.
Nonviral sequences at the termini of Δ3′ NCR transcripts inhibit replication.
We hypothesized that the increase in deletion mutant virion RNA infectivity over that of mutated transcripts is not due to sequence changes that had accumulated in the Δ3′ NCR PV1 genome but rather to fundamental differences between in vitro-derived transcripts and encapsidated viral RNAs. Recognized sequence differences between virion and transcribed deletion-mutant RNAs are located at the 5′ and 3′ termini. The 5′ end of the PV RNA genome is covalently linked to viral protein VPg (18, 32). In contrast, full-length genomic transcripts are generated in vitro using a bacteriophage T7 RNA polymerase that initiates transcription optimally when transcripts initiate with two G residues. The two-G start is known to reduce the infectivity of in vitro-derived transcripts (27, 66) and accounts, in part, for the lower specific infectivity of wild-type transcripts compared to wild-type virion RNA (Fig. 3). To examine the contribution of the covalently linked VPg to the deletion mutation virion RNA, virion RNA was subjected to proteinase K treatment prior to testing the specific infectivity. As has been reported previously for wild-type virion RNA (41), digestion of VPg had no effect on the specific infectivity of deletion mutant virion RNA (data not shown). A construct containing a hammerhead ribozyme 5′ of the poliovirus deletion mutation genome was generated (pT7RzΔ3′MluI), enabling us to generate in vitro-derived genomic transcripts that have authentic 5′ termini. Analysis of the specific infectivity of these transcripts revealed that an authentic 5′ terminus enhances the specific infectivity of the deletion mutation transcripts modestly, but no more than would be predicted based on similar studies with wild-type transcripts (data not shown).
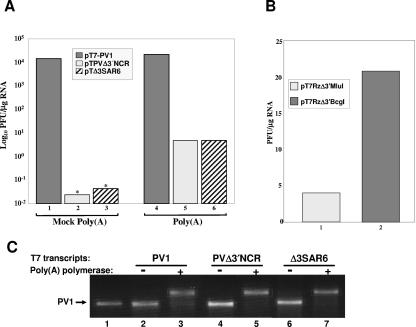
At the 3′ end of the PV genome is a genetically encoded poly(A) tract that is necessary for viral RNA infectivity (61, 72, 73). In vitro-derived transcripts contain a poly(A) tract but are flanked at the 3′ end by 4 nucleotides (pT7RzΔ3′MluI) or 7 nucleotides (pT7PVΔ3′NCR and pT7Δ3SAR6) of nonviral vector sequences. These sequences arise as a consequence of linearizing the full-length cDNA constructs by restriction enzyme digestion in preparation for in vitro transcription reactions. We reasoned that, while the 3′ nonviral sequences may pose little challenge to genomic replication in the presence of a 3′ NCR, the nonviral sequences may inhibit replication of genomic RNAs that have a complete 3′ NCR deletion. To address this possibility, we used yeast poly(A) polymerase to add poly(A) tracts of >200 residues to the ends of our in vitro-derived deletion mutation transcripts, thereby generating authentic 3′ termini. The addition of 3′ poly(A) tracts to wild-type transcripts had essentially no effect on wild-type RNA infectivity (Fig. 4A, compare bars 1 and 4). In contrast, when poly(A) polymerase was used to add a poly(A) tract to the 3′ ends of in vitro-derived deletion mutation transcripts, the infectivity increased by at least 2 orders of magnitude (100- to 200-fold) over mock poly(A) polymerase-treated transcripts (Fig. 4A, compare bars 2 and 5 and 3 and 6, respectively). Furthermore, the addition of an authentic 3′ poly(A) terminus to pT7Δ3SAR6 transcripts resulted in RNAs that had no increase in specific infectivity over that of pT7PVΔ3′NCR transcripts also treated with poly(A) polymerase (Fig. 4A, compare bars 5 and 6). While the specific infectivity of poly(A) polymerase-treated over mock poly(A) polymerase-treated transcripts was 2 orders of magnitude higher, this difference may be reflective of the fact that mock-treated transcripts had lower specific infectivities than transcripts that were untreated. In addition, the specific infectivities of the treated RNAs were still below those of the isolated virion RNAs. To address the possibility that both the 5′ and 3′ termini need be authentic to generate transcripts with specific infectivities matching virion RNA, we generated a construct that encoded both a 5′ hammerhead ribozyme and a 3′ BcgI linearization restriction enzyme site. By using the BcgI site at the 3′ end of the poly(A) tract, we were able to use the linearized cDNA genomic templates to generate transcripts with authentic 3′ poly(A) termini. Transcripts containing authentic 5′ and 3′ termini were approximately fivefold more infectious than transcripts with an authentic 5′ terminus alone (Fig. 4B). While these deletion mutation transcripts had the highest specific infectivities that we measured, they are still several orders of magnitude lower than those of the deletion mutant virion RNAs.
FIG. 4.
Specific infectivity analysis of nonviral terminal sequences. In vitro-derived RNAs were generated from the indicated cDNA templates. Specific infectivities (PFU/μg RNA) were determined by plaque assay following DEAE-dextran-mediated transfection of transcripts into HeLa (R19) cells and are reported as the averages of at least two experiments. (A) The 3′ termini of transcripts generated from the indicated cDNA constructs either were modified posttranscriptionally by the addition of an elongated poly(A) tract using poly(A) polymerase (bars 4 to 6) or remained unmodified in mock poly(A) polymerase reactions (bars 1 to 3). The specific infectivities reported by bars 2 and 3 (asterisks) were below the level of detection and therefore represent the maximum possible specific infectivities that could have been measured. (B) Both 5′ and 3′ nonviral sequences were eliminated from in vitro-derived transcripts by the use of a Klenow-treated BcgI-linearized cDNA template encoding a 5′ hammerhead ribozyme (bar 2). The infectivity of these transcripts is contrasted to transcripts that have an authentic 5′ terminus but contain nonviral sequences at the 3′ terminus (bar 1). (C) The ethidium bromide-stained agarose gel shows a shift in the sizes of poly(A) polymerase-treated RNA transcripts (lanes 3, 5, and 7) compared to those of the mock-treated transcripts (lanes 2, 4, and 6), demonstrating that poly(A) polymerase added additional poly(A) tracts to the 3′ ends of these transcripts. Purified PV1 virion RNA is shown as a marker in lane 1.
In vitro translation kinetics of in vitro-derived transcript RNAs.
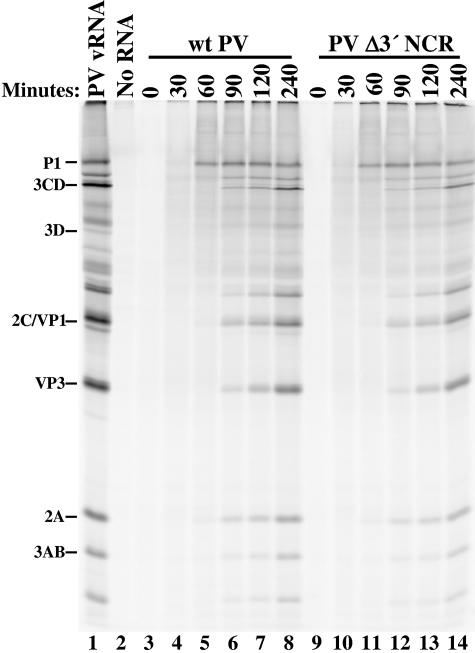
It was possible that the addition of an authentic 3′-terminal poly(A) did not affect genomic RNA replication directly but rather corrected an alternate deficiency of the deletion mutation RNA. In addition to stabilizing mRNAs, 3′ poly(A) tails have been shown to mediate increased translation efficiency. Sheets et al. demonstrated that the 3′ NCRs of cyclin A1, B1, B2, and c-mos mRNAs contain signals that lead to cytoplasmic poly(A) elongation, resulting in an increase in the translation efficiency of these messages (58). In addition, the 3′ NCRs of certain mRNAs have been shown to contain elements that augment or repress translation (30, 44). Previously, we demonstrated that deletion mutation and wild-type transcripts yield similar levels of accumulated viral translation products in vitro (63). These data represented the final accumulation of translation products and did not address the kinetics of translation and viral protein processing. To determine if the deletion mutation affects translation or processing rates, we analyzed the accumulation of PV translation products during in vitro translation. As shown in Fig. 5, HeLa S10 extracts incubated with wild-type or deletion mutation transcripts translated and processed viral polyprotein with equal efficiencies (Fig. 5, compare lanes 3 to 8 with 9 to 14). These data demonstrated that the deletion of the 3′ NCR did not affect the ability of these transcripts to serve as templates for translation.
FIG. 5.
In vitro translation kinetics of wild-type (wt) and Δ3′ NCR transcript RNAs. Translation and processing were carried out in HeLa cell extracts at 30°C in the presence of [35S]methionine. Extracts were incubated with PV1 virion RNA (lane 1) or no RNA (lane 2) for 4 h. At the indicated times, 10-μl portions of the extracts incubated with pT7-PV1-derived transcripts (lanes 3 to 8) or pT7PVΔ3′NCR-derived transcripts (lanes 9 to 14) were removed and translation was terminated by the addition of Laemmli sample buffer (31) and boiling for 3 to 5 min. These products were analyzed on an SDS-2.5% polyacrylamide gel and visualized by phosphorimager (Personal Molecular Imager FX; Bio-Rad, Hercules, California).
Stability of in vitro-derived Δ3′ NCR transcripts.
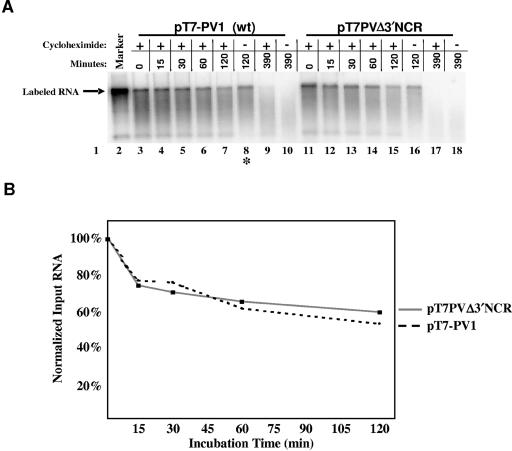
The half-lives of eukaryotic mRNAs vary from a few minutes to many hours or even days. Elements within the 3′ NCR of mRNAs have been shown to act as destabilization signals for short-lived mRNAs, such as c-fos mRNA (59, 71), or as stabilization signals for long-lasting mRNAs, such as for the mRNA encoding α-globin (69, 70). To evaluate a possible RNA instability effect caused by the deletion of the PV 3′ NCR, we analyzed the stability of the deletion mutation transcripts. Although our previous in vitro translation data (Fig. 5) (63) suggest that there is not a significant decrease in the stability of genomic RNA lacking a 3′ NCR, it is possible that actively translated RNAs are protected from ribonucleases. Because there is evidence suggesting that an elongating ribosome translating the PV genome interferes with negative-strand RNA synthesis (6, 20), there may be a kinetic transition in the viral life cycle when viral translation must be limited to allow the synthesis of negative-strand RNA intermediates. Thus, we analyzed RNA stability in the absence or presence of cycloheximide, a potent translation elongation inhibitor. Radiolabeled transcripts were incubated in S10 extracts at 37°C in the presence or absence of cycloheximide. At specific times, portions of the incubation mixture were removed and the RNA was phenol-chloroform extracted. The RNA was analyzed on an agarose gel by ethidium bromide staining (data not shown) and phosphorimager analysis (Fig. 6A). When data were normalized to the input RNA, there were no significant differences between the stability of radiolabeled deletion mutation transcripts and wild-type PV transcripts in the HeLa S10 extract (Fig. 6B). In addition, there was little or no effect on RNA stability during ongoing protein synthesis (Fig. 6A, compare lanes 7 to 8 and 15 to 16). Our data demonstrated that, in the presence or absence of translating ribosomes, there are no RNA stability effects produced by the deletion of the poliovirus 3′ NCR that could account for the dramatic specific-infectivity differences observed between in vitro-derived wild-type and Δ3′ NCR transcripts.
FIG. 6.
Wild-type and deletion mutation transcript stability in HeLa S10 extracts. Transcript stability was determined as described in Materials and Methods. (A) HeLa S10 extracts were incubated with radiolabeled wild-type (lanes 3 to 10) or deletion mutation (lanes 11 to 18) transcripts in the presence (lanes 3 to 7, 9, 11 to 15, and 17) or absence (lanes 8, 10, 16, and 18) of cycloheximide. PV virion RNA (lane 1) or a radiolabeled PV transcript (lane 2) was loaded as the marker. At the indicated times, portions of the extract mixture were removed and the RNA was phenol-chloroform extracted. Total RNA was separated by electrophoresis on a 1.1% agarose gel and visualized by ethidium bromide staining and UV illumination (data not shown). The gel was dried, and radiolabeled products were visualized and quantitated by phosphorimager analysis (Personal Molecular Imager FX; Bio-Rad, Hercules, California). *, lane containing a sample overloaded on the gel by ∼20% compared to other samples and rRNA loading controls. (B) Products correlating to full-length genomic RNA incubated in the presence of cycloheximide were normalized to input (zero time) levels and plotted on a graph.
DISCUSSION
The 3′ NCR is thought to contain a cis-acting element(s) responsible for the specific and efficient synthesis of negative-strand RNA intermediates during PV genomic replication. Support for this notion comes from the fact that, in the absence of coding selection pressure, the 3′ NCR is highly conserved among the three serotypes of PV. In addition, the 3′ NCR is proximal to the site of negative-strand RNA synthesis initiation at or near the end of the 3′ poly(A) tract. Finally, while multiple cis-acting replication element sequences have been described for PV, the lack of specific replication complex targeting sequences suggests that an additional element(s) exists to target the polyadenylated viral RNA for replication in the presence of an abundance of polyadenylated cellular mRNAs. Although numerous studies have implicated the 3′ NCR of picornaviruses as having a role in RNA replication (8, 16, 28, 37, 38, 47-49, 55, 57, 63), an understanding of a functional mechanism and a clearly defined minimal element within the 3′ NCR is lacking from these studies.
Our previous studies with PV transcripts lacking a 3′ NCR had raised the possibility that, during initial low-level RNA replication following transfections, compensatory mutations had accumulated in the deletion-mutant genome (63). Sequencing through the 3′ NCR lesion did not identify reversion mutations, indicating that putative compensatory mutations may have arisen elsewhere in the genome. Complete nucleotide sequencing of the genomic RNA from a third-passage virus stock revealed two nucleotide changes from the parental genomic sequence. These nucleotide changes resulted in amino acid changes in both the 2C and 3D proteins. When transcripts from a pT7Δ3SAR6 expression clone, containing both of the identified mutations, were transfected into HeLa cells, there was no reduction in the time to observed CPE, nor was there an increase in RNA specific infectivity. However, there was an increase in the specific infectivities of deletion mutation transcripts when the 5′ and/or the 3′ termini of the Δ3′ NCR transcripts were modified to mimic those of virion RNA. While wild-type poliovirus transcripts containing an authentic 5′ end are known to have a higher specific infectivity than genomic transcripts that contain nonviral sequences at their 5′ termini (25, 27, 66), the generation of wild-type transcripts with authentic 3′ poly(A) termini does not significantly increase their specific infectivities. An increase in infectivity due to an authentic 3′ terminus was unique to the deletion mutation RNAs and suggested that efficient initiation of deletion mutant negative-strand RNA synthesis is mediated through an authentic poly(A) 3′ terminus. Although both the 3′ poly(A) tract and the 3′ NCR of cellular mRNAs have been shown to be involved in translation and stability modulation, the deletion mutation transcripts were shown to be nearly identical to wild-type transcripts in both genome stability and translation efficiencies. Thus, while the addition of an authentic 3′ poly(A) terminus resulted in an increase in specific infectivity of deletion mutation transcripts, this increase was not due to a compensatory effect on RNA stability or translation and viral polypeptide processing. The equal specific infectivities of pT7PVΔ3′NCR- and pT7Δ3SAR6-derived transcripts, terminating in adenosine nucleotides or not, confirmed that Δ3′ NCR PV viral RNA does not contain compensatory mutations.
The reason for the much higher infectivity of deletion mutant virion RNA over that of the deletion mutation transcript RNA remains unclear. Based the our data showing that deletion mutant virion RNAs had specific infectivities within 1 log unit of those observed for wild-type virion RNAs (Fig. 3), we conclude that the mode of entry of the RNA into the cell (i.e., transfection versus infection) cannot account for this difference. It may be that the poly(A) tract needs to be of a precise length for optimal infectivity. However, the specific infectivities of deletion mutation RNAs with 16 (data not shown), 27, and 51 (this study) adenosine residues are essentially identical, while pT7RzΔ3′BcgI-derived transcripts have 46 adenosine residues but are more infectious than the original deletion mutant transcripts. Another possibility for the increased infectivity of virion RNA is that epigenetic information may be contained within the deletion mutant virion RNA. Such epigenetic information might take the form of an RNA structural element that requires a chaperone-like factor(s) to properly form (during RNA replication in the cytoplasm of infected cells) or perhaps the inclusion of modified nucleotide moieties that may be found within the infected cell. Alternatively, there could be an inhibitory RNA structure(s) or component(s) associated with generating genomic RNAs in vitro in the absence of host cell components. This putative epigenetic component would appear to be of no consequence in wild-type RNAs; however, in the absence of the RNA sequences and structures contained within the 3′ NCR, such a component may take on added importance.
Previous reports have provided speculation on the severity of the replication defect of our mutant virus harboring a complete 3′ NCR deletion during infection of HeLa cells (1, 46, 51, 67). Although the specific steps in RNA replication affected by the deletion are still under investigation, it is now clear that growth of this virus does not require second-site compensatory mutations (i.e., PVΔ3′NCR is infectious in contrast to quasi-infectious). While the 3′ NCR is not absolutely necessary for PV genomic RNA replication, data presented in this study showed that the 3′ NCR has a functional role in RNA replication since the deletion-mutant virus does not achieve wild-type replication levels. Previous studies examining the role of picornavirus 3′ NCRs in viral replication have come to similar conclusions (16, 28, 37, 38, 47-49, 55, 57). Moreover, we have recently shown that the poliovirus genomic 3′ NCR has a cell-dependent role in the initiation of positive-strand RNA synthesis (8). Apart from these previous findings, this study implicates the 3′ NCR in the role of nonterminal synthesis initiation of negative-strand RNA intermediates during PV RNA replication. We speculate that, when the 3′ NCR is intact, negative-strand RNA synthesis can initiate at a number of sites along the poly(A) tract, possibly by maintaining the position of the VPg protein primer. However, when the 3′ NCR contains a severe lesion or is completely deleted, initiation may be confined to the very 3′ terminus of the poly(A) tract. Therefore, we propose that the PV genomic 3′ NCR plays a role in multiple steps of RNA replication, affecting both positive-strand and negative-strand RNA synthesis. Point mutations and small deletions within the 3′ NCRs of enterovirus RNAs have been reported to be lethal (37, 39). We suggest that such lesions within the 3′ NCR disrupt a functional element that is recognized by factors involved in RNA replication complex formation, resulting in the misdirection of such factors to generate nonfunctional RNA replication complexes.
The deletion of the 3′ NCR of poliovirus RNA may only partially disrupt a cis-acting replication element that is constructed from multiple RNA structures. When the 3′ NCR is completely removed, the remaining element is sufficient to function, albeit at slightly less than wild-type levels. We examined computer-predicted structures of the 3′ ends of PV and Δ3′ NCR RNAs, including the 3′-most nucleotides of the 3Dpol coding sequence (Mfold V3.1) (34, 75). When we compared the predicted RNA secondary structures of wild-type and 3′ NCR deletion mutation PV RNAs, we found that the complete deletion of the 3′ NCR leads to a precise removal of predicted stem-loops X and Y but leaves a larger stem-loop structure in the 3D coding region unaffected (Fig. 7, compare wild type to Δ3′ NCR). This predicted 3D stem-loop structure is conserved among the three serotypes of PV (data not shown). Such a structure may represent a minimal replication element that, in concert with the 3′ NCR, forms a larger tertiary structure required for maximum levels of RNA replication.
It has been proposed that a cis-acting targeting element may not be necessary to assure accurate replication of the viral genome (12, 63). Novak and Kirkegaard showed that genomic RNA must be translated in order for it to be replicated (42). Furthermore, nascent nonstructural (P2 and P3) gene products are proximal to the 3′ terminus of RNAs from which the proteins are translated (50). Given the tight membrane association of RNA replication functions (62), it is not hard to imagine that an important cis signal for negative-strand RNA synthesis may be the proximal translation of viral nonstructural proteins. Furthermore, positive-strand RNA synthesis may require the completion of a negative-strand intermediate as a cis signal for replication. Finally, it has been shown that genomic replication and packaging are coupled (4, 43, 65), suggesting that each step in virus replication may be a signal that mediates the onset of the next step in the virus life cycle.
Acknowledgments
We acknowledge the DNA Core Automated DNA Sequencing Facility and its support from the UCI Cancer Center and the UCI School of Biological Sciences. D.M.B. and C.T.C. were predoctoral trainees supported by Public Health Service training grant AI 07319. This work was supported by Public Health Service grant AI 22693 from the National Institutes of Health.
REFERENCES
- 1.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 4.Baltimore, D., M. Girard, and J. E. Darnell. 1966. Aspects of synthesis of poliovirus RNA and formation of virus particles. Virology 29:179-189. [DOI] [PubMed] [Google Scholar]
- 5.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunner, J. E., J. H. C. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos, R., and L. P. Villarreal. 1982. An SV40 deletion mutant accumulates late transcripts in a paranuclear extract. Virology 119:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Charini, W. A., C. C. Burns, E. Ehrenfeld, and B. L. Semler. 1991. trans rescue of a mutant poliovirus RNA polymerase function. J. Virol. 65:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta, A. 1983. Purification of host factor required for in vitro transcription of poliovirus RNA. Virology 128:245-251. [DOI] [PubMed] [Google Scholar]
- 14.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in a poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 61:2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dildine, S. L., and B. L. Semler. 1989. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5′ noncoding region of genomic RNA. J. Virol. 63:847-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duque, H., and A. C. Palmenberg. 2001. Phenotypic characterization of three phylogenetically conserved stem-loop motifs in the mengovirus 3′ untranslated region. J. Virol. 75:3111-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanegan, J. B., and D. Baltimore. 1977. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc. Natl. Acad. Sci. USA 74:3677-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanegan, J. B., R. F. Pettersson, V. Ambros, N. J. Hewlett, and D. Baltimore. 1977. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl. Acad. Sci. USA 74:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harber, J., and E. Wimmer. 1993. Aspects of the molecular biology of picornaviruses, p. 189-224. In L. Carrasco, N. Sonenberg, and E. Wimmer (ed.), Regulation of gene expression in animal viruses. Plenum Press, New York, N.Y.
- 25.Harmon, S. A., O. C. Richards, D. F. Summers, and E. Ehrenfeld. 1991. The 5′-terminal nucleotides of hepatitis-A virus-RNA, but not poliovirus RNA, are required for infectivity. J. Virol. 65:2757-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, T. J. R., J. J. Dunn, and E. Wimmer. 1978. Identification of specific fragments containing 5′ end of poliovirus RNA after ribonuclease-III digestion. Nucleic Acids Res. 5:4039-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson, S. J., D. A. Konings, and P. Sarnow. 1993. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J. Virol. 67:2961-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 30.Kruys, V., M. Wathelet, P. Poupart, R. Contreras, W. Fiers, J. Content, and G. Huez. 1987. The 3′ untranslated region of the human interferon-beta messenger-RNA has an inhibitory effect on translation. Proc. Natl. Acad. Sci. USA 84:6030-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 35.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melchers, W. J., J. G. Hoenderop, H. J. B. Slot, C. W. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meredith, J. M., J. B. Rohll, J. W. Almond, and D. J. Evans. 1999. Similar interactions of the poliovirus and rhinovirus 3D polymerases with the 3′ untranslated region of rhinovirus 14. J. Virol. 73:9952-9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirmomeni, M. H., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 74:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 43.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostareck-Lederer, A., D. H. Ostareck, N. Standart, and B. J. Thiele. 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 13:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 46.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierangeli, A., M. Bucci, P. Pagnotti, A. M. Degener, and R. Perez Bercoff. 1995. Mutational analysis of the 3′-terminal extra-cistronic region of poliovirus RNA: secondary structure is not the only requirement for minus strand RNA replication. FEBS Lett. 374:327-332. [DOI] [PubMed] [Google Scholar]
- 48.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 20:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilipenko, E. V., K. Poperechny, S. V. Maslova, W. J. G. Melchers, H. J. B. Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (-) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (′kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 50.Richards, O. C., and E. Ehrenfeld. 1990. Poliovirus RNA replication. Curr. Top. Microbiol. Immunol. 161:89-119. [DOI] [PubMed] [Google Scholar]
- 51.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 53.Roehl, H. H., and B. L. Semler. 1994. In vitro biochemical methods for investigating RNA-protein interactions in picornaviruses, p. 169-182. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 4. Academic Press, Inc., Orlando, Fla. [Google Scholar]
- 54.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohll, J. B., N. Percy, R. Ley, D. J. Evans, J. W. Almond, and W. S. Barclay. 1994. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68:4384-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarnow, P., H. D. Bernstein, and D. Baltimore. 1986. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc. Natl. Acad. Sci. USA 83:571-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheets, M. D., C. A. Fox, T. Hunt, W. G. Vande, and M. Wickens. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 8:926-938. [DOI] [PubMed] [Google Scholar]
- 59.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 60.Simoes, E. A., and P. Sarnow. 1991. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J. Virol. 65:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spector, D. H., and D. Baltimore. 1975. Polyadenylic acid on poliovirus RNA. II. Poly(A) on intracellular RNAs. J. Virol. 15:1418-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tershak, D. R. 1984. Association of poliovirus proteins with the endoplasmic reticulum. J. Virol. 52:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 229:90-97. [DOI] [PubMed] [Google Scholar]
- 65.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Werf, S., J. Bradley, E. Wimmer, F. W. Studier, and J. J. Dunn. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, X., M. Kiledjian, I. M. Weiss, and S. A. Liebhaber. 1995. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol. Cell. Biol. 15:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss, I. M., and S. A. Liebhaber. 1994. Erythroid cell-specific determinants of α-globin mRNA stability. Mol. Cell. Biol. 14:8123-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson, T., and R. Treisman. 1988. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336:396-399. [DOI] [PubMed] [Google Scholar]
- 72.Yogo, Y., and E. Wimmer. 1972. Polyadenylic acid at the 3′-terminus of poliovirus RNA. Proc. Natl. Acad. Sci. USA 69:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yogo, Y., and E. Wimmer. 1975. Sequence studies of poliovirus RNA. III. Polyuridylic acid and polyadenylic acid as components of the purified poliovirus replicative intermediate. J. Mol. Biol. 92:467-477. [DOI] [PubMed] [Google Scholar]
- 74.Zell, R., and A. Stelzner. 1997. Application of genome sequence information to the classification of bovine enteroviruses: the importance of 5′- and 3′-nontranslated regions. Virus Res. 51:213-229. [DOI] [PubMed] [Google Scholar]
- 75.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. RNA Biochem. Biotechnol. 70:11-43. [Google Scholar]