Abstract
The development of antiviral drug resistance is an important problem in the treatment of human immunodeficiency virus type 1 (HIV-1) infection. Potent antiretroviral therapy is currently used for treatment, and typically consists of at least two reverse transcriptase (RT) inhibitors. We have previously reported that both drugs and drug-resistant RT mutants can increase virus mutation frequencies. To further assess the contributions of nucleoside RT inhibitors (NRTIs), nonnucleoside RT inhibitors (NNRTIs), and drug-resistant RTs to HIV mutagenesis, a new high-throughput assay system was developed. This assay system was designed to specifically detect frameshift mutations in the luciferase gene in a single virus replication cycle. New drug-resistant RTs were identified that significantly altered virus mutation frequencies. Consistent with our previous observations of NRTIs, abacavir, stavudine, and zalcitabine increased HIV-1 mutation frequencies, supporting the general hypothesis that the NRTIs currently used in antiviral drug therapy increase virus mutation frequencies. Interestingly, similar observations were made with NNRTIs. This is the first report to show that NNRTIs can influence virus mutation frequencies. NNRTI combinations, NRTI-NNRTI combinations, and combinations of drug and drug-resistant RTs led to significant changes in the virus mutation frequencies compared to virus replication of drug-resistant virus in the absence of drug or wild-type virus in the presence of drug. This indicates that combinations of RT drugs or drugs and drug-resistant virus created during the evolution of drug resistance can act together to increase HIV-1 mutation frequencies, which would have important implications for drug therapy regimens. Finally, the influence of drug-resistant RT mutants from CRF01_AE viruses on HIV-1 mutation frequencies was analyzed and it was found that only a highly drug resistant RT led to altered virus mutation frequencies. The results further suggest that high-level drug-resistant RT can significantly influence virus mutation frequencies. A structural model that explains the mutation frequency data is discussed.
Potent antiretroviral therapy of human immunodeficiency virus type 1 (HIV-1) infection with antiretroviral drugs consists of nucleoside RT inhibitors (NRTIs), nonnucleoside RT inhibitors (NNRTIs), and protease inhibitors. Antiretroviral drugs have been previously shown to influence HIV-1 mutation frequencies and the HIV-1 mutation rate. The first study of the impact of drugs on HIV-1 mutation frequencies was investigating how the NRTIs 3′-azido-3′-deoxythymidine (zidovudine) and (−)2′,3′-dideoxy-3′-thiacytidine (lamivudine) influence HIV-1 mutation frequencies (24). These analyses used the lacZα peptide gene as a mutation target that has been used in previous mutation rate studies of HIV-1.
Zidovudine increased the HIV-1 mutation frequency by 7.6-fold in a single round of replication, while lamivudine led to a 3.4-fold increase in virus mutation frequency. How NRTIs increase HIV-1 mutagenesis is presently not known, but the NRTIs currently used in therapy may have a similar mechanism to influence HIV-1 mutation frequencies. This is supported by the observation that HIV-1 mutation frequencies increased in an additive manner during virus replication in the presence of two NRTIs (i.e., zidovudine and lamivudine, zidovudine and dideoxyinosine, and lamivudine and dideoxyinosine) (23).
Zidovudine-resistant RT was also found to increase the virus mutation frequency by 4.3-fold, but the replication of lamivudine-resistant HIV-1 had no significant influence on the mutation frequency (24). Furthermore, it was observed that only high-level zidovudine-resistant RT mutants could influence the in vivo mutation frequency, such as those containing mutations M41L/T215Y and M41L/D67N/K70R/T215Y. These observations suggested that when virus replication occurs in the presence of suboptimal concentrations of drug, drug-resistant virus is selected and that replication of drug-resistant virus in the presence of drug could further increase the virus mutation rate.
To test this hypothesis, the combined effects of drug and drug-resistant virus were investigated (26). It was found that replication of zidovudine-resistant virus in the presence of zidovudine led to a multiplicative 24-fold increase in the virus mutation frequency compared to that observed with wild-type virus in the absence of drug. In addition, it was found that replication of a zidovudine/lamivudine dual-resistant virus in the presence of both zidovudine and lamivudine also led to a multiplicative 22.5-fold increase in the virus mutation frequency. These results indicated that when drug failure occurs due to the evolution of drug resistance, replication of the drug-resistant virus in the presence of drug could significantly increase HIV-1 mutagenesis.
Previous in vitro studies using purified HIV-1 RT showed that single base substitutions and single base frameshift mutations were predominant mutations in the HIV-1 mutational spectrum and were nonrandomly distributed (3). Most of these mutations were found at mutation hot spots, typically homopolymeric runs. It was observed that many single base substitutions occurred at either the 5′ end or the 3′ end of homopolymeric runs, indicating many single base substitutions, as well as frameshift mutations, are initiated by template-primer slippage (3, 4). Consistent with these observations, the homopolymeric runs were found to be hot spots for spleen necrosis virus RT to initiate frameshift mutations (most common mutations were +1 and −1) in a single round of viral replication (7). The mutation rate for runs of T's was the highest compared to rates for runs of A's, C's, and G's.
Moreover, the analysis of the HIV-1 mutation rate in a single round of replication also demonstrated that both base substitutions and frameshift mutations were common mutations during HIV-1 reverse transcription; the most common frameshift mutations were +1 mutations at a run of T's (28). Further study of mutations in HIV-1 proviruses following treatment with antiretroviral drugs showed that the mutation spectra of HIV-1 after drug treatment was comparable to the spectrum of mutants observed in the absence of drugs, indicating that the mechanisms by which mutations occurred were similar but that the rate had increased (24).
In order to extend our current knowledge of how antiretroviral drugs and drug-resistant RTs influence HIV-1 mutation frequencies, a new high-throughput assay system using the luc gene as a mutational target was developed to measure HIV-1 mutation frequencies based on previous observations (3, 4, 7, 24, 28). Using this new assay system several issues were addressed. First, specific mutations in HIV-1 RT that conferred resistance to antiretroviral drugs were tested to determine if they could influence the rate of HIV-1 mutation. Second, the hypothesis that the NRTIs currently used in drug therapy could increase HIV-1 mutation frequencies was tested. Third, it was tested whether NNRTIs could influence HIV-1 mutation frequencies. Fourth, the combined effects of drugs and of drugs and drug-resistant RTs on virus mutation frequencies were found to alter HIV-1 mutation frequencies. Finally, it was observed that high-level drug-resistant RT mutants from CRF01_AE viruses could significantly influence HIV-1 mutation frequencies.
MATERIALS AND METHODS
Retroviral vectors and expression plasmids.
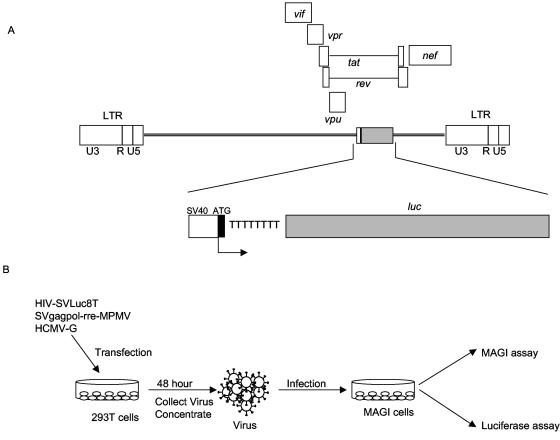
HIV-SVLuc8T was developed to measure the reversion mutation rate of HIV-1 using a mutated luc gene as the reporter. This vector was designed to specifically detect frameshift mutations in the luciferase gene in a single virus replication cycle. Eight T residues were inserted after the luc gene start codon by two-step PCR mutagenesis using the pGL3 control vector as the template. This insertion causes the complete loss of luciferase activity due to the loss of the open reading frame. A 1.5-kb deletion (from the SalI to NheI sites) within the open reading frame of the env gene was made in the HIV vector KP97 (graciously provided by Michael Emerman, Fred Hutchinson Cancer Research Center, Seattle, Wash.), which is an HIV vector derivative of pBRU2 with a 3.1-kb deletion (SphI to BglI) of the gag and pol genes. This env deletion was replaced with a 1.9-kb XhoI to XbaI fragment from pGL3, which contains the simian virus 40 promoter and the mutated luc gene (Fig. 1A).
FIG. 1.
Assay system used for analysis of virus mutation frequencies. (A) Schematic representation of the HIV-1 vector used to analyze HIV-1 mutation frequencies. The proviral DNA form of the vector is shown. The large rectangular boxes are long terminal repeats, which include U3, R, and U5. The simian virus 40 promoter (SV40), luc gene, and luc gene start codon are indicated. (B) Single round of replication assay for mutation frequencies. Virus-producing cells were transfected with HIV-1 reporter vector and helper plasmids, and the viruses produced were concentrated and used to infect fresh MAGI cells. The MAGI assay and luciferase assay were used to determine the reverse mutation frequencies.
The HIV-1 gag-pol expression plasmid used was pSVgagpol-rre-MPMV, which has been described previously (27). This expression vector contains the simian virus 40 promoter driving expression of the HIV-1 gag-pol genes. The vector used for expression of vesicular stomatitis virus glycoprotein envelope, pHCMV-G, was obtained J. Burns (University of California at San Diego, San Diego, CA). To be packaged into virus particles, HIV-SVLuc8T was complemented in trans with the HIV-1 gag-pol expression plasmid and pseudotyped with the vesicular stomatitis virus G envelope expression plasmid.
The HIV-1 RT mutants (subtype B) analyzed in these experiments were constructed by introducing mutations coding for RT amino acid substitutions into pSVgagpol-rre-MPMV by site-directed mutagenesis (Quick Change, Stratagene, La Jolla, CA). The helper vectors containing HIV-1 CRF01_AE RT variants were created by deletion of full-length CRF01_AE clones (p93JP-NH1 and its variants). The mutant RT genes confer different levels of NRTI resistance in the genetic backbone of 93JP-NH1 virus (38). A 2.4-kb deletion was made in each of the full-length CRF01_AE clones between the two AvrII sites. This deletion removed the vpr, tat, and env genes of CRF01_AE, so that these gene products will be supplied by the other plasmid constructs.
Cell culture and antiretroviral drugs.
293T cells were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL, Gaithersburg, MD), supplemented with 10% of fetal clone III serum (HyClone, Logan, UT). MAGI cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% of fetal clone III serum, G418 (0.2 mg/ml), and hygromycin (0.1 mg/ml). Virus-infected cells were scored by staining MAGI cells with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Antiretroviral drugs were obtained from the National Institutes of Health AIDS Research Reagents Program (Bethesda, MD) except for lamivudine, which was purchased from Sigma (St. Louis, MO).
Transfections and infections.
The experimental protocol developed to obtain a single round of HIV-1 vector replication is shown in Fig. 1B, and described in detail elsewhere (28). Briefly, HIV-SVLuc8T (9.5 μg) was cotransfected with pSVgagpol-rre-MPMV (9.5 μg) and vesicular stomatitis virus G (1 μg) into 293T cells in a 100-mm petri dish using the calcium phosphate precipitation method. Viruses were harvested 48 h posttransfection and concentrated using a Centricon Plus-20 filter (Millipore, Billerica, MA). For infections, concentrated virus was mixed with Polybrene (8 μg/ml) (Sigma, St. Louis, MO) and the virus-Polybrene mixture was used to infect two sets of fresh MAGI target cells (2.5 × 105 cells per dish). After 24 h, cells were washed once with phosphate-buffered saline (GIBCO BRL), and fresh medium was added. The cells were then incubated at 37C for an additional 48 h. Supernatants were removed, and one set of cells was used to perform the MAGI assay to determine virus titer (17). The other set of cells was lysed and used for the luciferase assay to detect the restored luciferase activity (Fig. 1B). Cell numbers at the time of these assays were typically 5 × 105 to 10 × 105 cells.
Reversion mutation detection.
Mutations that restored luciferase activity were determined in MAGI cells, which were treated or not with antiretroviral drugs. Drug treatments were typically done by maintaining MAGI target cells in medium supplemented with drug at the 50% inhibitory concentration (IC50) from 2 h prior to infection and until 24 h after infection. Seventy-two hours postinfection, infected MAGI cells were counted using the trypan blue dye exclusion method, harvested, and lysed in 1× luciferase assay lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N′,N′,N′,N′-tetracetic acid, 10% glycerol, 1% Triton X-100). Luciferase activity was quantified using the Promega Luciferase Assay System (Promega, Madison, WI). Mutation frequencies were calculated based on luciferase activity reading, viral titer, and cell numbers. The relative mutation frequency of wild-type viruses that were not treated with drug was defined as 1. The equation used to calculate the relative mutation frequencies (RMF) is RMF = {[(number of cells with no drug treatment/number of cells with drug treatment) × luciferase reading of drug-treated cells]/luciferase reading of cells with no drug treatment} × (viral titer of virus with no drug treatment/viral titer of drug-treated virus).
Determination of 50% inhibitory concentrations.
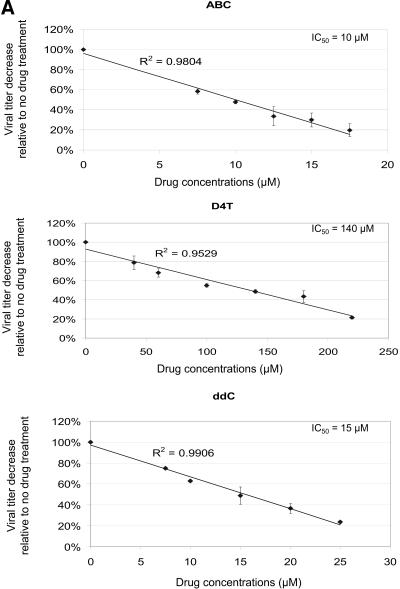
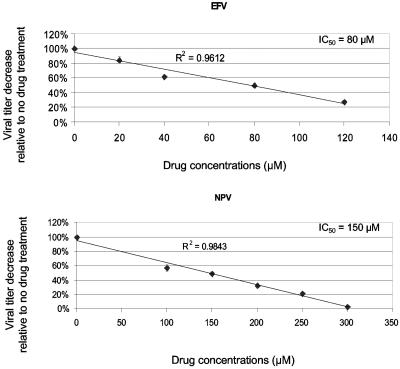
The MAGI assay was used to determine the IC50 value for each drug. Briefly, 2.5 × 105 fresh MAGI target cells were treated with antiretroviral drug at different concentrations 2 h prior to infection and continued until 24 h postinfection. Virus-Polybrene mixture was diluted to 1:1,000 and used to infect MAGI target cells. Seventy-two hours postinfection, infected MAGI cells were stained with X-Gal and positive blue cells were counted to determine virus titer at each concentration. The virus titer was plotted as a function of the drug concentration used, generating linear curves for all the drugs. IC50 values for each drug were calculated based on the linear plots (Fig. 2).
FIG. 2.
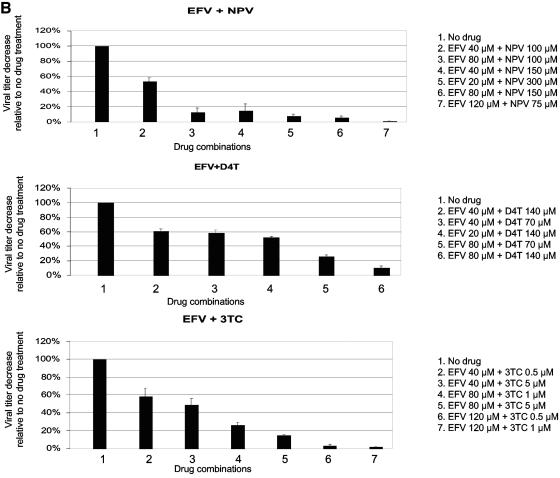
Drug susceptibility assays. (A) Determination of IC50 values. Data from three independent experiments are shown. Virus titers from drug-treated cells are shown relative to virus titer from untreated cells (set at 100%). The absolute virus titer from untreated cells was 8.2 × 105 ± 3.5 × 105 infected cells/ml of cell-free virus stock. The IC50 value for lamivudine was determined previously (24). ABC, abacavir; D4T, stavudine; ddC, zalcitabine; EFV, efavirenz; NVP, nevirapine. (B) Effects of drug combinations on virus titers. The concentrations of individual drugs used in the following experiments were chosen based on the effects of the individual drugs and combined drugs on virus infectivity. Data from three independent experiments are shown. Virus titers from drug-treated cells are shown relative to the virus titer from untreated cells (set at 100%). The absolute virus titer from untreated cells was 11.5 × 105 ± 1.6 × 105 infected cells/ml of cell-free virus stock.
Analysis of mutation spectra.
Cellular DNA was prepared from 105 cells infected by wild-type HIV-1 in the absence of drug. Following infection, the infected MAGI cells were washed once with phosphate-buffered saline, trypsinized and lysed in 500 μl of PCR lysis buffer (50 mM KCl, 10 mM Tris, pH 8.3, 1.8 mM MgCl2, 0.45% IGEPAL CA-630, 0.45% Tween 20). The cell lysate was treated with 3 μl of proteinase K (20 mg/ml) (Roche, Indianapolis, IN) at 50°C for 1 h. The 5′ end of the luc gene was amplified from cell lysate using nested PCR. The PCR products were then cloned into pCR2.1 TA cloning vector (Invitrogen, Calsbad, CA). The plasmid DNAs were isolated and sequenced.
Statistical analysis.
To investigate the effects of antiretroviral drugs and RT variants on mutation frequency, we fit a linear model to the normalized logarithms of the fold changes relative to the wild type. Normalization was conducted as described above. The model pooled data from all experiments and replicates to estimate effects for each drug and RT variant in addition to all estimable interactions. The model supposes that assay variability is constant across all conditions and replicates, an assumption that is consistent with the data. The model has a term for all drugs and RT variants in addition to interactions. Standard diagnostic techniques failed to reveal any major shortcomings of the model. Ninety-five percent confidence intervals for all fold changes were constructed by sampling parameters from their joint sampling distribution and then transforming the parameters (i.e., exponentiating) to obtain samples from the joint sampling distribution of the fold changes. We summarize the results with the upper and lower endpoints of these intervals in addition to the estimate of the fold change.
Structural analysis.
To investigate the initial binding sites of deoxynucleoside triphosphates and NRTIs with HIV-1 RT, we used the crystal structure of the RT-template-primer complex at a resolution of 3.1Å from the Protein Data Bank (PDB code 1N5Y) (37) as a template for substrate docking simulations. This structure represents an open configuration at the posttranslocation stage during the catalytic cycle of RT, which theoretically is competent for binding of the incoming-substrate. The template was docking with deoxynucleoside triphosphate and deoxynucleoside triphosphate analogs by using the automated ligand docking program AS_Dock (Ryoka Systems Inc., Chiba, Japan) operated in the Molecular Operating Environment. The precision of docking results with the AS_Dock is generally equivalent to the experimental error (i.e., a few angstroms). In fact, a result of dTTP docking with the AS_Dock at the catalytic site in an RT closed configuration had a root-mean-square deviation of ∼1.6 Å compared to that determined by x-ray crystallography (1RTD) (11), which was the range within the resolution of the crystal structure.
RESULTS
Development of a new high-throughput assay system to measure HIV-1 mutation frequencies.
Previous studies of HIV-1 mutation frequencies used the lacZα gene as a reporter gene (28). However, using this system is laborious and time-consuming. In order to more rapidly assess the ability of drugs and/or drug-resistant RTs to influence HIV-1 mutation frequencies, a new high-throughput assay system was developed. This system was designed to be a reversion assay that would specifically detect frameshift mutations in the luciferase reporter gene in a single round of replication (3, 4, 7, 24, 28) (Fig. 1).
The HIV-1 vector developed contained a mutated luc gene, which was inserted into the env gene. Specifically, eight T residues were inserted after the start codon of the luc gene (Fig. 1A). To be packaged into virus particles, the vector was complemented in trans with an HIV-1 gag-pol expression plasmid and a vesicular stomatitis virus glycoprotein envelope expression plasmid. Vector virus produced from 293T cells was used to infect fresh MAGI target cells (Fig. 1B). Reversion mutations were detected by measuring the restored luciferase activity in the infected MAGI cells, which were either untreated or treated with antiretroviral drug. The relative mutation frequency of wild-type virus was defined as 1. The relative mutation frequencies of viruses treated with antiretroviral drugs and of viruses expressing drug-resistant RT variants were compared to this value (Materials and Methods).
Several approaches were used to validate this assay system. First, a control HIV-1 vector was constructed in which two T residues were inserted after the ATG of the luc gene. A mutation hot spot has been defined as a homopolymeric run of three or more nucleotides (3, 7). Therefore, the reversion mutations that occurred in this 2T vector was expected to be much lower than that when the 8T vector was used. As expected, significantly lower reversion mutations were observed when the 2T vector was used (data not shown), providing indirect evidence that the observed reversion mutations occurred during the reverse transcription process. Second, analysis of the mutation spectra at the hot spot revealed plus one, minus two, and minus one frameshift mutations. In addition, a G-to-T base substitution was also observed (Table 1). This G-to-T mutation was located at the 3′ end of the run of T's, indicating that this mutation was likely initiated by dislocation mutagenesis (4).
TABLE 1.
Analysis of mutation spectra
| Mutational class | No. of mutants/total | Mutant frequencya (10−3) |
|---|---|---|
| +1 | 1/116 | 1.1 |
| −2 | 1/116 | 1.1 |
| −1 | 1/116 | 1.1 |
| G-to-T | 1/116 | 1.1 |
| Total | 4/116 | 4.4 |
Mutant frequency is in units of mutations/bp/cycle. The average number of relative light units for control experiments (virus replication in the presence of wild-type RT) was 6.1 × 102 + 35.
The mutation frequency calculated from this analysis, 4.4 × 10−3 mutations/cycle/base pair, is consistent with the previously determined HIV-1 frameshift mutation rate in a homopolymeic sequence, which is on the order of 10−3 (3, 28). Because only the +1 and −2 mutations can restore luciferase activity in this assay system, the relative mutation frequency of 1 correlates to 2.2 × 10−3 mutations/cycle/base pair. Based on the statistical analysis of data pooled from all control experiments, mutation frequencies are similar among all replicates, indicating that the calculated mutation frequency is representative.
Third, a control vector was constructed in which nine T residues were inserted after the start codon of the luc gene. This vector was designed to test whether the insertion of three amino acids would affect luciferase expression. The results revealed that the insertion of three amino acids did not significantly influence luciferase expression (data not shown). Fourth, it was observed that the detection of luciferase activity caused by reversion mutations was in the linear detection range (data not shown). Dilution of the cell lysate used in the luciferase assay resulted in a luciferase reading in the linear detection range even at high dilution points. Moreover, when the infected cell number used in the luciferase assay was increased, luciferase activity increased in a linear manner (data not shown), indicating that the increase in luciferase activity was correlated with the increase in reversion mutations that occurred during HIV-1 replication. No luciferase expression was detected in transfected cells (data not shown), indicating that the rate of reversion was very low and did not significantly contribute to the reversion frequencies observed in infected cells.
Finally, the influence of zidovudine and zidovudine-resistant RT on virus mutation frequencies was examined using this system. As shown in Table 2, zidovudine led to a 2.6-fold increase in virus mutation frequency and a zidovudine-resistant RT increased the virus mutation frequency by 2.2-fold. Replication of zidovudine-resistant virus in the presence of zidovudine led to a 6.5-fold increase in virus mutation frequencies. In comparison, it was reported that zidovudine increased the rate of +1 frameshift mutations at the run of T's in the lacZα gene by twofold (24). These data provide further evidence that this new assay system behaves in a predictable manner based on previously published data using the lacZα gene as a mutation target (24, 26) (Table 2).
TABLE 2.
Influence of zidovudine and zidovudine-resistant reverse transcriptase on HIV-1 mutant frequenciesa
| Drug | RT variant | Avg relative mutation frequencyb
|
|
|---|---|---|---|
| luc | lacZα | ||
| None | Wild type RT | 1 | 1 |
| Zidovudine | 2.6 ± 0.4 | 7.6 | |
| None | Zidovudine resistant RTc | 2.2 ± 0.5 | 4.3 |
| Zidovudine | 6.5 ± 0.3 | 24 | |
Data represent the average of three independent experiments. The average number of relative light units detected from infected cells for virus replication in wild-type RT was 7.0 × 102 ± 45.
Data were obtained by using the luc gene or lacZα (26) as the mutation target.
The resistant RT variant contained the mutations M41L, D67N, K70R, and T215Y.
Influence of drug-resistant RT mutants on HIV-1 mutation frequencies.
In order to analyze the effects of antiretroviral drugs on virus mutation frequencies, the IC50 values for each drug were first determined (Fig. 2A). Typically, cells were treated at the IC50 concentration of each drug to determine their influence on virus mutation frequencies. Previous studies have shown that HIV-1 replication with zidovudine-resistant RTs increased the mutation rate by as much as 4.3-fold, while replication of HIV-1 with a lamivudine-resistant RT had no significant influence on the mutation rate (24).
It was observed that only high-level zidovudine-resistant RT variants could influence the in vivo mutation rate (i.e., those containing the mutations M41L/T215Y and M41L/D67N/K70R/T215Y) (24). Moreover, it was found that combined drugs and drug-resistant RTs could further increase virus mutation frequencies (24-26). To further examine if virus mutation frequencies are influenced by drug-resistant RT variants in the presence of drugs, a series of drug-resistant RT mutants were analyzed (1, 10, 12, 20, 31, 33, 45). The mutant enzyme V75T confers resistance to the drug stavudine; Y115F confers resistance to abacavir; L74V/Y115F/M184V confers high-level resistance to abacavir and mild resistance to zalcitadine; G190A, Y318F, and K103N/Y318F confer resistance to the drugs efavirenz and nevirapine; and K103N is the most frequently observed mutation in patients treated with drug combination therapy, which includes efavirenz.
V75T was previously observed as a novel resistance mutation in cell cultures treated with stavudine. It was observed that the effect of V75T relative to wild-type RT was close to being statistically significant (P = 0.06). The amino acid residues that interact with the incoming deoxynucleoside triphosphate and form the deoxynucleoside triphosphate-binding site have been identified in structural studies. One substitution in the deoxynucleoside triphosphate binding site, Y115A, has been previously reported to decrease fidelity by a factor of 4 using the lacZα gene (14). We previously observed that the Y115A RT variant significantly increased (2.3-fold) virus mutation frequencies using one round of HIV-1 vector replication (25). Moreover, the Y115F and Y115V RT variants were found in lacZα cell-free fidelity assays to have slightly lower error rates than that of wild-type RT (6). In this study, we found that the Y115F RT mutant increased virus mutation frequency by only 1.20-fold, which is not significantly different from wild-type RT (Table 3) (see also below).
TABLE 3.
Influence of HIV-1 RT variants, NRTIs, and NNRTIs on virus mutation frequencies
| Class and RT variant | Druga (concn, μM) | Avg relative mutation frequencyb |
|---|---|---|
| Drug-resistant RT variantsc | ||
| Wild type | 1 | |
| V75T | 0.62, 0.80, 1.02 | |
| Y115F | 0.85, 1.20, 1.73 | |
| L74V/Y115F/M184V | 1.50, 1.91, 2.44 | |
| K103N | 0.89, 1.09, 1.31 | |
| G190A | 0.53, 0.83, 1.41 | |
| Y318F | 0.60, 0.74, 0.91 | |
| K103N/Y318F | 0.33, 0.46, 0.66 | |
| NRTIs and NRTI-resistant RT variantsd | ||
| Wild type | ABC | 1.68, 2.14, 2.72 |
| Y115F | ABC | 2.15, 3.01, 4.24 |
| L74V/Y115F/M184V | ABC | 2.77, 3.91, 5.47 |
| Wild type | d4T | 2.60, 3.46, 4.72 |
| V75T | d4T | 1.21, 1.65, 2.18 |
| Wild type | ddC | 0.95, 1.36, 1.89 |
| L74V/Y115F/M184V | ddC | 2.37, 3.39, 4.78 |
| Wild type | 3TC | 0.84, 1.20, 1.70 |
| Wild type | ddC (15) | 0.95, 1.36, 1.89 |
| ddC (25) | 1.98, 2.21, 2.43 | |
| 3TC (1) | 0.84, 1.20, 1.70 | |
| 3TC (2) | 1.82, 2.13, 2.41 | |
| NNRTIs and NNRTI-resistant RT variantse | ||
| Wild type | EFV | 2.25, 2.69, 3.18 |
| G190A | EFV | 2.25, 3.22, 4.54 |
| Y318F | EFV | 1.70, 2.43, 3.43 |
| K103N/Y318F | EFV | 1.05, 1.47, 2.07 |
| Wild type | NPV | 2.56, 3.12, 3.80 |
| G190A | NPV | 1.88, 2.63, 3.73 |
| Y318F | NPV | 2.21, 3.09, 4.45 |
| NNRTI and NNRTI combinationsf | ||
| Wild type | EFV (40) | 0.98, 1.37, 1.93 |
| NPV (100) | 1.85, 2.62, 3.76 | |
| EFV & NPV | 0.82, 2.71, 9.15 | |
| Y318F | EFV | 0.95, 1.36, 1.93 |
| NPV | 1.80, 2.64, 3.83 | |
| EFV & NPV | 0.56, 2.01, 6.99 | |
| NRTI, NNRTI, and drug-resistant RT variantsg | ||
| Wild type | EFV (80) | 2.25, 2.69, 3.18 |
| d4T (140) | 2.60, 3.48, 4.72 | |
| 3TC (1) | 0.84, 1.20, 1.70 | |
| EFV (80) & d4T (140) | 7.28, 9.33, 11.82 | |
| EFV (80) & 3TC (1) | 2.59, 3.64, 5.21 | |
| K103N | EFV (80) | 2.03, 2.83, 3.97 |
| d4T (140) | 4.78, 7.44, 11.96 | |
| 3TC (1) | 0.67, 0.97, 1.37 | |
| EFV (80) & d4T (140) | 8.28, 10.75, 12.53 | |
| EFV (80) & 3TC (1) | 1.44, 4.40, 13.11 |
ABC, abacavir; d4T, stavudine; ddC, zalcitabine; 3TC, lamivudine; EFV, efavivenz; NPV, nevirapine.
The estimated fold change (middle value) and the endpoints of the 95% confidence intervals for the fold changes are shown.
The average luciferase reading for virus replication in the presence of wild-type RT was 5.5 × 102 ± 25.
The influence of NRTI and NRTI-resistant RTs on HIV-1 mutant frequencies. The average luciferase reading for virus replication in the presence of wild-type RT was 5.8 × 102 ± 26.
The influence of NNRTI and NNRTI-resistant RTs on HIV-1 mutant frequencies. The average luciferase reading for virus replication in the presence of wild-type RT 5.9 × 102 ± 32.
The influence of EFV and NPV combination and drug-resistant RT on HIV-1 mutant frequencies. Data represent the average of three to twelve independent experiments. The average luciferase reading for virus replication in the presence of wild-type RT was 5.7 × 102 ± 33.
The effects of combinations of NRTI and NNRTI and drug-resistant RT on HIV-1 mutant frequencies. The average luciferase reading for virus replication in the presence of wild-type RT was 5.9 × 102 ± 34.
Cell culture selection of resistant mutants has shown that multiple mutations were required to create high levels of resistance to abacavir. One mutant, L74V/Y115F/M184V, was isolated during an in vitro passage experiment. This mutant showed a 10-fold increase in IC50 of abacavir, while it had a 4-fold decrease in susceptibility to zalcitabine (45). We observed that the L74V/Y115F/M184V RT mutations led to a 1.91-fold increase in HIV-1 mutation frequency and this increase is statistically significant.
Crystallographic analyses of HIV-1 RT and nonnucleoside RT inhibitor complexes have suggested that all NNRTIs occupy a hydrophobic binding pocket that is located in the palm subdomain of p66 and proximal to the polymerase active site (8, 34, 40, 41). NNRTI resistance is associated with mutations within the NNRTI binding pocket. Three single NNRTI-resistant RT mutations, K103N, G190A, and Y318F, which are all located in the NNRTI binding pocket, and one double mutation, K103N/Y318F, were tested to determine their influence on virus mutation frequencies. Clinically, the K103N mutation is the most frequently observed mutation in patients treated with efavirenz-containing therapies (1). The G190A mutation is also observed in patients treated with NNRTIs (12). The Y318F mutation is also associated with a decrease in susceptibility to NNRTIs, and viruses containing both Y318F and K103N have higher levels of drug resistance (10). As indicated in Table 3, the single RT variants K103N and G190A did not have a statistically significant impact. However, the Y318F and K103N/Y318F RT variants significantly decreased HIV-1 mutation frequencies, the latter quite substantially.
Influence of NRTIs and NRTI-resistant RT variants on virus mutation frequencies.
Previous studies using the lacZα gene as a mutational target have indicated that NRTIs could increase virus mutation frequencies (23, 24, 26). Zidovudine increased the HIV-1 mutation rate by as much as 7.6-fold (0.4 μM) in a single round of replication, while lamivudine increased the virus mutation rate by as much as 3.4-fold (0.3 μM) (26). A dose-dependent relationship between increased drug concentration and increased virus mutation frequencies has been reported for zidovudine, lamivudine, and dideoxyinosine (23, 26, 27). The maximum increase in virus mutation frequencies in the presence of dideoxyinosine was sixfold higher than the virus mutation frequency observed during replication in the absence of drug (23).
To further investigate the impact of NRTIs on HIV-1 mutation frequencies, virus mutation frequencies were determined in the presence of other NRTIs (i.e., abacavir, stavudine, and zalcitabine) at IC50 concentrations. Like the other NRTIs studied previously, abacavir and stavudine led to 2.1-fold and 3.5-fold increases in virus mutation frequencies, respectively, and these effects are statistically significant. Unexpectedly, the dCTP analog zalcitabine only increased HIV-1 mutation frequencies by 1.36-fold at its IC50 concentration and was not statistically significant (Table 3). Based on this observation, we also tested lamivudine at its IC50 concentration and found that it led to a 1.20-fold increase (not significant different from no drug). At the IC90 concentrations of zalcitabine and lamivudine, virus mutation frequencies increased by 2.21- and 2.13-fold, respectively (Table 3), indicating that higher concentrations of zalcitabine and lamivudine are needed for these drugs to significantly influence HIV-1 mutation frequencies. This indicates that increased virus mutation frequencies occur when drug concentrations increase.
It has been shown that both RT variants and drugs together could increase virus mutation frequencies (24, 26). To further test this, selected NRTIs and drug-resistant RT variants were analyzed for their influence on virus mutation frequencies. Mutant viruses containing either the Y115F or the L74V/Y115F/M184V RT mutations were grown in the presence of abacavir at its IC50 concentration. Both mutations were associated with a statistically significant increase in virus mutation frequencies compared to those observed during virus replication with the wild-type RT in the presence of abacavir (Table 3). Interestingly, it was also observed that HIV-1 replication with the L74V/Y115F/M184V RT variant in the presence of zalcitabine significantly influenced HIV-1 mutation frequencies compared to that observed during virus replication with wild-type RT in the presence of zalcitabine (3.39-fold versus 1.36-fold). Similarly, in the presence of stavudine, the V75T RT variant affected virus mutation frequencies compared to wild-type RT (3.46-fold versus 1.65-fold) (Table 3).
Influence of NNRTIs and NNRTI-resistant RT variants on virus mutation frequencies.
NNRTIs inhibit reverse transcription by binding to a hydrophobic pocket that is proximal to the active site of HIV-1 RT (8, 18, 34, 35, 40, 41, 43). Three NNRTIs are currently used for treatment of HIV-1 as part of combination antiretroviral therapy (39). Nothing is known about how NNRTIs affect HIV-1 mutation frequencies.
In this study, efavirenz and nevirapine and NNRTI-resistant RT variants were used to investigate how NNRTIs and NNRTI-resistant RT variants influence HIV-1 mutation frequencies. We found that efavirenz and nevirapine could increase HIV-1 mutation frequencies by 2.69 and 3.12-fold, respectively, and that these increases are statistically significant. This is the first report of NNRTIs being able to influence (increase) HIV-1 mutation frequencies. This observation surprisingly suggests that both NRTIs and NNRTIs have a similar influence on HIV-1 mutation frequencies (Table 3).
To determine the effects of NNRTI-resistant mutants on HIV-1 mutation, virus replication with NNRTI-resistant RT mutants was analyzed in the presence of these drugs. As shown in Table 3, the G190A and Y318F RT variants did not influence HIV-1 mutation frequencies compared to virus replication with wild-type RT in the presence of drug. The K103N/Y318F mutant led to an approximately 1.47-fold increase in the virus mutation frequencies in the presence of drugs compared to the 2.69-fold increase observed during virus replication with the wild-type RT in the presence of drugs.
Influence of combined drugs and HIV-1 RT variants on virus mutation frequencies.
Potent antiretroviral therapy regimens typically include drugs from two of the three classes of antiretroviral drugs (NRTIs, NNRTIs, and protease inhibitors). Four two-NRTI combinations are typically used in antiretroviral therapy, i.e., zidovudine plus lamivudine, zidovudine plus dideoxyinosine, stavudine plus dideoxyinosine, and stavudine plus lamivudine (39). It has been reported that an additive increase in virus mutation frequencies was observed during virus replication in the presence of NRTI combinations (i.e., zidovudine plus dideoxyinosine, zidovudine plus lamivudine, and lamivudine plus dideoxyinosine) (23).
In this study, various combinations of drugs (NNRTI plus NNRTI and NNRTI plus NRTI) were studied for their ability to act together to influence HIV-1 mutation frequencies during virus replication. First, the drug combination of efavirenz plus nevirapine (NNRTI plus NNRTI) was tested. Based upon the effects of the individual drugs and combined drugs on virus infectivity (Fig. 2A and B) and a lack of cytotoxicity (data not shown), an efavirenz concentration of 40 μM was used along with a nevirapine concentration of 100 μM. For each drug alone, these concentrations inhibited virus replication by one-fourth. It was observed that efavirenz and nevirapine together led to an increase in the virus mutation frequency of 2.71-fold, while virus replication in the presence of efavirenz or nevirapine alone led to 1.37-fold and 2.62-fold increases in the virus mutation frequencies, respectively (Table 3).
The clinically used drug combinations of NNRTI and NRTI were then analyzed, which included efavirenz plus stavudine and efavirenz plus lamivudine. The 80 μM concentration of efavirenz was used in combination with 140 μM of stavudine or 1 μM of lamivudine, based upon the effects of the individual drugs and combined drugs on virus infectivity (Fig. 2A and B) and a lack of cytotoxicity (data not shown). As shown in Table 3, efavirenz and stavudine together could increase the HIV-1 mutation frequency by 9.33-fold, while virus replication in the presence of efavirenz or stavudine alone led to 2.69-fold and 3.46-fold increases in the virus mutation frequencies, respectively. Interestingly, the virus mutation frequency was only increased by 3.64-fold during virus replication in the presence of both efavirenz and lamivudine, which is not significantly different from the virus mutation frequency caused by efavirenz alone (Table 3). This suggests that the concentration of lamivudine was low enough to not significantly affect virus mutation frequencies.
Since there are HIV-1-infected individuals undergoing antiviral therapy who harbor viruses with drug resistance mutations, the effects of drug-resistant mutants and combined drugs on HIV-1 mutation frequencies were analyzed. It has been shown that the K103N mutant can emerge in patients treated with efavirenz-containing therapy (1), and the Y318F mutant was associated with a decrease in susceptibility to all NNRTIs (10). Therefore, the abilities of K103N and Y318F to affect the virus mutation frequencies in the presence of combined drugs were tested. In the presence of combined drugs, the K103N mutant did not have a significant influence on virus mutation frequencies compared with the mutation frequency observed for virus replication with wild-type RT in the presence of drugs except for the combination of efavirenz and stavudine, and stavudine alone (Table 3). Furthermore, virus replication with the Y318F RT variant in the presence of both efavirenz and nevirapine did not have a significant influence on HIV-1 mutation frequencies compared to virus replication with wild-type RT in the presence of both efavirenz and nevirapine (Table 3).
Influence of CRF01_AE drug-resistant RT variants on virus mutation frequencies.
HIV-1 is classified into groups and subtypes based on sequences within the gag and env genes. Three separate groups, M (main), O (outlier), and N (non-M, non-O), exist. The most prevalent strains belong to group M. Group M also contains at least nine distinct subtypes or clades (A to D, F to H, and J and K), as well as several circulating recombinant forms (i.e., CRF01_AE virus) (22, 36). Much of our current understanding of HIV-1 drug resistance is derived from the studies of subtype B virus, which is the major subtype that circulates in North America and Europe. However, other subtypes, such as A, C, and E, are rapidly expanding worldwide. These variants may differ in rates of transmission, ability to cause progression to AIDS, and drug resistance profiles compared to subtype B virus (9, 15). Recently, the analysis of drug resistance profiles of recombinant RTs from subtypes CRF01_AE, B, and C demonstrated that each of these RTs possessed similar baseline sensitivity to NRTIs and NNRTIs (32).
To assess the influence of CRF01_AE mutants on HIV-1 mutation frequencies, a CRF01_AE molecular clone was used. This molecular clone was constructed from the 93JP-NH1 virus isolate (p93JP-NH1). Based upon previous studies, RT variants that confer multiple-drug resistance were introduced by site-directed mutagenesis (38). All CRF01_AE RT variants except CRF01_AE-mt1 have a β3-β4-loop-insertion mutation at a similar position seen in subtype B viruses with multiple NRTI resistance. The HIV-1 isolates with the insertion mutation predominated in a patient treated with multiple NRTIs. The RT variants and their drug resistance profiles are listed in Table 4. As shown in Table 4, the p93JP-NH1 RT influenced the virus mutation frequencies at a level comparable to wild-type subtype B RT. Only one CRF01_AE mutation that confers high levels of multi-nucleoside analog drug resistance had a significant influence on HIV-1 mutation frequencies (i.e., CRF01_AE mt-7).
TABLE 4.
Influence of HIV-1 CRF01_AE RT variants on virus mutation frequencies
| RT variant | Mutation(s)a | Drug resistance phenotypeb | Avg relative mutation frequencyc |
|---|---|---|---|
| 93JP-NH1 | None | None | 1 |
| CRF01_AE mt-1 | M41L, L210W, T215Y | AZT (mild) | 0.80, 0.88, 0.96 |
| CRF01_AE mt-2 | Insertiond | 3TC (weak) | 0.68, 0.92, 1.16 |
| CRF01_AE mt-5 | Insertion + AZTe | AZT (high), 3TC (mild), d4T (weak), ddI (weak) | 0.76, 0.86, 0.96 |
| CRF01_AE mt-6 | Insertiond + AZTe + T69l | AZT (high), 3TC (high), d4T (mild), ddI (mild), ddC (weak) | 0.81, 0.87, 0.93 |
| CRF01_AE mt-7 | Insertiond + AZTe + T69l and othersf | AZT (high), 3TC (high), d4T (mild), ddI (mild), ddC (weak) | 0.59, 0.69, 0.79 |
The insertion is an 11-amino-acid insertion located between codons 67 and 68 in the β3-β4 loop coding region of the RT gene.
See Table 3, footnote a.
Endpoints of 95% confidence intervals and the sample mean are presented. The average number of relative light units for virus replication in the presence of wild-type RT was 6.3 × 102 ± 38.
Reverse transcriptase variants had an 11-amino-acid insertion between codons 67 and 68 in the β3-β4 loop coding region. The amino acid insertion was NIHGGRDQGPA.
Reverse transcriptase variants with the zidovudine (AZT) resistance mutations M41L, D67N, K70R, L210W, and T215Y.
The other mutations in reverse transcriptase included D6E, K39E, E43K, T69I, G196E, T200R, and L228R.
Structural analysis.
To help determine the underlying molecular mechanisms by which drugs and drug resistance mutations alter the mutation frequency of HIV-1 subtype B and CRF01_A/E, we have conducted structural analyses by integrating HIV-1 RT structure-function information into the following two basic models for mutation. One basic model explains how misalignments of the primer-template are initiated, in which nucleotide misinsertion into the catalytic site plays a key role in inducing template-primer slippage for −1, −2, and +1 frameshift mutations and dislocation mutagenesis of HIV-1 RT (5). Studies of frameshift error rates with various polymerases, including HIV-1 RT, have supported this model (19). The other model explains how high levels of DNA replication fidelity are generated during the catalytic cycle of the various polymerases, in which there are at least two critical check points for the nucleotide selection in the early phase: check of base pair geometry at initial binding of substrate and check of steric hindrance during finger-domain rotation that locates the substrate at the catalytic site (19). Although the previous structural (11, 37) and kinetic (16, 35) studies with HIV-1 RT are consistent with this induced-fit model, the initial substrate binding site on RT to evaluate nucleotide selection fidelity remains to be determined.
To assess the initial binding site of deoxynucleoside triphosphates or NRTIs with HIV-1 RT, we have conducted computer-assisted docking simulations by using the crystal structure of the RT-template-primer complex (PDB code 1N5Y) (37) as a template for nucleotide docking. The crystal structure represents a fingers-open configuration at the posttranslocation stage during the catalytic cycle of RT, which is theoretically competent for de novo substrate binding. As shown in Fig. 3, dATP, which is the complementary substrate for the 1N5Y template DNA, was predicted to bind the site along the β3-β4 loops of the p66 fingers subdomain. Other deoxynucleoside triphosphate and deoxynucleoside triphosphate analogs such as zidovudine triphosphate and stavudine triphosphate shared the same binding site with the dATP (data not shown). Interestingly, the predicted initial binding position of the nucleotides was distinct from the position of dTTP at the catalytic site in the RT crystal structure of the fingers-closed configuration (11) and is consistent with the position to initiate effectively the base pair formation and fingers rotation in the induced-fit model. The position was also in agreement with the biochemical mode of NRTI inhibition, competitive inhibition, shown by kinetics studies. Furthermore, the position was analogous to that of other polymerases determined by x-ray crystallography (13, 21, 44, 46).
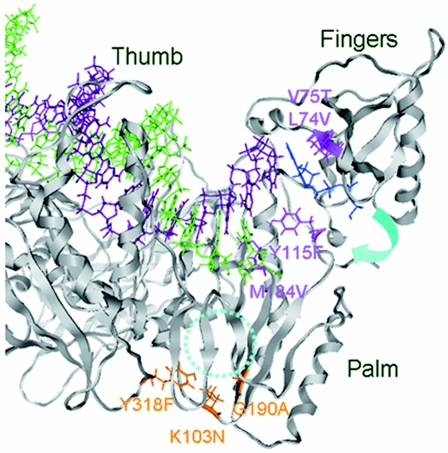
FIG. 3.
Structural model of the active site of the HIV-1 reverse transcriptase p66 subunit open configuration with newly bound dATP, primer, and template. The three-dimensional model that simulates de novo substrate binding to RT at the posttranslocation stage during the catalytic cycle was constructed by docking dATP onto the crystal structure (PDB code 1N5Y) (37) by using the molecular operating environment. The incoming dATP (blue sticks) in the open ternary complex is bound to the initial binding site along the β3-β4 loops in the p66 fingers subdomain. The ribbon represents the backbone of HIV-1 RT. The DNA template and primer are shown in magenta sticks and green sticks, respectively. The NRTI resistance mutations and NNRTI resistance mutations analyzed in this study are shown in pink and orange, respectively. A dotted cyan circle indicates the site for NNRTI binding (18, 43). The cyan arrow indicates the rotation of the fingers subdomain following substrate incorporation at the catalytic site of the enzyme (11).
In the crystal structure, positively charged amino acids are responsible for the initial binding of substrates (13, 21, 44, 46). Consistently, K65 and R72 in the β3 and β4 loops of HIV-1 RT correspond to those amino acids that are interacting with the deoxynucleoside triphosphate. The NRTI docking model with the information of the NNRTI binding site (18, 43) and the two basic models described above help explain the changes in mutation frequencies observed in the present study and in previous studies (25).
DISCUSSION
Previous studies using the lacZα peptide gene as a mutation target have indicated that both antiretroviral drugs and drug-resistant RTs can increase HIV-1 mutation frequencies (24-26). In this study, we have analyzed virus mutation frequencies in a single round of replication with an HIV-1 vector containing the luc gene. This vector was designed to specifically detect frameshift mutations based on previous in vitro and in vivo studies (3, 4, 7, 24, 28). The advantages of this new high-throughput assay system are that it allows the rapid assessment of the influence of drugs and drug-resistant RT variants on HIV-1 mutation frequencies and the sensitivity of the luciferase assay allows detection of reversion mutations in relatively small samples.
The virus mutation frequencies measured are representative based on previous observations made of frameshift mutations created in the presence and absence of drugs or drug-resistant HIV-1 using lacZα (26) and provide a good model for identifying mutations that occur during virus replication. However, in certain instances, this new assay may underestimate mutation frequency change because this HIV-1 vector was designed to detect only frameshift mutations and not the entire spectra of mutations. One case in point is when the influence of zidovudine and zidovudine-resistant RT mutants on virus mutation frequencies was compared. In this comparison, zidovudine and zidovudine-resistant RT led to a lower mutation frequency using luc versus the lacZα reporter (Table 2), indicating that there was an underestimate of the effects of drugs and drug-resistant RTs on virus mutation frequencies. However, this new system can quickly assess the influence of drugs and drug-resistant RT mutations on HIV-1 mutation frequencies during virus replication that could subsequently be analyzed in greater detail using the lacZα system.
Drug-resistant RTs influence HIV-1 mutation frequencies.
Several drug-resistant RT variants were observed to significantly alter HIV-1 mutation frequencies. First, the RT variant L74V/Y115F/M184V, which confers high resistance to abacavir, led to a significant increase in virus mutation frequencies, which is in contrast to the individual mutations, which do not lead to significant differences compared to wild-type RT. Second, the NNRTI-resistant RT variants Y318F and K103N/Y318F led to significant decreases in virus mutation frequencies. This is the first report that NNRTI-resistant RT can alter HIV mutation frequencies. Third, the CRF01_AE RT variant CRF01_AE mt-7, which confers high levels of multi-nucleoside analog drug resistance, led to a significant decrease in virus mutation frequencies. Our previous work found that only high-level zidovudine-resistant RT variants could influence the in vivo mutation rate. Therefore, our data suggests that increased drug resistance correlates with altered virus mutation frequencies.
It is not clear how the mutations that confer NNRTI resistance influence virus mutation frequencies. These residues are located within the hydrophobic NNRTI-binding pocket, which could cause conformational changes in the pocket. Therefore, it is possible that NNRTI-resistant RT mutants could indirectly affect the conformation of the HIV-1 RT active site and subsequently influence HIV-1 mutation frequencies. Biochemical studies of how NNRTI-resistant RT mutants influence misincorporation, mismatch extension, and processivity of RT would also help us to understand how NNRTI-resistant RTs affect viral mutagenesis.
NRTIs and NNRTIs can increase HIV-1 mutation frequencies.
The observation that abacavir, stavudine, and zalcitabine influence HIV-1 mutation frequencies indicates that the approved NRTIs currently used in drug therapy can increase virus mutation frequencies. Current studies are being directed at understanding the molecular basis for how NRTIs influence HIV-1 mutagenesis. These data suggest that NNRTIs and NRTIs have similar effects on HIV-1 mutagenesis. The mechanism by which NNRTIs increase HIV-1 mutation frequencies is presently unknown. One hypothesis is that the conformational change caused by NNRTIs binding noncatalytically to RT may affect enzyme fidelity.
Crystallographic studies have shown that NNRTIs cause a repositioning of the three-stranded β-sheet in the p66 subunit (containing the catalytic aspartic acid residues 110, 185, and 186), and there is a striking similarity between the actual conformations of the three-stranded β-sheet in the drug-bound p66 conformation and in the inactive p51 conformation (8). This suggests that the NNRTIs inhibit HIV-1 RT by locking the active catalytic site in an inactive conformation, reminiscent of the conformation observed in the inactive p51 subunit (8). This conformational change has a dramatic effect on the rate of the chemical step (transfer of deoxynucleoside monophosphate to the end of the primer molecule) of polymerization (35). Hence, the conformational change of the RT active site caused by NNRTIs could influence either nucleotide selectivity or RT processivity, which would lead to lower fidelity of NNRTI-bound RT compared to wild-type RT.
The effects of combined drugs (NNRTI plus NNRTI) on HIV-1 mutation frequencies were analyzed. In general, the virus mutation frequencies observed in the presence of combined drugs were significantly increased compared to that observed in the presence of the individual drugs. This indicates that these two NNRTIs can act together in an additive manner to further increase HIV-1 mutation frequencies. The clinical use of drug combinations of NNRTI plus NRTI, such as efavirenz plus stavudine or efavirenz plus lamivudine, were also examined and found to act together in an additive manner. This is the first report which shows that an NRTI-NNRTI combination could further increase HIV-1 mutation frequencies. Given that potent antiretroviral therapy typically includes two or three RT inhibitors, the combined effects of RT inhibitors on HIV-1 mutation frequencies may be clinically relevant.
Potential mechanisms for NRTI resistance mutations and NRTI-mediated changes in HIV-1 mutation frequency.
First, the docking model in Fig. 3 predicts that mutations near the β3-β4 loop alter the base pair geometry upon initial substrate binding, leading to changes in the frequency of correct substrate incorporation at the catalytic site. Thus, NRTI resistance mutations on the fingers subdomain that had evolved to play roles in better rejection of incorrect substrate will generally decrease the frequency of misinsertion and thereby frameshift mutation, as observed in this study (Table 3 and Fig. 3, V75T) and previous studies (25). The L74V mutation on the β3-β4 loop is unique in that it causes virus hypersensitivity to zidovudine (42). Such a mutation may decrease the fidelity of geometric selection of the correct substrate, leading to an increase in mutation frequency.
Second, the model predicts that mutations along the interface of the palm subdomain of RT alter the nature of steric hindrance during the movement of the fingers and subsequent proper positioning of substrate at the catalytic site, resulting in an altered mutation frequency. For example, the Y115A mutations will induce less restricted rotation of the fingers subdomain because of the loss of the aromatic ring and the hydroxyl group at the side chain of the phenylalanine, respectively, leading to an increase in the probability of misinsertion-mediated mutations, as was observed in the previous study (25). Third, mutations around the catalytic site that reduce the rate of polymerization will cause increased susceptibility to the ATP-mediated excision reaction for the removal of a residue at the primer 3′ end (29, 30), leading to an increase in fidelity of DNA replication.
The docking model also explains why various NRTIs can increase HIV-1 mutation frequencies. The experimental conditions used in the present studies result in relatively high concentrations of the intracellular triphosphate form of the NRTI (NRTI-triphosphate), which is a potent competitive inhibitor of deoxynucleoside triphosphate as suggested in both the present and previous studies. This in turn will result in the reduction of frequency in correct deoxynucleoside triphosphate incorporation into the catalytic site, provided that the NRTI-triphosphate-template geometry is similar to that of the correct deoxynucleoside triphosphate-template base pair, leading to an increase in misinsertion and frameshift mutation frequencies. For example, abacavir-triphosphate, as a dGTP analogue, will become a competitor of the correct substrate dATP in the present system, because abacavir-triphosphate has structural similarity to dATP in that it has a purine base, as dATP has. Similarly, stavudine triphosphate will become a competitor of dATP incorporation, because the stavudine triphosphate can make two hydrogen bonds with the T template after initial binding. However, dCTP analogs such as lamivudine and zalcitabine will be less efficient competitors because of the lack of favorable hydrogen bond formation with thymidine and because of the similarity of base structure. This differential competition for each NRTI-triphosphate in turn will lead to the differential reduction in frequency of correct substrate (dATP) incorporation into the catalytic site, leading to a differential increase in mutation frequency, as seen in the present study (Table 3).
Potential mechanisms for NNRTI and NNRTI resistance mutation-mediated changes in HIV-1 mutation frequency.
The induced-fit model and the activity of ATP-mediated excision (29, 30) suggest a potential mechanism to explain the NNRTI-mediated increase in mutation frequency. NNRTIs, as allosteric inhibitors of RT activity, inhibit the above ATP-mediated excision reaction, probably by locking the structure of the RT active center and inhibiting ATP binding to RT (2). Therefore it is possible that the binding of an NNRTI to RT suppresses ATP binding to RT, leading to suppression of ATP-mediated RT changes for increasing the fidelity of substrate selection, such as in the excision reaction. This in turn will increase misinsertion-mediated mutation frequencies. On the other hand, NNRTI resistance mutations will reverse the suppression effects of NNRTI in a resistance level-dependent manner, which will partially restore the ATP-binding activity and its function for higher fidelity (as observed in Table 3). Further studies using this newly developed high-throughput assay along with structure-function analysis of HIV-1 RT will help in our understanding of what influences HIV-1 mutagenesis in cells.
Acknowledgments
This work was supported by National Institutes of Health grant GM56615.
REFERENCES
- 1.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremske. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basavapathruni, A., C. M. Bailey, and K. S. Anderson. 2004. Defining a molecular mechanism of synergy between nucleoside and nonnucleoside AIDS drugs. J. Biol. Chem. 279:6221-6224. [DOI] [PubMed] [Google Scholar]
- 3.Bebenek, K., J. Abbotts, J. D. Roberts, S. H. Wilson, and T. A. Kunkel. 1989. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J. Biol. Chem. 264:16948-16956. [PubMed] [Google Scholar]
- 4.Bebenek, K., J. Abbotts, S. H. Wilson, and T. A. Kunkel. 1993. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 268:10324-10334. [PubMed] [Google Scholar]
- 5.Bebenek, K., J. D. Roberts, and T. A. Kunkel. 1992. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J. Biol. Chem. 267:3589-3596. [PubMed] [Google Scholar]
- 6.Boyer, P. L., and S. H. Hughes. 2000. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 74:6494-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns, D. P., and H. M. Temin. 1994. High rates of frameshift mutations within homo-oligomeric runs during a single cycle of retroviral replication. J. Virol. 68:4196-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnouf, R., J. Ren, C. Ross, Y. Jones, D. Stammers, and D. Stuart. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Struct. Biol. 2:303-308. [DOI] [PubMed] [Google Scholar]
- 9.Essex, M. 1999. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 53:71-88. [DOI] [PubMed] [Google Scholar]
- 10.Harrigan, P. R., M. Salim, D. K. Stammers, B. Wynhoven, Z. L. Brumme, P. McKenna, B. Larder, and S. D. Kemp. 2002. A mutation in the 3′ region of the human immunodeficiency virus type 1 reverse transcriptase (Y318F) associated with nonnucleoside reverase transcriptase inhibitor resistance. J. Virol. 76:6836-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 12.Huang, W., A. Gamarnik, K. Limoli, C. J. Petropoulos, and J. M. Whitcomb. 2003. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J. Virol. 77:1512-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, S. J., J. S. Taylor, and L. S. Beese. 2003. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. USA 100:3895-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonckheere, H., E. De Clercq, and J. Anne. 2000. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, Tyr183 and Met184. Eur. J. Biochem. 267:2658-2665. [DOI] [PubMed] [Google Scholar]
- 15.Kanki, P. J., D. J. Hamel, J. L. Sankale, C. Hsieh, I. Thior, F. Barin, S. A. Woodcock, A. Gueye-Ndiaye, E. Zhang, M. Montano, T. Siby, R. Marlink, I. NDoye, M. E. Essex, and, S. MBoup. 1999. Human immunodeficiency virus type 1 subtypes differ in disease progression. J. Infect. Dis. 179:68-73. [DOI] [PubMed] [Google Scholar]
- 16.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 17.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 20.Lacey, S. F., and B. A. Larder. 1994. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob. Agents Chemother. 38:1428-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., Y. Kong, S. Korolev, and G. Waksman. 1998. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 7:1116-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansky, L. M. 2003. Mutagenic outcome of combined antiviral drug treatment during human immunodeficiency virus type 1 replication. Virology 307:116-121. [DOI] [PubMed] [Google Scholar]
- 24.Mansky, L. M., and L. C. Bernard. 2000. 3′-Azido-3′-deoxythymidine (zidovudine) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency type 1. J. Virol. 74:9532-9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansky, L. M., E. Le Rouzic, S. Benichou, and L. C. Gajary. 2003. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutation frequencies. J. Virol. 77:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansky, L. M., D. K. Pearl, and L. C. Gajary. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutation frequencies. J. Virol. 76:9253-9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansky, L. M., S. Preveral, L. Selig, R. Benarous, and S. Benichou. 2000. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J. Virol. 74:7039-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelemans, H., R. M. Esnouf, H. Jonckheere, E. De Clercq, and J. Balzarini. 1998. Mutational analysis of Tyr-318 within the non-nucleoside reverse transcriptase inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 273:34234-34239. [DOI] [PubMed] [Google Scholar]
- 32.Quan, Y., B. G. Brenner, R. G. Marlink, M. Essex, T. Kurimura, and M. A. Wainberg. 2003. Drug resistance profiles of recombinant reverse transcriptases from human immunodeficiency virus type 1 subtypes A/E, B, and C. AIDS Res. Hum. Retroviruses 19:743-753. [DOI] [PubMed] [Google Scholar]
- 33.Ray, A. S., A. Basavapathruni, and K. S. Anderson. 2002. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J. Biol. Chem. 277:40479-40490. [DOI] [PubMed] [Google Scholar]
- 34.Ren, J., R. Esnouf, E. Garman, D. Somers, C. Ross, I. Kirby, J. Keeling, G. Darby, Y. Jones, D. Stuart, and D. Stammers. 1995. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Struct. Biol. 2:293-302. [DOI] [PubMed] [Google Scholar]
- 35.Rittinger, K., G. Divita, and R. S. Goody. 1995. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA 92:8046-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 37.Sarafianos, S. G., A. D. Clark, Jr., K. Das, S. Tuske, J. J. Birktoft, P. Ilankumaran, A. R. Ramesha, J. M. Sayer, D. M. Jerina, P. L. Boyer, S. H. Hughes, and E. Arnold. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 21:6614-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato, H., Y. Tomita, K. Ebisawa, A. Hachiya, K. Shibamura, T. Shiino, R. Yang, M. Tatsumi, K. Gushi, H. Umeyama, S. Oka, Y. Takebe, and Y. Nagai. 2001. Augmentation of human immunodeficiency virus type 1 subtype E (CRF01_AE) multiple-drug resistance by insertion of a foreign 11-amino-acid fragment into the reverse transcriptase. J. Virol. 75:5604-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shafer, R. W., and D. A. Vuitton. 1999. Highly active antiretroviral therapy (HAART) for the treatment of infection with human immunodeficiency virus type 1. Biomed. Pharmacother. 53:73-86. [DOI] [PubMed] [Google Scholar]
- 40.Smerdon, S. J., J. Jager, J. Wang, L. A. Kohlstaedt, A. J. Chirino, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1994. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 91:3911-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spence, R. A., W. M. Kati, K. S. Anderson, and K. A. Johnson. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St Clair, M. H., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 43.Tantillo, C., J. Ding, A. Jacobo-Molina, R. G. Nanni, P. L. Boyer, S. H. Hughes, R. Pauwels, K. Andries, P. A. Janssen, and E. Arnold. 1994. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J. Mol. Biol. 243:369-387. [DOI] [PubMed] [Google Scholar]
- 44.Temiakov, D., V. Patlan, M. Anikin, W. T. McAllister, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for substrate selection by t7 RNA polymerase. Cell 116:381-391. [DOI] [PubMed] [Google Scholar]
- 45.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin, Y. W., and T. A. Steitz. 2004. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116:393-404. [DOI] [PubMed] [Google Scholar]