Abstract
Murine models have suggested that CD8+ T-cell responses peak early in acute viral infections and are not sustained, but no evidence for humans has been available. To address this, we longitudinally analyzed the CD8+ T-cell response to human parvovirus B19 in acutely infected individuals. We observed striking CD8+ T-cell responses, which were sustained or even increased over many months after the resolution of acute disease, indicating that CD8+ T cells may play a prominent role in the control of parvovirus B19 and other acute viral infections of humans, including potentially those generated by live vaccines.
The emergence of new tools for the ex vivo analysis of cellular immune responses, especially CD8+ T-cell responses, has revealed an important role for such cells in a range of viral infections. In studies with mice, most work on acute infection has been focused on influenza and lymphocytic choriomeningitis virus (6, 11, 29). However, very little is known about CD8+ T-cell responses in acute infections in humans; most work has focused on latent and persistent infections, such as human immunodeficiency virus, cytomegalovirus (CMV), Epstein-Barr virus, and hepatitis B and C virus (1, 16, 17) infections. In infections which are truly cleared, such as influenza, responses may return to a low-level resting memory state (20). In contrast, in infections with very-low-level persistence, such as CMV infection, strong immune responses may be sustained, and indeed, may increase over time (12, 13). Such responses typically possess “mature” effector characteristics indicative of repetitive antigen exposure (2, 26). Parvovirus B19 (B19) is a common virus with significant pathology (7). As B19 is regarded as a typical “hit-and-run” virus, the humoral response plays a well-documented role for viral neutralization, but there is also evidence that low-level persistence can occur in certain cases (21, 27). Cellular immune responses have also recently been described, both CD4+ proliferative- and CD8+ cytotoxic-T-cell responses, with one HLA-B35-restricted epitope characterized so far (4, 5, 28). The small B19 genome is very stable and encodes only three major proteins, which makes it suitable for extensive study without compromise due to incoming antigenic variability. Here, we describe the first assessment of the breadth, specificity, and kinetics of the acute CD8+ T-cell responses in this infection.
B19 immunoglobulin M (IgM)-positive samples from five previously healthy adult females (S1 through S5; age range, 32 to 51 years) were prospectively identified at the Clinical Virology Laboratory at the Karolinska Hospital, Stockholm, Sweden, after local ethical approval of the study. All presented with symptoms of fever, arthralgia, fatigue, and rashes, and were diagnosed within 11 days. Medical history gave no indication of susceptibility to infections. Serum and heparinized blood samples were collected at intervals for 48 to 108 weeks after diagnosis. Serum was analyzed for B19 DNA by nested PCR with a sensitivity of 103 DNA copies/ml and for B19 IgM and IgG by using an enzyme immunoassay (Biotrin International, Dublin, Ireland) (28). Peripheral blood mononuclear cells (PBMC) were separated by Ficoll-Paque (Amersham, Uppsala, Sweden). DNA was extracted from PBMC by using the QIAamp DNA minikit (VWR, Stockholm, Sweden) and analyzed by PCR to enable B19 DNA detection in escaped cells from bone marrow. HLA class I genotyping was performed by ABC SSP Unitray (Dynal, Oslo, Norway). Published sequences were used for synthesizing 210 peptides covering the nonstructural protein (NS), the unique region of the minor (VP1ur) structural protein, and the major (VP2) structural proteins (Table 1) (24). Gamma interferon (IFN-γ) responses were measured by ELISpot (15), using biotinylated IFN-γ antibodies (Mabtech, Stockholm, Sweden) and by intracellular staining (ICS) (18). PBMC was depleted of CD8+ T cells by using microbeads (Miltenyi Biotec, Gladbach, Germany). Nonamer-mediated cytotoxicity was tested by 51Cr-release assay (22, 25). HLA restrictions were estimated by using the BIMAS algorithm (http://bimas.cit.nih.gov) and T2-cell assays (14) and by matching single HLA alleles of targets and effectors in 51Cr experiments.
TABLE 1.
Peptide specifications
| Protein
|
Peptide
|
Pool
|
||||
|---|---|---|---|---|---|---|
| Name | Length (aa)a | No. of peptides | Length (aa)b | Amino acid overlap | No. of pools | No. of peptides in pool |
| NS1 | 671 | 133 | 15 (11) | 10 | 14 | 10 (3) |
| VP1ur | 227 | 22 | 20 (17) | 10 | 3 | 10 (2) |
| VP2 | 554 | 55 | 20 (14) | 10 | 6 | 10 (5) |
aa, amino acid.
The length of the last peptide for the respective protein is given in parentheses.
The number of peptides in the last pool of the respective peptide set is given in parentheses.
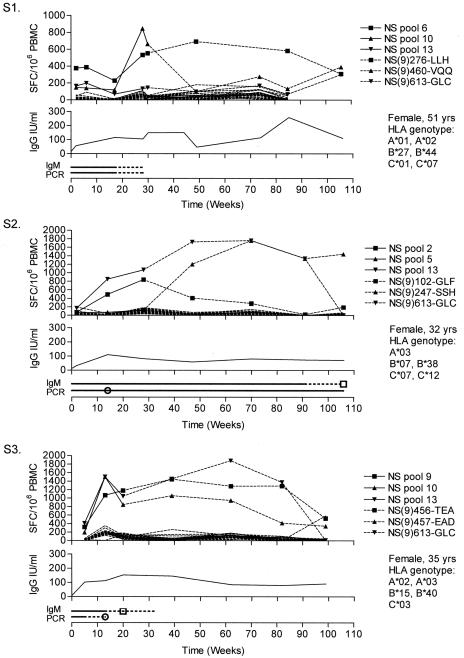
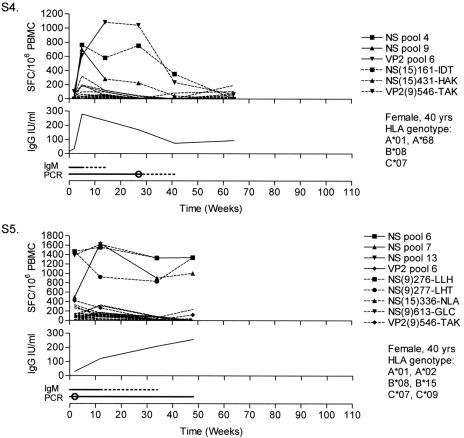
All individuals showed normal mitogen-induced IFN-γ responses and proliferation in vitro (data not shown). Fever, rashes, and fatigue resolved within 6 weeks. Responses to 8 of the 14 NS pools and 1 of the 6 VP2 pools were shown. No responses to the VP1ur pools were shown. All individuals responded to NS, whereas a VP2 response was present in only two individuals. Responses peaked at around 1 year in S1, S2, and S3, with a decline at about 2 years, whereas S4 showed a more rapid course, with a peak at 15 weeks and a decline at 1 year (Fig. 1). S5 was followed for 48 weeks with stable response levels. In S2, IgM antibodies were detected for more than 90 weeks, whereas in all others, they were lost between weeks 15 and 35. From the second week, IgG levels were raised and maintained above 6 IU/ml (23). In S2 and S5, B19 DNA was detected in PBMC throughout the entire follow-up, while this was lost between weeks 13 and 41 in the other individuals. Pool responses were fine mapped to five 15-mers and nine nonamers, of which seven nonamers were considered to represent novel CD8+ T-cell epitopes (Table 2; Fig. 2). By combined methods, it was possible to suggest HLA restrictions for four of these epitopes. When stimulating peptides of different lengths around the sequence of NS(9)456-TEA (see Table 2 for peptide nomenclature) were used in ICS, an additional B*44-restricted 11-mer epitope was detected (data not shown).
FIG. 1.
Longitudinal ELISpot, serology, and PCR data. Each graph is composed of three panels. The upper panel shows the IFN-γ responses over time expressed as SFC/106 PBMC. Solid lines with symbols represent positive peptide pool responses that were further mapped and are replaced at later time points by the responses to single peptides or nonamers, represented by dashed lines with the corresponding symbols. Dashed lines without symbols represent responses to the other peptide pools. For plotted responses to overlapping 15-mers (S4), responses to both peptides were equal, and only the second is plotted for clarity. For readability, error bars are not plotted, but mean standard deviations ± 95% confidence interval of positive triplicates were 58 ± 22, 53 ± 27, 46 ± 22, 37 ± 14, and 30 ± 18 SFC/106 PBMC for S1 through S5, respectively. The middle panel shows the IgG levels over time expressed in international units (IU) per ml. In the lower panel, the IgM serology is shown above the PCR results. A solid line was drawn between positive samples, and a dashed line represents time between a positive and negative result (or intermediate). Squares mark intermediate IgM serology results. Circles mark the time points when B19 DNA was lost in serum; further positivity then represents detectable viral DNA in PBMC. SFC, spot-forming cells.
TABLE 2.
Epitopes and peptides defined by mapping of pool responses
| Peptide namea | Amino acid sequenceb | HLA restriction | Specific lysis (%)c | Response after CD8+ depletion (%)d |
|---|---|---|---|---|
| NS (9) 102-GLF | GLFNNVLYH | NAf | 55 | 31 |
| NS (9) 247-SSH | SSHSGSFQI | NA | 65 | 0 |
| NS (9) 276-LLH | LLHTDFEQV | A*02 | 50 | 0 |
| NS (9) 277-LHTe | LHTDFEQVM | NA | 15 | 1 |
| NS (9) 456-TEA | TEADVQQWL | B*40 | 60 | 16 |
| NS (9) 457-EADe | EADVQQWLT | NA | 25 | 20 |
| NS (9) 460-VQQ | VQQWLTWCN | NA | 10 | 4 |
| NS (9) 613-GLC | GLCPHCINV | A*02 | 15 | 4 |
| VP2 (9) 546-TAK | TAKSRVHPL | B*08 | 25 | 0 |
| NS (11) 456-TEA | TEADVQQWLTW | B*44 | NA | NA |
| NS (15) 156-NID | NIDGYIDTCISATFR | |||
| NS (15) 161-IDT | IDTCISATFRRGACH | |||
| NS (15) 336-NLA | NLAMAIAKSVPVYGM | |||
| NS (15) 426-TTT | TTTTVHAKALKERMV | |||
| NS (15) 431-HAK | HAKALKERMVKLNFT |
Nomenclature used for peptide naming is as follows: B19 protein abbreviation and then the amino acid length in parentheses, followed by the position of the first amino acid in the protein according to the sequence published by Shade et al. (24). After the hyphen are the first three amino acids in the peptide sequence given.
Sequences in boldface are plausible to contain a shorter epitope due to the overlap of reactive 15-mers.
Percent specific lysis of pulsed autologous target cells in 51Cr-release assays at 50:1. NS(11)456-TEA was defined using ICS and was not tested in the cytotoxic assay.
IFN-γ response in ELISpot relative to PBMC.
Regarded as suboptimal epitopes with one amino acid shift.
NA, not available.
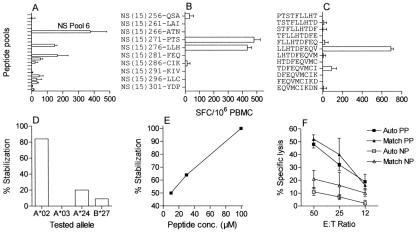
FIG. 2.
Example of the mapping and characterization of an epitope. The mapping and characterization of the epitope NS(9)247-LLH in S1 are shown. (A) The ELISpot IFN-γ responses to all NS peptide pools at week 2 are plotted. (B) Further mapping of the response to NS pool 6 revealed responses directed to two overlapping single 15-mers. SFC, spot-forming cells. (C) Testing of nonamers covering the sequence of the two reactive 15-mers revealed the epitope located in the overlap. (D) HLA restriction was confirmed by showing the epitope capable of stabilizing major histocompatibility complex (MHC) of T2 cells expressing HLA-A*02 but not MHC of cells with different expression. Peptide concentration was 30 μM. (E) An increasing peptide concentration (conc.) resulted in increasing stabilization of MHC of HLA-A*02-positive T2 cells. (F) Specific lysis in a 51Cr-release assay in which target cells were either pulsed (PP) or not pulsed (NP) with the epitope. In addition to autologous (Auto) targets, the HLA-A*02 restriction was confirmed by showing lysis of A*02 single matched target cells (Match). Error bars equal 1 standard deviation of triplicates where available. E:T ratio, effector-to-target cell ratio.
Thus, adults presenting with symptomatic B19 infection rapidly develop cellular immune responses with multiple specificities, which rise to high levels and are maintained for many months. The responses do not decay as anticipated but are kept at high levels for a long time, sometimes more than 2 years. This could be the result of continuous antigenic stimulation, analogous to truly persistent low-level CMV infection. Indeed, for three out of five individuals, B19 DNA was detected in peripheral blood for over 6 months. It is possible that B19 persists beyond this time in the bone marrow, in which prolonged replication has been observed (8, 19). Most responses were directed to NS, while two individuals also responded to an epitope in VP2. In contrast, neutralizing antibodies and a majority of CD4+ T-cell responses are reported to be primarily directed to the structural proteins (4, 5, 21). Thus, there is evidently a division in structural and nonstructural preference for the different arms of the immune system. Importantly, we found that S3, S4, and S5 all made three roughly equal responses at 5 to 10 weeks after symptom onset. All but one of these responses were maintained at equal high levels over at least a further 20 weeks. Thus, in B19 infection, multiple “equidominant” CD8+ T-cell immune responses occur simultaneously, with no clear pattern of changing dominant responses over time as, for example, seen for Epstein-Barr virus or murine CMV (9, 26). This may reflect the structural and replicative simplicity of B19, in that no division in early and late proteins exists. Other responses may have been missed either because they occurred very early or were not detected using 15- or 20-mer peptides. B19 is not classically persistent in normal infection and is present only in about 2% of bone marrow samples from healthy subjects (8). However, it seems clear that B19 infection is not to be immunologically described by the paradigm of lytic nonpersistent viruses that postulates rapid viral clearance and contraction of the initial cytotoxic-T-cell burst in the absence of antigenic drive (3, 10). B19 assumes characteristics of lytic viruses, both of lytic nonpersistent, and persistent, viruses, and elicits a striking pattern of immunological response in human primary infection. Harnessing such CD8+ T-cell responses to generate vaccine responsiveness is an exciting possibility. In further comprehensive approaches to characterization of the human CD8+ T-cell response, B19 may serve as a key model for dissecting the complexities of long-term virus-host relationships.
Acknowledgments
This study was supported by the Tobias Foundation, the Swedish Children's Cancer Foundation, the Swedish Cancer Foundation, the Swedish Research Council, the Wellcome Trust, the Medical Research Council, United Kingdom, and the Commission of the European Communities (QLK2-CT-2001-00877).
We thank Victor Levitsky, Kiyotaka Kuzushima, and Lopez de Castro for generously providing the T2 cell lines.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. [DOI] [PubMed] [Google Scholar]
- 3.Badovinac, V. P., B. B. Porter, and J. T. Harty. 2002. Programmed contraction of CD8(+) T cells after infection. Nat. Immunol. 3:619-626. [DOI] [PubMed] [Google Scholar]
- 4.Franssila, R., and K. Hedman. 2004. T-helper cell-mediated interferon-gamma, interleukin-10 and proliferation responses to a candidate recombinant vaccine for human parvovirus B19. Vaccine 22:3809-3815. [DOI] [PubMed] [Google Scholar]
- 5.Franssila, R., K. Hokynar, and K. Hedman. 2001. T helper cell-mediated in vitro responses of recently and remotely infected subjects to a candidate recombinant vaccine for human parvovirus b19. J. Infect. Dis. 183:805-809. [DOI] [PubMed] [Google Scholar]
- 6.Gourley, T. S., E. J. Wherry, D. Masopust, and R. Ahmed. 2004. Generation and maintenance of immunological memory. Semin. Immunol. 16:323-333. [DOI] [PubMed] [Google Scholar]
- 7.Heegaard, E. D., and K. E. Brown. 2002. Human parvovirus B19. Clin. Microbiol. Rev. 15:485-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heegaard, E. D., B. L. Petersen, C. J. Heilmann, and A. Hornsleth. 2002. Prevalence of parvovirus B19 and parvovirus V9 DNA and antibodies in paired bone marrow and serum samples from healthy individuals. J. Clin. Microbiol. 40:933-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hislop, A. D., N. E. Annels, N. H. Gudgeon, A. M. Leese, and A. B. Rickinson. 2002. Epitope-specific evolution of human CD8(+) T cell responses from primary to persistent phases of Epstein-Barr virus infection. J. Exp. Med. 195:893-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou, S., L. Hyland, K. W. Ryan, A. Portner, and P. C. Doherty. 1994. Virus-specific CD8+ T-cell memory determined by clonal burst size. Nature 369:652-654. [DOI] [PubMed] [Google Scholar]
- 11.Kagi, D., B. Ledermann, K. Burki, P. Seiler, B. Odermatt, K. J. Olsen, E. R. Podack, R. M. Zinkernagel, and H. Hengartner. 1994. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369:31-37. [DOI] [PubMed] [Google Scholar]
- 12.Khan, N., N. Shariff, M. Cobbold, R. Bruton, J. A. Ainsworth, A. J. Sinclair, L. Nayak, and P. A. Moss. 2002. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 169:1984-1992. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu, H., S. Sierro, A. V. Cuero, and P. Klenerman. 2003. Population analysis of antiviral T cell responses using MHC class I-peptide tetramers. Clin. Exp. Immunol. 134:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzushima, K., N. Hayashi, H. Kimura, and T. Tsurumi. 2001. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8(+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98:1872-1881. [DOI] [PubMed] [Google Scholar]
- 15.Larsson, M., X. Jin, B. Ramratnam, G. S. Ogg, J. Engelmayer, M. A. Demoitie, A. J. McMichael, W. I. Cox, R. M. Steinman, D. Nixon, and N. Bhardwaj. 1999. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS 13:767-777. [DOI] [PubMed] [Google Scholar]
- 16.Lauer, G. M., E. Barnes, M. Lucas, J. Timm, K. Ouchi, A. Y. Kim, C. L. Day, G. K. Robbins, D. R. Casson, M. Reiser, G. Dusheiko, T. M. Allen, R. T. Chung, B. D. Walker, and P. Klenerman. 2004. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127:924-936. [DOI] [PubMed] [Google Scholar]
- 17.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 76:6104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundqvist, A., T. Tolfvenstam, J. Bostic, M. Soderlund, and K. Broliden. 1999. Clinical and laboratory findings in immunocompetent patients with persistent parvovirus B19 DNA in bone marrow. Scand. J. Infect. Dis. 31:11-16. [DOI] [PubMed] [Google Scholar]
- 20.Maczek, C., T. G. Berger, B. Schuler-Thurner, E. S. Schultz, A. Hamann, P. R. Dunbar, V. Cerundolo, A. Steinkasserer, and G. Schuler. 2005. Differences in phenotype and function between spontaneously occurring melan-A-, tyrosinase- and influenza matrix peptide-specific CTL in HLA-A*0201 melanoma patients. Int. J. Cancer 115:450-455. [DOI] [PubMed] [Google Scholar]
- 21.Modrow, S., and S. Dorsch. 2002. Antibody responses in parvovirus B19 infected patients. Pathol. Biol. (Paris) 50:326-331. [DOI] [PubMed] [Google Scholar]
- 22.Nixon, D. F., A. R. Townsend, J. G. Elvin, C. R. Rizza, J. Gallwey, and A. J. McMichael. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484-487. [DOI] [PubMed] [Google Scholar]
- 23.Searle, K., C. Guilliard, and G. Enders. 1997. Parvovirus B19 diagnosis in pregnant women-quantification of IgG antibody levels (IU/ml) with reference to the international parvovirus B19 standard serum. Infection 25:32-34. [DOI] [PubMed] [Google Scholar]
- 24.Shade, R. O., M. C. Blundell, S. F. Cotmore, P. Tattersall, and C. R. Astell. 1986. Nucleotide sequence and genome organization of human parvovirus B19 isolated from the serum of a child during aplastic crisis. J. Virol. 58:921-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehy, M. E., A. B. McDermott, S. N. Furlan, P. Klenerman, and D. F. Nixon. 2001. A novel technique for the fluorometric assessment of T lymphocyte antigen specific lysis. J. Immunol. Methods 249:99-110. [DOI] [PubMed] [Google Scholar]
- 26.Sierro, S., R. Rothkopf, and P. Klenerman. 2005. Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur. J. Immunol. 35:1113-1123. [DOI] [PubMed] [Google Scholar]
- 27.Soderlund-Venermo, M., K. Hokynar, J. Nieminen, H. Rautakorpi, and K. Hedman. 2002. Persistence of human parvovirus B19 in human tissues. Pathol. Biol. (Paris) 50:307-316. [DOI] [PubMed] [Google Scholar]
- 28.Tolfvenstam, T., A. Oxenius, D. A. Price, B. L. Shacklett, H. M. Spiegel, K. Hedman, O. Norbeck, M. Levi, K. Olsen, M. Kantzanou, D. F. Nixon, K. Broliden, and P. Klenerman. 2001. Direct ex vivo measurement of CD8+ T-lymphocyte responses to human parvovirus B19. J. Virol. 75:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wherry, E. J., and R. Ahmed. 2004. Memory CD8 T-cell differentiation during viral infection. J. Virol. 78:5535-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]