Abstract
The potential threat of smallpox bioterrorism has made urgent the development of lower-virulence vaccinia virus vaccines. An attenuated LC16m8 (m8) vaccine was developed in 1975 from the Lister strain used in the World Health Organization smallpox eradication program but was not used against endemic smallpox. Today, no vaccines can be tested with variola virus for efficacy in humans, and the mechanisms of immune protection against the major intracellular mature virion (IMV) and minor extracellular enveloped virion (EEV) populations of poxviruses are poorly understood. Here, we determined the full-genome sequences of the m8, parental LC16mO (mO), and grandparental Lister (LO) strains and analyzed their evolutionary relationships. Sequence data and PCR analysis indicated that m8 was a progeny of LO and that m8 preserved almost all of the open reading frames of vaccinia virus except for the disrupted EEV envelope gene B5R. In accordance with this genomic background, m8 induced 100% protection against a highly pathogenic vaccinia WR virus in mice by a single vaccination, despite the lack of anti-B5R and anti-EEV antibodies. The immunogenicity and priming efficacy with the m8 vaccine consisting mainly of IMV were as high as those with the intact-EEV parental mO and grandparental LO vaccines. Thus, mice vaccinated with 107 PFU of m8 produced low levels of anti-B5R antibodies after WR challenge, probably because of quick clearance of B5R-expressing WR EEV by strong immunity induced by the vaccination. These results suggest that priming with m8 IMV provides efficient protection despite undetectable levels of immunity against EEV.
Variola virus (VAR), a member of the orthopoxvirus (OPV) family, is the causative agent of smallpox and caused millions of deaths before its eradication. Today, smallpox is again becoming a potential threat to humans, with abuse of VAR as a bioterrorist weapon (10, 15, 20, 26, 30, 37, 40). The World Health Organization (WHO) program for smallpox eradication indicated that vaccinia virus (VV) vaccination is the most effective preventive measure against the disease. However, WHO recommended discontinuing the vaccination in 1980 (55) due to rare (around 20 cases/106 vaccinees) but severe complications, such as postvaccinial encephalitis, progressive vaccinia, and eczema vaccinatum with the primary vaccination (4, 17, 34, 57). Thus, after a lag time of more than 20 years, serious attempts have been urged to restart the development of lower-virulence vaccine strains (2, 3, 9, 43, 45, 50). A vaccinia ACAM1000 clone has recently been established using cell cultures from the Dryvax (NYBH strain) vaccine (50), but it may induce myocarditis (4, 11). Modified vaccinia virus Ankara (MVA) and NYVAC (modified Copenhagen strain) replication-incompetent viruses are certainly safer but may require high vaccine doses or boosting with replication-competent vaccines (2, 9).
One of the safest replication-competent vaccines, a vaccinia virus LC16m8 strain (m8), was developed and established in the early 1970s with cell culture systems (24, 25) through a temperature-sensitive and low-virulence LC16mO intermediate clone (mO) from the Lister (Elstree) original strain (LO) that was used worldwide in the WHO program. The m8 virus exhibited the lowest levels of neurovirulence and the mildest adverse events among several vaccine strains, such as NYBH, CV1, and EM63, in monkeys, rabbits, and cortisone-induced immunocompromised mice (24, 38, 39). Its antigenicity was as high as that of the LO vaccine, not only in animals, but also in approximately 50,000 Japanese children vaccinated from 1973 to 1974 (over 90,000 doses in 1974 and 1975) with no reports of severe complications (24, 57). Based on these studies, cell culture-derived m8 was licensed in 1975 in Japan as a second-generation smallpox vaccine, but it has never been confronted with VAR.
Recent progress in molecular genetics has demonstrated that m8 has a single-nucleotide deletion creating a termination codon at amino acid (aa) position 93 in the B5R envelope (env) gene (47). Several papers have indicated that the destruction of B5R contributes to attenuation of poxviruses (12, 36, 44, 46, 47, 54). In turn, the B5R Env protein was suggested to function as an antigen that induces neutralizing antibodies (NAbs) to the extracellular enveloped virion (EEV) form of poxviruses (12, 19, 44). EEVs are free virions released from infected cells and may cause long-range dissemination of infection, although they comprise less than 1% of the virus population, the majority being the intracellular mature virion (IMV) form (12, 41, 44). In addition, B5R is also a component of viral particles on the cell surface termed cell-associated enveloped virions, which are more abundant than EEV and are important for cell-to-cell spread (44). Consequently, the spread of these VVs seems to be prevented by anti-B5R NAbs.
However, little is as yet understood regarding the mechanisms of immune protection against EEVs, cell-associated enveloped virions, and IMVs of poxviruses. Thus, a concern has arisen that the B5R truncation and other possible mutations introduced into m8 during processes of attenuation of the LO vaccine reduce the generation of the enveloped virions and therefore might make the attenuated m8 vaccine less protective or nonprotective against VAR (5, 44, 45). No vaccines, however, can be tested for efficacy against VAR in humans. Alternatively, intranasal infection with a mouse-adapted and highly pathogenic vaccinia virus Western Reserve (WR) strain provides a mouse model well suited for evaluating protective efficacy (2, 32, 50, 51).
Here, we determined and compared the full-genome sequences of the licensed m8, parental mO, and grandparental LO strains to examine whether m8 has inherited the intact genome of LO or acquired other alterations in the EEV-related genes. We also examined antibody responses to B5R, EEV, and IMV in mice after a single vaccination with m8, mO, and LO and evaluated the protective efficacy against intranasal WR challenge in vaccinated mice. The results suggest that the genes, except for B5R, of m8 are similar to those of LO and that consequently, the immunogenicity and protective efficacy of m8 are similar to those of LO.
MATERIALS AND METHODS
Cells and viruses.
RK13 cells were grown in Eagle's minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). HeLa cells were cultured in Dulbecco's modified MEM containing 5% FBS. High five (Tn5) insect cells were cultured at 26°C in TC100 medium (JRH Bioscience, Inc.) supplemented with 10% FBS. LO, mO, m8, and WR strains of VV (kind gifts from S. Hashizume) were propagated and titrated on RK13 cell monolayers (58). The WR virus used was selected by sensitivity to 5-bromo-2-deoxyuridine before propagation. When a VV IHD-J strain was used as a high producer of EEV, the virus was freshly prepared, titrated, and inoculated into cells (41).
Purification of viral DNA.
RK13 cells infected with m8, mO, or LO virus were harvested and disrupted by sonication in 10 mM Tris (pH 8.0)-1 mM EDTA buffer. Cell debris and nuclei were removed from cell lysates by low-speed centrifugation, and viruses were recovered by centrifugation at 15,000 × g for 40 min. Virions suspended in 0.1× Tris-EDTA were purified by centrifugation on 36% sucrose cushions and then on 20 to 40% linear sucrose density gradients, as described previously (29). After each centrifugation step, virion precipitates were resuspended by sonication to avoid virion aggregate formation. Genomic virus DNA was extracted from purified virions with sodium dodecyl sulfate-proteinase K and then with phenol-chloroform as described previously (42).
Sequence analysis of the complete viral DNA genomes.
Purified viral DNA was fragmented with a HydroShear recirculating point-sink flow system (GeneMachines). DNA fragments of 1.5 to 2.5 kbp were recovered by 0.8% agarose gel electrophoresis, blunt ended, and cloned into pUC18. The inserts of the shotgun clones were amplified by PCR with primers (5′-CAGTCACGACGTTGTAAAACGAC-3′ and 5′-GTGTGGAATTGTGAGCGGATAAC-3′) and Ex Taq polymerase (TaKaRa Bio. Inc.). The amplified DNAs were sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit on PRISM 3700 automated DNA sequencers (Applied Biosystems). The net virus nucleotide sequences were collected with PHRED/PHRAP software and then assembled and edited with Sequencher 4.0 software (GeneCodes Corp.) (13, 14). Primer walking was done for filling gaps and for confirming the order and lengths of the preassembled contigs, as well as the approximately 6-kbp inverted terminal repeats (ITRs) of both genome ends. As the terminal hairpin loops were not sequenced, the leftmost nucleotide of the assembled sequences was arbitrarily designated base number 1. The final DNA sequences of m8, mO, and LO were represented at more than 9.2-, 7.8-, and 8.9-fold redundancy, respectively, at each base position. Open reading frames (ORFs) were identified using National Center for Biotechnology Information BLAST and compared to the GenBank files of the nonredundant protein sequence database, including OPVs and the vaccinia Copenhagen (CPN) strain (21). When there was a large gap between ORFs, mini-ORFs (more than 33 aa) were tentatively predicted for m8 and mO. Noncoding regions were examined for putative early, intermediate, and late promoters with MEME version 3.0 and MAST version 3.0.
PCR analysis.
DNAs from LO and mO viruses were analyzed by PCR at six randomly selected loci of LO diversity, numbers L0202, L0403, L0638, L0640, L1000, and L1100, using combinations of the LO- or mO-specific forward primers and the common reverse primers. PCR mixtures were heat denatured at 95°C for 3 min and subjected to 30 cycles of 94°C for 20 s, 63°C for 40 s, and 72°C for 1 min. When the loci L0403 and L1000 were amplified, annealing was done at 61°C. The primers used were as follows: LO-0202 (5′-AGCTATTCTACCATAGCAAAT-3′), mO-0202 (5′-AGCTATTCTACCATAGCAGAA-3′), and R-0202 (5′-CTTGGTTGGTAGAAATGCGG-3′); LO-0403 (5′-TCTAGATAAAATCACTGACTTTC-3′), mO-0403 (5′-TCTAGATAAAATCACTGACTTTT-3′), and R-0403 (5′-AGGAATATGTATAAATGCGGG-3′); LO-0638 (5′-CATATTAGTAGTTCTGCGCAAT-3′), mO-0638 (5′-CATATTAGTAGTTCTGCGTAAG-3′), and R-0638 (5′-CATTATGGTGGCTAGTGATG-3′); LO-0640 (5′-CACCTCTACCGAATAGAGTA-3′), mO-0640 (5′-CACCTCTACCGAATAAAGTT-3′), and R-0630 (5′-GATCTAAATAGAATGCCGACC-3′); LO-1000 (5′-TTAATAGTTGATAGATACGCATTT-3′), mO-1000 (5′-AATAGTTGATAGATACGCGTTC-3′), and R-1000 (5′-CATTTATAACACTGTACTAAC-3′); and LO-1100 (5′-GAACTTCAGGCTGGTGAATC-3′), mO-1100 (5′-AGAACTTCAGGCTGGTAAATT-3′), and R-1100 (5′-CCATTAGTATCCATATACCATG-3′).
Comparison of EEV env-related genes.
The B5R gene and other EEV env-related genes, A33R, A34R, A36R, A56R, and F13L, of a calf lymph Lister vaccine (ListerVAX), mO, and IHD-J were amplified by PCR, sequenced, and compared in amino acid alignment with the VV CPN (GenBank M35027), WR (GenBank AY243312), and MVA (GenBank, U94848) strains and also with other OPVs: VAR (strain Bangladesh-1975; GenBank L22579), monkeypox virus (MPV) (strain Zaire-96-I-16; GenBank AF380138), and cowpox virus (CPV) (strain GRI-90; GenBank X94355).
Preparation of B5R and vaccinia virus antigens.
The ectodomain of B5R was amplified from ListerVAX DNA by PCR using primers B5R-Hisf-Bgl (5′-AGATCTACATGTACTGTACCCAC-3′) and B5R-ECTr-Bgl (5′-AGATCTATTCTAACGATTCTATTTCTTG-3′) and cloned into pGEM-Teasy (Promega). The B5R-ect insert was excised from the resultant pTe-Lis-B5R-ect and ligated into a pAcYM1 baculovirus transfer plasmid, pAcMel-His, modified with the melitin signal sequence and a six-His tag. A recombinant AcHis-Lister-B5R-ect baculovirus was constructed as described previously (33). Lysates of Tn5 insect cells were prepared with 1% NP-40 4 days after AcHis-Lister-B5R-ect infection. The lysates were clarified by centrifugation, and the recombinant B5R protein was purified by Ni column (Invitrogen) chromatography. For VV antigens, HeLa cells were infected with LO, harvested 4 days after infection, and lysed with 1% NP-40. The lysates were clarified by centrifugation.
Tests for immunogenicity and protective efficacy.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the National Institute of Infectious Diseases. Groups of 15 6-week-old female BALB/c mice were vaccinated with 105 or 107 PFU of m8, mO, or LO or with PBS. On day 21, five mice from each group were sacrificed to test for prechallenge antibody responses, and the other mice were challenged intranasally with 106 PFU of WR in 20 μl PBS (51). The mice were observed for clinical signs, examined for bodyweight, and sacrificed 14 days after WR challenge to test for postchallenge antibody responses. The immunogenicity of the recombinant B5R protein was confirmed by subcutaneous injection of BALB/c mice three times each with mixed-in aluminum adjuvant and with the B5R antigen adsorbed to Ni-agarose beads. The immunized mice were challenged with WR as described above 12 days after the last booster injection.
Anti-B5R and anti-vaccinia virus antibody ELISA.
Enzyme-linked immunosorbent assay (ELISA) plates were coated with B5R or VV antigen and blocked with 5% skim milk. Dilutions of serum samples were reacted to the plates, and bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Zymed Laboratory), followed by a substrate (ABTS; Roche Diagnostics). The cutoff optical density at 405 nm (OD405) value of 0.2 was calculated from the average OD, plus three times the standard deviation, for five mock-immunized mouse sera.
Virus neutralization and comet inhibition assays.
LO virus (100 PFU/100 μl determined on HeLa cells) was mixed with serially diluted mouse serum at 37°C for 1 h and then overnight at 4°C. HeLa cells in 24-well plates were infected with the serum-treated virus, cultured for 4 days, and stained with 0.1% crystal violet. The serum dilutions yielding a 50% plaque reduction were defined as IMV-neutralizing antibody titers. Comet-inhibiting activity in serum was examined as an indication of anti-EEV antibody responses (1). RK13 cells in 12-well plates were infected with IHD-J virus (100 PFU/well), incubated for 2 days in 2% FBS-Dulbecco's modified MEM containing mouse serum dilutions, and stained with crystal violet. The lengths of comets formed from primary plaques were measured under a microscope.
Histopathology and immunohistochemistry (IHC).
The mouse nasal tissues were fixed in 10% buffered formalin and embedded in paraffin. Paraffin block sections were stained with hematoxylin and eosin (HE). VV antigens were immunohistochemically detected with a labeled-streptavidin-biotin complex staining system (DAKO). Rabbit polyclonal antibodies raised by LO infection were used as a primary antibody. A catalyzed signal amplification method (DAKO) was also used to detect VV antigens with enhanced sensitivity.
Nucleotide sequence accession numbers.
The complete sequences of the vaccinia virus m8, mO, and LO strains have been deposited in GenBank under accession numbers AY678275, AY678277, and AY678276, respectively. The env gene sequences of IHD-J were deposited in DDBJ: A33R-A34R (accession no. AB191187), A36R (accession no. AB191188), A56R (accession no. AB191189), B5R (accession no. AB191190), and F13L (accession no. AB191191). As there were slight differences between the ListerVAX and compiled shotgun LO sequences, ListerVAX virus sequences were deposited in DDBJ as follows: B5R (accession no. AB191251), A56R (clone 1) (accession no. AB191252), and A56R (clone 3) (accession no. AB191253).
RESULTS
Complete genome sequences of m8, mO, and LO.
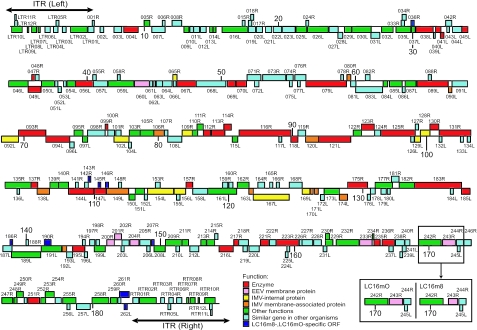
Genomic DNA was prepared from purified m8, mO, and LO virions, shotgun sequenced, and confirmed by primer walking. As m8 and mO are clonal viruses, their genome sequences were easily assembled to 189,158 and 189,157 bp, respectively, and were analyzed with reference to the GenBank files, including the vaccinia virus CPN strain (21). Comparison of the m8 and mO genomes indicated that their gene structures and organizations were almost the same (Fig. 1 and Table 1). Notably, there were only six point mutations between m8 and mO (Fig. 2A). Three of them were in noncoding regions, probably in promoter regions. A single-amino-acid substitution was found in 4 ORFs out of 286 putative major, minor, and mini-ORFs: a T-to-G mutation caused the change from Ile to Leu in the LC16M098L (F12L for CPN) gene, and an A-to-T mutation caused the replacements of Thr with Ser in the LC16M105R (A ORF T for CPN) gene and Ser with Arg in the LC16M012L (A54L for CPN) gene. The most remarkable change was a deletion of G in the LC16M243R (B5R for CPN) gene, which generated a termination codon and truncated the B5R Env protein of m8 EEV at amino acid position 93 (Fig. 2B), as described previously (47).
FIG. 1.
ORF map of the LC16m8 and LC16mO strains. The ORFs transcribed rightward and leftward are presented above and below the horizontal centerlines, respectively. The major difference between the two strains is boxed. Putative functions of ORFs were evaluated or predicted by a BLAST search of the GenBank database and are expressed in different colors. The double-headed arrows indicate the regions of the ITRs of the left and right ends.
TABLE 1.
ORF locations and features of the LC16m8 and LC16mO genomes
| ORF | Position in LC16m8 (aa length) | Position in LC16m0 | Promoter typea | Putative function | Category | Best-matching ORFb
|
ORF corresponding to CPN | ||
|---|---|---|---|---|---|---|---|---|---|
| Name | BLASTP Score | Source | |||||||
| LC16MLTR12R | 300-503 (67) | -c | Hypothetical protein | Similar gene in other organisms | C ORF H | 2e-36 | CPN | C ORF H (2e-36) | |
| LC16MLTR11R | 307-420 (37) | - | Hypothetical protein | Similar gene in other organisms | C ORF G | 4e-09 | CPN | C ORF G (4e-09) | |
| LC16MLTR10L | 860-84 (258) | - | Major secreted protein | Other functions | VACWR001 | e-113 | WR | B29R (e-112) | |
| LC16MLTR09L | 1353-1249 (34) | - | Tumor necrosis factor receptor II fragment | Other functions | PredictadbyGaneMark | 3e-17 | CPN | PredictedbyGeneMark11 (3e-17) | |
| LC16MLTR08L | 1940-1572 (122) | - | L? | Tumor necrosis factor receptor II homologue | Other functions | VACWR004 | 4e-73 | WR | C22L (3e-72) |
| LC16MLTR07L | 2204-2058 (48) | - | K1R protein fragment | Other functions | VACWR005 | 4e-24 | WR | PredictedbyGeneMark02 (5e-24) | |
| LC16MLTR06L | 2954-2568 (128) | - | Hypothetical protein | Similar gene in other organisms | VACWR007 | 4e-59 | WR | C20L (1e-55) | |
| LC16MLTR05R | 3387-3599 (70) | - | L? | Hypothetical protein | Similar gene in other organisms | C ORF F | 1e-29 | CPN | C ORF F (1e-29) |
| LC16MLTR04L | 3533-3204 (109) | - | L?,E | Hypothetical protein | Similar gene in other organisms | VACWR008 | 1e-62 | WR | C19L (5e-57) |
| LC16MLTR03L | 4141-3860 (93) | - | Hypothetical protein | Similar gene in other organisms | D4L | 3e-41 | Cowpox | PredictedbyGeneMark09 (3e-18) | |
| LC16MLTR02L | 5725-4475 (416) | - | L? | Host range protein | Other functions | C17L | 0.0 | CPN | C17L (0.0) |
| LC16M001R | 6087-6242 (51) | - | Hypothetical protein | Similar gene in other organisms | TC18R | 3e-65 | Tian Tan | ||
| LC16MLTR01L | 6215-5772 (147) | - | Hypothetical protein | Similar gene in other organisms | C16L | 4e-85 | CPN | C16L (4e-85) | |
| LC16M002L | 6938-6669 (89) | - | L? | Hypothetical protein | Similar gene in other organisms | C15L | 1e-35 | CPN | C15L (1e-35) |
| LC16M003L | 8281-7709 (190) | - | Hypothetical protein | Similar gene in other organisms | VACWR206 | e-108 | WR | C14L (3e-37) | |
| LC16M004L | 9505-8444 (353) | - | L? | Serine protease | Enzyme | C12L | 0.0 | CPN | C12L (0.0) |
| LC16M005R | 9950-10372 (140) | - | L? | Growth factor | Other functions | MVA005R | 3e-72 | MVA | C11R (8e-69) |
| LC16M006R | 11315-11512 (65) | - | Hypothetical protein | Similar gene in other organisms | C ORF E | e-14 | CPN | C ORF E (e-14) | |
| LC16M007L | 11520-10525 (331) | - | L? | Hypothetical protein | Similar gene in other organisms | C10L | 0.0 | CPN | C10L (0.0) |
| LC16M008R | 12034-12753 (239) | - | L? | Hypothetical protein | Similar gene in other organisms | C7R | e-105 | Cowpox | |
| LC16M009L | 13300-12826 (124) | - | L? | Interleukin 18 binding protein | Other functions | MVA008L | 5e-64 | MVA | |
| LC16M010L | 13631-13359 (90) | - | E | Hypothetical protein | Similar gene in other organisms | ACAM3000_MVA_009 | 5e-50 | ACAM3000 | |
| LC16M011L | 14072-13644 (142) | - | L? | Hypothetical protein | Similar gene in other organisms | ACAM3000_MVA_010 | 9e-80 | ACAM3000 | |
| LC16M012L | 14574-14161 (137) | - | L? | Hypothetical protein | Similar gene in other organisms | VACWR015 | 5e-71 | WR | |
| LC16M013L | 15074-14841 (77) | - | Host range protein | Other functions | VACWR016 | 6e-41 | WR | ||
| LC16M014L | 15311-15096 (11) | - | L? | Host range protein | Other functions | ACAM3000_MVA_013 | 9e-41 | ACAM3000 | |
| LC16M015R | 17265-17477 (70) | - | Hypothetical protein | Similar gene in other organisms | C ORF D | 8e-23 | CPN | C ORF D (8e-23) | |
| LC16M016L | 17671-15767 (634) | - | L?,E | Host range protein | Other functions | C9L | 0.0 | CPN | C9L (0.0) |
| LC16M017R | 17724-17972 (82) | - | L? | Hypothetical protein | Similar gene in other organisms | C ORF C | 7e-33 | CPN | C ORF C (7e-33) |
| LC16M018R | 17697-18121 (74) | - | Hypothetical protein | Similar gene in other organisms | C ORF B | 2e-37 | CPN | C ORF B (2e-37) | |
| LC16M019L | 18247-17714 (177) | - | L? | Hypothetical protein | Similar gene in other organisms | VACWR020 | e-102 | WR | C8L (6e-99) |
| LC16M020L | 18771-18319 (150) | - | L? | Hypothetical protein | Similar gene in other organisms | MVA018L | 1e-88 | MVA | C7L (2e-88) |
| LC16M021L | 19455-19000 (151) | - | L? | Hypothetical protein | Similar gene in other organisms | MVA019L | 6e-85 | MVA | C6L (7e-85) |
| LC16M022L | 20196-19582 (204) | - | Hypothetical protein | Similar gene in other organisms | C5L | e-120 | CPN | C5L (e-120) | |
| LC16M023L | 21209-20259 (316) | - | L?,E | Hypothetical protein | Similar gene in other organisms | C4L | 0.0 | CPN | C4L (0.0) |
| LC16M024R | 22010-22219 (69) | - | L? | Hypothetical protein | Similar gene in other organisms | C ORF A | 2e-36 | CPN | C ORF A (2e-36) |
| LC16M025L | 22067-21276 (263) | - | L? | Complement regulatory protein | Other functions | C3L | e-159 | CPN | C3L (e-159) |
| LC16M026L | 23672-22134 (512) | - | Kelch-like protein | Other functions | C2L | 0.0 | CPN | C2L (0.0) | |
| LC16M027L | 24413-23739 (224) | - | Hypothetical protein | Similar gene in other organisms | C1L | e-120 | CPN | C1L (e-120) | |
| LC16M028L | 24753-24400 (117) | - | L? | Hypothetical protein | Similar gene in other organisms | N1L | 5e-66 | CPN | N1L (5e-66) |
| LC16M029L | 25416-24889 (175) | - | Putative alpha amanitin-sensitive protein | Other functions | N2L | e-100 | CPN | N2L (e-100) | |
| LC16M030L | 26876-25458 (472) | - | Putative ankyrin isoform | Other functions | M1L | 0.0 | CPN | M1L (0.0) | |
| LC16M031L | 27516-26854 (220) | - | L? | Hypothetical protein | Similar gene in other organisms | M2L | e-132 | CPN | M2L (e-132) |
| LC16M032L | 28505-27651 (284) | - | E | Host range protein | Other functions | VACWR032 | e-155 | WR | K1L (e-153) |
| LC16M033R | 29114-29359 (81) | - | Hypothetical protein | Similar gene in other organisms | K ORF A | 4e-45 | CPN | K ORF A (4e-45) | |
| LC16M034R | 29181-29483 (100) | - | Hypothetical protein | Similar gene in other organisms | K ORF B | 1e-40 | CPN | K ORF B (1e-40) | |
| LC16M035L | 29836-28727 (369) | - | L?,E | Serine protease inhibitor 3 | Other functions | K2L | 0.0 | CPN | K2L (0.0) |
| LC16M036R | 29843-30079 (78) | - | L? | Hypothetical protein | LC16m8, LC16mO specific gene | ||||
| LC16M037L | 30154-29888 (88) | - | L?,E | elF-2 alpha protein | Other functions | MVA024L | 2e-50 | MVA | K3L (1e-49) |
| LC16M038L | 31488-30214 (424) | - | L?,E | Phospholipase D-like protein | Enzyme | K4L | 0.0 | CPN | K4L (0.0) |
| LC16M039L | 31649-31515 (44) | - | Hypothetical protein | Similar gene in other organisms | ACAM3000_MVA_026 | 9e-24 | ACAM3000 | ||
| LC16M040L | 32068-31664 (134) | - | Putative monoglyceride lipase | Enzyme | VACWR037 | 1e-72 | WR | K5L (2e-60) | |
| LC16M041L | 32291-32037 (84) | - | Lysophospholipase-like protein | Enzyme | K6L | 1e-45 | CPN | K6L (1e-45) | |
| LC16M042R | 32430-32879 (149) | - | L? | Hypothetical protein | Similar gene in other organisms | K7R | 2e-86 | CPN | K7R (2e-86) |
| LC16M043L | 32708-32514 (64) | - | Hypothetical protein | Similar gene in other organisms | K8 | 2e-21 | WR | ||
| LC16M044L | 33624-32944 (226) | - | Hypothetical protein | Similar gene in other organisms | F1L | e-122 | CPN | F1L (e-122) | |
| LC16M045L | 34079-33638 (147) | - | L? | dUTP pyrophosphatase | Enzyme | MVA030L | 3e-76 | MVA | F2L (4e-76) |
| LC16M046L | 35545-34103 (480) | - | L | Kalch-like protein | Other functions | F3L | 0.0 | CPN | F3L (0.0) |
| LC16M047R | 35827-36063 (78) | - | Ribonucleoside-diphosphate reductase | Enzyme | F ORF B | 3e-40 | CPN | F ORF B (3e-40) | |
| LC16M048R | 36075-36365 (96) | - | Hypothetical protein | Similar gene in other organisms | F ORF C | 3e-55 | CPN | F ORF C (3e-55) | |
| LC16M049L | 36515-35556 (318) | - | E | Ribonucleoside-diphosphate reductase | Enzyme | F4L | 0.0 | CPN | F4L (0.0) |
| LC16M050L | 37512-36547 (321) | - | L?,E | Major membrane protein | Other functions | F5L | e-168 | CPN | F5L (e-168) |
| LC16M051L | 37766-37542 (74) | - | L? | Hypothetical protein | Similar gene in other organisms | MVA035L | 5e-40 | MVA | F6L (7e-40) |
| LC16M052L | 38024-37782 (80) | - | E | Hypothetical protein | Similar gene in other organisms | MVA036L | 3e-46 | MVA | F7L (6e-43) |
| LC16M053L | 38387-38190 (65) | - | L? | Hypothetical protein | Similar gene in other organisms | ACAM3000_MVA_037 | 9e-25 | ACAM3000 | F8L (3e-24) |
| LC16M054L | 39085-38447 (212) | - | L | Putative membrane protein | Other functions | F9L | e-121 | CPN | F9L (e-121) |
| LC16M055R | 40370-40627 (85) | - | L? | Hypothetical protein | Similar gene in other organisms | F ORF D | 1e-44 | CPN | F ORF D (1e-44) |
| LC16M056L | 40391-39072 (439) | - | L | Putative ser/thr protein kinase | Enzyme | F10L | 0.0 | CPN | F10L (0.0) |
| LC16M057L | 41478-40414 (354) | - | L?,E | Hypothetical protein | Similar gene in other organisms | F11L | 0.0 | CPN | F11L (0.0) |
| LC16M058R | 42203-42418 (71) | - | L? | Hypothetical protein | Similar gene in other organisms | F ORF E | 2e-37 | CPN | F ORF E (2e-37) |
| LC16M059L | 43428-41521 (635) | - | L? | Putative EEV maturation protein | Other functions | F12L | 0.0 | CPN | F12L (0.0) |
| LC16M060L | 44588-43470 (372) | - | L | Major envelope protein | EEV membrane protein | F13L | 0.0 | CPN | F13L (0.0) |
| LC16M061L | 44827-44606 (73) | - | L?,E | Hypothetical protein | Similar gene in other organisms | MVA044L | 3e-28 | MVA | F14L (2e-27) |
| LC16M062L | 45026-44877 (49) | - | L | Hypothetical protein | Similar gene in other organisms | PredictedbyGeneMark | 7e-22 | CPN | PredictedbyGeneMark04 (7e-22) |
| LC16M063L | 45575-45099 (158) | - | L?,E | Hypothetical protein | Similar gene in other organisms | MVA045L | 1e-78 | MVA | F15L (6e-79) |
| LC16M064L | 46277-45582 (231) | - | L?,E | Hypothetical protein | Similar gene in other organisms | MVA046L | e-122 | MVA | F16L (e-121) |
| LC16M065R | 46339-46644 (101) | - | L | Putative DNA-binding virion core protein | IMV internal protein | ACAM3000_MVA_047 | 8e-44 | ACAM3000 | F17R (2e-43) |
| LC16M066L | 48586-46374 (70) | - | Hypothetical protein | Similar gene in other organisms | E ORF A | 2e-27 | CPN | E ORF A (2e-27) | |
| LC16M067L | 48080-46641 (479) | - | L? | Poly(A) polymerase large subunit | Enzyme | E1L | 0.0 | CPN | E1L (0.0) |
| LC16M068L | 50290-48077 (737) | - | Hypothetical protein | Similar gene in other organisms | E2L | 0.0 | CPN | E2L (0.0) | |
| LC16M069L | 50989-50417 (190) | - | Double-stranded RNA-specific adenosine | Enzyme | MVA050L | 2e-99 | MVA | E3L (3e-99) | |
| LC16M070L | 51824-51045 (259) | - | L,E | DNA-directed RNA polymerese | Enzyme | E4L | e-139 | CPN | E4L (e-139) |
| LC16M071R | 51873-52898 (341) | - | Hypothetical protein | Similar gene in other organisms | E5R | 0.0 | CPN | E5R (0.0) | |
| LC16M072L | 52750-52430 (106) | - | Hypothetical protein | Similar gene in other organisms | E ORF B | 4e-43 | CPN | E ORF B (4e-43) | |
| LC16M073R | 53035-54738 (567) | - | L? | Hypothetical protein | Similar gene in other organisms | E6R | 0.0 | CPN | E6R (0.0) |
| LC16M074R | 54805-55305 (166) | - | L | Hypothetical protein | Similar gene in other organisms | MVA054R | 6e-89 | MVA | E7R (7e-89) |
| LC16M075L | 55236-55026 (70) | - | Hypothetical protein | Similar gene in other organisms | E ORF C | 3e-38 | CPN | E ORF C (3e-38) | |
| LC16M076R | 55430-56251 (273) | - | L? | Hypothetical protein | Similar gene in other organisms | MVA055R | e-161 | MVA | E8R (e-160) |
| LC16M077L | 55830-55630 (66) | - | Hypothetical protein | Similar gene in other organisms | E ORF D | 5e-36 | CPN | E ORF D (5e-36) | |
| LC16M078R | 58856-59053 (65) | - | Hypothetical protein | Similar gene in other organisms | E ORF E | 2e-36 | CPN | E ORF E (2e-36) | |
| LC16M079L | 59278-56258 (1006) | - | L,E | DNA-directed DNA polymerase | Enzyme | E9L | 0.0 | CPN | E9L (0.0) |
| LC16M080R | 59310-59597 (95) | - | L | Putative redox protein | IMV membrane associated protein | MVA057R | 2e-54 | MVA | E10R (3e-53) |
| LC16M081L | 59981-59592 (129) | - | L | Hypothetical protein | Similar gene in other organisms | MVA058L | 3e-73 | MVA | E11L (4e-73) |
| LC16M082R | 60686-61033 (115) | - | Hypothetical protein | Similar gene in other organisms | E ORF F | 3e-59 | CPN | E ORF F (3e-59) | |
| LC16M083L | 61968-59968 (655) | - | E | Hypothetical protein | Similar gene in other organisms | O1L | 0.0 | CPN | O1L (0.0) |
| LC16M084L | 62342-62016 (108) | - | L? | Glutaredoxin | Other functions | ACAM3000_MVA_061 | 8e-61 | ACAM3000 | O2L (1e-60) |
| LC16M085L | 63426-62488 (312) | - | L,E | Putative DNA-binding virion care protein | Other functions | I1L | e-147 | CPN | I1L (e-147) |
| LC16M086L | 63654-63433 (73) | - | L | Hypothetical protein | Similar gene in other organisms | MVA063L | 3e-28 | MVA | I2L (4e-28) |
| LC16M087L | 64464-63655 (269) | - | I | DNA binding phosphoprotein | Other functions | MVA064L | e-139 | MVA | I3L (e-138) |
| LC16M088R | 65372-65605 (77) | - | Hypothetical protein | Similar gene in other organisms | I ORF A | 9e-34 | CPN | I ORF A (9e-34) | |
| LC15M089L | 66862-64547 (771) | - | L?,E | Ribonucleoside-diphosphate reductase large subunit | Enzyme | I4L | 0.0 | CPN | I4L (0.0) |
| LC16M090L | 67128-66889 (79) | - | L | Hypothetical protein | IMV membrane associated protein | I5L | 3e-40 | CPN | I5L (3e-40) |
| LC16M091L | 68295-67147 (382) | - | L? | Hypothetical protein | Similar gene in other organisms | I6L | 0.0 | CPN | I6L (0.0) |
| LC16M092L | 69559-68288 (423) | - | L | Hypothetical protein | IMV internal protein | I7L | 0.0 | CPN | I7L (0.0) |
| LC16M093R | 69565-71595 (676) | - | I,L? | RNA helicase/NPH-I/NTPase II | Enzyme | I8R | 0.0 | CPN | I8R (0.0) |
| LC16M094L | 73374-71599 (591) | - | L | Metalloprotease | Enzyme | G1L | 0.0 | CPN | G1L (0.0) |
| LC16M095R | 73700-74362 (220) | - | L? | Putative transcriptional elongation factor | Other functions | G2R | e-127 | CPN | G2R (e-127) |
| LC16M096L | 73706-73371 (111) | - | L | Hypothetical protein | Similar gene in other organisms | G3L | 2e-54 | CPN | G3L (2e-54) |
| LC16M097L | 74706-74332 (124) | - | L | Putative glutaredoxin | Other functions | MVA073L | 3e-68 | MVA | G4L (9e-69) |
| LC16M098R | 74709-76013 (434) | - | Hypothetical protein | Similar gene in other organisms | G5R | 0.0 | CPN | G5R (0.0) | |
| LC16M099R | 76021-76212 (63) | - | L?,E | RNA polymerase | Enzyme | MVA075R | 3e-26 | MVA | Predicted by Gene Mark05 (5e-26) |
| LC16M100R | 76214-76711 (165) | - | I,L? | Hypothetical protein | Similar gene in other organisms | VACWR084 | 2e-96 | WR | G6R (3e-95) |
| LC16M101R | 76806-77204 (132) | - | L?,E | Hypothetical protein | Similar gene in other organisms | G ORF A | 1e-60 | CPN | G ORF A (1e-60) |
| LC16M102L | 77791-76676 (371) | - | L | Putative virion core protein | IMV internal | G7L | 0.0 | CPN | G7L (0.0) |
| LC16M103R | 77822-78604 (250) | - | I,L? | Late transcription factor | Other functions | G8R | e-151 | CPN | G8R (e-151) |
| LC16M104L | 77970-77752 (72) | - | Hypothetical protein | Similar gene in other organisms | G ORF B | 3e-38 | CPN | G ORF B (3e-38) | |
| LC16M105R | 78624-79646 (340) | - | L? | Myristytprotein | Other functions | G9R | 0.0 | CPN | G9R (0.0) |
| LC16M106R | 79647-80399 (250) | - | L | Myristytated membrane protein | IMV membrane associated protein | L1R | e-142 | CPN | L1R (e-142) |
| LC16M107R | 80431-80688 (85) | - | E | Hypothetical protein | Similar gene in other organisms | MVA081R | 2e-29 | MVA | L2R (3e-29) |
| LC16M108L | 81730-80678 (350) | - | L | Hypothetical protein | Similar gene in other organisms | L3L | 0.0 | CPN | L3L (0.0) |
| LC16M109R | 81755-82510 (251) | - | L | Putative DNA-binding virion core protein | IMV internal protein | MVA083R | e-143 | MVA | L4R (e-142) |
| LC16M110R | 82520-82906 (128) | - | L | Putative membrane protein | Other functions | MVA084R | 1e-60 | MVA | L5R (2e-60) |
| LC16M111R | 82863-83324 (153) | - | L | Dimeric Virion protein | Other functions | MVA085R | 3e-82 | MVA | J1R (9e-83) |
| LC16M112R | 83340-83873 (177) | - | E | Thymidine kinase | Enzyme | J2R | 2e-95 | CPN | J2R (2e-95) |
| LC16M113R | 83939-84940 (333) | - | L?,E | Poly(A) polymerase subunit | Enzyme | MVA087R | e-172 | MVA | J3R (e-171) |
| LC16M114R | 84855-85412 (185) | - | L?,E | DNA-directed RNA polymerase | Enzyme | J4R | e-104 | CPN | J4R (e-104) |
| LC16M115L | 85895-85494 (133) | - | L? | Membrane protein | Other functions | J5L | 4e-69 | CPN | J5L (4e-69) |
| LC16M116R | 86002-89862 (1286) | - | L?,E | DNA-directed RNA polymerase subunit | Enzyme | J6R | 0.0 | CPN | J6R (0.0) |
| LC16M117L | 89180-88965 (71) | - | L? | Hypothetical protein | Similar gene in other organisms | H ORF A | 8e-36 | CPN | H ORF A (8e-36) |
| LC16M118L | 90374-89859 (171) | - | L | Tyrosine phosphatase | Enzyme | MVA091L | 1e-91 | MVA | H1L (6e-91) |
| LC16M119R | 90388-90957 (189) | - | Hypothetical protein | Similar gene in other organisms | H2R | e-109 | CPN | H2R (e-109) | |
| LC16M120L | 91934-90960 (324) | - | L | IMV membrane associated protein | IMV membrane associated protein | MVA093L | e-172 | MVA | H3L (e-171) |
| LC16M121L | 94322-91935 (795) | - | L | RNA polymerase-associated protein | Enzyme | H4L | 0.0 | CPN | H4L (0.0) |
| LC16M122R | 94508-95119 (203) | - | L? | Late transcription factor | Other functions | MVA095R | 1e-83 | MVA | H5R (4e-83) |
| LC16M123R | 95120-96064 (314) | - | L | DNA topoisomerase | Enzyme | H6R | 0.0 | CPN | H6R (0.0) |
| LC16M124R | 96101-96541 (146) | - | L | Hypothetical protein | Similar gene in other organisms | MVA097R | 6e-82 | MVA | H7R (7e-82) |
| LC16M125R | 96585-99119 (844) | - | L?,E | mRNA capping enzyme, large subunit | Enzyme | D1R | 0.0 | CPN | D1R (0.0) |
| LC16M126L | 99049-98795 (84) | - | Hypothetical protein | Similar gene in other organisms | D ORF A | 7e-43 | CPN | D ORF A (7e-43) | |
| LC16M127R | 99133-99375 (80) | - | L? | Hypothetical protein | Similar gene in other organisms | D ORF B | 1e-24 | CPN | D ORF B (1e-24) |
| LC16M128R | 99511-100224 (237) | - | L? | Structural protein | IMV Internal protein | VACWR108 | e-141 | WR | D3R (e-140) |
| LC16M129L | 89518-99078 (146) | - | L? | Putative Virion protein | IMV internal protein | MVA099L | 1e-81 | MVA | D2L (2e-81) |
| LC16M130R | 100224-100850 (218) | - | E | Uracil DNA glycosytase | Enzyme | MVA101R | e-124 | MVA | D4R (e-123) |
| LC16M131R | 100912-103269 (785) | - | L,E | Putative NTPase | Enzyme | D5R | 0.0 | CPN | D5R (0.0) |
| LC16M132L | 101117-100908 (69) | - | L? | Hypothetical protein | Similar gene in other organisms | D ORF C | 8e-26 | CPN | D ORF C (8e-26) |
| LC16M133L | 102713-102495 (72) | - | Hypothetical protein | Similar gene in other organisms | D ORF D | 7e-38 | CPN | D ORF D (7e-38) | |
| LC16M134L | 103247-103005 (80) | - | L? | Hypothetical protein | Similar gene in other organisms | D ORF E | 3e-45 | CPN | D ORF E (3e-45) |
| LC16M135R | 103310-105223 (637) | - | L | Early transcription factor | Other functions | D6R | 0.0 | CPN | D6R (0.0) |
| LC16M136L | 104388-104197 (63) | - | Hypothetical protein | Similar gene in other organisms | F-53 | 2e-21 | WR | ||
| LC16M137R | 105250-105735 (161) | - | L | DNA-directed RNA polymerase subunit | Enzyme | MVA104R | 2e-90 | MVA | D7R (6e-91) |
| LC16M138L | 106612-105698 (304) | - | Cell surface-binding protein | IMV membrane associated protein | VACWR113 | e-161 | WR | D8L (e-158) | |
| LC16M139R | 106654-107295 (213) | - | E | MutT-like protein | Other functions | D9R | e-121 | CPN | D9R (e-121) |
| LC16M140R | 107292-108038 (248) | - | L | MutT-like protein | Other functions | VACWR115 | e-144 | WR | D10R (e-142) |
| LC16M141R | 108556-108765 (69) | - | L? | Hypothetical protein | Similar gene in other organisms | D ORF F | 4e-36 | CPN | D ORF F (4e-36) |
| LC16M142R | 109234-109506 (90) | - | Hypothetical protein | Similar gene in other organisms | D ORF G | 8e-51 | CPN | D ORF G (8e-51) | |
| LC15M143R | 109503-109688 (61) | - | Hypothetical protein | LC16m8, LC16mO specific gene | |||||
| LC16M144L | 109934-108039 (631) | - | L | Nucleoside triphosphate phosphohydrolase I, DNA helicase | Enzyme | D11L | 0.0 | CPN | D11L (0.0) |
| LC16M145R | 110249-110437 (62) | - | L? | Hypothetical protein | LC16m8, LC16mO specific gene | ||||
| LC16M146R | 110794-111012 (72) | - | L? | Hypothetical protein | LC16m8, LC16mO specific gene | ||||
| LC16M147L | 110832-109969 (287) | - | L,E | mRNA capping enzyme, small subunit | Enzyme | VACWR117 | e-166 | WR | D12L (e-165) |
| LC16M148R | 111759-111993 (74) | - | L? | Hypothetical protein | Similar gene in other organisms | D ORF I | 2e-43 | CPN | D ORF I (2e-43) |
| LC16M149L | 112518-110863 (551) | - | L? | Rifampicin resistance protein | IMV membrane associated protein | D13L | 0.0 | CPN | D13L (0.0) |
| LC16M150L | 112994-112542 (150) | - | I,L | Late gene transactivator | Other functions | MVA111L | 1e-84 | MVA | A1L (5e-85) |
| LC16M151L | 113689-113015 (224) | - | I,L? | Late gene transactivator | Other functions | A2L | e-131 | CPN | A2L (e-131) |
| LC16M152L | 113916-113586 (76) | - | L | Hypothetical protein | Similar gene in other organisms | MVA113L | 6e-42 | MVA | |
| LC16M153R | 114510-114869 (119) | - | Hypothetical protein | Similar gene in other organisms | A ORF A | 2e-69 | CPN | A ORF A (2e-69) | |
| LC16M154L | 115865-113931 (644) | - | L? | Major care protein | IMV internal protein | A3L | 0.0 | CPN | A3L (0.0) |
| LC16M155L | 116348-116088 (86) | - | Hypothetical protein | Similar gene in other organisms | A ORF B | e-24 | CPN | A ORF B (e-24) | |
| LC16M156L | 116763-115918 (281) | - | L | Membrane associated core protein | IMV internal protein | A4L | e-116 | CPN | A4L (e-116) |
| LC16M157R | 116801-117295 (164) | - | L | DNA-directed RNA polymerase subunit | Enzyme | MVA116R | 5e-72 | MVA | A5R (6e-72) |
| LC16M158L | 118410-117292 (372) | - | I,L?,E | Hypothetical protein | Similar gene in other organisms | A6L | 0.0 | CPN | A6L (0.0) |
| LC16M159R | 119518-119904 (128) | - | L? | Hypothetical protein | Similar gene in other organisms | A ORF C | 1e-68 | CPN | A ORF C (1e-68) |
| LC16M160R | 119986-120291 (101) | - | L? | Hypothetical protein | Similar gene in other organisms | A ORF D | 3e-35 | CPN | A ORF D (3e-35) |
| LC16M161L | 120566-118434 (710) | - | L? | Early transcription factor | Other functions | A7L | 0.0 | CPN | A7L (0.0) |
| LC16M162R | 120620-121486 (288) | - | E | Putative intermediate transcription factor | Other functions | MVA119R | e-165 | MVA | A8R (e-164) |
| LC16M163L | 121805-121479 (108) | - | L | Hypothetical protein | IMV membrane associated protein | VACWR128 | 6e-42 | WR | A9L (3e-40) |
| LC16M164R | 122149-122649 (166) | - | Hypothetical protein | Similar gene in other organisms | A ORF E | 2e-82 | CPN | A ORF E (2e-82) | |
| LC16M165R | 123031-123258 (75) | - | Hypothetical protein | Similar gene in other organisms | A ORF F | 8e-39 | CPN | A ORF F (8e-39) | |
| LC16M166R | 123525-123752 (75) | - | Hypothetical protein | Similar gene in other organisms | A ORF G | 5e-43 | CPN | A ORF G (5e-43) | |
| LC16M167L | 124481-121806 (891) | - | L | Major core protein | IMV internal protein | A10L | 0.0 | CPN | A10L (0.0) |
| LC16M168R | 124496-125452 (318) | - | L | Hypothetical protein | Similar gene in other organisms | VACWR130 | e-160 | WR | A11R (e-159) |
| LC16M169L | 126032-125454 (192) | - | L | Virion protein | IMV Internal protein | A12L | 2e-79 | CPN | A12L (2e-79) |
| LC16M170L | 126268-126056 (70) | - | L | Putative IMV membrane protein | IMV membrane associated protein | A13L | 2e-20 | CPN | A13L (2e-20) |
| LC16M171L | 126648-126376 (90) | - | L | Putative IMV membrane protein | IMV membrane associated protein | MVA125L | 5e-44 | MVA | A14L (6e-44) |
| LC16M172L | 127100-126816 (94) | - | L,E | Hypothetical protein | Similar gene in other organisms | MVA126L | 2e-52 | MVA | A15L (3e-52) |
| LC16M173L | 128217-127084 (377) | - | L? | Myristylprotein | Other functions | A16L | 0.0 | CPN | A16L (0.0) |
| LC16M174L | 128831-128220 (203) | - | L | Putative phosphorylated IMV membrane protein | IMV membrane associated protein | A17L | 6e-86 | CPN | A17L (6e-86) |
| LC16M175R | 128846-130327 (493) | 128845-130326 | L? | DNA helicase | Enzyme | A18R | 0.0 | CPN | A18R (0.0) |
| LC16M176L | 130541-130308 (77) | 130540-130307 | L | Hypothetical protein | Similar gene in other organisms | MVA130L | 3e-42 | MVA | A19L (4e-42) |
| LC16M177R | 130894-132174 (426) | 130893-132173 | E | Putative DNA polymerase processivity factor | Other functions | A20R | 0.0 | CPN | A20R (0.0) |
| LC16M178L | 130895-130542 (117) | 130894-130541 | L? | Hypothetical protein | Similar gene in other organisms | MVA131L | 6e-57 | MVA | A21L (7e-57) |
| LC16M179L | 131714-131328 (128) | 131713-131327 | L? | Hypothetical protein | Similar gene in other organisms | A ORF H | 6e-52 | CPN | A ORF H (6e-52) |
| LC16M180L | 132017-131796 (73) | 132016-131795 | Hypothetical protein | Similar gene in other organisms | A ORF I | 2e-39 | CPN | A ORF I (2e-39) | |
| LC16M181R | 132104-132667 (187) | 132103-132668 | L?,E | Hypothetical protein | Similar gene in other organisms | VACWR142 | e-100 | WR | A22R (1e-99) |
| LC16M182R | 132687-133835 (382) | 132686-133834 | L? | Putative intermediate transcription factor | Other functions | A23R | 0.0 | CPN | A23R (0.0) |
| LC16M183R | 133832-137326 (1164) | 133831-137325 | L? | DNA-directed RNA polymerase subunit | Enzyme | A24R | 0.0 | CPN | A24R (0.0) |
| LC16M184L | 136716-138495 (73) | 136715-136494 | Hypothetical protein | Similar gene in other organisms | A ORF J | 2e-28 | CPN | A ORF J (2e-28) | |
| LC16M185L | 137963-137331 (210) | 137962-137330 | E | DNA-directed RNA polymerase subunit | Enzyme | A26L | 1e-64 | Cowpox | A26L (4e-45) |
| LC16M186R | 138773-138958 (61) | 138772-138957 | Hypothetical protein | LC16m8, LC16mO specific gene | |||||
| LC16M187L | 138918-138235 (227) | 138517-138234 | E | Hypothetical protein | Similar gene in other organisms | VACWR147 | e-128 | WR | |
| LC16M188R | 139964-140146 (60) | 139963-140145 | Hypothetical protein | Similar gene in other organisms | TA30R | 3e-18 | Tian Tan | ||
| LC16M189L | 141055-138878 (725) | 141054-138877 | L | A-type inclusion protein | Other functions | VACWR148 | 0.0 | WR | |
| LC16M190R | 141327-141827 (166) | 141326-141826 | Hypothetical protein | LC16m8, LC16mO specific gene | |||||
| LC16M191L | 142607-141099 (502) | 142606-141098 | L | Structural protein | Other functions | VACWR149 | 0.0 | WR | A26L (e-115) |
| LC16M192L | 142989-142657 (110) | 142988-142656 | L | Cell fusion protein | IMV membrane associated protein | MVA138L | 2e-52 | MVA | A27L (5e-52) |
| LC16M193L | 143430-142990 (146) | 143429-142989 | L | Hypothetical protein | Similar gene in other organisms | VACWR151 | 2e-84 | WR | A28L (7e-84) |
| LC16M194R | 144164-144376 (70) | 144163-144375 | L? | Hypothetical protein | Similar gene in other organisms | A ORF K | 1e-38 | CPN | A ORF K (1e-38) |
| LC16M195L | 144348-143431 (305) | 144347-143430 | L? | DNA-directed RNA polymerase subunit | Enzyme | A29L | e-178 | CPN | A29L (e-178) |
| LC16M196L | 144544-144311 (77) | 144543-144310 | L | Hypothetical protein | Similar gene in other organisms | A30L | 2e-28 | CPN | A30L (2e-28) |
| LC16M197R | 144704-145087 (127) | 144703-145086 | L? | Hypothetical protein | Similar gene in other organisms | MVA142R | 1e-61 | MVA | A31R (2e-61) |
| LC16M198R | 145175-145441 (88) | 145174-145440 | L? | Hypothetical protein | Similar gene in other organisms | A ORF L | 1e-46 | CPN | A ORF L (1e-46) |
| LC16M199L | 145866-145054 (270) | 145865-145053 | L?,E | ATP/GTP-binding protein | Other functions | A32L | e-151 | CPN | A32L (e-151) |
| LC16M200R | 145984-146541 (185) | 145983-146540 | L? | EEV glycoprotein | EEV membrane protein | A33R | 5e-96 | CPN | A33R (5e-96) |
| LC16M201R | 146565-147071 (158) | 146564-147070 | L,E | EEV glycoprotein | EEV membrane protein | VACWR157 | 2e-85 | WR | A34R (8e-85) |
| LC16M202R | 147115-147645 (176) | 147114-147644 | E | Hypothetical protein | Similar gene in other organisms | MVA146R | 1e-93 | MVA | A35R (2e-93) |
| LC16M203L | 147275-147045 (76) | 147274-147044 | L? | Hypothetical protein | Similar gene in other organisms | A ORF M | 7e-40 | CPN | A ORF M (7e-40) |
| LC16M204R | 147712-148377 (221) | 147711-148376 | L?,E | EEV membrane protein | EEV membrane protein | A36R | e-106 | CPN | A38R (e-106) |
| LC16M205R | 148441-149232 (263) | 148440-149231 | L? | Hypothetical protein | Similar gene in other organisms | VACWR150 | e-143 | WR | A37R (e-141) |
| LC16M206L | 149213-148962 (83) | 149212-148961 | Hypothetical protein | Similar gene in other organisms | A ORF O | 1e-41 | CPN | A ORF O (1e-41) | |
| LC16M207R | 149322-149510 (62) | 149321-149509 | L?,E | Hypothetical protein | LC16m8, LC16mO specific gene | ||||
| LC16M208L | 150340-149507 (277) | 150339-149506 | L? | CD47 antigen/integrin-associated protein | Other functions | A38L | e-149 | CPN | A38L (e-149) |
| LC16M209R | 150357-151568 (403) | 150356-151567 | L? | Semaphorin | Other functions | A39R | 0.0 | CPN | A39R (0.0) |
| LC16M210L | 151402-151133 (89) | 151401-151132 | Hypothetical protein | Similar gene in other organisms | A ORF P | 3e-51 | CPN | A ORF P (3e-51) | |
| LC16M211R | 151594-152073 (159) | 151593-152072 | L?,E | Natural killer cell receptor homologue | Other functions | VACWR165 | 4e-86 | WR | A40R (5e-70) |
| LC16M212L | 152830-152171 (219) | 152829-152170 | L? | Hypothetical protein | Similar gene in other organisms | MVA153L | e-131 | MVA | A41L (e-129) |
| LC16M213R | 152994-153395 (133) | 152993-153394 | L? | Profilin-like protein | Other functions | A42R | 1e-75 | CPN | A42R (1e-75) |
| LC16M214R | 153433-154017 (194) | 153432-154016 | L,E | Membrane glycoprotein | Other functions | A43R | e-112 | CPN | A43R (e-112) |
| LC16M215R | 154025-154261 (78) | 154024-154260 | E | Hypothetical protein | Similar gene in other organisms | MVA156R | 6e-23 | MVA | PredictedbyGeneMark06 (1e-15) |
| LC16M216L | 155397-154357 (346) | 155396-154356 | L? | Hydroxysteroid dehydrogenase | Enzyme | A44L | 0.0 | CPN | A44L (0.0) |
| LC16M217R | 155444-155821 (125) | 155443-155820 | L? | Superoxide dismutase (Cu-Zn)-related protein | Enzyme | VACWR171 | 1e-70 | WR | A45R (5e-69) |
| LC16M218R | 155811-156533 (240) | 155810-156532 | L?,E | Hypothetical protein | Similar gene in other organisms | MVA159R | e-127 | MVA | A46R (e-105) |
| LC16M219L | 155454-156137 (105) | 156453-156136 | Hypothetical protein | Similar gene in other organisms | A ORF Q | 6e-39 | CPN | A ORF Q (6e-39) | |
| LC16M220L | 157339-156581 (252) | 157337-156579 | L? | Hypothetical protein | Similar gene in other organisms | VACWR173 | e-129 | WR | A47L (e-125) |
| LC16M221R | 157439-158053 (204) | 157437-158051 | L? | Thymidytate kinase | Enzyme | A48R | e-115 | CPN | A48R (e-115) |
| LC16M222R | 158101-158589 (162) | 158099-158587 | L,E | Hypothetical protein | Similar gene in other organisms | A49R | 2e-90 | CPN | A49R (2e-90) |
| LC16M223R | 158622-160280 (552) | 158620-160278 | L | ATP-dependent DNA ligase | Enzyme | A50R | 0.0 | CPN | A50R (0.0) |
| LC16M224L | 159491-159291 (66) | 159489-159289 | Hypothetical protein | Similar gene in other organisms | A ORF R | 7e-38 | CPN | A ORF R (7e-38) | |
| LC16M225L | 159610-159407 (67) | 159608-159405 | Hypothetical protein | Similar gene in other organisms | A ORF S | 3e-36 | CPN | A ORF S (3e-36) | |
| LC16M226R | 160333-160554 (73) | 160331-160552 | L? | Hypothetical protein | Similar gene in other organisms | A51R | 3e-40 | CPN | A51R (3e-40) |
| LC16M227R | 160533-161333 (266) | 160531-161331 | Hypothetical protein | Similar gene in other organisms | A51R | e-150 | CPN | A51R (e-150) | |
| LC16M228R | 161403-161975 (190) | 161401-161973 | Hypothetical protein | Similar gene in other organisms | VACWR178 | 4e-92 | WR | A52R (3e-91) | |
| LC16M229R | 162275-162835 (186) | 162273-162833 | Tumor necrosis factor receptor | Other functions | A53R | 1e-50 | VV | A53R (1e-50) | |
| LC16M230R | 162291-162587 (98) | 162289-162585 | Tumor necrosis factor receptor | Other functions | A ORF T | 5e-40 | CPN | A ORF T (5e-40) | |
| LC16M231L | 162383-162111 (90) | 162381-162109 | Hypothetical protein | Similar gene in other organisms | A54L | 8e-49 | CPN | A54L (8e-49) | |
| LC16M232R | 163083-164777 (584) | 163081-164775 | L?,E | Kelch-like protein | Other functions | A55R | 0.0 | CPN | A55R (0.0) |
| LC16M233R | 164827-165759 (310) | 164825-165757 | L? | Hemagglutinin | EEV membrane protein | A56R | e-142 | CPN | A56R (e-142) |
| LC16M234R | 165777-165890 (37) | 165775-165888 | L | Guanylate kinase fragment | Other functions | PredictedbyGeneMark | 2e-18 | CPN | PredictedbyGeneMark07 (2e-18) |
| LC16M235R | 165904-166359 (151) | 165902-166357 | Guanylate kinase | Enzyme | A57R | 1e-82 | CPN | A57R (1e-82) | |
| LC16M236R | 166510-167412 (300) | 166508-167410 | L?,E | Putative ser/thr protein kinase | Enzyme | MVA167R | e-178 | MVA | B1R (e-177) |
| LC16M237L | 167333-167010 (107) | 167331-167008 | Hypothetical protein | Similar gene in other organisms | B ORF A | 2e-60 | CPN | B ORF A (2e-60) | |
| LC16M238R | 167502-168161 (219) | 167500-168159 | L? | Hypothetical protein | Similar gene in other organisms | B2R | e-130 | CPN | B2R (e-130) |
| LC16M239L | 168029-167829 (66) | 168027-167827 | Hypothetical protein | Similar gene in other organisms | B ORF B | 1e-35 | CPN | B ORF B (1e-35) | |
| LC16M240R | 168197-168571 (124) | 168195-168569 | Hypothetical protein | Similar gene in other organisms | B3R | 2e-62 | CPN | B3R (2e-62) | |
| LC16M241L | 168292-168005 (95) | 168290-168003 | Hypothetical protein | Similar gene in other organisms | B ORF C | 1e-52 | CPN | B ORF C (1e-52) | |
| LC16M242R | 169227-170903 (558) | 169225-170901 | L?,E | Ankyrin repeat protein | Other functions | B4R | 0.0 | CPN | B4R (0.0) |
| LC16M243R | 171004-171957d | L? | Plaque-size/Host range protein precursor | EEV membrane protein | MVA173R | 0.0 | MVA | B5R (e-179) | |
| 171293-171958 (221)d | Plaque-size/Host range protein precursor | EEV membrane protein | MVA173R | e-123 | MVA | B5R (e-122) | |||
| LC16M244R | 172040-172561 (173) | 172039-172560 | I,L?,E | Hypothetical protein | Similar gene in other organisms | MVA174R | 2e-99 | MVA | B5R (3e-99) |
| LC16M245L | 172317-172102 (71) | 172316-172101 | E | Hypothetical protein | Similar gene in other organisms | B ORF D | 4e-37 | CPN | B ORF D (4e-37) |
| LC16M246R | 172599-173147 (182) | 172598-173146 | L | Hypothetical protein | Similar gene in other organisms | B7R | e-107 | CPN | B7R (e-107) |
| LC16M247R | 173202-174020 (272) | 173201-174019 | L? | Interferon-gamma receptor | Other functions | VACWR190 | e-163 | WR | B8R (e-161) |
| LC16M248R | 174107-174340 (77) | 174106-174339 | L? | Putative ER-localized apoptosis regulator | Other functions | VACWR191 | 1e-42 | WR | B9R (3e-42) |
| LC16M249R | 174303-174803 (166) | 174302-174802 | Kelch-like protein | Other functions | B10R | 5e-82 | CPN | B10R (5e-82) | |
| LC16M250R | 174875-175093 (72) | 174874-175092 | L? | Hypothetical protein | Similar gene in other organisms | VACWR193 | 5e-25 | WR | B11R (3e-23) |
| LC16M251R | 175160-176011 (283) | 175159-176010 | Protein kinase | Enzyme | B12R | e-160 | CPN | B12R (e-160) | |
| LC16M252R | 176116-176466 (116) | 176115-176465 | Serine protease inhibitor | Other functions | ACAM3000_MVA_161 | 2e-63 | ACAM3000 | B13R (1e-61) | |
| LC16M253R | 175441-177109 (222) | 176440-177108 | Serine protease inhibitor | Other functions | B14R | e-127 | CPN | B14R (e-127) | |
| LC16M254R | 177186-177635 (149) | 177185-177634 | Hypothetical protein | Similar gene in other organisms | B15R | 4e-89 | CPN | B15R (4e-89) | |
| LC16M255R | 177748-178728 (326) | 177747-178727 | L? | Interleukin-1 binding protein precursor | Other functions | VACWR197 | 0.0 | WR | B16R (e-166) |
| LC16M256L | 178289-178062 (75) | 178288-178061 | Hypothetical protein | Similar gene in other organisms | B ORF F | 4e-29 | CPN | B ORF F (4e-29) | |
| LC16M257L | 179796-178774 (340) | 179795-178773 | L? | Hypothetical protein | Similar gene in other organisms | B17L | 0.0 | CPN | B17L (0.0) |
| LC16M258R | 179936-181177 (413) | 179935-181176 | Ankyrin-like protein | Other functions | B18R | 0.0 | CPN | B18R (0.0) | |
| LC16M259R | 181307-181810 (187) | 181306-181809 | L? | CrmE protein | Other functions | crmE | 2e-74 | USSR strain | |
| LC16M260R | 181859-182080 (73) | 181858-182079 | L? | Hypothetical protein | Similar gene in other organisms | CMP6L | 1e-80 | Camalpox | |
| LC16M261R | 181978-182691 (237) | 181977-182690 | L? | Hypothetical protein | LC16m8, LC16mO specific gene | ||||
| LC16M262L | 182555-182328 (75) | 182554-182327 | Hypothetical protein | LC16m8, LC16mO specific gene | |||||
| LC16MRTR01R | 182972-183415 (147) | 182971-183414 | Hypothetical protein | Similar gene in other organisms | B22R | 4e-85 | CPN | B22R (4e-85) | |
| LC16MRTR02R | 183462-184712 (418) | 183461-184711 | L? | Host range protein | Other functions | B23R | 0.0 | CPN | B23R (0.0) |
| LC16MRTR03R | 185046-185327 (93) | 185045-185326 | Hypothetical protein | Similar gene in other organisms | D4L | 3e-41 | Cowpox | PredictedbyGeneMark09 (3e-18) | |
| LC16MRTR04R | 185654-185983 (109) | 185653-185982 | L?,E | Hypothetical protein | Similar gene in other organisms | VACWR211 | 1e-62 | WR | B25R (5e-57) |
| LC16MRTR05L | 185800-185588 (70) | 185799-185587 | L? | Hypothetical protein | Similar gene in other organisms | B ORF G | 1e-29 | CPN | B ORF G (1e-29) |
| LC16MRTR06R | 186233-185619 (128) | 186232-186618 | Hypothetical protein | Similar gene in other organisms | VACWR212 | 4e-59 | WR | B26R (1e-55) | |
| LC16MRTR07R | 186983-187129 (48) | 186982-187128 | K1R protein fragment | Other functions | VACWR214 | 4e-24 | WR | PredictedbyGeneMark02 (5e-24) | |
| LC16MRTR08R | 187247-187615 (122) | 187246-187614 | L? | Tumor necrosis factor receptor II homologue | Other functions | VACWR215 | 4e-73 | WR | B26R (3e-72) |
| LC16MRTR09R | 167834-187938 (34) | 187833-187937 | Tumor necrosis factor receptor II fragment | Other functions | PredictedbyGeneMark | 3e-17 | CPN | PredictedbyGeneMark11 (3e-17) | |
| LC15MRTR10R | 188327-189103 (258) | 188326-189102 | Major secreted protein | Other functions | VACWR218 | e-113 | WR | B29R (e-112) | |
| LC16MRTR11L | 188880-188767 (37) | 188879-188766 | Hypothetical protein | Similar gene in other organisms | B ORF H | e-10 | CPN | B ORF H (e-10) | |
| LC16MRTR12L | 188887-188684 (67) | 188886-188683 | Hypothetical protein | Similar gene in other organisms | B ORF I | 2e-36 | CPN | B ORF I (2e-36) | |
Regulatory sequences upstream of the ORFs were classified into early (E), intermediate (I), late (L) and putative late (L?) promoters.
Best-matching ORF from BLASTP analysis of nonredundant protein database.
Broken lines indicate that LC16mO ORFs were in the same positions and had the same amino acid lengths as those of LC16m8.
LC16M243R ORF was full-size (317 aa) in LC16mO but was truncated (221 aa) in LC16m8.
FIG. 2.
Differences in nucleotide sequences between the LC16m8 and LC16mO strains. (A) The locations (1 to 6) of nucleotide point mutations in the genomes are shown schematically. (B) The nucleotide changes are shown in boldface lowercase letters. The resultant amino acid changes in ORFs are indicated by shaded boldface italics in loci (2, 5, and 6). Putative gene functions and the ORFs corresponding to the CPN strain are also shown.
Almost all of the m8 ORFs best matched those of OPV, mainly the vaccinia virus CPN strain. Therefore, m8 and CPN were strikingly similar in their genomic organizations and ORF orientations (Fig. 1 and Table 1) (21). The m8 virus retained 192 out of 198 major CPN ORFs (60 out of 65 minor CPN ORFs), including other EEV env-related genes, A33R, A34R, A36R, A56R, F12L, and F13L. Only a few differences were observed. CPN C21L/B27R and C19L/B24R were absent in the ITR regions of m8, although they appear to be nonessential and presumably do not represent functional genes (21). The m8 genome lacked nonessential ORFs C13L, B19R, and B20R of unknown function in the regions neighboring the ITR termini and A25L in the central coding region, which encodes a short fragment (65 aa) (21) homologous to an A-type inclusion protein of CPV (1,284 aa) (18). ORF LC16M191L (502 aa), however, corresponded to CPN A26L, also encoding a truncated homologue (322 aa) of the CPV inclusion protein (18, 21).
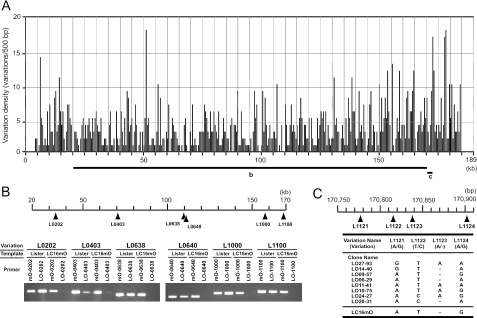
As LO had no history of virus cloning, nucleotide polymorphisms were observed at 1,264 sites in the genome putatively assembled by 4,913 sequencing reactions. This diversity was mapped from L0001 to L1264 along the whole genome (Fig. 3A; see Table S1 in the supplemental material). Sequences of the only marginal region spanning the diversity numbers from L1121 to L1124 (150 bp) revealed at least eight genotypes in LO, whereas mO possessed the “AT-G” genotype, which was the same as the LO09-57 clone in the region (Fig. 3C). Furthermore, PCR analysis of other randomly selected loci demonstrated that mO-specific primers amplified template LO DNA, but not vice versa (Fig. 3B). These results indicate that LO consists of a huge divergent virus population but likely contains the ancestors of mO. Because of the diversity of LO, however, it was impossible to exactly assign its consensus full-genome sequence and all ORFs. Thus, the LO shotgun sequences with major hits were tentatively assembled, compiled as an artificial genome sequence, and deposited in GenBank.
FIG. 3.
Polymorphism of the Lister strain genome. (A) Nucleotide sequence variations are presented in each 500-bp length along the central coding region of the Lister genome. (B) Six divergent loci, L0202, L0403, L0638, L0640, L1000, and L1100, were randomly selected. LO and mO genomic DNAs were amplified at the selected sites by PCR with the forward primers specific for LO or mO and the common reverse primers. (C) The marginal (150-bp) region spanning diversity numbers L1121 to -1124 of LO virus DNA were cloned, sequenced, and classified into eight genotypes. The genotype of LC16mO is also shown.
Analysis of the EEV env-related genes.
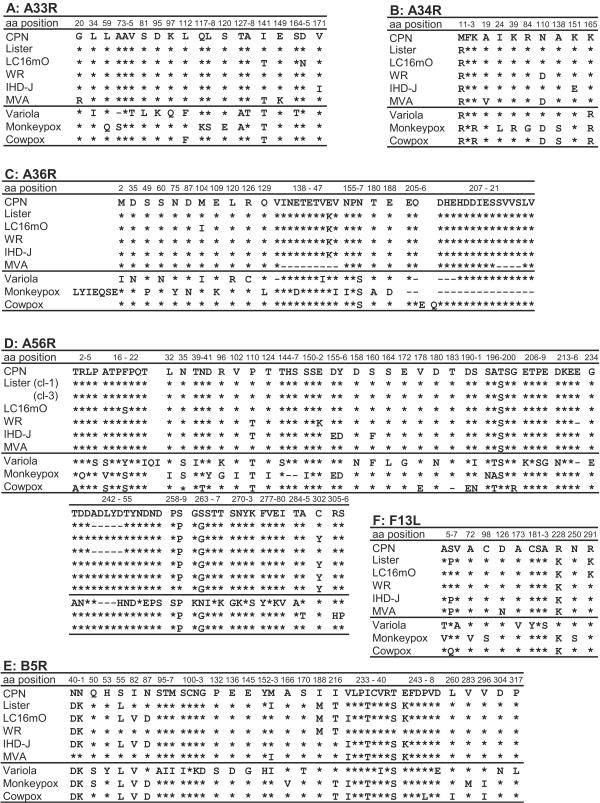
The evolutionary relationships of the EEV env-related genes in Lister-related viruses were further analyzed by sequencing of PCR amplicons from ListerVAX, another batch of mO and m8, and WR and IHD-J, which were stored in our laboratory. Because the mO and m8 sequences were identical except for B5R, the resultant amino acid alignments of A33R, A34R, A36R, A56R, F13L, and B5R of ListerVAX and mO were presented with reference to those of CPN and compared to other VV strains and OPVs deposited in GenBank (Fig. 4). ListerVAX had the same amino acid alignment in A33R as wild-type (wt) VV CPN or WR. On the other hand, mO A33R had two amino acid substitutions: Asn at amino acid position 165 (Asn165) was unique to mO, but Thr141 was found in mO and MVA, and also in VAR, MPV, and CPV of OPV (Fig. 4A). A34R was rather conserved in OPV, and no substitution was observed between ListerVAX and mO. Interestingly, however, Lys165 seems to be specific to VV (Arg165 for VAR, MPV, and CPV), and aa 110 (Asn or Asp) may classify OPV into two groups (Fig. 4B). Similarly, A36R was almost conserved in VV strains but divergent in other OPVs. ListerVAX, mO, WR, and IHD-J strains of VV, however, had a common Glu146-to-Lys146 substitution from CPN. An additional Met104-to-Ile104 change occurred in mO, although this was also the case in VAR (Fig. 4C).
FIG. 4.
Comparison of amino acid alignments of the EEV Env-related proteins in six vaccinia virus strains and other OPVs. The numbers at the top of each panel indicate the amino acid positions of the EEV proteins of vaccinia virus CPN strain. The asterisks and dashes show conserved and deleted amino acids, respectively, with reference to CPN. The vaccinia viruses compared are CPN, Lister (calf lymph Lister vaccine), LC16mO, WR, IHD-J, and MVA strains. Variola, monkeypox, and cowpox viruses shown for reference are Bangladesh-1975, Zaire-96-I-16, and GRI-90 strains, respectively.
As for A56R, ListerVAX was a mixture of wt-like VV (clone 3) and an mO-type mutant (clone 1) that possessed a 5-aa deletion from Ala245 to Asp249 and a conversion of Tyr302 to Cys302, which may be an ancestor clone of mO. Another difference between ListerVAX and mO was aa 19, which was Phe and Ser in ListerVAX and mO, respectively (Fig. 4D). Lys291 in F13L was unique to the Lister family viruses, whereas it was Arg291 in other VVs and OPVs, supporting the Lister lineage of mO. F13L Pro6 and Ser6 of ListerVAX and mO, respectively, seem to be within the divergence of OPV, because there was Pro6 in MVA and IHD-J and Ser6 in CPN, WR, VAR, and MPV (Fig. 4F). B5R is located close to the right-terminal end, and therefore, it was most divergent among the EEV env genes. ListerVAX differed from the compiled shotgun LO sequence in 3 nucleotides. However, the differences resulted in one amino acid substitution, from Ile82 to Val82, which also occurred in other OPVs. There were four amino acid changes in B5R between ListerVAX (Ile82, Asn87, Ile153, and Val233) and mO (Val82, Asp87, Met153, and Ile233) (Fig. 4E).
Altogether, these results confirm the notion that mO, and consequently m8, are the progeny of LO and not so divergent from LO, wt VV, or OPV, except for B5R.
Antibody responses by vaccination.
The truncated m8 and intact LO B5R proteins were compared for antigenic activity in initial experiments. BALB/c mice were subcutaneously immunized six times with the recombinant B5R proteins adsorbed to aluminum adjuvant or Ni-agarose beads. The mice were challenged by intranasal infection with 106 PFU of mouse-pathogenic WR virus 20 weeks after the first immunization and 12 days after the last booster injection. The LO B5R protein partially protected mice from death, with a survival rate of 78% after the appearance of severe clinical symptoms, such as ruffled fur, hunched posture, and weight loss, peaking at around 7 to 9 days after challenge. However, mice receiving the truncated m8 protein similarly developed symptoms, lost bodyweight, and died (100%) within 9 days (data not shown). These results confirm the immunogenicity of the intact B5R protein and also suggest a lack of antigenic activity of the truncated B5R protein.
Thus, B5R-defective m8 was compared with B5R-intact mO and LO for the ability to prime or induce anti-B5R and anti-EEV antibody responses before and after pathogenic-WR infection. BALB/c mice were vaccinated subcutaneously with a low (105 PFU) or high (107 PFU) dose of the vaccine strains. On day 21 after vaccination, one-third of the mice were bled to determine prechallenge antibody levels, and the other mice were challenged intranasally with 106 PFU of WR. Sera were collected 14 days later to test for postchallenge antibodies. Representative ELISA antibody levels in individual mice are shown in Fig. 5A, and the results of antibody responses examined are summarized in Table 2. ELISA antibody levels at prechallenge were low against VV antigens and undetectable against the B5R protein in all vaccinated mice. The titers and seroprevalences, if any were present, tended to be higher in 107 PFU vaccination groups than in those vaccinated with 105 PFU. Comet inhibition activity in sera, which is an indicator of anti-EEV antibodies, was negative in each of the vaccinated groups. NAb titers to VV, that is, IMV, were also low or undetectable; titers as low as 1:4 and 1:16 were detected only in groups of mice immunized with 107 PFU of mO and LO, respectively (Table 2).
FIG. 5.
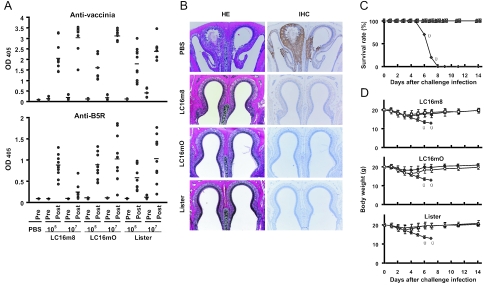
Protection against lethal WR challenge by vaccination with LC16m8. Groups of 6-week-old BALB/c mice were subcutaneously vaccinated and intranasally challenged as for Table 2. (A) Levels of antibodies in pre- and postchallenge sera of individual mice. Sera were examined by ELISA for vaccinia virus- and B5R-specific antibodies, and the results are shown with OD405 values at 1:400 and 1:100 dilutions, respectively. The horizontal bars indicate the averages. (B) Histopathology by HE staining and IHC by peroxidase staining of the nasal tissue collected from nonimmunized and vaccinated mice 9 and 14 days after challenge infection, respectively. (C) Survival and (D) bodyweights of mice after WR challenge. The mice had been vaccinated with 105 (open symbols) or 107 (solid symbols) PFU of LC16m8 (□ and ▪), LC16mO (○ and •), or Lister (▵ and ▴) strain or PBS (⧫). To avoid confusion, the average bodyweight ± standard deviation is shown in separate panels in comparison with the PBS group. The crosses indicate the deaths of mice.
TABLE 2.
Antibody responses in vaccinated mice at pre- and postchallenge infectiona
| Vaccination (day 0)
|
Prechallenge (day 21)
|
Postchallenge (day 35)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | Dose (PFU) | IgG ELISA (positive/total)
|
NAb | Comet inhibition | IgG ELISA (positive/total)
|
NAb | Comet inhibition | ||
| Anti-vaccinia virusb | Anti-B5Rb | Anti-vaccinia virusc | Anti-B5Rb | ||||||
| PBS | 0.10 (0/5) | 0.08 (0/5) | <4d | <10d | NDe | ND | ND | ND | |
| Lister | 105 | 0.20 (3/5) | 0.09 (0/5) | <4 | <10 | 1.78 (10/10) | 0.56 (10/10) | 4 | <10 |
| 107 | 1.00 (5/5) | 0.11 (0/5) | 16 | <10 | 2.42 (10/10) | 1.06 (10/10) | 64 | <10 | |
| LC16mO | 105 | 0.19 (2/5) | 0.09 (0/5) | <4 | <10 | 1.60 (10/10) | 0.83 (10/10) | 16 | <10 |
| 107 | 0.52 (4/5) | 0.10 (0/5) | 4 | <10 | 3.18 (10/10) | 1.03 (9/10) | 64 | <10 | |
| LC16m8 | 105 | 0.39 (2/5) | 0.08 (0/5) | <4 | <10 | 2.08 (10/10) | 0.85 (10/10) | 64 | <10 |
| 107 | 0.53 (4/5) | 0.08 (0/5) | <4 | <10 | 3.14 (10/10) | 0.21 (3/10) | 64 | <10 | |
Mice vaccinated with a single dose were challenged intranasally with 106 PFU of WR strain on day 21 and sacrificed on day 35.
Averages of OD405 values at a 1:100 dilution.
Averages of OD405 values at a 1:400 dilution.
The highest serum dilutions yielding a 50% plaque reduction or inhibitory comet formation.
ND, not determined.
Upon lethal challenge with virulent WR, however, high levels of anti-vaccinia virus ELISA antibodies were induced in all groups of mice vaccinated with m8, mO, and LO. Substantial levels of anti-B5R antibodies were also detected in all groups, except for that receiving 107 PFU of m8, where only 3 out of 10 mice developed anti-B5R antibodies (Fig. 5A and Table 2). Therefore, mice immunized with 107 PFU of m8 produced significantly (P < 0.0008) lower levels of anti-B5R antibodies after WR infection than did those immunized with 105 PFU of m8, 107 PFU of mO, or 107 PFU of LO (Fig. 5A), when compared by an unpaired Student's t test. The lethal challenge with WR did not elicit comet inhibition activity against EEV in vaccinated mice but induced and/or augmented NAb titers to IMV ranging from 1:4 to 1:64 (Table 2). Levels of antibodies after WR challenge were higher in mice immunized with 107 PFU than in those immunized with 105 PFU, indicating that mice were effectively primed with a higher dose of vaccine and boosted by WR infection. The exception was anti-B5R antibody titers in groups receiving B5R-defective m8 (Table 2 and Fig. 5A), probably because B5R-expressing EEV of WR was more quickly cleared before eliciting anti-B5R antibodies by stronger immunity induced with 107 PFU of m8 than with 105 PFU of m8.
Pathological findings.
The immunogenicities of the m8, mO, and LO vaccines were evaluated by histopathological and immunohistochemical analyses of the nasal tissue of mice, the primary infection site for pathogenic WR. The specimens from mice mock vaccinated with PBS demonstrated massive destruction and necrosis of the mucosal epithelium of the nasal cavity. The severe necrosis of olfactory epithelial cells was widespread in the nasal-cavity tissue (Fig. 5B, HE). VV antigens were distributed widely and intensively, colocalizing at the damaged areas of the epithelium (Fig. 5B, IHC). In contrast to nonimmune mice, severe epithelial destruction was rarely observed in the nasal cavities of mice vaccinated with a lower dose (105 PFU) of m8, mO, or LO. Their nasal specimens showed intact tissue morphology without evidence of recovery from tissue necrosis. In addition, no VV antigens were detected in nasal mucosal epithelial cells when examined by enhanced immunohistochmical staining (Fig. 5B, IHC). Similarly, no pathological changes were detectable after intranasal WR challenge in mice vaccinated with a higher dose (107 PFU) of m8, mO, or LO (data not shown).
Protection by m8, mO, and LO vaccines.
The immunological and histopathological studies described above suggest that m8 is as effective as mO and LO against pathogenic-OPV infection. Therefore, the protective efficacies of the m8, mO, and LO vaccine strains were further estimated in additional WR challenge experiments. Groups of 10 BALB/c mice vaccinated as for immunogenicity studies were examined for survival rate (Fig. 5C) and bodyweight loss (Fig. 5D) after intranasal inoculation with 106 PFU of WR. As this WR dose represented 10 LD50 for 6-week-old BALB/c mice (data not shown), the nonimmunized mice receiving PBS developed clinical symptoms, lost bodyweight, and died within 9 days after WR challenge. In contrast, none of the mice in the m8, mO, or LO vaccination group died (Fig. 5C). Vaccinated mice developed only a transient and slight loss of bodyweight, peaking at 3 or 4 days after challenge, but looked healthy without ruffled fur, inactivity, or respiratory distress and promptly gained weight thereafter (Fig. 5D). Notably, there were no significant differences in bodyweight between the low-dose (105 PFU) and high-dose (107 PFU) vaccination groups nor among the m8, mO, and LO vaccination groups (Fig. 5D).
DISCUSSION
In this study, we suggest that an attenuated vaccinia virus m8 strain that was licensed in 1975 in Japan as the second-generation smallpox vaccine is as efficacious as the first-generation LO vaccine that was used worldwide in the WHO smallpox eradication program.
The m8 vaccine was not used in a large population in areas of endemicity because smallpox was almost eradicated when it was developed. Today, no vaccines under development or in human trials can be tested for protective efficacy against smallpox by infection of humans with the causative virus, VAR. However, a pathogenic vaccinia virus WR strain provides an alternative small-animal model suited for evaluating protective immunization (2, 32, 50, 51). VV has rather low infectivity for mice, but WR is an exception, because it is adapted to mice by repeated passages in the mouse brain (27). Intranasal inoculation with as little as 105 PFU of WR elicited severe illness and 50% death in BALB/c mice, although they were less susceptible to VV infection than C57BL/6 and C3H/He mice (unpublished data). Thus, BALB/c mice vaccinated with the LO and LO-derived vaccine strains failed to develop definite erythema or pustules at the inoculated skin sites, which is classified as a “take” that is indicative of viral replication and therefore successful immunization in other vaccinia virus-sensitive hosts, such as humans, cows, and rabbits. Anti-B5R, -EEV, or -IMV antibodies were certainly undetectable or at low levels in vaccinated BALB/c mice. Nevertheless, the m8, mO and LO vaccines all protected mice comparably and completely against challenge with 106 PFU of WR. Notably, a single subcutaneous vaccination with m8 primed mice to render them as protective as vaccination with mO and LO, even at a low dose (105 PFU). Furthermore, with an increased WR challenge dose (107 PFU), 100% of mice vaccinated percutaneously with m8 (105 PFU) survived, while they lost significant weight temporarily and comparably to those vaccinated with the LO or NYBH strains (unpublished data) that had been used in humans.
OPVs are known to be highly cross-reactive among themselves in immune protection. Indeed, the m8 vaccine protected monkeys against MPV challenge (unpublished data), as recently described for the MVA vaccine (9). On the basis of these historical and experiential facts, CPV is thought to have been used in 1798 as the first human vaccine against VAR, and VV became the smallpox vaccine in the modern era. Similarly, OPVs are genetically highly conserved. Complete OPV genome sequences from VV, VAR, CPV, MPV, ectromelia virus, and camelpox virus have recently been investigated for phylogenetic analyses, with results indicating that CPV (strain GRI) is closely related to VV and that the genetic distances from VAR were lowest for camelpox virus (<0.0155), next lowest for VV (<0.0259), high for MPV (<0.0307), and highest for ectromelia virus (<0.0354) (22). These analyses may lead to the prediction that complete genome sequence data from VVs or OPVs will provide insight into the efficacy of smallpox vaccine strains.
Therefore, we determined the complete genome sequences of the licensed m8, parental mO, and grandparental LO strains. Our data may be interpreted to mean that the LO-related vaccines have similar abilities that would induce immune protection, supporting the above-mentioned prediction. Only four missense mutations occurred among the >280 deduced ORFs of m8 during evolution from the parental mO strain. The major change was a truncating mutation of the B5R gene. It is therefore noted that B5R was the only destroyed gene in m8 compared to mO. Furthermore, m8 and mO possessed almost all ORFs corresponding to the vaccinia virus CPN strain (21). As the grandparental LO strain has never been plaque cloned, its genome sequence exhibited huge polymorphisms, which were previously suggested by analyses of restriction enzyme fragments and pock or plaque size (46, 52, 53). However, our PCR sequencing of the EEV env-related genes indicated that they were all preserved in mO, and in LO as well, and that m8 was probably derived from a low-virulence clone of divergent LO. This genomic background of m8 suggests that it functions like LO as a smallpox vaccine, except for B5R.
B5R is the only NAb-inducing antigen of EEV so far identified (19). EEVs are extracellular free virions released from infected cells and seem to be prevented by NAbs (12, 19, 44). Destruction of B5R reduced the formation of EEV 5- to 10-fold (36, 44, 54), although they comprise less than 1% of the total virus population (41). In light of these findings, a concern has arisen that the m8 vaccine seems to contain reduced amounts of EEV that lacks the B5R antigen and might not be protective against long-range spread of VAR EEV (5, 44, 45). Our study of multiple immunizations with recombinant B5R proteins adsorbed to adjuvant showed that antigenic activity was absent in the truncated B5R protein of m8 but present in the intact protein of LO. In addition, infection or vaccination with live VV induced very few anti-EEV NAbs, and repeated inoculations were required to induce moderate NAb levels (19, 44), probably because of the small EEV population. Alternatively, low levels of the antibodies may be due to the low sensitivity of conventional assay systems. Wyatt et al. recently reported that NAbs can be produced after a single percutaneous vaccination (56). They recently developed and used a highly sensitive system, a semiautomated flow cytometric assay with recombinant VV expressing enhanced green fluorescent protein (8).
It was therefore important to examine the levels of protection against virulent WR infection in m8-vaccinated mice, irrespective of the absence of EEV B5R-specific antibody responses. Our results confirmed that a single vaccination with m8, mO, and LO failed to induce detectable levels of anti-EEV and anti-B5R antibodies. Nevertheless, mice immunized with these vaccines were 100% protected against pathogenic WR challenge as early as 3 weeks after vaccination. Moreover, m8 with the whole B5R gene deleted protected mice from lethal WR challenge (32). These findings suggest that many viral antigens other than B5R are also involved in protective immunity to EEV. In this regard, antibodies to the A33R Env antigen did not neutralize EEV but provided mice with 100% protection (19). Anti-A33R might disrupt fragile EEV Env and convert to IMV, which is easily neutralized by anti-vaccinia antibodies (19, 28). Alternatively, A33R-specific cellular immunity may be crucial for protective immunity.
We have only limited knowledge about the protective immune mechanisms against smallpox. Experience with worldwide vaccination, however, has suggested that the protective mechanisms involve innate immunity, including interferons, natural killer cells, and complements, and also acquired immunity, including specific antibody- and T-cell-mediated immune responses (12). Indeed, recent papers have revealed the involvement of gamma interferon-expressing CD8 and CD4 T cells, vaccinia-specific cytotoxic T cells, and T-helper type 1 memory in humans (6, 7, 31, 48) and mice (16, 35, 49). Several studies conducted out of urgency in the last few years using smallpox vaccine candidates came to similar conclusions with regard to the contribution of overall immunity to smallpox protection (2, 9, 50, 56). Moreover, priming effects in vaccinated persons were recently shown to be long-lived or long-lasting, for as long as 75 years after vaccination (23). These historical and most recent studies imply that vaccine priming for immunological memory is important so that effecter components, such as NAbs, CD4+ or CD8+ T cells, and various cytokines can promptly be induced or boosted to protective levels by VAR infection, regardless of whether they are above measurable levels before infection. In support of this hypothesis, we found that mice that received a single dose of LO-related vaccines could not fully develop antibody responses as early as 3 weeks after vaccination but could produce enhanced levels of antibodies and complete immune protection after pathogenic-virus infection.
The need to produce safer and more effective vaccines may increase in the future. Here, we determined the nucleotide sequences of the whole genomes from the m8, intermediate mO, and original LO vaccine strains. The accumulating information on complete genome sequences of attenuated or pathogenic VVs and other OPVs will provide a basis for producing new genetically engineered vaccines. The double-stranded DNA genomes of OPVs are known to be highly stable. However, a single nucleotide insertion just upstream of the m8 B5R mutation site has recently been reported to restore the ORF to the parental mO phenotype after repeated (10 or more) virus passages. Although the repaired viruses were a marginal population, attenuation that is achieved by a deletion of the whole B5R gene prevented the reversion of m8- to mO-type viruses (32), which have, however, much lower virulence than LO and NYBH (24, 25, 39). In turn, the genetic manipulation of m8 to replace genes related to protective immunity, but not to pathogenicity, with the counterpart genes of VAR may make m8 more efficacious. It will be necessary to study in detail the correlation between individual gene functions and antigenicity of the gene products for inducing protective immunity in the future.
Supplementary Material
Acknowledgments
We thank S. Hashizume for smallpox vaccine strains of vaccinia virus, LC16m8, LC16mO, and Lister Original (Elstree); Y. Sato for technical assistance; and N. Fujita, A. Kikuchi, M. Kudo, Y. Kuroda, S. Mimaki, M. Ohsawa, N. Okada, R. Sasaki, and S. Shinohara for assistance in sequencing and data processing.
This work was supported in part by grants from the Ministry of Health, Labor, and Welfare.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 2.Belyakov, I. M., P. Earl, A. Dzutsev, V. A. Kuznetsov, M. Lemon, L. S. Wyatt, J. T. Snyder, J. D. Ahlers, G. Franchini, B. Moss, and J. A. Berzofsky. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc. Natl. Acad. Sci. USA 100:9458-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birmingham, K., and G. Kenyon. 2001. Smallpox vaccine development quickened. Nat. Med. 7:1167. [DOI] [PubMed] [Google Scholar]
- 4.Chen, R. T., and J. M. Lane. 2003. Myocarditis: the unexpected return of smallpox vaccine adverse events. Lancet 362:1345-1346. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, J. 2002. Looking for vaccines that pack a wallop without the side effects. Science 298:2314. [DOI] [PubMed] [Google Scholar]
- 6.Combadiere, B., A. Boissonnas, G. Carcelain, E. Lefranc, A. Samri, F. Bricaire, P. Debre, and B. Autran. 2004. Distinct time effects of vaccination on long-term proliferative and IFN-producing T cell memory to smallpox in humans. J. Exp. Med. 199:1585-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drexler, I., C. Staib, W. Kastenmüller, S. Stevanovi, B. Schmidt, F. A. Lemonnier, H.-G. Rammensee, D. H. Busch, H. Bernhard, V. Erfle, and G. Sutter. 2003. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc. Natl. Acad. Sci. USA 100:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Earl, P. L., J. L. Americo, and B. Moss. 2003. Development and use of a vaccinia virus neutralization assay based on flow cytometric detection of green fluorescent protein. J. Virol. 77:10684-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earl, P. L., J. L. Americo, L. S. Wyatt, L. A. Eller, J. C. Whitbeck, G. H. Cohen, R. J. Eisenberg, C. J. Hartmann, D. L. Jackson, D. A. Kulesh, M. J. Martinez, D. M. Miller, E. M. Mucker, J. D. Shamblin, S. H. Zwiers, J. W. Huggins, P. B. Jahrling, and B. Moss. 2004. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428:182-185. [DOI] [PubMed] [Google Scholar]
- 10.Enserink, M. 2002. How devastating would a smallpox attack really be? Science 296:1592-1595. [DOI] [PubMed] [Google Scholar]
- 11.Enserink, M. 2004. Smallpox vaccines: looking beyond the next generation. Science 304:809. [DOI] [PubMed] [Google Scholar]
- 12.Esposito, J. J., and F. Fenner. 2001. Poxviruses, p. 2885-2921. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 13.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 15.Ferguson, N. M., M. J. Keeling, W. J. Edmunds, R. Gani, B. T. Grenfell, R. M. Anderson, and S. Leach. 2003. Planning for smallpox outbreaks. Nature 425:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg, C., S. Lustig, J. C. Whitbeck, R. J. Eisenberg, G. H. Cohen, and B. Moss. 2004. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J. Virol. 78:10230-10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulginiti, V. A., A. Papier, J. M. Lane, J. M. Neff, and D. A. Henderson. 2003. Smallpox vaccination: a review, part II. Adverse events. Clin. Infect. Dis. 37:251-271. [DOI] [PubMed] [Google Scholar]
- 18.Funahashi, S., T. Sato, and H. Shida. 1988. Cloning and characterization of the gene encoding the major protein of the A-type inclusion body of cowpox virus. J. Gen. Virol. 69:35-47. [DOI] [PubMed] [Google Scholar]
- 19.Galmiche, M. C., J. Goenaga, R. Wittek, and L. Rindisbacher. 1999. Neutralizing and protective antibodies directed against vaccinia virus envelope antigens. Virology 254:71-80. [DOI] [PubMed] [Google Scholar]
- 20.Gani, R., and S. Leach. 2001. Transmission potential of smallpox in contemporary populations. Nature 414:748-751. [DOI] [PubMed] [Google Scholar]
- 21.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266. [DOI] [PubMed] [Google Scholar]
- 22.Gubser, C., S. Hue, P. Kellam, and G. L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85:105-117. [DOI] [PubMed] [Google Scholar]
- 23.Hammarlund, E., M. W. Lewis, S. G. Hansen, L. I. Strelow, J. A. Nelson, G. J. Sexton, J. M. Hanifin, and M. K. Slifka. 2003. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9:1131-1137. [DOI] [PubMed] [Google Scholar]
- 24.Hashizume, S., H. Yoshizawa, M. Morita, and K. Suzuki. 1985. Properties of attenuated mutant of vaccinia virus, LC16m8, derived from Lister strain, p. 421-428. In G. V. Quinnan (ed.), Vaccinia viruses as vectors for vaccine antigens. Elsevier Science Publishing Co., Amsterdam, The Netherlands.
- 25.Hashizume, S., M. Morita, H. Yoshizawa, S. Suzuki, M. Arita, T. Komatsu, H. Amano, and I. Tagaya. 1973. Internal symposium on smallpox vaccines, Bilthoven. Symp. Ser. Immunobiol. Stand. 19:325-331. [Google Scholar]
- 26.Henderson, D. A. 1999. The looming threat of bioterrorism. Science 283:1279-1282. [DOI] [PubMed] [Google Scholar]
- 27.Henderson, D. A., and B. Moss. 1999. Smallpox and vaccinia, p. 74-97. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines. Saunders, Philadelphia, Pa.
- 28.Ichihashi, Y. 1996. Extracellular enveloped vaccinia virus escapes neutralization. Virology 217:478-485. [DOI] [PubMed] [Google Scholar]
- 29.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan, E. H., D. L. Craft, and L. M. Wein. 2002. Emergency response to a smallpox attack: the case for mass vaccination. Proc. Natl. Acad. Sci. USA 99:10935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy, J. S., S. E. Frey, L. Yan, A. L. Rothman, J. Cruz, F. K. Newman, L. Orphin, R. B. Belshe, and F. A. Ennis. 2004. Induction of human T cell mediated immune responses after primary and secondary smallpox vaccination. J. Infect. Dis. 190:1286-1294. [DOI] [PubMed] [Google Scholar]
- 32.Kidokoro, M., M. Tashiro, and H. Shida. 2005. Genetically stable and fully effective smallpox vaccine strain constructed from highly attenuated vaccinia LC16m8. Proc. Natl. Acad. Sci. USA 102:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitts, P. A., M. D. Ayres, and R. D. Possee. 1990. Linearization of baculovirus DNA enhances the recovery of recombinant virus expression vectors. Nucleic Acids Res. 18:5667-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane, J. M., F. L. Ruben, J. M. Neff, and J. D. Millar. 1969. Complications of smallpox vaccination, 1968. National surveillance in the United States. N. Engl. J. Med. 281:1201-1208. [DOI] [PubMed] [Google Scholar]
- 35.Legrand, F. A., P. H. Verardi, K. S. Chan, Y. Peng, L. A. Jones, and T. D. Yilma. 2005. Vaccinia viruses with a serpin gene deletion and expressing IFN-γ induce potent immune responses without detectable replication in vivo. Proc. Natl. Acad. Sci. USA 102:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Pomares, L., R. J. Stern, and R. W. Moyer. 1993. The ps/hr gene (B5R open reading frame homolog) of rabbitpox virus controls pock color, is a component of extracellular enveloped virus, and is secreted into the medium. J. Virol. 67:5450-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meltzer, M. I., I. Damon, J. W. LeDuc, and J. D. Millar. 2001. Modeling potential responses to smallpox as a bioterrorist weapon. Emerg. Infect. Dis. 7:959-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita, M., K. Suzuki, A. Yasuda, A. Kojima, M. Sugimoto, K. Watanabe, H. Kobayashi, K. Kajima, and S. Hashizume. 1987. Recombinant vaccinia virus LC16m0 or LC16m8 that expresses hepatitis B surface antigen while preserving the attenuation of the parental virus strain. Vaccine 5:65-70. [DOI] [PubMed] [Google Scholar]
- 39.Morita, M., Y. Aoyama, M. Arita, H. Amano, H. Yoshizawa, S. Hashizume, T. Komatsu, and I. Tagaya. 1977. Comparative studies of several vaccinia virus strains by intrathalamic inoculation into cynomolgus monkeys. Arch. Virol. 53:197-208. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, T., M. Mair, and T. V. Inglesby. 2002. Shining light on “Dark Winter.” Clin. Infect. Dis. 34:972-983. [DOI] [PubMed] [Google Scholar]
- 41.Payne, L. G. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 42.Rice, C. M., C. A. Franke, J. H. Strauss, and D. E. Hruby. 1985. Expression of Sindbis virus structural proteins via recombinant vaccinia virus: synthesis, processing, and incorporation into mature Sindbis virions. J. Virol. 56:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal, S. R., M. Merchlinsky, C. Kleppinger, and K. L. Goldenthal. 2001. Developing new smallpox vaccines. Emerg. Infect. Dis. 7:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 45.Smith, G. L., and G. McFadden. 2002. Smallpox: anything to declare? Nat. Rev. Immunol. 2:521-527. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi-Nishimaki, F., K. Suzuki, M. Morita, T. Maruyama, K. Miki, S. Hashizume, and M. Sugimoto. 1987. Genetic analysis of vaccinia virus Lister strain and its attenuated mutant LC16m8: production of intermediate variants by homologous recombination. J. Gen. Virol. 68:2705-2710. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi-Nishimaki, F., S. Funahashi, K. Miki, S. Hashizume, and M. Sugimoto. 1991. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology 181:158-164. [DOI] [PubMed] [Google Scholar]
- 48.Terajima, M., J. Cruz, G. Raines, E. D. Kilpatrick, J. S. Kennedy, A. L. Rothman, and F. A. Ennis. 2003. Quantitation of CD8+ T cell responses to newly identified HLA-A*0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J. Exp. Med. 197:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tscharke, D. C., G. Karupiah, J. Zhou, T. Palmore, K. R. Irvine, S. M. M. Haeryfar, S. Williams, J. Sidney, A. Sette, J. R. Bennink, and J. W. Yewdell. 2005. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 201:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weltzin, R., J. Liu, K. V. Pugachev, G. A. Myers, B. Coughlin, P. S. Blum, R. Nichols, C. Johnson, J. Cruz, J. S. Kennedy, F. A. Ennis, and T. P. Monath. 2003. Clonal vaccinia virus grown in cell culture as a new smallpox vaccine. Nat. Med. 9:1125-1130. [DOI] [PubMed] [Google Scholar]
- 51.Williamson, J. D., R. W. Reith, L. J. Jeffrey, J. R. Arrand, and M. Mackett. 1990. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J. Gen. Virol. 71:2761-2767. [DOI] [PubMed] [Google Scholar]
- 52.Wittek, R., A. Menna, D. Schümperli, S. Stoffel, H. K. Müller, and R. Wyler. 1977. HindIII and SstI restriction sites mapped on rabbit poxvirus and vaccinia virus DNA. J. Virol. 23:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittek, R., H. K. Müller, A. Menna, and R. Wyler. 1978. Length heterogeneity in the DNA of vaccinia virus is eliminated on cloning the virus. FEBS Lett. 90:41-46. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.World Health Organization. 1980. The global eradication of smallpox. Final report of the Global Commission for the Certification of Smallpox Eradication. World Health Organization, Geneva, Switzerland.
- 56.Wyatt, L. S., P. L. Earl, L. A. Eller, and B. Moss. 2004. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proc. Natl. Acad. Sci. USA 101:4590-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi, M., M. Kimura, and M. Hirayama. 1975. Report of the National Smallpox Vaccination Research Committee: study of side effects, complications and their treatments. Clin. Virol. 3:269-278. (In Japanese.) [Google Scholar]
- 58.Yasuda, A., J. Kimura-Kuroda, M. Ogimoto, M. Miyamoto, T. Sata, C. Takamura, T. Kurata, A. Kojima, and K. Yasui. 1990. Induction of protective immunity in animals vaccinated with recombinant vaccinia viruses that express PreM and E glycoprotein of Japanese encephalitis virus. J. Virol. 64:2788-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.