Abstract
Successful inhibition of human papillomavirus type 16 (HPV-16) late gene expression early in the life cycle is essential for persistence of infection, the highest risk factor for cervical cancer. Our study aimed to locate regulatory RNA elements in the early region of HPV-16 that influence late gene expression. For this purpose, subgenomic HPV-16 expression plasmids under control of the strong human cytomegalovirus immediate early promoter were used. An exonic splicing enhancer that firmly supported the use of the E4 3′ splice site at position 3358 in the early region of the HPV-16 genome was identified. The enhancer was mapped to a 65-nucleotide AC-rich sequence located approximately 100 nucleotides downstream of the position 3358 3′ splice site. Deletion of the enhancer caused loss of both splicing at the upstream position 3358 3′ splice site and polyadenylation at the early polyadenylation signal, pAE. Direct splicing occurred at the competing L1 3′ splice site at position 5639 in the late region. Optimization of the position 3358 3′ splice site restored splicing to that site and polyadenylation at pAE. Additionally, a sequence of 40 nucleotides with a negative effect on late mRNA production was located immediately downstream of the enhancer. As the E4 3′ splice site is employed by both early and late mRNAs, the enhancer constitutes a key regulator of temporal HPV-16 gene expression, which is required for early mRNA production as well as for the inhibition of premature late gene expression.
Human papillomavirus type 16 (HPV-16) is the major cause of cervical cancer (41, 60) and is one of the most common cancer-causing infections in the world. Cervical cancer accounts for 6% of all malignancies in women in developed countries but 26% in developing countries (37). While the majority of HPV-16 infections are cleared in less than a year, in rare cases, despite immunosurveillance, persistence is established. Persistence of HPV-16 infection is itself the highest risk factor for the development of cervical cancer (28) and requires successful inhibition of late gene expression early in the life cycle to avoid the immune response. As cervical cancer cells never express the late viral mRNAs or proteins, one can speculate that inhibition of late gene expression is a prerequisite for cancer progression.
Expression of HPV-16 genes is complex and is tightly regulated by differentiation-dependent transcription and RNA processing events (2, 40). Production of the late mRNAs encoding the antigenic capsid proteins L1 and L2 is restricted to terminally differentiated keratinocytes (41). The inhibition of the late genes may be partly attributed to RNA elements in the late region, suggested to regulate various RNA processing events such as mRNA stability, splicing, and translation (2, 40). Regulatory RNA elements have been identified in HPV-16 coding regions (42, 45) and also in the late untranslated region of HPV-1 (46), HPV-16 (31), HPV-31 (13), and bovine papillomavirus type 1 (BPV-1) (23), while differentiation-dependent splicing of the late genes has been studied in various HPV types (3, 19, 22, 30, 36). Premature induction of late gene expression by intervening in the regulation of posttranscriptional steps such as splicing could result in detection by the host and the clearance of infection. Identification of critical RNA sequences in the HPV-16 mRNAs implicated in these regulatory steps is therefore highly worthwhile.
Alternative splicing is a key mechanism in the control of gene expression of both eukaryotic and viral genes (6, 27). Splicing of pre-mRNA requires consensus cis-acting elements; a 5′ and 3′ splice site, a branchpoint sequence (BPS) and a polypyrimidine tract (PPT). Specific exonic or intronic sequences known as splicing enhancers and suppressors can promote or suppress the recognition of suboptimal 5′ and 3′ splice sites, thus regulating gene expression (5, 7, 54). Cellular protein factors such as serine-arginine-rich (SR) proteins and heterogenous ribonucleoproteins (hnRNPs), known to bind to these sequences, are thought to stabilize interactions between splice sites and the spliceosome, thereby regulating the selection of splice sites (6, 25).
In HPV-16, the late mRNAs are initiated and spliced at both 5′ and 3′ splice sites in the early region (1). All late mRNAs must be processed upstream of the early polyadenylation signal at an early 3′ splice site at position 3358. As both early and late messages require the use of this site, tight regulation must be necessary to avoid premature late gene expression during the life cycle. In fact, suppression of a suboptimal 3′ splice site in the late region by a splicing silencer in L1 (53) in proliferating cells allowed alternative selection of early-specific 3′ splice sites, thereby favoring early gene expression. In support of splicing regulation in papillomavirus, it has been previously demonstrated that late gene expression of BPV-1 is partly regulated by alternative 3′ splice site selection, controlled by at least five cis-acting RNA elements (55-58). We therefore proposed that alternative splice site selection of HPV-16 and consequently late gene expression must be under the control of regulatory cis-acting sequences in the early region and aimed to investigate this.
MATERIALS AND METHODS
Plasmid construction.
pBEL, pBELM, and pBS have been described previously (53), and pBS was referred to as pC16L2L1splice (53). To generate pBSM, PCR mutagenesis was performed using oligonucleotides L1MspliceFS and L1stopX or L2KPN1S and L1MspliceFA (described in reference 53), which was followed by insertion of this KpnI- and BamHI-digested PCR fragment into pBS.
pBN and pBNM, both containing a unique NsiI site, were created by cleavage of pBEL and pBELM, respectively, by NsiI, followed by religation. pBNB was generated by digestion of pBEL with NsiI and BssHII. Overhangs were filled in and religated using blunt-end ligation. pBNBM was generated by excision and insertion into pBNB of an ApaI-BamHI fragment from pBELM. pD1 and pD2 were created by digestion of pBELDU and pBELDP, respectively (52), by BssHII and MluI, followed by religation, made possible by the compatibility of the overhangs from these sites. BssHII and MluI were introduced at position 3396 and immediately upstream of pAE at position 4212, respectively, as described previously (52, 53). pDM1 and pDM2 were created by digestion of pBELMDU and pBELMDP, respectively (52), by BssHII and MluI, followed by religation, as described above.
The pT1 to pT10 constructs, with various deletions between positions 3396 and 4212 of the HPV-16 genome, required the use of the plasmid pBELDP (52). In this plasmid, the early polyadenlyation signal of pBEL has been specifically destroyed by mutations, converting the AAUAAA site to an MluI site necessary for further cloning. PCR was carried out on pBELDP using oligonucleotides 3705s (Table 1) and L2D (52), and the amplified fragment corresponding to positions 3652 to 4327 of HPV-16 was subcloned into PCR-II-TOPO (Invitrogen). A fragment cleaved by BssHII and StuI was subcloned into pBEL, from which a SalI-to-MluI fragment was cloned into pBELMDU (52), a construct containing an MluI site and a functional polyadenylation signal. The pT1 plasmid generated therefore contained a deletion from positions 3396 to 3651 of HPV-16 and a functional polyadenylation signal.
TABLE 1.
Oligonucleotides used to generate plasmids
| Oligonucleotide | Sequencea |
|---|---|
| 3705s | GCGCGCTCTAGAATGTTTAAGATATAGATTT |
| 3455s | GCGCGCTCTAGACCAACCACCCCGCCGCGACC |
| 3705a | ACGCGTTTTTAAAGTATTAGCATCAC |
| 3902a | ACGCGTATAGACATAAATCCAGTAG |
| 3903s | GCGCGCTCTAGAATGACAAATCTTGATACTS |
| 3640s | GCGCGCTCTAGACGGATTAACTGTAATAGTAAC |
| 3575s | GCGCGCTCTAGATGTTGCACAGAGACTCAGTG |
| 3515s | GCGCGCTCTAGACGACTATCCAGCGACCAAG |
| 3555s | GCGCGCTCTAGACCCCTGCCACACCACTAAGT |
| 3580a | TCTAGATTTAAAGTATTAGCATCACCTTTCTGTCCACTGAGTCTCTGTG |
| 3620a | TCTAGATTTAAAGTATTAGCATCACCTTTTCCTTTGTGTGAGCTGTTAA |
| E4SalBssHIIs | GTCGACGCGCGCCCAACCACCCCGCCGCGACC |
| 3579a | TCTAGACAACAACTTAGTGGTGTGGC |
| 3681s | GCGCGCTCTAGAAAAAGGTGATGCTAATACTTT |
| 3515BssHIIs | GCGCGCCGACTATCCAGCGACCAAG |
| 3599a | TCTAGACTGTCCACTGAGTCTCTGTG |
| 2530s | GTCGACCCATGCATTGGATGGAAATTTAGTTT |
| 3190a | CAGTTTGTATAGTGCATTGTATTGCATATGT |
| 3190s | ACATATGCAATACAATGCACTATACAAACTG |
| E5startantiXbaI | GATGCAGTATCTAGATTTGTCAT |
| E4OPSAa | ATTTCAGGAGAGGATACTTCGTTGCTGCTAAAAAAAAAAAAAAAAAGAGTTAGTGGCTGACCACCCG |
| E4OPSAs | CGGGTGGTCAGCCACTAACTCTTTTTTTTTTTTTTTTTAGCAGCAACGAAGTATCCTCTCCTGAAAT |
| E5startSalI/XbaI | GTCGACATGACAAATCTAGATACTGCATC |
| OPSDa | CATTTTAAAGTATTATACTTACCTGTTAAATGTAC |
| OPSDs | GTACATTTAACAGGTAAGTATAATACTTTAAAATG |
| K1 | ACGCGTGGTACCCTAGACGCTGTCCAATGCCATG |
Restriction sites are underlined.
To generate pT2 and pT3, PCR with pBELDP (52) using oligonucleotide 3455s with oligonucleotide 3705a or 3902a created PCR fragments corresponding to positions 3402 to 3652 and positions 3402 to 3849 of HPV-16, respectively. These were subcloned into PCR-II-TOPO (Invitrogen), cleaved using XbaI and MluI, and cloned into the pT1 construct, generating pT2 and pT3. To generate pT4, PCR was performed using oligonucleotides 3903s and L2D on pBELDP (52). The fragment, comprising nucleotides 3652 to 4327 of the HPV-16 genome, was subcloned into PCR-II-TOPO (Invitrogen), followed by cleavage by XbaI and MluI and insertion into pT1, generating the pT4 construct.
pT5, pT6, pT7, and pT8 were constructed as follows. PCR with the template pBELDP was carried out using the oligonucleotides 3640s and L2D, 3575s and L2D, 3515s and L2D, and 3555s and L2D. The resultant fragments corresponding to genomic HPV-16 positions 3587 to 4327, 3522 to 4327, 3462 to 4327, and 3502 to 4327, respectively, were subcloned into PCR-II-TOPO (Invitrogen). The fragments were subsequently cloned using XbaI and MluI into pT1, generating pT5, pT6, pT7, and pT8, respectively. To generate pT9 and pT10, two fragments were produced by PCR on the template pBELDP (52) using the oligonucleotide E4S (53) with either oligonucleotide 3580a or 3620a. The two fragments corresponded to genomic sequences between positions 3402 and 3651 but with the use of tails on the antisense oligonucleotides allowed deletions of different sizes within this sequence to be introduced. Both fragments were cloned into PCR-II-TOPO (Invitrogen), followed by cleavage using BssHII and XbaI and insertion into pT1. This process generated pT9 and pT10 with deletions corresponding to sequences between positions 3547 and 3628 and positions 3587 to 3628 of the HPV-16R sequence (2), respectively. pBELT9 was generated by excision and insertion of an ApaI-BamHI fragment from pBEL into pT9. To create pT11, PCR was performed with pBEL using E4SalBssHIIs and 3579a, and the fragment, subcloned into PCR-II-TOPO (Invitrogen), was transferred to pT1SD (see below) using the BssHII and XbaI sites.
To create pT1SD, PCR was carried out on pBELDP (52) using oligonucleotides 3681s and L2D, and the amplified fragment corresponding to positions 3628 to 4327 of HPV-16 was subcloned into PCR-II-TOPO (Invitrogen). This fragment was cleaved by XbaI and MluI and inserted into pT1, creating pT1SD, with a deletion similar to that of pT1 but containing the E4 5′ splice site. To insert the enhancer sequence into this deletion sequence, a sequence was PCR amplified from pBEL using oligonucleotides 3515BssHIIs and 3599a and subcloned into PCR-II-TOPO (Invitrogen). This fragment was cleaved using BssHII and XbaI and cloned into pT1SD, generating pT1SDE with 85 nucleotides inserted into the pT1 deletion.
To create pT1OPSA, PCR mutagenesis was first performed to destroy NsiI at position 3140 to render the NsiI site at position 2477 unique. PCR fragments produced using oligonucleotides 2530s and 3190a on the HPV-16R sequence and 3190s and E5startantiXbaI on pBELM were annealed and cloned into PCR-II-TOPO (Invitrogen), making PCRMUT-TOPO. This was used as a template for a second PCR mutagenesis step with oligonucleotides 2530s and E4OPSAa or E4OPSAs and E5startantiXbaI in order to optimize the E4 3′ splice site. This PCR mutagenesis product was subcloned into PCR-II-TOPO and subsequently cloned into pT1 using NsiI and BssHII, creating pT1OPSA.
To generate pOPSD, pBEL with an XbaI site at the start of E5 was used and is referred to as pBEX. To construct pBEX, PCR was carried out on the HPV-16R genome using oligonucleotides E5startSalI/XbaI and L1stop(XhoI) (9). The amplified fragment was cloned into pL0806 (9) using SalI and XhoI. A second PCR-amplified fragment was transferred to the resultant plasmid described above using BamHI and XhoI. This PCR product contained L1 sequences from positions 6152 to 7157 in which a cryptic polyadenylation site at position 6208 had been inactivated (39). Finally, the early region was amplified from HPV-16R using 757s (53) and E5startantiXbaI and cloned using SalI and XbaI, generating pBEX. pBEXM was generated by transfer of an ApaI-BamHI L1 fragment from pBELM. To create the OPSD plasmids, PCR mutagenesis using oligonucleotides 2530s and OPSDa or OPSDs and E5startantiXbaI on PCRMUT-TOPO was carried out, and the resulting fragment was cloned using NsiI and XbaI into pBEX and pBEXM to create pOPSD and pOPSDM, respectively.
Transfection and cell culture.
HeLa cells were cultured in Dulbecco's modified Eagle medium containing 10% heat-inactivated fetal bovine calf serum and penicillin-streptomycin. Transfections were carried out using Fugene 6 acccording to the manufacturer's instructions (Roche Molecular Biochemicals). A mixture of 3 μl of Fugene and 200 μl of Dulbecco's modified Eagle medium without serum was added to 1 μg of DNA and incubated for 15 min. This mixture was then added to 60-mm plates of HeLa cells at a 60% confluence in a drop-wise fashion. Cells were harvested at 24 h posttransfection.
RNA extraction, Northern blotting, and radiolabeled DNA probe synthesis.
Total cellular RNA was prepared 24 h posttransfection using a QiaShredder and an RNeasy minikit as instructed by the manufacturer (QIAGEN) and then treated with DNase I. Northern blot analysis was carried out by the size separation of 10 μg total RNA on a 1% agarose gel containing 2.2 M formaldehyde, followed by overnight transfer to nitrocellulose and hybridization. DNA probes were made with [α-32P]dCTP incorporated by random priming by use of a Decaprime kit (Ambion). The cytomegalovirus (CMV) probe was made as described previously (9), as was the L1 probe (53). The E4 probe was generated by digestion of pBELM (53) with BssHII and MluI, and the released fragment was purified and used as a template for probe synthesis using the Decaprime system. A second E4 probe used was also previously described (53) and is referred to here as the E4* probe.
RESULTS
The deletion of 817 nucleotides of the early region of HPV-16 between positions 3396 and 4212 caused a dramatic increase in late gene expression.
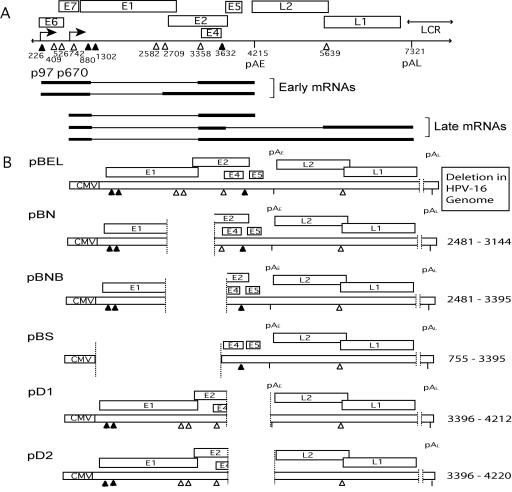
Deletional analysis was carried out to determine cis-acting sequences in the early region that are important for regulation of HPV-16 splicing. The genome of HPV-16 and representative mRNA transcripts are illustrated in Fig. 1A. The HPV-16 expression plasmids used were based on the previously described CMV-driven constructs pBEL (Fig. 1B) and pBELM (see Fig. 3A) (53), which contained the E1, E2, E4, and E5 early genes and early polyadenylation signal (pAE), followed by the L1 and L2 late genes and late polyadenylation signal (pAL). A series of deletions were made using available restriction enzyme sites in pBEL (Fig. 1B) and tested for their effect on production of late mRNAs in transfection and Northern blot experiments. The same deletions were made using pBELM (see Fig. 3A), which contains a previously reported mutated sequence of the first 514 nucleotides of the L1 coding region that inactivates a splicing silencer in L1 (53).
FIG. 1.
(A) Schematic diagram of the HPV-16 genome, with potential mRNA transcripts representing groups of early and late transcripts, transcribed from promoters p97 and p670, respectively, shown below the diagram. The boxes represent the coding regions of the genes and the positions of 5′ splice sites (filled triangles), 3′ splice sites (empty triangles), and polyadenylation sites are indicated. (B) Schematic representation of expression plasmids used to identify regulatory sequences in the early region affecting late gene expression. Numbers given are positions in the HPV-16R sequence. The position of the CMV promoter is indicated. Dotted lines indicate where deletions were made, and the nucleotides deleted are displayed on the right. (Dashed lines indicate deletion of a late untranslated region sequence in all plasmids [53].) Plasmids with L1 mutated not represented here are identical except for a mutant L1 sequence from position 1 to 514 of the L1 gene. pAE, early polyadenylation signal; pAL, late polyadenylation signal.
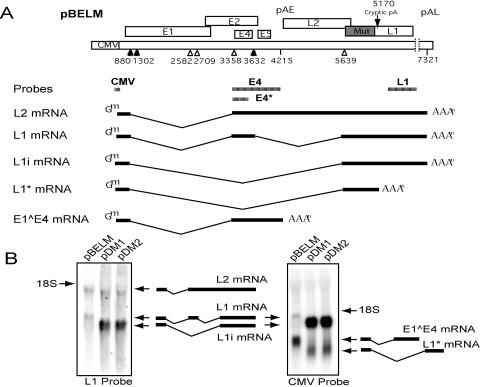
FIG. 3.
A deletion in the early region of HPV-16 induces a shift from early to late mRNA production. (A) Schematic diagram of pBELM with the mutated L1 sequence illustrated as a shaded box. Late mRNAs produced by the HPV-16 expression system used here are shown, with the positions of probes indicated by shaded bars. (B) Northern blots of total RNA extracted from HeLa cells transfected with pBEL, pDM1, and pDM2, hybridized to L1 and CMV probes. Arrows indicate the positions of mRNA species shown in the adjacent schematics and in panel A. L1* mRNA represents truncated L1 mRNA that is polyadenylated at an identified cryptic polyadenylation site at position 5170 in the HPV-16R genome. L1i mRNA represents a “mutation-induced” late message spliced from the position 880 5′ splice site directly to the position 5639 3′ splice site.
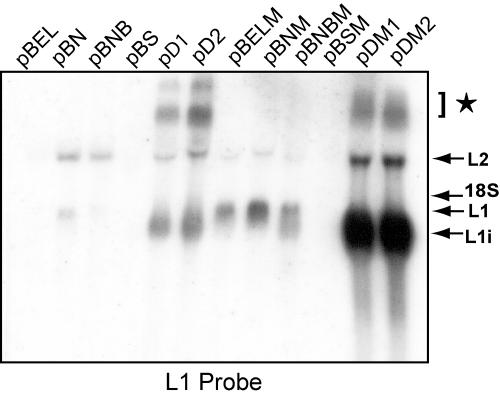
pBEL did not produce late mRNA levels detectable by the L1 probe (Fig. 2), while pBELM produced high levels of spliced L1 mRNA (Fig. 2), as reported previously (53). Deletion of almost the entire early region from pBEL in pBS did not induce late gene expression. Removal of the sequence between natural NsiI sites at genomic positions 2477 and 3140 caused a slight increase in both L2 and L1 mRNA levels, whereas deletion of the sequence between the 3′ NsiI site (position 3140) and BssHII (position 3396) did not enhance spliced L1 mRNA expression considerably (Fig. 2; compare pBN and pBNB to pBEL). However, removal of the sequence downstream of the position 3358 3′ splice site, between BssHII (3396) and MluI (4212) at pAE as seen in pD1 and pD2 (Fig. 2), dramatically increased late gene expression (Fig. 2). This striking effect could be seen in the presence and absence of pAE, as shown by a comparison of pD1 and pD2 (Fig. 2). All deletion constructs were made in combination with both wild-type and mutant L1 sequences. Effects on late gene expression were identical but were amplified greatly in the presence of the L1 mutant sequence, with the exception of pBSM, which abolished the production of late mRNA seen in the parental plasmid pBELM (Fig. 2). These results demonstrated that sequences in the early region were required for late gene expression, most likely including the 5′ splice site at position 880 or 1302. The various deletions in the early region caused an increase in late mRNA levels. When an 817-nucleotide deletion in the early region was combined with the L1 mutant sequence in pBELM, as in pDM1, a 280-fold increase in late mRNA levels was induced (Fig. 2; compare the first and last lanes of the Northern blot). These results established that the sequence between positions 3396 and 4212 contained RNA cis-acting sequences capable of blocking late gene expression in proliferating cells.
FIG. 2.
Identification of HPV-16 early region sequences that induce late gene expression. A Northern blot of total RNA extracted from HeLa cells transfected with the plasmids indicated was hybridized with L1 probe. Plasmids with a mutated L1 ORF are designated with an M in the plasmid name and are identical to plasmids represented in Fig. 1 except for a mutant L1 sequence from position 1 to 514 of the L1 gene. Bands representing late mRNA transcripts are indicated by arrows. A star indicates late mRNAs with differential splicing events in the early region. L1i mRNA represents a “mutation-induced” late message spliced from the position 880 5′ splice site directly to the position 5639 3′ splice site. Late mRNAs are illustrated schematically in Fig. 3A.
The deletion comprising positions 3396 to 4212 in the early region of HPV-16 induced a complete shift from early to L1 mRNA production.
To determine the effect of the deletion comprising positions 3396 to 4212 on early mRNAs, two plasmids illustrating the induced late mRNA effect, pDM1 and pDM2, were analyzed by Northern blot analysis using L1 and CMV probes (Fig. 3B). Short exposures of the filters with the L1 probe in repeated experiments convincingly highlighted a qualitative difference between the results with pBELM and those with pDM1 and pDM2 (Fig. 3B). pBELM produced high levels of spliced L1 mRNA (Fig. 3B), while in contrast the L1 mRNA produced from pDM1 and pDM2 migrated faster than pBELM L1 mRNA (Fig. 3B). This faster-migrating L1 mRNA did not hybridize to the E4 probe as expected, as this sequence is not present in pDM1 or pDM2 (data not shown; see Fig. 3A for location of the probe) and represents the L1 mRNA that is referred to here as mutation-induced L1 mRNA (L1i mRNA) (Fig. 3A).
To determine the major mRNA produced from pDM1 and pDM2, a probe named CMV (Fig. 3A), detecting immediate 5′ ends of all mRNAs produced, was used to detect both early and late mRNAs. The major mRNA species in pBELM was identified as the early E1∧E4 mRNA, while the major species in pDM1 and pDM2 was the late L1i mRNA (Fig. 3B). Low levels of an additional L1 mRNA species were seen with longer exposures of the CMV-probed filter (Fig. 3B, right). These mRNAs were referred to as L1* mRNA and represent a previously described L1 mRNA prematurely polyadenylated at position 5170 in the L1 open reading frame (ORF) (35, 39). In conclusion, these results established that a switch from early to late L1 mRNA production had been induced by the deletion of nucleotides 3396 to 4212 and prompted further analysis of this region of interest.
Deletion of 255 nucleotides between the E4 3′ splice site at position 3358 and the 5′ splice site at position 3632 upstream of HPV-16 pAE activated mutation-induced late gene expression.
The region between positions 3396 and 4212 was divided into four parts to locate the sequence responsible for the efficient inhibition of HPV-16 late gene expression (Fig. 4A). As shown on Northern blots probed for L1 (Fig. 4B), pT1 and pT4 deletions, which removed the 5′ splice site at nucleotide 3632, increased late gene expression to a level similar to that of pDM1 and expressed L1i mRNA, while pT2 and pT3 expressed L1 mRNA to a level similar to that of pBELM. These results demonstrated that sequences inhibiting late gene expression were still present in pT2 and pT3 but were destroyed or deleted in pT1 and pT4.
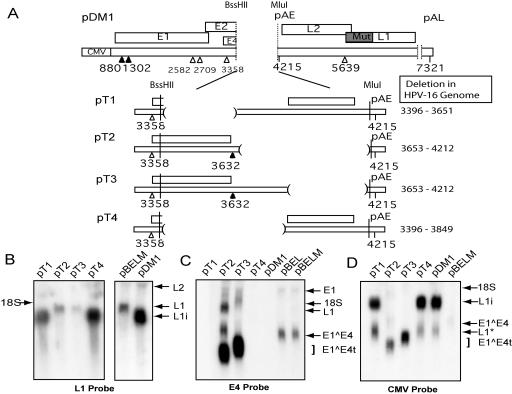
FIG. 4.
Deletion of 255 nucleotides in pBELM increased late gene expression. (A) Schematic illustrations of expression plasmids with further deletions of the E4 region are shown, as well as pDM1, a control plasmid. The nucleotide positions deleted in the HPV-16R genome are displayed on the right. The locations of the BssHII and MluI sites are indicated. (B) Northern blot of total RNA extracted from the indicated plasmids hybridized with the L1 probe. Arrows are labeled with the names of the mRNA species. L1* mRNA represents truncated L1 mRNA that is polyadenylated at an identified cryptic polyadenylation site at position 5170 in the HPV-16R genome. L1i mRNA represents a “mutation-induced” late message spliced from the position 880 5′ splice site directly to the position 5639 3′ splice site. (C, D) Northern blot of total RNA extracted from the indicated plasmids, probed with the E4 (C) or CMV (D) probe. Arrows indicate the positions of the mRNA species produced. A square bracket specifies the position of the truncated E1∧E4 species (E1∧E4) produced by pT2 and pT3.
Probing for the presence of the E4 exon on L1 mRNA using the E4 probe revealed that pT1 and pT4 did not express any mRNA containing the E4 exon (Fig. 4C). It was established that the L1i mRNA produced by pT1 and pT4 lacked the internal E4 exon. As expected, the L1i mRNAs produced from control plasmid pDM1 were also not detected by the E4 probe (Fig. 4C). We concluded that the L1i mRNA lacked the E4 exon and represented splicing from the 5′ splice site at position 880 to the L1 3′ splice site at position 5639. In contrast, the L1 mRNAs produced by pT2 and pT3 were detected by the E4 probe (Fig. 4C), as were the mRNAs produced by pBELM, and therefore contained the internal E4 exon. Moreover, the major mRNAs produced by pT2 and pT3 were the early mRNAs. These included truncated E1∧E4 messages, of a size relative to their deletions, containing parts of the E4 exon detectable by the E4 probe. Northern blot analysis with the CMV probe (Fig. 4D) confirmed that pT2 and pT3, like pBELM, produced primarily early mRNAs, while the majority of mRNAs produced by pT1, pT4, and pDM1 were late mRNAs. In conclusion, the largest deletion that still permitted production of E1∧E4 mRNAs, seen in pT2, established that the element required for production of early mRNAs lies upstream of nucleotide 3652 and is disrupted by a deletion from positions 3396 to 3651.
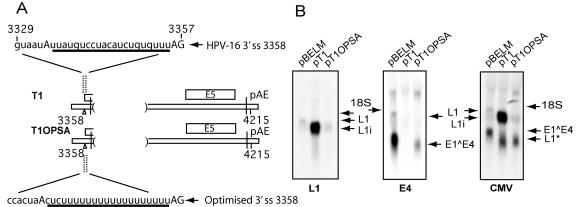
Optimization of the E4 3′ splice site at nucleotide 3358 overcame the absence of an enhancer and prevented mutation-induced late gene expression.
The results described above, which demonstrated that the E4 3′ splice site was not used in the presence of the deletions in pDM1, pT1, and pT4, implied that this 3′ splice site was under the control of a downstream splicing enhancer. If this was correct, it would be possible to reverse the effects of the deletion by optimizing the E4 3′ splice site in pT1 that lacked the enhancing sequence. Thus, we could investigate whether optimization would inhibit L1 mRNA production and promote E1∧E4 mRNA production, despite the absence of an enhancer. The E4 3′ splice site deviated from the consensus 3′ splice site sequence and had a brief and interrupted PPT (Fig. 5A). An optimized BPS, YNYURAC, and a PPT of U17 were introduced into the pT1 plasmid. The resultant plasmid, pT1OPSA, was transfected into HeLa cells and compared to pT1. Both of these plasmids lack the 5′ splice site at nucleotide 3632. Northern blots showed that pT1OPSA and pBELM produced similar levels of L1 mRNA, in contrast to pT1 (Fig. 5B, left). The L1 mRNA species of pT1OPSA was smaller with respect to the T1 deletion of 255 nucleotides in the E4 exon than that of the mutant pBELM (Fig. 5B, left). This finding was confirmed by probing for early mRNAs (Fig. 5B, middle), which showed that pT1OPSA produced an E1∧E4 mRNA detectable by the E4 probe, in contrast to pT1. Probing the same RNA with the CMV probe (Fig. 5B right) confirmed that pBELM and pT1OPSA produced primarily early mRNAs, while pT1 produced predominantly late mRNAs. These data supported the hypothesis of an enhancer-dependent 3′ splice site at position 3358. We concluded that an exonic enhancer element supported the use of the position 3358 3′ splice site and was located in a region upstream of position 3652. As this 3′ splice site is used in both early and late mRNAs in HPV-16, we continued to map this element, which may be significant in the switch between early and late mRNA production.
FIG. 5.
Optimization of the 3′ splice site at position 3358 overcame the absence of an enhancer. (A) The position 3358 3′ splice site and its optimized counterpart, OPSA, are shown in a schematic diagram. Capital letters represent the branch point and the 3′ splice site. The polypyrimidine tract is underlined, and genomic positions are indicated. (B) Northern blots of total RNA extracted from HeLa cells after transfection of pT1, pT1OPSA, and pBELM, hybridized to the L1, E4, and CMV probes. Arrows indicate the positions of the mRNA species produced.
The 65-nucleotide enhancer element was located between positions 3462 and 3527 of the HPV-16 genome.
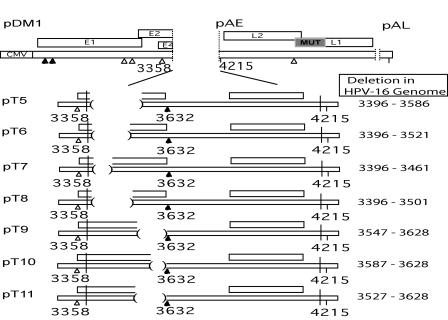
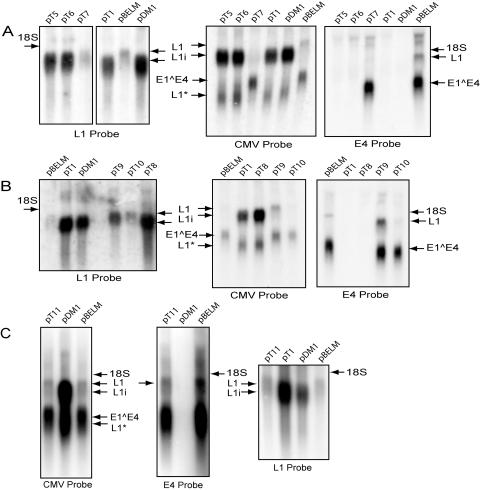
Three deletion constructs named pT5, pT6, and pT7 (Fig. 6) were analyzed in transfection and Northern blot experiments to deduce the 5′ end of the enhancer element in the T1 deletion region. A deletion of 65 nucleotides from positions 3396 to 3461 in pT7 did not activate late gene expression (Fig. 7A, left) but rather produced L1 mRNA levels similar to those produced by pBELM, as shown using the L1 probe. However, larger deletions of 125 and 190 nucleotides, in pT6 and pT5, respectively, activated late gene expression and produced L1i mRNA at a level similar to that of pDM1 (Fig. 7A, left). After probing for early mRNAs using CMV and E4 probes (Fig. 7A, middle and right), pT5 and pT6 revealed a phenotype equal to that of pDM1 and pT1. These did not produce E4 exon-containing mRNAs as detected by the E4 probe, whereas pT7 expressed E1∧E4 mRNA strongly (Fig. 7A, right). We concluded that the larger deletions in pT5 and pT6 interrupted the exonic enhancer element, locating the 5′ border between positions 3461 and 3651. A deletion of 105 nucleotides between positions 3396 and 3501 in pT8 (Fig. 6) was designed to further examine the 5′ border of the enhancer as determined by pT6 and pT7 as described above. The pT8 deletion activated mutation-induced late gene expression and produced L1i mRNA levels similar to that of control plasmid pT1 (Fig. 7B, left). Similar to pT1, pT8 did not produce any early mRNAs detectable by the E4 probe, and the E4 exon was no longer included in L1 mRNA splicing (Fig. 7B, right). This result confirmed that the pT8 deletion interrupted the enhancer, and thus the 5′ border of the enhancer sequence remained at position 3461.
FIG. 6.
Schematic illustrations of further deletion constructs made with expression plasmids pT5 to pT11 are shown, as well as pDM1, a control plasmid. Deletion sites are indicated by parentheses, and 5′ and 3′ splice sites are represented by filled and empty triangles, respectively. The nucleotide positions deleted in the HPV-16R sequence are displayed on the right.
FIG. 7.
Further deletions mapped the enhancer element between positions 3462 and 3547. (A) Northern blots of total RNA from transfection of plasmids pT5 to pT7 and controls, hybridized to the L1, CMV, and E4 probes. (B) Northern blots of total RNA extracted from HeLa cells transfected with plasmids pT8 to pT10 and controls, hybridized to the L1, CMV, and E4 probes. (C) Northern blots of total RNA extracted from HeLa cells transfected with plasmids pT11 and controls pBELM and pDM1, hybridized to the CMV, E4, and L1 probes. For all panels, arrows indicate the positions of the mRNA species produced.
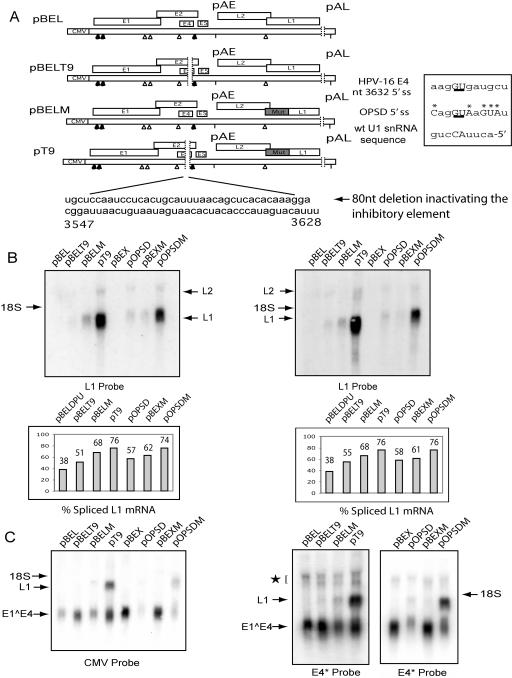
Two more deletions were made in pBELM to investigate the 3′ border of the enhancer element. pT9 lacked 81 nucleotides 5′ of the 3632 5′ splice site, whereas pT10 lacked 41 nucleotides 5′ of the 3632 5′ splice site (Fig. 6). pT10 produced L1 mRNA at levels similar to that of pBELM and also had the same phenotype as pBELM when it was probed for early mRNAs using the CMV and E4 probes (Fig. 7B, middle and right). These results mapped the 3′ border of the enhancer to immediately upstream of position 3587.
The deletion in pT9 produced higher levels of L1 mRNA than did pBELM and pT10 when the L1 probe was used (Fig. 7B, left). However, both the L1 and CMV probes highlighted the slower migration pattern of the pT9 L1 mRNA relative to that of the L1i mRNA produced by pT1 and pT8, which suggested that pT9 contained the E4 exon (Fig. 7B, left and middle). Indeed, the E4 probe confirmed that the pT9 L1 mRNA contained the internal E4 exon (Fig. 7B, right). The results of the L1, CMV, and E4 probes demonstrated that pT9 displayed all the properties of a plasmid containing a functional enhancer element of the 3′ splice site at position 3358 but induced an increase in spliced L1 mRNA. These results indicated that the overall increase in L1 mRNA produced from pT9 in comparison to that produced from pBELM could be explained by the deletion of a sequence inhibitory to the downstream position 3632 5′ splice site, while the enhancer element of the upstream 3′ splice site at position 3358 was still intact.
By use of the CMV and E4 probes (Fig. 7B, middle and right), early mRNAs were detected at high levels in pT9 but were undetectable in pT1 and pT8. Early mRNAs produced by pT9 were similar in size and abundance to those produced by pBELM and pT10. In summary, these results confirmed that the position 3358 3′ splice site was in fact used in pT9 and demonstrated that the enhancer was functional. The 3′ border was now situated at position 3547, thereby locating the enhancer to an 85-nucleotide sequence between positions 3462 and 3547. A final deletion construct named pT11 made to map the 3′ border removed an additional 20 nucleotides from the proposed enhancer (Fig. 6). Using the E4 probe, early mRNAs were detected at high levels in pT11 and pBELM but were undetectable in pDM1 (Fig. 7C, middle). The CMV and L1 probes (Fig. 7C, left and right) revealed that L1 mRNA produced by pT11 was similar in size and abundance to those produced by pBELM. The 3′ border was moved to position 3527, thereby locating the enhancer to a 65-nucleotide sequence between positions 3462 and 3527.
The use of the position 3358 3′ splice site was restored when the enhancer element was inserted into the pT1 deletion.
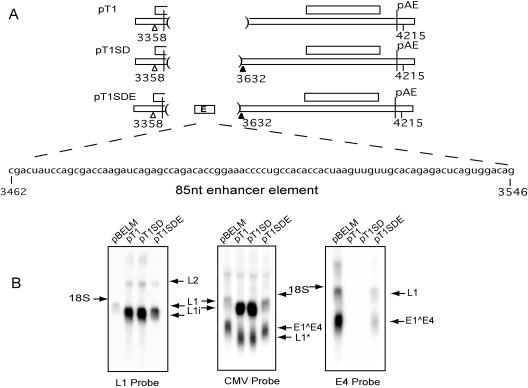
To confirm that a functional enhancer element was present downstream of the E4 3′ splice site at position 3358, the sequence between positions 3462 and 3547 was inserted into the pT1 deletion construct, which lacked the enhancer and did not produce mRNAs containing the E4 exon (Fig. 8A). For this purpose, a derivative of pT1 (pT1SD) was also used; in pT1SD, the pT1 sequence was extended to contain the position 3632 5′ splice site and the enhancer could be readily inserted (Fig. 8A). pT1SD had an mRNA-expressing phenotype similar to that of pT1 when tested in parallel on Northern blots probed for late and early mRNAs (Fig. 8B). pT1SDE, with the enhancer sequence inserted between the position 3358 3′ splice site and the position 3632 5′ splice site, expressed its respective L1 mRNA at a higher level than that of control plasmid pBELM but at a lower level than the L1 mRNA produced by pT1 (Fig. 8B, left). This result illustrated that the inserted fragment promoted the use of the 3′ splice site at position 3358 but at a slightly lower efficiency than the full wild-type sequence did. The pT1SDE L1 mRNA was smaller than the pBELM L1mRNA (Fig. 8B, left and middle,) indicating that it contained a truncated E4 exon. This result was confirmed with the E4 probe (Fig. 8B, right). Thus, pT1SDE exhibited a phenotype similar to that exhibited by pBELM, in which E1∧E4 mRNAs were expressed and polyadenylated at pAE due to the use of the position 3358 3′ splice site, which was dependent on the enhancer element. The higher L1 mRNA levels produced by pT1SDE relative to pBELM (Fig. 8B, left) are best explained by the absence of the same sequence inhibitory to late mRNA splicing at the downstream 5′ splice site at position 3632, which caused an increase in L1 mRNA from pT9 (Fig. 7B, left). This may have been responsible for the increase in L1 mRNA expression over that produced by pBELM, at the expense of the early mRNAs polyadenylated at pAE, pointing towards the similarity of the pT1SDE and pT9 phenotypes. We concluded that an exonic splicing enhancer (ESE) was located in the inserted fragment spanning positions 3462 to 3527.
FIG. 8.
Use of the position 3358 3′ splice site was restored when the enhancer was inserted into the pT1 deletion. (A) Representative illustrations of the deletion regions of pT1, pT1SD, and pT1SDE. A box represents the inserted enhancer, and the sequence is presented below. (B) Northern blots of total RNA extracted from HeLa cells after transfection of the respective plasmids hybridized to the L1, CMV, and E4 probes. Arrows indicate the positions of the mRNA species produced.
Optimization of the suboptimal position 3632 5′ splice site or deletion of a sequence upstream of the same 5′ splice site increased late gene expression in both wild-type and mutant L1-expressing plasmids.
Having established that the use of the 3′ splice site at nucleotide 3358 was dependent on a 65-nucleotide enhancer situated between positions 3462 and 3527, we wished to gain further evidence for an adjacent inhibitory sequence with an effect on the use of the E4 5′ splice site (Fig. 7B, lane pT9). The pT9 and pT10 deletion constructs lacked a common 40 nucleotides, in the 5′-most direction of the 5′ splice site at position 3632 (Fig. 6). The T10 deletion, representing the 3′-most 40 nucleotides of pT9, did not increase late gene expression in comparison to that of pBELM (Fig. 7B, left and middle). From this we deduced that the 3′-most 40 nucleotides did not contribute to the inhibitory effect on the position 3632 5′ splice site seen in pT9. Thus, the 3′ border of a sequence with a negative effect on the use of the E4 5′ splice site was mapped to between positions 3547 and 3587 of the HPV-16 genome.
In order to determine the relative importance in late mRNA splicing of the inhibitory sequence located upstream of the 5′ splice site at position 3632 and the splicing silencer downstream of the L1 3′ splice site at position 5639 (53), the T9 deletion that inactivates the inhibitory sequence was introduced into pBEL in the context of wild-type L1 (Fig. 9A). The T9 deletion had a smaller effect on late gene expression in pBEL than mutation of the splicing silencer in pBELM, as shown by the slight increase in spliced L1 mRNA levels in the case of pBELT9 (Fig. 9B). The increase in late gene expression by the T9 deletion was also confirmed by use of the CMV and E4* probes (Fig. 9C). However, the combined effect of the T9 deletion and the L1 mutation increased the level of spliced L1 mRNA from 53% for the T9 deletion alone (51%, 55%) (Fig. 9B) or 68% for the L1 mutation alone to 76% (Fig. 9B). These results confirmed that the T9 sequence inhibited splicing of L1 mRNA. An increase in late mRNA splicing predicts a decrease in early mRNA levels since early polyadenylation and late mRNA splicing are mutually exclusive events. Surprisingly, the early probes revealed that the 80-nucleotide deletion increased E1∧E4 mRNA (Fig. 9C, left and middle) in both wild-type (compare pBEL and pBELT9 results) and mutant L1-expressing plasmids (compare pBELM and pT9 results). These results implied that the inhibitory sequence suppressed the 5′ splice site and reduced early mRNA stability or had an inhibitory effect on both the position 3632 5′ and position 3358 3′ splice sites, the latter possibly through the enhancer.
FIG. 9.
Disruption of an inhibitory sequence and optimization of the “weak” position 3632 5′ splice site increased late gene expression. (A)Schematic diagrams of plasmids tested to study the inhibitory sequence are shown on the left, and the location and sequence of the 80-nucleotide deletion disrupting the inhibitor is indicated. 5′ and 3′ splice sites are represented by filled and empty triangles, respectively. The position 3632 5′ splice site, its optimized counterpart OPSD, and the complementary wild-type U1 snRNA sequence are shown on the right. Uppercase letters below an asterisk represent base changes from the HPV-16 sequence, while the underlined uppercase letters represent the 5′ splice site. Genomic positions are indicated. (B) Two Northern blots representing independent experiments of total RNA extracted from HeLa cells after transfection of the indicated plasmids hybridized to the L1 probe are shown at the top, with the quantified percentages of spliced L1 mRNAs displayed in graphs belowl. (C) A Northern blot of total RNA extracted after transfection of the indicated plasmids hybridized to the CMV probe is shown on the left. Northern blots of total RNA extracted after transfection of the respective plasmids hybridized to E4* probe are shown on the right. Arrows indicate the positions of the mRNA species produced. A star indicates unspliced early mRNAs.
The position 3632 5′ splice site ought to play a pivotal role in the switch to L1 mRNA production but prior to this must avoid recognition to allow early polyadenylation. The presence of an inhibitory sequence suggested that the position 3632 5′ splice site was a suboptimal splice site. To investigate this regulated splice site, the position 3632 5′ splice site was optimized by replacement with an optimal U1 snRNA binding site (Fig. 9A, right) in the context of both wild-type and mutant splicing silencer sequences in L1, generating plasmids pOPSD and pOPSDM, respectively. pBELDPU, in which the early polyadenylation signal had been deactivated (52), produced 38% L1 mRNA (53). By comparison, the OPSD mutation caused an increase in spliced late mRNA to 58% (57%, 58%) (Fig. 9B), while the OPSD mutation in combination with the mutant L1 sequence, pOPSDM, produced 75% spliced L1 mRNA (74%, 76%) (Fig. 9B), compared to 62% from the parental plasmid pBEXM (62%, 61%) (Fig. 9B). The increase in late mRNA splicing in pOPSD and the additive effect on late mRNA splicing in pOPSDM established that the 5′ splice site at position 3632 is suboptimal. The enhanced splicing of L1 mRNAs was accompanied by a predicted decrease in early mRNA levels (Fig. 9C, CMV and E4* probes). In summary, the decrease seen in early mRNAs to the benefit of the late mRNAs produced by OPSD plasmids demonstrated that the position 3632 5′ splice site is suboptimal. In contrast, the T9 deletion, which caused an increase in both spliced early and spliced late mRNAs, demonstrated the existence of an RNA element that inhibits splicing of L1 mRNAs and inhibits production of E1∧E4 mRNAs, either by inhibiting early mRNA splicing or by reducing early mRNA stability.
DISCUSSION
This study identified a 65-nucleotide ESE that supported the use of the position 3358 3′ splice site in the E4 coding region of HPV-16. The E4 ESE is located approximately 100 nucleotides downstream of the E4 3′ splice site on the E4 exon, present in many early HPV-16 mRNAs as well as the late mRNAs (1). The 65-nucleotide ESE is 66% adenosine and cytosine rich (AC rich) and when a uridine-rich sequence of 10 nucleotides at the 3′ end is disregarded, this becomes 75% AC rich. Thus, the HPV-16 E4 ESE resembles a class of previously described enhancers, AC-rich enhancers (ACE), the role of which has been recognized in the regulated splicing of viral and cellular genes (8, 24, 43, 48, 50, 58). Although they were initially characterized using in vivo selection experiments (12), ACEs have since been shown to enhance splicing at an upstream 3′ splice site in an NS2-specific exon of parvovirus minute virus of mice (24) and at a differentiation-specific 3′ splice site in BPV-1 (58), both in bipartite elements. ACEs have also been found in exon v4 of the human CD44 gene (43), in exon 6B of the α1(XI) collagen gene (8), and in exon 4 of the calcitonin/calcitonin gene-related peptide gene (50). Motifs common in some ACEs (8, 43, 50) are repeats of CAA, first seen in the Drosophila doublesex (dsx) gene AC-rich repeat element (49). The E4 ESE identified here contains only one such CAA but interestingly contains both of the AC-rich motifs (CACCA and ACACC) present in the parvovirus ACE (24).
While many examples of SR proteins interacting with purine-rich enhancers have been described previously (32, 49), binding of the heterologous ACEs has not been attributed to one such single class of proteins. However, the Drosophila splicing regulator Tra2, an SR-related protein, is known to regulate exon inclusion by interaction with the CAA-containing dsx splicing enhancer (44). In addition, strong evidence that the related human transformer 2 (hTra2β) of 40 kDa interacts with the calcitonin/calcitonin gene-related peptide exon 4 ACE has been provided (50). Also, a non-SR protein of 50 kDa, human YB-1 binding protein, was demonstrated to bind ACE elements in exon v4 of the human CD44 gene and to induce exon inclusion in vivo (43). However, in support of SR protein involvement in papillomavirus gene regulation, SR proteins have been previously linked to the regulated splicing of BPV-1. BPV-1 purine-rich enhancers SE1 and SE2, regulating an upstream early-specific 3′ splice site (55), demonstrated strong affinity for the SR proteins ASF/SF2, SRp55, and SRp75 (56), while the BPV-1 silencer exonic splicing suppressor immediately downstream of SE1 bound SC35 and SRp54/55, among other proteins (57).
The ESE-dependent E4 3′ splice site lacks a consensus BPS and has a suboptimal PPT. In fact, the longest run of uninterrupted pyrimidines in the PPT was four residues, UCCU. Despite the absence of the E4 ESE, optimization of the E4 3′ splice site PPT and replacement of the putative BPS with a consensus BPS, YNYURAC, restored early mRNA production and polyadenylation efficiency in an induced late mRNA-producing phenotype, suggesting that the E4 ESE functions to facilitate the recruitment of U2AF65 to the weak PPT, in keeping with previous reports (32, 59). This also exemplifies the coupling of polyadenylation to an upstream 3′ splice site in the processing of the terminal exon (4, 33, 34). Terminal exon recognition depends on the intrinsic strength of the processing signals at either end of the terminal exon (4). In support of this definition, an upstream 3′ splice site has been shown to increase polyadenylation efficiency (10, 11), while cleavage/polyadenylation sites can enhance U2AF65 binding to the PPT of upstream 3′ splice sites via interaction between the carboxy terminus of poly(A) polymerase and U2AF65 (51). Our results suggest that the E4 ESE supports the processing of the E4 terminal exon, despite a lengthy terminal exon of 854 nucleotides, by strengthening E4 3′ splice site recognition as the most 3′ splice site to the early polyadenylation signal. In the absence of the enhancer, the optimized E4 3′ splice site PPT presumably resulted in improved binding of U2AF65, thus reestablishing recognition of the terminal exon. This implies a key role for the E4 ESE in the precise and efficient recognition of a weak 3′ splice site to allow the production of early mRNAs terminated at pAE.
One interesting question to ask is why an upstream 3′ splice site is not selected when the E4 3′ splice site is no longer supported by the ESE. In pD1 and pD2, which lack the enhancer but not the splicing silencer, which is still present at the L1 3′ splice site, splicing occurs at high levels at the L1 3′ splice site rather than at upstream splice sites in the early region. For example, the upstream E2 3′ splice site at position 2709 has an uninterrupted pyrimidine tract of 11 residues and is by comparison superior to the E4 and the L1 3′ splice site sequences, the latter consisting of nine consecutive pyrimidines. The resulting E2 terminal exon, polyadenylated at pAE, would be 1,500 nucleotides in length, slightly shorter than the L1 terminal exon, which is approximately 1,680 nucleotides long. The length of the terminal exon in each case described above, however, is far greater than the optimal exon length for enhancing recognition of the last exon (4). This argues for competition for recognition by the E2 and L1 3′ splice sites, each facilitated by enhancer and suppressor sequences. In this study, the inactivation of the E4 splice site led to splicing at the L1 3′ splice site with an intact splicing silencer. One can therefore speculate that splicing silencers or enhancers must regulate recognition of the E2 3′ splice site and may require additional factors in order to be utilized.
In order to polyadenylate early mRNAs, the late-specific E4 5′ splice site must not be recognized. In support of this, a 5′ splice site directly upstream of a polyadenylation signal has been shown to inhibit polyadenylation (26). Cross-exon interaction of the E4 3′ splice site with the downstream 5′ splice site must also be avoided at all costs early in the viral life cycle to prevent production of L1 mRNA. Optimization of the E4 5′ splice site inhibited early polyadenylation and early mRNA production to some extent. Interestingly, the E1∧E4 mRNAs produced from plasmids with an optimized 5′ splice site were larger than the wild-type mRNAs. This effect on the processing of the terminal exon remains to be resolved by reverse transcription-PCR and sequencing of the 5′ and 3′ termini but may be a result of activation of a downstream cryptic polyadenylation site in the L2 coding region due to reduced pAE efficiency caused by the optimized 5′ splice site. Optimization had a moderate effect in contrast to the dramatic late-to-early switch caused by optimization of the 3′ splice site in an enhancer-deleted construct. In addition to highlighting the central role of the E4 3′ splice site, this difference prompts speculation that the upstream 40-nucleotide sequence still present had a negative effect on the processing of the E4 exon or the optimized 5′ splice site. That optimization of the 5′ splice site had a weak effect supports the idea of a late-specific 5′ splice site with overall low efficiency and the need for a regulated late-specific L1 3′ splice site (53). The difference in magnitude of effect caused by optimization of 3′ and 5′ splice sites suggests that while potential exonic sequences inhibitory to the 5′ splice site and its suboptimal nature may represent important hindrances, it is the coupling of the enhancer-supported E4 3′ splice site to polyadenylation that is paramount in preventing premature L1 production. However, the recognition or suppression of the weak E4 5′ splice site must also be regulated differentially during the life cycle to avoid or permit L1 mRNA processing. In this study, the deletion of the inhibitory sequence upstream of the 5′ splice site may have resulted in a reduction in cellular protein binding and, consequently, the stabilization of the late mRNAs, because elimination of the inhibitory sequence increased late mRNA expression. This increase was lesser than that produced in combination with deactivation of a splicing silencer in L1, which suggests that stronger regulation of L1 mRNA production occurs at the L1 3′ splice site. However, a moderate effect on L1 mRNA splicing, most likely at the 5′ splice site, suggests that the identification of factors binding to the inhibitor is still of interest. Control of the E4 5′ splice site may yet be complemented by regulatory sequences in the downstream intron. Evidence to support this is the slightly higher level of L1 mRNA expressed by pT2 and T3 than that expressed by pBELM, suggesting that there may be inhibitory sequences in the final intron of L1 pre-mRNA. Regulation of 3′ splice sites is, however, more commonly recognized than that at 5′ splice sites. Therefore, adding to the key task of the E4 ESE in supporting early rather than late mRNA production, suppression of the downstream L1 3′ splice site by a hnRNP A1-binding silencer (53) may play a larger role in L1 mRNA splicing regulation than the inhibitory sequence upstream of the E4 5′ splice site.
In both HPV-16 and BPV-1, a 3′ splice site in the early region plays a central role in regulated gene expression. In BPV-1, alternative 3′ splice site selection and subsequently late mRNA production are controlled by at least five separate splicing elements (55-58). A suboptimal 3′ splice site is used in the processing of most early BPV-1 mRNAs, from which a switch to a downstream late-specific 3′ splice site allows the production of L1 mRNA. In HPV-16, the suboptimal E4 3′ splice site is required throughout the virus life cycle for expression of the E1∧E4 protein, the most abundant viral protein in HPV-infected epithelia (18), and presumably the E5 protein from the spliced E1∧E4 transcript, as well as for expression of L2 and L1 late mRNAs. Unlike BPV-1, the suboptimal HPV-16 3′ splice site in question cannot be silenced early in the life cycle to prevent late gene expression; in fact, it must always be active to produce E1∧E4 mRNA throughout the life cycle. The use of a downstream differentiation-specific 3′ splice site along with the increased use of a weak downstream L1 5′ splice site occurs in BPV-1 to induce late gene expression. In HPV-16, expression of the late genes requires coordinated regulation of the downstream 5′ splice site, the early polyadenylation signal, and the L1 3′ splice site. In common with HPV-16, BPV-1 has a strong ESE (SE1) that regulates the utilization of an early-specific 3′ splice site, in conjunction with a second ESE downstream (SE2) (55). To add to the complexity of regulation, an exonic splicing suppressor (ESS) directly downstream of SE1 modulates the use of the early-specific suboptimal 3′ splice site. Of special interest is an AC-rich enhancer, identified in BPV-1 as part of an exonic bipartite element controlling a second downstream 3′ splice site essential for L1 expression (58). In summary, BPV-1 and HPV-16 differ in the regulated splicing of late gene expression due to a different arrangement of splicing signals that may therefore bind different factors. In the case of HPV-16, in which a common 3′ splice site is used for early and late mRNA processing, we provide evidence here that a strong coupling to the early polyadenylation signal conferred by the E4 ESE is necessary to block premature late gene expression.
The HPV-16 E1∧E4 protein has been considered a late protein because its expression coincides with vegetative viral DNA replication prior to the late structural capsid proteins (17). Additionally, late in infection it is abundantly expressed from the differentiation-dependent promoter and accumulates in the differentiated cell layers of the epithelium (18). HPV-16 E1∧E4 has been shown to cause the collapse of the cytoskeletal network (16) and to associate with mitochondria (38). Despite its late association, E1∧E4 is first expressed earlier, concomitant with the early proteins E6, E7, E1, and E2, and is detectable in the basal layers (29). A G2 arrest function was recently described for HPV-16 E1∧E4 in HeLa cells (14), suggesting that HPV-16 E1∧E4 has a role early in the life cycle, possibly to counteract E7-mediated proliferation. The E2 protein also affects the mitotic checkpoint in human keratinocytes (21) and causes G2 arrest in the fission yeast Schizosaccharomyces pombe (20). Given the effect of E1∧E4 on cell cycle progression and that the E4 ORF is embedded in that of E2, it is appealing to suggest that expression of E2 and E4 must be coordinated for this common function in the life cycle. Mutation of the E1 5′ splice site of HPV-11 in a GFP-E1E2 replicon abolished E1∧E4 splicing and increased E1 protein expression (15). In fact, the expression of E1∧E4 must be regulated over time to support its various functions and to allow the expression of E2 and E1 in parallel. Our hypothesis supports a model in which the E4 3′ splice site is efficiently used but subject to modulation throughout the life cycle. The E4 ESE then plays a key regulatory role in the HPV-16 life cycle by supporting the use of the E4 3′ splice site, allowing the modulated expression of the abundant E1∧E4 transcript required throughout infection. Furthermore, the E4 ESE represents a barrier for late gene expression. When the strong 3′ splice site at position 3358 competes with the L1 3′ splice site at position 5639, the E4 ESE enables the recognition of the 3358 3′ splice site and the RNA processing machinery is directed to the pAE. Thus, splicing is coupled to polyadenylation at the early pAE, and the late mRNAs are not produced. A previous study shows that recombinant HPV-31 lacking the early polyadenylation signal increases late gene expression in keratinocytes differentiated in methylcellulose, suggesting a link between early polyadenylation usage and a block in late gene expression (47). However, use of the E4 3′ splice site must be flexible as upstream 3′ splice site usage is required for the expression of E2 early in the life cycle. Therefore, the E4 ESE must be constitutively active to allow expression and polyadenylation of early mRNA but flexible to allow differential early mRNA expression. We conclude that in addition to its role in early gene expression, the HPV-16 E4 ESE plays a crucial role as an inhibitor of premature late gene expression. In this capacity, the E4 ESE may influence the ability of HPV-16 to be concealed from the immune response and to establish persistent infections that may progress to high-grade lesions or cancer. Therefore, we speculate that HPV regulatory elements may affect the pathogenesis of the HPVs.
Acknowledgments
We thank Daniel Öberg for pBEX and pBEXM and helpful discussions.
This study was supported by the Swedish Research Council (Medicine) and the Swedish Cancer Society.
REFERENCES
- 1.Baker, C., and C. Calef. 1996. Maps of papillomavirus mRNA transcripts. In S. R. Billakanti, C. E. Calef, A. D. Farmer, A. L. Halpern, and G. L. Myers (ed.), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical biology and biophysics. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 2.Baker, C. C. 1997. Posttranscriptional regulation of papillomavirus gene expression. In S. R. Billakanti, C. E. Calef, A. D. Farmer, A. L. Halpern, and G. L. Myers (ed.), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical biology and biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 3.Barksdale, S., and C. C. Baker. 1995. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J. Virol. 69:6553-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 5.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 7.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., H. Sumiyoshi, J. T. Oxford, H. Yoshioka, F. Ramirez, and N. P. Morris. 2001. Cis-acting elements regulate alternative splicing of exons 6A, 6B and 8 of the alpha1(XI) collagen gene and contribute to the regional diversification of collagen XI matrices. Matrix Biol. 20:589-599. [DOI] [PubMed] [Google Scholar]
- 9.Collier, B., D. Öberg, X. Zhao, and S. Schwartz. 2002. Specific inactivation of inhibitory sequences in the 5′ end of the human papillomavirus type 16 L1 open reading frame results in production of high levels of L1 protein in human epithelial cells. J. Virol. 76:2739-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke, C., and J. C. Alwine. 1996. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol. Cel.l Biol. 16:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke, C., H. Hans, and J. C. Alwine. 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 19:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulter, L. R., M. A. Landree, and T. A. Cooper. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cumming, S. A., C. E. Repellin, M. McPhilips, J. C. Redford, J. B. Clements, and S. V. Graham. 2002. The human papillomavirus type 31 untranslated region contains a complex bipartite negative regulatory element. J. Virol. 76:5993-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. Millar, and J. Doorbar. 2002. Identification of a G2 arrest domain in the E1∧E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, W., G. Jin, B. Y. Lin, B. A. Van Tine, T. R. Broker, and L. T. Chow. 2003. mRNA splicing regulates human papillomavirus type 11 E1 protein production and DNA replication. J. Virol. 77:10213-10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 17.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 18.Doorbar, J., and G. Myers. 1996. The E4 protein, p. 58-80. In G. Myers, H. Delius, J. Icenogel, H.-U. Bernard, H. Halpern, C. Baker, and C. Wheeler (ed.), Human papillomaviruses 1996, vol. III. Los Alamos National Laboratory, Los Alamos, N.Mex. [Google Scholar]
- 19.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 20.Fournier, N., K. Raj, P. Saudan, S. Utzig, R. Sahli, V. Simanis, and P. Beard. 1999. Expression of human papillomavirus 16 E2 protein in Schizosaccharomyces pombe delays the initiation of mitosis. Oncogene 18:4015-4021. [DOI] [PubMed] [Google Scholar]
- 21.Frattini, M. G., S. D. Hurst, H. B. Lim, S. Swaminathan, and L. A. Laimins. 1997. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 16:318-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 65:5806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gersappe, A., and D. J. Pintel. 1999. CA- and purine-rich elements form a novel bipartite exon enhancer which governs inclusion of the minute virus of mice NS2-specific exon in both singly and doubly spliced mRNAs. Mol. Cell. Biol. 19:364-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunderson, S. I., M. Polycarpou-Schwarz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 27.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 28.Ho, G. Y., R. D. Burk, S. Klein, A. S. Kadish, C. J. Chang, P. Palan, J. Basu, R. Tachezy, R. Lewis, and S. Romney. 1995. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J. Natl. Cancer Inst. 87:1365-1371. [DOI] [PubMed] [Google Scholar]
- 29.Howley, P. M., and D. R. Lowy. 2001. Papillomaviridae and their replication, p. 2197-2229. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 30.Hummel, M., H. B. Lim, and L. A. Laimins. 1995. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J. Virol. 69:3381-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1991. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 65:2093-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavigueur, A., H. La Branche, A. R. Kornblihtt, and B. Chabot. 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 7:2405-2417. [DOI] [PubMed] [Google Scholar]
- 33.Niwa, M., and S. M. Berget. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 5:2086-2095. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 35.Öberg, D., B. Collier, X. Zhao, and S. Schwartz. 2003. Mutational inactivation of two distinct negative RNA elements in the human papillomavirus type 16 L2 coding region induces production of high levels of L2 in human cells. J. Virol. 77:11674-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozbun, M. A., and C. Meyers. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkin, D. M., F. I. Bray, and S. S. Devesa. 2001. Cancer burden in the year 2000. The global picture. Eur. J. Cancer 37(Suppl. 8):24-66. [DOI] [PubMed] [Google Scholar]
- 38.Raj, K., S. Berguerand, S. Southern, J. Doorbar, and P. Beard. 2004. E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 78:7199-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rollman, E., L. Arnheim, B. Collier, D. Öberg, H. Hall, J. Klingström, J. Dillner, D. V. Pastrana, C. B. Buck, J. Hinkula, B. Wahren, and S. Schwartz. 2004. HPV-16 L1 genes with inactivated negative RNA elements induce potent immune responses. Virology 322:182-189. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz, S., X. Zhao, D. Öberg, and M. Rush. 2004. Regulation of papillomavirus late gene expression. Recent Res. Dev. Virol. 6:29-45. [Google Scholar]
- 41.Shah, K. V., and P. M. Howley. 1996. Papillomaviruses, p. 2077-2109. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 42.Sokolowski, M., W. Tan, M. Jellne, and S. Schwartz. 1998. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 72:1504-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stickeler, E., S. D. Fraser, A. Honig, A. L. Chen, S. M. Berget, and T. A. Cooper. 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tacke, R., M. Tohyama, S. Ogawa, and J. L. Manley. 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93:139-148. [DOI] [PubMed] [Google Scholar]
- 45.Tan, W., B. K. Felber, A. S. Zolotukhin, G. N. Pavlakis, and S. Schwartz. 1995. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol. 69:5607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, W., and S. Schwartz. 1995. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 69:2932-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terhune, S. S., W. G. Hubert, J. T. Thomas, and L. A. Laimins. 2001. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. J. Virol. 75:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]
- 49.Tian, M., and T. Maniatis. 1993. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74:105-114. [DOI] [PubMed] [Google Scholar]
- 50.Tran, Q., T. P. Coleman, and J. R. Roesser. 2003. Human transformer 2beta and SRp55 interact with a calcitonin-specific splice enhancer. Biochim. Biophys. Acta 1625:141-152. [DOI] [PubMed] [Google Scholar]
- 51.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, X., D. Öberg, M. Rush, J. Fay, H. Lambkin, and S. Schwartz. 2005. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 79:4270-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, X., M. Rush, and S. Schwartz. 2004. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 78:10888-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, Z. M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11:278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, Z. M., P. He, and C. C. Baker. 1996. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J. Virol. 70:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Z. M., P. J. He, and C. C. Baker. 1997. Structural, functional, and protein binding analyses of bovine papillomavirus type 1 exonic splicing enhancers. J. Virol. 71:9096-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, Z. M., M. Huynen, and C. C. Baker. 1998. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc. Natl. Acad. Sci. USA 95:14088-14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng, Z. M., E. S. Reid, and C. C. Baker. 2000. Utilization of the bovine papillomavirus type 1 late-stage-specific nucleotide 3605 3′ splice site is modulated by a novel exonic bipartite regulator but not by an intronic purine-rich element. J. Virol. 74:10612-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10:1356-1368. [DOI] [PubMed] [Google Scholar]
- 60.zur Hausen, H. 1999. Papillomaviruses in human cancers. Proc. Assoc. Am. Physicians 111:581-587. [DOI] [PubMed] [Google Scholar]