The challenge
Complete sequencing of genomes for human, mouse, and several other species was a technological breakthrough that identified and mapped thousands of genes and non-coding regions, much of which had heretofore been unknown. But it soon became apparent that significant knowledge gaps existed in understanding the in vivo function of most of these genes. Scientific research progress to address this deficiency was painstakingly slow and arduous, resulting in only partial functional annotation of a small number of well-characterized genes and gene sets. This self-fulfilling research paradigm overlooked genes with little to no known function, leaving in its wake a neglected “dark” genome. To accelerate progress and reveal gene function and insights into genetic associations and causes of disease, a fundamental shift from incremental steps to transformative change was needed. In response, a collaborative, global initiative emerged to systematically generate and phenotype a comprehensive collection of genetically modified “knockout” mouse models. This mandate was adopted and implemented by the International Mouse Phenotyping Consortium (IMPC) (Brown and Moore 2012a, b), a network of 21 academic research institutions across 15 countries on 5 continents, including leading laboratories from Europe, North America, Asia, and Africa. IMPC members agreed a mission to “create a comprehensive catalog of mammalian gene function that is freely available for researchers” by producing mouse models with targeted disruptions of every human orthologous protein-coding gene in the mouse genome. These knockout models have been and continue to be subjected to a standardized series of phenotyping assays across multiple body systems (Brown and Moore 2012a, b; Brown et al. 2005), allowing for identification of key biological processes and functional pleiotropy (Brown and Lad 2019), sexual dimorphism (Karp et al. 2017; Wilson et al. 2022), and essentiality for each gene (Cacheiro et al. 2020). Depositing mice and data into publicly accessible repositories are making these resources available for researchers around the world to extend this new knowledge into studies of the genetic effects on specific disease mechanisms. These efforts aim to accelerate disease diagnoses, identify new druggable targets, develop novel therapeutic interventions, and enact effective disease prevention strategies (Groza et al. 2023).
The impact
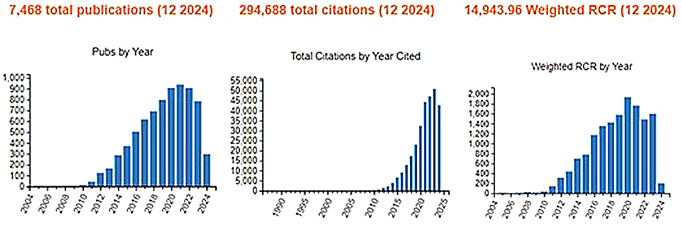
To date, data emerging from the study of IMPC mice has become an invaluable scientific resource for the biomedical research community, facilitating the study of gene function and the identification of novel therapeutic targets for human diseases. The vast phenotypic data generated not only in the project consortium itself but also by the greater biomedical research community using IMPC-generated mouse models and data has substantially enhanced our understanding of gene-disease relationships and genetic influences on mechanisms of disease. A publication tracking system using natural language processing methods, followed by annotator reviews through an IMPC-specific literature monitoring and curation tool (Cacheiro et al. 2024), identified nearly 7,500 papers that have used IMPC mice, data, and/or biomaterials to expand preclinical knowledge on a variety of diseases and disorders, including cardiac dysfunction (Guo et al. 2018; Wang et al. 2019; Spielmann et al. 2022), schizophrenia (Mihali et al. 2012; Lago and Bahn 2022; Garrett et al. 2024), Alzheimer’s (Rao et al. 2020; Cheng et al. 2021; Wang et al. 2021), ciliopathies (Wang et al. 2020; Higgins et al. 2022), osteoporosis (Swan et al. 2020; Formosa et al. 2021; Stein et al. 2023), metabolic syndrome (Ng and Gloyn 2013; Rozman et al. 2018; Andersen et al. 2022), hearing loss (Bowl et al. 2017; Trpchevska et al. 2022), developmental conditions(Dickinson et al. 2016; Dhombres et al. 2022), ophthalmic disorders (Khaled et al. 2019; Chee et al. 2023; Fritsche et al. 2016), dermatopathologies (Morell et al. 2022), and others. As shown in Fig. 1, according to NIH iCite, a digital tracking tool for citations (Hutchins et al. 2019a, b), influence (Hutchins et al. 2016, 2017) and the prediction of translational progress (Hutchins et al. 2019a, b), the exact number of 7468 IMPC-related publications from 2005 to 2024 have resulted in a total of 294,688 citations. These publications originated either from consortia contributing to the development of IMPC like EUMORPHIA (Brown et al. 2005), EUCOMM (Friedel et al. 2007), EUMODIC and Sanger MGP (Hrabe et al. 2015; Ayadi et al. 2012), from the IMPC itself, or from researchers using IMPC resources. Altogether they resulted in a weighted relative citation ratio (RCR) of 14943.96, more than double the number of total publications, indicating a highly influential set of articles. These achievements, and surely more to come, will fuel successful research grant applications and publication of more scientific papers on an even greater variety of disease topics in the future, especially as newer data are added to the growing catalogue of mouse models for more genes in the remaining years of the project.
Fig. 1.
Influence of IMPC publications according to NIH iCite (https://icite.od.nih.gov) on 5 December 2024
An expanding community of users and growing numbers of papers indicate that the IMPC is fulfilling expectations and delivering new scientific knowledge about genes and gene function that is useful for the scientific community. While this demonstrates the IMPC has been effectively illuminating the previously dark genome, these metrics do not reflect the translational impact of IMPC. For example, how has IMPC inspired clinical insights, catalyzed the accuracy and speed of disease diagnoses, accelerated the identification of novel druggable targets, led to the development of new or repurposing of existing therapeutics, or validated effective preventative strategies. These are very difficult measures to assess. An analysis using the translation module of NIH iCite that predicts the translation of scientific knowledge into clinical studies (Hutchins et al. 2019a, b) reveals that 789 of the 7468 IMPC publications mentioned above have been cited in clinical publications. Indeed, by identifying and characterizing disease-associated genes, IMPC resources are contributing to the development of preclinical models with predictive value for eliminating diagnostic odysseys, enhancing drug discovery, and reducing disease incidence. As a platform for translating newly revealed molecular mechanisms underlying diseases to impacts on human health, IMPC resources have contributed significantly to improvements in the diagnosis, treatment, and prevention of a wide range of conditions, from rare genetic disorders to common diseases like cancer, cardiovascular diseases, cognitive decline, and metabolic disorders.
Moreover, phenotype observations from IMPC knockout mice have directly impacted clinical medical practice, especially in personalized medicine. Our recent analysis highlighting the value of the IMPC for human genetic studies found that the resource has been implicated in at least 109 validated rare disease–gene associations over the last decade (Cacheiro et al. 2024). In addition, knockout models of human disease-associated genes have been crucial for evaluating the efficacy and safety of new therapies and improving the precision of treatments tailored to individual genetic profiles of patients. The public health impact of IMPC is also substantial in that it fosters the development of new diagnostic tools and therapies that can address unmet clinical needs of patient populations. Specific examples for this usage of the IMPC resource in preclinical research are described in the following paragraphs.
Specific examples
The IMPC has facilitated the development of new tools for diagnosing, managing, and preventing complex diseases, as studies using IMPC mice have led to discovery and characterization of potential biomarkers that could be used for early detection and diagnosis, monitoring disease progression, prognostication, and the development of personalized therapeutic strategies. For example, studies using Prkcb knockout mice support PKC-beta as a potential new drug target for the treatment of high-fat induced non-alcoholic fatty liver disease (Shu et al. 2021). In addition, mice with low Tgfbr2 expression are predisposed to spontaneous gastrointestinal tract tumors, suggesting that TGFBR2 could be a potential biomarker for early detection of colorectal cancer in humans (Gough et al. 2021). Experiments using Bcl2l11 knockout mice showing premature neuronal apoptosis have led to investigations into using BCL2L11 as a biomarker to assess neurodegenerative disorders and cognitive decline in Alzheimer’s and Parkinson’s diseases (Sionov et al. 2015). Further, Fto knockout mice showing metabolic dysfunction have prompted studies of FTO gene variants as biomarkers for obesity risk in humans (Najd-Hassan-Bonab et al. 2022). Also, altered immune responses and increased susceptibility to lupus-like phenotypes in Ifnar1 knockout mice have highlighted interferon (IFN) signaling pathways and IFN-induced biomarkers as potential diagnostic markers of autoimmune disease (Ban et al. 2021).
As well as being used for investigating novel biomarkers, IMPC knockout mouse models have also uncovered fundamental biological mechanisms that have accelerated drug discovery pipelines by helping researchers identify new drug targets, validate existing ones, and assess drug efficacy and safety. Experiments using IMPC mice have supported research studies targeting Tgfbr2 for cancer drug discovery (Gough et al. 2021), S1pr1 for neurological diseases (Kandjani et al. 2023), Fto for obesity and metabolic diseases (Azzam et al. 2022), Sirt6 for aging and cancer (Akter et al. 2021), and Prokr2 for obesity and circadian rhythm disorders (Sarfati et al. 2010; Martinez-Mayer and Perez-Millan 2023). Studies on these models have provided preclinical validation of drug targets and help optimize therapeutic strategies.
The IMPC has also provided critical insights into the genetic basis of diseases, identifying key molecular pathways and potential therapeutic targets for precision medicine and pharmacogenomics. Research using IMPC mouse models have demonstrated how genetic alterations influence disease phenotypes, guiding the design of safe and effective treatments that are tailored to individual genetic profiles. For example, drug efficacy in gene-targeted therapeutics have been predicted in studies on Tgfbr2 knockout mice (Zhao et al. 2024), drug safety associated with genetic variability has been assessed in Cpd2d6 mice (Taylor et al. 2020), and genetic factors that make individuals susceptible to type 2 diabetes, obesity, and cardiovascular diseases have been identified using Pparg mice (Lefterova et al. 2014). With respect to brain research, Slc20a2 mice have provided a solid preclinical model to study the development and treatment of a rare neurodegenerative disorder of brain calcification (Jensen et al. 2018), P2rx7 mice can be used to study immunological features and subtypes of depression (Urbina-Trevino et al. 2022), and several IMPC lines are suitable models for drug repurposing studies in schizophrenia (Lago and Bahn 2022). These models have helped researchers better understand the genetic underpinnings of diseases, predicted drug responses, optimized therapeutic strategies, and identified at-risk populations who may benefit from personalized treatments.
For example, Kmo (kynurenine 3-monooxygenase), an enzyme in the kynurenine pathway, plays a role in the excessive inflammatory response to pancreatitis which involves release of pro-inflammatory cytokines, oxidative stress, and activation of immune cells (Mole et al. 2016). Kmo knockout mice produced by the KOMP2 project reduced levels of inflammatory cytokines like quinolinic acid and prevented the excessive activation of macrophages and neutrophils in an experimental model of acute pancreatitis. Not only did this study provide compelling evidence that the Kmo knockout mouse was a valuable tool for understanding genetic regulation of acute pancreatitis, but it also demonstrated that Kmo inhibition could modulate metabolic pathways related to inflammation. This finding may be especially relevant in the context of critical illness and organ failure. In this way, these studies have laid the groundwork for exploring Kmo as a druggable target for the development of KMO inhibitors to control inflammation, prevent organ damage, and improve survival in acute pancreatitis and other diseases.
Notably, in recent years IMPC has also facilitated research exploring the feasibility and efficacy of gene therapy approaches in a broad range of disease areas. Examples range from genetic forms of deafness or hearing loss and associated vestibular deficits (Michalski and Petit 2022; Ding et al. 2021; Maudoux et al. 2022), restoration of visual function for blue cone monochromacy and retinal degeneration (Deng et al. 2018, 2019; Beryozkin et al. 2021; Qian et al. 2022; Hsu et al. 2023; Lu et al. 2023; Abu-Diab et al. 2023), managing hereditary spastic paraplegia (Hauser et al. 2019; Chen et al. 2023; Lim et al. 2024) and other upper motor neuron diseases like amyotrophic lateral sclerosis (Genc et al. 2022), and treating mitochondrial myopathy (Pereira et al. 2020), muscular dystrophies (Li et al. 2023; Hindi et al. 2023), and cardiac dysfunction (Li et al. 2020; Magadum et al. 2021; Martin et al. 2021; Wingert et al. 2024). Other examples include evaluation of new therapies for lysosomal and glycogen storage diseases (Koeberl et al. 2024; Gardin et al. 2024; Chen et al. 2022; Lim et al. 2020; Vidal et al. 2018; Goodman et al. 2021), neurodegenerative disorders (Jaillard et al. 2021; Lee et al. 2023; Jonquieres et al. 2018), metabolic dysfunctions (Khoja et al. 2018; Pontoizeau et al. 2022, 2024; Sonaimuthu et al. 2021) and mucopolysaccharidosis (Roca et al. 2017). These preclinical studies prompted by IMPC-generated data and resources have inspired the development of treatments for rare genetic diseases for which no treatments currently exist and which are often fatal, with the possibility to significantly impact patients’ lives. The examples above also show that IMPC provides insights not only into rare monogenetic diseases where only limited patient cohorts exist, but also for more common and age-related conditions like blindness, hearing loss, and heart failure. In the latter cases a mouse model can be a useful tool for the development of a therapeutic intervention in preclinical research if it features an aspect of the pathological mechanism that is targeted.
In summary, the IMPC is a cornerstone of translating functional genomics into clinical impact despite its limitations. An obvious limitation is the choice of a single inbred strain to produce all IMPC knockout mouse lines for feasibility reasons. It is well known that phenotypes can change depending on genetic modifiers in different genetic backgrounds, as e.g. already shown for social behaviour (Arbogast et al. 2016), vision (Hoelter et al. 2008) and hearing (Newton et al. 2023), which also implies that the IMPC programme should be considered a sensitized screen. Nevertheless, the mice, data, and biomaterials it has and continues to produce are shaping the future of biomedical research, drug development, and medical practice. These contributions are crucial for making real impacts to advance precision medicine, improve patient outcomes, and enhance public health worldwide.
Author contributions
All authors contributed to the conception and design of the manuscript. The first draft was written by KCKL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
This work was supported in part by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung [BMBF]) and the Bavarian State Ministry of Science and the Arts (Staatsministerium für Wissenschaft und Kunst) within the initial phase of the German Center for Mental Health (DZPG), grant: 01EE2303E, and by NIH grants (UM1OD023221) to the Davis-Toronto-Collaborative-Consortium (DTCC) and UM1HG006370 to the Mouse Phenotying Informatics (MPI2) consortium.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
SMH is a member of the EBRAINS Science and Technology Committee and IMPC Vice Chair.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abu-Diab A et al (2023) Homozygous knockout of Cep250 leads to a relatively late-onset Retinal Degeneration and Sensorineural hearing loss in mice. Transl Vis Sci Technol 12(3):3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter R et al (2021) A Comprehensive Analysis into the Therapeutic Application of Natural Products as SIRT6 Modulators in Alzheimer’s Disease, Aging, Cancer, Inflammation, and Diabetes. Int J Mol Sci, 22(8) [DOI] [PMC free article] [PubMed]
- Andersen MK et al (2022) Loss of sucrase-isomaltase function increases acetate levels and improves metabolic health in Greenlandic cohorts. Gastroenterology 162(4):1171–1182e3 [DOI] [PubMed] [Google Scholar]
- Arbogast T et al (2016) Reciprocal effects on Neurocognitive and metabolic phenotypes in mouse models of 16p11.2 deletion and duplication syndromes. PLoS Genet 12(2):e1005709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A et al (2012) Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome 23(9–10):600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam SK, Alsafar H, Sajini AA (2022) FTO m6A demethylase in obesity and Cancer: implications and underlying molecular mechanisms. Int J Mol Sci, 23(7) [DOI] [PMC free article] [PubMed]
- Ban T et al (2021) Genetic and chemical inhibition of IRF5 suppresses pre-existing mouse lupus-like disease. Nat Commun 12(1):4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beryozkin A et al (2021) A new mouse model for retinal degeneration due to Fam161a deficiency. Sci Rep, 11(1): p. 2030 [DOI] [PMC free article] [PubMed]
- Bowl MR et al (2017) A large scale hearing loss screen reveals an extensive unexplored genetic landscape for auditory dysfunction. Nat Commun 8(1):886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDM, Lad HV (2019) The dark genome and pleiotropy: challenges for precision medicine. Mamm Genome 30(7–8):212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW (2012a) Towards an encyclopaedia of mammalian gene function: the International mouse phenotyping Consortium. Dis Model Mech 5(3):289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW (2012b) The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome 23(9–10):632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Chambon P, de Angelis MH (2005) EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet 37(11):1155 [DOI] [PubMed] [Google Scholar]
- Cacheiro P et al (2020) Human and mouse essentiality screens as a resource for disease gene discovery. Nat Commun 11(1):655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheiro P et al (2024) Computational identification of disease models through cross-species phenotype comparison. Dis Model Mech, 17(6) [DOI] [PMC free article] [PubMed]
- Chee JM et al (2023) Genome-wide screening reveals the genetic basis of mammalian embryonic eye development. BMC Biol 21(1):22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X et al (2022) AAV9/MFSD8 gene therapy is effective in preclinical models of neuronal ceroid lipofuscinosis type 7 disease. J Clin Invest, 132(5) [DOI] [PMC free article] [PubMed]
- Chen X et al (2023) Intrathecal AAV9/AP4M1 gene therapy for hereditary spastic paraplegia 50 shows safety and efficacy in preclinical studies. J Clin Invest, 133(10) [DOI] [PMC free article] [PubMed]
- Cheng B et al (2021) Triggering receptor expressed on myeloid Cells-2 (TREM2) interacts with colony-stimulating factor 1 receptor (CSF1R) but is not necessary for CSF1/CSF1R-Mediated microglial survival. Front Immunol 12:633796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hrabe M et al (2015) Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet 47(9):969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WT et al (2018) Human L- and M-opsins restore M-cone function in a mouse model for human blue cone monochromacy. Mol Vis 24:17–28 [PMC free article] [PubMed] [Google Scholar]
- Deng WT et al (2019) Rescue of M-cone function in aged Opn1mw-/- mice, a model for late-stage Blue Cone Monochromacy. Invest Ophthalmol Vis Sci 60(10):3644–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhombres F et al (2022) Prenatal phenotyping: a community effort to enhance the human phenotype ontology. Am J Med Genet C Semin Med Genet 190(2):231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME et al (2016) High-throughput discovery of novel developmental phenotypes. Nature 537(7621):508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N et al (2021) Advances in genome editing for genetic hearing loss. Adv Drug Deliv Rev 168:118–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa MM et al (2021) A Roadmap to Gene discoveries and Novel therapies in monogenic low and high bone Mass disorders. Front Endocrinol (Lausanne) 12:709711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel RH et al (2007) EUCOMM–the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic 6(3):180–185 [DOI] [PubMed] [Google Scholar]
- Fritsche LG et al (2016) A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 48(2):134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin A et al (2024) A functional mini-GDE transgene corrects impairment in models of glycogen storage disease type III. J Clin Invest, 134(2) [DOI] [PMC free article] [PubMed]
- Garrett L et al (2024) Co-expression of prepulse inhibition and schizophrenia genes in the mouse and human brain. Neurosci Appl 3:104075 [Google Scholar]
- Genc B et al (2022) Upper motor neurons are a target for gene therapy and UCHL1 is necessary and sufficient to improve cellular integrity of diseased upper motor neurons. Gene Ther 29(3–4):178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S et al (2021) Deficiency of the sedoheptulose kinase (Shpk) does not alter the ability of hematopoietic stem cells to rescue cystinosis in the mouse model. Mol Genet Metab 134(4):309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough NR, Xiang X, Mishra L (2021) TGF-beta signaling in liver, pancreas, and gastrointestinal diseases and Cancer. Gastroenterology 161(2):434–452e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groza T et al (2023) The International mouse phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res 51(D1):D1038–D1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R et al (2018) Cardiomyocyte-specific disruption of Cathepsin K protects against doxorubicin-induced cardiotoxicity. Cell Death Dis 9(6):692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser S et al (2019) mRNA as a Novel Treatment Strategy for Hereditary Spastic Paraplegia Type 5. Mol Ther Methods Clin Dev 15:359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins K et al (2022) Analysis of genome-wide knockout mouse database identifies candidate ciliopathy genes. Sci Rep 12(1):20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindi SM et al (2023) Enveloped viruses pseudotyped with mammalian myogenic cell fusogens target skeletal muscle for gene delivery. Cell 186(16):3520 [DOI] [PubMed] [Google Scholar]
- Hoelter SM et al (2008) Sighted C3H mice–a tool for analysing the influence of vision on mouse behaviour? Front Biosci 13:5810–5823 [DOI] [PubMed] [Google Scholar]
- Hsu Y et al (2023) Subretinal gene therapy delays vision loss in a Bardet-Biedl syndrome type 10 mouse model. Mol Ther Nucleic Acids 31:164–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins BI et al (2016) Relative citation ratio (RCR): a New Metric that uses Citation Rates to measure influence at the article level. PLoS Biol 14(9):e1002541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins BI et al (2017) Additional support for RCR: a validated article-level measure of scientific influence. PLoS Biol 15(10):e2003552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins BI et al (2019a) The NIH Open Citation Collection: a public access, broad coverage resource. PLoS Biol 17(10):e3000385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins BI et al (2019b) Predicting translational progress in biomedical research. PLoS Biol 17(10):e3000416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard C et al (2021) The metabolic signaling of the nucleoredoxin-like 2 gene supports brain function. Redox Biol 48:102198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen N et al (2018) Mice knocked out for the primary brain calcification-Associated Gene Slc20a2 Show Unimpaired prenatal survival but retarded growth and nodules in the brain that grow and calcify over Time. Am J Pathol 188(8):1865–1881 [DOI] [PubMed] [Google Scholar]
- Kandjani OJ et al (2023) S1PR1 modulators in multiple sclerosis: efficacy, safety, comparison, and chemical structure insights. Eur J Med Chem 250:115182 [DOI] [PubMed] [Google Scholar]
- Karp NA et al (2017) Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat Commun 8:15475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled ML et al (2019) PPIP5K2 and PCSK1 are Candidate Genetic Contributors to familial Keratoconus. Sci Rep 9(1):19406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoja S et al (2018) Conditional disruption of hepatic carbamoyl phosphate synthetase 1 in mice results in hyperammonemia without orotic aciduria and can be corrected by liver-directed gene therapy. Mol Genet Metab 124(4):243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeberl DD et al (2024) Gene therapy for glycogen storage diseases. J Inherit Metab Dis 47(1):93–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago SG, Bahn S (2022) The druggable schizophrenia genome: from repurposing opportunities to unexplored drug targets. NPJ Genom Med 7(1):25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J et al (2023) ANKS1A regulates LDL receptor-related protein 1 (LRP1)-mediated cerebrovascular clearance in brain endothelial cells. Nat Commun 14(1):8463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI et al (2014) PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol Metab 25(6):293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y et al (2020) gp130 controls cardiomyocyte proliferation and heart regeneration. Circulation 142(10):967–982 [DOI] [PubMed] [Google Scholar]
- Li H et al (2023) Systemic AAV9.BVES delivery ameliorates muscular dystrophy in a mouse model of LGMDR25. Mol Ther 31(2):398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JA et al (2020) A Novel Gene Therapy Approach for GSD III using an AAV Vector Encoding a bacterial glycogen debranching enzyme. Mol Ther Methods Clin Dev 18:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH et al (2024) ARL6IP1 gene delivery reduces neuroinflammation and neurodegenerative pathology in hereditary spastic paraplegia model. J Exp Med, 221(1) [DOI] [PMC free article] [PubMed]
- Lu J et al (2023) Gene augmentation therapy to rescue degenerative photoreceptors in a Cwc27 mutant mouse model. Exp Eye Res 234:109596 [DOI] [PubMed] [Google Scholar]
- Magadum A et al (2021) Therapeutic delivery of Pip4k2c-Modified mRNA attenuates Cardiac Hypertrophy and Fibrosis in the failing heart. Adv Sci (Weinh) 8(10):2004661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TG et al (2021) Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun 12(1):2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mayer J, Perez-Millan MI (2023) Phenotypic and genotypic landscape of PROKR2 in neuroendocrine disorders. Front Endocrinol (Lausanne) 14:1132787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux A, Vitry S, El-Amraoui A (2022) Vestibular deficits in deafness: clinical presentation, animal modeling, and treatment solutions. Front Neurol 13:816534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski N, Petit C (2022) Central auditory deficits associated with genetic forms of peripheral deafness. Hum Genet 141(3–4):335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihali A et al (2012) Modeling resilience to schizophrenia in genetically modified mice: a novel approach to drug discovery. Expert Rev Neurother 12(7):785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole DJ et al (2016) Kynurenine-3-monooxygenase inhibition prevents multiple organ failure in rodent models of acute pancreatitis. Nat Med 22(2):202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M et al (2022) SIDT1 plays a key role in type I IFN responses to nucleic acids in plasmacytoid dendritic cells and mediates the pathogenesis of an imiquimod-induced psoriasis model. EBioMedicine 76:103808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najd-Hassan-Bonab L et al (2022) The role of FTO variant rs1421085 in the relationship with obesity: a systematic review and meta-analysis. Eat Weight Disord 27(8):3053–3062 [DOI] [PubMed] [Google Scholar]
- Newton S et al (2023) Absence of Embigin accelerates hearing loss and causes sub-viability, brain and heart defects in C57BL/6 N mice due to interaction with Cdh23(ahl). iScience 26(10):p108056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HJ, Gloyn AL (2013) Bridging the gap between genetic associations and molecular mechanisms for type 2 diabetes. Curr Diab Rep 13(6):778–785 [DOI] [PubMed] [Google Scholar]
- Pereira CV et al (2020) Myopathy reversion in mice after restauration of mitochondrial complex I. EMBO Mol Med 12(2):e10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoizeau C et al (2022) Neonatal gene therapy achieves sustained disease rescue of maple syrup urine disease in mice. Nat Commun 13(1):3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoizeau C et al (2024) Successful treatment of severe MSUD in Bckdhb(-/-) mice with neonatal AAV gene therapy. J Inherit Metab Dis 47(1):41–49 [DOI] [PubMed] [Google Scholar]
- Qian X et al (2022) AAV8-Mediated gene therapy rescues retinal degeneration phenotype in a Tlcd3b knockout mouse model. Invest Ophthalmol Vis Sci 63(3):11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV et al (2020) Amyloid-beta accumulation cycle as a prevention and/or therapy target for Alzheimer’s disease. Aging Cell 19(3):e13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca C et al (2017) Disease correction by AAV-mediated gene therapy in a new mouse model of mucopolysaccharidosis type IIID. Hum Mol Genet 26(8):1535–1551 [DOI] [PubMed] [Google Scholar]
- Rozman J et al (2018) Identification of genetic elements in metabolism by high-throughput mouse phenotyping. Nat Commun 9(1):288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfati J, Dode C, Young J (2010) Kallmann syndrome caused by mutations in the PROK2 and PROKR2 genes: pathophysiology and genotype-phenotype correlations. Front Horm Res 39:121–132 [DOI] [PubMed] [Google Scholar]
- Shu Y et al (2021) Hepatocyte-specific PKCbeta deficiency protects against high-fat diet-induced nonalcoholic hepatic steatosis. Mol Metab 44:101133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov RV, Vlahopoulos SA, Granot Z (2015) Regulation of Bim in Health and Disease. Oncotarget 6(27):23058–23134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonaimuthu P et al (2021) Gene delivery corrects N-acetylglutamate synthase deficiency and enables insights in the physiological impact of L-arginine activation of N-acetylglutamate synthase. Sci Rep 11(1):3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann N et al (2022) Extensive identification of genes involved in congenital and structural heart disorders and cardiomyopathy. Nat Cardiovasc Res 1(2):157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M et al (2023) Why animal experiments are still indispensable in Bone Research: A Statement by the European Calcified Tissue Society. J Bone Min Res 38(8):1045–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan AL et al (2020) Mouse mutant phenotyping at scale reveals novel genes controlling bone mineral density. PLoS Genet 16(12):e1009190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C et al (2020) A review of the important role of CYP2D6 in Pharmacogenomics. Genes (Basel), 11(11) [DOI] [PMC free article] [PubMed]
- Trpchevska N et al (2022) Genome-wide association meta-analysis identifies 48 risk variants and highlights the role of the stria vascularis in hearing loss. Am J Hum Genet 109(6):1077–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina-Trevino L, von Mucke-Heim IA, Deussing JM (2022) P2X7 Receptor-Related Genetic Mouse Models - Tools for Translational Research in Psychiatry. Front Neural Circuits 16:876304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal P et al (2018) Rescue of GSDIII phenotype with gene transfer requires liver- and muscle-targeted GDE expression. Mol Ther 26(3):890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jonquieres G et al (2018) Uncoupling N-acetylaspartate from brain pathology: implications for Canavan disease gene therapy. Acta Neuropathol 135(1):95–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT et al (2019) Cardioprotection by the mitochondrial unfolded protein response requires ATF5. Am J Physiol Heart Circ Physiol 317(2):H472–H478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y et al (2020) Impacts of ciliary neurotrophic factor on the retinal transcriptome in a mouse model of photoreceptor degeneration. Sci Rep 10(1):6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H et al (2021) Genome-scale metabolic network reconstruction of model animals as a platform for translational research. Proc Natl Acad Sci U S A, 118(30) [DOI] [PMC free article] [PubMed]
- Wilson LAB et al (2022) Sex differences in allometry for phenotypic traits in mice indicate that females are not scaled males. Nat Commun 13(1):7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert J et al (2024) Cardiomyocyte-specific RXFP1 overexpression protects against pressure overload-induced cardiac dysfunction independently of relaxin. Biochem Pharmacol 225:116305 [DOI] [PubMed] [Google Scholar]
- Zhao G et al (2024) TGF-betaR2 signaling coordinates pulmonary vascular repair after viral injury in mice and human tissue. Sci Transl Med 16(732):eadg6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.