Abstract
Cryptococcus neoformans var. neoformans strains have historically been divided into serotypes A and D on the basis of reactivity with rabbit sera. Previously, we noted that two murine immunoglobulin M monoclonal antibodies (MAbs) to the capsular glucuronoxylomannan produced different indirect immunofluorescence (IF) patterns, described as annular and punctate, when bound to C. neoformans cells from different strains. In this study, we examined the reactivity of these two MAbs, known as 12A1 and 13F1, with 20 C. neoformans var. neoformans strains, of which 13 were serotype A and 7 were serotype D. For all strains, MAb binding was studied by IF and agglutination assays. In addition, we blindly tested the IF patterns of 22 C. neoformans var. neoformans strains. For selected strains, MAb binding was studied by flow cytometry (FACScan) and phagocytosis assays. The epitopes recognized by MAbs 12A1 and 13F1 were found in all of the strains. MAb 12A1 binding produced an annular IF pattern with all of the strains, irrespective of the serotype classification. MAb 13F1 binding produced annular binding with all of the serotype A strains and punctate binding with 19 of 20 serotype D strains. In general, the punctate IF pattern was associated with lower fluorescence intensity, a requirement for higher antibody concentrations to produce yeast cell agglutination, and lower opsonic efficacy. Our results provide strong support for the existing classification of two serological types for strains assigned to variety neoformans and indicate qualitative and quantitative antigenic differences among serotype A and D strains.
Cryptococcus neoformans is unique among the pathogenic fungi in that it has a polysaccharide capsule which is a major virulence factor (15). Structural differences in the capsular polysaccharide result in antigenic differences that have been used to classify strains into four serotypes, known as A, B, C, and D (6). C. neoformans strains have also been classified into two varieties on the basis of several genetic and biochemical differences. C. neoformans var. neoformans comprises serotypes A and D, whereas C. neoformans var. gattii comprises serotypes B and C. The serotype classification for C. neoformans was originally developed in the 1940s by using reciprocally absorbed rabbit immune sera (11, 12). The usefulness of the serotype classification scheme has been limited by the fact that most var. neoformans strains have been grouped as serotype A, despite considerable evidence for structural variation in the glucuronoxylomannan (GXM) of strains assigned to this serotype (21). The relationship between serotype A and D strains is uncertain. Detailed structures for the GXMs of all of the serotypes have been proposed, but the molecular structures responsible for the antigenic differences which allow classification into particular serotypes are not understood (6).
Monoclonal antibody (MAb) technology provides a potential alternative to rabbit sera for generating reagents for the study of the antigenic composition of the capsule. Several groups have generated MAbs to the capsular polysaccharide of C. neoformans (1, 2, 4, 10, 14, 21, 22). Unfortunately, most of the MAbs studied to date are not specific for a given serotype (1). An exception is MAb E1, which binds distinctly to serotype A strains and can be useful for classifying strains (9). Recently, a MAb with specificity for serotype D strains has been described (14). The availability of MAbs that can consistently discriminate between C. neoformans var. neoformans strains may assist in the study of capsular structure.
Previously, we reported that two immunoglobulin M (IgM) MAbs derived from the same progenitor B cell bound to spatially different epitopes on the C. neoformans capsule (17, 20). In this study, we evaluated the binding of these MAbs to a larger set of well-characterized strains and correlated immunofluorescence (IF) binding patterns with agglutination, phagocytosis, and flow cytometry studies. The results indicate that IF patterns correlate with serotype classification and other serological assays.
(The data in this report are from a thesis to be submitted by W. Cleare in partial fulfillment of the requirements for the degree of doctor of philosophy at the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, N.Y.)
MATERIALS AND METHODS
Strains.
C. neoformans 24067, 34874, 28958, 34873, and 34870 were obtained from the American Type Culture Collection (Rockville, Md.). Strains J11A, SB4, SB6, J22, and J9A were isolated from patients with cryptococcal meningitis in New York City. Strains CN 6, CN 15, CN 98, CN 110, and CN 145 were provided by Stuart Levitz (Boston, Mass.); 184A was provided by Juneanne Murphy (Oklahoma City, Okla.). Strains 371, 62066, and H99 were obtained from J. E. Bennett (National Institutes of Health, Bethesda, Md.), Robert Cherniak (Atlanta, Ga.), and John Perfect (Durham, N.C.), respectively. The serotype classification of the strains listed in Table 1 was derived by classical rabbit serological methods or typed by nuclear magnetic resonance analysis, and the serotype assignment was confirmed by serology. Twenty-two additional C. neoformans var. neoformans isolates (9 serotype A and 13 serotype D) for blind IF analysis were provided by Mary Brandt (Atlanta, Ga.). All strains were maintained on Sabouraud dextrose agar slants (Difco Laboratories, Detroit, Mich.) at 4°C and grown in Sabouraud dextrose broth (Difco) at 30°C prior to use.
TABLE 1.
Serotypes, IF patterns, and agglutination endpoints of 20 C. neoformans strains
| Strain | Serotype | IF pattern
|
Agglutination endpoint (μg/ml)
|
||
|---|---|---|---|---|---|
| 12A1 | 13F1 | 12A1 | 13F1 | ||
| J11A | A | Annular | Annular | 0.98 | 1.95 |
| H99 | A | Annular | Annular | 3.9 | 3.8 |
| SB4 | A | Annular | Annular | 3.9 | 7.8 |
| 184A | A | Annular | Annular | 1.95 | 0.98 |
| CN15 | A | Annular | Annular | 1.95 | 7.8 |
| CN98 | A | Annular | Annular | 0.24 | 0.98 |
| CN145 | A | Annular | Annular | 7.8 | 7.8 |
| CN110 | A | Annular | Annular | 1.95 | 31.3 |
| CN6 | A | Annular | Annular | 7.8 | 62.5 |
| 62066 | A | Annular | Annular | 7.8 | 62.5 |
| SB6 | A | Annular | Annular | NDa | ND |
| 34870 | A | Annular | Annular | ND | ND |
| 371 | A | Annular | Annular | ND | ND |
| 24067 | D | Annular | Punctate | 7.8 | 62.5 |
| 34874 | D | Annular | Punctate | 0.48 | 15.6 |
| 34873 | D | Annular | Punctate | 3.9 | 31.3 |
| J9A | D | Annular | Punctate | 0.98 | 31.3 |
| 28957 | D | Annular | Punctate | 0.49 | 62.5 |
| 28958 | D | Annular | Punctate | 0.49 | 15.6 |
| J22 | D | Annular | Annular | 0.49 | 0.49 |
ND, not done.
MAbs.
MAbs 12A1 and 13F1 have already been described (2, 17, 20). Both are IgM and were generated from splenocytes of BALB/c mice immunized with the GXM-tetanus toxoid vaccine. Ascitic fluid containing hybridoma protein was generated by injecting 107 hybridoma cells into the peritoneal cavities of pristane-primed BALB/c mice and obtaining the fluid by paracentesis. Antibody concentration was determined by enzyme-linked immunosorbent assay relative to isotype-matched standards. For phagocytosis experiments, the antibodies were purified by mannose protein affinity chromatography in accordance with the manufacturer’s (Pierce, Rockford, Ill.) instructions.
Indirect IF.
Stationary-phase (3 to 5 days) yeast cells were washed three times in phosphate-buffered saline (PBS; 0.137 M NaCl, 0.003 M sodium phosphate, pH 7.4), and 106 cells were placed into microcentrifuge tubes (liquid method) or mounted onto poly-l-lysine-coated slides (Sigma, St. Louis, Mo.) (dry method). MAb (10 μg/ml) was added to cryptococcal cells, and the mixture was incubated for 2 h at room temperature. Yeast cells were washed with PBS and incubated with 10 μg of fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgM (Southern Biotechnology, Birmingham, Ala.) per ml for 1 h at room temperature in the dark. Cells were washed again with PBS and suspended in 50 μl of mounting medium consisting of a solution of 50% glycerol–0.1 M n-propyl gallate (Sigma) in PBS. The cells were mounted on glass slides and viewed with an Axiophot microscope (Zeiss, Thornwood, N.Y.) equipped with an FITC filter. Confocal microscopy was performed with an MRC 600 laser scanning confocal microscope (Bio-Rad, Hercules, Calif.). Single optical sections were collected at a magnification of ×4 with Nikon 60×N.A. 1.4 phase 4 optics.
Agglutination assay.
Agglutination assays were done with 96-well microtiter plates by using a modification of previously described protocols (7). Briefly, serial dilutions of MAb which corresponded to MAb concentrations of 500 to 0.0019 μg/ml in a solution of 75 mM NaCl–10 mM sodium phosphate buffer (pH 7.2)–0.5% bovine serum albumin–1 mM azide were done in 96-well polystyrene microtiter plates (no. 25801-96; Corning Glass Works, Corning, N.Y.). Approximately 5 × 104 yeast cells were added to each well. The plate was shaken for 15 min on a rotating platform (Eberbach Corp., Ann Arbor, Mich.) at moderate speed and incubated without shaking at 4°C overnight. Agglutination occurred rapidly, but the endpoint was read after overnight incubation because this resulted in more reproducible measurements. The agglutination endpoint was the highest dilution of MAb at which agglutination was observed.
Phagocytosis assay.
The J774.16 macrophage cell line originated from a murine reticulum cell sarcoma (8) and was used to study the opsonic efficacy of these MAbs for a selected set of strains as previously described (7). The J774.16 cells used were obtained from Barry Bloom (Bronx, N.Y.). Briefly, J774.16 cells were plated on tissue culture plates (no. 3695; Costar, Cambridge, Mass.) and stimulated with 500 U of murine recombinant gamma interferon (Genzyme, Cambridge, Mass.) and 3 μg of lipopolysaccharide (Sigma, St. Louis, Mo.) per ml. C. neoformans cells were washed and suspended in feeding medium (Dulbecco’s modified Eagle medium [Mediatech, Washington, D.C.] with 10% heat-inactivated fetal calf serum [Bioproducts for Science, Indianapolis, Ind.], 10% NCTC-109 [Gibco Laboratories, Life Technologies, Inc., Grand Island, N.Y.], and 1% nonessential amino acids [Cellgro; Mediatech]) and then added to the J774.16 monolayer in a macrophage-to-yeast ratio of 1:1 with either 20% fresh mouse serum, 20% heat-inactivated mouse serum, or MAb 12A1 or 13F1. Serum was obtained from BALB/c mice immediately before the assay. Whole blood was separated from serum by centrifugation. The serum used was either heat inactivated (56°C for 30 min) or fresh. The suspension was incubated for 2 h at 37°C. The J774.16 cell monolayer was then washed several times with PBS to remove nonadherent organisms, fixed with cold methanol, and stained with Giemsa (Sigma). The phagocytic index (PI) is the number of internalized yeast cells per number of macrophages per field. Internalized cells were differentiated from attached cells by their presence in a well-defined phagocytic vacuole. Previous studies using Uvitex B fluorescence staining have shown that one can easily discriminate between attached and internalized C. neoformans by microscopic examination of Giemsa-stained cultures (18, 19). PIs were determined by light microscopy (Nikon Diaphot; Nikon, Inc., Instrument Division, Garden City, N.J.) at a magnification of ×600. For each experiment, at least five fields were counted with approximately 200 macrophages per field.
Flow cytometry.
Flow cytometry (fluorescence-activated cell sorting [FACS]), performed on a FACScan (Becton Dickinson Immunocytometry Systems, San Jose, Calif.), was used to study the IF of individual cells. A single colony of each strain was grown in Sabouraud dextrose broth for 72 h. Cells were washed twice in sterile PBS and prepared as described above for IF studies. The suspension used for FACS consisted of 107 cells incubated for 1 h with 100 μg of either MAb 12A1 or 13F1 per ml, followed by two washes in sterile PBS. After incubation with FITC-labeled goat anti-mouse IgM, samples were washed and analyzed by using the FACScan equipped with an argon laser. Green fluorescence in C. neoformans cells was measured at a wavelength of 530 ± 15 nm. A total of 10,000 cells were analyzed for each colony. Cells without MAb gave the same results as cells incubated with an irrelevant IgM murine MAb and were therefore used as negative controls. Frequency histograms of fluorescence distribution were generated by plotting the number of cells (y axis) versus log fluorescence intensity (x axis) with the LYSIS II data analysis program (Becton Dickinson). The size distribution of cells with and without MAb staining was uniform when examined by forward scatter and side scatter parameters. Furthermore, there was no difference in light scatter between stained and unstained cells, indicating that the average particle size for both populations was the same.
Statistical analysis.
The Mann-Whitney rank sum test was used to determine the statistical significance of agglutination endpoints. Chi-square analysis and the Fisher exact test were used to determine the significance of the correlation between the binding pattern and the established serotype. The Student t test was used to compare fluorescence intensities measured by FACS analysis. Statistical analysis was done by using Primer of Statistics—The Program (McGraw Hill Co., New York, N.Y.).
RESULTS
Indirect IF.
To determine the ability of MAbs 12A1 and 13F1 to discriminate between serotype A and D strains, 20 C. neoformans var. neoformans strains whose serotypes are known (Table 1) were examined by IF assay. Twenty-two C. neoformans var. neoformans isolates (9 serotype A and 13 serotype D) were tested blindly. IF with MAb 12A1 produced an annular pattern with all of the strains, regardless of serotype classification. IF with MAb 13F1 produced an annular pattern with all of the serotype A strains and a punctate pattern with 19 (95%) of 20 serotype D strains. Photographs of annular and punctate IF patterns have been previously published (17, 20). These results indicate a statistically significant difference in the ability of MAb 13F1 to produce annular or punctate IF patterns on serotype A and D strains, respectively (P ≤ 0.001 by either chi square or Fisher exact test analysis). To determine whether the IF pattern was affected by the method used to prepare the sample, we compared the pattern obtained with cells suspended in liquid versus that obtained with cells attached to a slide by drying for five strains. Both methods produced consistent results. To determine if the MAb 13F1 IF pattern was dependent on the age of the culture, we analyzed IF patterns for the log (24-h) and stationary (72-h) growth phases of five strains. The same IF pattern was observed for both growth phases.
Agglutination studies.
Addition of MAbs 12A1 and 13F1 to each of the C. neoformans strains listed in Table 1 produced cell agglutination, but there were significant differences in the antibody concentration required for agglutination to occur. For strains with annular binding by MAb 12A1, the agglutination endpoints ranged from 0.24 to 7.8 μg/ml (geometric mean of 1.6 μg/ml). For strains with annular binding by MAb 13F1, the agglutination endpoints ranged from 0.49 to 62.5 μg/ml (geometric mean of 5.7 μg/ml). For strains with punctate binding by MAb 13F1, the agglutination endpoints ranged from 15.6 to 62.5 μg/ml (geometric mean of 31.2 μg/ml). Hence, C. neoformans agglutination required significantly higher concentrations of MAb 13F1 than MAb 12A1 and punctate binding was associated with higher agglutination endpoints (P ≤ 0.05).
Phagocytosis assay.
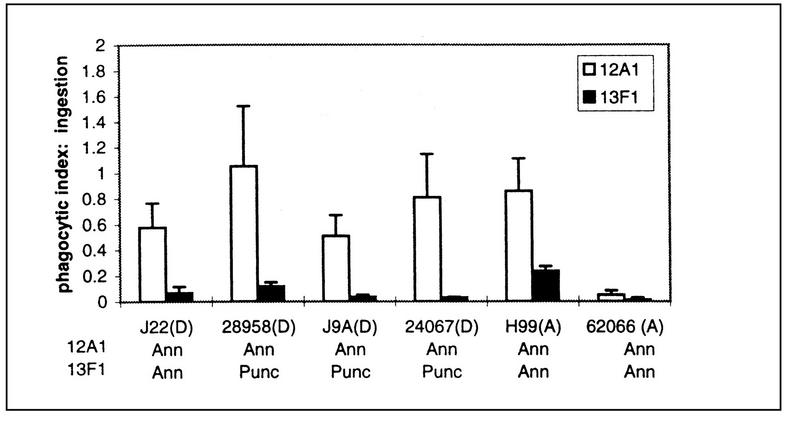
The PI was measured as the number of internalized C. neoformans cells divided by the number of macrophages. The PIs for six of the strains with MAbs 12A1 and 13F1 were determined. There were significant strain differences in the ability of MAbs 12A1 and 13F1 to promote ingestion, as well as differences in the ability of MAbs 12A1 and 13F1 to promote ingestion for a given strain. For all strains, the addition of MAb 12A1 to J774.16 and C. neoformans suspensions resulted in a higher PI than that measured for MAb 13F1 (Fig. 1).
FIG. 1.
PIs resulting from the incubation of several strains of C. neoformans with J774.16 cells in the presence of either MAb 12A1 or 13F1. PIs were measured by counting ingested cells. The x axis also denotes the fluorescence pattern obtained with MAbs 12A1 and 13F1 for each strain shown. The abbreviations Ann and Punc refer to annular and punctate IF patterns, respectively. Serotypes are indicated in parentheses. In the absence of a MAb, the PIs were essentially zero (see Fig. 2).
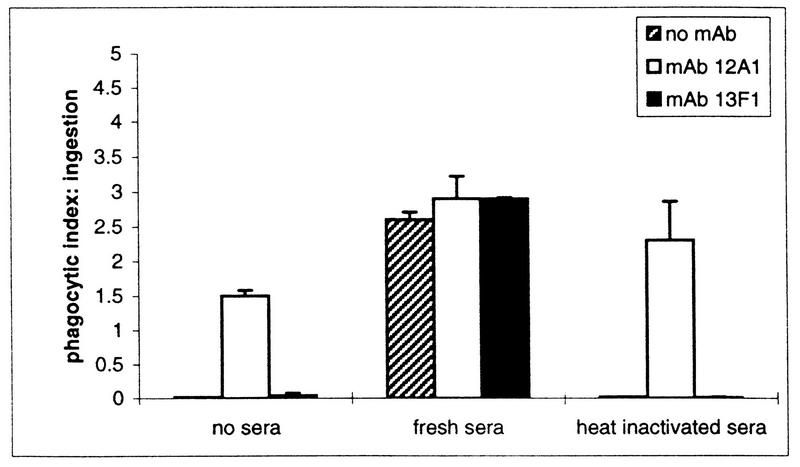
The effect of complement on MAb 12A1- and 13F1-mediated phagocytosis was studied with strain 24067 (Fig. 2). This strain was selected because it demonstrates the prototypical annular and punctate binding with MAbs 12A1 and 13F1, respectively. In the presence of complement, PI was significantly higher for all measurements. Hence, addition of complement produced quantitative changes in the opsonization of strain 24067 whether or not MAb 12A1 or 13F1 was present.
FIG. 2.
PIs resulting from the incubation of strain 24067 with J774.16 cells in the presence of either MAb 12A1 or 13F1 and fresh or heat-inactivated mouse serum.
Flow cytometry.
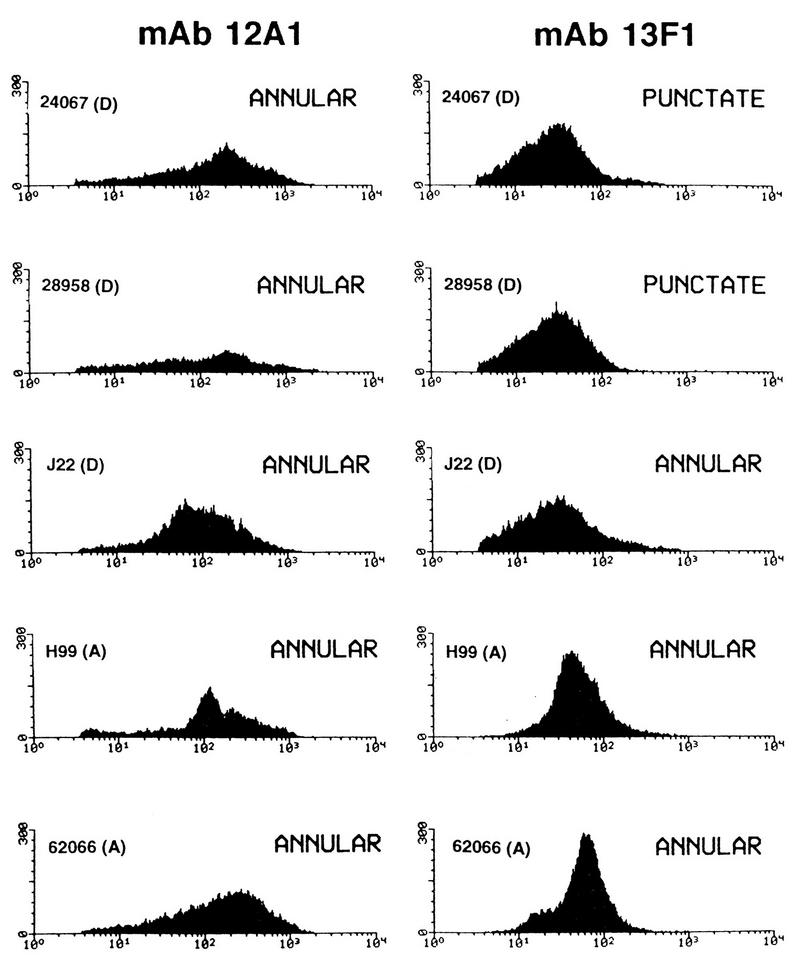
The patterns of MAb 12A1 and 13F1 binding to five strains was analyzed by FACS (Fig. 3). For all strains, there was considerable cell-to-cell variation in fluorescence intensity. For all strains, the fluorescence intensity observed with MAb 12A1 was higher than that observed with MAb 13F1 (Table 2). Punctate binding by MAb 13F1 produced the lowest fluorescence intensities observed among the samples studied.
FIG. 3.
FACS profiles of five strains of C. neoformans after incubation with MAb 12A1 or 13F1 and an FITC-labeled goat anti-mouse IgM antibody. For each measurement, 10,000 cells were counted. On the y axis is the number of cells, and on the x axis is the log fluorescence intensity. Serotypes are shown in parentheses.
TABLE 2.
Indirect IF intensity measured by FACS
| Strain (serotype) | Mean relative fluorescence intensity ± SDa
|
P valueb | |
|---|---|---|---|
| MAb 12A1 | MAb 13F1 | ||
| 24067 (D) | 241 ± 261 | 41 ± 51 | <0.001 |
| 28959 (D) | 558 ± 1,172 | 35 ± 36 | <0.001 |
| J22 (D) | 141 ± 148 | 49 ± 71 | <0.001 |
| 62066 (A) | 241 ± 248 | 65 ± 46 | <0.001 |
| H99 (A) | 333 ± 550 | 66 ± 67 | <0.001 |
Relative fluorescence intensity was measured by FACS.
The P value was calculated by the Student t test for 5,000 events per group.
Reproducibility and consistency of measurements.
IF and FACS studies were done multiple times for some strains on different days, and the results were always consistent. Relative fluorescence intensities with MAbs 12A1 and 13F1 were measured by FACS at least seven times on different days. For all measurements, the intensity observed with MAb 12A1 was greater than that observed with MAb 13F1. Agglutination endpoints were measured for several strains on different days, and the endpoints were always within 1 dilution.
DISCUSSION
IgM MAbs 12A1 and 13F1 are closely related in molecular structure but differ in epitope specificity, as established by differences in serotype reactivity (3), competition assays (16), and differences in binding to peptide mimetopes (23). Here we demonstrate differences in MAb 12A1 and 13F1 binding among C. neoformans var. neoformans strains. For MAb 12A1, an annular IF binding pattern was observed with all of the strains studied, irrespective of the serotype classification. For MAb 13F1, annular and punctate IF binding patterns were associated with serotypes A and D, respectively. A significant correlation was demonstrated for MAb 13F1 punctate binding and a serotype D classification. On the basis of the strains studied, we calculate that a punctate IF pattern with MAb 13F1 has a predictive value for serotype D of 95%.
One exception to the expected punctate binding by MAb 13F1 on serotype D strains was J22. This strain is unusual in that it has a novel GXM triad structure (5). Whether this characteristic contributed to the unusual 13F1 binding pattern relative to other serotype D strains is unclear. The finding of the J22 exception to the association between MAb 13F1 punctate binding and a serotype D classification has several possible explanations. First, J22 may not be a classical serotype D strain, as suggested by its unusual GXM triad structure. Second, different antigenic determinants may be responsible for MAb 13F1 binding and serotype classification based on rabbit sera. In this scenario, serotype classification could be associated with a pattern of MAb 13F1 binding but each phenomenon could depend on different GXM structural components. Alternatively, the same antigenic determinants could be recognized by MAb 13F1 and the rabbit serotype-specific sera but structural heterogeneity in GXM could result in occasional strains that exhibit unusual serological characteristics. We cannot distinguish between these possibilities.
Previous IF studies of the binding of other MAbs to C. neoformans var. neoformans strains have reported annular patterns with serotype A and D organisms. MAb 13F1 is unusual in that it produces punctate IF on most serotype D strains. The physical basis for the punctate pattern is unknown. The IF pattern results from fluorescence emitted by the secondary antibody, which is labeled with fluorescein. One possibility is that the epitope recognized by 13F1 is found in discrete regions of the capsule. Alternatively, MAb 13F1 may bind in a manner that allows cross-linking of MAb 13F1 molecules by the secondary antibody with the result that aggregates form in the capsule. In any case, the IF differences in MAb 12A1 and 13F1 patterns indicate binding to different epitopes and also imply differences in epitope location in the capsule structure. This observation is consistent with and supports the report by Vogel in 1966 that specific antigens differ in location among strains, depending on the serotype classification (24).
To determine whether the IF binding phenomena observed with MAbs 12A1 and 13F1 correlate with other serological techniques, additional studies were done with agglutination and FACS assays. Punctate binding by MAb 13F1 was associated with a requirement for a higher antibody concentration to produce agglutination. This suggests that epitopes which elicit punctate IF binding are relatively infrequent in the surface of the capsule and/or that the interaction between the MAb and this type of antigenic determinant is weak. Since the apparent affinities of MAbs 12A1 and 13F1 for GXM are very similar (16), it is likely that the higher agglutination endpoints for punctate binding reflect a difference in epitope distribution. FACS revealed that MAb 13F1 produced a lower mean cell fluorescence intensity than MAb 12A1. Furthermore, punctate binding produced a lower mean cell fluorescence intensity than annular binding. Since the affinities of MAbs 12A1 and 13F1 for GXM are similar (16) and the affinity of the secondary, FITC-labeled reagent for each IgM should be the same, these results suggest that the epitope recognized by MAb 13F1 is found at either a lower density or in a different distribution in the capsule. Other explanations for the lower fluorescence intensity include lower epitope accessibility for MAb 13F1 or the secondary antibody. FACS analysis revealed considerable variation in the fluorescence intensities of individual cells for all of the strains studied. This variation was surprising since we used cultures derived from single colonies grown to early stationary phase. This variation in individual cell IF intensity presumably implies cell-to-cell differences in epitope density and/or content.
To determine if the differences in binding between MAbs 12A1 and 13F1 correlate with a functional effect, we studied the ability of these antibodies to promote phagocytosis of C. neoformans by J774.16 murine macrophage-like cells. Phagocytosis assays revealed that the annular binding pattern was associated with more effective opsonization than the punctate binding pattern. Furthermore, MAb 12A1 was a more effective opsonin than MAb 13F1 for all of the strains studied, irrespective of the IF binding pattern. The differences in phagocytic efficacy between MAbs 12A1 and 13F1 may reflect differences in epitope distribution as described for other systems (13).
The epitopes recognized by MAbs 12A1 and 13F1 were found in all of the C. neoformans var. neoformans strains studied. The observation that MAb 13F1 discriminates between most serotype A and D strains provides strong support for the present serotype classification scheme devised with rabbit sera. MAb 13F1 binding to serotype A and D strains revealed differences in IF pattern and intensity, strongly suggesting that the antigenic differences between these serotypes are both qualitative and quantitative. In this regard, MAb 13F1 differs from the other MAbs used in serotyping schemes, which discriminate between serotype A and D strains solely on the basis of quantitative differences in binding.
In summary, we have demonstrated an association between the IF binding patterns of two IgM MAbs and the serotype classification of C. neoformans. Determination of whether the association is causal must await structural information about the epitopes recognized by these MAbs and the polysaccharide structures responsible for serotype classification. Our results suggest that this MAb pair may be useful for serological studies of C. neoformans strains. The results highlight the structural complexity of the capsule, the antigenic heterogeneity of C. neoformans strains, and the usefulness of MAbs in serological studies.
ACKNOWLEDGMENTS
We thank Mary Brandt for generously providing several C. neoformans strains. We also thank David Gebhard for assistance in flow cytometry and Michael Cammer for assistance in confocal microscopy.
A.C. is supported by National Institutes of Health grants RO1-AI33774 and RO1-AI13342 and a Burroughs Wellcome Fund Development Therapeutics Award. W.C. was supported by a minority graduate student supplement to grant RO1-AI33774. The flow cytometry facility of the Chanin Cancer Center is supported by National Cancer Institute support grant 5P30-CA13330. This support is gratefully acknowledged.
REFERENCES
- 1.Belay T, Cherniak R, Kozel T R, Casadevall A. Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect Immun. 1997;65:718–728. doi: 10.1128/iai.65.2.718-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Mukherjee J, Scharff M D. Monoclonal antibody ELISAs for cryptococcal polysaccharide. J Immunol Methods. 1992;154:27–35. doi: 10.1016/0022-1759(92)90209-c. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall A, Mukherjee J, Devi S, Schneerson R, Robbins J, Scharff M D. Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;165:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 4.Casadevall A, Scharff M D. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–160. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherniak R, Morris L C, Belay T, Spitzer E D, Casadevall A. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect Immun. 1995;63:1899–1905. doi: 10.1128/iai.63.5.1899-1905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherniak R, Sundstrom J B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect Immun. 1994;62:1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleare W, Mukherjee S, Spitzer E D, Casadevall A. Prevalence in Cryptococcus neoformans strains of a polysaccharide epitope which can elicit protective antibodies. Clin Diagn Lab Immunol. 1994;1:737–740. doi: 10.1128/cdli.1.6.737-740.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damiani G, Kiyotaki C, Soeller W, Sasada M, Peisach J, Bloom B R. Macrophage variants in oxygen metabolism. J Exp Med. 1980;152:808–822. doi: 10.1084/jem.152.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dromer F, Gueho E, Ronin O, Dupont B. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J Clin Microbiol. 1993;31:359–363. doi: 10.1128/jcm.31.2.359-363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dromer F, Salamero J, Contrepois A, Carbon C, Yeni P. Production, characterization, and antibody specificity of a mouse monoclonal antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect Immun. 1987;55:742–748. doi: 10.1128/iai.55.3.742-748.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans E D, Kessel J F. The antigenic composition of Cryptococcus neoformans. J Immunol. 1951;67:109–114. [PubMed] [Google Scholar]
- 12.Evans E E. An immunologic comparison of twelve strains of Cryptococcus neoformans (Torula histolytica) Proc Soc Exp Biol Med. 1949;71:644–646. doi: 10.3181/00379727-71-17283. [DOI] [PubMed] [Google Scholar]
- 13.Griffin F M, Jr, Griffin J A, Leider J E, Silverstein S C. Studies on the mechanism of phagocytosis. J Exp Med. 1975;142:1263–1282. doi: 10.1084/jem.142.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda R, Nishimura S, Nishikawa A, Shinoda T. Production of agglutinating monoclonal antibody against antigen 8 specific for Cryptococcus neoformans serotype D. Clin Diagn Lab Immunol. 1996;3:89–92. doi: 10.1128/cdli.3.1.89-92.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee J, Casadevall A, Scharff M D. Molecular characterization of the antibody responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J Exp Med. 1993;177:1105–1106. doi: 10.1084/jem.177.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee J, Nussbaum G, Scharff M D, Casadevall A. Protective and non-protective monoclonal antibodies to Cryptococcus neoformans originating from one B-cell. J Exp Med. 1995;181:405–409. doi: 10.1084/jem.181.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Feldmesser M, Casadevall A. J774 murine macrophage-like cell interactions with Cryptococcus neoformans in the presence and absence of opsonins. J Infect Dis. 1996;173:1222–1231. doi: 10.1093/infdis/173.5.1222. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee S, Lee S C, Casadevall A. Antibodies to Cryptococcus neoformans glucuronoxylomannan enhance antifungal activity of murine macrophages. Infect Immun. 1995;63:573–579. doi: 10.1128/iai.63.2.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussbaum G, Cleare W, Casadevall A, Scharff M D, Valadon P. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J Exp Med. 1997;185:685–697. doi: 10.1084/jem.185.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiropulu C, Eppard R A, Otteson E, Kozel T R. Antigenic variation within serotypes of Cryptococcus neoformans detected by monoclonal antibodies specific for the capsular polysaccharide. Infect Immun. 1989;57:3240–3242. doi: 10.1128/iai.57.10.3240-3242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todaro-Luck F, Reiss E, Cherniak R, Kaufman L. Characterization of Cryptococcus neoformans capsular glucuronoxylomannan polysaccharide with monoclonal antibodies. Infect Immun. 1989;57:3882–3887. doi: 10.1128/iai.57.12.3882-3887.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadon P, Nussbaum G, Boyd L F, Margulies D H, Scharff M D. Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 24.Vogel R A. The indirect fluorescent antibody test for the detection of antibody in human cryptococcal disease. J Infect Dis. 1966;116:573–580. doi: 10.1093/infdis/116.5.573. [DOI] [PubMed] [Google Scholar]