Abstract
The SWI-SNF complex in yeast and related complexes in higher eukaryotes have been implicated in assisting gene activation by overcoming the repressive effects of chromatin. We show that the ability of the transcriptional activator GAL4 to bind to a site in a positioned nucleosome is not appreciably impaired in swi mutant yeast cells. However, chromatin remodeling that depends on a transcriptional activation domain shows a considerable, although not complete, SWI-SNF dependence, suggesting that the SWI-SNF complex exerts its major effect at a step subsequent to activator binding. We tested this idea further by comparing the SWI-SNF dependence of a reporter gene based on the GAL10 promoter, which has an accessible upstream activating sequence and a nucleosomal TATA element, with that of a CYC1-lacZ reporter, which has a relatively accessible TATA element. We found that the GAL10-based reporter gene showed a much stronger SWI-SNF dependence than did the CYC1-lacZ reporter with several different activators. Remarkably, transcription of the GAL10-based reporter by a GAL4-GAL11 fusion protein showed a nearly complete requirement for the SWI-SNF complex, strongly suggesting that SWI-SNF is needed to allow access of TFIID or the RNA polymerase II holoenzyme. Taken together, our results demonstrate that chromatin remodeling in vivo can occur by both SWI-SNF-dependent and -independent avenues and suggest that the SWI-SNF complex exerts its major effect in transcriptional activation at a step subsequent to transcriptional activator-promoter recognition.
Transcriptional activation in eukaryotes requires activator proteins and the preinitiation complex, including TATA-binding protein (TBP) and RNA polymerase, to recognize their respective binding sites in a gene promoter. Each of these interactions can be strongly inhibited in vitro by incorporating the relevant recognition site into a nucleosome (20, 26, 37, 44, 51, 53, 59, 78, 84). Since eukaryotic DNA is packaged into nucleosomes, this creates a potential problem. One solution would be for eukaryotic promoters to be constitutively free of nucleosomes, but this is not the case (79). Another would be for the cell to have evolved chromatin remodeling activities which would allow activators and other components of the preinitiation complex to recognize their sites in chromatin. Several candidate activities have been identified (9, 12, 21, 28, 35, 73–75, 80), but the roles they play in vivo are poorly understood.
One candidate for helping transcriptional activation occur in the context of chromatin in vivo is the SWI-SNF complex, first identified in the yeast Saccharomyces cerevisiae. This large complex, of about 2 × 106 Da, contains at least 11 distinct polypeptides (8, 61, 72) and is conserved across most of the eukaryotic kingdom (60, 80). Transcriptional induction of a variety of genes is impaired in yeast cells lacking components of the SWI-SNF complex (62, 87), and genetic and biochemical evidence suggests that the SWI-SNF complex may aid transcriptional induction by helping to counteract the repressive effects of packaging DNA into chromatin. Suppressors of the swi phenotype in yeast include deletion of one of the two sets of genes encoding histones H2A and H2B, as well as point mutations in histones H3 and H4 that increase the accessibility of nucleosomal DNA (25, 38, 63, 81, 87). Furthermore, the chromatin structure of the active SUC2 promoter, whose activity is strongly SWI-SNF dependent, is altered in swi cells but is partially restored in suppressor mutants (18a, 25, 86). Finally, in vitro studies have shown that the yeast SWI-SNF complex or its mammalian homolog can alter nucleosomal structure in a way that facilitates the binding of derivatives of the transcriptional activator GAL4 to nucleosomal sites (14, 80). Although the ratio of SWI-SNF to nucleosomes in these experiments was vastly greater than that found in vivo, this work nonetheless demonstrated that the SWI-SNF complex can alter nucleosome structure in a way that facilitates protein binding. A key finding was that this alteration was ATP dependent (14), since SWI2-SNF2, a critical component of the complex, has ATPase activity that is essential for its in vivo activity (34, 43).
The SWI-SNF complex could support transcriptional induction in chromatin by facilitating factor binding, as suggested by the in vitro experiments in which SWI-SNF assists the binding of GAL4 derivatives to nucleosomal sites (14, 80). Alternatively, it could assist subsequent steps such as recruitment of TATA-binding protein (26) or RNA polymerase, as suggested by a report that SWI-SNF proteins are associated with the RNA polymerase II holoenzyme (82). It could even assist in elongation through nucleosomes (5), although if this were its only point of action, it is difficult to understand how a tethered SWI-SNF component could activate transcription (41, 42).
We have previously shown that GAL4 can bind to its recognition sequence in a nucleosome in yeast with concomitant nucleosome disruption (55, 69). We have also shown that perturbation of nucleosome positioning can depend on the presence of an activation domain (69). Here we assess whether either of these events requires participation by the SWI-SNF complex. Our results suggest that in genes requiring SWI-SNF for maximal activation, the SWI-SNF complex exerts its effect principally at a step that occurs subsequent to transcriptional activator binding. By inference, the results also suggest that some other chromatin-remodeling activity is likely to assist in the initial binding of transcriptional activators to their recognition sites in at least some promoters.
MATERIALS AND METHODS
Strains, media, and genetic methods.
The S. cerevisiae strains used are derivatives of S288C and are listed in Table 1. Strain CY297b was derived from CY297 (7) by two-step deletion of the TRP1 gene (1). The yeast strains were grown at 30°C in complete synthetic dropout media (Bio 101) (60) containing 2% glucose, 1.5% raffinose, or 2% galactose and were transformed by a modification (24) of the method of Ito et al. (27).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| FY24 | MATα ura3-52 trp1Δ63 leu2Δ1 | 83 (isogenic to FY23) |
| YNN282 | MATα trp1-Δ his3-Δ200 ura3-52 lys-801a ade2-10 | 65 |
| CY296 | MATa gal4Δ::LEU2 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 ura3-Δ99 trp1-Δ99 | 7 |
| CY297 | MATα gal4Δ::LEU2 swi1Δ::LEU2 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 ura3-Δ99 | 7 |
| CY297b | MATα gal4Δ::LEU2 swi1Δ::LEU2 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 ura3-Δ99 trp1 | This work |
Plasmids.
The episome TA17Δ80 was introduced into yeast by first excising bacterial sequences from two pUC19-based plasmids carrying complementary pieces of TA17Δ80 and ligating those pieces, as described previously (55). TALS was introduced after excision of the complete yeast sequence from a pUC19 vector and religation (65).
To create the lacZ reporter plasmid 314-17Δ80lacZΔNco (bearing the reporter gene GAL4-CYC1-lacZ), a PstI fragment encompassing the lacZ gene and CYC1 TATA box from plasmid pLG669-Z (23) was ligated into the PstI site of pRS104-17Δ80 (55) at 823 map units of the TRP1ARS1 episome; the KpnI-HindIII fragment of the resulting plasmid containing the lacZ gene and TRP1ARS1 sequences was then inserted into the polylinker of the CEN-containing shuttle vector pRS314 (67) to create pRS314-17Δ80lacZ. Finally, the URA3 gene contained in this plasmid was removed by excision of a 1.7-kb NcoI fragment to yield pRS314-17Δ80lacZΔNco. The GAL4 binding site is 400 bp upstream of the first lacZ ATG.
The GAL10-MEL1 fusion gene was carried on plasmid pBM150SKMEL1. The MEL1 gene coding sequences were amplified with primers 5′-ATAATTTCTTACTGGATCCTAGGAGAGCAACGGAATTCAAAGCAA-3′ and 5′-AAAATTGAAGAGAATTCGGGCAAAAATTGGTACCAATGCATCCAA-3′. Restriction sites (BamHI and EcoRI or EcoRI and KpnI; underlined bases) were introduced to facilitate cloning. The PCR product was digested with EcoRI, which removes the MEL1 transcription start site, and the fragment was cloned into pBluescript (SK−) to generate SKMEL1. To confirm that MEL1 coding sequences in SKMEL1 were functional, a BamHI-KpnI fragment from SKMEL1 was subcloned into a yeast expression vector and the resulting plasmid was transformed into yeast. SKMEL1 plasmids having functional MEL1 coding sequences were isolated as EcoRI fragments and cloned into pBM150 (29) to generate pBM150SKMEL1.
The reporter plasmids carrying GAL4-CYC1-lacZ and GAL10-MEL1 were introduced into CY296 and CY297b (Table 1) along with expression vectors for GAL4·ftz(3–413) (17), GAL4·ER·VP16 (48, 69), or GAL4·GAL11. The GAL4 gene was introduced as a 3.6-kb BamHI fragment (40) carried on the CEN-containing shuttle vector pRS416 (13) in cells harboring the GAL4-CYC1-lacZ reporter, or on the 2μm plasmid pRS426 (13) in cells harboring the GAL10-MEL1 reporter. To construct the expression plasmid for GAL4·GAL11, the coding sequence for amino acids 1 to 93 of GAL4 (which includes the DNA-binding and dimerization domains) fused to the ADH1 promoter was amplified from pRS313GAL4·ER·VP16 (48) with primers 5′-ACACTTGAGCTCGTCGACTGACCCGGGCAATGCTTTTATATC-3′ and 5′-CAAGGTACCAATAAATGATGGTAAATG-3′. The PCR product was digested with KpnI and SacI (underlined) and ligated into pRS416 (13) to yield pRS416-GAL4DBD. The coding sequence for GAL11 amino acids 799 to 1081 (4) was amplified with primers 5′-AGGTCTGAGCTCAATACCGCTAAGTCAACC-3′ and 5′-AGGTTCACCGAGCTCTCAGCACTAGCTAACCGG-3′. The resulting fragment was cut with SacI (underlined sites) and ligated with SacI-cut pRS416-GAL4DBD. Clones with the correct orientation of the GAL11-encoding sequences were identified and subcloned into pRS413 (13).
β-Galactosidase and α-galactosidase (MEL1) assays.
Cells harboring lacZ reporters were grown to an absorbance at 600 nm (A600) between 0.5 and 1.5 in galactose medium for GAL4 activation or in glucose medium for activation by GAL4·ER·VP16, GAL4·ftz, and GAL4·GAL11; for activation by GAL4·ER·VP16, 0.1 μM β-estradiol was added and growth was continued for 4 h before assaying for β-galactosidase activity as described previously (52, 64). To assay for α-galactosidase (the MEL1 gene product) activity, we combined features of an assay involving cell extracts (30, 33) and the commonly used permeabilized cell assay used to measure β-galactosidase activity (64). Yeast cultures (5 to 10 ml) were grown in galactose (for GAL4) or raffinose (for GAL4·ER·VP16, GAL4· ftz, and GAL4·GAL11) to an A600 between 0.5 and 1.5. A 1-ml volume of cells was centrifuged, and the pellet was taken up in 200 μl of 20 mM HEPES (pH 7.5)–10 mM dithiothreitol–0.002% sodium dodecyl sulfate. One or two drops of chloroform was added with a Pasteur pipette, and the sample was vortexed for 10 s. After a 5-min preequilibration at 30°C, 800 μl of a 7 mM solution of p-nitrophenyl-α-galactose (Sigma) in 61 mM citric acid–77 mM Na2HPO4 (pH 4) was added and incubation at 30°C was continued. At various times, 100-μl aliquots were added to 900 μl of 0.1 M Na2CO3 to stop the reaction and develop the yellow color characteristic of the cleaved p-nitrophenyl-α-galactose (which remains colorless at pH 4). The terminated reaction products were centrifuged for 5 min in a microcentrifuge, and the A400 was determined. Units of α-galactosidase activity are given as [A400/(A600 × time in minutes)] × 1,000, using samples taken at time points such that the A400 <3.0.
Indirect end-label analysis of plasmid chromatin.
Yeast nuclei (66) were prepared from 1 liter of cells grown to an A600 between 0.6 and 1.6, and 300-μl aliquots were digested with micrococcal nuclease (MNase) for 10 min at 37°C at concentrations varying from 0 to 50 U/ml. Alternatively, spheroplast lysates (32) were prepared from 100 to 200 ml of cells and treated for 5 min at 37°C with 0 to 400 U of MNase per ml. The cleavage patterns did not vary within this range of concentrations. For digestion of naked DNA, samples were treated with proteinase K, extracted with phenol-chloroform, taken up in 300 μl of 150 mM NaCl–5 mM KCl–1 mM EDTA–20 mM Tris.HCl (pH 8.0)–2 mM CaCl2–5 mM MgCl2 or in 300 μl of 10 mM HEPES (pH 7.5)–2 mM CaCl2–5 mM MgCl2, and digested as above. The digestions were stopped by the addition of 55 μl of 5% sodium dodecyl sulfate–5 mg of proteinase K per ml to the 300-μl reaction mixtures and processed as described previously (55, 69). MNase cleavage patterns were analyzed by indirect end labeling as previously described (55, 69). The probes used were the 231-bp EcoRV-HindIII fragment (see Fig. 1) and the 200-bp EcoRV-XbaI fragment (see Fig. 2) from TRP1ARS1, both prepared by PCR; the 276-bp BamHI-SalI fragment from pBM150SKMEL1 (see Fig. 4); and a 252-bp PvuII-ClaI fragment from pRS314 (see Fig. 5). The probes were labeled by random priming. Marker lanes contained 0.5 ng of a ΦX/HaeIII digest, and labeled ΦX/HaeIII DNA was included with the hybridization probe. Chromatin was prepared from at least two independent clones for all MNase analyses.
FIG. 1.
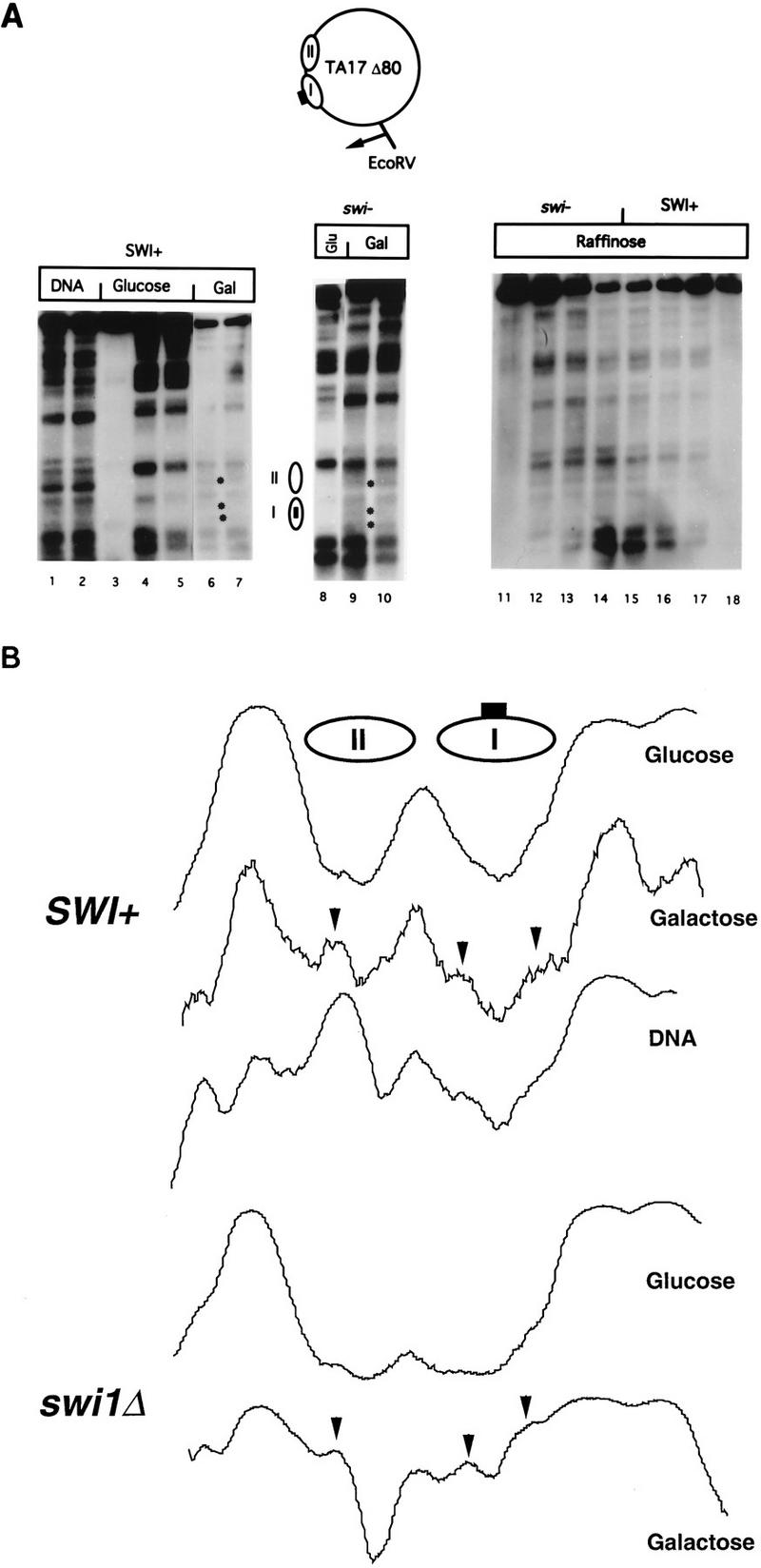
Perturbation of positioned nucleosomes in the yeast episome TA17Δ80 by GAL4 binding in SWI+ and swi1Δ yeast cells. (A) MNase cleavage sites in chromatin from SWI+ and swi1Δ cells grown in glucose, raffinose, or galactose medium were mapped relative to the EcoRV site as indicated. Samples were digested with MNase at 4 U/ml (lane 1), 10 U/ml (lane 2), 0 U/ml (lanes 3, 11, and 18), 2 U/ml (lanes 4, 6, 12, and 17), 5 U/ml (lanes 5, 7, 8, 10, 13, and 16), or 20 U/ml (lanes 9, 14, and 15). Bands cleaved in chromatin from cells grown in galactose are marked by asterisks to the right of lanes 6 and 9. Locations of nucleosomes I and II in cells grown in glucose are indicated; the box in nucleosome I represents the GAL4-binding site. (B) Densitometric scans of MNase cleavage patterns in the vicinity of nucleosomes I and II. Scans represent, in descending order, lanes 4, 6, 1, 8, and 10 in panel A. Arrowheads indicate cleavage sites induced in galactose. Note that the cleavage sites in the region of nucleosome I, which are cleaved weakly in chromatin from cells grown in galactose, are also weakly cut in naked DNA.
FIG. 2.
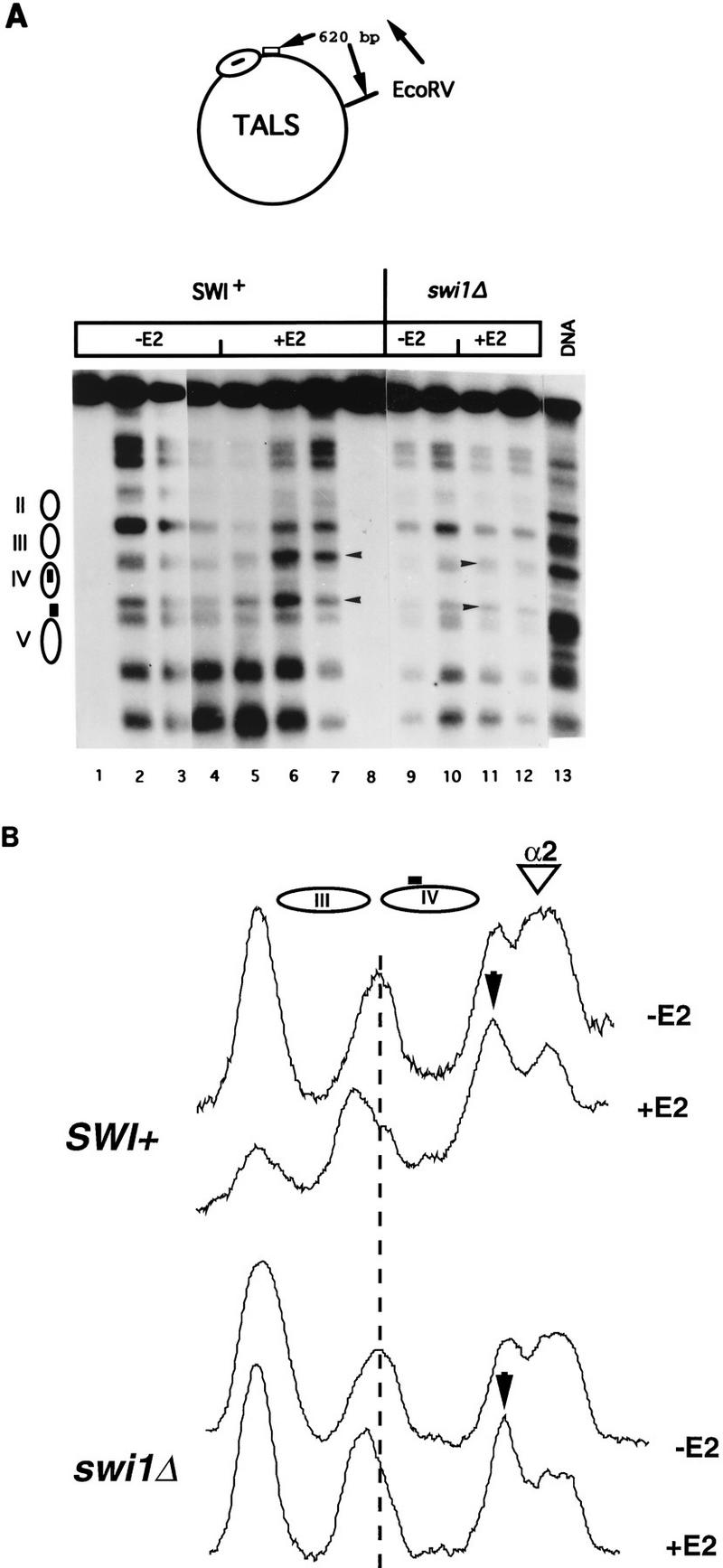
(A) Perturbation of TALS chromatin by the GAL4·ER·VP16 activation domain in SWI+ and swi1Δ yeast haploid α cells. MNase cleavage sites in cells harboring TALS and expressing GAL4· ER·VP16, either incubated for 4 h with 0.1 μM β-estradiol (+E2 lanes) or not (−E2 lanes), were mapped relative to the EcoRV site as indicated. Samples were digested with MNase at 0 U/ml (lanes 1 and 8), 5 U/ml (lanes 2 and 7), 20 U/ml (lanes 3, 6, 9, and 12), 50 U/ml (lanes 4, 5, 10, and 11), or 10 U/ml (lane 13). Control samples (0 U/ml) for the swi1Δ samples were identical in appearance to those shown for the SWI+ samples (data not shown). Arrowheads indicate bands cleaved preferentially under activating (+E2) conditions. The locations of nucleosomes II to V are indicated; the box in nucleosome IV represents the GAL4-binding site, and the box between nucleosomes IV and V represents the α2/MCM1 operator. (B) Densitometric scans of MNase cleavage patterns from lanes 4, 5, 10, and 11. The arrowhead indicates the lower of the two hormone-induced enhanced cleavage sites seen in the panel A, and the dotted line allows visualization of the slight shift in position of the cleavage site corresponding to the upper arrowhead in panel A.
FIG. 4.
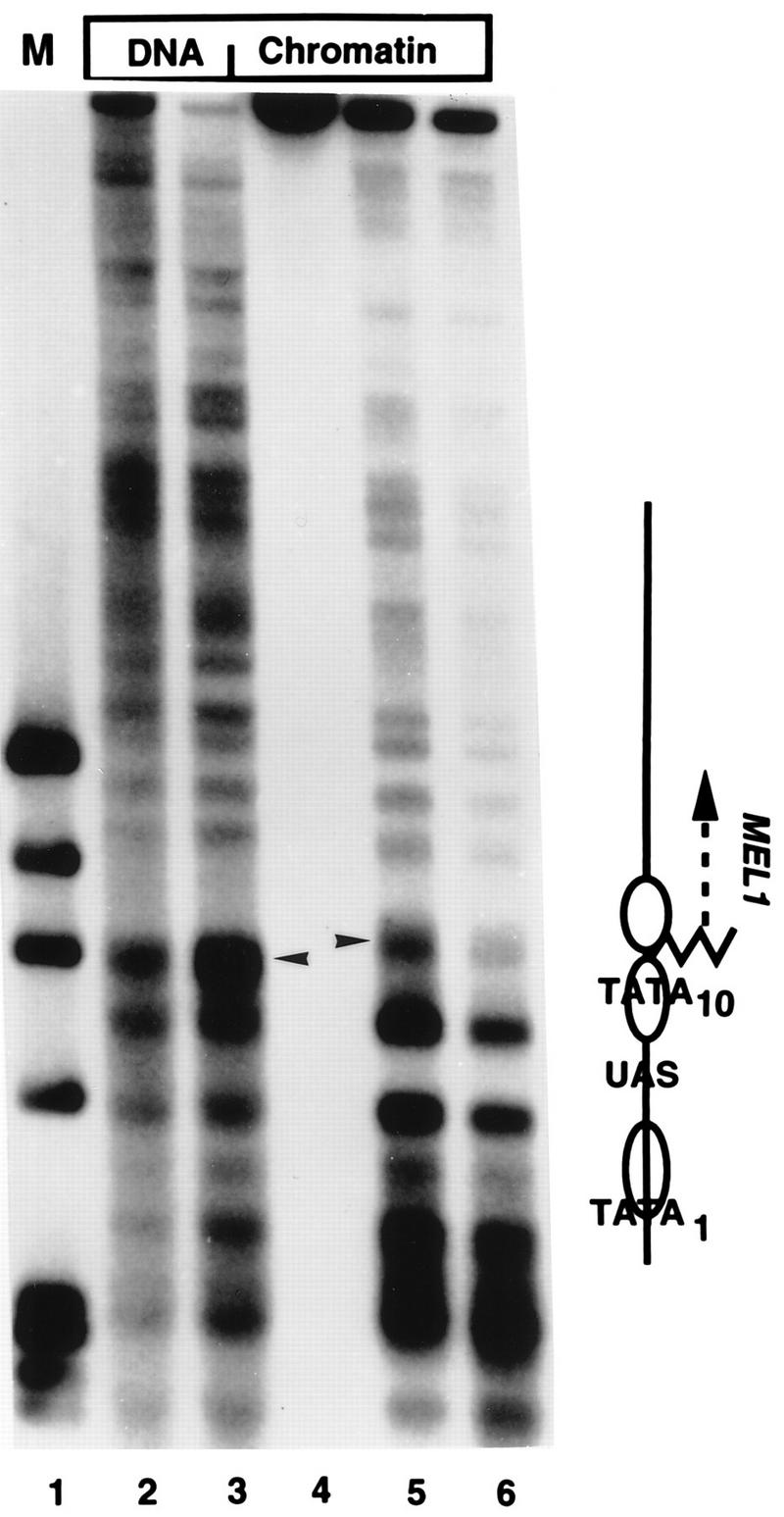
Indirect end-label analysis of the chromatin structure of the GAL10-MEL1 gene fusion. MNase cleavage sites in naked DNA or chromatin, as indicated, were mapped relative to a SalI site 830 bp upstream of the GAL10 TATA element, on the GAL1 side of the promoter in plasmid pBM150SKMEL1. Samples were digested with 4 (lane 2), 10 (lane 3), 0 (lane 4), 150 (lane 5) or 300 (lane 6) U of MNase per ml. Note the strong cleavage in naked DNA (arrowhead, lanes 2 and 3), which is protected in chromatin; a new cleavage (arrowhead, lanes 5 and 6) is present slightly higher on the gel, corresponding to the nucleosome-sized protected region containing the GAL10 TATA element. The overall pattern of cleavages in the vicinity of the UAS and GAL10 TATA is identical to that seen in the endogenous GAL1-10 promoter (data not shown). The locations of relevant promoter elements are indicated on the right; the jagged line represents the site of the fusion between the GAL10 promoter and the MEL1 coding sequence. Lane 1 contains ΦX/HaeIII markers.
FIG. 5.
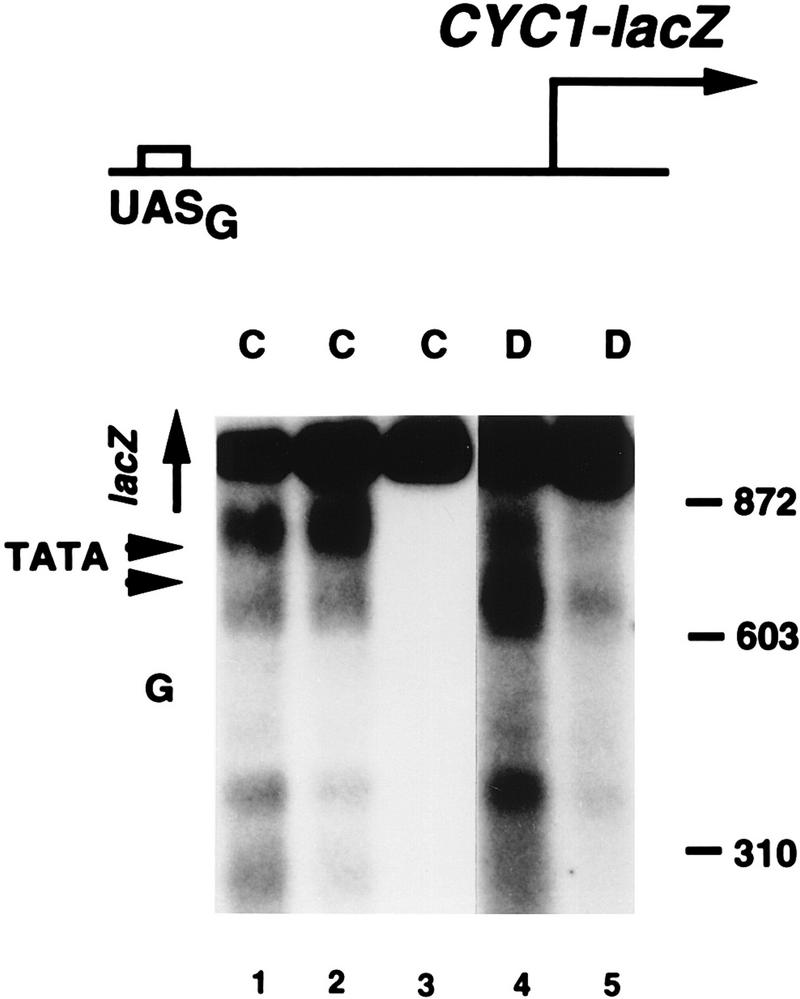
Indirect end-label analysis of chromatin structure of 314-17Δ80lacZΔNco, which bears the GAL4-CYC1-lacZ reporter gene. MNase cleavage sites in chromatin (lanes C) and DNA (lanes D) were mapped relative to a PvuII site that is 400 bp upstream of the GAL4-binding site (UASG). Cell lysates (32) or DNA was digested with MNase at 20 U/ml (lane 1), 5 U/ml (lane 2), 0 U/ml (lane 3), 4 U/ml (lane 4), or 10 U/ml (lane 5). The location of the GAL4 site is indicated by G, and the locations of the two major TATA elements in the CYC1 promoter (45) and the lacZ gene are also indicated. Lanes 1 to 3 and lanes 4 and 5 were taken from separate gels which ran identically (as shown by size markers).
Topoisomer analysis.
DNA from three independent clones was prepared as described previously (54) from 10-ml cultures grown to an A600 of 0.6 to 1.2 by rapid glass bead lysis. Purified DNA was electrophoresed at 2.5 V/cm for 18 to 20 h on 1.5% agarose gels containing 40 μg of chloroquine diphosphate (Sigma) per ml, transferred to nylon membranes, and hybridized with probes specific for TRP1ARS1 sequences as described above. The Gaussian centers of the topoisomer distributions were calculated as described previously (54) with images scanned on a Molecular Dynamics PhosphorImager. Topological changes in swi1Δ cells were measured in strain CY297b; changes in SWI+ cells were measured in FY24 cells for GAL4-induced changes and in YNN282 cells for GAL4·ER·VP16-induced changes. Topological changes induced by GAL4 and GAL4·ER·VP16 are identical between YNN282 and FY24 cells (reference 69 and data not shown).
RESULTS
Activator binding to nucleosomal sites in swi yeast cells.
To test whether the SWI-SNF complex is required for binding of a transcriptional activator to a nucleosomal site in yeast, we examined the disruption of nucleosome positioning by GAL4 binding in the yeast episome TA17Δ80. This plasmid contains a TRP1 marker and a single strong GAL4-binding site of 17 bp, which is located near the middle of a strongly positioned nucleosome when GAL4 is absent or present in very small amounts. The growth of cells in galactose, which induces GAL4 expression, or exogenous expression of GAL4 or its derivatives results in perturbation of this nucleosome by GAL4 (55).
We introduced TA17Δ80 into SWI+ and swi1Δ yeast strains, both derived from the parent strain S288C. SWI1 is essential to SWI-SNF complex integrity, since the complex cannot be isolated intact from swi1Δ cells (61); furthermore, swi1Δ, swi2Δ, and swi3Δ cells have very similar phenotypes, indicating that loss of any of these proteins inactivates the complex (62). Nucleosome positioning in TA17Δ80 was examined in SWI+ and swi1Δ yeast cells grown in glucose or galactose by MNase cleavage followed by indirect end labeling (56, 85). Two regions of about 150 bp are cleaved by MNase as naked DNA but protected against cleavage in chromatin from yeast cells grown in glucose, indicating the presence of two positioned nucleosomes, one of which includes the GAL4-binding site (Fig. 1A, lanes 1 to 5) (55). These regions show enhanced cleavage by MNase in SWI+ cells grown in galactose (lanes 6 and 7), indicating that GAL4 is able to compete successfully against the histones for occupancy of its site, as observed previously (55). Protection against MNase cleavage is also seen in swi1Δ cells grown in glucose, indicating that nucleosome positioning is not generally affected by the SWI-SNF complex (lane 8; we note that the site between nucleosomes I and II is cleaved very weakly in this sample, but this site shows some variability in its cleavage from sample to sample and in general is not cleaved more weakly in swi1Δ yeast cells than in wild-type cells). In galactose, MNase cleavages in the regions protected by nucleosomes I and II in glucose are observed (lanes 9 and 10). The MNase cleavage pattern from cells grown in galactose is very similar between SWI+ and swi1Δ cells (Fig. 1B), and we have found this to be reproducibly the case. We conclude that the binding of GAL4 to a nucleosomal binding site, with concomitant chromatin remodeling, can occur independently of the SWI-SNF complex in vivo.
GAL4 synthesis is also stimulated by growth in raffinose (by relief of glucose repression [22, 57]); however, in the absence of galactose, the GAL4 activation domain is masked by GAL80 and it cannot activate transcription (31, 50). GAL4 produced by growth in raffinose can also perturb nucleosome positioning in TA17Δ80 in both wild-type and swi1Δ yeast cells (lanes 11 to 18). Perturbation of TA17Δ80 chromatin by GAL4 in raffinose is somewhat weaker than in galactose for both SWI+ and swi1Δ yeast cells, consistent with previous results obtained with SWI+ yeast cells (55). Thus, binding of the transcription factor GAL4 in both its activating and nonactivating configurations can occur at a nucleosomal site, with concomitant nucleosome perturbation, in yeast cells lacking a functional SWI-SNF complex.
Role of the SWI-SNF complex in activation domain-dependent chromatin remodeling.
Although the SWI-SNF complex is not needed for GAL4 to gain access to a strong binding site in chromatin in vivo (Fig. 1), this does not imply that chromatin remodeling by the SWI-SNF complex is not important for transcriptional activation. Other proteins, including TBP and RNA polymerase, must also bind their sites in chromatin, and they could require chromatin remodeling mediated by the SWI-SNF complex to do so. To examine whether chromatin perturbation occurring subsequent to GAL4 binding is assisted by the SWI-SNF complex, we took advantage of our recent observations of activator-dependent changes in chromatin structure in another yeast episome, TALS (69).
The TALS episome is packaged into strongly positioned nucleosomes in yeast haploid α cells (36, 65, 66). TALS also contains a GAL4-binding site derived from the GAL3 promoter, although the region of the GAL3 promoter downstream of this site is absent (3); this GAL4-binding site is packaged into a nucleosome in yeast α cells grown in glucose (36, 65, 66). Using the chimeric, hormone-dependent activator, GAL4·ER· VP16, we have shown that the chromatin structure of the episome is only minimally affected in the absence of hormone (69). The addition of β-estradiol in the presence of GAL4· ER·VP16 results in remodeling of TALS chromatin, as assessed by MNase cleavage, restriction endonuclease accessibility, and topological analysis. Similarly, the endogenous activator GAL4 only slightly affects TALS chromatin structure under nonactivating conditions but has considerable effect under activating conditions (69). Remodeling depends on the presence of both the activator and the GAL4-binding site.
We detected remodeling of TALS chromatin by GAL4· ER·VP16 upon addition of hormone in both SWI+ and swi1Δ cells as determined by alterations in the MNase cleavage pattern (Fig. 2). Perturbation is seen in the regions flanking nucleosome IV, which contains the GAL4-binding site. First, the relative intensity of the two cleavage sites between nucleosomes IV and V changes upon hormone addition, with cleavage at the site proximal to nucleosome IV being relatively enhanced in the presence of hormone (Fig. 2A, compare lanes 5 to 7 with lanes 2 to 4 for SWI+ cells and compare lanes 11 and 12 with lanes 9 and 10 for swi1Δ cells; Fig. 2B). Second, a new site is cleaved at the edge of nucleosome III in the presence of hormone at the expense of the cleavage site between nucleosomes III and IV (Fig. 2). Similar changes were observed in both SWI+ and swi1Δ cells when GAL4 was used as the activator (data not shown). Since the GAL4-binding site in TALS is nucleosomal in the absence of GAL4 (69), these results again indicate that GAL4 (and GAL4·ER·VP16) can bind to a nucleosomal site without help from the SWI-SNF complex, corroborating the results of Fig. 1. Furthermore, these results indicate that activation domain-dependent changes in chromatin structure can occur in yeast in the absence of a functional SWI-SNF complex.
Although TALS chromatin is clearly perturbed by GAL4·ER·VP16 in the presence of β-estradiol in swi1Δ cells, there were some indications that this perturbation was less dramatic than in SWI+ cells. For example, we often observed nearly complete replacement of the cleavage site between nucleosomes III and IV with the new site at the edge of nucleosome III in SWI+ cells in the presence of hormone-activated GAL4· ER·VP16, whereas this shift was reproducibly seen only partially in swi1Δ cells (Fig. 2B, compare the shift in the peak indicated by the dotted line in traces derived from SWI+ and swi1Δ cells). However, these are subtle changes and are certainly not subject to quantitative analysis. Therefore, to obtain a more quantitative comparison of activator-dependent changes in TALS chromatin structure in swi and SWI+ cells, we examined plasmid topology.
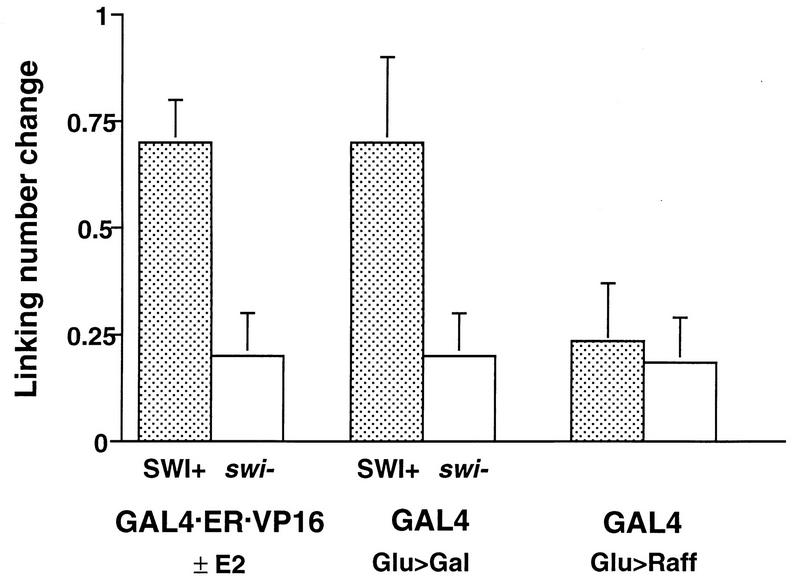
Since each nucleosome confers one negative supercoil on closed circular DNA, nucleosome loss results in an increased linking number (19, 68). We previously demonstrated that both GAL4 and GAL4·ER·VP16, in their activate configurations, cause an average increase in linking number (loss of negative supercoiling) of 0.7 per molecule of TALS (69). To compare this effect on TALS topology in SWI+ and swi1Δ cells, DNA was rapidly harvested from the appropriate cells under activating and nonactivating conditions (i.e., glucose versus galactose for GAL4 and plus or minus hormone for GAL4·ER·VP16) and treated to inactivate topoisomerases (so that the distribution of supercoiled plasmids reflected the in vivo distribution). The DNA was electrophoresed in agarose containing 40 μg of chloroquine diphosphate per ml, conditions under which the plasmids migrate as positively supercoiled topoisomers. These topoisomer distributions conform to Gaussian distributions, which allows their centers to be measured precisely from the relative intensities of individual topoisomers (54). Consequently, differences in topology can also be measured with precision. Quantitation of several independent experiments yielded values for the loss of negative supercoiling in TALS in SWI+ and swi1Δ cells (summarized in Fig. 3). The hormone-dependent alteration in TALS topology in the presence of GAL4·ER·VP16 is substantially reduced in swi1Δ cells (Fig. 3, P <0.001 for the null hypothesis that the difference between SWI+ and swi1Δ cells is not significant [49]). Similarly, alteration of TALS topology by GAL4 under activating conditions is reduced in swi1Δ compared to SWI+ cells (Fig. 3, P <0.01). Under nonactivating conditions (cells grown in raffinose), a small change in topology is seen in both SWI+ and swi1Δ cells (Fig. 3), possibly indicating that this effect is due to SWI-SNF-independent binding of nonactivating GAL4 complexed with GAL80. Taken together, the results of Fig. 2 and 3 indicate that SWI-SNF contributes to but is not absolutely required for activator-dependent remodeling of TALS chromatin. This conclusion is consistent with the partial dependence upon the SWI-SNF complex for transcriptional activation observed in previous studies (7, 18, 25, 42, 62) as well as in the present work (see Fig. 6).
FIG. 3.
Topological perturbation of TALS chromatin in SWI+ and swi1Δ yeast cells by GAL4 and by GAL4 · ER · VP16. Linking-number changes induced by hormone addition in the presence of GAL4·ER·VP16 or by growth in galactose (Gal) or raffinose (Raff) compared to glucose (Glu) in the presence of the endogenous GAL4 gene are indicated along with standard errors.
FIG. 6.
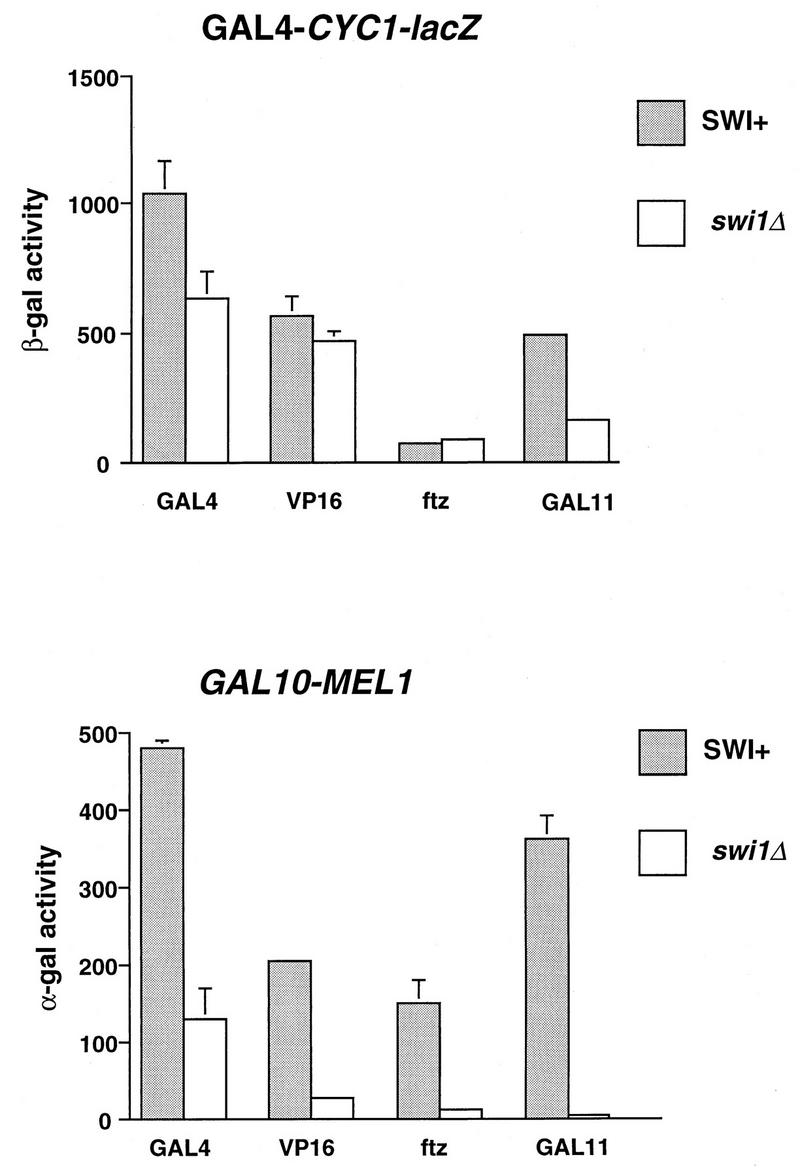
Activity of the two reporter genes, GAL4-CYC1-lacZ and GAL10-MEL1, induced by different activation domains in SWI+ (strain CY296) and swi1Δ (strain CY297b) cells. The activators used were GAL4, GAL4·ER·VP16, GAL4·ftz, and GAL4·GAL11. Activities were measured for at least three independent clones for each sample. Some standard errors were too small for the error bars to be seen in the graphs.
SWI-SNF dependence of two promoters differing in chromatin structure at proximal elements.
The results presented so far suggest that an activator can bind to a strong binding site in chromatin independently of the SWI-SNF complex, whereas chromatin remodeling occurring subsequent to transcriptional activator binding depends at least partly on SWI-SNF. We would therefore predict that a promoter with an organized chromatin structure that includes the TATA element and transcription start site would show a relatively strong SWI-SNF dependence compared to a promoter having more accessible proximal elements. We would also predict that a promoter having an accessible upstream activating sequence (UAS) could show a strong SWI-SNF dependence if its proximal elements were present in positioned nucleosomes. In contrast, if the primary function of the SWI-SNF complex were to allow activator access to chromatin, such a promoter should not show any SWI-SNF dependence at all.
To test this idea, we compared transcription from two promoters in SWI+ and swi1Δ yeast cells. One reporter gene was a GAL4-CYC1-lacZ fusion, having a single strong binding site for GAL4 upstream of the CYC1 promoter sequence and a lacZ coding sequence, and the other was a fusion of the GAL10 promoter to the MEL1 coding sequence. The native GAL10 promoter contains four GAL4-binding sites in its UAS (which is shared with the GAL1 promoter) in a nucleosome-free region, but downstream sequences, including the TATA element, are packaged in a highly organized array of positioned nucleosomes (10, 46, 47). We fused the GAL10 promoter with the MEL1 coding sequence, which encodes α-galactosidase. The activity of this enzyme can be quantified by a simple colorimetric assay (see Materials and Methods), similar to that commonly used for β-galactosidase. An indirect end-label assay of MNase cleavage sites revealed that the chromatin structure of the GAL1-10 promoter is well preserved in the MEL1 fusion (Fig. 4). The accessible UAS is not cut by MNase in either chromatin or naked DNA (the GAL1-10 UAS is cut by DNase I in chromatin and naked DNA [46]) and is flanked by protected regions of about 150 bp, characteristic of positioned nucleosomes. The GAL10 TATA element is near the center of such a protected region, and a BanII site 23 bp from the TATA element is also strongly protected in chromatin (data not shown). In contrast, one of the two major TATA elements of the CYC1 promoter (45) in the GAL4-CYC1-lacZ reporter gene is in a region highly accessible to MNase (Fig. 5).
We examined transcription from the GAL10-MEL1 and GAL4-CYC1-lacZ reporter genes in SWI+ and swi1Δ cells by using several different activation domains fused to the GAL4 DNA-binding domain as activators, including the native activator GAL4. Figure 6 shows that neither GAL4, GAL4·ER·VP16, nor GAL4·ftz, which differ substantially in their ability to activate transcription, exhibit strong SWI-SNF dependence at the GAL4-CYC1-lacZ promoter. In contrast, the ability of GAL4 to activate transcription from the GAL10-MEL1 fusion gene was reduced about fourfold in swi1Δ relative to SWI+ yeast cells, consistent with previous work showing that GAL10 mRNA levels are reduced several fold in swi yeast cells (62). Even greater (8- to 12-fold) reduction in α-galactosidase activity from GAL10-MEL1 was seen in swi1Δ cells with GAL4·ER·VP16 and GAL4·ftz, indicating that the reduction in activity in swi1Δ cells seen with GAL4 was not due to indirect effects on genes required for galactose induction (2). These results indicate that a promoter having an accessible UAS and highly ordered chromatin structure around the TATA element and transcription start site (GAL10-MEL1) shows a stronger SWI-SNF dependence than one having an accessible proximal promoter region.
We also examined transcription from these two promoters by recruiting the RNA polymerase II holoenzyme via a GAL4·GAL11 fusion protein (4, 15). Transcriptional activation of the GAL4-CYC1-lacZ reporter by GAL4·GAL11 was comparable to activation by GAL4, consistent with the results of earlier work (4, 15, 18), and was reduced about threefold in swi1Δ compared to SWI+ cells, indicating that the SWI-SNF dependence of transcription from this promoter is slightly increased when normal activator function is bypassed by holoenzyme recruitment (Fig. 6). Transcription of GAL10-MEL1 by GAL4·GAL11 in SWI+ cells was also robust (Fig. 6). Remarkably, the transcription of this reporter by GAL4·GAL11 in swi1Δ cells was almost abolished (Fig. 6). Consistent with this result, GAL4·GAL11 strongly supports the growth of gal4 SWI+ yeast in galactose medium but does not support the growth of gal4 swi1Δ yeast in galactose (data not shown). The nearly complete SWI-SNF dependence of GAL10-MEL1 transcription by GAL4·GAL11 supports the idea that the SWI-SNF complex exerts its effect at a step subsequent to transcriptional activator binding by two lines of reasoning. First, if SWI-SNF was needed principally to allow activator binding, the same SWI-SNF dependence should be observed for a given promoter no matter which activation domain is tethered to the GAL4 DNA-binding domain, which is not consistent with the very different dependencies on the SWI-SNF complex seen for GAL4 (about 4-fold) and GAL4·GAL11 (about 50-fold). Second, if GAL4·GAL11 activates transcription by directly recruiting the RNA polymerase II holoenzyme (15), the near-complete SWI-SNF dependence seen at the GAL10-MEL1 promoter with this protein suggests a stringent requirement for the SWI-SNF complex in order for the basal transcription machinery (holoenzyme or TFIID) to gain access to the proximal promoter. This would be consistent with the SWI-SNF complex being associated with the holoenzyme (82), although this is at present somewhat controversial (9).
DISCUSSION
The SWI-SNF complex is required in vivo for maximal transcriptional induction of a number of promoters and for chromatin remodeling at the SUC2 promoter in yeast (see Introduction). In vitro, the SWI-SNF complex can enable the binding of GAL4 to a single site in a nucleosomal template (14). These findings suggest a model in which the SWI-SNF complex assists transcriptional activation in vivo by helping activator proteins bind to their sites in chromatin (60). Alternatively, the SWI-SNF complex could be recruited by activators, for example in association with the RNA polymerase II holoenzyme (82), and could assist in steps subsequent to activator binding, such as binding of TBP and RNA polymerase II to their sites in chromatin.
We have tested these ideas by using templates with defined chromatin structures to examine transcription and chromatin perturbation induced by GAL4 and derivatives thereof in vivo. Our results indicate that activator binding to nucleosomal sites can occur essentially independently of SWI-SNF and that the SWI-SNF complex is likely to play its major role in overcoming the repressive effects of chromatin at steps subsequent to activator binding during transcriptional activation. First, perturbation of a positioned nucleosome containing a single strong GAL4-binding site by GAL4 occurs essentially independently of SWI-SNF (Fig. 1). Second, activation domain-dependent chromatin remodeling showed substantial SWI-SNF dependence (Fig. 3), although it was not eliminated entirely (Fig. 2), consistent with the partial dependence on SWI-SNF observed for transcriptional activation (7, 18, 25, 42, 62) (Fig. 6). Finally, a GAL10-MEL1 fusion, which has a nonnucleosomal UAS and a highly organized chromatin structure in the region of the TATA element and transcription start site, shows a strong SWI-SNF dependence with GAL4, GAL4·ER·VP16, and GAL4·ftz and a nearly complete SWI-SNF dependence with GAL4·GAL11 (Fig. 6), consistent with a requirement for SWI-SNF activity at a step subsequent to activator binding.
GAL4 binding to a nucleosomal site in swi1Δ yeast cells.
We have shown that GAL4 can bind to a nucleosomal site with concomitant nucleosome perturbation in swi yeast cells. We cannot rule out that the SWI-SNF complex enhances GAL4 binding to TA17Δ80 to a minor degree which our assays are not sensitive enough to measure; nor can we exclude that examination of other transcription factors, or weaker GAL4-binding sites, might show an increased dependence on the SWI-SNF complex. In fact, GAL4 binding to a pair of weak sites is impaired in swi yeast cells in at least one instance (7). One way this could be reconciled with the present work is if stable binding to weak sites were especially dependent on the activation domain. Indeed, evidence exists that transcription factor binding and chromatin perturbation depends on activation domains in vivo (6, 69–71). Furthermore, stable binding of GAL4 to low- and moderate-affinity binding sites has been shown to be strengthened by the presence of a nearby TATA element (76). A likely explanation for this effect, which requires the presence of an activation domain, is that binding to low- and moderate-affinity sites is enhanced by cooperative interactions between GAL4 and general transcription factors such as TFIID. If interactions involving the latter were diminished in swi cells, GAL4 binding could be affected. This explanation might also help to account for factor occupancy being diminished only about 2-fold in swi cells while transcription from the same reporter was decreased by 25-fold (7).
Recently, it has been shown that chromatin remodeling of the PHO5 promoter by the transcription factor PHO4 can occur in swi yeast cells (18). This example differs from the work described here in that PHO4 binding first occurs at a site between two positioned nucleosomes, which then allows nucleosome disruption and binding to the nucleosomal site (77). It seems likely that this disruption may differ mechanistically from that of nucleosome I of TA17Δ80 by GAL4 (Fig. 1), in which binding to a nucleosomal site occurs directly. Different mechanisms for the two systems are also suggested by the different results obtained with PHO4-GAL11 with the PHO5 promoter (weak SWI-SNF dependence, similar to that seen with PHO4) from those obtained with GAL4·GAL11 with the GAL10-MEL1 reporter (nearly complete dependence, and much stronger dependence than seen with GAL4). However, the conclusion that chromatin remodeling by activators can occur by a pathway independent of SWI-SNF in vivo is consistent with our findings. Further work is required to elucidate the mechanism by which GAL4 can access a nucleosomal site in vivo and to determine whether any of several other candidate chromatin-remodeling activities (9, 12, 28, 73, 75) contribute to chromatin remodeling during transcription factor binding in vivo.
A role for SWI-SNF in activation domain-dependent remodeling of chromatin.
We found that activation domain-dependent perturbation of TALS chromatin, especially as measured by a change in topology, was reduced in swi1Δ yeast (Fig. 3) but not eliminated (Fig. 2 and 3). This perturbation occurs outside of the context of a natural promoter (69), suggesting that the differences seen in swi cells are unlikely to be a consequence of defects in transcription. Similarly, the chromatin structure of the derepressed SUC2 promoter is altered by mutations in SNF2-SWI2 or SNF5, and the differences between SWI+ and swi cells are observed even when the TATA element has been mutated (25), although a direct effect of an activator on the chromatin structure of this locus has not been established. These findings suggest that transcriptional activators have a general capability to remodel chromatin in the vicinity of their binding sites and that this ability is impaired but not abolished in the absence of the SWI-SNF complex.
We have examined the changes in the chromatin structure of the GAL10-MEL1 promoter which accompany activation in SWI+ yeast cells in the hope of learning more about the specific remodeling which occurs at this strongly SWI-SNF-dependent promoter. However, in agreement with observations made with the GAL10 promoter (11, 16, 47), we found only subtle changes in chromatin structure in the GAL10 promoter upon activation, although changes in the MNase cleavage pattern of the transcribed MEL1 coding sequence were seen (data not shown). Nevertheless, the apparent incorporation of the TATA element into a positioned nucleosome in the repressed promoter (10, 14, 43) (Fig. 4), in contrast to the accessible TATA element in the much less SWI-SNF-dependent GAL4-CYC1-lacZ reporter (Fig. 5), suggests that TBP recruitment could be an important step requiring chromatin remodeling by the SWI-SNF complex. Consistent with this notion, the SUC2 promoter, which also displays a strong SWI-SNF dependence (25, 58), appears to have its TATA element incorporated into a nucleosome under repressed conditions (18a, 25, 86). The PHO5 promoter, which has a nucleosomal TATA element and which shows little SWI-SNF dependence for its activation (18), appears to deviate from this rule. Perhaps in this gene the disruption that accompanies PHO4 binding to its nucleosomal site near the TATA element is sufficient to allow TBP access, or it may be that PHO4 is able to recruit other chromatin-remodeling activities that obviate the need for the SWI-SNF complex. Alternatively, chromatin remodeling by the SWI-SNF complex may be required for the binding of general transcription factors or RNA polymerase subsequent to TFIID binding.
Participation of the SWI-SNF complex in transcriptional activation in a step that depends on both the activation domain and the promoter is consistent with previous work (39, 42, 62). One mechanism which could account for this dependency would be recruitment by activation domains, directly or indirectly, of an RNA polymerase II holoenzyme which includes the SWI-SNF complex (82). This would also account for the complete SWI-SNF dependence of GAL10-MEL1 transcription by GAL4·GAL11. However, the extent to which the SWI-SNF complex is associated with the RNA polymerase II holoenzyme in vivo remains to be resolved (9).
If the role of the SWI-SNF complex is entirely to remodel chromatin to allow access to DNA by proteins involved in transcriptional activation, the nearly complete SWI-SNF dependence of GAL10-MEL1 transcription by GAL4·GAL11 also implies a strong requirement for chromatin remodeling for transcriptional activation at this promoter. Since GAL4 showed only partial SWI-SNF dependence at this promoter (Fig. 6), we infer that GAL4 must be able to remodel chromatin by some alternative pathway. It is possible that activators are generally capable of recruiting more than one chromatin-remodeling activity via more than one pathway. For example, SWI-SNF might be recruited along with the RNA polymerase holoenzyme (82) and the SAGA complex via adaptor complexes (21). Other identified chromatin-remodeling activities (9, 28, 73, 75) might be recruited by presently undefined pathways. Whether these many candidate activities are partly redundant or have evolved to operate at specific promoters or during specific steps of transcriptional activation or are involved in other processes which must contend with nucleosomal templates, such as replication and repair, remains to be determined.
ACKNOWLEDGMENTS
We thank R. T. Simpson for support and encouragement in the initial phase of this work; S. Hanes, M. Johnston, and D. Picard for gifts of plasmids; C. Peterson for yeast strains, many helpful discussions, and communication of unpublished results; and M. J. Curcio for helpful discussions. We are also grateful to Stephen Johnston for providing a plasmid carrying the MEL1 gene and for help in developing the α-galactosidase assay. We gratefully acknowledge the use of the Wadsworth Center molecular genetics core facility.
This work was supported by NIH grant GM51993 to R.H.M.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajwa W, Torchia T E, Hopper J E. Yeast regulatory gene GAL3: carbon regulation; UASGAL elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol Cell Biol. 1988;8:3439–3447. doi: 10.1128/mcb.8.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barberis A J, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 6.Bunker C A, Kingston R E. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc Natl Acad Sci USA. 1996;93:10820–10825. doi: 10.1073/pnas.93.20.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalli G, Bachmann D, Thoma F. Inactivation of topoisomerases affects transcription-dependent chromatin transitions in rDNA but not in a gene transcribed by RNA polymerase II. EMBO J. 1996;15:590–597. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Li B, Workman J L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 14.Coté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 15.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 16.Fedor M J, Kornberg R D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989;9:1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzpatrick V D, Ingles C J. The Drosophila fushi tarazu polypeptide is a DNA-binding transcriptional activator in yeast cells. Nature. 1989;337:666–668. doi: 10.1038/337666a0. [DOI] [PubMed] [Google Scholar]
- 18.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 18a.Gavin I M, Simpson R T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Germond J E, Hirt B, Oudet P, Gross-Bellard M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP-TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant P A, Duggan L, Coté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 22.Griggs D W, Johnston M. Regulated expression of the GAL4 gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarante L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill J, Ian K A, Donald G, Griffiths D E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 26.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Marata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 29.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston S A, Hopper J E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston S A, Salmeron J M, Jr, Dincher S S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 32.Kent N A, Bird L E, Mellor J. Chromatin analysis in yeast using NP-40 permeabilized spheroplasts. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kew O M, Douglas H C. Genetic coregulation of galactose and melibiose utilization in Saccharomyces. J Bacteriol. 1976;125:33–41. doi: 10.1128/jb.125.1.33-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 35.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 36.Kladde M P, Simpson R T. Positioned nucleosomes inhibit Dam methylation in vivo. Proc Natl Acad Sci USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 38.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 39.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 40.Laughon A, Gesteland R F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984;4:260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurent B C, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 43.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 44.Laybourn P J, Kadonaga J T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 45.Li W-Z, Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984;12:8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohr D, Lopez J. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem. 1995;270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 48.Louvion J-F, Havaux-Copf B, Picard D. Fusion of GAL4-VP16 to a steroid binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene. 1993;131:129–134. doi: 10.1016/0378-1119(93)90681-r. [DOI] [PubMed] [Google Scholar]
- 49.Lukacs E. Probability and mathematical statistics: an introduction. London, United Kingdom: Academic Press, Ltd.; 1972. [Google Scholar]
- 50.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 51.Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987;7:1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 53.Morse R H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989;8:2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morse R H. Topoisomer heterogeneity of plasmid chromatin in living cells. J Mol Biol. 1991;222:133–137. doi: 10.1016/0022-2836(91)90198-f. [DOI] [PubMed] [Google Scholar]
- 55.Morse R H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 56.Nedospasov S A, Georgiev G P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980;92:532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- 57.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcriptional control. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 60.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interaction? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 61.Peterson C L, Dingwall A, Scott Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 63.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 65.Roth S Y, Dean A, Simpson R T. Yeast α2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990;10:2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimizu M, Roth S Y, Szent-Gyorgyi C, Simpson R T. Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simpson R T, Thoma F, Brubaker J M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 69.Stafford G A, Morse R H. Chromatin remodeling by transcriptional activation domains in a yeast episome. J Biol Chem. 1997;272:11526–11534. doi: 10.1074/jbc.272.17.11526. [DOI] [PubMed] [Google Scholar]
- 70.Svaren J, Schmitz J, Hörz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka M. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci USA. 1996;93:4311–4315. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Treich I, Cairns B R, de los Santos T, Brewster E, Carlson M. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 74.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 75.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 76.Vashee S, Kodadek T. The activation domain of GAL4 protein mediates cooperative promoter binding with general transcription factors in vivo. Proc Natl Acad Sci USA. 1995;92:10683–10687. doi: 10.1073/pnas.92.23.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venter U, Svaren J, Schmitz J, Schmid A, Hörz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vettese-Dady M, Walter P, Chen H, Juan L J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wallrath L L, Lu Q, Granok H, Elgin S C R. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 80.Wang W, Coté J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 81.Wechsler M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin-versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF transcriptional activators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 83.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 84.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 85.Wu C. The 5′ ends of Drosophila heat-shock genes in chromatin are sensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 86.Wu L, Winston F. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4230–4234. doi: 10.1093/nar/25.21.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]