Abstract
The lactogenic hormones, i.e., prolactin and glucocorticoids, act in concert to stimulate transcription factors responsible for hormone-dependent milk protein gene expression. In the mammary gland, prolactin activates Stat5a and Stat5b and glucocorticoids activate the glucocorticoid receptor (GR). Immunoprecipitation experiments revealed that in mammary cells, Stat5a, Stat5b, and the GR are physically associated in vivo. The association is not dependent on lactogenic hormone treatment and is evident at all stages of mammary gland development. Immunodepletion experiments indicated that a fraction of GR and Stat5 proteins are not associated, suggesting that there are different intracellular pools of these proteins. Lactogenic hormone treatment of HC11 mammary cells resulted in tyrosine phosphorylation of Stat5a and Stat5b, dimerization, and rapid nuclear translocation of both Stat5 proteins. Following hormone treatment, Stat5a-Stat5b heterodimers were detected by their coimmunoprecipitation. In addition, immunodepletion experiments followed by gel shift analyses revealed the presence of active Stat5a and Stat5b homodimers. In mammary cells, Stat5b homodimers are less abundant than Stat5a homodimers. Although the GR does not bind the Stat5 DNA binding site directly, it could be detected with the Stat5-DNA complex. These results suggest that glucocorticoids affect milk protein gene expression via association of the GR with Stat5. Thus, there is a functional coupling between Stat-dependent and nuclear hormone receptor-dependent gene transcription.
Mammary gland differentiation requires the coordinated action of growth factors and hormones that promote morphological development and milk protein production in the lactating gland (44). In order to understand the molecular events contributing to mammary cell differentiation, in vitro cell culture systems have proven invaluable. The HC11 mouse mammary epithelial cell line is a useful model system for studying mammary cell differentiation (3, 6, 7, 43), since treatment of the cells with the lactogenic hormones, i.e., prolactin (PRL) and glucorticoids, leads to the production of several milk proteins (29). The PRL intracellular target is MGF/Stat5 (16, 46), a member of the Stat family of transcription factors (signal transducer and activator of transcription) (37). PRL binding to its receptor leads to Janus kinase 2 (Jak2)-mediated phosphorylation of Stat5 on tyrosine (11, 36). Activated Stat5 dimers translocate to the nucleus where they bind to gamma interferon (IFN-γ)-activated (GAS)-like sites leading to transcriptional activation of target genes, including β-casein. Stat5a and Stat5b, the products of two closely related genes (2, 23, 30), are constitutively expressed in HC11 cells (48) and in mammary glands at all stages of development (18, 25). Stat5a and Stat5b are able to act independently as homodimers or in combination as heterodimers (28, 34). Despite their homology, recent reports indicate that they can be differentially activated (28), suggesting that they might have functional difference. In fact, mice deficient in Stat5a (24) or in Stat5b (45) have impaired mammary gland development and do not lactate, showing that the two Stat5 proteins have distinct functions in the gland. Here, we report the existence of active Stat5a and Stat5b homodimers and heterodimers in both HC11 cells and in lactating mammary gland tissue. Interestingly, despite the similar levels of Stat5a and Stat5b in the mammary gland, Stat5b homodimers are less abundant than Stat5a homodimers.
Interaction of the glucocorticoid receptor (GR) with its steroid ligand results in an allosteric change that allows the complex to bind specific DNA sequences, the glucocorticoid response element (GRE), and to modulate transcription of target genes (4). In HC11 cells, optimal β-casein gene expression is achieved only in the presence of both lactogenic hormones, i.e., glucocorticoids and PRL (9). The β-casein gene promoter does not have a consensus GRE. However, half-palindromic GREs have been detected. Mutation of the Stat5 binding site in the β-casein gene promoter inhibits both glucocorticoid- and PRL-induced transcription (40), suggesting that the GAS-like site influences glucocorticoid-mediated transcription. It has recently been shown that there is a functional interaction between Stat5 and the GR in transient cotransfected COS cells (42). Here, we show by coimmunoprecipitation that a portion of the cellular pools of Stat5 and GR are physically associated in mammary cells. The GR-Stat5 association may be more general, since it was also found in liver extracts and in NIH 3T3 mouse fibroblasts. The interaction appears specific for Stat5, since Stat3 is not associated with the GR in any of the cells tested. In lactogenic hormone-treated mammary cells, the GR is part of the Stat5-DNA complex, suggesting that there is a functional coupling between Stat- and nuclear hormone receptor-dependent gene transcription.
MATERIALS AND METHODS
Materials.
[32P]ATP was purchased from Amersham. The following antibodies were used: rabbit polyclonal antiserum raised against a purified rat GR fragment (amino acids 440 to 795) (17), anti-Stat5a and -Stat5b sera raised in rabbits against specific C-terminal peptides (Stat5a [LDARLSPPAGLFTSARSSLS] and Stat5b [MDSQWIPHAQS] [48]), phosphotyrosine-specific monoclonal antibody (MAb) (10), rabbit anti-Raf-1 serum 1558 (47), and anti-Stat3, from Santa Cruz Biotechnology (sc-482).
Cell culture and lactogenic induction.
NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. HC11 mammary epithelial cells were grown to confluency and maintained for 3 days in RPMI 1640 supplemented with 10% fetal calf serum and 10 ng of epidermal growth factor and 5 μg of insulin per ml. These competent cells (43) were washed and incubated for 18 h in serum-free medium (RPMI 1640 containing 1-mg/ml fetuin and 10-μg/ml transferrin) and then treated for the indicated times with serum-free medium supplemented with 10−6 M dexamethasone, 5-μg/ml insulin, and 5-μg/ml ovine prolactin (luteotropic hormone; Sigma).
Extracts of whole cells and tissues.
Cells were washed with phosphate-buffered saline and scraped into 700 μl of a buffer containing 10 mM NaHPO4 (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol (DTT), 400 mM KCl, 10% glycerol, 5-μg/ml aprotinin, 5-μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 μM NaF, 2 mM Na2VO4, and 50 μM Na-glycerophosphate. After three cycles of freezing and thawing, extracts were centrifuged for 15 min at 17,000 × g and supernatants were recovered and kept at −70°C (51). Livers and mammary glands isolated from different stages of development were taken from BALB/c mice and frozen in liquid nitrogen. Whole-cell lysates were prepared as described above, with the help of a Polytron.
Subcellular fractionation.
Cells were lysed in 700 μl of hypotonic buffer (10 mM KCl, 20 mM HEPES [pH 7.0], 1 mM MgCl2, 0.1% Triton X-100, and 20% glycerol, protease, and phosphatase inhibitors [as described above]) in a Potter homogenizer (25 strokes) and centrifuged for 5 min at 800 × g (Eppendorf centrifuge). Supernatants (cytoplasmic fraction) were frozen at −70°C. Pelleted nuclei were resuspended in hypertonic buffer (hypotonic buffer plus 300 mM NaCl), and protein extracts were prepared by constant agitation for 20 min at 4°C. Debris was removed by centrifugation, and nuclear extracts were frozen at −70°C (41). Nuclear fraction of lactating mammary glands was prepared based on a published method (12, 41). Minced tissue (about 2 g) was increased to 7 to 10 ml with homogenization buffer (10 mM HEPES [pH 7.6], 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, 2 M sucrose, 10% glycerol), and cells were broken with a Teflon glass homogenizer. Cell disruption and nucleus integrity were monitored under the microscope. The homogenate was layered on a cushion of homogenizer buffer and centrifuged at 120,000 × g for 30 min at 4°C (35,000 rpm; Beckmann TL-100 ultracentrifuge). The pellet was resuspended in extraction buffer (10 mM HEPES [pH 7.6], 400 mM KCl, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 2 mM Na2VO4, 10% glycerol), and the buffer was incubated for 1 h at 4°C under agitation. The lysate was centrifuged for 5 min at 10,000 × g and dialyzed against NED buffer (10 mM HEPES [pH 7.6], 40 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 10% glycerol) twice (250 ml each time) for 90 min at 4°C. Nuclear protein extracts were aliquoted and frozen at −70°C.
Immunoprecipitation and Western blot analysis.
Protein concentrations were determined by the Bradford method. A 500-μg amount of protein was incubated with 1 μl of Stat5 and GR antisera for 1 h on ice. Protein A-Sepharose-coupled beads (Sigma) were added for 30 min at 4°C under constant agitation. The beads were pelleted and washed three times with extraction buffer and once with TNE buffer (140 mM NaCl, 50 mM Tris [pH 7 to 8], 5 mM EDTA). Immunoprecipitated proteins were boiled in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [8 to 10% polyacrylamide]). Immunodepletion was performed by two consecutive incubations of the extracts with the specific antiserum. Proteins were electroblotted onto polyvinylidene difluoride membranes, and specific protein detection was performed in TTBS solution (50 mM Tris [pH 7.5], 150 mM NaCl, 0.05% Tween 20, 20% normal horse serum) with specific antisera against phosphotyrosine (1:30,000), Stat5a (1:5,000), Stat5b (1:2,000), GR (1:2,000), raf (1:1,000), and Stat3 (1:1,000). Proteins were visualized with peroxidase-coupled second antibody by the ECL detection system (Amersham). Quantification of specific bands was performed with different times of exposure and ImageQuant software (Molecular Dynamics). For reprobing, membranes were stripped in 62.5 mM Tris (pH 6.7)–2% SDS–100 mM β-mercaptoethanol at 40°C for 40 min.
EMSA.
A 6-μg amount of nuclear protein extract was incubated with the Stat5 binding site from the bovine β-casein promoter (5′-AGATTTCTAGGAATTCAATCC-3′) (46) (30,000 cpm, 5 fmol) for 30 min on ice in 20 μl of electrophoretic mobility shift assay (EMSA) buffer [10 mM HEPES (pH 7.6), 2 mM NaHPO4, 0.25 mM EDTA, 1 mM DTT, 5 mM MgCl2, 80 mM KCl, 2% glycerol, 100-μg/ml poly(dI-dC)]. Supershifting was performed by adding anti-Stat5a or anti-Stat5b serum during the last 15 min of the binding reaction at a dilution of 1:500. Specific binding was analyzed on a native 4% polyacrylamide gel that was prerun for 2 h at 200 V in 0.25× TBE buffer (22.5 mM Tris-borate [pH 8.0], 0.5 mM EDTA). The samples were loaded and electrophoresed in the same buffer for 1 h at 200 V. The gels were dried and exposed to X-ray film. Specific bands were quantified with a PhosphorImager and ImageQuant software (Molecular Dynamics).
Immunoprecipitation of the GR-Stat5-DNA complex.
The method used for immunoprecipitation of the complex was basically as previously described (1). A 6-μg amount of nuclear extract was incubated with [γ32P]ATP-labeled wild-type (Col 7) (5′-TGTGGACTTCTTGGAATTAAGGAACTTTTG-3′) or mutated (Col 7.3) 5′-TGTGGACTTATTTTAATTAAGGAACTTTTG-3′ (38) Stat5 DNA binding site in 20 μl of EMSA buffer on ice for 30 min (approximately 15,000 cpm, 3 fmol). GR antiserum or preimmune serum was added at a dilution of 1:800 for 30 min, followed by 10 μl of Sepharose-coupled protein A for another 30 min. Samples were washed once with 500 μl of hypotonic extraction buffer and twice with TNE buffer. Radiolabeled probe associated with immunoprecipitated material was measured and compared with total added radioactivity.
RESULTS
Kinetics of Stat5 DNA binding following lactogenic induction and identification of both Stat5a and Stat5b in the DNA complex.
Activation of the PRL receptor in HC11 mammary epithelial cells leads to phosphorylation of Stat5a and Stat5b on tyrosine (48). The activated Stat5 proteins bind to a GAS-like site in the promoter of the β-casein gene and stimulate its transcription (40). To monitor the activation of Stat5, we investigated the formation of Stat5-DNA complexes by EMSAs with nuclear extracts from lactogenic hormone-treated cells and the Stat5 binding site from the β-casein promoter as a probe. Binding was maximal within 7 min and declined by 3 h to a level which remained constant for 24 h (Fig. 1A). In order to analyze the composition of the Stat5-DNA complex, nuclear protein extracts of HC11 cells treated for 7 min with lactogenic hormones were analyzed by EMSA in the presence of anti-Stat5a or anti-Stat5b serum. Addition of either anti-Stat5a or anti-Stat5b serum resulted in a supershift of part of the complex (Fig. 1B, lanes 3 and 4, respectively), which was not observed with preimmune serum (Fig. 1B, lane 2). Simultaneous addition of Stat5a and Stat5b antisera caused a supershift of the entire complex (Fig. 1B, lane 5). Two distinct complexes were observed in the anti-Stat5a but not the anti-Stat5b supershifted material (Fig. 1B; compare supershifts in lanes 3 and 4). This point will be discussed further with reference to Fig. 3. A similar analysis was done with nuclear extracts taken from the mammary glands of lactating mice, in which the highest levels of Stat5a and Stat5b activation and dimerization have been detected (5, 25) (Fig. 1C). As for HC11 cells, addition of anti-Stat5a or anti-Stat5b serum resulted in a supershift of part of the complex (Fig. 1C, lanes 2 and 3), while simultaneous addition of both antisera resulted in a complete supershift (Fig. 1C, lane 4). These results demonstrate that only Stat5a and Stat5b interact with this site in lactogenic hormone-treated HC11 cells and in lactating mammary gland.
FIG. 1.
Stat5a and Stat5b DNA binding in mammary cells (EMSA). (A) A 6-μg amount of HC11 nuclear protein extract prepared from cell cultures treated for the indicated times with lactogenic hormones (lanes 2 to 7) or noninduced cells (lane 1) was incubated with radiolabeled oligonucleotide containing the Stat5 binding site. Complexes were electrophoresed in a 4% polyacrylamide native gel. (B and C) EMSA performed in the presence of the indicated antisera by using 6 μg of nuclear protein extracts from HC11 cells treated with lactogenic hormones for 7 min or 1 μg of nuclear protein extracts from lactating mammary gland, respectively. Prolactin, insulin, and dexamethasone were at concentrations of 5 μg/ml, 5 μg/ml, and 1 μM, respectively.
FIG. 3.
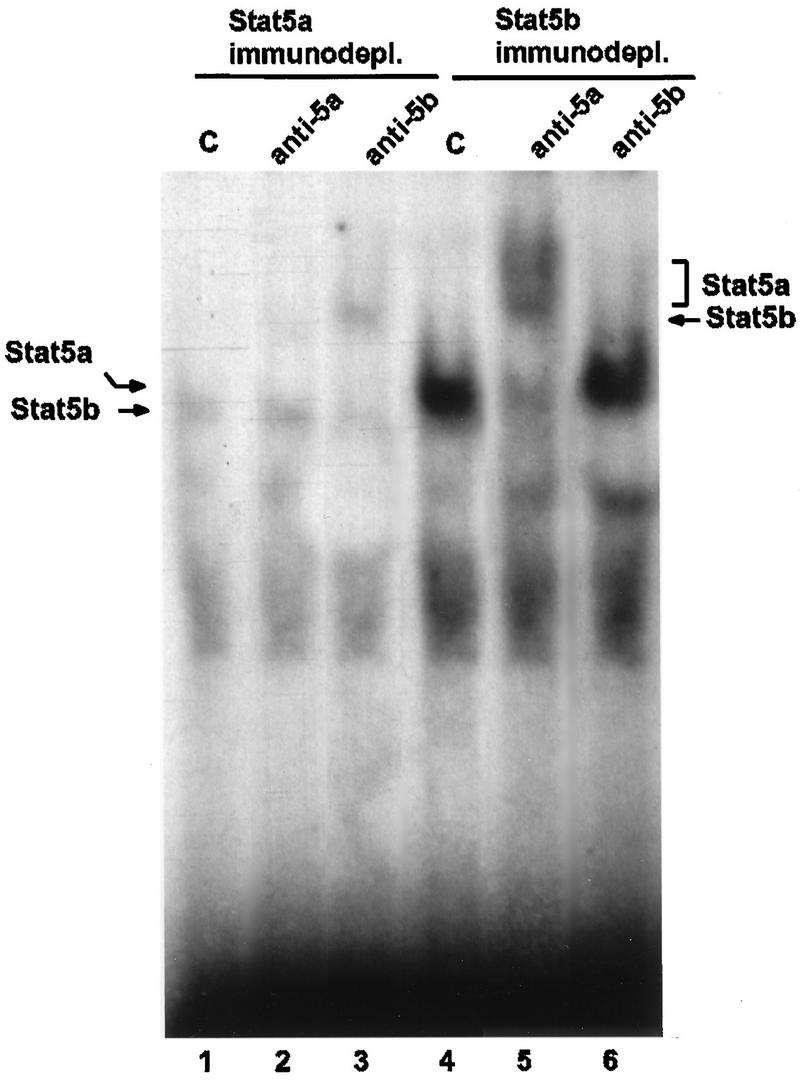
Active Stat5a and Stat5b homodimers in HC11 cells. A 200-μg amount of nuclear protein extracts from HC11 cells treated for 7 min with lactogenic hormones was immunodepleted (immunodepl.) of Stat5a or Stat5b as indicated; 6 μg of immunodepleted extracts was analyzed by EMSA either in the absence (control [C]) or in the presence of Stat5a or Stat5b antiserum as indicated. Complexes were resolved in a 4% polyacrylamide native gel. Complexes containing Stat5a or Stat5b and their supershifts are indicated. Prolactin, insulin, and dexamethasone were at concentrations of 5 μg/ml, 5 μg/ml, and 1 μM, respectively.
Stat5 tyrosine phosphorylation, nuclear translocation, and dimerization.
Homodimers of both Stat5a and Stat5b are independently able to activate transcription from the β-casein gene promoter in transiently transfected COS cells (23). Figure 1 shows that both Stat5a and Stat5b are able to bind to the DNA but does not provide information on the identity of the dimers, i.e., whether they are homodimers or heterodimers. Two approaches were taken to identify the Stat5 dimers. To probe for heterodimers, coimmunoprecipitation of the Stat5 proteins was examined. Identification of homodimers was determined by immunodepletion combined with EMSAs (see below). Stat5a and Stat5b were immunoprecipitated from nuclear and cytoplasmic fractions of lactogenic hormone-treated HC11 cells. Equal amounts of protein representing approximately one-eighth as much cytoplasmic material relative to nuclear material were used. Stat5a-Stat5b coimmunoprecipitation was detected only in the nuclear fraction (Fig. 2A and B, lanes 4 to 6) in which the highest level of association was found after 7 min of hormone treatment (Fig. 2A and B, lane 5). In the noninduced nuclear fraction, there was a small amount of Stat5a-Stat5b association (Fig. 2, lane 4).
FIG. 2.
Stat5a-Stat5b activation, nuclear translocation, and dimerization in HC11 cells. HC11 cells were treated with lactogenic hormones for the indicated times. A 500-μg amount of the nuclear and cytoplasmic protein fractions was immunoprecipitated (ip) with anti-Stat5a (A)- or anti-Stat5b (B)-specific antiserum. Immunocomplexes were resolved by SDS-8% PAGE. Filters were sequentially probed for phosphotyrosine (P-Y), Stat5a, and Stat5b by using specific antisera. Following each incubation, the membrane was stripped as described in Materials and Methods. b*, tyrosine-phosphorylated form of Stat5b. Molecular mass markers (in kilodaltons) are indicated on the right side. In the Western analysis for Stat5b, the upper bands in panels A (top panel) and B (lowest panel) are due to a nonspecific protein. Prolactin, insulin, and dexamethasone were at concentrations of 5 μg/ml, 5 μg/ml, and 1 μM, respectively. WB, Western blot; d, days.
Activation of Stat5 proteins was also investigated with phosphotyrosine-specific antiserum. The strongest Stat5 tyrosine phosphorylation correlated with the highest level of heterodimers (7 min) (Fig. 2A and B, lane 5) and DNA binding activity (Fig. 1A, lane 2). Due to the coimmunoprecipitation of Stat5a and Stat5b, the tyrosine phosphorylation results reflect the levels in both Stat5 proteins. A portion of the tyrosine-phosphorylated Stat5a (but not of Stat5b) was still in the cytoplasm after 7 min of induction (Fig. 2A and B, middle panels, lane 2). Active Stat5a was also found in the nuclei of nonstimulated cells (Fig. 2A, middle panel, lane 4). Active, nuclear Stat proteins have also been observed under other nonstimulated conditions (34). The phosphotyrosine contents of both nuclear Stat5 proteins decrease within 3 to 4 h (not shown) and remain at the level shown for 1 day (Fig. 2A and B, lane 6). As we have previously shown (48), Stat5b displays a slower electrophoretic mobility upon hormone induction. This shift was partially reversed after 1 day of induction, and only the upper band had detectable tyrosine phosphorylation (Fig. 2B, lower panel, lane 6). In the nuclear fraction from cells treated for 1 day with hormones, both Stat5b forms coimmunoprecipitated with Stat5a (Fig. 2A, lane 6), which suggests that inactive Stat5a-Stat5b heterodimers may exist at this time of induction.
Homodimers of Stat5a and Stat5b in mammary cells.
To identify active homodimers of Stat5a and Stat5b, specific immunodepletion followed by EMSA analyses was performed. Nuclear extracts from cells treated for 7 min with lactogenic hormones were immunodepleted of Stat5a or Stat5b by two consecutive immunoprecipitations with each Stat5 antiserum. Each serum recognizes the specific Stat5 protein as a monomer and as part of a homodimer or heterodimer. This procedure led to depletion of each Stat5 protein, as determined by Western analyses (data not shown). The immunodepleted supernatants were analyzed by EMSA with the Stat5 site of the β-casein promoter (Fig. 3). Stat5a-immunodepleted supernatants had weak DNA binding activity (Fig. 3, lane 1), which was completely supershifted by the anti-Stat5b serum (Fig. 3, lane 3). Addition of Stat5a-specific antiserum had no effect on the migration of the Stat-DNA complex (Fig. 3, lane 2), confirming the Stat5a immunodepletion. Stat5b-immunodepleted supernatants had strong DNA binding activity (Fig. 3, lane 4) which could be supershifted only by the anti-Stat5a serum. The shifted complex migrated as a doublet (Fig. 3, lane 5) in a manner similar to that observed in a previous publication (25) and in Fig. 1B (lane 3). This doublet was not observed in extracts from 10-day-lactating mammary glands (Fig. 1C, lane 2), suggesting that in HC11 cells there are two forms of Stat5a homodimers. Quantification of the complexes revealed that there are five times as many Stat5a homodimers as Stat5b homodimers in HC11 cells treated with lactogenic hormone.
Heterodimers of Stat5a and Stat5b were detected via their coimmunoprecipitation in extracts from lactating mammary gland tissue, which is the developmental stage with the highest level of Stat5 activation (25). The question of Stat5 homodimers was not addressed in that study (25). Immunodepletion experiments combined with EMSA analyses were used to identify the Stat5 homodimers present in lactating mammary glands. Each Stat5 protein was immunodepleted from nuclear extracts prepared from the glands of lactating mice. The results of the EMSA performed with the immunodepleted extracts are shown in Fig. 4. The complex formed by Stat5a-immunodepleted extracts could be shifted completely by anti-Stat5b serum (Fig. 4, lane 4), while the complex formed by Stat5b-immunodepleted extracts could be shifted completely by anti-Stat5a serum (Fig. 4, lane 6). The supershifted complexes were masked by a background band, which appeared in all of the immunodepleted extracts (Fig. 4, lanes 2 to 7) but not in normal extracts (lane 1). Quantification of the complexes revealed that the ratio of Stat5a homodimers to Stat5b homodimers in lactating mammary glands was 3:2.
FIG. 4.
Active Stat5a and Stat5b homodimers in lactating mammary glands. Nuclear protein extracts from mammary glands lactating for 10 days were immunodepleted of Stat5a or Stat5b as indicated. A 6-μg amount of immunodepleted (immunodepl.) extracts was analyzed by EMSA either in the absence (control [C]) or in the presence of Stat5a or Stat5b antiserum as indicated. Stat5a- or Stat5b-DNA complexes are indicated by arrows on the right side. Immunodepleted extracts gave a background band which masked the Stat5a and Stat5b supershifts. The electrophoretic mobility of the Stat-DNA complex from nonimmunodepleted nuclear extract is shown in lane 1.
Stat5 and the GR are physically associated.
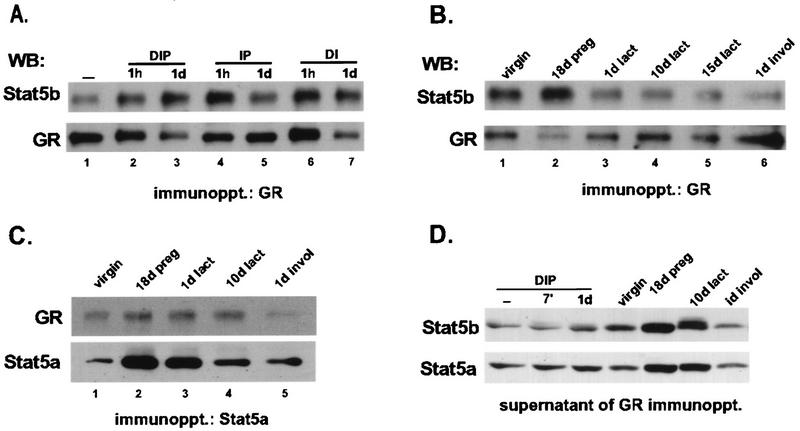
In HC11 cells, transcription of the β-casein gene is synergistically induced by PRL and glucocorticoids (5, 9). It has been shown that there is a physical interaction between GR and Stat5 in transient cotransfected COS cells (42); however, their association in a natural setting has not been reported. We examined the interaction Stat5 and the GR in mammary cells after immunoprecipitation of GR from whole-cell extracts. Coimmunoprecipitation of the GR and Stat5 was observed in HC11 cells treated with all combinations of lactogenic hormones (Fig. 5A) and in mammary gland extracts made from all developmental stages (Fig. 5B, lanes 1 to 6). The GR from dexamethasone-treated cells has a reduced electrophoretic mobility, likely reflecting ligand-induced changes in phosphorylation (17) (Fig. 5A, lanes 2, 3, 6, and 7). Association of the GR and Stat5a throughout mammary gland development was also observed when the precipitating serum was Stat5a specific (Fig. 5C). In the mammary gland, there appears to be a developmental-stage variation in the amounts of GR molecules that are associated with Stat5 (e.g., compare extracts from mammary glands from mice in involuting to pregnant stages). These differences were detected in independent assays; however, analyses of additional animals taken from each stage of development would be required in order to confirm the biological significance of this observation. Analysis of GR-immunodepleted extracts from both HC11 and mammary gland extracts revealed uncomplexed Stat5a and Stat5b proteins (Fig. 5D). Immunodepletion of Stat5a and Stat5b also left free GR in the supernatant (data not shown). These results indicate that only part of the cellular GR and Stat5 proteins is associated, perhaps due to specific modifications.
FIG. 5.
Stat5-GR association in mammary cells. (A) HC11 cells were treated for the indicated times with different combinations of lactogenic hormones (dexamethasone [D], insulin [I], and prolactin [P]). Control cells (−) were left in serum-free medium. A 500-μg amount of whole-cell protein extracts was immunoprecipitated (immunoppt.) with GR antiserum and subjected to SDS-10% PAGE. The membrane was successively incubated with Stat5b and GR antisera. (B) A 500-μg amount of whole-cell protein extracts of mammary gland tissues prepared from glands of virgin, 18-day-pregnant, 1-day-lactating, 10-day-lactating, and 15-day-lactating mice and from 1-day-involuting glands was immunoprecipitated with GR antiserum and analyzed as described in above. (C) A 500-μg amount of whole-cell protein extracts of mammary gland tissues prepared from glands of virgin, 18-day-pregnant, 1-day-lactating, and 10-day-lactating mice and from 1-day-involuting glands were immunoprecipitated with Stat5a antiserum. The membrane was successively incubated with GR and Stat5a antisera. (D) A 80-μg amount of GR-immunodepleted extracts was resolved by SDS-8% PAGE and incubated with Stat5a and Stat5b antisera. Prolactin, insulin, and dexamethasone were at concentrations of 5 μg/ml, 5 μg/ml, and 1 μM, respectively. WB, Western blot; d, days; preg, pregnant; lact, lactating; invol, involuted.
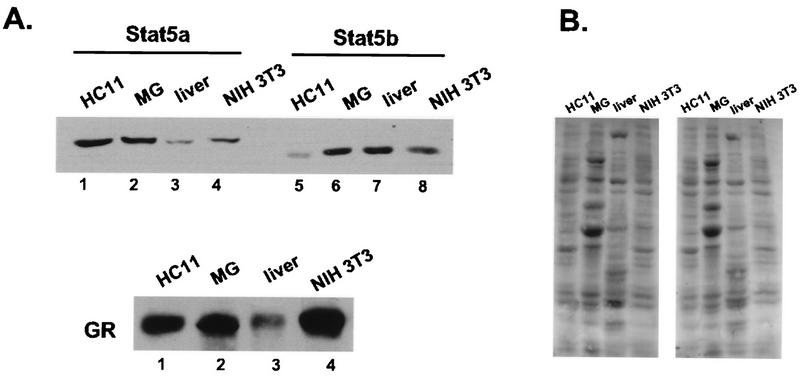
In order to determine if the GR-Stat5 association also occurs in other cells and organs, we examined the interaction of the two proteins in the liver and in NIH 3T3 mouse fibroblasts. A Western analysis showing the level of each of the three proteins is shown in Fig. 6A. Coimmunoprecipitation of the GR with both Stat5a and Stat5b was observed in all cases (Fig. 7A and B), suggesting that association of the GR with Stat5 may be widespread. In contrast, there was no coimmunoprecipitation of the GR and Stat3, which was also abundantly expressed in each extract (Fig. 7C).
FIG. 6.
Protein levels of Stat5a, Stat5b, and GR in in vitro-cultured cells and organs. (A) An 80-μg amount of whole-cell extracts made from HC11 cells, lactating mammary gland (MG), liver, and NIH 3T3 cells was resolved by SDS-8% PAGE. The membrane was probed for Stat5a (left side) or Stat5b (right side) (upper panel), stripped, and reprobed for GR (lower panel) with specific antisera. (B) The membranes were stained with 0.1% amido black to control for protein loading.
FIG. 7.

Stat5-GR association in in vitro-cultured cells and organs. A 500-μg amount of whole-cell extracts made from HC11 cells, lactating mammary gland (MG), liver, and NIH 3T3 cells was immunoprecipitated (ip) with Stat5a antiserum (A) or Stat5b antiserum (B) and resolved by SDS-10% PAGE. The membrane was incubated sequentially with GR and Stat5a (A) or Stat5b (B) antiserum. (C) An 80-μg amount of the protein extracts described above (lanes 1 to 4) or 500 μg of the same extracts immunoprecipitated with GR antiserum (lanes 5 to 8) was resolved by SDS-10% PAGE and probed for Stat3 with specific antiserum. Arrows indicate the proteins shown on the Western Western blot (WB). Ig, immunoglobulin.
Stat5 and GR nuclear translocation.
The observation that Stat5 and the GR are physically associated in mammary cells, irrespective of their activation states, raises the question of whether independent activation of Stat5 or the GR would result in nuclear translocation of both molecules. To examine this, HC11 cells were treated with various combinations of hormones and nuclear and cytoplasmic fractions were probed for Stat5a and the GR by Western analyses (Fig. 8A). There was approximately one-eighth as much cytoplasmic material relative to nuclear material in this Western blot. The purity of the extracts was controlled by a Western analysis for the cytoplasmic Ser/Thr kinase Raf-1, which is easily detectable in HC11 cells (48) (Fig. 8C). In control nonstimulated cells, there is a low level of both the GR and Stat5a in the nucleus (Fig. 8A, lane 8). Quantification of the bands indicates that in cells treated with the lactogenic hormones, there are 5 and 3.5 times more, respectively, GR and Stat5 in the nucleus compared to nonstimulated cells (Fig. 8A, lanes 9 versus 8). Quantification of the GR in the nuclei of cells treated with PRL indicated that there is approximately 1.5 times more GR compared to nonstimulated cells (Fig. 8A, upper panel, lanes 11 and 12 versus lane 8). A similar increase in nuclear Stat5a is observed in cells treated with dexamethasone compared to control cells (Fig. 8A, lower panel, lanes 13 and 14 versus lane 8). These results show that nuclear translocation is mainly dependent on ligand-induced activation. The levels of nuclear GR and Stat5 detected under conditions in which the cognate ligand was not present were much lower than the levels detected in the nuclei of cells treated with both lactogenic hormones. The mechanism underlying the ligand-independent translocation is at present unknown. Interestingly, we observed that there is 1.5 times more Stat5a in the nuclear fraction of cells treated with both lactogenic hormones compared to the same fraction from cells treated with PRL only (Fig. 8A, lanes 9 and 10 versus 11 and 12). This result suggests that more Stat5 may be recruited to the nucleus in the presence of ligand-activated GR.
FIG. 8.
Stat5 and GR nuclear translocation. (A) HC11 cells were treated for the indicated times with different combinations of lactogenic hormones (dexamethasone [D], insulin (I), and prolactin [P]). Control cells (−) were left in serum-free medium. Nuclear and cytoplasmic fractions were prepared, and 80 μg of each was subjected to SDS-8% PAGE and probed with Stat5a and GR antisera. (B) HC11 cells were treated for the indicated times with lactogenic hormones; 80 μg of whole-cell protein extracts was subjected to SDS-8% PAGE and incubated with Stat5a and Stat5b antisera. (C) Cellular fractionation was controlled by probing two independent preparations of each fraction with a Raf-1-specific antiserum. Prolactin, insulin, and dexamethasone were at concentrations of respectively, 5 μg/ml, 5 μg/ml, and 1 μM, respectively.
It is well documented that glucocorticoid treatment leads to an increase in the turnover of the GR, which is reflected by a decrease in the amount of protein (17). This is seen in the GR immunoprecipitates from HC11 cells treated for 1 day with dexamethasone (Fig. 5A, lower panel, lanes 3 and 7). The level of nuclear Stat5 increases following 7 min of PRL treatment and decreases slightly after 1 h, suggesting that activated Stat5 might also turn over more rapidly than its nonactivated counterpart (Fig. 8A, lower panel, lanes 10 and 12). However, Western analyses carried out with whole-cell extracts show that the levels of Stat5a and Stat5b remain constant between 1 h and 1 day of lactogenic hormone treatment (Fig. 8B), suggesting that PRL does not cause a dramatic change in Stat5 turnover.
GR is in the Stat5-DNA complex.
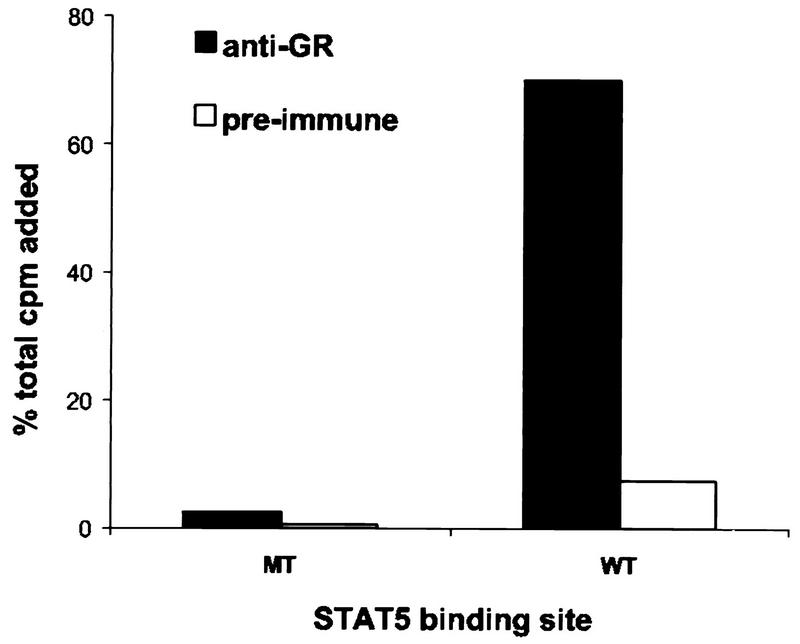
Although a physical association between the GR and Stat5 is easily detectable by coimmunoprecipitation experiments, we failed to demonstrate this interaction in an EMSA followed by a supershift with GR antiserum. This might be due to disassembly of the complex during electrophoresis. These results contrast with those obtained for COS cells, in which it was possible to perform supershifts of the Stat5-DNA complex with GR antiserum (42). This difference is likely to be due to the high level of protein expression which can be achieved in COS cells, which might not mimic the stoichiometry of the endogenous molecules. In order to determine whether the GR is a part of the Stat5-DNA complex in mammary cells, another experimental approach was employed (1). Nuclear extracts from HC11 cells treated for 7 min with lactogenic hormones were incubated in an EMSA binding reaction mixture with the radiolabeled wild-type Stat5-DNA binding site. Anti-GR or preimmune serum was added, and the complexes were immunoprecipitated. Labeled probe associated with immunoprecipitated material was counted and expressed as a percentage of initially added probe (Fig. 9). A total of 70% of the wild-type binding site was immunoprecipitated when the GR antiserum was added to the reaction mixture. In contrast, GR antiserum was unable to immunoprecipitate the radiolabeled, mutated Stat5 binding site. This result shows that the Stat5 binding site can be immunoprecipitated by the GR antiserum, indicating that GR participates in the Stat5-DNA complex.
FIG. 9.
GR antiserum immunoprecipitates the Stat5-DNA complex. HC11 cells were treated for 7 min with lactogenic hormones, and 6 μg of nuclear protein extract was incubated in EMSA buffer with radiolabeled mutated (MT) or wild-type (WT) Stat5 binding site. Anti-GR or preimmune serum was added to the reaction and immunoprecipitated with protein A-Sepharose beads. Immunocomplexes were collected, and radioactivity was quantified in a scintillation counter. Values were expressed as percentages of initially added probe. Prolactin, insulin, and dexamethasone were at concentrations of 5 μg/ml, 5 μg/ml, and 1 μM, respectively.
DISCUSSION
The intracellular mediators of the lactogenic hormones, i.e., glucocorticoids and PRL, are the GR and Stat5. The pathways activating these two transcription factors are distinct but converge at the promoter of the β-casein milk protein gene. In this paper, we describe the specific association of a portion of the Stat5 and GR proteins in mammary epithelial cells, which might provide a mechanism for the synergistic action of PRL and glucocorticoids in the induction of β-casein gene expression. In addition, we show that following lactogenic hormone treatment, both Stat5 proteins rapidly translocate to the nucleus and interact with the GAS-like site of the β-casein gene promoter. We demonstrate that active Stat5a and Stat5b homodimers as well as Stat5 heterodimers are present in mammary cells and that the Stat5a homodimers are more abundant than Stat5b homodimers.
Stat5 is activated by numerous cytokines and growth factors in diverse cell types (14, 34, 35), suggesting that Stat5 proteins have specific roles in various cell types. Differences in the activation of Stat5a and Stat5b have been previously observed. In COS cells transfected with the β-casein promoter-luciferase construct, Stat5b (but not Stat5a) stimulates basal β-casein gene transcription in the absence of PRL (23). In mammary cells, Stat5a and Stat5b have different binding affinities for the three different Stat5 binding sites mapped to the β-lactoglobulin gene promoter (31). Similarly, the two Stat5 binding sites mapped in the murine β-casein gene promoter (40) interact with homodimers of Stat5a and Stat5b with different affinities (13). Interestingly, occupancy of both sites was observed only in extracts from lactating mammary glands but not in extracts from the glands of pregnant mice (39). These data suggest that during the development of the mammary gland, selective gene activation might be attributed to the activation of different Stat5 dimers, making it important to characterize the dimers present in the gland. In addition to Stat5a-Stat5b heterodimers, we observed that Stat5a and Stat5b homodimers are present and that the Stat5a homodimers are more abundant than the Stat5b homodimers in both HC11 cells treated with lactogenic hormones and in lactating mammary glands. Our experiments do not allow a direct comparison between the levels of Stat5a-Stat5b heterodimers and the levels of Stat5 homodimers. The Stat5-DNA complex contains all three species; however, due to the similar sizes of Stat5a and Stat5b, it has not been possible to separate the complexes during EMSAs. In addition, during the immunodepletion experiments, the heterodimers as well as one of the specific homodimers precipitate together. However, coimmunoprecipitation of Stat5a and Stat5b is readily detectable (24, 25) (Fig. 3), suggesting that in the mammary gland the heterodimers are also quite abundant.
The mechanism underlying the formation of active Stat dimers (either heterodimers or homodimers) and their translocation to the nucleus has not been elucidated. In the mammary gland, all three dimeric Stat5 species are found. However, the mere presence of the two Stat5 proteins does not ensure that all three dimers will be formed. In HeLa cells and U937 cells, which express high amounts of both Stat5 proteins, IFN-α activates Stat5b but not Stat5a in the former, while it activates Stat5a but not Stat5b in the latter (28). In B cells, Stat5 and Stat1 simultaneously activated with, respectively, interleukin 3 and IFN-γ, interact with the same site. However, no Stat5-Stat1 heterodimers have been observed, even when different ratios of the cotransfected proteins were expressed (50). In addition, the level of Stat protein does not appear to determine the ratio of dimers formed. As we show here, lactating mammary glands have similar levels of Stat5a and Stat5b, yet Stat5a homodimers are more abundant than Stat5b homodimers. The SH2 domain could influence Stat activation and dimerization by showing a preference for docking sites on the upstream activator and/or on other Stats. This situation is illustrated by growth hormone (GH) activation of Stat5b and Stat3. Although both Stats are activated by GH, Stat5b preferentially interacts with the GH receptor while Stat3 docks to JAK2 (53). Furthermore, Stat1 has a higher affinity for Stat2 phosphopeptides compared to other Stat phosphopeptides (15). In analogy, the Stat5b-SH2 domain might preferentially interact with activated Stat5a, favoring Stat5a-Stat5b heterodimers over Stat5 homodimers. The sequences of the SH2 domain of the two Stat5 proteins differ by five residues (22), which might support this hypothesis.
Maximal Stat5 DNA binding activity is observed 7 to 15 min following addition of lactogenic hormones and then decreases to a low level which remains constant for several days. These binding kinetics do not mimic the rate of β-casein gene transcription, which increases steadily between 3 and 24 h (3). Stat5a and Stat5b are both phosphorylated on tyrosine following short-term or long-term lactogenic hormone treatment, and despite the decrease in their levels of phosphotyrosine after long-term treatment, Stat5a-Stat5b heterodimers can still be detected (Fig. 2). It is possible that the composition of the homodimers changes, something which we are currently analyzing. These results suggest that strong Stat5 DNA binding activity is necessary to initiate but may not be necessary to sustain transcription. It is known that other transcription factors are important in milk protein gene expression and that they act synergistically in this process (20, 33). These proteins may potentiate long-term maintenance of the transcription rate.
Stat5a-deficient mice (24) and Stat5b-deficient mice (45) have both impaired mammary gland development, and the females do not lactate. Milk proteins are present in these animals, with the exception of the WAP protein, the level of which is significantly lower in Stat5a-deficient mice. These observations suggest that homodimers of Stat5a and Stat5b, probably in combination with other transcription factors, are generally able to promote milk protein gene expression but that only the expression of the three different dimers can promote full development of the mammary gland. In Stat5a-deficient mice, the level of Stat5b phosphorylation is lower than that in wild-type mice (24), suggesting that activation of Stat5a and of Stat5b does not occur independently. A similar observation has been reported for IFN-α-stimulated cells, in which Stat1 tyrosine phosphorylation is dependent on Stat2 (32).
Activation of both Stat5 and the GR is essential for optimal β-casein gene expression. Mutation of the Stat5 binding site in the β-casein gene promoter abolishes both PRL and glucocorticoid responsiveness (40), suggesting that Stat5 is involved in both PRL- and glucocorticoid-mediated responses. Recently, an association of the two proteins was reported for transfected COS cells, in which glucocorticoids enhance Stat5-mediated β-casein promoter activity (43). In addition, half-palindromic GREs have been mapped in this promoter (52), and mutation of some of these sites caused a decrease in lactogenic hormone-mediated β-casein gene expression (19). We show here that Stat5 and the GR are complexed in vivo in the mammary gland and liver. In contrast, we could not detect an association between the GR and Stat3, which was expressed in all cell types analyzed (Fig. 7). While preliminary, these results suggest that the GR interacts only with a defined set of Stat proteins. Alternatively, a particular GR-Stat interaction may be exclusive and tissue specific. Analysis of this association in mammary cells revealed that in contrast to the results seen in COS cells, ligand-induced activation of Stat5 and the GR is not necessary for this association. These data suggest that endogenous Stat5 and GR undergo posttranslational modifications, perhaps phosphorylation, which allow their specific association. Immunodepletion experiments revealed that part of the Stat5 and GR molecules are not associated, suggesting that there are different intracellular pools of the two transcription factors. These observations agree with the finding that they translocate independently to the nucleus. Interestingly, quantification of the nuclear Stat5 showed that cells treated with both glucocorticoids and PRL have approximately 1.5 more nuclear Stat5 than cells treated only with PRL (Fig. 8A). This observation suggests that although activation is not a prerequisite for Stat5 and GR association, it might strengthen the association, resulting in increased nuclear translocation of Stat5. We could not detect any difference in the level of nuclear GR in cells treated with glucocorticoids in the presence or absence of PRL. However, there is more GR than Stat5 in the nucleus, and a difference between these two conditions might not be detectable. Alternatively, the presence of PRL-activated Stat5 might influence the GR-Stat5 complex in a different manner. The role of GR phosphorylation on the receptor function is poorly understood. Previous studies indicate that ligand-dependent GR phosphorylation has little effect on its ability to activate transcription from a complex (murine mammary tumor virus) promoter (26). However, a recent study showed that the phosphorylation status of the GR had a strong effect on hormone-mediated transcription from a half-palindromic GRE (49), similar to the one described for milk protein gene promoters and other glucocorticoid-responsive genes (8). The role of Stat5 and GR phosphorylation in milk protein gene expression is currently being analyzed.
Transcription of the WAP gene also depends on the synergistic effects of glucocorticoids and PRL (21). In this case, the mechanism may be slightly different, since GR-mediated WAP expression depends on cooperation among the nuclear factor I (NF-I) site, the Stat5 site, and clustered GRE half-sites (21). These observations suggest that Stat5-GR cooperation can be extended to other milk protein genes, suggesting that this cooperativity might be a common mechanism for glucocorticoid-mediated milk gene expression. GR regulates the activity of many other transcription factors through direct protein-protein interactions (for a review, see reference 27). Our demonstration of a GR-Stat5 interaction in the mammary gland is the first described cooperation between a Stat and a member of the nuclear receptor family of transcription factors.
ACKNOWLEDGMENTS
We thank Patrick Matthias for his many useful discussions and suggestions, Heidi Lane and Diana Graus-Porta for their suggestions on the manuscript, and John Daly for help with Fig. 9.
REFERENCES
- 1.Alonso C R, Pesce C G, Kornblihtt A R. The CCAAT-binding proteins CP1 and NF-I cooperate with ATF-2 in the transcription of the fibronectin gene. J Biol Chem. 1996;271:22271–22279. doi: 10.1074/jbc.271.36.22271. [DOI] [PubMed] [Google Scholar]
- 2.Azam M, Erdjument-Bromage H, Kreider B L, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle J N, Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball R K, Friis R R, Schoenenberger C A, Doppler W, Groner B. Prolactin regulation of β-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J. 1988;7:2089–2095. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 5.Cella, N. Unpublished data.
- 6.Cella N, Cornejo-Uribe R R, Montes G S, Hynes N E, Chammas R. The lysosomal-associated membrane protein LAMP-1 is a novel differentiation marker for HC11 mouse mammary epithelial cells. Differentiation. 1996;61:113–120. doi: 10.1046/j.1432-0436.1996.6120113.x. [DOI] [PubMed] [Google Scholar]
- 7.Chammas R, Taverna D, Cella N, Santos C, Hynes N E. Laminin and tenascin assembly and expression regulates HC11 mouse mammary cell differentiation. J Cell Sci. 1994;107:1031–1040. doi: 10.1242/jcs.107.4.1031. [DOI] [PubMed] [Google Scholar]
- 8.Dean D M, Sanders M M. Ten years after: reclassification of steroid-responsive genes. Mol Endocrinol. 1996;10:1489–1495. doi: 10.1210/mend.10.12.8961259. [DOI] [PubMed] [Google Scholar]
- 9.Doppler W, Groner B, Ball R K. Prolactin and glucocorticoid hormones synergistically induce expression of transfected rat beta-casein gene promoter constructs in a mammary epithelial cell line. Proc Natl Acad Sci USA. 1989;86:104–108. doi: 10.1073/pnas.86.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druker B J, Mamon H J, Roberts T M. Oncogenes, growth factors, and signal transduction. N Engl J Med. 1989;21:1383–1391. doi: 10.1056/NEJM198911163212007. [DOI] [PubMed] [Google Scholar]
- 11.Dusanter-Fourt I, Muller O, Ziemiecki A, Mayeux P, Druker B, Djiane J, Wilks A, Harpur A G, Fisher S, Gisselbrecht S. Identification of JAK protein tyrosine kinases as signaling molecules for prolactin:functional analysis of prolactin receptor and prolactin-erythropoietin receptor chimera expressed in lymphoid cells. EMBO J. 1994;13:2583–2591. doi: 10.1002/j.1460-2075.1994.tb06548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 13.Gouilleux, F. Personal communication.
- 14.Gouilleux F, Pallard C, Dusanter-Fourt I, Wakao H, Haldosen L-A, Norstedt G, Levy D, Groner B. Prolactin, growth hormone, erythropoietin and granulocyte-macrophage colony stimulating factor induce MGF-Stat5 DNA binding activity. EMBO J. 1995;14:2005–2013. doi: 10.1002/j.1460-2075.1995.tb07192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenlund A, Morales M O, Viviano B L, Yan H, Krolewski J, Schreiber R D. STAT recruitment by tyrosine phosphorylated receptors: an ordered reversible affinity driven process. Immunity. 1995;2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 16.Groner B, Gouilleux F. Prolactin-mediated gene activation in mammary epithelial cells. Curr Opin Genet Dev. 1995;5:587–594. doi: 10.1016/0959-437x(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 17.Hoeck W, Rusconi S, Groner B. Down-regulation and phosphorylation of glucocorticoid receptors in cultured cells. J Biol Chem. 1989;264:14396–14402. [PubMed] [Google Scholar]
- 18.Kazansky A V, Raught B, Lindsey S M, Wang A-F, Rosen J M. Regulation of mammary gland factor/Stat5a during mammary gland development. Mol Endocrinol. 1995;9:1598–1609. doi: 10.1210/mend.9.11.8584036. [DOI] [PubMed] [Google Scholar]
- 19.Lechner J, Welte T, Tomasi J K, Bruno P, Cairns C, Gustafsson J-A, Doppler W. Promoter-dependent synergy between glucocorticoid receptor and Stat5 in the activation of β-casein gene transcription. J Biol Chem. 1997;272:20954–20960. doi: 10.1074/jbc.272.33.20954. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Rosen J M. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in trangenic mice. Mol Cell Biol. 1995;15:2063–2070. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Rosen J M. Glucocorticoid regulation of rat whey acidic protein gene expression involves hormone-induced alterations of chromatin structure in the distal promoter region. Mol Endocrinol. 1994;8:1328–1335. doi: 10.1210/mend.8.10.7854350. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, Mietz J, Modi W S, John S, Leonard W J. Cloning of human Stat5B. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 23.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Robinson G W, Wagner K-U, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Robinson G W, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinol. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 26.Manson S A, Housley P R. Site-directed mutagenesis of the phosphorylation sites in the mouse glucocorticoid receptor. J Biol Chem. 1993;268:21501–21504. [PubMed] [Google Scholar]
- 27.McEwan I J, Wright A P H, Gustafsson J. Mechanism of gene expression by the glucocorticoid receptor: role of protein-protein interactions. BioEssays. 1997;2:153–160. doi: 10.1002/bies.950190210. [DOI] [PubMed] [Google Scholar]
- 28.Meinke A, Barahmand-Pour F, Wöhrl S, Stoiber D, Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol Cell Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merlo G R, Graus-Porta D, Cella N, Marte B M, Taverna D, Hynes N E. Growth, differentiation and survival of HC11 mammary epithelial cells: diverse effects of receptor tyrosine kinase-activating peptide growth factors. Eur J Cell Biol. 1996;70:97–105. [PubMed] [Google Scholar]
- 30.Mui A L F, Wakao H, O’Farrell A M, Harada N, Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO J. 1995;14:1116–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philp J A C, Burdon T G, Watson C J. Differential activation of Stats 3 and 5 during mammary gland development. FEBS Lett. 1996;396:77–80. doi: 10.1016/0014-5793(96)01069-1. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi S A, Leung S, Kerr I M, Stark G R, Darnell J E., Jr Function of Stat2 protein in transcription activation by alpha interferon. Mol Cell Biol. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raught B, Warren S-L L, Rosen J M. Developmentally and hormonally regulated CCAAT/enhancer-binding factor isoforms influence β-casein gene expression. Mol Endocrinol. 1995;9:1223–1232. doi: 10.1210/mend.9.9.7491114. [DOI] [PubMed] [Google Scholar]
- 34.Ripperger J A, Fritz S, Richter K, Hocke G M, Lottspeich F, Fey G H. Transcription factors Stat3 and Sta5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- 35.Ruff-Jamison S, Chen K, Cohen S. Epidermal growth factor induces the tyrosine phosphorylation and nuclear translocation of Stat5 in mouse liver. Proc Natl Acad Sci USA. 1995;92:4215–4218. doi: 10.1073/pnas.92.10.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rui H, Kirken R A, Farrar W L. Activation of receptor-associated tyrosine kinase JAK2 by prolactin. J Biol Chem. 1994;269:5364–5368. [PubMed] [Google Scholar]
- 37.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Ann Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt-Ney M, Happ B, Hofer P, Hynes N E, Groner B. Mammary gland-specific nuclear factor activity is positively regulated by lactogenic hormones and negatively by milk stasis. Mol Endocrinol. 1992;6:1988–1997. doi: 10.1210/mend.6.12.1491685. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt-Ney M, Happ B, Ball R K, Groner B. Developmental and environmental regulated of a mammary gland-specific nuclear factor essential for transcription of the gene encoding β-casein. Proc Natl Acad Sci USA. 1992;89:3130–3134. doi: 10.1073/pnas.89.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt-Ney M, Doppler W, Ball R K, Groner B. β-Casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol Cell Biol. 1991;11:3745–3755. doi: 10.1128/mcb.11.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Standke G J R, Meier V S, Groner B. Mammary gland factor activated by prolactin in mammary epithelial cells and acute-phase response factor activated by the interleukin-6 and liver cells share DNA binding and transactivation potential. Mol Endocrinol. 1994;8:469–477. doi: 10.1210/mend.8.4.7519723. [DOI] [PubMed] [Google Scholar]
- 42.Stöcklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 43.Taverna D, Groner B, Hynes N E. Epidermal growth factor receptor, platelet-derived growth factor receptor, and c-erbB-2 receptor activation all promote growth but have distinctive effects upon mouse mammary epithelial cell differentiation. Cell Growth Differ. 1991;2:14–154. [PubMed] [Google Scholar]
- 44.Topper Y, Freeman C S. Multile hormone interactions in the development of the mammary gland. Physiol Rev. 1980;60:1049–1106. doi: 10.1152/physrev.1980.60.4.1049. [DOI] [PubMed] [Google Scholar]
- 45.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S-H, Ram P A, Waxman D J, Davey H W. Requirement of Stat5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wartmann M, Davis R J. The native structure of the activated Raf protein kinase is a membrane bound multi-subunit complex. J Biol Chem. 1994;269:6695–6701. [PubMed] [Google Scholar]
- 48.Wartmann M, Cella N, Hofer P, Groner B, Liu X, Hennighausen L, Hynes N E. Lactogenic hormone activation of Stat5 and transcription of the β-casein gene in mammary epithelial cells is independent of p42 ERK2 mitogen-activated protein kinase activity. J Biol Chem. 1996;271:31863–31868. doi: 10.1074/jbc.271.50.31863. [DOI] [PubMed] [Google Scholar]
- 49.Webster J C, Jewell C M, Bodwell J E, Munck A, Sar M, Cidlowski J A. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J Biol Chem. 1997;272:9287–9293. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]
- 50.Wehinger J, Gouilleux F, Groner B, Finke J, Mertelsmann R, Weber-Nordt R M. IL-10 induces DNA binding activity of the three Stat proteins (Stat1, Stat3 and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett. 1996;394:365–370. doi: 10.1016/0014-5793(96)00990-8. [DOI] [PubMed] [Google Scholar]
- 51.Welte T, Garimorth K, Philipp S, Doppler W. Prolactin-dependent activation of a tyrosine phosphorylated DNA binding factor in mouse mammary epithelial cells. Mol Endocrinol. 1994;8:1091–1102. doi: 10.1210/mend.8.8.7527899. [DOI] [PubMed] [Google Scholar]
- 52.Welte T, Philipp S, Cairns C, Gustafsson J-A, Doppler W. Glucocorticoid receptor binding sites in the promoter region of milk protein genes. J Steroid Biochem Mol Biol. 1993;47:75–81. doi: 10.1016/0960-0760(93)90059-6. [DOI] [PubMed] [Google Scholar]
- 53.Yi W, Kim S-O, Jiang J, Park S-H, Kraft A S, Waxman D J, Frank S J. Growth hormone receptor cytoplasmic domain differentially promotes tyrosine phosphorylation of signal transducers and activators of transcription 5b and 3 by activated JAK2 kinase. Mol Endocrinol. 1996;10:1425–1443. doi: 10.1210/mend.10.11.8923468. [DOI] [PubMed] [Google Scholar]