Abstract
Spt3 and Mot1 are two transcription factors of Saccharomyces cerevisiae that are thought to act in a related fashion to control the function of TATA-binding protein (TBP). Current models suggest that while Spt3 and Mot1 do not directly interact, they do function in a related fashion to stabilize the TBP-TATA interaction at particular promoters. Consistent with this model, certain combinations of spt3 and mot1 mutations are inviable. To identify additional proteins related to Spt3 and Mot1 functions, we screened for high-copy-number suppressors of the mot1 spt3 inviability. This screen identified a previously unstudied gene, MOT3, that encodes a zinc finger protein. We show that Mot3 binds in vitro to three sites within the retrotransposon Ty long terminal repeat (δ) sequence. One of these sites is immediately 5′ of the δ TATA region. Although a mot3 null mutation causes no strong phenotypes, it does cause some mild phenotypes, including a very modest increase in Ty mRNA levels, partial suppression of transcriptional defects caused by a mot1 mutation, and partial suppression of an spt3 mutation. These results, in conjunction with those of an independent study of Mot3 (A. Grishin, M. Rothenberg, M. A. Downs, and K. J. Blumer, Genetics, in press), suggest that this protein plays a varied role in gene expression that may be largely redundant with other factors.
RNA polymerase II (pol II) transcription involves a balance of positive and negative regulators to properly control gene expression. The study of pol II transcription has identified several general factors that are required to initiate and transcribe mRNA from most promoters (77). Other studies have focused on gene-specific activators, such as the Saccharomyces cerevisiae activators Gal4 or Gcn4, or coactivators, such as the TATA-binding protein (TBP)-associated factors, and the mechanisms by which these two groups of proteins help the general transcription apparatus (31, 34). Repression of transcription, by both gene-specific repressors and general repressors, is also important for the proper regulation of pol II transcription. The mechanisms and targets of repression appear as varied as the mechanisms that lead to activation of transcription (19, 63). One target of repression is TBP, whose binding to the promoter TATA sequence represents one of the early rate-limiting steps in the formation of a pol II preinitiation complex (11, 12).
To understand the complex interplay of the many diverse proteins and mechanisms of transcriptional regulation, we have isolated and analyzed a number of mutations that affect pol II transcription initiation in vivo. The mutations were isolated as suppressors of insertion mutations caused by Ty elements or Ty long terminal repeats, δ sequences (29, 71, 72). This analysis has identified a number of genes, called SPT genes, for suppressor of Ty (70). Although identified by this specific selection, most SPT genes have now been shown to be generally important or essential for growth and transcription in vivo. One related group of SPT genes includes SPT3 (37, 74, 75), SPT7 (30), SPT8 (24), SPT20/ADA5 (48, 59), and SPT15, which encodes TBP (23, 25). These genes are related by common mutant phenotypes that include suppression of a common set of insertion mutations, mating defects, and sporulation defects. Spt3, Spt7, Spt8, and Spt20/Ada5 have recently been shown to be part of the large multifunctional complex, SAGA, that may interact with both TBP and transcriptional activators (8, 23, 32, 39, 58).
In a genetic screen for factors functionally related to Spt3, a mutation in MOT1 was isolated that is inviable in combination with an spt3Δ mutation (47). MOT1 is an essential gene that was originally identified by selections for mutations that increase transcription from a number of pol II-dependent genes (20, 54). Biochemical analysis has demonstrated that Mot1 exists in a complex with TBP (55, 56) and that in vitro, Mot1 can disrupt the TBP-TATA interaction in an ATP-dependent fashion (1, 4, 5). More recent work has demonstrated that Mot1 also plays a positive role in transcription (18, 47, 50). Our previous analysis showed that spt3 and mot1 mutants have certain common phenotypes, including decreased levels of Ty mRNA (47). It was proposed that Spt3 and Mot1 could both be positive factors in transcription by helping TBP to bind to functional TATA boxes, with Mot1 functioning by removing TBP from nonfunctional TATA sequences and Spt3 functioning by stabilizing TBP on certain functional TATA sequences. Neither Mot1 or Spt3 has been shown to bind to DNA, and there is no evidence that Mot1 and Spt3 interact directly with each other.
To understand better how Mot1 and Spt3 control TBP function, we have further investigated the synthetic lethal interaction in the spt3Δ mot1 double mutant. We hypothesized that mot1-24, the mot1 allele that is synthetically lethal with spt3Δ, may impair a protein-protein interaction with another Mot1- or Spt3-related factor. Thus, overexpression of such a factor might restore its functional interaction with Mot1 and suppress the inviability of the spt3Δ mot1-24 double mutant. We therefore isolated high-copy-number suppressors of the spt3Δ mot1-24 synthetic lethality and identified a single gene called MOT3. This gene encodes a zinc-finger protein that binds to three sites in the Ty δ sequence in vitro. Analysis of a mot3Δ mutation showed that it causes a modest increase in Ty mRNA levels, partially suppresses a transcription defect of mot1 mutants, and partially suppresses an spt3 mutant phenotype. Results of an independent study of Mot3 (33) demonstrate that mot3Δ also has a wide variety of effects, both positive and negative, at several promoters. Thus, we have identified a previously unstudied zinc finger protein, Mot3, that appears to regulate some aspect of transcription, perhaps by controlling TATA box function.
MATERIALS AND METHODS
Yeast strains and genetic methods.
The yeast strains used in this study (Table 1), unless otherwise stated, were derived from an S288C GAL2+ derivative (73) and were constructed by standard methods (62). All spt, mot1, and toa1 mutations used in this study have been described previously (23, 36, 72). The Ty912Δ44-lacZ fusion is integrated at HIS4 at the normal position of Ty912 (21). This fusion contains 388 bp of Ty912 fused, in frame, with the Escherichia coli lacZ gene (72). Yeast strains were transformed by a lithium acetate procedure (26). Standard methods of mating, sporulation, and tetrad analysis were used (62).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| FY2 | MATα ura3-52 |
| FY104 | MATa his3Δ200 ura3-52 trp1Δ63 |
| FY119 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 |
| FY126 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 ade8 suc2ΔUAS |
| FY294 | MATa spt3-202 ura3-52 leuΔ1 lys2-173R2 his4-917δ trp1Δ63 |
| FY252 | MATα ura3-52 his3Δ200 leu2Δ1 ade8 |
| FY630 | MATa his4-917δ lys2-173R2 leu2δ1 ura3-52 trp1Δ63 |
| FY925 | MATα Ty912Δ44-lacZ lys2-173R2 ura3-52 |
| FY1472 | MATα Ty912Δ44-lacZ lys2-173R2 ura3-52 trp1Δ63 |
| FY1473 | MATα Ty912Δ44-lacZ-15 lys2-173R2 ura3-52 trp1Δ63 |
| FY1474 | MATα Ty912Δ44-lacZ-15 lys2-173R2 ura3-52 trp1Δ63 |
| FY1475 | MATα Ty912Δ44-lacZ-16 lys2-173R2 ura3-52 trp1Δ63 |
| FY1475 | MATα Ty912Δ44-lacZ-16 lys2-173R2 ura3-52 trp1Δ63 |
| FY1483 | MATa Ty912Δ44-lacZ ura3-52 mot3Δ2::HIS3 lys2-173R2 |
| FY1484 | MATa Ty9125Δ44-lacZ ura3-52 mot3Δ2::HIS3 lys2-173R2 trp1Δ63 |
| FY1485 | MATα Ty912Δ44-lacZ-15 ura3-52 mot3Δ2::HIS3 lys2-173R2 |
| FY1486 | MATα Ty912Δ44-lacZ-15 ura3-52 mot3Δ2::HIS3 trp1Δ63 his3Δ200 |
| FY1487 | MATa Ty912Δ44-lacZ-16 ura3-52 mot3Δ2::HIS3 lys2-173R2 |
| FY1488 | MATα Ty912Δ44-lacZ-16 ura3-52 mot3Δ2::HIS3 trp1Δ63 |
| FY1625 | MATα mot1-24 toa1-18 ade8 leu2Δ1 ura3-52 his4-912δ pRS7.1BglII (MOT1 pRS316) |
| FY1626 | MATa mot1-24 spt6-14 leu2Δ1 ade8 his4-912δ or his4-917δ lys2-128δ pCC11 (SPT6 YCp50) |
| FY1627 | MATa toa1-18 spt3-202 ura3-52 leu2Δ1 pCC1 (SPT3 YCp50) |
| FY1628 | MATα mot3Δ2::HIS3 ura3-52 his3Δ200 |
| FY1629 | MATα spt7Δ::LEU2 mot1-24 trp1Δ63 leu2Δ1 ura3-52 pFW127 (SPT7 YCp50) |
| FY1630 | MATα mot1-24 mot3Δ2::HIS3 his3Δ200 ura3-52 |
| FY1631 | MATa mot3Δ2::HIS3 his3Δ200 lys2-173R2 ura3-52 |
| FY1632 | MATα spt3-202 mot3Δ2::HIS3 his3Δ200 lys2-173R2 ura3-52 |
| FY1633 | MATa mot1-24 his3Δ200 trp1Δ63 ade8 ura3-52 |
| L641 | MATa mot1-24 spt3Δ203::TRP1 ura3-52 leu2Δ1 trp1Δ63 pCC1 (SPT3 YCp50) |
Media.
Rich (YPD), minimal (SD), synthetic complete (SC), 5-fluoroorotic acid (5-FOA), and sporulation media were prepared as described previously (62). Suppression of insertion mutations was scored on SD medium supplemented with the required nutrients or on SC medium lacking appropriate nutrients. Yeast transformants were selected on the appropriate SC medium.
DNA preparation and analysis.
E. coli HB101 and DH5α were used as hosts for plasmids (64). The plasmids were constructed, maintained, and isolated by standard methods (64). They were recovered from yeast as described previously (60). Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) and Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and used as recommended by the manufacturer.
RNA isolation and Northern blot analysis.
Cells for RNA isolation were grown at 30°C in supplemented SD medium to a density of 1 × 107 to 2 × 107 cells/ml. Total RNA was isolated by a hot-phenol method (7). Northern transfer and hybridizations were performed as described previously (66). 32P-labeled probes were generated with a Boehringer Mannheim Biochemicals nick translation kit or by random hexamer labeling (7). The Northern blots were quantitated on a Molecular Dynamics PhosphorImager.
Sequence analysis of MOT3.
For sequencing MOT3, the appropriate restriction fragments were subcloned into pRS425. Sequencing was performed with the U.S. Biochemical Corp. Sequenase version 2.0 kit. Synthetic primers and m13 universal and reverse primers were used to determine the sequence on both strands. The sequence was compared against known sequences and proteins in the GenBank, EMBL, and PIR databases with the BLAST program (2).
Plasmids.
The pRS series of vectors is described previously (17, 65). The vector used to express Mot3 in E. coli, pET-His has been described previously (16). pKA1 (3) and pFW32 (75) have been described previously. pJM162 is the XhoI-NotI fragment containing MOT1 from pRS7.1BglII (20) subcloned into the XhoI-NotI sites of pRS425.
The original plasmid isolate of MOT3 is pJM189 and is a Sau3A S. cerevisiae genomic fragment in the BamHI site of Yep13. pJM141 is a 6-kb BamHI fragment from pJM189 subcloned into the BamHI site of pRS425. pJM142 is a XhoI-BamHI fragment from pJM141 subcloned into the XhoI-BamHI sites of pRS425. pJM187, which contains MOT3 cloned under the control of the MET25 promoter, was generated by digesting pJM182 with BamHI and XhoI, treating the fragments with the Klenow fragment and deoxynucleotide triphosphates, gel purifying the insert, and ligating into the SmaI site of p425MET25. pJM188 is the XhoI-BamHI fragment containing MOT3 from pJM142 subcloned into the XhoI-BamHI sites of pRS315.
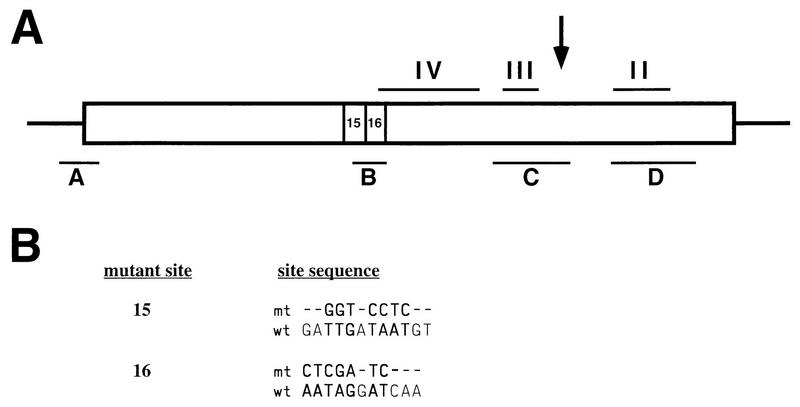
To generate mutant alleles of Ty912Δ44-lacZ, pAD1 was mutagenized by oligonucleotide-directed mutagenesis as described previously (21, 43). Mutagenized regions were sequenced on both strands and subcloned into an unmutagenized pAD1. The oligonucleotides used for mutagenesis of pAD1 were LGO15 (GAAACGCAAGGAGGTACCTCGTAATAGGATC) and LGO16 (GGATTGATAATGTCTCGAGTACAATGAATATAAA). Plasmid pAD1 was mutagenized with the above-described oligonucleotides to generate plasmids pAD15 (LGO15) and pAD16 (LGO16).
The plasmids used for nick translation of Northern probes were pB161 (Ty1) and pHB59 (TPI1; generously provided by H. Baker).
A screen for high-copy-number suppressors of mot1 spt3Δ synthetic lethality.
Strain L641 (mot1-24 spt3Δ) with pCC1 (SPT3 CEN URA3) is temperature sensitive because of mot1-24 and 5-FOA sensitive because of the mot1-24 spt3Δ inviability. This strain was transformed with a YEp13 (2μm LEU2) library (76), and approximately 12,000 Leu+ colonies were screened for the ability to grow on 5-FOA and the ability to grow on SC−Leu at 37°C. No high-copy-number suppressors of the temperature sensitivity (Ts−) phenotype were isolated. This was not surprising, since the Ts− phenotype is partially dominant, although the other mot1-24 phenotypes are recessive (46, 47). Plasmids were rescued from 5-FOA resistant strains as described previously and transformed into E. coli DH5α. Ninety-seven plasmids that, when retransformed into L641, reconferred the high-copy-number suppression of the 5-FOA sensitivity phenotype were obtained.
Construction and analysis of a mot3Δ null mutation.
A mot3Δ mutation that deletes the entire MOT3 open reading frame was generated by PCR (9). Primers JMH15 (ACTAATAGGCAACAGTAGGCAAATAGTAAAGGGACATATCATATTGGCCTCCTCTAGTACACC) and JMH16 (AAATGAGTGGGAAGGGATATTTTGTGTGTCTATAAAGTCTATCTAGCGCGCCTCGTTCAGAATG) were used to PCR amplify the HIS3 gene from plasmid pRS313 under the following conditions: 95°C for 3 min, 1 cycle; 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min, 30 cycles; 72°C, for 5 min, 1 cycle. This PCR product replaces nucleotides 117 to 1623 of the GenBank sequence with a functional HIS3 gene. The resulting PCR product, which has 40 bp of homology to sequences flanking the MOT3 open reading frame on either side, was transformed into a his3Δ200/his3Δ200 diploid constructed from FY104 and FY252. His+ transformants were selected on SC−His, sporulated, and analyzed by tetrad dissection. Four-spore tetrads were analyzed by Southern blotting to confirm the correct deletion of the MOT3 open reading frame. This null is designated mot3Δ2::HIS3.
Affinity purification of Mot3.
A 6-histidine-tagged Mot3 (6His-Mot3) was generated by PCR amplifying (94°C for 3 min; 94°C for 30 s, 55°C for 1 min, and 72°C, for 2 min for 30 cycles; 72°C for 3 min) the MOT3 open reading frame with Pfu polymerase and primers JMP30 (CCGCTCGAGATGAATGCGGACCATC) and JMP31 (CGGGATCCCTATTTGTTGTGAC). A BamHI site was incorporated into JMP30 and an XhoI site was incorporated into JMP31 for subcloning. Following amplification, the PCR product was digested with BamHI and XhoI and subcloned into the BamHI-XhoI sites of the pET-His vector to create pJM182. The pET-His vector contains a polylinker that has been modified to express a protein in frame with six histidines. pJM182 was transformed into strain BL21/De3. 6His-Mot3 was produced in E. coli by growing cells to an optical density at 600 nm of 0.7 and inducing with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C. Following induction, the cells were harvested by centrifugation at 4,000 rpm for 10 min, resuspended in buffer A (50 mM NaH2PO4 [pH 8.0], 1 M NaCl, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, and quick-frozen. The cells were thawed, refrozen, and disrupted by sonication. Sonicated cells were centrifuged at 9,000 rpm for 20 min. The supernatant was bound, for 2 h at 4°C with constant rocking, to Ni2+-nitrotriacetic acid (NTA) agarose (Qiagen) equilibrated in buffer A–25 mM imidazole. 6His-Mot3 bound to the Ni2+-NTA agarose was washed with 40 column volumes of buffer A–25 mM imidazole, 10 column volumes of buffer A–50 mM imidazole, and 5 column volumes of buffer A–75 mM imidazole. 6His-Mot3 was found to elute in the 75 mM imidazole eluate. The 75 mM imidazole Mot3 eluate was then dialyzed against 25 mM HEPES (pH 8.0)–0.1 M KCl–25 μM ZnSO4–1 mM dithiothreitol (DTT)–12.5 mM MgCl2–20% glycerol–1 mM phenylmethylsulfonyl fluoride for 6 h with two changes of buffer. Mock extracts in which strain BL21/DE3 contained the vector alone were prepared in the same manner as for the 6His-Mot3 protein. Protein concentrations were determined by a Bradford protein assay (Bio-Rad, Richmond, Calif.). Before aliquoting and freezing were performed, insulin was added to 0.2 mg/ml to both 6His-Mot3 and the mock protein preparation. The addition of insulin was required to preserve Mot3 activity upon freeze-thawing (46). The 6His-Mot3 protein was judged to be greater than 90% pure by silver staining.
Mot3 mobility shift assay.
Mot3 mobility shift assays were carried out in the following DNA-binding buffer: 25 mM HEPES (pH 8.0)–5 mM MgCl2–0.1 mM EDTA–50 mM KCl–10% glycerol. Reactions mixtures (10 to 20 μl) contained 1× binding buffer, 0.5 mM DTT, 500 μg of bovine serum albumin per ml, 6His-Mot3 protein, and 2,500 cpm of 32P-labeled probe (approximately 0.1 to 1 ng of DNA). The probes were quantitated by an ethidium bromide spot test (7). The reaction mixtures were incubated for 20 min at room temperature and loaded directly onto a 4% polyacrylamide gel (40:1 cross-linking) during electrophoresis. The gels contained 1× TGE (25 mM Tris, 190 mM glycine, 1 mM EDTA [pH 8.3]) plus 0.5 mM DTT. Electrophoresis buffer consisted of 1× TGE. The gels were preelectrophoresed for 30 min at 100 mV and electrophoresed at room temperature for 1.5 h at 120 mV. Following electrophoresis, the gels were vacuum dried onto Whatman filter paper and then exposed to Kodak XAR X-OMAT autoradiographic film at −70°C overnight.
Gel shift experiments in which 1,10-o-phenanthroline was used were carried out as described above with the following exceptions. A concentration of 10 mM 1,10-o-phenanthroline in 20% ethanol was added to the reaction mixtures to a final concentration of 1 mM, and the mixtures were incubated for 20 min and loaded onto the gel. Mot3 DNA binding was reconstituted by adding back divalent cations at 2 mM to reaction mixtures in which 1,10 o-phenanthroline had been added and then incubating the mixtures for 20 min at room temperature before loading them onto the running gel.
his4-912δ gel shift probes.
Wild-type probes, unless otherwise noted, were generated by cutting pKA1 with SpeI-SspI and α-32P-end labeling with the Klenow fragment.
Mutant Ty912δ gel shift probes were generated by digesting mutant Ty912Δ44-containing plasmids with SpeI-SspI and α-32P-end labeling with the Klenow fragment. Mutant gel shift probes were isolated from the following plasmids pAD15 (mutant 15) and pAD16 (mutant 16).
Mot3 DNase I footprint assays.
To footprint the antisense strand of his4-912δ, plasmid pKA1 was cut with SpeI and HaeIII and end labeled with [α32P]dATP by filling in the recessed SpeI site with Klenow fragment. End-labeled probe was separated on a native polyacrylamide gel, excised from the gel, and eluted either by electroelution or by a crush-and-soak method that has been described previously (64). The probe was ethanol precipitated and quantitated by an ethidium bromide spot test as described previously (7). DNase I (Sigma) was prepared in DNase I buffer (50% glycerol, 20 mM Tris-HCl [pH 7.5], 1 mM MgCl2) to 1 mg/ml and stored at −20°C. A DNase I concentration at which 30 to 50% of the probe remained uncut was chosen for the experiments and was determined for each probe used. Footprinting reaction mixtures contained the following components: 1× gel shift buffer, 1 μg of poly(dG-dC) (Pharmacia) per ml, 500 μg of bovine serum albumin per ml, 0.5 mM DTT, 6His-Mot3, and 20,000 cpm of end-labeled his4-912δ (approximately 4 to 5 ng of DNA) probe in a 25-μl volume. The reactions were initiated by adding 5 μl of DNase I and incubating for 1 min at room temperature. The reactions were then stopped by adding an equal volume of stop solution containing 200 mM NaCl, 20 mM EDTA, 1% sodium dodecyl sulfate, and 250 μg of yeast tRNA per ml. The reaction products were extracted with 25:24 phenol-chloroform, ethanol precipitated, and resuspended in 6 μl of formamide loading buffer consisting of deionized formamide with 0.1 N NaOH (2:1, vol/vol), 0.25% bromophenol blue, and 0.25% xylene cyanol. Chemical sequencing reactions were carried out with the same probe as described previously (49). Footprints were run on 7% acrylamide–8.3 M urea gels. The gels were preelectrophoresed for 1 h in 1× Tris-borate-EDTA (TBE). Following the preelectrophoresis, the bottom buffer was brought to 1 M with unbuffered sodium acetate to generate an electrolyte gradient (7). These gel conditions allowed resolution of the complete his4-912δ probe on one gel. Samples were loaded onto the gel, and the gel was electrophoresed at 60 W until the bromophenol blue reached the bottom. Following electrophoresis, the gels were vacuum dried on Whatman filter paper and exposed to autoradiography film at −70°C.
To footprint the sense strand of his4-912δ, pKA1 was cut with AccI and HpaII and [α-32P]dATP end labeled with the Klenow fragment. Probe isolation, footprint assays, and gel electrophoresis were carried out exactly as described above for the antisense strand.
β-Galactosidase assays.
Cells were grown to 1 × 107 to 2 × 107 cells/ml in SD medium supplemented with the appropriate amino acids. Crude extracts were prepared and assayed as described previously (62). β-Galactosidase levels are expressed in Miller units and were normalized to the total protein concentration as determined by the Bradford protein assay (Bio-Rad).
Nucleotide sequence accession number.
The accession number of the MOT3 nucleotide sequence is U25279.
RESULTS
Isolation of MOT3, a high-copy-number suppressor of the mot1-24 spt3Δ synthetic lethality.
Previous analysis has shown that Mot1 and Spt3 are functionally related and that the combination of spt3Δ and mot1-24 causes lethality (47). The mot1-24 mutation encodes a single amino acid change (K1507R) in a residue conserved among members of the Snf2/Swi2 family of putative helicases (20, 22). If this amino acid impairs protein-protein interactions between Mot1 and another factor, overexpression of such a factor might suppress either the mot1-24 Ts− phenotype or the spt3Δ mot1-24 synthetic lethality. We screened for such suppressors as described in Materials and Methods and identified 97 plasmids that suppress the spt3Δ mot1-24 double-mutant lethality. Two of these plasmids contained MOT1, 3 plasmids contained SPT3, and 80 other plasmids shared overlapping restriction fragments. Suppression of the double-mutant lethality by a member of this class is shown in Fig. 1. The remaining 12 plasmids were found to suppress the mot1-24 spt3Δ synthetic lethality only weakly and were not studied further. In the following studies, we have focused on the gene identified in the set of 80 plasmids. The gene was named MOT3 for modulator of transcription.
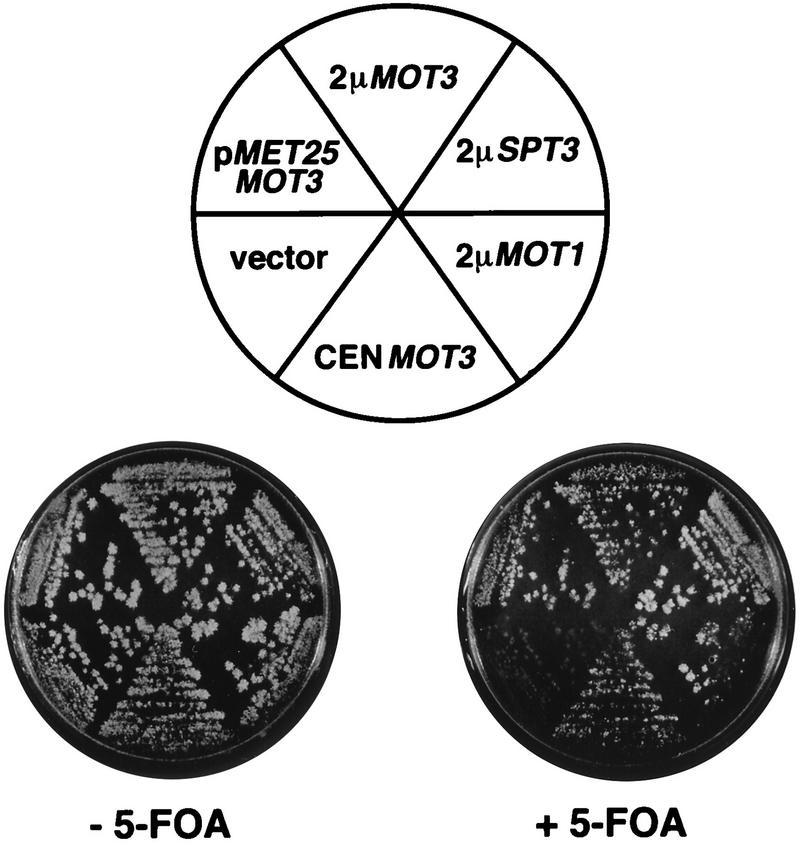
FIG. 1.
MOT3 suppresses the synthetic lethality of the spt3Δ mot1-24 double mutant. Strain L641 (mot1-24 spt3Δ pCC1) was transformed with the following plasmids: 2μm MOT3 (pJM142), 2μm MOT3 expressed from the MET25 promoter (pJM182), 2μm SPT3 (pFW32), 2μm MOT1 (pRS7.1BglII), CEN MOT3 (pJM188), and the vector alone (p425MET25). Leu+ transformants were colony purified on SC-Leu and replica plated to Sc-Leu-Met plates containing or lacking 5-FOA. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer.
Since this work was done before the completion of the S. cerevisiae genome sequence, we took several steps to identify and analyze MOT3. To determine the map position of MOT3, a fragment of a MOT3 genomic clone was used to probe a filter containing an ordered set of λ clones containing S. cerevisiae genomic DNA. This analysis placed MOT3 on chromosome XIII near ILV2 (46). To identify the open reading frame responsible for the high-copy-number suppression of the mot1-24 spt3Δ synthetic lethality, subclones of the original plasmid were constructed and tested for high-copy-number suppression by observing growth on 5-FOA (Fig. 1). A single open reading frame was identified and completely sequenced. The sequence, described below, is identical to that subsequently determined by the S. cerevisiae genome project (10). To verify that overexpression of this open reading frame suppressed the spt3Δ-mot1 lethality, MOT3 was cloned and expressed under the control of the MET25 promoter. Expression of MOT3 under conditions of MET25 induction suppressed the mot1-24 spt3Δ synthetic lethality (Fig. 1).
The MOT3 open reading frame is predicted to encode a novel protein of 490 amino acids (Fig. 2A). Mot3 has a striking amino acid composition, consisting of several stretches of asparagine, glutamine, alanine, proline, and serine. These residues account for 60% of the Mot3 protein. The presence of polyglutamine, polyalanine, and polyproline stretches is reminiscent of domains that have been found in several transcriptional repressors and have been determined to be involved in transcriptional repression (35). In addition to this striking amino acid composition, two C2-H2 zinc finger DNA-binding motifs were identified in the C terminus. These zinc fingers are homologous to other known zinc finger DNA-binding domains (Fig. 2B). Taken together, these results show that overexpression of a newly identified zinc finger protein, Mot3, suppresses mot1-24 spt3Δ synthetic lethality.
FIG. 2.

The MOT3 protein is arginine, glutamine, proline, serine, and alanine rich. (A) The predicted translation product of the MOT3 open reading frame. Arginine (N), proline (P), serine (S), alanine (A), and glutamine (Q) residues are shown in boldface and a larger font. The two Cys2-His2 zinc finger motifs are underlined. (B) The Mot3 zinc fingers most closely related to the yeast proteins Msn2, Msn4, and YPL230W. The consensus residues shown on the top line are identical in all four yeast zinc finger regions. These proteins are primarily homologous over the zinc finger DNA-binding domain and overall have less than 15% identity. The shaded letters indicate residues that are found in at least two of the four zinc finger sequences. The numbers to the right and the left of the alignment are the amino acid numbers for the published sequence of each protein. The alignment was performed with the MEGALIGN alignment program of the DNASTAR sequence analysis programs.
Suppression by MOT3 overexpression is specific for spt3Δ mot1-24 lethality.
One model for MOT3 high-copy-number suppression of mot1-24 spt3Δ synthetic lethality is that it suppresses either mot1-24 or spt3Δ individually. We therefore tested whether MOT3 overexpression would suppress any of the mutant phenotypes caused by either mot1-24 or spt3Δ single mutations. mot1-24 has been shown previously to cause several mutant phenotypes, including Spt−, slow growth, Ts−, and a decreased level of Ty mRNA (20, 47, 54). We found that MOT3 overexpression had no effect on any of these phenotypes in a mot1-24 mutant (46). spt3Δ mutations have been shown previously to cause slow growth, an Spt− phenotype, and decreased levels of Ty mRNA (23, 72). As with mot1-24, MOT3 overexpression did not suppress these spt3Δ phenotypes (46). Consistent with these results, when high-copy-number MOT3 is present in a mot1-24 spt3Δ mutant, it suppresses the double-mutant lethality but not any of the other phenotypes conferred by either mot1-24 or spt3Δ. Finally, we also tested if MOT3 overexpression would cause any mutant phenotypes in a MOT1+ SPT3+ strain. We found that MOT3 overexpression caused no detectable mutant phenotypes, including effects of growth or the Spt phenotype (46).
In addition to spt3Δ mot1-24 synthetic lethality, we previously observed synthetic lethality for other double-mutant combinations among spt, mot1-24, and toa1 mutations (47). TOA1 encodes the large subunit of the general transcription factor TFIIA (57). To determine if MOT3 overexpression could suppress any of these other double-mutant lethalities, we overexpressed MOT3 in the different double-mutant combinations and tested them for viability (Table 2). As expected, high-copy-number MOT3 strongly suppressed spt3Δ mot1-24 inviability. The spt7Δ mot1-24 and spt6-14 mot1-24 double-mutant inviabilities were only weakly suppressed, and the mot1-24 toa1-18 and spt3Δ toa1-18 double-mutant lethalities were not detectably suppressed. These results suggest that high-copy-number suppression by MOT3 is relatively specific for the spt3Δ mot1-24 combinations.
TABLE 2.
High-copy-number suppression of double-mutant inviability of toa1-18, mot1-24, and spt mutations
| Strain genotype | Plasmid genotype | High-copy-number suppressiona
|
|
|---|---|---|---|
| Vector | 2μm MOT3 | ||
| toa1-18 | None | −b | −b |
| mot1-24 | None | −b | −b |
| spt3Δ mot1-24 | SPT3 URA3 (pCC1) | − | + |
| spt3Δ toa1-18 | SPT3 URA3 (pCC1) | − | − |
| mot1-24 toa1-18 | MOT1 URA3 (pRS7.1BglII) | − | − |
| mot1-24 spt7Δ | SPT7 URA3 (pFW127) | − | ± |
| mot1-24 spt6-14 | SPT6 URA3 (pCC11) | W | ± |
High-copy-number suppression was determined by transforming the strains containing the plasmid of the indicated genotype with pRS425 (LEU2 2μm) or pJM142 (LEU2 2μm MOT3) and observing growth on plates containing and lacking 5-FOA. +, strong growth; ±, weak growth; W, very weak growth; −, no growth.
These strains were tested for temperature-sensitive growth at 37°C.
A mot3Δ mutation causes some mild mutant phenotypes.
To analyze the mot3 null phenotype, we constructed a mot3Δ mutation, mot3Δ2, that replaces the entire MOT3 open reading frame with the HIS3 gene. This mutation was recombined into an S. cerevisiae strain, replacing the wild-type MOT3 gene. The null mutant was found to be viable and to have a wild-type growth rate. mot3Δ2 mutants were examined for a number of possible phenotypes. Of those tested, only one additional phenotype was discovered: mot3Δ mutants are approximately twice as sensitive to UV light as are wild-type strains (38). Those phenotypes unaffected by the mot3Δ mutation included growth on sucrose, raffinose, glucose, and galactose as carbon sources, the Spt− phenotype; suppression of a SUC2 upstream activation sequence deletion; temperature sensitivity; pseudohyphal growth; and haploid invasive growth (46).
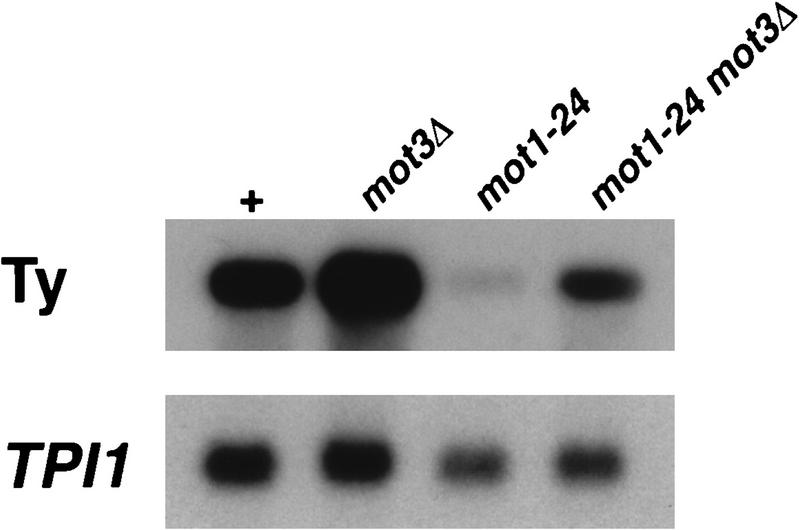
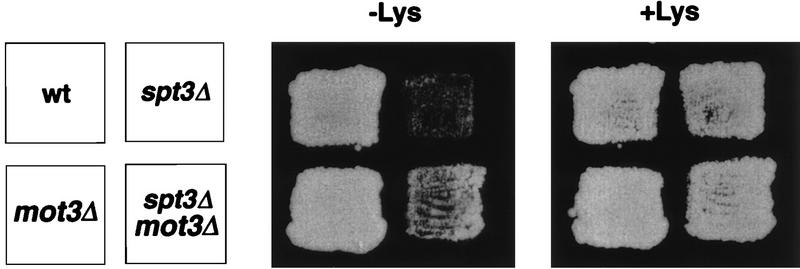
mot3Δ2 mutants were further analyzed in combination with either mot1-24 or spt3Δ. The mot3Δ mot1-24 and mot3Δ spt3Δ double mutants were viable and could therefore be tested for all known mot1-24 and spt3Δ mutant phenotypes. In the mot3Δ mot1-24 mutant, there was a detectable effect on one mot1-24 phenotype, the Ty mRNA level (Fig. 3). In the mot3Δ2 mot1-24 double mutant, Ty mRNA levels were elevated compared to the levels in the mot1-24 single mutant (Fig. 3). Thus, mot3Δ partially suppresses the mot1-24 decrease in Ty mRNA levels. In the mot3Δ spt3Δ double mutant, mot3Δ also weakly suppressed one spt3Δ phenotype: the Spt− phenotype with respect to the lys2-173R2 insertion mutation (Fig. 4). However, the slow growth of spt3Δ mutants was not suppressed and the decrease in Ty mRNA levels was only weakly suppressed by mot3Δ (46).
FIG. 3.
Ty transcript levels are slightly elevated in a mot3Δ mutant. Ty mRNA levels were examined in wild-type, mot3Δ2, mot1-24, and mot1-24 mot3Δ2 strains. The strains used from left to right are FY2, JMY519, JMY613, and JMY572. Ty mRNA levels were normalized to TPI1 mRNA levels. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer.
FIG. 4.
mot3Δ2 weakly suppresses the Spt− phenotype of spt3Δ. Strains patched on a YPD plate were replica plated to plates containing (+) or lacking (−) lysine. The strains used were JMY607, JMY612, FY630, and FY294. wt, wild type. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer.
Since both mot1-24 and spt3Δ are weakly suppressed by mot3Δ2, it seemed possible that mot3Δ2 would weakly suppress the mot1-24 spt3Δ synthetic lethality. To test this possibility, we constructed a spt3Δ mot1-24 mot3Δ2 triple mutant containing a wild-type SPT3 plasmid. The viability of the triple mutant in the absence of the SPT3 plasmid was determined by its ability to grow on 5-FOA medium. The triple mutant was as 5-FOA sensitive as the spt3Δ mot1-24 double mutant (46). Thus, a mot3Δ2 mutation does not suppress the spt3Δ mot1-24 double mutant lethality.
A number of other spt mutations, spt7Δ, spt15-21, and spt20Δ, cause phenotypes similar to those due to spt3Δ, including suppression of the insertion mutations lys2-173R2 and his4-917δ (25, 30, 32, 58, 59, 72, 74). Therefore, to determine if mot3Δ would suppress these spt mutations as it does spt3Δ, we examined suppression of these insertion mutations in mot3Δ spt7Δ, mot3Δ spt15-21, and mot3Δ spt20Δ double mutants. For the lys2-173R2 insertion, mot3Δ2 was found to weakly suppress spt15-21 but not spt7Δ or spt20Δ (46). The suppression of the Spt− phenotype of spt15-21 is of similar strength to the suppression observed for spt3Δ (46). The Spt− phenotype with respect to his4-917δ was unaffected by mot3Δ in combination with any of these spt mutations (46).
Since a mot3Δ2 mutation was found to weakly suppress the Ty transcriptional defect caused by mot1-24, we analyzed Ty mRNA levels in MOT3+ and mot3Δ strains. As shown in Fig. 3, a reproducible 1.6-fold increase in Ty mRNA levels was observed in mot3Δ mutants as compared to the level in a MOT3+ strain. This very modest increase in transcription is opposite to the effect observed for spt3 and mot1 mutants, which have greatly reduced Ty mRNA levels (74). Thus, a modest increase in Ty transcript levels, suppression of the decrease in transcription of a mot1 mutation, and weak suppression of an spt3Δ mutation suggest that Mot3 may play a role in repressing Ty transcription.
Mot3 binds to the Ty912δ sequence.
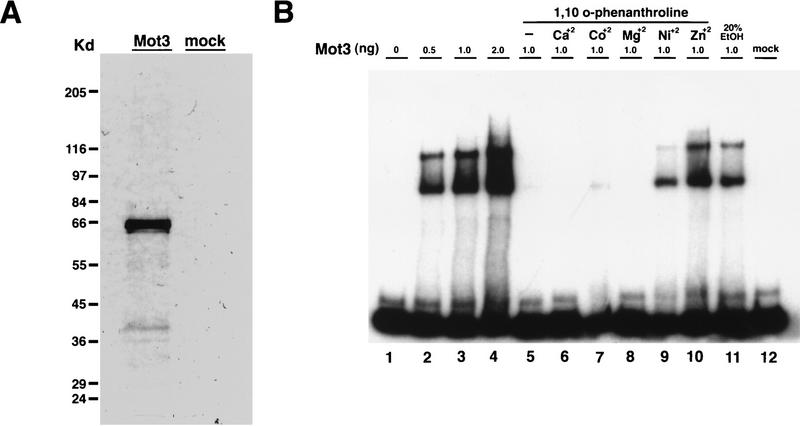
The two zinc finger DNA-binding motifs of Mot3 strongly suggested that it is a sequence-specific DNA-binding protein. To assay DNA binding, we first overexpressed and purified full-length Mot3 with a 6-histidine tag (6His-Mot3) from E. coli, using Ni2+ affinity chromatography (see Materials and Methods). By this method, the 6His-Mot3 was judged to be more than 90% pure by silver staining (Fig. 5A). In addition, we performed the same purification steps on a mock extract in which the expression vector contained no insert. 6His-Mot3 migrates at a position corresponding to a molecular mass of approximately 66 kDa, larger than its predicted molecular mass of 49 kDa. Highly charged proteins have previously been observed to migrate at positions significantly greater than their predicted molecular masses (30, 67).
FIG. 5.
Mot3 binds to the Ty912δ promoter in a zinc-dependent fashion. (A) Purification of Mot3 from an E. coli-overexpressing strain. 6His-Mot3 and mock extracts were affinity purified by Ni2+-NTA chromatography. Mot3 (150 ng) and an equivalent amount of mock extract were run on a sodium dodecyl sulfate–12.5% polyacrylamide gel and silver stained. The positions of protein molecular mass markers are shown on the left. Mot3 is judged to be >90% pure. The approximately 37-kDa band in the Mot3 lane appears to be a proteolytic product of Mot3 that consistently copurifies with 6His-Mot3. (B) Mot3 binding to the Ty912δ promoter is zinc dependent. Electrophoretic mobility shift assays were performed with an end-labeled Ty912δ DNA probe as described in Materials and Methods. The following amounts of Mot3 were added to each lane: 1, no protein; 2 to 4, 0.5, 1.0, and 2.0 ng, respectively; 5 to 10, 1 ng; 11, 1 ng plus 20% ethanol; 12, mock extract was added. Where used, either 2 mM indicated cation or 1 mM 1,10-o-phenanthroline was added. EtOH, ethanol. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer.
To assay DNA binding by Mot3, we performed gel mobility shift assays with the Ty912δ promoter DNA. Ty912δ is the long terminal repeat sequence from a Ty1 element integrated at the HIS4 promoter (61). The δ promoter was chosen because MOT3 was isolated as a high-copy-number suppressor of spt3 mot1 lethality and both Spt3 and Mot1 are required for δ promoter function (40, 72, 74). In addition, as described above, mot3Δ slightly increases Ty mRNA levels in both MOT1 and mot1-24 backgrounds. The results of these experiments (Fig. 5B, lanes 2 to 4) show that Mot3 causes a mobility shift of a Ty912δ promoter fragment. The presence of two major gel-shifted bands suggests that more than one Mot3 protein can bind per probe.
To determine if these gel shifts were zinc dependent, we added the zinc chelator, 1,10-o-phenanthroline to the gel shift reaction mixtures. 1,10-o-Phenanthroline is a high-affinity zinc chelator that can disrupt the DNA binding of zinc-dependent DNA-binding proteins (41). As predicted, Mot3-dependent gel shifts are sensitive to 1,10-o-phenathroline (Fig. 5B, lane 5), and they can be reconstituted by adding back either zinc or nickel to the gel shift reaction mixture (lanes 9 and 10). It is unclear if nickel ions can replace zinc ions to reconstitute Mot3 binding or if the nickel sulfate is contaminated by zinc. Thus, Mot3 binds to the Ty912δ promoter in a zinc-dependent fashion.
To determine if Mot3 bound to the Ty912δ probe in a sequence-specific manner, we carried out DNase I footprint analysis. Purified Mot3 protein was titrated onto two Ty912δ probes, 32P labeled to visualize binding to each strand. The combination of these probes allowed us to examine binding over the complete Ty912δ element. Between the two probes, four footprints (A to D) are apparent (Fig. 6). Sites B, C, and D are within the δ sequences, while site A lies at the HIS4-δ junction. In addition, there are a number of DNase I-hypersensitive cleavages at the boundaries of sites A, B and D. Site C appears to be less well protected than sites A, B, and D, as evidenced by the apparent cleavages that occur in this region when other sites appear to be saturated for binding. The footprints are summarized schematically in Fig. 6B and are compared to previously described TBP footprints in this region (3).
FIG. 6.
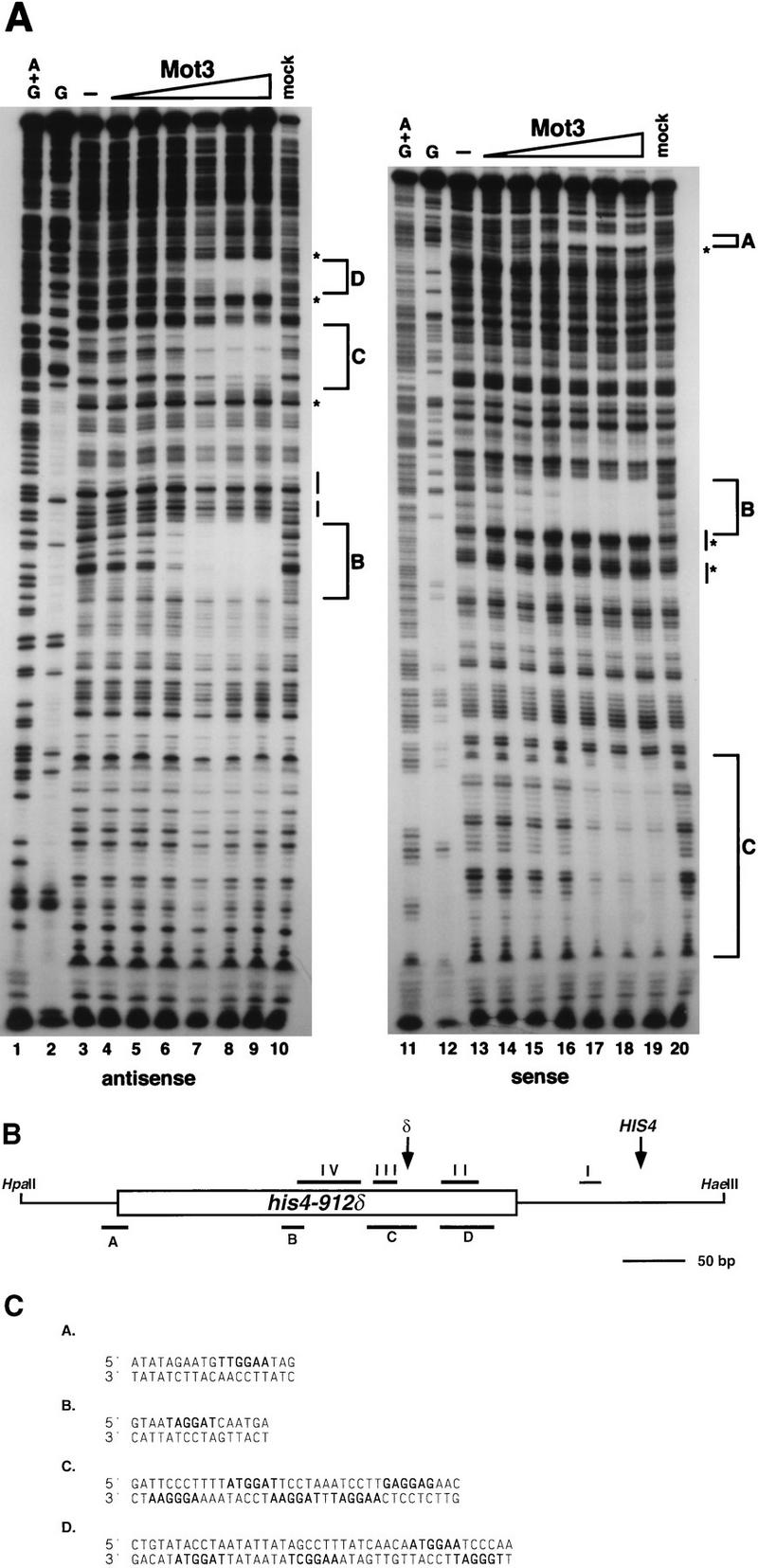
Mot3 binds to four sites in his4-912δ DNase I footprint analysis of Mot3 binding to the his4-912δ 5′ region was carried out as described in Materials and Methods. (A) DNase I footprint analysis was performed with increasing amounts of purified Mot3 protein on the sense (gel on the right) and antisense (gel on the left) strands of his4-912δ. Reaction mixtures contained the following amounts of purified Mot3 protein: lanes 3 and 13, 0 ng; lanes 4 and 14, 5 ng; lanes 5, and 15, 10 ng; lanes 6 and 16, 25 ng; lanes 7 and 17, 50 ng; lanes 8 and 18, 75 ng; lanes 9 and 19, 100 ng. Lanes 10 and 20 contained mock extract. Lanes 1 and 11 contained chemical sequencing reactions for adenine and guanine (A+G), and lanes 2 and 12 contained chemical sequencing reactions for guanines (G). Lettered brackets indicate the regions protected by Mot3. Asterisks indicate DNase I-hypersensitive sites. Lines drawn parallel to the gel indicate the positions of the two TATA elements of the Ty912δ promoter. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer. (B) Schematic diagram of the his4-912δ region. The box represents the 334-bp δ sequence, and the thin lines represent HIS4 sequences. The restriction sites HpaII and HaeIII indicate the 5′ and 3′ boundaries of the footprint probes, respectively. Footprints for TBP described previously are indicated by horizontal lines and roman numerals above the δ. Site I is the HIS4 TATA. The tandem TATA boxes are located in site IV. Footprints for Mot3 are indicated by horizontal lines and letters below the promoter. Arrows labeled δ or HIS4 indicate the δ promoter and HIS4 transcription start sites, respectively, and transcription for both is from left to right. Relative to the HIS4 transcription start site at +1, the extents of protection by Mot3 are as follows: site A, −423 to −442; site B, −278 to −293; site C, −186 to −223; site D, −132 to −173. The δ is inserted at position −98 relative to the HIS4 +1. (C) Sequences protected in panel A are shown. Sequences in boldface are potential binding sites. Due to resolution at the ends of the gels, the extent of protection in sites A and D is approximate.
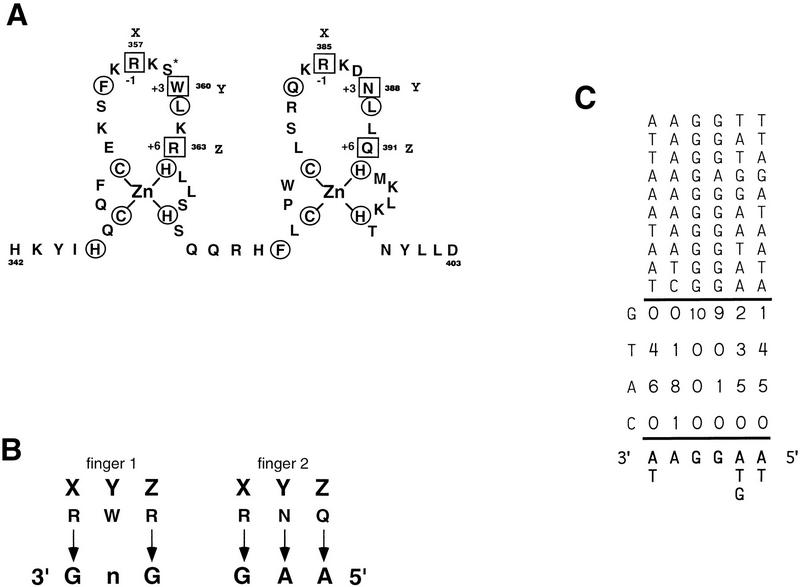
Analysis of other zinc finger proteins suggests that each zinc finger DNA-binding domain recognizes a 3-bp subsite (42, 51–53). Because Mot3 contains two zinc finger domains, a 6-bp recognition sequence might be expected to be common to each of these footprint regions. A comparison of these sequences suggests that the sequence important for Mot3 DNA binding contains a GGA sequence (Fig. 6C). Crystal structure analysis of other zinc finger proteins suggests that arginine, histidine, and lysine residues in conserved positions of the zinc finger are involved in base pairing with guanines (53). Thus, a G-rich sequence might be expected for the Mot3-binding site. The exact composition of the binding sequence seems to be more loosely determined at other positions, and further experiments are necessary to address this issue.
Analysis of Ty912δ promoter mutations that mutate a Mot3-binding site.
One of the Mot3-binding sites identified in vitro is immediately 5′ of the two TATA regions in the Ty912δ promoter. To determine if this site plays a Mot3-dependent role in Ty912δ promoter function, we measured the expression from both wild-type and mutant promoters, using a Ty912δ-lacZ fusion gene. For this analysis, the putative Mot3-binding site B was disrupted by introducing two different sets of clustered point mutations into the Ty912δ-lacZ promoter (21) (summarized in Fig. 7). The wild-type and mutant Ty912δ-lacZ fusions were then integrated at the HIS4 locus in both MOT3 and mot3Δ strains, and expression was measured by β-galactosidase assays. The results (Table 3) show that for a wild-type Ty912δ promoter, a mot3Δ2 mutation causes a modest increase (1.6-fold) in β-galactosidase activity, consistent with the observed increase in Ty mRNA levels (Fig. 3). Similarly, analysis of the two different mutations that overlap with Mot3 site B (mutations 15 and 16) also caused a slight increase (1.4-fold) in β-galactosidase activity compared to that of a wild-type Ty912δ-lacZ fusion. In a mot3Δ2 background, these two promoter mutations did not cause a further increase in β-galactosidase activity, suggesting that Mot3 confers weak repression via this site. We also used this Ty912δ-lacZ fusion to test if increased levels of Mot3 affect Ty expression. We found that there was no significant effect on β-galactosidase levels, which were 1,597 ± 33 U under overexpression conditions and 1,796 ± 41 U under normal conditions.
FIG. 7.
Ty912Δ44 promoter mutations. (A) Schematic diagram of Ty912δ-lacZ promoter mutations and their relationship to Mot3- and TBP-binding sites in the Ty912δ promoter. The large box represents the Ty912Δ44 promoter region. The small boxes numbered 15 and 16 are site-directed mutations whose mutant sequences are shown in panel B. The horizontal bars above the large box represent previously described TBP-binding sites II to IV, and the horizontal bars below the large box represent Mot3-binding sites. These site-directed mutants Ty912Δ44-15 and Ty912Δ44-16 are integrated at the normal his4-912δ position and are fused to the lacZ gene. The δ transcription start site is indicated by an arrow. (B) Mutant (mt) and wild-type (wt) sequences are indicated for the his4-912δ promoter mutations Ty912Δ44-15 and Ty912Δ44-16. The mutation number is indicated at the left, and the mutant and wild-type sequences for the indicated mutations are indicated at the right. For each mutation, the mutant sequence is indicated at the top and the wild-type sequence is indicated at the bottom. Absence of a mutation is indicated by a dash, and mutations are indicated by the nucleotide of the new mutation.
TABLE 3.
Effect of a mot3Δ mutation on Ty912Δ44::lacZ fusion alleles
| Mutationa | β-Galactosidase activity of wtb | Mutant/wt fold change | β-Galactosidase activity (U) of mot3Δb | mot3Δ/wt fold change |
|---|---|---|---|---|
| None (wt) | 1,999 ± 89 | 1.0 | 3,181 ± 322 | 1.6 |
| 15 | 2,783 ± 213 | 1.4 | 3,044 ± 187 | 1.1 |
| 16 | 2,790 ± 254 | 1.4 | 2,810 ± 94 | 1.0 |
Ty912Δ44::lacZ promoter mutations are described in Materials and Methods and in Results.
β-Galactosidase values are presented as described in Materials and Methods. Each value represents the average β-galactosidase units from two strains, each assayed three times. A negative control lacking Ty912Δ44::lacZ fusion produced 1 U of β-galactosidase activity. wt, wild type.
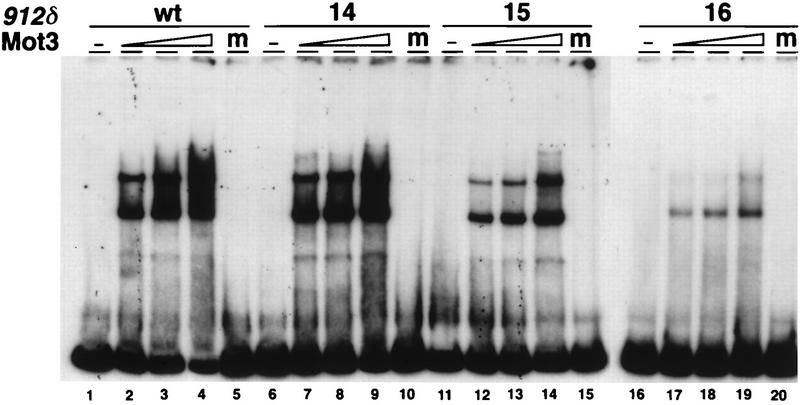
To determine if the Ty912δ mutations that alter Mot3-binding site B actually disrupt Mot3 binding in vitro, we carried out Mot3 gel shift assays with wild-type and mutant Ty912δ gel shift probes. Our results (Fig. 8) show that the site 16 mutation causes a drastic decrease in Mot3 binding. The site 15 mutation also impairs binding, but less severely than the site 16 mutation. The fact that mutations in sites 15 and 16 lead to slightly increased levels of expression from a Ty912δ-lacZ fusion and that mot3Δ mutations lead to increased Ty mRNA levels suggest that Mot3 binds to site B in vivo and plays a modest role in repression of Ty transcription.
FIG. 8.
Mutations in the Ty912δ promoter disrupt Mot3 DNA binding. Electrophoretic mobility shift experiments were carried out with wild-type (wt) and three mutant (panels 14, 15, and 16) end-labeled Ty912δ promoter fragments as described in Materials and Methods. The promoter fragments are labeled on the top line. Mutations 15 and 16 are site-directed mutants of the Ty912δ promoter described in the legend to Fig. 7. Mutation 14 is a set of clustered base pair changes immediately 5′ to mutation 15 but not affecting the putative Mot3 binding site (21). Each probe was incubated with increasing amounts of purified Mot3 protein. The reaction mixtures contained the following amounts of Mot3: lanes 1, 6, 11, and 16, 0 ng of protein; lanes 2, 7, 12, and 17, 0.5 ng; lanes 3, 8, 13, and 18, 1.0 ng; lanes 4, 9, 14, and 19, 2.0 ng. Reaction mixtures in lanes 5, 10, 15, and 20 were incubated with mock extract. Lanes 1 to 15 and 16 to 20 were run on separate gels and were combined in Adobe Photoshop for the purpose of this figure. The two gels were run in the same gel box, the Mot3 prep used for both gels was the same, and the proteins had identical DNA-binding activities. This figure was produced with Adobe Photoshop and a Fujix Pictography 3000 printer.
DISCUSSION
In this study, we sought to investigate further the functional relationship between the transcription factors Mot1 and Spt3 by isolating high-copy-number suppressors of mot1-24 spt3Δ synthetic lethality. This analysis identified the MOT3 gene, which encodes a previously unstudied zinc finger DNA-binding protein. We have shown that purified Mot3 binds to multiple sites in the Ty912δ sequence in a zinc-dependent manner. One binding site is immediately 5′ of the two δ TATA elements. A mot3Δ mutant shows a modest (1.6-fold) increase in the level of Ty transcripts and of a Ty912δ-lacZ reporter. These data suggest that Mot3 may contribute to repression of the Ty912δ promoter, possibly by controlling TATA box function. Mot3 has also been identified in two other studies. Results reported by Grishin et al. (33) suggest that Mot3 can have both positive and negative effects on transcription at many promoters. In a third study, Mot3 was identified as a high-copy-number suppressor of an mpk1Δ/slt2Δ mutation (44, 45). MPK1 is mitogen-activated protein kinase important for cell integrity, and mpk1Δ mutants lyse at 37°C. Thus, Mot3 is a DNA-binding protein that appears to affect the expression of many genes both when it is missing and when it is overproduced.
Our analysis of mot3Δ mutants has demonstrated that mot3Δ partially suppresses some of the phenotypes of mot1-24 and spt3Δ. In addition, mot3Δ causes a mild increase in sensitivity to UV light (38). Overexpression of MOT3 suppresses spt3Δ mot1-24 synthetic lethality. Grishin et al. (33) have also shown that both mot3Δ and MOT3 overexpression affect gene expression. However, none of the phenotypes of a mot3Δ mutant are especially strong, suggesting that Mot3 may be functionally redundant with one or more additional factors. An example of redundancy of zinc finger proteins has been observed for the yeast proteins Msn2 and Msn4, which were both isolated as high-copy-number suppressors of a snf1Δ mutation (27). Snf1 is a kinase involved in glucose repression of the SUC2 gene (13–15). Neither msn2Δ nor msn4Δ alone causes a detectable phenotype, but msn2Δ msn4Δ double mutants have modest defects in invertase expression and in growth on galactose media (27). Msn2 and Msn4 have significant homology outside of their predicted zinc finger domains (27). However, Mot3 has no significant sequence homolog in the S. cerevisiae genome (46). Thus, if proteins functionally redundant with Mot3 exist in S. cerevisiae, they do not have significant sequence similarity. Alternatively, Mot3 may not be redundant with other functions. Rather, it may play only minor roles in transcription, or it may play a significant role in some aspect of S. cerevisiae growth that has not yet been examined. Analysis of additional phenotypes might lead to the identification of other defects, and mutant screens for proteins that interact with Mot3 could identify redundant factors.
Based on our in vitro results and those of Grishin et al. (33), Mot3 appears capable of binding to many sites in the S. cerevisiae genome. Mot3 binds to three sites in the Ty912δ sequence and one site at the His4/Ty912δ 5′ boundary. The significance of these Mot3-binding sites for regulation of the Ty912δ promoter is unclear. Mutation of the Ty912δ Mot3-binding site 5′ to the TATA sequences caused a small increase in Ty912δ-lacZ expression, similar to the effect seen in a mot3Δ mutant with the wild-type Ty912δ-lacZ fusion. Conceivably, Mot3 binding to one or more sites in the Ty912δ sequence may depend on cooperative binding to high- and low-affinity sites within this sequence. The Drosophila repressor eve has been shown in vitro to have multiple high-affinity and low-affinity sites within a promoter; the high-affinity sites are important for cooperative binding of eve to its low-affinity sites and to its ability to repress (6, 68). As yet, our experiments do not fully address the issue of high- and low-affinity Mot3-binding sites. From footprinting data in Fig. 6A, site B appears to be better protected and to be protected at lower concentrations than the other three sites (sites A, C, and D). Site B may therefore represent a higher-affinity binding site than sites A, C, and D. Further mutational analysis of the Mot3-binding sites will be necessary to determine the affinity of Mot3 for different binding sites and to determine the contribution of these sites to δ promoter activity. In vivo binding analysis will be necessary to determine if Mot3 actually binds to as many sites in vivo as indicated by the in vitro data.
Crystallographic analysis of other zinc finger proteins has resulted in a number of empirical rules for DNA recognition by zinc finger DNA-binding domains. These rules suggest that residues at three conserved positions (usually lysines, histidines, and arginines) in the fingers (denoted in Fig. 9 as X, Y, and Z) interact with guanines in the DNA-binding site while asparagines and glutamines at these positions interact with adenines (42, 52, 53). Application of these empirical rules to Mot3 suggests that the Mot3 DNA-binding domain might recognize 3′ GNGGAA 5′ (where N indicates any nucleotide) (Fig. 9B). The GGA sequence appears in the Mot3 footprinted sites of the Ty912δ sequence. If this GGA site is used to anchor the Ty912δ-binding sites determined in vitro (Fig. 6), an alignment of these sequences predicts the following binding site: 3′ (A/T)AGG(A/T/G)(A/T) 5′ (Fig. 9). However, analysis of other zinc finger proteins suggests that there are exceptions to these emperical rules. For example, in the case of the yeast Adr1 zinc finger protein, none of the predicted subsites are found in the true binding site (69), and in the case of the Drosophila Tramtrack protein, a nonconserved serine is observed to make a base pair contact while a conserved histidine is observed to make a phosphate contact (28). The presence of a similar serine in the Mot3 zinc finger suggests that Mot3 might show side chain interactions similar to those of Tramtrack (Fig. 9A). Future analyses of Mot3 DNA-binding will help test these predictions.
FIG. 9.
Analysis of the Mot3-binding site. (A) Schematic diagram of the zinc finger motifs of Mot3 are shown. Residues that are part of the zinc finger consensus are circled, and residues that have been shown previously to participate in base contacts are boxed. The serine with the asterisk is referred to in the Discussion and is predicted to make additional base pair contacts. (B) Predicted Mot3 site based on previous analysis of zinc fingers. Residues labeled X, Y, and Z are shown with the nucleotides that are predicted to be contacted by those residues. The sites are read 3′ to 5′. (C) The sites predicted to be bound in sites A to D in Fig. 6C are lined up, and a consensus is generated for the prefered base(s) at each position.
Another interesting feature of the in vitro footprint experiments is the presence of a number of hypersensitive sites. In all sites except site D, these hypersensitive sites show a polarity that suggests that Mot3 is inducing a conformational change in DNA unidirectionally, making the DNA more susceptible to cleavage. In site D, where hypersensitive sites appear to occur on both sides, Mot3 might be recognizing sites on both strands, which could explain the large region of protection. Conformational alterations of DNA might represent a mechanism by which Mot3 regulates gene expression.
There are several possible mechanisms by which overexpression of MOT3 might suppress spt3Δ mot1-24 inviability. We proposed previously that both Mot1 and Spt3 help TBP to bind to functional TATA boxes at particular promoters (47). When Mot1 and Spt3 are both mutated, the cumulative defect in TBP binding might be too severe to support viability. Alternatively, the inviability could arise because one or more essential genes are not adequately expressed in the spt3 mot1-24 double mutant. The suppression of this double-mutant defect by overexpression of MOT3 could occur by suppression of either the mot1-24 or spt3Δ mutation alone. The original rationale for this high-copy-number screen was to find factors that might interact with Mot1. However, this possibility is unlikely, since MOT3 overexpression does not suppress any of the phenotypes caused by either mot1-24 or spt3Δ (46). In a second model, Mot3 could be a positive regulator of an essential gene that is underexpressed in the spt3Δ mot1-24 double mutant; in this model, MOT3 overexpression restores enough of the expression of this essential gene to allow viability. Consistent with this idea, Grishin et al. (33) have shown that overexpression of MOT3 causes overexpression of a number of genes in vivo. Whatever the mechanism of MOT3 high-copy-number suppression, its specificity for the spt3Δ mot1-24 double mutant strongly suggests that MOT3 overexpression is compensating for a transcriptional defect found specifically in this double mutant. Consistent with this model is the observation that even though a mot3Δ mutation is capable of at least partially suppressing phenotypes of both spt3Δ and mot1-24 single mutants, it is unable to suppress the inviability of the double mutant.
In summary, we have discovered a previously unstudied zinc finger protein as a high-copy-number suppressor of the spt3Δ mot1-24 synthetic lethality. We have shown that Mot3 binds to the Ty912δ sequence in vitro. Our results, combined with those of Grishin et al. (33), suggest that Mot3 may act as either a repressor or an activator. Our analysis of Ty912δ shows that Mot3 appears to be acting as a weak repressor. In light of the weak phenotypes of a mot3Δ, further genetic experiments are necessary to determine the in vivo relevance of Mot3 binding to δ sequences. Future studies of Mot3 function will help illuminate the function of zinc finger-binding proteins in transcription regulation and the possible role of Mot3 in the regulation of transcription in vivo.
ACKNOWLEDGMENTS
We thank Bob Kingston for helpful discussions during the course of this work. We thank Kendall Blumer and David Levin for communicating unpublished results.
This work was supported by NIH grant GM45720 to F.W. J.M. was supported in part by a grant from the Lucille P. Markey Trust.
REFERENCES
- 1.Abmayr S M, Workman J L, Roeder R G. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 1988;2:542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arndt K M, Ricupero S L, Eisenmann D M, Winston F. Biochemical and genetic characterization of a yeast TFIID mutant that alters transcription in vivo and DNA binding in vitro. Mol Cell Biol. 1992;12:2372–2382. doi: 10.1128/mcb.12.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 5.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP-binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 6.Austin R J, Biggin M. A domain of the even skipped protein represses transcription by preventing TFIID binding to a promoter: repression by cooperative binding. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley Interscience; 1988. [Google Scholar]
- 8.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, Ada2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 9.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman S, Churcher C, Badcock K, Brown D, Chillingworth T, Connor R, Dedman K, Devlin K, Gentles S, Hamlin N, Hunt S, Jagels K, Lye G, Moule S, Odell C, Pearson D, Rajandream M, Rice P, Skelton J, Walsh S, Whitehead S, Barrell B. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIII. Nature. 1997;387:90–93. [PubMed] [Google Scholar]
- 11.Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 12.Buratowski S, Hahn S, Guarente L, Sharp P. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 13.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 14.Celenza J L, Carlson M. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celenza J L, Carlson M. Structure and expression of the SNF1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:54–60. doi: 10.1128/mcb.4.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B P C, Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–75. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 17.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 18.Collart M. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 20.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudley, A., and F. Winston. Unpublished data.
- 22.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. Spt3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 24.Eisenmann D M, Chapon C, Roberts S M, Dollard C, Winston F. The Saccharomyces cerevisae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenmann D M, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA-binding factor, TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 26.Elble R. A simple and efficient procedure for the transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 27.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a snf1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairall L, Schwabe J W R, Chapman L, Finch J T, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 29.Fassler J S, Winston F. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics. 1988;118:203–212. doi: 10.1093/genetics/118.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gansheroff L J, Dollard C, Tan P, Winston F. The Saccharomyces cerevisae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich J, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 32.Grant P, Duggan L, Cote J, Roberts S, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis C, Winston F, Berger S, Workman J. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 33.Grishin, A., M. Rothenberg, M. A. Downs, and K. J. Blumer. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in S. cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 34.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 35.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressor. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 36.Happel A, Winston F. A mutant tRNA affects δ-mediated transcription in Saccharomyces cerevisae. Genetics. 1992;132:361–374. doi: 10.1093/genetics/132.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirschlorn J N, Winston F. SPT3 is required for normal levels of α-factor and a-factor expression in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:822–827. doi: 10.1128/mcb.8.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hongay, C., and F. Winston. Unpublished results.
- 39.Horiuchi J, Silverman N, Pina B, Marcus G A, Guarente L. Ada1, a novel component of the Ada/Gcn5 complex, has broader effects than Gcn5, Ada2, or Ada3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Y W, Stillman D J. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 41.Kadonaga J, Carner K, Masiarz F, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 42.Klevit R. Recognition of DNA by Cys2-His2 zinc fingers. Science. 1991;253:1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- 43.Kunkel T, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 44.Lee K S, Hines L K, Levin D E. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol Cell Biol. 1993;9:5843–5853. doi: 10.1128/mcb.13.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin, D. E. Personal communication.
- 46.Madison, J., and F. Winston. Unpublished results.
- 47.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TBP to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcus G, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 50.Moqtaderi, Z., and K. Struhl. Personal communication.
- 51.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 52.Pavletich N, Pabo C. Zinc-finger DNA recognition: crystal structure of a Zif28-DNA complex at 2.1A. Science. 1991;252:809. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 53.Pavletich N P, Pabo C O. Crystal structure of a five-finger GLI-DNA complex: new perspective on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 54.Piatti S, Tazzi R, Pizzagalli A, Plevani P, Lucchini G. Control of DNA synthesis genes in budding yeast: involvement of the transcriptional modulator MOT1 in the expression of the DNA polymerase α gene. Chromosoma. 1992;102:107–113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 55.Poon D, Campbell A M, Bai Y, P W A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 56.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 57.Ranish J A, Lane W S, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 58.Roberts S, Winston F. Essential function of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robzyk K, Kassir Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roeder G S, Farabaugh P J, Chaleff D T, Fink G R. The origins of gene instability in yeast. Science. 1980;209:1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- 62.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 63.Roth S Y. Chromatin-mediated transcriptional repression in yeast. Curr Opin Genet Dev. 1995;5:168–173. doi: 10.1016/0959-437x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 65.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takano E, Maki M, Mori H, Hatanaka M, Marti T, et al. Pig heart calpastatin: identification of repetitive domain structure and anomolous behavior in polyacrylamide gel electrophoresis. Biochemistry. 1988;27:1964–1972. doi: 10.1021/bi00406a024. [DOI] [PubMed] [Google Scholar]
- 68.Tenharmsel A, Austin R J, Savenelli N, Biggin M D. Cooperative binding at a distance by even skipped protein correlates with repression and suggests a mechanism of silencing. Mol Cell Biol. 1993;13:2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thukral S, Eisen A, Young E T. Two monomers of yeast transcription factor Adr1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991;11:1566. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winston F. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast. In: McKnight S, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1271–1293. [Google Scholar]
- 71.Winston F, Chaleff D T, Valent B, Fink G R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winston F, Dollard C, Malone E A, Clare J, Kapakos J G, Farabaugh P, Minehart P L. Three genes are required for trans-activation of Ty transcription in yeast. Genetics. 1987;115:649–656. doi: 10.1093/genetics/115.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winston F, Dollard K, Ricupero-Hovasse S. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 74.Winston F, Durbin K J, Fink G. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 75.Winston F, Minehart P L. Analysis of the yeast SPT3 gene and identification of its product, a positive regulator of Ty transcription. Nucleic Acids Res. 1986;14:6885–6900. doi: 10.1093/nar/14.17.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshihisa T, Anraku Y. Nucleotide sequence of AMS1, the structure gene of vacuolar α mannosidase of S. cerevisiae. Biochem Biophys Res Commun. 1989;163:908–915. doi: 10.1016/0006-291x(89)92308-5. [DOI] [PubMed] [Google Scholar]
- 77.Zawel L, Reinberg D. Common themes in assembly of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–566. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]