Abstract
1,25-Dihydroxyvitamin D3 [1,25(OH)2D3], the hormonal ligand for vitamin D3, is a potent inducer of myeloid-leukemic-cell differentiation. Such cells differentiate exclusively into monocytes/macrophages in response to this ligand. Since 1,25(OH)2D3 transduces its hormone signal through the vitamin D3 receptor (VDR), a ligand-modulated transcription factor and member of the nuclear hormone receptor superfamily, we sought to identify direct VDR target genes induced during this differentiation process. To do so, we applied a modified differential screen with a nascent-RNA purification strategy using biases for immediate-early-response genes induced by 1,25(OH)2D3 in the myelomonocytic cell line U937. Using this screen, we had previously identified p21Waf1/Cip1 as a gene transcriptionally induced by 1,25(OH)2D3 and demonstrated that this induction facilitates the differentiation of U937 cells into monocytes/macrophages (24). Here, we describe in detail our differential screen strategy and the identification and isolation of 20 1,25(OH)2D3-inducible genes or unknown cDNAs by means of this screen. One gene newly identified as a target of VDR regulation in myeloid cells is the homeobox HoxA10 gene. HoxA10 protein may act as a general regulator of cell growth, since overexpression of HoxA10 facilitated the differentiation of U937 cells into monocytes/macrophages independent of 1,25(OH)2D3 and acted to strongly inhibit the growth of the breast cancer cell line MCF-7 by arresting these cells in G1.
The processes of cellular proliferation and the progressive acquisition of a specialized phenotype show a remarkable degree of coordination. The essentially hierarchical nature of development and tissue maintenance is elegantly displayed in the replenishment of the hematopoietic system of the adult vertebrate. All the various types of blood and lymph cells are derived during fetal and adult life from a common pluripotent hematopoietic stem cell. These stem cells are rare in bone marrow (0.01% of bone marrow cells), and a large proportion of primitive stem cells are quiescent. A small number of multipotent stem cells laid down during embryogenesis give rise to a much larger population of more developmentally restricted progenitor cells. These cells then proliferate further to produce the functional, mature, postmitotic cells required to replace those cells lost through natural processes (e.g., apoptosis, postterminal differentiation). The stem cells themselves are capable of self-renewal to replace those that become committed to differentiation. It is clear that a balance in cell types and numbers is maintained throughout this progression from a less to a more differentiated state.
Leukemia is defined as the uncontrolled proliferation or expansion of hematopoietic cells that do not retain the capacity to differentiate normally into mature blood cells. Some hematologic disorders are not, strictly speaking, leukemias because they display only part of the full leukemic phenotype—either growth expansion (myeloproliferative syndromes and chronic-phase chronic myelogenous leukemia) or differentiation block (myelodysplasia syndrome); yet both of these conditions can progress to acute leukemia (26). This observation suggests that full leukemic transformation requires defects in both growth and differentiation. Several compounds and hormones, however, are capable of reversing this transformation by inducing leukemic cells to undergo differentiation (in some cases by growth arresting cells in G1; see below). For example, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], the active metabolite of vitamin D3, is a potent inducer of myeloid-leukemic-cell differentiation. Abe et al. (1) first reported that in vitro, murine myeloid leukemia M1 cells could be induced to differentiate into cells that were functionally and morphologically similar to macrophages by using 10−10 to 10−8 M 1,25(OH)2D3. The same group showed that treatment with this secosteroid in vivo considerably prolonged the survival of mice inoculated with M1 cells (15). Subsequent studies have established that 1,25(OH)2D3 could induce monocytic differentiation in human myeloid leukemic cell lines (27) and in blasts from patients with acute myeloid leukemia (14) or myelodysplasia syndrome (16). In animals and patients, however, differentiation occurs only at high concentrations of 1,25(OH)2D3 (10 to 100 nM), thus limiting the potential therapies as a result of the toxicity of this hormone (it induces hypercalcemia by stimulating calcium absorption) at nanomolar concentrations (17).
1,25(OH)2D3 transduces its signal through the vitamin D3 receptor (VDR), a member of a large group of related, transcriptional regulatory proteins that comprise the nuclear receptor superfamily. 1,25(OH)2D3-mediated effects on myeloid-cell differentiation ought therefore to be initiated through the transcriptional regulation of specific VDR target genes. To isolate such genes, we carried out a modified differential screen with a nascent-RNA purification strategy (6, 35) that biases for immediate-early-response genes that are induced by 1,25(OH)2D3 in the myelomonocytic cell line U937. The first gene isolated by means of this screen was p21Waf1/Cip1, which was subsequently shown in fact to be transcriptionally induced by 1,25(OH)2D3; this induction facilitates the differentiation of U937 cells into monocytes/macrophages (24). This strongly suggested that our differential screen satisfied two important criteria: it enriched for genes directly under the control of VDR, and the genes in question encoded proteins that play key roles in the induction of myeloid-cell differentiation.
Here, we describe our differential-screen strategy in detail, identify 20 1,25(OH)2D3-inducible genes or unknown cDNAs that we have isolated with this screen, and characterize one such gene as a newly identified target of VDR regulation in myeloid cells. The product of this gene, HoxA10, may act as a general regulator of cell growth, since its overexpression facilitates the differentiation of U937 cells into monocytes/macrophages independent of 1,25(OH)2D3 and acts to strongly inhibit the growth of the breast cancer cell line MCF-7 by arresting these cells in G1.
MATERIALS AND METHODS
Cell culture and plasmids.
U937 human myelomonocytic cells (clone 4; provided by K. Nilsson [29]) were routinely maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 5 mM l-glutamine. MCF-7 human breast adenocarcinoma cells were propagated in Eagle’s minimum essential medium with nonessential amino acids and 10% fetal bovine serum. pCMV5-HoxA10 was produced by inserting full-length HoxA10 as an EcoRI fragment generated from pBS-HoxA10 (a generous gift of C. Largman) into the EcoRI site of pCMV5.
Total nascent RNA preparation.
The isolation of newly synthesized mRNA during 1,25(OH)2D3 induction was based on a previously described protocol (6, 35). U937 cells at a density of 0.5 × 106 cells/ml were treated with 1.0 × 10−7 M 1,25(OH)2D3 (a gift from M. Uskokovic, Hoffman LaRoche) or ethanol, together with 10 μg of cycloheximide/ml, 200 mM 4-thiouridine (Sigma Chemicals), and 2.5 μCi of [3H]uridine/ml for 4 h. Total cellular RNA (400 μg) was then resuspended in 1 ml of pyrocarbonic acid diethyl ester-H2O. One milliliter of 2× buffer A (0.1 M sodium acetate [pH 5.5], 0.2% sodium dodecyl sulfate (SDS), 0.3 M NaCl, and 8 mM EDTA) was added, heat denatured for 5 min at 65°C, and then cooled on ice. A 6.0-ml volume of Affi-gel 501 (Bio-Rad) matrix was washed with 10 column volumes of buffer A. RNA was loaded onto the column and incubated for 10 min at room temperature, and the flowthrough was collected. Three column volumes of buffer A and 3 column volumes of buffer A plus 0.5 M NaCl were used to wash the column. Thiol-labeled RNA was then eluted with 18 ml of buffer A plus 2-mercaptoethanol by collecting 12 fractions of 1.5 ml each. Aliquots (40 μl) of each fraction were then counted to monitor recovery of RNA. Peak fractions of RNA were precipitated by adding sodium acetate and ethanol, and RNA was quantitated by absorbance at 260 and 280 nm. Peak RNA recovered in the thiouridine fractions was approximately 5 to 8% of the input RNA. Poly(A)+ RNA was generated from the total nascent RNA by one round of oligo(dT) chromatography with the Fast-Track Kit (Invitrogen), following the manufacturer’s specifications.
cDNA library construction and differential screening.
Five micrograms of pBluescript II SK+ (Stratagene) plasmid DNA was digested by EcoRI and XhoI and then dephosphorylated by calf intestine phosphatase (Boehringer Mannheim) and gel purified. The recovered vector DNA was aliquoted and stored at −20°C. Five micrograms of the 1,25(OH)2D3-treated nascent mRNA from U937 cells was used as the source of cDNA for library construction following the manufacturer’s instructions. The distribution of radioactive tracer cDNA was from 300 to 7,000 bp, and the yield of cDNA was determined by trichloroacetic acid precipitation. EcoRI linkers containing non-self-complementary overhangs were ligated into the cDNA, which was subsequently size fractionated to recover fragments over 500 bp in length. cDNA-plasmid ligation was set up with 50 to 100 ng of cDNA and 10 to 20 ng of vector. Following incubation at 16°C overnight, the ligation reaction was diluted fourfold with sterile H2O, and a small portion of the library (one tenth) was transformed into Epicurian Escherichia coli XL2-blue MRF′ ultracompetent cells (Stratagene) by means of the supplier’s recommended protocol. Eighteen clones were randomly picked and examined by EcoRI-XhoI digestion; 100% of the clones had inserts, ranging in size from 500 to 2,000 bp.
For differential colony screening, transformation mixtures were plated directly onto nitrocellulose filters overlaid onto Luria-Bertani agar–ampicillin plates, each 150-mm-diameter dish containing approximately 5,000 to 6,000 clones. Two carefully marked replicate filters were prepared by direct transfer of bacterial colonies onto a second prewetted nitrocellulose filter. A master plate and two filters were prepared in this manner from each plate. The initial screen was made up of about 100,000 unamplified clones—approximately one-fifth of the cDNA library. The colonies were grown directly on the nitrocellulose for 8 h at 37°C. Each filter was then transferred to an agar plate containing 100 μg of chloramphenicol per ml to amplify the plasmid. After 3 h at 37°C, the colonies were lysed. The remainder of the transformation mix was plated out and grown overnight. The following morning, the bacterial colonies were scraped in a batch with 30 ml of Luria-Bertani agar–ampicillin, and 1.0-ml aliquots were stored at −70°C. The filters were treated with 0.5 M NaOH–1.5 M NaCl, neutralized in 0.5 M Tris-HCl (pH 7.5), rinsed in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), and air dried. The DNA was immobilized on the filters by UV cross-linking (Strategene) and then baked at 80°C in a vacuum oven for 2 h.
To generate positive and negative probes, 2 μg of nascent poly(A)+ RNA from each treatment was used to synthesize a single-stranded cDNA probe with a high specific activity by using Superscript Reverse Transcriptase II (BRL) and [α-32P]dCTP. The size distribution of the total 32P-cDNA was examined by electrophoresis on a 1% alkaline agarose gel. The filters were prehybridized for at least 2 h at 42°C in 50% formamide, 6× SSPE, 5× Denhardt’s solution, 1% SDS, and 100 μg of poly(A) · poly(C). The hybridization was carried out for 72 h at 42°C under the same conditions except with a final probe concentration of 2.0 × 106 cpm/ml. One of the duplicate filters from each dish was hybridized to the positive probe [+1,25(OH)2D3], and the other duplicate was hybridized to the negative probe [−1,25(OH)2D3]. The filters were washed twice at room temperature for 30 min per wash with 2× SSPE–0.1% SDS and then once at room temperature for 30 min with 0.2× SSPE–0.1% SDS. The final wash was carried out at 50°C with 0.1× SSPE and 0.1% SDS for 30 min. After autoradiography, the positive and negative films were superimposed and compared at each spot. From the primary screen (0.1 × 106 unamplified clones), approximately 4,000 colonies that exhibited differential hybridization to the stimulated and unstimulated probes were picked and grown in 96-well plates. Duplicate dot blots were prepared with a replicator-beaded lid (TSP; Nunc) on HATF membranes (Millipore) from each plate and hybridized again with the positive and negative 1,25(OH)2D3 cDNA probes as in the primary screen. Candidates (approximately 200) which still appeared to be differentially expressed after the secondary screen were isolated for Northern blot analysis.
Northern blot analysis.
Total RNA (20 μg) from 0-, 4-, and 12-h treatments of U937 cells with 1,25(OH)2D3 were subjected to electrophoresis on a 1.2% formaldehyde-agarose gel and transferred to nylon membranes (NEN Genescreen Plus; New England Biolabs) by capillary action. Hybridization was carried out in a solution of 50% formamide, 2× Denhardt’s solution, 5× SSPE, 5% dextran sulfate, and 0.1% SDS at 42°C for 24 h, followed by washes: twice in 2× SSPE plus 0.1% SDS at room temperature for 15 min, once in 0.1× SSPE plus 0.1% SDS at room temperature for 15 min, and once in 0.1× SSPE plus 0.1% SDS at 50 to 65°C for 30 min. Probes from all of the unknown clones were prepared by PCR from the pBluescript vector with T3 and T7 primers. The amplified products were then gel purified and radioactively labeled with [α-32P]dCTP by random priming.
Clones whose RNA was induced in the Northern blots were partially sequenced with the T3 primer from pBluescript (pBluescript II SK+ was used to create the original cDNA plasmid library from 1,25(OH)2D3-treated U937 cells). Sequence data was analyzed by means of the BLAST algorithm, and both the DNA and the translated sequences were assessed for identities and/or homologies to known genes or proteins.
Nuclear run-on assay.
Uninduced and induced [10−7 M 1,25(OH)2D3] U937 cells were harvested, washed twice in phosphate-buffered saline, and lysed in Nonidet P-40 lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.5% [vol/vol] Nonidet P-40). The lysed cells were spun for 5 min at 50 × g, after which the nuclei were resuspended in 100 μl of glycerol storage buffer (40% [vol/vol] glycerol, 50 mM Tris-HCl [pH 8.3], 5 mM MgCl2, and 0.1 mM EDTA). To 100 μl of nuclei, 100 μl of 2× reaction buffer (10 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.3 M KCl), 5 mM concentrations of each nucleotide (ATP, CTP, and GTP), and 10 μl of 10-mCi/ml [α-32P]UTP (3,000 Ci/mmol; Amersham) were added. The reaction mixture was incubated for 30 min at 30°C, the reaction was terminated by the addition of 40 U of RNase-free DNase I in HSB buffer (0.5 M NaCl, 50 mM MgCl2, 2 mM CaCl2, and 10 mM Tris-HCl [pH 7.4]), and the mixture was further incubated at 30°C for 5 min. After the addition of 200 μl of SDS-Tris buffer and 10 μl of proteinase K (20 mg/ml), the reaction was incubated for another 30 min at 42°C. RNA was extracted, precipitated, and dissolved in N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) solution (10 mM TES [pH 7.4], 10 mM EDTA, and 0.2% [wt/vol] SDS). The RNA was hybridized to HoxA10 cDNA immobilized on a nitrocellulose membrane for 36 h at 65°C. Strips were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dried, and exposed to X-ray film. β-Actin cDNA was used as a control.
Immunoblots.
Total cell extracts were resolved by 15 to 7.5% SDS-polyacrylamide gel electrophoresis and transferred to a polyscreen polyvinylidene difluoride transfer membrane (NEN). Blots were incubated with a polyclonal anti-HoxA10 rabbit antibody (Babco) and developed by the use of enhanced chemiluminescence (Amersham).
Transfection and flow cytometry.
A total of 107 early-log-phase U937 cells were transiently cotransfected with 10 μg of pCMV5 vector with or without specific insert cDNAs and 8 μg of pGreen Lantern-1, a plasmid containing a modified version of the reporter gene Gene Fluorescent Protein (GFP) (Gibco-BRL) (for clarity, this plasmid is called pCMV-GFP throughout this work). Cells were harvested, washed twice, resuspended in 400 μl of RPMI 1640, electroporated (2,800 μF and 250 V; BTX) in 4-mm cuvettes, and diluted in 20 ml of RPMI 1640 complete medium containing 10% fetal calf serum. Forty-eight hours later, cells were harvested, filtered through cotton to remove dead cells, and analyzed by a fluorescence-activated cell sorter (FACS) (Becton Dickinson) with phycoerythrin-conjugated CD11b (Caltag). Only GFP-positive cells were taken into account for the analysis. For MCF-7 transfections, 107 MCF-7 cells (60 to 80% confluent) were cotransfected with 2 μg of pCMV-GFP and 8 μg of empty pCMV5 vector or pCMV5-HoxA10 by means of electroporation (1,000 μF, 100 V) as just described. Forty-eight hours after electroporation, cells were harvested, washed two times with phosphate-buffered saline, and resuspended in minimum essential medium without additions. GFP-positive cells were sorted (Becton Dickinson), and the DNA contents of their isolated nuclei were then determined by flow cytometry.
RESULTS
A differential screen to enrich for 1,25(OH)2D3-inducible genes.
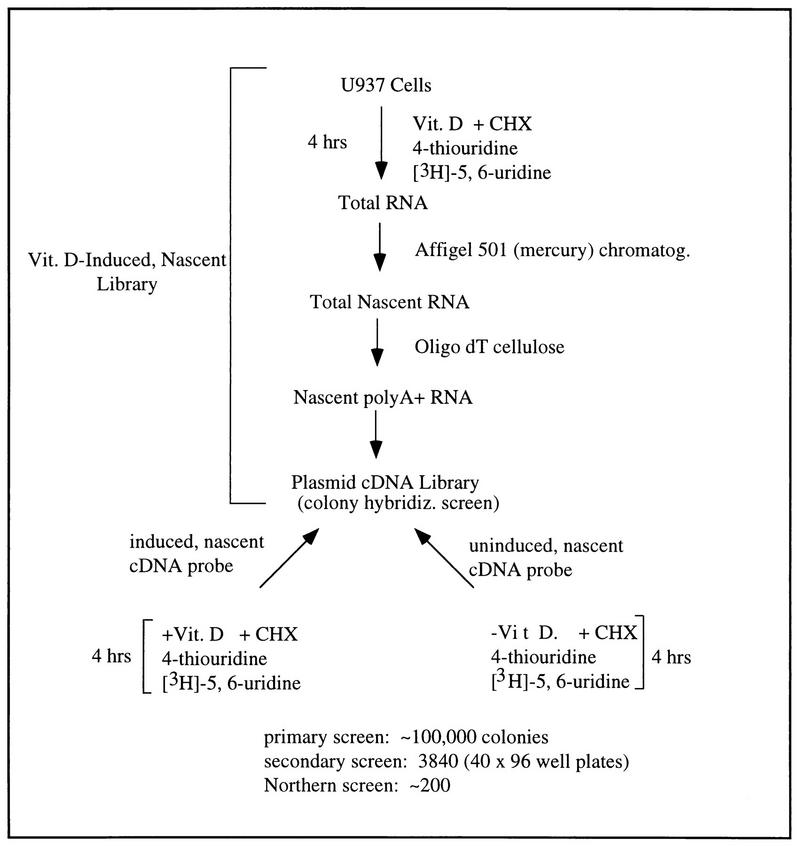
We sought to isolate and clone 1,25(OH)2D3-regulated genes that might initiate the differentiation of U937 cells in response to the ligand. To do so, we designed a modified differential screen that would bias for direct-transcriptional-target genes of VDR. As outlined in Fig. 1, three strategies were jointly employed to increase the likelihood of having an immediate-early transcript induced by 1,25(OH)2D3 represented in the probes and the cDNA library, as well as to increase the sensitivity of differential hybridization. First, since a 4-h pulse of the ligand was sufficient to commit U937 cells to differentiate along a monocyte/macrophage pathway (23a), cells that were harvested to generate the cDNA library and as sources of the induced positive probe were treated with 1.0 × 10−7 M 1,25(OH)2D3 for no more than 4 h, thus limiting the number of activated downstream genes (i.e., those not directly regulated by VDR) that would be represented in the library or probes. Second, in both the 1,25(OH)2D3-treated and the untreated cell populations, cycloheximide was included to inhibit protein synthesis and thereby preclude indirect effects of the ligand. Because cycloheximide was used in the library and in the preparation of both the induced and the uninduced probes, differential expression of putative clones observed upon colony screening should not be due to the effects of this drug, and therefore the possibility of cloning only cycloheximide-inducible genes is eliminated. Third, 4-thiouridine, together with a [3H]uridine tracer, was added to cells during the 4-h 1,25(OH)2D3 treatment to enrich for nascent-mRNA transcripts that could be selectively purified by organo-mercury chromatography (35). In this way, newly synthesized transcripts [e.g., those made during the 4-h 1,25(OH)2D3 pulse] could be enriched by separating them from the preexisting, housekeeping-type gene transcripts which could lower the relative abundances of the induced genes.
FIG. 1.
A differential screening strategy for the isolation of VDR target genes upregulated during the induced differentiation of U937 cells. See text for details. CHX, cycloheximide; Vit. D, vitamin D3.
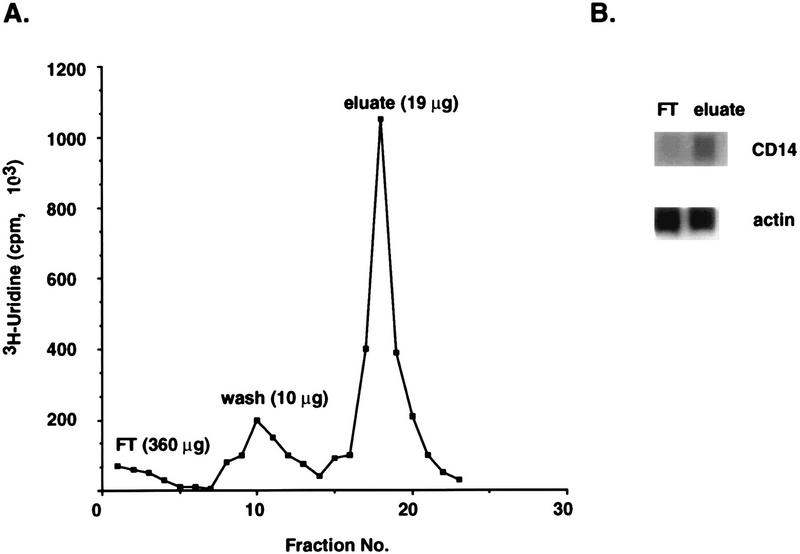
To this end, total cellular RNA accumulated in either the presence or absence of 1,25(OH)2D3 was fractionated by passage through an affinity phenyl-mercury agarose column such that thiol-labeled transcripts were covalently bound to the column, while unlabeled RNA was collected in the flowthrough (Fig. 2A). Nonspecifically bound RNA was removed from the column by low- and high-salt washes, and the thiol-labeled RNA was subsequently eluted in the presence of 50 mM 2-mercaptoethanol. Typically, about 95% of the total RNA input was recovered in the flowthrough and nonspecifically bound column fractions (Fig. 2A). When the nonselected total RNA was compared with the thiol-selected RNA by Northern hybridization with a probe encoding CD14, a gene known to be induced by 1,25(OH)2D3 in U937 cells, a dramatic increase in the relative abundance of CD14 transcripts was observed in the thiol-selected RNA (Fig. 2B). This result demonstrates our method to be an effective enrichment of newly synthesized U937 transcripts during a 4-h 1,25(OH)2D3 pulse.
FIG. 2.
Purification of thiol-labeled, nascent mRNA. (A) Affinity column profile of the nascent-RNA selection. A total of 400 μg of cellular RNA was isolated from U937 cells treated with 1,25(OH)2D3 and cycloheximide for 4 h. This RNA was applied to a phenyl-mercury agarose column (Affi-gel 501). The column was washed extensively, and thiol- and [3H]uridine-labeled transcripts were eluted as described in Materials and Methods. (B) Enrichment of 1,25(OH)2D3-induced nascent mRNA. RNA samples were fractionated as described for panel A, except that cycloheximide was excluded. The total nonselected RNA (column flowthrough [FT]) and eluted RNA (eluate) were analyzed by Northern blot hybridization with CD14 cDNA as a probe. An actin probe was included as a control for loading variability.
Full-length or partial cDNAs were generated from U937 cells treated with 1,25(OH)2D3 for 4 h to create a plasmid library. Over 100,000 colonies were initially screened. After a secondary screen, 162 candidate clones were selected to generate probes for Northern analysis of cells treated for 0, 4, and 12 h with 1,25(OH)2D3. Of these, induction of 20 clones appeared to be increased fourfold or more within 4 h of treatment (Fig. 3). After obtaining partial DNA sequences, we assigned the clones to four different categories (Table 1). Class I clones correspond to known genes. The first clone sequenced turned out to have identity to the cyclin-dependent kinase (CDK) inhibitor p21Cip1/Waf1. We have established that the p21 gene is transcriptionally induced by 1,25(OH)2D3 and that this induction facilitates the differentiation of U937 cells into monocytes (24). Other genes include those encoding the ribosomal proteins S4 and L21, Ki antigen, and cyclin A, HoxA10, Mad1, and CD14. Class II clones have no homology to known proteins at the DNA level but do contain isolated regions with similarity to known proteins at the amino acid level. Such clones include clone 340, which has weak homology to TAN-1, the human homolog of the Drosophila Notch protein (10), and clone 26, which has weak homology to NF-κB. Class III clones appear in the database as expressed sequence tags. Class IV clones are clones that do not appear in any database and are not homologous to any known gene. Examples of the typical inductions observed with Northern blots for members of each class are shown in Fig. 3.
FIG. 3.
1,25(OH)2D3 induction of candidate target gene RNAs in U937 cells. Shown are Northern blots of RNA from U937 cells treated with 1,25(OH)2D3 for 0, 4, and 12 h. Clone numbers correspond to the nomenclature in Table 1. Equal amounts of RNA were loaded in each lane, as indicated by the representative membrane stained with methylene blue and showing identical intensities of 28S and 18S rRNAs at each of three time points.
TABLE 1.
1,25(OH)2D3-induced clones from U937 differential screen
| Class | Clone | Extent of induction | Description from BLAST searcha |
|---|---|---|---|
| I | 42 | ++ | Gene encoding ribosomal protein L21 |
| 116 | ++++ | p21Cip1/Waf1 | |
| 124 | ++ | Gene encoding ribosomal protein S4 | |
| 316 | + | CD14 | |
| 322 | ++ | HoxA10 | |
| 344 | ++ | Gene encoding cyclin A | |
| 346 | ++++ | Gene encoding Ki antigen | |
| 347 | ++ | Mad1 | |
| II | 19 | + | Low similarity to dopamine receptor |
| 26 | ++ | Similarity to NF-κB | |
| 174 | + | Similarity to microtubule-associated protein and triacylglyceride lipase | |
| 303 | + | Similarity to respiratory syncytial virus gag | |
| 309 | ++ | Similarity to human immunodeficiency virus env | |
| 310 | + | Low similarity to Fos-related antigen | |
| 340 | +++++ | Low similarity to TAN-1 (Drosophila Notch homolog) | |
| III | 63 | + | EST |
| 168 | +++ | EST | |
| IV | 31 | + | Unknown |
| 304 | + | Unknown | |
| 308 | ++ | Unknown |
Nucleotide sequences obtained by sequencing the 5′ ends of the cDNA inserts were compared with those present in GenBank databases. EST, expressed sequence tag, which is from the random sequencing of cDNA libraries. Unknown, clones whose sequences find no match or significant homology in the GenBank or the EST database.
HoxA10 induction by 1,25(OH)2D3.
On the basis of the known or implied functions of some of the isolated 1,25(OH)2D3-inducible genes, we proceeded to investigate the possible roles of these genes in facilitating the differentiation of myeloid leukemic cells. For example, our previous studies of p21 responsiveness to 1,25(OH)2D3 in U937 cells established this gene as a direct transcriptional target of VDR leading to growth arrest at the G1 phase of the cell cycle and differentiation towards a monocyte/macrophage lineage. Likewise, owing to its identity as a homeobox gene, HoxA10 was also an intriguing candidate for study both as a VDR target gene and as a trigger of differentiation.
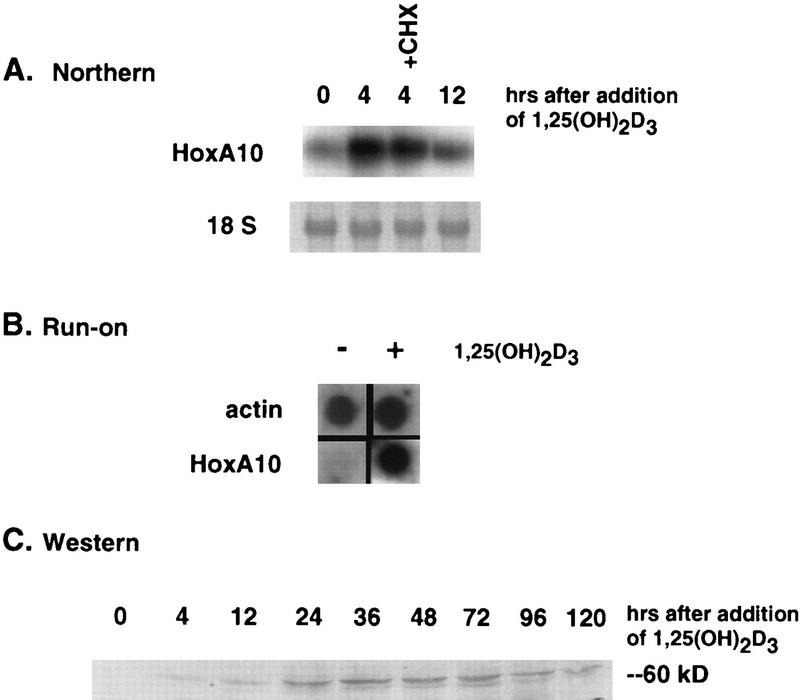
To confirm that HoxA10 is transcriptionally regulated by 1,25(OH)2D3 and VDR, HoxA10 mRNA accumulation in response to 1,25(OH)2D3 in the presence and absence of cycloheximide was examined by Northern blotting. Consistent with the strategy used by the differential screen, HoxA10 RNA was induced after 4 h in the absence or presence of cycloheximide (Fig. 4A), after which its levels decreased. To determine whether induction was at the level of transcription, nuclear run-on assays were performed with nuclei prepared from 1,25(OH)2D3-treated or untreated U937 cells. As shown in Fig. 4B, hybridization with a HoxA10 probe indicated strong induction of transcription after 4 h of ligand treatment. The kinetics of HoxA10 protein expression in response to 1,25(OH)2D3 reveal detectable but low levels at 4 and 12 h following the addition of ligand to U937 cells, peaking at 36 h and gradually diminishing (but detectable) through 120 h of exposure to ligand (Fig. 4C). The transient kinetics of HoxA10 expression may relate to its putative role in the differentiation of myeloid cells (see Discussion).
FIG. 4.
1,25(OH)2D3 induces HoxA10 expression in U937 cells. (A) Northern blot analysis of mRNA isolated from exponentially growing U937 cells treated with 1,25(OH)2D3 (10−7 M), with or without cycloheximide (CHX), for the indicated times. The methylene blue-stained 18S band was used as a control for RNA loading. (B) Nuclear run-on analysis of nuclear extracts isolated from cells grown in the presence or absence of 1,25(OH)2D3. Cells were harvested and nuclei were prepared 4 h after hormone or ethanol addition. β-Actin was used as a loading control. (C) HoxA10 protein levels following 1,25(OH)2D3 treatment. HoxA10 protein was assayed by immunoblotting with anti-HoxA10 antibody (Babco) with 20 μg of whole-cell extracts from cells treated with 1,25(OH)2D3 (10−7 M) for the indicated times.
Transient HoxA10 overexpression facilitates differentiation of U937 cells.
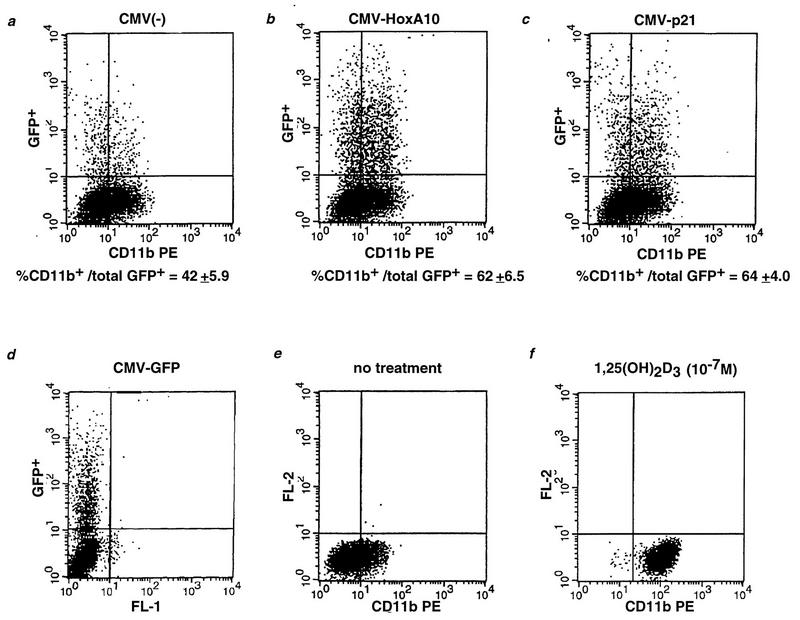
The early 1,25(OH)2D3 induction of HoxA10 expression and the role of the latter as a putative transcription factor suggested that this event may, in fact, initiate differentiation of U937 cells into monocytes/macrophages. We tested this hypothesis by expressing full-length HoxA10 from a cytomegalovirus (CMV) promoter following transient transfection of U937 cells with this plasmid. To identify transfected cells and to normalize for fluctuations in transfection efficiency, a CMV construct expressing GFP was included in the same transfection. After 48 h, GFP-positive cells were assayed for the appearance of the monocyte/macrophage-specific cell surface marker CD11b. As a positive control, cells were transfected with GFP and CMV-p21. As a negative control, an empty CMV expression vector was cotransfected with CMV-GFP and similarly assayed for both GFP and CD11b expression. Figure 5 demonstrates that the number of double-positive cells was significantly greater when CMV-HoxA10 was introduced into cells (Fig. 5B), compared to the CMV vector (Fig. 5A). That is, the percentage of CD11b-positive cells, normalized as a fraction of GFP-positive cells, increased specifically when HoxA10 was overexpressed relative to the empty overexpression vector. As expected, p21 also elicited expression of the differentiation marker (Fig. 5C); however, coexpression of p21 and HoxA10 did not appear to confer an additive or cooperative induction of CD11b (data not shown). This suggests that while both proteins can confer an effect on myeloid differentiation, they may do so through independent pathways, requiring the cooperation of additional, distinct factors.
FIG. 5.
Induction of differentiation of U937 cells by transient overexpression of HoxA10 in the absence of 1,25(OH)2D3. U937 cells were transiently cotransfected with an empty CMV vector [CMV(−)] (A) or with CMV expressing HoxA10 (B) or p21 (C), together with CMV-Green Lantern, a reporter plasmid constitutively expressing GFP. Forty-eight hours posttransfection, cells were harvested and extracts were prepared and labeled with PE-coupled CD11b. FACS analysis was then carried out to quantitate GFP (vertical axis) and CD11b fluorescent staining (horizontal axis). Double-positive cells are shown in the upper right quadrant; the fraction of CD11b-positive cells relative to the total GFP-positive population in each experiment is shown as a percentage below panels A to C. Each value is the mean of three independent experiments. GFP was used to mark transfected cells (D) and to normalize for fluctuations in transfection efficiencies in the empty CMV vector, CMV-HoxA10, and CMV-p21 transfections. Results for untreated and 1,25(OH)2D3-treated cells (48-h treatment) were included in panels E and F as negative and positive controls, respectively, for CD11b induction. FL, fluorescence.
HoxA10 is induced in MCF-7 cells by 1,25(OH)2D3 and facilitates cell cycle arrest.
The human HoxA10 gene was originally cloned from a U937 cell library (25) and found to be expressed exclusively in myelomonocytic cell types (22). Using an antibody raised against the C terminus of the HoxA10 protein (34) (Fig. 4), we examined the expression of the protein in several different cell lines and surprisingly detected it to varying degrees in seven lines tested, ranging from HaCaT, an immortalized keratinocyte cell line, to the MCF-7 line (data not shown). Apart from U937 cells, MCF-7 was the only other cell type in which we found HoxA10 to be responsive to 1,25(OH)2D3 induction (Fig. 6A).
FIG. 6.
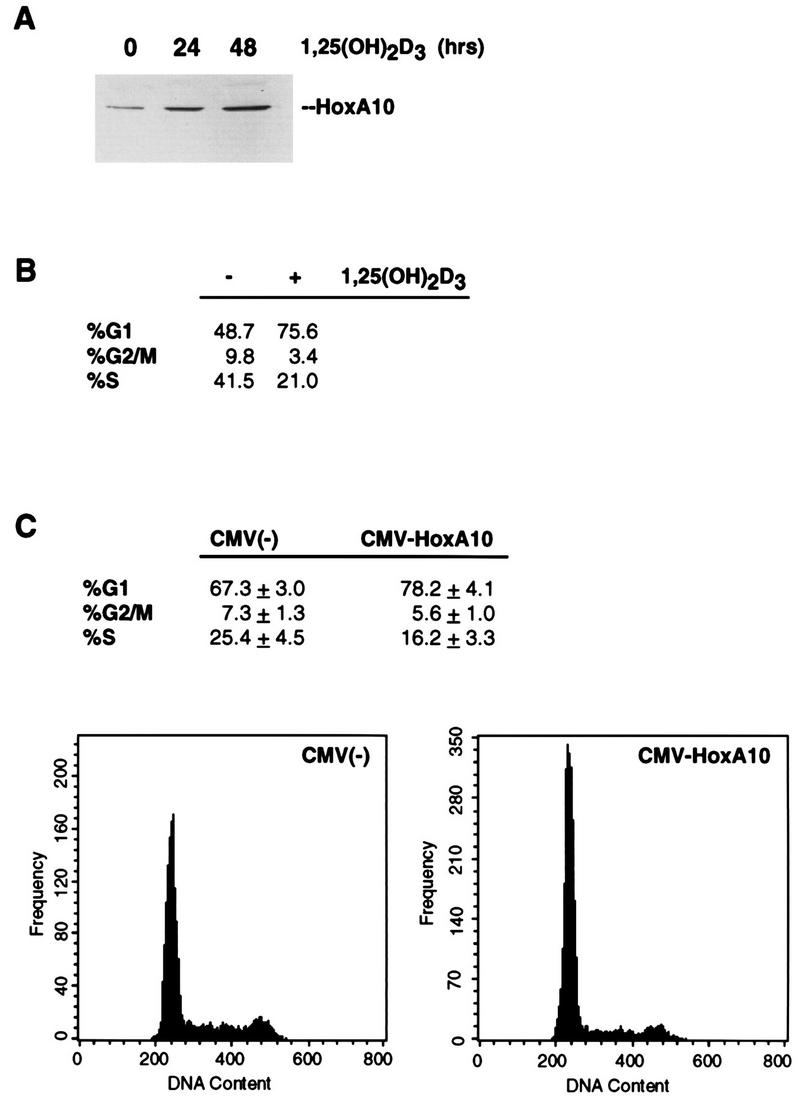
HoxA10 is induced in MCF-7 cells by 1,25(OH)2D3 and facilitates cell cycle arrest. (A) HoxA10 is induced by 1,25(OH)2D3 in MCF-7 cells. Whole-cell extracts were isolated from MCF-7 cells 0, 24, and 48 h after the addition of 10−8 M 1,25(OH)2D3. Twenty micrograms of total protein was separated by SDS-polyacrylamide gel electrophoresis, transferred to a polyvinylidine difluoride membrane, and probed with an anti-HoxA10 antibody. (B) MCF-7 cells are arrested in G1 by 1,25(OH)2D3. The DNA content of nuclei isolated from MCF-7 cells that were either untreated or treated for 24 h with 10−8 M 1,25(OH)2D3 was determined by FACS analysis after staining with propidium iodide. (C) Transient ectopic overexpression of HoxA10 induces G1 arrest in MCF-7 cells. The cell cycle distribution of MCF-7 cells was determined for cells that were cotransfected with an empty vector [CMV(−)] or CMV-HoxA10 together with CMV-GFP. GFP-positive cells were sorted, cell cycle distribution based on DNA content was determined by propidium iodide staining, and data were processed by using the Multicycle program (Phoenix Flow Systems). Shown are the averages of three separate experiments, and below are the DNA profiles. Note that the frequency scales are different on the two graphs.
Treatment of MCF-7 cells with 1,25(OH)2D3 resulted in a strong G1-phase growth arrest after 24 h (Fig. 6B). We therefore tested HoxA10’s ability to arrest MCF-7 cell growth by using a strategy similar to that carried out for U937 cells (described in the legend to Fig. 5). Since MCF-7 cells are already differentiated, we asked whether the ectopic overexpression of HoxA10 could induce cell cycle arrest. CMV-HoxA10 and CMV-GFP were therefore used to cotransfect MCF-7 cells; GFP-positive cells were sorted and analyzed for DNA content by propidium iodide staining. Expression of HoxA10 indeed conferred a significant decrease in the number of cells in S phase and an accompanying higher G1-phase content relative to cells transfected with the CMV vector alone (Fig. 6C). Thus, HoxA10 expression and its action as a growth inhibitor do not appear to be restricted to myeloid cell types.
DISCUSSION
We have described here a strategy for enriching for immediate-early genes that are induced by 1,25(OH)2D3 in myeloid leukemic cells. The crux of our strategy has been to bias for isolating the earliest targets of this regulation, i.e., direct, transcriptionally regulated genes of 1,25(OH)2D3 and VDR. We incorporated into our screen three components that most likely increased the probability of obtaining such targets: (i) a very short (4-h) pulse of the ligand prior to isolating RNA; (ii) cycloheximide to block de novo protein synthesis and thereby reduce the chance of isolating inducible genes downstream; and (iii) the addition of 4-thiouridine during the 4-h ligand treatment to permit the enrichment and isolation of only those mRNA species synthesized during the 4-h exposure to 1,25(OH)2D3. We then screened by using a standard differential approach with probes generated from 1,25(OH)2D3-treated and untreated cells. However, in principle, this approach could easily be combined with any of a number of current methods for isolating and cloning regulated genes: differential display, subtraction, or suppressive subtractive hybridization.
As is typical for any of these approaches, a number of the isolated cDNAs are unknown and novel. Of these, some have intriguing but quite weak homologies to known genes. For example, clone 340, which is strongly induced by 1,25(OH)2D3, has some partial amino acid sequence homology to TAN-1, the human homolog of the Drosophila Notch protein (10), and clone 310 has some similarity to Fos-related antigen. Some of the inducible cDNAs contain Alu repeat sequences or transposable elements. But a significant number of the isolated cDNAs are known genes encoding proteins with interesting functions. Four of these genes, encoding ribosomal proteins L21 and S4, Ki antigen, and cyclin A (Table 1), might not be predicted to be upregulated during differentiation. The cellular function of Ki antigen is unknown. It was originally identified as a nuclear protein recognized by systemic lupus erythematosus patient antisera (28, 29). Its sequence is between 33 and 41% identical to PA28, an activator of the 20S proteasome; both PA28 and Ki antigen are induced by gamma interferon at the mRNA level (2, 13). What is surprising about the induction of Ki antigen in myeloid cells by 1,25(OH)2D3 is that in mouse fibroblasts, its expression mirrors that of c-myc, which is generally associated with proliferation (28).
Similarily, increases in cyclin A are typically associated with cell growth, not differentiation. In proliferating cancer cells in culture, there is a striking positive correlation between the amount of cyclin A mRNA and protein and the number of cells in S and G2/M phase, so much so that cyclin A might be considered a marker for tumor cell proliferation (9). To our knowledge, there is no report that cyclin A is upregulated by an external signal during cell growth arrest or differentiation. However, in our differentiation assay, we consistently observed a transient burst of proliferation following 1,25(OH)2D3 treatment of U937 cells that precedes growth inhibition and differentiation. This short increase in proliferation is accompanied by increases in the levels of cyclin A, cyclin D1, and cyclin E proteins (31a). We do not as yet understand why differentiation might require an initial increase in proliferation, but it may explain why a target like cyclin A was isolated in the screen.
Three other known genes, p21Waf1/Cip1, Mad1, and HoxA10, were scored as positive in the differential screen and, based on their known functions, provide varying degrees of insight into how differentiation is induced by 1,25(OH)2D3. We previously described the identification of p21 as a 1,25(OH)2D3-inducible target gene by means of this screen, and we subsequently established that the p21 gene is, in fact, transcriptionally induced by 1,25(OH)2D3 and that this induction facilitates the differentiation of U937 cells into monocytes/macrophages. Overexpression of p27 also leads to U937 differentiation (24). Thus, cell cycle arrest in G1 gives way to differentiation, at least in this cell type.
Mad1 is a heterodimeric partner of Max (5); Max in turn is a requisite DNA binding partner of Myc (8). These two dimer species appear to have opposing biological effects: Myc-Max heterodimers transactivate target genes and are associated with the proliferative state (but also, paradoxically, with apoptosis) and oncogenesis. Myc overexpression is sufficient to drive cells through the cell cycle, and the Myc gene family has been implicated in many types of human malignancies (3, 11). These effects of Myc presumably occur through the action of Myc-Max heterodimers. In contrast, a switch in Max dimer partners from Myc to Mad results in a complex that has been associated with growth inhibition and differentiation in a number of cell lines (4, 20, 36). Ayer and Eisenman have shown that in U937 cells induced to differentiate with phorbol esters, Mad protein levels are rapidly induced and are accompanied by a switch from Myc-Max to Mad-Max heterodimers (4). Thus, the induction of Mad1 by 1,25(OH)2D3 may be initiating a similar switch that is required to alter the proliferative effects of Myc toward a growth-inhibitory state.
The human homeobox gene HoxA10 was originally cloned in U937 cells and reported to have a myeloid-restricted expression pattern. Surveying normal and leukemic marrow samples, Largman and coworkers found that this gene is expressed in CD34+ normal marrow cells but not in CD34− marrow cells or in mature neutrophils, monocytes, and lymphocytes (22). Moreover, when HoxA10 was overexpressed in murine bone marrow cells with a retroviral vector, a significant increase in the number of colonies formed containing megakaryocytes and blast cells with an absence of macrophage and pre-B-lymphoid progenitor cells (34) was observed. Taken together, these results suggest that the HoxA10 gene is active in the early stages of myelopoiesis and is downregulated as myeloid cells mature and differentiate. It is perhaps, then, somewhat surprising that when we transiently overexpressed HoxA10 in U937 cells independent of 1,25(OH)2D3, a significant degree of macrophage-specific CD11b expression was detectable in the transfected cells relative to the expression vector alone (Fig. 5). The upregulation of HoxA10 in response to 1,25(OH)2D3 may serve to increase the basal levels normally found in myeloid cells to further drive differentiation, but this may require only a transient, early increase in HoxA10 levels. Consistent with this, the kinetics of HoxA10 mRNA induction in response to 1,25(OH)2D3 peak at 4 h following addition of the ligand and subside thereafter (Fig. 4A).
The notion of central regulatory roles played by Hox genes during hematopoiesis is not new. Homeobox gene products function as key developmental switches in the determination of cell fate and tissue identity during Drosophila embryogenesis (12, 23). Since hematopoiegenesis in many ways parallels embryogenesis, it is reasonable to hypothesize that homeobox genes are involved in hematopoietic differentiation. In fact, in surveys of human leukemic cell lines, lineage-specific expression patterns of Hox genes have been observed (21). Moreover, there are suggestions that mutations of homeobox genes might be leukemogenic (33). That a ligand like 1,25(OH)2D3 is regulating a homeobox gene also has a precedent in the well-characterized effects of retinoic acid on Hox genes in the mouse, whereby the genes are induced in a linear cascade that reflects their arrayed organization on particular chromosomes (18). Langston and Gudas have identified retinoic-acid-responsive elements in the Hoxa-1 gene promoter (19), and they also showed that retinoic acid failed to induce Hoxa-1 expression in a murine differentiation-defective embryonal carcinoma cell line carrying a mutant gene for RARα (31). A retinoic-acid-responsive element has also been characterized upstream of the mouse Hoxd-4 gene (30). To our knowledge, our finding here that HoxA10 is induced by 1,25(OH)2D3 in what appears to be a direct transcriptional induction is the first report of a Hox gene regulated by this ligand. We recently sequenced over 4 kb of the human HoxA10 upstream region from genomic YACs (3a), and we will determine whether functional vitamin D-responsive elements reside in this region and are responsible for mediating 1,25(OH)2D3 induction.
HoxA10 may play a more generalized role in cellular regulation beyond the myeloid lineage. Its mouse homolog, Hoxa-10, is expressed in the developing limb bud and in the gut and urogenital tract; its expression patterns are very similar to that observed for the Drosophila homeotic complex, Abdominal B (AbdB) (7). Recently, Hoxa-10-deficient mice were generated (32). Male homozygotes have severe defects in spermatogenesis, resulting in sterility, and female homozygotes, which ovulate normally, are sterile due to early embryonic death at day 3 postcoitus. In the mouse, then, this gene appears to play an important role in both male and female sterility and extends its role beyond hematopoiesis. Along these lines, we found that the human HoxA10 protein is expressed in numerous cell types and is induced by 1,25(OH)2D3 in breast tumor MCF-7 cells, in addition to U937 cells. The overexpression of HoxA10 in MCF-7 cells results in G1 arrest and growth inhibition. Thus the HoxA10 protein may be playing a central, early role in growth control. The molecular mechanism whereby this putative transcription factor regulates cell growth and differentiation generally, and in response to hormonal ligands such as 1,25(OH)2D3 in particular, will undoubtedly be an area of increasing attention.
ACKNOWLEDGMENTS
N.Y.R. and M.L. contributed equally to this work.
We thank T. Delohery, J. Wrana, D. Tenen, and A. Koff for important suggestions and discussions and V. Bromleigh, G. Farmer, and C. Rachez for comments on the manuscript. We are indebted to J. Massagué, C. Largman, D. Tenen, K. Nilsson, and M. Uskokovic for various reagents used here.
This work was supported by NIH grants DK-45460 and DK-52621 to L.P.F. and MSKCC Support Grant CA-08748. N.Y.R. was supported in part by a fellowship from the Dutch NWO Talent Stipendium. L.P.F. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Abe E, Miyaura C, Sakagami H, Takeda M, Konno K, Yamazaki T, Yoshiki S, Suda T. Differentiation of mouse myeloid leukemia cells induced by 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci USA. 1981;78:4990–4994. doi: 10.1073/pnas.78.8.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J Y, Tanahashi N, Akiyama K, Hisamatsu H, Noda C, Tanaka K, Chung C H, Shibmara N, Willy P J, Mott J D, Slaughter C A, DeMartino G N. Primary structures of two homologous subunits of PA28, a γ-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 1995;366:37–42. doi: 10.1016/0014-5793(95)00492-r. [DOI] [PubMed] [Google Scholar]
- 3.Amati B, Land H. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation, and death. Curr Opin Genet Dev. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 3a.Anderson, E. C., N. R. Rots, and L. P. Freedman. Unpublished data.
- 4.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 5.Ayer D E, Jretzner L, Eisenman D E. Mad: a heterodimeric partner of Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 6.Beadling C, Johnson K W, Smith K A. Isolation of interleukin 2-induced immediate early genes. Proc Natl Acad Sci USA. 1993;90:2719–2723. doi: 10.1073/pnas.90.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson G V, Nguyen T-H E, Maas R L. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol. 1995;15:1591–1601. doi: 10.1128/mcb.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 9.Brechot C. Oncogenic activation of cyclin A. Curr Opin Genet Dev. 1993;3:11–18. doi: 10.1016/s0959-437x(05)80335-1. [DOI] [PubMed] [Google Scholar]
- 10.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 11.Evan G I, Littlewood T D. The role of c-myc in cell growth. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 12.Gehring W J, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 13.Gray C W, Slaughter C A, DeMartino G N. PA28 activator protein forms regulatory caps on proteasome stacked rings. J Mol Biol. 1994;236:7–15. doi: 10.1006/jmbi.1994.1113. [DOI] [PubMed] [Google Scholar]
- 14.Hellstrom E, Robert K H, Soppi E, Putkoken P O, Gahrton G. Effects of retinoic acid, 1,25-dihydroxyvitamin D3, cytosine arabinoside, and interferon α on bone marrow cells from patients with myelodysplastic syndrome. Leuk Res. 1989;13:113–120. doi: 10.1016/0145-2126(89)90157-4. [DOI] [PubMed] [Google Scholar]
- 15.Honma Y, Huzumi M, Abe E, Konno K, Fukushima M, Hata S, Nishii Y, DeLuca H F, Suda T. 1α,25-dihydroxyvitamin D3 and 1α-hydroxyvitamin D3 prolong the survival time of mice inoculated with myeloid leukemia cells. Proc Natl Acad Sci USA. 1983;80:201–204. doi: 10.1073/pnas.80.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell A L, Satukel T A, Bloomfield C D, Davey F R, Ball E D. High dose vitamin D for treatment of myelodysplasia: a pilot study. Br J Haematol. 1991;77:30–35. [Google Scholar]
- 17.Kizaki M, Koeffler H P. Differentiation-inducing agents in the treatment of myelodysplastic syndromes. Semin Oncol. 1992;19:95–111. [PubMed] [Google Scholar]
- 18.Langston A W, Gudas L. Retinoic acid and homeobox gene regulation. Curr Opin Genet Dev. 1994;4:550–555. doi: 10.1016/0959-437x(94)90071-a. [DOI] [PubMed] [Google Scholar]
- 19.Langston A W, Gudas L J. Identification of a retinoic responsive enhancer 3′ of the murine gene Hox-1.6. Mech Dev. 1992;38:217–228. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 20.Larsson L G, Pettersson M, Oberg F, Nillson K, Luscher B. Expression of mad, mxi1, max, and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 21.Lawrence H J, Largman C. Homeobox genes in normal hematopoiesis and leukemia. Blood. 1992;80:2445–2453. [PubMed] [Google Scholar]
- 22.Lawrence H J, Sauvageau G, Ahmadi N, Lopez A R, LeBeau M M, Link M, Humphries K, Largman C. Stage- and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp Hematol. 1995;23:1160–1166. [PubMed] [Google Scholar]
- 23.Levine M, Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988;55:537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 23a.Liu M. Ph.D. thesis. Ithaca, N.Y: Cornell University School of Medical Sciences; 1996. [Google Scholar]
- 24.Liu M, Lee M-H, Cohen M, Freedman L P. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 25.Lowney P, Corral J, Detmer K, LeBeau M M, Deaven L, Lawrence H J, Largman C. A human Hox 1 homeobox gene exhibits myeloid-specific expression of alternative transcripts in human hematopoietic cells. Nucleic Acids Res. 1991;19:3443–3449. doi: 10.1093/nar/19.12.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore M A S. Growth and maturation factors in leukemia. In: Oldham R K, editor. Principles of cancer biotherapy. New York, N.Y: Raven Press; 1987. pp. 399–445. [Google Scholar]
- 27.Munker R, Norman A, Koeffler H P. Vitamin D compounds: effect on clonal proliferation and differentiation of human myeloid cells. J Clin Invest. 1986;78:474–480. doi: 10.1172/JCI112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido T, Shimada K, Nishida Y, Lee R S, Pardee A B, Nishizuka Y. Loss in transformed cells of cell cycle regulation of expression of nuclear protein recognized by SLE patient antisera. Exp Cell Res. 1989;182:284–289. doi: 10.1016/0014-4827(89)90299-1. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido T, Shimada K, Shibata M, Hata M, Sakamoto M, Takasaki Y, Sato C. Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Exp Immunol. 1990;79:209–214. doi: 10.1111/j.1365-2249.1990.tb05180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Ollson I, Gullberg U, Ivhed I, Nilsson K. Induction of differentiation of the human histiocytic lymphoma cell line U-937 by 1,25-dihydroxyvitamin D3. Cancer Res. 1983;43:5862–5870. [PubMed] [Google Scholar]
- 30.Popperl H, Featherstone M S. Identification of a retinoic acid response element upstream of the murine Hox-42 homeobox gene. Mol Cell Biol. 1993;13:217–265. doi: 10.1128/mcb.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt M A C, Langston A W, Gudas L J, McBurney M W. Retinoic acid fails to induce the expression of Hox genes in differentiation-defective murine embryonal carcinoma cells carrying a mutant gene for alpha retinoic acid receptor. Development. 1993;53:105–113. doi: 10.1111/j.1432-0436.1993.tb00650.x. [DOI] [PubMed] [Google Scholar]
- 31a.Rots, N. Y., and L. P. Freedman. Unpublished data.
- 32.Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–463. doi: 10.1038/374460a0. [DOI] [PubMed] [Google Scholar]
- 33.Shen W-F, Detmer K, Morgan D A, Largman C, Lawrence H J. Modulation of homeobox gene expression alters the phenotype of human hematopoietic cell lines. EMBO J. 1992;11:2445–2453. doi: 10.1002/j.1460-2075.1992.tb05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorsteindottir U, Sauvageau G, Hough M R, Dragowska W, Landsorp P M, Lawrence H J, Largman C, Humphries R K. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid differentiation. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodford T A, Schlegel R, Pardee A B. Selective isolation of newly synthesized mammalian mRNA after in vivo labelling with 4-thiouridine or 6-thiouridine. Anal Biochem. 1988;171:166–172. doi: 10.1016/0003-2697(88)90138-8. [DOI] [PubMed] [Google Scholar]
- 36.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]