Abstract
Mating and virulence of the human fungal pathogen Cryptococcus neoformans are controlled by calcineurin, a serine-threonine-specific calcium-activated phosphatase that is the target of the immunosuppressive drugs cyclosporine A and FK506. In previous studies, a calcineurin binding protein (Cbp1, Rcn1, Dscr1/Csp1-3/MCIP1-3) that is conserved from yeasts to humans has been identified, but whether this protein functions to regulate calcineurin activity or facilitate calcineurin function as a signaling effector has been unclear. Here we show that, like calcineurin, Cbp1 is required for mating in C. neoformans. By contrast, Cbp1 plays no role in promoting calcineurin-dependent growth at 37°C and is not essential for haploid fruiting. Site-directed mutagenesis studies provide evidence that tandem phosphorylation and dephosphorylation of two serine residues in the conserved SP repeat motif are critical for Cbp1 function. Epistasis analysis supports models in which Cbp1 functions coordinately with calcineurin to direct hyphal elongation during mating. Taken together, these findings provide insights into the roles of Cbp1 as an accessory subunit or effector of calcineurin-specific signaling pathways, which may be features conserved among the calcipressins to govern calcineurin signaling in immune cells, cardiomyocytes, and neurons of multicellular eukaryotes.
Signal transduction cascades are utilized by all organisms to facilitate the transmission of signals perceived at the cell surface to effectors within the cell. Protein kinases and phosphatases are the primary components of signal transduction cascades and mediate signaling via reversible phosphorylation. Signal transduction cascades are employed by many pathogenic fungi to allow rapid adaptation to changing environmental conditions within the host.
Cryptococcus neoformans is a significant human fungal pathogen and a leading cause of meningoencephalitis and respiratory disease among immunosuppressed individuals. The highly conserved Ca2+-calmodulin-activated serine/threonine-specific protein phosphatase calcineurin is necessary for the pathogenesis of several medically important fungi, including the human fungal pathogen C. neoformans, and regulates many physiological processes necessary for life, including morphogenesis, cell wall biosynthesis, and ion homeostasis (reviewed in reference 10). Calcineurin is composed of two subunits, a catalytic A subunit and a regulatory B subunit, and is the target of the immunosuppressive compounds FK506 and cyclosporine A, which inhibit its phosphatase activity (26). In C. neoformans, calcineurin activity is required for cell viability at 37°C and for the regulation of hyphal elongation and basidium formation, a prerequisite for the generation of infectious spores (4, 9, 31).
The life cycle of C. neoformans is well characterized and includes two distinct filamentous stages, mating (dikaryotic filamentation) and monokaryotic fruiting (1). Mating involves fusion between haploid cells of opposite mating types, a and α, in response to pheromone and nutrient deprivation, and leads to the production of dikaryotic filaments that differentiate at the tips to form basidia, the site of karyogamy and meiosis and subsequent basidiospore release (22). Monokaryotic fruiting occurs in haploid cells of either mating type, in the absence of a mating partner, generating monokaryotic filaments that differentiate at the tips to form basidia, which produce only α or a basidiospores (44, 48).
To study their implications for virulence and as drug targets, effectors of calcineurin that regulate hyphal elongation during mating and monokaryotic fruiting have been sought. This analysis revealed three candidates: the novel calcineurin-binding protein Cbp1, the calcineurin mutant multicopy suppressor Cts1, and the β-1,3 glucan synthase Fks1 (8, 13, 20). Using the yeast two-hybrid system, C. neoformans var. grubii Cbp1 was originally identified as a novel protein that physically interacts with calcineurin (13). This calcineurin binding protein, Cbp1, contains a conserved serine proline repeat, which serves as a substrate for the phosphatase activity of calcineurin. Cbp1 is a founding member of a family of calcineurin-binding proteins, called the calcipressins, which are present in organisms as diverse as model yeasts, pathogenic fungi, and humans. The yeast and human functional homologs, Rcn1 and DSCR1/MCIP1, respectively, similarly interact with calcineurin and modulate its activity and physiological functions via their actions as potential effectors or feedback regulators of calcineurin signaling (reviewed in references 10, 14, 27, 32, 37, and 49).
All members of the calcipressin family possess a conserved central peptide region, termed the serine-proline (SP) repeat, that serves as a mitogen-activated protein kinase substrate for a priming phosphorylation event that facilitates a subsequent phosphorylation at an adjacent site by glycogen synthase kinase-3 (GSK-3) kinases, with the GSK-3 phosphorylation event subject to reversal by the phosphatase action of calcineurin (47). Recent studies have shown that phosphorylation within this region is important, as the mutation of the conserved serines results in the loss of nuclear localization of DSCR1 (34). Additionally, recent studies by Hilioti et al. have shown that preservation of the GSK-3 phosphorylation event within the SP repeat of Rcn1 is necessary for calcineurin activity in S. cerevisiae (15). Thus, it is likely that the phosphorylation state of this region is involved in the modulation of calcineurin function, either via allosteric regulation of calcineurin activity or through subcellular targeting.
To examine the role of the calcineurin-binding protein Cbp1 as a modulator of calcineurin activity in C. neoformans, and to elucidate the importance of the highly conserved serine-proline repeat region for Cbp1 function, we isolated and disrupted the C. neoformans var. neoformans CBP1 gene. cbp1 mutant strains were assessed for phenotypes associated with loss of calcineurin function, including growth at high temperature, hyphal elongation during mating and monokaryotic fruiting, and calcium ion sensitivity. Additionally, to determine the role of the highly conserved SP repeat region on Cbp1 and calcineurin function, site-directed mutations were generated within CBP1 to mimic the phosphoserine or nonphosphorylated states of the SP repeat, and the influence of each substitution on calcineurin-specific signaling events was analyzed. Our studies provide compelling evidence that Cbp1 functions as a specificity modulator or targeting subunit of calcineurin-dependent hyphal elongation during mating. Additionally, we demonstrate that the phosphorylation state of the conserved SP repeat of Cbp1 does not significantly influence calcineurin association, but does direct the activity of calcineurin during hyphal elongation.
MATERIALS AND METHODS
Strains, media, and reagents.
All C. neoformans strains used in this study are derived from the congenic serotype D (var. neoformans) strains JEC21 (MATα) and JEC20 (MATa). The genotype of strain MCC3 is MATa cna1::ADE2 ura5 ade2. Strains JEC43 (MATα ura5) and JEC34 (MATa ura5) are 5-FOA resistant derivatives of JEC21 and JEC20, respectively. The genotypes of the cbp1 deletion strains derived from JEC20, JEC43, or JEC21 are as follows: DSF64 (MATa cbp1::NAT), DSF67 (MATα cbp1::NAT), DSF58 (MATα cbp1::NAT ura5) and DSF57 (MATa cbp1::NAT ura5). C. neoformans strains were grown on standard yeast medium except where otherwise indicated.
Oligonucleotide primers and sequencing.
Oligonucleotide primers for PCR and sequencing were synthesized by Integrated DNA Technologies, Inc. Sequencing was performed by the Davis Sequencing (Davis, CA) and the Duke University DNA analysis facilities using the ABI 377, version 3.3 (PE Applied Biosystems). Sequence data were analyzed with MacVector version 7.2 and the DNASTAR software suite version 4.0. Primers used in this study are listed in Table 1.
TABLE 1.
Primer list
| Primer | Description | Sequence (5′ to 3′)a |
|---|---|---|
| 8949 | CBP1 5′ UTR primer (−500) | CGTTCTGATATCGTGGGCGG |
| 7868 | CBP1 5′ pcr expression seroD | CTGGAAAAAATGACCATAAGCC |
| 7869 | CBP1 3′ pcr expression seroD | CCAAGACTCGGATGATGGCG |
| 8480 | CBP1 overlap primer 1 | ACACTCGCCCTCCTTCTTCCTCAC |
| 8163 | CBP1 overlap primer 2 | CGATTATATTACCTTCCACCGGCTGCGAGGATGTGAGCT |
| 8164 | CBP1 overlap primer 3 | AGCTGACATCCTCGCAGCCGGTGGAAGGTAATATAATCG |
| 8739 | CBP1 overlap primer 4 | GGCTCCTTGTCTCTGAAACACAACTTTCACATCTCCCCG |
| 8740 | CBP1 overlap primer 5 | CGGGGAGATGAGAAAGTTGTGTTTCAGAGACAAGGAGCC |
| 8167 | CBP1 overlap primer 6 | GCGGTAGTAGGCTCAGAACC |
| 8481 | CBP1 reverse primer | TCATCTGCCTTGCTCCCACCTGTCC |
| 8933 | CBP1 S138A forward | CCACACAACTTTCTCATCGCCCCGCCAGGTTCTCCGCC |
| 8934 | CBP1 S138A reverse | GGCGGAGAACCTGGCGGGGCGATGAGAAAGTTGTGTGG |
| 8935 | CBP1 S142A forward | CTCATCTCCCCGCCAGGTGCTCCGCCTGAAGGCTGGGAACC |
| 8936 | CBP1 S142A reverse | GGTTCCCAGCCTTCAGGCGGAGCACCTGGCGGGGAGATGAG |
| 8941 | CBP1 S138A, S142A forward | CCACACAACTTTCTCATCGCCCCGCCAGGTGCTCCGCCTGAAGGCTGGGAACC |
| 8942 | CBP1 S138A, S142A reverse | GGTCCCCAGCCTTCAGGCGGAGCACCTGGCGGGGCGATGAGAAAGTTGTGTGG |
| 8945 | CBP1 C-terminal FLAG forward | GCAATGCCTCCTTTAGATTATAAGGATGATGATGATAAGTGATCCGCCATCATCC G |
| 8946 | CBP1 C-terminal FLAG reverse | CGGATGATGGCGGATCACTTATCATCATCATCCTTATAATCTAAAGGAGGCATTGC |
| 8947 | CBP1 delta FLISPPGSPP forward | CCTCTTCCACACACCGATTATAAGGATGATGATGATAAGGAAGGCTGGGAACC |
| 8948 | CBP1 delta FLISPPGSPP reverse | GGTTCCCAGCCTTCCTTATCATCATCATCCTTATAATCGTTGTGTGGAAGAGG |
| 9394 | CBP1 S138E forward | CCACACAACTTTCTCATCGAGCCGCCAGGTTCTCCGCC |
| 9395 | CBP1 S138E reverse | GGCGGAGAACCTGGCGGCTCGATGAGAAAGTTGTGTGG |
| 9396 | CBP1 S142E forward | CTCATCTCCCCGCCAGGTGAACCGCCTGAAGGCTGGGAACC |
| 9397 | CBP1 S142E reverse | GGTTCCCAGCCTTCAGGCGGTTCACCTGGCGGGGAGATGAG |
| 9398 | CBP1 S138A, S142E forward | CCACACAACTTTCTCATCGCCCCGCCAGGTGAACCGCCTGAAGGCTGGGAACC |
| 9399 | CBP1 S138A, S142E reverse | GGTCCCCAGCCTTCAGGCGGTTCACCTGGCGGGGCGATGAGAAAGTTGTGTGG |
| 9400 | CBP1 S138E, S142A forward | CCACACAACTTTCTCATCGAGCCGCCAGGTGCTCCGCCTGAAGGCTGGGAACC |
| 9401 | CBP1 S138E, S142A reverse | GGTCCCCAGCCTTCAGGCGGAGCACCTGGCGGCTCGATGAGAAAGTTGTGTGG |
| 9402 | CBP1 S138E, S142E forward | CCACACAACTTTCTCATCGAGCCGCCAGGTGAACCGCCTGAAGGCTGGGAACC |
| 9403 | CBP1 S138E, S142E reverse | GGTCCCCAGCCTTCAGGCGGTTCACCTGGCGGCTCGATGAGAAAGTTGTGTGG |
| 9674 | CBP1 5′ EcoRI for pGAD424 | GATCTGGAATTCATGACCATAAGCC |
| 9675 | CBP1 3′ BglII for pGAD424 | CTCCCAAGATCTGGATGATGGCG |
Underlines indicate codon changes to introduce substitutions.
Disruption of the C. neoformans CBP1 gene.
The CBP1 gene was disrupted by homologous recombination with a cassette containing the nourseothricin dominant drug resistance gene from Streptomyces noursei, nat1, fused to the C. neoformans ACT1 promoter and TRP1 terminator and flanked on either side by CBP1 gene sequence (28). The cassette was introduced by a sequential overlap PCR approach as follows: the left arm of the cassette (340 bp) was generated by amplification of CBP1 genomic DNA with primers 8480 and 8164; the right arm of the cassette (425 bp) was generated by amplification of CBP1 genomic DNA with primers 8739 and 8167; the drug resistance module was amplified from the cloned NAT cassette (1700 bp) with primers 8163 and 8740; and the final 2.5-kbp overlap product was generated by PCR with the flanking primers 8480 and 8167 (5). Disruption was confirmed by PCR of genomic DNA from wild-type and cbp1::nat disruptants with a primer pair, 8480 and 8481, flanking the disruption cassette insertion site (positions 421 to 474) within the CBP1 gene.
Northern analysis.
Total RNA was isolated from strains grown at 25°C in YPD or synthetic minimal medium to mid-log phase using the Trizol reagent, as previously described (20). Total RNA (20 μg) was separated by formaldehyde-agarose (1.2%) electrophoresis, visualized by ethidium bromide staining, and transferred to nylon (Magnacharge, Osmotics, Inc.). PCR-generated fragments of the GPI10, CNB1, and CBP1 genes were labeled with [α-32P]dCTP and used as probes during Northern hybridization of immobilized RNA.
Filamentation assays.
Mating assays were performed by growing strains of opposite mating type on YPD solid medium for 48 to 72 h at 25°C prior to being cocultured on V8 or filamentation agar medium at 25°C for 7 days in the dark. Monokaryotic fruiting assays were performed similarly, with strains grown on YPD solid medium prior to spotting onto filamentation agar followed by incubation at 25°C for 14 days in the dark. Filamentation for both mating and fruiting assays was scored microscopically, as previously described (4). Quantitative mating (cell fusion) assays were performed as previously described (16, 40).
FLAG-tagging and site-directed mutagenesis of Cbp1.
The FLAG epitope (DYKDDDDK) was introduced into the CBP1 coding region by amplification of the CBP1 cDNA with primers 8949 and 8946 to generate a C-terminal Cbp1-FLAG fusion. Overlap PCR with the CBP1::FLAG template was subsequently used to generate the site-directed mutants of conserved serine residues within the serine-proline repeat (SPR). Briefly, a set of overlapping primers (shown in Table 1) incorporating the serine to alanine or serine to glutamic acid substitution(s) was used in combination with flanking primers 8949 and 7869 to generate full-length overlap products containing the C-terminal FLAG fusion. Deletion of the SP repeat of Cbp1 was generated by replacement of the region with the FLAG epitope. The SP repeat (amino acids 135 to 144) was replaced by an in-frame substitution with the FLAG epitope using a PCR overlap method as described above with primers 8947 and 8948 paired with the flanking primer set to generate the SP repeat deletion product. All overlap products were then ligated into the TA cloning vector pCR2.1 (Invitrogen) and introduced into Escherichia coli TOP10 cells (Invitrogen). Plasmids generated in this study are listed in Table 2.
TABLE 2.
Plasmid list
| Plasmid | Descriptiona | Construction | Reference |
|---|---|---|---|
| pDSF100 | CBP1 cDNA in pCRT7-NT | cDNA PCR (7868, 7869) | This study |
| pDSF111 | CBP1::FLAG in pCR2.1 | PCR (8949, 8946) | This study |
| pDSF112 | S138A in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 8933, 8934) | This study |
| pDSF114 | S142A in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 8935, 8936) | This study |
| pDSF116 | S138A + S142A in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 8941, 8942) | This study |
| pDSF118 | CBP1ΔSPR in pCR2.1 | overlap PCR (8949, 7869, 8947, 8948) | This study |
| pDSF122 | CBP1::FLAG in pPM8 | BamHI-XbaI from pDSF111 | This study |
| pDSF123 | S138A in pPM8 | BamHI-XbaI from pDSF112 | This study |
| pDSF125 | S142A in pPM8 | BamHI-XbaI from pDSF114 | This study |
| pDSF127 | S138A + S142A in pPM8 | BamHI-XbaI from pDSF116 | This study |
| pDSF129 | CBP1ΔSPR in pPM8 | BamHI-XbaI from pDSF118 | This study |
| pDSF135 | S138E in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 9394, 9395) | This study |
| pDSF136 | S142E in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 9396, 9397) | This study |
| pDSF137 | S138A + S142E in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 9398, 9399) | This study |
| pDSF138 | S138E + S142A in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 9400, 9401) | This study |
| pDSF139 | S138E + S142E in pCR2.1 | pDSF111, overlap PCR (8949, 7869, 9402, 9403) | This study |
| pDSF141 | S138E in pPM8 | BamHI-XbaI from pDSF135 | This study |
| pDSF142 | S142E in pPM8 | BamHI-XbaI from pDSF136 | This study |
| pDSF143 | S138A + S142E in pPM8 | BamHI-XbaI from pDSF137 | This study |
| pDSF144 | S138E + S142A in pPM8 | BamHI-XbaI from pDSF138 | This study |
| pDSF145 | S138E + S142E in pPM8 | BamHI-XbaI from pDSF139 | This study |
| pDSF146 | S138A cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 8933, 8934) | This study |
| pDSF147 | S142A cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 8935, 8936) | This study |
| pDSF149 | S138A + S142A cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 8941, 8942) | This study |
| pDSF150 | S138E cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 9394, 9395) | This study |
| pDSF151 | S142E cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 9396, 9397) | This study |
| pDSF152 | S138A + S142E cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 9398, 9399) | This study |
| pDSF153 | S138E + S142A cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 9400, 9401) | This study |
| pDSF154 | S138E + S142E cDNA in pCRT7NT | pDSF100, overlap PCR (7868, 7869, 9402, 9403) | This study |
| pDSF155 | S138A* in pCR2.1 | pDSF146, PCR (9674, 9675) | This study |
| pDSF156 | S138A + S142A* in pCR2.1 | pDSF149, PCR (9674, 9675) | This study |
| pDSF157 | S138E + S142A* in pCR2.1 | pDSF153, PCR (9674, 9675) | This study |
| pDSF158 | S138E + S142E* in pCR2.1 | pDSF154, PCR (9674, 9675) | This study |
| pDSF159 | CBP1* in pCR2.1 | pDSF100, PCR (9674, 9675) | This study |
| pDSF160 | S142A* in pCR2.1 | pDSF147, PCR (9674, 9675) | This study |
| pDSF161 | S138E* in pCR2.1 | pDSF150, PCR (9674, 9675) | This study |
| pDSF162 | S138A + S142E* in pCR2.1 | pDSF152, PCR (9674, 9675) | This study |
| pDSF163 | S142E* in pCR2.1 | pDSF151, PCR (9674, 9675) | This study |
| pDSF164 | S138A in pGAD424 | EcoRI-BglII from pDSF155 | This study |
| pDSF165 | CBP1 in pGAD424 | EcoRI-BglII from pDSF159 | This study |
| pDSF166 | S142E in pGAD424 | EcoRI-BglII from pDSF163 | This study |
| pDSF167 | CBP1 genomic in pCR2.1 | PCR (8949, 7869) | This study |
| pDSF173 | S138E in pGAD424 | EcoRI-BglII from pDSF161 | This study |
| pDSF174 | S142A in pGAD424 | EcoRI-BglII from pDSF160 | This study |
| pJMM119 | CNA1 in pGBT9 | Görlach |
Asterisks denote cDNA-pCR2.1 constructs.
Expression and detection of Cbp1 fusion proteins in C. neoformans.
To generate the Cbp1-FLAG, Cbp1S138A-FLAG, Cbp1S138E-FLAG, Cbp1S142A-FLAG, Cbp1S142E-FLAG, Cbp1S138A+S142A-FLAG, Cbp1S138A+S142E-FLAG, Cbp1S138E+S142A-FLAG, Cbp1S138E+S142E-FLAG, and Cbp1ΔSRR-FLAG protein fusions, the CBP1::FLAG coding regions were isolated as BamHI-XbaI fragments from the pCR2.1 constructs described above and ligated into the BamHI and XbaI sites of the pPM8 shuttle vector. The resulting constructs were linearized with I-SceI and introduced into C. neoformans by electroporation (7). Transformants were grown on medium lacking uracil to maintain the pPM8 plasmid. Total protein was isolated from transformants grown at 25°C with complete yeast extraction buffer (Calbiochem). Total protein (10 μg) was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (Bio-Rad) prior to immunoblotting with rabbit anti-FLAG polyclonal antibody (Sigma). Following hybridization with the secondary anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (Sigma), fusion proteins were detected with a chemiluminescent substrate (Pierce).
Two-hybrid plasmid constructions and assays.
The coding region of CBP1 was obtained by amplification of a JEC21-derived cDNA library with primers 7868 and 7869, generating a 780-bp product that was TOPO TA cloned into pCRT7-NT (Invitrogen) and subcloned as an EcoRI-BglII fragment into the EcoRI and BamHI sites of pGAD424. The serine to alanine and serine to glutamic acid substitutions of the SP repeat region of Cbp1 were generated by overlap PCR of the original CBP1 cDNA construct, followed by subcloning into pGAD424 (see Tables 1 and 2). The CNA1 gene in pGBT9 plasmid pJMM119 was generated as previously described (13). Yeast two-hybrid and chlorophenol red-beta-d-galactopyranoside assays were performed as described previously (9).
Nucleotide sequence accession number.
The genomic and coding sequences for CBP1 from C. neoformans var. neoformans (serotype D) have been deposited in GenBank under accession number AF230799.
RESULTS
Cbp1 does not mediate calcineurin-dependent high temperature growth.
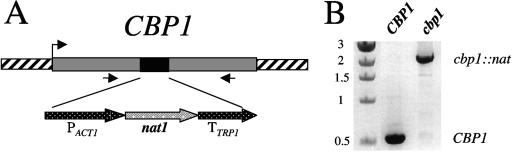
To further characterize the functions of Cbp1 in calcineurin-dependent processes in C. neoformans, the CBP1 gene from C. neoformans var. neoformans (serotype D) was isolated by PCR with primers designed from a sequence trace obtained from the C. neoformans genome database (http://www.tigr.org) queried with the C. neoformans var. grubii (serotype A) CBP1 coding region (13). The CBP1 gene was disrupted by homologous recombination with a nourseothricin dominant drug resistance cassette generated by a PCR overlap approach, as described in Materials and Methods (Fig. 1A). Disruption was detected by PCR amplification of genomic DNA from wild type and cbp1::nat disruptants with a primer pair flanking the disruption cassette insertion site within the CBP1 gene (Fig. 1B). The disruption frequency observed was 45% (9 of 20) for both mating types. The cbp1::nat disruption was confirmed by Northern and Southern hybridization (Fig. 2D and data not shown).
FIG. 1.
Disruption of the CBP1 gene by homologous recombination. (A) The CBP1 gene was disrupted by the insertion of the 1.7-kb nourseothricin dominant drug resistance cassette into the coding region of CBP1, as described in Materials and Methods. (B) Disruptants were identified by PCR with primers flanking the cassette insertion site within the CBP1 coding region. A representative PCR result is shown for both wild-type (CBP1) and disrupted (cbp1) alleles. Sizes are indicated at left in kilobases.
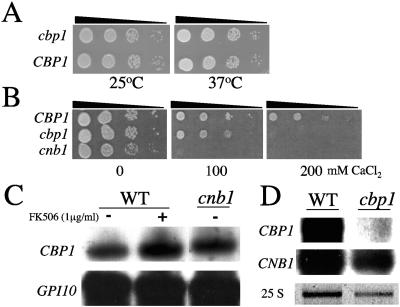
FIG. 2.
Growth of cbp1 mutant is sensitive to exogenous calcium but not elevated temperature and regulation of CBP1 transcription is calcineurin-independent. (A). The isogenic wild-type CBP1 (JEC21) and the cbp1 mutant strain were serially diluted and spotted onto YPD medium for 72 h at 25°C or 37°C. (B) The isogenic wild-type CBP1 (JEC21) and cbp1 and cnb1 mutant strains were serially diluted and spotted onto YPD medium with 0, 100, or 200 mM CaCl2 for 72 h at 25°C. (C and D) Northern blot analysis of total RNA isolated from isogenic wild-type (WT, strain JEC21), cnb1, and cbp1 mutant strains grown: (C) at 37°C in the presence or absence of 1 μg/ml FK506 for 4 h, or (D) at 25°C. Following hybridization with the CBP1 probe, the blots were reprobed with GPI10 (C) or CNB1 (D) for additional loading and hybridization controls. The 25 S rRNA stained with ethidium bromide serves as a loading control for D.
The importance of Cbp1 for growth at elevated temperature in the serotype D background was assessed, based on previous studies in which both the calcineurin A catalytic (CNA1) and regulatory (CNB1) subunits were demonstrated to be essential for growth at 37°C. Viability of the cbp1 mutant strain at both 25°C and 37°C was equivalent to that of the wild type strain (Fig. 2A). Because calcineurin mutants also have morphological defects at both permissive and restrictive temperatures, we also sought to examine the role of Cbp1 in morphogenesis. Wild-type and cbp1 mutant strains were grown to mid-log phase at both 25°C and 37°C in YPD, fixed, and visualized by differential interference contrast optics. In contrast to calcineurin mutants, cbp1 mutant cells did not demonstrate any alterations in morphology and were indistinguishable from wild type at either growth temperature (data not shown). In addition, overexpression of the CBP1 gene from an episomal shuttle plasmid, under the control of its endogenous promoter, in either the cna1 or cnb1 mutant strain was insufficient to restore growth at 37°C (data not shown). These results demonstrate that Cbp1 does not function downstream of calcineurin to mediate growth at elevated temperature.
In Saccharomyces cerevisiae, calcineurin signaling mediates calcium tolerance (6, 14, 42). To determine the role of calcineurin and the calcineurin-modulator Cbp1 with respect to calcium homeostasis in C. neoformans, we compared the abilities of cbp1 and cnb1 mutants to grow in the presence of exogenous calcium at 25°C. While growth of the calcineurin mutant strain (cnb1) was inhibited in the presence of 100 mM or higher levels of calcium ions (CaCl2), the cbp1 mutant strain was viable at 100 mM, but not at 200 mM, CaCl2, and was not hypersensitive to similar concentrations of NaCl or LiCl (Fig. 2B and data not shown). This result indicates that both calcineurin and Cbp1 are necessary for calcium homeostasis in C. neoformans and suggests that Cbp1 may function in a calcineurin-dependent manner.
CBP1 transcript levels are independent of calcineurin function.
Because functional homologs of Cbp1 in both S. cerevisiae (Rcn1) and multicellular eukaryotes (MCIP1/DSCR1) are transcriptionally regulated by calcineurin, we examined the possible influence of calcineurin on CBP1 transcript levels in C. neoformans (11, 14, 19, 51). Northern analysis was performed with total RNA isolated from wild-type and cnb1 mutant strains. Cells were grown to mid log phase in YPD at 25°C prior to a 4 h shift to 37°C in the presence or absence of the calcineurin inhibitor FK506. CBP1 transcripts were present at equivalent levels in wild-type cells compared to those in which calcineurin was mutated or inhibited (Fig. 2C).
In S. cerevisiae, Rcn1 has also been shown to regulate the expression of the calcineurin catalytic and regulatory subunits (19). We next examined whether Cbp1 plays a similar role in transcription of the calcineurin genes in C. neoformans. Expression of both CBP1 and the calcineurin B gene CNB1 was assayed by Northern blot in both wild-type and cbp1 mutant cells grown to mid-log phase in YPD at 25°C. There was no significant difference in CNB1 expression between wild-type and cbp1 mutant cells, and, as expected, the CBP1 transcript was absent in the cbp1 mutant strains (Fig. 2D). Taken together, these results show that calcineurin is not required to promote expression of the CBP1 gene, nor does Cbp1 appear to regulate the expression of calcineurin in C. neoformans, suggesting that Cbp1 does not participate in a negative feedback circuit to regulate calcineurin activity, in contrast to calcipressin family members in other organisms.
Cbp1 is required for hyphal elongation during mating.
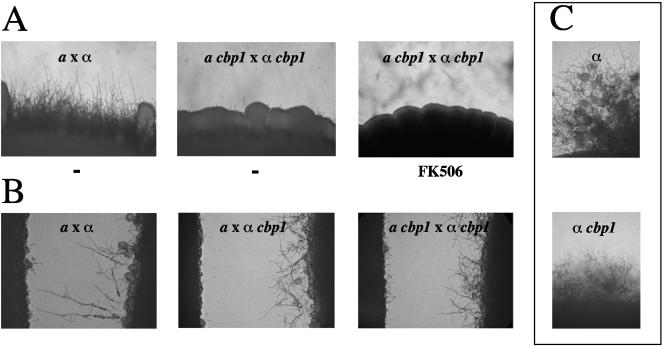
Calcineurin is required for monokaryotic (fruiting) hyphal elongation and for the production of the heterokaryotic filamentous mycelium resulting from cell fusion (mating) in C. neoformans (4). As calcineurin mediates growth at elevated temperature and filamentation by distinct mechanisms, we hypothesized that Cbp1 could serve as a calcineurin effector or regulator during hyphal elongation (4, 8). When the role of Cbp1 in mating was examined, we found that cbp1 mutant strains were unable to filament in a bilateral mutant (a cbp1 x α cbp1) cross, indicating that Cbp1, in addition to calcineurin, is also required for mating (Fig. 3A). No apparent filaments were produced, even with prolonged incubation in the dark to promote high efficiency mating, although a few basidia could be found embedded in the mating mix. The absence of obvious filaments, despite the presence of basidia, was suggestive of defective filamentation, but because the basidia were embedded in the mating mix, it could not be determined whether the basidial heads were barren or with spores. While a pronounced mating defect was observed in the cbp1 × cbp1 bilateral mutant cross, mating occurred at a wild-type level in unilateral cbp1 × cna1 and cbp1 × cnb1 mutant crosses, similar to previous observations with cna1 × cnb1 mutant crosses (Fig. 3A and data not shown). To examine the influence of the cbp1 mutation on cell fusion efficiencies, we performed quantitative mating assays and found no reduction in the fusion efficiency of cbp1 mutants compared to that exhibited by wild-type cells (data not shown).
FIG. 3.
Cbp1 is necessary for mating. (A) Deletion of the CBP1 gene results in bilateral sterility. Isogenic a and α CBP1 wild-type (JEC20 and JEC21) and cbp1 strains were coincubated on V8 agar at 25°C for 7 days, in the presence or absence of 1 μg/ml FK506. (B) Isogenic a and α CBP1 wild-type and cbp1 mutant strains were grown in confrontation on filament agar for 72 h at 25°C to evaluate the role of Cbp1 in responses to pheromone. (C) The α CBP1 wild-type and cbp1 mutant strains were grown on filament agar for 72 h at 25°C to evaluate the role of Cbp1 in monokaryotic fruiting. For each coculture and confrontation, the colony edge was photographed at 100× magnification.
To test if the bilateral mating defect observed with the cbp1 mutants is attributable to increased, rather than impaired calcineurin function, we tested and found that the calcineurin inhibitor FK506 (1 μg/ml) did not restore mating of cbp1 mutant cells (Fig. 3A). To verify that any potential inhibition of calcineurin activity by Cbp1 was not masked by the inhibitory effects of FK506, a range of FK506 concentrations were used to examine the influence of calcineurin inhibition on filamentation in wild-type, and cnb1 and cbp1 mutant unilateral and bilateral crosses. While concentrations of FK506 higher than 0.01 μg/ml inhibited all filamentation, subinhibitory concentrations, which dampen calcineurin activity but preserve minimal mating function, did not restore mating in cbp1 deficient bilateral crosses (data not shown). Additionally, while CBP1 overexpression in cbp1 mating partners restored filamentation in bilateral crosses, the overexpression of CBP1 in cnb1 mating partners failed to suppress the mating defect of calcineurin-deficient cells (Fig. 4B and C). These findings are in agreement with our previous results that support a positive role for calcineurin during mating and suggest that Cbp1 plays a stimulatory, rather than inhibitory, signaling role with respect to calcineurin function (4).
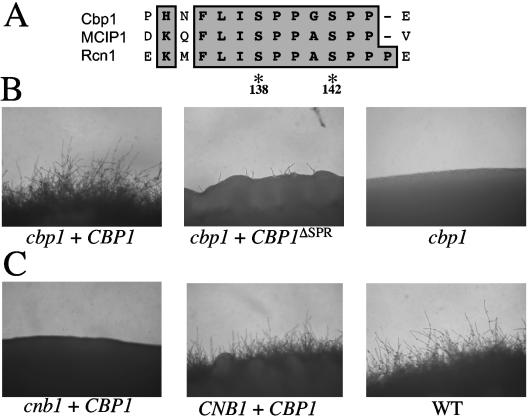
FIG. 4.
SP repeat of Cbp1 is required for hyphal elongation. (A) Alignment of the central peptide sequence motif containing the SP repeat for Cbp1 (C. neoformans), Rcn1 (S. cerevisiae), and DSCR1/MCIP1 (human). Amino acid positions for the conserved serines within the serine-proline repeat of C. neoformans Cbp1 are indicated. (B) Expression of Cbp1 complements to restore mating in a cbp1 mutant. Isogenic MATa and MATα cbp1 ura5 strains were transformed with constructs containing the CBP1 alleles in the pPM8 shuttle vector. Transformants were coincubated on nitrogen-limiting agar (V8) and photographed after incubation at 25°C for 7 days. (ΔSPR denotes the allele with deletion of the serine-proline repeat.) (C) Overexpression of Cbp1 fails to restore mating in a calcineurin mutant. Isogenic MATa and MATα wild-type (CNB1) and cnb1 ura5 strains were transformed with a construct containing the CBP1 allele in the pPM8 shuttle vector and cocultured on V8 medium for 7 days at 25°C. A wild-type cross (WT) is shown for comparison. Colony edges were photographed at 100× magnification.
To determine the importance of Cbp1 in responses to pheromone during mating, we examined the ability of α cbp1 mutants to undergo monokaryotic fruiting in a confrontation assay, in which α cells are monitored for the ability to produce filaments in response to pheromone from adjacent a cells (40). In contrast to calcineurin-deficient α cells, the cbp1 mutant strains were competent to produce both conjugation tubes and filaments in response to pheromone from adjacent a cells (wild-type or cbp1), although the extent of filamentation was significantly reduced compared to the wild-type control (Fig. 3B).
Interestingly, unlike filaments from a pheromone-responsive wild-type α cells, the filaments produced by the α cbp1 cells were not oriented toward the a mating partner, resulting in the absence of fusion with confronting a cells, although the filaments produced by the α cbp1 partner were viable and no fusion defect was observed when cbp1 α and a cells were cocultured in direct contact (Fig. 3B and data not shown). Mating factor α pheromone production was not impaired in α cbp1 strains, as typical morphological changes in a cells, either wild-type or cbp1, were induced following confrontation with α cbp1 cells and reverse transcription-PCR analysis of RNA isolated from confronting α cbp1 cells revealed no reduction in MFα1 transcript levels (Fig. 3B and data not shown). In addition, α cbp1 mutant strains generated monokaryotic filaments in the absence of a mating partner and the extent of filamentation resembled that observed for α cbp1 during a pheromone confrontation and was reduced compared to wild-type (Fig. 3C). These results suggest that calcineurin may differentially regulate monokaryotic and dikaryotic filamentation, and that, while not required for haploid fruiting, Cbp1 may be necessary for calcineurin-dependent a pheromone-directed morphological response.
Cbp1 function is calcineurin-dependent.
Members of the calcipressin family of calcineurin-regulatory proteins each possess a central peptide sequence motif, FLISPPxSPP, that is important for the modulatory action of the calcipressins on the activity of calcineurin (Fig. 4A). This motif also serves as a substrate for calcineurin (10, 13, 19). The phosphorylation state of the serines within the SP repeat is regulated via sequential phosphorylation initiated by phosphorylation of the second serine (serine 142) by a mitogen-activated protein kinase, which creates a consensus recognition site for phosphorylation of the first serine (serine 138) by glycogen synthase kinase-3 (15, 47).
To examine the function of the SP repeat of Cbp1 in mating, FLAG-tagged versions of wild-type Cbp1 and a mutant in which the SP repeat was replaced with the FLAG epitope were expressed from the CBP1 promoter in the pPM8 shuttle vector in the wild-type, cbp1, and cnb1 strains (Fig. 4B and C). Wild-type Cbp1 restored filamentation in a bilateral cbp1 mutant cross. In contrast, the CBP1ΔSPR allele failed to restore filamentation. These data could indicate that the SP repeat is important for Cbp1 function during mating. However, we found that loss of the SP repeat results in transcript instability (see Fig. 6B). Additionally, overexpression of wild-type CBP1 did not alter mating in wild-type cells or restore mating in cells lacking calcineurin (Fig. 4C and data not shown). Therefore, our data support a model in which Cbp1 activity requires the presence of functional calcineurin, as overexpression of Cbp1 did not restore mating in cells lacking active calcineurin.
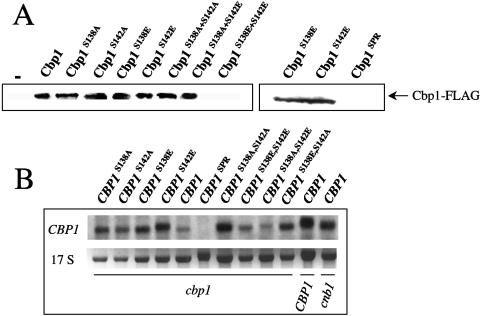
FIG. 6.
Influence of SP repeat phosphorylation on Cbp1 stability. (A) Phosphorylation of SP repeat regulates Cbp1 stability. Immunoblot analysis to detect FLAG-tagged Cbp1 fusions from total protein (10 μg) isolated from bilateral crosses of transformants containing CBP1::FLAG alleles cocultured on V8 medium for 48 h at 25°C. (B) Northern blot analysis of total RNA isolated from isogenic cbp1 ura5 transformants containing CBP1::FLAG alleles and grown to mid-log phase in selection medium at 25°C. Total RNA from wild-type and cnb1 ura5 strains were included as controls. Hybridization with the CBP1 probe as described for Fig. 2. The 17S rRNA stained with ethidium bromide serves as a loading control.
Phosphorylation status of the SP repeat of Cbp1 regulates hyphal elongation and protein stability.
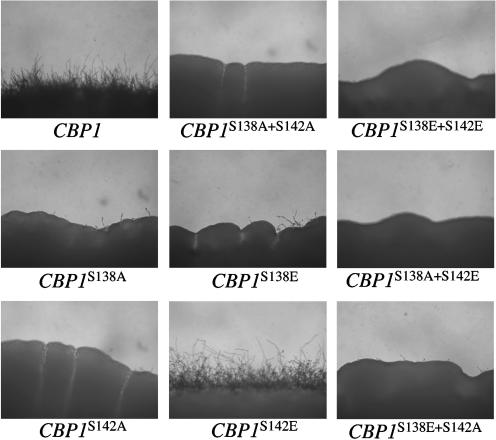
The importance of the phosphorylation status of each serine within the SP repeat of Cbp1 in mating was examined. FLAG-tagged versions of the wild-type CBP1 allele and a series of mutant alleles in which the serine residues at positions 138 and/or 142 were replaced with either alanine (A), to mimic the nonphosphorylated form, or glutamic acid (E), to mimic the phosphorylated form were generated. Each allele was expressed from the CBP1 promoter in the pPM8 shuttle vector in both mating partners of the cbp1 mutant and the ability to restore filamentation in a bilateral cross of cbp1 strains was examined. Substitution of serine for alanine at either or both positions, or combinations of serine to alanine and serine to glutamic acid (S138A, S142A, S138A+S142A, S138A+S142E, and S138E+S142A) failed to complement the cbp1 mutation, as there was no filamentation compared to the wild-type control (Fig. 5). However, the phospho-mimic serine to glutamic acid substitution at position 142 (S142E) was able to provide Cbp1 function and restored filamentation to near wild-type levels, whereas the serine to glutamic acid substitution at position 138 (S138E) did not (Fig. 5). Interestingly, the combined substitution of both serines to glutamic acid (S138E+S142E) resulted in a loss of Cbp1 function and a complete abrogation of filamentation.
FIG. 5.
Phosphorylation of S142, and tandem phosphorylation/dephosphorylation of S138, is necessary for Cbp1 function. Transformation with the S142E CBP1 allele, but not the S138E or S138E+S142E allele, restores mating in a bilateral cbp1 cross. Transformants were coincubated on mating medium (V8) and photographed at 100× magnification after incubation at 25°C for 7 days.
To examine the influence of serine substitution on Cbp1 protein stability, the FLAG-tagged Cbp1 fusions were affinity-purified from total protein lysates isolated from bilateral crosses of cells expressing the various CBP1::FLAG alleles. After SDS-PAGE and transfer to polyvinylidene difluoride membranes, blots were exposed to a polyclonal anti-FLAG antibody and relevant conjugated secondary antibody to detect the Cbp1 fusions. As expected, the wild-type and S142E alleles of CBP1 were stably expressed, as were several other alleles (S138A, S142A, S138E, etc). Thus, the failure of these mutants to complement the cbp1 mutation is not attributable to a loss of protein stability (Fig. 6A). However, the dual phospho-mimic S138E+S142E allele did result in a loss of protein stability, and no epitope-tagged Cbp1 was detected (Fig. 6A, left panel). In addition, the replacement of the serine-proline repeat also resulted in a loss of detectable protein (Fig. 6A, right panel). However, in this case (CBP1SPR), Northern analysis revealed the message was unstable, whereas for all the other mutants the CBP1 transcript was detected (Fig. 6B). That the Cbp1 S142E substitution is active, while the S138A+S142E substitution is not, suggests that the function of Cbp1 in filamentation is regulated via phosphorylation of serine 142, and that a sequential initial phosphorylation and subsequent dephosphorylation of serine 138 is important for stability, and possibly also activity, of the Cbp1 protein.
SP repeat phosphorylation does not mediate calcineurin binding.
Because the phosphorylation status of the conserved SP repeat controls Cbp1 function, we used the yeast two-hybrid assay to test whether phosphorylation promotes Cbp1-calcineurin interactions. Wild-type and serine-substituted Cbp1 mutants were fused to the Gal4 activation domain (GAD), the C. neoformans calcineurin A subunit (Cna1) was fused to the Gal4 DNA-binding domain, and the two were coexpressed in a yeast two-hybrid reporter strain (13). As shown previously for the serotype A Cbp1 protein, the serotype D Cbp1 protein also binds calcineurin (13) (Fig. 7A).
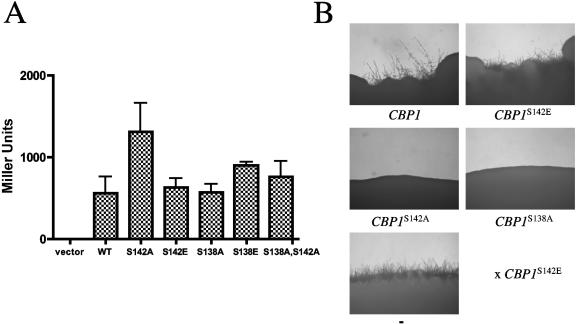
FIG. 7.
Influence of SP repeat phosphorylation on Cbp1-calcineurin interaction. (A) Phosphorylation status of SP repeat does not mediate the interaction of Cbp1 with calcineurin. Cotransformation of two-hybrid yeast reporter strain expressing calcineurin B (Y187) with plasmids expressing the Gal4 DNA-binding domain alone or fused to calcineurin A and the Gal4 activation domain alone or fused to Cbp1 (alleles listed on x-axis). Interaction values are shown in Miller units. (B) Mixed-allele mating assay. Isogenic MATα and MATa cbp1 ura5 strains were transformed with constructs containing the CBP1 alleles in the pPM8 shuttle vector. Transformants were coincubated in mixed-allele pairs on V8 agar and photographed after incubation at 25°C for 3 days. +, filamentation in mating assay; −, absence of filamentation.
Surprisingly, the interaction of Cbp1S142A with the calcineurin A subunit was noticeably stronger than that observed for the wild-type fusion, while all of the other serine substitutions examined, including Cbp1S142E and Cbp1S138A+S142A, bound calcineurin at levels equivalent to wild-type (Fig. 7A). Because the calcineurin-binding potential of the wild-type Cbp1 fusion was equivalent to that of the Cbp1S142E phospho-mimic, and both bound to a lesser extent compared to Cbp1S142A, we infer that the wild-type Cbp1 fusion may be phosphorylated by an endogenous mitogen-activated protein kinase at serine 142 and that either phosphorylation or the glutamic acid substitution reduces binding compared to Cbp1S142A. However, the phosphorylation state of the wild-type Cbp1 could not be determined from this assay. As previously observed for the Cbp1-calcineurin association (13), the calcineurin B subunit was also required for binding, demonstrating that the phosphorylation state of Cbp1 does not influence the necessity of the intact calcineurin heterodimer for binding (data not shown).
To corroborate the two-hybrid assays with in vivo activity in C. neoformans, a mixed-allele assay was developed based on the role of Cbp1 in mating. The CBP1 serine substitution alleles were expressed in one cbp1 mating partner and crossed to an opposite cbp1 mating partner expressing the functional CBP1S142E allele. Filamentation in the mixed allele assay was observed when cells expressing the CBP1S142E allele were crossed to wild-type (as expected) and CBP1S142E, CBP1S138E, CBP1S138A+S142E, or CBP1S138E+S142A alleles or to cells expressing no Cbp1 (Fig. 7B and data not shown). However, filamentation was noticeably absent for combinations of the CBP1S142E allele with mating partners expressing the CBP1S142A, CBP1S138A, and CBP1S138A+S142A mutant alleles. That Cbp1 S142E provides wild-type Cbp1 function when the mating partner lacks Cbp1 but not when the S138A, S142A, or S138A+S142A substitutions are expressed suggests a potential competition among phosphorylated and nonphosphorylated forms of Cbp1 for access to calcineurin. Together, these findings demonstrate that while phosphorylation of serine 142 is necessary for Cbp1 function in mating, the association of Cbp1 with calcineurin may be increased in the absence of phosphorylation at the SP repeat.
Overexpression of activated calcineurin partially overcomes loss of Cbp1 function.
These findings support a model in which phosphorylated Cbp1 promotes mating via the regulation of calcineurin activity, but do not distinguish the point of action of Cbp1 with respect to calcineurin. To address this issue, epistasis analysis was performed to examine the effect of overexpression of a constitutively active allele of the calcineurin catalytic subunit on mating in wild-type and cbp1 mutant cells. A constitutively active allele of CNA1 (CNA1-ΔCam-AI) was generated by truncation of the distal portion of the gene containing the calmodulin binding and autoinhibitory domains, cloned into the pPM8 shuttle vector, and expressed from the CNA1 promoter in wild-type, and cnb1, cna1, and cbp1 mutant strains (21, 33).
The extent of mating observed when the CNA1-ΔCam-AI allele was overexpressed in wild-type cells was indistinguishable from cells bearing the control plasmid. Mating and filamentation were restored by expression of the constitutively active CNA1-ΔCam-AI allele in cna1 mutant strains (data not shown) (21). Overexpression of the CNA1-ΔCam-AI allele failed to restore filamentation in the cnb1 mutant strain lacking the calcineurin B regulatory subunit, demonstrating that both subunits are required for calcineurin function even when the catalytic subunit is liberated from the requirement for calmodulin binding for activation. In the cbp1 bilateral cross, overexpression of the CNA1-ΔCam-AI allele in both mating partners partially restored mating, but not to the extent observed in the cna1 mutant strain background (Fig. 8A, and data not shown). Thus, when calcineurin is rendered constitutively active, the requirement for Cbp1 in mating can be partially bypassed.
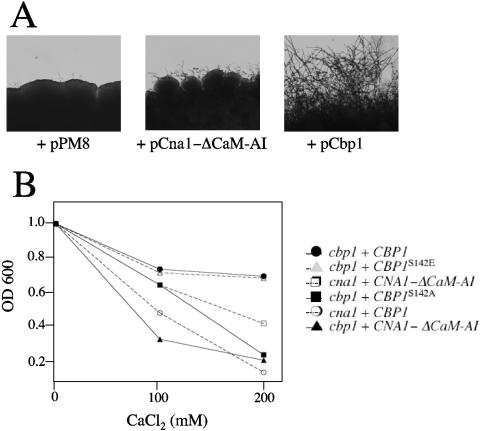
FIG. 8.
Cbp1 is a positive regulator/effector of calcineurin. (A) Overexpression of activated calcineurin partially overcomes loss of Cbp1 function. Isogenic a and α cbp1 strains were transformed with plasmids expressing the CNA1ΔCam-AI calcineurin A constitutive truncation mutant or CBP1 and coincubated in bilateral crosses on V8 agar at 25°C for 7 days. For each coculture and confrontation, the colony edge was photographed at 100× magnification. (B) Overexpression of Cbp1 or Cbp1S142E restores growth of calcium-sensitive cbp1 mutants. Transformants (cna1 or cbp1) harboring plasmids expressing Cbp1, Cbp1S142E, or Cbp1S142A, or constitutive calcineurin A, Cna1-ΔCam-AI, were inoculated at low density (OD600 = 1) into YPD with or without CaCl2 (100 or 200 mM) and incubated for 18 h at 25°C prior to culture density determination at OD600, each in duplicate.
In addition to mating, Cbp1 and calcineurin are required for calcium tolerance (Fig. 2). To determine whether Cbp1 cooperates with calcineurin to regulate calcium homeostasis, the ability of the CBP1, CBP1S142A, CBP1S142E, and CNA1-AIΔ alleles to restore calcium tolerance in wild-type and cbp1 and cna1 mutant strains was monitored. Both Cbp1 and Cbp1S142E, but not Cbp1S142A or CNA1-ΔCam-AI, restored calcium tolerance to cbp1 mutants (Fig. 8B). Expression of CBP1 was not sufficient to restore calcium tolerance in cna1 mutants (Fig. 8B), similar to the finding that increased Cbp1 also fails to restore mating of cna1 mutants. In addition, cna1 mutant strains were only minimally restored for growth in exogenous calcium by the constitutively active CNA1-ΔCam-AI, likely due to the loss of calmodulin responsiveness, and no effect was observed with this allele in cbp1 mutant cells (Fig. 8B). In all cases, wild-type cells were not altered in calcium sensitivity (data not shown). Taken together, these epistasis findings provide evidence to support a model in which Cbp1 modulates the specificity of calcineurin activity to promote mating and calcium homeostasis by facilitating calcineurin substrate recognition and/or subcellular targeting (Fig. 9).
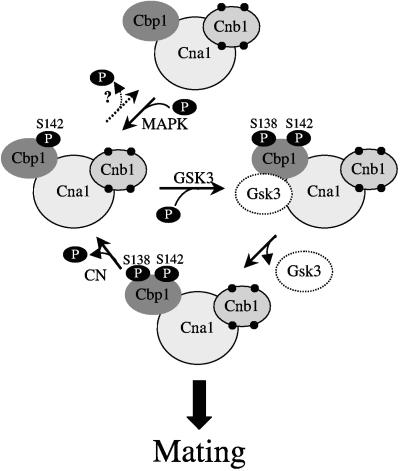
FIG. 9.
Model of calcineurin regulation by Cbp1 to promote mating-specific hyphal elongation. Calcineurin-associated Cbp1 is phosphorylated at serine 142 by mitogen-activated protein kinase, creating a priming site for phosphorylation at serine 138 by GSK-3. Similar to the model for protein phosphatase 1/I-2, GSK-3 associates with the calcineurin/Cbp1 complex and phosphorylates serine 138, leading to the activation of calcineurin function in mating-specific filamentation via substrate targeting of the complex. After phosphorylation of serine 138, GSK-3 loses binding affinity to the calcineurin/Cbp1 complex and dissociates. Calcineurin reverses phosphorylation at serine 138, leading to the return to the inactive state. The specific kinases involved in the phosphorylation of serines 138 and 142, and the putative subcellular targeting compartment and substrate(s) are not yet known. This model incorporates observations of Cbp1 and calcineurin function in C. neoformans and closely resembles the mode of regulation of the mammalian and yeast protein phosphatase 1/I-2(Glc8)/GSK-3 complexes, with significant similarity to the proposed model for regulation of calcineurin by Rcn1 in S. cerevisiae (15, 25, 39, 45).
DISCUSSION
The goals of this study were to examine the role of the calcineurin-binding protein Cbp1 as a modulator of calcineurin function in C. neoformans and to elucidate the importance of the highly conserved serine proline repeat region for Cbp1 function. Cbp1 is a member of a family of proteins conserved from fungi to humans that function as both effectors and regulators of calcineurin signaling pathways. In C. neoformans, calcineurin, a highly conserved serine/threonine-directed phosphatase, is required for filamentation during mating and monokaryotic fruiting, as well as for growth at 37°C and virulence (4, 9, 31).
Our earlier characterization of Cbp1 function was performed in the C. neoformans var. grubii (serotype A) background, whereas the congenic C. neoformans var. neoformans (serotype D) strains provide an optimal system for genetic studies of mating and monokaryotic fruiting. Therefore, to more comprehensively determine the contribution of Cbp1 to calcineurin function, we isolated and disrupted the C. neoformans var. neoformans CBP1 gene. In contrast to calcineurin, Cbp1 is not essential for growth at 37°C or monokaryotic fruiting. However, similar to calcineurin mutants, cbp1 mutants display a bilateral mating defect, indicating that Cbp1 also promotes hyphal elongation during mating. Interestingly, Cbp1 is required for mating, but not for α pheromone-induced morphogenesis. Because a pheromone-responsiveness is important for mating and a pheromone enhances, but is not necessary for, haploid fruiting of α cells, our findings suggest that Cbp1 is a mating-specific effector or governs calcineurin substrate specificity during mating-specific hyphal elongation (16, 40).
Calcineurin and calcium ion gradients play an important role in the regulation of hyphal morphogenesis in many fungi (reviewed in references 10, 18, and 43). In both Neurospora crassa and C. neoformans, calcineurin is required for heterokaryon viability and hyphal elongation following cell fusion, while in S. cerevisiae, calcineurin is required for survival following pheromone-induced growth arrest in haploid cells (4, 30, 35, 50). Recent evidence suggests a role for the calcineurin-dependent modulation of localized vesicle fusion, enzyme activation, and cytoskeletal organization during both hyphal elongation and neurite extension via the regulation of polarized calcium gradients (2, 23, 29, 35, 41). In this study, we demonstrated that Cbp1 functions in a calcineurin-dependent manner to promote the regulation of calcium ion tolerance and hyphal elongation. As localized calcium ion homeostasis is necessary for hyphal elongation, Cbp1 may act to promote hyphal elongation by restricting calcineurin activity to sites of tip growth to promote the establishment and maintenance of calcium ion-responsive hyphal growth during mating. Alternatively, Cbp1 may function to stabilize calcineurin protein levels during calcium signaling conditions (19).
Existing models propose that members of the calcipressin family serve as both activators and inhibitors of calcineurin, similar to the action of inhibitor-2 (I-2) on protein phosphatase 1 (14, 15, 38, 46). To determine the nature of the influence of Cbp1 on calcineurin function in C. neoformans, we performed epistasis analyses with combinations of calcineurin inhibition, and Cbp1 or activated calcineurin overexpression and monitored the influence of each to restore filamentation to mating defective cbp1 and calcineurin deficient strains. Our previous studies demonstrated that calcineurin is required for filamentation in C. neoformans, suggesting a positive role for calcineurin during hyphal elongation (4). Therefore, if Cbp1 negatively regulates filamentation via calcineurin inhibition, the loss of Cbp1 could promote filamentation, and conversely, if Cbp1 promotes the activation of calcineurin, the loss of Cbp1 would abrogate calcineurin-dependent filamentation.
Our analyses revealed that the overexpression of CBP1 failed to restore mating in calcineurin-deficient mating reactions, addition of calcineurin inhibitors did not restore mating in cbp1 mutant strains, even at subinhibitory concentrations which dampen calcineurin activity but preserve minimal mating function, overexpression of wild-type and phospho-mimic (S142E) alleles of Cbp1 restored mating in cbp1 deficient mating reactions, even in the presence of subinhibitory concentrations of FK506, and overexpression of an activated calcineurin catalytic allele restored filamentation in both cbp1 and cna1, but not cnb1, backgrounds. As neither loss nor overexpression of Cbp1 blocked calcineurin-dependent functions during high temperature growth or monokaryotic fruiting, our findings suggest that Cbp1 functions as a targeting subunit or chaperone to specifically promote, and potentially limit, events leading to mating-dependent filamentation.
The specificity of Cbp1 function in calcineurin-dependent mating argues against a general role for calcipressins as simple allosteric regulators of calcineurin, and provides a contrasting opinion to recent models proposed from studies with the Cbp1 functional homolog, Rcn1 (15). In addition, the recent elucidation of the importance of DSCR1/MCIP1 in endothelial cell morphogenesis and the identification of the MEK kinase Raf-1 as a DSCR1/MCIP1 binding partner provide added support for our findings regarding the role of Cbp1 as a calcineurin-dependent targeting subunit (3, 17). Furthermore, unlike Rcn1 and DSCR1/MCIP1, Cbp1 does not stimulate or inhibit calcineurin expression in C. neoformans, and thus does not participate in a feedback loop to indirectly regulate calcineurin activity (11, 15, 19, 51).
One of the most striking features of the calcipressin family of calcineurin binding proteins is that they all share a central highly conserved peptide region that contains a SP repeat (SPPxSPP) implicated in the regulation of calcipressin localization, as well as calcineurin function (12, 15, 24, 34, 36). A variety of studies, including our own, have demonstrated that the two invariant serine residues contained within the SP repeat are phosphorylated, and this occurs first via a priming event on the distal serine (S142) followed by modification of the more N-terminal serine residue (S138) (13, 15, 47). However, there exists some discordance among the current models regarding the role of the phosphorylation status of the calcipressin SP repeat on the regulation of calcineurin activity. While one model supports the activation of calcineurin by calcipressin in its phosphoprotein form, with subsequent inhibition following dephosphorylation and continued occupancy of the active site, another model endorses the inhibition of calcineurin function as a result of calcipressin SP repeat phosphorylation.
These studies provide evidence that the conserved serine residues of the SP repeat motif are critical for both Cbp1 and calcineurin biological function. That neither the S138A nor the S142A mutant protein is functional suggests that phosphorylation of both residues is necessary for activity, in support of the model of calcineurin activation via a SP repeat phosphorylation. That the S138E allele is also nonfunctional suggests that both phosphorylation and dephosphorylation of S138 is involved, whereas the finding that S142E is functionally wild-type supports models in which this residue only requires phosphorylation and not dephosphorylation. Because the overexpression of the dual phospho-mimic S138E+S142E resulted in the loss of Cbp1 protein stability, we speculate that Cbp1 normally exists in this form only transiently and must be bound to calcineurin to maintain stability.
We accommodate these findings in a model (Fig. 9) in which Cbp1 is phosphorylated initially at S142 by a mitogen-activated protein kinase, either before or after binding to calcineurin, facilitating the subsequent phosphorylation of calcineurin-bound Cbp1 at S138 by GSK-3. This cooperative phosphorylation event generates a dually phosphorylated Cbp1 that activates calcineurin function to promote hyphal elongation during mating. The subsequent reversal of phosphorylation at S138 by calcineurin then produces a monophosphorylated species that potentially serves to limit calcineurin activity, either directly by down-regulating calcineurin phosphatase activity, or indirectly via relocation of the complex to alternate sites.
These findings suggest that the phosphorylation state of the SP repeat of Cbp1 does not preclude binding to calcineurin, but is important for Cbp1, as well as calcineurin, function during events leading to mating-specific filamentation. Our studies have addressed the importance of Cbp1 and the phosphorylation status of the SP repeat during mating-specific hyphal elongation in C. neoformans and provide a novel approach to analyze the roles of Cbp1 and other calcipressins as regulators of calcineurin specificity, with applications toward the elucidation of mechanisms for the prevention of calcineurin-associated human disorders.
Acknowledgments
We thank Jill Blankenship, Ping Wang, and Jim Cutler for comments and discussions, Richard Kleinschmidt and James R. Young for technical support, and Peter Kraus for providing the constitutively active CNA1 (CNA1ΔCam-AI) allele construct and unpublished results. We also thank Fujisawa for providing FK506 and members of the C. neoformans genome project at the Stanford Genome Technology Center and Nagasaki University (NIH U01AI47087) and the Institute for Genomic Research (NIH U01AI48594) for sequence data.
This work was supported by NIAID (K22 Career Development Award) AI055302 (to D. Fox) and NIAID R01 grants AI39115, AI42159, and AI50438 (to J. Heitman). Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans, p. 217-238. In J. F. Ernst and A. Schmidt (ed.), Dimorphism in human pathogenic and apathogenic yeasts, vol. 5. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 2.Chang, H. Y., K. Takei, A. M. Sydor, T. Born, F. Rusnak, and D. G. Jay. 1995. Asymmetric retraction of growth cone filopodia following focal inactivation of calcineurin. Nature 376:686-690. [DOI] [PubMed] [Google Scholar]
- 3.Cho, Y. J., M. Abe, S. Y. Kim, and Y. Sato. 2005. Raf-1 is a binding partner of DSCR1. Arch. Biochem. Biophys. 439:121-128. [DOI] [PubMed] [Google Scholar]
- 4.Cruz, M. C., D. S. Fox, and J. Heitman. 2001. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 20:1020-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 6.Degand, I., P. Catty, E. Talla, D. Thines-Sempoux, A. de Kerchove d'Exaerde, A. Goffeau, and M. Ghislain. 1999. Rabbit sarcoplasmic reticulum Ca2+-ATPase replaces yeast PMC1 and PMR1 Ca2+-ATPases for cell viability and calcineurin-dependent regulation of calcium tolerance. Mol. Microbiol. 31:545-556. [DOI] [PubMed] [Google Scholar]
- 7.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell Biol. 10:4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, D. S., G. M. Cox, and J. Heitman. 2003. Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot. Cell 2:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, D. S., M. C. Cruz, R. A. Sia, H. Ke, G. M. Cox, M. E. Cardenas, and J. Heitman. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39:835-849. [DOI] [PubMed] [Google Scholar]
- 10.Fox, D. S., and J. Heitman. 2002. Good fungi gone bad: the corruption of calcineurin. Bioessays 24:894-903. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes, J. J., L. Genesca, T. J. Kingsbury, K. W. Cunningham, M. Perez-Riba, X. Estivill, and S. de la Luna. 2000. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Genet. 9:1681-1690. [DOI] [PubMed] [Google Scholar]
- 12.Genesca, L., A. Aubareda, J. J. Fuentes, X. Estivill, S. De La Luna, and M. Perez-Riba. 2003. Phosphorylation of calcipressin 1 increases its ability to inhibit calcineurin and decreases calcipressin half-life. Biochem. J. 374:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görlach, J., D. S. Fox, N. S. Cutler, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilioti, Z., and K. W. Cunningham. 2003. The RCN family of calcineurin regulators. Biochem. Biophys. Res. Commun. 311:1089-1093. [DOI] [PubMed] [Google Scholar]
- 15.Hilioti, Z., D. A. Gallagher, S. T. Low-Nam, P. Ramaswamy, P. Gajer, T. J. Kingsbury, C. J. Birchwood, A. Levchenko, and K. W. Cunningham. 2004. GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev. 18:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsueh, Y. P., and W. C. Shen. 2005. A homolog of Ste6, the a-factor transporter in Saccharomyces cerevisiae, is required for mating but not for monokaryotic fruiting in Cryptococcus neoformans. Eukaryot. Cell 4:147-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka, M., M. Abe, K. Shiiba, I. Sasaki, and Y. Sato. 2004. Down syndrome candidate region 1, a downstream target of VEGF, participates in endothelial cell migration and angiogenesis. J. Vasc. Res. 41:334-344. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, S. L., and I. B. Heath. 1993. Roles of calcium ions in hyphal tip growth. Microbiol. Rev. 57:367-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsbury, T. J., and K. W. Cunningham. 2000. A conserved family of calcineurin regulators. Genes Dev. 14:1595-1604. [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus, P. R., D. S. Fox, G. M. Cox, and J. Heitman. 2003. The Cryptococcus neoformans mitogen-activated protein kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus, P. R., C. Nichols, and J. Heitman. 2005. Calcium- and calcineurin-independent roles for calmodulin in Cryptococcus neoformans morphogenesis and high-temperature growth. Eukaryot. Cell 4:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821-833. [PubMed] [Google Scholar]
- 23.Lautermilch, N. J., and N. C. Spitzer. 2000. Regulation of calcineurin by growth cone calcium waves controls neurite extension. J. Neurosci. 20:315-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, H. Y., H. J. Michtalik, S. Zhang, T. T. Andersen, D. A. Van Riper, K. K. Davies, G. Ermak, L. M. Petti, S. Nachod, A. V. Narayan, N. Bhatt, and D. R. Crawford. 2003. Oxidative and calcium stress regulate DSCR1 (Adapt78/MCIP1) protein. Free Radic. Biol. Med. 35:528-539. [DOI] [PubMed] [Google Scholar]
- 25.Lin, T. H., Y. C. Chen, C. L. Chyan, L. H. Tsay, T. C. Tang, H. H. Jeng, F. M. Lin, and H. B. Huang. 2003. Phosphorylation by glycogen synthase kinase of inhibitor-2 does not change its structure in free state. FEBS Lett. 554:253-256. [DOI] [PubMed] [Google Scholar]
- 26.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J. O. 2003. Endogenous protein inhibitors of calcineurin. Biochem. Biophys. Res. Commun. 311:1103-1109. [DOI] [PubMed] [Google Scholar]
- 28.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 29.Meberg, P. J., S. Ono, L. S. Minamide, M. Takahashi, and J. R. Bamburg. 1998. Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell Motil. Cytoskeleton 39:172-190. [DOI] [PubMed] [Google Scholar]
- 30.Moser, M. J., J. R. Geiser, and T. N. Davis. 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16:4824-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parry, R. V., and C. H. June. 2003. Calcium-independent calcineurin regulation. Nat. Immunol. 4:821-823. [DOI] [PubMed] [Google Scholar]
- 33.Parsons, J. N., G. J. Wiederrecht, S. Salowe, J. J. Burbaum, L. L. Rokosz, R. L. Kincaid, and S. J. O'Keefe. 1994. Regulation of calcineurin phosphatase activity and interaction with the FK506-FKBP12 binding protein complex. J. Biol. Chem. 269:19610-19616. [PubMed] [Google Scholar]
- 34.Pfister, S. C., G. M. Machado-Santelli, S. W. Han, and F. Henrique-Silva. 2002. Mutational analyses of the signals involved in the subcellular location of DSCR1. BMC Cell Biol. 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prokisch, H., O. Yarden, M. Dieminger, M. Tropschug, and I. B. Barthelmess. 1997. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet. 256:104-114. [DOI] [PubMed] [Google Scholar]
- 36.Rothermel, B., R. B. Vega, J. Yang, H. Wu, R. Bassel-Duby, and R. S. Williams. 2000. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 275:8719-8725. [DOI] [PubMed] [Google Scholar]
- 37.Rothermel, B. A., R. B. Vega, and R. S. Williams. 2003. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc. Med. 13:15-21. [DOI] [PubMed] [Google Scholar]
- 38.Ryeom, S., R. J. Greenwald, A. H. Sharpe, and F. McKeon. 2003. The threshold pattern of calcineurin-dependent gene expression is altered by loss of the endogenous inhibitor calcipressin. Nat. Immunol. 4:874-881. [DOI] [PubMed] [Google Scholar]
- 39.Sakashita, G., H. Shima, M. Komatsu, T. Urano, A. Kikuchi, and K. Kikuchi. 2003. Regulation of type 1 protein phosphatase/inhibitor-2 complex by glycogen synthase kinase-3beta in intact cells. J. Biochem. (Tokyo) 133:165-171. [DOI] [PubMed] [Google Scholar]
- 40.Shen, W. C., R. C. Davidson, G. M. Cox, and J. Heitman. 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1:366-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman-Gavrila, L. B., and R. R. Lew. 2003. Calcium gradient dependence of Neurospora crassa hyphal growth. Microbiology 149:2475-2485. [DOI] [PubMed] [Google Scholar]
- 42.Tanida, I., A. Hasegawa, H. Iida, Y. Ohya, and Y. Anraku. 1995. Cooperation of calcineurin and vacuolar H+-ATPase in intracellular Ca2+ homeostasis of yeast cells. J. Biol. Chem. 270:10113-10119. [DOI] [PubMed] [Google Scholar]
- 43.Torralba, S., and I. B. Heath. 2001. Cytoskeletal and Ca2+ regulation of hyphal tip growth and initiation. Curr. Top. Dev. Biol. 51:135-187. [DOI] [PubMed] [Google Scholar]
- 44.Tscharke, R. L., M. Lazera, Y. C. Chang, B. L. Wickes, and K. J. Kwon-Chung. 2003. Haploid fruiting in Cryptococcus neoformans is not mating type alpha-specific. Fungal Genet. Biol. 39:230-237. [DOI] [PubMed] [Google Scholar]
- 45.Tung, H. Y., W. Wang, and C. S. Chan. 1995. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell. Biol. 15:6064-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vega, R. B., B. A. Rothermel, C. J. Weinheimer, A. Kovacs, R. H. Naseem, R. Bassel-Duby, R. S. Williams, and E. N. Olson. 2003. Dual roles of modulatory calcineurin-interacting protein 1 in cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 100:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vega, R. B., J. Yang, B. A. Rothermel, R. Bassel-Duby, and R. S. Williams. 2002. Multiple domains of MCIP1 contribute to inhibition of calcineurin activity. J. Biol. Chem. 277:30401-30407. [DOI] [PubMed] [Google Scholar]
- 48.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, R. S., and P. Rosenberg. 2002. Calcium-dependent gene regulation in myocyte hypertrophy and remodeling. Cold Spring Harb. Symp. Quant. Biol. 67:339-344. [DOI] [PubMed] [Google Scholar]
- 50.Withee, J. L., J. Mulholland, R. Jeng, and M. S. Cyert. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, J., B. Rothermel, R. B. Vega, N. Frey, T. A. McKinsey, E. N. Olson, R. Bassel-Duby, and R. S. Williams. 2000. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 87:E61-68. [DOI] [PubMed] [Google Scholar]