Abstract
Chromatin disassembly and reassembly, mediated by histone chaperones such as anti-silencing function 1 (Asf1), are likely to accompany all nuclear processes that occur on the DNA template. In order to gain insight into the functional conservation of Asf1 across eukaryotes, we have replaced the budding yeast Asf1 protein with Drosophila Asf1 (dAsf1) or either of the two human Asf1 (hAsf1a and hAsf1b) counterparts. We found that hAsf1b is best able to rescue the growth defect of Saccharomyces cerevisiae lacking Asf1. Moreover, dAsf1 and hAsf1b but not hAsf1a can replace the role of yeast Asf1 in protecting against replicational stress and activating the PHO5 gene, while only hAsf1a can replace the role of Asf1 in protecting against double-stranded-DNA-damaging agents. Furthermore, it appears that the interaction between Asf1 and the DNA damage checkpoint protein Rad53 is not required for Asf1's role in maintaining genomic integrity. In addition to indicating the functional conservation of the Asf1 proteins across species, these studies suggest distinct roles for the two human Asf1 proteins.
Chromatin is made up of a basic repeating unit called the nucleosome. The nucleosome consists of approximately 147 base pairs of DNA wrapped around a histone octamer comprising two molecules each of histones H2A, H2B, H3, and H4 (12, 13). The deposition of histone proteins onto the DNA to form chromatin mainly occurs following DNA replication. This chromatin assembly process is mediated (at least in vitro) by histone H3-H4 chaperones, including chromatin assembly factor 1 (CAF-1) and anti-silencing function 1 (Asf1) (30). The role of Asf1-mediated chromatin assembly in cellular processes is not limited to DNA replication, as budding yeast deleted for ASF1 have phenotypes that implicate Asf1 in transcriptional silencing, transcriptional regulation, protection against DNA-damaging agents, genomic instability, normal cell cycle progression, and DNA replication (1, 2, 5, 8-10, 14, 19, 20, 22-25, 28, 30). Although Asf1 is highly conserved through evolution, it is unclear whether Asf1 from higher eukaryotes also plays a role in these processes.
Asf1 functions as an H3-H4 histone chaperone to regulate transcription. Saccharomyces cerevisiae Asf1 was originally identified by its ability to derepress transcriptional silencing when overexpressed (10, 27). Transcriptional silencing is mediated by a specialized repressive chromatin structure akin to heterochromatin in higher eukaryotes. While deletion of ASF1 results in a nominal decrease in transcriptional silencing (10, 27), this defect is exacerbated by inactivation of CAF-1, suggesting a role for Asf1 in silencing (30). In addition to a role in transcriptional silencing, Asf1 has also been shown to have a direct role in transcriptional activation. For example, degradation of Asf1 results in the immediate deregulation of many genes without the requirement for passage through S phase (33). As such, the transcriptional role of Asf1 is independent of its role during DNA replication-coupled chromatin assembly. A molecular mechanism by which Asf1 may directly influence gene expression has become evident at the yeast PHO5 and PHO8 promoters, where Asf1-mediated nucleosome disassembly is required for transcriptional activation of these genes (1).
Asf1 also appears to play an important, although poorly understood, role in maintaining genomic stability. For example, yeast cells deleted for Asf1 grow slowly, due to activation of the DNA damage checkpoint as a result of an increased level of spontaneous DNA damage (24). Furthermore, Asf1 mutants have an increased rate of genomic instability (19, 22, 24). Spontaneous DNA damage and genomic instability are usually the result of problems during DNA replication, suggesting that Asf1-mediated chromatin assembly influences DNA replication. Yeast cells deleted for ASF1 are also highly sensitive to DNA-damaging agents (31) and it is possible that Asf1 assembles and/or disassembles chromatin during DNA repair.
Asf1 has also been implicated as a target of the DNA damage checkpoint response. Specifically, Asf1 binds to the yeast DNA damage checkpoint kinase Rad53 (5, 8). Upon activation of the DNA damage checkpoint, Rad53 is phosphorylated, causing Asf1 to be released and allowing it to bind histones (5, 8). This dynamic interaction may activate Asf1 to bring histones to DNA damage in order to repair the chromatin structure.
While most of the cellular processes affected by Asf1 have been discovered in yeast, it is unclear what role Asf1 plays in higher eukaryotes. Drosophila melanogaster Asf1 has been shown to be essential and also appears to influence chromatin structure, as measured by transcriptional assays of heterochromatin structure in flies (16). The human homologs of Asf1, hAsf1a and hAsf1b, both bind to replication-specific and transcription-specific histone variants, H3.1 and H3.3, respectively, suggesting that both human Asf1 proteins may also assemble/disassemble chromatin during DNA replication and transcription (29). Indeed, RNA interference depletion of human Asf1a and Asf1b results in an elongated S phase, suggesting a role for human Asf1 proteins during DNA replication (7). It is important to functionally compare higher eukaryotic and yeast Asf1 in order to gain insight into the role of Asf1 in Drosophila and humans.
In order to determine whether Asf1 is functionally conserved across eukaryotic species and to examine whether human Asf1a and Asf1b have distinct functions, we tested the ability of the Drosophila and human Asf1 proteins to functionally replace S. cerevisiae Asf1. Using genetic and biochemical approaches, we show that there is remarkable conservation and interesting distinctions between the functions and protein-protein interactions mediated by the yeast, Drosophila, and human Asf1 proteins that provide novel insight into the function of Asf1 in higher eukaryotes. Indeed, we find that the functions of the yeast Asf1 protein have been divided between the two human Asf1 proteins, with hAsf1a playing a primary role in DNA repair and hAsf1b contributing to genomic stability during DNA replication, transcription, and replication.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used were as follows. ACN026, MATα ade2-1 leu2-3,112, lys5, ura3-52, ade3::Gal10::HO ΔHO::Ade1, Δhml::ADE1 Δhmr::ade1 ASF1-13myc:KAN (this study); BAT011, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1::LEU2 TEVIIL::URA HMRa::ADE2 asf1::hAsf1b-13myc KAN (this study); BAT013, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1::LEU2 TEVIIL::URA HMRa::ADE2 asf1::dAsf1-13myc KAN (this study); BAT014, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 TEVIIL::URA HMRa::ADE2 asf1::dAsf1-13myc KAN (this study); BAT015, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 cac1::LEU2 TEVIIL::URA HMRa::ADE2 asf1::hAsf1a-13myc KAN (this study); BAT016, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 TEVIIL::URA HMRa::ADE2 asf1::hAsf1a-13myc KAN (this study); JKT0010, MATa his3-11 leu2-3,112 lys2 trp-1 ura3-1 bar1::LEU2 (1); JKT0049, MATα ade2-1 his3-11 leu2-3,112 trp1-1 ura301 cac1::LEU2 TELVIIL::URA3 (this study); KDY006, MATα ade2-1 leu2-3,112 his3-11 trp1-1 ura3-1 TEVIIL::URA HMRa::ADE2 asf1::hAsf1b-13myc KAN (this study); MPY0042, MATa his3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1:::LEU2 asf1::KAN (1); ROY1169, MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 cac1::LEU2 asf1::his5+ TELVIIL::URA3 HMRa::ADE2 can1-100 (30); ROY1170, MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 asf1::his5+ TELVIIL::URA3 HMRa::ADE2 can1-100 (30); ROY1171, MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 cac1::LEU2 TELVIIL::URA3 HMRa::ADE2 can1-100 (30); and ROY1172, MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 TELVIIL::URA3 HMRa::ADE2 can1-100 (30).
Immunoprecipitation.
Immunoprecipitation of proteins was performed as described previously (11) with the following alterations. Yeast cells were grown to an optical density at 600 nm (OD600) of 1.0 to 1.5. Cells were washed twice with phosphate-buffered saline (PBS). One half of the culture was then incubated for 30 min at room temperature with 3 mM of the thiol-cleavable homobifunctional cross-linker dithiobis(succinimidyl propionate) (DSP) in PBS made from a 0.1 M stock diluted in dimethyl sulfoxide (4). The other half of the culture was left untreated.
Cells were washed with PBS and then resuspended in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.2% Triton X-100, phenylmethylsulfonyl fluoride, benzamidine, aprotinin, leupeptin) at 4°C. Total protein was measured and equivalent amounts of protein were immunoprecipitated for each sample, between 6 mg/ml and 8 mg/ml. Asf1-Myc was immunoprecipitated using a 1:50 dilution of anti-rabbit polyclonal c-Myc (A-14) (Santa Cruz Biotechnology). The anti-Myc antibody was captured on protein A-Sepharose beads, which was washed in lysis buffer and then the beads were resuspended in 20 μl of protein sample buffer, boiled, and 10 μl was resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and detected by Western blot analysis. Nitrocellulose membranes were blocked in 3% milk in Tris-buffered saline (TBS) plus 0.05% Tween 20 and primary antibodies H3AcK9/14 (1:500) (Upstate), Rad53 (1:100) (yC-19) (Santa Cruz Biotechnology), and c-Myc (1:100) were incubated in 3% milk in TBS plus 0.05% Tween 20 overnight and washed with TBS plus 0.05% Tween 20. Protein was detected using anti-rabbit or anti-goat secondary antibodies conjugated to horseradish peroxidase and ECL reagents (Amersham Biosciences). Each immunoprecipitation was repeated at least 3 independent times with similar results.
DNA damage sensitivity and silencing assays.
Yeast strains were grown to log phase and adjusted to an OD600 of 1.0 and plated in 10-fold serial dilutions onto YEPD or YEPD containing hydroxyurea, bleomycin, camptothecin, methane methyl sulfonate, 5′fluoroorotic acid, or low adenine. Yeast were grown for 2 to 3 days at 30°C on plates.
Growth analysis.
Yeast cultures were grown to log phase and diluted to OD600 of 0.1 and optical density was measured every hour and graphed using Excel. Strains were streaked onto YEPD to visualize colony size compared to the wild type.
Phosphatase assay.
The phosphatase assay was performed as described previously (1).
RESULTS
Human Asf1b best compensates for the growth defect of yeast deleted for ASF1.
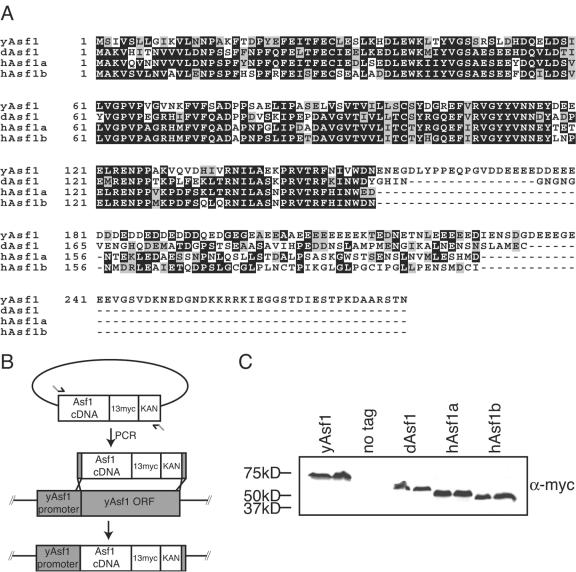
We sought to gain insight into the functional conservation of the Asf1 proteins across eukaryotes and the potential functional differences between the two human Asf1 proteins. In contrast to the single Asf1 protein in budding yeast and Drosophila, there are two forms of Asf1 in humans, hAsf1a and hAsf1b. The sequence of the C terminus of Asf1 is divergent across species (Fig. 1A). The C terminus is dispensable for the role of Asf1 in silencing, resistance to DNA-damaging agents and replicational stress, interaction with histones, Rad53 and HIRA, and Asf1's ability to deposit histones onto DNA in vitro (3, 32). The conserved N-terminal region of Drosophila Asf1 is 59% identical to the yeast protein, while hAsf1a and hAsf1b are 57% and 59% identical to yeast Asf1, respectively, and are 84% identical to each other.
FIG. 1.
A. Alignment of yeast, Drosophila, and human Asf1 proteins, showing conserved and nonconserved regions of the Asf1 gene, where black represents identical residues and gray represents conserved residues. B. Schematic of insertion of Asf1 homologs into yeast. PCR fragments carrying the open reading frames of human and Drosophila Asf1 were inserted by homologous recombination downstream of the endogenous yeast ASF1 promoter. Yeast DNA is represented by gray boxes and homolog DNA is represented by white boxes. A C-terminal 13-Myc epitope tag and kanamycin resistance gene were included for detection and selection on medium, respectively. C. Equivalent expression of the yeast, Drosophila, and human Asf1 proteins in yeast; 6 μg of total protein extracts from strains ACN026 (yAsf1), ROY1172 (no tag), BAT014 (dAsf1), BAT016 (hAsf1a), and KDY006 (hAsf1b) were loaded on a 7.5% SDS gel and Western blotted using a Myc antibody.
Due to the similarities between the Asf1 proteins, we asked whether the Drosophila or human Asf1 proteins could functionally compensate for the deletion of budding yeast ASF1. We integrated the Drosophila gene (dASF1) and the human gene (hASF1a and hASF1b) open reading frames in place of the endogenous yeast ASF1 open reading frame (Fig. 1B). In this manner, the Drosophila or either human Asf1 protein was the sole Asf1 protein within the cell and under the transcriptional control of the yeast ASF1 promoter. A C-terminal 13-Myc epitope was included to verify equal expression levels of the yeast, Drosophila, and human Asf1 proteins (Fig. 1C).
In order to compare the functions of Asf1 proteins across eukaryotes, we tested the ability of the Drosophila and human Asf1 homologs to replace yeast Asf1 for growth. Yeast deleted for ASF1 grow slowly due to an elongated metaphase of the cell cycle (24). This metaphase elongation is due to activation of the DNA damage checkpoint in response to an elevated level of spontaneous DNA damage during DNA replication (24). We compared the growth of wild-type yeast, yeast deleted for ASF1, and those containing only hAsf1a, hAsf1b, or dAsf1. We found that human Asf1b best complemented the slow growth rate of yeast deleted for ASF1, while hAsf1a and dAsf1 only partially complemented the growth rate (Fig. 2A). In support of this, the colony size for yeast containing hAsf1b was closest to that of wild-type Asf1 yeast, while colonies of yeast strains containing hAsf1a and dAsf1 were only slightly bigger than yeast deleted for ASF1 (Fig. 2B). Furthermore, yeast containing hAsf1b accumulated in the G2/M phase of the cell cycle to a much lesser degree than yeast deleted for ASF1 or yeast containing hAsf1a or dAsf1, as determined by flow cytometry analysis (data not shown). These results indicate that hAsf1b can mimic the role of yeast Asf1 in preventing slow growth, suggesting that hAsf1b is more functionally similar to yeast Asf1.
FIG. 2.
hAsf1b best complements the growth defect of yeast deleted for ASF1. A. Exponentially growing cultures of strains JKT049 (yAsf1), BAT015 (hAsf1a), BAT011 (hAsf1b), BAT013 (dAsf1), and ROY1169 (asf1Δ) were diluted to an OD of 0.1, and OD readings were taken over time and plotted. B. The same strains used in panel A were plated and grown for 3 days before images were photographed.
Drosophila Asf1 and human Asf1b protect against replicational stress, while human Asf1a protects against double-strand DNA-damaging agents.
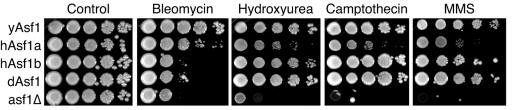
In order to further examine the role of the Asf1 homologs in maintaining genomic integrity, we tested their ability to provide resistance to agents that induce DNA damage and replicational stress. Yeast lacking ASF1 are highly sensitive to the radiomimetic drug bleomycin, which induces double-strand DNA damage (5, 30). We found that human Asf1a, but not human Asf1b or Drosophila Asf1, is able to restore resistance to bleomycin (Fig. 3).
FIG. 3.
Ability of homologs of Asf1 to provide resistance to DNA-damaging agents. Exponentially growing cultures of yeast strains ROY1172 (yAsf1), BAT016 (hAsf1a), KDY006 (hAsf1b), BAT014 (dAsf1), and ROY1170 (asf1Δ) were plated in 10-fold serial dilutions on rich medium (control), rich medium plus 1.5 mU bleomycin, 100 mM hydroxyurea, 10 μM camptothecin, and 0.01% methyl methane sulfonate and grown for 2 to 3 days at 30°C.
Yeast lacking ASF1 are also highly sensitive to agents that induce DNA damage during DNA replication, such as the DNA-alkylating agent methyl methane sulfonate, the topoisomerase I religation inhibitor camptothecin, and hydroxyurea, which results in replicational stalling (10, 24, 27, 30). We found that human Asf1b and Drosophila Asf1 are able to fully restore resistance to hydroxyurea, camptothecin, and methyl methane sulfonate, while human Asf1a only partially restores resistance to these agents (Fig. 3). The sensitivity of strains carrying the homologs of Asf1 to DNA-damaging agents was not a result of changes in Asf1 transcription, because protein levels of epitope-tagged yeast Asf1 do not change upon treatment of DNA-damaging agents (data not shown) and all the homologs were under the control of the yeast Asf1 transcriptional regulatory sequences.
These results allow us to separate the functions of the human Asf1 proteins, where hAsf1a functions in maintaining viability in response to double-strand DNA damage, and hAsf1b function is more important during DNA replication. We suggest that hAsf1a contributes to the repair of DNA damage in human cells by disassembling and reassembling the chromatin structure at a DNA break, while hAsf1b acts in disassembling and reassembling histones onto DNA at replication forks. This is an interesting divergence of species, since there is only one copy of yeast Asf1 to perform both of these functions. Furthermore, Drosophila Asf1 appears to be more functionally similar to human Asf1b than Asf1a during the maintenance of genomic integrity in response to exogenous DNA-damaging agents and replicational stress.
Human Asf1a and Asf1b and Drosophila Asf1 partially complement the role of yeast Asf1 in transcriptional silencing.
In order to gain further insight into the function of the human and Drosophila Asf1 proteins, we examined whether they could mediate transcriptional silencing in yeast. Asf1 contributes to transcriptional silencing at the yeast telomeres and the HMR and HML silent mating type loci (30). We used a URA3 marker inserted proximal to the telomere of chromosome VII and an ADE2 marker inserted into the HMR locus in order to measure silencing by the different homologs of Asf1. These experiments were performed in yeast deleted for the CAC1 gene, encoding a subunit of the chromatin assembly factor CAF-1 complex, as this deletion allows the role of Asf1 in silencing to be measured (30). Silencing of the telomere-proximal URA3 gene is measured by growth on the drug 5′ fluoroorotic acid (5′FOA), where wild-type silencing of URA3 allows growth on 5′FOA and a silencing defect causes death on 5′FOA. We found that hAsf1b and dAsf1 allowed more growth on 5′FOA than yeast deleted for ASF1 or yeast containing hAsf1a (Fig. 4). This result indicates that hAsf1b and dAsf1 can best mimic the role of yeast Asf1 for telomeric silencing, suggesting that hAsf1b may play a more prominent role in heterochromatin formation in human cells than hAsf1a.
FIG. 4.
Ability of homologs of Asf1 to mediate transcriptional silencing. Yeast strains ROY1171 (yAsf1), BAT015 (hAsf1a), BAT011 (hAsf1b), BAT013 (dAsf1), and ROY1169 (asf1Δ) were plated in 10-fold serial dilutions onto rich medium, medium containing the indicated amounts of 5′FOA per 250 ml, or medium containing low adenine and grown for 2 to 3 days at 30°C.
Silencing of the HMR-inserted ADE2 gene is measured by colony color, as silencing of ADE2 leads to the build-up of a red pigment from the adenine biosynthesis pathway, yielding red colonies. A silencing defect at HMR::ADE2 results in white colonies, as seen for the yeast deleted for ASF1 (Fig. 4). hAsf1a, hAsf1b, and dAsf1 yielded colonies that were more pink than yeast deleted for ASF1 but less pink than colonies containing yeast Asf1 (Fig. 4). Slight variations in the colony color were observed in the strains containing hAsf1a, hAsf1b, and dAsf1, presumably due to the defect in maintenance of transcriptional silencing that results from the deletion of CAC1 in these strains (15). This result shows that hAsf1a, hAsf1b, and dAsf1 can partially mimic the role of yeast Asf1 for silencing of the mating type locus. Taken together, these data indicate that the human and Drosophila counterparts of Asf1 can partially mimic the silencing function of yeast Asf1, although this appears to be in a locus-specific manner for hAsf1a.
Human Asf1b and dAsf1 complement the role of yeast Asf1 in activation of the PHO5 gene.
Next, we tested the ability of the human and Drosophila Asf1 homologs to activate the PHO5 gene. Asf1-mediated disassembly of the nucleosomes over the PHO5 promoter is required for activation of the PHO5 gene (1). We assayed the ability of the Asf1 homologs to activate PHO5 by measuring the acid phosphatase activity of the product of the PHO5 gene (1). We found that while hAsf1b and dAsf1 were able to activate the PHO5 gene to the same level as yeast Asf1, there was a slight delay in PHO5 activation (Fig. 5). By contrast, hAsf1a was unable to activate the PHO5 gene (Fig. 5). The defect in PHO5 activation in strains carrying the homologs of Asf1 was not a result of changes in Asf1 transcription, because protein levels of epitope-tagged yeast Asf1 do not change in different phosphate media (data not shown) and all the homologs were under the control of the yeast Asf1 transcriptional regulatory sequences. These data show that hAsf1b and dAsf1 but not hAsf1a are able to replace yeast Asf1 for its role in activation of the PHO5 promoter. Activation of the PHO5 gene is due to a direct role of Asf1 in chromatin disassembly at the promoter (1), suggesting, because hAsf1b but not hAsf1a can activate PHO5, that human Asf1b but not hAsf1a is likely to be directly involved in transcriptional regulation in human cells.
FIG. 5.
Ability of homologs of Asf1 to activate PHO5. Yeast strains JKT010 (yAsf1), BAT014 (dAsf1), BAT016 (hAsf1a), KDY006 (hAsf1b), and MPY0042 (asf1Δ) were grown overnight, diluted in phosphate-depleted medium to OD600 of 0.25, and phosphatase activity was measured at the time points indicated.
Human and Drosophila counterparts of yeast Asf1 have different histone-binding abilities.
To further increase our understanding of the functional conservation of the Asf1 proteins in yeast, humans, and Drosophila, we asked whether the Drosophila and human Asf1 proteins could interact with some of the binding partners of yeast Asf1. Asf1 binds to histone H3 (30), and presumably this interaction is essential for its histone chaperone function during chromatin assembly and disassembly. We measured the ability of the Asf1 homologs to bind endogenous yeast histones by coimmunoprecipitation analysis. We were barely able to detect the interaction between the endogenous yeast Asf1 protein and endogenous histone H3 by immunoprecipitation analysis (Fig. 6). Therefore, we treated cells with the 12-angstrom cross-linker dithiobis(succinimidyl propionate) prior to immunoprecipitation to stabilize existing protein-protein interactions (4).
FIG. 6.
Asf1 homologs bind differentially to yeast histone H3 and Rad53. Yeast strains ROY1172 (no tag), ACN026 (yAsf1), BAT014 (dAsf1), BAT016 (hAsf1a), and KDY006 (hAsf1b) were grown to OD600 of ∼1.0, lysed, and immunoprecipitated with a Myc antibody. Equal amounts of protein were loaded onto an SDS-PAGE gel and Western blotted for either Myc, acetylated lysine 9/14 of H3 or Rad53. All soluble histones are modified on H3 lysine 9, and therefore the signal with the acetylated lysine 9/14 antibody reflects the amount of soluble histones. The presence (+) or absence (−) of treatment with the DSP cross-linker is indicated. Equal amounts of Asf1, H3K9/14, and Rad53 were detected in the input samples from all strains (data not shown).
Addition of the cross-linker enabled detection of coprecipitating histone H3 with yeast Asf1 (Fig. 6). By contrast, interaction between human Asf1a and yeast histones was apparent even without the cross-linker, indicating that the interaction between hAsf1a and yeast histones is more stable than the interaction between yeast Asf1 and histones. The interaction between hAsf1b and yeast histones appeared to be less stable than the interaction between yeast Asf1 and histones, since less histone H3 coprecipitated with hAsf1b compared to yAsf1 in the presence of cross-linker (Fig. 6). The interaction between dAsf1 and yeast histones appeared to be the least stable, as coprecipitating histone H3 was not detectable even with the cross-linker. It is important to note that the absence of detectable coprecipitating histone H3 is very unlikely to indicate that the Asf1 protein does not interact with histone H3, but is rather a reflection of the relative stability of the Asf1-H3 interaction during the process of protein extraction and immunoprecipitation.
We used anti-acetylated histone H3 antibodies for this experiment, as they were more sensitive than anti-H3 antibodies. The different affinities of the Asf1 homologs for histones are unlikely to reflect different affinities for acetylated histones, as Asf1 binds to the C terminus of histone H3, not the N terminus (18). These data show that the relative order of affinity of the Asf1 proteins for the yeast histones, from highest to lowest, is hAsf1a, yAsf1, hAsf1b, dAsf1.
Human and Drosophila counterparts of yeast Asf1 fail to bind Rad53.
The interaction between Asf1 and the DNA checkpoint protein Rad53 may be important for the role of Asf1 in protecting against genomic instability (5, 8). Using coimmunoprecipitation analysis, we were able to detect the interaction between yeast Asf1 and Rad53 both with and without the DSP cross-linker (Fig. 6). By contrast, neither hAsf1a, hAsf1b, nor dAsf1 detectably bound Rad53 in this assay. Given that hAsf1a can functionally replace the role of yeast Asf1 for protection from DNA-damaging agents and that hAsf1b and dAsf1 can functionally replace the role of yeast Asf1 during replicational stress (Fig. 3), these Rad53 binding results indicate that the interaction between Asf1 and Rad53 is unlikely to be important for the function of Asf1 in protecting against genomic insults and replicational stress.
DISCUSSION
Mechanistic insight into the function of Asf1 has come largely from studies in yeast. Showing that Drosophila Asf1 and the two human Asf1 proteins can function in yeast demonstrates that the function of Asf1 is highly similar in higher eukaryotes and yeast. The eukaryotic Asf1 proteins have been highly conserved through evolution, with Drosophila and the two human Asf1 proteins being able to perform different aspects of the function of budding yeast Asf1. Furthermore, although the two human Asf1 proteins are highly similar in sequence, they are likely to have different functions within human cells because they complement distinct functions of yeast Asf1.
We conclude from our data that hAsf1a and hAsf1b have unique functions. Indeed, it appears that the functions of yeast Asf1 have been divided between the two human Asf1 proteins. Human Asf1b can replace yeast Asf1's role in growth (likely a consequence of Asf1 protecting against endogenous genomic instability during DNA replication) (24), protection against exogenous insults during DNA replication and PHO5 activation, while hAsf1a can replace yeast Asf1's role in protecting against double-strand DNA-damaging agents. Consistent with our observed role for hAsf1b but not hAsf1a during replication, quiescent human cells (nonreplicating) contain hAsf1a but have no detectable hAsf1b (21). Furthermore, human Asf1b partially compensates for the function of yeast Asf1 in transcriptional silencing at the telomere and HMR locus, while hAsf1a partially compensates for the function of yeast Asf1 in transcriptional silencing only at the HMR locus. The molecular role that Asf1 plays in each of these processes has yet to be determined.
Asf1 is required to mediate H3 and H4 removal from the promoter of the PHO5 gene to allow PHO5 activation (1). Although it has yet to be proven, the functions of Asf1 during DNA replication, silencing, repair, and cell growth are also likely to reflect Asf1-mediated histone H3-H4 disassembly and/or reassembly accompanying DNA replication and DNA repair. The functional distinctions between the two human Asf1 proteins in yeast are likely to indicate that they perform distinct functions in human cells.
We propose that hAsf1a assembles and disassembles chromatin during DNA repair, while hAsf1b assembles and disassembles chromatin during DNA replication in human cells. Presumably, these specialized functions of the human Asf1 proteins would be mediated through differential protein-protein interactions. Yeast Asf1 has recently been shown to be recruited to DNA by replication protein C (RFC) (6), and it would be insightful to determine whether hAsf1b but not hAsf1a can bind to human RFC. Furthermore, given that we have recently localized Drosophila Asf1 to DNA replication foci (Schulz and Tyler, submitted), it will be interesting to determine whether human Asf1b but not Asf1a localizes to DNA replication foci in human cells.
Drosophila Asf1 is more functionally similar to hAsf1b (and yeast Asf1) than hAsf1a, as determined by their abilities to complement deletion of ASF1 in yeast. This would have not been predicted from the sequence identity between the proteins alone, because the identity between the conserved regions of hAsf1b and dAsf1 is 71%, while that between the conserved regions of hAsf1a and dAsf1 is 75%. Furthermore, analysis of the protein sequences does not identify regions of Asf1 that could account for the functional similarities between hAsf1b and dAsf1, as there are only three residues (82S, 86E, and 120P) that are conserved between dAsf1 and hAsf1b but absent from hAsf1a. It is conceivable that these residues are important for mediating differential protein interactions between hAsf1a and hAsf1b, but these residues do not map to any of the known interaction interfaces (3, 17). Furthermore, there is only one residue (154D) that is present in hAsf1b, dAsf1, and yeast Asf1 but absent from hAsf1a (154E). It is unlikely that this one residue is responsible for the functional similarity between dAsf1, yAsf1, and hAsf1b and the many functional differences to hAsf1a. The recent nuclear magnetic resonance structure of human Asf1a showed several loops that were shifted relative to their positions in the yeast Asf1 crystal structure (3, 17). Although the hAsf1b structure is not yet solved, it is possible that these loops dictate the functional specificity of the human Asf1 proteins by mediating differential protein-protein interactions.
The ability of the human and Drosophila Asf1 proteins to function in yeast indicates a striking degree of conservation of the Asf1 proteins through evolution. This functional complementation demonstrates not only that the functions of the Asf1 proteins are similar across eukarotyes, but also that the human and Drosophila proteins can interact with the yeast counterparts of their binding partners. As such, the regions of the Asf1 proteins that mediate these protein-protein interactions must fall within the highly conserved regions of Asf1 (Fig. 1A) and the binding partners of Asf1 must also have been highly conserved through evolution.
It is noteworthy that human Asf1a and Asf1b and Drosophila Asf1 have quite different affinities for the yeast histones, with hAsf1a very strongly binding and hAsf1b and dAsf1 weakly binding yeast histones. This does not necessarily reflect the affinities of the Asf1 proteins for the histones from their own species, but may reflect diversification of the histone binding region of the Asf1 proteins through evolution. However, the differential affinity of the human and Drosophila Asf1 proteins for yeast histones allows us to draw correlations with their functional abilities in yeast. It is quite striking that a stronger affinity for histones does not correlate with greater functional competence of the human Asf1 proteins in yeast. For example, hAsf1a binds yeast histones very strongly (stronger than yAsf1), but only complements the DNA damage sensitivity phenotype, not the growth defect, the sensitivity to replicational stress, or the defect in PHO5 activation or transcriptional silencing in cells lacking yeast Asf1.
It is possible that a highly dynamic interaction with histones is required in order for Asf1 to function during transcription and replication. For example, if all the hAsf1a in yeast is already bound to histones due to its high affinity for yeast histones, one could imagine that there would not be any free hAsf1a available to remove the histones from the PHO5 promoter to facilitate PHO5 activation. Similarly, an Asf1-histone interaction that is too strong could prevent the histones being released onto the DNA during chromatin assembly. We can conclude that a stronger interaction is not always a better interaction in the case of highly dynamic functions like those of the histone chaperone Asf1.
It is possible that the different abilities of hAsf1a and hAsf1b to bind to yeast histones determines how well they mimic the yeast Asf1 protein. In this case, it is possible that the observed different functions of hAsf1a and hAsf1b are an artifact of placing these proteins in yeast and that hAsf1a and hAsf1b may be fully redundant in human cells. However, the available evidence suggests that this is not the case, and that hAsf1a and hAsf1b are likely to have distinct functions in vivo. RNA interference inactivation of hAsf1a leads to a moderate delay in S phase, while RNA interference of hAsf1b leads to no delay (7). RNA interference of both hAsf1a and hAsf1b leads to a much greater delay in S phase (7). Similarly, hAsf1a but not hAsf1b is required to promote the formation of senescence-associated heterochromatin foci in primary fibroblasts (34). While studies of hAsf1a and hAsf1b in human cells are limited, the information that is available supports our data showing that hAsf1a and hAsf1b have some specific and redundant functions in vivo.
Asf1 binds to the DNA damage checkpoint kinase Rad53 in yeast (5, 8). While this interaction is intriguing and strongly suggests that Asf1 and Rad53 function together to facilitate DNA repair and replication, we find that this is unlikely to be the case. Our data indicate that the interaction between Rad53 and Asf1 is unlikely to be important for Asf1's function in protecting against spontaneous genomic instability, DNA replicational stress and double-strand DNA-damaging agents. Furthermore, we have been unable to detect an interaction between dAsf1 and the Drosophila equivalent of Rad53, Dmchk2, by coimmunoprecipitation and in vitro binding analyses (data not shown). Similarly, an interaction between human Chk2 and human Asf1 has not been detected (7). Furthermore, we found that Dmchk2 is not able to complement the loss of Rad53 in budding yeast (data not shown). It therefore appears that the interaction between Rad53 and Asf1 is specific to yeast and probably explains why Drosophila and human Asf1 failed to bind to Rad53 (Fig. 6). Nonetheless, the fact that human and Drosophila Asf1 can complement the role of yeast Asf1 in protecting against DNA-damaging agents and replicational stress indicates that the interaction between Rad53 and Asf1 is not required for these functions of Asf1.
These studies also demonstrate that phosphorylation of human and Drosophila Asf1 is unlikely to be essential for its function. Human Asf1a is phosphorylated by Tousled-like kinases (Tlks) during S phase, while hAsf1b is phosphorylated by Tlks in M phase (26). There are no obvious Tlk homologs in yeast, although it is impossible to rule out whether there are functionally analogous kinases in yeast. However, phosphorylation of yeast Asf1 has not been observed, suggesting that phosphorylation similar to that mediated by Tlks does not occur on the human and Drosophila Asf1 proteins when they are present in yeast. The fact that human and Drosophila Asf1 proteins are at least partially functional in yeast indicates that Tlk-mediated phosphorylation is not essential for their function in yeast.
It is possible that phosphorylation of Asf1 in human and Drosophila cells may play a regulatory role that is not conserved in yeast. It is noteworthy that human Asf1a but not Asf1b is phosphorylated in an S-phase-specific manner by Tlks in human cells (26). Perhaps phosphorylation of hAsf1a is required for it to mediate chromatin assembly and disassembly during replication, and the absence of the S-phase-specific Tlk-mediated phosphorylation of hAsf1a in yeast may be responsible for the failure of hAsf1a to protect against genomic instability and replication stress-inducing drugs in yeast. Future studies are required to determine whether Tlk-mediated phosphorylation of Asf1 activates, represses, or regulates the function of Asf1.
Acknowledgments
We thank Christine English, Laura Schulz, and Jeffrey Linger for critical reading of the manuscript. We thank Kim Dao Dang and Andy Nelson for technical assistance with strain construction. We are grateful to Tin-Tin Su and Wei Du for reagents to examine the Dmchk2-dAsf1 interaction.
This work was supported by a grant from the NIH (CA95641) to J.K.T. J.K.T. is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Chimura, T., T. Kuzuhara, and M. Horikoshi. 2002. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 99:9334-9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daganzo, S. M., J. P. Erzberger, W. M. Lam, E. Skordalakes, R. Zhang, A. A. Franco, S. J. Brill, P. D. Adams, J. M. Berger, and P. D. Kaufman. 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13:2148-2158. [DOI] [PubMed] [Google Scholar]
- 4.de Gunzburg, J., R. Riehl, and R. A. Weinberg. 1989. Identification of a protein associated with p21ras by chemical crosslinking. Proc. Natl. Acad. Sci. USA 86:4007-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emili, A., D. M. Schieltz, J. R. Yates 3rd, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 6.Franco, A. A., W. M. Lam, P. M. Burgers, and P. D. Kaufman. 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groth, A., D. Ray-Gallet, J. P. Quivy, J. Lukas, J. Bartek, and G. Almouzni. 2005. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17:301-311. [DOI] [PubMed] [Google Scholar]
- 8.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krawitz, D. C., T. Kama, and P. D. Kaufman. 2002. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 11.Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman, J. D. Boeke, and D. Young. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276:24068-24074. [DOI] [PubMed] [Google Scholar]
- 12.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 13.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol 119:1-16. [DOI] [PubMed] [Google Scholar]
- 14.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monson, E. K., D. de Bruin, and V. A. Zakian. 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. USA 94:13081-13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mousson, F., A. Lautrette, J. Y. Thuret, M. Agez, R. Courbeyrette, B. Amigues, E. Becker, J. M. Neumann, R. Guerois, C. Mann, and F. Ochsenbein. 2005. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. USA 102:5975-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munakata, T., N. Adachi, N. Yokoyama, T. Kuzuhara, and M. Horikoshi. 2000. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells 5:221-233. [DOI] [PubMed] [Google Scholar]
- 19.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osada, S., A. Sutton, N. Muster, C. E. Brown, J. R. Yates 3rd, R. Sternglanz, and J. L. Workman. 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15:3155-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polo, S. E., S. E. Theocharis, J. Klijanienko, A. Savignoni, B. Asselain, P. Vielh, and G. Almouzni. 2004. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 64:2371-2381. [DOI] [PubMed] [Google Scholar]
- 22.Prado, F., F. Cortes-Ledesma, and A. Aguilera. 2004. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 5:497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramey, C. J., S. Howar, M. Adkins, J. Linger, J. Spicer, and J. K. Tyler. 2004. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 24:10313-10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 26.Sillje, H. H., and E. A. Nigg. 2001. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11:1068-1073. [DOI] [PubMed] [Google Scholar]
- 27.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 30.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 31.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Biol. Cell 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umehara, T., T. Chimura, N. Ichikawa, and M. Horikoshi. 2002. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells 7:59-73. [DOI] [PubMed] [Google Scholar]
- 33.Zabaronick, S. R., and J. K. Tyler. 2005. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol. Cell. Biol. 25:652-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8:19-30. [DOI] [PubMed] [Google Scholar]