The cocrystal of 5-fluorocytosine and 4-hydroxybenzaldehyde (1/1), C4H4FN3O·C7H6O2, crystallizes in the monoclinic P21/c space group. The crystal structure features a robust supramolecular network stabilized by N—H⋯O, N—H⋯N, O—H⋯O, C—H⋯O and C–H⋯F hydrogen bonds, forming diverse ring motifs including R22(8), R44(22), R66(32), and R88(34). Additional C—F⋯π interactions contribute to the crystal cohesion. Hirshfeld surface analysis reveals that O⋯H/H⋯O contacts dominate the intermolecular interactions, emphasizing the key role of hydrogen bonding in the crystal packing.
Keywords: cocrystal; supramolecular network; dimeric motif; tetrameric motif; Hirshfeld surface analysis,fingerprint plots
Abstract
The 1:1 cocrystal of 5-fluorocytosine (5FC) and 4-hydroxybenzaldehyde (4HB), C4H4FN3O·C7H6O2 has been synthesized and its structure characterized by single-crystal X-ray diffraction and Hirshfeld surface analysis. The compound crystallizes in the monoclinic P21/c space group. A robust supramolecular architecture is stabilized by N—H⋯O, N—H⋯N, C—H⋯O and C—H⋯F hydrogen bonds, forming R22(8), R44(22), R66(32), and R88(34) ring motifs. The N—H⋯O and N—H⋯N hydrogen bonds form strong directional interactions, contributing to the R22(8) and R88(34) motifs through dimeric and extended ring structures. O—H⋯O interactions link 5FC and 4HB molecules, generating an R66(32) ring that enhances the packing. Weaker C—H⋯F bonds help form the R44(22) tetrameric motif, supporting the overall three-dimensional supramolecular framework. Additionally, C—F⋯π interactions between the fluorine atom and the aromatic ring add further to the crystal cohesion. Hirshfeld surface analysis and two-dimensional fingerprint plots confirm that O⋯H/H⋯O contacts are the most significant, highlighting the central role of hydrogen bonding in the stability and organization of the crystal structure.
1. Chemical context
Cocrystals have gained considerable attention in supramolecular chemistry for their ability to improve the physical and chemical properties of active pharmaceutical ingredients (APIs) and functional materials without altering the molecular structure of the drug. They are defined as crystalline, single-phase solids composed of two or more distinct molecular and/or ionic compounds, typically in a stoichiometric ratio, which are neither simple salts nor solvates (Aitipamula et al., 2012 ▸; Almarsson & Zaworotko, 2004 ▸). Cocrystals are stabilized through non-covalent interactions such as hydrogen bonding, π–π stacking, halogen bonding, and van der Waals forces. Their design is guided by the principles of crystal engineering, involving the careful selection of suitable coformers and the application of supramolecular synthons, such as the R22(8) hydrogen-bonded motif (Etter, 1990 ▸; Etter et al., 1990 ▸; Desiraju, 1995 ▸). In the pharmaceutical industry, cocrystallization offers a promising strategy for enhancing the solubility, stability, and bioavailability of poorly soluble drugs. (Alvani & Shayanfar, 2022 ▸; Shi et al., 2024 ▸). Compared to conventional techniques such as salt formation, micronization, solid dispersion, amorphous forms, and encapsulation, cocrystals offer the advantage of maintaining a stable crystalline structure, which facilitates detailed characterization by X-ray diffraction (Bolla & Nangia, 2016 ▸; Bolla et al., 2022 ▸).
2. Structural commentary
Single-crystal X-ray diffraction analysis reveals that the title compound crystallizes in the monoclinic P21/c space group with one molecule each of 5-fluorocytosine (5FC) and 4-hydroxybenzaldehyde (4HB) present in the asymmetric unit. An ellipsoid plot of the compound is shown in Fig. 1 ▸. Proton transfer does not occur between the hydroxyl group of benzaldehyde and the pyrimidine ring nitrogen atom of 5FC. The C—O bond length in the hydroxyl group of the 4HB molecule is 1.3520 (13) Å, with the corresponding internal bond angle [C2A—N1A—C3A = 120.00 (8)°] in agreement with reported literature values (Louis et al., 1982 ▸; Mohana et al., 2016 ▸, 2023 ▸; Sangavi et al., 2024 ▸).
Figure 1.
The molecular structure of the title cocrystal with displacement ellipsoids drawn at the 50% probability level. Hydrogen bonds are shown as dashed lines.
3. Supramolecular features and Hirshfeld surface analysis
The primary interaction motif is formed via N—H⋯O and C—H⋯F hydrogen bonds (Table 1 ▸). The N4A amino group and F1A atom of the 5FC molecule interact with the O2B and C7B atoms of the 4HB molecule, resulting in an  (8) heterodimeric synthon. Heterodimers are further linked through a weak C—H⋯Oiii [symmetry code: (iii) −x + 2, −y + 1, −z + 1] hydrogen bond involving the C4A atom of 5FC and the O1B atom of 4HB. The interaction leads to the formation of an
(8) heterodimeric synthon. Heterodimers are further linked through a weak C—H⋯Oiii [symmetry code: (iii) −x + 2, −y + 1, −z + 1] hydrogen bond involving the C4A atom of 5FC and the O1B atom of 4HB. The interaction leads to the formation of an 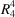 (22) tetrameric synthon. The tetrameric motif is further extended through a homodimeric
(22) tetrameric synthon. The tetrameric motif is further extended through a homodimeric  (8) synthon, formed by N—H⋯Ni [symmetry code: (i) −x, y +
(8) synthon, formed by N—H⋯Ni [symmetry code: (i) −x, y +  , −z +
, −z +  ] and N—H⋯Oii [symmetry code: (ii) −x, y −
] and N—H⋯Oii [symmetry code: (ii) −x, y −  , −z +
, −z +  ] hydrogen bonds. These interactions involve atoms N1A, N2A, N3A and O1A of the 5-fluorocytosine (5FC) molecule. The formation of this homodimeric synthon bridges adjacent tetrameric units, resulting in a large
] hydrogen bonds. These interactions involve atoms N1A, N2A, N3A and O1A of the 5-fluorocytosine (5FC) molecule. The formation of this homodimeric synthon bridges adjacent tetrameric units, resulting in a large  (34) ring motif. The alternating arrangement of
(34) ring motif. The alternating arrangement of 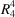 (22) and
(22) and  (34) rings leads to the development of a three-dimensional supramolecular cage-like architecture. This network is further consolidated by O—H⋯O hydrogen-bonding interactions between the O1A atom of the 5FC molecule and the hydroxyl (–OH) group of the 4-hydroxybenzaldehyde (4HB) molecule. The hydrogen bonding occurs via an O—H⋯Oiv [symmetry code: (iv) x + 1, −y +
(34) rings leads to the development of a three-dimensional supramolecular cage-like architecture. This network is further consolidated by O—H⋯O hydrogen-bonding interactions between the O1A atom of the 5FC molecule and the hydroxyl (–OH) group of the 4-hydroxybenzaldehyde (4HB) molecule. The hydrogen bonding occurs via an O—H⋯Oiv [symmetry code: (iv) x + 1, −y +  , z −
, z −  ] interaction, forming an
] interaction, forming an  (32) ring motif (Fig. 2 ▸). This interaction strengthens the packing and adds complexity to the supramolecular network. In addition to hydrogen bonding, the crystal structure is further consolidated by weak C—H⋯F and C—F⋯π interactions. The C—F⋯π interaction (Fig. 3 ▸) is observed between 5FC molecules [C1A⋯Cgv = 3.2676 (9) Å, C1A—F1A⋯Cg = 89.41 (6)°, where Cg is the centroid of the 5FC ring; symmetry code: (v) 1 + x, y, z]. The observed angle is consistent with values reported in the literature (Sikorski et al., 2005 ▸; Vangala et al., 2002 ▸).
(32) ring motif (Fig. 2 ▸). This interaction strengthens the packing and adds complexity to the supramolecular network. In addition to hydrogen bonding, the crystal structure is further consolidated by weak C—H⋯F and C—F⋯π interactions. The C—F⋯π interaction (Fig. 3 ▸) is observed between 5FC molecules [C1A⋯Cgv = 3.2676 (9) Å, C1A—F1A⋯Cg = 89.41 (6)°, where Cg is the centroid of the 5FC ring; symmetry code: (v) 1 + x, y, z]. The observed angle is consistent with values reported in the literature (Sikorski et al., 2005 ▸; Vangala et al., 2002 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2A—H1⋯N1Ai | 0.88 (1) | 2.06 (1) | 2.9354 (12) | 175 (1) |
| N3A—H1CC⋯O2B | 0.89 (1) | 2.10 (1) | 2.9848 (13) | 170 (1) |
| N3A—H1A⋯O1Aii | 0.90 (1) | 2.04 (1) | 2.9328 (12) | 176 (1) |
| C4A—H4A⋯O1Biii | 0.93 | 2.48 | 3.2905 (14) | 145 |
| O1B—H1B⋯O1Aiv | 0.86 (2) | 1.85 (2) | 2.6934 (13) | 166 (2) |
| C6B—H6B⋯F1Aiii | 0.93 | 2.56 | 3.3446 (14) | 143 |
| C7B—H7B⋯F1A | 0.93 | 2.51 | 2.9886 (14) | 112 |
Symmetry codes: (i)  ; (ii)
; (ii) 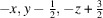 ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Figure 2.
Three-dimensional supramolecular cage-like architecture formed via N—H⋯O, N—H⋯N, O—H⋯O, C—H⋯F and C—H⋯O hydrogen bonds. [Symmetry codes: (i) −x, y +  , −z +
, −z +  ; (ii) −x, y −
; (ii) −x, y −  , −z +
, −z +  ; (iii) −x + 2, −y + 1, −z + 1; (iv) x + 1, −y +
; (iii) −x + 2, −y + 1, −z + 1; (iv) x + 1, −y +  , z −
, z −  .]
.]
Figure 3.
A view of the C—F⋯π interaction (symmetry operation 1 + x, y, z).
Hirshfeld surface (HS) analysis was performed for the title compound to visualize and quantify its intermolecular interactions. Fig. 4 ▸ presents the van der Waals interactions using a Hirshfeld surface mapped over dnorm (Spackman & Jayatilaka, 2009 ▸), generated with Crystal Explorer 21 (Spackman et al., 2021 ▸). This analysis reveals significant intermolecular hydrogen bonds of the types N—H⋯O, N—H⋯N and O—H⋯O interactions. In the surface representation, red areas indicate strong hydrogen bonding, blue regions correspond to contacts close to the sum of the van der Waals radii, and white regions represent weaker interactions.
Figure 4.
The Hirshfeld surface mapped over dnorm showing the N—H⋯O, N—H⋯N and O—H⋯O interactions as dashed gray lines.
To analyze the relative contributions of different intermolecular interactions, two-dimensional fingerprint plots were generated (McKinnon et al., 2007 ▸) and these are shown in Fig. 5 ▸. These plots indicate that the most prominent contacts are O⋯H/H⋯O (26.6%), followed by H⋯H (25.5%), C⋯H/H⋯C (16.7%), N⋯H/H⋯N (10.0%) and F⋯H/H⋯F (6.2%). The crystallographic analysis reveals a robust supramolecular network in the title compound, stabilized by hydrogen bonds (N—H⋯O, N—H⋯N, O—H⋯O and C—H⋯F) and C—F⋯π interactions, forming a three-dimensional cage-like supramolecular architecture. Hirshfeld surface analysis highlights prominent O⋯H/H⋯O interactions, alongside other significant contacts, contributing to crystal stability. The study demonstrates how non-covalent interactions, including hydrogen-bonding and π interactions, govern the molecular packing and cohesion, supporting the principles of supramolecular chemistry in crystal engineering.
Figure 5.
Fingerprint plots showing the total contribution of individual interactions and those delineated into O⋯H/H⋯O, H⋯H, C⋯H/H⋯C, N⋯H/H⋯N and F⋯H/H⋯F interactions.
4. Database survey
5-Fluorocytosine (5FC) is a synthetic antimycotic compound, first synthesized in 1957 and widely used as an antitumor agent. It is also active against fungal infection (Portalone & Colapietro, 2007 ▸; Vermes et al., 2000 ▸). It becomes active by deamination of 5FC into 5-fluorouracil by the enzyme cytosine deaminase (CD) and inhibits RNA and DNA synthesis (Morschhauser, 2003 ▸). The Cambridge Structural Database (CSD, v5.45, June 2024; Groom et al., 2016 ▸) reference codes for the monohydrate are BIRMEU, BIRMEU01, BIRMEU02, BIRMEU03, MEBQUG, MEBQIU, MEBQOA and GATMUL (Louis et al., 1982 ▸; Portalone & Colapietro, 2006 ▸; Hulme & Tocher, 2006 ▸; Portalone, 2011 ▸), and for the polymorphs: DUKWIQ, DUKWAI and DUKWEM (Tutughamiarso et al., 2009 ▸). A wide range of cocrystals has also been documented, such as XOQQUS, MECTUL, MECVEX, MECVIB, MECVOH, MECVUN, MECWAU, MECWEY, MECWOI, MECWUO, MECXEZ, MECXID, MECXOJ, GIFWIF, UJUJAM, and POCWUD (Souza et al., 2019 ▸;Tutughamiarso et al., 2012 ▸; Tutughamiarso & Egert, 2012 ▸; Mohana et al., 2016 ▸, 2023 ▸; Sangavi et al., 2024 ▸). Salts include WEWZAA01, SIJXAM, SIJXIU, SIJXUG, EDATOS, GIFWEB, POCXAK, ZAPFEE and ROLTUJ WEWZAA01, SIJXAM, SIJXIU, SIJXUG, EDATOS, GIFWEB, POCXAK, ZAPFEE and ROLTUJ (Perumalla et al., 2013a ▸,b ▸; Prabakaran et al., 2001 ▸; Mohana et al., 2017 ▸; Karthikeyan et al., 2014 ▸) have been reported in the literature. 4-Hydroxybenzaldehydes are potential therapeutic agents for the treatment of human angiostrongyliasis. The crystal structure of 4-hydroxybenzaldehyde (Jasinski et al., 2008 ▸), as well as its cocrystal (Nowak & Sikorski, 2023 ▸) and polymorphic forms (Simões et al., 2013 ▸) have also been reported. 5FC contains multiple hydrogen-bond donors and acceptors, including amino and carbonyl groups, and 4-HBA offers hydroxyl and aldehyde functionalities capable of forming hydrogen bonds, along with an aromatic ring that can engage in π–π interactions. The present work focuses on the supramolecular hydrogen bonding interactions in the crystal structure of 1:1 cocrystals of 5-fluorocytosine-4-hydroxybenzaldehyde.
5. Synthesis and crystallization
The title compound was synthesized by mixing a hot ethanolic solution of 5-fluorocytosine with 4-hydroxybenzaldehyde in a 1:1 molar ratio. The solution was heated in a water bath at 333 K for 30 minutes and then allowed to cool slowly to room temperature. After a few days, colorless crystals had separated out of the mother liquor.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The H atoms of the N—H, –NH2 and OH groups were located in difference-Fourier maps and refined freely. Other H atoms were placed geometrically (C—H = 0.93 Å) and refined using a riding model with Uiso(H) = 1.2Ueq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C4H4FN3O·C7H6O2 |
| M r | 251.22 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 297 |
| a, b, c (Å) | 4.2126 (1), 9.6687 (1), 26.8628 (5) |
| β (°) | 94.186 (1) |
| V (Å3) | 1091.21 (3) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.07 |
| Crystal size (mm) | 0.27 × 0.21 × 0.17 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, HyPix |
| Absorption correction | Analytical (CrysAlis PRO; Rigaku OD, 2023 ▸) |
| Tmin, Tmax | 0.782, 0.840 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 19824, 2243, 2127 |
| R int | 0.019 |
| (sin θ/λ)max (Å−1) | 0.630 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.034, 0.101, 1.04 |
| No. of reflections | 2243 |
| No. of parameters | 180 |
| No. of restraints | 4 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.19 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004463/oi2017sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004463/oi2017Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989025004463/oi2017Isup3.cml
CCDC reference: 2452037
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Crystal data
| C4H4FN3O·C7H6O2 | F(000) = 520 |
| Mr = 251.22 | Dx = 1.529 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 4.2126 (1) Å | Cell parameters from 18043 reflections |
| b = 9.6687 (1) Å | θ = 3.3–76.2° |
| c = 26.8628 (5) Å | µ = 1.07 mm−1 |
| β = 94.186 (1)° | T = 297 K |
| V = 1091.21 (3) Å3 | Block, colorless |
| Z = 4 | 0.27 × 0.21 × 0.17 mm |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Data collection
| XtaLAB Synergy, Dualflex, HyPix diffractometer | 2243 independent reflections |
| Radiation source: micro-focus sealed X-ray tube | 2127 reflections with I > 2σ(I) |
| Detector resolution: 10.0000 pixels mm-1 | Rint = 0.019 |
| ω scans | θmax = 76.2°, θmin = 3.3° |
| Absorption correction: analytical (CrysAlisPro; Rigaku OD, 2023) | h = −3→5 |
| Tmin = 0.782, Tmax = 0.840 | k = −12→12 |
| 19824 measured reflections | l = −33→33 |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.034 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.101 | w = 1/[σ2(Fo2) + (0.0563P)2 + 0.2241P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 2243 reflections | Δρmax = 0.20 e Å−3 |
| 180 parameters | Δρmin = −0.19 e Å−3 |
| 4 restraints | Extinction correction: SHELXL2019/2 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: dual | Extinction coefficient: 0.0088 (13) |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. The data collection, cell refinement, and data reduction were performed using CrysAlisPro (Rigaku OD, 2023). Structure solution was carried out with SHELXT 2014/5 (Sheldrick, 2015a) and refinement was done using SHELXL-2016/6 (Sheldrick, 2015b). Molecular graphics were prepared using PLATON (Spek, 2020), Mercury (Macrae et al., 2020) and POVRay (Cason 2004). |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1A | 0.6004 (2) | 0.39753 (7) | 0.63425 (3) | 0.0557 (2) | |
| O1A | −0.1067 (2) | 0.40145 (8) | 0.79019 (3) | 0.0439 (2) | |
| N1A | 0.1299 (2) | 0.27556 (9) | 0.73165 (3) | 0.0331 (2) | |
| N2A | 0.1501 (2) | 0.51932 (9) | 0.73260 (3) | 0.0344 (2) | |
| H1 | 0.076 (3) | 0.5966 (14) | 0.7444 (6) | 0.050 (4)* | |
| N3A | 0.3795 (3) | 0.15570 (10) | 0.67141 (4) | 0.0401 (3) | |
| H1CC | 0.492 (3) | 0.1523 (16) | 0.6446 (5) | 0.049 (4)* | |
| H1A | 0.294 (3) | 0.0798 (14) | 0.6845 (5) | 0.045 (4)* | |
| C1A | 0.4154 (3) | 0.40215 (11) | 0.67318 (4) | 0.0351 (3) | |
| C2A | 0.3072 (2) | 0.27452 (11) | 0.69223 (4) | 0.0313 (2) | |
| C3A | 0.0531 (2) | 0.39737 (10) | 0.75270 (4) | 0.0323 (2) | |
| C4A | 0.3331 (3) | 0.52236 (11) | 0.69305 (4) | 0.0356 (3) | |
| H4A | 0.399417 | 0.605918 | 0.680151 | 0.043* | |
| O1B | 1.1638 (2) | 0.27251 (10) | 0.35919 (3) | 0.0520 (3) | |
| H1B | 1.074 (4) | 0.2080 (18) | 0.3414 (7) | 0.076 (5)* | |
| O2B | 0.6793 (3) | 0.13585 (11) | 0.57435 (3) | 0.0605 (3) | |
| C1B | 1.0816 (3) | 0.25581 (12) | 0.40653 (4) | 0.0383 (3) | |
| C2B | 0.8831 (3) | 0.14938 (12) | 0.42006 (4) | 0.0421 (3) | |
| H2B | 0.798860 | 0.087622 | 0.396053 | 0.050* | |
| C3B | 0.8117 (3) | 0.13552 (13) | 0.46892 (4) | 0.0450 (3) | |
| H3B | 0.677170 | 0.064757 | 0.477749 | 0.054* | |
| C4B | 0.9386 (3) | 0.22637 (12) | 0.50533 (4) | 0.0401 (3) | |
| C5B | 1.1361 (3) | 0.33238 (13) | 0.49140 (4) | 0.0440 (3) | |
| H5B | 1.222274 | 0.393527 | 0.515464 | 0.053* | |
| C6B | 1.2064 (3) | 0.34827 (13) | 0.44238 (5) | 0.0471 (3) | |
| H6B | 1.336473 | 0.420430 | 0.433386 | 0.056* | |
| C7B | 0.8656 (3) | 0.21566 (14) | 0.55746 (4) | 0.0474 (3) | |
| H7B | 0.972343 | 0.275878 | 0.579901 | 0.057* |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1A | 0.0776 (5) | 0.0414 (4) | 0.0534 (5) | −0.0025 (3) | 0.0416 (4) | 0.0020 (3) |
| O1A | 0.0648 (5) | 0.0344 (4) | 0.0352 (4) | 0.0058 (4) | 0.0229 (4) | 0.0028 (3) |
| N1A | 0.0453 (5) | 0.0265 (4) | 0.0287 (4) | −0.0013 (3) | 0.0106 (4) | 0.0013 (3) |
| N2A | 0.0456 (5) | 0.0250 (4) | 0.0338 (5) | 0.0007 (3) | 0.0103 (4) | −0.0009 (3) |
| N3A | 0.0588 (6) | 0.0295 (5) | 0.0341 (5) | −0.0013 (4) | 0.0177 (4) | −0.0018 (4) |
| C1A | 0.0421 (6) | 0.0337 (6) | 0.0310 (5) | −0.0024 (4) | 0.0131 (4) | 0.0025 (4) |
| C2A | 0.0383 (5) | 0.0296 (5) | 0.0265 (5) | −0.0006 (4) | 0.0053 (4) | 0.0009 (4) |
| C3A | 0.0416 (5) | 0.0285 (5) | 0.0274 (5) | 0.0006 (4) | 0.0065 (4) | 0.0018 (4) |
| C4A | 0.0423 (6) | 0.0290 (5) | 0.0364 (5) | −0.0035 (4) | 0.0096 (4) | 0.0047 (4) |
| O1B | 0.0734 (6) | 0.0513 (5) | 0.0327 (4) | −0.0134 (4) | 0.0137 (4) | 0.0001 (4) |
| O2B | 0.0838 (7) | 0.0605 (6) | 0.0404 (5) | 0.0017 (5) | 0.0255 (5) | 0.0006 (4) |
| C1B | 0.0477 (6) | 0.0370 (5) | 0.0311 (5) | 0.0040 (4) | 0.0082 (4) | 0.0018 (4) |
| C2B | 0.0536 (7) | 0.0389 (6) | 0.0343 (6) | −0.0031 (5) | 0.0075 (5) | −0.0055 (4) |
| C3B | 0.0549 (7) | 0.0419 (6) | 0.0397 (6) | −0.0053 (5) | 0.0140 (5) | −0.0004 (5) |
| C4B | 0.0464 (6) | 0.0422 (6) | 0.0324 (5) | 0.0101 (5) | 0.0080 (4) | −0.0005 (4) |
| C5B | 0.0528 (7) | 0.0425 (6) | 0.0367 (6) | 0.0017 (5) | 0.0026 (5) | −0.0072 (5) |
| C6B | 0.0595 (7) | 0.0406 (6) | 0.0418 (6) | −0.0090 (5) | 0.0083 (5) | −0.0015 (5) |
| C7B | 0.0569 (7) | 0.0521 (7) | 0.0342 (6) | 0.0110 (6) | 0.0102 (5) | −0.0031 (5) |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Geometric parameters (Å, º)
| F1A—C1A | 1.3498 (12) | O1B—H1B | 0.857 (15) |
| O1A—C3A | 1.2521 (13) | O2B—C7B | 1.2122 (17) |
| N1A—C2A | 1.3397 (13) | C1B—C6B | 1.3889 (17) |
| N1A—C3A | 1.3558 (13) | C1B—C2B | 1.3907 (16) |
| N2A—C4A | 1.3578 (14) | C2B—C3B | 1.3746 (16) |
| N2A—C3A | 1.3715 (13) | C2B—H2B | 0.9300 |
| N2A—H1 | 0.877 (13) | C3B—C4B | 1.3920 (17) |
| N3A—C2A | 1.3231 (14) | C3B—H3B | 0.9300 |
| N3A—H1CC | 0.891 (13) | C4B—C5B | 1.3887 (18) |
| N3A—H1A | 0.900 (12) | C4B—C7B | 1.4594 (15) |
| C1A—C4A | 1.3353 (15) | C5B—C6B | 1.3791 (17) |
| C1A—C2A | 1.4235 (14) | C5B—H5B | 0.9300 |
| C4A—H4A | 0.9300 | C6B—H6B | 0.9300 |
| O1B—C1B | 1.3520 (13) | C7B—H7B | 0.9300 |
| C2A—N1A—C3A | 120.00 (8) | O1B—C1B—C2B | 122.30 (10) |
| C4A—N2A—C3A | 121.95 (9) | C6B—C1B—C2B | 119.97 (10) |
| C4A—N2A—H1 | 120.1 (10) | C3B—C2B—C1B | 119.96 (11) |
| C3A—N2A—H1 | 117.8 (10) | C3B—C2B—H2B | 120.0 |
| C2A—N3A—H1CC | 121.8 (10) | C1B—C2B—H2B | 120.0 |
| C2A—N3A—H1A | 115.6 (9) | C2B—C3B—C4B | 120.64 (11) |
| H1CC—N3A—H1A | 122.4 (14) | C2B—C3B—H3B | 119.7 |
| C4A—C1A—F1A | 121.33 (9) | C4B—C3B—H3B | 119.7 |
| C4A—C1A—C2A | 120.77 (10) | C5B—C4B—C3B | 118.92 (10) |
| F1A—C1A—C2A | 117.90 (9) | C5B—C4B—C7B | 118.91 (11) |
| N3A—C2A—N1A | 119.97 (9) | C3B—C4B—C7B | 122.16 (11) |
| N3A—C2A—C1A | 120.74 (9) | C6B—C5B—C4B | 120.93 (11) |
| N1A—C2A—C1A | 119.29 (9) | C6B—C5B—H5B | 119.5 |
| O1A—C3A—N1A | 121.43 (9) | C4B—C5B—H5B | 119.5 |
| O1A—C3A—N2A | 118.86 (9) | C5B—C6B—C1B | 119.58 (11) |
| N1A—C3A—N2A | 119.71 (9) | C5B—C6B—H6B | 120.2 |
| C1A—C4A—N2A | 118.21 (9) | C1B—C6B—H6B | 120.2 |
| C1A—C4A—H4A | 120.9 | O2B—C7B—C4B | 126.30 (12) |
| N2A—C4A—H4A | 120.9 | O2B—C7B—H7B | 116.9 |
| C1B—O1B—H1B | 107.7 (13) | C4B—C7B—H7B | 116.9 |
| O1B—C1B—C6B | 117.73 (11) | ||
| C3A—N1A—C2A—N3A | −179.36 (10) | O1B—C1B—C2B—C3B | 178.79 (11) |
| C3A—N1A—C2A—C1A | 0.55 (16) | C6B—C1B—C2B—C3B | −0.23 (19) |
| C4A—C1A—C2A—N3A | 177.61 (11) | C1B—C2B—C3B—C4B | −0.64 (19) |
| F1A—C1A—C2A—N3A | −1.78 (16) | C2B—C3B—C4B—C5B | 0.72 (18) |
| C4A—C1A—C2A—N1A | −2.30 (17) | C2B—C3B—C4B—C7B | 179.39 (11) |
| F1A—C1A—C2A—N1A | 178.31 (9) | C3B—C4B—C5B—C6B | 0.07 (18) |
| C2A—N1A—C3A—O1A | −178.50 (10) | C7B—C4B—C5B—C6B | −178.65 (11) |
| C2A—N1A—C3A—N2A | 1.84 (16) | C4B—C5B—C6B—C1B | −0.9 (2) |
| C4A—N2A—C3A—O1A | 177.67 (10) | O1B—C1B—C6B—C5B | −178.06 (11) |
| C4A—N2A—C3A—N1A | −2.66 (16) | C2B—C1B—C6B—C5B | 1.01 (19) |
| F1A—C1A—C4A—N2A | −179.10 (9) | C5B—C4B—C7B—O2B | 173.98 (12) |
| C2A—C1A—C4A—N2A | 1.53 (17) | C3B—C4B—C7B—O2B | −4.7 (2) |
| C3A—N2A—C4A—C1A | 0.92 (17) |
4-Amino-5-fluoro-1H-pyrimidin-2-one–4-hydroxybenzaldehyde (1/1) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2A—H1···N1Ai | 0.88 (1) | 2.06 (1) | 2.9354 (12) | 175 (1) |
| N3A—H1CC···O2B | 0.89 (1) | 2.10 (1) | 2.9848 (13) | 170 (1) |
| N3A—H1A···O1Aii | 0.90 (1) | 2.04 (1) | 2.9328 (12) | 176 (1) |
| C4A—H4A···O1Biii | 0.93 | 2.48 | 3.2905 (14) | 145 |
| O1B—H1B···O1Aiv | 0.86 (2) | 1.85 (2) | 2.6934 (13) | 166 (2) |
| C6B—H6B···F1Aiii | 0.93 | 2.56 | 3.3446 (14) | 143 |
| C7B—H7B···F1A | 0.93 | 2.51 | 2.9886 (14) | 112 |
Symmetry codes: (i) −x, y+1/2, −z+3/2; (ii) −x, y−1/2, −z+3/2; (iii) −x+2, −y+1, −z+1; (iv) x+1, −y+1/2, z−1/2.
Funding Statement
We thank Howard University and the National Science Foundation Major Research Instrumentation program (NSF DMR-2117502) for financially supporting the acquisition of the Rigaku Synergy single-crystal X-ray diffractometer used in this study.
References
- Aitipamula, S., Banerjee, R., Bansal, A. K., Biradha, K., Cheney, M. L., Choudhury, A. R., Desiraju, G. R., Dikundwar, A. G., Dubey, R., Duggirala, N., Ghogale, P. P., Ghosh, S., Goswami, P. K., Goud, N. R., Jetti, R. R. K. R., Karpinski, P., Kaushik, P., Kumar, D., Kumar, V., Moulton, B., Mukherjee, A., Mukherjee, G., Myerson, A. S., Puri, V., Ramanan, A., Rajamannar, T., Reddy, C. M., Rodriguez-Hornedo, N., Rogers, R. D., Row, T. N. G., Sanphui, P., Shan, N., Shete, G., Singh, A., Sun, C. C., Swift, J. A., Thaimattam, R., Thakur, T. S., Kumar Thaper, R., Thomas, S. P., Tothadi, S., Vangala, V. R., Variankaval, N., Vishweshwar, P., Weyna, D. R. & Zaworotko, M. J. (2012). Cryst. Growth Des.12, 2147–2152.
- Almarsson, O. & Zaworotko, M. J. (2004). Chem. Commun. pp. 1889–1896. [DOI] [PubMed]
- Alvani, A. & Shayanfar, A. (2022). Cryst. Growth Des.22, 6323–6337.
- Bolla, G. & Nangia, A. (2016). Chem. Commun.52, 8342–8360. [DOI] [PubMed]
- Bolla, G., Sarma, B. & Nangia, A. K. (2022). Chem. Rev.122, 11514–11603. [DOI] [PubMed]
- Cason, C. J. (2004). POV-RAY for Windows. Persistence of Vision Raytracer Pvt Ltd, Victoria, Australia. http://www.povray.org
- Desiraju, G. R. (1995). Angew. Chem. Int. Ed. Engl.34, 2311–2327.
- Etter, M. C. (1990). Acc. Chem. Res.23, 120–126.
- Etter, M. C., Urbanczyk-Lipkowska, Z., Zia-Ebrahimi, M. & Panunto, T. W. (1990). J. Am. Chem. Soc.112, 8415–8426.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hulme, A. T. & Tocher, D. A. (2006). Cryst. Growth Des.6, 481–487.
- Jasinski, J. P., Butcher, R. J., Narayana, B., Swamy, M. T. & Yathirajan, H. S. (2008). Acta Cryst. E64, o187. [DOI] [PMC free article] [PubMed]
- Karthikeyan, A., Thomas Muthiah, P. & Perdih, F. (2014). Acta Cryst. E70, 328–330. [DOI] [PMC free article] [PubMed]
- Louis, T., Low, J. N. & Tollin, P. (1982). Cryst. Struct. Commun.11, 1059–1064.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst.53, 226–235. [DOI] [PMC free article] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Mohana, M., Muthiah, P. T., Sanjeewa, L. D. & McMillen, C. D. (2016). Acta Cryst. E72, 552–555. [DOI] [PMC free article] [PubMed]
- Mohana, M., Thomas Muthiah, P. & McMillen, C. D. (2017). Acta Cryst. E73, 361–364. [DOI] [PMC free article] [PubMed]
- Mohana, M., Thomas Muthiah, P., McMillen, C. D. & Butcher, R. J. (2023). Acta Cryst. C79, 61–67. [DOI] [PubMed]
- Morschhäuser, J. (2003). Pharm. Unserer Zeit32, 124–129. [DOI] [PubMed]
- Nowak, P. & Sikorski, A. (2023). RSC Adv.13, 20105–20112. [DOI] [PMC free article] [PubMed]
- Perumalla, S. R., Pedireddi, V. R. & Sun, C. C. (2013a). Cryst. Growth Des.13, 429–432.
- Perumalla, S. R., Pedireddi, V. R. & Sun, C. C. (2013b). Mol. Pharm.10, 2462–2466. [DOI] [PubMed]
- Portalone, G. (2011). Chem. Cent. J.5, 51. [DOI] [PMC free article] [PubMed]
- Portalone, G. & Colapietro, M. (2006). Acta Cryst. E62, o1049–o1051.
- Portalone, G. & Colapietro, M. (2007). J. Chem. Crystallogr.37, 141–145.
- Prabakaran, P., Murugesan, S., Muthiah, P. T., Bocelli, G. & Righi, L. (2001). Acta Cryst. E57, o933–o936. [DOI] [PubMed]
- Rigaku OD. (2023). CrysAlis PRO. Rigaku Oxford Diffraction Ltd, Yarnton, England.
- Sangavi, M., Kumaraguru, N., Butcher, R. J. & McMillen, C. D. (2024). Acta Cryst. C80, 30–36. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. C71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. A71, 3–8.
- Shi, J., Zhang, Y., An, Q., Li, Y. & Liu, L. (2024). J. Solid State Chem.331, 124545.
- Sikorski, A., Krzymiński, K., Niziołek, A. & Błażejowski, J. (2005). Acta Cryst. C61, o690–o694. [DOI] [PubMed]
- Simões, R. G., Bernardes, C. E. S. & da Piedade, M. E. M. (2013). Cryst. Growth Des.13, 2803–2814.
- Souza, M. S., Diniz, L. F., Alvarez, N., da Silva, C. C. P. & Ellena, J. (2019). New J. Chem.43, 15924–15934.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm11, 19–32.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst.54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Tutughamiarso, M. & Egert, E. (2012). Acta Cryst. B68, 444–452. [DOI] [PubMed]
- Tutughamiarso, M., Bolte, M. & Egert, E. (2009). Acta Cryst. C65, o574–o578. [DOI] [PubMed]
- Tutughamiarso, M., Wagner, G. & Egert, E. (2012). Acta Cryst. B68, 431–443. [DOI] [PubMed]
- Vangala, V. R., Nangia, A. & Lynch, D. E. (2002). Chem. Commun. pp. 1304–1305. [DOI] [PubMed]
- Vermes, A., Guchelaar, H. J. & Dankert, J. (2000). J. Antimicrob. Chemother.46, 171–179. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004463/oi2017sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004463/oi2017Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989025004463/oi2017Isup3.cml
CCDC reference: 2452037
Additional supporting information: crystallographic information; 3D view; checkCIF report







