The previously unknown molecular SCXRD structure and crystal-packing pattern of a diclofenac derivative were assessed in detail including intra- and intermolecular interactions such as hydrogen bonds and halogen bonds. The validity of these interactions was further evaluated computationally using QM calculations.
Keywords: crystal structure, diclofenac derivative, hydrogen bonding, halogen bonds, halogen-π-interactions
Abstract
The title compound, C15H15Cl3NO2, was synthesized from diclofenac and chloromethyl chlorosulfate under phase-transfer conditions, and crystallizes in the monoclinic space group P21/c. As a result of steric strain, the two adjacent aromatic six-membered rings cannot be co-planar, while the terminal ring on one side of the molecule and the methyl acetate moiety atoms on the other reside roughly in the same plane. The angle between the planes of the two aromatic rings is rather wide at 64.27 (8)°. The crystal is tightly packed and consolidated by a large number and notable range of intermolecular contacts, including relatively strong classical hydrogen bonds but also halogen bonds and even short contacts between chlorine atoms and π-bonds. The intermolecular interactions were further analysed using DFT methods, the results of which are discussed in comparison to the experimental X-ray data.
1. Chemical context
Chloromethyl esters and their chemical relatives bromomethyl and iodomethyl esters are of great synthetic interest, as they easily react with carboxylic acids in the presence of a base to form acylals (Keydel & Link, 2024 ▸), which serve as prodrug motifs both for drugs already on the market, e.g. sultamicillin (Betrosian & Douzinas, 2009 ▸), pivmecillinam (Burchette et al., 2024 ▸) and clevidipine (Nordlander et al., 2004 ▸), and for experimental compounds of different potential indications (Gao et al., 2022 ▸; Dalaijargal et al., 2022 ▸; Zheng et al., 2022 ▸). During our ongoing investigations into the optimization of the synthesis of acylals, we used the chloromethyl ester of the well-known and widely used NSAID (non-steroidal anti-inflammatory drug) diclofenac as one model substance. It was synthesized from diclofenac in a transesterification reaction with chloromethyl chlorosulfate under phase-transfer conditions according to known methods (Binderup & Hansen, 1984 ▸; Harada et al., 1994 ▸).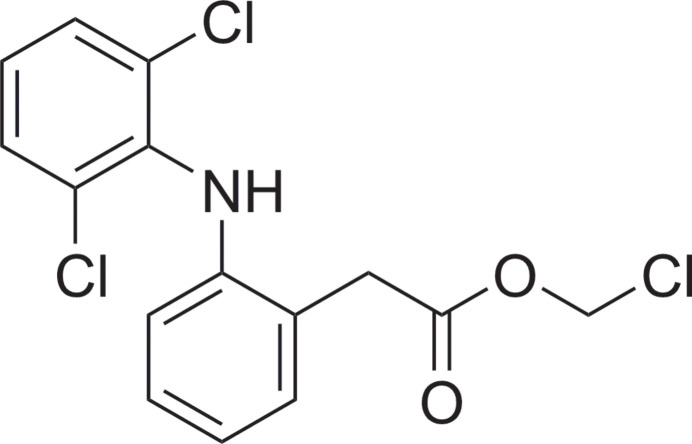
Preparations of the title compound by very similar but not identical procedures were reported before (Dugar et al., 2012 ▸; Xu et al., 2015 ▸), while no characterization data other than low-resolution mass data (m/z = 344) were reported. The magnificent crystals obtained were characterized and the previously unknown crystal structure is reported herein.
2. Structural commentary
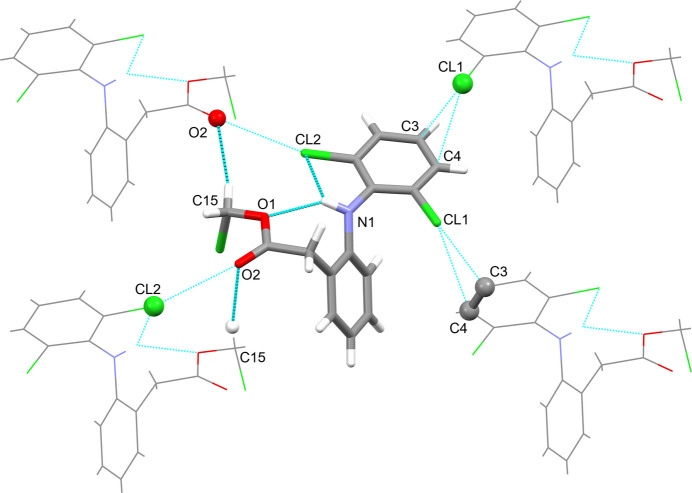
The molecular structure of the title compound is shown in Fig. 1 ▸. All atoms are in general positions and are all refined in the asymmetric unit. The metrical parameters of bond lengths and angles are inconspicuous as they all fall into the commonly reported ranges. The two aromatic rings are not co-planar but arranged with an angle between the planes (through all six atoms of each ring) of 64.27 (8)°, which is notably wide. This is also evident in the torsion angles involving N1 and C13 as second or third atom (Table 1 ▸). An in-plane situation is hindered by the two chlorine ortho substituents of the aniline moiety, while the actually observed torsion between the rings and between the central ring and the ester group is stabilized by two intramolecular hydrogen bonds with N1—H1N as donor to the two acceptors, which are the methylester oxygen atom O1 and the chlorine substituent Cl2 (Table 2 ▸). Fig. 2 ▸ shows all contacts of the asymmetric unit, including the intramolecular hydrogen bonds. The non-hydrogen atoms of the methylacetate chain and the entire aniline moiety lie roughly in the same plane with an angle between the aniline’s six-membered ring and the C13/C14/O1/O2 plane of only 9.78 (8)°. The central phenyl ring (C7–C12) and the terminal chlorine Cl3 point away from the common aniline/acetate plane into the same direction. The nitrogen atom N1 is slightly pyramidalized with a sum of all three angles of 349.6 ° (see Table 1 ▸ for the angles), which is significantly lower than for a planar nitrogen with an angle sum of 360°. Again, this geometry is likely supported by the intramolecular hydrogen bonds (Table 2 ▸).
Figure 1.
The molecular structure of the title compound chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Ellipsoids of the anisotropic atoms are shown at the 50% probability level while the hydrogen atoms have arbitrary radii.
Table 1. Selected bond and torsion angles (°).
| C7—N1—H1N | 113.6 (12) | C2—C1—Cl2 | 118.39 (12) |
| C2—C1—C6 | 122.66 (13) | ||
| C7—N1—C6—C1 | 124.89 (15) | C11—C12—C13—C14 | −90.93 (16) |
| C7—N1—C6—C5 | −59.6 (2) | C7—C12—C13—C14 | 87.17 (16) |
| C6—N1—C7—C8 | −13.4 (2) | C12—C13—C14—O2 | 104.07 (17) |
| C6—N1—C7—C12 | 166.62 (13) | C12—C13—C14—O1 | −73.22 (15) |
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1 | 0.87 (2) | 2.217 (19) | 2.9568 (15) | 142.2 (16) |
| N1—H1N⋯Cl2 | 0.87 (2) | 2.536 (19) | 2.9890 (13) | 113.1 (15) |
| C15—H15A⋯O2i | 0.95 (2) | 2.59 (2) | 3.521 (2) | 168.2 (15) |
| C2—H2⋯N1ii | 0.96 (2) | 2.93 (2) | 3.886 (2) | 173.6 (16) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 2.
The intra- and intermolecular interactions that support or define the molecule’s geometry and the three-dimensional packing in the crystal. Hydrogen bonds are shown as thick blue lines, halogen bonds and halogen–π interactions are shown as thin blue lines. Atoms of the asymmetric unit that are engaged in contacts are labelled.
3. Supramolecular features
The molecules of the title compound in the crystal are engaged in three notable intermolecular interactions (Fig. 2 ▸). These are, firstly, a non-classical hydrogen bond between one methylene hydrogen of the chloromethyl acetate moiety and the carbonyl oxygen atom of the same moiety of an adjacent molecule (Table 2 ▸; C15—H15A⋯O2; symmetry operator: −x + 1, y −  , −z +
, −z +  ). This interaction is repeated one-directionally so that it protrudes throughout the crystal. It can be classified as a C(5) hydrogen-bonding motif (Bernstein et al., 1995 ▸). Secondly, there is a short contact between the terminal chlorine and the carbonyl oxygen atom of a neighbouring molecule, which has a C15—Cl3⋯O1 angle that is sufficiently close to linear at 169.06° (Table 3 ▸; symmetry operator: x,
). This interaction is repeated one-directionally so that it protrudes throughout the crystal. It can be classified as a C(5) hydrogen-bonding motif (Bernstein et al., 1995 ▸). Secondly, there is a short contact between the terminal chlorine and the carbonyl oxygen atom of a neighbouring molecule, which has a C15—Cl3⋯O1 angle that is sufficiently close to linear at 169.06° (Table 3 ▸; symmetry operator: x,  − y, −
− y, − + z). This contact may be considered a halogen bond in which the chlorine’s σ-hole opposite to its pivot carbon atom C15 interacts with the electron density of a free electron pair of the oxygen (Metrangolo et al., 2008 ▸). Thirdly, there are strikingly close contacts between the aniline substituent Cl1 and the aniline ring carbon atoms C3 and, in particular, C4 of an adjacent molecule (symmetry operator: −x, y +
+ z). This contact may be considered a halogen bond in which the chlorine’s σ-hole opposite to its pivot carbon atom C15 interacts with the electron density of a free electron pair of the oxygen (Metrangolo et al., 2008 ▸). Thirdly, there are strikingly close contacts between the aniline substituent Cl1 and the aniline ring carbon atoms C3 and, in particular, C4 of an adjacent molecule (symmetry operator: −x, y +  , −z +
, −z +  ). The Cl—C distances and C5—Cl1⋯C3/4 angles are 3.38 Å/177.53° and 3.34 Å/154.79°, respectively. Interactions between iodine and aromatic systems were identified spectroscopically as early as 1949 (Benesi & Hildebrand, 1949 ▸). What is observed in the crystal structure of the title compound may be characterized as semi-localized halogen–π bond (Schollmeyer et al., 2008 ▸).
). The Cl—C distances and C5—Cl1⋯C3/4 angles are 3.38 Å/177.53° and 3.34 Å/154.79°, respectively. Interactions between iodine and aromatic systems were identified spectroscopically as early as 1949 (Benesi & Hildebrand, 1949 ▸). What is observed in the crystal structure of the title compound may be characterized as semi-localized halogen–π bond (Schollmeyer et al., 2008 ▸).
Table 3. Non-covalent intermolecular contacts (Å, °).
| Contact | Operator | Distance | Geometry | Angle |
|---|---|---|---|---|
| Cl3⋯O2 |
x, −y +  , z − , z − 
|
3.066 (12) | C15—Cl3⋯O2 | 169.1 (07) |
| Cl1⋯C3 | −x, y +  , −z + , −z + 
|
3.385 (18) | C5—Cl1⋯C3 | 177.6 (06) |
| Cl1⋯C4 | −x, y +  , −z + , −z + 
|
3.340 (16) | C5—Cl1⋯C4 | 154.8 (06) |
| C14⋯O2 | −x + 1, −y + 1, −z + 2 | 3.044 (18) | O2—C14⋯O2 | 82.3 (09) |
| Cl3⋯C15 | −x + 1, y +  , −z + , −z + 
|
3.577 (17) | C15—Cl3⋯C15 | 115.8 (03) |
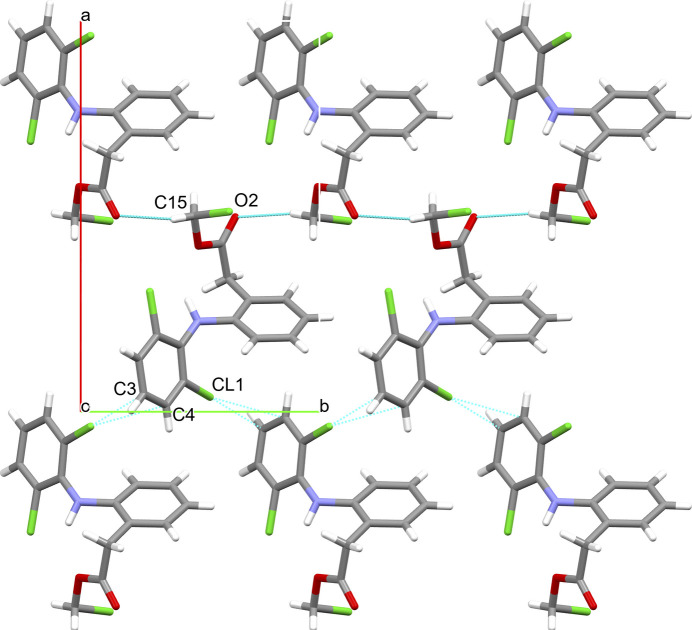
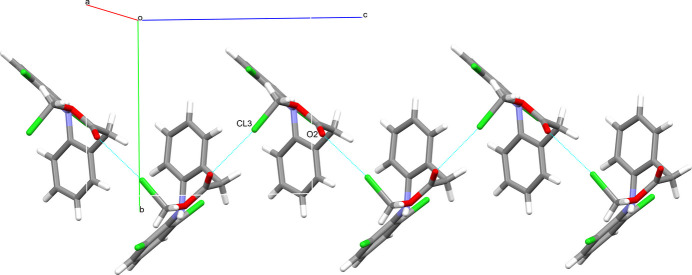
The intermolecular hydrogen bond and the halogen–π contact both form infinite zigzag chains that protrude in the direction of the crystallographic b axis (Fig. 3 ▸, top). The hydrogen bond connects molecules that point alternately up and down parallel to the crystallographic a axis. The same is true for the halogen–π contact (Fig. 3 ▸, bottom). Together they thereby form a layer structure in the ab plane with an overall thickness of half of the crystallographic c axis (Fig. 4 ▸). The interactions between the layers, which are stacked in the c-axis direction, comprise the halogen bonds between the terminal chlorine Cl3 and the carbonyl oxygen atom of the ester O3, which also protrude in a zigzag pattern (Fig. 5 ▸).
Figure 3.
The C(5) hydrogen-bonding motif involving the methyl group and the carbonyl oxygen of the methylacetate moiety (top) and the halogen–π interaction (bottom). Both intermolecular interactions protrude parallel to the crystallographic b-axis. Labelled atoms belong to the respective asymmetric unit. Hydrogen bonds are shown as thick blue lines, halogen–π interactions as thin blue lines. Only atoms of the asymmetric unit are labelled.
Figure 4.
The layer structure formed with a combination of hydrogen bonding and halogen–π interaction contacts (shown in Fig. 3 ▸) in the crystallographic ab plane with a thickness of approximately 0.5 times c.
Figure 5.
The halogen bond between the terminal chlorine Cl3 and the carbonyl oxygen atom, which protrudes parallel to the crystallographic c-axis (contacts shown in blue). This interaction thereby connects the layer structure shown in Fig. 4 ▸ to the layers above and below, forming a three-dimensional network of contacts that stabilize or define the packing pattern. Only atoms of the asymmetric unit are labelled.
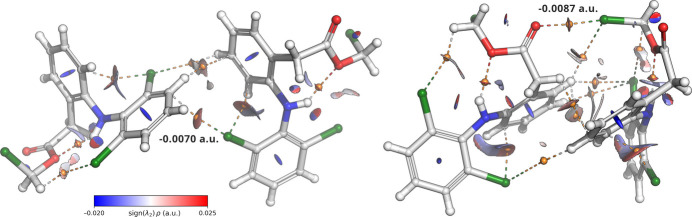
To support the crystallographically identified intermolecular contacts, quantum chemical calculations were performed at the B3LYP-D4/def2-TZVPP level of theory using non-covalent interaction (NCI) analysis based on the reduced density gradient (Fig. 6 ▸). The halogen bond between the terminal chlorine atom Cl3 and the carbonyl oxygen O1 shows a moderately attractive interaction [sin (λ2)ρ = −0.0087 a.u.], consistent with a σ-hole-driven halogen bond. Similarly, the interaction between Cl1 and the adjacent ring atoms C3 and C4, interpreted as a halogen–π contact, exhibits a weak, dispersion-driven attraction [sin (λ2)ρ = −0.0070 a.u.]. These results align with the crystallographic findings and support the role of these non-covalent interactions in stabilizing the supramolecular assembly.
Figure 6.
Non-covalent interaction (NCI) isosurface plot (isovalue = 0.4) of dimers based on the reduced density gradient (RDG) computed from the electron density at the B3LYP-D4/def2-TZVPP level (Schrödinger, 2025 ▸). The orange spheres highlight the bond critical points (BCP) of the identified non-covalent interactions.
In addition to the stronger and more apparent non-covalent interactions observed there are or may be three weaker contacts present (Table 3 ▸). The first potential interaction involves C15 and Cl3 and could be interpreted as a so-called tetrel bond into a π-hole on carbon (Varadwaj et al., 2023 ▸). This runs jointly with the hydrogen bond between O2 and C15 along the crystallographic b axis. The second one comprises a short distance between two carbonyl moieties (C14, O2) of adjacent molecules, which are aligned in an inverted parallel arrangement. This may be regarded as a dispersion effect of the two π-systems, since the geometry of the four atoms excludes a tetrel bond-type interaction. Considering O2 is also involved in relatively stronger hydrogen bonding and a halogen bond, the carbonyl arrangement is likely an effect of the packing rather than that it defines it. The third potential interaction would be a non-classical hydrogen bond C2—H2⋯N1 (Table 2 ▸) protruding along the crystallographic c axis. Considering the distances between the respective C, H and N atoms and the fact that the adjacent halogen – contact also runs along the c-axis direction in the exact same zigzag manner, it is more likely that the former arrangement is a consequence of the more prominent and stronger latter one. We do not consider these three weaker contacts to be of particular importance for the overall packing.
Despite the presence of two aromatic six-membered rings, no aromatic π–π interactions are observed with metrical parameters (distance, slippage etc.) that would suggest any relevance for the packing pattern.
4. Database survey
The CSD (Groom et al., 2016 ▸) was searched using the ConQuest programme version 2024.2.0 (Bruno et al., 2002 ▸), as part of the CSD software suite. A search for the entire backbone (aniline, phenyl ring, and acetate moiety) resulted in 186 hits. Limiting the search by the introduction of the two chlorine substituents on the aniline ring, which are characteristic for diclofenac, reduced the hits by only 6 to 180, which is quite notable. To further narrow down the hit list to molecules that would be well comparable to the title compound, the methyl carbon of the ester group was included. This search returned 29 hits of which 7 are repeatedly reported, i.e. 22 distinct molecular structures in the database. From these were excluded metal-bearing hits, those with relatively large substituents such as sugars, and those with another not covalently bound component in the crystal, e.g., solvent. This resulted in 14 hits and 7 individual structures distinct in the substituents on the methylacetate moiety, which were assessed in comparison with the title compound. One general defining structural feature is the torsion of the central phenyl ring relative to the aniline moiety. A change in torsion direction results in the aniline and the acetate moiety swapping places when looking at the central ring as leaning away from the viewer, while the aniline and acetate moieties lie in the horizontal plane. In the title compound, the torsion angle C6—N1—C7—C12 is 166.62 (13)° and the dichloroaniline ring appears on the right-hand side of the reclining central phenyl ring. This is also the case for the acetoxyacetic acid derivative with a torsion of 178.8° involving the analogous four atoms (BOXMUZ; Jelsch et al., 2020 ▸), for the ethylacetate derivative with ca. 164.5° analogous torsion in all six reported structures (DUBWAB and DUBWAB01–05; Nugrahani et al., 2019 ▸; Chodkiewicz et al., 2022 ▸), and the methylacetate derivative with 165.1° (XEYZIL01; Saleem et al., 2008 ▸). For the latter, a second structure is published in a different crystal system and space group for which the torsion of analogous atoms occurs in opposite direction with −178.3° (XEYZIL; Nawaz et al., 2007 ▸) and the dichloroaniline moiety appears to the left-hand side of the viewer looking at the reclining central phenyl ring. That is also the case for a mixed structure of diclofenac together with its ethylacetate derivative with both molecules having negative analogous torsions of −161.5 and −173.6° (DUBWEF; Nugrahani et al., 2019 ▸), respectively, the 2-propylacetate derivative with −167.0° (MISBEW; Nawaz et al., 2008 ▸), the 4-nitro-aniline and 3,5-dinitrophenyl derivative with a torsion of −148.0° (UNUDEN; Tariq et al., 2011 ▸), and another acetoxyacetic acid derivative, which is a respective torsional isomer of BOXMUZ with a torsion angle of −172.8° and −173.2° in the two reported structures (VUGCUV and VUGCUV01; Alvarez-Larena et al., 1992 ▸; Goud et al., 2013 ▸). All observed absolute torsion angle values fall into a relatively narrow range of 160–180°, which emphasizes a certain joint geometric rigidity of the two aromatic rings, likely enforced by steric strain from the ortho-chlorine substituents and the intramolecular hydrogen bonding involving the NH group. In the title compound and in XEYZIL it is the substituted oxygen that points toward the nitrogen atom for an intramolecular hydrogen bond. In all other cases it is the carbonyl oxygen. This orientation is therefore somehow unusual and as a result, these two are the only examples in which the carbonyl oxygen atom is free to engage in intermolecular hydrogen bonding, which the substituted oxygen does not in any of the other assessed examples. As a result, the intermolecular interactions of the title compound chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate are particularly rich and three-dimensional, which facilitates a unique packing pattern amongst these derivatives.
5. Synthesis and crystallization
To a solution of diclofenac (1.481 g, 5.0 mmol, 1.00 equiv.), sodium hydrogen carbonate (1.260 g, 15.0 mmol, 3.00 equiv.) and tetrabutylammonium hydrogen sulfate (162 mg, 0.5 mmol, 0.10 equiv.) in water (50 mL) and dichloromethane (10 mL), a solution of chloromethyl chlorosulfate (0.759 mL, 7.5 mmol, 1.50 equiv.) in dichloromethane (40 mL) was added dropwise at 273 K over a period of 1 h. The ice bath was removed, and the reaction was allowed to warm to room temperature and left to stir for 16 h. After completion of the reaction (monitored by TLC analysis of the organic phase), the organic phase was separated and washed with saturated sodium hydrogen carbonate solution (3×) and brine. After drying over MgSO4, the solvent was removed under reduced pressure and the residue was purified by dry column vacuum chromatography (0–20% ethyl acetate in hexane) yielding a white solid. Crystallization was accomplished from methanol yielding the title compound as colourless rhomboid crystals (yield 1.325 g, 3.845 mmol, 77%): Rf = 0.24 (hexane); m.p. 359 K; 1H-NMR (400 MHz, DMSO-d6) δ 7.79–7.72 (m, 2H), 7.62 (dd, J = 7.5 Hz, 1H), 7.39 (d, J = 7.4 Hz, 1H), 7.24–7.18 (m, 1H), 7.09 (td, J = 7.5 Hz, 1H), 6.39 (d, J = 7.3 Hz, 1H), 3.89 (s, 2H).; 13C NMR (101 MHz, DMSO-d6) δ 169.70, 142.98, 137.00, 131.28, 131.05, 129.12, 127.99, 126.23, 121.88, 120.41, 115.60, 69.74, 36.33; ESI-HRMS: m/z calculated for [C15H1235Cl3NO2 + H]+ 344.0006, found: 344.0001. Purity: 99.4% (254 nm). General experimental information such as the employed instruments as well as the NMR spectra can be found in the supporting information.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All hydrogen atoms were located and refined freely.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C15H12Cl3NO2 |
| M r | 344.61 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 299 |
| a, b, c (Å) | 15.1487 (1), 9.2389 (1), 10.8661 (1) |
| β (°) | 94.242 (1) |
| V (Å3) | 1516.62 (2) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 5.50 |
| Crystal size (mm) | 0.18 × 0.13 × 0.10 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Single source at home/near, HyPix |
| Absorption correction | Numerical (face indexed; CrysAlis PRO, Rigaku OD, 2024 ▸) |
| Tmin, Tmax | 0.507, 0.856 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 54733, 3301, 3145 |
| R int | 0.031 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.031, 0.081, 1.07 |
| No. of reflections | 3301 |
| No. of parameters | 238 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.46 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004074/nu2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004074/nu2009Isup2.hkl
General experimental information such as the employed instruments as well as the NMR spectra can be found in the Supplementary Information file. DOI: 10.1107/S2056989025004074/nu2009sup3.pdf
Supporting information file. DOI: 10.1107/S2056989025004074/nu2009Isup4.cml
CCDC reference: 2449037
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We acknowledge support by the Open Access Publication Fund of the University of Greifswald.
supplementary crystallographic information
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Crystal data
| C15H12Cl3NO2 | F(000) = 704 |
| Mr = 344.61 | Dx = 1.509 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 15.1487 (1) Å | Cell parameters from 37256 reflections |
| b = 9.2389 (1) Å | θ = 5.1–79.6° |
| c = 10.8661 (1) Å | µ = 5.50 mm−1 |
| β = 94.242 (1)° | T = 299 K |
| V = 1516.62 (2) Å3 | Block, colourless |
| Z = 4 | 0.18 × 0.13 × 0.10 mm |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Data collection
| XtaLAB Synergy, Single source at home/near, HyPix diffractometer | 3301 independent reflections |
| Radiation source: micro-focus sealed X-ray tube | 3145 reflections with I > 2σ(I) |
| Detector resolution: 10.0000 pixels mm-1 | Rint = 0.031 |
| ω scans | θmax = 80.1°, θmin = 5.6° |
| Absorption correction: numerical (face indexed; CrysAlisPro, Rigaku OD, 2024) | h = −19→19 |
| Tmin = 0.507, Tmax = 0.856 | k = −11→11 |
| 54733 measured reflections | l = −12→13 |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.031 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.081 | All H-atom parameters refined |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0387P)2 + 0.4573P] where P = (Fo2 + 2Fc2)/3 |
| 3301 reflections | (Δ/σ)max = 0.001 |
| 238 parameters | Δρmax = 0.34 e Å−3 |
| 0 restraints | Δρmin = −0.46 e Å−3 |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.03768 (2) | 0.54114 (4) | 0.78340 (4) | 0.05292 (12) | |
| Cl2 | 0.30724 (3) | 0.28951 (5) | 0.56128 (5) | 0.06261 (13) | |
| Cl3 | 0.51214 (4) | 0.62138 (5) | 0.63287 (4) | 0.06918 (15) | |
| O1 | 0.42456 (7) | 0.49277 (11) | 0.80019 (10) | 0.0451 (2) | |
| O2 | 0.49799 (7) | 0.64736 (13) | 0.93006 (11) | 0.0539 (3) | |
| N1 | 0.23090 (8) | 0.49080 (13) | 0.74080 (12) | 0.0430 (3) | |
| C1 | 0.19495 (9) | 0.31935 (15) | 0.57362 (14) | 0.0424 (3) | |
| C2 | 0.13490 (11) | 0.24256 (17) | 0.49680 (15) | 0.0491 (3) | |
| C3 | 0.04534 (11) | 0.26343 (18) | 0.50614 (16) | 0.0520 (4) | |
| C4 | 0.01648 (10) | 0.35706 (17) | 0.59320 (16) | 0.0479 (3) | |
| C5 | 0.07738 (9) | 0.43110 (15) | 0.67077 (13) | 0.0408 (3) | |
| C6 | 0.16882 (9) | 0.41802 (14) | 0.66173 (13) | 0.0385 (3) | |
| C7 | 0.23400 (8) | 0.64393 (14) | 0.75110 (13) | 0.0387 (3) | |
| C8 | 0.18753 (10) | 0.73305 (17) | 0.66555 (16) | 0.0468 (3) | |
| C9 | 0.18976 (11) | 0.88217 (18) | 0.67993 (19) | 0.0559 (4) | |
| C10 | 0.23924 (13) | 0.94408 (18) | 0.7780 (2) | 0.0611 (5) | |
| C11 | 0.28691 (11) | 0.85629 (17) | 0.86162 (17) | 0.0529 (4) | |
| C12 | 0.28521 (9) | 0.70601 (15) | 0.85000 (13) | 0.0408 (3) | |
| C13 | 0.34086 (10) | 0.61469 (18) | 0.94222 (14) | 0.0451 (3) | |
| C14 | 0.43033 (9) | 0.58920 (15) | 0.89501 (12) | 0.0400 (3) | |
| C15 | 0.50149 (11) | 0.47487 (18) | 0.73558 (16) | 0.0496 (3) | |
| H2 | 0.1567 (13) | 0.178 (2) | 0.4372 (19) | 0.060 (5)* | |
| H3 | 0.0032 (14) | 0.212 (2) | 0.4547 (19) | 0.066 (6)* | |
| H4 | −0.0433 (14) | 0.370 (2) | 0.6010 (18) | 0.060 (5)* | |
| H8 | 0.1548 (13) | 0.687 (2) | 0.5951 (19) | 0.060 (5)* | |
| H9 | 0.1590 (14) | 0.939 (2) | 0.621 (2) | 0.065 (6)* | |
| H10 | 0.2419 (15) | 1.045 (3) | 0.785 (2) | 0.081 (7)* | |
| H11 | 0.3225 (13) | 0.897 (2) | 0.9322 (19) | 0.060 (5)* | |
| H13A | 0.3497 (12) | 0.666 (2) | 1.0206 (17) | 0.051 (5)* | |
| H13B | 0.3129 (12) | 0.522 (2) | 0.9566 (16) | 0.050 (5)* | |
| H15A | 0.4927 (12) | 0.390 (2) | 0.6869 (18) | 0.054 (5)* | |
| H15B | 0.5529 (14) | 0.474 (2) | 0.7902 (18) | 0.056 (5)* | |
| H1N | 0.2831 (13) | 0.450 (2) | 0.7451 (17) | 0.053 (5)* |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0467 (2) | 0.0523 (2) | 0.0613 (2) | 0.00247 (15) | 0.01461 (16) | −0.00421 (16) |
| Cl2 | 0.0417 (2) | 0.0615 (3) | 0.0856 (3) | 0.00329 (16) | 0.01117 (18) | −0.0195 (2) |
| Cl3 | 0.0967 (4) | 0.0564 (2) | 0.0574 (2) | 0.0089 (2) | 0.0263 (2) | 0.00548 (18) |
| O1 | 0.0401 (5) | 0.0443 (5) | 0.0507 (6) | 0.0000 (4) | 0.0028 (4) | −0.0075 (4) |
| O2 | 0.0447 (6) | 0.0593 (7) | 0.0565 (6) | −0.0093 (5) | −0.0037 (5) | −0.0064 (5) |
| N1 | 0.0365 (6) | 0.0359 (6) | 0.0552 (7) | 0.0023 (5) | −0.0060 (5) | −0.0036 (5) |
| C1 | 0.0393 (7) | 0.0378 (7) | 0.0502 (8) | 0.0005 (5) | 0.0035 (6) | 0.0008 (6) |
| C2 | 0.0536 (8) | 0.0431 (7) | 0.0502 (8) | −0.0010 (6) | 0.0009 (6) | −0.0058 (6) |
| C3 | 0.0491 (8) | 0.0467 (8) | 0.0584 (9) | −0.0069 (7) | −0.0087 (7) | −0.0021 (7) |
| C4 | 0.0365 (7) | 0.0447 (8) | 0.0616 (9) | −0.0029 (6) | −0.0017 (6) | 0.0051 (7) |
| C5 | 0.0390 (7) | 0.0358 (6) | 0.0476 (7) | 0.0003 (5) | 0.0037 (5) | 0.0037 (5) |
| C6 | 0.0372 (6) | 0.0333 (6) | 0.0447 (7) | −0.0003 (5) | −0.0002 (5) | 0.0032 (5) |
| C7 | 0.0333 (6) | 0.0353 (6) | 0.0481 (7) | 0.0000 (5) | 0.0075 (5) | 0.0001 (5) |
| C8 | 0.0380 (7) | 0.0456 (8) | 0.0567 (9) | 0.0012 (6) | 0.0041 (6) | 0.0069 (7) |
| C9 | 0.0456 (8) | 0.0446 (8) | 0.0785 (11) | 0.0067 (7) | 0.0120 (8) | 0.0160 (8) |
| C10 | 0.0606 (10) | 0.0349 (8) | 0.0899 (13) | 0.0011 (7) | 0.0189 (9) | −0.0002 (8) |
| C11 | 0.0522 (8) | 0.0423 (8) | 0.0654 (10) | −0.0045 (6) | 0.0131 (7) | −0.0112 (7) |
| C12 | 0.0368 (6) | 0.0400 (7) | 0.0466 (7) | −0.0012 (5) | 0.0098 (5) | −0.0039 (5) |
| C13 | 0.0459 (7) | 0.0500 (8) | 0.0398 (7) | −0.0042 (6) | 0.0050 (6) | −0.0016 (6) |
| C14 | 0.0433 (7) | 0.0374 (6) | 0.0386 (6) | −0.0010 (5) | −0.0016 (5) | 0.0035 (5) |
| C15 | 0.0484 (8) | 0.0429 (8) | 0.0582 (9) | 0.0090 (6) | 0.0082 (7) | −0.0009 (7) |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Geometric parameters (Å, º)
| Cl1—C5 | 1.7325 (15) | C5—C6 | 1.4012 (19) |
| Cl2—C1 | 1.7381 (14) | C7—C8 | 1.393 (2) |
| Cl3—C15 | 1.7693 (17) | C7—C12 | 1.401 (2) |
| O1—C14 | 1.3600 (17) | C8—C9 | 1.387 (2) |
| O1—C15 | 1.4141 (18) | C8—H8 | 0.98 (2) |
| O2—C14 | 1.1945 (17) | C9—C10 | 1.381 (3) |
| N1—C6 | 1.3983 (17) | C9—H9 | 0.92 (2) |
| N1—C7 | 1.4197 (17) | C10—C11 | 1.381 (3) |
| N1—H1N | 0.87 (2) | C10—H10 | 0.94 (2) |
| C1—C2 | 1.384 (2) | C11—C12 | 1.394 (2) |
| C1—C6 | 1.400 (2) | C11—H11 | 0.98 (2) |
| C2—C3 | 1.381 (2) | C12—C13 | 1.517 (2) |
| C2—H2 | 0.96 (2) | C13—C14 | 1.503 (2) |
| C3—C4 | 1.377 (2) | C13—H13A | 0.974 (19) |
| C3—H3 | 0.95 (2) | C13—H13B | 0.974 (19) |
| C4—C5 | 1.384 (2) | C15—H15A | 0.95 (2) |
| C4—H4 | 0.92 (2) | C15—H15B | 0.94 (2) |
| C14—O1—C15 | 116.33 (12) | C10—C9—C8 | 120.39 (16) |
| C6—N1—C7 | 122.99 (12) | C10—C9—H9 | 120.8 (13) |
| C6—N1—H1N | 113.0 (12) | C8—C9—H9 | 118.8 (13) |
| C7—N1—H1N | 113.6 (12) | C9—C10—C11 | 119.47 (15) |
| C2—C1—C6 | 122.66 (13) | C9—C10—H10 | 119.8 (14) |
| C2—C1—Cl2 | 118.39 (12) | C11—C10—H10 | 120.7 (15) |
| C6—C1—Cl2 | 118.93 (11) | C10—C11—C12 | 121.33 (17) |
| C3—C2—C1 | 119.30 (15) | C10—C11—H11 | 121.4 (12) |
| C3—C2—H2 | 121.8 (12) | C12—C11—H11 | 117.3 (12) |
| C1—C2—H2 | 118.9 (12) | C11—C12—C7 | 118.86 (14) |
| C4—C3—C2 | 120.11 (15) | C11—C12—C13 | 119.23 (14) |
| C4—C3—H3 | 119.3 (13) | C7—C12—C13 | 121.88 (13) |
| C2—C3—H3 | 120.6 (13) | C14—C13—C12 | 109.44 (12) |
| C3—C4—C5 | 119.86 (14) | C14—C13—H13A | 107.7 (11) |
| C3—C4—H4 | 120.6 (12) | C12—C13—H13A | 110.0 (11) |
| C5—C4—H4 | 119.6 (12) | C14—C13—H13B | 109.3 (11) |
| C4—C5—C6 | 122.19 (14) | C12—C13—H13B | 111.6 (11) |
| C4—C5—Cl1 | 118.02 (11) | H13A—C13—H13B | 108.7 (15) |
| C6—C5—Cl1 | 119.79 (11) | O2—C14—O1 | 122.81 (14) |
| N1—C6—C1 | 121.46 (12) | O2—C14—C13 | 126.51 (14) |
| N1—C6—C5 | 122.61 (13) | O1—C14—C13 | 110.62 (12) |
| C1—C6—C5 | 115.79 (12) | O1—C15—Cl3 | 109.92 (10) |
| C8—C7—C12 | 119.59 (13) | O1—C15—H15A | 106.6 (12) |
| C8—C7—N1 | 121.61 (13) | Cl3—C15—H15A | 107.2 (12) |
| C12—C7—N1 | 118.80 (12) | O1—C15—H15B | 111.2 (12) |
| C9—C8—C7 | 120.34 (16) | Cl3—C15—H15B | 107.3 (12) |
| C9—C8—H8 | 121.7 (12) | H15A—C15—H15B | 114.6 (16) |
| C7—C8—H8 | 118.0 (12) | ||
| C6—C1—C2—C3 | 0.2 (2) | C12—C7—C8—C9 | −1.9 (2) |
| Cl2—C1—C2—C3 | 179.24 (13) | N1—C7—C8—C9 | 178.14 (14) |
| C1—C2—C3—C4 | −1.6 (3) | C7—C8—C9—C10 | 1.1 (2) |
| C2—C3—C4—C5 | 0.5 (2) | C8—C9—C10—C11 | 0.3 (3) |
| C3—C4—C5—C6 | 2.1 (2) | C9—C10—C11—C12 | −0.8 (3) |
| C3—C4—C5—Cl1 | −177.06 (12) | C10—C11—C12—C7 | 0.1 (2) |
| C7—N1—C6—C1 | 124.89 (15) | C10—C11—C12—C13 | 178.24 (15) |
| C7—N1—C6—C5 | −59.6 (2) | C8—C7—C12—C11 | 1.3 (2) |
| C2—C1—C6—N1 | 177.97 (14) | N1—C7—C12—C11 | −178.74 (13) |
| Cl2—C1—C6—N1 | −1.03 (19) | C8—C7—C12—C13 | −176.84 (13) |
| C2—C1—C6—C5 | 2.1 (2) | N1—C7—C12—C13 | 3.2 (2) |
| Cl2—C1—C6—C5 | −176.86 (10) | C11—C12—C13—C14 | −90.93 (16) |
| C4—C5—C6—N1 | −179.08 (13) | C7—C12—C13—C14 | 87.17 (16) |
| Cl1—C5—C6—N1 | 0.05 (19) | C15—O1—C14—O2 | −6.3 (2) |
| C4—C5—C6—C1 | −3.3 (2) | C15—O1—C14—C13 | 171.15 (12) |
| Cl1—C5—C6—C1 | 175.83 (10) | C12—C13—C14—O2 | 104.07 (17) |
| C6—N1—C7—C8 | −13.4 (2) | C12—C13—C14—O1 | −73.22 (15) |
| C6—N1—C7—C12 | 166.62 (13) | C14—O1—C15—Cl3 | −77.80 (14) |
Chloromethyl 2-[2-(2,6-dichlorophenylamino)phenyl]acetate. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1 | 0.87 (2) | 2.217 (19) | 2.9568 (15) | 142.2 (16) |
| N1—H1N···Cl2 | 0.87 (2) | 2.536 (19) | 2.9890 (13) | 113.1 (15) |
| C15—H15A···O2i | 0.95 (2) | 2.59 (2) | 3.521 (2) | 168.2 (15) |
| C2—H2···N1ii | 0.96 (2) | 2.93 (2) | 3.886 (2) | 173.6 (16) |
Symmetry codes: (i) −x+1, y−1/2, −z+3/2; (ii) x, −y+1/2, z−1/2.
Funding Statement
Funding for this research was provided by: Deutsche Forschungsgemeinschaft (grant No. 463304797 to Andreas Link).
References
- Alvarez-Larena, A., Piniella, J. F., Carrasco, E., Ginebreda, A., Julia, S. & Germain, G. (1992). J. Crystallogr. Spectrosc. Res.22, 323–328.
- Benesi, H. A. & Hildebrand, J. H. (1949). J. Am. Chem. Soc.71, 2703–2707.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Betrosian, A. P. & Douzinas, E. E. (2009). Expert Opin. Drug Metab. Toxicol.5, 1099–1112. [DOI] [PubMed]
- Binderup, E. & Hansen, E. (1984). Synth. Commun.14, 857–864.
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. [DOI] [PubMed]
- Burchette, J. E., Covert, K., Tu, P., White, B. P., Chastain, D. B. & Cluck, D. B. (2024). Ann. Pharmacother. https://doi.org/10.1177/10600280241288554. [DOI] [PubMed]
- Chodkiewicz, M., Pawlędzio, S., Woińska, M. & Woźniak, K. (2022). IUCrJ, 9, 298–315. [DOI] [PMC free article] [PubMed]
- Dalaijargal, B., Mi, L., Wu, Z., Yin, Y., Liang, H., Qiu, Y., Yan, Y.-J., Jin, H. & Chen, Z.-L. (2022). Med. Chem. Res.31, 1003–1010.
- Dugar, S., Mahajan, D. & Hollinger Peter, F. (2012). Sphaera Pharma Pvt. Ltd WO2012137225.
- Gao, R.-D., Maeda, M., Tallon, C., Feinberg, A. P., Slusher, B. S. & Tsukamoto, T. (2022). ACS Med. Chem. Lett.13, 1892–1897. [DOI] [PMC free article] [PubMed]
- Goud, N. R., Suresh, K. & Nangia, A. (2013). Cryst. Growth Des.13, 1590–1601.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Harada, N., Hongu, M., Tanaka, T., Kawaguchi, T., Hashiyama, T. & Tsujihara, K. (1994). Synth. Commun.24, 767–772.
- Jelsch, C., Devi, R. N., Noll, B. C., Guillot, B., Samuel, I. & Aubert, E. (2020). J. Mol. Struct.1205, 127600.
- Keydel, T. & Link, A. (2024). Molecules29, 4451. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst.53, 226–235. [DOI] [PMC free article] [PubMed]
- Metrangolo, P., Meyer, F., Pilati, T., Resnati, G. & Terraneo, G. (2008). Angew. Chem. Int. Ed.47, 6114–6127. [DOI] [PubMed]
- Nawaz, H., Khawar Rauf, M., Ebihara, M. & Badshah, A. (2008). Acta Cryst. E64, o334. [DOI] [PMC free article] [PubMed]
- Nawaz, H., Khawar Rauf, M., Fuma, Y., Ebihara, M. & Badshah, A. (2007). Acta Cryst. E63, o1228–o1229. [DOI] [PMC free article] [PubMed]
- Nordlander, M., Sjöquist, P.-O., Ericsson, H. & Rydén, L. (2004). Cardiovasc. Drug Rev.22, 227–250. [DOI] [PubMed]
- Nugrahani, I., Utami, D., Nugraha, Y. P., Uekusa, H., Hasianna, R. & Darusman, A. A. (2019). Heliyon5, e02946. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2024). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Saleem, R., Shabir, G., Hanif, M., Qadeer, G. & Wong, W.-Y. (2008). Acta Cryst. E64, o2400. [DOI] [PMC free article] [PubMed]
- Schollmeyer, D., Shishkin, O. V., Rühl, T. & Vysotsky, M. O. (2008). CrystEngComm10, 715–723.
- Schrödinger (2025). Schrödinger Release 2025.1: Jaguar, Schrödinger, LLC, New York, NY.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Tariq, M. I., Jameel, M., Tahir, M. N., Ali, T. & Rizwan, M. (2011). Acta Cryst. E67, o791–o792. [DOI] [PMC free article] [PubMed]
- Varadwaj, P. R., Varadwaj, A., Marques, H. M. & Yamashita, K. (2023). CrystEngComm25, 1411–1423.
- Westrip, S. P. (2010). J. Appl. Cryst.43, 920–925.
- Xu, Z., Wu, J., Zheng, J., Ma, H., Zhang, H., Zhen, X., Zheng, L. T. & Zhang, X. (2015). Int. Immunopharmacol.25, 528–537. [DOI] [PubMed]
- Zheng, W., Tu, B., Zhang, Z., Li, J., Yan, Z., Su, K., Deng, D., Sun, Y., Wang, X., Zhang, B., Zhang, K., Wong, W.-L., Wu, P., Hong, W. D. & Ang, S. (2022). Front. Chem.10, 1094841. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004074/nu2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004074/nu2009Isup2.hkl
General experimental information such as the employed instruments as well as the NMR spectra can be found in the Supplementary Information file. DOI: 10.1107/S2056989025004074/nu2009sup3.pdf
Supporting information file. DOI: 10.1107/S2056989025004074/nu2009Isup4.cml
CCDC reference: 2449037
Additional supporting information: crystallographic information; 3D view; checkCIF report