In the crystal, molecules are linked by C—H⋯O interactions and C—H⋯F interactions to form sheets parallel to the (002) plane. In addition, S—O⋯π and π–π interactions link molecules along the a-axis direction. van der Waals interactions between molecular sheets consolidate the packing.
Keywords: crystal structure, disorder, acylation, furan, sulfonamide, 4+2 cycloaddition, weak interactions, Hirshfeld surface analysis
Abstract
The molecular conformation of the title compound, C29H22F3NO7S, is stable due to the intramolecular C—H⋯O hydrogen bonds. The central seven-membered ring adopts a distorted chair form. In the 7-oxabicyclo[2.2.1]hepta-2,5-diene unit, the five-membered rings adopt envelope conformations. In the crystal, the molecules are linked by C—H⋯O and C—H⋯F interactions, forming sheets parallel to the (002) plane. Additionally, S—O⋯π and π–π interactions [centroid-to-centroid distance = 3.6159 (7) Å] connect the molecules along the a-axis direction. van der Waals interactions between the molecular sheets reinforce the molecular packing. A Hirshfeld surface analysis was conducted to visualize the various intermolecular interactions, indicating that the largest contribution to the surface contacts is from H⋯H interactions (37.3%), followed by O⋯H/H⋯O (24.1%), F⋯H/H⋯F (19.0%), and C⋯H/H⋯C (10.3%) interactions.
1. Chemical context
7-Oxabicyclo[2.2.1]heptenes, products of the thermic reaction between furans and alkenes or alkynes, have great synthetic potential as a useful tool for the design of a broad diversity of substances with various practical properties. For example, these scaffolds can be used in the synthesis of polycyclic arenes – fragments of graphene – and serve as models for new carbon-based electronic materials (Eda et al., 2015 ▸; Criado et al., 2013 ▸; Furrer et al., 2013 ▸). The 7-oxabicyclo[2.2.1]heptane moiety annelated with other rings serves as a scaffold for the preparation of molecular tweezers (Murphy et al., 2016 ▸; Warrener et al., 1999 ▸), supramolecular systems (Chou et al., 2015 ▸; Oh et al., 2010 ▸; Eckert-Maksić et al., 2005 ▸), bridging donor–acceptor molecules (Chakrabarti et al., 2007 ▸), various bioactive and natural products (Roscalesa et al., 2017 ▸; Enev et al., 2012 ▸; Gromov et al., 2009 ▸; Schindler et al., 2009 ▸; Vogel et al., 1999 ▸), high-molecular-weight materials (Margetić et al., 2010 ▸; Warrener et al., 2001 ▸; Vogel et al., 1999 ▸), etc. Under acid catalysis and temperature, cycloaddition intermediates can be converted into phenols, cyclohexenoles, or substituted aromatic hydrocarbons (Zaytsev et al., 2019 ▸; Zubkov et al., 2012a ▸,b ▸; Guliyeva et al., 2024 ▸). Continuing our research into the chemistry of furyl-substituted sulfonamides (Burkin et al., 2024 ▸; Mammadova et al., 2023a ▸,b ▸), a new approach toward the cycloaddition of dimethyl but-2-ynedioate (DMAD) with substituted furans (Zubkov et al., 2009 ▸; Borisova et al., 2018a ▸,b ▸) has been developed. In particular, in the course of the thermic [4 + 2] cycloaddition between DMAD and sulfamide 2, an interesting sequence of reaction steps was observed; [4 + 2] cycloaddition, cleavage of the epoxy bridge, and a subsequent aromatization of the cyclohexene ring (Fig. 1 ▸).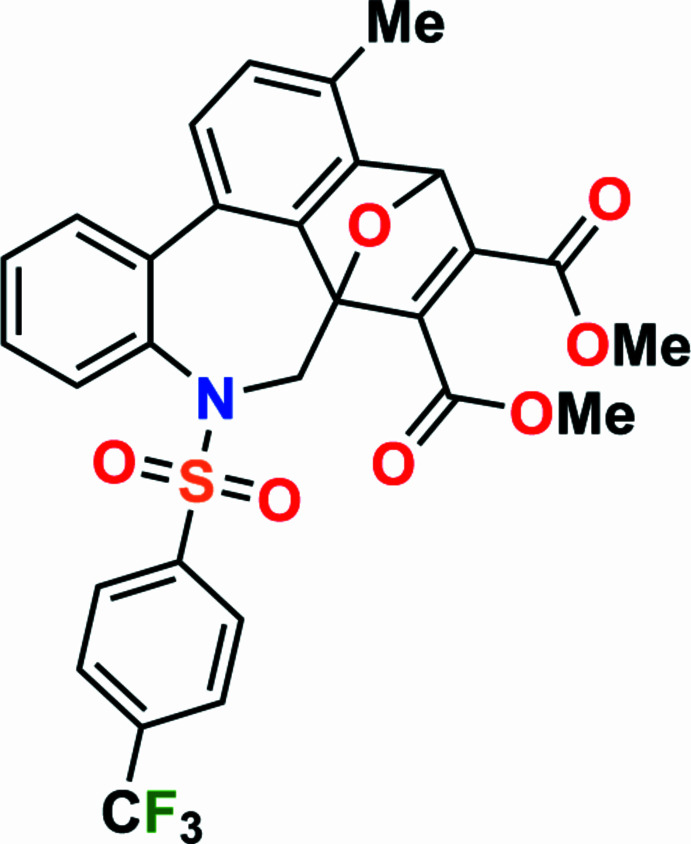
Figure 1.
Synthesis of dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate.
2. Structural commentary
Fig. 2 ▸ shows the molecular structure of the title compound, intramolecular C—H⋯O hydrogen bonds, and naming of the rings in the molecule. The molecular conformation is stable due to the intramolecular hydrogen bonds C7—H7B⋯O17, C7—H7A⋯O1 and C19—H19⋯O2, which form S(6), S(5) and S(5) ring motifs, respectively (Fig. 2 ▸; Table 1 ▸; Bernstein et al., 1995 ▸). Fig. 3 ▸ shows a detailed view of the central rings of the molecule. The central ring A (C6A/C7/N8/C8A/C12A/C12B/C12C) exhibits a distorted chair form [puckering parameters: q2 = 0.708 (1), q3 = 207 (1) Å, φ(2) = −29.76 (9), φ(3) = −138.1 (4) °, QT = 0.738 (1) Å, and spherical polar angle θ(2) = 73.70 (9)°]. Ring A (r.m.s. deviation of fitted atoms = 0.2783 Å) subtends dihedral angles of 20.58 (5), 50.46 (5), 30.64 (5) and 28.18 (5)°, respectively, with rings D (C1–C3/C3A/C12C/C12B), E (C3A/C4–C6/C6A/C12C), F (C8A/C9–C12/C12A) and G (C18–C23). In the 7-oxabicyclo[2.2.1]hepta-2,5-diene unit, the five-membered rings B (O13/C4/C3A/C12C/C6A) and C (O13/C4–C6/C6A) show envelope conformations on atom O13 [B: q(2) = 0.5436 (12) Å, φ(2) = 0.35 (14)° and C: q(2) = 0.5395 (12) Å, φ(2) = 179.95 (14)°]. The bond lengths and angles in the title compound are in good agreement with those reported for related compounds (see Database survey section).
Figure 2.
Molecular structure of the title compound showing atom labelling and ellipsoids at the 30% probability level. The minor disorder component has been omitted for clarity.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C4—H4⋯O16i | 1.00 | 2.49 | 3.3893 (16) | 149 |
| C7—H7A⋯O1 | 0.99 | 2.35 | 2.8481 (15) | 111 |
| C7—H7B⋯O17 | 0.99 | 2.46 | 3.0071 (14) | 115 |
| C12—H12⋯O2ii | 0.95 | 2.55 | 3.1357 (15) | 120 |
| C15—H15A⋯O13ii | 0.98 | 2.47 | 3.3741 (17) | 153 |
| C17—H17B⋯F1iii | 0.98 | 2.54 | 3.3033 (17) | 135 |
| C19—H19⋯O2 | 0.95 | 2.52 | 2.9003 (17) | 104 |
| C22—H22⋯O14i | 0.95 | 2.37 | 3.2698 (19) | 158 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 3.
A detailed view of the central rings of the title molecule.
3. Supramolecular features and Hirshfeld surface analysis
In the crystal, molecules form  (17) ring motifs by C—H⋯O interactions and are linked by C—H⋯F interactions to form sheets parallel to the (002) plane (Figs. 4 ▸ and 5 ▸; Table 1 ▸). Additionally, S—O⋯π (Fig. 5 ▸; Table 1 ▸) and π–π interactions [Fig. 6 ▸; Cg3⋯Cg6 = 3.6159 (7) Å, slippage = 0.804 Å; where Cg3 and Cg6 are the centroids of rings D (C1–C3/C3A/C12C/C12B) and G (C18–C23), respectively] link the molecules along the a-axis direction. van der Waals interactions between the molecular sheets reinforce the molecular packing.
(17) ring motifs by C—H⋯O interactions and are linked by C—H⋯F interactions to form sheets parallel to the (002) plane (Figs. 4 ▸ and 5 ▸; Table 1 ▸). Additionally, S—O⋯π (Fig. 5 ▸; Table 1 ▸) and π–π interactions [Fig. 6 ▸; Cg3⋯Cg6 = 3.6159 (7) Å, slippage = 0.804 Å; where Cg3 and Cg6 are the centroids of rings D (C1–C3/C3A/C12C/C12B) and G (C18–C23), respectively] link the molecules along the a-axis direction. van der Waals interactions between the molecular sheets reinforce the molecular packing.
Figure 4.
A view along the a axis of the title compound, showing the crystal packing. C—H⋯O and C—H⋯F hydrogen bonds are shown as dashed lines; H atoms not involved in hydrogen bonding have been omitted.
Figure 5.
A view along the b axis of the title compound, showing the crystal packing. C—H⋯O and C—H⋯F hydrogen bonds are shown as dashed lines; H atoms not involved in hydrogen bonding have been omitted.
Figure 6.
A partial packing diagram showing S—O⋯π and π–π interactions as dashed lines. H atoms not involved in hydrogen bonding have been omitted.
Hirshfeld surfaces and the corresponding two-dimensional fingerprint plots were created using CrystalExplorer17.5 (Spackman et al., 2021 ▸) in order to visualize the intermolecular interactions (Tables 1 ▸ and 2 ▸). Fig. 7 ▸ shows the full two-dimensional fingerprint plot and those delineated into the major contacts: H⋯H (37.3%), O⋯H/H⋯O (24.1%), F⋯H/H⋯F (19.0%) and C⋯H/H⋯C (10.3%). Smaller contributions are made by O⋯C/C⋯O(4.9%), O⋯O(1.6%), C⋯C (1.5%), F⋯C/C⋯F (0.7%), F⋯O/O⋯F (0.4%), N⋯H/H⋯N (0.2%), S⋯C/C⋯S (0.1%) and S⋯H/H⋯S (0.1%) interactions.
Table 2. Summary of short interatomic contacts (Å) in the title compound.
| Contact | distance | Symmetry operation |
|---|---|---|
| F3⋯H17C | 2.72 | x, −1 + y, z |
| F1⋯H17B | 2.54 | −1 + x, −1 + y, z |
| H12⋯H1 | 2.54 | 1 − x, 1 − y, 1 − z |
| O13⋯H15A | 2.47 | −1 + x, y, z |
| O14⋯H22 | 2.37 | 1 − x,  + y, + y,  − z − z
|
| O14⋯H15B | 2.64 | 2 − x,  + y, + y,  − z − z
|
| H22⋯O14 | 2.37 | 1 − x, − + y, + y,  − z − z
|
| H19⋯H19 | 2.27 | −x, 1 − y, 1 − z |
| H17A⋯H10 | 2.51 | 1 − x, 2 − y, 1 − z |
| H10⋯H9 | 2.58 | 1 − x, 2 − y, 1 − z |
Figure 7.
(a) The full two-dimensional fingerprint plot for the title compound and those delineated into (b) H⋯H, (c) O⋯H/H⋯O, (c) F⋯H/H⋯F and (c) C⋯H/H⋯C contacts.
4. Database survey
A search of the Cambridge Structural Database (Version 5.41, last update November 2019; Groom et al., 2016 ▸) for 11-oxatricyclo[6.2.1.02,7]undecanes gave 739 hits, while a search for 3-methyl-11-oxatricyclo[6.2.1.02,7]undecanes gave zero hits. In these searches, the most related compounds are CSD refcode COKHAP (Sadikhova et al., 2024 ▸) and POYBEL (Zubkov et al., 2009 ▸). In COKHAP, two hexane rings and one oxane ring are fused together. The two hexane rings tend toward a distorted boat conformation, while the tetrahydrofuran and dihydrofuran rings adopt envelope conformations. The oxane ring is puckered. In the crystal, C—H⋯O hydrogen bonds connect the molecules into a three-dimensional network. POYBEL comprises a fused pentacyclic system containing two five-membered (cyclopentane and tetrahydrofuran) and three six-membered (tetrahydropyridinone, tetrahydropyridine and benzene) rings. Both five-membered rings of the bicyclic fragment have the usual envelope conformations, and the two central six-membered rings adopt sofa and non-symmetrical half-chair conformations.
In addition, three related compounds containing the O=S=O group are YIKROD (Mammadova et al., 2023a ▸), KETGID (Schinke et al., 2022 ▸) and LUJKUA (Yakuth et al., 2024 ▸). In YIKROD, intramolecular interactions are observed between the furan and benzene rings of the 4-cyanophenyl group. In the crystal, molecules are connected via C—H⋯O and C—H⋯N hydrogen bonds, forming layers parallel to the (100) plane. These layers are interconnected by C⋯H interactions and weak van der Waals interactions. In KETGID, the 1,2-oxazole and methanone fragments form an almost coplanar unit. The crystal structure features three short intermolecular C—H⋯O contacts involving the methanesulfonyl-O atoms. In LUJKUA, the asymmetric unit contains two distinct molecules, which exhibit differences in conformation resulting from a variation in key torsion angles. These distinctions influence the molecular orientation and intermolecular interactions, with strong N—H⋯N and N—H⋯O hydrogen bonds forming a centrosymmetric tetramer stabilized by π–π stacking.
5. Synthesis and crystallization
Dimethyl but-2-ynedioate (133.2 µL, 1.1 mmol) was added to a solution of N-(furan-2-ylmethyl)-N-[2-(5-methylfuran-2-yl)phenyl]-4-(trifluoromethyl)benzenesulfonamide 2 (100 mg, 0.22 mmol) in o-xylene (5 mL). The mixture was refluxed for 5 h. After cooling of the reaction to r.t, the solvent was evaporated under reduced pressure and the crude product was purified by column chromatography (eluent: from hexane to ethyl acetate). The title compound was obtained as colourless powder, yield 27%, 35 mg (0.059 mmol); m.p. 486–487 K. A single crystal of the title compound was grown from ethanol. IR (KBr), ν (cm−1): 1753 (CO2), 1325 (νas SO2), 1169 (νs SO2). 1H NMR (700.2 MHz, CDCl3) (J, Hz): δ 7.71 (dd, J = 7.6, 1.7, 1H, H Ar), 7.50–7.44 (m, 5H, H Ar), 7.20 (d, J = 8.1, 2H, H Ar), 6.69 (d, J = 7.9, 1H, H Ar), 6.61 (d, J = 8.1, 1H, H Ar), 5.91 (s, 1H, H Ar), 5.15 (d, J = 16.7, 1H, NCH), 4.47 (d, J = 16.7, 1H, NCH), 3.76 (s, 3H, OCH3), 3.47 (s, 3H, OCH3), 2.29 (s, 3H, CH3). 13C{1H} NMR (176.1 MHz, CDCl3): δ there are no signal of CF3 163.1, 162.4, 151.2, 150.6, 145.3, 144.1, 142.7, 137.4, 137.0, 133.3 (q, J = 32.4, 1 C), 132.4, 130.9, 130.0, 129.7, 129.2 (2C), 128.3, 127.9 (2C), 126.1, 124.5 (q, J = 4.1, 2 C), 96.9, 81.3, 54.8, 52.5, 52.2, 17.4. 19F{1H} NMR (658.8 MHz, CDCl3): −63.27. MS (ESI) m/z: [M + H]+ 586. Elemental analysis calculated (%) for C29H22F3NO7S: C 59.49, H 3.79, N 2.39, S 5.48; found: C 59.81, H 3.48, N 2.19, S 5.33.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All C-bound H atoms were positioned geometrically (C—H = 0.95 and 1.00 Å) and refined using a riding model with Uiso(H) = 1.2 or 1.5Ueq(C). The methyl group (C13) attached to the benzene ring was found to be disordered over two positions with a refined occupancy ratio of 0.53 (2): 0.47 (2). A SADI instruction was used to restrain the C3—C13 and C3—C13′ bonds. The anisotropic displacement parameters of both parts of the carbon atom of the disordered methyl group were restrained to be similar with EADP instruction.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C29H22F3NO7S |
| M r | 585.53 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 7.6375 (5), 11.0324 (6), 30.2019 (8) |
| β (°) | 93.983 (1) |
| V (Å3) | 2538.7 (2) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.79 |
| Crystal size (mm) | 0.35 × 0.18 × 0.17 |
| Data collection | |
| Diffractometer | Rigaku XtaLAB Synergy-S, HyPix-6000HE area-detector |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2021 ▸) |
| Tmin, Tmax | 0.713, 0.737 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 31570, 5526, 5187 |
| R int | 0.049 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.037, 0.103, 1.05 |
| No. of reflections | 5526 |
| No. of parameters | 379 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.45, −0.43 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004426/nx2026sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004426/nx2026Isup2.hkl
CCDC reference: 2451675
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
GMB and SA thank to Common Use Center ‘Physical and Chemical Research of New Materials, Substances and Catalytic Systems’. This publication has been supported by the RUDN University Scientific Projects Grant System, project No. 021408–2-000, as well as by the Baku Engineering University (Azerbaijan) and Azerbaijan Medical University. The author’s contributions are as follows. Conceptualization, MA and GMM; synthesis, GMB and SA; AGK NMR analysis; X-ray analysis, VNK, NAG; writing (review and editing of the manuscript) MA and GMM; funding acquisition KIH; supervision, MA and GMM.
supplementary crystallographic information
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Crystal data
| C29H22F3NO7S | F(000) = 1208 |
| Mr = 585.53 | Dx = 1.532 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54184 Å |
| a = 7.6375 (5) Å | Cell parameters from 20964 reflections |
| b = 11.0324 (6) Å | θ = 2.9–79.9° |
| c = 30.2019 (8) Å | µ = 1.79 mm−1 |
| β = 93.983 (1)° | T = 100 K |
| V = 2538.7 (2) Å3 | Prism, colourless |
| Z = 4 | 0.35 × 0.18 × 0.17 mm |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Data collection
| Rigaku XtaLAB Synergy-S, HyPix-6000HE area-detector diffractometer | 5187 reflections with I > 2σ(I) |
| Radiation source: micro-focus sealed X-ray tube | Rint = 0.049 |
| φ and ω scans | θmax = 80.1°, θmin = 2.9° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2021) | h = −9→8 |
| Tmin = 0.713, Tmax = 0.737 | k = −14→13 |
| 31570 measured reflections | l = −38→38 |
| 5526 independent reflections |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.037 | H-atom parameters constrained |
| wR(F2) = 0.103 | w = 1/[σ2(Fo2) + (0.0633P)2 + 0.8141P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 5526 reflections | Δρmax = 0.45 e Å−3 |
| 379 parameters | Δρmin = −0.43 e Å−3 |
| 1 restraint | Extinction correction: SHELXL-2019/2 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: difference Fourier map | Extinction coefficient: 0.00089 (14) |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | −0.08268 (3) | 0.68046 (3) | 0.60331 (2) | 0.01536 (10) | |
| F1 | 0.03672 (12) | 0.08285 (8) | 0.60261 (3) | 0.0324 (2) | |
| F2 | 0.22478 (16) | 0.13678 (9) | 0.55695 (5) | 0.0501 (3) | |
| F3 | 0.29014 (14) | 0.14333 (10) | 0.62756 (5) | 0.0539 (3) | |
| O1 | −0.16062 (12) | 0.70213 (9) | 0.64434 (3) | 0.0216 (2) | |
| O2 | −0.17402 (12) | 0.70996 (9) | 0.56162 (3) | 0.0225 (2) | |
| C1 | 0.48862 (15) | 0.49339 (11) | 0.57852 (4) | 0.0171 (2) | |
| H1 | 0.496038 | 0.479440 | 0.547665 | 0.021* | |
| C2 | 0.55556 (16) | 0.40746 (11) | 0.60859 (4) | 0.0187 (2) | |
| H2 | 0.607100 | 0.336071 | 0.597644 | 0.022* | |
| C3 | 0.54990 (15) | 0.42217 (11) | 0.65483 (4) | 0.0172 (2) | |
| C3A | 0.47063 (15) | 0.52723 (11) | 0.66816 (4) | 0.0156 (2) | |
| C4 | 0.44151 (15) | 0.58212 (12) | 0.71373 (4) | 0.0171 (2) | |
| H4 | 0.444763 | 0.524274 | 0.739264 | 0.021* | |
| C5 | 0.56803 (16) | 0.69088 (11) | 0.71852 (4) | 0.0165 (2) | |
| C6 | 0.50225 (15) | 0.77414 (11) | 0.68956 (4) | 0.0149 (2) | |
| C6A | 0.33411 (15) | 0.71572 (11) | 0.66698 (4) | 0.0141 (2) | |
| C7 | 0.18668 (15) | 0.79745 (11) | 0.64834 (4) | 0.0158 (2) | |
| H7A | 0.096592 | 0.803245 | 0.670287 | 0.019* | |
| H7B | 0.234316 | 0.879823 | 0.644221 | 0.019* | |
| N8 | 0.10246 (13) | 0.75554 (9) | 0.60592 (3) | 0.0148 (2) | |
| C8A | 0.19749 (15) | 0.76703 (11) | 0.56657 (4) | 0.0144 (2) | |
| C9 | 0.14221 (16) | 0.85518 (12) | 0.53567 (4) | 0.0186 (2) | |
| H9 | 0.041411 | 0.902688 | 0.540357 | 0.022* | |
| C10 | 0.23335 (17) | 0.87407 (12) | 0.49810 (4) | 0.0217 (3) | |
| H10 | 0.195141 | 0.934034 | 0.477015 | 0.026* | |
| C11 | 0.38118 (17) | 0.80441 (12) | 0.49158 (4) | 0.0202 (3) | |
| H11 | 0.445235 | 0.817581 | 0.466155 | 0.024* | |
| C12 | 0.43538 (16) | 0.71573 (12) | 0.52212 (4) | 0.0171 (2) | |
| H12 | 0.535760 | 0.668175 | 0.517074 | 0.020* | |
| C12A | 0.34499 (15) | 0.69485 (11) | 0.56029 (4) | 0.0143 (2) | |
| C12B | 0.40963 (14) | 0.60123 (11) | 0.59260 (4) | 0.0144 (2) | |
| C12C | 0.40171 (14) | 0.61400 (11) | 0.63793 (4) | 0.0137 (2) | |
| O13 | 0.27786 (11) | 0.64524 (8) | 0.70381 (3) | 0.01706 (19) | |
| C13 | 0.6235 (19) | 0.3275 (12) | 0.6870 (4) | 0.0238 (3) | 0.53 (2) |
| H13A | 0.732679 | 0.294903 | 0.676576 | 0.036* | 0.53 (2) |
| H13B | 0.538176 | 0.261703 | 0.689067 | 0.036* | 0.53 (2) |
| H13C | 0.647483 | 0.364271 | 0.716356 | 0.036* | 0.53 (2) |
| C13' | 0.625 (2) | 0.3275 (13) | 0.6867 (5) | 0.0238 (3) | 0.47 (2) |
| H13D | 0.747394 | 0.348027 | 0.695799 | 0.036* | 0.47 (2) |
| H13E | 0.621140 | 0.248114 | 0.672104 | 0.036* | 0.47 (2) |
| H13F | 0.556513 | 0.324858 | 0.712874 | 0.036* | 0.47 (2) |
| C14 | 0.74252 (17) | 0.68539 (12) | 0.74318 (4) | 0.0187 (3) | |
| O14 | 0.82551 (14) | 0.77048 (10) | 0.75805 (4) | 0.0298 (2) | |
| O15 | 0.79316 (12) | 0.56938 (9) | 0.74709 (3) | 0.0221 (2) | |
| C15 | 0.96586 (17) | 0.54877 (14) | 0.76935 (5) | 0.0252 (3) | |
| H15A | 1.051895 | 0.601521 | 0.756353 | 0.038* | |
| H15B | 0.999568 | 0.463854 | 0.765660 | 0.038* | |
| H15C | 0.962354 | 0.567077 | 0.801032 | 0.038* | |
| C16 | 0.58807 (15) | 0.88141 (11) | 0.67119 (4) | 0.0160 (2) | |
| O16 | 0.68066 (13) | 0.95432 (9) | 0.69113 (3) | 0.0247 (2) | |
| O17 | 0.54422 (11) | 0.88538 (8) | 0.62730 (3) | 0.01773 (19) | |
| C17 | 0.63324 (19) | 0.97498 (13) | 0.60257 (5) | 0.0244 (3) | |
| H17A | 0.585829 | 0.974193 | 0.571579 | 0.037* | |
| H17B | 0.759006 | 0.956514 | 0.603902 | 0.037* | |
| H17C | 0.615553 | 1.055289 | 0.615390 | 0.037* | |
| C18 | −0.02522 (15) | 0.52580 (11) | 0.60113 (4) | 0.0161 (2) | |
| C19 | −0.01841 (17) | 0.47033 (12) | 0.56005 (4) | 0.0213 (3) | |
| H19 | −0.055255 | 0.512804 | 0.533666 | 0.026* | |
| C20 | 0.04285 (18) | 0.35202 (13) | 0.55788 (5) | 0.0241 (3) | |
| H20 | 0.049166 | 0.312688 | 0.530058 | 0.029* | |
| C21 | 0.09484 (17) | 0.29210 (12) | 0.59721 (5) | 0.0220 (3) | |
| C22 | 0.08252 (17) | 0.34703 (12) | 0.63835 (5) | 0.0224 (3) | |
| H22 | 0.115329 | 0.303757 | 0.664827 | 0.027* | |
| C23 | 0.02216 (17) | 0.46516 (12) | 0.64049 (4) | 0.0195 (3) | |
| H23 | 0.013327 | 0.503979 | 0.668321 | 0.023* | |
| C24 | 0.16202 (19) | 0.16488 (13) | 0.59579 (6) | 0.0296 (3) |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.01088 (15) | 0.01765 (16) | 0.01767 (16) | 0.00027 (9) | 0.00183 (11) | 0.00049 (10) |
| F1 | 0.0335 (5) | 0.0180 (4) | 0.0464 (5) | −0.0065 (3) | 0.0085 (4) | −0.0001 (4) |
| F2 | 0.0591 (7) | 0.0245 (5) | 0.0717 (8) | 0.0035 (4) | 0.0407 (6) | −0.0050 (5) |
| F3 | 0.0374 (5) | 0.0263 (5) | 0.0941 (10) | 0.0095 (4) | −0.0236 (6) | −0.0078 (5) |
| O1 | 0.0165 (4) | 0.0240 (5) | 0.0252 (5) | 0.0012 (3) | 0.0086 (4) | −0.0018 (4) |
| O2 | 0.0159 (4) | 0.0257 (5) | 0.0250 (5) | −0.0014 (3) | −0.0049 (3) | 0.0034 (4) |
| C1 | 0.0160 (5) | 0.0185 (6) | 0.0170 (5) | −0.0007 (4) | 0.0020 (4) | −0.0035 (4) |
| C2 | 0.0174 (5) | 0.0157 (5) | 0.0231 (6) | 0.0007 (4) | 0.0027 (4) | −0.0026 (4) |
| C3 | 0.0137 (5) | 0.0166 (6) | 0.0213 (6) | −0.0015 (4) | 0.0013 (4) | 0.0031 (4) |
| C3A | 0.0129 (5) | 0.0186 (6) | 0.0154 (5) | −0.0025 (4) | 0.0017 (4) | 0.0016 (4) |
| C4 | 0.0153 (5) | 0.0215 (6) | 0.0148 (5) | 0.0005 (4) | 0.0025 (4) | 0.0028 (4) |
| C5 | 0.0169 (6) | 0.0218 (6) | 0.0110 (5) | 0.0010 (4) | 0.0028 (4) | −0.0018 (4) |
| C6 | 0.0143 (5) | 0.0193 (6) | 0.0114 (5) | 0.0005 (4) | 0.0023 (4) | −0.0036 (4) |
| C6A | 0.0137 (5) | 0.0174 (5) | 0.0116 (5) | −0.0014 (4) | 0.0035 (4) | 0.0002 (4) |
| C7 | 0.0142 (5) | 0.0178 (5) | 0.0155 (5) | 0.0011 (4) | 0.0013 (4) | −0.0029 (4) |
| N8 | 0.0123 (4) | 0.0175 (5) | 0.0144 (5) | −0.0002 (4) | 0.0012 (4) | −0.0006 (4) |
| C8A | 0.0133 (5) | 0.0157 (5) | 0.0141 (5) | −0.0026 (4) | 0.0011 (4) | −0.0011 (4) |
| C9 | 0.0182 (5) | 0.0183 (6) | 0.0192 (6) | 0.0011 (4) | 0.0003 (4) | 0.0015 (5) |
| C10 | 0.0244 (6) | 0.0218 (6) | 0.0188 (6) | −0.0007 (5) | 0.0002 (5) | 0.0054 (5) |
| C11 | 0.0210 (6) | 0.0258 (6) | 0.0141 (5) | −0.0032 (5) | 0.0033 (4) | 0.0017 (5) |
| C12 | 0.0150 (5) | 0.0217 (6) | 0.0146 (5) | −0.0011 (4) | 0.0011 (4) | −0.0027 (4) |
| C12A | 0.0139 (5) | 0.0161 (5) | 0.0128 (5) | −0.0021 (4) | −0.0010 (4) | −0.0022 (4) |
| C12B | 0.0117 (5) | 0.0165 (5) | 0.0151 (5) | −0.0014 (4) | 0.0012 (4) | −0.0010 (4) |
| C12C | 0.0108 (5) | 0.0148 (5) | 0.0155 (5) | −0.0007 (4) | 0.0018 (4) | −0.0009 (4) |
| O13 | 0.0149 (4) | 0.0225 (4) | 0.0142 (4) | 0.0008 (3) | 0.0043 (3) | 0.0036 (3) |
| C13 | 0.0248 (7) | 0.0199 (6) | 0.0267 (7) | 0.0026 (5) | 0.0027 (6) | 0.0074 (5) |
| C13' | 0.0248 (7) | 0.0199 (6) | 0.0267 (7) | 0.0026 (5) | 0.0027 (6) | 0.0074 (5) |
| C14 | 0.0192 (6) | 0.0258 (6) | 0.0112 (5) | 0.0013 (5) | 0.0007 (4) | 0.0012 (4) |
| O14 | 0.0306 (5) | 0.0287 (5) | 0.0282 (5) | −0.0025 (4) | −0.0117 (4) | −0.0016 (4) |
| O15 | 0.0185 (4) | 0.0259 (5) | 0.0216 (4) | 0.0025 (4) | −0.0020 (3) | 0.0031 (4) |
| C15 | 0.0177 (6) | 0.0360 (7) | 0.0217 (6) | 0.0042 (5) | −0.0010 (5) | 0.0066 (5) |
| C16 | 0.0151 (5) | 0.0168 (5) | 0.0164 (5) | 0.0021 (4) | 0.0024 (4) | −0.0020 (4) |
| O16 | 0.0266 (5) | 0.0233 (5) | 0.0236 (5) | −0.0069 (4) | −0.0016 (4) | −0.0053 (4) |
| O17 | 0.0190 (4) | 0.0192 (4) | 0.0150 (4) | −0.0035 (3) | 0.0011 (3) | 0.0015 (3) |
| C17 | 0.0274 (6) | 0.0215 (6) | 0.0246 (6) | −0.0037 (5) | 0.0051 (5) | 0.0066 (5) |
| C18 | 0.0127 (5) | 0.0180 (6) | 0.0179 (6) | −0.0022 (4) | 0.0028 (4) | −0.0001 (4) |
| C19 | 0.0243 (6) | 0.0231 (6) | 0.0167 (6) | −0.0038 (5) | 0.0035 (5) | −0.0010 (5) |
| C20 | 0.0276 (7) | 0.0220 (6) | 0.0235 (6) | −0.0045 (5) | 0.0069 (5) | −0.0050 (5) |
| C21 | 0.0169 (6) | 0.0171 (6) | 0.0325 (7) | −0.0035 (5) | 0.0045 (5) | −0.0018 (5) |
| C22 | 0.0218 (6) | 0.0198 (6) | 0.0250 (6) | −0.0031 (5) | −0.0016 (5) | 0.0030 (5) |
| C23 | 0.0210 (6) | 0.0199 (6) | 0.0175 (6) | −0.0025 (5) | 0.0010 (4) | −0.0003 (4) |
| C24 | 0.0227 (7) | 0.0199 (6) | 0.0469 (9) | −0.0026 (5) | 0.0067 (6) | −0.0021 (6) |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Geometric parameters (Å, º)
| S1—O1 | 1.4314 (9) | C10—H10 | 0.9500 |
| S1—O2 | 1.4338 (10) | C11—C12 | 1.3877 (18) |
| S1—N8 | 1.6359 (10) | C11—H11 | 0.9500 |
| S1—C18 | 1.7642 (13) | C12—C12A | 1.4034 (16) |
| F1—C24 | 1.3434 (17) | C12—H12 | 0.9500 |
| F2—C24 | 1.334 (2) | C12A—C12B | 1.4819 (17) |
| F3—C24 | 1.343 (2) | C12B—C12C | 1.3816 (16) |
| C1—C2 | 1.3857 (18) | C13—H13A | 0.9800 |
| C1—C12B | 1.4129 (17) | C13—H13B | 0.9800 |
| C1—H1 | 0.9500 | C13—H13C | 0.9800 |
| C2—C3 | 1.4096 (18) | C13'—H13D | 0.9800 |
| C2—H2 | 0.9500 | C13'—H13E | 0.9800 |
| C3—C3A | 1.3805 (17) | C13'—H13F | 0.9800 |
| C3—C13' | 1.507 (10) | C14—O14 | 1.2022 (18) |
| C3—C13 | 1.509 (9) | C14—O15 | 1.3400 (17) |
| C3A—C12C | 1.4001 (17) | O15—C15 | 1.4562 (15) |
| C3A—C4 | 1.5340 (17) | C15—H15A | 0.9800 |
| C4—O13 | 1.4439 (15) | C15—H15B | 0.9800 |
| C4—C5 | 1.5411 (17) | C15—H15C | 0.9800 |
| C4—H4 | 1.0000 | C16—O16 | 1.2045 (16) |
| C5—C6 | 1.3416 (18) | C16—O17 | 1.3452 (15) |
| C5—C14 | 1.4820 (17) | O17—C17 | 1.4378 (15) |
| C6—C16 | 1.4795 (17) | C17—H17A | 0.9800 |
| C6—C6A | 1.5517 (16) | C17—H17B | 0.9800 |
| C6A—O13 | 1.4466 (13) | C17—H17C | 0.9800 |
| C6A—C7 | 1.5198 (16) | C18—C19 | 1.3876 (17) |
| C6A—C12C | 1.5358 (16) | C18—C23 | 1.3899 (18) |
| C7—N8 | 1.4680 (15) | C19—C20 | 1.390 (2) |
| C7—H7A | 0.9900 | C19—H19 | 0.9500 |
| C7—H7B | 0.9900 | C20—C21 | 1.393 (2) |
| N8—C8A | 1.4405 (15) | C20—H20 | 0.9500 |
| C8A—C9 | 1.3928 (17) | C21—C22 | 1.391 (2) |
| C8A—C12A | 1.4031 (17) | C21—C24 | 1.4961 (19) |
| C9—C10 | 1.3877 (18) | C22—C23 | 1.3854 (19) |
| C9—H9 | 0.9500 | C22—H22 | 0.9500 |
| C10—C11 | 1.3910 (19) | C23—H23 | 0.9500 |
| O1—S1—O2 | 121.06 (6) | C12—C12A—C12B | 119.67 (11) |
| O1—S1—N8 | 106.49 (6) | C12C—C12B—C1 | 115.67 (11) |
| O2—S1—N8 | 107.06 (5) | C12C—C12B—C12A | 123.09 (11) |
| O1—S1—C18 | 108.27 (6) | C1—C12B—C12A | 121.22 (11) |
| O2—S1—C18 | 107.08 (6) | C12B—C12C—C3A | 122.41 (11) |
| N8—S1—C18 | 105.97 (5) | C12B—C12C—C6A | 132.85 (11) |
| C2—C1—C12B | 121.64 (11) | C3A—C12C—C6A | 104.66 (10) |
| C2—C1—H1 | 119.2 | C4—O13—C6A | 96.88 (8) |
| C12B—C1—H1 | 119.2 | C3—C13—H13A | 109.5 |
| C1—C2—C3 | 122.36 (11) | C3—C13—H13B | 109.5 |
| C1—C2—H2 | 118.8 | H13A—C13—H13B | 109.5 |
| C3—C2—H2 | 118.8 | C3—C13—H13C | 109.5 |
| C3A—C3—C2 | 115.46 (11) | H13A—C13—H13C | 109.5 |
| C3A—C3—C13' | 123.5 (7) | H13B—C13—H13C | 109.5 |
| C2—C3—C13' | 121.1 (7) | C3—C13'—H13D | 109.5 |
| C3A—C3—C13 | 123.0 (6) | C3—C13'—H13E | 109.5 |
| C2—C3—C13 | 121.5 (6) | H13D—C13'—H13E | 109.5 |
| C3—C3A—C12C | 122.45 (11) | C3—C13'—H13F | 109.5 |
| C3—C3A—C4 | 133.39 (11) | H13D—C13'—H13F | 109.5 |
| C12C—C3A—C4 | 104.11 (10) | H13E—C13'—H13F | 109.5 |
| O13—C4—C3A | 100.46 (9) | O14—C14—O15 | 124.85 (12) |
| O13—C4—C5 | 99.91 (10) | O14—C14—C5 | 126.04 (12) |
| C3A—C4—C5 | 105.22 (9) | O15—C14—C5 | 109.10 (11) |
| O13—C4—H4 | 116.3 | C14—O15—C15 | 115.88 (11) |
| C3A—C4—H4 | 116.3 | O15—C15—H15A | 109.5 |
| C5—C4—H4 | 116.3 | O15—C15—H15B | 109.5 |
| C6—C5—C14 | 129.68 (12) | H15A—C15—H15B | 109.5 |
| C6—C5—C4 | 105.54 (11) | O15—C15—H15C | 109.5 |
| C14—C5—C4 | 123.47 (11) | H15A—C15—H15C | 109.5 |
| C5—C6—C16 | 129.54 (11) | H15B—C15—H15C | 109.5 |
| C5—C6—C6A | 105.26 (10) | O16—C16—O17 | 124.64 (12) |
| C16—C6—C6A | 122.82 (10) | O16—C16—C6 | 127.35 (12) |
| O13—C6A—C7 | 110.59 (9) | O17—C16—C6 | 108.01 (10) |
| O13—C6A—C12C | 100.14 (9) | C16—O17—C17 | 116.09 (10) |
| C7—C6A—C12C | 119.46 (10) | O17—C17—H17A | 109.5 |
| O13—C6A—C6 | 99.57 (9) | O17—C17—H17B | 109.5 |
| C7—C6A—C6 | 119.05 (10) | H17A—C17—H17B | 109.5 |
| C12C—C6A—C6 | 104.71 (9) | O17—C17—H17C | 109.5 |
| N8—C7—C6A | 113.89 (10) | H17A—C17—H17C | 109.5 |
| N8—C7—H7A | 108.8 | H17B—C17—H17C | 109.5 |
| C6A—C7—H7A | 108.8 | C19—C18—C23 | 121.92 (12) |
| N8—C7—H7B | 108.8 | C19—C18—S1 | 119.01 (10) |
| C6A—C7—H7B | 108.8 | C23—C18—S1 | 118.95 (10) |
| H7A—C7—H7B | 107.7 | C18—C19—C20 | 119.34 (12) |
| C8A—N8—C7 | 118.49 (9) | C18—C19—H19 | 120.3 |
| C8A—N8—S1 | 119.23 (8) | C20—C19—H19 | 120.3 |
| C7—N8—S1 | 121.75 (8) | C19—C20—C21 | 118.86 (12) |
| C9—C8A—C12A | 120.98 (11) | C19—C20—H20 | 120.6 |
| C9—C8A—N8 | 117.87 (11) | C21—C20—H20 | 120.6 |
| C12A—C8A—N8 | 121.12 (10) | C22—C21—C20 | 121.45 (13) |
| C10—C9—C8A | 120.45 (12) | C22—C21—C24 | 118.61 (13) |
| C10—C9—H9 | 119.8 | C20—C21—C24 | 119.93 (13) |
| C8A—C9—H9 | 119.8 | C23—C22—C21 | 119.65 (12) |
| C9—C10—C11 | 119.40 (12) | C23—C22—H22 | 120.2 |
| C9—C10—H10 | 120.3 | C21—C22—H22 | 120.2 |
| C11—C10—H10 | 120.3 | C22—C23—C18 | 118.72 (12) |
| C12—C11—C10 | 120.20 (12) | C22—C23—H23 | 120.6 |
| C12—C11—H11 | 119.9 | C18—C23—H23 | 120.6 |
| C10—C11—H11 | 119.9 | F2—C24—F3 | 107.37 (13) |
| C11—C12—C12A | 121.37 (11) | F2—C24—F1 | 106.36 (13) |
| C11—C12—H12 | 119.3 | F3—C24—F1 | 105.22 (13) |
| C12A—C12—H12 | 119.3 | F2—C24—C21 | 112.84 (13) |
| C8A—C12A—C12 | 117.59 (11) | F3—C24—C21 | 112.32 (13) |
| C8A—C12A—C12B | 122.72 (10) | F1—C24—C21 | 112.23 (11) |
| C12B—C1—C2—C3 | −0.27 (19) | C12—C12A—C12B—C12C | 143.46 (12) |
| C1—C2—C3—C3A | 0.97 (18) | C8A—C12A—C12B—C1 | 146.80 (12) |
| C1—C2—C3—C13' | −179.4 (8) | C12—C12A—C12B—C1 | −34.76 (17) |
| C1—C2—C3—C13 | 180.0 (7) | C1—C12B—C12C—C3A | 1.32 (17) |
| C2—C3—C3A—C12C | −0.53 (17) | C12A—C12B—C12C—C3A | −176.99 (11) |
| C13'—C3—C3A—C12C | 179.9 (8) | C1—C12B—C12C—C6A | 177.79 (11) |
| C13—C3—C3A—C12C | −179.5 (7) | C12A—C12B—C12C—C6A | −0.5 (2) |
| C2—C3—C3A—C4 | −177.38 (12) | C3—C3A—C12C—C12B | −0.65 (18) |
| C13'—C3—C3A—C4 | 3.0 (8) | C4—C3A—C12C—C12B | 177.00 (11) |
| C13—C3—C3A—C4 | 3.6 (7) | C3—C3A—C12C—C6A | −177.97 (11) |
| C3—C3A—C4—O13 | −148.68 (13) | C4—C3A—C12C—C6A | −0.32 (11) |
| C12C—C3A—C4—O13 | 34.06 (11) | O13—C6A—C12C—C12B | 149.70 (12) |
| C3—C3A—C4—C5 | 107.93 (15) | C7—C6A—C12C—C12B | 28.95 (18) |
| C12C—C3A—C4—C5 | −69.34 (11) | C6—C6A—C12C—C12B | −107.51 (14) |
| O13—C4—C5—C6 | −33.53 (11) | O13—C6A—C12C—C3A | −33.39 (11) |
| C3A—C4—C5—C6 | 70.27 (12) | C7—C6A—C12C—C3A | −154.14 (10) |
| O13—C4—C5—C14 | 158.45 (10) | C6—C6A—C12C—C3A | 69.41 (11) |
| C3A—C4—C5—C14 | −97.75 (13) | C3A—C4—O13—C6A | −54.37 (10) |
| C14—C5—C6—C16 | 4.6 (2) | C5—C4—O13—C6A | 53.28 (10) |
| C4—C5—C6—C16 | −162.42 (11) | C7—C6A—O13—C4 | −179.14 (10) |
| C14—C5—C6—C6A | 167.07 (12) | C12C—C6A—O13—C4 | 53.93 (10) |
| C4—C5—C6—C6A | 0.07 (12) | C6—C6A—O13—C4 | −53.03 (10) |
| C5—C6—C6A—O13 | 33.30 (11) | C6—C5—C14—O14 | 36.1 (2) |
| C16—C6—C6A—O13 | −162.73 (10) | C4—C5—C14—O14 | −158.94 (13) |
| C5—C6—C6A—C7 | 153.40 (10) | C6—C5—C14—O15 | −144.93 (13) |
| C16—C6—C6A—C7 | −42.62 (15) | C4—C5—C14—O15 | 20.01 (16) |
| C5—C6—C6A—C12C | −69.93 (11) | O14—C14—O15—C15 | −3.06 (18) |
| C16—C6—C6A—C12C | 94.05 (12) | C5—C14—O15—C15 | 177.98 (10) |
| O13—C6A—C7—N8 | −105.16 (11) | C5—C6—C16—O16 | −44.8 (2) |
| C12C—C6A—C7—N8 | 10.18 (15) | C6A—C6—C16—O16 | 155.42 (12) |
| C6—C6A—C7—N8 | 140.52 (10) | C5—C6—C16—O17 | 135.61 (13) |
| C6A—C7—N8—C8A | −72.75 (13) | C6A—C6—C16—O17 | −24.19 (14) |
| C6A—C7—N8—S1 | 98.88 (11) | O16—C16—O17—C17 | 8.00 (17) |
| O1—S1—N8—C8A | −170.14 (9) | C6—C16—O17—C17 | −172.38 (10) |
| O2—S1—N8—C8A | −39.32 (10) | O1—S1—C18—C19 | 152.89 (10) |
| C18—S1—N8—C8A | 74.73 (10) | O2—S1—C18—C19 | 20.83 (11) |
| O1—S1—N8—C7 | 18.29 (11) | N8—S1—C18—C19 | −93.20 (11) |
| O2—S1—N8—C7 | 149.11 (9) | O1—S1—C18—C23 | −30.97 (11) |
| C18—S1—N8—C7 | −96.85 (10) | O2—S1—C18—C23 | −163.03 (10) |
| C7—N8—C8A—C9 | −107.68 (13) | N8—S1—C18—C23 | 82.94 (11) |
| S1—N8—C8A—C9 | 80.47 (13) | C23—C18—C19—C20 | −2.14 (19) |
| C7—N8—C8A—C12A | 70.01 (15) | S1—C18—C19—C20 | 173.88 (10) |
| S1—N8—C8A—C12A | −101.83 (12) | C18—C19—C20—C21 | 0.4 (2) |
| C12A—C8A—C9—C10 | −0.56 (19) | C19—C20—C21—C22 | 1.6 (2) |
| N8—C8A—C9—C10 | 177.14 (11) | C19—C20—C21—C24 | −179.74 (12) |
| C8A—C9—C10—C11 | −0.2 (2) | C20—C21—C22—C23 | −1.9 (2) |
| C9—C10—C11—C12 | 0.9 (2) | C24—C21—C22—C23 | 179.43 (12) |
| C10—C11—C12—C12A | −0.7 (2) | C21—C22—C23—C18 | 0.18 (19) |
| C9—C8A—C12A—C12 | 0.70 (17) | C19—C18—C23—C22 | 1.85 (19) |
| N8—C8A—C12A—C12 | −176.93 (10) | S1—C18—C23—C22 | −174.17 (9) |
| C9—C8A—C12A—C12B | 179.17 (11) | C22—C21—C24—F2 | −158.19 (13) |
| N8—C8A—C12A—C12B | 1.54 (17) | C20—C21—C24—F2 | 23.13 (19) |
| C11—C12—C12A—C8A | −0.05 (18) | C22—C21—C24—F3 | −36.65 (18) |
| C11—C12—C12A—C12B | −178.57 (11) | C20—C21—C24—F3 | 144.67 (14) |
| C2—C1—C12B—C12C | −0.87 (17) | C22—C21—C24—F1 | 81.66 (17) |
| C2—C1—C12B—C12A | 177.47 (11) | C20—C21—C24—F1 | −97.02 (16) |
| C8A—C12A—C12B—C12C | −34.98 (17) |
Dimethyl 3-methyl-8-{[4-(trifluoromethyl)phenyl]sulfonyl}-7,8-dihydro-4H-4,6a-epoxybenzo[b]naphtho[1,8-de]azepine-5,6-dicarboxylate . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C4—H4···O16i | 1.00 | 2.49 | 3.3893 (16) | 149 |
| C7—H7A···O1 | 0.99 | 2.35 | 2.8481 (15) | 111 |
| C7—H7B···O17 | 0.99 | 2.46 | 3.0071 (14) | 115 |
| C12—H12···O2ii | 0.95 | 2.55 | 3.1357 (15) | 120 |
| C15—H15A···O13ii | 0.98 | 2.47 | 3.3741 (17) | 153 |
| C17—H17B···F1iii | 0.98 | 2.54 | 3.3033 (17) | 135 |
| C19—H19···O2 | 0.95 | 2.52 | 2.9003 (17) | 104 |
| C22—H22···O14i | 0.95 | 2.37 | 3.2698 (19) | 158 |
Symmetry codes: (i) −x+1, y−1/2, −z+3/2; (ii) x+1, y, z; (iii) x+1, y+1, z.
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Borisova, K., Nikitina, E., Novikov, R., Khrustalev, V., Dorovatovskii, P., Zubavichus, Y., Kuznetsov, M., Zaytsev, V., Varlamov, A. & Zubkov, F. (2018a). Chem. Commun.54, 2850–2853. [DOI] [PubMed]
- Borisova, K. K., Kvyatkovskaya, E. A., Nikitina, E. V., Aysin, R. R., Novikov, R. A. & Zubkov, F. I. (2018b). J. Org. Chem.83, 4840–4850. [DOI] [PubMed]
- Burkin, G. M., Kvyatkovskaya, E. A., Khrustalev, V. N., Hasanov, K. I., Sadikhova, N. D., Akkurt, M. & Bhattarai, A. (2024). Acta Cryst. E80, 418–422. [DOI] [PMC free article] [PubMed]
- Chakrabarti, S., Liu, M., Waldeck, D. H., Oliver, A. M. & Paddon-Row, M. N. (2007). J. Am. Chem. Soc.129, 3247–3256. [DOI] [PubMed]
- Chou, T.-C. & Li, Y.-J. (2015). Tetrahedron71, 5620–5633.
- Criado, A., Vilas-Varela, M., Cobas, A., Peréz, D., Pena, D. & Guitián, E. (2013). J. Org. Chem.78, 12637–12649. [DOI] [PubMed]
- Eckert-Maksić, M., Margetić, D., Kirin, S., Milić, D. & Matković-Čalogović, D. (2005). Eur. J. Org. Chem. pp. 4612–4620.
- Eda, S., Eguchi, F., Haneda, H. & Hamura, T. (2015). Chem. Commun.51, 5963–5966. [DOI] [PubMed]
- Enev, V. S., Felzmann, W., Gromov, A., Marchart, S. & Mulzer, J. (2012). Chem. Eur. J.18, 9651–9668. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst.45, 849–854.
- Furrer, F., Linden, A. & Stuparu, M. C. (2013). Chem. Eur. J.19, 13199–13206. [DOI] [PubMed]
- Gromov, A., Enev, V. & Mulzer, J. (2009). Org. Lett.11, 2884–2886. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Guliyeva, N. A., Burkin, G. M., Annadurdyyeva, S., Khrustalev, V. N., Atioğlu, Z., Akkurt, M. & Bhattarai, A. (2024). Acta Cryst. E80, 62–66. [DOI] [PMC free article] [PubMed]
- Mammadova, G. Z., Annadurdyyeva, S., Burkin, G. M., Khrustalev, V. N., Akkurt, M., Yıldırım, S. Ö. & Bhattarai, A. (2023b). Acta Cryst. E79, 499–503. [DOI] [PMC free article] [PubMed]
- Mammadova, G. Z., Yakovleva, E. D., Burkin, G. M., Khrustalev, V. N., Akkurt, M., Çelikesir, S. T. & Bhattarai, A. (2023a). Acta Cryst. E79, 747–751. [DOI] [PMC free article] [PubMed]
- Margetić, D., Eckert-Maksić, M., Trošelj, P. & Marinić, Z. (2010). J. Fluorine Chem.131, 408–416.
- Murphy, R. B., Norman, R. E., White, J. M., Perkins, M. V. & Johnston, M. R. (2016). Org. Biomol. Chem.14, 8707–8720. [DOI] [PubMed]
- Oh, C. H., Yi, H. J. & Lee, K. H. (2010). Bull. Korean Chem. Soc.31, 683–688.
- Rigaku OD (2021). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Roscalesa, S. & Plumet, J. (2017). Nat. Prod. Commun.12, 713–732. [PubMed]
- Sadikhova, N. D., Atioğlu, Z., Guliyeva, N. A., Podrezova, A. G., Nikitina, E. V., Akkurt, M. & Bhattarai, A. (2024). Acta Cryst. E80, 83–87. [DOI] [PMC free article] [PubMed]
- Schindler, C. S. & Carreira, E. M. (2009). Chem. Soc. Rev.38, 3222–3241. [DOI] [PubMed]
- Schinke, J., Gelbrich, T. & Griesser, U. J. (2022). Acta Cryst. E78, 979–983. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst.54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Vinaya, Yakuth, S. A., Mohan Kumar, T. M., Bhaskar, B. L., Divakara, T. R., Yathirajan, H. S., Basavaraju, Y. B. & Parkin, S. (2024). Acta Cryst. E80, 1354–1358. [DOI] [PMC free article] [PubMed]
- Vogel, P., Cossy, J., Plumet, J. & Arjona, O. (1999). Tetrahedron55, 13521–13642.
- Warrener, R. N., Margetic, D., Amarasekara, A. S., Butler, D. N., Mahadevan, I. B. & Russell, R. A. (1999). Org. Lett.1, 199–202.
- Warrener, R. N., Margetić, D., Foley, P. J., Butler, D. N., Winling, A., Beales, K. A. & Russell, R. A. (2001). Tetrahedron57, 571–582.
- Zaytsev, V. P., Mertsalov, D. F., Chervyakova, L. V., Krishna, G., Zubkov, F. I., Dorovatovskii, P. V., Khrustalev, V. N. & Zarubaev, V. V. (2019). Tetrahedron Lett.60, 151204.
- Zubkov, F. I., Airiyan, I. K., Ershova, J. D., Galeev, T. R., Zaytsev, V. P., Nikitina, E. V. & Varlamov, A. V. (2012b). RSC Adv.2, 4103–4109.
- Zubkov, F. I., Ershova, J. D., Orlova, A. A., Zaytsev, V. P., Nikitina, E. V., Peregudov, A. S., Gurbanov, A. V., Borisov, R. S., Khrustalev, V. N., Maharramov, A. M. & Varlamov, A. V. (2009). Tetrahedron65, 3789–3803.
- Zubkov, F. I., Zaytsev, V. P., Puzikova, E. S., Nikitina, E. V., Khrustalev, V. N., Novikov, R. A. & Varlamov, A. V. (2012a). Chem. Heterocycl. Compd.48, 514–524.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989025004426/nx2026sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989025004426/nx2026Isup2.hkl
CCDC reference: 2451675
Additional supporting information: crystallographic information; 3D view; checkCIF report









