Abstract
IME1 encodes a transcriptional activator required for the transcription of meiosis-specific genes and initiation of meiosis in Saccharomyces cerevisiae. The transcription of IME1 is repressed in the presence of glucose, and a low basal level of IME1 RNA is observed in vegetative cultures with acetate as the sole carbon source. Upon nitrogen depletion a transient induction in the transcription of IME1 is observed in MATa/MATα diploids but not in MAT-insufficient strains. In this study we demonstrate that the transcription of IME1 is controlled by an extremely unusual large 5′ region, over 2,100 bp long. This area is divided into four different upstream controlling sequences (UCS). UCS2 promotes the transcription of IME1 in the presence of a nonfermentable carbon source. UCS2 is flanked by three negative regions: UCS1, which exhibits URS activity in the presence of nitrogen, and UCS3 and UCS4, which repress the activity of UCS2 in MAT-insufficient cells. UCS2 consists of alternate positive and negative elements: three distinct constitutive URS elements that prevent the function of any upstream activating sequence (UAS) under all growth conditions, a constitutive UAS element that promotes expression under all growth conditions, a UAS element that is active only in vegetative media, and two discrete elements that function as UASs in the presence of acetate. Sequence analysis of IME1 revealed the presence of two almost identical 30- to 32-bp repeats. Surprisingly, one repeat, IREd, exhibits constitutive URS activity, whereas the other repeat, IREu, serves as a carbon-source-regulated UAS element. The RAS-cyclic AMP-dependent protein kinase cAPK pathway prevents the UAS activity of IREu in the presence of glucose as the sole carbon source, while the transcriptional activators Msn2p and Msn4p promote the UAS activity of this repeat in the presence of acetate. We suggest that the use of multiple negative and positive elements is essential to restrict transcription to the appropriate conditions and that the combinatorial effect of the entire region leads to the regulated transcription of IME1.
In the budding yeast Saccharomyces cerevisiae the choice between meiosis-sporulation and alternative developmental pathways such as the mitotic cell cycle, pseudohyphae growth, or G1 arrest depends on the expression and activity of a master regulator, Ime1p. This is deduced from the observation that cells deleted for IME1 are sporulation deficient and arrest in meiosis at G1 prior to any meiotic event, i.e., transcription of meiosis-specific genes, premeiotic DNA replication, meiotic recombination, and nuclear divisions (15, 49). IME1 encodes a transcriptional activator (23, 48) that is recruited to the promoters of early meiosis-specific genes by interacting with a sequence-specific DNA binding protein, Ume6p (41).
Initiation of meiosis depends on two signals: starvation for nutrients and the presence of MATa1 and MATα2 gene products (17). The nutrient signal is required at several levels: for the transcription of IME1 (15), for the translation of IME1 mRNA (47), for the association of Ime1p with its meiotic target, Ume6p (41), and for entry into the first meiotic division (21). The MAT signal is also required in more than one step: for the transcription of IME1 and for efficient meiosis (15, 47). The purpose of this study has been to identify the elements in IME1 that are required for its regulated transcription and to determine the role of the RAS-cyclic AMP-dependent protein kinase (cAPK) pathway in the activity of the regulated upstream activating sequence (UAS) elements. Therefore, we shall summarize below the known information concerning the transcription of IME1.
Transcripts of IME1 are not detected in the presence of glucose, and a low basal level is present in vegetative acetate media (15). Upon nitrogen depletion the level of IME1 mRNA increases in MATa/MATα diploids, reaching a peak at about 6 to 8 h and then declining (15, 49). The transcription of IME1 is not induced in cells that do not carry both the MATa1 and MATα2 alleles (MAT-insufficient cells) (15, 49).
Very little is known about the organization of the IME1 locus. The sequence of IME1 identifies three putative TATA boxes: TATATTA at −353, TATTTAA at −330, and TATAAAT at −158. Deletions of these TATA boxes revealed that the functional TATA is at −330 (1). Accordingly, the main transcription initiation site of IME1 RNA was mapped to −229 (1, 47). The complete genomic sequence of S. cerevisiae reveals that upstream of IME1 there is an extremely large region, 4,122 bp long, that is devoid of open reading frames, tRNA, or rRNA. This suggests the possibility that a large region may be involved in the transcriptional regulation of IME1. Indeed, previous reports have pointed to this phenomenon (5, 12). Covitz and Mitchell reported that the region between −2243 and −1743 upstream of IME1 ATG carries a negative element that prevents the expression of IME1 in MATa/MATa cells (5). Furthermore, a 21-bp element (RRE) located at base pair −2024 to −2044 binds Rme1p (5), a zinc finger protein that represses the transcription of IME1 in MAT-insufficient cells (15, 16, 37). The regulated region may extend even further, since multiple copies of IME1 sequences from −3166 to −3762 promote sporulation in both the presence of nutrients and in MATa/MATa diploids (12).
Except for Rme1p, the transcriptional activators and repressors that directly affect the transcription of IME1 are unknown. Nonetheless, several genes that affect the transcription of IME1 have been identified. IME4 encodes a positive regulator that is absolutely required for the transcription of IME1 (44). The transcription of IME4 is induced only in MATa/MATα diploids that are shifted to nitrogen-depleted medium (44), suggesting that Ime4p transmits both MAT and nitrogen signals. IME4 does not encode a DNA binding protein, and its mode of action is not known. The third gene that mediates MAT regulation to IME1 is RES1; a dominant mutation, RES1-1, promotes sporulation of MAT-insufficient diploids (14). RES1 has yet to be cloned, but epistasis tests suggest that it acts in a pathway distinct from either Ime4p or Rme1p (14, 44). The nitrogen depletion signal seems to be transmitted to IME1 via the RAS-cAPK pathway: mutations that cause lower activity of cAPK, such as cdc25, ras2, and cyr1, lead to the expression of IME1 and to meiosis in the presence of nitrogen (references 26, 27, and 49 and references therein). On the other hand, mutations that cause constitutive activity of cAPK, such as RAS2-val19 and bcy1, are sporulation deficient and are suppressed by overexpression of IME1 (27). Mutations in several genes lower the level of IME1 RNA; these include the serine-threonine protein kinase MCK1 (34) and the DNA binding protein RIM1 (50) and its proteolytic cleavage regulators RIM8, RIM9, and RIM13 (22, 51).
In this paper we report a systematic analysis of the 5′ untranslated region of IME1 and identify the elements that are required for its regulated transcription. We show that IME1 is regulated by an unusually large region that is composed of alternate negative and positive elements. Our analysis reveals the presence of distinct elements responding to MAT, carbon, and nitrogen regulation. We demonstrate that the RAS-cAPK pathway transmits a glucose signal to one of the regulated UAS elements, IREu. Moreover, gel-shift and expression assays show that Msn2p and Msn4p (Msn2/4p) function as the transcription factors mediating the UAS activity of IREu in the presence of acetate as the sole carbon source.
MATERIALS AND METHODS
Plasmids.
Since in many cases multiple steps were involved in plasmid construction, here we describe only the structure of the plasmids, and details are available upon request. YEp1636 carries MSN2 on a 2μm URA3 vector. In this paper we have used two types of single-copy shuttle vectors: ARS CEN and 2μm CEN. As previously reported, both vectors are relatively stable and are maintained at a copy number of about one per genome (53).
(i) IME1.
The following plasmids carry the entire IME1 gene with various portions of its 5′ region on the yeast shuttle vector YCp50 (40): YCp51, IME1 (−621 to +2132); YCp214, IME1 (−4401 to +2132); YCpAS169, IME1 (−915 to +2132); and YCpAS210, IME1 (−1444 to +2132). The following plasmids carry the entire IME1 gene with various portions of its 5′ region on the yeast shuttle vector YCpLac33 (10): YCpAS174, IME1 (−1641 to +2132); YCp1348, IME1 (−1369 to +2132); and YCp1704, IME1 (−2112 to +2132). The following plasmids carry portions of IME1 on pUC119: YIpAS360, IME1 (−4401 to +2132 with a deletion between −1641 to −1369 that was replaced by the URA3 gene); and P2010, IME1 (−1122 to −789).
(ii) ime1-lacZ.
The following plasmids carry the ime1-lacZ gene with various portions of its 5′ region. In these chimeric genes IME1 at bp +202 was in-frame fused to the eighth amino acid of the Escherichia coli lacZ gene. The following constructs are carried on a CEN derivative of E357 (2μm vector) (30): YCpAS128, IME1 (−4401 to +202); YCpAS133, IME1 (−621 to +202); YCpAS134, IME1 (−3015 to +202); YCpAS135, IME1 (−1369 to +202); YCpAS146, IME1 (−2112 to +202); YCpAS148, IME1 (−915 to +202); YCpAS149, IME1 (−1641 to +202); YCpAS152, IME1 (−1444 to +202); YCpAS198, IME1 (−915 to 621 and −346 to +202); YCpAS340, IME1 (−1369 to −1202 and −621 to +202); YCpAS341, IME1 (−1369 to −1122 and −621 to +202); YCpAS344, IME1 (−915 to −621 and −480 to +202); and YCp1457, IME1 (−4401 to −1641 and −1369 to +202). The following constructs are carried on pUN75 (7): YCp1333, IME1 (−756 to +202); YCp1376, IME1 (−1122 to +202); YCp1379, IME1 (−1369 to +202); YCp1427, IME1 (−1153 to +202); YCp1476, IME1 (−1202 to +202); YCp1477, IME1 (−621 to +202); and YCp1956, IME1 (−788 to +202).
(iii) HIS4uas-ime1-his4-lacZ.
In the following plasmids various portions of the IME1 5′ region were inserted in the his4-lacZ chimeric gene by using the X-1 construct carrying the HIS4uas-his4-lacZ gene with a deletion from −181 to −202, leaving 2 Gcn4p binding sites upstream of a XhoI site (31). The following chimeric genes are carried on the shuttle vector YCp50: YCp1138, carries no IME1 information; and YCpAS334, IME1 (−449 to −227). The following plasmids are carried on YIplac128 (10): YIp2006, carries no IME1 information; YIp2032, IME1 (−915 to −788); YIp2055, IME1 (−788 to −756); and YIp2067, IME1 (−1202 to −1153).
(iv) ime1-his4-lacZ.
In the following plasmids various portions of the IME1 5′ region were inserted in the his4-lacZ chimeric gene by using the X-52 construct carrying the his4-lacZ gene (without a UAS) with a deletion from −144 to −316 and a XhoI linker at −144 (31). The following chimeric genes are carried on the shuttle vector YCp50: YCp1139, carries no IME1 information; YCp336, IME1 (−915 to −621); YCp1497, IME1 (−1641 to −1369); YCp1689, IME1 (−1639 to −915); YCp1692, IME1 (−1641 to −1202); YCp1696, IME1 (−1641 to −1122); YCp1975, IME1 (−1641 to −1153); and YCp1981, IME1 (−1122 to −915). The following construct is carried on pUN75 (7): YCp1391, IME1 (−756 to −621). The following plasmids are carried on YIpLac128 (10): YIp2007, carries no IME1 information; YIp1979, IME1 (−1641 to −1122); YIp1980, IME1 (−1641 to −1202); YIp1990, IME1 (−1641 to −1153); YIp1994, IME1 (−1153 to −1122); YIp2020, IME1 (−1122 to −788); YIp2023, IME1 (−915 to −788); and YIp2083, IME1 (−788 to −756).
Yeast strains.
The following yeast strains were used: Y419G, MATa/MATα ura3-52/ura3-52 leu2-3,112/leu2-3,112 ade2/ade2-R8 lys2/LYS2 his7/HIS7 can1-11/CAN1 trp5-Δ/TRP5; Y419G1, isogenic to Y419G but MATa/MATa; Y419G-ΔUCS3 and Y419G1-ΔUCS3, isogenic to Y419G and Y419G1, respectively, but heterozygous for a deletion of upstream controlling sequence 3 (UCS3) (a one-step replacement of a portion of the IME1 5′ region was accomplished following transformation of parental strains with an EcoRI-ClaI fragment carrying IME1 [−4401 to −1641]-URA3-IME1 [−1369 to −621] from YIpAS360); Y422, MATa/MATα ura3-52/ura3-52 trp1-Δ/trp1-Δ leu2-3,112/leu2-3,112 ade2-1/ade2-R8 his4-519/HIS4 his6-1/HIS6 gal/GAL+ can1/CAN1; Y424, isogenic to Y422 but ime1::TRP1/ime1::TRP1 (9); Y1065, MATα ura3-52 trp1-Δ leu2-3,112 his3::hisG ade2-R8 GAL+ CANs gal80::hisG gal4::hisG; Y1093, isogenic to Y1065 but cdc25-2::URA3 (a one-step replacement of CDC25 was accomplished following transformation of Y1065 with a SalI-PvuII fragment carrying cdc25-2::URA3 [27]); Y1132, isogenic to Y1065 but msn2::HIS3 msn4::URA3 (a one-step replacement of MSN2 and MSN4 was accomplished by transformation with a BamHI-SphI fragment from pt32-ΔXB::HIS3 [8] and EcoRI fragment from pZfh45-GEX3X [8], respectively); Y1133, isogenic to Y1065 but bcy1::URA3 (a one-step replacement of BCY1 was accomplished by transformation with a BamHI fragment carrying sra1-20::URA3 [51a]). Correct replacements were checked by Southern blotting (data not shown). In addition, various ime1-his4-lacZ chimeric genes were integrated at the LEU2 loci in various haploid and diploid strains. Integrative plasmids used were digested with PpuMI prior to transformation.
Media and genetic techniques.
PSP2 (minimal acetate medium), and SPM (sporulation medium) have been described previously (18). Synthetic dextrose (SD) medium has also been described previously (46). Meiosis was induced as follows: cells were grown in PSP2 supplemented with the required amino acids to 107 cells/ml, washed once with water, and resuspended in SPM (time zero; also designated SA). Yeast transformation with lithium acetate was done as described by Ito et al. (13). Proteins were extracted from at least three independent transformants and assayed for β-galactosidase activity as previously described (28, 39). Results are given in Miller units.
Gel-shift assays.
The following complementary oligonucleotides were annealed to create the 38-bp IREd double-stranded DNA: UROdsal, 5′ AATTCTTTCCGTCTTCGAGGGAAAGGATCAAAGGCGCG, and UROdrI, 5′ GAAAGGCAGAAGCTCCCTTTCCTAGTTTCCGCGCAGCT. The following complementary oligonucleotides were annealed to create the 38-bp IREu double-stranded DNA: IREulr1, 5′ AATTCTTTTCGTCTTCGAGGGGAAGGATCAAAGGCGCG, and IREuls 5′ TCGACGCGCCTTTGATCCTTCCCCTCGAAGACGAAAAG. The UASrm DNA was isolated as a 127-bp XhoI-BglII fragment from P2010. These DNA fragments were end labeled with [α-32P]dATP by using the Klenow enzyme. Gel-shift assays were performed essentially as described previously (2). The probe (300 ng) was incubated with 10 μg of whole yeast cell extracts (prepared as described in references 2 and 44). The reaction mixture was applied to a 5% polyacrylamide gel. The gel was dried and exposed to both a phosphorimager and to X-ray film.
RESULTS
At least four UCS (UCS1 to UCS4) mediate the regulated transcription of IME1.
In a previous report (47) we compared the expression in β-galactosidase units of an ime1-lacZ chimeric gene present on a single-copy vector to the expression determined by Northern blot analysis of a genomic copy of IME1. The reported analysis showed that both methods gave similar results, i.e., the same pattern of regulation. This allowed us to make use of this ime1-lacZ chimeric gene for a deletion analysis aimed at identifying positive and negative elements that regulate the transcription of IME1. The validity of this approach, i.e., using lacZ chimeric genes on a single-copy vector rather than integrating the chimeric genes at a specific place in the genome, was further examined by additional methods. First, using Northern blot analysis we found that the transcription of an ime1-lacZ chimeric gene present on a single-copy vector was regulated in the same manner as the genomic copy of IME1, although the level of the IME1 RNA was higher when expressed from the genomic copy (reference 1 and data not shown). Second, the levels of expression of various lacZ chimeric genes present on a single-copy vector were compared to the levels of expression of the same genes integrated in the genome. Although in the genome the chimeric genes gave rise to higher levels of expression, their regulation was identical in both cases (see Fig. 6 and data not shown). We assume that the lower levels of expression observed when the chimeric genes are placed on a single-copy vector is due to either plasmid loss or repression effects from the centromere.
FIG. 6.
Effects of various elements present in IME1 UCS2 on the expression of his4-lacZ. Various elements from UCS2 were inserted into the his4-lacZ chimeric gene. Y422 strains carrying this gene on either a CEN vector (YCp plasmid) or integrated in the genomic LEU2 gene (YIp plasmid) were grown as described in the legends for Fig. 1 and 4. The levels of β-galactosidase are given in Miller units. The results are averages of three or four independent transformants. Standard deviations were less than 10%.
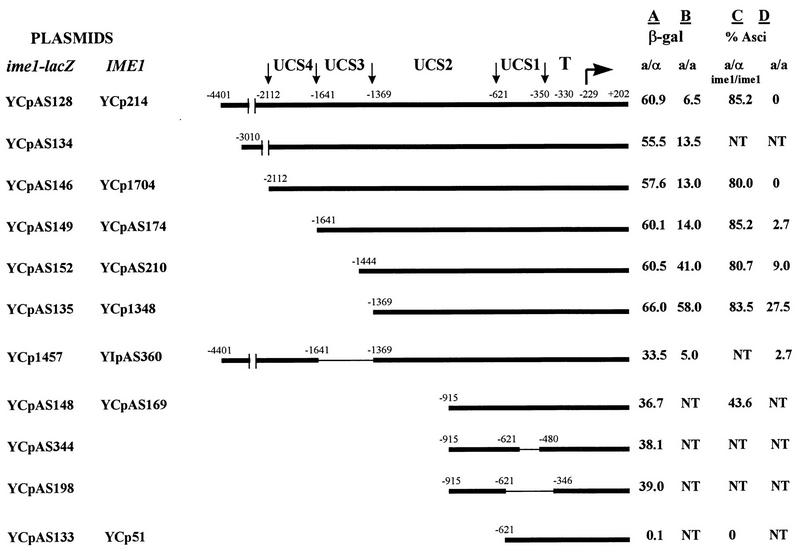
Various portions of the 5′ upstream region of IME1 were fused in-frame to lacZ by using naturally occurring restriction sites. In these chimeric genes IME1 was fused at amino acid 69 to the eighth amino acid of the E. coli lacZ gene. Proteins were extracted from MATa/MATα diploids (derivatives of strain 419G) carrying these fusion genes, at 0 and 6 h in SPM, and from cells grown in SD to 107 cells/ml. The activity of β-galactosidase was determined. Figure 1 shows a schematic representation of the various plasmids used. In vegetative medium with either glucose or acetate as the sole carbon source the chimeric genes were not expressed, giving rise to less than 0.1 Miller units of β-galactosidase (data not shown). At 6 h in SPM, an ime1-lacZ fusion that extends to −1369 gave rise to essentially the same level of β-galactosidase as the one that extends to −4401 (Fig. 1, column A, compare YCpAS135 to YCpAS128, YCpAS134, YCpAS146, YCpAS149, and YCpAS152). On the other hand, an ime1-lacZ fusion that extends to −621 (Fig. 1, column A, YCpAS133) was not expressed. These results suggest that the region between −621 to −1369, designated UCS2, exhibits UAS activity. In order to confirm this conclusion we measured the abilities of plasmids carrying the entire IME1 gene with various portions of the 5′ upstream region to complement an ime1 disruption allele, i.e., to promote sporulation to a MATa/MATα ime1-0/ime1-0 diploid. An IME1 gene that extends to −1369 and one which carries IME1 information up to −4401 bp gave rise to almost identical levels of sporulation (Fig. 1, column C, YCp214, YCp1704, YCpAS174, YCpAS210, and YCp1348), whereas an IME1 gene that extends to −621 did not promote sporulation (Fig. 1, column C, YCp51). Partial deletion of UCS2 led to a decrease in both the level of expression and the efficiency of sporulation (Fig. 1, columns A and C, YCpAS148 and YCpAS169).
FIG. 1.
The transcription of IME1 is mediated by at least four UCSs (UCS1 to UCS4). Yeast strains carrying on a CEN plasmid either ime1-lacZ (columns A and B) or IME1 (columns C and D) with various portions of the 5′ upstream region were grown in PSP2 to 107 cells/ml. Cells were washed once in water and were resuspended in SPM. At 6 h in SPM proteins were extracted and β-galactosidase activities were determined. The levels of β-galactosidase are given in Miller units. The results are averages of three or four independent transformants. Standard deviations were less than 10%. At 72 h the percentages of asci were determined. NT, not tested. The following strains were used: Y419G (MATa/MATα) (column A); Y419G1 (MATa/MATa, isogenic to Y419G (columns B and D); and Y424 (MATa/MATα ime1-0/ime1-0) (column C). The borders of the designated UCS and the TATA box are designated UCS1 to UCS4 and T, respectively. The transcription initiation site is indicated.
The induced level of expression of IME1 that occurs upon nitrogen depletion depends on the presence of both MATa1 and MATα2 gene products and is absent in MATa/MATa diploids (15). In order to identify the site(s) that mediates this regulation, the above-described ime1-lacZ and IME1 plasmids were transformed into an isogenic MATa/MATa strain. The level of β-galactosidase at 6 h in SPM and the percentage of asci following 72 h of incubation in SPM were measured. MATa/MATa diploids that carry the entire IME1 gene with 5′ regions extending up to at least −2112 are properly regulated, i.e., they do not sporulate (Fig. 1, column D, YCp214 and YCp1704). Lack of sporulation in these diploids correlates with a low level of expression of the corresponding ime1-lacZ chimeric gene in this MATa/MATa strain in comparison to the MATa/MATα diploid (Fig. 1, column B, YCpAS128, YCpAS134, and YCpAS146). However, MATa/MATa cells that carry an IME1 or an ime1-lacZ gene with a 5′ region extending to −1369 do sporulate, and they show almost the same level of β-galactosidase as their isogenic MATa/MATα diploids (Fig. 1, columns B and D, compare YCpAS128 to YCpAS135 and YCp214 to YCp1348). The discrepancy between an almost complete level of expression of IME1 and a low efficiency of sporulation (66 versus 60 U of β-galactosidase and 27.5 versus 83.5% asci) suggests that the MATa1 and MATα2 gene products might be required in meiosis not only for the expression of IME1 but also for an additional meiotic event. A similar conclusion was drawn when the level of IME1 RNA was compared in MATa/MATα and MATa/MATa rme1Δ/rme1Δ diploids or when Ime1p was overexpressed in MATa/MATa diploids. In both cases a high level of expression did not lead to a high percentage of asci (15).
The results presented in Fig. 1 suggest that upstream of −1369 reside a site or sites that repress the transcription of IME1 in MAT-insufficient diploids. Further deletion analysis revealed the existence of two such sites, UCS3, which resides between −1369 and −1641, and UCS4, which resides between −1641 and −2112. Deletion of either UCS3 or UCS4 promotes low, inefficient sporulation in MATa/MATa diploids (Fig. 1, column D, compare YCp1704 to YCpAS174, YCpAS210, and YIpAS360). Note that deletion of UCS4 was checked in a MATa/MATa diploid carrying IME1 on a CEN plasmid, whereas deletion of UCS3 was determined in a MATa/MATa diploid heterozygote for a genomic deletion of this element. A MATa/MATα diploid heterozygote for the same deletion (strain Y419GΔUCS3) sporulated with high efficiency (80.1% asci), suggesting that the low levels of sporulation obtained in the MATa/MATa diploid (Y419G1ΔUCS3) are specific. Interestingly, the effect of UCS3 or UCS4 deletion could be observed only when sporulation was assayed, and the levels of β-galactosidase from MATa/MATa diploids carrying comparable ime1-lacZ genes were not affected (Fig. 1, column B compare YCpAS146 to YCpAS149 and YCp1457). These results indicate that very low levels of Ime1p suffice for initiation of meiosis and that deletion of both UCS3 and UCS4 elements relieves the requirement for MATa and MATα for the expression of IME1 and for meiosis.
Identification and function of UCS1.
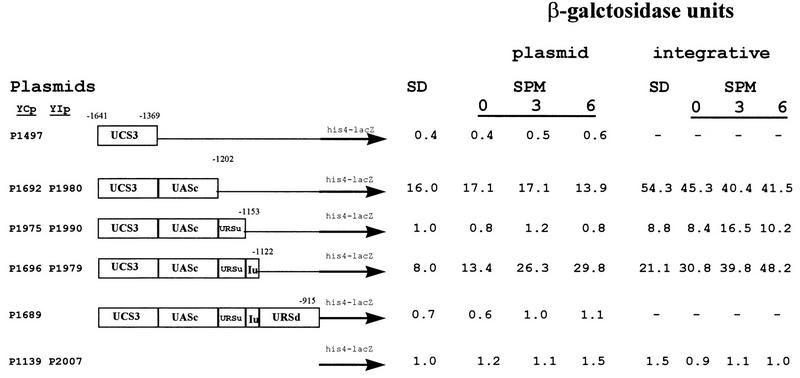
Figure 1 demonstrates that the region between the TATA box at −330 (1) and −621 does not carry any UAS element. Accordingly, two deletions in this region, between −480 and −621 and between −346 and −621, did not reduce the level of expression of these fusion genes in comparison to that of the control plasmid (Fig. 1, column A, compare YCpAS148 to YCpAS344 and YCpAS198). However, it is possible that this region is required to repress rather than activate the transcription of IME1. Indeed, MATa/MATα diploids carrying an ime1-lacZ gene with a deletion between −346 and −621 (YCpAS198) gave rise to low, but elevated levels of β-galactosidase in vegetative cultures: 2.5 U when glucose was the sole carbon source and 15.4 U in the presence of acetate as the sole carbon source. On the other hand, plasmids carrying the entire region gave less than 0.1 U in both glucose and acetate vegetative media. This region, designated UCS1, appears to play a role in nutrient repression of IME1.
In order to confirm the proposed negative role of UCS1, we inserted it downstream of HIS4uas in a his4-lacZ fusion and determined the level of lacZ in MATa/MATα diploids (Y419G) carrying either this plasmid (YCpAS334) or the parental plasmid bearing the HIS4uas-his4-lacZ chimeric gene (YCp1138). Figure 2 shows that the level of expression of UAShis4-lacZ is increased in the presence of acetate as the sole carbon source in comparison to that in the presence of glucose as the sole carbon source. Moreover, as previously reported (6), the transcription of HIS4 is further induced upon nitrogen depletion. Insertion of UCS1 upstream of a HIS4 TATA box prevents the transcriptional activation function of HIS4 UAS in vegetative culture with either glucose or acetate as the sole carbon source (Fig. 2, SD or SA, respectively) but has no effect upon nitrogen depletion (Fig. 2, SPM). UCS1 was also inserted upstream of a his4-lacZ fusion (plasmid YCp1139) that lacked its own UAS. The resulting chimeric gene (on plasmid YCpAS331) remained silent under all growth conditions, confirming that UCS1 does not possess any UAS activity and that it serves only as an upstream repression sequence.
FIG. 2.
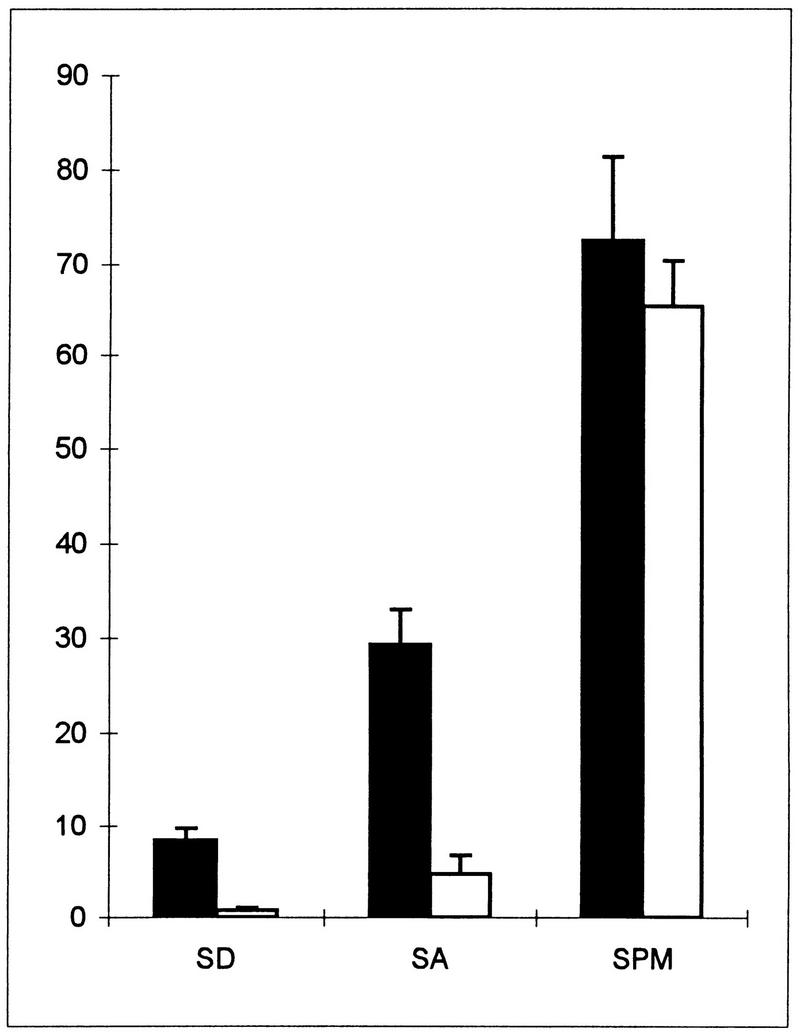
UCS1 possesses URS activity in the presence of nitrogen. Strain Y419G carrying the UAShis4-lacZ (filled bars) or UAShis4-UCS1-lacZ (open bars) chimeric gene on a CEN vector (YCp1138 or YCpAS334, respectively) was grown as described in the legend for Fig. 1, and at 0 (SA) and 6 (SPM) h in SPM samples were taken to extract proteins and measure lacZ levels. In addition, proteins were extracted from cells grown in glucose medium (SD) to 107 cells/ml. The levels of β-galactosidase are given in Miller units. The results are averages of three or four independent transformants. Error bars indicate standard deviations.
UCS2 contains positive and negative elements.
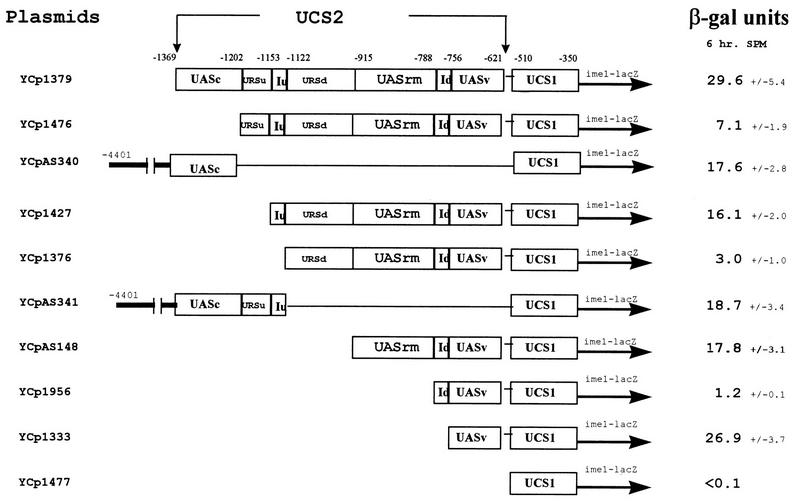
In order to identify the elements within UCS2 that are responsible for its regulated expression, 5′ serial deletions of UCS2 were constructed in the ime1-lacZ fusion. The level of β-galactosidase following 6 h in SPM was determined for MATa/MATα diploids carrying these chimeric genes. Figure 3 demonstrates that UCS2 is made of alternate positive and negative elements. YCp1379 gave rise to 29.6 U of β-galactosidase. Deletion of 167 bp in YCp1476 led to only 7.1 U and a further deletion of 50 bp, as in YCp1427, led to 16.1 U, and so on.
FIG. 3.
UCS2 consists of positive and negative elements. MATa/MATα diploids (Y422) carrying on a CEN vector various ime1-lacZ chimeric genes were grown in PSP2 to 107 cells/ml. Cells were washed once in water and were resuspended in SPM. At 6 h in SPM proteins were extracted and β-galactosidase activities were determined. The levels of β-galactosidase are given in Miller units. The results are averages of three or four independent transformants, and standard deviations are indicated.
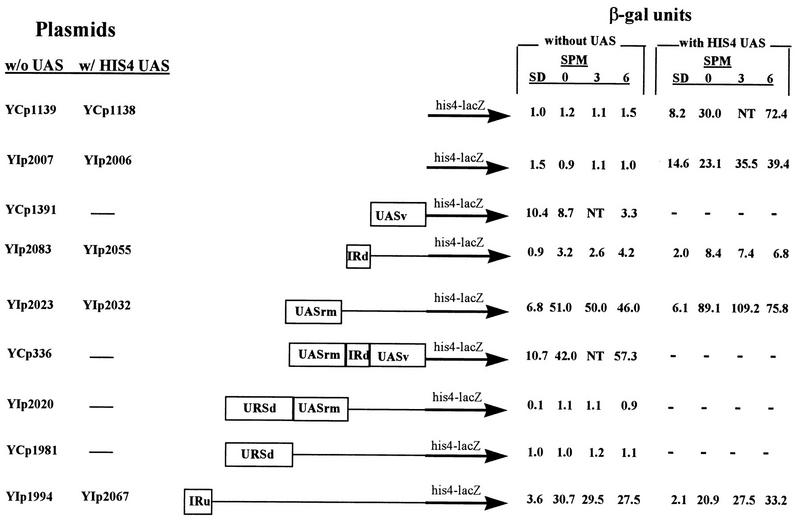
The ime1-lacZ chimeric genes cannot be used to determine whether the UAS or URS activity of the various elements is subject to nutrient regulation. This is due to both the presence of UCS1 and translational regulation (47) that prevents expression in the presence of nitrogen. Therefore, characterization of UCS2 was achieved following the insertion of these elements in a his4-lacZ gene lacking or carrying its own UAS. The level of β-galactosidase under various growth conditions was determined for MATa/MATα diploids carrying these chimeric genes either integrated at the LEU2 locus or on a CEN plasmid (Fig. 4 and 6). For simplicity, the various identified elements will be separately described, starting from the 3′ element.
FIG. 4.
UCS2 contains positive and negative elements that mediate expression of his4-lacZ and HIS4uas-his4-lacZ. Various elements from UCS2 were inserted into either the HIS4uas-his4-lacZ or his4-lacZ chimeric gene. Y422 strains carrying these genes on either a CEN vector (YCp plasmid) or integrated in the genomic LEU2 gene (YIp plasmid) were grown as described in the legend for Fig. 1. Samples were taken to extract proteins and measure lacZ levels at 0, 3, and 6 h in SPM. In addition, proteins were extracted from cells grown in SD medium to 107 cells/ml. The levels of β-galactosidase are given in Miller units. The results are averages of three or four independent transformants. Standard deviations were less than 10%.
(i) The UASv element.
An ime1-lacZ gene that extends to −756 was almost completely expressed (Fig. 3, compare YCp1333 to YCp1379), whereas an ime1-lacZ gene that extends to −621 (YCp1477) was not expressed. These results suggest that the region between −621 to −756 exhibits a UAS activity, designated UASv (for UAS activity in vegetative cultures). Figure 4 demonstrates that insertion of this element upstream of the his4-lacZ chimeric gene promotes its expression in vegetative culture with either glucose or acetate as the sole carbon source (compare YCp1391 to YCp1139). Upon nitrogen depletion (6 h in SPM) lower levels of β-galactosidase are observed, suggesting that either UASv is less active in this condition or that it is totally inactive in this condition and that the activity observed is due to the stability of the β-galactosidase protein that is transcribed and translated in the vegetative cultures.
(ii) The IRE elements.
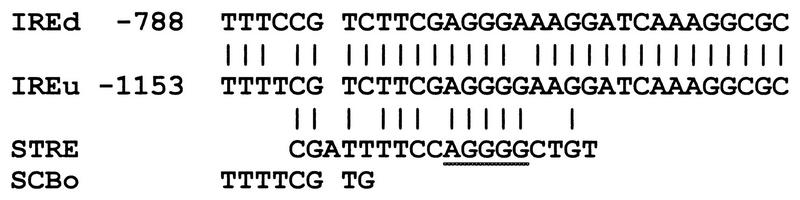
Computer analysis of UCS2 revealed the presence of two almost identical 32-bp repeats designated IRE (IME1 repeated element) (Fig. 5). The presence of such a large repeat suggests that it may recruit identical regulators. Therefore, using designed oligonucleotides and PCR, we constructed ime1-lacZ fusion genes whose 5′ ends terminated either upstream to each one of the IRE repeats or downstream to these elements. Figure 3 demonstrates that the upstream element, IREu, serves as a UAS element: an ime1-lacZ chimeric gene that extends to the IREu element shows an about fivefold increase in expression compared to that of a chimeric gene that does not include this element (Fig. 3, YCp1427 YCp1376). Insertion of the IREu element upstream of the silent his4-lacZ gene promotes its expression (Fig. 4, compare YIp2007 to YIp1994, and Fig. 6, compare YIp1990 to YIp1979 and YCp1975 to YCp1696). Moreover, the UAS activity of the IREu element is subject to nutrient regulation: low UAS activity is observed in vegetative medium with glucose as the sole carbon source, and an increase in UAS activity is observed in vegetative medium with acetate as the sole carbon source and in SPM upon nitrogen depletion. The lower level of expression in the presence of glucose may be due either to a lack of UAS activity or to specific repression of the UAS activity in this medium. In order to distinguish between these two possibilities we inserted the IREu element downstream of a HIS4 UAS in a his4-lacZ chimera and determined its effect on the function of the HIS4 UAS under various growth conditions. Figure 4 shows that the resulting HIS4uas-IREu-his4-lacZ chimeric gene (YIp2067) gives levels of β-galactosidase similar to that given by the IREu-his4-lacZ chimeric gene (YIp1994). Thus, under all growth conditions the HIS4 UAS seems silent, suggesting that the regulatory protein(s) that binds to the IREu element interferes with the binding of Gcn4p to HIS4 UAS. This analysis could not determine, therefore, if the IREu element also serves as a URS.
FIG. 5.
Sequence alignment of the two IRE elements in IME1 to known UAS elements. The sequence of the STRE element is underlined. Given is the sequence used in band-shift assays as a probe for the binding of Msn2/4p (25). The SCB element (3) is in opposite orientation.
Surprisingly, the downstream element, IREd, exhibits URS activity: a chimeric gene carrying UASv and UCS1 is expressed, whereas the addition of the IREd element causes a 22-fold reduction in expression (Fig. 3, compare plasmid YCp1956 to YCp1333). Insertion of this element upstream of the silent his4-lacZ gene promotes low levels of expression, about 10-fold less than the ones observed for the IREu-his4-lacZ chimera (Fig. 4, compare YIp2083 to YIp1994). Insertion of IREd downstream to HIS4 UAS in a his4-lacZ fusion results in a three- to sevenfold reduction in its activity under all growth conditions (Fig. 4, compare YIp2055 and YIp2006). However, since the positive element, IREu, also prevents the UAS activity of HIS4, further analysis is required to determine if IREd can repress any UAS or, specifically, UASv.
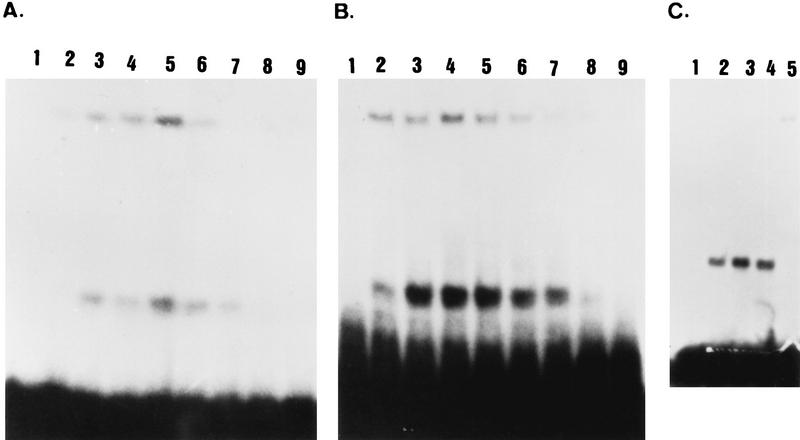
Gel-shift assays using either the IREu or IREd elements and total proteins extracted from yeast cells grown under the same conditions described above for the lacZ experiments, revealed the formation of two specific DNA-protein complexes (Fig. 7). Nonspecific competition with 10× and 100× plasmid pUC19 DNA did not reduce the formation of these complexes (data not shown), whereas specific competition showed such a decrease (Fig. 7A, compare lanes 8 and 9 to lane 3, and Fig. 7B, compare lanes 6 and 7 to lane 3). Moreover, since the addition of the nonspecific competitor increases the level of these complexes (data not shown), in the experiment reported in Fig. 7 each lane also included cold plasmid pUC19 DNA. Figure 7A demonstrates that the formation of the IREu-protein complexes is subject to nutrient regulation: in the presence of glucose very low levels are observed, and about a twofold increase is observed in vegetative medium with acetate as the sole carbon source (phosphorimager calculations). Upon nitrogen depletion the intensity of the complexes is further increased. Thus, the regulated UAS activity of IREu, i.e., low in glucose and high in acetate and upon nitrogen depletion, mirrors the formation of the two DNA-protein complexes on this element. This analysis suggests that IREu serves only as a UAS element (but see below).
FIG. 7.
Two protein-DNA complexes are formed on the IRE elements. Gel retardation assays were performed with 300 ng of 32P-end-labeled 38-bp double-stranded oligonucleotides IREulr1 and IREuls, creating IREu (A and C), or UROdsal and UROdr1, creating IREd (B), and 30 ng of cold plasmid pUC19 DNA. Lane 1 is free probe. Proteins were extracted from cells grown in minimal glucose medium to 107 cells/ml (panels A and B, lane 2; panel C, lanes 2 and 4) or from cells pregrown in PSP2 to 107 cells/ml, washed once in water, and resuspended in SPM for 0 (panels A and B, lanes 3 and 6 to 9; panel C, lanes 3 and 5), 3 (panels A and B, lane 4), and 6 (panels A and B, lane 5) h. For lanes 6 and 7 cold IREd was added in excess molar ratios of 0.1 and 0.5, respectively. For lanes 8 and 9 cold IREu was added in excess molar ratios of 0.1 and 0.5, respectively. The strains used were as follows: Y419G, a wild-type diploid (panels A and B); Y1065, a wild-type haploid (panel C, lanes 2 and 3), and Y1132, an msn2Δ msn4Δ haploid (panel C, lanes 4 and 5).
Figure 7B demonstrates that the same DNA-protein complexes that are formed on IREu are also formed on IREd. However, for IREd, the lower-molecular-weight complex seems constitutive in the presence of acetate (compare intensities in lanes 3 to 5). The different affinity of the regulatory proteins to each element can be revealed by competition analysis: Fig. 7 demonstrates that the IREu element is a better competitor than the IREd element when the probe is made either of IREu (Fig. 7A, compare lanes 6 and 7 to 8 and 9) or IREd (Fig. 7B, compare lanes 6 and 7 to 8 and 9). The difference in binding affinity pinpoints the two mismatched regions as the binding sites for the regulatory proteins and explains the lower UAS activity of this element compared to that of the IREu element (Fig. 4 and 5).
(iii) The UASrm element.
Figure 3 identifies the presence of a UAS element, designated UASrm (regulated middle), from bp −788 to −915. An ime1-lacZ gene whose 5′ end extends to this element is expressed, whereas a chimeric gene ending prior to this element is not expressed (Fig. 3, compare YCpAS148 to YCp1956). The nature of this element, i.e., regulated or constitutive, was revealed by inserting it in the his4-lacZ reporter gene. Figure 4 demonstrates that UASrm exhibits nutrient-regulated UAS activity: it promotes low expression in the presence of glucose and exhibits high activity in acetate and sporulation media (Fig. 4, compare YIp2023 to YIp2007). In vegetative medium with acetate as the sole carbon source, and upon nitrogen starvation, the presence of UASrm does not interfere with the activity of HIS4 UAS, and additive levels of β-galactosidase are observed (Fig. 4, compare YIp2032 to YIp2023 and YIp2006). Gel-shift assays confirm these conclusions. Figure 8 shows the binding of a specific protein(s) to UASrm. The level of the bound protein is regulated by nutrients: a low level is present in the presence of glucose, and an eightfold increase (calculated by phosphorimager) is observed in acetate-containing medium. These results reflect the regulated UAS activity of this element. Figure 4 shows a slight reduction in the activity of UASrm at 6 h in SPM (46 versus 50 U); however, a more pronounced reduction (fourfold) is observed for the formation of the DNA-protein complex (Fig. 8, compare SPM to SA). We suggest that this discrepancy is due to the stability of the lacZ protein.
FIG. 8.
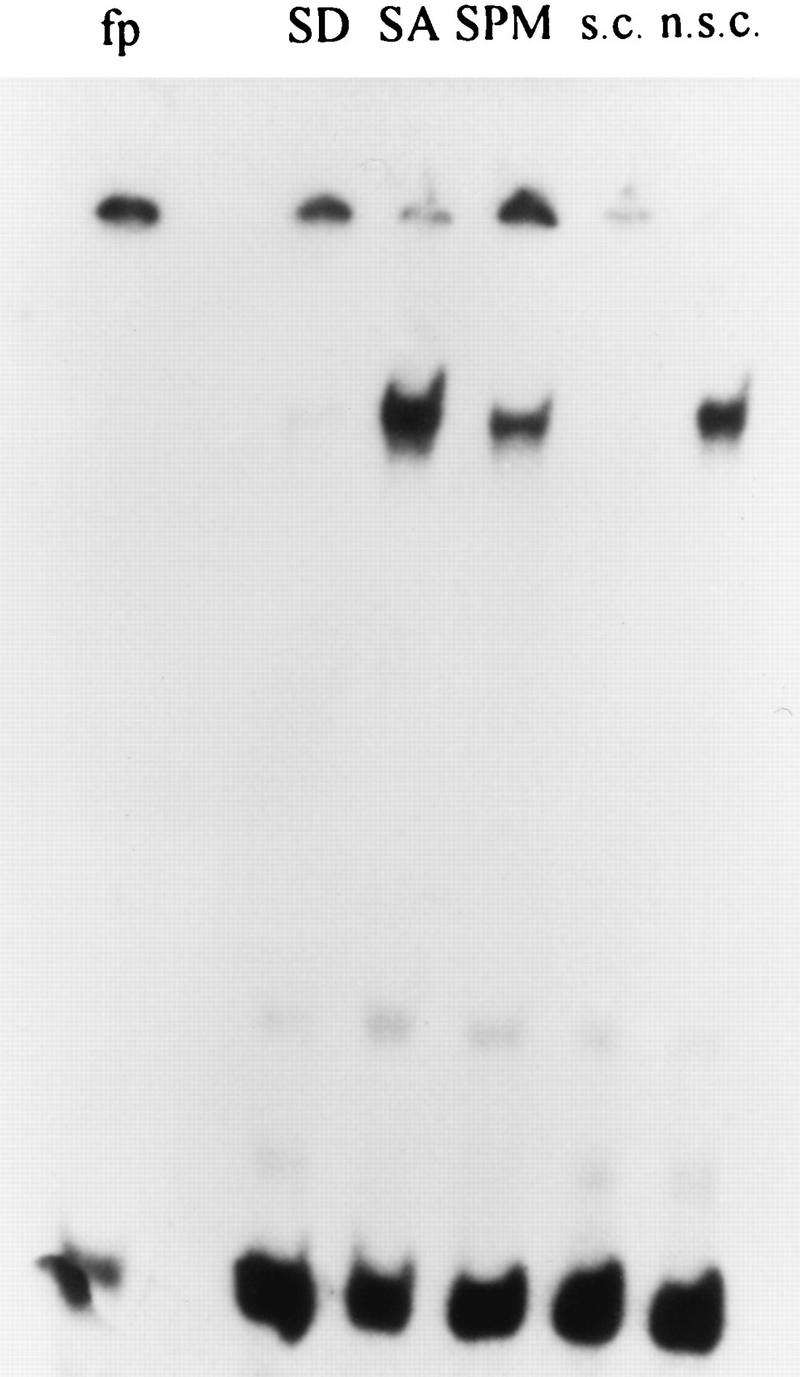
A single protein-DNA complex is formed on UASrm. Gel retardation was performed with 300 ng of a 32P-end-labeled UASrm element present on a 127-bp XhoI-BglII fragment derived from p2010. Proteins were extracted from cells of strain Y419G grown in minimal glucose medium (SD) to 107 cells/ml or from cells pregrown in PSP2 to 107 cells/ml, washed once in water, and resuspended in SPM for 0 (SA) and 5 h (SPM). For competition, cold plasmid pUC19 DNA (n.s.c) or UASrm DNA (s.c) was added in excess molar ratios of 10 to protein samples extracted at 5 h in SPM. fp, free probe.
(iv) URSd.
Upstream of UASrm, the region between −915 and −1122 possesses URS activity. An ime1-lacZ chimeric gene extending to −915 is expressed, whereas an ime1-lacZ chimeric gene extending to −1122 shows about sixfold reduction in expression (Fig. 3, compare YCpAS148 to YCp1376). An his4-lacZ gene carrying this element is not expressed (Fig. 4, YCp1981), suggesting that it exhibits no UAS activity. The URS activity of URSd is not subject to regulation and is observed under all growth conditions: it lowers the UAS activity of IME1 UASs in glucose and acetate media and in SPM (Fig. 4, compare YIp2020 to YIp2023, and Fig. 6, compare YCp1689 to YCp1696).
(v) URSu.
Figure 3 indicates that the region between −1153 and −1202 exhibits URS activity. Addition of this element 5′ to an ime1-lacZ gene that extends to −1153 caused twofold repression (Fig. 3, compare expression from YCp1476 to that from YCp1427). Addition of this element 3′ to IME1 UASc in an his4-lacZ chimeric gene caused between 2- and 20-fold repression under all growth conditions (Fig. 6, compare expression from YIp1990 to that from YIp1980 and expression from YCp1975 to that from YCp1692).
(vi) UASc.
Figure 3 indicates that the region between −1202 to −1369 exhibits UAS activity. A fourfold increase in expression was observed when this element was added to a plasmid lacking it (Fig. 3, compare expression from YCp1379 to that from YCp1476 and expression from YCpAS340 to that from YCp1477). The UAS activity of this element is constitutive under all growth conditions. This is demonstrated when UASc (constitutive) is inserted upstream of his4-lacZ (Fig. 6, compare YCp1692 and YIp1980 to YCp1497 and YIp2007).
The glucose signal is transmitted to IREu via the RAS-cAPK pathway and the Msn2/4p transcription factors.
An active RAS-cAPK pathway represses the transcription of IME1 (27). We determined if this effect is mediated via any of the regulated UAS elements of IME1 by examining the effect of a temperature-sensitive mutation in CDC25 (RAS exchange factor [4]) on their UAS activity. Table 1 demonstrates that the level of expression of a UASrm-his4-lacZ chimeric gene is identical in wild-type and cdc25-2 haploids grown at either the permissive or restrictive temperature. Thus, UASrm is not the target for the RAS-cAPK pathway. On the other hand, the UAS activity of IREu is increased in cdc25-2 isogenic strains; in SD medium, a 6- to 10-fold increase is observed. This increase is already evident at the permissive temperature, and higher levels are observed at the nonpermissive temperature. At 34°C the level of expression of IREu-his4-lacZ is higher than the level observed for the wild-type strain in SA medium (30.8 versus 10.4 U), suggesting that Cdc25p transmits a glucose signal that prevents the UAS activity of IREu. Cdc25p is required to activate Rasp; however, Rasp activates two signal pathways, that of the cAPK and the pheromone-induced mitogen-activated protein kinase cascade (8a, 11). In order to establish that for IREu Cdc25p modulates the activity of cAPK, we examined the effect of deletion of the regulatory subunit of cAPK, BCY1, on the level of expression of IREu-his4-lacZ. Table 1 demonstrates that in bcy1Δ strains IREu does not activate transcription. We conclude that the RAS-cAPK pathway transmits a glucose signal to IREu.
TABLE 1.
The RAS/cAPK pathway prevents the UAS activity of IREu in the presence of glucose
| Strain genotypea | Temp (°C) | Level of β-galactosidase inb:
|
|||
|---|---|---|---|---|---|
| IREu
|
UASrm
|
||||
| SD | SA | SD | SA | ||
| Wild type | 25 | 0.8 ± 0.14 | 8.2 ± 0.56 | 10.7 ± 1.18 | 46.1 ± 4.69 |
| 30 | 0.88 ± 0.22 | 8.55 ± 0.25 | 7.4 ± 2.34 | 51.2 ± 9.0 | |
| 34 | 3.4 ± 0.42 | 10.4 ± 0.85 | 21.3 ± 6.71 | 54.4 ± 6.69 | |
| cdc25-2 | 25 | 5.4 ± 0.57 | 55.3 ± 3.89 | 8.1 ± 1.48 | 64.2 ± 11.5 |
| 34 | 30.8 ± 2.19 | 163.0 ± 8.41 | 4.2 ± 0.8 | 56.0 ± 7.07 | |
| bcy1Δ::URA3 | 30 | 0.4 ± 0.18 | 0.82 ± 0.28 | ||
| msn2Δ msn4Δ | 30 | 0.15 ± 0.03 | 0.1 ± 0.01 | ||
| [MSN2] | 30 | 1.82 ± 0.13 | 8.21 ± 1.77 | ||
The following isogenic strains were used: Y1065 (wild type), Y1093 (cdc25-2), Y1133 (bcy1Δ::URA3), Y1132 (msn2Δmsn4Δ), Y1065 carrying YEP1636, ([MSN2]).
Levels of β-galactosidase (± standard deviations) are given in Miller units.
What DNA binding protein serves as a target for the RAS-cAPK pathway and binds to and activates IREu? In order to identify positive regulators, we screened for multicopy plasmids that increase the expression of an IREu-cyc1-lacZ chimeric gene (38). Several genes were identified, including MSN2. Table 1 demonstrates that when MSN2 is carried on a multicopy plasmid a twofold increase in the UAS activity of IREu is observed in glucose medium. Deletion of MSN2 has no effect (data not shown). Nonetheless, this negative result may be due to the presence of its close homolog, MSN4 (8). Indeed, in the isogenic, msn2Δ msn4Δ double mutant, IREu shows no UAS activity (Table 1). MSN2-MSN4 encode a transcriptional activator (8) that binds to and activates STRE elements (25, 43). Sequence analysis reveals that IREu carries such an element (Fig. 5). We used gel-shift assays to determine if Msn2/4p bind to IREu. As described above, Fig. 7 demonstrates that two DNA-protein complexes are formed on IREu. The levels of these DNA-protein complexes are higher in Fig. 7A and B than in Fig. 7C. This is probably due to the fact that for the results shown in the former panels proteins were extracted from diploid cells, whereas for the results in the latter panel protein were extracted from haploid cells. In vegetative medium with acetate as the sole carbon source the lower-molecular-weight DNA-protein complex disappears when proteins are extracted from the msn2Δ msn4Δ double mutant (Fig. 7C, compare lanes 3 to lane 5). Interestingly, in glucose medium deletion of both MSN2 and MSN4 has no effect (Fig. 7C, compare lane 2 to 3), suggesting that in glucose and acetate media this band consists of different proteins. We assume that, normally, in SA medium only Msn2/4p bind the STRE element, whereas the protein that binds IREu DNA in glucose medium may be either degraded or excluded from binding. Further work is required to establish if this protein functions in glucose medium as a transcriptional activator or repressor.
DISCUSSION
Using systematic deletion analysis of an ime1-lacZ fusion we demonstrate that the transcription of IME1 is regulated by a remarkably large region, over 2,100 bp long, which is the largest region known in S. cerevisiae. In comparison, the HO gene, another gene with a long and complex promoter, is made of only 1,400 bp (32). Preliminary data suggest that 5′ of the 2,100-bp upstream region of IME1 there are yet additional elements that control its transcription: Granot et al. (12) showed that multiple copies of an 0.5-kb XhoI-BglII fragment from positions −3166 to −3762 of IME1 promote low levels of sporulation to a MATa/MATa diploid. They proposed that a titration of a negative regulator that binds to this region is responsible for derepression of the transcription of IME1 in this mater diploid (12). In agreement, in this report we demonstrate that deletion of this region promotes a twofold increase in the expression of ime1-lacZ in MATa/MATa diploids (Fig. 1 column B, compare YCpAS128 and YCpAS134).
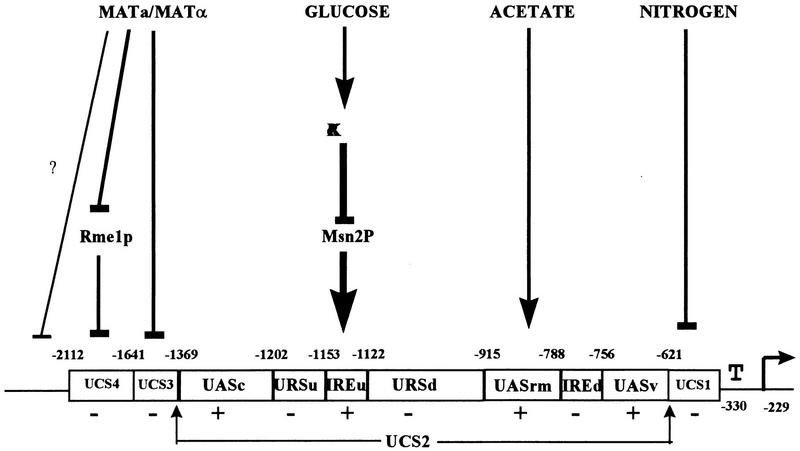
A schematic map of IME1 is illustrated in Fig. 9. For simplicity, the 5′ region of IME1 was divided into four UCS, UCS1 to UCS4. UCS1, UCS3, and UCS4 function as negative elements, whereas UCS2 functions as a positive element that is absolutely required for the transcription of IME1. UCS1 and UCS2 mediate nutrient regulation: UCS1 prevents the transcription of IME1 in the presence of nitrogen, whereas UCS2 promotes its transcription in the presence of a nonfermentable carbon source, i.e., acetate versus glucose. UCS3 and UCS4 repress the transcription of IME1 in MAT-insufficient cells. This organization of the IME1 5′ region explains its mode of transcription. In vegetative medium with glucose as the sole carbon source, IME1 is silent, since two of its UAS elements, IREu and UASrm, are not active. On the other hand in vegetative medium with acetate as the sole carbon source, the UAS activity of these elements leads to the low levels of IME1 RNA. Complete activation requires lack of repression from UCS1, UCS3, and UCS4, a condition that occurs only in MATa/MATα diploids upon nitrogen depletion.
FIG. 9.
Schematic structure of IME1 5′ untranslated region. The elements that respond to the various meiotic signals, MAT, glucose, acetate, and nitrogen, are illustrated. A positive role is marked by an arrow, whereas a negative role is marked by a line. ?, preliminary result; +, UAS; −, URS.
UCS4 (−1641 to −2112) represses the transcription of IME1 in cells that do not carry both MATa1 and MATα2 gene products. This result is in agreement with that reported by Covitz and Mitchell (5) showing that a region between −2243 and −1743 consists of a negative element that transmits the MAT signal. UCS4 carries the binding site for Rme1p (5), a negative regulator that mediates MAT repression to IME1 (15). In this report we show that the MAT signal is also transmitted via UCS3. Sequence analysis reveals that UCS3 does not carry the Rme1p binding site. Further analysis is required to identify the gene(s) whose product(s) binds to and regulates this element. Here we demonstrate that deletion of both UCS3 and UCS4 is required for complete derepression (Fig. 1). In agreement, Covitz and Mitchell (5) have shown that an IME1 gene that extends to −2001 (deletion of only UCS4) gives rise to 18% asci in MATa/MATa cells, whereas an IME1 gene that extends to −1202 (deletion of both UCS3 and UCS4) gives rise to a higher level, i.e., 34%. Covitz and Mitchell (5) suggested that only the UCS4 region transmitted a MAT signal, since an IME1 gene that extends to −2243 and carried a deletion between −1122 and −1743 gave only 1.3% asci in MATa/MATa cells. Two reasons may explain their inability to identify UCS3. (i) The IME1 gene used in that study was not efficiently expressed, since two positive regulators, designated here as UASc and IREu, were also deleted. (ii) Deletion of only UCS3 is not sufficient for complete derepression.
Deletion analysis reveals that UCS2 consists of alternate positive and negative elements (Fig. 9). Previously, a less detailed deletion analysis divided UCS2 into only two positive elements, UASd and UASu (45), which consist of UASv, IREd, and UASrm and of URSd, IREu, and UASc, respectively. The presence of UCS1 prevents the characterization of these elements in the intact IME1 or ime1-lacZ gene. Moreover, since nutrients also regulate the translation of IME1 (47), the expression of the various ime1-lacZ chimeric genes could be determined only under sporulation conditions. Therefore, the various elements were inserted in a heterologous reporter gene, his4-lacZ, carrying or lacking its own UAS. This analysis reveal that UCS2 is made of the following elements: a constitutive UAS element, UASc, that promotes expression of a reporter gene under all growth conditions; three constitutive URS elements, URSu, URSd, and IREd; a UAS element, UASv, that promotes expression in vegetative media; and two regulated UAS elements, IREu and UASrm, that promote expression of reporter genes in the presence of acetate. Accordingly, in the presence of acetate as the sole carbon source the abundance of specific DNA-protein complexes, formed on these regulated UASs, is higher than in the presence of glucose (Fig. 7 and 8).
One of the striking features of the analysis reported here is the presence of two, almost identical repeats, IREu and IREd (Fig. 5), that in the context of IME1 sequences are not identical in function. This difference is also reflected by both the UAS activity of these elements in the heterologous his4-lacZ gene (Fig. 4) and the different affinities in the binding of specific proteins to these elements (Fig. 7). Figure 5 shows that an A to G mutation in the IREu element creates an AGGGG STRE. STRE elements function as UAS that respond to various stresses including nitrogen depletion (24, 42). STRE elements are activated by the binding of the transcriptional activator Msn2p or its homolog Msn4p (25, 43), suggesting that these proteins also bind to and regulate the function of IREu. Indeed, cells deleted for both MSN2 and MSN4 show the elimination of a specific DNA-protein band from IREu (Fig. 7C) and a substantial reduction in the UAS activity of IREu (Table 1). The IREd element carries the AGGGA PDS element to which in vitro-made Msn2p and Msn4p do not bind (25), explaining why in vivo, by itself, the IREd element serves as only a weak UAS.
The second mismatch between the IRE elements creates, in IREu, the sequence TTTTCGTC , which is almost identical (7 of 8 nucleotides) to the known cell cycle box (SCB) CACGAAAA (3) (Fig. 5). SCBs serve as UAS elements upon exit from G1 arrest and are present in the HO gene as well as in the G1 cyclins CLN1 and CLN2 (3, 33, 35). We suggest that the upper DNA-protein complex on the IRE elements (Fig. 7) is formed on this sequence. Transcription mediated by SCB elements is accomplished by the binding of the Swi4p and Swi6p transcription factors (20, 33, 35). We determined, therefore, the role of these proteins in regulating the expression of both the ime1-lacZ and IREu-his4-lacZ chimeric genes. Deletion of either SWI4 or SWI6 resulted in a three- to fourfold increase in the expression of both genes (38). The Swi4p-Swi6p complex activates transcription only upon progression from G1 to the mitotic S phase (19, 20), explaining why the transcription of IME1 is induced upon G1 arrest (47). Thus, the Swi4p-Swi6p complex is a positive regulator for initiation of the mitotic cell cycle and a negative regulator for initiation of meiosis. Similarly, Rme1p is a positive regulator for the transcription of the G1 cyclin CLN2 and a negative regulator for the transcription of IME1 (15, 29, 52).
In this report we demonstrate that the RAS-cAPK pathway prevents the UAS activity of IREu in the presence of glucose (Table 1), whereas the transcriptional activators Msn2/4p are required for both the formation of a specific DNA-protein complex on IREu and its UAS activity in acetate medium (Fig. 7C and Table 1). Since in vitro-made Msn2p binds to the STRE element (25, 43) that is also present in IREu (Fig. 5), we suggest that cAPK mediates its effect on IREu via Msn2/4p (Fig. 9). Accordingly, in a recent meeting it was reported (12a) that nuclear localization of Msn2p is modulated by cAPK: in the presence of high cAPK activity, or in nonstress conditions, Msn2p is confined to the cytoplasm, whereas upon stress it is translocated to the nucleus.
In this report we show that the RAS-cAPK pathway transmits a glucose signal to IREu. On the other hand, previous reports have shown that this pathway transmits a nitrogen rather than a glucose signal to both IME1 and meiosis (references 26 and 27 and references therein). Since in the latter case the entire IME1 5′ region was examined, it is possible that this pathway, with a different element (UCS1?), also transmits a nitrogen signal to IME1. However, biochemical evidence suggests that the RAS-cAPK pathway transmits a glucose rather than a nitrogen signal (54) and that the level of cyclic AMP is not affected by nitrogen (36). We suggest, therefore, that the complex organization of the IME1 5′ region is responsible for this discrepancy. We propose that nitrogen repression through UCS1 can be relieved only upon full activation of both IREu and UASrm. Thus, in the presence of glucose, activation of the IREu element by low cAPK activity does not suffice to overcome UCS1 repression and cells do not initiate meiosis in glucose media. However, in acetate media, when UASrm is also activated, UCS1 is neutralized and IME1 is expressed.
IME1 encodes the master regulator of the developmental pathway, meiosis. It is not surprising, therefore, that the transcription of IME1 is mediated by an extremely large and complex region. The IME1 5′ region consists of constitutive as well as regulated positive and negative elements. The combinatorial effect of the entire region leads to the regulated transcription of IME1, i.e., silent in vegetative media with glucose as the sole carbon source, low levels of mRNA in vegetative acetate media, and increased levels upon nitrogen depletion. We suggest that the use of many negative elements is essential to restrict transcription to the appropriate conditions, since any deviation may lead to the initiation of meiosis at the wrong time or condition, resulting in cell death.
ACKNOWLEDGMENTS
We thank R. Davis, G. Fink, A. Sugino, and A. Tzagaloff for kindly providing plasmids. We thank D. Cassel and D. Kornitzer for critical reading of the manuscript.
This work was supported by grants from the Israel Academy of Sciences (Y.K) and the U.S.-Israel Binational Science Foundation (G.S.).
S.S, A.S., and G.S contributed equally to this article.
REFERENCES
- 1.Ben-Dov, N., and Y. Kassir. Unpublished data.
- 2.Bram R J, Kornberg R D. Specific protein binding to far upstream activating sequences in polymerase II promoter. Proc Natl Acad Sci USA. 1985;82:43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 4.Broach J R. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- 5.Covitz P A, Mitchell A P. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 6.Donahue T F, Daves R S, Lucchini G, Fink G R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983;32:89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- 7.Elledge S J, Davis R W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 8.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Fink, G. Personal communication.
- 9.Foiani M, Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. Meiosis specific protein kinase, Ime2, is required for the correct timing of DNA replication and nuclear divisions in yeast meiosis. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- 10.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 11.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 12.Granot D, Margolskee J P, Simchen G. A long region upstream of the IME1 gene regulates meiosis in yeast. Mol Gen Genet. 1989;218:308–314. doi: 10.1007/BF00331283. [DOI] [PubMed] [Google Scholar]
- 12a.Griffioen, G., C. Schuller, W. Gorner, and H. Ruis. Personal communication.
- 13.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao G, Shah J C, Clancy M J. An RME1-independent pathway for sporulation control in Saccharomyces cerevisiae acts through IME1 transcript accumulation. Genetics. 1990;126:823–835. doi: 10.1093/genetics/126.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassir Y, Granot D, Simchen G. IME1, a positive regulator of meiosis in the yeast Saccharomyces cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 16.Kassir Y, Simchen G. Regulation of mating and meiosis in yeast by the mating-type region. Genetics. 1976;82:187–206. doi: 10.1093/genetics/82.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassir Y, Simchen G. Pathways leading to meiotic differentiation in the yeast Saccharomyces cerevisiae. Curr Genet. 1989;15:167–170. [Google Scholar]
- 18.Kassir Y, Simchen G. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:94–109. doi: 10.1016/0076-6879(91)94009-2. [DOI] [PubMed] [Google Scholar]
- 19.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 20.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 21.Lee R H, Honigberg S M. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol Cell Biol. 1996;16:3222–3232. doi: 10.1128/mcb.16.6.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Mitchell A P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandel S, Robzyk K, Kassir Y. The IME1 gene encodes a potent transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev Genet. 1994;15:139–147. doi: 10.1002/dvg.1020150204. [DOI] [PubMed] [Google Scholar]
- 24.Marchler G, Schuller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Pastor M T, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto K, Uno I, Ishikawa T. Initiation of meiosis in yeast mutants defective in adenylate cyclase and cyclic AMP-dependent protein kinase. Cell. 1983;32:417–423. doi: 10.1016/0092-8674(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura A, Treinin M, Mitsuzawa H, Kassir Y, Uno I, Simchen G. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. EMBO J. 1990;9:3225–3232. doi: 10.1002/j.1460-2075.1990.tb07521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Mitchell A P, Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature. 1986;319:738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- 30.Myers A M, Tzagaloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 31.Nagawa F, Fink G R. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasmyth K. At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell. 1985;42:213–223. doi: 10.1016/s0092-8674(85)80117-3. [DOI] [PubMed] [Google Scholar]
- 33.Nasmyth K, Dirick L. The role of SW14 and SW16 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 34.Neigeborn L, Mitchell A P. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- 35.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 36.Olempska-Beer Z, Freese E. Initiation of meiosis and sporulation in Saccharomyces cerevisiae does not require a decrease in cyclic AMP. Mol Cell Biol. 1987;7:2141–2147. doi: 10.1128/mcb.7.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rine J, Sprague G F, Jr, Herskowitz I. rme1 mutation of Saccharomyces cerevisiae: map position and bypass of mating type locus control of sporulation. Mol Cell Biol. 1981;1:958–960. doi: 10.1128/mcb.1.10.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robzyk K. Transcriptional regulation of the IME1 gene in yeast. Ph.D. thesis. Haifa, Israel: Technion; 1997. [Google Scholar]
- 39.Rose M, Botstein D. Construction and use of gene fusions to LacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 40.Rose M D, Broach J R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 41.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruis H, Schuller C. Stress signaling in yeast. Bioassays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah J C, Clancy M J. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shefer-Vaida M, Sherman A, Ashkenzi T, Robzyk K, Kassir Y. Positive and negative feedback loops affect the transcription of IME1, a positive regulator of meiosis in Saccharomyces cerevisiae. Dev Genet. 1995;16:219–228. doi: 10.1002/dvg.1020160302. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 47.Sherman A, Shefer M, Sagee S, Kassir Y. Post-transcriptional regulation of IME1 determines initiation of meiosis in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:375–384. doi: 10.1007/BF00279441. [DOI] [PubMed] [Google Scholar]
- 48.Smith H E, Driscoll S E, Sia R A L, Yuan H E, Mitchell A P. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–784. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith H E, Mitchell A P. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su S S, Mitchell A P. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 1993;21:3789–3797. doi: 10.1093/nar/21.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su S S, Mitchell A P. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics. 1993;133:67–77. doi: 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Tatchell, K. Personal communication.
- 52.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Penalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toone W M, Johnson A L, Banks G R, Toyn J H, Stuart D, Wittenberg C, Johnston L H. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tschumper G, Carbon J. Copy number control by a yeast centromere. Gene. 1983;23:221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- 55.Van Aelst L, Boy-Marcotte E, Camonis J H, Thevelein J M, Jacquet M. The C-terminal part of the CDC25 gene product plays a key role in signal transduction in the glucose-induced modulation of cAMP level in Saccharomyces cerevisiae. Eur J Biochem. 1990;193:675–680. doi: 10.1111/j.1432-1033.1990.tb19386.x. [DOI] [PubMed] [Google Scholar]