Abstract
Repair of a double-strand break (DSB) in yeast can induce very frequent expansions and contractions in a tandem array of 375-bp repeats. These results strongly suggest that DSB repair can be a major source of amplification of tandemly repeated sequences. Most of the DSB repair events are not associated with crossover. Rearrangements appear in 50% of these repaired recipient molecules. In contrast, the donor template nearly always remains unchanged. Among the rare crossover events, similar rearrangements are found. These results cannot readily be explained by the gap repair model of Szostak et al. (J. W. Szostak, T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl, Cell 33:25–35, 1983) but can be explained by synthesis-dependent strand annealing (SDSA) models that allow for crossover. Support for SDSA models is provided by a demonstration that a single DSB repair event can use two donor templates located on two different chromosomes.
Tandem repeat instability is implicated in several human genetic diseases. The best-documented examples of deleterious rearrangements in tandem repeats are the massive amplifications of microsatellite DNA, known to be responsible for a dozen diseases, including fragile X syndrome and Huntington’s disease (for reviews, see references 62 and 74). Rearrangements affecting minisatellites (repeats of 10 to 50 nucleotides) can be harmful, too (4). For example, expansions of a minisatellite are associated with epilepsy (30, 31, 73). During meiosis, minisatellites can display a very high rate of modification, including intra-allele duplications and deletions, and nonreciprocal interallelic transfer of information (2, 24). Recently, a human minisatellite was also found to display massive amplification (78). Rearrangements in tandem repeats are not specific to micro- and minisatellites. Expansions and contractions of larger tandem repeats have been observed in Drosophila melanogaster and yeast (48, 49, 69, 75, 76).
While replication slippage can easily account for small changes in microsatellite copy number (63), the origin of massive amplifications remains a mystery. Since the predominant rearrangement events observed in minisatellites are nonreciprocal interallelic transfers of information, the meiotic instability affecting those sequences is thought to result from gene conversions rather than replication (24). Tandem repeat rearrangements observed in Drosophila are linked to P-M dysgenesis and have also been supposed to be the consequence of genetic recombination, because P-element excision is known to induce gene conversion (9, 29, 48, 49, 69).
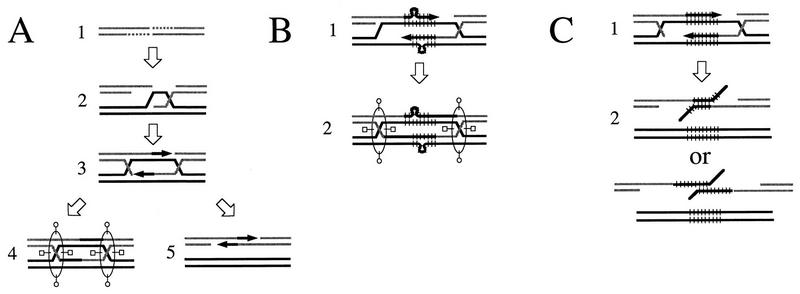
Gene conversions are most often explained by the double-strand break (DSB) repair model, proposed by Resnick and Martin (54) and Szostak et al. (68) to account for recombination events in yeast and other fungi. Many of the features of this model, or of its revised version (67), have been experimentally verified in Saccharomyces cerevisiae. The initial observation that a DSB in the DNA double helix induced a gene conversion in mitotic cells (45) was corroborated by results showing that some site-specific gene conversions, such as mating-type switching and intron homing, are initiated by site-specific DSBs (3, 52, 64), as are most meiotic gene conversions and chromosomal exchanges (33, 66). DSB formation is followed by resection of the ends (Fig. 1A, step 1) (65, 67, 77). The resulting 3′ ends can then invade the template (Fig. 1A, step 2) and provide a primer for new DNA synthesis, resulting in the restoration of the degraded single strands.
FIG. 1.
The SDSA model, and how it could explain rearrangements in tandem arrays. (A) Two alternative DSB repair models. The ends of the break are first resected (step 1), and the resulting 3′ ends can invade a homologous template and prime DNA synthesis (steps 2 and 3). Szostak et al. (68) proposed that resolution would require cutting of two Holliday junctions, resulting in crossover or noncrossover products (step 4), but to explain the lack or low frequency of crossover events accompanying mitotic gene conversion, other authors (10, 22, 42) proposed that the newly synthesized strands could be unwound from the template and anneal together (step 5). (B) A model of tandem repeat rearrangements derived from the DSB repair model of Szostak et al. (68). Expansions and contractions could be due to slippage events during semiconservative DNA synthesis (step 1). The resulting loop could be corrected in either direction, or not be corrected, but anyway, after resolution of the Holliday junctions (step 2), the rearrangements should be found on both donor and recipient molecules. (C) Origin of tandem repeat rearrangements according to a simple SDSA model. Both 3′ ends of the broken molecule invade the homologous template and prime DNA synthesis (step 1). Then, the newly synthesized strands are unwound from the template and anneal together. Two of the many possibilities of annealing are represented (step 2).
As gene conversion in yeast was observed to be frequently associated with crossover, the DSB repair model postulates that resolution of the gene conversion occurs through the cutting of two Holliday junctions (Fig. 1A, step 4) (58, 68). Holliday junctions are symmetrical structures, and it was assumed that they could be resolved in two different ways, resulting either in crossover or in no crossover (23). In the original experiment supporting the DSB repair model, half of the gene conversion events were accompanied by crossover (46). However, in most subsequent studies, the rate of crossover associated with gene conversion was found to be much less than 50%, and many types of gene conversion are rarely associated with crossover, in Saccharomyces (26, 27, 51), Drosophila (10, 18), Ustilago maydis (11), bacteria (41), and humans (24). Thus, a series of models which do not require Holliday junctions have been proposed. Their basic assumption is that after strand invasion and new DNA synthesis, the newly synthesized DNA strands are unwound from the template, allowing the annealing of the two free 3′ ends surrounding the DSB (Fig. 1A, step 5) (4, 10, 11, 22, 38, 41–44). Because this model of gene conversion includes an event of single-strand annealing, it was termed synthesis-dependent strand annealing (SDSA) by Nassif et al. (43).
The distinctive feature of SDSA models is that the recipient locus receives two newly synthesized strands of DNA and the donor template remains unchanged. DNA synthesis is thus conservative and fundamentally different from the semiconservative genome replication that occurs during the S phase of the cell cycle. As McGill et al. (38) noted, this could be the result of an unwinding, by a topoisomerase, of the two duplex DNAs created during gene conversion, restoring the original template strands to one duplex and the newly made strands to the other. An alternative model is that newly synthesized strands are continuously displaced from the template, analogous to the bubble migration mechanism proposed by Formosa and Alberts (14). In this view, newly synthesized DNA maintains only a short base-paired contact near the point of synthesis, as in RNA transcription, and the newly synthesized DNA is largely unpaired with its template and is free to anneal with a complementary strand.
SDSA models were initially proposed to explain a lack of crossover, but they can also explain the observation that during yeast mating-type switching and P-element excision, gene conversion events are unidirectional (18, 38), in other words, that there is no transfer of information from the repaired molecule to the template. In addition, they can explain the P-induced rearrangements in tandem repeats in Drosophila (9, 29, 48, 49, 69) and the meiotic rearrangements in human minisatellites (2, 4, 24) and in yeast CUP1 sequences (75, 76) as the consequences of DSB repair. Figure 1C shows how unwinding of newly synthesized sequences can lead to tandem repeat rearrangements. Although recombination has been implicated in these systems, there are limitations to a complete description of the process: the hypothesis of an initiating DSB cannot be checked, and the donor and recipient molecules cannot both be recovered (in the Drosophila and human systems) or cannot be identified (in the yeast CUP1 system). Moreover, to explain P-induced rearrangements, one had to suppose that the template for gene conversion was the sister chromatid, which is impossible to demonstrate.
In this paper, we present an experimental system in which both the donor and recipient molecules can be recovered and identified after recombination induced by a known DSB at a specific site. We demonstrate that when a repeated array is transferred by DSB repair to a broken molecule in yeast, the transferred sequence displays various expansions and contractions in half of the recombinants. The template itself is not modified. The expansions sometimes involve the duplication of more than 4 kb. To explain such events by “classical” replication slippage, one would have to hypothesize that the newly synthesized DNA strands are unwound from their template over several kilobases, which is the distinctive feature of conservative DNA synthesis (14). Furthermore, we formally demonstrate that DSB repair can be achieved from two different templates, which also requires the detachment of newly synthesized DNA. These results support the SDSA model for gene conversion and the idea that the DNA synthesis associated with DNA repair is fundamentally different from the semiconservative replication of the genome. We suggest that DSB repair can be a major source of amplification of tandemly repeated sequences. Thus, the massive amplifications of microsatellites responsible for many genetic diseases could have their origin in DNA repair rather than in genome replication.
MATERIALS AND METHODS
Plasmids.
A series of five plasmids, diagrammed in Fig. 2, were derived from Ted, a centromeric plasmid marked by the URA3 gene (provided by W. Kramer). In pFP14, a genomic XhoI/SalI fragment including the LEU2 gene was inserted into the polylinker of Ted. In the other four Ted derivatives, an extra sequence was inserted in the KpnI site of LEU2. In pFP5, the extra sequence contains two 5S arrays of seven 5S genes surrounding the white gene from D. melanogaster and was taken from pCA422 (49). When pFP5 is transformed into yeast, the repeated 5S genes undergo deletions similar to those described in this study, but the plasmid is stable after transformation (50). The two plasmids containing 5S genes and diagrammed in Fig. 2 are transformant derivatives of pFP5. In pFP23, the insert is a EcoRI fragment containing white, and in pFP24, it is a EcoRI/SmaI fragment containing a truncated white copy. pFP13, the plasmid shown in Fig. 6, is a centromeric plasmid marked by TRP1 and contains an XhoI/SalI fragment corresponding to the LEU2 gene, where the 400-bp KpnI/EcoRI fragment has been replaced by a 40-bp synthetic HO cut site.
FIG. 2.
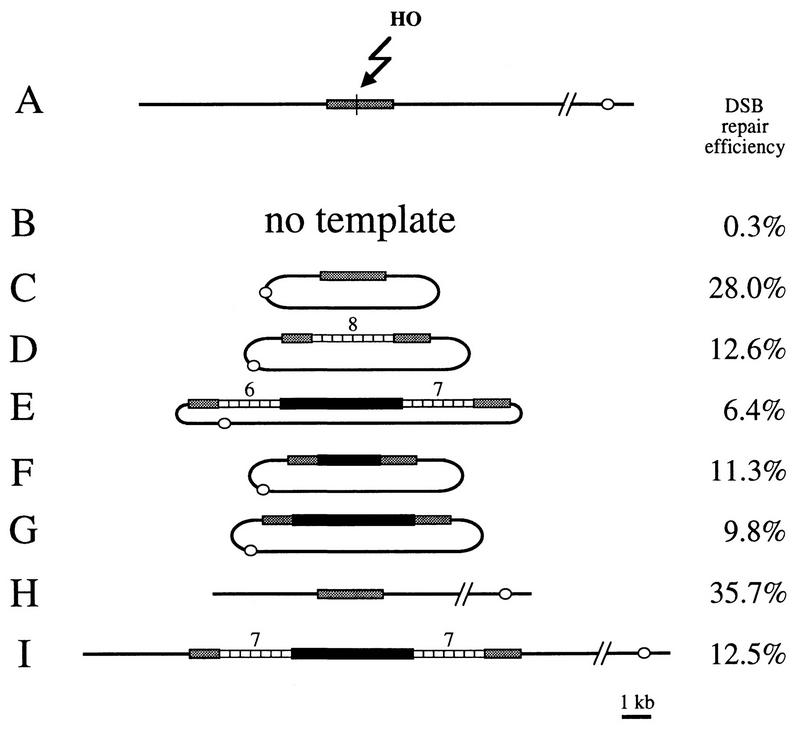
Experimental system to study DSB repair involving tandem arrays. An HO cut site is introduced into a KpnI site of the chromosomal endogenous LEU2 gene. The HO gene can be expressed from an inducible promoter and will cut the LEU2 gene (A). The structures of the templates are shown as follows: no homologous template (B); plasmid LEU2 template (C); plasmid LEU2 templates containing eight D. melanogaster 5S genes in a tandem array (D), two 5S arrays surrounding the D. melanogaster white gene (E), the 5′ part of the white gene (F), or the entire white gene (G); chromosomal LEU2 template (H); and chromosomal leu2 template with a 9.1-kb insert corresponding to two 5S arrays surrounding the white gene (I). Shaded box, LEU2 gene; open box, one D. melanogaster 5S gene (375 bp); solid box, D. melanogaster white gene; open circle, centromere. The number of repeats in each 5S array is given above the array.
FIG. 6.
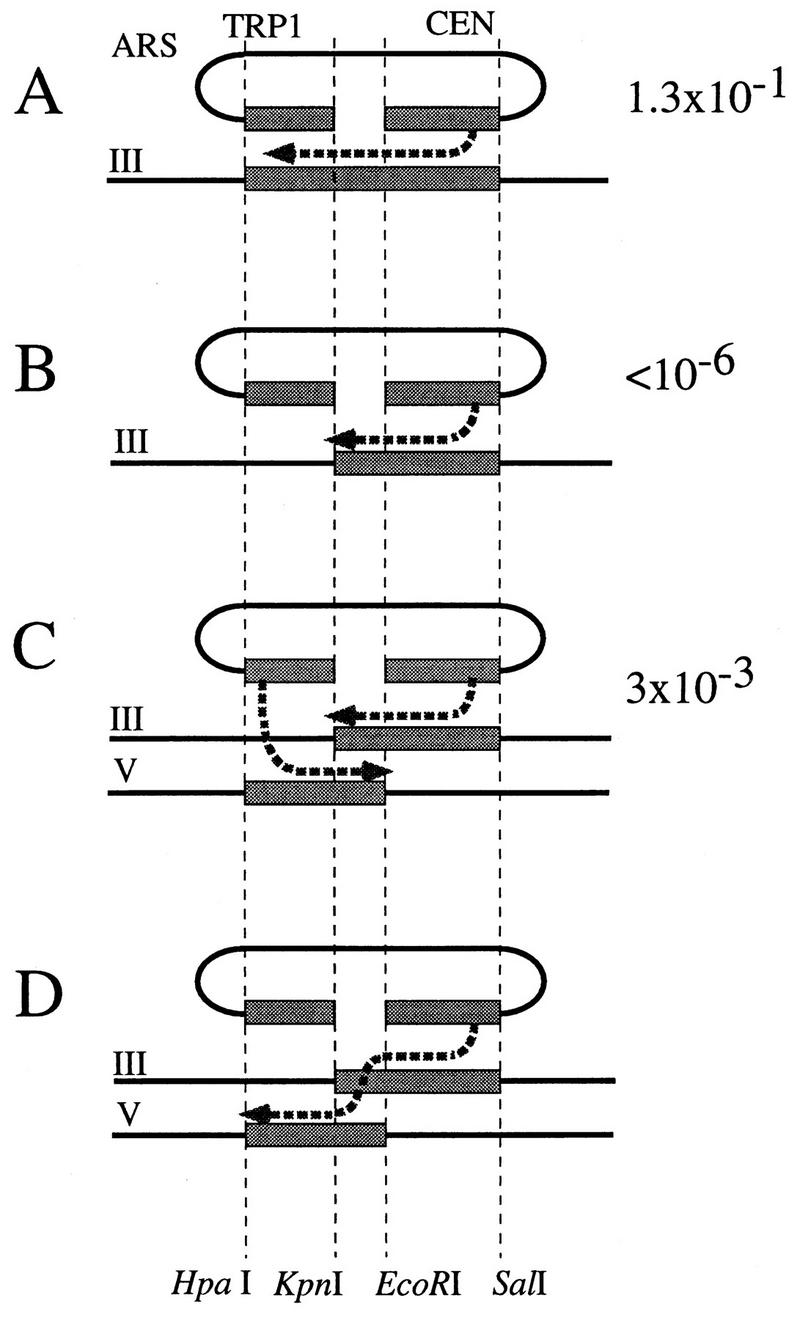
Direct evidence for the SDSA model. (A) A plasmid with a gapped leu2 gene can be repaired on an uninterrupted LEU2 template. This event minimally requires one strand invasion. The newly synthesized strand can then anneal to the other 3′ end of the DSB. (B) A truncated template with no overlap with one side of the gap does not allow for efficient repair. (C and D) Two truncated templates also allow for gap repair. Each template is lacking an overlap on one side of the gap but is overlapping with the other template. On each panel, an arrow(s) represents the invading 3′ end(s) after it has been extended by DNA synthesis. Panels C and D represent two different mechanisms to account for the same events: both 3′ ends invade and prime DNA synthesis before the newly synthesized DNA strands are unwound from the templates and anneal together (C), or DNA synthesis from a single 3′ end can switch from one template to the other (D). In both cases, two strand invasion steps and one annealing step are required.
We used the following plasmids to modify the chromosomal genome. pLS27 (provided by Laurence Signon) contains the URA3 marker and a LEU2 gene with a 117-bp HO cut site inserted into the KpnI site. pFP18 contains a MATa locus where the BglII/BsaAI fragment containing the HO cut site is replaced by a hisG::URA3::hisG cassette (1). pJH18 contains the URA3 and LEU2 genes in the same orientation, and pFP20, a derivative of pJH18, contains the same insert in LEU2 as pFP5. pFP10 contains the URA3 marker and a truncated leu2 gene, missing the EcoRI/SalI fragment embracing the 3′ end of the gene.
Strains.
The S. cerevisiae strains studied in this study all derive from JKM111 (40), which contains a GAL::HO fusion inserted into the chromosomal ADE3 locus (57), and G304 (20). All transformations were performed as described by Chen et al. (7). One-step disruption (55) and two-step replacement (59) were used to modify the genome, and the corresponding transformed strains were checked by Southern blotting. The MAT locus of JKM111 was disrupted with pFP18, and an HO cut site was introduced into its LEU2 gene with the pLS27 plasmid, resulting in the YFP17 strain. To provide a chromosomal LEU2 donor template, pJH18 or pFP20 was integrated into the ura3-52 gene, on chromosome V of YFP17. To provide a plasmid LEU2 template, pFP5, pFP14, pFP23, or pFP24 was transformed into YFP17.
G556, a G304 derivative, contains a GAL::HO fusion integrated into the chromosomal ADE3 gene. In the related G522 strain, an XhoI/KpnI fragment in the 5′ part of LEU2 has been replaced by ADE1. pFP10 was integrated in G522, into the ura3-52 gene on chromosome V, to produce the G520 strain, with two truncated leu2 donor templates. G520, G522, and G558 were transformed with pFP13 to obtain the strains diagrammed in Fig. 6.
DSB induction and characterization of recombinants.
Yeast was grown for 24 h in yeast extract-peptone (YEP)-dextrose, or in uracil or tryptophan dropout medium if plasmid selection was required. This culture was then used to inoculate 50 ml of YEP-glycerol at an initial concentration of 106 cells per ml. The YEP-glycerol culture was grown overnight, to a final concentration of 1 × 107 to 5 × 107 cells per ml, to prepare the cells for galactose induction. Then cells were plated on YEP-dextrose and YEP-galactose plates at a concentration of about 200 cells per plate. In the absence of any DSB, colonies appear on YEP-dextrose and YEP-galactose with the same efficiency (data not shown). For strains with an HO cut site on the chromosomal LEU2 gene, DSB repair efficiency was scored as the ratio of the number of colonies on YEP-galactose to that on YEP-dextrose. Independent colonies were then subcloned, and independent subclones were molecularly characterized as described by Pâques et al. (49). For the strains with an HO cut site on a plasmid LEU2 gene, we used a different procedure. For the strains with a single truncated leu2 template (Fig. 6B) or with two truncated leu2 templates (Fig. 6C), colonies were replicated onto leucine dropout plates, and the efficiency of gap repair was scored as the frequency of Leu+ colonies. Twenty Leu+ recombinants from the strain with two templates were checked molecularly and were shown to have gap repaired the plasmid LEU2 gene. For the strain with a single continuous LEU2 template (Fig. 6A), the LEU2 marker cannot be used to monitor homologous repair because all the cells already have a functional LEU2 copy. Therefore, colonies were replicated onto tryptophan dropout plates, and the efficiency of gap repair was scored as the frequency of Trp+ colonies among all colonies. Twenty Trp+ recombinants were checked molecularly, and in these strains, the plasmid LEU2 gene had been gap repaired.
Identification of crossover events.
A leu2 donor was inserted on chromosome V of YFP17 by targeted integration of pFP20 in ura3-52, so that the leu2 donor is surrounded by ura3-52 and URA3. Spontaneous “pop-out” events recombining the functional URA3 gene and the mutated ura3-52 in a single ura3-52 gene occur with a frequency of about 10−4 and result in papillations when the strain is replicated onto 5-fluoro-orotic acid (5-FOA) medium (21). Crossover occurring between the two leu2 copies on chromosomes III and V will give rise to a reciprocal translocation. The URA3 and ura3-52 gene will then be separated, pop-out will be impossible, and the papillations will disappear (21). Recombinants were patched onto 5-FOA plates, and if no papillation was observed, the strain was analyzed molecularly by Southern blotting. Crossover structure was tested with the diagnostic SphI and EagI restriction enzymes (see Fig. 4). However, because of coconversions of an adjacent delta sequence, only 20% of total crossover events could be identified by this screen. Nevertheless, these coconversions do not seem to affect the nature and frequency of the rearrangements we observe (50).
FIG. 4.
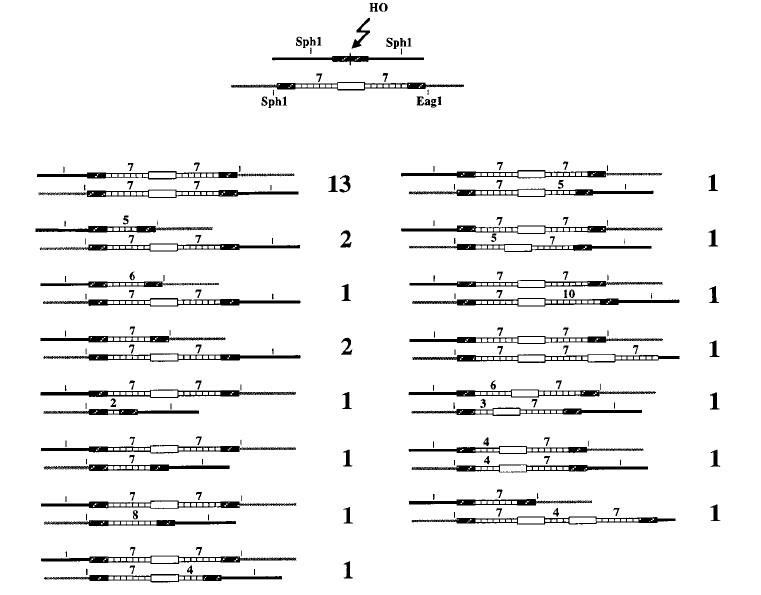
Crossover events. The diagram at the top represents the donor and the recipient. The cutting of the donor by HO endonuclease, and genetic and molecular identification of crossover events (with SphI and EagI diagnostic restriction enzymes) allowed us to characterize the 29 events whose structures are shown here.
Media and growth conditions.
YEP-dextrose and synthetic dropout media used for the growth of S. cerevisiae were made according to the method of Sherman et al. (60). YEP-galactose contains 2% galactose (wt/vol) instead of glucose to replace glucose as a carbon source. YEP-glycerol contains 2% glycerol (wt/vol) instead of glucose.
RESULTS
P-induced rearrangements in tandem repeats in Drosophila and meiotic rearrangements in the yeast CUP1 locus and in human minisatellites led to the hypothesis that copying of a repeated array during DSB repair could occur with deletions and duplications of the repeated sequence (4, 9, 24, 29, 48, 49, 69, 75, 76). To test this hypothesis, we designed an assay with the yeast S. cerevisiae (Fig. 2). We used the site-specific HO endonuclease under the control of an inducible promoter to deliver one DSB per cell within the LEU2 gene. In S. cerevisiae, DSBs are repaired mainly by homologous recombination, so when no homologous template is available, DSB repair is very inefficient (0.3% of the cells). However, a homologous LEU2 template, either on a centromeric plasmid (Fig. 2C) or inserted on a chromosome (Fig. 2H), allows DSB healing in 28 to 36% of the cells. Various additional sequences were inserted in the template, at the exact same location as the HO cut site, so that in order to repair the break by homologous recombination, the broken molecules would have to copy the insert. Three of these inserts have a complex repeated structure: they carry a single D. melanogaster 5S RNA gene array containing eight repeats (Fig. 2D) or the D. melanogaster white gene surrounded by two 5S gene arrays (Fig. 2E and 2I). These same constructs were made and used previously to characterize P-induced rearrangement in Drosophila (49). Repair occurred in 6 to 13% of the cells, depending on the size of the insert and the location (plasmid or chromosome) of the template. When repair occurred on a template containing a repeated array, we checked the structure of a sample of repaired molecules to look for DSB-induced tandem repeat rearrangements.
Copying of a repeated array during DSB repair is accompanied by expansions and contractions of this array.
After galactose induction of DSBs, independent recombinants were recovered. With a plasmid template, only noncrossover events can be recovered, because crossover would result in a dicentric chromosome. With a chromosomal template (Fig. 2I), molecular analysis showed that a crossover event created a pair of reciprocal translocations in 5 of 80 cases (6.3%). We first focused on the noncrossover events.
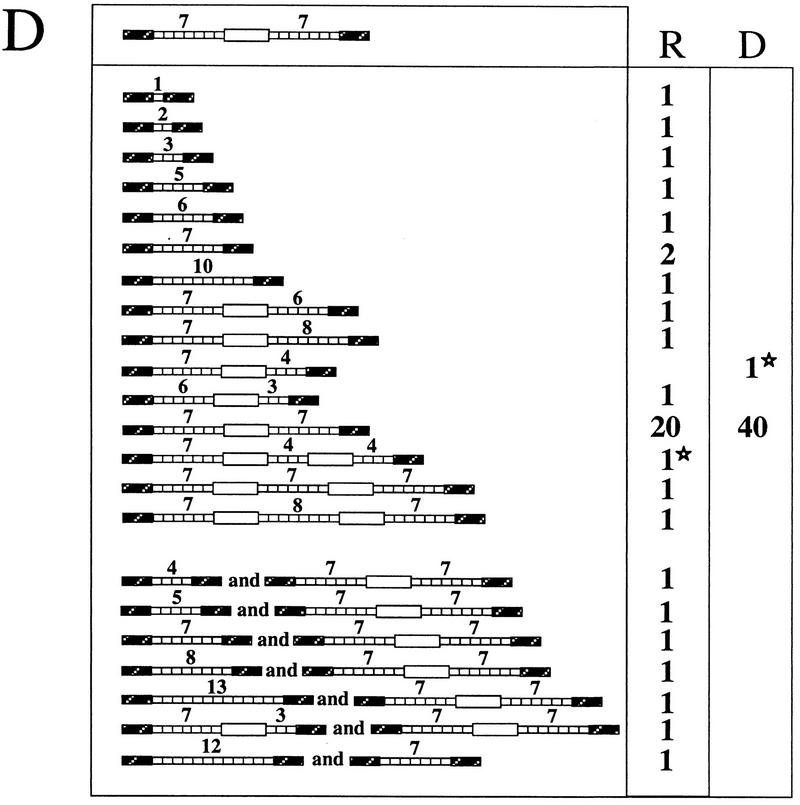
Analysis of the noncrossover-repaired molecules showed that 36 to 48% of them had assimilated a rearranged repeated insert. The types of tandem repeat rearrangements are shown in Fig. 3. With a plasmid template containing a 2.9-kb insert corresponding to eight 5S genes (Fig. 2D), 36% of the repaired molecules were rearranged, including 10 deletions and 6 duplications (Fig. 3A). With a plasmid template containing two 5S arrays and the white gene (8.7 kb; Fig. 2E), 45% of the repair events were associated with rearrangements, nearly all of them deletions, with a single large duplication including white (Fig. 3B, column R). In most of the rearrangements that did not remove the white gene, only one of the two 5S arrays was modified. The same distribution of events was observed when this template was integrated in chromosome V (Fig. 2I and 3C).
FIG. 3.
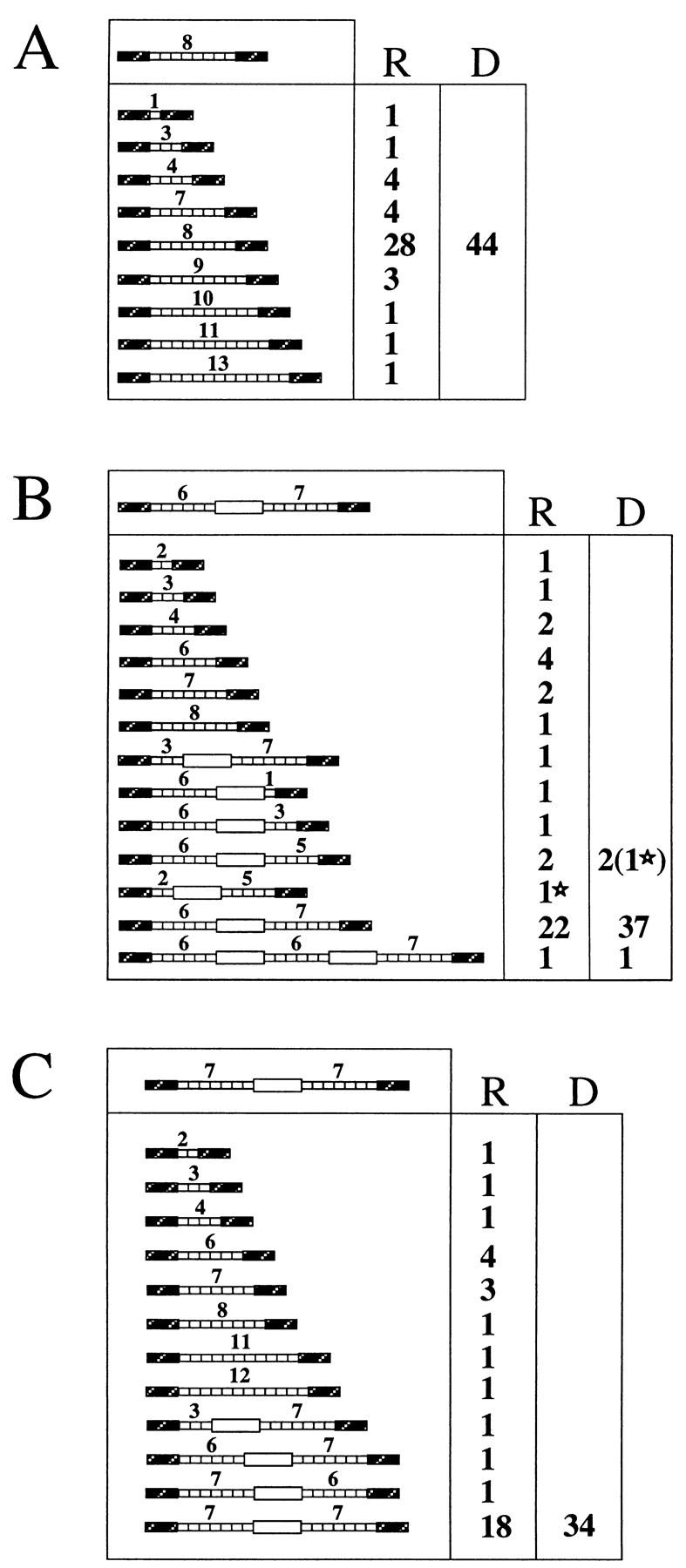
Recombinants obtained by DSB repair involving tandem repeats. For each panel, the structure of the template is diagrammed in the upper box, and the numbers of recipient (R) and donor (D) molecules with each structure are given on the right. (A) Forty-four recombinants obtained after DSB repair using the plasmid template diagrammed in Fig. 2D. (B) Forty recombinants obtained after DSB repair using the plasmid template diagrammed in Fig. 2E. (C) Thirty-four noncrossover recombinants obtained after repair using the template diagrammed in Fig. 2I. (D) Forty-one noncrossover recombinant lines obtained after repair in G1 of the template diagrammed in Fig. 2I. In seven cases (bottom), two populations of cells differing in the structure of the repaired molecule, were obtained. In panels B and D, one recombinant displays tandem repeat rearrangement in both donor and recipient. In these cases, the corresponding donor and recipient molecules are labeled with stars.
In these experiments, random colonies appearing after galactose induction were picked and subcloned before characterization. One could argue that if induction occurred in G2, we did not necessarily recover the donor and recipient molecules from the same recombination event. Therefore, we separated unbudded (G1) and large-budded (G2) cells by micromanipulation and induced them separately. The survival was 13.0% for G1 cells (47 of 360) and 25.6% for G2 cells (45 of 176). The entire colony derived from each of 44 G1 cells was analyzed. The structures of the 41 noncrossover events are shown in Fig. 3D. Again, various rearrangements are found on a recipient molecule in about half of the cases (21 of 41). Interestingly, we sometimes recovered a mixture of two different structures of the recipient locus (Fig. 3D, seven bottom rows). In six cases, one recipient molecule had received a contracted tandem array while the other had received an unrearranged array. In one case, the recipient molecules had two different contracted tandem arrays. These composite colonies would not have been obtained in the former experiment, as the recombinants were subcloned before characterization. Here, however, we recovered and analyzed the entire colony derived from a G1 cell, so that a rearranged molecule (recipient or donor) could not have segregated away from the recipient before analysis.
While the repeated sequences that were transferred into the recipient molecule were frequently rearranged, the template was modified in only 4 of 159 cases (Fig. 3B and D, columns D). In two of these four cases, the recipient molecule also displayed a complex rearrangement. We conclude that the rearrangements are almost always confined to the newly synthesized sequences.
These results cannot readily be explained by the DSB repair model proposed by Szostak et al. (68), which assumes that semiconservative DNA synthesis is initiated from both 3′ ends of the DSB (Fig. 1A, step 3). Rearrangements could occur on either of the newly synthesized strands (Fig. 1B, step 1) and, after resolution of the Holliday junctions, would be found on both donor and recipient molecules. Our results support SDSA models, which imply a conservative DNA synthesis and predict that whatever rearrangement occurs during DNA synthesis will be found on the recipient molecule only.
Crossovers and tandem repeat rearrangements are compatible.
In its simplest form, the SDSA model of DSB repair predicts that rearrangements should occur on the recipient molecule without altering the donor, and without any associated crossover (10, 22, 42). As illustrated in Fig. 1C, both ends would invade the template and prime new DNA synthesis. Rearrangements would appear during the resolution step, because of the multiple possibilities of annealing offered by the repetitive structure. In this version of SDSA, rearrangements and crossovers would be mutually exclusive.
To test this hypothesis, we looked for crossover events and checked if they were associated with 5S gene deletions or duplications. Only five crossovers (6.3%) had been detected among 80 characterized recombinants (the noncrossover events are diagrammed in Fig. 3C and D). Therefore, we used a genetic screen to identify more events (see Materials and Methods). Figure 4 diagrams the structures of 29 crossover recombinants. Thirteen of them were not associated with tandem repeat rearrangement. Of the 16 recombinants with rearrangements, 13 had a rearrangement in only one of the repeated arrays. Two large duplications encompassing white were found among the rearrangement events. In noncrossover events, we can define a donor and a recipient molecule, but in crossover events, both are split by the reciprocal translocation. This is probably why the crossover events we characterized display rearrangement on either of the two 5S loci found after DNA repair. Thus, rearrangements occur in about 55% of crossovers. This ratio is not significantly different from the 48% of rearrangements found among noncrossover events. Both deletions and duplications appear to be perfectly compatible with crossovers. This rules out the simple SDSA hypothesis cited above (Fig. 1C).
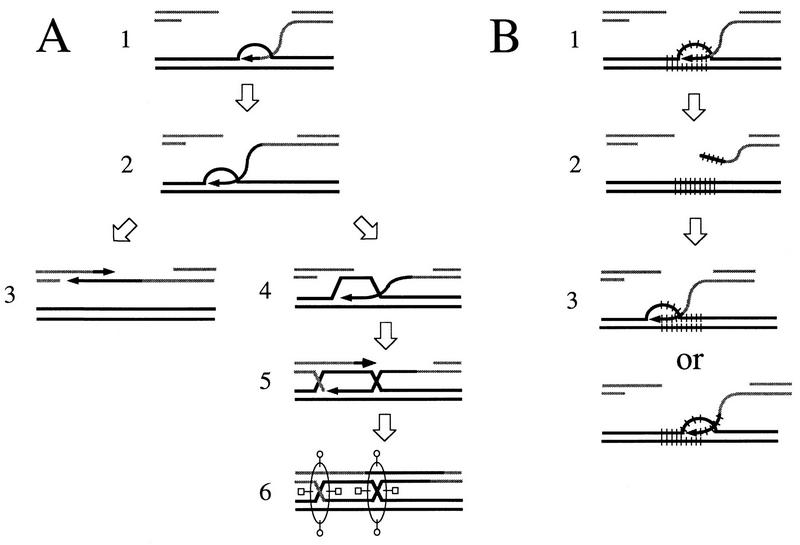
Thus, we favor an alternative model of SDSA (11, 41, 44), which assumes that most of the time, only one 3′ end invades the template donor, and it is extended by bubble migration (Fig. 5A). As pointed out by Ferguson and Holloman (11), this SDSA model is compatible with crossovers. Invasion or pairing of the second 3′ end may stabilize the strand displaced by the first one, making unwinding unlikely. The gene conversion would thus resemble the branched intermediates proposed by Szostak et al. (68), and resolution of the Holliday junctions would occur with or without crossover.
FIG. 5.
A model of SDSA compatible with crossovers. (A) A single 3′ end invades the template and primes synthesis (step 1) and is extended by bubble migration (step 2). Most of the time, resolution can occur by annealing (step 3). However, invasion of the template by the second 3′ end may sometimes stabilize the strand displaced by the first 3′ end (step 4). DNA synthesis would become semiconservative (step 5), and Holliday junctions could be formed (step 6) and subsequently cut. (B) Creation of tandem repeat rearrangements by reinvasion. A single 3′ end would invade the donor template and initiate DNA synthesis (step 1). It would then be unwound from the template (step 2) and invade this template a second time. Because repeated sequences have been added to the 3′ end, this second invasion can be initiated on any of the tandem repeats on the template. Two different possibilities are shown here (step 3). The conversion could then be accomplished by annealing or by formation and cutting of Holliday junctions, as shown in panel A.
This SDSA model can also account for the tandem repeat rearrangements we observe (Fig. 5B). After a newly synthesized strand is unwound, it may reinvade the template duplex. If newly synthesized DNA strands from both sides of the break do not overlap yet, it may even be the only possibility, for no annealing would be possible. Rearrangements could then occur because of the multiple possibilities of strand reinvasion (Fig. 5B, step 3) allowed by the redundant structure, and they would thus be the consequence of “slippage-like” events. Subsequently, resolution could occur by annealing (Fig. 5A, step 3) or by formation and cutting of Holliday junctions (Fig. 5A, step 4), and thus, crossovers and tandem repeat rearrangements would be compatible.
Use of a split template during DSB repair.
In Drosophila, many complex P-induced gene conversion events have been found, where sequences were supposed to be copied from two different templates (18, 19, 43). However, in these studies, one of the two templates was presumed to be a sister chromatid, which could not be demonstrated. Similar observations have been reported with S. cerevisiae in a transformation experiment (61). The origin of such events could rely on the ability of newly synthesized DNA to unwind from its template during DNA repair: DNA synthesis could be initiated on two different templates, and the two different new DNA molecules could then be annealed together (18, 43, 61). This corresponds to the model of SDSA diagrammed in Fig. 1A, except that the two 3′ ends invade sequences on different chromosomes. Another possibility is that template switching would occur during DNA synthesis (19), which corresponds to the SDSA model diagrammed in Fig. 5A, the one we favor (see above).
We designed a system to test if DSB repair can occur when it must use two different templates (Fig. 6). A plasmid with a gapped leu2 gene containing an HO cut site was transformed into three yeast strains differing only in their LEU2 donor sequences. Gap repair resulting in a functional plasmid LEU2 gene was induced by HO endonuclease. It is well documented that gap repair of a plasmid can occur by gene conversion from a chromosomal template (44–46), and in our assay, a complete LEU2 donor template allowed for gap repair in 13% of the cells (Fig. 6A). As predicted by current models, gap repair was very inefficient (<10−6) with a truncated leu2 template that shares no common sequence with one side of the DSB in the plasmid leu2 (Fig. 6B). However, a significantly higher level of gap repair (0.3%) was observed with two overlapping truncated templates located on different chromosomes (Fig. 6C). We confirmed genetically and molecularly that the gap-repaired, functional LEU2 gene was plasmid borne and was not the result of a chromosomal rearrangement.
This result clearly shows that during DSB repair, sequences can be recruited from two different overlapping templates. Our assay does not allow us to decide between the two models proposed above, i.e., (i) the two 3′ ends each invade one of the templates (Fig. 6C), and resolution occurs by annealing of the newly synthesized DNA, or (ii) one of the 3′ ends invades one of the donors, and then switches to the other donor, in the overlapping region (Fig. 6D), and then anneals to the second end of the plasmid.
Gap repair efficiency decreases as the size of the gap increases.
When we induced a DSB in a chromosomal leu2 gene to obtain gap repair events that would have copied 5S genes, we noticed that as the size of the repeated array inserted in the leu2 template increased, gap repair efficiency decreased. With a plasmid template, repair occurred in 28% of the cells when the LEU2 donor contained no insert (Fig. 2C), but repair efficiency dropped to 12.6% with a 2.9-kb insert of eight 5S genes (Fig. 2D) and to 6.4% with a 9.1-kb insert containing two 5S arrays surrounding the white gene (Fig. 2E). When the template was on chromosome V, repair efficiencies were quite similar, dropping from 35.7% with no insert (Fig. 2H) to 12.5% with the 9.1-kb insert (Fig. 2I). This effect on gap repair could be specific to repeated sequences or to the 5S gene; therefore, we checked the effects of two other inserts. When we introduced a truncated 2.1-kb white gene into the LEU2 gene (Fig. 2F), DSB repair efficiency was 11.3%, very similar to the 12.6% efficiency obtained with a 2.9-kb 5S array. With a complete 4.1-kb white gene as an insert, repair efficiency reproducibly dropped further, to 9.8% (Fig. 2G). These results indicate that the decrease in DSB repair efficiency observed with inserts of increasing size is not 5S specific or specific to a redundant structure: increasing amounts of sequences from the white gene have the same effect. Our conclusion is that during double-strand gap repair, the size of the DNA segment to be copied limits the efficiency of repair. The most likely difficulty in gap repair appears to be in the processivity of DNA synthesis.
DISCUSSION
We describe here rearrangements of a repeated array during DSB-induced gene conversion. These rearrangements are very similar to changes in tandem repeats that have previously been observed in Drosophila by using the same substrates (48, 49), in human minisatellites (24), and in the yeast CUP1 repeated locus (75, 76) and which have been explained as the consequences of gene conversion. However, this is the first study in which we know not only where but also when the DSB appears, so that we can determine how often DSB repair results in tandem repeat rearrangements. One of our most striking results is that half of the repair events are associated with a deletion or a duplication.
This is also the first study in which we could recover and identify both donor and recipient sequences. This is impossible in the Drosophila and human systems, as mentioned above. Although Welch et al. (75, 76), who characterized meiotic contractions and expansions of the CUP1 locus in yeast, could recover all the recombination products, they could not identify donor and recipient molecules, nor could they tell how often repair occurred without rearrangement. This advantage allowed us to examine the mechanism of DSB repair in detail. We made two key observations.
First, when gene conversion is not associated with crossing-over, the rearrangements are almost always at the recipient site while the donor template remains unchanged. Second, while crossing-over associated with gene conversion is rare, about half of these events also contain changes in the number of repeats, but most of the time on only one of the two participating chromosomes. As we discuss below, these events are not readily explained by the DSB gap repair model proposed by Szostak et al. (68) or by an SDSA model that does not permit crossing-over (10, 22, 42). We can account for all these events by a modified form of the SDSA model that allows crossovers to occur (11).
DSB-induced rearrangements in tandem repeats are best explained in terms of an SDSA model.
The gap repair model of Szostak et al. (68) envisions that the two 3′ ends of a DSB will each invade the donor template and copy a new strand of DNA. This DNA synthesis is assumed to be semiconservative, like genome replication. Thus, the recipient locus as well as the donor locus become heteroduplex, with one original and one newly synthesized strand. Therefore, in the discussion below, we will refer to the Szostak et al. (68) model for DSB gap repair as a semiconservative gene conversion (SCGC) model. It might be imagined that during this semiconservative replication process, there was replication slippage, leaving a loop on either the newly synthesized or the template strand, to add or subtract repeats (5, 36, 63, 71, 72). However, such events should be just as likely to occur at the donor locus as at the recipient locus, since each is the product of an equivalent semiconservative replication process (Fig. 1B). But we find such rearrangements almost always at the recipient site. Moreover, although replication slippage can explain small duplications, such as the 100-bp duplications observed in a rad27 mutant (70), they can hardly account for the events we observe, involving sometimes more than 4 kb. To explain these duplications by replication slippage, one has to assume that the newly synthesized strands can be unwound from their template over several thousand base pairs, so that they can reinitiate DNA synthesis far upstream from the point they reached.
One might also hypothesize that rearrangements at the recipient locus are the consequence of a secondary event, after gap repair had produced a donor and a recipient with identical numbers of repeats. We think this unlikely for two reasons. First, the initiating lesion again would have to be confined to the recipient locus, and there is no evident asymmetry in the intermediate structures postulated by the SCGC gap repair model that would account for such bias. Second, while one might imagine a nick or DSB stimulating deletions between flanking repeats, as in models of single-strand annealing (6, 12, 34, 35, 47, 56, 65), it is difficult to explain how these lesions would produce increases in copy number.
Thus, we believe that these results are better explained in terms of SDSA. Here, newly synthesized DNA does not remain base paired to the template, as in normal DNA replication; rather, newly synthesized strands are displaced, allowing them to anneal. Because alignment of complementary newly synthesized strands can occur in different registers, the total number of repeats in the recipient site can easily have fewer or more copies than the donor template. The distinctive feature of SDSA is that the recipient locus receives two newly synthesized strands of DNA, while the donor template remains unchanged, so that DNA synthesis is conservative rather than semiconservative. This, of course, accounts for the observation that rearrangements are almost always found at the recipient and not at the donor site.
When we analyzed whole colonies resulting from a single G1 cell, we sometimes obtained mixed populations of recipient molecules (Fig. 3D). These events can be explained if repair of the DSB did not occur until after replication of the broken chromosome, at which time two independent repair events occurred. Alternatively, the DNA synthesis initiated from the second 3′ end after annealing (Fig. 5A, step 3) could also be prone to slippage-like events, resulting in heteroduplexes in the recipient molecule that would, after the first cell division, yield two different recipient molecules. However, we cannot rule out the possibility that in these cases, induction actually occurred after the cell had entered the S or G2 phase.
Direct evidence of SDSA.
The most compelling evidence for SDSA comes from our demonstration of gap repair when the template consists of two unlinked overlapping sequences, each of which has homology to only one end of the DSB (Fig. 6). Reconstitution of the intact LEU2 gene on a centromeric plasmid was accomplished 0.3% of the time, approximately 40 times less often than when the two ends of the DSB were both homologous to a single template strand. The difference in efficiency most likely reflects the fact that repair from two templates requires an additional strand invasion step. Gap repair from a single, intact template minimally requires a single strand invasion producing a displaced new strand of DNA that can anneal with the other 3′ end of the DSB (Fig. 6A). In contrast, repair from two overlapping unlinked templates requires two independent strand invasion steps, followed by an annealing step (Fig. 6C and D). Once one end of the DSB interacts with a donor, the interaction of the second end with the same template could be greatly enhanced.
We envision two alternative versions of this model. In the first (Fig. 6C), each end invades a template and each spins out a strand, which then anneals to repair the DSB. Alternatively, only one end may invade the first template, then detach and invade the second template (Fig. 6D) to complete the missing region, and finally anneal to the second end of the DSB, which need never have invaded the donor duplex. The latter mechanism resembles copy choice models (32), and a similar model has been proposed recently to account for P-induced events that seem to require the use of two different templates (19). In either case, there are two strand invasions and an annealing step.
The two models proposed in Fig. 6 can both account for the change in copy number during the copying of a template containing tandem repeats, as shown in Fig. 1C and 5B. However, the reinvasion (or template switching) mechanism can also account for gap repair events associated with crossing-over.
A modified SDSA model can account for crossover-associated events.
About half of the gap repair events yield the same number of repeats as the donor. One explanation for this distinctive clustering of “perfect” events is that many of them arise by a mechanism (i.e., SCGC) that is different from that which produces the gene conversions with different repeat numbers (SDSA). This possibility cannot be excluded, and it led us to examine gene conversion events with crossing-over. If all SDSA occurred without crossover, then one might conjecture that the crossover-associated events might derive from SCGC. In this case, one might expect all such events to have the same number of repeats as the donor; but we showed that this was not so (Fig. 4). Moreover, 13 of the 16 crossover-associated events showed a rearrangement on only one of the two arrays. This is not consistent with an unequal reciprocal exchange event occurring between two initially identical regions, although it could occur if some repeats were looped out in heteroduplex DNA (75). Consequently, although we cannot rule out the possibility that some gap repair events occur via SCGC, the crossover outcome predicted by such a model was not found.
Crossover-associated rearrangements can be explained by the reinvasion of one end during the copying of the template-containing repeats. Most of the time, the conversion event could be accomplished by an annealing with the second end of the DSB (Fig. 5A, step 3). However, if the second end also paired with the template, it might sometimes stabilize the displaced strand (Fig. 5A, step 4). DNA synthesis on both strands could then become semiconservative (Fig. 5A, step 5), and Holliday junctions could arise (Fig. 5A, step 6) and be cut, as predicted by Szostak et al. (68). This would account for the events shown in Fig. 4.
It must be pointed out that once DNA synthesis becomes semiconservative, no further change in copy number is likely. If a rearrangement has been generated by a reinvasion event (Fig. 5B, step 3) prior to the invasion of the second 3′ end (Fig. 5A, step 4) and the switch to a semiconservative mode of DNA synthesis (Fig. 5A, step 5), it will remain outside the region where the Holliday junctions are formed (Fig. 5A, step 6). Branch migration across the expanded or contracted repeated array would be difficult because it would have to bypass a loop. Thus, the rearrangement will not be affected by the crossover and will be clustered on one chromosome, which is what is observed in 13 of 16 cases. Invasion and reinvasion happening simultaneously on both sides of the DSB could result in two rearranged arrays, as happens in three cases.
We therefore propose that during DNA repair in vegetative cells, DNA synthesis is mostly not semiconservative but can sometimes switch to the semiconservative mode (11), which would allow for the appearance of crossover events. Given the low frequency of crossovers, switching to a semiconservative mode of DNA synthesis should be a rare event, and gene conversion would be accomplished by annealing most of the time. In this case, the two processes diagrammed in Fig. 1B and 5B (and the two processes suggested in Fig. 6, which are their analogs) would include exactly the same number of events, i.e., two strand invasions priming synthesis and one annealing, and would be two different versions of the same mechanism.
Meiotic gene conversions are associated with crossover more often than are mitotic conversions, which could indicate that semiconservative DNA synthesis would occur more frequently during DNA repair in meiotic cells. However, an overall deficit in crossover still appears (13), and some data indicate that nonsemiconservative DNA synthesis might be important in meiotic cells as well (17, 53).
DNA synthesis as a rate-limiting step.
If DNA repair synthesis by bubble migration is prone to be interrupted, it could also explain another feature of our results: gap repair efficiency decreases when the size of the gap increases. During genome replication, DNA polymerases are apparently very processive, but PolI, the bacterial DNA polymerase associated with DNA repair, is not (28). Nothing is known about the DNA polymerase(s) associated with DSB repair in eukaryotes, but our results suggest that they are not processive. When long sequences have to be synthesized before DNA repair can be accomplished, repair fails most of the time. An insert of 9 kb decreases repair efficiency fourfold. In addition, half of the repair events apparently require a reinitiation of DNA synthesis, resulting in a rearrangement. It is only the other half, the perfect repair events with no deletion or duplication, which may reflect uninterrupted DNA synthesis across the repeated template.
Altogether, our data strongly suggest that the DNA synthesis associated with DNA repair is fundamentally different from the genome replication occurring during the S phase: it is neither semiconservative nor processive.
DSB repair, a major cause of tandem repeat amplification?
Fu et al. (15) proposed that the massive amplifications of microsatellites could be the consequence of reinitiation of DNA synthesis. Our result show that DNA synthesis resulting from DSB repair is more likely to induce tandem repeat rearrangements than DNA synthesis associated with genome replication. Thus, the origin of the massive amplifications of microsatellites in fragile X syndrome or Huntington’s disease may be linked more to DNA repair than to genome replication. Thus, in the case of fragile X syndrome, amplifications appear during oogenesis or shortly after fertilization (15, 37), possibly during meiosis, when DSBs may frequently cut the genome. Some microsatellites have been shown to have the potential to form very stable hairpins (8, 16, 39), which could pause the replication machinery (25) and be responsible for small slippage events. The effect of such structures would be much more severe on the synthesis associated with DNA repair, because any pause of the polymerase could favor dissociation of the elongating strand from its template. One can imagine that with microsatellites, several events of dissociation and reinvasion of the elongating strand could occur during a single repair event. Very large amplifications could thus appear as a consequence of repairing a single DSB.
ACKNOWLEDGMENTS
We thank Susan Lovett, Michael Lichten, and members of the Haber laboratory for their helpful comments on the manuscript.
This work was supported by National Institutes of Health grant GM20056. F.P. was a fellow of the Association pour la Recherche contre le Cancer, and then of the Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armour J A, Jeffreys A J. Biology and applications of human minisatellite loci. Curr Opin Genet Dev. 1992;2:850–856. doi: 10.1016/s0959-437x(05)80106-6. [DOI] [PubMed] [Google Scholar]
- 3.Bremer M C, Gimble F S, Thorner J, Smith C L. VDE endonuclease cleaves Saccharomyces cerevisiae genomic DNA at a single site: physical mapping of the VMA1 gene. Nucleic Acids Res. 1992;20:5484–5490. doi: 10.1093/nar/20.20.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buard J, Jeffreys A J. Big, bad minisatellites. Nat Genet. 1997;15:327–328. doi: 10.1038/ng0497-327. [DOI] [PubMed] [Google Scholar]
- 5.Canceill D, Ehrlich S D. Copy-choice recombination mediated by DNA polymerase III holoenzyme from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6647–6652. doi: 10.1073/pnas.93.13.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll D, Wright S H, Wolff R K, Grzesiuk E, Maryon E B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986;6:2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Mariappan S V S, Catasti P, Ratliff R, Moyzis R K, Laayoun A, Smith S S, Bradbury E M, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc Natl Acad Sci USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delattre M, Anxolabéhère D, Coen D. Prevalence of localized rearrangements vs. transposition among events induced by Drosophila transposase on a P transgene. Genetics. 1996;141:1407–1424. doi: 10.1093/genetics/141.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engels W R, Johnson-Schlitz D M, Eggleston W B, Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 13.Fogel S, Mortimer R K, Lusnack K. Mechanisms of meiotic gene conversion, or “wanderings on a foreign standard,”. In: Strathern J N, Jones E W, Broach J R, editors. The molecular analysis of the yeast Saccharomyces cerevisiae: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 289–339. [Google Scholar]
- 14.Formosa T, Alberts B M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y-H, Kuhl D P A, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J M H, Holden J J A, Fenwick R G, Warren S T, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 16.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 17.Gilbertson L A, Stahl F W. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1996;144:27–41. doi: 10.1093/genetics/144.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloor G B, Nassif N A, Johnson-Schlitz D M, Preston C R, Engels W R. Targeted gene replacement in Drosophila via P element-induced gap-repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 19.Gonzy-Treboul G, Lepesant J A, Deutsch J. Enhancer-trap targeting at the Broad-Complex locus of Drosophila melanogaster. Genes Dev. 1995;9:1137–1148. doi: 10.1101/gad.9.9.1137. [DOI] [PubMed] [Google Scholar]
- 20.Haber J E, Leung W-Y. Lack of chromosome territoriality: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris S, Rudnicki K S, Haber J E. Gene conversion and crossing-over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings P J. Recombination in the eukaryotic nucleus. Bioessays. 1988;9:61–64. doi: 10.1002/bies.950090206. [DOI] [PubMed] [Google Scholar]
- 23.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 24.Jeffreys A J, Tamaki K, MacLeod A, Monckton D G, Neil D L, Armour J A L. Complex gene conversion events in germline mutation at human minisatellites. Nat Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Ohshima K, Shimizu M, Amirhaeri S, Wells R D. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease gene. J Biol Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 26.Klar A J, Strathern J N. Resolution of recombination intermediates generated during yeast mating type switching. Nature. 1984;310:744–748. doi: 10.1038/310744a0. [DOI] [PubMed] [Google Scholar]
- 27.Klein H L. Lack of association between intrachromosomal gene conversion and reciprocal exchange. Nature. 1984;310:748–753. doi: 10.1038/310748a0. [DOI] [PubMed] [Google Scholar]
- 28.Kornberg A, Baker T. DNA replication. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 29.Kurkulos M, Weinberg J M, Roy D, Mount S M. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics. 1994;136:1001–1011. doi: 10.1093/genetics/136.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafrenière R G, Rochefort D L, Chrétien N, Rommens J M, Cochius J I, Kälviäinen R, Nousiainen U, Patry G, Farrell K, Söderfeldt K, et al. Unstable insertion of the 5′ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat Genet. 1997;15:298–302. doi: 10.1038/ng0397-298. [DOI] [PubMed] [Google Scholar]
- 31.Laliotti M D, Scott H S, Buresi C, Rossier C, Bottani A, Morris M A, Malafosse A, Antonorakis S E. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature. 1997;386:847–851. doi: 10.1038/386847a0. [DOI] [PubMed] [Google Scholar]
- 32.Lederberg J. Recombination mechanisms in bacteria. J Cell Comp Physiol. 1955;45:75–107. doi: 10.1002/jcp.1030450506. [DOI] [PubMed] [Google Scholar]
- 33.Lichten M, Goldman A S H. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 34.Lin F-L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin F-L, Sperle K, Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990;10:113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovett S T, Feschenko V V. Stabilization of diverged tandem repeats by mismatch repair: evidence for deletion formation via a misaligned replication intermediate. Proc Natl Acad Sci USA. 1996;93:7120–7124. doi: 10.1073/pnas.93.14.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malter H E, Iber J C, Willemsen R, de Graaff E, Tarleton J C, Leisti J, Warren S T, Oostra B A. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet. 1997;15:165–169. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- 38.McGill C, Shafer B, Strathern J N. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- 39.Miltas M, Yu A, Dill J, Kamp T J, Chambers E J, Haworth I S. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller J E, Clyman J, Huang Y-J, Parker M M, Belfort M. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 42.Nasmyth K A. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 43.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson H H, Sweetser D B, Nickoloff J A. Effect of terminal nonhomology and homeology on double-strand-break-induced gene conversion tract directionality. Mol Cell Biol. 1996;16:2951–2957. doi: 10.1128/mcb.16.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orr-Weaver T L, Szostack J W, Rothstein R. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orr-Weaver T L, Szostack J W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozenberger B A, Roeder G S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pâques F, Wegnez M. Deletions and amplifications of tandemly arranged ribosomal 5S genes internal to a P element occur at a high rate in a dysgenic context. Genetics. 1993;135:469–476. doi: 10.1093/genetics/135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pâques F, Bucheton B, Wegnez M. Rearrangements involving repeated sequences within a P element preferentially occur between units close to the transposon extremities. Genetics. 1996;142:459–470. doi: 10.1093/genetics/142.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pâques, F., and J. E. Haber. Unpublished data.
- 51.Plessis A, Dujon B. Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene. 1993;134:41–50. doi: 10.1016/0378-1119(93)90172-y. [DOI] [PubMed] [Google Scholar]
- 52.Plessis A, Perrin A, Haber J E, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porter S E, White M A, Petes T D. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics. 1993;134:5–19. doi: 10.1093/genetics/134.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resnick M A, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 55.Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 56.Rudin N, Haber J E. Efficient gap repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1991;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 58.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 59.Sherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 61.Silberman R, Kupiec M. Plasmid-mediated induction of recombination in yeast. Genetics. 1994;137:41–48. doi: 10.1093/genetics/137.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinden R R, Wells R D. DNA structure, mutation, and human genetic disease. Curr Opin Biotechnol. 1992;3:612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- 63.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 64.Strathern J N, Klar A J, Hicks J B, Abraham J A, Ivy J M, Nasmyth K A, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 65.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-strand DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 67.Sun H, Treco D, Szostak J W. Extensive 3′-overhanging, single-stranded DNA associated with meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 68.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 69.Thompson-Stewart D, Karpen G H, Spradling A C. A transposable element can drive the concerted evolution of tandemly repetitious DNA. Proc Natl Acad Sci USA. 1994;91:9042–9046. doi: 10.1073/pnas.91.19.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 71.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trinh T Q, Sinden R R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 73.Virtaneva K, D’Amato E, Miao J, Koskiniemi M, Norio R, Avanzini G, Franceschetti S, Michelucci R, Tassinari C A, Omer S, et al. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat Genet. 1997;15:393–396. doi: 10.1038/ng0497-393. [DOI] [PubMed] [Google Scholar]
- 74.Warren S T. The expanding world of trinucleotide repeats. Science. 1996;271:1374–1375. doi: 10.1126/science.271.5254.1374. [DOI] [PubMed] [Google Scholar]
- 75.Welch J W, Maloney D H, Fogel S. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol Gen Genet. 1990;222:304–310. doi: 10.1007/BF00633833. [DOI] [PubMed] [Google Scholar]
- 76.Welch J W, Maloney D H, Fogel S. Gene conversions within the Cup1r region from heterologous crosses in Saccharomyces cerevisiae. Mol Gen Genet. 1990;229:261–266. doi: 10.1007/BF00272164. [DOI] [PubMed] [Google Scholar]
- 77.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–674. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre H J, Lapsys N, Le Paslier D, Doggett N A, Sutherland G R, Richards R I. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]