Abstract
A genetic screen was devised to identify Saccharomyces cerevisiae splicing factors that are important for the function of the 5′ end of U2 snRNA. Six slt (stands for synthetic lethality with U2) mutants were isolated on the basis of synthetic lethality with a U2 snRNA mutation that perturbs the U2-U6 snRNA helix II interaction. SLT11 encodes a new splicing factor and SLT22 encodes a new RNA-dependent ATPase RNA helicase (D. Xu, S. Nouraini, D. Field, S. J. Tang, and J. D. Friesen, Nature 381:709–713, 1996). The remaining four slt mutations are new alleles of previously identified splicing genes: slt15, previously identified as prp17 (slt15/prp17-100), slt16/smd3-1, slt17/slu7-100, and slt21/prp8-21. slt11-1 and slt22-1 are synthetically lethal with mutations in the 3′ end of U6 snRNA, a region that affects U2-U6 snRNA helix II; however, slt17/slu7-100 and slt21/prp8-21 are not. This difference suggests that the latter two factors are unlikely to be involved in interactions with U2-U6 snRNA helix II but rather are specific to interactions with U2 snRNA. Pairwise synthetic lethality was observed among slt11-1 (which affects the first step of splicing) and several second-step factors, including slt15/prp17-100, slt17/slu7-100, and prp16-1. Mutations in loop 1 of U5 snRNA, a region that is implicated in the alignment of the two exons, are synthetically lethal with slu4/prp17-2 and slu7-1 (D. Frank, B. Patterson, and C. Guthrie, Mol. Cell. Biol. 12:5179–5205, 1992), as well as with slt11-1, slt15/prp17-100, slt17/slu7-100, and slt21/prp8-21. These same U5 snRNA mutations also interact genetically with certain U2 snRNA mutations that lie in the helix I and helix II regions of the U2-U6 snRNA structure. Our results suggest interactions among U2 snRNA, U5 snRNA, and Slt protein factors that may be responsible for coupling and coordination of the two reactions of pre-mRNA splicing.
Precursor-mRNA (pre-mRNA) splicing takes place in the spliceosome through a two-step transesterification reaction. At least 40 splicing factors have been identified by genetic means in the yeast Saccharomyces cerevisiae. Most have been implicated in specific steps of the splicing pathway (31, 41). During the process of spliceosome assembly, small nuclear RNAs (snRNAs) and the pre-mRNA substrate, in association with protein factors, undergo extensive conformational changes which establish RNA-RNA interactions that are important for both splicing reactions. Among these factors are the DExD or DExH proteins: RNA-dependent ATPases (possibly RNA helicases), which include Prp2p, Prp5p, Prp16p, Prp22p, Prp28p (26, 31), Prp43p (2), and Slt22p (also called Brr2p) (22, 34, 53). Their functions are essential for the formation and maintenance of RNA-RNA interactions in the spliceosome.
With few exceptions (28, 29, 36), Watson-Crick base pairing is important for most RNA-RNA interactions in the formation of the spliceosome (26). In particular, intermolecular base-pairing interactions that occur between U2 and U6 snRNAs, forming helices I and II (Fig. 1A), are likely to be involved in bringing the 5′ splice site (through a U6–5′-splice-site interaction [21, 23, 42]) and the branchpoint site (through a U2-branchpoint interaction [37]) into proximity. This is necessary for the first-step reaction (Fig. 1A). Residues in both U2 and U6 snRNAs that are important for either or both steps of splicing (11, 30) form part of these interactions or lie nearby. This finding has led to the suggestion that these RNA structures may form the so-called active center for catalysis of the splicing reactions (27, 30). The helix II portion of human U6 snRNA has also been shown to influence dissociation of the U4-U6 snRNA duplex in vitro, perhaps by stabilizing it (5).
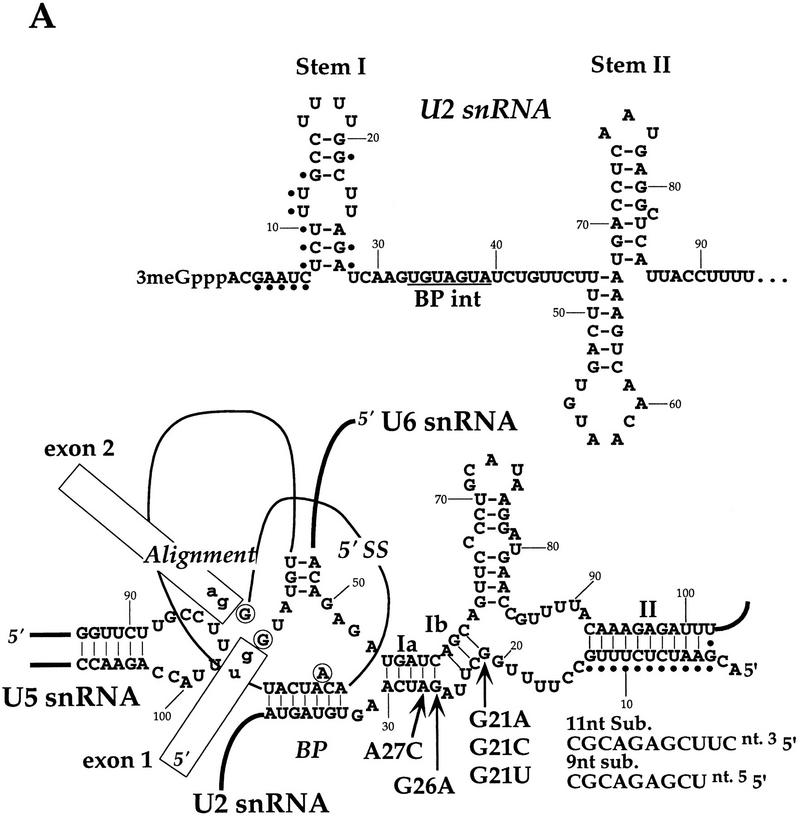
FIG. 1.
(A) Yeast U2 snRNA. The top diagram shows the structure of yeast U2 snRNA with the 5′-end region, including stems I and II. BP int, the region that interacts through base pairing with the branchpoint (BP) site in pre-mRNA. The bottom diagram represents U2-U6 snRNA interactions (intermolecular helices Ia, Ib, and II), illustrated in the context of BP-U2 (BP) and 5′-splice-site–U6 (5′ SS) interactions, and alignment of exons by U5 snRNA (Alignment). Nucleotides involved directly in the transesterification reaction are circled. Dots indicate the U2-U6 snRNA helix II region that is mutated in the 11-nt substitution (Sub.) mutation of U2 snRNA. The 11-nt substitution (12) used in the genetic screen and other U2 snRNA mutations used to test allele specificity are shown under U2 snRNA in the context of U2-U6 snRNA interactions. (B) Growth phenotypes of yeast strains carrying the U2 snRNA mutations shown in panel A. (C) Yeast strain used in the genetic screen. The strain (12) contains a chromosomal deletion of the U2 snRNA gene (SNR20), which was replaced with HIS3 (SNR20Δ::HIS3), and two plasmids carrying wt SNR20 (URA3 CEN-ARS, i.e., a maintenance plasmid) and mutant snr20-11nt (TRP1 CEN-ARS). Following EMS mutagenesis, cells harboring extragenic mutants (slt’s) that became synthetically lethal with the mutant U2 snRNA were also sensitive to 5-FOA. (D) Growth phenotypes of six slt mutants. In addition to conferring synthetic lethality, these slt mutants all confer growth defects at various elevated temperatures. Shown are the 2-day growth phenotypes of the slt strains obtained following a series of back-crosses with a wt strain (carrying SNR20) grown on yeast extract-peptone-dextrose medium. Note that the slt15, slt16, and slt22 strains all grow slowly at 30°C.
The highly conserved loop 1 region of U5 snRNA is important for the juxtaposition of the two exons, which is essential for the second step of splicing (35). However, canonical base pairing is not obviously involved in this interaction, suggesting that additional factors are required for its establishment and maintenance. In fact, two splicing factors, Prp8p (45, 47, 48) and Slu7p (6, 13), have been implicated in the alignment of the two exons and/or in 3′-splice-site selection. In addition, a functional role for U2 snRNA in the alignment of the two exons has been suggested by site-specific cross-linking between the first nucleotide of the 3′ exon and the U23 and A30 nucleotides of U2 snRNA (33). Despite the available information, it is not clear how different RNA structures in the active center are coordinated, either spatially or temporally, prior to and after the first splicing step.
The yeast U2 snRNA contains all conserved elements found in other eukaryotic U2 snRNAs, despite its unusually large size (approximately 1,200 nucleotides [nt]) (Fig. 1A). The stem II region of U2 snRNA is important for recognition of the branchpoint site and association of the U2 snRNP with the pre-mRNA. Several factors, including Prp5p (a DEAD-box protein), SF3a (Prp9p Prp11p Prp21p), and SF3b, have been implicated in this function (40, 51, 54). In contrast, relatively little is known about the factors involved in subsequent steps of spliceosomal function during which the stem I region of U2 snRNA undergoes extensive conformational rearrangements to form the U2-U6 snRNA interactions (see above and Fig. 1A).
We have devised a synthetic-lethality genetic screen to search for such factors. In this screen, a mutant U2 snRNA carrying an 11-nt substitution in the stem I region (12) was used as the starting mutation. Although this mutation can potentially perturb the U2-U6 snRNA helix II interaction (Fig. 1A), it confers only a mild growth defect, which is likely due to functional redundancy of the helix Ib and helix II regions in U2-U6 snRNA (12). Our genetic screen yielded six slt (stands for synthetic lethality with U2) mutants. We have characterized and cloned all the SLT genes, two of which encode new splicing factors: Slt11p (a possible RNA-binding protein), Slt22p (an RNA-dependent ATPase [53]), and four previously characterized factors, Slt15p (previously named Prp17p [Slt15p/Prp17p]), Slt16p/Smd3p, Slt17p/Slu7p, and Slt21p/Prp8p. These factors are required for either or both steps of splicing. Our genetic and biochemical characterizations suggest that the functions of these factors and of U2 snRNA may overlap that of invariant loop 1 of U5 snRNA in the alignment of the two exons in the spliceosome.
MATERIALS AND METHODS
Yeast strains and plasmids.
All yeast strains used in this study were derived from W303-1A or -1B (MATa or MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100). The original strain used in the genetic screen, containing a chromosomal deletion of SNR20 (the U2 snRNA gene), has been described previously (12). A new ΔSNR20 disruption was made by deleting the ClaI-HpaI region of ∼810 nt containing the 5′-end half of the U2 snRNA gene (∼750 nt) and by replacing it with the yeast HIS3 gene. The wild-type (wt) SNR20 gene was carried on pRS316 (URA3 CEN-ARS). Two resultant haploids, YDX2299A (MATa) and YDX22100A (MATα), were used in the subsequent genetic experiments. Chromosomal deletion of SNR6 (the U6 snRNA gene) was constructed by deleting the entire coding region and replacing it with the yeast HIS3 gene. Two haploid ΔSNR6 strains, YXU91 (MATα) and YXU92 (MATa), were obtained. Strains containing slt11, slt17, slt21, and slt22 mutations in the ΔSNR20 background were crossed to both ΔSNR6 strains in order to generate haploid strains carrying double deletions as well as slt mutations. SNR20 and SNR6 were carried on the same maintenance plasmid. These strains were used to test genetic interaction among slt, U2, and U6 mutations. U6 snRNA mutations (3-nt substitutions near the 3′ end: designated a, b, c, and d) have been described (12).
Strains containing slu mutations and U5 snRNA constructs (U98A, U98C, and the U97C U99C double mutation [U97C/U99C] [14]) and U5 snRNA constructs were provided by C. Guthrie (University of California, San Francisco). PCR was used to subclone mutant U5 snRNA fragments into pRS315 (LEU2 CEN-ARS). The ΔSNR7 disruption was constructed by deleting the entire coding region and replacing it with the yeast HIS3 gene. Two disruption strains, YXU37A (MATa) and YXU37B (MATα), were used in subsequent experiments. Strains containing chromosomal deletions of both SNR20 and SNR7 were created by crossing the two single-deletion haploid strains; a maintenance plasmid carrying both the SNR20 and SNR7 genes was then introduced into the resultant heterozygous diploid before sporulation. The double-deletion strains (YXU53 and YXU54) were obtained from the progenies following sporulation and tetrad dissection. A strain containing the prp2-1 mutation and the PRP8 gene were provided by J. Beggs (University of Edinburgh). prp2-1 and prp28-102 plasmids were obtained from R.-J. Lin (Beckman Research Institute of the City of Hope) and T.-H. Chang (Ohio State University), respectively. For the testing of synthetically lethal interactions between slu (or prp) mutations and U2 snRNA mutations, the slu (or prp) mutations were segregated genetically into the ΔSNR20 strain through at least two consecutive crosses with YDX2299A or YDX22100A. Following tetrad dissection, progeny haploids containing the slu (or prp) mutation and ΔSNR20 were selected and used to test synthetic lethality with the U2 snRNA mutations. In order to exclude differences in genetic background, at least four independent ΔSNR20 slu (or prp) isolates were used in these experiments. Genomic fragments containing the prp2-1 and prp28-102 mutations were introduced into W303-1A by two-step gene replacement. Both mutations were then segregated to the ΔSNR20 background as described above.
EMS mutagenesis, screening, and genetic characterization.
In the initial genetic screen, a yeast strain with a chromosomal deletion of SNR20 (12) containing SNR20 and snr20-11nt carried on URA3 and TRP1 plasmids, respectively, was subject to ethyl methanesulfonate (EMS) mutagenesis to yield a survival rate of 10 to 20%. The surviving cells were screened for sensitivity to 5-fluoroorotic acid (5-FOA) at 30°C. This indicates dependence of viability on SNR20 (carried on a URA3 plasmid), which reflects the lethality generated by snr20-11nt in combination with extragenic mutations (i.e., synthetic lethality) (Fig. 1C). In order to eliminate artifacts (e.g., mutations which affect the uracil pathway), the snr20-11nt/TRP1 plasmids in 5-FOA-sensitive cells were replaced by another SNR20 plasmid. The resultant cells that were sensitive to 5-FOA were discarded. A scheme for genetic characterization was designed to ensure that the slt mutants were phenotypically and genetically appropriate (they should have arisen from mutation at a single locus which also confers a recessive growth defect). Briefly, the temperature-sensitive phenotype of each original slt strain was first segregated into the chromosomal SNR20 background by crossing the slt strain with the wt strain (W303-1A). The resulting temperature-sensitive haploid was then back-crossed with the wt strain at least three times to ensure that the temperature sensitivity phenotype was due to mutation at a single locus. The final temperature-sensitive haploid was crossed with the newly constructed SNR20 deletion strain (YDX2299A or YDX22100A). Synthetic lethality was then tested in strains containing both the temperature sensitivity mutation and ΔSNR20 derived from the above-described cross. These strains were also used to generate the slt ΔSNR20 plus ΔSNR6 strains described above.
Splicing extract preparation and in vitro splicing.
Yeast whole-cell extracts were prepared from wt and slt cells according to the method of Lin et al. (24) with modifications. The substrate—32P-labeled yeast pre-actin RNA—was synthesized by runoff transcription with T7 RNA polymerase and 32P[UTP] as described previously (24). For in vitro splicing assays, equal volumes (10 μl) of whole-cell extract and buffer component, containing labeled substrate, were mixed and incubated at the temperatures indicated in the figures. Splicing intermediates and products were resolved in a 5% polyacrylamide gel.
RNA isolation and primer extension.
All slt mutant strains used for RNA analysis contained SNR20 on the chromosome (see above). Total yeast RNA isolation and primer extension were performed as described previously (17).
Cloning by complementation.
In order to clone the wt SLT genes, a YCp50-borne yeast genomic DNA library, CENBANK A (38), was introduced into slt cells and the transformants were selected for temperature resistance at 37°C. Plasmids containing complementing genomic DNA fragments were recovered from the positives. A minilibrary approach was used to define the minimal complementing regions. The original complementing plasmid was first digested with a variety of restriction enzymes, and the resulting fragments were ligated to another vector with a different selective marker. Total plasmid DNA prepared from these mini-libraries was introduced into the slt strains, followed by selection for full complementation. The overlapping region in plasmids rescued from different mini-libraries contain slt-complementing open reading frames. Nucleotide sequences of both ends of these fragments were determined and then used to search the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/) for a match. DNA sequences of complementing regions were retrieved, and fragments containing only single open reading frames were tested for complementation of both growth defect and synthetic lethality.
Genetic analysis of synthetic lethality.
In order to examine potential synthetic lethality of two mutations, a heterozygous diploid was first generated from two parental slt (slu or prp) haploid strains and was then subjected to sporulation and tetrad dissection. If haploids containing both mutations are viable, three types of tetrads should be obtained: parental ditype (PD; each spore inherits either of the original mutations), nonparental ditype (NPD; two spores inherit both mutations, while the other two inherit neither mutation, i.e., it has the wt phenotype), and tetratype (T; two spores inherit either of the original mutations, one inheriting both mutations and the remaining one inheriting neither mutation, i.e., it has the wt phenotype). However, if the combination of both mutations is lethal, only PD tetrads can be obtained from such a diploid. T tetrads should contain only three viable spores (one wt spore and two spores with either mutation), and NPD tetrads should contain only two wt spores; the lack of complete (i.e., with four viable spores) NPD and T tetrads indicates that the two mutations in question are synthetically lethal. In most cases, at least 20 scoreable tetrads from each heterozygous diploid were analyzed phenotypically to determine synthetic lethality.
RESULTS
Genetic screening, characterization, and molecular cloning of SLT genes.
The U2 snRNA mutation (snr20-11nt) used in the search for synthetic-lethality mutants contains an 11-nt substitution in the stem I region (Fig. 1A) (12). Although such a substitution may potentially disrupt the proposed U2 snRNA stem structure and perturb the helix II interaction of U2-U6 snRNA, it confers only a mild growth defect at both 37 and 16°C (12) (Fig. 1B). Cells containing the wt U2 snRNA gene (SNR20) carried on a URA3-marked plasmid and snr20-11nt on a TRP1-marked plasmid were mutagenized with EMS. Surviving cells were screened for extragenic mutations that failed to allow growth at 30°C in the absence of SNR20 (as indicated by sensitivity to 5-FOA). These extragenic mutations result from lethality generated by snr20-11nt in combination with a second mutation (Fig. 1C), i.e., they produce synthetic lethality, which suggests functional interaction between the two gene products. These mutations were designated slt mutations (for synthetic lethality with U2 snRNA).
On the basis of 5-FOA sensitivity, nine candidate slt mutant strains were isolated from approximately 8,000 colonies that survived EMS mutagenesis. In the presence of SNR20, all mutant strains displayed a temperature-sensitive growth defect (see below). They were crossed with prp strains in our collection to test for complementation. slt21 failed to complement prp8-1 at 37°C, indicating that it might correspond to a new allele of PRP8. The complementation test involving slt15 and prp17-1 yielded an ambiguous result. However, following the cloning of SLT15 gene, it became clear that slt15 is allelic to prp17-1. Each slt mutation occurs at a single locus and confers a recessive growth defect (Fig. 1D and see Materials and Methods).
The splicing defect associated with five slt mutations, slt11, -15, -16, -17, and -22 (Fig. 1D), representing five different genes, was determined. In vitro splicing assays were performed with extracts prepared from wt, slt11, slt22, and slt17 cells. At 25°C the splicing activities of slt11 and slt22 extracts were lower than that of the wt extract (Fig. 2A, lanes 2 and 3), and at 33°C neither extract had detectable splicing activity (Fig. 2A, lanes 5 and 6). No preferential accumulation of intermediates was observed, suggesting that both mutations affect pre-mRNA splicing prior to or at the first step. A first-step splicing defect was also observed in vivo for both mutations (data not shown). When the slt17 extract was assayed at 25 or 33°C, reduced splicing activity was detected (Fig. 2A, lanes 11 to 14). Accumulation of splicing intermediates was observed following a 20-min incubation at either temperature (Fig. 2A, lanes 12 and 14), suggesting that the slt17 mutation affects primarily the efficiency of the second-step reaction. This observation is consistent with molecular cloning data which show that slt17 is a new allele of SLU7.
FIG. 2.
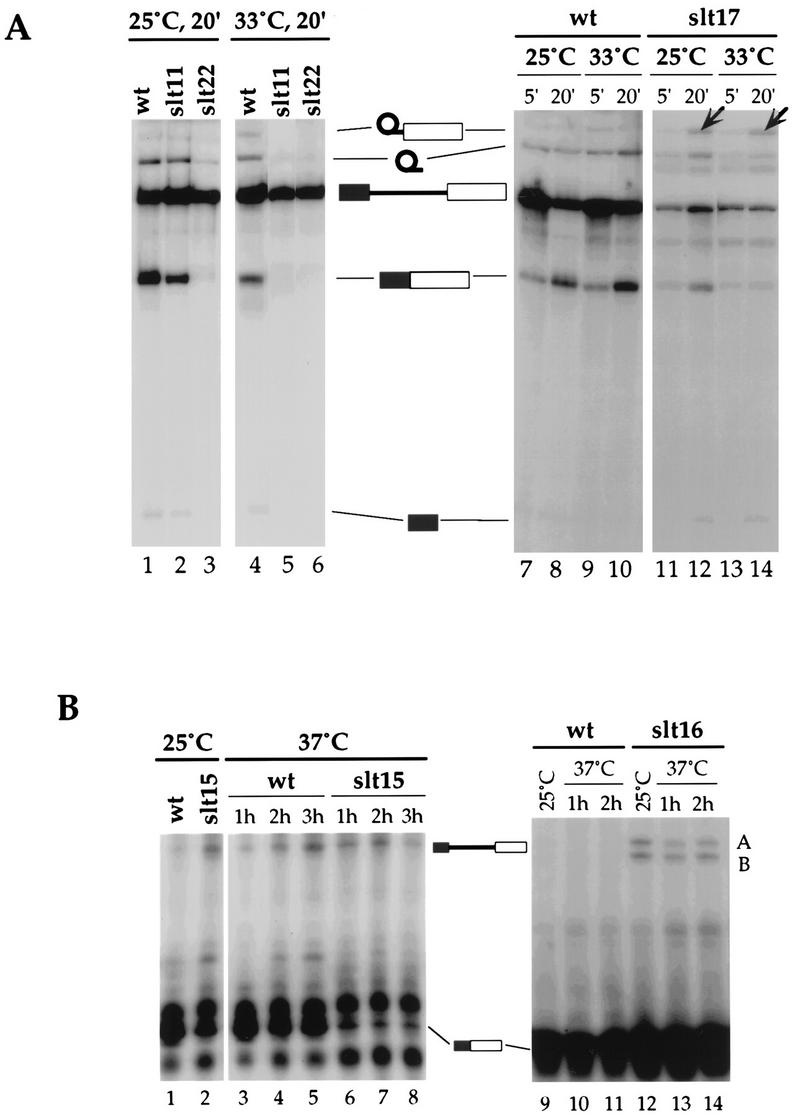
Splicing defects associated with slt mutations. (A) In vitro splicing defects associated with the slt11, slt22, and slt17 mutations. Splicing reactions with 32P-labeled actin pre-mRNA substrate were performed at 25 and 33°C for 20 min with whole-cell extracts prepared from wt, slt11, slt22, and slt17 cells. Precursor, intermediates (free 5′ exon and lariat intron–3′ exon), and final products (5′ exon–3′ exon and lariat intron) of the splicing reaction are indicated between the gels. Arrows in lanes 12 and 14 indicate preferential accumulation of lariat-intron–3′-exon intermediate in slt17 extract. (B) Primer-extension analyses of inhibition of splicing in vivo. The left gels show reduced levels of mature actin RNA in slt15 cells. The right gels show inhibition of pre-U3 splicing in slt16 cells. The two upper bands labeled A and B correspond to pre-U3A and pre-U3B, respectively. Cells were grown at 25°C for several generations and then were shifted to 37°C. Total yeast RNA was isolated before (grown at 25°C) and following a shift to 37°C for the times indicated. Levels of spliced and unspliced RNAs were measured by primer extension with labeled oligonucleotide complementary to the second exon. Precursor and mature RNAs are indicated.
In vivo splicing defects in slt15 and slt16 mutants were detected by primer-extension assays. Total yeast RNA was isolated from these mutant cells before and following a shift from 25°C to a nonpermissive temperature (37°C). The levels of spliced and unspliced actin and U3 RNAs were measured by primer extension with labeled oligonucleotides complementary to the second exons of each transcript. Inhibition of pre-mRNA splicing at 25°C was noted for both mutant strains (Fig. 2B, lanes 2 and 12). This inhibition is consistent with the slow growth conferred at 25 and 30°C by both mutations (Fig. 1D and data not shown). The level of mature mRNA was reduced in slt15 cells following the shift (Fig. 2B, lanes 6 to 8), while accumulation of unspliced precursors was observed in slt16 cells at 37°C (Fig. 2B, lanes 13 and 14). These data indicate that both slt15 and slt16 mutations affect pre-mRNA splicing in vivo.
The wt genes of slt11, -15, -16, -17, and -22 were cloned by complementation with a low-copy-number yeast genomic library. The minimum complementing fragments for each SLT gene were identified by a mini-library method (see Materials and Methods). Table 1 summarizes the results of molecular cloning of SLT genes. Among the six SLT genes identified in our genetic screen, two (SLT11 and SLT22) encode new splicing factors and the remainder correspond to splicing genes isolated previously.
TABLE 1.
Summary of SLT genes and proteins
| SLT | Gene sequence name | Other name(s) for gene | Protein size (aa)a | Protein motif, homology, and/or function | Splicing step(s) | Source or reference(s) |
|---|---|---|---|---|---|---|
| SLT11 | YBR065c | 364 | Zn-2+ fingers, Rpl25 homology | First step | This study | |
| SLT15 | YDR364c | PRP17/SLU4/CDC40 | 455 | WD repeats | Second step | 14, 20 |
| SLT16 | YLR147c | SMD3 | 101 | Sm core protein | snRNP biogenesis | 39 |
| SLT17 | YDR088c | SLU7 | 382 | Zn2+ knuckle | Second step | 6, 13, 14, 20 |
| SLT21 | YHR165c | PRP8/DBF3 | 2,413 | Proline-rich repeats | First and second steps | 45–47 |
| SLT22 | YER172c | BRR2/SNU246/RSS1 | 2,163 | RNA-dependent ATPase | First step | 22, 34, 53 |
aa, amino acid.
Characterization of the slt22-1 mutation and the RNA-dependent ATPase activity associated with Slt22p have been reported (53). The interaction between Slt21p/Prp8p and U2 snRNA and the role of Slt21p/Prp8p in formation of the active spliceosome will be reported elsewhere.
SLT11.
SLT11, a new splicing-related gene, corresponds to YBR065c. It encodes a 41-kDa protein of 364 amino acids. Two putative zinc fingers (CX2CX17CX2C and CX2CX6CX2C) are present in the N-terminal region, while the central region (amino acids 151 to 299) shows homology (27% identity and 40% similarity) to yeast ribosomal protein L25. The C terminus (approximately 30 amino acids) of Slt11p is highly charged due to the presence of blocks of lysine residues. The original slt11-1 mutation blocks splicing prior to the first step (Fig. 2A, lane 5) without an apparent effect on spliceosome assembly (unpublished data), suggesting that Slt11p exerts its function immediately prior to the first-step splicing reaction.
SLT15/SLU4/PRP17.
SLT15/SLU4/PRP17 was first isolated in a genetic screen for temperature sensitivity mutations that also affect pre-mRNA splicing (prp17 [49]) and in another screen independently (slu4 [14]). The encoded protein contains WD repeats, which are also present in another splicing factor, Prp4p (4, 17). Prp17p is involved exclusively in the second step of splicing (20). The new allele is named prp17-100. The primary defect of the slt15/prp17-100 mutation was associated with the reduction of mature RNA, and no preferential accumulation of unspliced precursor was observed (Fig. 2B, lanes 6 through 8), suggesting that the mutation affects the stability of mature RNA and/or, more likely, the second step of splicing.
SLT17/SLU7.
SLT17/SLU7 encodes a second-step splicing factor (6, 13, 20) (Fig. 2A, lanes 11 through 14). The gene product contains a so-called zinc knuckle that is similar to one in retroviral nucleocapsid protein and that is required for RNA-protein interaction (13). The slt17 allele is renamed slu7-100. Both slu4-1/prp17-2 and slu7-1 were isolated in a genetic screen for splicing mutations that are synthetically lethal with mutations in invariant loop 1 of U5 snRNA (14), a region that has been implicated in alignment of the two exons in the second step of splicing (35). However, Slu7p and Prp17p exert their functions at different steps, with respect to that of hydrolysis of ATP by Prp16p (20), a putative RNA helicase, whose function is required for the second step (7, 8, 43).
SLT16/SMD3.
SLT16/SMD3 encodes a yeast core Sm protein with homology to human SmD3 (39). slt16 is the first reported temperature-sensitive allele of this gene and is renamed smd3-1. The protein is associated with all five spliceosomal snRNAs and is thought to be involved in the biogenesis of snRNPs. In an extract depleted of Smd3p, splicing is blocked before or at the first step (39). A similar in vivo splicing defect was observed for the slt16/smd3-1 mutation (Fig. 2B, lanes 12 through 14). The human SmD3 protein has been shown to be cross-linked to the 5′ splice site in a site-specific manner (25, 52). To our knowledge, this study is the first time that an Sm protein has been shown genetically to be important for the function of a particular snRNA in splicing. However, it remains to be determined whether this genetic interaction between Smd3p and U2 snRNA is direct (affects the function of U2 snRNA) or indirect (affects the stability of snRNAs).
Genetic interactions of slt, slu, and prp mutations with U2 snRNA.
We determined whether the phenotypes of the slt mutations are specific for the 11-nt substitution in the stem I region of U2 snRNA, with which they were originally identified. We included in our genetic analysis three sets of U2 snRNA mutations that lie in regions that are involved in U2-U6 snRNA helices: helix Ia (G26A and A27C), helix Ib (substitutions at G21), and helix II (11- and 9-nt substitutions) (Fig. 1A). In addition to the six slt mutations identified in this study, four slu (14) and four prp mutations were also included (Table 2).
TABLE 2.
Genetic interaction of slt, slu, and prp mutations with U2 snRNA mutations
| Mutation | U2 snRNA mutationa
|
||||||
|---|---|---|---|---|---|---|---|
| Helix Ia
|
Helix Ib
|
Helix II
|
|||||
| A27C | G26A | G21A | G21C | G21U | 11 nt | 9 nt | |
| slt | |||||||
| slt11-1b | + | −− | − | −− | − | −− | −− |
| slt15/prp17-100c | −− | −− | −− | −− | −− | −− | −− |
| slt16/smd3-1b | + | −− | + | −− | + | −− | −− |
| slt17/slu7-100c | + | − | + | −− | + | − | − |
| slt21/prp8-21d | + | + | + | −− | + | −− | −− |
| slt22-1b | + | + | + | −− | + | −− | −− |
| slu | |||||||
| slu2-1 | + | + | + | + | + | + | + |
| slu3-1 | + | + | + | + | + | + | + |
| slu4/prp17-2c | −− | −− | −− | −− | −− | −− | −− |
| slu5-1 | + | −− | + | −− | + | + | + |
| slu7-1c | −− | + | − | −− | − | −− | − |
| prp | |||||||
| prp2-1b | + | + | + | + | + | + | + |
| prp16-1c | + | −− | + | + | + | + | + |
| prp28-1b | + | + | + | + | + | + | + |
| prp28-102b | + | + | + | + | + | + | + |
Helices Ia, Ib, and II refer to intermolecular structures of U2-U6 snRNAs. Yeast strains carrying slt, slu, or prp mutations and SNR20Δ::HIS3 were transformed with plasmids carrying wt or mutant U2 snRNA genes, and the resultant transformants were then grown on medium containing 5-FOA at 30°C (if the slu or prp mutant in question was viable) and/or 25°C to test for viability of cells containing both mutations. −−, no growth of double-mutant strains, i.e., synthetically lethal at 25 and 30°C; −, no growth of double-mutant strains at 30°C but poor growth at 25°C (≥4 days); +, no additive growth defects of double-mutant strains.
Mutations block splicing prior to the first step.
Required for the second step.
Required for both steps of the splicing.
Three kinds of genetic interactions were observed between slt and U2 snRNA mutations (Table 2): (i) slt11-1 and slt15/prp17-100 showed virtually no allele specificity, as they were synthetically lethal with all the U2 snRNA substitutions tested (with the exception of A27C for slt11-1); (ii) slt16/smd3-1 and slt17/slu7-100 showed synthetic lethality with specific substitutions in the U2 snRNA part of the U2-U6 helix Ia and Ib regions and with the helix II region; and (iii) slt21/prp8-21 and slt22-1 were allele specific and lethal only with G21C in helix Ib and with the 11- and 9-nt substitutions in helix II but not with other U2 snRNA mutations tested.
Among the slu mutations, slu2 and slu3 showed no synthetic lethality with any of the U2 snRNA mutations tested (Table 2). slu4/prp17-2 had the same allele specificity as slt15/prp17-100. slu5 was lethal only with two U2 snRNA mutations, G21C and G26A, but not with the 11-nt substitution used in the genetic screen for slt mutants. slu7-1 showed broader allele specificity than slt17/slu7-100 in that it was lethal with both G21A and G21U in addition to G21C, which may affect the U2-U6 snRNA helix Ib interaction.
The four prp mutations correspond to three RNA-dependent ATPases involved in events which occur after the formation of prespliceosome (Prp28p), concomitantly with U2-U6 snRNA interactions (Prp2p), or in the second step (Prp16p). With the exception of only one prp16-1 interaction, none of these mutations showed synthetic lethality (Table 2). These results provided genetic evidence that the RNA conformational rearrangements that are affected by the original U2 snRNA mutation (11-nt substitution) is specific for Slt22p (53).
Thus, the synthetically lethal interactions of most slt and slu mutants are rather general with respect to mutations in U2 snRNA. This may reflect the possibility that a large portion of the U2 snRNA and/or the U2-U6 snRNA structure, rather than individual structural elements, is involved in interactions with these factors.
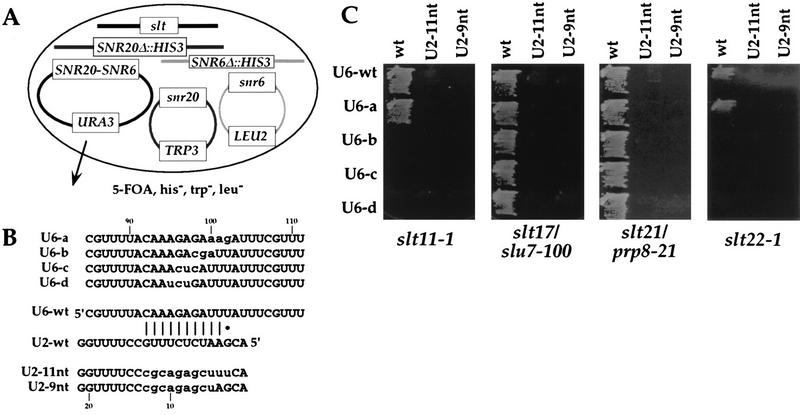
Genetic interactions of slt mutations with U6 mutations in U2-U6 snRNA helix II.
The original 11-nt substitution in U2 snRNA used in our genetic screen has the potential to perturb U2-U6 snRNA helix II (12). Consequently, we determined whether this genetic interaction extended to nucleotides on the U6 snRNA side of helix II. Yeast strains which contain slt mutations in combination with double deletions of SNR20 and SNR6 were constructed (Fig. 3A). These were used to determine whether specific U6 snRNA mutations (12) were synthetically lethal with the slt mutations and whether partial restoration of a mutationally disrupted helix II could suppress the synthetic lethality that is observed with the original slt mutations. (Note that we were unable to determine whether a corresponding 9-nt U6 snRNA substitution conferred synthetic lethality, since that mutation is lethal [12].) Two slt mutations that encoded new splicing factors (slt11-1 and slt22-1) and two that encoded existing ones (slt17/slu7-100 and slt21/prp8-21) were tested.
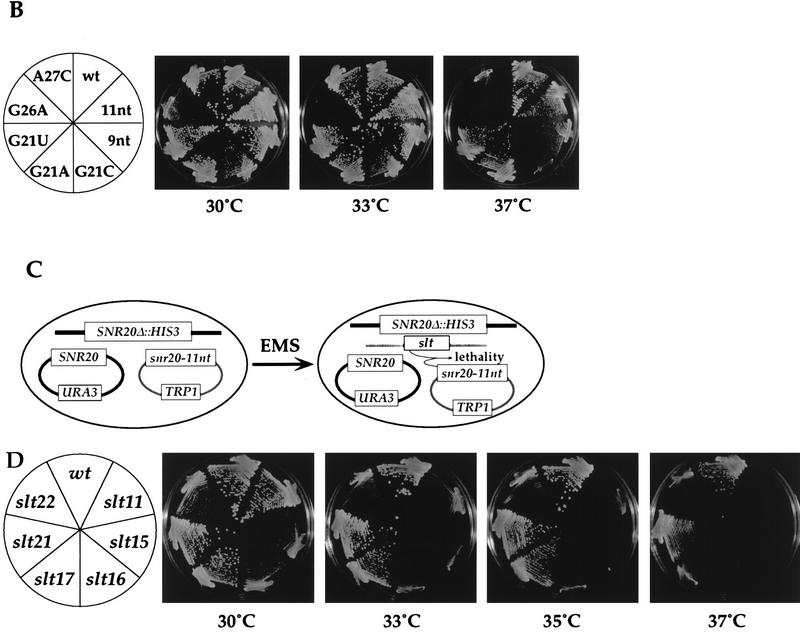
FIG. 3.
Genetic interactions between slt mutations and the U2-U6 snRNA helix II. (A) Representative yeast strain used in the genetic tests. (B) U2 and U6 snRNA mutations used in the genetic tests. The U6 snRNA mutations and their growth phenotypes in a wt (SLT) background are described in reference 12. (C) Growth for 2 days at 30°C of four slt strains containing various combinations of U2 and U6 snRNA mutations. Mutant U2 snRNA (TRP1-marked) and U6 snRNA (LEU2-marked) plasmids were introduced into the yeast strains shown in panel A. The resultant transformants were grown on 5-FOA-containing selective medium. Note that the slt22-1 mutation confers slow growth at 30°C.
slt22-1 showed synthetic lethality with three of the U6 snRNA mutations in the helix II region (U6-b, -c, and -d) (Fig. 3C). This is consistent with the observation that the RNA-dependent ATPase activity of this factor is related to U2-U6 snRNA helix II and that a disrupted helix II (because of the U2 snRNA 11-nt substitution) is not an efficient in vitro substrate for Slt22p (53). slt11-1 showed similar synthetic lethality, which at the current state of knowledge of this splicing factor we are not yet able to explain. None of these synthetically lethal interactions was suppressed by partial restoration of helix II through the inclusion of the U2 snRNA 9- or 11-nt mutations (Fig. 3C). For slt22-1, this lack of suppression suggests that the helix II structures formed by the U6-b, -c, and -d mutations are not efficient substrates for the mutant protein.
Neither slt17/slu7-100 nor slt21/prp8-21 was synthetically lethal with any of the U6 snRNA mutations tested (Fig. 3C). This result suggests that these two factors are unlikely to be involved in interactions with U2-U6 snRNA helix II but that they are specific to interaction with U2 snRNA.
Genetic interactions among slt and slu mutations.
The fact that the slt mutations are all synthetically lethal with certain U2 snRNA mutations suggested that at least some of them might form a functional unit(s). We assessed this possibility by testing the synthetic lethality of pairs of slt mutations, as well as of selected prp mutations, by attempting to create double-mutant haploids (Table 3 and Materials and Methods). Failure to obtain such haploids indicates that the two mutations in question are synthetically lethal. The results of these experiments are summarized in Tables 3 and 4 and Fig. 4.
TABLE 3.
Summary of tetrad-dissection analysis
| Tetrad typea | Spore phenotypea | No. of spores of each phenotype in crossb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| slt11 × slt15 | slt11 × slu4 | slt11 × slt17 | slt11 × slu7 | slt11 × prp16 | slt15 × slt17 | slt15 × slu7 | slt15 × prp16 | slt17 × slu4 | slt17 × prp16 | ||
| PD | 4 (2ts:2ts) | 5 | 4 | 8 | 3 | 3 | 7 | 3 | 1 | 3 | 3 |
| 3 (2ts:1ts) | 1 | 3 | 2 | 4 | 2 | 0 | 4 | 2 | 3 | 1 | |
| T | 4 (1wt:2ts:1ts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 (1wt:2ts) | 27 | 25 | 39 | 23 | 17 | 23 | 19 | 11 | 14 | 12 | |
| NPD | 4 (2wt:2ts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 (2wt:1ts) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2 (2wt) | 5 | 8 | 9 | 5 | 4 | 4 | 3 | 2 | 2 | 3 | |
| Total | 38 | 40 | 58 | 35 | 26 | 34 | 29 | 16 | 22 | 19 | |
The three types of tetrads were determined as follows: (i) the PD tetrads contained four or three viable spores, all with the original temperature sensitivity (ts) phenotype; (ii) the T tetrads contained four viable spores with only one with the wt phenotype (the one with double mutants [ts]) or three viable spores with only one with the wt phenotype and two with the original temperature sensitivity phenotype; and (iii) NPD tetrads contained four viable spores with two with the wt and two with the temperature sensitivity phenotype, three viable spores with two with the wt and one with the temperature sensitivity phenotype, or two viable wt spores.
slt15 is prp17-100, slt16 is smd3-1, slt17 is slu7-100, and slu4 is prp17-2. The slt11-1 mutation (underlined) affects the first step of splicing, while the other mutations affect the second step of splicing. Results of tetrad dissection of heterozygous diploids that generate viable double-mutation spores are not shown but are summarized in Table 4.
TABLE 4.
Summary of synthetic lethality among slt and slu mutations
| Mutation | Growth phenotype of double mutant containinga:
|
|||||
|---|---|---|---|---|---|---|
| slt11b | slt15c | slt16b | slt17c | slt21d | slt22b | |
| slt11b | NA | |||||
| slt15c | − | NA | ||||
| slt16b | + | + | NA | |||
| slt17c | − | − | + | NA | ||
| slt21d | + | + | + | +/− | NA | |
| slt22b | + | + | ND | + | + | NA |
| slu4c | − | NA | + | −* | + | ND |
| slu7c | − | −* | + | NA | +/− | ND |
| prp16c | − | −* | NA | −* | +/− | ND |
| prp2-1b | + | ND | ND | + | + | + |
slt15 is prp17-100, slt16 is smd3-1, slt17 is slu7-100, slu4 is prp17-2, and slt21 is prp8-21. Symbols: +, the double mutant is viable and without additive defect; −, the double mutant is lethal (i.e., it has synthetic lethality); +/−, the double mutant is viable but confers a severe growth defect (slow growth at 25°C, i.e., it has partial synthetic lethality); and *, see reference 14. NA, not applicable; ND, not determined.
Mutations block splicing prior to the first step.
Required for the second step.
Required for both steps.
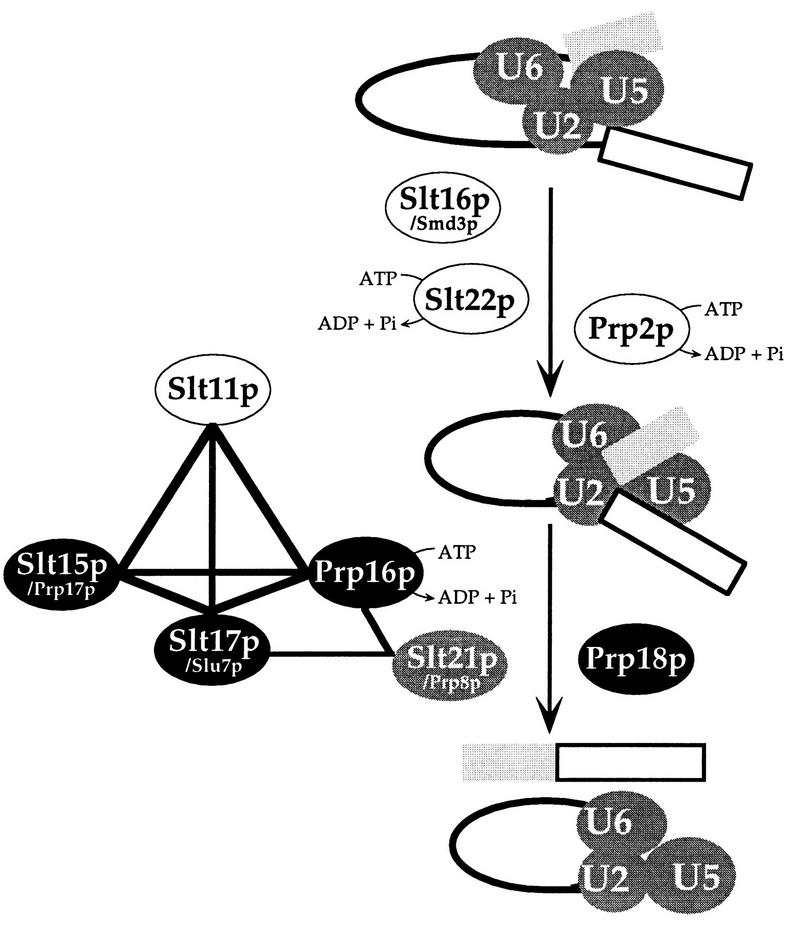
FIG. 4.
Summary of genetic interactions among factors involved in either or both steps of splicing. Thick lines indicate synthetic lethality at all temperatures. Thin lines indicate partial synthetic lethality (Table 4). None of the slt16/smd3-1, slt22-1, and prp2-1 mutations show synthetic lethality with other mutations tested. Genetic interactions between second-step mutations, including prp18 and prp8, have also been described elsewhere (14, 20, 48). Pi, inorganic phosphate.
Pairwise synthetic lethality was observed in strains carrying the following mutations: slt11-1 (first-step mutation), slt15/prp17-100, slu4/prp17-2, slt17/slu7-100, slu7-1, and prp16-1 (all second-step mutations). Two members of this group, slt17/slu7-100 and prp16-1, also showed weak but significant synthetic lethality with slt21/prp8-21 (the gene product is required for both splicing steps). Although not identified in our screen, prp16-1 is also synthetically lethal with one particular mutation, G26A, in U2 snRNA (Table 2) and with four slt mutations (Table 4; Fig. 4). All other combinations showed no significant synthetic lethality.
We suggest that Slt11p, Prp17p, Slu7p, Prp16p, and Prp8p form two overlapping functional units. The fact that this group of factors affects both splicing steps might indicate hitherto-unsuspected connections between the two reactions of pre-mRNA splicing.
Genetic interactions among slt and U5 snRNA mutations.
All of our slt mutations were isolated as synthetically lethal with a U2 snRNA mutation, yet two of them (slt15 and slt17) are in the same genes (slu4 and slu7, respectively) as mutations that were identified originally as being synthetically lethal with U5 snRNA mutations (14). Furthermore, three (possibly four) Slt factors may act in functional groups (see above), which suggests the possibility of an interaction involving some of the Slt factors and U5 snRNA.
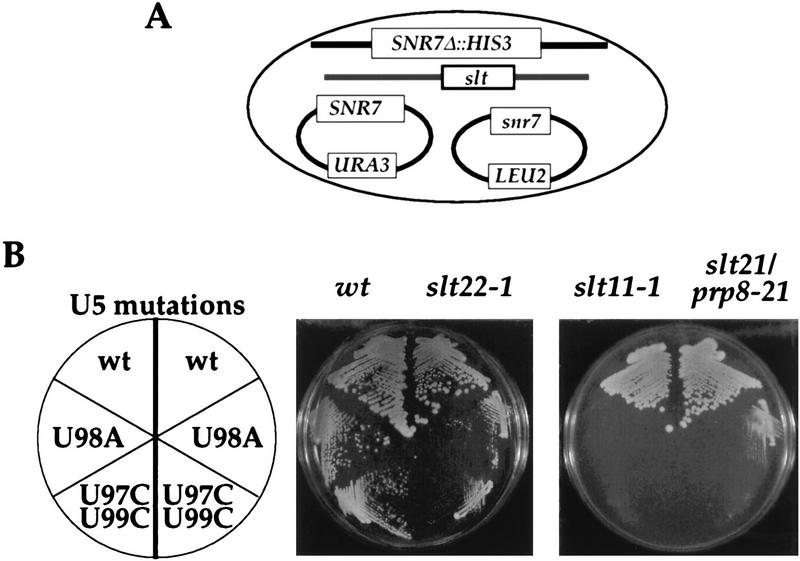
This possibility was tested by constructing strains in which each of five slt mutations, slt11-1, slt15/prp17-100, slt17/slu7-100, slt21/prp8-21, and slt22-1, were segregated to a ΔSNR7 (SNR7 the U5 snRNA gene) background. A plasmid carrying either of two U5 snRNA mutations, U98A and the U97C/U99C double mutation, was then introduced into these strains to test by plasmid shuffling for synthetic lethality (Fig. 5A).
FIG. 5.
Synthetic lethality of slt mutants with loop 1 mutations of U5 snRNA. (A) Yeast strains carrying slt mutations and a chromosomal deletion of the U5 snRNA gene (SNR7Δ::HIS3), with SNR7 on a URA3 CEN-ARS plasmid, were transformed with wt and mutant U5 snRNA plasmids (LEU2 CEN-ARS). (B) Results of 5-day growth at 30°C of the resultant transformants on medium containing 5-FOA. Note that slt22-1 and U5 snRNA double mutants grew significantly slower than strains carrying either mutation alone.
slt11-1 and slt21/prp8-21 showed synthetic lethality at 30°C with both U5 snRNA mutations (Fig. 5B), although slt21/prp8-21 showed some growth with the U5 U98A mutation at 25°C (Fig. 5B and data not shown). Weak synthetic lethality was observed between slt22-1 and both U5 mutations at 25 and 30°C (Fig. 5B and data not shown). It has been reported that the original slu4/prp17-2 and slu7-1 mutations are synthetically lethal with the U5 snRNA U98A mutation (14); we found that slt15/prp17-100 is lethal with both the U98A mutation and the U97C/U99C double mutations and that slt17/slu7-100 is lethal with U97C/U99C (data not shown).
Thus, four slt mutations that were identified on the basis of synthetic lethality with U2 snRNA mutations are also synthetically lethal with mutations in U5 snRNA loop 1. Two of these, slt11-1 and slt21/prp8-21, affect the first splicing step, while the tethering function of U5 snRNA is required only for the second step (35). We conclude that these factors, including the new one, Slt11p, may be involved in the interactions involving both U2 and U5 snRNAs and in both splicing steps.
Genetic interactions between U2 and U5 snRNAs.
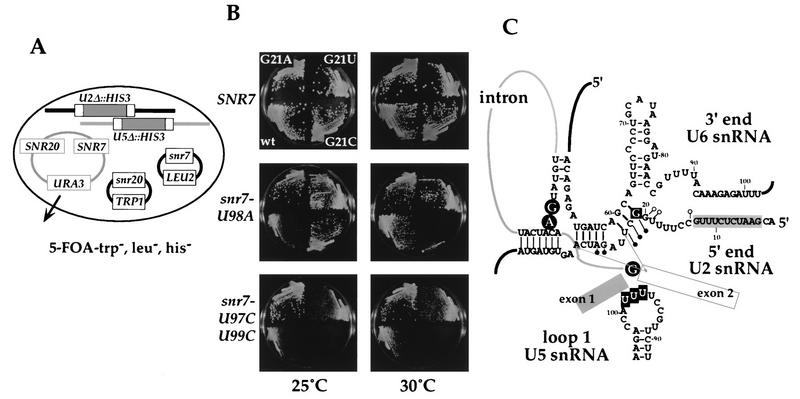
The data presented above indicate that members of a group of splicing factors, which were identified originally on the basis of their genetic interaction with U2 snRNA, also interact with one another and with U5 snRNA. It is possible that these factors act together in functions related to both U2 and U5 snRNAs, such as coordination of the two steps of splicing and/or tethering of the two exons following the first splicing step (35). This suggests that there may be genetic interactions between U2 and U5 snRNAs. The two U5 snRNA mutations used in the genetic screen for slu mutants (14), U98A/U99C, and a series of U2 snRNA mutations were tested for genetic interaction with the yeast strain shown in Fig. 6A.
FIG. 6.
Genetic interactions (synthetic lethality and suppression) between substitutions at the G21 position in U2 snRNA and loop 1 mutations of U5 snRNA. (A) Yeast strain containing both SNR20Δ::HIS3 and SNR7Δ::HIS3. Mutant U2 snRNA (TRP1 CEN-ARS) and U5 snRNA (LEU2 CEN-ARS) plasmids were introduced into this strain to test for genetic interactions. The resultant transformants were first grown on 5-FOA-containing medium at both 25 and 30°C. Cells containing snr20-G21C and snr7-U97C/U99C failed to grow on this medium; i.e., they are synthetically lethal. Other 5-FOA resistant cells were then grown on selective medium at the temperatures indicated. (B) Four-day growth at 25 and 30°C of U5 snRNA-wt, -U98A, -U97C/U99C in combination with U2 snRNA-wt, -G21A, -G21C, and -G21U in the absence of a maintenance plasmid. Note that U2-G21C is synthetically lethal with U5-U97C/U99C. (C) Summary of genetic interactions between U2 and U5 snRNAs and the locations of U2 and U5 snRNA mutations in the context of other demonstrated RNA-RNA interactions in the spliceosome. Positions 97, 98, and 99 in U5 snRNA loop 1 are enclosed in filled squares. Filled circles indicate mutations at positions in U2 snRNA that are synthetically lethal with the U5 snRNA mutations tested (see Table 5 for a summary). G21 of U2 snRNA is enclosed in a filled square; mutations at this position were able to suppress the U5 snRNA mutations tested. The U2 snRNA part of the U2-U6 snRNA helix II is shaded. Substitutions in this region (i.e., the 11-nt substitution) cause synthetic lethality with slt mutations but not with U5 snRNA mutations. Lines from U23 and A30 in U2 snRNA to exon 2 indicate site-specific cross-linking (33).
In the helix II region, the original 11- or 9-nt substitutions of U2 snRNA showed no synthetic lethality with either U5 snRNA mutation (Table 5), suggesting that U2-U6 snRNA helix II may not be involved directly in interaction with U5 snRNA.
TABLE 5.
Growth phenotypes of U2 and U5 snRNA double mutants
| U2 region or nucleotide | Mutation | Growth phenotype of double mutant containing the indicated U5 mutationa
|
||||||
|---|---|---|---|---|---|---|---|---|
| wt | U98A at °C:
|
U97C/U99C at °C:
|
||||||
| 25 | 30 | 33 | 25 | 30 | 33 | |||
| wt | ++++ | +/− | ++ | +/− | +/− | + | +/− | |
| Helix II | 11-nt substitution | ++++c | +/− | ++ | ND | +/− | + | ND |
| 9-nt substitution | ++++c | +/− | ++ | ND | +/− | + | ND | |
| G21b | G21A | ++++c | +++ | ++ | − | +++ | +++ | − |
| G21C | +++c | +++ | − | − | − | − | − | |
| G21U | ++++c | +++ | +++ | − | +/− | +/− | − | |
| C22 | C22A | ++++ | − | − | − | − | − | − |
| C22G | ++++ | − | − | − | − | − | − | |
| C22U | ++++ | +/− | +/− | − | +/− | +/− | − | |
| U23 | U23A | ++++ | − | − | − | − | − | − |
| U23G | ++++ | − | − | − | − | − | − | |
| U23C | ++++ | − | − | − | − | − | − | |
| G26 | G26A | ++c | − | − | ND | − | − | ND |
| A27 | A27C | ++++c | − | − | − | − | − | − |
However, all substitutions at C22 and U23 of U2 snRNA in the helix Ib region of U2-U6 snRNA were synthetically lethal or debilitating with either of the U5 snRNA mutations tested, even though these U2 snRNA mutations themselves conferred little or no growth defect at these temperatures (Table 5). It is noteworthy that U23 is in close contact with the first nucleotide of the 3′ exon, which is tethered to the 5′ exon through interactions with loop 1 of U5 snRNA (33).
Mutations at the adjacent G21 position of U2 snRNA showed a mixed phenotype (Fig. 6B). At 25°C all three substitutions at G21 suppressed the slow growth conferred by the U5 U98A mutation, at 33°C all three were synthetically lethal, and at 30°C (the intermediate temperature) G21A and G21U had little effect while C21C was synthetically lethal. With the U5 U97C/U99C double mutation, on the other hand, only the G21A substitution suppressed at 25 and 30°C while the G21C substitution was synthetically lethal at both temperatures (Table 5; Fig. 6B).
G26A and A27C, the only two viable substitutions in the U2 part of U2-U6 snRNA helix Ia (11, 30), were synthetically lethal with either U5 snRNA mutation tested (Table 5). No synthetic lethality was observed with mutations at other positions in U2 snRNA (e.g., substitutions G20C, U19G, C14G, and C14A [unpublished data]).
These results suggest an interaction of U5 snRNA loop 1 with the U2 portion of U2-U6 snRNAs helix I (Fig. 6B) but not with helix II. While there is no biochemical evidence for a direct RNA-RNA interaction between loop 1 of U5 snRNA and nt 21, 22, and 23 of U2 snRNA, the genetic results presented here (Fig. 5B and Table 5) suggest that an interaction may nevertheless exist. Given the role of U5 snRNA loop 1 in alignment of the two exons (35), it may be that the helix I region of U2 snRNA also plays a role in this function. In support of this suggestion, we note that cross-linking experiments (33) indicate that the helix I region of U2 snRNA is in close contact with exon 2. This idea is consistent with the finding that Slt17p/Slu7p and Slt2p/Prp8p are involved in 3′-splice-site selection (6, 13, 20, 45, 47, 48).
DISCUSSION
The genetic screen described here has identified six yeast splicing factors (called slt factors). Mutations in genes encoding these factors become synthetically lethal with mutations in the stem I region of U2 snRNA (corresponding to helix II of the U2-U6 snRNA composite structure). Each of these splicing factors affects either or both steps of splicing. One of these factors, Slt22p, is likely to be involved with U2-U6 snRNA helix II (53). Three factors, Slt15p/Prp17p, Slt17p/Slu7p, and Slt21p/Prp8p, are linked to the tethering of U5 snRNA loop 1 (14, 45). On the basis of genetic interactions among some members of this group of factors and other prp mutations, we suggest that Slt11p, Prp17p, Slu7p, Prp16p, and Prp8p form two overlapping functional units. Four slt mutations in this group are also synthetically lethal with mutations in U5 snRNA loop 1. From this we conclude that these factors, including a new one, Slt11p, may be involved in the functions of both U2 and U5 snRNAs, perhaps in alignment of the two exons in the yeast spliceosome and/or coordination of the two steps of splicing. Genetic tests indicated an interaction between mutations in loop 1 of U5 snRNA and those near the 5′ end of U2 snRNA. Given the role of U5 snRNA loop 1 in the tethering of exons (35), it may be that U2 snRNA also plays a role in this function. Our findings suggest a mechanism for coupling and coordination of the two steps of splicing.
The functions of U2-U6 snRNA helix I and helix II regions.
During maturation of the spliceosome, snRNAs and the pre-mRNA substrate undergo conformational changes in order to juxtapose the splice sites and to form structures that are important for the subsequent steps of splicing (26). For both helix I and helix II of U2-U6 snRNA, there is evidence that the U6 snRNA components may have functions that are independent of their interaction in the helical structures. For example, mutations in the U6 snRNA components of yeast helix II are lethal and cannot be suppressed by base-compensatory mutations in the corresponding U2 snRNA portion of U2-U6 snRNA (12). Similar genetic asymmetry was also observed for nucleotides involved in the helix Ib interaction (11, 27, 28). Indeed, the 5′ end of U2 snRNA itself may be important for the second step of splicing. The genetic interactions of both Slt17p/Slu7p and Slt21p/Prp8p with U2 snRNA are specific to the U2 portion of U2-U6 snRNA helix II (Fig. 3). This observation differentiates the function of the 5′-end region of U2 snRNA from its role in U2-U6 snRNA helix II.
There is evidence that the role of U2-U6 snRNA helix II may be exerted prior to the splicing reactions. In vitro dissociation of the human U4-U6 snRNA duplex appears to be regulated by two elements in U6 snRNA: the center region (upstream of U4-U6 snRNA stem I, including the conserved ACAGAG motif that recognizes the 5′ splice site) and the 3′-end region, which forms U2-U6 snRNA helix II (5). It has been suggested that U2-U6 snRNA helix II may stabilize the U4-U6 snRNA duplex until the spliceosome is fully assembled (5). It has also been suggested that the energy released from the unwinding of helix II is sufficient to disassemble the U4-U6 snRNA duplex (5). If this is so, one possible function of helix II is to hold indirectly the U4-U6 snRNA structure in place to antagonize the premature formation of structures that are important for the splicing reactions (i.e., U2-U6 snRNA helix Ia and the 3′-end stem-loop of U6 snRNA). The unwinding of U2-U6 snRNA helix II by a possible RNA helicase, such as Slt22p (53), may be a crucial regulatory step in the initiation of splicing.
Interactions between the U2 and U5 snRNPs.
The genes encoding the Slt factors were isolated on the basis of genetic interaction with U2 snRNA, yet four of them are related functionally to U5 snRNA. This finding suggests that the U2 snRNA portion of U2-U6 snRNA helix II (the 5′-end region), following resolution of this helix by Slt22p, may interact with U5 snRNA. Three Slt factors are related to the tethering function of U5 snRNA loop 1. Slt21p/Prp8p, which has been shown to be cross-linked to both the 5′ and 3′ splice sites in human (52) and yeast (45, 47, 48), may act to stabilize the exon-U5 snRNA loop 1 interaction (45). This factor is also involved in the recognition of the polypyrimidine tract (46, 47) and 3′-splice-site selection (46). The other two factors, Prp17p and Slu7p, are linked genetically to the function of U5 snRNA loop 1 (14) and are in close contact with the 3′ splice site (1, 6, 13, 20, 48).
Further observations support the idea of interaction between the U2 and U5 snRNPs. The failure of mutant slt22-1p to function, possibly because of an inability to unwind U2-U6 snRNA helix II, results in the accumulation of an unusual, “dead-end” splicing complex that lacks the U5 snRNP (53). This suggests that the U2 snRNA part of helix II may be involved in holding the U5 snRNP to the spliceosome. A related experiment with a HeLa cell nuclear extract demonstrated that an oligoribonucleotide complementary to U5 snRNA (a region in the 3′ side of loop 1) can induce the formation of a U1-U4-U5 snRNP complex (3). This unusual U1-U4-U5 snRNP complex may represent a transient step in spliceosome assembly during which the U1–5′-splice-site and U4-U6 snRNA interactions are disrupted and displaced by U6–5′-splice-site and U2-U6 snRNA interactions, respectively. In certain circumstances the U5 snRNP may associate preferentially with the U1 and U4 snRNPs, which are in the process of dissociating from the spliceosome (3). It is known that residues in loop 1 of U5 snRNA can be cross-linked to nucleotides of both pre-mRNA exons before and after the first step of splicing (33, 44, 50, 52). We suggest that U5 snRNA may become associated tightly with the spliceosome concomitantly with the unwinding of U2-U6 snRNA helix II and that the 5′ end of U2 snRNA may provide an RNA interaction(s) to anchor the U5 snRNA and/or to assist the U5 snRNA in the alignment of the two exons.
It has been proposed that the stem-loop structure of the U5 snRNA loop 1 region is analogous to subdomain ID3 of autocatalytic group II introns, which is essential for 5′-splice-site recognition and tethering of the free 5′ exon (32). However, unlike the loop region of group II ID3, which contains the exon-binding site (EBS1) complementary to the 3′ end of the 5′ exon (19), uridine-rich U5 snRNA loop 1 may interact with the two exons by noncanonical base pairing, since the exon sequences at both splice sites are not conserved in pre-mRNAs. An additional RNA interaction(s) may be necessary to stabilize the U5-exon interaction. In group II introns, a bulged region, α′, at the bottom of the subdomain ID3 stem-loop, interacts with a region in subdomain IB, α (15). Recently, an anchoring function for the tertiary α-α′ interaction has been suggested (16). Our data (reference 53 and this study) may suggest a similar role for the 5′ end of U2 snRNA in binding the U5 snRNP to the spliceosome, possibly in association with protein factors identified in our genetic screen.
The observed genetic interactions between mutations in U2 and U5 snRNAs (Fig. 4) suggest a role for the helix Ib region of U2 snRNA in the second step: (i) this region of U2 snRNA may interact with U5 snRNA to assist the tethering function of loop 1 and/or (ii) the 5′ end of U2 snRNA (particularly nt 21, 22, and 23), in addition to that of U5 snRNA loop 1, may interact with either or both exons to provide tethering. Since the U23 position of U2 snRNA is in close contact with the exon region of the 3′ splice site (33), the second possibility is more likely and is supported by the synthetic lethality of mutations in two second-step factors (Prp17p and Slu7) with mutations in the 5′ end of U2 snRNA (Table 2).
Recently, Chiara et al. (9) have shown in the HeLa system that cross-linking of the U2 snRNP protein, U2AF65, to the polypyrimidine tract is replaced by cross-linking of three U5 snRNP proteins, p110, p116, and p220 (the latter is an Slt21p/Prp8p ortholog) prior to the second splicing step. This observation provides evidence that an interaction (direct or indirect) between the U2 snRNP, bound to the branchpoint site, and the U5 snRNP is important for positioning the latter on the 3′ splice site and thus for selection of the 3′ splice site. Our genetic data led to a similar conclusion.
Interactions among Slt factors: coordination of the two steps of splicing.
Four slt mutations, slt11-1, slt15/prp17-100, slt17/slu7-100, and slt21/prp8-21, show pairwise synthetic lethality with each other and with prp16-1 (Fig. 4). prp16-1 was identified initially as a suppressor of a mutation at the branchpoint site; this mutation resulted in a second-step block due to the use of a cryptic branchpoint site (7). McPheeters (29) has shown genetic suppression of nonadenosine branchpoints by mutations in two regions of U6 snRNA that are either part of U2-U6 snRNA helix Ia (U57) or of the intramolecular stem-loop adjacent to helix Ib (nt 63 to 65, 71, and 82 to 84) and that an allele of prp16, prp16-302, is synthetically lethal with one of these U6 snRNA suppressors, U57C (29). Our results demonstrate that prp16-1 is synthetically lethal with U2 substitutions in the U2-U6 snRNA helix Ia region (G26A).
The RNA-dependent ATPase activity of Prp16p is involved in remodeling the spliceosome in order that the second-step reaction occurs (43) and, analogously to what occurs with group II self-splicing (18), that the lariat intermediate is proofread (8). This suggestion of proofreading was supported by the observation that replacement of the adenosine residue at the branchpoint can block the second-step reaction (37). These observations have led to the suggestion that kinetic coordination of the two steps of pre-mRNA splicing is achieved by a mechanism in which the second step serves as a trap for intermediates that have undergone productive branching, thus driving the full splicing reaction to completion (10).
Four Slt factors (Slt11p, Slt15p/Prp17p, Slt17p/Slu7p, and Slt21p/Prp8p) as well as Prp16p may act as two units with overlapping functions (Fig. 4). Through these potential interactions, four components or functions of the spliceosome that are important for the two steps of splicing are connected: U2 snRNA (interacting with U6 snRNA and juxtaposing the 5′ splice site and branchpoint site), U5 snRNA (tethering the two exons), Prp16p (proofreading the first-step reaction and remodelling the spliceosome), and Slt17p/Slu7p plus Slt21p/Prp8p (selection of the 3′ splice site). Slt11p, required for the first step of splicing, also interacts genetically with three second-step factors (Fig. 4). These interactions may reflect a mechanism that couples and coordinates the two steps of splicing.
ACKNOWLEDGMENTS
We thank B. Funnell, S. Nouraini, and V. Lay for reading the manuscript; J. D. Beggs, A. J. Newman, A. M. MacMillan, and D. Jansma for comments and constructive discussion; R. W. van Nues, J. D. Beggs, T.-H. Chang, P. Raghunathan, C. Guthrie, D.-H. Kim, and R.-J. Lin for providing constructs and/or yeast strains; and U. Vijayraghavan for verifying the PRP17 sequence.
This research was supported by the National Cancer Institute of Canada and the Medical Research Council of Canada.
REFERENCES
- 1.Ansari A, Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1994;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenas J E, Abelson J N. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ast G, Weiner A M. A U1/U4/U5 snRNP complex induced by a 2′-O-methyl-oligoribonucleotide complementary to U5 snRNA. Science. 1996;272:881–884. doi: 10.1126/science.272.5263.881. [DOI] [PubMed] [Google Scholar]
- 4.Ayadi L, Miller M, Banroques J. Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA. 1997;3:197–209. [PMC free article] [PubMed] [Google Scholar]
- 5.Brow D A, Vidaver R M. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA. 1995;1:122–131. [PMC free article] [PubMed] [Google Scholar]
- 6.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess S, Couto J R, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 8.Burgess S M, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 9.Chiara M D, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin K, Pyle A M. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- 11.Fabrizio P, Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- 12.Field D J, Friesen J D. Functionally redundant interactions between U2 and U6 spliceosomal snRNAs. Genes Dev. 1996;10:489–501. doi: 10.1101/gad.10.4.489. [DOI] [PubMed] [Google Scholar]
- 13.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 14.Frank D, Patterson B, Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris-Kerr C L, Zhang M, Peebles C L. The phylogenetically predicted base-pairing interaction between alpha and alpha′ is required for group II splicing in vitro. Proc Natl Acad Sci USA. 1993;90:10658–10662. doi: 10.1073/pnas.90.22.10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetzer M, Wurzer G, Schweyen R J, Mueller M W. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature. 1997;386:417–420. doi: 10.1038/386417a0. [DOI] [PubMed] [Google Scholar]
- 17.Hu J, Xu Y, Schappert K, Harrington T, Wang A, Braga R, Mogridge J, Friesen J D. Mutational analysis of the PRP4 protein of Saccharomyces cerevisiae suggests domain structure and snRNP interactions. Nucleic Acids Res. 1994;22:1724–1734. doi: 10.1093/nar/22.9.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquier A, Jacquesson-Breuleux N. Splice site selection and role of the lariat in a group II intron. J Mol Biol. 1991;219:415–428. doi: 10.1016/0022-2836(91)90183-7. [DOI] [PubMed] [Google Scholar]
- 19.Jacquier A, Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987;50:17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- 20.Jones M H, Frank D N, Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc Natl Acad Sci USA. 1995;92:9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandels-Lewis S, Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 22.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Luhrmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 23.Lesser C F, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 24.Lin R J, Newman A J, Cheng S-C, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 25.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 26.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 27.Madhani H D, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 28.Madhani H D, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- 29.McPheeters D S. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- 30.McPheeters D S, Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of Group I introns. Cell. 1992;171:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- 31.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 32.Newman A J, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 33.Newman A J, Teigelkamp S, Beggs J D. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 34.Noble S M, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Keefe R T, Norman C, Newman A J. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 36.Parker R, Siliciano P G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 37.Query C C, Moore M J, Sharp P A. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- 38.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 39.Roy J, Zheng B, Rymond B C, Woolford J L., Jr Structurally related but functionally distinct yeast SmD core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruby S W, Chang T H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 41.Rymond B, Rosbash M. Yeast pre-mRNA splicing. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 42.Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci USA. 1992;89:11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sontheimer E J, Steitz J A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 45.Teigelkamp S, Newman A J, Beggs J D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umen J G, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umen J G, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 48.Umen J G, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 49.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 50.Wassarman D A, Steitz J A. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 51.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 52.Wyatt J R, Sontheimer E J, Steitz J A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 53.Xu D, Nouraini S, Field D, Tang S J, Friesen J D. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 54.Yan D, Ares M J. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]